- 1College of Agriculture and Biological Science, Dali University, Dali, Yunnan, China
- 2School of Life Sciences, Guizhou Normal University, Guiyang, China
- 3Co-Innovation Center for Cangshan Mountain and Erhai Lake Integrated Protection and Green Development of Yunnan Province, Dali University, Dali, Yunnan, China
Junewangiaceae (Sordariomycetes) is a family with a relatively recent taxonomic history and a small number of described species. However, a major challenge within this family is the inability to distinguish between various genera based solely on the phylogenetic analysis. In this study, we introduced two new species, Junewangia guangxiensis and J. synnematica, which formed independent clades in phylogenetic analysis and displayed characteristics that were easily distinguishable from other species within this family. Additionally, three previously known species, viz., Dictyosporella aquatica, D. thailandensis, and J. thailandensis, are reported from China for the first time. Furthermore, D. bambusicola is documented from a freshwater habitat. The results enhance our understanding of Junewangiaceae and provide some suggestions for addressing the taxonomic problems of this family in the future.
Introduction
Biodiversity is one of the most significant topics globally in current times (Díaz, 2022). However, our current understanding of biodiversity remains limited, especially in the fungal realm (Wanasinghe et al., 2023). In freshwater ecosystems, many fungi form an integral part of the material cycle and energy flow, making them an integral part of freshwater ecosystems (Hyde et al., 2016; Grossart et al., 2019). Over the past decade, more than 200 new species of Ascomycota, including members of the family Junewangiaceae, have been discovered in freshwater habitats in China (Luo et al., 2019; Bao et al., 2021; Dong et al., 2020; Calabon et al., 2022; Shen et al., 2022; Yang et al., 2023). However, these discoveries still remain insufficient compared to the forecasted number of species (Bánki et al., 2023; Calabon et al., 2023). Therefore, relevant research should continue to be conducted on a broader scale in the future.
Ariyawansa et al. (2015) introduced Dictyosporella within the family Annulatascaceae. The asexual morph of Dictyosporella is characterized by reduced conidiophores, monoblastic, terminal, thin-walled conidiogenous cells with or without separating cells, and muriform, broadly ellipsoidal to subglobose conidia (Yuan et al., 2020; Dong et al., 2021; Hyde et al., 2023). Later, the phylogenetic analysis by Xia et al. (2017) established the Junewangiaceae family to accommodate the acrodictys-like genus Junewangia. However, subsequent phylogenetic analysis by Luo et al. (2019) showed that Dictyosporella, Junewangia, and Sporidesmiella clustered within Junewangiaceae, a conclusion that is supported by follow-up studies (Dong et al., 2021; Li et al., 2021; Hyde et al., 2023). Additionally, Goh and Kuo (2020) introduced Jennwenomyces, which was segregated from Belemnospora. Although phylogenetic analysis positioned Jennwenomyces within Junewangiaceae, Goh and Kuo (2020) classified it as a Sordariomycetes genus insertae sedis.
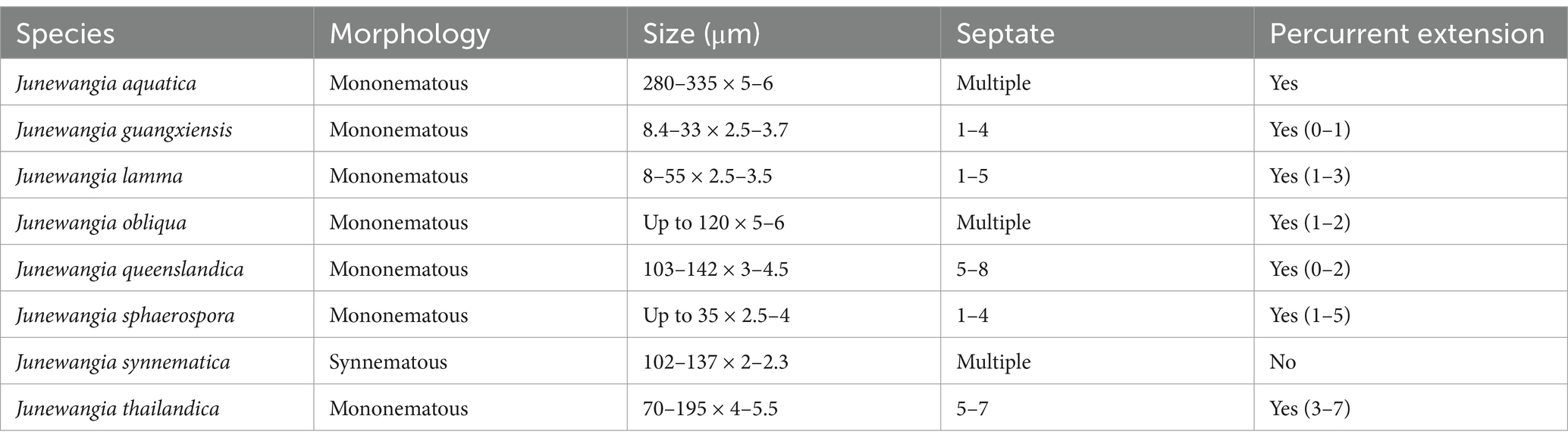
Currently, the phylogenetic boundaries between Dictyosporella, Junewangia, Jennwenomyces, and Sporidesmiella remain unclear (Goh and Kuo, 2020; Hyde et al., 2023). These genera exhibit distinct morphological differences. Both Dictyosporella and Junewangia have muriform, broadly ellipsoidal to subglobose conidia, but Junewangia is differentiated by macronematous, mononematous, erect conidiophores with percurrent proliferations (Figure 1; Ariyawansa et al., 2015; Xia et al., 2017; Song et al., 2018a; Yuan et al., 2020; Dong et al., 2021; Hyde et al., 2023). Jennwenomyces resembles Sporidesmiella in having macronematous, mononematous, cylindrical, thick-walled conidiophores (Goh and Kuo, 2020). However, Jennwenomyces differs in having navicular, euseptate conidia, whereas Sporidesmiella produces obovoid or clavate, distoseptate conidia (Figure 1; Luo et al., 2019; Goh and Kuo, 2020; Li et al., 2021; Liu et al., 2024; Tian et al., 2024).

Figure 1. Asexual morph of Junewangiaceae. (a–e) Dictyosporella spp.; (f–k) Junewangia spp.; (l–n) Sporidesmiella spp.; (o–s) Jennwenomyces fusiformis.
Materials and methods
Samples collection
Specimens of submerged decaying wood were collected from freshwater habitats (stream and river) in Yunnan Province (July 2023 and February 2024) and Guangxi Zhuang Autonomous Region (February 2024), China. The specimens were brought to the laboratory in plastic bags to preserve their integrity. The sample processing followed the method described by Shen et al. (2023): the samples were cut to the appropriate length, numbered, and placed in a disinfected plastic crisper for incubated culture at room temperature.
Isolation and morphological examination
Fungal colonies on natural substrates were observed using a Guiguang GL-99BI compound stereomicroscope (Guilin Guiguang Instrument Co., Ltd., Guilin, China) and photographed with a Nikon SMZ1000 stereo zoom microscope (Nikon Corporation, Tokyo, Japan). Fungal structures were photographed using a Nikon ECLIPSE Ni-U compound microscope (Nikon Corporation, Tokyo, Japan) fitted with a Nikon DS-Ri2 digital camera (Nikon Corporation, Tokyo, Japan), according to the guidelines provided in the study by Luo et al. (2018). Fungal species were isolated using single spore isolation following the method outlined in the study by Senanayake et al. (2020). Germinating ascospores and conidia were transferred to fresh potato dextrose agar (PDA) media and incubated at room temperature. Herbarium specimens (dry woody branches with fungal material) were deposited in the Herbarium of Cryptogams at the Kunming Institute of Botany, Academia Sinica (HKAS), Kunming, China. The isolates obtained in this study were deposited in the China General Microbiological Culture Collection Center (CGMCC), Beijing, China and the Kunming Institute of Botany Culture Collection Center (KUNCC), Kunming, China. Names of the new taxa were registered in Fungal Names (FN).1 We followed the suggestions provided in the study by Thines et al. (2020) and italicized all the Latin names that appeared in the text.
DNA extraction, PCR amplification, and sequencing
The genomic DNA was extracted from fungal mycelium. A Trelief™ Hi-Pure Plant Genomic DNA Kit (Beijing TsingKe Biotech Co., Ltd., Beijing, China) was used to extract total genomic DNA following the manufacturer’s instructions. DNA amplification was performed by polymerase chain reaction (PCR). the large subunit of nuclear ribosomal RNA gene (LSU), the nuclear ribosomal internal ranscribed spacer (ITS), the small subunit of nuclear ribosomal RNA gene (SSU), the translation elongation factor 1-alpha (tef1-α), and the second-largest subunit of RNA polymerase II (rpb2) gene regions were amplified using the primer pairs LR0R/LR5 (Vilgalys and Hopple, 1990), ITS5/ITS4 (White, 1990), NS1/NS4 (White, 1990), 983F/2218R (Rehner and Buckley, 2005), and fRPB2-5F/fRPB2-7cR (Liu et al., 1999). The amplifications were performed in a 25 μL reaction volume containing 9.5 μL ddH2O, 12.5 μL 2 × Taq PCR Master Mix with blue dye (Shanghai Sangon Biological Engineering Technology and Services Co., Shanghai, China), 1 μL DNA template, and 1 μL of each primer (10 μM). PCR products were checked on 1% agarose electrophoresis gels stained with GelRed (Beijing TsingKe Biotech Co., Ltd., Beijing, China). The sequencing reactions were carried out using the primers mentioned above by Shanghai Sangon Biological Engineering Technology and Services Co., Shanghai, China.
Phylogenetic analyses
The Basic Local Alignment Search Tool (BLAST) searches in the National Center of Biotechnology Information (NCBI) preliminarily screened out strains of Junewangiaceae. Five gene markers, LSU, ITS, SSU, tef1-α, and rpb2, were used for the multigene analyses, with the whole or part of them concatenated for different fungal groups. Single-locus sequences were aligned using the online multiple alignment program MAFFT version 7 (Rozewicki et al., 2019), and this alignment was manually optimized in BioEdit version 7.0.5.3 (Hall, 1999). The concatenated sequence alignments were obtained from SequenceMatrix version 1.7.8 (Vaidya et al., 2011).
Maximum likelihood (ML) analysis was performed using Randomized Axelerated Maximum Likelihood High-Performance Computing 2 (RAxML-HPC2) on ACCESS (Stamatakis, 2006; Stamatakis et al., 2008) on the Extreme Science and Engineering Discovery Environment (XSEDE) TeraGrid of the CIPRES Science Gateway online platform (Miller et al., 2010) with rapid bootstrap analysis, which was followed by 1,000 bootstrap replicates. The final tree was selected among the suboptimal trees from each run by comparing the likelihood scores under the general time-reversible gamma (GTRGAMMA) parameter substitution model.
Bayesian inference (BI) analysis was performed in a likelihood framework implemented in MrBayes version 3.1.2 (Ronquist et al., 2012). The Markov Chain Monte Carlo (MCMC) sampling approach was used to calculate posterior probabilities (PP) (Rannala and Yang, 1996). A Bayesian analysis of six simultaneous Markov chains was run for 10,000,000 generations, with trees sampled at intervals of every 1,000 generations. The sequences generated in this study have been deposited in GenBank and are listed in Table 1.
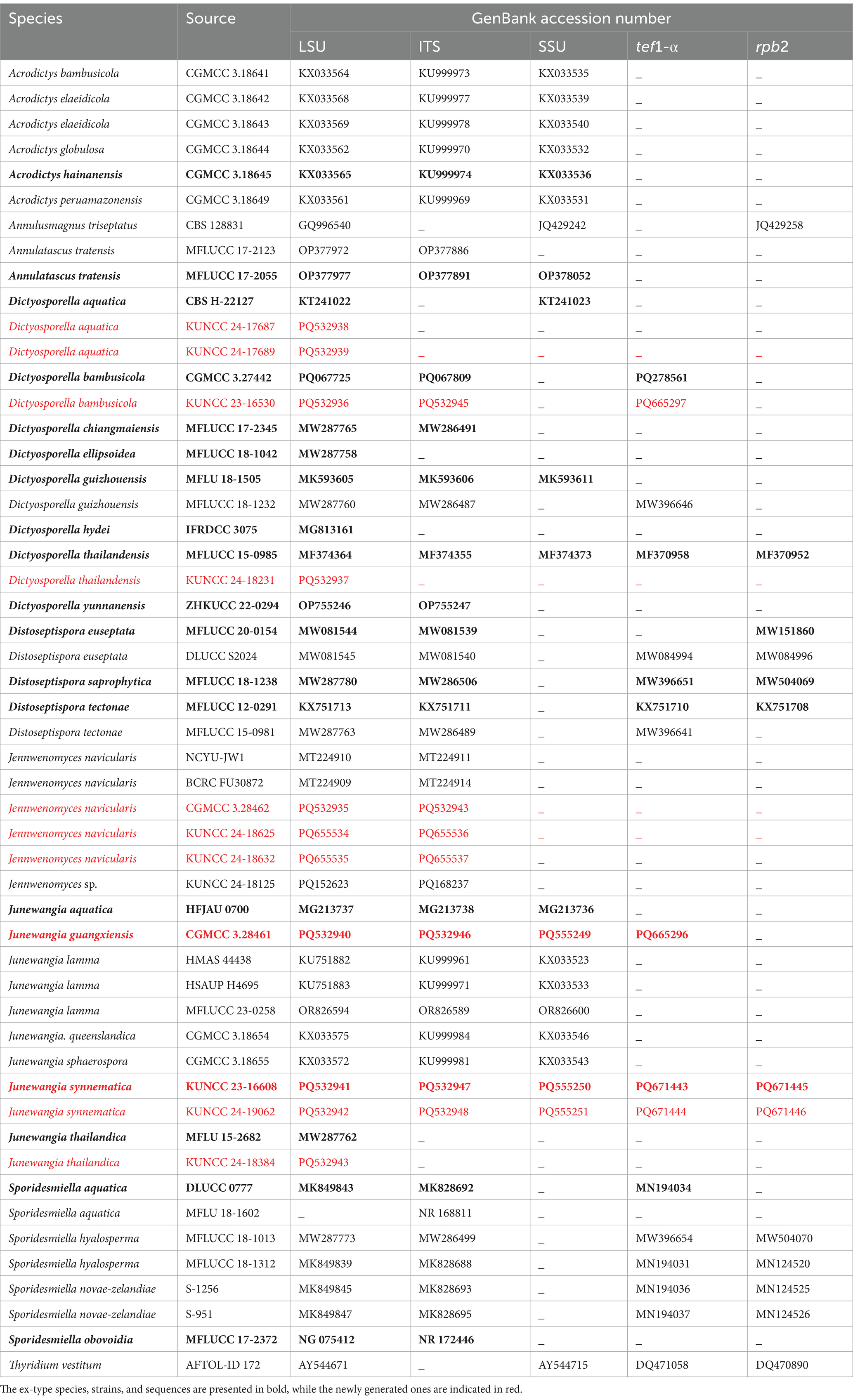
Table 1. Strains/specimens used for phylogenetic analysis along with their GenBank accession numbers.
Results
Phylogenetic analyses
The dataset that includes combined LSU, ITS, SSU, tef1-α, and rpb2 sequence data comprises 52 strains with 4,092 characters, including gaps (LSU: 1–802 bp, ITS: 803–1,310 bp, SSU: 1,311–2,178 bp, tef1-α: 2,179–3,042 bp, and rpb2: 3,043–4,092 bp). Thyridium vestitum (AFTOL-ID 172) was selected as the outgroup taxon. RAxML and Bayesian analyses were conducted and resulted in generally congruent topologies. The best RAxML tree with a final likelihood value of −21,682.843109 is presented. The matrix contained 1,439 distinct alignment patterns, with 53.62% undetermined characters or gaps. The estimated base frequencies were as follows: A = 0.241236, C = 0.253535, G = 0.284886, T = 0.220342; the substitution rates were as follows: AC = 1.104233, AG = 2.702435, AT = 1.264711, CG = 0.798496, CT = 6.288150, and GT = 1.000000; the gamma distribution shape parameter was α = 0.205348.
In the phylogenetic tree, 11 newly obtained strains are nested within the family Junewangiaceae (Figure 2). The species Junewangia guangxiensis (CGMCC 3.28461) and J. synnematica (KUNCC 23-16608 and KUNCC 24-19062) cluster together with Dictyosporella hydei (IFRDCC 3075), J. aquatica (HFJAU 0700), and J. thailandica (MFLU 15-2682 and KUNCC 24-18384) in the same clade with 97% ML/1.00 PP support. Three new collections of Jennwenomyces navicularis (CGMCC 3.28462, KUNCC 24-18625, and KUNCC 24-18632) grouped with the clade of Je. navicularis with 88% ML/1.00 PP support. Other four known species D. aquatica (KUNCC 24-17687 and KUNCC 24-17689), D. bambusicola (KUNCC 23-16530), D. thailandensis (KUNCC 24-18231), and J. thailandica (KUNCC 24-18384) cluster with their holotype or ex-type strains with 100% ML/1.00 PP, 100% ML/1.00 PP, 99% ML/0.98 PP, and 100% ML/1.00 PP support, respectively (Figure 2).
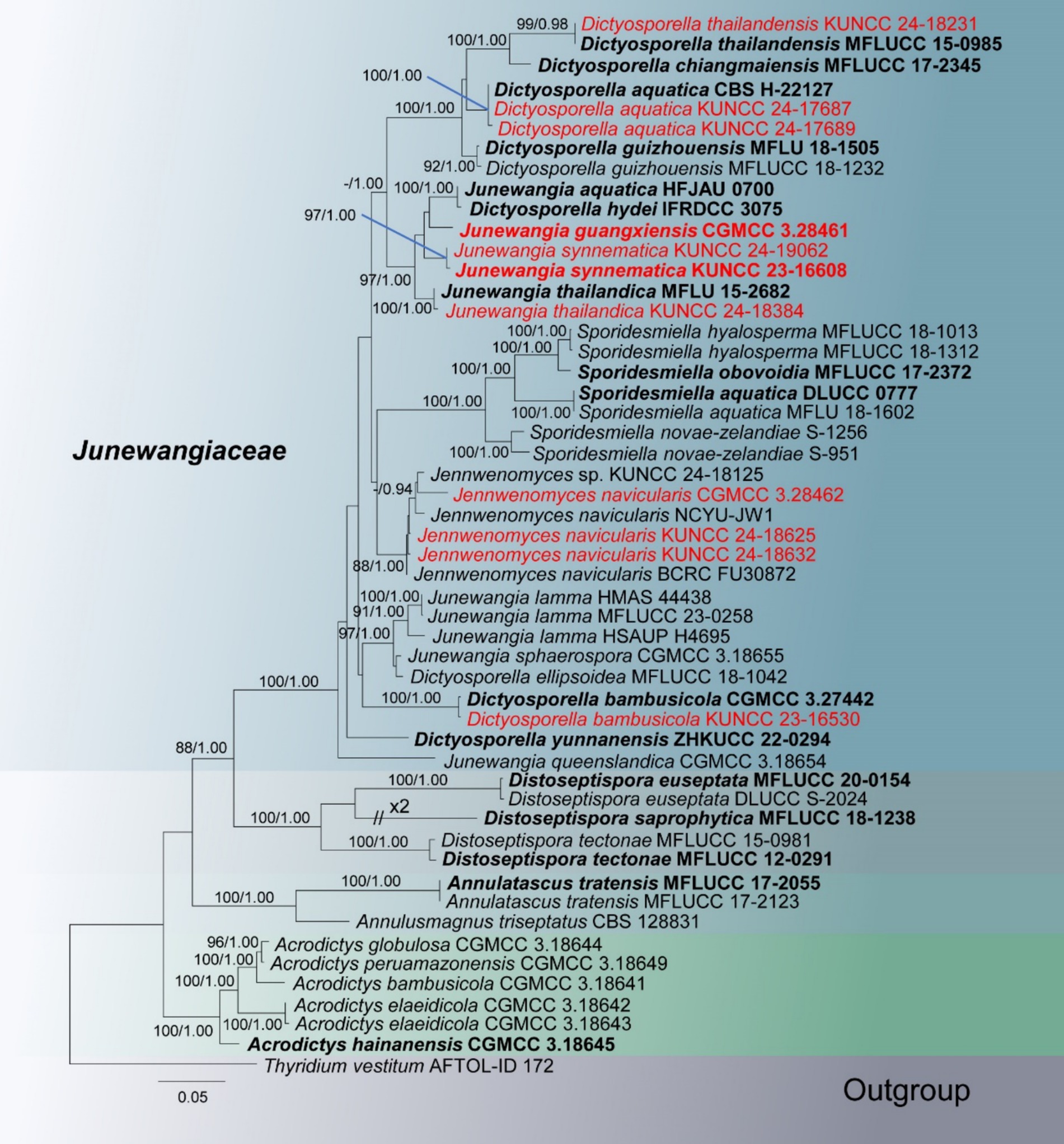
Figure 2. RAxML tree based on combined LSU, ITS, SSU, tef1-α, and rpb2 sequence date of Junewangiaceae. Bootstrap support values for maximum likelihood (ML) greater than 80% and Bayesian posterior probabilities (PP) greater than 0.90 are given as ML/PP above the nodes. The newly obtained sequences are indicated in red and ex-type strains are indicated in bold.
Taxonomy
Dictyosporella aquatica Abdel-Aziz, Fungal Diversity 75: 119 (2015), Figure 3.
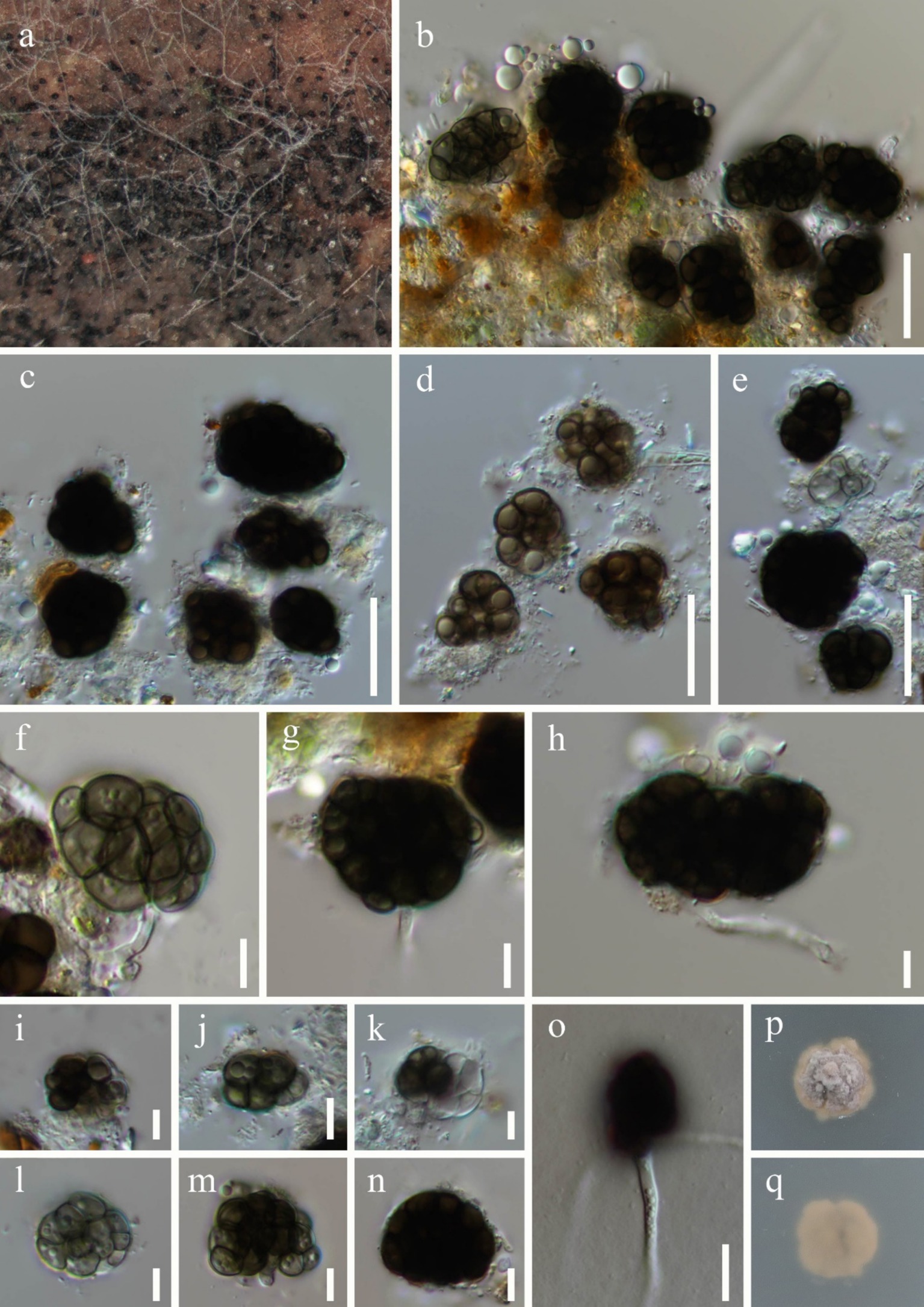
Figure 3. Dictyosporella aquatica (HKAS 144562). (a) Colonies on the substratum; (b–e,i–n) Conidia; (f–h) Conidiophores with conidia; (o) Germinating conidium; (p) Colony on PDA on the surface; (q) Colony on PDA on reverse. Scale bars: (b–e) 20 μm; (f–n) 5 μm; (o) 10 μm.
Fungal names number: FN551481.
Saprobic on submerged stems of Bambusoideae sp. Asexual morph: Colonies – superficial, effuse, gathered or scattered, dark brown to black, glistening. Mycelium – partly immersed, partly superficial, composed of aseptate, smooth, hyaline hyphae. Conidiophores – mostly reduced, mononematous, semi-macronematous, cylindrical, hyaline. Conidiogenous cells – monoblastic, integrated, terminal, hyaline. Conidia 16–29 × 9.8–24 μm ( = 22.1 × 16 μm, n = 30), acrogenous, solitary, helicoid when young, later becoming irregularly shaped, composed of subglobose to globose or irregular cells, guttulate, olive or brown when immature, dark brown when mature. Sexual morph: Undetermined.
Culture characteristics: Conidia germinating on PDA within 36 h and germ tubes. Colonies on PDA reaching 10 mm diameter after 8 weeks at room temperature. Colonies on the surface of PDA, protruding, with regular edges, dry, surface rough, grey to pale brown; pale brown, smooth from reverse.
Material examined: China, Guangxi Zhuang Autonomous Region, Nanning City (22°49′48″N; 108°13′32″E), on submerged Bambusoideae sp. in a freshwater river, 17 November 2023, Qiu-Xia Yang, S-5975 (HKAS 144562), living culture, KUNCC 24-17687; ibid., S-5977 (HKAS 144563), living culture, KUNCC 24-17689.
Notes: In the phylogenetic tree, two new collections (KUNCC 24-17687 and KUNCC 24-17689) were clustered with the ex-type strain of Dictyosporella aquatica (CBS H-22127) with 100% ML and 1.00 PP support (Figure 2). Our new collections resemble D. aquatica in having solitary conidia that are helicoid when young, later becoming irregular in shape, composed of subglobose to globose cells (Ariyawansa et al., 2015). We therefore identify our new collections as D. aquatica, a species introduced by Ariyawansa et al. (2015) on submerged decayed stems of Phragmites australis (Poaceae) in Egypt. This is the first report of this species in China.
Dictyosporella bambusicola X.D. Yu & Jian K. Liu, Mycosphere 15(1): 5104 (2024), Figure 4.

Figure 4. Dictyosporella bambusicola (HKAS 144564). (a) Colonies on the substratum; (b–e) Conidiophores with conidia; (f–k) Conidia; (l) Germinating conidium. (m) Colony on the surface of PDA; (n) Colony on the reverse of PDA. Scale bars: (b–l) 10 μm.
Fungal names number: FN854894.
Saprobic on submerged stems of Bambusoideae sp. Asexual morph: Colonies – superficial, effuse, sporodochia, brown. Mycelium – immersed, composed of aseptate, smooth, hyaline hyphae. Conidiophores – mostly reduced, semi-macronematous to micronematous, mononematous, unbranched, cylindrical, hyaline, aseptate, smooth and thin-walled. Conidiogenous cells 2.5–5.9 × 1.3–2.5 μm ( = 3.3 × 1.8 μm, n = 10), monoblastic, integrated, determinate, terminal, cylindrical, hyaline. Conidia 12–23 × 9.3–18 μm ( = 18 × 12.7 μm, n = 40), acrogenous, solitary, ellipsoidal, muriform, complanate, longitudinal or oblique and transverse separation, slightly constricted at the septum, composed of 3–4 rows of cells with a paler basal cell, hyaline when immature and becoming brown when mature, guttulate. Sexual morph: Undetermined.
Culture characteristics: Conidia germinating on PDA within 24 h and germ tubes produced from the base. Colonies on PDA reaching 8 mm diameter after 4 weeks at room temperature. Colonies on the surface of PDA, protruding, dry, with regular edges, surface rough, dark brown with a light brownish-yellow, gelatinous edge, light brownish-yellow, smooth from reverse.
Material examined: China, Yunnan Province, Honghe Hani and Yi Autonomous Prefecture, Mile City (24°42′69.75″N; 103°48′34.68″E), on submerged Bambusoideae sp. in a freshwater stream, 14 July 2023, Xing-Ya Zeng, S-5433 (HKAS 144564), living culture, KUNCC 23-16530.
Notes: Phylogenetic analysis revealed that our new collection clustered with the ex-type strain of Dictyosporella bambusicola with 100% ML and 1.00 PP support (Figure 2). Morphologically, our new collection exhibits micronematous, cylindrical conidiophores and ellipsoidal, muriform conidia, which are similar to D. bambusicola (Yu et al., 2024). A comparison of the ITS, LSU, and tef1-α sequence similarity between the new collection and D. bambusicola showed that 99.77% (434/435 bp), 100% (814/814 bp), and 99.45% (905/910 bp) similarity, respectively. We therefore identify our new collection as D. bambusicola based on phylogeny and morphological characteristics.
Dictyosporella thailandensis W. Dong, H. Zhang & K.D. Hyde, Fungal Diversity 85: 33 (2017), Figure 5.

Figure 5. Dictyosporella thailandensis (HKAS 144552). (a) Freshwater habitat; (b) and (c) Ascomata on the substratum; (d) Vertical section of ascoma; (e) Structure of peridium; (f) Paraphyses; (g–i) Asci; (j) Apex of ascus; (k–m) Ascospores; (n) Germinating ascospore; (o) Colony on the surface of PDA; (q) Colony on the reverse of PDA. Scale bars: (d) 30 μm; (e,f,n) 10 μm; (g–i) 20 μm; (j,k–m) 5 μm.
Fungal names number: FN553770.
Saprobic on submerged decaying wood. Asexual morph: Undetermined. Sexual morph: Ascomata 102–200 μm height and 101–197 μm diameter, scattered, solitary or gregarious, semi-immersed, with neck erumpent through host surface, uniloculate, ellipsoidal to subglobose, dark brown to black. Neck – cylindrical, central or lateral, hyaline to pale brown, periphysate, with multiple fluff appendages. Peridium 5.9–33 μm thickness, coriaceous, two-layered, outer layer dark brown to black, inner layer comprising multiple rows of hyaline to pale brown, thick-walled, irregular cells taxtura prismatica. Paraphyses 2.6–6 (−10) μm wide, hyaline, septate, slightly constricted at the septum, unbranched, tapering toward the apex, guttulate. Asci 113–167 (−180) × 9–14 μm ( = 142.4 × 11.5 μm, n = 20), eight-spored, unitunicate, cylindrical, with up to 43 μm long, tapering or dilated at the base pedicellate, rounded at the apex, with a refractive, wedge-shaped, J-, apical ring. Ascospores 15–19 × 5.6–7.3 μm ( = 17.5 × 6.4 μm, n = 30), oblique uniseriate, ellipsoidal, straight, hyaline, tri-septate, slightly constricted at the septum, guttulate, with dull and filamentous appendages at both ends.
Culture characteristics: Ascospores germinating on PDA within 12 h and germ tubes produced from one end. Colonies on PDA reaching 10 mm diameter after 4 weeks at room temperature. Colonies on the surface of PDA, flat surface with a small protrusion in the center, irregular edges, dry, white; white to pale brown, smooth from reverse
Material examined: China, Guangxi Zhuang Autonomous Region, Wuzhou City (23°43′47.98″N; 110°85′96.75″E), on unknown submerged decaying wood in a freshwater stream, 23 February 2024, Fa-Li Li, S-6304 (HKAS 144552), living culture, KUNCC 24-18231.
Notes: Dictyosporella thailandensis was introduced by Zhang et al. (2017) from a freshwater habitat in Thailand. This species is characterized by subglobose or ellipsoidal ascomata with hyaline to pale yellow neck erumpent through the host surface, unitunicate, long cylindrical, pedicellate asci with an apical ring, and straight, 3-septate ascospores with filamentous bipolar appendages (Zhang et al., 2017). Our new collection matches the characteristics of D. thailandensis, and the phylogenetic analysis also supports identifying our new collection as D. thailandensis. We therefore identify our new collection as D. thailandensis, and this is the first report of this species in China.
Jennwenomyces navicularis (R.F. Castañeda & Heredia) Goh &C.H. Kuo, Mycological Progress 19: 874 (2020), Figure 6.

Figure 6. Jennwenomyces navicularis (HKAS 144559). (a) Freshwater habitat; (b) Colonies on the substratum; (c–g) Conidiophores with conidium; (h) Conidia; (i) Germinating conidium; (j) Colony on the surface of PDA; (k) Colony on the reverse of PDA. Scale bars: (c–i) 20 μm.
Fungal names number: FN835023.
Saprobic on submerged stems of Bambusoideae sp. Asexual morph: Colonies – superfical, scattered, effuse, brown. Mycelium – mostly immersed, composed of aseptate, smooth, hyaline hyphae. Conidiophores 66–133 × 3.2–6.1 μm ( = 102.5 × 4.4 μm, n = 20), macronematous, mononematous, unbranched, cylindrical, 2–5-septate, straight or slightly flexuous, reddish-brown, paler toward the apex, smooth and thick-walled. Conidiogenous cells 36–56 × 3.3–5 μm ( = 42.4 × 4 μm, n = 20), polyblastic, integrated, terminal, determinate, sympodial. Conidia 33–64 × 5.9–10 μm ( = 49.6 × 8.6 μm, n = 20), acrogenous, solitary, smooth-walled, clavate when immature, becoming navicular when mature, tapering at both ends, with a rounded apex and a narrow hilum at the base, straight, 4(-5)-euseptate, partly with an inconspicuous septate at the apex cell, pale reddish-brown. Sexual morph: Undetermined.
Culture characteristics: Conidia germinating on PDA within 12 h and germ tubes produced from both ends. Colonies on PDA reaching 8 mm diameter after 6 weeks at room temperature. Colonies on the surface of PDA, surface rough, dry, regular edges, reddish-brown; dark brown in the center with reddish-brown edges from reverse.
Material examined: China, Guangxi Zhuang Autonomous Region, Guigang City (23°82′91.20″N; 110°26′68.92″E), on submerged Bambusoideae sp. in a freshwater stream, 23 February 2024, Tian-Tian Zhao, S-6457 (HKAS 144559), living culture, CGMCC 3.28462 = KUNCC 24-18389; Hechi City (24°55′70.58″N; 107°21′44.02″E), on unknown submerged decaying wood in a freshwater stream, 19 February 2024, Tian-Tian Zhao, S-6135 (HKAS 144551), living culture, KUNCC 24-18625; Baise City (24°08′90.39″N; 106°64′83.78″E), on unknown submerged decaying wood in a freshwater stream, 18 February 2024, Tian-Tian Zhao, S-6358 (HKAS 144553), living culture, KUNCC 24-18632.
Notes: Goh and Kuo (2020) established Jennwenomyces to accommodate J. navicularis which was transferred from Belemnospora. In the phylogenetic analysis, our new collections grouped with J. navicularis with 88% and 1.00 PP support (Figure 2). Comparison of the internal transcribed spacer (ITS) sequence of the new collection (CGMCC 3.28462) with J. navicularis (BCRC FU30872 and NCYU-JW1) and Jennwenomyces sp. (KUNCC 24-18125) revealed differences of 4.20% (22/524 bp, eight gaps), 5.15% (27/524 bp, eight gaps), and 4.58% (24/524 bp, eight gaps). However, our new collections have unbranched, cylindrical conidiophores and navicular, euseptate conidia with rounded apex, which fits well with the description of J. navicularis. Although there are significant differences between the ITS sequences of the new collection (CGMCC 3.28462) and J. navicularis (BCRC FU30872 and NCYU-JW1), we can identify our new collections as J. navicularis based on phylogenetic analysis and morphological characteristics. Moreover, studying this genus requires the discovery of more species in the future.
Junewangia guangxiensis W.P. Wang & Z.L. Luo, sp. nov., Figure 7.

Figure 7. Junewangia guangxiensis (HKAS 144567, holotype). (a) Freshwater habitat; (b) Colonies on the substratum; (c–g) Conidiophores with conidia; (h–k) Conidia; (l) Germinating conidium; (m) Colony on the surface of PDA; (n) Colony on the reverse of PDA. Scale bars: (c–g) 15 μm; (h–l) 10 μm.
Fungal names number: FN572298.
Etymology: Referring to the Guangxi Zhuang Autonomous Region, China, where the species was collected.
Holotype: HKAS 144567.
Saprobic on submerged decaying wood. Asexual morph: Colonies – effuse, scattered, gathered in small groups, and brown. Mycelium – mostly immersed, composed of aseptate, smooth, and pale brown hyphae. Conidiophores 8.4–33 × 2.5–3.7 μm ( = 18.1 × 3 μm, n = 30), macronematous, mononematous, erect, cylindrical, straight or slightly flexuous, 1–4-septate, unbranched, brown, smooth, thick-walled, with 0–1 percurrent proliferations. Conidiogenous cells – monoblastic, integrated, terminal, cuneiform to doliiform, pale brown. Conidia – 17–23 × 8.5–12 μm ( = 20.1 × 10.5 μm, n = 40), acrogenous, solitary, ellipsoidal, muriform, composed of two columns cells with a cuneiform basal cell, longitudinal or oblique, and transverse separation, slightly constricted at the septum, partly rows without longitudinal septate, 1–2 cells at the apex row, rounded apical, brown, guttulate. Sexual morph: Undetermined.
Culture characteristics: Conidia germinating on PDA within 24 h and germ tubes produced from the base. Colonies on PDA reaching 8 mm diameter after 9 weeks at room temperature. Colonies on the surface of PDA, regular edges, dry, with a small, punctate protrusion in the center, reddish-brown to brown; brown and smooth from reverse.
Material examined: China, Guangxi Zhuang Autonomous Region, Nanning City (23°02′84.57″N; 107°52′41.73″E), on unknown submerged decaying wood in a freshwater stream, 28 February 2024, Zheng-Quan Zhang, S-6434 (HKAS 144567, holotype), ex-type culture, CGMCC 3.28461 = KUNCC 24-18376.
Notes: Junewangia guangxiensis has macronematous, erect, thick-walled conidiophores, which align well with the generic concept of Junewangia (Baker et al., 2002; Xia et al., 2017; Dong et al., 2021). Junewangia guangxiensis has short conidiophores that resemble J. sphaerospora (Table 2). However, J. guangxiensis differs from J. sphaerospora by having fewer percurrent proliferations (0–1 vs. 1–5 in J. sphaerospora), cuneiform to doliiform conidiogenous cells, and ellipsoidal, muriform, narrower conidia (8.5–12 vs. 12–18 μm wide) (Xia et al., 2017). Phylogenetic analysis also supports J. guangxiensis as a distinct species (Figure 2). We therefore recognize J. guangxiensis as a new species.
Junewangia synnematica W.P. Wang & Z.L. Luo, sp. nov., Figure 8.

Figure 8. Junewangia synnematica (HKAS 144554, holotype). (a) Colony on the substratum; (b) Synnema; (c,d) Synnemata with conidia; (e) Conidiogenous cells; (f) Conidiogenous cell with conidia; (g–l) Conidia; (m) Germinating conidium; (n) Colony on the surface of PDA; (o) Colony on the reverse of PDA. Scale bars: (b–d) 50 μm; (e–m) 10 μm.
Fungal names number: FN572299.
Etymology: Referring to synnematous conidiophores of this fungus.
Holotype: HKAS 144554.
Saprobic on submerged decaying wood. Asexual morph: Colonies – erect, scattered, and brown to dark brown conidia gathered at the apex of synnemata. Mycelium - immersed, composed of aseptate, smooth, hyaline, unbranched hyphae. Conidiophores 102–137 × 2–2.3 μm ( = 122.8 × 2.1 μm, n = 10), macronematous, synnematous, cylindrical, straight or slightly flexuous, multiseptate, unbranched, brown, slightly paler toward the apex, smooth and thick-walled. Synnemata 192–213 × 21–40 μm ( = 201.5 × 29.7 μm, n = 5), erect, rigid, cylindrical, and dark brown. Conidiogenous cells 7.2–14.4 × 2.6–3.9 μm ( = 11.3 × 3 μm, n = 10), monoblastic, integrated, determinate, terminal, cylindrical, brown. Conidia 21–28 × 13–17 μm ( = 26.1 × 14.7 μm, n = 40), acrogenous, solitary, broadly ellipsoidal to pyriform, muriform, smooth-walled, with longitudinal and transverse separation; oblique septate at the apex row, slightly constricted at septum, composed of 3 columns and 4 (−5) rows of cells; broadly rounded apical, brown, guttulate; with a globose to subglobose, 6.3–7.7 μm in diameter, hyaline, and thin-walled separating cell connect to conidiogenous cells, becoming cuneiform base when mature. Sexual morph: Undetermined.
Culture characteristics: Conidia germinating on PDA within 24 h and germ tubes produced from the base. Colonies on PDA reaching 10 mm diameter after 6 weeks at room temperature. Colonies on the surface of PDA, irregular edges, umbellate, dry, reddish-brown to brown; smooth, reddish-brown to brown from reverse.
Material examined: China, Yunnan Province, Qujing City, Luoping County (25°01′52.57″N; 104°42′47.40″E), on unknown submerged decaying wood in a freshwater river, 15 July 2023, Fa-Li Li, S-5643 (HKAS 144554, holotype), ex-type culture, KUNCC 23-16608; Wenshan Zhuang and Miao Autonomous Prefecture, Malipo County (23°49′99.99″N; 104°97′74.49″E), on unknown submerged decaying wood in Dahe River, 28 February 2024, Ting-Xiang Liu, S-6470 (HKAS 144568, paratype), living culture, KUNCC 24-19062.
Notes: Junewangia synnematica is the first species in the genus Junewangia which has synnematous conidiophores (Table 2); it is distinguished from other species in this genus (Baker et al., 2002; Xia et al., 2017; Song et al., 2018a; Dong et al., 2021). In the phylogenetic analysis, J. synnematica (KUNCC 23-16608 and KUNCC 24-19062) clustered with Dictyosporella hydei, J. aquatica, J. guangxiensis, and J. thailandica in the same clade with 97% ML and 1.00 PP support (Figure 2). A comparison of the ITS sequence of the ex-type strain of J. synnematica with J. aquatica (HFJAU 0700) and J. guangxiensis (KUNCC 24-18376) (as D. hydei and J. thailandica lack ITS sequence in GenBank) showed differences of 6.73% (37/550 bp, nine gaps) and 4.93% (27/548 bp, seven gaps), respectively. We therefore introduced J. synnematica as a new species based on morphological characteristics and phylogenetic analysis.
Junewangia thailandica W. Dong, H. Zhang & K.D. Hyde, Mycosphere 12(1): 53 (2021), Figure 9.

Figure 9. Junewangia thailandica (HKAS 144560). (a) Freshwater habitat; (b,c) Colonies on the substratum; (d–g) Conidiophores with conidia; (h) Conidiogenous cells with conidia; (i,j) Conidia; (k) Germinating conidium; (l) Colony on the surface of PDA; (m) Colony on the reverse of PDA. Scale bars: (d–g) 30 μm; (h) 20 μm; (i–k) 10 μm.
Fungal names number: FN558045.
Saprobic on submerged decaying wood. Asexual morph: Colonies - erect, scattered, brown, globose conidia at the apex of conidiophores. Mycelium – partly immersed, partly superficial, composed of septate, smooth, branched, brown hyphae. Conidiophores 112–177 × 4.7–6.3 μm ( = 148.3 × 5.4 μm, n = 20), macronematous, mononematous, cylindrical, straight or slightly flexuous, 5–7-septate, unbranched, brown, paler toward the apex, smooth and thick-walled, with 2–6 percurrent proliferations. Conidiogenous cells – monoblastic, integrated, terminal, cylindrical, pale brown, smooth-walled. Conidia 24–37 × 18–28 μm ( = 29.4 × 22.7 μm, n = 50), acrogenous, solitary, broadly ellipsoidal to subglobose, muriform, smooth-walled, longitudinal or oblique and transverse separation, constricted at septum, composed 3 (−4) rows cells, broadly rounded apical, brown, guttulate, with a cuneiform basal cell. Sexual morph: Undetermined.
Culture characteristics: Conidia germinating on PDA within 24 h and germ tubes produced from surface. Colonies on PDA reaching 8 mm diameter after 8 weeks at room temperature. Colonies semi-immersed in PDA, irregular edges, surface flat with a small protrusion in the center, dry, dark brown with a layer of gray hyphae covering the surface; dark brown from reverse.
Material examined: China, Guangxi Zhuang Autonomous Region, Yulin City (22°24′48.70″N; 109°70′42.37″E), on unknown submerged decaying wood in a freshwater stream, 25 February 2024, Wen-Peng Wang, S-6449 (HKAS 144560), living culture, KUNCC 24-18384.
Notes: Phylogenetic analysis showed that our new collection (KUNCC 24-18384) clustered with the holotype of Junewangia thailandica (MFLU 15-2682) with 100% ML and 1.00 PP support (Figure 2). Morphologically, our new collection fits well with the conception of J. thailandica in having macronematous, mononematous, cylindrical, and unbranched conidiophores with several percurrent proliferations and broadly ellipsoidal to subglobose, muriform conidia with similar size (24–37 × 18–28 vs. 22–32.5 × 16.5–23 μm) (Dong et al., 2021). We therefore identify our new collection as J. thailandica, a species first described by Dong et al. (2021) from a freshwater habitat in Thailand, and this is a new geographical record in China.
Discussion
In this study, we introduced two new species of the genus Junewangia based on morphological characteristics and phylogenetic analysis. Phylogenetic analysis showed that the four genera, Dictyosporella, Jennwenomyces, Junewangia, and Sporidesmiella were chaotically clustered, especially Dictyosporella and Junewangia (Luo et al., 2019; Dong et al., 2021; Hyde et al., 2023; Figure 2). This finding suggests that species identification within Junewangiaceae cannot be solely based on the phylogenetic analysis. As shown in Figure 1, the primary difference between Dictyosporella and Junewangia lies in the conidiophores, whereas their muriform conidia are not significantly different (Xia et al., 2017; Song et al., 2018a,b; Dong et al., 2021). Some genera with closely phylogenetic relationships have similar conidia produced from different conidiophores, for example, Dendryphion and Torula (Su et al., 2016, 2018). Of course, some taxa exhibit highly variable conidiophores, even within the same genus or species, for example, Phaeoiseria and Pleurotheciella (Crous et al., 2015, 2017; Boonmee et al., 2021; Shi et al., 2021; Wang et al., 2024).
When discussing these issues, we cannot overlook the influence of factors, such as altitude, latitude, temperature, and host, on fungal morphology. Some studies have found that the host can influence the fungal morphology, particularly on conidiophores (Réblová et al., 2016; Jayawardena et al., 2022; Li et al., 2025), and this phenomenon is also observed in Junewangia (Xia et al., 2017). Understanding the host’s influence could be a way to resolve the taxonomic issue between Dictyosporella and Junewangia, but there are still too few relevant species for reference. In addition, studying the sexual morph is another potential approach to solving the taxonomic problem of Junewangiaceae. Currently, the sexual morph is only reported in Dictyosporella (Zhang et al., 2017; Dong et al., 2021).
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Author contributions
W-PW: Data curation, Formal analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing. C-GL: Conceptualization, Writing – review & editing. T-XL: Formal analysis, Investigation, Writing – review & editing. H-WS: Software, Validation, Writing – review & editing. Z-LL: Conceptualization, Data curation, Formal analysis, Funding acquisition, Methodology, Resources, Software, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. We would like to thank the National Natural Science Foundation of China (Project ID: 32060005) and the Yunnan Fundamental Research Project (202201AW070001) for financial support. This work was also supported by the Foundation of Yunnan Province Science and Technology Department (202305AM070003).
Acknowledgments
We would like to thank Dr. Shaun Pennycook (Landcare Research, Auckland, New Zealand) for checking the fungal nomenclature. Wen-Peng Wang extends thanks to Qiu-Xia Yang, Zheng-Quan Zhang, and Xing-Ya Zeng for sample collection. Fa-Li Li, Ming-Chuan Li, Tian-Tian Zhao, and Si-Rui Yu (College of Agriculture and Biological Science, Dali University, Yunnan, China) are acknowledged for helping with DNA extraction.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
1. ^https://nmdc.cn/fungalnames/, accessed on 2 January 2025.
References
Ariyawansa, H. A., Hyde, K. D., Jayasiri, S. C., Buyck, B., Chethana, K. W. T., Dai, D. Q., et al. (2015). Fungal diversity notes 111–252—taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 75, 27–274. doi: 10.1007/s13225-015-0346-5
Baker, W. A., Partridge, E. C., and Morgan-Jones, G. (2002). Notes on hyphomycetes LXXXV. Junewangia, a genus in which to classify four Acrodictys species and a new taxon. Mycotaxon 81, 293–319.
Bánki, O., Roskov, Y., Döring, M., Ower, G., Vandepitte, L., Hobern, D., et al. (2023). Catalogue of life checklist (version 2023-01-12). Catalogue Life. doi: 10.48580/dfqz
Bao, D. F., Hyde, K. D., McKenzie, E. H. C., Jeewon, R., Su, H. Y., Nalumpang, S., et al. (2021). Biodiversity of lignicolous freshwater hyphomycetes from China and Thailand and description of sixteen species. J. Fungi 7:669. doi: 10.3390/jof7080669
Boonmee, S., Wanasinghe, D. N., Calabon, M. S., Huanraluek, N., Chandrasiri, S. K. U., Jones, E. B. G., et al. (2021). Fungal diversity notes 1387–1511: taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Divers. 111, 1–335. doi: 10.1007/s13225-021-00489-3
Calabon, M. S., Hyde, K. D., Jones, E. B. G., Bao, D. F., Bhunjun, C. S., Phukhamsakda, C., et al. (2023). Freshwater fungal biology. Mycosphere 14, 195–413. doi: 10.5943/mycosphere/14/1/4
Calabon, M. S., Hyde, K. D., Jones, E. B. G., Luo, Z. L., Dong, W., Hurdeal, V. G., et al. (2022). Freshwater fungal numbers. Fungal Divers. 114, 3–235. doi: 10.1007/s13225-022-00503-2
Crous, P. W., Schumacher, R. K., Wingfield, M. J., Lombard, L., Giraldo, A., Christensen, M., et al. (2015). Fungal systematics and evolution: FUSE 1. Sydowia 67, 81–118. doi: 10.12905/0380.sydowia67-2015-0081
Crous, P. W., Wingfield, M. J., Burgess, T. I., Carnegie, A. J., Hardy, G. E., Smith, D., et al. (2017). Fungal planet description sheets: 625–715. Persoonia 39, 270–467. doi: 10.3767/persoonia.2017.39.11
Díaz, S. (2022). COP15 biodiversity plan risks being alarmingly diluted. Nature 612:9. doi: 10.1038/d41586-022-04154-w
Dong, W., Hyde, K. D., Jeewon, R., Doilom, M., Yu, X. D., Wang, G. N., et al. (2021). Towards a natural classification of annulatascaceae-like taxa II: introducing five new genera and eighteen new species from freshwater. Mycosphere 12, 1–88. doi: 10.5943/mycosphere/12/1/1
Dong, W., Wang, B., Hyde, K. D., McKenzie, E. H. C., Raja, H. A., Tanaka, K., et al. (2020). Freshwater Dothideomycetes. Fungal Divers. 105, 319–575. doi: 10.1007/s13225-020-00463-5
Goh, T. K., and Kuo, C. H. (2020). Jennwenomyces, a new hyphomycete genus segregated from Belemnospora, producing versicolored phragmospores from percurrently extending conidiophores. Mycol. Prog. 19, 869–883. doi: 10.1007/s11557-020-01602-7
Grossart, H., den Wyngaert, S. V., Kagami, M., Wurzbacher, C., Cunliffe, M., and Rojas-Jimenez, K. (2019). Fungi in aquatic ecosystems. Nat. Rev. Microbiol. 17, 339–354. doi: 10.1038/s41579-019-0175-8
Hall, T. A. (1999). “BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT” in Nucleic acids symposium series (Oxford, UK: Oxford University Press), 95–98.
Hyde, K. D., Fryar, S., Tian, Q., Bahkali, A. H., and Xu, J. C. (2016). Lignicolous freshwater fungi along a north-south latitudinal gradient in the Asian/Australian region; can we predict the impact of global warming on biodiversity and function? Fungal Ecol. 19, 190–200. doi: 10.1016/J.FUNECO.2015.07.002
Hyde, K. D., Norphanphoun, C., Ma, J., Yang, H. D., Zhang, J. Y., Du, T. Y., et al. (2023). Mycosphere notes 387–412 – novel species of fungal taxa from around the world. Mycosphere 14, 663–744. doi: 10.5943/mycosphere/14/1/8
Jayawardena, R. S., Hyde, K. D., Wang, S., Sun, Y. R., Suwannarach, N., Sysouphanthong, P., et al. (2022). Fungal diversity notes 1512–1610: taxonomic and phylogenetic contributions on genera and species of fungal taxa. Fungal Divers. 117, 1–272. doi: 10.1007/s13225-022-00513-0
Li, L., Bhat, D. J., Jiang, H. B., Li, J. F., Dawoud, D. M., Sun, F. Q., et al. (2025). New insights into freshwater ascomycetes: discovery of novel species in diverse aquatic habitats. Front. Cell. Infect. Microbiol. 14:1515972. doi: 10.3389/fcimb.2024.1515972
Li, X. H., Liu, Y. L., Song, H. Y., Hu, D. M., Gao, Y., Hu, H. J., et al. (2021). Sporidesmiella lignicola sp. nov., a new hyphomycetous fungus from freshwater habitats in China. Biodivers. Data J. 9:e77414. doi: 10.3897/BDJ.9.e77414
Liu, N. G., Hyde, K. D., Sun, Y. R., Bhat, D. J., Jones, E. B. G., Jumpathong, J., et al. (2024). Notes, outline, taxonomy and phylogeny of brown-spored hyphomycetes. Fungal Divers. 129, 1–281. doi: 10.1007/s13225-024-00539-6
Liu, Y. J., Whelen, S., and Hall, B. D. (1999). Phylogenetic relationships among ascomycetes: evidence from an RNA polymerase II subunit. Mol. Biol. Evol. 16, 1799–1808. doi: 10.1093/OXFORDJOURNALS.MOLBEV.A026092
Luo, Z. L., Hyde, K. D., Liu, J. K., Bhat, D. J., Bao, D. F., Li, W. L., et al. (2018). Lignicolous freshwater fungi from China II: novel Distoseptispora (Distoseptisporaceae) species from northwestern Yunnan Province and a suggested unified method for studying lignicolous freshwater fungi. Mycosphere 9, 444–461. doi: 10.5943/mycosphere/9/3/2
Luo, Z. L., Hyde, K. D., Liu, J. K., Maharachchikumbura, S. S. N., Jeewon, R., Bao, D. F., et al. (2019). Freshwater Sordariomycetes. Fungal Divers. 99, 451–660. doi: 10.1007/s13225-019-00438-1
Miller, M. A., Pfeiffer, W., and Schwartz, T. (2010). Creating the CIPRES science gateway for inference of large phylogenetic trees. In Proceedings of the 2010 gateway computing environments workshop (GCE), New Orleans, LA, USA, 14 November 2010.
Rannala, B., and Yang, Z. (1996). Probability distribution of molecular evolutionary trees: a new method of phylogenetic inference. J. Mol. Evol. 43, 304–311. doi: 10.1007/BF02338839
Réblová, M., Seifert, K. A., Fournier, J., and Štěpánek, V. (2016). Newly recognised lineages of perithecial ascomycetes: the new orders Conioscyphales and Pleurotheciales. Persoonia 37, 57–81. doi: 10.3767/003158516X689819
Rehner, S. A., and Buckley, E. (2005). A Beauveria phylogeny inferred from nuclear ITS and EF1-α sequences: evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 97, 84–98. doi: 10.3852/mycologia.97.1.84
Ronquist, F., Teslenko, M., Van Der Mark, P., Ayres, D. L., Darling, A., Höhna, S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Rozewicki, J., Li, S. L., Amada, K. M., Standley, D. M., and Katoh, K. (2019). MAFFT-DASH: integrated protein sequence and structural alignment. Nucleic Acids Res. 47, W5–W10. doi: 10.1093/nar/gkz342
Senanayake, I. C., Rathnayaka, A. R., Marasinghe, D. S., Calabon, M. S., Gentekaki, E., Lee, H. B., et al. (2020). Morphological approaches in studying fungi: collection, examination, isolation, sporulation and preservation. Mycosphere 11, 2678–2754. doi: 10.5943/mycosphere/11/1/20
Shen, H. W., Bao, D. F., Bhat, D. J., Su, H. Y., and Luo, Z. L. (2022). Lignicolous freshwater fungi in Yunnan Province, China: an overview. Mycology 13, 119–132. doi: 10.1080/21501203.2022.2058638
Shen, H. W., Bao, D. F., Boonmee, S., Su, X. J., Tian, X. G., Hyde, K. D., et al. (2023). Lignicolous freshwater fungi from plateau lakes in China (I): morphological and phylogenetic analyses reveal eight species of Lentitheciaceae, including new genus, new species and new records. J. Fungi 9:962. doi: 10.3390/jof9100962
Shi, L., Yang, H., Hyde, K. D., Wijayawardene, N. N., Wang, G. N., Yu, X. D., et al. (2021). Freshwater Sordariomycetes: new species and new records in Pleurotheciaceae, Pleurotheciales. Phytotaxa 518, 143–166. doi: 10.11646/phytotaxa.518.2.4
Song, H. Y., Huo, G. H., and Hu, D. M. (2018b). Dictyosporella hydei sp. nov., an asexual species from freshwater habitats in China. Phytotaxa 358, 181–188. doi: 10.11646/phytotaxa.358.2.5
Song, H. Y., Zhong, P. A., Liao, J. L., Wang, Z. H., Hu, D. M., and Huang, Y. J. (2018a). Junewangia aquatica (Junewangiaceae), a new species from freshwater habitats in China. Phytotaxa 336, 272–278. doi: 10.11646/phytotaxa.336.3.5
Stamatakis, A. (2006). RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690. doi: 10.1093/bioinformatics/btl446
Stamatakis, A., Hoover, P., and Rougemont, J. (2008). A rapid bootstrap algorithm for the RAxML web servers. Syst. Biol. 57, 758–771. doi: 10.1080/10635150802429642
Su, H. Y., Hyde, K. D., Maharachchikumbura, S. S. N., Ariyawansa, H. A., Luo, Z. L., Promputtha, I., et al. (2016). The families Distoseptisporaceae fam. Nov., Kirschsteiniotheliaceae, Sporormiaceae and Torulaceae, with new species from freshwater in Yunnan Province, China. Fungal Divers. 80, 375–409. doi: 10.1007/s13225-016-0362-0
Su, X. J., Luo, Z. L., Jeewon, R., Bhat, D. J., Bao, D. F., Li, W. L., et al. (2018). Morphology and multigene phylogeny reveal new genus and species of Torulaceae from freshwater habitats in northwestern Yunnan, China. Mycol. Prog. 17, 531–545. doi: 10.1007/s11557-018-1388-3
Thines, M., Aoki, T., Crous, P. W., Hyde, K. D., Lücking, R., Malosso, E., et al. (2020). Setting scientific names at all taxonomic ranks in italics facilitates their quick recognition in scientific papers. IMA Fungus 11:25. doi: 10.1186/s43008-020-00048-6
Tian, W. H., Jin, Y., Liao, Y. C., Faraj, T. K., Guo, X. Y., and Maharachchikumbura, S. S. N. (2024). New and interesting pine-associated hyphomycetes from China. J. Fungi 10:546. doi: 10.3390/jof10080546
Vaidya, G., Lohman, D. J., and Meier, R. (2011). Cladistics multi-gene datasets with character set and codon information. Cladistics 27, 171–180. doi: 10.1111/j.1096-0031.2010.00329.x
Vilgalys, R., and Hopple, M. (1990). Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J. Bacteriol. 172, 4238–4246. doi: 10.1128/jb.172.8.4238-4246.1990
Wanasinghe, D. N., Nimalrathna, T. S., Xian, L. Q., Faraj, T. K., Xu, J. C., and Mortimer, P. E. (2023). Taxonomic novelties and global biogeography of Montagnula (Ascomycota, Didymosphaeriaceae). MycoKeys 101, 191–232. doi: 10.3897/mycokeys.101.113259
Wang, W. P., Bhat, D. J., Yang, L., Shen, H. W., and Luo, Z. L. (2024). New species and records of Pleurotheciaceae from karst landscapes in Yunnan Province, China. J. Fungi 10:516. doi: 10.3390/jof10080516
White, T. J. (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protocols 38, 315–322. doi: 10.1016/B978-0-12-372180-8.50042-1
Xia, J. W., Ma, Y. R., Li, Z., and Zhang, X. G. (2017). Acrodictys-like wood decay fungi from southern China, with two new families Acrodictyaceae and Junewangiaceae. Sci. Rep. 7:7888. doi: 10.1038/s41598-017-08318-x
Yang, J., Liu, L. L., Jones, E. B. G., Hyde, K. D., Liu, Z. Y., Bao, D. F., et al. (2023). Freshwater fungi from karst landscapes in China and Thailand. Fungal Divers. 119, 1–212. doi: 10.1007/s13225-023-00514-7
Yu, X. D., Zhang, S. N., Liang, X. D., Zhu, J. T., Hyde, K. D., and Liu, J. K. (2024). Bambusicolous Fungi from southwestern China. Mycosphere 15, 5038–5145. doi: 10.5943/mycosphere/15/1/24
Yuan, H. S., Lu, X., Dai, Y. C., Hyde, K. D., Kan, Y. H., Kušan, I., et al. (2020). Fungal diversity notes 1277–1386: taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 104, 1–266. doi: 10.1007/s13225-020-00461-7
Keywords: two new taxa, aquatic fungi, morphology, phylogeny, taxonomy
Citation: Wang W-P, Lin C-G, Liu T-X, Shen H-W and Luo Z-L (2025) Additions to the family Junewangiaceae (Sordariomycetes): novel species and new records from freshwater habitats in Southwestern China. Front. Microbiol. 16:1566263. doi: 10.3389/fmicb.2025.1566263
Edited by:
Baokai Cui, Beijing Forestry University, ChinaReviewed by:
Jing Si, Beijing Forestry University, ChinaHuang Zhang, Kunming University of Science and Technology, China
Copyright © 2025 Wang, Lin, Liu, Shen and Luo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zong-Long Luo, bHVvem9uZ2xvbmdmdW5naUAxNjMuY29t
 Wen-Peng Wang
Wen-Peng Wang Chuan-Gen Lin
Chuan-Gen Lin Ting-Xiang Liu1
Ting-Xiang Liu1 Hong-Wei Shen
Hong-Wei Shen Zong-Long Luo
Zong-Long Luo