- 1Key Laboratory of Ministry of Education for Genetics, Breeding and Multiple Utilization of Crops, College of Horticulture, Fujian Agriculture and Forestry University, Fuzhou, China
- 2State Key Laboratory of Agricultural and Forestry Biosecurity, College of Life Sciences, Fujian Agriculture and Forestry University, Fuzhou, China
- 3Department of Biological Sciences, University of Illinois, Chicago, IL, United States
- 4Guangxi Institute of Botany, Chinese Academy of Sciences, Guilin, China
- 5College of Plant Protection, Gansu Agricultural University, Lanzhou, China
- 6College of Life Science and Technology, Xinjiang University, Urumqi, China
- 7Institute of Tropical Bioscience and Biotechnology, Chinese Academy of Tropical Agricultural Sciences, Hainan Key Laboratory of Tropical Microbe Resources, Haikou, China
- 8Fuzhou Technology and Business University, Fuzhou, China
Introduction: Bamboo is the largest member of the family Poaceae, and these fast-growing plants have many beneficial ecological effects. Both mutualistic and pathogenic fungi associate with bamboo, however, only a limited number of such fungal species have been identified, and information concerning the diversity of fungi that parasitizes bamboo leaves is sparse.
Methods: Fungi were isolated from diseased leaves of bamboo from Fujian Province, China. Nucleotide sequences of four genetic loci were used for taxonomic and phylogenetic reconstruction. In addition, the morphological characteristics of fungal structures as well as growth parameters were characterized.
Results: We report two new fungal species, one each within the Didymosphaeriaceae and Magnaporthaceae, both diverse genera that include saprophytes, endophytes, and pathogens. Based on combined morphological and phylogenetic analyses, including the nucleotide sequences of the internal transcribed spacer (ITS), the 28S large subunit of ribosomal RNA (LSU), the 18S small subunit of nuclear ribosomal RNA gene (SSU), the large subunit of RNA polymerase I (rpb1), and the translation elongation factor 1-α gene (tef1-α), the two new species described herein have been named as Bifusisporella magnum sp. nov., and Letendraea bambusae sp. nov. Detailed descriptions and morphological features of the species are provided.
Discussion: These data identify and describe new species within the Didymosphaeriaceae and Magnaporthaceae, expanding the diversity and distribution of fungi associating with bamboo.
1 Introduction
Bamboo (Bambusoideae, Poales) is one of the most important forms of wood in the world (Wysocki et al., 2015), with rich species diversity. It is found in tropical, subtropical, and temperate regions on several continents, albeit rare in Europe (Phookamsak et al., 2022). Global bamboo resources are abundant, with a total of 140 genera and over 1,750 species. In China, there are ~770 species of bamboo, along with five hybrids and 203 forms or varieties, spread across 42 genera (Yang et al., 2024). China boasts the richest bamboo resources, the highest diversity, and the largest total cultivation area, accounting for more than 50% of the world's bamboo species (Liu et al., 2018; Dlamini et al., 2022). Bamboo species are widely distributed from cold mountain regions to hot tropical regions, form beneficial associations with a variety of fungi, but can also be vulnerable to fungal infection. Plant-associated fungi can be endophytic, living within plant tissues, exist on the surface of the plant as epiphytes, function as saprobes, and/or form parasitic interactions, leading to plant disease (Mu et al., 2024). Currently, more than 1,100 species of fungi associated with bamboo have been documented or described globally (Dai et al., 2018; Jiang et al., 2022). Intriguingly, several “bambusicolous” fungi have been used in Chinese medicine to treat various human diseases and are known to produce a variety of bioactive functional metabolites (Yan et al., 2023).
Studies of bambusicolous fungi have been carried out since the 18th century, the earliest of which was initiated by Léveillé (1845), and aspects of their molecular phylogeny developed by Tanaka et al. (2009), who established a new family, Tetraplosphaeriaceae, and assigned it within the order Pleosporales. A second, diverse family of bamboo associating fungi, Didymosphaeriaceae, also within Pleosporales, has further been defined (Ariyawansa et al., 2014b; Dai et al., 2016). Species within these families are widely distributed globally, can grow on and/or parasitize a variety of hosts, including plants and occasionally other fungi and animals including human. These families encompass endophytes, epiphytes, saprophytes, and pathogens typically found on woody branches, herb stems, leaves, pods, and soil (Ren et al., 2022).
Munk (1953) introduced Didymosphaeriaceae in the order Melanommatales, with the type genus Didymosphaeria Fuckel (Barr, 1990). The sexual morph of these fungi is characterized by one-septal ascospores and trabeculate pseudoparaphyses which anastomose mostly above the asci (Ariyawansa et al., 2014a). The asexual morphs of Didymosphaeriaceae are diverse (Wijayawardene et al., 2022). Members of the genus of Montagnulaceae display one to multi-septate ascospores and generally cellular pseudoparaphyses. However, Ariyawansa et al. (2014a) compared the new species Didymosphaeria rubi-ulmifolii with the type species, D. futilis (Berk. & Broome) Rehm, based on multilocus phylogenetic analyses, demonstrating that D. rubi-ulmifolii clustered as a separate genus in Montagnulaceae, indicating that Montagnulaceae and Didymosphaeriaceae are synonymous (Ariyawansa et al., 2014b). Letendraea belongs to the Didymosphaeriaceae of the Pleosporales. Some species in the genus Letendraea, such as Letendraea sp. WZ07, are considered endophytic fungi that can colonize healthy plant tissues without causing obvious disease symptoms, and these fungi have been shown to produce secondary metabolites with anti-inflammatory activity (Qiao et al., 2022; Yan et al., 2024).
The order Magnaporthales, belongs to the class Sordariomycetes within the phylum Ascomycota. Wijayawardene et al. (2022) proposed five families and 39 genera within this order. Magnaporthaceae is the most diverse family within the Magnaporthales (Feng et al., 2024). Cannon (1994) proposed the Magnaporthaceae family to accommodate Magnaporthe and its related genus Buergenerula. As early as 2009, Magnaporthaceae was independently classified as a new order, Magnaporthales, based on morphological characteristics and phylogenetic analysis of LSU and SSU genes (Thongkantha et al., 2009). Asexual forms appear as simple, unbranched or branched conidia, conidia straight or curved, transparent to light brown, septate, conidiogenous cells have pigment (Klaubauf et al., 2014). Members of the Magnaporthaceae include important plant pathogens of rice, grasses, and cereals. During an investigation of endophytic fungi in Brazilian Sorghum bicolor leaves, two isolates were discovered that exhibited morphological and phylogenetic differences from previously known members within the Magnaporthaceae. Consequently, Silva et al. (2019) established a new genus, Bifusisporella to accommodate these isolates.
In this study, we report on the identification and characterization of two species isolated from diseased bamboo plants, namely: Bifusisporella magnum sp. nov. (Magnaporthaceae) and Letendraea bambusae sp. nov. (Didymosphaeriaceae), based on phylogenetic (molecular) and morphological analyses. Phylogenetic reconstruction involved a multi-locus approach, combining data from five genetic loci: the intervening 5.8S nrRNA gene (ITS), the 28S large subunit of nuclear ribosomal RNA gene (LSU), the 18S small subunit of nuclear ribosomal RNA gene (SSU), the large subunit of RNA polymerase I (rpb1), and the translation elongation factor 1-α gene (tef1-α) nucleotide sequences. These results expand the diversity of fungi infecting bamboo plants and provide distinguishing characteristics of bambusicolous fungi to better elucidate relationships between species.
2 Materials and methods
2.1 Sampling and isolation
Fungal specimens isolated from diseased leaves of bamboo were collected in Fujian Province, China. Diseased leaves with obvious fungal necrosis and/or typical blight-spot symptoms were cut into ~25 mm2 pieces. The fragments were soaked in 75% ethanol solution for 60 s, washed once, then soaked in sterile deionized water for 20 s, then transferred to 5% sodium hypochlorite for 30–40 s, and then washed in sterile deionized water for three times for 45–60 s. The washed fragments were placed on sterilized filter paper to dry, then transferred to PDA plates and incubated at 25°C for 7 days (Su et al., 2021). Fungal hyphae on the growing edges of colonies were transferred to a new PDA plate until pure fungal cultures were obtained. Spore production and visual appearance of the colonies were examined on SNA plates amended with sterilized pine needles or bamboo leaf fragments, and plates were incubated at 25°C under a regimen of 12 h of near-ultraviolet light and 12 h of darkness (Zhang et al., 2023).
2.2 Morphological observation
After 7–14 days of incubation, fungal colonies were photographed with a digital camera (Canon EOS 6D MarkII, Tokyo, Japan) to detail the morphology of the conidiomata, conidiophores, and conidiogenous cells using a stereomicroscope (Nikon SMZ745, Tokyo, Japan) and biological microscope (Ni-U, Tokyo, Japan) with a digital camera (Olympus, Tokyo, Japan). Images were analyzed using Digimizer 5.4.4 software for various size parameters of fungal structures. A single colony derived from the purified culture was cut into 1 cm2 pieces and preserved in 10% glycerol and sterile water at 4°C for further detailed study.
2.3 DNA extraction, PCR amplification and sequencing
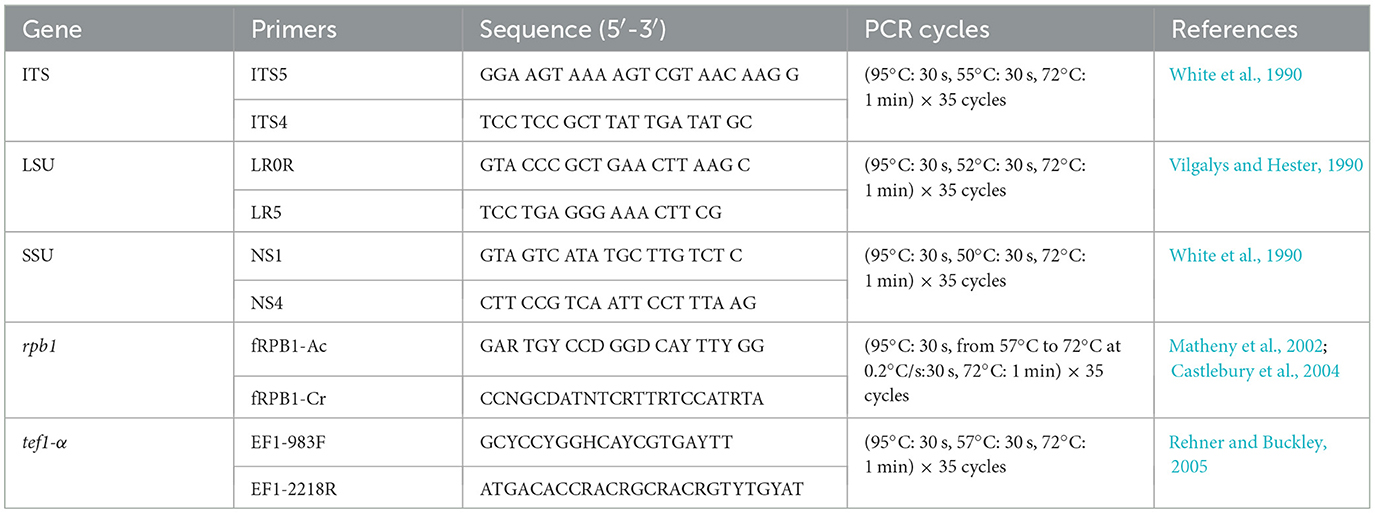
A fungal DNA Mini-kit (OMEGA-D3390, Feiyang Biological Engineering Corporation, Guangzhou, China) was used to extract fungal genomic DNA from growing hyphae. Genetic loci corresponding to the ITS region were amplified by polymerase chain reaction (PCR) using primers ITS5 and ITS4 (White et al., 1990; Zou et al., 2022), with primers LR0R and LR5 used to amply the LSU region (Vilgalys and Hester, 1990), primers NS1 and NS4 for the SSU region (White et al., 1990), and primers fRPB1-Ac and fRPB1-Cr for amplification of the rpb1 gene (Matheny et al., 2002; Castlebury et al., 2004), The tef1-α locus was amplified using EF1-983F and EF1-2218R (Rehner and Buckley, 2005). Primer sequences are given in Table 1.
PCR amplification was performed using a Bio-Rad thermal cycler (Hercules, CA, USA). Amplification reactions were performed in 25 μl reaction volumes consisting of 12.5 μl 2 × Rapid Taq Master Mix (Vazyme, Nanjing, China), 1 μl forward and reverse primers (10 μM) (Sangon, Shanghai, China), and 1 μl template genomic DNA, adjusted with sterile deionized water to a total of 25 μl. The integrity and sizes of PCR products were estimated by 1% agarose gel electrophoresis. All PCR products were purified, and their nucleotide sequences were determined by a commercial service (Tsingke Company Limited, Fuzhou, China). Sequence assembly was performed using MEGA 7.0 (Kumar et al., 2016). The new sequences generated in this study have been uploaded to GenBank (https://www.ncbi.nlm.nih.gov, accession numbers given in Table 2).
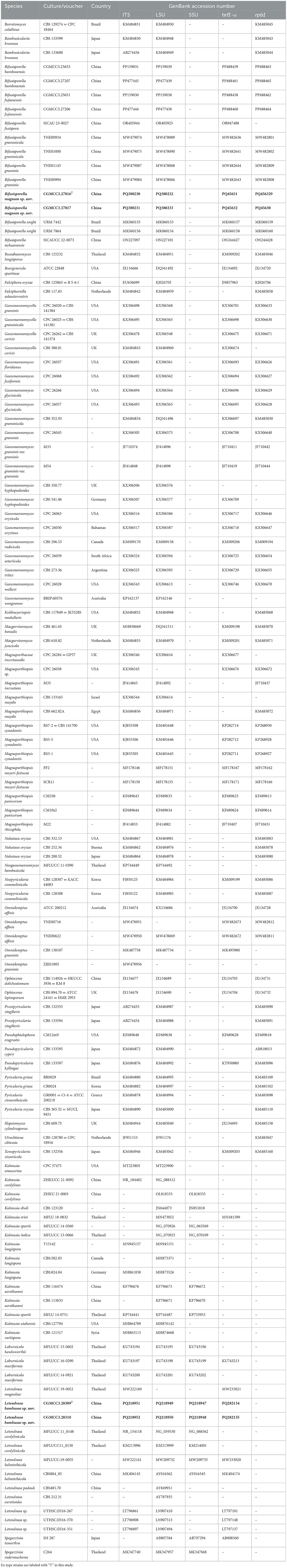
Table 2. Species and GenBank accession numbers of DNA sequences used in this study, with new sequences indicated in bold.
2.4 Sequence alignment and phylogenetic analysis
Sets of orthologous sequences for all loci were downloaded from GenBank. All sequences were aligned using the MAFFT v. 7 online service (http://mafft.cbrc.jp/alignment/server/, accessed on 27 September 2024) (Katoh et al., 2019) and manually adjusted in BioEdit v .7.2.6.1 (He et al., 2022) and MEGA 7.0 (Kumar et al., 2016). Using Maximum Likelihood (ML) and Bayesian Inference (BI) analyses, phylogenetic trees were constructed to explore the evolutionary relationships of fungi within the Magnaporthaceae and Didymosphaeriaceae families, respectively. Spegazzinia tessarthra (SH 287) and Spegazzinia radermacherae (C264) were selected as outgroup taxa of Didymosphaeriaceae. Ophioceras dolichostomum (CBS 114926) and Ophioceras leptosporum (CBS 894.70) (Ophioceraceae, Magnaporthales) were used as outgroup taxa of Magnaporthaceae (Zhao et al., 2024).
Bayesian Posterior Probabilities (BPP) was assessed using Markov Chain Monte Carlo (MCMC) methods (Rannala and Yang, 1996; Zhaxybayeva and Gogarten, 2002). Two parallel runs were conducted, each with four chains, starting from 2,000,000 generations of random trees, sampling every 100 generations, resulting in a total of 20,000 trees, the average standard deviation of split frequencies was set to be < 0.01, starting from random trees (Ronquist and Huelsenbeck, 2003). The first 25% of the trees were discarded as burn-in, and the remaining 75% were used to calculate the Bayesian Posterior Probabilities (BPP) for the majority rule consensus tree (Ji et al., 2022). The consensus trees were visualized in FigTree v.1.4.4 (https://tree.bio.ed.ac.uk/software/figtree; Rambaut, 2018) and were embellished using Adobe Illustrator CS 6.0 (Adobe Systems Inc., USA). Maximum likelihood bootstrap support values (≥70%) and Bayesian Posterior Probabilities (≥0.90) were considered as significantly supported branches. The new sequences generated in this study have been deposited in GenBank (https://www.ncbi.nlm.nih.gov; accessed on 12 October 2024).
3 Results
3.1 Phylogenetic analysis
A total of two samples were collected from diseased bamboo leaves. Of these, four purified fungal colonies were obtained. Genomic DNA was extracted from all specimens and the nucleotide sequences of the internal transcribed spacer (ITS), the 28S large subunit of ribosomal RNA (LSU), the 18S small subunit of nuclear ribosomal RNA gene (SSU), the large subunit of RNA polymerase I (rpb1), the translation elongation factor 1-α gene (tef1-α) gene were determined as detailed in the Materials and Methods section.
For construction of the phylogenetic tree of isolates matching Magnaporthaceae, the concatenated sequence dataset of ITS, LSU, tef1-α, and rpb1 was used in analyses which included 83 taxa with Ophioceras dolichostomum (CBS 114926) and Ophioceras leptosporum (CBS 894.70) as the outgroup (Figure 1). For each fungal isolate, 2,869 bp of nucleotide sequences corresponding to portions of the ITS, LSU, rpb1, and tef1-α loci (ITS: 1–433; LSU: 434–1,213; rpb1: 1,214–1,981; tef1-α: 1,982–2,869) were isolated. Based on these and morphological data, a new species, Bifusisporella magnum, was identified, related to Bifusisporella bambooensis (CGMCC3.25653), with high support (ML-BS: 98% and BYPP: 1). For Bifusisporella magnum, 1,346 distinct and 398 variable nucleotide sequences, including gaps were identified, with 909 patterns parsimony informative. ModelFinder (Kalyaanamoorthy et al., 2017) was used to select the best-fit model using AIC criterion, maximum likelihood phylogenies were inferred using IQ-TREE (Nguyen et al., 2015) under the GTR+R4+F model for 5000 ultrafast (Minh et al., 2013) bootstraps, as well as the Shimodaira–Hasegawa–like approximate likelihood-ratio test (Guindon et al., 2010) (Lsetnst = 1, rates = Invgamma). Average standard deviation of split frequencies was 0.004924. Based on these analyses, the two isolates of Bifusisporella described herein represent one novel species.
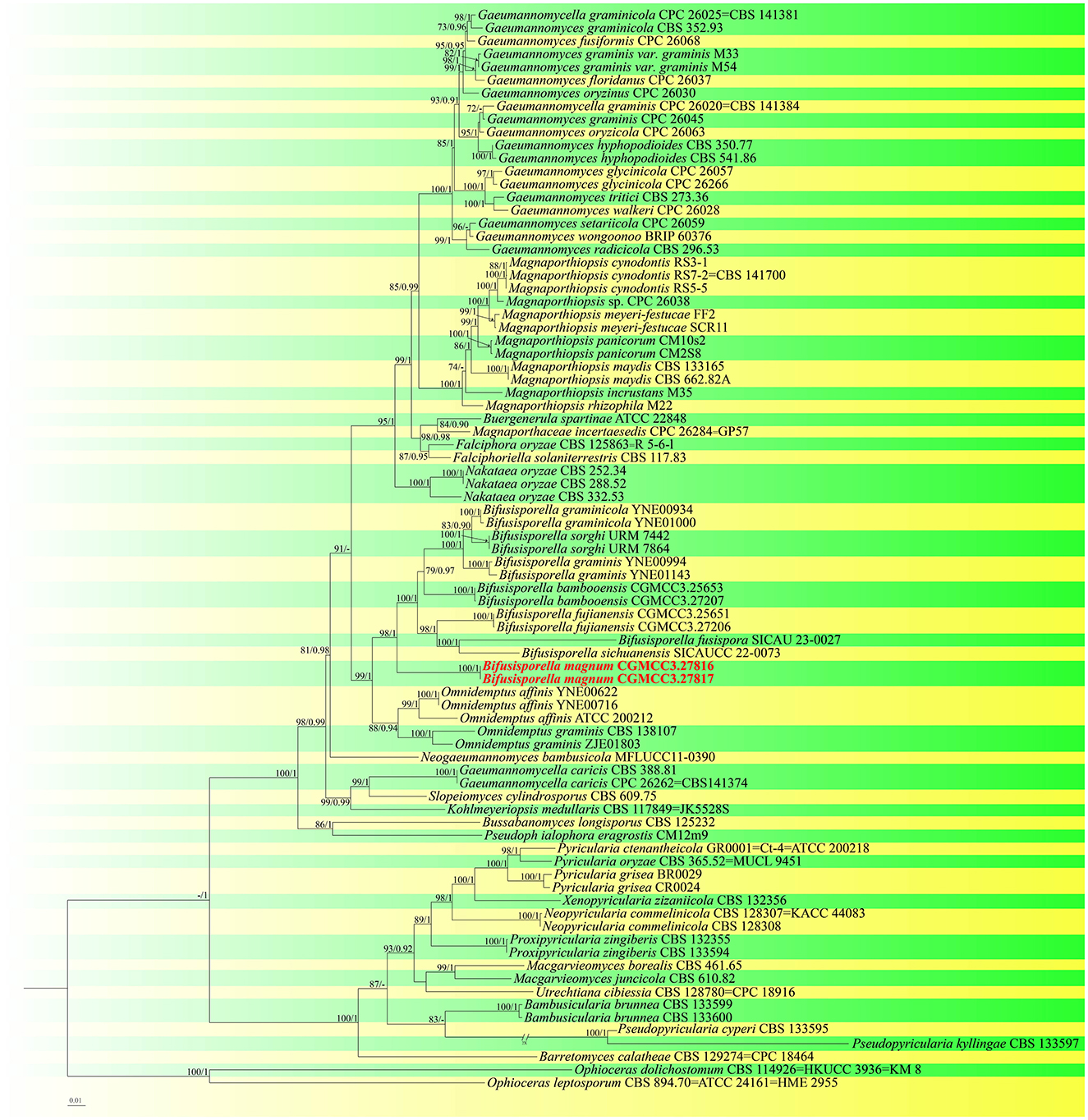
Figure 1. Molecular phylogenetic analysis of Magnaporthaceae and Pyriculariaceae based on a combined ITS, LSU, tef1-α, and rpb1 sequences. IQ-TREE ultra-fast bootstrap (BS) (UFBoot) values ≥70% and MrBayes posterior probability (PP) ≥0.90 are displayed above or below the respective branches (ML/BI). The newly described species is marked red.
For the Letendraea isolates, in our dataset, the ITS + LSU + SSU + tef1-α concatenated sequence dataset had an aligned length of 2,654 total characters (ITS: 1–405, LSU: 406–1,176, SSU: 1,177–1,941, tef1-α: 1,942–2,654) including gaps, of which 2,282 characters were constant, 151 were variable and parsimony-uninformative, and 221 bp were parsimony-informative. ModelFinder (Kalyaanamoorthy et al., 2017) was employed to choose the best-fit model based on AIC criterion, maximum likelihood phylogenies were inferred using IQ-TREE (Nguyen et al., 2015) under the TIM2e+R2 model for 5,000 ultrafast (Minh et al., 2013) bootstraps, as well as the Shimodaira–Hasegawa–like approximate likelihood-ratio test (Lsetnst = 1, rates = Equal) (Guindon et al., 2010). Bayesian analyses resulted in an average standard deviation of split frequencies = 0.003168. The topology shown by ML and BI were almost identical, although there were slight differences in statistical support for some branches (Figure 2). Based on phylogenetic resolution and morphological analysis, we report a new species of Didymosphaeriaceae: Letendraea bambusae, with the two strains studied herein representing one novel species. The new species Letendraea bambusae clustered alongside Letendraea sp. (UTHSC: DI16-267) and formed a clade sister to Letendraea sp. (UTHSC: DI16-267) with a posterior probability of 1.
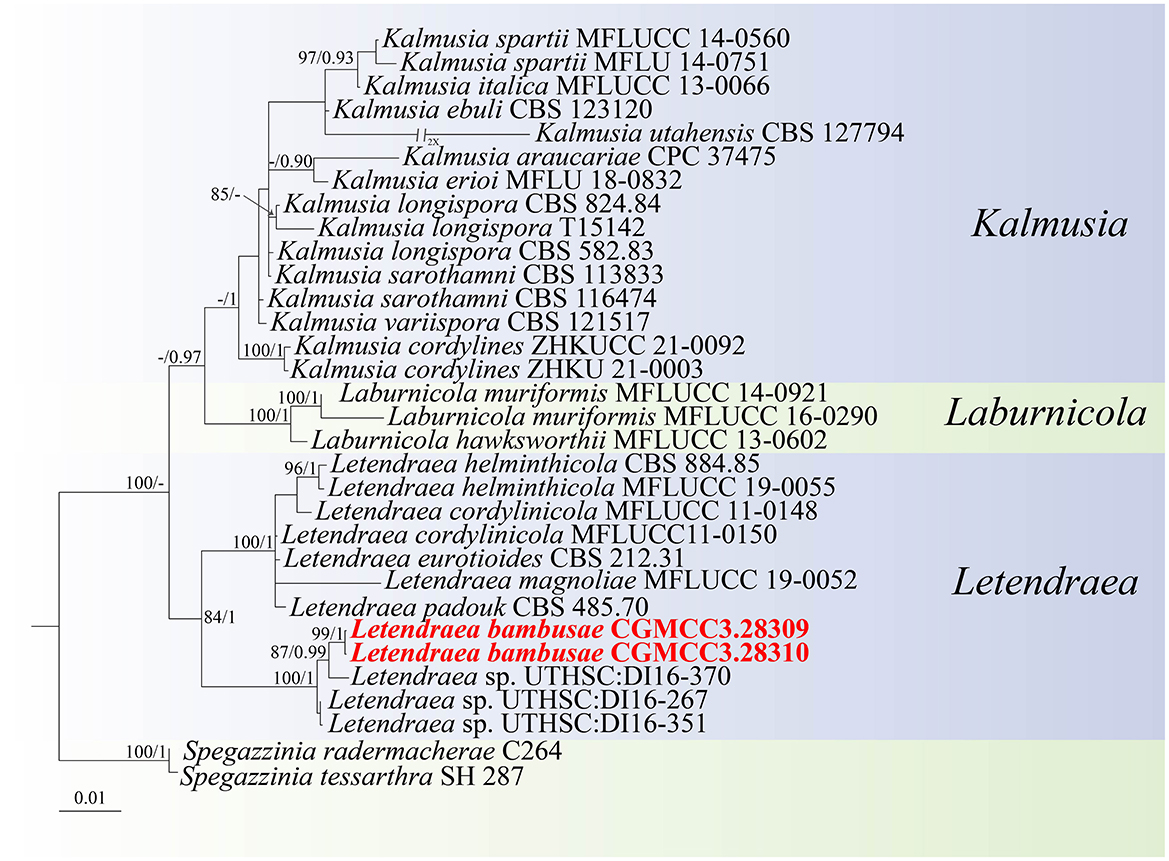
Figure 2. Phylogenetic tree of Didymosphaeriaceae inferred from Bayesian inference analyses base on a combined ITS, LSU, SSU, and tef1-α sequence dataset, with Spegazzinia tessarthra (SH 287) and Spegazzinia radermacherae (C264) as outgroup. IQ-TREE ultra-fast bootstrap (BS; UFBoot) values ≥70% and MrBayes posterior probability (PP) ≥0.90 are displayed above or below the respective branches (ML/BI). The newly described species is marked in red.
3.2. Taxonomy
Bifusisporella magnum sp. nov. Z.Y. Zhao, X.Y. Guan and J.Z. Qiu (Figure 3).
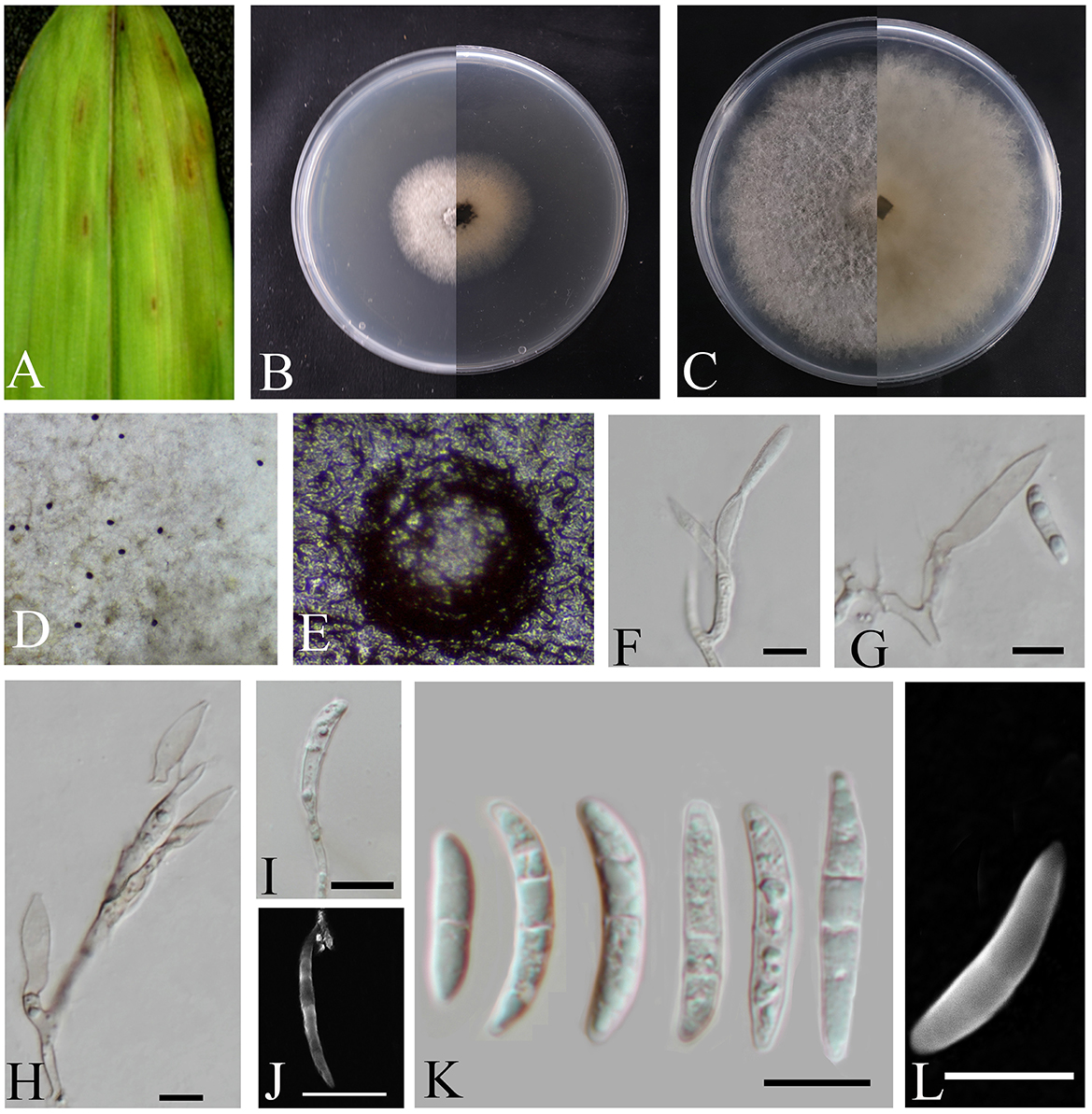
Figure 3. Bifusisporella magnum (HMAS353209). (A) Leaves of host plant. (B, C) Upper and reverse view of colony after incubation for 7 days on PDA and 14 days. (D, E) Conidiomata sporulating on PDA. (F–J) Conidiogenous cells and conidia. (K, L) Conidia. (F–L) Scale bars: 10 μm.
MycoBank: MB856574
Holotype: China, Fujian Province, Fujian Agriculture and Forestry University (119°14′35.14″E, 26°5′2.55″N), from diseased leaves of bamboo in China, March 2023, Z.Y. Zhao (holotype HMAS353209; ex-type living culture CGMCC3.27816).
Etymology: From Latin magnus (“great”). In reference to the large sized conidiogenous cells.
Description: The oval-shaped leaf spots feature a gradient of color, shifting from white at the periphery to brown at the center. Conidiomata emits droplets that form distinct, black, spherical protrusions on the agar surface. Conidiophores reduced to conidiophores cells. Conidiogenous cells are cylindrical or pot-shaped, solitary or clustered, curved, and elongated in shape. 18.8–30.4 × 4.6–6.4 μm, mean ± SD = 22.5 ± 4.7 × 5.4 ± 0.9 μm, n = 20. Conidia are sickle-shaped or crescent-shaped, with a smooth or cracked surface, and are transparent. 0–4 septa, 11.8–28.9 × 2.1–4.3 μm, mean ± SD = 21.2 ± 4.4 × 3.5 ± 0.6 μm, n = 30.
Culture characteristics: Colonies on PDA are fluffy, velvety, and grayish-white, calculated growth rate of 0.6 cm/day at 25°C.
Material examined: China, Fujian Province, Fujian Agriculture and Forestry University (119°14′35.14″E, 26°5′2.55″N), from diseased leaves of bamboo in China, March 2023, Z.Y. Zhao (paratype HMAS353210; ex-paratype living culture CGMCC3.27817).
Notes: In this study, two isolates corresponding to the genus Bifusisporella was collected from the Bambusoideae. The strain of the genus Bifusisporella was identified as a new species, with the two isolates representing one species. Phylogenetic analysis utilizing four genomic loci revealed that B. magnum constitutes a distinct clade, closely related to B. bambooensis (CGMCC3.25653), and this classification is strongly supported by statistical evidence (98% ML/1 pp). Compared to B. bambooensis, B. magnum sp. nov. has larger conidiogenous cells and smaller conidia (18.8–30.4 × 4.6–6.4 vs. 7.2–21.0 × 4.2–6.4 μm; 11.8–28.9 × 2.1–4.3 vs. 10.8–45.0 × 2.8–4.9 μm); Molecularly, an unnamed sequence that is at least 97% similar to a fully annotated type-derived sequence in a global ITS alignment can be annotated at the family level and often at the genus level (Nilsson et al., 2019), the nucleotide comparison of ITS, LSU, tef1-α, and rpb1 (CGMCC3.27816) showed differences with the sequences of B. bambooensis (CGMCC3.25653), similarities are 16.1% (86/534), 2.8% (22/779), 3.7% (32/869), and 11.6% (73/628), therefore, the strain of the genus Bifusisporella was identified as a new species.
Letendraea bambusae sp. nov. Z.Y. Zhao, X.Y. Guan and J.Z. Qiu (Figure 4).
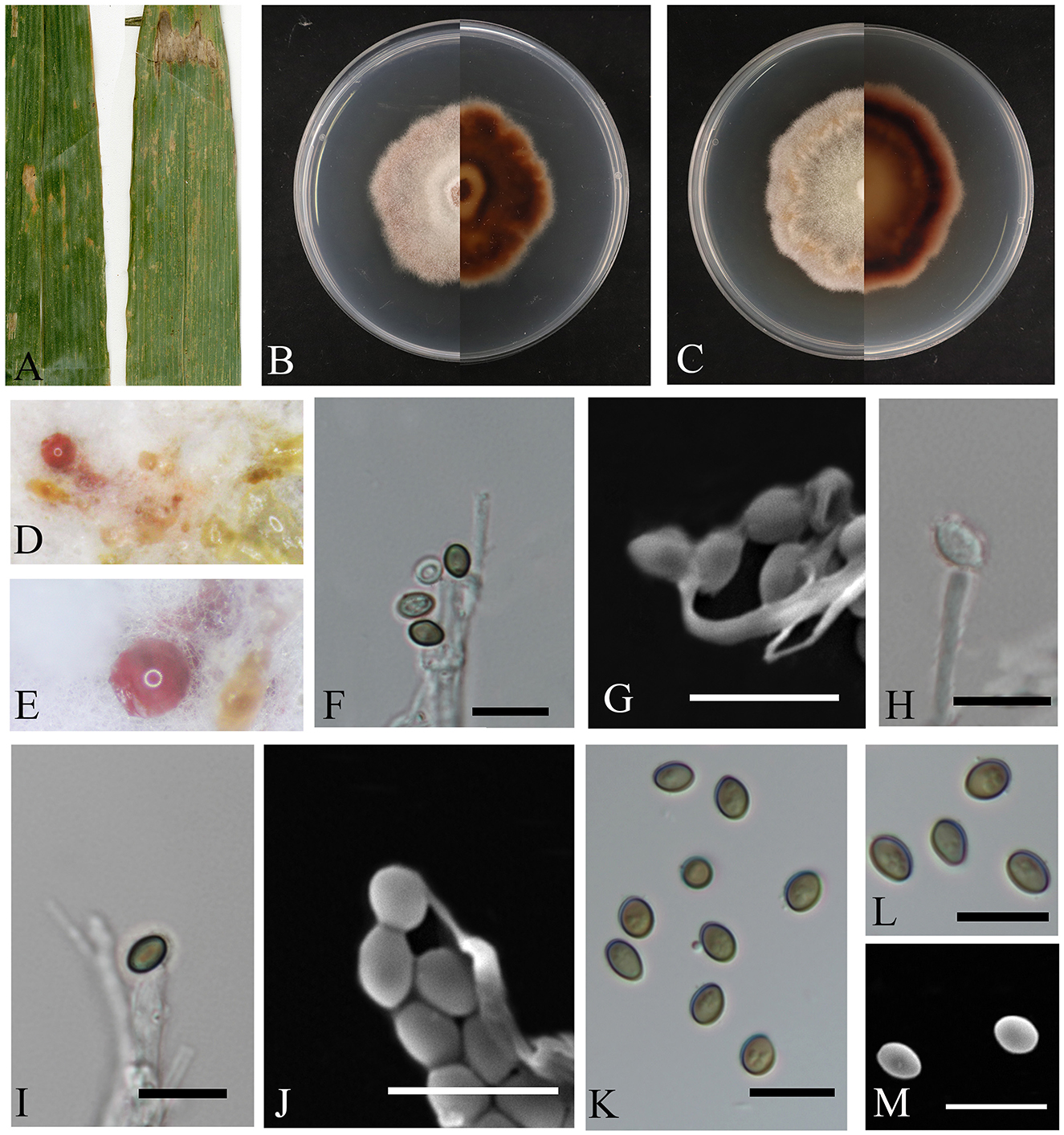
Figure 4. Letendraea bambusae (HMAS353152). (A) Leaves of host plant. (B, C) Upper and reverse view of colony after incubation for 7 days on PDA and 14 days. (D, E) Conidiomata sporulating on PDA. (F–J) Conidiogenous cells and conidia. (K–M) Conidia. (F–M) Scale bars: 10 μm.
MycoBank: MB856575
Holotype: China, Fujian Province, Hua'an Bamboo Garden (117°32′23.35″E, 25°0′29.47″N), from diseased leaves of bamboo. June 2023, Z.Y. Zhao (holotype HMAS353152; ex-type living culture CGMCC3.28309).
Etymology: The epithet “bambusae” refers to the host, which is bamboo.
Description: The leaf spots are irregular, with colors transitioning from reddish-brown on the outer edges to white at the center. Conidiomata bulging on agar, transparent to pink, spherical in aggregate; conidiophores hyaline, smooth, subcylindrical, conidia are round or oval, with smooth surface, initially transparent and becoming olive or brown upon maturation, 3.6–5.9 × 2.1–3.9 μm, mean ± SD = 4.7 ± 0.6 × 3.0 ± 0.4 μm, n = 40.
Culture characteristics: the colonies on PDA are fluffy and velvety, with the colony surface appearing white, the reverse side of the colony transitions from a nearly circular, pale-yellow center to a reddish-brown color as it extends outwards. The growth rate was 0.7 cm/day.
Material examined: China, Fujian Province, Hua'an Bamboo Garden (117°32′23.35″E, 25°0′29.47″N), from diseased leaves of bamboo. June 2023, Z.Y. Zhao (paratype HMAS353153; ex-paratype living culture CGMCC3.28310).
Notes: In this study, the two isolates originated from diseased bamboo leaves. Phylogenetic analysis revealed that these isolates represent the same species and cluster together with Letendraea sp. (UTHSC: DI16-267) on a single clade, with high statistical support for their differences (BYPP = 1, ML-BS = 84%), the nucleotide comparison of ITS, LSU, SSU, and tef1-α (CGMCC3.28309) showed differences with the sequences of a closely related species Letendraea helminthicola (MFLUCC 19-0055), similarities were 19.0% (104/548), 57.5% (454/790), 1.0% (10/954), and 73.6% (559/759).
4 Discussion
The subtropical region of China is considered as an important area rich in biodiversity and a significant nature reserve for endemic plants (Dai et al., 2018). Fujian Province is located in the eastern part of the subtropical region, and rainfall comes from the East Asian monsoon, which determines the vegetation characteristics of East Asia (Webster et al., 1998). The main vegetation are evergreen broad-leaved forests, and the main tree species in Fujian include Masson's pine, bamboo forests, and willow trees (Zhu and Tan, 2024). The diversity of tree species in Fujian, combined with a warm and humid climate, is an important factor in promoting the proliferation of related fungi (Liu et al., 2024). In this study, two new species (Bifusisporella magnum sp. nov., and Letendraea bambusae sp. nov.) from Fujian Province, China are identified and described.
Members of the Magnaporthaceae family exists in all trophic levels of plants, including roots, stems, and leaves (Shearer et al., 1999). Many of these fungi display host preferences, especially those parasitic on the plants of Poaceae and Cyperaceae (Luo and Zhang, 2013; Yuan et al., 2010). In a study of endophytic fungi in Brazilian two-color sorghum (Sorghum bicolor) leaves, Bifusisporella was proposed as a new genus based on asexual morphological characteristics such as macroconidia and microconidia (Silva et al., 2019). Currently, four Bifusisporella species have been reported isolated healthy sorghum leaves (Silva et al., 2019). B. sichuanensis was proposed by Zeng et al. (2022) for strains isolated from the leaves of poplar trees in Sichuan Province, China, while B. fujianensis and B. bambooensis were proposed by Zhao et al. (2024) for strains isolated from diseased bamboo leaves. Here, B. magnum sp. nov. is also proposed for strains isolated from diseased bamboo leaves. It is likely that these fungi evolved from endophytic predecessors, as evidenced by conserved traits of appressorium development and lignocellulose-degrading enzyme secretion (Bhunjun et al., 2024). Many endophytic fungi within the Magnaporthales order possess the enzymatic machinery for cellulose and lignin decomposition, despite these functions being non-essential for their endophytic existence. The majority of Bifusisporella species generate melanized appressoria that closely resemble those observed in pathogenic fungi in both morphology and structure. Phylogenetic conservation of infection structures (appressoria) and lignocellulolytic machinery among Magnaporthales endophytes indicates evolutionary predisposition for niche flexibility, including possible pathogenic/saprobic conversions under ecological pressures (Konta et al., 2023; Feng et al., 2024).
Saccardo (1880) introduced the genus Letendraea, with L. eurotioides as its type species. Species of Letendraea often cause infections in various terrestrial habitats and have been associated with leaf spot diseases in the asparagus (Cordyline sp.) family. These fungi exhibit a similar morphology to Wilmia, a monotypic genus, containing the single species Wilmia brasiliensis. However, Wilmia (formerly known as Phaeosphaeriaceae) has been demonstrated to be synonymous with Letendraea (Didymosphaeriaceae) (Ariyawansa et al., 2014b). Currently, Letendraea contains four strains with L. eurotioides (CBS 212.31), L. padouk (CBS 485.70), L. helminthicola (CBS 884.85), L. cordylinicola (MFLUCC11-0148T, MFLUCC 11-0150). Our data add one new species to this group, L. bambusae sp. nov., along with a detailed description and its morphological illustration.
The taxonomic study of bambusicolous fungi has become an important research topic due to the increasing cultivation and use of bamboo worldwide (Dai et al., 2018; Phookamsak et al., 2022). East Asia, as the global center of bamboo distribution (especially China, Japan, and Korea), has rich bamboo species resources (Bambusoideae), but in-depth studies on the phylogenetic classification of bambusicolous fungi remain scarce (Hyde et al., 2002), resulting in a lack of molecular data that could help elucidate a significantly greater proportion of the diversity of bamboo associating fungi. Our data describe two new species and provides additional resources in the exploration of the ecology of bambusicolous fungi in East Asia. These results expand our knowledge of bambusicolous fungi, and also provides new insights into the range and potential specificity of their interactions in East Asia.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article.
Author contributions
XG: Conceptualization, Data curation, Investigation, Writing – original draft. ZZ: Conceptualization, Data curation, Formal analysis, Investigation, Writing – original draft. NK: Conceptualization, Formal analysis, Writing – review & editing. TM: Conceptualization, Data curation, Investigation, Writing – original draft, Writing – review & editing. MZhe: Data curation, Formal analysis, Investigation, Writing – original draft. LY: Data curation, Formal analysis, Investigation, Writing – original draft. YM: Data curation, Formal analysis, Investigation, Writing – original draft. JS: Data curation, Formal analysis, Software, Writing – original draft. JY: Data curation, Formal analysis, Methodology, Writing – original draft. HP: Conceptualization, Data curation, Formal analysis, Investigation, Writing – review & editing. YL: Conceptualization, Data curation, Formal analysis, Software, Writing – original draft. MZhu: Conceptualization, Data curation, Formal analysis, Investigation, Writing – original draft. HLv: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Writing – original draft. ZH: Conceptualization, Data curation, Formal analysis, Investigation, Writing – original draft. HLi: Data curation, Formal analysis, Writing – review & editing. LF: Data curation, Formal analysis, Writing – review & editing. XM: Data curation, Formal analysis, Methodology, Writing – review & editing. HM: Data curation, Formal analysis, Methodology, Writing – review & editing. ZQ: Data curation, Formal analysis, Writing – review & editing. JQ: Conceptualization, Data curation, Funding acquisition, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. The research was financed by the National Natural Science Foundation of China (Nos. 32270029, U1803232, and 31670026), the National Key R&D Program of China (No. 2017YFE0122000), a Social Service Team Support Program Project (No. 11899170165), Science and Technology Innovation Special Fund (Nos. KFB23084, CXZX2019059S, and CXZX2019060G) of Fujian Agriculture and Forestry University, a Fujian Provincial Major Science and Technology Project (No. 2022NZ029017), an Investigation and evaluation of biodiversity in the Jiulong River Basin (No. 082·23259-15), Macrofungal and microbial resource investigation project in Longqishan Nature Reserve (No. SMLH2024(TP)-JL003#), and an Investigation of macrofungal diversity in Junzifeng National Nature Reserve, Fujian Province (No. Min Qianyu Sanming Recruitment 2024-23).
Acknowledgments
We would like to thank Weibin Zhang, Jinhui Chen, Chenjie Yang, Sen Liu at Fujian Agriculture and Forestry University, China, for their help with collecting samples. The authors would also like to thank China General Microbiological Culture Collection Center and Fungarium (HMAS), Institute of Microbiology, CAS for strain and sample storage help.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Gen AI was used in the creation of this manuscript.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ariyawansa, H. A., Camporesi, E., Thambugala, K. M., Mapook, A., Kang, J. C., Alias, S. A., et al. (2014a). Confusion surrounding Didymosphaeria — phylogenetic and morphological evidence suggest Didymosphaeriaceae is not a distinct family. Phytotaxa 176, 102–119. doi: 10.11646/phytotaxa.176.1.12
Ariyawansa, H. A., Tanaka, K., Thambugala, K. M., Phookamsak, R., Tian, Q., Camporesi, E., et al. (2014b). A molecular phylogenetic reappraisal of the Didymosphaeriaceae (= Montagnulaceae). Fungal Divers. 68, 69–104. doi: 10.1007/s13225-014-0305-6
Bhunjun, C. S., Phukhamsakda, C., Hyde, K. D., McKenzie, E. H. C., Saxena, R. K., Li, Q., et al. (2024). Do all fungi have ancestors with endophytic lifestyles? Fungal Divers. 125, 73–98. doi: 10.1007/s13225-023-00516-5
Cannon, P. F. (1994). The newly recognized family Magnaporthaceae and its interrelationships. Syst. Ascomycetum 13, 25–42.
Castlebury, L. A., Rossman, A. Y., Sung, G. H., Hyten, A. S., and Spatafora, J. W. (2004). Multigene phylogeny reveals new lineage for Stachybotrys chartarum, the indoor air fungus. Mycol. Res. 108, 864–872. doi: 10.1017/S0953756204000607
Dai, D. Q., Bahkali, A. H., Ariyawansa, H. A., Li, W. J., Bhat, J. D., Zhao, R. L., et al. (2016). Neokalmusia didymospora (Didymosphaeriaceae), a new species from bamboo. Sydowia 68, 17–25. doi: 10.1016/j.sjbs.2015.01.020
Dai, D. Q., Tang, L. Z., and Wang, H. B. (2018). A review of Bambusicolous Ascomycetes. Bamboo Curr. Future Prospect. 2018, 165–183. doi: 10.5772/intechopen.76463
Dlamini, L. C., Fakudze, S., Makombe, G. G., Muse, S., and Zhu, J. (2022). Bamboo as a valuable resource and its utilization in historical and modern-day China. BioResources 17, 1926–1938. doi: 10.15376/biores.17.1.Dlamini
Feng, J. W., Chen, X. Y., Chen, K. Y., Druzhinina, I. S., Voglmayr, H., Crous, P. W., et al. (2024). A reappraisal of families within the order Magnaporthales and description ofnew endophytic taxa associated with Poaceae plants in China. Mycosphere 15, 6240–6346. doi: 10.5943/mycosphere/15/1/26
Guindon, S., Dufayard, J. F., Lefort, V., Anisimova, M., Hordijk, W., Gascuel, O., et al. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. doi: 10.1093/sysbio/syq010
He, J., Han, X., Luo, Z. L., Xian, L., Tang, S. M., Luo, H. M., et al. (2022). Species diversity of Ganoderma (Ganodermataceae, Polyporales) with three new species and a key to Ganoderma in Yunnan Province, China. Front. Microbiol. 13:1035434. doi: 10.3389/fmicb.2022.1035434
Hyde, K. D., Zhou, D. Q., and Dalisayl, T. (2002). Bambusicolous fungi: a review. Fungal Divers. 9, 1–14.
Ji, X., Zhou, J. L., Song, C. G., Xu, T. M., Wu, D. M., Cui, B. K., et al. (2022). Taxonomy, phylogeny and divergence times of Polyporus (Basidiomycota) and related genera. Mycosphere 13, 1–52. doi: 10.5943/mycosphere/13/1/1
Jiang, H. B., Phookamsak, R., Hongsanan, S., Bhat, D. J., Mortimer, P. E., Suwannarach, N., et al. (2022). A review of bambusicolous Ascomycota in China with an emphasis on species richness in southwest China. Studi. Fungi 7:20. doi: 10.48130/SIF-2022-0020
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., von Haeseler, A., and Jermiin, L. S. (2017). ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14, 587–589. doi: 10.1038/nmeth.4285
Katoh, K., Rozewicki, J., and Yamada, K. D. (2019). MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 20, 1160–1166. doi: 10.1093/bib/bbx108
Klaubauf, S., Tharreau, D., Fournier, E., Groenewald, J. Z., Crous, P. W., De Vries, R. P., et al. (2014). Resolving the polyphyletic nature of Pyricularia (Pyriculariaceae). Stud. Mycol. 79, 85–120. doi: 10.1016/j.simyco.2014.09.004
Konta, S., Tibpromma, S., Karunarathna, S. C., Samarakoon, M. C., Steven, L. S., Mapook, A., et al. (2023). Morphology and multigene phylogeny reveal ten novel taxa in Ascomycota from terrestrial palm substrates (Arecaceae) in Thailand. Mycosphere 14, 107–152. doi: 10.5943/mycosphere/14/1/2
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Liu, S., Zhu, M. J., Keyhani, N. O., Wu, Z. Y., Lv, H. J., Heng, Z. A., et al. (2024). Three new species of Russulaceae (Russulales, Basidiomycota) from Southern China. J. Fungi. 10:70. doi: 10.3390/jof10010070
Liu, W. Y., Hui, C. M., Wang, F., Wang, M., and Liu, G. L. (2018). Review of the resources and utilization of bamboo in China. Bamboo Curr. Future Prospect. 8, 133–142. doi: 10.5772/intechopen.76485
Luo, J., and Zhang, N. (2013). Magnaporthiopsis, a new genus in Magnaporthaceae (Ascomycota). Mycologia 105, 1019–1029. doi: 10.3852/12-359
Matheny, P. B., Liu, Y. J., Ammirati, J. F., and Hall, B. D. (2002). Using RPB1 sequences to improve phylogenetic inference among mushrooms (Inocybe, Agaricales). Am. J. Bot. 89, 688–698. doi: 10.3732/ajb.89.4.688
Minh, B. Q., Nguyen, M. A., and von Haeseler, A. (2013). Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 30, 1188–1195. doi: 10.1093/molbev/mst024
Mu, T. C., Lin, Y. S., Keyhani, N. O., Pu, H. L., Lv, Z. Y., Lan, C. H., et al. (2024). Phylogenetic and morphological evidence for three new species of Diaporthales (Ascomycota) from Fujian Province, China. J. Fungi. 10:383. doi: 10.3390/jof10060383
Munk, A. (1953). The system of the pyrenomycetes. A contribution to avnatural classification of the group Sphaerialessensu Lindau. vDansk Bot Ark 15, 1–163.
Nguyen, L. T., Schmidt, H. A., von Haeseler, A., and Minh, B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Nilsson, R. H., Larsson, K. H., Taylor, A. F. S., Bengtsson-Palme, J., Jeppesen, T. S., Schigel, D., et al. (2019). The UNITE database for molecular identification of fungi: handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res. 47, D259–D264. doi: 10.1093/nar/gky1022
Phookamsak, R., Jiang, H. B., Suwannarach, N., Lumyong, S., Xu, J. C., Xu, S., et al. (2022). Bambusicolous Fungi in Pleosporales: introducing four novel taxa and a new habitat record for Anastomitrabeculia didymospora. J. Fungi. 8:630. doi: 10.3390/jof8060630
Qiao, Z. P., Wang, M. Y., Liu, J. F., and Wang, Q. Z. (2022). Green synthesis of silver nanoparticles using a novel endophytic fungus Letendraea sp. WZ07: characterization and evaluation of antioxidant, antibacterial and catalytic activities (3-in-1 system). Inorg. Chem. Commun. 138:109301. doi: 10.1016/j.inoche.2022.109301
Rambaut, A. (2018). Molecular Evolution, Phylogenetics and epidemiology. FigTree ver.1.4.4 software. Available online at: http://tree.bio.ed.ac.uk/software/figtree (accessed October 2022).
Rannala, B., and Yang, Z. H. (1996). Probability distribution of molecular evolutionary trees: a new method of phylogenetic inference. J. Mol. Evol. 43, 304–311. doi: 10.1007/BF02338839
Rehner, S. A., and Buckley, E. (2005). A Beauveria phylogeny inferred from nuclear ITS and EF1-α sequences: evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 97, 84–98. doi: 10.3852/mycologia.97.1.84
Ren, G., Wanasinghe, D. N., de Farias, A. R. G., Hyde, K. D., Yasanthika, E., Xu, J., et al. (2022). Taxonomic novelties of woody litter fungi (Didymosphaeriaceae, Pleosporales) from the greater Mekong subregion. Biology 11:1660. doi: 10.3390/biology11111660
Ronquist, F., and Huelsenbeck, J. P. (2003). MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574. doi: 10.1093/bioinformatics/btg180
Saccardo, P. A. (1880). Fungi Gallicilecti a cl. viris P. Brunaud, Abb. Letendre, A. Malbranche, J. Therry, velediti in Mycotheca Gallica C. Roumeguèri. Series II. Michelia. 2, 39–135.
Shearer, C. A., Crane, J. L., and Chen, W. (1999). Freshwater Ascomycetes: Ophioceras species. Mycologia 91, 145–156. doi: 10.1080/00275514.1999.12061004
Silva, R. M. F., Oliveira, R. J. V., Bezerra, J. D. P., Bezerra, J. L., Souza-Motta, C. M., Silva, G. A., et al. (2019). Bifusisporellasorghi gen. et sp. nov. (Magnaporthaceae) to accommodate an endophytic fungus from Brazil. Mycol. Prog. 18, 847–854. doi: 10.1007/s11557-019-01494-2
Su, D., Zhang, W. H., Sun, R., Zhang, Z. T., and Lyu, G. (2021). First report of Botryosphaeria dothidea causing leaf spot on Kadsura coccinea in China. Plant Dis. 105:2714. doi: 10.1094/PDIS-01-21-0150-PDN
Tanaka, K., Hirayama, K., Yonezawa, H., Hatakeyama, S., Harada, Y., Sano, T., et al. (2009). Molecular taxonomy of bambusicolous fungi, Tetraplosphaeriaceae, a new pleosporalean family with Tetraploa-like anamorphs. Stud. Mycol. 64, 175–209. doi: 10.3114/sim.2009.64.10
Thongkantha, S., Jeewon, R., Vijaykrishna, D., Lumyong, S., McKenzie, E. H. C., Hyde, K. D., et al. (2009). Molecular phylogeny of Magnaporthaceae (Sordariomycetes) with a new species, Ophioceras chiangdaoense from Dracaena loureiroi in Thailand. Fungal Divers. 34, 157–173. doi: 10.1002/yea.1657
Vilgalys, R., and Hester, M. (1990). Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 172, 4238–4246. doi: 10.1128/jb.172.8.4238-4246.1990
Webster, P. J., Magana, V. O., Palmer, T. N., Shukla, J., Tomas, R. A., Yanai, M., et al. (1998). Monsoons: processes, predictability, and the prospects for prediction. J. Geophys. Res. 103, 14451–14510. doi: 10.1029/97JC02719
White, T. J., Bruns, T. D., Lee, S. B., and Taylor, J. W. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics” in PCR Protocols: A Guide to Methods and Applications, eds. M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (San Diego, NY: Academic PRess), 315–322. doi: 10.1016/B978-0-12-372180-8.50042-1
Wijayawardene, N. N., Hyde, K. D., Dai, D. Q., Sánchez-García, M., Goto, B. T., Saxena, R. K., et al. (2022). Outline of fungi and fungus-like taxa – 2021. Mycosphere 13, 53–453. doi: 10.5943/mycosphere/13/1/2
Wysocki, W. P., Clark, L. G., Attigala, L., Ruiz-Sanchez, E., and Duvall, M. R. (2015). Evolution of the bamboos (Bambusoideae; Poaceae): a full plastome phylogenomic analysis. BMC Evol. Biol. 15:50. doi: 10.1186/s12862-015-0321-5
Yan, H. J., Zhang, J. P., Tan, Z. L., Chen, Y. C., Li, C. N., Li, S., et al. (2024). Secondary metabolites from the endophytic fungus Letendraea helminthicola A820 with Anti-inflammatory activity. Chem Biodivers. 22:e202402114. doi: 10.1002/cbdv.202402114
Yan, K., Zhang, J., Cai, Y., Cao, G., Meng, L., Soaud, S. A., et al. (2023). Comparative analysis of endophytic fungal communities in bamboo species Phyllostachys edulis, Bambusa rigida, and Pleioblastus amarus. Sci. Rep. 13:20910. doi: 10.1038/s41598-023-48187-1
Yang, C. L., Xu, X. L., Zeng, Q., Liu, L. J., Liu, F., Deng, Y., et al. (2024). Bambusicolous mycopathogens in China with an update on taxonomic diversity, novel species, pathogenicity, and new insights. Mycosphere 15, 4788–4918. doi: 10.5943/mycosphere/15/1/21
Yuan, Z. L., Lin, F. C., and Zhang, C. L. Kubicek, C.P. (2010). A new species of Harpophora (Magnaporthaceae) recovered from healthy wild rice (Oryza granulata) roots, representing a novel member of beneficial dark septate endophyte. FEMS Microbiol. Lett. 307, 94–101. doi: 10.1111/j.1574-6968.2010.01963.x
Zeng, Q., Lv, Y. C., Xu, X. L., Deng, Y., Wang, F. H., Liu, S. Y., et al. (2022). Morpho-molecular characterization of microfungi associated with Phyllostachys (Poaceae) in Sichuan, China. J. Fungi. 8:702. doi: 10.3390/jof8070702
Zhang, Z. X., Liu, X. Y., Tao, M. F., Liu, X. Y., Xia, J. W., Zhang, X. G., et al. (2023). Taxonomy, phylogeny, divergence time estimation, and biogeography of the Family Pseudoplagiostomataceae (Ascomycota, Diaporthales). J. Fungi. 9:82. doi: 10.3390/jof9010082
Zhao, Z. Y., Mu, T. C., Keyhani, N. O., Pu, H. L., Lin, Y., Lv, Z., et al. (2024). Diversity and new species of ascomycota from bamboo in China. J. Fungi. 10:454. doi: 10.3390/jof10070454
Zhaxybayeva, O., and Gogarten, J. P. (2002). Bootstrap, Bayesian probability and maximum likelihood mapping: exploring new tools for comparative genome analyses. BMC Genomics 3, 1–15. doi: 10.1186/1471-2164-3-4
Zhu, H., and Tan, Y. (2024). The origin of evergreen broad-leaved forests in East Asia from the evidence of floristic elements. Plants 13:1106. doi: 10.3390/plants13081106
Keywords: Didymosphaeriaceae, Magnaporthaceae, fungal phylogenetics, taxonomy, fungal morphology
Citation: Guan X, Zhao Z, Keyhani NO, Mu T, Zheng M, Yang L, Mao Y, Shang J, Yang J, Pu H, Lin Y, Zhu M, Lv H, Heng Z, Liang H, Fan L, Ma X, Ma H, Qiu Z and Qiu J (2025) Two new species of Ascomycota on bamboo leaves in Fujian, China. Front. Microbiol. 16:1555501. doi: 10.3389/fmicb.2025.1555501
Received: 04 January 2025; Accepted: 22 May 2025;
Published: 25 June 2025.
Edited by:
Wen-Jun Li, Sun Yat-sen University, ChinaCopyright © 2025 Guan, Zhao, Keyhani, Mu, Zheng, Yang, Mao, Shang, Yang, Pu, Lin, Zhu, Lv, Heng, Liang, Fan, Ma, Ma, Qiu and Qiu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhenxing Qiu, cWl1emhlbnhpbmcyMDA2QDEyNi5jb20=; Junzhi Qiu, anVuemhpcWl1QDEyNi5jb20=
 Xiayu Guan1
Xiayu Guan1 Taichang Mu
Taichang Mu Mengjia Zhu
Mengjia Zhu Longfei Fan
Longfei Fan Xiaoli Ma
Xiaoli Ma Junzhi Qiu
Junzhi Qiu