- 1College of Life Sciences, Hebei University, Baoding, China
- 2Key Laboratory of Microbial Diversity Research and Application of Hebei Province, Baoding, China
- 3Engineering Research Center of Microbial Breeding and Conservation, Baoding, China
This study is aimed at the actual demand for exploring new species resources of Streptomyces, and aims to solve the technical bottleneck of Streptomyces isolation and culture. A new method was established based on the resuscitation function of the RpfA protein from Streptomyces coelicolor CGMCC 4.1658T to isolate unculturable or difficult-to-culture Streptomyces species, and it was applied to explore Streptomyces species resources in special habitats in the frozen soils of the Qinghai-Tibet Plateau. The RpfA protein of S. coelicolor was heterologously expressed and validated for its in vitro activity. The purified RpfA protein was then used to isolate Streptomyces from soil samples in the frozen soils of the Qinghai-Tibet Plateau, followed by an investigation into the impact of the RpfA protein on the cultivability of Streptomyces species. The results showed that the RpfA protein had a significant promoting effect on the germination of spores of both S. coelicolor itself and other species of the Streptomyces genus, and when a suitable concentration of RpfA protein was added to the culture medium, it could significantly improve the culturability of members of phylum Actinomycetota, especially Streptomyces species. In addition, many new species of the genus Streptomyces and other genera of phylum Actinomycetota were discovered. This study provides a new approach for further exploring Streptomyces species resources in special environments such as the Qinghai-Tibet Plateau and developing new biologically active substances produced by Streptomyces.
1 Introduction
Streptomyces are known for producing a wide variety of antibiotics and are considered to be an endless treasure trove of natural product biosynthesis diversity. Approximately two-thirds of the antibiotics used clinically are produced by Streptomyces (Barka et al., 2016; Bérdy, 2012; Gao et al., 2018; Olanrewaju and Babalola, 2019). However, in recent decades, a large number of Streptomyces species have been repeatedly isolated, leading to a decreasing chance of discovering new antibiotics. Moreover, most Streptomycetes cannot be cultured under laboratory conditions because they are in a state of being “uncultivable” or “not yet cultivated,” and thus have not been discovered so far, and they are the source of many unusual bioactive natural products (Lackner et al., 2017; van Dorst et al., 2016). Research has shown that the production of antibiotics by Streptomyces is species-specific (Liu et al., 2013), therefore, exploring new Streptomyces species resources is an effective way to solve the difficulty of discovering antibiotics. For the Streptomyces isolation and cultivation technology, past research has usually designed suitable culture media for Streptomyces growth (Kuester and Williams, 1964; Stewart, 2012; Suutari et al., 2002), but these traditional strategies limited the discovery of new species. Therefore, it is urgent to develop new isolation strategies and achieve breakthroughs in isolation and cultivation techniques to explore new resources of Streptomyces species.
The next-generation sequencing technology reveals that there remains an extremely high level of unknown (currently “uncultivable”) microbial diversity in nature, particularly in extreme environments (Li et al., 2021; Locey and Lennon, 2016; Pham and Kim, 2012). Meanwhile, when Streptomyces is confronted with adverse growth conditions, it forms dormant spores with conspicuously thicker cell wall (Cava and de Pedro, 2014; Sexton et al., 2020; Vollmer et al., 2008) and no significant metabolic activity (Kell and Young, 2000), which has led to “unculturable” or difficult-to-culture state in laboratory cultures (Lopez Marin et al., 2021). The permafrost of the Qinghai-Tibet Plateau serves as a representative of extreme environments, which it features long-term low temperatures, a severe deficiency of liquid water, and limited available nutrients, making Streptomyces strains within it more prone to form dormant spores with low metabolic activity. These challenges have significantly hindered the isolation of microorganisms.
Resuscitation-promoting factor (Rpf) was first discovered by Mukamolova et al. (1998) in the supernatant of Micrococcus luteus. This protein can stimulate the resuscitation of dormant M. luteus cells at picomolar concentrations (pmol/L), was named as Resuscitation-promoting factor (Rpf) (Mukamolova et al., 1999). Studies have shown that proteins in the Rpf family belong to the class of hydrolytic glycosyltransferases, similar to lysozymes and lytic transglycosylases, and are able to cleave the β-1,4-glycosidic bond between N-acetylglucosamine (GlcNAc) residues and N-acetylmuramic acid (MurNAc) residues in bacterial peptidoglycan (Maione et al., 2015). Previous researches have demonstrated that Rpfs can promote the germination and growth of dormant spores (Nikitushkin et al., 2016; Sexton et al., 2020; Sexton et al., 2015). Rpfs proteins are widely distributed among Gram-positive bacteria within the phylum Actinomycetota. Bioinformatics analysis has revealed that rpf genes in different species vary in length, genetic structure, host specificity, and activity (Flärdh and Buttner, 2009; Ravagnani et al., 2005; Tantivitayakul et al., 2020). However, all rpf genes encode proteins that contain a conserved domain with lysozyme-like activity (the Rpf-like domain), which is approximately 70 amino acids (Ravagnani et al., 2005). S. coelicolor can encode five Rpfs proteins (RpfA - RpfE), all Rpf proteins structurally possess a C-type lysozyme-like domain, and their active sites have a conserved Glu, all of which are secreted proteins, unlike the Rpfs proteins of Mycobacterium tuberculosis (Sexton et al., 2015). It has been 22 years since the Rpf protein in M. luteus was first reported to be able to stimulate the recovery of dormant bacteria. During this period, the application of Rpf has been extended from the aspects of pathogen containment (Coletta-Filho et al., 2006), vaccine development (Romano et al., 2012) and diagnostic testing (Arroyo et al., 2018) to the research field of introducing Rpf protein into polluted environment remediation and sewage biological treatment system (Su et al., 2021; Su et al., 2013). The introduction of Rpf offers a novel approach for reviving difficult-to-culture microorganisms. However, there are few systematic studies on the effect of Rpf for cultivation of soil Streptomyces.
The RpfA protein of S. coelicolor can promote the resuscitation of high G + C Gram-positive bacteria, making it possible to successfully isolate dormant Actinomycetes in the environment. In this experiment, the RpfA protein of S. coelicolor was heterologously expressed in Escherichia coli, and the recombinant Rpf protein was verified for in vitro activity and added to the isolation medium to evaluate its culture effect on soil Streptomyces. The aim is to explore a new method for mining Streptomyces strain resources. Therefore, this study established a new method for isolating unculturable or difficult-to-culture Streptomyces species based on the resuscitation function of the RpfA protein from S. coelicolor, and applied it to excavate Streptomyces species resources in extreme environments of frozen soils of the Qinghai-Tibet Plateau. The results showed that the RpfA protein from S. coelicolor has a stimulatory effect on the spore germination of both itself and other Streptomyces species. Furthermore, the RpfA protein can significantly improve the cultivability of members of phylum Actinomycetota, especially Streptomyces species, of frozen soils of the Qinghai-Tibet Plateau, and some potential new species were isolated. This study provides a new approach for further exploring Streptomyces species resources in unique environments of the Qinghai-Tibet Plateau and developing new biologically active substances derived from Streptomyces.
2 Materials and methods
2.1 Strains and plasmids
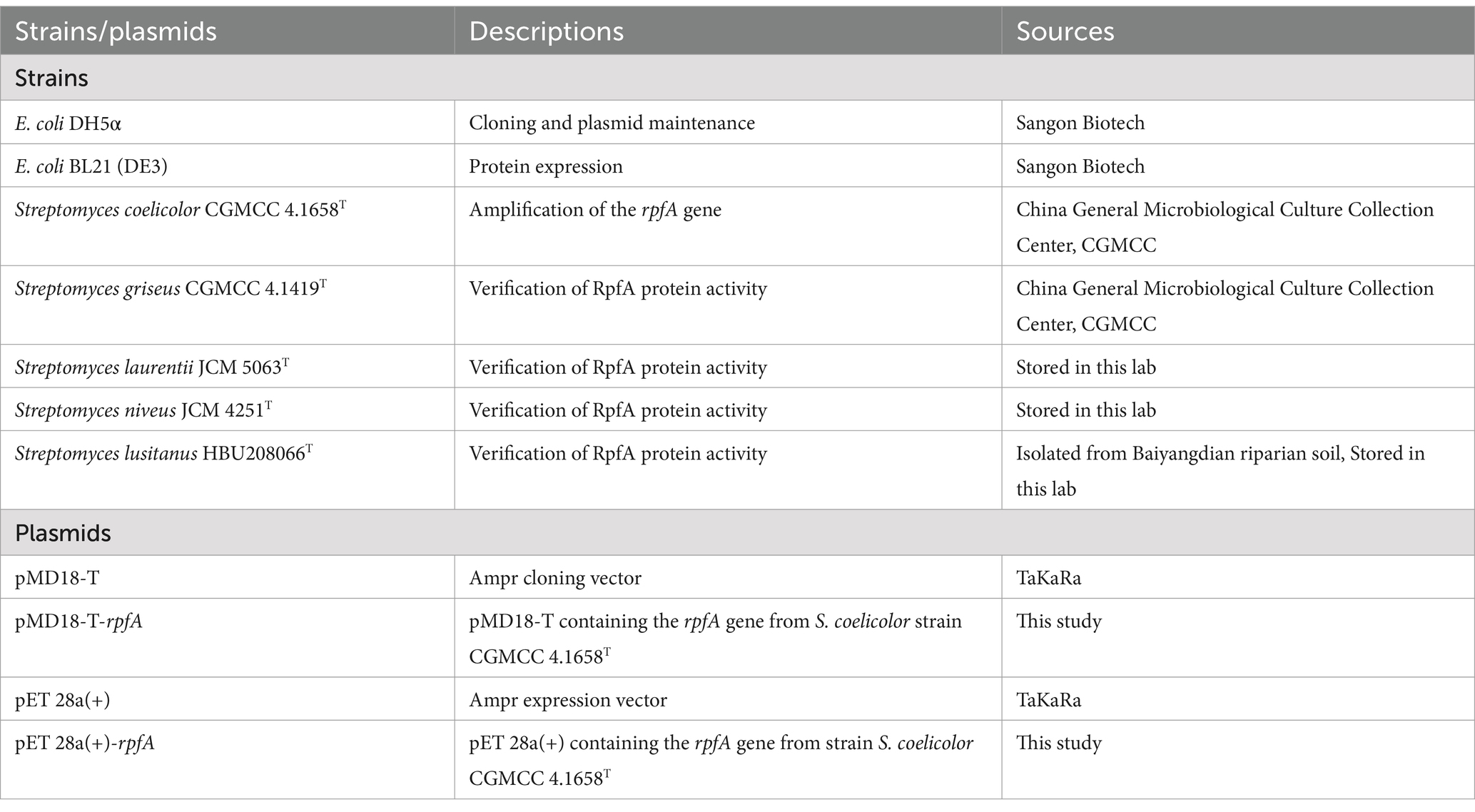
The experimental strains and plasmids used in this study are listed in Table 1.
2.2 Medium and growth conditions
S. coelicolor CGMCC 4.1658T, S. griseus CGMCC 4.1419T, S. laurentii JCM 5063T, S. niveus JCM 4251T, S. lusitanus HBU208066 were cultured in R2A medium (Reasoner and Geldreich, 1985) at 28°C; E. coli DH5α and E. coli BL21 (DE3) were cultured in LB medium at 37°C. R2A and Glucose-Tryptone culture media (Chen et al., 2007) were used to isolate Actinomycetota strains from frozen soils of the Qinghai-Tibet Plateau.
2.3 Preparation of recombinant RpfA protein from Streptomyces coelicolor
The rpfA gene and its encoded protein from S. coelicolor were subjected to multiple sequence alignment and protein domain prediction using the online software UniProt (accessed on January 29, 2024)1 and HMMER (accessed on January 29, 2024).2 Genomic DNA of S. coelicolor CGMCC 4.1658T was extracted using the “Chelex100” method (de Lamballerie et al., 1992), and PCR amplification of rpfA gene was performed using the primers rpf-F 5′-CAGTAC CATATG GCCACCGCGTCCGAGTGGGAC-3′ and rpf-R 5′-CGAAGT GGATCC TTACTTCAGGTGCAGCTGCTG-3′ (restriction sites Nde I/BamH I used for cloning purposes are indicated in bold and underlined). The purified PCR product was ligated to the Nde I/BamH I-digested pET-28a (+) plasmid and transformed into E. coli BL21(DE3) competent cells. The recombinant cells were cultivated in LB medium containing kanamycin (50 μg/mL), induced with 1 mM/L IPTG, collected, and washed twice with phosphate-buffered saline (PBS). After sonication to disrupt the cells, centrifugation was performed at 4°C 10,000 rpm for 20 min, and both the supernatant and precipitate were collected, and then analyzed by SDS-PAGE, respectively. The RpfA target protein was purified using Ni-NTA-agarose column (Suzhou Biodragon Technology Co., Ltd., BDTL0011-10). The concentration of RpfA protein was determined using the Lowry method (Waterborg and Matthews, 1994). Finally, the purified recombinant RpfA protein was stored at −20°C for subsequent experiments.
2.4 Verification of the spore germination-promoting activity of the RpfA protein
Sixty 16S rRNA gene sequences of type strains belonging to species of the genus Streptomyces were retrieved from the GenBank database.3 Phylogenetic clustering analysis was conducted using MEGA 11.0 software. According to the formation of distinct phylogenetic groups, one species from each group was selected as a representative to assess the promotion of the RpfA protein of S. coelicolor on spore germination activity in its own and other Streptomyces species. Spore suspensions of the different species were prepared and coated on R2A plates. Spore germination rates were determined by adding 0.1, 1, 10% (v/v) RpfA protein in above R2A plates as experimental group and without adding RpfA protein as control group, respectively. In addition, in order to verify the resuscitation and stimulation function of RpfA protein, sterilized inactivated RpfA protein was used as the inactivated control group. These R2A plates were incubated at 28°C for 72 h. Colony counts on the R2A solid medium were enumerated every 12 h, and the spore germination rate was calculated as follows: spore germination rate = [number of germinated spores (CFU/mL)/total number of spores in the spore suspension (CFU/mL)] × 100%. The above experiments were repeated 3 times.
To further verify the stimulatory effect of the RpfA protein on spore germination, the BacTiter-Glo™ Microbial Cell Viability Assay Kit (BacTiter-Glo™, Catalog No. G82300, purchased from Madison, Wisconsin, United States) was used to measure the cellular viability during spore germination of different Streptomyces species, following the manufacturer’s instructions (Sun et al., 2016). This method detects cellular viability by measuring ATP content. According to the colony counting results of different species of Streptomyces during spore germination, RpfA protein of optimal concentration was added into spore suspension of optimal dilution gradient of the strain to be tested as the experimental group, and no RpfA protein was added as the control group, with three replicates each group. The cultures were incubated at 28°C with shaking at 160 rpm for 72 h. Luminescence intensity was measured every 12 h. Simultaneously, D2O isotope labeling combined with Single-Cell Raman Spectroscopy (D2O-SCRS) (Wang et al., 2022) was utilized to assess the metabolic activity of spore germination with and without the addition of RpfA protein.
2.5 Isolation and identification of Streptomyces species in frozen soils of the Qinghai-Tibet Plateau
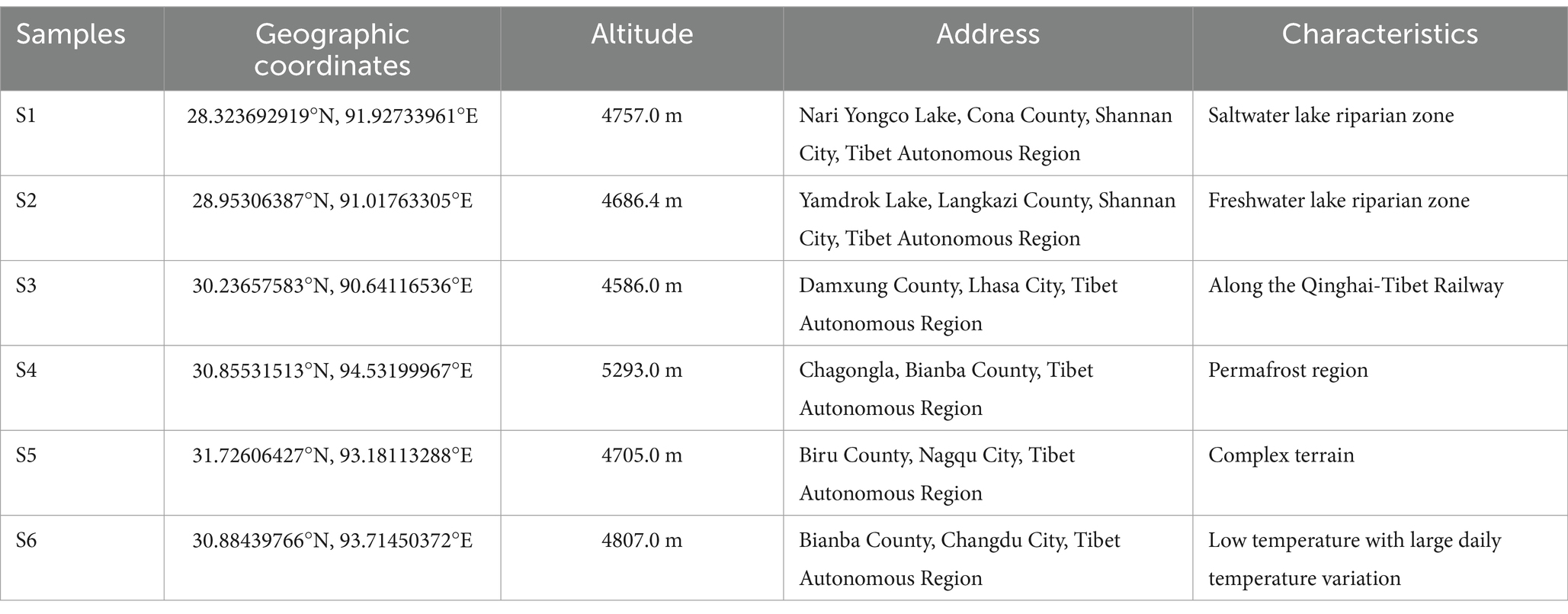
Soil samples were collected from six locations in the frozen soils of the Qinghai-Tibet Plateau (Table 2). The processed soil samples were gradient-diluted to 10−3, and 100 μL of each gradient was plated onto R2A and glucose-tryptone solid media, respectively. The media were pre-supplemented with sterile-filtered S. coelicolor RpfA protein at concentrations of 10, 1, and 0.1% (v/v), as well as final concentrations of 50 μg/mL of cycloheximide, nalidixic acid, and nystatin, and 40 μg/mL of kanamycin. Groups without RpfA protein served as controls. Each group had three replicates, and they were incubated at 28°C for more than 2 weeks. The colonies on the plates were isolated, purified, and preserved.
The Genomic DNA of the isolates were extracted using “chelex 100” method (de Lamballerie et al., 1992), and the 16S rRNA gene fragments were PCR-amplified using the universal bacterial primers 27F (5′-AGAGTTTGATCCTGGCTCAG-3′) and 1492R (5′-GGTTACCTTGTTACGACTT-3′). The amplification products were sent to Sangon Biotech (Shanghai) Co., Ltd. for sequencing. The sequencing results were analyzed and compared using the EzTaxon database on the EzBioCloud online website4 (Kim et al., 2012). Multiple alignments were performed using MEGA 11.0 (Kumar et al., 2016), and a phylogenetic tree was constructed using the Neighbor-Joining method (Saitou and Nei, 1987). The 16S rRNA gene trees diagram were drawn using the iTOL online tool5 (Lefebvre et al., 2009) to determine the taxonomic status and affiliation of the strains.
2.6 Statistical analysis
Statistical analysis of the data was performed using GraphPad Prism (version 9.4.1) and Origin 8.0. The mean ± standard deviation (SD) for each parameter was calculated using data from three replicates. The significance level for analysis of variance (ANOVA) was set at p < 0.05.
3 Results
3.1 Characteristics of the RpfA protein from Streptomyces coelicolor CGMCC 4.1658T
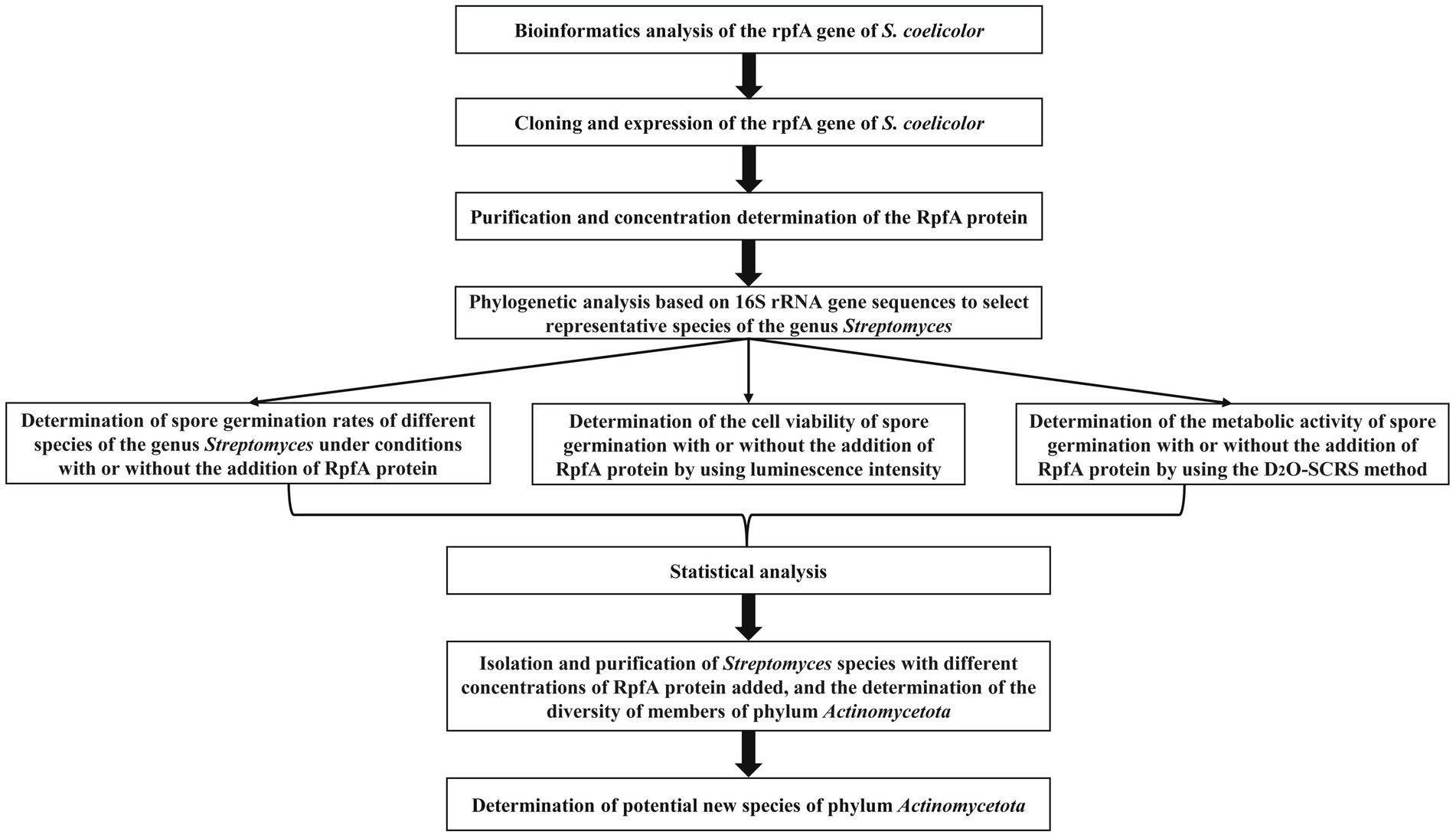
The experimental scheme of this study is shown in Figure 1. The physicochemical properties of the amino acid sequence encoded by the rpfA gene of S. coelicolor were predicted and analyzed using the Expasy online software.6 The protein consists of 244 amino acids, including 20 types of amino acids, with alanine (Ala) and glycine (Gly) being the most abundant. The Grand average of hydropathicity (GRAVY) is −0.519. The II value of this protein is 25.39. The hydrophilicity and hydrophobicity of the RpfA protein amino acid sequence were analyzed using the ProtScale tool. The hydrophilic amino acids in the RpfA protein amino acid sequence are slightly more abundant than the hydrophobic amino acids, and the amino acid hydrophilicity index is around −1 (Figure 2A). Prediction and analysis of the signal peptide of the RpfA protein encoded by the rpfA gene was conducted using the online software Signal P-5.0.7 The possibility of the cleavage site of the signal peptide of the RpfA protein being located between the 41st and 42nd amino acids (ASA-AT) is 0.1830, and the possibility of the presence of a Sec/SPI signal peptide is 0.5828 (Figure 2B). The amino acid sequence of the RpfA protein was analyzed for potential phosphorylation sites using the online tool NetPhos 3.1.8 The analysis results indicated that the RpfA protein has 23 serine (Ser) phosphorylation sites, 10 threonine (Thr) phosphorylation sites, and 4 tyrosine (Tyr) phosphorylation sites (Figure 2C). The conserved domains of the protein sequence encoded by the rpfA gene were predicted and analyzed using HM-MER.9 The RpfA protein-encoding chain has two domains: one Rpf catalytic domain for hydrolyzing β-1,4-glycosidic bonds and one Lysin (LysM, C121525) domain (Figure 2D). The three-dimensional structure of the RpfA protein was predicted using SWISS-MODEL (Waterhouse et al., 2018). The results showed that its key catalytic site was glutamic acid, the substrate-binding residues were threonine and tryptophan. The reliability score of the RpfA protein is 57.69%. The template sequence method used X-ray with a resolution of 1.17 Å. The alignment covered amino acids 45–122, with a coverage of 0.32 (Figure 2E).
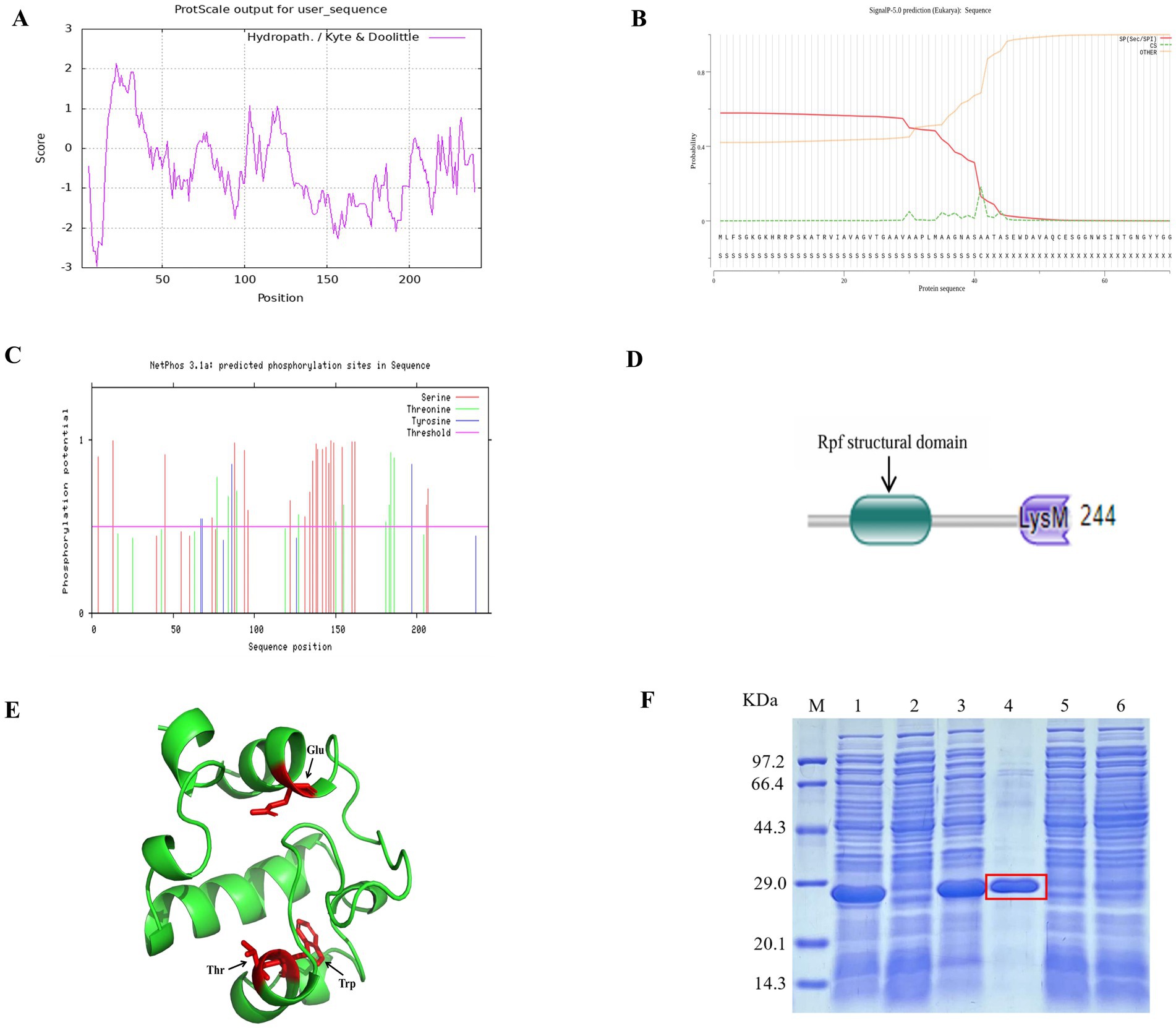
Figure 2. Characteristic information of the RpfA protein from S. coelicolor. (A) Hydrophilicity and hydrophobicity prediction diagram; (B) Signal peptide prediction diagram; (C) Phosphorylation site prediction diagram; (D) Structural domains; (E) Tertiary structure prediction diagram, Glu is the key catalytic site of the RpfA protein in S.coelicolor, Thr and Try are its substrate-binding residues; (F) SDS-PAGE electrophoresis for detection of rpfA gene expression and protein purification. M: Protein molecular weight markers; Lane 1 and Lane 3: lysed proteins of E. coli BL21(DE3) containing pET-28a (+)-rpf; Lane 2 and Lane 5: lysed proteins of E. coli BL21(DE3) containing pET-28a (+); Lane 4: purified RpfA protein (marked with a red frame); Lane 6: lysed proteins of E. coli BL21(DE3) containing pET-28a (+)-rpf before induction.
3.2 Purification and identification of the RpfA protein from Streptomyces coelicolor CGMCC 4.1658T
The prokaryotic expression vector pET-28a (+)-rpfA for the rpfA gene of S. coelicolor was constructed and efficiently expressed heterologously in E. coli BL21 (DE3). The heterologously expressed RpfA protein was purified using Ni-NTA-agarose affinity chromatography, yielding RpfA protein with a molecular weight of approximately 24.9 kDa and a concentration of 1.19 mg/mL (Figure 2F).
3.3 Phylogenetic analysis of some species within the genus Streptomyces and selection of representative species
Sixty type strains of Streptomyces species (Supplementary Table S1) formed five major phylogenetic groups (Supplementary Figure S1). One representative species was selected from each group (indicated in bold), and they were S. coelicolor CGMCC 4.1658T, S. griseus CGMCC 4.1419T, S. laurentii JCM 5063T, S. niveus JCM 4251T, and S. lusitanus HBU208066T, respectively. These strains were used as test strains in subsequent experiments to verify the ability of the RpfA protein from S. coelicolor to promote spore germination activity in itself and other Streptomyces species.
3.4 Effect of RpfA protein on spore germination rates among different Streptomyces species
The addition of different concentrations of RpfA protein had varying effects on the spore germination rates of S. coelicolor, S. griseus, S. laurentii, S. niveus, and S. lusitanus (Figure 3). When a final concentration of 10% (v/v) RpfA protein was added to R2A medium, the number of colonies formed by the spores of S. coelicolor and S. niveus was significantly higher than that in the experimental groups with 0.1 and 1% (v/v) RpfA protein added and the control group without RpfA protein. The addition of 1% (v/v) RpfA protein had the most significant promoting effect on spore germination of S. griseus. In contrast, 0.1% (v/v) RpfA protein had the most significant promoting effect on spore germination of S. laurentii and S. lusitanus. After adding different concentrations of RpfA protein, the spore germination rates of different Streptomyces species varied (Supplementary Table S2). Compared with the control group, when cultured for 72 h and after adding the optimal concentration of RpfA protein, the spore germination rates of S. coelicolor, S. niveus, S. griseus, S. laurentii, and S. lusitanus increased by 45, 48.9, 22.2, 64, and 48.9%, respectively (Table 3). Furthermore, it was found that inactivated RpfA protein did not promote spore germination (Figure 4).
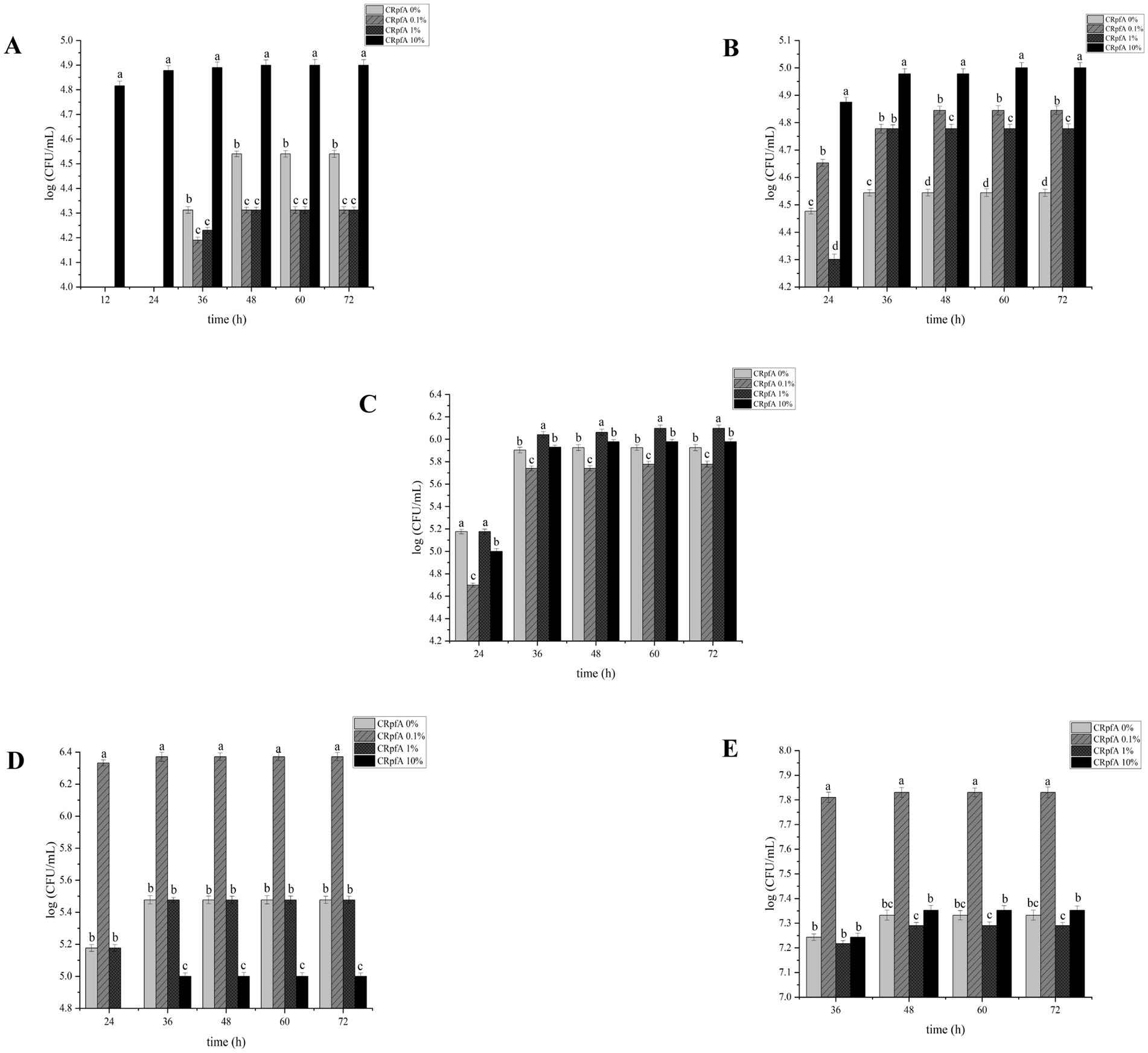
Figure 3. Effect of different concentrations of RpfA protein on spore germination of Streptomyces strain. Different letters above the bar graph indicate statistically significant differences at p ≤ 0.05 (n = 3). (A) S. coelicolor; (B) S. niveus; (C) S. griseus; (D) S. laurentii; (E) S. lusitanus; CRpfA represents the concentration of RpfA protein.
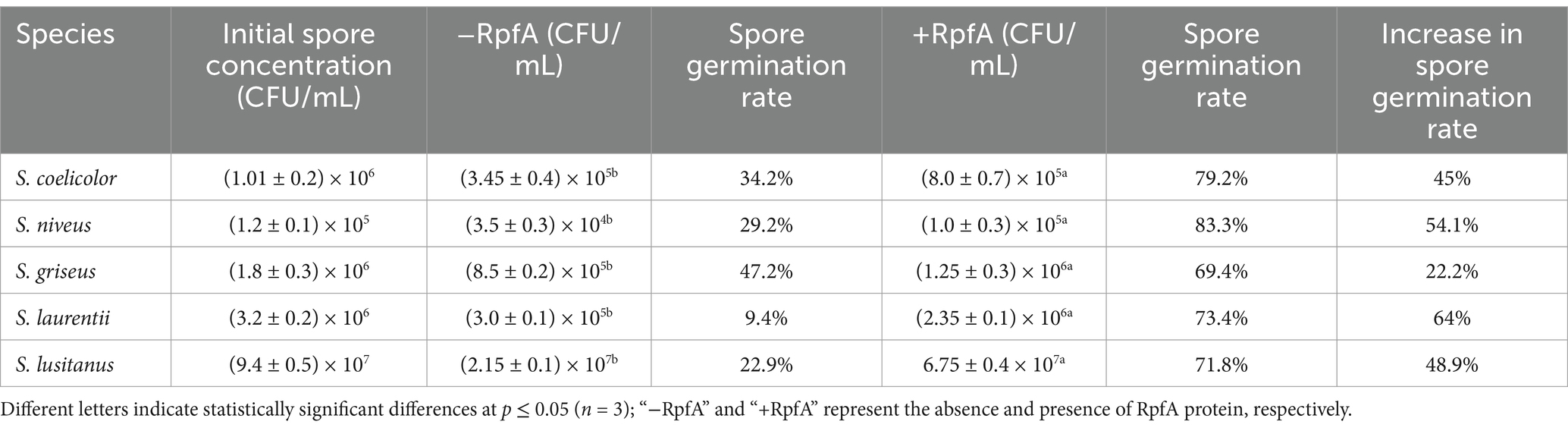
Table 3. Spore germination rates of different Streptomyces species in the experimental group with the optimal concentration of Rpf protein and the control group at 72 h of cultivation.
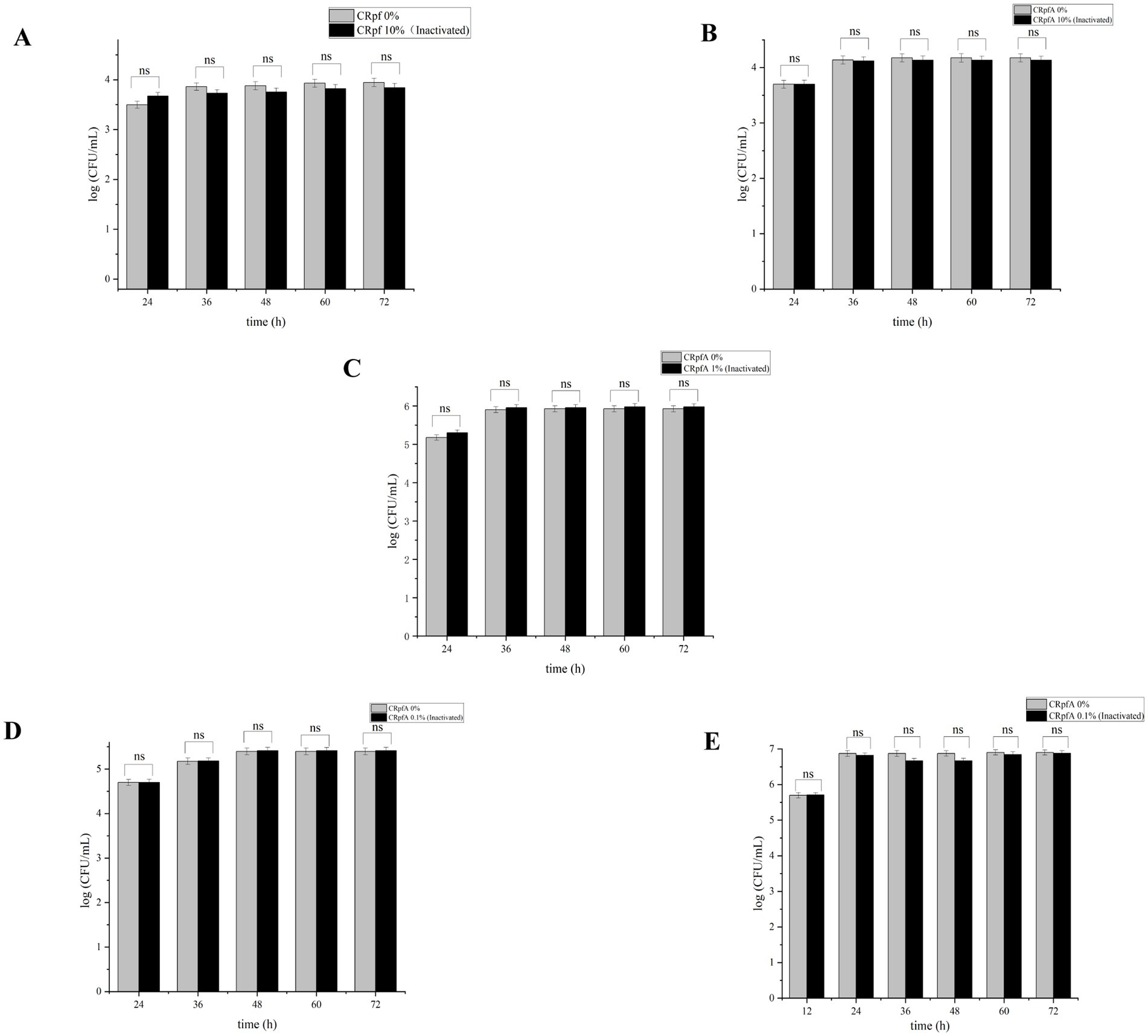
Figure 4. Effect of inactivated RpfA protein on the number of colonies formed by Streptomyces spore germination. Significance level of ANOVA, *(0.01 < =p < 0.05), **(0.001 < =p < 0.01), ***(p < 0.001), ns(p > 0.05), (n = 3). (A) S. coelicolor; (B) S. niveus; (C) S. griseus; (D) S. laurentii; (E) S. lusitanus; CRpfA represents the concentration of RpfA protein.
3.5 Cell viability during spore germination measured by luminescence intensity
To further verify the promoting effect of RpfA protein on spore germination, the BacTiter-Glo™ Microbial Cell Viability Assay Kit was used to measure the cell viability during spore germination of S. coelicolor CGMCC 4.1658T, S. niveus JCM 4251T, S. griseus CGMCC 4.1419T, S. laurentii JCM 5063T, and S. lusitanus HBU208066T in the presence or absence of RpfA protein (Figure 5). The BacTiter-Glo™ reagent relies on the properties of thermostable luciferase (Ultra-Glo recombinant luciferase) and a patented formula for extracting ATP from bacteria. The BacTiter-Glo™ assay produces a luminescent signal, with the intensity of the signal being directly proportional to the amount of ATP present, which in turn is proportional to the cellular viability in the culture. Bioluminescent sensors based on luciferin have been proven to effectively detect ATP concentrations, with a linear relationship between analyte concentration and bioluminescent intensity (Abbasi et al., 2024; Karimi et al., 2022; Luo et al., 2009). The assay results showed that, compared with the control group, the experimental group with the optimal concentration of RpfA protein had continuously increasing Relative Light Unit (RLU) values over time, which were significantly higher than those of the control group.
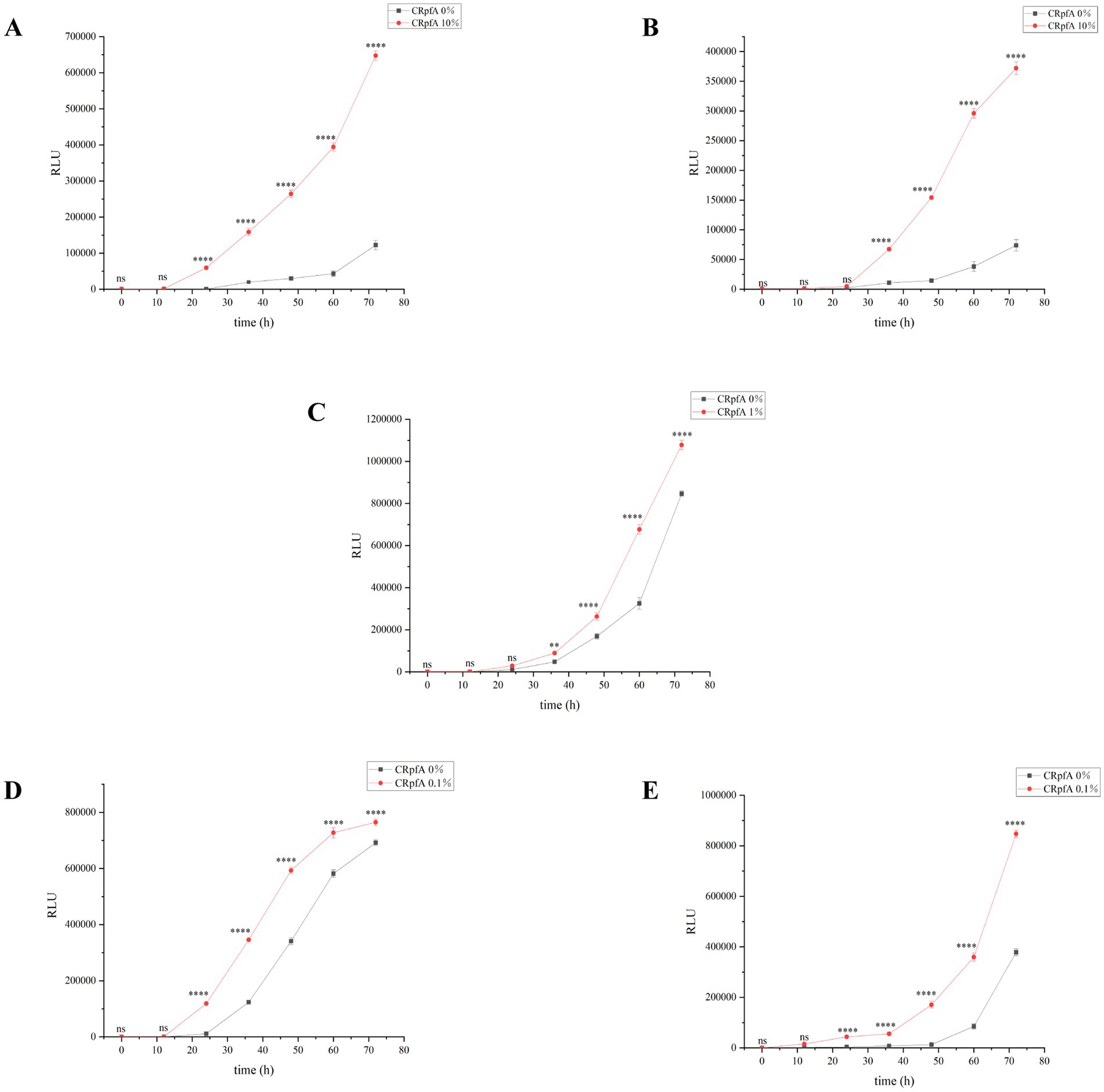
Figure 5. Verification of the effect of RpfA protein on cellular viability during Streptomyces spore germination using BacTiter-Glo™. Significance level of ANOVA, *(0.01 < =p < 0.05), **(0.001 < =p < 0.01), ***(p < 0.001), ns(p > 0.05), (n = 3). (A) S. coelicolor; (B) S. niveus; (C) S. griseus; (D) S. laurentii; (E) S. lusitanus; CRpfA represents the concentration of RpfA protein.
3.6 Metabolic activity of spores during germination determined by D2O isotope labeling combined with single-cell Raman spectroscopy (D2O-SCRS)
Raman spectroscopy is a fingerprint spectrum that reflects the structure of materials, arising from the interaction between light and material. When high-intensity incident light scatters on material molecules, a portion of the scattered light differs in frequency from the incident light, with the frequency shifting based on the structure of the tested material. This portion of the scattered light is known as Raman scattering (Huang et al., 2010; Smith et al., 2016). The use of heavy water (D2O) isotope labeling combined with single-cell Raman spectroscopy (SCRS) allows for the identification and classification of viable microorganisms at the single-cell level in complex samples. The carbon-deuterium (C-D) characteristic peak in single-cell Raman spectroscopy can clearly detect the incorporation of D, derived from D2O, into the biomass of microorganisms, thereby reflecting their metabolic activity (Berry et al., 2015; Hua et al., 2021; Ni et al., 2021).
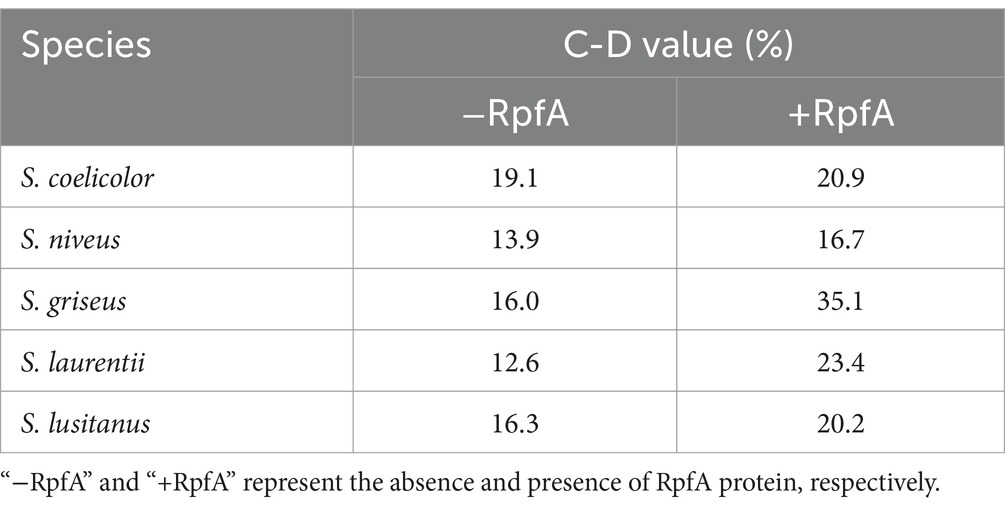
Raman spectroscopy was adopted to further verify the effect of RpfA protein on the metabolic activity during spore germination of S. coelicolor CGMCC 4.1658T, S. niveus JCM 4251T, S. griseus CGMCC 4.1419T, S. laurentii JCM 5063T, and S. lusitanus HBU208066T (Figure 6). After 72 h of culturing, the intensities of the C-D and C-H peaks of the prepared samples were detected by a single-cell Raman phenotypic monitoring system, and their C-D values were calculated (Table 4). The results showed that in the experimental groups with the optimal concentration of RpfA protein added, the C-D values of all five Streptomyces strains were higher than those in the control group to varying degrees, indicating that the cellular metabolic activity in the experimental groups with RpfA protein during spore germination was higher. Additionally, both the C-D and C-H peak intensities were higher in the experimental groups.
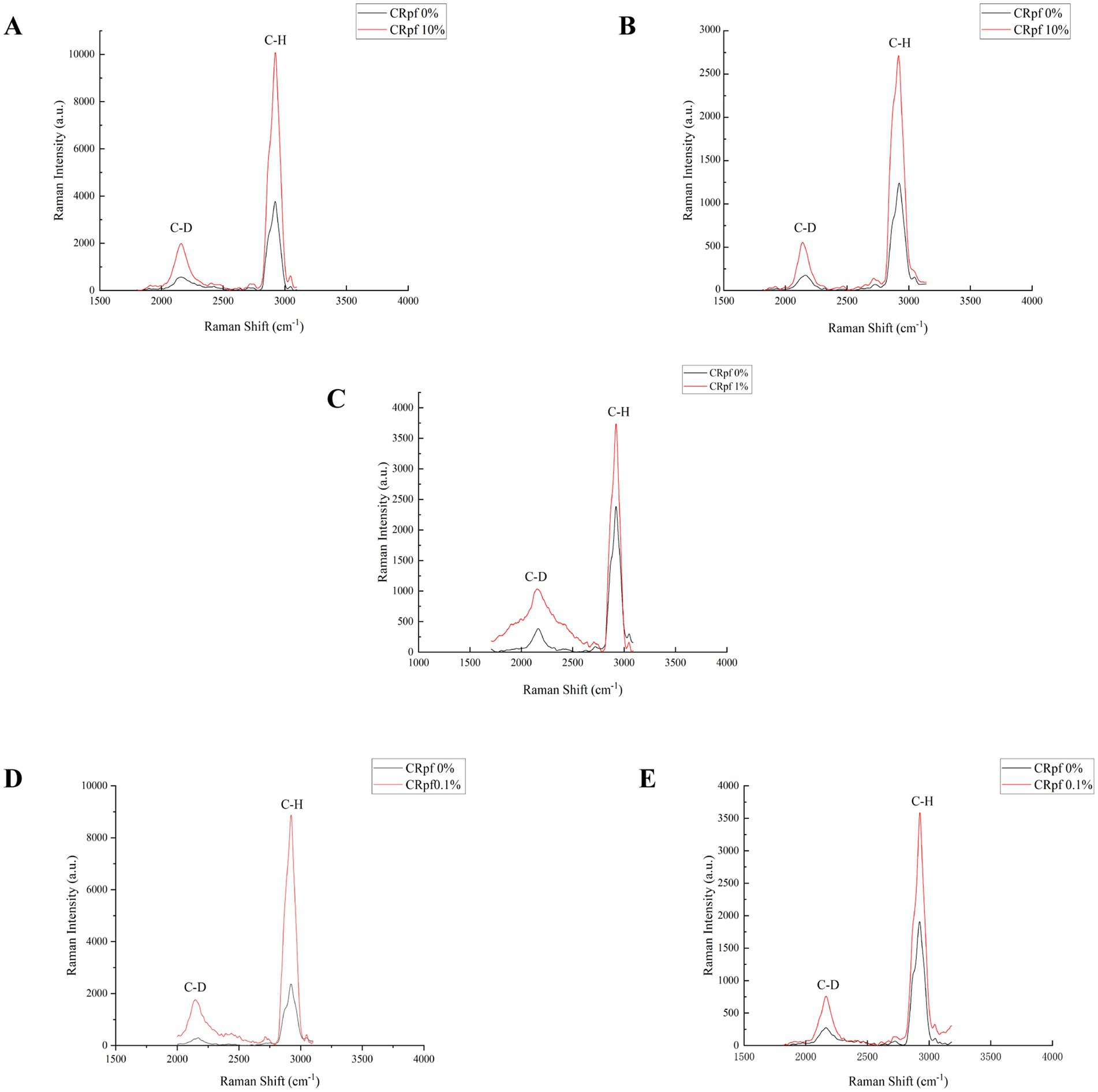
Figure 6. Peak of single-cell Raman spectroscopy during spore germination after D2O isotope labeling. (A) S. coelicolor; (B) S. niveus; (C) S. griseus; (D) S. laurentii; (E) S. lusitanus; CRpfA represents the concentration of RpfA protein.
3.7 Streptomyces strains isolated from six soil samples after adding RpfA protein
From soil samples collected at six sampling sites in the frozen soils of the Qinghai-Tibet Plateau, a total of 137 strains of Streptomyces were isolated using R2A and glucose-tryptone media (including experimental groups with different concentrations of RpfA protein added and control groups without RpfA protein added). Specifically, 80, 27, and 25 strains of Streptomyces were obtained when the concentrations of RpfA protein added were 10, 1, and 0.1%, respectively; whereas in the control groups without RpfA protein added, only 5 strains of Streptomyces were isolated (Table 5).
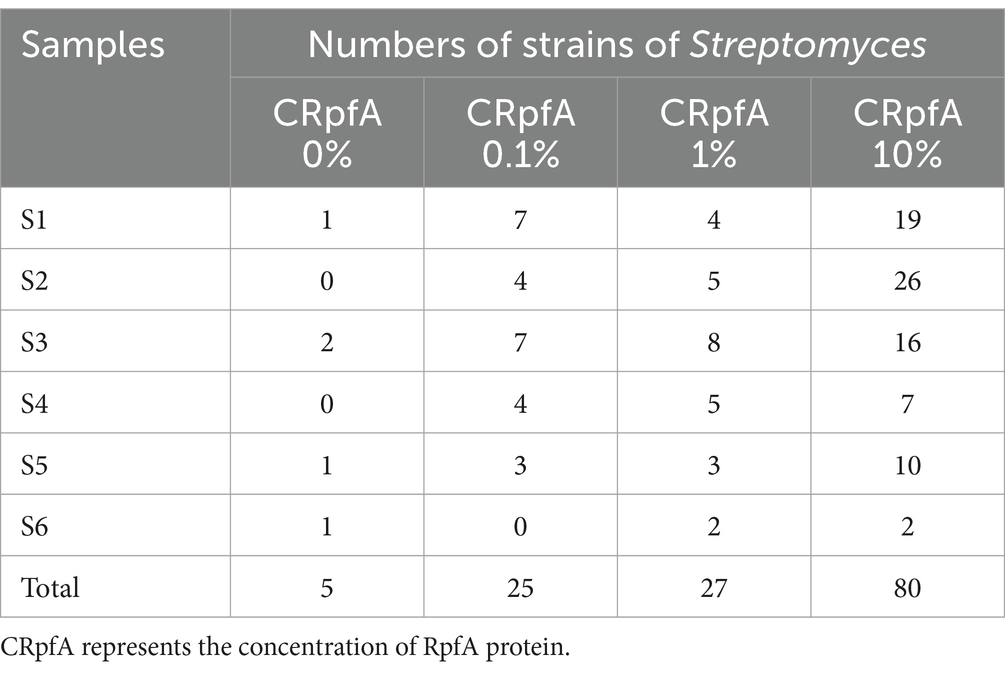
Table 5. The quantities of strains of genus Streptomyces isolated by adding different the concentrations of RpfA protein in six soil samples.
When 10, 1, and 0.1% RpfA protein were added to the media, 19, 4, and 7 strains of genus Streptomyces were isolated from sample S1, respectively; while only 1 strain was isolated in the control group. In sample S2, 26, 5, and 4 strains were obtained, respectively, with no Streptomyces strains isolated in the control group. For sample S3, 16, 8, and 7 strains were isolated, respectively, while 2 strains were isolated in the control group. In sample S4, 7, 5, and 4 strains were obtained, respectively, with no Streptomyces strains isolated in the control group. For sample S5, 10, 3, and 3 strains were isolated, respectively, while only 1 strain was isolated in the control group. In sample S6, 2 strains of Streptomyces were isolated when both 10 and 1% RpfA protein were added, but no Streptomyces strains were isolated when 0.1% RpfA protein was added; 1 strain was isolated in the control group (Table 5).
16S rRNA sequence analysis was conducted on all Streptomyces strains isolated from the six samples to determine their species (Supplementary Table S3). The results revealed that the addition of RpfA protein in the six samples increased the diversity and quantity of isolated Streptomyces to varying degrees (Supplementary Tables S4–S9; Figure 7). Among the five Streptomyces strains isolated from the control groups without RpfA protein, they belonged to four species: S. tateyamensis, S. olivoviridis, S. liangshanensis, and S. cyaneofuscatus. Except for S. liangshanensis, the other three species were also isolated in the experimental groups with RpfA protein added.
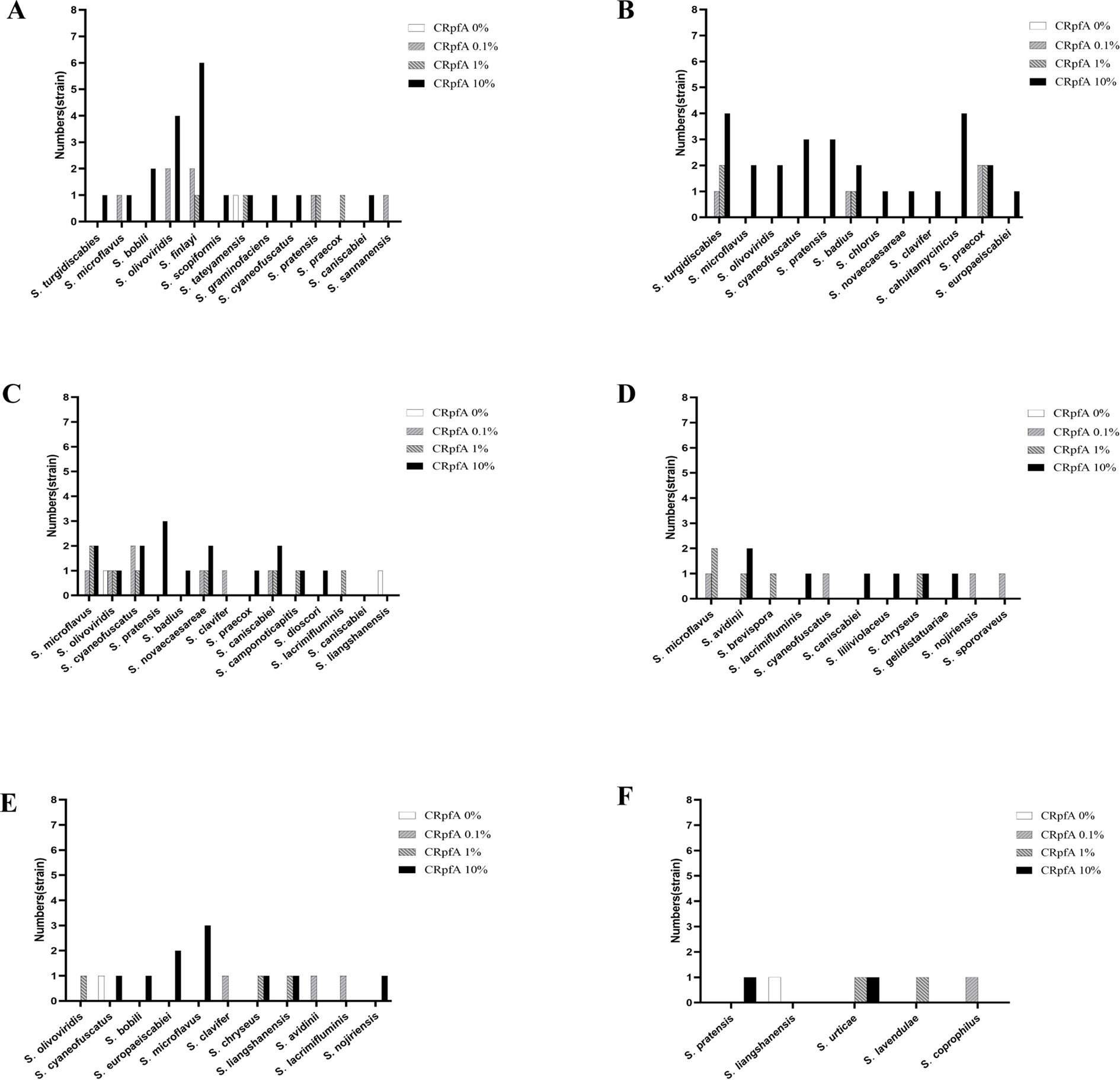
Figure 7. Statistical graphs of the quantities of Streptomyces strains isolated from six soil samples adding different the concentrations of RpfA protein. (A) S1; (B) S2; (C) S3; (D) S4; (E) S5; (F) S6; CRpfA represents the concentration of RpfA protein.
Based on the 16S rRNA gene sequences of these 137 Streptomyces strains, a phylogenetic tree was constructed (Figure 8). It was found that the five Streptomyces strains isolated from the control groups without RpfA protein addition clustered into two small groups.
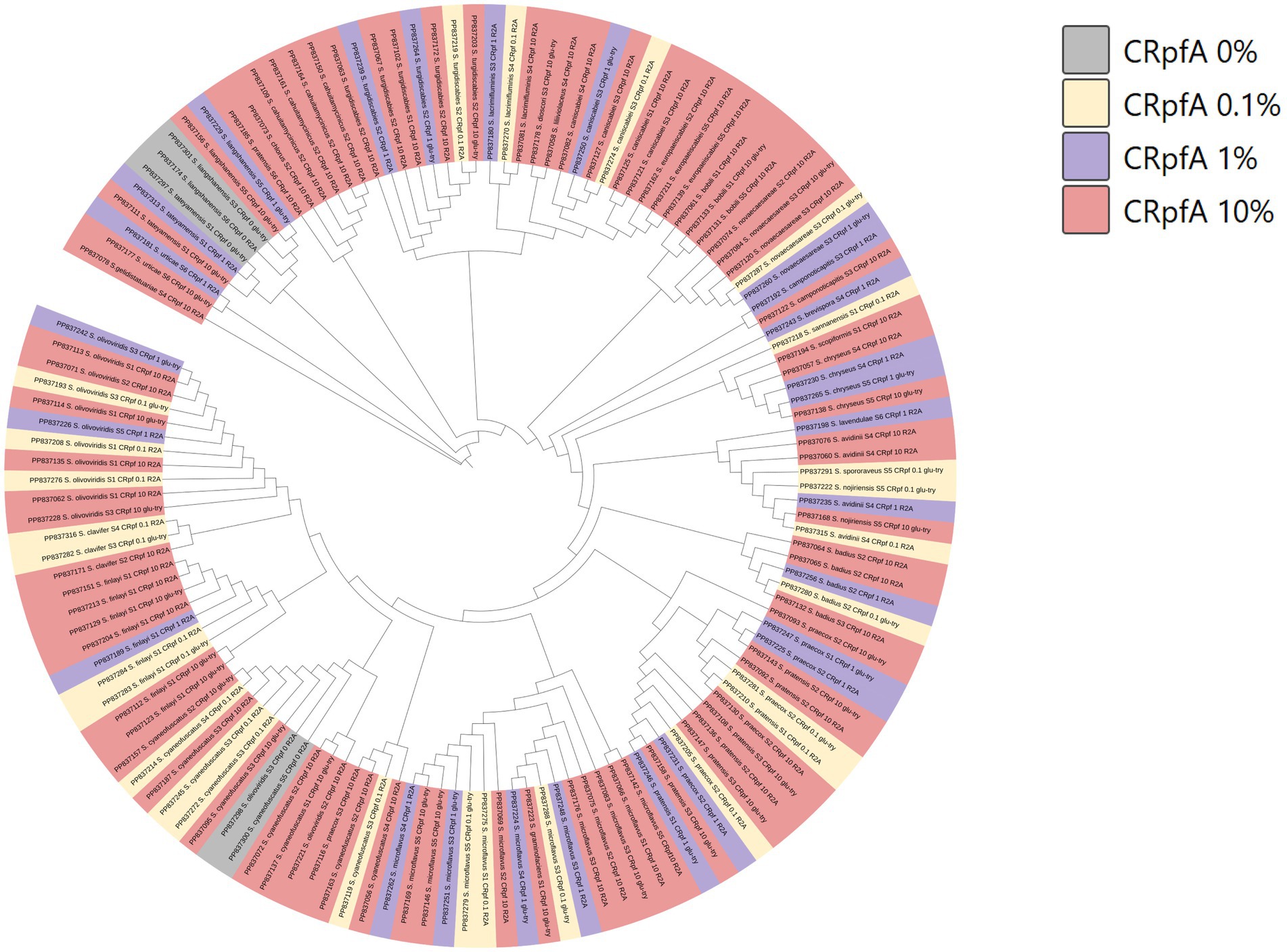
Figure 8. Phylogenetic tree constructed based on Streptomyces strains isolated from six samples. CRpfA represents the concentration of RpfA protein.
3.8 Diversity of members of phylum Actinomycetota obtained by adding RpfA protein
A total of 264 strains (Supplementary Tables S3, S10) of phylum Actinomycetotas were isolated from six soil samples collected from the frozen soils of the Qinghai-Tibet Plateau (Figure 9). The quantities and proportions different genera of phylum Actinomycetota obtained by adding different concentrations of RpfA protein are shown in Supplementary Table S11, and a phylogenetic tree was constructed based on these strains (Figure 10). When 10% RpfA protein was added, a total of 139 strains of Actinomycetota, belonging to 15 genera, were isolated, with Streptomyces being the dominant genus, accounting for 80 strains and 57.55% of the total. When 1% RpfA protein was added, 56 strains of phylum Actinomycetota, belonging to 11 genera, were isolated, with Streptomyces also being the dominant genus, accounting for 27 strains and 48.21% of the total. When 0.1% RpfA protein was added, 45 strains of phylum Actinomycetota, belonging to 9 genera, were isolated, with Streptomyces again being the dominant genus, accounting for 25 strains and 55.56% of the total. However, in the control group without RpfA protein, 24 strains of phylum Actinomycetota, belonging to 8 genera, were isolated, with Nocardia being the dominant genus, accounting for 9 strains and 37.50% of the total, while only 5 Streptomyces strains were isolated. The diversity of the members of phylum Actinomycetota isolated from the 6 samples with the addition of different concentrations of RpfA protein using R2A and glucose-tryptone media at the generic level was shown in Supplementary Figure S2.
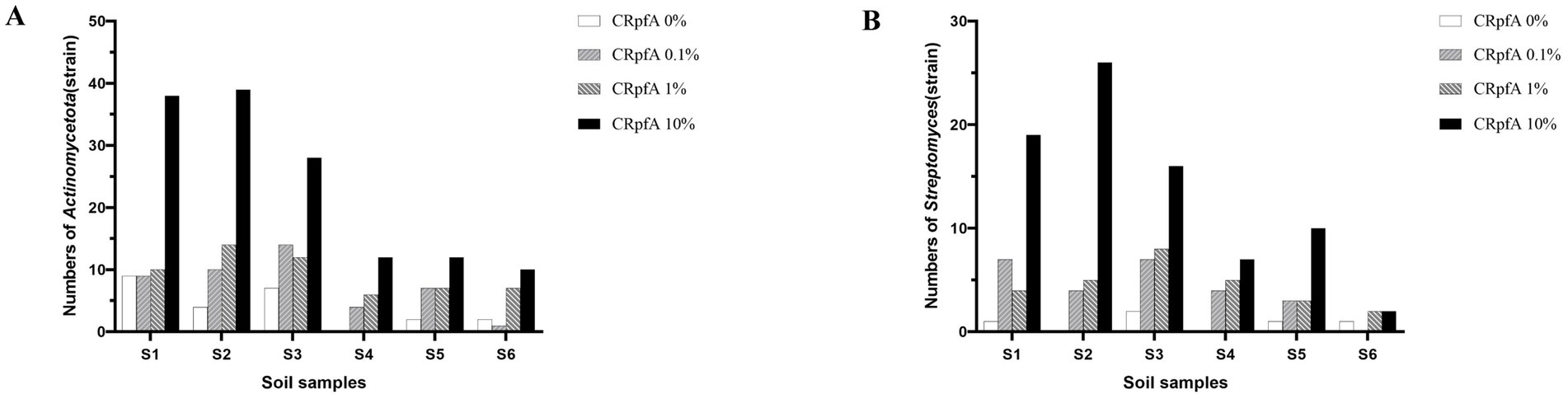
Figure 9. Statistical graphs of the quantities of strains of phylum Actinomycetota (A) and genus Streptomyces (B) strains isolated from six soil samples. CRpfA represents the concentration of RpfA protein.
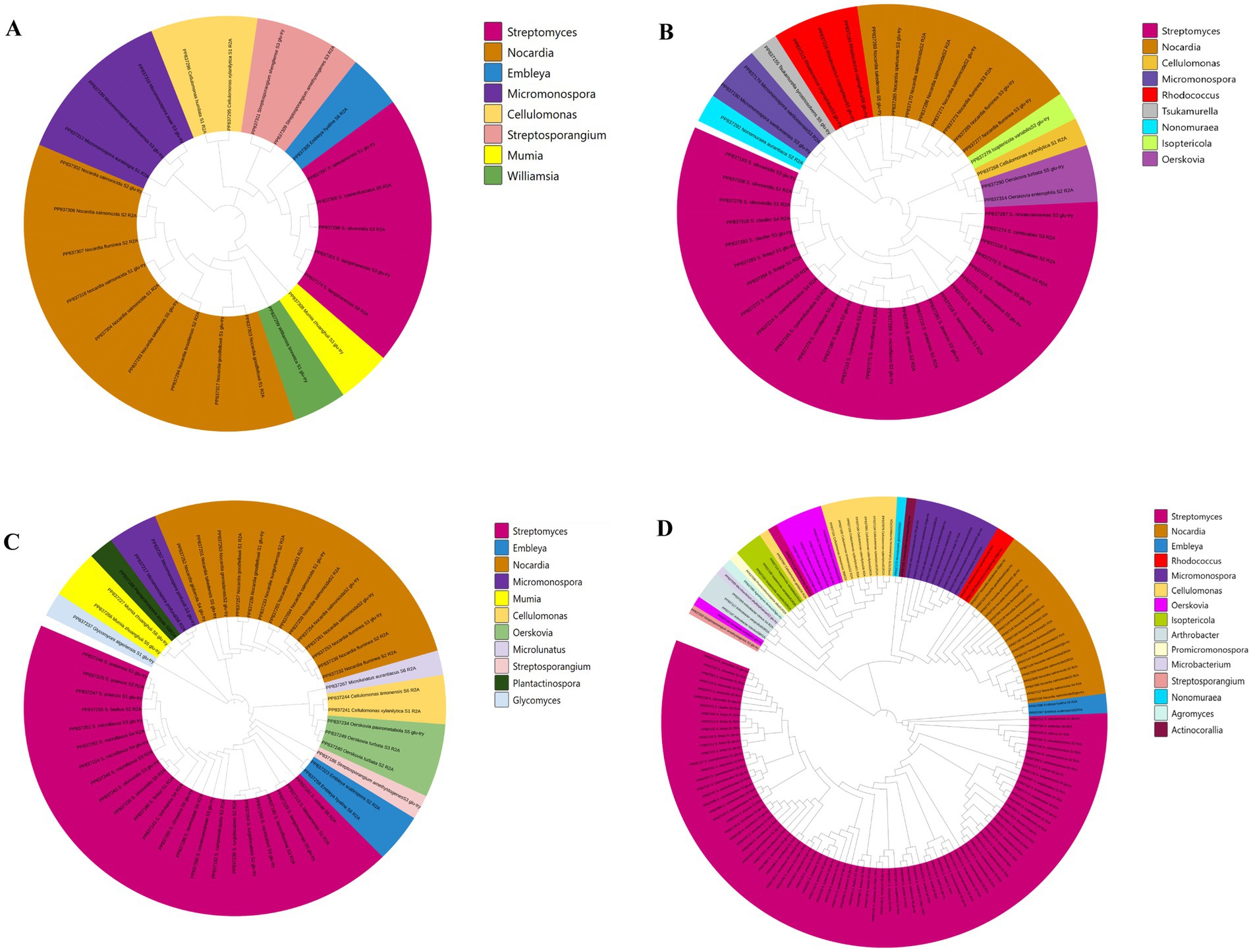
Figure 10. Phylogenetic tree constructed based on phylum Actinomycetota strains isolated from the frozen soils of the Qinghai-Tibet Plateau. (A) CRpfA 0%; (B) CRpfA 0.1%; (C) CRpfA 1%; (D) CRpfA 10%; CRpfA represents the concentration of RpfA protein.
Furthermore, it was also found that five genera were only isolated when 10% RpfA protein was added, namely Arthrobacter (2.16%), Actinocorallia (0.72%), Agromyces (0.72%), Microbacterium (0.72%) and Promicromonospora (0.72%). Three genera were only isolated when 1% RpfA protein was added, namely Glycomyces (1.79%), Microlunatus (1.79%), and Plantactinospora (1.79%). One genus, Tsukamurella (2.22%), was only isolated when the concentration of RpfA protein added was 0.1%. In contrast, Williamsia (4.17%) was only isolated in the absence of RpfA protein.
3.9 Potential new species of phylum Actinomycetota
Among the isolated strains of phylum Actinomycetota, a total of 21 strains exhibited <98.65% similarity in their 16S rRNA gene sequences to the closest type strains of their relatives, falling below the threshold for species delineation in prokaryotes (Kim et al., 2014). These were considered potential new species, accounting for 15.3% of the isolated strains, including 11 within the genus Streptomyces and 10 in other genera (Supplementary Table S12). In the R2A medium, when 10% RpfA protein is added, 6 potential new species of the Streptomyces genus, 2 potential new species of the Cellulomonas genus, and 1 potential new species of the Embleya genus are isolated; when 1% RpfA protein is added, 2 potential new species of the Streptomyces genus and 1 potential new species of the Embleya genus are isolated; when 0.1% RpfA protein is added, 1 potential new species of the Streptomyces genus, 1 potential new species of the Cellulomonas genus, and 1 potential new species of the Nonomuraea genus are isolated; when no RpfA protein is added, 1 potential new species of the Nocardia genus and 1 potential new species of the Embleya genus are isolated. In the glucose-tryptone medium, when 10% RpfA protein is added, 1 potential new species of the Streptomyces genus and 1 potential new species of the Micromonospora genus are isolated; when no RpfA protein is added, 1 potential new species of the Streptomyces genus and 1 potential new species of the Williamsia genus are isolated.
4 Discussion
In this study, bioinformatics method was used to predict the structure and related information of the RpfA protein of S. coelicolor. The instability index (II) value of this protein is 25.39. Generally, a protein with an II value less than 40 is considered to be stable (Gamage et al., 2019), indicating that this protein is relatively stable. According to the analysis results of the physicochemical properties of the amino acid sequence encoded by the rpfA gene of S. coelicolor, the hydrophilic amino acids in the RpfA protein amino acid sequence are slightly more than the hydrophobic amino acids, and the amino acid hydrophilicity index is around −1, indicating that the RpfA protein is a hydrophilic protein. The prediction and analysis by the online software Signal P-5.0 show that RpfA has a signal peptide. The SP (Sec/SPI) curve indicates that RpfA is a typical secretory protein. The signal peptide binds to receptors on the cell membrane, guiding the transport of proteins and closely related to the soluble expression of proteins (Gong et al., 2022). Therefore, it is speculated that the RpfA protein can be expressed in a soluble form. The RpfA protein has 23 serine (Ser) phosphorylation sites, 10 threonine (Thr) phosphorylation sites, and 4 tyrosine (Tyr) phosphorylation sites (Figure 1C). It is speculated that the RpfA protein may participate in cell wall activities through reversible phosphorylation, which is thought to further promote to cell resuscitation (Cohen-Gonsaud et al., 2005; Cohen-Gonsaud et al., 2004). The RpfA protein-encoding chain has two domains: one Rpf catalytic domain for hydrolyzing β-1,4-glycosidic bonds and one Lysin (LysM, C121525) domain related to the properties of lysozyme (Buist et al., 2008). The homologous modeling results of S. coelicolor rpfA gene showed that the RpfA protein had the highest homology with M. tuberculosis rpfB, and the sequence matching degree was 49% (<70% was identified as a new protein), which contained a complete domain (Nikitushkin et al., 2015). The RpfA protein of S. coelicolor has a glycosylase - like domain in structure, with glutamic acid as its key catalytic site and threonine and tryptophan as the substrate - binding residues. These results lay a foundation for the subsequent research work and are helpful to further reveal the biological function of this protein in S. coelicolor.
In this study, the rpfA gene from S. coelicolor was heterologously expressed in E. coli, resulting in the production of recombinant RpfA protein with a molecular weight of approximately 24.9 kDa. The effect of the purified RpfA protein on the spore germination rate of different species of the genus Streptomyces was determined. The RpfA protein can promote the germination of its own spores as well as those of other Streptomyces species. Furthermore, it was found that inactivated RpfA protein did not promote spore germination (Figure 4), further proving that RpfA protein promotes spore germination not as a nutrient but as a protein with resuscitation function. It has also been shown that the products encoded by the five rpf genes (rpfA-E) of S. coelicolor can promote the rapid germination of dormant spores and affect the vegetative cgrowth and sporulation (Haiser et al., 2009; Sexton et al., 2015), which is consistent with the above results. The effect of RpfA protein on the viability of spore germination cells was determined using the BacTiter-Glo™ Microbial Cell Viability Assay Kit, and the assay results showed that, compared with the control group, the experimental group with the optimal concentration of RpfA protein had continuously increasing RLU values over time, which were significantly higher than those of the control group. This indicated that after adding RpfA protein, the cell viability during spore germination was more vigorous and the metabolic level was higher, further proving that the RpfA protein of S. coelicolor can promote the germination of its own spores as well as those of other Streptomyces species. The optimal concentration of this RpfA protein for promoting spore germination varies among species, and not always is a higher concentration of RpfA protein associated with a better effect on spore germination. It is speculated that this difference in activity and concentration may be related to differences in the genome and growth cycle of different Streptomyces species (Flärdh and Buttner, 2009), as well as the copy number, length of the rpfs gene, and genetic diversity of Rpf proteins (Tantivitayakul et al., 2020). This hypothesis awaits further in-depth analysis at the genetic level and more exploration of the Rpf protein family in the future.
It was reported that D2O had no significant effect on spore germination and initial growth (Öberg et al., 2023; Zhang et al., 2020). Therefore, the method combining D2O isotope labeling with single-cell Raman spectroscopy was adopted to further verify the effect of RpfA protein on the metabolic activity during spore germination of S. coelicolor CGMCC 4.1658T, S. niveus JCM 4251T, S. griseus CGMCC 4.1419T, S. laurentii JCM 5063T, and S. lusitanus HBU208066T (Figure 6). The experimental group with added RpfA protein had a higher cellular metabolic activity during spore germination compared to the control group, and the intensities of the C-D and C-H peaks in the experimental group were also higher, suggesting that due to the higher metabolic level, more intracellular substances were synthesized, resulting in a higher concentration of chemical bonds. The aforementioned studies fully demonstrate that the RpfA protein of S. coelicolor can promote the spore germination of its own as well as other Streptomyces species. Additionally, our previous work validated the activity of the Rpf protein from Nocardiosis salophila CGMCC 4.1195T, confirming its significant resuscitation effect on N. salophila in the viable but non-culturable (VBNC) state (Zhang et al., 2023). These findings have laid the foundation for establishing new methods to isolate unculturable or difficult-to-culture Actinomycetes based on the resuscitation function of Rpfs proteins.
A total of 264 strains (Supplementary Tables S3, S10) of phylum Actinomycetotas were isolated from six soil samples collected from the frozen soils of the Qinghai-Tibet Plateau. The result showed that the RpfA protein from S. coelicolor also promotes the isolation and cultivation of members of phylum Actinomycetota, with the most significant promotion effect observed on Streptomyces species, accounting for more than 50% of the total. There is a correlation between the quantity of strains of genus Streptomyces obtained and the concentration of RpfA protein added. In this study, the highest diversity and quantity of strains of genus Streptomyces were obtained when 10% RpfA protein was added. Based on the 16S rRNA gene sequences of these 137 Streptomyces strains, a phylogenetic tree was constructed (Figure 8). It was found that the five Streptomyces strains isolated from the control groups without RpfA protein addition clustered into two small groups, indicating that these species, which do not rely on the presence of RpfA protein for growth, have relatively close genetic relationships. This further suggests that the RpfA protein from S. coelicolor exhibits species selectivity in enhancing the cultivability of strains of genus Streptomyces, which is consistent with the previous observation that RpfA protein has differential effects on promoting spore germination activity among different Streptomyces species. The reason of why S. liangshanensis was only isolated from sample S3 and S6 without RpfA protein, but not from two samples with RpfA protein, might be that the species were not sensitive to the resuscitation effect of RpfA protein, or RpfA protein might have inhibitory effect on it to some extent. And different species of phylum Actinomycetota have varying sensitivities to Rpf protein. Most species of phylum Actinomycetota require higher concentrations of RpfA protein to promote their resuscitation and growth, while a few species only need low concentrations of RpfA protein to stimulate their growth, and high concentrations of RpfA protein may even be not conducive to their resuscitation growth. In general, the heterologously expressed RpfA protein from S. coelicolor can effectively enhance the cultivability of members of phylum Actinomycetotas, especially genus Streptomyces, of frozen soil samples on the Qinghai-Tibet Plateau.
The introduction of Rpf offers a novel approach for reviving difficult-to-culture microorganisms. Previous studies have also indicated that Rpf proteins derived from M. luteus and other species can enhance the cultivability of members of phylum Actinomycetota (Puspita et al., 2012; Wang et al., 2021). Furthermore, numerous studies have demonstrated that the addition of Rpfs proteins can resuscitate many strains of phylum Actinomycetota in a viable but non-culturable (VBNC) state that possess pollutant-degrading capabilities in the environment, thereby enhancing their biodegradation capacity for pollutants (Lothigius et al., 2010; Luo et al., 2019; Su et al., 2021; Su et al., 2018; Ye et al., 2020; Yu et al., 2020). However, there are few systematic studies on the effect of Rpf for cultivation of soil Streptomyces.
Among the isolates of phylum Actinomycetota, there were 21 potential new species, of which 8 belonged to genus Streptomyces, indicating that the RpfA protein of S. coelicolor could promote the isolation of new species of Actinomycetota, especially Streptomyces. The isolation rate of new species was also related to the type of medium used. Lopez et al. reported that Rpf proteins facilitated the isolation of 51 potential novel bacterial species (Lopez Marin et al., 2021). Wang et al. also isolated two new species of Actinomycetota from soil samples treated with Rpf (Wang et al., 2021). These studies provided a methodological reference for using Rpf proteins to excavate new species resources of phylum Actinomycetota.
5 Conclusion
Through this investigation, the purified RpfA protein of S. coelicolor was obtained by heterologous expression in E. coli. The RpfA protein of S. coelicolor significantly promotes the spore germination of itself and other Streptomyces species, and can significantly improve the cultivability of members of phylum Actinomycetota, especially genus Streptomyces and its potential new species in the frozen soil samples of the Qinghai-Tibet Plateau, the results support and verify the conclusion that RpfA protein can effectively increase the “cultivability” of actinobacteria under laboratory conditions. This provides a new method for exploring and preserving Streptomyces species resources in the unique environment of the Qinghai-Tibet Plateau and further developing new bioactive substances derived from Streptomyces.
In this study, the concentration of RpfA protein from S. coelicolor required to promote spore germination in itself and across species was observed to vary depending on the species. It is not the case that higher concentrations of RpfA protein leading to better spore germination promotion. This variation in activity and concentration may be related to differences in the genomes of various Streptomyces species, including the copy number, length, genetic structure of rpf genes (Tantivitayakul et al., 2020), host specificity, and differences in growth cycles (Flärdh and Buttner, 2009). For instance, S. coelicolor has five copies of rpf genes, while the other four Streptomyces strains tested have only four copies (Gong et al., 2022). In this study, RpfA protein was added at the early growth stage, leading to the speculation that different species exhibit varying sensitivities to RpfA protein at different growth stages. This is reflected in the significant differences in the concentration of RpfA protein required to promote spore germination in different Streptomyces species. This hypothesis awaits further analysis at the genetic level and more in-depth exploration of the Rpf protein family in the future.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
YX: Data curation, Writing – original draft, Writing – review & editing. JL: Data curation, Methodology, Resources, Software, Writing – review & editing. JM: Writing – review & editing. NS: Writing – review & editing. XZ: Funding acquisition, Methodology, Project administration, Supervision, Validation, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was funded by the National Natural Science Foundation of China (Grant No. 32270020).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1557511/full#supplementary-material
Footnotes
2. ^https://www.ebi.ac.uk/Tools/hmmer/
3. ^https://www.ncbi.nlm.nih.gov
4. ^https://www.ezbiocloud.net/
7. ^https://services.healthtech.dtu.dk/services/SignalP-5.0/
8. ^https://services.healthtech.dtu.dk/services/NetPhos-3.1/
References
Abbasi, R., Imanbekova, M., and Wachsmann-Hogiu, S. (2024). On-chip bioluminescence biosensor for the detection of microbial surface contamination. Biosens. Bioelectron. 254:116200. doi: 10.1016/j.bios.2024.116200
Arroyo, L., Marín, D., Franken, K., Ottenhoff, T. H. M., and Barrera, L. F. (2018). Potential of DosR and Rpf antigens from Mycobacterium tuberculosis to discriminate between latent and active tuberculosis in a tuberculosis endemic population of Medellin Colombia. BMC Infect. Dis. 18:26. doi: 10.1186/s12879-017-2929-0
Barka, E. A., Vatsa, P., Sanchez, L., Gaveau-Vaillant, N., Jacquard, C., Meier-Kolthoff, J. P., et al. (2016). Taxonomy, physiology, and natural products of actinobacteria. Microbiol. Mol. Biol. Rev. 80, 1–43. doi: 10.1128/mmbr.00019-15
Bérdy, J. (2012). Thoughts and facts about antibiotics: where we are now and where we are heading. J. Antibiot. (Tokyo) 65, 385–395. doi: 10.1038/ja.2012.27
Berry, D., Mader, E., Lee, T. K., Woebken, D., Wang, Y., Zhu, D., et al. (2015). Tracking heavy water (D2O) incorporation for identifying and sorting active microbial cells. Proc. Natl. Acad. Sci. USA 112, E194–E203. doi: 10.1073/pnas.1420406112
Buist, G., Steen, A., Kok, J., and Kuipers, O. P. (2008). LysM, a widely distributed protein motif for binding to (peptido)glycans. Mol. Microbiol. 68, 838–847. doi: 10.1111/j.1365-2958.2008.06211.x
Cava, F., and de Pedro, M. A. (2014). Peptidoglycan plasticity in bacteria: emerging variability of the murein sacculus and their associated biological functions. Curr. Opin. Microbiol. 18, 46–53. doi: 10.1016/j.mib.2014.01.004
Chen, Y. G., Jiang, Y., Li, W. J., Cui, X. L., and Xu, L. H. (2007). Micirobial diversity and screening of antitumor activity of Actinomycete strains from saline and alkaline environments in the Qinghai Province, P. R. China. Wei Sheng Wu Xue Bao 47, 757–762. doi: 10.13343/j.cnki.wsxb.2007.05.001
Cohen-Gonsaud, M., Barthe, P., Bagnéris, C., Henderson, B., Ward, J., Roumestand, C., et al. (2005). The structure of a resuscitation-promoting factor domain from Mycobacterium tuberculosis shows homology to lysozymes. Nat. Struct. Mol. Biol. 12, 270–273. doi: 10.1038/nsmb905
Cohen-Gonsaud, M., Keep, N. H., Davies, A. P., Ward, J., Henderson, B., and Labesse, G. (2004). Resuscitation-promoting factors possess a lysozyme-like domain. Trends Biochem. Sci. 29, 7–10. doi: 10.1016/j.tibs.2003.10.009
Coletta-Filho, H. D., Takita, M. A., Souza, A. A., Neto, J. R., Destéfano, S. A., Hartung, J. S., et al. (2006). Primers based on the rpf gene region provide improved detection of Xanthomonas axonopodis pv. Citri in naturally and artificially infected citrus plants. J. Appl. Microbiol. 100, 279–285. doi: 10.1111/j.1365-2672.2005.02787.x
de Lamballerie, X., Zandotti, C., Vignoli, C., Bollet, C., and de Micco, P. (1992). A one-step microbial DNA extraction method using "Chelex 100" suitable for gene amplification. Res. Microbiol. 143, 785–790. doi: 10.1016/0923-2508(92)90107-y
Flärdh, K., and Buttner, M. J. (2009). Streptomyces morphogenetics: dissecting differentiation in a filamentous bacterium. Nat. Rev. Microbiol. 7, 36–49. doi: 10.1038/nrmicro1968
Gamage, D. G., Gunaratne, A., Periyannan, G. R., and Russell, T. G. (2019). Applicability of instability index for in vitro protein stability prediction. Protein Pept. Lett. 26, 339–347. doi: 10.2174/0929866526666190228144219
Gao, H., Li, G., and Lou, H. X. (2018). Structural diversity and biological activities of novel secondary metabolites from endophytes. Molecules 23:646. doi: 10.3390/molecules23030646
Gong, X., Lu, H., Wu, J., Zhou, Y., Yang, L., Wang, Y., et al. (2022). Enzymatic properties and biological activity of resuscitation-promoting factor B of Rhodococcus sp. (GX12401). Front. Microbiol. 13:965843. doi: 10.3389/fmicb.2022.965843
Haiser, H. J., Yousef, M. R., and Elliot, M. A. (2009). Cell wall hydrolases affect germination, vegetative growth, and sporulation in Streptomyces coelicolor. J. Bacteriol. 191, 6501–6512. doi: 10.1128/jb.00767-09
Hua, X., He, J., Wang, J., Zhang, L., Zhang, L., Xu, Q., et al. (2021). Novel tigecycline resistance mechanisms in Acinetobacter baumannii mediated by mutations in adeS, rpoB and rrf. Emerg. Microbes Infect. 10, 1404–1417. doi: 10.1080/22221751.021.1948804
Huang, W. E., Li, M., Jarvis, R. M., Goodacre, R., and Banwart, S. A. (2010). Shining light on the microbial world the application of Raman microspectroscopy. Adv. Appl. Microbiol. 70, 153–186. doi: 10.1016/s0065-2164(10)70005-8
Karimi, E., Nikkhah, M., and Hosseinkhani, S. (2022). Label-free and bioluminescence-based nano-biosensor for ATP detection. Biosensors (Basel) 12:918. doi: 10.3390/bios12110918
Kell, D. B., and Young, M. (2000). Bacterial dormancy and culturability: the role of autocrine growth factors. Curr. Opin. Microbiol. 3, 238–243. doi: 10.1016/s1369-5274(00)00082-5
Kim, O. S., Cho, Y. J., Lee, K., Yoon, S. H., Kim, M., Na, H., et al. (2012). Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int. J. Syst. Evol. Microbiol. 62, 716–721. doi: 10.1099/ijs.0.038075-0
Kim, M., Oh, H. S., Park, S. C., and Chun, J. (2014). Towards a taxonomic coherence between average nucleotide identity and 16S rRNA gene sequence similarity for species demarcation of prokaryotes. Int. J. Syst. Evol. Microbiol. 64, 346–351. doi: 10.1099/ijs.0.059774-0
Kuester, E., and Williams, S. T. (1964). Selection of media for isolation of Streptomycetes. Nature 202, 928–929. doi: 10.1038/202928a0
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lackner, G., Peters, E. E., Helfrich, E. J., and Piel, J. (2017). Insights into the lifestyle of uncultured bacterial natural product factories associated with marine sponges. Proc. Natl. Acad. Sci. USA 114, E347–e356. doi: 10.1073/pnas.1616234114
Lefebvre, T., Miambi, E., Pando, A., Diouf, M., and Rouland-Lefèvre, C. (2009). Gut-specific actinobacterial community structure and diversity associated with the wood-feeding termite species, Nasutitermes corniger (Motschulsky) described by nested PCR-DGGE analysis. Insect. Soc. 56, 269–276. doi: 10.1007/s00040-009-0020-6
Li, S., Dong, L., Lian, W. H., Lin, Z. L., Lu, C. Y., Xu, L., et al. (2021). Exploring untapped potential of Streptomyces spp. in Gurbantunggut Desert by use of highly selective culture strategy. Sci. Total Environ. 790:148235. doi: 10.1016/j.scitotenv.2021.148235
Liu, G., Chater, K. F., Chandra, G., Niu, G., and Tan, H. (2013). Molecular regulation of antibiotic biosynthesis in Streptomyces. Microbiol. Mol. Biol. Rev. 77, 112–143. doi: 10.1128/mmbr.00054-12
Locey, K. J., and Lennon, J. T. (2016). Scaling laws predict global microbial diversity. Proc. Natl. Acad. Sci. USA 113, 5970–5975. doi: 10.1073/pnas.1521291113
Lopez Marin, M. A., Strejcek, M., Junkova, P., Suman, J., Santrucek, J., and Uhlik, O. (2021). Exploring the potential of Micrococcus luteus culture supernatant with resuscitation-promoting factor for enhancing the culturability of soil bacteria. Front. Microbiol. 12:685263. doi: 10.3389/fmicb.2021.685263
Lothigius, A., Sjöling, A., Svennerholm, A. M., and Bölin, I. (2010). Survival and gene expression of enterotoxigenic Escherichia coli during long-term incubation in sea water and freshwater. J. Appl. Microbiol. 108, 1441–1449. doi: 10.1111/j.1365-2672.2009.04548.x
Luo, D., Chen, J., Xie, G., Yue, L., and Wang, Y. (2019). Enzyme characterization and biological activities of a resuscitation promoting factor from an oil degrading bacterium Rhodococcus erythropolis KB1. PeerJ 7:e6951. doi: 10.7717/peerj.6951
Luo, J., Liu, X., Tian, Q., Yue, W., Zeng, J., Chen, G., et al. (2009). Disposable bioluminescence-based biosensor for detection of bacterial count in food. Anal. Biochem. 394, 1–6. doi: 10.1016/j.ab.2009.05.021
Maione, V., Ruggiero, A., Russo, L., De Simone, A., Pedone, P. V., Malgieri, G., et al. (2015). NMR structure and dynamics of the resuscitation promoting factor RpfC catalytic domain. PLoS One 10:e0142807. doi: 10.1371/journal.pone.0142807
Mukamolova, G. V., Kaprelyants, A. S., Young, D. I., Young, M., and Kell, D. B. (1998). A bacterial cytokine. Proc. Natl. Acad. Sci. USA 95, 8916–8921. doi: 10.1073/pnas.95.15.8916
Mukamolova, G. V., Kormer, S. S., Kell, D. B., and Kaprelyants, A. S. (1999). Stimulation of the multiplication of Micrococcus luteus by an autocrine growth factor. Arch. Microbiol. 172, 9–14. doi: 10.1007/s002030050733
Ni, H., Jing, X., Xiao, X., Zhang, N., Wang, X., Sui, Y., et al. (2021). Microbial metabolism and necromass mediated fertilization effect on soil organic carbon after long-term community incubation in different climates. ISME J. 15, 2561–2573. doi: 10.1038/s41396-021-00950-w
Nikitushkin, V. D., Demina, G. R., and Kaprelyants, A. S. (2016). Rpf proteins are the factors of reactivation of the dormant forms of actinobacteria. Biochemistry (Mosc) 81, 1719–1734. doi: 10.1134/s0006297916130095
Nikitushkin, V. D., Demina, G. R., Shleeva, M. O., Guryanova, S. V., Ruggiero, A., Berisio, R., et al. (2015). A product of RpfB and RipA joint enzymatic action promotes the resuscitation of dormant mycobacteria. FEBS J. 282, 2500–2511. doi: 10.1111/febs.13292
Öberg, R., Dahlberg, T., Malyshev, D., and Andersson, M. (2023). Monitoring bacterial spore metabolic activity using heavy water-induced Raman peak evolution. Analyst 148, 2141–2148. doi: 10.1039/d2an02047e
Olanrewaju, O. S., and Babalola, O. O. (2019). Streptomyces: implications and interactions in plant growth promotion. Appl. Microbiol. Biotechnol. 103, 1179–1188. doi: 10.1007/s00253-018-09577-y
Pham, V. H., and Kim, J. (2012). Cultivation of unculturable soil bacteria. Trends Biotechnol. 30, 475–484. doi: 10.1016/j.tibtech.2012.05.007
Puspita, I. D., Kamagata, Y., Tanaka, M., Asano, K., and Nakatsu, C. H. (2012). Are uncultivated bacteria really uncultivable? Microbes Environ. 27, 356–366. doi: 10.1264/jsme2.me12092
Ravagnani, A., Finan, C. L., and Young, M. (2005). A novel firmicute protein family related to the actinobacterial resuscitation-promoting factors by non-orthologous domain displacement. BMC Genomics 6:39. doi: 10.1186/1471-2164-6-39
Reasoner, D. J., and Geldreich, E. E. (1985). A new medium for the enumeration and subculture of bacteria from potable water. Appl. Environ. Microbiol. 49, 1–7. doi: 10.1128/aem.49.1.1-7.1985
Romano, M., Aryan, E., Korf, H., Bruffaerts, N., Franken, C. L., Ottenhoff, T. H., et al. (2012). Potential of Mycobacterium tuberculosis resuscitation-promoting factors as antigens in novel tuberculosis sub-unit vaccines. Microbes Infect. 14, 86–95. doi: 10.1016/j.micinf.2011.08.011
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425. doi: 10.1093/oxfordjournals.molbev.a040454
Sexton, D. L., Herlihey, F. A., Brott, A. S., Crisante, D. A., Shepherdson, E., Clarke, A. J., et al. (2020). Roles of LysM and LytM domains in resuscitation-promoting factor (Rpf) activity and Rpf-mediated peptidoglycan cleavage and dormant spore reactivation. J. Biol. Chem. 295, 9171–9182. doi: 10.1074/jbc.RA120.013994
Sexton, D. L., St-Onge, R. J., Haiser, H. J., Yousef, M. R., Brady, L., Gao, C., et al. (2015). Resuscitation-promoting factors are cell wall-lytic enzymes with important roles in the germination and growth of Streptomyces coelicolor. J. Bacteriol. 197, 848–860. doi: 10.1128/jb.02464-14
Smith, R., Wright, K. L., and Ashton, L. (2016). Raman spectroscopy: an evolving technique for live cell studies. Analyst 141, 3590–3600. doi: 10.1039/c6an00152a
Stewart, E. J. (2012). Growing unculturable bacteria. J. Bacteriol. 194, 4151–4160. doi: 10.1128/jb.00345-12
Su, X., Li, S., Xie, M., Tao, L., Zhou, Y., Xiao, Y., et al. (2021). Enhancement of polychlorinated biphenyl biodegradation by resuscitation promoting factor (Rpf) and Rpf-responsive bacterial community. Chemosphere 263:128283. doi: 10.1016/j.chemosphere.2020.128283
Su, X., Shen, H., Yao, X., Ding, L., Yu, C., and Shen, C. (2013). A novel approach to stimulate the biphenyl-degrading potential of bacterial community from PCBs-contaminated soil of e-waste recycling sites. Bioresour. Technol. 146, 27–34. doi: 10.1016/j.biortech.2013.07.028
Su, X., Wang, Y., Xue, B., Zhang, Y., Mei, R., Zhang, Y., et al. (2018). Resuscitation of functional bacterial community for enhancing biodegradation of phenol under high salinity conditions based on Rpf. Bioresour. Technol. 261, 394–402. doi: 10.1016/j.biortech.2018.04.048
Sun, W., Weingarten, R. A., Xu, M., Southall, N., Dai, S., Shinn, P., et al. (2016). Rapid antimicrobial susceptibility test for identification of new therapeutics and drug combinations against multidrug-resistant bacteria. Emerg. Microbes Infect. 5:e116, 1–11. doi: 10.1038/emi.2016.123
Suutari, M., Lignell, U., Hyvärinen, A., and Nevalainen, A. (2002). Media for cultivation of indoor Streptomycetes. J. Microbiol. Methods 51, 411–416. doi: 10.1016/s0167-7012(02)00100-8
Tantivitayakul, P., Juthayothin, T., Ruangchai, W., Smittipat, N., Disratthakit, A., Mahasirimongkol, S., et al. (2020). Identification and in silico functional prediction of lineage-specific SNPs distributed in DosR-related proteins and resuscitation-promoting factor proteins of Mycobacterium tuberculosis. Heliyon 6:e05744. doi: 10.1016/j.heliyon.2020.e05744
van Dorst, J. M., Hince, G., Snape, I., and Ferrari, B. C. (2016). Novel culturing techniques select for heterotrophs and hydrocarbon degraders in a subantarctic soil. Sci. Rep. 6:36724. doi: 10.1038/srep36724
Vollmer, W., Blanot, D., and de Pedro, M. A. (2008). Peptidoglycan structure and architecture. FEMS Microbiol. Rev. 32, 149–167. doi: 10.1111/j.1574-6976.2007.00094.x
Wang, C., Chen, R., Xu, J., and Jin, L. (2022). Single-cell Raman spectroscopy identifies Escherichia coli persisters and reveals their enhanced metabolic activities. Front. Microbiol. 13:936726. doi: 10.3389/fmicb.2022.936726
Wang, Y., Shi, J., Tang, L., Zhang, Y., Zhang, Y., Wang, X., et al. (2021). Evaluation of Rpf protein of Micrococcus luteus for cultivation of soil actinobacteria. Syst. Appl. Microbiol. 44:126234. doi: 10.1016/j.syapm.2021.126234
Waterborg, J. H., and Matthews, H. R. (1994). The Lowry method for protein quantitation. Methods Mol. Biol. 32, 1–4. doi: 10.1385/0-89603-268-x:1
Waterhouse, A., Bertoni, M., Bienert, S., Studer, G., Tauriello, G., Gumienny, R., et al. (2018). SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–w303. doi: 10.1093/nar/gky427
Ye, Z., Li, H., Jia, Y., Fan, J., Wan, J., Guo, L., et al. (2020). Supplementing resuscitation-promoting factor (Rpf) enhanced biodegradation of polychlorinated biphenyls (PCBs) by Rhodococcus biphenylivorans strain TG9T. Environ. Pollut. 263:114488. doi: 10.1016/j.envpol.2020.114488
Yu, C., Liu, Y., Jia, Y., Su, X., Lu, L., Ding, L., et al. (2020). Extracellular organic matter from Micrococcus luteus containing resuscitation-promoting factor in sequencing batch reactor for effective nutrient and phenol removal. Sci. Total Environ. 727:138627. doi: 10.1016/j.scitotenv.2020.138627
Zhang, M., Hong, W., Abutaleb, N. S., Li, J., Dong, P. T., Zong, C., et al. (2020). Rapid determination of antimicrobial susceptibility by stimulated Raman scattering imaging of D2O metabolic incorporation in a single bacterium. Adv. Sci. (Weinh.) 7:2001452. doi: 10.1002/advs.202001452
Keywords: Streptomyces coelicolor , resuscitation-promoting factor, RpfA protein, frozen soils of the Qinghai-Tibet Plateau, resources of Streptomyces species
Citation: Xie Y, Liu J, Ma J, Shi N and Zhang X (2025) Excavation of resources of Streptomyces species in frozen soils of the Qinghai-Tibet Plateau based on RpfA protein of Streptomyces coelicolor. Front. Microbiol. 16:1557511. doi: 10.3389/fmicb.2025.1557511
Edited by:
Tatiana A. Vishnivetskaya, The University of Tennessee, Knoxville, United StatesReviewed by:
Sergii Krysenko, University of Tübingen, GermanyAli Moridi, Shahid Beheshti University, Iran
Copyright © 2025 Xie, Liu, Ma, Shi and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiumin Zhang, emh4aXVtaW4xMTA2QDEyNi5jb20=
†These authors have contributed equally to this work
 Yuxiao Xie1†
Yuxiao Xie1† Nan Shi
Nan Shi Xiumin Zhang
Xiumin Zhang