- 1School of Pharmacy and Life Sciences, Jiujiang University, Jiujiang, China
- 2Key Laboratory of Molecular Biophysics, Ministry of Education, College of Life Science and Technology, Huazhong University of Science and Technology, Wuhan, China
Microbial lipases constitute the primary source of commercialized and industrial lipases, and they are extensively utilized across numerous industrial sectors. Compared to fungal lipases, bacterial lipases catalyze a broader spectrum of reactions with higher activity, enhanced stability, and improved stress resistance. Among them, lipases from Pseudomonas and Burkholderia cepacia are among the most widely employed microbial lipases. Furthermore, bacterial extracellular lipases act as crucial virulence factors, playing a significant role in the pathogenesis of bacteria. However, the production of bacterial lipases is typically low, rendering them expensive in the market and insufficient to meet the substantial demand for industrial production. To achieve large-scale production of bacterial lipases, stable and efficient homologous expression has proven to be an effective strategy. However, elucidating how bacterial lipase genes are regulated is the initial step for developing stable and efficient homologous expression, and a pressing scientific challenge. To date, the regulatory mechanisms governing the expression of bacterial lipase genes remain unclear, significantly impeding the construction of robust and high-yield homologous expression systems. Concurrently, understanding these regulatory mechanisms can facilitate early diagnosis of lipase-related pathogenic bacterial infections, and aid in the development of novel antibacterial drugs. In this review, we summarized the advancements in understanding the expression regulation of bacterial lipase genes, including direct regulators, the quorum sensing (QS) system, the Gac/Rsm system and its related regulators, as well as other regulators. Additionally, based on our ongoing research, we also discussed potential research directions in this field, aiming to provide valuable insights for the construction of homologous expression systems with high-yield lipases.
1 Introduction
Lipases (triacylglycerol acylhydrolases, EC 3.1.1.3), belonging to the α/β hydrolase superfamily, are ubiquitous in various animals, plants and microorganisms. They can catalyze the hydrolysis of long-chain acylglycerols at the oil–water interface, and also various synthetic reactions in micro-aqueous or non-aqueous phases, such as transesterification, esterification, aminolysis and alcoholysis (Jaeger et al., 1999; Ali et al., 2023; Kim et al., 2023; Zhao et al., 2024). They are the third most important industrial enzymes after proteases and amylases (Filho et al., 2019). Microbial lipases are the primary source of commercialized and industrial lipases, widely used in numerous fields such as food, beverages, oils and fats, detergents, feed, textiles, leather, novel materials, fine chemicals, pharmaceuticals, cosmetics, papermaking, environmental treatment, bioenergy, etc. (Jaeger et al., 1999; Enespa and Singh, 2023; Ali et al., 2023; Kim et al., 2023). Compared to fungal lipases, bacterial lipases catalyze a wider range of reactions with higher activity, better stability and stress resistance. Among them, lipases form Pseudomonas and Burkholderia cepacia are one of the most widely used microbial lipases (Arpigny and Jaeger, 1999; Jaeger et al., 1999; Gupta et al., 2004; Sánchez et al., 2018). In addition, bacterial extracellular lipases are also important virulence factors that play a significant role in the pathogenesis of bacteria (Stehr et al., 2003).
However, the production of bacterial lipases is generally low, and various strategies such as conventional breeding, fermentation condition optimization and heterologous expression have failed to effectively address this issue. As a result, bacterial lipases are costly and hard to meet the large industrial demand. Rosenau and Jaeger proposed that the large-scale production of bacterial lipases could be achieved through the following three steps: elucidating the molecular mechanisms on the expression regulation of lipase genes, constructing genetically engineered strains for stable and efficient homologous expression of lipases, and optimizing the fermentation conditions for these genetically engineered strains (Rosenau and Jaeger, 2000). However, to date, the above regulatory mechanisms remain unclear, which significantly hinders the construction of these genetically engineered strains. Additionally, elucidating these mechanisms can also facilitate early diagnosis of lipase-related pathogenic bacterial infections and the development of new antibacterial drugs. Therefore, studying the above mechanisms holds significant theoretical and practical importance.
Currently, research hotspots in the field of lipases mainly include gene resource mining, enzymatic property characterization, immobilization technology and industrial applications. However, research progress in gene expression regulation has been slow, which is unfavorable for elucidating the regulatory mechanisms and severely obstructs the construction of genetically engineered strains. Up to now, the expression of bacterial lipase genes is primarily regulated by direct regulators, the QS system, the Gac/Rsm system and its related regulators, as well as other regulators (Figure 1 and Table 1). The regulatory mechanisms of these regulators are detailed below.
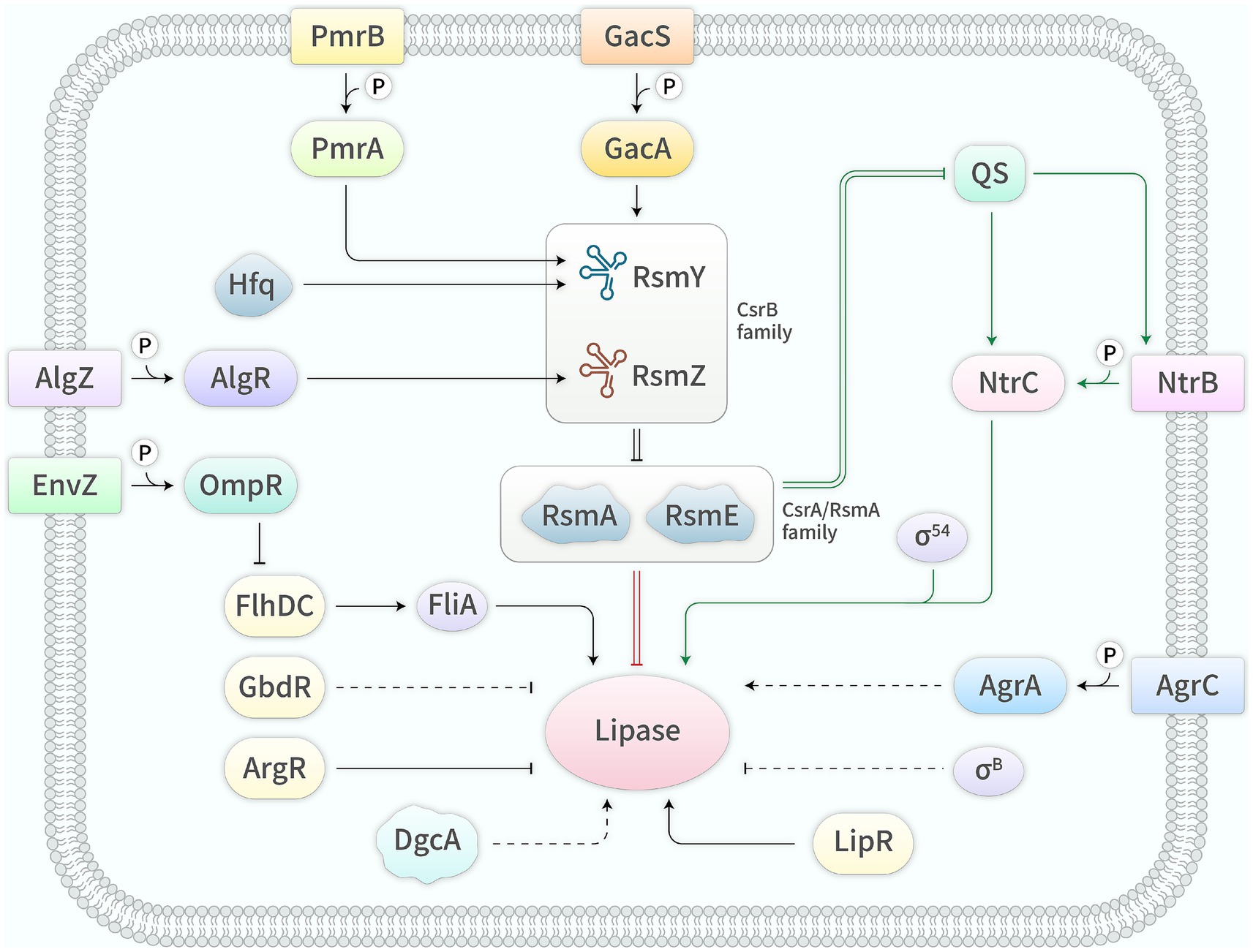
Figure 1. Regulators and their mechanisms in the expression of bacterial lipase genes. →, activation; —|, inhibition; =, interaction; —, known mechanism; ┄, unknown mechanism.
2 Direct regulators
Several studies have shown that the transcription of bacterial lipase genes is directly regulated by the two-component system NtrB/C superfamily (Jaeger et al., 1999; Rosenau and Jaeger, 2000; Krzeslak et al., 2008; Krzeslak et al., 2012). The NtrB superfamily is a class of sensor kinases that can sense extracellular environmental signals, although the nature of the signaling molecules remains unclear. In contrast, the NtrC superfamily, being a class of response regulators, can bind to the upstream activating sequence (UAS) of the promoter, thereby activating the transcription of lipase genes. Upon sensing environmental signaling molecules, the NtrB superfamily undergoes autophosphorylation and transfers the phosphate group to the NtrC superfamily, then the phosphorylated NtrC superfamily acts as a class of transcriptional activators (Monteagudo-Cascales et al., 2022). To date, two members of this protein superfamily, i.e., CbrA/B in P. aeruginosa and LipQ/R in P. alcaligenes, have been reported to directly regulate the transcription of lipase genes (Jaeger et al., 1999; Rosenau and Jaeger, 2000; Krzeslak et al., 2008; Abdou et al., 2011; Krzeslak et al., 2012). It is worth noting that the NtrC superfamily requires the assistance of the alternative sigma factor σ54 to activate gene transcription, and σ54-defective strains produce little or no lipase (Jaeger et al., 1999; Rosenau and Jaeger, 2000; Cox et al., 2001; Krzeslak et al., 2012). Moreover, the lipase operon lip/lif from P. aeruginosa contains two promoters, P1 and P2, and its transcription initiated by P1 requires σ54, while the physiological function of P2 remains unclear and requires further investigation (Jaeger et al., 1999; Rosenau and Jaeger, 2000). Similarly, in B. glumae, two putative σ54-dependent promoters were identified ahead of the lipase operon lipAB, which suggests that the transcription initiation of this operon may involve the NtrC superfamily and σ54. Also in this bacterium, a putative cAMP response protein (CRP) binding site was also found upstream of the lipAB-operon, implying that lipAB transcription is subjected to catabolite repression and induced by the CRP-cAMP complex in the absence of glucose (Beselin, 2005).
Furthermore, in Streptomyces the lipase gene lipA and its contiguous and downstream gene lipR, encoding a transcriptional activator that belongs to the LuxR family, form an operon and their transcription is directly activated by LipR binding to conserved sequences upstream of the −35 region (Servin-Gonzalez et al., 1997; Valdez et al., 1999). In Xenorhabdus nematophila, the flagella sigma factor, FliA (σ28), directly enhances the transcription of the lipase gene xlpA by binding to the consensus binding sequence contained in the xlpA promoter (Park and Forst, 2006). In P. protegens, RsmE, a member of the CsrA/RsmA family, was reported to directly inhibit lipA translation through binding to the ACAAGGAUGU sequence overlapping the Shine-Dalgarno (SD) sequence of lipA mRNA, thereby hindering the access of the 30S ribosomal subunit to the SD sequence (Zha et al., 2014). ArgR, a transcriptional regulator belonging to the AraC/XylS family, plays a key role in arginine metabolism regulation, and in P. protegens it has also been confirmed to directly repress lipA transcription by binding to the ArgR binding site (TGTCGCCAAAGCGTCATGGGG) located in the lipA promoter to produce steric hindrance. In addition, lipA expression in both wild-type and argR mutant is inhibited by arginine, and arginine exhibits a synergistic inhibitory effect with ArgR (Ying et al., 2019). Similarly, ArgR in P. aeruginosa was reported to also restrain lipA transcription, and deletion of argR significantly increases lipA transcript by up to 2.3 times, meanwhile the addition of arginine further enhances this effect, increasing lipA transcript by 9 times. The specific mechanism underlying this regulation requires further investigation (Yang and Lu, 2007). The differential regulatory mechanism of ArgR between P. protegens and P. aeruginosa may result from variations in the promoter sequences of their respective target genes.
3 QS system
Numerous studies have demonstrated that the expression of bacterial lipase genes exhibits cell density-dependence, known as QS (Jaeger et al., 1999; Lewenza et al., 1999; Rosenau and Jaeger, 2000; Christensen et al., 2003; Devescovi et al., 2007; Su et al., 2019). QS is a process by which bacteria communicate using self-produced autoinducers as signaling molecules. The QS system is involved in regulating numerous biological functions, including the synthesis of various enzymes and antibiotics, the production of virulence factors, the synthesis of extracellular polysaccharides, and the formation of biofilms and spores (Miller and Bassler, 2001; Boyer and Wisniewski-Dye, 2009; Juszczuk-Kubiak, 2024). To date, the molecular mechanism by which the QS system activates the expression of lipase genes has only been elucidated in P. aeruginosa, where it transcriptionally activates lipA mediated by directly enhancing the transcription of cbrA/B (Jaeger et al., 1999; Rosenau and Jaeger, 2000; Duan and Surette, 2007; Boyer and Wisniewski-Dye, 2009; Williams and Cámara, 2009). In the QS system, bacteria sense population density by producing diffusible small molecule substances known as autoinducers. Most Gram-positive bacteria use small peptides as QS signal molecules, while Gram-negative bacteria utilize various small molecule substances as QS signal molecules, with acyl-homoserine lactones (acyl-HSLs) being the most representative (Lewenza et al., 1999; Juszczuk-Kubiak, 2024). The majority of Gram-negative bacteria employ acyl-HSLs as their QS signal molecules, which are produced by LuxI-type signal synthases and gradually accumulate as the population density increases. When the concentration reaches a certain threshold, they bind to the LuxR-type receptors, enabling the latter to function as transcription activators (Williams and Cámara, 2009; Parsek and Greenberg, 2000; Majdura et al., 2023).
The acyl-HSLs mediated QS system is considered the classical QS system, with the QS system of P. aeruginosa being the most representative. It possesses two acyl-HSL signaling systems, namely the Las and Rhl systems. The Las system includes LasI, the signal synthase that produces N-(3-oxo-dodecanoyl)-homoserine lactone (3-oxo-C12-HSL), and LasR, the signal receptor that, upon binding to 3-oxo-C12-HSL, activates the transcription of certain target genes. This activation requires 3-oxo-C12-HSL dependent LasR polymerization. The Rhl system is the second QS system, comprising RhlI, the signal synthase that synthesizes N-butanoyl-homoserine lactone (C4-HSL), and RhlR, the signal receptor that, upon binding to C4-HSL, induces the transcription of certain genes, such as promoting the transcription of cbrA/B in the regulation network of lipase gene expression. While the transcriptional activity of RhlR requires C4-HSL, its dimerization does not. LasR and RhlR can also induce the transcription of their respective homologous signal synthase genes, forming a positive feedback loop, the process also known as self-induction, which leads to rapid increase and diffusion of signal molecules. These two QS systems form a cascade relationship, where the Las system activates the Rhl system, meaning that LasR-3-oxo-C12-HSL activates the transcription of rhlI/R (Parsek and Greenberg, 2000; Fuqua and Greenberg, 2002; Schuster and Greenberg, 2006; Williams and Cámara, 2009; Majdura et al., 2023).
4 Gac/Rsm system
The Gac/Rsm system is highly conserved in Gram-negative bacteria and consists of the two-component system GacS/A, the CsrB family and the CsrA/RsmA family (Lapouge et al., 2008). The GacS/A system has been proven to play a crucial role in the expression regulation of bacterial lipase genes. Deletion of gacS, gacA, or both gacS/A inhibits the expression of lipase genes (Reimmann et al., 1997; Jaeger et al., 1999; Rosenau and Jaeger, 2000; Williams and Cámara, 2009; Lalaouna et al., 2012; Zha et al., 2014). In P. aeruginosa, deletion of gacA reduces lipase production by 3-fold, whereas overexpression of gacA increases lipase production by 1.4-fold (Reimmann et al., 1997). The GacS/A system is composed of the membrane-bound sensor kinase GacS and the response regulator GacA. GacS contains the HAMP domain (histidine kinase, adenylyl cyclase, methyl-accepting protein, and phosphatase), the HisKA domain (histidine kinase), the HATPase-c domain (ATPase), the REC domain (response regulator), and the Hpt domain (phosphotransfer). The HAMP domain is responsible for GacS-GacS interaction, while the HisKA/HATPase-c/REC domains are responsible for GacS-GacA interaction (Workentine et al., 2009; Song et al., 2023). GacS senses unknown signal molecules and undergoes autophosphorylation. It interacts with GacA to transfer the phosphate group to the latter, and phosphorylated GacA functions as a transcription activator, initiating the transcription of the CsrB family of small regulatory RNAs (sRNAs) (Sonnleitner and Haas, 2011; Gallegos et al., 2024). In P. aeruginosa, GacA directly regulates only the transcription of two sRNA genes rsmY and rsmZ (Brencic et al., 2009), while in P. syringae, it also directly regulates the transcription of other genes (Cha et al., 2012). The CsrB family of sRNAs are rich in GGA motifs and form multiple stem-loop structures, enabling specific binding to CsrA/RsmA family proteins and alleviating the translational inhibition of target genes (Humair et al., 2010; Sonnleitner and Haas, 2011; Marzi and Romby, 2012). The CsrA/RsmA family is a class of RNA-binding proteins that can bind to multiple specific motifs (often GGA or ANGGA) on the 5′ untranslated region (UTR) of target mRNAs. Since one of these specific motifs is close to or overlaps with the SD sequence, which prevents the recruitment of the 30S ribosomal subunit to the ribosome-binding site (RBS), ultimately inhibiting the translation of the target mRNAs (Sonnleitner and Haas, 2011).
In P. aeruginosa, RsmA, a member of the CsrA/RsmA family, which specifically binds to the 5’UTR of lasI and rhlI mRNAs, leading to their translational inhibition and ultimately resulting in the inhibition of lipA expression at the transcriptional level (Rosenau and Jaeger, 2000; Williams and Cámara, 2009). In addition, RsmA also transcriptionally promotes lipA expression through another unknown pathway. Deletion of rsmA reduces lipase production by 1.9-fold, while overexpression of rsmZ lowers lipase production by 5.6-fold but its deletion has almost no effect on lipase production, which may be related to the redundancy of the CsrB family of sRNAs. The stimulative effect of RsmA may be indirect, achieved by repressing inhibitors of lipA expression, with the specific inhibitors yet to be further identified (Heurlier et al., 2004). However, in P. protegens lacking the classical QS system, RsmA and RsmE, the members of the CsrA/RsmA family, have been shown to regulate lipA expression through different pathways. RsmA mainly activates lipA translation by inhibiting rsmE translation and secondarily inhibits lipA transcription through an unknown pathway, while RsmE directly inhibits lipA translation by binding to the SD sequence of lipA mRNA (Zha et al., 2014).
In summary, in P. aeruginosa, the Gac/Rsm system exhibits duality in regulating lipA expression, primarily promoting lipA transcription through the Gac/RsmA/QS/CbrA/B/LipA pathway, and secondarily inhibiting lipA transcription through the Gac/RsmA/unknown regulator(s)/LipA pathway (Figure 2, L). However, in P. protegens, the system regulates lipA expression through other pathways, which primarily and directly activates lipA translation through the Gac/RsmE/LipA pathway, and secondarily and indirectly activates lipA transcription through the Gac/RsmA/unknown regulator(s)/LipA pathway (Figure 2, R). P. protegens chooses different regulatory pathways to activate lipA expression by the Gac/Rsm system, which may be due to the direct binding of RsmE to the SD sequence of lipA mRNA and this bacterium without the classical QS system.
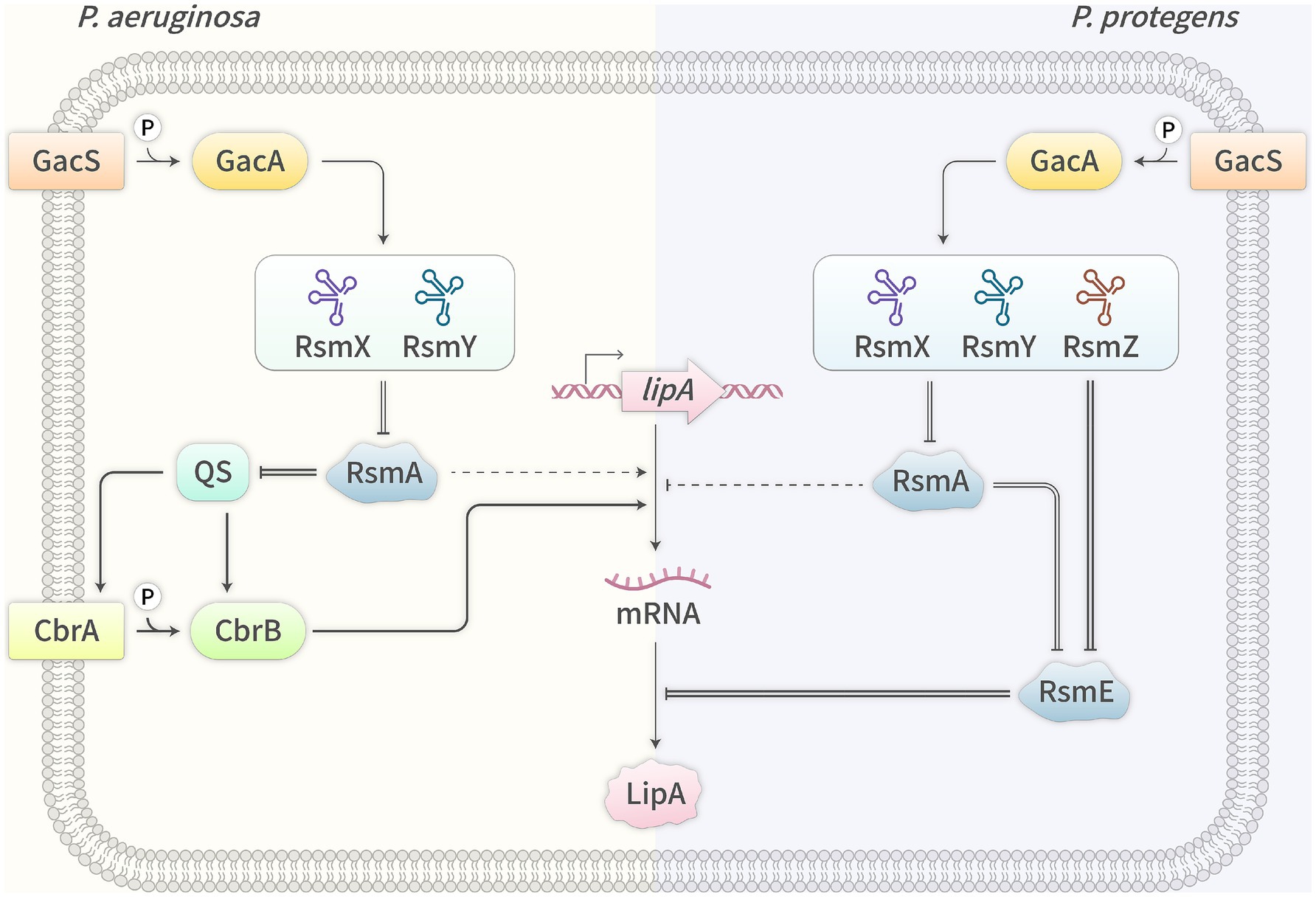
Figure 2. Expression regulation of lipase genes by Gac/Rsm system in P. aeruginosa (L) and P. protegens (R). →, activation; —|, inhibition; =, interaction; —, known mechanism; ┄, unknown mechanism.
5 Regulators mediated by Gac/Rsm system
Each component of the Gac/Rsm system is finely controlled by multiple regulators to adapt to complex and diverse environments (Lapouge et al., 2008). These regulators may modulate the expression of lipase genes mediated by the Gac/Rsm system. In P. protegens, AlgR mainly promotes lipA transcription by directly binding to the promoter sequence of rsmZ, but the mechanism of RsmZ regulating lipA expression is unknown. Besides, AlgR also enhances lipA expression at translational level with an unknown mechanism (Li et al., 2017). Also in this bacterium, Hfq mainly strengthens lipA translation through increasing the transcript of rsmY by an unknown mechanism and enhancing the stability of RmsY by directly binding to the sequence of RsmY, but the mechanism by which RsmY regulates lipA expression remains unclear. Moreover, Hfq also boosts lipA transcription via an unknown mechanism (Liu et al., 2017). In P. aeruginosa, the two-component system PmrB/A mainly activates lipA translation through the PmrB/A/RsmY/RsmA/LipA pathway by adopting the mechanism of PmrA directly binding to the promoter sequence of rsmY. Furthermore, PmrB/A also promotes lipA expression at the transcriptional level through an unknown mechanism (Liu et al., 2018). It is worth noting that the Gac/Rsm system is also controlled by other regulators, such as RetS, LadS, PA1611, SuhB, Lon, HptB, PsrA, IHF, MvaT/U, BfiS/R, CafA, GidA and TrmE (Zha and Yan, 2015), suggesting that they may also regulate the expression of lipase genes through the pathway mediated by the Gac/Rsm system or other pathways.
6 Other regulators
In Staphylococcus aureus, lipase production is repressed by the alternative sigma factor σB (Kullik et al., 1998) and enhanced by the two-component system AgrC/A (McNamara and Iandolo, 1998), but their regulatory mechanisms are still unknown. In P. fluorescens, the two-component system EnvZ/OmpR inhibits lipA expression, and the mutant of envZ::Tn5 increases lipase production by 2-4-fold and the overexpression of envZ depresses lipase production by 10-20-fold. Of note, EnvZ/OmpR synergistically inhibits lipase production with NaCl, and NaCl acts through a mechanism independent of EnvZ/OmpR, but their regulatory mechanisms are also unclear (McCarthy et al., 2004). It is encouraging to note that the mechanism by which EnvZ/OmpR regulates lipase gene expression in X. nematophila has been elucidated, it represses xlpA transcription through the EnvZ/OmpR/FlhDC/FliA/XlpA pathway (Park and Forst, 2006). In P. aeruginosa, lipA expression is activated by the dimethylglycine demethylase DgcA and suppressed by the AraC/XylS family transcriptional regulator GbdR being necessary for choline catabolism, but their regulatory mechanisms remain unclear (Hampel et al., 2014). It is worth mentioning that the QS system is also controlled by other regulators in addition to the Gac/Rsm system, these regulators control the QS system either by affecting the activity or expression of signal receptors or by influencing the production of signal molecules (Juhas et al., 2005; Schuster and Greenberg, 2006; Boyer and Wisniewski-Dye, 2009; Williams and Cámara, 2009; Zha and Yan, 2015), ultimately, may regulate the expression of lipase genes.
Moreover, the expression of bacterial lipase genes is also regulated by various physiological factors, such as carbon sources, nitrogen sources, fatty acyl esters, iron, temperature, etc. Among carbon sources, organic acids such as pyruvate and succinate enhance lipase production, whereas most sugars, especially glucose, inhibit lipase production in P. fluorescens (Makhzoum et al., 1995). The inhibition of lipase production by most sugars may occur because lipase gene expression is subject to catabolite repression mediated by the CRP-cAMP complex. In the case of nitrogen sources, amino acids (e.g., arginine, threonine and lysine) and inorganic nitrogen sources (e.g., ammonium salts of mineral acids) support good lipase production in P. fluorescens (Makhzoum et al., 1995). In Acinetobacter calcoaceticus, casamino-acids and tryptone improve lipase yield, and the addition of ammonium further increases it. The increase in yield caused by nitrogen sources is unrelated to lipA transcription, suggesting that post-transcriptional processes, including enzyme protection, inactivation and secretion, must be considered important factors affecting lipase production (Cordenons et al., 1996). However, the transcription of lipA is repressed by high amino acid concentrations in Bacillus subtilis (Eggert et al., 2003). Among fatty acyl esters, many triglycerides and Spans and Tweens are strong inducers for lipase production in P. aeruginosa, P. fluorescens and Thermus thermophilus (Gilbert et al., 1991; Makhzoum et al., 1995; Deive et al., 2009), but triolein is a strong inhibitor in P. fluorescens (Makhzoum et al., 1995). Interestingly, the hydrolysis products of lipase, long-chain fatty acids like oleic acid, significantly repress lipase production in P. aeruginosa, P. fluorescens and A. calcoaceticus (Gilbert et al., 1991; Makhzoum et al., 1995; Kok et al., 1996), but a short-chain fatty acid, caproic acid, enhances considerably lipase production in P. fluorescens (Makhzoum et al., 1995). The feedback inhibition of long-chain fatty acids implies the involvement of a fatty acyl-responsive DNA-binding protein that, upon fatty acid binding, acts as a transcriptional repressor to downregulate the expression of lipase genes (Kok et al., 1996). In P. fluorescens, iron is found to strongly repress lipA transcription, this repression may be mediated by the iron-sensing Fur repressor (Woods et al., 2001). In some psychrotrophic P. fluorescens stains, temperature has also been reported to regulate the expression of lipase genes, with the maximum lipase production occurring below the optimal growth temperature, suggesting a cold-adaptation strategy (Merieau et al., 1993; Guillou et al., 1995; Makhzoum et al., 1995; Woods et al., 2001). The low-temperature regulation may be achieved at the post-transcriptional or post-translational level (Woods et al., 2001). All in all, the mechanisms by which these physiological factors regulate the expression of lipase genes remain unclear, but these results indicate that there are still many unidentified regulators in the expression regulatory network of lipase genes.
7 Conclusion and perspectives
Although some progress has been made in the expression regulation of bacterial lipase genes, there are still some challenges to be faced. For instance, there are difficulties in efficiently screening and identifying key regulators, constructing genetically engineered strains for stable and efficient homologous expression of lipases, and optimizing fermentation processes to enhance lipases production. Fortunately, the continuous development of biotechnology and the application of novel technologies have presented new opportunities for the research on the expression regulation of bacterial lipase genes. For example, synthetic biology can be leveraged to construct bacterial lipases with entirely new functions, new gene editing technologies like CRISPR allow for precise modification and optimization of bacterial lipase genes, and artificial intelligence and big data analytics based on multi-omics can be utilized to predict and dissect the complex networks involved in the expression regulation of bacterial lipase genes. In summary, researches on the expression regulation of bacterial lipase genes hold vast prospects and significant applied value. Future researches will continue to propel advancements in this field, offering more possibilities for the production and application of industrial lipases.
Author contributions
D-mZ: Conceptualization, Funding acquisition, Supervision, Writing – original draft, Writing – review & editing. Y-jY: Conceptualization, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was supported by the National Natural Science Foundation of P. R. China (32360146), the Natural Science Foundation of Jiangxi Province of P. R. China (20202BAB213022) and the Open Project of Key Laboratory of Molecular Biophysics, Ministry of Education of P. R. China (20230110) to D-mZ.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abdou, L., Chou, H. T., Haas, D., and Lu, C. D. (2011). Promoter recognition and activation by the global response regulator CbrB in Pseudomonas aeruginosa. J. Bacteriol. 193, 2784–2792. doi: 10.1128/JB.00164-11
Ali, S., Khan, S. A., Hamayun, M., and Lee, I. J. (2023). The recent advances in the utility of microbial lipases: a review. Microorganisms 11:510. doi: 10.3390/microorganisms11020510
Arpigny, J. L., and Jaeger, K. E. (1999). Bacterial lipolytic enzymes: classification and properties. Biochem. J. 343, 177–183. doi: 10.1042/bj3430177
Beselin, A. Optimization of lipase production in Burkholderia glumae. PhD thesis, Ruhr-Universität Bochum (2005)
Boyer, M., and Wisniewski-Dye, F. (2009). Cell-cell signalling in bacteria: not simply a matter of quorum. FEMS Microbiol. Ecol. 70, 1–19. doi: 10.1111/j.1574-6941.2009.00745.x
Brencic, A., McFarland, K. A., McManus, H. R., Castang, S., Mogno, I., Dove, S. L., et al. (2009). The GacS/GacA signal transduction system of Pseudomonas aeruginosa acts exclusively through its control over the transcription of the RsmY and RsmZ regulatory small RNAs. Mol. Microbiol. 73, 434–445. doi: 10.1111/j.1365-2958.2009.06782.x
Cha, J. Y., Lee, D. G., Lee, J. S., Oh, J. I., and Baik, H. S. (2012). GacA directly regulates expression of several virulence genes in Pseudomonas syringae pv. Tabaci 11528. Biochem. Biophys. Res. Commun. 417, 665–672. doi: 10.1016/j.bbrc.2011.11.124
Christensen, A. B., Riedel, K., Eberl, L., Flodgaard, L. R., Molin, S., Gram, L., et al. (2003). Quorum-sensing-directed protein expression in Serratia proteamaculans B5a. Microbiology 149, 471–483. doi: 10.1099/mic.0.25575-0
Cordenons, A., Gonzalez, R., Kok, R., Hellingwerf, K. J., and Nudel, C. (1996). Effect of nitrogen sources on the regulation of extracellular lipase production in Acinetobacter calcoaceticus strains. Biotechnol. Lett. 18, 633–638. doi: 10.1007/BF00130756
Cox, M., Gerritse, G., Dankmeyer, L., and Quax, W. J. (2001). Characterization of the promoter and upstream activating sequence from the Pseudomonas alcaligenes lipase gene. J. Biotechnol. 86, 9–17. doi: 10.1016/S0168-1656(00)00397-7
Deive, F. J., Carvalho, E., Pastrana, L., Rúa, M. L., Longo, M. A., and Sanroman, M. A. (2009). Strategies for improving extracellular lipolytic enzyme production by Thermus thermophilus HB27. Bioresour. Technol. 100, 3630–3637. doi: 10.1016/j.biortech.2009.02.053
Devescovi, G., Bigirimana, J., Degrassi, G., Cabrio, L., LiPuma, J. J., Kim, J., et al. (2007). Involvement of a quorum-sensing-regulated lipase secreted by a clinical isolate of Burkholderia glumae in severe disease symptoms in rice. Appl. Environ. Microbiol. 73, 4950–4958. doi: 10.1128/AEM.00105-07
Duan, K., and Surette, M. G. (2007). Environmental regulation of Pseudomonas aeruginosa PAO1 las and Rhl quorum-sensing systems. J. Bacteriol. 189, 4827–4836. doi: 10.1128/JB.00043-07
Eggert, T., Brockmeier, U., Dröge, M. J., Quax, W. J., and Jaeger, K. E. (2003). Extracellular lipases from Bacillus subtilis: regulation of gene expression and enzyme activity by amino acid supply and external pH. FEMS Microbiol. Lett. 225, 319–324. doi: 10.1016/S0378-1097(03)00536-6
Enespa, C. P., and Singh, D. P. (2023). Sources, purification, immobilization and industrial applications of microbial lipases: an overview. Crit. Rev. Food Sci. Nutr. 63, 6653–6686. doi: 10.1080/10408398.2022.2038076
Filho, D. G., Silva, A. G., and Guidini, C. Z. (2019). Lipases: sources, immobilization methods, and industrial applications. Appl. Microbiol. Biotechnol. 103, 7399–7423. doi: 10.1007/s00253-019-10027-6
Fuqua, C., and Greenberg, E. P. (2002). Listening in on bacteria: acyl-homoserine lactone signalling. Nat. Rev. Mol. Cell Biol. 3, 685–695. doi: 10.1038/nrm907
Gallegos, M. T., Garavaglia, M., and Valverde, C. (2024). Small regulatory RNAs of the Rsm clan in Pseudomonas. Mol. Microbiol. 122, 563–582. doi: 10.1111/mmi.15313
Gilbert, E. J., Drozd, J. W., and Jones, C. W. (1991). Physiological regulation and optimization of lipase activity in Pseudomonas aeruginosa EF2. J. Gen. Microbiol. 137, 2215–2221. doi: 10.1099/00221287-137-9-2215
Guillou, C., Merieau, A., Trebert, B., and Guespin, M. J. (1995). Growth temperature is involved in the regulation of extracellular lipase at two different levels in Pseudomonas fluorescens strain MF0. Biotechnol. Lett. 17, 377–382. doi: 10.1007/BF00130793
Gupta, R., Gupta, N., and Rathi, P. (2004). Bacterial lipases: an overview of production, purification and biochemical properties. Appl. Microbiol. Biotechnol. 64, 763–781. doi: 10.1007/s00253-004-1568-8
Hampel, K. J., LaBauve, A. E., Meadows, J. A., Fitzsimmons, L. F., Nock, A. M., and Wargo, M. J. (2014). Characterization of the GbdR regulon in Pseudomonas aeruginosa. J. Bacteriol. 196, 7–15. doi: 10.1128/JB.01055-13
Heurlier, K., Williams, F., Heeb, S., Dormond, C., Pessi, G., Singer, D., et al. (2004). Positive control of swarming, rhamnolipid synthesis, and lipase production by the posttranscriptional RsmA/RsmZ system in Pseudomonas aeruginosa PAO1. J. Bacteriol. 186, 2936–2945. doi: 10.1128/JB.186.10.2936-2945.2004
Humair, B., Wackwitz, B., and Haas, D. (2010). GacA-controlled activation of promoters for small RNA genes in Pseudomonas fluorescens. Appl. Environ. Microbiol. 76, 1497–1506. doi: 10.1128/AEM.02014-09
Jaeger, K. E., Dijkstra, B. W., and Reetz, M. T. (1999). Bacterial biocatalysts: molecular biology, three-dimensional structures, and biotechnological applications of lipases. Ann. Rev. Microbiol. 53, 315–351. doi: 10.1146/annurev.micro.53.1.315
Juhas, M., Eberl, L., and Tümmler, B. (2005). Quorum sensing: the power of cooperation in the world of Pseudomonas. Environ. Microbiol. 7, 459–471. doi: 10.1111/j.1462-2920.2005.00769.x
Juszczuk-Kubiak, E. (2024). Molecular aspects of the functioning of pathogenic bacteria biofilm based on quorum sensing (QS) signal-response system and innovative non-antibiotic strategies for their elimination. Int. J. Mol. Sci. 25:2655. doi: 10.3390/ijms25052655
Kim, B. H., Hwang, J., and Akoh, C. C. (2023). Liquid microbial lipase—recent applications and expanded use through immobilization. Curr. Opin. Food Sci. 50:100987. doi: 10.1016/j.cofs.2023.100987
Kok, R. G., Nudel, C. B., Gonzalez, R. H., Nugteren-Roodzant, I. M., and Hellingwerf, K. J. (1996). Physiological factors affecting production of extracellular lipase (LipA) in Acinetobacter calcoaceticus BD413: fatty acid repression of lipA expression and degradation of LipA. J. Bacteriol. 178, 6025–6035. doi: 10.1128/jb.178.20.6025-6035.1996
Krzeslak, J., Gerritse, G., van Merkerk, R., Cool, R. H., and Quax, W. J. (2008). Lipase expression in Pseudomonas alcaligenes is under the control of a two-component regulatory system. Appl. Environ. Microbiol. 74, 1402–1411. doi: 10.1128/AEM.01632-07
Krzeslak, J., Papaioannou, E., van Merkerk, R., Paal, K. A., Bischoff, R., Cool, R. H., et al. (2012). Lipase a gene transcription in Pseudomonas alcaligenes is under control of RNA polymerase σ54 and response regulator LipR. FEMS Microbiol. Lett. 329, 146–153. doi: 10.1111/j.1574-6968.2012.02516.x
Kullik, I., Giachino, P., and Fuchs, T. (1998). Deletion of the alternative sigma factor σB in Staphylococcus aureus reveals its function as a global regulator of virulence genes. J. Bacteriol. 180, 4814–4820. doi: 10.1128/JB.180.18.4814-4820.1998
Lalaouna, D., Fochesato, S., Sanchez, L., Schmitt-Kopplin, P., Haas, D., Heulin, T., et al. (2012). Phenotypic switching in Pseudomonas brassicacearum involves GacS-and GacA-dependent Rsm small RNAs. Appl. Environ. Microbiol. 78, 1658–1665. doi: 10.1128/AEM.06769-11
Lapouge, K., Schubert, M., Allain, F. H. T., and Haas, D. (2008). Gac/Rsm signal transduction pathway of γ-proteobacteria: from RNA recognition to regulation of social behaviour. Mol. Microbiol. 67, 241–253. doi: 10.1111/j.1365-2958.2007.06042.x
Lewenza, S., Conway, B., Greenberg, E. P., and Sokol, P. A. (1999). Quorum sensing in Burkholderia cepacia: identification of the LuxRI homologs CepRI. J. Bacteriol. 181, 748–756. doi: 10.1128/JB.181.3.748-756.1999
Li, M., Yan, J., and Yan, Y. (2017). The Pseudomonas transcriptional regulator AlgR controls LipA expression via the noncoding RNA RsmZ in Pseudomonas protegens Pf-5. Biochem. Biophys. Res. Commun. 487, 173–180. doi: 10.1016/j.bbrc.2017.04.034
Liu, W., Li, M., Jiao, L., Wang, P., and Yan, Y. (2018). PmrA/PmrB two-component system regulation of lipA expression in Pseudomonas aeruginosa PAO1. Front. Microbiol. 8:2690. doi: 10.3389/fmicb.2017.02690
Liu, W., Li, M., Yan, J., and Yan, Y. (2017). The role of Hfq in regulation of lipA expression in Pseudomonas protegens Pf-5. Sci. Rep. 7:10356. doi: 10.1038/s41598-017-10808-x
Majdura, J., Jankiewicz, U., Gałązka, A., and Orzechowski, S. (2023). The role of quorum sensing molecules in bacterial-plant interactions. Meta 13:114. doi: 10.3390/metabo13010114
Makhzoum, A., Knapp, J. S., and Owusu, R. K. (1995). Factors affecting growth and extracelluar lipase production by Pseudomonas fluorescens 2D. Food Microbiol. 12, 277–290. doi: 10.1016/S0740-0020(95)80108-1
Marzi, S., and Romby, P. (2012). RNA mimicry, a decoy for regulatory proteins. Mol. Microbiol. 83, 1–6. doi: 10.1111/j.1365-2958.2011.07911.x
McCarthy, C. N., Woods, R. G., and Beacham, I. R. (2004). Regulation of the aprX-lipA operon of Pseudomonas fluorescens B52: differential regulation of the proximal and distal genes, encoding protease and lipase, by ompR-envZ. FEMS Microbiol. Lett. 241, 243–248. doi: 10.1016/j.femsle.2004.10.027
McNamara, P. J., and Iandolo, J. J. (1998). Genetic instability of the global regulator agr explains the phenotype of the xpr mutation in Staphylococcus aureus KSI9051. J. Bacteriol. 180, 2609–2615. doi: 10.1128/JB.180.10.2609-2615.1998
Merieau, A., Gugi, B., Guespin-Michel, J. F., and Orange, N. (1993). Temperature regulation of lipase secretion by Pseudomonas fluorescens strain MFO. Appl. Microbiol. Biotechnol. 39, 104–109. doi: 10.1007/BF00166857
Miller, M. B., and Bassler, B. L. (2001). Quorum sensing in bacteria. Ann. Rev. Microbiol. 55, 165–199. doi: 10.1146/annurev.micro.55.1.165
Monteagudo-Cascales, E., Santero, E., and Canosa, I. (2022). The regulatory hierarchy following signal integration by the CbrAB two-component system: diversity of responses and functions. Genes 13:375. doi: 10.3390/genes13020375
Park, D., and Forst, S. (2006). Co-regulation of motility, exoenzyme and antibiotic production by the EnvZ-OmpR-FlhDC-FliA pathway in Xenorhabdus nematophila. Mol. Microbiol. 61, 1397–1412. doi: 10.1111/j.1365-2958.2006.05320.x
Parsek, M. R., and Greenberg, E. P. (2000). Acyl-homoserine lactone quorum sensing in gram-negative bacteria: a signaling mechanism involved in associations with higher organisms. PNAS 97, 8789–8793. doi: 10.1073/pnas.97.16.8789
Reimmann, C., Beyeler, M., Latifi, A., Winteler, H., Foglino, M., Lazdunski, A., et al. (1997). The global activator GacA of Pseudomonas aeruginosa PAO positively controls the production of the autoinducer N-butyryl-homoserine lactone and the formation of the virulence factors pyocyanin, cyanide, and lipase. Mol. Microbiol. 24, 309–319. doi: 10.1046/j.1365-2958.1997.3291701.x
Rosenau, F., and Jaeger, K. E. (2000). Bacterial lipases from Pseudomonas: regulation of gene expression and mechanisms of secretion. Biochimie 82, 1023–1032. doi: 10.1016/S0300-9084(00)01182-2
Sánchez, D. A., Tonetto, G. M., and Ferreira, M. L. (2018). Burkholderia cepacia lipase: a versatile catalyst in synthesis reactions. Biotechnol. Bioeng. 115, 6–24. doi: 10.1002/bit.26458
Schuster, M., and Greenberg, E. P. (2006). A network of networks: quorum-sensing gene regulation in Pseudomonas aeruginosa. Int. J. Med. Microbiol. 296, 73–81. doi: 10.1016/j.ijmm.2006.01.036
Servin-Gonzalez, L., Castro, C., Perez, C., Rubio, M., and Valdez, F. (1997). bldA-dependent expression of the Streptomyces exfoliatus M11 lipase gene (lipA) is mediated by the product of a contiguous gene, lipR, encoding a putative transcriptional activator. J. Bacteriol. 179, 7816–7826. doi: 10.1128/jb.179.24.7816-7826.1997
Song, H., Li, Y., and Wang, Y. (2023). Two-component system GacS/GacA, a global response regulator of bacterial physiological behaviors. Eng Microbiol 3:100051. doi: 10.1016/j.engmic.2022.100051
Sonnleitner, E., and Haas, D. (2011). Small RNAs as regulators of primary and secondary metabolism in Pseudomonas species. Appl. Microbiol. Biotechnol. 91, 63–79. doi: 10.1007/s00253-011-3332-1
Stehr, F., Kretschmar, M., Kröger, C., Hube, B., and Schäfer, W. (2003). Microbial lipases as virulence factors. J. Mol. Catal. B Enzym. 22, 347–355. doi: 10.1016/S1381-1177(03)00049-3
Su, Y., Tang, K., Liu, J., Wang, Y., Zheng, Y., and Zhang, X. H. (2019). Quorum sensing system of Ruegeria mobilis Rm01 controls lipase and biofilm formation. Front. Microbiol. 9:3304. doi: 10.3389/fmicb.2018.03304
Valdez, F., González-Cerón, G., Kieser, H. M., and Servı́n-González, L. (1999). The Streptomyces coelicolor A3(2) lipAR operon encodes an extracellular lipase and a new type of transcriptional regulator. Microbiol 145, 2365–2374. doi: 10.1099/00221287-145-9-2365
Williams, P., and Cámara, M. (2009). Quorum sensing and environmental adaptation in Pseudomonas aeruginosa: a tale of regulatory networks and multifunctional signal molecules. Curr. Opin. Microbiol. 12, 182–191. doi: 10.1016/j.mib.2009.01.005
Woods, R. G., Burger, M., Beven, C. A., and Beacham, I. R. (2001). The aprX-lipA operon of Pseudomonas fluorescens B52: a molecular analysis of metalloprotease and lipase production. Microbiology 147, 345–354. doi: 10.1099/00221287-147-2-345
Workentine, M. L., Chang, L., Ceri, H., and Turner, R. J. (2009). The GacS-GacA two-component regulatory system of Pseudomonas fluorescens: a bacterial two-hybrid analysis. FEMS Microbiol. Lett. 292, 50–56. doi: 10.1111/j.1574-6968.2008.01445.x
Yang, Z., and Lu, C. D. (2007). Functional genomics enables identification of genes of the arginine transaminase pathway in Pseudomonas aeruginosa. J. Bacteriol. 189, 3945–3953. doi: 10.1128/JB.00261-07
Ying, W., Wang, X., Shi, H., Yan, L., Zhang, B., Li, H., et al. (2019). ArgR directly inhibits lipA transcription in Pseudomonas protegens Pf-5. Biochimie 167, 34–41. doi: 10.1016/j.biochi.2019.08.018
Zha, D., Xu, L., Zhang, H., and Yan, Y. (2014). The two-component GacS-GacA system activates lipA translation by RsmE but not RsmA in Pseudomonas protegens Pf-5. Appl. Environ. Microbiol. 80, 6627–6637. doi: 10.1128/AEM.02184-14
Zha, D., and Yan, Y. (2015). Progress in expression regulation of bacterial lipase genes-a review. Acta Microbiol. Sin. 55, 1378–1384. doi: 10.13343/j.cnki.wsxb.20150117
Keywords: lipase, expression regulation, Gac/Rsm system, QS system, two-component system
Citation: Zha D-m and Yan Y-j (2025) Expression regulation of bacterial lipase genes: a review. Front. Microbiol. 16:1592059. doi: 10.3389/fmicb.2025.1592059
Edited by:
Rodolfo García-Contreras, National Autonomous University of Mexico, MexicoReviewed by:
Prassan Choudhary, Amity University, Chhattisgarh, IndiaBruno De Oliveira, São Paulo State University, Brazil
Shaymaa Yusuf, Assiut University, Egypt
Copyright © 2025 Zha and Yan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dai-ming Zha, ZG16aGFAamp1LmVkdS5jbg==; ZG16aGEyMDE1QDEyNi5jb20=; Yun-jun Yan, eWFueXVuanVuQGh1c3QuZWR1LmNu
 Dai-ming Zha
Dai-ming Zha Yun-jun Yan
Yun-jun Yan