- 1Department of Microbiology, School of Medicine, University of Alabama at Birmingham, Birmingham, AL, United States
- 2Regional Biocontainment BSL-3 Research Laboratory (SEBLAB), University of Alabama at Birmingham, Birmingham, AL, United States
The coronavirus disease (COVID-19) pandemic has unveiled a complex spectrum of disease manifestations, from acute respiratory distress to persistent health effects. In its acute phase, COVID-19 unleashes a storm of lung injury, marked by diffuse alveolar damage, thrombosis, and inflammation (Bussani et al., 2020). Yet, for many, the battle doesn't end with recovery from acute symptoms. A significant number of patients experience prolonged health issues known as “long COVID-19” (Davis et al., 2023) or post-acute sequelae of SARS-CoV-2 (PASC; Swank et al., 2023), challenging our understanding of viral infections and recovery. This condition is also referred to as “persistent SARS-CoV-2 infection,” where the virus persists beyond 2 months post-initial onset (Furie et al., 2023). Another phenomenon, termed “walking pneumonia,” encompasses a range of persistent viral respiratory infections symptoms, such as those of SARS-CoV-2 infection, rather than a specific diagnosis (Walking Pneumonia: What You Should Know, n.d.). The burning question remains: does this persistence represent an extended infectious cycle, or does the virus truly transitions into a persistent state? We propose the hypothesis that SARS-CoV-2 transitions to persistent infection, facilitated by syncytia formation, which may be key to unlocking the unknowns of long COVID-19.
RNA viruses (i.e., alphaviruses) employ sophisticated strategies to avoid host defenses and develop persistent infection. Two fascinating mechanisms have emerged for RNA viruses (Schlesinger and Schlesinger, 1986)—both begin with a cytopathic viral assault, but their paths later diverge (Figure 1). In one path, the virus transitions to formation of defective interfering (DI) particles, a defective viral genome that is capable of replication and propagation in the presence of helper genome or wild type virus (Girgis et al., 2022; Weaver et al., 1999). The alternative strategy occurs when the virus overcomes the interferon defense, leaving only a small fraction of infected cells as survivors (Schlesinger and Schlesinger, 1986). These observations come from in vitro experiments, have now been confirmed in the more complex realm of coronaviruses. Syncytium, sometimes also called a “giant cell,” is a large, multi-nucleated cell that is born from cell-cell fusions (Zimmerberg et al., 1993). While syncytia naturally occur in some tissues, such as muscles, as a part of normal growth (Zimmerberg et al., 1993), their appearance can also be a sign of a more pathological outcome (Rajah et al., 2022). This cellular fusion is orchestrated by an ensemble of transmembrane and membrane-associated proteins of diverse origin (Ashorn et al., 1993; Mohan, 1992). Apart from DI particles, syncytia also serve as another hallmark of SARS-CoV-2′s persistence. Syncytia formation is a process that SARS-CoV-2 has mastered with outstanding efficiency. In vitro research on SARS-CoV-2 evolution has revealed that the spike protein that contains furin cleavage site (FCS) drives virus toward utilization of Angiotensin-Converting Enzyme 2 (ACE2)/Transmembrane serine protease 2 (TMPRSS2) receptors and syncytia formation (Frolova et al., 2022; Shiliaev et al., 2021). In the case of viral infection-induced syncytia formation, it requires two key players: cellular receptors/co-receptors and viral spike proteins (Yin et al., 2024; Zimmerberg et al., 1993). Together, they transform the cellular landscape into a playground for viral persistence.
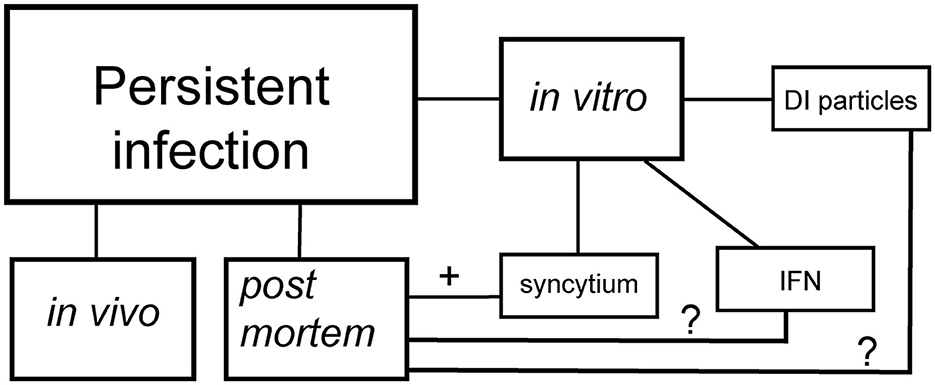
Figure 1. Schematic presentation of persistent SARS-CoV-2 infection. DI particles—the establishment of persistent infection via the emergence of defective interfering particles. IFN—the establishment of persistent infection via the interferon selection. Syncytium—the establishment of persistent infection via formation of syncytia.
The virus-induced syncytia formation phenomenon has been observed across a diverse phylogenetic spectrum, from influenzas to HIV-1 and HIV-2, from the highly contagious measles to respiratory syncytial virus (Zimmerberg et al., 1993). The SARS and MERS coronaviruses, closely related to SARS-CoV-2, have also demonstrated this cellular merger in infected tissues (Bussani et al., 2020; Chan et al., 2013; Franks et al., 2003; Hoffmann et al., 2020; Matsuyama et al., 2010; Qian et al., 2013; Tian et al., 2020), proving that syncytia formation is not merely a laboratory curiosity observed in vitro (Buchrieser et al., 2020). For viruses, this cellular fusion ability confers an outstanding advantage, helping them evade antibody neutralization and avoid the broader immune response (Frolova et al., 2022; Rajah et al., 2022). Moreover, as demonstrated in HIV-1 and HIV-2, it can serve as an alternative highway for infection spread (Pearce-Pratt and Phillips, 1993).
In the intricate dance of viral infection, TMPRSS2 emerges as a key choreographer. While not essential for host cell fusion in vitro, its presence significantly enhances the process (Frolova et al., 2022; Shiliaev et al., 2021). This leads us to an intriguing hypothesis: could TMPRSS2 expression in the population be a limiting factor for syncytia formation and, by extension, SARS-CoV-2 persistence in vivo? This could shed light on the puzzling presence of SARS-CoV-2 N protein in some endothelial cells of postmortem human lungs (Swank et al., 2023). Furthermore, several groups have demonstrated that the syncytia formed during SARS-CoV-2 infection are formidable, as they are resistant to both humoral immunity and most anti-COVID drugs. These fused cells may also act as viral factories, producing S protein that sheds, fueling the inflammatory fire (Frolova et al., 2022; Swank et al., 2023). Yet, the question of whether these viruses can use syncytia as intercellular bridges for spread in vivo remains a mystery.
In the ongoing saga of long COVID-19, one compelling theory points to persistent viral reservoirs lurking in certain tissues. These viral hideouts could nestle in neuronal tissue, brain glia, the vagus nerve, endothelium, or bronchi, potentially arising from survival selection under interferon pressure.
Autopsy of postmortem lung tissue from confirmed SARS-CoV-2 cases has revealed a landscape dotted with abnormal pneumocytes and syncytia, both pseudo and genuine (Bussani et al., 2020). These findings are invaluable, offering a frozen snapshot of syncytia formation in vivo, almost impossible to observe in real-time in living subjects. Intriguingly, affected tissues are often those less accessible to immune cells or hidden by barriers in the central nervous system (CNS; Daneman and Prat, 2015; Gopallawa et al., 2023).
In a twist of viral persistence, some COVID-19 cases have shown a delayed viral presence, with particles detected beyond 3 weeks post-infection, coinciding with significant lung fibrosis (Bussani et al., 2020; Swank et al., 2023). The authors suggested the hypothesis of a remote viral influence, where the pathogen's effects linger long after its clearance, possibly through the action of shed S1 protein (Frolova et al., 2022).
Unlike its syncytia-forming viral cousins such as HIV or CMV, SARS-CoV-2 hasn't yet been caught causing latent or persistent infection, meaning it cannot integrate its genetic material into host cell genomes. However, as a member of the RNA+ group of viruses, SARS-CoV-2 might have a few tricks for establishing persistence. Three potential strategies emerge: the development of DI particles, the selection of specific mutations, or the modulation of interferon production (Weiss et al., 1980). While interferon modulation seems to align best with observations from acute COVID-19 autopsies (Swank et al., 2023), we can't rule out the DI particle strategy. These viral particles have been spotted in vitro (Girgis et al., 2022), tempting us to wonder if they might also be playing a similar, low frequency game in vivo. Reports of SARS-CoV-2 lingering in pneumocytes and endothelial cells (Bussani et al., 2020) have been attributed to slow viral replication, but could syncytia-mediated spread be buying the virus extra time to evade clearance?
In conclusion, we find ourselves facing a gap in our understanding of how SARS-CoV-2 outmaneuvers the immune response to establish persistent infection. The possibility that syncytia serve as viral “escape rooms” requires deeper investigation. Additionally, the potential role of DI particles in vivo remains an open question, calling for further research to unravel these viral mysteries.
Author contributions
OP: Writing – original draft, Writing – review & editing, Conceptualization, Investigation, Visualization. FD: Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was funded by NIAID Regional Biocontainment Lab (RBL) awards UC6AI058599, G20AI167409, G20AI167409, and UC7AI180255.
Acknowledgments
The authors thank Professor Scott Ballinger for valuable suggestions and critical reading of the draft. Authors also thanks the University of Alabama and Heersink School of Medicine for the opportunity to develop their perspective in virology.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Gen AI was used in the creation of this manuscript.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ashorn, P., Berger, E. A., and Moss, B. (1993). Vaccinia virus vectors for study of membrane fusion mediated by human immunodeficiency virus envelope glycoprotein and CD4. Methods Enzymol. 221, 12–18. doi: 10.1016/0076-6879(93)21004-R
Buchrieser, J., Dufloo, J., Hubert, M., Monel, B., Planas, D., Rajah, M. M., et al. (2020). Syncytia formation by SARS-CoV-2-infected cells. EMBO J. 39:e106267. doi: 10.15252/embj.2020106267
Bussani, R., Schneider, E., Zentilin, L., Collesi, C., Ali, H., Braga, L., et al. (2020). Persistence of viral RNA, pneumocyte syncytia and thrombosis are hallmarks of advanced COVID-19 pathology. EBioMedicine 61:103104. doi: 10.1016/j.ebiom.2020.103104
Chan, J. F.-W., Chan, K.-H., Choi, G. K.-Y., To, K. K.-W., Tse, H., Cai, J.-P., et al. (2013). Differential cell line susceptibility to the emerging novel human betacoronavirus 2c EMC/2012: implications for disease pathogenesis and clinical manifestation. J. Infect. Dis. 207, 1743–1752. doi: 10.1093/infdis/jit123
Daneman, R., and Prat, A. (2015). The blood–brain barrier. Cold Spring Harb. Perspect. Biol. 7:a020412. doi: 10.1101/cshperspect.a020412
Davis, H. E., McCorkell, L., Vogel, J. M., and Topol, E. J. (2023). Long COVID: major findings, mechanisms and recommendations. Nat. Rev. Microbiol. 21, 133–146. doi: 10.1038/s41579-022-00846-2
Franks, T. J., Chong, P. Y., Chui, P., Galvin, J. R., Lourens, R. M., Reid, A. H., et al. (2003). Lung pathology of severe acute respiratory syndrome (SARS): a study of 8 autopsy cases from Singapore. Hum. Pathol. 34, 743–748. doi: 10.1016/S0046-8177(03)00367-8
Frolova, E. I., Palchevska, O., Lukash, T., Dominguez, F., Britt, W., and Frolov, I. (2022). Acquisition of furin cleavage site and further SARS-CoV-2 evolution change the mechanisms of viral entry, infection spread, and cell signaling. J. Virol. 96:e0075322. doi: 10.1128/jvi.00753-22
Furie, N., Mandelboim, M., Zuckerman, N., Belkin, A., Seluk, L., Shafran, I., et al. (2023). Persistent severe acute respiratory syndrome coronavirus 2 pneumonia in patients treated with anti-CD20 monoclonal antibodies. Open Forum Infect. Dis. 10:ofad464. doi: 10.1093/ofid/ofad464
Girgis, S., Xu, Z., Oikonomopoulos, S., Fedorova, A. D., Tchesnokov, E. P., Gordon, C. J., et al. (2022). Evolution of naturally arising SARS-CoV-2 defective interfering particles. Commun. Biol. 5:1140. doi: 10.1038/s42003-022-04058-5
Gopallawa, I., Dehinwal, R., Bhatia, V., Gujar, V., and Chirmule, N. (2023). A four-part guide to lung immunology: invasion, inflammation, immunity, and intervention. Front. Immunol. 14:1119564. doi: 10.3389/fimmu.2023.1119564
Hoffmann, M., Kleine-Weber, H., and Pöhlmann, S. (2020). A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol. Cell 78, 779–784.e5. doi: 10.1016/j.molcel.2020.04.022
Matsuyama, S., Nagata, N., Shirato, K., Kawase, M., Takeda, M., and Taguchi, F. (2010). Efficient activation of the severe acute respiratory syndrome coronavirus spike protein by the transmembrane protease TMPRSS2. J. Virol. 84, 12658–12664. doi: 10.1128/JVI.01542-10
Mohan, P. (1992). Anti-AIDS drug development—challenges and strategies. Pharm. Res. 9, 703–714. doi: 10.1023/A:1015882901078
Pearce-Pratt, R., and Phillips, D. M. (1993). Studies of adhesion of lymphocytic cells: implications for sexual transmission of human immunodeficiency virus1. Biol. Reprod. 48, 431–445. doi: 10.1095/biolreprod48.3.431
Qian, Z., Dominguez, S. R., and Holmes, K. V. (2013). Role of the spike glycoprotein of human middle east respiratory syndrome coronavirus (MERS-CoV) in virus entry and syncytia formation. PLoS ONE 8:e76469. doi: 10.1371/journal.pone.0076469
Rajah, M. M., Bernier, A., Buchrieser, J., and Schwartz, O. (2022). The mechanism and consequences of SARS-CoV-2 spike-mediated fusion and syncytia formation. J. Mol. Biol. 434:167280. doi: 10.1016/j.jmb.2021.167280
Schlesinger, S., and Schlesinger, M. J. (1986). The Togaviridae and Flaviviridae, eds. S. Schlesinger and M. J. Schlesinger (Springer: New York). doi: 10.1007/978-1-4757-0785-4
Shiliaev, N., Lukash, T., Palchevska, O., Crossman, D. K., Green, T. J., Crowley, M. R., et al. (2021). Natural and recombinant SARS-CoV-2 isolates rapidly evolve in vitro to higher infectivity through more efficient binding to heparan sulfate and reduced S1/S2 cleavage. J. Virol. 95:e0135721. doi: 10.1128/JVI.01357-21
Swank, Z., Senussi, Y., Manickas-Hill, Z., Yu, X. G., Li, J. Z., Alter, G., et al. (2023). Persistent circulating severe acute respiratory syndrome coronavirus 2 spike is associated with post-acute coronavirus disease 2019 sequelae. Clin. Infect. Dis. 76, e487–e490. doi: 10.1093/cid/ciac722
Tian, S., Hu, W., Niu, L., Liu, H., Xu, H., and Xiao, S.-Y. (2020). Pulmonary pathology of early-phase 2019 novel coronavirus (COVID-19) pneumonia in two patients with lung cancer. J. Thorac. Oncol. 15, 700–704. doi: 10.1016/j.jtho.2020.02.010
Walking Pneumonia: What You Should Know. (n.d.). Available online at: https://www.yalemedicine.org/news/walking-pneumonia (accessed October 13, 2024).
Weaver, S. C., Brault, A. C., Kang, W., and Holland, J. J. (1999). Genetic and fitness changes accompanying adaptation of an arbovirus to vertebrate and invertebrate cells. J. Virol. 73, 4316–4326. doi: 10.1128/JVI.73.5.4316-4326.1999
Weiss, B., Rosenthal, R., and Schlesinger, S. (1980). Establishment and maintenance of persistent infection by sindbis virus in BHK cells. J. Virol. 33, 463–474. doi: 10.1128/jvi.33.1.463-474.1980
Yin, P., Martin, C. K., and Kielian, M. (2024). Virus stealth technology: tools to study virus cell-to-cell transmission. PLoS Pathogens 20:e1012590. doi: 10.1371/journal.ppat.1012590
Keywords: persistent infection, SARS-CoV-2, syncytia, cell-cell fusion, long COVID
Citation: Palchevska O and Dominguez F (2025) Syncytium: the viral escape room secret to persistent infection of SARS-CoV-2. Front. Microbiol. 16:1561274. doi: 10.3389/fmicb.2025.1561274
Received: 15 January 2025; Accepted: 14 May 2025;
Published: 04 June 2025.
Edited by:
Subhash C. Verma, University of Nevada, Reno, United StatesReviewed by:
Christian Barbato, National Research Council (CNR), ItalyErica Diani, University of Verona, Italy
Copyright © 2025 Palchevska and Dominguez. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Oksana Palchevska, b3BhbGNoZXZAdWFiLmVkdQ==
 Oksana Palchevska
Oksana Palchevska Francisco Dominguez2
Francisco Dominguez2