- 1College of Biology and Food Engineering, Anhui Polytechnic University, Wuhu, China
- 2Anhui Zhang Hengchun Pharmaceutical Co., Ltd, Wuhu, China
- 3Wuhu Green Food Industry Research Institute Co., Ltd, Wuhu, China
- 4College of Chemistry and Environmental Engineering, Anhui Polytechnic University, Wuhu, China
- 5Anhui Engineering Laboratory for Industrial Microbiology Molecular Breeding, Wuhu, China
Menaquinone-7 (MK-7) production from renewable feedstocks using Bacillus subtilis natto provides a promising pathway toward sustainability. However, MK-7 yields are often limited by poor microbial cell viability. In this study, a novel fermentation strategy aimed at prolonging the lifespan of B. subtilis natto was investigated to enhance MK-7 biosynthesis. The results showed that the combination of soy protein hydrolysate (SPH) and Span 80 (SP80) increased intracellular and extracellular MK-7 yields by 5.6- and 7.2-fold, respectively, compared to the SP-based medium. This enhancement was associated with an extended lifespan of B. subtilis natto, as evidenced by increased optical density and cell length, and a reduced cell death rate—13.5% in the stationary phase compared to 39.5% in the SP-based medium. Moreover, a significant correlation was observed between key MK-7 biosynthetic genes and lifespan-related genes. The expression of growth-related autolysis genes skfF and sdp, which contribute to reduced cannibalism, was downregulated by 4.89- and 5.19-fold, respectively. In contrast, expression of the cell division genes ftsZ and ftsL, which promote cell division, was upregulated by 7.52- and 6.31-fold, respectively. Improved fermentation performance was attributed to a 2.26-fold increase in oligopeptides (Mw < 1 kDa) and moderate levels of amino acids, particularly Phe, Arg, and Glu, derived from the enzymatic hydrolysis of soy protein by Protamex 1.6, present in the SPH+SP80 medium. Meanwhile, the prolonged lifespan also promoted MK-7 biosynthesis by upregulating key membrane-associated genes such as menA and menD, which showed 1.43- and 1.47-fold increases in expression, respectively. This contributed to improved MK-7 precursor availability and enhanced MK-7 assembly efficiency in B. subtilis natto. Collectively, these results indicate that prolonging the lifespan of B. subtilis natto using SPH+SP80 represents a promising and effective strategy for improving industrial MK-7 production.
1 Introduction
Menaquinone-7 (MK-7), a highly valuable lip-soluble form of vitamin K2, features a ring structure comprising a 2-methyl-1,4-naphthoquinone core and a side chain of seven isoprene units. MK-7 has been identified as a component of microbial plasma membranes, where it plays an important role in electron transport and oxidative phosphorylation (Wu et al., 2021). In the current era of heightened health awareness, MK-7 has gained increasing attention for its efficacy in preventing and treating various diseases, especially those affecting bone and blood health (Liu et al., 2021). Interestingly, recent studies have shown that MK-7 acts as a potent anti-ferroptotic compound (Mishima et al., 2022), which is associated with the prevention of numerous age-related diseases, including osteoporosis, cardiovascular disease, cancer, Alzheimer’s disease, and diabetes. With the global rise in aging populations, the demand for MK-7 has grown substantially due to its broad therapeutic potential.
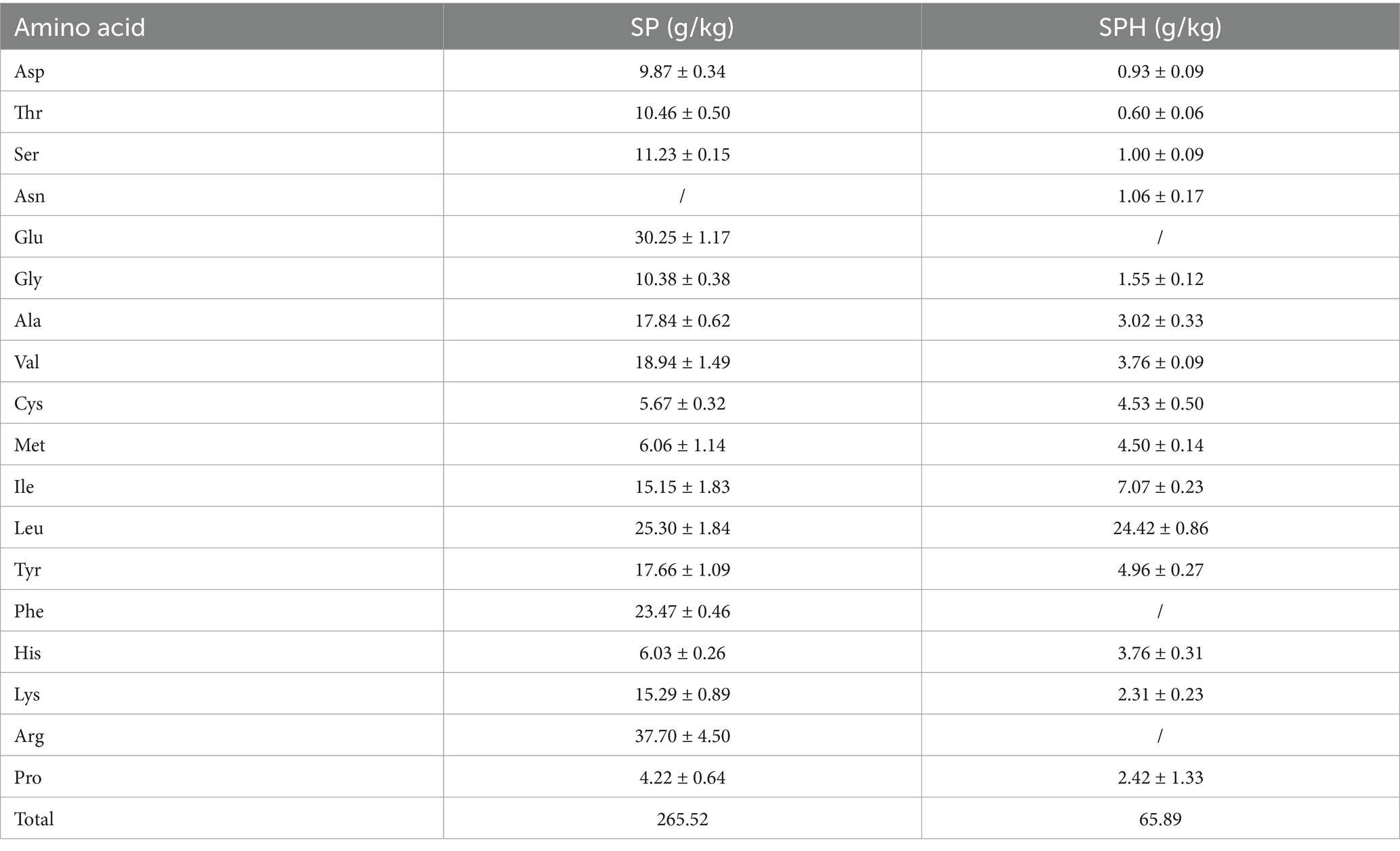
Several bacteria, including Escherichia coli, Lactobacillus, B. subtilis natto, and Bacillus amyloliquefaciens, have been employed to produce MK-7. B. subtilis natto became the dominant strain due to its safety, short growth cycle, and relatively higher MK-7 yield (Table 1). These bacteria are commercially available worldwide as nutritional supplements in food industrial production. However, the high production cost caused by the low yield has been the greatest challenge in the biosynthesis of MK-7.
Lifespan engineering, which regulates the growth performance of cell factories by extending cellular longevity and productivity, represents a promising strategy to increase metabolic production. The lifespan of microorganisms is divided into the chronological (defined as the length of time that cells in stationary phase culture remain viable) and replicative lifespan (defined by the number of daughter cells produced before the decline phase) (Lindner et al., 2008). Inhibition of cell autolysis was a viable approach to extend the chronological lifespan.
For example, Zhao et al. combined knockout of peptidoglycan hydrolase-related genes, including sigD, lytE, lytF, lytC, lytD, and lytG, which significantly increased cell growth rate and alpha-amylase production (Zhao et al., 2018). Regulation of the replicative lifespan in B. subtilis not only promoted rapid cell growth and altered cellular morphology but also modulated cell division dynamics. The rate of mass transfer depended on the specific surface area of the cell, while the cell morphology could be partially regulated by the rate of cell division (Jiang et al., 2015). For example, a previous study inhibited cell filopodia by overexpressing the genes ftsZ and ftsA associated with the D phase of cytokinesis, resulting in 57.1% higher growth density, 30% higher specific growth rate, and 227% higher production of human leptin protein in E. coli (Jeong and Lee, 2003). Although there had been many successful cases of modifying the chronological lifespan (CLS) and replicative lifespan (RLS) of microorganisms to increase metabolite content, it was particularly important to find a convenient and fast way to regulate cell lifespan due to the unavailability of gene editing tools for B. subtilis natto.
As a cost-effective and easily accessible nitrogen source, plant protein hydrolysates have been applied to markedly enhance the activity and improve the lifespan of bacterial cells. The mechanisms may be that the addition of small molecular weight proteins, polypeptides, or amino acids in the growth medium changed membrane lipid composition, cell membrane fluidity, membrane permeability, and intracellular homeostasis, and thus impacted cell growth (Jin et al., 2022). In addition, surfactant was applied to increase microbial metabolite production, remove target products inside the cells, and avoid production inhibition (Liu et al., 2014). To the best of our knowledge, no detailed investigation has been conducted on the impacts of SPH and surfactants in improving bacterial lifespan and enhancing MK-7 synthesis. The inter-relationships between SPH and surfactant prolonging the cell lifespan and the relationship between lifespan prolongation and MK-7 biosynthesis were not elucidated thoroughly. These results will provide a novel perspective on the prolongation of lifespan in B. subtilis natto and better elucidate the mechanism of SPH and surfactant supplementation in MK-7 synthesis from multiple dimensions.
2 Materials and methods
2.1 The preparation of soy protein hydrolysates
Soy peptone (SP), soy protein, and Protamex1.6 were purchased from Jinan Weiduofeng Biotechnology Co., Ltd. (Jinan, China), Jiangsu Ruiduo Bioengineering Co., Ltd. (Jiangsu, China), and Keppel Biotechnology Co., Ltd. (Qingdao, China), respectively. The preparation of soy protein hydrolysate (SPH) is referred to as the previous method (Yi et al., 2024). The soy protein and distilled water with a mass ratio of 1:15 were pretreated at 80°C for 10 min, and then Protamex 1.6 (1.5%, w/w) was added for 24 h of hydrolysis at pH 7.0, 50°C. The hydrolysates were heated to 100°C for 10 min to inactivate the enzyme and then centrifuged at 8,000 g and 4°C for 10 min. The supernatants were designated as SPH.
2.2 Strains and culture conditions
Bacillus subtilis natto strain CICC No. 25137 was screened by nitrosoguanidine and low-energy ion beam implantation (Song et al., 2014) and stored in the China Center of Industrial Culture Collection. The bacterial cells were first cultivated in seed medium (3% soy peptone, 0.1% yeast extract, 5% glycerol, and 0.9% NaCl) for 15 h at 37°C with 150 rpm. Then, the bacterial cells were harvested and inoculated into the fermentation medium at a 3% inoculum size. The base medium consisted of 5% glycerol, 1% yeast extract, 0.4% K2HPO4, and 0.2% KH2PO4. To form different experimental groups, additional components were included as follows: 3% SP (SP group), 0.3% Span 80 (SP80 group), 3% SPH (SPH group), and a combination of 3% SPH and 0.3% SP80 (SPH+SP80 group). Fermentation was conducted at 37°C for 84 h, with the medium occupying 30% of the total volume of the fermentation flask.
2.3 Degree of hydrolysis, molecular weight (mw) distribution, and free amino acids
The SP, SPH, SP80, or SPH+SP80 and distilled water with a mass ratio of 1:15 were pretreated at 80°C for 10 min, and then protamex1.6 (1.5%, w/w) was added for 24 h of hydrolysis at pH 7.0, 50°C. The hydrolysates were heated to 100°C for 10 min to inactivate the enzyme and then centrifuged at 8,000 g and 4°C for 10 min. The supernatant was obtained and tested for degree of hydrolysis, Mw distribution, and free amino acids.
The o-phthalaldehyde method (Nielsen et al., 2001) was used to determine the degree of hydrolysis for SP and SPH. Serine was used for the standard curve. The degree of hydrolysis was measured using the following formula:
where Cserine, Wserine, X, V, P, N, α, and β were the concentration of serine per gram of protein (mmol/g), the total number of peptide bonds (mmol/g), the sample mass (g), the volume of hydrolysate (L), the protein mass fraction of the sample (%), the dilution ratio, and the correction factors of 0.97 and 0.34, respectively.
The high Mw distribution (>10 kDa) of the sample was analyzed by SDS-PAGE. Separation gels (1.5 mm thick, 81 × 74 mm) consisted of a 12% polyacrylamide resolving gel and a 4% polyacrylamide stacking gel. 20 μL of each sample, protein marker ZD101 (14.4 kDa-116.0 kDa), and 5 μL of loading buffer were loaded onto the gel and run at 150 V in a Bio-Rad Mini-Protean Tetra system. Finally, the gel was stained using Coomassie brilliant blue fast-stain solution and photographed.
The low Mw distribution (<10 kDa) of the sample was analyzed by high-performance liquid chromatography (Agilent, USA) equipped with a UV detector and a TSK gel G2000SWXL (7.8 × 300 mm, 250 mm). The mobile phase was water: acetonitrile (4:1, v/v) and flowed at 0.5 mL/min. The wavelength of 220 nm was used for detection and analysis.
The free amino acids were measured by an L-8900 amino acid analyzer (Hitachi, Tokyo, Japan). The sample pretreatment was performed using the previous method (Li et al., 2022).
2.4 Determination of biomass, MK-7 yield, cell viability, and membrane potential
The cell biomass of fermentation broth was measured by determining the optical density at 600 nm. The MK-7 detection method was the same as in our previous research (Zhou et al., 2023).
The cell viability was assessed by flow cytometry (Guava EasyCyte, America). The bacterial solution cultured in 84 h of fermentation was centrifuged at 4°C and 8,000 g for 3 min, washed 3 times with 0.2 M PBS buffer, and resuspended in buffer (the bacterial density was 107–108 CFU/mL). 100 μL of solution from each tube was transferred to new 1.5 mL microcentrifuge tubes, vortexed, and stained with Propidium Iodide (PI).
The membrane potential was measured by flow cytometry (Novo et al., 2000). Firstly, B. subtilis natto was incubated in SP, SPH, SP80, and SPH+SP80 fermentation medium at 37°C for 84 h. Then, the cells were harvested by centrifugation (5,723 g, 10 min, 4°C). The competent cells were prepared by resuspending in 1 mL of ice-cold buffer solution [distilled deionized water (DDW), 0.9% NaCl, or 1 mM MgCl2, 30 mM DiOC2(3)]. The mixtures were incubated in the dark at room temperature for 4 min after being mixed vigorously by vortex. Finally, the cells were centrifuged (8,000 g, 10 min, 4°C), washed three times with ice-cold DDW, and analyzed by flow cytometry.
2.5 Cell morphology observation
The cells were centrifuged after 48 h of fermentation and washed twice with 0.1 M phosphate buffer. The dried cells were then fixed with 2.5% glutaraldehyde at 4°C for 6–12 h. The cells were then washed three times with 0.1 M phosphate buffer (20 min each time) and dehydrated by gradient ethanol (30, 50, 70, 80, 95, and 100% ethanol once, 20 min each time). Finally, the cells were coated with gold powder and observed using a scanning electron microscope (Thermo Fisher Scientific Corp., USA).
2.6 Detection of surface tension, NADH and NAD+, and maintenance energy coefficient (MEC)
The Wilhelmy plate method was applied to test the surface tension of a solution containing SP, SPH, SP80, and SPH+SP80 (Ma et al., 2023). A liquid film formed because of surface tension when the specific plate with known length and height was fixed on the tensiometer and contacted with the gas–liquid interface. The surface tension was measured by both the liquid film’s gravitational force and the contact line’s length. The results for SP, SPH, SP80, and SPH+SP80 solutions were calibrated using ethanol and pure water. For each example, three measurements were performed at 25°C, and the average was obtained.
NADH/NAD+ levels of B. subtilis natto were measured referring to the previous method (Wang et al., 2019). NAD+ total (NAD+ and NADH) or NADH levels were quantified by a colorimetric assay at 450 nm using a SpectraMax i3x (Molecular Devices, USA). The NADH/NAD+ ratio was calculated using the following formula:
The MEC of B. subtilis natto was measured using the previous method (Ren et al., 2024). The strains were incubated continuously in a 500 mL fermenter containing a 50 mL fermentation medium. The culture was switched from batch to chemostat mode when the glycerol was almost consumed. The agitation rate, pH, and temperature were controlled at 400 rpm, 6.6, and 37°C. Glycerol consumption was continuously monitored until the growth of cells reached a steady state. The value of the MEC was expressed as the rate of glycerol consumption (mgly).
2.7 RNA isolation, library construction, sequencing, and data analyses
The samples of B. subtilis natto were obtained by centrifuging the fermentation broth after 6 days of fermentation. RNA was extracted by the RNAlock reagent, and total RNA was collected by the TRIzol reagent (Invitrogen, USA). The extracted RNA was detected with an Agilent Bioanalyzer 2,100 (Agilent Technologies, Palo Alto, CA, USA). The RNA library construction from samples was completed by the Shanghai Human Genome Research Center (Wu et al., 2021).
After obtaining the data, the SeqPrep/Sickle software first selected the clean data by removing the adapter reads and low-quality and short fragments from the original data. Second, the differentially expressed genes (DEGs) between SP and SPH+SP80 were identified by mapping the clean data to reference sequences. Finally, the DEGs were mapped to terms in the gene ontology (GO) and KEGG databases for analyzing the functions and pathways, which were judged through the log2 ratio ≥ 1 and FDR of ≤ 0.05.
3 Results and discussion
3.1 Effect of MK-7 on microbial cell viability
Previous studies proved that the protein complex released MK-7 in the cell membrane of B. subtilis natto to the external environment (Chatake et al., 2018). To detect whether an MK-7-producing strain has a good product tolerance, 0, 100, 200, and 300 mg/L MK-7 were added to the medium at the beginning of fermentation, and the cell growth and morphology of B. subtilis natto BN-P15-11-1 were investigated. As shown in Figures 1A,E,F, with the increase of MK-7 concentration in the medium, the OD of B. subtilis natto at 48 h showed a decreasing trend. The addition of 100, 200, and 300 mg/L MK-7 to the culture broth led to a 12, 34, and 56% decrease in optical density at 600 nm (OD600) compared to the control, indicating that the addition of extracellular MK-7 had a certain inhibitory effect on the growth of B. subtilis natto. The intracellular and extracellular MK-7 showed an increasing trend (Figures 1C,D). However, the increase of intracellular and extracellular MK-7 content produced by B. subtilis natto was not significant because extracellular MK-7 included MK-7 added in the medium.
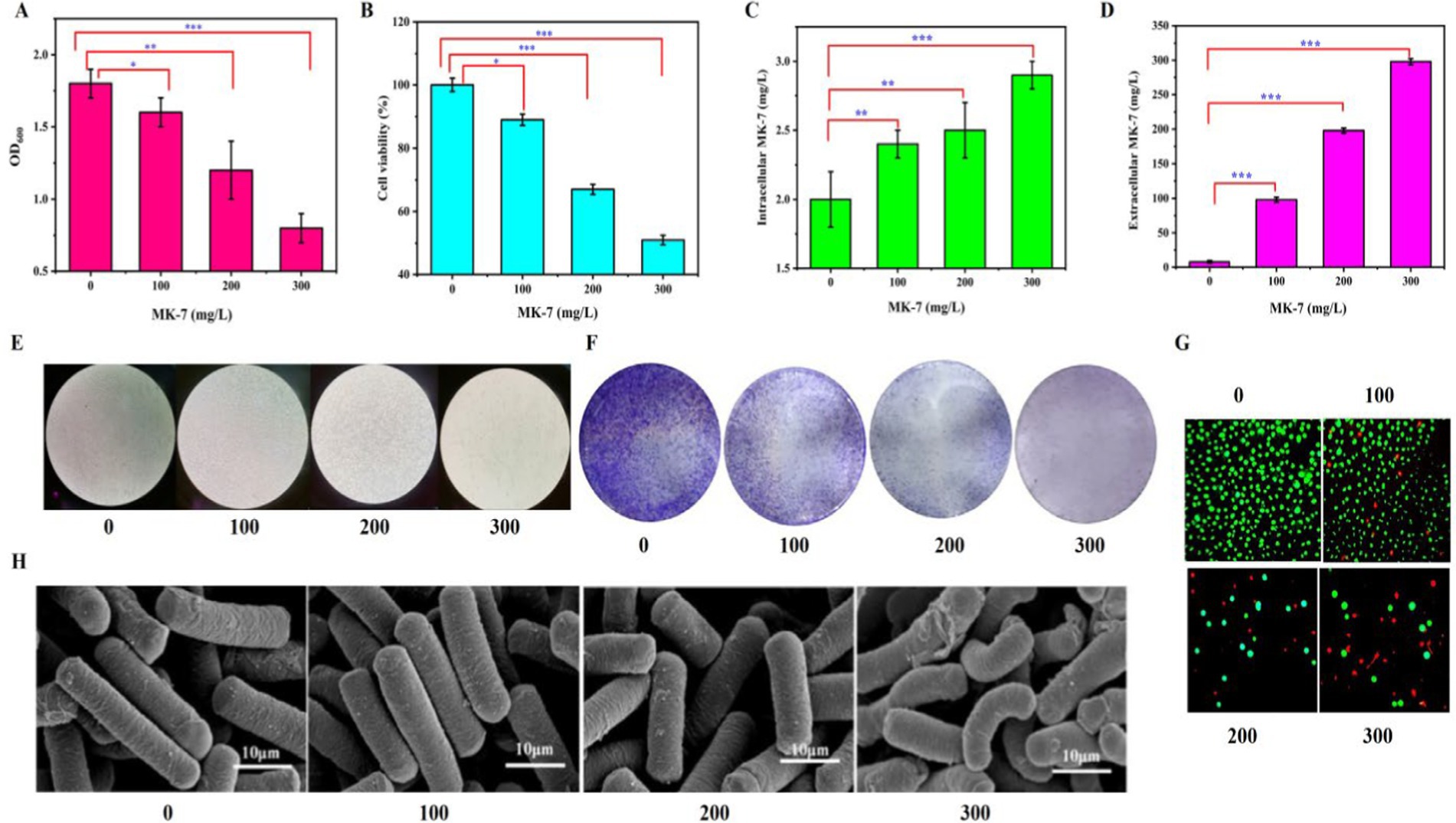
Figure 1. Effect of MK-7 on Bacillus subtilis natto. (A) Effect of MK-7 on B. subtilis natto biomass in 48 h of fermentation. (B) Effect of MK-7 on cell viability in 84 h of fermentation. (C) Effect of MK-7 on intracellular MK-7 production in 84 h of fermentation. (D) Effect of MK-7 on extracellular MK-7 production in 84 h of fermentation. Values and error bars represent the mean values and standard deviations of biological triplicates. (E,F) Ultrastructural changes to cell morphology under a regular optical microscope without (E) and with (F) crystalline violet dyeing in cells cultivated with 0, 100, 200, and 300 mg/L MK-7 in 84 h of fermentation. (G) Fluorescence micrographs of different concentrations of MK-7 on B. subtilis natto (green for living cells, red for apoptotic cells) in 84 h of fermentation. (H) Scanning electron microscopic images of B. subtilis cultivated with 0, 100, 200, and 300 mg/L MK-7 in 48 h of fermentation.
Representative phase contrast images of cells cultivated with 0, 100, 200, and 300 mg/L MK-7 were applied to measure the effect of the target metabolite on cell viability (Figure 1G). When MK-7 was not added to the medium, the cells exhibited abundant green fluorescence with no red fluorescence. After the addition of 100, 200, and 300 mg/L MK-7 to B. subtilis natto, the number of cells with green fluorescence reduced significantly, while the number of cells with red fluorescence increased. Furthermore, the total number of cells in the field of view was reduced as the concentration of MK-7 increased. The addition of 100, 200, and 300 mg/L MK-7 to the medium led to a 21, 33, and 49% decrease in cell viability compared to the control (Figure 1B), indicating that higher concentrations of MK-7 induced apoptosis in B. subtilis natto.
The control cells, which were not cultivated with MK-7, appeared smooth, rod-shaped, and exhibited intact surface morphology. More damaged cells with broken cell walls were observed when B. subtilis natto was cultivated with 300 mg/L MK-7 when detected by scanning electron microscopy (Figure 1H), indicating that MK-7 could induce cellular shortening and breakage in B. subtilis natto.
These results demonstrated that the addition of MK-7 caused damage to cell viability in B. subtilis natto. Because this fat-soluble vitamin is synthesized on the cell membrane, extracellular secretion is a crucial approach for its large-scale synthesis due to the intracellular space and feedback inhibition (Wu et al., 2021). However, from the above experimental results, it can be seen that extracellular secretion exceeding 100 mg/L will negatively impact cell replication and temporal lifespan. Therefore, it is necessary to adjust the lifespan of B. subtilis natto to further improve the synthetic ability of MK-7.
3.2 Combination of SPH and SP80 could prolong the lifespan and decrease the death rate of Bacillus subtilis natto
SP was chosen as the control to evaluate the impact of the SPH-based medium for MK-7 production by B. subtilis natto in submerged fermentation because it was the most commonly used nitrogen source for MK-7 production (Berenjian et al., 2011). Previous studies also found that the addition of SP could adjust the cell cycle (Yi et al., 2024), and it was speculated that SPH, as a hydrolysate of SP, also played a very important role in prolonging the cell cycle. We added SP and SPH separately to the culture medium to verify this speculation and examined their effects on the RLS and CLS. It was found that the optical density at 600 nm (OD600) increased by 33% in the SPH group compared to the SP group (Figure 2A), indicating that the addition of SPH was more effective in increasing bacterial biomass during the stable growth period. The death rate was 39.5% when SPH was added, which was 58.1% when SP was added after 84 h of fermentation (Figures 2B,C), suggesting the bacterial mortality rate also decreased when SPH was added. Moreover, the length of individual bacterial cells significantly increased after the addition of SPH through scanning electron microscopy observation (Figure 2D). The results above suggested that SPH prolonged the RLS of cells. By examining the changes in intracellular and extracellular MK-7 content after SPH addition, it was found that the intracellular MK-7 content increased 7.2-fold, while the extracellular content just increased 1.3-fold compared to the control (Figures 2E,F). The reason may be that the significant accumulation of MK-7 within the cell inhibited bacterial metabolism (Cui et al., 2020). Previous studies found that surfactants promoted the efflux of intracellular MK-7, ultimately leading to its extensive synthesis (Liu et al., 2014). Therefore, we investigated the effects of various surfactants on the synthesis of MK-7 and, ultimately, SP80, which could be widely used in the food industry and had the greatest promoting effect on MK-7 (see Supplementary Figure S1).
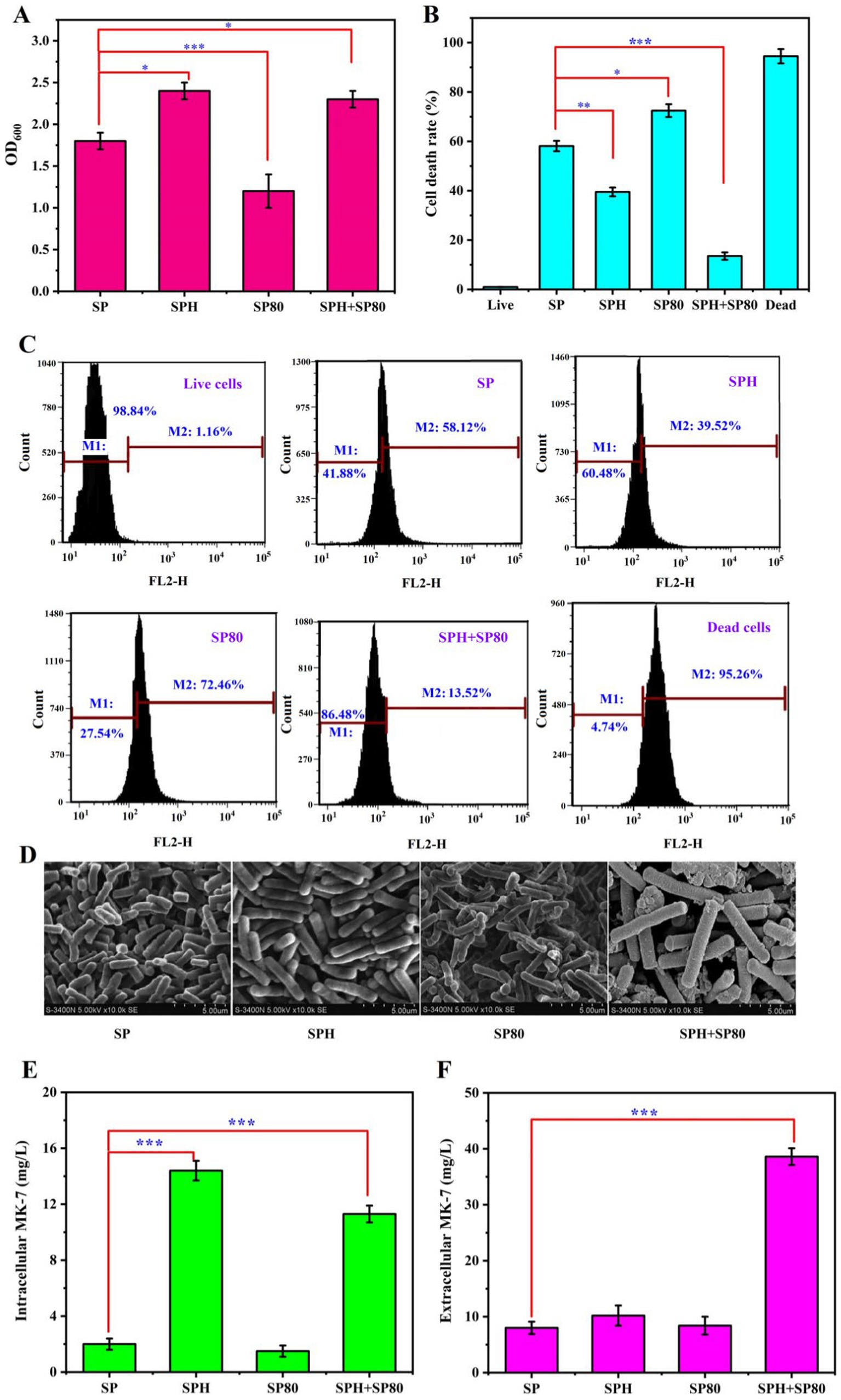
Figure 2. Effect of SP, SPH, SP80, and SPH+SP80 on B. subtilis natto. (A) Effect of SP, SPH, SP80, and SPH+SP80 on B. subtilis natto biomass in 48 h of fermentation. (B) Effect of SP, SPH, SP80, and SPH+SP80 on mortality rate in 84 h of fermentation. Live: negative control, which means cells were cultivated in basal fermentation medium and were not stained by PI; dead: positive control, which means cells cultivated in basal fermentation medium, destroyed by ultrasonic waves at a frequency of 20 kHz for 5 min, and stained by PI. (C) Survival of B. subtilis natto cultivated with SP, SPH, SP80, and SPH+SP80 in 84 h of fermentation, the number of apoptotic cells detected by PI-stained flow cytometry. PI is impermeable to live cells and is commonly used to detect dead cells in a population; relative cell viability is determined based on the fluorescence intensity of PI. (D) Scanning electron microscopic images of B. subtilis natto cultivated with SP, SPH, SP80, and SPH+SP80 in 48 h of fermentation. (E) Effect of SP, SPH, SP80, and SPH+SP80 on intracellular MK-7 production in 84 h of fermentation. (F) Effect of SP, SPH, SP80, and SPH+SP80 on extracellular MK-7 production in 84 h of fermentation. Values and error bars represent the mean values and standard deviations of biological triplicates.
By comparing the effects of SP80 and SPH+SP80 on biomass, survival rate, cell morphology, and MK-7 synthesis, it was found that the OD value of bacterial cells decreased by 34%, and bacterial length had no significant change compared to SP+SP80 (Figures 2A,D). These results indicated that SP80 itself was not conducive to bacterial RLS. However, in the SPH+SP80 group, the optical density at 600 nm (OD600) increased by 28% in 48 h of fermentation, and bacterial mortality decreased to 13.5% after 84 h of fermentation. Notably, the length of the B. subtilis natto increased significantly compared to the control (Figure 2D).
Intracellular MK-7, especially extracellular MK-7, had an obvious increase, which was 5.6-fold and 7.2-fold compared to the control (Figures 2E,F). The result was consistent with our previous study, which found that the addition of surfactant led to MK secretion into the extracellular space in Escherichia sp. (Liu et al., 2014). It was mentioned that the extracellular productivity was 0.46 mg/L·h−1 in this study, which was much higher than 0.33 mg/L·h−1 in that study. The reason may be that different surfactants had varying secretion abilities toward different bacteria. The total of MK-7 reached 52.9 mg/L, the highest among all studies except those involving the engineered B. subtilis 168 strain (Table 1). Due to the frequent loss of plasmids during the passage of genetically engineered strains in industrial production, non-genetically engineered strains were more favored. It was worth mentioning that the productivity of MK-7 reached 0.629 mg/L·h−1 when SPH and SP80 were added to the medium, which was the highest compared with other strategies to regulate MK-7 synthesis (Table 1). In other words, the production rate obtained under this strategy was 8.6% higher than that of the B. subtilis 168 engineering strain (Yang et al., 2019), indicating an increase in the MK-7 content obtained in a shorter period. Taken together, SPH+SP80 could greatly prolong B. subtilis natto lifespan and increase extracellular and total MK-7 content. Thus, clarifying the underlying mechanisms between nutrients and lifespan was important to achieve more stable and efficient MK-7 production in B. subtilis natto.
3.3 SPH+SP80 was more conducive for nutrient absorption in the culture medium and prolonged the Bacillus subtilis natto lifespan
The free amino acid composition, hydrolysis degree, surface tension, Mw distribution, and membrane integrity of SP, SP80, SPH, and SPH+SP80 were measured to analyze the mechanism for SPH+SP80 promoting the cell lifespan. Table 2 showed that the level of total amino acids in SP (265.52 g/kg) was significantly higher than that in SPH (65.89 g/kg). Especially, Phe, Arg, and Glu content, which were 23.47 ± 0.46 g/kg, 37.70 ± 4.50 g/kg, and 30.25 ± 1.17 g/kg in the SP group, could not be tested in the SPH group. As for Phe, it involved the negative feedback in MK-7 synthesis by inhibiting the.
3-deoxy-arabino-heptulonate 7-phosphate synthase (AroA) is responsible for the MK-7 precursor, naphthoquinone ring, synthesis (Yang et al., 2019; Figure 3A). The concentrations of both Arg and Glu could influence global regulators between carbon and nitrogen metabolism in Bacillus subtilis (Sonenshein, 2007), further affecting the synthesis of metabolites, including the MK-7. In addition, Arg also showed negative effects on MK-7 production. This corresponded to the results of Arg showing a potential antimicrobial role in the dissociation of the gingivalis biofilm (Li et al., 2020).
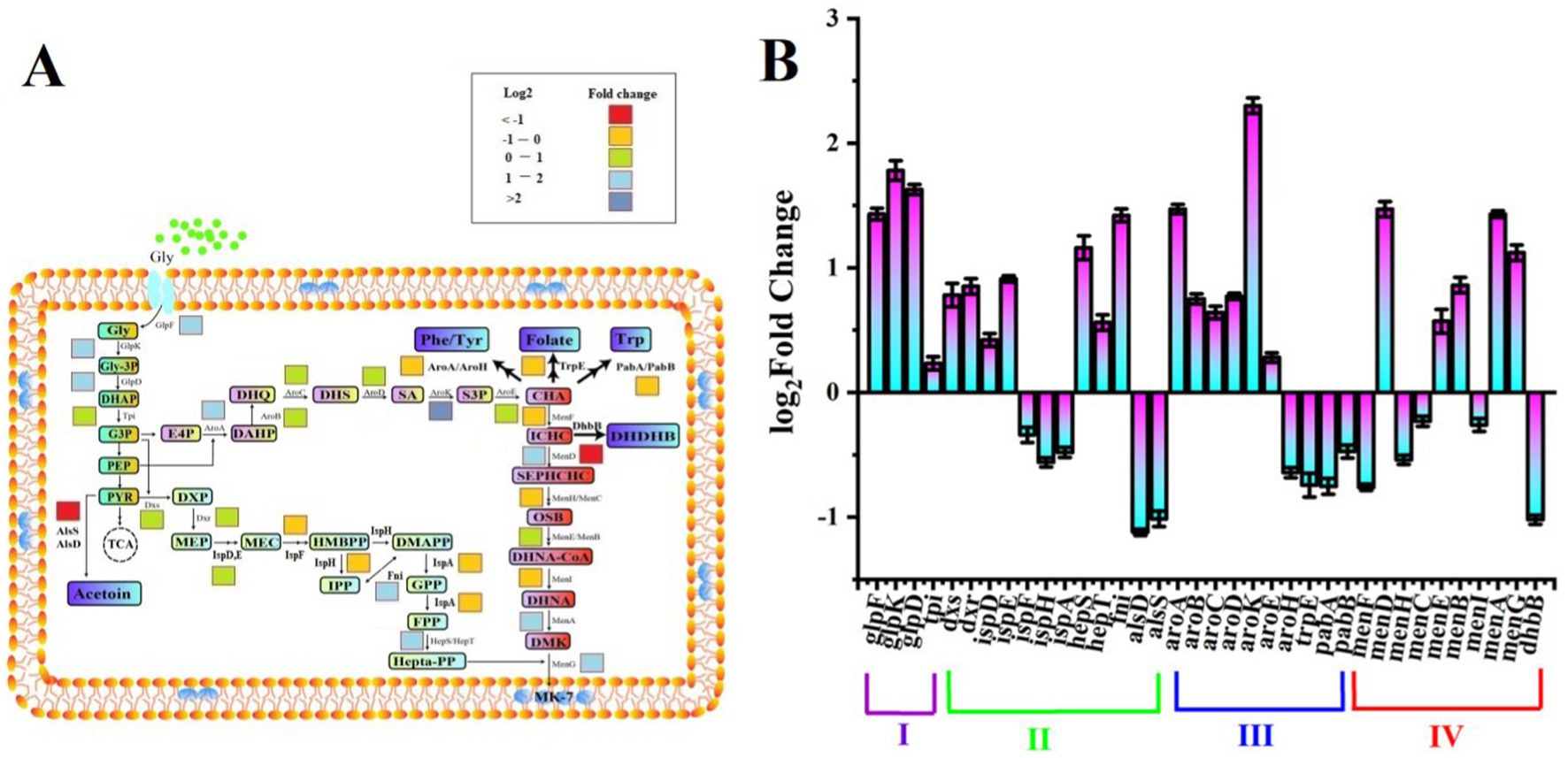
Figure 3. Changes in the gene expression level related to MK-7 biosynthesis between SPH+SP80 and SP groups. Biosynthetic pathway of MK-7 in Bacillus subtilis natto (A). The level of gene expression related to MK-7 biosynthesis of SPH+SP80 was higher than that of the SP group (B).
The hydrolysis degrees of SP, SP80, SPH, and SPH+SP80 were 15.2, 0.0, 22.3, and 22.1%, respectively (Figure 4A). More small peptides or amino acids produced by hydrolysis were conducive to fermentation (Yang et al., 2018). The surface tension of SP80 and SPH+SP80 medium decreased noticeably (34.56 mN/m, 32.78 mN/m) compared to SP and SPH medium (68.43 mN/m, 67.94 mN/m). Reduced surface tension was more conducive for nutrients to enter cells.
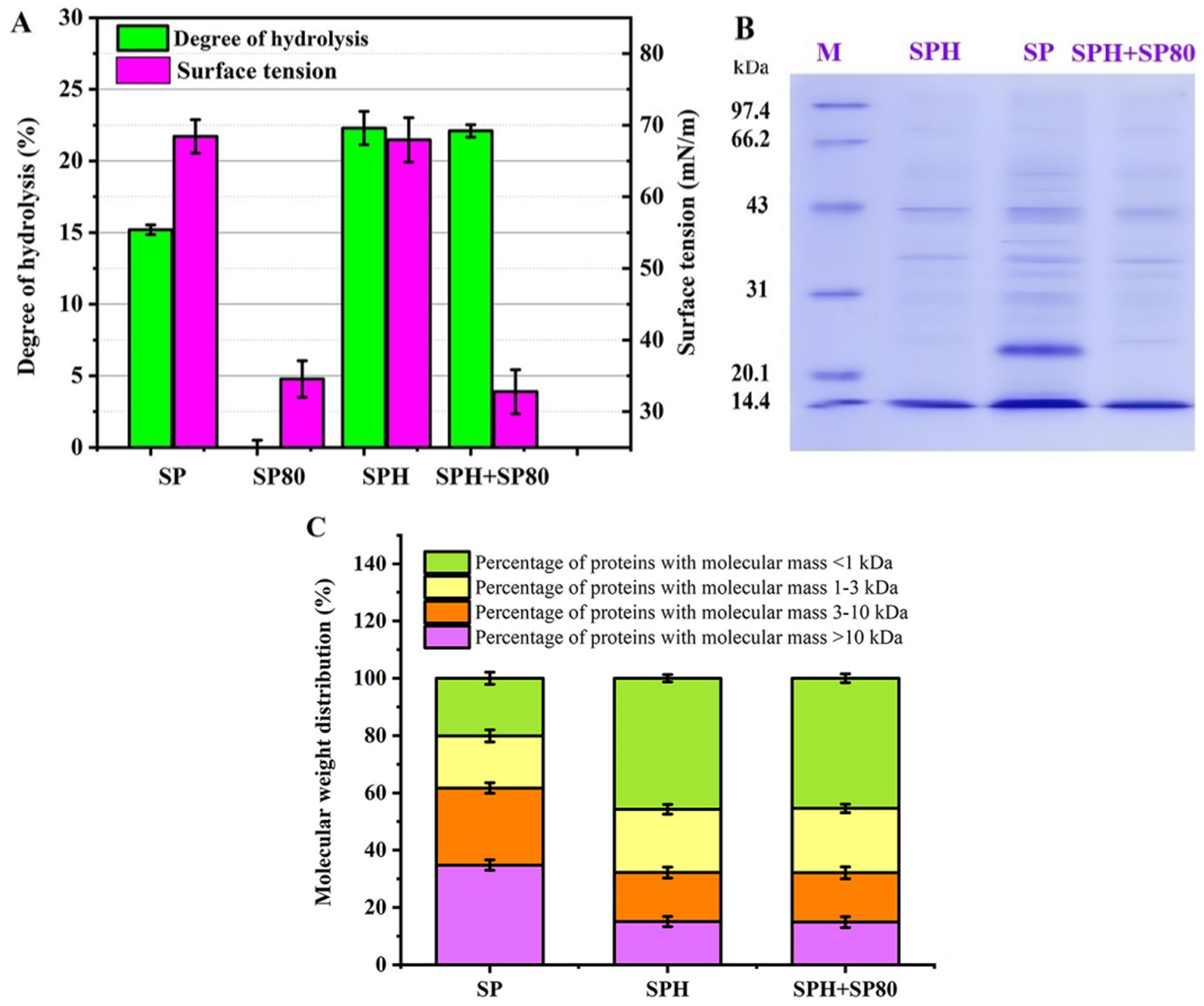
Figure 4. The degree of hydrolysis, surface tension, Mw distribution, and membrane permeability of SP, SPH, SP80, and SPH+SP80. (A) The degree of hydrolysis and surface tension for SP, SPH, SP80, and SPH+SP80. The SP, SPH, SP80, or SPH+SP80, and distilled water with a mass ratio of 1:15 were pretreated at 80°C for 10 min, and then protamex1.6 (1.5%, w/w) was added for 24 h of hydrolysis at pH 7.0, 50°C. The hydrolysates were heated to 100°C for 10 min to inactivate the enzyme and then centrifuged at 8000 g and 4°C for 10 min. The supernatant was obtained and tested. (B,C) The Mw distribution of SP, SPH, SP80, and SPH+SP80. SP, cells cultivated in basal fermentation medium supplemented with 30 g/L SP; SPH, cells cultivated in basal fermentation medium supplemented with 30 g/L SPH; SP80, cells cultivated in basal fermentation medium supplemented with 3.0 g/L SP80; SPH+SP80, cells cultivated in basal fermentation medium supplemented with 30 g/L SPH and 3.0 g/L SP80.
The Mw of SP, SP80, SPH, and SPH+SP80 groups was measured by SDS-PAGE and HPLC. More bands were visible in the SDS-PAGE gel from the SP group than from the other three groups (Figure 4B), meaning SP was composed of more high-Mw proteins (Mw > 10 kDa), which was further confirmed in Figure 4C (a 34.8% proportion of higher Mw proteins). As for SPH and SPH+SP80, the proportion of small peptides (Mw < 1 kDa) in SPH (45.7%) and SPH+SP80 (45.4%) were prominently higher than those in SP (20.1%) in the medium (Figure 4C). Because the small peptides (Mw < 1 kDa) could be directly transported into cells through a specific peptide transport system without competing with amino acids, promoting growth and stress resistance to the external environment (Jin et al., 2022), it was more efficient for cell growth and prolonging cell lifespan.
Taken together, SPH+SP80 performed better on the cell lifespan since SPH produced 2.26-fold small peptides (Mw < 1 kDa), which could be directly transported into cells through a specific peptide transport system. SP80 addition reduced surface tension and was more conducive to nutrients entering cells. Furthermore, the content of adverse amino acid components in SPH, such as Phe, Arg, and Glu, for the synthesis of MK-7 was reduced (Table 2). Further investigation was needed to elucidate the underlying mechanisms behind this phenomenon.
3.4 SPH+SP80 regulated the lifespan of Bacillus subtilis natto
To explore why SPH+SP80 regulated lifespan and MK-7 production in B. subtilis natto, the gene expression levels in related metabolic pathways affected by SPH and SP80 were detected by the Illumina RNAseq. The results showed significant changes in expression levels in 1269 identical genes (Figure 5A). The functional differences were mainly in “metabolism,” “cellular processes,” “cellular microenvironment,” and “environmental information processed” (Figure 5B). The top 50 genes with significant changes that belonged to these pathways included growth lifespan, ubiquinone and another terpenoid-quinone biosynthesis, oxidative phosphorylation, bacterial secretion system, microbial metabolism in diverse environments, transporter activity, and nitrogen metabolism (Figures 5B,C).
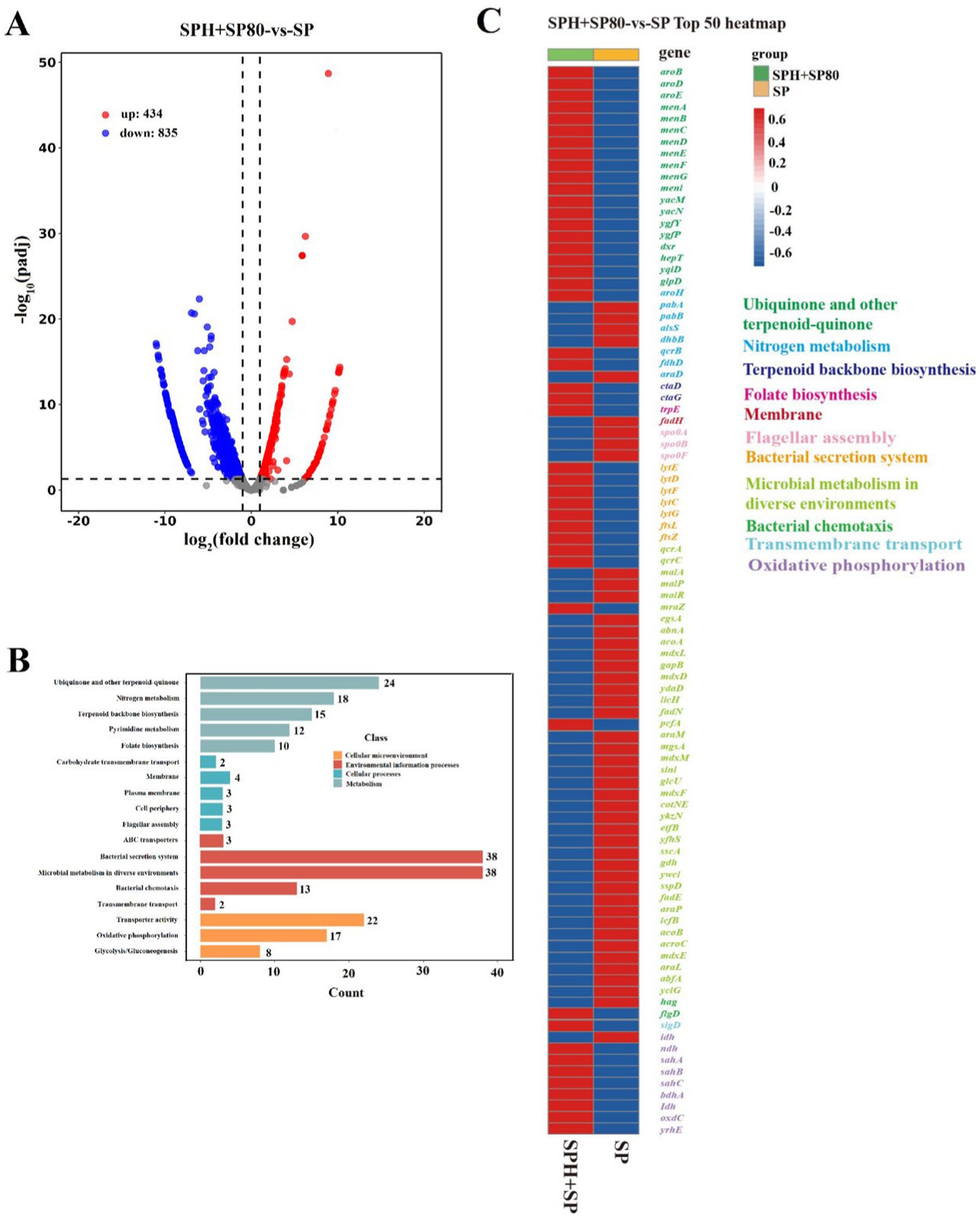
Figure 5. Summary of draft reads of samples by Illumina deep sequencing. (A) Visualization of differential genes between SP and SPH+SP80 samples by volcano plots. (B) Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis of functions of differential genes. (C) Top 50 of upregulated and downregulated genes of SP and SPH+SP80 samples. All experiments were independently conducted at least three times, and the results were expressed as mean ± standard deviation (SD).
The different expressions of lifespan-related genes were studied to explore how SPH+SP80 regulated external morphology in B. subtilis natto. The first category of CLS-related genes included those involved in cannibalism (i.e., spo0A, skfA, skfE, skfF, and sdp). When spo0A was not expressed, the cells would be killed, and their contents would provide nutrients that feed their siblings and delay the sporulation of the entire bacterial population (Wang et al., 2014). The skf and sdp operons were regulated by spo0A. skfA, the first gene of the skf operon, encodes a small peptide that kills siblings. The product encoded by skfE and skfF was used to export this peptide. The sdp operon, which produced a signaling protein, was used to enhance the killing effect. It could be seen from Figure 6 that the expression levels of growth-related autolysis genes spo0A, skfA, skfE, skfF, and sdp were downregulated 1.31-, 2.08-, 3.23-, 4.89-, and 5.19-fold, respectively, indicating that SPH+SP80 decreased cannibalism and thus prolonged CLS.
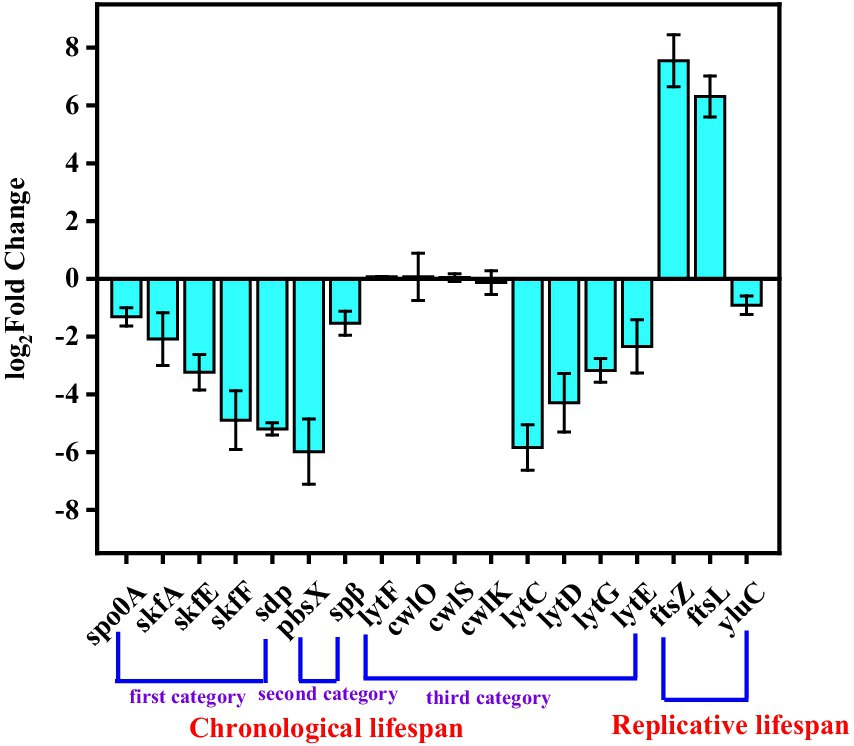
Figure 6. Expression level of genes related to the lifespan of cells between the SP and SPH+SP80 groups. All experiments were independently carried out at least three times, and the results were expressed as mean ± standard deviation (SD).
The second category of CLS-related genes included prophages that could induce cell lysis and thus influence the lifespan of B. subtilis natto. Although the B. subtilis genome harbored at least ten putative prophages, only pbsx and spβ were lysogenic (Kunst et al., 2007). Generally, the prophage PBSX was induced to express only when treated with factors that caused survival-oriented behavior responses. Spβ was difficult to induce the gene expression in wild-type B. subtilis natto because the c gene encoded a strong repressor of spβ induction. It could be seen from Figure 6 that the expression levels of pbsx and spβ were downregulated 5.96- and 1.53-fold, respectively. Thus, SPH+SP80 eliminated the survival pressure of bacterial cells and then decreased pbsx expression.
The third category of CLS-related genes encoded peptidoglycan hydrolases that could also mediate cell lysis. Peptidoglycan hydrolases were encoded by the vegetative cell wall hydrolases lytC, lytD, lytE, lytF, lytG, cwlO, cwlS, and cwlK. It can be seen from Figure 6 that the expression levels of lytF, cwlO, cwlS, and cwlK had no obvious change. However, the expression levels of lytC, lytD, lytG, and lytE were downregulated 5.84-, 4.29-, 3.31-, and 2.51-fold, respectively. The reason may be that lytC, lytD, and lytG possess amidase or N-acetylglucosaminidase activities, respectively (Horsburgh et al., 2003). Meanwhile, lytE encoded an endopeptidase, which degrades isolated cell wall preparations.
The cell division proteins, FtsZ and FtsL, were essential cytokines associated with Z-loop maturation, facilitated the division process, and finally determined the RLS of bacteria. The zinc metalloprotease YluC was required to turn over genes mraZ and ftsL (Hashimoto et al., 2012). In our study, the expression levels of ftsZ and ftsL were upregulated by 7.52-, 6.31-fold, and the expression level of yluC was downregulated by 0.91-fold (Figure 6). Additionally, previous studies discovered that deletion of ftsL resulted in the decondensation of the FtsZ loop (Z-loop), preventing cells from producing more daughters (Kodama et al., 2007), and deletion of yluC prolonged the length of E. coli by approximately 33% compared to the WT (Bisicchia et al., 2007), which also verified the important role of FtsZ, FtsL, and YluC in regulating RLS of Bacillus subtilis natto.
Taken together, the expression level of genes that encode prophages, peptidoglycan hydrolases, the bacterial dcw cluster, and those playing roles in cannibalism during sporulation changed noticeably in the SPH+SP80 group. It not only promoted rapid cell growth, accelerated cell division, and partially delayed cell senescence of B. subtilis natto but also altered cellular morphology.
3.5 SPH+SP80 regulated the MEC, membrane potential, and electron generation system of Bacillus subtilis natto
The MEC was a physiological parameter that reflected the energy required to maintain intracellular homeostasis. In cells, a high MEC testified to a low energy utilization efficiency. The MEC of four strains selected from the SP, SPH, SP80, and SPH+SP80 groups was tested, and strains in the SPH+SP80 group showed the lowest MEC (0.33 mmol·g (cdw)−1 h−1) compared to the strain of the SP group, while the SP group strain had the highest MEC of 0.39 mmol·g (cdw)−1 h−1 (Table 3). Therefore, the strains of SPH+SP80 had a higher energy utilization efficiency due to their low maintenance energy metabolism. Additionally, the OD600 of SP, SPH, SP80, and SPH+SP80 groups also changed, with strains in the SPH+SP80 group exhibiting a rise in OD600 by 54.1%.
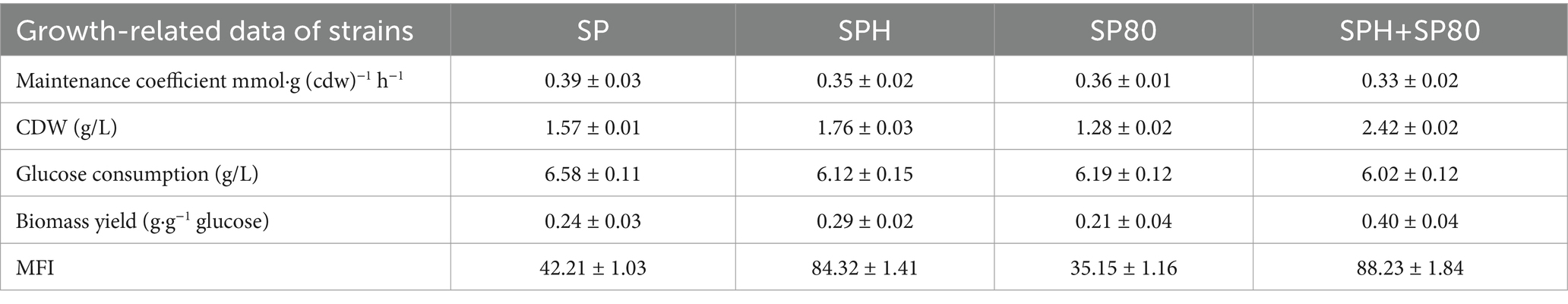
Table 3. Comparison of growth-related data of strains from SP, SPH, SP80, SPH+SP80 group under aerobic conditions.
MK-7 and cytochrome c are important components of the cell membrane and play key roles in electron transfer (Figure 7A). To investigate the effects of SP, SPH, SP80, and SPH+SP80 groups on electron transfer in the cell membrane, the membrane potential of the strain cultured for 6 days was detected. The mean fluorescence intensity (MFI) of the SPH+SP80 group (88.23 ± 1.84) increased by 2.09 times compared to the SP group (42.21 ± 1.03) (Table 3), indicating the SPH+SP80 group had electrical hyperpolarization. To explore which process causes electrical hyperpolarization, the expression level of a gene involved in encoding oxalate-decarboxylase (OxdC, catalyzing the conversion of oxalate to form electrons) was detected. Unexpectedly, the expression level of oxdC in the SPH+SP80 group was reduced by 65% compared to the SP group. Furthermore, the expression levels of yrhE and fdhD, which encode formate dehydratase (oxidizing formate to form electrons), were downregulated by 64 and 94% (Figure 7B), indicating that electrical hyperpolarization of the SPH+SP80 group was not due to these two processes. Therefore, we explored the other processes that could donate large amounts of electrons.
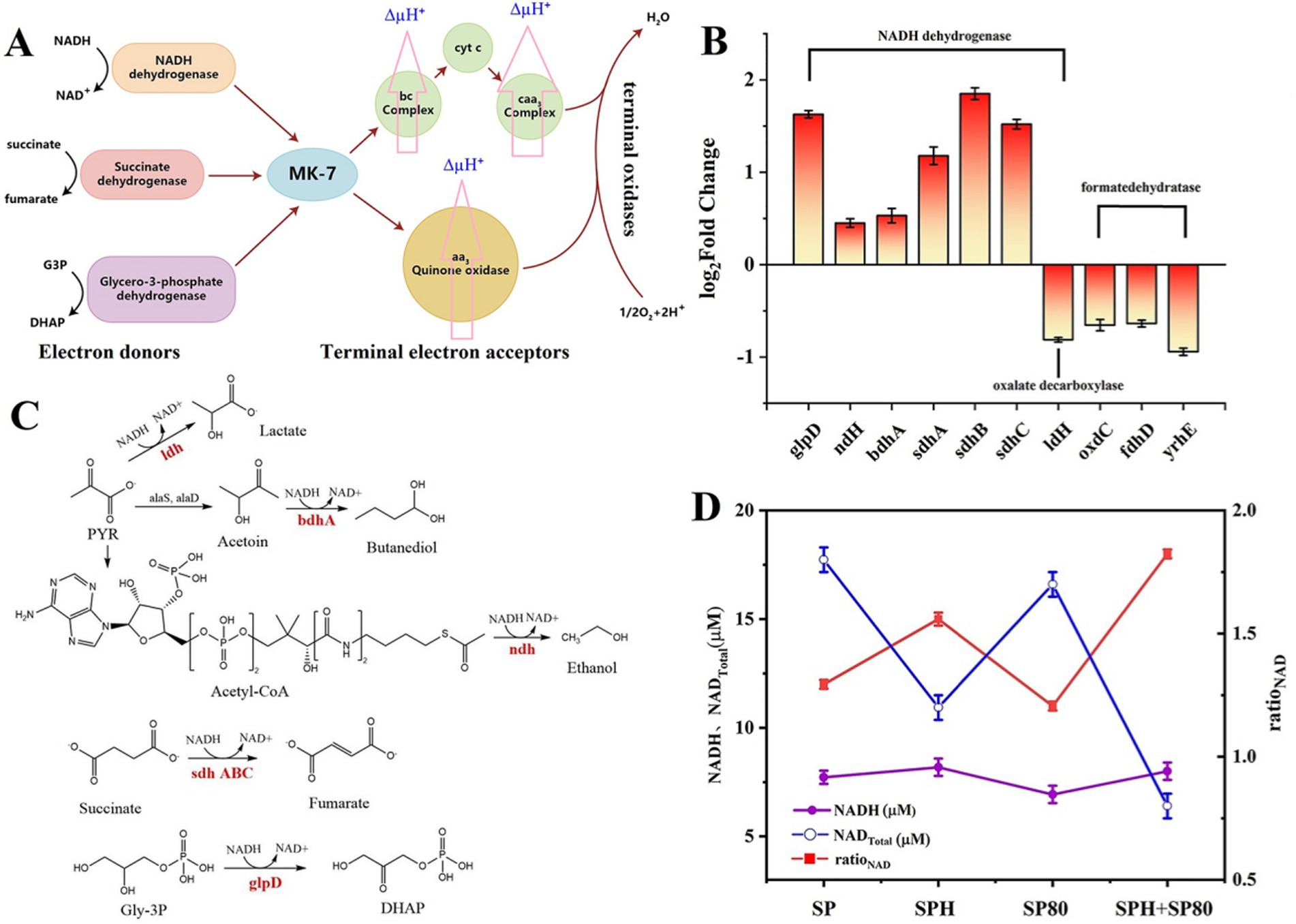
Figure 7. Schematic diagram of the electron transfer chain in B. subtilis natto. Electrons are extracted under the action of NADH dehydrogenase, succinate dehydrogenase, and glycerol 3 3-phosphate dehydrogenase. MK-7 and cytochrome c (Cyt c) act as electron transport carriers, and finally, electrons are delivered to oxygen to form water (A). Changes in the membrane potential were evaluated by total NAD content, NADH content yield, and NADH/NAD+ ratio in SP, SPH, SP80, and SPH+SP80 groups (B). Catalytic reaction of NADH reduction in cells, including l-lactate dehydrogenase (ldH), (R, R)-butanediol dehydrogenase (bdhA), NADH dehydrogenase (ndH), aerobic glycerol-3-phosphate dehydrogenase (glpD), and succinate dehydrogenase flavoprotein (sdhABC) (C). The expression level of NADH dehydrogenase of SPH+SP80 is compared with the SP group; positive numbers mean upregulation, and negative numbers mean downregulation (D). All experiments were independently carried out at least three times, and the results were expressed as mean±standard deviation (SD). G3P, glyceraldehyde-3-phosphate. DHAP, dihydroxyacetonephosphate.
The expression levels of NADH dehydrogenases that catalyze NADH to form electrons were detected. The process of these reactions was presented in Figure 7C. Most NADH dehydrogenases were upregulated, especially glpD, sdhA, sdhB, and sdhC, which had a growth rate of 1.62-, 1.17-, 1.85-, and 1.52-fold, respectively (Figure 7B). The level of NADH, NAD+ total (NAD+ and NADH), and the ratio of NADH/NAD+ were also detected. The level of NAD+ total and NADH increased by 3.6 and 50% in the SPH+SP80 group compared to the SP group, respectively. However, the ratio of NADH/NAD+ reduced to 44% (Equations 1–3) (Figure 7D). The ratio of NADH/NAD+ has been proven to be inversely proportional to MK-7 content in B. subtilis (Wang et al., 2019).
3.6 SPH+SP80 affected the pathway related to the biosynthesis of MK-7
Four modules named glycerol metabolism pathway (Module I), methylerythritol phosphate (MEP) pathway (Module II), shikimate (SA) pathway (Module III), and MK-7 pathway (Module IV) were involved in the biosynthesis pathway of MK-7 in B. subtilis natto (Figure 3A; Yang et al., 2019). The different expression changes of genes involved in four modules were detected (Supplementary Table S1). In Module I, three enzymes, such as glycerol kinase (GlpK), Gly-3P dehydrogenase (GlpD), and glycerol facilitator (GlpF), played crucial roles in the SPH+SP80 group because the expression of glpK, glpD, and glpF was upregulated 1.78-, 1.62-, and 1.43-fold, respectively.
The change of most of the enzymes in Module II was not significant in the SPH+SP80 group (Figures 3A,B). Except for membrane proteins like Fni and HepS, which catalyzed a reversible reaction of isopentenyl diphosphate (IPP) to form dimethylallyl diphosphate (DMAPP), the reaction of farnesyl diphosphate (FPP) to heptaprenyl diphosphate (HDP) was upregulated 1.42-fold, and 1.16-fold, respectively.
As for Module III, the expression of aroA, aroB, aroC, aroD, aroK, and aroE was upregulated 1.47-, 0.75-, 0.64-, 0.77-, 2.30-, and 0.28-fold, respectively (Figure 3B), which verified a previous study showing that AroA and AroK played a crucial role in the SA pathway (Cui et al., 2019). The genes encoding PabA, PabB, AroH, and TrpE, which were used to synthesize aromatic amino acids such as tryptophan (Trp), tyrosine (Tyr), and phenylalanine (Phe) from chorismate, were downregulated by 36, 26, 25, and 53%, respectively.
Nine enzymes (MenA, B, C, D, E, F, G, H, and I) were involved in the synthesis of 1,4-dihydroxy-2-naphthoyl-CoA (DHNA-CoA) in Module IV. Interestingly, only genes encoding membrane proteins (menA, menB, menD, menE, and menG) were upregulated, especially menA, menG, and menD, whose expression levels were highly regulated by 1.43-, 1.12-, and 1.47-fold, respectively, in the SPH+SP80. These results provide clues for further modification of MK-7 biosynthesis at the transcriptional level.
Taken together, an active electron transfer system stimulated the expression of related genes on the cell membrane, such as ispE, hepS, aroA, aroK, menA, menD, and menG, which were key enzyme-encoding genes for synthesizing MK-7, and finally promoted the synthesis of MK-7.
4 Conclusion
In the fermentation process, B. subtilis natto cultivated with different nutrients exhibited significant differences in lifespan and MK-7 production. The value of OD600 increased by 28%, and the mortality rate decreased to 13.5% in SPH+SP80, while the death rate was 58.1% when SP was added after 84 h of fermentation. Intracellular MK-7, especially extracellular MK-7, showed a noticeable increase of 5.6-fold and 7.2-fold compared to the control (Figures 2E,F). The total MK-7 content in SPH+SP80 reached 52.9 mg/L, which was 5.3-fold compared to that in SP. The biomass and cell size were markedly enhanced by SPH+SP80, which were associated with the enhancement of cell viability, downregulation of CLS-related genes (spo0A, skfA, skfE, skfF, sdp, pbsx, spβ, lytC, lytD, lytG, and lytE), and the upregulation of the RLS-related genes (ftsZ and ftsL). Low Mw (<1 kDa) components and moderate amino acids, particularly Phe, Arg, and Glu in SPH+SP80, might be the predominant factors contributing to these differences. Moreover, the prolongation of the lifespan induced the MK-7 biosynthesis by upregulating the key genes on the cell membrane, such as ispE, hepS, aroA, aroK, menA, menD, and menG, related to MK-7 synthesis, therefore enhancing MK-7 precursor, isoprene side chain supply, and MK-7 assembly efficiency in B. subtilis natto. Briefly, SPH+SP80 reduced surface tension, enhanced nutrient absorption, and finally extended the lifespan of B. subtilis natto. The prolongation of the lifespan induced the MK-7 biosynthesis by upregulating the key genes on the cell membrane. For the majority of industrial microorganisms, it was difficult to produce genetically engineered bacteria due to the lack of suitable gene editing tools.
The addition of SPH and SP80 provided a potential strategy for controlling the physiological state and metabolic synthesis capabilities of industrial strains.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
Author contributions
L-xH: Conceptualization, Resources, Software, Writing – original draft. YC: Data curation, Methodology, Validation, Writing – review & editing. H-pZ: Investigation, Resources, Supervision, Writing – review & editing. WT: Data curation, Investigation, Methodology, Validation, Writing – review & editing. X-lG: Methodology, Software, Writing – original draft. C-cW: Data curation, Investigation, Methodology, Writing – review & editing. H-mZ: Formal analysis, Visualization, Writing – review & editing. R-mH: Investigation, Validation, Writing – review & editing. Y-qL: Methodology, Software, Writing – review & editing. YL: Conceptualization, Project administration, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. The study was supported by the National Natural Science Foundation of China (No. 32372295), the Outstanding Youth Research Project in Anhui Province Universities (No. 2023AH020013), and Anhui Provincial Undergraduate Innovation and Entrepreneurship Program (No. 202310363254).
Conflict of interest
L-xH was employed by Anhui Zhang Hengchun Pharmaceutical Co., Ltd. YC, C-cW, H-mZ, R-mH, and YL were employed by Wuhu Green Food Industry Research Institute Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1578160/full#supplementary-material
References
Berenjian, A., Mahanama, R., Talbot, A., Biffin, R., Regtop, H., Valtchev, P., et al. (2011). Efficient media for high menaquinone-7 production: response surface methodology approach. Nat. Biotechnol. 28, 665–672. doi: 10.1016/j.nbt.2011.07.007
Bisicchia, P., Noone, D., Lioliou, E., Howell, A., Quigley, S., Jarmer, T. J. H., et al. (2007). The essential YycFG two-component system controls cell wall metabolism in Bacillus subtilis. Mol. Microbiol. 65, 180–200. doi: 10.1111/j.1365-2958.2007.05782.x
Bøe, A. C., and Holo, H. (2020). Engineering Lactococcus lactis for increased bitamin K2 production. Front. Bioeng. Biotech. 8, 1–4. doi: 10.3389/fbioe.2020.00191
Chatake, T., Yanagisawa, Y., Inoue, R., Sugiyama, M., Matsuo, T., Fujiwara, S., et al. (2018). Purification and structural characterization of water-soluble menaquinone-7 produced by Bacillus subtilis natto. J. Food Biochem. 42:e12630. doi: 10.1111/jfbc.12630
Cui, S., Lv, X., Wu, Y., Li, J., Du, G., Rodrigo, L. A., et al. (2019). Engineering a bifunctional Phr60-Rap60-Spo0A quorum-sensing molecular switch for dynamic fine-tuning of menaquinone-7 synthesis in Bacillus subtilis. ACS Synth. Biol. 8, 1826–1837. doi: 10.1021/acssynbio.9b00140
Cui, S., Xia, H., Chen, T., Gu, Y., Lv, X., Liu, Y., et al. (2020). Cell membrane and electron transfer engineering for improved synthesis of menaquinone-7 in Bacillus subtilis. iScience 23:100918. doi: 10.1016/j.isci.2020.100918
Gao, Q., Chen, H., Wang, G., Yang, W., Zhong, X., Liu, J., et al. (2021). Highly efficient production of menaquinone-7 from glucose by metabolically engineered Escherichia coli. ACS Synth. Biol. 10, 756–765. doi: 10.1021/acssynbio.0c00568
Goodman, S. R., Marrs, B. L., Narconis, R. J., and Olson, R. E. (1976). Isolation and description of a menaquinone mutant from Bacillus licheniformis. J. Bacteriol. 125, 282–289. doi: 10.1128/jb.125.1.282-289.1976
Hashimoto, M., Ooiwa, S., and Sekiguchi, J. (2012). Synthetic lethality of the lytE cwlO genotype in Bacillus subtilis is caused by lack of D, L-endopeptidase activity at the lateral cell wall. J. Bacteriol. 194, 796–803. doi: 10.1128/jb.05569-11
Horsburgh, G. J., Atrih, A., Williamson, M. P., and Foster, S. J. (2003). Lyt G of Bacillus subtilis is a novel peptidoglycan hydrolase: the major active glucosaminidase. Biochemistry 42, 257–264. doi: 10.1021/bi020498c
Jeong, K. J., and Lee, S. Y. (2003). Enhanced production of recombinant proteins in Escherichiacoli by filamentation suppression. Appl. Environ. Microbiol. 69, 1295–1298. doi: 10.1128/AEM.69.2.1295-1298.2003
Jiang, X. R., Wang, H., Shen, R., and Chen, G. Q. (2015). Engineering the bacterial shapes for enhanced inclusion bodies accumulation. Metab. Eng. 29, 227–237. doi: 10.1016/j.ymben.2015.03.017
Jin, X., Yang, H., Coldea, T. E., Andersen, M. L., and Zhao, H. (2022). Wheat gluten peptides enhance ethanol stress tolerance by regulating the membrane lipid composition in yeast. J. Agric. Food Chem. 70, 5057–5065. doi: 10.1021/acs.jafc.2c00236
Kodama, T., Endo, K., Ara, K., Ozaki, K., Kakeshita, H., Yamane, K., et al. (2007). Effect of Bacillus subtilis spo0A mutation on cell wall lytic enzymes and extracellular proteases, and prevention of cell lysis. J. Biosci. Bioeng. 103, 13–21. doi: 10.1263/jbb.103.13
Kunst, F., Ogasawara, N., Moszer, I., Albertini, A. M., Alloni, G. O., Azevedo, V., et al. (2007). The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature 390, 249–256. doi: 10.1038/36786
Li, Y. Y., Li, B. S., Liu, W. W., Cai, Q., Wang, H. Y., Liu, Y. Q., et al. (2020). Effects of D-arginine on Porphyromonas gingivalis biofilm. J. Oral Sci. 62, 57–61. doi: 10.2334/josnusd.19-0075
Li, C., Wang, H., Ma, G., Wang, L., Zheng, Z., Zhao, G., et al. (2022). Enhancement of vitamin K2 efflux in Bacillus subtilis natto via a potential protein receptor for increased yield. J. Food Qual. 2022, 1–9. doi: 10.1155/2022/8407829
Lindner, A. B., Madden, R., Demarez, A., Stewart, E., and Taddei, F. (2008). Asymmetric segregation of protein aggregates is associated with cellular aging and rejuvenation. Proc. Natl. Acad. Sci. USA 105, 3076–3081. doi: 10.1073/pnas.0708931105
Liu, Y., Ding, X. M., Xue, Z. L., Hu, L. X., Cheng, Q., Chen, M. H., et al. (2017a). Site-directed mutagenesis of Ubi a to promote menaquinone biosynthesis in Elizabethkingia meningoseptica. Process Biochem. 58, 186–192. doi: 10.1016/j.procbio.2017.05.002
Liu, Y., Ding, X. M., Xue, Z. L., Hu, L. X., Zhang, N. J., Wang, Z., et al. (2017b). The change of the state of cell membrane can enhance the synthesis of menaquinone in Escherichia coli. World J. Microbiol. Biotechnol. 33:52. doi: 10.1007/s11274-017-2222-9
Liu, S. X., Li, S., Shen, G. M., Sukumar, N., Krezel, A. M., and Li, W. K. (2021). Structural basis of antagonizing the vitamin K catalytic cycle for anticoagulation. Science 371:652401. doi: 10.1126/science.abc5667
Liu, Y., Zheng, Z. M., Qiu, H. W., Zhao, G. H., Liu, H., Li, Z. M., et al. (2014). Surfactant supplementation to enhance the production of vitamin K2 metabolites in shake flask cultures using Escherichia Sp. mutant FM3-1709. Food Technol Biotech 52, 269–275.
Ma, X. T., Li, M. B., Xu, X. F., and Sun, C. (2023). On the role of surface charge and surface tension tuned by surfactant in stabilizing bulk nanobubbles. Appl. Surf. Sci. 608:155232. doi: 10.1016/j.apsusc.2022.155232
Ma, Y. W., McClure, D. D., Somerville, M. V., Proschogo, N. W., Dehghani, F., Kavanagh, J. M., et al. (2019). Metabolic engineering of the MEP pathway in Bacillus subtilis for increased biosynthesis of menaquinone-7. ACS Synth. Biol. 8, 1620–1630. doi: 10.1021/acssynbio.9b00077
Mishima, E., Ito, J., Wu, Z. J., Nakamura, T., Wahida, A., Doll, S., et al. (2022). A non-canonical vitamin K cycle is a potentferroptosis suppressor. Nature 608, 778–783. doi: 10.1038/s41586-022-05022-3
Nielsen, P., Petersen, D., and Dambmann, C. (2001). Improved method for determining food protein degree of hydrolysis. J. Food Sci. 66, 642–646. doi: 10.1111/j.1365-2621.2001.tb04614.x
Novo, D. J., Perlmutter, N. G., Hunt, R. H., and Shapiro, H. M. (2000). Multiparameter fow cytometric analysis of antibiotic effects on membrane potential, membrane permeability, and bacterial counts of Staphylococcus aureus and Micrococcus luteus. Antimicrob. Agents Chemother. 44, 827–834. doi: 10.1128/aac.44.4.827-834.2000
Puri, A., Iqubal, M., Zafar, R., and Panda, B. P. (2015). Influence of physical, chemical and inducer treatments on menaquinone-7 biosynthesis by Bacillus subtilis MTCC 2756. Songklanakarin J. Sci. Technol. 37, 283–289.
Ren, K. X., Wang, Q., Chen, J. H., Zhang, H. W., Guo, Z. L., Xu, M. J., et al. (2024). Design-build-test of recombinant Bacillus subtilis chassis cell by lifespan engineering for robust bioprocesses. Synthetic Syst. Biotechnol. 9, 470–480. doi: 10.1016/j.synbio.2024.04.004
Sonenshein, A. L. (2007). Control of key metabolic intersections in Bacillus subtilis. Nat. Rev. Microbiol. 5, 917–927. doi: 10.1038/nrmicro1772
Song, J. Y., Liu, H. X., Wang, L., Dai, J., Liu, Y., Liu, H., et al. (2014). Enhanced production of vitamin K2 from Bacillus subtilis (natto) by mutation and optimization of the fermentation medium. Braz. Arch. Biol. Technol. 57, 606–612. doi: 10.1590/S1516-8913201402126
Tsukamoto, Y., Kasai, M., and Kakuda, H. (2001). Construction of a Bacillus subtilis (natto) with high productivity of vitamin K2 (menaquinone-7) by analog resistance. Biosci. Biotechnol. Biochem. 65, 2007–2015. doi: 10.1271/bbb.65.2007
Wang, Y., Chen, Z., Zhao, R., Jin, T., Zhang, X., and Chen, X. (2014). Deleting multiple lytic genes enhances biomass yield and production of recombinant proteins by Bacillus subtilis. Microb. Cell Factories 13:129. doi: 10.1186/s12934-014-0129-9
Wang, H., Liu, H., Wang, L., Zhao, G., Tang, H., Sun, X., et al. (2019). Improvement of menaquinone-7 production by Bacillus subtilis natto in a novel residue-free medium by increasing the redox potential. Appl. Microbiol. Biotechnol. 103, 7519–7535. doi: 10.1007/s00253-019-10044-5
Wu, J., Li, W., Zhao, S. G., Qian, S. H., Wang, Z., Zhou, M. J., et al. (2021). Site-directed mutagenesis of the quorum sensing transcriptional regulator SinR affects the biosynthesis of menaquinone in Bacillus subtilis. Microb. Cell Factories 20, 1–19. doi: 10.1186/s12934-021-01603-5
Xu, J. Z., Yan, W. L., and Zhang, W. G. (2017). Enhancing menaquinone-7 production in recombinant Bacillus amyloliquefaciens by metabolic pathway engineering. RSC Adv. 7, 28527–28534. doi: 10.1039/C7RA03388E
Yang, S., Cao, Y., Sun, L., Li, C., Lin, X., Cai, Z., et al. (2019). Modular pathway engineering of Bacillus subtilis to promote de novo biosynthesis of menaquinone-7. ACS Synth. Biol. 8, 70–81. doi: 10.1021/acssynbio.8b00258
Yang, H., Zong, X., Xu, Y., Zeng, Y., and Zhao, H. (2018). Improvement of multiple-stress tolerance and ethanol production in yeast during very-high-gravity fermentation by supplementation of wheat-gluten hydrolysates and their ultrafiltration fractions. J. Agric. Food Chem. 66, 10233–10241. doi: 10.1021/acs.jafc.8b04196
Yi, Y. X., Chen, M. T., Coldea, T. E., Yang, H. R., and Zhao, H. F. (2024). Soy protein hydrolysates induce menaquinone-7 biosynthesis by enhancing the biofilm formation of Bacillus subtilis natto. Food Microbiol. 124:104599. doi: 10.1016/j.fm.2024.104599
Zhao, L., Jing, J. W., and Chen, G. Q. (2018). Engineering peptidoglycan degradation related genes of Bacillus subtilis for better fermentation processes. Bioresour. Technol. 248, 238–247. doi: 10.1016/j.biortech.2017.05.134
Zhou, M. J., Wu, J., Hu, L. X., Hu, W. S., Huang, J. B., Huang, X. L., et al. (2023). Enhanced vitamin K2 production by engineered Bacillus subtilis during leakage fermentation. World J. Microbiol. Biotechnol. 39:224. doi: 10.1007/S11274-023-03671-8
Appendix
Abbreviations of enzymes
Module I: GlpF, glycerol uptake facilitator; GlpK, glycerol kinase; GlpD, glycerol-3-phosphate dehydrogenase; Tpi, triosephosphate isomerase.
Module II: Dxs, 1-deoxyxylulose-5-phosphate synthase; Dxr, 1-deoxyxylulose-5-phosphate reductoisomerase; IspD, 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase; IspE, 4-(cytidine 5′ -diphospho)-2-C-methyl-D-erythritol kinase; IspF, 2-C-methyl-D-erythritol 2,4-cyclodiphosphate synthase; IspG, (E)-4-hydroxy-3-methylbut-2-enyldiphosphate synthase; IspH, 4-hydroxy-3-methylbut-2- enyl diphosphate reductase; Idi, isopentenyldiphosphate delta-isomerase; IspA, which acts as geranyl pyrophosphate synthase and farnesyl pyrophosphate synthase; HepS/HepT, heptaprenyl diphosphate synthase component I/II; AlsS, acetolactate synthase; AlsD, acetolactate decarboxylase.
Module III: AroA, 3-deoxy-7-phosphoheptulonate synthase; AroB, 3-dehydroquinate synthase; AroC, 3-dehydroquinate dehydratase; AroD, shikimate dehydrogenase; AroK, shikimate kinase; AroE, 3-phosphoshikimate-1-carboxyvinyltransferase; AroF, chorismate synthase; AroH, chorismate mutase; TrpE, anthranilate synthase; PabB/PabA, para-aminobenzoate synthase component I/II.
Module IV: MenF, isochorismate synthase; MenD, 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylate synthase; MenH, 2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylate synthase; MenC, o-succinylbenzoate synthase; MenE, o-succinylbenzoic acid-CoA ligase; MenB, 1,4-dihydroxy-2-naphthoyl-CoA synthase; MenI, 1,4-dihydroxy-2-naphthoyl-CoA hydrolase; MenA, 1,4-dihydroxy-2-naphthoate heptaprenyltransferase; MenG, demethylmenaquinone methyltransferase; DhbB, bifunctional isochorismate lyase/aryl carrier protein.
Abbreviations of metabolites
Module I: Gly, glycerol; Gly-3P, glycerol-3-phosphate; DAHP, 3-deoxy-arabino-heptulonate 7-phosphate; G3P, glyceraldehyde-3-phosphate.
Module II: PEP, phosphoenolpyruvate; PYR, pyruvate; DXP, 1-deoxyxylulose-5-phosphate; MEP, methyl-eryth ritol-4-diphosphate; HMBPP, 1-hydroxy-2-methyl-2-butenyl 4-diphosphate; DMAPP, dimethylallyl diphosphate; IPP, isopentenyl diphosphate; GPP, geranyl diphosphate; FPP, farnesyl diphosphate.
Module III: E4P, erythrose 4-phosphate; CHA, chorismate; DHQ, 3-dehydroquinate; DHS, 3-dehydroshikimate; SA, shikimate; S3P, shikimate 3-phosphate; Phe, phenylalanine; Tyr, tyrosine; Trp, tryptophan.
Module IV: ICHA, isochorismate; SEPHCHC, 2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene-1-carboxylate; OSB, 2-succinylbenzoate; DHNA-CoA, 1,4-dihydroxy-2-naphthoyl-CoA; DHNA, 1,4-dihydroxy-2-naphthoate; DMK, 2-demethylmenaquinone; MK-7, menaquinone-7; DHDHB, (2S,3S)-2,3-dihydro-2,3-dihydroxybenzoate.
Keywords: menaquinone-7, lifespan, soy protein hydrolysate, span 80, Bacillus subtilis natto
Citation: Hu L-x, Chen Y, Zhou H-p, Tao W, Gao X-l, Wu C-c, Zhang H-m, Han R-m, Li Y-q and Liu Y (2025) Combination of SPH and SP80 prolongs the lifespan of Bacillus subtilis natto to enhance industrial menaquinone-7 biosynthesis. Front. Microbiol. 16:1578160. doi: 10.3389/fmicb.2025.1578160
Edited by:
Masahiro Ito, Toyo University, JapanCopyright © 2025 Hu, Chen, Zhou, Tao, Gao, Wu, Zhang, Han, Li and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hong-ping Zhou, emhwemhwQDI2My5uZXQ=; Yan Liu, bGl1eWFuQGFocHUuZWR1LmNu
†These authors have contributed equally to this work and share first authorship
 Liu-xiu Hu1,2†
Liu-xiu Hu1,2† Chuan-chao Wu
Chuan-chao Wu Yan Liu
Yan Liu