- 1College of Agriculture and Biology, Liaocheng University, Liaocheng, China
- 2College of Smart Agriculture, Yulin Normal University, Yulin, China
- 3College of Pharmaceutical Sciences and Food Engineering, Liaocheng University, Liaocheng, China
Background: The global spread of carbapenem-resistant Escherichia coli is a major public health concern. An investigation of their presence in the human and food chain products would facilitate the elucidation of the route of their food-borne transmission. Thus, the aim of this study was to investigate the prevalence of NDM-positive E. coli isolates in chicken at retail markets in Shandong, China.
Methods: A total of 60 NDM-positive isolates were recovered from 531 E. coli isolates obtained from chickens at the retail market in Shandong. Antimicrobial susceptibility testing and polymerase chain reaction screening were performed to investigate the phenotype and genotype of carbapenemase resistance. Genomic characteristics of the -producing isolates were determined by WGS and bioinformatic analysis.
Results: All of these isolates were multidrug-resistant (MDR), with a majority exhibiting resistance to meropenem, ampicillin, ceftazidime, cefotaxime, florfenicol, sulfamethoxazole/trimethoprim, and tetracycline. Whole genome sequencing (WGS) analysis indicated that these isolates were belonged to 18 distinct sequence types (STs), with the most prevalent STs being ST515 (17/60) and ST69 (11/60). Additionally, WGS analysis revealed that clonal spread of NDM-positive ST69 and ST515 E. coli isolates at markets in different cities in Shandong. Phylogenomic analysis showed that NDM-positive E. coli isolates from chickens were closely related to those of human origin.
Conclusion: This study provides a new insight into the spread of NDM-positive E. coli isolates from retail chicken, and offers essential data for public health management.
Introduction
The overuse of antibiotics can lead to the emergence of antimicrobial-resistant microorganisms in food-producing animals and in products derived from them, such as meat, eggs, and milk (Hassan et al., 2021; Chu et al., 2024; Sun et al., 2024). Consuming or coming into contact with food containing antimicrobial-resistant microorganisms can lead to the development of foodborne diseases that are challenging to treat, posing a significant threat to anti-infective treatment (Peng et al., 2024). Global annual deaths from foodborne illnesses caused by bacteria and other microorganisms exceed 400,000 (Todd, 2020). Therefore, the prevalence of carbapenem-resistant isolates is a public health concern that needs to be addressed on a global scale.
The main mechanism of carbapenem resistance is the production of carbapenemases. New Delhi metallo-β-lactamase (NDM) is one of the most common carbapenemases and has the strongest hydrolytic activity, hydrolyzing all β-lactam antibiotics except aztreonam (Kaewnirat et al., 2022). Since the discovery of NDM in clinical Escherichia coli in 2008, NDM-positive E. coli isolates have been shown to be globally distributed, with NDM-positive strains detected in virtually all countries conducting epidemiological surveys (Yong et al., 2009). To date, NDM-positive E. coli isolates have been recovered from hospitals and hospital sewage in major provinces/municipalities in China (Li et al., 2024; Liu et al., 2024). In addition, carbapenem antibiotics have never been approved for use in food-producing animals in the world; however, there have been sporadic reports of NDM-positive E. coli isolates from various food-producing animals and the downstream meat production chain (Bai et al., 2022; Wang et al., 2022; Wen et al., 2023). It is worth noting that NDM-positive E. coli isolates were highly prevalent among the Chinese poultry production chain, including commercial broiler farms, slaughterhouses, and supermarkets (Fu et al., 2024). In recent years, sporadic cases of NDM-positive E. coli isolates from chickens have been detected in retail markets in some local areas (Lv et al., 2022), suggesting that chickens in retail markets may be a “reservoir” for NDM-positive E. coli isolates. However, a large-scale survey of the NDM-positive E. coli isolates from retail market chickens remains unexplored.
In this study, we conducted a large-scale investigation on the prevalence of NDM-positive E. coli isolates in chickens from retail markets in Shandong, China. The antibiotic resistance phenotype and genotype, genetic diversity, phylogenetic relationships, and genetic environments of blaNDM were evaluated. The findings of this study will provide fundamental data to help guide the development of food safety policies and ensure public health.
Materials and methods
Isolates collection
A total of 531 chicken samples were randomly collected from retail markets (5–8 chicken samples in each retail market) across 8 cities [Liaocheng (n = 75), Jinan (n = 93), Dezhou (n = 47), Taian (n = 52), Yantai (n = 87), Linyi (n = 74), Qingdao (n = 39), and Weifang (n = 64)] of Shandong Province from April 2018 to October 2020. In brief, all samples were added to 1 mL of lysogeny broth (LB) and incubated for 16–18 h at 37°C, followed by inoculation onto MacConkey plates for 12 h. Then, red clones were selected for identification using MALDI-TOF MS Axima™ (Shimadzu-Biotech Corp., Kyoto, Japan) and 16S rRNA sequencing. Finally, all the E. coli isolates were stored in 30% glycerol broth at −80°C.
All isolates were resuscitated on LB broth, and for 16–18 h at 37°C, followed by inoculation onto MacConkey plates containing 1.0 mg/L meropenem. Five major carbapenemase resistance genes (blaKPC, blaNDM, blaIMP, blaOXA-48-like, and blaVIM) were detected in carbapenem-resistant isolates using polymerase chain reaction (PCR) with previously described primers (Lv et al., 2022).
Antimicrobial susceptibility testing
The MICs of 16 antibiotics (ampicillin, cefotaxime, ceftazidime, aztreonam, amikacin, gentamicin, ciprofloxacin, nalidixic acid, tetracycline, tigecycline, doxycycline, fosfomycin, sulfamethoxazole/trimethoprim, meropenem, colistin, and florfenicol) for all recovery isolates were determined using agar dilution and interpreted according to the Clinical and Laboratory Standards Institute (CLSI) guidelines (CLSI, 2020). The breakpoints of colistin and tigecycline for Enterobacteriaceae were interpreted according to the EUCAST criteria (European Committee on Antimicrobial Susceptibility Testing, 2019). E. coli ATCC 25922 served as a quality control strain for susceptibility testing.
Conjugation assay testing
To determine the transferability of the resistance genes, streptomycin-resistant E. coli strain C600 was used as the recipient, and the conjugation assay was performed using a filter mating method. Transconjugants were selected using MacConkey agar plates containing 1.0 mg/L meropenem and 1,500 mg/L streptomycin.
WGS and phylogenetic analysis
The genomic DNA of all NDM-positive isolates was subjected to 250 bp paired-end WGS using the Illumina MiSeq system (Illumina, San Diego, CA, United States), and the paired-end Illumina reads were assembled using SPAdes v3.6.2.18 MLST (Bankevich et al., 2012). Antibiotic resistance genes (ARGs) and plasmid replicon types were analyzed using the Center for Genomic Epidemiology server.1
The hosts and countries of 196 E. coli isolates were retrieved from the NCBI database,2 and the assembly genomes of the 196 isolates were downloaded from the NCBI database (as of September 2024). All assembly genomes were used for core-genome alignments to produce a phylogenetic tree using the Parsnp software of the Harvest suite (Todd et al., 2014). In this pipeline, bases that have likely undergone recombination are removed using PhiPack (Bruen et al., 2006), and only columns passing a set of filters based on these criteria are considered reliable core-genome SNPs. The final set of core-genome SNPs was submitted to FastTree 2 for reconstructing a maximum likelihood phylogenetic tree using default parameters (Poon et al., 2010). The VCF file of all variants identified by Parsnp was then used to determine pair-wise single-nucleotide variant distances between the core genomes of all strains. For the phylogenetic tree, a reference genome was randomly selected using the ‘-r!’ switch. The lineages of the phylogenetic tree were defined using rhierbaps version 6.0 (Cheng et al., 2013). The heat map was generated using R 3.3.2 (R Foundation for Statistical Computing) and was used to construct the tree that was visualized using FigTree v1.4.2 and iTOL v4 (Letunic and Bork, 2019). Plasmid maps were generated using the BRIG.
Results
Prevalence of carbapenemase-producing Escherichia coli isolates
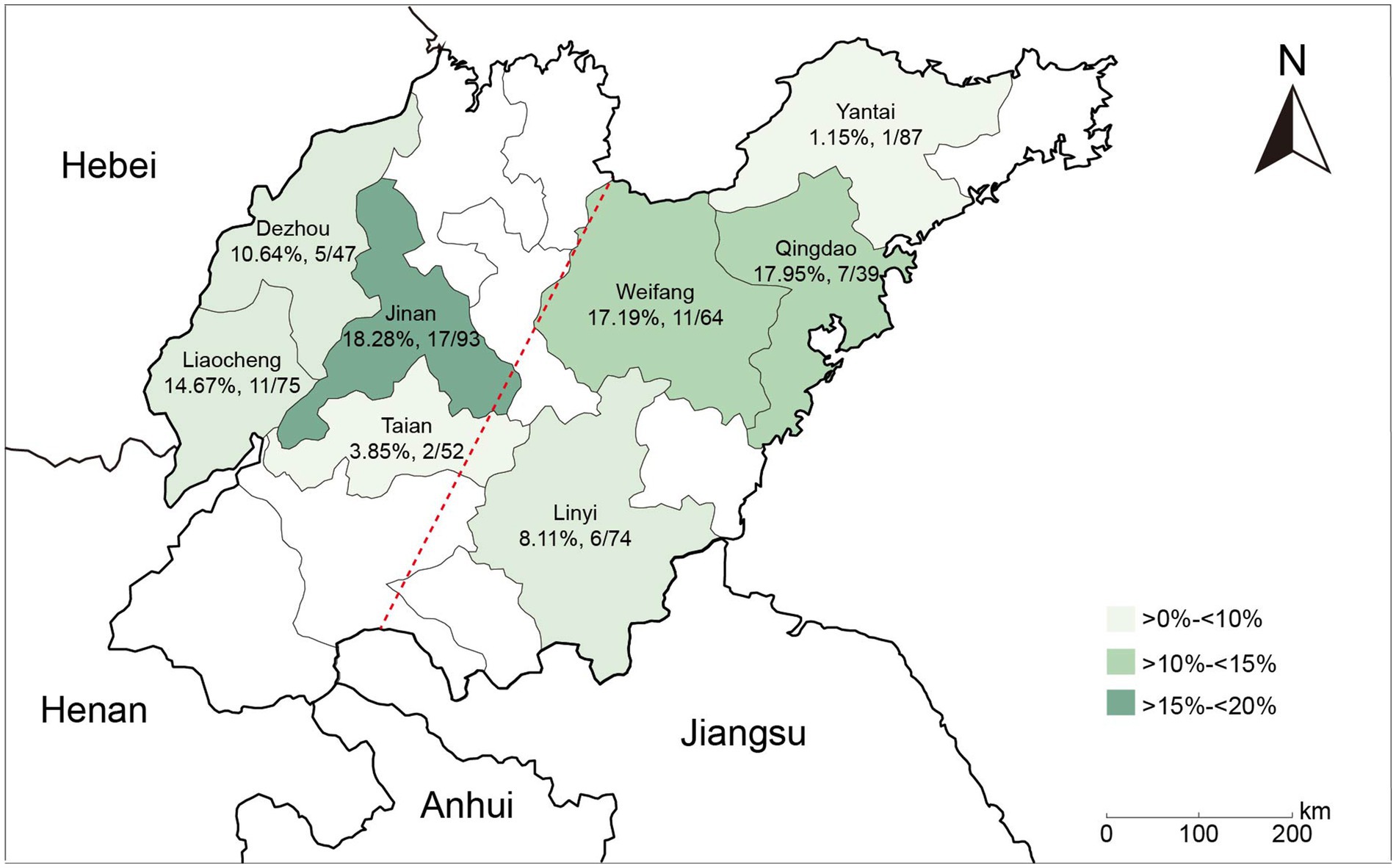
In this study, a total of 60 (11.30%) NDM-positive isolates were recovered from 531 E. coli isolates obtained from retail market chickens in Shandong, China. No other carbapenemase-encoding genes were detected among these carbapenem-resistant isolates. Among NDM-positive isolates, the most predominant variants were blaNDM-5 (86.67%, 52/60), followed by blaNDM-9 (10.00%, 6/60), blaNDM-1 (1.67%, 1/60), and blaNDM-20 (1.67%, 1/60) (Supplementary Figure S1a). The highest detection rate of NDM-positive E. coli isolates was observed in Jinan (18.28%, 17/93), followed by Qingdao (17.95%, 7/39), Weifang (17.19%, 11/64), and Liaocheng (14.67%, 11/75). In contrast, Taian (3.85%, 2/52) and Yantai (1.15%, 1/87) displayed the lowest detection rates (Figure 1). Moreover, the prevalence of NDM-positive E. coli isolates was higher in northwestern Shandong (13.11%, 35/267) than in southeastern Shandong (9.47%, 25/264) (Supplementary Figure S1b).
Antibiotic resistance phenotypes
All 60 NDM-positive E. coli isolates exhibited resistance to ampicillin, cefotaxime, ceftazidime, and trimethoprim–sulfamethoxazole (Figure 2). In addition, the majority of these isolates remained resistant to meropenem (83.33%, 50/60), tetracycline (83.33%, 50/60), florfenicol (73.33%, 44/60), doxycycline (65.00%, 39/60), gentamicin (53.33%, 32/60), ciprofloxacin (90%, 54/60), and nalidixic acid (53.33%, 32/60). In contrast, a lower prevalence of resistance phenotypes was observed among aztreonam (21.67%, 13/60), amikacin (3.33%, 2/60), fosfomycin (13.33%, 8/60), and colistin (6.67%, 4/60) (Figure 2). Of note, none of the isolates showed resistance to tigecycline.
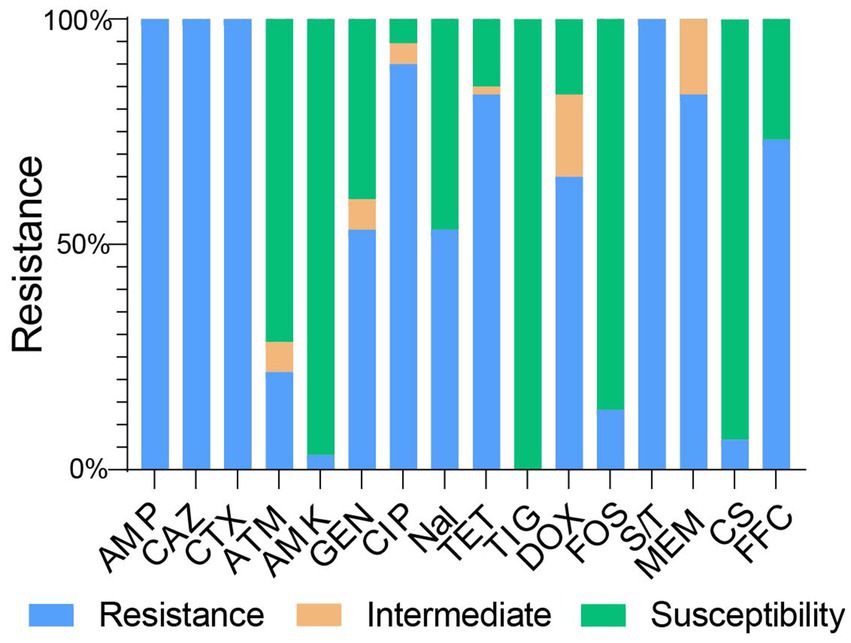
Figure 2. Minimum inhibitory concentrations of tested antimicrobial agents for the studied bacterial isolates. AMP, ampicillin; CTX, cefotaxime; CAZ, ceftazidime; ATM, aztreonam; AMK, amikacin; GEN, gentamicin; CIP, ciprofloxacin; Nal, Nalidixic acid; TET, tetracycline; TIG, tigecycline; DOX, doxycycline; FOS, fosfomycin; S/T, sulfamethoxazole/trimethoprim; MEM, meropenem; CS, colistin; FFC, florfenicol.
Phylogenetic analysis of NDM-positive Escherichia coli isolates
Whole-genome sequencing data were generated for 60 NDM-positive E. coli isolates, and the WGS results revealed that these isolates were categorized into 18 distinct STs, with 6 isolates classified as unclassified STs. Overall, ST515 (28.3%, 17/60) was the most prevalent isolate in Jinan and Weifang, followed by ST69 (18.3%, 11/60) in Qingdao and Liaocheng. These findings indicate a distinct geographic distribution preference (Figure 3). A phylogenetic tree was established using 60 NDM-positive E. coli isolates. Phylogenomic analysis revealed that all the E. coli isolates were classified into five distinct lineages. It is worth noting that all isolates in the lineages I belonged to ST515 and were sourced from Jinan and Weifang, with these isolates sharing only 0 ~ 24 core-genome SNPs (cgSNP) among themselves. In addition, isolates in lineage III belonged to ST69 and originated from Qingdao and Liaocheng, sharing only 0 ~ 13 SNPs among them (Figure 3). These findings indicated the clonal transmission of NDM-positive ST69 and ST515 E. coli isolates across various regions. Furthermore, our phylogenomic analysis demonstrated that the majority of NDM-positive E. coli isolates exhibited a significant degree of variation in core-genome sequences, suggesting a high genetic diversity of NDM-positive E. coli isolates from chicken at retail markets in Shandong Province, China.
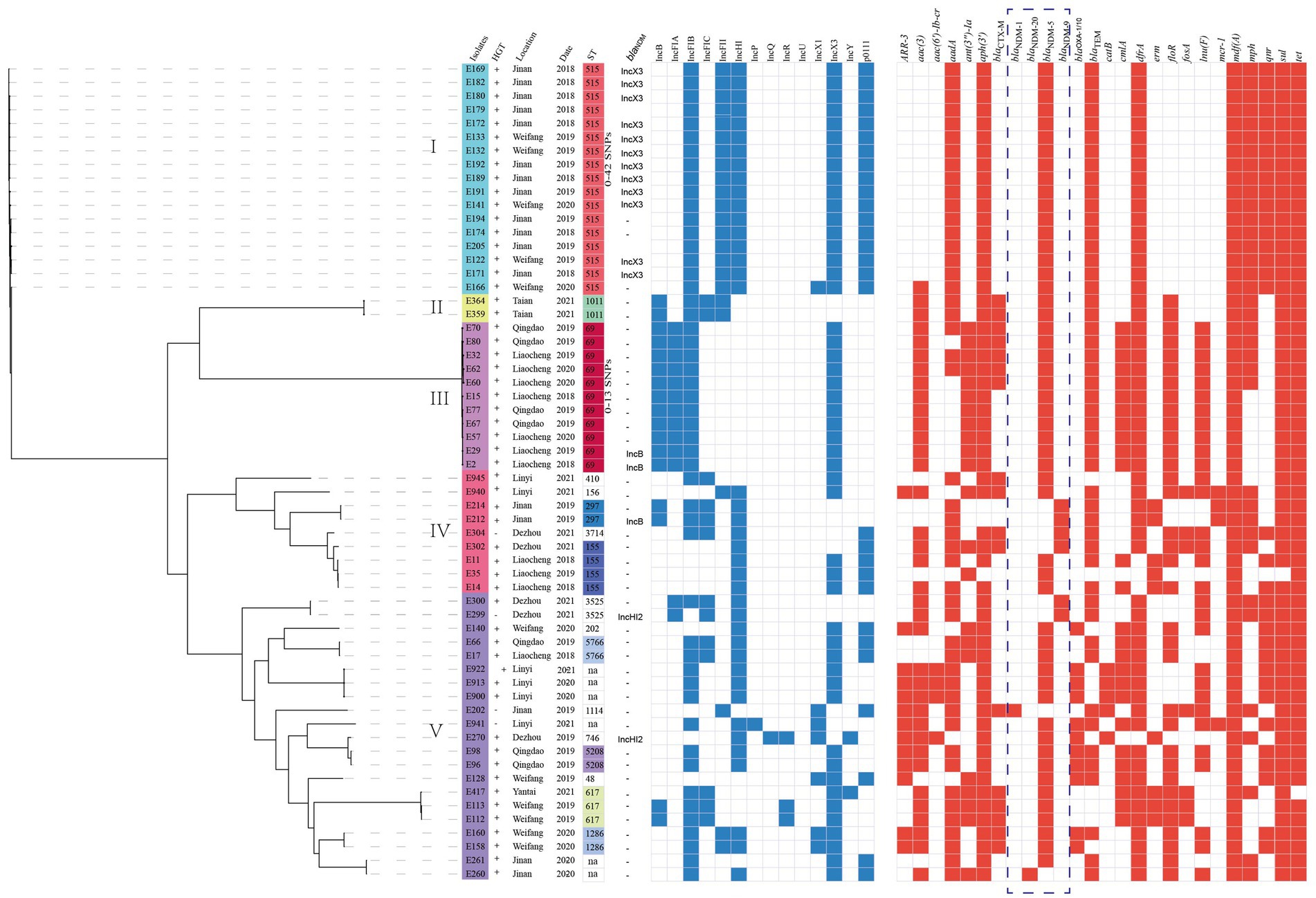
Figure 3. Phylogenetic analysis of NDM-positive E. coli isolates in this study (n = 60). Bayesian evolutionary tree was constructed using core-genome SNPs. Each isolate is labeled with the city of isolation year and ST. The red-filled squares indicate the possession of the indicated antimicrobial resistance genes (ARGs).
To further assess the relationship between the isolates from the current study and other resources in Shandong, China. A total of 196 NDM-positive E. coli isolates were collected in Shandong from the NCBI database (as of 2024). Then, a maximum likelihood phylogenetic tree was constructed using these 256 NDM-positive E. coli isolates. The 256 NDM-positive E. coli isolates were grouped into 8 clades and shared 216,099 core-genome SNPs (cgSNPs). It should be noted that 11 NDM-positive ST746 E. coli isolates from humans in Shandong (GenBank assembly GCA_001893215.1, GCA_001894425.1, GCA_001893995.1, GCA_001894025.1, GCA_001893225.1, GCA_001893375.1, GCA_001893305.1, GCA_001894035.1, GCA_001894065.1, GCA_001893765.1, and GCA_001894315.1) shared only 69–94 SNPs with NDM-positive ST746 E. coli isolates from a retail chicken in Shandong in this study (E270) (Figure 4), revealing a strong correlation of NDM-positive E. coli isolates among animals and foods.
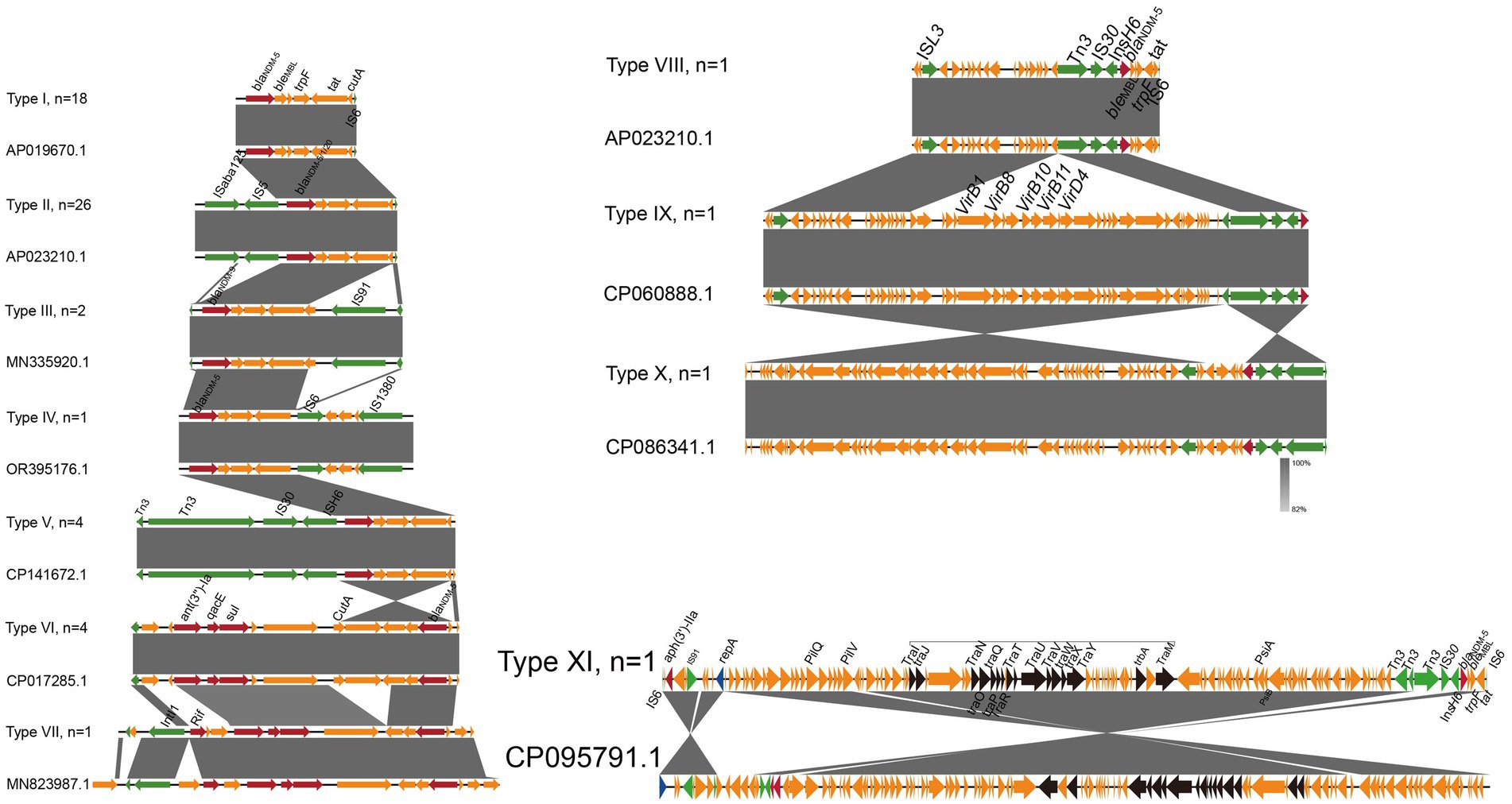
Figure 4. Phylogenetic structures of the NDM-positive E. coli isolates from this study and the GenBank database. The maximum likelihood tree shows the relationships among the 256 NDM-positive E. coli isolates. Isolate hosts from which isolates were obtained are indicated in the outer ring.
Plasmid analysis
A total of 14 incompatible group plasmid replicon types were detected among the 60 blaNDM-positive E. coli isolates including IncB (28%, 17/60), IncFIA (22%, 13/60), IncFIB (83%, 50/60), IncFIC (22%, 13/60), IncFII (38%, 23/60), IncX3 (82%, 49/60), IncHI (67%, 40/60), and other plasmids (Figure 3). Of note, WGS and BRIG analyses revealed that the blaNDM genes may be located in IncX3 (n = 12), IncHI2 (n = 2), and IncB (n = 3) plasmids (Figure 3; Supplementary Figure S2). In addition, all of the IncX3 plasmid carrying blaNDM genes were successfully transferred to recipients (E. coli C600str) by conjugation. These findings suggest that IncX3 plasmids may have undergone backbone fusion events with other plasmids, potentially driving horizontal gene transfer of resistance determinants.
Analysis of antibiotic resistance genes
We conducted a comprehensive antimicrobial analysis on E. coli isolates that revealed the presence of the β-lactam resistance genes (blaNDM-1, blaNDM-20, blaNDM-5, blaNDM-9, blaOXA-1/10, blaCTX-M, and blaTEM). Other important resistance determinants that confer resistance to quinolones (qnr), aminoglycosides (aadA, aph, armA and aac), fosfomycin (fosA), chloramphenicol/florfenicol (floR), sulfonamides (sul), macrolide (mdf(A) and mph(A)), rifampicin (ARR-3), tetracycline (tet), and trimethoprim (dfrA). In addition, we identified two isolates (E212 and E214) that co-harbored blaNDM-9 and mcr-1, as well as two isolates (E940 and E941) that co-harbored blaNDM-5 and mcr-1 (Figure 3).
The genetic environments of blaNDM
A total of 11 genetic contexts (type I to type XI) were found in 60 isolates (Figure 5). All of the type VII and XI genetic contexts were novel in the GenBank database. Notably, blaNDM-5 was found in 10 of the 11 genetic context types. In all types, various variants of blaNDM (blaNDM-1, blaNDM-5, blaNDM-9, and blaNDM-20) were directly associated with the bleMBL-trpF-tat genes. The type I arrangement was blaNDM-bleMBL-trpF-tat-cutA and was inserted upstream of InsH6. blaNDM-1 and blaNDM-20 were located in type II genetic contexts, which are identical to the type II context of blaNDM-5. In addition, we found that the type II genomic context ISAba125-IS5-blaNDM-bleMBL-trpF-tat-IS6 was the most prevalent NDM genetic environment in this study (43.3%, 26/60), which is identical to a blaNDM-5-carrying plasmid of carbapenem-resistant Enterobacteriaceae isolates from humans in China (Yang et al., 2022). Meanwhile, the backbone structure of blaNDM-5 including the ISAba125-IS5-blaNDM-bleMBL-trpF-tat-IS6 was usually carrying the IncX3-type plasmid and highly conserved. In the type VIII contexts, the IS91 was inserted downstream of InsH6, and blaNDM-9 was uniquely associated with the single distinct genetic context. Compared with type VIII, the type IV context revealed deletion of IS91, but the IS1380 was inserted upstream of In6. Type V contains the highest number of mobile genetic elements, including Tn3, IS30, and ISH6. In type VI, the ant(3″)-Ia, qacE, and sul resistance genes were located upstream of blaNDM. Type VII was similar to type III, with the exception that a rif resistance gene was found upstream of ant(3″)-Ia. In the type VIII contexts, the ISL3 was inserted upstream of type V. In contrast, type IX inserts a virulence gene (vir) cluster at the Tn3 of type VIII. The sequence of the type IX plasmid, when compared to the IncX3 plasmids (GenBank: CP060888.1), from an E. coli strain isolated from a patient in Zhejiang province, showed 100% nucleotide identity and 100% coverage (Yang et al., 2022). Based on type IV, type X reverses the upstream of blaNDM with the action of ISL3 and ISH6. Type XI reverses the upstream of blaNDM through the action of Tn3, with the plasmid transfer gene tra cluster positioned upstream of blaNDM.
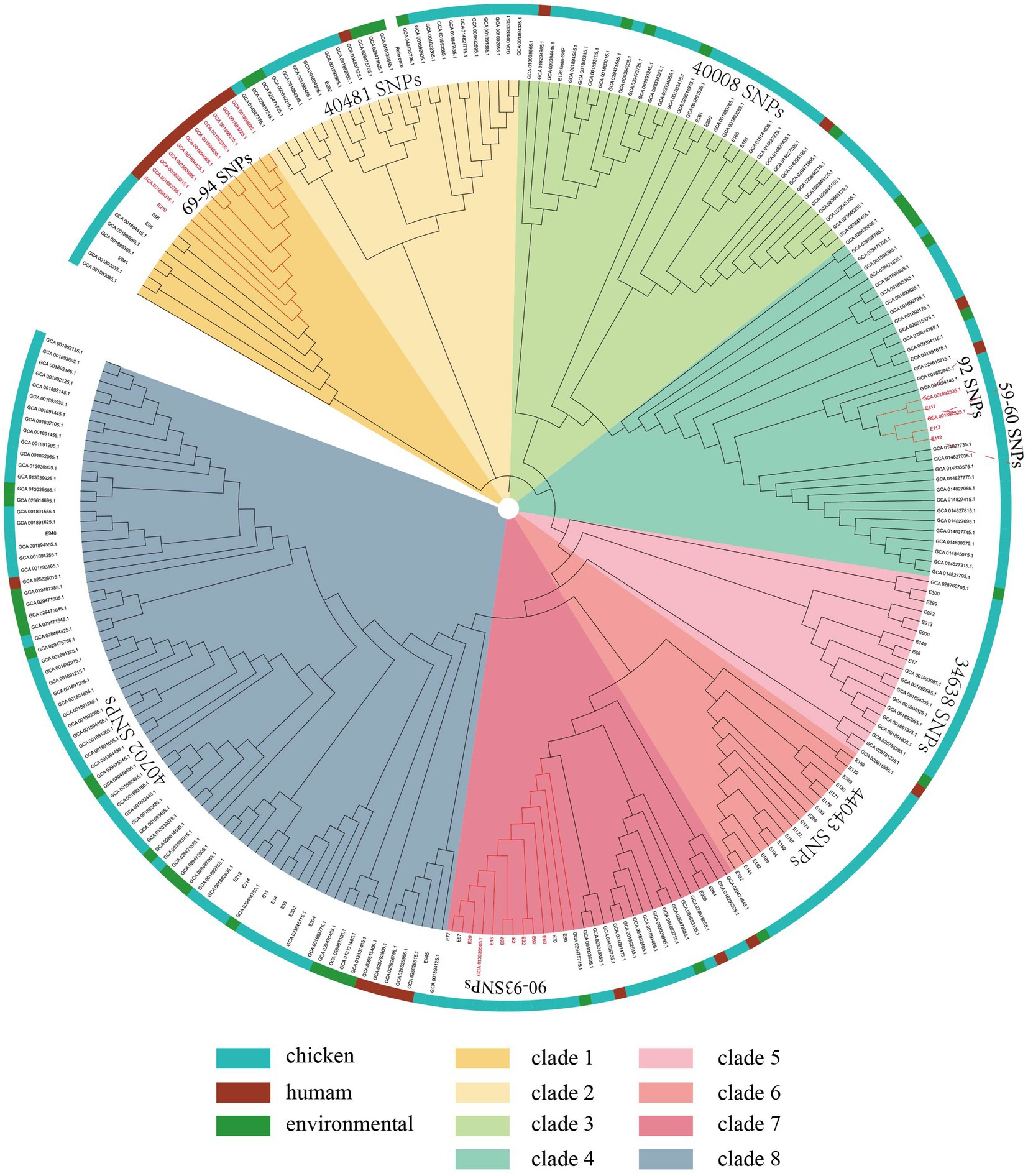
Figure 5. Genomic environments of blaNDM of E. coli isolates. The figure was generated using Easyfig. Regions of homology are marked by shading, and regions of ≥ 99.0% nucleotide sequence identity are shaded gray. Arrows indicate the direction of transcription of the genes. In addition, red represents resistance gene, blue represents mobile genetic elements, black represents transfer gene tra clusters, and yellow represents other coding sequence (CDS).
Discussion
In the present study, we investigated the prevalence of NDM-positive E. coli isolates from chickens in the retail markets in Shandong, China, during 2018–2020. Recent studies have revealed that blaNDM was the most frequently identified mobile carbapenem resistance gene found in various Enterobacteriaceae species, especially in Asia (Wu et al., 2023). However, NDM-positive E. coli isolates from retail chicken were rarely detected. In this study, we obtained 11.30% of NDM-positive E. coli isolates from retail chicken in markets in Shandong, China. Furthermore, our previous study also found that 15.42% of NDM-positive E. coli were highly represented in ducks in the coastal areas of China, which is similar to our result (Wang et al., 2021a,b). In contrast, the overall incidence of NDM-positive strains in animals was lower in pigs (6.05%) (Wen et al., 2023), suggesting that the blaNDM gene is highly prevalent in poultry. Furthermore, a recent study found that the abattoir is a hotspot for cross-contamination, amplifying blaNDM (Fu et al., 2024). The blaNDM gradually increases along the chicken (4.70%)–slaughterhouse (7.60%)–retail (65.56%) chain (Fu et al., 2024), suggesting that NDM-positive E. coli reaches the dinner table via the farm–slaughterhouse–retail route, thereby endangering human health.
WGS analysis demonstrated the predominance of NDM-5 in this study. In fact, blaNDM-5 was first reported in an E. coli strain isolated from a patient in the UK in 2011 (Hornsey et al., 2011). Since then, blaNDM-5 has been detected in more than 40 countries, especially in China and Southeast Asia (Lv et al., 2022). Of further concern, NDM-5-producing Enterobacterales have been recovered from a variety of other sources worldwide, including food, livestock, companion animals, wildlife, and the environment (Hans et al., 2023). Plasmid profiling demonstrated that blaNDM genes were predominantly harbored by IncX3-type plasmids, with significantly lower carriage rates observed in IncFII and IncHI plasmid types. In fact, IncX3 plasmids are the most common type of plasmid carrying blaNDM-5 in China (Ma et al., 2020). The IncX3 plasmids found in this study further highlight the importance of the epidemic IncX3 plasmid in the spread of the blaNDM-5 gene within the chicken market. MLST analysis revealed that these isolates belonged to 18 distinct STs, indicating a high diversity of STs in NDM-positive E. coli isolates from Chicken at retail markets. Our previous study found that the NDM-positive E. coli isolates from duck farms had 30 distinct STs and showed obvious distinctive diversities in geographical distribution (Wang et al., 2021a,b). In addition, it was also found that NDM-positive E. coli from raw meat in retail markets were classified into 14 different STs (Nisa et al., 2024); these studies showed that the ST diversity of NDM-positive E. coli from retail market chickens was lower than that from farms and higher than that from raw meat. In this study, ST69 and ST515 are the most prevalent E. coli isolates. Previous studies have also identified ST69 E. coli isolates as emerging and high-risk clones and were one of the most frequent clinical isolates in urinary tract infections (Cuénod et al., 2023; Mattioni Marchetti et al., 2023), while ST515 E. coli isolates have only been prevalent in vegetable markets in Northern Thailand (Chotinantakul et al., 2022). This study showed that there was cross-regional clonal transmission of ST69 between Liaocheng and Qingdao, and of ST515 between Jinan and Weifang. This pattern of cross-regional dissemination is particularly evident in the coastal areas of China and other developed regions, such as the grass carp were recovered from different markets and different sample booths in Guangdong, and the ducks were recovered from different farms in Anhui (Guo et al., 2024; Lv et al., 2022). In addition, NDM-positive ST69 and ST515 E. coli isolates persist in their host strain during 3 years of ongoing investigation, suggesting that NDM-positive ST69 and ST515 E. coli isolates may pose challenges in their treatment (Wang et al., 2018).
A total of 11 genetic contexts (types I to XIII) were found in 60 NDM-carrying plasmids. In type VI, a fusion plasmid was found that was recombined from an IncX3 plasmid carrying blaNDM and an IncF plasmid carrying ant(3ʹ)-Ia, qacE, sul. This indicated that the formation of IncX3-FIB hybrid plasmids through the integration of IncF plasmids into IncX3 plasmid backbones probably facilitated the transmission of IncX3-F plasmids and ARGs (Bai et al., 2023). For type IX, it was confirmed that the blaNDM-carrying plasmid could mediate the transmission of the virulence plasmid through the formation of a fusion plasmid by Tn3. The mobile genetic element Tn3 plays an important role in the transmission of the NDM gene (Li et al., 2024). These results indicated a diversity of genetic environments forblaNDM in the Enterobacteriaceae.
WGS analysis also revealed that blaNDM coexisted with other 30 other types of ARGs, 15 of these ARGs were highly prevalent with detection rates >50%. Previous research has shown that plasmid fusion can expand the host range of plasmids, accelerating the dissemination of ARGs among various bacteria (Lv et al., 2022), and can further promote the spread and persistence of carbapenem-resistant microbes in retail chicken at market. In this study, we first describe an E. coli isolate of coexistence of the mcr-1 and blaNDM genes. The co-occurrence of blaNDM and mcr-1 positive plasmids may have occurred, and several studies have been reported (Liu et al., 2024; Nisa et al., 2024). Of note, mcr-1—which confers resistance to the last-resort antibiotic colistin (Liu et al., 2023)—was detected in 4 NDM-positive E. coli isolates (2 blaNDM-5 and mcr-1 and 2 blaNDM-9 and mcr-1). blaNDM and mcr-1 antimicrobial resistance genes confer resistance to colistin and carbapenems, which are antimicrobials often used as last-resort antibiotics in hospitals (Bai et al., 2023). Some recent studies have reported that blaNDM-5 and mcr-1 co-producing E. coli isolates were recovered from food animals (Wang et al., 2021a,b). The results highlight that the food animals in markets serve as an important reservoir of E. coli isolates carrying blaNDM and mcr-1, posing a serious public health threat via food chain transmission.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
J-XM: Software, Writing – original draft. S-CB: Validation, Writing – original draft. J-QX: Formal analysis, Writing – original draft. Z-QH: Investigation, Writing – original draft. Y-XQ: Resources, Writing – original draft. J-xW: Validation, Writing – review & editing. Y-XS: Validation, Writing – review & editing. Y-BL: Project administration, Writing – original draft, Writing – review & editing. M-GW: Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was funded by the National Natural Science Foundation of China (32202859); The results of the Research Initiation Project for High-level Talents of Yulin Normal University in 2024 (G2024ZK13).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1550742/full#supplementary-material
Footnotes
References
Bai, S. C., Fang, L. X., Xiao, H. L., Zhang, Y., Guo, W. Y., Zhang, J. X., et al. (2023). Genomics analysis of KPC-2 and NDM-5-producing Enterobacteriaceae in migratory birds from Qinghai Lake, China. Appl. Microbiol. Biotechnol. 107, 7531–7542. doi: 10.1007/s00253-023-12746-3
Bai, S., Yu, Y., Kuang, X., Li, X., Wang, M., Sun, R., et al. (2022). Molecular characteristics of antimicrobial resistance and virulence in Klebsiella pneumoniae strains isolated from goose farms in Hainan, China. Appl. Environ. Microbiol. 88:e0245721. doi: 10.1128/aem.02457-21
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Bruen, T. C., Philippe, H., and Bryant, D. (2006). A simple and robust statistical test for detecting the presence of recombination. Genetics 172, 2665–2681. doi: 10.1534/genetics.105.048975
Cheng, L., Connor, T. R., Siren, J., Aanensen, D. M., and Corander, J. (2013). Hierarchical and spatially explicit clustering of DNA sequences with BAPS software. Mol. Biol. Evol. 30, 1224–1228. doi: 10.1093/molbev/mst028
Chotinantakul, K., Woottisin, S., and Okada, S. (2022). The emergence of CTX-M-55 in extended-Sectrum beta-lactamase-producing Escherichia coli from vegetables sold in local Markets of Northern Thailand. Jpn. J. Infect. Dis. 75, 296–301. doi: 10.7883/yoken.JJID.2021.139
Chu, Y., Ruan, Y. X., Liu, J. Q., Zhang, Y., Wang, M. G., Liao, X. P., et al. (2024). Population genomics of emerging multidrug-resistant Salmonella derby from pork and human in Guangdong, China. LWT Food Sci. Technol. 205:116535. doi: 10.1016/j.lwt.2024.116535
CLSI. (2020). Performance Standards for Antimicrobial Susceptibility Testing. Available online at: https://clsi.org/
Cuénod, A., Agnetti, J., Seth-Smith, H. M. B., Roloff, T., Wälchli, D., Shcherbakov, D., et al. (2023). Bacterial genome-wide association study substantiates papGII of Escherichia coli as a major risk factor for urosepsis. Genome Med. 15:89. doi: 10.1186/s13073-023-01243-x
European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters. (2019). Available online at: http://www.eucast.org/clinical_breakpoints/
Fu, B., Xu, J., Yin, D., Sun, C., Liu, D., Zhai, W., et al. (2024). Transmission of blaNDM in Enterobacteriaceae among animals, food and human. Emerg. Microb. Infect. 13:2337678. doi: 10.1080/22221751.2024.2337678
Guo, C.-H., Chu, M.-J., Liu, T., Wang, J., Zou, M., and Liu, B.-T. (2024). High prevalence and transmission of blaNDM-positive Escherichia coli between farmed ducks and slaughtered meats: an increasing threat to food safety. Int. J. Food Microbiol. 424:110850. doi: 10.1016/j.ijfoodmicro.2024.110850
Hans, J. B., Pfennigwerth, N., Neumann, B., Pfeifer, Y., Fischer, M. A., Eisfeld, J., et al. (2023). Molecular surveillance reveals the emergence and dissemination of NDM-5-producing Escherichia coli high-risk clones in Germany, 2013 to 2019. Eur. Secur. 28:2200509. doi: 10.2807/1560-7917.ES.2023.28.10.2200509
Hassan, M. M., El Zowalaty, M. E., Lundkvist, Å., Järhult, J. D., Khan Nayem, M. R., Tanzin, A. Z., et al. (2021). Residual antimicrobial agents in food originating from animals. Trends Food Sci. Technol. 111, 141–150. doi: 10.1016/j.tifs.2021.01.075
Hornsey, M., Phee, L., and Wareham, D. W. (2011). A novel variant, NDM-5, of the New Delhi metallo-β-lactamase in a multidrug-resistant Escherichia coli ST648 isolate recovered from a patient in the United Kingdom. Antimicrob. Agents Chemother. 55, 5952–5954. doi: 10.1128/AAC.05108-11
Kaewnirat, K., Chuaychob, S., Chukamnerd, A., Pomwised, R., Surachat, K., Phoo, M. T. P., et al. (2022). In vitro synergistic activities of Fosfomycin in combination with other antimicrobial agents against Carbapenem-resistant Escherichia coli harboring blaNDM-1 on the IncN2 plasmid and a study of the genomic characteristics of these pathogens. Infect. Drug Resist. 15, 1777–1791. doi: 10.2147/IDR.S357965
Letunic, I., and Bork, P. (2019). Interactive tree of life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47, W256–w259. doi: 10.1093/nar/gkz239
Li, X., Li, C., Zhou, L., Wang, Q., Yao, J., Zhang, X., et al. (2024). Global phylogeography and genomic characterization of blaKPC and blaNDM-positive clinical Klebsiella aerogenes isolates from China, 2016-2022. Sci. Total Environ. 923:171560. doi: 10.1016/j.scitotenv.2024.171560
Liu, K. D., Jin, W. J., Li, R. B., Zhang, R. M., Sun, J., Liu, Y. H., et al. (2023). Prevalence and molecular characteristics of mcr-1-positive Escherichia coli isolated from duck farms and the surrounding environments in coastal China. Microbiol. Res. 270:127348. doi: 10.1016/j.micres.2023.127348
Liu, L., Zhao, M., Tang, Y., Shen, A., Yang, X., Yao, L., et al. (2024). Dissemination of clinical Escherichia coli strains harboring mcr-1, blaNDM-7 and siderophore-producing plasmids in a Chinese hospital. Antimicrob. Resist. Infect. Control 13:66. doi: 10.1186/s13756-024-01423-3
Lv, L. C., Lu, Y. Y., Gao, X., He, W. Y., Gao, M. Y., Mo, K. B., et al. (2022). Characterization of NDM-5-producing Enterobacteriaceae isolates from retail grass carp (Ctenopharyngodon idella) and evidence of blaNDM-5-bearing IncHI2 plasmid transfer between ducks and fish. Zool. Res. 43, 255–264. doi: 10.24272/j.issn.2095-8137.2021.426
Ma, T., Fu, J., Xie, N., Ma, S., Lei, L., Zhai, W., et al. (2020). Fitness cost of blaNDM-5-carrying p3R-IncX3 plasmids in wild-type NDM-free Enterobacteriaceae. Microorganisms 8:377. doi: 10.3390/microorganisms8030377
Mattioni Marchetti, V., Hrabak, J., and Bitar, I. (2023). Fosfomycin resistance mechanisms in Enterobacterales: an increasing threat. Front. Cell. Infect. Microbiol. 13:1178547. doi: 10.3389/fcimb.2023.1178547
Nisa, T. T., Sugawara, Y., Hamaguchi, S., Takeuchi, D., Abe, R., Kuroda, E., et al. (2024). Genomic characterization of carbapenemase-producing Enterobacterales from Dhaka food markets unveils the spread of high-risk antimicrobial-resistant clones and plasmids co-carrying blaNDM and mcr-1.1. JAC Antimicrob. Resist. 6:dlae124. doi: 10.1093/jacamr/dlae124
Peng, J., Xiao, R., Wu, C., Zheng, Z., Deng, Y., Chen, K., et al. (2024). Characterization of the prevalence of Salmonella in different retail chicken supply modes using genome-wide and machine-learning analyses. Food Res. Int. 191:114654. doi: 10.1016/j.foodres.2024.114654
Poon, A. F. Y., Price, M. N., Dehal, P. S., and Arkin, A. P. (2010). FastTree 2-approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490
Sun, Q., Dyar, O. J., Zhao, L., Tomson, G., Nilsson, L. E., Grape, M., et al. (2024). Overuse of antibiotics for the common cold - attitudes and behaviors among doctors in rural areas of Shandong Province, China. BMC Pharmacol Toxicol. 16:6. doi: 10.1186/s40360-015-0009-x
Todd, E. (2020). Food-borne disease prevention and risk assessment. Int. J. Environ. Res. Public Health 17:5129. doi: 10.3390/ijerph17145129
Todd, J., Treangen, B. D. O., Koren, S., and Phillippy, A. M. (2014). The harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 15:524. doi: 10.1186/s13059-014-0524-x
Wang, M. G., Fang, C., Liu, K. D., Wang, L. L., Sun, R. Y., Zhang, R. M., et al. (2022). Transmission and molecular characteristics of blaNDM-producing Escherichia coli between companion animals and their healthcare providers in Guangzhou, China. J. Antimicrob. Chemother. 77, 351–355. doi: 10.1093/jac/dkab382
Wang, Q., Wang, X., Wang, J., Ouyang, P., Jin, C., Wang, R., et al. (2018). Phenotypic and genotypic characterization of Carbapenem-resistant Enterobacteriaceae: data from a longitudinal large-scale CRE study in China (2012-2016). Clin. Infect. Dis. 67, S196–S205. doi: 10.1093/cid/ciy660
Wang, M. G., Yu, Y., Wang, D., Yang, R. S., Jia, L., Cai, D. T., et al. (2021b). The emergence and molecular characteristics of New Delhi Metallo beta-lactamase-producing Escherichia coli from ducks in Guangdong, China. Front. Microbiol. 12:677633. doi: 10.3389/fmicb.2021.677633
Wang, M. G., Zhang, R. M., Wang, L. L., Sun, R. Y., Bai, S. C., Han, L., et al. (2021a). Molecular epidemiology of carbapenemase-producing Escherichia coli from duck farms in south-east coastal China. J. Antimicrob. Chemother. 76, 322–329. doi: 10.1093/jac/dkaa433
Wen, R., Wei, H., Zhang, T., Ma, P., Wang, Q., Li, C., et al. (2023). Epidemiological characterisation of blaNDM-positive Enterobacterales from food-producing animal farms in Southwest China. Microorganisms 11:2304. doi: 10.3390/microorganisms11092304
Wu, J.-W., Quyen, T. L. T., Hsieh, Y.-C., Chen, Y.-Y., Wu, L.-T., and Pan, Y.-J. (2023). Investigation of carbapenem-resistant Klebsiella pneumoniae in Taiwan revealed strains co-harbouring blaNDM and blaOXA-48-like and a novel plasmid co-carrying blaNDM-1 and blaOXA-181. Int. J. Antimicrob. Agents 62:106964. doi: 10.1016/j.ijantimicag.2023.106964
Yang, C., Han, J., Berglund, B., Zou, H., Gu, C., Zhao, L., et al. (2022). Dissemination of blaNDM-5 and mcr-8.1 in carbapenem-resistant Klebsiella pneumoniae and Klebsiella quasipneumoniae in an animal breeding area in eastern China. Front. Microbiol. 13:1030490. doi: 10.3389/fmicb.2022.1030490
Yong, D., Toleman, M. A., Giske, C. G., Cho, H. S., Sundman, K., Lee, K., et al. (2009). Characterization of a new Metallo-β-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 53, 5046–5054. doi: 10.1128/AAC.00774-09
Keywords: carbapenemase, Escherichia coli, blaNDM, MLST, ARGs
Citation: Ma J-X, Bai S-C, Xu J-Q, He Z-Q, Qi Y-X, Wang J-x, Shi Y-X, Li Y-B and Wang M-G (2025) Molecular epidemiology of New Delhi metallo-β-lactamase-producing Escherichia coli in retail market chickens, Shandong, China. Front. Microbiol. 16:1550742. doi: 10.3389/fmicb.2025.1550742
Edited by:
Yasser Mahmmod, Long Island University, United StatesReviewed by:
Vittoria Mattioni Marchetti, University of Pavia, ItalyFabrice Compain, L’Institut Mutualiste Montsouris, France
Copyright © 2025 Ma, Bai, Xu, He, Qi, Wang, Shi, Li and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Min-Ge Wang, d2FuZ21pbmdlQGxjdS5lZHUuY24=
†These authors have contributed equally to this work
 Jing-Xian Ma1†
Jing-Xian Ma1† Yu-Bao Li
Yu-Bao Li Min-Ge Wang
Min-Ge Wang