- 1Institute of Plant and Microbial Biology, Academia Sinica, Taipei, Taiwan
- 2Department of Tropical Agriculture and International Cooperation, National Pingtung University of Science and Technology, Pingtung City, Taiwan
- 3Bioinformatics Program, Taiwan International Graduate Program, National Taiwan University, Taipei, Taiwan
- 4Bioinformatics Program, Institute of Statistical Science, Taiwan International Graduate Program, Academia Sinica, Taipei, Taiwan
- 5Genome and Systems Biology Degree Program, National Taiwan University and Academia Sinica, Taipei, Taiwan
- 6Department of Life Science, National Taiwan University, Taipei, Taiwan
- 7Institute of Plant Biology, National Taiwan University, Taipei, Taiwan
Lignocellulose biomass is one of the most abundant resources for sustainable biofuels. However, scaling up the biomass-to-biofuels conversion process for widespread usage is still pending. One of the main bottlenecks is the high cost of enzymes used in key process of biomass degradation. Current research efforts are therefore targeted at creative solutions to improve the feasibility of lignocellulosic-degrading enzymes. One way is to engineer multi-enzyme complexes that mimic the bacterial cellulosomal system, known to increase degradation efficiency up to 50-fold when compared to freely-secreted enzymes. However, these designer cellulosomes are instable and less efficient than wild type cellulosomes. In this review, we aim to extensively analyze the current knowledge on the lignocellulosic-degrading enzymes through three aspects. We start by reviewing and comparing sets of enzymes in bacterial and fungal lignocellulose degradation. Next, we focus on the characteristics of cellulosomes in both systems and their feasibility to be engineered. Finally, we highlight three key strategies to enhance enzymatic lignocellulose degradation efficiency: discovering novel lignocellulolytic species and enzymes, bioengineering enzymes for improved thermostability, and structurally optimizing designer cellulosomes. We anticipate these insights to act as resources for the biomass community looking to elevate the usage of lignocellulose as biofuel.
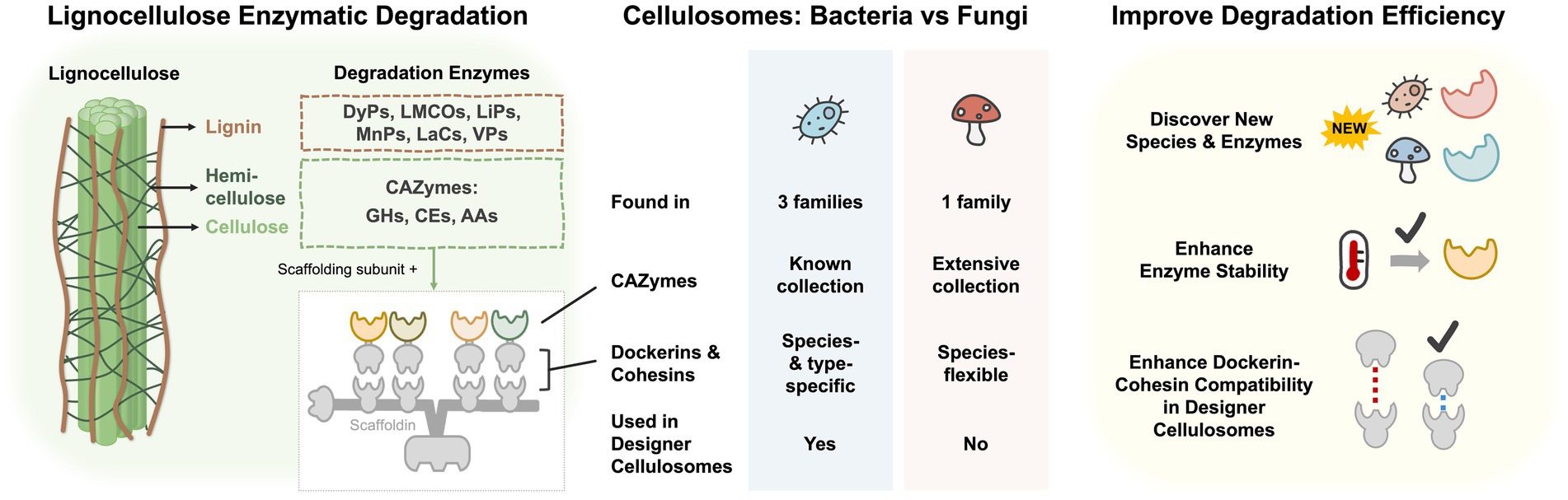
Graphical Abstract. Overview of lignocellulose enzymatic degradation in bacteria and fungi. The left panel shows the degradation enzymes of lignocellulose. Cellulosome is composed of multiple CAZymes and the scaffolding subunit containing scaffoldin, dockerins, cohesins, and carbohydrate-binding module. The middle panel indicates the differences between cellulosomes of bacteria and fungi. The right panel is the potential methods to improve lignocellulose degradation efficiency. DyPs, dye-depolymerizing peroxidases; LMCOs, laccase and laccase-like multi-copper oxidases; LiPs, lignin peroxidases; MnPs, manganese-dependent peroxidases; Lacs, laccases; VPs, versatile peroxidases; CAZymes, carbohydrate-active enzymes; GH, glycoside hydrolase; CE, carbohydrate esterase; AA, auxiliary activity families.
1 Introduction
Plant biomass is a resource of renewable energy that is made up of byproducts from agriculture, forestry and the food supply chain. Some examples include wheat straw, sugarcane bagasse, cereal husk and tree bark (Talebnia et al., 2010; Binod et al., 2012; Elmekawy et al., 2013; Jablonsky et al., 2017). With its abundant availability and regenerative nature, plant biomass is a promising candidate to replace fossil fuels as an energy source. Global land biomass was estimated at 1.8 trillion tons in 2018, including 140 billion tons of plant wastes produced annually (Tursi, 2019), altogether enough for world’s annual consumption over 80 times (Anonymous, 2018). However, this resource remains under-exploited. In 2023, biomass energy only made up of about 5% of total energy production globally (Anonymous, 2023).
The main barrier for widespread use of plant biomass energy is the low economic feasibility of upscaling the energy conversion process. Two factors contribute to the cost economics of biomass degradation:
1. The need for pre-treatment, a process to reduce recalcitrance of lignocellulose through thermochemical or physical methods, contributes to about 20% of total cost of production (Yang and Wyman, 2008). Acid hydrolysis and pyrolysis are two common pre-treatments, where concentrated acids and high temperatures ranging from 300 to 800°C are used. Both of these approaches have been proven to be highly efficient, for example, the use of acid hydrolysis pre-treatment prior to enzymatic hydrolysis increases raw poplar wood degradation by 14 times (Antczak et al., 2018). However, these methods impose high costs as they are energy-intensive and may require downstream neutralization of toxic byproducts (Chen et al., 2014).
2. Lignocellulosic-degrading enzymes which perform the key hydrolysis process in breaking down lignocellulose components to yield glucose are highly priced (Singh et al., 2016; Musa and Elnour, 2024). Industrial-grade enzymes have low availability as large amounts of substrates are needed for their production (Bhatia et al., 2021). Although the cost of cellulase enzymes depends on factors like the type of feedstock and enzyme dosage, they can contribute up to 48% of the total cost of producing ethanol from lignocellulose (Liu et al., 2016). In addition, effective biomass degradation requires high concentration of enzymes due to prolonged hydrolysis time (Singhvi and Kim, 2020).
Besides economic feasibility, issues such as substrate specificity of enzymes and the need for optimization of enzymatic cocktails require labor-intensive solutions, further limiting the potential for upscaling biomass usage (Siqueira et al., 2020; Wingren et al., 2003; Hu et al., 2023; Méndez Arias et al., 2016).
1.1 The need for cost-effective enzymatic systems
Extensive ongoing research in multiple fields is aimed at designing a cost-effective enzymatic system which requires minimum pre-treatment and lowering the costs of degrading enzymes. For example, developing new pre-treatment methods for recalcitrant components (Wang et al., 2013), simplifying enzyme purification methods and logistics (Takimura et al., 2013; Nhim et al., 2024), and modification of enzyme properties such as activity and thermostability through protein engineering methods (Bloom et al., 2005).
The graphical abstract depicts three aspects of lignocellulosic-degrading enzymes that we are focusing on in this review. First, we characterized the sets of enzymes naturally involved in bacterial and fungal lignocellulose degradation as shown in the left panel of the graphical abstract. In the middle panel, we explore the characteristics of a highly-efficient multi-enzyme complex termed cellulosome in bacterial and fungal systems, and their feasibility to be engineered. Thirdly, as shown in the right panel, we describe three main directions in enhancing enzymatic degradation efficiency in biomass usage.
2 Enzymatic degradation of lignocellulose
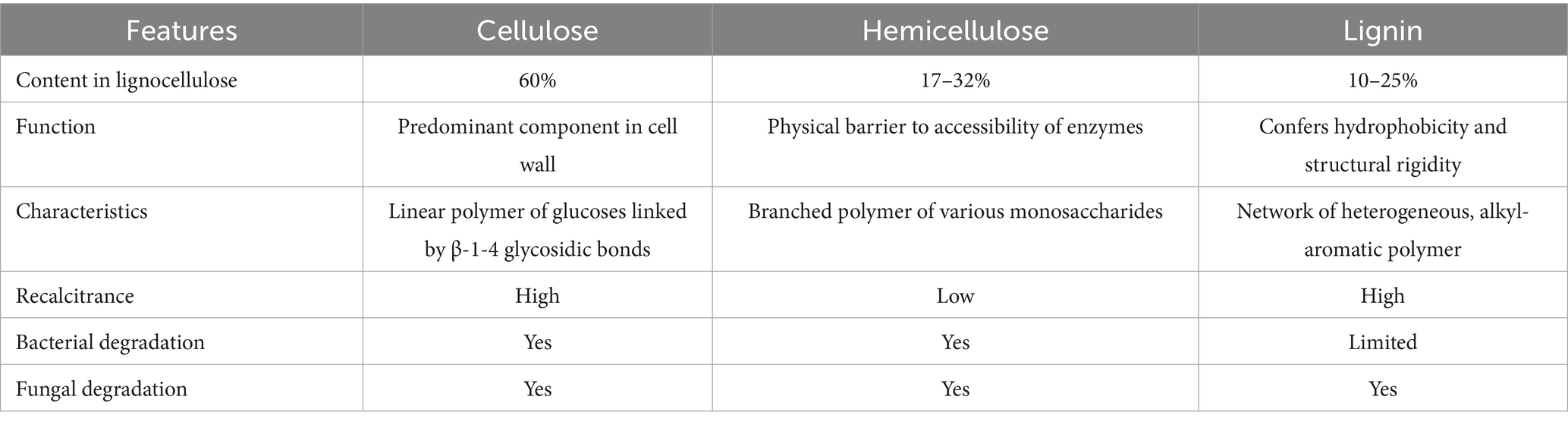
Lignocellulosic biomass consists of three main components, cellulose (60%), hemicellulose (17–32%) and lignin (10–25%) (Payne et al., 2015; Sankaran et al., 2021; Akinosho et al., 2014). Cellulose is composed of a linear polymer chain of 7,000–15,000 glucoses connected by β-1-4-glycosidic bonds in a cross-linked structure, which prevents enzyme access (Rosenau et al., 2018), forming one of the most stable polymers on earth. Hemicellulose is a branched heteropolysaccharide composed of various sugar monomers, such as xylose, arabinose, mannose, and glucose, and its structure varies across plant species (Gao et al., 2023; Paz-Cedeno et al., 2022). It exhibits a lower degree of polymerization and reduced molecular weight in comparison to cellulose. Hemicellulose functions as a linking agent between lignin and cellulose fibers, as well as a physical barrier that obstructs access to cellulose, contributing to increased rigidity within the composite structure (Syguła et al., 2024). Lignin, the compound that contributes to biomass recalcitrance, i.e., degree of resistance against degradation, consists of cross-linked phenyl propane polymers (Gibson, 2012). It contributes to cell wall integrity and forms an outer barrier for degrading enzymes to access cellulose and hemicellulose (Boerjan et al., 2003).
The enzymatic degradation of lignocellulose can be achieved by diverse bacterial and fungal species, each exhibiting differing efficiencies and enzyme sets. Table 1 summarizes the functions and characteristics of cellulose, hemicellulose and lignin, as well as whether bacterial and fungal enzymatic degradation is possible. Enzymes that are involved in breaking down complex carbohydrates and polysaccharides into smaller products are known as carbohydrate-active enzymes (CAZymes), which include Glycoside Hydrolase (GH), Carbohydrate Esterase (CE) and Auxiliary Activity (AA) families (Kameshwar et al., 2019). These enzymes act in tandem to target different components of lignocellulose. For example, the degradation of hemicellulose requires backbone-cleaving GHs and ancillary enzymes that attack the functional groups.
3 Cellulose and hemicellulose degradation
Cellulose and hemicellulose degrading bacteria can be found in the environment as well as in the gastrointestinal system of ruminant animals (Weimer, 2022; Datta, 2024). Both aerobic and anaerobic bacteria are capable of degrading cellulose and hemicellulose. For fungi, they are mainly identified within Ascomycetes and Basidiomycetes (Li et al., 2022). These fungi are divided into brown-rot, white-rot, and soft-rot fungi based on their ability to decompose each type of lignocellulose component (Fukasawa, 2021). Here we discuss and compare the degradation mechanisms of cellulose and hemicellulose in bacteria and fungi.
3.1 Bacteria and fungi share similar degradation enzymes
The degradation of cellulose is achieved by GHs, which function to break the glycosidic linkage between two carbohydrates. There are three types of GH families characterized by their modes of activity and location of cleavage (Akinosho et al., 2014; Datta, 2024; Bhardwaj et al., 2019):
1. Endoglucanases cleave β-1,4-glycosidic bonds from the internal structure and are more active on the soluble region of the substrate. GH5 to GH12, GH26, GH44, GH45, GH48, GH51, GH74, and GH124 are regarded as endoglucanases (Alnoch et al., 2023).
2. Exoglucanases cleave bonds from the terminals of substrates and release cellobiose molecules. GH3, GH43 and few members from other GH families belong to this category (Hirano et al., 2016).
3. β-glucosidases are responsible for degrading the cellobiose released from endoglucanases and exoglucanases. β-glucosidases remove the non-reducing end glucosyl residues from saccharides and glycosides to produce glucose (Ketudat Cairns and Esen, 2010; Puchart et al., 2023). GH1-GH3, GH5, GH30, GH39 and GH116 members are key β-glucosidases.
Due to the diverse nature of hemicellulose composition, multiple sets of enzymes are involved in its degradation. To degrade xylans, the xylanase endoenzymes, such as those from GH10, function by hydrolyzing the β-D-xylopyranosyl bonds to release xylobiose, xylooligosaccharides, and xylose (Puchart et al., 2021). As for mannans, the β-mannanase, β-mannosidase, acetyl mannan esterase, β-glucosidase and α-galactosidase belonging to GH1, GH2, GH3, GH5, GH26, GH27, and GH113 (Dawood and Ma, 2020) are involved. The GH74, GH29 and GH95 members exhibit the xyloglucan-specific endo-β-1,4-glucanase activity to degrade the backbone chain of xyloglucan (Zerillo et al., 2013).
CEs and AAs are accessory enzymes that facilitate the actions of GHs in degrading cellulose and hemicellulose. CEs target pectin oligosaccharides and arabinoxylans to cleave ester linkages and release acyl or alkyl, while AA9 to AA17 families consist of lytic polysaccharide monooxygenases (LPMOs) that cleave the crystalline polysaccharide chain of cellulose and other carbohydrates via direct oxidative reactions (Ward et al., 2020).
In general, bacteria and fungi share similar sets of enzymes listed above to degrade cellulose and hemicellulose (Haitjema et al., 2017; Floudas et al., 2012). However, certain groups of fungi possess greater variety of enzymes than others, therefore contributing to different degree of degradation ability. For example, a genome-wide search revealed that white-rot fungi generally possess more GHs compared to brown-rot fungi, suggesting a divergence in the GH family between these two types of fungi (Riley et al., 2014). White-rot fungi encode GH6 and GH7 enzymes that can degrade crystalline cellulose, which are missing in brown-rot fungi likely due to gene loss events (Floudas et al., 2012).
In addition, endoglucanases of certain fungal species have been shown to be more effective in targeting amorphous domains, most likely because fungi species have more “retaining” type endoglucanases such as GH7, GH5 and GH12 (Bharadwaj et al., 2020) than the bacterial species. “Retaining” type endoglucanases are known to target cellulose domains that are disordered and therefore have lowered recalcitrance (Ioelovich, 2021; Wang et al., 2016).
4 Lignin degradation
Lignin degradation involves the breaking down of alkyl-aromatic polymers into simpler compounds. It occurs in two steps, depolymerization and aromatic ring cleavage. Depolymerization occurs via the oxidative degradation mechanism, while the aromatic ring cleavage step involves long-range electron transfer process and phenolic moieties oxidation (Higuchi, 2004). In both bacteria and fungi, the cleavage of the aromatic ring is catalyzed by non-heme iron-containing intradiol and extradiol dioxygenases (Harayama and Rekik, 1989; Semana and Powlowski, 2019). Intradiol dioxygenases cleave catecholic substrates between the two hydroxyl groups using a non-heme Fe (III) center, while extradiol dioxygenases cleave catecholic substrates adjacent to the hydroxyl groups and typically contain a non-heme Fe (II) center.
Both bacterial and fungal species exhibit lignin degradation ability, for example the fungus Aspergillus flavus has a lignin degradation rate of 44.6% within 3 days under optimized conditions (Li et al., 2020) while the bacterium Bacillus cereus shows a lignin degradation rate of 89% (Kumar et al., 2022). The enzymes involved in lignin degradation of fungi and bacteria are presented in the following sections.
4.1 Fungi possess a diverse set of lignin degradation enzymes
The white-rot fungi are well-known to degrade the lignin. A diverse set of lignin degradation enzymes is found in white-rot fungi, including dye-depolymerizing peroxidases (DyPs), laccase and laccase-like multi-copper oxidases (LMCOs), lignin peroxidases (LiP), manganese-dependent peroxidases (MnP), laccases (LaC), versatile peroxidases (VP) and unspecific peroxygenases (UPO) (Yadav et al., 2022). DyPs belong to a superfamily of heme peroxidases (Sugano, 2009), whereas LMCOs belong to the multicopper oxidase superfamily (Leonowicz et al., 2001). Both DyPs and LCMOs form two major groups of lignin-degrading enzymes in fungi.
DyPs exhibit broad substrate specificity, oxidizing typical peroxidase substrates such as 2,2′-Azino-bis(3-ethylbenzothiazoline-6-sulfonic acid) (ABTS), 2,6-dimethoxyphenol (DMP) and various dyes, and potentially demonstrate additional hydrolase or oxygenase activities (Sugano, 2009). The LMCOs demonstrate oxidase activity, facilitating the oxidation of diverse phenolic compounds as well as certain non-phenolic substrates, while simultaneously reducing molecular oxygen to water (Ausec et al., 2011). The LiPs degrade the aromatic compounds and phenolic compounds in lignin with the presence of H2O2 via oxidation processes (Kumar and Chandra, 2020). MnP facilitates the oxidation of Mn2+ to Mn3+ using hydrogen peroxide, forming a Mn2+/Mn3+ redox pair. In the presence of a chelating agent, such as oxalate or malate, the Mn3+ ion is released from the enzyme’s active site (Kumar and Chandra, 2020; Janusz et al., 2017). These chelators stabilize Mn3+, preventing its disproportionation into Mn2+ and insoluble Mn4+. The stabilized Mn3+ complex can then diffuse into the lignified cell wall, where it oxidizes phenolic and nonphenolic lignin components. Versatile peroxidases possess a unique substrate specificity that combines the characteristics of LiP and MnP (Yadav et al., 2022). Unlike LiPs and MnPs, VPs can oxidize a broader range of phenolic and nonphenolic substrates, including veratryl alcohol, methoxybenzenes, and various lignin model compounds. UPOs are multifunctional oxidoreductases with wide substrate versatility as they are able to act both as oxygen donors and reductants (Ullrich et al., 2004; Pandya et al., 2014). This dual-functionality enables UPOs to emerge as research targets for process development (Tieves et al., 2019).
Some brown-rot fungi, like Gloeophyllum trabeum also exhibit lignin degradation ability (Arimoto et al., 2015). Though both brown-rot and white-rot fungi break down lignin by releasing acyl or alkyl groups (Wilhelm et al., 2019; de Gonzalo et al., 2016; Grgas et al., 2023; Christopher et al., 2014), their mechanisms differ significantly (Janusz et al., 2017; Cragg et al., 2015; Embacher et al., 2023). Brown-rot fungi use a non-enzymatic mechanism known as the Fenton reaction, an energy-saving process and relies on Fe (II) ions and hydrogen peroxide (Yadav et al., 2022; Embacher et al., 2023). This reaction reduces Fe (III) to Fe (II) in plant cells, producing-OH radicals that depolymerize lignin polymers (Yadav et al., 2022; Embacher et al., 2023) and break down cellulose and hemicellulose, making them more accessible to other lignocellulose degradation enzymes. Some Ascomycetes, including soft-rot fungi, secrete oxidative enzymes such as laccases and peroxidases, which help degrade lignin by oxidizing its phenolic units and breaking down its structure using hydrogen peroxide, respectively. For example, Podospora anserina demonstrates its ability to break down lignin through enhanced synthesis of laccases (Ferrari et al., 2021; van Erven et al., 2020).
4.2 Bacterial lignin-degrading enzymes: low efficiency, high adaptability
Lignin-degrading enzymes have been identified genetically (Janusz et al., 2017; Bugg et al., 2011) and isolated from many bacteria species. However, the degradation strategies in bacteria are less understood, and are believed to be generally less efficient compared to fungi (He et al., 2017). Bacteria capable of lignin modification and degradation belong to class Actinomycetes which mainly live in the soil (Bugg et al., 2011), as well as Gammaproteobacteria and Alphaproteobacteria (Xu et al., 2019). Only two classes of lignin-degrading enzymes are identified in bacteria, DyPs and LMCOs (Kamimura et al., 2017; Levy-Booth et al., 2022; Wilhelm et al., 2019). Bacterial laccases and DyPs have relatively lower oxidizing power than fungal enzymes, and therefore target lignin polymers of lower molecular weight (de Gonzalo et al., 2016).
Despite their low efficiency, bacterial lignin-degrading enzymes can function in a broader range of environmental conditions (Grgas et al., 2023). With better thermal stability and higher optimal pH (Levy-Booth et al., 2022; Christopher et al., 2014), bacterial laccases are attractive candidates for industrial usage. Another advantage unique to bacterial lignin-degradation is their capability of modifying or converting lignin into valuable bioproducts. Instead of rapidly degrading it in order to access cellulose and hemicellulose, bacteria utilize lignin as a substrate. After lignin monomers are converted into phenolic compounds, aromatic ring cleavage will occur for cellular uptake while producing bioproducts that are of industrial interest such as muconic acid and polyhydroxyalkanoates (Li and Zheng, 2020).
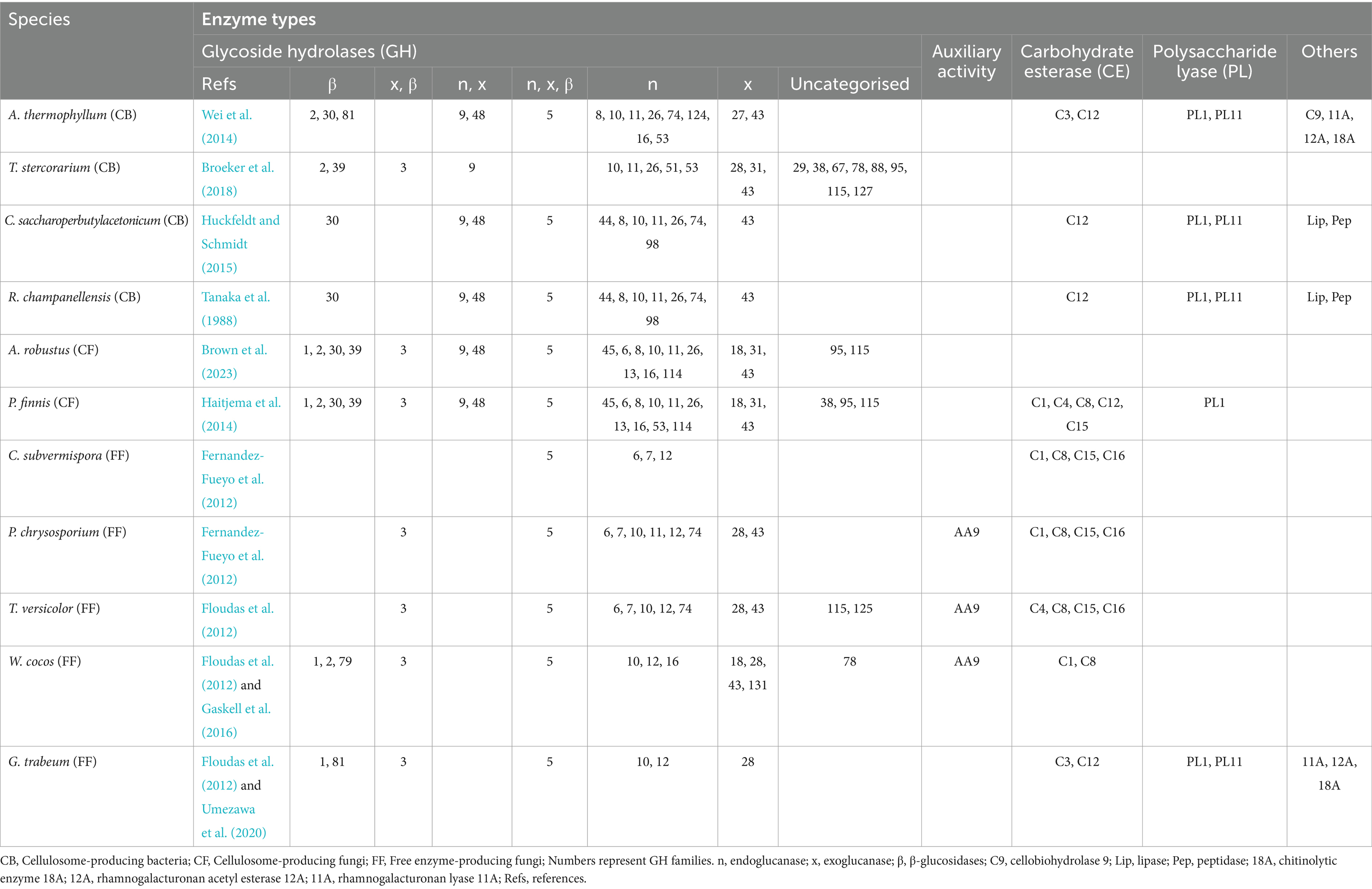
As examples, Table 2 details the extensive sets of carbohydrate-active enzyme families identified in four bacteria and seven representative fungal species.
5 The assembly of cellulosome enhances degradation efficiency
The enzymes described above can either be freely-secreted or assembled into a multi-enzyme complex known as cellulosome. Majority of the lignocellulolytic bacteria and fungi that are aerobic belong to the freely-secreting category. As shown in Figure 1A, secreted enzymes diffuse as individual catalytic units and function independently to degrade lignocellulose (Cragg et al., 2015). Trichoderma reesei and Aspergillus niger are the two main fungi species that are being extensively exploited for the production of cellulolytic cocktails (Xu et al., 2009). Bacterial strains are generally less favored in industrial settings due to their lower secretion capacity although species such as Streptomyces have been identified as versatile candidates (Kashiwagi et al., 2017).
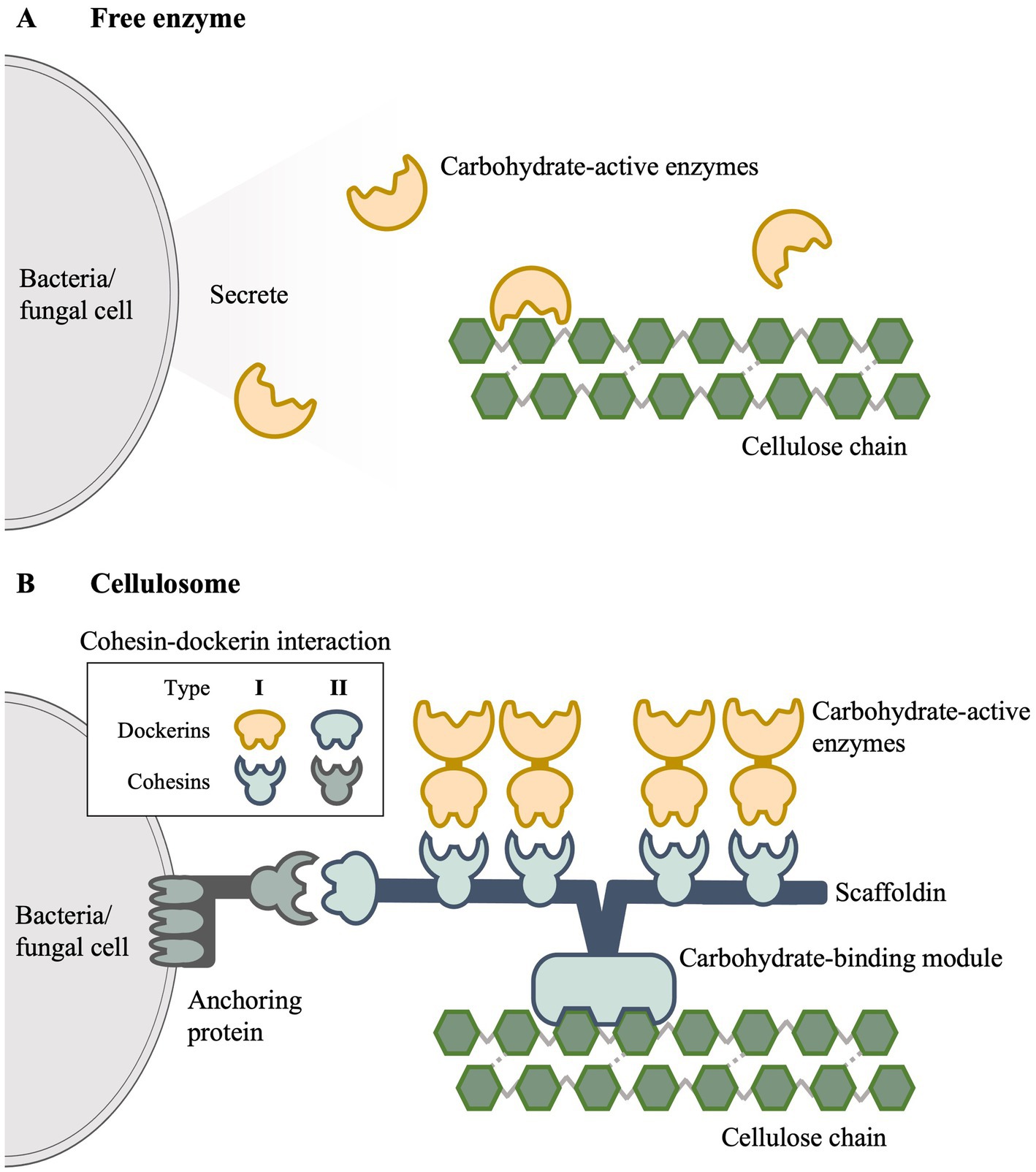
Figure 1. Overview of enzymatic mechanisms of lignocellulose degradation. (A) Free enzyme mechanism. (B) Cellulosome.
On the other hand, the cellulosome, first discovered in Acetivibrio thermocellum (Bayer et al., 2004; Lamed and Bayer, 1988), is a multiprotein complex in which lignocellulolytic enzymes are tethered together for enhanced efficient degradation and systematic hydrolytic activity (Artzi et al., 2017). A simplified cellulosome is depicted in Figure 1B, where it is predominantly situated on the cell wall surface of microorganisms with enzymes attached to a large non-catalytic subunit known as scaffoldin. This building-block-like structure is assembled through a flexible mechanism involving specialized domains called cohesins and dockerins. The type I cohesin-dockerin interaction allows binding of carbohydrate-active enzymes to scaffoldin (Fan et al., 2012), while type II interaction allows the scaffoldin to anchor to the cell surface (Doi et al., 2003). The carbohydrate-binding module (CBM) serves to attach the cellulosome structure to the cellulose substrate (Akinosho et al., 2014; Doi et al., 2003), while the connection of cellulosome to the cell surface is facilitated by surface (S)-layer proteins (Bayer et al., 2008; Fontes and Gilbert, 2010).
The efficiency of bacterial cellulosome has been demonstrated through comparisons of cellulose hydrolysis rate, with A. thermocellum achieving a 50-fold increase over free enzymes in T. ressei (Demain et al., 2005) and a 15-fold decrease when A. thermocellum’s cellulosome formation is impaired (Zverlov Vladimir et al., 2008). It has been proposed that the spatial enzyme proximity offered by cellulosome structure enhances synergistic interactions between catalytic units (Bayer et al., 2004), while anchoring to the cellulosome increases enzyme stability (Fontes and Gilbert, 2010). Cellulosomes can be found in anaerobic bacteria of the Clostridia class and Neocallimastigomycetes fungi (Artzi et al., 2017; Doi et al., 2003; Xu et al., 2003; Xu et al., 2004; Sabathé et al., 2002; Zhivin et al., 2017; Morrison and Miron, 2000; Moraïs et al., 2016; Ding et al., 2001; Cai et al., 2010; Levi Hevroni et al., 2020; Ben David et al., 2015). While none of the cellulosomal-producing species are currently used in industrial applications, they are key research targets in optimizing the biofuel production process (Vodovnik and Lindic, 2025).
5.1 The building blocks of cellulosomes: cohesins and dockerins
Components of cellulosomes are held together by high-affinity interactions (Pagès et al., 1997; Fierobe et al., 2002) between cohesins and dockerins. Three types of cohesin-dockerin interactions were identified so far based on sequence similarity: type I which is responsible for attaching enzymes to scaffoldin, type II which attaches scaffoldin to the cell, and type III which consists of those that are sequentially divergent from the other types (Karpol et al., 2013). Note that this classification is based on bacterial cellulosomes, and the more recently discovered fungal cohesins and dockerins are currently unclassified (Alves et al., 2021).
Cohesin-dockerin has type-and species-specificity, where cohesins of Type I do not interact with dockerins of Type II (Fontes and Gilbert, 2010; Miras et al., 2002; Schaeffer et al., 2002), and cohesins of one species do not interact with dockerins of another species (Fendri et al., 2009). Analysis of binding sites showed that low sequence similarities and differences in structural site topology explain these specificities in cohesins and dockerins (Carvalho et al., 2005). The combination of strict species-and type-specificity with module-promiscuity enables accurate assembly of cellulosome with enough heterogeneity of enzyme content, a factor that has been identified to increase degradation efficiency (Fendri et al., 2009).
As cellulosome is an inherently dynamic structure (Barth et al., 2018), the precise arrangement of the components, including cohesins and dockerins, remains poorly understood in most species. Firstly, the amino acid residues that are responsible for binding specificity and promiscuity as described above is not well understood, as mutational studies only attained conclusive results in limited species (Slutzki et al., 2015). Secondly, cohesins and dockerins have been found to play a role in maintaining the plasticity and stability of the whole cellulosome, but the mechanism remains unclear. Alternative binding modes have been found in certain types of cohesin (Alves et al., 2021; Carvalho et al., 2007) which may enable flexibility in the spatial organization of the scaffoldin subunits (Lamote et al., 2023). These binding modes had been hypothesized to be enzymatically regulated by an intramolecular structure located on dockerins. Also, the precise spatial arrangement of enzymatic subunits is found to rely on cohesin-cohesin interactions (Barth et al., 2018). These findings suggest that cohesins and dockerins are the basis of the regulation of cellulosome stability and plasticity, but quantitative evidence to these hypotheses is still lacking (Lamote et al., 2023).
Thorough understanding of the regulation and organization of cohesin-dockerin interactions may be able to improve the process of lignocellulose degradation, such as development of methods to manipulate the specificity of cohesin-dockerin complexes, as well as to enhance stability and flexibility of artificial designer cellulosomes.
5.2 Characterization of cellulosomes based on bacterial systems
The architecture of cellulosomes can vary from simple to complex, depending on the amount of interacting scaffoldins and the intricacy of the complex (Bayer et al., 2004). A simple cellulosome is composed of a single scaffoldin that contains up to nine enzymatic subunits, according to the number of cohesin modules on the scaffoldin, like in Clostridium josui (Ichikawa et al., 2014). Highly structured cellulosomes contain several scaffoldins and many enzymes. For example, eight scaffoldins and up to 63 enzymes are identified from A. thermocellum (Fontes and Gilbert, 2010).
Cellulosomes display selective enzyme synthesis based on the substrate. For instance, when grown on differently pretreated yellow poplar and pine-made pulp, A. thermocellum’s cellulosomes show varied enzyme compositions (Wei et al., 2014; Leis et al., 2018). Pine pulp, with its higher lignin content, leads to the production of enzymes like xylanase and mannanase for lignin breakdown, while pretreated yellow poplar prompts the synthesis of endoglucanases, exoglucanases, and β-glucosidases (Leis et al., 2018). Similarly, Clostridium cellulovorans produces different sets of enzymes, when grown on cellulose, xylan, galactomannan and pectin (Aburaya et al., 2019).
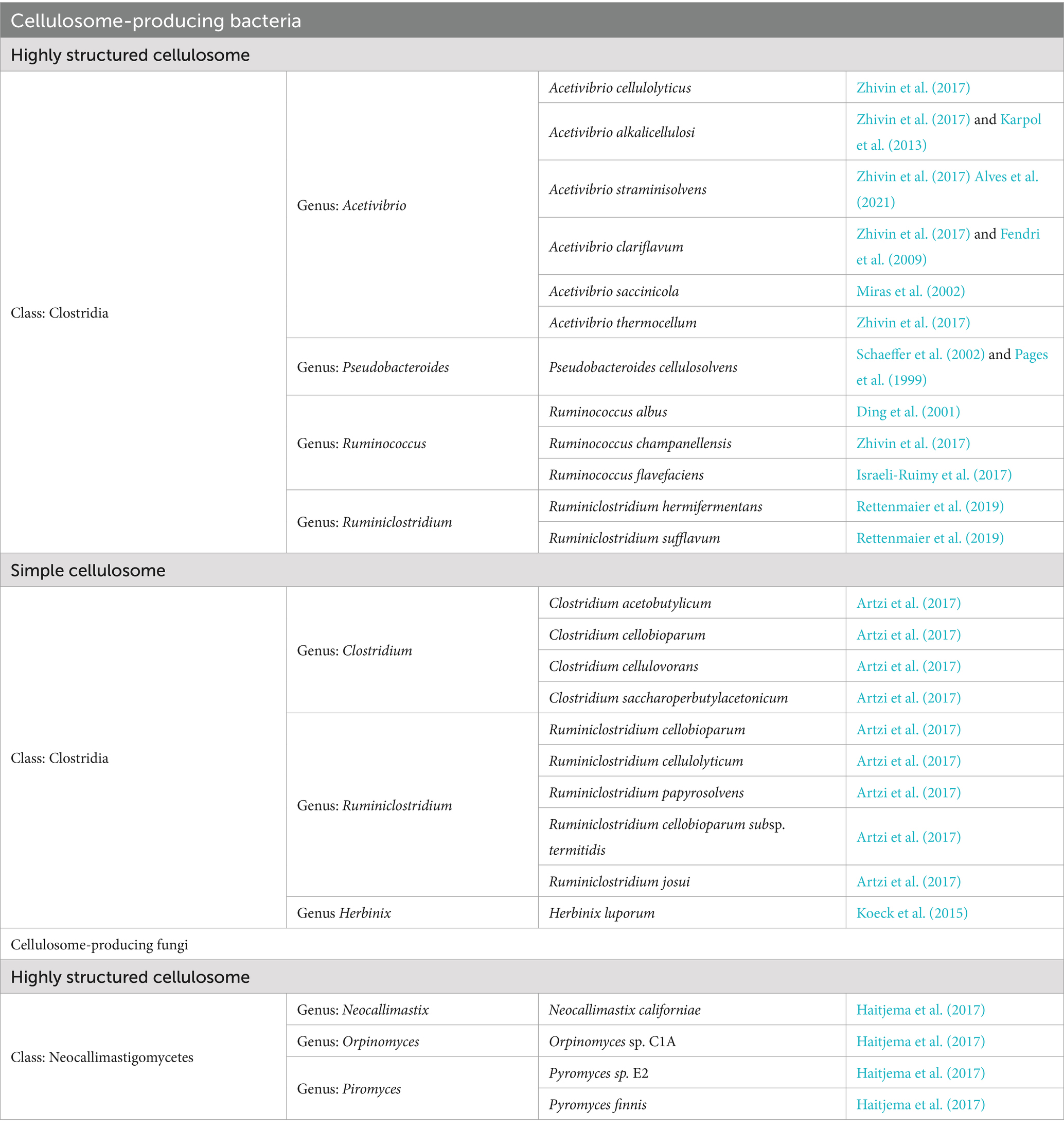
Current understandings of cellulosomes are mainly derived from bacterial species, as only limited fungal cellulosomes are discovered to date. Table 3 lists 22 bacteria and 4 fungi species that were commonly reported in the literature to produce cellulosomes. Cellulosome is presently exclusive to anaerobic bacteria and fungi of Clostridia and Neocallimastigomycetes members (Artzi et al., 2017; Doi et al., 2003; Xu et al., 2003; Xu et al., 2004; Sabathé et al., 2002; Zhivin et al., 2017; Morrison and Miron, 2000; Moraïs et al., 2016; Ding et al., 2001; Cai et al., 2010; Levi Hevroni et al., 2020; Ben David et al., 2015). Class Clostridia consists of anaerobic bacteria that mainly colonize the human gastrointestinal system and soil, whereas Neocallimastigomycetes is a phylum of anaerobic fungi that colonizes the herbivore gastrointestinal system. The strict anaerobic environment and limited energy sources of anaerobic bacteria are two factors that are hypothesized to impose selective pressures for the evolution of cellulosomes. This adaptation allows for efficient lignocellulosic degradation to obtain cellular energy (Fontes and Gilbert, 2010).
5.3 Fungal cellulosomes are poorly characterized due to genetic and isolation challenges
Fungal cellulosomes remain poorly understood due to the limited number of identified cellulosome-producing fungi and the low genetic similarity of their cellulosomal genes to bacterial counterparts (Youssef Noha et al., 2013). This means that the knowledge from bacterial cellulosomes might not be directly transferable onto fungal cellulosomes. To date, fungal cellulosomes have been isolated from anaerobic fungi residing in gastrointestinal systems of herbivores (Haitjema et al., 2017; Lillington et al., 2021; Brown et al., 2023; Gomez-Flores et al., 2017). Despite the early 20th-century discovery of fungal cellulosomes in the anaerobic rumen fungus Neocallimastix frontalis (Wilson and Wood, 1992), it was not until recently that their biological characteristics and gene-level components were unveiled (Haitjema et al., 2017; Lillington et al., 2021; Brown et al., 2023). This delay was due to the challenges in isolating these organisms, attributed to their anaerobic nature and complex nutritional needs (Haitjema et al., 2014). The biological features of the fungal cellulosome were initially explored through the herbivore gut-isolated Piromyces finnis (Lillington et al., 2021) and the presence of dockerin domains, partial ScaA scaffolding, and GH48 indicate that fungal cellulosomes consist of similar compounds as bacterial cellulosomes.
5.4 Key differences between bacteria and fungi cellulosomes
There are two key differences in cellulosomes between bacteria and fungi:
1. The genetic sequences of fungal dockerin and cohesin domains have no similarity to bacterial counterparts (Haitjema et al., 2017). The fungal dockerins form tandem repeats, which are believed to enhance cellulosome binding more than singular, non-repeated domains (Raghothama et al., 2001). Contrary to the precise dockerin-cohesin interaction observed in bacteria (Bayer et al., 2004), fungal versions have low species-specificity. Dockerins from one fungal species can attach to cellulosome components of another (Fanutti et al., 1995), suggesting the potential for various fungal species within the gut to aid in cellulosome structure formation, thereby boosting its efficiency (Haitjema et al., 2017). This cross-binding capability, so far confirmed only in species from the early-diverging fungal phylum Neocallimastigomycota, suggests a possibly widespread conservation of the scaffoldin and dockerin/cohesin system.
2. The second key difference is that fungi with cellulosomes possess the most extensive collection of carbohydrate-active enzymes discovered to date (Solomon et al., 2016). Of all the dockerin-domain-containing proteins in five strains of anaerobic fungi, 13% are GH enzymes that are absent in bacterial cellulosomes (Haitjema et al., 2017), therefore contributing enzymatic functions that are unique to fungi. For example, using β-glucosidases GH3, fungal cellulosome is able to convert cellulose into monosaccharides, instead of the more complex oligosaccharides produced by bacterial cellulosome (Steenbakkers et al., 2003). Fungal cellulosomes also prefer to degrade more recalcitrant lignocellulose components compared to bacterial cellulosomes (Hagen et al., 2021).
Even though the understanding for fungal cellulosome is still based on pieces of evidence, unique features that stood out from bacterial cellulosome may suggest fungal cellulosome to be a potential candidate for industrial applications.
6 Ways to improve lignocellulose degradation for industrial applications
Currently, most industrial applications depend on enzymatic cocktails derived from aerobic fungi such as T. reesei and Aspergillus sp. (Seppälä et al., 2017). The significant expense of these mixtures is a major barrier to making biofuels economically viable (Klein-Marcuschamer et al., 2012). Moreover, optimizing these mixtures based on the substrate is necessary, as the generic “one-size-fits-all” approach has been proven ineffective (Chylenski et al., 2017).
Bioengineering and genomic advancement can help to improve the process by shortening the reaction time of enzymes, reducing the enzyme concentration needed, and enhancing enzyme stability in extreme heat or pH conditions. Here we discuss three main focuses in optimizing lignocellulose degradation for industrial applications.
6.1 Discovery of novel lignocellulolytic species and enzymes
With more and more species being isolated and sequenced, genomic (such as NCBI) and proteomic [such as CAZy (Drula et al., 2022)] databases are exponentially increasing in size. Discovering and adding novel lignocellulose-degrading species and enzymes in various ecosystems to the existing repertoire is a highly valuable effort to improve lignocellulose degradation for industrial use.
One example is the aforementioned gut anaerobic fungi. While genomic studies of these gut fungi species showed that their enzyme repertoires possess four times more carbohydrate-active enzymes than fungi that utilize free enzyme mechanisms, a large portion of open reading frames in their genomes remain unannotated (Seppälä et al., 2017), therefore opening the possibility of discovering novel, and efficient enzymes. Similar findings have been obtained through metagenomic analysis. For example, highly complexed microbial communities with a diversified set of cellulases and accessory enzymes were found in three naturally-degraded energy crops (Montella et al., 2017). A recent genomic scan of bacterial genomes for scaffoldins, cohesins, dockerins and enzymes revealed 9 new bacterial species that are likely to produce simple cellulosomes (Minor et al., 2024).
6.2 Engineering lignocellulolytic enzymes for thermostability
Enzyme engineering is a common practice to modify enzymatic properties through altering the amino acid sequences (Bloom et al., 2005). One of the more popular procedures is through random mutagenesis directed evolution, where a target gene in a microorganism is randomly mutated followed by high-throughput screening approaches (Liu et al., 2009). For example, thermostability is one of the highly desired traits, as besides maintaining reactivity in high temperature, thermostable enzymes can be recycled more efficiently (Skovgaard and Jorgensen, 2013). A quadruple mutant of A. thermocellum endoglucanase Cel8A was produced through consensus-guided mutagenesis and molecular dynamics simulation, where its hydrolysis activity was increased by 14-fold at 85°C compared to wild type enzyme (Anbar et al., 2012). Similarly, thermostable exoglucanase (Smith et al., 2012) and β-glucosidase (Yoav et al., 2019) were also successfully generated in the recent years. These results demonstrated bioengineering as a reliable tool in producing enzymes of desired qualities for industrial use.
6.3 Improving structural stability of designer cellulosomes
Designer cellulosomes constructed with bacteria A. thermocellum and A. cellulolyticus have shown to be two to three times more effective at breaking down crystalline lignocellulose than free enzymes (Fierobe et al., 2002; Morais et al., 2011; Tsai et al., 2013; Gefen et al., 2012). Designer cellulosomes offer a key advantage by enabling the integration of diverse enzymes from multiple species, including both cellulosomal and non-cellulosomal enzymes, which has been shown to increase degradation efficiencies. Reconstituted A. thermocellum cellulosome with “external” enzymes that is from other species are more effective in degrading crystalline cellulosome (Hirano et al., 2016). Incorporation of non-cellulosomal enzymes such as LPMOs and expansins also showed similar effects (Chen et al., 2016; Arfi et al., 2014). Cross-species combination of cellulosomal enzymes with dockerin domains has also been shown to have a positive effect on degradation efficiency as glucose production is enhanced when β-glucosidases from A. thermocellum was tagged with the dockerin from Ruminococcus flavefaciens (Tsai et al., 2009). The benefit of cross-species combination of enzymes is also proven in fungi, as different types of fungi follow a succession in natural wood decay. The process begins with early colonizers consuming residual sap carbohydrates, followed by intermediate decay by brown-rot and white-rot fungi, and ends with soft-rot fungi thriving in nutrient-rich conditions, ultimately leading to lignin-degraded wood dominated by soft-rot fungi (Fukasawa, 2021; Fukasawa et al., 2009). This shows that in order to effectively degrade the lignocellulose, a combination of species is required. Structural stability of designer cellulosomes can be enhanced via cohesins-dockerins affinity, and improving the thermostability of other components.
6.3.1 Improving the compatibility between cohesins and dockerins
Designer cellulosomes have issues of low mechanical stability, which is mainly caused by the compatibility of selected cohesins and dockerins (Levasseur et al., 2004). It is necessary to ensure that the selected cohesin-dockerin pair is only able to interact with each other and not with any other modules that are incorporated in the design (Lamote et al., 2023). However, often times the molecular characterization of cohesins and dockerins available are not thorough enough to verify their specificities. For example, differing concentration and pH have been shown to affect binding specificities. The species-specific interaction between cohesins and dockerins typically occurs at low concentrations; however, at higher concentrations of dockerin-containing enzymes, dockerins can also interact with non-cognate but closely related cohesin domains (Barak et al., 2005). In addition, pH value is identified as a factor affecting cohesins-dockerins affinity, which regulate the amounts of protein complex and the structural stability of designer cellulosome (Barbosa et al., 2010; Duarte et al., 2023; Yao et al., 2020).
To optimize the selection of cohesin-dockerin pairs, deciphering the interactions in natural cellulosome is a key step. Through structural predictions of protein interactions, such as the highly accurate prediction model AlphaFold 3 (Abramson et al., 2024), cohesin-dockerin affinities can be estimated without the need for laborious experiments. Future studies may leverage recent advances in protein structure prediction, such as AlphaFold 3, to explore cohesin-dockerin interactions in silico. In addition to predicting the 3D structures of protein complexes, AlphaFold 3 provides confidence metrics, including the predicted template modeling (pTM) score and predicted aligned error (PAE), which may help assess the reliability of predicted interactions (Abramson et al., 2024; Zhang and Skolnick, 2004; Varadi et al., 2022).
In addition, unique traits of fungal cellulosomes might be key to solving this. Low species-specificity of cohesins and dockerins can enhance compatibility between components, and additional C-terminal fusions on fungal dockerins allow capacity for more enzymes to be added to the cellulosome (Dorival et al., 2022).
6.3.2 Improving thermostability of cellulosomal components
Due to the mesophilic origin of most cellulosomal components, the use of cellulosomes is often limited to temperatures up to 50°C (Bayer et al., 2004). To create a thermostable designer cellulosome with maintaining functional activity at elevated temperatures, one approach is to incorporate a scaffoldin composed of mesophilic modules with thermophilic enzymes (Galanopoulou et al., 2016). A tetravalent scaffoldin containing three mesophilic cohesins and one cohesin with a cellulose-binding module from Clostridium thermocellum was paired with thermophilic enzymes from Geobacillus and Caldicellulosiruptor species, each linked to dockerins matching the cohesins. This designer cellulosome maintained stability and functionality for at least 6 h at 60°C, achieving over 50% greater hydrolysis efficiency compared to free enzymes. The findings demonstrate that mesophilic scaffoldin components can be utilized at elevated temperatures, offering a viable strategy for designing cellulosomes for high-temperature biorefinery applications. Techniques such as thermostability enhancements and modular scaffoldin designs integrating mesophilic and thermophilic elements have enabled the creation of heat-resistant cellulosomes.
7 Conclusion
The utilization of lignocellulose biomass is a sustainable alternative to fossil carbon resources in producing biofuels to contribute to the circular economy. It is therefore of great interest to develop robust and efficient procedures to degrade lignocellulose. In this review we present the diverse lignocellulose degradation mechanisms found in nature, with the intention of using them as guidance for the development of more efficient degradation of biomass. Additionally, we highlight three key optimization focuses, including discovery of new species and enzymes, enzyme bioengineering and enhancing structural stability of designer cellulosomes. We expect these insights to serve as resources for the biomass community to develop more streamlined and sustainable solutions for utilizing lignocellulose as biofuel.
7.1 Future prospects
By drawing conclusions on lignocellulose degradation systems existing in nature, here a model favorable for industrial usage could be raised. After increasing the surface area of biomass using mechanical pre-treatment, lignin degradation is performed by bacterial AA enzymes to take advantage of their heat tolerance and ability to produce economical byproducts. Compared to costly thermochemical methods, pairing mechanical pretreatment and bacterial lignin degrading enzymes to remove recalcitrance could be more cost efficient. Recombinant expression of DyPs and laccase in E. coli have yielded high levels of expression (Lambertz et al., 2016; Ihssen et al., 2015), and bacterial cultivation benefits from fast growth rate and high tolerance to growth conditions. After lignin is removed, fungal endoglucanase should be used to degrade amorphous cellulose, followed by the application of designer cellulosomes to hydrolyze the remaining crystallized cellulose and hemicellulose. To capitalize on their cross-binding ability, designer cellulosomes are recommended to be assembled with fungal scaffoldin backbone and cohesin/dockerin modules. To ensure that the enzymes work synergistically, the arrangement of the enzymes on the cellulosome should take the natural succession of microorganism communities into consideration. For example, enzymes from white rot fungi should come before intermediate decay fungi followed by soft rot fungi. One of the challenges of designer cellulosome engineering lies in the large diversity of cellulosome components, creating exponential number of combinatorial possibilities that are costly to troubleshoot and optimize one by one (Lamote et al., 2023). Through advancements such as combinatorial assembly method via VersaTile platform (Vanderstraeten et al., 2022) and neural network models in protein–protein interaction prediction via AlphaFold 3 (Abramson et al., 2024), more economically efficient pipelines can be developed.
In the future, we anticipate answers to biological questions such as what is the mechanism behind the secretion of a selected choice of substrate-specific synergistic enzymes, how and when is the cellulosome assembled, what selection factors contribute to the repeated evolution of cellulosomal structure in both bacteria and fungi, etc. These insights, together with a wider collection of synthetic biology tools, will help actualize the usage of biomass as a sustainable energy.
Author contributions
K-TH: Writing – original draft, Writing – review & editing. HL: Visualization, Writing – original draft, Writing – review & editing. Y-CH: Visualization, Writing – review & editing. G-JL: Writing – review & editing. P-YL: Writing – review & editing. Y-CJL: Writing – review & editing. P-YC: Conceptualization, Funding acquisition, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by Academia Sinica (NTU-AS Innovative Joint Program: AS-NTU-112-12) and the National Science and Technology Council Taiwan Ministry of Science and Technology (112-2311-B-001-007 and 111-2927-I-001-505 to P-YC).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abramson, J., Adler, J., Dunger, J., Evans, R., Green, T., Pritzel, A., et al. (2024). Accurate structure prediction of biomolecular interactions with alpha fold 3. Nature 630, 493–500. doi: 10.1038/s41586-024-07487-w
Aburaya, S., Aoki, W., Kuroda, K., Minakuchi, H., and Ueda, M. (2019). Temporal proteome dynamics of Clostridium cellulovorans cultured with major plant cell wall polysaccharides. BMC Microbiol. 19:118. doi: 10.1186/s12866-019-1480-0
Akinosho, H., Yee, K., Close, D., and Ragauskas, A. (2014). The emergence of Clostridium thermocellum as a high utility candidate for consolidated bioprocessing applications. Front. Chem. 2:66. doi: 10.3389/fchem.2014.00066
Alnoch, R. C., Salgado, J. C. S., Alves, G. S., de Andrades, D., Meleiro, L. P., Segato, F., et al. (2023). Biochemical characterization of an endoglucanase GH7 from thermophile Thermothielavioides terrestris expressed on aspergillus nidulans. Catalysts 13:582. doi: 10.3390/catal13030582
Alves, V. D., Fontes, C., and Bule, P. (2021). Cellulosomes: highly efficient cellulolytic complexes. Subcell. Biochem. 96, 323–354. doi: 10.1007/978-3-030-58971-4_9
Anbar, M., Gul, O., Lamed, R., Sezerman, U. O., and Bayer, E. A. (2012). Improved thermostability of Clostridium thermocellum endoglucanase Cel 8A by using consensus-guided mutagenesis. Appl. Environ. Microbiol. 78, 3458–3464. doi: 10.1128/AEM.07985-11
Antczak, A., Marchwicka, M., Szadkowski, J., Drozdzek, M., Gawron, J., Radomski, A., et al. (2018). Sugars yield obtained after acid and enzymatic hydrolysis of fast-growing poplar Wood species. Bioresources 13, 8629–8645. doi: 10.15376/biores.13.4.8629-8645
Arfi, Y., Shamshoum, M., Rogachev, I., Peleg, Y., and Bayer, E. A. (2014). Integration of bacterial lytic polysaccharide monooxygenases into designer cellulosomes promotes enhanced cellulose degradation. Proc. Natl. Acad. Sci. USA 111, 9109–9114. doi: 10.1073/pnas.1404148111
Arimoto, M., Yamagishi, K., Wang, J., Tanaka, K., Miyoshi, T., Kamei, I., et al. (2015). Molecular breeding of lignin-degrading brown-rot fungus Gloeophyllum trabeum by homologous expression of laccase gene. AMB Express 5:81. doi: 10.1186/s13568-015-0173-9
Artzi, L., Bayer, E. A., and Morais, S. (2017). Cellulosomes: bacterial nanomachines for dismantling plant polysaccharides. Nat. Rev. Microbiol. 15, 83–95. doi: 10.1038/nrmicro.2016.164
Ausec, L., van Elsas, J. D., and Mandic-Mulec, I. (2011). Two-and three-domain bacterial laccase-like genes are present in drained peat soils. Soil Biol. Biochem. 43, 975–983. doi: 10.1016/j.soilbio.2011.01.013
Barak, Y., Handelsman, T., Nakar, D., Mechaly, A., Lamed, R., Shoham, Y., et al. (2005). Matching fusion protein systems for affinity analysis of two interacting families of proteins: the cohesin-dockerin interaction. J. Mol. Recognit. 18, 491–501. doi: 10.1002/jmr.749
Barbosa, L. R. S., Ortore, M. G., Spinozzi, F., Mariani, P., Bernstorff, S., and Itri, R. (2010). The importance of protein-protein interactions on the pH-induced conformational changes of bovine serum albumin: a small-angle X-ray scattering study. Biophys. J. 98, 147–157. doi: 10.1016/j.bpj.2009.09.056
Barth, A., Hendrix, J., Fried, D., Barak, Y., Bayer, E. A., and Lamb, D. C. (2018). Dynamic interactions of type I cohesin modules fine-tune the structure of the cellulosome of Clostridium thermocellum. Proc. Natl. Acad. Sci. USA 115, E11274–E11283. doi: 10.1073/pnas.1809283115
Bayer, E. A., Belaich, J. P., Shoham, Y., and Lamed, R. (2004). The cellulosomes: multienzyme machines for degradation of plant cell wall polysaccharides. Ann. Rev. Microbiol. 58, 521–554. doi: 10.1146/annurev.micro.57.030502.091022
Bayer, E. A., Lamed, R., White, B. A., and Flint, H. J. (2008). From cellulosomes to cellulosomics. Chem. Rec. 8, 364–377. doi: 10.1002/tcr.20160
Ben David, Y., Dassa, B., Borovok, I., Lamed, R., Koropatkin, N. M., Martens, E. C., et al. (2015). Ruminococcal cellulosome systems from rumen to human. Environ. Microbiol. 17, 3407–3426. doi: 10.1111/1462-2920.12868
Bharadwaj, V. S., Knott, B. C., Ståhlberg, J., Beckham, G. T., and Crowley, M. F. (2020). The hydrolysis mechanism of a GH45 cellulase and its potential relation to lytic transglycosylase and expansin function. J. Biol. Chem. 295, 4477–4487. doi: 10.1074/jbc.RA119.011406
Bhardwaj, N., Kumar, B., and Verma, P. (2019). A detailed overview of xylanases: an emerging biomolecule for current and future prospective. Bioresour. Bioprocess. 6:276. doi: 10.1186/s40643-019-0276-2
Bhatia, S. K., Vivek, N., Kumar, V., Chandel, N., Thakur, M., Kumar, D., et al. (2021). Molecular biology interventions for activity improvement and production of industrial enzymes. Bioresour. Technol. 324:124596. doi: 10.1016/j.biortech.2020.124596
Binod, P., Satyanagalakshmi, K., Sindhu, R., Janu, K. U., Sukumaran, R. K., and Pandey, A. (2012). Short duration microwave assisted pretreatment enhances the enzymatic saccharification and fermentable sugar yield from sugarcane bagasse. Renew. Energy 37, 109–116. doi: 10.1016/j.renene.2011.06.007
Bloom, J. D., Meyer, M. M., Meinhold, P., Otey, C. R., Mac Millan, D., and Arnold, F. H. (2005). Evolving strategies for enzyme engineering. Curr. Opin. Struct. Biol. 15, 447–452. doi: 10.1016/j.sbi.2005.06.004
Boerjan, W., Ralph, J., and Baucher, M. (2003). Lignin biosynthesis. Annu. Rev. Plant Biol. 54, 519–546. doi: 10.1146/annurev.arplant.54.031902.134938
Broeker, J., Mechelke, M., Baudrexl, M., Mennerich, D., Hornburg, D., Mann, M., et al. (2018). The hemicellulose-degrading enzyme system of the thermophilic bacterium Clostridium stercorarium: comparative characterisation and addition of new hemicellulolytic glycoside hydrolases. Biotechnol. Biofuels 11:229. doi: 10.1186/s13068-018-1228-3
Brown, J. L., Gierke, T., Butkovich, L. V., Swift, C. L., Singan, V., Daum, C., et al. (2023). High-quality RNA extraction and the regulation of genes encoding cellulosomes are correlated with growth stage in anaerobic fungi. Front. Fungal Biol. 4:1171100. doi: 10.3389/ffunb.2023.1171100
Bugg, T. D. H., Ahmad, M., Hardiman, E. M., and Rahmanpour, R. (2011). Pathways for degradation of lignin in bacteria and fungi. Nat. Prod. Rep. 28, 1883–1896. doi: 10.1039/c1np00042j
Cai, S., Li, J., Hu, F. Z., Zhang, K., Luo, Y., Janto, B., et al. (2010). Cellulosilyticum ruminicola, a newly described rumen bacterium that possesses redundant fibrolytic-protein-encoding genes and degrades lignocellulose with multiple carbohydrate-borne fibrolytic enzymes. Appl. Environ. Microbiol. 76, 3818–3824. doi: 10.1128/AEM.03124-09
Carvalho, A. L., Dias, F. M., Nagy, T., Prates, J. A., Proctor, M. R., Smith, N., et al. (2007). Evidence for a dual binding mode of dockerin modules to cohesins. Proc. Natl. Acad. Sci. USA 104, 3089–3094. doi: 10.1073/pnas.0611173104
Carvalho, A. L., Pires, V. M., Gloster, T. M., Turkenburg, J. P., Prates, J. A., Ferreira, L. M., et al. (2005). Insights into the structural determinants of cohesin-dockerin specificity revealed by the crystal structure of the type II cohesin from Clostridium thermocellum Sdb a. J. Mol. Biol. 349, 909–915. doi: 10.1016/j.jmb.2005.04.037
Chen, C., Cui, Z., Song, X., Liu, Y. J., Cui, Q., and Feng, Y. (2016). Integration of bacterial expansin-like proteins into cellulosome promotes the cellulose degradation. Appl. Microbiol. Biotechnol. 100, 2203–2212. doi: 10.1007/s00253-015-7071-6
Chen, R., Zhu, S., Chen, C., Cheng, B., Chen, J., and Wu, Y.-x. (2014). Reviving the acid hydrolysis process of lignocellulosic material in biorefinery. Bioresources 9, 1824–1827. doi: 10.15376/biores.9.2.1824-1827
Christopher, L. P., Yao, B., and Ji, Y. (2014). Lignin biodegradation with laccase-mediator systems. Front. Energy Res. 2:12. doi: 10.3389/fenrg.2014.00012
Chylenski, P., Forsberg, Z., Stahlberg, J., Varnai, A., Lersch, M., Bengtsson, O., et al. (2017). Development of minimal enzyme cocktails for hydrolysis of sulfite-pulped lignocellulosic biomass. J. Biotechnol. 246, 16–23. doi: 10.1016/j.jbiotec.2017.02.009
Cragg, S. M., Beckham, G. T., Bruce, N. C., Bugg, T. D., Distel, D. L., Dupree, P., et al. (2015). Lignocellulose degradation mechanisms across the tree of life. Curr. Opin. Chem. Biol. 29, 108–119. doi: 10.1016/j.cbpa.2015.10.018
Datta, R. (2024). Enzymatic degradation of cellulose in soil: a review. Heliyon 10:e24022. doi: 10.1016/j.heliyon.2024.e24022
Dawood, A., and Ma, K. (2020). Applications of microbial beta-Mannanases. Front. Bioeng. Biotechnol. 8:598630. doi: 10.3389/fbioe.2020.598630
de Gonzalo, G., Colpa, D. I., Habib, M. H., and Fraaije, M. W. (2016). Bacterial enzymes involved in lignin degradation. J. Biotechnol. 236, 110–119. doi: 10.1016/j.jbiotec.2016.08.011
Demain, A. L., Newcomb, M., and Wu, J. H. (2005). Cellulase, clostridia, and ethanol. Microbiol. Mol. Biol. Rev. 69, 124–154. doi: 10.1128/MMBR.69.1.124-154.2005
Ding, S. Y., Rincon, M. T., Lamed, R., Martin, J. C., McCrae, S. I., Aurilia, V., et al. (2001). Cellulosomal scaffoldin-like proteins from Ruminococcus flavefaciens. J. Bacteriol. 183, 1945–1953. doi: 10.1128/JB.183.6.1945-1953.2001
Doi, R. H., Kosugi, A., Murashima, K., Tamaru, Y., and Han, S. O. (2003). Cellulosomes from mesophilic bacteria. J. Bacteriol. 185:5907-14. doi: 10.1128/JB.185.20.5907-5914.2003
Dorival, J., Moraïs, S., Labourel, A., Rozycki, B., Cazade, P.-A., Dabin, J., et al. (2022). Mapping the deformability of natural and designed cellulosomes in solution. Biotechnol. Biofuels Bioprod. 15:68. doi: 10.1186/s13068-022-02165-3
Drula, E., Garron, M.-L., Dogan, S., Lombard, V., Henrissat, B., and Terrapon, N. (2022). The carbohydrate-active enzyme database: functions and literature. Nucleic Acids Res. 50, D571–D577. doi: 10.1093/nar/gkab1045
Duarte, M., Alves, V. D., Correia, M., Caseiro, C., Ferreira, L. M. A., Romão, M. J., et al. (2023). Structure-function studies can improve binding affinity of cohesin-dockerin interactions for multi-protein assemblies. Int. J. Biol. Macromol. 224, 55–67. doi: 10.1016/j.ijbiomac.2022.10.102
Elmekawy, A., Diels, L., De Wever, H., and Pant, D. (2013). Valorization of cereal based biorefinery byproducts: reality and expectations. Environ. Sci. Technol. 47, 9014–9027. doi: 10.1021/es402395g
Embacher, J., Zeilinger, S., Kirchmair, M., Rodriguez-R, L. M., and Neuhauser, S. (2023). Wood decay fungi and their bacterial interaction partners in the built environment – a systematic review on fungal bacteria interactions in dead wood and timber. Fungal Biol. Rev. 45:100305. doi: 10.1016/j.fbr.2022.100305
Fan, L. H., Zhang, Z. J., Yu, X. Y., Xue, Y. X., and Tan, T. W. (2012). Self-surface assembly of cellulosomes with two miniscaffoldins on Saccharomyces cerevisiae for cellulosic ethanol production. Proc. Natl. Acad. Sci. USA 109, 13260–13265. doi: 10.1073/pnas.1209856109
Fanutti, C., Ponyi, T., Black, G. W., Hazlewood, G. P., and Gilbert, H. J. (1995). The conserved noncatalytic 40-residue sequence in Cellulases and Hemicellulases from anaerobic Fungi functions as a protein docking domain (*). J. Biol. Chem. 270, 29314–29322. doi: 10.1074/jbc.270.49.29314
Fendri, I., Tardif, C., Fierobe, H. P., Lignon, S., Valette, O., Pagès, S., et al. (2009). The cellulosomes from Clostridium cellulolyticum: identification of new components and synergies between complexes. FEBS J. 276, 3076–3086. doi: 10.1111/j.1742-4658.2009.07025.x
Fernandez-Fueyo, E., Ruiz-Duenas, F. J., Ferreira, P., Floudas, D., Hibbett, D. S., Canessa, P., et al. (2012). Comparative genomics of Ceriporiopsis subvermispora and Phanerochaete chrysosporium provide insight into selective ligninolysis. Proc. Natl. Acad. Sci. USA 109, 5458–5463. doi: 10.1073/pnas.1119912109
Ferrari, R., Gautier, V., and Silar, P. (2021). “Chapter three-lignin degradation by ascomycetes, p 77-113” in Advances in botanical research. eds. M. Morel-Rouhier and R. Sormani (New York, NY: Academic Press).
Fierobe, H. P., Bayer, E. A., Tardif, C., Czjzek, M., Mechaly, A., Belaich, A., et al. (2002). Degradation of cellulose substrates by cellulosome chimeras. Substrate targeting versus proximity of enzyme components. J. Biol. Chem. 277, 49621–49630. doi: 10.1074/jbc.M207672200
Floudas, D., Binder, M., Riley, R., Barry, K., Blanchette, R. A., Henrissat, B., et al. (2012). The Paleozoic origin of enzymatic lignin decomposition reconstructed from 31 fungal genomes. Science 336, 1715–1719. doi: 10.1126/science.1221748
Fontes, C. M., and Gilbert, H. J. (2010). Cellulosomes: highly efficient nanomachines designed to deconstruct plant cell wall complex carbohydrates. Annu. Rev. Biochem. 79, 655–681. doi: 10.1146/annurev-biochem-091208-085603
Fukasawa, Y. (2021). Ecological impacts of fungal wood decay types: a review of current knowledge and future research directions. Ecol. Res. 36, 910–931. doi: 10.1111/1440-1703.12260
Fukasawa, Y., Osono, T., and Takeda, H. (2009). Microfungus communities of Japanese beech logs at different stages of decay in a cool temperate deciduous forest. Can. J. For. Res. 39, 1606–1614. doi: 10.1139/X09-080
Galanopoulou, A. P., Moraïs, S., Georgoulis, A., Morag, E., Bayer, E. A., and Hatzinikolaou, D. G. (2016). Insights into the functionality and stability of designer cellulosomes at elevated temperatures. Appl. Microbiol. Biotechnol. 100, 8731–8743. doi: 10.1007/s00253-016-7594-5
Gao, Y., Guo, M., Wang, D., Zhao, D., and Wang, M. (2023). Advances in extraction, purification, structural characteristics and biological activities of hemicelluloses: a review. Int. J. Biol. Macromol. 225, 467–483. doi: 10.1016/j.ijbiomac.2022.11.099
Gaskell, J., Blanchette, R. A., Stewart, P. E., Bon Durant, S. S., Adams, M., Sabat, G., et al. (2016). Transcriptome and Secretome analyses of the Wood decay fungus Wolfiporia cocos support alternative mechanisms of lignocellulose conversion. Appl. Environ. Microbiol. 82, 3979–3987. doi: 10.1128/AEM.00639-16
Gefen, G., Anbar, M., Morag, E., Lamed, R., and Bayer, E. A. (2012). Enhanced cellulose degradation by targeted integration of a cohesin-fused beta-glucosidase into the Clostridium thermocellum cellulosome. Proc. Natl. Acad. Sci. USA 109, 10298–10303. doi: 10.1073/pnas.1202747109
Gibson, L. J. (2012). The hierarchical structure and mechanics of plant materials. J. R. Soc. Interface 9, 2749–2766. doi: 10.1098/rsif.2012.0341
Gomez-Flores, M., Nakhla, G., and Hafez, H. (2017). Hydrogen production and microbial kinetics of Clostridium termitidis in mono-culture and co-culture with Clostridium beijerinckii on cellulose. AMB Express 7:84. doi: 10.1186/s13568-016-0256-2
Grgas, D., Rukavina, M., Bešlo, D., Štefanac, T., Crnek, V., Šikić, T., et al. (2023). The bacterial degradation of lignin—a review. Water 15:272. doi: 10.3390/w15071272
Hagen, L. H., Brooke, C. G., Shaw, C. A., Norbeck, A. D., Piao, H., Arntzen, M. Ø., et al. (2021). Proteome specialization of anaerobic fungi during ruminal degradation of recalcitrant plant fiber. ISME J. 15, 421–434. doi: 10.1038/s41396-020-00769-x
Haitjema, C. H., Gilmore, S. P., Henske, J. K., Solomon, K. V., de Groot, R., Kuo, A., et al. (2017). A parts list for fungal cellulosomes revealed by comparative genomics. Nat. Microbiol. 2:17087. doi: 10.1038/nmicrobiol.2017.87
Haitjema, C. H., Solomon, K. V., Henske, J. K., Theodorou, M. K., and O'Malley, M. A. (2014). Anaerobic gut fungi: advances in isolation, culture, and cellulolytic enzyme discovery for biofuel production. Biotechnol. Bioeng. 111, 1471–1482. doi: 10.1002/bit.25264
Harayama, S., and Rekik, M. (1989). Bacterial aromatic ring-cleavage enzymes are classified into two different gene families*. J. Biol. Chem. 264, 15328–15333. doi: 10.1016/S0021-9258(19)84830-5
He, K., Chen, G., Zeng, G., Huang, Z., Guo, Z., Huang, T., et al. (2017). Applications of white rot fungi in bioremediation with nanoparticles and biosynthesis of metallic nanoparticles. Appl. Microbiol. Biotechnol. 101, 4853–4862. doi: 10.1007/s00253-017-8328-z
Higuchi, T. (2004). Microbial degradation of lignin: role of lignin peroxidase, manganese peroxidase, and laccase. Proc. Jpn. Acad. B 80, 204–214. doi: 10.2183/pjab.80.204
Hirano, K., Kurosaki, M., Nihei, S., Hasegawa, H., Shinoda, S., Haruki, M., et al. (2016). Enzymatic diversity of the Clostridium thermocellum cellulosome is crucial for the degradation of crystalline cellulose and plant biomass. Sci. Rep. 6:35709. doi: 10.1038/srep35709
Hu, Y., Priya, A., Chen, C., Liang, C., Wang, W., Wang, Q., et al. (2023). Recent advances in substrate-enzyme interactions facilitating efficient biodegradation of lignocellulosic biomass: a review. Int. Biodeterior. Biodegradation 180:105594. doi: 10.1016/j.ibiod.2023.105594
Huckfeldt, T., and Schmidt, O. (2015). Hausfäule-und Bauholzpilze. Bausanierung: Erkennen und Beheben von Bauschäden.
Ichikawa, S., Karita, S., Kondo, M., and Goto, M. (2014). Cellulosomal carbohydrate-binding module from Clostridium josui binds to crystalline and non-crystalline cellulose, and soluble polysaccharides. FEBS Lett. 588, 3886–3890. doi: 10.1016/j.febslet.2014.08.032
Ihssen, J., Reiss, R., Luchsinger, R., Thöny-Meyer, L., and Richter, M. (2015). Biochemical properties and yields of diverse bacterial laccase-like multicopper oxidases expressed in Escherichia coli. Sci. Rep. 5:10465. doi: 10.1038/srep10465
Ioelovich, M. (2021). Preparation, characterization and application of Amorphized cellulose—a review. Polymers 13:4313. doi: 10.3390/polym13244313
Israeli-Ruimy, V., Bule, P., Jindou, S., Dassa, B., Moraïs, S., Borovok, I., et al. (2017). Complexity of the Ruminococcus flavefaciens FD-1 cellulosome reflects an expansion of family-related protein-protein interactions. Sci. Rep. 7:42355. doi: 10.1038/srep42355
Jablonsky, M., Nosalova, J., Sladkova, A., Haz, A., Kreps, F., Valka, J., et al. (2017). Valorisation of softwood bark through extraction of utilizable chemicals. A review. Biotechnol. Adv. 35, 726–750. doi: 10.1016/j.biotechadv.2017.07.007
Janusz, G., Pawlik, A., Sulej, J., Swiderska-Burek, U., Jarosz-Wilkolazka, A., and Paszczynski, A. (2017). Lignin degradation: microorganisms, enzymes involved, genomes analysis and evolution. FEMS Microbiol. Rev. 41, 941–962. doi: 10.1093/femsre/fux049
Kameshwar, A. K. S., Ramos, L. P., and Qin, W. (2019). CAZymes-based ranking of fungi (CBRF): an interactive web database for identifying fungi with extrinsic plant biomass degrading abilities. Bioresour. Bioprocess. 6:51. doi: 10.1186/s40643-019-0286-0
Kamimura, N., Takahashi, K., Mori, K., Araki, T., Fujita, M., Higuchi, Y., et al. (2017). Bacterial catabolism of lignin-derived aromatics: new findings in a recent decade: update on bacterial lignin catabolism. Environ. Microbiol. Rep. 9, 679–705. doi: 10.1111/1758-2229.12597
Karpol, A., Jobby, M. K., Slutzki, M., Noach, I., Chitayat, S., Smith, S. P., et al. (2013). Structural and functional characterization of a novel type-III dockerin from Ruminococcus flavefaciens. FEBS Lett. 587, 30–36. doi: 10.1016/j.febslet.2012.11.012
Kashiwagi, N., Ogino, C., and Kondo, A. (2017). Production of chemicals and proteins using biomass-derived substrates from a Streptomyces host. Bioresour. Technol. 245, 1655–1663. doi: 10.1016/j.biortech.2017.06.001
Ketudat Cairns, J. R., and Esen, A. (2010). β-Glucosidases. Cell. Mol. Life Sci. 67, 3389–3405. doi: 10.1007/s00018-010-0399-2
Klein-Marcuschamer, D., Oleskowicz-Popiel, P., Simmons, B. A., and Blanch, H. W. (2012). The challenge of enzyme cost in the production of lignocellulosic biofuels. Biotechnol. Bioeng. 109, 1083–1087. doi: 10.1002/bit.24370
Koeck, D. E., Ludwig, W., Wanner, G., Zverlov, V. V., Liebl, W., and Schwarz, W. H. (2015). Herbinix hemicellulosilytica gen. Nov., sp. nov., a thermophilic cellulose-degrading bacterium isolated from a thermophilic biogas reactor. Int. J. Syst. Evol. Microbiol. 65, 2365–2371. doi: 10.1099/ijs.0.000264
Kumar, A., and Chandra, R. (2020). Ligninolytic enzymes and its mechanisms for degradation of lignocellulosic waste in environment. Heliyon 6:e03170. doi: 10.1016/j.heliyon.2020.e03170
Kumar, R., Singh, A., Maurya, A., Yadav, P., Yadav, A., Chowdhary, P., et al. (2022). Effective bioremediation of pulp and paper mill wastewater using Bacillus cereus as a possible Kraft lignin-degrading bacterium. Bioresour. Technol. 352:127076. doi: 10.1016/j.biortech.2022.127076
Lambertz, C., Selin, E., Rainer, F., and Commandeur, U. (2016). Progress and obstacles in the production and application of recombinant lignin-degrading peroxidases. Bioengineered 7, 145–154. doi: 10.1080/21655979.2016.1191705
Lamed, R., and Bayer, E. A. (1988). The Cellulosome of Clostridium thermocellum. Adv. Appl. Microbiol. 33, 1–46. doi: 10.1016/S0065-2164(08)70203-X
Lamote, B., da Fonseca, M. J. M., Vanderstraeten, J., Meert, K., Elias, M., and Briers, Y. (2023). Current challenges in designer cellulosome engineering. Appl. Microbiol. Biotechnol. 107, 2755–2770. doi: 10.1007/s00253-023-12474-8
Leis, B., Held, C., Andreessen, B., Liebl, W., Graubner, S., Schulte, L. P., et al. (2018). Optimizing the composition of a synthetic cellulosome complex for the hydrolysis of softwood pulp: identification of the enzymatic core functions and biochemical complex characterization. Biotechnol. Biofuels 11:220. doi: 10.1186/s13068-018-1220-y
Leonowicz, A., Cho, N. S., Luterek, J., Wilkolazka, A., Wojtas-Wasilewska, M., Matuszewska, A., et al. (2001). Fungal laccase: properties and activity on lignin. J. Basic Microbiol. 41, 185–227. doi: 10.1002/1521-4028(200107)41:3/4<185::AID-JOBM185>3.0.CO;2-T
Levasseur, A., Pages, S., Fierobe, H. P., Navarro, D., Punt, P., Belaich, J. P., et al. (2004). Design and production in Aspergillus niger of a chimeric protein associating a fungal feruloyl esterase and a clostridial dockerin domain. Appl. Environ. Microbiol. 70, 6984–6991. doi: 10.1128/AEM.70.12.6984-6991.2004
Levi Hevroni, B., Moraïs, S., Ben-David, Y., Morag, E., and Bayer, E. A. (2020). Minimalistic Cellulosome of the Butanologenic bacterium Clostridium saccharoperbutylacetonicum. MBio 11:443. doi: 10.1128/mBio.00443-20
Levy-Booth, D. J., Navas, L. E., Fetherolf, M. M., Liu, L. Y., Dalhuisen, T., Renneckar, S., et al. (2022). Discovery of lignin-transforming bacteria and enzymes in thermophilic environments using stable isotope probing. ISME J. 16, 1944–1956. doi: 10.1038/s41396-022-01241-8
Li, T., Cui, L., Song, X., Cui, X., Wei, Y., Tang, L., et al. (2022). Wood decay fungi: an analysis of worldwide research. J. Soils Sediments 22, 1688–1702. doi: 10.1007/s11368-022-03225-9
Li, S. F., Wang, H., Chen, J. L., Zhu, H. X., Yao, R. S., and Wu, H. (2020). Degradation and transformation of lignin by a fungus aspergillus Flavus strain F-1. Iran. J. Biotechnol. 18:e2461. doi: 10.30498/IJB.2020.155690.2461
Li, X., and Zheng, Y. (2020). Biotransformation of lignin: mechanisms, applications and future work. Biotechnol. Prog. 36:e2922. doi: 10.1002/btpr.2922
Lillington, S. P., Chrisler, W., Haitjema, C. H., Gilmore, S. P., Smallwood, C. R., Shutthanandan, V., et al. (2021). Cellulosome localization patterns vary across life stages of anaerobic Fungi. MBio 12:e0083221. doi: 10.1128/mBio.00832-21
Liu, W., Hong, J., Bevan, D. R., and Zhang, Y. H. (2009). Fast identification of thermostable beta-glucosidase mutants on cellobiose by a novel combinatorial selection/screening approach. Biotechnol. Bioeng. 103, 1087–1094. doi: 10.1002/bit.22340
Liu, G., Zhang, J., and Bao, J. (2016). Cost evaluation of cellulase enzyme for industrial-scale cellulosic ethanol production based on rigorous Aspen plus modeling. Bioprocess Biosyst. Eng. 39, 133–140. doi: 10.1007/s00449-015-1497-1
Méndez Arias, J., Modesto, L. F. A., Polikarpov, I., and Pereira, N. Jr. (2016). Design of an enzyme cocktail consisting of different fungal platforms for efficient hydrolysis of sugarcane bagasse: optimization and synergism studies. Biotechnol. Prog. 32, 1222–1229. doi: 10.1002/btpr.2306
Minor, C. M., Takayesu, A., Ha, S. M., Salwinski, L., Sawaya, M. R., Pellegrini, M., et al. (2024). A genomic analysis reveals the diversity of cellulosome displaying bacteria. Front. Microbiol. 15:1473396. doi: 10.3389/fmicb.2024.1473396
Miras, I., Schaeffer, F., Béguin, P., and Alzari, P. M. (2002). Mapping by site-directed mutagenesis of the region responsible for cohesin-dockerin interaction on the surface of the seventh cohesin domain of Clostridium thermocellum Cip a. Biochemistry 41, 2115–2119. doi: 10.1021/bi011854e
Montella, S., Ventorino, V., Lombard, V., Henrissat, B., Pepe, O., and Faraco, V. (2017). Discovery of genes coding for carbohydrate-active enzyme by metagenomic analysis of lignocellulosic biomasses. Sci. Rep. 7:42623. doi: 10.1038/srep42623
Morais, S., Barak, Y., Hadar, Y., Wilson, D. B., Shoham, Y., Lamed, R., et al. (2011). Assembly of xylanases into designer Cellulosomes promotes efficient hydrolysis of the Xylan component of a natural recalcitrant cellulosic substrate. MBio 2:233. doi: 10.1128/mBio.00233-11
Moraïs, S., Ben David, Y., Bensoussan, L., Duncan, S. H., Koropatkin, N. M., Martens, E. C., et al. (2016). Enzymatic profiling of cellulosomal enzymes from the human gut bacterium, Ruminococcus champanellensis, reveals a fine-tuned system for cohesin-dockerin recognition. Environ. Microbiol. 18, 542–556. doi: 10.1111/1462-2920.13047
Morrison, M., and Miron, J. (2000). Adhesion to cellulose by Ruminococcus albus: a combination of cellulosomes and Pil-proteins? 1. FEMS Microbiol. Lett. 185, 109–115. doi: 10.1111/j.1574-6968.2000.tb09047.x
Musa, K. H., and Elnour, A. A. M. (2024). Advances and future perspectives in biotechnological and bioconversional of dates byproducts. J. Agric. Food Res. 16:101145. doi: 10.1016/j.jafr.2024.101145
Nhim, S., Baramee, S., Tachaapaikoon, C., Pason, P., Ratanakhanokchai, K., Uke, A., et al. (2024). Effective semi-fed-batch saccharification with high lignocellulose loading using co-culture of Clostridium thermocellum and Thermobrachium celere strain A9. Front. Microbiol. 15:1519060. doi: 10.3389/fmicb.2024.1519060
Pagès, S., Bélaïch, A., Bélaïch, J. P., Morag, E., Lamed, R., Shoham, Y., et al. (1997). Species-specificity of the cohesin-dockerin interaction between Clostridium thermocellum and Clostridium cellulolyticum: prediction of specificity determinants of the dockerin domain. Proteins 29, 517–527. doi: 10.1002/(SICI)1097-0134(199712)29:4<517::AID-PROT11>3.0.CO;2-P
Pages, S., Belaich, A., Fierobe, H. P., Tardif, C., Gaudin, C., and Belaich, J. P. (1999). Sequence analysis of scaffolding protein Cip C and ORFXp, a new cohesin-containing protein in Clostridium cellulolyticum: comparison of various cohesin domains and subcellular localization of ORFXp. J. Bacteriol. 181, 1801–1810. doi: 10.1128/JB.181.6.1801-1810.1999
Pandya, C., Farelli, J. D., Dunaway-Mariano, D., and Allen, K. N. (2014). Enzyme promiscuity: engine of evolutionary innovation. J. Biol. Chem. 289, 30229–30236. doi: 10.1074/jbc.R114.572990
Payne, C. M., Knott, B. C., Mayes, H. B., Hansson, H., Himmel, M. E., Sandgren, M., et al. (2015). Fungal cellulases. Chem. Rev. 115, 1308–1448. doi: 10.1021/cr500351c
Paz-Cedeno, F. R., Solorzano-Chavez, E. G., Dias, L. M., Otaviano, C. A., Bustamante, L. J. A., Monti, R., et al. (2022). Composition and chemical structure of hemicelluloses and polysaccharides with capability of gel formation. Hemicellulose Biorefinery Sust. Solution Value Addition Bio-Based Products Bioenergy 2022, 111–137. doi: 10.1007/978-981-16-3682-0_4
Puchart, V., Šuchová, K., and Biely, P. (2021). Xylanases of glycoside hydrolase family 30- an overview. Biotechnol. Adv. 47:107704. doi: 10.1016/j.biotechadv.2021.107704
Puchart, V., Šuchová, K., and Biely, P. (2023). “Chapter 6- importance of accessory enzymes in hemicellulose degradation” in Polysaccharide-degrading biocatalysts. eds. R. Goldbeck and P. Poletto (New York, NY: Academic Press), 139–176.
Raghothama, S., Eberhardt, R. Y., Simpson, P., Wigelsworth, D., White, P., Hazlewood, G. P., et al. (2001). Characterization of a cellulosome dockerin domain from the anaerobic fungus Piromyces equi. Nat. Struct. Biol. 8, 775–778. doi: 10.1038/nsb0901-775
Rettenmaier, R., Kowollik, M. L., Klingl, A., Liebl, W., and Zverlov, V. (2019). Ruminiclostridium herbifermentans sp. nov., a mesophilic and moderately thermophilic cellulolytic and xylanolytic bacterium isolated from a lab-scale biogas fermenter fed with maize silage. Int. J. Syst. Evol. Microbiol. 71:4692. doi: 10.1099/ijsem.0.004692
Riley, R., Salamov, A. A., Brown, D. W., Nagy, L. G., Floudas, D., Held, B. W., et al. (2014). Extensive sampling of basidiomycete genomes demonstrates inadequacy of the white-rot/brown-rot paradigm for wood decay fungi. Proc. Natl. Acad. Sci. USA 111, 9923–9928. doi: 10.1073/pnas.1400592111
Rosenau, T., Potthast, A., Hofinger, A., Bacher, M., Yoneda, Y., Mereiter, K., et al. (2018). Toward a better understanding of cellulose swelling, dissolution, and regeneration on the molecular level. Cellulose Sci. Technol. 22, 99–125. doi: 10.1002/9781119217619.ch5
Sabathé, F., Bélaïch, A., and Soucaille, P. (2002). Characterization of the cellulolytic complex (cellulosome) of Clostridium acetobutylicum. FEMS Microbiol. Lett. 217, 15–22. doi: 10.1111/j.1574-6968.2002.tb11450.x
Sankaran, R., Markandan, K., Khoo, K. S., Cheng, C. K., Ashokkumar, V., Deepanraj, B., et al. (2021). The expansion of lignocellulose biomass conversion into bioenergy via Nanobiotechnology. Front. Nanotechnol. 3:793528. doi: 10.3389/fnano.2021.793528
Schaeffer, F., Matuschek, M., Guglielmi, G., Miras, I., Alzari, P. M., and Béguin, P. (2002). Duplicated dockerin subdomains of Clostridium thermocellum endoglucanase Cel D bind to a cohesin domain of the scaffolding protein Cip a with distinct thermodynamic parameters and a negative cooperativity. Biochemistry 41, 2106–2114. doi: 10.1021/bi011853m
Semana, P., and Powlowski, J. (2019). Four aromatic Intradiol ring cleavage dioxygenases from Aspergillus niger. Appl. Environ. Microbiol. 85:1786. doi: 10.1128/AEM.01786-19
Seppälä, S., Wilken, S. E., Knop, D., Solomon, K. V., and O'Malley, M. A. (2017). The importance of sourcing enzymes from non-conventional fungi for metabolic engineering and biomass breakdown. Metab. Eng. 44, 45–59. doi: 10.1016/j.ymben.2017.09.008
Singh, R., Kumar, M., Mittal, A., and Mehta, P. K. (2016). Microbial enzymes: industrial progress in 21st century. 3 Biotech 6:174. doi: 10.1007/s13205-016-0485-8
Singhvi, M., and Kim, B. S. (2020). Current developments in lignocellulosic biomass conversion into biofuels using Nanobiotechology approach. Energies 13:5300. doi: 10.3390/en13205300
Siqueira, J. G. W., Rodrigues, C., Vandenberghe, L. P. S., Woiciechowski, A. L., and Soccol, C. R. (2020). Current advances in on-site cellulase production and application on lignocellulosic biomass conversion to biofuels: a review. Biomass Bioenergy 132:105419. doi: 10.1016/j.biombioe.2019.105419
Skovgaard, P. A., and Jorgensen, H. (2013). Influence of high temperature and ethanol on thermostable lignocellulolytic enzymes. J. Ind. Microbiol. Biotechnol. 40, 447–456. doi: 10.1007/s10295-013-1248-8
Slutzki, M., Reshef, D., Barak, Y., Haimovitz, R., Rotem-Bamberger, S., Lamed, R., et al. (2015). Crucial roles of single residues in binding affinity, specificity, and promiscuity in the cellulosomal cohesin-dockerin interface. J. Biol. Chem. 290, 13654–13666. doi: 10.1074/jbc.M115.651208
Smith, M. A., Rentmeister, A., Snow, C. D., Wu, T., Farrow, M. F., Mingardon, F., et al. (2012). A diverse set of family 48 bacterial glycoside hydrolase cellulases created by structure-guided recombination. FEBS J. 279, 4453–4465. doi: 10.1111/febs.12032
Solomon, K. V., Haitjema, C. H., Henske, J. K., Gilmore, S. P., Borges-Rivera, D., Lipzen, A., et al. (2016). Early-branching gut fungi possess a large, comprehensive array of biomass-degrading enzymes. Science 351, 1192–1195. doi: 10.1126/science.aad1431
Steenbakkers, P. J., Harhangi, H. R., Bosscher, M. W., van der Hooft, M. M., Keltjens, J. T., van der Drift, C., et al. (2003). beta-glucosidase in cellulosome of the anaerobic fungus Piromyces sp. strain E2 is a family 3 glycoside hydrolase. Biochem. J. 370, 963–970. doi: 10.1042/bj20021767
Sugano, Y. (2009). DyP-type peroxidases comprise a novel heme peroxidase family. Cell. Mol. Life Sci. 66, 1387–1403. doi: 10.1007/s00018-008-8651-8
Syguła, E., Ciolkosz, D., and Białowiec, A. (2024). The significance of structural components of lignocellulosic biomass on volatile organic compounds presence on biochar - a review. Wood Sci. Technol. 58, 859–886. doi: 10.1007/s00226-024-01557-y
Takimura, O., Yanagida, T., Fujimoto, S., and Minowa, T. (2013). Estimation of bioethanol production Cost from Rice straw by on-site enzyme production. J. Japan Petroleum Institute 56, 150–155. doi: 10.1627/jpi.56.150
Talebnia, F., Karakashev, D., and Angelidaki, I. (2010). Production of bioethanol from wheat straw: an overview on pretreatment, hydrolysis and fermentation. Bioresour. Technol. 101, 4744–4753. doi: 10.1016/j.biortech.2009.11.080
Tanaka, H., Enoki, A., Fuse, G., and Nishimoto, K. (1988). Interactions in successive exposure of Wood to varying Wood-inhabiting Fungi 42, 29–35. doi: 10.1515/hfsg.1988.42.1.29
Tieves, F., Willot, S. J.-P., van Schie, M. M. C. H., Rauch, M. C. R., Younes, S. H. H., Zhang, W., et al. (2019). Formate oxidase (FOx) from aspergillus oryzae: one catalyst enables diverse H2O2-dependent biocatalytic oxidation reactions. Angew. Chem. Int. Ed. 58, 7873–7877. doi: 10.1002/anie.201902380
Tsai, S. L., DaSilva, N. A., and Chen, W. (2013). Functional display of complex cellulosomes on the yeast surface via adaptive assembly. ACS Synth. Biol. 2, 14–21. doi: 10.1021/sb300047u
Tsai, S. L., Oh, J., Singh, S., Chen, R., and Chen, W. (2009). Functional assembly of minicellulosomes on the Saccharomyces cerevisiae cell surface for cellulose hydrolysis and ethanol production. Appl. Environ. Microbiol. 75, 6087–6093. doi: 10.1128/AEM.01538-09
Tursi, A. (2019). A review on biomass: importance, chemistry, classification, and conversion. Biofuel Research J. 6, 962–979. doi: 10.18331/BRJ2019.6.2.3
Ullrich, R., Nüske, J., Scheibner, K., Spantzel, J., and Hofrichter, M. (2004). Novel Haloperoxidase from the agaric basidiomycete Agrocybe aegerita oxidizes aryl alcohols and aldehydes. Appl. Environ. Microbiol. 70, 4575–4581. doi: 10.1128/AEM.70.8.4575-4581.2004
Umezawa, K., Niikura, M., Kojima, Y., Goodell, B., and Yoshida, M. (2020). Transcriptome analysis of the brown rot fungus Gloeophyllum trabeum during lignocellulose degradation. PLoS One 15:e0243984. doi: 10.1371/journal.pone.0243984
van Erven, G., Kleijn, A. F., Patyshakuliyeva, A., Di Falco, M., Tsang, A., de Vries, R. P., et al. (2020). Evidence for ligninolytic activity of the ascomycete fungus Podospora anserina. Biotechnol. Biofuels 13:75. doi: 10.1186/s13068-020-01713-z
Vanderstraeten, J., da Fonseca, M. J. M., De Groote, P., Grimon, D., Gerstmans, H., Kahn, A., et al. (2022). Combinatorial assembly and optimisation of designer cellulosomes: a galactomannan case study. Biotechnol. Biofuels Bioprod. 15:60. doi: 10.1186/s13068-022-02158-2
Varadi, M., Anyango, S., Deshpande, M., Nair, S., Natassia, C., Yordanova, G., et al. (2022). Alpha fold protein structure database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 50, D439–D444. doi: 10.1093/nar/gkab1061
Vodovnik, M., and Lindic, N. (2025). Towards the application of nature's catalytic nanomachines: cellulosomes in 2nd generation biofuel production. Biotechnol. Adv. 79:108523. doi: 10.1016/j.biotechadv.2025.108523
Wang, Y., Song, X., Zhang, S., Li, J., Shu, Z., He, C., et al. (2016). Improving the activity of Trichoderma reesei cel 7B through stabilizing the transition state. Biotechnol. Bioeng. 113, 1171–1177. doi: 10.1002/bit.25887
Wang, Z. J., Zhu, J. Y., Fu, Y. J., Qin, M. H., Shao, Z. Y., Jiang, J. G., et al. (2013). Lignosulfonate-mediated cellulase adsorption: enhanced enzymatic saccharification of lignocellulose through weakening nonproductive binding to lignin. Biotechnol. Biofuels 6:156. doi: 10.1186/1754-6834-6-156
Ward, L., Islam, M. S., Kao, N., and Ahmed-Haras, M. R. (2020). An overview of recent developments in hetero-catalytic conversion of cellulosic biomass. Res. Eng. Sci. Technol. 4, 43–54. doi: 10.22597/rcest.v4.65
Wei, H., Fu, Y., Magnusson, L., Baker, J. O., Maness, P. C., Xu, Q., et al. (2014). Comparison of transcriptional profiles of Clostridium thermocellum grown on cellobiose and pretreated yellow poplar using RNA-Seq. Front. Microbiol. 5:142. doi: 10.3389/fmicb.2014.00142
Weimer, P. J. (2022). Degradation of cellulose and hemicellulose by ruminal microorganisms. Microorganisms 10:2345. doi: 10.3390/microorganisms10122345
Wilhelm, R. C., Singh, R., Eltis, L. D., and Mohn, W. W. (2019). Bacterial contributions to delignification and lignocellulose degradation in forest soils with metagenomic and quantitative stable isotope probing. ISME J. 13, 413–429. doi: 10.1038/s41396-018-0279-6
Wilson, C. A., and Wood, T. M. (1992). The anaerobic fungus Neocallimastix frontalis: isolation and properties of a cellulosome-type enzyme fraction with the capacity to solubilize hydrogen-bond-ordered cellulose. Appl. Microbiol. Biotechnol. 37, 125–129. doi: 10.1007/BF00174216
Wingren, A., Galbe, M., and Zacchi, G. (2003). Techno-economic evaluation of producing ethanol from softwood: comparison of SSF and SHF and identification of bottlenecks. Biotechnol. Prog. 19, 1109–1117. doi: 10.1021/bp0340180
Xu, Q., Bayer, E. A., Goldman, M., Kenig, R., Shoham, Y., and Lamed, R. (2004). Architecture of the Bacteroides cellulosolvens cellulosome: description of a cell surface-anchoring scaffoldin and a family 48 cellulase. J. Bacteriol. 186, 968–977. doi: 10.1128/JB.186.4.968-977.2004
Xu, Q., Gao, W., Ding, S. Y., Kenig, R., Shoham, Y., Bayer, E. A., et al. (2003). The cellulosome system of Acetivibrio cellulolyticus includes a novel type of adaptor protein and a cell surface anchoring protein. J. Bacteriol. 185, 4548–4557. doi: 10.1128/JB.185.15.4548-4557.2003
Xu, Z., Lei, P., Zhai, R., Wen, Z., and Jin, M. (2019). Recent advances in lignin valorization with bacterial cultures: microorganisms, metabolic pathways, and bio-products. Biotechnol. Biofuels 12:32. doi: 10.1186/s13068-019-1376-0
Xu, Q., Singh, A., and Himmel, M. E. (2009). Perspectives and new directions for the production of bioethanol using consolidated bioprocessing of lignocellulose. Curr. Opin. Biotechnol. 20, 364–371. doi: 10.1016/j.copbio.2009.05.006
Yadav, V. K., Gupta, N., Kumar, P., Dashti, M. G., Tirth, V., Khan, S. H., et al. (2022). Recent advances in synthesis and degradation of lignin and lignin nanoparticles and their emerging applications in nanotechnology. Materials 15:953. doi: 10.3390/ma15030953
Yang, B., and Wyman, C. E. (2008). Pretreatment: the key to unlocking low-cost cellulosic ethanol. Biofuels Bioproducts Biorefining 2, 26–40. doi: 10.1002/bbb.49
Yao, X., Chen, C., Wang, Y., Dong, S., Liu, Y. J., Li, Y., et al. (2020). Discovery and mechanism of a pH-dependent dual-binding-site switch in the interaction of a pair of protein modules. Sci. Adv. 6:eabd7182. doi: 10.1126/sciadv.abd7182
Yoav, S., Stern, J., Salama-Alber, O., Frolow, F., Anbar, M., Karpol, A., et al. (2019). Directed evolution of Clostridium thermocellum beta-glucosidase a towards enhanced Thermostability. Int. J. Mol. Sci. 20:4701. doi: 10.3390/ijms20194701
Youssef Noha, H., Couger, M. B., Struchtemeyer Christopher, G., Liggenstoffer Audra, S., Prade Rolf, A., Najar Fares, Z., et al. (2013). The genome of the anaerobic fungus Orpinomyces sp. strain C1A reveals the unique evolutionary history of a remarkable plant biomass degrader. Appl. Environ. Microbiol. 79, 4620–4634. doi: 10.1128/AEM.00821-13
Zerillo, M. M., Adhikari, B. N., Hamilton, J. P., Buell, C. R., Lévesque, C. A., and Tisserat, N. (2013). Carbohydrate-active enzymes in Pythium and their role in plant Cell Wall and storage polysaccharide degradation. PLoS One 8:e72572. doi: 10.1371/journal.pone.0072572
Zhang, Y., and Skolnick, J. (2004). Scoring function for automated assessment of protein structure template quality. Proteins Structure Funct. Bioinf. 57, 702–710. doi: 10.1002/prot.20264
Zhivin, O., Dassa, B., Moraïs, S., Utturkar, S. M., Brown, S. D., Henrissat, B., et al. (2017). Unique organization and unprecedented diversity of the Bacteroides (Pseudobacteroides) cellulosolvens cellulosome system. Biotechnol. Biofuels 10:211. doi: 10.1186/s13068-017-0898-6
Zverlov Vladimir, V., Klupp, M., Krauss, J., and Schwarz, W. H. (2008). Mutations in the Scaffoldin gene, cip a, of Clostridium thermocellum with impaired Cellulosome formation and cellulose hydrolysis: insertions of a new transposable element, IS 1447, and implications for Cellulase synergism on crystalline cellulose. J. Bacteriol. 190, 4321–4327. doi: 10.1128/JB.00097-08
Keywords: cellulosome, lignocellulose, enzymatic degradation, carbohydrate-active enzymes, biomass, biofuel
Citation: Hsin K-T, Lee H, Huang Y-C, Lin G-J, Lin P-Y, Lin Y-CJ and Chen P-Y (2025) Lignocellulose degradation in bacteria and fungi: cellulosomes and industrial relevance. Front. Microbiol. 16:1583746. doi: 10.3389/fmicb.2025.1583746
Edited by:
Anandharaj Marimuthu, Joint Genome Institute, United StatesReviewed by:
Willem J. H. Van Berkel, Wageningen University and Research, NetherlandsYuridia Mercado-Flores, Universidad Politécnica de Pachuca, Mexico
Copyright © 2025 Hsin, Lee, Huang, Lin, Lin, Lin and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pao-Yang Chen, cGFveWFuZ0BnYXRlLnNpbmljYS5lZHUudHc=
†These authors have contributed equally to this work
 Kuan-Ting Hsin
Kuan-Ting Hsin HueyTyng Lee
HueyTyng Lee Yu-Chun Huang
Yu-Chun Huang Guan-Jun Lin
Guan-Jun Lin Pei-Yu Lin
Pei-Yu Lin Ying-Chung Jimmy Lin
Ying-Chung Jimmy Lin Pao-Yang Chen
Pao-Yang Chen