- Independent Researcher, Seattle, WA, United States
More than three decades ago multidrug-resistant (MDR) clones of the pathogens: Staphylococcus aureus, Escherichia coli, Klebsiella pneumoniae, Clostridioides difficile, Enterococcus faecium, Pseudomonas aeruginosa and Acinetobacter baumannii have started to disseminate across wide geographical areas. A characteristic feature of all these MDR lineages is the carriage of some mutations in the quinolone resistance-determining regions (QRDRs) of DNA gyrase and topoisomerase IV which besides conferring resistance to fluoroquinolones are associated with a fitness benefit. Several lines of evidence strongly suggest that extra fitness conferred by these mutations facilitated the dissemination of the international MDR lineages. MDR pathogens require extra energy to cover the fitness cost conferred by the excess antibiotic resistance gene cargo. However, extra energy generated by upgraded metabolic activity was demonstrated to increase the uptake of antibiotics enhancing susceptibility. Accordingly, MDR bacteria need additional positive fitness schemes which, similarly to the QRDR advantage, will not compromise resistance. Some of these, not clone-specific effects are large genomes, the carriage of low-cost plasmids, the transfer of plasmid genes to the chromosome, the application of weak promoters in integrons and various techniques for the economic control of the activity of the integrase enzyme including a highly sophisticated system in A. baumannii. These impacts – among others – will confer a fitness advantage promoting the spread of MDR pathogens. However, even the potential of extra fitness generated by the combined effect of various schemes is not without limit and virulence-related genes or less relevant antibiotic resistance gene cargoes will often be sacrificed to permit the acquisition of high-priority resistance determinants. Accordingly major MDR clone strains are usually less virulent than susceptible isolates. In summary, a fitness approach to the research of antibiotic resistance is very useful since the fitness status of MDR bacteria seem to profoundly impact the capacity to disseminate in the healthcare setting.
1 Introduction
Resistance to antibiotics in bacterial pathogens constitutes a serious challenge worldwide endangering patients mostly in the healthcare setting and resulting in considerable excess mortality. Though antibiotic resistance had appeared shortly after the introduction of the first antibiotics (Acar et al., 2012) it became a major issue in the 1990s when some genetically related groups of various pathogenic MDR bacteria started to disseminate across wide geographical swathes and have ever since remained with us in many places. Interestingly the spread of these MDR strains proved exclusively clonal and was observed primarily in a handful of pathogens: Staphylococcus aureus, Escherichia coli, Klebsiella pneumoniae, Clostridioides difficile, Enterococcus faecium, Pseudomonas aeruginosa, and Acinetobacter baumannii (Hawkey and Jones, 2009; Fuzi, 2016; Fuzi et al., 2017; Fuzi et al., 2020; Fuzi and Sokurenko, 2023). The question arises what could have triggered the dissemination of these MDR lineages?
Several lines of strong circumstantial evidence suggest a “fluoroquinolone connection.” Conspicuously, the emergence of the MDR international clones commenced in the 1990s when the consumption of fluoroquinolone type antibiotics, and primarily that of ciprofloxacin, rose substantially (Linder et al., 2005). Moreover, fluoroquinolone resistant pathogens were demonstrated to carry significantly bigger and more diverse antibiotic resistance gene cargoes than fluoroquinolone susceptible strains (Coluzzi et al., 2023). Furthermore, the onset of mutations in the quinolone resistance-determining regions (QRDRs) in E. coli was shown to always precede the subsequent acquisition of the excess resistance gene loads (Coluzzi et al., 2023). Among these early onset QRDR mutations the most common alterations preceding the evolution of an MDR phenotype usually affect the gyrA S83 and parC S80 residues (positions for E. coli) (Coluzzi et al., 2023). However, how can all of these observations be linked to the success of the major international MDR clones of human pathogenic bacteria?
2 An excess gene cargo involves fitness cost
MDR bacteria inescapably carry an extra cargo of genes that will provide adequate protection in an antibiotic environment like the healthcare setting. However, the carriage and expression of most of the antibiotic resistance genes are well-established to compromise fitness (Andersson and Hugnes, 2010). Moreover, many resistance-associated mutations or changes in the target molecules of antibiotics exert additional negative fitness effects (Melnyk et al., 2015; Vanacker et al., 2023). Accordingly, the successful handling of the fitness issue is crucial for MDR pathogens which need not only protection against antibiotics used in the healthcare setting but also have to retain an appropriate growth rate (fitness) permitting dissemination. Several groups emphasized that the fitness status of pathogens plays a salient role in governing dissemination in the healthcare setting (Martínez and Baquero, 2002; Shin and Ko, 2015; Bartke et al., 2021; Baquero et al., 2021; Dunn et al., 2021).
The efficient regulation of gene acquisition and the level of expression is of utmost importance not just for the reduction of fitness cost associated with resistance to antibiotics but also for retaining a proper fitness balance across the entire genome during adaptation to diverse environmental conditions (Holden and Webber, 2020; Elg et al., 2024). When bacteria need to increase the expression of a particular set of genes they will reduce the expression of some other groups of genes which are less relevant under existing conditions (Holden and Webber, 2020). This trade-off can involve both the antibiotic resistance genes and virulence genes: bacteria were observed to increase the expression of some resistance genes upon exposure to antibiotics, while, concurrently reduced the expression of virulence genes (Holden and Webber, 2020). Accordingly, bacteria to preserve fitness may become less pathogenic when showing an MDR phenotype (Holden and Webber, 2020).
These observations suggest that successful MDR bacteria should command better fitness than less prevalent MDR pathogens allowing their extensive dissemination in the healthcare setting often with the maintenance of some virulence features. The question arises why can not bacteria simply enhance their metabolic activity to cover the fitness cost conferred by the extra antibiotic resistance gene cargo? The reason is that increased metabolic activity will not only generate extra energy. The import of more nutrients will also result in a substantial rise in antibiotic uptake which will greatly enhance susceptibility (Stokes et al., 2019; Lopatkin et al., 2021; Tao et al., 2023). This is the reason why metabolic activity is well-established to be scaled back during antibiotic exposure in bacteria even though the pathogens would need extra energy (Lopatkin et al., 2021).
Consequently, we presume, that MDR bacteria should pursue alternative fitness generating/saving schemes which will not seriously compromise resistance to antibiotics. We suggest that bacteria possess/employ a variety of devices to attain this goal. The international MDR clone strains of pathogenic bacteria command a specific beneficial fitness feature. As we shall see these agents proved capable of evolving some energetically favorable QRDR mutations which besides providing resistance to fluoroquinolones confer a fitness advantage onto the isolates without compromising antibiotic resistance.
In the following the paper will provide an up-to-date analysis of the QRDR impact and, for the first time, will review several additional energetically favorable fitness effects which support the acquisition and expression of an antibiotic resistance gene cargo without enhancing susceptibility. It is supposed that the combined effect of these features govern a bacterial pathogen’s ability to economically handle an MDR phenotype and assist in preserving sufficient fitness permiting dissemination in the healthcare setting.
3 Two energetically favorable QRDR mutations
3.1 Two QRDR mutations carried by most major clone MDR pathogens
It is a characteristic feature of all the major MDR clones in various bacterial species that they carry at least one but usually both of the so-called double-serine QRDR mutations affecting the gyrA S83 and parC S80 positions in. E. coli or equivalent positions in other species (Fuzi et al., 2017; Fuzi et al., 2020; Fuzi and Sokurenko, 2023). The onset of these mutations, as stated above, precedes the acquisition of excess genetic material associated with resistance to diverse groups of antibiotics (Coluzzi et al., 2023). An abundance of papers demonstrated that these mutations, but especially those affecting the gyrA S83 position, will confer a fitness benefit onto the pathogens, measured by the growth rate under laboratory conditions. This was observed in E. coli (Komp Lindgren et al., 2005; Marcusson et al., 2009; Machuca et al., 2014; Huseby et al., 2017), Salmonella Typhimurium (Giraud et al., 2003), Salmonella Typhi (Baker et al., 2013), C. difficile (Vernon et al., 2019), and Campylobacter jejuni (Luo et al., 2005).
The fitness impact of the parC S80 mutation proved more dependent on the genetic background of the tested isolates. It was investigated in only two pathogens: E. coli and Salmonella spp. The parC S80 mutation was observed to confer a slight fitness cost in two studies (Marcusson et al., 2009; Huseby et al., 2017), while an additional study reported a fitness gain (Baker et al., 2013).
Similarly, the combined fitness effect of the double-serine gyrA S83 plus parC S80 mutations (positions in E. coli) was strongly influenced by epistasis. The double-serine mutations were linked to a small fitness cost in two studies (Marcusson et al., 2009; Huseby et al., 2017), however, a third study demonstrated a highly significant fitness benefit in a distinct E. coli isolate (Machuca et al., 2014). Moreover, in P. aeruginosa and Burkholderia cepacia alterations equivalent to the double QRDR mutations were shown to enhance fitness influenced also by epistasis (Pope et al., 2008; Agnello et al., 2016).
Nevertheless, the question arises why have nearly all bacterial species evolutionarily preserved the double-serine QRDR residues if they ab ovo compromise (or may compromise in a strain-specific fashion) the activity of the enzymes?
3.2 Common natural compounds are toxic to QRDR mutants
Interestingly the penetrance of the serine residue in positions equivalent to that of gyrA 83 in E. coli is not complete across the kingdom of bacteria. Some Ehrlichia spp. and most Mycobacteria spp. were shown to harbor non-serine residues in the equivalent positions (Maurin et al., 2001; Guillemin et al., 1998). Since Ehrlichia spp. virtually spend their lifetime in their insect hosts where they remain fairly protected from environmental impacts and Mycobacteria command a hardly penetrable cell wall, this observation may hint that the serine residue might confer protection against some naturally occurring noxa to which bacteria are exposed in the environment.
Japanese scientists showed that fluoroquinolone resistant strains of S. aureus carrying the double-serine mutations were all susceptible to a natural antibiotic (nybomycin, a quinolone-dione) produced by several Streptomyces species (Hiramatsu et al., 2012) and a very common herbal alkaloid, apigenin (Morimoto et al., 2015). Moreover, nybomycin also showed activity against fluoroquinolone resistant Enterococci (Hiramatsu et al., 2012). Though the effect of nybomycin was recently shown to be non-mutation specific in E. coli (Shiriaev et al., 2023), resistance to apigenin seems to be unequivocally linked to the gyrA 84 serine residue (position in S. aureus) (Hiramatsu et al., 2012). Moreover, a recent study has shown that multiple additional herbal flavonoids inhibit fluoroquinolone resistant S. aureus and E. faecium strains in a similar fashion (Morimoto et al., 2023). Likewise to apigenin these alkaloids are very common in nature (Sen et al., 2016). It is tempting to speculate that due to extensive exposition to herbal flavonoids and in the absence of quinolones, ancient bacteria compromised the activity of their DNA gyrase enzymes by evolving the gyrA S83 (E. coli) and equivalent residues (Hiramatsu et al., 2012).
Though apigenin was reported to be more effective against gram-positive than gram-negative bacteria (Osonga et al., 2019; Shiriaev et al., 2023), under natural conditions bacteria are exposed to the combined effect of an abundance of substances and some (perhaps many) of them could facilitate the penetration of flavonoids into the gram-negative cell wall. Obviously exposure to antibiotics constitute a much greater threat for modern bacteria in the healthcare setting than environmental noxa. Accordingly bacterial pathogens nowadays can reliably mutate their double-serine QRDR residues under the antibiotic pressure despite they being impacted by the naturally toxic compounds.
In the following chapters we are going to review both the genetic and epidemiological evidence supporting a role for both the energetically favorable QRDR mutations and some other factors in the dissemination of the international MDR clones of high priority bacterial pathogens (Table 1; Tables S1, S2). Although every human pathogenic bacterial species exposed to fluoroquinolones was impacted a review of all affected species lies beyond the scope of this paper. Similarly, the influence of slow-degrading fluoroquiunolones in the environment on the acquisition of antibiotic resistance (Frade et al., 2014; Bungau et al., 2021) can not be covered.
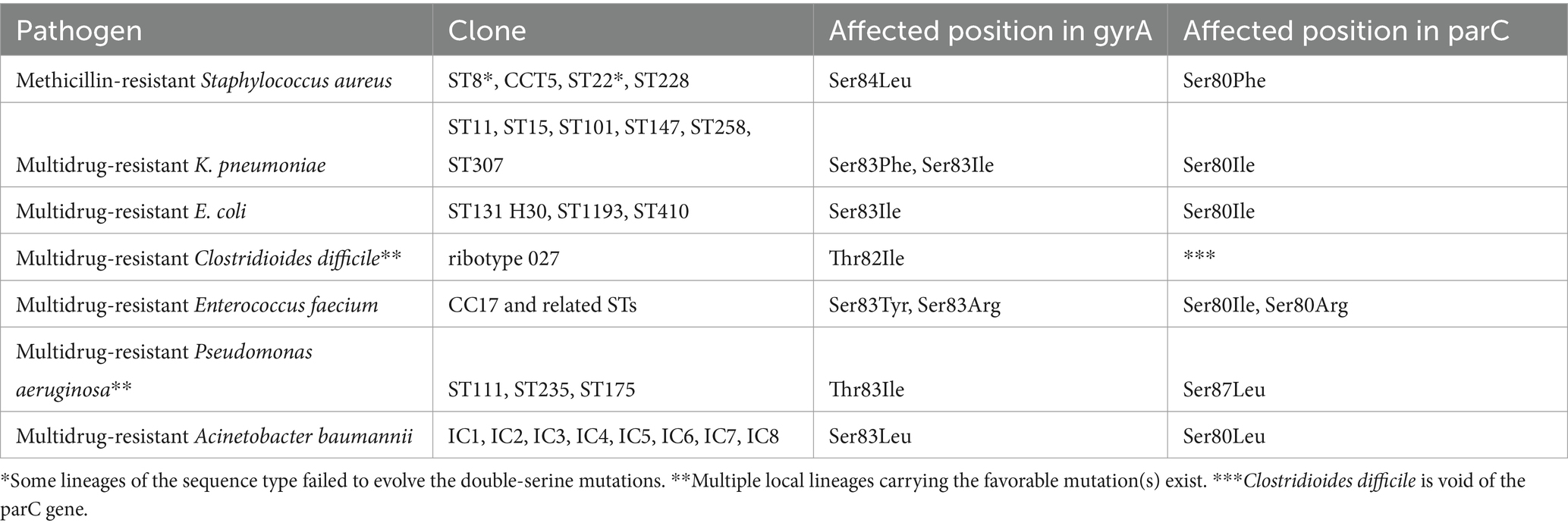
Table 1. Characteristic double-serine/threonine QRDR mutations in the international clones/lineages of various MDR pathogens (references in text).
4 International MDR clone pathogens command epidemiological advantage in a fluoroquinolone environment
4.1 Methicillin-resistant Staphylococcus aureus
It was our group that demonstrated for the first time that fitness advantage commanded by the major international clones of an MDR pathogen potentially promoted their dissemination (Table 2). The global spread of the international lineages of health care-associated (HA)-MRSA about three decades ago (Nübel et al., 2011; Grundmann et al., 2014; Lawal et al., 2022) resulted in clonal shifts worldwide. In Hungary initially the ST5 and ST228 clones achieved dominance over previous minor STs (Conceição et al., 2007) what was followed by an invasion of ST22 isolates (Grundmann et al., 2014) (Table S1).
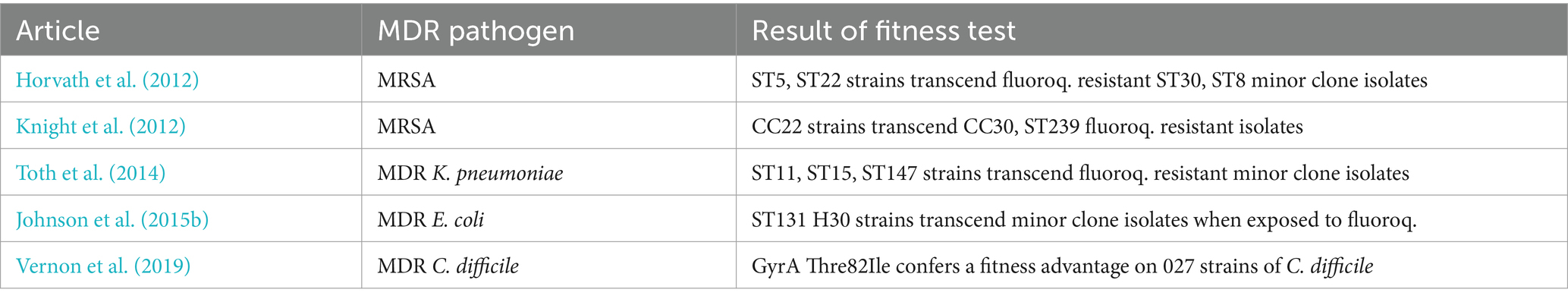
Table 2. Superior fitness of international MDR clone strains vis-a-vis minor clone isolates demonstrated by propagation assays.
Since the epidemiological shifts were associated with a rise in the incidence of HA-MRSA in Hungary (Horvath et al., 2012; Fuzi, 2016) we investigated whether or not distinct fitness commanded by the various clones could have contributed to their success. We demonstrated that fitness costs across clones elicited by resistance to fluoroquinolones were highly diverse and could have contributed to the observed clonal replacements. Propagation assays conducted by fluoroquinolone resistant major clone strains, minor clone isolates in which resistance to ciprofloxacin was induced in vitro clearly showed that the major clones commanded superior fitness (Horvath et al., 2012). The significantly faster growth rate detected in the fluoroquinolone resistant major clone isolates might have accounted for both the clonal shifts and the rise in the incidence of HA-MRSA (Horvath et al., 2012). Trends in the consumption of fluoroquinolones also proved closely associated with the increase in rates for HA-MRSA in Hungary (Fuzi, 2016). Concurrently, Knight et al. (2012) investigating the fitness of strains from diverse HA-MRSA clones in the United Kingdom reported identical findings with our results. Subsequently, Holden et al. (2013) emphasized that the success of the ST22 clone was associated with the acquisition of fluoroquinolone resistance (Table S1).
Moreover, a plethora of papers have been published on clonal shifts where major clones of HA-MRSA strains (primarily CC5 and ST22) replaced diverse minor clones/STs in adult wards in the healthcare setting or at country level supporting the notion of a fluoroquinolone impact (Velazquez-Meza et al., 2004; Ma et al., 2006; Amorim et al., 2007; Conceição et al., 2007; Aires-De-Sousa et al., 2008; Knight et al., 2012; Espadinha et al., 2013; Lim et al., 2013; Lawes et al., 2015; Hsu et al., 2015; De Oliveira et al., 2019; Junaidi et al., 2023; Coombs et al., 2023) (Table S1). The findings of De Oliveira et al. (2019) are especially interesting: they observed that MRSA strains carrying the double-serine mutations replaced isolates with single-serine alterations even if they belonged to an identical sequence type. Furthermore, multiple groups reported a significant association between fluoroquinolone use and the incidence of HA-MRSA (LeBlanc et al., 2006; Couderc et al., 2014; Fuzi, 2016; Wang et al., 2019; Bolla et al., 2020; Corcione et al., 2021; Tan et al., 2022; Aldeyab et al., 2022; Chen et al., 2022; Baede et al., 2023) that obviously implies a “major-clone-effect” (Table S2).
The most important determinant of the success of the major clones of HA-MRSA (ST5, ST22) – as it was mentioned previously – should have been the characteristic carriage of the energetically favorable double-serine QRDR mutations affecting the gyrA S84 and grlA S80 residues which are mostly absent from minor lineages which often harbor additional QRDR alterations that might be associated with fitness cost (review: Fuzi et al., 2017) (Tables 1, 3).
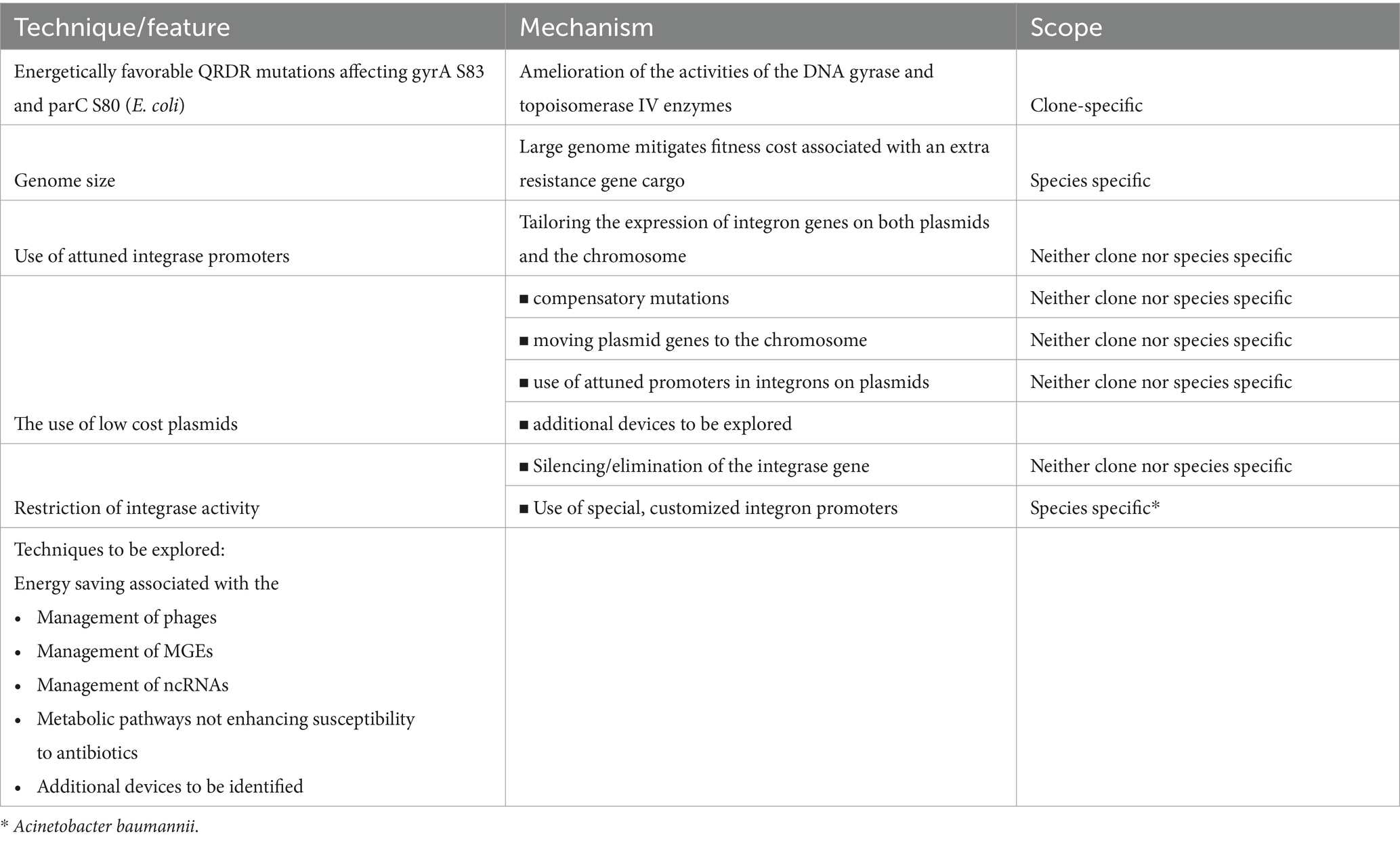
Table 3. Energy saving/generating techniques/features in MDR bacteria mitigating fitness cost associated with the carriage of an antibiotic resistance gene cargo (see references in text).
Interestingly the American ST8 clone (USA300) carrying gyrA S84; grlA S80 mutations proved capable of disseminating in US hospitals at an earlier time period when the consumption of fluoroqinolones used to be higher (Planet, 2017; Challagundla et al., 2018) and subsequently evolved into a global MDR HA-MRSA clone (Glaser et al., 2016). In contrast, the Hungarian ST8 strains developed just one favorable mutation (in position grlA S80) and proved unable to disseminate in the healthcare setting (Horvath et al., 2012; Fuzi, 2016). Similarly, while the ST22 strains carrying the double-serine mutations disseminated extensively in Western countries (Grundmann et al., 2014) some ST22 isolates in China that failed to develop these genetic traits showed lower incidence in the healthcare setting (Zhao et al., 2023).
4.2 Multidrug-resistant Klebsiella pneumoniae
The clonal shift with HA-MRSA was followed by a conspicuous clonal reduction observed with ESBL-producing K. pneumoniae in Hungary (Damjanova et al., 2006; Damjanova et al., 2008). Interestingly, in contrast to the previous polyclonal scene, all of the novel strains belonged to just three major STs (ST11, ST15, ST147) and carried the CTX-M-15 ESBL gene (Damjanova et al., 2006; Damjanova et al., 2008). The new major clones disseminated exclusively in adult hospital wards where fluoroquinolones were in extensive use but not in the perinatal intensive care units where these antibiotics were not considered an appropriate choice of therapy (Szilagyi et al., 2010). Interestingly in the perinatal intensive care units the MDR K. pneumoniae isolates remained polyclonal and primarily produced SHV type ESBL enzymes (Damjanova et al., 2008; Szilagyi et al., 2010) (Table S1). Similarly to MRSA the observed clonal change was associated with a rise in the incidence for ESBL-producing K. pneumoniae in Hungary (Szilagyi et al., 2010) (Table S2).
With regard to previous findings obtained with MRSA (Horvath et al., 2012) we performed propagation assays with major and minor ST ESBL-producing and fluoroquinolone resistant strains of K. pneumoniae. We found that the major clone/ST strains displayed significantly faster growth rates than minor clone isolates (Toth et al., 2014). In addition, some of our minor clone Klebsiella strains, originally susceptible to fluoroquinolones, suffered substantial fitness cost during in vitro induction of resistance to ciprofloxacin while other minor lineage strains ab ovo failed to assume high-level resistance (Toth et al., 2014). In contrast major ST MDR strains retained growth rates just slightly compromised relative to that of fluoroquinolone susceptible isolates (Toth et al., 2014) (Table 2).
All major clone MDR K. pneumoniae strains carried three QRDR mutations, including the double-serine alterations, in the gyrA S83, D87 and parC S80 positions. In contrast, the minor ST strains either failed to evolve any of these mutations or acquired fewer of them (Toth et al., 2014) (Table 1). These findings could account for the clonal dynamics observed with ESBL-producing K. pneumoniae in Hungary. Moreover, they may clarify the background of the widespread international dissemination of the CTX-M-15 enzyme.
Subsequently two additional major international lineages of MDR K. pneumoniae emerged and disseminated extensively worldwide. The ST258 clone, a close relative of the ST11 lineage, that played a significant role in the global spread of KPC-type carbapenemases and the ST307 clone (Bowers et al., 2015; Peirano et al., 2020) (Table S1). The available data shows that the carriage of QRDR mutations affecting primarily the double-serine (gyrA S83, parC S80) positions, but sometimes also an additional alteration in the gyrA D87 position, is a general feature of ST11, ST15, ST147; ST101, ST258 and ST307 MDR K. pneumoniae lineages (Bowers et al., 2015; Park et al., 2015; Roe et al., 2019; Zeng et al., 2020; Peirano et al., 2020; Núñez-Samudio et al., 2022; Cai and Wang, 2022; Rodrigues et al., 2022; Rodrigues et al., 2023) (Table 1). Accordingly, these strains command an energy benefit when acquiring an MDR phenotype advancing their dissemination in a fluoroquinolone environment allowing for the replacement of minor clone isolates (Damjanova et al., 2008; Chen et al., 2014; Wang Q. et al., 2024; Chatzidimitriou et al., 2024; Peirano and Pitout, 2025) (Table S1).
Moreover, similarly to HA-MRSA multiple epidemiological studies demonstrated that besides β-lactam antibiotics, the consumption of fluoroquinolons influences the incidence of ESBL-producing K. pneumoniae that mostly involves the rates of strains from major STs (Szilagyi et al., 2010; Forde et al., 2017; Wang et al., 2019; Corcione et al., 2021; Tan et al., 2022) (Table S2). In addition, Logan et al. (2019) observed that in pediatric wards, which are supposed to be low-fluoroquinolone areas the incidence of minor ST strains of K. pneumoniae producing carbapenemases was higher than that of major ST isolates. Authors emphasized that the findings are in contrast with that reported by multiple groups for adult departments.
4.3 Multidrug-resistant Escherichia coli
It is well-established that strains of both global lineages of MDR E. coli, the ST131 H30 and ST1193 clones, carry, among others, the energetically favorable double-serine QRDR mutations affecting the positions gyrA S83 and parC S80 (Colpan et al., 2013; Pitout and Finn, 2020; Pitout et al., 2022). Moreover, an emerging clone – ST410 – may be on the way of becoming another major international MDR lineage of E. coli (Pitout et al., 2023). ST410 isolates also characteristically carry the favorable double-serine mutations besides two additional alterations (Roer et al., 2018) (Table 1). Pitout et al. (2024) demonstrated that the novel clades of ST410, in contrast to previous ones, acquired the favorable QRDR mutations relatively recently and that might have contributed to the clone’s success with an MDR phenotype.
American scientists experimentally demonstrated a positive fitness effect associated with resistance to fluoroquinolones, similarly to that observed with the international STs of MRSA and MDR K. pneumoniae, in the global E. coli MDR lineage of ST131 H30 (Johnson et al., 2015a; Johnson et al., 2015b) (Table 2). This “fitness advantage” was linked to a number of favorable mutations in the gyrA, parC, and parE genes and a significantly weaker efflux activity relative to isolates from other lineages (Johnson et al., 2015a; Johnson et al., 2015b). This fitness benefit should have contributed to the extraordinary success of the ST131 H30 lineage and that of other major MDR clones of E coli (Nicolas-Chanoine et al., 2008, 2014; Price et al., 2013; Stoesser et al., 2016; Tchesnokova et al., 2019; Pitout and Finn, 2020; Pitout et al., 2022, 2024) (Table S1).
Moreover, subsequently Stoesser et al. (2016) analyzing ST131 H30 strains with WGS technique concluded that “strong selection pressure exerted by the widespread use of fluoroquinolones and extended-spectrum cephalosporins”…“most likely” played a major role in the selection of the H30 and H30R lineages of E. coli (Table S2). In addition, several groups suggested that there was an association between the success of the ST131 MDR lineage and fluoroquinolone resistance (QRDR mutations). Van der Donk et al. (2013) showed that the increased use of fluoroquinolones resulted in a rise in the incidence of ST131 strains. Banerjee and Johnson (2013) suggested that fluoroquinolone use was associated with the spread of ST131 E. coli. Li et al. (2021) linked the dissemination of the ST131 MDR clone to DNA gyrase and topoisomerase IV mutations. Cummins et al. (2021) showed that there was a link between resistance to fluoroquinolones, increased fitness and the prevalence of ST131-H30 and ST1193 isolates (Table S2).
Moreover, the fitness benefit commanded by the ST131 H30 and ST1193 clones might have contributed not only to a rise in the incidence of infections caused by these bacteria but also to the successful enteric colonization of healthy individuals by these lineages and by strains from other STs carrying one or two favorable QRDR mutations (Tchesnokova et al., 2023; Fuzi and Sokurenko, 2023).
4.4 Multidrug-resistant Clostridioides difficile
In C. difficile and some other species—P. aeruginosa, B. cepacia, C. jejuni—the residue playing a prominent role in the binding of fluoroquinolones to DNA gyrase (position gyrA 83 in E. coli) is not serine but threonine. Moreover, C. difficile, like some additional slow-growing bacteria — C. jejuni and the Mycobacteria — are void of the topoisomerase IV enzyme. Consequently QRDR resistance in these species will primarily reside with alterations in the DNA gyrase gene (Dridi et al., 2002).
The most renowned international MDR lineage of C. difficle is the 027 ribotype. Ribotype 027 isolates are characteristically resistant to fluoroquinolones and carry the gyrA T82I mutation (He et al., 2013; Spigaglia, 2016) (Table 1). Wasels et al. (2015) reported that resistance to fluoroquinolones in ribotype 027 strains was associated with a very slight fitness cost which was linked to the presence of the gyrA T82I mutation. However, Vernon et al. (2019) subsequently unequivocally demonstrated that the gyrA T82I mutation confers a fitness advantage on many 027 isolates in a strain-specific fashion (Table 2). Some additional DNA gyrase mutations were also associated with greater fitness in C. difficile, however, these alterations conferred significantly lower MIC values to fluoroquinolones than the gyrA T82I mutation (Vernon et al., 2019).
These findings strongly argue for a mechanism similar to that observed with HA-MRSA, ESBL-producing K. pneumoniae and MDR E. coli since mutations affecting the gyrA T82 position have also been shown to be a feature of isolates of additional major-non-027-international ribotypes of C. difficile (Carman et al., 2009; Saxton et al., 2009; Spigaglia et al., 2010; Lin et al., 2011; Solomon et al., 2011; Lee et al., 2014; Baldan et al., 2015; Kuwata et al., 2015; Spigaglia, 2016; Imwattana et al., 2021; Shu et al., 2023; Plankaova et al., 2023) (Table 1).
In contrasts to the rather restricted clonal landscape observed with HA-MRSA, ESBL-producing K. pneumoniae and MDR E. coli where the capacity of developing favorable gyrA and parC mutations seems to be restricted to just a few international ST strains (see above), multiple lineages of C. difficile proved capable of evolving the gyrA T82I mutation which resulted in a relative diversity in the clonal spectrum (Bauer et al., 2011; Tickler et al., 2014; Freeman et al., 2015; Liu et al., 2023) (Table 1).
The extraordinary expansion of the 027 ribotype and that of multiple additional gyrA T82I-positive lineages (Goorhuis et al., 2008; Freeman et al., 2015; Liu et al., 2023) (Table S1) plus a well-documented clonal replacement attest to the superior dissemination capacity of international C. difficle clones in a fluroquinolone environment.
A comprehensively investigated clonal shift with C. difficile was reported from a Korean hospital. The previously prevalent ribotype 001 strains were replaced by isolates from the 014, 017, and 018 ribotypes within a couple of years (Lee et al., 2014) (Table S1). Interestingly, all of the novel ribotype strains carried the energetically favorable T82I gyrA mutation while the Korean ribotype 001 isolates harbored a different mutation in the same position (T82V gyrA) which was previously demonstrated to be associated with a high fitness cost (Wasels et al., 2015).
Moreover, the proposed “fluoroquinolone mechanism” for the selection of the major clones of C. difficile is supported by the observation that the proportion of ribotype 027 strains is significantly greater in adult wards than in pediatric units (McFarland et al., 2016). Furthermore, an abundance of studies demonstrated that a decrease in the consumption of fluoroquinolones results in a decline in the incidence of C. difficile infections in general and of those caused by major clone (ribotype) strains in particular (Pepin et al., 2004; McDonald et al., 2005; LeBlanc et al., 2006; Sarma et al., 2015; Dingle et al., 2017; Rizzo et al., 2019; Redmond et al., 2019; Kazakova et al., 2020; Guh et al., 2020; Slimings and Riley, 2021; Couture et al., 2024) (Table S2).
4.5 Multidrug-resistant Enterococcus faecium
MDR E. faecium showing resistance to various antibiotics including vancomycin (VRE) poses a serious challenge for antibiotic therapy worldwide. A few lineages related to a single international clonal complex of VRE (CC17) dominate the hospital setting in many geographic areas (Willems et al., 2005; Freitas et al., 2016).
VRE disseminated in Europe in the 1990s initially as an enteric colonizer due to the extensive use of avoparcin, a veterinary glycopeptide antibiotic (Van den Bogaard et al., 1997). However, no major clone of VRE was observed in the healthcare setting during this period (Clark et al., 1993; Van den Braak et al., 1998). Then surprisingly major MDR lineages of VRE all linked to a single clonal complex, CC17, started to expand across large geographic areas (Willems et al., 2005; Cattoir and Leclercq, 2013) (Table S1). Most of these strains were characteristically resistant to fluoroquinolones and carried – sometimes among others – the double-serine QRDR alterations (Leavis et al., 2006; López et al., 2011; Valdezate et al., 2012; Troscianczyk et al., 2022) (Table 1). Moreover, the use of fluoroquinolones was reported to be associated with the incidence of strains from the CC17 VRE lineage (Top et al., 2007). Furthermore, fluoroquinolone consumption was observed to be more closely associated with VRE rates than the consumption of glycopeptide antibiotics (Forstner et al., 2015) (Table S2). In addition, CC17 strains were demonstrated to start to extensively disseminate when having acquired resistance to fluoroquinolones by evolving the favorable QRDR mutations and only subsequently developed resistance to vancomycin (López et al., 2011; Cattoir and Leclercq, 2013). Moreover, this clone was recently reported to spread as a vancomycin-susceptible but fluoroquinolone-resistant lineage featuring the double-serine mutations (Aung et al., 2023) (Table S1). Finally, it is certainly not accidental that all MDR (VRE) E faecium lineages are associated with a single clonal complex: it was demonstrated that the capacity to evolve the double-serine QRDR mutations is extremely rare among E. faecium isolates (De Lastours et al., 2017).
4.6 Multidrug-resistant Pseudomonas aeruginosa
The dissemination of MDR P. aeruginosa, similarly to many other resistant pathogens, is well-established to be clonal. Besides the three main international STs – ST235, ST111 and ST175 (Cabot et al., 2012; Del Barrio-Tofiño et al., 2017; Pérez et al., 2019; Recio et al., 2021; Torrens et al., 2022) (Table S1)—multiple local high-risk clones were also observed (Pérez et al., 2019; Kocsis et al., 2019; Silveira et al., 2020; Torrens et al., 2022; Hu et al., 2021; Kiyaga et al., 2022).
The carriage of the energetically favorable double QRDR mutations—gyrA T83I and parC S87L in P. aeruginosa—is a characteristic feature of all three major international MDR clones of ST235, ST111 and ST175 (Cabot et al., 2012; Kos et al., 2015; Recio et al., 2021; Torrens et al., 2022) (Table 1). Moreover, the ST175 strains usually carry an additional gyrA D87N QRDR alteration (Cabot et al., 2012; Kos et al., 2015; Del Barrio-Tofiño et al., 2017; Torrens et al., 2022).
Some of the local high-risk clones (ST773, ST654; ST357, ST463) were also reported to often carry the favorable double-threonine/serine QRDR mutations (Kocsis et al., 2019; Torrens et al., 2022; Kiyaga et al., 2022; Zhang et al., 2023; Jung et al., 2024). However, many minor clone strains were observed to harbor just one of the energetically favorable mutations which is often combined with additional, diverse, less common DNA gyrase and topoisomerase IV alterations (Kos et al., 2015; Del Barrio-Tofiño et al., 2017; Kiyaga et al., 2022).
The significance of the double threonine/serine QRDR mutations in P. aeruginosa was demonstrated by Treepong et al. (2018) who showed that the epidemiological success of the ST235 clone commenced when the strains assumed resistance to fluoroquinolones which was only followed by the acquisition of an MDR phenotype. Moreover, Brüggemann et al. (2018) demonstrated that the ST235 strains of P. aeruginosa, despite a shared clonal ancestry, “possessed individual evolutionary histories” when having acquired an MDR phenotype. This observation raises the question: if not the genetically related evolution, then what is the common feature in the successful isolates of the ST that permitted/facilitated dissemination? The answer seems obvious, the carriage of the double-threonine/serine QRDR mutations. Furthermore, Hakki et al. (2019) demonstrated that the use of fluoroquinolones results in an increase in the rate of MDR ST111 P. aeruginosa with strains displaying resistance to antibiotics the patients were not exposed to (Table S2).
4.7 Multidrug-resistant Acinetobacter baumannii
The dissemination of MDR A. baumannii – similarly to other MDR pathogens – remains largely clonal. Strains from a few global clonal complexes (“international clones”) comprising multiple STs are responsible for the majority of infections (Karah et al., 2012; Hamidian and Nigro, 2019; Valcek et al., 2022; Castillo-Ramírez, 2023) (Table S1). The major ST MDR strains characteristically carry double-serine QRDR mutations affecting positions: gyrA S81 and parC S80 (Warner et al., 2016; Shen et al., 2016; Nodari et al., 2020; Wiradiputra et al., 2023) (Table 1) which are equivalent to those observed in the global clones of other international MDR lineages.
The crucial role of fluoroquinolone use in the dissemination of MDR A. baumannii was confirmed by Matsui et al. (2018) conducting a large epidemiological study in Japan. The authors concluded that the judicious use of “…fluoroquinolones may hold the key to overcoming epidemic clones” of MDR A. baumannii.
5 Not-clone-specific positive fitness schemes associated with MDR phenotype
Besides the favorable double-serine QRDR fitness effect MDR pathogens command/employ a variety of not-clone-specific energy saving/generating schemes to support the carriage and expression of an extra antibiotic resistance gene cargo which will not compromise antibiotic resistance.
5.1 Genome size and fitness
It has been suggested more than fifteen years ago that the size of the bacterial genome impacts the capacity of a particular pathogen to assume an MDR phenotype (Projan, 2007). Bacterial species with large genomes seem capable of harboring a greater cargo of antibiotic resistance genes without suffering serious fitness cost than species with smaller genomes (Projan, 2007; Iranzo et al., 2017). A newly acquired excess block of genes will constitute a much smaller proportion in a large bacterial genome than in a small bacterial genome and will confer a smaller fitness cost onto the agent (Iranzo et al., 2017).
It is certainly not accidental that the pathogens comprising the major international MDR clones mentioned above command the biggest genomes among human pathogenic bacteria (Projan, 2007). In addition, it should not be a contingency that human pathogenic bacteria featuring the largest genomes, P. aeruginosa and K. pneumoniae (Projan, 2007), are well-known to be capable of acquiring a pandrug-resistant phenotype (Kryzhevskyi et al., 2023; de Man et al., 2016). Interestingly, A. baumannii featuring a somewhat smaller genome (Duan et al., 2024) was also observed to assume pandrug-resistance (Fernández-Vázquez et al., 2023). However, this exploit apart from the considerable size of the genome, could be the consequence of some additional energy saving schemes in A. baumannii: a highly sophisticated management of plasmids (Maslova et al., 2022), the application of special metabolic pathways (Djahanschiri et al., 2022) and, most importantly, an unique technique for the judicious control of its integrase enzymes (Couvé-Deacon et al., 2019) (the latter see in detail in chapter: Plasmids and integrons). Interestingly, E. coli and Salmonella spp. which command somewhat bigger genomes than A. baumannii but smaller than those of P. aeruginosa and K. pneumoniae (Projan, 2007) have not been reported to have acquired pandrug-resistance to date hinting at the relevance of some unique energy saving devices observed in A. baumannii.
S. aureus and E. faecium command smaller genomes than any of the gram-negative pathogens mentioned above (Projan, 2007). Accordingly, pandrug-resistance has not been observed in these gram-positive agents. Moreover, serious limitations remain even as to the MDR phenotypes that S. aureus and E. faecium can assume. S. aureus was demonstrated to suffer highly significant fitness cost when acquiring resistance to both daptomycin and vancomycin (Roch et al., 2017; McGuinness et al., 2017). Furthermore, vancomycin resistant S. aureus (VRSA) strains usually remain susceptible to a variety of non-glycopeptide antibiotics (Cong et al., 2019) which is most certainly a fitness effect. In addition, the incidence of VRSA remains very low despite several reports of individual infections occurring across large geographical areas (McGuinness et al., 2017). The failure of VRSA to disseminate is certainly linked to a serious compromise in fitness (McGuinness et al., 2017). Moreover, similarly to S. aureus, resistance to daptomycin results in a serious fitness cost in vancomycin resistant E. faecium (VRE) that compromises even the strains’ resistance to glycopeptides (Zeng et al., 2022).
5.2 Plasmids and integrons
Bacteria evolved several well-defined and not clone-specific energy-saving/generating schemes – independent of the size of genome—which assist in the preservation of fitness and allow proper growth rates subsequent to the acquisition of extra gene cargoes without seriously compromising antibiotic resistance.
One of the most important vehicles bacteria employ in their defensive strategy against exposure to harmful substances, including antibiotics, are plasmids. Plasmids allow bacteria to transiently acquire helpful genes that can be subsequently eliminated when conditions rehabilitate permitting for the restoration of fitness. The carriage of plasmids is associated with fitness cost (Modi and Adams, 1991; Adler et al., 2014; Dunn et al., 2021) that is primarily related to the metabolic cost and the molecular conflicts emanating from the “interactions of the plasmid and host machineries” (Modi and Adams, 1991).
However, the plasmid-related fitness cost can subsequently be attenuated mainly by the evolution of compensatory mutations (Yano et al., 2016; Hall et al., 2021; Ares-Arroyo et al., 2022; Carrilero et al., 2023). Moreover, some plasmids were reported to exert just a minimal adverse effect onto host bacteria. The fitness effects of some plasmids were thoroughly investigated in the major MDR clones of E. coli and were established to be usually minimal rendering their carriage economical (San Millan and MacLean, 2017; Johnson et al., 2019; Dunn et al., 2021; Palkovicova et al., 2022; Vanacker et al., 2023; Chen et al., 2024).
The main source of fitness cost associated with the carriage of plasmids was demonstrated to be the number of genes expressed (Sandegren et al., 2012; Adler et al., 2014; Vogwill and MacLean, 2015; Wein et al., 2019; Vanacker et al., 2023) although the energy loss proved very diverse across individual genes (Rajer and Sandegren, 2022). However, it is not just the expression of genes but also the level of expression that is associated with fitness cost during both plasmid-, and chromosomal adaptation (Kloos et al., 2021). The extensive dissemination of the IncP-1 plasmid in a variety of bacterial species was linked to its sophisticated machinery capable of decreasing (“fine-tuning”) gene expression and minimize fitness cost (Adamczyk and Jagura-Burdzy, 2003). The successful horizontal spread of the mcr-1 colistin resistance gene was similarly established to have been permitted by the scaling back (“optimization”) of its expression (Ogunlana et al., 2023). Moreover, the substantial decrease of transcription in multiple genes of the broad-host range plasmid pBP136 was observed to “drastically” improve its maintenance in host bacteria (Elg et al., 2024).
Consequently, the optimization/fine-tuning of the level of gene expression in both the chromosome and plasmids is a salient fitness maintaining exercise for bacteria. The ability to collectively control and optimize the expression of multiple genes, including many of those associated with resistance to antibiotics, bacteria employ the genetic elements called integrons. Integrons are ubiquitos in bacteria and can be located on both plasmids and the chromosome (Noel et al., 2022). Various genes recruited to an integron gene cassette have a common promoter which allows for maintaining a collective level of expression. Integron genes with strong promoters will be expressed usually at a much higher level than integron genes with weak promoters (Fonseca and Vicente, 2022). Strains of the major lineages of MDR E. coli (ST131 H30; ST1193) harbor mostly class 1 integrons (Li et al., 2020; Li et al., 2021) featuring generally weak promoters (Vinué et al., 2011; Wei et al., 2013) allowing for a considerable economization.
Some data are available on the promoter types of integrons in major MDR clone isolates in some additional pathogens. The carriage of type 1 integrons was reported common also in MDR strains of K. pneumoniae (Derakhshan et al., 2016; Bakr and Zaki, 2019; Wang et al., 2023). Moreover, Wang et al. (2023) detected mostly the “relative weak” PcH1 promoter in the integrons of their MDR K. pneumoniae isolates. Weak promoters remain dominant also in class 1 integrons in MDR Proteus spp. and were demonstrated to be associated with a favorable fitness effect (Xiao et al., 2019). Moreover, similarly to MDR E. coli and MDR K. pneumoniae it was recently established that the carriage of class 1 integrons is characteristic for MDR high-risk clones of P. aeruginosa (Lee et al., 2023). The relevance of energy conservation by the judicious expression of integron genes in P. aeruginosa was recently demonstrated by Kikuchi et al. (2024) who showed that a novel integron acquired years earlier by the species had originally harbored a strong promoter but started to globally disseminate only after its expression has been attenuated.
The application of weak promoters is not the only means for saving energy with integron function. A. baumannii uses an exceptional mechanism for the control of its costly integrase enzymes. Integrons are associated with the integrase enzyme, a crucial element of the integron system, catalyzing both insertion and excision of gene cassettes. The expression of the integrase must be strictly controlled to prevent substantial DNA damage, a consequence of inadvertant activity (Cambray et al., 2011). Most bacteria restrict the unintended expression of integrase by employing the LexA SOS system (Cambray et al., 2011). However, this type of integrase control was demonstrated in E. coli to result in an enormous fitness cost especially if the integrase has to be regularly blocked by an SOS response due to steady stress effects like continuous exposure to antibiotics (Kozuch et al., 2020; Jones and Uphoff, 2021). Moreover, MDR bacteria often carry multiple integrons whose integrases must be kept in check individually to prevent substantial damage to the genetic makeup (Couvé-Deacon et al., 2019; Xiu et al., 2024).
A. baumannii lack the LexA system and use instead the “umuDAb” protein for stress control (Aranda et al., 2013; Gregg-Jolly, 2022) that was reported to facilitate the acquisition of some antibiotic resistance determinants (Norton et al., 2013). However, A. baumannii basically employs a quite different, and highly energy conserving strategy for the control of its integrase enzymes: it uses special integron promoters which will ab ovo restrict integrase activity (Couvé-Deacon et al., 2019; Fonseca and Vicente, 2022; Xiu et al., 2024). Integrons in A. baumannii usually carry structurally strong casette promoters which are attenuated, however, will exert sufficient transcriptional interference with the integrase promoter to limit the expression of the enzyme and conserving thereby substantial energy (Couvé-Deacon et al., 2019; Xiu et al., 2024). This unique mechanism of integrase control might have contributed to the ability of A. baumannii to acquire a pandrug-resistant phenotype with a relatively modest size of genome compared with those of P. aeruginosa and K. pneumoniae.
Energy conservation by the “silencing” of integrase genes was observed also in the international MDR clones of E. coli (ST131, ST1193). In these lineages a considerable proportion of the integrons were reported to have been truncated and the deletions mostly affected the integrase genes preventing the enzyme’s transcription (Li et al., 2021; Wyrsch et al., 2022).
Furthermore, as mentioned above, A. baumannii commands some additional specific energy generating/saving schemes which besides the restriction of integrase transcription should support the carriage of excess gene cargoes: the employment of some special metabolic pathways (Djahanschiri et al., 2022) and the highly efficient conduct of the plasmidome (Maslova et al., 2022).
Besides compensatory mutations, the fitness cost associated with plasmids can be further mitigated by the transfer of long-standing antibiotic resistance genes from the plasmid to the chromosome which also results in some fitness advantage that may support the maintenance of an MDR phenotype. Plasmid genes which come from an alien agent command a codon-usage scheme that is different from that of the host bacterium to which the tRNA gene pool of the cell has been tailored (López et al., 2019; López et al., 2020). However, plasmid genes transferred to the chromosome will adapt with time and gradually assume the codon usage scheme of the host which should be associated with some energy conservation (López et al., 2020). It is certainly not accidental that the transfer of antibiotic resistance genes from plasmids to the chromosome was observed in some major MDR clone strains of E. coli (Price et al., 2013), K. pneumoniae (Mathers et al., 2017); P. aeruginosa (Hong et al., 2016; Papagiannitsis et al., 2020) and A. baumannii (Valcek et al., 2022; Wang W. et al., 2024). The gene transfer from plasmid to chromosome may involve the whole integron which will then be often further economized by the elimination of the integrase gene (Cury et al., 2016; Goswami et al., 2020).
6 Virulence, antibiotic resistance and fitness: always a trade-off
Although the fitness benefit/conservation conferred by the double serine QRDR mutations, low cost plasmids, integrons with weak promoters and other fitness schemes permit the international MDR bacteria to carry an extensive extra gene cargo, the energy gain has its limits. As we have seen with S. aureus and E. faecium, sometimes less relevant antibiotic resistance genes have to be sacrificed to maintain appropriate fitness with the carriage of an indispensible resistance gene cargo (Roch et al., 2017; McGuinness et al., 2017; Zeng et al., 2022).
In addition, the “fitness limit” warrants that virulence genes, which are not essential for survival in the antibiotic environment of the healthcare setting, will often be deleted in a trade-off in the major MDR clones of bacteria. The “fluoroquinolone-related” major MDR clone strains are usually less virulent than the antibiotic susceptible isolates from the same species and it is most certainly the command of appropriate fitness that primarily supports their dissemination in the healthcare setting.
Martínez and Baquero (2002) emphasized more than twenty years ago the precedence of antibiotic resistance over virulence in the selection of international clones in their classic paper. They observed: “If antibiotics are almost ubiquitously present in the hosts, as in an intensive care unit…the spread of the resistant bacteria is favored”.
We previously reviewed the virulence features of MDR clones in S. aureus, E. coli, K. pneumoniae, E. faecium, C. difficile and P. aeruginosa (Fuzi et al., 2020). Although some virulence factors – primarily the formation of biofilm – contribute to the dissemination of multiple international MDR clones we showed that these pathogens usually harbor fewer virulence determinants than more susceptible lineages from the same species (Fuzi et al., 2020). In the following we are going to review some well-documented examples demonstrating that the carriage of virulence features is often a function of available fitness subsequent to the acquisition of an indispensible antibiotic resistance gene cargo.
Strains from the major MDR ST8 clone of HA-MRSA were reported to carry fewer virulence factors than more susceptible HA-MRSA isolates (Dauwalder et al., 2008). Moreover, the SCCmec II, III elements which are typically harbored by HA-MRSA strains and are bigger and confer significantly higher MIC values to β-lactam antibiotics than the SCCmec IV/V casettes often include a mobile genetic element (psm-mec) whose product compromises the accessory gene regulator system (agr) attenuating the virulence of HA-MRSA isolates (Kaito et al., 2013; McCarthy et al., 2015).
Moreover, Duprilot et al. (2020) showed that ST131 strains of MDR E. coli sacrificed multiple virulence genes to retain appropriate fitness that should have contributed to the clone’s success. The ST410 clone of E. coli shows a relatively low virulence-associated factor score and has started to emerge as a major international MDR lineage not after acquiring more virulence genes but only subsequent to having evolved the double-serine mutations plus two additional QRDR alterations (Pitout et al., 2024). This set of mutations should have conferred both the extra fitness benefit and the appropriate level of fluoroquinolone resistance that was required for dissemination in adult hospital wards.
The attenuation of virulence in some of the so-called hypervirulent MDR major STs of K. pneumoniae is well-reflected in the observation that the plasmid responsible for the introduction of multiple virulence genes into many of these isolates proved capable of dissemination exclusively after having significantly lowered the associated fitness cost by eliminating the relevant virulence gene cluster of iroBCDN (Jia et al., 2024).
Dingle et al. (2023) demonstrated that β-lactam resistance conferred by PBP mutations were just as characteristic for 027 strains of C. difficile as the carriage of the energetically favorable gyrA T82I and gyrB D426N/V alterations. In addition, they observed that two fluoroquinolone susceptible lineages commanding similar PBP mutations (010 and 039) proved quite successful in non-fluoroquinolone environments. Interestingly the 010 and 039 lineages are both non-toxigenic which might have conferred a fitness benefit onto the isolates. Since PBP mutations were demonstrated to have been associated with fitness cost in Clostridium perfringens (Park and Rafii, 2017), Dingle et al.’s (2023) observation may suggest that the gyrA T82I and gyrB D426N/V mutations might have conferred a fitness benefit onto the 027 strains that could have permitted the clone to collectively acquire PBP mutations and the toxin gene cargo. In contrast the 010 and 039 lineages proved unable to evolve the favorable QRDR mutations that might have prevented these clones from carrying both the mutated PBP proteins and an extra toxin gene load.
Though MDR strains of P. aeruginosa often display an attenuated virulence profile relative to susceptible isolates (Peña et al., 2015) many high-risk clone strains from the sequence types: ST235; ST111; ST773; ST357 were observed to show considerable virulence which was predominantly linked to the production of the exoU toxin (Peña et al., 2015; Sánchez-Diener et al., 2017; Papagiannitsis et al., 2020; Recio et al., 2021; Kiyaga et al., 2022; Stoikov et al., 2023; Wu et al., 2024; Jung et al., 2024). Interestingly, in contrast to these clones, strains from the ST175 MDR major lineage, one of the biggest MDR groups in P. aeruginosa, are mostly less virulent and remain void of the exoU gene (Sánchez-Diener et al., 2017; Recio et al., 2021; Silva et al., 2023). The question arises, why are many high-risk clones of P. aeruginosa capable of harboring a greater virulence gene cargo, involving primarily the exoU toxin-related genes, than ST175 strains?
The exoU+ TTSS type III genetic background was demonstrated to be conducive for the evolution of QRDR mutations in general and for the development of the threonine/serine alterations in particular in P. aeruginosa (Agnello and Wong-Beringer, 2012). Most of the virulent high-risk clone MDR strains characteristically carry the double threonine/serine QRDR mutations (Cabot et al., 2012; Kos et al., 2015; Recio et al., 2021; Torrens et al., 2022) conferring a fitness advantage onto the isolates. Alternatively, the ST175 strains are well-established to typically carry an additional gyrA D87 substitution (Cabot et al., 2012; Kos et al., 2015; Recio et al., 2021; Torrens et al., 2022) that should compromise the fitness gain associated with the double-threonine/serine mutations and prevent the carriage of a greater virulence gene cargo, like exoU. The mutations affecting an equivalent position to gyrA D87 in P. aeruginosa were demonstrated to be associated with a fitness cost in various Enterobacterales spp. by several groups of investigators (Komp Lindgren et al., 2005; Marcusson et al., 2009; Baker et al., 2013).
Finally, in contrast to other pathogens, major clone strains of MDR A. baumannii can often display greater virulence than minor clone isolates (Kumkar et al., 2022; Park et al., 2023). This interesting characteristic of the MDR major clone strains is most certainly permitted by the specific energy generating/saving schemes commanded by A. baumannii, primarily the efficient control of its integrase enzymes (Couvé-Deacon et al., 2019; Xiu et al., 2024) that may also allow for the acquisition of pandrug-resistance with a relatively modest size of genome.
7 Perspective
Several lines of strong circumstantial evidence suggest that bacteria need extra energy when they acquire an excess gene cargo to be able to cope with the surplus load. However, enhancement of the basic energy generating metabolic activity is well-established to increase susceptibility to antibiotics. Accordingly bacterial pathogens need to command/employ other energy generating/saving schemes that will confer a fitness benefit without compromising susceptibility. Some QRDR mutations associated with fitness benefit have been demonstrated to support the acquisition and expression of antibiotic resistance genes contributing substantially to the dissemination of the major international MDR clones in various bacterial species (Fuzi et al., 2017; Fuzi and Sokurenko, 2023). A large genome also promotes the physiologically sustainable carriage of acquired antibiotic resistance genes rendering some bacterial species more prone to assume MDR phenotypes (Projan, 2007). Moreover, additional general and species-specific energy saving/generating schemes contribute to the ability of pathogens to acquire diverse quantities of excess gene cargoes (Table 3). Bacteria commanding more abundant/efficient fitness schemes will prove more capable of assuming an MDR phenotype and/or retaining virulence genes.
The fitness schemes reviewed in this paper are certainly relevant for MDR bacteria, however, comprise only a fraction of the miscellaneous techniques pathogens employ to save/generate extra energy without compromising antibiotic resistance. The impact of the efficient management of various mobile genetic elements, phages, ncRNAs and the effect of distinct metabolic pathways – among others – remain to be investigated. Accordingly, future studies should be aimed at exploring additional beneficial fitness schemes assisting the acquisition of an MDR phenotype and applying a fitness approach to any research associated with antibiotic resistance.
Data availability statement
Publicly available datasets were analyzed in this study. This data can be found at PubMed.
Author contributions
MF: Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Acknowledgments
The author greatly appreciates the expert counsel given by Prof. Evgeni Sokurenko (Department of Microbiology, University of Washington, Seattle, United States).
Conflict of interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The author(s) declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1556656/full#supplementary-material
References
Acar, J. F., Moulin, G., Page, S. W., and Pastoret, P. P. (2012). Antimicrobial resistance in animal and public health: introduction and classification of antimicrobial agents. Rev. Sci. Tech. 31, 15–21. doi: 10.20506/rst.31.1.2093
Adamczyk, M., and Jagura-Burdzy, G. (2003). Spread and survival of promiscuous IncP-1 plasmids. Acta Biochim. Pol. 50, 425–453. doi: 10.18388/abp.2003_3696
Adler, M., Anjum, M., Berg, O. G., Andersson, D. I., and Sandegren, L. (2014). High fitness costs and instability of gene duplications reduce rates of evolution of new genes by duplication-divergence mechanisms. Mol. Biol. Evol. 31, 1526–1535. doi: 10.1093/molbev/msu111
Agnello, M., Finkel, S. E., and Wong-Beringer, A. (2016). Fitness cost of fluoroquinolone resistance in clinical isolates of Pseudomonas aeruginosa differs by type III secretion genotype. Front. Microbiol. 7:1591. doi: 10.3389/fmicb.2016.01591
Agnello, M., and Wong-Beringer, A. (2012). Differentiation in quinolone resistance by virulence genotype in Pseudomonas aeruginosa. PLoS One 7:e42973. doi: 10.1371/journal.pone.0042973
Aires-De-Sousa, M., Correia, B., and De Lencastre, H.Multilaboratory Project Collaborators (2008). Changing patterns in frequency of recovery of five methicillin-resistant Staphylococcus aureus clones in Portuguese hospitals: surveillance over a 16-year period. J. Clin. Microbiol. 46, 2912–2917. doi: 10.1128/JCM.00692-08
Aldeyab, M. A., Bond, S. E., Conway, B. R., Lee-Milner, J., Sarma, J. B., and Lattyak, W. J. (2022). A threshold logistic modelling approach for identifying thresholds between antibiotic use and methicillin-resistant Staphylococcus aureus incidence rates in hospitals. Antibiotics (Basel) 11:1250. doi: 10.3390/antibiotics11091250
Amorim, M. L., Faria, N. A., Oliveira, D. C., Vasconcelos, C., Cabeda, J. C., Mendes, A. C., et al. (2007). Changes in the clonal nature and antibiotic resistance profiles of methicillin-resistant Staphylococcus aureus isolates associated with spread of the EMRSA-15 clone in a tertiary care Portuguese hospital. J. Clin. Microbiol. 45, 2881–2888. doi: 10.1128/JCM.00603-07
Andersson, D. I., and Hugnes, D. (2010). Antibiotic resistance and its cost: is it possible to reverse resistance? Nat. Rev. Microbiol. 8, 260–271. doi: 10.1038/nrmicro2319
Aranda, J., Poza, M., Shingu-Vazquez, M., Cortes, P., Boyce, J. D., and Adler, B. (2013). Identification of a DNA-damage-inducible regulon in Acinetobacter baumannii. J. Bacteriol. 195, 5577–5582. doi: 10.1128/JB.00853-13
Ares-Arroyo, M., Fernández-García, M., Wedel, E., Montero, N., Barbas, C., Rey-Stolle, M. F., et al. (2022). Genomics, transcriptomics, and metabolomics reveal that minimal modifications in the host are crucial for the compensatory evolution of ColE1-like plasmids. mSphere 7:e0018422. doi: 10.1128/msphere.00184-22
Aung, M. S., Urushibara, N., Kawaguchiya, M., Ohashi, N., Hirose, M., Kudo, K., et al. (2023). Antimicrobial resistance, virulence factors, and genotypes of Enterococcus faecalis and Enterococcus faecium clinical isolates in northern Japan: identification of optrA in ST480 E. faecalis. Antibiotics (Basel) 12:108. doi: 10.3390/antibiotics12010108
Baede, V. O., Gupta, A., Knight, G. M., Schouls, L. M., Laing, K., Tavakol, M., et al. (2023). Markers of epidemiological success of methicillin-resistant Staphylococcus aureus isolates in European populations. Clin. Microbiol. Infect. 29, 1166–1173. doi: 10.1016/j.cmi.2023.05.015
Baker, S., Duy, P. T., Nga, T. V., Dung, T. T., Phat, V. V., Chau, T. T., et al. (2013). Fitness benefits in fluoroquinolone-resistant Salmonella Typhi in the absence of antimicrobial pressure. eLife 2:e01229. doi: 10.7554/eLife.01229
Bakr, A. O., and Zaki, M. E. S. (2019). Molecular study of Klebsiella pneumoniae virulence genes from patients with hospital acquired sepsis. Clin. Lab. 65. doi: 10.7754/Clin.Lab.2018.180709
Baldan, R., Trovato, A., Bianchini, V., Biancardi, A., Cichero, P., Mazzotti, M., et al. (2015). Clostridium difficile PCR ribotype 018, a successful epidemic genotype. J. Clin. Microbiol. 53, 2575–2580. doi: 10.1128/JCM.00533-15
Banerjee, R., and Johnson, J. R. (2013). Escherichia coli ST131: variations on a theme of clonal expansion. Enferm. Infecc. Microbiol. Clin. 31, 355–356. doi: 10.1016/j.eimc.2013.01.004
Baquero, F., Martínez, J. L. F., Lanza, V., Rodríguez-Beltrán, J., Galán, J. C., et al. (2021). Evolutionary pathways and trajectories in antibiotic resistance. Clin. Microbiol. Rev. 34:e0005019. doi: 10.1128/CMR.00050-19
Bartke, K., Garoff, L., Huseby, D. L., Brandis, G., and Hughes, D. (2021). Genetic architecture and fitness of bacterial interspecies hybrids. Mol. Biol. Evol. 38, 1472–1481. doi: 10.1093/molbev/msaa307
Bauer, M. P., Notermans, D. W., van Benthem, B. H., Brazier, J. S., Wilcox, M. H., Rupnik, M., et al. (2011). Clostridium difficile infection in Europe: a hospital-based survey. Lancet 377, 63–73. doi: 10.1016/S0140-6736(10)61266-4
Bolla, C., Di Pietrantonj, C., Ferrando, E., Pernecco, A., Salerno, A., D'Orsi, M., et al. (2020). Example of antimicrobial stewardship program in a community hospital in Italy. Med. Mal. Infect. 50, 342–345. doi: 10.1016/j.medmal.2019.11.008
Bowers, J. R., Kitchel, B., Driebe, E. M., MacCannell, D. R., Roe, C., Lemmer, D., et al. (2015). Genomic analysis of the emergence and rapid global dissemination of the clonal group Klebsiella pneumoniae pandemic. PLoS One 10:e0133727. doi: 10.1371/journal.pone.0133727
Brüggemann, H., Migliorini, L. B., Sales, R. O., Koga, P. C. M., Souza, A. V., Jensen, A., et al. (2018). Comparative genomics of nonoutbreak Pseudomonas aeruginosa strains underlines genome plasticity and geographic relatedness of the global clone ST235. Genome Biol. Evol. 10, 1852–1857. doi: 10.1093/gbe/evy139
Bungau, S., Tit, D. M., Behl, T., Aleya, L., and Zaha, D. C. (2021). Aspects of excessive antibiotic consumption and environmental influences correlated with the occurrence of resistance to antimicrobial agents. Curr Opin Environ Sci Health 19:100224. doi: 10.1016/j.coesh.2020.10.012
Cabot, G., Ocampo-Sosa, A. A., Domínguez, M. A., Gago, J. F., Juan, C., Tubau, F., et al. (2012). Genetic markers of widespread extensively drug-resistant Pseudomonas aeruginosa high-risk clones. Antimicrob. Agents Chemother. 56, 6349–6357. doi: 10.1128/AAC.01388-12
Cai, G., and Wang, Y. (2022). Molecular patterns of clinically important fluoroquinolone resistance in multidrug-resistant Klebsiella pneumoniae isolates during nosocomial outbreaks in Shanghai, PR China. J. Med. Microbiol. 71. doi: 10.1099/jmm.0.001583
Cambray, G., Sánchez-Alberola, N., Campoy, S., Guerin, É., Da Re, S., González-Zorn, B., et al. (2011). Prevalence of SOS-mediated control of integron integrase expression as an adaptive trait of chromosomal and mobile integrons. Mob. DNA 2:6. doi: 10.1186/1759-8753-2-6
Carman, R., Genheimer, C., Rafii, F., Park, M., Hiltonsmith, M., and Lyerly, D. (2009). Diversity of moxifloxacin resistance during a nosocomial outbreak of a predominantly ribotype ARU 027 Clostridium difficile diarrhea. Anaerobe 15, 244–248. doi: 10.1016/j.anaerobe.2009.09.009
Carrilero, L., Dunn, S. J., Moran, R. A., McNally, A., and Brockhurst, M. A. (2023). Evolutionary responses to acquiring a multidrug resistance plasmid are dominated by metabolic functions across diverse Escherichia coli lineages. mSystems 8:e0071322. doi: 10.1128/msystems.00713-22
Castillo-Ramírez, S. (2023). Genomic epidemiology of Acinetobacter baumannii goes global. MBio 14:e0252023. doi: 10.1128/mbio.02520-23
Cattoir, V., and Leclercq, R. (2013). Twenty-five years of shared life with vancomycin-resistant enterococci: is it time to divorce? J. Antimicrob. Chemother. 68, 731–742. doi: 10.1093/jac/dks469
Challagundla, L., Luo, X., Tickler, I. A., Didelot, X., Coleman, D. C., Shore, A. C., et al. (2018). Range expansion and the origin of USA300 north American epidemic methicillin-resistant Staphylococcus aureus. MBio 9, e02016–e02017. doi: 10.1128/mBio.02016-17
Chatzidimitriou, M., Kavvada, A., Kavvadas, D., Kyriazidi, M. A., Eleftheriadis, K., Varlamis, S., et al. (2024). Carbapenem-resistant Klebsiella pneumoniae in the Balkans: clonal distribution and associated resistance determinants. Acta Microbiol. Immunol. Hung. 71, 10–24. doi: 10.1556/030.2024.02230
Chen, B., Li, S., Lin, S., Huang, M., and Dong, H. (2022). The association between antibiotics and community-associated Staphylococcus aureus colonization in the United States population: analysis of the National Health and nutrition examination survey (NHANES). Medicine (Baltimore) 101:e31702. doi: 10.1097/MD.0000000000031702
Chen, L., Mathema, B., Pitout, J. D., DeLeo, F. R., and Kreiswirth, B. N. (2014). Epidemic Klebsiella pneumoniae ST258 is a hybrid strain. MBio 5, e01355–e01314. doi: 10.1128/mBio.01355-14
Chen, L., Peirano, G., Yen, K., Wang, B., Terlecky, A., and DeVinney, R. (2024). CRISPR-Cas9-mediated IncF plasmid curing in extraintestinal pathogenic Escherichia coli. Microbiol. Spectr. 12:e0369223. doi: 10.1128/spectrum.03692
Clark, N. C., Cooksey, R. C., Hill, B. C., Swenson, J. M., and Tenover, F. C. (1993). Characterization of glycopeptide-resistant enterococci from U.S. hospitals. Antimicrob. Agents Chemother. 37, 2311–2317. doi: 10.1128/AAC.37.11.2311
Colpan, A., Johnston, B., Porter, S., Clabots, C., Anway, R., Thao, L., et al. (2013). Escherichia coli sequence type 131 (ST131) subclone H30 as an emergent multidrug-resistant pathogen among US veterans. Clin. Infect. Dis. 57, 1256–1265. doi: 10.1093/cid/cit503
Coluzzi, C., Guillemet, M., Mazzamurro, F., Touchon, M., Godfroid, M., Achaz, G., et al. (2023). Chance favors the prepared genomes: horizontal transfer shapes the emergence of antibiotic resistance mutations in core genes. Mol. Biol. Evol. 40:msad217. doi: 10.1093/molbev/msad217
Conceição, T., Aires-de-Sousa, M., Fuzi, M., Tóth, A., Pászti, J., Ungvári, E., et al. (2007). Replacement of methicillin-resistant Staphylococcus aureus clones in Hungary over time: a 10-year surveillance study. Clin. Microbiol. Infect. 13, 971–979. doi: 10.1111/j.1469-0691.2007.01794.x
Cong, Y., Yang, S., and Rao, X. (2019). Vancomycin resistant Staphylococcus aureus infections: a review of case updating and clinical features. J. Adv. Res. 21, 169–176. doi: 10.1016/j.jare.2019.10.005
Coombs, G. W., Daley, D. A., Shoby, P., and Mowlaboccus, S.Australian Group on Antimicrobial Resistance (AGAR) (2023). Australian Staphylococcus aureus surveillance outcome program (ASSOP) bloodstream infection annual report 2022. Commun. Dis. Intell. 47:47. doi: 10.33321/cdi.2023.47.67
Corcione, S., Shbaklo, N., Vicentini, C., Corradi, A., Scabini, S., Pinna, S. M., et al. (2021). Impact of an empiric antimicrobial therapy manual on antimicrobial usage and multidrug resistant organism trends in a large Italian teaching hospital. Infect. Prev. Pract. 4:100187. doi: 10.1016/j.infpip.2021.100187
Couderc, C., Jolivet, S., Thiébaut, A. C., Ligier, C., Remy, L., Alvarez, A. S., et al. (2014). Fluoroquinolone use is a risk factor for methicillin-resistant Staphylococcus aureus acquisition in long-term care facilities: a nested case-case-control study. Clin. Infect. Dis. 59, 206–215. doi: 10.1093/cid/ciu236
Couture, S., Frenette, C., Schiller, I., Alfaro, R., Dendukuri, N., Thirion, D., et al. (2024). The changing epidemiology of Clostridioides difficile infection and the NAP1/027 strain in two Quebec hospitals: a 17-year time-series study. Antimicrob Steward Healthc. Epidemiol. 4:e99. doi: 10.1017/ash.2024.95
Couvé-Deacon, E., Jové, T., Afouda, P., Barraud, O., Tilloy, V., Scaon, E., et al. (2019). Class 1 integrons in Acinetobacter baumannii: a weak expression of gene cassettes to counterbalance the lack of LexA-driven integrase repression. Int. J. Antimicrob. Agents 53, 491–499. doi: 10.1016/j.ijantimicag.2018.11.012
Cummins, E. A., Snaith, A. E., McNally, A., and Hall, R. J. (2021). The role of potentiating mutations in the evolution of pandemic Escherichia coli clones. Eur. J. Clin. Microbiol. Infect. Dis. 17. doi: 10.1007/s10096-021-04359-3
Cury, J., Jové, T., Touchon, M., Néron, B., and Rocha, E. P. (2016). Identification and analysis of integrons and cassette arrays in bacterial genomes. Nucleic Acids Res. 44, 4539–4550. doi: 10.1093/nar/gkw319
Damjanova, I., Toth, Á., Paszti, J., Bauernfeind, A., and Fuzi, M. (2006). Nationwide spread of clonally related CTX-M-15-producing multidrug-resistant Klebsiella pneumoniae strains in Hungary. Eur. J. Clin. Microbiol. Infect. Dis. 25, 275–278. doi: 10.1007/s10096-006-0120-4
Damjanova, I., Toth, A., Paszti, J., Hajbel-Vekony, G., Jakab, M., Berta, J., et al. (2008). Expansion and countrywide dissemination of ST11, ST15 and ST147 ciprofloxacin-resistant CTX-M-15-type β-lactamase-producing Klebsiella pneumoniae epidemic clones in Hungary in 2005–the new‘MRSAs?’. J. Antimicrob. Chemother. 62, 978–985. doi: 10.1093/jac/dkn287
Dauwalder, O., Lina, G., Durand, G., Bes, M., Meugnier, H., Jarlier, V., et al. (2008). Epidemiology of invasive methicillin-resistant Staphylococcus aureus clones collected in France in 2006 and 2007. J. Clin. Microbiol. 46, 3454–3458. doi: 10.1128/JCM.01050-08
De Lastours, V., Maugy, E., Mathy, V., Chau, F., Rossi, B., Guérin, F., et al. (2017). Ecological impact of ciprofloxacin on commensal enterococci in healthy volunteers. J. Antimicrob. Chemother. 72, 1574–1580. doi: 10.1093/jac/dkx043
De Man, T. J. B., Lutgring, J. D., Lonsway, D. R., Anderson, K. F., Kiehlbauch, J. A., Chen, L., et al. (2016). Genomic analysis of a pan-resistant isolate of Klebsiella pneumoniae, United States 2016. MBio 9, e00418–e00440. doi: 10.1128/mBio.00440-18
De Oliveira, T. L. R., Cavalcante, F. S., Chamon, R. C., Ferreira, R. B. R., and Dos Santos, K. R. N. (2019). Genetic mutations in the quinolone resistance-determining region are related to changes in the epidemiological profile of methicillin-resistant Staphylococcus aureus isolates. J. Glob. Antimicrob. Resist. 19, 236–240. doi: 10.1016/j.jgar.2019.05.026
Del Barrio-Tofiño, E., López-Causapé, C., Cabot, G., Rivera, A., Benito, N., Segura, C., et al. (2017). Genomics and susceptibility profiles of extensively drug-resistant Pseudomonas aeruginosa isolates from Spain. Antimicrob. Agents Chemother. 61, e01589–e01517. doi: 10.1128/AAC.01589-17
Derakhshan, S., Najar Peerayeh, S., and Bakhshi, B. (2016). Association between presence of virulence genes and antibiotic resistance in clinical Klebsiella pneumoniae isolates. Lab. Med. 47, 306–311. doi: 10.1093/labmed/lmw030
Dingle, K. E., Didelot, X., Quan, T. P., Eyre, D. W., Stoesser, N., Golubchik, T., et al. (2017). Effects of control interventions on Clostridium difficile infection in England: an observational study. Lancet Infect. Dis. 17, 411–421. doi: 10.1016/S1473-3099(16)30514-X
Dingle, K. E., Freeman, J., Didelot, X., Quan, T. P., Eyre, D. W., Swann, J., et al. (2023). Penicillin binding protein substitutions cooccur with fluoroquinolone resistance in epidemic lineages of multidrug-resistant Clostridioides difficile. MBio 14:e0024323. doi: 10.1128/mbio.00243-23
Djahanschiri, B., Di Venanzio, G., Distel, J. S., Breisch, J., Dieckmann, M. A., Goesmann, A., et al. (2022). Evolutionarily stable gene clusters shed light on the common grounds of pathogenicity in the Acinetobacter calcoaceticus-baumannii complex. PLoS Genet. 18:e1010020. doi: 10.1371/journal.pgen.1010020
Dridi, L., Tankovic, J., Burghoffer, B., Barbut, F., and Petit, J. C. (2002). GyrA and gyrB mutations are implicated in cross-resistance to ciprofloxacin and moxifloxacin in Clostridium difficile. Antimicrob. Agents Chemother. 46, 3418–3421. doi: 10.1128/AAC.46.11.3418-3421.2002
Duan, Z., Li, X., Li, S., Zhou, H., Hu, L., Xia, H., et al. (2024). Nosocomial surveillance of multidrug-resistant Acinetobacter baumannii: a genomic epidemiological study. Microbiol Spectr 12:e0220723. doi: 10.1128/spectrum.02207-23
Dunn, S., Carrilero, L., Brockhurst, M., and McNally, A. (2021). Limited and strain-specific transcriptional and growth responses to acquisition of a multidrug resistance plasmid in genetically diverse Escherichia coli lineages. mSystems 6, e00021–e00083. doi: 10.1128/mSystems.00083-21
Duprilot, M., Baron, A., Blanquart, F., Dion, S., Pouget, C., Lettéron, P., et al. (2020). Success of Escherichia coli O25b:H4 sequence type 131 clade C associated with a decrease in virulence. Infect. Immun. 88, e00520–e00576. doi: 10.1128/IAI.00576-20
Elg, C. A., Mack, E., Rolfsmeier, M., McLean, T. C., Kosterlitz, O., Soderling, E., et al. (2024). Evolution of a plasmid regulatory circuit ameliorates plasmid fitness cost. bioRxiv. doi: 10.1101/2024.02.05.579024
Espadinha, D., Faria, N. A., Miragaia, M., Lito, L. M., Melo-Cristino, J., de Lencastre, H., et al. (2013). Extensive dissemination of methicillin-resistant Staphylococcus aureus (MRSA) between the hospital and the community in a country with a high prevalence of nosocomial MRSA. PLoS One 8:e59960. doi: 10.1371/journal.pone.0059960
Fernández-Vázquez, J. L., Hernández-González, I. L., Castillo-Ramírez, S., Jarillo-Quijada, M. D., Gayosso-Vázquez, C., Mateo-Estrada, V. E., et al. (2023). Pandrug-resistant Acinetobacter baumannii from different clones and regions in Mexico have a similar plasmid carrying the Bla(OXA-72) gene. Front. Cell. Infect. Microbiol. 13:1278819. doi: 10.3389/fcimb.2023.1278819
Fonseca, É. L., and Vicente, A. C. (2022). Integron functionality and genome innovation: an update on the subtle and smart strategy of integrase and gene cassette expression regulation. Microorganisms 10:224. doi: 10.3390/microorganisms10020224
Forde, C., Stierman, B., Ramon-Pardo, P., Dos Santos, T., and Singh, N. (2017). Carbapenem-resistant Klebsiella pneumoniae in Barbados: driving change in practice at the national level. PLoS One 12:e0176779. doi: 10.1371/journal.pone.0176779
Forstner, C., Diab-Elschahawi, M., Kivaranovic, D., Graninger, W., Mitteregger, D., Macher, M., et al. (2015). Non-linear significant relationship between use of glycopeptides and isolation of vancomycin-resistant Enterococcus species in a university hospital setting. Antimicrob. Resist. Infect. Control 4:25. doi: 10.1186/s13756-015-0064-5
Frade, M. F. F., Dias, M., Teixeira, C. S. C., and Palma, M. S. A. (2014). Environmental contamination by fluoroquinolones. Braz. J. Pharm. Sci. 50, 41–54. doi: 10.1590/S1984-82502011000100004
Freeman, J., Vernon, J., Morris, K., Nicholson, S., Todhunter, S., Longshaw, C., et al. (2015). Pan-european longitudinal surveillance of antibiotic resistance among prevalent Clostridium difficile ribotypes. Clin. Microbiol. Infect. 21, 248.e9–248.e16. doi: 10.1016/j.cmi.2014.09.017
Freitas, A. R., Tedim, A. P., Francia, M. V., Jensen, L. B., Novais, C., Peixe, L., et al. (2016). Multilevel population genetic analysis of vanA and vanB Enterococcus faecium causing nosocomial outbreaks in 27 countries (1986-2012). J. Antimicrob. Chemother. 71, 3351–3366. doi: 10.1093/jac/dkw312
Fuzi, M. (2016). Dissimilar fitness associated with resistance to fluoroquinolones influences clonal dynamics of various multiresistant bacteria. Front. Microbiol. 7:1017. doi: 10.3389/fmicb.2016.01017
Fuzi, M., Rodriguez Baño, J., and Toth, A. (2020). Global evolution of pathogenic bacteria with extensive use of fluoroquinolone agents. Front. Microbiol. 11:271. doi: 10.3389/fmicb.2020.00271
Fuzi, M., and Sokurenko, E. (2023). Commensal fitness advantage may contribute to the global dissemination of multidrug-resistant lineages of bacteria. The case of uropathogenic E. coli. Pathogens 12:1150. doi: 10.3390/pathogens12091150
Fuzi, M., Szabo, D., and Csercsik, R. (2017). Double-serine fluoroquinolone resistance mutations advance major international clones and lineages of various multi-drug resistant bacteria. Front. Microbiol. 8:2261. doi: 10.3389/fmicb.2017.02261
Giraud, E., Cloeckaert, A., Baucheron, S., Mouline, C., and Chaslus-Dancla, E. (2003). Fitness cost of fluoroquinolone resistance in Salmonella enterica serovar Typhimurium. J. Med. Microbiol. 52, 697–703. doi: 10.1099/jmm.0.05178-0
Glaser, P., Martins-Simões, P., Villain, A., Barbier, M., Tristan, A., Bouchier, C., et al. (2016). Demography and intercontinental spread of the USA300 community-acquired methicillin-resistant Staphylococcus aureus lineage. MBio 7, e02115–e02183. doi: 10.1128/mBio.02183-15
Goorhuis, A., Bakker, D., Corver, J., Debast, S. B., Harmanus, C., Notermans, D. W., et al. (2008). Emergence of Clostridium difficile infection due to a new hypervirulent strain, polymerase chain reaction ribotype 078. Clin. Infect. Dis. 47, 1162–1170. doi: 10.1086/592257
Goswami, C., Fox, S., Holden, M. T. G., Connor, M., Leanord, A., and Evans, T. J. (2020). Origin, maintenance and spread of antibiotic resistance genes within plasmids and chromosomes of bloodstream isolates of Escherichia coli. Microb Genom 6:e000353. doi: 10.1099/mgen.0.000353
Gregg-Jolly, L. A. (2022). Locking down SOS mutagenesis repression in a dynamic pathogen. J. Bacteriol. 204:e0022022. doi: 10.1128/jb.00220-22
Grundmann, H., Schouls, L. M., Aanensen, D. M., Pluister, G. N., Tami, A., Chlebowicz, M., et al. (2014). The dynamic changes of dominant clones of Staphylococcus aureus causing bloodstream infections in the European region: results of a second structured survey. Euro Surveill. 19:20987. doi: 10.2807/1560-7917.ES2014.19.49.20987
Guh, A. Y., Mu, Y., Winston, L. G., Johnston, H., Olson, D., Farley, M. M., et al. (2020). Trends in U.S. burden of Clostridioides difficile infection and outcomes. N. Engl. J. Med. 382, 1320–1330. doi: 10.1056/NEJMoa1910215
Guillemin, I., Jarlier, V., and Cambau, E. (1998). Correlation between quinolone susceptibility patterns and sequences in the a and B subunits of DNA gyrase in mycobacteria. Antimicrob. Agents Chemother. 42, 2084–2088. doi: 10.1128/AAC.42.8.2084
Hakki, M., Humphries, R. M., Hemarajata, P., Tallman, G. B., Shields, R. K., Mettus, R. T., et al. (2019). Fluoroquinolone prophylaxis selects for meropenem-nonsusceptible Pseudomonas aeruginosa in patients with hematologic malignancies and hematopoietic cell transplant recipients. Clin. Infect. Dis. 68, 2045–2052. doi: 10.1093/cid/ciy825
Hall, J. P. J., Wright, R. C. T., Harrison, E., Muddiman, K. J., Wood, A. J., Paterson, S., et al. (2021). Plasmid fitness costs are caused by specific genetic conflicts enabling resolution by compensatory mutation. PLoS Biol. 19:e3001225. doi: 10.1371/journal.pbio.3001225
Hamidian, M., and Nigro, S. J. (2019). Emergence, molecular mechanisms and global spread of carbapenem-resistant Acinetobacter baumannii. Microb Genom 5:e000306. doi: 10.1099/mgen.0.000306
Hawkey, P. M., and Jones, A. M. J. (2009). The changing epidemiology of resistance. Antimicrob Chemother 64, i3–i10. doi: 10.1093/jac/dkp256
He, M., Miyajima, F., Roberts, P., Ellison, L., Pickard, D. J., Martin, M. J., et al. (2013). Emergence and global spread of epidemic healthcare-associated Clostridium difficile. Nat. Genet. 45, 109–113. doi: 10.1038/ng.2478
Hiramatsu, K., Igarashi, M., Morimoto, Y., Baba, T., Umekita, M., and Akamatsu, Y. (2012). Curing bacteria of antibiotic resistance: reverse antibiotics, a novel class of antibiotics in nature. Int. J. Antimicrob. Agents 39, 478–485. doi: 10.1016/j.ijantimicag.2012.02.007
Holden, M. T., Hsu, L. Y., Kurt, K., Weinert, L. A., Mather, A. E., Harris, S. R., et al. (2013). A genomic portrait of the emergence, evolution, and global spread of a methicillin-resistant Staphylococcus aureus pandemic. Genome Res. 23, 653–664. doi: 10.1101/gr.147710.112
Holden, E. R., and Webber, M. A. (2020). MarA, RamA, and SoxS as mediators of the stress response: survival at a cost. Front. Microbiol. 11:828. doi: 10.3389/fmicb.2020.00828
Hong, J. S., Yoon, E. J., Lee, H., Jeong, S. H., and Lee, K. (2016). Clonal dissemination of Pseudomonas aeruginosa sequence type 235 isolates carrying blaIMP-6 and emergence of blaGES-24 and blaIMP-10 on novel genomic islands PAGI-15 and -16 in South Korea. Antimicrob. Agents Chemother. 60, 7216–7223. doi: 10.1128/AAC.01601-16
Horvath, A., Dobay, O., Kardos, S., Ghidan, A., Toth, A., Paszti, J., et al. (2012). Varying fitness cost associated with resistance to fluoroquinolones governs clonal dynamic of methicillin-resistant Staphylococcus aureus. Eur. J. Clin. Microbiol. Infect. Dis. 31, 2029–2036. doi: 10.1007/s10096-011-1536-z
Hsu, L. Y., Harris, S. R., Chlebowicz, M. A., Lindsay, J. A., Koh, T. H., Krishnan, P., et al. (2015). Evolutionary dynamics of methicillin-resistant Staphylococcus aureus within a healthcare system. Genome Biol. 16:81. doi: 10.1186/s13059-015-0643-z
Hu, Y., Liu, C., Wang, Q., Zeng, Y., Sun, Q., Shu, L., et al. (2021). Emergence and expansion of a carbapenem-resistant Pseudomonas aeruginosa clone are associated with plasmid-borne Bla (KPC-2) and virulence-related genes. mSystems 6, e00121–e00154. doi: 10.1128/mSystems.00154-21
Huseby, D. L., Pietsch, F., Brandi, S. G., Garoff, L., Tegehall, A., and Hughes, D. (2017). Mutation supply and relative fitness shape the genotypes of ciprofloxacin-resistant Escherichia coli. Mol. Biol. Evol. 34, 1029–1039. doi: 10.1093/molbev/msx052
Imwattana, K., Rodríguez, C., Riley, T. V., and Knight, D. R. (2021). A species-wide genetic atlas of antimicrobial resistance in Clostridioides difficile. Microb. Genom. 7:000696. doi: 10.1099/mgen.0.000696
Iranzo, J., Cuesta, J. A., Manrubia, S., Katsnelson, M. I., and Koonin, E. V. (2017). Disentangling the effects of selection and loss bias on gene dynamics. Proc. Natl. Acad. Sci. USA 114, E5616–E5624. doi: 10.1073/pnas.1704925114
Jia, X., Zhu, Y., Jia, P., Li, C., Chu, X., Sun, T., et al. (2024). The key role of iroBCDN-lacking pLVPK-like plasmid in the evolution of the most prevalent hypervirulent carbapenem-resistant ST11-KL64 Klebsiella pneumoniae in China. Drug Resist Update 77:101137. doi: 10.1016/j.drup.2024.101137
Johnson, J. R., Johnston, B., Kuskowski, M. A., Sokurenko, E. V., and Tchesnokova, V. (2015a). Intensity and mechanisms of fuoroquinolone resistance within the H30 and H30Rx subclones of Escherichia coli sequence type 131 compared with other fluoroquinolone resistant E. coli. Antimicrob. Agents Chemother. 59, 4471–4480. doi: 10.1128/AAC.00673-15
Johnson, J. R., Johnston, B. D., Porter, S. B., Clabots, C., Bender, T. L., Thuras, P., et al. (2019). Rapid emergence, subsidence, and molecular detection of Escherichia coli sequence type 1193-fimH64, a new disseminated multidrug-resistant commensal and extraintestinal pathogen. J. Clin. Microbiol. 57, 1664–1718. doi: 10.1128/JCM.01664-18
Johnson, J. R., Porter, S. B., Thuras, P., Johnson, T. J., Price, L. B., Tchesnokova, V., et al. (2015b). Greater ciprofloxacin tolerance as a possible selectable phenotype underlying the pandemic spread of the H30 subclone of Escherichia coli sequence type 131. Antimicrob. Agents Chemother. 59, 7132–7135. doi: 10.1128/AAC.01687-15
Jones, E. C., and Uphoff, S. (2021). Single-molecule imaging of LexA degradation in Escherichia coli elucidates regulatory mechanisms and heterogeneity of the SOS response. Nat. Microbiol. 6, 981–990. doi: 10.1038/s41564-021-00930-y
Junaidi, N. S. S. A., Shakrin, N. N. S. M., Desa, M. N. M., and Yunus, W. M. Z. W. (2023). Dissemination pattern of hospital-acquired methicillin-resistant Staphylococcus aureus and community-acquired MRSA isolates from Malaysian hospitals: a review from a molecular perspective. Malays J. Med. Sci. 30, 26–41. doi: 10.21315/mjms2023.30.2.3
Jung, H., Pitout, J. D. D., Matsumura, Y., Strydom, K. A., Kingsburgh, C., Ehlers, M. M., et al. (2024). Genomic epidemiology and molecular characteristics of Bla(NDM-1)-positive carbapenem-resistant Pseudomonas aeruginosa belonging to international high-risk clone ST773 in the Gauteng region, South Africa. Eur. J. Clin. Microbiol. Infect. Dis. 43, 627–640. doi: 10.1007/s10096-024-04763-5
Kaito, C., Saito, Y., Ikuo, M., Omae, Y., Mao, H., Nagano, G., et al. (2013). Mobile genetic element SCCmec-encoded psm-mec RNA suppresses translation of agrA and attenuates MRSA virulence. PLoS Pathog. 9:e1003269. doi: 10.1371/journal.ppat.1003269
Karah, N., Sundsfjord, A., Towner, K., and Samuelsen, Ø. (2012). Insights into the global molecular epidemiology of carbapenem non-susceptible clones of Acinetobacter baumannii. Drug Resist. Updat. 15, 237–247. doi: 10.1016/j.drup.2012.06.001
Kazakova, S. V., Baggs, J., McDonald, L. C., Yi, S. H., Hatfield, K. M., Guh, A., et al. (2020). Association between antibiotic use and hospital-onset Clostridioides difficile infection in US acute care hospitals, 2006–2012: an ecologic analysis. Clin. Infect. Dis. 70, 11–18. doi: 10.1093/cid/ciz169
Kikuchi, Y., Yoshida, M., Kuwae, A., Asami, Y., Inahashi, Y., and Abe, A. (2024). Correlation between the spread of IMP-producing bacteria and the promoter strength of blaIMP gene. J. Antibiot. (Tokyo) 77, 315–323. doi: 10.1038/s41429-024-00715-5
Kiyaga, S., Kyany'a, C., Muraya, A. W., Smith, H. J., Mills, E. G., Kibet, C., et al. (2022). Genetic diversity, distribution, and genomic characterization of antibiotic resistance and virulence of clinical Pseudomonas aeruginosa strains in Kenya. Front. Microbiol. 13:835403. doi: 10.3389/fmicb.2022.835403
Kloos, J., Gama, J. A., Hegstad, J., Samuelsen, Ø., and Johnsen, P. J. (2021). Piggybacking on niche adaptation improves the maintenance of multidrug-resistance plasmids. Mol. Biol. Evol. 38, 3188–3201. doi: 10.1093/molbev/msab091
Knight, G. M., Budd, E. L., Whitney, L., Thornley, A., Al-Ghusein, H., Planche, T., et al. (2012). Shift in dominant hospital-associated methicillin-resistant Staphylococcus aureus (HA-MRSA) clones over time. J. Antimicrob. Chemother. 67, 2514–2522. doi: 10.1093/jac/dks245
Kocsis, B., Toth, A., Gulyas, D., Ligeti, B., Katona, K., Rokusz, L., et al. (2019). Acquired qnrVC1 and blaNDM-1 resistance markers in an international high-risk Pseudomonas aeruginosa ST773 clone. J. Med. Microbiol. 68, 336–338. doi: 10.1099/jmm.0.000927
Komp Lindgren, P., Marcusson, L. L., Sandvang, D., Frimodt-Møller, N., and Hughes, D. (2005). Biological cost of single and multiple norfloxacin resistance mutations in Escherichia coli implicated in urinary tract infections. Antimicrob. Agents Chemother. 49, 2343–2351. doi: 10.1128/AAC.49.6.2343-2351.2005
Kos, V. N., Déraspe, M., McLaughlin, R. E., Whiteaker, J. D., Roy, P. H., Alm, R. A., et al. (2015). The resistome of Pseudomonas aeruginosa in relationship to phenotypic susceptibility. Antimicrob. Agents Chemother. 59, 427–436. doi: 10.1128/AAC.03954-14
Kozuch, B. C., Shaffer, M. G., and Culyba, M. J. (2020). The parameter-fitness landscape of lexA autoregulation in Escherichia coli. mSphere 5, e00718–e00720. doi: 10.1128/mSphere.00718-20
Kryzhevskyi, V., Strokous, V., Lifshyts, Y., Rybianets, Y., Oberniak, A., Krikunov, A., et al. (2023). Case report: azithromycin-meropenem combination therapy as a low-cost approach to combat PDR gram-negative infections of war wounds in Ukraine. Front. Med. (Lausanne) 10:1264492. doi: 10.3389/fmed.2023.1264492
Kumkar, S. N., Kamble, E. E., Chavan, N. S., Dhotre, D. P., and Pardesi, K. R. (2022). Diversity of resistant determinants, virulence factors, and mobile genetic elements in Acinetobacter baumannii from India: a comprehensive in silico genome analysis. Front. Cell. Infect. Microbiol. 12:997897. doi: 10.3389/fcimb.2022.997897
Kuwata, Y., Tanimoto, S., Sawabe, E., Shima, M., Takahashi, Y., Ushizawa, H., et al. (2015). Molecular epidemiology and antimicrobial susceptibility of Clostridium difficile isolated from a university teaching hospital in Japan. Eur. J. Clin. Microbiol. Infect. Dis. 34, 763–772. doi: 10.1007/s10096-014-2290-9
Lawal, O. U., Ayobami, O., Abouelfetouh, A., Mourabit, N., Kaba, M., Egyir, B., et al. (2022). A 6-year update on the diversity of methicillin-resistant Staphylococcus aureus clones in Africa: a systematic review. Front. Microbiol. 13:860436. doi: 10.3389/fmicb.2022.860436
Lawes, T., López-Lozano, J. M., Nebot, C., Macartney, G., Subbarao-Sharma, R., Dare, C. R., et al. (2015). Turning the tide or riding the waves? Impacts of antibiotic stewardship and infection control on MRSA strain dynamics in a Scottish region over 16 years: non-linear time series analysis. Br. Med. J. Open 5:e006596. doi: 10.1136/bmjopen-2014-006596
Leavis, H. L., Willems, R. J. L., Top, J., Marc, J., and Bonten, M. (2006). Highlevel ciprofloxacin resistance from point mutations in gyrA and parC confined to global hospital-adapted clonal lineage CC17 of Enterococcus faecium. J. Clin. Microbiol. 44, 1059–1064. doi: 10.1128/JCM.44.3.1059-1064.2006
LeBlanc, L., Pépin, J., Toulouse, K., Ouellette, M. F., Coulombe, M. A., Corriveau, M. P., et al. (2006). Fluoroquinolones and risk for methicillin-resistant Staphylococcus aureus, Canada. Emerg. Infect. Dis. 12, 1398–1405. doi: 10.3201/eid1209.060397
Lee, J. H., Kim, N. H., Jang, K. M., Jin, H., Shin, K., Jeong, B. C., et al. (2023). Prioritization of critical factors for surveillance of the dissemination of antibiotic resistance in Pseudomonas aeruginosa: a systematic review. Int. J. Mol. Sci. 24:15209. doi: 10.3390/ijms242015209
Lee, J. H., Lee, Y., Lee, K., Riley, T. V., and Kim, H. (2014). The changes of PCR ribotype and antimicrobial resistance of Clostridium difficile in a tertiary care hospital over 10 years. J. Med. Microbiol. 63:819823, 819–823. doi: 10.1099/jmm.0.072082-0
Li, D., Reid, C. J., Kudinha, T., Jarocki, V. M., and Djordjevic, S. P. (2020). Genomic analysis of trimethoprim-resistant extraintestinal pathogenic Escherichia coli and recurrent urinary tract infections. Microb Genom 6:mgen000475. doi: 10.1099/mgen.0.000475
Li, D., Wyrsch, E. R., Elankumaran, P., Dolejska, M., Marenda, M. S., Browning, G. F., et al. (2021). Genomic comparisons of Escherichia coli ST131 from Australia. Microb Genom 7:000721. doi: 10.1099/mgen.0.000721
Lim, K. T., Hanifah, Y. A., Mohd Yusof, M. Y., Ito, T., and Thong, K. L. (2013). Comparison of methicillin-resistant Staphylococcus aureus strains isolated in 2003 and 2008 with an emergence of multidrug resistant ST22: SCCmec IV clone in a tertiary hospital, Malaysia. J. Microbiol. Immunol. Infect. 46, 224–233. doi: 10.1016/j.jmii.2013.02.001
Lin, Y. C., Huang, Y. T., Tsai, P. J., Lee, T. F., Lee, N. Y., Liao, C. H., et al. (2011). Antimicrobial susceptibilities and molecular epidemiology of clinical isolates of Clostridium difficile in Taiwan. Antimicrob. Agents Chemother. 55, 1701–1705. doi: 10.1128/AAC.01440-10
Linder, J. A., Huang, E. S., Steinman, M. A., Gonzales, R., and Stafford, R. S. (2005). Fluoroquinolone prescribing in the United States: 1995 to 2002. Am. J. Med. 118, 259–268. doi: 10.1016/j.amjmed.2004.09.015
Liu, C., Monaghan, T., Yadegar, A., Louie, T., and Kao, D. (2023). Insights into the evolving epidemiology of Clostridioides difficile infection and treatment: a global perspective. Antibiotics (Basel). 12:1141. doi: 10.3390/antibiotics12071141
Logan, L. K., Nguyen, D. C., Scaggs Huang, F. A., Qureshi, N. K., Charnot-Katsikas, A., Bartlett, A. H., et al. (2019). A multi-centered case-control study of factors associated with Klebsiella pneumoniae carbapenemase-producing Enterobacteriaceae infections in children and young adults. Pediatr. Infect. Dis. J. 38, 490–495. doi: 10.1097/INF.0000000000002176
Lopatkin, A. J., Bening, S. C., Manson, A. L., Stokes, J. M., Kohanski, M. A., Badran, A. H., et al. (2021). Clinically relevant mutations in core metabolic genes confer antibiotic resistance. Science 371:eaba0862. doi: 10.1126/science.aba0862
López, J. L., Lozano, M. J., Fabre, M. L., and Lagares, A. (2020). Codon usage optimization in the prokaryotic tree of life: how synonymous codons are differentially selected in sequence domains with different expression levels and degrees of conservation. MBio 11, e00720–e00766. doi: 10.1128/mBio.00766-20
López, J. L., Lozano, M. J., Lagares, A., Fabre, M. L., Draghi, W. O., Del Papa, M. F., et al. (2019). Codon usage heterogeneity in the multipartite prokaryote genome: selection-based coding bias associated with gene location, expression level, and ancestry. MBio 10, e00505–e00519. doi: 10.1128/mBio.00505-19
López, M., Tenorio, C., Del Campo, R., Zarazaga, M., and Torres, C. (2011). Characterization of the mechanisms of fluoroquinolone resistance in vancomycin-resistant enterococci of different origins. J. Chemother. 23, 87–91. doi: 10.1179/joc.2011.23.2.87
Luo, N., Pereira, S., Sahin, O., Lin, J., Huang, S., Michel, L., et al. (2005). Enhanced in vivo fitness of fluoroquinolone-resistant Campylobacter jejuni in the absence of antibiotic selection pressure. Proc. Natl. Acad. Sci. USA 102, 541–546. doi: 10.1073/pnas.0408966102
Ma, X. X., Ito, T., Chongtrakool, P., and Hiramatsu, K. (2006). Predominance of clones carrying Panton-valentine leukocidin genes among methicillin-resistant Staphylococcus aureus strains isolated in Japanese hospitals from 1979 to 1985. J. Clin. Microbiol. 44, 4515–4527. doi: 10.1128/JCM.00985-06
Machuca, J., Briales, A., Labrador, G., Díaz-de-Alba, P., López-Rojas, R., Docobo-Pérez, F., et al. (2014). Interplay between plasmid mediated and chromosomalmediated fluoroquinolone resistance and bacterial fitness in Escherichia coli. J. Antimicrob. Chemother. 69:320315, 3203–3215. doi: 10.1093/jac/dku308
Marcusson, L. L., Frimodt-Møller, N., and Hughes, D. (2009). Interplay in the selection of fluoroquinolone resistance and bacterial fitness. PLoS Pathog. 5:e1000541. doi: 10.1371/journal.ppat.1000541
Martínez, J. L., and Baquero, F. (2002). Interactions among strategies associated with bacterial infection: pathogenicity, epidemicity, and antibiotic resistance. Clin. Microbiol. Rev. 15, 647–679. doi: 10.1128/CMR.15.4.647-679.2002
Maslova, O., Mindlin, S., Beletsky, A., Mardanov, A., and Petrova, M. (2022). Plasmids as key players in Acinetobacter adaptation. Int. J. Mol. Sci. 23:10893. doi: 10.3390/ijms231810893
Mathers, A. J., Stoesser, N., Chai, W., Carroll, J., Barry, K., Cherunvanky, A., et al. (2017). Chromosomal integration of the Klebsiella pneumoniae carbapenemase gene, Bla(KPC), in Klebsiella species is elusive but not rare. Antimicrob. Agents Chemother. 61, e01823–e01816. doi: 10.1128/AAC.01823-16
Matsui, M., Suzuki, M., Suzuki, M., Yatsuyanagi, J., Watahiki, M., Hiraki, Y., et al. (2018). Distribution and molecular characterization of Acinetobacter baumannii international clone II lineage in Japan. Antimicrob. Agents Chemother. 62, e02190–e02117. doi: 10.1128/AAC.02190-17
Maurin, M., Abergel, C., and Raoult, D. (2001). DNA gyrase-mediated natural resistance to fluoroquinolones in Ehrlichia spp. Antimicrob. Agents Chemother. 45, 2098–2105. doi: 10.1128/AAC.45.7.2098-2105.2001
McCarthy, H., Rudkin, J. K., Black, N. S., Gallagher, L., O’Neill, E., and O’Gara, J. P. (2015). Methicillin resistance and the biofilm phenotype in Staphylococcus aureus. Front. Cell. Infect. Microbiol. 5:1. doi: 10.3389/fcimb.2015.00001
McDonald, L. C., Killgore, G. E., Thompson, A., Owens, R. C. Jr., Kazakova, S. V., Sambol, S. P., et al. (2005). An epidemic, toxin gene-variant strain of Clostridium difficile. N. Engl. J. Med. 353, 2433–2441. doi: 10.1056/NEJMoa051590
McFarland, L. V., Ozen, M., Dinleyici, E. C., and Goh, S. (2016). Comparison of pediatric and adult antibiotic-associated diarrhea and Clostridium difficile infections. World J. Gastroenterol. 22, 3078–3104. doi: 10.3748/wjg.v22.i11.3078
McGuinness, W. A., Malachowa, N., and DeLeo, F. R. (2017). Vancomycin resistance in Staphylococcus aureus. Yale J. Biol. Med. 90, 269–281
Melnyk, A. H., Wong, A., and Kassen, R. (2015). The fitness costs of antibiotic resistance mutations. Evol. Appl. 8, 273–283. doi: 10.1111/eva.12196
Modi, R. I., and Adams, J. (1991). Coevolution in bacterial-plasmid populations. Evolution 45, 656–667. doi: 10.1111/j.1558-5646.1991.tb04336.x
Morimoto, Y., Aiba, Y., Miyanaga, K., Hishinuma, T., Cui, L., Baba, T., et al. (2023). CID12261165, a flavonoid compound as antibacterial agents against quinolone-resistant Staphylococcus aureus. Sci. Rep. 13:1725. doi: 10.1038/s41598-023-28859-8
Morimoto, Y., Baba, T., Sasaki, T., and Hiramatsu, K. (2015). Apigenin as an anti-quinolone-resistance antibiotic. Int. J. Antimicrob. Agents 46, 666–673. doi: 10.1016/j.ijantimicag.2015.09.006
Nicolas-Chanoine, M. H., Bertrand, X., and Madec, J. Y. (2014). Escherichia coli ST131, an intriguing clonal group. Clin. Microbiol. Rev. 27, 543–574. doi: 10.1128/CMR.00125-13
Nicolas-Chanoine, M. H., Blanco, J., Leflon-Guibout, V., Demarty, R., Alonso, M. P., Caniça, M. M., et al. (2008). Intercontinental emergence of Escherichia coli clone O25:H4-ST131 producing CTX-M-15. J. Antimicrob. Chemother. 61, 273–281. doi: 10.1093/jac/dkm464
Nodari, C. S., Cayô, R., Streling, A. P., Lei, F., Wille, J., Almeida, M. S., et al. (2020). Genomic analysis of carbapenem-resistant Acinetobacter baumannii isolates belonging to major endemic clones in South America. Front. Microbiol. 11:584603. doi: 10.3389/fmicb.2020.584603
Noel, H. R., Petrey, J. R., and Palmer, L. D. (2022). Mobile genetic elements in Acinetobacter antibiotic-resistance acquisition and dissemination. Ann. N. Y. Acad. Sci. 1518, 166–182. doi: 10.1111/nyas.14918
Norton, M. D., Spilkia, A. J., and Godoy, V. G. (2013). Antibiotic resistance acquired through a DNA damage-inducible response in Acinetobacter baumannii. J. Bacteriol. 195, 1335–1345. doi: 10.1128/JB.02176-12
Nübel, U., Strommenger, B., Layer, F., and Witte, W. (2011). From types to trees: reconstructing the spatial spread of Staphylococcus aureus based on DNA variation. Int. J. Med. Microbiol. 301, 614–618. doi: 10.1016/j.ijmm.2011.09.007
Núñez-Samudio, V., Pimentel-Peralta, G., Herrera, M., Pecchio, M., Quintero, J., and Landires, I. (2022). Molecular enetic epidemiology of an emerging antimicrobial-resistant Klebsiella pneumoniae clone (ST307) obtained from clinical isolates in Central Panama. Antibiotics (Basel) 11:1817. doi: 10.3390/antibiotics11121817
Ogunlana, L., Kaur, D., Shaw, L. P., Jangir, P., Walsh, T., Uphoff, S., et al. (2023). Regulatory fine-tuning of mcr-1 increases bacterial fitness and stabilises antibiotic resistance in agricultural settings. ISME J. 17, 2058–2069. doi: 10.1038/s41396-023-01509-7
Osonga, F. J., Akgul, A., Miller, R. M., Eshun, G. B., Yazgan, I., Akgul, A., et al. (2019). Antimicrobial activity of a new class of phosphorylated and modified flavonoids. ACS Omega 4, 12865–12871. doi: 10.1021/acsomega.9b00077
Palkovicova, J., Sukkar, I., Delafuente, J., Valcek, A., Medvecky, M., Jamborova, I., et al. (2022). Fitness effects of blaCTX-M-15-harbouring F2:A1:B- plasmids on their native Escherichia coli ST131 H30Rx hosts. J. Antimicrob. Chemother. 77, 2960–2963. doi: 10.1093/jac/dkac250
Papagiannitsis, C. C., Verra, A., Galani, V., Xitsas, S., Bitar, I., Hrabak, J., et al. (2020). Unravelling the features of success of VIM-producing ST111 and ST235 Pseudomonas aeruginosa in a Greek hospital. Microorganisms 8:1884. doi: 10.3390/microorganisms8121884
Park, M., and Rafii, F. (2017). Exposure to beta-lactams results in the alteration of penicillin-binding proteins in Clostridium perfringens. Anaerobe 45, 78–85. doi: 10.1016/j.anaerobe.2017.02.004
Park, S. M., Suh, J. W., Ju, Y. K., Kim, J. Y., Kim, S. B., Sohn, J. W., et al. (2023). Molecular and virulence characteristics of carbapenem-resistant Acinetobacter baumannii isolates: a prospective cohort study. Sci. Rep. 13:19536. doi: 10.1038/s41598-023-46985-1
Park, D. J., Yu, J. K., Park, K. G., and Park, Y. J. (2015). Genotypes of ciprofloxacin-resistant Klebsiella pneumoniae in Korea and their characteristics according to the genetic lineages. Microb. Drug Resist. 21, 622–630. doi: 10.1089/mdr.2015.0001
Peirano, G., Chen, L., Kreiswirth, B. N., and Pitout, J. D. D. (2020). Emerging antimicrobial-resistant high-risk Klebsiella pneumoniae clones ST307 and ST147. Antimicrob. Agents Chemother. 64, e01148–e01120. doi: 10.1128/AAC.01148-20
Peirano, G., and Pitout, J. D. D. (2025). Rapidly spreading Enterobacterales with OXA-48-like carbapenemases. J. Clin. Microbiol. 63:e0151524. doi: 10.1128/jcm.01515-24
Peña, C., Cabot, G., Gómez-Zorrilla, S., Zamorano, L., Ocampo-Sosa, A., Murillas, J., et al. (2015). Influence of virulence genotype and resistance profile in the mortality of Pseudomonas aeruginosa bloodstream infections. Clin. Infect. Dis. 60, 539–548. doi: 10.1093/cid/ciu866
Pepin, J., Valiquette, L., Alary, M. E., Villemure, P., Pelletier, A., Forget, K., et al. (2004). Clostridium difficile-associated diarrhea in a region of Quebec from 1991 to 2003: a changing pattern of disease severity. CMAI 171, 466–472. doi: 10.1503/cmaj.1041104
Pérez, A., Gato, E., Pérez-Llarena, J., Fernández-Cuenca, F., Gude, M. J., Oviaño, M., et al. (2019). High incidence of MDR and XDR Pseudomonas aeruginosa isolates obtained from patients with ventilator-associated pneumonia in Greece, Italy and Spain as part of the MagicBullet clinical trial. J. Antimicrob. Chemother. 74, 1244–1252. doi: 10.1093/jac/dkz030
Pitout, J. D. D., and Finn, T. J. (2020). The evolutionary puzzle of Escherichia coli ST131. Infect. Genet. Evol. 81:104265. doi: 10.1016/j.meegid.2020.104265
Pitout, J. D. D., Peirano, G., Chen, L., DeVinney, R., and Matsumura, Y. (2022). Escherichia coli ST1193: following in the footsteps of E. coli ST131. Antimicrob. Agents Chemother. 66:e0051122. doi: 10.1128/aac.00511-22
Pitout, J. D., Peirano, G., and DeVinney, R. (2023). The contributions of multidrug resistant clones to the success of pandemic extra-intestinal pathogenic Escherichia coli. Expert Rev. Anti-Infect. Ther. 21, 343–353. doi: 10.1080/14787210.2023.2184348
Pitout, J. D. D., Peirano, G., Matsumura, Y., DeVinney, R., and Chen, L. (2024). Escherichia coli sequence type 410 with carbapenemases: a paradigm shift within E. coli toward multidrug resistance. Antimicrob. Agents Chemother. 68:e0133923. doi: 10.1128/aac.01339-23
Planet, P. J. (2017). Life after USA300: the rise and fall of a superbug. J. Infect. Dis. 215, S71–S77. doi: 10.1093/infdis/jiw444
Plankaova, A., Brajerova, M., Capek, V., Balikova Novotna, G., Kinross, P., Skalova, J., et al. (2023). Clostridioides difficile infections were predominantly driven by fluoroquinolone-resistant Clostridioides difficile ribotypes 176 and 001 in Slovakia in 2018-2019. Int. J. Antimicrob. Agents 62:106824. doi: 10.1016/j.ijantimicag.2023.106824
Pope, C. F., Gillespie, S. H., Pratten, J. R., and McHugh, T. D. (2008). Fluoroquinolone-resistant mutants of Burkholderia cepacia. Antimicrob. Agents Chemother. 52, 1201–1203. doi: 10.1128/AAC.00799-07
Price, L. B., Johnson, J. R., Aziz, M., Clabots, C., Johnston, B., Tchesnokova, V., et al. (2013). The epidemic of extended-spectrum-beta-lactamase-producing Escherichia coli ST131 is driven by a single highly pathogenic subclone, H30-Rx. MBio 4, e00377–e00313. doi: 10.1128/mBio.00377-13
Projan, S. J. (2007). (Genome) size matters. Antimicrob. Agents Chemother. 51, 1133–1134. doi: 10.1128/AAC.01370-06
Rajer, F., and Sandegren, L. (2022). The role of antibiotic resistance genes in the fitness cost of multiresistance plasmids. MBio 13:e0355221. doi: 10.1128/mbio.03552-21
Recio, R., Viedma, E., González-Bodí, S., Villa, J., Orellana, M. Á., Mancheño-Losa, M., et al. (2021). Clinical and bacterial characteristics of Pseudomonas aeruginosa affecting the outcome of patients with bacteraemic pneumonia. Int. J. Antimicrob. Agents 58:106450. doi: 10.1016/j.ijantimicag.2021.106450
Redmond, S. N., Silva, S. Y., Wilson, B. M., Cadnum, J. L., and Donskey, C. J. (2019). Impact of reduced fluoroquinolone use on Clostridioides difficile infections resulting from the fluoroquinolone-resistant ribotype 027 strain in a veterans affairs medical center. Pathog. Immun. 4, 251–259. doi: 10.20411/pai.v4i2.327
Rizzo, K. R., Yi, S. H., Garcia, E. P., Zahn, M., and Epson, E. (2019). Reduction in Clostridium difficile infection rates following a multifacility prevention initiative in Orange County, California: a controlled interrupted time series evaluation. Infect. Control Hosp. Epidemiol. 40, 872–879. doi: 10.1017/ice.2019.135
Roch, M., Gagetti, P., Davis, J., Ceriana, P., Errecalde, L., Corso, A., et al. (2017). Daptomycin resistance in clinical MRSA strains is associated with a high biological fitness cost. Front. Microbiol. 8:2303. doi: 10.3389/fmicb.2017.02303
Rodrigues, C., Desai, S., Passet, V., Gajjar, D., and Brisse, S. (2022). Genomic evolution of the globally disseminated multidrug-resistant Klebsiella pneumoniae clonal group 147. Microb Genom 8:000737. doi: 10.1099/mgen.0.000737
Rodrigues, C., Lanza, V. F., Peixe, L., Coque, T. M., and Novais, Â. (2023). Phylogenomics of globally spread clonal groups 14 and 15 of Klebsiella pneumoniae. Microbiol Spectr 11:e0339522. doi: 10.1128/spectrum.03395-22
Roe, C. C., Vazquez, A. J., Esposito, E. P., Zarrilli, R., and Sahl, J. W. (2019). Diversity, virulence, and antimicrobial resistance in isolates from the newly emerging Klebsiella pneumoniae ST101 lineage. Front. Microbiol. 10:542. doi: 10.3389/fmicb.2019.00542
Roer, L., Overballe-Petersen, S., Hansen, F., Schønning, K., Wang, M., Røder, B. L., et al. (2018). Escherichia coli sequence type 410 is causing new international high-risk clones. mSphere 3, e00337–e00318. doi: 10.1128/mSphere.00337-18
San Millan, A., and MacLean, R. C. (2017). Fitness costs of plasmids: a limit to plasmid transmission. Microbiol. Spectr. 5. doi: 10.1128/microbiolspec.MTBP-0016-2017
Sánchez-Diener, I., Zamorano, L., López-Causapé, C., Cabot, G., Mulet, X., Peña, C., et al. (2017). Interplay among resistance profiles, high-risk clones, and virulence in the Caenorhabditis elegans, Pseudomonas aeruginosa infection model. Antimicrob. Agents Chemother. 61, e01586–e01517. doi: 10.1128/AAC.01586-17
Sandegren, L., Linkevicius, M., Lytsy, B., Melhus, Å., and Andersson, D. I. (2012). Transfer of an Escherichia coli ST131 multiresistance cassette has created a Klebsiella pneumoniae-specific plasmid associated with a major nosocomial outbreak. J. Antimicrob. Chemother. 67, 74–83. doi: 10.1093/jac/dkr405
Sarma, J. B., Marshall, B., Cleeve, V., Tate, D., Oswald, T., and Woolfrey, S. (2015). Effects of fluoroquinolone restriction (from 2007 to 2012) on Clostridium difficile infections: interrupted time-series analysis. J. Hosp. Infect. 91, 74–80. doi: 10.1016/j.jhin.2015.05.013
Saxton, K., Baines, S. D., Freeman, J., O'Connor, R., and Wilcox, M. H. (2009). Effects of exposure of Clostridium difficile PCR ribotypes 027 and 001 to fluoroquinolones in a human gut model. Antimicrob. Agents Chemother. 53, 412–420. doi: 10.1128/AAC.00306-08
Sen, P., Kumar Sahu, P., Haldar, R., Kumar Sahu, K., Prasad, P., and Ro, A. (2016). Apigenin naturally occurring flavonoids: occurrence and bioactivity. UK J Pharmac Biosci 4, 56–68. doi: 10.20510/ukjpb/4/i6/134666
Shen, M., Luan, G., Wang, Y., Chang, Y., Zhang, C., Yang, J., et al. (2016). Coexistence of blaOXA-23 with armA in quinolone-resistant Acinetobacter baumannii from a Chinese university hospital. Diagn. Microbiol. Infect. Dis. 84, 230–231. doi: 10.1016/j.diagmicrobio.2015.10.009
Shin, J., and Ko, S. K. (2015). Effect of plasmids harbouring blaCTX-M on the virulence and fitness of Escherichia coli ST131 isolates. Int. J. Antimicrob. Agents 46, 214–218. doi: 10.1016/j.ijantimicag.2015.04.012
Shiriaev, D. I., Sofronova, A. A., Berdnikovich, E. A., Lukianov, D. A., Komarova, E. S., Marina, V. I., et al. (2023). Nybomycin inhibits both types of E. coli DNA gyrase—fluoroquinolone-sensitive and fluoroquinolone-resistant. Antimicrob. Agents Chemother. 95, e00777–e00720. doi: 10.1128/AAC.00777-20
Shu, C., Zhou, J., Yu, H., Fu, W., Shen, J., Liang, L., et al. (2023). Genomic epidemiology and antimicrobial resistance profiles of Clostridioides difficile from multi-hospitals in a city in eastern China. Infect Drug Resist 16, 3379–3388. doi: 10.2147/IDR.S407497
Silva, A., Silva, V., López, M., Rojo-Bezares, B., Carvalho, J. A., Castro, A. P., et al. (2023). Antimicrobial resistance, genetic lineages, and biofilm formation in Pseudomonas aeruginosa isolated from human infections: an emerging one health concern. Antibiotics (Basel) 12:1248. doi: 10.3390/antibiotics12081248
Silveira, M. C., Rocha-de-Souza, C. M., Albano, R. M., de Oliveira Santos, I. C., and Carvalho-Assef, A. P. D. (2020). Exploring the success of Brazilian endemic clone Pseudomonas aeruginosa ST277 and its association with the CRISPR-Cas system type I-C. BMC Genomics 21:255. doi: 10.1186/s12864-020-6650-9
Slimings, C., and Riley, T. V. (2021). Antibiotics and healthcare facility-associated Clostridioides difficile infection: systematic review and meta-analysis 2020 update. J. Antimicrob. Chemother. 76, 1676–1688. doi: 10.1093/jac/dkab091
Solomon, K., Fanning, S., McDermott, S., Murray, S., Scott, L., Martin, A., et al. (2011). PCR ribotype prevalence and molecular basis of macrolide-lincosamide-streptogramin B (MLSB) and fluoroquinolone resistance in Irish clinical Clostridium difficile isolates. J. Antimicrob. Chemother. 66, 1976–1982. doi: 10.1093/jac/dkr275
Spigaglia, P. (2016). Recent advances in the understanding of antibiotic resistance in Clostridium difficile infection. Ther. Adv. Infect. Dis. 3, 23–42. doi: 10.1177/2049936115622891
Spigaglia, P., Barbanti, F., Dionisi, A., and Mastrantonio, P. (2010). Clostridium difficile isolates resistant to fluoroquinolones in Italy: emergence of PCR ribotype 018. J. Clin. Microbiol. 48, 2892–2896. doi: 10.1128/JCM.02482-09
Stoesser, N., Sheppard, A. E., Pankhurst, L., De Maio, N., Moore, C. E., Sebra, R., et al. (2016). Evolutionary history of the global emergence of the Escherichia coli epidemic clone ST131. MBio 7:e02162. doi: 10.1128/mBio.02162-15
Stoikov, I., Ivanov, I. N., Donchev, D., Teneva, D., Dobreva, E., Hristova, R., et al. (2023). Genomic haracterization of IMP-producing Pseudomonas aeruginosa in Bulgaria reveals the emergence of IMP-100, a novel plasmid-mediated variant coexisting with a chromosomal VIM-4. Microorganisms 11:2270. doi: 10.3390/microorganisms11092270
Stokes, J. M., Lopatkin, A. J., Lobritz, M. A., and Collins, J. J. (2019). Bacterial metabolism and antibiotic efficacy. Cell Metab. 30, 251–259. doi: 10.1016/j.cmet.2019.06.009
Szilagyi, E., Fuzi, M., Damjanova, I., Böröcz, K., Szonyi, K., Tóth, A., et al. (2010). Investigation of extended-spectrum beta-lactamase-producing Klebsiella pneumoniae outbreaks in Hungary between 2005 and 2008. Acta Microbiol. Immunol. Hung. 57, 43–53. doi: 10.1556/AMicr.57.2010.1.4
Tan, S. Y., Khan, R. A., Khalid, K. E., Chong, C. W., and Bakhtiar, A. (2022). Correlation between antibiotic consumption and the occurrence of multidrug-resistant organisms in a Malaysian tertiary hospital: a 3-year observational study. Sci. Rep. 12:3106. doi: 10.1038/s41598-022-07142-2
Tao, J. J., Li, S. H., Wu, J. H., Peng, X. X., and Li, H. (2023). Pts promoter influences antibiotic resistance via proton motive force and ROS in Escherichia coli. Front. Microbiol. 14:1276954. doi: 10.3389/fmicb.2023.1276954
Tchesnokova, V., Larson, L., Basova, I., Sledneva, Y., Choudhury, D., Heng, J., et al. (2023). Increase in the community carriage of fluoroquinolone-resistant Escherichia coli despite a reduction in antibiotic prescriptions. Commun. Med. 3:110. doi: 10.1038/s43856-023-00337-2
Tchesnokova, V., Radey, M., Chattopadhyay, S., Larson, L., Weaver, J. L., Kisiela, D., et al. (2019). Pandemic fluoroquinolone resistant Escherichia coli clone ST1193 emerged via simultaneous homologous recombinations in 11 gene loci. Proc. Natl. Acad. Sci. USA 116, 14740–14748. doi: 10.1073/pnas.1903002116
Tickler, I. A., Goering, R. V., Whitmore, J. D., Lynn, A. N., Persing, D. H., Tenover, F. C., et al. (2014). Strain types and antimicrobial resistance patterns of Clostridium difficile isolates from the United States, 2011 to 2013. Antimicrob. Agents Chemother. 58, 4214–4218. doi: 10.1128/AAC.02775-13
Top, J., Willems, R., Blok, H., de Regt, M., Jalink, K., Troelstra, A., et al. (2007). Ecological replacement of Enterococcus faecalis by multiresistant clonal complex 17 Enterococcus faecium. Clin. Microbiol. Infect. 13, 316–319. doi: 10.1111/j.1469-0691.2006.01631.x
Torrens, G., van der Schalk, T. E., Cortes-Lara, S., Timbermont, L., Del Barrio-Tofiño, E., Xavier, B. B., et al. (2022). Susceptibility profiles and resistance genomics of Pseudomonas aeruginosa isolates from European ICUs participating in the ASPIRE-ICU trial. J. Antimicrob. Chemother. 77, 1862–1872. doi: 10.1093/jac/dkac122
Toth, A., Kocsis, B., Damjanova, I., Kristof, K., Janvari, L., Paszti, J., et al. (2014). Fitness cost associated with resistance to fluoroquinolones is diverse across clones of Klebsiella pneumoniae and may select for CTX-M-15 type extendedspectrum β-lactamase. Eur. J. Clin. Microbiol. Infect. Dis. 33, 837–843. doi: 10.1007/s10096-013-2022-6
Treepong, P., Kos, V. N., Guyeux, C., Blanc, D. S., Bertrand, X., Valot, B., et al. (2018). Global emergence of the widespread Pseudomonas aeruginosa ST235 clone. Clin. Microbiol. Infect. 24, 258–266. doi: 10.1016/j.cmi.2017.06.018
Troscianczyk, A., Nowakiewicz, A., Osinska, M., Łagowski, D., Gnat, S., and Chudzik-Rząd, B. (2022). Comparative characteristics of sequence types, genotypes and virulence of multidrug-resistant Enterococcus faecium isolated from various hosts in eastern Poland. Spread of clonal complex 17 in humans and animals. Res. Microbiol. 173:103925. doi: 10.1016/j.resmic.2022.103925
Valcek, A., Nesporova, K., Whiteway, C., De Pooter, T., De Coster, W., Strazisar, M., et al. (2022). Genomic analysis of a strain collection containing multidrug-, extensively drug-, pandrug-, and carbapenem-resistant modern clinical isolates of Acinetobacter baumannii. Antimicrob. Agents Chemother. 66:e0089222. doi: 10.1128/aac.00892-22
Valdezate, S., Miranda, C., Navarro, A., Freitas, A. R., Cabrera, J. J., Carrasco, G., et al. (2012). Clonal outbreak of ST17 multidrug-resistant Enterococcus faecium harbouring an Inc18-like::Tn1546 plasmid in a haemo-oncology ward of a Spanish hospital. J. Antimicrob. Chemother. 67, 832–836. doi: 10.1093/jac/dkr545
Van den Bogaard, A. E., Mertens, P., London, N. H., and Stobberingh, E. E. (1997). High prevalence of colonization with vancomycin- and pristinamycin-resistant enterococci in healthy humans and pigs in the Netherlands: is the addition of antibiotics to animal feeds to blame? J. Antimicrob. Chemother. 40, 454–456. doi: 10.1093/jac/40.3.454
Van den Braak, N., van Belkum, A., van Keulen, M., Vliegenthart, J., Verbrugh, H. A., and Endtz, H. P. (1998). Molecular characterization of vancomycin-resistant enterococci from hospitalized patients and poultry products in the Netherlands. J. Clin. Microbiol. 36, 1927–1932. doi: 10.1128/JCM.36.7.1927-1932.1998
Van der Donk, C., van de Bovenkamp, J., Bamelis, H., Driessen, C., Feldhoff, K. H., Kalka-Moll, W., et al. (2013). Prevalence and spread of multidrug-resistant Escherichia coli including ST131 in different patient populations in the Euroregion Meuse-Rhine. Future Microbiol. 8, 1027–1037. doi: 10.2217/fmb.13.61
Vanacker, M., Lenuzza, N., and Rasigade, J. P. (2023). The fitness cost of horizontally transferred and mutational antimicrobial resistance in Escherichia coli. Front. Microbiol. 14:1186920. doi: 10.3389/fmicb.2023.1186920
Velazquez-Meza, M. E., Aires de Sousa, M., Echaniz-Aviles, G., Solórzano Santos, F., Miranda-Novales, G., Silva-Sanchez, J., et al. (2004). Surveillance of methicillin-resistant Staphylococcus aureus in a pediatric hospital in Mexico City during a 7-year period (1997 to 2003): clonal evolution and impact of infection control. J. Clin. Microbiol. 42, 3877–3880. doi: 10.1128/JCM.42.8.3877-3880.2004
Vernon, J. J., Wilcox, M. H., and Freeman, J. (2019). Effect of fluoroquinolone resistance mutation Thr-82Ile on Clostridioides difficile fitness. J. Antimicrob. Chemother. 74, 877–884. doi: 10.1093/jac/dky535
Vinué, L., Jové, T., Torres, C., and Ploy, M. C. (2011). Diversity of class 1 integron gene cassette pc promoter variants in clinical Escherichia coli strains and description of a new P2 promoter variant. Int. J. Antimicrob. Agents 38, 526–529. doi: 10.1016/j.ijantimicag.2011.07.007
Vogwill, T., and MacLean, R. C. (2015). The genetic basis of the fitness costs of antimicrobial resistance: a meta-analysis approach. Evol. Appl. 8, 284–295. doi: 10.1111/eva.12202
Wang, Q., Wang, R., Wang, S., Zhang, A., Duan, Q., Sun, S., et al. (2024). Expansion and transmission dynamics of high risk carbapenem-resistant Klebsiella pneumoniae subclones in China: an epidemiological, spatial, genomic analysis. Drug Resist. Updat. 74:101083. doi: 10.1016/j.drup.2024.101083
Wang, W., Weng, J., Wei, J., Zhang, Q., Zhou, Y., He, Y., et al. (2024). Whole genome sequencing insight into carbapenem-resistant and multidrug-resistant Acinetobacter baumannii harboring chromosome-borne Bla(OXA-23). Microbiol Spectr 12:e0050124. doi: 10.1128/spectrum.00501-24
Wang, R., Yang, Q., Zhang, S., Hong, Y., Zhang, M., and Jiang, S. (2019). Trends and correlation of antibiotic susceptibility and antibiotic consumption at a large teaching hospital in China (2007-2016): a surveillance study. Ther. Clin. Risk Manag. 15, 1019–1027. doi: 10.2147/TCRM.S210872
Wang, T., Zhu, Y., Zhu, W., Cao, M., and Wei, Q. (2023). Molecular characterization of class 1 integrons in carbapenem-resistant Enterobacterales isolates. Microb. Pathog. 177:106051. doi: 10.1016/j.micpath.2023.106051
Warner, W. A., Kuang, S. N., Hernandez, R., Chong, M. C., Ewing, P. J., Fleischer, J., et al. (2016). Molecular characterization and antimicrobial susceptibility of Acinetobacter baumannii isolates obtained from two hospital outbreaks in Los Angeles County, California, USA. BMC Infect. Dis. 16:194. doi: 10.1186/s12879-016-1526-y
Wasels, F., Kuehne, S. A., Cartman, S. T., Spigaglia, P., Barbanti, F., Minton, N. P., et al. (2015). Fluoroquinolone resistance does not impose a cost on the fitness of Clostridium difficile in vitro. Antimicrob. Agents Chemother. 59, 1794–1796. doi: 10.1128/AAC.04503-14
Wei, Q., Jiang, X., Li, M., Li, G., Hu, Q., Lu, H., et al. (2013). Diversity of gene cassette promoter variants of class 1 integrons in uropathogenic Escherichia coli. Curr. Microbiol. 67, 543–549. doi: 10.1007/s00284-013-0399-1
Wein, T., Hülter, N. F., Mizrahi, I., and Dagan, T. (2019). Emergence of plasmid stability under non-selective conditions maintains antibiotic resistance. Nat. Commun. 10:2595. doi: 10.1038/s41467-019-10600-7
Willems, R. J., Top, J., van Santen, M., Robinson, D. A., Coque, T. M., Baquero, F., et al. (2005). Global spread of vancomycin-resistant Enterococcus faecium from distinct nosocomial genetic complex. Emerg. Infect. Dis. 11, 821–828. doi: 10.3201/1106.041204
Wiradiputra, M. R. D., Thirapanmethee, K., Khuntayaporn, P., Wanapaisan, P., and Chomnawang, M. T. (2023). Comparative genotypic characterization related to antibiotic resistance phenotypes of clinical carbapenem-resistant Acinetobacter baumannii MTC1106 (ST2) and MTC0619 (ST25). BMC Genomics 24:689. doi: 10.1186/s12864-023-09734-2
Wu, T., Zhang, Z., Li, T., Dong, X., Wu, D., Zhu, L., et al. (2024). The type III secretion system facilitates systemic infections of Pseudomonas aeruginosa in the clinic. Microbiol Spectr 12:e0222423. doi: 10.1128/spectrum.02224-23
Wyrsch, E. R., Bushell, R. N., Marenda, M. S., Browning, G. F., and Djordjevic, S. P. (2022). Global phylogeny and F virulence plasmid carriage in pandemic Escherichia coli ST1193. Microbiol Spectr 10:e0255422. doi: 10.1128/spectrum.02554
Xiao, L., Wang, X., Kong, N., Cao, M., Zhang, L., Wei, Q., et al. (2019). Polymorphisms of gene cassette promoters of the class 1 integron in clinical Proteus isolates. Front. Microbiol. 10:790. doi: 10.3389/fmicb.2019.00790
Xiu, Y., Dai, Y., Yin, S., and Wei, Q. (2024). Analysis of the class 1 integrons, carbapenemase genes and biofilm formation genes occurrence in Acinetobacter baumannii clinical isolates. Pol. J. Microbiol. 73, 189–197. doi: 10.33073/pjm-2024-017
Yano, H., Wegrzyn, K., Loftie-Eaton, W., Johnson, J., Deckert, G. E., Rogers, L. M., et al. (2016). Evolved plasmid-host interactions reduce plasmid interference cost. Mol. Microbiol. 101, 743–756. doi: 10.1111/mmi.13407
Zeng, W., Feng, L., Qian, C., Chen, T., Wang, S., Zhang, Y., et al. (2022). Acquisition of daptomycin resistance by Enterococcus faecium confers collateral sensitivity to glycopeptides. Front. Microbiol. 13:815600. doi: 10.3389/fmicb.2022.815600
Zeng, L., Zhang, J., Li, C., Fu, Y., Zhao, Y., Wang, Y., et al. (2020). The determination of gyrA and parC mutations and the prevalence of plasmid-mediated quinolone resistance genes in carbapenem resistant Klebsiella pneumonia ST11 and ST76 strains isolated from patients in Heilongjiang Province, China. Infect. Genet. Evol. 82:104319. doi: 10.1016/j.meegid.2020.104319
Zhang, X., Tang, M., Xu, Y., Xu, M., Qian, C., Zheng, X., et al. (2023). Characteristics of rare ST463 carbapenem-resistant Pseudomonas aeruginosa clinical isolates from blood. J. Glob. Antimicrob. Resist. 32, 122–130. doi: 10.1016/j.jgar.2023.01.011
Keywords: fluoroquinolones, resistance, clones, energy saving schemes, fitness
Citation: Fuzi M (2025) The fitness connection of antibiotic resistance. Front. Microbiol. 16:1556656. doi: 10.3389/fmicb.2025.1556656
Edited by:
José Manuel Rodriguez-Martínez, Sevilla University, SpainReviewed by:
Simona Gabriela Bungau, University of Oradea, RomaniaBela Kocsis, Semmelweis University, Hungary
Copyright © 2025 Fuzi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Miklos Fuzi, bWlrbG9zZnV6aUBnbWFpbC5jb20=
 Miklos Fuzi
Miklos Fuzi