- 1Department of Microbiology and Immunology, School of Medicine, Xi'an Jiaotong University, Xi'an, China
- 2Shaanxi Center for Disease Control and Prevention, Xi’an, China
- 3Department of Rehabilitation Medicine, Zhujiang Hospital, Southern Medical University, Guangzhou, China
- 4Department of Public Health, Xi’an Medical University, Xi’an, China
- 5Xi’an Center for Disease Control and Prevention, Xi’an, China
- 6Yanta Center for Disease Control and Prevention, Xi’an, China
- 7Hanzhong Center for Disease Control and Prevention, Hanzhong, China
- 8Ankang Center for Disease Control and Prevention, Ankang, China
- 9Weinan Center for Disease Control and Prevention, Weinan, China
- 10Yulin Center for Disease Control and Prevention, Yulin, China
Introduction: Monophasic Salmonella Typhimurium, characterized by the absence of phase II flagellar antigens, has become increasingly prevalent as a foodborne pathogen, raising significant public health concerns due to its multidrug resistance. This study investigated the genomic characteristics and antibiotic resistance profiles of the monophasic Salmonella Typhimurium strains isolated from patients and food sources in Shaanxi Province, China.
Methods: A total of 58 strains were collected between 2020 and 2021, with 4 strains isolated from food and 54 from patients. Whole genome sequencing was performed to assess genomic features. Antimicrobial susceptibility was tested against 17 antimicrobial agents using the broth dilution method, while pulsed-field gel electrophoresis (PFGE) and multi-locus sequence typing were employed for genetic characterization and epidemiological analysis. Phylogenetic analysis was conducted using single nucleotide polymorphism clustering.
Results and discussion: Our results revealed that all the strains belonged to the ST34 and did not carry virulence genes on pSLT (NC_003277). There were 12 strains carrying the STM2757 gene. The isolates exhibited a considerable diversity in PFGE subtypes. Phenotypic antimicrobial resistance showed that the strains were most resistant to tetracycline (94.34%; 50/53) and ampicillin (94.34%; 50/53), followed by streptomycin (88.68%; 47/53) and ampicillin/sulbactam (64.15%; 34/53). Resistance gene prediction highlighted the presence of 64 distinct genes, with aac(6’)-Iaa found in all strains (100%) and tet(B) in 93.1% of strains. Notably, the floR gene, relevant for resistance to phenicols, was observed in 44.83% of isolates. Genomic analysis revealed that 96.55% of strains were positive for the sodC1 virulence gene, whereas only 10.34% carried the sopE gene. The most plasmid replicon was IncQ1 (84.48%; 49/58), followed by IncHI2 (32.76%; 19/58) and IncHI2A (32.76%; 19/58). Single nucleotide polymorphism analysis showed that 2 strains were clustered together with SRR17830210 (UK outbreak isolate) with a bootstrap value of 0.949. There were only 12 allelic differences between SNXiAn21SAL011 and the reference strain. Conclusively, the monophasic Salmonella Typhimurium ST34 strains in Shaanxi Province demonstrated unique genomic and antimicrobial resistance traits. This study may help to prevent outbreaks and rationalize salmonellosis antimicrobial therapeutics.
1 Introduction
Salmonella is among the most prevalent foodborne pathogens. Specifically, nontyphoidal Salmonella remains an important etiology of diarrhea, accounting for an estimated 197.35 million episodes (95% uncertainty interval: 127.37 million to 299.20 million) and resulting in approximately 84,799 deaths (95% uncertainty interval: 46,201 to 144,935) annually worldwide (Woh et al., 2021). The Salmonella genus includes two species and six subspecies, and there are approximately 2,659 serotypes (Monte and Sellera, 2020). Among these serotypes, Salmonella enterica serovar Typhimurium and Enteritidis are most frequently associated with human diseases (Ritter et al., 2019).
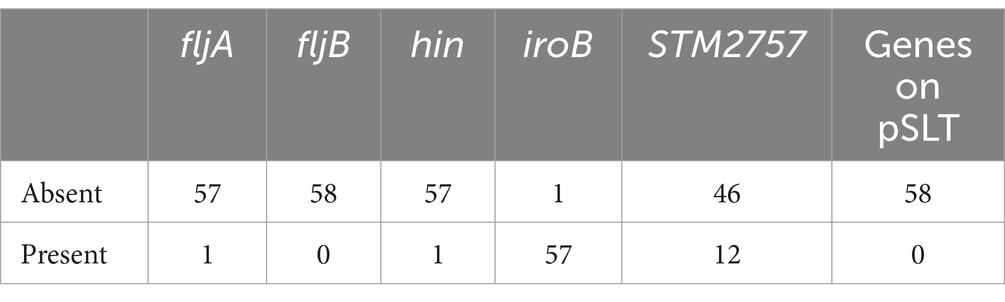
The antigenic formula of monophasic Salmonella Typhimurium is 4,[5],12:i:-, representing a variant of Salmonella Typhimurium. This variant was first isolated from poultry in Portugal during the late 1980s and has since disseminated extensively across Europe, America, Australia, and Asia (Ingle et al., 2021). The primary characteristic of monophasic variants is the absence of phase II flagellar antigens. Potential molecular mechanisms for this absence include the deletion of the fljB gene, which encodes the phase II flagellar antigen, or the deletion of related regulatory genes such as fljA and hin. These deletions result in a serotype change from 4,[5],12:i:1,2 to 4,12:i:- or 4,5,12:i:- (Bugarel et al., 2012). Multiple-locus sequence typing (MLST) analysis has revealed that this serotype predominantly consists of ST19, ST34, and ST313, with ST34 being the most significant type (Van Puyvelde et al., 2019).
In recent years, monophasic strains of Salmonella enterica have rapidly spread, resulting in worldwide outbreaks and a concomitant increase in multidrug resistance, which poses significant public health concerns (Larkin et al., 2022; Lund et al., 2022). This serotype currently ranks third among Salmonella infections in the European Union and fifth in the United States (National Center for Emerging and Zoonotic Infectious Diseases (U.S.), n.d.; European Food Safety Authority and European Centre for Disease Prevention and Control, 2021). Since December 2021, several European countries have reported suspected clusters of infections with ST34 monophasic Salmonella enterica Typhimurium, which have been associated with the consumption of contaminated chocolate products (World Health Organization, 2022). Notably, monophasic Salmonella Typhimurium has emerged as the third most prevalent serotype in China (Bai et al., 2022), thereby raising significant public health concerns. It has been reported that monophasic Salmonella Typhimurium has become the dominant serotype of human Salmonella isolates in Guangdong and Guangxi province in China (Ke et al., 2018; Zeng et al., 2020). However, research specifically focusing on monophasic Salmonella Typhimurium in Shaanxi Province remains limited.
This study investigated the genomic characteristics and antibiotic resistance profiles of 58 strains of monophasic Salmonella Typhimurium, which were collected from various sources, including patients with diarrhea-related illnesses and food samples. Whole genome sequencing (WGS), bioinformatics analysis, pulsed-field gel electrophoresis (PFGE), single nucleotide polymorphism (SNP) analysis, and antimicrobial susceptibility testing were performed on these strains. Our findings elucidate the molecular epidemiological characteristics of local monophasic Salmonella Typhimurium, which may aid in preventing outbreaks and optimizing antimicrobial therapies for salmonellosis.
2 Methods
2.1 Sample collection
This study collected 58 strains of monophasic Salmonella Typhimurium isolated from 6 cities (Xi’an, Xianyang, Weinan, Hanzhong, Ankang, and Yulin) during the years 2020 and 2021. Among them, 4 strains were obtained from food sources (one cooked meat product and three samples of raw pork), while 54 strains were isolated from patients with diarrhea-related illnesses. This study uses strains obtained from Xi’an Center for Disease Control and Prevention (CDC), Yanta CDC, Hanzhong CDC, Ankang CDC, Weinan CDC, and Yulin CDC. These institutions did not require the study to be reviewed or approved by an ethics committee because these strains were gifted from these institutions.
2.2 PFGE
According to the China CDC Pathogen Identification Net (CCPIN) protocol, a standardized PFGE typing method was used for the molecular subtyping of Salmonella. In brief, Salmonella isolates were plated onto Columbia agar supplemented with 5% sheep blood (HuanKai Microbial, Guangzhou, China) and incubated at 37°C for 24 h. The concentration of the Salmonella suspension was adjusted to 4.5 McFarland units. The Salmonella enterica serotype Braenderup strain (H9812) served as a molecular weight marker (Ribot et al., 2006). Plugs were created by mixing 400 μL of 1% Seakem Gold (Lonza, Rockland, United States) with 400 μL of 1% SDS (Solarbio, Beijing, China) in the bacterial suspension. This mixture was digested with proteinase K at a concentration of 20 mg/mL (Merck, Darmstadt, Germany) at 54°C for 2 h. Digestion was subsequently performed at 37°C for 2 h using the XbaI enzyme (50 U) (Takara, Dalian, China). Additionally, 1.5 g of Seakem Gold agarose was dissolved in 150 mL of 0.5 × TBE (TianGen, Beijing, China). Gel electrophoresis was performed on the CHEF Mapper (Bio-Rad Laboratories, Hercules, United States), with molecular weight parameters set to 30–700 kb and an electrophoresis time of 18 h at 14°C. Other parameters remained at default settings. After electrophoresis, the gel was stained with GelRed (Bosunlife, Beijing, China) for 20 min. The dendrogram was automatically generated online by the CCPIN platform. The software deployment and background parameters were uniformly set by the China CDC, ensuring consistency in the analysis.
2.3 Antimicrobial susceptibility testing
The antimicrobial resistance of the isolated Salmonella strains was phenotypically evaluated using the broth dilution method. The minimum inhibitory concentration of a panel of 17 antimicrobial agents from 8 antimicrobial classes was determined. The results were interpreted according to the most recent Clinical Laboratory Standard Institute guidelines (2024) (CLSI, 2024). The tested antimicrobial agents included phenicols (chloramphenicol, 4–32 μg/mL), sulphonamide and trimethoprim (trimethoprim-sulfamethoxazole, 0.5–8 μg/mL), polymyxins (colistin, 0.25–8 μg/mL), beta-lactamase inhibitors (ertapenem, 0.25–8 μg/mL; meropenem, 0.125–2 μg/mL; cefotaxime, 0.25–16 μg/mL; ceftazidime, 0.25–16 μg/mL; ceftazidime/avibactam, 0.25/4–8/4 μg/mL; ampicillin, 2–32 μg/mL; ampicillin/sulbactam, 2–32 μg/mL), tetracyclines (tetracycline, 1–16 μg/mL; tigecycline, 0.25–8 μg/mL), fluoroquinolones (ciprofloxacin, 0.015–2 μg/mL; nalidixic acid, 4–32 μg/mL), macrolides (azithromycin, 2–64 μg/mL), and aminoglycosides (amikacin, 4–64 μg/mL; streptomycin, 4–32 μg/mL). Escherichia coli ATCC 25922 and Klebsiella pneumoniae ATCC700603 served as the quality control strains. Heatmap visualization was performed using the R (version 4.4.2) programming language.
2.4 WGS
Genomic DNA was extracted using the QIAamp DNA Mini Kit (QIAGEN, Hilden, Germany). WGS was conducted on the Illumina MiniSeq platform (Illumina, San Diego, CA, United States) using a 150-base pair paired-end strategy. The genomic DNA library was constructed using the Nextera XT DNA Library Preparation Kit (MicroFuture, Beijing, China). Subsequently, genomic sequencing was performed using the MiniSeq High Output Kit (300 cycles) (MicroFuture, Beijing, China).
2.5 Bioinformatic analysis
The raw sequence reads were checked for quality and assembled using SPAdes v3.15.2 (Bankevich et al., 2012). The gene sequences of fljA, fljB, hin, iroB, and STM2757 were retrieved from the National Center for Biotechnology Information, and sequence alignment was performed using BLAST 2.12.0+. Virulence gene prediction was conducted based on the virulence factors database (Wu et al., 2005). In silico serotyping of Salmonella strains and antimicrobial resistance genes were detected using pipelines on the CCPIN platform. The MLST and the plasmid replicons of the strains were screened and determined by using the MLST 2.0 server and Plasmid Finder 2.1 at the Center for Genomic Epidemiology (Larsen et al., 2012; Carattoli et al., 2014), respectively.
2.6 Phylogenetic relationship analysis
The genome sequences of four representative strains of Salmonella enterica (UK outbreak isolates), specifically SRR17830210, SRR18021617, SRR18488397, and SRR18590198, were retrieved from the European Bioinformatics Institute/European Nucleotide Archive database. Using SRR17830210 as the reference genome and the CSI Phylogeny 1.4 tool at the Center for Genomic Epidemiology (Kaas et al., 2014), SNP clustering analysis was performed on 30 Salmonella enterica strains isolated in 2021. These strains were selected because of their relevance to the European multi-country outbreak of monophasic Salmonella Typhimurium ST34 linked to chocolate products that occurred between the end of 2021 and the beginning of 2022, allowing for a more focused analysis of epidemiologically significant isolates. Finally, the resulting phylogenetic tree was visualized using the iTOL software for better interpretation and communication of the evolutionary relationships. The Ridom SeqSphere+ (version 10.0.4) was used for core genome MLST (Junemann et al., 2013).
3 Results
3.1 Basic genetic characteristics
To determine the genetic characterization, basic gene analysis was performed. As shown in Table 1, the 58 strains of monophasic Salmonella Typhimurium did not carry the fljB gene or virulence genes on the virulence plasmid of Salmonella Typhimurium (pSLT) (NC_003277). One strain carried fljA, one strain carried hin, 57 strains carried iroB, and 12 strains carried the STM2757 gene (Table 1).
3.2 PFGE and MLST sequence types
To determine the genotype, PFGE and MLST were performed. The results revealed that the 58 strains of monophasic Salmonella Typhimurium exhibited significant diversity in PFGE subtypes (Figure 1). These findings suggest the possibility of multiple independent emergence events. However, MLST typing results showed that both the food isolates and the patient isolates belonged to the ST34 type.
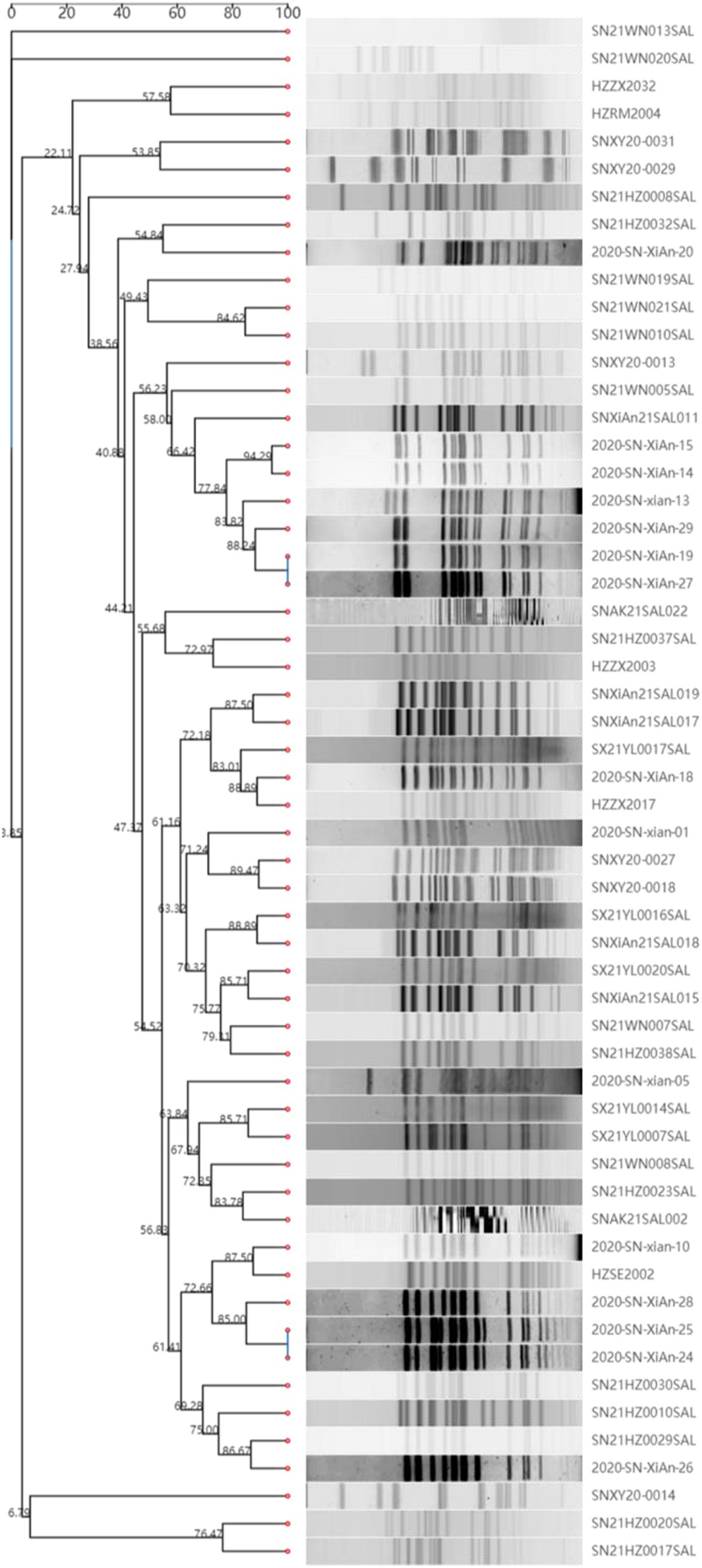
Figure 1. PFGE sequence types of 58 strains of monophasic Salmonella Typhimurium. The Salmonella serotype Braenderup strain (H9812) served as a molecular weight marker. The dendrogram was automatically generated online using the CCPIN platform.
3.3 Phenotypic antimicrobial resistance
To assess the most common patterns of antibiotic resistance, the phenotypic antimicrobial resistance of 53 successfully resuscitated monophasic Salmonella Typhimurium strains was evaluated against 17 antimicrobial agents belonging to 8 different classes. Antimicrobial resistance was classified as resistant, susceptible, or intermediate according to the guidelines established by the Clinical Laboratory Standard Institute. As presented in Table 2, the highest resistance rates were found in tetracycline and ampicillin (94.34%; 50/53), followed by streptomycin (88.68%; 47/53) and ampicillin/sulbactam at 64.15% (34/53). Additionally, the resistance rate to chloramphenicol and trimethoprim-sulfamethoxazole was both 47.17% (25/53). Additionally, we found three strains exhibiting resistance to colistin; however, none of these strains carried the mcr resistance gene.
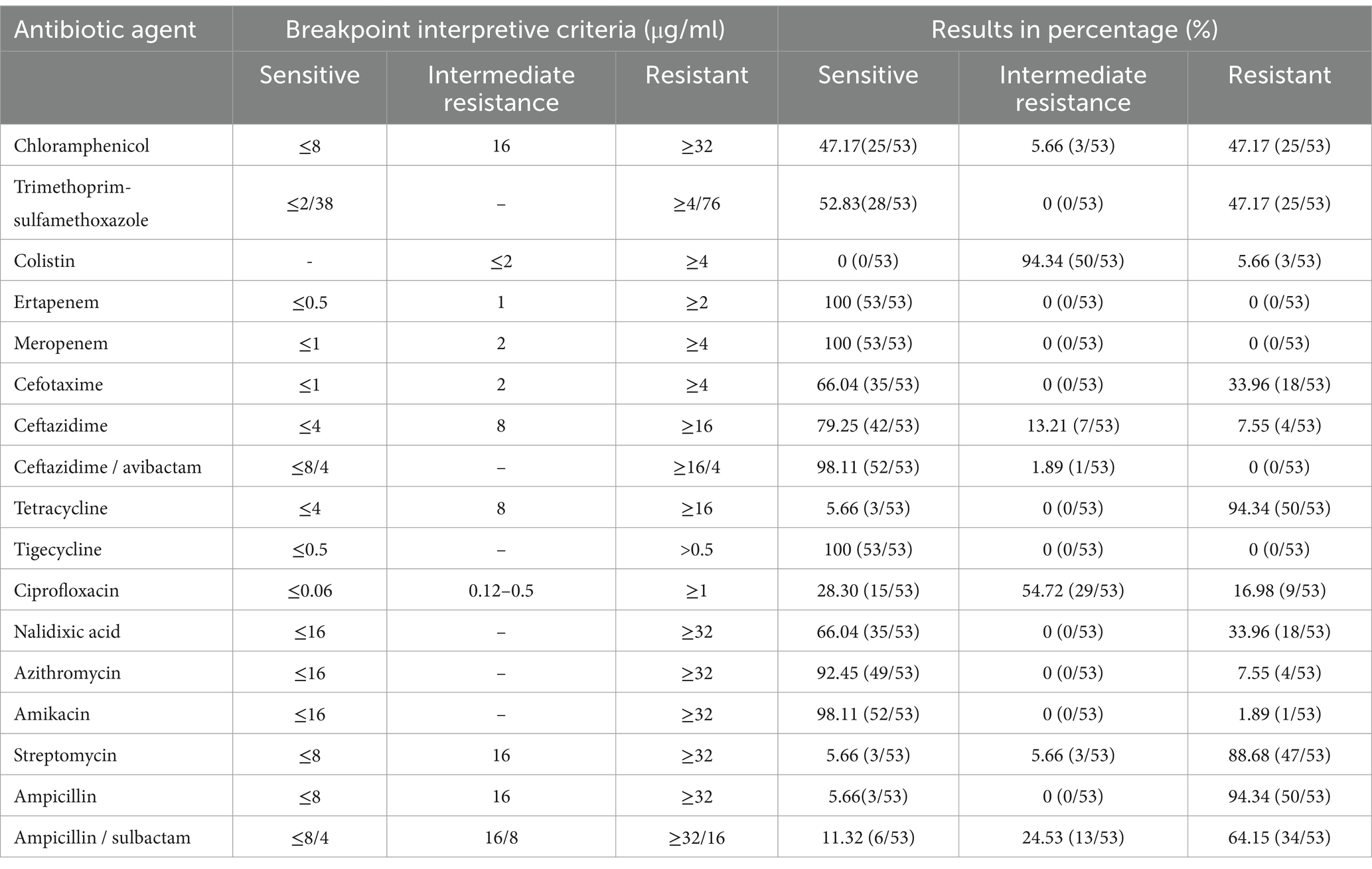
Table 2. Antimicrobial susceptibility interpretation of the 53 isolated monophasic Salmonella Typhimurium strains.
When considering intermediate strains as susceptible, all strains were classified as resistant, as illustrated in Figure 2. Among the isolated strains, 26.42% (14/53) were resistant to five antimicrobial agents, while 24.53% (13/53) showed resistance to three agents, and 20.75% (11/53) were resistant to four agents.
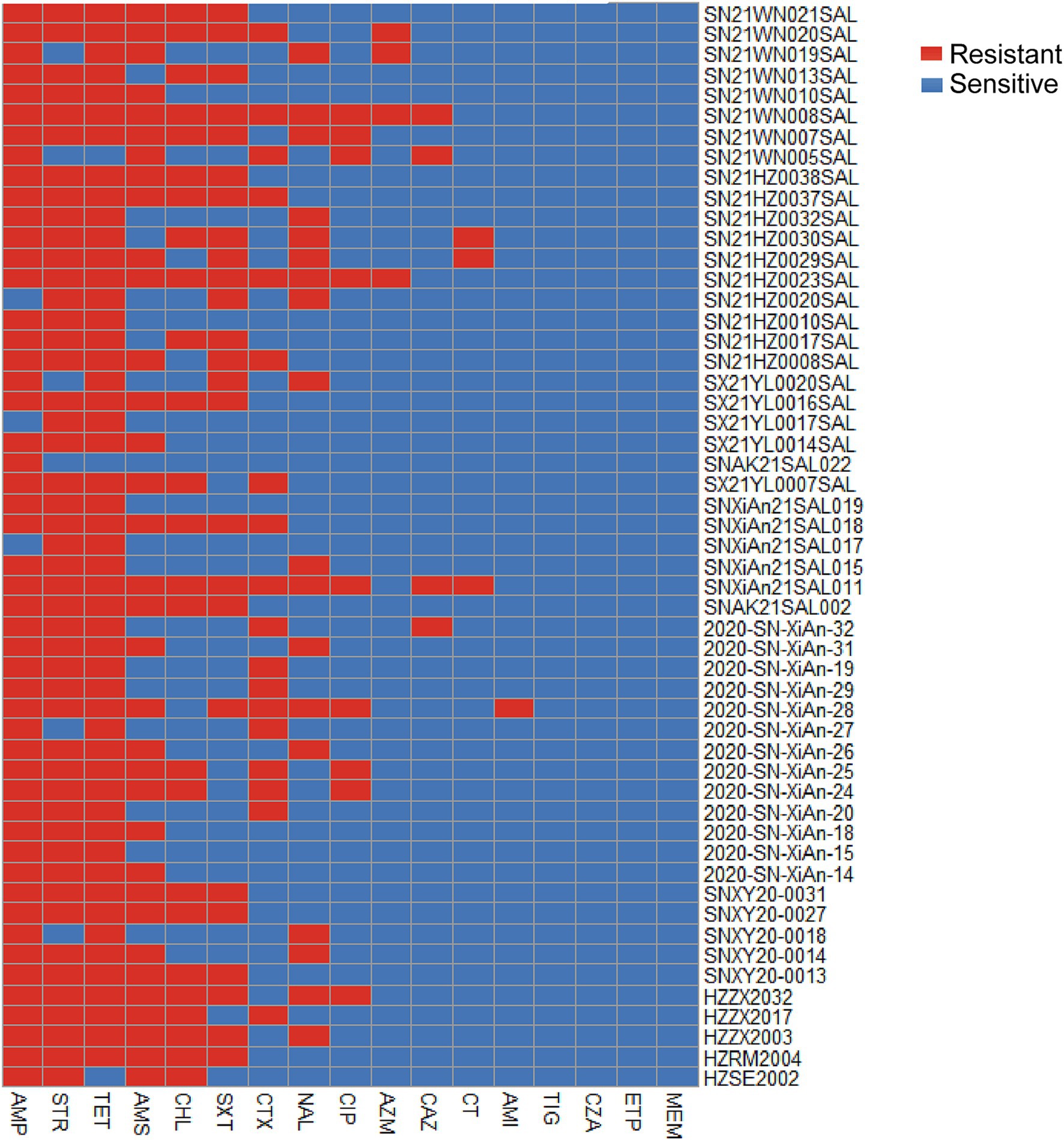
Figure 2. Heatmap of the phenotypic antimicrobial resistance of 53 isolated Salmonella strains. Red represents resistance, while blue indicates sensitivity. The heatmap was constructed based on the resistance similarity of each strain to 17 antimicrobial agents. CHL, Chloramphenicol; SXT, Trimethoprim-sulfamethoxazole; CT, Colistin; ETP, Ertapenem; MEM, Meropenem; CTX, Cefotaxime; CAZ, Ceftazidime; CZA, Ceftazidime / avibactam; TET, Tetracycline; TIG, Tigecycline; CIP, Ciprofloxacin; NAL, Nalidixic acid; AZM, Azithromycin; AMI, Amikacin; STR, Streptomycin; AMP, Ampicillin; AMS, Ampicillin / sulbactam.
3.4 Prediction of the antimicrobial resistance genes
The genomes of 58 strains were uploaded to the CCPIN platform for the prediction of antimicrobial resistance genes. The results indicated the presence of 64 distinct genes associated with resistance to 7 antimicrobial classes. Notably, aac(6′)-Iaa was found to be the most prevalent gene conferring resistance to aminoglycosides, detected in 100.00% (58/58) of the strains. This was followed by tet(B), which conferred resistance to tetracycline in 93.10% (54/58) of the strains, blaTEM-1B, responsible for beta-lactam resistance, found in 74.14% (43/58), and sul2, which was resistant to trimethoprim-sulfamethoxazole (74.14%; 43/58). Additionally, the floR gene was present in 44.83% (26/58) of the strains, while qnrS1 was detected in 41.38% (24/58) (Figure 3). We did not find mutations in the gyrA, gyrB, parC, and parE genes among the ciprofloxacin-resistant strains (data not shown).
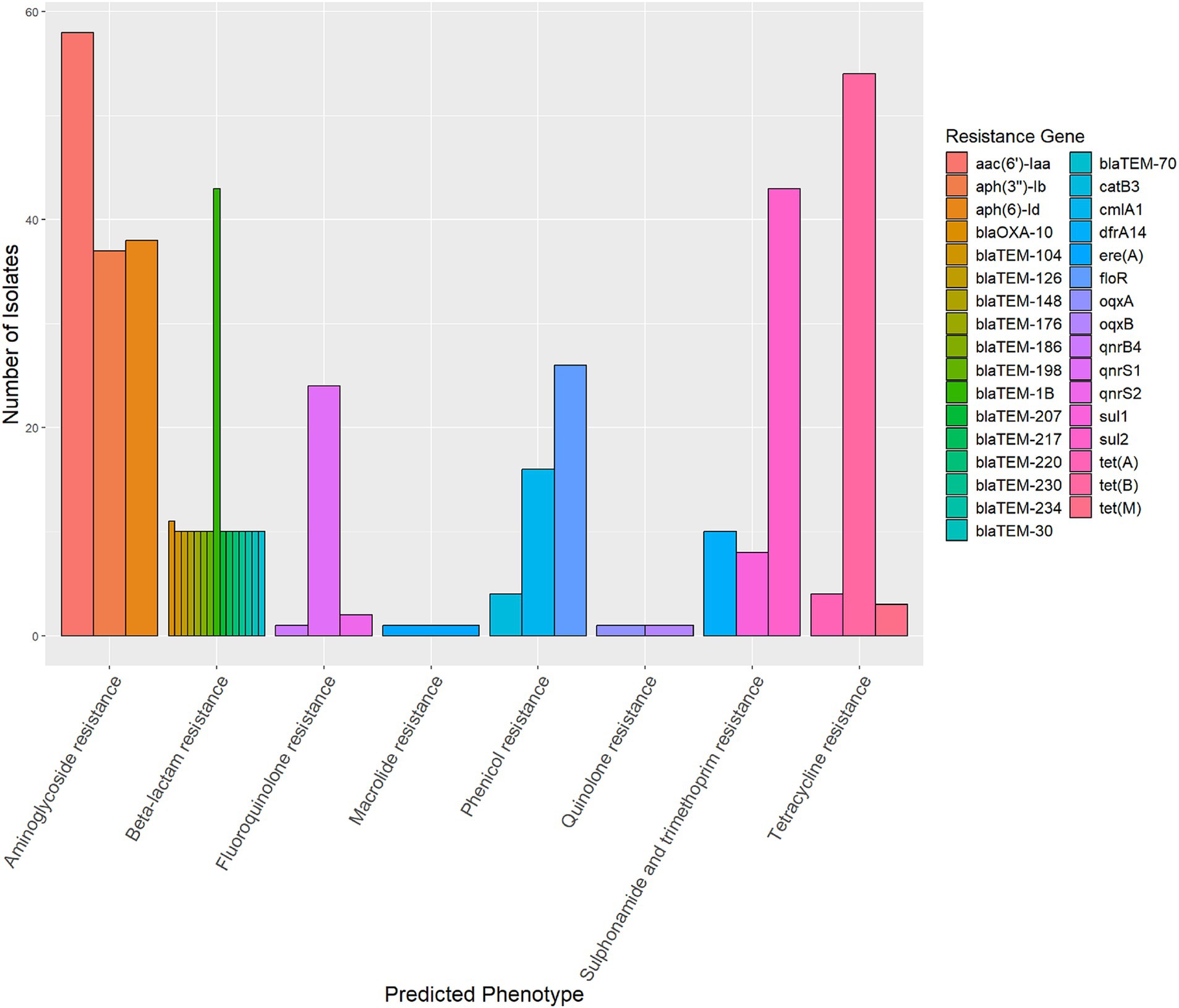
Figure 3. The prediction of antimicrobial resistance genes of 58 strains of monophasic Salmonella Typhimurium. There were 64 distinct genes associated with resistance to seven antimicrobial classes. The diagram illustrates the top three genes for each antimicrobial class.
3.5 Prediction of the virulence genes
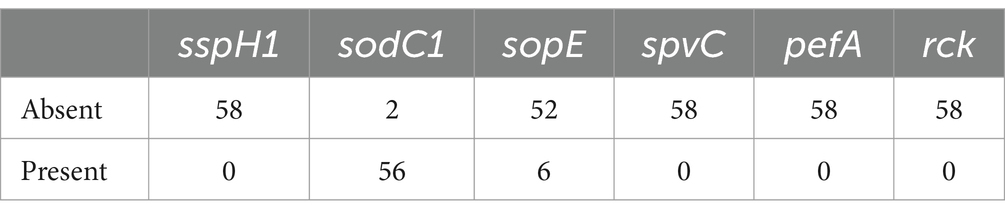
To determine the virulence genes of Salmonella, the genomes of the 58 isolates were assessed with the virulence factors database. Among these, the sodC1 gene was positive in 96.55% (56/58) strains, while the sopE gene was positive in 10.34% (6/58) strains (Table 3). No strains were found to carry the genes sspH1, spvC, pefA, or rck.
3.6 Plasmid replicon predictions
The most prevalent plasmid replicon identified was IncQ1, detected in 84.48% (49/58) of the isolates, followed by IncHI2 and IncHI2A, each found in 32.76% (19/58) of the isolates. The strains carried between one and four types of plasmid replicons, and 8 of 58 isolates (13.79%) were found to harbor no plasmid replicons.
3.7 The phylogenetic relationship of isolates
In this study, SRR17830210 served as the reference genome. The SNP analysis of 30 monophasic Salmonella Typhimurium strains isolated in 2021 and 4 representative strains revealed that there were 3 distinct clusters (Figure 4). The SN21WN005SAL, SNXiAn21SAL011, SNXiAn21SAL017, SX21YL0014SAL, and 4 representative strains were clustered together in the same branch with a bootstrap value of 0.750 (Figure 4). Meanwhile, SN21WN005SAL and SNXiAn21SAL011 were clustered together with the reference strain (SRR17830210) with a bootstrap value of 0.949 (not within five SNPs).
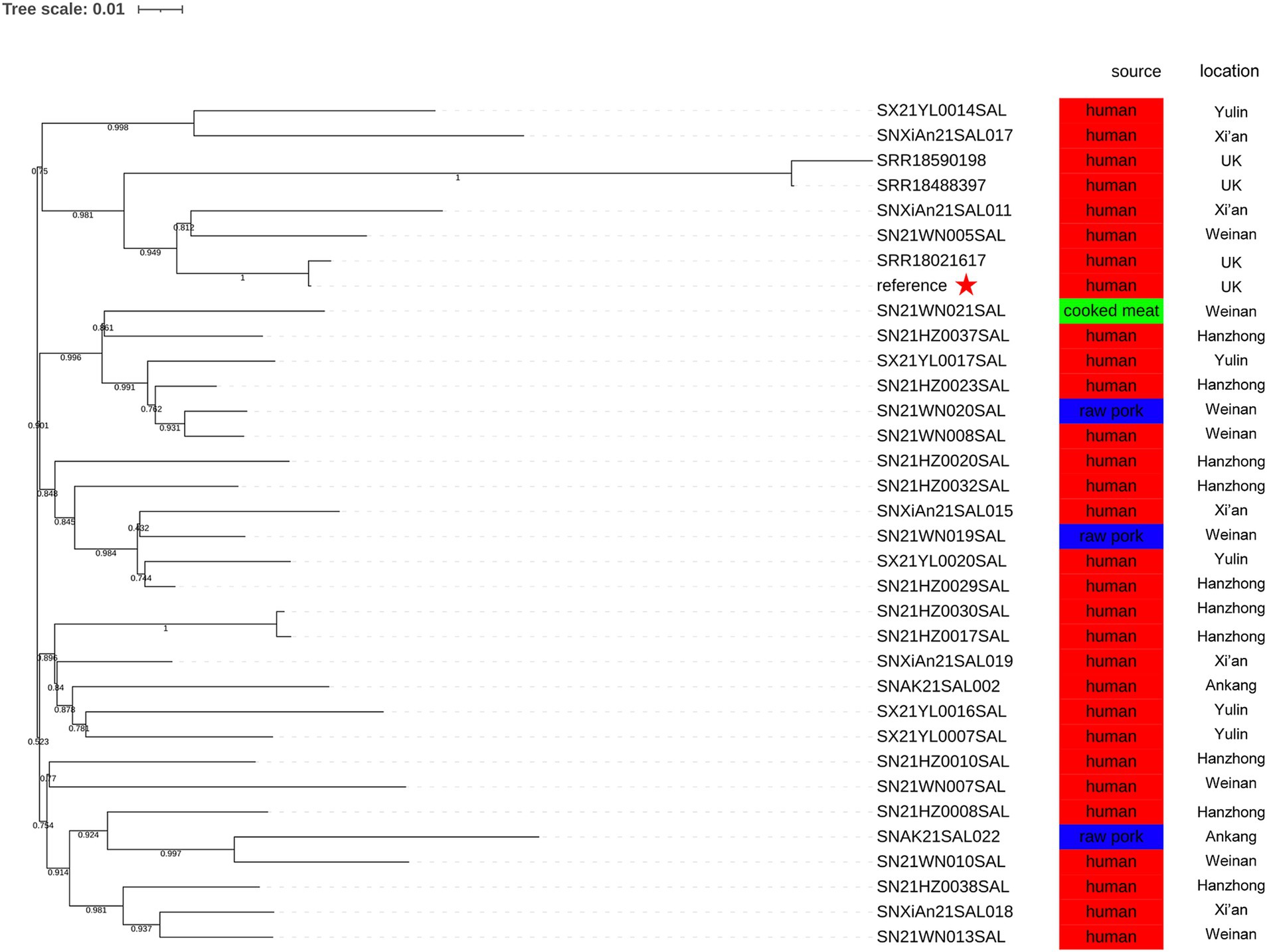
Figure 4. Phylogenetic relationship between 4 representative strains (SRR17830210, SRR18021617, SRR18488397, and SRR18590198) and 30 monophasic Salmonella Typhimurium strains isolated in 2021. The reference genome was SRR17830210. The geographical locations and sources are presented.
Using core genome MLST analysis, the difference between SNXiAn21SAL011, SRR17830210, and SRR18021617 was only 12 allelic differences (ADs), while SN21WN005SAL had a difference of 10 ADs compared to SNXiAn21SAL011 (Figure 5).
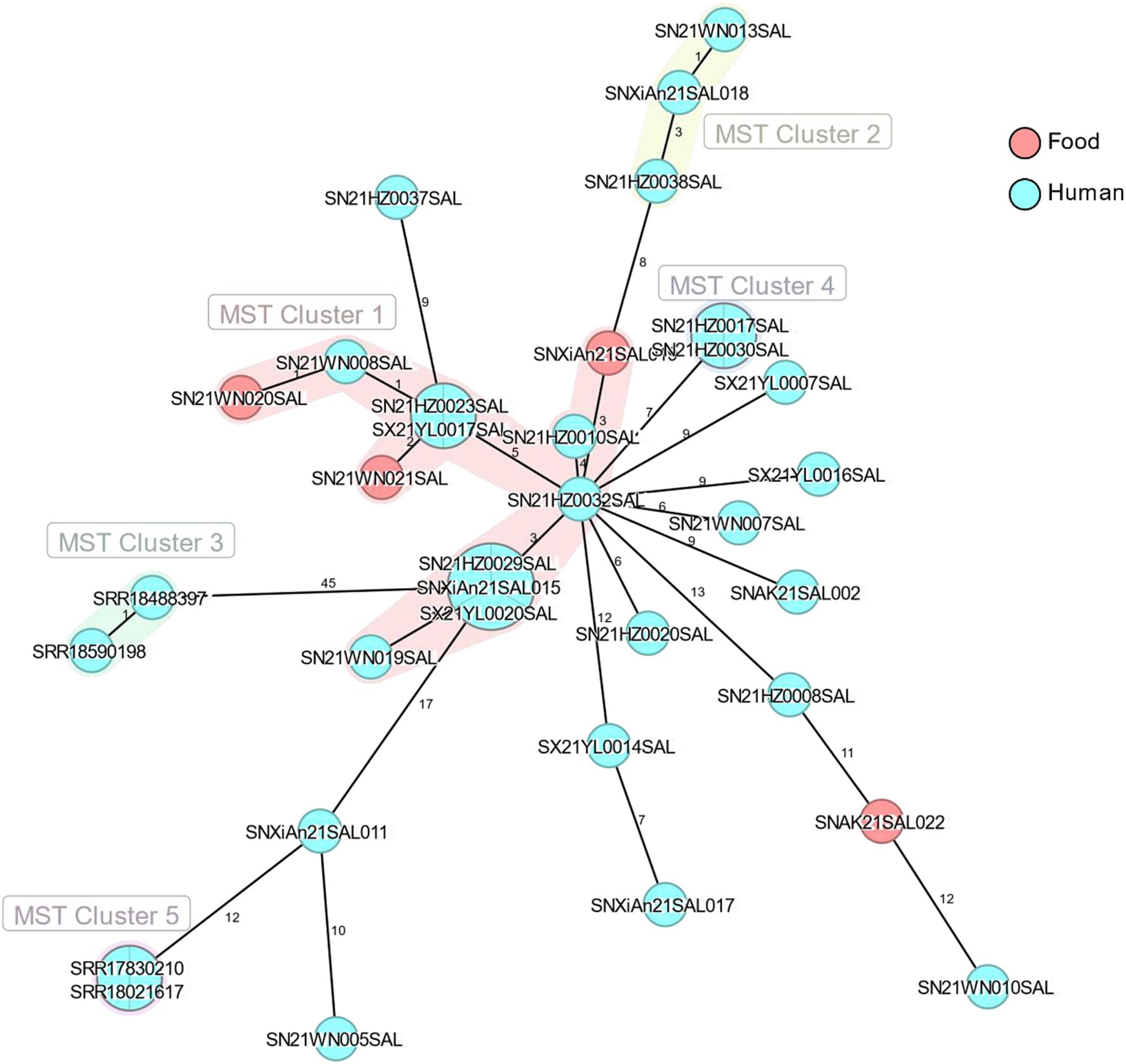
Figure 5. The minimum spanning tree of 4 representative strains (SRR17830210, SRR18021617, SRR18488397, and SRR18590198) and 30 monophasic Salmonella Typhimurium strains isolated in 2021. The numbers between strains represented allelic differences by using core genome MLST analysis. Light red circles represent strains from food sources, and cyan circles represent strains from human sources.
4 Discussion
There are multiple distinct clones of monophasic Salmonella Typhimurium, initially categorized into three geographical clones: the Spanish clone, the U.S. clone, and the major European clone. The Spanish clone, which is associated with pigs and pork products, is characterized by the deletion of the fljA, fljB, hin, and iroB genes, while the STM2757 gene is conserved (Garaizar et al., 2002). The U.S. clone exhibits a unique genomic deletion pattern surrounding the fljBA operon but retains the hin and iroB genes (Soyer et al., 2009). The major European clone identified in 2005 is distinguished by the absence of the fljA, fljB, and hin genes, as well as the loss of the typical pSLT; however, it harbors conserved iroB and STM2757 genes (Lucarelli et al., 2012). This clone primarily corresponds to ST34. Currently, the prevalence of the European clone has gradually surpassed that of the Spanish and U.S. clones (Elnekave et al., 2018). This study analyzed 58 strains of monophasic Salmonella. Among them, 4 strains originated from food, including 1 from cooked meat products and 3 from raw pork. The remaining 54 strains were isolated from patients. All 58 strains belonged to the ST34 type and lacked the fljB gene and virulence genes typically found on pSLT. However, only 12 strains possessed the STM2757 gene. Additionally, we utilized both PFGE and WGS to characterize the isolates. While WGS offers a comprehensive genomic understanding of relatedness, PFGE is a traditional molecular typing method that is widely used for outbreak strain tracing. The inclusion of PFGE allows us to compare our findings with a vast database of previous PFGE gel images, providing valuable context and validation for our results. Furthermore, the integration of these methodologies enhances our ability to detect genetic variations that could impact the epidemiological relationships between strains.
Previous research has indicated that Salmonella 4,[5],12:i:– isolates have a considerable diversity of subtypes, even among samples from a single country (Zamperini et al., 2007). It is suggested that this increased heterogeneity implies greater adaptability, which may contribute to the success of this serotype (Sun et al., 2020). In our study, the 58 strains of monophasic Salmonella Typhimurium demonstrated significant diversity in PFGE subtypes, suggesting that these isolates may differ considerably in their genetic backgrounds. While the lack of fljB expression is a notable trait among monophasic variants, the observed differences in PFGE patterns indicate that these variations could also arise from a range of genetic factors, including other gene deletions, mutations, or insertions unrelated to fljB. These insights highlight the necessity for further comprehensive genomic analyses to better understand the relationship between genetic deletions and the phenotypic characteristics of monophasic Salmonella Typhimurium.
The antibiotic resistance pattern of Salmonella serves as an important indicator of clonal evolution and lineage classification. The Spanish clone is resistant to up to 7 antimicrobial agents, including ampicillin, chloramphenicol, gentamicin, streptomycin/spectinomycin, sulfonamides, tetracyclines, and trimethoprim (ACGSSuTTp type). The ASSuT pattern, commonly observed in monophasic Salmonella Typhimurium in European strains from humans and pig products (Petrovska et al., 2016; Seixas et al., 2016), is also prevalent in swine and pork products in the United States, contributing significantly to the increase in multidrug-resistant Salmonella strains. This study revealed that tetracycline (94.34%; 50/53) and ampicillin (94.34%; 50/53) were the most resistant antimicrobial agents in Shaanxi Province, followed closely by streptomycin (88.68%; 47/53). This antimicrobial resistance pattern was similar to that of the ASSuT pattern. The antimicrobial resistance in the European clone may be attributed to the primary antibiotics used in pig breeding (Sun et al., 2020). Moreover, tetracycline and ampicillin are less effective antimicrobial agents against Salmonella isolates from the pig slaughtering process (Geng et al., 2021). These findings emphasize how antimicrobial use in livestock, particularly in pork production, may drive these patterns of resistance.
The European clones carry the same antimicrobial resistance genes, including blaTEM, strA-strB, sul2, and tet(B) (Hopkins et al., 2010; Seixas et al., 2016). In this study, our results showed that aac(6′)-Iaa was the most prevalent gene conferring resistance to aminoglycosides (100.00%; 58/58), followed by tet(B), which expressed resistance to tetracycline (93.10%; 54/58), blaTEM-1B, which conferred resistance to beta-lactam antibiotics (74.14%; 43/58), and sul2, which was resistant to sulfonamides (74.14%; 43/58). Notably, floR (44.83%; 26/58) was identified as the gene with the highest prevalence of resistance to phenicols. Phenotypic resistance analysis revealed that the resistance levels for chloramphenicol and trimethoprim-sulfamethoxazole were comparable (47.17%; 25/53). It has been reported that the detection rates of blaTEM-1B, sul2, and floR are high in the pig slaughtering process (Geng et al., 2021). Additionally, the multidrug-resistant Salmonella DT104 also carries the floR gene (Switt et al., 2009). While our analysis has drawn comparisons to these three recognized clones, it is essential to recognize that the emergence of monophasic Salmonella Typhimurium is dynamic and multifaceted, with evidence of widespread genetic diversity. Numerous strains now exist that do not fit neatly into the original categories. Our study identified unique characteristics in the isolates from Shaanxi Province. This finding highlights the ongoing evolution of monophasic Salmonella Typhimurium and underscores the necessity for continuous surveillance and investigation of emerging strains.
Quinolones are essential antibiotics for the treatment of invasive Salmonella infections in humans (Cito et al., 2016). In He et al.’s (2020) study, 40.4% of monophasic Salmonella Typhimurium isolates were found to carry at least one plasmid-mediated quinolone resistance gene in China. The predominant genes identified were oqxAB and aac(6′)-Ib-cr, while the overall positive rate for the qnr gene was 7.8%. In this study, the prevalence of the qnrS1 gene, which conferred reduced susceptibility to fluoroquinolones rather than full resistance, was found to be 41.38% (24/58). Additionally, one strain carrying the blaCTX-M-14 gene was detected, which has been previously reported in a monophasic isolate collected from swine in Portugal (Clemente et al., 2013), and recovered from a human clinical sample (Fernandez et al., 2016). Notably, given that fluoroquinolones are generally contraindicated in children and pregnant women, ceftriaxone is often administered instead. This practice has implications for the treatment of infections, especially considering the presence of the blaCTX-M gene, which indicates potential resistance to cephalosporins.
Among the analyzed strains in our study, 96.55% (56/58) were positive for the virulence gene sodC1, while 10.34% (6/58) tested positive for sopE. The sodC1 and sopE genes, located on prophages, exhibited comparable virulence capacities in comparison with Salmonella Typhimurium, causing human salmonellosis (Ngoi et al., 2018). The sodC1 gene encodes a putative Cu/Zn superoxide dismutase (Ammendola et al., 2008), while SopE serves as one of the effector proteins that promote efficient bacterial entry into host cells (Rudolph et al., 1999). The virulence gene sopE was described in monophasic Salmonella isolates from Europe (Petrovska et al., 2016) and the United States (Elnekave et al., 2018).
The prediction results of plasmid replicons in 58 Salmonella isolates indicated that the predominant plasmid replicon was IncQ1 (84.48%; 49/58), followed by IncHI2 (32.76%; 19/58) and IncHI2A (32.76%; 19/58). It has been reported that IncQ1 is strongly associated with African multidrug-resistant Salmonella Enteritidis isolates (Cao et al., 2023). Wu et al. (2005) reported that IncHI2A_1 and IncHI2_1 were predominant in Salmonella Typhimurium ST34 isolates throughout the pig slaughtering process in China (Geng et al., 2021). It has been demonstrated that these plasmids were associated with resistance to different antimicrobial classes, including β-lactams, aminoglycosides, sulfonamides, tetracyclines, and polymyxins (Elbediwi et al., 2020a; Elbediwi et al., 2020b; Gu et al., 2020; McMillan et al., 2020). The virulence and antimicrobial resistance profiles of the isolates associated with these plasmids will be further studied.
Different pathogens show variations in SNP numbers and distributions due to differences in their genomic compositions. The SNP differences between outbreak strains may be related to the mutation rates and different outbreak patterns (such as geographic extent and period). The United Kingdom monophasic Salmonella Typhimurium outbreak reported a cluster of representative outbreak isolates within five ADs or five SNPs (European Centre for Disease Prevention and Control, European Food Safety Authority, 2022). In the present study, although the difference between SNXiAn21SAL011, SRR17830210, and SRR18021617 was 12 ADs, the SNP phylogenetic analysis showed that SN21WN005SAL, SNXiAn21SAL011, SRR17830210, and SRR18021617 were closely related, with a bootstrap value of 0.949. This result indicates the genetic similarities of the monophasic Salmonella Typhimurium strains. The metadata analysis revealed that all these isolates were from Shaanxi Province, with 4 strains sourced from food and the remaining from human cases. Notably, the genetic similarities of isolates from the same province could be attributed to shared environmental factors, local agricultural practices, and the transmission dynamics of Salmonella. Further research should explore these correlations to understand better the epidemiological aspects of the strains examined.
In this study, we investigated the genomic characterization and antibiotic resistance of monophasic Salmonella Typhimurium in Shaanxi Province, China. Although all strains belonged to sequence type ST34 and did not harbor virulence genes on the pSLT plasmid, 12 strains carried the STM2757 gene. The phenotypic assessment of antimicrobial resistance demonstrated that tetracycline, ampicillin, streptomycin, and trimethoprim-sulfamethoxazole were less effective antimicrobial agents. Moreover, the resistance levels of chloramphenicol and trimethoprim-sulfamethoxazole were found to be comparable. Gene prediction indicated that aac(6′)-Iaa was the most prevalent resistance gene, followed by tet(B), blaTEM-1B, and sul2. Furthermore, floR exhibited the highest resistance among the phenicol-related genes. Though SNXiAn21SAL011, SRR17830210, and SRR18021617 were not within five ADs, the SNP phylogenetic relationships of SN21WN005SAL, SNXiAn21SAL011, SRR17830210, and SRR18021617 were closely related. This study not only contributes essential local molecular epidemiological insights but also underscores the urgent need for enhanced surveillance and strategic interventions to combat foodborne pathogens and optimize treatment options for salmonellosis in humans.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author contributions
YS: Data curation, Investigation, Resources, Software, Validation, Writing – original draft. YuL: Investigation, Resources, Writing – review & editing. SL: Funding acquisition, Writing – review & editing. SW: Formal analysis, Resources, Writing – review & editing. GM: Investigation, Resources, Writing – review & editing. YaL: Investigation, Writing – review & editing. JZ: Investigation, Resources, Writing – review & editing. YC: Investigation, Writing – review & editing. WL: Investigation, Resources, Writing – review & editing. TS: Investigation, Resources, Writing – review & editing. CW: Investigation, Resources, Writing – review & editing. JX: Conceptualization, Project administration, Resources, Validation, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the Shaanxi Province Key Research and Development Plan (Grant no. 2023-YBSF-633).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ammendola, S., Pasquali, P., Pacello, F., Rotilio, G., Castor, M., Libby, S. J., et al. (2008). Regulatory and structural differences in the cu,Zn-superoxide dismutases of Salmonella enterica and their significance for virulence. J. Biol. Chem. 283, 13688–13699. doi: 10.1074/jbc.M710499200
Bai, L., Wang, J., Liu, L., Cui, S., Guo, Y., Li, N., et al. (2022). Implications for risk management of foodborne pathogens in China from the outbreak of monophasic Salmonella enterica serovar typhimurium contaminated chocolate products. Chin. J. Prev. Med. 11, 1648–1656. doi: 10.3760/cma.j.cn112150-20220712-00711
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Bugarel, M., Vignaud, M. L., Moury, F., Fach, P., and Brisabois, A. (2012). Molecular identification in monophasic and nonmotile variants of Salmonella enterica serovar typhimurium. Microbiology 1, 481–489. doi: 10.1002/mbo3.39
Cao, G., Zhao, S., Kuang, D., Hsu, C. H., Yin, L., Luo, Y., et al. (2023). Geography shapes the genomics and antimicrobial resistance of Salmonella enterica Serovar Enteritidis isolated from humans. Sci. Rep. 13:1331. doi: 10.1038/s41598-022-24150-4
Carattoli, A., Zankari, E., Garcia-Fernandez, A., Voldby Larsen, M., Lund, O., Villa, L., et al. (2014). In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–3903. doi: 10.1128/AAC.02412-14
Cito, F., Baldinelli, F., Calistri, P., Di Giannatale, E., Scavia, G., Orsini, M., et al. (2016). Outbreak of unusual Salmonella enterica serovar typhimurium monophasic variant 1,4 [5],12:i:-, Italy, June 2013 to September 2014. Euro Surveill. 21, 1–10. doi: 10.2807/1560-7917.ES.2016.21.15.30194
Clemente, L., Manageiro, V., Ferreira, E., Jones-Dias, D., Correia, I., Themudo, P., et al. (2013). Occurrence of extended-spectrum beta-lactamases among isolates of Salmonella enterica subsp. enterica from food-producing animals and food products, in Portugal. Int. J. Food Microbiol. 167, 221–228. doi: 10.1016/j.ijfoodmicro.2013.08.009
CLSI (2024). “Performance standards for antimicrobial susceptibility testing” in CLSI supplement M100. 34th. eds. F. R. Cockerill, K. Bush, M. N. Dudley, G. M. Eliopoulos, D. J. Hardy, and D. W. Hecht, et al. (Berwyn, Pennsylvania, USA: Clinical and Laboratory Standards Institute).
Elbediwi, M., Beibei, W., Pan, H., Jiang, Z., Biswas, S., Li, Y., et al. (2020a). Genomic characterization of mcr-1-carrying Salmonella enterica Serovar 4,[5],12:i:- ST 34 clone isolated from pigs in China. Front. Bioeng. Biotechnol. 8:663. doi: 10.3389/fbioe.2020.00663
Elbediwi, M., Pan, H., Biswas, S., Li, Y., and Yue, M. (2020b). Emerging colistin resistance in Salmonella enterica serovar Newport isolates from human infections. Emerg. Microbes Infect. 9, 535–538. doi: 10.1080/22221751.2020.1733439
Elnekave, E., Hong, S., Mather, A. E., Boxrud, D., Taylor, A. J., Lappi, V., et al. (2018). Salmonella enterica serotype 4,[5],12:i:- in swine in the United States Midwest: an emerging multidrug-resistant clade. Clin. Infect. Dis. 66, 877–885. doi: 10.1093/cid/cix909
European Centre for Disease Prevention and Control, European Food Safety Authority (2022). Multi-country outbreak of monophasic Salmonella Typhimurium sequence type 34 linked to chocolate products – first update – 18 may 2022. EFSA Support. Publ. 19:7352E. doi: 10.2903/sp.efsa.2022.EN-7352
European Food Safety Authority and European Centre for Disease Prevention and Control (2021). The European Union one health 2020 Zoonoses report. EFSA J. 19:e06971. doi: 10.2903/j.efsa.2021.6971
Fernandez, J., Garcia, V., Bances, M., and Rodicio, M. R. (2016). CTX-M-14 production by a clinical isolate of the European clone of Salmonella enterica 4,[5],12:i. J. Glob. Antimicrob. Resist. 7, 130–131. doi: 10.1016/j.jgar.2016.08.009
Garaizar, J., Porwollik, S., Echeita, A., Rementeria, A., Herrera, S., Wong, R. M., et al. (2002). DNA microarray-based typing of an atypical monophasic Salmonella enterica serovar. J. Clin. Microbiol. 40, 2074–2078. doi: 10.1128/JCM.40.6.2074-2078.2002
Geng, Y., Wu, W., Zhou, L., Li, J., Geng, Y., and Yang, Y. (2021). Synergistic effects of LY294002 and ABT199 on the cell cycle in K562, HL60 and KG1a cells. Oncol. Rep. 45:97. doi: 10.3892/or.2021.8048
Gu, D., Wang, Z., Tian, Y., Kang, X., Meng, C., Chen, X., et al. (2020). Prevalence of salmonella isolates and their distribution based on whole-genome sequence in a chicken slaughterhouse in Jiangsu, China. Front Vet Sci 7:29. doi: 10.3389/fvets.2020.00029
He, J., Sun, F., Sun, D., Wang, Z., Jin, S., Pan, Z., et al. (2020). Multidrug resistance and prevalence of quinolone resistance genes of Salmonella enterica serotypes 4,[5],12:i:- in China. Int. J. Food Microbiol. 330:108692. doi: 10.1016/j.ijfoodmicro.2020.108692
Hopkins, K. L., Kirchner, M., Guerra, B., Granier, S. A., Lucarelli, C., Porrero, M. C., et al. (2010). Multiresistant Salmonella enterica serovar 4,[5],12:i:- in Europe: a new pandemic strain? Euro Surveill. 15:19580. doi: 10.2807/ese.15.22.19580-en
Ingle, D. J., Ambrose, R. L., Baines, S. L., Duchene, S., Goncalves da Silva, A., Lee, D. Y. J., et al. (2021). Evolutionary dynamics of multidrug resistant Salmonella enterica serovar 4,[5],12:i:- in Australia. Nat. Commun. 12:4786. doi: 10.1038/s41467-021-25073-w
Junemann, S., Sedlazeck, F. J., Prior, K., Albersmeier, A., John, U., Kalinowski, J., et al. (2013). Updating benchtop sequencing performance comparison. Nat. Biotechnol. 31, 294–296. doi: 10.1038/nbt.2522
Kaas, R. S., Leekitcharoenphon, P., Aarestrup, F. M., and Lund, O. (2014). Solving the problem of comparing whole bacterial genomes across different sequencing platforms. PLoS One 9:e104984. doi: 10.1371/journal.pone.0104984
Ke, B., Zeng, H., He, D., Tan, H., Li, B., Liang, Y., et al. (2018). Circulation and etiological characterization of Salmonella enterica serotype in human in Guangdong province, 2007-2016. Chin. J. Epidemiol. 39, 63–66. doi: 10.3760/cma.j.issn.0254-6450.2018.01.013
Larkin, L., Pardos de la Gandara, M., Hoban, A., Pulford, C., Jourdan-Da Silva, N., de Valk, H., et al. (2022). Investigation of an international outbreak of multidrug-resistant monophasic Salmonella Typhimurium associated with chocolate products, EU/EEA and United Kingdom, February to April 2022. Euro Surveill. 27:2200314. doi: 10.2807/1560-7917.ES.2022.27.15.2200314
Larsen, M. V., Cosentino, S., Rasmussen, S., Friis, C., Hasman, H., Marvig, R. L., et al. (2012). Multilocus sequence typing of total-genome-sequenced bacteria. J. Clin. Microbiol. 50, 1355–1361. doi: 10.1128/JCM.06094-11
Lucarelli, C., Dionisi, A. M., Filetici, E., Owczarek, S., Luzzi, I., and Villa, L. (2012). Nucleotide sequence of the chromosomal region conferring multidrug resistance (R-type ASSuT) in salmonella typhimurium and monophasic Salmonella Typhimurium strains. J. Antimicrob. Chemother. 67, 111–114. doi: 10.1093/jac/dkr391
Lund, S., Tahir, M., Vohra, L. I., Hamdana, A. H., and Ahmad, S. (2022). Outbreak of monophasic Salmonella Typhimurium sequence type 34 linked to chocolate products. Ann. Med. Surg. (Lond) 82:104597. doi: 10.1016/j.amsu.2022.104597
McMillan, E. A., Jackson, C. R., and Frye, J. G. (2020). Transferable plasmids of Salmonella enterica associated with antibiotic resistance genes. Front. Microbiol. 11:562181. doi: 10.3389/fmicb.2020.562181
Monte, D. M., and Sellera, F. P. (2020). Salmonella. Emerg. Infect. Dis. 26:2955. doi: 10.3201/eid2612.et2612
National Center for Emerging and Zoonotic Infectious Diseases (U.S.). National Enteric Disease Surveillance: Salmonella Annual Report, 2016. Available online at: https://www.cdc.gov/nationalsurveillance/pdfs/2016-Salmonella-report-508.pdf (Accessed April 19, 2022).
Ngoi, S. T., Yap, K. P., and Thong, K. L. (2018). Genomic characterization of endemic Salmonella enterica serovar Typhimurium and Salmonella enterica serovar I 4,[5],12:i:- isolated in Malaysia. Infect. Genet. Evol. 62, 109–121. doi: 10.1016/j.meegid.2018.04.027
Petrovska, L., Mather, A. E., AbuOun, M., Branchu, P., Harris, S. R., Connor, T., et al. (2016). Microevolution of monophasic Salmonella Typhimurium during epidemic, United Kingdom, 2005-2010. Emerg. Infect. Dis. 22, 617–624. doi: 10.3201/eid2204.150531
Ribot, E. M., Fair, M. A., Gautom, R., Cameron, D. N., Hunter, S. B., Swaminathan, B., et al. (2006). Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, salmonella, and Shigella for PulseNet. Foodborne Pathog. Dis. 3, 59–67. doi: 10.1089/fpd.2006.3.59
Ritter, A. C., Tondo, E. C., Siqueira, F. M., Soggiu, A., Varela, A. P. M., Mayer, F. Q., et al. (2019). Genome analysis reveals insights into high-resistance and virulence of Salmonella Enteritidis involved in foodborne outbreaks. Int. J. Food Microbiol. 306:108269. doi: 10.1016/j.ijfoodmicro.2019.108269
Rudolph, M. G., Weise, C., Mirold, S., Hillenbrand, B., Bader, B., Wittinghofer, A., et al. (1999). Biochemical analysis of SopE from Salmonella typhimurium, a highly efficient guanosine nucleotide exchange factor for RhoGTPases. J. Biol. Chem. 274, 30501–30509. doi: 10.1074/jbc.274.43.30501
Seixas, R., Santos, T. R., Machado, J., Tavares, L., Bernardo, F., Semedo-Lemsaddek, T., et al. (2016). Phenotypic and molecular characterization of salmonella 1,4,[5],12:i:- R-type ASSuT isolates from humans, animals, and environment in Portugal, 2006-2011. Foodborne Pathog. Dis. 13, 633–641. doi: 10.1089/fpd.2016.2156
Soyer, Y., Moreno Switt, A., Davis, M. A., Maurer, J., McDonough, P. L., Schoonmaker-Bopp, D. J., et al. (2009). Salmonella enterica serotype 4,5,12:i:-, an emerging salmonella serotype that represents multiple distinct clones. J. Clin. Microbiol. 47, 3546–3556. doi: 10.1128/JCM.00546-09
Sun, H., Wan, Y., Du, P., and Bai, L. (2020). The epidemiology of monophasic Salmonella Typhimurium. Foodborne Pathog. Dis. 17, 87–97. doi: 10.1089/fpd.2019.2676
Switt, A. I., Soyer, Y., Warnick, L. D., and Wiedmann, M. (2009). Emergence, distribution, and molecular and phenotypic characteristics of Salmonella enterica serotype 4,5,12:i. Foodborne Pathog. Dis. 6, 407–415. doi: 10.1089/fpd.2008.0213
Van Puyvelde, S., Pickard, D., Vandelannoote, K., Heinz, E., Barbe, B., de Block, T., et al. (2019). An African Salmonella Typhimurium ST313 sublineage with extensive drug-resistance and signatures of host adaptation. Nat. Commun. 10:4280. doi: 10.1038/s41467-019-11844-z
Woh, P. Y., Yeung, M. P. S., Goggins, W. B. 3rd, Lo, N., Wong, K. T., Chow, V., et al. (2021). Genomic epidemiology of multidrug-resistant Nontyphoidal salmonella in young children hospitalized for gastroenteritis. Microbiol. Spectr. 9:e0024821. doi: 10.1128/Spectrum.00248-21
World Health Organization (2022). Disease Outbreak News; Multi-country outbreak of Salmonella Typhimurium linked to chocolate products - Europe and the UnitedStates of America. Available at: https://www.who.intemergencies/disease-outbreak-news/item/2022.DON369
Wu, L. X., La Rose, J., Chen, L., Neale, C., Mak, T., Okkenhaug, K., et al. (2005). CD28 regulates the translation of Bcl-xL via the phosphatidylinositol 3-kinase/mammalian target of rapamycin pathway. J. Immunol. 174, 180–194. doi: 10.4049/jimmunol.174.1.180
Zamperini, K., Soni, V., Waltman, D., Sanchez, S., Theriault, E. C., Bray, J., et al. (2007). Molecular characterization reveals Salmonella enterica serovar 4,[5],12:i:- from poultry is a variant typhimurium serovar. Avian Dis. 51, 958–964. doi: 10.1637/7944-021507-REGR.1
Zeng, X., Lv, S., Qu, C., Lan, L., Tan, D., Li, X., et al. (2020). Serotypes, antibiotic resistance, and molecular characterization of non-typhoidal salmonella isolated from dianrheic patients in Guangxi Zhuang autonomous region, China, 2014--2017. Food Control 120:107478. doi: 10.1016/j.foodcont.2020.107478
Keywords: salmonella, antimicrobial resistance, virulence factors, plasmid replicons, WGS, SNP, PFGE, MLST
Citation: Shi Y, Liu Y, Li S, Wu S, Ma G, Luan Y, Zhang J, Chen Y, Liu W, Shen T, Wang C and Xu J (2025) Genomic characteristics and antibiotic resistance profiles of monophasic Salmonella Typhimurium in Shaanxi Province, China. Front. Microbiol. 16:1565631. doi: 10.3389/fmicb.2025.1565631
Edited by:
Mohamed K. Fakhr, University of Tulsa, United StatesReviewed by:
Keri N. Norman, Texas A&M University, United StatesDaniel F. M. Monte, North Carolina State University, United States
Copyright © 2025 Shi, Liu, Li, Wu, Ma, Luan, Zhang, Chen, Liu, Shen, Wang and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiru Xu, OTEzNDM2M0BxcS5jb20=
 Yi Shi1,2
Yi Shi1,2 Jiru Xu
Jiru Xu