- 1College of Food Science, South China Agricultural University, Guangzhou, China
- 2Guangdong Provincial Key Laboratory of Microbial Safety and Health, State Key Laboratory of Applied Microbiology Southern China, Institute of Microbiology, Guangdong Academy of Sciences, Guangzhou, China
by Wang, C., Ye, Q., Jiang, A., Zhang, J., Shang, Y., Li, F., Zhou, B., Xiang, X., Gu, Q., Pang, R., Ding, Y., Wu, S., Chen. M., Wu, Q., and Wang, J. (2022). Front. Microbiol. 13:820431. doi: 10.3389/fmicb.2022.820431
In the published article, there were errors in Figures 2, 3, page 8 as published.
The purpose of both Figures 2, 3 was to explore the sensitivity of the novel identified target detection between genomic DNA and pure culture of P. aeruginosa. Unfortunately, during the final uploading of the data, the Figures 2, 3 were pasted incorrectly.
The corrected Figures 2, 3 and their captions appear below.
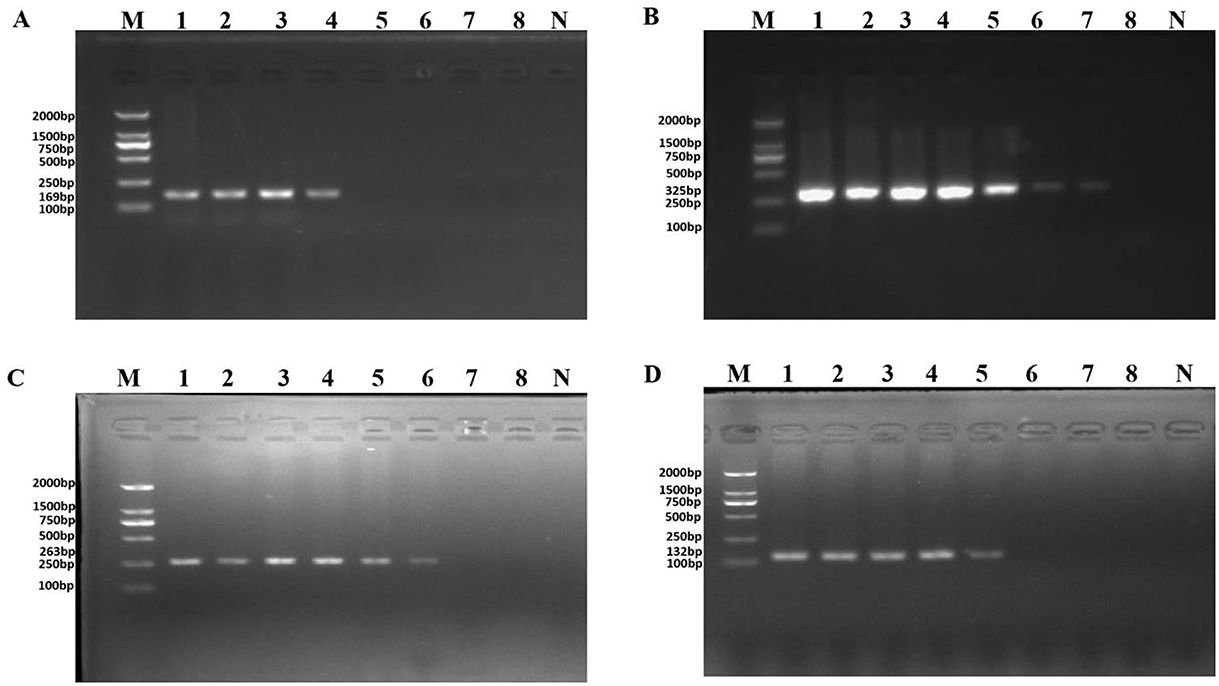
Figure 2. PCR detection sensitivity using dilutions of genomic DNA from Pseudomonas aeruginosa ATCC 15442. Lane M = DSTM 2000 marker (Dongsheng Biotechnology, Guangdong, China); lane N = negative control (double-distilled H2O); lanes 1–8 = 65.4 ng/μl, 6.54 ng/μl, 654 pg/μl, 65.4 pg/μl, 6.54 pg/μl, 654 fg/μl, and 65.4 fg/μl, 6.54 fg/μl, respectively. (A) Primer set PA1 (169 bp); (B) primer set PA2 (325 bp); (C) primer set PA3 (263 bp); and (D) primer set PA4 (132 bp).
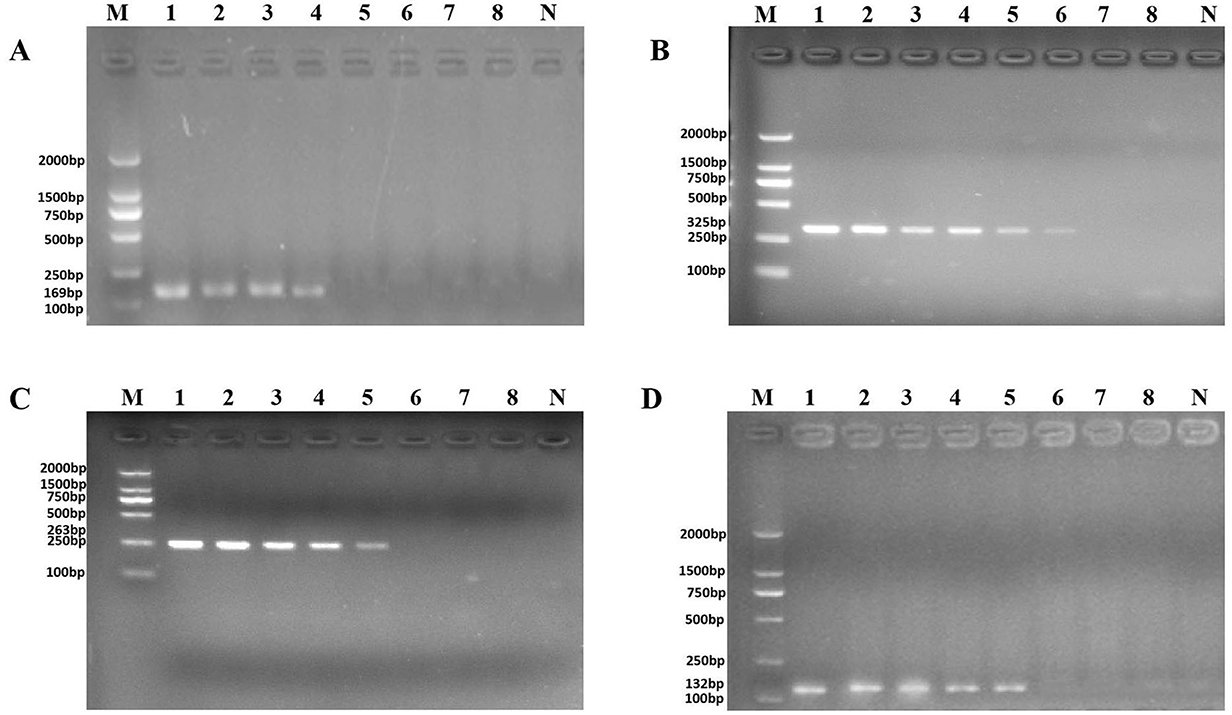
Figure 3. PCR detection sensitivity using dilutions of a pure culture of P. aeruginosa ATCC 15442. Lane M = DSTM 2000 marker (Dongsheng Biotechnology, Guangdong, China); lane N = negative control (double-distilled H2O); and lanes 1–8 = 2.07 × 108 CFU/ml, 2.07 × 107 CFU/ml, 1.85 × 106 CFU/ml, 4.15 × 105 CFU/ml, 4.3 × 104 CFU/ml, 9.7 × 103 CFU/ml, 1.4 × 102 CFU/ml, and 2 × 101 CFU/ml, respectively. (A) Primer set PA1 (169 bp); (B) primer set PA2 (325 bp); (C) primer set PA3 (263 bp); and (D) primer set PA4 (132 bp).
In the published article, there was an error in Supplementary Figure 1. During the assembling of different Figures, we mistakenly pasted Figure S1.d at the position of Figure S1.h.
The authors apologize for this error and state that this does not change the scientific conclusions of the article in any way. The original article has been updated.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Keywords: novel target gene, Pseudomonas aeruginosa, pangenome analysis, PCR, ready-to-eat vegetables
Citation: Wang C, Ye Q, Jiang A, Zhang J, Shang Y, Li F, Zhou B, Xiang X, Gu Q, Pang R, Ding Y, Wu S, Chen M, Wu Q and Wang J (2025) Corrigendum: Pseudomonas aeruginosa detection using conventional PCR and quantitative real-time PCR based on species-specific novel gene targets identified by pangenome analysis. Front. Microbiol. 16:1583946. doi: 10.3389/fmicb.2025.1583946
Received: 26 February 2025; Accepted: 24 March 2025;
Published: 24 April 2025.
Edited and reviewed by: Filipa Grosso, University of Porto, Portugal
Copyright © 2025 Wang, Ye, Jiang, Zhang, Shang, Li, Zhou, Xiang, Gu, Pang, Ding, Wu, Chen, Wu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qingping Wu, d3VxcDIwM0AxNjMuY29t; Juan Wang, d2FuZ2p1YW5Ac2NhdS5lZHUuY24=
†These authors have contributed equally to this work
 Chufang Wang1,2†
Chufang Wang1,2† Jumei Zhang
Jumei Zhang Yuting Shang
Yuting Shang Fan Li
Fan Li Qihui Gu
Qihui Gu Rui Pang
Rui Pang Yu Ding
Yu Ding Shi Wu
Shi Wu Moutong Chen
Moutong Chen Qingping Wu
Qingping Wu Juan Wang
Juan Wang