- 1Faculty of Agriculture, Forestry and Food Engineering, Yibin University, Yibin, China
- 2College of Veterinary Medicine, Qingdao Agricultural University, Qingdao, China
Introduction: Papillomaviruses have been previously identified in mammals, avians and fish. However, few numbers of reptiles’ PVs have been characterized.
Methods: This study investigated the oral cavity of the splendid japalure (Japalura splendida) in southwestern China using high-throughput sequencing. The presence of papillomavirus strain JsPV in oral samples was confirmed using PCR with consensus primers.
Results and discussion: In this study, a papillomavirus strain, designated JsPV, in the oral cavity of the splendid japalure (Japalura splendida) in southwestern China. The complete JsPV genome was sequenced, comprising 222 bp. Phylogenetic analysis based on the L1 protein revealed that JsPV clustered closely with gecko-derived strains (HfrePV1 and HfrePV2) and other sauropsid-associated papillomaviruses, while remaining distinct from mammalian- and fish-associated lineages. These findings provide insights into the evolutionary origins of papillomaviruses in reptiles.
1 Introduction
Papillomaviruses (PVs) have been detected across all vertebrate taxa, including reptiles, and constitute a diverse family of non-enveloped, double-stranded DNA viruses with genomes ranging from 7 to 8 kb in length. To date, only a limited number of non-mammalian PVs have been characterized, including yellow-necked francolin (Francolinus leucoscepus), common chaffinch (Fringilla coelebs), northern fulmar (Fulmar glacialis), African grey parrot (Psittacus erithacus), Adélie penguin (Pygoscelis adeliae), green sea turtle (Chelonia mydas), loggerhead sea turtle (Caretta caretta), gilt-head bream fish (Sparus aurata), and Asian house gecko (Hemidactylus frenatus) (Agius et al., 2019). Papillomaviridae genomes typically encode early regulatory proteins (E1 and E2) and late structural proteins (L1 and L2) (Agius et al., 2019). E1 serves as an adenosine triphosphate (ATP)-dependent DNA helicase, playing a key role in viral genome replication and episomal amplification, thought to be essential throughout the viral life cycle (Bergvall et al., 2013). To date, over 230 types of human PVs (HPVs) and 159 types of non-human PVs have been identified (Van Doorslaer et al., 2017). HPVs account for an estimated 27.9 to 30.0% of cancer cases globally (Zur Hausen, 2009; Bravo et al., 2010). PVs are implicated not only in human malignancies but also in neoplasms affecting various animal species. Notable examples include bovine papillomavirus, cottontail rabbit papillomavirus, rodent papillomavirus, feline papillomavirus, and canine oral papillomavirus, all of which have been recognized for their oncogenic potential in their respective hosts (Gil da Costa and Medeiros, 2014; Uberoi and Lambert, 2017; Medeiros-Fonseca et al., 2023). In reptiles, PVs are associated with mucocutaneous and cutaneous epithelial proliferative lesions (Herbst et al., 2009); however, no clear link has been established between PV infection and tumor development in these species.
The splendid japalure (Japalura splendida) is native to southwestern China, including Yunnan, Sichuan, Chongqing, and Hubei provinces, where it primarily inhabits forest edges (Huang et al., 2019). It is an insectivorous species and is commonly maintained in captivity as a pet (Tian et al., 2022). In this study, a papillomavirus was identified for the first time in the oral cavity of splendid japalure using high-throughput sequencing technology and virus-specific polymerase chain reaction (PCR) analysis. This discovery provides insights into the characteristics and evolutionary history of PVs in lizards.
2 Materials and methods
2.1 Sample preparation, DNA sequencing, and sequence analysis
Samples were obtained from a splendid japalure near the Jinsha River (28°64′12″N, 104°27′22″E) in the absence of overt clinical symptoms. Oral samples were collected using sterile swabs, which were placed in RNase-free tubes and transported on dry ice to Novogene Bioinformatics Technology Co., Ltd. (Beijing, China). High-throughput sequencing technology, including DNA sequencing and subsequent bioinformatic analyses, was performed as described previously (Liu et al., 2023). Briefly, DNA was extracted from oral samples using the E.Z.N.A.® Stool DNA Kit (Omega BioTek, Norcross, GA, United States). A commercial library preparation kit [Rapid Plus DNA Lib Prep Kit for Illumina (RK20208)] was employed to construct the DNA library (ABclonal, Wuhan, China) according to the manufacturer’s protocol (Wang et al., 2024). Subsequently, library quality was assessed on the Agilent 5400 system (AATI) and quantified by real-time PCR (1.5 nM). The qualified libraries were pooled and sequenced on Illumina platforms with PE150 strategy in Novogene Bioinformatics Technology Co., Ltd. (Beijing, China), according to effective library concentration and the data amount required. Raw reads for clonal reads and low sequencing quality tails were processed using Trimmomatic (Bolger et al., 2014). Clean reads were assembled using the MEGAHIT software (Li et al., 2016) and were taxonomic classification using VIRify pipeline (Rangel-Pineros et al., 2023). All the contigs were split into high confidence (HC), low confidence (LC) and putative prophage (PP) sets. The assigned taxonomy was based on the informative ViPhOG hits per contig and performed on genus, family, subfamily (Moreno-Gallego and Reyes, 2021). The open reading frames (ORFs) in the viral genome were predicted by the BLASTx search results. The protein domains were identified and annotated using the NCBI conserved domain search (E-value <10−5) (Marchler-Bauer and Bryant, 2004).
2.2 Detection of papillomavirus in oral samples by PCR
Papillomavirus-specific primers for complete genome sequencing were designed based on known sequences (Supplementary Table S1). PCRs were conducted in a total of volume of 50 μL containing 5 μL of 10× buffer, 3 μL of dNTPs mixture (2.5 mM), 5 μL (10 ng) of DNA, 1 μL of forward primer (10 μM), 1 μL of reverse primer (10 μM), 0.5 μL (5 U) of LA Taq polymerase (TaKaRa, Tokyo, Japan), and 34.5 μL of sterile water. PCR was conducted with the cycling parameters: 94°C for 5 min, followed by 35 cycles at 94°C for 0.5 min, 56°C for 0.5 min, and 72°C for 2 min, and a final extension at 72°C for 10 min (Hu et al., 2015). Prior to sequencing, amplified products were cloned into the pMD18-T vector (TaKaRa). Three independent clones were sequenced using Sanger method with universal primers (M13F: AGGGTTTTCCCAGTCACG; M13R: CAGGAAACAGCTATGAC) to confirm sequence integrity. Complete sequences were manually assembled and alignment using Vector 10, and DNASTAR, respectively.
2.3 Phylogenetic and homology model analyses
The amino acid sequence of L1 protein was aligned with Pairwise nucleotide sequence similarity using Needleman–Wunsch algorithm of global alignment.2 Phylogenetic trees were constructed based on L1 protein sequences using the maximum-likelihood (ML) method with the LG + I + G + F model in MEGA v7.0. Bootstrap values were estimated for 1,000 replicates. Homology modeling of papillomavirus proteins was conducted using Phyre2.2 (PHYRE2 Protein Fold Recognition Server, ic.ac.uk).
3 Results and discussion
PVs are widely distributed across vertebrate and invertebrate species, including mammals (Varga et al., 2014; Bravo and Félez-Sánchez, 2015; Brücher and Jamall, 2014; Herbster et al., 2012), birds (Terai et al., 2002; Tachezy et al., 2002; Varsani et al., 2014; Gaynor et al., 2015; Varsani et al., 2015), fish (López-Bueno et al., 2016), and reptiles (Agius et al., 2019; Herbst et al., 2009; Lange et al., 2011). Among reptiles, previous studies have reported papillomavirus infections only in turtles and geckos, with viral strains distinct from those identified in other hosts (Agius et al., 2019; Herbst et al., 2009). In this study, a papillomavirus was identified for the first time in the oral cavity of a splendid japalure in the absence of visible lesions. The complete genome of this strain, designated JsPV, was amplified by PCR, revealing a genome length of 7,222 bp with a GC content of 43%. Conserved early proteins E1 (706 amino acids) and E2 (386 amino acids), along with late structural proteins L1 (512 amino acids) and L2 (444 amino acids), were identified (Figure 1A). A 612 bp region (8.5% of the genome) located between the stop codon of L1 and the start of E7 was identified, corresponding to the long control region (LCR) observed in other PVs (Figure 1B). It contained a polyadenylation (PolyA) site [257–262 nucleotides (nt)], a TATA box (254–258 nt) within an AT-rich region, one E2-binding site (ACC-N20-GGT) located at 177–202 nt and two modified putative E2-binding sites (AAC-N7-32-GGT) were found at 177–189 nt and 581–618 nt. No Nf1 or Sp1 binding sites matching the motifs identified in JsPV were detected. The putative E7 oncoprotein consisted of 110 amino acids (nucleotides (nt) 613–942), with a zinc-binding motif (CxxC) located in the C-terminal region (84–99 amino acids) and non-folded CR1 and CR2 motifs, although it lacked the conserved retinoblastoma protein (pRb) binding motif (L-x-C-x-E) (Figure 1A). Structural analysis indicated that the E7 protein of JsPV shared similarity with those of gecko-derived PVs (Supplementary Figure S1). Multiple sequence alignment indicated that L1 protein of JsPV shared 37.6–57% identity with mammalian- and sauropsid-associated PVs (Figure 2). Compared to the genomic architecture of other known sauropsid PVs, JsPV, along with HfrePV1 and HfrePV2, encoded only five proteins (E7, E1, E2, L2, and L1), lacking the E6 gene—the fewest reported among lacertilian PVs. E6 is a small oncoprotein (Boulet et al., 2007) that can form a trimeric complex with the E6AP ubiquitin ligase and p53 (Tan et al., 2012), leading to cell transformation (Liu et al., 2002) and immortalization (Boon et al., 2015), suggesting that lacertilian PVs may lack the molecular capacity to induce tumorigenesis. Previous studies have identified PVs in the genera Hemidactylus, Gehyra, and Lepidodactylus in Australia (Agius et al., 2019). However, the present study provides the first evidence of PVs in the genus Japalura, suggesting that these viruses may have originated from a distinct ancestral lineage in lizards and may have adapted to specific ecological niches within sympatric Lacertidae. Furthermore, the splendid japalure is endemic to China and inhabits faunal assemblages distinct from those of gecko-derived PVs. This finding suggests that there is a potential pathogenic risk of papillomavirus transmission to other reptiles. Further investigations are needed to explore the broader distribution of these viruses across China.
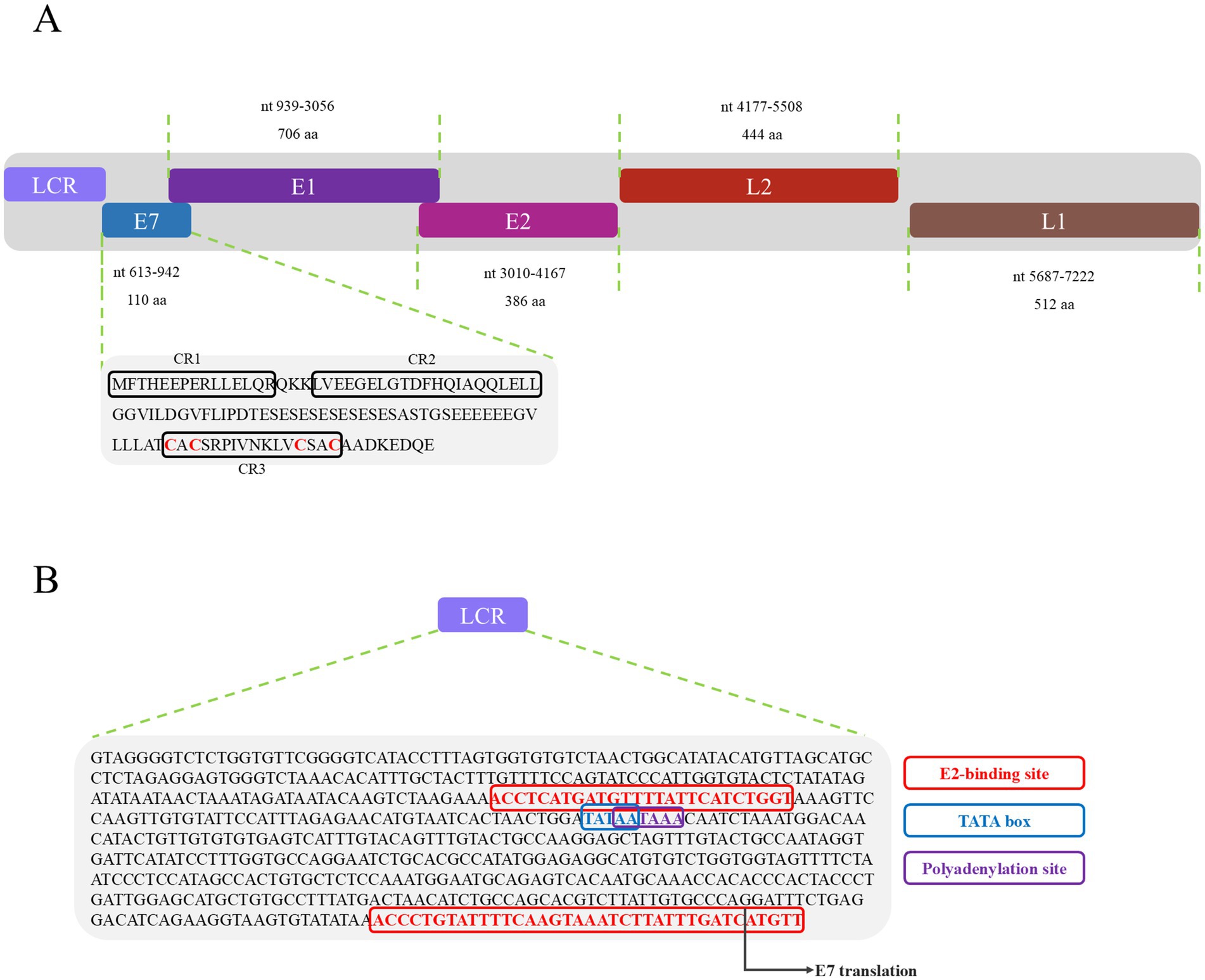
Figure 1. Genomic organization of JsPV. (A) Illustration of the genomic organization of JsPV, including LCR, E7, E1, E2, L2, and L1. The sequence and motif annotation of E7 protein, black boxes indicate conserved regions referred to CR1, CR2, and CR3, and red letters indicate putative Zn-binding domain. (B) Representation of the LCR motifs identified in JsPV. Coloured boxes represent different regulatory elements as per key provided, including E2-binding site, TATA box, polyadenylation site.
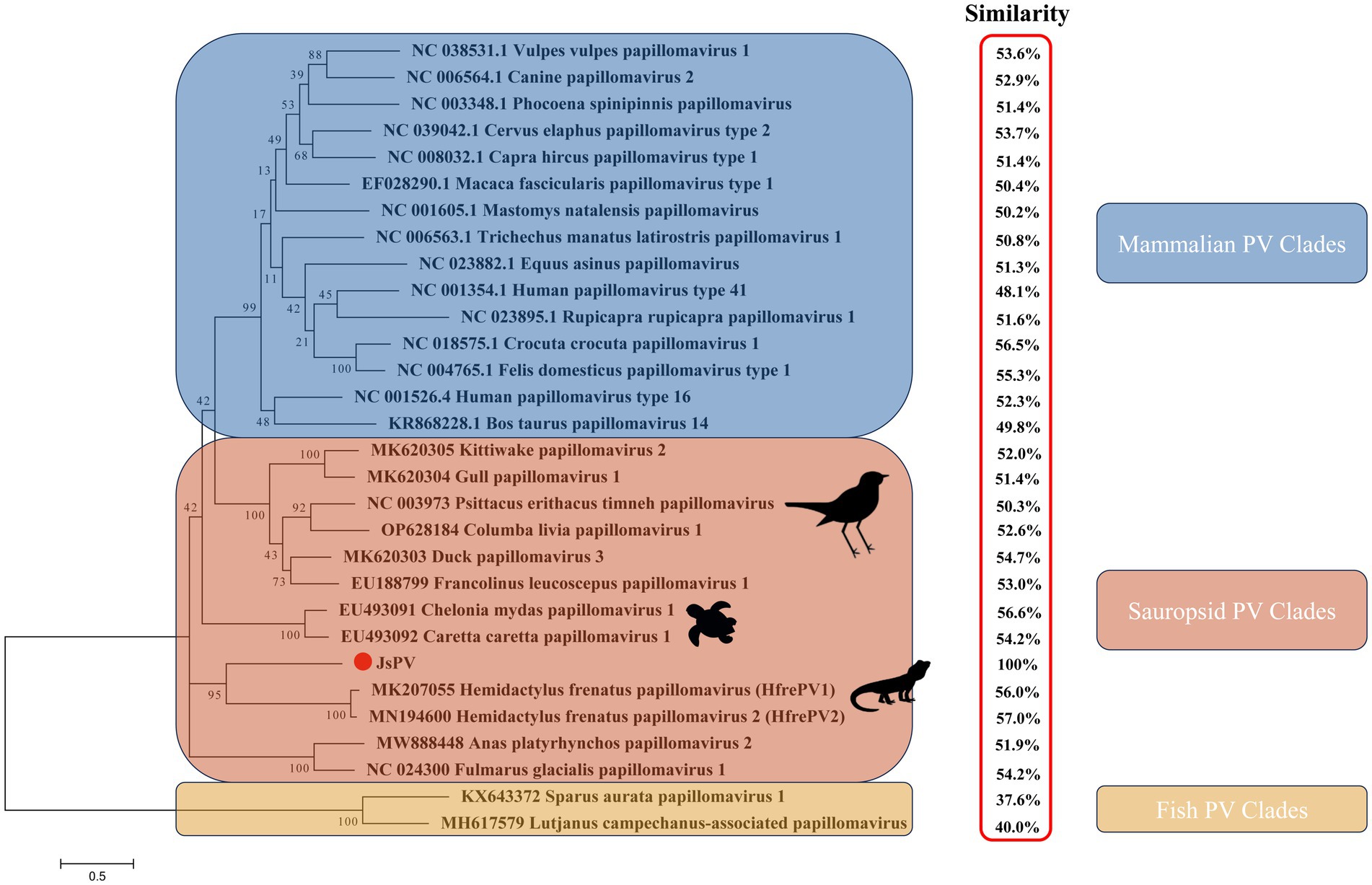
Figure 2. Maximum likelihood phylogenetic tree showing the relationship between JsPV and other papillomaviruses based on the L1 protein. The results of the L1 protein similarity comparison are showed in the red box.
Phylogenetic analysis demonstrated that the lizard-derived JsPV clustered closely with gecko-associated strains (HfrePV1 and HfrePV2), forming a distinct clade separate from those associated with snakes, birds, turtles, and mammals (Figure 2). This finding suggests that species from geographically distinct regions may have originated from ancestral lizards and their associated PVs. Additionally, turtle-associated PVs formed a unique cluster within the sauropsid clade, supporting the hypothesis that PVs in reptiles have evolved independently with limited interspecific host transmission.
Overall, this study provides evidence of PV infection in lizards without obvious clinical symptom, the same as avian papillomaviruses. Whether the JsPV could develop a persist infection or a simply present in the oral cavity was unknown in lizard. Future research should focus on elucidating the evolutionary origins of reptilian PVs and expanding our current understanding of sauropsid viral genomes.
Data availability statement
The data presented in the study are deposited in the online repository. The names of the repository/repositories and accession number(s) can be found below: NCBI–PRJNA1233463, SRR32623559, PV259886.
Ethics statement
The manuscript presents research on animals that do not require ethical approval for their study.
Author contributions
ZT: Data curation, Writing – original draft, Writing – review & editing. TL: Data curation, Writing – original draft. YF: Data curation, Writing – original draft. JL: Methodology, Writing – original draft. SL: Methodology, Writing – original draft. WC: Data curation, Writing – original draft. QP: Data curation, Methodology, Supervision, Writing – original draft, Writing – review & editing. XH: Formal analysis, Resources, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the Doctor Launch Project of Yibin University (2019QD09 and 2019QD10) and Foundation of Key Laboratory of Southwest China Wildlife Resources Conservation, China West Normal University, Ministry of Education (XNYB24-04).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1590538/full#supplementary-material
SUPPLEMENTARY FIGURE S1 | Structural models of putative E7 ORFs of HPV-45 (A), CcPV1 (B), HfrePV1 (C), and JsPV (D). Homology models for JsPV based on the experimentally derived sequence for HPV45 E7.
Footnotes
References
Agius, J. E., Phalen, D. N., Rose, K., and Eden, J. S. (2019). New insights into Sauropsid Papillomaviridae evolution and epizootiology: discovery of two novel papillomaviruses in native and invasive Island geckos. Virus Evol. 5:vez051. doi: 10.1093/ve/vez051
Bergvall, M., Melendy, T., and Archambault, J. (2013). The E1 proteins. Virology 445, 35–56. doi: 10.1016/j.virol.2013.07.020
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Boon, S. S., Tomaić, V., Thomas, M., Roberts, S., and Banks, L. (2015). Cancer-causing human papillomavirus E6 proteins display major differences in the phospho-regulation of their PDZ interactions. J. Virol. 89, 1579–1586. doi: 10.1128/JVI.01961-14
Boulet, G., Horvath, C., Vanden Broeck, D., Sahebali, S., and Bogers, J. (2007). Human papillomavirus: E6 and E7 oncogenes. Int. J. Biochem. Cell Biol. 39, 2006–2011. doi: 10.1016/j.biocel.2007.07.004
Bravo, I. G., de Sanjosé, S., and Gottschling, M. (2010). The clinical importance of understanding the evolution of papillomaviruses. Trends Microbiol. 18, 432–438. doi: 10.1016/j.tim.2010.07.008
Bravo, I. G., and Félez-Sánchez, M. (2015). Papillomaviruses: viral evolution, cancer and evolutionary medicine. Evol. Med. Public Health 2015, 32–51. doi: 10.1093/emph/eov003
Brücher, B. L., and Jamall, I. S. (2014). Epistemology of the origin of cancer: a new paradigm. BMC Cancer 14:331. doi: 10.1186/1471-2407-14-331
Gaynor, A. M., Fish, S., Duerr, R. S., Cruz, F. N. Jr., and Pesavento, P. A. (2015). Identification of a novel papillomavirus in a Northern Fulmar (Fulmarus glacialis) with viral production in cartilage. Vet. Pathol. 52, 553–561. doi: 10.1177/0300985814542812
Gil da Costa, R. M., and Medeiros, R. (2014). Bovine papillomavirus: opening new trends for comparative pathology. Arch. Virol. 159, 191–198. doi: 10.1007/s00705-013-1801-9
Herbst, L. H., Lenz, J., Van Doorslaer, K., Chen, Z., Stacy, B. A., Wellehan, J. F. Jr., et al. (2009). Genomic characterization of two novel reptilian papillomaviruses, Chelonia mydas papillomavirus 1 and Caretta caretta papillomavirus 1. Virology 383, 131–135. doi: 10.1016/j.virol.2008.09.022
Herbster, S., Ferraro, C. T., Koff, N. K., Rossini, A., Kruel, C. D., Andreollo, N. A., et al. (2012). HPV infection in Brazilian patients with esophageal squamous cell carcinoma: interpopulational differences, lack of correlation with surrogate markers and clinicopathological parameters. Cancer Lett. 326, 52–58. doi: 10.1016/j.canlet.2012.07.018
Hu, X., Li, N., Tian, Z., Yin, X., Qu, L., and Qu, J. (2015). Molecular characterization and phylogenetic analysis of transmissible gastroenteritis virus HX strain isolated from China. BMC Vet. Res. 11:72. doi: 10.1186/s12917-015-0387-8
Huang, W., Luo, H., Luo, S., Huang, A., Ni, Q., Yao, Y., et al. (2019). The complete mitogenome of the splendid japalure Japalura splendida (Squamata, Agamidae). Mitochondrial DNA B 4, 2641–2642. doi: 10.1080/23802359.2019.1643797
Lange, C. E., Favrot, C., Ackermann, M., Gull, J., Vetsch, E., and Tobler, K. (2011). Novel snake papillomavirus does not cluster with other non-mammalian papillomaviruses. Virol. J. 8:436. doi: 10.1186/1743-422X-8-436
Li, D., Luo, R., Liu, C. M., Leung, C. M., Ting, H. F., Sadakane, K., et al. (2016). MEGAHIT v1.0: a fast and scalable metagenome assembler driven by advanced methodologies and community practices. Methods 102, 3–11. doi: 10.1016/j.ymeth.2016.02.020
Liu, Z., Liu, Y., Hong, Y., Rapp, L., Androphy, E. J., and Chen, J. J. (2002). Bovine papillomavirus type 1 E6-induced sensitization to apoptosis is distinct from its transforming activity. Virology 295, 230–237. doi: 10.1006/viro.2001.1351
Liu, A., Tian, Z., Yin, C., Zou, J., Wu, S., Luo, Y., et al. (2023). The analysis of oral and fecal virome detects multiple novel emerging viruses in snakes. Transbound. Emerg. Dis. 2023:4214812. doi: 10.1155/2023/4214812
López-Bueno, A., Mavian, C., Labella, A. M., Castro, D., Borrego, J. J., Alcami, A., et al. (2016). Concurrence of iridovirus, polyomavirus, and a unique member of a new group of fish papillomaviruses in lymphocystis disease-affected gilthead sea bream. J. Virol. 90, 8768–8779. doi: 10.1128/JVI.01369-16
Marchler-Bauer, A., and Bryant, S. H. (2004). CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 32, W327–W331. doi: 10.1093/nar/gkh454
Medeiros-Fonseca, B., Faustino-Rocha, A. I., Medeiros, R., Oliveira, P. A., and Gil da Costa, R. M. (2023). Canine and feline papillomaviruses: an update. Front. Vet. Sci. 10:1174673. doi: 10.3389/fvets.2023.1174673
Moreno-Gallego, J. L., and Reyes, A. (2021). Informative regions in viral genomes. Viruses 13:1164. doi: 10.3390/v13061164
Rangel-Pineros, G., Almeida, A., and Beracochea, M. (2023). VIRify: an integrated detection, annotation and taxonomic classification pipeline using virus-specific protein profile hidden Markov models. PLoS Comput. Biol. 19:e1011422. doi: 10.1371/journal.pcbi.1011422
Tachezy, R., Rector, A., Havelkova, M., Wollants, E., Fiten, P., Opdenakker, G., et al. (2002). Avian papillomaviruses: the parrot Psittacus erithacus papillomavirus (PePV) genome has a unique organization of the early protein region and is phylogenetically related to the chaffinch papillomavirus. BMC Microbiol. 2:19. doi: 10.1186/1471-2180-2-19
Tan, M. J., White, E. A., Sowa, M. E., Harper, J. W., Aster, J. C., and Howley, P. M. (2012). Cutaneous β-human papillomavirus E6 proteins bind mastermind-like coactivators and repress notch signaling. Proc. Natl. Acad. Sci. U.S.A. 109, E1473–E1480. doi: 10.1073/pnas.1205991109
Terai, M., DeSalle, R., and Burk, R. D. (2002). Lack of canonical E6 and E7 open reading frames in bird papillomaviruses: Fringilla coelebs papillomavirus and Psittacus erithacus timneh papillomavirus. J. Virol. 76, 10020–10023. doi: 10.1128/JVI.76.19.10020-10023.2002
Tian, Z., Pu, H., Cai, D., Luo, G., Zhao, L., Li, K., et al. (2022). Characterization of the bacterial microbiota in different gut and oral compartments of splendid japalure (Japalura sensu lato). BMC Vet. Res. 18:205. doi: 10.1186/s12917-022-03300-w
Uberoi, A., and Lambert, P. F. (2017). Rodent papillomaviruses. Viruses 9:362. doi: 10.3390/v9120362
Van Doorslaer, K., Li, Z., Xirasagar, S., Maes, P., Kaminsky, D., Liou, D., et al. (2017). The Papillomavirus Episteme: a major update to the papillomavirus sequence database. Nucleic Acids Res. 45, D499–d506. doi: 10.1093/nar/gkw879
Varga, J., De Oliveira, T., and Greten, F. R. (2014). The architect who never sleeps: tumor-induced plasticity. FEBS Lett. 588, 2422–2427. doi: 10.1016/j.febslet.2014.06.019
Varsani, A., Kraberger, S., Jennings, S., Porzig, E. L., Julian, L., Massaro, M., et al. (2014). A novel papillomavirus in Adélie penguin (Pygoscelis adeliae) faeces sampled at the Cape Crozier colony, Antarctica. J. Gen. Virol. 95, 1352–1365. doi: 10.1099/vir.0.064436-0
Varsani, A., Porzig, E. L., Jennings, S., Kraberger, S., Farkas, K., Julian, L., et al. (2015). Identification of an avian polyomavirus associated with Adélie penguins (Pygoscelis adeliae). J. Gen. Virol. 96, 851–857. doi: 10.1099/vir.0.000038
Wang, N., Yu, H. J., Han, X. Y., Li, C., Ye, R. Z., Du, L. F., et al. (2024). Genomic characterization of an emerging Rickettsia barbariae isolated from tick eggs in northwestern China. Emerg. Microbes Infect. 13:2396870. doi: 10.1080/22221751.2024.2396870
Keywords: evolution, Japalura splendida , lizard, papillomavirus, reptile
Citation: Tian Z, Li T, Fan Y, Li J, Luo S, Cao W, Pan Q and Hu X (2025) First detection of a lizard-associated papillomavirus in the splendid japalure (Japalura splendida) from southwestern China. Front. Microbiol. 16:1590538. doi: 10.3389/fmicb.2025.1590538
Edited by:
Marta Canuti, University of Copenhagen, DenmarkReviewed by:
Jinxin Xie, Xinjiang Agricultural University, ChinaAndré Santos, Federal University of Rio de Janeiro, Brazil
Copyright © 2025 Tian, Li, Fan, Li, Luo, Cao, Pan and Hu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qing Pan, cGFucWluZzIwMDUwMTAxQDEyNi5jb20=; Xiaoliang Hu, bGlhbmc2NzlAMTYzLmNvbQ==
 Zhige Tian
Zhige Tian Tingjie Li1
Tingjie Li1 Jiayi Li
Jiayi Li Qing Pan
Qing Pan Xiaoliang Hu
Xiaoliang Hu