- 1School of Basic Medicine, Youjiang Medical University for Nationalities, Baise, China
- 2College of Animal Science & Technology, Guangxi University, Nanning, China
- 3College of Animal Science, Anhui Science and Technology University, Chuzhou, China
- 4Laboratory Animal Center, Youjiang Medical University for Nationalities, Baise, China
- 5Library, Youjiang Medical University for Nationalities, Baise, China
Obesity is increasingly becoming a challenge with China’s economic development. There is an urgent need to identify more affordable methods to combat this condition. Paederia scandens (PS), a cost-effective herbal remedy widely used in China for treating inflammation and pain, shows potential in this regard. To investigate its anti-obesity mechanisms, we established a high-fat diet (HFD)-induced obesity model in mice. The obese mice subsequently received daily oral gavage of PS extract for 21 consecutive days. Upon the completion of the experiment, blood samples were collected to analyze lipid profiles, including total cholesterol (TC), triglycerides (TG), high-density lipoprotein (HDL-C), and low-density lipoprotein (LDL-C). Abdominal adipose tissue was subjected to hematoxylin–eosin (HE) staining for histological analysis, while fecal samples underwent 16S rRNA sequencing to assess gut microbiota composition. Our findings revealed that PS supplementation significantly reduced body weight, lipid metabolism biomarkers, and adipocyte size. PS treatment also restored gut microbial diversity, with 19 specific microbial taxa and 25 KEGG pathways identified as potential mediators of its anti-obesity effects. Notably, PS modulated key obesity-associated gut microbiota, including Alistipes, Lachnoclostridium, Odoribacter, Prevotellaceae UCG-001, Rikenellaceae RC9-gut group, and norank_g Bacteroidales S24-7 group. Serum metabolomics analysis implicated L-ascorbic acid, stevioside, allopurinol, and gingerol, along with amino acid and energy metabolism pathways, in the anti-obesity mechanism of PS. These results provide novel theoretical insights into the therapeutic potential of PS for obesity prevention and treatment.
1 Introduction
Metabolic diseases have emerged as a critical global health challenge (Wu Y. et al., 2024), with obesity representing a central contributor to this epidemic (Zhang et al., 2023). Recent data reveal that 50% of Chinese adults and 25% of children meet the criteria for obesity risk (Wang et al., 2021), a condition projected to incur global economic costs exceeding USD 4.32 trillion annually by 2035 (Mahase, 2023). This escalating crisis has intensified demands for innovative interventions targeting obesity-related mechanisms, including gut microbiota modulation and metabolic regulation (The Lancet Diabetes Endocrinology, 2021).
Paederia scandens (PS), a cost-effective botanical deeply rooted in Chinese traditional medicine, presents unique therapeutic potential (Wang et al., 2014), and is widely consumed as both a medicinal herb and a dietary component in Hainan Province (China). Phytochemical analyses have identified multiple bioactive constituents in PS, including 6’-O-E-feruloylmonotropein (Trung et al., 2023), iridoid glycosides (Xu et al., 2023), and paederosidic acid (Li et al., 2023). Experimental studies demonstrate its multifunctional properties: ameliorating non-alcoholic fatty liver disease through antioxidant enhancement in chickens (Wu et al., 2019), suppressing inflammatory pathways to alleviate diarrhea in mice (Ai et al., 2023), and restoring gut microbial balance in arthritis models (Xiao et al., 2018). These findings collectively position PS as a promising candidate for metabolic regulation, though its anti-obesity mechanisms remain unexplored.
Growing evidence implicates gut microbiota dysbiosis in obesity pathogenesis (Gomes et al., 2018). Specific microbial taxa exhibit distinct roles: Megamonas rupellensis promotes lipid absorption (Wu C. et al., 2024), Fusimonas intestine and Lachnospiraceae facilitate diet-induced obesity (Takeuchi et al., 2023), and Bacteroidetes demonstrates anti-obesity correlations (Ley et al., 2006). Therapeutic agents such as spermidine (Ma et al., 2020) and ginsenoside Rg1 (Liu et al., 2023) mitigate obesity via microbiota modulation, suggesting microbial targets for PS intervention. Concurrently, serum metabolomic studies reveal obesity-associated perturbations in glutamate (Liu et al., 2017) and branched-chain amino acids (Wahl et al., 2012), though PS’s metabolic regulatory effects remain uncharacterized.
To address these knowledge gaps, we established a high-fat diet-induced murine obesity model to systematically investigate PS’s anti-obesity efficacy. PS was orally administered to investigate its effects on body weight, biochemical indicators, and histopathological changes in obese mice. 16S rRNA and metabolomic sequencing were used to determine whether PS supplementation ameliorated fecal microbial dysregulation and serum metabolite disorders in the obesity model. Finally, we identified gut microbes and serum metabolites associated with the underlying protective mechanisms of PS against obesity. This multidisciplinary approach provides critical insights into PS’s therapeutic potential while advancing strategies for combating metabolic disorders.
2 Materials and methods
2.1 Animals and groups
In this study, specific pathogen-free (SPF) male C57BL/6 J mice (aged 5 weeks; body weight 19.97 ± 0.93 g) were obtained from RUIYE laboratories (Guangzhou, China; Production License for Experimental Animals: SCXK(YUE)2022–0063; Certificate No.44829700021758). A total of 22 mice were randomly assigned to 3 groups: normal control (NC, n = 6), obesity model (OB, n = 8), and PS extract treatment (PS, n = 8). All animals were housed at the Experimental Animal Center of Youjiang Medical University for Nationalities (Laboratory Animal Use License: SYXK(GUI)2022–0004) under controlled conditions: temperature 22 ± 2°C, relative humidity 55 ± 5%, and 12-h light/dark cycles. The NC group received a standard diet throughout the 13-week experiment, while the OB and PS groups were fed a high-fat diet (HFD) (4.73 kcal/g digestible energy, Charles River Co., Ltd., Jiaxing, China). From weeks 10 to 13, the NC and OB groups were administered normal saline (1 mL/kg/day), whereas the PS group received the PS extraction solution. All animal feeding, management, and experiment procedures strictly comply with the Regulations on the Management of Experimental Animals (China National Standard GB/T 35823–2018).
2.2 PS extract preparation
PS extract was prepared by boiling 200 g of dried PS raw material (Zexintang Pharmaceutical Co., Ltd., Bozhou, China) in 1000 mL of double-distilled water. The solution was concentrated to 100 mL under reduced pressure, yielding extract with a final PS concentration of 2 g/mL.
2.3 Physiological, biochemical indexes, and sample collection
The weekly body weight was recorded for all mice. Feces samples were collected at week 13. Following isoflurane anesthesia, blood and abdominal adipose tissue were harvested. Blood samples were centrifuged (3,000 rpm, 15 min, 4°C) to isolate serum. Serum total cholesterol (TC) (A111-1-1), triglycerides (TG) (A110-1-1), high-density lipoprotein cholesterol (HDL-C) (A112-1-1), and low-density lipoprotein cholesterol (LDL-C) (A113-1-1) were quantified using ELISA kits (Jiancheng Bioengineering Institute, Nanjing, China). Abdominal fat was fixed in 4% paraformaldehyde and embedded in paraffin blocks (4 μm thickness). Feces samples were immediately snap-frozen in liquid nitrogen and stored at −80°C for subsequent analysis.
2.4 16s rRNA amplicon sequencing
Fecal microbial genomic DNA was extracted using the MagPure Soil DNA LQ Kit (Magen, Cat. No. D6356-02; Guangzhou, China), following the manufacturer’s protocol. DNA quality was verified by NanoDrop 2000 spectrophotometry and 1.5% agarose gel electrophoresis. The V3–V4 region of 16S rRNA was amplified via PCR using barcoded primers 343F (5’-TACGGRAGGCAGCAG-3′) and 798R (5’-AGGGTATCTAATCCT-3′). Libraries were constructed and sequenced on the Illumina MiSeq 6000 platform (OE Biotech, Shanghai, China). The analysis pipeline for 16S rRNA was completed according to a previous study (Mo et al., 2022). Briefly, the raw sequences were analyzed using EasyAmplicon v1.0 with the minimum unique size in the dereplication, and the sintax_cutoff in the removal of plastids and non-bacteria was 12 and 0.1, respectively. After quality control, all the samples were subsampled to 57735 sequences per sample and aligned using the silva_16s_v123 database. Beta diversity was calculated using unweighted UniFrac distances and visualized via principal coordinate analysis (PCoA). The LDA score ≥4 was defined as significant for differential microbes using linear discriminant analysis effect size (Lefse) software. The different KEGG pathways were identified by STAMP software with Bonferroni corrections (p < 0.05).
2.5 Serum untargeted metabolomics
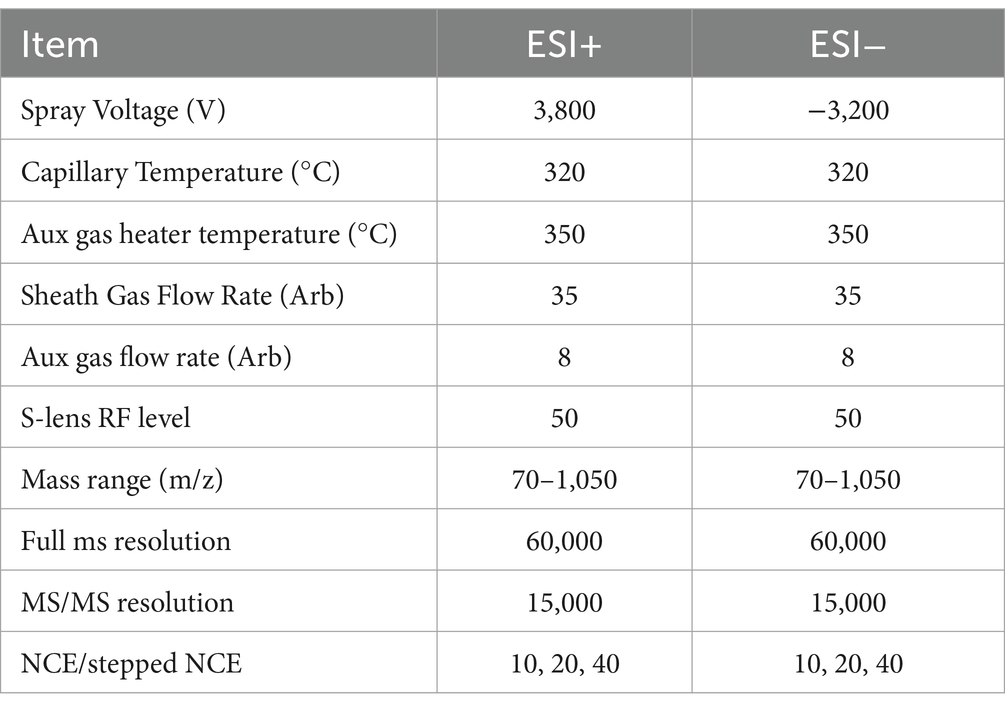
After thawing on ice, each serum sample (80 μL) was transferred to 2 mL Eppendorf (EP) tubes containing 400 μL of protein precipitator solvent (ACN:methanol = 1:2, V/V) spiked with internal standards. Samples were sonicated (10 min) and allowed to equilibrate at −40°C overnight. Following centrifugation (12,000 rpm, 20 min, 4°C), 150 μL of supernatant was transferred to sample vials for liquid chromatography-mass spectrometry (LC–MS) analysis at Oebiotech Company (Shanghai, China). LC–MS analysis was performed using an ACQUITY UPLC HSS T3 column (100 mm × 2.1 mm, 1.8 μm, Waters, Milford, MA, USA). Mass spectrometry parameters for positive/negative ion modes are detailed in Table 1. Chromatographic gradients (mobile phase composition) and instrumental parameter profiles are provided in Tables 2, 3, respectively.
Raw LC–MS data were processed through peak filtration, peak extraction, alignment, and retention time correction using XCMS software (v4.5.1). Metabolite annotation was performed by querying the Human Metabolome Database (HMDB), LipidMaps (v2.3), METLIN, and LuMet-Animal 3.0 databases. Then the principal component analysis (PCA), supervised orthogonal partial least squares discriminant analysis (OPLS-DA), permutation test (200 permutations), and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis were conducted following established protocols (Mo et al., 2021). Metabolites with specific variable importance in projection (VIP) scores (VIP > 2), fold changes (FC) (FC > 4 or FC < 0.25), and p-values (p < 0.001) were defined as significantly differential metabolites. Time-series clustering of metabolite profiles was performed using STEM software (see Table 4).
3 Results
3.1 Addition of PS reduced obesity related indices
During the first and second weeks, the mean body weight of mice in the NC, OB, and PS groups showed no significant difference (p > 0.05) (Figure 1A). Notably, from weeks 3 to 12, HFD-fed mice (OB and PS groups) showed significantly elevated body weights compared to the NC group (p < 0.01) (Figure 1A). By week 13, the body weight in the PS group was significantly higher than in the NC group (p < 0.05; Figure 1A). Intriguingly, no significant differences were observed between the OB and PS groups from weeks 1 to 10 (p > 0.05; Figure 1A). Following PS administration (weeks 11–13), the PS group exhibited progressive weight reduction, culminating in significantly lower body weight versus the OB group at week 13 (p < 0.01; Figure 1A). These findings suggest that PS supplementation ameliorates HFD-induced weight gain. Serum TC, TG, HDL-C, and LDL-C in the OB group were markedly elevated compared to the NC and PS groups (p < 0.05; Figure 1B). Similarly, LDL-C levels in the NC group were significantly suppressed relative to the PS group (p < 0.05; Figure 1B). Histological analysis of inguinal adipose tissue revealed that the size of fat cells in the OB group was larger than those in the NC and PS groups (p > 0.05; Figure 1C). Critically, adipocyte sizes in the PS group did not statistically match those in the NC group (p > 0.05), indicating PS-mediated normalization of adipocyte morphology (Figure 1C). Thus, our results show that HFD exacerbates obesity-related metabolic indices and adipocyte hypertrophy, and PS intervention attenuates HFD-induced dyslipidemia and adipose remodeling.
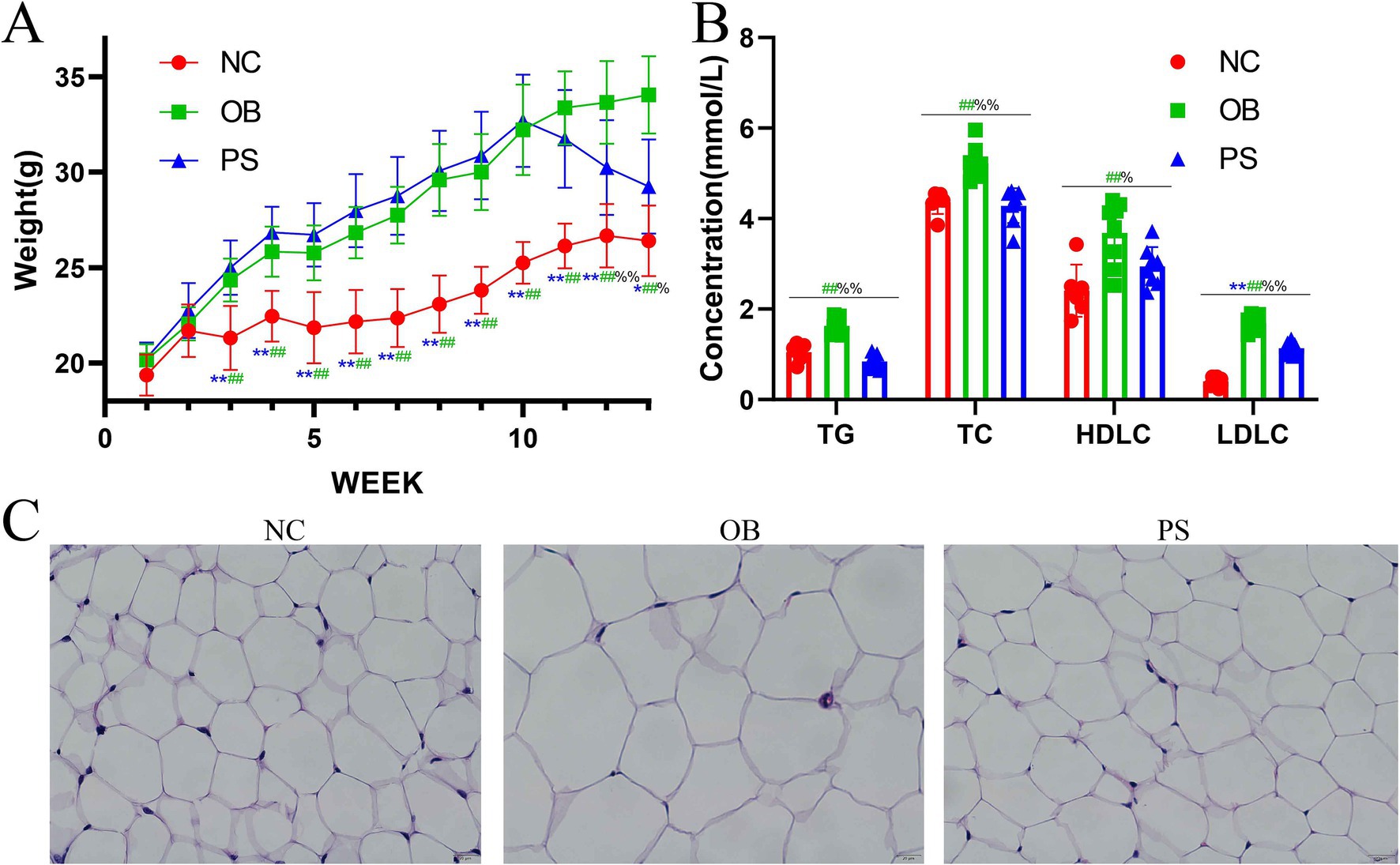
Figure 1. Indices and hematoxylin–eosin (HE)-stained sections from obesity. (A) body weight; (B) TG, TC, HDL-C, and LDL-C concentrations in blood; (C) HE-stained section of abdominal fat in three groups. Note: The ** was the extremely significant difference between the NC group and the PS group (p < 0.01); the ## was the extremely significant difference between the NC group and the OB group (p < 0.01); the %% was the extremely significant difference between the OB group and the PS group (p < 0.01); the * was the significant difference between the NC group and the PS group (p < 0.05); the # was the significant difference between the NC group and the OB group (p < 0.05); and the % was the significant difference between the OB group and the PS group (p < 0.05).
3.2 Addition of PS changes the microbe diversity
The difference in fecal microbiota among the NC, OB, and PS groups was identified using 16S rRNA sequencing. After quality control and dereplication, a total of 1,397,098 reads were obtained, with the number of available sequences ranging from 78,153 to 81,920. The rarefaction curve analysis showed that the data met the requirements for bioinformatics (Figure 2A). A total of 794 amplicon sequence variants (ASVs) were identified, with 169 ASVs (21.3%) shared by all samples and 316 ASVs (39.8%) present in over 90% of the samples. After normalization to 57,735 sequences per sample, both the ACE and Chao1 indices in the OB group were significantly lower than those in the NC group (p < 0.05; Figures 2B,C). However, no significant differences were observed between the PS and NC groups in these indices (p > 0.05; Figures 2B,C). β-diversity analysis revealed structure differences in microbiota composition among the three groups (Figure 2D). Principal coordinates (PCo) 1 and 2 accounted for 54.66 and 19.3% of the total variation, respectively (Figure 2D). Notably, the NC group exhibited closer clustering with the PS group than with the OB group (Figure 2D).
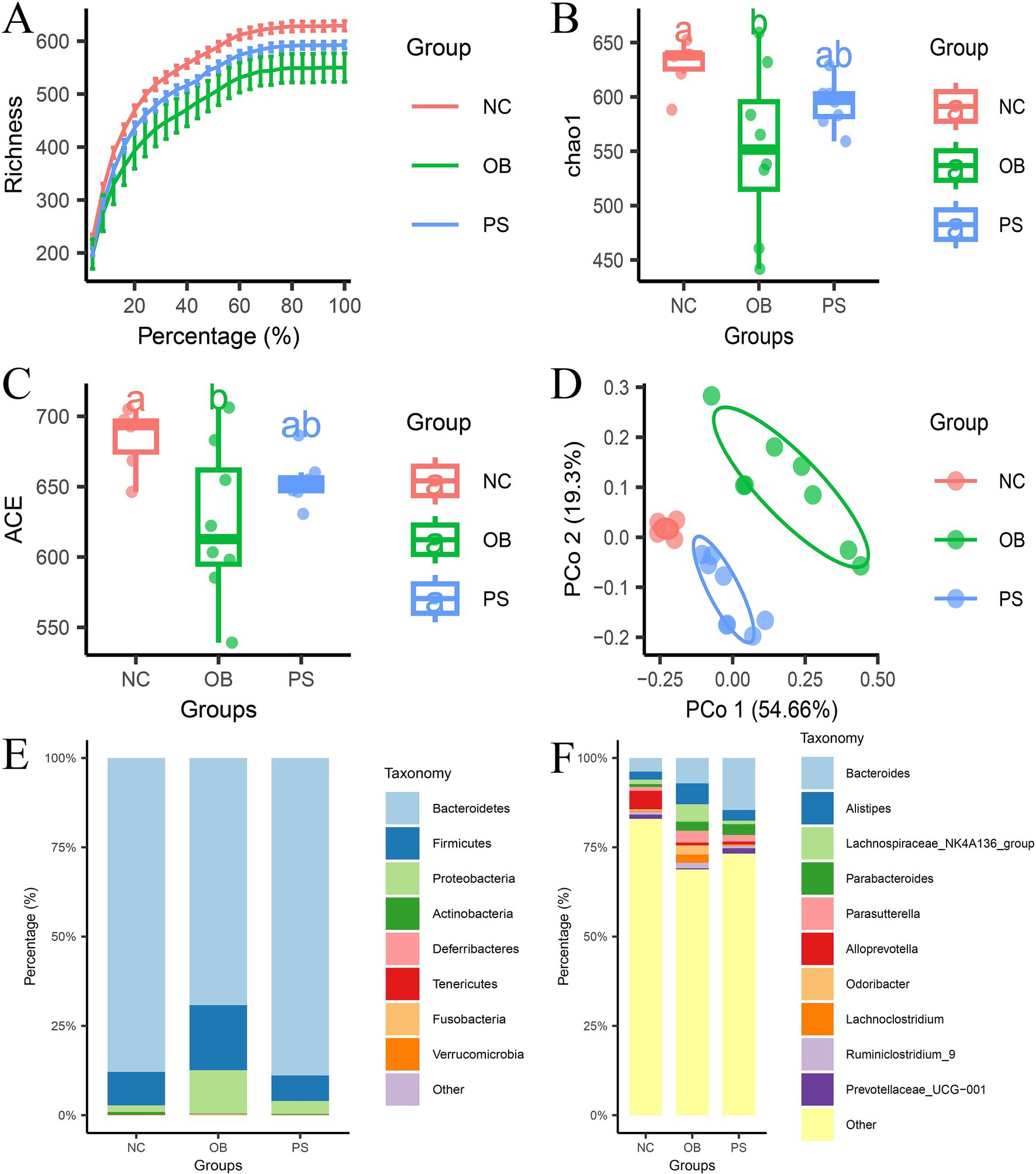
Figure 2. Microbial composition analysis. (A) the rarefaction curve in three groups; (B) the Chao1 index in three groups; (C) the ACE index in three groups; (D) β-diversity in the three groups; (E) the microbial composition at the phylum level; and F: the microbial composition in the genus level.
3.3 Addition of PS changes the microbe composition
The relative abundances of bacteria are shown at the phylum level (Figure 2E) and the genus level (Figure 2F). A total of 8 phyla, 17 classes, 21 orders, 38 families, and 92 genera were identified in this study. The major bacteria in the phyla were Bacteroidetes, Firmicutes, and Proteobacteria, accounting for over 95.7% of the total in each sample. The relative abundance of Bacteroidetes in the NC, OB, and PS groups was 87.87, 69.22, and 88.89%, respectively. The relative abundance of Firmicutes in the NC, OB, and PS groups was 9.41, 18.23, and 7.14%, respectively. The relative abundance of Proteobacteria in the NC, OB, and PS groups was 1.84, 12.07, and 3.64%, respectively. These results showed that the microbial composition at the phylum level in the PS group was closer to that of the NC group than to that of the OB group. The main bacterial genera in the NC group at the genus level were Alloprevotella, Bacteroides, Alistipes, Ruminococcaceae UCG-014, and the Lachnospiraceae NK4A136 group. The main bacterial genera in the OB group at the genus level were Bacteroides, Alistipes, Lachnospiraceae NK4A136 group, Parasutterella, Odoribacter, Parabacteroides, and Lachnoclostridium. The main bacterial genera in the PS group at the genus level were Bacteroides, Parabacteroides, Alistipes, Parasutterella, and Rikenellaceae RC9-gut group. These results indicate that the addition of PS changed the dominant microbial composition in feces, and these changes may be related to the protective mechanism of PS against HFD-induced obesity in mice.
3.4 Difference in microbes and pathways between the NC and OB groups
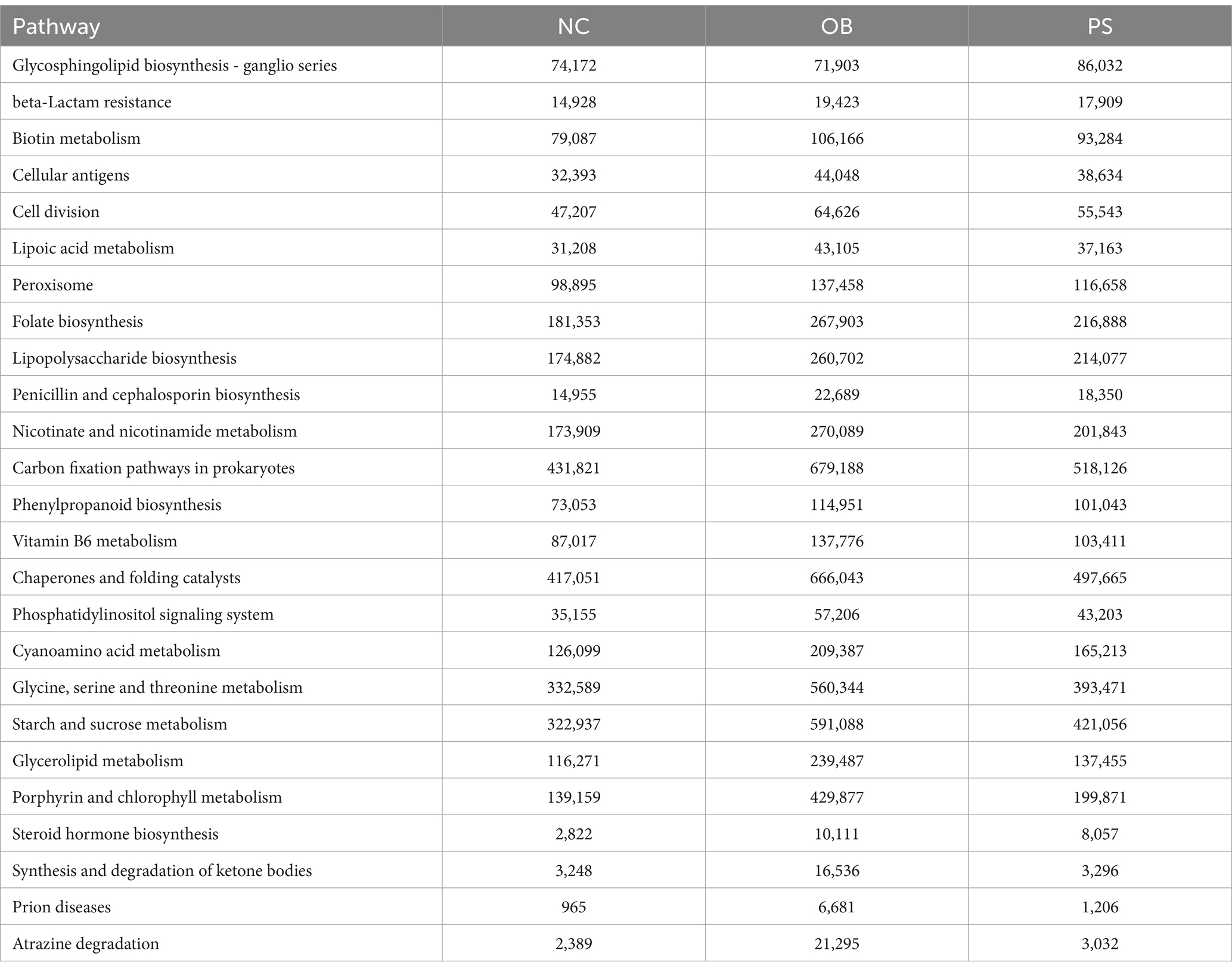
The relative abundances of 19 microbes showed significant differences between the NC and OB groups (p < 0.05), including Alloprevotella, norank_g Bacteroidales S24-7 group, Bacteroidales S24-7 group, Prevotellaceae, Bacteroidales, Bacteroidia, and Bacteroidetes, which were significantly higher in the NC group than in the OB group (Figure 3A). The relative abundance of Alistipes, Bacteroides, Lachnoclostridium, norank_g Enterobacteriaceae, Bacteroidaceae, Lachnospiraceae, Porphyromonadaceae, Rikenellaceae, Clostridiales, Clostridia, Firmicutes, and Proteobacteria was significantly lower in the NC group than in the OB group (Figure 3A). The KEGG pathway analysis revealed that 21 KEGG pathways in the NC group were significantly more abundant than those in the OB group, including glycosphingolipid biosynthesis - ganglio series, protein digestion and absorption, biotin metabolism, lipoic acid metabolism, lipopolysaccharide biosynthesis, folate biosynthesis, nicotinate and nicotinamide metabolism, carbon fixation pathways in prokaryotes, vitamin B6 metabolism, and glycine, serine and threonine metabolism pathways, among others (Figure 3B). Additionally, nine KEGG pathways in the NC group were significantly less abundant than those in the OB group, including glycerolipid metabolism, steroid hormone biosynthesis, and synthesis and degradation of ketone bodies pathways, among others (Figure 3B).
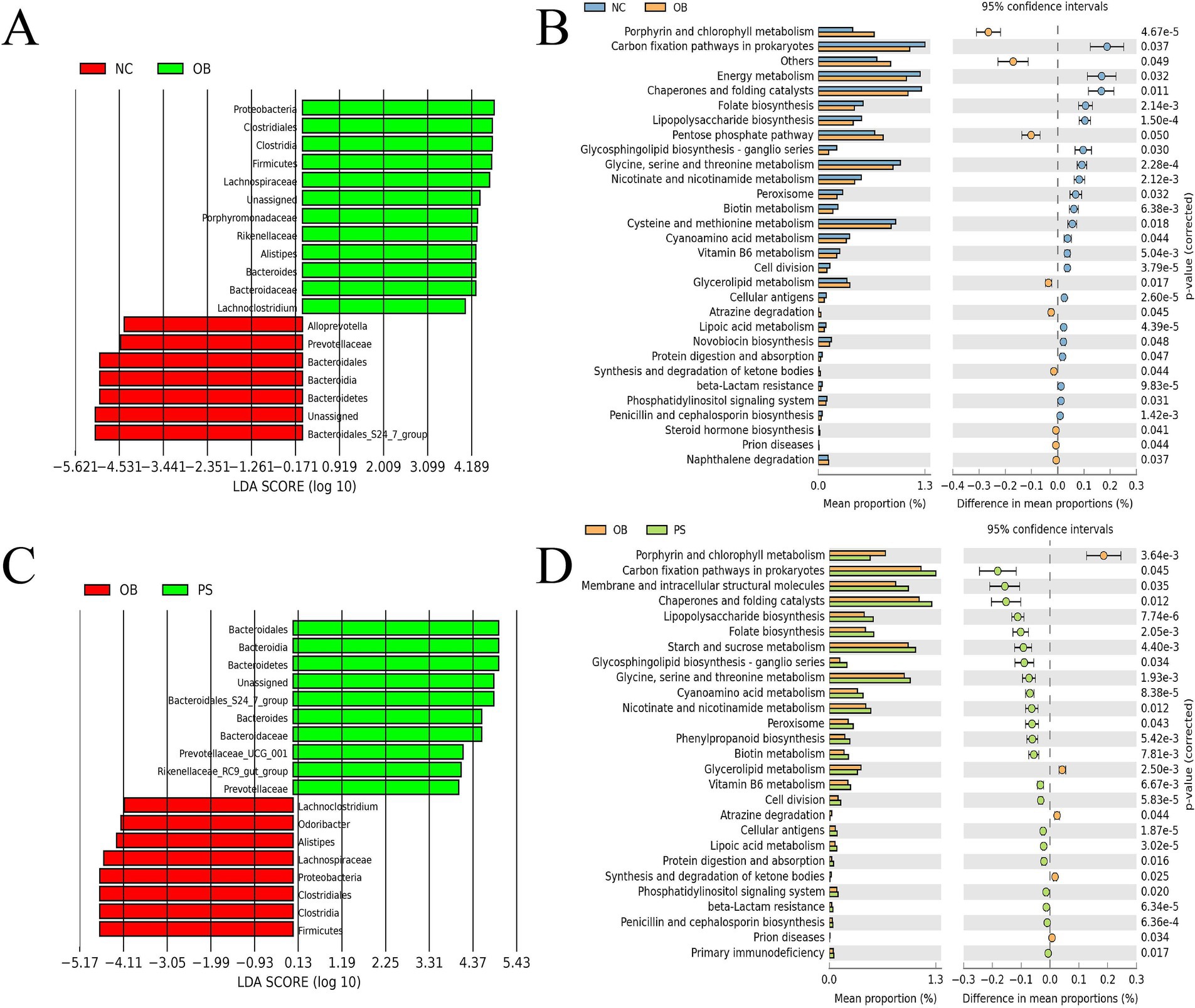
Figure 3. Difference in microbes and pathways. (A) difference in microbes between the NC and OB groups. (B) the difference in pathways between the NC and OB groups. (C) difference in microbes between the OB and PS groups. (D) difference in pathways between the OB and PS groups.
3.5 Difference in microbes and pathways between the OB and PS groups
The relative abundances of 18 microbes showed significant differences between the OB and PS groups (p < 0.05), including Alistipes, Lachnoclostridium, Odoribacter, Lachnospiraceae, Clostridiales, Clostridia, Firmicutes, and Proteobacteria, which were significantly higher in the OB group than in the PS group (Figure 3C). The relative abundance of Bacteroides, Prevotellaceae UCG-001, Rikenellaceae RC9-gut group, norank_g Bacteroidales S24-7 group, Bacteroidaceae, Bacteroidales S24-7 group, Prevotellaceae, Bacteroidales, Bacteroidia, and Bacteroidetes was significantly lower in the OB group than in the PS group (Figure 3C). The KEGG pathway analysis revealed that five KEGG pathways in the OB group were significantly more abundant than those in the PS group, including synthesis and degradation of ketone bodies and glycerolipid metabolism pathways (Figure 3D). Additionally, 22 KEGG pathways in the OB group were significantly less abundant than those in the PS group, including glycine, serine, and threonine metabolism; starch and sucrose metabolism; nicotinate and nicotinamide metabolism; vitamin B6 metabolism; carbon fixation pathways in prokaryotes; folate biosynthesis; lipopolysaccharide biosynthesis; lipoic acid metabolism; biotin metabolism; glycosphingolipid biosynthesis—ganglio series; and protein digestion and absorption pathways, among others (Figure 3D).
3.6 Addition of PS restored the disorder of the feces microbe
Based on microbial differences among the NC, OB, and PS groups, our study identified seven microbial taxa in the OB group with significantly higher relative abundance than those in both the NC and PS groups, including Lachnoclostridium, Alistipes, Proteobacteria, Firmicutes, Clostridiales, Lachnospiraceae, and Clostridia (p < 0.05). Meanwhile, the relative abundance of these taxa in the PS group was higher than that in the NC group, though not statistically significant (p > 0.05). Additionally, the relative abundance of Rikenellaceae was significantly higher in the OB and PS groups compared to the NC group (p < 0.05), with the OB group showing a non-significantly higher abundance than the PS group (p > 0.05). Thus, eight differential microbial taxa exhibited the highest abundance in the OB group, followed by the PS group, while the NC group showed the lowest levels (Figure 4A).
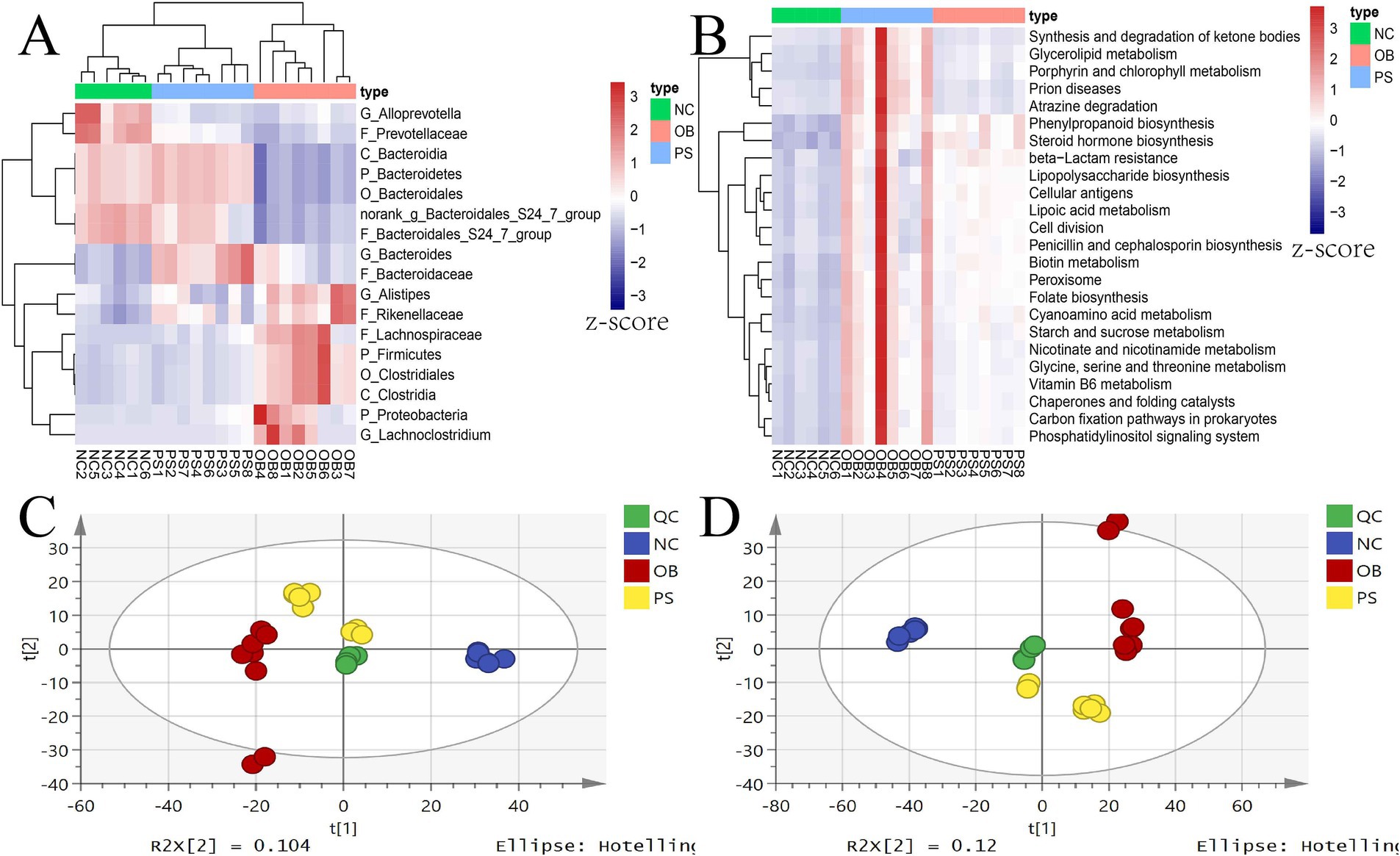
Figure 4. Microbial taxa and pathways related to PS anti-obesity and the PCA plot of metabolites. (A) the microbial taxa related to PS anti-obesity; (B) the microbial pathways related to PS anti-obesity; (C) the PCA plot in the positive ion model; and (D) the PCA plot in the negative ion model.
Three distinct microbial taxa in the OB group exhibited significantly lower relative abundances than those in both the NC and PS groups, including Bacteroidetes, Bacteroidales, and Bacteroidia (p < 0.05). Meanwhile, the relative abundance of these taxa in the PS group was higher than that in the NC group, though not statistically significant (p > 0.05). Thus, these three taxa showed the highest abundance in the PS group, followed by the NC group, and the lowest in the OB group. Additionally, three differential taxa in the NC group were significantly more abundant than those in the OB and PS groups (p < 0.05), with the PS group also showing significantly higher levels than the OB group (p < 0.05), including the Bacteroidales S24-7 group, Prevotellaceae, and norank_g Bacteroidales S24-7 group. Therefore, their abundance was highest in the NC group, intermediate in the PS group, and lowest in the OB group (Figure 4A). In summary, PS supplementation alleviated fecal microbial dysbiosis by reducing the abundance of Lachnoclostridium, Alistipes, Proteobacteria, Firmicutes, Clostridiales, Lachnospiraceae, Rikenellaceae, and Clostridia while enhancing the abundance of Bacteroidetes, Bacteroidales, Bacteroidia, Bacteroidales S24-7 group, Prevotellaceae, and norank_g Bacteroidales S24-7 group.
Meanwhile, the dysregulation of 25 significantly altered KEGG pathways was normalized by PS supplementation, such as the decrease of the glycosphingolipid biosynthesis—ganglio series pathway in the OB group was restored by PS intervention (Figure 4B). Additionally, the abnormally elevated levels of biotin metabolism, lipoic acid metabolism, folate biosynthesis, lipopolysaccharide biosynthesis, nicotinate and nicotinamide metabolism, carbon fixation pathways in prokaryotes, vitamin B6 metabolism, glycine, serine and threonine metabolism, starch and sucrose metabolism, glycerolipid metabolism, steroid hormone biosynthesis, and synthesis and degradation of ketone body pathways in the OB group were also restored by PS (Figure 4B). These findings suggest that PS may counteract obesity induced by HFDs by modulating these 25 microbial KEGG pathways.
3.7 Addition of PS changes the serum metabolites profiles
A total of 3,084 metabolites were identified in the serum metabolome, with 1,413 and 1,671 metabolites in the positive and negative ion modes, respectively (Supplementary Table S1). PCA results indicated that the model interpretation rates of X (R2X) for the positive and negative ion modes were 0.456 and 0.564, respectively. In both PCA score plots, the four quality control (QC) samples clustered tightly, confirming data reproducibility (Figures 4C,D). Notably, the PS group was positioned between the NC and OB groups in both the positive and negative ion modes.
3.8 Difference in metabolites and pathways between the NC and OB groups
OPLS-DA analysis revealed a clear separation between the NC and OB groups in both ion modes (Figures 5A,B). The model interpretation rates of Y (R2Y) in the positive and negative ion modes were 0.662 and 0.761, respectively. The prediction ability (Q2) in the positive and negative ion modes was over 0.99 in both modes. Permutation test intercepts for both modes were below −0.45, confirming model validity. Among 136 significantly differential metabolites, 56 were upregulated and 80 were downregulated in the NC group compared to the OB group (Supplementary Table S2). MetaboAnalyst 4.0 analysis identified seven enriched metabolic pathways, including D-amino acid metabolism, citrate cycle (TCA cycle), pyruvate metabolism, glycolysis/gluconeogenesis, alanine, aspartate and glutamate metabolism, glyoxylate and dicarboxylate metabolism, and the arachidonic acid metabolism pathway (Figure 5C).
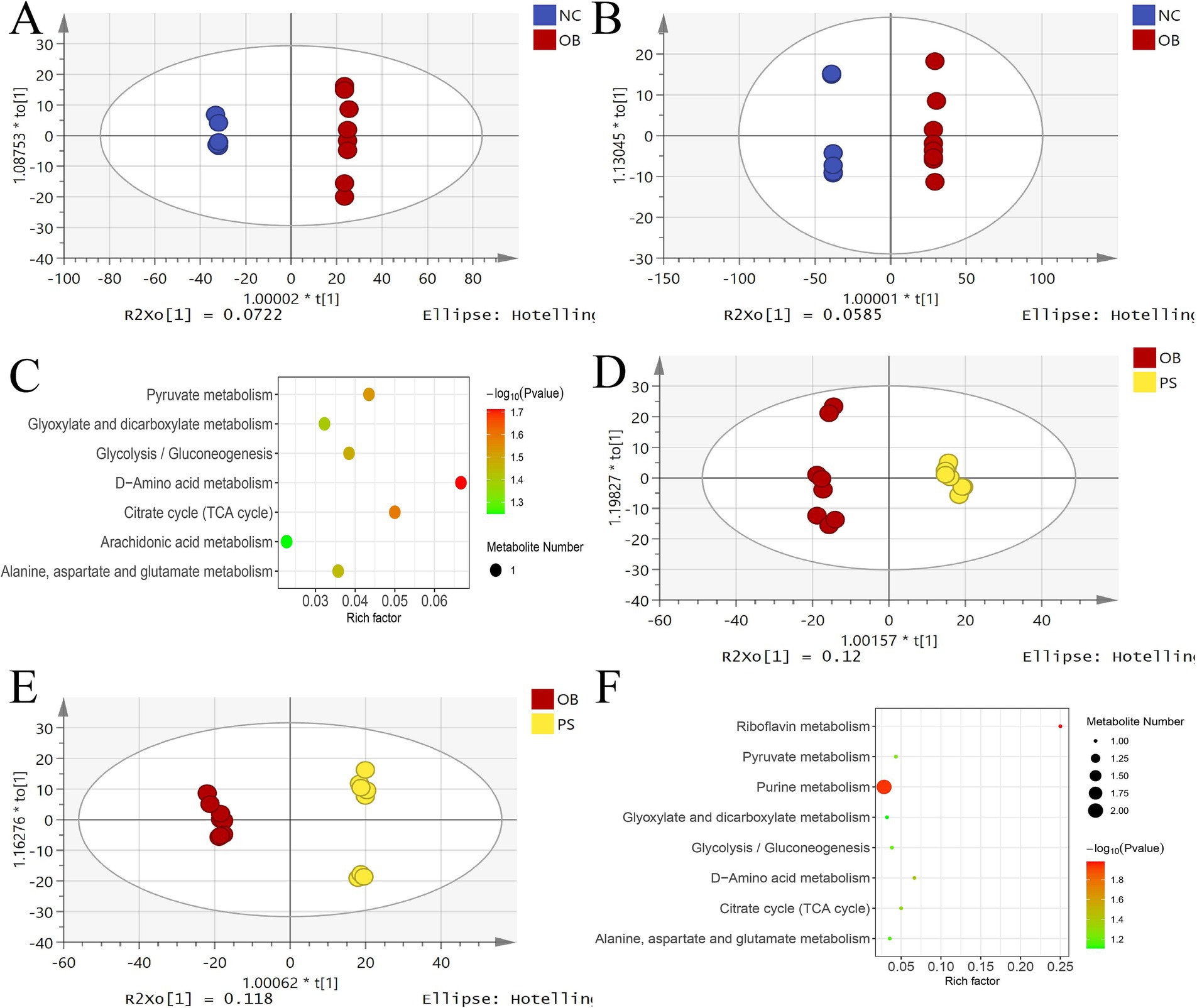
Figure 5. OPLS-DA analysis in different groups. (A) the OPLS-DA score plot in positive between the NC and OB groups; (B) the OPLS-DA score plot in negative between the NC and OB groups; (C) the KEGG plot of significantly different metabolites between the NC and OB groups; (D) the OPLS-DA score plot in positive between the PS and OB groups; (E) the OPLS-DA score plot in negative between the PS and OB groups; and (F) the KEGG plot of significantly different metabolites between the PS and OB groups.
3.9 Difference in metabolites and pathways between the OB and PS groups
OPLS-DA analysis revealed a clear separation between the OB and PS groups in both ion modes (Figures 5D,E). The R2Y values for the positive and negative ion modes were 0.437 and 0.488, respectively. The Q2 in both the positive and negative ion modes was over 0.93 in both modes. Permutation test intercepts for both modes were below −0.28, confirming model reliability. Among 60 significantly differential metabolites, 6 were upregulated and 54 downregulated in the OB group compared to the PS group (Supplementary Table S3). MetaboAnalyst 4.0 analyzes eight enriched metabolic pathways, including riboflavin metabolism, purine metabolism, D-amino acid metabolism, citrate cycle (TCA cycle), pyruvate metabolism, glycolysis/gluconeogenesis, alanine, aspartate and glutamate metabolism, and glyoxylate and dicarboxylate metabolism pathway (Figure 5F).
3.10 Addition of PS restored the disorder of serum metabolites
Hierarchical cluster analysis of all significant differential metabolites was performed using the Stem software. From the NC to PS and then OB groups, 189 significant difference metabolites were stratified into three distinct clusters (Figure 6A). Clusters 2, 3, and 11 contained 46, 28, and 18 metabolites, respectively (Supplementary Table S4; Figures 6B–D). Notably, metabolite concentration in clusters 2 and 3 of the PS group was intermediate between the NC and OB groups, suggesting that these 74 metabolites (cluster 2 + 3) may be associated with PS-mediated alleviation of HFD-induced obesity in mice. These metabolites were enriched in seven metabolic pathways, including D-amino acid metabolism, citrate cycle (TCA cycle), pyruvate metabolism, glycolysis/gluconeogenesis, alanine, aspartate and glutamate metabolism, glyoxylate and dicarboxylate metabolism, and the arachidonic acid metabolism pathway (Figure 6E).
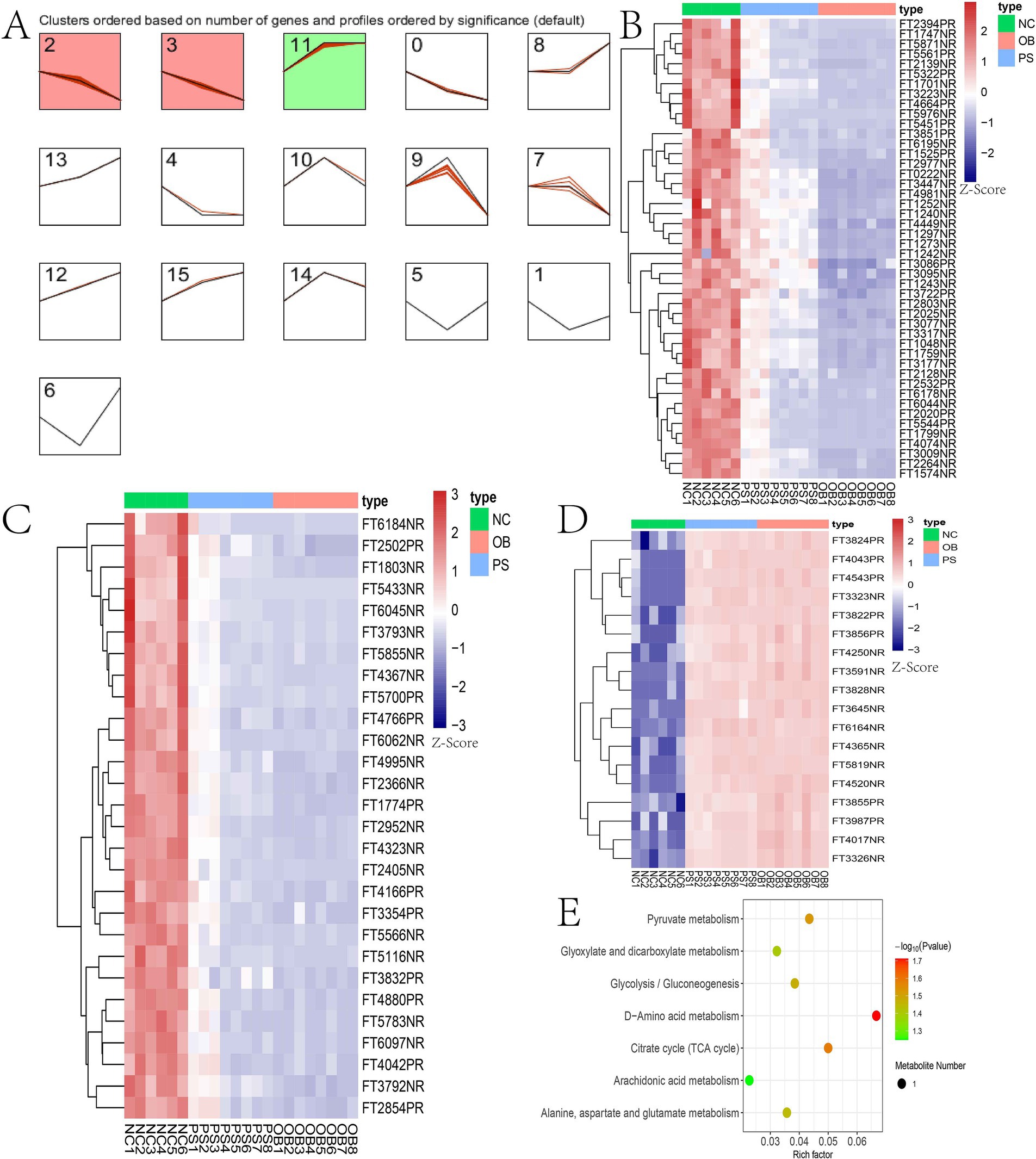
Figure 6. Hierarchical cluster analysis results. (A) the hierarchical cluster of 189 metabolites; (B) the metabolites in cluster 2; (C) the metabolites in cluster 3; (D) the metabolites in cluster 11; (E) the KEGG plot of clusters 2 and 3.
4 Discussion
To investigate the protective mechanisms of PS against obesity, we established an HFD-induced obesity model in mice and administered PS via oral gavage to obese subjects. Our study demonstrated that HFD-fed mice exhibited significantly higher body weight than those on a standard diet, consistent with previous findings (Le Roy et al., 2022). Following PS intervention starting at week 10, the body weight of the PS group became lower than the OB group, with statistically significant differences emerging by week 13. These results indicate that PS mitigates high-fat diet-induced weight gain in mice. Similar to the effects of semaglutide (Ren et al., 2022), PS treatment significantly reduced serum lipid levels, including TC, TG, HDL-C, and LDL-C. Notably, TC, TG, and HDL-C levels in the PS group returned to normal ranges compared to the OB group. Additionally, the adipocyte size in the PS group was markedly smaller than in the OB group. Thus, our results showed that the oral administration of PS restored body weight, normalized blood lipid indices, and reduced adipocyte hypertrophy in obese mice. These results confirm that oral PS supplementation is an effective strategy for alleviating obesity in murine models.
Our microbial analysis revealed that HFD decreased α-diversity in mice, while PS intervention restored it to baseline levels. β-diversity metrics demonstrated closer clustering between the NC and PS groups compared to the OB group, indicating PS-mediated restoration of gut microbiota composition in obesity. Differential abundance analysis identified seven microbes that were decreased, and 12 microbes were enhanced in the OB group compared to the NC group. These findings align with prior studies of HFD-induced depletion of Alloprevotella and Prevotellaceae, alongside enrichment of Alistipes (Kong et al., 2019). Wei et al. (2020) indicated that the HFD decreased the abundance of the Bacteroidales S24-7 group and Bacteroidetes and increased the abundance of Firmicutes and Proteobacteria. Gong et al. (2022) reported that the abundance of Lachnoclostridium in the obesity group was enhanced. Duan et al. (2024) found that the abundance of Lachnospiraceae and Bacteroides in HFD mice was enhanced. Yang et al. (2021) evidenced that the abundance of Porphyromonadaceae in HFD mice was improved, and Daniel et al. (2014) declared that the abundance of Rikenellaceae in HFD mice was enhanced. Notably, PS treatment reversed 14 of 18 OB-associated microbial dysregulations, including eight microbes [Alistipes (Kong et al., 2019), Lachnoclostridium (Gong et al., 2022), Lachnospiraceae (Duan et al., 2024), Rikenellaceae (Daniel et al., 2014), Clostridiales, Clostridia, Firmicutes, and Proteobacteria] that were decreased by the PS addition, and six microbes (norank_g Bacteroidales S24-7 group (Wei et al., 2020), Bacteroidales S24-7 group (Wei et al., 2020), Prevotellaceae (Kong et al., 2019), Bacteroidales, Bacteroidia, and Bacteroidetes) that were increased by the PS addition. Meanwhile, the abundance of Odoribacter, Prevotellaceae UCG-001, and Rikenellaceae RC9-gut group in the PS group was significantly higher than those in the OB group. Lachnoclostridium (Dicks, 2024) and Alistipes (Chanda and De, 2024) were the short-chain fatty acid (SCFA) producers, and the SCFAs can affect adipose tissue metabolism, lipid oxidation capacity, β cell function, and insulin secretion via the SCFA-sensing G protein-coupled receptors (GPCRs), SCFA transporter (SLC16A and SLC5A), and other SCFA receptor-independent pathways (You et al., 2022). The higher abundances of Lachnoclostridium and Alistipes in obesity samples in our study and previous studies may be related to the negative feedback mechanism of the body. The norank_g Bacteroidales S24-7 group was a butyrate-producing bacterium (Fang et al., 2020), and the butyrate can resist inflammation and obesity (Liu et al., 2018) through GPCR signaling pathways (Saban Güler et al., 2025). The relationship between the Prevotellaceae UCG-001 and obesity in the body (Han et al., 2024) may be related to the butyrate because the butyrate can enhance the abundance of Prevotellaceae UCG-001 in the gut (Wang et al., 2024). As a potentially obesity-related biomarker (Liu et al., 2022; Vallianou et al., 2023), the Odoribacter improves glucose control and inflammatory profile in obese mice by depleting circulating succinate (Huber-Ruano et al., 2022). A previous study showed that the Rikenellaceae RC9-gut group was related to HFD-induced obesity (Fu et al., 2022), and the Rikenellaceae RC9-gut group was positively correlated with HFD-induced “harmful indicators” and negatively correlated with “beneficial indicators” (Gao et al., 2020). Thus, PS might alleviate obesity in mice by restoring the abundance at the genus levels of Alistipes, Lachnoclostridium, Odoribacter, Prevotellaceae UCG-001, Rikenellaceae RC9-gut group, and norank_g Bacteroidales S24-7 group.
Our metabolomic profiling revealed that the PS group clustered intermediately between the NC and OB groups in both positive and negative ion modes, which was the same as the β-diversity results in the microbe analysis. This spatial distribution suggests the partial restoration of serum metabolite profiles in HFD-induced obese mice by PS intervention. Differential metabolite analysis identified 136 serum metabolites significantly altered by HFD, including cholestanetriol, gingerol, isovaleric acid, and stevioside. Zhu et al. (2021) illustrated that the HFD enhanced the serum cholestanetriol in mice, which was consistent with our study. Same as our study, isovaleric acid was an SCFA in the body and regulated obesity by decreasing lipogenesis (Heimann et al., 2016). Saravanan et al. (2014) reported that the gingerol could decrease the glucose level, body weight, leptin, insulin, amylase, lipase plasma, and tissue lipids in obesity mice. Brahma Naidu et al. (2016) proposed that it might modulate the lipid metabolism through decreasing activity in lipogenesis as well as increasing fatty-acid oxidation, which is contrary to our study. Wang et al. (2012) found that the stevioside may ameliorate insulin resistance in HFD mice by attenuating adipose tissue inflammation and inhibiting the NF-κB pathway. These results may be due to the negative feedback mechanism of the body. The KEGG analysis showed that two amino acid metabolism and three energy metabolism-related pathways were enriched in the different metabolites, which was consistent with the different pathways in the microbe analysis. Thus, the HFD may induce obesity in mice through enhancing the levels of cholestanetriol, gingerol, and stevioside and decreasing the levels of isovaleric acid.
A total of 60 metabolites were identified as significantly different between the PS group and the OB group, including allopurinol, inosine, and L-ascorbic acid. Consistent with our study, allopurinol not only alleviates HFD-induced hypertension and proteinuria (El-Bassossy and Shaltout, 2015) but also ameliorates the HFD-induced hepatic steatosis through modulation of lipid metabolism, inflammation, and the ER stress pathway in mice (Cho et al., 2021). Niemann et al. (2022) demonstrated that inosine can combat obesity by regulating thermogenic fat, which is consistent with our findings. Lee et al. (2019) reported that ascorbic acid suppresses HFD-induced obesity and non-alcoholic fatty liver disease is activation of PPARα. KEGG analysis revealed that two amino acid metabolism and three energy metabolism-related pathways were enriched among the differential metabolites, aligning with the pathways identified in microbial analysis. Cluster analysis indicated that 74 metabolites may be associated with the PS-mediated alleviation of HFD-induced obesity in mice, including allopurinol (El-Bassossy and Shaltout, 2015), cholestanetriol (Zhu et al., 2021), gingerol (Saravanan et al., 2014), inosine (Niemann et al., 2022), L-ascorbic acid (Lee et al., 2019), and stevioside (Wang et al., 2012). Notably, the disrupted concentrations of inosine, L-ascorbic acid, stevioside, allopurinol, and gingerol in the OB group were restored by PS treatment. Furthermore, these 74 metabolites were enriched in two amino acid metabolism and three energy metabolism-related pathways, consistent with the microbial analysis. Collectively, PS might alleviate obesity in mice by restoring serum concentrations of inosine, L-ascorbic acid, stevioside, allopurinol, and gingerol and by regulating amino acid and energy metabolism-related pathways.
However, there were some limitations to our study. For instance, due to the short experimental period, the long-term effect might not be observed; the PS extraction method might have a possible influence on its active ingredients; the number of individuals in each group was small, and the PS administration groups with different doses were absent. We will improve these problems through further study.
5 Conclusion
In this study, an HFD was used to successfully establish a mice obesity model. Oral administration of PS significantly alleviated obesity, as evidenced by reductions in TG, TC, LDL-C, and HDL-C levels, as well as decreased adipocyte size. 16S rRNA sequencing analysis revealed that PS modulated the abundance of 17 microbial taxa associated with obesity mitigation, including Alistipes, Lachnoclostridium, Odoribacter, Prevotellaceae UCG-001, Rikenellaceae RC9-gut group, and norank_g Bacteroidales S24-7 group. Metabolomic analysis identified serum metabolites such as L-ascorbic acid, stevioside, allopurinol, and gingerol, along with amino acid metabolism and energy metabolism-related pathways, as key mechanisms underlying the anti-obesity effects of PS. These findings provide novel theoretical and experimental support for the potential application of PS in obesity prevention and treatment.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://ngdc.cncb.ac.cn/, CRA021056.
Ethics statement
The animal study was approved by Youjiang Medical University for Nationalities. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
YY: Conceptualization, Data curation, Writing – original draft. JS: Conceptualization, Data curation, Writing – original draft. JM: Methodology, Software, Writing – original draft. JL: Investigation, Supervision, Writing – review & editing. BP: Supervision, Writing – review & editing. YP: Validation, Writing – review & editing. LJ: Visualization, Writing – review & editing. DW: Project administration, Resources, Writing – review & editing. XF: Funding acquisition, Resources, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was funded by Youjiang Medical University for Nationalities high-level talent research project (grant number RZ2300001266); Baise City science and technology bureau research topic (grant number 20221428).
Acknowledgments
We would like to thank the State Key Laboratory for Conservation and Utilization of Subtropical Agro-bioresources for its instrument support.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1554537/full#supplementary-material
References
Ai, L., Guo, L., Liu, W., Xue, X., Li, L., Sheng, Z., et al. (2023). Determination and mechanism of antidiarrheal chemical constituents of Paederia scandens determined by HPLC-ESI-MS integrated with network pharmacology. ACS Omega 8, 28834–28845. doi: 10.1021/acsomega.3c03887
Brahma Naidu, P., Uddandrao, V. V. S., Ravindar Naik, R., Suresh, P., Meriga, B., Begum, M. S., et al. (2016). Ameliorative potential of gingerol: promising modulation of inflammatory factors and lipid marker enzymes expressions in HFD induced obesity in rats. Mol. Cell. Endocrinol. 419, 139–147. doi: 10.1016/j.mce.2015.10.007
Chanda, D., and De, D. (2024). Meta-analysis reveals obesity associated gut microbial alteration patterns and reproducible contributors of functional shift. Gut Microbes 16:2304900. doi: 10.1080/19490976.2024.2304900
Cho, I.-J., Oh, D.-H., Yoo, J., Hwang, Y.-C., Ahn, K. J., Chung, H.-Y., et al. (2021). Allopurinol ameliorates high fructose diet induced hepatic steatosis in diabetic rats through modulation of lipid metabolism, inflammation, and ER stress pathway. Sci. Rep. 11:9894. doi: 10.1038/s41598-021-88872-7
Daniel, H., Gholami, A. M., Berry, D., Desmarchelier, C., Hahne, H., Loh, G., et al. (2014). High-fat diet alters gut microbiota physiology in mice. ISME J. 8, 295–308. doi: 10.1038/ismej.2013.155
Dicks, L. M. T. (2024). How important are fatty acids in human health and can they be used in treating diseases? Gut Microbes 16:2420765. doi: 10.1080/19490976.2024.2420765
Duan, X., Zhang, L., Liao, Y., Lin, Z., Guo, C., Luo, S., et al. (2024). Semaglutide alleviates gut microbiota dysbiosis induced by a high-fat diet. Eur. J. Pharmacol. 969:176440. doi: 10.1016/j.ejphar.2024.176440
El-Bassossy, H. M., and Shaltout, H. A. (2015). Allopurinol alleviates hypertension and proteinuria in high fructose, high salt and high fat induced model of metabolic syndrome. Transl. Res. 165, 621–630. doi: 10.1016/j.trsl.2014.11.008
Fang, S., Chen, X., Pan, J., Chen, Q., Zhou, L., Wang, C., et al. (2020). Dynamic distribution of gut microbiota in meat rabbits at different growth stages and relationship with average daily gain (ADG). BMC Microbiol. 20:116. doi: 10.1186/s12866-020-01797-5
Fu, S., Dang, Y., Xu, H., Li, A., Zhou, X., Gao, X., et al. (2022). Aloe Vera-fermented beverage ameliorates obesity and gut dysbiosis in high-fat-diet mice. Food Secur. 11:3728. doi: 10.3390/foods11223728
Gao, X., Chang, S., Liu, S., Peng, L., Xie, J., Dong, W., et al. (2020). Correlations between α-linolenic acid-improved multitissue homeostasis and gut microbiota in mice fed a high-fat diet. mSystems 5, e00391–e00320. doi: 10.1128/mSystems.00391-20
Gomes, A. C., Hoffmann, C., and Mota, J. F. (2018). The human gut microbiota: metabolism and perspective in obesity. Gut Microbes 9, 1–18. doi: 10.1080/19490976.2018.1465157
Gong, J., Shen, Y., Zhang, H., Cao, M., Guo, M., He, J., et al. (2022). Gut microbiota characteristics of people with obesity by Meta-analysis of existing datasets. Nutrients 14:2993. doi: 10.3390/nu14142993
Han, Y.-H., Cui, X.-W., Li, Y.-X., Chen, X., Zhang, H., Zhang, Y., et al. (2024). Bacterial cellulose is a desirable biological macromolecule that can prevent obesity via modulating lipid metabolism and gut microbiota. Int. J. Biol. Macromol. 283:137522. doi: 10.1016/j.ijbiomac.2024.137522
Heimann, E., Nyman, M., Pålbrink, A.-K., Lindkvist-Petersson, K., and Degerman, E. (2016). Branched short-chain fatty acids modulate glucose and lipid metabolism in primary adipocytes. Adipocytes 5, 359–368. doi: 10.1080/21623945.2016.1252011
Huber-Ruano, I., Calvo, E., Mayneris-Perxachs, J., Rodríguez-Peña, M.-M., Ceperuelo-Mallafré, V., Cedó, L., et al. (2022). Orally administered Odoribacter laneus improves glucose control and inflammatory profile in obese mice by depleting circulating succinate. Microbiome 10:135. doi: 10.1186/s40168-022-01306-y
Kong, C., Gao, R., Yan, X., Huang, L., and Qin, H. (2019). Probiotics improve gut microbiota dysbiosis in obese mice fed a high-fat or high-sucrose diet. Nutrition 60, 175–184. doi: 10.1016/j.nut.2018.10.002
Le Roy, T., Moens de Hase, E., Van Hul, M., Paquot, A., Pelicaen, R., Régnier, M., et al. (2022). Dysosmobacter welbionis is a newly isolated human commensal bacterium preventing diet-induced obesity and metabolic disorders in mice. Gut 71, 534–543. doi: 10.1136/gutjnl-2020-323778
Lee, H., Ahn, J., Shin, S. S., and Yoon, M. (2019). Ascorbic acid inhibits visceral obesity and nonalcoholic fatty liver disease by activating peroxisome proliferator-activated receptor α in high-fat-diet-fed C57BL/6J mice. Int. J. Obes. 43, 1620–1630. doi: 10.1038/s41366-018-0212-0
Ley, R. E., Turnbaugh, P. J., Klein, S., and Gordon, J. I. (2006). Microbial ecology: human gut microbes associated with obesity. Nature 444, 1022–1023. doi: 10.1038/4441022a
Li, Y., Li, Y., Zhu, Y., Ji, W., Wang, Y., Dong, X., et al. (2023). Structure-based virtual screening for discovery of paederosidic acid from Paederia scandens as novel P2Y14R antagonist. Phytomedicine 115:154851. doi: 10.1016/j.phymed.2023.154851
Liu, W., Fang, X., Zhou, Y., Dou, L., and Dou, T. (2022). Machine learning-based investigation of the relationship between gut microbiome and obesity status. Microbes Infect. 24:104892. doi: 10.1016/j.micinf.2021.104892
Liu, R., Hong, J., Xu, X., Feng, Q., Zhang, D., Gu, Y., et al. (2017). Gut microbiome and serum metabolome alterations in obesity and after weight-loss intervention. Nat. Med. 23, 859–868. doi: 10.1038/nm.4358
Liu, Y., Jin, Z.-Y., Wang, J.-X., Wang, D., Liu, H., Li, D., et al. (2023). Ginsenoside Rg1 activates brown adipose tissue to counteract obesity in high-fat diet-fed mice by regulating gut microbes and bile acid composition. Food Funct. 14, 4696–4705. doi: 10.1039/d2fo03142f
Liu, H., Wang, J., He, T., Becker, S., Zhang, G., Li, D., et al. (2018). Butyrate: a double-edged sword for health? Adv. Nutr. 9, 21–29. doi: 10.1093/advances/nmx0009
Ma, L., Ni, Y., Wang, Z., Tu, W., Ni, L., Zhuge, F., et al. (2020). Spermidine improves gut barrier integrity and gut microbiota function in diet-induced obese mice. Gut Microbes 12, 1832857–1832819. doi: 10.1080/19490976.2020.1832857
Mahase, E. (2023). Global cost of overweight and obesity will hit $4.32tn a year by 2035, report warns. BMJ 380:523. doi: 10.1136/bmj.p523
Mo, J., Lu, Y., Jiang, S., Yan, G., Xing, T., Xu, D., et al. (2022). Effects of the probiotic, Lactobacillus delbrueckii subsp. bulgaricus, as a substitute for antibiotics on the gastrointestinal tract microbiota and metabolomics profile of female growing-finishing pigs. Animals (Basel) 12:1778. doi: 10.3390/ani12141778
Mo, J., Sun, L., Cheng, J., Lu, Y., Wei, Y., Qin, G., et al. (2021). Non-targeted metabolomics reveals metabolic characteristics of porcine Atretic follicles. Front. Vet. Sci. 8:679947. doi: 10.3389/fvets.2021.679947
Niemann, B., Haufs-Brusberg, S., Puetz, L., Feickert, M., Jaeckstein, M. Y., Hoffmann, A., et al. (2022). Apoptotic brown adipocytes enhance energy expenditure via extracellular inosine. Nature 609, 361–368. doi: 10.1038/s41586-022-05041-0
Ren, Q., Chen, S., Chen, X., Niu, S., Yue, L., Pan, X., et al. (2022). An effective glucagon-like Peptide-1 receptor agonists, Semaglutide, improves Sarcopenic obesity in obese mice by modulating skeletal muscle metabolism. Drug Des. Devel. Ther. 16, 3723–3735. doi: 10.2147/DDDT.S381546
Saban Güler, M., Arslan, S., Ağagündüz, D., Cerqua, I., Pagano, E., Berni Canani, R., et al. (2025). Butyrate: a potential mediator of obesity and microbiome via different mechanisms of actions. Food Res. Int. 199:115420. doi: 10.1016/j.foodres.2024.115420
Saravanan, G., Ponmurugan, P., Deepa, M. A., and Senthilkumar, B. (2014). Anti-obesity action of gingerol: effect on lipid profile, insulin, leptin, amylase and lipase in male obese rats induced by a high-fat diet. J. Sci. Food Agric. 94, 2972–2977. doi: 10.1002/jsfa.6642
Takeuchi, T., Kameyama, K., Miyauchi, E., Nakanishi, Y., Kanaya, T., Fujii, T., et al. (2023). Fatty acid overproduction by gut commensal microbiota exacerbates obesity. Cell Metab. 35, 361–375.e9. doi: 10.1016/j.cmet.2022.12.013
The Lancet Diabetes Endocrinology (2021). Obesity in China: time to act. Lancet Diabetes Endocrinol. 9:407. doi: 10.1016/S2213-8587(21)00150-9
Trung, N. Q., Thu Thanh, N. T., Hoa, N. T., Mechler, A., and Vo, Q. V. (2023). Feruloylmonotropeins: promising natural antioxidants in Paederia scandens. RSC Adv. 13, 6153–6159. doi: 10.1039/d3ra00458a
Vallianou, N. G., Kounatidis, D., Tsilingiris, D., Panagopoulos, F., Christodoulatos, G. S., Evangelopoulos, A., et al. (2023). The role of next-generation probiotics in obesity and obesity-associated disorders: current knowledge and future perspectives. Int. J. Mol. Sci. 24:6755. doi: 10.3390/ijms24076755
Wahl, S., Yu, Z., Kleber, M., Singmann, P., Holzapfel, C., He, Y., et al. (2012). Childhood obesity is associated with changes in the serum metabolite profile. Obes. Facts 5, 660–670. doi: 10.1159/000343204
Wang, N., Dilixiati, Y., Xiao, L., Yang, H., and Zhang, Z. (2024). Different short-chain fatty acids unequally modulate intestinal homeostasis and reverse obesity-related symptoms in Lead-exposed high-fat diet mice. J. Agric. Food Chem. 72, 18971–18985. doi: 10.1021/acs.jafc.4c04193
Wang, L., Jiang, Y., Han, T., Zheng, C., and Qin, L. (2014). A phytochemical, pharmacological and clinical profile of Paederia foetida and P. scandens. Nat. Prod. Commun. 9, 879–886. doi: 10.1177/1934578X1400900640
Wang, Z., Xue, L., Guo, C., Han, B., Pan, C., Zhao, S., et al. (2012). Stevioside ameliorates high-fat diet-induced insulin resistance and adipose tissue inflammation by downregulating the NF-κB pathway. Biochem. Biophys. Res. Commun. 417, 1280–1285. doi: 10.1016/j.bbrc.2011.12.130
Wang, Y., Zhao, L., Gao, L., Pan, A., and Xue, H. (2021). Health policy and public health implications of obesity in China. Lancet Diabetes Endocrinol. 9, 446–461. doi: 10.1016/S2213-8587(21)00118-2
Wei, G., Ye, Y., Yan, X., Chao, X., Yang, F., Wang, M., et al. (2020). Effect of banana pulp dietary fibers on metabolic syndrome and gut microbiota diversity in high-fat diet mice. J. Food Biochem. 44:e13362. doi: 10.1111/jfbc.13362
Wu, Y., Mo, J., Liang, J., Pu, X., Dong, Y., Zhu, X., et al. (2024). Multiomic study of the protective mechanism of Persicaria capitata (Buch.-ham. Ex D.Don) H.Gross against streptozotocin-induced diabetic nephropathy in Guizhou miniature pigs. Phytomedicine 128:155499. doi: 10.1016/j.phymed.2024.155499
Wu, Q., Tang, H., and Wang, H. (2019). The anti-oxidation and mechanism of essential oil of Paederia scandens in the NAFLD model of chicken. Animals (Basel) 9:850. doi: 10.3390/ani9100850
Wu, C., Yang, F., Zhong, H., Hong, J., Lin, H., Zong, M., et al. (2024). Obesity-enriched gut microbe degrades myo-inositol and promotes lipid absorption. Cell Host Microbe 32, 1301–1314.e9. doi: 10.1016/j.chom.2024.06.012
Xiao, M., Fu, X., Ni, Y., Chen, J., Jian, S., Wang, L., et al. (2018). Protective effects of Paederia scandens extract on rheumatoid arthritis mouse model by modulating gut microbiota. J. Ethnopharmacol. 226, 97–104. doi: 10.1016/j.jep.2018.08.012
Xu, Y., Zeng, J., Wang, L., Xu, J., He, X., and Wang, Y. (2023). Anti-inflammatory iridoid glycosides from Paederia scandens (Lour.) Merrill. Phytochemistry 212:113705. doi: 10.1016/j.phytochem.2023.113705
Yang, Y., Fukui, R., Jia, H., and Kato, H. (2021). Amaranth supplementation improves hepatic lipid dysmetabolism and modulates gut microbiota in mice fed a high-fat diet. Food Secur. 10:1259. doi: 10.3390/foods10061259
You, H., Tan, Y., Yu, D., Qiu, S., Bai, Y., He, J., et al. (2022). The therapeutic effect of SCFA-mediated regulation of the intestinal environment on obesity. Front. Nutr. 9:886902. doi: 10.3389/fnut.2022.886902
Zhang, X., Ha, S., Lau, H. C.-H., and Yu, J. (2023). Excess body weight: novel insights into its roles in obesity comorbidities. Semin. Cancer Biol. 92, 16–27. doi: 10.1016/j.semcancer.2023.03.008
Zhu, T., Zhao, J., Zhuo, S., Hu, Z., Ouyang, S., Wunier, n., et al. (2021). High fat diet and high cholesterol diet reduce hepatic vitamin D-25-hydroxylase expression and serum 25-Hydroxyvitamin D3 level through elevating circulating cholesterol, glucose, and insulin levels. Mol. Nutr. Food Res. 65:e2100220. doi: 10.1002/mnfr.202100220
Keywords: Paederia scandens , obesity, high-fat induced, microbes, metabolites
Citation: Yang Y, Si J, Mo J, Li J, Pan B, Pan Y, Jiang L, Wang D and Feng X (2025) Paederia scandens extract alleviates obesity via modulating the gut microbiota and serum metabolome disorder. Front. Microbiol. 16:1554537. doi: 10.3389/fmicb.2025.1554537
Edited by:
Mohammad Altamimi, An-Najah National University, PalestineReviewed by:
Fengjie Sun, Georgia Gwinnett College, United StatesXiaokang Ma, Hunan Agricultural University, China
Copyright © 2025 Yang, Si, Mo, Li, Pan, Pan, Jiang, Wang and Feng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xueping Feng, ZnhwMjAwMjA2MDVAeW11bi5lZHUuY24=; Decai Wang, NTYwMjMwOTBAeW11bi5lZHUuY24=
†These authors have contributed equally to this work
 Yuanyuan Yang1†
Yuanyuan Yang1† Jiayuan Mo
Jiayuan Mo Jin Li
Jin Li Lihe Jiang
Lihe Jiang Decai Wang
Decai Wang Xueping Feng
Xueping Feng