- State Key Laboratory of Microbial Metabolism, Joint International Research Laboratory of Metabolic and Developmental Sciences, School of Life Sciences and Biotechnology, Shanghai Jiao Tong University, Shanghai, China
Introduction: Long-term studies on the dynamic changes in nitrogen metabolism and functional microbial communities under anaerobic conditions, particularly those driven by organic amendments, remain scarce.
Methods: We conducted a year-long anaerobic microcosm experiment using three organic amendments—aerobically fermented pig-manure digestate (ACM), compost (ACP) and straw powder (ACS)—alongside an inorganic fertilizer-only control (ACN).
Results: Temporal shifts revealed that organic amendments drove distinct nitrogen metabolism pathways. Amendments of digestate and compost promoted the proliferation of nitrogen-mineralizing bacteria such as Ramlibacter and Lysobacter, leading to significant ammonium accumulation. After 12-month incubation, the ACM treatment caused a 75.6-fold increase in ammonium, a 43.4% rise in total nitrogen (TN), and a 27.0% increase in total organic carbon (TOC). In contrast, the ACS treatment exhibited superior nitrogen fixation, with an average of 1.69-fold higher rate than ACM and 5.30 fold higher than ACP The ACS treatment enriched cellulolytic nitrogen-fixing bacteria, including Clostridium, and nitrogen-fixing archaea.
Discussion: This study provides profound insights in to the unique nitrogen metabolism pathways influenced by organic amendments under anoxic conditions, ultimately offering valuable insights into improved soil fertility and sustainable nitrogen management practices in agricultural systems.
1 Introduction
Nitrogen fertilizers have been widely applied in recent decades to support high agricultural crop yields (Ladha et al., 2016; Guo et al., 2022). However, excess fertilization and low utilization lead to several environmental problems, such as nitrous oxide (N2O) emissions (Li et al., 2024), ammonia volatilization (Ju and Zhang, 2017), and nitrate leaching (Zhao et al., 2022). Various management strategies to overcome the above issues have been evaluated (Young et al., 2021; Hassan et al., 2022; Gao and Cabrera Serrenho, 2023). Deeper application of fertilizers reduces ammonia volatilization by minimizing nitrogen exposure to the atmosphere (Ju and Zhang, 2017; Li et al., 2022), and simultaneously promote the formation of anaerobic micro-zones within the soil (Li et al., 2016; Schlüter et al., 2025). Additionally, combining organic amendments, such as manure (Wang et al., 2022), manure compost (Oyetunji et al., 2022), crop residue (Fu et al., 2021), and digestate (Samoraj et al., 2022; Chozhavendhan et al., 2023), with nitrogen fertilizers can improve soil fertility, enhance nitrogen use efficiency, and contribute to stable crop yields and environmental benefits (Selim, 2020; Liu et al., 2021a; Zhai et al., 2022; Liu et al., 2023).
Long-term field trials have revealed that organic amendment inputs contribute to soil organic carbon (SOC) stabilization by increasing the abundance of stable aliphatic and aromatic carbon compounds, as evidenced by solid-state nuclear magnetic resonance (solid-state NMR) spectroscopy (Guo et al., 2019; Yuan et al., 2025). Importantly, such carbon-fraction diversity is increasingly recognized as a key factor influencing soil microbial community composition, with potential consequences for biogeochemical cycling processes (Fierer, 2017). Material-driven heterogeneity, particularly variations in carbon fraction composition, has also been increasingly recognized as a critical driver of microbial community assembly and biogeochemical processes (Kallenbach et al., 2016; Liang et al., 2017). Changes in the soil carbon-to-nitrogen ratios (C/N) may regulate nitrogen immobilization and mineralization by influencing soil microbial community composition and the activities of microorganisms in decomposing organic matter fractions (Lin et al., 2019; Cui et al., 2022). In addition, exogenous organic matter can stimulate soil organic matter (SOM) mineralization through positive priming effects, enhancing nutrient availability in soil (Hicks et al., 2019). Several long-term field experiments have demonstrated that manure application promotes organic nitrogen accumulation and mineralization (Abubaker et al., 2012; Wu et al., 2017; Xia et al., 2021).
Deeper placement of fertilizers helps to create anaerobic zones, which consequently enhance denitrification and increase N₂O emissions (Meng et al., 2022). However, inconsistent findings regarding the impacts of combined inorganic and organic fertilization on N₂O emissions have been reported in literature (Reddy and Crohn, 2014; Akhtar et al., 2020; Zhang et al., 2023; Zuo et al., 2023; Wu et al., 2024). These discrepancies are likely due to variations in the types of organic amendments and differences in nitrogen-cycling microbial communities (He et al., 2019; Zhou et al., 2022). Thus, it is critical to investigate how organic amendments with distinct carbon compositions modulate the structure and function of nitrogen-transforming microbiomes under oxygen-limited conditions. Such insights are essential for unraveling the microbial mechanisms underlying nitrogen cycling and for informing fertilization strategies that improve nitrogen use efficiency while reducing environmental risks.
Previous studies have investigated the effects of organic amendments on soil microbial community compositions and nitrogen cycling functions (Liu et al., 2021b; Cui et al., 2022; Chen et al., 2023). However, most investigations have relied on short-term incubations or endpoint sampling, which provide limited insights into the long-term succession dynamics of nitrogen-transforming microbiomes (Ouyang et al., 2020; Bossolani et al., 2023; Pastorelli et al., 2024). While a few efforts have introduced a dynamic perspective, such as a 16-week anaerobic incubation tracking microbial community shifts under straw-amended soils (Yang et al., 2023). In that study, 16S rRNA gene-based taxonomic profiling was employed, but without any functional analysis.
Yet nitrogen transformation processes, such as denitrification, nitrogen fixation, and organic matter decomposition, typically involve slow and progressive shifts in microbial community composition and functional activity. Localized anaerobic conditions frequently arise in agricultural soils following deep fertilizer placement or intense microbial respiration, and the biogeochemical processes occurring within these zones unfold over extended periods. Consequently, short-term studies may fail to capture the cumulative functional succession of nitrogen-transforming microbiomes under oxygen-limited conditions, underscoring the need for long-term incubation experiments that better simulate these ecological dynamics. Nevertheless, the mechanisms by which amendment-derived carbon fraction differences drive the functional succession of nitrogen-transforming microbiomes under prolonged oxygen limitation remain poorly understood. To address these gaps, we designed a one-year-long anaerobic incubation experiment using γ-ray irradiated, carbon-normalized organic amendments with standardized nitrate additions, coupled with multi-time-point shotgun metagenomics and nitrogenous gas flux monitoring, aiming to elucidate the microbial mechanisms by which the discrepancy of amendment-derived carbon fractions drives nitrogen cycling under sustained oxygen limitation.
2 Materials and methods
2.1 Experimental materials
Soil samples were collected from the Shangzhuang Experimental Station (39°48′N, 116°28′E) of China Agricultural University, which was initiated in October 2006. The experimental site followed a winter wheat–summer maize rotation, with fertilizers applied based on basal and follow-up applications. The original soil used in this study was topsoil (0–20 cm) collected from the conventional fertilizer straw-no-return treatment group (Abbreviated C for original soil), passed through a 2 mm sieve, and stored at 4°C before subsequent experiments. The main properties of the soil were displayed in Supplementary Table S1.
Three types of organic amendments were used in this study: digestate remaining after anaerobic fermentation of swine manure for methane production (abbreviated as M), compost from aerobic fermentation of pig manure (abbreviated as P), and a 1:1 (w:w) mixture of wheat straw and corn straw powder (abbreviated as S). These amendments were selected for their relevance to agricultural practices and their distinct impacts on nitrogen cycling and microbial dynamics. Straw was chosen to simulate in-situ straw return, representing crop residues from the experimental field. Compost and digestate, both derived from pig manure, were selected due to their widespread use in agricultural practices. The differences in fermentation methods influence nitrogen release patterns and microbial community impacts.
The organic amendments were sterilized to exclude the introduction of exogenous bacteria before being added to the soil. Sterilization was conducted with cobalt 60 irradiation using a sterilization dose of 50 kGy by the Shanghai Radiant Technology Company, following established laboratory protocols and methods validated in previous studies (Wu et al., 2019). The main properties of the organic amendments were displayed in Supplementary Table S1.
2.2 Experimental design
To investigate the effects of adding organic matter with different carbon and nitrogen fractions on soil nitrogen metabolism, four experimental groups were established. The experimental groups consisted of anaerobically fermented pig manure digestate (ACM), aerobically fermented pig manure compost (ACP), straw powder (ACS), and a control group without organic matter amendment (ACN). In these group designations, ‘A’ represents anaerobic conditions, and ‘C’ refers to conventional nitrogen-fertilized soil.
Organic matters were added to the soil based on equivalent total carbon input to ensure that any observed effects were caused by differences in the composition of the amendments rather than differences of carbon content. The total amount of aerobically fermented pig manure compost, which had the highest nitrate content, was set to be added to 10% (w:w) of the soil (with an initial total carbon content of 8.992 g per kg of dry soil). The amounts of anaerobically fermented pig manure digestate and straw powder were calculated to match the total carbon content applied in the ACP treatment, ensuring consistency in carbon input across all treatments.
To control the unexpected effects of variations in carbon and nitrate content, we ensured consistency in the total carbon content and nitrate levels before the start of the incubation. This would ensure that observed differences in nitrogen metabolism and microbial community composition were solely due to variations in the carbon components of the amendments.
Organic amendment was introduced in two batches: the first at the start of the incubation and the second after six months of incubation. The total amount of organic matters addition was evenly divided between these two batches, maintaining equivalent carbon input across all treatments, as described above. This two-batch application was designed to simulate a basal and follow-up fertilization pattern commonly used in agricultural management. Potassium nitrate solution was added to maintain consistent nitrate levels across all treatments, accounting for the nitrate already present in the organic amendments.
The organic matters and potassium nitrate solution were added to 25 g of soil in serum bottles, and the headspace was replaced with pure helium (99.99%) to maintain anaerobic conditions. Anaerobic conditions were achieved by performing multiple rounds of gas replacement with helium, and the anaerobic level was confirmed through monitoring, which showed oxygen concentrations remained below detection limits throughout the experiment. Incubation was carried out at 25°C in the dark, with soil moisture maintained at 75% water holding capacity (WHC). Additional nitrate and water were injected every three months to sustain the experimental conditions. Periodic supplementation every 3 months mimicked the fertilization pulses typical in agricultural settings, allowing for a more realistic assessment of the impact of organic amendments on nitrogen metabolism.
The experiment followed a parallel replicated design with 18 replicates per group. Triplicate samples were destructively collected from each group at time point of 7 days (D7), 3 months (M3), 6 months (M6), 9 months (M9), and 1 year (M12), for high-throughput sequencing of the 16S rRNA gene (V3-V4 regions) and for physicochemical analysis, including total carbon (TC), total nitrogen (TN), total organic carbon (TOC), pH, nitrate, nitrite, and ammonium. Before each sampling, headspace gas in the system was measured using the ROBOT (Supplementary Figure S2), an instrument capable of continuously monitoring the gas concentrations in the headspace of the bottles (Molstad et al., 2007). Bacterial community composition and functional gene profiles were analyzed through metagenomic shotgun sequencing at 7 days, 6 months, and 1 year.
The detailed information of experimental design is provided in Supplementary Figures S1, S2 and Supplementary Table S2.
2.3 Physical and chemical analysis of soils and organic amendment
TC, TOC, and TN were analyzed with an elemental analyzer (Vario EL III/Isoprime). TOC was determined after removing inorganic carbon by hydrochloric acid fumigation (Ramnarine et al., 2011). Inorganic nitrogen contents were determined with the cadmium reduction column method for nitrate and nitrite (Huffman and Barbarick, 1981) and with the indophenol blue method for ammonium (Dorich and Nelson, 1983). Total organic nitrogen (TON) was calculated as the total nitrogen minus the inorganic nitrogen (nitrate, nitrite, and ammonium). pH was determined in a solution with a distilled water–soil ratio of 2.5:1 using a pH meter (Mettler Toledo, Switzerland).
The organic carbon fractions of the four experimental groups were determined at Day 0 following the addition of the corresponding organic matter using the 13C cross-polarization/total sideband suppression (CP/TOSS) method of solid-state NMR (Bruker Avance Neo 600 MHz). The NMR integrals of each functional group in the organic carbon fractions were calculated using the Topspin 4.13 program (Chen et al., 2018).
To compare the denitrifying potential of soil before and after 1-year incubation, the denitrification gas kinetics with addition of 150 mg N/kg of nitrate were assessed using the ROBOT system (Supplementary Figure S2) for multiple sampling during 3 day period (Molstad et al., 2007).
2.4 Calculation of nitrogen fixation rates
To quantify nitrogen fixation, we developed a calculation approach based on the difference between the total amount of added nitrate and the remaining nitrogen gas (N₂) concentration in each closed system. The calculation of nitrogen fixation was conducted separately for two stages, as the seal of the vials was opened in between. They are stages of the first 6 months (1) and the second 6 months (2), as indicated below.
where (1) F1 represents the amount of nitrogen fixation of each group in the first stage, A1 represents the total amount of nitrate nitrogen added to each sample during the D0–M6 period (210 mg/kg), and NG1 represents the nitrogen content measured in each closed system at M6. (2) F2 represents the amount of nitrogen fixation of each group during the second stage, and A2 represents the total amount of nitrate nitrogen added to each sample during the M6–M12 period (120 mg/kg), while NG2 represents the nitrogen content measured in each closed system at M12.
2.5 16S rRNA gene amplicon sequencing and bioinformatics analysis
DNA was extracted from 0.5 g of soil samples as described previously (Griffiths et al., 2000; Paulin et al., 2013). Two rounds of polymerase chain reaction (PCR) were used to amplify the V3-V4 hypervariable regions of 16S rRNA genes from the soil DNA to construct high-throughput sequencing libraries (Zhang et al., 2016; Wu et al., 2019). The first round of PCR was performed using the universal primers B341F/B785R and the PCR products were purified by magnetic bead adsorption. The second round of PCR was performed using purified products as the template and primers with sequencing index tags added to the DNA amplicons obtained by the second round of PCR. The constructed sequencing libraries were then mixed in equal concentrations and subjected to high-throughput sequencing on the Illumina Miseq sequencing platform.
The obtained sequences were processed using the QIIME2 platform (Bolyen et al., 2019), and the “cutadapt” plug-in was used to excise junctions and primers, followed by the use of the DADA2 program to perform quality control of sequences (trimming, filtering, denoising, and chimera removal) and to merge amplicon sequence variants (ASVs) to obtain the original abundance matrix and ASV sequence information. A total of 1,949,178 high-quality sequences were obtained from 63 samples after quality control and filtration, and 9,561 ASVs were obtained after clustering (Supplementary Table S3). The sequence numbers of all samples were normalized to 17,000 (the number of sequences in the sample with the fewest sequences) to enable direct comparisons among samples. The subsequent analyses conducted in this study were based on normalized data. The ASV classifications were then taxonomically annotated using the SILVA138.2 database (Bolyen et al., 2019). The rarefaction curves (Supplementary Figure S7A) eventually flattened, indicating that the sequencing depth was sufficient to meet the requirements of the subsequent data analysis.
2.6 Metagenome shotgun sequencing and bioinformatics analysis
Soil metagenomic DNA was used for library construction and sequenced on an Illumina NovaSeq high-throughput sequencing platform (Personal Co. Ltd., Shanghai). A total of 39 samples were collected in triplicates from initial samples, and from four incubation groups in D7, M6 and M12 for metagenomic sequencing. Clean metagenomic sequencing reads were obtained after quality control using FastQC (Andrews, 2010), followed by assembly with the MEGAHIT (Li et al., 2015) to obtain contigs that were assembled into scaffolds using IDBA (Peng et al., 2010). Basic sequencing and quality control metrics for all samples, including raw data, GC content, Q30, and assembly statistics, are summarized in Supplementary Table S4. Gene prediction was then performed using the MetaGeneMark (Hyatt et al., 2012), followed by clustering of proteins using CD-HIT (Fu et al., 2012) into groups with >90% sequence similarity to enable removal of redundancy. The longest sequence was then used as the representative sequence to obtain non-redundant protein sets. The htseq-count was used to count the number of reads mapped to the non-redundant genes and assess gene abundances using transcripts per million (TPM) values. Nitrogen-related functional proteins were then annotated using the NcycDB based on the non-redundant protein datasets (Tu et al., 2019). Furthermore, carbohydrate degradation-related functional proteins (referred to as CAZymes) were annotated using the CAZy database (Drula et al., 2022), while taxonomic annotation was conducted using the NCBI non-redundant (NR) protein database.
Each sample was subjected to generate metagenome-assembled genome (MAG) using MetaBAT2 (Kang et al., 2019). The obtained MAGs were then quality-filtered using the CheckM (Parks et al., 2015). “High-quality draft genomes” with estimates of completeness >90% and contamination <5%, in addition to “medium-quality draft genomes” with estimates of completeness ≥50% and contamination <10%, were retained, for a total of 651 MAGs (Bowers et al., 2017). The genome datasets were subjected to de-redundancy using the dRep (Olm et al., 2017) based on 99% average nucleotide identity (ANI), leading to a total of 470 non-redundant MAGs. The relative abundances of non-redundant MAGs were calculated using CoverM (https://github.com/wwood/CoverM/releases/tag/v0.6.1). Taxonomic annotations of the non-redundant MAGs were then assigned using the GTDB-Tk (release 207) (Chaumeil et al., 2022). Phylophlan v.3.0 was subsequently used for phylogenetic tree construction (Asnicar et al., 2020), followed by visualization of trees using iTOL (Letunic and Bork, 2019). Nitrogen cycling gene annotations within MAGs were assigned using NcycDB, while annotations of lignin, cellulose, and hemicellulose degrading enzymes were assigned with the CAZy database. Kyoto Encyclopedia of Genes and Genomes (KEGG) Orthology (KO) annotation of MAGs for the KEGG database was conducted using kofam_scan v.1.3.0 (ver. 2023-03-02 KEGG release 105.0) (Aramaki et al., 2020). The KEGGDecoder was then used to parse the kofam_scan module outputs to determine the completeness of various metabolic pathways (Graham et al., 2018). Detailed sequencing and analysis parameters were provided in Supplementary Table S12.
2.7 Statistical analysis
Repeated measures Two-way analysis of variance (ANOVA) was applied to analyze the denitrification gas data (e.g., N₂O and N₂ dynamics) to assess the effects of organic amendment type and incubation duration across multiple time points. For other physicochemical indicators (e.g., ammonium concentration, nitrogen fixation rate), standard Two-way ANOVA was used. To meet ANOVA assumptions, logarithmic transformation was applied to the data where necessary, based on assessments of normality (Shapiro–Wilk test) and homogeneity of variances (Brown-Forsythe test). Post hoc comparisons were conducted using Tukey’s honestly significant difference (HSD) test to identify significant differences between group means (Ratsiatosika et al., 2024). All analyses were performed using GraphPad Prism 10 (Swift, 1997), with the significance level set at α = 0.05 for all tests. Full details of the statistical analysis are provided in Supplementary Tables S8–S11.
Principal coordinates analysis (PCoA) was calculated using R vegan package for Bray-Curtis distances and plotted using ggplot2 (Wickham, 2011) package. The overall statistical significance of group separation was assessed using permutational multivariate analysis of variance (PERMANOVA).
We constructed six random forest regression models using the R package ‘randomForest’ (Breiman, 2001) to identify MAGs associated with changes in ammonium content (Figure 1G), nitrogen fixation (Figure 1I), N₂O accumulation (Figures 1A,D), and total denitrification (N₂O + N₂, Figures 1B,E) across incubation stages. The significance of selected MAGs was assessed using the percentage increase in mean square error (MSE%), with higher values indicating greater importance (Jiao et al., 2018). Model performance was evaluated using the ‘A3’ package for cross-validation R2 and permutation significance tests, while predictor variable significance was assessed via ‘rfPermute’ (Jiao et al., 2018). Full model details and the importance and significance of selected MAGs are provided in Supplementary Tables S6, S7, respectively.
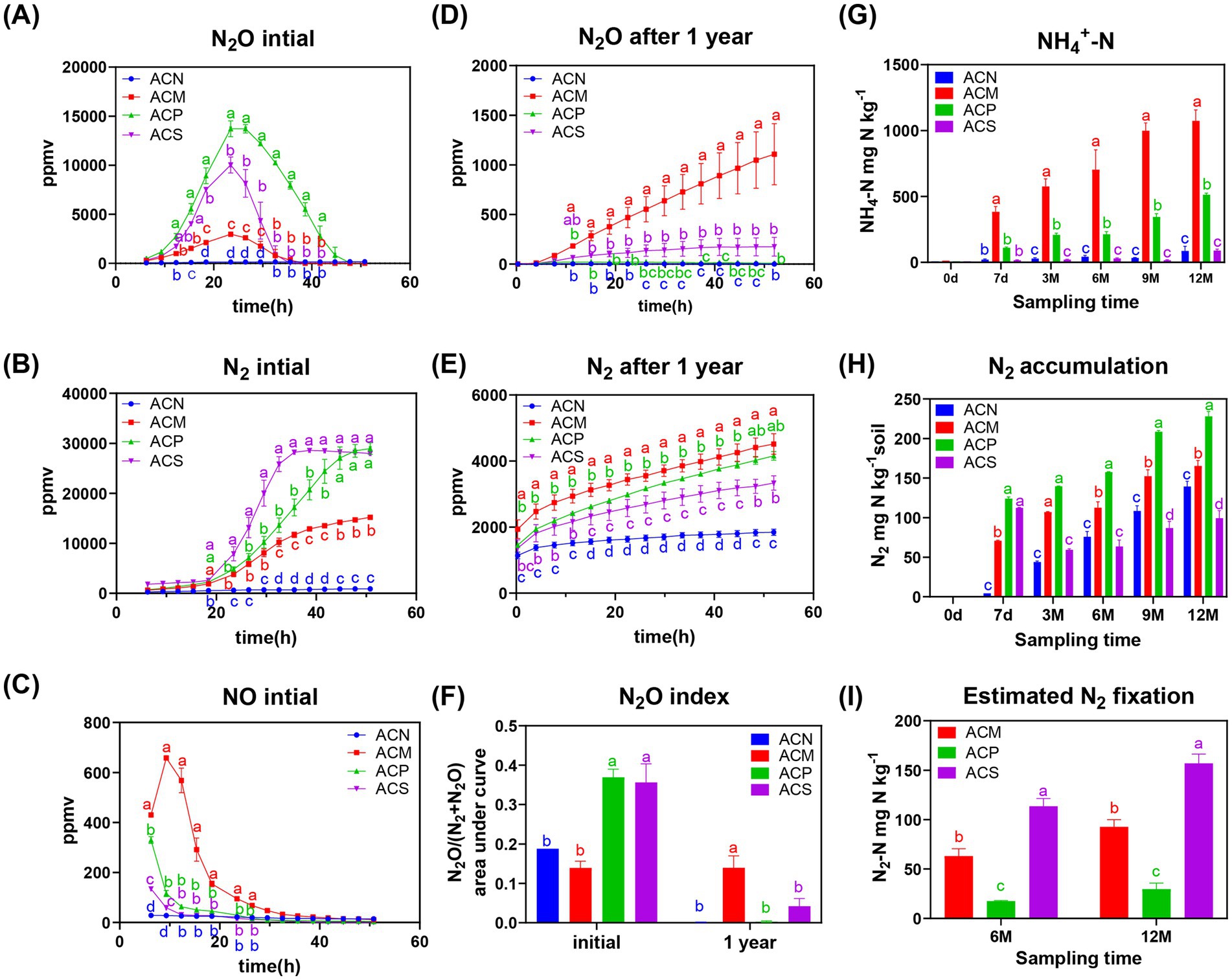
Figure 1. Dynamic variations in N2O, NH4+-N, N2, NO and nitrogen fixation activity during the one - year incubation. N2O (A), N2 (B), and NO (C) dynamics measured in soil sample before the1-year incubation. N2O (D) and N2 (E) dynamics measured in soil after the 1-year incubation. (F) N2O index before and after 1-year incubation. (G) Accumulation of ammonium during the incubation. (H) Accumulation of nitrogen gas during the incubation. (I) Amount of fixed nitrogen after 6 and 12 months of incubation. Error bars indicate the standard error of the means (n = 3). Significance was determined using repeated measures Two-way ANOVA for (A–E) and standard Two-way ANOVA for (F–I), followed by Tukey’s HSD post hoc test. Different letters indicate significant differences between groups at each time period (p < 0.05). Global and inter-time point significance results are provided in Supplementary Tables S8, S10, and Supplementary Figure S6.
3 Results
3.1 Effects of different organic amendments on nitrogen metabolic processes
The three types of organic amendments differentially affected soil physicochemical parameters in Fluvo-aquic soils (Supplementary Figures S3–S5). Solid-state NMR measurement of composition of carbon fractions in the four experimental groups at day 0 of incubation indicated that all three organic matter additions increased the percentage of labile carbon fractions (Supplementary Figure S3B), as the TOC content of the ACM, ACP, and ACS groups were equally and significantly higher than that of ACN (p < 0.05, Supplementary Figure S4B). The N-alkyl functional group with a chemical shift of 45–65 ppm (Supplementary Figure S3C) was higher in the ACM than ACP.
The three organic amendments differentially affected nitrogen metabolism, specially, all of them enhanced soil denitrification capacity (Figures 1A–C). Furthermore, organic amendment significantly increased the production and accumulation of N2O and N2 at the beginning of incubation (p < 0.05, Figures 1A,B). After one year of incubation, N2 concentrations were high in all of the organic matter addition groups (Figure 1E), while only minimal N2O accumulation was detected in the ACM group (Figures 1D,F). At the beginning of the incubations, the ACP and ACS group treatments accumulated greater N2O, while the ACS group consumed nitrate more rapidly through denitrification (Figures 1A,B). Among the three organic treatments, ACM addition accumulated the least amount of N2O and consumed nitrate through denitrification most slowly (Figures 1A,B).
During the incubation period, ammonium significantly accumulated in the ACM and ACP groups with incubation time (p < 0.05, Figure 1G). Moreover, nitrogen balance calculations revealed that the amount of accumulated ammonium significantly exceeded the initial nitrate input (Supplementary Figure S5B), suggesting that the primary source of ammonium was the mineralization of organic nitrogen from the added organic amendments. In contrast, the ACS group exhibited an initial increase followed by a decrease in nitrogen levels, implying the presence of nitrogen fixation within the incubation systems (Figure 1H). The calculated nitrogen fixation suggested the highest levels in the ACS compared with the ACM and ACP groups, with a increasing trend observed within all treatments over time (Figure 1I).
3.2 Effects of different organic amendments on microbial communities
After one year of incubation, the Shannon diversity index was consistently lower in all experimental groups with organic amendments (ACM, ACP and ACS) than in the control group (ACN) (p < 0.05, Supplementary Figure S7B). Furthermore, PCoA analysis based on Bray–Curtis distances (Figure 2A) indicated that the type of organic amendment and incubation time significantly affected microbial community composition (p < 0.001, Supplementary Table S11). Consistently, differences in phylum-level compositions were observed among groups (Figure 2B and Supplementary Table S13). For example, the organic amendments notably increased Firmicutes abundance (p < 0.05, Supplementary Table S13). Microbial community compositions in different groups also showed observable changes with incubation time, with the exception of the ACN group in which the majority of phyla remained relatively stable. Overall, ACS communities exhibited different variation tendency compared to the communities of two manure-based groups, ACM and ACP.
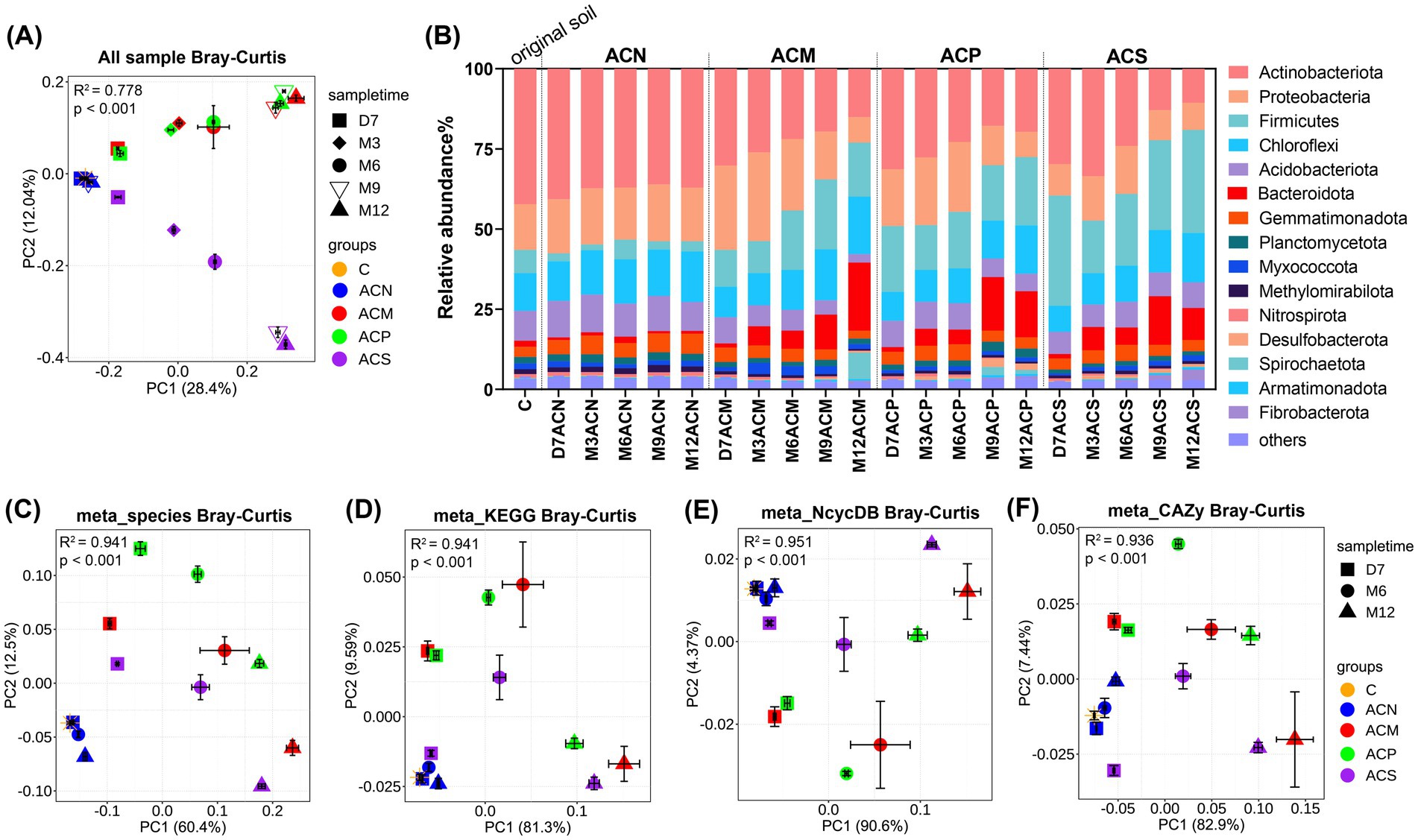
Figure 2. Microbial community compositional variation among treatments. (A) PCoA analysis of all samples based on Bray–Curtis distances of 16S rRNA gene sequence data. (B) Bacterial composition at the phylum level of samples. (C) PCoA analysis of microbial community composition based on species-level Bray–Curtis distances of metagenomic sequence data. (D) PCoA analysis of all KOs annotated based on kegg database. (E) PCoA analysis of functional genes related to the nitrogen cycle annotated based on NcycDB. (F) PCoA analysis of functional genes related to carbohydrate metabolism annotated based on CAZY database. All PCoA analyses were based on Bray–Curtis distances. The coordinates of each point in each PCoA plot were the mean of each group of samples, and the error bars indicate the standard error of the means (n = 3). Differences among groups were calculated via PERMANOVA (A,C–F).
3.3 Temporal effects of organic amendments on nitrogen metabolism genes
From samples of this study, all sixty-eight nitrogen cycling genes annotated in NcycDB and associated with eight nitrogen cycling pathways were successfully identified in the metagenomic data (Supplementary Table S5). Among these pathways, Organic degradation and synthesis (ODAS), denitrification, Dissimilatory nitrate reduction (DNRA), and Assimilatory nitrate reduction (ANRA) showed consistently higher relative abundances across all samples (Supplementary Figure S8). Among these, two nitrogen mineralization functional genes (gdh_K00262 and gdh_K15371) were identified in the ODAS pathway, which is associated with organic nitrogen degradation and ammonium production. Specifically, gdh_K00262 was enriched over whole peroid in the experimental group with organic amendment, while gdh_K15371 was enriched only in the early stage of incubation in ACM and ACP groups. In contrast, DNRA-related functional genes (nirB, nirD, nrfA, nrfC) and ANRA-related genes (nirA), which are associated with ammonium production, were consistently less abundant in the ACM and ACP groups compared to the ACN group (Figure 3). Furthermore, the total abundances of DNRA and ANRA pathway genes showed declining trends over time compared to the original soil (Supplementary Figures S8D,E). The nitrogen fixation genes nifD, nifK, and nifH were significantly enriched with incubation time in the ACS group (p < 0.05, Figure 3). In the organic amendment groups, enrichment of the denitrification functional genes napA, nirK, and nosZ decreased with incubation time (Supplementary Figure S9), while the relative abundances of norB genes significantly increased in these three groups (p < 0.05, Figure 3).
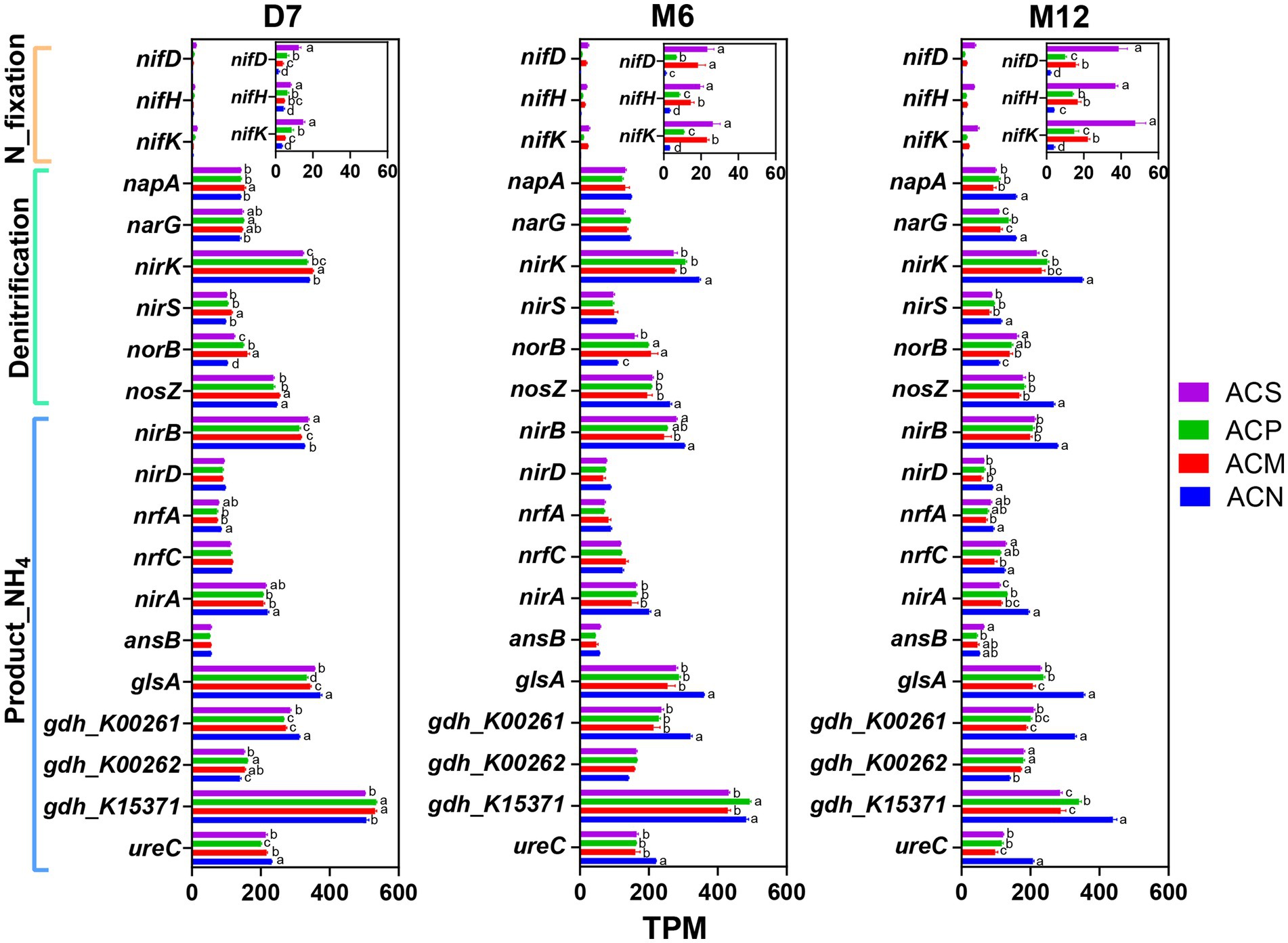
Figure 3. The abundances of functional genes for nitrogen fixation, denitrification and catalytic ammonium production function at different time points (D7: 7 days; M6: 6 months; M12: 12 months). Gene abundance in the metagenomic data was normalized and expressed as transcripts per million (TPM). Error bars indicate the standard error of the means (n = 3). Differences in the values were calculated based on analysis of variance (ANOVA) and Tukey’s HSD post hoc test. Different letters denote significant differences among groups at a significance level of p < 0.05.
3.4 Effects of organic amendments on the functional microbiome of nitrogen mineralization and denitrification
Considering the microorganisms that encoded nitrogen mineralization functional genes (Figures 4A–C), organic amendment enriched some specific taxa in all communities including Lysobacter and Luteimonas, in addition to the enrichment of Anaeromyxobacter and Lentimicrobium harboring the gdh_K00262 genes at the later stage of cultivation. Organic matter addition also enriched Anaeromyxobacter that contains the denitrifying genes of narG, napA, and norB (Figures 4D,E,H). Lysobacter and Luteimonas harbor genes of nirK, norB, and nosZ (Figures 4F,H,I). In the microbial communities carrying the functional genes gdh_K00262, gdh_K15371, ureC, nirK and nosZ, compared with the ACN group, the abundances of Luteitalea and Solirubrobacter decreased across the incubation in the ACM, ACP, and ACS groups. Among the microorganisms carrying nifH, nifD and nifK, Anaeromyxobacter was enriched in organic amendment groups, while Clostridium, Methanocella, and Methanosarcina were only enriched in the ACS group (Figures 4J–L).
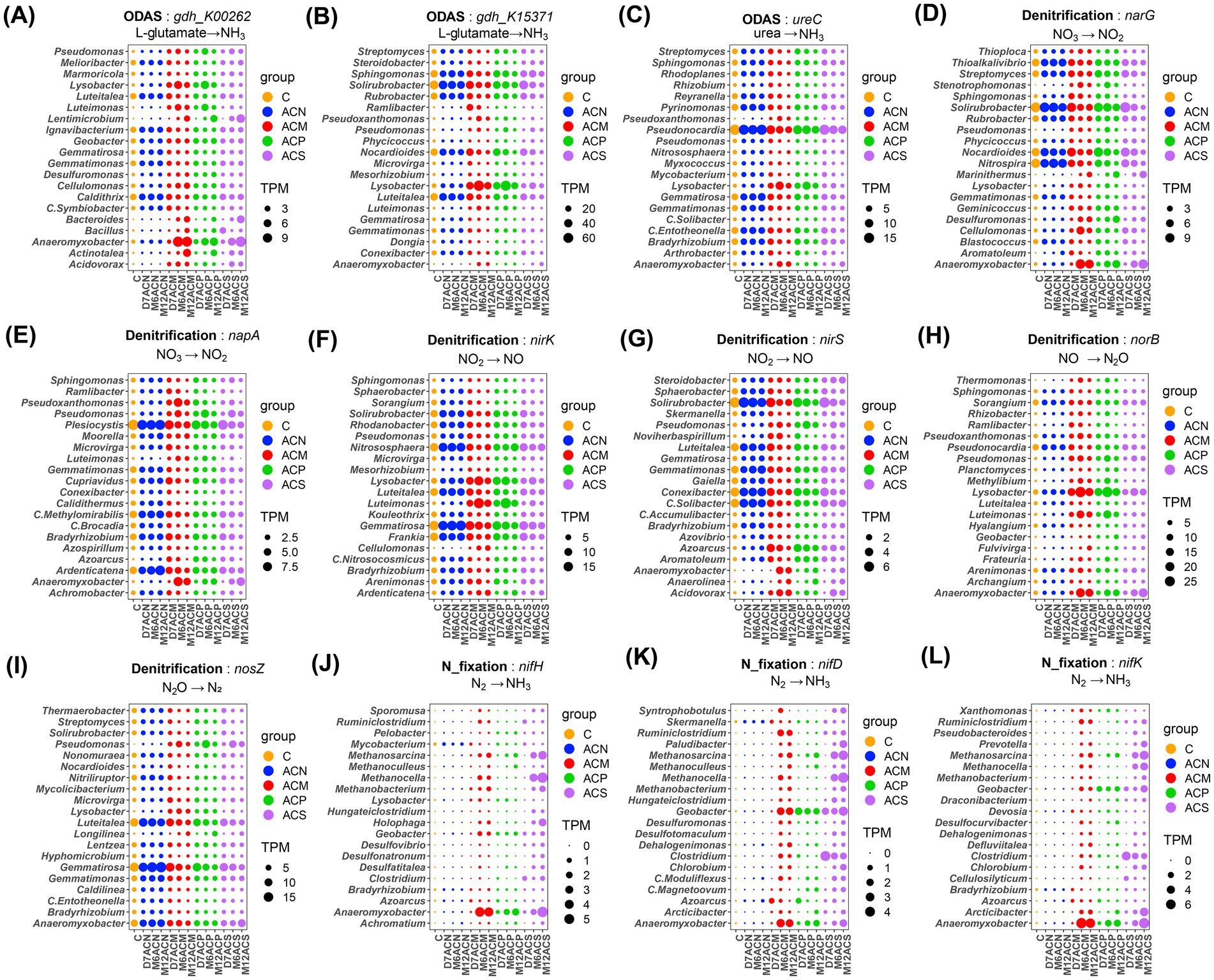
Figure 4. Top 20 dominant bacterial genera involved in nitrogen mineralization, denitrification and nitrogen fixation pathways. Dominant bacteria contain gdh_K00262 (A), gdh_K15371 (B), ureC (C), narG (D), napA (E), nirK (F), nirS (G), norB (H), nosZ (I), nifH (J), nifD (K), nifK (L), respectively. Circle size is the TPM value. Different colors represent different groups.
3.5 Diversity and nitrogen functional annotations of MAGs
To better understand the links between microbial species and nitrogen cycling processes, we conducted metagenome binning to reconstruct MAGs. A total of 651 MAGs were reconstructed, of which 208 were high-quality draft genomes and 443 were medium-quality draft genomes (Bowers et al., 2017). 470 MAGs were retained after de-redundancy based on 99% ANI. The MAGs were assigned to 29 phyla, 47 classes, 84 orders (464 MAGs), 107 families (453 MAGs), and 105 genera (369 MAGs) based on annotation by the GTDB database (Figure 5). NcycDB-based functional annotation of nitrogen cycling genes indicated that 373 MAGs contained one or more functional genes for denitrification, while 443 MAGs contained genes related to ammonium production from organic nitrogen mineralization and 79 MAGs encoded functional genes for nitrogen fixation (Figure 5). Among all the 79 MAGs predicted for nitrogen fixation, 33 MAGs contained multiple cellulolytic and hemicellulolytic enzyme-encoding genes (Supplementary Figure S12), which were primarily derived from Bacteroidota, Fibrobacterota, and Firmicutes.
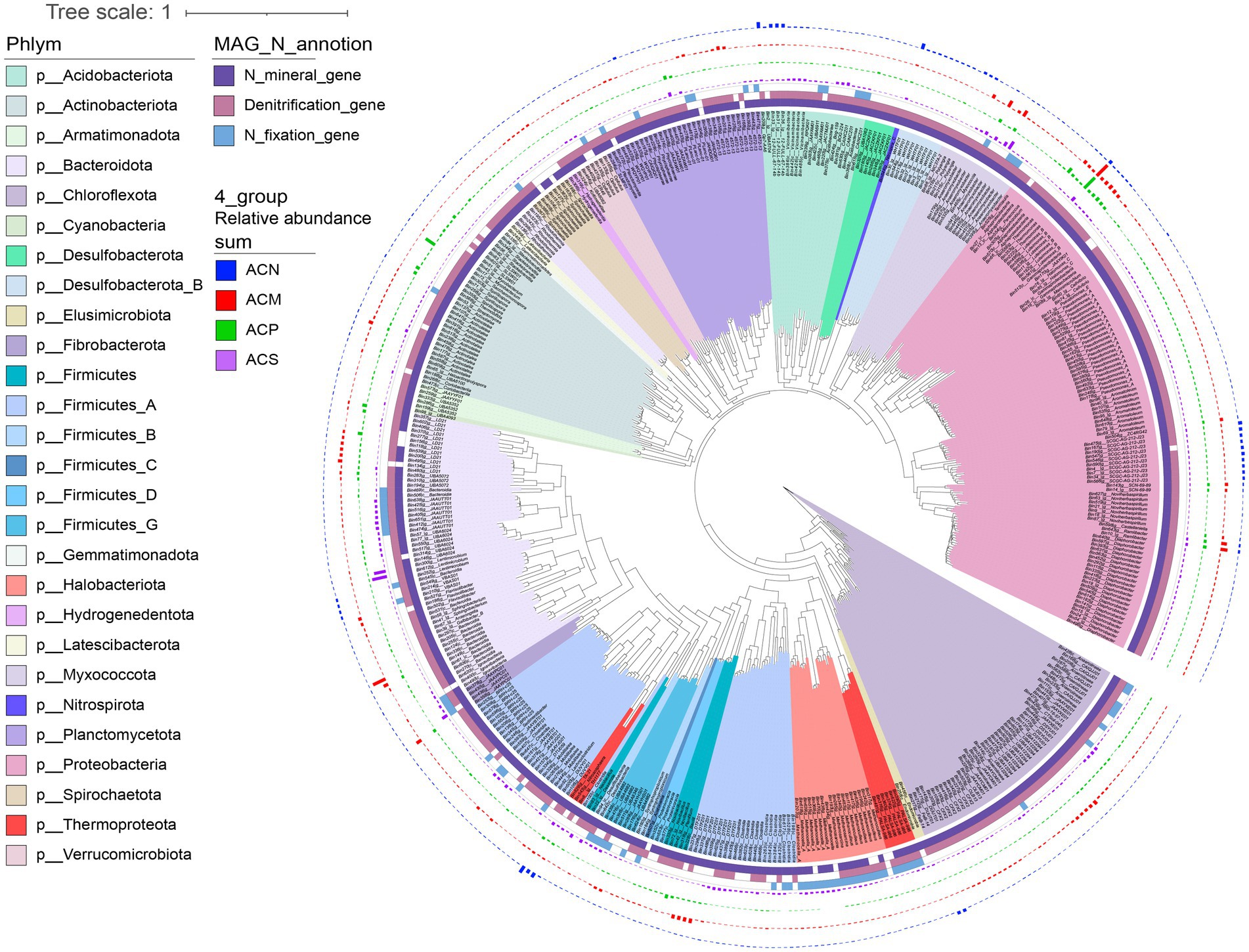
Figure 5. Phylogenetic tree showing the diversity, total abundances, and annotated nitrogen-related function of 470 reconstructed metagenome-assembled genomes (MAGs). Different colors on the phylogenetic tree branches represent different bacterial or archaeal phyla. The four outer circles of different colored bars represent the relative abundance of MAGs in the four experimental groups, with ACN in blue, ACM in red, ACP in green, and ACS in purple. The 3 inner circles indicate the types of nitrogen functional genes annotated in the MAG. The dark purple color indicates that a nitrogen mineralization gene is annotated, the light purple color indicates that a denitrification gene is annotated, and the light blue color indicates that a nitrogen fixation gene is annotated.
Twenty-six MAGs associated with changes in ammonium content were enriched in the ACM and ACP groups (Figure 6). In particular, MAGs primarily annotated as Burkholderiaceae, Xanthomonadaceae, and Anaeromyxobacteraceae were identified by the random forest regression model as important predictors contributing to ammonium accumulation. Among these, Ramlibacter exhibited higher relative abundance in the early cultivation stages, while Luteimonas_B had a higher relative abundance in the middle cultivation stage, and Anaeromyxobacter was more abundant in both the middle and late culture stages (p < 0.05, Supplementary Figure S13). Twelve MAGs that were important for changes in nitrogen fixation exhibited higher relative abundance in the late culture stage of the ACS group, with some also showing higher relative abundance at the late culture stage of the ACM group (Figure 6). Furthermore, MAGs with higher relative abundance at the late culture stage were primarily affiliated with the JAAUTT01, Methanocella_A, Methanosarcina, and Methanoculleus genera (p < 0.05, Supplementary Figure S13). Among the 25 MAGs associated with the denitrification process, 10 MAGs contained only N2O production genes and lacked nosZ for N2O reduction; these MAGs were primarily found in the ACM group (Figure 6). Additionally, Neobacillus MAG93 was identified as a complete denitrifier capable of converting nitrate to N2 and had high abundance in the ACS group at day 7 (p < 0.05, Supplementary Figure S13).

Figure 6. Random forest-selected metagenome-assembled genomes (MAGs) associated with the function of ammonium accumulation, nitrogen fixation, and denitrification. Heatmap indicates the relative abundance of the selected important MAGs. The bubble map represents the carbon and nitrogen-related functional genes annotated in the selected MAGs, different colors represent different modules. The cross-validation R2 values for the random forest regression models are as follows: ammonium accumulation (R2 = 0.73), nitrogen fixation (R2 = 0.81), and denitrification (R2 = 0.64 to 0.85 across different models). The details of the random forest regression model are displayed in Supplementary Table S6.
4 Discussion
4.1 Organic amendment and organic matter mineralization
During the incubation period, the ACM and ACP treatments resulted in significant ammonium accumulation, far exceeding the initial nitrate input (Supplementary Figure S5B). This results suggests that the primary source of ammonium derived from organic matter mineralization. The low abundances of DNRA and ANRA pathway-related genes (e.g., nirB, nirD, nrfA, nirA; Figure 3) and the overall declining trends of these pathways (Supplementary Figures S8D,E) further support their minimal contributions to ammonium accumulation. The increased gross nitrogen mineralization rates in these treatments were primarily attributed to one or more of several mechanisms, which are explored in the following sections.
First, the differences in the added organic matter fractions drove distinct nitrogen transformation pathways and fluxes. Solid-state NMR analysis indicated that the digestate and compost addition groups contained higher N-alkyl C fractions compared with the straw group, and this fraction primarily originated from proteins and amino acids (Chen et al., 2018). The rapid turnover of amino acids in soil (Jones and Kielland, 2012) may be one of the reasons for quick accumulation of mineralization-derived ammonium nitrogen in the ACM and ACP samples. By contrast, the higher percentage of the N-alkyl C component of ACM compared with the ACP group resulted in greater accumulation of ammonium in the ACM group. The stability index (Chen et al., 2018) relating to the ratio of alkyl-C to O-alkyl-C (A/O-A ratio) of the ACM group was lower than within the ACP group (Supplementary Figure S3D), indicating that its components were more easily degraded.
Second, manure additions greatly increased the abundance of the glutamate dehydrogenase encoding gene gdh_k15371, in addition to the abundances of the nitrogen-acquiring enzyme-encoding genes K01255 (leucine aminopeptidase, LAP) and K01207 (β-1,4-n-acetylglucosaminidase, NAG) during the early incubation stage (Supplementary Figure S10). The higher abundance of these functional genes during the initial stage suggests that the ACM and ACP groups had higher potential activities of organic nitrogen mineralizing enzymes and extracellular nitrogen-acquiring enzymes (Hu et al., 2020; Chen and Sinsabaugh, 2021). Nitrogen-acquiring enzyme activities have been shown to be positively correlated with nitrogen mineralization rates (Wang et al., 2023). These changes would increase the supply of dissolved organic nitrogen (DON) required for microbial nitrogen uptake and mineralization, as well as improve the ability to utilize organic nitrogen (Jian et al., 2016). Consistently with Wang et al. (2024), field applications of manure and digestate increased nitrogen-acquiring enzyme activities. This also aligns with the microbial nitrogen mining hypothesis (Craine et al., 2007), which suggests that excess exogenous carbon promotes microbial secretion of extracellular enzymes to acquire the nitrogen needed for growth.
Third, compost and digestate pig manure additions led to the enrichment of key bacterial taxa involved in nitrogen mineralization. MAGs associated with ammonium nitrogen production were enriched in the ACM and ACP groups. Among these, MAG10 and MAG643 of the genus Ramlibacter, which annotated contain the nitrogen-acquiring enzyme LAP encoding gene K01255, were most abundant in the early stage of incubation when ammonium accumulation rate was at its peak. Random forest regression analysis identified these MAGs as important predictors of ammonium levels. The abundance of genus Ramlibacter was also significantly and positively correlated with ammonium accumulation in another study (Hu et al., 2023).
4.2 Nitrogen fixation triggered by straw addition
The significant amount of nitrogen fixed in the ACS group could be due to the combined involvement of cellulolytic nitrogen-fixing bacteria (CNFB) and methanogenic archaea.
Straw addition enriched CNFB (Harindintwali et al., 2020), which produce cellulases and nitrogen-fixing enzymes, facilitating the degradation of cellulose into simple sugars and promoting nitrogen fixation. CNFB have been widely studied in agricultural composting due to their cellulose-degrading and nitrogen-enriching functions (Harindintwali et al., 2020; Harindintwali et al., 2022). Lignocellulose is a carbon-rich component of crop residues such as straw that primarily comprises cellulose, hemicellulose, and lignin (Harindintwali et al., 2020). Bacteria encoding nifH, nifD, and nifK were enriched in the ACS group, as represented by the anaerobic cellulose-degrading Clostridium (Figures 4J–L), with species having cellulolytic and nitrogen-fixing functions (McDonald et al., 2012; Poehlein et al., 2017). These bacteria may play a key role in both straw degradation and nitrogen fixation in the ACS group during early incubation. The D7ACS group was enriched in CAZymes involved in cellulose and hemicellulose catabolism (Bredon et al., 2018) (Supplementary Figure S11). Consequently, straw addition was hypothesized to rapidly promote the abundance of bacteria encoding cellulose and hemicellulose catabolic enzyme, such as MAG92 (Figure 6).
Nitrogen-fixing taxa enriched in the ACS group in mid-to late-culture periods comprised several methanogenic archaea, including Methanocella and Methanosarcina. The end products of the anaerobic degradation of lignocellulose are primarily carbon dioxide and methane (Nguyen and Khanal, 2018; Song et al., 2023). Methanogenic archaea in confined anaerobic systems use the products from the final step of lignocellulose fermentation, such as acetic acid, hydrogen, and carbon dioxide, to produce methane (Evans et al., 2019). These organisms were enriched over time in this study. Nitrogen fixation abilities had been documented in some methanogenic archaea (Belay et al., 1984; Murray and Zinder, 1984; Bomar et al., 1985; Scherer, 1988), although it is not a common trait among all methanogenic archaea. It is consistent with the result that the methanogen MAGs from the ACS group encoded genes for nitrogen fixation. The enrichment of these organisms should promote nitrogen fixation and methanogenesis by using fermentation products such as short fatty acids.
However, although this study provides insights into the regulation of soil N-functional communities by exogenous organic matter additions, we recognize that there are some limitations in the study. Our study did not adequately consider all factors that may affect the soil microbial community and N cycling. The factors, such as aeration in soil and plant cultivation and so on, deserve to be further explored in future studies for a more comprehensive understanding of the complexity of soil ecosystems. Despite these limitations, our findings offer promising insights for practical applications. The rapid accumulation of ammonium from anaerobically fermented pig manure digestate highlights its potential as a readily available nitrogen source to meet the early-stage nutrient demands of plants in agricultural cropping systems. Conversely, the enrichment of nitrogen-fixing CNFB and methanogenic archaea in straw-amended soils promoted long-term nitrogen fixation under anaerobic conditions. These results highlight the opportunity to optimize organic amendment strategies, balancing immediate nutrient availability with sustained nitrogen-fixing capacity in soils.
5 Conclusion
Adding compost, digestate, or straw to soil differentially shapes nitrogen (N) cycling via microbial mechanisms. Compost/digestate boosted microbial enzymes for nitrogen acquisition and ammonium accumulation via mineralization, while straw enriched methanogenic nitrogen-fixing archaea. These organic inputs create distinct nitrogen metabolism pathways by altering microbial communities and functional genes, ultimately affecting soil N availability and greenhouse gases. Strategically selecting organic amendments could synchronize nutrient release with crop demands while suppressing nitrous oxide emissions. This microbial-driven nitrogen partitioning offers a framework to optimize organic input timing/location, balancing soil fertility and environmental sustainability in intensive agriculture. Digestate provides a readily available nitrogen source for early-stage plant growth, while straw promotes long-term nitrogen fixation under anaerobic conditions. By selecting appropriate organic amendments based on the actual soil conditions, farmers may benefit from optimizing nitrogen cycling and improve soil sustainability.
Data availability statement
The data presented in the study are deposited in the NCBI Sequence Read Archive repository, accession number PRJNA1013701.
Author contributions
YM: Conceptualization, Investigation, Methodology, Writing – original draft, Writing – review & editing. QW: Investigation, Writing – review & editing. XW: Formal analysis, Software, Writing – review & editing. WS: Investigation, Writing – review & editing. XZ: Conceptualization, Funding acquisition, Supervision, Writing – review & editing, Writing – original draft.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the National Natural Science Foundation of China (NSFC project 31971526) and the National Key Research and Development Program of China (2017YFD0200102).
Acknowledgments
The authors would like to express their gratitude to Shanshan Zhang and Bona Dai (Instrumental Analysis Center of Shanghai Jiao Tong University, IAC-SJTU) for their help in the pretreatment method of soil total organic carbon determination and in the detection method of soil solid-state NMR, respectively.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1588169/full#supplementary-material
References
Abubaker, J., Risberg, K., and Pell, M. (2012). Biogas residues as fertilisers – effects on wheat growth and soil microbial activities. Appl. Energy 99, 126–134. doi: 10.1016/j.apenergy.2012.04.050
Akhtar, K., Wang, W., Ren, G., Khan, A., Enguang, N., Khan, A., et al. (2020). Straw mulching with inorganic nitrogen fertilizer reduces soil CO(2) and N(2)O emissions and improves wheat yield. Sci. Total Environ. 741:140488. doi: 10.1016/j.scitotenv.2020.140488
Andrews, S. (2010). FastQC: A quality control tool for high throughput sequence data. Cambridge, United Kingdom: Babraham Bioinformatics, Babraham Institute.
Aramaki, T., Blanc-Mathieu, R., Endo, H., Ohkubo, K., Kanehisa, M., Goto, S., et al. (2020). KofamKOALA: KEGG Ortholog assignment based on profile HMM and adaptive score threshold. Bioinformatics 36, 2251–2252. doi: 10.1093/bioinformatics/btz859
Asnicar, F., Thomas, A. M., Beghini, F., Mengoni, C., Manara, S., Manghi, P., et al. (2020). Precise phylogenetic analysis of microbial isolates and genomes from metagenomes using PhyloPhlAn 3.0. Nature. Communications 11:2500. doi: 10.1038/s41467-020-16366-7
Belay, N., Sparling, R., and Daniels, L. (1984). Dinitrogen fixation by a thermophilic methanogenic bacterium. Nature 312, 286–288. doi: 10.1038/312286a0
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9
Bomar, M., Knoll, K., and Widdel, F. (1985). Fixation of molecular nitrogen by Methanosarcina barkeri. FEMS Microbiol. Ecol. 31, 47–55. doi: 10.1016/0378-1097(85)90046-1
Bossolani, J. W., Leite, M. F. A., Momesso, L., ten Berge, H., Bloem, J., and Kuramae, E. E. (2023). Nitrogen input on organic amendments alters the pattern of soil–microbe-plant co-dependence. Sci. Total Environ. 890:164347. doi: 10.1016/j.scitotenv.2023.164347
Bowers, R. M., Kyrpides, N. C., Stepanauskas, R., Harmon-Smith, M., Doud, D., Reddy, T. B. K., et al. (2017). Minimum information about a single amplified genome (MISAG) and a metagenome-assembled genome (MIMAG) of bacteria and archaea. Nat. Biotechnol. 35, 725–731. doi: 10.1038/nbt.3893
Bredon, M., Dittmer, J., Noël, C., Moumen, B., and Bouchon, D. (2018). Lignocellulose degradation at the holobiont level: teamwork in a keystone soil invertebrate. Microbiome 6:162. doi: 10.1186/s40168-018-0536-y
Chaumeil, P. A., Mussig, A. J., Hugenholtz, P., and Parks, D. H. (2022). GTDB-Tk v2: memory friendly classification with the genome taxonomy database. Bioinformatics 38, 5315–5316. doi: 10.1093/bioinformatics/btac672
Chen, J., and Sinsabaugh, R. L. (2021). Linking microbial functional gene abundance and soil extracellular enzyme activity: implications for soil carbon dynamics. Glob. Chang. Biol. 27, 1322–1325. doi: 10.1111/gcb.15506
Chen, M., Xu, J., Li, Z., Li, D., Wang, Q., Zhou, Y., et al. (2023). Long-term nitrogen fertilization-induced enhancements of acid hydrolyzable nitrogen are mainly regulated by the most vital microbial taxa of keystone species and enzyme activities. Sci. Total Environ. 874:162463. doi: 10.1016/j.scitotenv.2023.162463
Chen, X., Xu, Y., Gao, H. J., Mao, J., Chu, W., and Thompson, M. L. (2018). Biochemical stabilization of soil organic matter in straw-amended, anaerobic and aerobic soils. Sci. Total Environ. 625, 1065–1073. doi: 10.1016/j.scitotenv.2017.12.293
Chozhavendhan, S., Karthigadevi, G., Bharathiraja, B., Praveen Kumar, R., Abo, L. D., Venkatesa Prabhu, S., et al. (2023). Current and prognostic overview on the strategic exploitation of anaerobic digestion and digestate: a review. Environ. Res. 216:114526. doi: 10.1016/j.envres.2022.114526
Craine, J. M., Morrow, C., and Fierer, N. (2007). Microbial nitrogen limitation increases decomposition. Ecology 88, 2105–2113. doi: 10.1890/06-1847.1
Cui, J., Zhu, R., Wang, X., Xu, X., Ai, C., He, P., et al. (2022). Effect of high soil C/N ratio and nitrogen limitation caused by the long-term combined organic-inorganic fertilization on the soil microbial community structure and its dominated SOC decomposition. J. Environ. Manag. 303:114155. doi: 10.1016/j.jenvman.2021.114155
Dorich, R. A., and Nelson, D. W. (1983). Direct colorimetric measurement of ammonium in potassium chloride extracts of soils. Soil Sci. Soc. Am. J. 47, 833–836. doi: 10.2136/sssaj1983.03615995004700040042x
Drula, E., Garron, M.-L., Dogan, S., Lombard, V., Henrissat, B., and Terrapon, N. (2022). The carbohydrate-active enzyme database: functions and literature. Nucleic Acids Res. 50, D571–D577. doi: 10.1093/nar/gkab1045
Evans, P. N., Boyd, J. A., Leu, A. O., Woodcroft, B. J., Parks, D. H., Hugenholtz, P., et al. (2019). An evolving view of methane metabolism in the Archaea. Nat. Rev. Microbiol. 17, 219–232. doi: 10.1038/s41579-018-0136-7
Fierer, N. (2017). Embracing the unknown: disentangling the complexities of the soil microbiome. Nat. Rev. Microbiol. 15, 579–590. doi: 10.1038/nrmicro.2017.87
Fu, B., Chen, L., Huang, H., Qu, P., and Wei, Z. (2021). Impacts of crop residues on soil health: a review. Environmental Pollutants Bioavailability 33, 164–173. doi: 10.1080/26395940.2021.1948354
Fu, L., Niu, B., Zhu, Z., Wu, S., and Li, W. (2012). CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28, 3150–3152. doi: 10.1093/bioinformatics/bts565
Gao, Y., and Cabrera Serrenho, A. (2023). Greenhouse gas emissions from nitrogen fertilizers could be reduced by up to one-fifth of current levels by 2050 with combined interventions. Nature Food 4, 170–178. doi: 10.1038/s43016-023-00698-w
Graham, E. D., Heidelberg, J. F., and Tully, B. J. (2018). Potential for primary productivity in a globally-distributed bacterial phototroph. ISME J. 12, 1861–1866. doi: 10.1038/s41396-018-0091-3
Griffiths, R. I., Whiteley, A. S., O’Donnell, A. G., and Bailey, M. J. (2000). Rapid method for Coextraction of DNA and RNA from natural environments for analysis of ribosomal DNA-and rRNA-based microbial community composition. Appl. Environ. Microbiol. 66, 5488–5491. doi: 10.1128/AEM.66.12.5488-5491.2000
Guo, C., Liu, X., and He, X. (2022). A global meta-analysis of crop yield and agricultural greenhouse gas emissions under nitrogen fertilizer application. Sci. Total Environ. 831:154982. doi: 10.1016/j.scitotenv.2022.154982
Guo, Z., Zhang, Z., Zhou, H., Wang, D., and Peng, X. (2019). The effect of 34-year continuous fertilization on the SOC physical fractions and its chemical composition in a vertisol. Sci. Rep. 9:2505. doi: 10.1038/s41598-019-38952-6
Harindintwali, J. D., Wang, F., Yang, W., Zhou, J., Muhoza, B., Mugabowindekwe, M., et al. (2022). Harnessing the power of cellulolytic nitrogen-fixing bacteria for biovalorization of lignocellulosic biomass. Ind. Crop. Prod. 186:115235. doi: 10.1016/j.indcrop.2022.115235
Harindintwali, J. D., Zhou, J., and Yu, X. (2020). Lignocellulosic crop residue composting by cellulolytic nitrogen-fixing bacteria: a novel tool for environmental sustainability. Sci. Total Environ. 715:136912. doi: 10.1016/j.scitotenv.2020.136912
Hassan, M. U., Aamer, M., Mahmood, A., Awan, M. I., Barbanti, L., Seleiman, M. F., et al. (2022). Management strategies to mitigate N2O emissions in agriculture. Life 12:439. doi: 10.3390/life12030439
He, T., Yuan, J., Luo, J., Wang, W., Fan, J., Liu, D., et al. (2019). Organic fertilizers have divergent effects on soil N2O emissions. Biol. Fertil. Soils 55, 685–699. doi: 10.1007/s00374-019-01385-4
Hicks, L. C., Meir, P., Nottingham, A. T., Reay, D. S., Stott, A. W., Salinas, N., et al. (2019). Carbon and nitrogen inputs differentially affect priming of soil organic matter in tropical lowland and montane soils. Soil Biol. Biochem. 129, 212–222. doi: 10.1016/j.soilbio.2018.10.015
Hu, X., Gu, H., Sun, X., Wang, Y., Liu, J., Yu, Z., et al. (2023). Distinct influence of conventional and biodegradable microplastics on microbe-driving nitrogen cycling processes in soils and plastispheres as evaluated by metagenomic analysis. J. Hazard. Mater. 451:131097. doi: 10.1016/j.jhazmat.2023.131097
Hu, R., Wang, X. P., Xu, J. S., Zhang, Y. F., Pan, Y. X., and Su, X. (2020). The mechanism of soil nitrogen transformation under different biocrusts to warming and reduced precipitation: from microbial functional genes to enzyme activity. Sci. Total Environ. 722:137849. doi: 10.1016/j.scitotenv.2020.137849
Huffman, S., and Barbarick, K. (1981). Soil nitrate analysis by cadmium reduction. Commun. Soil Sci. Plant Anal. 12, 79–89. doi: 10.1080/00103628109367129
Hyatt, D., LoCascio, P. F., Hauser, L. J., and Uberbacher, E. C. (2012). Gene and translation initiation site prediction in metagenomic sequences. Bioinformatics 28, 2223–2230. doi: 10.1093/bioinformatics/bts429
Jian, S., Li, J., Chen, J., Wang, G., Mayes, M. A., Dzantor, K. E., et al. (2016). Soil extracellular enzyme activities, soil carbon and nitrogen storage under nitrogen fertilization: a meta-analysis. Soil Biol. Biochem. 101, 32–43. doi: 10.1016/j.soilbio.2016.07.003
Jiao, S., Chen, W., Wang, J., Du, N., Li, Q., and Wei, G. (2018). Soil microbiomes with distinct assemblies through vertical soil profiles drive the cycling of multiple nutrients in reforested ecosystems. Microbiome 6:146. doi: 10.1186/s40168-018-0526-0
Jones, D. L., and Kielland, K. (2012). Amino acid, peptide and protein mineralization dynamics in a taiga forest soil. Soil Biol. Biochem. 55, 60–69. doi: 10.1016/j.soilbio.2012.06.005
Ju, Xt., and Zhang, C. (2017). Nitrogen cycling and environmental impacts in upland agricultural soils in North China: a review. J. Integr. Agric. 16, 2848–2862. doi: 10.1016/S2095-3119(17)61743-X
Kang, D. D., Li, F., Kirton, E., Thomas, A., Egan, R., An, H., et al. (2019). MetaBAT 2: an adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ 7:e7359. doi: 10.7717/peerj.7359
Kallenbach, C. M., Frey, S. D., and Grandy, A. S. (2016). Direct evidence for microbial-derived soil organic matter formation and its ecophysiological controls. Nature Communications 7:13630. doi: 10.1038/ncomms13630
Ladha, J. K., Tirol-Padre, A., Reddy, C. K., Cassman, K. G., Verma, S., Powlson, D. S., et al. (2016). Global nitrogen budgets in cereals: a 50-year assessment for maize, rice and wheat production systems. Sci. Rep. 6:19355. doi: 10.1038/srep19355
Letunic, I., and Bork, P. (2019). Interactive tree of life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47, W256–W259. doi: 10.1093/nar/gkz239
Li, L., Hong, M., Zhang, Y., and Paustian, K. (2024). Soil N2O emissions from specialty crop systems: a global estimation and meta-analysis. Glob. Chang. Biol. 30:e17233. doi: 10.1111/gcb.17233
Li, Y., Kronzucker, H. J., and Shi, W. (2016). Microprofiling of nitrogen patches in paddy soil: analysis of spatiotemporal nutrient heterogeneity at the microscale. Sci. Rep. 6:27064. doi: 10.1038/srep27064
Li, D., Liu, C. M., Luo, R., Sadakane, K., and Lam, T. W. (2015). MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics 31, 1674–1676. doi: 10.1093/bioinformatics/btv033
Li, L., Wu, T., Li, Y., Hu, X., Wang, Z., Liu, J., et al. (2022). Deep fertilization improves rice productivity and reduces ammonia emissions from rice fields in China; a meta-analysis. Field Crop Res. 289:108704. doi: 10.1016/j.fcr.2022.108704
Liang, C., Schimel, J. P., and Jastrow, J. D. (2017). The importance of anabolism in microbial control over soil carbon storage. Nature Microbiol. 2:17105. doi: 10.1038/nmicrobiol.2017.105
Lin, Y., Ye, G., Kuzyakov, Y., Liu, D., Fan, J., and Ding, W. (2019). Long-term manure application increases soil organic matter and aggregation, and alters microbial community structure and keystone taxa. Soil Biol. Biochem. 134, 187–196. doi: 10.1016/j.soilbio.2019.03.030
Liu, P., Guo, X., Zhou, D., Zhang, Q., Ren, X., Wang, R., et al. (2023). Quantify the effect of manure fertilizer addition and optimal nitrogen input on rainfed wheat yield and nitrogen requirement using nitrogen nutrition index. Agric. Ecosyst. Environ. 345:108319. doi: 10.1016/j.agee.2022.108319
Liu, J., Shu, A., Song, W., Shi, W., Li, M., Zhang, W., et al. (2021b). Long-term organic fertilizer substitution increases rice yield by improving soil properties and regulating soil bacteria. Geoderma 404:115287. doi: 10.1016/j.geoderma.2021.115287
Liu, B., Wang, X., Ma, L., Chadwick, D., and Chen, X. (2021a). Combined applications of organic and synthetic nitrogen fertilizers for improving crop yield and reducing reactive nitrogen losses from China’s vegetable systems: a meta-analysis. Environ. Pollut. 269:116143. doi: 10.1016/j.envpol.2020.116143
McDonald, J. E., Rooks, D. J., and McCarthy, A. J. (2012). Methods for the isolation of cellulose-degrading microorganisms. Methods Enzymol.510, 349–74. doi: 10.1016/B978-0-12-415931-0.00019-7
Meng, X., Ma, C., and Petersen, S. O. (2022). Sensitive control of N2O emissions and microbial community dynamics by organic fertilizer and soil interactions. Biol. Fertil. Soils 58, 771–788. doi: 10.1007/s00374-022-01662-9
Molstad, L., Dorsch, P., and Bakken, L. R. (2007). Robotized incubation system for monitoring gases (O2, NO, N2O N2) in denitrifying cultures. J. Microbiol. Methods 71, 202–211. doi: 10.1016/j.mimet.2007.08.011
Murray, P. A., and Zinder, S. H. (1984). Nitrogen fixation by a methanogenic archaebacterium. Nature 312, 284–286. doi: 10.1038/312284a0
Nguyen, D., and Khanal, S. K. (2018). A little breath of fresh air into an anaerobic system: how microaeration facilitates anaerobic digestion process. Biotechnol. Adv. 36, 1971–1983. doi: 10.1016/j.biotechadv.2018.08.007
Olm, M. R., Brown, C. T., Brooks, B., and Banfield, J. F. (2017). dRep: a tool for fast and accurate genomic comparisons that enables improved genome recovery from metagenomes through de-replication. ISME J. 11, 2864–2868. doi: 10.1038/ismej.2017.126
Ouyang, Y., Norton, J. M., and Parales, R. E. (2020). Short-term nitrogen fertilization affects microbial community composition and nitrogen mineralization functions in an agricultural soil. Appl. Environ. Microbiol. 86:e02278-19. doi: 10.1128/aem.02278-19
Oyetunji, O., Bolan, N., and Hancock, G. (2022). A comprehensive review on enhancing nutrient use efficiency and productivity of broadacre (arable) crops with the combined utilization of compost and fertilizers. J. Environ. Manag. 317:115395. doi: 10.1016/j.jenvman.2022.115395
Parks, D. H., Imelfort, M., Skennerton, C. T., Hugenholtz, P., and Tyson, G. W. (2015). CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 25, 1043–1055. doi: 10.1101/gr.186072.114
Pastorelli, R., Casagli, A., Rocchi, F., Tampio, E., Laaksonen, I., Becagli, C., et al. (2024). Effects of anaerobic Digestates and biochar amendments on soil health, greenhouse gas emissions, and microbial communities: a Mesocosm study. Appl. Sci. 14:1917. doi: 10.3390/app14051917
Paulin, M. M., Nicolaisen, M. H., Jacobsen, C. S., Gimsing, A. L., Sørensen, J., and Bælum, J. (2013). Improving Griffith’s protocol for co-extraction of microbial DNA and RNA in adsorptive soils. Soil Biol. Biochem. 63, 37–49. doi: 10.1016/j.soilbio.2013.02.007
Peng, Y., Leung, H. C. M., Yiu, S. M., and Chin, F. Y. L. (2010). IDBA - a practical iterative de bruijn graph de novo assembler. Lecture Notes Comput. Sci. 426–440. doi: 10.1007/978-3-642-12683-3-28
Poehlein, A., Funkner, K., Schüler, M. A., and Daniel, R. (2017). First insights into the genome sequence of the cellulolytic bacterium Clostridium hungatei DSM 14427. Genome Announc. 5:e00363-17. doi: 10.1128/genomea.00363-17
Ramnarine, R., Voroney, R. P., Wagner-Riddle, C., and Dunfield, K. E. (2011). Carbonate removal by acid fumigation for measuring the δ13C of soil organic carbon. Can. J. Soil Sci. 91, 247–250. doi: 10.4141/cjss10066
Ratsiatosika, O., Trap, J., Herinasandratra, V., Razafimbelo, T., Bernard, L., and Blanchart, E. (2024). Earthworms enhance the performance of organic amendments in improving rice growth and nutrition in poor ferralsols. Soil Biol. Biochem. 195:109477. doi: 10.1016/j.soilbio.2024.109477
Reddy, N., and Crohn, D. M. (2014). Effects of soil salinity and carbon availability from organic amendments on nitrous oxide emissions. Geoderma 235-236, 363–371. doi: 10.1016/j.geoderma.2014.07.022
Samoraj, M., Mironiuk, M., Izydorczyk, G., Witek-Krowiak, A., Szopa, D., Moustakas, K., et al. (2022). The challenges and perspectives for anaerobic digestion of animal waste and fertilizer application of the digestate. Chemosphere 295:133799. doi: 10.1016/j.chemosphere.2022.133799
Scherer, P. (1988). Vanadium and molybdenum requirement for the fixation of molecular nitrogen by two Methanosarcina strains. Arch. Microbiol. 151, 44–48. doi: 10.1007/BF00444667
Schlüter, S., Lucas, M., Grosz, B., Ippisch, O., Zawallich, J., He, H., et al. (2025). The anaerobic soil volume as a controlling factor of denitrification: a review. Biol. Fertil. Soils 61, 343–365. doi: 10.1007/s00374-024-01819-8
Selim, M. M. (2020). Introduction to the integrated nutrient management strategies and their contribution to yield and soil properties. Int. J. Agronomy 2020, 1–14. doi: 10.1155/2020/2821678
Song, Y., Pei, L., Chen, G., Mu, L., Yan, B., Li, H., et al. (2023). Recent advancements in strategies to improve anaerobic digestion of perennial energy grasses for enhanced methane production. Sci. Total Environ. 861:160552. doi: 10.1016/j.scitotenv.2022.160552
Swift, M. L. (1997). GraphPad prism, data analysis, and scientific graphing. J. Chem. Inf. Comput. Sci. 37, 411–412. doi: 10.1021/ci960402j
Tu, Q., Lin, L., Cheng, L., Deng, Y., and He, Z. (2019). NCycDB: a curated integrative database for fast and accurate metagenomic profiling of nitrogen cycling genes. Bioinformatics 35, 1040–1048. doi: 10.1093/bioinformatics/bty741
Wang, C., Ma, Y., He, W., Kuzyakov, Y., Bol, R., Chen, H., et al. (2024). Soil quality and ecosystem multifunctionality after 13-year of organic and nitrogen fertilization. Sci. Total Environ. 931:172789. doi: 10.1016/j.scitotenv.2024.172789
Wang, C., Ma, X., Shen, J., Chen, D., Zheng, L., Ge, T., et al. (2022). Reduction in net greenhouse gas emissions through a combination of pig manure and reduced inorganic fertilizer application in a double-rice cropping system: three-year results. Agric. Ecosyst. Environ. 326:107799. doi: 10.1016/j.agee.2021.107799
Wang, J., Wu, L., Xiao, Q., Huang, Y., Liu, K., Wu, Y., et al. (2023). Long-term manuring enhances soil gross nitrogen mineralization and ammonium immobilization in subtropical area. Agric. Ecosyst. Environ. 348:108439. doi: 10.1016/j.agee.2023.108439
Wu, G., Liang, F., Wu, Q., Feng, X.-G., Shang, W. D., Li, H. W., et al. (2024). Soil pH differently affects N2O emissions from soils amended with chemical fertilizer and manure by modifying nitrification and denitrification in wheat-maize rotation system. Biol. Fertil. Soils 60, 101–113. doi: 10.1007/s00374-023-01775-9
Wu, Y., Shaaban, M., Deng, C., Peng, Q., and Hu, R. (2017). Changes in the soil N potential mineralization and nitrification in a rice paddy after 20 years application of chemical fertilizers and organic matter. Can. J. Soil Sci. 97, 290–299. doi: 10.1139/cjss-2016-0065
Wu, X., Wang, Y., Zhu, Y., Tian, H., Qin, X., Cui, C., et al. (2019). Variability in the response of bacterial community assembly to environmental selection and biotic factors depends on the immigrated Bacteria, as Revealed by a Soil Microcosm Experiment. mSystems 4:e00496-19. doi: 10.1128/msystems.00496-19
Xia, Y., Chen, X., Zheng, S., Gunina, A., Ning, Z., Hu, Y., et al. (2021). Manure application accumulates more nitrogen in paddy soils than rice straw but less from fungal necromass. Agric. Ecosyst. Environ. 319:107575. doi: 10.1016/j.agee.2021.107575
Yang, Y., Shen, L., Zhao, X., Agathokleous, E., Wang, S., Ren, B., et al. (2023). Long-term fertilization enhances the activity of anaerobic oxidation of methane coupled to nitrate reduction and associated microbial abundance in paddy soils. Soil Biol. Biochem. 185:109130. doi: 10.1016/j.soilbio.2023.109130
Young, M. D., Ros, G. H., and de Vries, W. (2021). Impacts of agronomic measures on crop, soil, and environmental indicators: a review and synthesis of meta-analysis. Agric. Ecosyst. Environ. 319:107551. doi: 10.1016/j.agee.2021.107551
Yuan, Y., Liang, Y., Cai, H., Yuan, J., Li, C., Liu, H., et al. (2025). Soil organic carbon accumulation mechanisms in soil amended with straw and biochar: entombing effect or biochemical protection? Biochar 7:33. doi: 10.1007/s42773-025-00431-9
Zhai, L., Wang, Z., Zhai, Y., Zhang, L., Zheng, M., Yao, H., et al. (2022). Partial substitution of chemical fertilizer by organic fertilizer benefits grain yield, water use efficiency, and economic return of summer maize. Soil Tillage Res. 217:105287. doi: 10.1016/j.still.2021.105287
Zhang, C., Ju, X., Zhang, J., Rees, R. M., and Müller, C. (2023). Soil pH and long-term fertilization affect gross N transformation and N2O production pathways in Chinese and UK croplands. Biol. Fertil. Soils 59, 527–539. doi: 10.1007/s00374-022-01695-0
Zhang, Q., Wu, Y., Wang, J., Wu, G., Long, W., Xue, Z., et al. (2016). Accelerated dysbiosis of gut microbiota during aggravation of DSS-induced colitis by a butyrate-producing bacterium. Sci. Rep. 6:27572. doi: 10.1038/srep27572
Zhao, J., Pullens, J. W. M., Sørensen, P., Blicher-Mathiesen, G., Olesen, J. E., and Børgesen, C. D. (2022). Agronomic and environmental factors influencing the marginal increase in nitrate leaching by adding extra mineral nitrogen fertilizer. Agric. Ecosyst. Environ. 327:107808. doi: 10.1016/j.agee.2021.107808
Zhou, W., Ma, Q., Wu, L., Hu, R., Jones, D. L., Chadwick, D. R., et al. (2022). The effect of organic manure or green manure incorporation with reductions in chemical fertilizer on yield-scaled N2O emissions in a citrus orchard. Agric. Ecosyst. Environ. 326:107806. doi: 10.1016/j.agee.2021.107806
Keywords: metagenome, functional genes, microcosm experiment, nitrogen cycling, organic amendments
Citation: Ma Y, Wu Q, Wang X, Sui W and Zhang X (2025) Carbon components in organic amendments drive nitrogen metabolism in one-year-long anaerobic soil microcosms. Front. Microbiol. 16:1588169. doi: 10.3389/fmicb.2025.1588169
Edited by:
Brahim Bouizgarne, Ibn Zohr University, MoroccoReviewed by:
Xiaomeng Chen, Northeast Agricultural University, ChinaAzim Khalid, National Institute for Agricultural Research, Morocco
Copyright © 2025 Ma, Wu, Wang, Sui and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaojun Zhang, eGp6aGFuZzY4QHNqdHUuZWR1LmNu
†Present address: Qiaoyu Wu, State Key Laboratory of Pollution Control and Resources Reuse, Shanghai Institute of Pollution Control and Ecological Security, College of Environmental Science and Engineering, Tongji University, Shanghai, China
‡ORCID: Xiaojun Zhang, https://orcid.org/0000-0002-4559-0177
 Yiming Ma
Yiming Ma Qiaoyu Wu†
Qiaoyu Wu† Xiaojun Zhang
Xiaojun Zhang