- 1Biology Department, School of Science, College of Science and Technology, University of Rwanda, Kigali, Rwanda
- 2Rwanda Biomedical Centre, Kigali, Rwanda
- 3School of Health Sciences, College of Medicine and Health Sciences, University of Rwanda, Kigali, Rwanda
- 4School of Veterinary Medicine, College of Agriculture, Animal Sciences, and Veterinary Medicine, University of Rwanda, Nyagatare, Rwanda
- 5Natural Product Informatics Research Center, KIST Gangneung Institute of Natural Products, Gangneung, Republic of Korea
- 6Tanzania Livestock Research Institute, Dar es Salaam, Tanzania
- 7Department of Clinical Sciences, College of Veterinary Medicine, Kansas State University, Manhattan, KS, Unites States
Globally, Campylobacter jejuni and C. coli have been associated with human gastroenteritis. More importantly, there are increasing reports of Campylobacter strains that are resistant to commonly used antimicrobials. In Rwanda, the prevalence and the antimicrobial susceptibility profiles of thermophilic Campylobacter strains remain underexplored. Since human campylobacteriosis is a foodborne disease with chicken and pigs being among their major reservoirs, this study aimed to determine the prevalence and antimicrobial susceptibility profiles of thermophilic Campylobacter species from human, chicken, and pig feces in Rwanda. A total of 385, 337, and 359 human, pig and chicken feces, respectively, were investigated for the presence of Campylobacter species. Isolation was done by culture and presumptive colonies were confirmed by the Polymerase Chain Reaction (PCR). The Kirby-Bauer disc diffusion method was employed to determine the susceptibility profiles of obtained isolates against six (06) antimicrobials, namely erythromycin (ERY), ciprofloxacin (CIP), streptomycin (STR), gentamicin (GEN), tetracycline (TET), and chloramphenicol (CHL). The used antimicrobials include drugs of choice or alternative treatment for human campylobacteriosis. The overall prevalence of thermophilic Campylobacter was 7.0% (27/385) in humans, 7.1% (24/337) in pigs, and 32.0% (115/359) in chicken. C. jejuni was the predominant species in all hosts with detection frequencies of 92.6%, 66.7%, and 73.9% in humans, pigs, and poultry, respectively. Increased resistance rates to ERY (70.1–92.4%) and STR (68.2–88.0%) were observed particularly among chicken isolates. Multi-drug resistance (MDR) was observed among the isolates, with the highest rates observed in chicken isolates (88.0%). Proportions of MDR among pig (40.9%) and human (40.7%) isolates were more or less similar. These findings highlight the presence of thermophilic Campylobacter strains in humans and livestock which are resistant to commonly used antimicrobials. Based on the potential of interspecies transmission, it is recommended to adopt a One Health approach to curb antimicrobial resistance. Further genomic analysis will shed more light on the transmission and drug resistance patterns.
Introduction
Campylobacter spp. are major foodborne pathogens responsible for zoonotic gastroenteritis worldwide (Scallan et al., 2011) with C. jejuni and C. coli accounting for 95% of human campylobacteriosis cases (Badjo et al., 2024; Acheson and Allos, 2001). Different reservoirs have been associated with campylobacteriosis but poultry has been linked to around 70% of human campylobacteriosis (Epps et al., 2013; Boes et al., 2005). Pigs are considered the primary host for C. coli which is the second most prevalent species linked to human campylobacteriosis after C. jejuni (Wagenaar et al., 2013; Georges-Courbot et al., 1986). Overall, Clinical symptoms of Campylobacter infections include abdominal pain, diarrhea, vomiting, chills, and fever (Same and Tamma, 2018). The transmission of Campylobacter to humans occurs through the consumption of raw or undercooked meat, especially poultry, contaminated milk and water, and direct contact with farm animals (Khairullah et al., 2024; Akase et al., 2024; Igwaran and Okoh, 2020).
The escalating incidence of campylobacteriosis, particularly in low- and middle-income countries (LMICs), presents a significant public health challenge. Africa is estimated to have the world's highest incidence of campylobacteriosis (French et al., 2024). The latter is hyper-endemic in sub-Saharan Africa (SSA) due to factors such as poor sanitation, limited access to clean water, and close contact with animals in domestic and agricultural settings (Olum et al., 2023). Human campylobacteriosis often presents symptoms similar to other gastrointestinal infections, which explains its frequent exclusion from the diagnostic and reporting processes (Paintsil et al., 2023; Gahamanyi et al., 2020a; Asuming-Bediako et al., 2019). Despite its significant occurrence in the food chain and strong association with diarrheal cases in children (Coker et al., 2002; Gölz et al., 2018), campylobacteriosis has generally received modest attention.
Indiscriminate use of antibiotics in human and veterinary medicine has led to an increased resistance of Campylobacter strains to the drugs of choice. Indeed, both macrolides (erythromycin or azythromycin) and fluoroquinolone (ciprofloxacin) were used as drugs of choice (Wieczorek and Osek, 2013; García-Fernández et al., 2018), but increasing resistance of Campylobacter to ciprofloxacin has led to the adoption of erythromycin as the best choice (Dai et al., 2020). Alternative treatment include aminoglycosides and tetracyclines (Koolman et al., 2015; Reddy and Zishiri, 2017). Furthermore, the World Health Organization (WHO) has classified fluoroquinolone-resistant Campylobacter as a priority pathogen requiring the development of new antibiotics (Gahamanyi et al., 2020b). We previously showed an increased resistance to ciprofloxacin and erythromycin in Ghana and Tanzania among Campylobacter of human origin (Gahamanyi et al., 2020a) while Kenya recorded an increased resistance against tetracycline and ciprofloxacin in Campylobacter from chicken (Asuming-Bediako et al., 2019). Also, a study reported a high prevalence of multidrug-resistant (MDR) Campylobacter isolates from humans and poultry in Tanzania and Kenya (French et al., 2024). Thus, antimicrobial-resistant Campylobacter strains are becoming more prevalent in the East African region (Gahamanyi et al., 2020a; Asuming-Bediako et al., 2019; Zachariah et al., 2021).
In Rwanda, Campylobacter species were identified in human stool (Kabayiza et al., 2014) and environmental samples (Ssemanda et al., 2018), but the prevalence in livestock is not fully documented. Moreover, studies on antimicrobial susceptibility profiles of Campylobacter isolates are scarce, possibly due to inherent difficulties associated with its culture (Mileng et al., 2021) or the fact that it is not on the list of commonly suspected bacteria in clinical settings. Therefore, the objective of the study was to determine the prevalence and antimicrobial susceptibility profiles of thermophilic Campylobacter species from humans, chicken, and pigs in Rwanda. The ultimate goal is to generate data that will be used to design a One Health control strategy for campylobacteriosis and associated antimicrobial resistance.
Materials and methods
Study area
The study was conducted in three (03) different districts of Rwanda, namely Nyarugenge (Kigali city), Huye, and Gisagara Districts (Southern Province). Clinical samples were collected from Kigali University Teaching Hospital (CHUK), Nyarugenge District Hospital, Muhima and Biryogo Health Centers, while pig and poultry samples were collected from the largest farms located in the Southern Province (Huye and Gisagara Districts).
Study design
This study adopted a cross-sectional design and was conducted from March 2024 to August 2024. The study used a purposive sampling strategy. The human aspect targeted individuals presenting at the hospital with abdominal pain and/or diarrhea.
Sample size determination
The sample size was determined using Cochran's Formula as follows:
Formula: n . Where n is the sample size; Z2 is the Z score (Z = 1.96); p is the expected prevalence; and e2 is the desired level of precision or margin of error. Since the confidence interval is 95%, the error margin is 0.05 or 5%. For humans, p = 0.5 used when the true prevalence is unknown was chosen and the sample size being n = 385. For pigs, the sample size of 337 was calculated based on previous prevalence (p = 0.325) (Kashoma et al., 2015). For chicken, the sample size was 376 based on previous prevalence (p = 0.425) (Chuma et al., 2016).
Sample collection and Campylobacter isolation
Human stool samples (n = 385) were collected from CHUK (n = 21), Nyarugenge DH (n = 63), Muhima HC (n = 58), and Biryogo HC (n = 243). Pig feces (n = 337) were collected from Tumba (n = 56), Ngoma (n = 56), Maraba (n = 56), Kigoma (n = 56), and Ruhashya (n = 56) sectors of Huye and Save sector of Gisagara district (n = 57). Chicken feces (n = 376) were collected from poultry farms located in Ruhashya (n = 130) and from one farm in Ngoma (n = 246) of Huye district. In all sites, the collected samples were placed into stool collection containers labeled with the site and geographical locations. Approximately five (05) grams of fecal samples were collected and transported in a cooler box to the Microbiology laboratory of the University of Rwanda, College of Science and Technology, for further processing.
Upon reaching the laboratory, human stool samples were inoculated into the Nutrient Broth No.2 (CM0067, Oxoid, Basingstoke, UK) supplemented with Preston Campylobacter selective Supplements SR0204 and SR0232E (Oxoid Ltd, Basingstoke, Hampshire, England), and 5% defibrinated Sheep Blood (SR0051B; Oxoid Ltd, Basingstoke, Hampshire, England). Tubes were incubated at 42 °C for 24 h under microaerophilic conditions generated with CampyGen™ sachets CN0025A (Oxoid Ltd, Basingstoke, Hampshire, England) before being subcultured onto modified Charcoal Cefoperazone Deoxycholate Agar (mCCDA; Oxoid Ltd, Basingstoke, Hampshire, England) as previously described (Srijan et al., 2013). Pig and chicken feces were immediately inoculated onto mCCDA supplemented with SR0155E (Oxoid Ltd, Basingstoke, Hampshire, England). Plates were incubated at 37 °C for 48 h (Guévremont et al., 2006). Presumptive colonies of Campylobacter (moistened, gray, flat, and tendency to spread) were sub-cultured onto blood agar supplemented with 5% (v/v) of defibrinated sheep blood and incubated at 37 °C for 48 h under microaerophilic conditions as previously described (Guévremont et al., 2006). Obtained pure colonies were further characterized with Gram staining, cellular morphology, catalase, and oxidase tests (Modi et al., 2015). Campylobacter isolates were preserved in Mueller Hinton Broth (MHB) supplemented with 25% glycerol and kept at −80 °C until further processing as previously described (Yildiz et al., 2024).
DNA extraction and Polymerase chain reaction (PCR)
Genomic DNA was extracted from pure colonies using the Quick-DNA TM Fecal/Soil Microbe Miniprep Kit (ZYMO Research, USA) according to the manufacturer's instructions. Extracted DNA was quantified using a quantusTM fluorometer while the quality of the extracted DNA was assessed with a nanodrop (EzDrop 1000 micro-volume spectrophotometer). Extracted DNA samples were of good quality and quantity for further processing. DNA samples were kept at −20 °C before PCR was performed.
After DNA extraction, a multiplex-PCR using genus-specific primers (C412F; C1228R), C. jejuni and C. coli primers was carried out as previously described (Yamazaki-Matsune et al., 2007). The choice of primers was made with a focus on their ability to specifically identify the genus and species of Campylobacter (Linton et al., 1997; Pajaniappan et al., 2008). The PCR mixture was 25 μl, and it comprised 12.5 μl of 2X Master Mix (New England Biolabs), 1 μl of each primer (10 μM), 1.5 μl of template DNA (20 μg/ml), and 9 μl of sterile deionized water. UltraPure™ DNase/RNase-Free Distilled Water (Invitrogen) was used as negative control for each PCR run. The amplification conditions involved one cycle at 95 °C for 5 min for denaturation, followed by 35 cycles, each consisting of 94 °C for 30 s, 60 °C for 45 s for annealing, and 72 °C for 45 s, with a final extension at 72 °C for 7 min (Gahamanyi et al., 2021). PCR products were kept at 4 °C before analyzed by gel electrophoresis. The bands of the amplification products were compared to the NEB 100 bp DNA ladder (New England Biolabs). Gel electrophoresis was performed for band visualization of PCR products. A 2% (v/v) agarose gel stained with gel red suspended in 1X TAE buffer was utilized. Samples were loaded into wells using 6X blue-orange loading dye, and bands of PCR products were observed with a UV transilluminator at 161 bp, 502 bp, 816 bp for C. jejuni, C. coli, and Campylobacter genus, respectively. For quality control, C. jejuni strain (ATCC® 33560™) and C. coli strains (ATCC® 33559™) were used.
Antimicrobial susceptibility testing
Antimicrobial susceptibility testing (AST) was conducted using the disc diffusion method on Muller Hinton Agar (MHA; Oxoid Ltd., Basingstoke, UK) in accordance with the guidelines set by the Clinical Laboratory Standards Institute (CLSI) (2024) and the European Committee for Antimicrobial Susceptibility Testing (EUCAST) for Campylobacter isolates from human samples and livestock1, respectively. EUCAST lacks Gentamicin and Chloramphenicol guidelines for C. jejuni and C. coli and therefore, Enterobacteriales breakpoints were used. For livestock, since Streptomycin was missing from the EUCAST guidelines, CLSI-Enterobacterales guidelines were used. Various antibiotics, including Ciprofloxacin (CIP, 5 μg), Erythromycin (ERY, 15 μg), Tetracycline (TET, 30 μg), Streptomycin (STR, 10 μg), Gentamicin (GEN, 10 μg), and Chloramphenicol (CHL, 30 μg) were used to assess the AST profiles of isolated Campylobacter. However, due to limited stock, TET was not used for Campylobacter isolates from chicken.
Briefly, preserved Campylobacter isolates were revived onto Blood Agar and sub-cultured onto the same media to obtain pure colonies free from glycerol. Obtained pure colonies were suspended in sterile normal saline, and turbidity was adjusted to 0.5 McFarland standard. Using sterile glass spreaders, bacterial suspension was spread over MHA supplemented with 5% defibrinated sheep blood and 20 mg/L β-NAD. Using forceps, antibiotic discs were evenly distributed over the plates ensuring a spacing of at least 24 mm. Plates were then incubated at 42 °C for 24 h in a microaerophilic environment as previously described (Strakova et al., 2024). After incubation, the inhibition zones were measured using a ruler. Resistance to three or more classes of antibiotics was referred to as multidrug resistance (MDR) as previously described (Ocejo et al., 2019).
Data analysis
Microsoft Excel was used for data entry and cleaning while SPSS version 27 was used for statistical analysis. Descriptive statistics including proportions of antibiotic resistance, and attribute frequencies, were computed. Antimicrobial susceptibility testing was done based on cultural characteristics. Samples harboring two species of Campylobacter (C. jejuni and C. coli) were removed from the final AST analysis. To assess the association between source of sample and Campylobacter species, a chi-square test of independence was performed. This test was performed at a 95% confidence level with a significance threshold set at α = 0.05.
Results
Prevalence of Campylobacter in different compartments
The prevalence of Campylobacter varied across different sampling sites and sample types. In humans, the overall positivity rate was 7.0%, with Biryogo HC recording the highest prevalence at 8.6%, followed by Nyarugenge DH (6.3%) and Muhima H.C. (3.4%). No positive cases were detected at CHUK. In pigs, the prevalence was 7.1%, and Save exhibited the highest positivity rate of 21.1%, followed by Kigoma and Ruhashya (8.9%) and Maraba (3.6%). No positive cases were detected in samples from Tumba and Ngoma. Chicken samples had the highest prevalence of Campylobacter at 32.0%, with positivity rates of 34.9% in Ngoma and 26.9% in Ruhashya (Table 1).
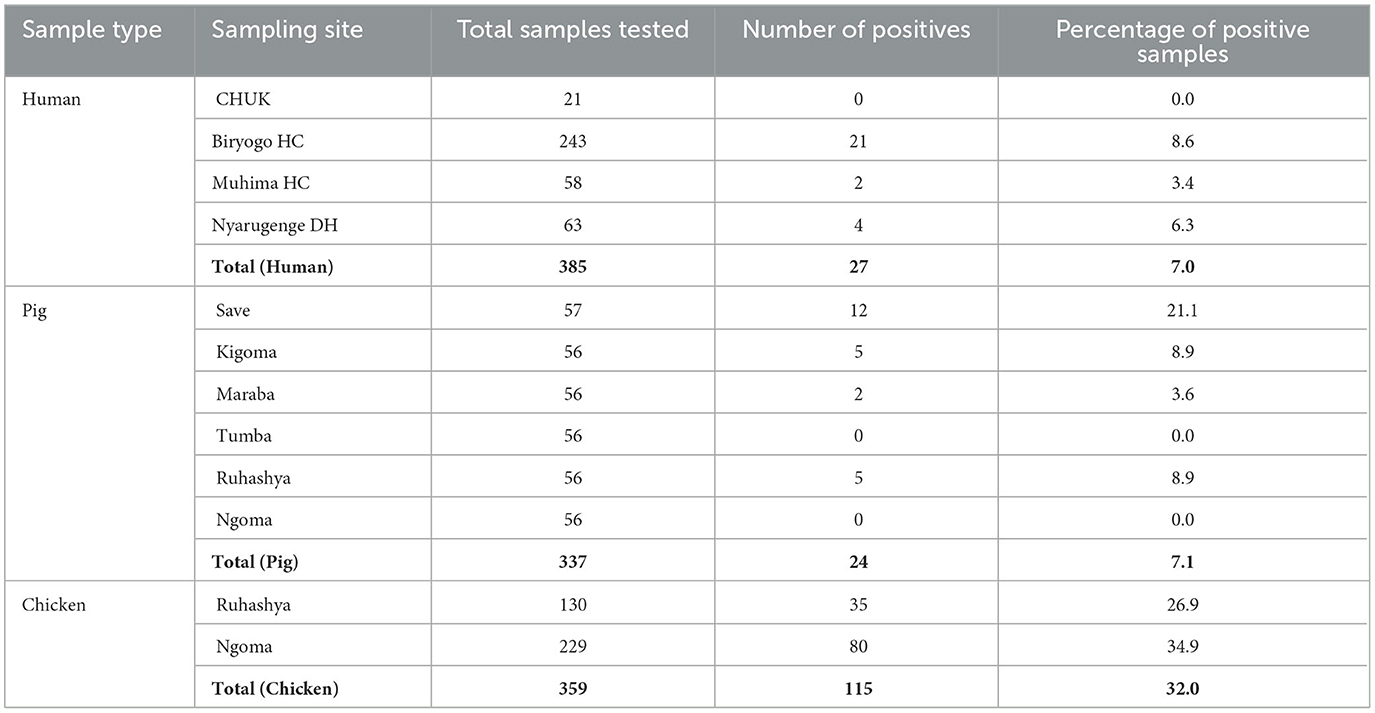
Table 1. Distribution of samples, and isolation frequencies of Campylobacter from humans, pigs, and chicken in Rwanda (2024).
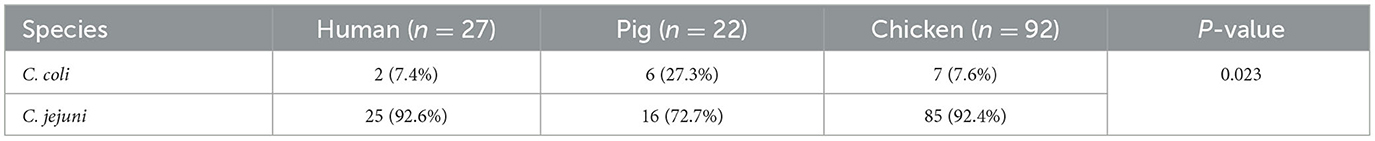
Campylobacter isolates were identified as C. jejuni and C. coli by the polymerase chain reaction. Findings showed that the predominant species was C. jejuni with detection rates varying between 66.7 and 92.6%, while C. coli was detected at rates varying between 7.4 and 27.3% (Table 2). The chi-square test of independence revealed a significant association between sample source and Campylobacter species (p-value of 0.023). We also identified both species (C. jejuni and C. coli) in two isolates (8.3%) from pigs and 23 (20.0%) from chicken.
Antimicrobial susceptibility profiles of Campylobacter species from human, pigs, and chicken
Antimicrobial susceptibility testing (AST) was subsequently performed on isolates from the two species (C. jejuni and C. coli) identified by PCR. Campylobacter isolates from all hosts showed higher resistance to ERY and STR. In chicken, C. coli demonstrated resistance rates of 100, 85.7, and 71.4% to CIP, ERY, and STR, respectively. Among pig isolates, C. coli exhibited high resistance to ERY (83.3%) and CIP (66.7%), while lower resistance rates were observed for gentamicin (GEN, 33.3%) and chloramphenicol (CHL, 16.7%). In human isolates, high resistance was noted against STR (100%) and ERY (50%), whereas no resistance was detected for GEN, CIP, CHL, and tetracycline (TET; Table 3).
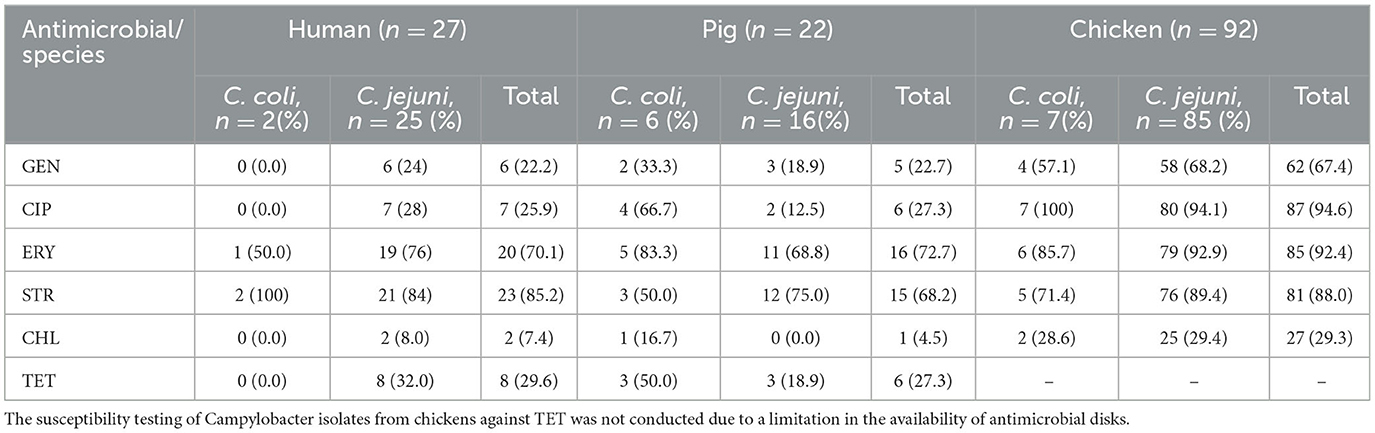
Table 3. Antimicrobial resistance profiles of identified C. jejuni and C. coli species from the three hosts.
For C. jejuni, high levels of resistance were observed in chicken isolates, with rates of 94.1, 92.9, and 89.4% for CIP, ERY, and STR, respectively. In pig isolates, notable resistance was recorded for STR (75%) and ERY (68.8%). Among human isolates, resistance was high for ERY (76%) and STR (84%; Table 3).
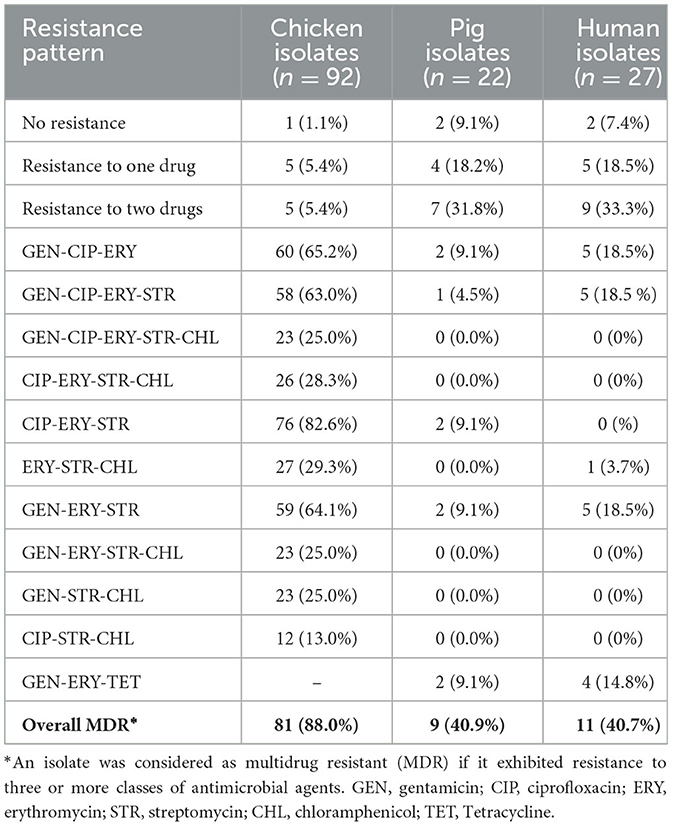
Of the 92 Campylobacter isolates from chicken, 91 (98.9%) were resistant to at least one antimicrobial agent. Similarly, 25 (92.6%) out of 27 Campylobacter isolates from humans and 20 (90.9%) out of 22 Campylobacter isolates from pigs exhibited resistance to at least one antimicrobial agent (Table 4).
The MDR profiles of Campylobacter isolates against tested antimicrobials were also evaluated. Overall, MDR was observed in 88.0% (n = 81), 40.9% (n = 9), and 40.7% (n = 11) isolates from chicken, pigs, and humans, respectively. In chicken, the highest MDR rate (82.6%) was observed against CIP-ERY-STR. In pigs, the highest MDR rate of 9.1% was observed against GEN-CIP-ERY, CIP-ERY-STR, GEN ERY-STR, and GEN-ERY-TET. For human isolates, the highest MDR rate (18.5%) was observed against GEN-CIP-ERY-STR, GEN-CIP-ERY, and GEN-ERY-STR (Table 4).
Discussion
The current study observed varying frequencies of isolation of thermophilic Campylobacter from human, pig, and chicken samples. The highest prevalence of Campylobacter was observed in chicken (32%), while the prevalence of the bacterium in humans (7%) and pigs (7.1%) was similar. The findings of this study suggest that the poultry-human transmission route could be more important than the pig-human transmission. Furthermore, this is in line with findings from our systematic review indicating that chicken had the highest prevalence of Campylobacter among known reservoirs in Africa (Gahamanyi et al., 2020a). Indeed, chicken are known to be the primary reservoir for Campylobacter (de Zoete et al., 2007) due to their high body temperature (42 °C) favorable for the bacteria's growth (Sibanda et al., 2018). A previous human study in Rwanda reported a higher Campylobacter prevalence of 15.5% (Kabayiza et al., 2014). The discrepancy may be a result of the diagnostic methods used (PCR vs. culture) and the age of patients (infants and younger children vs. adults). The PCR is more sensitive than culture and children are more at risk of Campylobacter of infections especially in LMICs. Nevertheless, our findings are comparable to findings reported in Ethiopia (Worku et al., 2024) but lower than the one reported in Tanzania (Komba et al., 2015). Contrarily, the prevalence recorded in this study was higher than the one reported in Burkina Faso (Sangaré et al., 2012). Since humans acquire Campylobacter through food/drink contamination from reservoir animals (poultry, pigs, cattle), the observed country-level differences may be due to the varying food chains, feeding habits, and differing sanitation levels. For pigs, the prevalence of Campylobacter obtained in this study was slightly higher than 2.3% reported in South Africa (Jonker and Picard, 2010), but significantly lower than 32.5% reported in Tanzania (Kashoma et al., 2015). These disparities likely reflect differences in production systems, dietary practices, and standards of hygiene. For instance, the Rwandan pig food chain is less developed compared to the one in Tanzania.
Campylobacter jejuni was the predominant species in all three hosts with 92.6, 72.7, and 66.7% in humans, chicken, and pigs, respectively. The prevalence of C. coli was higher in pigs (27.3%), while co-occurrence of both species was recorded in 20 and 8.3% of chicken, and pig, respectively. Generally, human campylobacteriosis is associated with C. jejuni at 80–85%, while the percentage of infection due to C. coli is between 10 and 15% (Bullman et al., 2012). Worldwide, C. jejuni is more common in poultry, while C. coli is predominant in pigs (Harvey et al., 1999). The increased prevalence of C. jejuni can be linked to the fact that it survives better at low temperatures than C. coli (Ortiz et al., 2024). It is important to note that C. jejuni is equipped with virulence genes and is associated with higher pathogenesis compared to C. coli (Thakur et al., 2010). It is also not surprising to obtain both C. jejuni and C. coli as previously described (Liao et al., 2022).
The current study assessed the antimicrobial susceptibility profiles of Campylobacter isolates. In general, over 90% of Campylobacter isolates from all three (03) hosts exhibited resistance to at least one antimicrobial agent. Higher resistance rates ranging from 70.8 to 85.2% against STR and 74.1 to 83.5% against ERY were observed in this study. The resistance of Campylobacter isolates from chicken to CIP was 93.9%. Both CIP and ERY are drugs of choice for the treatment of human campylobacteriosis (Harvey et al., 1999), but raising resistance levels call for alternative treatment strategies (Dai et al., 2020). Erythromycin and azithromycin belong to the macrolide class which have been used as first line drugs for treating camplyobacteriosis (García-Fernández et al., 2018) but azithromycin showed lower MIC values than those of erythromycin against Gram-negative bacteria including Campylobacter (Retsema et al., 1987; Wei and Kang, 2018). The WHO has classified fluoroquinolone-resistant Campylobacter strains among the pathogens requiring the development of new antibiotics (Zainol et al., 2024). The resistance to STR was higher than 50% reported in Ethiopia (Hagos et al., 2021). Increased resistance to STR has been associated with mutations in the rpsL gene coding for a ribosomal protein (RpsL) or the expression of aminoglycoside-modifying enzyme [ANT(6)-I; Retsema et al., 1987; Wei and Kang, 2018; Dahl et al., 2021; Hormeño et al., 2018]. Resistance of Campylobacter isolates from humans and pigs to CIP (25.9–27.0%) was higher than 16% reported in Sub-Saharan Africa (Hlashwayo et al., 2021) and 22.1% reported in Tanzania (Komba et al., 2015). Resistance to CIP has been associated with its misuse in treating diarrhea of unknown etiology and the persistence of its resistance in the community once acquired (Sproston et al., 2018). Resistance to GEN was lower (22.2–25%) among human and pig-derived isolates but higher in chicken isolates (66.9%). Resistance to GEN has been relatively low because it is limited to treating systemic infections (Lynch et al., 2020). Resistance to CHL was relatively low, ranging from 7.4 to 26.9% across the isolates. This suggests that CHL remains a viable option for the treatment of Campylobacter infections. Similarly, GEN also exhibited relatively low resistance rates, making it another potential alternative for managing human campylobacteriosis. The use of these antimicrobials could provide effective options, particularly in cases where resistance to first-line treatments such as CIP and ERY is high.
The AMR profiles of C. jejuni and C. coli isolates were compared. For ERY, C. jejuni exhibited higher resistance than C. coli, except among isolates from pigs. Conversely, resistance to CIP and STR was higher in C. coli than in C. jejuni, except for isolates from human stool. Both C. coli and C. jejuni showed lower resistance to CHL. Resistance to GEN was generally below 50%, except for isolates from chickens. These findings suggest that CHL and GEN could serve as alternative antimicrobials for treating human campylobacteriosis. However, their prudent use is essential to mitigate the risk of increasing resistance.
The present study detected MDR Campylobacter isolates with the frequencies of MDR being higher among isolates from chicken. The MDR rates in isolates from pigs and humans were comparable. The frequently occurring MDR combinations were GEN-CIP-ERY and GEN-ERY-STR. Notably, 25% of isolates from chickens, 4.5% from pigs, and 14.8% from humans were resistant to five of the tested antimicrobials. A previous study revealed higher resistance of Campylobacter isolates to a combination of fluoroquinolone, tetracycline, and macrolide (Tang et al., 2020). A different study on chicken reported higher resistance to quinolones, tetracycline, and sulfamethoxazole-trimethoprim (Giacomelli et al., 2014). These findings underscore the growing concern over MDR in Campylobacter strains across different hosts, highlighting the need for strengthening antimicrobial stewardship programs and the development of alternative treatment options, including phage therapy and the use of natural products.
This study faced a number of limitations. First, the cross-sectional approach could not capture potential changes in prevalence over different seasons or the influence of hygiene and other interventions. Second, the Kirby-Bauer disk diffusion method used is qualitative in nature and could not determine the level of AMR against tested antimicrobials. Last, different protocols were used for Campylobacter isolation in human vs. pig and chicken feces. For human feces, enrichment with Preston broth was used while for pig and chicken feces, direct plating onto mCCDA was preferred. This was due to the low intensity of Campylobacter in humans compared to animals. Despite the highlighted shortcomings, this study is the first in Rwanda to isolate Campylobacter from animal feces by culture and to report antimicrobial profiles. The findings highlight a significant public health threat that should be tackled in a One Health approach.
Conclusion
The current study highlights higher prevalence of Campylobacter in chicken feces when compared to pig and human feces. Campylobacter jejuni was the predominant species across the three hosts, but C. coli was also identified in a considerable number of fecal samples, mainly from pigs and chicken. An increased resistance of Campylobacter isolates to different antimicrobials, including drugs of choice (ERY and CIP) was observed. The study showed that GEN and CHL could be used as alternative treatment options. Further research and development toward non-antibiotic control measures are highly recommended. Genomic studies will provide insights of transmission patterns and specific resistance genes.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding authors.
Ethics statement
This study was approved by Institutional Review Board (IRB) of the College of Medicine and Health Sciences (CMHS) at the University of Rwanda with reference number 244/CMHS IRB/2023. The study was conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation in this study was provided by the participants or the legal guardian for children and farm owners.
Author contributions
NG: Conceptualization, Data curation, Formal analysis, Funding acquisition, Methodology, Supervision, Validation, Writing – original draft, Writing – review & editing. AH: Formal analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing. JH: Investigation, Methodology, Project administration, Validation, Writing – original draft. JM: Conceptualization, Funding acquisition, Methodology, Project administration, Validation, Writing – original draft. SN: Data curation, Formal analysis, Investigation, Methodology, Software, Validation, Writing – original draft. AU: Methodology, Project administration, Resources, Supervision, Writing – review & editing. EN: Conceptualization, Formal analysis, Methodology, Supervision, Validation, Writing – review & editing. EI: Conceptualization, Funding acquisition, Investigation, Methodology, Supervision, Validation, Writing – review & editing. JB: Investigation, Project administration, Supervision, Writing – original draft. C-HP: Conceptualization, Funding acquisition, Supervision, Writing – review & editing. EK: Conceptualization, Funding acquisition, Supervision, Writing – review & editing. CM: Conceptualization, Project administration, Supervision, Writing – review & editing. NR: Conceptualization, Funding acquisition, Methodology, Supervision, Writing – review & editing. RA: Conceptualization, Funding acquisition, Methodology, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was funded by the PASET Regional Scholarship and Innovation Grant under its Junior Investigator Research Award (JIRA) with grant number RSIF/JIRA/002.
Acknowledgments
Authors acknowledge the support from laboratory staff and farmers during sample collection. We also thank patients who consented to be part of this study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Gen AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
References
Acheson, D., and Allos, B. M. (2001). Campylobacter jejuni infections: update on emerging issues and trends. Clin. Infect. Dis. 32, 1201–1206. doi: 10.1086/319760
Akase, S., Obata, H., Okada, W., Saiki, D., Konishi, N., Yokoyama, K., et al. (2024). A case of food poisoning caused by Campylobacter jejuni after the ingestion of undercooked chicken meal with subsequent development of Guillain–Barré syndrome. Jpn. J. Infect. Dis. 77, 353–355. doi: 10.7883/yoken.JJID.2024.108
Asuming-Bediako, N., Parry-Hanson Kunadu, A., Abraham, S., and Habib, I. (2019). Campylobacter at the human–food interface: the African perspective. Pathogens 8:87. doi: 10.3390/pathogens8020087
Badjo, A. O. R., Kabore, N. F., Zongo, A., Gnada, K., Ouattara, A., Muhigwa, M., et al. (2024). Burden and epidemiology of Campylobacter species in acute enteritis cases in Burkina Faso. BMC Infect. Dis. 24:808. doi: 10.1186/s12879-024-09709-y
Boes, J., Nersting, L., Nielsen, E. M., Kranker, S., Enøe, C., Wachmann, H. C., et al. (2005). Prevalence and diversity of Campylobacter jejuni in pig herds on farms with and without cattle or poultry. J. Food Prot. 68, 722–727. doi: 10.4315/0362-028X-68.4.722
Bullman, S., O'leary, J., Corcoran, D., Sleator, R. D., and Lucey, B. (2012). Molecular-based detection of non-culturable and emerging campylobacteria in patients presenting with gastroenteritis. Epidemiol. Infect. 140, 684–688. doi: 10.1017/S0950268811000859
Chuma, I. S., Nonga, H. E., Mdegela, R. H., and Kazwala, R. R. (2016). Epidemiology and RAPD-PCR typing of thermophilic campylobacters from children under five years and chickens in Morogoro Municipality, Tanzania. BMC Infect. Dis. 16, 1–11. doi: 10.1186/s12879-016-2031-z
Clinical and Laboratory Standards Institute (CLSI), Z. Z. (2024). M100 Performance Standards for Antimicrobial Susceptibility Testing, 34th Edn. Wayne, PA: Clinical and Laboratory Standards Institute. Available at: https://www.sid.ir/en/Journal/ViewPaper.aspx?ID=450165 (Accessed December 18, 2024).
Coker, A. O., Isokpehi, R. D., Thomas, B. N., Amisu, K. O., and Obi, C. L. (2002). Human campylobacteriosis in developing countries. Emerg. Infect. Dis. 8, 237–243. doi: 10.3201/eid0803.010233
Dahl, L. G., Joensen, K. G., Østerlund, M. T., Kiil, K., and Nielsen, E. M. (2021). Prediction of antimicrobial resistance in clinical Campylobacter jejuni isolates from whole-genome sequencing data. Eur. J. Clin. Microbiol. Infect. Dis. 40, 673–682. doi: 10.1007/s10096-020-04043-y
Dai, L., Sahin, O., Grover, M., and Zhang, Q. (2020). New and alternative strategies for the prevention, control, and treatment of antibiotic-resistant Campylobacter. Transl. Res. 223, 76–88. doi: 10.1016/j.trsl.2020.04.009
de Zoete, M. R., van Putten, J. P. M., and Wagenaar, J. A. (2007). Vaccination of chickens against Campylobacter. Vaccine 25, 5548–5557. doi: 10.1016/j.vaccine.2006.12.002
Epps, S. V., Harvey, R. B., Hume, M. E., Phillips, T. D., Anderson, R. C., and Nisbet, D. J. (2013). Foodborne Campylobacter: infections, metabolism, pathogenesis and reservoirs. Int. J. Environ. Res. Public. Health 10, 6292–6304. doi: 10.3390/ijerph10126292
French, N. P., Thomas, K. M., Amani, N. B., Benschop, J., Bigogo, G. M., Cleaveland, S., et al. (2024). Population structure and antimicrobial resistance in Campylobacter jejuni and C. coli isolated from humans with diarrhea and from poultry, East Africa. Emerg. Infect. Dis. 30, 2079–2089. doi: 10.3201/eid3010.231399
Gahamanyi, N., Mboera, L. E. G., Matee, M. I., Mutangana, D., and Komba, E. V. G. (2020a). Prevalence, risk factors, and antimicrobial resistance profiles of thermophilic Campylobacter species in humans and animals in Sub-Saharan Africa: a systematic review. Int. J. Microbiol. 2020:2092478. doi: 10.1155/2020/2092478
Gahamanyi, N., Song, D. G., Cha, K. H., Yoon, K. Y., Mboera, L. E. G., Matee, M. I., et al. (2020b). Susceptibility of campylobacter strains to selected natural products and frontline antibiotics. Antibiotics 9:790. doi: 10.3390/antibiotics9110790
Gahamanyi, N., Song, D. G., Yoon, K. Y., Mboera, L. E. G., Matee, M. I., Mutangana, D., et al. (2021). Genomic characterization of fluoroquinolone-resistant thermophilic campylobacter strains isolated from layer chicken feces in Gangneung, South Korea by whole-genome sequencing. Genes 12:1131. doi: 10.3390/genes12081131
García-Fernández, A., Dionisi, A. M., Arena, S., Iglesias-Torrens, Y., Carattoli, A., and Luzzi, I. (2018). Human Campylobacteriosis in Italy: emergence of multi-drug resistance to ciprofloxacin, tetracycline, and erythromycin. Front. Microbiol. 9:1906. doi: 10.3389/fmicb.2018.01906
Georges-Courbot, M. C., Baya, C., Beraud, A. M., Meunier, D. M., and Georges, A. J. (1986). Distribution and serotypes of Campylobacter jejuni and Campylobacter coli in enteric Campylobacter strains isolated from children in the Central African Republic. J. Clin. Microbiol. 23, 592–594. doi: 10.1128/jcm.23.3.592-594.1986
Giacomelli, M., Salata, C., Martini, M., Montesissa, C., and Piccirillo, A. (2014). Antimicrobial Resistance of Campylobacter jejuni and Campylobacter coli from poultry in Italy. Microb. Drug Resist. 20, 181–188. doi: 10.1089/mdr.2013.0110
Gölz, G., Kittler, S., Malakauskas, M., and Alter, T. (2018). Survival of Campylobacter in the food chain and the environment. Curr. Clin. Microbiol. Rep. 5, 126–134. doi: 10.1007/s40588-018-0092-z
Guévremont, E., Nadeau, É., Sirois, M., and Quessy, S. (2006). Antimicrobial susceptibilities of thermophilic Campylobacter from humans, swine, and chicken broilers. Can. J. Vet. Res. 70, 81–86.
Hagos, Y., Gugsa, G., Awol, N., Ahmed, M., Tsegaye, Y., Abebe, N., et al. (2021). Isolation, identification, and antimicrobial susceptibility pattern of Campylobacter jejuni and Campylobacter coli from cattle, goat, and chicken meats in Mekelle, Ethiopia. PLoS ONE 16:e0246755. doi: 10.1371/journal.pone.0246755
Harvey, R. B., Young, C. R., Ziprin, R. L., Hume, M. E., Genovese, K. J., Anderson, R. C., et al. (1999). Prevalence of Campylobacter spp isolated from the intestinal tract of pigs raised in an integrated swine production system. J. Am. Vet. Med. Assoc. 215, 1601–1604. doi: 10.2460/javma.1999.215.11.1601
Hlashwayo, D. F., Sigaúque, B., Noormahomed, E. V., Afonso, S. M. S., Mandomando, I. M., and Bila, C. G. (2021). A systematic review and meta-analysis reveal that Campylobacter spp. and antibiotic resistance are widespread in humans in sub-Saharan Africa. PLOS ONE 16:e0245951. doi: 10.1371/journal.pone.0245951
Hormeño, L., Ugarte-Ruiz, M., Palomo, G., Borge, C., Florez-Cuadrado, D., Vadillo, S., et al. (2018). ant(6)-I genes encoding aminoglycoside O-nucleotidyltransferases are widely spread among streptomycin resistant strains of Campylobacter jejuni and Campylobacter coli. Front. Microbiol. 9:2515. doi: 10.3389/fmicb.2018.02515
Igwaran, A., and Okoh, A. I. (2020). Occurrence, virulence and antimicrobial resistance-associated markers in Campylobacter species isolated from retail fresh milk and water samples in two district municipalities in the Eastern Cape Province, South Africa. Antibiot. Basel 9:426. doi: 10.3390/antibiotics9070426
Jonker, A., and Picard, J. A. (2010). Antimicrobial susceptibility in thermophilic Campylobacter species isolated from pigs and chickens in South Africa. J. S. Afr. Vet. Assoc. 81, 228–236. doi: 10.4102/jsava.v81i4.153
Kabayiza, J. C., Andersson, M. E., Nilsson, S., Bergström, T., Muhirwa, G., and Lindh, M. (2014). Real-time PCR identification of agents causing diarrhea in rwandan children less than 5 years of age. Pediatr. Infect. Dis. J. 33, 1037–1042. doi: 10.1097/INF.0000000000000448
Kashoma, I. P., Kassem, I. I., Kumar, A., Kessy, B. M., Gebreyes, W., Kazwala, R. R., et al. (2015). Antimicrobial resistance and genotypic diversity of Campylobacter isolated from pigs, dairy, and beef cattle in Tanzania. Front. Microbiol. 6:1240. doi: 10.3389/fmicb.2015.01240
Khairullah, A. R., Yanestria, S. M., Effendi, M. H., Moses, I. B., Jati Kusala, M. K., Fauzia, K. A., et al. (2024). Campylobacteriosis: a rising threat in foodborne illnesses. Open Vet. J. 14, 1733–1750. doi: 10.5455/OVJ.2024.v14.i8.1
Komba, E. V. G., Mdegela, R. H., Msoffe, P. L. M., Nielsen, L. N., and Ingmer, H. (2015). Prevalence, antimicrobial resistance and risk factors for thermophilic Campylobacter infections in symptomatic and asymptomatic humans in Tanzania. Zoonoses Public Health 62, 557–568. doi: 10.1111/zph.12185
Koolman, L., Whyte, P., Burgess, C., and Bolton, D. (2015). Distribution of virulence-associated genes in a selection of Campylobacter isolates. Foodborne Pathog. Dis. 12, 424–432. doi: 10.1089/fpd.2014.1883
Liao, Y. S., Chen, B. H., Teng, R. H., Wang, Y. W., Chang, J. H., Liang, S. Y., et al. (2022). Antimicrobial resistance in Campylobacter coli and Campylobacter jejuni from human campylobacteriosis in Taiwan, 2016 to 2019. Antimicrob. Agents Chemother. 66, e01736–e01721. doi: 10.1128/AAC.01736-21
Linton, D., Lawson, A. J., Owen, R. J., and Stanley, J. (1997). PCR detection, identification to species level, and fingerprinting of Campylobacter jejuni and Campylobacter coli direct from diarrheic samples. J. Clin. Microbiol. 35, 2568–2572. doi: 10.1128/jcm.35.10.2568-2572.1997
Lynch, C. T., Lynch, H., Burke, S., Hawkins, K., Buttimer, C., Mc Carthy, C., et al. (2020). Antimicrobial resistance determinants circulating among thermophilic Campylobacter isolates recovered from broilers in Ireland over a one-year period. Antibiotics 9:308. doi: 10.3390/antibiotics9060308
Mileng, K., Ramatla, T. A., Ndou, R. V., Thekisoe, O. M. M., and Syakalima, M. (2021). Isolation and antibiotic sensitivity of Campylobacter species from fecal samples of broiler chickens in North West Province, South Africa. Vet. World 14, 2929–2935. doi: 10.14202/vetworld.2021.2929-2935
Modi, S., Brahmbhatt, M. N., Chatur, Y. A., and Nayak, J. B. (2015). Prevalence of Campylobacter species in milk and milk products, their virulence gene profile and anti-bio gram. Vet. World 8, 1–8. doi: 10.14202/vetworld.2015.1-8
Ocejo, M., Oporto, B., and Hurtado, A. (2019). Occurrence of Campylobacter jejuni and Campylobacter coli in cattle and sheep in Northern Spain and changes in antimicrobial resistance in two studies 10-years apart. Pathogens 8:98. doi: 10.3390/pathogens8030098
Olum, M. O., Masila, E., Muhoma, V. A., Too, E., Mungube, E. O., and Maichomo, M. (2023). “Campylobacteriosis in Sub-Saharan Africa,” in Bacterial Infectious Diseases Annual, eds. G. Katarzyna and J. Tomas (London: IntechOpen), 1–17.
Ortiz, B. T., Rodríguez, D., and Restrepo, S. (2024). Prevalence and risk factors of Campylobacter jejuni and Campylobacter coli in fresh chicken carcasses from retail sites in Bogotá, Colombia. Heliyon 10:e26356. doi: 10.1016/j.heliyon.2024.e26356
Paintsil, E. K., Masanta, W. O., Dreyer, A., Ushanov, L., Smith, S. I., Frickmann, H., et al. (2023). Campylobacter in Africa – a specific viewpoint. Eur. J. Microbiol. Immunol. 13, 107–124. doi: 10.1556/1886.2023.00043
Pajaniappan, M., Hall, J. E., Cawthraw, S. A., Newell, D. G., Gaynor, E. C., Fields, J. A., et al. (2008). A temperature-regulated Campylobacter jejuni gluconate dehydrogenase is involved in respiration-dependent energy conservation and chicken colonization. Mol. Microbiol. 68, 474–491. doi: 10.1111/j.1365-2958.2008.06161.x
Reddy, S., and Zishiri, O. T. (2017). Detection and prevalence of antimicrobial resistance genes in Campylobacter spp. isolated from chickens and humans. Onderstepoort J. Vet. Res. 84, 1–6. doi: 10.4102/ojvr.v84i1.1411
Retsema, J., Girard, A., Schelkly, W., Manousos, M., Anderson, M., Bright, G., et al. (1987). Spectrum and mode of action of azithromycin (CP-62,993), a new 15-membered-ring macrolide with improved potency against gram-negative organisms. Antimicrob. Agents Chemother. 31, 1939–1947. doi: 10.1128/AAC.31.12.1939
Same, R. G., and Tamma, P. D. (2018). Campylobacter infections in children. Pediatr. Rev. 39, 533–541. doi: 10.1542/pir.2017-0285
Sangaré, L., Nikiéma, A., Zimmermann, S., Sanou, I., Congo-Ouédraogo, M., Diabaté, A., et al. (2012). Campylobacter spp. epidemiology and antimicrobial susceptibility in a developing country, Burkina Faso (West Africa). Afr. J. Clin. Exp. Microbiol. 13, 110–117. doi: 10.4314/ajcem.v13i2.9
Scallan, E., Hoekstra, R. M., Angulo, F. J., Tauxe, R. V., Widdowson, M. A., Roy, S. L., et al. (2011). Foodborne illness acquired in the United States-major pathogens. Emerg. Infect. Dis. 17, 7–15. doi: 10.3201/eid1701.P11101
Sibanda, N., McKenna, A., Richmond, A., Ricke, S. C., Callaway, T., Stratakos, A. C., et al. (2018). A review of the effect of management practices on Campylobacter prevalence in poultry farms. Front. Microbiol. 9:2002. doi: 10.3389/fmicb.2018.02002
Sproston, E. L., Wimalarathna, H. M. L., and Sheppard, S. K. (2018). Trends in fluoroquinolone resistance in Campylobacter. Microb. Genom. 4:e000198. doi: 10.1099/mgen.0.000198
Srijan, A., Bodhidatta, L., Mason, C. J., Bunyarakyothin, G., Jiarakul, W., and Vithayasai, N. (2013). Field evaluation of a transport medium and enrichment broth for isolation of Campylobacter species from human diarrheal stool samples. Open J. Med. Microbiol. 3, 48–52. doi: 10.4236/ojmm.2013.31007
Ssemanda, J. N., Reij, M. W. G., an Middendorp, E., Bouw, R., van der Plaats, and Franz, E.. (2018). Foodborne pathogens and their risk exposure factors associated with farm vegetables in Rwanda. Food Control 89, 86–96. doi: 10.1016/j.foodcont.2017.12.034
Strakova, N., Michova, H., Shagieva, E., Ovesna, P., Karpiskova, R., and Demnerova, K. (2024). Genotyping of Campylobacter jejuni and prediction tools of its antimicrobial resistance. Folia Microbiol. 69, 207–219. doi: 10.1007/s12223-023-01093-5
Tang, M., Zhou, Q., Zhang, X., Zhou, S., Zhang, J., Tang, X., et al. (2020). Antibiotic resistance profiles and molecular mechanisms of campylobacter from chicken and pig in China. Front. Microbiol. 11:592496. doi: 10.3389/fmicb.2020.592496
Thakur, S., Zhao, S., McDermott, P. F., Harbottle, H., Abbott, J., English, L., et al. (2010). Antimicrobial resistance, virulence, and genotypic profile comparison of Campylobacter jejuni and Campylobacter coli isolated from humans and retail meats. Foodborne Pathog. Dis. 7, 835–844. doi: 10.1089/fpd.2009.0487
Wagenaar, J. A., French, N. P., and Havelaar, A. H. (2013). Preventing Campylobacter at the source: why is it so difficult?. Clin. Infect. Dis. 57, 1600–1606. doi: 10.1093/cid/cit555
Wei, B., and Kang, M. (2018). Molecular basis of macrolide resistance in Campylobacter strains isolated from poultry in South Korea. BioMed Res. Int. 2018:4526576. doi: 10.1155/2018/4526576
Wieczorek, K., and Osek, J. (2013). Antimicrobial resistance mechanisms among Campylobacter. BioMed Res. Int. 2013, 1–12. doi: 10.1155/2013/340605
Worku, M., Tessema, B., Ferede, G., Ochieng, L., Leliso, S. A., Mutua, F., et al. (2024). Campylobacter jejuni and Campylobacter coli infection, determinants and antimicrobial resistance patterns among under-five children with diarrhea in Amhara National Regional State, Northwest Ethiopia. PLoS ONE 19:e0304409. doi: 10.1371/journal.pone.0304409
Yamazaki-Matsune, W., Taguchi, M., Seto, K., Kawahara, R., Kawatsu, K., Kumeda, Y., et al. (2007). Development of a multiplex PCR assay for identification of Campylobacter coli, Campylobacter fetus, Campylobacter hyointestinalis subsp. hyointestinalis, Campylobacter jejuni, Campylobacter lari and Campylobacter upsaliensis. J. Med. Microbiol. 56, 1467–1473. doi: 10.1099/jmm.0.47363-0
Yildiz, M., Sahin, O., and Adiguzel, M. C. (2024). Prevalence and antimicrobial resistance of Campylobacter species in shelter-housed healthy and diarrheic cats and dogs in Turkey. Vet. Med. Sci. 10:e1327. doi: 10.1002/vms3.1327
Zachariah, O. H., Lizzy, M. A., Rose, K., and Angela, M. M. (2021). Multiple drug resistance of Campylobacter jejuni and Shigella isolated from diarrhoeic children at Kapsabet County referral hospital, Kenya. BMC Infect. Dis. 21:109. doi: 10.1186/s12879-021-05788-3
Keywords: thermophilic Campylobacter, antimicrobial resistance, livestock, One Health, Rwanda
Citation: Gahamanyi N, Habimana AM, Harerimana JP, Mukayisenga J, Ntwali S, Umuhoza A, Nsengiyumva E, Irimaso E, Bosco Shimirwa J, Pan C-H, Komba EV, Muvunyi CM, Rujeni N and Amachawadi RG (2025) Antimicrobial susceptibility profiles of thermophilic Campylobacter species from human, pig, and chicken feces in Rwanda. Front. Microbiol. 16:1570290. doi: 10.3389/fmicb.2025.1570290
Received: 06 February 2025; Accepted: 05 August 2025;
Published: 25 August 2025.
Edited by:
Fábio Sellera, Universidade Metropolitana de Santos, BrazilReviewed by:
Muhammad Akbar Shahid, Bahauddin Zakariya University, PakistanPhil Giffard, Charles Darwin University, Australia
Rita Barata, National Institute for Agricultural and Veterinary Research (INIAV), Portugal
Copyright © 2025 Gahamanyi, Habimana, Harerimana, Mukayisenga, Ntwali, Umuhoza, Nsengiyumva, Irimaso, Bosco Shimirwa, Pan, Komba, Muvunyi, Rujeni and Amachawadi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Noel Gahamanyi, bm9lbC5nYWhhbWFueWlAcmJjLmdvdi5ydw==; Raghavendra G. Amachawadi, YWdyYWdoYXZAdmV0Lmstc3RhdGUuZWR1
†These authors have contributed equally to this work
 Noel Gahamanyi
Noel Gahamanyi Arsene Musana Habimana
Arsene Musana Habimana Jean Paul Harerimana1
Jean Paul Harerimana1 Salomon Ntwali
Salomon Ntwali Emmanuel Nsengiyumva
Emmanuel Nsengiyumva Emmanuel Irimaso
Emmanuel Irimaso Cheol-Ho Pan
Cheol-Ho Pan Erick Vitus Komba
Erick Vitus Komba Claude Mambo Muvunyi
Claude Mambo Muvunyi Nadine Rujeni
Nadine Rujeni