- 1Engineering Research Center of Tropical Medicine Innovation and Transformation of Ministry of Education, International Joint Research Center of Human-machine Intelligent Collaborative for Tumor Precision Diagnosis and Treatment of Hainan Province, Hainan Provincial Key Laboratory of Research and Development on Tropical Herbs, School of Pharmacy, Hainan Medical University, Haikou, China
- 2Analysis and Test Center, Chinese Academy of Tropical Agricultural Sciences, Haikou, China
- 3Hainan Academy of Inspection and Testing, Haikou, China
Inocybe is a cosmopolitan genus of ectomycorrhizal fungi within the family Inocybaceae. Members of this genus are recognized as a group of toxic mushrooms linked to poisoning incidents worldwide. Clarifying this species diversity and toxin profiles within this genus is critical for both taxonomy and public health. In this study, we describe two newly identified species, I. bicystidiata sp. nov. and I. microcarpa sp. nov., based on phylogenetic analysis and morphological evidence. Phylogenetically, the two new species belong to I. sect. Leptocybe. Inocybe bicystidiata is characterized by nodulose basidiospores with saddle-shaped projections and the coexistence of thick-walled pleurocystidia and thin-walled paracystidia on the lamellar side. Inocybe microcarpa is characterized by very small basidiomata, spinose basidiospores with forked projections, absence of pleurocystidia, and thin-walled cheilocystidia. Ecologically, both of these new species occur in tropical rainforests dominated by Parashorea chinensis, which is considered as their presumed host. In addition, new geographic data are reported for three previously documented species of I. sect. Leptocybe, I. acutata, I. juji, and I. peppa, based on newly obtained specimens. Comprehensive ultra-performance liquid chromatography-mass spectrometry (UPLC–MS)/MS toxin screening revealed no detectable levels of muscarine, psilocybin, psilocin, bufotenine, or baeocystin in 10 examined species of sect. Leptocybe. This contrasts with the known toxin-producing Inocybe lineages, suggesting a divergent secondary metabolism in this clade.
1 Introduction
Inocybaceae Jülich (1982: 374) is a family of ectomycorrhizal fungi (Agaricales) commonly found in temperate and tropical forests, with over 1,050 described species across Africa, Asia, Europe, North America, Oceania, and South America (Matheny et al., 2020). The family has been revised based on the results of a recent phylogenetic study that divides the family into seven genera, with Inocybe (Fr.) Fr. (1863: 346) is the largest genus in the family (Matheny et al., 2020). Many species in the family contain fungal toxins such as muscarine, psilocybin, and phalloidin (Li et al., 2022; Zhang Y. Z. et al., 2022; Kosentka et al., 2013; Matheny et al., 2023), which usually lead to poisoning events (Li, H. J. et al., 2020, 2021, 2022, 2023, 2024; Xu et al., 2020; Deng et al., 2022; Chandrasekharan et al., 2020; Parnmen et al., 2021). Accordingly, the basic data on species diversity, geographical distribution, and their toxins are crucial for poison prevention and the utilization of this group of fungi (Deng et al., 2022). However, only a small proportion of Inocybaceae species have been assigned for toxin detection (Kosentka et al., 2013), and the toxin status of many taxa in the family remains under-recognized.
The genus Inocybe represents the most evolutionarily successful group, with over 1,000 documented species worldwide (Matheny and Kudzma, 2019). The members of Inocybe are characterized by small basidiomata, fibrillose to rimose pileus, smooth, nodulose to spinose basidiospores with a distinct apiculus, and usually thick-walled hymenial cystidia. However, Inocybe species collected from tropical forests have some peculiar features: thin-walled cheilocystidia or thin-walled pleurocystidia (Horak, 1979; Pradeep et al., 2016; Latha and Manimohan, 2017; Gao et al., 2024). The I. alienospora group was initially recovered in a LSU phylogeny with three species (I. hydrocybiformis, I. lasseri, and I. stellata), but the topography was not well supported (Horak et al., 2015). Subsequent studies expanded this clade to encompass Australian taxa (I. alienospora, I. lasseroides) and Indian taxa (I. barbruka, I. kuruvensis, I. papiliformis), formally designated as the I. alienospora clade (Pradeep et al., 2016; Latha and Manimohan, 2017; Matheny and Bougher, 2017). Our recent work has refined the phylogenetic framework of this group through the inclusion of specimens from China, leading to its formal classification as I. sect. Leptocybe (Gao et al., 2024). Concurrently, eight new species from temperate to tropical China and new distributions for certain species were reported (He et al., 2022; Gao et al., 2024). At present, 17 species are recognized within the sect. Leptocybe; however, several phylogenetically distinct lineages remain unresolved and necessitate taxonomic revision.
During a 2024 mycological survey in Xishuangbanna Tropical Rainforest National Park (Yunnan Province, China), two previously undescribed species of I. sect. Leptocybe were discovered. Integrated morphological examination and molecular phylogenetic analyses confirmed their taxonomic novelty. We provide detailed descriptions of these new species, supplemented with diagnostic illustrations and comparative discussions with allied taxa. Additionally, new geographic distributions of the previously documented species I. acutata, I. juji, and I. peppa were reported based on the recently obtained specimens. To better understand the toxic profiles of I. sect. Leptocybe in China, a targeted screening of toxins and quantitative analysis were performed using a comprehensive method of ultra-performance liquid chromatography-mass spectrometry (UPLC–MS/MS).
2 Materials and methods
2.1 Chemicals and reagents
Standards: Muscarine (catalog No. 0000045268) was purchased from Merck. Psilocybin (catalog No. 017013022YD04020200507) and psilocin (catalog No. 002005025TR00920200417) were obtained from Shanghai Yuansi Standard Science and Technology Co., Ltd. Bofotenine (catalog No. CFN91165) was sourced from Shanghai Keshun Biotechnology Co., Ltd. (Shanghai, China), and baeocystin (catalog No. B115315) was purchased from Toronto Research Chemicals. All standard solutions were stored at −20°C and protected from light. HPLC-grade acetonitrile and methanol were obtained from HuBei FTSCI BioTech Co., Ltd. (Wuhan, China), Acetic Acid and Ammonium acetate were obtained from Xilong Scientific Co., Ltd. (Shantou, China), and Shanghai Aladdin Biochemical Technology Co., Ltd. (Shanghai, China) Ultrapure water with electrical resistivity of 18.2 MΩ/cm and total organic carbon (TOC) < 3 ppb used in all experiments was produced by a Milli-Q water purification system (Millipore, Billerica, MA, United States).
2.2 Field sampling and morphological studies
Specimens were collected from Wangtianshu Scenic Area, a national nature reserve in Mengla County, Xishuangbanna Prefecture, Yunnan Province, China, with a tropical climate. In the field, ecological images were taken using a digital camera. The Basidiomata were documented while fresh, with color assignments based on the criteria set by Kornerup and Wanscher (1978). Subsequently, the specimens were dried overnight in an electric oven at 45°C and then sealed in plastic bags (Yu et al., 2020; Deng et al., 2021a, 2021b, 2022; Zhao et al., 2022; Hu et al., 2023). Following the study, the specimens were deposited in the Herbarium of the Changbai Mountain National Natural Reserve (ANTU), along with their corresponding FCAS (Fungarium of Changbai Mountain Academy of Sciences, FCAS) numbers.
Macromorphological features were documented from field notes and color photographs. Microscopic examinations were carried out using a light microscope. Mushroom tissues from the pileus, lamellae, and stipes were cut into thin sections by hand with the aid of a stereoscope (AV100–240 V). Dried specimens were sliced and rehydrated in a 5% potassium hydroxide (KOH) solution, and a 1% Congo Red solution was used when necessary. Basidiospores, basidia, hymenophoral trama, pleurocystidia, paracystidia, cheilocystidia, pileipellis/pileal trama, stipitipellis/stipe trama, and oily hyphae were examined and measured. For each specimen, side views of at least 100 mature basidiospores were measured, excluding the apiculus, using the format [a/b/c] to denote the measurement of “a” basidiospores from “b” individuals across “c” collections. Measurement data were presented as (d) e−h−f (g), where “d” and “g” represent the minimum and maximum values, respectively; “e” and “f” correspond to the values at the 5th and 95th percentiles when the data are ordered from smallest to highest; and “h” signifies the average value (Ge et al., 2021; Liu et al., 2021; Na et al., 2022). Furthermore, the roundness of spores was quantified using the length-to-width ratio (Q), which effectively differentiated between species. Qm denotes the average Q-value, and Qm ± SD indicates the average plus or minus the sample standard deviation (Ge et al., 2021).
2.3 DNA extraction, polymerase chain reaction, and sequencing
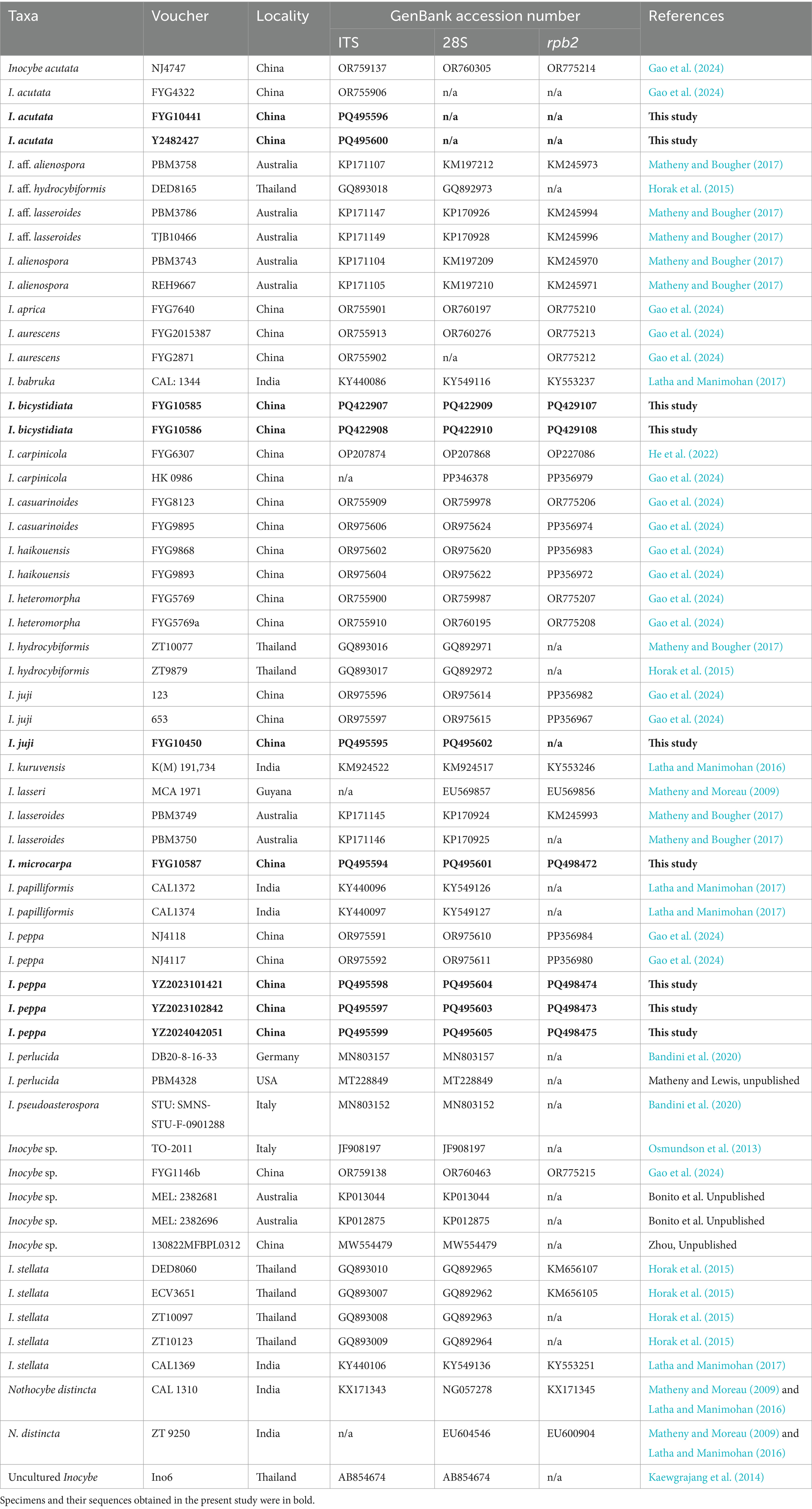
Three loci were identified from the samples in this study, including the rDNA internal transcribed spacer (ITS) region, 28S (LSU, large subunit of ribosomal DNA), and the second largest subunit of DNA-directed RNA polymerase II (rpb2). The genomic DNA was extracted using the NuClean Plant Genomic DNA Kit (ComWin Biotech, Beijing, China) and stored at −20°C. Polymerase chain reaction (PCR) was performed using the primer pairs ITS1-F/ITS4 for the ITS region (Gardes and Bruns, 1993), LR0R/LR7 for the 28S region (Vilgalys and Hester, 1990), and rpb2-6F/rpb2-7.1R for the rpb2 region (Matheny, 2005). The standard PCR reaction mixture consisted of 9.5 μL water, 12.5 μL 2 × Taq Plus Master Mix (Dye) (CW0690L, ComWin Biotech, Beijing, China), 1 μL of each primer, and 1 μL template DNA. The PCR program consisted of an initial heating step of 5 min at 95°C for 4 min; then 35 cycles of denaturation at 94°C for 1 min, annealing at 53°C for 1 min and extension at 72°C for 1 min, with a final extension at 72°C for 8 min (Zhang M. et al., 2022). After amplification, the PCR products were sent to Sangon Biotech (Guangdong and Hainan) Ltd. for purification and sequencing. The sequencing results were analyzed using BioEdit v7.0.9.0 software (Hall, 1999) and assembled using SeqMan v7.1.0 within DNASTAR v7.1.0 (44.1) software (Burland, 2000). The newly generated DNA sequences were submitted to GenBank sequence database.1
2.4 Sequence alignment and phylogenetic analyses
For the phylogenetic analysis, validated sequences of I. sect. Leptocybe were retrieved from GenBank (Matheny and Moreau, 2009; Osmundson et al., 2013; Kaewgrajang et al., 2014; Horak et al., 2015; Latha and Manimohan, 2016, 2017; Matheny and Bougher, 2017; Bandini et al., 2020; He et al., 2022; Gao et al., 2024). Nothocybe distincta (K.P.D. Latha & Manim.) Matheny & K.P.D. Latha was used to root the phylogenetic tree (Latha and Manimohan, 2016). The alignment of the three partitions was performed using the MAFFT online tool2 with the E-INS-i iterative refinement strategy (Katoh et al., 2019). Sequence alignments were manually refined using BioEdit v7.0.9.0 (Hall, 1999). The three individual partitions (ITS, 28S, and rpb2) were concatenated into a single multiple sequence alignment using MEGA v5.02 (Tamura et al., 2011). Maximum likelihood (ML) analyses were performed using the IQ-TREE web server with 1,000 bootstrap replicates of ultrafast bootstrap resampling (Trifinopoulos et al., 2016). The optimal models for each partition in Bayesian Inference (BI) analyses were determined using MrModeltest v2.3 (Nylander, 2004). Finally, BI analyses were performed using MrBayes v3.2.7a (Ronquist et al., 2012), with the selected models applied to each partition. Four Markov chains were run, sampling every 100 generations. The first 25% of the trees were discarded after confirming that the average standard deviation of partition frequencies was less than 0.009 (Ronquist et al., 2012). Results were processed using FigTree v1.4.3 software3, with support values (with ML bootstrap proportions ≥70% and BI posterior probabilities ≥95 or < 95%) displayed on each branch. Phylogenetic results are ultimately displayed and annotated on the tvBOT website4 (Xie et al., 2023).
2.5 Sample preparation for toxin detection
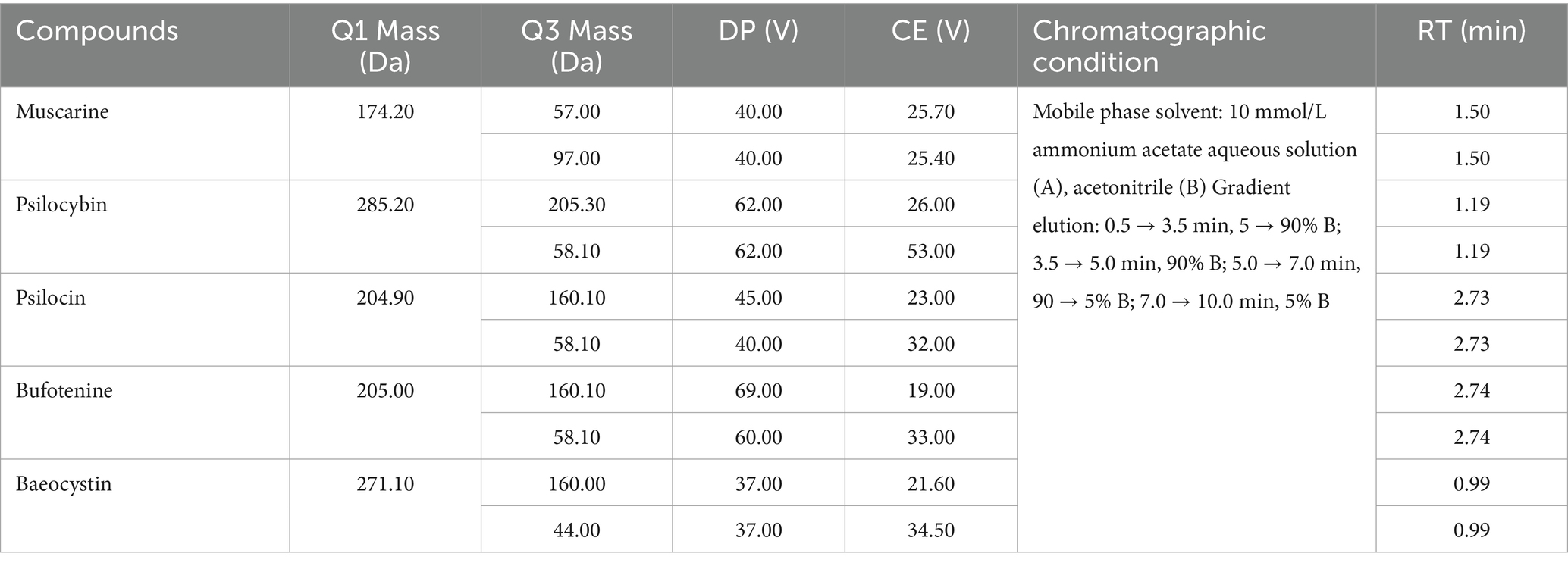
Dried mushroom samples (10 mg) were ground into powder and transferred to a 2 mL centrifuge tube. Then, 2 mL methanol/water (70:30, v/v) was added to the mixture. The tubes were then vortexed for 30 min, placed in an ultrasonic bath (33 Hz, 25°C) for 30 min, and centrifuged at 12,000 rpm for 5 min at 4°C. Extraction solution was filtered through a 0.22 μm microporous membrane. Targeted screening was performed under the optimized UPLC-MS/MS detection conditions (Table 1). Extracts were prepared in the same way as described above and subjected to UPLC-MS/MS analysis. Working standards were prepared by mixing the stock standards with an acetonitrile/water solution (5,95, v/v). Calibration curves were generated with 1.0, 2.0, 5.0, 10.0, 20.0, 50,0, 100.0, and 200.0 ng/mL of the working standards solution using UPLC-MS/MS. Limit of detection (LOD) and limit of quantitation (LOQ) were calculated as the concentrations corresponding to signals that are 3 and 10 times the baseline noise’s standard deviation, respectively (Table 2).
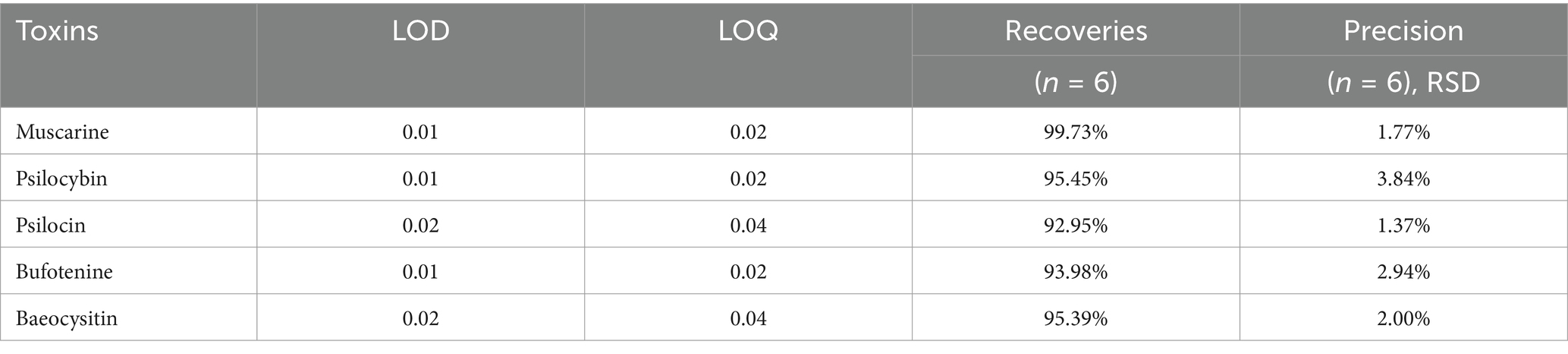
Table 2. The LOD, LOQ, recoveries, relative standard deviations (RSD), and precision of five mushroom toxins.
2.6 UPLC-MS/MS conditions
The liquid chromatography system consisted of two LC-30 AD pumps and a Shimadzu SIL-30 AC autosampler. An AB Sciex Triple Quad 6,500+ system (Applied Biosystems/MDS Analytical Technologies, Foster City, CA, United States) equipped with an electrospray ionization interface was used for mass spectrometric detection. An HSS T3 column (2.1 mm × 100 mm, 1.8 μm) (Waters, United States) was used as the separation column. The flow rate was 0.3 mL/min. The temperature of the analytical column was set at 40°C. The injection volume was 2 μL. Analyst software (v1.6) was used for detection, data acquisition, and processing. The specific UPLC parameters and mass spectral parameters of the fungal toxins are presented in Table 1.
3 Results
3.1 Molecular phylogeny
This study generated 22 new sequences (9 from ITS, 7 from 28S, and 6 from rpb2) and submitted them to GenBank (Table 3). The dataset included 53 taxa and 682 sites for the ITS partition, 52 taxa and 1,265 sites for the 28S partition, and 36 taxa and 613 sites for the rpb2 partition. For the ML analyses, IQTREE automatically selected the following DNA substitution models: ITS (TVM + F + G4), 28S (HKY + F + I + G4), and rpb2 (TNe + G4). ML phylogenetic analysis yielded a final log-likelihood value of −11828.001. MrModeltest identified the best-fitting models as GTR + G for the ITS and GTR + I + G for both the 28S and rpb2 partitions. The BI phylogenetic analysis was completed after 120,000 generations, at which point the mean standard deviation of the split frequencies converged to 0.008840, and the adequate sample size (ESS) reached 365.42. The average potential scale reduction factor (PSRF) was 1.000.
The phylogenetic results generated by ML and BI analyses have a similar topography, so only the ML tree is shown here. As shown in Figure 1, the two new species were placed in separate lineages in I. sect. Leptocybe. Inocybe bicystidiata, nested within the alienospora subclade, and was sister to most species except the basal taxon I. kuruvensis in this subclade. Inocybe microcarpa clustered with several tropical Asian taxa, I. hydrocybiformis, I. aff. hydrocybiformis, I. barbruka, and I. papilliformis in the hydrocybiformis subclade. Newly obtained specimens of I. acutata, I. juji, and I. peppa have been placed with their holotypes or authentic material in their respective lineages.
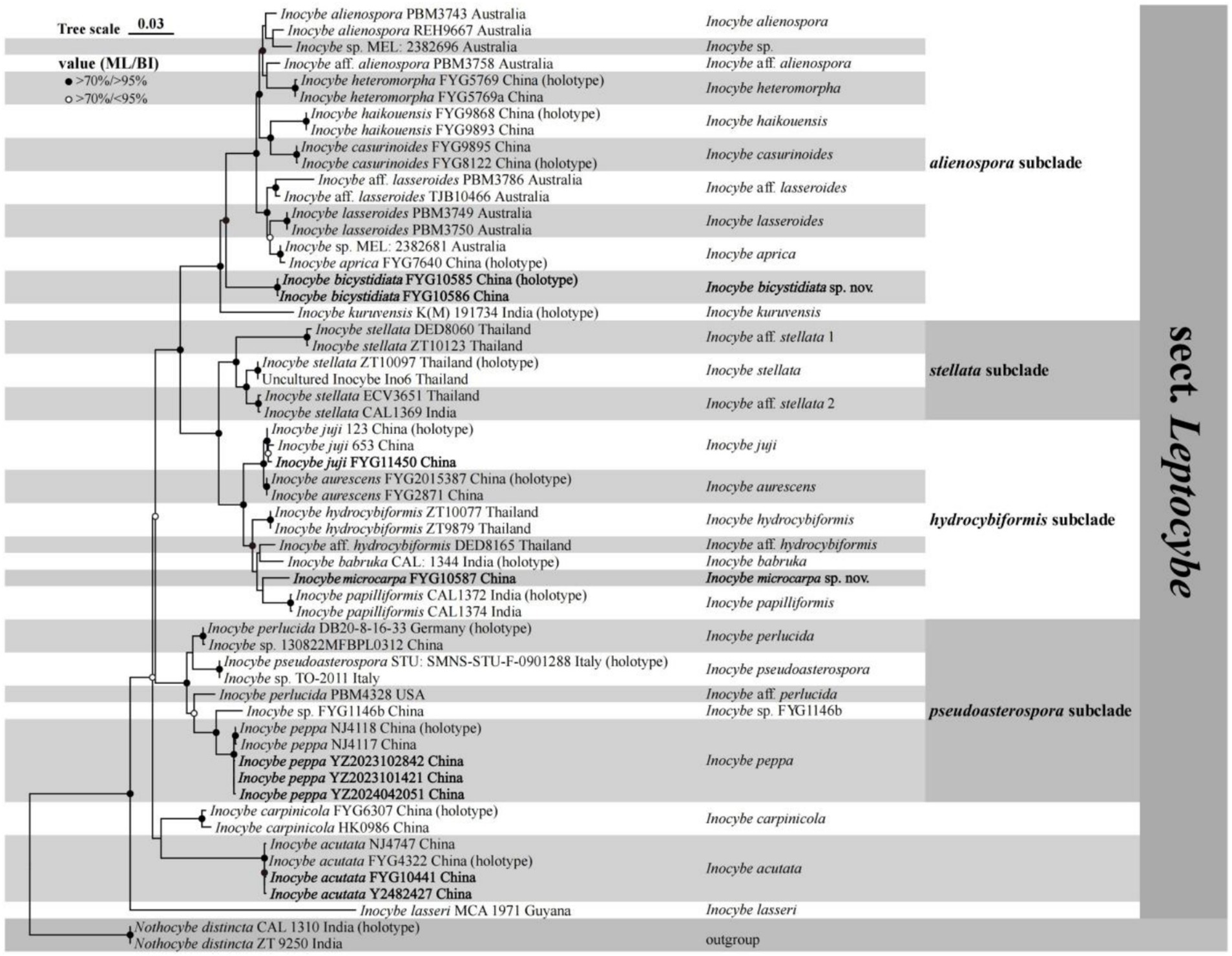
Figure 1. Phylogram generated by maximum likelihood (ML) and Bayesian inference (BI) analysis based on a combined dataset of nuclear genes (ITS, 28S, and rpb2). The tree is rooted with Nothocybe distincta (CAL1310 and ZT9250). Support values (ML-bp ≥ 70% and BI-pp ≥ 95% indicated by black circles; ML-bp ≥ 70% and BI-pp < 95% indicated by white circles centered in black) are shown at the nodes.
3.2 Taxonomy
Inocybe acutata Takah. Kobay. & Nagas., Mycotaxon 48: 461 (1993)
Remarks: Inocybe acutata was originally described from Tottori, subtropical Japan, and subsequently found in China (Anhui, Zhejiang, and Jiangsu provinces). In 2024, we obtained three additional specimens from Jilin (temperate climate), Hubei (subtropical climate), and Guizhou (subtropical climate) provinces. Inocybe acutata is characterized by small and slender basidiomata, spinose basidiospores without saddle-shaped projections, absence of metuloid pleurocystidia, and thin-walled cheilocystidia. A detailed description and line drawings/color plates of I. acutata can be found in Kobayashi (1993) and Gao et al. (2024).
Habitat and ecology: Found in subtropical evergreen deciduous forests or temperate mixed deciduous and coniferous forests.
Distribution: China (Anhui, Zhejiang, Jiangsu, Jilin, Guizhou, and Hubei) and Japan (holotype).
Newly collected specimens: China. Jilin Province: Baishan City, Fusong County, Lushuihe Town, at 42°32′59″N, 128°00′39″E, alt. 649 m, 26 August 2024, leg. Tolgor Bau, Y2482427 (FCAS4073); Guizhou Province: Tongren City, Jiangkou County, Fanjingshan Nature Reserve, at 27°54′40″N, 108°28′32″E, alt. 1,600 m, 21 July 2024, leg. Y.-G. Fan and W.-J. Yu, FYG10441 (FCAS4074); and Hubei Province: Yichang City, Yiling District, in deciduous forest, alt. 1,086 m, 11 June 2024, Y.-P. Ge, L.-J. Wang, J.-W. Guo, G.-Y. Qiu, NJ 5065 (FCAS4078).
Inocybe bicystidiata W.J. Yu, Y.G. Fan & J.L. Gao, sp. nov. Figures 2, 3 Chinese name: 双囊丝盖伞 (Double-cystidium Fiber Cap)

Figure 2. Basidiomata of Inocybe bicystidiata. (a–c) FYG10585 (FCAS4069, holotype); (d) FYG10586 (FCAS4070). Scale bars: (a) and (c–d) = 10 mm; (b) = 1 mm. Photos by Y.-G. Fan.
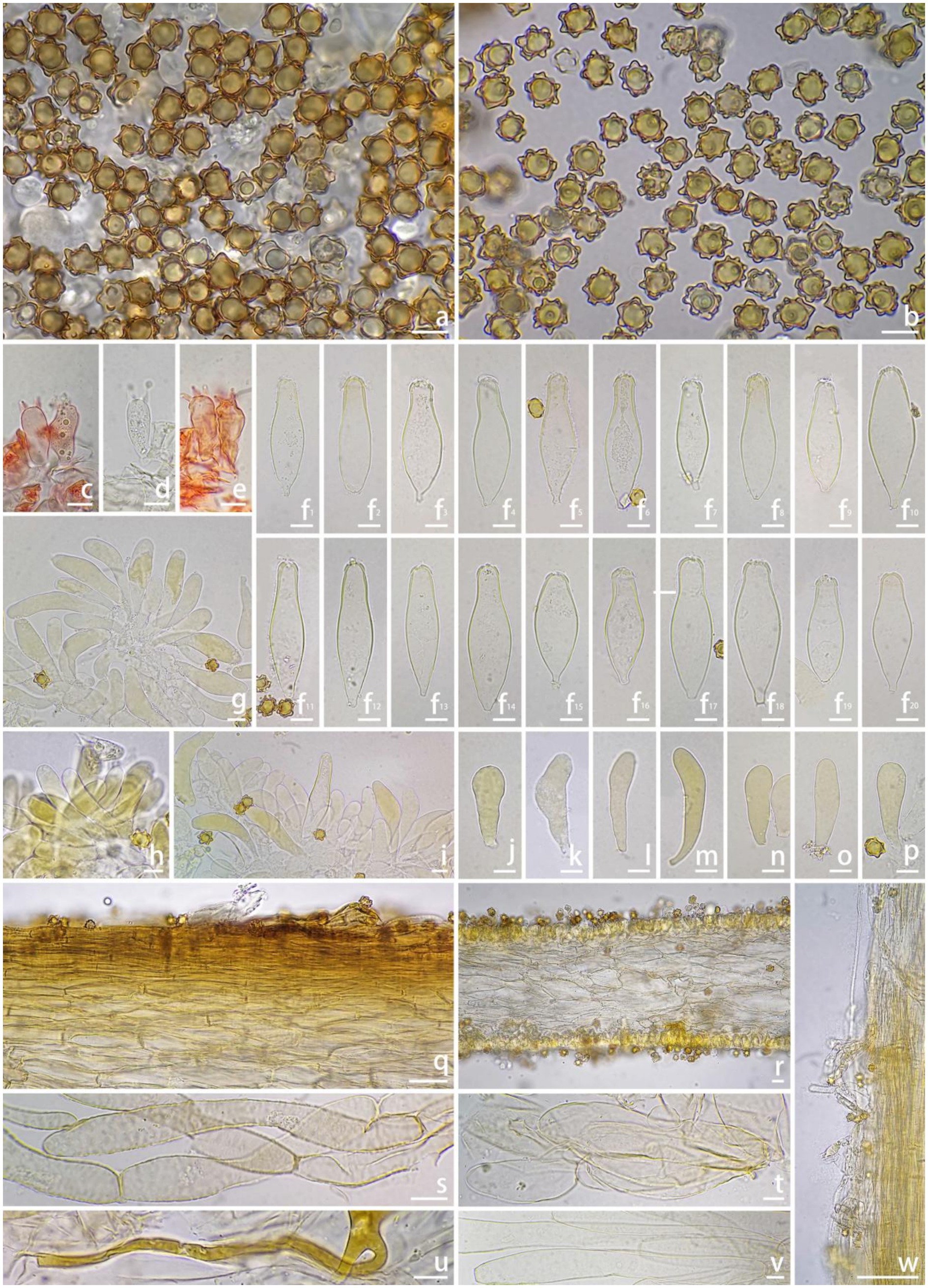
Figure 3. Microscopic features of Inocybe bicystidiata (FCAS4069, holotype). (a,b) Basidiospores; (c–e) Basidia; (f 1 –f 20 ) Pleurocystidia; (g–i) Cheilocystidia; (j–p) Paracystidia in side of lamellae; (q) Pileipellis; (r) Hymenophoral trama; (s) Pileipellis hyphae; (t) Hymenophoral trama hyphae; (u) Oily hyphae; (v) Stipe trama hyphae; (w) Stipitipellis in the apex of stipe. Scale bars: (a–v) = 10 μm; (w) = 100 μm. Images by J.-L. Gao.
Mycobank: MB856828
Etymology: bicystidiata (L.), referring to the coexistence of thick-walled pleurocystidia and scattered thin-walled paracystidia on the lamellar side.
Diagnosis: Inocybe bicystidiata has slender basidiomata, thin- to thick-walled pleurocystidia, thin-walled yellowish-pigmented cheilocystidia, and a fungoid odor. Most similar to I. kuruvensis, but differs in the presence of thick-walled pleurocystidia and thin-walled paracystidia on the lamellar side.
Holotype: China. Yunnan Province: Xishuangbanna Dai Autonomous Prefecture, Mengla County, Xishuangbanna Tropical Rainforest National Park, Wangtianshu Scenic Spot, 21°37′21″N, 101°35′15″E, alt. 711 m, 28 July 2024, in tropical rainforest dominated by Parashorea chinensis Wang Hsie (Dipterocarpaceae), leg. Y.-G. Fan & W.-J. Yu, FYG10585 (FCAS4069), GenBank accession no: ITS (PQ422907); LSU (PQ422909) and rpb2 (PQ429107).
Description: Basidiomata small and slender. Pileus 9–19 mm wide, hemispherical when young, then convex to planoconvex with a small pointed umbo or at times non-umbonate when matured; margin at first inrolled, then depressed to straight; cortina present in young specimens, often with brown analoid remnants from spore deposition; surface dry, dotted scaly around the disc, squamulose with finely recurved fibrils to mid-radius, radially fibrillose to rimulose with uneven streaks elsewhere; uniformly yellowish (4A2–4A4) when young, then yellowish (4B5–4C5) or yellowish-brown (5C4–5C5), brownish (5C6–5D6) to dark brown (5D6–5E6) toward the center when mature; veil remnants yellow (4C6–4D6) to pale brown (5C4–5C5). Lamellae adnexed, subdistant, 2–3 mm wide, alternately distributed with 4–6 tiers of lamellulae; white (4A1) or pale grayish-white (4A4–4B5) at first, yellowish-brown (5C4–5D5) to brown (5D5–5D6), edge pallid, indistinctly fimbriate. Stipe 40–52 × 1.2–2 mm, terete, central, solid, equal with a slightly enlarged base; surface dry, covered with a layer of veil remnants, appressed-fibrillose to silky smooth; yellow (4C6–4D6) to brown (5C3–5D4) when young, brown (5E4–5E5) when mature. Context fleshy in pileus, slightly yellow (4A4–4B4) with a pale brownish (5D6) tinge near the cuticle, 0.3 mm thick at mid-radius, up to 1.5 mm thick under the umbo; fibrillose and striate in stipe, yellowish (4C6–4D6) or slightly brownish (5D6). Odor fungoid.
Basidiospores [100/4/2], (7.0) 8.0–9.44–11.1 (12.2) × (5.9) 7.0–8.31–9.7 (11.2) μm, Q = (1.00) 1.00–1.14–1.29 (1.50), Qm ± SD = 1.14 ± 0.099 with spines, (4.8) 5.0–6.29–7.2 (9.0) × (3.9) 4.2–5.36–6.7 (8.0) μm, Q = (1.00) 1.00–1.18–1.38 (1.55), Qm ± SD = 1.18 ± 0.121 without spines; nodulose with spines that sometimes saddle-shaped projections or bifurcate, apiculus distinct, yellowish in 5% KOH, thick-walled, with yellow ovoid contents. Basidia 18–35 × 9–11 μm, subclavate to clavate, apex obtuse, bases usually tapered, with 4- or 2-sterigmata 2–9 μm length, colorless to slightly yellowish. Pleurocystidia 43–62 × 14–22 μm, abundant, mostly fusiform to utriform, apices rounded or obtuse, base usually tapering to a small pedicel, thick-walled, walls pale yellowish, up to 1.5 μm thick. Paracystidia on the lamellar side 32–61 × 9–15 μm, mostly clavate to broadly clavate, apices rounded or obtuse, base usually tapering to a small pedicel, colorless to slightly yellowish. Cheilocystidia 32–95 × 7–18 μm, abundant, subclavate to clavate, sometimes cylindrical or broadly clavate, apices rounded or obtuse, base tapered, thin-walled, colorless to pale yellowish. Hymenophoral trama 50–125 μm thick, subregular to regular, consisting of subinflated to inflated hyphae measuring 17–33 μm wide, colorless, smooth, thin-walled, wall slightly yellowish. Pileipellis a cutis, 35–48 μm wide, subregular to regular, yellowish-brown in mass, consisting of cylindrical hyphae measuring 12–15 μm wide, pale yellowish, walls slightly yellowish. Pileal trama 100–200 μm wide, regular, hyphae subinflated, colorless, 18–37 μm wide. Stipitipellis regular, hyphae cylindrical, 4–15 μm wide, encrusted, colorless. Stipe trama regularly arranged, composed of colorless, thin-walled, cylindrical hyphae 13–27 μm wide. Caulocystida not observed. Oily hyphae 3–6 μm wide, cylindrical, pale yellow to yellow, smooth, diverticulate, in hymenophoral trama. Clamp connections present in all tissues.
Habitat and ecology: Scattered on mosses in tropical rainforests dominated by P. chinensis (Dipterocarpaceae).
Distribution: Known from the type locality in Yunnan Province of China.
Additional specimens examined: CHINA. Yunnan Province: Xishuangbanna Dai Autonomous Prefecture, Xishuangbanna Tropical Rainforest National Park, Wangtianshu Scenic Spot, 21°37′21″N, 101°35′15″E, alt. 711 m, 28 July 2024, in tropical rainforest dominated by P. chinensis Wang Hsie (Dipterocarpaceae), leg. Y.-G. Fan & W.-J. Yu, FYG10586 (FCAS4070).
Remarks: Inocybe bicystidiata was recently found in Yunnan’s P. chinensis-dominated tropical rainforest. The non-umbonate pileus, featuring dotted, appressed to more or less raised squamulose, along with the slender continuate stipes, makes the new species impressive in the field. Microscopically, it has subglobose basidiospores with subconical or saddle-shaped nodules typically protruding about 2 μm long, thin-walled cheilocystidia, and thick-walled pleurocystidia together with scattered thin-walled paracystidia on the sides of the lamellae. The new species is phylogenetically placed in the alienospora subclade of I. sect. Leptocybe (Gao et al., 2024). This subclade includes I. lasseroides, I. alienospora, I. kuruvensis, and four recently described taxa from China, namely, I. aprica, I. casuarinoides, I. haikouensis, I. heteromorpha. Species in this subclade share thick-walled pleurocystidia and thin-walled cheilocystidia, usually with yellow pigments. In contrast to I. bicystidiata, the four recently described Chinese species exhibit brown to umber-brown and smaller basidiomata, appressed-fibrillose to appressed scaly pileus, apparently different outlines in the basidiospores, different associated plants (Casuarina or Fagaceae trees) and a geographical distribution in the Hainan province of China (Gao et al., 2024). The remaining three species are similar to the new species in having raised scales in the pileus, but I. alienospora has subumbonate pileus, more flanged or saddle-shaped nodules in the basidiospores and thicker walled pleurocystidia (Horak, 1979; Matheny and Bougher, 2017); I. lasseroides has umbonate pileus, ovoid–fusoid pleurocystidia with thicker walls (Matheny and Bougher, 2017); I. kuruvensis has dark brown pileus with erect scales, broadly fusiform pleurocystidia with thicker walls (Latha and Manimohan, 2017).
Inocybe juji Y.G. Fan, Y.P. Ge & J.L. Gao, Mycology 15(4): 28 (2024)
Remarks: Inocybe juji was originally described from Anhui province, subtropical China. In 2024, we obtained an additional specimen from Hainan province (tropical climate). Inocybe juji is characterized by a dirty-yellow to brownish-yellow pileus, spinose basidiospores with saddle-shaped projections, and thin-walled and yellowish reflecting cheilo- and pleurocystidia. A detailed description and color plates of I. juji can be found in Gao et al. (2024).
Habitat and ecology: Scattered in subtropical evergreen broad-leaved forests dominated by fagaceous trees or in tropical cloud forests dominated by fagaceous trees.
Distribution: Anhui and Hainan Provinces in China.
Newly collected specimens: China. Hainan Province: Wuzhishan City, Nansheng Town, Wuzhishan station of Hainan Tropical Rain Forest National Park, at 109°40′43″E, 18°51′53″N, alt. 690 m, 2 August 2024, leg. J.-L. Gao, G.-H. Liu, and X. Chen, FYG10450 (FCAS4072).
Inocybe microcarpa W.J. Yu, Y.G. Fan & L.J. Gao, sp. nov. Figure 4

Figure 4. Basidiomata and microscopic features of Inocybe microcarpa (FCAS4071, holotype). (a,b) Basidiomata; (c,d) Basidiospores; (e,f) Basidia; (g–i) Cheilocystidia; (j) Pileipellis hyphae; (k) Stipitipellis hyphae; (l) Stipitipellis in the apex of stipe. Scale Bars: (a,b) = 10 mm; (c–l) = 10 μm. Images: (a,b) by Y.-G. Fan; (c–l) by J.-L. Gao.
Chinese name: 小果丝盖伞 (Tiny Fiber Cap)
Mycobank: MB856831
Etymology: microcarpa (L.), in reference to the conspicuously small basidiomata.
Diagnosis: Inocybe microcarpa has very small and slender basidiomata; grooved pileus, ginger-yellow veil remnants on stipe surface, subglobose to ovoid, spinose with simple or sometimes branched spines reaching up to 3.1 μm on basidiospores, and thin-walled, yellowish pigmented cheilocystidia. Most similar to I. hydrocybiformis, but differs in conspicuously smaller basidiomata, ginger-colored stipes, larger basidiospores, and shorter cheilocystidia.
Holotype: China. Yunnan Province: Xishuangbanna Dai Autonomous Prefecture, Xishuangbanna Tropical Rainforest National Park, Wangtianshu Scenic Spot, 21°37′21″N, 101°35′15″E, alt. 711 m, in tropical rainforest dominated by P. chinensis (Dipterocarpaceae), 28 July 2024, leg. Y.-G. Fan & W.-J. Yu, FYG10587 (FCAS4071), GenBank accession no: ITS (PQ495594); LSU (PQ495601); and rpb2 (PQ498472).
Description: Basidiomata very small and slender. Pileus 4–5 mm wide, initially obtusely conical to campanulate, hemispherical to convex or broadly convex with a small subacute umbo, margin incurved when young, depressed to straight when mature, cortina present in young specimens; surface dry, glabrous or with finely fibrils toward the disc, radically fibrillose with grooves outward; uniformly brownish (5D4–5D5) to brown (6C4–6C5) when young, then yellowish brown (4C4–4D5) to brown (6C4–6C5), brownish (5C4–5C5) or yellowish brown (4C4–4D5) to yellowish (4B5–4B6) toward the center when mature. Lamellae adnexed, distant, 0.9–1 mm wide, alternately distributed with 2–3 tiers lamellulae; color initially yellowish brown (4C4–4D5) to yellow (4B4–4C5), then yellowish brown (4C4–4D5), yellowish (4A5–4B6) when mature, edge pale yellowish (4A3–4A4), not fimbriate. Stipe 10–12 × 0.8–1 mm, terete, central, solid, equal with a slightly enlarged base; surface dry, covered with a layer of yellowish (4B5–4C5) fibrils that from veil remnants at the apex; pale yellowish (4B3–4C4) to yellowish brown (4C4–4D5) when young, brownish (5C4–5C5) to darkly brownish (5E5–5E6) when mature. Context thin in pileus, translucent and pale yellowish in stipes. Odor not recorded.
Basidiospores [100/2/1], (9.9) 10.2–11.55–12.7 (13.1) × (8.7) 9.5–10.60–11.9 (12.2) μm, Q = (1.01) 1.02–1.09–1.20 (1.24), Qm ± SD = 1.09 ± 0.052, subglobose to ovoid, spinose with simple or sometimes bifurcate spines up to 3.1 μm, pale yellowish in 5% KOH, thick-walled, sometimes with pale yellow ovoid contents. Basidia 27–38 × 10–15 μm, subclavate to clavate, apex obtuse, bases usually tapered, with 2- or 4-sterigmata 3–9 μm long, colorless to pale yellowish, sometimes golden yellowish. Cheilocystidia 28–47 × 8–12 μm, abundant, narrowly clavate to clavate, sometimes broadly clavate, apices rounded or obtuse, base tapered, thin-walled, colorless to pale yellowish, sometimes with golden yellowish pigments. Hymenophoral trama subregular to regular, consisting of cylindrical to subinflated hyphae 9–23 μm wide, colorless, smooth, thin-walled, walls slightly yellowish. Pileipellis a cutis, regular, yellowish to yellowish-brown in mass, consisting of cylindrical hyphae measured 5–10 μm wide, pale yellowish, smooth, thin-walled, walls pale yellowish. Pileal trama regular, hyphae cylindrical to subinflated, colorless, 10–15 μm wide. Stipitipellis regular, hyphae cylindrical, 4–9 μm wide, encrusted, smooth, colorless. Caulocystida not observed. Oily hyphae not observed. Clamp connections present in all tissues.
Habitat and ecology: Scattered on mosses in tropical rainforests dominated by P. chinensis (Dipterocarpaceae).
Distribution: Known from the type locality in Yunnan Province of China.
Remarks: Inocybe microcarpa is easily overlooked because of its very small basidiomata. It occurs on moss beds in tropical rainforests dominated by P. chinensis. The striped pileus and the ginger-colored fibrils on the stipes make this species conspicuous in the field. Microscopically, it has spinose basidiospores with bifurcate projections and thin-walled, yellow-pigmented cheilocystidia, and no pleurocystidia. Phylogenetically, I. microcarpa is placed in the subclade hydrocybiformis and tends to cluster with the lineage formed by I. babruka, I. papilliformis, and I. hydrocybiformis. These three species have similar spinose basidiospores with forked or saddle-shaped projections. However, I. babruka, described from Kerala (tropical India), has larger basidiomata, shorter projections in basidiospores, and longer cheilocystidia described as “gloeocystidia,” and a habitat near Hopea ponga trees (Dipterocarpaceae) (Latha and Manimohan, 2017); I. hydrocybiformis, described from Singapore and Malaysia and subsequently found in India, has larger basidiomata, shorter projections on average (up to 2.5 μm), longer cheilocystidia, and the presence of caulocystidia (Horak, 1979; Horak et al., 2015; Pradeep et al., 2016); I. papilliformis described from tropical India has larger basidiomata, an acute umbo in the pileus, significantly larger basidiospores measuring 15–19.5 × 14–18 μm, thick-walled pleurocystidia as metuloids, and a habitat on sandy soil under H. parviflora and Vateria indica (Dipterocarpaceae) (Pradeep et al., 2016; Latha and Manimohan, 2017).
Inocybe peppa Y.G. Fan, Y.P. Ge, J.L. Gao & W.J. Yu, Mycology 15(4): 32 (2024)
Remarks: Inocybe peppa was originally described from Zhejiang, subtropical China. Here, we obtained four additional specimens from Sichuan (subtropical climate) and Shandong (warm temperate climate) provinces in 2023 and 2024. Inocybe peppa is characterized by small basidiomata, campanulate pileus, stellate basidiospores, and fusoid to broadly fusoid cheilo- and pleurocystidia. A detailed description and color plates of Inocybe peppa can be found in Gao et al. (2024).
Habitat and ecology: Scattered in subtropical evergreen broad-leaved forests.
Distribution: Zhejiang, Sichuan, and Shandong Provinces in China.
Newly collected specimens: China. Sichuan Province: Dazhou City, Xuanhan County, at 31°23′49″N, 107°34′42″E, alt. 500 m, 14 October 2023, leg. X.-M. Yang, YZ2023101421 (FCAS4076), 28 October 2023, leg. X.-M. Yang, YZ2023102842 (FCAS4075), 20 April 2024, leg. X.-M. Yang, YZ2024042051 (FCAS4077); Shandong Province, Tai’an, Dajinkou Town, Mount Tai, Yuquan Temple, at 36°18′15″N, 117°05′12″E, alt. 554 m, 20 July 2023, leg. Y.-P. Ge & Q. Na, HK1152 (FCAS4079), GenBank: ITS (PQ676244).
3.3 Method validation and toxin detection results
UPLC-MS/MS analyses demonstrated robust method performance for toxin identification. Calibration curves of muscarine, psilocybin, baeocystin, psilocin, and bufotenine exhibited excellent linearity (R2 > 0.99; Figure 5). The precision of the analytical method was satisfactory, with a relative standard deviation of less than 5% for replicate measurements (Table 2). Method validation using Lentinula edodes spiked samples showed recoveries of 92.95%–99.73% and repeatability (RSD) within 2.60%–6.13% (Table 2), confirming the protocol’s suitability for quantifying these five toxins. All examined specimens of I. sect. Leptocybe (n = 32) underwent UPLC-MS/MS profiling. Targeted screening revealed no detectable levels of muscarine, psilocybin, psilocin, bufotenine, or baeocystin across 10 species within the section.
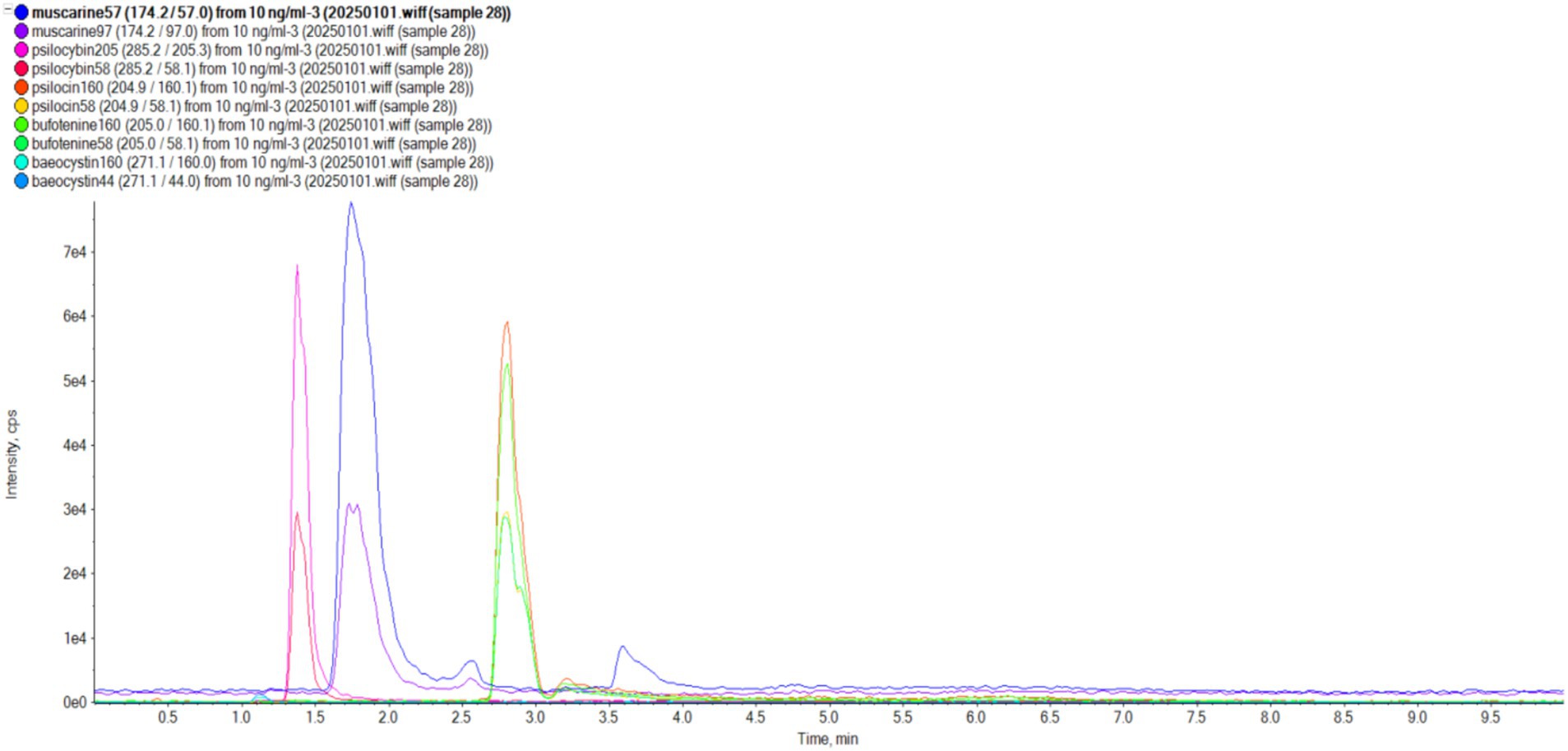
Figure 5. Standards of five mushroom toxins (muscarine, psilocybin, psilocin, bufotenine, and baeocystin).
4 Discussion
The two new species, I. bicystidiata and I. microcarpa, were both discovered in tropical forests dominated by P. chinensis, a tree species of the Dipterocarpaceae family. Parashorea chinensis is also recognized as an iconic species in the tropical rainforests of China, but has a limited distribution in Yunnan and Guangxi provinces (Han et al., 2020). The tree species faces survival challenges due to changes in climate and native site conditions caused by human activities (Dai et al., 2017), and has been listed as a Class I endangered plant in China and by the International Union for Conservation of Nature (IUCN) (Dai et al., 2017; Li et al., 2004). The two new Inocybe species are hypothesized to be mycorrhizal partners of P. chinensis, but require further verification. In addition, new geographical distributions of I. acutata, I. juji, and I. peppa are reported based on recently collected specimens. Notably, I. acutata was discovered in Jilin Province, extending its range northward from subtropical southern China to temperate northeast China. Similarly, I. peppa was recorded in Shandong Province, a region characterized by a warm temperate monsoon climate. Additionally, I. juji was confirmed in the tropical montane cloud forests of Hainan.
Species of the I. sect. Leptocybe generally have small brown basidiomata, cortinatae and slender stipes, moderately crowded lamellae, thin-walled cheilocystidia and nodose basidiospores, usually with forked or saddle-shaped projections (Gao et al., 2024). However, notable exceptions occur within the section. For instance, the type species I. acutata exhibits simple spinose basidiospores and exclusively thin-walled pleurocystidia/cheilocystidia (Kobayashi, 1993); while I. juji and I. aurescens lack metuloids but retain thin-walled pleurocystidia and cheilocystidia. In this study, I. bicystidiata displays a unique combination: thick-walled pleurocystidia coexisting with thin-walled paracystidia on lamellar sides (Figure 3). While its paracystidia resemble those of I. acutata, the latter species completely lacks lamellar metuloids. Re-examination of Chinese I. acutata specimens revealed that its purported “paracystidia” structurally resemble basidioles, being nearly confluent with the hymenium layer. These elements can nevertheless be distinguished from true basidioles by their oily cytoplasmic inclusions and distinct subapical pigmentation in 1% Congo Red (Gao et al., 2024). Contrasting cystidial patterns are observed in the hydrocybiformis subclade: I. hydrocybiformis was originally described as lacking pleurocystidia (Horak, 1979), yet Thai specimens showed scattered thin-walled pleurocystidia resembling cheilocystidia (Horak et al., 2015); I. papilliformis initially reported both metuloid cheilo- and pleurocystidia, but subsequent studies found no pleurocystidia (Latha and Manimohan, 2017). Inocybe microcarpa in our study showed no discernible thin-walled elements distinct from basidia/basidioles on lamellar sides.
Muscarine and psilocybin are the primary toxins in Inocybe fruiting bodies (Stijve et al., 1985; Stijve and Kuyper, 1985; Kosentka et al., 2013; Xu et al., 2020; Li, S. N. et al. 2021). Secondary psychotropic compounds (e.g., baeocystin and psilocin) and amatoxins have been sporadically reported in select species (Semerdzieva et al., 1986; Gartz, 1987; Kosentka et al., 2013; Ren et al., 2016). Despite the genus’ high species diversity, toxin profiles remain understudied, with only a limited subset of taxa analyzed to date. A literature review identified 13 Inocybe species testing negative for both muscarine and psilocybin: I. appendiculata, I. fraudans, Inocybe aff. fraudans, I. godeyi, I. grammata (= I. albodisca), I. granulosipes, I. incarnata, I. luteifolia, I. nigrescens, I. subexilis, I. tahquamenonsis, I. viscata, and I. xanthomelas (Kosentka et al., 2013).
Notably, our UPLC-MS/MS analyses detected none of the five targeted toxins (muscarine, psilocybin, psilocin, bufotenine, baeocystin) across the 10 examined species of I. sect. Leptocybe (Table 4). These species represent five of the seven major phylogenetic lineages within the section (Figure 1), though materials from the neotropical I. lasseri and the I. stellata subclade were unavailable for testing. The newly described I. microcarpa was excluded from toxin screening due to insufficient biomass.
Muscarine production was considered an ancestral trait in the Inocybe s.s.-Pseudosperma-Nothocybe clade, but exhibits multiple evolutionary losses (Kosentka et al., 2013). This phylogenetic plasticity complicates definitive toxin status attribution at the sectional level. Consequently, the absence of detected toxins in I. sect. Leptocybe requires validation through expanded sampling, particularly for unscreened lineages (e.g., stellata subclade) and chemically uncharacterized species.
Data availability statement
The datasets presented in this study can be found in the online repository https://www.ncbi.nlm.nih.gov/genbank/ and the accession numbers are mentioned in Table 3.
Author contributions
J-LG: Data curation, Formal analysis, Funding acquisition, Methodology, Resources, Writing – original draft. X-PW: Conceptualization, Data curation, Formal analysis, Methodology, Supervision, Writing – review & editing. Y-LZ: Formal analysis, Methodology, Writing – review & editing. W-JY: Funding acquisition, Resources, Supervision, Writing – review & editing. Y-GF: Conceptualization, Funding acquisition, Resources, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the Scientific Research Project of Hainan Higher Education Institutions (Hnky2023ZD-8), the Hainan Province Science and Technology Special Fund (ZDYF2024SHFZ129), the National Science Foundation of China (32260005 and 32470008), the Hainan Provincial Natural Science Foundation (323MS044) (Y-GF and W-JY), and the Postgraduate Innovation Fund Project of the Hainan Medical University (HYYS2022B09 and Qhys2023-483).
Acknowledgments
We thank Prof. Tolgor Bau (Jilin Agricultural University), Yu-Peng Ge (Ludong University), and Mrs. Xianmei Yang for providing valuable specimens. We also thank Dr. Chunying Deng (Guizhou Academy of Sciences), Mr. Jing Wang (Guizhou Institute of Biology), and Prof. Jiang Xu (Guangdong Academy of Agricultural Sciences) for their kind help in fieldwork.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
1. ^https://www.ncbi.nlm.nih.gov/genbank/
2. ^https://mafft.cbrc.jp/alignment/server/
References
Bandini, D., Oertel, B., Schüssler, C., and Eberhardt, U. (2020). Noch mehr risspilze: fünfzehn neue und zwei wenig bekannte arten der gattung Inocybe. Mycol. Bavar. 20, 13–101.
Burland, T. G. (2000). DNASTAR’s lasergene sequence analysis software. Methods Mol. Biol. 132, 71–91. doi: 10.1385/1-59259-192-2:71
Chandrasekharan, B., Pradeep, C., and Vrinda, B. (2020). Inocybe poisoning from Kerala-a case study. J. Mycopathol. Res. 57, 255–258.
Dai, W., Zhou, L., and Yang, M. (2017). Research and utilization of Dipterocarpaceae plants in China (in chinese). World Forestry Res. 30, 46–51. doi: 10.13348/j.cnki.sjlyyj.2017.0056.y
Deng, L. S., Kang, R., Zeng, N. K., Yu, W. J., Chang, C., Xu, F., et al. (2021a). Two new Inosperma (Inocybaceae) species with unexpected muscarine contents from tropical China. Myco Keys 85, 87–108. doi: 10.3897/mycokeys.85.71957
Deng, L. S., Yu, W. J., Zeng, N. K., Liu, L. J., Liu, L. Y., and Fan, Y. G. (2021b). Inosperma subsphaerosporum (Inocybaceae), a new species from Hainan, tropical China. Phytotaxa 502, 169–178. doi: 10.11646/phytotaxa.502.2.5
Deng, L. S., Yu, W. J., Zeng, N. K., Zhang, Y. Z., Wu, X. P., Li, H. J., et al. (2022). A new muscarine-containing Inosperma (Inocybaceae, Agaricales) species discovered from one poisoning incident occurring in tropical China. Front. Microbiol. 13:923435. doi: 10.3389/fmicb.2022.923435
Gao, J. L., Ge, Y. P., Matheny, P. B., He, P. M., Wu, X. P., Bau, T., et al. (2024). A phylogeny of the Inocybe alienospora group (Agaricales) with emphasis on seven new species from China and emendation of sect. Leptocybe. Mycology 15, 1–44. doi: 10.1080/21501203.2024.2380069
Gardes, M., and Bruns, T. D. (1993). ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Mol. Ecol. 2, 113–118. doi: 10.1111/j.1365-294X.1993.tb00005.x
Gartz, J. (1987). Variation der Alkaloidmengen in Fruchtkörpern von Inocybe aeruginascens. Planta Med. 53, 539–541. doi: 10.1055/s-2006-962805
Ge, Y. P., Liu, Z. W., Zeng, H., Cheng, X. H., and Na, Q. (2021). Updated description of Atheniella (Mycenaceae, Agaricales), including three new species with brightly coloured pilei from Yunnan Province, Southwest China. Myco Keys 81, 139–164. doi: 10.3897/mycokeys.81.67773
Han, X., Huang, Z., Cheng, F., and Yang, M. (2020). Physiochemical properties and microbial community characteristics of izosphere soil in Parashorea chinensis plantatio (in chinese). Chin. J. Appl. Ecol. 31, 3365–3375. doi: 10.13287/j.1001-9332.202010.019
Hall, T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. Nucleic Acids Symp. Ser. 41, 95–98.
He, P. M., Fan, Y. G., Deng, L. S., and Yu, W. J. (2022). Inocybe carpinicola (Inocybaceae, Agaricales), a new nodulose-spored species from Hainan Province, China. Phytotaxa 575, 79–88. doi: 10.11646/phytotaxa.575.1.5
Horak, E., Matheny, P. B., Desjardin, D. E., and Soytong, K. (2015). The genus Inocybe (Inocybaceae, Agaricales, Basidiomycota) in Thailand and Malaysia. Phytotaxa 230, 201–238. doi: 10.11646/phytotaxa.230.3.1
Hu, J. H., Yu, W. J., Deng, L. S., Fan, Y. G., Bau, T., Tang, L. P., et al. (2023). The detection of major clades and new species of Mallocybe (Inocybaceae, Agaricales) from China with elongate cheilocystidia. Mycol. Prog. 22:15. doi: 10.1007/s11557-022-01854-5
Kaewgrajang, T., Sangwanit, U., Kodama, M., and Yamato, M. (2014). Ectomycorrhizal fungal communities of Dipterocarpus alatus seedlings introduced by soil inocula from a natural forest and a plantation. J. Fungal Res. 19, 260–267. doi: 10.1007/s10310-013-0408-z
Katoh, K., Rozewicki, J., and Yamada, K. D. (2019). MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 20, 1160–1166. doi: 10.1093/bib/bbx108
Kornerup, A., and Wanscher, J. H. (1978). Methuen handbook of colour. 3rd Edn. London: Eyre Methuen.
Kosentka, P., Sprague, S. L., Ryberg, M., Gartz, J., May, A. L., Campagna, S. R., et al. (2013). Evolution of the toxins muscarine and psilocybin in a family of mushroom-forming fungi. PLoS One 8:e64646. doi: 10.1371/journal.pone.0064646
Latha, K. P. D., and Manimohan, P. (2016). Five new species of Inocybe (Agaricales) from tropical India. Mycologia 108, 110–122. doi: 10.3852/14-358
Li, Q., He, T., and Xu, Z. (2004). Generic relationships of Parashorea chinensis Wang Hsie (Dipterocarpaceae) based on cpDNA sequences. Taxon. 53, 461–466. doi: 10.2307/4135622
Li, S. N., Xu, F., Jiang, M., Liu, F., Wu, F., Zhang, P., et al. (2021). Two new toxic yellow Inocybe species from China: morphological characteristics, phylogenetic analyses and toxin detection. MycoKeys 81, 185–204. doi: 10.3897/mycokeys.81.68485
Li, H. J., Zhang, H. S., Zhang, Y. Z., Zhang, K. P., Zhou, J., Yin, Y., et al. (2020). Mushroom poisoning outbreaks-China, 2019. China CDC Wkly. 2, 19–24. doi: 10.46234/ccdcw2020.005
Li, H. J., Zhang, Y. Z., Zhang, H. S., Zhou, J., Chen, Z. H., Liang, J. Q., et al. (2024). Mushroom poisoning outbreaks-China, 2023. China CDC Wkly. 6, 64–68. doi: 10.46234/ccdcw2024.014
Li, H. J., Zhang, Y. Z., Zhang, H. S., Zhou, J., Liang, J. Q., Yin, Y., et al. (2023). Mushroom poisoning outbreaks-China, 2022. China CDC Wkly. 5, 45–50. doi: 10.46234/ccdcw2023.009
Li, H. J., Zhang, H. S., Zhang, Y. Z., Zhou, J., Yin, Y., He, Q., et al. (2021). Mushroom poisoning outbreaks-China, 2020. China CDC Wkly. 3, 41–45. doi: 10.46234/ccdcw2021.014
Li, H. J., Zhang, H. S., Zhang, Y. Z., Zhou, J., Yin, Y., He, Q., et al. (2022). Mushroom poisoning outbreaks-China, 2021. China CDC Wkly. 4, 35–40. doi: 10.46234/ccdcw2022.010
Liu, Z. W., Na, Q., Cheng, X. H., Wu, X. M., and Ge, Y. P. (2021). Mycena yuezhuoi sp. nov. (Mycenaceae, Agaricales), a purple species from the peninsula areas of China. Phytotaxa 511, 148–162. doi: 10.11646/phytotaxa.511.2.3
Matheny, P. B. (2005). Improving phylogenetic inference of mushrooms with RPB1 and RPB2 nucleotide sequences (Inocybe; Agaricales). Mol. Phylogenet. Evol. 35, 1–20. doi: 10.1016/j.ympev.2004.11.014
Matheny, P. B., and Bougher, N. L. (2017). Fungi of Australia: Inocybaceae. Canberra: CSIRO Publishing.
Matheny, P. B., Hobbs, A. M., and Esteve-Raventós, F. (2020). Genera of Inocybaceae: new skin for the old ceremony. Mycologia 112, 83–120. doi: 10.1080/00275514.2019.1668906
Matheny, P. B., and Kudzma, L. V. (2019). New species of Inocybe (Agaricales) from eastern North America. J. Torrey. Bot. Soc. 146, 213–235. doi: 10.3159/TORREY-D-18-00060.1
Matheny, P. B., Kudzma, L. V., Graddy, M. G., Mardini, S. M., Noffsinger, C. R., Swenie, R. A., et al. (2023). A phylogeny for north American Mallocybe (Inocybaceae) and taxonomic revision of eastern north American taxa. Fungal Syst. Evol. 12, 153–202. doi: 10.3114/fuse.2023.12.09
Matheny, P. B., and Moreau, P. (2009). A rare and unusual lignicolous species of Inocybe (Agaricales) from eastern North America. Brittonia 61, 163–171. doi: 10.1007/s12228-008-9066-4
Na, Q., Hu, Y. P., Zeng, H., Song, Z. Z., Ding, H., Cheng, X. H., et al. (2022). Updated taxonomy on Gerronema (Porotheleaceae, Agaricales) with three new taxa and one new record from China. MycoKeys 89, 87–120. doi: 10.3897/mycokeys.89.79864
Nylander, J. (2004). MrModeltest V2. Program distributed by the author. Bioinformatics 24, 581–583. doi: 10.1093/bioinformatics/btm388
Osmundson, T. W., Robert, V. A., Schoch, C. L., Baker, L. J., Smith, A., Robich, G., et al. (2013). Filling gaps in biodiversity knowledge for macrofungi: contributions and assessment of an herbarium collection DNA barcode sequencing project. PLoS One 8:e62419. doi: 10.1371/journal.pone.0062419
Parnmen, S., Nooron, N., Leudang, S., Sikaphan, S., Polputpisatkul, D., Pringsulaka, O., et al. (2021). Foodborne illness caused by muscarine-containing mushrooms and identification of mushroom remnants using phylogenetics and LC-MS/MS. Food Control 128:108182. doi: 10.1016/j.foodcont.2021.108182
Pradeep, C. K., Vrinda, K. B., Varghese, S. P., Korotkin, H. B., and Matheny, P. B. (2016). New and noteworthy species of Inocybe (Agaricales) from tropical India. Mycol. Prog. 15:24. doi: 10.1007/s11557-016-1174-z
Ren, J. L., Bau, T., and Bao, H. Y. (2016). Distribution characteristics of amatoxins in fungi excluding Amanita from Northeast China. Mycosystema 35, 1080–1098. doi: 10.13346/j.mycosystema.150140
Ronquist, F., Teslenko, M., Mark, P., Ayres, D. L., Darling, A., Höhna, S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Semerdzieva, M., Wurst, M., Koza, T., and Gartz, J. (1986). Psilocybin in Fruchtkörpern von Inocybe aeruginascens. Planta Med. 52, 83–85. doi: 10.1055/s-2007-969085
Stijve, T., Klán, J., and Kuyper, T. W. (1985). Occurrence of psilocybin and baeocystin in the genus Inocybe (Fr.) Fr. Persoonia 12, 469–473.
Stijve, T., and Kuyper, T. W. (1985). Occurrence of psilocybin in various higher fungi from several European countries. Planta Med. 51, 385–387. doi: 10.1055/s-2007-969526
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Trifinopoulos, J., Nguyen, L. T., Haeseler, A., and Minh, B. Q. (2016). W-IQ-TREE: a fast online phylogenetic tool for maximum like lihood analysis. Nucleic Acids Res. 44, W232–W235. doi: 10.1093/nar/gkw256
Vilgalys, R., and Hester, M. (1990). Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 172, 4238–4246. doi: 10.1128/jb.172.8.4238-4246.1990
Xie, J. M., Chen, Y. R., Cai, G. J., Cai, R. L., Hu, Z., and Wang, H. (2023). Tree visualization by one Table (tv BOT): a web application for visualizing, modifying and annotating phylogenetic trees. Nucleic Acids Res. 51, W587–W592. doi: 10.1093/nar/gkad359
Xu, F., Zhang, Y. Z., Zhang, Y. H., Guan, G. Y., Zhang, K. P., Li, H. J., et al. (2020). Mushroom poisoning from Inocybe serotina: a case report from Ningxia, Northwest China with exact species identification and muscarine detection. Toxicon 179, 72–75. doi: 10.1016/j.toxicon.2020.03.003
Yu, W. J., Chang, C., Qin, L. W., Zeng, N. K., Wang, S. X., and Fan, Y. G. (2020). Pseudosperma citrinostipes (Inocybaceae), a new species associated with keteleeria from southwestern China. Phytotaxa 450, 8–16. doi: 10.11646/phytotaxa.450.1.2
Zhang, M., Xie, D. C., Wang, C. Q., Deng, W. Q., and Li, T. H. (2022). New insights into the genus Gyroporus (Gyroporaceae, Boletales), with establishment of four new sections and description of five new species from China. Mycology 13, 223–242. doi: 10.1080/21501203.2022.2094012
Zhang, Y. Z., Yan, Y. Y., Li, H. J., Fan, Y. G., and Xu, F. (2022). Toxin screening of Pseudosperma umbrinellum (Agaricals, Basidiomycota): first report of phalloidin in Inocybaceae mushroom. Toxicon 217, 155–161. doi: 10.1016/j.toxicon.2022.08.005
Keywords: Inocybaceae, new taxa, molecular phylogeny, taxonomy, toxin detection
Citation: Gao J-L, Wu X-P, Zhou Y-L, Yu W-J and Fan Y-G (2025) Additions to the Inocybe sect. Leptocybe (Agaricales) in China: new species from tropical rainforests, new geographical distributions, and toxin detection. Front. Microbiol. 16:1540570. doi: 10.3389/fmicb.2025.1540570
Edited by:
Xiancan Zhu, Anhui Normal University, ChinaReviewed by:
Wei Li, Shantou University, ChinaJing Si, Beijing Forestry University, China
Fei Xu, Ningxia Center for Disease Control and Prevention, China
Copyright © 2025 Gao, Wu, Zhou, Yu and Fan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wen-Jie Yu, aW5vY3liZUBxcS5jb20=; Yu-Guang Fan, bXljZW5hQHFxLmNvbQ==
†These authors have contributed equally to this work
 Jia-Long Gao1†
Jia-Long Gao1† Wen-Jie Yu
Wen-Jie Yu Yu-Guang Fan
Yu-Guang Fan