- 1Department of Biochemistry, Indian Institute of Science, Bangalore, India
- 2Department of Bioengineering, Indian Institute of Science, Bangalore, India
Lipopolysaccharide (LPS) O-antigen and enterobacterial common antigen (ECA) play crucial roles in maintaining the outer membrane in Gram-negative bacteria. Mutations in the biosynthetic pathways of LPS and ECA may lead to accumulation of intermediates, resulting in morphological changes and activation of stress responses. However, the functional consequences of abrogation of both O-antigen and ECA synthesis in Salmonella enterica serovar Typhimurium (S. Typhimurium) are not well investigated. In this study, we generated single and double-deletion mutants of rfbB and rffG, encoding dTDP-glucose 4,6-dehydratase paralogs that are important in the synthesis of both O-antigen and ECA. Importantly, mutations in the dTDP-D-glucose 4,6-dehydratase encoding gene in humans are known to cause Catel-Manzke syndrome, a rare genetic disease. All four strains, i.e., wild type (WT), ΔrfbB, ΔrffG and ΔrfbBΔrffG, grew well in rich Luria Bertani (LB) liquid media at 37°C; however, the functional loss of both rfbB and rffG, but not in single-deletion strains, resulted in round cell morphology and smaller colony size in LB agar plates. There was no significant differences in the growth of the four strains in minimal media at 37°C (nutritional deficiency), in LB at 42°C (high temperature), acidic pH or LB with 3–4% NaCl (high osmolarity; however the ΔrfbBΔrffG strain was hypersensitive to bile and cell wall-targeting antibiotics). These results demonstrated that the ΔrfbBΔrffG strain was sensitive to some stress conditions. Interestingly, the ΔrfbBΔrffG strain displayed an altered LPS profile, autoaggregated rapidly compared to the WT and the single mutant strains and showed high N-phenylnaphthylamine (NPN) fluorescence indicating greater surface hydrophobicity. Furthermore, transcriptomic analysis identified flagellar and SPI-1 pathways to be highly downregulated in ΔrfbBΔrffG which led to impaired swimming as well as swarming motility, lower adhesion and invasion of HeLa cells. Importantly, the ΔrfbBΔrffG strain was less proficient in colonizing Peyer’s patches, spleen and liver, was unable to induce pro-inflammatory cytokines and was attenuated in both the oral and intraperitoneal models of S. Typhimurium infection in mice. Overall, this study highlights the importance of rfbB and rffG in maintaining cell wall and cell membrane integrity, colony and cellular morphology, motility and virulence in S. Typhimurium.
Introduction
Salmonellosis is a global public health concern, especially with its rise as a food borne pathogen, increase in the number of antimicrobial resistant strains and high mortality by non-typhoidal strains due to co-infections in Africa (Kumar et al., 2025). The treatment of infections caused by Salmonella with the current generation of antibiotics continues to be a challenge, owing to the presence of the outer membrane. The outer membrane of Gram-negative bacteria is made up of lipopolysaccharide (LPS) and outer membrane proteins (OMPs). The presence of the LPS molecules provide a barrier which excludes toxic substances such as large hydrophobic antibiotics (Farrag et al., 2019). Another feature that is present on the outer membrane of species from the order Enterobacterales is the enterobacterial common antigen (ECA). ECA comprises repeating trisaccharide units composed of 4-acetamide-4,6 dideoxy D-galactose, N-acetyl-D-mannosaminuronic acid and N-acetyl-D-glucosamine (Gilbreath et al., 2012). The LPS, along with ECA, provides structural integrity to the outer membrane which is vital for the bacteria as their loss is associated with significant fitness cost (Spöring et al., 2018; Ilg et al., 2009).
Salmonella enterica serovar Typhimurium (S. Typhimurium) must overcome a wide array of host-mediated antimicrobial responses to establish successful infection in the host. LPS along with ECA form essential components of immunodominant molecules for these Gram-negative pathogens. Therefore, studying bacterial mutants deficient in LPS and ECA biosynthesis pathway offers a promising area of research. The enzyme, dTDP-glucose 4,6-dehydratase (RfbB) is involved in the dTDP-rhamnose biosynthesis pathway and is crucial in the formation of the LPS O-antigen, exopolysaccharides, capsular polysaccharides, and glycolipids (Supplementary Figure S1). This enzyme catalyzes the conversion of dTDP-α-D-glucose into dTDP-4-keto-6-deoxyglucose. The genes encoding the enzymes involved in the biosynthesis of O-specific polysaccharides are clustered in the rfb region (Supplementary Figure S2). However, another gene, rffG, is a paralog of rfbB, (Yao and Valvano, 1994) the product of which is another dTDP-glucose 4,6-dehydratase that catalyzes the same reaction as RfbB, but functions in the enterobacterial common antigen (ECA) pathway (Marolda and Valvano, 1995; Stevenson et al., 1994) (Supplementary Figure S1). A double deletion of these genes has been proposed to be a potential live vaccine candidate (Huang et al., 2016).
RfbB (also known as RmlB) functions as a homodimer and each monomer exhibits an α/β structure that is divided into two domains: the larger N-terminal domain that binds the nucleotide cofactor NAD+ while the C-terminal domain which is responsible for binding the sugar substrate, dTDP-D-glucose (Allard et al., 2002; Beis et al., 2003). The highly conserved active site, Tyr-X-X-X-Lys catalytic couple and the Gly-X-Gly-X-X-Gly motif at the N terminus characterize RfbB/RmlB as a member of the short-chain dehydrogenase/reductase (SDR) family. During glycan biosynthesis, TDP-dehydratase generates dTDP-rhamnose from dTDP-glucose. Undecaprenyl phosphate (UndP) and the UndP-linked oligosaccharides are important in LPS, capsular polysaccharides, peptidoglycan and additional pathways (Supplementary Figure S1). Consequently, disruptions in the biosynthetic pathways of LPS and ECA biosynthesis can lead accumulation of intermediates, resulting in morphological changes and activation of stress pathways (Castelli and Véscovi, 2011; Jorgenson and Young, 2016; Jorgenson et al., 2016; Castelli et al., 2008; Jorgenson and Bryant, 2021). Importantly, mutations in the dTDP-D-glucose 4,6-dehydratase encoding gene in humans are known to cause Catel-Manzke syndrome, a rare bone disease often characterized by abnormalities in the index finger (Ehmke et al., 2014).
Previously, our laboratory was involved in a collaborative study on the functional roles of UDP-glucose 4,6 dehydratase in Candida albicans (Sen et al., 2011). Studies conducted with the single-deletion mutants of either O-antigen or ECA have shown the importance of these two pathways on the physiology of S. Typhimurium (Rai Ashutosh and Mitchell Angela, 2020; Marshall and Gunn, 2015; Park et al., 2018; Huang et al., 2016). One study has shown that a strain harboring double deletion of rffG and rfbB has a potential to be a live vaccine candidate (Huang et al., 2016). However, there is paucity of information on the detailed characterization of the direct as well as indirect effects of the loss of both O-antigen and ECA in S. Typhimurium. With this objective in mind, we initiated this study and generated single and double-deletion mutants of rffG and rfbB to study the functional loss of dTDP-glucose 4,6-dehydratase. The broad objective of this study was to understand the consequences of the combinatorial loss of O-antigen and the ECA and its effects on the physiology and virulence of S. Typhimurium.
Materials and methods
Bacterial strains and growth conditions
The bacterial strains used in this study are listed in Supplementary Table S1. All bacterial cultures were grown in Luria-Bertani (LB) medium consisting of 10 g/L tryptone (HiMedia Laboratories, Mumbai, India), 10 g/L NaCl (Merck, Darmstadt, Germany), and 5 g/L yeast extract (HiMedia Laboratories) at 37°C. Strains containing pKD46 were cultured at 30°C with constant shaking at 160 rpm. LB containing antibiotics were used at the following concentrations: kanamycin, 50 μg/mL; chloramphenicol, 30 μg/mL; tetracycline, 10 μg/mL and/or ampicillin, 100 μg/mL.
Generation of knockout strains and complementation
All single-deletion mutants used in this study were generated using the one-step gene disruption strategy (Datsenko and Wanner, 2000). S. Typhimurium 14028s (WT) was used as the parent strain for all experiments. For the construction of ΔrffG, primers listed in Supplementary Table S2 (Sigma, Bangalore, India) were designed to amplify the kanamycin cassette from the template, pKD4. The resulting PCR-amplified product was purified and electroporated into the WT strain harboring pKD46, which expresses the λ Red recombinase. A similar methodology was followed for generating the ΔrfbB strain, where the gene was replaced with the chloramphenicol cassette (pKD3 template). The double deletion strain, ΔrfbBΔrffG was constructed by amplifying rfbB gene fragment and electroporating the amplicon into ΔrffG strain harboring pKD46. All the mutants were confirmed by PCR amplification using primers designed to anneal ~100 bp upstream and downstream of the gene (Supplementary Table S2).
Cloning of genes for complementation
Salmonella Typhimurium 14028s genomic DNA was used as a template for the PCR amplification of genes rffG and rfbB with the specific primers (Supplementary Table S2) using the Phusion DNA polymerase. The genes, rffG and rfbB were cloned between the sites, NcoI and HindIII in the pACDH plasmid (Rao and Varshney, 2001). Positive clones were confirmed by Sanger sequencing (Aggrigenome, India) and transformed into appropriate strains for complementation.
Stress assays
Salmonella Typhimurium WT, ∆rfbB, ∆rffG and ∆rfbB∆rffG strains were grown in 3 mL LB or LB with appropriate concentration of antibiotics overnight at 37°C with 160 rpm. The optical density at 600 nm (OD600) of the pre-inoculum was normalized to 2.0, and 50 μL of the culture (normalized pre-inoculum) was added to 5 mL of LB without or with different concentrations of either bile salts (Sigma Aldrich, USA), NaCl or different pH range. The cultures were incubated at 37°C under shaking conditions at 160 rpm for 6–8 h and OD600 were measured in an UV–Visible spectrophotometer (Shimadzu) (Ray et al., 2019).
RNA isolation and cDNA synthesis
For total RNA preparation, bacterial cultures were grown for 3 h in LB, pelleted and stabilized with RNAprotect Bacteria Reagent (Qiagen) and stored at −80°C. Total RNA was extracted using TRIzol (Sigma, St. Louise, Missouri, USA). Subsequently, 2–5 μg DNase-treated RNA was reversed transcribed to cDNA using the RevertAid First Strand cDNA synthesis kit (Thermo Fisher Scientific, Walter, MA, USA) (Ray et al., 2019).
Quantitative real-time PCR
The cDNA was diluted to 1:20 and analysed using iQ5 Real time PCR detection system (BioRad, Hercules, California, USA) with SYBR Green detection system. Each sample was kept in triplicates in a 96-well plate (BioRad, Hercules, California, USA) in total reaction mixture of 10 μL containing 2X SYBR iQ SYBR Green supermix and 10 μM primer mix. PCR conditions were as followed, 95°C for 5 min, followed by 39 cycles of 95°C for 30s, 57°C for 30s and 72°C for 30s. The amplification specificity and the primer dimer were calculated by the melt curve acquired after 81 cycles of heating PCR products from 55°C to 95°C for 20s, with 5°C increase per cycle. The untreated WT cells, at the respective time points were normalized using the reference control gene, gmk, and all other samples were calculated as fold change to this reference value, using the 2-ΔΔCt method (Livak and Schmittgen, 2001).
To estimate qPCR primer efficiency, RNA was isolated from WT 14028s at the indicated time point using the TRIzol method. cDNA was synthesized as described previously and serially diluted at 1:10, 1:100, 1:1000, and 1:10,000. Real-time PCR was performed under the conditions mentioned above. The Ct values were plotted against the cDNA concentration, and the slope of the standard curve was calculated. Primer efficiency and amplification factor was determined using the formula: Efficiency = 10(−1/slope)-1 (Rogers-Broadway and Karteris, 2015). The primer efficiency was calculated to be in the range of 1.7–1.8, as described in Supplementary Figure S4.
Atomic force microscopy
The bacterial cells were pelleted down after growth for indicated time by centrifugation at 6000 rpm and washed with double distilled water thrice. Around 5 μL was placed on top of a glass coverslip and the images were obtained by using NX-10 AFM (Park systems, South Korea) (Varghese et al., 2020; Verma et al., 2021). Cellular width was measured using the image processing software XEI (Park Systems, South Korea). For the ΔrfbBΔrffG strain, which are spherical, although not perfect spheres, the cellular width was measured considering the smaller dimension to be the width of the cell.
MIC determination
The MIC was determined by E-tests using Ezy-MIC strips (HiMedia, Mumbai, India) according to the manufacturer’s protocol. Briefly, the overnight grown cultures were normalized to 2.0 OD at 600 nm and spread-plated onto the Mueller Hinton agar plates by spread plate method. Pre-coated MIC strips with antibiotics were placed in the middle of the agar plates with the help of a sterile swab and the plates were left in an incubator maintained at 37°C for 18 h. The MIC was read at the point of the strip at which zone of clearance coincided with the strip (Bhaskarla et al., 2016).
Analysis of the LPS profile
Bacterial cells were grown for 16 h, and the OD was normalized to 2.0 at 600 nm. Cells were centrifuged and the pellets were suspended in 150 mL of lysis buffer containing proteinase K (Thermo Scientific, Walter, MA, USA) followed by hot phenol extraction and a subsequent extraction of the aqueous phase with diethyl ether. LPS was separated on 12% (w/v) acrylamide gels using a Tricine-SDS buffer system and visualized by silver staining (Hitchcock and Brown, 1983).
NPN assay
The outer membrane permeability was measured with the help of NPN assay as previously described (Spöring et al., 2018). Briefly, the cells were grown till OD of 0.5 at 600 nm and the cells were harvested by centrifugation. The pellet was washed with 5 mM HEPES (pH 7.2) and adjusted to an OD of 0.5 at 600 nm. NPN was added to each well at the final concentration of 10 μM. 200 μL was added to a flat bottom 96-well plate and fluorescence excitation and emission was measured at 350 nm and 420 nm, respectively.
Auto-aggregation assay
The bacterial growth for every strain was normalized to OD 2.0 at 600 nm and the cultures were kept in a static, upright position for 30 min. At regular intervals of 10 min, 100 μL of the culture was collected from the top and the absorbance was measured at 600 nm. Absorbance at 600 nm versus time was plotted over a 30-min time interval (Raghunathan et al., 2011).
Motility assays
Bacterial motility assays were performed as described previously (Ray et al., 2020; Thakur et al., 2020). Briefly, swimming motility assay was performed on a freshly prepared 0.3% agar whereas for swarming motility assays, 0.5% agar was used along with 0.5% glucose (HiMedia, Mumbai, India). For both types of motility assays, 2 μL of overnight culture normalized to 1 OD600 were inoculated in the center of the plates, and the plates were incubated at 37°C for 8 h and then imaged using the ImageQuant LAS4000 (GE Healthcare).
HeLa cell and mice infections
The detailed protocol followed for HeLa cell infections are in the Supplementary methods. Six to eight-week old, male, C57BL/6 mice (n = 4–6 mice per group) were orally infected with ~1 × 108 CFU/mouse or intra-peritoneally (i.p.) with ~1 × 103 CFU/mouse of either the WT or the ΔrfbBΔrffG strain (Majumdar et al., 2017). Organ bacterial burden was estimated 4 days after infection. The organs were harvested, weighed, and homogenized in 1 mL sterile PBS. Appropriate dilutions were plated on LB agar plates to enumerate CFU as log10(CFU/gram tissue weight).
Measurement of cytokines in sera
The amounts of TNFα, IL6 and IFNγ in sera were quantified using sandwich ELISA (Yadav et al., 2018) using the appropriate kits (Thermo Fisher Scientific, USA). Sera were collected at day 4–5 post oral gavage or day 3 post i.p. injection. 3,3′,5,5’-Tetramethylbenzidine (TMB) was used as the chromogenic substrate, and absorbance was recorded at 450 nm using VersaMax Microplate Reader (Molecular Devices, USA).
Other assays
The detailed protocols involved in multiple sequence alignment (MSA), RNA sequencing and data analysis, isolation of outer membrane proteins (OMP) and adhesion and invasion assays are described in detail in the Supplementary methods section.
Statistical analysis
All graphs were plotted, and the statistical analyses performed using GraphPad Prism 8 (v 8.0.2) software (GraphPad, La Jolla, CA). For most experiments, statistical analyses were performed using either one-way or two-way ANOVA. Data are represented as mean ± SEM, where * p < 0.05, ∗∗ p < 0.01, ∗∗∗ p < 0.001 and ∗∗∗∗ p < 0.0001. For in vivo infection studies, statistical analyses of mice survival curves post infection were performed using the log-rank (Mantel-Cox) test. For estimation of cytokines, statistical analysis was performed using the Kruskal Wallis test, where * p < 0.05; ** p < 0.01 and *** p < 0.001.
Results
Multiple sequence alignment of RffG and RfbB protein homologs indicate presence of a conserved YXXXK motif
The genes under investigation, rffG and rfbB are paralogs, (Yao and Valvano, 1994) each encoding the enzyme dTDP-glucose 4,6-dehydratase (Supplementary Figure S1) and are organized as part of distinct operons (Supplementary Figure S2). A sequence analysis was performed to investigate the protein sequence conservation from lower to higher organisms and to also analyze whether the known functional motifs are conserved. To obtain homologs, the protein FASTA sequences of RfbB (361aa, locus id: STM14_2591) and RffG (355aa, locus id, STM14_4720) from S. Typhimurium 14028s were retrieved from NCBI. These sequences were then independently used as query sequences to perform protein BLAST against the target non-redundant protein sequence database. The hits obtained were further filtered. Multiple sequence alignment and representation of the aligned sequences were performed using CLUSTAL Omega and JalView, respectively. The crystal structure of dTDP-D-Glucose 4,6-dehydratase (RfbB or RmlB) from S. Typhimurium, shows two regions in the active site: the cavity created by the NAD+ binding region and the dTDP-D-glucose-binding regions (Allard et al., 2001). Mechanistic roles of the residues Thr133, Tyr167 and Lys171 in the catalytic motif of the enzyme have been studied through mutagenesis in Escherichia coli (Gerratana et al., 2001). We observed that eukaryotic homologs have a very low degree of sequence (25–40%) similarity. However, the Tyr (residue 167), part of the YXXXK motif is strictly conserved in this extended SDR family and is also the substrate binding site. Thr133 and Lys171 in RfbB are also highly, but not absolutely, conserved. Other highly conserved residues include the three glycine residues, with motif (Gly-X-Gly-X-X-Gly) located close to the N terminus (at positions 8, 10 and 13 in RfbB) are involved in NAD+ binding. Although the SDR signature motif (LftettayapSspYSASKASSdHLVrAWR) slightly varied in the RffG sequence, it did retain the active site Tyr and Lys residues (Supplementary Figure S3A). The other residues known to make contact with the nucleotide sugar are Thr133, Asp134, Glu135, Asn196, Arg231 and Asn266. Notably, amino acid variations were observed in Shigella, Mycobacterium, Drosophila and Saccharomyces species (Supplementary Figure S3). Overall, it was interesting to observe the presence of homologs of this enzyme from E. coli to humans.
S. Typhimurium ΔrfbBΔrffG strain shows higher optical density and a delayed lag phase compared to the WT and single-deletion mutants
To better understand the functional roles of rffG and rfbB in S. Typhimurium 14028s, single and double-deletion mutants were generated, and growth assays were performed. The WT and the single-deletion strains displayed similar growth kinetics; however, the ΔrfbBΔrffG strain attained a slightly higher optical density (OD) than the other strains beginning from the mid-exponential phase till the stationary phase (Supplementary Figure S5A). Notably, introducing the WT copy of either rffG or rfbB restored the OD to WT levels (Supplementary Figure S5B). To further investigate whether the higher OD displayed by the ΔrfbBΔrffG strain corresponded with the actual number of viable cells, appropriate dilutions were plated on agar plates at various time points. Cellular viability between the WT and the single-deletion mutants were not significantly different; however, the ΔrfbBΔrffG strain at 3 h showed lesser CFU when compared to other strains (Supplementary Figure S5C). Apart from demonstrating a prolonged lag phase in the liquid medium wrt CFU, there were no other observable growth differences in the ΔrfbBΔrffG strain compared to the WT and the single-deletion mutants.
S. Typhimurium ΔrfbBΔrffG forms smaller colonies and a distinct colony morphology on LB agar plates
Interestingly, we observed that the colonies formed by ΔrfbBΔrffG strain on LB agar plates were considerably smaller in size when compared to the WT or the single-deletion mutants (Figure 1A). Complementing with the WT copy of either of the genes restored the colony size of the ΔrfbBΔrffG to that of the WT (Figure 1C). This indicated that the ΔrfbBΔrffG strain was more compromised in growth on solid media than in liquid media. Also, we noted that the colony texture of the ΔrfbBΔrffG strain appeared much smoother compared to the WT and the single-deletion mutants (Figure 1B).
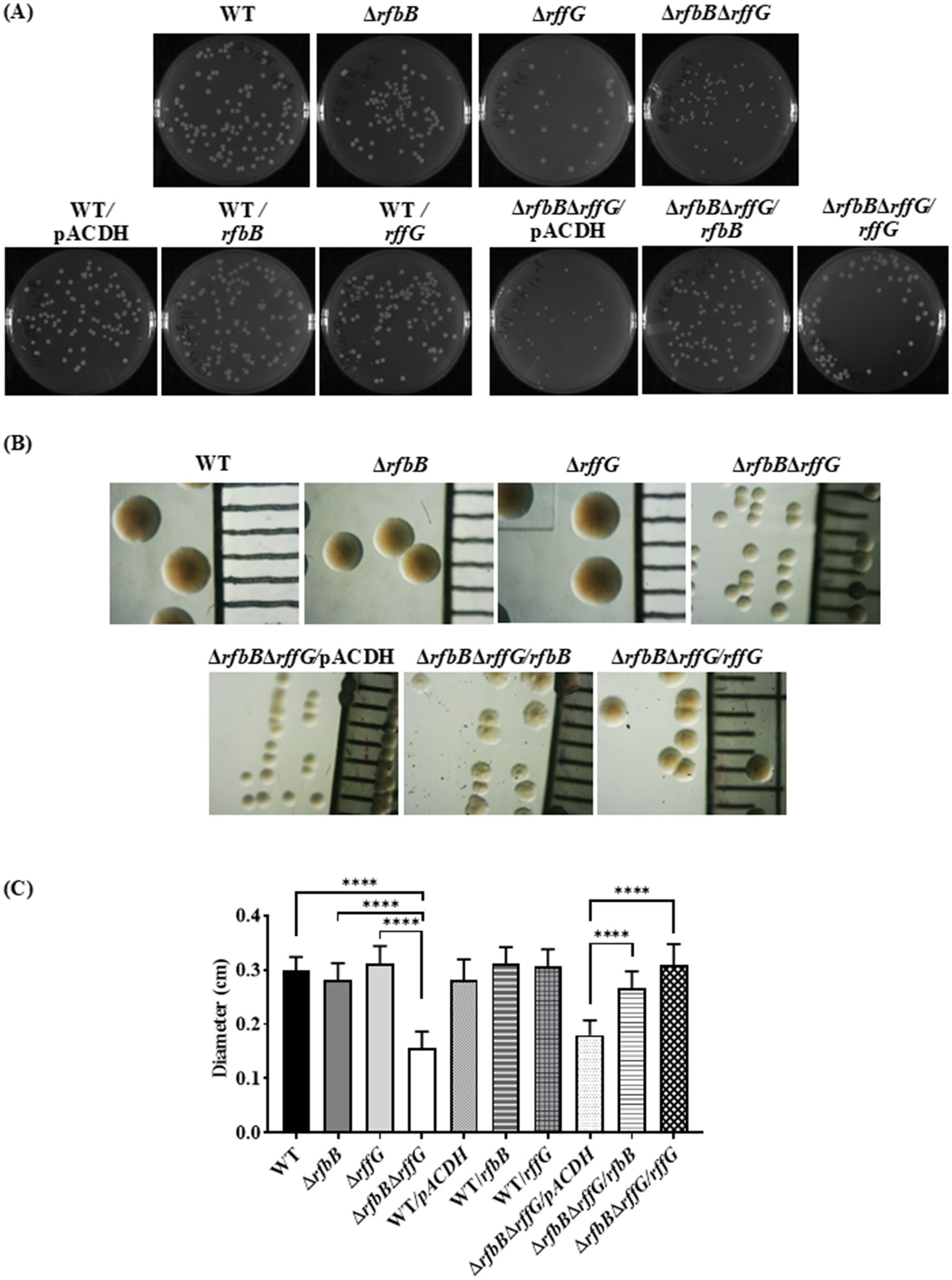
Figure 1. Salmonella Typhimurium ΔrfbBΔrffG strain forms smaller colonies on LB agar. (A) S. Typhimurium WT, ΔrfbB, ΔrffG, ΔrfbBΔrffG and complemented strains were plated on LB agar plates, and their images were captured after 16 h of incubation at 37°C. (B) Representative images depicting individual colony morphology of the above-mentioned strains obtained through a light microscope. (C) Diameter of the colony of WT, ΔrfbB, ΔrffG and ΔrfbBΔrffG and the complemented strains on LB agar plates. Statistical analysis was performed using one-way ANOVA, where ****p < 0.0001. Data are representative of three independent experiments plotted as mean ± SEM. Scale Bar for (B): 1 mm per unit.
S. Typhimurium ΔrfbBΔrffG have distinct cell morphology as revealed by atomic force microscopy
Furthermore, differences in the cellular morphology among the different strains were examined using AFM imaging in a kinetic manner (Figure 2 and Supplementary Figure S6). The cells of the ΔrfbBΔrffG strain were wider than those of the WT strain and single-deletion mutants. At 6 h and 12 h of growth, most of the cells of the ΔrfbBΔrffG strain exhibited a round or spherical morphology (Figures 2A,B). Quantification of the cellular width revealed that the ΔrfbBΔrffG strain was significantly wider than the WT strain. This phenotype was completely restored upon the expression of the WT copy of either rfbB or rffG in trans (Figure 2C).
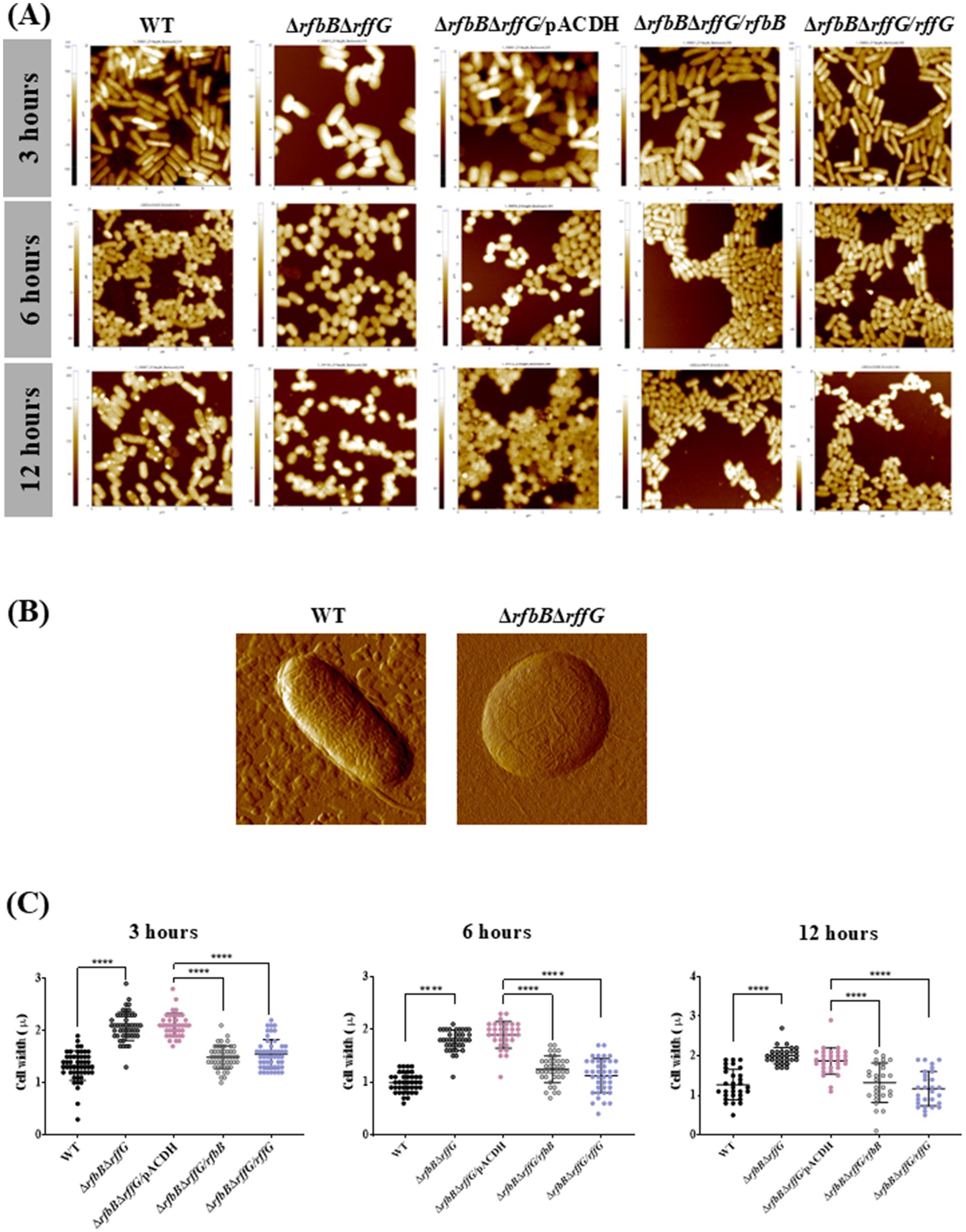
Figure 2. Complementation of S. Typhimurium ΔrfbBΔrffG strain with WT copy of rfbB or rffG restores cellular width. (A) The bacterial strains were grown for the indicated time points (3, 6 or 12 h) and AFM images were acquired in the non-contact mode. (B) Single cell images of the WT and the ΔrfbBΔrffG at 6 h. (C) Quantification of the cellular width was performed and for each condition, the width of at least 50 cells was determined. Data are representative of 3 independent experiments plotted as mean ± SEM. Statistical analysis was performed using two-way ANOVA, where * p < 0.05; ** p < 0.01; *** p < 0.001 and **** p < 0.0001.
S. Typhimurium ΔrfbBΔrffG is highly susceptible to bile stress and cell wall targeting antibiotics
The susceptibility of these strains to common stresses encountered by S. Typhimurium was evaluated. The four strains were grown in LB at 37°C, high temperature (42°C) and minimal media at 37°C; however, no significant growth alterations among the strains were noted (Figures 3A,B). Furthermore, no differences between strains were observed in pH and osmolar stresses (Figures 3C,D). Enteric pathogens such as S. Typhimurium are resistant to high concentrations of bile (van Velkinburgh and Gunn, 1999); therefore, a growth susceptibility assay was conducted by exposing the strains to different concentrations of bile. No growth differences were observed between the WT and the single-deletion mutants (Figure 3E). Strikingly, the ΔrfbBΔrffG strain exhibited a significant growth defect in a dose-dependent manner. As depicted in Figure 3F, the bile-sensitive phenotype of the ΔrfbBΔrffG strain was rescued upon complementation with the WT copy of either rffG or rfbB.
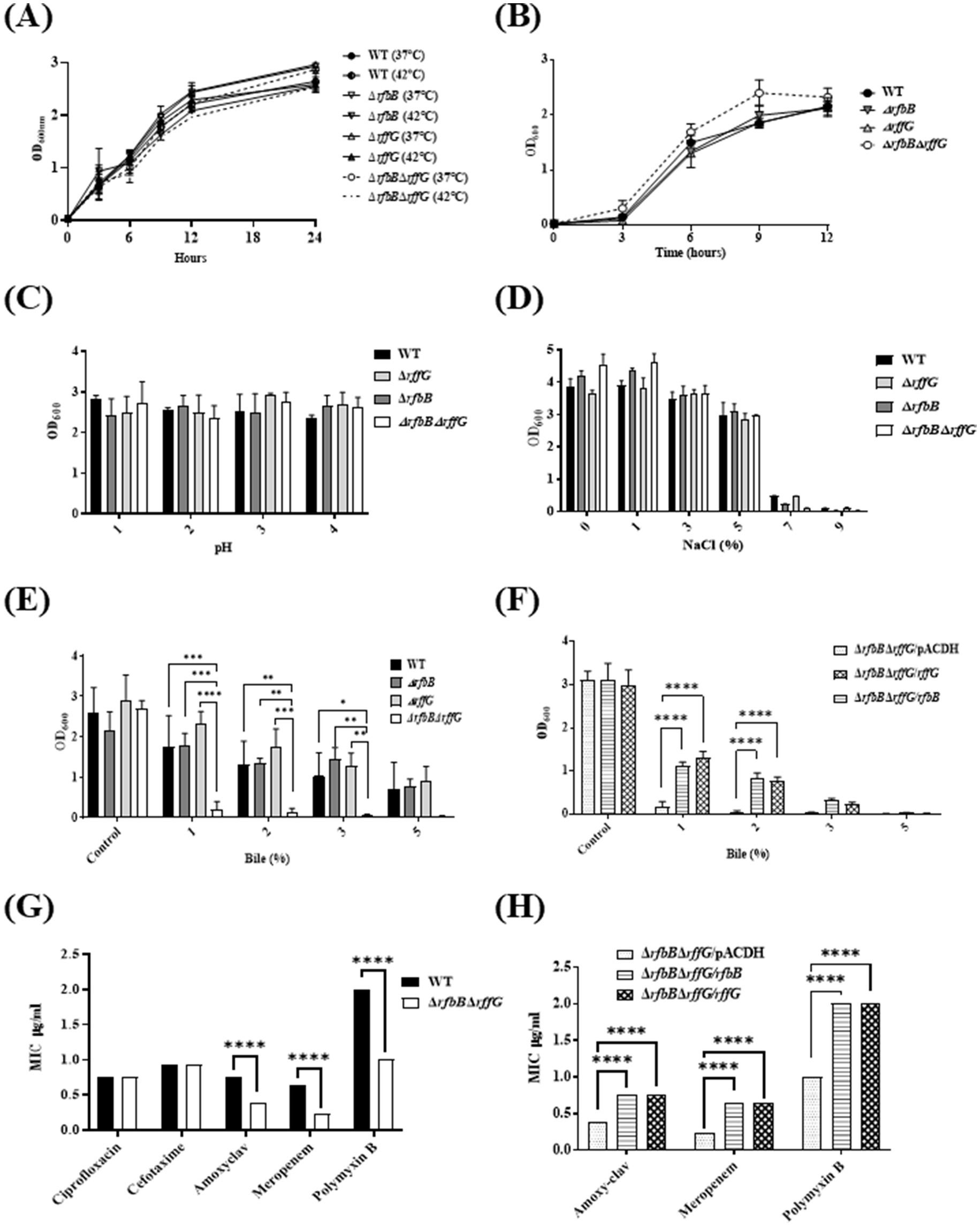
Figure 3. Salmonella Typhimurium ΔrfbBΔrffG strain is susceptible to bile and cell wall targeting antibiotics. S. Typhimurium WT and ΔrfbBΔrffG were grown (A) in LB at 37°C and 42°C (B) in minimal media. To test the effects of growth at high temperature, S. Typhimurium WT, ΔrfbB, ΔrffG, and ΔrfbBΔrffG were grown in LB and media at 37°C and 42°C overnight. Post incubation, the cultures were normalized to an optical density (OD) of 2. Subsequently, 0.2% (v/v) of normalized cultures were inoculated into LB medium and incubated for 3 h. Following this, 0.2% (v/v) of the culture was transferred to respective tubes containing LB media and incubated at 37°C and 42°C with shaking at 160 RPM. Optical density measurements were recorded at specified time points. S. Typhimurium WT, ΔrfbB, ΔrffG and ΔrfbBΔrffG growth in 5 mL LB broth (C) of different pH, and (D) with different concentrations of NaCl. (E) S. Typhimurium WT, ΔrfbB, ΔrffG and ΔrfbBΔrffG and (F) complemented strains, ΔrfbBΔrffG/pACDH, ΔrfbBΔrffG/rfbB, ΔrfbBΔrffG/rffG strains were treated with the indicated concentrations of bile and grown for 8 h at 37°C and 160 rpm. Bacterial growth was quantified by measuring OD at 600 nm. Epsilometer Test (E-Test) results to determine MIC of the (G) WT and the ΔrfbBΔrffG strains as well as the (H) complemented strains. Briefly, overnight grown cultures were normalized to OD 2 at 600 nm and plated onto the Mueller Hinton agar plates by spread plate method. Pre-coated Ezy-MIC strips with selected antibiotics were placed in the middle of the agar plates with the help of a sterile swab and the plates were incubated at 37°C for 18 h. Data are representative of three independent experiments and plotted as mean ± SEM. Statistical analysis was performed using two-way ANOVA, where * p < 0.05; ** p < 0.01; *** p < 0.001 and **** p < 0.0001.
Antimicrobial susceptibility of these strains was also tested by exposing the four strains to commonly used antibiotics, ciprofloxacin (fluroquinolone), amoxyclav, meropenem (β-lactam antibiotics), and polymyxin B (cationic antimicrobial polypeptide). Although there were no notable differences in the minimum inhibitory concentration (MIC) among the strains treated with ciprofloxacin, the ΔrfbBΔrffG strain exhibited lower MIC values for amoxyclav, meropenem, and polymyxin B (Figure 3G). This phenotype could also be restored by complementing with the WT copy of either rffG or rfbB in trans (Figure 3H).
S. Typhimurium ΔrfbBΔrffG shows a truncated O-antigen profile, displays auto-aggregation behavior and binds more to NPN dye
The susceptibility to cell wall targeting antibiotics and bile indicated that the cell wall and membrane integrity may be compromised in the ΔrfbBΔrffG strain. To investigate any alterations in the outer membrane protein (OMP) profile, the LPS and the OMPs of these strains were analyzed. WT and the single–deletion mutants did not display any difference in the LPS profile. However, ΔrfbBΔrffG strain showed the absence of the repeating O-antigen structure (Figure 4A). The OMP profile of the ΔrfbBΔrffG was also monitored and compared to the isogenic WT strain. However, no observable difference between the WT and the ΔrfbBΔrffG strain was noted (Supplementary Figure S7). The proteins clustered around 35–45 kDa are likely to be OmpA, OmpD, and OmpC (Ray et al., 2019).
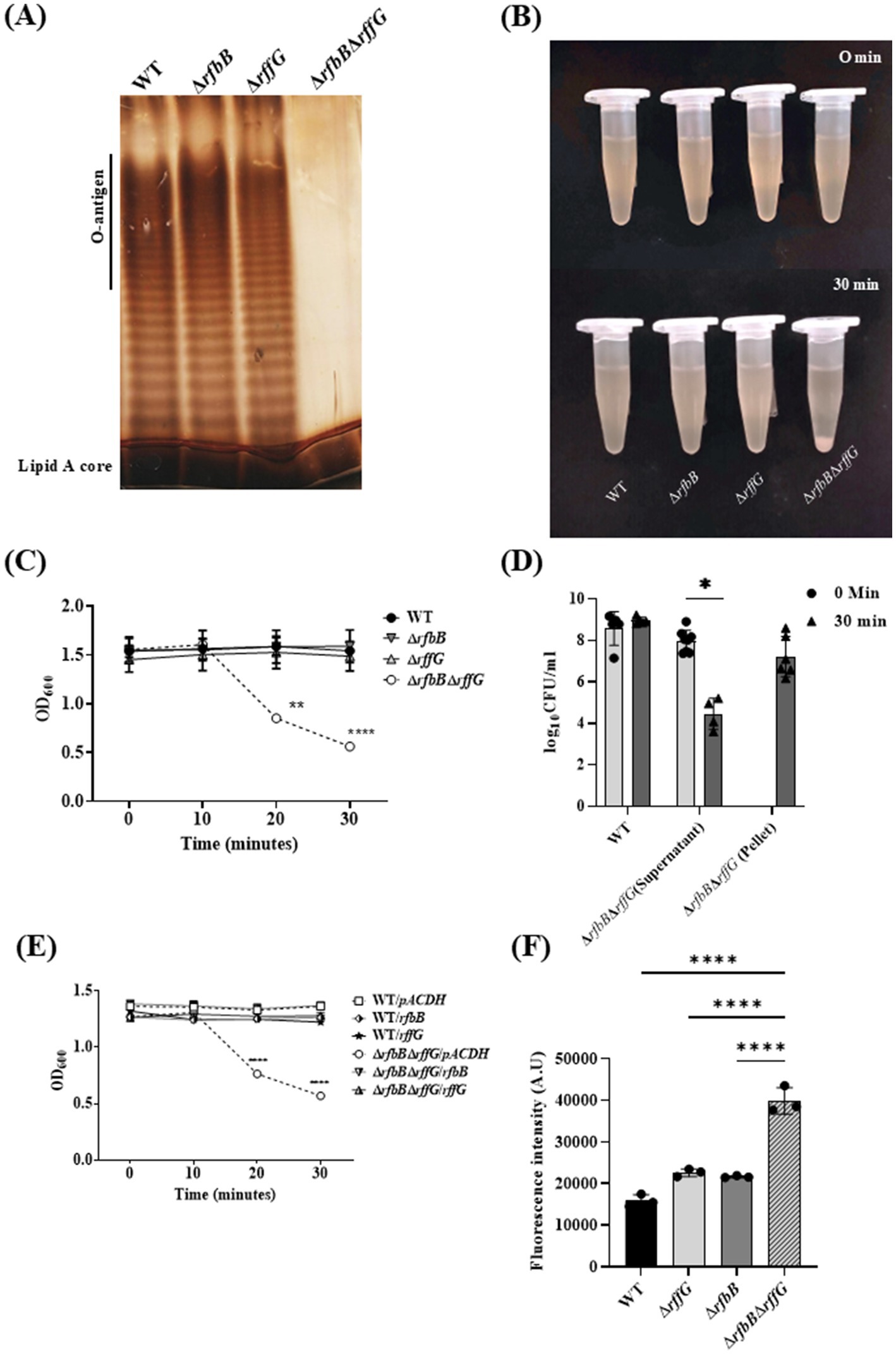
Figure 4. Salmonella Typhimurium ΔrfbBΔrffG strain displays a truncated O-antigen profile, rapid auto-aggregation and binds more to NPN dye. (A) S. Typhimurium WT, ΔrfbB, ΔrffG and ΔrfbBΔrffG strains were grown in LB broth at 37°C for 15 h. LPS was isolated and resolved on a 12% SDS PAGE. (B,C) Auto-aggregation behavior of the WT and the gene-deleted strains. Briefly, 1 mL of 1.5 OD normalized, S. Typhimurium WT, ΔrfbB, ΔrffG and ΔrfbBΔrffG cultures in LB broth were kept in an upright position for 30 min. At every 10-min interval, 100 μL was aspirated from the top of the solution, and the OD was measured at 600 nm. (D) Post 30 min, the supernatant was aspirated from the top of the culture, and the residual pellet was resuspended in 1 mL of sterile LB. Appropriate dilutions were then plated, and colony-forming units (CFU) were recorded. (E) Complementation with the WT copy of the gene(s) show a rescue in the phenotype. (F) S. Typhimurium ΔrfbBΔrffG accumulates higher amounts of the NPN dye. Briefly, the representative bacterial strains were grown to an OD of 0.5 at 600 nm. Cells were harvested, and the bacterial pellet was washed with 5 mM HEPES (pH 7.2) and adjusted to an OD of 0.5 at 600 nm. NPN was added to each well at a concentration of 10 μM. Fluorescence excitation and emission were measured at 350 nm and 420 nm, respectively. Statistical analysis for this assay was performed using one-way ANOVA, where ** p < 0.01. Data are representative of three independent experiments and plotted as mean ± SEM. Statistical analysis was performed using two-way ANOVA, where ** p < 0.01 and **** p < 0.0001.
During these experiments, we also observed that the ΔrfbBΔrffG strain quickly formed aggregates. When it was kept static at room temperature (Figures 4B,C). CFU analysis demonstrated that cell viability was unaffected during the rapid aggregation of cells (Figure 4D). This auto-aggregation behavior was optimal at 1% NaCl concentration, whereas a modulation in the concentration of NaCl (either higher or lower than 1%) reduced the auto-aggregation rate (data not shown). The auto-aggregation phenotype of the ΔrfbBΔrffG strain could also be complemented by introducing a WT copy of either rffG or rfbB expressed in trans (Figure 4E). This auto-aggregation behavior of the ΔrfbBΔrffG strain could be due to the absence of O-antigen repeating units which may lead to greater surface hydrophobicity as seen in higher NPN binding (Figure 4F), culminating in higher cell-to-cell aggregation.
RNA-seq reveals differential gene expression between the WT and the ΔrfbBΔrffG strain
A global transcriptomic analysis of 22 distinct infection-related condition in S. Typhimurium was published as a Salmonella compendium (Kröger et al., 2013). We investigated the expression of rffG, rfbB as well as other well-known genes from the Salmonella compendium v2.0 (Supplementary Table S3). Further, the phenotypic characterization of the S. Typhimurium WT, ΔrffG, ΔrfbB and ΔrfbBΔrffG strains in this study revealed that major differences occurred only in the ΔrfbBΔrffG strain, whereas the single-deletion strains did not display any observable differences upon comparision to the isogenic WT strain. Therefore, a transcriptomics experiment was conducted to identify differentially regulated genes in the WT and the ΔrfbBΔrffG strains. RNA-seq analysis revealed considerable gene expression differences between the WT and the ΔrfbBΔrffG strain. Those genes found to be significantly modulated were mapped to pathways using KEGG Mapper (Supplementary Figure S8).
Firstly, it was observed that more number of pathways were downregulated in the ΔrfbBΔrffG strain as compared to pathways that were upregulated (Supplementary Figure S8). The genes belonging to flagellar assembly pathway as well as genes involved in the infection process, specifically invasion-related genes, were among the most significantly downregulated genes in the ΔrfbBΔrffG strain. Other pathways which play indispensable roles during the pathogenesis of S. Typhimurium such as chemotaxis, quorum sensing, O-antigen biosynthesis, and LPS biosynthesis were also found to be downmodulated in the ΔrfbBΔrffG strain as compared to the isogenic WT strain. Among the prominent pathways which were upregulated in the ΔrfbBΔrffG strain as compared to the WT were the glycerophospholipid biosynthesis, cationic antimicrobial resistance, and nitrogen metabolism.
The flagellar assembly pathway is downregulated and the ΔrfbBΔrffG strain displays severe motility defects
The observation from the RNA-seq was validated by focusing on the flagellar assembly pathway as well as Salmonella infection-related genes. The flagellar assembly pathway in S. Typhimurium is hierarchically organized into three classes: Class I promoter includes flhDC which is the master regulator, Class II and Class III gene products comprise the basal body, hook, filament, and the motor force generator, respectively, (Das et al., 2018; Ray et al., 2020). The genes, flhD, fliC and fljB were expressed in lower amounts in the single-deletion mutants but highly downregulated in the ΔrfbBΔrffG strain (Figures 5A–C). However, fliC was not significantly downregulated in the ΔrffG strain.
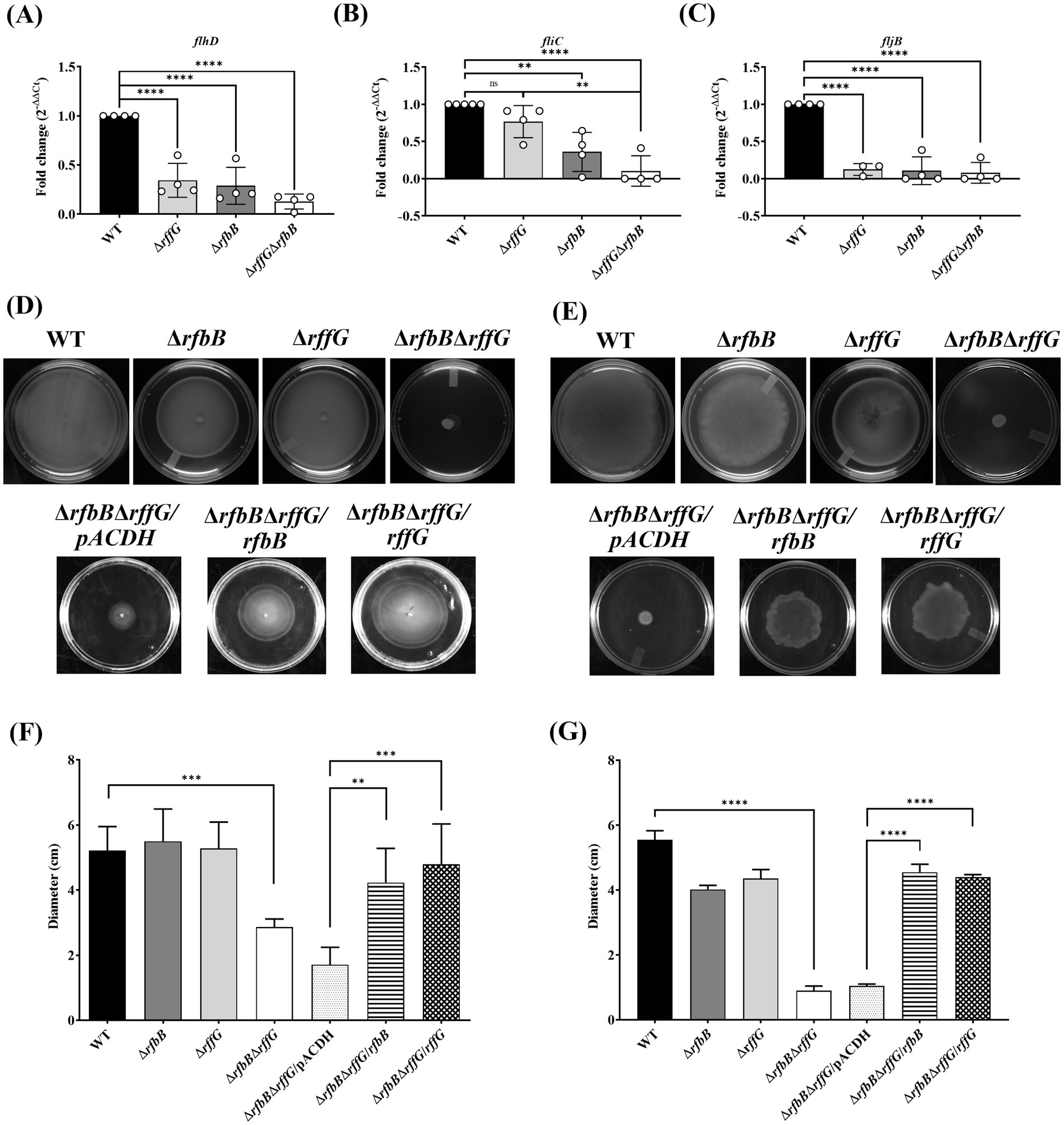
Figure 5. Salmonella Typhimurium ΔrfbBΔrffG strain is compromised in swimming and swarming motility. Total RNA was extracted from the bacterial samples after 3 h of growth in LB broth at 37°C and 160 rpm. A qRT-PCR analysis was performed to monitor the expression of the genes, (A) flhD, (B) fliC and (C) fljB and the expression normalized with the reference gene, gmk. The (D) swimming and (E) swarming motility assays were performed on 0.3 and 0.5% agar, respectively. Equal amounts of cultures were inoculated at the center of the motility-agar plates and the plates were incubated at 37°C for 8 h in an upright condition. The distance covered by each strain after 8 h post inoculation for (F) swimming and (G) swarming motility was measured and analyzed using ImageJ software. Multiple measurements were obtained for each strain. Data are representative of three independent experiments and plotted as mean ± SEM. Statistical analysis was performed using one-way ANOVA, where * p < 0.05; ** p < 0.01; *** p < 0.001 and **** p < 0.0001.
When motility assays were performed in these strains, there were no distinguishable differences in swimming motility between the WT and the single-deletion strains (Figures 5D,F). However, the ΔrfbBΔrffG strain was highly compromised in swimming motility and complementation with the WT copy of either rffG or rfbB in ΔrfbBΔrffG background restored motility. The single-deletion strains appeared to be slightly but significantly compromised in swarming behavior as compared to the WT strain. The ΔrfbBΔrffG strain, on the other hand, was found to be completely non-motile on swarming agar plates (Figures 5E,G). Complementation with the WT copy of either rffG or rfbB in ΔrfbBΔrffG background restored the swarming defect.
The ΔrfbBΔrffG strain displays compromised ability to infect HeLa cells
S. Typhimurium can infect and replicate within epithelial cells and macrophages and genes present in the Salmonella Pathogenicity Islands 1 and 2 play key roles (Hurley et al., 2014). There are three steps in the infection process: adhesion, invasion, and intracellular replication. Initially, the expression of a few genes belonging to the SPI-1 pathway was measured. The expression of hilA, hilD and sipC, all of which regulate the expression of other key downstream effectors of the SPI-1 pathway, were downregulated in the single-deletion strains but two-fold downregulated in the ΔrfbBΔrffG as compared to the WT strain (Figures 6A–C).
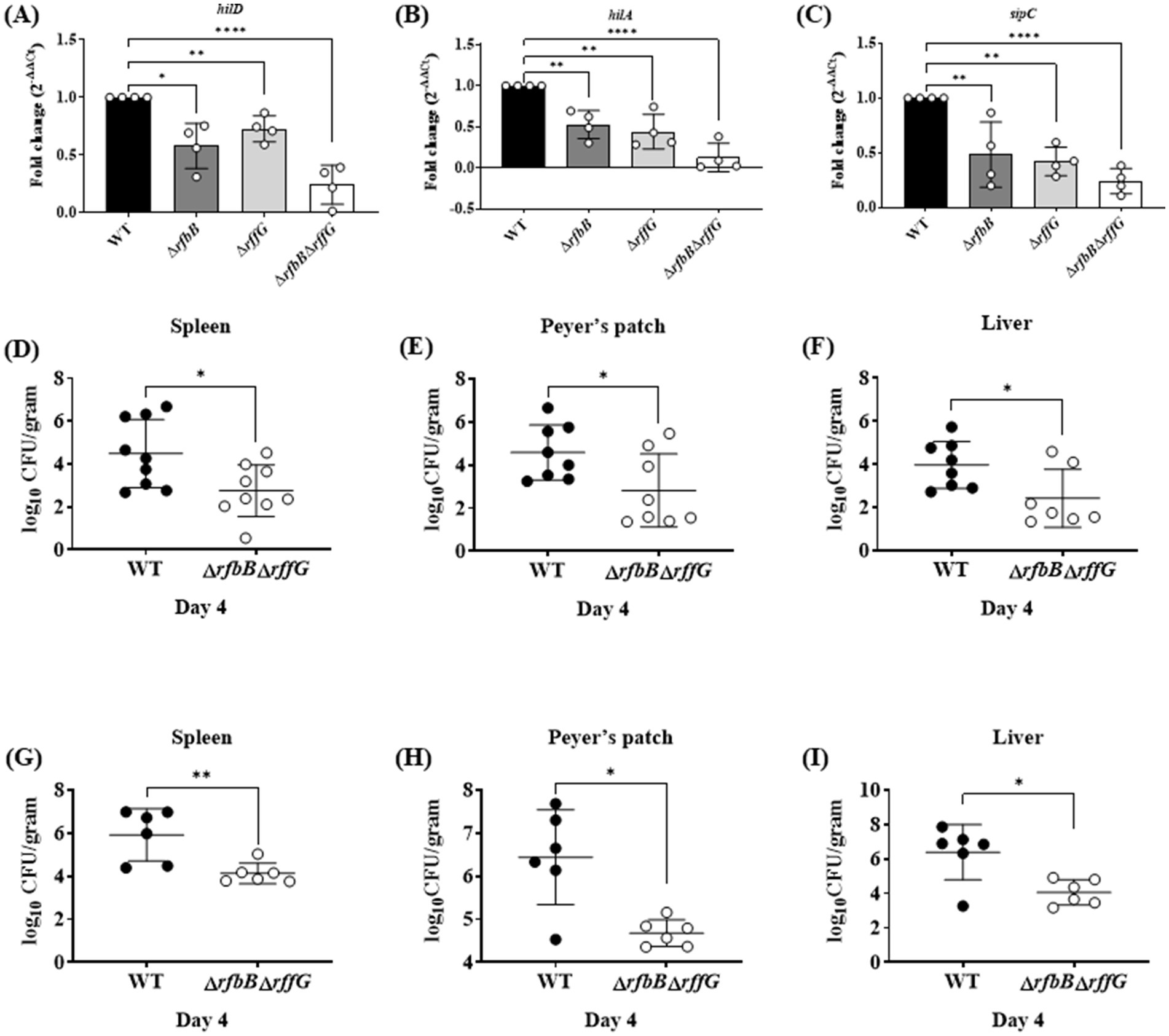
Figure 6. Salmonella Typhimurium ΔrfbBΔrffG strain is less proficient in colonizing different organs. Total RNA was extracted from the bacterial samples after 3 h of growth in LB broth at 37°C and 160 rpm. A qRT-PCR analysis was performed to monitor the expression of the SPI-1 genes, (A) hilD (B) hilA and (C) sipC, and the expression normalized with the reference gene, gmk. Bacterial burden in the organs was estimated after (D–F) oral and (G-I) intraperitoneal infection of C57BL/6 mice with the WT and the ΔrfbBΔrffG strains. Infected mice were sacrificed on day 4 post infection, organs were harvested, and the bacterial burden in the spleen, Peyer’s Patch and the liver was estimated by plating serial dilutions of the tissue homogenate on LB agar plate. Gene expression analysis data are representative of three independent experiments plotted as mean ± SEM and statistical analysis was performed using one-way ANOVA, where * p < 0.05; ** p < 0.01; *** p < 0.001 and **** p < 0.0001. For estimating bacterial burden in organs, statistical analysis was performed using the Mann Whitney U test.
The three steps in the cellular infection process were studied in HeLa cells using the gentamicin protection assay (Supplementary Figures S9A–C). The single-deletion strains were slightly compromised than the WT in adhering to the HeLa cells, whereas the ΔrfbBΔrffG strain was significantly compromised compared to the other strains. Additionally, the single-deletion mutants also showed a slight, although significant reduction in the number of viable bacteria at 2 h post infection compared to the WT strain. Interestingly, the ΔrfbBΔrffG strain showed a significant reduction of the number of viable bacteria at 2 h post infection. Overall, these observations indicate that the ΔrfbBΔrffG strain is compromised in comparison to the other strains in its ability to adhere and invade HeLa cells.
The ΔrfbBΔrffG strain is less proficient in colonizing different organs and induces lower pro-inflammatory cytokine response
To determine if the ΔrfbBΔrffG strain would exhibit any noticeable defects during in vivo infection. C57BL/6 mice were infected with the WT and the ΔrfbBΔrffG strain through oral as well as intraperitoneal (i.p.) routes. Subsequently, the organ CFU burden and other responses were estimated days 4 post infection (Figures 6D–I). In the oral infection model, the pathogens must traverse the intestinal epithelial barrier, whereas in the i.p. infection model, pathogens directly enter the systemic circulation. After 4-days post oral infection, the ΔrfbBΔrffG strain displayed two log-fold lower infection burden in the Peyer’s patches, liver, and spleen (Figures 6D–F). It is possible that the lowered CFU burden observed in the different organs might be due to the reduced expression of SPI-1 genes needed to traverse the epithelial barrier. To address this question, C57BL/6 mice were intraperitoneally infected with the WT and the ΔrfbBΔrffG strains as intraperitoneal infection would not require active breach of the intestinal epithelial barrier. After 4 days post infection via the intraperitoneal route, the CFU burden of the ΔrfbBΔrffG strain was two log-fold lower compared to the WT strain (Figures 6G–I). This clearly suggested that the low proficiency to infect and colonize different organs was not solely due to the reduced expression of the SPI-1 genes in the ΔrfbBΔrffG strain but could be due to multiple and varied mechanisms. In addition, the ΔrfbBΔrffG strain induced lesser amounts of pro-inflammatory cytokines such as TNF-α, IL6, and IFN-γ than the WT strain in both the oral (Figures 7A–C) and the i.p. (Figures 7D–F) modes of infection.
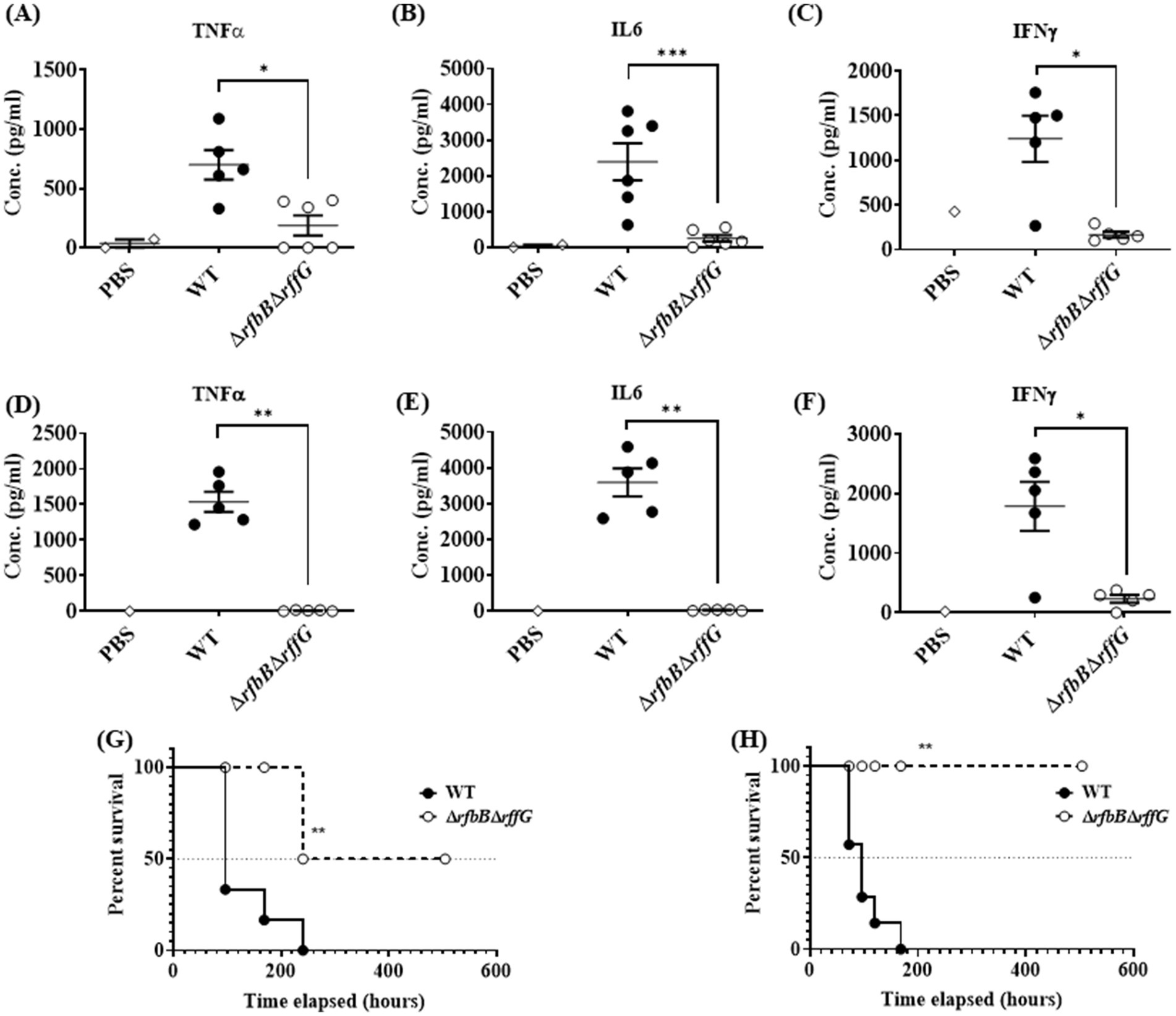
Figure 7. Salmonella Typhimurium ΔrfbBΔrffG strain is highly attenuated in oral and intraperitoneal mice models of infection. The levels of pro-inflammatory cytokines (A) TNFα (B) IL6, and (C) IFNγ at day 4 post infection were estimated from the serum collected from mice infected orally with 1×108 CFU of the WT and ΔrfbBΔrffG strains. Similarly, mice infected intraperitoneally with 1×103 CFU of S. Typhimurium WT and ΔrfbBΔrffG were sacrificed at day 4 post infection and the serum levels of cytokines, (D) TNFα (E) IL6 and (F) IFNγ were quantified. Kaplan–Meier survival analysis comparing mice survival for 21 days upon infection with the WT and the ΔrfbBΔrffG strains, infected either, (G) orally or (H) intraperitoneally. Data are representative of three independent experiments plotted as mean ± SEM. Statistical analysis was performed using Log-rank (Mantel-Cox) test, where **p < 0.005 and ***p < 0.0001. For cytokine estimation, statistical analysis was performed using Kruskal Wallis test, where *p < 0.05; **p < 0.01 and ***p < 0.001.
ΔrfbBΔrffG strain is attenuated in both oral and intraperitoneal model of S. Typhimurium infection
Finally, survival of the mice after oral and i.p. infection was monitored for 21 days post infection. Mice infected with the WT strain succumbed to infection as early as 4 days post infection and 100% mortality was observed by day 12. In contrast, infection with the ΔrfbBΔrffG strain resulted in prolonged mice survival. By day 21, only 20% mortality was observed (Figure 7G). Similarly, in experiments where the mice were infected via the i.p. route, all the WT-infected mice succumbed by day 7 post infection whereas the ΔrfbBΔrffG strain showed no mortality till 21 days post infection (Figure 7H). Overall, these results suggest that the ΔrfbBΔrffG strain was highly attenuated in causing systemic disease in C57BL/6 mice compared to the WT strain.
Discussion
In this study, we sought to investigate how the absence of the genes, rffG and rfbB affect the physiology of S. Typhimurium. Disruptions in the biosynthetic pathways of O-antigen and ECA can lead to significant cell shape abnormalities and activate stress response systems in various bacterial species. These defects primarily result due to limiting the availability of Und-P, which is important in the synthesis of key molecules (Supplementary Figure S1), leading to morphological alterations. E.coli (Danese et al., 1998; Jorgenson and Young, 2016; Jorgenson et al., 2016; Jorgenson and Bryant, 2021) and S. flexneri (Qin et al., 2025) show different tolerance to UndP sequestrations, where limited production of ECA renders S. flexneri more susceptible to cell death. In E. coli, mutations in the O-antigen flippase gene (wzxB) or ligase gene (waaL) result in the buildup of Und-PP-linked intermediates, causing cells to become swollen, filamentous, or form chains (Jorgenson and Young, 2016). Similarly, in the ECA pathway, mutations in the wecE and rfbA lead to the accumulation of ECA-lipid II intermediates, sequestering Und-P and causing cell shape defects and sensitivity to detergents (Jorgenson and Young, 2016; Jorgenson et al., 2016). In Serratia marcescens, mutants defective in ECA biosynthesis trigger activation of the regulator of capsule synthesis (Rcs) stress response system (Castelli and Véscovi, 2011; Castelli et al., 2008). In Bacillus subtilis, depletion of undecaprenyl pyrophosphate phosphatase activity leads to morphological defects and strongly activates the σM-dependent cell envelope stress response (Zhao et al., 2016). Recent studies reveal that UDP-glucose acts as a sensing molecule regulating cell size and division in E. coli (Hill et al., 2013). It links nutrient availability with cell division by activating OpgH, an inner-membrane glycosyltransferase involved in bacterial envelope biogenesis. In nutrient-rich conditions, UDP-glucose promotes OpgH-mediated inhibition of FtsZ ring formation by direct interaction with its N-terminal domain, delaying cell division. Also, peptidoglycan biosynthesis is important in maintaining the cellular shape in S. Typhimurium (van Teeseling et al., 2017).
Initial studies revealed that the ΔrfbBΔrffG strain demonstrated a higher OD in the stationary phase as compared to the WT and the single deletion strains but did not display any difference in the CFU in stationary phase (Supplementary Figure S5). AFM studies conducted with the various strains revealed that the ΔrfbBΔrffG strain displayed a predominantly round morphology as compared to other strains (Figure 2). Interestingly, Salmonella mutants lacking ECA (Ramos-Morales et al., 2003) or LPS (Spöring et al., 2018) do not show the round phenotype as noted in ΔrfbBΔrffG strain. Our double mutants displays a change in cell morphology as it shows increased cellular width but no filamentation observed as compared to WT (Figure 2). Mutations in mre and rod, which are components of cytoskeletal arrangements, in S. Typhimurium give rise to a round cellular morphology (Costa and Antón, 1993). MinC and MinD are required for proper cell division. In the absence of MinC or MinD, the FtsZ ring fails to locate to the middle of a cell, leading to abnormal cell division and morphological aberrations (Wu et al., 2016). It is possible that the change in cell shape in the ΔrfbBΔrffG strain may account for an increased OD (Yoon et al., 2015; Stevenson et al., 2016) compared to the WT or single deletion strains. Further work is required to shed more insight into the mechanisms at play that affects the distinct cellular morphology in the ΔrfbBΔrffG strain.
We subjected these strains to various stresses that S. Typhimurium commonly encounters during the host colonization process. Although no growth difference was detected among the strains in many stress conditions, the ΔrfbBΔrffG strain was found to be highly susceptible to bile-induced stress, with, cell wall targeting antibiotics and cationic polypeptides (Figure 3). LPS acts as a membrane barrier; consequently, LPS mutants are susceptible to bile (Urdaneta and Casadesús, 2017). Salmonella strains lacking ECA do not show alterations in morphology, LPS profile or motility, although they are attenuated in mice models of infection (Gilbreath et al., 2012). The outer membrane of Gram-negative bacteria is unique among all biological membranes by virtue of their properties to exclude hydrophobic substances, primarily due to LPS. The probe 1-N-phenylnaphthylamine (NPN) is used to study membranes as it gives strong fluorescence in phospholipid environments but fluoresces weakly in aqueous environments (Helander and Mattila-Sandholm, 2000). The dye accumulated to a similar extent in the WT and the single deletion strains. However, an increased amount of fluorescence was observed in the ΔrfbBΔrffG strain (Figure 4F), indicating enhanced surface hydrophobicity which may be responsible for greater aggregation (Figures 4B–E). In fact, the susceptibility to cell wall targeting antibiotics and bile indicated that the outer membrane integrity may be compromised in the ΔrfbBΔrffG strain (Figures 3E–H).
A comparative transcriptomic analysis (RNA-seq) between the WT and the ΔrfbBΔrffG strains showed major alterations in several pathways: motility, invasion, chemotaxis (Supplementary Figure S8). The flagellar assembly pathway was one, among the many prominent pathways that was found to be highly downregulated in the ΔrfbBΔrffG strain. In fact, motility studies revealed that the ΔrfbBΔrffG strain was significantly compromised in swimming as well as swarming motility as compared to the WT and the single deletion strains (Figure 5). Swimming motility represents individual movement in liquid environments using flagella, whereas swarming motility is also flagella-driven, multicellular and on solid or semi-solid surfaces (Thakur et al., 2020). Mutations in genes of the O antigen and ECA biosynthesis pathways can activate the Rcs phosphorelay system, a key envelope stress response mechanism in bacteria belonging to the Enterobacteriaceae family (Meng et al., 2021; Liu et al., 2022). Sequestration of Und-P in O-antigen and ECA mutants due to the accumulated biosynthetic intermediates compromises peptidoglycan synthesis and cell wall integrity, leading to morphological defects and triggering stress responses, including the Rcs system (Castelli and Véscovi, 2011; Spöring et al., 2018; Castelli et al., 2008). The Rcs phosphorelay is a critical envelope stress-sensing signal transduction pathway that plays a major role in motility. Through its transcriptional regulator RcsB, the phosphorelay directly represses flhDC expression, which encodes the master transcriptional regulator, FlhD4C2. Reduced flhDC levels leads to decreased flagellum production and inhibits motility (Francez-Charlot et al., 2003). In E. coli, the Rcs two component signaling pathway plays a role in repression of motility in LPS mutants (Girgis et al., 2007). In S., Typhimurium Rcs activation represses flagellar gene expression via flhDC, leading to impaired swarming motility (Wang et al., 2007). Similarly, in E. coli, the RcsC-YojN-RcsB phosphorelay modulates capsular synthesis and swarming behavior (Takeda et al., 2001). The lack of proper LPS perturbs the outer membrane, leading to activation of signaling pathways, including RpoE and Rcs phosphor relay system, and degradation of FlhDC, the class I motility regulator (Spöring et al., 2018). In Proteus mirabilis, disruption of cell envelope-associated genes impairs swarmer cell development and motility through various mechanisms (Morgenstein and Rather, 2011). RcsB also promotes the expression of several fimbrial genes, including paralogs of the fimbrial transcriptional regulator MrpJ (Bode et al., 2015). Collectively, these findings reinforce the role of the Rcs system in coordinating bacterial envelope stress responses with motility regulation. It is possible that in the ΔrfbBΔrffG strain, the absence of the O-antigen and ECA leads to loss of surface lubrication and aberrant signaling events, leading to compromised motility (Figure 5). Further studies are required to investigate the Rcs activation and other pathways to understand their functional roles in these mutants strains.
Another, pathway that was found to be highly downmodulated in the ΔrfbBΔrffG strain was the SPI-1 pathway (Supplementary Figure S8 and Figure 6). The significant difference in the invasion for the ΔrfbBΔrffG strain observed in HeLa, could be due to less SPI-1 effector proteins (Supplementary Figure S9). Both SPI-1 and the flagellar biosynthesis pathway are essential for successful colonization of mice (Dieye et al., 2009; Stecher et al., 2004). Studies have shown that inv mutants of S. Typhimurium, which do not have functional SPI-1, when administered perorally to BALB/c mice, results in higher 50% lethal doses (LD50). In addition, inv mutants were compromised in their ability to colonize the Peyer’s patches, small intestinal wall, and the spleen when administered perorally (Porter and Curtiss, 1997). However, no such differences were observed when these strains were administered intraperitoneally. Although the ΔrfbBΔrffG strain had the ability to colonize different organs, it was less proficient than the parent, WT strain (Figure 6) and induced lower pro-inflammatory cytokine responses (Figure 7). Importantly, the ΔrfbBΔrffG strain did not cause lethal infection when administered via oral as well as the i.p. routes of infection (Huang et al., 2016; Zhou et al., 2023). This observation is consistent with previous findings in Candida albicans, where it was observed that a GAL102p mutant (a homolog of dTDP-glucose 4,6-dehydratase) was unable to grow in resident peritoneal macrophages. The mutant also elicited a weak pro-inflammatory cytokine response in vitro as well as in an in vivo mouse model of systemic candidiasis (Sen et al., 2011). Both O-antigen, ECA and the flagella are important antigens recognized by the immune system (Hajam et al., 2017; Kintz et al., 2017). The absence or down regulation of these components would cause aberrant immune responses in mice infected with the ΔrfbBΔrffG strain. Earlier studies have also reported that the LPS or ECA mutant are highly attenuated in S. Typhimurium oral infection model (Kong et al., 2011; Gilbreath et al., 2012). The absence of the O-antigen and ECA in the ΔrfbBΔrffG strain may make the infection cycle non-optimal and/or these strains may be more susceptible to complement mediated lysis (Murray et al., 2006). Despite the lowered virulence of the ΔrfbBΔrffG strain, its ability to grow within host organs may be a concern for its proposed use as a live attenuated vaccine (Huang et al., 2016). Previously, we have demonstrated that another attenuated strain, ΔrpoS, can colonize host organs (Rananware et al., 2021). It is likely that mice can co-exist or eventually clear the infection when infected with attenuated S. Typhimurium strains.
Finally, we present a model that accounts for the key observations (Figure 8). Briefly, the genes, rffG and rfbB are paralogs that encode the enzyme dTDP-glucose 4,6- dehydratase, which is known to catalyze steps of O-antigen and ECA biosynthesis (Supplementary Figure S1). Consequently, the functional loss of dTDP-glucose 4,6-dehydratase activity leads to profound physiological differences, a reduced ability to colonize different organs and lowered virulence in the mouse model of infection. The LPS biosynthesis pathway has generated a lot of optimism as a drug target (Zhang et al., 2018). To the best of our knowledge, this is one of the primary reports to demonstrate in detail the physiological consequences of the loss of dTDP-glucose 4,6- dehydratase activity in S. Typhimurium. This study, therefore, lays the foundation for further research and better understanding of the combinatorial roles of O-antigen and ECA, either directly or indirectly, on S. Typhimurium physiology and pathogenesis.
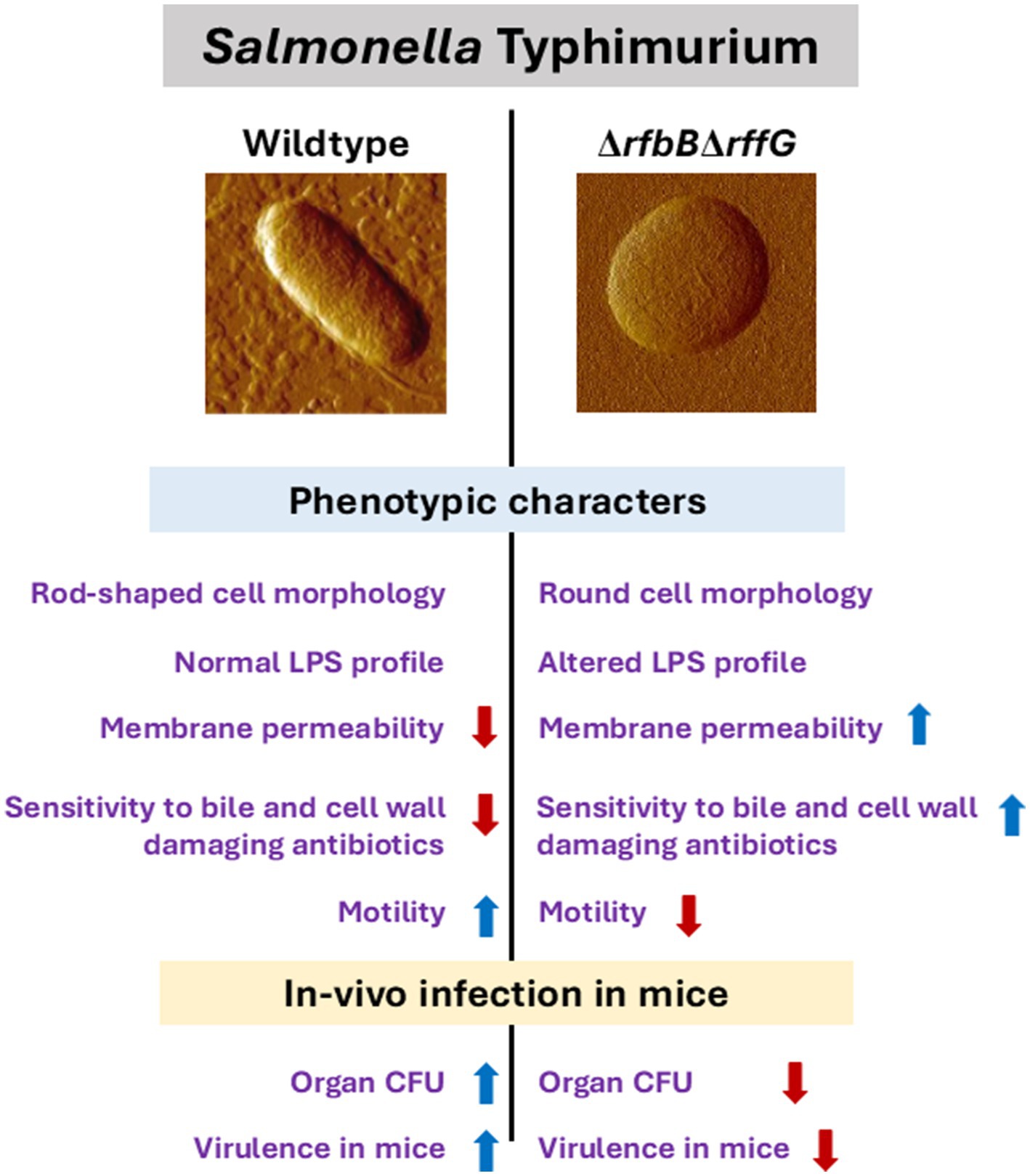
Figure 8. Salmonella Typhimurium WT and ∆rfbB∆rffG show distinct physiological and phenotypic differences. The genes, rfbB and rffG, encode the enzyme dTDP-glucose 4,6- dehydratase, which catalyzes intermittent steps of O-antigen and ECA biosynthesis. Functional loss of dTDP-glucose 4,6-dehydratase renders S. Typhimurium strains incapable of synthesizing both O-antigen and ECA. This leads to profound physiological differences between the S. Typhimurium, WT and the ΔrfbBΔrffG strains. The ΔrfbBΔrffG strain exhibits distinct cellular morphology, altered LPS profile, increased outer membrane permeability and susceptibility to bile and cell wall targeting antibiotics. Functional loss of dTDP-glucose 4,6-dehydratase also led to inhibition of motility and a reduced ability to colonize different organs in the mouse model of infection. Consequently, the ΔrfbBΔrffG strain displayed significant virulence attenuation as compared to the WT.
Data availability statement
The RNAseq dataset accession number presented in this article is not readily available because the experiment was performed with one biological replicate to obtain a preliminary idea regarding the DEGs in the ΔrfbBΔrffG strain compared to the isogenic WT control after growth in a nutrient rich medium (LB) at 3 hours. The findings from this preliminary analysis were further validated using qPCR (Figure 5 and Figure 6). Requests to access this dataset and all other data should be directed to Dipankar Nandi (bmFuZGlAaWlzYy5hYy5pbg==).
Ethics statement
The animal study was approved by the Institutional Animal Ethics Committee. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
SC: Formal analysis, Investigation, Methodology, Writing – original draft. PB: Data curation, Formal analysis, Investigation, Methodology, Visualization, Writing – original draft. JJ: Formal analysis, Investigation, Writing – review & editing. SP: Formal analysis, Investigation, Writing – review & editing. TV: Formal analysis, Investigation, Writing – review & editing. AK: Investigation, Writing – review & editing. DC: Investigation, Writing - review & editing. MD: Investigation, Writing – review & editing. PS: Investigation, Writing - review & editing. DN: Conceptualization, Funding acquisition, Project administration, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by core grants from IISc and the DBT-IISc partnership program. In addition, the infrastructural support from DST-FIST and UGC CAS/SAP is greatly appreciated.
Acknowledgments
Imaging studies were performed at the Bioengineering Imaging and Advanced Facility for Microscopy and Microanalysis (AFMM) facilities in IISc. We thank the help of the Central Animal Facility, IISc, for all the experiments involving mice. The support from all members of the DpN lab is greatly appreciated. This study is dedicated to the memory of Prof. Parag Sadhale who inspired this work. We recall fondly his contributions as a scientist and as a fine human being.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1572117/full#supplementary-material
Abbreviations
LPS, Lipopolysaccharide; S. Typhimurium, Salmonella enterica serovar Typhimurium; ECA, enterobacterial common antigen; WT, Wild-type; LB, Luria-Bertani; OMP, outer membrane protein; SDR, short-chain dehydrogenase/reductase; AFM, atomic force microscopy; SPI, Salmonella Pathogenicity Island; RNA-seq, RNA-sequencing; NPN, 1-N-phenylnaphthylamine; TMB, 3,3′,5,5’-Tetramethylbenzidine.
References
Allard, S. T. M., Beis, K., Giraud, M.-F., Hegeman, A. D., Gross, J. W., Wilmouth, R. C., et al. (2002). Toward a structural understanding of the dehydratase mechanism. Structure 10, 81–92. doi: 10.1016/S0969-2126(01)00694-3
Allard, S. T. M., Giraud, M.-F., Whitfield, C., Graninger, M., Messner, P., and Naismith, J. H. (2001). The crystal structure of dTDP-d-glucose 4,6-dehydratase (RmlB) from Salmonella enterica serovar typhimurium, the second enzyme in the dTDP-l-rhamnose pathway. J. Mol. Biol. 307, 283–295. doi: 10.1006/jmbi.2000.4470
Beis, K., Allard, S. T. M., Hegeman, A. D., Murshudov, G., Philp, D., and Naismith, J. H. (2003). The structure of NADH in the enzyme dTDP-d-glucose dehydratase (RmlB). J. Am. Chem. Soc. 125, 11872–11878. doi: 10.1021/ja035796r
Bhaskarla, C., Das, M., Verma, T., Kumar, A., Mahadevan, S., and Nandi, D. (2016). Roles of Lon protease and its substrate MarA during sodium salicylate-mediated growth reduction and antibiotic resistance in Escherichia coli. Microbiology 162, 764–776. doi: 10.1099/mic.0.000271
Bode, N. J., Debnath, I., Kuan, L., Schulfer, A., Ty, M., and Pearson, M. M. (2015). Transcriptional analysis of the MrpJ network: modulation of diverse virulence-associated genes and direct regulation of mrp fimbrial and flhDC flagellar operons in Proteus mirabilis. Infect Immun. 83:2542–56.
Castelli, M. E., Fedrigo, G. V., Clementín, A. L., Ielmini, M. V., Feldman, M. F., and García, V. E. (2008). Enterobacterial common antigen integrity is a checkpoint for flagellar biogenesis in Serratia marcescens. J. Bacteriol. 190, 213–220. doi: 10.1128/JB.01348-07
Castelli, M. E., and Véscovi, E. G. (2011). The Rcs signal transduction pathway is triggered by enterobacterial common antigen structure alterations in Serratia marcescens. J. Bacteriol. 193, 63–74. doi: 10.1128/JB.00839-10
Costa, C. S., and Antón, D. N. (1993). Round-cell mutants of Salmonella typhimurium produced by transposition mutagenesis: lethality of rodA and mre mutations. Mol. Gen. Genet. 236, 387–394
Danese, P. N., Oliver, G. R., Barr, K., Bowman, G. D., Rick, P. D., and Silhavy, T. J. (1998). Accumulation of the enterobacterial common antigen lipid II biosynthetic intermediate stimulates degP transcription in Escherichia coli. J. Bacteriol. 180, 5875–5884. doi: 10.1128/JB.180.22.5875-5884.1998
Das, C., Mokashi, C., Mande, S. S., and Saini, S. (2018). Dynamics and control of flagella assembly in Salmonella typhimurium. Front. Cell. Infect. Microbiol. 8:36. doi: 10.3389/fcimb.2018.00036
Datsenko, K. A., and Wanner, B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. 97, 6640–6645. doi: 10.1073/pnas.120163297
Dieye, Y., Ameiss, K., Mellata, M., and Curtiss, R. (2009). The Salmonella Pathogenicity Island (SPI) 1 contributes more than SPI2 to the colonization of the chicken by Salmonella enterica serovar Typhimurium. BMC Microbiol. 9:3. doi: 10.1186/1471-2180-9-3
Ehmke, N., Caliebe, A., Koenig, R., Kant, S. G., Stark, Z., Cormier-Daire, V., et al. (2014). Homozygous and compound-heterozygous mutations in TGDS cause Catel-Manzke syndrome. Am. J. Hum. Genet. 95, 763–770. doi: 10.1016/j.ajhg.2014.11.004
Farrag, H. A., Abdallah, N., Shehata, M. M. K., and Awad, E. M. (2019). Natural outer membrane permeabilizers boost antibiotic action against irradiated resistant bacteria. J. Biomed. Sci. 26:69. doi: 10.1186/s12929-019-0561-6
Francez-Charlot, A., Laugel, B., Van Gemert, A., Dubarry, N., Wiorowski, F., Castanié-Cornet, M. P., et al. (2003). RcsCDB His-Asp phosphorelay system negatively regulates the flhDC operon in Escherichia coli. Mol Microbiol. 49:823–32.
Gerratana, B., Cleland, W. W., and Frey, P. A. (2001). Mechanistic roles of Thr134, Tyr160, and Lys 164 in the reaction catalyzed by dTDP-glucose 4,6-dehydratase. Biochemistry 40, 9187–9195. doi: 10.1021/bi0108249
Gilbreath, J. J., Colvocoresses Dodds, J., Rick, P. D., Soloski, M. J., Merrell, D. S., and Metcalf, E. S. (2012). Enterobacterial common antigen mutants of Salmonella enterica serovar Typhimurium establish a persistent infection and provide protection against subsequent lethal challenge. Infect. Immun. 80, 441–450. doi: 10.1128/IAI.05559-11
Girgis, H. S., Liu, Y., Ryu, W. S., and Tavazoie, S. (2007). A comprehensive genetic characterization of bacterial motility. PLoS Genet. 3, 1644–1660. doi: 10.1371/journal.pgen.0030154
Hajam, I. A., Dar, P. A., Shahnawaz, I., Jaume, J. C., and Lee, J. H. (2017). Bacterial flagellin-a potent immunomodulatory agent. Exp. Mol. Med. 49:e373. doi: 10.1038/emm.2017.172
Helander, I. M., and Mattila-Sandholm, T. (2000). Fluorometric assessment of gram-negative bacterial permeabilization. J. Appl. Microbiol. 88, 213–219. doi: 10.1046/j.1365-2672.2000.00971.x
Hill, N. S., Buske, P. J., Shi, Y., and Levin, P. A. (2013). A moonlighting enzyme links Escherichia coli cell size with central metabolism. PLoS Genet. 9:1003663. doi: 10.1371/journal.pgen.1003663
Hitchcock, P. J., and Brown, T. M. (1983). Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J. Bacteriol. 154, 269–277. doi: 10.1128/jb.154.1.269-277.1983
Huang, C., Liu, Q., Luo, Y., Li, P., Liu, Q., and Kong, Q. (2016). Regulated delayed synthesis of lipopolysaccharide and enterobacterial common antigen of Salmonella Typhimurium enhances immunogenicity and cross-protective efficacy against heterologous Salmonella challenge. Vaccine 34, 4285–4292. doi: 10.1016/j.vaccine.2016.07.010
Hurley, D., Mccusker, M. P., Fanning, S., and Martins, M. (2014). Salmonella-host interactions - modulation of the host innate immune system. Front. Immunol. 5:481. doi: 10.3389/fimmu.2014.00481
Ilg, K., Endt, K., Misselwitz, B., Stecher, B., Aebi, M., and Hardt, W. D. (2009). O-antigen-negative Salmonella enterica serovar Typhimurium is attenuated in intestinal colonization but elicits colitis in streptomycin-treated mice. Infect. Immun. 77, 2568–2575. doi: 10.1128/IAI.01537-08
Jorgenson, M. A., and Bryant, J. C. (2021). A genetic screen to identify factors affected by undecaprenyl phosphate recycling uncovers novel connections to morphogenesis in Escherichia coli. Mol. Microbiol. 115, 191–207. doi: 10.1111/mmi.14609
Jorgenson, M. A., Kannan, S., Laubacher, M. E., and Young, K. D. (2016). Dead-end intermediates in the enterobacterial common antigen pathway induce morphological defects in Escherichia coli by competing for undecaprenyl phosphate. Mol. Microbiol. 100, 1–14. doi: 10.1111/mmi.13284
Jorgenson, M. A., and Young, K. D. (2016). Interrupting biosynthesis of O antigen or the lipopolysaccharide Core produces morphological defects in Escherichia coli by sequestering Undecaprenyl phosphate. J. Bacteriol. 198, 3070–3079. doi: 10.1128/JB.00550-16
Kintz, E., Heiss, C., Black, I., Donohue, N., Brown, N., Davies, M. R., et al. (2017). Salmonella enterica Serovar Typhi lipopolysaccharide O-antigen modification impact on serum resistance and antibody recognition. Infect. Immun. 85, e01021–e01016. doi: 10.1128/IAI.01021-16
Kong, Q., Yang, J., Liu, Q., Alamuri, P., Roland, K. L., and Curtiss, R. (2011). Effect of deletion of genes involved in lipopolysaccharide core and O-antigen synthesis on virulence and immunogenicity of Salmonella enterica serovar typhimurium. Infect. Immun. 79, 4227–4239. doi: 10.1128/IAI.05398-11
Kröger, C., Colgan, A., Srikumar, S., Händler, K., Sivasankaran, S. K., Hammarlöf, D. L., et al. (2013). An infection-relevant transcriptomic compendium for Salmonella enterica Serovar Typhimurium. Cell Host Microbe 14, 683–695. doi: 10.1016/j.chom.2013.11.010
Kumar, G., Kumar, S., Jangid, H., Dutta, J., and Shidiki, A. (2025). The rise of non-typhoidal Salmonella: an emerging global public health concern. Front. Microbiol. 16:1524287. doi: 10.3389/fmicb.2025.1524287
Liu, T., Liu, Y., Bu, Z., Yin, F., Zhang, Y., Liu, J., et al. (2022). The Rcs System Contributes to the Motility Defects of the Twin-Arginine Translocation System Mutant of Extraintestinal Pathogenic Escherichia coli. J Bacteriol. 19:e0061221.
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Majumdar, S., Deobagkar-Lele, M., Adiga, V., Raghavan, A., Wadhwa, N., Ahmed, S. M., et al. (2017). Differential susceptibility and maturation of thymocyte subsets during Salmonella Typhimurium infection: insights on the roles of glucocorticoids and interferon-gamma. Sci. Rep. 7:40793. doi: 10.1038/srep40793
Marolda, C. L., and Valvano, M. A. (1995). Genetic analysis of the dTDP-rhamnose biosynthesis region of the Escherichia coli VW187 (O7:K1) rfb gene cluster: identification of functional homologs of rfbB and rfbA in the rff cluster and correct location of the rffE gene. J. Bacteriol. 177, 5539–5546. doi: 10.1128/jb.177.19.5539-5546.1995
Marshall, J. M., and Gunn, J. S. (2015). The O-antigen capsule of Salmonella enterica serovar Typhimurium facilitates serum resistance and surface expression of FliC. Infect. Immun. 83, 3946–3959. doi: 10.1128/IAI.00634-15
Meng, J., Young, G., and Chen, J. (2021). The Rcs system in Enterobacteriaceae: envelope stress responses and virulence regulation. Front. Microbiol. 15:627104. doi: 10.3389/fmicb.2021.627104
Morgenstein, R. M., and Rather, P. N. (2011). Role of the Umo proteins and the Rcs phosphorelay in the swarming motility of the wild type and an O-antigen (waaL) mutant of Proteus mirabilis. J Bacteriol. 194:669–76.
Murray, G. L., Attridge, S. R., and Morona, R. (2006). Altering the length of the lipopolysaccharide O antigen has an impact on the interaction of Salmonella enterica serovar Typhimurium with macrophages and complement. J. Bacteriol. 188, 2735–2739. doi: 10.1128/JB.188.7.2735-2739.2006
Park, S., Won, G., Kim, J., Kim, H. B., and Lee, J. H. (2018). Potent O-antigen-deficient (rough) mutants of Salmonella Typhimurium secreting Lawsonia intracellularis antigens enhance immunogenicity and provide single-immunization protection against proliferative enteropathy and salmonellosis in a murine model. Vet. Res. 49:57. doi: 10.1186/s13567-018-0552-8
Porter, S. B., and Curtiss, R. (1997). Effect of inv mutations on Salmonella virulence and colonization in 1-day-old white Leghorn chicks. Avian Dis. 41, 45–57. doi: 10.2307/1592442
Qin, J., Hong, Y., Maczuga, N. T., Morona, R., and Totsika, M. (2025). Tolerance mechanisms in polysaccharide biosynthesis: implications for undecaprenol phosphate recycling in Escherichia coli and Shigella flexneri. PLoS Genet. 21:1011591. doi: 10.1371/journal.pgen.1011591
Raghunathan, D., Wells Timothy, J., Morris Faye, C., Shaw Robert, K., Bobat, S., Peters Sarah, E., et al. (2011). SadA, a trimeric autotransporter from Salmonella enterica Serovar Typhimurium, can promote biofilm formation and provides limited protection against infection. Infect. Immun. 79, 4342–4352. doi: 10.1128/IAI.05592-11
Rai Ashutosh, K., and Mitchell Angela, M. (2020). Enterobacterial common antigen: synthesis and function of an enigmatic molecule. MBio 11:20. doi: 10.1128/mbio.01914-20
Ramos-Morales, F., Prieto, A. I., Beuzón, C. R., Holden, D. W., and Casadesús, J. (2003). Role for Salmonella enterica enterobacterial common antigen in bile resistance and virulence. J. Bacteriol. 185, 5328–5332. doi: 10.1128/JB.185.17.5328-5332.2003
Rananware, S., Pathak, S., Chakraborty, S., Bisen, R. Y., Chattopadhyay, A., and Nandi, D. (2021). Autoimmune-prone lpr mice exhibit a prolonged but lethal infection with an attenuated Salmonella typhimurium strain. Microb. Pathog. 150:104684. doi: 10.1016/j.micpath.2020.104684
Rao, A. R., and Varshney, U. (2001). Specific interaction between the ribosome recycling factor and the elongation factor G from Mycobacterium tuberculosis mediates peptidyl-tRNA release and ribosome recycling in Escherichia coli. EMBO J. 20, 2977–2986. doi: 10.1093/emboj/20.11.2977
Ray, S., Da Costa, R., Das, M., and Nandi, D. (2019). Interplay of cold shock protein E with an uncharacterized protein, YciF, lowers porin expression and enhances bile resistance in Salmonella typhimurium. J. Biol. Chem. 294, 9084–9099. doi: 10.1074/jbc.RA119.008209
Ray, S., Da Costa, R., Thakur, S., and Nandi, D. (2020). Salmonella Typhimurium encoded cold shock protein E is essential for motility and biofilm formation. Microbiology 166, 460–473. doi: 10.1099/mic.0.000900
Rogers-Broadway, K. R., and Karteris, E. (2015). Amplification efficiency and thermal stability of qPCR instrumentation: current landscape and future perspectives. Exp. Ther. Med. 10, 1261–1264. doi: 10.3892/etm.2015.2712
Sen, M., Shah, B., Rakshit, S., Singh, V., Padmanabhan, B., Ponnusamy, M., et al. (2011). UDP-glucose 4, 6-dehydratase activity plays an important role in maintaining cell wall integrity and virulence of Candida albicans. PLoS Pathog. 7:e1002384. doi: 10.1371/journal.ppat.1002384
Spöring, I., Felgner, S., Preuße, M., Eckweiler, D., Rohde, M., Häussler, S., et al. (2018). Regulation of flagellum biosynthesis in response to cell envelope stress in Salmonella enterica Serovar Typhimurium. mBio 9, e00736–e00717. doi: 10.1128/mBio.00736-17
Stecher, B., Hapfelmeier, S., Müller, C., Kremer, M., Stallmach, T., and Hardt, W. D. (2004). Flagella and chemotaxis are required for efficient induction of Salmonella enterica serovar Typhimurium colitis in streptomycin-pretreated mice. Infect. Immun. 72, 4138–4150. doi: 10.1128/IAI.72.7.4138-4150.2004
Stevenson, K., Mcvey, A. F., Clark, I. B. N., Swain, P. S., and Pilizota, T. (2016). General calibration of microbial growth in microplate readers. Sci. Rep. 6:38828. doi: 10.1038/srep38828
Stevenson, G., Neal, B., Liu, D., Hobbs, M., Packer, N. H., Batley, M., et al. (1994). Structure of the O antigen of Escherichia coli K-12 and the sequence of its rfb gene cluster. J. Bacteriol. 176, 4144–4156. doi: 10.1128/jb.176.13.4144-4156.1994
Takeda, S., Fujisawa, Y., Matsubara, M., Aiba, H., and Mizuno, T. (2001). A novel feature of the multistep phosphorelay in Escherichia coli: a revised model of the RcsC --> YojN --> RcsB signalling pathway implicated in capsular synthesis and swarming behaviour. Mol Microbiol. 40:440–50.
Thakur, S., Ray, S., Jhunjhunwala, S., and Nandi, D. (2020). Insights into coumarin-mediated inhibition of biofilm formation in Salmonella Typhimurium. Biofouling 36, 479–491. doi: 10.1080/08927014.2020.1773447
Urdaneta, V., and Casadesús, J. (2017). Interactions between Bacteria and bile salts in the gastrointestinal and hepatobiliary tracts. Front Med (Lausanne) 4:163. doi: 10.3389/fmed.2017.00163
van Teeseling, M. C. F., De Pedro, M. A., and Cava, F. (2017). Determinants of bacterial morphology: from fundamentals to possibilities for antimicrobial targeting. Front. Microbiol. 8:1264. doi: 10.3389/fmicb.2017.01264
van Velkinburgh, J. C., and Gunn, J. S. (1999). PhoP-PhoQ-regulated loci are required for enhanced bile resistance in Salmonella spp. Infect. Immun. 67, 1614–1622. doi: 10.1128/IAI.67.4.1614-1622.1999
Varghese, A., Ray, S., Verma, T., and Nandi, D. (2020). Multicellular string-like structure formation by Salmonella Typhimurium depends on cellulose production: roles of Diguanylate Cyclases, YedQ and YfiN. Front. Microbiol. 11:613704. doi: 10.3389/fmicb.2020.613704
Verma, T., Annappa, H., Singh, S., Umapathy, S., and Nandi, D. (2021). Profiling antibiotic resistance in Escherichia coli strains displaying differential antibiotic susceptibilities using Raman spectroscopy. J. Biophotonics 14:e202000231. doi: 10.1002/jbio.202000231
Wang, Q., Zhao, Y., McClelland, M., and Harshey, R. M. (2007). The RcsCDB signaling system and swarming motility in Salmonella enterica serovar typhimurium: dual regulation of flagellar and SPI-2 virulence genes. J Bacteriol. 189:8447–57.
Wu, H., Fan, Z., Jiang, X., Chen, J., and Chen, G.-Q. (2016). Enhanced production of polyhydroxybutyrate by multiple dividing E. coli. Microb. Cell Factories 15:128. doi: 10.1186/s12934-016-0531-6
Yadav, S., Pathak, S., Sarikhani, M., Majumdar, S., Ray, S., Chandrasekar, B. S., et al. (2018). Nitric oxide synthase 2 enhances the survival of mice during Salmonella typhimurium infection-induced sepsis by increasing reactive oxygen species, inflammatory cytokines and recruitment of neutrophils to the peritoneal cavity. Free Radic. Biol. Med. 116, 73–87. doi: 10.1016/j.freeradbiomed.2017.12.032
Yao, Z., and Valvano, M. A. (1994). Genetic analysis of the O-specific lipopolysaccharide biosynthesis region (rfb) of Escherichia coli K-12 W3110: identification of genes that confer group 6 specificity to Shigella flexneri serotypes Y and 4a. J. Bacteriol. 176, 4133–4143. doi: 10.1128/jb.176.13.4133-4143.1994
Yoon, J. H., Shin, J.-H., Park, J. H., and Park, T. H. (2015). Effect of light intensity on the correlation between cell mass concentration and optical density in high density culture of a filamentous microorganism. Korean J. Chem. Eng. 32, 1842–1846. doi: 10.1007/s11814-015-0012-3
Zhang, G., Baidin, V., Pahil, K. S., Moison, E., Tomasek, D., Ramadoss, N. S., et al. (2018). Cell-based screen for discovering lipopolysaccharide biogenesis inhibitors. Proc. Natl. Acad. Sci. USA 115, 6834–6839. doi: 10.1073/pnas.1804670115
Zhao, H., Sun, Y., Peters, J. M., Gross, C. A., Garner, E. C., and Helmann, J. D. (2016). Depletion of Undecaprenyl pyrophosphate phosphatases disrupts cell envelope biogenesis in Bacillus subtilis. J. Bacteriol. 198, 2925–2935. doi: 10.1128/JB.00507-16
Keywords: lipopolysaccharide, enterobacterial common antigen, Salmonella Typhimurium, rfbB , rffG , dTDP-D-glucose 4, 6-dehydratase
Citation: Chakraborty S, Banerjee P, Joseph JP, Pathak S, Verma T, Karhale AK, Chandra D, Das M, Saha P and Nandi D (2025) Functional loss of rffG and rfbB, encoding dTDP-glucose 4,6-dehydratase, alters colony morphology, cell shape, motility and virulence in Salmonella Typhimurium. Front. Microbiol. 16:1572117. doi: 10.3389/fmicb.2025.1572117
Edited by:
Axel Cloeckaert, Institut National de recherche pour l’agriculture, l’alimentation et l’environnement (INRAE), FranceReviewed by:
Sara Petrin, Experimental Zooprophylactic Institute of the Venezie (IZSVe), ItalyRuchika Dehinwal, Harvard University, United States
Paul Ugalde Silva, Rhode Island Hospital, United States
Chamith Hewawaduge, Sri Lanka Institute of Biotechnology, Sri Lanka
Copyright © 2025 Chakraborty, Banerjee, Joseph, Pathak, Verma, Karhale, Chandra, Das, Saha and Nandi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dipankar Nandi, bmFuZGlAaWlzYy5hYy5pbg==
†These authors have contributed equally to this work and share first authorship
 Subhashish Chakraborty
Subhashish Chakraborty Pip Banerjee
Pip Banerjee Joel P. Joseph
Joel P. Joseph Sanmoy Pathak
Sanmoy Pathak Taru Verma
Taru Verma Aagosh Kishor Karhale
Aagosh Kishor Karhale Deepti Chandra1
Deepti Chandra1 Mrinmoy Das
Mrinmoy Das Dipankar Nandi
Dipankar Nandi