- 1Department of Nephrology and Endocrinology, Dongzhimen Hospital, Beijing University of Chinese Medicine, Beijing, China
- 2College of Pharmacy, Baotou Medical College, Baotou, China
- 3College of Traditional Chinese Medicine, Beijing University of Chinese Medicine, Beijing, China
- 4Graduate School, Heilongjiang University of Chinese Medicine, Harbin, China
- 5Changzhi People’s Hospital Affiliated to Changzhi Medical College, Changzhi, China
Background: Liver injury (LI) is responsible for a significant number of fatalities each year. In the context of Mongolian medicine, Rhododendron molle (Blume) G. Don (RM) is utilized for its properties to treatment of hepatic disorders. However, the underlying mechanisms of its action remain poorly understood.
Objectives: Clarifying the process through which RM enhances LI.
Methods: The chemical constituents were subjected to analysis, and network pharmacology alongside molecular docking studies were conducted. Additionally, ELISA, staining techniques, metabolomic analyses, and 16S rDNA sequencing were performed.
Results: A total of 17 components have been identified from RM, including liver disease-related compounds such as kaempferol, emodin, quercetin. Network pharmacology has identified notable genes that exhibit a strong binding affinity to active compounds, including emodin, which interacts with IL6 and PPARG, and aloeemodin, which binds to IL6 and AKT1. In a rat model of LI induced by CCL4, low dose (0.07875 g/kg) of RM demonstrated a reduction in ALT and γ-GT levels (p < 0.05). Metabolomic analysis indicated that RM has an impact on the concentrations of 13-OxoODE, morphine, and niacinamide in rat models exhibiting LI, simultaneously several metabolic pathways, including steroid biosynthesis, linoleic acid metabolism, and tryptophan metabolism. By integrating the findings from metabolomics with KEGG pathways, it was determined that RM may ameliorate LI by activating specific pathways and modulating fatty acid metabolic processes, particularly linoleic acid and arachidonic acid metabolism. Furthermore, low-dose RM (RML) was found to enhance beneficial gut microbiota such as Lactobacillus, suggesting its potential role in the regulation of intestinal homeostasis and barrier integrity.
Conclusion: RML has the potential to enhance the composition of intestinal microbiota by through the differential regulation of various metabolized components, including 13-OxoODE, morphine, and niacinamide, it influences several metabolic pathways, notably steroid biosynthesis, lysine degradation, interconversions of pentose and glucuronate, as well as the metabolism of linoleic acid. Additionally, it may promote the proliferation of HT002 and Lactobacillus probiotics, thereby contributing to the amelioration of LI. It establishes a robust foundation for future applications and the development of associated pharmaceuticals.
1 Introduction
Liver injury (LI) represents a significant global health issue, exhibiting a range of epidemiological characteristics that differ across regions and populations. A report published in 2023 indicates that chronic liver disease and LI rank among the foremost causes of mortality globally, resulting in an estimated 2 million deaths annually (Devarbhavi et al., 2023). The causes of LI are complex and varied, including factors such as viral infections and alcoholic liver disease (Hackstein et al., 2023), drug-induced hepatotoxicity (Zhang et al., 2024), and non-alcoholic fatty liver disease (Chen et al., 2024). These elements can initiate a series of pathological processes characterized by hepatic inflammation, fibrosis, and cirrhosis, which may eventually lead to liver failure or the development of hepatocellular carcinoma (Lin et al., 2023).
Under typical physiological conditions, the gut microbiota is involved in various physiological processes and is essential for the support and regulation of liver functions (Zhang et al., 2022). The disruption of microbial homeostasis may result in the impairment of the liver’s metabolic, detoxification, and immune regulatory functions (Zhang et al., 2024). The disruption of gut microbiota homeostasis is characterized by a decline in the population of beneficial bacterial taxa, including Bacteroidota, Lactobacillus, Actinobacteriota, Proteobacteria, and Clostridium. Concurrently, there is an overgrowth of pathogenic bacteria, such as Erysipelotrichaceae_UCG-003, Lachnoclostridium, Enterococcus, and Dorea. This dysbiosis further compromises the integrity of the intestinal barrier, leading to increased intestinal permeability. As a result, a greater number of bacteria and their metabolites can translocate into the bloodstream, thereby exacerbating the inflammatory response and the severity of intestinal inflammation (Li et al., 2022). Conversely, the immune disorder induced by LI will have an impact on the intestinal microecological environment, altering the habitat of gut microbiota. This disruption may inhibit the proliferation of beneficial bacteria while facilitating the growth of opportunistic pathogens (Zhang et al., 2024). Consequently, a detrimental cycle is established, perpetually advancing the progression and decline of LI.
The medicinal application of RM is derived from the desiccated flowers of Rhododendron molle (Guo et al., 2021a). It possesses properties that facilitate the expulsion of wind and the elimination of dampness, as well as the dispersion of blood stasis and the alleviation of pain. This treatment is frequently employed for conditions such as rheumatic arthralgia, unilateral or bilateral headaches, and swelling and pain resulting from trauma, in addition to persistent tinea (Chinese Pharmacopoeia Commission, 2020). “Bashaga” is a frequently utilized medicinal substance within the realm of Mongolian medicine, boasting a rich historical background and holding significant importance in this field. In clinical practice, this substance functions as a substitute for “Bashaga” medicinal herbs within the framework of Mongolian traditional medicine, specifically in certain areas of Hulunbuir City, Chifeng City, and Tongliao City in Inner Mongolia (Xiao et al., 2020). The “Compendium of Chinese Herbal Medicine-Mongolian Medicine Volume” documents that RM possesses therapeutic properties for the treatment of various conditions, including dermatological disorders, cardiovascular diseases, and hepatic diseases, among others.
Nevertheless, the rationale for its application and the mechanisms that underpin its therapeutic effects remain to be clarified (Zhao et al., 2017). Recent pharmacological investigations into RM have identified its various pharmacological properties, including anti-inflammatory, analgesic, anesthetic, hypotensive, insecticidal, antiviral, and anticancer activities (Cai et al., 2018; Guo et al., 2021b). The primary constituents of RM encompass diterpenoids, flavonoids, triterpenoids, and coumarins (Guo et al., 2020). Diterpenoids, including grayanotoxin I, rhodojaponin VI, and rhodojaponin V, are identified as the toxic constituents of RM. Administration of these compounds at elevated doses may result in neurotoxicity and cardiotoxicity, with the potential for fatal outcomes due to respiratory failure in extreme cases. Conversely, the administration of RM at lower doses has been associated with anti-inflammatory and analgesic properties (Liu et al., 2018). Recent research has indicated that the fingerprint of RM contains flavonoid constituents, including kaempferol and quercetin (Hu et al., 2024), both components play crucial roles within the paradigm of traditional Chinese medicine. They exhibit pharmacological properties that positively influence human health, including antioxidant, anti-inflammatory, anti-obesity, and hepatoprotective effects (Gu et al., 2023; Singh et al., 2023). In light of the therapeutic efficacy of “Bashaga”-type Mongolian medicines in the treatment of liver diseases observed in clinical practice, we aim to conduct further investigations into the mechanisms by which RM mitigates LI.
Consequently, this study aimed to investigate the ameliorative effects of RM on LI by employing a CCl4-induced LI model. Initially, we conducted a comprehensive analysis of the components of RM, followed by network pharmacology analysis and molecular docking of the identified constituents to establish a RM-target network and a “component-target-metabolite” network. This approach allowed for a thorough elucidation of the in vivo targets and signaling pathways associated with RM. Furthermore, we compared the pharmacodynamics of three different doses of RM through pathological assessments and ELISA, performed fecal metabolomics on both high and low dose groups, and analyzed the gut microbiota associated with the optimal dose. This study provides preliminary insights into the dosing of RM and investigates the mechanisms through which RM mitigates LI.
2 Result
2.1 Analysis of the components of RM
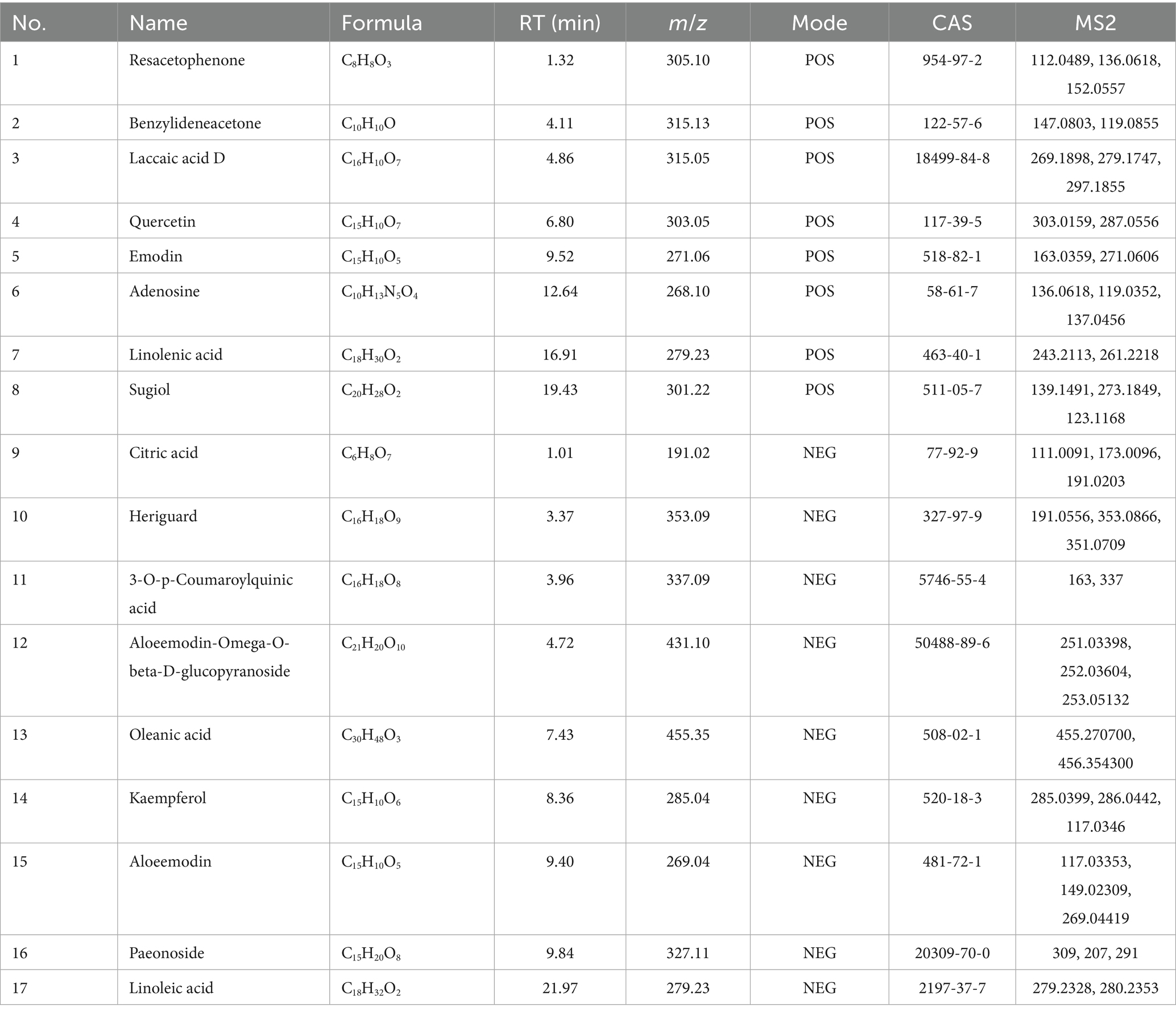
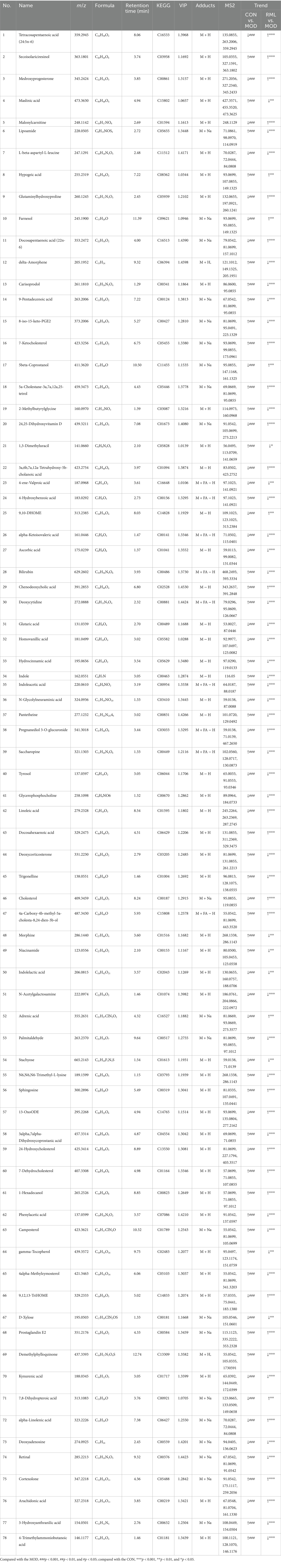
Total ion chromatograms of the RM were obtained in both negative and positive ion modes. An analysis of the mass spectrometric data, coupled with database comparisons and secondary identification methods, led to the identification of 17 components within the RM across both ionization modes (Figure 1 and Table 1). The identified components comprised citric acid, heriguard, oleanic acid (Wang et al., 2014), emodin, quercetin, kaempferol (Liu and Kong, 2009), among others.
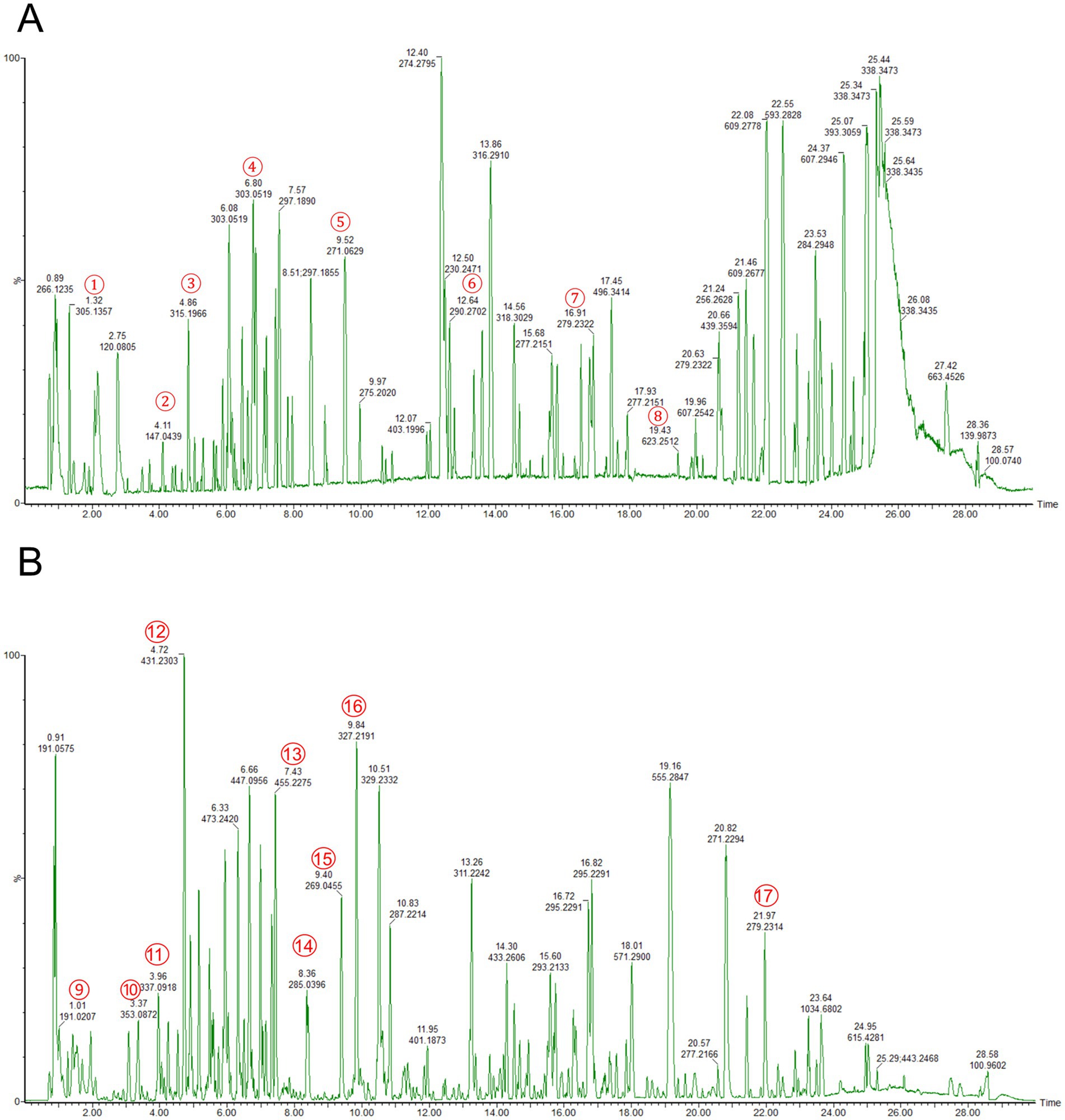
Figure 1. TIC of the components of RM in positive and negative ion modes. (A) TIC of the components of RM in positive ion mode. (B) TIC of the components of RM in negative ion mode. ① Resacetophenone. ② Benzylideneacetone. ③ Laccaic acid D. ④ Quercetin. ⑤ Emodin. ⑥ Adenosine. ⑦ Linolenic acid. ⑧ Sugiol. ⑨ Citric acid. ⑩ Heriguard. ⑪ 3-O-p-Coumaroylquinic acid. ⑫ Aloeemodin-Omega-O-beta-D-glucopyranoside. ⑬ Oleanic acid. ⑭ Kaempferol. ⑮ Aloeemodin. ⑯ Paeonoside. ⑰ Linoleic acid.
2.2 Network pharmacology
2.2.1 Screening of targets of RM components
The 17 identified chemical constituents were systematically entered into the Swiss Target Prediction database, resulting in the identification of 16 potential active constituents, such as isorhamnetin, kaempferol, andromedotoxin along with their corresponding action targets. Subsequently, all targets underwent a screening and deduplication process, yielding a total of 370 unique constituent targets.
2.2.2 Related targets of RM acting on LI
A total of 1,378 disease targets were identified. By utilizing the microsignature platform to analyze intersections, 125 overlapping targets associated with potential active constituents related to RM diseases were identified (Figure 2A).
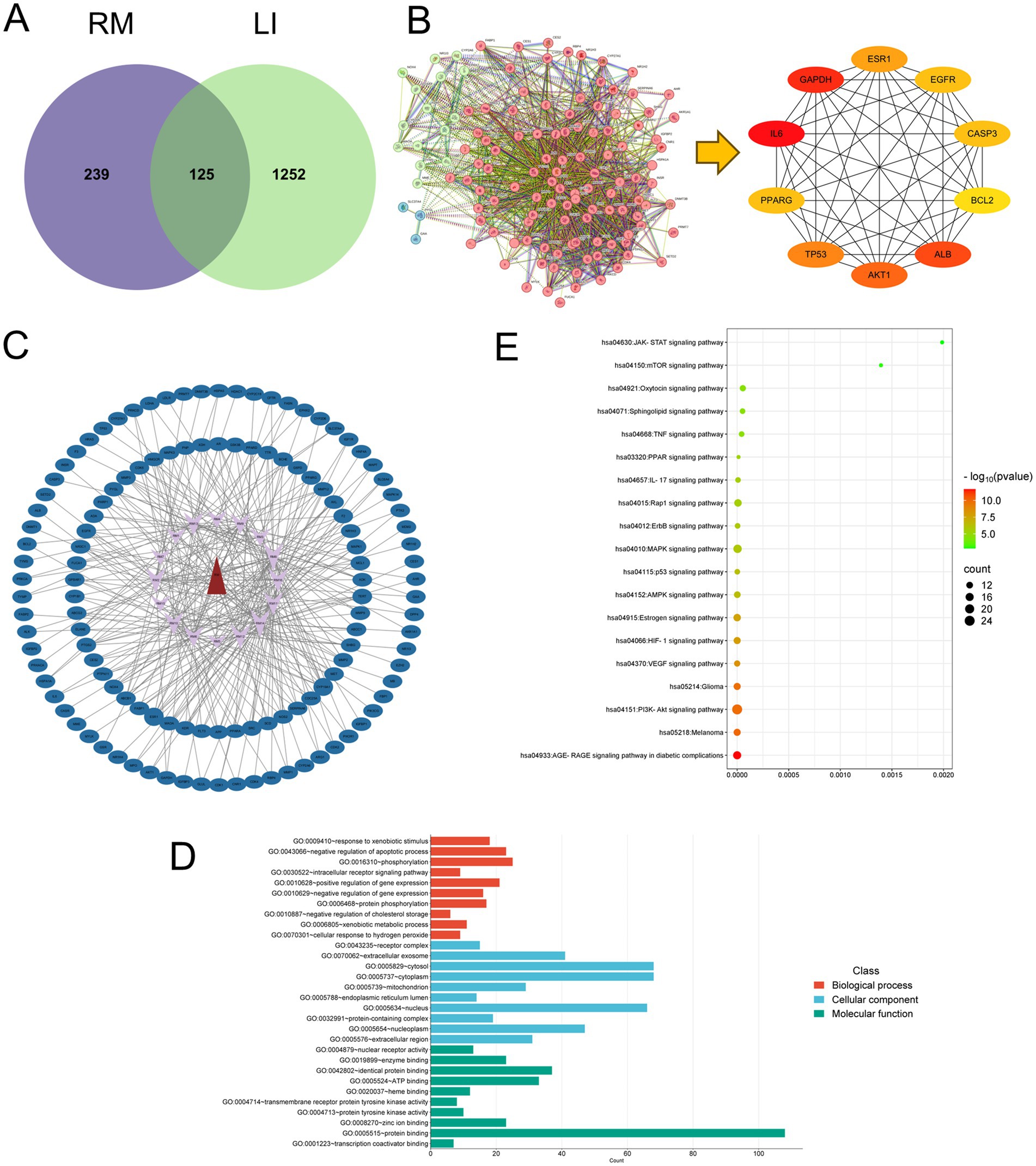
Figure 2. Network pharmacology research on the treatment of LI with RM. (A) The intersection targets between the potential active ingredients of RM and LI. (B) The core targets of RM for improving LI. (C) The “drug-ingredient-target” network. (D) GO enrichment analysis. (E) KEGG enrichment analysis.
2.2.3 Construction of PPI network
The analysis of the protein–protein interaction (PPI) network has revealed 10 principal targets: IL-6, PPARG, TP53, AKT1, ALB, BCL2, CASP3, EGFR, ESR1, and GAPDH. These targets exhibit a high degree of interconnectivity and are likely to be integral to the therapeutic effects of RM in the context of LI. Importantly, these targets are implicated in critical biological processes, including inflammation (IL-6, AKT1), apoptosis regulation (TP53, BCL2, CASP3), and metabolic modulation (PPARG, ESR1). This suggests that RM may provide multi-targeted protective effects in LI (Figure 2B).
2.2.4 “Drug-component-target” interaction network and topological analysis
Network analysis has revealed six principal bioactive components in RM, namely adenosine, aloeemodin, emodin, kaempferol, paeonoside, and quercetin, which were identified due to their significant network connectivity (Figure 2C). These compounds demonstrated the most robust interactions with targets associated with liver injury, indicating their potential role as key active constituents responsible for mediating the therapeutic effects of RM.
2.2.5 GO and KEGG pathway analysis
Following the importation of 125 intersection targets into the DAVID online database, a comprehensive screening yielded a total of 1,200 entries. Among these, 472 were categorized under biological processes, predominantly encompassing the negative regulation of apoptotic processes, intracellular receptor signaling pathways, positive regulation of gene expression, and negative regulation of gene expression, among others. Additionally, 62 entries were classified as cellular components, primarily involving receptor complexes, extracellular exosomes, cytosol, and mitochondria. Furthermore, 130 entries were identified under molecular functions, with a focus on protein binding, identical protein binding, zinc ion binding, and ATP binding, among other functions. The top 10 entries with the lowest p-values in each category were selected for visual analysis using MicroBioinformatics (Figure 2D). The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis identified 103 pathways. After filtering out irrelevant pathways, such as those related to “cancer,” “lipids and atherosclerosis,” and “cytomegalovirus infection,” the top 20 pathways with the highest number of enriched targets were selected based on ascending p-values to create a bubble chart. The KEGG enrichment analysis indicated that Rhododendron molle is significantly associated with pathways pertinent to liver injury treatment, particularly involving signaling pathways such as the PI3K-Akt signaling pathway, VEGF signaling pathway, HIF-1 signaling pathway, and estrogen signaling pathway (Figure 2E).
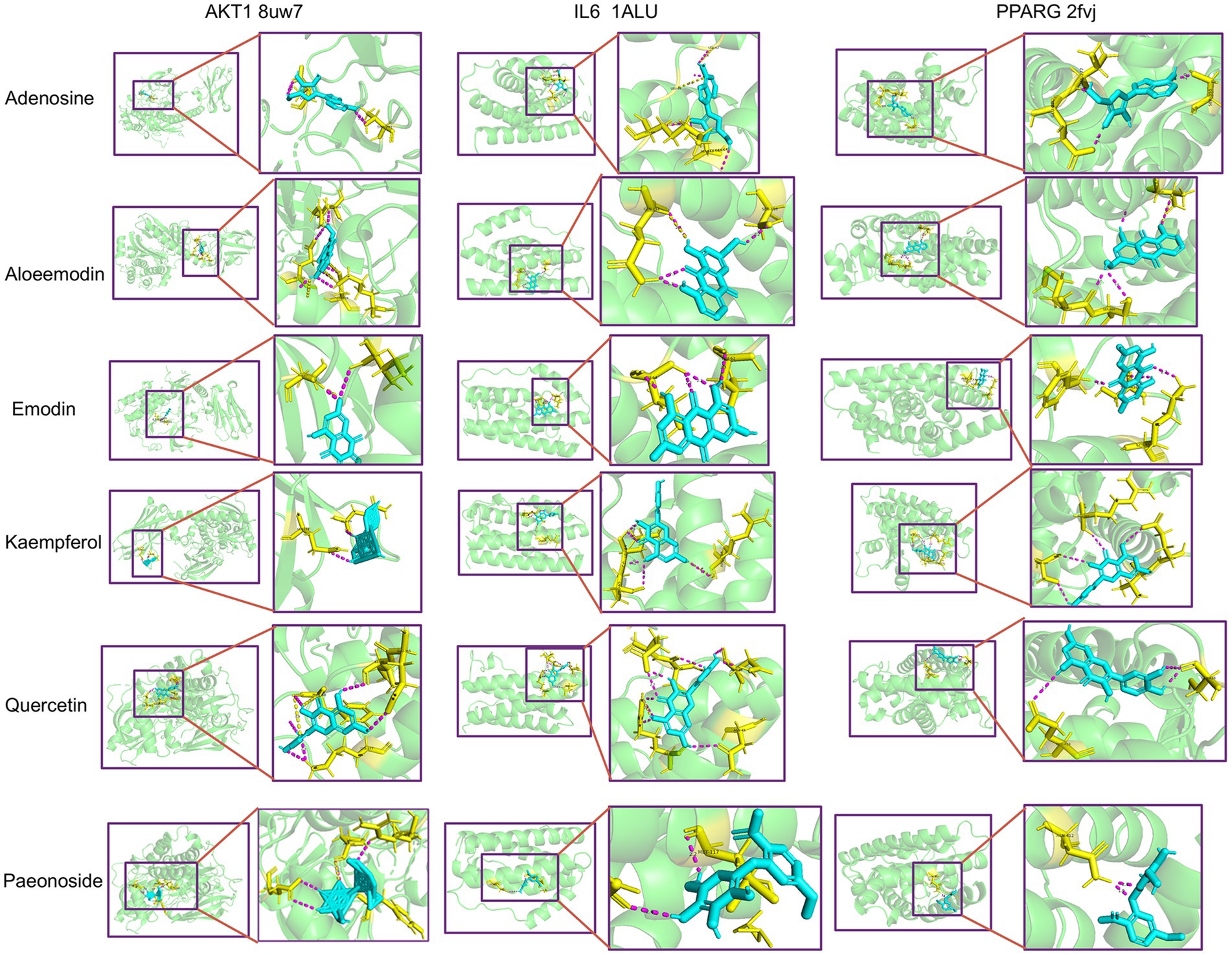
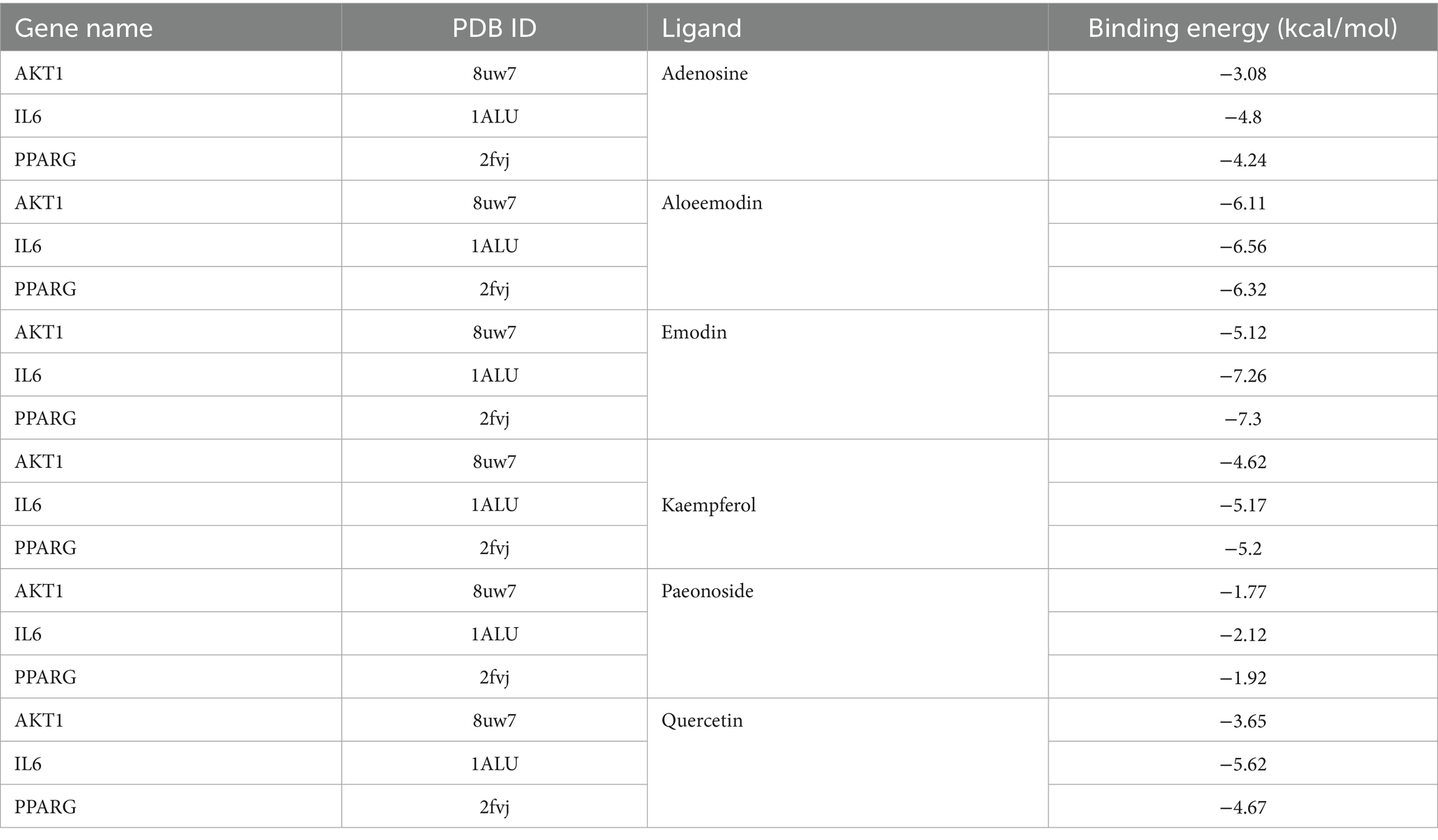
2.2.6 Molecular docking
In order to assess the binding affinity of the primary constituents of Rhododendron molle (Blume) G. Don with significant biological targets, we conducted molecular docking analyses. An analysis was conducted on the top 10 core targets identified by the “CytoHubba” plugin within the Cytoscape framework. From this analysis, six compounds—quercetin, kaempferol, emodin, aloe-emodin, adenosine, and paeonoside were selected for molecular docking studies with the key targets IL6, AKT1, and PPARG to assess their binding affinity and stability. The results of the molecular docking indicated a significant inverse relationship between the binding free energy and the stability of the ligand-receptor complexes. Specifically, conformations characterized by lower binding energies demonstrated enhanced thermodynamic stability. Comprehensive docking results are illustrated in Figure 3 and detailed in Table 2. The results show that the components aloeemodin and emodin have a high binding energy with the target IL-6, and aloeemodin also has a high binding energy with PPARG.
2.3 The effects of RM on the pathological changes of liver tissue in rats with LI
Histological examination of liver tissue using hematoxylin and eosin (HE) staining (Figure 4A) revealed distinct differences between the control group (CON) and the experimental groups. In the CON group, the liver lobules of the rats exhibited a uniform distribution, with liver cords arranged in a regular or striped configuration. Conversely, the model group (MOD) displayed significant lipid depletion within the liver cells, accompanied by areas of degeneration and necrosis, as well as extensive infiltration of granulocytes. In comparison to the MOD group, both the low-dose Rhododendron molle (Blume) G. Don group (RML) and the Silymarin group (Sily) demonstrated a reduction in inflammatory cell infiltration within the liver tissue. Additionally, there was a partial alleviation of lipid vacuolation in the liver cells, leading to a notable improvement in the extent of tissue damage. Notably, the pathological morphology of liver tissue in both the RML and Sily groups closely resembled that of the CON group.
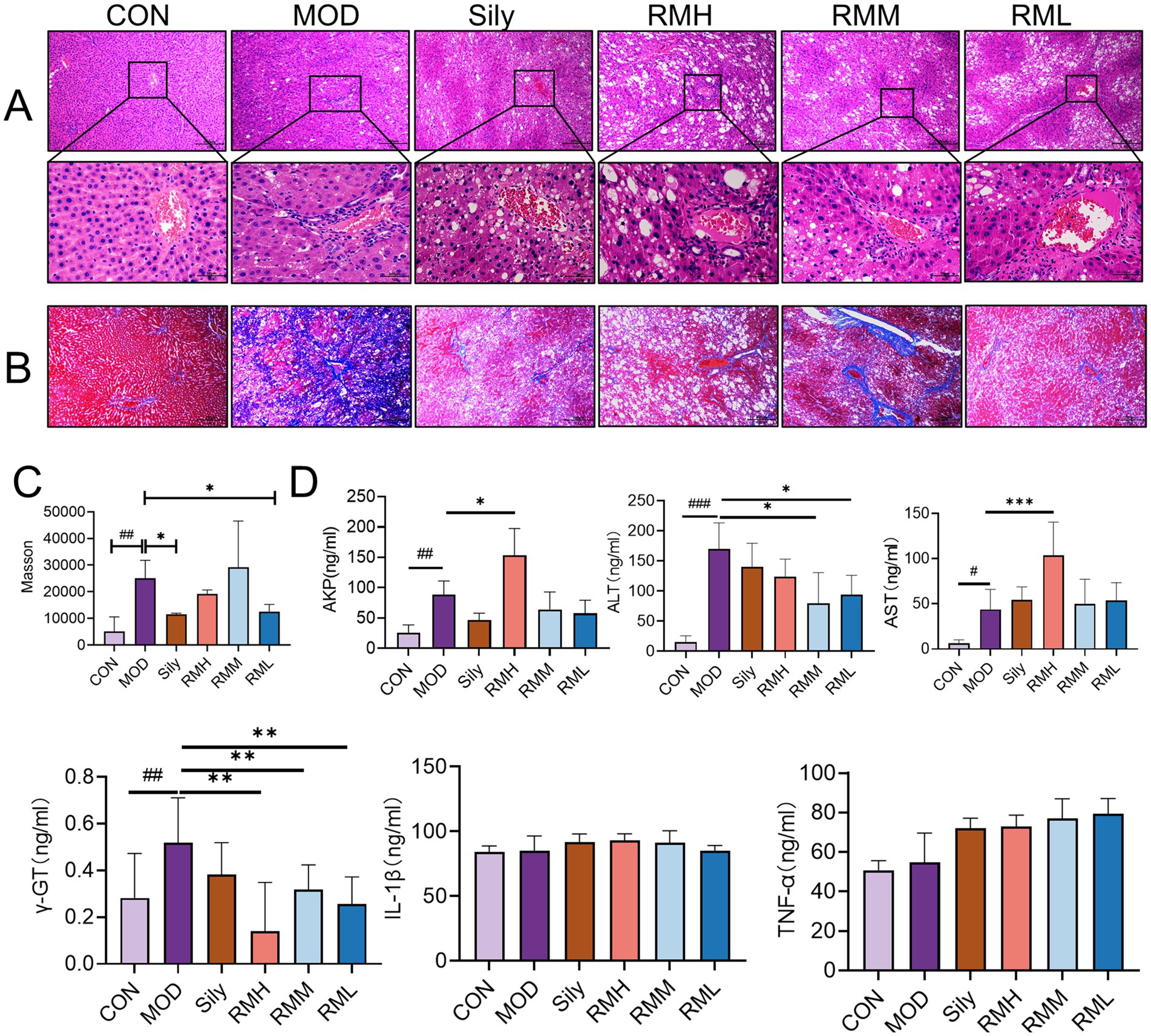
Figure 4. The effects of RM on the biochemical indexes of rats with LI. (A) HE staining of liver tissue under 100× and 400× microscopy. (B) Masson staining of liver tissue under 100× microscopy. (C) Scoring of Masson staining of liver tissue. (D) ELISA analysis of AKP, γ-GT, AST, ALT, TNF-α, and IL-1β in liver tissue. Compared with the MOD, ###p < 0.001, ##p < 0.01, and #p < 0.05; compared with the CON, ***p < 0.001, **p < 0.01, and *p < 0.05.
Masson staining of liver tissue revealed (Figure 4B) that the CON group exhibited no significant fibrosis, while the MOD group displayed pronounced collagen fiber deposition within the liver tissue. In comparison to the MOD group, the liver tissue fibrosis in rats from the RML group, as well as those receiving medium-dose (RMM) and high-dose (RMH) Rhododendron molle (Blume) G. Don, and the Sily group, showed varying degrees of alleviation. The collagen deposition score in the MOD group was significantly elevated compared to the CON group (p < 0.05) (Figure 4C). Furthermore, the collagen deposition scores in the RML and Sily groups were both significantly lower than those in the MOD group (p < 0.05), with the RML group demonstrating a lesser degree of fibrosis and a collagen deposition score that approached that of the CON group.
2.4 The effects of RM on the biochemical indexes of liver tissue in rats with LI
To investigate the impact of RM on liver function and inflammatory markers in rats with LI, we performed enzyme-linked immunosorbent assays on liver tissue samples. The findings indicated (Figure 4D) that, in comparison to the CON group, the levels of alkaline phosphatase (AKP), gamma-glutamyl transpeptidase (γ-GT), aspartate aminotransferase (AST), and alanine aminotransferase (ALT) were significantly elevated in the MOD group. Conversely, the inflammatory markers tumor necrosis factor-alpha (TNF-α) and interleukin-1 beta (IL-1β) did not exhibit significant variations. Following the administration of three doses of RM to the rats with LI, the RMH demonstrated an increase in AKP levels compared to the MOD group, while no significant changes were observed in the RMM and RML groups. Additionally, the RMH group showed increased levels of AST and ALT in the liver tissue relative to the MOD group, and all three doses of RM resulted in a reduction of γ-GT levels in the liver tissue. However, the inflammatory markers TNF-α and IL-1β no significant changes across the LI and treatment groups. Based on the biochemical index results, it was observed that the RML group led to a reduction in ALT and γ-GT levels in rats with LI, and to a certain extent, also decreased AST and AKP levels. Consequently, we deduced that RML may have a more favorable effect on the repair of LI.
2.5 The effects of RM on the fecal metabolomics of rats with LI
2.5.1 Analysis of the fecal metabolic profile of rats with LI by RM
Based on the analysis of pathological sections and biochemical indices, we determined that the RML dosage was optimal. We hypothesized that the RMH dosage might exacerbate LI. Consequently, we undertook a non-targeted fecal metabolomics investigation comparing RMH and RML. The results of the principal component analysis (PCA) (Figure 5A) and partial least squares discrimination analysis (PLS-DA) (Figure 5B) indicated that metabolites within each group exhibited intra-group clustering and inter-group differentiation, the test results show (Figure 5C) that the R2 value is 0.623, indicating that the PLS-DA model has good predictive ability. To further evaluate the distinctions between the two groups, we employed orthogonal partial least squares discriminant analysis (OPLS-DA). The OPLS-DA model (Figure 5D) and the corresponding test results for each group (Figure 5E) revealed a notable distinction in metabolic products between the CON and MOD groups across both positive and negative ion modes. Furthermore, the Q2 values for the comparisons between the CON and MOD groups, as well as the MOD and RML groups, exceeded 0.9, suggesting that the predictive capacity of the OPLS-DA model is highly robust, thereby indicating notable differences in fecal metabolites between the two groups.
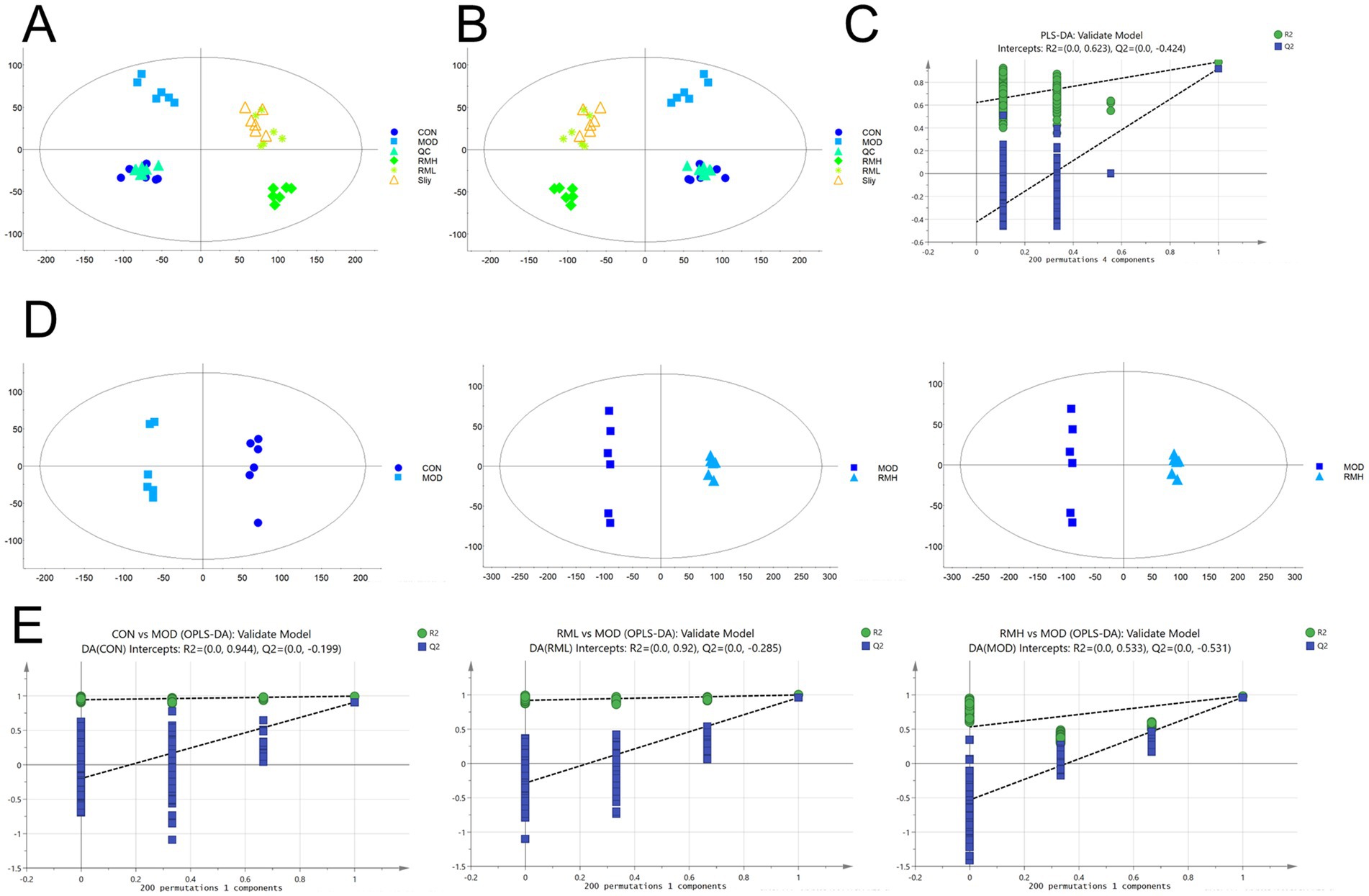
Figure 5. The metabolic profile analysis of RMH and RML. (A) PCA analysis of the each group under positive and negative ion modes. (B) PLSDA analysis of the each group under positive and negative ion modes. (C) PLS-DA test plots for each group. (D) OPLS-DA analysis of each group under positive and negative ion modes. (E) OPLS-DA tests for each group under positive and negative ion modes.
2.5.2 Screening of differential metabolic components and cluster analysis
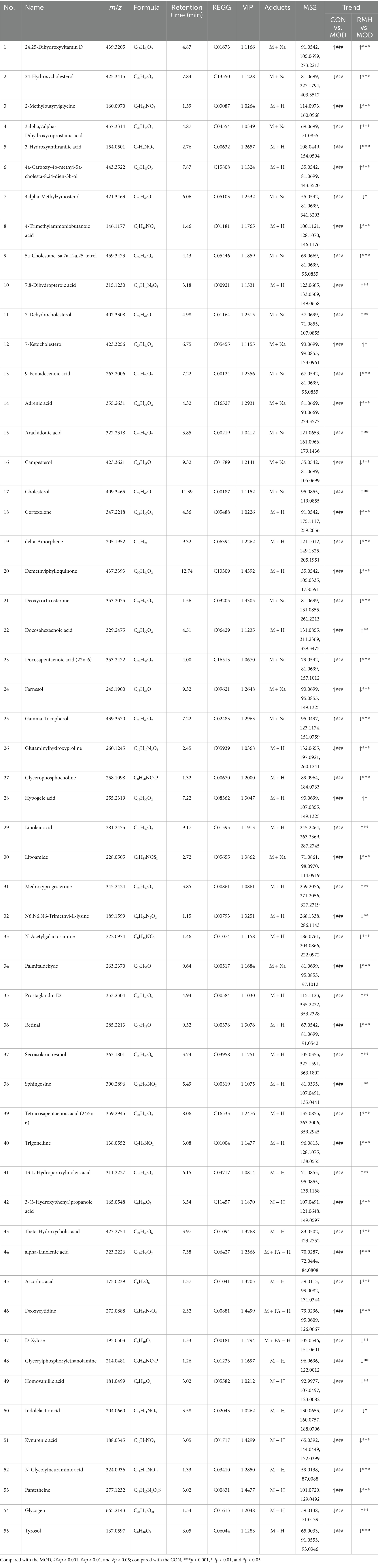
Utilizing a VIP >1 and a p < 0.05, differential metabolic components were identified among the CON and MOD groups, the MOD and RMH groups, as well as the MOD and RML groups. Specifically, a total of 84 differential metabolic components were detected in both positive and negative ion modes between the CON and MOD groups, among these, based on VIP >1 and a p < 0.05 while 55 differential metabolic components were identified in the MOD and RMH groups (Table 3), and 78 differential metabolic components were recognized in the MOD and RML groups (Table 4).
Cluster heatmap analysis was conducted based on the aforementioned differential metabolic components, The clustering analysis indicated that certain components of both the RML and RMH groups exhibited similarities to those of the CON group, with a greater number of components in the RML group demonstrating trends that aligned more closely with those of the CON group (Figures 6B,C). Furthermore, there were 34 common differential metabolic components identified across the four groups: CON, MOD, RMH, and RML (Figure 6A). A subsequent cluster analysis was performed on these 34 components (Figure 6D). RML and RMH exhibit distinct influences on the metabolic trends of various compounds, including 4a-Carboxy-4b-methyl-5a-cholesta-8,24-dien-3b-ol, 7-Dehydrocholesterol, cholesterol, and kynurenic acid. The observed variations in these metabolites may account for the differing effects associated with RMH and RML.
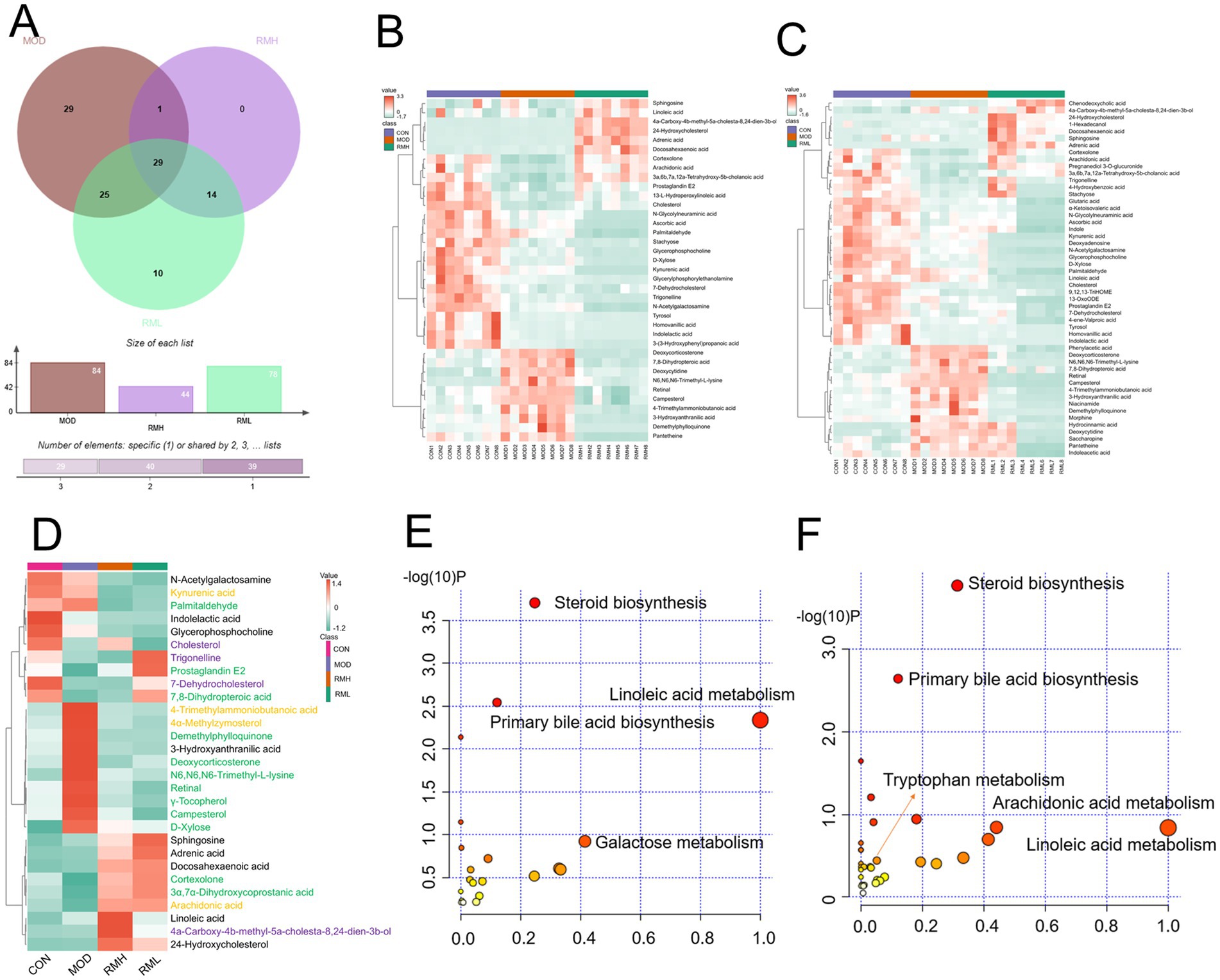
Figure 6. Analysis of fecal differential metabolic components. (A) OPLS-DA analysis of feces in positive and negative ion modes. (B) Venn analysis of differential metabolic components in each group. (C) Clustering heatmap of RMH of the CON and MOD. (D) Clustering heatmap analysis of RML group of the CON and MOD. (E) Clustering heatmap of common differential metabolic components of RMH and RML groups of the CON and MOD. (F) Metabolic pathways of the RMH group. (G) Metabolic pathways of the RML group. The color green signifies a notable upregulation or downregulation within the model group, which is subsequently reversed following treatment administration. The color purple denotes significant upregulation or downregulation in the model group, characterized by contrasting trends observed at both high and low dosage levels. Conversely, the color yellow represents significant upregulation or downregulation in the model group, exhibiting a consistent trend but varying degrees of change at high and low doses.
2.5.3 Metabolic pathway analysis
To further investigate the effects of the selected metabolites and to identify the potential biochemical pathways impacted by LI, MetPA1 was utilized to elucidate the most pertinent pathways influenced by LI. As illustrated in Figures 6E,F, the metabolic pathways in the RMH group were predominantly enriched in steroid biosynthesis, linoleic acid metabolism, and arachidonic acid metabolism, among others. Conversely, the RML group was primarily associated with steroid biosynthesis, lysine degradation, pentose and glucuronate interconversions, linoleic acid metabolism, primary bile acid biosynthesis, and tryptophan metabolism, among other pathways. The variations in metabolites that exert reciprocal effects predominantly influence steroid biosynthesis, linoleic acid metabolism, and tryptophan metabolism, among other processes.
2.6 Integrated analysis of metabolomics and network pharmacology
We conducted an analysis of the intersection between 30 differential metabolites identified from the RML and their associated targets, alongside 125 drug-disease-metabolite targets predicted through network pharmacology (Figure 7A). This analysis yielded a total of 76 intersecting targets. The top 10 genes were identified using the Cytohubba plugin (Figure 7B), which included IL6, PPARG, AKT1, TP53, BCL2, CASP3, ESR1, PTGS2, MAPK3, and EGFR. Subsequently, we employed the “CytoNCA” plugin to identify the highest-ranked differential metabolic components (Figure 7C), which comprised 3α,7α-Dihydroxycoprostanic acid, arachidonic acid, cortexolone, deoxycorticosterone, pantetheine, and 13-OxoODE. The 76 core targets were then analyzed using the Metascape platform to perform Gene Ontology (GO) (Figure 7D) and KEGG (Figure 7E) analyses. The GO analysis encompassed three primary categories: biological process (BP), cellular component (CC), and molecular function (MF), which included a total of 209 BP, 42 CC, and 87 MF terms. The KEGG enrichment analysis indicated that RM positively influenced LI through 146 signaling pathways modulated by the differential metabolic components. Following the exclusion of pathways associated with cancer and those irrelevant to liver function, the most significant pathways identified were the PI3K-Akt signaling pathway, TNF signaling pathway, HIF-1 signaling pathway, IL-17 signaling pathway, PPAR signaling pathway, and mTOR signaling pathway, among others.
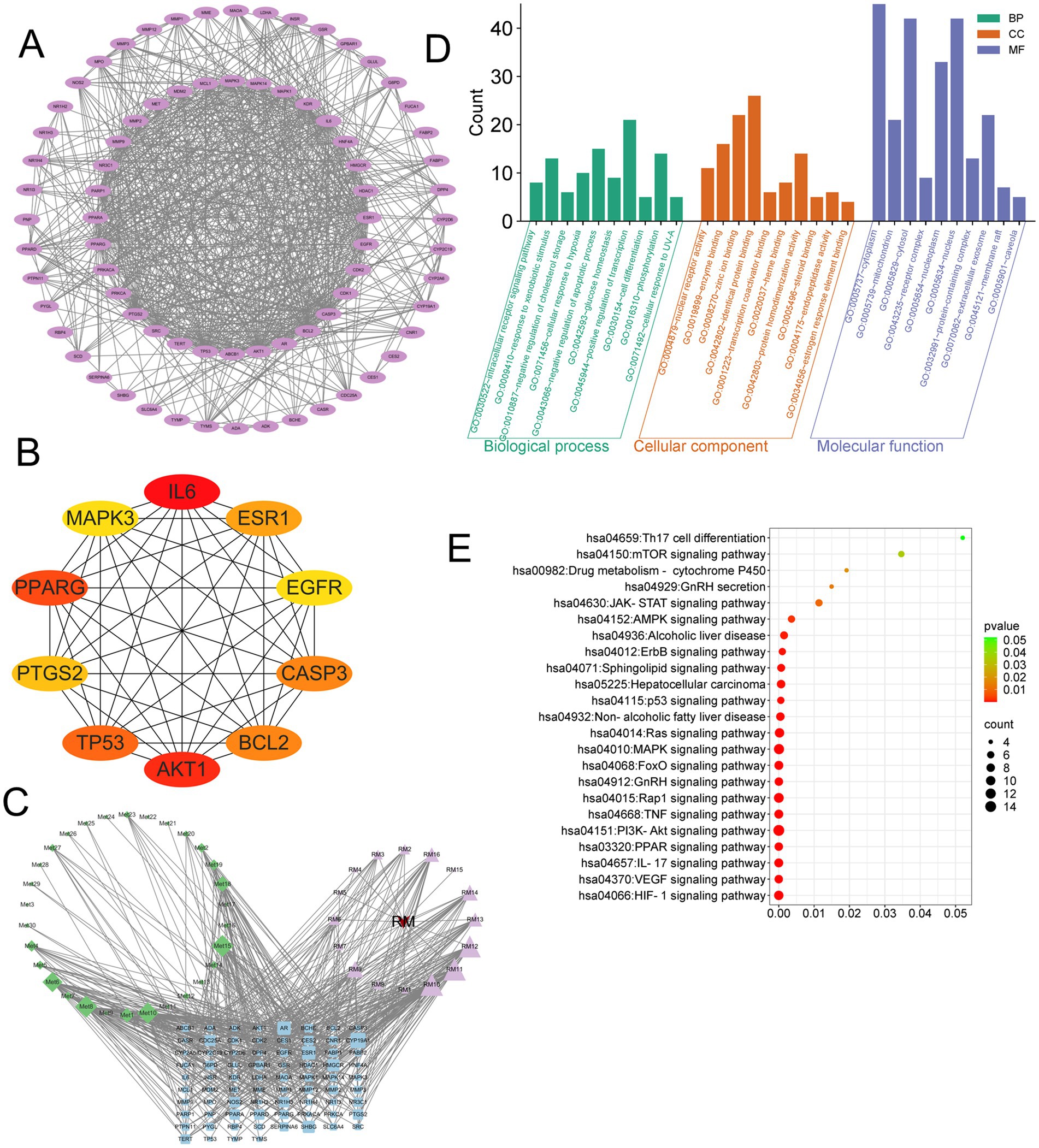
Figure 7. Integrated analysis of network pharmacology and metabolomics. (A) PPI network of RM components-disease-metabolite targets. (B) Top 10 targets after screening of core targets. (C) Network interaction of RM components-metabolites-disease. (D) GO enrichment analysis. (E) KEGG enrichment analysis.
2.7 The impact of RML on the intestinal microbiota of rats with LI
2.7.1 The impact of RML on the abundance of the intestinal microbiota of rats with LI
In light of the findings from pharmacodynamics and metabolomics, a comprehensive examination of the gut microbiota was performed for the group receiving the optimal RML. Alpha diversity analysis was utilized to assess the species diversity within individual samples, employing indices such as Chao1, Peilou_e, Shannon, and Simpson. Our findings (Figure 8A) indicate that RML did not significantly affect the richness and evenness of the intestinal microbiota in rats with LI. Following this, we performed a principal coordinate analysis (PCoA) on the operational taxonomic unit abundance matrices derived from the four experimental groups to evaluate the similarity among the samples and groups. The PCoA scores (Figure 8B) revealed a distinct separation of the MOD group from both the CON and RML groups, while showing a relative proximity to the Sily group.
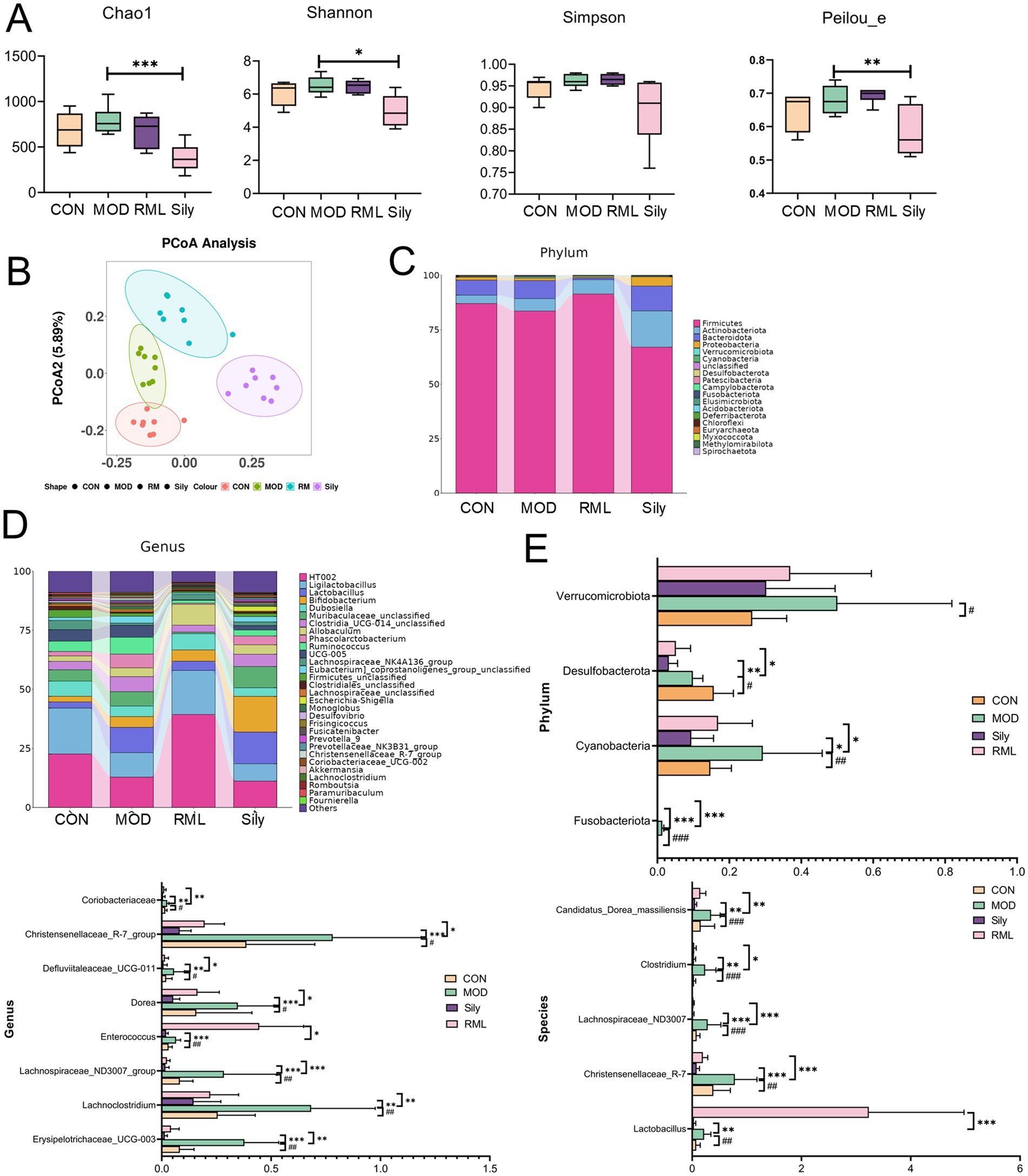
Figure 8. The impact of RML on the intestinal microbiota of rats with LI. (A) Alpha diversity analysis (Chao1 index, Simpson index, Shannon index, Peilou_e index). (B) Beta diversity analysis. (C) Stacked chart of differential microbiota at the phylum level. (D) Stacked chart of differential microbiota at the genus level. (E) Significance analysis of differential microbiota at the phylum, genus, and species levels. Compared with the MOD, ###p < 0.001, ##p < 0.01, and #p < 0.05; compared with the CON, ***p < 0.001, **p < 0.01, and *p < 0.05.
2.7.2 Analysis of intestinal microbial composition
The findings presented in the species annotation stacked charts (Figures 8C,D) indicate notable variations in the composition of microbial species across different taxa at both the phylum and genus levels.
We conducted an analysis of the microbial community changes at the phylum, genus, and species levels (Figure 8E). At the phylum level, a significant increase in the abundances of Fusobacteriota, Cyanobacteria, and Desulfobacterota was observed in comparison to the CON group. Following the administration of the Sily and RML groups, a marked reduction in the abundances of these phyla was noted. At the genus level, the administration of CCl4 resulted in a significant increase in the abundances of Erysipelotrichaceae_UCG-003, Lachnoclostridium, Lachnospiraceae_ND3007_group, Enterococcus, Dorea, Defluviitaleaceae_UCG-011, Christensenellaceae_R-7_group, and Coriobacteriaceae, among others. Conversely, both Sily and RML treatments led to a significant decrease in the abundances of these genera. To investigate the impact of RM on the intestinal microbiota at the species level, we conducted an analysis of the top 30 microbiota and performed a significance analysis on those that exhibited notable changes. The microbiota that demonstrated significant alterations are as follows: Lactobacillus, Christensenellaceae_R-7, Lachnospiraceae_ND3007, Clostridium, and Candidatus_Dorea_massiliensis. Subsequent to the administration of RM, there was a significant increase in the abundance of Lactobacillus, whereas the levels of Christensenellaceae_R-7, Lachnospiraceae_ND3007, Clostridium, and Candidatus_Dorea_massiliensis were restored to levels that approached normalcy.
In this investigation, we employed the Linear discriminant analysis Effect Size (LEfSe) method to elucidate the significant variations in the composition of intestinal microbiota across different treatment groups, as illustrated in Figure 9A. By establishing a significance threshold of p < 0.05, we were able to identify a range of biomarkers that exhibited statistical significance. Within the CON group, we identified a total of seven microbiota types that were significantly enriched, including Firmicutes, Oscillospiraceae, and Clostridiales, which encompassed various taxonomic levels such as classes, orders, families, genera, and species within the Firmicutes phylum. The MOD group exhibited enrichment of 23 microbiota types, including Clostridia, Ruminococcaceae, Negativicutes, Acidaminococcaceae, Phascolarctobacterium, UCG_005, Monoglobales, and Christensenellaceae. In the RML group, 33 microbiota types were predominantly enriched, including Bacteroidota, Lactobacillus, Actinobacteriota, Proteobacteria, Clostridium, and Atopobiaceae. Lastly, the Sily group demonstrated enrichment of nine microbiota types, which included Bacilli, Lactobacillaceae, HT002, and Enterorhabdus.
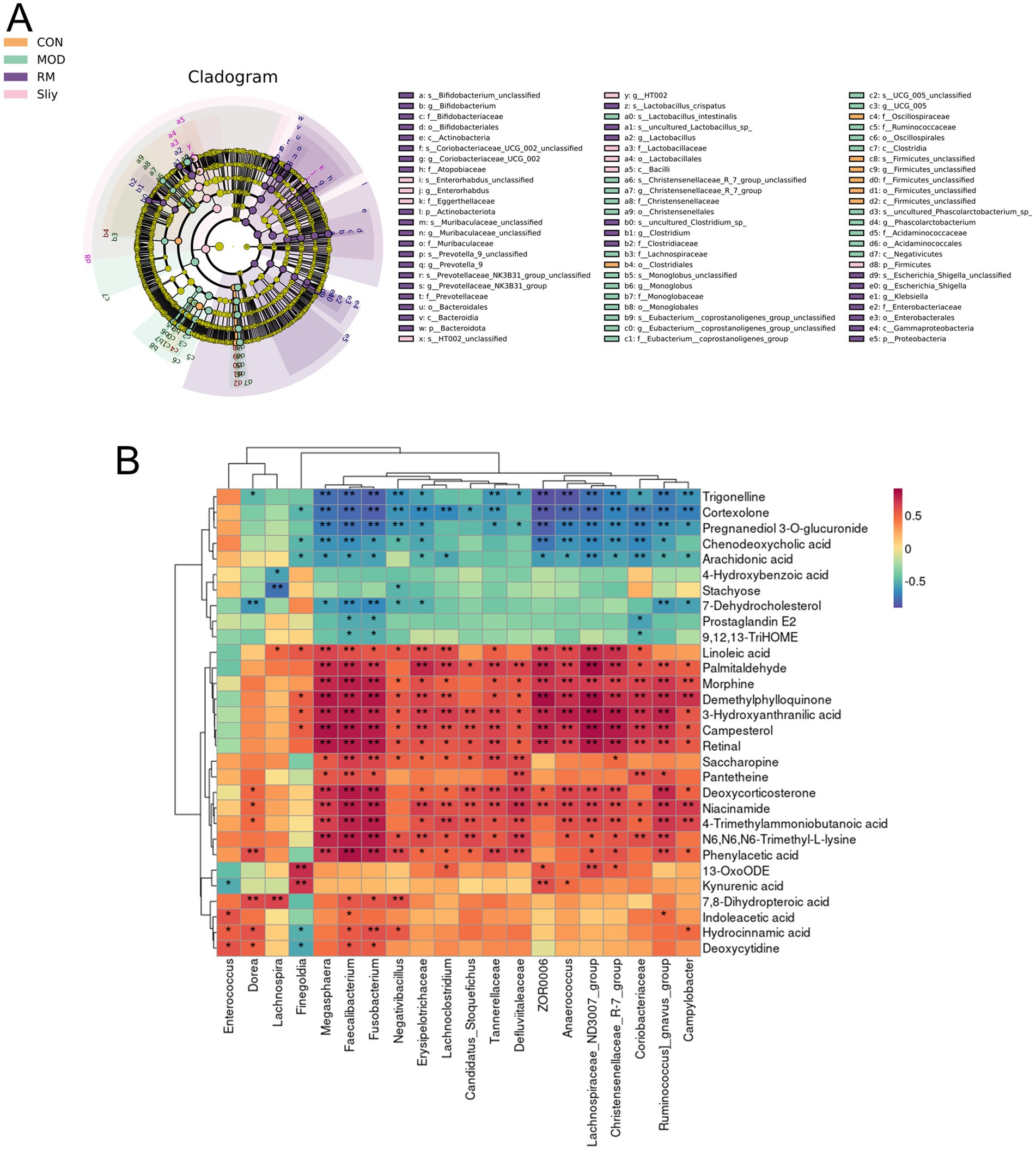
Figure 9. LEfSe analysis and correlation between differential metabolic components and gut microbiota. (A) LEfSe analysis of the intestinal microbiota. (B) Correlation analysis of the intestinal microbiota and the top 30 differential metabolic components in the RML group.
2.8 Analysis of the correlation between differential metabolic components and gut microbiota
The association between intestinal microbiota and host metabolic processes was examined through the calculation of the Spearman correlation coefficient (Figure 9B). The analysis revealed that specific bacterial taxa exhibited significant correlations with a range of metabolites (p < 0.05). Notably, kynurenic acid demonstrated a positive correlation with Finegoldia, ZOR0006, and Anaerococcus. Conversely, 7-Dehydrocholesterol was found to have a negative correlation with Erysipelotrichaceae. Additionally, Deoxycytidine showed positive correlations with Enterococcus, Dorea, Fusobacterium, and Faecalibacterium. Furthermore, 7,8-Dihydropteroic acid was positively correlated with Dorea, Lachnospira, Fusobacterium, Faecalibacterium, and Negativibacillus. Lastly, palmitaldehyde exhibited positive correlations with a diverse array of bacterial groups, including Fusobacterium, Faecalibacterium, Ruminococcus_gnavus_group, Anaerococcus, Tannerellaceae, Erysipelotrichaceae, Lachnoclostridium, Lachnospiraceae_ND3007_group, Campylobacter, Megasphaera, Defluviitaleaceae, Christensenellaceae_R-7_group, ZOR0006, and Coriobacteriaceae.
3 Discussion
The liver, serving as the primary organ responsible for the metabolism of pharmaceuticals and toxic compounds within the human body, is particularly susceptible to injury from drugs, toxins, and viral infections (Su et al., 2023). In light of this context, the investigation of natural liver-protective compounds has emerged as a prominent area of research. This field is essential for the preservation of liver health and the effective regulation of various physiological processes, including metabolism, synthesis, and detoxification (Yang et al., 2019; Lu et al., 2016; Yang et al., 2015). RM is recognized as “Bashaga” within the context of Mongolian medicine and possesses a longstanding tradition of use in the treatment of liver ailments. Notably, the most pronounced hepatotoxic effects are observed following administration at dosages of 1.5 and 3 g/kg (Yao et al., 2011), consequently, we initiated a comprehensive investigation into the potential of RM to enhance LI, as well as the underlying mechanisms through which it exerts this effect. Our findings indicate that the RML, particularly at a dosage of 0.75 g/kg, exhibited the most pronounced efficacy in mitigating LI. Consequently, to assess the potential hepatotoxic effects of a higher dosage of RM, specifically 3 g/kg, we conducted subsequent fecal metabolomics analyses on both RMH and RML.
Through an analysis utilizing network pharmacology, the primary active constituents of RM that contribute to the amelioration of LI have been identified as adenosine, aloeemodin, emodin, kaempferol, paeonoside, and quercetin. The pivotal targets implicated in the therapeutic action of these components include AKT1, IL6, and PPARG. Prior research has demonstrated that in a model of acetaminophen-induced LI, kaempferol is capable of activating the Nrf2 signaling pathway, leading to the upregulation of Gpx4, inhibition of ferroptosis, and subsequent mitigation of LI. Furthermore, kaempferol may exert its effects through the modulation of targets such as AKT1. This study suggests that kaempferol may collaboratively contribute to the protective effects against LI by activating the Nrf2 signaling pathway and regulating targets like AKT1 (Mohamed et al., 2024).
At present, ALT, AST, AKP and γ-GT and are the primary biomarkers utilized for the clinical diagnosis of LI. In our investigation, the levels of these enzymes in the MOD group were significantly elevated in comparison to the CON group, thereby confirming the successful establishment of our experimental model. Additionally, the administration of RML resulted in a notable reduction in the levels of ALT and γ-GT in rats with LI, as well as a partial decrease in AKP levels. RM exhibits bidirectional regulation with a narrow therapeutic window. Low doses predominantly activate hepatic protective mechanisms, yielding significant efficacy. However, medium-to-high doses allow inherent hepatotoxic components to prevail, diminishing or reversing protection and reducing efficacy. At moderate doses, protective and toxic effects counterbalance, resulting in non-dose-dependent changes in classic liver enzymes.
Metabolomics serves as a significant methodology for the identification of novel biomarkers associated with LI (Gaggini et al., 2019). Based on the fecal metabolomics study of rats with LI, we screened the differential metabolic components of RMH and RML. Research has indicated that 13-OxoODE is directly linked to LI, this compound appears to facilitate the progression of LI by exacerbating oxidative stress, inducing endoplasmic reticulum stress, disrupting lipid metabolism, and promoting apoptosis (Zhang et al., 2017). In CECs, the synthesis of 13-Oxo-ODE can be stimulated through the metabolism of linoleic acid. Findings suggest 13-Oxo-ODE protects intestinal barrier integrity by suppressing inflammation. LPS-induced inflammation compromises barrier function, triggering dysbiosis, bacterial/metabolite translocation, immune activation, chronic inflammation, and potential systemic metabolic disturbances (Chopyk and Grakoui, 2020). The concentration of this metabolite was markedly reduced in the MOD group and exhibited a significant increase following the administration of RML, aligning with our findings.
Our investigation further demonstrated that RML markedly decreased bilirubin concentrations in LI rats. In the context of liver fibrosis, bilirubin metabolism frequently displays considerable irregularities, with increased serum bilirubin levels serving as a critical indicator of hepatic functional impairment. As liver fibrosis advances, the functionality of hepatocytes progressively declines, diminishing their capacity to uptake, conjugate, and eliminate bilirubin, which consequently leads to the accumulation of unconjugated bilirubin in the bloodstream (Surendran et al., 2020).
13-L-Hydroperoxylinoleic acid, an oxidative metabolite derived from linoleic acid, is integral to the process of hepatic fibrogenesis. In the context of nonalcoholic fatty liver disease and nonalcoholic steatohepatitis, increased concentrations of 13-L-Hydroperoxylinoleic acid contribute to the exacerbation of hepatocyte injury and the acceleration of fibrotic progression (Kasahara et al., 2023). The levels of 13-L-Hydroperoxylinoleic acid were markedly increased in the context of MOD, with a further significant elevation observed following the administration of RMH. Therefore, metabolomics research indicates that RML downregulated differential metabolic components such as 13-OxoODE, morphine, and niacinamide, while RMH showed the opposite trend.
This research incorporated the mechanisms of action associated with network pharmacology alongside a metabolomic analysis of RM in the context of treating LI. The findings indicated that the principal components of RM facilitated the amelioration of LI by modulating differential metabolites and engaging specific genes, including IL6, PPARG, and AKT1. These components exerted their therapeutic effects through various signaling pathways, notably the HIF-1 signaling pathway, IL-17 signaling pathway, PI3K-Akt signaling pathway, and TNF signaling pathway. Notably, during the process of LI amelioration, IL-6 emerged as a pivotal factor in the protection and repair of hepatic tissue, actively promoting the dedifferentiation and regenerative processes of hepatocytes (Li et al., 2023). The function of PPARG in enhancing LI is primarily demonstrated through its regulation of lipid metabolism, suppression of inflammatory responses, mitigation of oxidative stress, and facilitation of hepatocyte repair and regeneration (Wu et al., 2021). Furthermore, the function of AKT1 in enhancing LI is primarily evident through its facilitation of the upregulation of hepatocyte growth factor and vascular endothelial growth factor. This process results in the increased release of anti-inflammatory cytokines and growth factors within an inflammatory context, thereby contributing to a protective effect against immune-mediated hepatitis (Wang et al., 2024).
Through network pharmacology association of common differential metabolic components combined with RM components, we identified several critical pathways associated with the enhancement of LI. Significantly, the PI3K-Akt signaling pathway is implicated in the enhancement of LI. Activation of this pathway has been shown to suppress the synthesis of ECM and the activation of HSCs, while simultaneously promoting hepatocyte proliferation and inhibiting apoptosis, among other physiological functions (Jackson et al., 2008; Shamsan et al., 2024; Wang et al., 2022). Fatty acid metabolism is a critical component of liver function. It is integral to both the synthesis and storage of hepatic lipids, and it is also significantly associated with various pathological processes, including inflammatory responses, oxidative stress, and mitochondrial dysfunction (Kumar et al., 2023). The cytochrome P450 (CYP450) metabolic pathway of arachidonic acid (AA) is significant in the context of non-alcoholic fatty liver disease. Arachidonic acid influences the inflammatory response of hepatocytes via the phosphoinositide 3-kinase (PI3K)/Akt signaling pathway (Wang et al., 2025). Simultaneously, research has demonstrated that the activation of the PI3K-Akt signaling pathway exerts diverse influences on linoleic acid metabolism in liver tissue, encompassing antioxidant properties, regulation of lipid metabolism, responses to endoplasmic reticulum stress, and modulation of the FoxO signaling pathway (Li et al., 2024). Through the integration of metabolomics data with metabolic pathways and KEGG pathways, our findings suggest that RM has the potential to enhance lipid index by activating specific pathways and modulating fatty acid metabolic processes, including those related to linoleic acid and arachidonic acid metabolism.
The impairment of intestinal barrier function enhances the translocation of intestinal bacteria, which subsequently results in inflammation and the development of inflammatory conditions (Paulraj and Hojun, 2018). At the phylum level, the relative abundances of microbiota, including Verrucomicrobiota, Desulfobacterota, Cyanobacteria, and Fusobacteriota, were found to be elevated in rats subjected to CCl4-induced LI. Notably, Fusobacteriota is classified as a Gram-negative bacterium. In the intestinal microbial communities of healthy individuals, it typically constitutes a specific proportion; however, its abundance can fluctuate markedly under certain pathological conditions, such as colitis (Yu et al., 2022). In our investigation, the RML intervention effectively mitigated the dysbiosis of Fusobacteriota observed in rats with LI, restoring it to baseline levels. At the genus level, notable increases were observed in Erysipelotrichaceae_UCG-003, Lachnoclostridium, Lachnospiraceae_ND3007_group, Enterococcus, Dorea, Defluviitaleaceae_UCG-011, Christensenellaceae_R-7_group, and Coriobacteriaceae among the LI-affected rats, with varying degrees of reduction noted in the RML-treated group. Notably, Erysipelotrichaceae has been significantly elevated in various pathological conditions, including hyperlipidemia, chronic hepatitis B-related liver fibrosis, and non-alcoholic fatty liver disease (Matsushita et al., 2015; Neuschwander-Tetri, 2017). In our research, we observed that RML resulted in a reduction of Erysipelotrichaceae abundance in rats exhibiting LI. Furthermore, the variation in the relative abundance of Lachnoclostridium demonstrated a strong correlation with the severity of liver diseases. Specifically, in individuals diagnosed with alcoholic fatty liver disease (AFLD), the relative abundance of Lachnoclostridium at the genus level was significantly elevated in the AFLD cohort compared to the healthy control group (Chi et al., 2023). Moreover, Lachnoclostridium exhibited a notable increase in abundance within the cholestatic liver disease cohort and demonstrated a correlation with various serological markers indicative of liver pathology (Wu et al., 2020). At the species level, CCl4 administration resulted in a significant elevation of Lactobacillus, Christensenellaceae_R-7, Lachnospiraceae_ND3007, Clostridium, and Candidatus_Dorea_massiliensis in rat subjects. Notably, Lactobacillus exhibited a marked increase in comparison to the MOD group. Lactobacillus is intricately associated with liver pathologies, as it has the capacity to enhance the population of beneficial bacteria while suppressing the proliferation of pathogenic bacteria, thus contributing to the modulation of the intestinal microbiota balance (Kim et al., 2022). It has the potential to enhance the integrity of the intestinal barrier (Liu et al., 2022), additionally, it may mitigate liver inflammation by modulating the expression of inflammatory mediators (Yamazaki et al., 2022). The concentration of Lactobacillus exhibited a significant increase in the MOD group, suggesting that RML may contribute to the regulation of intestinal microbiota, enhancement of the intestinal barrier, and attenuation of inflammation levels in the large intestine.
The experimental findings indicated that the RML group exhibited significantly greater protective effects in alleviating LI compared to the RMH group. Biochemical assessments revealed that RML not only effectively diminished the area of collagen fiber deposition in the liver tissues of LI rats but also led to a substantial reduction in serum levels of ALT and γ-GT. Furthermore, metabolomic analysis elucidated that RML treatment resulted in a notable upregulation of 13-Oxo-ODE levels, alongside a downregulation of morphine and bilirubin expression. Importantly, the abnormal increase in 13-L-Hydroperoxylinoleic acid levels observed in the RMH group may correlate with the exacerbation of LI symptoms. Collectively, these results suggest that RML demonstrates superior efficacy in ameliorating LI. Nonetheless, the mechanisms by which potentially toxic components in RM influence LI, particularly regarding their role in exacerbating LI through specific pathways, necessitate further investigation in future studies. Initially, our research did not include validation of the RM reference standard. However, although 17 constituents were identified, their ADME properties lack pharmacokinetic validation. Importantly, predicted core targets (e.g., AKT1, IL-6, PPARG) require functional confirmation through cellular or genetic models. Finally, observed microbiota modulation necessitates causal verification via fecal transplantation to establish direct mechanistic links.
4 Materials and methods
Rhododendron molle (Blume) G. Don (RM) was purchased from Anguo Qi’ao Traditional Chinese Medicine Drinking Tablets Co. IL-1β, TNF-α, AST, ALT, AKP, γ-GT were purchased from Kote Biologicals, Mass spectinol, and acetonitrile were purchased from Thermo Fisher.
4.1 Drug extraction
Weigh 200 g of the RM. Subsequently, immerse it in 70% ethanol at a solid-to-liquid ratio of 1:10 for a duration of 30 min. Following this, perform reflux extraction under gentle boiling conditions for three cycles, each lasting 2 h. The resulting extract should then be subjected to suction filtration, followed by concentration and drying processes. The final extract is to be freeze-dried and subsequently ground into a powder for future applications.
4.2 Component analysis of RM
4.2.1 Sample pretreatment
In the sample pretreatment procedure, 0.25 g of the powdered RM was accurately measured and subsequently combined with 10 milliliters of methanol. Following this, ultrasonic extraction was performed for a duration of 30 min. The resulting mixture was then subjected to centrifugation and filtration. Ultimately, the supernatant was collected for sample injection in subsequent analytical processes.
4.2.2 Chromatographic conditions
The detection system was an Obitrap Exploris 120 (Thermo Fisher Scientific). The chromatographic column used was a Waters Acquity UPLC BEH with dimensions of 2.1100 mm and a particle size of 1.7 μm. The mobile phase consisted of two components: Mobile phase A was water containing 0.1% formic acid, and mobile phase B was acetonitrile. Gradient elution was adopted as follows: From 0 to 25 min, the proportion of mobile phase A changed from 99 to 1%, while that of mobile phase B changed from 1 to 99%. From 25 to 27 min, the proportion of mobile phase A was 1% and that of mobile phase B was 99%. From 27 to 28 min, the proportion of mobile phase A changed from 1 to 99%, and that of mobile phase B changed from 99 to 1%. From 28 to 30 min, the proportion of mobile phase A was 99% and that of mobile phase B was 1%. The injection volume was 2.0 μL, the flow rate was set at 0.3 mL/min, and the column temperature was maintained at 35.0°C.
4.2.3 Mass spectrometry conditions
The ion source was a heated electrospray ionization (HESI). Both positive and negative ion detection modes were adopted. The sheath gas pressure was set at 35 Arb, the auxiliary gas volumetric flow rate was 10 Arb, the spray voltage was 3,500 V, the ion transfer tube temperature was 320°C, and the auxiliary gas temperature was 300°C. The scanning mode was Fullscan-ddMS2. The resolution of the full MS was 60,000, while the resolution of the dd-MS2 was 15,000. The scanning range was from 100 to 1,500, and the collision energy was 30%.
4.2.4 Data processing
Progenesis QI was employed for baseline filtering, peak identification, integration, retention time correction, and peak alignment. The Xcalibur 4.0 software was utilized to analyze the data pertaining to the molecular ions and fragment ions associated with each component peak. By integrating the precise mass numbers of the compounds, the secondary fragmentation spectra, and referencing the HMDB database2 alongside the PubChem database,3 the primary chemical constituents of Rhododendron molle were identified.
4.3 Network pharmacology studies
4.3.1 Screening of the targets of RM components
We performed a network-pharmacological analysis on the components of RM that were identified through MS/MS techniques. Following the retrieval of the Canonical SMILE sequences of these compounds from the PubChem database, we employed the SwissTargetPrediction database4 to forecast the potential targets associated with RM.
4.3.2 Acquisition of LI-related disease targets and intersection target genes
Conduct a search for genes associated with liver injury (LI) in the GeneCards database5 using “Liver Injury” as the search term. Subsequently, compile a summary of the retrieved results to identify the complete set of genes related to LI. Employ Venny 2.1.0 (csic.es) to determine the intersection of genes between the target genes of the active components of RM and the genes associated with LI, and create a Venn diagram to visually represent these findings.
4.3.3 Construction of the “drug-active component-potential target” network
In the Cytoscape 3.10.1 software, a network was developed to represent the interactions between drug-active components and their potential targets, which was then subjected to analysis. The significance of each node in relation to others, as well as the extent of each node’s influence, was evaluated using the Degree value. As a result, the primary components involved in the treatment of LI by RM were identified through a screening process that employed Degree values.
4.3.4 Construction of the protein–protein interaction network
The genes that overlap between RM and LI were uploaded to the STRING database,6 with the organism specified as “Homo sapiens” This process generated a PPI network diagram, which was subsequently saved in “tsv” file format. The resulting data were then imported into Cytoscape version 3.10.1, where the top 10 core targets were identified utilizing the “Cytohubba” plugin for network analysis.
4.3.5 Enrichment analysis of GO biological processes and KEGG pathways
Enrichment analyses for GO biological processes and KEGG pathways were performed using the DAVID database.7 Upon entering the database, users are prompted to select the “Functional Annotation” option. The predicted critical targets were subsequently uploaded to the DAVID database to facilitate the KEGG enrichment analysis. Users must designate “GENE SYMBOL” and specify “Human” to ensure the selection of “Homo sapiens” prior to data submission. Following this, the “KEGG Pathways” option is selected to download the relevant pathway files. The screening criteria established a significance threshold of p < 0.05, simultaneously, it is essential to eliminate extraneous pathways, including but not limited to “cancer,” “atherosclerosis,” and “proteoglycan signaling pathway in cancer,” with results organized by p-value to identify the top 20 pathways. The enrichment results were ultimately visualized using the online platform Bioinformatics,8 which offers tools for the graphical representation of the data.
4.3.6 Molecular docking validation
Utilizing the Degree values from the network analysis, core components and high-ranking targets were identified for molecular docking studies, employing software tools such as AutoDock Tools and PyMOL. The two-dimensional structures of the small molecules were sourced from the PubChem database,9 and the Open Babel GUI software was employed to convert these structures into mol2 file format for subsequent applications. Following this, the three-dimensional structural files of the core targets were retrieved from the Protein Data Bank (PDB).10 Preliminary procedures, including the extraction of small molecules and the removal of hydrogen atoms, were conducted using PyMOL, with the resultant files saved for future reference. The small molecule and target protein data were then imported into AutoDockTools for the molecular docking process. Finally, PyMOL was utilized once more for the visual analysis of the docking outcomes.
4.4 Animal experimentation
4.4.1 Materials
Silymarin purchased from Rigalon; Rhododendron molle (Blume)w G. Don was purchased from Anguo Qi’ao Traditional Chinese Medicine Drinking Tablets Co.; AKP, γ-GT, AST, ALT, TNF-α, IL-1β were purchased from Kurt Bio; Methanol and acetonitrile were purchased from Thermo Fisher.
4.4.2 Animal experimentation and grouping
Sixty specific pathogen-free (SPF) male SD rats with a body weight of 180 ± 20 g (provided by (Beijing) Biotechnology Co., Ltd.) were acclimated for 7 days and then randomly divided into six groups: CON with 10 rats, MOD with 10 rats, RMH, RMM, RML each with 10 rats, and a Sily group with 10 rats. The license number for the experimental animals is: SCXK (Jing) 2019–0010. The housing conditions were maintained at a temperature of 22–26°C, approximately 33% humidity, and a 12-h light/dark cycle. The animal experiments were approved by the Medical Ethics Committee of Baotou Medical College, Inner Mongolia University of Science and Technology (approval number: Baotou Medical College Lunjian Animal 2021 No. 018). The experiment lasted for 14 days. The MOD group, RM group, and Sily group were subjected to modeling twice weekly (40% CCl4 + corn oil) on Mondays and Thursdays each week. The initial injection dose was 5 mg/kg, followed by subsequent injections of 3 mg/kg. The CON and MOD groups were given 0.1% sodium carboxymethyl cellulose solution by gavage daily; the Sily group was dissolved in 0.1% sodium carboxymethyl cellulose solution at a dosage of 50 mg/kg. According to the “Chinese Pharmacopoeia,” the recommended human oral dose of RM is 0.6–1.5 g. In this study, 1.5 g was designated as the medium dose, while doses 2-fold lower and 2-fold higher than the medium dose were defined as the low and high doses, respectively. Based on a standard human body weight of 60 kg, the equivalent doses for rats were calculated as follows: low dose = (0.75/60) × 6.3, medium dose = (1.5/60) × 6.3, and high dose = (3/60) × 6.3, which translates to crude drug dosages of 0.07875 g/kg, 0.0575 g/kg, and 0.315 g/kg, respectively. These were dissolved in 0.1% sodium carboxymethyl cellulose and administered by gavage.
4.4.3 Treatment and sample collection
On the fourteenth day of the experimental protocol, following a period of fasting and water deprivation, fresh fecal samples were collected from the rats utilizing the extrusion method. A total of three samples were obtained and subsequently stored at −80°C in a freezer. Anesthesia was administered to the rats via intraperitoneal injection of 3% pentobarbital sodium at a dosage of 30 mg/kg. Following anesthesia, blood samples were extracted from the abdominal aorta of each group of rats. These blood samples were then subjected to centrifugation at 4°C at a speed of 3,500 r/min for a duration of 15 min to facilitate the separation of serum. The resulting supernatant serum was transferred into 250 μL centrifuge tubes for storage. Subsequently, the rats were euthanized through cervical dislocation, and the liver tissues were excised, rinsed with normal saline to eliminate residual blood, and sectioned into multiple portions, which were wrapped in aluminum foil. A segment of the liver tissue, approximately 2 cm from the hepatic portal region, was preserved in paraformaldehyde for future histological analysis, including HE staining and Masson’s trichrome staining.
4.5 Observation of hepatic tissue pathological changes using HE and Masson’s staining
After the liver tissues from each group of rats were fixed for a duration of 48 h, they underwent processing in an automated dehydration apparatus for gradient dehydration, followed by embedding. Subsequent to sectioning and dewaxing, the tissue sections were stained utilizing the HE and Masson’s staining kits in accordance with the manufacturer’s protocols. The HE staining technique was employed to examine the pathological morphology of the liver lobules, while Masson’s staining was utilized to assess the deposition of liver fibrosis and the distribution of fibrotic tissue. The percentage of collagen fiber area was quantified using Image-Pro Plus 6.0 software.
4.6 ELISA for the detection of liver function and inflammatory markers
Liver tissues were obtained from a − 80°C freezer and subsequently thawed on ice. A weight ratio of 1:9 was established, comprising liver tissue and physiological saline, and the tissue was homogenized while maintained on ice. Post-homogenization, the resultant mixture underwent centrifugation at 3,000 rpm for a duration of 10 min utilizing a centrifuge. The supernatant was then meticulously collected and processed in accordance with the protocols outlined in the ELISA kits designed for the assessment of liver function and inflammatory markers.
4.7 Fecal untargeted metabolomics analysis
4.7.1 Sample preparation
Fecal samples were obtained from a −80°C freezer. Approximately 100 mg of fecal matter was transferred into a 1.5 mL centrifuge tube, to which 1,000 μL of 75% methanol was added. The resulting mixture was homogenized and agitated for 30 s, followed by sonication for a duration of 10 min. Subsequently, the mixture was centrifuged at 13,000 rpm, and the supernatant was filtered through a membrane to prepare the sample for fecal analysis. An injection volume of 4 μL was utilized for the analytical procedure.
4.7.2 QC sample preparation
Carefully measure 100 mg of fecal samples from each group and combine them thoroughly to produce a homogenized composite sample.
4.7.3 Chromatographic conditions
Chromatographic column: ACQUITY UPLCTM C18 column (100 mm × 2.1 mm i.d., 1.7 μm) (Waters Corporation, United States); Mobile phase: Mobile phase A is 0.1% acetonitrile, and mobile phase B is 0.1% formic acid in water; Column temperature: 40°C; Flow rate: 0.4 mL/min; Injection volume: 4 μL; Gradient elution method: 0–2 min, 99–50% A, 1–50% B; 3–10 min, 50% A to 1% A, 50% B to 99% B; 11–15 min, 1–99% A, 99% B to 1% B.
4.7.4 Mass spectrometry conditions
Ion source: Electrospray ionization (ESI) source; Switching between positive and negative ion modes; Operating mode: Full scan-data dependent secondary scanning (Full MS-ddMS2); Mass spectrometry parameters are as follows: Sheath gas flow rate: 30 arb (ESI+), 35 arb (ESI−); Auxiliary gas flow rate: 10 arb; Spray voltage: 3.5 kV (ESI+), 3.0 kV (ESI−); Column temperature set to 35°C.
4.7.5 Data processing
The raw data were analyzed using Progenesis QI software to obtain the normalized label-free results of metabolic peaks for each sample. The samples were normalized based on the total ion intensity of each chromatogram. The data matrix consisting of sample codes, RT-m/z pairs, and peak areas was imported into MassLynx V4.1 software for PCA, PLS-DA, and OPLS-DA. Based on the OPLS-DA analysis results of the control and model groups, a VIP list file was created to identify metabolic variables with VIP >1 and p < 0.05 from independent sample t-tests for further analysis. By combining public databases such as HMDB (see text footnote 2), KEGG,11 mbrole2,12 and relevant literature, the structures of potential biomarkers for LI were identified. PCA, PLS-DA, and OPLS-DA analyses were conducted using EZinfo 3.0 for Waters, and the validation analysis was performed using SIMCA 14.1. The names of the identified biomarkers were imported into the Metaboanalyst 6.0 website13 to analyze and determine the disrupted metabolic pathways. Metabolic pathways closely related to liver injury were obtained based on the criterion of impact >0.
4.8 Component-gene-disease-metabolite network
4.8.1 Network construction
The differential metabolites identified between RM and LI are regarded as potential targets for RM in its interaction with LI. Following this, the overlapping targets are incorporated into Cytoscape version 3.10.3 to construct a “component-target-metabolite” interaction network.
4.8.2 PPI analysis and core target screening
An interaction analysis of potential targets was performed utilizing the STRING database to examine proteins at a systemic level. We identified targets associated with RM components, LI components, and metabolic components, subsequently importing the intersecting targets into Cytoscape version 3.10.3. Employing the “Cytohubba” plugin, we identified the top 10 core targets and conducted a visual analysis of these targets.
4.8.3 GO and KEGG enrichment analysis
The overlapping targets were uploaded to the Metascape database14 for GO enrichment and KEGG pathway analysis, in order to identify representative biological processes and pathways.
4.9 Fecal 16S rDNA analysis
4.9.1 Sample preparation
In accordance with the protocols for 16S rDNA gene sequencing, Hangzhou Lianchuan Biotechnology Co., Ltd. employed the E.Z.N.A.® Stool DNA Kit to extract DNA from rat intestinal content samples. Subsequent to the extraction process, agarose gel electrophoresis was utilized to assess the quality of the extracted DNA, while a UV spectrophotometer was employed for quantification purposes, thereby ensuring the accuracy and reliability of the obtained results.
4.9.2 High-throughput 16S rDNA sequencing
The V3-V4 region of the prokaryotic small subunit (16S) rRNA gene was amplified using primers 341F (5′-CCTACGGGNGGCWGCAG-3′) and 805R (5′-GACTACHVGGGTATCTAATCC-3′). The total volume of the PCR amplification reaction mixture was 25 μL, which contained 25 ng of template DNA, 12.5 μL of a specific reagent, 2.5 μL of each primer, and ddH₂O (deionized distilled water) to adjust the volume. Subsequently, the PCR products were separated by 2% agarose gel electrophoresis to confirm the obtained DNA fragments. Throughout the DNA extraction process, ultrapure water was utilized instead of the sample solution. This was done to eliminate the possibility of false-positive PCR results and served as a negative control. The PCR products were purified by AMPure XT beads (Beckman Coulter Genomics, Danvers, MA, United States) and quantified using Qubit (Invitrogen, United States). The amplicon pool was then prepared for sequencing. The size and quantity of the amplicon library were, respectively, evaluated on an Agilent 2100 Bioanalyzer (Agilent, United States) and with a library quantification kit from Illumina (Kapa Biosciences, Woburn, MA, United States). Sequencing of the library was carried out on the NovaSeq PE250 platform.
4.9.3 Data processing
The gut microbiota data were processed utilizing the QIIME2 software. Alpha diversity analysis was performed using Graphpad version 9.5. Beta diversity and species composition analyses were conducted on the Lianchuan Bioinformatics Cloud Platform.15 Additionally, the LEfSe analysis was executed on the Biosciences Cloud Platform,16 with the linear discriminant analysis (LDA) score established at 3.5.
4.10 Statistical methods
Data analysis was conducted using the SPSS 22.0 statistical software. Measurement data were expressed as the mean ± standard deviation ( ). For comparisons among multiple groups, one-way analysis of variance (one-way ANOVA) was employed. When pairwise comparisons were needed, the t-test was utilized. A p-value less than 0.05 was considered to indicate a statistically significant difference.
5 Conclusion
This investigation systematically examined the mechanisms by which RM addresses LI through the integration of metabolomics, network pharmacology, and 16S rRNA methodologies. The findings indicated that RM, particularly the RML variant, demonstrated a protective effect against CCl4-induced LI by modulating various metabolic pathways, including Arachidonic acid metabolism, Linoleic acid metabolism, and Steroid biosynthesis. These results underscore the multi-target and multi-pathway nature of RM’s action in mitigating CCl4-induced LI. In contrast, RMH did not demonstrate any advantageous effects on LI as evidenced by biochemical markers and histopathological examinations. These findings suggest that RM may represent a promising therapeutic candidate for the treatment of CCl4-induced LI, with the identified targets potentially serving as viable candidates for future diagnostic or therapeutic interventions. According to our findings, the suggested clinical dosage is 0.07875 g/kg. Furthermore, the insights gained from the network pharmacology analysis of RM will inform the objectives of subsequent research endeavors.
Data availability statement
The original contributions presented in the study are publicly available. All raw Illumina sequencing data have been deposited in the NCBI SRA under BioProject accession number PRJNA1294298. Mass spectrometry data for the metabolomics metabolites are available in the article and Supplementary material.
Ethics statement
The animal study was approved by the Ethical Committee of Baotou Medical College (No. 2021018). The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
XJ: Formal analysis, Investigation, Methodology, Validation, Writing – original draft. YZ: Formal analysis, Methodology, Writing – original draft. TiancM: Methodology, Validation, Writing – original draft. TianwM: Data curation, Investigation, Methodology, Writing – original draft. XL: Formal analysis, Software, Writing – original draft. DH: Formal analysis, Writing – original draft. HM: Data curation, Software, Supervision, Validation, Visualization, Writing – review & editing. HC: Conceptualization, Data curation, Funding acquisition, Project administration, Supervision, Validation, Visualization, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was financially supported by the National Natural Science Foundation of China (No. 82060784); Inner Mongolia Natural Science Foundation (No. 2023MS08029); Inner Mongolia Autonomous Region Higher Education Institutions “Young Scientific and Technological Talent Support Program” (No. NJYT24079); and the 2024 Baotou Medical College Research and Innovation Program (BYKYCX202464).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1570229/full#supplementary-material
Footnotes
1. ^https://www.metaboanalyst.ca/MetaboAnalyst/home.xhtml
3. ^https://pubchem.ncbi.nlm.nih.gov/
4. ^http://www.swisstargetprediction.ch
5. ^https://genealacart.genecards.org/
7. ^https://david.ncifcrf.gov/
8. ^https://www.bioinformatics.com.cn/login/
9. ^https://pubchem.ncbi.nlm.nih.gov/
11. ^http://www.genome.jp/kegg/
12. ^http://csbg.cnb.csic.es/mbrole2/index.php
13. ^http://www.metaboanalyst.ca/
References
Cai, Y. Q., Hu, J. H., Qin, J., Sun, T., and Li, X. L. (2018). Rhododendron molle (Ericaceae): phytochemistry, pharmacology, and toxicology. Chin. J. Nat. Med. 16, 401–410. doi: 10.1016/S1875-5364(18)30073-6
Chen, X., Shi, L., Ran, X.-Y., Zhang, L.-N., Xie, K.-P., Zhao, Y., et al. (2024). Rational design of an intramolecular hydrogen bond enhanced fluorescent probe for diagnosis of drug-induced liver injury. ACS Mater. Lett. 6, 1059–1068. doi: 10.1021/acsmaterialslett.4c00002
Chi, X., Sun, X., Xu, X., Chen, D., Liu, S., Chen, J., et al. (2023). Characteristics and clinical significance of intestinal microbiota in patients with alcoholic fatty liver disease. Chin. J. Liver Dis. 15, 19–27. doi: 10.3969/j.issn.1674-7380.2023.01.004
Chinese Pharmacopoeia Commission (2020). Pharmacopoeia of the People’s Republic of China: a part. Beijing: China Medical Science and Technology Press.
Chopyk, D. M., and Grakoui, A. (2020). Contribution of the intestinal microbiome and gut barrier to hepatic disorders. Gastroenterology 159, 849–863. doi: 10.1053/j.gastro.2020.04.077
Devarbhavi, H., Asrani, S. K., Arab, J. P., Nartey, Y. A., Pose, E., and Kamath, P. S. (2023). Global burden of liver disease: 2023 update. J. Hepatol. 79, 516–537. doi: 10.1016/j.jhep.2023.03.017
Gaggini, M., Carli, F., Rosso, C., Younes, R., D’Aurizio, R., Bugianesi, E., et al. (2019). Altered metabolic profile and adipocyte insulin resistance mark severe liver fibrosis in patients with chronic liver disease. Int. J. Mol. Sci. 20:6333. doi: 10.3390/ijms20246333
Gu, C. Q., Tang, L. D., Hao, Y. H., Dong, S., Shen, J., Xie, F. M., et al. (2023). Network pharmacology and bioinformatics were used to construct a prognostic model and immunoassay of core target genes in the combination of quercetin and kaempferol in the treatment of colorectal cancer. J. Cancer 14, 1956–1980. doi: 10.7150/jca.85517
Guo, X. H., Feng, J. W., Wang, Y. D., Chen, H. P., Leng, J., Liu, Y. P., et al. (2021a). Analysis of the herbal verification and historical evolution of the preparation of Rhododendron molle (Blume) G. Don. Chin J. Exp. Formul. 27, 172–180. doi: 10.13422/j.cnki.syfjx.20211559
Guo, X. H., Feng, J. W., You, Q., Chen, H. P., and Liu, Y. P. (2020). Modern research progress on the toxic Chinese herb Rhododendron molle (Blume) G Don. China Pharm Ind 29, 96–104. doi: 10.3969/j.issn.1006-4931.2020.23.027
Guo, X. H., Liu, X., Wu, W. H., Wang, Y.D., and Leng, J. (2021b). Simultaneous determination of four components in Ephedra sinica herbal medicine by HPLC-ELSD method. J. Tradit. Chin. Med. 36, 2922–2925. doi: 10.1016/j.jtcme.2021.07.010
Hackstein, C. P., Spitzer, J., Symeonidis, K., Horvatic, H., Bedke, T., Steglich, B., et al. (2023). Interferon-induced IL-10 drives systemic T-cell dysfunction during chronic liver injury. J. Hepatol. 79, 150–166. doi: 10.1016/j.jhep.2023.02.026
Hu, H., Wang, Y., Li, J. H., et al. (2024). Research on fingerprint spectrum and content determination of Mongolian medicine Rhododendron molle (Blume) G. Don formula granules. Zhongnan Pharm. 22, 2962–2968.
Jackson, L. N., Larson, S. D., Silva, S. R., Rychahou, P. G., Chen, L. A., Qiu, S., et al. (2008). PI3K/Akt activation is critical for early hepatic regeneration after partial hepatectomy. Am. J. Physiol. Gastrointest. Liver Physiol. 294:G1401-10. doi: 10.1152/ajpgi.00062.2008
Kasahara, N., Imi, Y., Amano, R., Shinohara, M., Okada, K., Hosokawa, Y., et al. (2023). A gut microbial metabolite of linoleic acid ameliorates liver fibrosis by inhibiting TGF-β signaling in hepatic stellate cells. Sci. Rep. 13:18983. doi: 10.1038/s41598-023-46404-5
Kim, H., Fugaban, J. I. I., Holzapfel, W. H., and Todorov, S. D. (2022). Selection of beneficial bacterial strains with potential as oral probiotic candidates. Probiotics. Antimicrob. Proteins 14, 1077–1093. doi: 10.1007/s12602-021-09896-z
Kumar, V., Sethi, B., Staller, D. W., Xin, X., Ma, J., Dong, Y., et al. (2023). Anti-miR-96 and Hh pathway inhibitor MDB5 synergistically ameliorate alcohol-associated liver injury in mice. Biomaterials 295:122049. doi: 10.1016/j.biomaterials.2023.122049
Li, Y., Cao, B., Lin, M., Xu, J., Qi, S., Wang, J., et al. (2024). An integrative lipidomics and transcriptomics study revealing Bavachin and Icariin synergistically induce idiosyncratic liver injury. Immunopharmacol. Immunotoxicol. 46:924. doi: 10.1080/08923973.2024.2424293
Li, D. K., Chaudhari, S. N., Lee, Y., Sojoodi, M., Adhikari, A. A., Zukerberg, L., et al. (2022). Inhibition of microbial deconjugation of micellar bile acids protects against intestinal permeability and liver injury. Sci. Adv. 8:eabo2794. doi: 10.1126/sciadv.abo2794
Li, L., Cui, L., Lin, P., Liu, Z., Bao, S., Ma, X., et al. (2023). Kupffer-cell-derived IL-6 is repurposed for hepatocyte dedifferentiation via activating progenitor genes from injury-specific enhancers. Cell Stem Cell 30, 283–299. doi: 10.1016/j.stem.2023.01.009
Lin, X., Wang, J. X., Li, M. G., Qiu, J., Li, X., Qi, L., et al. (2023). Farnesoid X receptor is an important target for the treatment of disorders of bile acid and fatty acid metabolism in mice with nonalcoholic fatty liver disease combined with cholestasis. J. Gastroenterol. Hepatol. 38, 1438–1446. doi: 10.1111/jgh.16279
Liu, Y., and Kong, L. (2009). Research on flavonoid components in Rhododendron molle (Blume) G. Don. Herbal Med. 40, 199–201. doi: 10.3321/j.issn:0253-2670.2009.02.010
Liu, Y., Liu, Q., Zhao, J., Zhang, H., Zhai, Q., and Chen, W. (2022). Strain-specific regulative effects of Lactobacillus plantarum on intestinal barrier dysfunction are associated with their capsular polysaccharides. Int. J. Biol. Macromol. 222, 1343–1352. doi: 10.1016/j.ijbiomac.2022.09.136
Liu, A. L., Zhang, L., and Du, G. H. (2018). Historical understanding and evaluation of the Rhododendron molle (Blume) G. Don poison incident. Pharmacol. Clin. Appl. Tradit. Chin. Med. 34, 161–163.
Lu, Y., Hu, D., Ma, S., Zhao, X., Wang, S., Wei, G., et al. (2016). Protective effect of wedelolactone against CCl₄-induced acute liver injury in mice. Int. Immunopharmacol. 34, 44–52. doi: 10.1016/j.intimp.2016.02.003
Matsushita, N., Osaka, T., Haruta, I., Ueshiba, H., Yanagisawa, N., Omori-Miyake, M., et al. (2015). Effect of lipopolysaccharide on the progression of non-alcoholic fatty liver disease in high caloric diet-fed mice. Scand. J. Immunol. 83, 109–118. doi: 10.1111/sji.12397
Mohamed, G. A., El-Agamy, D. S., Abdallah, H. M., Sindi, I. A., Almogaddam, M. A., Alzain, A. A., et al. (2024). Kaempferol sophoroside glucoside mitigates acetaminophen-induced hepatotoxicity: role of Nrf 2/NF-κB and JNK/ASK-1 signaling pathways. Heliyon 10:e31448. doi: 10.1016/j.heliyon.2024.e31448
Neuschwander-Tetri, B. A. (2017). Non-alcoholic fatty liver disease. BMC Med. 15:45. doi: 10.1186/s12916-017-0806-8
Paulraj, K., and Hojun, K. (2018). Protective effects of lactic acid bacteria against TLR4 induced inflammatory response in hepatoma Hep G2 cells through modulation of Toll-like receptor negative regulators of mitogen-activated protein kinase and NF-κB signaling. Front. Immunol. 9:1537. doi: 10.3389/fimmu.2018.01537
Shamsan, E., Almezgagi, M., Gamah, M., Khan, N., Qasem, A., Chuanchuan, L., et al. (2024). The role of PI3k/AKT signaling pathway in attenuating liver fibrosis: a comprehensive review. Front. Med. 11:1389329. doi: 10.3389/fmed.2024.1389329
Singh, J., Rasane, P., Kaur, R., Kaur, H., Garg, R., Kaur, S., et al. (2023). Valorization of grape (Vitis vinifera) leaves for bioactive compounds: novel green extraction technologies and food-pharma applications. Front. Chem. 11:1290619. doi: 10.3389/fchem.2023.1290619
Su, R., He, H., Sun, C., Wang, X., and Liu, X. (2023). Prediction of drug-induced hepatotoxicity based on histopathological whole slide images. Methods 212, 31–38. doi: 10.1016/j.ymeth.2023.01.005
Surendran, S. P., Thomas, R. G., Moon, M. J., Park, R., Lee, J. H., and Jeong, Y. Y. (2020). A bilirubin-conjugated chitosan nanotheranostics system as a platform for reactive oxygen species stimuli-responsive hepatic fibrosis therapy. Acta Biomater. 116, 356–367. doi: 10.1016/j.actbio.2020.09.014
Wang, X., Hu, Y. W., Yuan, D., and Fu, H. Z. (2014). Chemical composition study of Rhododendron molle (Blume) G. Don’s milk. J. Chin. Pharm. Sci. 23, 94–98. doi: 10.1016/j.cjps.2014.03.003
Wang, W., Liu, M., Fu, X., Qi, M., Zhu, F., Fan, F., et al. (2024). Hydroxysafflor yellow A ameliorates alcohol-induced liver injury through PI3K/Akt and STAT3/NF-κB signaling pathways. Phytomedicine 132:155814. doi: 10.1016/j.phymed.2024.155814
Wang, X., Yang, X., Liu, C., Yin, Z., Zhang, Z., Feng, T., et al. (2025). Serum pharmacochemistry combined with network pharmacology reveals the hepatotoxicity mechanism of Alangium chinense (Lour.) Harms. J. Ethnopharmacol. 340:119312. doi: 10.1016/j.jep.2024.119312
Wang, M., Zhang, J., and Gong, N. (2022). Role of the PI3K/Akt signaling pathway in liver ischemia reperfusion injury: a narrative review. Ann. Palliat. Med. 11, 10567–10574. doi: 10.21037/apm-21-3286
Wu, L. Y., Tang, L., Zhang, J., Wu, Y. B., Tang, B., Yang, G. D., et al. (2020). Characterization of intestinal flora and its clinical relevance in patients with cholestatic liver disease. J. Third Mil. Med. Univ. 42, 2251–2258. doi: 10.16016/j.1000-5404.202005245
Wu, L. W., Yu, Q., Cheng, P., and Guo, C. Y. (2021). PPAγ plays an important role in acute hepatic ischemia-reperfusion injury via AMPK/mTOR pathway. PPAR Res. 2021:6626295. doi: 10.1155/2021/6626295
Xiao, H., Zhu, X. H., Ji, M. Y., Yi, L. T., and Li, M. H. (2020). A survey on the application of “bashaga”-type drugs in compound preparations in Inner Mongolia. Chin. J. Exp. Formul. 26, 190–198. doi: 10.13422/j.cnki.syfjx.20191812
Yamazaki, T., Yamada, S., Ohshio, K., Sugamata, M., and Morita, Y. (2022). Lactobacillus paracasei KW3110 prevents inflammatory-stress-induced mitochondrial dysfunction in mouse macrophages. Int. J. Mol. Sci. 23:1443. doi: 10.3390/ijms23031443
Yang, B.-Y., Zhang, X.-Y., Guan, S.-W., and Hua, Z.-C. (2015). Protective effect of procyanidin B2 against CCl4-induced acute liver injury in mice. Molecules 20, 12250–12265. doi: 10.3390/molecules200712250
Yang, X., Zhu, X., Tang, X., Liu, M., Zheng, H., and Zheng, L. (2019). Astragalus polysaccharides meliorate cardiovascular dysfunction in iron-overloaded thalassemic mice. Exp. Biol. Med. 244, 1202–1209. doi: 10.1177/1535370219876540
Yao, M., Jin, L. Y., Dai, W. Y., He, L. M., Gao, W. Q., and Bao, Y. M. (2011). Animal studies on the detoxifying effects of gardenia on the liver toxicity of Rhododendron molle (Blume) G. Don. Shizhen J. Tradit. Chin. Med. 22, 2183–2185. doi: 10.3969/j.issn.1008-0805.2011.09.060
Yu, J., Zhao, J., Xie, H., Cai, M., Yao, L., Li, J., et al. (2022). Dendrobium huoshanense polysaccharides ameliorate ulcerative colitis by improving intestinal mucosal barrier and regulating gut microbiota. J. Funct. Foods 96:105231. doi: 10.1016/j.jff.2022.105231
Zhang, Y., Dong, R., Zhou, H., Wang, J., Shi, J., Ye, S., et al. (2024). The flavonoid glycoside from Abrus cantoniensis Hance alleviates alcoholic liver injury by inhibiting ferroptosis in an AMPK-dependent manner. J. Agric. Food Chem. 72, 16323–16333. doi: 10.1021/acs.jafc.4c02912
Zhang, X.-t., Yang, Y., Ji, C., Fu, Y., Pu, X., and Xu, G. (2024). Ganoderma lucidum polysaccharides reduce the severity of acute liver injury by improving the diversity and function of the gut microbiota. Heliyon 10:e35559. doi: 10.1016/j.heliyon.2024.e35559
Zhang, P., Zheng, L., Duan, Y., Gao, Y., Gao, H., Mao, D., et al. (2022). Gut microbiota exaggerates triclosan-induced liver injury via gut-liver axis. J. Hazard. Mater. 421:126707. doi: 10.1016/j.jhazmat.2021.126707
Zhang, W. L., Zhong, W., Sun, Q., Sun, X., and Zhou, Z. (2017). Hepatic overproduction of 13-HODE due to ALOX15 upregulation contributes to alcohol-induced liver injury in mice. Sci. Rep. 7:8976. doi: 10.1038/s41598-017-02759-0
Keywords: Rhododendron molle (Blume) G. Don, network pharmacology, molecular docking, metabolomics, intestinal flora
Citation: Jiang X, Zhuang Y, Meng T, Meng T, Li X, He D, Meng H and Chang H (2025) Integrative network pharmacology, metabolomics and gut flora studies reveal mechanisms of action of Rhododendron molle (Blume) G. Don to ameliorate liver injury. Front. Microbiol. 16:1570229. doi: 10.3389/fmicb.2025.1570229
Edited by:
Jun Zhou, Shandong First Medical University, ChinaReviewed by:
Yina Huang, Anhui Polytechnic University, ChinaJian Yang, Shanghai University of Traditional Chinese Medicine, China
Copyright © 2025 Jiang, Zhuang, Meng, Meng, Li, He, Meng and Chang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hongyu Meng, bWVuZ2h5aHlAMTI2LmNvbQ==; Hong Chang, Y2hhbmdob25nX2Nvb2xAMTYzLmNvbQ==
 Xiaolei Jiang
Xiaolei Jiang Yafeng Zhuang2
Yafeng Zhuang2 Tianwei Meng
Tianwei Meng Xinghua Li
Xinghua Li Hong Chang
Hong Chang