- 1School of Light Industry Science and Engineering, Beijing Technology and Business University (BTBU), Beijing, China
- 2Beijing Advanced Innovation Center for Food Nutrition and Human Health, Beijing Technology and Business University (BTBU), Beijing, China
- 3The Key Laboratory of Molecular Microbiology and Technology, Ministry of Education, College of Life Sciences, Nankai University, Tianjin, China
- 4College of Life Sciences, Shanxi University, Taiyuan, China
Caffeic acid, a plant-derived phenolic compound, has attracted much attention in the fields of medicines and cosmetics due to its remarkable physiological activities including antioxidant, anti-inflammation, antibacteria, antivirus and hemostasis. However, traditional plant extraction and chemical synthesis methods exist some problems such as high production costs, low extraction efficiency and environmental pollution. In recent years, the construction of microbial cell factories for the biosynthesis of caffeic acid has attracted much attention due to its potential to offer an efficient and environmentally-friendly alternative for caffeic acid production. This review introduces the caffeic acid biosynthesis pathway first, after which the characteristics of microbial hosts for caffeic acid production are analyzed. Then, the main strategies for caffeic acid production in microbial hosts, including selection and optimization of heterologous enzymes, enhancement of the metabolic flux to caffeic acid, supply and recycling of cofactor, and optimization of the production process, are summarized and discussed. Finally, the future prospects and perspectives of microbial caffeic acid production are discussed. Recent breakthroughs have achieved caffeic acid titers of up to 6.17 g/L, demonstrating the potential of microbial biosynthesis. Future research can focus on the enhancement of metabolic flux to caffeic acid biosynthesis pathway, the development of robust microbial hosts with improved tolerance to caffeic acid and its precursors, and the establishment of cost-effective industrial production processes.
1 Introduction
Caffeic acid (3,4-dihydroxycinnamic acid) is a natural phenolic compound widely existed in plants such as Crataegus pinnatifida, Thymian, Solidago virgaurea, and Eucommia ulmoides (Touaibia et al., 2011; Yang et al., 2013; Espindola et al., 2019). It has attracted much attention in the fields of medicines and cosmetics due to its demonstrated pharmacological actions including antioxidant, anti-inflammation, antibacteria, antivirus, hemostasis, analgesis, prevention of cardiovascular disease, protection of brain tissue damage, and promotion of wound healing (Touaibia et al., 2011; Khan et al., 2016; Alson et al., 2018; Teng et al., 2020; Pavlikova, 2022). Moreover, its derivatives like caffeic acid phenethyl ester and chlorogenic acid also have important pharmacological activities (Wang et al., 2017; Xiao et al., 2022).
According to statistics and forecasts by QYResearch, the global market sales of caffeic acid reached 0.13 billion USD in the year 2024 and is expected to reach 0.16 billion USD by the year 2030, with a compound annual growth rate of 3.8% from 2024 to 2030. Caffeic acid is commonly extracted from plant sources, but its low accumulation in plants makes the separation process both difficult and expensive to some extent (Ye et al., 2010). Additionally, extraction methods involve high-temperature treatments, the use of petroleum ether, and solvent extraction, which are energy-intensive and environmentally unfriendly (Zhang et al., 2014; Rodrigues et al., 2015; Han et al., 2024; Qian et al., 2024). Caffeic acid can also be produced by chemical synthesis, but the application of this technique is greatly limited by the requirement for toxic solvents, the complex production process and the production of byproducts (Zhang et al., 2014). In recent years, the development of synthetic biology techniques and metabolic engineering make it possible to produce plant-specific secondary metabolites in heterologous microbial hosts. The use of microbial hosts has made significant contributions to the biosynthesis of industrial and pharmaceutical compounds due to its advantages of low production cost, high product purity, high efficiency, environmental friendliness, and the simplicity of the genetic manipulation process (Marienhagen and Bott, 2013; Davy et al., 2017; Cravens et al., 2019). Heterologous production of caffeic acid in microbial hosts provides a feasible way to produce caffeic acid and has attracted much attention of researchers. The first report on microbial production of caffeic acid was achieved in Streptomyces fradiae XKS (Berner et al., 2006). Subsequently, more and more studies have been carried out, and the concentration of caffeic acid has reached 6.17 g/L through the introduction of its heterologous biosynthetic pathway and the metabolic engineering of the microbial host (Sakae et al., 2023).
This review summarizes the recent advances in the heterologous production of caffeic acid in microbial hosts, discusses the tools and strategies employed at the gene, protein, and pathway levels, as well as the optimization of the fermentation process to achieve efficient biosynthesis of caffeic acid. Furthermore, the future opportunities and challenges in the production of caffeic acid is also discussed.
2 Caffeic acid biosynthesis pathway
The biosynthetic pathway of caffeic acid and its metabolic regulation are shown in Figure 1. Glucose is converted into phosphoenolpyruvate and erythrose-4-phosphate via the glycolytic pathway and the pentose phosphate pathway, respectively. Biosynthesis of aromatic amino acids starts with shikimate pathway. The 3-deoxy-D-arabino-heptulosonate-7-phosphate (DAHP) synthase, which is regulated by feedback-inhibition allosteric mechanism, catalyzes the condensation of erythrose-4-phosphate and phosphoenolpyruvate to DAHP. Six enzymatic reactions convert DAHP to chorismite, which is the precursor of aromatic amino acids. These aromatic amino acids are the final products of biosynthetic pathways in most microbial hosts, such as Escherichia coli and S. cerevisiae. However, in plants and some microorganisms, L-phenylalanine and L-tyrosine are function as intermediates products to synthesize secondary metabolites such as phenylpropanoids. Caffeic acid is mainly produced by the universal route of phenylpropanoid (Lin and Yan, 2012; Rodrigues et al., 2015). First, L-phenylalanine and L-tyrosine are converted to cinnamic acid and p-coumaric acid, respectively, by the removal of ammonia under the catalysis of phenylalanine ammonia lyase (PAL) and tyrosine ammonia lyase (TAL). Cinnamic acid is then hydroxylated to p-coumaric acid under the action of cinnamate-4-hydroxylase (C4H). Then p-coumaric acid is further hydroxylated by coumarate-3-hydroxylase (C3H) to form caffeic acid. Both C4H and C3H are plant-specific cytochrome P450 dependent monooxygenases, and thus, cytochrome P450 reductase (CPR) is required for efficient electron transfer (Kim et al., 2011; Lin and Yan, 2012; Cheniany and Ganjeali, 2016).
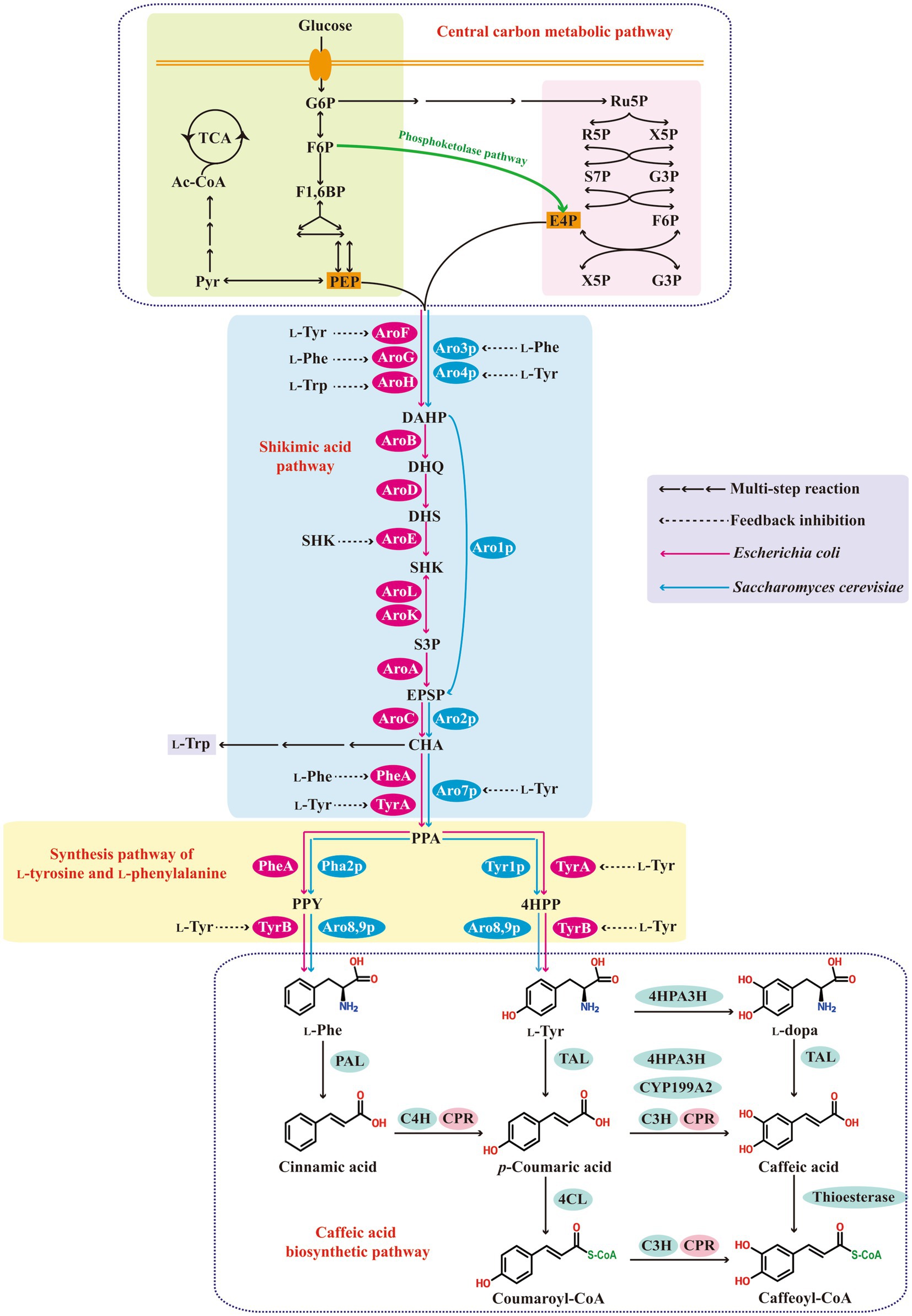
Figure 1. The biosynthetic pathway of caffeic acid and its metabolic regulation. G6P, glucose-6-phosphate; F6P, fructose-6-phosphate; F1,6BP, fructose 1,6-biphosphate; PEP, phosphoenolpyruvate; Pyr, pyruvate; Ac-CoA, acetyl-CoA; TCA tricarboxylic acid cycle; Ru5P, ribulose 5-phosphate; R5P, ribose-5-phosphate; X5P, xylulose-5-phosphate; S7P, sedoheptulose 7-phosphate; G3P, glyceraldehyde 3-phosphate; E4P, erythrose-4-phosphate; DAHP, 3-deoxy-D-arabinoheptulosonic acid-7-phosphate; DHQ, 3-dehydroquinate; DHS, 3-dehydroshikimate; SHIK, shikimate; S3P, shikimate 3-phosphate; EPSP, 5-enolpyruvyl-shikimate 3-phosphate; CHA, chorismic acid; PPA, prephenate; 4HPP, p-hydroxyphenylpyruvate; L-Phe, L-phenylalanine; L-Tyr, L-tyrosine; L-Trp, L-tryptophan; PAC, hydroxy-acetaldehyde; AroF, AroG, AroH, Aro3p, and Aro4p, 3-deoxy-D-arabinoheptulosonic acid-7-phosphate synthetase; AroB, 3-dehydroquinate synthetase; AroD, 3-dehydroquinate dehydratase; AroE, shikimate dehydrogenase; AroL and AroK, shikimate kinase; AroA, 5-enolpyruvyl-shikimate 3-phosphate synthetase; Aro1p, pentafunctional enzyme; AroC and Aro2p, chorismic acid synthetase; Aro7p, prephenate mutase; TyrA and Tyr1p, prephenate dehydrogenase; PheA and Pha2p, prephenate dehydratase; TyrB, Aro8p and Aro9p, aromatic aminotransferase; PAL, phenylalanine ammonia-lyase; C4H, cinnamate-4-hydroxylase; TAL, tyrosine ammonia-lyase; C3H, p-coumarate-3-hydroxylase; CPR, cytochrome P450 reductase; 4HPA3H, 4-hydroxyphenylacetate 3-hydroxylase; CYP199A2, cytochrome P450 CYP199A2; 4CL, 4-coumarate-CoA ligase.
3 Microbial hosts for caffeic acid biosynthesis
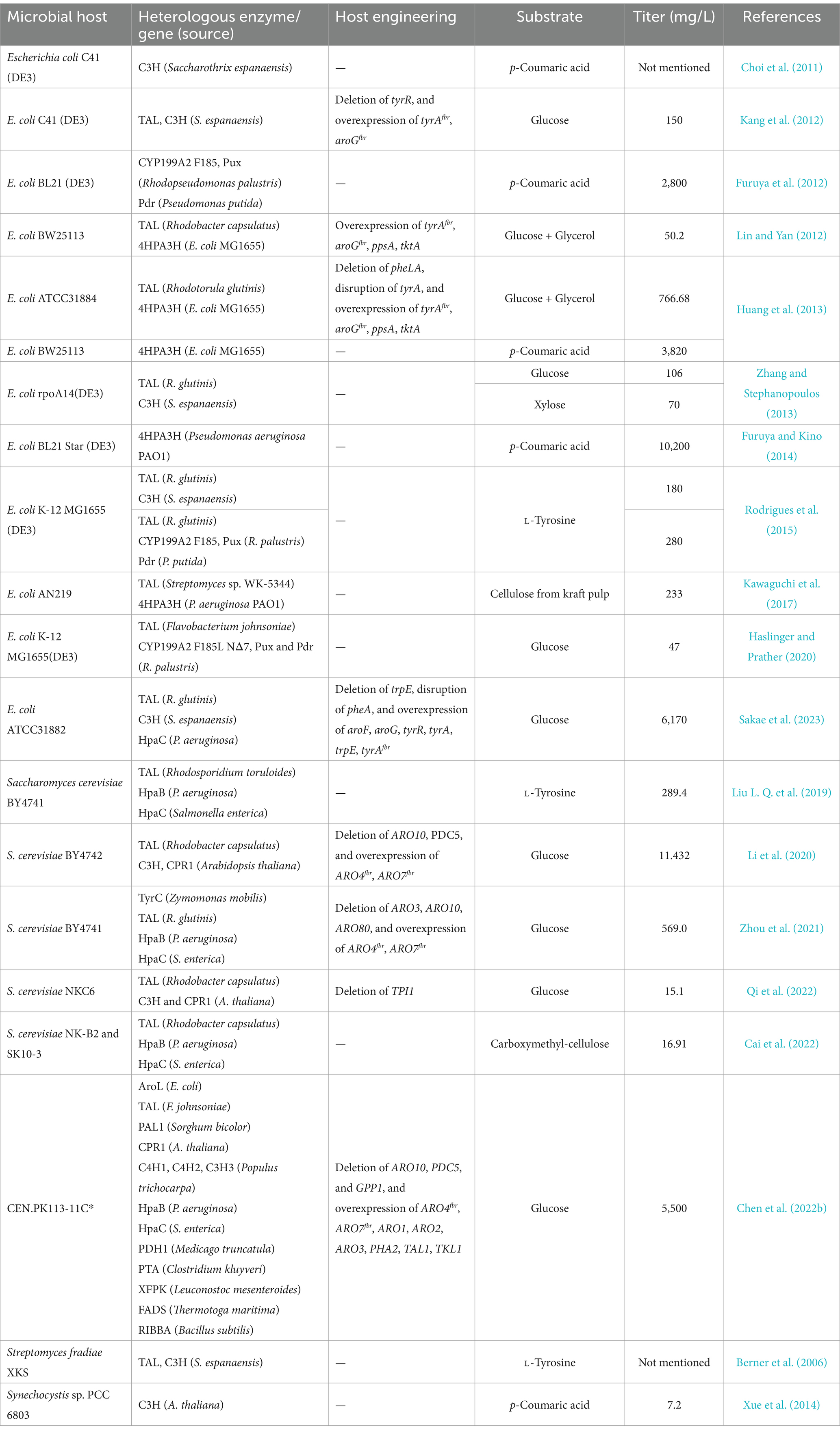
Caffeic acid biosynthesis refers to the implantation of caffeic acid biosynthetic pathway into useful microbial hosts for heterologous expression to produce caffeic acid by metabolic engineering and synthetic biology. The heterologous genes from eukaryotes, prokaryotes, and plants are incorporated into the microbial hosts, which convert cheap renewable carbon sources to caffeic acid through their own metabolic processes. It is very important to select a suitable host for the efficient production of caffeic acid. The commonly used hosts for caffeic acid production are E. coli as prokaryotes and Saccharomyces cerevisiae as eukaryotes. The main research of caffeic acid biosynthesis in microbial host are shown in Table 1, detailing the microbial hosts utilized, the enzymes or genes introduced, the substrates added to the culture medium, and the yield of caffeic acid.
3.1 E. coli host
Escherichia coli is taken as an ideal host for the engineering and production of different biomolecules due to its rapid growth rate, short life cycle, low cultivation cost, clear genetic background, easy genetic manipulation, and good ability for enzyme overexpression (Makrides, 1996; Wu et al., 2013; Liu et al., 2016; Wang et al., 2016). Moreover, L-tyrosine and p-coumaric acid, the key precursors of caffeic acid, are crucial for increasing caffeic acid production and easy to be improved in E. coli (Shrestha et al., 2019). In addition, E. coli is better suited for caffeic acid production than yeast due to its superior tolerance to p-coumaric acid (Shin et al., 2011; Zhang and Stephanopoulos, 2013; Li et al., 2020). The first attempt to produce caffeic acid in E. coli was described by Choi et al. (2011), who introduced C3H from Saccharothrix espanaensis into E. coli C41 (DE3) strain and achieved caffeic acid with the supplementation of precursor p-coumaric acid. However, a significant limitation of using E. coli as an expression host is its lack of capacity for post-translational modifications, which are typically necessary for the correct folding and functional activity of recombinant proteins.
3.2 Saccharomyces cerevisiae host
Like E. coli, S. cerevisiae is also taken as an ideal host for the engineering and production of different biomolecules due to its easy growth, low cultivation cost, clear genetic background, and easy genetic manipulation (Yesilirmak and Sayers, 2009; Borodina and Nielsen, 2014). Furthermore, S. cerevisiae is a generally recognized as safe (GRAS) host and widely used in food markets and pharmaceutical products (Krivoruchko and Nielsen, 2015; Pereira et al., 2019). One major advantage of using S. cerevisiae rather than E. coli as a host is that its subcellular compartmentation is similar to that of plant cells, which make it has ability to perform post-translational modifications (Rainha et al., 2020). Thus S. cerevisiae can adequately express membrane proteins such as cytochrome P450 (Kim et al., 2011; Koopman et al., 2012; Sadeghi et al., 2013; Bostick et al., 2016). Li et al. (2020) determined that the codon optimized C3H gene from Arabidopsis thaliana can successfully express and hydroxylate the substrate p-coumaric acid to caffeic acid in S. cerevisiae strain. Nevertheless, S. cerevisiae is more sensitive to high concentration of p-coumaric acid (Shrestha et al., 2019).
3.3 Other hosts
Apart from E. coli and S. cerevisiae, S. fradiae XKS and cyanobacterium Synechocystis PCC 6803 have also been engineered to produce caffeic acid. Berner et al. (2006) have successfully expressed TAL and C3H from S. espanaensis in S. fradiae XKS, and achieved caffeic acid with the supplementation of L-tyrosine. Xue et al. (2014) introduced C3H from A. thaliana into cyanobacterium Synechocystis PCC 6803 to obtain 7.2 mg/L caffeic acid, when supplemented with p-coumaric acid.
4 Metabolic engineering to enhance caffeic acid production
Metabolic engineering of caffeic acid production in microbial hosts has achieved great progress in recent years. The main strategies for increasing caffeic acid productivity using microbial cell factories included selecting and optimizing heterologous enzymes via enzyme engineering, increasing the metabolic flux to caffeic acid via pathway engineering, enhancing the supply and recycling of cofactor via cofactor engineering, and optimizing the production process via fermentation engineering.
4.1 Enzyme selection and engineering
The heterologous enzymes needed for the synthesis of caffeic acid in microorganisms mainly include ammonia-lyase and hydroxylase.
4.1.1 Ammonia-lyase
TAL catalyzes the deamination of L-tyrosine to produce p-coumaric acid. PAL catalyzes the deamination of L-phenylalanine to produce cinnamic acid, which is then hydroxylated at the 4-position of the benzene ring to form p-coumaric acid by the action of C4H and its associated CPR. Jendresen et al. (2015) screened and expressed 22 wild type or artificially modified TAL and PAL genes for p-coumaric acid production, and found that enzymes from Flavobacterium johnsoniae and Herpetosiphon aurantiacus resulted in high p-coumaric acid production in E. coli, S. cerevisiae, and Lactococcus lactis strains. TAL from S. espanaensis can also result in high production of p-coumaric acid in E. coli strain. Kawaguchi et al. (2017) found that TAL (fevV) from Streptomyces sp. WK-5344 can be successfully expressed and convert L-tyrosine to p-coumaric acid in E. coli strain.
4.1.2 Hydroxylase
C3H catalyzes the hydroxylation of p-coumaric acid at 3-position of the benzene ring to form caffeic acid. The hydroxylase which can function in E. coli mainly contains S. espanaensis C3H, Rhodopseudomonas palustris cytochrome P450 CYP199A2, and E. coli endogenous 4-hydroxyphenylacetate 3-hydroxylase (4HPA3H). Choi et al. (2011) used an E. coli strain C41 (DE3) harboring C3H from S. espanaensis and achieved caffeic acid in the presence of p-coumaric acid. Furuya et al. (2012) achieved the caffeic acid with an E. coli strain BL21 (DE3) harboring cytochrome P450 CYP199A2 from R. palustris and its associated putidaredoxin reductase gene (pdR) from Pseudomonas putida and palustrisredoxin gene (pux) from R. palustris with the supplementation of p-coumaric acid. To enhance the activities of CYP199A2, the F185 residue of CYP199A2 was substituted with Tyr (F185Y), Trp (F185W), Val (F185V), Leu (F185L), Ile (F185I), Gly (F185G), Ala (F185A), Ser (F185S), or Thr (F185T), respectively. The F185L mutant was confirm to have the highest hydroxylation activity for p-coumaric acid, which was 5.5 times higher than that of wild-type CYP199A2, and the production of caffeic acid from p-coumaric acid using the F185L whole-cell catalyst reached 2.8 g/L (Furuya et al., 2012). Further, Haslinger and Prather (2020) enhanced the efficiency of the electron transfer step from the two redox partner proteins to CYP199A2 F185L through using the natural redox system, tethering the redox complex by creating genetic fusions with high-affinity tethering domains, and supplying extra gene copies coding for pux. About 47 mg/L caffeic acid was obtained from the E. coli MG1655 (DE3) strain incorporating TAL from F. johnsoniae, CYP199A2 F185L, tethered redox complex, and extra gene copies coding for pux with glucose as the only carbon source (Haslinger and Prather, 2020). It has been demonstrated that CYP199A2 F185L with its redox partners allowed a higher caffeic acid titer when compared with C3H from S. espanaensis (Rodrigues et al., 2015). 4HPA3H complex, which is composed of 4-hydroxyphenylacetate 3-monooxygenase (HpaB) and NADPH-flavin oxidoreductase (HpaC), can hydroxylate a broad range of substrates at the 3-position, including L-tyrosine, tyrosol, and p-hydroxyphenylacetate (Galan et al., 2000). Considering the structural similarity of p-coumaric acid to L-tyrosine, tyrosol, and p-hydroxyphenylacetate, researchers hypothesized that 4HPA3H should be able to catalyze p-coumaric acid as well (Furuya and Kino, 2014). Lin and Yan (2012) first identified that the E. coli native 4HPA3H could convert p-coumaric acid to caffeic acid efficiently. Huang et al. (2013) overexpressed HpaBC with a high-copy plasmid pZE12-luc in E. coli BW25113 strain, and produced 3.82 g/L caffeic acid from 4 g/L p-coumaric acid in shake flasks after 24 h cultivation.
The hydroxylase which can function in S. cerevisiae mainly contains 4HPA3H derived from different species and C3H from A. thaliana. Li et al. (2020) overexpressed A. thaliana-derived C3H and CPR1 with a high-copy plasmid pLC41 in yeast BY4742, and produced 18.1 mg/L caffeic acid from 600 mg/L p-coumaric acid in shake flasks after 120 h cultivation, indicating the A. thaliana-derived C3H can be expressed in S. cerevisiae strain and catalyze the conversion of p-coumaric acid to caffeic acid. Liu L. Q. et al. (2019) constructed functional 4HPA3H in S. cerevisiae strain by recruiting HpaB and HpaC from several bacteria, and obtained the highest production of caffeic acid (289.4 mg/L) from 500 mg/L L-tyrosine by co-overexpressing of TAL from Rhodosporidium toruloides and the enzyme combination of HpaB from Pseudomonas aeruginosa and HpaC from Salmonella enterica in shake flasks after 96 h cultivation.
4.2 Pathway engineering
There is an endogenous aromatic amino acid biosynthesis pathway in microbial hosts, such as E. coli and S. cerevisiae (Figure 1). However, this pathway is mostly regulated by feedback inhibition of allosterically controlled key enzymes and the accumulation of aromatic amino acid is very low, thus the enhancement of metabolic flux to L-phenylalanine and L-tyrosine is vitally important for caffeic acid production. Meanwhile, microbial hosts, such as E. coli and S. cerevisiae, lack a phenylpropanoid metabolism pathway, thus the construction of heterologous synthesis pathway of caffeic acid is essential for bioconversion or de novo biosynthesis of caffeic acid. The two main goals within this step include the enhancement of metabolic flux to L-phenylalanine and L-tyrosine and the functional introduction of heterologous synthesis pathway of caffeic acid.
4.2.1 The enhancement of metabolic flux to L-phenylalanine and L-tyrosine
The mainly strategies to enhance the endogenous metabolic flux to L-phenylalanine and L-tyrosine biosynthesis contain the enhancement of precursor supply, elimination of enzyme feedback inhibition regulation, blocking of competitive pathways, and balance of the ratio between phosphoenolpyruvate and erythrose 4-phosphate.
In E. coli strain, Kang et al. (2012) constructed a L-tyrosine high-producing strain by knocking out tyrR to remove the repression of TyrR protein to L-tyrosine metabolic pathway, overexpressing aroGfbr to relieve the feedback inhibition of L-phenylalanine on AroG protein, and overexpressing tyrAfbr mutant to relieve the feedback inhibition of L-tyrosine on TyrA protein, and achieved a 2.6-fold higher caffeic acid production (150 mg/L) from glucose by overexpressing TAL and C3H from S. espanaensis in this L-tyrosine high-producing strain. Furthermore, the production of L-tyrosine was increased by overexpressing ppsA and tktA to enhance the supply of precursors phosphoenolpyruvate and erythrose 4-phosphate, disrupting pheA to block the competitive pathway of L-phenylalanine, and disrupting tyrA to eliminate feedback inhibition of L-tyrosine (Lin and Yan, 2012; Huang et al., 2013).
In S. cerevisiae strain, Li et al. (2020) constructed a L-tyrosine high-producing strain by overexpressing the mutated feedback-resistant ARO4K229L and ARO7G141S to alleviate feedback inhibition by L-tyrosine on DAHP synthase and chorismite mutase, and knocking out ARO10 and PDC5 to avoid production of aromatic alcohols and direct the pathway flux to L-tyrosine, and resulted in a 14.1-fold higher caffeic acid production from glucose by overexpressing TAL from Rhodobacter capsulatus and C3H from A. thaliana in this L-tyrosine high-producing strain. The activity of endogenous prephenate dehydrogenase Tyr1p in S. cerevisiae is feedback-inhibited by L-phenylalanine, while the cyclohexadienyl dehydrogenase TyrC from Zymomonas mobilis can also catalyze the conversion of prephenic acid to 4-hydroxyphenylpyruvate without being feedback-inhibited by L-phenylalanine (Gold et al., 2015; Mao et al., 2017). Zhou et al. (2021) engineered a high-throughput caffeic acid synthetic pathway by knocking out ARO3 and ARO10, and inducibly overexpressing ARO4K229L, ARO7G141S, TyrC, TAL from Rhodotorula glutinis, hpaB from P. aeruginosa, and hpaC from S. enterica, achieving 569.0 mg/L of caffeic acid from glucose in shake flask fermentation, with a 1.6-fold increase in production due to the enhanced L-tyrosine metabolic flux. Liu Q. L. et al. (2019) engineered the aromatic amino acid biosynthesis pathway by introducing a phosphoketolase-based pathway to divert glycolytic flux toward erythrose 4-phosphate formation, and optimizing the carbon distribution between aromatic amino acid biosynthesis pathway and glycolysis pathway through replacing the promoters of several important genes at key nodes between these two pathways. Furthermore, they simultaneously introduced a single heterologous enzymatic step from L-tyrosine (TAL from F. johnsoniae) and two enzymatic steps from L-phenylalanine (PAL2, C4H and CPR2 from A. thaliana, a cytochrome B5 from S. cerevisiae) in this strain to produce a maximum p-coumaric acid titer of 12.5 g/L, providing a new strategy for constructing platform strain for high L-phenylalanine and L-tyrosine yields (Liu Q. L. et al., 2019).
4.2.2 The introduction of heterologous synthesis pathway of caffeic acid
In prokaryotic microorganisms, caffeic acid is commonly synthesized by a L-tyrosine derived pathway due to the difficult expression of plant-specific cytochrome P450 enzymes in prokaryotic microbial system, whereas in eukaryotic microorganisms, caffeic acid can be synthesized by a L-tyrosine derived pathway or a L-phenylalanine derived pathway. L-Phenylalanine or L-tyrosine are first converted to p-coumaric acid. Then, p-coumaric acid is further converted to caffeic acid by sam5-encoded C3H from the S. espanaensis, or cytochrome P450 CYP199A2 from R. palustris, or 4HPA3H from different microbial sources (Furuya et al., 2012; Lin and Yan, 2012; Li et al., 2020). The L-phenylalanine derived pathway was far more efficient compared with the L-tyrosine derived pathway. Liu Q. L. et al. (2019) introduced a L-phenylalanine derived pathway containing PAL2, C4H, and CPR2 from A. thaliana and cytochrome B5 (CYB5) from S. cerevisiae, as well as a L-tyrosine derived pathway containing highly specific TAL from F. johnsoniae in S. cerevisiae strain, respectively, and found that the L-phenylalanine derived pathway can produced 337.6 mg/L p-coumaric acid under glucose-limited conditions, which was 26.17-fold higher than that of the L-tyrosine derived pathway. Chen et al. (2022b) introduced a L-phenylalanine derived pathway containing Sorghum bicolor PAL1, A. thaliana CPR1, and an identified Cyp complex (containing C4H2, C4H1, and C3H3 from Populus trichocarpa) into S. cerevisiae strain expressing TAL from F. johnsoniae, and resulted in a 5.66-fold increase in p-coumaric acid accumulation in shake-flask cultivation. In addition, L-tyrosine can also be first converted to L-dopa by 4HPA3H, and then TAL converts L-dopa to caffeic acid. A potential caffeic acid biosynthesis pathway is to convert L-tyrosine to p-coumaric acid, then to coumaroyl-CoA, to caffeoyl-CoA, and finally to caffeic acid by TAL, 4-coumarate-CoA ligase, C3H, and thioesterase, respectively (Zhang and Stephanopoulos, 2013) (Figure 1).
4.3 Cofactor engineering
At present, researchers mainly improve the microbial synthesis of caffeic acid products by selecting and optimizing heterologous enzymes of caffeic acid synthesis pathway and enhancing the metabolic flux of L-tyrosine and L-phenylalanine in host cells, while neglecting the role of cofactors in the biosynthesis of natural products. In the biosynthetic pathway of caffeic acid, HpaBC is a two-component monooxygenase. HpaB performs the hydroxylation reaction with the aid of FADH2 cofactor, whereas HpaC, serving as a flavin reductase, is responsible for the conversion of FAD to FADH2 through the consumption of NADH (Galan et al., 2000; Xun and Sandvik, 2000; Phillips and Shephard, 2008). This interplay ensures the seamless progression of the monooxygenase reaction. Meanwhile, the regeneration of NADPH is crucial for the function of cytochrome P450 enzymes in the synthesis of the precursor p-coumaric acid (Yang et al., 2020). Chen et al. (2022a) systematically optimized the supply and recycling pathways of cofactors NADPH and FADH2 in the S. cerevisiae strain, increasing the titer of caffeic acid by 31% to 5.5 g/L, which is the highest titer reported for caffeic acid. Fre, a flavin reductase enzyme in E. coli, catalyzes the reduction of flavin to FADH2 by utilizing NAD (P) H as the electron donor (Fontecave et al., 1987; Fieschi et al., 1995). Zhou et al. (2022) increased the hydroxylation efficiency of HpaBC by 810% in E. coli strain through the fusion expression of Fre and HpaBC, which accelerate the regeneration of FADH2. Effendi and Ng (2023) enriched FADH2 cofactor pools via deploying fre gene in E. coli strain, which improve ferulic acid titer by 37.14%.
4.4 Fermentation engineering
4.4.1 Microbial co-culture strategy
Microorganism co-cultivation can not only fully utilize the advantages of various microorganisms but also alleviate the metabolic stress on single microbial cells, and is being increasingly applied to the biosynthesis of natural products (Nakayama et al., 2011; Goers et al., 2014; Zhang et al., 2017; Rodrigues et al., 2020). Cai et al. (2022) co-cultured engineered S. cerevisiae strain SK10-3 harboring the carboxymethyl cellulose hydrolysis pathway composed of Talaromyces emersonii CBHI, Trichoderma reesei EGII, and Aspergillus aculeatus BGLI with engineered S. cerevisiae strain NK-B2b harboring p-coumaric acid synthesis pathway composed of R. capsulatus TAL and produced 71.71 mg/L p-coumaric acid from carboxymethyl cellulose, which is 155.9-fold higher than the mono-culture of SK10-3b harboring both carboxymethyl cellulose hydrolysis pathway and p-coumaric acid synthesis pathway. Then, the de novo biosynthesis of caffeic acid from carboxymethyl-cellulose was achieved through the co-culture of engineered S. cerevisiae strains SK10-3, NK-B2b, and NK-B2d harboring caffeic acid synthesis pathway composed of P. aeruginosa hpaB and S. enterica hpaC. This strategy provides a novel approach for the low-cost and high-efficiency synthesis of caffeic acid (Cai et al., 2022).
4.4.2 Optimization of the fermentation medium
Research has found that the composition of culture medium, especially the carbon source, greatly affect the yield of caffeic acid. Furuya et al. (2012) confirmed that glycerol was more effective than glucose in supporting the hydroxylation carried out by E. coli cells expressing the F185L mutant of CYP199A2. By using the F185L whole-cell catalyst in the presence of glycerol, the production of caffeic acid from p-coumaric acid reached 2.8 g/L, 1.65-fold higher than in the presence of glucose (Furuya et al., 2012). Zhang and Stephanopoulos (2013) evaluated the synthesis of caffeic acid by recombinant E. coli strain under various conditions and discovered that glucose was more suitable as a carbon source than xylose. Furthermore, when comparing the MOPS medium, rich medium, and synthetic medium, it was found that the rich medium yielded the highest caffeic acid titer, whereas the synthetic medium resulted in the highest specific titer (the ratio of caffeic acid production to cell density) (Zhang and Stephanopoulos, 2013). Li et al. (2020) found that engineered strains of S. cerevisiae exhibited greater growth density and 4.1-fold higher caffeic acid production in the rich medium YPD containing 4% glucose compared to the synthetic medium SC-Ura-His containing 2% glucose. Kawaguchi et al. (2017) produce 233 mg/L of caffeic acid from inedible cellulose through simultaneous saccharification and fermentation (SSF) reactions using a recombinant E. coli strain. Sakae et al. (2023) produce 6.17 g/L of caffeic acid using a modified M9Y medium with 50 g/L glucose in a jar fermenter.
5 Conclusion and future prospect
Caffeic acid, an important natural compound of plants, is not only widely used in the pharmaceutical and cosmetic industries but also frequently serves as an intermediate in the biosynthesis of many high-value natural compounds, such as caffeic acid phenethyl ester and chlorogenic acid, thus possessing significant commercial value. In recent years, the demand for caffeic acid has been growing continuously with the improvement of people’s living standards. The construction of cell factories for the synthesis of caffeic acid provides a green and sustainable alternative to traditional plant extraction and chemical synthesis methods. By utilizing renewable resources, avoiding the use of toxic solvents and complex chemical reactions, and minimizing waste generation, the microbial production of caffeic acid not only reduces environmental pollution but also provides significant support for the development of a circular bioeconomy. In recent years, microbial biosynthesis of caffeic acid has achieved remarkable success, with the highest reported de novo production titer reaching 6.17 g/L (Sakae et al., 2023). Microbial synthesis of caffeic acid is expected to make further progress in the following areas.
5.1 Enhancement of metabolic flux to caffeic acid biosynthesis pathway
The low activity of heterologous enzymes in microbial hosts and the insufficient supply of precursor molecules by microbial metabolism are the main factors limiting the concentration of caffeic acid. Therefore, it is necessary to combine the regulation of aromatic amino acid metabolic pathways, the screening of high-activity caffeic acid synthetase, and the enhancement of cofactors supply and recycling together to strengthen the metabolic flux of caffeic acid biosynthesis in microbial cells. In recent years, the application of machine learning algorithms in the learning phase of the design-build-test-learn cycle has been increasing. Through machine learning, it is possible to design and optimize biosynthetic pathways, analyze the relationship between protein structure and function, and optimize protein design (Kim et al., 2020; Cuperlovic-Culf et al., 2023; Sieow et al., 2023). To improve the activity of enzymes, protein engineering, particularly directed evolution, can be employed. Luo et al. (2021) reported a deep-learning algorithm named evolutionary context-integrated neural network, which leverages evolutionary contexts to predict functional fitness for protein engineering, enabling accurate mapping from sequence to function and generalization from low-order to higher-order mutants. The optimization of each step in the biosynthetic pathway should be carried out to ensure that every metabolite and precursor is directed toward the final product. Additionally, a comprehensive understanding of the synthetic and molecular biology of each component involved in caffeic acid biosynthesis, including the genome, transcriptome, proteome, and metabolome, can significantly enhance the efficiency of production from microbial platforms.
5.2 Screening and construction of suitable microbial hosts
Currently, the common microbial host for caffeic acid biosynthesis are E. coli and S. cerevisiae. On the one hand, C4H, which is required for the synthesis of caffeic acid, belongs to the cytochrome P450 dependent monooxygenases and is difficult to express in E. coli. Therefore, caffeic acid can only be synthesized through the L-tyrosine derived pathway in E. coli. As a eukaryote, S. cerevisiae has certain advantages in expressing plant-derived P450 enzymes. In S. cerevisiae, caffeic acid can be synthesized through both the L-tyrosine derived pathway and the L-phenylalanine derived pathway, which greatly increases the yield of caffeic acid. On the other hand, high concentrations of caffeic acid and p-coumaric acid have certain inhibitory effects on the growth of E. coli and S. cerevisiae. With the continuous improvement of caffeic acid production, the tolerance of microbial host to caffeic acid and p-coumaric acid will become a restrictive factor. Research indicates that E. coli has a higher tolerance to caffeic acid and p-coumaric acid than S. cerevisiae. In the future, the mechanisms by which caffeic acid and p-coumaric acid inhibit the growth of microbial hosts can be deeply analyzed, and the microbial hosts can be modified in a “top-down” manner or engineered through “bottom-up” synthetic biology design to construct strains with high tolerance to caffeic acid and p-coumaric acid. Some approaches, such as adaptive laboratory evolution and multi-functional genome-wide CRISPR system, can be designed to enhance the tolerance of microbial host to caffeic acid and p-coumaric acid while enhancing the yield of caffeic acid. Wang et al. (2021) employed an adaptive laboratory evolution approach with increasing concentrations of ferulic acid to significantly enhance the tolerance of the Yarrowia lipolytica strain XYL+ to ferulic acid and identified the tolerance-related genes via transcriptomic analysis of the evolved strain. Lian et al. (2019) enhanced the furfural tolerance of yeast using the multi-functional genome-wide CRISPR system, which can identify genetic determinants of complex phenotypes that have not been previously characterized, especially those genes that interact synergistically when perturbed to different expression levels. In addition, a co-culture system of strains can also be established to fully leverage the advantages of different microbial hosts for the production of caffeic acid.
5.3 Establishment of a low cost, high efficiency industrial production process of caffeic acid
At present, the biosynthesis of caffeic acid is still at the laboratory level. The carbon source for microbial fermentation is mostly glucose. Compared to glucose, cellulose is mainly sourced from low-cost agricultural residues, such as straw and wood chips. With the advancement of cellulase technology and the optimization of pretreatment processes, utilizing cellulose as a carbon source for the industrial-scale production of bio-products may not only reduce production costs but also provide support for the development of a circular economy. In the future, it is possible to develop cheap fermentation raw materials such as cellulose, optimize and scale up the fermentation process, reduce the difficulty and cost of separation and purification, and establish an industrial production process for caffeic acid that is low-cost and highly efficient.
Author contributions
YLi: Funding acquisition, Project administration, Writing – original draft, Writing – review & editing, Investigation. JL: Investigation, Writing – review & editing. MZ: Investigation, Writing – review & editing. YLia: Supervision, Writing – review & editing. FW: Project administration, Supervision, Writing – review & editing. MQ: Project administration, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the National Natural Science Foundation of China (32100068) and the General Projects of Science and Technology Program of Beijing Municipal Education Commission (KM202210011009).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Alson, S. G., Jansen, O., Cieckiewicz, E., Rakotoarimanana, H., Rafatro, H., Degotte, G., et al. (2018). In-vitro and in-vivo antimalarial activity of caffeic acid and some of its derivatives. J. Pharm. Pharmacol. 70, 1349–1356. doi: 10.1111/jphp.12982
Berner, M., Krug, D., Bihlmaier, C., Vente, A., Muller, R., and Bechthold, A. (2006). Genes and enzymes involved in caffeic acid biosynthesis in the actinomycete Saccharothrix espanaensis. J. Bacteriol. 188, 2666–2673. doi: 10.1128/JB.188.7.2666-2673.2006
Borodina, I., and Nielsen, J. (2014). Advances in metabolic engineering of yeast Saccharomyces cerevisiae for production of chemicals. Biotechnol. J. 9, 609–620. doi: 10.1002/biot.201300445
Bostick, C. D., Hickey, K. M., Wollenberg, L. A., Flora, D. R., Tracy, T. S., and Gannett, P. M. (2016). Immobilized cytochrome P 450 for monitoring of P 450-P 450 interactions and metabolism. Drug Metab. Dispos. 44, 741–749. doi: 10.1124/dmd.115.067637
Cai, M., Liu, J. Y., Song, X. F., Qi, H., Li, Y. Z., Wu, Z. Z., et al. (2022). De novo biosynthesis of p-coumaric acid and caffeic acid from carboxymethyl-cellulose by microbial co-culture strategy. Microb. Cell Factories 21:81. doi: 10.1186/s12934-022-01805-5
Chen, R. B., Gao, J. Q., Yu, W., Chen, X. H., Zhai, X. X., Chen, Y., et al. (2022a). Author correction: engineering cofactor supply and recycling to drive phenolic acid biosynthesis in yeast. Nat. Chem. Biol. 18:1032. doi: 10.1038/s41589-022-01129-w
Chen, R. B., Gao, J. Q., Yu, W., Chen, X. H., Zhai, X. X., Chen, Y., et al. (2022b). Engineering cofactor supply and recycling to drive phenolic acid biosynthesis in yeast. Nat. Chem. Biol. 18, 520–529. doi: 10.1038/s41589-022-01014-6
Cheniany, M., and Ganjeali, A. (2016). Developmental role of phenylalanine-ammonia-lyase (PAL) and cinnamate 4-hydroxylase (C4H) genes during adventitious rooting of Juglans regia L. microshoots. Acta Biol. Hung. 67, 379–392. doi: 10.1556/018.67.2016.4.4
Choi, O., Wu, C. Z., Kang, S. Y., Ahn, J. S., Uhm, T. B., and Hong, Y. S. (2011). Biosynthesis of plant-specific phenylpropanoids by construction of an artificial biosynthetic pathway in Escherichia coli. J. Ind. Microbiol. Biotechnol. 38, 1657–1665. doi: 10.1007/s10295-011-0954-3
Cravens, A., Payne, J., and Smolke, C. D. (2019). Synthetic biology strategies for microbial biosynthesis of plant natural products. Nat. Commun. 10:2142. doi: 10.1038/s41467-019-09848-w
Cuperlovic-Culf, M., Nguyen-Tran, T., and Bennett, S. A. L. (2023). Machine learning and hybrid methods for metabolic pathway modeling. Methods Mol. Biol. 2553, 417–439. doi: 10.1007/978-1-0716-2617-7_18
Davy, A. M., Kildegaard, H. F., and Andersen, M. R. (2017). Cell factory engineering. Cell Syst 4, 262–275. doi: 10.1016/j.cels.2017.02.010
Effendi, S. S. W., and Ng, I. S. (2023). High value ferulic acid biosynthesis using modular design and spent coffee ground in engineered Escherichia coli chassis. Bioresour. Technol. 384:129262. doi: 10.1016/j.biortech.2023.129262
Espindola, K. M. M., Ferreira, R. G., Narvaez, L. E. M., Silva Rosario, A. C. R., da Silva, A. H. M., Silva, A. G. B., et al. (2019). Chemical and pharmacological aspects of caffeic acid and its activity in hepatocarcinoma. Front. Oncol. 9:541. doi: 10.3389/fonc.2019.00541
Fieschi, F., Niviere, V., Frier, C., Decout, J. L., and Fontecave, M. (1995). The mechanism and substrate specificity of the NADPH: flavin oxidoreductase from Escherichia coli. J. Biol. Chem. 270, 30392–30400. doi: 10.1074/jbc.270.51.30392
Fontecave, M., Eliasson, R., and Reichard, P. (1987). NAD (P)H: flavin oxidoreductase of Escherichia coli. A ferric iron reductase participating in the generation of the free radical of ribonucleotide reductase. J. Biol. Chem. 262, 12325–12331. doi: 10.1016/S0021-9258(18)45356-2
Furuya, T., Arai, Y., and Kino, K. (2012). Biotechnological production of caffeic acid by bacterial cytochrome P 450 CYP199A2. Appl. Environ. Microbiol. 78, 6087–6094. doi: 10.1128/AEM.01103-12
Furuya, T., and Kino, K. (2014). Catalytic activity of the two-component flavin-dependent monooxygenase from Pseudomonas aeruginosa toward cinnamic acid derivatives. Appl. Microbiol. Biotechnol. 98, 1145–1154. doi: 10.1007/s00253-013-4958-y
Galan, B., Diaz, E., Prieto, M. A., and Garcia, J. L. (2000). Functional analysis of the small component of the 4-hydroxyphenylacetate 3-monooxygenase of Escherichia coli W: a prototype of a new flavin: NAD (P) H reductase subfamily. J. Bacteriol. 182, 627–636. doi: 10.1128/JB.182.3.627-636.2000
Goers, L., Freemont, P., and Polizzi, K. M. (2014). Co-culture systems and technologies: taking synthetic biology to the next level. J. R. Soc. Interface 11:20140065. doi: 10.1098/rsif.2014.0065
Gold, N. D., Gowen, C. M., Lussier, F. X., Cautha, S. C., Mahadevan, R., and Martin, V. J. J. (2015). Metabolic engineering of a tyrosine-overproducing yeast platform using targeted metabolomics. Microb. Cell Factories 14:73. doi: 10.1186/s12934-015-0252-2
Han, X. H., Li, S. M., Sun, X. X., Zhang, J. Y., Zhang, X. R., and Bi, X. D. (2024). Preparation of imidazolium ionic liquid functionalized paper membrane for selective extraction of caffeic acid and its structural and functional analogues from Taraxaci herba. Biomed. Chromatogr. 38:e5953. doi: 10.1002/bmc.5953
Haslinger, K., and Prather, K. L. J. (2020). Heterologous caffeic acid biosynthesis in Escherichia coli is affected by choice of tyrosine ammonia lyase and redox partners for bacterial cytochrome P 450. Microb. Cell Factories 19:26. doi: 10.1186/s12934-020-01300-9
Huang, Q., Lin, Y. H., and Yan, Y. J. (2013). Caffeic acid production enhancement by engineering a phenylalanine over-producing Escherichia coli strain. Biotechnol. Bioeng. 110, 3188–3196. doi: 10.1002/bit.24988
Jendresen, C. B., Stahlhut, S. G., Li, M. J., Gaspar, P., Siedler, S., Förster, J., et al. (2015). Highly active and specific tyrosine ammonia-lyases from diverse origins enable enhanced production of aromatic compounds in bacteria and Saccharomyces cerevisiae. Appl. Environ. Microbiol. 81, 4458–4476. doi: 10.1128/AEM.00405-15
Kang, S. Y., Choi, O., Lee, J. K., Hwang, B. Y., Uhm, T. B., and Hong, Y. S. (2012). Artificial biosynthesis of phenylpropanoic acids in a tyrosine overproducing Escherichia coli strain. Microb. Cell Factories 11:153. doi: 10.1186/1475-2859-11-153
Kawaguchi, H., Katsuyama, Y., Danyao, D., Kahar, P., Nakamura-Tsuruta, S., Teramura, H., et al. (2017). Caffeic acid production by simultaneous saccharification and fermentation of Kraft pulp using recombinant Escherichia coli. Appl. Microbiol. Biotechnol. 101, 5279–5290. doi: 10.1007/s00253-017-8270-0
Khan, F. A., Maalik, A., and Murtaza, G. (2016). Inhibitory mechanism against oxidative stress of caffeic acid. J. Food Drug Anal. 24, 695–702. doi: 10.1016/j.jfda.2016.05.003
Kim, G. B., Kim, W. J., Kim, H. U., and Lee, S. Y. (2020). Machine learning applications in systems metabolic engineering. Curr. Opin. Biotechnol. 64, 1–9. doi: 10.1016/j.copbio.2019.08.010
Kim, Y. H., Kwon, T., Yang, H. J., Kim, W., Youn, H., Lee, J. Y., et al. (2011). Gene engineering, purification, crystallization and preliminary X-ray diffraction of cytochrome P 450 p-coumarate-3-hydroxylase (C3H), the Arabidopsis membrane protein. Protein Expr. Purif. 79, 149–155. doi: 10.1016/j.pep.2011.04.013
Koopman, F., Beekwilder, J., Crimi, B., van Houwelingen, A., Hall, R. D., Bosch, D., et al. (2012). De novo production of the flavonoid naringenin in engineered Saccharomyces cerevisiae. Microb. Cell Factories 11:155. doi: 10.1186/1475-2859-11-155
Krivoruchko, A., and Nielsen, J. (2015). Production of natural products through metabolic engineering of Saccharomyces cerevisiae. Curr. Opin. Biotechnol. 35, 7–15. doi: 10.1016/j.copbio.2014.12.004
Li, Y. Z., Mao, J. W., Liu, Q. L., Song, X. F., Wu, Y. Z., Cai, M., et al. (2020). De novo biosynthesis of caffeic acid from glucose by engineered Saccharomyces cerevisiae. ACS Synth. Biol. 9, 756–765. doi: 10.1021/acssynbio.9b00431
Lian, J. Z., Schultz, C., Cao, M. F., Hamedi Rad, M., and Zhao, H. M. (2019). Multi-functional genome-wide CRISPR system for high throughput genotype-phenotype mapping. Nat. Commun. 10:5794. doi: 10.1038/s41467-019-13621-4
Lin, Y. H., and Yan, Y. J. (2012). Biosynthesis of caffeic acid in Escherichia coli using its endogenous hydroxylase complex. Microb. Cell Factories 11:42. doi: 10.1186/1475-2859-11-42
Liu, X. L., Lin, J., Hu, H. F., Zhou, B., and Zhu, B. Q. (2016). De novo biosynthesis of resveratrol by site-specific integration of heterologous genes in Escherichia coli. FEMS Microbiol. Lett. 363:fnw061. doi: 10.1093/femsle/fnw061
Liu, L. Q., Liu, H., Zhang, W., Yao, M. D., Li, B. Z., Liu, D., et al. (2019). Engineering the biosynthesis of caffeic acid in Saccharomyces cerevisiae with heterologous enzyme combinations. Engineering 5, 287–295. doi: 10.1016/j.eng.2018.11.029
Liu, Q. L., Yu, T., Li, X. W., Chen, Y., Campbell, K., Nielsen, J., et al. (2019). Rewiring carbon metabolism in yeast for high level production of aromatic chemicals. Nat. Commun. 10:4976. doi: 10.1038/s41467-019-12961-5
Luo, Y., Jiang, G. D., Yu, T. H., Liu, Y., Vo, L., Ding, H. T., et al. (2021). ECNet is an evolutionary context-integrated deep learning framework for protein engineering. Nat. Commun. 12:5743. doi: 10.1038/s41467-021-25976-8
Makrides, S. C. (1996). Strategies for achieving high-level expression of genes in Escherichia coli. Microbiol. Rev. 60, 512–538. doi: 10.1128/mr.60.3.512-538.1996
Mao, J. W., Liu, Q. L., Song, X. F., Wang, H. S. Y., Feng, H., Xu, H. J., et al. (2017). Combinatorial analysis of enzymatic bottlenecks of l-tyrosine pathway by p-coumaric acid production in Saccharomyces cerevisiae. Biotechnol. Lett. 39, 977–982. doi: 10.1007/s10529-017-2322-5
Marienhagen, J., and Bott, M. (2013). Metabolic engineering of microorganisms for the synthesis of plant natural products. J. Biotechnol. 163, 166–178. doi: 10.1016/j.jbiotec.2012.06.001
Nakayama, S., Kiyoshi, K., Kadokura, T., and Nakazato, A. (2011). Butanol production from crystalline cellulose by cocultured Clostridium thermocellum and Clostridium saccharoperbutylacetonicum N1-4. Appl. Environ. Microbiol. 77, 6470–6475. doi: 10.1128/AEM.00706-11
Pavlikova, N. (2022). Caffeic acid and diseases-mechanisms of action. Int. J. Mol. Sci. 24:588. doi: 10.3390/ijms24010588
Pereira, R., Wei, Y. J., Mohamed, E., Radi, M., Malina, C., Herrgard, M. J., et al. (2019). Adaptive laboratory evolution of tolerance to dicarboxylic acids in Saccharomyces cerevisiae. Metab. Eng. 56, 130–141. doi: 10.1016/j.ymben.2019.09.008
Phillips, I. R., and Shephard, E. A. (2008). Flavin-containing monooxygenases: mutations, disease and drug response. Trends Pharmacol. Sci. 29, 294–301. doi: 10.1016/j.tips.2008.03.004
Qi, H., Yu, L., Li, Y. Z., Cai, M., He, J. Z., Liu, J. Y., et al. (2022). Developing multi-copy chromosomal integration strategies for heterologous biosynthesis of caffeic acid in Saccharomyces cerevisiae. Front. Microbiol. 13:851706. doi: 10.3389/fmicb.2022.851706
Qian, S. Q., Lu, M. Q., Zhou, X. R., Sun, S. S., Han, Z. L., and Song, H. W. (2024). Improvement in caffeic acid and ferulic acid extraction by oscillation-assisted mild hydrothermal pretreatment from sorghum straws. Bioresour. Technol. 396:130442. doi: 10.1016/j.biortech.2024.130442
Rainha, J., Gomes, D., Rodrigues, L. R., and Rodrigues, J. L. (2020). Synthetic biology approaches to engineer Saccharomyces cerevisiae towards the industrial production of valuable polyphenolic compounds. Life 10:56. doi: 10.3390/life10050056
Rodrigues, J. L., Araujo, R. G., Prather, K. L. J., Kluskens, L. D., and Rodrigues, L. R. (2015). Heterologous production of caffeic acid from tyrosine in Escherichia coli. Enzym. Microb. Technol. 71, 36–44. doi: 10.1016/j.enzmictec.2015.01.001
Rodrigues, J. L., Gomes, D., and Rodrigues, L. R. (2020). A combinatorial approach to optimize the production of Curcuminoids from tyrosine in Escherichia coli. Front. Bioeng. Biotechnol. 8:59. doi: 10.3389/fbioe.2020.00059
Sadeghi, M., Dehghan, S., Fischer, R., Wenzel, U., Vilcinskas, A., Kavousi, H. R., et al. (2013). Isolation and characterization of isochorismate synthase and cinnamate 4-hydroxylase during salinity stress, wounding, and salicylic acid treatment in Carthamus tinctorius. Plant Signal. Behav. 8:e27335. doi: 10.4161/psb.27335
Sakae, K., Nonaka, D., Kishida, M., Hirata, Y., Fujiwara, R., Kondo, A., et al. (2023). Caffeic acid production from glucose using metabolically engineered Escherichia coli. Enzym. Microb. Technol. 164:110193. doi: 10.1016/j.enzmictec.2023.110193
Shin, S. Y., Han, N. S., Park, Y. C., Kim, M. D., and Seo, J. H. (2011). Production of resveratrol from p-coumaric acid in recombinant Saccharomyces cerevisiae expressing 4-coumarate: coenzyme a ligase and stilbene synthase genes. Enzym. Microb. Technol. 48, 48–53. doi: 10.1016/j.enzmictec.2010.09.004
Shrestha, A., Pandey, R. P., and Sohng, J. K. (2019). Biosynthesis of resveratrol and piceatannol in engineered microbial strains: achievements and perspectives. Appl. Microbiol. Biotechnol. 103, 2959–2972. doi: 10.1007/s00253-019-09672-8
Sieow, B. F., Sotto, R. D., Seet, Z. R. D., Hwang, I. Y., and Chang, M. W. (2023). Synthetic biology meets machine learning. Methods Mol. Biol. 2553, 21–39. doi: 10.1007/978-1-0716-2617-7_2
Teng, Y. N., Wang, C. C. N., Liao, W. C., Lan, Y. H., and Hung, C. C. (2020). Caffeic acid attenuates multi-drug resistance in cancer cells by inhibiting efflux function of human P-glycoprotein. Molecules 25:247. doi: 10.3390/molecules25020247
Touaibia, M., Jean-Francois, J., and Doiron, J. (2011). Caffeic acid, a versatile pharmacophore: an overview. Mini Rev. Med. Chem. 11, 695–713. doi: 10.2174/138955711796268750
Wang, J., Guleria, S., Koffas, M. A., and Yan, Y. J. (2016). Microbial production of value-added nutraceuticals. Curr. Opin. Biotechnol. 37, 97–104. doi: 10.1016/j.copbio.2015.11.003
Wang, J., Mahajani, M., Jackson, S. L., Yang, Y. P., Chen, M. Y., Ferreira, E. M., et al. (2017). Engineering a bacterial platform for total biosynthesis of caffeic acid derived phenethyl esters and amides. Metab. Eng. 44, 89–99. doi: 10.1016/j.ymben.2017.09.011
Wang, Z. D., Zhou, L. L., Lu, M. R., Zhang, Y. W., Perveen, S., Zhou, H. R., et al. (2021). Adaptive laboratory evolution of Yarrowia lipolytica improves ferulic acid tolerance. Appl. Microbiol. Biotechnol. 105, 1745–1758. doi: 10.1007/s00253-021-11130-3
Wu, J. J., Liu, P. R., Fan, Y. M., Bao, H., Du, G. C., Zhou, J. W., et al. (2013). Multivariate modular metabolic engineering of Escherichia coli to produce resveratrol from L-tyrosine. J. Biotechnol. 167, 404–411. doi: 10.1016/j.jbiotec.2013.07.030
Xiao, F., Lian, J. Z., Tu, S., Xie, L. L., Li, J., Zhang, F. M., et al. (2022). Metabolic engineering of Saccharomyces cerevisiae for high-level production of chlorogenic acid from glucose. ACS Synth. Biol. 11, 800–811. doi: 10.1021/acssynbio.1c00487
Xue, Y., Zhang, Y., Grace, S., and He, Q. F. (2014). Functional expression of an Arabidopsis P450 enzyme, p-coumarate-3-hydroxylase, in the cyanobacterium Synechocystis PCC 6803 for the biosynthesis of caffeic acid. J. Appl. Phycol. 26, 219–226. doi: 10.1007/s10811-013-0113-5
Xun, L., and Sandvik, E. R. (2000). Characterization of 4-hydroxyphenylacetate 3-hydroxylase (HpaB) of Escherichia coli as a reduced flavin adenine dinucleotide-utilizing monooxygenase. Appl. Environ. Microbiol. 66, 481–486. doi: 10.1128/AEM.66.2.481-486.2000
Yang, S. Y., Hong, C. O., Lee, G. P., Kim, C. T., and Lee, K. W. (2013). The hepatoprotection of caffeic acid and rosmarinic acid, major compounds of Perilla frutescens, against t-BHP-induced oxidative liver damage. Food Chem. Toxicol. 55, 92–99. doi: 10.1016/j.fct.2012.12.042
Yang, J. Z., Liang, J. C., Shao, L., Liu, L. H., Gao, K., Zhang, J. L., et al. (2020). Green production of silybin and isosilybin by merging metabolic engineering approaches and enzymatic catalysis. Metab. Eng. 59, 44–52. doi: 10.1016/j.ymben.2020.01.007
Ye, J. C., Hsiao, M. W., Hsieh, C. H., Wu, W. C., Hung, Y. C., and Chang, W. C. (2010). Analysis of caffeic acid extraction from Ocimum gratissimum Linn. By high performance liquid chromatography and its effects on a cervical cancer cell line. Taiwan. J. Obstet. Gynecol. 49, 266–271. doi: 10.1016/S1028-4559(10)60059-9
Yesilirmak, F., and Sayers, Z. (2009). Heterelogous expression of plant genes. Int. J. Plant Genomics 2009:296482. doi: 10.1155/2009/296482
Zhang, W., Liu, H., Li, X., Liu, D., Dong, X. T., Li, F. F., et al. (2017). Production of naringenin from D-xylose with co-culture of E. coli and S. cerevisiae. Eng. Life Sci. 17, 1021–1029. doi: 10.1002/elsc.201700039
Zhang, H., and Stephanopoulos, G. (2013). Engineering E. coli for caffeic acid biosynthesis from renewable sugars. Appl. Microbiol. Biotechnol. 97, 3333–3341. doi: 10.1007/s00253-012-4544-8
Zhang, P. X., Tang, Y. P., Li, N. G., Zhu, Y., and Duan, J. A. (2014). Bioactivity and chemical synthesis of caffeic acid phenethyl ester and its derivatives. Molecules 19, 16458–16476. doi: 10.3390/molecules191016458
Zhou, P. P., Yue, C. L., Shen, B., Du, Y., Xu, N. N., and Ye, L. D. (2021). Metabolic engineering of Saccharomyces cerevisiae for enhanced production of caffeic acid. Appl. Microbiol. Biotechnol. 105, 5809–5819. doi: 10.1007/s00253-021-11445-1
Keywords: caffeic acid, microbial production, pathway engineering, metabolic engineering, synthetic biology
Citation: Li Y, Li J, Zhang M, Liao Y, Wang F and Qiao M (2025) Heterologous production of caffeic acid in microbial hosts: current status and perspectives. Front. Microbiol. 16:1570406. doi: 10.3389/fmicb.2025.1570406
Edited by:
Karthik Loganathan, Salem Microbes Pvt. Ltd., IndiaReviewed by:
Yiwen Zhang, Dalian University of Technology, ChinaA. Sai Ramesh, Prathyusha Engineering College (PEC), India
Copyright © 2025 Li, Li, Zhang, Liao, Wang and Qiao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fenghuan Wang, d2FuZ2ZlbmdodWFuQHRoLmJ0YnUuZWR1LmNu; Mingqiang Qiao, cWlhb21xQG5hbmthaS5lZHUuY24=
 Yuanzi Li
Yuanzi Li Jiaxin Li
Jiaxin Li Miao Zhang1,2
Miao Zhang1,2 Fenghuan Wang
Fenghuan Wang Mingqiang Qiao
Mingqiang Qiao