- 1Department of Biochemistry, Maharshi Dayanand University, Rohtak, Haryana, India
- 2INtegrative GENomics of HOst-PathogEn (INGEN-HOPE) Laboratory, CSIR-Institute of Genomics and Integrative Biology (CSIR-IGIB), New Delhi, India
- 3Academy of Scientific and Innovative Research (AcSIR), Ghaziabad, India
Background: Delftia species have attracted significant interest for their biofertilizer and biocontrol capabilities, particularly in promoting the growth of crops such as Oryza sativa, Brassica campestris, and Solanum lycopersicum. However, their potential in supporting wheat cultivation remains largely unexplored.
Methods: A culture-dependent approach was employed to isolate a Delftia strain from the wheat rhizosphere. The biofertilizer potential of the isolate was systematically evaluated through a series of physiological, biochemical, and molecular assays, as well as field trials to assess its efficacy under agronomic conditions.
Results and discussion: Culture-dependent investigation of the wheat rhizosphere led to the isolation of a multifunctional plant growth-promoting bacterium, designated as strain NSC. Morphological and physiological characterization identified NSC as a gram-negative, rod-shaped, motile bacterium with optimal growth at pH 7.0 and 35°C. Phylogenetic and phylogenomic analyses confirmed its taxonomic identity as Delftia lacustris. In vitro assays revealed its ability to solubilize phosphate (0.325 IU), reduce nitrate (0.401 IU), produce indole-3-acetic acid (0.485 IU), and exhibit ACC deaminase activity (0.512 IU) and siderophore production. The strain demonstrated strong antifungal activity against Fusarium oxysporum and Rhizoctonia solani. Strain NSC exhibited significant tolerance to abiotic stresses, including drought [up to 40% PEG (w/v)], heavy metals, and high salinity [up to 11.69% NaCl (w/v), 11.18% KCl (w/v), and 4.24% LiCl (w/v)]. Genome analysis identified key genes associated with phosphate solubilization (PhoR, PhoB, PhoU, PstABCD), nitrogen fixation (nifC, nifU), auxin and siderophore biosynthesis, rhizosphere colonization, and antifungal mechanisms (chitinase, PhnZ). In planta studies showed significantly enhanced seed germination (93.33% ± 0.23), seedling growth, and biomass accumulation under stress conditions (p < 0.05). Field trials further validated the strain’s efficacy, showing marked improvements in plant growth and yield parameters (p = 0.0001). These results underscore the potential of D. lacustris NSC as an effective biofertilizer and biocontrol agent for sustainable agriculture.
Conclusion: Delftia lacustris strain NSC exhibits multifunctional plant growth-promoting and biocontrol activities, including enhanced nutrient mobilization, pathogen suppression, and abiotic stress tolerance. Its demonstrated efficacy under field conditions and environmentally benign profile highlight its potential as a sustainable bioinoculant for wheat production systems.
Introduction
The increasing global emphasis on sustainable agriculture has spurred interest in plant growth-promoting rhizobacteria as environmentally friendly alternatives to chemical fertilizers and pesticides (Chandran et al., 2021). Among various genera, Delftia has gained attention due to its metabolic versatility (Braña et al., 2016), environmental adaptability (Yin et al., 2022), and plant-associated beneficial traits (Yin et al., 2022). Delftia species are motile, aerobic, non-fermentative, gram-negative rods classified under Betaproteobacteria (Shigematsu et al., 2003). These bacteria thrive in diverse ecological niches, including wastewater, contaminated soils, clinical samples, and both the rhizosphere and endosphere of plants, highlighting their resilience and functional diversity (Fan et al., 2023). Several Delftia strains have demonstrated plant growth-promoting and biocontrol properties.
Delftia sp. Lp-1 and D. acidovorans DSM 39 possess nitrogen-fixation abilities, allowing them to function as free-living diazotrophs that enhance soil nitrogen levels (Han et al., 2005; Agafonova et al., 2017). D. tsuruhatensis, Delftia sp. JD2, and Delftia sp. BTL-M2, have been reported to colonize the roots of important crops like Solanum lycopersicum, Arabidopsis thaliana, Medicago sativa, and Pisum sativum (Morel et al., 2015; Brambilla et al., 2022; Islam et al., 2023). These root-colonizing strains enhance plant development by promoting nutrient availability, synthesizing phytohormones, facilitating root elongation, and lateral root formation (Morel et al., 2016). Field studies have further validated the plant growth-promoting effects of Delftia strains. Notably, Paraburkholderia fungorum BRRh-4 and Delftia sp. BTL-M2 improved growth and yield in Oryza sativa, highlighting their agricultural potential (Islam et al., 2023). Similarly, D. tsuruhatensis promotes rice growth under nitrogen-deficient conditions by enhancing nutrient solubilization and stimulating root system architecture (Harahap et al., 2023). These strains are also known to produce siderophores, iron-chelating compounds that facilitate iron uptake while restricting iron availability to plant pathogens, thereby serving as indirect biocontrol agents (Guo et al., 2016).
Beyond growth promotion, Delftia species are increasingly recognized for their stress-mitigation capabilities (Pattnaik et al., 2021). Many strains exhibit resilience to extreme environmental conditions, particularly heavy metal contamination (Sazykin et al., 2023). D. lacustris MS3 has been isolated from lead- and chromium-polluted soils, while D. acidovorans SPH-1 mitigates cadmium, zinc, and lead toxicity (Bautista-Hernandez et al., 2012; Samimi and Shahriari-Moghadam, 2021). D. lacustris and Delftia sp. JD2, have demonstrated resistance to copper, nickel, and hexavalent chromium, making them promising candidates for bioremediation applications (Braña et al., 2016; Morel et al., 2016). Given their well-documented plant growth-promotion, biocontrol activities, and stress tolerance, Delftia species hold significant promise as multifunctional bioinoculants. However, most research has focused on horticultural and model crops, such as tomato, rice, and Arabidopsis, with limited investigation into their role in cereal crops, particularly wheat.
Wheat is a globally cultivated staple, and enhancing its productivity in a sustainable manner is critical for food security. Modern agricultural practices based on excessive agrochemical use have led to soil degradation, nutrient imbalances, and increased disease susceptibility, reinforcing the need for biological alternatives (Sharma et al., 2024b). The rhizosphere of wheat plants represents a rich microbial niche with untapped potential for discovering novel PGP strains (Sharma et al., 2024a). Given the metabolic traits observed in other Delftia isolates (Islam et al., 2023), it is believable that wheat-associated Delftia strains may possess similar or enhanced biofertilizer and biocontrol attributes. However, these need to be experimentally validated. Hereby, the present study was planned to assess the biofertilizer and biocontrol properties of the wheat rhizospheric Delftia isolate. The impact of Delftia isolate needs to be checked on wheat seed germination in the presence of phytopathogens and saline conditions, followed by its impact on plant growth and yield to confirm biofertilizer and biocontrol properties. Moreover, Delftia’s ability to tolerate abiotic stresses makes it an ideal candidate for deployment in marginal or degraded agricultural lands. However, the stress response physiology of the wheat rhizosphere Delftia isolate needs to be checked through experimentation. In this context, the present study was designed to comprehensively characterize the isolate through physiological, biochemical, and genomic analyses. The central objective was to validate the Delftia strain’s potential to promote wheat growth, enhance nutrient uptake, and improve resilience to both biotic and abiotic stresses. Ultimately, these observations would confirm the biocontrol and biofertilizer potential of wheat rhizospheric Delftia isolate before its employment to sustainably enhance wheat crop yield.
Materials and methods
Study design
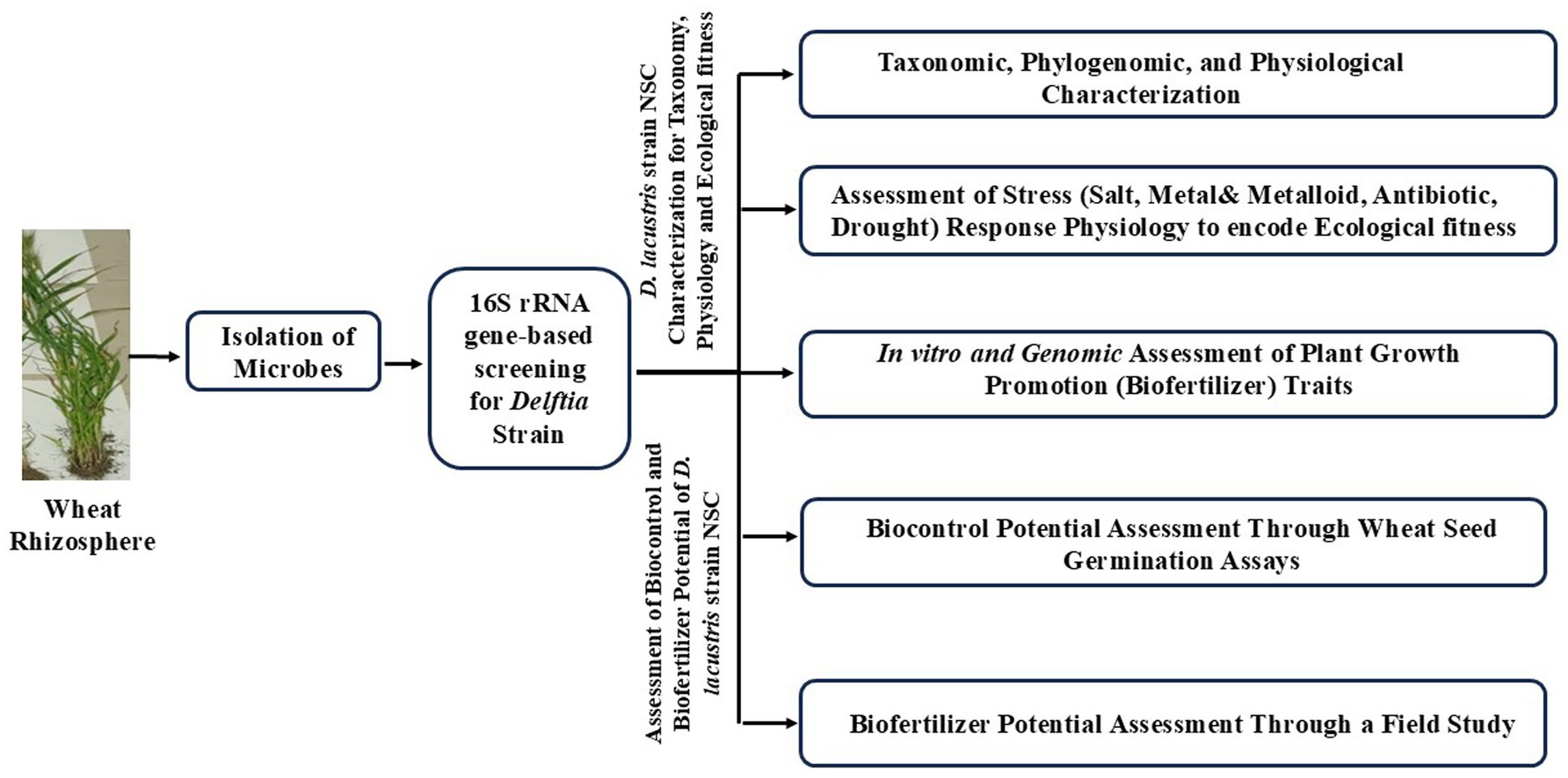
The present study was designed to assess the ecological fitness, biofertilizer, and biocontrol potential of wheat rhizospheric D. lacustris strain NSC. A workflow was developed to assess these characteristics of D. lacustris strain NSC (Figure 1).
Isolation of wheat rhizosphere microbes (WRMs)
Wheat rhizospheric microbes were isolated using a previously standardized method (Supplementary Material 1) (Sharma et al., 2024a). Briefly, rhizospheric soil was collected from randomly selected wheat plants (05) cultivated in an experimental field at Maharshi Dayanand University, Rohtak (28° 52′44” N, 76° 37′19″E), Haryana, India. Wheat rhizosphere samples were harvested with a sterile scoop and immediately transferred to a sterile bag, which was transported immediately to the laboratory for isolation of microbes. Roots were mechanically shaken in the sterile bag to collect the rhizospheric soil. Rhizospheric soil was transferred aseptically in a sterile tube for isolation of rhizospheric microbes following standardized methodology (Supplementary Material 1) (Sharma et al., 2024a). Phylogenetic affiliation of the isolated microbial strains was assessed through 16S rRNA gene-based phylotype identification (Sharma et al., 2024b).
Molecular, physiological, biochemical, and genomic characterization of Delftia strain NSC
Gram staining was performed with a Gram staining kit (K001-1KT, Himedia). The bacterial growth was observed at different pH ranges (3, 4, 5, 7, 8, 9, 10, 11, and 12) and temperature ranges (10°C, 20°C, 30°C, 40°C, 50°C, 60°C) to identify its optimal growth conditions. Delftia strain NSC growth pattern was observed in LB broth for 48 h at 37°C with constant shaking at 200 rpm to check its growth pattern and generation time (Supplementary Material 2) (Sharma et al., 2024a). Substrate utilization preference was assessed with a Hi-Carbo kit (Himedia, KB009A-1KT, KB009B-1KT, and KB009C-1KT) (Supplementary Material 3). Biochemical properties were performed using assays for amylase (Swain et al., 2006), catalase (Iwase et al., 2013), pectinase (Oumer and Abate, 2018), cellulase (Kasana et al., 2008), esterase (Ramnath et al., 2017), and protease (Vijayaraghavan et al., 2017) (Supplementary Materials 4–9). The stress response physiology was assessed by performing specific assays for salt stress tolerance, metal stress tolerance, and oxidative stress (Supplementary Material 10) (Sharma et al., 2024a). DNA was extracted from the microbial isolate using the alkali lysis method (Chauhan et al., 2009). The qualitative and quantitative analysis of the DNA was performed with agarose gel electrophoresis and Qubit HS DNA estimation kits (Invitrogen, United States), respectively. Genomic DNA was sequenced using Illumina MiSeq platform using Nextera XT DNA Library Prep kit (Yadav et al., 2022) to unveil its genomic architecture. Quality filtration, genome assembly, and genome completeness were checked using standard methodology (Yadav et al., 2022). Genomic characterization and comparative genomics were performed against other Delftia using a standardized methodology (Supplementary Material 11) (Yadav et al., 2022).
Assessment of biofertilizer-related features in D. lacustris strain NSC
Delftia lacustris strain NSC was characterized for its biofertilization potential through a series of functional assays, including phosphate solubilization (Behera et al., 2017), nitrate reductase activity (Kim and Seo, 2018), indole-3-acetic acid (IAA) production (Ehmann, 1977), ammonia production (Bhattacharyya et al., 2020), and siderophore biosynthesis (Himpsl and Mobley, 2019) (Supplementary Materials 2–16). Its drought stress tolerance property was also evaluated (Supplementary Material 17) (Mustamu et al., 2023). Its efficiency to mitigate salt-induced oxidative stress was evaluated by measuring its 1-aminocyclopropane-1-carboxylic acid (ACC) deaminase activity (Maheshwari et al., 2020) (Supplementary Material 18). The RAST software assessed the genetic features related to plant growth promotion.1
Assessment of Delftia lacustris strain NSC biofertilizer and biocontrol properties in laboratory conditions
The biocontrol potential was evaluated against Rhizoctonia solani, and Fusarium oxysporum (Supplementary Material 19). Both of these phytopathogens are the most prevalent and economically damaging soil-borne fungi affecting wheat, known to cause root rot, damping-off, and vascular wilt, which significantly reduce crop yield and quality (Wille et al., 2019; Rhouma et al., 2024). Pre-inoculation of seeds with biofertilizer strains has been known to improve plant growth parameters like shoot and root length, etc. (O'Callaghan, 2016; Kumar et al., 2023). The assessment focused on their effects on seed germination efficiency and wheat seedlings’ root and shoot lengths (Singh and Kayastha, 2014; Sharma et al., 2024a). To investigate the role of D. lacustris strain NSC in seed germination under saline conditions, seeds were soaked in an overnight-grown microbial culture (cell density of 1011 cells/mL), supplemented with NaCl concentrations ranging from 0 to 1 M, for 16 h at 37°C. Control seeds were soaked directly in NaCl solutions of equivalent concentrations for 16 h at 37°C. Following the soaking treatment, seeds were wrapped in germination sheets and placed in 50 mL culture tubes containing 5 mL of Hoagland solution. The tubes were incubated in the dark at ambient temperature (25°C) for 7 days. After incubation, wheat seed germination percentage, alpha-amylase activity, and root and shoot lengths were measured (Singh and Kayastha, 2014; Sharma et al., 2024a).
Assessment of D. lacustris strain NSC biofertilizer properties under field conditions
Roots of wheat cultivar (WC)-306 plants treated with D. lacustris strain NSC were harvested at various Feeks and assessed for their phosphate-solubilizing ability (Behera et al., 2017), nitrate reductase activity (Kim and Seo, 2018), reducing sugar content (Khatri and Chhetri, 2020), and total sugar content (Ludwig and Goldberg, 1956) (Supplementary Materials 20–25). These results were compared to those of untreated WC-306 plants. In addition, several growth and yield parameters were evaluated in both treated and untreated plants, including seed germination percentage, number of tillers per plant, number of leaves per plant, number of spikes per plant, spike length, number of spikelets per spike, and grain yield, to assess the impact of D. lacustris strain NSC on wheat growth and productivity (Supplementary Material 26).
Statistical analysis
All experiments were performed with replicates (a total of six replicates for microbial physiological and biochemical studies, and 10 replicates for all plant-based studies). Statistical analyses and data visualization were conducted using SIGMA Plot 15.0. Significant difference between microbial-treated and control groups for growth parameters (number of tillers, number of leaves per plant, spike length, number of spikes per plant, number of spikelets per plant, grain weight per 1,000 grains, and grain yield was determined using One-Way ANOVA using Systat Software, Sigma Plot 15.0). Data distribution was checked both visually (histogram analysis) and statistically (Shapiro–Wilk Test) before performing ANOVA analysis.
Results
Identification of microbial isolate NSC
Wheat rhizosphere has a pH of 7.3 ± 0.0025, a temperature of 22.6°C ± 0.01, and a moisture content of 11.5 ± 1.10014. A total of 12 microbial isolates were obtained from the rhizosphere. The 16S rRNA gene-based taxonomic characterization revealed that only NSC isolate’s 16S rRNA gene shared 99.86% homology with Delftia lacustris strain 332 (NR 116495) in the NCBI Nr database. This identification was further confirmed through phylogenetic analysis (Figure 2A). Based on this observation, the isolated NSC was used for further characterization.
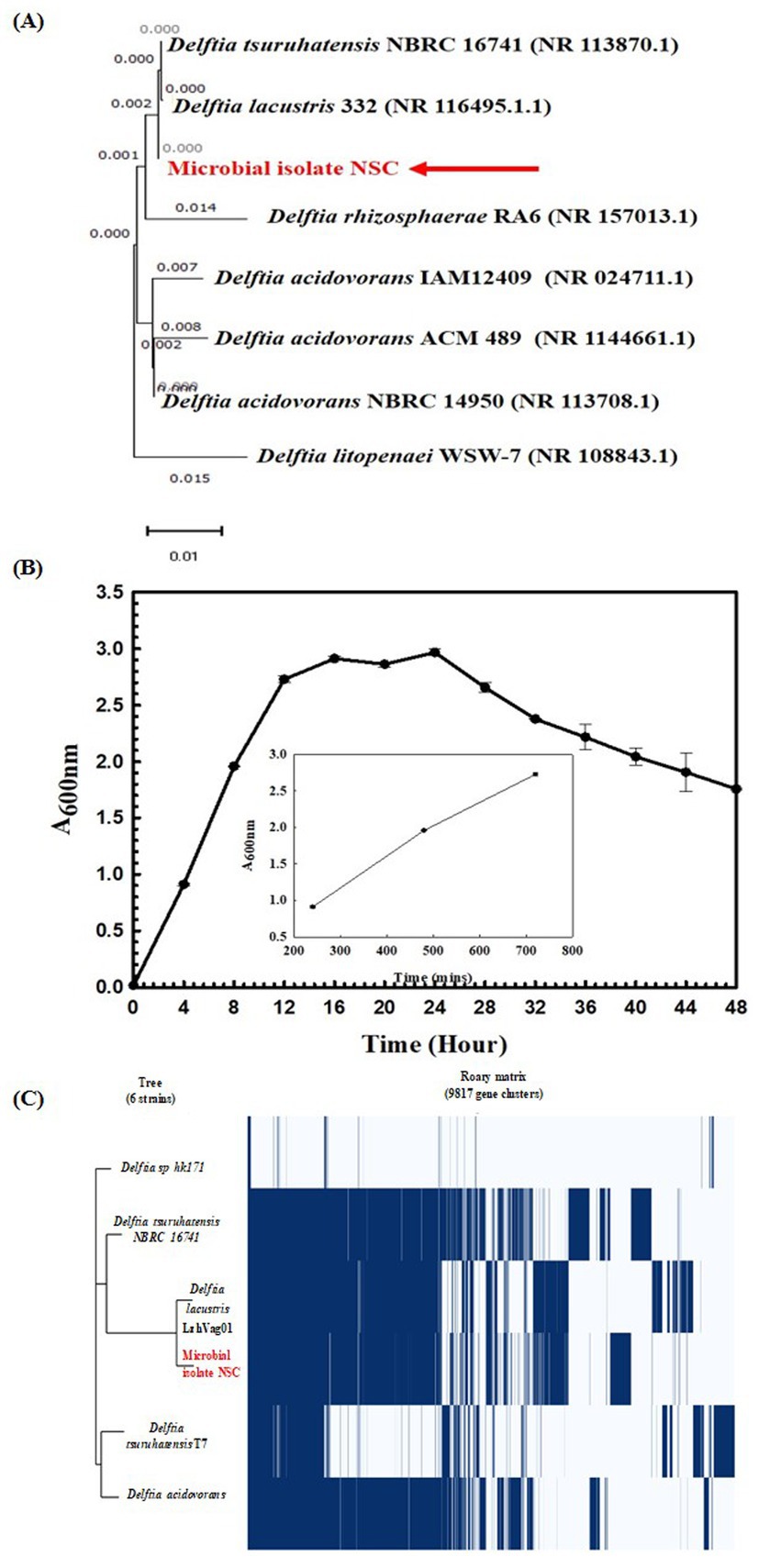
Figure 2. Phylogenetic and Physiological characterization of Delftia lacustris strain NSC. Phylogenetic affiliation of microbial isolates NSC with the other Delftia species (A). The phylogenetic tree was constructed using the Neighbor-joining method of phylogenetics with 1,000 bootstrap replications using MEGA-X software. The out-group was represented by Delftia litopenaei WSW-7 SSU rRNA gene sequence. Growth pattern analysis of Delftia lacustris strain NSC was observed after incubating the cultures for 48 h in LB broth with constant shaking at 200 rpm. The inset figure represents the linear growth phase used to calculate generation time (B). Phylogenomic analysis of D. lacustris strain NSC with other Delftia strains (C). The phylogenomic tree of the microbial isolate was generated using the FastTree v2.1.10 tool through Roary. The left panel illustrates the phylogenetic relationship of D. lacustris NSC with other Delftia species, while the right panel shows the core and accessory genes shared among different D. lacustris strains.
Molecular, physiological, biochemical, and genomic characterization of D. lacustris strain NSC
Microscopic examination identified the isolate as a Gram-negative, rod-shaped, motile bacterium. Physiologically, it exhibited optimal growth at pH 7.0 and 35°C temperature, reaching the logarithmic phase after 12 h with a generation (doubling) time of approximately 54.31 min (Figure 2B). Growth profile analysis at various temperatures and pH conditions indicated its sustained growth between 10°C and 50°C and pH 5 and 8. It also demonstrated facultative anaerobic growth, with an optical density (O. D.) of 0.402 at 600 nm after 24 h under anaerobic conditions. Biochemical testing showed it was positive for amylase, esterase, lipase, protease, and catalase activities, and its substrate utilization profile was consistent with that of other Delftia species (Supplementary Table S1). Antibiotic susceptibility testing revealed that it was resistant to amoxicillin, bacitracin, cephalothin, vancomycin, ceftazidime, and ofloxacin, but remained sensitive to oxytetracycline, novobiocin, and lincomycin, resembling the antibiotic resistance profile of other Delftia strains (Supplementary Table S2).
Stress response assays demonstrated that it could tolerate saltine stress [up to 11.69% NaCl (w/v), 11.18% KCl (w/v), 4.24% LiCl (w/v)], metal stress [0.068% CdCl2 (w/v), up to 0.1% Na3AsO4 (w/v), 0.15% NaAsO2 (w/v)], and oxidizing agents stress [up to 4.25% H2O2 (v/v)]. These properties were found similar to other Delftia strains (Supplementary Table S3). These findings highlight the adaptability of the D. lacustris strain NSC under stress conditions. Genome sequencing has generated 805,929 paired-end raw reads, which were assembled into 6,235,469 base pairs represented through 220 contigs. Functional annotation of the genome revealed 5,466 protein-coding sequences, 8 rRNA genes, 82 tRNA genes, and one tmRNA gene. The Average Nucleotide Identity (ANI) among different species of Delftia sp. ranged from 74 to 98%, reflecting significant genomic variation between species. Notably, the ANI between D. lacustris NSC and Delftia lacustris LzhVag01 was 98.28%, higher than those observed with other species within the genus (Supplementary Table S4). The classification of D. lacustris strain NSC as part of the Delftia species was further confirmed using tetra correlation. Delftia sp. LMG 24775 showed a z-score of 0.99989 when compared to D. lacustris strain NSC, confirming its close genetic relationship. Other Delftia species exhibited high similarity, with z-scores ranging from approximately 0.95–0.99 (Supplementary Table S5). Following ANIb and tetra confirmation, the genome of D. lacustris strain NSC was compared to those of other Delftia species to assess genome-wide similarities and differences. A matrix generated using the Roary tool highlighted the extensive nature of the genome, showing that D. lacustris strain NSC exhibited the greatest similarity to D. lacustris LzhVag01 (Figure 2C). The Roary analysis also revealed that all Delftia genomes shared a significant number of genes. Notably, the D. lacustris strain NSC genome lacks pathogenic genes or virulence-related genes, supporting its non-pathogenic nature.
Genomic analysis also identified genes involved in phosphate solubilization and transport (Table 1). Beyond phosphate solubilization, its genome includes genes associated with plant growth promotion activities, such as auxin biosynthesis, nitrogen assimilation, and siderophore biosynthesis (Table 1). Presence of genes for arsenic resistance, oxidative stress tolerance, metal stress tolerance, and salt tolerance, confirming the genome-derived stress-mitigating physiology of D. lacustris strain NSC. A detailed examination of the D. lacustris strain NSC genomes also revealed genes encoding proteins for the synthesis of Type 1 and IV pili and exopolysaccharides (Table 1), crucial for plant surface adhesion, auto-aggregation, and early biofilm formation.

Table 1. Genetic features identified within Delftia lacustris strain NSC genome encoding various proteins involved in nutrient assimilation, solubilization, colonization, and antifungal properties.
Assessment of biofertilizer-related features in D. lacustris strain NSC
Delftia lacustris strain NSC demonstrated phosphate solubilizing activity (0.325 IU), nitrate reductase activity (0.401 IU), and produced IAA (0.485 IU). The qualitative analysis showed that D. lacustris strain NSC could also generate ammonia. Siderophore biosynthesis assays revealed the formation of an orange color and the development of a clearance zone (19.5 ± 0.02 mm), indicating the siderophore production ability. It has also demonstrated strong growth even in the presence of up to 40% polyethylene glycol (PEG). This result suggests that D. lacustris strain NSC possesses traits that can help plants withstand drought by enhancing growth in challenging environments. Additionally, it was found to produce the enzyme ACC deaminase, with an activity level of 0.512 IU. ACC deaminase activity suggests that it may play a crucial role in promoting plant health under stressful conditions, such as drought, by reducing oxidative stress and supporting plant growth.
Assessment of D. lacustris strain NSC biofertilizer and biocontrol properties in laboratory conditions
Plant growth promotion properties of D. lacustris strain NSC were sequentially assessed at different plant growth stages, starting from seed germination to plant grain yield. Wheat seed germination assay reflected that the untreated control group had a seed germination rate of 75.66 ± 0.57735%. In contrast, seeds pre-treated with D. lacustris strain NSC exhibited a germination efficiency of 93.33 ± 0.23% (Figure 3A), reflecting approximately a 123.35-fold increase (p = 0.0001) in germination percentage.
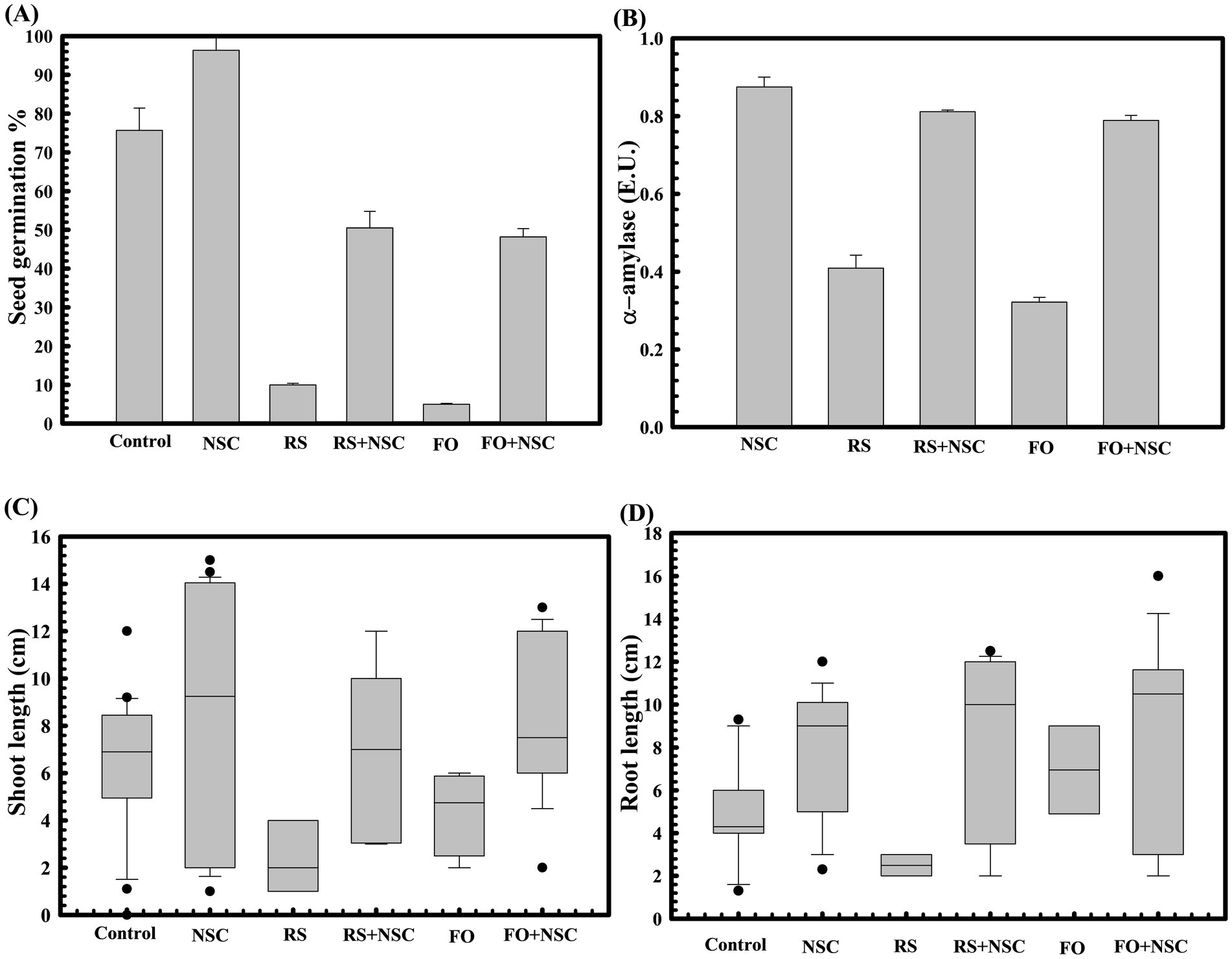
Figure 3. Effect of D. lacustris strain NSC on wheat seed germination parameters. The seed germination percentage in D. lacustris strain NSC pretreated seed in reference to the control (untreated) (A). The alpha amylase activity of D. lacustris strain NSC pretreated wheat seeds in the presence and absence of plant pathogenic fungi Rhizoctonia solani and Fusarium oxysporum (B). The impact of seeds pretreatment with D. lacustris strain NSC on shoot (C) and root length (D) of WC-306 seedling in the presence of R. solani and F. oxysporum. Plotted values are the mean of triplicates along with the observed standard deviation. Here NSC, D. lacustris NSC; FO, Fusarium oxysporum; RS, Rhizoctonia solani.
The protective effect of D. lacustris strain NSC was also assessed against phytopathogen infestation during wheat seed germination. In the presence of Rhizoctonia solani and Fusarium oxysporum, seed germination was substantially reduced in untreated seeds to 10 ± 1% (p = 0.024) and 5 ± 0.57735% (p = 0.032), respectively. However, pre-treatment with D. lacustris strain NSC led to improved germination efficiencies of 52.25 ± 0.23% and 48.19 ± 0.55% in the presence of R. solani and F. oxysporum, respectively (Figure 3A). Notably, it has significantly enhanced seed germination in the presence of these phytopathogens by approximately 520-fold (p = 0.0001) and 963.8-fold (p = 0.0001) compared to untreated control seeds. These findings strongly suggest the biocontrol potential of the D. lacustris strain NSC, as it improved seed germination in the presence of phytopathogens and significantly outperformed untreated seeds (p = 0.0025) (Figure 3). Moreover, R. solani and F. oxysporum exposure during wheat seed germination significantly reduced alpha-amylase activity to 0.42 IU and 0.22 IU, respectively. Pre-treatment with D. lacustris strain NSC significantly restored alpha-amylase activity to 0.792 IU (p = 0.0001) and 0.735 IU (p = 0.0001) in the presence of R. solani and F. oxysporum, respectively (Figure 3B).
Accordingly, the impact of seeds inoculation with strain NSC on plantlet growth parameters was also assessed. Pre-treatment with D. lacustris strain NSC also increased wheat plantlets’ shoot and root length. Seeds treated with D. lacustris NSC exhibited significantly increased shoot length (p = 0.033) (Figure 3C) and root length (p = 0.002) (Figure 3D) relative to untreated seeds. Furthermore, it has significantly improved the root (p = 0.020, p = 0.032) and shoot length (p = 0.031, p = 0.129) of plantlets derived from wheat seeds exposed to R. solani and F. oxysporum, respectively. Even after exposure to these pathogens, the average shoot and root lengths of plantlets treated with D. lacustris isolate NSC remained significantly higher (p = 0.0297 and p = 0.010) compared to the untreated control, highlighting its biocontrol potential.
The role of D. lacustris strain NSC in enhancing wheat seed germination under abiotic stress, such as high salinity, was also evaluated. Germination rates significantly decreased with increasing salt concentrations, up to 500 mM NaCl (Figure 4). However, seeds pre-treated with D. lacustris strain NSC exhibited improved germination under high salinity conditions (Figure 4A). Specifically, seed germination increased by approximately 10.11-fold (p = 0.0001) at 1 M NaCl, with corresponding increases in alpha-amylase activity (0.412 IU at 1 M NaCl) (Figure 4B) compared to controls. Pre-treatment with it enhanced seed germination and promoted wheat plantlet growth. At the highest NaCl concentration (1 M), seed pre-treatment exhibited a ~4.02-fold increase in shoot length (p = 0.0003) (Figure 4E) and a ~2.34-fold increase in root length (p = 0.0001) (Figure 4F) compared to untreated seeds under saline conditions (Figures 4C,D).
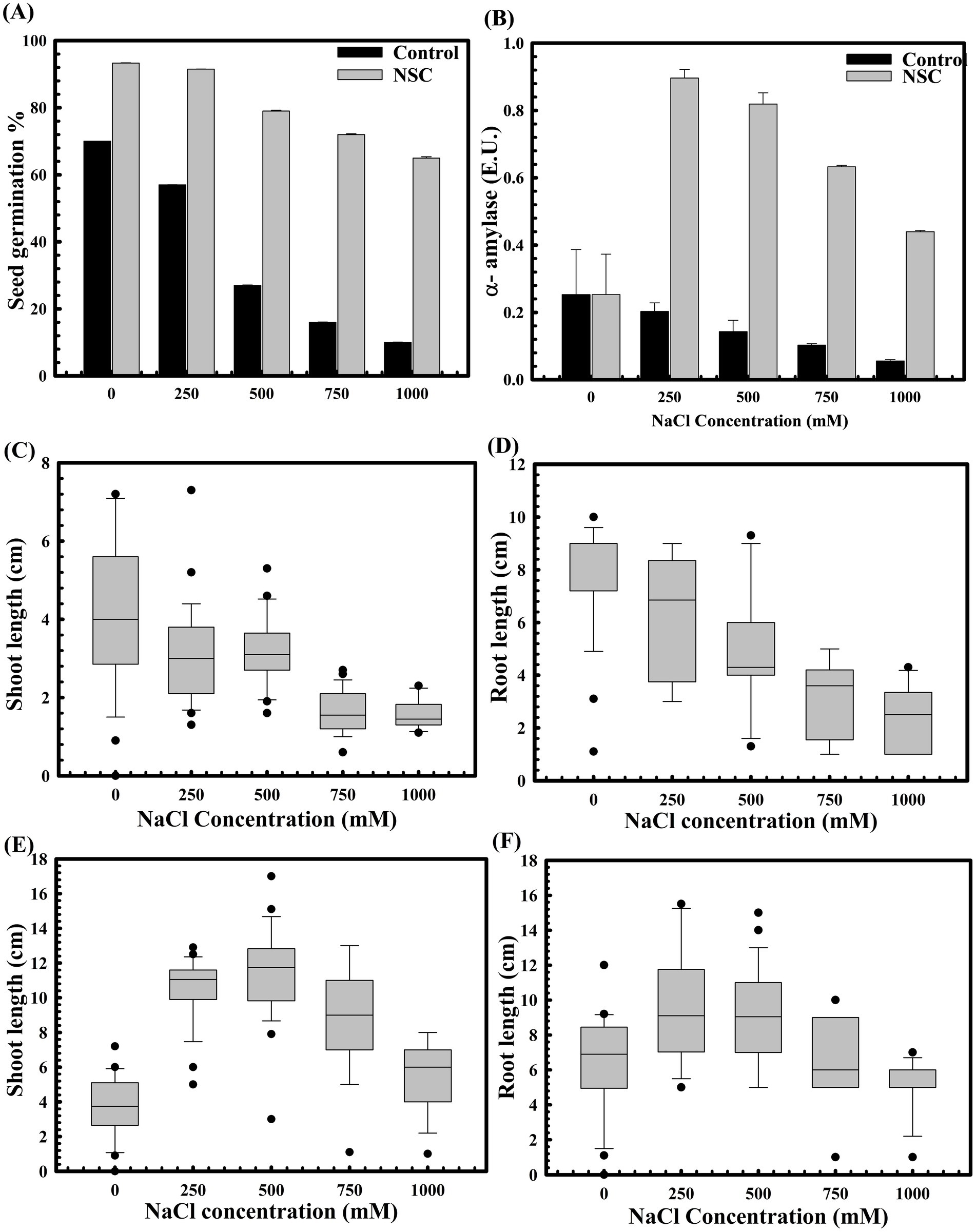
Figure 4. The impact of D. lacustris strain NSC pre-treatment on wheat seed germination in saline conditions. The seed germination assays were performed to compare the effect of D. lacustris strain NSC on seed germination under saline conditions (A). The alpha amylase activity of D. lacustris strain NSC pretreated wheat seeds under saline stress conditions (B). Shoot and root lengths of WC-306 seedlings were assessed under both untreated (C,D) and D. lacustris strain NSC pre-treated group (E,F) under NaCl-induced salinity stress. All assays were performed in triplicate.
Assessment of D. lacustris isolate NSC biofertilizer properties under field conditions
The physiological parameters were assessed at wheat plant growth stages, also known as Feeks. The rhizosphere of wheat plants derived from seeds pre-treated with D. lacustris strain NSC exhibited substantial enhancements in various physiological parameters such as phosphate solubilization (p = 0.0025), nitrogen assimilation (p = 0.0005), the release of total sugar (p = 0.0022), and reducing sugar (p = 0.0011) relative to untreated controls. Across multiple growth stages (Feeks 1, 2, 3, 6, 9, and 10.5), the concentrations of total sugars and reducing sugars in the rhizosphere were significantly elevated in the pre-treated plants, with total sugars increasing by ~2.1 fold increase (Figure 5A), and reducing sugars by ~2.03 fold (Figure 5B). Likewise, extracellular alkaline phosphatase activity was markedly higher in the pre-treated plants, showing increases ranging from ~ 3.48-fold at different stages (Figure 5C). Additionally, pre-treatment resulted in enhanced nitrogen assimilation in the rhizosphere, as evidenced by a substantial increase in nitrate reductase activity. This activity was elevated by ~2-fold at Feeks 3, with consistent increases observed at other stages, ranging from ~6.01-fold higher than that in untreated seeds (Figure 5D). These findings indicate that the pre-treatment facilitated improved nutrient cycling, particularly in relation to sugars and phosphorus, and promoted greater nitrogen uptake, thereby supporting enhanced growth and development of the wheat plants. Wheat plants originated from seeds treated with D. lacustris strain NSC showed a significant increase in the number of tillers (p = 0.0042), number of leaves per plant (p = 0.0002), spike length (p = 0.0025), number of spikes per plant (p = 0.0033), number of spikelets per plant (p = 0.0001), grain weight per 1,000 grains (p = 0.0011) and grain yield (p = 0.0001) (Table 2). These findings suggested that D. lacustris strain NSC enhanced wheat productivity by promoting both vegetative growth (e.g., more tillers and leaves) and reproductive success (e.g., longer spikes, more spikelets, and higher grain weight), leading to higher overall grain yields. This could have important implications for sustainable agriculture, particularly as these microbial isolates may reduce the need for chemical fertilizers and enhance crop performance under varying environmental conditions.
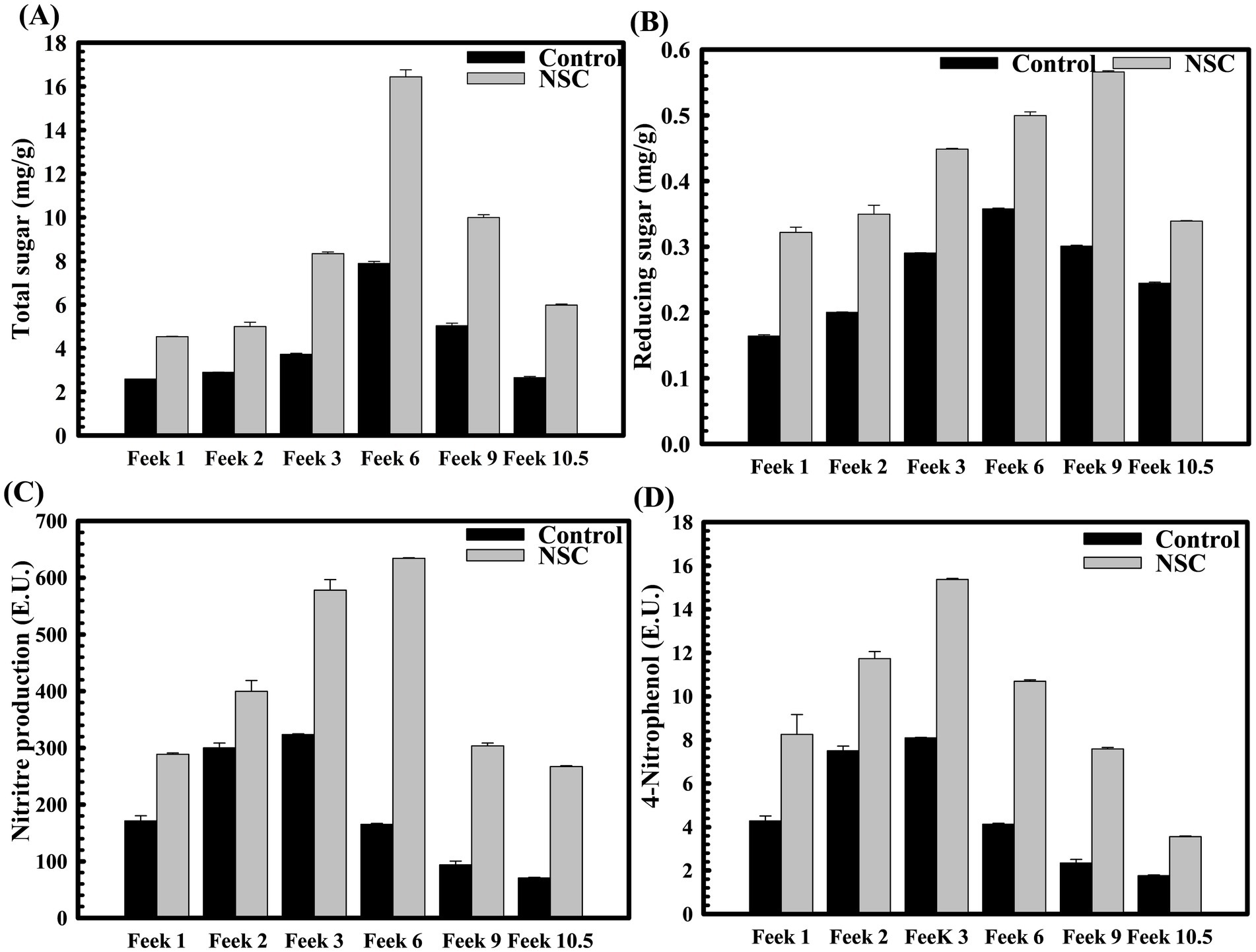
Figure 5. The impact of D. lacustris strain NSC pre-treatment on rhizosphere sugar content, phosphatase, and nitrate reduction activity across various wheat growth stages. Total sugar content was observed at different Feeks (1.0, 2.0, 3.0, 6.0, 9.0, and 10.5) in the presence and the absence of microbial inoculant NSC (A). Reducing sugar was estimated using DNS assay at different Feeks (1.0, 2.0, 3.0, 6.0, 9.0, and 10.5) was assessed in the presence and the absence of microbial inoculant NSC (B). Assessment of nitrate reductase activity at different Feeks (1.0, 2.0, 3.0, 6.0, 9.0, and 10.5) in the presence or absence of D. lacustris strain NSC (C). Assessment of alkaline phosphatase activity at different Feeks (1.0, 2.0, 3.0, 6.0, 9.0, 10.5) in the presence or absence of D. lacustris strain NSC (D). The experiment was carried out in triplicate. Plotted values are the mean of triplicate readings and their observed standard deviation.
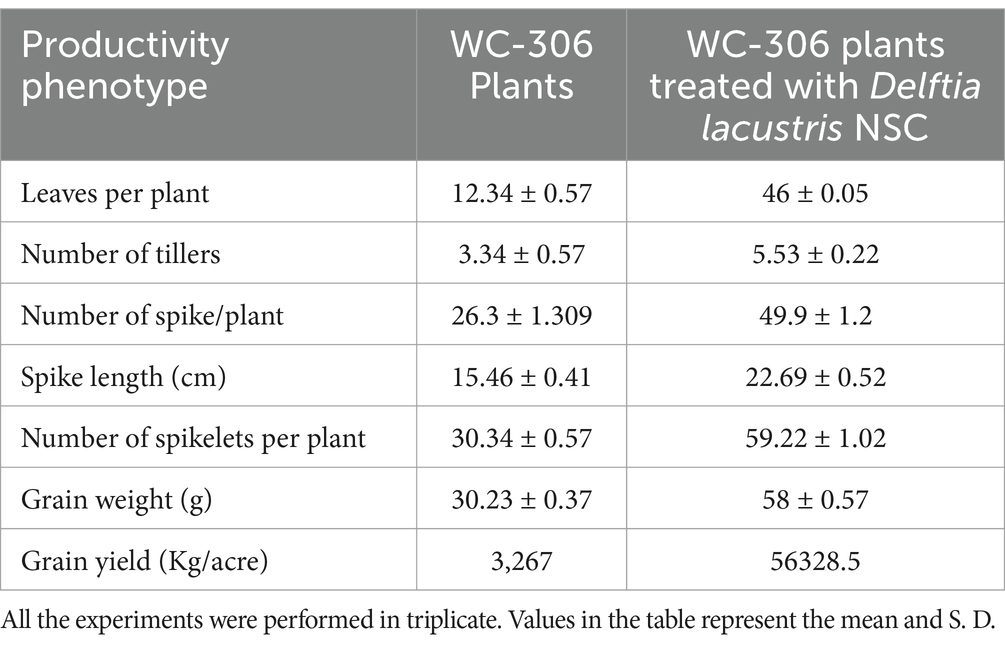
Table 2. Assessment of plant productivity features in D. lacustris strain NSC treated seeds in comparison to untreated.
Discussion
In the present study, a bacterial isolate (designated NSC) was obtained from the wheat rhizosphere, a microecological niche known for harboring diverse beneficial microbes (Han et al., 2005; Ubalde et al., 2012). Each microbial isolate must be taxonomically and phylogenetically characterized prior to its employment for any applications (Keswani et al., 2019). Taxonomic and phylogenetic analyses confirmed the affiliation of isolate NSC with D. lacustris strain 332. This identification was further supported by phylogenetic clustering and genome-wide similarity metrics, including average nucleotide identity (ANI) and tetra correlation values. Microscopic examination revealed that the isolate is a Gram-negative, rod-shaped, and motile bacterium, consistent with the established morphology of Delftia spp. (Grady et al., 2016). All these results ensured the isolation of the target bacterium, i.e., D. lacustris, from the wheat rhizosphere. Physiological assessments showed that isolated NSC grows optimally at neutral pH and under mesophilic conditions, but it also tolerates a wide range of temperatures and pH levels. Given that soil is a dynamic ecosystem with fluctuating environmental parameters (Mehta et al., 2021; Morigasaki et al., 2024), microbial adaptability to such changes is crucial. The ability of D. lacustris NSC to thrive under varying conditions highlights its ecological fitness and survivability in the soil environment.
Biochemical assays demonstrated the production of several hydrolytic enzymes, including amylase, lipase, esterase, and protease, which play roles in nutrient mobilization and pathogen suppression (Figueiredo et al., 2021). In nutrient-poor soil environments, a microbe’s metabolic versatility supports its survival and functional performance (Hopkins and Dungait, 2010; Wang et al., 2015). Isolate NSC exhibited a broad substrate utilization profile and robust metabolic machinery, positioning it as a strong candidate for soil application as a plant growth-promoting bacterium (PGPB).
Soils are increasingly contaminated by anthropogenic pollutants, particularly heavy metals and metalloids such as cadmium and arsenic (Vareda et al., 2019). These contaminants disrupt soil ecology and impair plant growth (Hou et al., 2025). Delftia strains are known for their ability to tolerate salinity and metal/metalloid stress, making them suitable for application in degraded soils (Bautista-Hernandez et al., 2012; Samimi and Shahriari-Moghadam, 2021). In line with this, isolate NSC demonstrated tolerance to multiple abiotic stresses, including salinity, cadmium, arsenic, and oxidative stress (H₂O₂), confirming its environmental resilience. These traits are consistent with previous reports of stress-tolerant Delftia strains from contaminated environments (Gulati et al., 2020; Kumar S. et al., 2022; Kumar M. et al., 2022).
Antibiotic accumulation in soil, resulting from manure, pharmaceuticals, sludge, or native microbial activity, poses additional ecological stress (Cycoń et al., 2019; Hashmi et al., 2017). Isolate NSC displayed resistance to multiple antibiotics, including amoxicillin, bacitracin, cephalothin, vancomycin, ceftazidime, netillin, and ofloxacin. Genome-wide analysis revealed the presence of antibiotic resistance genes, suggesting the isolate’s ability to survive in antibiotic-contaminated environments. The combination of an extended substrate range, metabolic richness, and stress tolerance underscores its rhizosphere fitness and colonization potential—key features for an effective biofertilizer (Compant et al., 2005).
This study primarily aimed to evaluate the biofertilizer and biocontrol potential of D. lacustris NSC through genomic, physiological, and field-based investigations. Genome analysis revealed numerous plant growth-promoting genes, including those involved in phosphate solubilization, nitrogen assimilation, indole-3-acetic acid (IAA) biosynthesis, siderophore production, and ACC deaminase activity—well-established PGP mechanisms (Glick, 2014; Egamberdieva et al., 2017; Sharma et al., 2024a). The genome also encodes genes for chitinase, proteases, phenazine, and siderophores—critical components for biocontrol via fungal cell wall degradation and iron sequestration (Nefzi et al., 2019; Karmegham et al., 2020). ACC deaminase activity helps mitigate ethylene-induced stress (Danhorn and Fuqua, 2007), while genes for exopolysaccharide biosynthesis and Type I/IV pili support root adhesion, biofilm formation, and rhizosphere competence (Zhang et al., 2024). While concerns exist about the pathogenic potential of some Delftia strains (Berg et al., 2005), genomic analysis of D. lacustris NSC revealed the absence of known virulence factors, suggesting it is safe for agricultural use.
Phosphate solubilizing activity, nitrate reductase activity, ammonia production, and IAA biosynthesis are a few essential properties of potential biofertilizer strains (Tang et al., 2020). Results have showcased that D. lacustris strain NSC harbors all these biofertilizer traits. It was also found to tolerate osmotic stress induced by polyethylene glycol (PEG). PEG is a well-known osmotic stress inducer (Zlobin et al., 2018) and is used as a selection marker for drought stress resistance plant growth-promoting microbes (Ashry et al., 2022). PEG induced stress tolerance property of D. lacustris strain NSC, indicating its potential to support plant growth even under drought conditions. ACC deaminase activity further strengthens its utility as a stress-alleviating PGPR (Glick, 2014). These properties are essential for any biofertilizer strain and could enhance nutrient assimilation to improve host growth (Rao et al., 2020). Collectively, these features position D. lacustris NSC as a promising biofertilizer for sustainable wheat production (Brambilla et al., 2022).
Although these traits are promising, they are based primarily on the strain’s intrinsic properties. Therefore, the present study also evaluated the actual impact of D. lacustris NSC on wheat seed germination, growth, and yield.
Wheat seed germination is a critical determinant of crop productivity (Reed et al., 2022). Phytopathogens such as R. solani and F. oxysporum negatively impact germination and reduce yield (Agha et al., 2023). Microbes with antifungal activity against these pathogens can serve as effective biocontrol agents (Reed et al., 2022; Agha et al., 2023). Genomic evidence of antifungal genes in isolate NSC, along with experimental results, confirmed its ability to enhance wheat seed germination both under normal and pathogen-challenged conditions. Additionally, pre-inoculation with isolate NSC significantly increased seed alpha-amylase activity, essential for mobilizing stored starch into sugars to support early seedling vigor (Kumar et al., 2023). This enzymatic activity may explain the improved germination rates and seedling establishment. Pre-treatment with NSC also significantly improved shoot and root lengths in wheat seedlings under both non-stress and stress conditions. Notably, the isolate remained effective under salt stress (up to 1 M NaCl), consistent with the genomic identification of stress-resistance genes. It underlines its potential application in saline-affected soils (Zhao et al., 2016). These results highlight the dual biofertilizer and biocontrol capabilities of D. lacustris NSC under varied environmental conditions.
Field evaluations further validated these findings. Across multiple wheat growth stages (Feeks 1–10.5), NSC-treated plants displayed higher rhizospheric nutrient activity, including increased levels of reducing sugars, total sugars, nitrate reductase, and alkaline phosphatase activity. These enhancements indicate improved microbial-driven nutrient cycling, benefiting plant nutrient uptake and metabolism (Richardson et al., 2009). Agronomic parameters such as tiller number, spikelet density, grain weight, and overall yield significantly improved in NSC-treated plants, highlighting its potential to boost vegetative growth and reproductive output. In summary, the genomic and functional characteristics of D. lacustris NSC demonstrate its potential as both a plant growth-promoting biofertilizer and biocontrol agent. Its resilience to environmental stressors and ability to enhance wheat growth and yield underscore its suitability for sustainable agriculture, particularly in marginal or degraded soils with reduced chemical input.
Data availability statement
The 16S rRNA gene sequence datasets generated in this study were deposited at NCBI, PRJNA1223623 (https://www.ncbi.nlm.nih.gov/sra/PRJNA1223623).
Author contributions
PS: Data curation, Investigation, Methodology, Writing – original draft, Writing – review & editing. RP: Formal analysis, Funding acquisition, Methodology, Project administration, Resources, Software, Validation, Writing – original draft, Writing – review & editing. NC: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. Authors acknowledge the funding support from Bill and Melinda Gates Foundation (BMGF), grant number -INV-033578 to RP.
Acknowledgments
The authors acknowledge CSIR-Institute of Genomics and Integrative Biology, New Delhi, India for DNA sequencing facility.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1576536/full#supplementary-material
Footnotes
References
Agafonova, N. V., Doronina, N. V., Kaparullina, E. N., Fedorov, D. N., Gafarov, A. B., Sazonova, O. I., et al. (2017). A novel Delftia plant symbiont capable of autotrophic methylotrophy. Microbiology 86, 96–105. doi: 10.1134/S0026261717010039
Agha, S. I., Ullah, M., Khan, A., Jahan, N., Ullah, S. M., Tabassum, B., et al. (2023). Biocontrol rhizobacteria enhances growth and yield of wheat (Triticum aestivum) under field conditions against Fusarium oxysporum. Bioengineered 14:2260923. doi: 10.1080/21655979.2023.2260923
Ashry, N. M., Alaidaroos, B. A., Mohamed, S. A., Badr, O. A. M., El-Saadony, M. T., and Esmael, A. (2022). Utilization of drought-tolerant bacterial strains isolated from harsh soils as a plant growth-promoting rhizobacteria (PGPR). Saudi J. Biol. Sci. 29, 1760–1769. doi: 10.1016/j.sjbs.2021.10.054
Bautista-Hernandez, D. A., Ramírez-Burgos, L. I., Duran-Páramo, E., and Fernández-Linares, L. (2012). Zinc and lead biosorption by Delftia tsuruhatensis: a bacterial strain resistant to metals isolated from mine tailings. JWARP 4, 207–216. doi: 10.4236/jwarp.2012.44023
Behera, B. C., Yadav, H., Singh, S. K., Mishra, R. R., Sethi, B. K., Dutta, S. K., et al. (2017). Phosphate solubilization and acid phosphatase activity of Serratia sp. isolated from mangrove soil of Mahanadi river delta, Odisha, India. JGEB 15, 169–178. doi: 10.1016/j.jgeb.2017.01.003
Berg, G., Eberl, L., and Hartmann, A. (2005). The rhizosphere as a reservoir for opportunistic human pathogenic bacteria. Environ. Microbiol. 7, 1673–1685. doi: 10.1111/j.1462-2920.2005.00891.x
Bhattacharyya, C., Banerjee, S., Acharya, U., Mitra, A., Mallick, I., Haldar, A., et al. (2020). Evaluation of plant growth promotion properties and induction of antioxidative defense mechanism by tea rhizobacteria of Darjeeling, India. Sci. Rep. 10:15536. doi: 10.1038/s41598-020-72439-z
Brambilla, S., Stritzler, M., Soto, G., and Ayub, N. (2022). A synthesis of functional contributions of rhizobacteria to growth promotion in diverse crops. Rhizosphere 24:100611. doi: 10.1016/j.rhisph.2022.100611
Braña, V., Cagide, C., and Morel, M. A. (2016). “The sustainable use of Delftia in agriculture, bioremediation, and bioproducts synthesis” in Microbial models: From environmental to industrial sustainability. ed. S. Castro-Sowinski (Singapore: Springer), 227–247.
Chandran, H., Meena, M., and Swapnil, P. (2021). Plant growth-promoting rhizobacteria as a green alternative for sustainable agriculture. Sustain. For. 13:10986. doi: 10.3390/su131910986
Chauhan, N. S., Ranjan, R., Purohit, H. J., Kalia, V. C., and Sharma, R. (2009). Identification of genes conferring arsenic resistance to Escherichia coli from an effluent treatment plant sludge metagenomic library: arsenic resistance genes from sludge metagenome. FEMS Microbiol. Ecol. 67, 130–139. doi: 10.1111/j.1574-6941.2008.00613.x
Compant, S., Duffy, B., Nowak, J., Clément, C., and Barka, E. A. (2005). Use of plant growth-promoting bacteria for biocontrol of plant diseases: principles, mechanisms of action, and future prospects. Appl. Environ. Microbiol. 71, 4951–4959. doi: 10.1128/AEM.71.9.4951-4959.2005
Cycoń, M., Mrozik, A., and Piotrowska-Seget, Z. (2019). Antibiotics in the soil environment—degradation and their impact on microbial activity and diversity. Front. Microbiol. 10:338. doi: 10.3389/fmicb.2019.00338
Danhorn, T., and Fuqua, C. (2007). Biofilm formation by plant-associated bacteria. Ann. Rev. Microbiol. 61, 401–422. doi: 10.1146/annurev.micro.61.080706.093316
Egamberdieva, D., Wirth, S. J., Alqarawi, A. A., Abd, E., and Hashem, A. (2017). Phytohormones and beneficial microbes: essential components for plants to balance stress and fitness. Front. Microbiol. 8:2104. doi: 10.3389/fmicb.2017.02104
Ehmann, A. (1977). The van URK-Salkowski reagent — a sensitive and specific chromogenic reagent for silica gel thin-layer chromatographic detection and identification of indole derivatives. J. Chromatogr. A 132, 267–276. doi: 10.1016/S0021-9673(00)89300-0
Fan, D., Schwinghamer, T., Liu, S., Xia, O., Ge, C., Chen, Q., et al. (2023). Characterization of endophytic bacteriome diversity and associated beneficial bacteria inhabiting a macrophyte Eichhornia crassipes. Front. Plant Sci. 14:1176648. doi: 10.3389/fpls.2023.1176648
Figueiredo, L., Santos, R. B., and Figueiredo, A. (2021). Defense and offense strategies: the role of aspartic proteases in plant-pathogen interactions. Biology 10:75. doi: 10.3390/biology10020075
Glick, B. R. (2014). Bacteria with ACC deaminase can promote plant growth and help to feed the world. Microbiol. Res. 169, 30–39. doi: 10.1016/j.micres.2013.09.009
Grady, E. N., MacDonald, J., Liu, L., Richman, A., and Yuan, Z. C. (2016). Current knowledge and perspectives of Paenibacillus: a review. Microb. Cell Factories 15:203. doi: 10.1186/s12934-016-0603-7
Gulati, M., Vyas, P., Rahi, P., and Gulati, A. (2020). Plant growth-promoting and rhizosphere-competent Delftia lacustris isolates with biocontrol potential. Microbiol. Res. 235:126436.
Guo, H., Yang, Y., Liu, K., Xu, W., Gao, J., Duan, H., et al. (2016). Comparative genomic analysis of Delftia tsuruhatensis MTQ3 and the identification of functional NRPS genes for Siderophore production. Biomed. Res. Int. 2016:3687619. doi: 10.1155/2016/3687619
Han, J., Sun, L., Dong, X., Cai, Z., Sun, X., Yang, H., et al. (2005). Characterization of a novel plant growth-promoting bacteria strain Delftia tsuruhatensis HR4 both as a diazotroph and a potential biocontrol agent against various plant pathogens. Syst. Appl. Microbiol. 28, 66–76. doi: 10.1016/j.syapm.2004.09.003
Harahap, R. T., Azizah, I. R., Setiawati, M. R., Herdiyantoro, D., and Simarmata, T. (2023). Enhancing upland rice growth and yield with indigenous plant growth-promoting rhizobacteria (PGPR) isolate at N-fertilizers dosage. Agriculture 13:1987. doi: 10.3390/agriculture13101987
Hashmi, M. Z., Strezov, V., and Varma, A. (2017). Antibiotics and antibiotics resistance genes in soils: Monitoring, toxicity, risk assessment and management. Cham: Springer International Publishing.
Himpsl, S. D., and Mobley, H. L. T. (2019). Siderophore Detection Using Chrome Azurol S and Cross-Feeding Assays, in Proteus mirabilis, ed. M. M. Pearson (New York, NY: Springer New York), 97–108. doi: 10.1007/978-1-4939-9601-8_10
Hopkins, D. W., and Dungait, J. A. J. (2010). “Soil microbiology and nutrient cycling” in Soil microbiology and sustainable crop production. eds. G. R. Dixon and E. L. Tilston (Dordrecht: Springer Netherlands), 59–80.
Hou, D., Jia, X., Wang, L., McGrath, S. P., Zhu, Y.-G., Hu, Q., et al. (2025). Global soil pollution by toxic metals threatens agriculture and human health. Science 388, 316–321. doi: 10.1126/science.adr5214
Islam, T., Fatema, n., Hoque, M. N., Gupta, D. R., Mahmud, N. U., Sakif, T. I, et al. (2023). Improvement of growth, yield, and associated bacteriome of rice by the application of probiotic Paraburkholderia and Delftia. Front. Microbiol. 14:1212505. doi: 10.3389/fmicb.2023.1212505
Iwase, T., Tajima, A., Sugimoto, S., Okuda, K., Hironaka, I., Kamata, Y., et al. (2013). A simple assay for measuring catalase activity: a visual approach. Sci. Rep. 3:3081. doi: 10.1038/srep03081
Karmegham, N., Vellasamy, S., Natesan, B., Sharma, M. P., Al Farraj, D. A., and Elshikh, M. S. (2020). Characterization of antifungal metabolite phenazine from rice rhizosphere fluorescent pseudomonads (FPs) and their effect on sheath blight of rice. Saudi J. Biol. Sci. 27, 3313–3326. doi: 10.1016/j.sjbs.2020.10.007
Kasana, R. C., Salwan, R., Dhar, H., Dutt, S., and Gulati, A. (2008). A rapid and easy method for the detection of microbial cellulases on agar plates using gram’s iodine. Curr. Microbiol. 57, 503–507. doi: 10.1007/s00284-008-9276-8
Keswani, C., Prakash, O., Bharti, N., Vílchez, J. I., Sansinenea, E., Lally, R. D., et al. (2019). Re-addressing the biosafety issues of plant growth promoting rhizobacteria. Sci. Total Environ. 690, 841–852. doi: 10.1016/j.scitotenv.2019.07.046
Khatri, D., and Chhetri, S. B. B. (2020). Reducing Sugar, Total Phenolic Content, and Antioxidant Potential of Nepalese Plants. Biomed Res. Int. 2020, 1–7. doi: 10.1155/2020/7296859
Kim, J., and Seo, H. (2018). In vitro nitrate reductase activity assay from Arabidopsis crude extracts. Bioprotocol 8:e2785. doi: 10.21769/BioProtoc.2785
Kumar, S., Diksha,, Sindhu, S., and Kumar, R. (2022). Biofertilizers: an ecofriendly technology for nutrient recycling and environmental sustainability. Curr. Res. Microb. Sci. 3:100094:100094. doi: 10.1016/j.crmicr.2021.100094
Kumar, M., Poonam,, Ahmad, S., and Singh, R. P. (2022). Plant growth promoting microbes: diverse roles for sustainable and ecofriendly agriculture. Energy Nexus 7:100133. doi: 10.1016/j.nexus.2022.100133
Kumar, A., Rani, R., and Sharma, S. (2023). Role of Delftia spp. in bioremediation and agriculture: an emerging PGPR. J. Environ. Manag. 330:117160.
Ludwig, T. G., and Goldberg, H. J. V. (1956). The Anthrone Method for the Determination of Carbohydrates in Foods and in Oral Rinsing. J Dent Res. 35, 90–94. doi: 10.1177/00220345560350012301
Maheshwari, R., Bhutani, N., and Suneja, P. (2020). Isolation and characterization of ACC deaminase producing endophytic Bacillus mojavensis PRN2 from Pisum sativum. Iran. J. Biotech. 18:2308. doi: 10.30498/ijb.2020.137279.2308
Mehta, P., Yadav, M., Ahmed, V., Goyal, K., Pandey, R., and Chauhan, N. S. (2021). Culture-independent exploration of the hypersaline ecosystem indicates the environment-specific microbiome evolution. Front. Microbiol. 12:686549. doi: 10.3389/fmicb.2021.686549
Morel, M. A., Cagide, C., Minteguiaga, M. A., Dardanelli, M. S., and Castro-Sowinski, S. (2015). The pattern of secreted molecules during the co-inoculation of alfalfa plants with Sinorhizobium meliloti and Delftia sp. strain JD2: an interaction that improves plant yield. MPMI 28, 134–142. doi: 10.1094/MPMI-08-14-0229-R
Morel, A. M., Iriarte, A., Jara, E., Musto, H., and Castro-Sowinski, S. (2016). Revealing the biotechnological potential of Delftia sp. JD2 by a genomic approach. AIMS Bioeng. 3, 156–175. doi: 10.3934/bioeng.2016.2.156
Morigasaki, S., Matsui, M., Ohtsu, I., Doi, Y., Kawano, Y., Nakai, R., et al. (2024). Temporal and fertilizer-dependent dynamics of soil bacterial communities in buckwheat fields under long-term management. Sci. Rep. 14:9896. doi: 10.1038/s41598-024-60655-w
Mustamu, E. N., Tampubolon, K., Alridiwirsah Basyuni, M., AL-Taey, D. K. A., Jawad Kadhim Al Janabi, H., and Mehdizadeh, M. (2023). Drought stress induced by polyethylene glycol (PEG) in local maize at the early seedling stage. Heliyon 9:e20209:e20209. doi: 10.1016/j.heliyon.2023.e20209
Nefzi, A., Abdallah, R. A. B., Jabnoun-Khiareddine, H., Ammar, N., and Daami-Remadi, M. (2019). Ability of endophytic fungi associated with Withania somnifera L. to control Fusarium crown and root rot and to promote growth in tomato. Braz. J. Microbiol. 50, 481–494. doi: 10.1007/s42770-019-00062-w
O'Callaghan, M. (2016). Microbial inoculation of seed for improved crop performance: issues and opportunities. Appl. Microbiol. Biotechnol. 100, 5729–5746. doi: 10.1007/s00253-016-7590-9
Oumer, O. J., and Abate, D. (2018). Screening and molecular identification of pectinase producing microbes from coffee pulp. Biomed. Res. Int. 2018, 1–7. doi: 10.1155/2018/2961767
Pattnaik, S., Mohapatra, B., and Gupta, A. (2021). Plant growth-promoting microbe mediated uptake of essential nutrients (Fe, P, K) for crop stress management: microbe–soil–plant continuum. Front. Agron. 3:689972. doi: 10.3389/fagro.2021.689972
Ramnath, L., Sithole, B., and Govinden, R. (2017). Identification of lipolytic enzymes isolated from bacteria indigenous to Eucalyptus wood species for application in the pulping industry. Biotechnol. Rep. 15, 114–124. doi: 10.1016/j.btre.2017.07.004
Rao, Y. H. C., Mohana, C., and Satish, S. (2020). “Biocommercial aspects of microbial endophytes for sustainable agriculture” in Microbial endophytes. Eds. K. Ajay and E. K. Radhakrishnan (United Kingdom: Woodhead Publishing), 323–347.
Reed, R. C., Bradford, K. J., and Khanday, I. (2022). Seed germination and vigor: ensuring crop sustainability in a changing climate. Heredity 128, 450–459. doi: 10.1038/s41437-022-00497-2
Rhouma, A., Hajji, H. L., Atallaoui, K., Kouadri, A. M., and Khrieba, M. (2024). Fusarium oxysporum f. sp. lycopersici, the causal agent of vascular wilt disease of tomatoes: from its taxonomy to disease management. Asian J. Mycol. 7, 21–36.
Richardson, A. E., Barea, J. M., McNeill, A. M., and Prigent-Combaret, C. (2009). Acquisition of phosphorus and nitrogen in the rhizosphere and plant growth promotion by microorganisms. Plant Soil 321, 305–339. doi: 10.1007/s11104-009-9895-2
Samimi, M., and Shahriari-Moghadam, M. (2021). Isolation and identification of Delftia lacustris strain-MS3 as a novel and efficient adsorbent for lead biosorption: kinetics and thermodynamic studies, optimization of operating variables. Biochem. Eng. J. 173:108091. doi: 10.1016/j.bej.2021.108091
Sazykin, I., Khmelevtsova, L., Azhogina, T., and Sazykina, M. (2023). Heavy metals influence on the bacterial community of soils: a review. Agriculture 13:653. doi: 10.3390/agriculture13030653
Sharma, P., Pandey, R., and Chauhan, N. S. (2024a). Biofertilizer and biocontrol properties of Stenotrophomonas maltophilia BCM emphasize its potential application for sustainable agriculture. Front. Plant Sci. 15:1364807. doi: 10.3389/fpls.2024.1364807
Sharma, P., Pandey, R., and Chauhan, N. S. (2024b). Unveiling wheat growth promotion potential of phosphate solubilizing Pantoea agglomerans PS1 and PS2 through genomic, physiological, and metagenomic characterizations. Front. Microbiol. 15:1467082. doi: 10.3389/fmicb.2024.1467082
Shigematsu, T., Yumihara, K., Ueda, Y., Numaguchi, M., Morimura, S., and Kida, K. (2003). Delftia tsuruhatensis sp. nov., a terephthalate-assimilating bacterium isolated from activated sludge. Int. J. Syst. Evol. Microbiol. 53, 1479–1483. doi: 10.1099/ijs.0.02285-0
Singh, K., and Kayastha, A. M. (2014). Α-amylase from wheat (Triticum aestivum) seeds: its purification, biochemical attributes, and active site studies. Food Chem. 162, 1–9. doi: 10.1016/j.foodchem.2014.04.043
Swain, M. R., Kar, S., Padmaja, G., and Ray, R. C. (2006). Partial characterization and optimization of production of extracellular alpha-amylase from Bacillus subtilis isolated from culturable cow dung microflora. Pol. J. Microbiol. 55, 289–296.
Tang, A., Haruna, A. O., Majid, N. M. A., and Jalloh, M. B. (2020). Potential PGPR properties of cellulolytic, nitrogen-fixing, phosphate-solubilizing Bacteria in rehabilitated tropical Forest soil. Microorganisms 8:442. doi: 10.3390/microorganisms8030442
Ubalde, M. C., Braña, V., Sueiro, F., Morel, M. A., Martínez-Rosales, C., Marquez, C., et al. (2012). The versatility of Delftia sp. isolates as tools for bioremediation and biofertilization technologies. Curr. Microbiol. 64, 597–603. doi: 10.1007/s00284-012-0108-5
Vareda, J. P., Valente, A. J. M., and Durães, L. (2019). Assessment of heavy metal pollution from anthropogenic activities and remediation strategies: a review. J. Environ. Manag. 246, 101–118. doi: 10.1016/j.jenvman.2019.05.126
Vijayaraghavan, P., Rajendran, P., Prakash Vincent, S. G., Arun, A., Abdullah Al-Dhabi, N., Valan Arasu, M., et al. (2017). Novel sequential screening and enhanced production of fibrinolytic enzyme by Bacillus sp. IND12 using response surface methodology in solid-state fermentation. Biomed. Res. Int. 2017:3909657. doi: 10.1155/2017/3909657
Wang, L., Fan, D., Chen, W., and Terentjev, E. M. (2015). Bacterial growth, detachment and cell size control on polyethylene terephthalate surfaces. Sci. Rep. 5:15159. doi: 10.1038/srep15159
Wille, L., Messmer, M. M., Studer, B., and Hohmann, P. (2019). Insights to plant–microbe interactions provide opportunities to improve resistance breeding against root diseases in grain legumes. Plant Cell Environ. 42, 20–40. doi: 10.1111/pce.13214
Yadav, M., Kumar, T., Kanakan, A., Maurya, R., Pandey, R., and Chauhan, N. S. (2022). Isolation and characterization of human intestinal Bacteria Cytobacillus oceanisediminis NB2 for probiotic potential. Front. Microbiol. 13:932795. doi: 10.3389/fmicb.2022.932795
Yin, Z., Liu, X., Qian, C., Sun, L., Pang, S., Liu, J., et al. (2022). Pan-genome analysis of Delftia tsuruhatensis reveals important traits concerning the genetic diversity, pathogenicity, and biotechnological properties of the species. Microbiol Spectr 10:e0207221. doi: 10.1128/spectrum.02072-21
Zhang, L., Zhang, X., Bai, H., Li, T., Zhang, Z., Zong, X., et al. (2024). Characterization and genome analysis of the Delftia lacustris strain LzhVag01 isolated from vaginal discharge. Curr. Microbiol. 81:232. doi: 10.1007/s00284-024-03758-x
Zhao, K., Penttinen, P., Guan, T., Xiao, J., Chen, Q., Lindström, K., et al. (2016). The diversity and antimicrobial traits of actinobacteria isolated from medicinal plants in tropical environments. Front. Microbiol. 7:2061.
Keywords: biocontrol agents, biofertilizer, Delftia , food security, rhizosphere microbiota, sustainable agriculture, wheat yield
Citation: Sharma P, Pandey R and Chauhan NS (2025) Assessment of the wheat growth-promoting potential of Delftia lacustris strain NSC through genomic and physiological characterization. Front. Microbiol. 16:1576536. doi: 10.3389/fmicb.2025.1576536
Edited by:
Selva Dhandapani, Agri-Food and Biosciences Institute, IrelandReviewed by:
Rajeshwari Negi, Eternal University, IndiaSam A. Masih, Sam Higginbottom University of Agriculture, Technology and Sciences, India
Copyright © 2025 Sharma, Pandey and Chauhan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nar Singh Chauhan, bnNjaGF1aGFuQG1kdXJvaHRhay5hYy5pbg==; Rajesh Pandey, cmFqZXNocEBpZ2liLnJlcy5pbg==
 Pinki Sharma
Pinki Sharma Rajesh Pandey
Rajesh Pandey Nar Singh Chauhan
Nar Singh Chauhan