- 1School of Traditional Chinese Medicine, Southern Medical University, Guangzhou, China
- 2Hong Kong University of Science and Technology, Guangzhou, China
- 3School of Chinese Materia Medica, Guangdong Pharmaceutical University, Guangzhou, China
- 4Department of Breast Cancer, Cancer Center, Guangdong Provincial People's Hospital, Southern Medical University, Guangzhou, China
- 5Center for Drug Research and Development, Guangdong Provincial Key Laboratory for Research and Evaluation of Pharmaceutical Preparations, Guangdong Pharmaceutical University, Guangzhou, China
- 6Key Laboratory of Glucolipid Metabolic Disorder, Ministry of Education, Guangdong Pharmaceutical University, Guangzhou, China
Introduction: Sepsis is a life-threatening condition that often leads to organ dysfunction and systemic inflammation, with gut microbiota dysbiosis playing a crucial role in its pathogenesis. The role of Desulfovibrio vulgaris (D. vulgaris), a potentially pathogenic bacterium, in sepsis remains unclear.
Methods: We first assessed the abundance of D. vulgaris in the feces of septic mice and patients using qPCR. Mice were then orally gavaged with D. vulgaris (2 × 108 CFU/mouse/day) for 7 consecutive days followed by cecal ligation and puncture (CLP) surgery. We monitored survival, assessed organ damage, and measured inflammation. Peritoneal macrophages were isolated to analyze the phosphorylation of key MAPK and NF-κB signaling pathways. Finally, oxidative stress levels in the liver, lungs, and kidneys were evaluated, measuring markers such as GSH, CAT, and SOD.
Results: The abundance of D. vulgaris was significantly increased in the feces of both septic mice and patients. Supplementation with D. vulgaris exacerbated sepsis in mice, resulting in lower survival rates, more severe organ damage, and heightened inflammation. Phosphorylation of MAPK and NF-κB pathways in peritoneal macrophages was significantly enhanced. Additionally, D. vulgaris amplified oxidative stress across multiple organs, as indicated by increased ROS levels and decreased antioxidant enzyme activity.
Conclusion: Our findings suggest that D. vulgaris exacerbates the progression of sepsis by enhancing inflammation, activating key immune signaling pathways, and increasing oxidative stress. These processes contribute to organ dysfunction and increased mortality, highlighting the potential pathogenic role of D. vulgaris in sepsis.
Introduction
Sepsis and septic shock are major global healthcare challenges, with mortality rates exceeding 25%, resulting in substantial morbidity and healthcare costs (Fleischmann et al., 2016). Timely intervention is essential to improve survival outcomes in sepsis patients, as delayed treatment increases mortality by 4% per hour (Han X. et al., 2021). Mortality rates among critically ill sepsis patients vary based on factors such as sepsis severity, patient demographics, and healthcare settings. Recent studies have reported intensive care unit mortality rates for sepsis patients ranging from approximately 26–41.9% (Sakr et al., 2018; Fleischmann-Struzek et al., 2020).
Sepsis, as a systemic inflammatory response syndrome, involves complex immune, metabolic, and inflammatory regulatory networks (van der Poll et al., 2017; Zhang and Ning, 2021; Chen et al., 2022). The onset of sepsis is intricately linked to multiple factors (Giamarellos-Bourboulis et al., 2024), with gut microbiota dysbiosis emerging as a critical contributor (Haak and Wiersinga, 2017; Sun et al., 2023). In sepsis, gut microbiota undergoes profound disruption, compromising the intestinal barrier and triggering aberrant immune responses (Lankelma et al., 2017; Piccioni et al., 2024). Gut microbiota dysbiosis influences systemic immune activation through the “gut-immune axis” contributing to the intensification of inflammation, organ dysfunction, and poor clinical outcomes in sepsis patients. This dysbiosis may drive sepsis progression through mechanisms such as the release of pro-inflammatory cytokines, increased oxidative stress, and immune cell dysfunction (Liu W. et al., 2020; Fang et al., 2022; Chancharoenthana et al., 2023; Chen et al., 2023; Kunst et al., 2023). As a result, understanding the role of gut microbiota in sepsis has become a prominent area of investigation in recent years.
Among the gut microbiota, Desulfovibrio species are anaerobic, Gram-negative, and sulfate-reducing bacteria that play an important role in host health by influencing metabolism and immune responses (Figliuolo et al., 2017; Lu et al., 2023). Desulfovibrio vulgaris (D. vulgaris), a prominent species within this group, produces hydrogen sulfide (H₂S) through sulfate reduction (Ueki and Lovley, 2022). While low concentrations of H₂S serve as signaling molecules, higher concentrations can be toxic, damaging intestinal epithelial cells, compromising the gut barrier, and amplifying systemic inflammation (Jin et al., 2024; Munteanu et al., 2024a). These properties suggest a potential role for D. vulgaris in exacerbating conditions like sepsis, where immune responses and intestinal barrier dysfunction are critical factors.
Desulfovibrio vulgaris has garnered significant attention due to its potential pathogenic role in various diseases. Its overgrowth has been linked to the development of ulcerative colitis, inflammatory bowel disease, Parkinson’s disease, and colorectal cancer (Murros et al., 2021; An et al., 2023; Huang et al., 2024; Xie et al., 2024; Dong et al., 2025). D. vulgaris abundance is increased in patients with ulcerative colitis and correlates with the severity of the disease (An et al., 2023; Xie et al., 2024). One study demonstrated that the flagellin of D. vulgaris exacerbated ulcerative colitis in mice by activating the NAIP/NLRC4 inflammasome and inducing macrophage pyroptosis (An et al., 2023). Another study revealed that D. vulgaris or its flagellin interacted with LRRC19, rather than TLR5, to activate related signaling pathways, promoting the production of pro-inflammatory cytokines and aggravating ulcerative colitis (Xie et al., 2024). Furthermore, transplantation of D. vulgaris into healthy mice can induce intestinal inflammation, disrupt the intestinal barrier, and reduce short-chain fatty acid levels, thereby significantly exacerbating DSS-induced colitis (Huang et al., 2024). A previous study suggested that inhibiting Desulfovibrio could mitigate acute gastrointestinal inflammation in sepsis (Han C. et al., 2021). Additionally, Desulfovibrio has been reported to promote the production of LPS, a key component of Gram-negative bacterial outer membranes, which plays a crucial role in triggering acute inflammation and sepsis (Weglarz et al., 2003; Novakovic et al., 2016). Moreover, an increased abundance of Desulfovibrio has been positively correlated with elevated serum levels of AST and ALT, both indicators of liver injury (Cao et al., 2020). Although the role of D. vulgaris in various diseases has been preliminarily explored, studies on the specific role of D. vulgaris in sepsis are limited. The changes in its abundance within the gut microbiota of septic patients and its direct impact on disease progression remain unclear.
To address this issue, we established a murine sepsis model to explore the role of D. vulgaris in sepsis. We compared the abundance of D. vulgaris in the gut microbiota of septic mice and patients. To further investigate the impact of D. vulgaris on sepsis progression, we examined the inflammatory responses and oxidative stress across multiple organs. Our findings revealed a significant increase in D. vulgaris abundance in septic mice and patients. Notably, D. vulgaris exacerbated systemic inflammation by activating the mitogen-activated protein kinase (MAPK) and nuclear factor kappa B (NF-κB) signaling pathways in macrophages, contributing to multi-organ damage. Additionally, D. vulgaris promoted oxidative stress in various tissues, intensifying the pathological damage associated with sepsis. These results highlight the pivotal role of D. vulgaris in sepsis progression and suggest that targeting its growth or function could provide a novel therapeutic approach for alleviating sepsis-induced organ damage.
Materials and methods
Human samples
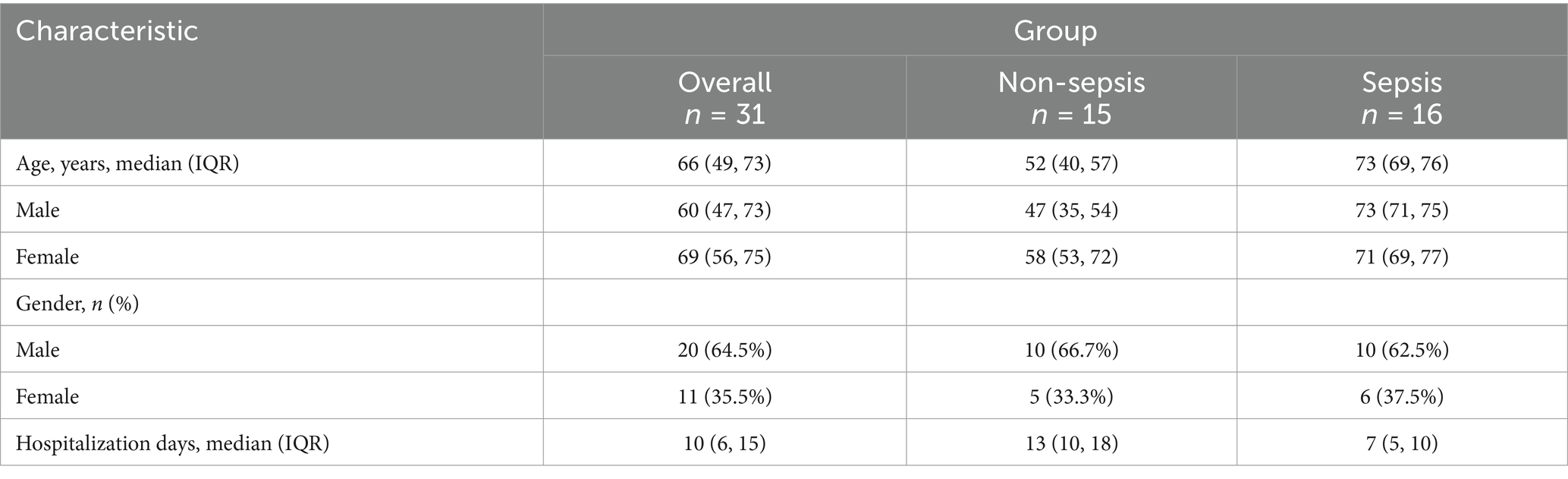
All human serum samples utilized in this study were procured from Nanfang Hospital. Patients diagnosed with sepsis in the ICU were selected based on the SEPSIS 3.0 criteria (Singer et al., 2016), while non-septic samples were collected from individuals undergoing trauma-related surgical procedures. Written informed consent was obtained from all participants prior to their inclusion. The study was approved by the Medical Ethics Committee of NanFang Hospital of Southern Medical University under the reference number NFEC-202403-K11. The baseline characteristics of the patients are summarized in Table 1.
Mouse cecal ligation and puncture model
Male C57BL/6 mice (6–8 weeks old, 20–22 g) were obtained from Beijing Sibefur Laboratory Animal Technology Co., Ltd. Mice were housed under a 12-h light/dark cycle with free access to food and water at 22–24°C. Anesthesia was induced with isoflurane and confirmed by the loss of the righting reflex. The abdominal area was disinfected with 75% ethanol, and the fur was shaved. A midline incision (~2 cm) was made to expose the cecum, taking care not to damage the mesenteric vessels. The cecum was ligated ~1 cm from the tip using 4–0 silk and punctured once with an 18G needle at the distal portion (~3/4 from the tip). A small amount of cecal content was extruded to keep the puncture site open. The cecum was returned to the abdominal cavity, and the incision was closed in layers. Post-surgery, mice received saline (50 mL/kg) for resuscitation and were kept on a heated pad. Survival was monitored for 36 h, and Kaplan–Meier survival curves were generated. Tissue samples for multi-organ injury analysis were collected 6 h or 12 h after CLP. The animal study was approved by the Animal Ethics Committee of the Animal Experiment Center of Southern Medical University. All procedures were conducted in accordance with local legislation and institutional guidelines.
Mouse endotoxemia model
Male C57BL/6 mice were intraperitoneally injected with 25 mg/kg lipopolysaccharide (LPS from E. coli O111:B4; Sigma, Cat# L2630) to induce endotoxemia. Cecal contents were harvested 12 h after LPS administration for further analysis.
Desulfovibrio vulgaris culture
Desulfovibrio vulgaris (Cat# bio-092027) was obtained from Biobw. The strain was cultured anaerobically for 48 h on Columbia blood agar plates at 37°C. After incubation, bacterial colonies were scraped from the plates, centrifuged, and resuspended for enumeration. Mice were orally gavaged with D. vulgaris at a dose of 2 × 108 CFU per mouse daily for 7 consecutive days. Following this treatment, CLP surgery was performed to induce sepsis.
Bacterial DNA extraction
Approximately 20–30 mg of cecal contents were mixed with 500 μL of PBS containing 0.5% Tween 20, homogenized, and vortexed. The sample underwent three freeze–thaw cycles (−80°C for 10 min, 60°C for 5 min, repeated three times) to disrupt the cell walls. Then, 5 μL of proteinase K (20 mg/mL), 10 μL of RNase (0.1 mg/μL), and 25 μL of 10% SDS were added, and the mixture was incubated at 55°C for 1 h. After incubation, 75 μL of phenol was added and mixed, followed by an equal volume of PCI solution. The mixture was centrifuged at 13,000 RPM for 10 min at 4°C, and the supernatant was collected. The extraction was repeated with PCI solution and chloroform (1:1 ratio) with 3 M sodium acetate (1/10th volume), followed by centrifugation. To precipitate the DNA, 2.5 volumes of pre-chilled ethanol were added, and the mixture was centrifuged again. The DNA pellet was washed with 80% ethanol, air-dried, and resuspended in 100 μL ultrapure water. The DNA was stored at −20°C for further analysis.
Desulfovibrio abundance analysis
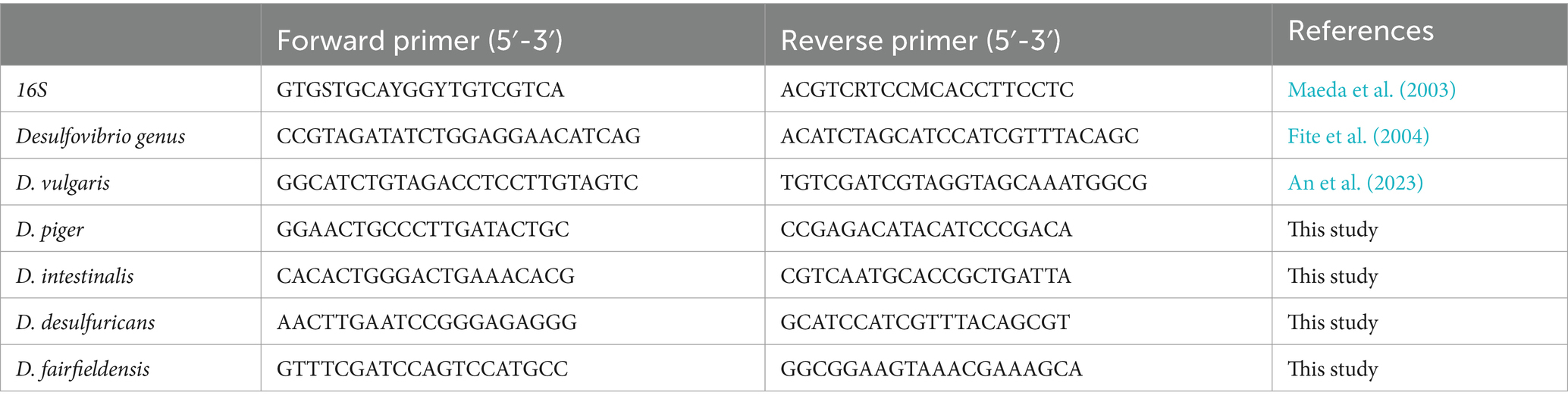
The relative abundance of the Desulfovibrio genus and key species, including D. vulgaris, D. piger, D. desulfuricans, and D. fairfieldensis, was quantified using real-time quantitative PCR (qPCR). Specific primers targeting these taxa are listed in Table 2. qPCR reactions were performed in triplicate using a SYBR Green-based master mix (TOYOBO, Cat# QPK-201). The cycling conditions were optimized to ensure high specificity and amplification efficiency. Relative gene expression levels were calculated using the 2−ΔΔCt method, with 16S rRNA serving as the internal control for normalization.
Real-time quantitative PCR analysis
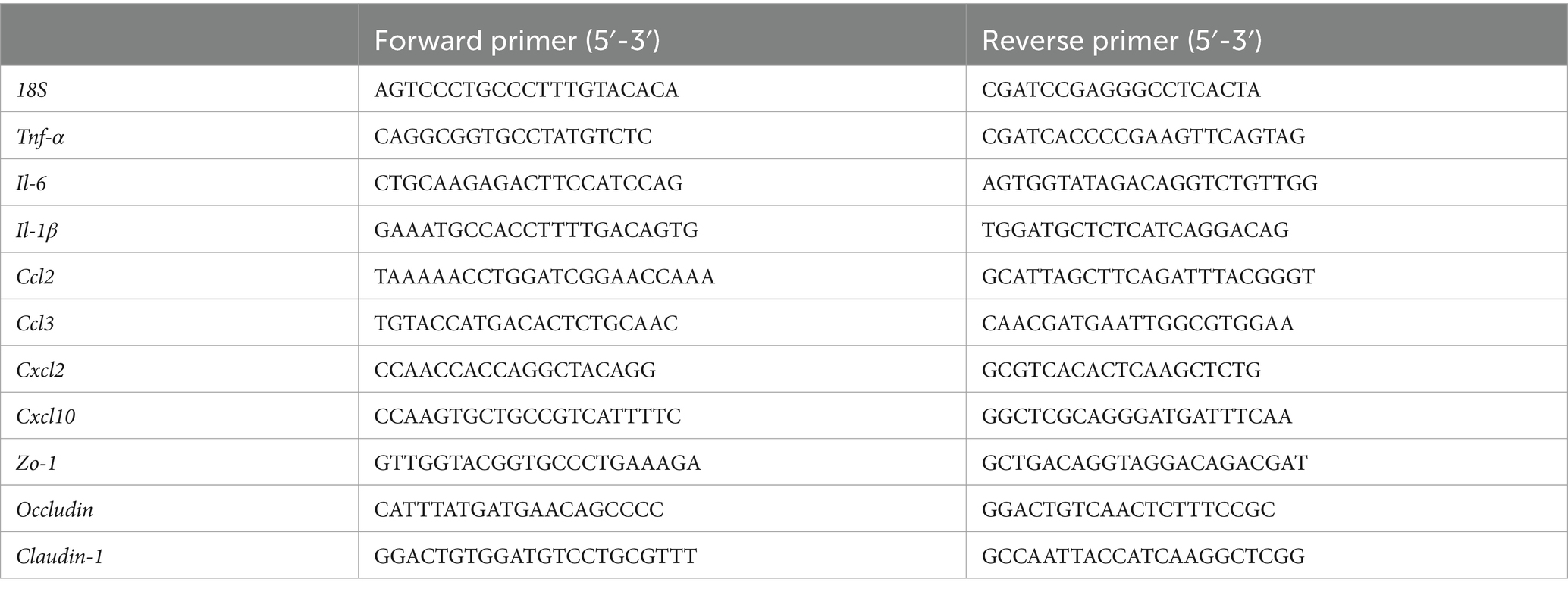
To evaluate the transcriptional levels of inflammatory cytokines and chemokines in mouse tissues, total RNA was isolated under RNase-free conditions using TRIzol Reagent (Invitrogen, Cat# 15596018). The RNA was then reverse-transcribed into complementary DNA (cDNA) using a reverse transcription kit (TOYOBO, Cat# FSQ-101). qPCR was performed on a Roche LightCycler® 96 Real-Time PCR System with SYBR Green as the fluorescent dye. Gene-specific primers for inflammatory cytokines and chemokines were designed and are listed in Table 3. The 18S ribosomal RNA (18S rRNA) served as an internal reference gene for normalization. Relative gene expression levels were calculated using the 2−ΔΔCt method.
ALT and AST assays
Serum ALT and AST levels were measured by a kinetic method using commercial assay kits (Nanjing Jiancheng Bioengineering Institute; ALT: Cat# C009-3-1; AST: Cat# C010-3-1) following the manufacturer’s instructions. Peripheral blood was collected and centrifuged at 12,000 rpm for 15 min at 4°C to isolate serum. In a 96-well plate, 200 μL of Reagent 1, 50 μL of Reagent 2, and 10 μL of serum were mixed per well. The absorbance at 340 nm was monitored at both time points of 0 and 1 min at a temperature of 37°C using a microplate reader (Epoch, BioTek). The activities of ALT and AST were then calculated based on the formulas according to the manufacturer.
Serum creatinine measurement
Serum creatinine (Cr) levels were determined using a creatinine assay kit (Nanjing Jiancheng Bioengineering Institute, Cat# C011-2-1) based on the sarcosine oxidase method. In a 96-well plate, 6 μL of serum samples, 6 μL of creatinine standard solution (442 μmol/L), and 6 μL of double-distilled water were added to the test wells, standard wells, and blank wells, respectively. Subsequently, 180 μL of Enzyme Solution A was added to each well. The plate was incubated at 37°C for 5 min, and absorbance at 546 nm (A1) was recorded using a microplate reader. Then, 60 μL of Enzyme Solution B was added to each well, followed by another 5-min incubation at 37°C. Absorbance at 546 nm (A2) was recorded, and creatinine levels were calculated according to the kit’s formula.
Blood urea nitrogen measurement
Blood urea nitrogen (BUN) was measured using a urea nitrogen assay kit (Cat# C013-2-1, Nanjing Jiancheng Bioengineering Institute) via the urease method. In brief, 20 μL of serum samples, 20 μL of urea nitrogen standard solution (10 mM), and 20 μL of double-distilled water were added to the test wells, standard wells, and blank wells, respectively. Each well was supplemented with 250 μL of buffer enzyme solution, mixed by vortexing for 10 s, and incubated in a water bath at 37°C for 10 min. Next, 1 mL of color reagent and 1 mL of alkaline sodium hypochlorite solution were added, vortexed for 10 s to ensure complete mixing, and incubated again at 37°C for 10 min. Subsequently, 200 μL of the solution was transferred to a 96-well plate, and the absorbance was measured at 640 nm using a microplate reader. BUN levels were calculated according to the kit’s formula.
Measurement of GSH and antioxidant enzyme activities
For the determination of GSH and antioxidant enzyme activities, 100 mg of fresh tissue samples were homogenized in 900 μL of PBS (pH 7.4, 0.1 M). The homogenate was then centrifuged at 3,500 × g for 10 min, and the resulting supernatant was used to measure the activities of GSH and antioxidant enzyme activities including superoxide dismutase (SOD) and catalase (CAT) using commercially available assay kits (Nanjing Jiancheng Bioengineering Institute). Protein concentration in the supernatant was quantified using a Pierce BCA Protein Assay Kit (Pierce, Cat# 23225).
The GSH content was determined using the Glutathione Assay Kit (Cat# A006-2-1) according to the DTNB [5,5′-dithiobis (2-nitrobenzoic acid)] method. Absorbance was measured at 405 nm, and the GSH content was expressed as μmol per g protein.
CAT activity was assessed using the CAT Assay Kit (Cat# A007-1-1), and results were expressed as U per mg protein. One unit of CAT activity is defined as the amount of enzyme that decomposes 1 μmol of H₂O₂ per minute per mg of fresh sample at 37°C.
SOD activity was measured using the SOD Assay Kit (Cat# A001-1-2) and expressed as U per mg protein. One unit of SOD activity corresponds to the amount of extract that causes 50% inhibition in the reduction of xanthine, as monitored at 550 nm.
Hematoxylin and eosin staining
Mouse liver, kidney, and lung tissues were fixed in 4% paraformaldehyde for 24 h, followed by dehydration, clearing, paraffin embedding, and sectioning. The embedded tissues were cut into 5 μm sections, baked at 60°C for 1 h, and stored for later use. For staining, sections were deparaffinized in xylene (twice for 10 min each), then sequentially rehydrated through a graded ethanol series (100, 95, 90, and 80%, each for 2 min) before rinsing in distilled water. Hematoxylin staining was performed for 8 min, followed by rinsing with running tap water for 10 min. Sections were then differentiated in 1% hydrochloric acid alcohol for 10 s and rinsed with running tap water for 10 min to restore the blue color. Eosin staining was performed for 3 min, followed by sequential dehydration in 80, 90, 95, and 100% ethanol, and clearing in xylene (twice for 5 min each). Histological evaluation was conducted by capturing at least six random fields per H&E-stained slide under a light microscope. Liver, kidney, and lung injuries were assessed and scored based on histopathological criteria described in a previous study published by our research group (Chen et al., 2023). In brief, liver injury was assessed based on necrosis, inflammation, ballooning degeneration, and hepatic cord disruption, with each parameter scored from 0 to 3. Lung injury was evaluated based on alveolar congestion, hemorrhage, inflammatory cell infiltration, and alveolar wall thickening, with each parameter scored from 0 to 3. Kidney injury was assessed based on the extent of tubular and glomerular damage, with a scoring range of 0 to 5.
Measurement of ROS production
ROS production in tissues was assessed using the dihydroethidium (DHE) oxidation assay. Frozen liver, lung, and kidney sections were incubated with 5 μM DHE (Thermo Scientific, Cat# D23107) at 37°C for 20 min. After incubation, the ROS levels were observed using a Leica microscope (Leica DMi8), and the average fluorescence intensity was measured and analyzed using ImageJ software.
TUNEL staining
To evaluate cell death in the liver, kidney, and lung tissues of CLP mice, paraffin-embedded tissue sections were subjected to terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) staining using a commercial kit (KeyGEN BioTECH, Cat# KGA1405-100) according to the manufacturer’s instructions. Briefly, tissue sections were deparaffinized in xylene, rehydrated through a graded ethanol series, and rinsed with PBS. Permeabilization was performed by incubating the sections with Proteinase K (20 μg/mL) at 37°C for 30 min, followed by PBS washes. The TdT enzyme reaction mixture was prepared by combining Equilibration Buffer, biotin-11-dUTP, and TdT Enzyme, and 50 μL of the mixture was applied to each section. The slides were incubated in a humidified chamber at 37°C in the dark for 60 min. After washing with PBS, a Streptavidin-TRITC labeling solution was applied, and the slides were incubated at 37°C in the dark for 30 min. Following another PBS wash, nuclei were counterstained with DAPI (1 μg/mL) for 10 min at room temperature. Excess DAPI was removed by rinsing with PBS, and the sections were mounted with an anti-fade medium and coverslipped. TUNEL-positive cells (apoptotic cells) were identified by red fluorescence under a fluorescence microscope, while DAPI stained all nuclei blue. The number of TUNEL-positive cells was quantified using image analysis software.
ELISA
Enzyme-linked immunosorbent assay (ELISA) was performed to determine the serum concentrations of TNF-α, IL-1β, and IL-6 using commercial ELISA kits from Neobioscience according to the manufacturer’s instructions (TNF-α: Cat# EMC102a, IL-1β: Cat# EMC001b, IL-6: Cat# EMC004). Briefly, serum samples were appropriately diluted. A 100 μL aliquot of each diluted sample or standard was added and incubated at 37°C for 90 min. Following incubation, the plate was washed, and a biotinylated detection antibody solution (100 μL) was added to each well and incubated at 37°C for 60 min. After washing, 100 μL of enzyme-conjugated streptavidin was added to each well and incubated at 37°C for 30 min. The plate was washed again, and 100 μL of TMB substrate solution was added to each well. The reaction was allowed to develop, then stopped using the stop solution. The absorbance was measured at 450 nm using a microplate reader. Cytokine concentrations in the samples were determined by comparing the absorbance values to the standard curve generated using the provided standards.
Peritoneal macrophage sorting
Peritoneal lavage fluid from septic mice was passed through a 70 μm nylon mesh and centrifuged. The cell pellet was resuspended in MACS buffer (PBS with 0.5% BSA and 2 mM EDTA). To label macrophages, 10 μL of F4/80 MicroBeads antibody (Miltenyi Biotec, Cat# 130–110-443) was added, followed by a 20-min incubation at 4°C. F4/80-positive macrophages were then isolated using LS columns (Miltenyi Biotec) according to the manufacturer’s instructions.
Western blot analysis
Total protein was extracted using RIPA buffer (Beyotime, Cat# P0013C), and protein concentrations were determined using a Pierce BCA Protein Assay Kit. Equal amounts of protein were separated by 10% SDS-PAGE and transferred onto nitrocellulose membranes. The membranes were blocked with 5% BSA and incubated overnight at 4°C with primary antibodies against SAPK/JNK (1:1,000, Proteintech, Cat# 66210-1-Ig), phospho-SAPK/JNK (1:1,000, CST, Cat# 9255S), ERK1/2 (1:1,000, CST, Cat# 4695S), phospho-ERK1/2 (1:1,000, CST, Cat# 9101S), NF-κB P65 (1:1,000, CST, Cat# 8242S), phospho-NF-κB P65 (1:1,000, CST, Cat# 3033S), P38 MAPK (1:1,000, Proteintech, Cat# 14064-1-AP), and phospho-P38 MAPK (1:1,000, CST, Cat# 9211S). ZO-1 (1:1,000, Affinity, Cat# AF5145), Occludin (1:1,000, Affinity, Cat# DF7504), and Claudin-1 (1:1,000, Affinity, Cat# AF0127). After washing, the membranes were incubated with HRP-conjugated secondary antibodies (RayBiotech; Goat anti-Mouse IgG(H + L)-HRP: Cat# RM3001, Goat anti-Rabbit IgG(H + L)-HRP: Cat# RM3002). Protein bands were visualized using an enhanced chemiluminescence system (Bio-Rad) and quantified with ImageJ software.
Immunohistochemistry
Paraffin-embedded tissue sections were deparaffinized and rehydrated, followed by antigen retrieval in citrate buffer using a microwave for 15 min. After cooling to room temperature, endogenous peroxidase activity was blocked with 3% hydrogen peroxide for 10 min, and nonspecific binding was blocked with 3% BSA for 30 min. Sections were incubated overnight at 4°C with a primary antibody against CD11b (1:100, Servicebio, Cat# GB11058). The following day, sections were washed with PBS and incubated with a secondary antibody at room temperature for 1 h. The signal was developed using 3,3′-diaminobenzidine (DAB), followed by hematoxylin counterstaining. Images were acquired using a microscope, capturing at least five fields of view per section. Quantification of CD11b positive area was performed using the IHC profile plugin in ImageJ software.
Statistical analysis
All statistical analyses were performed using GraphPad Prism software. Data were first tested for normality using the Shapiro–Wilk test and for homogeneity of variances using Levene’s test. If the data met the assumptions of normality and homogeneity of variance, comparisons between the two groups were conducted using an unpaired Student’s t-test. Survival analyses were conducted using the Kaplan–Meier method with log-rank tests. Data were expressed as mean ± SEM. Statistical significance was indicated as follows: *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
Results
Increased abundance of Desulfovibrio vulgaris in the gut microbiota of septic mice and patients
Desulfovibrio species are Gram-negative, anaerobic bacteria that are prevalent in the human gut microbiome (Singh et al., 2023). We collected cecal contents from septic mice 12 h after CLP surgery and extracted bacterial DNA (Figure 1A). Then, we assessed the abundance of Desulfovibrio species and several of its strains. The results showed a trend toward increased Desulfovibrio abundance in the cecum of septic mice, although this did not reach statistical significance (Figure 1B). Notably, D. vulgaris abundance in the cecum of septic mice was significantly elevated (Figure 1C), whereas the abundance of other Desulfovibrio species, including D. piger, D. intestinalis, D. fairfieldensis, and D. desulfuricans, remained unchanged (Figure 1D). To confirm whether this effect was observed in endotoxemia, we induced endotoxemia in mice by intraperitoneal injection of lipopolysaccharide (LPS; 25 mg/kg) and collected cecal contents for DNA extraction 12 h later (Figure 1E). qPCR analysis revealed a significant increase in D. vulgaris abundance in the cecum of endotoxemic mice (Figure 1F). Furthermore, in sepsis patients, the abundance of D. vulgaris was significantly higher compared to non-septic controls (Figure 1G). Collectively, these findings indicate a significant increase in D. vulgaris abundance in the gut microbiota of both septic mice and patients, suggesting that D. vulgaris may play a pivotal role in the pathogenesis of sepsis.
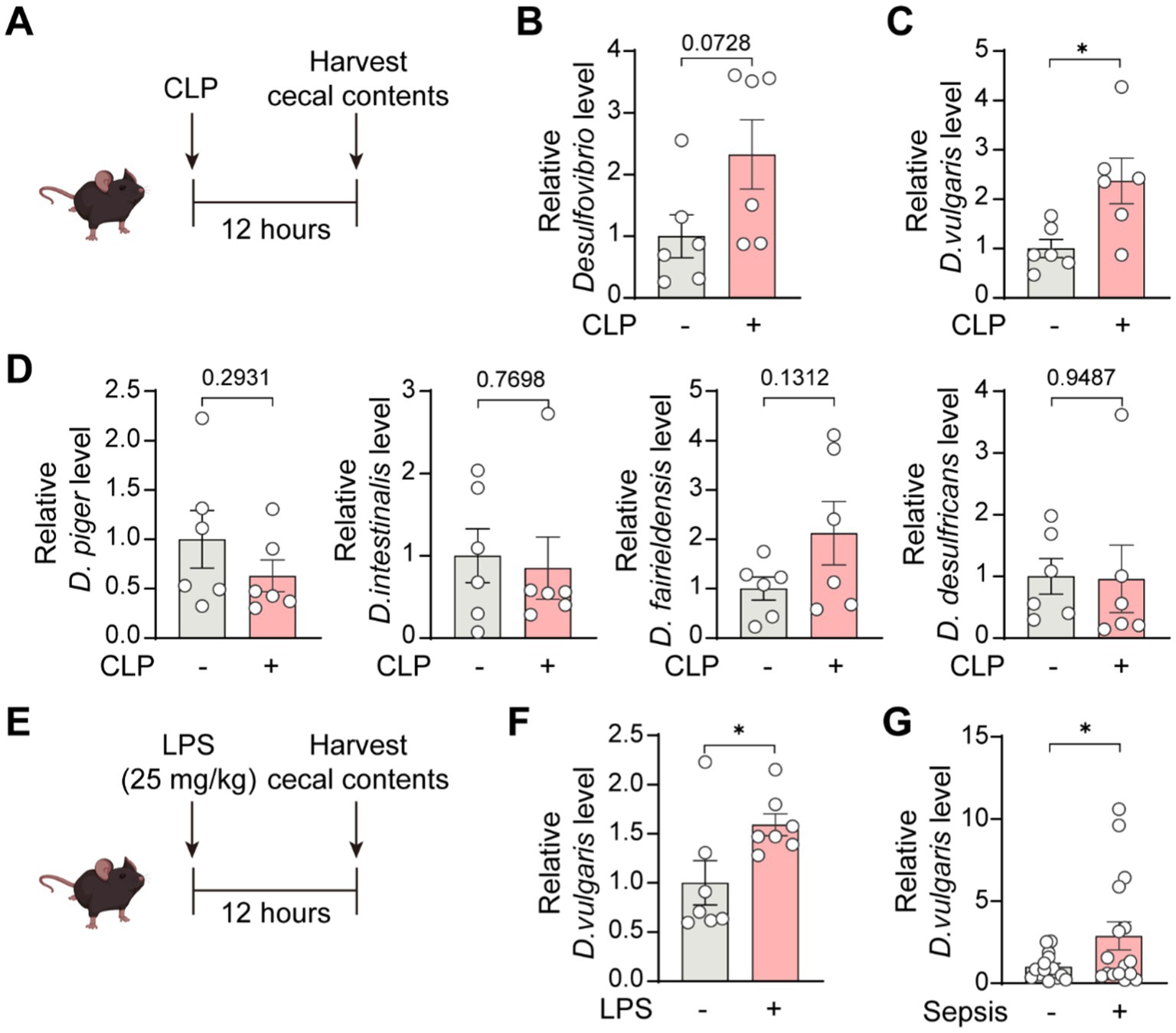
Figure 1. Increased abundance of D. vulgaris in the gut microbiota of septic mice and patients. (A) Cecal contents were collected from septic mice 12 h post-CLP surgery, and bacterial DNA was extracted. (B) qPCR analysis of the abundance of Desulfovibrio genus in the cecal microbiota of septic mice. n = 6. (C) qPCR analysis revealing increased D. vulgaris abundance in septic mouse cecal microbiota. n = 6. (D) qPCR analysis of the abundance of D. piger, D. intestinalis, D. fairfieldensis, and D. desulfuricans in the cecal microbiota of septic mice. n = 6. (E) Cecal contents were collected 12 h post-intraperitoneal LPS injection (25 mg/kg), with bacterial DNA extraction. (F) qPCR analysis showing increased D. vulgaris abundance in the cecal microbiota of endotoxemic mice. n = 7. (G) Comparison of D. vulgaris abundance in the cecal microbiota of septic versus non-septic patients. n = 15–16. Data are presented as mean ± SEM. Statistical comparisons were performed using two-tailed unpaired Student’s t-test. *p < 0.05.
Desulfovibrio vulgaris exacerbates mortality and organ damage in septic mice
To further explore the effects of D. vulgaris in sepsis, mice were gavaged with D. vulgaris (2 × 108 CFU/mouse/day) for 7 consecutive days, followed by CLP surgery. Survival rates were monitored for 36 h post-surgery, and tissues were collected for analysis (Figure 2A). D. vulgaris successfully colonized the gut, as evidenced by its significantly increased abundance in mouse feces (Supplementary Figure S1A). The results showed that D. vulgaris significantly reduced the survival rate of mice with sepsis compared to controls (Figure 2B), indicating that D. vulgaris exacerbates sepsis-induced mortality. Additionally, D. vulgaris treatment markedly elevated serum levels of ALT, AST, Cr, and BUN in septic mice (Figure 2C), suggesting damage to the liver and kidney tissues. However, D. vulgaris did not elevate ALT, AST, Cr, or BUN levels in healthy mice (Supplementary Figure S1B). Furthermore, D. vulgaris aggravated intestinal mucosal barrier damage in septic mice (Figures 2D,E), whereas it had no impact on the intestinal mucosal barrier in healthy mice (Supplementary Figure S1C). To investigate the direct impact of D. vulgaris on organ tissues, histological analysis was performed. Treatment with D. vulgaris markedly aggravated pathological damage in the liver, kidney, and lung tissues of septic mice. (Figure 3A). However, the proliferation of D. vulgaris did not cause pathological damage in the tissues of healthy mice (Supplementary Figure S2). TUNEL staining revealed a significant increase in apoptosis in these organs following D. vulgaris treatment (Figure 3B). These results suggest that D. vulgaris exacerbates multi-organ pathological damage in sepsis through induction of tissue injury and cellular apoptosis.
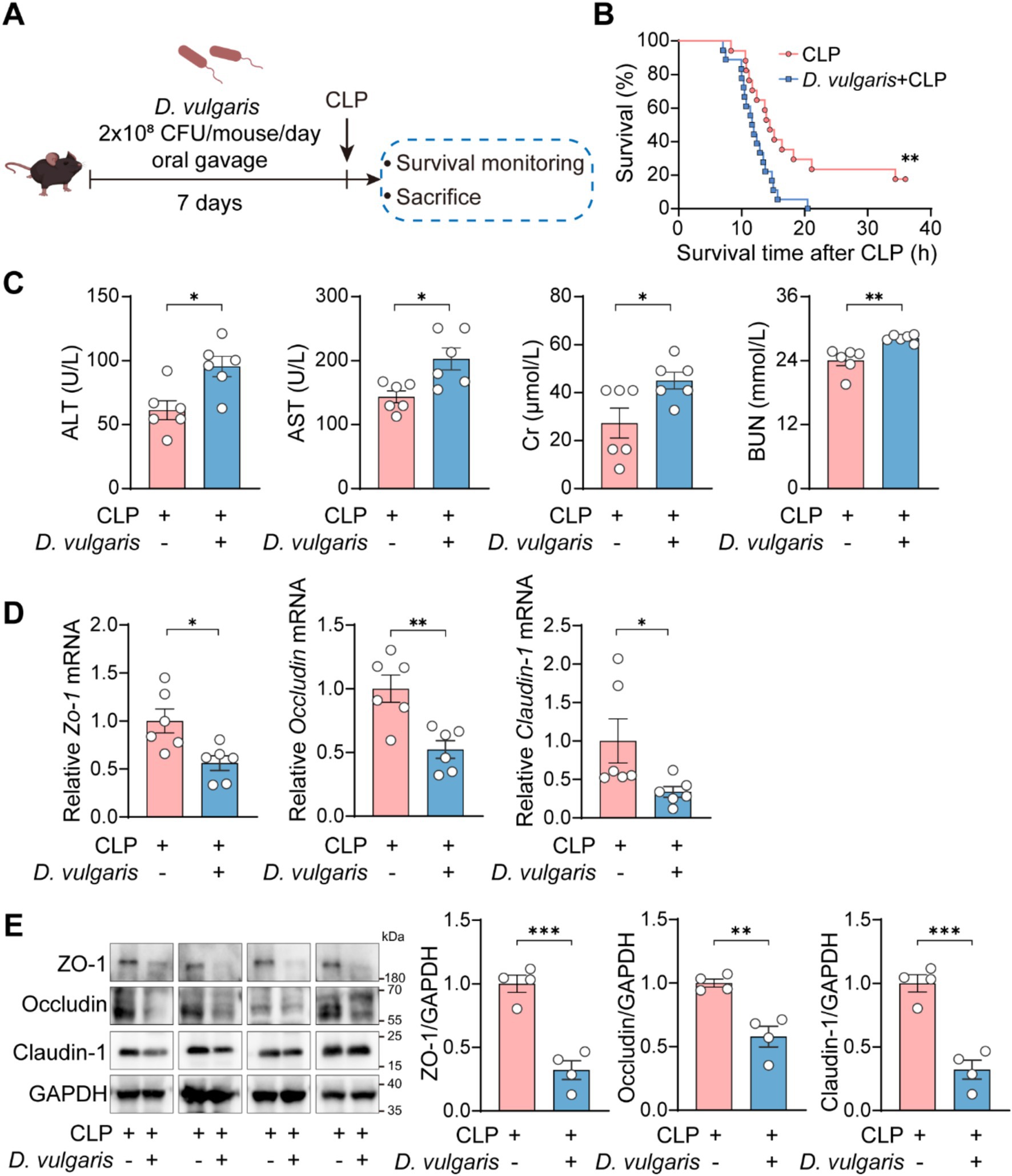
Figure 2. D. vulgaris exacerbates mortality and organ dysfunction in septic mice. (A) Experimental design for D. vulgaris administration: mice were gavaged with D. vulgaris (2 × 108 CFU/mouse) daily for 7 days before CLP surgery. Survival was monitored for 36 h, and tissues were collected 12 h post-CLP. (B) Survival rates of D. vulgaris-treated mice after CLP. n = 17–18. (C) Serum ALT, AST, Cr, and BUN levels in septic mice after D. vulgaris gavage. n = 6. (D) The relative mRNA levels of Zo-1, Occludin, and Claudin-1 in the colon. n = 6. (E) The protein expression levels of ZO-1, Occludin, and Claudin-1 in the colon. n = 4. Data are presented as mean ± SEM. Survival rates were analyzed using the Kaplan–Meier method with the log-rank test (B). Statistical comparisons were performed using two-tailed unpaired Student’s t-tests (C–E). *p < 0.05, **p < 0.01, ***p < 0.001.
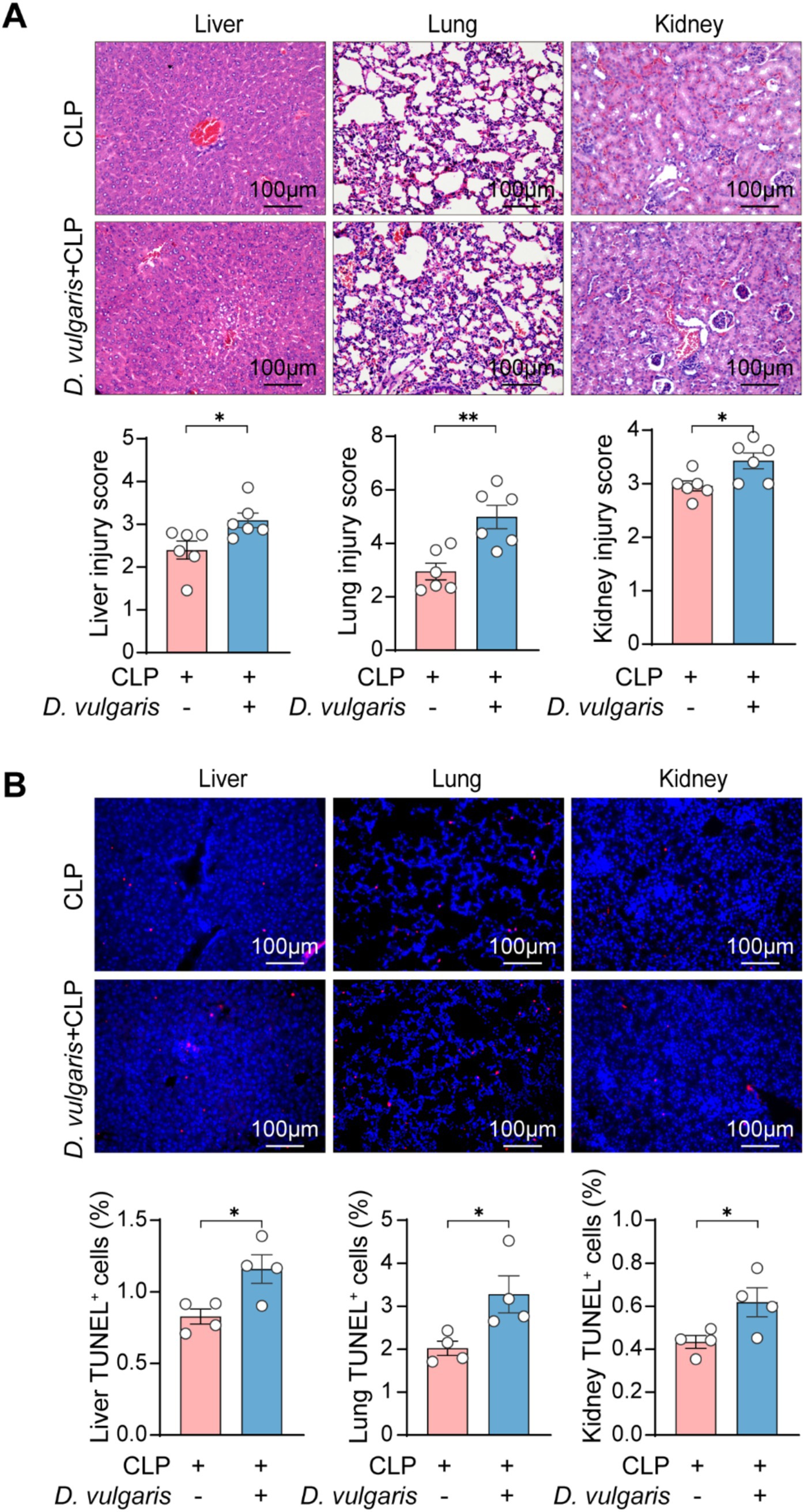
Figure 3. D. vulgaris promotes multi-organ pathological damage in septic mice. (A) Representative H&E-stained histological images and quantitative analysis of liver, kidney, and lung tissue damage in septic mice after D. vulgaris administration. n = 6. (B) TUNEL staining (red: TUNEL-positive cells, blue: cell nuclei) showing apoptosis in liver, kidney, and lung tissues of septic mice after D. vulgaris gavage. n = 4. Data are presented as mean ± SEM. Statistical comparisons were performed using two-tailed unpaired Student’s t-test. *p < 0.05, **p < 0.01. Scale bars, 100 μm.
Desulfovibrio vulgaris enhances inflammatory responses in septic mice
To examine the effect of D. vulgaris on inflammation, we measured levels of inflammatory cytokines in both serum and tissues. ELISA analysis showed that D. vulgaris treatment significantly elevated serum levels of the pro-inflammatory cytokines TNF-α, IL-1β, and IL-6 in septic mice (Figure 4A). Immunohistochemical analysis further demonstrated a marked increase in the infiltration of CD11b+ cells in the liver, kidneys, and lungs of septic mice treated with D. vulgaris (Figures 4B,C). Additionally, qPCR analysis revealed that D. vulgaris treatment significantly upregulated the transcriptional levels of inflammatory cytokines and chemokines in these organs (Figure 4D). These findings collectively suggest that D. vulgaris treatment exacerbates inflammation in septic mice, as evidenced by increased levels of pro-inflammatory cytokines, enhanced immune cell infiltration, and elevated gene expression of inflammatory mediators in key organs.
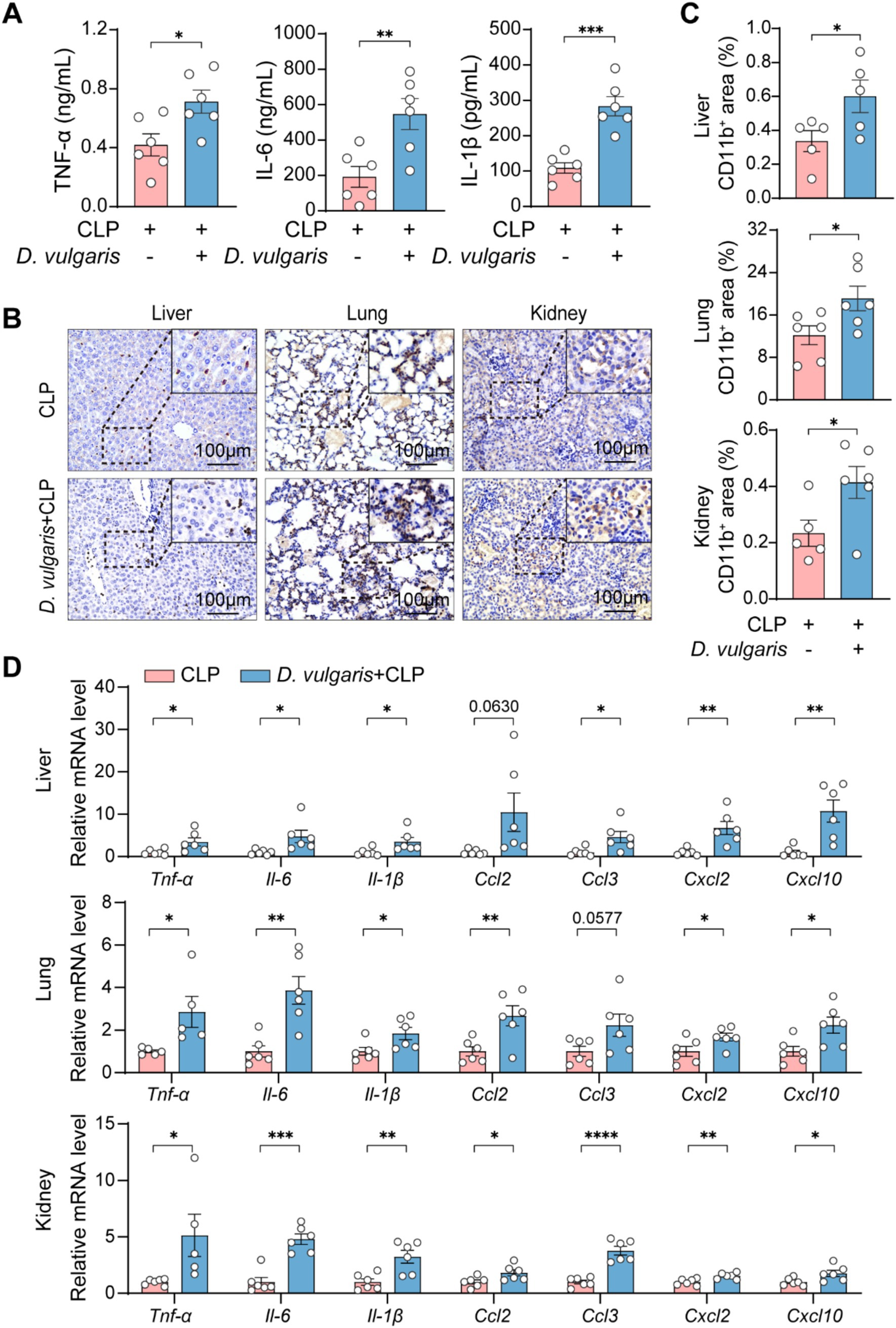
Figure 4. D. vulgaris induces inflammation in septic mice. (A) ELISA measurement of TNF-α, IL-1β, and IL-6 levels in the serum of D. vulgaris-treated septic mice. n = 6. (B,C) Immunohistochemical staining showing CD11b+ cell infiltration in liver, kidney, and lung tissues. n = 5–6. (D) qPCR analysis of inflammatory cytokines and chemokines in liver, kidney, and lung tissues. Data are presented as mean ± SEM. Statistical comparisons were performed using two-tailed unpaired Student’s t-test. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. Scale bars, 100 μm.
Desulfovibrio vulgaris promotes the activation of MAPK and NF-κB pathways in septic mice
To investigate the molecular mechanisms of D. vulgaris-induced inflammation, we focused on key signaling pathways involved in immune activation. The MAPK and NF-κB pathways are central regulators of inflammation and are commonly activated in conditions like sepsis (Gottschalk et al., 2016; Zhang and Ning, 2021). MAPK signaling regulates gene expression, cell survival, and cytokine production and the MAPK family includes three major types of kinases: ERK (Extracellular signal-Regulated Kinase), JNK (c-Jun N-terminal Kinase), and p38 MAPK (Cargnello and Roux, 2011). NF-κB, a transcription factor family, controls immune responses and pro-inflammatory cytokine expression (Guo et al., 2024). Both pathways are activated by external stimuli, such as bacteria, virus, or injury (Rahman and McFadden, 2011; Dong et al., 2015; Lane et al., 2019; Liu S. Q. et al., 2020; Guo et al., 2023), and can amplify inflammatory responses by regulating each other’s effects (Nyati et al., 2017; Bansal et al., 2022). Given their critical roles in inflammation, we specifically investigated the effects of D. vulgaris on peritoneal macrophages from septic mice. Western blot analysis revealed significant phosphorylation of JNK, ERK, and p38 MAPK, as well as pronounced phosphorylation of NF-κB p65 in peritoneal macrophages from septic mice following D. vulgaris treatment (Figures 5A,B). These findings suggest that D. vulgaris induces inflammation through the activation of both MAPK and NF-κB signaling pathways, potentially driving its pro-inflammatory effects.
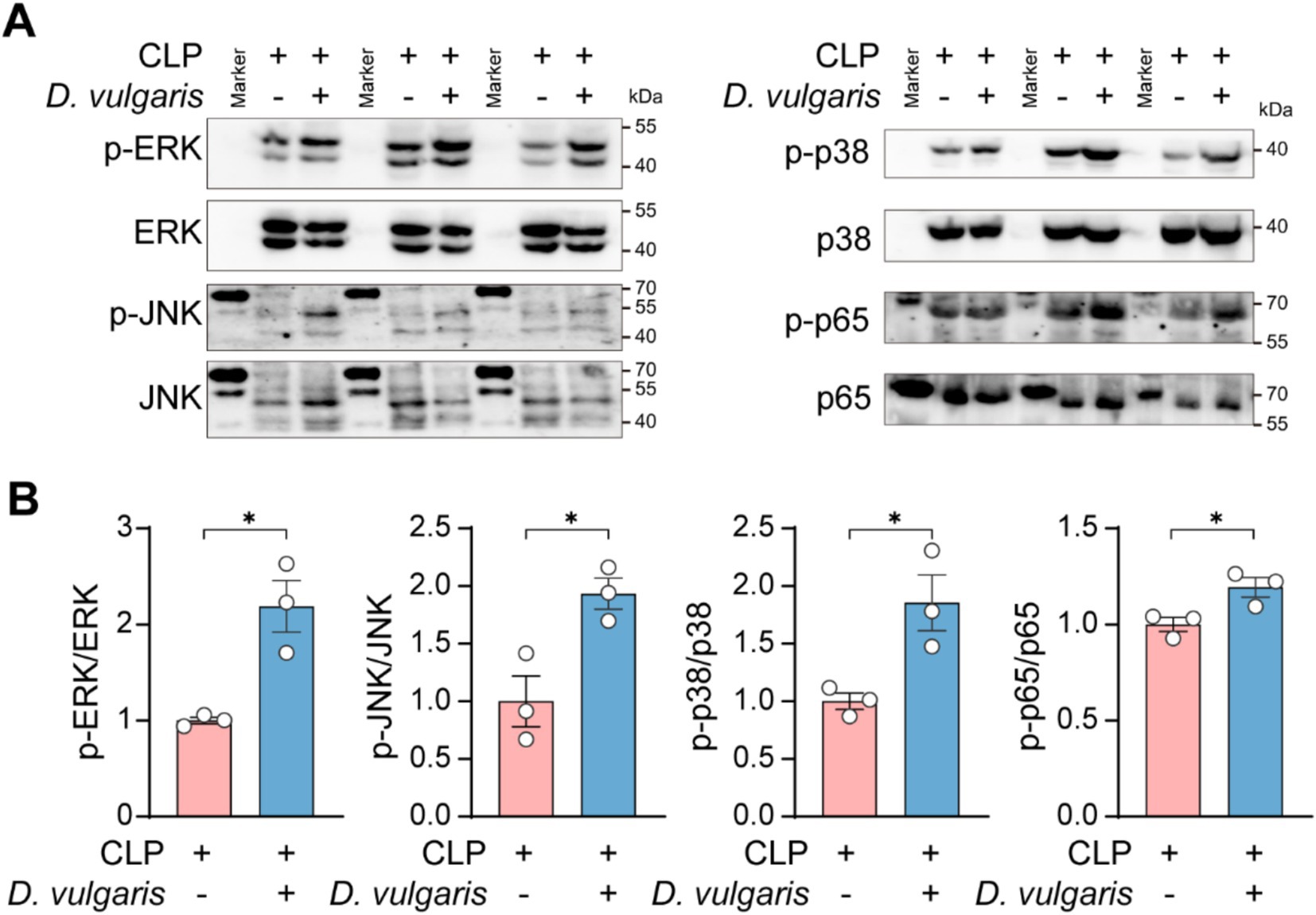
Figure 5. D. vulgaris promotes the activation of MAPK and NF-κB pathways in peritoneal macrophages of septic mice. (A,B) Western blot analysis of phosphorylated MAPK and NF-κB in peritoneal macrophages isolated from septic mice treated with D. vulgaris. n = 3. Data are presented as mean ± SEM. Statistical comparisons were performed using two-tailed unpaired Student’s t-test. *p < 0.05.
Desulfovibrio vulgaris exacerbates oxidative stress injury in sepsis mice
Oxidative stress plays a central role in the pathophysiology of sepsis, driving inflammatory responses and contributing to organ dysfunction (Victor et al., 2009; McCreath et al., 2016). To assess the impact of D. vulgaris on oxidative stress during sepsis, we measured ROS levels in the liver, kidney, and lung tissues of septic mice. D. vulgaris treatment resulted in a significant elevation of ROS in all three tissues (Figure 6A), indicating a marked exacerbation of oxidative stress. Concomitantly, we observed a significant reduction in the levels of key antioxidant enzymes, including GSH, CAT, and SOD (Figure 6B), which are critical for maintaining redox homeostasis. These findings suggest that D. vulgaris disrupts the delicate balance between ROS and antioxidant defenses, thereby amplifying oxidative damage in septic tissues. This redox imbalance may further exacerbate tissue injury, promoting the progression of sepsis-related organ dysfunction. Collectively, our data underscore the pivotal role of oxidative stress in D. vulgaris-mediated exacerbation of sepsis pathophysiology and suggest potential therapeutic strategies targeting oxidative pathways to mitigate sepsis-associated organ damage.
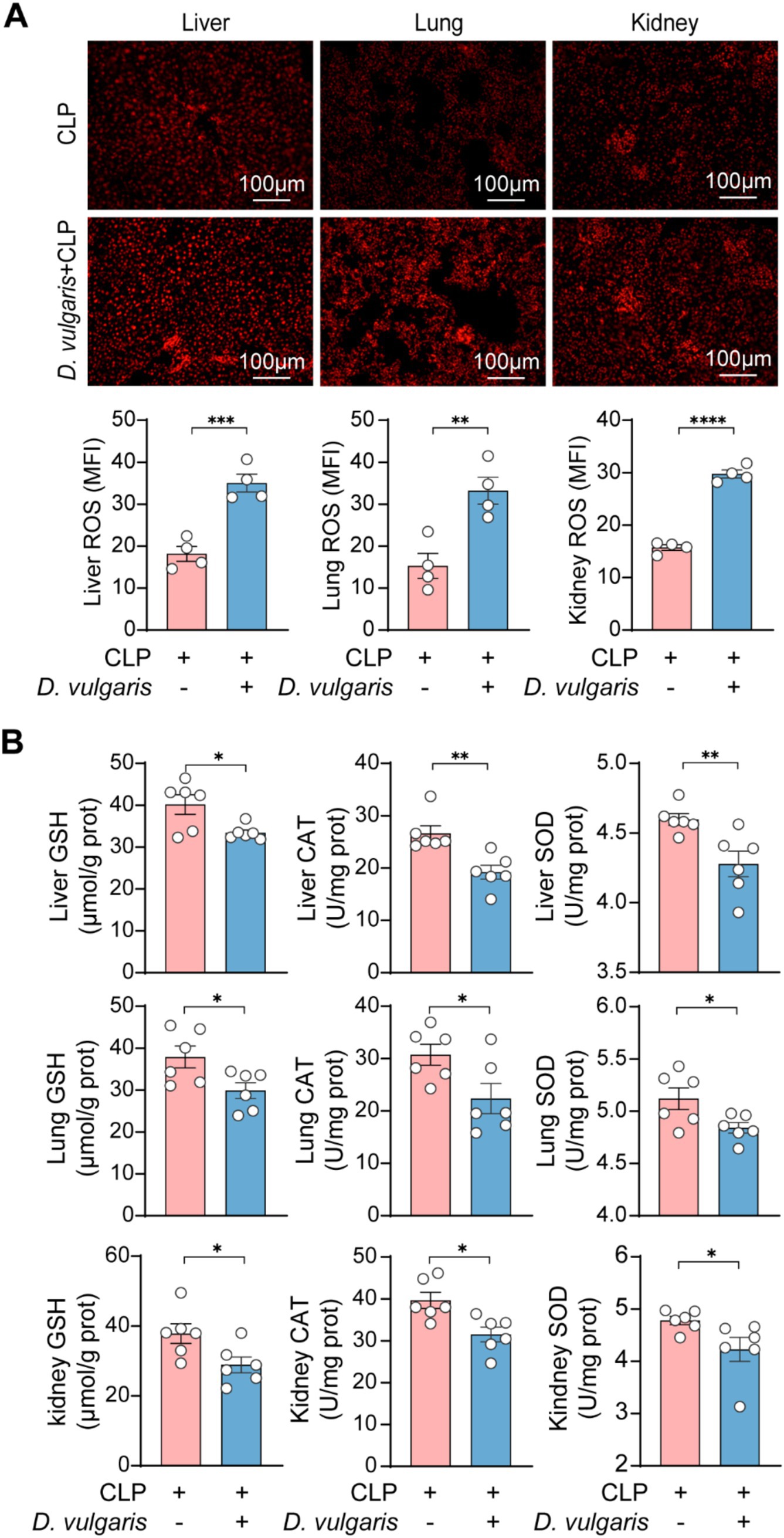
Figure 6. D. vulgaris induces oxidative stress in septic mice. (A) ROS levels in liver, kidney, and lung tissues of septic mice treated with D. vulgaris. n = 4. (B) Antioxidant levels (GSH, CAT, and SOD) in liver, kidney, and lung tissues of septic mice. n = 6. Data are presented as mean ± SEM. Statistical comparisons were performed using two-tailed unpaired Student’s t-test. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. Scale bars, 100 μm.
Based on our findings, we propose the following model (Figure 7): Sepsis-induced dysbiosis increases the abundance of D. vulgaris in the gut. This shift activates MAPK and NF-κB signaling pathways in macrophages, leading to enhanced inflammation. Additionally, D. vulgaris promotes the production of ROS, exacerbating oxidative stress. Together, these processes worsen sepsis-related organ damage, increase tissue injury, and contribute to higher mortality.
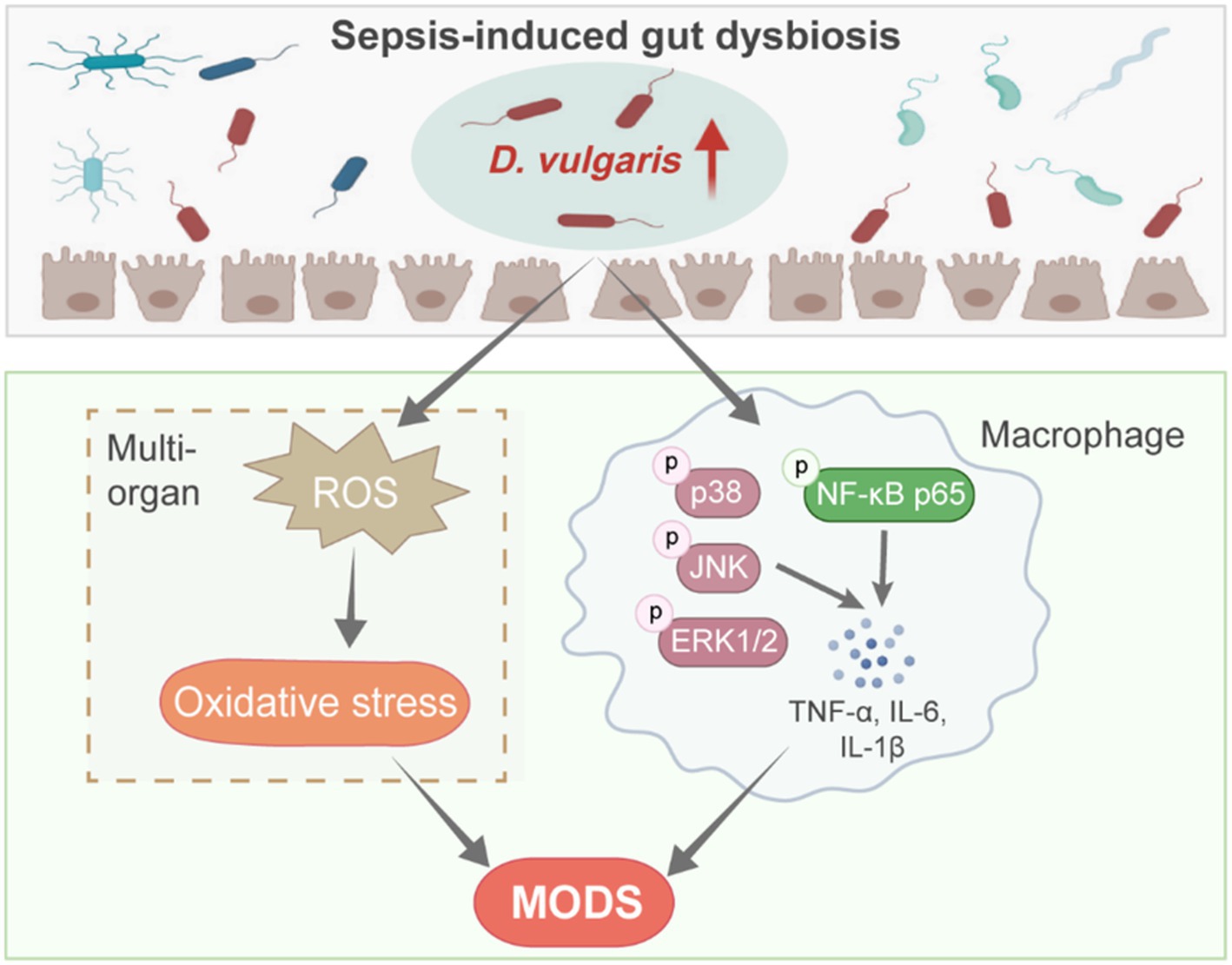
Figure 7. D. vulgaris promotes inflammation and oxidative stress in septic mice. Model illustrating the effects of increased D. vulgaris abundance in the gut during sepsis, leading to activated macrophage MAPK and NF-κB pathways, heightened inflammation, and increased oxidative stress in multiple organs.
Discussion
The gut microbiota plays a central role in regulating immune responses and has recently been identified as a key factor in the pathogenesis of sepsis. Dysbiosis of the gut microbiota is widely recognized as a key pathological mechanism in sepsis (Haak and Wiersinga, 2017). Recent studies have shown that the gut microbiome not only plays a crucial role in immune system modulation but also significantly influences the progression of sepsis (Sun et al., 2022). Previous studies have shown that gut microbiota dysbiosis contributes to sepsis-induced liver injury (Gong et al., 2019), highlighting its significant impact on organ function and disease progression.
In our study, we observed an increase in the abundance of Desulfovibrio species, as a conditional pathogenic bacterium, in the cecum of septic mice. This finding suggests that Desulfovibrio may be in a delicate regulatory state within the gut microbiota, with its abundance potentially influenced by specific pathological conditions. The rise in Desulfovibrio abundance may be associated with gut microbiota dysbiosis, diet (high-fat, low-fiber), antibiotic use, and increased intestinal sulfur compounds (Han C. et al., 2021; Singh et al., 2023; Zhou H. et al., 2024). Previous studies show that Desulfovibrio displays variable behavior across different disease models, including alcohol-induced liver injury, inflammatory bowel disease, ulcerative colitis, liver fibrosis, colorectal cancer, and stroke (Loubinoux et al., 2002; Rowan et al., 2010; Yang et al., 2019; Wang et al., 2021; Li et al., 2024; Qi et al., 2024; Zhang et al., 2024). These observations reflect the complex role of Desulfovibrio in host–microbe interactions across different pathological contexts. Additionally, previous studies have linked Desulfovibrio species such as D. fairfieldensis and D. desulfuricans to the onset of bacteremia (Pimentel and Chan, 2007; Urata et al., 2008; Yamaizumi et al., 2024). These findings support the notion that Desulfovibrio acts as a potential pathogenic factor during sepsis.
Although the overall abundance of Desulfovibrio did not show significant changes in our sepsis models, we observed a marked increase in D. vulgaris abundance in the cecum of CLP-induced septic mice, which was confirmed in the LPS-induced endotoxemia model. Previous research has shown that D. vulgaris proliferation exacerbates intestinal inflammation in ulcerative colitis, with the extent of proliferation positively correlating with the severity of intestinal disease (Rowan et al., 2010; An et al., 2023; Xie et al., 2024). This reinforces the key role of D. vulgaris in gut barrier function, immune responses, and disease progression.
In our septic mouse model, D. vulgaris-treated mice had significantly lower survival rates compared to controls. Histological analysis revealed that D. vulgaris exacerbated pathological damage to the liver, kidney, and lung tissues in septic mice. Furthermore, TUNEL staining revealed that D. vulgaris increased apoptosis in these tissues. In addition to its effects on these tissues, D. vulgaris also exacerbated damage to the intestinal mucosal barrier. The disruption of the intestinal barrier further contributed to the systemic inflammatory response, which is a critical factor in the progression of sepsis (Haak and Wiersinga, 2017). These findings underscore the multi-organ damage caused by D. vulgaris and its potential to worsen the overall clinical outcomes of sepsis. This aligns with previous research on the pathogenic role of Desulfovibrio species in other inflammatory diseases, particularly inflammatory bowel diseases, where D. vulgaris proliferation has been shown to worsen intestinal barrier damage and the secretion of pro-inflammatory cytokines (IL-1β, iNOS, and TNF-α), ultimately exacerbating DSS-induced colitis (Huang et al., 2024). Furthermore, previous studies have demonstrated that outer membrane vesicles (OMVs) from D. fairfieldensis can impair intestinal epithelial integrity and activate intrinsic inflammatory responses (Nie et al., 2022). D. desulfuricans has been reported to aggravate atherosclerosis by increasing intestinal permeability and activating the endothelial TLR4/NF-κB pathway in Apoe−/− mice (Zhang et al., 2023). Additionally, excessive H₂S production, often associated with gut dysbiosis, has been shown to compromise the intestinal barrier and exacerbate systemic inflammation (Munteanu et al., 2024b).
Beyond local tissue damage, D. vulgaris also significantly promotes the systemic inflammatory response in septic mice. We observed a marked elevation in pro-inflammatory cytokines such as TNF-α, IL-1β, and IL-6 in the serum of septic mice, suggesting that D. vulgaris may activate the intestinal immune system, thereby enhancing systemic inflammation. Additionally, we found that D. vulgaris significantly activated MAPK and NF-κB signaling pathways in peritoneal macrophages of septic mice. Previous study shows that D. vulgaris flagellin can activate LRRC19 to trigger NF-κB and MAPK pathways, and subsequently induces the production of pro-inflammatory chemokines and cytokines in vivo (Xie et al., 2024). However, an in vitro study suggests that D. vulgaris may induce the expression of pro-inflammatory cytokines such as TNF-α and iNOS via the PI3K/Akt pathway in a TLR2-dependent manner (Singh et al., 2024), which warrants further investigation.
Another key pathological feature of sepsis is oxidative stress, which exacerbates the pathological progression of the disease (Forman and Zhang, 2021). In our study, D. vulgaris significantly increased the levels of ROS in the liver, kidneys, and lungs of septic mice, while significantly reducing antioxidant levels (GSH, CAT, and SOD). We speculate that D. vulgaris exacerbates oxidative stress in septic mice by increasing free radical production and inhibiting antioxidant defense mechanisms, which in turn accelerates the progression of multi-organ dysfunction in sepsis. Interestingly, D. vulgaris contains genes for encoding superoxide reductase and SOD, which enable it to neutralize superoxide anions and survive in oxygen-rich environments (Fournier et al., 2004; Heidelberg et al., 2004; Zhou et al., 2010). However, in the context of sepsis, D. vulgaris may exacerbate oxidative stress. Elevated H₂S levels exhibit cytotoxic effects by inhibiting mitochondrial respiration or increasing ROS generation, thereby amplifying inflammation (Qi et al., 2024). We hypothesize that although D. vulgaris can survive in an oxidative environment, certain factors produced by D. vulgaris, such as LPS, flagellin, and H2S metabolites, could increase the oxidative burden, promoting ROS production and impairing antioxidant defenses, thereby worsening oxidative stress in septic tissues.
Although our study provides valuable insights into the role of D. vulgaris in sepsis, several limitations should be acknowledged. We have not fully elucidated the specific pathogenic mechanisms through which D. vulgaris exacerbates sepsis, particularly in terms of its effects on immune cell regulation and interactions with other signaling pathways. Additionally, while we focused primarily on D. vulgaris, the complexity of gut dysbiosis in sepsis involves a shift in microbial balance, including reductions in commensal bacteria and overgrowth of potential pathogens. D. vulgaris-specific bacteriophages could offer a more precise approach, selectively targeting D. vulgaris while preserving beneficial gut bacteria (Walker et al., 2006). Dietary interventions may also play a role, as D. vulgaris relies on sulfate for metabolism. Reducing sulfur-rich dietary components, such as high animal protein intake, thereby suppressing D. vulgaris growth and potentially mitigating organ injury caused by its overgrowth (Zhou L. et al., 2024). Interactions between D. vulgaris and other gut microbes may also play a significant role in the progression of sepsis and require further investigation.
Conclusion
The present study reveals that the increased abundance of D. vulgaris in septic mice may exacerbate sepsis through multiple mechanisms, including increased mortality, enhanced inflammation, oxidative stress, and multi-organ dysfunction. D. vulgaris likely contributes to systemic inflammation and tissue damage by activating critical signaling pathways such as MAPK and NF-κB. These findings provide a new perspective on the role of the gut microbiome in sepsis and offer potential therapeutic targets for sepsis treatment. Targeting both inflammation and oxidative stress may provide potential therapeutic avenues to mitigate the harmful effects of D. vulgaris in sepsis.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Ethics statement
The studies involving humans were approved by the Medical Ethics Committee of Nanfang Hospital of Southern Medical University. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study. The animal study was approved by the Animal Ethics Committee of the Animal Experiment Center of Southern Medical University. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
RW: Data curation, Methodology, Validation, Writing – original draft, Writing – review & editing. ZY: Formal analysis, Methodology, Writing – review & editing. PG: Formal analysis, Methodology, Validation, Writing – review & editing. XX: Formal analysis, Methodology, Validation, Writing – review & editing. YZ: Data curation, Methodology, Validation, Writing – original draft, Writing – review & editing. SF: Formal analysis, Methodology, Validation, Writing – review & editing. MY: Data curation, Formal analysis, Funding acquisition, Writing – review & editing. XH: Formal analysis, Methodology, Writing – review & editing. ZW: Formal analysis, Methodology, Writing – review & editing. AC: Data curation, Supervision, Writing – review & editing. YG: Conceptualization, Data curation, Project administration, Supervision, Writing – review & editing. XZ: Conceptualization, Data curation, Project administration, Supervision, Visualization, Writing – review & editing. WX: Conceptualization, Data curation, Funding acquisition, Project administration, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was funded by the National Natural Science Foundation of China (Nos. 82174322, 82474501, and 82305059), the Natural Science Foundation of Guangdong Province (Nos. 2023A1515011483 and 2021A1515110711), and the Guangzhou Science and Technology Program key projects (No. 202206010110).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1574998/full#supplementary-material
References
An, Y., Zhai, Z., Wang, X., Ding, Y., He, L., Li, L., et al. (2023). Targeting Desulfovibrio vulgaris flagellin-induced NAIP/NLRC4 inflammasome activation in macrophages attenuates ulcerative colitis. J. Adv. Res. 52, 219–232. doi: 10.1016/j.jare.2023.08.008
Bansal, A., Mostafa, M. M., Kooi, C., Sasse, S. K., Michi, A. N., Shah, S. V., et al. (2022). Interplay between nuclear factor-kappaB, p38 MAPK, and glucocorticoid receptor signaling synergistically induces functional TLR2 in lung epithelial cells. J. Biol. Chem. 298:101747. doi: 10.1016/j.jbc.2022.101747
Cao, Y., Zou, L., Li, W., Song, Y., Zhao, G., and Hu, Y. (2020). Dietary quinoa (Chenopodium quinoa Willd.) polysaccharides ameliorate high-fat diet-induced hyperlipidemia and modulate gut microbiota. Int. J. Biol. Macromol. 163, 55–65. doi: 10.1016/j.ijbiomac.2020.06.241
Cargnello, M., and Roux, P. P. (2011). Activation and function of the MAPKs and their substrates, the MAPK-activated protein kinases. Microbiol. Mol. Biol. Rev. 75, 50–83. doi: 10.1128/MMBR.00031-10
Chancharoenthana, W., Kamolratanakul, S., Schultz, M. J., and Leelahavanichkul, A. (2023). The leaky gut and the gut microbiome in sepsis - targets in research and treatment. Clin. Sci. 137, 645–662. doi: 10.1042/CS20220777
Chen, Q., Liang, X., Wu, T., Jiang, J., Jiang, Y., Zhang, S., et al. (2022). Integrative analysis of metabolomics and proteomics reveals amino acid metabolism disorder in sepsis. J. Transl. Med. 20:123. doi: 10.1186/s12967-022-03320-y
Chen, X., Wu, R., Li, L., Zeng, Y., Chen, J., Wei, M., et al. (2023). Pregnancy-induced changes to the gut microbiota drive macrophage pyroptosis and exacerbate septic inflammation. Immunity 56, 336–352.e9. doi: 10.1016/j.immuni.2023.01.015
Dong, Z. W., Chen, J., Ruan, Y. C., Zhou, T., Chen, Y., Chen, Y., et al. (2015). CFTR-regulated MAPK/NF-kappaB signaling in pulmonary inflammation in thermal inhalation injury. Sci. Rep. 5:15946. doi: 10.1038/srep15946
Dong, Y., Meng, F., Wang, J., Wei, J., Zhang, K., Qin, S., et al. (2025). Desulfovibrio vulgaris flagellin exacerbates colorectal cancer through activating LRRC19/TRAF6/TAK1 pathway. Gut Microbes 17:2446376. doi: 10.1080/19490976.2024.2446376
Fang, H., Wang, Y., Deng, J., Zhang, H., Wu, Q., He, L., et al. (2022). Sepsis-induced Gut Dysbiosis mediates the susceptibility to Sepsis-associated encephalopathy in mice. mSystems 7:e0139921. doi: 10.1128/msystems.01399-21
Figliuolo, V. R., Dos Santos, L. M., Abalo, A., Nanini, H., Santos, A., Brittes, N. M., et al. (2017). Sulfate-reducing bacteria stimulate gut immune responses and contribute to inflammation in experimental colitis. Life Sci. 189, 29–38. doi: 10.1016/j.lfs.2017.09.014
Fite, A., Macfarlane, G. T., Cummings, J. H., Hopkins, M. J., Kong, S. C., Furrie, E., et al. (2004). Identification and quantitation of mucosal and faecal desulfovibrios using real time polymerase chain reaction. Gut 53, 523–529. doi: 10.1136/gut.2003.031245
Fleischmann, C., Scherag, A., Adhikari, N. K. J., Hartog, C. S., Tsaganos, T., Schlattmann, P., et al. (2016). Assessment of global incidence and mortality of hospital-treated sepsis. Current estimates and limitations. Am. J. Respir. Crit. Care Med. 193, 259–272. doi: 10.1164/rccm.201504-0781OC
Fleischmann-Struzek, C., Mellhammar, L., Rose, N., Cassini, A., Rudd, K. E., Schlattmann, P., et al. (2020). Incidence and mortality of hospital- and ICU-treated sepsis: results from an updated and expanded systematic review and meta-analysis. Intensive Care Med. 46, 1552–1562. doi: 10.1007/s00134-020-06151-x
Forman, H. J., and Zhang, H. (2021). Targeting oxidative stress in disease: promise and limitations of antioxidant therapy. Nat. Rev. Drug Discov. 20, 689–709. doi: 10.1038/s41573-021-00233-1
Fournier, M., Dermoun, Z., Durand, M. C., and Dolla, A. (2004). A new function of the Desulfovibrio vulgaris Hildenborough [Fe] hydrogenase in the protection against oxidative stress. J. Biol. Chem. 279, 1787–1793. doi: 10.1074/jbc.M307965200
Giamarellos-Bourboulis, E. J., Aschenbrenner, A. C., Bauer, M., Bock, C., Calandra, T., Gat-Viks, I., et al. (2024). The pathophysiology of sepsis and precision-medicine-based immunotherapy. Nat. Immunol. 25, 19–28. doi: 10.1038/s41590-023-01660-5
Gong, S., Yan, Z., Liu, Z., Niu, M., Fang, H., Li, N., et al. (2019). Intestinal microbiota mediates the susceptibility to polymicrobial sepsis-induced liver injury by granisetron generation in mice. Hepatology 69, 1751–1767. doi: 10.1002/hep.30361
Gottschalk, R. A., Martins, A. J., Angermann, B. R., Dutta, B., Ng, C. E., Uderhardt, S., et al. (2016). Distinct NF-kappaB and MAPK activation thresholds uncouple steady-state microbe sensing from anti-pathogen inflammatory responses. Cell Syst. 2, 378–390. doi: 10.1016/j.cels.2016.04.016
Guo, Q., Jin, Y., Chen, X., Ye, X., Shen, X., Lin, M., et al. (2024). NF-kappaB in biology and targeted therapy: new insights and translational implications. Signal Transduct. Target. Ther. 9:53. doi: 10.1038/s41392-024-01757-9
Guo, Y., Zhang, H., Lv, Z., Du, Y., Li, D., Fang, H., et al. (2023). Up-regulated CD38 by daphnetin alleviates lipopolysaccharide-induced lung injury via inhibiting MAPK/NF-kappaB/NLRP3 pathway. Cell Commun. Signal 21:66. doi: 10.1186/s12964-023-01041-3
Haak, B. W., and Wiersinga, W. J. (2017). The role of the gut microbiota in sepsis. Lancet Gastroenterol. Hepatol. 2, 135–143. doi: 10.1016/S2468-1253(16)30119-4
Han, C., Guo, N., Bu, Y., Peng, Y., Li, X., Ma, X., et al. (2021). Intestinal microbiota and antibiotic-associated acute gastrointestinal injury in sepsis mice. Aging (Albany NY) 13, 10099–10111. doi: 10.18632/aging.202768
Han, X., Spicer, A., Carey, K. A., Gilbert, E. R., Laiteerapong, N., Shah, N. S., et al. (2021). Identifying high-risk subphenotypes and associated harms from delayed antibiotic orders and delivery*. Crit. Care Med. 49, 1694–1705. doi: 10.1097/CCM.0000000000005054
Heidelberg, J. F., Seshadri, R., Haveman, S. A., Hemme, C. L., Paulsen, I. T., Kolonay, J. F., et al. (2004). The genome sequence of the anaerobic, sulfate-reducing bacterium Desulfovibrio vulgaris Hildenborough. Nat. Biotechnol. 22, 554–559. doi: 10.1038/nbt959
Huang, G., Zheng, Y., Zhang, N., Huang, G., Zhang, W., Li, Q., et al. (2024). Desulfovibrio vulgaris caused gut inflammation and aggravated DSS-induced colitis in C57BL/6 mice model. Gut Pathog 16:39. doi: 10.1186/s13099-024-00632-w
Jin, Y. Q., Yuan, H., Liu, Y. F., Zhu, Y. W., Wang, Y., Liang, X. Y., et al. (2024). Role of hydrogen sulfide in health and disease. MedComm 5:e661. doi: 10.1002/mco2.661
Kunst, C., Schmid, S., Michalski, M., Tumen, D., Buttenschon, J., Muller, M., et al. (2023). The influence of gut microbiota on oxidative stress and the immune system. Biomedicines 11:1388. doi: 10.3390/biomedicines11051388
Lane, K., Andres-Terre, M., Kudo, T., Monack, D. M., and Covert, M. W. (2019). Escalating threat levels of bacterial infection can be discriminated by distinct MAPK and NF-kappaB signaling dynamics in single host cells. Cell Syst. 8, 183–196.e4. doi: 10.1016/j.cels.2019.02.008
Lankelma, J. M., van Vught, L. A., Belzer, C., Schultz, M. J., van der Poll, T., de Vos, W. M., et al. (2017). Critically ill patients demonstrate large interpersonal variation in intestinal microbiota dysregulation: a pilot study. Intensive Care Med. 43, 59–68. doi: 10.1007/s00134-016-4613-z
Li, G., Liu, H., Yu, Y., Wang, Q., Yang, C., Yan, Y., et al. (2024). Desulfovibrio desulfuricans and its derived metabolites confer resistance to FOLFOX through METTL3. EBioMedicine 102:105041. doi: 10.1016/j.ebiom.2024.105041
Liu, W., Cheng, M., Li, J., Zhang, P., Fan, H., Hu, Q., et al. (2020). Classification of the gut microbiota of patients in intensive care units during development of sepsis and septic shock. Genomics Proteomics Bioinformatics 18, 696–707. doi: 10.1016/j.gpb.2020.06.011
Liu, S. Q., Xie, Y., Gao, X., Wang, Q., and Zhu, W. Y. (2020). Inflammatory response and MAPK and NF-kappaB pathway activation induced by natural street rabies virus infection in the brain tissues of dogs and humans. Virol. J. 17:157. doi: 10.1186/s12985-020-01429-4
Loubinoux, J., Bronowicki, J. P., Pereira, I. A., Mougenel, J. L., and Faou, A. E. (2002). Sulfate-reducing bacteria in human feces and their association with inflammatory bowel diseases. FEMS Microbiol. Ecol. 40, 107–112. doi: 10.1111/j.1574-6941.2002.tb00942.x
Lu, G., Zhang, Y., Ren, Y., Shi, J. S., Xu, Z. H., and Geng, Y. (2023). Diversity and comparison of intestinal desulfovibrio in patients with liver cirrhosis and healthy people. Microorganisms 11:276. doi: 10.3390/microorganisms11020276
Maeda, H., Fujimoto, C., Haruki, Y., Maeda, T., Kokeguchi, S., Petelin, M., et al. (2003). Quantitative real-time PCR using TaqMan and SYBR green for Actinobacillus actinomycetemcomitans, Porphyromonas gingivalis, Prevotella intermedia, tetQ gene and total bacteria. FEMS Immunol. Med. Microbiol. 39, 81–86. doi: 10.1016/S0928-8244(03)00224-4
McCreath, G., Scullion, M. M., Lowes, D. A., Webster, N. R., and Galley, H. F. (2016). Pharmacological activation of endogenous protective pathways against oxidative stress under conditions of sepsis. Br. J. Anaesth. 116, 131–139. doi: 10.1093/bja/aev400
Munteanu, C., Onose, G., Postaru, M., Turnea, M., Rotariu, M., and Galaction, A. I. (2024a). Hydrogen sulfide and gut microbiota: their synergistic role in modulating sirtuin activity and potential therapeutic implications for neurodegenerative diseases. Pharmaceuticals (Basel) 17:1480. doi: 10.3390/ph17111480
Munteanu, C., Onose, G., Rotariu, M., Postaru, M., Turnea, M., and Galaction, A. I. (2024b). Role of microbiota-derived hydrogen sulfide (H(2)S) in modulating the gut-brain axis: implications for Alzheimer's and Parkinson's disease pathogenesis. Biomedicines 12:2670. doi: 10.3390/biomedicines12122670
Murros, K. E., Huynh, V. A., Takala, T. M., and Saris, P. E. J. (2021). Desulfovibrio bacteria are associated with Parkinson's disease. Front. Cell. Infect. Microbiol. 11:652617. doi: 10.3389/fcimb.2021.652617
Nie, Y., Xie, X. Q., Zhou, L., Guan, Q., Ren, Y., Mao, Y., et al. (2022). Desulfovibrio fairfieldensis-derived outer membrane vesicles damage epithelial barrier and induce inflammation and pyroptosis in macrophages. Cells 12:89. doi: 10.3390/cells12010089
Novakovic, B., Habibi, E., Wang, S. Y., Arts, R. J. W., Davar, R., Megchelenbrink, W., et al. (2016). beta-glucan reverses the epigenetic state of LPS-induced immunological tolerance. Cell 167, 1354–1368.e14. doi: 10.1016/j.cell.2016.09.034
Nyati, K. K., Masuda, K., Zaman, M. M., Dubey, P. K., Millrine, D., Chalise, J. P., et al. (2017). TLR4-induced NF-kappaB and MAPK signaling regulate the IL-6 mRNA stabilizing protein Arid5a. Nucleic Acids Res. 45, 2687–2703. doi: 10.1093/nar/gkx064
Piccioni, A., Spagnuolo, F., Candelli, M., Voza, A., Covino, M., Gasbarrini, A., et al. (2024). The gut microbiome in sepsis: from dysbiosis to personalized therapy. J. Clin. Med. 13:6082. doi: 10.3390/jcm13206082
Pimentel, J. D., and Chan, R. C. (2007). Desulfovibrio fairfieldensis bacteremia associated with choledocholithiasis and endoscopic retrograde cholangiopancreatography. J. Clin. Microbiol. 45, 2747–2750. doi: 10.1128/JCM.00969-07
Qi, Q., Zhang, H., Jin, Z., Wang, C., Xia, M., Chen, B., et al. (2024). Hydrogen sulfide produced by the gut microbiota impairs host metabolism via reducing GLP-1 levels in male mice. Nat. Metab. 6, 1601–1615. doi: 10.1038/s42255-024-01068-x
Rahman, M. M., and McFadden, G. (2011). Modulation of NF-kappaB signalling by microbial pathogens. Nat. Rev. Microbiol. 9, 291–306. doi: 10.1038/nrmicro2539
Rowan, F., Docherty, N. G., Murphy, M., Murphy, B., Calvin Coffey, J., and O'Connell, P. R. (2010). Desulfovibrio bacterial species are increased in ulcerative colitis. Dis. Colon Rectum 53, 1530–1536. doi: 10.1007/DCR.0b013e3181f1e620
Sakr, Y., Jaschinski, U., Wittebole, X., Szakmany, T., Lipman, J., Ñamendys-Silva, S. A., et al. (2018). Sepsis in intensive care unit patients: worldwide data from the intensive care over nations audit. Open Forum Infect. Dis. 5:ofy313. doi: 10.1093/ofid/ofy313
Singer, M., Deutschman, C. S., Seymour, C. W., Shankar-Hari, M., Annane, D., Bauer, M., et al. (2016). The third international consensus definitions for sepsis and septic shock (Sepsis-3). JAMA 315, 801–810. doi: 10.1001/jama.2016.0287
Singh, S. B., Braun, C. A., Carroll-Portillo, A., Coffman, C. N., and Lin, H. C. (2024). Sulfate-reducing bacteria induce pro-inflammatory TNF-alpha and iNOS via PI3K/Akt pathway in a TLR 2-dependent manner. Microorganisms 12:1833. doi: 10.3390/microorganisms12091833
Singh, S. B., Carroll-Portillo, A., and Lin, H. C. (2023). Desulfovibrio in the gut: the enemy within? Microorganisms 11:1772. doi: 10.3390/microorganisms11071772
Sun, S., Wang, D., Dong, D., Xu, L., Xie, M., Wang, Y., et al. (2023). Altered intestinal microbiome and metabolome correspond to the clinical outcome of sepsis. Crit. Care 27:127. doi: 10.1186/s13054-023-04412-x
Sun, T., Wang, L., and Zhang, H. (2022). Intestinal microbiota in sepsis. Intensive Care Res. 2, 1–7. doi: 10.1007/s44231-022-00001-8
Ueki, T., and Lovley, D. R. (2022). Desulfovibrio vulgaris as a model microbe for the study of corrosion under sulfate-reducing conditions. mLife 1, 13–20. doi: 10.1002/mlf2.12018
Urata, T., Kikuchi, M., Hino, T., Yoda, Y., Tamai, K., Kodaira, Y., et al. (2008). Bacteremia caused by Desulfovibrio fairfieldensis. J. Infect. Chemother. 14, 368–370. doi: 10.1007/s10156-008-0629-9
van der Poll, T., van de Veerdonk, F. L., Scicluna, B. P., and Netea, M. G. (2017). The immunopathology of sepsis and potential therapeutic targets. Nat. Rev. Immunol. 17, 407–420. doi: 10.1038/nri.2017.36
Victor, V. M., Espulgues, J. V., Hernandez-Mijares, A., and Rocha, M. (2009). Oxidative stress and mitochondrial dysfunction in sepsis: a potential therapy with mitochondria-targeted antioxidants. Infect. Disord. Drug Targets 9, 376–389. doi: 10.2174/187152609788922519
Walker, C. B., Stolyar, S. S., Pinel, N., Yen, H. C., He, Z., Zhou, J., et al. (2006). Recovery of temperate Desulfovibrio vulgaris bacteriophage using a novel host strain. Environ. Microbiol. 8, 1950–1959. doi: 10.1111/j.1462-2920.2006.01075.x
Wang, Y., Zhang, Y., Wang, Z., Tang, J., Cao, D. X., Qian, Y., et al. (2021). A clinical nomogram incorporating salivary Desulfovibrio desulfuricans level and oral hygiene index for predicting colorectal cancer. Ann. Transl. Med. 9:754. doi: 10.21037/atm-20-8168
Weglarz, L., Dzierzewicz, Z., Skop, B., Orchel, A., Parfiniewicz, B., Wisniowska, B., et al. (2003). Desulfovibrio desulfuricans lipopolysaccharides induce endothelial cell IL-6 and IL-8 secretion and E-selectin and VCAM-1 expression. Cell. Mol. Biol. Lett. 8, 991–1003
Xie, R., Gu, Y., Li, M., Li, L., Yang, Y., Sun, Y., et al. (2024). Desulfovibrio vulgaris interacts with novel gut epithelial immune receptor LRRC19 and exacerbates colitis. Microbiome 12:4. doi: 10.1186/s40168-023-01722-8
Yamaizumi, K., Kyotani, M., and Kenzaka, T. (2024). Bacteremia caused by Desulfovibrio desulfuricans with the intestinal tract as the portal of entry: two case reports and a literature review. BMC Infect. Dis. 24:725. doi: 10.1186/s12879-024-09623-3
Yang, J., McDowell, A., Kim, E. K., Seo, H., Lee, W. H., Moon, C. M., et al. (2019). Development of a colorectal cancer diagnostic model and dietary risk assessment through gut microbiome analysis. Exp. Mol. Med. 51, 1–15. doi: 10.1038/s12276-019-0313-4
Zhang, Y. Y., and Ning, B. T. (2021). Signaling pathways and intervention therapies in sepsis. Signal Transduct. Target. Ther. 6:407. doi: 10.1038/s41392-021-00816-9
Zhang, K., Qin, X., Qiu, J., Sun, T., Qu, K., Din, A. U., et al. (2023). Desulfovibrio desulfuricans aggravates atherosclerosis by enhancing intestinal permeability and endothelial TLR4/NF-kappaB pathway in Apoe (−/−) mice. Genes Dis. 10, 239–253. doi: 10.1016/j.gendis.2021.09.007
Zhang, Y., Wang, H., Sang, Y., Liu, M., Wang, Q., Yang, H., et al. (2024). Gut microbiota in health and disease: advances and future prospects. MedComm 5:e70012. doi: 10.1002/mco2.70012
Zhou, A., He, Z., Redding-Johanson, A. M., Mukhopadhyay, A., Hemme, C. L., Joachimiak, M. P., et al. (2010). Hydrogen peroxide-induced oxidative stress responses in Desulfovibrio vulgaris Hildenborough. Environ. Microbiol. 12, 2645–2657. doi: 10.1111/j.1462-2920.2010.02234.x
Zhou, H., Huang, D., Sun, Z., and Chen, X. (2024). Effects of intestinal Desulfovibrio bacteria on host health and its potential regulatory strategies: a review. Microbiol. Res. 284:127725. doi: 10.1016/j.micres.2024.127725
Keywords: sepsis, macrophage, Desulfovibrio vulgaris, inflammation, oxidative stress
Citation: Wu R, Yu Z, Guo P, Xiang X, Zeng Y, Fu S, Yang M, Huang X, Wang Z, Chen A, Ge Y, Zhao X and Xiao W (2025) Desulfovibrio vulgaris exacerbates sepsis by inducing inflammation and oxidative stress in multiple organs. Front. Microbiol. 16:1574998. doi: 10.3389/fmicb.2025.1574998
Edited by:
Huan Li, Lanzhou University, ChinaCopyright © 2025 Wu, Yu, Guo, Xiang, Zeng, Fu, Yang, Huang, Wang, Chen, Ge, Zhao and Xiao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Xiao, eHc3Njg4QHNtdS5lZHUuY24=; Xiaoshan Zhao, emhhb3hzQHNtdS5lZHUuY24=; Yuewei Ge, Z2V5dWV3ZWlAZ2RwdS5lZHUuY24=
†These authors have contributed equally to this work
 Rong Wu1†
Rong Wu1† Yunong Zeng
Yunong Zeng Yuewei Ge
Yuewei Ge Wei Xiao
Wei Xiao