- 1School of Marine Sciences, Ningbo University, Ningbo, China
- 2Southern Ocean Science and Engineering Guangdong Laboratory (Zhanjiang), Zhanjiang, China
- 3Ningbo Institute of Oceanography, Ningbo, Zhejiang, China
- 4Shandong Freshwater Fisheries Research Institute, Jinan, China
As the aquaculture capacity of S. paramamosain in southern China nears saturation, northern coastal regions, which are characterized by abundant water resources, ample feed availability, and favorable climatic conditions, have emerged as ideal areas for aquaculture expansion. This study investigates the aquatic environment of S. paramamosain cultured in seawater and saline-alkali ponds in northern China. Over the course of a five-month aquaculture experiment, water samples were collected from seawater and saline-alkali ponds and subsequently analyzed using 16S rRNA gene sequencing technology to examine the bacterial community composition and its relationship with physicochemical water quality parameters. Sensitive bacterial species were identified as well. The results revealed that seawater ponds exhibited higher salinity and dissolved oxygen levels, but lower pH, ammonia nitrogen, and nitrite nitrogen concentrations. In contrast, saline-alkali ponds exhibited elevated pH, ammonia nitrogen, and nitrite nitrogen levels, accompanied by reduced salinity and dissolved oxygen. Bacterial communities in seawater ponds demonstrated greater species richness, evenness, and diversity indices, whereas those in saline-alkali ponds were characterized by reduced diversity and distinct dominant bacterial groups. Redundancy analysis (RDA) identified salinity, pH, and dissolved oxygen as the principal environmental factors influencing bacterial community structure. Using the IndVal method, we identified strong associations between specific bacterial species and pond types, such as Sphingoaurantiacus and Cobetia in seawater ponds, and Roseivivax, Tropicimonas, and Thiobacillus in saline-alkali ponds. Environmental factors exerted distinct effects on bacterial communities in the two pond types, with sensitive bacterial species demonstrating significant specificity and strong correlations with water quality parameters. Functional predictions indicated that microbes in saline-alkali ponds prioritized resource acquisition and stress resistance, whereas those in seawater ponds emphasized nitrogen metabolism and protein synthesis. This study demonstrated significant differences in bacterial community characteristics between seawater and saline-alkali ponds, which were strongly influenced by water quality parameters. These findings are crucial for optimizing the growth environment of S. paramamosain, providing essential data for improving aquaculture conditions and promoting the development of northern S. paramamosain farming.
1 Introduction
The mud crab (Scylla paramamosain) is an important commercial crustacean species distributed throughout the Indo-Pacific region (Lin et al., 2012; Zhang et al., 2024). In China, the distribution and commercial production of S. paramamosain are primarily concentrated in the coastal regions south of the Yangtze River estuary (Ma et al., 2014; Zhou et al., 2023). Currently, the aquaculture capacity of S. paramamosain in southern China has reached saturation, necessitating the development of additional cultivation areas to meet increasing demand. The northern coastal regions of China are characterized by abundant water resources, ample feed availability, and a suitable climate, making them the preferred areas for the expanded cultivation of S. paramamosain. For instance, the Yellow River Delta region (37°35′–38°12’N, 118°33′–119°20′E), a recently formed landmass resulting from sediment deposition carried by the Yellow River (Cui et al., 2009), is a promising candidate for the expansion of S. paramamosain aquaculture. However, it is important to note that the Yellow River Delta (YRD) region exhibits distinctive salinization patterns characterized by complex salt compositions and spatial heterogeneity. The predominant salt types include NaCl, Na2SO4, and NaHCO3, with sodium ions constituting 70–85% of exchangeable cations in surface soils (Huang et al., 2015). Soil salinity typically ranges from 0.3 to 3.0%, showing significant seasonal variation due to the monsoon climate – higher in dry seasons (spring) and lower after summer rainfall (Komiyama et al., 2020). These factors may pose challenges to the cultivation of S. paramamosain in the region and should be carefully considered in the development of aquaculture practices.
Aquaculture ponds are complex ecosystems in which microorganisms in both water and sediment interact (Lahiri et al., 2018). Microorganisms influence the entire biogeochemical cycle and are crucial to productivity, nutrient cycling, water quality, and the health of farmed animals (Falkowski et al., 2008). Microbial communities are well-known to respond rapidly to environmental changes, such as pH, oxygen concentration, nitrogen concentration, and salinity (Xiong et al., 2015). The close relationship between water quality and bacterial communities in aquaculture ponds has been well-documented. Bacterial communities help maintain the aquatic ecosystem and can serve as indicators of its wellbeing (Paerl et al., 2003). Furthermore, bacteria are an integral part of the diet of various fish and crustaceans in aquaculture environments, including rotifers, copepods, shrimp, and fish (Troch et al., 2012). Bacteria are occasionally discussed as potential sources of essential nutrients and are considered viable food sources within the aquaculture food chain (Nichols, 2003). Clearly, bacteria represent a critical component of pond ecosystems, and the structure and diversity of their communities significantly impact the overall health of these ecosystems (Vincent et al., 2008). However, current studies on bacterial communities in S. paramamosain aquaculture ponds are primarily limited to the southern coastal regions of China (Wei et al., 2019; Kasan et al., 2021), with bacterial communities in northern China remaining largely unexplored.
Water temperature and salinity are key physicochemical parameters that significantly influence bacterial community structure and diversity, which in turn affect the growth of S. paramamosain (Deng et al., 2020). For example, studies on Litopenaeus vannamei aquaculture ponds have shown that temperature fluctuations between 25 and 35°C promote the proliferation of potential pathogenic bacteria, such as Vibrio species (Cheng et al., 2005). The excessive growth of Vibrio bacteria can degrade water quality by accumulating harmful substances, including ammonia nitrogen and nitrite, and depleting dissolved oxygen levels (Qiu et al., 2023; Zhang et al., 2022). Regarding salinity, deviations from the optimal range—whether excessively high or low—can disrupt bacterial community composition, altering the balance between beneficial and harmful bacteria (Gorrasi et al., 2021; Wang et al., 2019). A decline in beneficial bacteria can destabilize essential ecological processes, such as the nitrogen cycle, thereby impairing the efficient transformation of metabolic byproducts like ammonia nitrogen. This degradation in water quality can hinder the growth and survival of crab species (Hastuti et al., 2021).
The pH is a critical physicochemical parameter that profoundly influences bacterial communities, thereby affecting the growth of S. paramamosain (Deng et al., 2020). Studies have shown that in acidic environments with reduced pH, acidophilic bacteria, such as Thiobacillus species, tend to dominate the bacterial community (Johnson and Quatrini, 2020). These bacteria produce acidic byproducts during their metabolic processes, further lowering the water’s pH and exacerbating acidification (Harrison, 1984; Crescenzi et al., 2006). Acidified aquatic environments can disrupt the osmoregulation and respiratory metabolism of S. paramamosain, resulting in impaired growth, molting difficulties, weakened immunity, and heightened susceptibility to pathogens (Yuan et al., 2024).
Dissolved oxygen (DO) levels are crucial for both bacterial communities and the growth of S. paramamosain (Jie et al., 2021). In low-DO environments, facultative anaerobic bacteria, such as Enterobacter species, tend to proliferate (Kato et al., 1997). The overgrowth of Enterobacter bacteria depletes oxygen and produces toxic byproducts, such as ammonia nitrogen and hydrogen sulfide, which deteriorate water quality (Wang et al., 2019). This decline in water quality can induce hypoxia in S. paramamosain, leading to diminished feeding, impaired growth, and even mortality (Wang et al., 2021).
Bacterial communities also influence physicochemical water quality parameters, thereby indirectly affecting the growth environment of S. paramamosain (Pati et al., 2023). For example, nitrifying bacteria play a crucial role in the nitrogen cycle by converting ammonia nitrogen into nitrate, thereby reducing ammonia levels and enhancing water quality (Spietz et al., 2015). Denitrifying bacteria, in contrast, convert nitrate into nitrogen gas, removing excess nitrogen from the water and preventing eutrophication (Holmes et al., 2019). However, an imbalance in the bacterial community, characterized by a decline in beneficial bacteria such as nitrifying and denitrifying bacteria and an increase in harmful bacteria, can negatively impact water quality. This imbalance may lead to elevated levels of ammonia nitrogen and nitrite, along with decreased dissolved oxygen (Fitzgerald et al., 2015; Guo et al., 2009). Such unfavorable conditions can impair the growth of S. paramamosain, compromise its immune system, and increase its susceptibility to diseases (Chen and Wang, 2019). To address this, particularly in the context of expanding aquaculture to northern regions, there is a critical need to fill the gaps in microbial data from saline-alkaline ponds in the north and analyze their ecological differences from seawater pond communities. This research will provide insights into how bacterial communities in these distinct environments influence water quality and, consequently, the health and growth of S. paramamosain. Therefore, studying the structure and diversity of bacterial communities in seawater and saline-alkaline ponds in northern regions, and their interactions with physicochemical water quality parameters, is essential for optimizing aquaculture conditions and ensuring the healthy growth of S. paramamosain.
This study focuses on a comparative analysis of bacterial communities in seawater and saline-alkali aquaculture ponds for S. paramamosain in northern China. Using 16S rRNA gene sequencing, we characterized the bacterial communities in both pond types and investigated their correlations with physicochemical water quality parameters. In northern China, seawater and saline-alkali ponds represent two dominant aquaculture environments, exhibiting marked differences in key parameters such as salinity, pH, and dissolved oxygen (DO). These differences may critically shape bacterial community structure and function. However, systematic comparisons of bacterial communities between these two environments remain limited, necessitating in-depth exploration of their interactions with aquaculture conditions. To address this gap, we analyzed the composition of bacterial communities in seawater and saline-alkali ponds and their interactions with environmental factors, aiming to elucidate the characteristics and dynamics of microbial communities across distinct aquaculture systems. Specifically, this study addresses the following questions: (1) how do bacterial community structure and diversity differ between seawater and saline-alkali ponds? (2) Which environmental factors predominantly drive these structural and functional differences? (3) Are specific bacterial taxa strongly associated with a particular pond type? (4) What functional differences exist in bacterial communities between the two environments? By addressing these questions, this work provides a scientific foundation for optimizing aquaculture environments for S. paramamosain in northern China, enhancing productivity and quality, and promoting sustainable development of regional aquaculture.
2 Materials and methods
2.1 Experimental settings and breeding management
From May 1 to September 30, 2022, a five-month breeding experiment on S. paramamosain was conducted in seawater ponds (118°58′E, 37°37’N) at Haiyue Aquatic Science and Technology Co., Ltd., Dongying City, and saline-alkali ponds (118°56′E, 37°40’N) at Zhengda Sangtian Agricultural Development Co., Ltd., Dongying City. Both pond types were rectangular, with a surface area of 2,240 m2 and a water depth of 1.2 m. Prior to the experiment, all ponds were disinfected using bleaching powder and enclosed with plastic film to prevent crab escape. Oxygen supplementation was provided as needed, based on weather conditions and the feeding and activity levels of the crabs.
Juvenile S. paramamosain crabs in the C4 stage, weighing 2 ± 0.2 g, were obtained from Ningbo Huada Haichang Co., Ltd., for experimental aquaculture. Only active, healthy juveniles with intact limbs were selected for the experiment. The stocking density in each pond was set at 1.2 crabs per square meter. Throughout the aquaculture period, one-third of the total water volume in each pond was replaced monthly. All crabs were fed once daily with artificial feed sourced from Ningbo Haiding Aquatic Feed Co., Ltd., at a feeding rate of 10–15%, which was consistent across all experimental groups. The formulated feed for S. paramamosain included fish meal, squid paste, soybean meal, shrimp meal, brewer’s yeast, phospholipids, refined fish oil, high-gluten flour, minerals, trace elements, vitamins, and polysaccharides.
2.2 Sample collection
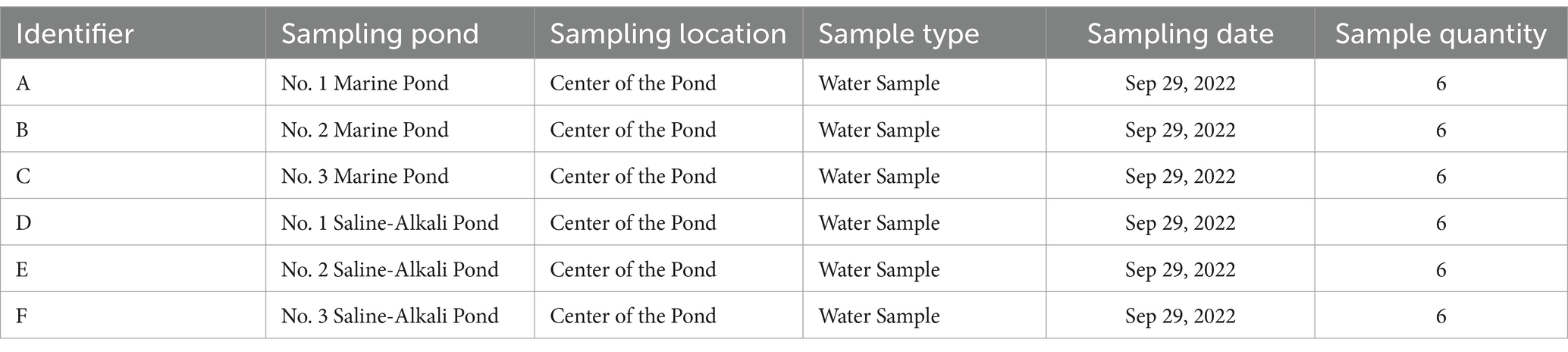
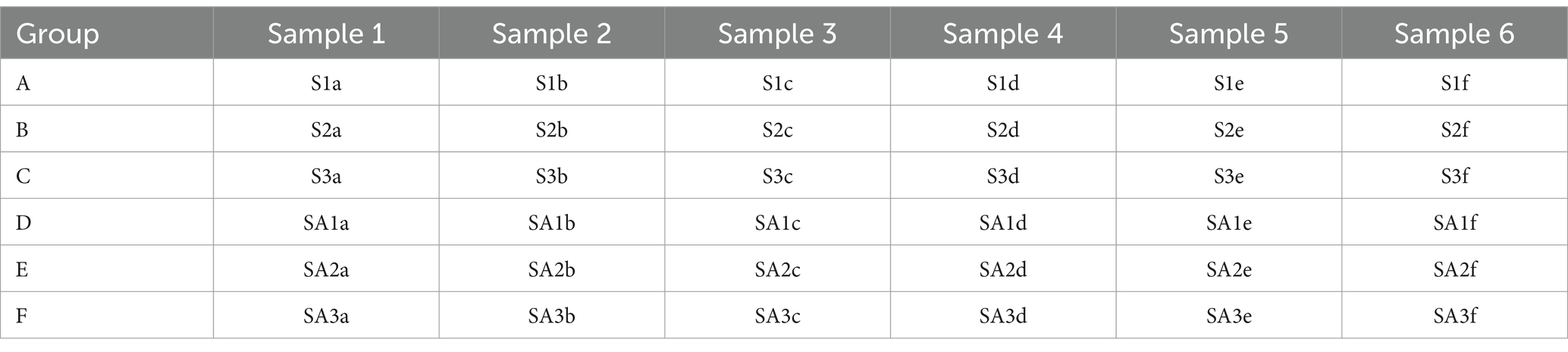
In this study, water samples were collected on September 29, 2022, from three seawater ponds and three saline-alkali ponds. The sampling locations were positioned at the center of each pond, and water was collected at a depth of 0.6 m. At each sampling site, six 100 mL water samples were collected using pre-sterilized polycarbonate sampling bottles, resulting in a total of 36 water samples. To prevent potential microbial contamination, all sampling equipment was sterilized by high-temperature autoclaving before use. The collected water samples were thoroughly homogenized on-site and then aliquoted into sterile centrifuge tubes. The samples were immediately stored at 4°C to minimize microbial activity and chemical alterations. Upon completion of sampling, all water samples were promptly transported to the laboratory. Sample processing was conducted in a biosafety cabinet under sterile conditions, and all aliquoting procedures were completed within 2 h. The processed samples were then immediately stored in an ultra-low temperature freezer at −80°C for further analysis. All water samples were classified according to the alphanumeric identifiers provided in Table 1 and further subdivided as specified in Table 2 to ensure traceability and accuracy in subsequent analyses.
2.3 Physicochemical indexes of the water samples
The temperature, salinity, pH, dissolved oxygen (DO), and ammonia (NH₄+-N) concentrations in the water samples were measured using a handheld multi-parameter instrument (YSI Pro 10,102,030, Yellow Springs Instrument, United States). Additionally, nitrate nitrogen (NO₃−-N) concentrations were determined using the “Water Quality—Determination of Nitrogen (Nitrite)—Spectrophotometric Method” in accordance with the Chinese standard GB 7493–87, the detailed procedure is as follows: Initially, sample pre-treatment is conducted. If the pH exceeds 11, phenolphthalein indicator should be added, and phosphoric acid titrated until the red color disappears, adjusting the pH to 1.8 ± 0.3. In cases where the sample contains suspended solids or coloration, aluminum hydroxide suspension should be added, followed by stirring and filtration. Subsequently, a series of standard nitrite nitrogen solutions (ranging from 0 to 0.05 mg/L) are prepared. After adding the coloring agent, absorbance is measured to construct a standard curve. Next, 50 mL of the sample is mixed with 1 mL of the coloring agent and left to stand for 20 min before measuring the absorbance at a wavelength of 540 nm using a 10 mm cuvette. The concentration is then calculated based on the standard curve. Interfering substances such as chloramines and chlorine can be eliminated by adjusting the pH or adding masking agents. Reagents used must be of analytical grade, and water should be nitrite-free distilled water obtained through double distillation. Coloring agents should be freshly prepared and stored away from light. Instrumentation requirements include a UV spectrophotometer, with glassware cleaned using hydrochloric acid. Quality control measures encompass blank tests, precision (with an RSD ≤ 0.07%), and spike recovery rates (92.7–96.7%). This method quantifies through the absorbance of azo dyes, necessitating strict adherence to standardized procedures to ensure accurate and reliable results.
2.4 DNA extraction and purification
DNA extraction was performed using the cetyltrimethylammonium bromide (CTAB) method, following the protocol provided by Beijing Novogene Bioinformatics Technology Co., Ltd. Initially, 1,000 μL of CTAB lysis buffer was added to a 2.0 mL EP tube, followed by the addition of lysozyme and the sample (Aboul-Maaty and Oraby, 2019). The mixture was incubated at 65°C with periodic gentle inversion to ensure complete sample lysis. After incubation, the lysate was centrifuged at 12,000 rpm for 10 min, and the supernatant was transferred to a clean tube. An equal volume of phenol (pH 8.0): chloroform: isoamyl alcohol (25:24:1, v/v/v) was added, mixed thoroughly by inversion, and centrifuged at 12,000 rpm for 10 min. The aqueous phase was collected and subjected to a second extraction with chloroform:isoamyl alcohol (24:1, v/v), followed by centrifugation under identical conditions. DNA was precipitated by adding isopropanol to the supernatant, gently mixing, and incubating at −20°C. The precipitate was collected by centrifugation at 12,000 rpm for 10 min. The DNA pellet was washed twice with 75% ethanol, air-dried on a sterile surface, and dissolved in nuclease-free water (ddH₂O). To ensure DNA quality, the concentration and purity of the extracted DNA were measured using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific), with absorbance ratios of A260/280 between 1.8 and 2.0 considered acceptable. Additionally, DNA integrity was verified by electrophoresis on a 1% agarose gel stained with ethidium bromide. To remove residual RNA, 1 μL of RNase A was added, and the mixture was incubated at 37°C for 15 min. The purified DNA was stored at −20°C for further analysis.
2.5 16S rDNA gene amplification and Illumina high-throughput sequencing
The V4 region of the 16S rDNA gene was amplified using the primer pair 515F (5’-GTGCCAGCMGCCGCGGTAA-3′) and 806R (5’-GGACTACHVGGGTWTCTAAT-3′) to assess bacterial diversity (Caporaso et al., 2011). Each polymerase chain reaction (PCR) was performed in a 25 μL reaction volume containing 15 μL of Phusion High-Fidelity PCR Master Mix, 0.2 μM of each primer, and 10 ng of genomic DNA as the template. The thermal cycling conditions consisted of an initial denaturation at 98°C for 1 min, followed by 30 cycles of denaturation at 98°C for 10 s, annealing at 50°C for 30 s, and extension at 72°C for 30 s. A final extension step was performed at 72°C for 5 min. PCR products were purified using magnetic beads, after which the amplicons were quantified. The purified PCR products were pooled equimolarity to generate a sequencing library. The quality and quantity of the prepared library were assessed using a Qubit fluorometer and quantitative PCR (qPCR). The sequencing was then performed on an Illumina platform to obtain paired-end reads.
2.6 Illumina data processing
Raw sequencing reads were demultiplexed based on unique barcode and primer sequences assigned to each sample. Paired-end reads were merged using FLASH (version 1.2.11) to generate raw tags, followed by quality filtering with fastp (version 0.23.1) to obtain high-quality clean tags (Magoc and Salzberg, 2011; Bokulich et al., 2013). Chimeric sequences were identified and removed using the UCHIME algorithm, with reference-based filtering performed against the Silva database, resulting in effective tags (Edgar et al., 2011). Amplicon sequence variants (ASVs) were generated using the DADA2 plugin in QIIME2 (Maruyama et al., 2020). Taxonomic classification of ASVs was carried out with QIIME2, utilizing the Silva 138.1 database for bacterial sequences. Data normalization was performed using the sample with the lowest sequence count to ensure consistency across datasets. Subsequent analyses included alpha diversity metrics (e.g., Shannon and Simpson indices) and beta diversity evaluations [e.g., principal coordinates analysis (PCoA) and unweighted pair group method with arithmetic mean (UPGMA) clustering]. Taxonomic profiles, heatmaps, and diversity indices were visualized using R packages such as ggplot2 and pheatmap. These analyses provided a comprehensive overview of bacterial community composition and structure across samples.
2.7 IndVal method
The Indicator Value (IndVal) method is grounded in the concept that a species’ association with a specific habitat is determined by its frequency of occurrence and abundance within that habitat (Leote et al., 2020). This method assesses the relationship between species and habitats by calculating a composite index that integrates both the species’ average abundance and its occurrence frequency across sample plots. The specificity of a species is represented by its occurrence rate within a particular habitat, reflecting how exclusive the species is to that habitat. Fidelity, in contrast, refers to the proportion of the species’ mean abundance within the habitat, indicating its consistency and prevalence in the sampled environment. In the R programming environment, the indicspecies package is used to compute the IndVal index, where the package’s functions automatically calculate both specificity and fidelity based on species’ occurrence and abundance data from the sample plots. The resulting IndVal value quantifies the strength of the species’ association with a given habitat, with higher values indicating a stronger relationship. A higher IndVal score suggests that a species is more indicative of the habitat, thereby enhancing its potential as an indicator species for that habitat.
2.8 Statistical analysis
All statistical analyses were performed using IBM SPSS Statistics 20, with independent Student’s t-tests applied to assess significant differences. Alpha diversity indices, including Chao1, Pielou_e, Shannon, and Simpson indices, were calculated using MetaParallel v3.5.2 (Jing et al., 2017). Relative abundance maps for the top 10 phyla and genera were created using Microsoft Excel 2010. The UPGMA cluster tree for water samples collected from seawater and saline-alkali ponds was constructed using the ape, phangorn, and ggtree packages in R. Principal coordinate analysis (PCoA) and non-metric multidimensional scaling (NMDS) were conducted at the phylum and genus levels using Bray–Curtis distance matrices, implemented with the WGCNA package in R v3.6.0. Environmental variables correlated with bacterial community composition were identified using the BioEnv procedure in the vegan package in R v3.6.0 (Li X. et al., 2024; Li Z. et al., 2024). Canonical correspondence analysis (CCA) was performed using the vegan and ggplot2 packages in R v3.6.0 to examine the contribution of environmental factors to microbial community shifts. Indicator species associated with seawater and saline-alkali ponds were identified using the labdsv package in R. Pearson correlation analyses were conducted in R v3.6.0 to assess the relationships between indicator species and physicochemical water quality parameters. Finally, functional annotation of microbial communities was performed using PICRUSt2, and clustering heatmaps of functional genes and sample groups were generated using the pheatmap package, with KEGG pathway annotations retrieved via the KEGGREST package in R.
3 Results
3.1 Analysis of physical and chemical indexes of water samples in seawater ponds and saline-alkali ponds
A comparative analysis was performed on the physicochemical parameters of water quality between seawater ponds (Groups A, B, C) and saline-alkali ponds (Groups D, E, F) used for S. paramamosain aquaculture (Table 3). The water temperature in the seawater ponds (27.30–27.67°C) was slightly higher than in the saline-alkali ponds (26.50–27.67°C), though the difference was not statistically significant (p > 0.05). In contrast, salinity levels in the seawater ponds (27.40–27.67 ppt) were significantly higher than in the saline-alkali ponds (7.33–7.80 ppt), with a statistically significant difference observed (p < 0.05). The pH in the seawater ponds (7.62–7.64) was slightly lower than in the saline-alkali ponds (8.67–8.72), and this difference was statistically significant (p < 0.05). Dissolved oxygen concentrations in the seawater ponds (8.83–8.91 mg/L) were significantly higher than in the saline-alkali ponds (5.89–6.25 mg/L), with a statistically significant difference (p < 0.05). Ammonia nitrogen concentrations were lower in the seawater ponds (0.30–0.37 mg/L), while higher levels were observed in the saline-alkali ponds (0.53–0.58 mg/L), with the difference being statistically significant (p < 0.05). Finally, nitrite nitrogen concentrations were significantly lower in the seawater ponds (0.027–0.031 mg/L) compared to the saline-alkali ponds (0.045–0.048 mg/L), with a significant difference (p < 0.05).
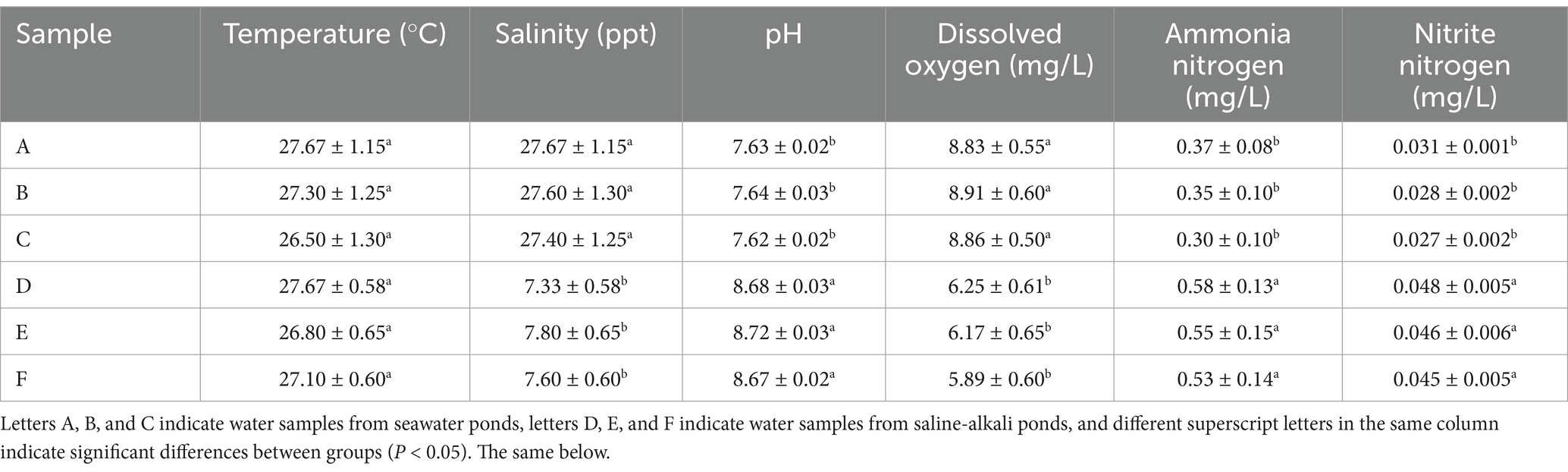
Table 3. Analysis of physical and chemical indexes of water samples in seawater ponds and saline-alkali ponds.
3.2 Diversity analysis of bacterial communities in seawater pond and saline-alkaline pond
A comparative analysis was performed on the microbial diversity indices of water samples from seawater ponds (Groups A, B, C) and saline-alkali ponds (Groups D, E, F). The indices included the Chao 1 index, Pielou_e evenness index, Shannon diversity index, and Simpson index (Figure 1). The Chao 1 index indicated that species richness was highest in Group C, significantly exceeding that of Groups A and B in the seawater ponds, as well as all groups in the saline-alkali ponds (p < 0.05). Among the saline-alkali ponds, Group D exhibited relatively high species richness, whereas Groups E and F had lower richness, with significant differences observed when compared to the seawater ponds (p < 0.05). Overall, species richness was higher in seawater ponds compared to saline-alkali ponds. The Pielou_e evenness index showed that the microbial community in Group C had the highest evenness, significantly higher than that in Groups D, E, and F (p < 0.05). No significant differences in evenness were observed among Groups A, B, and C in the seawater ponds (p > 0.05). Evenness was generally lower in the saline-alkali ponds, with Group F showing the lowest evenness, indicating a more uneven microbial community distribution. The Shannon diversity index revealed that Group C had the highest diversity, significantly surpassing that of all saline-alkali pond groups (p < 0.05). Groups A and B followed, while Groups D, E, and F exhibited progressively lower diversity, with Group F being the lowest. Microbial diversity in the saline-alkali ponds was notably lower than in the seawater ponds. The Simpson index further confirmed these findings. Group C had a Simpson index close to 1, suggesting low dominance of specific microbial species and a more balanced community structure. Groups A and B showed slightly lower Simpson indices. In contrast, Groups D, E, and F had even lower Simpson indices, with Group F being the lowest, indicating fewer dominant microbial species and a less balanced community structure.
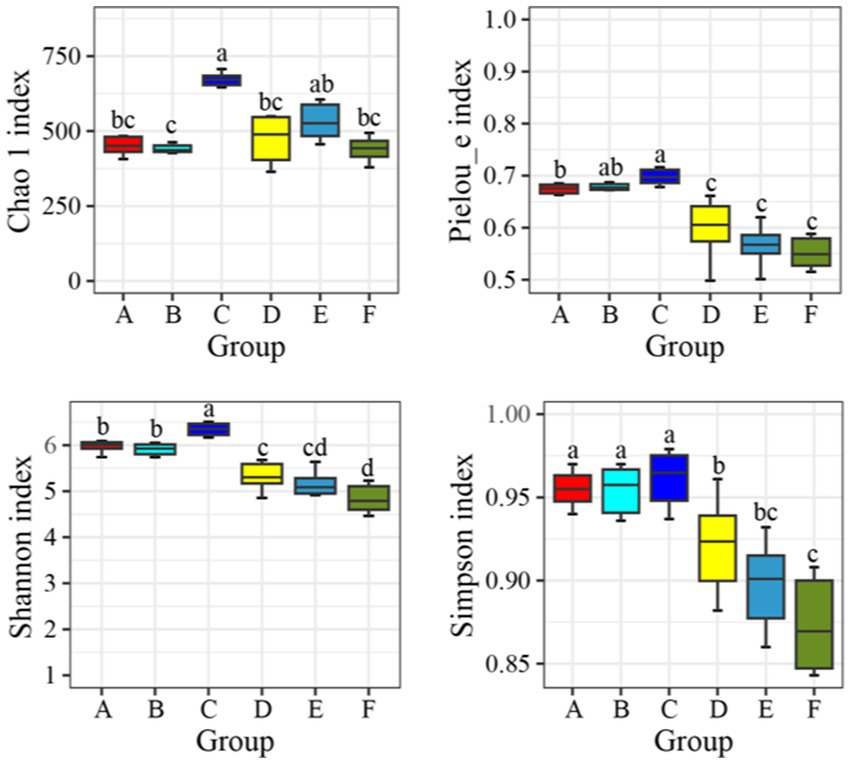
Figure 1. Diversity index of bacteria and microorganisms in seawater ponds and saline-alkali ponds. The data indicates the mean standard deviation, and different letters indicate significant differences between groups (p < 0.05).
3.3 Composition analysis of bacterial communities in seawater ponds and saline-alkali ponds
At the phylum level, there was minimal variation in the dominant bacterial communities between the seawater and saline-alkali ponds, which were primarily composed of Cyanobacteria, Proteobacteria, Actinobacteriota, and Bacteroidota (Figure 2). However, the relative abundances of Verrucomicrobiota and Bdellovibrionota were significantly higher in the seawater ponds, while Cyanobacteria and Firmicutes were more abundant in the saline-alkali ponds. At the genus level, significant differences were observed in the relative abundances of bacterial communities between the two pond types. In the seawater ponds, the dominant genera included Cyanobium_PCC-6307, Synechococcus_CC9902, DS001, and Marivita. In contrast, the dominant genera in the saline-alkali ponds were unidentified_Chloroplast, Cyanobium_PCC-6307, Synechococcus_CC9902, and Candidatus_Aquiluna.
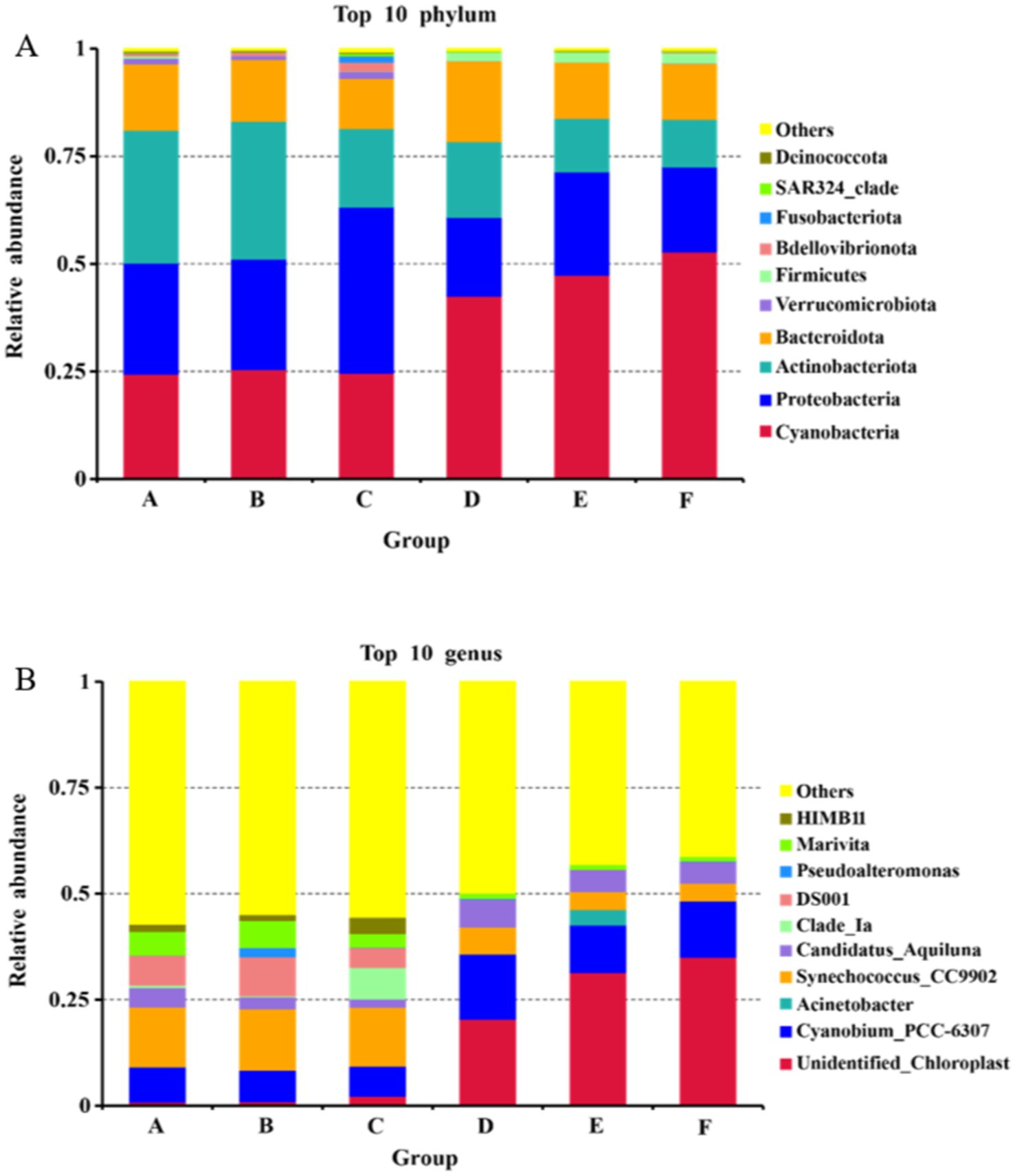
Figure 2. The relative abundance maps of the top 10 bacteria in seawater ponds and saline-alkali ponds are (A) at the level of phylum, and (B) at the level of genus.
3.4 Structural analysis of bacterial communities in seawater ponds and saline-alkali ponds
From Figure 3A, it is evident that Groups A, B, and C cluster together, as do Groups D, E, and F, forming two distinct branches. This clustering indicates a notable difference in the bacterial community structure at the phylum level between the seawater and saline-alkali ponds. In the seawater ponds, the bacterial communities are primarily dominated by Cyanobacteria, Proteobacteria, Actinobacteriota, and Bacteroidetes, with each phylum contributing approximately 25% to the overall relative abundance. The close clustering of samples from the seawater ponds suggests a high degree of similarity, indicating that the microbial community structure is stable and uniformly distributed across these ponds. In contrast, the bacterial community structure in the saline-alkali ponds shows greater variability. Dominant phyla in these ponds include Cyanobacteria, Proteobacteria, Actinobacteriota, and Bacteroidetes, along with smaller proportions of Firmicutes and other bacterial groups. Notably, Cyanobacteria accounts for a substantial 50% of the relative abundance in the saline-alkali ponds, reflecting a distinct microbial composition compared to the seawater ponds. When examining the clustering tree, the branch distances between groups within the seawater ponds are relatively short, which suggests that the microbial communities in these ponds are similar and tightly clustered. Similarly, groups within the saline-alkali ponds show short distances, indicating comparable microbial structures within these ponds. However, the distances between the seawater and saline-alkali pond groups are markedly larger, underscoring significant differences in the microbial community composition between the two pond types.
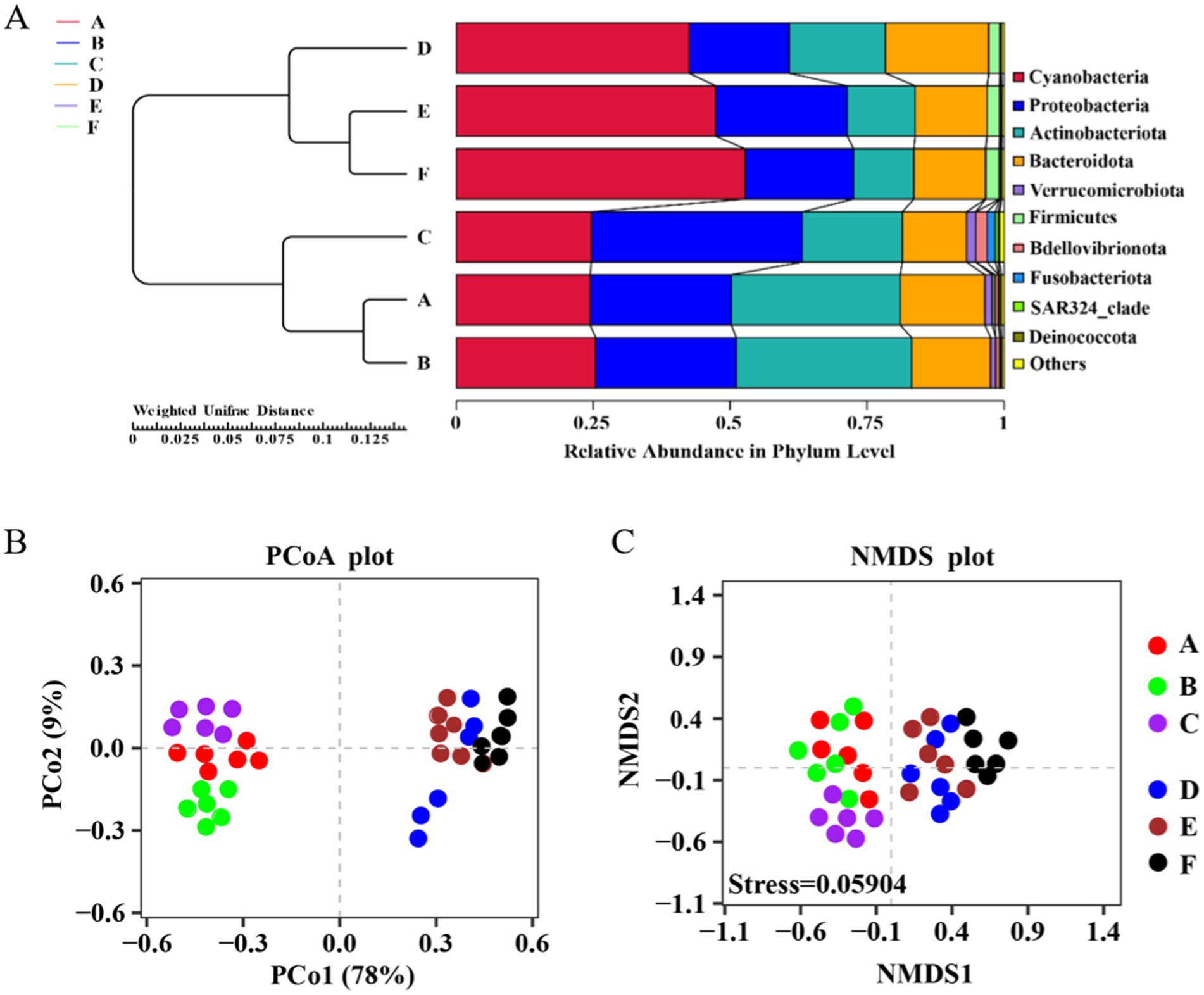
Figure 3. (A) UPGMA cluster tree of seawater pond and saline-alkali pond water samples, (B) PCoA map and (C) NMDS map of bacterial communities in the aquaculture ponds.
The PCoA plot (Figure 3B) illustrates the distribution of different samples in the principal coordinate space. The first principal coordinate (PCo1) accounts for 78% of the variation, while the second (PCo2) explains an additional 9%. The plot clearly reveals a distinct clustering pattern, with samples from Groups A, B, and C (seawater ponds) forming a cluster on the left side, and samples from Groups D, E, and F (saline-alkali ponds) grouping together on the right. This separation indicates significant differences in bacterial community structure between the seawater and saline-alkali ponds. The NMDS plot (Figure 3C) further supports this observation. With a stress value of 0.05904, the NMDS analysis demonstrates good reliability of the results. Similar to the PCoA plot, the NMDS plot shows a clear separation between the seawater pond and saline-alkali pond samples, with Groups A, B, and C clustering together, while Groups D, E, and F form a distinct cluster. This segregation highlights the significant differences in bacterial community composition between the two pond types, reinforcing the findings from the PCoA analysis.
3.5 Relationship between bacterial communities and environmental factors
The environmental factors of water samples from the seawater ponds were standardized to eliminate dimensional differences. BioEnv analysis revealed that the most influential combination of environmental factors driving bacterial community variations was salinity, pH, and dissolved oxygen, with a correlation of r = 0.76. These factors were then used in the redundancy analysis (RDA). Salinity (positively correlated, r = 0.83), dissolved oxygen (positively correlated, r = 0.94), and pH (negatively correlated, r = −0.61) emerged as the key environmental factors along the first (RDA1) and second (RDA2) axes, collectively explaining 96.9% of the variation in the bacterial community (Figure 4A). Similarly, the environmental factors of water samples from the saline-alkali ponds were standardized to address dimensional differences. BioEnv analysis identified salinity, pH, and ammonia nitrogen as the most relevant factors (correlation, r = 0.83). These were subsequently used in RDA. Salinity (positively correlated, r = 0.91), pH (positively correlated, r = 0.61), and ammonia nitrogen (negatively correlated, r = −0.83) were the primary environmental factors along RDA1 and RDA2. Together, they explained 93.8% of the variation in the bacterial community (Figure 4B).
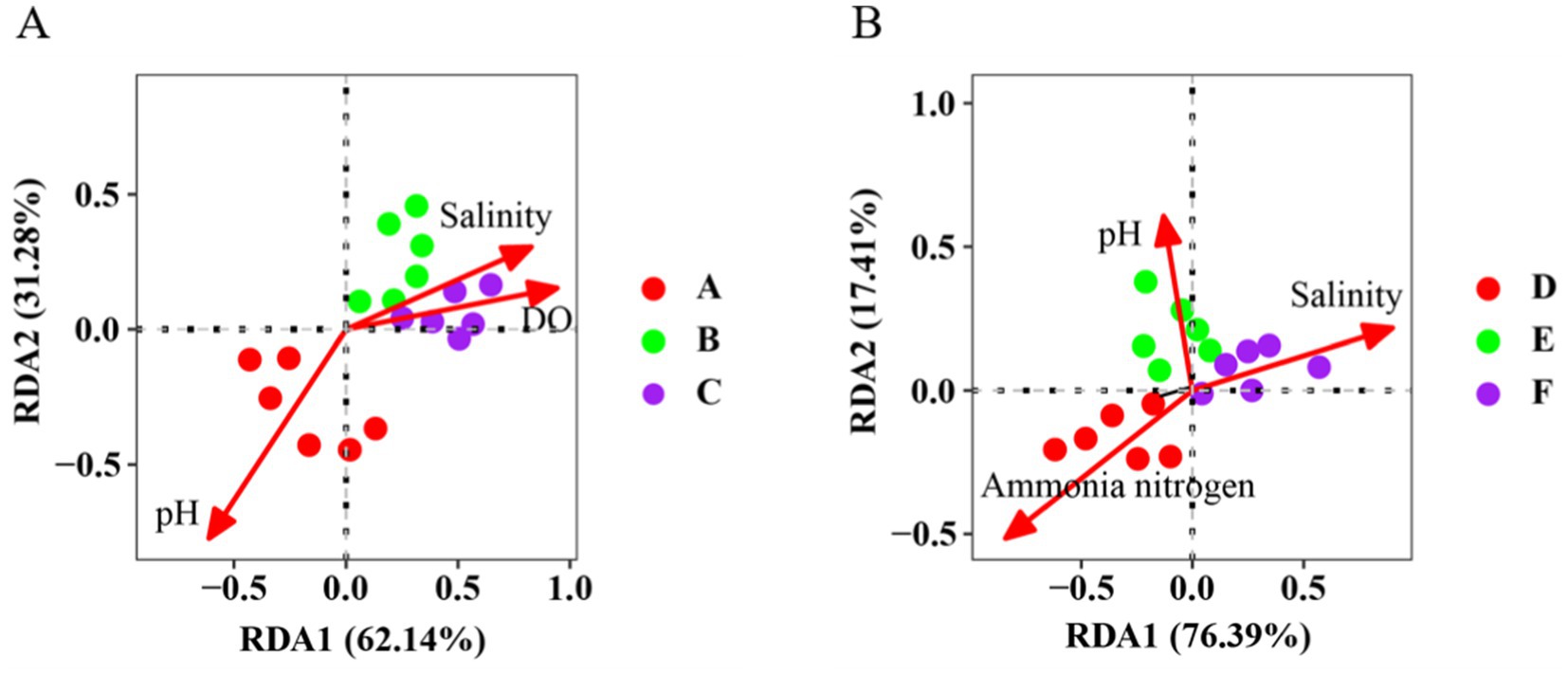
Figure 4. Bidirectional RDA of bacterial communities in (A) the Seawater pond water sample and (B) the Saline-alkali water pond water sample and their main environmental factors (DO, dissolved oxygen).
3.6 Correlation analysis between physical and chemical indexes of water samples and bacterial communities
The IndVal method was applied at the genus level to identify sensitive bacterial species, with an indicator value threshold set at 0.65 (p < 0.01). The distribution characteristics of these sensitive species are summarized in Table 4. Several sensitive bacterial species were identified in the seawater ponds. Sphingoaurantiacus and Cobetia were strongly associated with Group B, with indicator values of 0.6714 and 0.6647, respectively, and low probability values (0.004 and 0.003). This indicates a clear specific distribution tendency in seawater ponds. Additionally, Propionigenium, unidentified_SAR116_clade, Clade_Ia, and unidentified_Clade_III were associated with Group C, exhibiting high indicator values of 0.9504, 0.9083, 0.8979, and 0.8777, respectively, all with a probability value of 0.001. These results underscore their strong indicative role in seawater ponds.
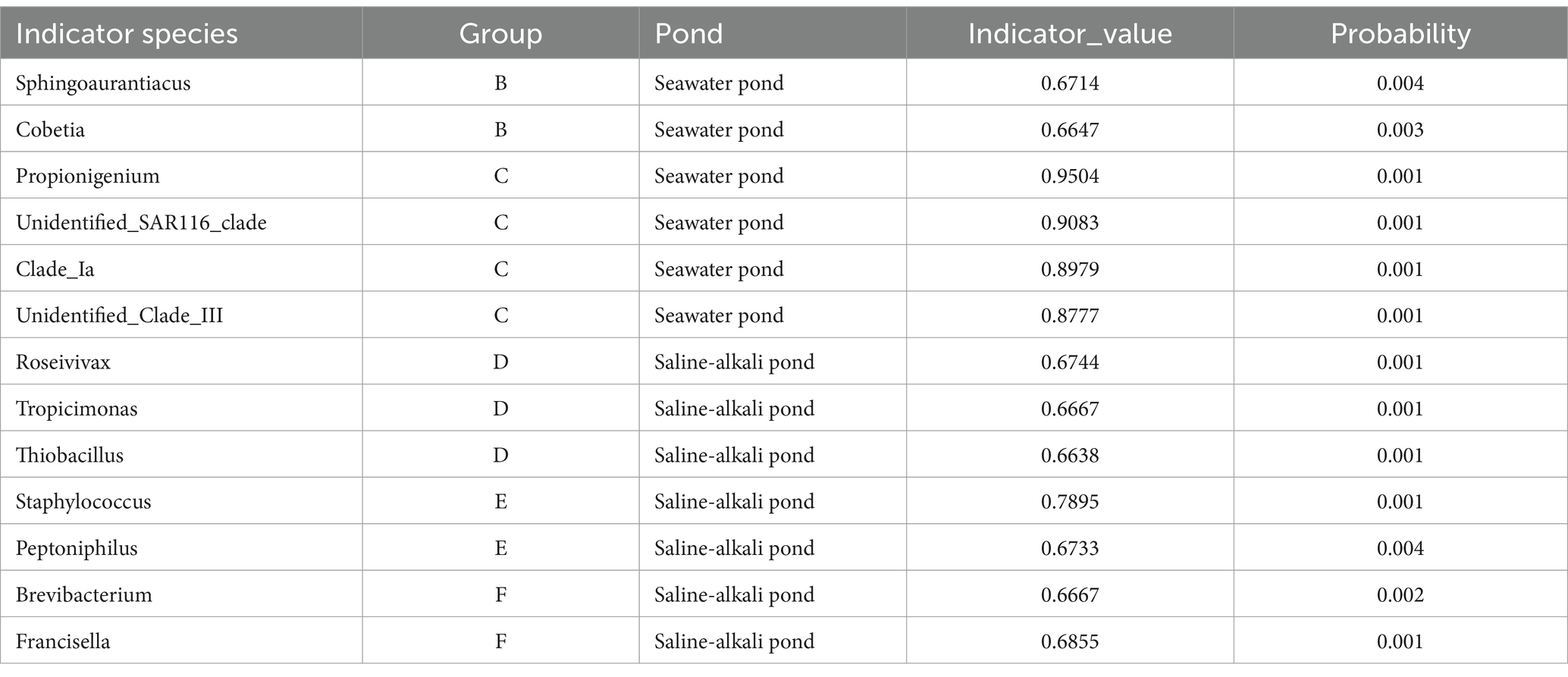
Table 4. Distribution characteristics of indicator bacteria in seawater ponds and saline-alkali ponds.
In the saline-alkali ponds, Roseivivax, Tropicimonas, and Thiobacillus were predominantly associated with Group D, exhibiting indicator values ranging from 0.6638 to 0.6744, with a probability of 0.001. These species are characteristic of saline-alkali ponds. Staphylococcus and Peptoniphilus, which were linked to Group E, showed indicator values of 0.7895 and 0.6733, with associated probabilities of 0.001 and 0.004, respectively. These values underscore the significant distribution patterns of these species within saline-alkali ponds. Additionally, Brevibacterium and Francisella, linked to Group F, displayed indicator values of 0.6667 and 0.6855, with probabilities of 0.002 and 0.001, confirming their role as sensitive bacterial species in saline-alkali ponds. These findings offer valuable insights into the distinct microbial community structures between seawater and saline-alkali ponds, forming a foundation for future ecological research and aquaculture applications.
The Pearson correlation heatmap (Figure 5) illustrates significant relationships between various sensitive bacterial species and water quality physicochemical parameters. The color intensity of the heatmap reflects the strength of these correlations, with red denoting positive correlations and blue indicating negative correlations. Specifically, salinity exhibited a positive correlation with Clade_Ia, unidentified_SAR116_clade, Propionigenium, Cobetia, and Sphingoaurantiacus in seawater pond samples, while it showed a negative correlation with Francisella and Thiobacillus in saline-alkali pond samples. pH demonstrated a negative correlation with unidentified_Clade_III and unidentified_SAR116_clade in seawater ponds, but a positive correlation with Thiobacillus and Francisella in saline-alkali ponds. Dissolved oxygen was positively correlated with unidentified_Clade_III, Propionigenium, and Cobetia in seawater ponds, whereas it was negatively correlated with Roseivivax, Tropicimonas, Thiobacillus, Staphylococcus, and Peptoniphilus in saline-alkali ponds. Ammonia nitrogen showed a positive correlation with Tropicimonas, Thiobacillus, and Staphylococcus in saline-alkali ponds, but a negative correlation with Sphingoaurantiacus in seawater ponds. Nitrite nitrogen was positively correlated with Thiobacillus, Staphylococcus, and Francisella in saline-alkali ponds, while it was negatively correlated with unidentified_Clade_III in seawater ponds.
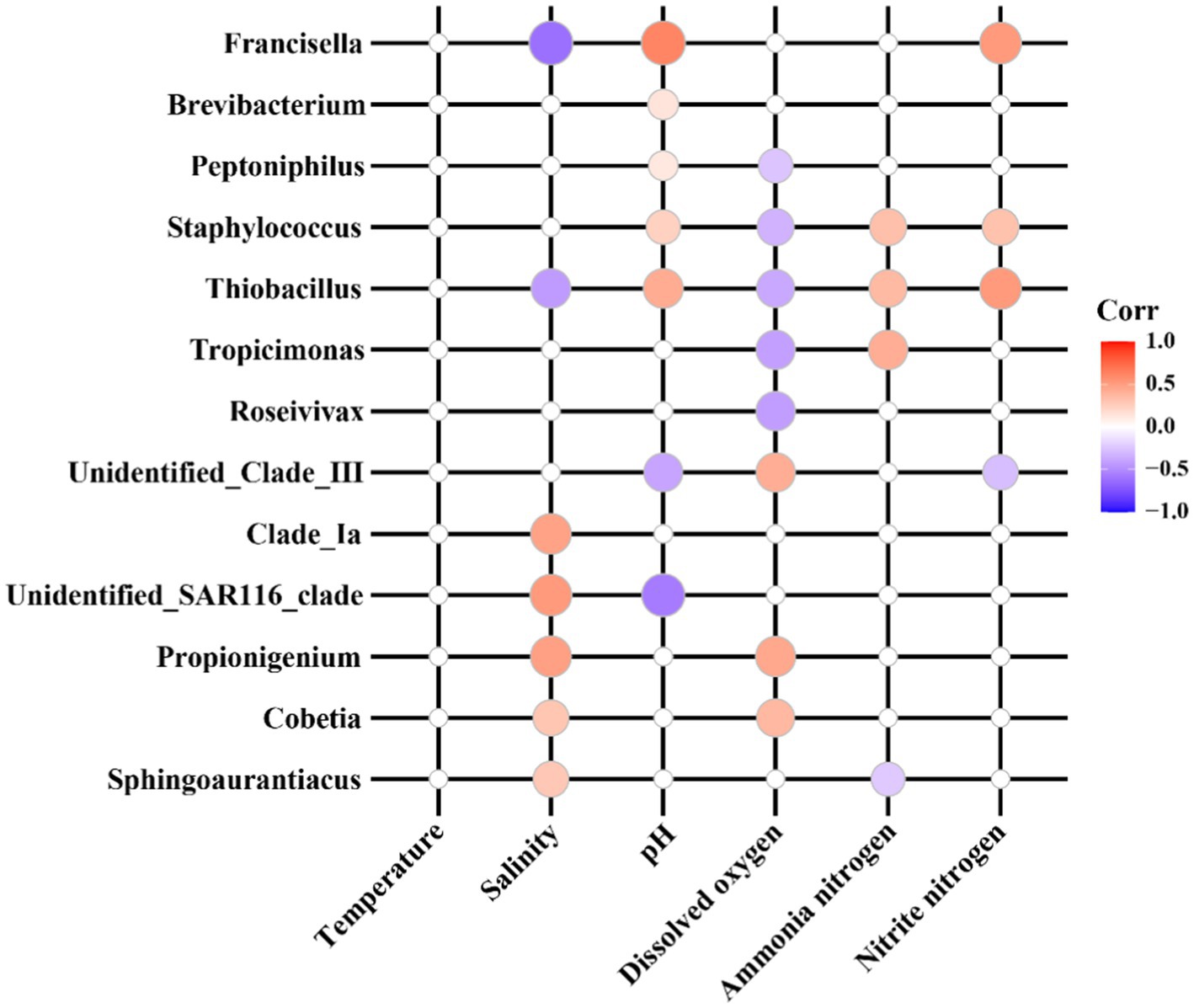
Figure 5. Correlation analysis between sensitive indicator flora and physical and chemical indexes of water quality.
3.7 Function prediction analysis
Using PICRUSt2, functional gene annotation and clustering analyses were performed on water samples from seawater and saline-alkali ponds, revealing substantial differences in the functional composition of microbial communities between the two environments. The functional gene data for this analysis were sourced from the KEGG Orthology (KO) database. The clustering heatmap results demonstrated environmental specificity in the distribution of functional genes across the pond water samples, reflecting microbial adaptive strategies to their respective ecosystems (Figure 6). In saline-alkali ponds, microbial functional genes identified from the KEGG Orthology (KO) database were predominantly associated with resource acquisition and adaptation to environmental stress. For instance, genes K01992 and K01990, which encode sugar and glutamate transport proteins, were more abundant in saline-alkali environments. This suggests that microbes in these ponds actively acquire sugars and amino acids to cope with nutrient scarcity and high osmotic pressure. Moreover, the high prevalence of phosphate transport protein genes, such as K02034, K02033, and K02032, indicates that phosphorus availability in saline-alkali ponds may be limited, prompting microbes to enhance phosphorus utilization through high-affinity phosphate transport systems. In contrast, the functional genes in seawater ponds, also sourced from the KEGG Orthology (KO) database, were primarily related to nitrogen metabolism and protein synthesis. Genes K00059 and K06147, which are associated with glutamate dehydrogenase and nitrate reductase, respectively, exhibited high nitrogen cycling activity. The presence of gene K03088, encoding ribosomal proteins, suggests an increased capacity for protein synthesis, potentially to meet the higher nutrient inputs and dynamic environmental conditions in seawater ponds. Additionally, the gene K08884 (ATP-binding cassette transport protein), identified through the KEGG Orthology (KO) database, was significantly abundant in both environments, indicating that active resource transport via ABC transport systems is a common metabolic strategy among microbes. In summary, the functional differences in microbial community composition between seawater and saline-alkali ponds reflect the impact of environmental factors on gene distribution, as demonstrated by KEGG Orthology (KO) database analyses. Microbes in saline-alkali ponds are adapted to stress through efficient resource utilization and osmotic pressure regulation, while those in seawater ponds primarily engage in nutrient cycling and protein metabolism. These findings underscore the ecological significance of microbial functional diversity in these two distinct aquatic ecosystems.
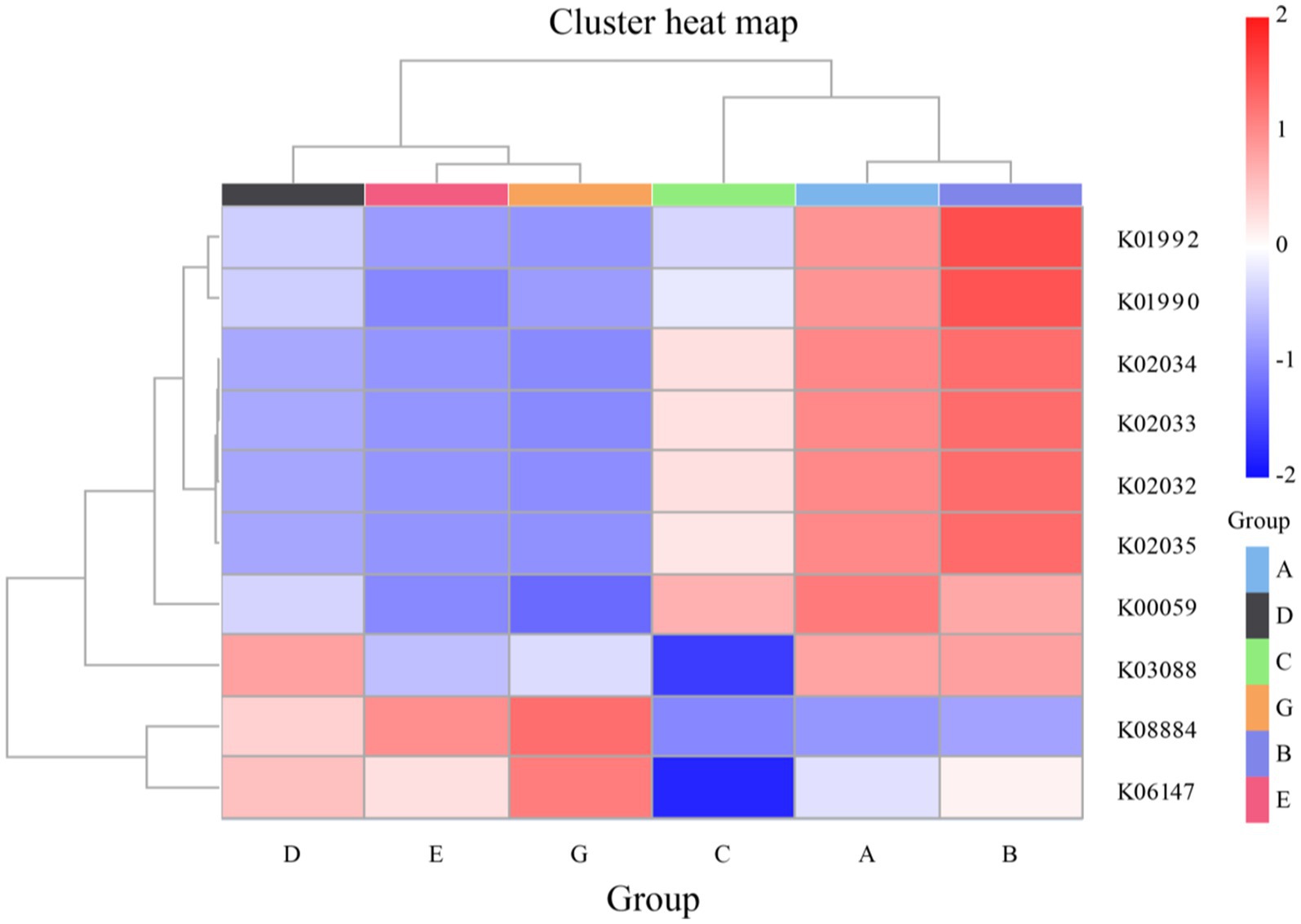
Figure 6. PICRUSt2 function annotation clustering heat map of seawater ponds and saline-alkali water ponds. The abscissa represents sample groups (D, E, G, C, A, B), and the ordinate displays KEGG orthologs (KOs) under positive selection (e.g., K01992, K01990). The color depth represents the relative abundance of functional genes.
4 Discussion
4.1 Diversity and structural differences of bacterial communities
The bacterial community diversity in seawater and saline-alkali ponds exhibits both similarities and significant differences. The Shannon and Simpson diversity indices reveal that while both pond types host a wide array of bacterial taxa, the diversity in the saline-alkali pond is marginally lower than in the seawater pond. This observation is consistent with studies by Liu et al. (2023) and others, who noted that extreme environments, such as saline-alkaline waters, tend to favor more specialized microbial communities (Niu et al., 2023). The dominance of halophilic and alkaliphilic bacteria in the saline-alkali pond further supports this notion (Valenzuela-Encinas et al., 2009). Contributing factors to the differences in bacterial diversity include variations in salinity, pH, and alkalinity, which exert selective pressure on microbial communities (Sorokin et al., 2012). Moreover, the more stable environmental conditions in seawater ponds, in contrast to the fluctuating conditions in saline-alkali ponds, may explain the higher diversity observed in the former (Paul and Mormile, 2017).
At the phylum level, both seawater and saline-alkali ponds are predominantly dominated by Cyanobacteria, Proteobacteria, and Actinobacteria, although their relative abundances differ significantly across the two environments. Seawater ponds exhibit a higher abundance of Proteobacteria, particularly the class Gammaproteobacteria, which aligns with the findings of Glaring et al. (2015) and reflects their adaptability to saline conditions. In contrast, Cyanobacteria and Firmicutes are more abundant in saline-alkali ponds, consistent with their reported tolerance to high alkalinity (Valenzuela-Encinas et al., 2009). At the genus level, Synechococcus_CC9902, DS001, and Marivita are highly abundant in seawater ponds, reflecting their salt tolerance and prevalence in marine environments (Ding et al., 2021). In contrast, unidentified_Chloroplast and Cyanobium_PCC-6307 dominate in saline-alkali ponds, indicating adaptation to high pH and ammonia nitrogen (Li X. et al., 2024; Li Z. et al., 2024). This finding is consistent with previous research by Kono and Spalding (2020), who observed alkaliphilic bacteria in high-pH environments.
The bacterial community structure was assessed using Principal Coordinates Analysis (PCoA), Non-metric Multidimensional Scaling (NMDS), and hierarchical clustering, revealing significant differences between the microbial communities of seawater and saline-alkali ponds. The seawater pond community exhibits a more even species distribution, consistent with Lyons and Schneider (1990), who observed balanced structures in stable, high-salinity environments. In contrast, saline-alkali ponds show pronounced clustering, indicating lower diversity but greater specialization. This pattern aligns with β-diversity indices and Kalwasińska et al. (2017), who linked extreme conditions to homogeneous communities dominated by specialized taxa. The observed differences in microbial community structure between the two pond types may be attributed to the unique combination of high alkalinity and pH in saline-alkali ponds, which impose stronger selective pressures on bacterial species (Yu et al., 2024; Sorokin et al., 2012).
4.2 Effects of water quality parameters on bacterial communities
This study reveals significant differences in the physicochemical water quality parameters between seawater and saline-alkali ponds, which have important implications for bacterial community structure. In high-salinity seawater ponds, bacterial community structure is relatively stable and exhibits higher diversity, with dominant groups like Cyanobacteria and Proteobacteria being evenly distributed. This is because salinity affects the osmotic pressure balance in bacterial cells, and high salinity selects for bacteria adapted to hyperosmotic environments, allowing them to dominate the community and influence its composition and diversity (Zahran, 1997). In contrast, the lower salinity in saline-alkali ponds leads to changes in microbial community structure, such as an increase in the relative abundance of Firmicutes, which may reflect adaptive strategies to lower salt concentrations (Sun et al., 2024).
pH, another critical factor, significantly impacts bacterial community composition. Seawater ponds, with lower pH (7.62–7.64), favor the dominance of Cyanobacteria and Proteobacteria, which play crucial roles in nutrient cycling (Landsman, 2019). Saline-alkali ponds, with higher pH (8.67–8.72), promote the proliferation of Cyanobacteria and Firmicutes, as these groups are better adapted to alkaline conditions (Belkin et al., 2024). The variation in pH alters bacterial metabolic activity and enzyme function, thereby affecting both community structure and functional diversity (Tu et al., 2022).
Dissolved oxygen (DO) concentrations differ markedly between the two pond types. Seawater ponds, with higher DO (8.83–8.91 mg/L), support aerobic bacteria and facilitate nitrogen cycling (Oren and Shilo, 1985), maintaining microbial stability and diversity. Saline-alkali ponds, with lower DO (5.89–6.25 mg/L), may promote anaerobic or facultative anaerobic bacteria, altering organic matter decomposition and nutrient cycling efficiency. These shifts are driven by differences in oxygen utilization and ecological niche distribution (Zhang et al., 2023; Pan et al., 2023).
4.3 Indicator bacteria and aquaculture environment stability
The differences in indicator bacteria between seawater and saline-alkali ponds can largely be attributed to the distinct environmental conditions in each ecosystem (Brauner et al., 2012; Heenatigala and Fernando, 2016), particularly variations in salinity, pH, and alkalinity. In seawater ponds, stable salinity and slightly alkaline pH favor indicator bacteria such as Sphingoaurantiacus and Cobetia, which are adapted to marine environments and sensitive to salinity fluctuations (Fernández-Juárez et al., 2022). In saline-alkali ponds, extreme alkalinity and pH select for alkaliphilic/halophilic species like Brevibacterium and Thiobacillus, reflecting adaptation to harsh conditions (Alvarado-López et al., 2023; Zhao et al., 2021).
Correlation analyses reveal contrasting relationships with physicochemical parameters. In seawater ponds, Sphingoaurantiacus and Cobetia positively correlate with salinity (Mu et al., 2020; Ganesan et al., 2023), indicating their role as salinity indicators. In saline-alkali ponds, Brevibacterium and Thiobacillus positively correlate with alkalinity/pH (Pei et al., 2021), while Roseivivax and Thiobacillus negatively correlate with dissolved oxygen and organic matter. This reflects adaptation to low-oxygen, nutrient-poor conditions (Cheng et al., 2024), aligning with previous studies on saline-alkaline environments (Ahmad et al., 2024).
Sphingoaurantiacus in seawater ponds negatively correlates with organic matter, likely due to competitive exclusion in nutrient-rich systems (Leite et al., 2023). These findings highlight the specificity of indicator bacteria as environmental health markers, underscoring the need to monitor salinity, pH, and dissolved oxygen to maintain microbial stability in S. paramamosain aquaculture.
4.4 The relationship between the prediction of bacterial community function and the culture of S. paramamosain
Functional prediction analysis reveals significant differences in the functional composition of microbial communities between seawater and saline-alkali ponds, highlighting the impact of environmental factors on the distribution of functional genes and microbial adaptation strategies. In saline-alkali ponds, microorganisms prioritize resource acquisition and stress resistance, as indicated by genes encoding glucose/glutamate transporters (K01992 and K01990) and high-affinity phosphate transport systems (K02034, K02033, and K02032), reflecting adaptive strategies to optimize phosphorus and nutrient utilization under limited availability (Knappe et al., 2003). In seawater ponds, microbial functions are dominated by nitrogen metabolism and protein synthesis, with genes such as glutamate dehydrogenase (K00059) and nitrate reductase (K06147), aligning with efficient nitrogen processing in nutrient-rich environments (Ahmed et al., 2021). Notably, ABC transporter genes (K08884) are highly expressed in both environments, indicating conserved strategies for active resource uptake. These functional differences suggest saline-alkali pond microbes rely on phosphorus optimization and osmotic regulation, while seawater pond microbes focus on nitrogen cycling and protein synthesis. For S. paramamosain aquaculture, these findings provide actionable insights: enhancing nitrogen metabolism in seawater ponds via optimized feed formulation and introducing phosphate-solubilizing microbes in saline-alkali ponds to leverage K02034/K02032 functions, thereby improving nutrient utilization and crab growth.
4.5 Limitations and future directions
This study has several limitations that warrant consideration. First, the reliance on a single sampling event (September 2022) may overlook seasonal variations in microbial dynamics, necessitating multi-seasonal sampling to comprehensively assess temporal shifts in bacterial communities. Second, while PICRUSt2 provided functional predictions of microbial metabolism, experimental validation through metatranscriptomic or enzymatic assays is critical to confirm the expression and activity of identified genes. Third, proposed management strategies, such as probiotic supplementation or phosphorus optimization, require field trials to evaluate their practical efficacy in enhancing S. paramamosain survival and growth under real-world aquaculture conditions. Addressing these limitations will strengthen the robustness of findings and facilitate actionable insights. Furthermore, indicator species in seawater and saline-alkali ponds, such as Sphingoaurantiacus, Cobetia, Roseivivax, Tropicimonas, and Thiobacillus etc., could be harnessed for synergistic regulation of harmful substances in water bodies through multiple mechanisms, which represents a promising future research direction. Future research should integrate multi-omics approaches (e.g., metagenomics and host-microbiome interaction studies), environmental modulation, and controlled aquaculture trials to optimize microbial management strategies. Such efforts will advance sustainable practices for S. paramamosain farming in northern China’s saline-alkali ecosystems, balancing productivity with ecological resilience.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found: https://www.ncbi.nlm.nih.gov/, PRJNA1232404.
Ethics statement
The animal studies were approved by Ethics committee of Ningbo University. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author contributions
DZ: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Software, Validation, Writing – original draft. YF: Conceptualization, Resources, Software, Validation, Visualization, Writing – review & editing. LL: Data curation, Formal analysis, Funding acquisition, Investigation, Project administration, Resources, Software, Supervision, Writing – original draft, Writing – review & editing. WL: Resources, Software, Visualization, Writing – original draft. ZL: Software, Validation, Writing – original draft. YY: Data curation, Software, Validation, Writing – review & editing. MZ: Data curation, Formal analysis, Investigation, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the National Key R&D Program of China (grant number 2023YFD2401005), the Dongying Science and Technology Innovation Major Project (grant number 2023ZDJH30), the Zhejiang Provincial Natural Science Foundation of China (grant number LY20C190005) and the K. C. Wong Magna Fund of Ningbo University.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Aboul-Maaty, N. A. F., and Oraby, H. A. S. (2019). Extraction of high-quality genomic DNA from different plant orders applying a modified CTAB-based method. Bull. Natl. Res. Cent. 43, 1–10. doi: 10.1186/s42269-019-0066-1
Ahmad, L., Shah, G. M. S., and Biswas, A. (2024). Fundamentals and applications of crop and climate science. Cham, Switzerland: Springer.
Ahmed, A., Khurshid, A., Tang, X., Wang, J., Khan, T. U., and Mao, Y. (2021). Structural and functional impacts of microbiota on pyropia yezoensis and surrounding seawater in cultivation farms along coastal areas of the yellow sea. Microorganisms 9:1291. doi: 10.3390/microorganisms9061291
Alvarado-López, M. J., Garrido-Hoyos, S. E., Raynal-Gutiérrez, M. E., El-Kassis, E. G., Luque-Almagro, V. M., and Rosano-Ortega, G. (2023). Cyanide biodegradation by a native bacterial consortium and its potential for goldmine tailing biotreatment. Water 15:1595. doi: 10.3390/w15081595
Belkin, P., Nechaeva, Y., Blinov, S., Vaganov, S., Perevoshchikov, R., and Plotnikova, E. (2024). Sediment microbial communities of a technogenic saline-alkaline reservoir. Heliyon. 10:e33640. doi: 10.1016/j.heliyon.2024.e33640
Bokulich, N. A., Subramanian, S., Faith, J. J., Gevers, D., Gordon, J. I., Knight, R., et al. (2013). Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat. Methods 10, 57–59. doi: 10.1038/nmeth.2276
Brauner, C. J., Gonzalez, R. J., and Wilson, J. M. (2012). Extreme environments: hypersaline, alkaline, and ion-poor waters. Fish Physiol. 32, 435–476. doi: 10.1016/B978-0-12-396951-4.00009-8
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Lozupone, C. A., Turnbaugh, P. J., et al. (2011). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. 108, 4516–4522. doi: 10.1073/pnas.1000080107
Chen, F., and Wang, K. (2019). Characterization of the innate immunity in the mud crab Scylla paramamosain. Fish Shellfish Immunol. 93, 436–448. doi: 10.1016/j.fsi.2019.07.076
Cheng, S., Meng, F., Wang, Y., Zhang, J., and Zhang, L. (2024). The potential linkage between sediment oxygen demand and microbes and its contribution to the dissolved oxygen depletion in the Gan River. Front. Microbiol. 15:1413447. doi: 10.3389/fmicb.2024.1413447
Cheng, W., Wang, L. U., and Chen, J. C. (2005). Effect of water temperature on the immune response of white shrimp Litopenaeus vannamei to Vibrio alginolyticus. Aquaculture 250, 592–601. doi: 10.1016/j.aquaculture.2005.04.060
Crescenzi, F., Crisari, A., D'Angel, E., and Nardella, A. (2006). Control of acidity development on solid sulfur due to bacterial action. Environ. Sci. Technol. 40, 6782–6786. doi: 10.1021/es0610131
Cui, S., Yang, C., Yang, F., and Zhang, J. (2009). Evaluating the ecological performance of wetland restoration in the Yellow River Delta, China. Ecol. Engineer. 35, 1090–1103. doi: 10.1016/j.ecoleng.2009.03.022
Deng, Y., Cheng, C., Feng, J., Liu, S., and Guo, Z. (2020). Rapid environmental change shapes pond water microbial community structure and function, affecting mud crab (Scylla paramamosain) survivability. Appl. Microbiol. Biotechnol. 104, 2229–2241. doi: 10.1007/s00253-019-10328-w
Ding, Y., Song, X., Cao, X., He, L., Liu, S., and Yu, Z. (2021). Healthier communities of phytoplankton and bacteria achieved via the application of modified clay in shrimp aquaculture ponds. Int. J. Environ. Res. Public Health 18:11569. doi: 10.3390/ijerph182111569
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C., and Knight, R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. doi: 10.1093/bioinformatics/btr381
Falkowski, P. G., Fenchel, T., and Delong, E. F. (2008). The microbial engines that drive Earth's biogeochemical cycles. Science 320, 1034–1039. doi: 10.1126/science.1153213
Fernández-Juárez, V., Jaén-Luchoro, D., Brito-Echeverría, J., Agawin, N. S., Bennasar-Figueras, A., and Echeveste, P. (2022). Everything is everywhere: physiological responses of the Mediterranean Sea and eastern Pacific Ocean epiphyte Cobetia sp. to varying nutrient concentration. Microb. Ecol. 83, 296–313. doi: 10.1007/s00248-021-01766-z
Fitzgerald, C. M., Camejo, P., Oshlag, J. Z., and Noguera, D. R. (2015). Ammonia-oxidizing microbial communities in reactors with efficient nitrification at low-dissolved oxygen. Water Res. 70, 38–51. doi: 10.1016/j.watres.2014.11.041
Ganesan, G., Balu, C., Ganesan, S., Arulmani, S. R. B., and Kandasamy, S. (2023). Antibiofilm activity of biosurfactant produced by a sponge-associated marine Cobetia sp. JCG-23. Biomass Convers. Biorefin., 1–15. doi: 10.1007/s13399-023-04808-3
Glaring, M. A., Vester, J. K., Lylloff, J. E., Abu Al-Soud, W., Sørensen, S. J., and Stougaard, P. (2015). Microbial diversity in a permanently cold and alkaline environment in Greenland. PLoS One 10:e0124863. doi: 10.1371/journal.pone.0124863
Gorrasi, S., Franzetti, A., Ambrosini, R., Pittino, F., Pasqualetti, M., and Fenice, M. (2021). Spatio-temporal variation of the bacterial communities along a salinity gradient within a thalassohaline environment (saline di tarquinia salterns, Italy). Molecules 26:1338. doi: 10.3390/molecules26051338
Guo, J., Peng, Y., Wang, S., Zheng, Y., Huang, H., and Wang, Z. (2009). Long-term effect of dissolved oxygen on partial nitrification performance and microbial community structure. Bioresour. Technol. 100, 2796–2802. doi: 10.1016/j.biortech.2008.12.036
Harrison, A. P. (1984). The acidophilic thiobacilli and other acidophilic bacteria that share their habitat. Ann. Rev. Microbiol. 38, 265–292. doi: 10.1146/annurev.mi.38.100184.001405
Hastuti, Y. P., Rusmana, I., Nirmala, K., Affandi, R., and Fatma, Y. S. (2021). Characterization of denitrifying and dissimilatory nitrate reduction to ammonium bacteria isolated from mud crab culture environment. Microbiol. Biotechnol. Lett. 49, 432–439. doi: 10.48022/mbl.2011.11011
Heenatigala, P. P. M., and Fernando, M. U. L. (2016). Occurrence of bacteria species responsible for vibriosis in shrimp pond culture systems in Sri Lanka and assessment of the suitable control measures. Sri Lanka J. Aquatic Sci. 21:7481. doi: 10.4038/sljas.v21i1.7481
Holmes, D. E., Dang, Y., and Smith, J. A. (2019). Nitrogen cycling during wastewater treatment. Adv. Appl. Microbiol. 106, 113–192. doi: 10.1016/bs.aambs.2018.10.003
Huang, S., Gao, W., Tang, J., and Li, C. (2015). Total salt content and ion composition in tillage layer of soils in the main vegetable production regions of China. Plant Nutr. Fert. Sci. 21, 10–18. doi: 10.11674/zwyf.15010
Jie, Y. K., Cheng, C. H., Wang, L. C., Ma, H. L., Deng, Y. Q., Liu, G. X., et al. (2021). Hypoxia-induced oxidative stress and transcriptome changes in the mud crab (Scylla paramamosain). Comp. Biochem. Physiol. C Toxicol. Pharmacol. 245:109039. doi: 10.1016/j.cbpc.2021.109039
Jing, G., Sun, Z., Wang, H., Gong, Y., Huang, S., Ning, K., et al. (2017). Parallel-META 3: comprehensive taxonomical and functional analysis platform for efficient comparison of microbial communities. Sci. Rep. 7:40371. doi: 10.1038/srep40371
Johnson, D. B., and Quatrini, R. (2020). Acidophile microbiology in space and time. Curr. Issues Mol. Biol. 39, 63–76. doi: 10.21775/cimb.039.063
Kalwasińska, A., Felföldi, T., Szabó, A., Deja-Sikora, E., Kosobucki, P., and Walczak, M. (2017). Microbial communities associated with the anthropogenic, highly alkaline environment of a saline soda lime, Poland. Antonie Leeuwenhoek. 110, 945–962. doi: 10.1007/s10482-017-0866-y
Kasan, N. A., Sze Yee, C., Manan, H., Ahmad Ideris, A. R., Kamaruzzan, A. S., Waiho, K., et al. (2021). Study on the implementation of different biofloc sedimentable solids in improving the water quality and survival rate of mud crab, Scylla paramamosain larvae culture. Aquac. Res. 52, 4807–4815. doi: 10.1111/are.15314
Kato, M. T., Field, J. A., and Lettinga, G. (1997). Anaerobe tolerance to oxygen and the potentials of anaerobic and aerobic cocultures for wastewater treatment. Braz. J. Chem. Eng. 14, 395–407. doi: 10.1590/S0104-66321997000400015
Knappe, S., Flugge, U. I., and Fischer, K. (2003). Analysis of the plastidic phosphate translocator gene family in Arabidopsis and identification of new phosphate translocator-homologous transporters, classified by their putative substrate-binding site. Plant Physiol. 131, 1178–1190. doi: 10.1104/pp.016519
Komiyama, A., Poungparn, S., Umnouysin, S., Rodtassana, C., Kato, S., Pravinvongvuthi, T., et al. (2020). Daily inundation induced seasonal variation in the vertical distribution of soil water salinity in an estuarine mangrove forest under a tropical monsoon climate. Ecol. Res. 35, 638–649. doi: 10.1111/1440-1703.12118
Kono, A., and Spalding, M. H. (2020). LCI1, a Chlamydomonas reinhardtii plasma membrane protein, functions in active CO2 uptake under low CO2. Plant J. 102, 1127–1141. |. doi: 10.1111/tpj.14761
Lahiri, S., Ghosh, D., and Sarkar, P. D. (2018). Biogeochemical cycling bacteria and nutrient dynamics in waste stabilization pond system. Wastewater Manage Through Aquacul., 29–52. doi: 10.1007/978-981-10-7248-2_2
Landsman, A. (2019). Shrimp production environment and the gut microbiome: effects of aquaculture practices and selective breeding on the gut microbiome of Pacific Whiteleg shrimp, Litopenaeus Vannamei : South Dakota State University. 22587247.
Leite, M. R. L., de Alcantara Neto, F., Dutra, A. F., Mendes, L. W., Antunes, J. E. L., Melo, V. M. M., et al. (2023). Silicon application influences the prokaryotic communities in the rhizosphere of sugarcane genotypes. Appl. Soil Ecol. 187:104818. doi: 10.1016/j.apsoil.2023.104818
Leote, P., Cajaiba, R. L., Cabral, J. A., Brescovit, A. D., and Santos, M. (2020). Are data-mining techniques useful for selecting ecological indicators in biodiverse regions? Bridges between market basket analysis and indicator value analysis from a case study in the neotropics. Ecol. Indic. 109:105833. doi: 10.1016/j.ecolind.2019.105833
Li, X., Che, W., Piao, J., Song, Y., Wang, X., Zhang, Y., et al. (2024). Biochar increases Rice yield in soda saline-alkali Paddy fields by improving saline-alkali stress and phosphorus use efficiency. Agronomy 14:2159. doi: 10.3390/agronomy14092159
Li, Z., Guo, X., Guo, Z., Shi, X., Zhou, J., Liu, Z., et al. (2024). 3D morphological scanning and environmental correlates of Bufo gargarizans in the Yellow River Basin. Animals 14:369. doi: 10.3390/ani14030369
Lin, Z., Qiao, J., Zhang, Y., Guo, L., Huang, H., Yan, F., et al. (2012). Cloning and characterisation of the sptoll gene from green mud crab, Scylla paramamosain. Dev. Comp. Immunol. 37, 164–175. doi: 10.1016/j.dci.2011.09.003
Liu, Q., Lei, X., Li, J., Chu, L., Wang, F., Shan, H., et al. (2023). Microbial communities and nitrogen cycling in Litopenaeus vannamei and Mercenaria mercenaria polyculture ponds. Aquacult. Rep. 33:101769. doi: 10.1016/j.aqrep.2023.101769
Lyons, J., and Schneider, D. W. (1990). Factors influencing fish distribution and community structure in a small coastal river in southwestern Costa Rica. Hydrobiologia 203, 1–14. doi: 10.1007/BF00005608
Ma, H., Jiang, W., Liu, P., Feng, N., Ma, Q., Ma, C., et al. (2014). Identification of transcriptome-derived microsatellite markers and their association with the growth performance of the mud crab (Scylla paramamosain). PLoS One 9:e89134. doi: 10.1371/journal.pone.0089134
Magoc, T., and Salzberg, S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
Maruyama, H., Masago, A., Nambu, T., Mashimo, C., and Okinaga, T. (2020). Amplicon sequence variant-based oral microbiome analysis using QIIME 2. J. Osaka Dent. Univ. 54, 273–281. doi: 10.18905/jodu.54.2_273
Mu, D. S., Wang, S., Liang, Q. Y., Du, Z. Z., Tian, R., Ouyang, Y., et al. (2020). Bradymonabacteria, a novel bacterial predator group with versatile survival strategies in saline environments. Microbiome 8, 126–115. doi: 10.1186/s40168-020-00902-0
Nichols, D. S. (2003). Prokaryotes and the input of polyunsaturated fatty acids to the marine food web. FEMS Microbiol. Lett. 219, 1–7. doi: 10.1016/S0378-1097(02)01200-4
Niu, M., Li, X., Chen, Y., Qin, K., Liang, G., Hu, Y., et al. (2023). Response of intestinal microbiota to saline-alkaline water in mud crab (Scylla paramamosain) based on multiple low salinity culture modes. Front. Mar. Sci. 10:1153326. doi: 10.3389/fmars.2023.1153326
Oren, A., and Shilo, M. (1985). Factors determining the development of algal and bacterial blooms in the Dead Sea: a study of simulation experiments in outdoor ponds. FEMS Microbiol. Ecol. 1, 229–237. doi: 10.1111/j.1574-6968.1985.tb01154.x
Paerl, H. W., Dyble, J., Moisander, P. H., Noble, R. T., Piehler, M. F., Pinckney, J. L., et al. (2003). Microbial indicators of aquatic ecosystem change: current applications to eutrophication studies. FEMS Microbiol. Ecol. 46, 233–246. doi: 10.1016/S0168-6496(03)00200-9
Pan, Y., She, D., Shi, Z., Cao, T., Xia, Y., and Shan, J. (2023). Salinity and high pH reduce denitrification rates by inhibiting denitrifying gene abundance in a saline-alkali soil. Sci. Rep. 13:2155. doi: 10.1038/s41598-023-29311-7
Pati, S. G., Paital, B., Panda, F., Jena, S., and Sahoo, D. K. (2023). Impacts of habitat quality on the physiology, ecology, and economical value of mud crab Scylla sp.: a comprehensive review. Water 15:2029. doi: 10.3390/w15112029
Paul, V. G., and Mormile, M. R. (2017). A case for the protection of saline and hypersaline environments: a microbiological perspective. FEMS Microbiol. Ecol. 93:fix091. doi: 10.1093/femsec/fix091
Pei, S., Niu, S., Xie, F., Wang, W., Zhang, S., and Zhang, G. (2021). Brevibacterium limosum sp. nov., Brevibacterium pigmenatum sp. nov., and Brevibacterium atlanticum sp. nov., three novel dye decolorizing actinobacteria isolated from ocean sediments. J. Microbiol. 59, 898–910. doi: 10.1007/s12275-021-1235-0
Qiu, Z., Zhao, J., Luo, Q., Qian, R., Lin, X., and Xu, Q. (2023). Effects of soybean oligosaccharides instead of glucose on growth, digestion, antioxidant capacity and intestinal flora of crucian carp cultured in biofloc system. Aquacult. Rep. 29:101512. doi: 10.1016/j.aqrep.2023.101512
Sorokin, D. Y., Janssen, A. J. H., and Muyzer, G. (2012). Biodegradation potential of halo (alkali) philic prokaryotes. Crit. Rev. Environ. Sci. Technol. 42, 811–856. doi: 10.1080/10643389.2010.534037
Spietz, R. L., Williams, C. M., Rocap, G., and Horner-Devine, M. C. (2015). A dissolved oxygen threshold for shifts in bacterial community structure in a seasonally hypoxic estuary. PLoS One 10:e0135731. doi: 10.1371/journal.pone.0135731
Sun, Z., Yao, Z., Gao, P., Zhou, K., Li, Y., Wei, Y., et al. (2024). Effects of fishery utilization on the physicochemical index and microbial community composition in saline-alkaline water. ACS Omega 9, 18872–18881. doi: 10.1021/acsomega.3c08437
Troch, M. D., Boeckx, P., Cnudde, C., Gansbeke, D. V., Vanreusel, A., Vincx, M., et al. (2012). Bioconversion of fatty acids at the basis of marine food webs: insights from a compound-specific stable isotope analysis. Mar. Ecol. Prog. Ser. 465, 53–67. doi: 10.3354/meps09920
Tu, D., Ke, J., Luo, Y., Hong, T., Sun, S., Han, J., et al. (2022). Microbial community structure and shift pattern of industry brine after a long-term static storage in closed tank. Front. Microbiol. 13:975271. doi: 10.3389/fmicb.2022.975271
Valenzuela-Encinas, C., Neria-González, I., Alcántara-Hernández, R. J., Estrada-Alvarado, I., Zavala-Díaz de la Serna, F. J., Dendooven, L., et al. (2009). Changes in the bacterial populations of the highly alkaline saline soil of the former Lake Texcoco (Mexico) following flooding. Extremophiles 13, 609–621. doi: 10.1007/s00792-009-0244-4
Vincent, W. F., Hobbie, J. E., and Laybourn-Parry, J. (2008). “Introduction to the limnology of high-latitude lake and river ecosystems” in Polar lakes and rivers: limnology of Arctic and Antarctic aquatic ecosystems, 1–24.
Wang, M., Chen, S., Chen, L., Wang, D., and Zhao, C. (2019). The responses of a soil bacterial community under saline stress are associated with cd availability in long-term wastewater-irrigated field soil. Chemosphere 236:124372. doi: 10.1016/j.chemosphere.2019.124372
Wang, Y., Guo, H., Gao, X., and Wang, J. (2021). The intratumor microbiota signatures associate with subtype, tumor stage, and survival status of esophageal carcinoma. Front. Oncol. 11:754788. doi: 10.3389/fonc.2021.754788
Wei, H., Wang, H., Tang, L., Mu, C., Ye, C., Chen, L., et al. (2019). High-throughput sequencing reveals the core gut microbiota of the mud crab (Scylla paramamosain) in different coastal regions of southern China. BMC Genomics 20:829. doi: 10.1186/s12864-019-6219-7
Xiong, J., Chen, H., Hu, C., Ye, X., Kong, D., and Zhang, D. (2015). Evidence of bacterioplankton community adaptation in response to long-term mariculture disturbance. Sci. Rep. 5:15274. doi: 10.1038/srep15274
Yu, K., Qiu, Z., He, S., Lian, C. A., Zhang, Q., Qiao, X., et al. (2024). Large scale exploration reveals rare taxa as key members shaping microbial assembly in alkaline lake sediments. npj Biofil. Microb. 10, 1–13. doi: 10.21203/rs.3.rs-3831384/v1
Yuan, Z., Niu, J., Qin, K., Wang, C., Mu, C., and Wang, H. (2024). Metabolic mechanism of Scylla paramamosain gill mitochondria in response to acute low salinity stress. Aquacult. Rep. 37:102234. doi: 10.1016/j.aqrep.2024.102234
Zahran, H. H. (1997). Diversity, adaptation and activity of the bacterial flora in saline environments. Biol. Fertil. Soils 25, 211–223. doi: 10.1007/s003740050306
Zhang, Y., Gao, W., Yuan, Y., Cui, W., Xiang, Z., Ye, S., et al. (2024). Impact and accumulation of calcium on soft-shell mud crab Scylla paramamosain in recirculating aquaculture system. Aquaculture 593:741323. doi: 10.1016/j.aquaculture.2024.741323
Zhang, P., Huang, Q., Peng, R., Jiang, X., Jiang, M., Zeng, G., et al. (2022). Environmental factors of rearing water and growth performance of shrimp (Penaeus vannamei) in a microalgal monoculture system. Aquaculture 561:738620. doi: 10.1016/j.aquaculture.2022.738620
Zhang, S., Rasool, G., Wang, S., Zhang, Y., Guo, X., Wei, Z., et al. (2023). Biochar and chlorella increase rice yield by improving saline-alkali soil physicochemical properties and regulating bacteria under aquaculture wastewater irrigation. Chemosphere 340:139850. doi: 10.1016/j.chemosphere.2023.139850
Zhao, Y., Wang, J., Liu, Y., Zheng, P., and Hu, B. (2021). Microbial interaction promotes desulfurization efficiency under high pH condition. Environ. Res. 200:111423. doi: 10.1016/j.envres.2021.111423
Keywords: Scylla paramamosain, bacterial communities, 16S rRNA gene sequencing, aquaculture, seawater pond, saline-alkali water pond
Citation: Zhou D, Fu Y, Liu L, Lin W, Lu Z, Ye Y and Zhang M (2025) Comparative analysis of bacterial communities and environmental interactions in seawater and saline-alkali aquaculture ponds for Scylla paramamosain in northern China. Front. Microbiol. 16:1589304. doi: 10.3389/fmicb.2025.1589304
Edited by:
Xianbiao Lin, Ocean University of China, ChinaReviewed by:
Hongyu Ma, Shantou University, ChinaYafei Duan, South China Sea Fisheries Research Institute, China
Copyright © 2025 Zhou, Fu, Liu, Lin, Lu, Ye and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lei Liu, bGl1bGVpMUBuYnUuZWR1LmNu Minglei Zhang, em1senp6cXpAMTYzLmNvbQ==
 Dongping Zhou1,2
Dongping Zhou1,2 Lei Liu
Lei Liu Weichuan Lin
Weichuan Lin Yangfang Ye
Yangfang Ye