- 1Evolutionary Ecology and Genetics, Zoological Institute, Kiel University, Kiel, Germany
- 2Antibiotic Resistance Group, Max-Planck-Institute for Evolutionary Biology, Ploen, Germany
Introduction: Antimicrobial resistance (AMR) is a critical global health issue caused by antibiotic overuse, leading to the rise of multi-resistant pathogens such as in bacteria of the ESKAPE group. Alternative or combination therapies, including bacteriophages and plaque-forming predatory bacteria, are being explored in response. Bdellovibrio bacteriovorus, a Gram-negative bacterial predator belonging to the Bdellovibrio and like organisms (BALOs), can kill other Gram-negative bacteria after the periplasmic invasion, including multidrug-resistant pathogens. However, a combined treatment of antibiotics and plaque-forming predatory bacteria requires the predatory bacteria to be resistant to the antibiotic. The predator’s unique growth requirements limit standardized AMR testing methods.
Materials and methods: We propose a streamlined three-step protocol to measure AMR in plaque-forming predatory bacteria. It requires the (i) cultivation of a dense Bdellovibrio culture with a suitable prey strain, followed by (ii) a double-layered agar plaque assay using a prey strain resistant to the antibiotic of interest, and (iii) the application of E-test strips for minimum inhibitory concentration (MIC) determination. We apply the method to the commonly used strain B. bacteriovorus HD100. We use P. aeruginosa H03 as prey for MIC determination for five antibiotics.
Results: Our results show consistent MICs for B. bacteriovorus HD100 across independent experiments. Reliable MIC determination for meropenem was limited by P. aeruginosa H03 susceptibility to this antibiotic. Further, we observed a positive association between MIC values and predator inoculum concentration for ceftazidime, ciprofloxacin, and gentamicin. Prolonged incubation time increased MIC values, notably for ciprofloxacin. While resistant to piperacillin, predator plaques were absent on plates with piperacillin-tazobactam combinations.
Conclusion: The streamlined approach described here to determine MICs in plaque-forming predatory bacteria proves effective and robust, when using a suitable (i.e., resistant) prey. It provides a starting point for the joint study of antibiotics and plaque-forming predatory bacteria.
1 Introduction
Antibiotics are indispensable to our healthcare system, but over time, their overuse has led bacteria to evolve resistance. The antimicrobial resistance (AMR) crisis has been classified as a global health concern by the World Health Organization (WHO) (Michael et al., 2014). In 2017, the WHO called for new antimicrobial development for a list of pathogens and designated “priority status” to the multi-resistant ESKAPE pathogens (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species), which cause the majority of nosocomial infections (De Oliveira et al., 2020).
In addition to the development of new antibiotics, there are approaches to adapt the application of antibiotics, e.g., combined or sequential therapy of different antibiotics (Andersson et al., 2020; Roemhild and Schulenburg, 2019; Sanz-García et al., 2023). Others examined the possibility of using bacteriophages to defeat bacterial infections (Dufour et al., 2016; Galtier et al., 2017; Jault et al., 2019; Yen et al., 2017). However, this is accompanied by several challenges, including the innate immune response to bacteriophages and the rapid evolution of bacterial resistance to phages during therapy (Kortright et al., 2019).
Because both novel antibiotics and phage therapy face practical hurdles, a third biological option—predatory bacteria—has attracted attention. Several studies have explored the therapeutic potential of predatory bacteria, i.e., Bdellovibrio bacteriovorus, with encouraging outcomes observed in vivo in poultry (Atterbury et al., 2011), rats (Shatzkes et al., 2016), mice (Findlay et al., 2019), and zebrafish (Willis et al., 2016).
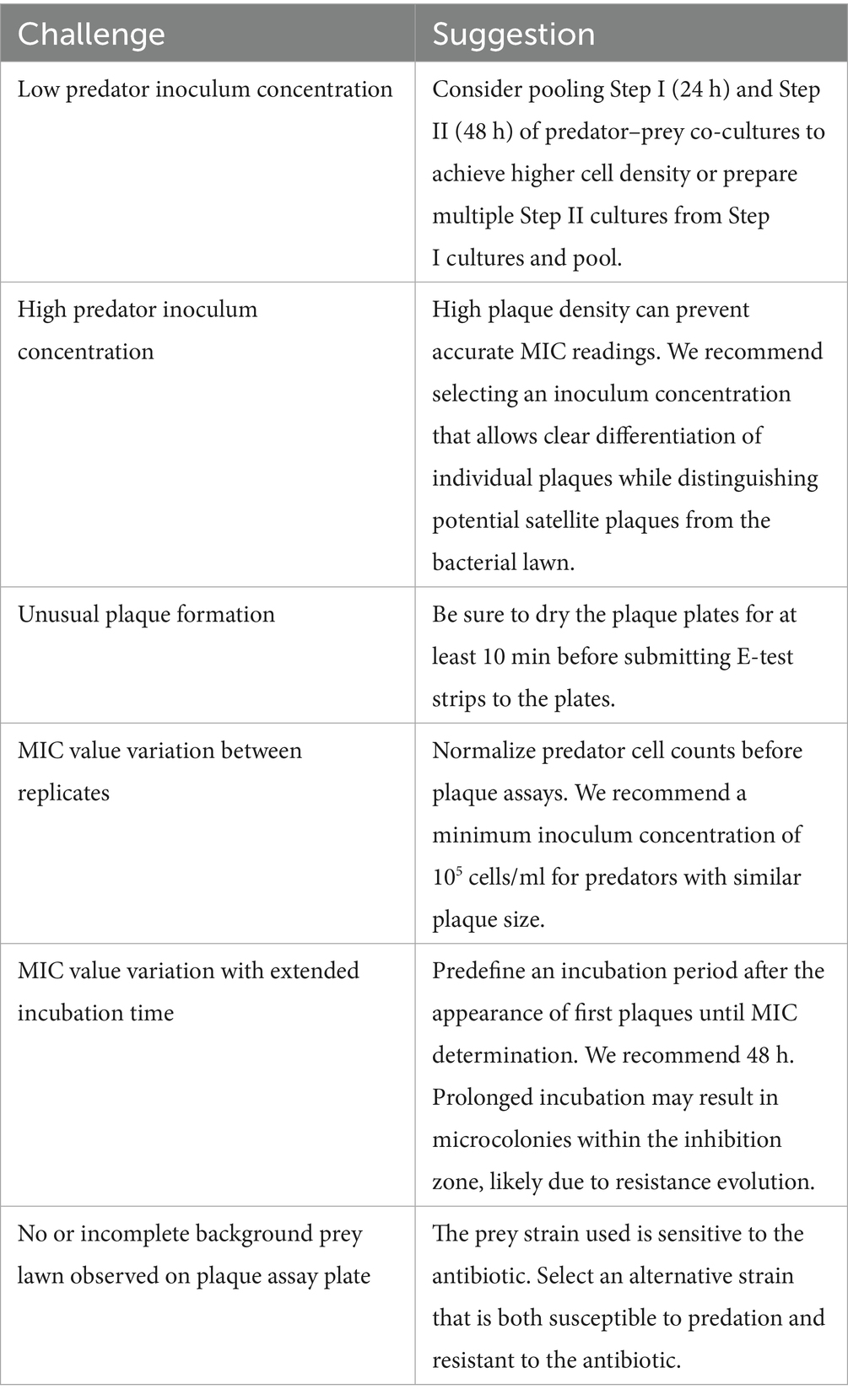
B. bacteriovorus is a highly motile, Gram-negative obligate predator that belongs to the group of Bdellovibrio and like organisms (BALOs) whose members are found within the phylum Bdellovibrionota (genera Bdellovibrio, Pseudobdellovibrio, Bacteriovorax, Peredibacter, Halobacteriovorax, Pseudobacteriovorax, and Micavibrio). All BALOs are characterized by a biphasic life cycle. The attack phase comprises free-swimming predator cells that actively seek out prey (Figures 1A,B). Upon locating Gram-negative bacteria, they enter a prey-associated phase, which starts with BALOs attaching to the prey cells. Periplasmic predators then invade the periplasm of the prey. Sealing the damaged peptidoglycan wall behind them, they form an osmotically stable bdelloplast, establishing and growing through elongation and non-binary fission. Once flagella have developed, the bdelloplast is lysed, releasing up to seven progenies that immediately enter the attack phase (Laloux, 2020). In contrast, epibiotic predators remain attached to the outside of the prey cell while consuming its cellular content. Various B. bacteriovorus strains demonstrate a broad host range, including ESKAPE pathogens (Negus et al., 2017). Unlike bacteriophages, B. bacteriovorus rapidly hydrolyzes all genomic DNA within its prey, reducing the risk of horizontal gene transfer (Matin and Rittenberg, 1972; Monnappa et al., 2013), a key mechanism in the spread of AMR (Djordjevic et al., 2024). Further, this predator has been shown to invade and destroy biofilms (Kadouri and O’Toole, 2005; Núñez et al., 2005), which can act as protective barriers for bacteria against antibiotics (Stewart and Costerton, 2001). Further, a recent study demonstrated that Bdellovibrio can effectively prey on antibiotic persister E. coli cells, which are typically tolerant to conventional antibiotics (Matan and Jurkevitch, 2022). Importantly, B. bacteriovorus is non-pathogenic to human serum and cell lines in vitro (Baker et al., 2017; Gupta et al., 2016; Monnappa et al., 2016; Raghunathan et al., 2019), highlighting its role as a promising “living antibiotic” against multi-resistant pathogens. However, B. bacteriovorus does not eliminate its prey. Although the exact mechanisms of prey resistance remain to be elucidated (Das and Negus, 2024), several studies have reported environmentally induced phenotypic prey resistance, where prey cells transition into non-susceptible physiological cell states (Kadouri and O’Toole, 2005; Shemesh and Jurkevitch, 2004; Youdkes et al., 2020). This implies that while effectively reducing bacterial load, B. bacteriovorus cannot fully clear the infection on its own (Cavallo et al., 2021). Therefore, combining B. bacteriovorus with a second agent, like an antibiotic, may be necessary to effectively treat infections and prevent the spread of new AMR genes. In this regard, Im et al. (2017) showed that combining violacein with B. bacteriovorus HD100 reduced pathogen viability by 2,970-fold (99.96%) in mixed bacterial cultures.
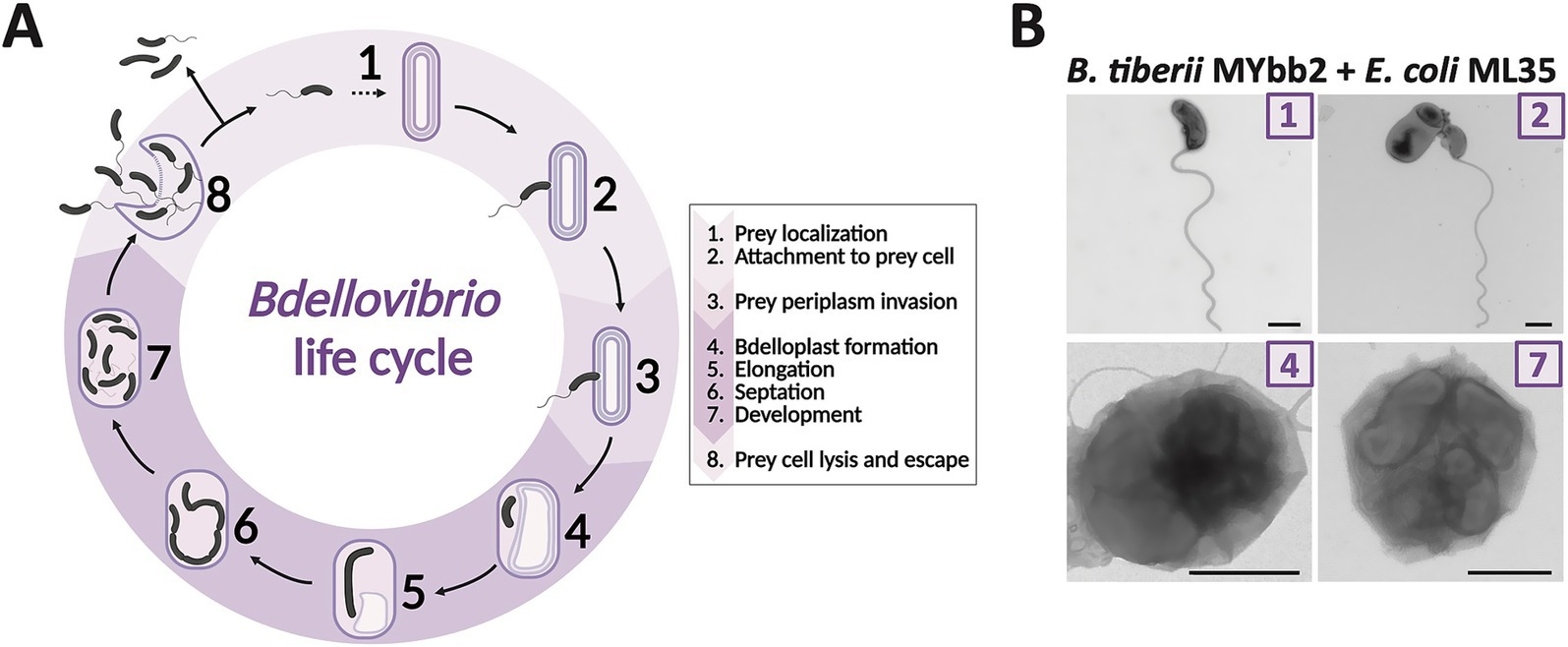
Figure 1. Periplasmic life cycle of Bdellovibrio. (A) In their attack phase, a free-swimming predator cell actively seeks out prey (1). Upon locating a Gram-negative cell, it attaches to the prey’s cell wall (2). Following attachment, the predator enters the prey-associated phase, invading the prey’s periplasm (3), and sealing the damaged cell wall behind them. Inside the resulting bdelloplast (4), the predator cell elongates (5), undergoes septation (6), and produces up to seven progeny cells. Once the progeny develops flagella (7), the prey cell is lysed, releasing the newly formed Bdellovibrio cells (8), which then seek new prey to continue the cycle. (B) B. tiberii MYbb2 with prey E. coli ML35 in different life cycle stages: Numbers in image corners correspond to the proposed life cycle stage in A. In stage (2), the predator is attached to an E. coli prey cell. Stages (4) and (7) demonstrate intraperiplasmic growth in the bdelloplast, with one or more Bdellovibrio cells confined within the outer membrane of a prey cell. Samples were imaged by TEM after negative staining. Scale bar is 500 nm. Life cycle illustration was created in BioRender. Wülbern (2025) https://BioRender.com/6ybxp6x, Figure based on Maher et al. (2025), modified by the authors.
To explore combinations of antibiotics and plaque-forming bacteria (such as B. bacteriovorus) against Gram-negative pathogens, a reliable method for determining predator antibiotic susceptibility is needed. As these predators only grow in the presence of prey and usually do not form colonies, the standard methods for determining the minimum inhibitory concentrations (MIC), such as the gradient method via E-test strips on bacterial lawns, cannot be applied.
Marine et al. (2020) proposed an assay to determine AMR in B. bacteriovorus. Specifically, the method requires a predator–prey co-culture in liquid medium to estimate B. bacteriovorus growth by measuring absorbance at 600 nm of the said co-culture with various concentrations of antibiotics. Due to their small size, B. bacteriovorus cells barely contribute to optical density measurements (OD600). Hence, measured absorbance indirectly reflects the decreasing cell density of the E. coli prey cells in co-cultures with B. bacteriovorus. This was followed by a plaque assay to verify the predator–prey ratio detected by absorbance at 600 nm (Marine et al., 2020). While this method yielded encouraging results, it involved extensive data analysis to determine the AMR profile of tested B. bacteriovorus strains. A key challenge in MIC testing for plaque-forming predatory bacteria remains the need for live prey, which are themselves susceptible to antibiotics and can confound results. While the Marine et al. (2020) approach is valid, MIC testing on plates with antibiotic test strips (e.g., E-tests) can simplify the process by allowing direct readout and eliminating the need for liquid culture transfers (Figure 2). To date, however, a method for MIC analysis of plaque-forming predatory bacteria on plates is not available.
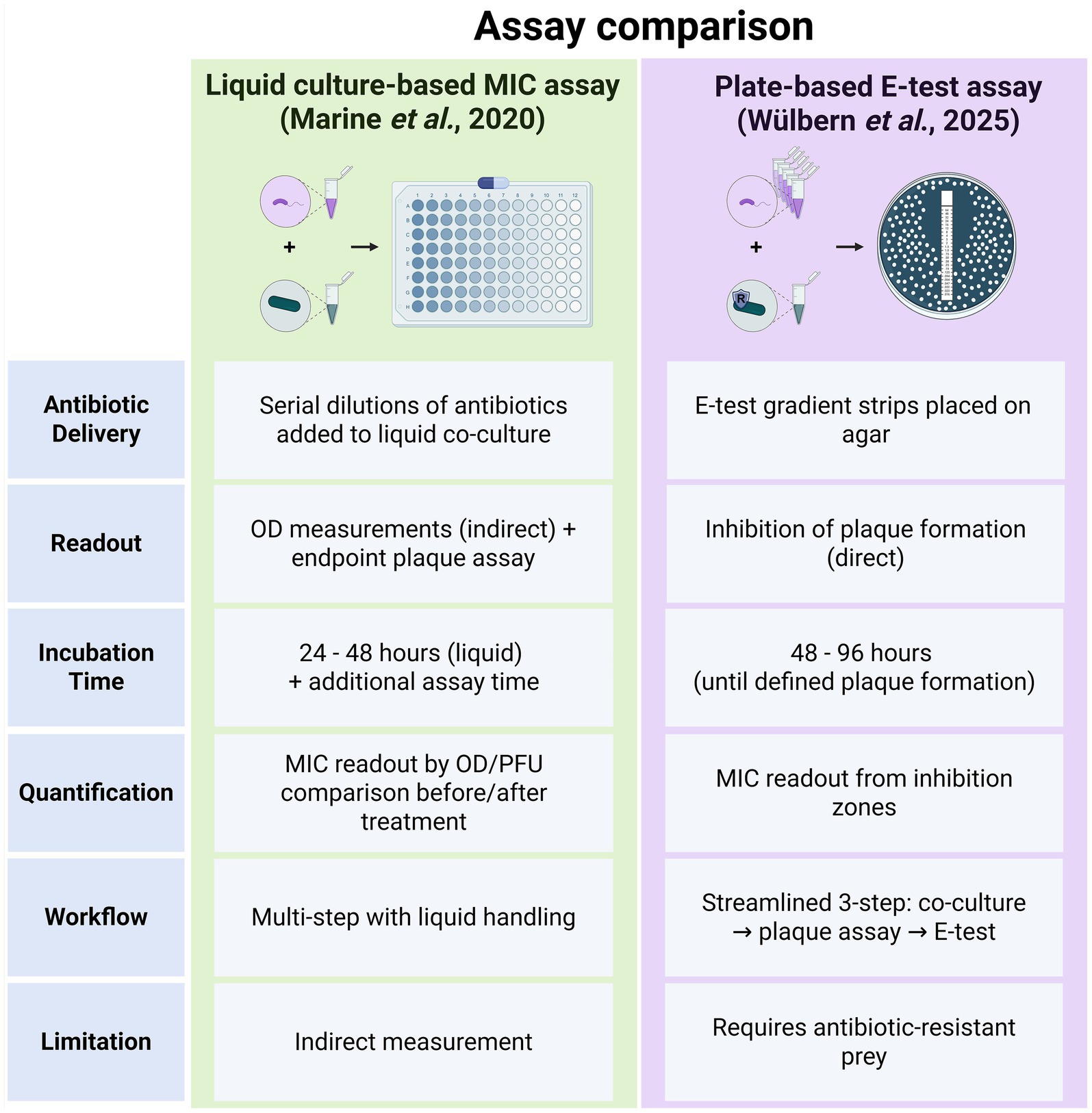
Figure 2. Comparison of two methods for MIC determination in plaque-forming predatory bacteria. The left panel summarizes the liquid culture-based MIC assay as described by Marine et al. (2020), which relies on indirect readouts (optical density) and multiple incubation steps. The right panel illustrates the streamlined E-test-based method introduced in this study, which directly measures inhibition of predator plaque formation on agar plates. Key differences include antibiotic delivery, readout type, incubation time, assay complexity, and methodological limitations. Created in BioRender. Wülbern (2025) https://BioRender.com/6ybxp6x.
This study proposes an alternative, streamlined three-step assay using E-test strips on plaque plates to determine the predator’s MIC. To ensure broad applicability across clinically relevant antibiotics, we evaluated the sensitivity of two different Bdellovibrio strains to antibiotics from five pharmaceutical classes, representing various mechanisms of action. We demonstrated that applying E-test strips to double-layered agar plates with a multi-resistant prey bacterium (e.g., P. aeruginosa strain H03) provides reliable MIC values for all measurable antibiotics. For this approach, selecting an appropriate prey strain is critical, as sensitivity of the prey to the tested antibiotic can prevent accurate determination of the MIC for the plaque-forming bacterium.
2 Materials and methods
2.1 Resources and reagents
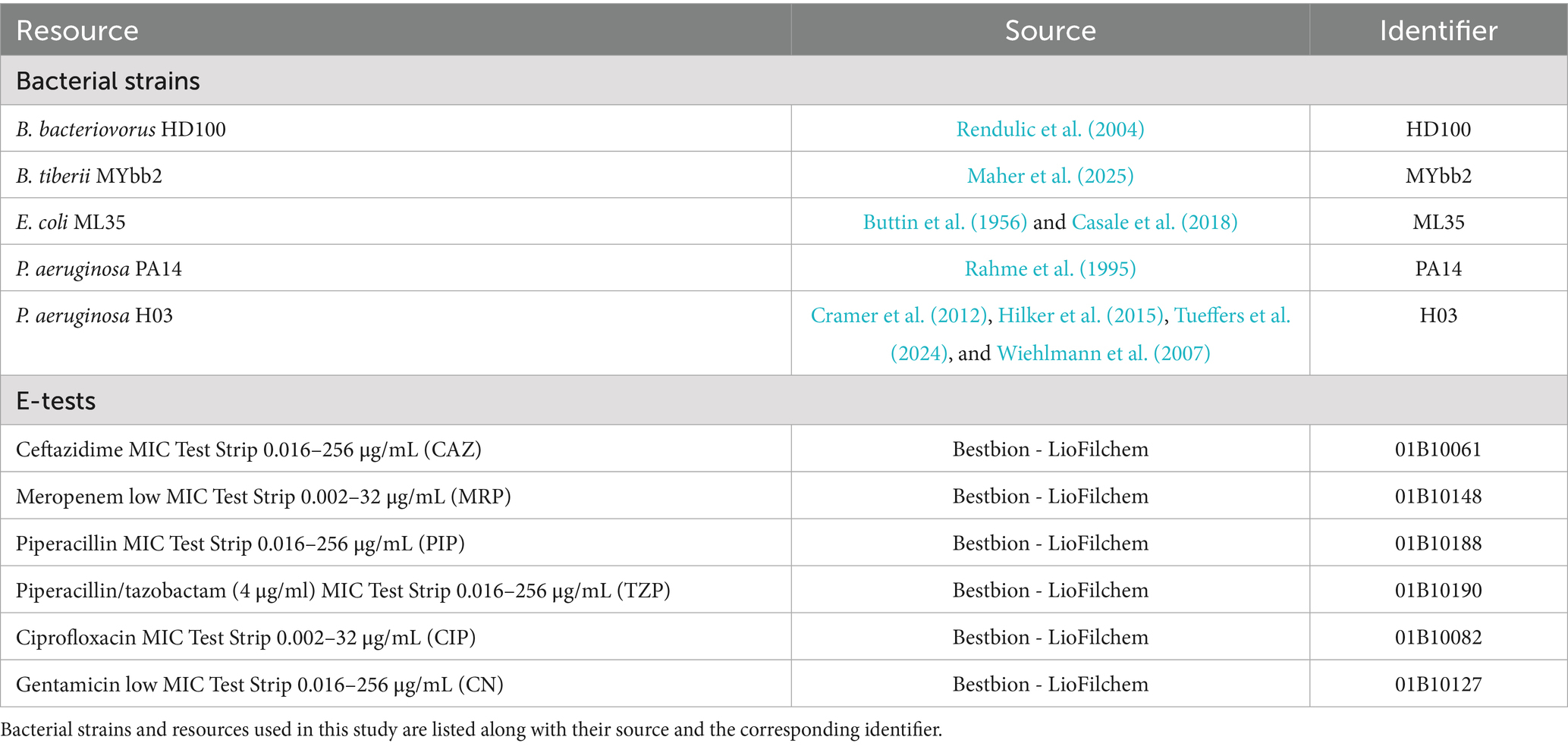
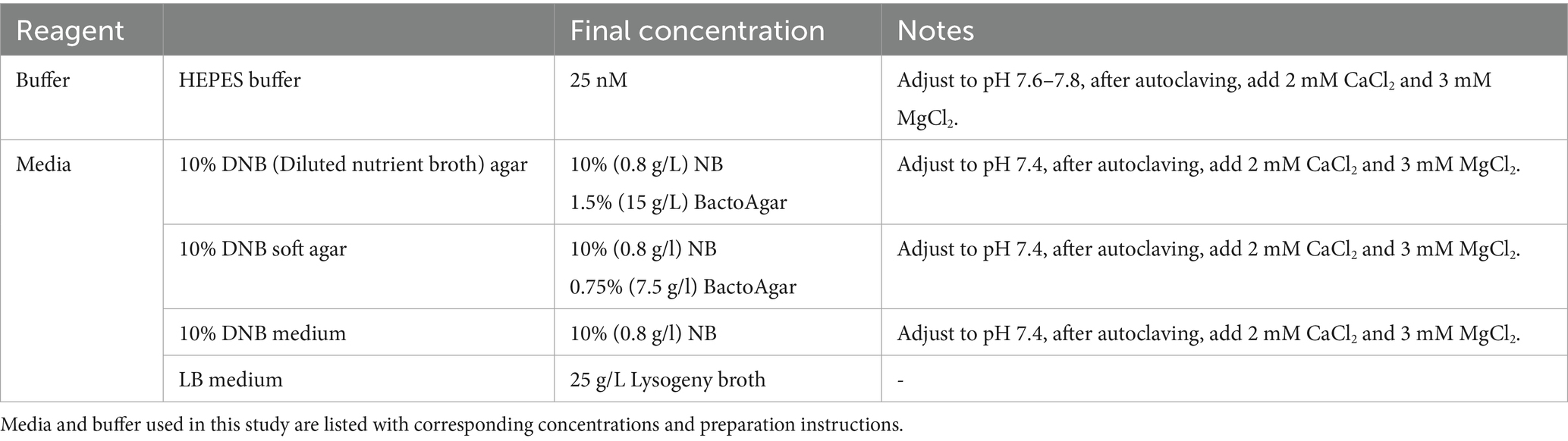
Bacterial strains and MIC test strips used in this study are detailed in Table 1. Table 2 lists all reagents, their respective concentrations, and preparation instructions.
2.2 Tested antibiotics
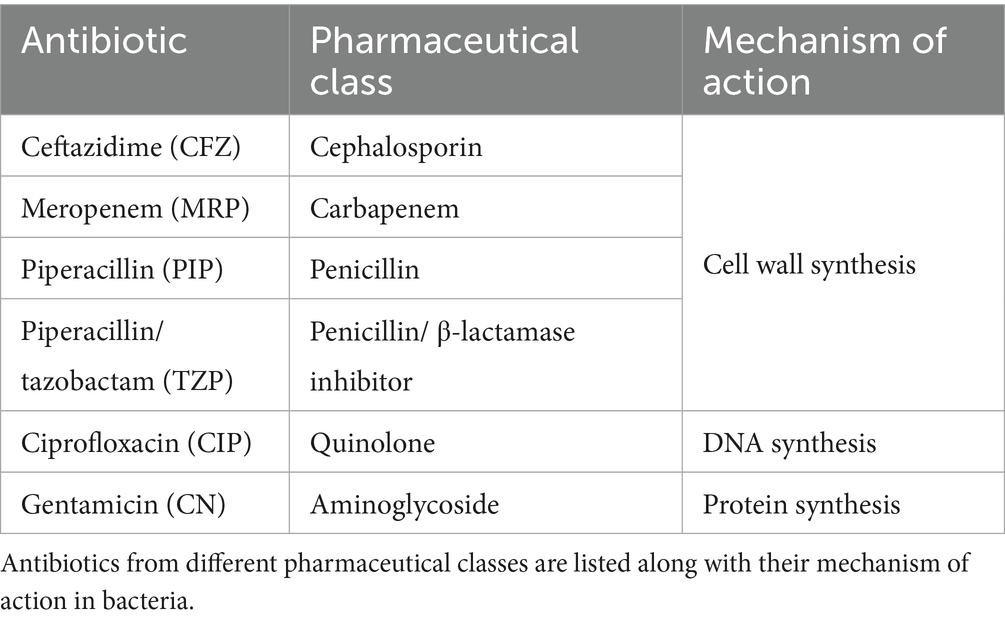
To determine Bdellovibrio AMR profiles, we utilized the multi-resistant strain P. aeruginosa H03 as background prey for plaque assays. We selected antibiotics from different pharmaceutical classes based on the resistance profile of H03 as determined in Tueffers et al. (2024).
2.3 Methods
To evaluate AMR profiles of plaque-forming predatory bacteria with the proposed method, it is necessary to (i) establish a dense culture of Bdellovibrio with a suitable prey strain. This culture is then used in a plaque assay (ii), using a prey strain resistant to the AB of interest, since the method relies on a background lawn of the prey to visualize lysis. And lastly, an E-test strip is added to this plaque assay plate (iii). In the following paragraphs, we will explain these steps in more detail. Thereafter, we give detailed methods used to identify factors that affect the robustness of the newly proposed method, i.e., inoculum concentration effects on MIC, and describe the statistical methods used to evaluate this effect.
Note: E-test-based MIC values may vary based on a variety of assay parameters, including cell density (Smith and Kirby, 2018; Udekwu et al., 2009), the growth medium, its pH, incubation time, and incubation temperature (Kowalska-Krochmal and Dudek-Wicher, 2021). To minimize variability and ensure reliable results, it is important to follow standardized protocols throughout the proposed assay.
i. Cultivation of Bdellovibrio with a suitable prey strain
We used the standard laboratory strain B. bacteriovorus HD100 to establish and evaluate the robustness of the proposed assay. Note: To test the desired Bdellovibrio AMR profile, it is crucial to ensure robust bacterial growth and high cell viability for the start culture. To achieve this, we co-cultivated Bdellovibrio cells with the common prey strain Escherichia coli ML35 in DNB liquid medium for maintenance, as liquid co-cultures with P. aeruginosa H03 yielded comparably lower cell counts (Supplementary Figure S1). The plaque assay and E-test were then performed using the highly AB-resistant strain P. aeruginosa H03 as prey.
As obligate predators, Bdellovibrio cells require co-cultivation with prey bacteria. E. coli strain ML35 and P. aeruginosa strain H03 (Cramer et al., 2012; Hilker et al., 2015; Tueffers et al., 2024; Wiehlmann et al., 2007) were used as prey cells for liquid co-cultures and plaque assays, respectively. Prey cell overnight cultures were started from glycerol stocks (80%) and grown in 50 mL LB (Lysogeny broth) medium, continuously shaking for 16 h at 37°C and 180 rpm. Thereafter, prey cells were adjusted to an OD600 of 10 after being washed in 25 mM HEPES (2-[4-(2-hydroxyethyl)piperazin-1-yl] ethane sulfonic acid solution, pH 7.6–7.8, supplemented with 2 mM CaCl2 and 3 mM MgCl2) (Jurkevitch, 2012).
For Bdellovibrio cultivation in liquid medium, we followed the protocol of Remy et al. (2022) with minor adaptations. In short, single plaques were aspirated from double-layered DNB (10% diluted nutrient broth, supplemented with 2 mM CaCl2 and 3 mM MgCl2) agar plates, released into a sterile glass tube with 2.7 mL DNB medium with 300 μL of E. coli ML35 (final OD600 = 1), and incubated overnight shaking (180 rpm) at 28°C (Step I). After 24 h, 100 μL of the co-cultivate was transferred to fresh 2.6 mL DNB medium with 300 μL of E. coli ML35 (final OD600 = 1) and incubated overnight, shaking (180 rpm) at 28°C (Step II).
Note: The method described here is intended to produce a dense Bdellovibrio culture. To estimate culture density, we propose a visual assessment. Bdellovibrio predation leads to the clearing of the culture, which can be readily observed by eye. Under the microscope, using 40 × or 100 × magnification, Bdellovibrio cells should appear small, fast-swimming, and should constitute the majority of cells in the co-culture after incubation. Since exact cell counts are difficult to determine prior to E-test application, it is important to use multiple dilutions of the Bdellovibrio culture to achieve optimal conditions (details provided below).
i. Plaque assay
Note: To assess the Bdellovibrio AMR profile in the proposed assay, the readout is reflected in the absence of plaque-forming units (PFUs) on double-layered agar plates. Hence, selecting a suitable prey strain is critical for establishing a reliable background to differentiate between areas where Bdellovibrio was able to predate and form PFUs and those where it was inhibited by the antibiotic tested. In this study, we chose P. aeruginosa H03, a multi-resistant and suitable prey strain for B. bacteriovorus HD100. Sensitivity of the prey strain to specific antibiotics may make it necessary to switch to a different strain that is then resistant to such specific antibiotics.
Before each plaque assay, Step I and Step II Bdellovibrio cells were pooled for higher cell yield and separated from the remaining ML35 prey cells and debris by double-filtration through 0.45 μm syringe filters (Figure 3A). The successful removal of prey cells and debris should be confirmed microscopically. Cells were concentrated by centrifugation for 10 min at 4000 rpm. Bdellovibrio cell pellets were then resuspended in 1 mL 25 mM HEPES. A ten-fold serial dilution of the resuspended Bdellovibrio cells (100–10−5) was prepared in 25 mM HEPES. Double-layered DNB agar plates were prepared according to standard protocol (Jurkevitch, 2012). In detail, prepared DNB agar plates (1.5% agarose) served as a nutrient layer. For the top DNB soft agar layer (0.75% agarose), 300 μL P. aeruginosa H03 prey cells (final OD600 = 0.6) and 100 μL concentrated Bdellovibrio cells were added to 5 mL molten DNB soft agar (58°C), mixed, and poured over the DNB agar plate, yielding a ~ 3 mm layer. Double-layered plates were allowed to dry for 30 min, before proceeding (Figure 3B).
Figure 3. Step-by-step guide for plaque-forming predator MIC determination. (A) After double-filtration (0.45 μm) of a predator–prey co-culture, prepare a ten-fold serial dilution (100–10−5) of the plaque-forming predatory bacteria. (B) Combine respective predator dilution with a multi-resistant prey of choice in DNB soft-agar and plate for plaque assay. Dry plates for 30 min. (C): Place E-test strips onto the soft agar layer of plaque assay plates of three replicates, and dry plates for another 10 min. (D): After incubation of inverted plates at 28°C for 4–7 days, evaluate the MIC of the plaque-forming predatory bacteria. Created in BioRender. Wülbern (2025) https://BioRender.com/6ybxp6x.
Note: For reliable readout, ensuring good quality plaque assay plates is important. Hence, it is important to pour the molten DNB soft agar evenly and to be sure to dry the plaque plates for at least 30 min, before submitting E-test strips to the plates.
i. Determination of Bdellovibrio MIC by E-test
For colony-forming bacteria, the MIC of an antimicrobial is routinely assessed using the gradient method (Kadeřábková et al., 2024). For this, E-test strips containing a predefined gradient of antibiotic concentrations are applied to inoculated agar plates. The MIC value is read as the intercept of the inhibition ellipse edge. The present study employed E-test strips (LioFilchem, Italy) to double-layered plaque assay plates seeded with P. aeruginosa H03 and a Bdellovibrio strain. In detail, following the preparation of dry plaque assay plates, sterile, fully thawed E-tests (RT) were placed onto the soft agar layer of the respective dilution plates (Figure 3C). This step was performed in triplicates.
Note: Determining optimal predator dilutions used in the plaque assay is essential to avoid complete prey lawn clearance at low dilutions or low plaque density at high dilutions, both of which hinder accurate quantification. Pre-evaluating optimal dilutions minimizes costs by preventing repeated trials with E-tests (see “Quantification of inoculum concentration”).
After E-test placement, plates were dried for 10 min and then sealed with parafilm. Inverted plates were incubated at 28°C for four to 7 days.
Note: When using MIC E-test strips for colony-forming bacteria, the standard incubation time ranges between 16 and 48 h at 37°C. However, these incubation periods are not applicable to plaque-forming predatory bacteria. Depending on parameters such as the prey used in assays, the incubation time for Bdellovibrio can range between 48 and 96 h before the appearance of the first plaques. Antibiotics from E-test strips typically diffuse into the agar and remain stable for extended periods—up to 10 days with the LioFilchem E-test strips used in this study (according to manufacturer information). To ensure accurate results during prolonged incubation, care must be taken to prevent the plates from drying out. We also tested for antibiotic degradation during prolonged incubation and found no evidence of such degradation after up to 8 days (Supplementary Figure S2; detailed methodology is provided below). Once plaques appear, plates should be incubated for a predetermined period to ensure sufficiently high cell densities for accurate MIC determination and reproducibility across experiments.
Plaque formation was monitored in 24-h intervals, and MICs were determined once plaques were sufficiently large, yet distinguishable and dense to allow for accurate MIC evaluation (Figure 3D). The lowest antibiotic concentration at which predator plaque growth was inhibited completely was interpreted as the intercept of the inhibition zone.
Note: We recommend consulting the Liofilchem® MIC Test Strip Photographic Guide for adequate MIC value interpretation.1
2.4 Quantification of inoculum concentration
To determine the inoculum concentration of Bdellovibrio cells, we used E. coli ML35 as prey, as preliminary experiments yielded higher predation efficiency with this strain compared to P. aeruginosa H03 (Supplementary Figure S1). We determine the inoculum concentration using plaque-forming units (PFUs)/ml of Bdellovibrio culture. For accurate PFU determination, it is essential to use plates where plaques are both distinguishable and sufficiently dense (on 9 cm plates: app. PFU > 30). Therefore, higher dilutions (10−4 and 10−5) from the ten-fold serial dilutions were utilized for cell density determination. Optimal cell density dilutions may need to be adjusted based on respective conditions, prey, and the plaque-forming predatory bacterial strain used. Double-layered DNB agar plates were prepared according to standard protocol (Jurkevitch, 2012) and as described before (see ii. Plaque assay). Cell numbers (PFU/ml) were determined by counting the total number of plaques per plate and adjusting the value based on the dilution factor and total plated volume.
2.5 Statistical analysis
Data analysis was performed using R (v. 4.3.2, R Core Team, 2023). Statistical analysis was performed using the “lme4” package (v.1.1.35.5, Bates et al., 2015). Plotting was performed using the “ggplot2” package (v.3.5.1, Wickham, 2016). A linear mixed model with the formula log2(MIC) ~ log2(PFU/ml) + (1|biological replicate) was used for the antibiotics ciprofloxacin (CIP) and gentamicin (CN) to examine the relationship between MIC values and PFU counts, but not for the other antibiotics as they were either lacking MIC variability (PIP/TAZ) or as they showed no linear trend (CFZ). To account for dependencies within the technical replicates associated with the same biological replicate, the biological replicate was included as a random factor. MIC and PFU/ml values were log-transformed to meet the assumptions of normality for the residuals.
2.6 Test for antibiotic degradation over prolonged incubation times
To assess potential antibiotic degradation during prolonged incubation, a standard E-test assay was performed using three biological replicate cultures of the P. aeruginosa reference strain PA14 for all antibiotics tested (CIP, CN, CAZ, TZP), following the manufacturer’s protocol (LioFilchem). Incubation was extended to 8 days, and MICs were recorded at both 24 h and 8 days.
2.7 Validation of MIC values obtained from E-tests
MICs for B. bacteriovorus HD100 and gentamycin, as determined via E-test, were validated using an E-test-independent method. Gentamycin was added at varying concentrations (0.032, 0.053, 0.106, 0.2, and 0.4 μg/mL) to DNB agar plates and DNB soft agar, followed by a standard plaque assay with three biological replicate cultures of B. bacteriovorus HD100 as described earlier. As in the E-test, the multi-resistant P. aeruginosa strain H03 was used as prey. To account for the observed inoculum effect, predator inoculum size was simultaneously determined by performing a parallel plaque assay without antibiotics, using E. coli ML35 as prey. All plates were incubated at 28°C for 7 days.
3 Results
This study aimed to establish a reliable method to test the antimicrobial resistance (AMR) of plaque-forming predatory bacteria such as Bdellovibrio bacteriovorus. We also examined the impact of two additional parameters, i.e., incubation duration and inoculum concentration.
3.1 Robust readouts for MIC values for plaque-forming predatory bacteria
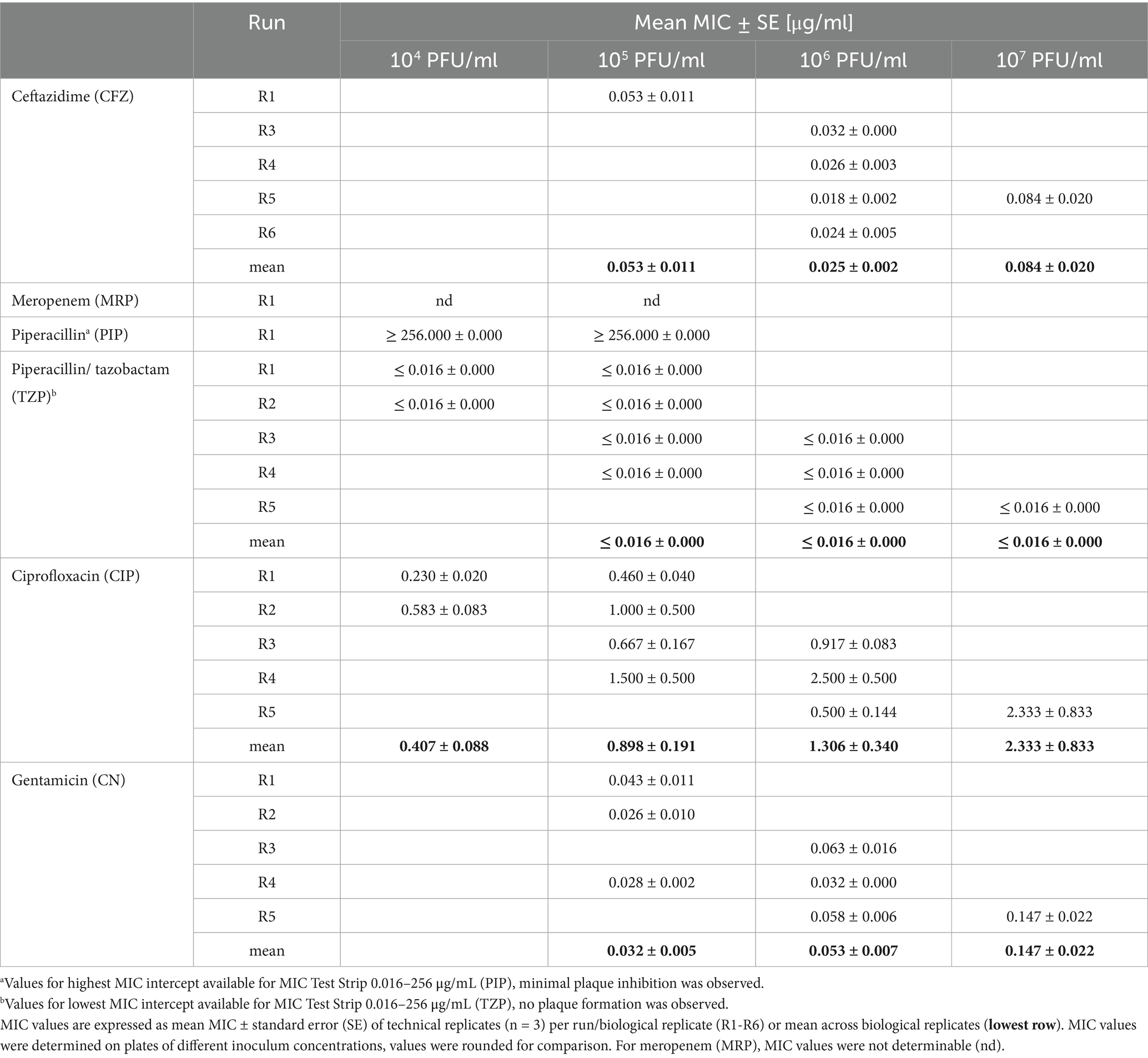
We evaluated the susceptibility of B. bacteriovorus HD100 to five antibiotics of various pharmaceutical classes: ceftazidime (CFZ), meropenem (MRP), piperacillin (PIP), piperacillin/tazobactam (TZP), ciprofloxacin (CIP), and gentamicin (CN) (Table 3). The assay was performed in five independent runs, each using a distinct Bdellovibrio culture and varying initial inoculum concentrations based on the respective culture densities. Across these runs, minimal variation in MIC values was observed among technical replicates (Table 4; Figure 4). Dilutions with an inoculum concentration in the range of 106 PFU/ml provided the most comprehensive data across antibiotics (Table 4; Supplementary Table S1). Supplementary Table S1 includes a detailed overview of individual MIC values per run and precise PFU/ml concentrations.
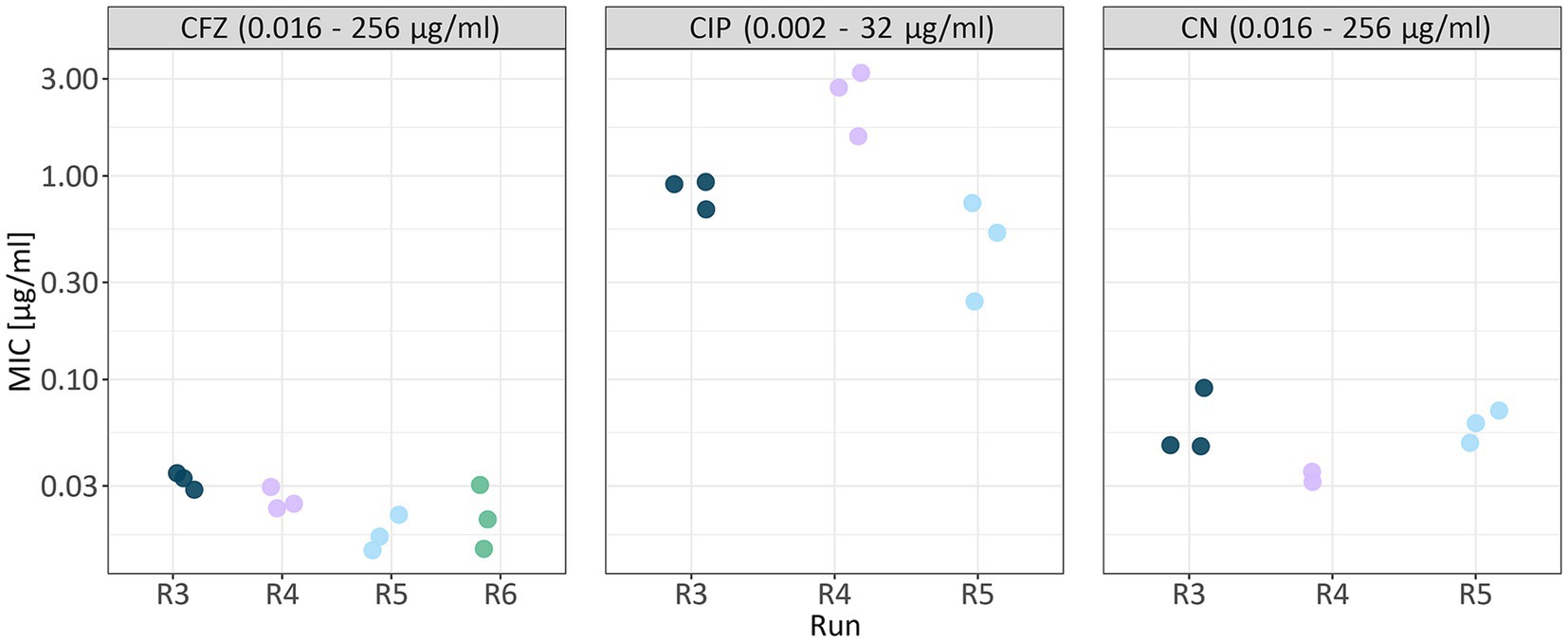
Figure 4. MIC values of technical replicates across runs for the most comprehensive dilution dataset. Only MIC values determined at a dilution of 106 PFU/ml are shown. Each antibiotic (CFZ, ceftazidime; CIP, ciprofloxacin; CN, gentamicin) is displayed in a separate panel. Data points are colored by biological replicate (run), and for each run, technical replicates are shown. Values given in the boxes on top of the plot indicate the antibiotic concentration range of the respective MIC test strip.
The inhibition of P. aeruginosa growth at elevated meropenem concentrations prevented reliable MIC determination for B. bacteriovorus HD100 with this antibiotic. Across all runs, plaque formation was observed at the highest antibiotic concentrations on B. bacteriovorus strain HD100 plates with E-test strips for piperacillin, indicating resistance to this β-lactam antibiotic (MIC ≥ 256 μg/mL). In contrast, adding a β-lactamase inhibitor, e.g., tazobactam, to render susceptibility (Cheng et al., 2017), led to no plaque formation, indicating that the MIC is below the lowest antibiotic concentration tested, ≤ 0.016 μg/mL.
We validated the observed MIC for gentamycin using an E-test-independent method by incorporating gentamycin directly into the agar and soft agar prior to performing a plaque assay. Using a B. bacteriovorus HD100 inoculum of approximately 108 PFU/mL, we determined MIC values of 0.2 μg/mL in two of three biological replicates, and 0.053 μg/mL in the remaining replicate. These MIC values align with those obtained from E-tests conducted with similar inoculum densities (e.g., 0.147 μg/mL for 107 PFU/mL).
In summary, the proposed assay produced robust MIC values for all considered antibiotics, in all cases dependent on the availability of a suitable (i.e., resistant) prey strain.
3.2 Relationship between MIC values and predator inoculum concentration
For colony-forming bacteria, several studies have shown a direct relationship between MIC values and inoculum density (inoculum effect) (García-Rivera et al., 2024; Smith and Kirby, 2018). In this context, we aimed to test different inoculum concentrations for B. bacteriovorus HD100 by applying E-test strips to different dilutions of the same biological replicates (Table 4).
We tested for a linear association between mean MIC values and inoculum concentration for ciprofloxacin (linear mixed model, p = 9.54e-05), and gentamicin (linear mixed model, p = 0.000654), but not ceftazidime (Figure 5; Supplementary Table S2; Supplementary Figure S3). Interestingly, for ceftazidime, we noted higher MIC values at inoculum concentrations in the range of 105 PFU/ml and 107 PFU/ml compared to 106 PFU/ml indicating no linear relationship. Especially for sensitive cells, E-test results may vary by 1 or 2 steps in the readable MIC between replicates due to a very small antibiotic gradient. However, for these dilutions data were obtained in one run each, showing the need for replication. In contrast, we saw little variation across four independent runs for inoculum concentrations of 106 PFU/ml (Figure 5; Table 4).
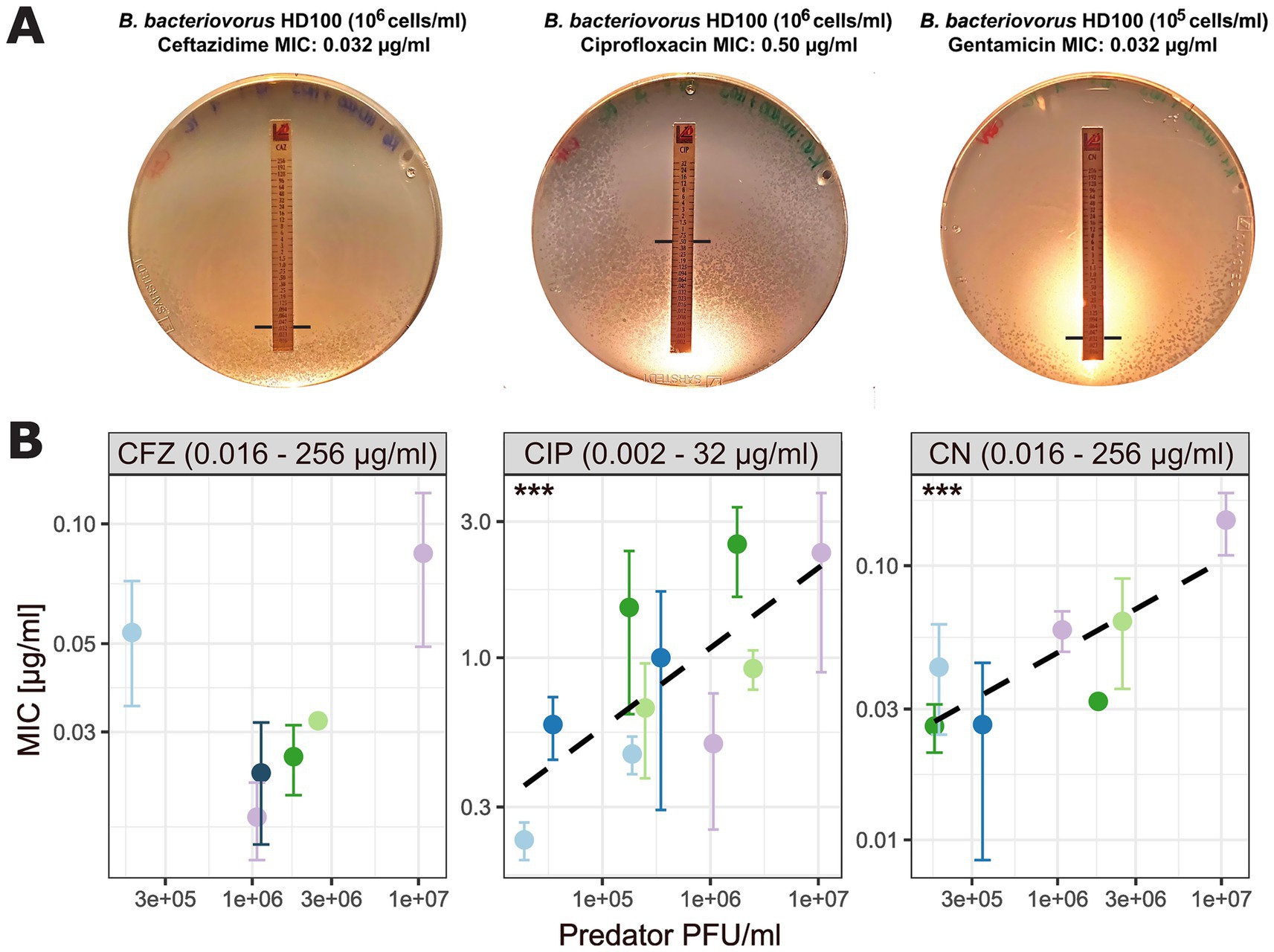
Figure 5. Relationship between predator inoculum concentration and MIC values. (A) Exemplary images of DNB agar plates showing B. bacteriovorus plaques with P. aeruginosa H03 as background lawn and E-test stripes. Black lines represent predator MIC intercept. (B) Association between MIC and predator inoculum concentration (measured as PFU/ml). Each antibiotic (CFZ, ceftazidime; CIP, ciprofloxacin; CN, gentamicin) is displayed in a separate panel with individual x- and y-axes. Black dashed lines for CIP and CN represent linear regression lines fitted to the data. Asterisks indicate p-values as calculated using a linear mixed model, with biological replicate as random variable to account for dependencies (Supplementary Table S2). Data points (colored by biological replicate) represent the mean MIC value of three technical replicates for biological replicates, with variability indicated by SD bars. Values given in the boxes on top of the plot indicate the antibiotic concentration range of the respective MIC test strip.
3.3 Relationship between MIC values and incubation time
First plaques of B. bacteriovorus HD100 appeared after four to 7 days. Plates were then incubated for another 48–72 h before MIC determination. In some cases, we observed small, scattered plaques in the periphery of inhibition zones. This may result from a reduced predator’s predation efficiency or, in general, slower growth due to increasing inhibitory concentrations. They can also be an indication of de novo resistant mutants, as they arise from a single cell, they will inherently be smaller. In cases where small, scattered plaques obscured the inhibition zone, the incubation time was extended in 24-h intervals until a clear MIC intercept could be reliably determined. Notably, we observed that longer incubation times influenced MIC values. Specifically, for ciprofloxacin, where the initial MIC was 0.5 μg/mL, an additional incubation time of 48 h yielded a four-fold increased MIC intercept (MIC = 2 μg/mL) (Figure 6).
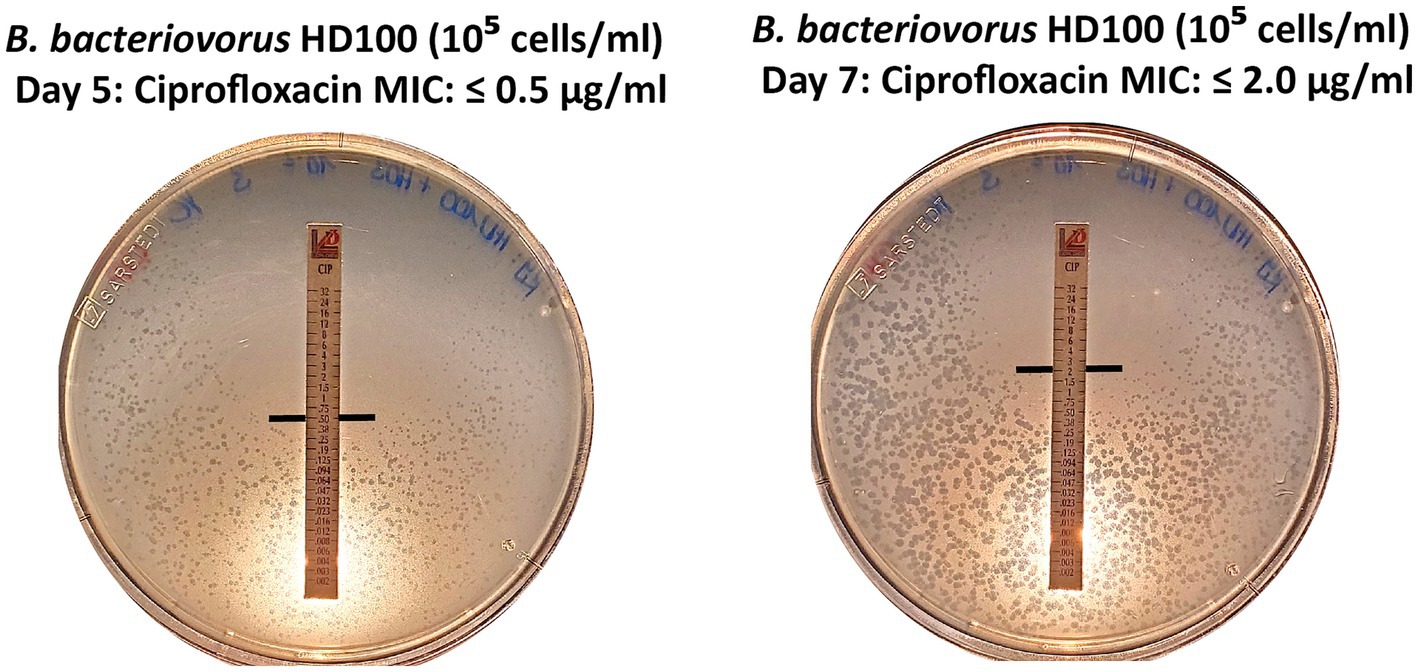
Figure 6. Extended incubation by 48 h yields a 4-fold increase in MIC value. For B. bacteriovorus HD100, the MIC intercept was determined at 0.5 μg/mL after five days and increased to 2 μg/mL after 7 days for ciprofloxacin.
3.4 Susceptible prey limits MIC determination for plaque-forming predatory bacteria
Meropenem is a carbapenem antibiotic frequently employed to treat bacteria resistant to other β-lactam antibiotics, such as the multidrug-resistant P. aeruginosa strain H03. This strain has been previously reported to exhibit susceptibility to meropenem at elevated concentrations (Tueffers et al., 2024). In line with this, the growth of P. aeruginosa H03 was visibly inhibited at higher antibiotic concentrations (> 0.25 μg/mL). Notably, B. bacteriovorus HD100 formed plaques throughout the lawn of P. aeruginosa H03. Therefore, a clear MIC intercept for B. bacteriovorus could not be determined using P. aeruginosa H03 (Figure 7).
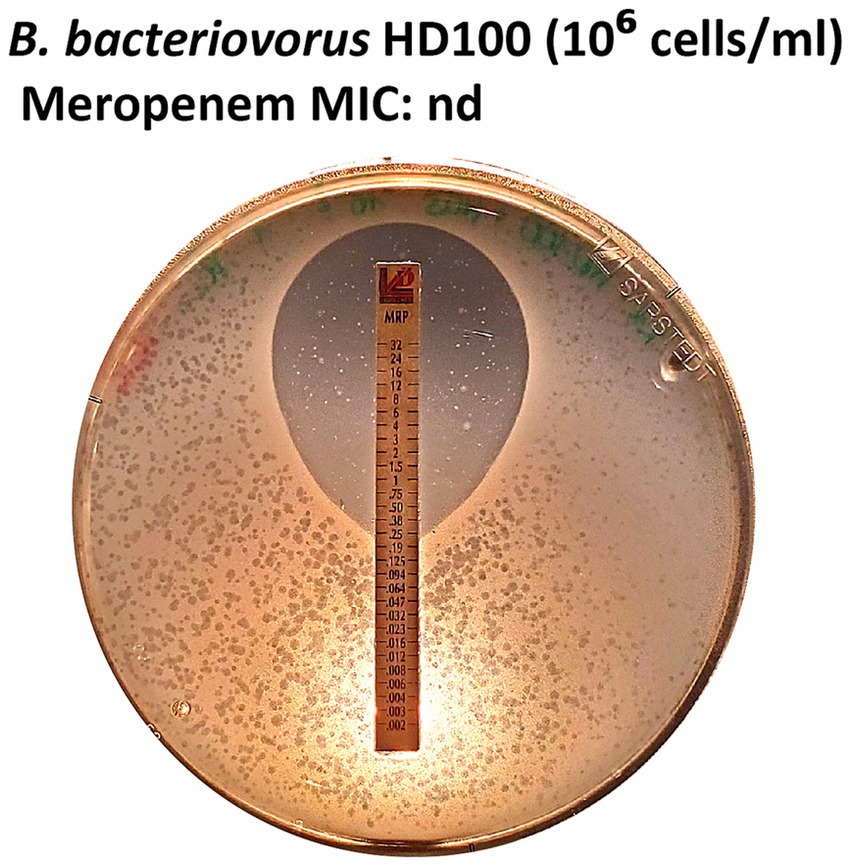
Figure 7. MIC not determinable for meropenem E-tests. The growth of P. aeruginosa H03 was inhibited at antibiotic concentrations > 0.25 μg/mL, while B. bacteriovorus HD100 formed plaques around the inhibition zone.
3.5 Challenges and suggestions
The proposed assay shows great potential for determining MIC values for plaque-forming predatory bacteria when accounting for a few challenges (Table 5). Small variations in determined MIC values were linked to predator inoculum concentration and plaque plate incubation time and are further discussed in the next sections.
4 Discussion
4.1 E-tests on plaque assay plates are a suitable method to determine the AMR of plaque-forming predatory bacteria
In this study, we propose a streamlined method to determine the AMR in plaque-forming predatory bacteria. We show that prey choice, inoculum density, and experimental conditions affect the determination of AMR in Bdellovibrio.
In detail, we used E. coli ML35 as prey for Bdellovibrio inoculum concentration determination, as plates seeded with this prey yielded higher numbers of Bdellovibrio plaques than those seeded with P. aeruginosa H03. This difference may be linked to the production of various antimicrobial compounds to evade predation. For instance, like many bacteria utilizing the quorum sensing (QS) system, P. aeruginosa produces diffusible signaling factors (DSF) (Twomey et al., 2012), that can delay predation and compromise membrane integrity in B. bacteriovorus (Dwidar et al., 2021). A recent study showed that the production of another QS molecule, 2-heptyl-3hydroxy-4(1H)-quinolone (PQS), was toxic to B. bacteriovorus (Hoshiko et al., 2021). Additionally, P. aeruginosa synthesizes toxic compounds such as pyocyanin, a phenazine pigment, and cyanides, which have been shown to reduce B. bacteriovorus motility rates and inhibit its predatory activity (Mun et al., 2017). Despite this potential multi-layered line of defense, predation through B. bacteriovorus is not inhibited, yet decreased for P. aeruginosa compared to E. coli (Supplementary Figure S1). While the effect of pyocyanin on Bdellovibrio motility is unlikely to influence Bdellovibrio MIC, intraperiplasmic Bdellovibrio cells may be affected. Mun et al. (2017) demonstrated that bdelloplasts exposed to cyanides exhibit inhibited development, making them likely less metabolically active and, hence, potentially taking in fewer antibiotics that require active transport.
Beyond antimicrobial defenses, inoculum density also impacts predation dynamics. Several studies link inoculum concentration to MIC values (inoculum effect) in colony-forming bacteria (García-Rivera et al., 2024; Smith and Kirby, 2018). Our findings indicate a similar effect in plaque-forming predatory bacteria, underscoring the need for standardized inoculum densities for reliable MIC results. For consistent, reproducible MIC readouts, it is recommended to use a standardized optical density for colony-forming bacteria when applying MIC E-tests. Accordingly, we strongly recommend testing respective plaque-forming predatory bacteria using standardized cell densities, for instance by fluorescence-based inoculum quantification, as suggested by Remy et al. (2022), or by PFU/ml determination using plaque assays. Ideally, the standardized cell densities should align with the requirements of subsequent experimental assays for which the MICs were determined.
Further, when selecting a prey strain, it is crucial to consider prey susceptibility to applied antibiotics, which can affect MIC determination for plaque-forming predatory bacteria, as seen for meropenem in this study, highlighting the importance of careful prey choice. P. aeruginosa strain H03 has been previously reported to exhibit susceptibility to meropenem at elevated concentrations (Tueffers et al., 2024) in line with the results of our study (> 0.25 μg/mL, Figure 7). In turn, meropenem was excluded from further testing, as no definite MIC intercept could be determined for the plaque-forming predatory bacteria. Prey strains should be selected for resistance to the antibiotics being tested, e.g., strains resistant to meropenem when assessing meropenem resistance in plaque-forming predatory bacteria. Representative Pseudomonas aeruginosa strain panels may be consulted to identify suitable candidates (Tueffers et al., 2024). One of the advantages of our proposed method is that this limitation is easily detectable, as prey lawn inhibition can be directly observed on double agar plates.
Importantly, prey cells may release enzymes that modify or degrade antibiotics, potentially leading to an underestimation of MIC values for the plaque-forming predatory bacteria applied. Therefore, we advise using multi-resistant prey bacteria, for which resistance does not rely on secreted enzymes, but on other resistance mechanisms (e.g., target modification or efflux). In addition, incubation conditions are crucial, particularly due to the extended periods required for plaque formation. Prolonged incubation can impact antibiotic stability, independent of prey activity (Lallemand et al., 2016). For instance, P. aeruginosa H03 encodes genes for aminoglycoside-modifying enzymes (aph(3′)-II, aph(6)-Ic, and aac(6’)Ib4) that may lower gentamicin concentration over prolonged exposure (Tueffers et al., 2024) and thus lead to an overestimation of the predator MIC value. Additionally, extended incubation under selective pressure increases the likelihood of resistant mutants. P. aeruginosa produces bactericidal compounds such as pyocyanin, DSF, and PQS (Hoshiko et al., 2021; Twomey et al., 2012), which may select for resistant Bdellovibrio cells. Hence, small, scattered plaques observed in the periphery of the inhibition zone should be interpreted cautiously. Plaques occurring at the edge of the inhibition zone after longer incubation may be more resistant cells. We observed these cases only for the drug ciprofloxacin which is a known SOS-response inducer that can lead to hypermutation, suggesting that these plaques are indeed resistant BALOs (Crane et al., 2021). In general, MICs determined with these assays should always be confirmed within agar dilution if precise MICs are necessary. We recommend to disregard microcolonies within the inhibition zone as these are likely resistant mutants and determine the MIC on the edge of the ellipse after a standardized incubation period.
4.2 Bdellovibrio AMR profiles reveal ciprofloxacin to be a suitable candidate for combined therapy
After establishing the robustness of the method, we applied it in a preliminary test on Bdellovibrio tiberii MYbb2, a close relative of B. bacteriovorus HD100 (Maher et al., 2025). Although only tested in a single biological replicate, MIC values for MYbb2 (CFZ: 0.021 μg/mL, TZP: ≤ 0.016 μg/mL, CIP: 1.667 μg/mL, CN: 0.032 μg/mL) were within the range observed for HD100. These initial results suggest that the method may be applicable to related species, though further testing is required to confirm reproducibility and species-specific differences. Both tested strains exhibit high resistance to ciprofloxacin while showing high susceptibility to piperacillin/tazobactam. Similarly, a previous study on the antibiotic resistance of another Bdellovibrio species, the epibiotic B. exovorus JSS, reported high resistance to ciprofloxacin. Interestingly, B. exovorus JSS presented higher resistance to piperacillin/tazobactam than to gentamicin (Pasternak et al., 2014). This indicates that distinct Bdellovibrio species have distinct susceptibility patterns. More information on the different species’ AMR profiles could provide valuable insights for effective combination therapies of plaque-forming predatory bacteria and antibiotics. Additionally, the proposed assay can be used to identify clinical breakpoints for Bdellovibrio. These thresholds, defined by the European Committee of Antimicrobial Susceptibility Testing (EUCAST), determine antibiotic resistance and are crucial in the context of combination therapy. We want to emphasize that the here proposed assay should also be adapted to other gradient-based MIC methods, such as the disc diffusion assay. While they do not yield precise MIC values, which are critical for experimental assays, they may offer a cost-effective alternative for high-throughput screening. Ideally, a suitable antibiotic for combined therapy would be one for which the pathogen is susceptible within clinical dosing limits but against which B. bacteriovorus is resistant.
In this context, piperacillin might be an interesting candidate antibiotic for combination therapy, as B. bacteriovorus HD100 demonstrated full resistance at the highest concentrations. However, if this resistance is mediated by excreted β-lactamases, as indicated by the loss of resistance after addition of β-lactamase inhibitors, these enzymes may function as a public good, potentially protecting the targeted pathogen as well. Future work should measure β-lactamase activity in predator supernatants to confirm this mechanism. Therefore, the full mechanism of resistance needs to be investigated to understand the potential of piperacillin-based combination therapies.
Thus, in this study, ciprofloxacin was determined as the most promising candidate for combination therapy with B. bacteriovorus HD100 against P. aeruginosa, as the predator showed high resistance to this antibiotic (MIC 1.306 ± 0.34 μg/mL for 106 PFU/ml and MIC 2.333 ± 0.833 μg/mL for 107 PFU/ml). However, effective therapy requires consideration of in vivo antibiotic concentrations. Pharmacokinetic studies of ciprofloxacin have shown maximum plasma concentrations of 2.5 ± 1.1 μg/mL and sputum concentrations of 1.8 ± 1.0 μg/mL during standard oral dosage (Begg et al., 2000). Given that these levels exceed the average MIC values observed for B. bacteriovorus HD100 (0.898 ± 0.191 μg/mL), high PFU/ml concentrations of B. bacteriovorus may be required to ensure compatibility with standard oral ciprofloxacin dosages. Alternatively, lower ciprofloxacin doses could still be effective when used in combination with Bdellovibrio. Finally, introducing plaque-forming predatory bacteria to the human body alongside antibiotics should prompt multiple considerations, such as their predation efficacy in physiologically complex conditions. While its clinical application potential in humans is still unknown, Bdellovibrio is non-pathogenic in human serum and epithelial cell cultures (Baker et al., 2017; Gupta et al., 2016; Monnappa et al., 2016; Raghunathan et al., 2019) and induces only mild inflammatory responses in human cell lines (Bonfiglio et al., 2019). Although ineffective in intravenous application (Shatzkes et al., 2016), studies have already shown promising results in this regard for B. bacteriovorus against pathogens upon oral administration in animal models (Atterbury et al., 2011; Findlay et al., 2019; Shatzkes et al., 2016; Willis et al., 2016) and in human serum cell lines (Baker et al., 2017; Gupta et al., 2016; Monnappa et al., 2016; Raghunathan et al., 2019), although without antibiotic co-administration. Future research could build on our new assay to investigate combined therapies in vivo, offering a deeper understanding of potential clinical applications.
5 Conclusion
The here proposed streamlined three-step protocol to measure AMR in plaque-forming predatory bacteria proved effective and robust when using a suitable prey. Our plate-based assay using antibiotic test strips (e.g., E-tests) enables direct readout and removes the need for liquid culture transfers (Figure 2). With this study, our goal was to help lay the groundwork for future clinical applications as the basis for combined treatment studies of antibiotics and plaque-forming predatory bacteria. While promising, these results are preliminary; in vivo validation and regulatory studies are necessary.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
JW: Conceptualization, Formal analysis, Investigation, Methodology, Supervision, Visualization, Writing – original draft, Writing – review & editing. YC: Data curation, Formal analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing. FB: Conceptualization, Funding acquisition, Methodology, Supervision, Writing – original draft, Writing – review & editing. HS: Conceptualization, Funding acquisition, Writing – original draft, Writing – review & editing. JJ: Conceptualization, Funding acquisition, Methodology, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. We are grateful for financial support from the German Research Foundation (Deutsche Forschungsgemeinschaft, DFG) within project JO1786/1-1 (to JJ) and the Research and Training Group (RTG) 2501 on Translational Evolutionary Research (project 4.2 to HS and to FB). We further acknowledge funding by the Max-Planck Society (fellowship to HS). We are grateful for funding from the Damp Foundation (Damp Stiftung) within the SKILLED project (to HS).
Acknowledgments
We thank Una Steinleitner for her valuable contributions to the initial experiments that guided the development of the methodology used in this study and the Schulenburg lab for general advice.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that Gen AI was used in the creation of this manuscript. AI-based tools were used solely for language editing (grammar and readability).
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1582371/full#supplementary-material
Footnotes
References
Andersson, D. I., Balaban, N. Q., Baquero, F., Courvalin, P., Glaser, P., Gophna, U., et al. (2020). Antibiotic resistance: turning evolutionary principles into clinical reality. FEMS Microbiol. Rev. 44, 171–188. doi: 10.1093/femsre/fuaa001
Atterbury, R. J., Hobley, L., Till, R., Lambert, C., Capeness, M. J., Lerner, T. R., et al. (2011). Effects of orally administered Bdellovibrio bacteriovorus on the well-being and Salmonella colonization of young chicks. Appl. Environ. Microbiol. 77, 5794–5803. doi: 10.1128/AEM.00426-11
Baker, M., Negus, D., Raghunathan, D., Radford, P., Moore, C., Clark, G., et al. (2017). Measuring and modelling the response of Klebsiella pneumoniae KPC prey to Bdellovibrio bacteriovorus predation, in human serum and defined buffer. Sci. Rep. 7:8329. doi: 10.1038/s41598-017-08060-4
Bates, D., Mächler, M., Bolker, B., and Walker, S. (2015). Fitting linear mixed-effects models using lme4. J. Stat. Softw. 67, 1–48. doi: 10.18637/jss.v067.i01
Begg, E. J., Robson, R. A., Saunders, D. A., Graham, G. G., Buttimore, R. C., Neill, A. M., et al. (2000). The pharmacokinetics of oral fleroxacin and ciprofloxacin in plasma and sputum during acute and chronic dosing. Br. J. Clin. Pharmacol. 49, 32–38. doi: 10.1046/j.1365-2125.2000.00105.x
Bonfiglio, G., Neroni, B., Radocchia, G., Pompilio, A., Mura, F., Trancassini, M., et al. (2019). Growth control of adherent-invasive Escherichia coli (AIEC) by the predator bacteria Bdellovibrio bacteriovorus: a new therapeutic approach for Crohn’s disease patients. Microorganisms 8:17. doi: 10.3390/microorganisms8010017
Buttin, G., Cohen, G. N., Monod, J., and Rickenberg, H. V. (1956). Galactoside-permease of Escherichia coli. Ann. Inst. Pasteur 91, 829–857.
Casale, A., Clark, S., Grasso, M., Kryschuk, M., Ritzer, L., Trudeau, M., et al. (2018). Complete genome sequence of Escherichia coli ML35. Genome Announc. 6:e00034-18. doi: 10.1128/genomeA.00034-18
Cavallo, F. M., Jordana, L., Friedrich, A. W., Glasner, C., and van Dijl, J. M. (2021). Bdellovibrio bacteriovorus: a potential ‘living antibiotic’ to control bacterial pathogens. Crit. Rev. Microbiol. 47, 630–646. doi: 10.1080/1040841X.2021.1908956
Cheng, L., Nelson, B. C., Mehta, M., Seval, N., Park, S., Giddins, M. J., et al. (2017). Piperacillin-tazobactam versus other antibacterial agents for treatment of bloodstream infections due to AmpC β-lactamase-producing Enterobacteriaceae. Antimicrob. Agents Chemother. 61:e00276-17. doi: 10.1128/AAC.00276-17
Cramer, N., Wiehlmann, L., Ciofu, O., Tamm, S., Høiby, N., and Tümmler, B. (2012). Molecular epidemiology of chronic Pseudomonas aeruginosa airway infections in cystic fibrosis. PLoS One 7:e50731. doi: 10.1371/journal.pone.0050731
Crane, J. K., Alvarado, C. L., and Sutton, M. D. (2021). Role of the SOS response in the generation of antibiotic resistance in vivo. Antimicrob. Agents Chemother. 65:21. doi: 10.1128/aac.00013-21
Das, S. K., and Negus, D. (2024). How do gram-negative bacteria escape predation by Bdellovibrio bacteriovorus? NPJ Antimicrobial Resistance 2, 1–8. doi: 10.1038/s44259-024-00048-1
De Oliveira, D. M. P., Forde, B. M., Kidd, T. J., Harris, P. N. A., Schembri, M. A., Beatson, S. A., et al. (2020). Antimicrobial resistance in ESKAPE pathogens. Clin. Microbiol. Rev. 33, e00181–e00119. doi: 10.1128/CMR.00181-19
Djordjevic, S. P., Jarocki, V. M., Seemann, T., Cummins, M. L., Watt, A. E., Drigo, B., et al. (2024). Genomic surveillance for antimicrobial resistance — a one health perspective. Nat. Rev. Genet. 25, 142–157. doi: 10.1038/s41576-023-00649-y
Dufour, N., Clermont, O., La Combe, B., Messika, J., Dion, S., Khanna, V., et al. (2016). Bacteriophage LM33_P1, a fast-acting weapon against the pandemic ST131-O25b:H4 Escherichia coli clonal complex. J. Antimicrob. Chemother. 71, 3072–3080. doi: 10.1093/jac/dkw253
Dwidar, M., Jang, H., Sangwan, N., Mun, W., Im, H., Yoon, S., et al. (2021). Diffusible signaling factor, a quorum-sensing molecule, interferes with and is toxic towards Bdellovibrio bacteriovorus 109J. Microb. Ecol. 81, 347–356. doi: 10.1007/s00248-020-01585-8
Findlay, J. S., Flick-Smith, H. C., Keyser, E., Cooper, I. A., Williamson, E. D., and Oyston, P. C. F. (2019). Predatory bacteria can protect SKH-1 mice from a lethal plague challenge. Sci. Rep. 9:7225. doi: 10.1038/s41598-019-43467-1
Galtier, M., De Sordi, L., Sivignon, A., de Vallée, A., Maura, D., Neut, C., et al. (2017). Bacteriophages targeting adherent invasive Escherichia coli strains as a promising new treatment for Crohn’s disease. J. Crohn's Colitis 11, 840–847. doi: 10.1093/ecco-jcc/jjw224
García-Rivera, C., Sánchez-Bautista, A., Parra-Grande, M., Ricart-Silvestre, A., Ventero, M. P., Tyshkovska, I., et al. (2024). Comparison of different methods for assaying the in vitro activity of cefiderocol against carbapenem-resistant Pseudomonas aeruginosa strains: influence of bacterial inoculum. Antibiotics 13:663. doi: 10.3390/antibiotics13070663
Gupta, S., Tang, C., Tran, M., and Kadouri, D. E. (2016). Effect of predatory bacteria on human cell lines. PLoS One 11:e0161242. doi: 10.1371/journal.pone.0161242
Hilker, R., Munder, A., Klockgether, J., Losada, P. M., Chouvarine, P., Cramer, N., et al. (2015). Interclonal gradient of virulence in the Pseudomonas aeruginosa pangenome from disease and environment. Environ. Microbiol. 17, 29–46. doi: 10.1111/1462-2920.12606
Hoshiko, Y., Nishiyama, Y., Moriya, T., Kadokami, K., López-Jácome, L. E., Hirano, R., et al. (2021). Quinolone signals related to Pseudomonas quinolone signal-quorum sensing inhibits the predatory activity of Bdellovibrio bacteriovorus. Front. Microbiol. 12:722579. doi: 10.3389/fmicb.2021.722579
Im, H., Choi, S. Y., Son, S., and Mitchell, R. J. (2017). Combined application of bacterial predation and violacein to kill polymicrobial pathogenic sommunities. Sci. Rep. 7:14415. doi: 10.1038/s41598-017-14567-7
Jault, P., Leclerc, T., Jennes, S., Pirnay, J. P., Que, Y.-A., Resch, G., et al. (2019). Efficacy and tolerability of a cocktail of bacteriophages to treat burn wounds infected by Pseudomonas aeruginosa (PhagoBurn): a randomised, controlled, double-blind phase 1/2 trial. Lancet Infect. Dis. 19, 35–45. doi: 10.1016/S1473-3099(18)30482-1
Jurkevitch, E. (2012). Isolation and classification of Bdellovibrio and like organisms. Curr. Protoc. Microbiol. 26:7B.1.1-7B.1.20. doi: 10.1002/9780471729259.mc07b01s26
Kadeřábková, N., Mahmood, A. J. S., and Mavridou, D. A. I. (2024). Antibiotic susceptibility testing using minimum inhibitory concentration (MIC) assays. NPJ Antimicrob Resist 2, 1–9. doi: 10.1038/s44259-024-00051-6
Kadouri, D., and O’Toole, G. A. (2005). Susceptibility of biofilms to Bdellovibrio bacteriovorus attack. Appl. Environ. Microbiol. 71, 4044–4051. doi: 10.1128/AEM.71.7.4044-4051.2005
Kortright, K. E., Chan, B. K., Koff, J. L., and Turner, P. E. (2019). Phage therapy: a renewed approach to combat antibiotic-resistant bacteria. Cell Host Microbe 25, 219–232. doi: 10.1016/j.chom.2019.01.014
Kowalska-Krochmal, B., and Dudek-Wicher, R. (2021). The minimum inhibitory concentration of antibiotics: methods, interpretation, clinical eelevance. Pathogens 10:165. doi: 10.3390/pathogens10020165
Lallemand, E. A., Lacroix, M. Z., Toutain, P.-L., Boullier, S., Ferran, A. A., and Bousquet-Melou, A. (2016). In vitro degradation of antimicrobials during use of broth microdilution method can increase the measured minimal inhibitory and minimal bactericidal concentrations. Front. Microbiol. 7:2051. doi: 10.3389/fmicb.2016.02051
Laloux, G. (2020). Shedding light on the cell biology of the predatory bacterium Bdellovibrio bacteriovorus. Front. Microbiol. 10:3136. doi: 10.3389/fmicb.2019.03136
Maher, R. L., Wülbern, J., Zimmermann, J., Yeh, E., Benda, L., Repnik, U., et al. (2025). Comparative analysis of novel Pseudobdellovibrionaceae genera and species yields insights into the genomics and evolution of bacterial predation mode. Biorxiv. doi: 10.1101/2025.02.19.638989
Marine, E., Milner, D. S., Lambert, C., Sockett, R. E., and Pos, K. M. (2020). A novel method to determine antibiotic sensitivity in Bdellovibrio bacteriovorus reveals a DHFR-dependent natural trimethoprim resistance. Sci. Rep. 10:5315. doi: 10.1038/s41598-020-62014-x
Matan, O., and Jurkevitch, E. (2022). Predation of antibiotic persister bacteria by the predatory bacterium Bdellovibrio bacteriovorus. Environ. Microbiol. Rep. 14, 239–244. doi: 10.1111/1758-2229.13054
Matin, A., and Rittenberg, S. C. (1972). Kinetics of deoxyribonucleic acid destruction and synthesis during growth of Bdellovibrio bacteriovorus strain 109D on Pseudomonas putida and Escherichia coli. J. Bacteriol. 111, 664–673. doi: 10.1128/jb.111.3.664-673.1972
Michael, C. A., Dominey-Howes, D., and Labbate, M. (2014). The antimicrobial resistance crisis: causes, consequences, and management. Frontiers. Public Health 2:145. doi: 10.3389/fpubh.2014.00145
Monnappa, A. K., Bari, W., Choi, S. Y., and Mitchell, R. J. (2016). Investigating the responses of human epithelial cells to predatory bacteria. Sci. Rep. 6:33485. doi: 10.1038/srep33485
Monnappa, A. K., Dwidar, M., and Mitchell, R. J. (2013). Application of bacterial predation to mitigate recombinant bacterial populations and their DNA. Soil Biol. Biochem. 57, 427–435. doi: 10.1016/j.soilbio.2012.09.010
Mun, W., Kwon, H., Im, H., Choi, S. Y., Monnappa, A. K., and Mitchell, R. J. (2017). Cyanide production by Chromobacterium piscinae shields it from Bdellovibrio bacteriovorus HD100 predation. MBio 8:e01370-17. doi: 10.1128/mBio.01370-17
Negus, D., Baker, M., Raghunathan, D., Tyson, J., and Sockett, R. E. (2017). Predator versus pathogen: how does predatory Bdellovibrio bacteriovorus interface with the challenges of killing gram-negative pathogens in a host setting? Ann. Rev. Microbiol. 71, 441–457. doi: 10.1146/annurev-micro-090816-093618
Núñez, M. E., Martin, M. O., Chan, P. H., and Spain, E. M. (2005). Predation, death, and survival in a biofilm: Bdellovibrio investigated by atomic force microscopy. Colloids Surf. B Biointerfaces 42, 263–271. doi: 10.1016/j.colsurfb.2005.03.003
Pasternak, Z., Njagi, M., Shani, Y., Chanyi, R., Rotem, O., Lurie-Weinberger, M. N., et al. (2014). In and out: an analysis of epibiotic vs periplasmic bacterial predators. ISME J. 8, 625–635. doi: 10.1038/ismej.2013.164
R Core Team (2023). R: A language and environment for statistical computing. Vienna: R Foundation for Statistical Computing.
Raghunathan, D., Radford, P. M., Gell, C., Negus, D., Moore, C., Till, R., et al. (2019). Engulfment, persistence and fate of Bdellovibrio bacteriovorus predators inside human phagocytic cells informs their future therapeutic potential. Sci. Rep. 9:4293. doi: 10.1038/s41598-019-40223-3
Rahme, L. G., Stevens, E. J., Wolfort, S. F., Shao, J., Tompkins, R. G., and Ausubel, F. M. (1995). Common virulence factors for bacterial pathogenicity in plants and animals. Science 268, 1899–1902. doi: 10.1126/science.7604262
Remy, O., Lamot, T., Santin, Y., Kaljević, J., De Pierpont, C., and Laloux, G. (2022). An optimized workflow to measure bacterial predation in microplates. STAR Protoc. 3:101104. doi: 10.1016/j.xpro.2021.101104
Rendulic, S., Jagtap, P., Rosinus, A., Eppinger, M., Baar, C., Lanz, C., et al. (2004). A predator unmasked: life cycle of Bdellovibrio bacteriovorus from a genomic perspective. Science 303, 689–692. doi: 10.1126/science.1093027
Roemhild, R., and Schulenburg, H. (2019). Evolutionary ecology meets the antibiotic crisis. Evol. Med. Public Health 2019, 37–45. doi: 10.1093/emph/eoz008
Sanz-García, F., Gil-Gil, T., Laborda, P., Blanco, P., Ochoa-Sánchez, L.-E., Baquero, F., et al. (2023). Translating eco-evolutionary biology into therapy to tackle antibiotic resistance. Nat. Rev. Microbiol. 21, 671–685. doi: 10.1038/s41579-023-00902-5
Shatzkes, K., Singleton, E., Tang, C., Zuena, M., Shukla, S., Gupta, S., et al. (2016). Predatory bacteria attenuate Klebsiella pneumoniae burden in rat lungs. MBio 7:e01847-16. doi: 10.1128/mBio.01847-16
Shemesh, Y., and Jurkevitch, E. (2004). Plastic phenotypic resistance to predation by Bdellovibrio and like organisms in bacterial prey. Environ. Microbiol. 6, 12–18. doi: 10.1046/j.1462-2920.2003.00530.x
Smith, K. P., and Kirby, J. E. (2018). The inoculum effect in the era of multidrug resistance: minor differences in inoculum have dramatic effect on MIC determination. Antimicrob. Agents Chemother. 62:e00433-18. doi: 10.1128/AAC.00433-18
Stewart, P. S., and Costerton, J. W. (2001). Antibiotic resistance of bacteria in biofilms. Lancet 358, 135–138. doi: 10.1016/S0140-6736(01)05321-1
Tueffers, L., Batra, A., Zimmermann, J., Botelho, J., Buchholz, F., Mendoza Mejía, N., et al. (2024). Variation in the response to antibiotics and life-history across the major Pseudomonas aeruginosa clone type (mPact) panel. Microbiol. Spectr. 12:e00143-24. doi: 10.1128/spectrum.00143-24
Twomey, K. B., O’Connell, O. J., McCarthy, Y., Dow, J. M., O’Toole, G. A., Plant, B. J., et al. (2012). Bacterial cis-2-unsaturated fatty acids found in the cystic fibrosis airway modulate virulence and persistence of Pseudomonas aeruginosa. ISME J. 6, 939–950. doi: 10.1038/ismej.2011.167
Udekwu, K. I., Parrish, N., Ankomah, P., Baquero, F., and Levin, B. R. (2009). Functional relationship between bacterial cell density and the efficacy of antibiotics. J. Antimicrob. Chemother. 63, 745–757. doi: 10.1093/jac/dkn554
Wiehlmann, L., Wagner, G., Cramer, N., Siebert, B., Gudowius, P., Morales, G., et al. (2007). Population structure of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. 104, 8101–8106. doi: 10.1073/pnas.0609213104
Willis, A. R., Moore, C., Mazon-Moya, M., Krokowski, S., Lambert, C., Till, R., et al. (2016). Injections of predatory bacteria work alongside host immune cells to treat Shigella infection in zebrafish larvae. Curr. Biol. 26, 3343–3351. doi: 10.1016/j.cub.2016.09.067
Wülbern, J. (2025). Figures 1–3. Created in BioRender. Available at: https://BioRender.com/6ybxp6x
Yen, M., Cairns, L. S., and Camilli, A. (2017). A cocktail of three virulent bacteriophages prevents Vibrio cholerae infection in animal models. Nat. Commun. 8:14187. doi: 10.1038/ncomms14187
Keywords: Bdellovibrio, antibiotic, plaque assay, E-tests, antibiotic resistance, BALOs, Pseudomonas aeruginosa
Citation: Wülbern J, Carstensen Y, Buchholz F, Schulenburg H and Johnke J (2025) A streamlined method to determine the antibiotic resistance of plaque-forming predatory bacteria. Front. Microbiol. 16:1582371. doi: 10.3389/fmicb.2025.1582371
Edited by:
John Osei Sekyere, University of Pretoria, South AfricaReviewed by:
Barathan Muttiah, University of Malaya, MalaysiaAhmed Elfadadny, Tokyo University of Agriculture and Technology, Japan
Esraa Halawa, Cairo University, Egypt
Copyright © 2025 Wülbern, Carstensen, Buchholz, Schulenburg and Johnke. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Julia Johnke, ampvaG5rZUB6b29sb2dpZS51bmkta2llbC5kZQ==
†These authors have contributed equally to this work and share first authorship
 Janna Wülbern
Janna Wülbern Yvonne Carstensen
Yvonne Carstensen Florian Buchholz
Florian Buchholz Hinrich Schulenburg
Hinrich Schulenburg Julia Johnke
Julia Johnke