- 1NASA Marshall Space Flight Center, Materials and Processes Laboratory, Huntsville, AL, United States
- 2Department of Biology, University of Mississippi, Oxford, MS, United States
- 3Genomics and Microbiome Core Facility, Rush University, Chicago, IL, United States
- 4Biotechnology and Planetary Protection Group, Jet Propulsion Laboratory, Pasadena, CA, United States
- 5Amentum Space Exploration Division, Huntsville, AL, United States
- 6GeoControl Systems, Huntsville, AL, United States
Introduction: Microorganisms can have major impacts on the success of NASA’s missions, including the integrity of materials, the protection of extraterrestrial environments, the reliability of scientific results, and maintenance of crew health. Robust cleaning and sterilization protocols for spacecraft and associated environments are currently in place in NASA facilities, but microbial contamination should be further controlled and its impact on NASA’s missions and science must be minimized. To address this, air and surfaces across cleanrooms and uncontrolled spaces at the Marshall Space Flight Center were sampled and microbial burden and diversity were analyzed.
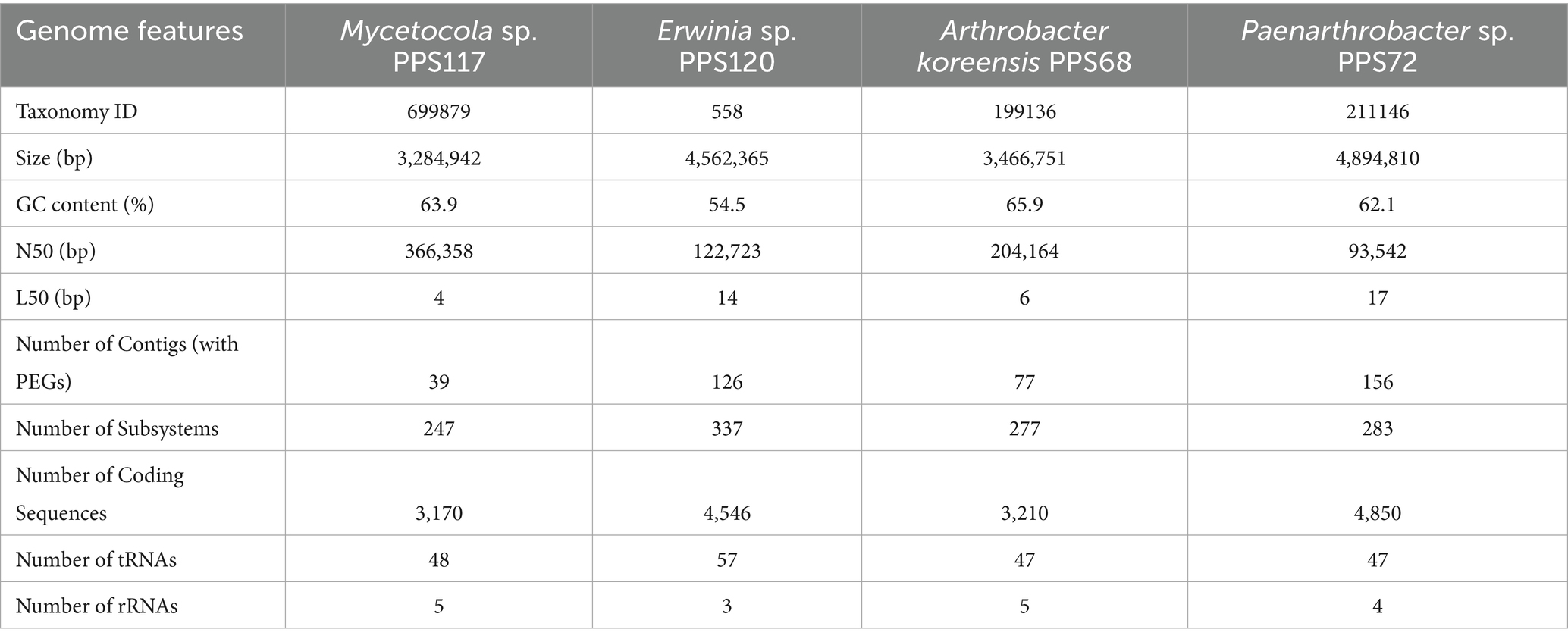
Methods: A library of 82 microbial strains was isolated, curated, characterized, and a subset (n = 24) was subjected to simulated space environmental stressors, including desiccation, vacuum, proton radiation, and ultraviolet radiation. Out of these, four non-spore-former species (Arthrobacter koreensis PPS68, Paenarthrobacter sp. PPS72, Mycetocola sp. PPS117, and Erwinia sp. PPS120) exhibiting the highest resistance to tested stressors were selected for whole genome sequencing and comparative genomic, pan-resistomics and functional analyses.
Results: The analysis revealed genomic features among these four species, encompassing genes critical for amino acid biosynthesis, carbohydrate metabolism, and stress response mechanisms. Erwinia sp. PPS120 had genomic features indicative of metabolic flexibility and stress response capabilities, particularly under oxidative stress conditions. Notably, strain A. koreensis PPS68 had unique genomic features predictive of resilience to desiccation and ionizing radiation, supported by genes for oxidative stress resistance, membrane stability, and nutrient acquisition. A. koreensis contains several genes which are also reported in established radioresistant strains, for predicted functions related to DNA-repair, osmoprotection, and efflux.
Discussion: NASA cleanrooms harbor hardy non-spore-forming bacteria capable of surviving vacuum, ionizing radiation, and UV. Their adaptations to space stressors suggest limitations of today’s spore-centric bioburden assays to explore expanded planetary-protection standards. The modular exposure assay and reference genomes are important resources for microbial risk assessment, decontamination design, and safeguarding both robotic missions and closed human habitats in space and earth where microbial presence and colonization could compromise life-support systems and crew health.
1 Introduction
Microbiology within National Aeronautics and Space Administration (NASA) typically falls into a handful of disciplines that include but are not limited to Space Biology, Environmental Control and Life Support Systems (ECLSS), Astrobiology, and Planetary Protection (PP). As NASA continues the exploration of our solar system, both robotically and in complex crewed missions, understanding the microbes that pose the greatest risk to mission success is of utmost importance. In robotic missions to certain target bodies of interest, like Mars, it is critical to prevent microbial contamination that could adversely impact scientific objectives (Olsson-Francis et al., 2023). Specifically, NASA PP policy requires stringent cleanliness protocols to be in place to remove most microbes from spacecraft surfaces and volumes, with the level of expected cleanliness increasing for missions landing on sensitive solar system bodies. However, the methods used to quantify cleanliness are largely based on the cultivation of bacterial spores [i.e., NASA Standard Assay (NSA, NASA-HDBK-6022, 2010) and provide information neither about the risk of contaminating target bodies (e.g., are putative extremophiles present?) nor appropriate mitigation strategies (e.g., is heat required, or will an ethanol wipe suffice?)].
Each cleanroom and spacecraft assembly facility is unique, and it is therefore important to characterize the microbial populations present in such environments to understand potential PP risks. Several studies have identified persistent microbial species in NASA cleanrooms and spacecraft assembly facilities, with some non-spore-forming species exhibiting resistance to radiation, desiccation, and cleaning protocols. For example, Tersicoccus phoenicis, was first identified in NASA’s Phoenix Mars Lander cleanroom in 2007 (Vaishampayan et al., 2013). This non-spore-forming bacteria thrives in nutrient-poor conditions and was found in two separate cleanrooms 2,500 miles apart, suggesting that cleanroom-adapted microbes can spread within these specialized environments. Similarly, Deinococcus phoenicis, a radiation-resistant bacterium isolated from the Phoenix Lander assembly facility at NASA’s Kennedy Space Center, has been shown to survive extreme gamma radiation (D10 > 8 kGy) and UV exposure [D10 > 1,000 Jm−2], demonstrating that non-spore-forming extremophiles may pose PP related risks (Vaishampayan et al., 2014). This is particularly concerning because PP protocols primarily target spore-forming bacteria, potentially overlooking hardy non-spore-formers like Deinococcus (Mattimore and Battista, 1996). Additional bacterial species have been repeatedly isolated in spacecraft assembly facilities, reinforcing the importance of understanding microbial persistence in these environment. Acinetobacter radioresistens, was cultured from the Mars Odyssey orbiter during assembly-facility (La Duc et al., 2007), while Brevundimonas diminuta, a Gram-negative bacterium emerged as a predominant species following intensified cleaning regimes during the Phoenix Lander assembly (Ghosh et al., 2010). The resilience of B. diminuta in nutrient-poor, disinfected environments suggests it may persist despite rigorous sterilization measures, has made it a model organism for PP studies (Kimura et al., 2023). Furthermore, spore-forming bacteria, particularly Bacillus and related genera (Paenibacillus, Geobacillus, Oceanobacillus), remain classic contaminants in cleanrooms, with numerous strains being recovered from both NASA and ESA facilities (La Duc et al., 2007; Moissl-Eichinger et al., 2012).
For crewed missions beyond low Earth orbit, microbial adaptations to high-radiation environments could impact microbial survival or physiology, which may impact crew health or other scientific objectives. It is important to investigate microbial survival and adaptation under space-like stressors, including desiccation, vacuum, and proton radiation. Previous studies have evaluated the effects of space stressors on human-associated microorganisms (Tarpley et al., 1953; Cao et al., 2015; Bruckbauer et al., 2020; Cortesão et al., 2020), liquid cultures (Beblo-Vranesevic et al., 2017; Verbeelen et al., 2024), and spore-forming bacteria (Schuerger et al., 2003; Khodadad et al., 2017; Cortesão et al., 2019; Deng et al., 2023). However, little is known about the effects of deep-space-like stressors on dried, non-spore-forming cleanroom isolates (Osman et al., 2007; Dance, 2025).
The scope of this project was to determine the effects of space-like stressors, including desiccation, vacuum, and proton radiation, on dried microbes isolated from various locations at the Marshall Space Flight Center (MSFC) with varying degrees of cleanliness. Given that each cleanroom is unique, investigating the microbial composition at MSFC is critical for assessing potential contamination risks. We performed multiple studies at several proton fluence levels and used the resultant data to narrow our test group for subsequent tests. Selected PP-relevant candidate strains were subjected to whole genome sequencing (WGS) and comparative genomic and functional analyses.
2 Materials and methods
2.1 Microbial collection and isolation
Four cleanrooms at MSFC were selected for sampling together with two uncontrolled lab spaces. One cleanroom is not maintained and certified as a cleanroom but still has controlled access. In all cases, air samples were collected into 10 mL sterile water using a Coriolis μ air sampler (Bertin, Montigny-le-Bretonneux, France). Subsequently, field control samples were collected by waving a swab through the air for approximately 5 s. Surface samples of small area (5″ by 3″) using either dry or sterile water-wetted swabs (806WC, Puritan) or large area (27″ by 27″) were collected using 9″ by 9″ sterile water-wetted wipes (TX3211; TexWipe, Kernersville, NC).
Air samples (300 L/min) were collected for 5 min into 10–15 mL sterile water. After completion, collection cones were capped and stored up to 7 days at 4°C until processed. Swabs were collected and then stored dry in sterile tubes at 4°C for up to 7 days until processed. Wipes were dampened in 10 mL sterile water within a sterile petri dish and following collection, stored in sterile glass jars at 4°C up to 27 days until processed.
Tryptic Soy Agar (TSA) (BD Biosciences, Franklin Lakes, NJ) and Tryptic Soy Broth (TSB) (BD Biosciences, Franklin Lakes, NJ) were used as media for cultivation of the bacteria. All swabs were streaked directly onto the surface of a TSA plate and then incubated at 32°C for at least 2 days. Wipes were submerged in 35 mL of sterile water and then sonicated. A volume of 250 μL was then pipetted onto TSA, spread using a sterile spreader and incubated at 32°C for at least 2 days. Last, 100 μL of liquid collected via Coriolis air sampler was spread on TSA using sterile spreaders and incubated at 32°C for at least 2 days. In most cases, if overgrowth was observed, or too numerous colonies to count (TNTC), dilutions were made of the liquid into sterile water and replated until countable colonies were observed.
Strains of Bacillus atrophaeus ATCC 9372 and Deinococcus radiodurans ATCC 13939 purchased from American Type Culture Collection (ATCC) were used as positive controls. Sterile water (n = 3) and DNA purification kit blanks (n = 3) were employed as negative controls.
2.2 Microbial library curation
For all plates, representative colonies for all morphologies observed were selected and given a unique identifier. Selected colonies were all streaked to confirm purity on TSA and incubated at either 25°C or 32°C depending on whether the colony was classified as fungi or bacteria, respectively, until sufficient growth and isolated colonies were observed for subsequent experiments. If multiple colony morphologies were detected, single colonies were selected and streaked again. If molds were observed and single colonies could not be obtained, the isolates were sub-cultured until only a single morphology was detected.
For the physical curation, when possible, strains were subcultured from one colony using a sterile loop into 1.5–3 mL TSB and grown stationary and incubated at either 25°C or 32°C. When turbid growth was observed, the strains were frozen with a 1:1 volume of 25% sterile glycerol in cryotubes and stored at −80°C. If an organism was unable to grow in liquid culture, colonies from the TSA plates were scraped with a sterile loop into a 1:1 mixture of TSB:25% glycerol and frozen. All frozen cultures were then re-streaked on TSA with a sterile loop and grown at 25°C or 32°C until growth was seen. If no growth, a mixed culture, or a culture with unmatched description was observed, the culture was discarded, and renewed attempts were made to obtain a pure culture from the original plates. It was only after a stock culture was frozen and the growth was verified that the original plates were discarded. All strains were classified as either suspected fungi or suspected bacteria based on colony morphology.
2.3 DNA extraction
DNA extraction was conducted using a Quick-DNA Fungal/Bacterial Miniprep Kit (Zymo Research, D6005, Irvine, CA). To collect pellets, pure cultures were scraped from TSA plates using sterile loops and deposited into a weighed microcentrifuge tube. The tubes were centrifuged at maximum speed for 1 min in an accuSpin Micro 17 Microcentrifuge (13-100-675; Thermo Fisher Scientific, Waltham, MA). The microcentrifuge tubes were weighed, and pellet weights were recorded. Pellet wet weights of 50–100 mg were targeted and multiple plates were used if necessary to get sufficient biomass. Pellets were stored in a −20°C freezer until ready for processing. For DNA extraction, pellets were thawed in a biosafety level 2 (BSL2) hood and the manufacturer’s instructions were followed for isolating genomic DNA. DNA concentration was measured using a Thermo Scientific Nanodrop Lite (Waltham, MA).
2.4 Sanger sequencing and DNA analysis
Microbial taxonomic characterization was performed using capillary electrophoresis (i.e., Sanger) sequencing of small subunit (SSU; 16S or 18S) ribosomal RNA (rRNA) gene amplicons generated from PCR amplification of genomic DNA using bacterial or fungal primer sets. Off-the-shelf primers for bacterial 16S rRNA gene amplification (1,500 bp amplicon) were purchased from Integrated DNA Technologies (IDT). Fungal primers ITS5F and ITS4R, custom synthesized by IDT (Coralville, IA), were used to amplify internal transcribed spacer (ITS) regions (500 bp amplicon) of suspected fungal isolates (Schoch et al., 2012).
PCR was conducted using an Applied Biosystems SimpliAmp Thermalcycler (A41192; Thermo Fisher Scientific, Waltham, MA). Detailed information can be found in the Supplementary methods.
Amplicons were sequenced by Azenta Life Sciences-Genewiz for Sanger sequencing. FASTQ sequences were downloaded, individually assessed and trimmed of unresolved bases (on average the first 20 to 40 bases and the last 20 to 150 bases depending on the length and quality of the sequence), and annotated using the rRNA/ITS database using the Targeted Loci Project Information from either the 16S ribosomal RNA sequence (Bacteria and Archaea) or ITS from Fungi and type reference material in the National Center for Biotechnology Information (NCBI) Basic Local Alignment Search Tool [BLAST; (e.g., Altschul et al., 1990)]. Genus-level annotation was only assigned when the closest taxonomically annotated reference sequence was at least 97% similar, and species-level annotation was only assigned when reference sequences were >99% similar, and there was a 0.2% difference between the top IDs (Supplementary Table 1).
2.5 Microbial culturing for exposure studies
Media used for culturing strains was TSA, Nutrient Agar with 1% glucose (BD Biosciences, Franklin Lakes, NJ), Lennox Broth (LB) agar, and TSB. Frozen stock cultures were scraped with a sterile loop and streaked across the surface of a TSA plate, a LB agar plate, or a nutrient agar plate, and then incubated at 32°C for at least 2 days. Once adequate growth was observed, cells from a single colony were collected with a sterile loop and inoculated into 3 mL of TSB. Cultures were grown either stationary or shaking at 250 rpm at 32°C for at least 2 days or until adequate turbidity was observed. Optical density was measured at 600 nm wavelength using a Genesys 50 UV/VIS spectrophotometer (Thermo Fisher Scientific, Waltham, MA) immediately following vigorous vortexing. Cultures were centrifuged and resuspended in sterile water to an OD600 of 0.1 (or approximately 1 × 106 cells/mL). Positive controls B. atrophaeus ATCC 9372 and D. radiodurans ATCC 13939 were included as needed.
2.6 Microbial application and radiation exposure
Kapton-HN film was cut into 1-in2 coupons and sterilized using a 3870ELP Heidolph Tuttnauer benchtop sterilizer. While each run was performed with independent cultures prepared specially for that experiment, the data from each run informed the next, meaning that the runs were all conducted differently and are outlined individually below and as described in Supplementary Table 2. Diluted cultures were mixed with a vortex and the specified volume was applied as a single droplet to the surface of a labeled Kapton coupon. All work was performed in a biosafety cabinet. Samples were allowed to dry for up to 3 days. Samples were then transferred to the radiation test facility for exposure. In all cases, media controls without bacteria were similarly exposed. Ambient coupons devoid of microbes but spotted with sterile water (negative control) were transferred between buildings along with the experimental coupons but were unexposed to vacuum or radiation. For the final run, a second set of controls, called vacuum controls, were also subjected to the ~1E-6 Torr vacuum, like the test specimens.
In a series of experiments, microbial cultures were deposited on sterile Kapton coupons and subjected to proton and ultraviolet (UV) radiation to assess their resilience under extreme conditions. The coupons were prepared in a biosafety cabinet, where microbial samples were dried before exposure. Proton radiation experiments were conducted at the Combined Environmental Effects Facility (CEEF) at MSFC. It is difficult to measure the dose of radiation received by such small targets (microbes), and pathfinder studies demonstrated the usefulness of relying on fluence for dosing (Cengel et al., 2010). Therefore, samples were exposed to 100 keV protons at fluences ranging from 2 × 1015 to 4 × 1015 p+/cm2 under high vacuum (~1E-6 Torr). Each run featured minor procedural variations. In the first run, 250 μL of microbial culture was applied, but all samples being irradiated, leaving no controls. Subsequent runs utilized 200 μL to ensure better drying and included ambient unexposed controls. The second, third, and fourth runs proceeded with increased fluences and adjusted exposure times, ranging from 10.7 to 36.5 h, under vacuum durations of 46 to 145 h (Supplementary Table 2).
Additionally, a UV radiation experiment exposed samples to 254 nm light at an intensity of 80 W/m2 at a distance of ~18 cm for 5 or 10 min using a Spectroline XL-1500 UV Crosslinker. These samples were processed on the same day to minimize contamination. Instances of contamination were seen only in the first run, and improved sample handling was employed in subsequent runs, improving outcomes.
2.7 Post irradiation processing
Following radiation experiments, samples were transferred to a BSL2 lab for further processing. In all cases, sample processing included the transfer of a single Kapton coupon into a sterile 50 mL conical tube. A volume of 5 mL of sterile water was added, and the sample was vortexed vigorously to release cells from the coupon surface. In the case of the first and second runs, 250 μL of eluate was pipetted on appropriate solid media and spread across the surface using a sterile L-shaped spreader. Plates were incubated at 32°C for 2 days. For the last two runs, spot plating was performed with 100 μL. All negative control samples were treated the same way as the experimental samples. An inoculum control was not included for the first run. For the inoculum growth control in the second run, 100 μL was spread plated to verify growth. For the remaining runs, 100 μL of inoculum were pipetted as spots on the agar surface and incubated at 32°C until sufficient growth was observed. Due to the high number of plates, quantitative data was not obtained, rather an arbitrary qualitative system was employed to describe the growth as growth (+), minor growth (minor), no growth (−), or contamination. Growth is defined as colonies observed in three replicate samples per microbe. Minor growth is defined as at least one colony observed in at least one replicate sample. No growth was defined as the lack of observable growth in any sample. Contamination was indicated if microbial colonies differing in appearance from those of the selected isolates were identified. Control coupons were processed identically. Finally, inocula were plated to ensure that adequate starting culture was present and to compare morphology and determine any contamination.
2.8 Bacterial whole genome sequencing (WGS)
WGS of strains PPS68, PPS72, PPS117, and PPS120 were carried out as previously described (Popovich et al., 2021). DNA quantity and quality were measured using fluorimetry (Qubit 4; ThermoFisher) and electrophoresis (TapeStation4150; Agilent). Library preparation was performed using a Nextera XT kit (#FC-131-1024; Illumina Inc. San Diego, CA) according to the manufacturer’s instructions with 1 ng template input and 12 cycles of indexing PCR. An equal-volume pool of all libraries was then prepared, and deep sequencing was performed using an Illumina NovaSeq X instrument targeting 5 million clusters per sample (paired-end, 2 × 150 base reads). Draft genomes were assembled using the software package CLC Genomics Workbench (v22; Qiagen). Library preparation was performed at the Genomics and Microbiome Core Facility (GMCF) at Rush University, IL, USA, and deep sequencing was performed at the DNA Services Facility at the University of Illinois at Urbana-Champaign.
2.9 Genome assembly and 16S rRNA gene identification and annotation
Bacterial genomes were de novo assembled using the CLC Genomics Workbench software (v24) using shotgun sequence data generated on an Illumina sequencer. Default settings were applied for the assembly process, except for the minimum contig length, which was set to 200 base pairs to ensure the inclusion of smaller but potentially important contigs. Using BLAST analysis (i.e., BLASTn-v2.12.0+) against the NCBI’s 16S ribosomal RNA sequences database, 16S rRNA genes were extracted from assembled genome sequences. In addition, the assembled genomes were annotated by Rapid Annotation using Subsystem Technology (RAST) as described previously (Chander et al., 2017). Data for each annotation was exported from the RAST server and further processed through Pan-ResistomeFinder. The “role” column in the RAST exported annotation data shows gene/protein while other columns assign these genes into larger groups based on their function. There may be multiple entries of a single gene/protein in the column “role” based on their multiple roles. To account for the functional diversity of each gene, uniqueness of genes in this column was ignored.
2.10 Strain selection for comparative resistome analysis with radioresistant strains of phylum Actinomycetota
Strains with experimentally validated D₁₀ values of 2 kGy or higher (for γ-irradiation or equivalent UV-C fluence) were designated as “resistant,” while those below this threshold served as “sensitive” controls (Table 1). The final panel comprises eleven resistant strains Deinococcus radiodurans R1 (Daly, 2009), Deinococcus radiomollis (Callegan et al., 2008), Kocuria rhizophila PT10 (Guesmi et al., 2021b), Kocuria rosea DSM 20447 (Asgarani et al., 2012), Kineococcus radiotolerans SRS30216 (Bagwell et al., 2008), Promicromonospora panici PT9 (Guesmi et al., 2021a), Rubrobacter xylanophilus DSM 9941 (Ferreira et al., 1999), Rubrobacter radiotolerans DSM 5868 (Ferreira et al., 1999), Rubrobacter radiotolerans RSPS-4 (Ferreira et al., 1999), Geodermatophilus obscurus DSM 43160, and Geodermatophilus dictyosporus DSM 44208 (Montero-Calasanz et al., 2015). The sensitive group includes Escherichia coli O157, E. coli K-12 MG1655, and Arthrobacter sp. PAMC 25486 (KOPRI Repository, n.d.). These additional set of strains provide a robust two-class framework for downstream comparative analysis of gene presence patterns related to radiation resistance.
2.11 Taxonomic annotation of the selected strain panel
NCBI taxonomy database confirms that, apart from the genus Deinococcus (order Deinococcales, phylum Deinococcota), all other reference strains in the radiation phenotype panel belong to the phylum Actinobacteriota. The resistant group includes Kocuria rhizophila PT10 and Kocuria rosea DSM 20447 from the order Micrococcales, Kineococcus radiotolerans SRS30216 from Kineosporiales, Promicromonospora panici PT-9 from Propionibacteriales, Geodermatophilus obscurus DSM 43160 and Geodermatophilus dictyosporus DSM 44208 from Geodermatophilales, and Rubrobacter radiotolerans DSM 5868, Rubrobacter radiotolerans RSPS-4, and Rubrobacter xylanophilus DSM 9941 from Rubrobacterales. Among the sensitive strains, Arthrobacter sp. PAMC 25486 (order Micrococcales) also belongs to Actinobacteriota, while the two Escherichia coli strains, O157 and K-12 MG1655, are members of the phylum Proteobacteria (order Enterobacterales). This updated composition emphasizes broad phylogenetic coverage while confirming that the majority of strains in the panel, particularly the resistant ones, are taxonomically affiliated with Actinobacteriota.
2.12 Automated resistome extraction using Pan-ResistomeFinder
For extraction and comparison of stress and resistance-associated genes from multiple microbial genomes, we used Pan-ResistomeFinder (https://github.com/atulchander/Pan-ResistomeFinder). This tool supports automated processing of genomic annotation files in.tsv,.csv,.xls, or.xlsx formats, each corresponding to a different strain. It ensures the presence of core functional descriptors (Category, Subcategory, Subsystem, and Role) and constructs a unified gene presence–absence matrix by merging input files on these shared descriptors. Genes were marked with a binary indicator (1 for presence) across strains, and missing annotations were imputed as 0. The merged matrix captures the distribution of functional genes across the strains. To focus specifically on stress and resistance-related functions, Pan-ResistomeFinder performs a keyword-guided resistome search. This produces a filtered output containing genes relevant to resilience, which is basis for subsequent resistomics. To characterize strain-specific resistome overlap, we implemented a focused extraction protocol using Pan-ResistomeFinder. First, the resistome output matrix filtered for stress and resistance-related genes was queried to identify total resistome in the dataset. In this total resistome dataset, a subset of data was created showing presence of genes in A. koreensis PPS68 and simultaneously found in at least one of the confirmed resistant strains. This subset was saved as the “shared resistome” dataset, representing genes potentially conserved in resistance-competent Actinobacteria. From this shared subset, we further excluded any genes that were also present in any known sensitive strains (E. coli O157, E. coli K-12 MG1655, and Arthrobacter sp. PAMC 25486). The resulting exclusive set, comprising genes unique to strain PPS68 and resistant strains only, was saved separately to capture PPS68-associated resistome signatures that may be selectively retained in radiation-adapted taxa.
2.13 Genome annotation using average nucleotide identity (ANI) analysis
We used Genome Taxonomy Database (GTDB)-Tk v2.4.0 to compute the Average Nucleotide Identity (ANI) between our query genomes and the reference genomes from the GTDB release r220. This analysis aimed to identify the closest representative genome for each query genome based on ANI. Four query genomes in.fna format were analyzed against the comprehensive GTDB reference dataset. The analysis was conducted using the ani_rep command in GTDB-Tk, with the genome directory, output directory, file extension (.fna). The results included two main output files: gtdbtk.ani_summary.tsv, which provided ANI values for all comparisons, and gtdbtk.ani_closest.tsv, which lists the closest representative genome for each query genome.
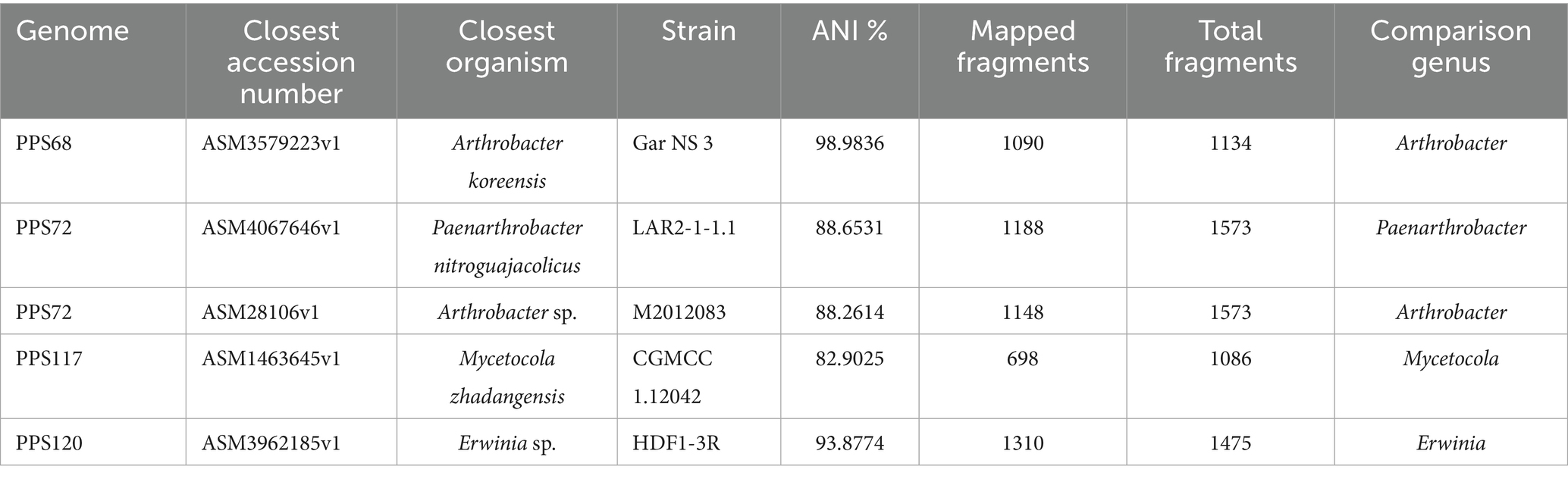
Further analysis was performed using the FastANI software with default parameters to calculate the Average Nucleotide Identity (ANI) for the four query genomes: strain PPS68, strain PPS72, strain PPS117, and strain PPS120. Output included ANI values, number of fragments mapped, and total fragments analyzed. FastANI analysis was performed by the Rush Research Bioinformatics Core (RRBC) at Rush University. These genomic comparisons were performed between the assembled draft genomes and reference genomes downloaded from the NCBI database. All available NCBI genomes from genera of interest, including GenBank (GCA) and RefSeq (GCF), were downloaded for comparison. Genus-level taxonomic identification was initially confirmed using 16S ribosomal RNA gene BLAST analysis (Altschul et al., 1997; Jain et al., 2018).
3 Results
In total, 82 microbial isolates were recovered from four sampling locations at the MSFC with varying levels of cleanliness, including ISO8 locations (A-C) and uncontrolled locations (D-F). As the aim of this study was to isolate and identify cleanroom extremotolerant with the ability to withstand harsh conditions, but not to evaluate cleanroom cleanliness, samples were collected and processed using multiple approaches that differed from the NSA (NSA, NASA-HDBK-6022, 2010). Furthermore, the collection of isolates was limited only to culturable aerobic microbes given that downstream exposure studies required that the strains be cultured. While this narrowed the diversity of microbes recovered, it was beyond the scope of this work to fully evaluate the composition of cleanroom microbes using cultivation and molecular approaches. Based on plate morphology, three were tentatively identified as fungi and the remaining 79 were presumptively identified as bacteria. Of the 82 isolates, 68 strains yielded PCR amplification with bacterial and fungal domain-level primer sets. The remaining 14 isolates were re-streaked on TSA and DNA was re-extracted, enabling genus-level or higher taxonomic identification for all. In total, 71 isolates provided high-quality sequencing data with annotations to the genus level or beyond (Supplementary Table 1). DNA amplification issues were primarily attributed to the misidentification of yeast/fungal strains as bacteria, as yeast lack the 16S rRNA gene required for 16S PCR amplification. For example, of the 14 isolates re-sequenced, five yeasts were initially misidentified as bacteria, but light microscopy confirmed yeast-like morphologies. Additionally, some isolates exhibited sequencing issues, such as high background noise, indicative of insufficient DNA quality or quantity. This may have been due to non-axenic cultures despite multiple purification attempts.
Several strains (n = 24) were selected based on their taxonomic identity, growth characteristics, and a targeted literature review conducted using PubMed. The search included keywords such as “radiation resistance,” “spore-forming,” “extremophile,” and the genus or species names of the isolates. Articles were screened for evidence of physiological traits associated with extremotolerance (e.g., resistance to desiccation, oxidative stress, or nutrient limitation), and strains with such attributes were prioritized for further characterization.
To assess their survival under space environmental conditions, a series of ionizing radiation experiments were conducted on a subset of the strains (n = 24). Five radiation exposure tests were performed, with the results summarized in Table 2. For the first run, microbial isolates were exposed to 100 keV protons to a fluence of 2 × 1015 p+/cm2. Of the 24 strains, 13 isolates demonstrated marked survival to this exposure, while seven showed minor growth. For the second run, 13 strains were tested again along with D. radiodurans (ATCC 13939) as a positive control. In this run, four strains survived with one showing minor growth and no contamination observed. For comparison, the D. radiodurans positive control exhibited only minor growth. In subsequent tests, B. atrophaeus (ATCC 9372) was added as a more resistant positive control. For the third run the radiation dose was increased by 50%, and five isolate strains were exposed in addition to D. radiodurans and spores of B. atrophaeus as positive controls. Three of five strains survived these exposure conditions, along with two instances of minor growth. In addition, D. radiodurans demonstrated minor growth, while B. atrophaeus spores survived. Similar results were observed in the fourth exposure experiment. In this fourth test, radiation conditions were identical to those of the third run but included two control sample sets: (1) ambient controls in which all taxa were maintained at normal pressure, and (2) vacuum which were subjected to the same level of vacuum as test specimens (Table 2).
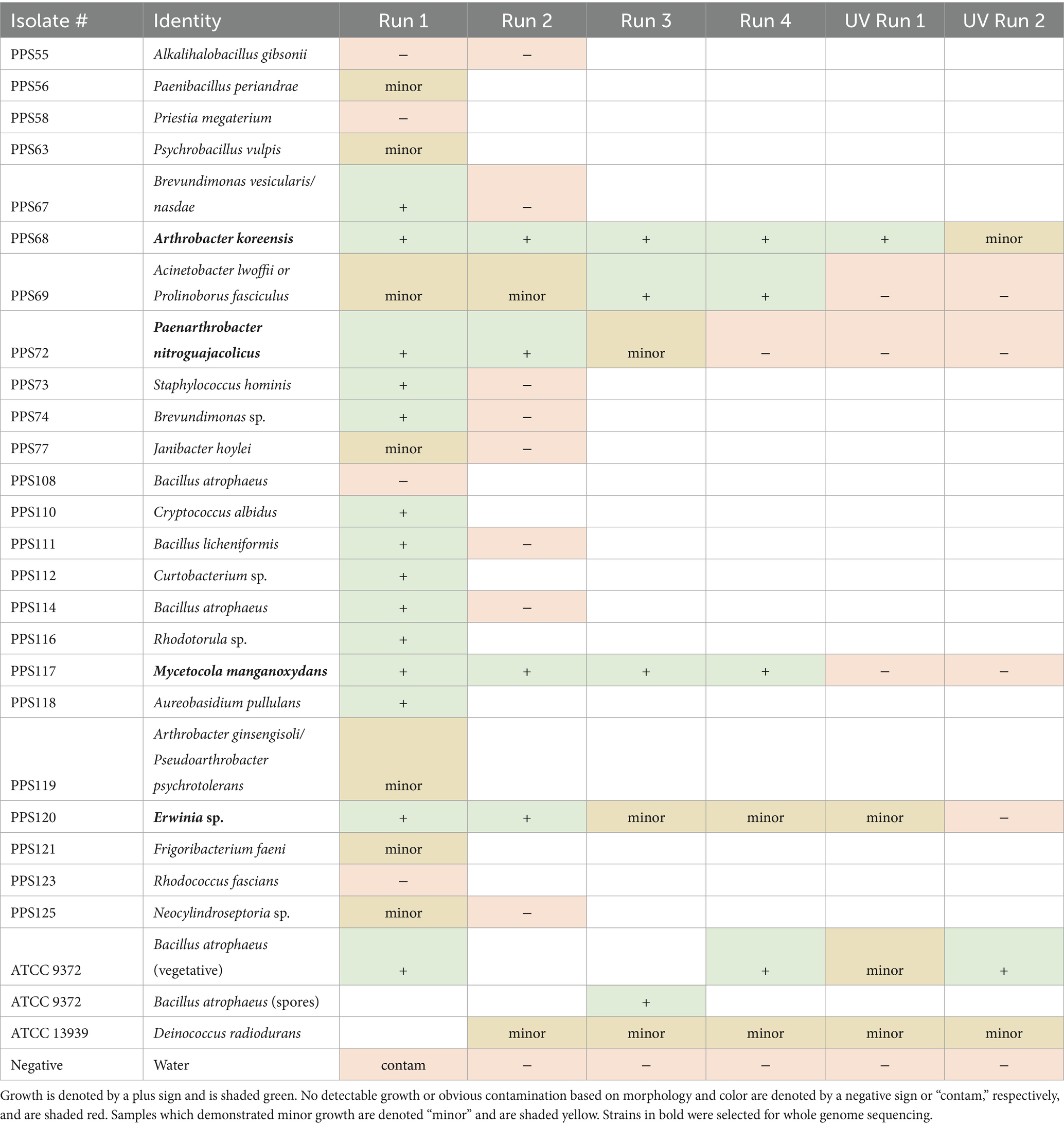
Table 2. Identification of strains isolated during this study based on 16S or ITS rRNA gene sequencing and survival against various simulated space environmental conditions.
After proton exposure, the resistant microbes were exposed to ultraviolet (UV254) radiation, representative of another type of space-relevant stressors. UV is not only relevant as a space environmental condition but has been considered as a sterilization procedure for “break-the-chain” PP workflows in the Mars Sample Return mission (Schuerger and Moores, 2023). In the UV experiment, the five most resistant microbes from the third run were exposed to 254 nm UV radiation at an intensity of 80 W/m2 for either UV Run 1 or 2 (5 or 10 min, respectively). While UV exposure was effective at killing most of the microbes tested, strain PPS120 (Erwinia sp.) exhibited minor survival. The minor growth was determined based on the results where one to three colonies were seen on one out of two samples exposed. In addition, survival of the strain PPS68 (A. koreensis) was recorded, when exposed to either 5 or 10 min of UV radiation. Positive controls (B. atrophaeus spores and D. radiodurans) tested exhibited minor growth.
Based on the resistance profiles observed, four strains were sequenced for WGS, including strains PPS68, PPS72, PPS117, and PPS120. Based on 16S rRNA gene sequencing these strains were identified as Arthrobacter koreensis, Paenarthrobacter nitroguajacolicus, Mycetocola manganoxydans, and Erwinia sp., respectively. However, WGS analysis identified the four isolates only to the genus level, except for strain PPS68. The WGS analysis suggests that three of the isolates may represent novel species, (Tables 3,4). Further taxonomic analysis was performed using ANI analysis.
3.1 Genome characteristics and relatedness in selected cleanroom strains
The 16S rRNA analysis identified the closest taxonomic affiliations of the query strains based on sequence similarity to known type strains. Strain PPS117 showed 99.03% similarity to Mycetocola zhadangensis ZD1-4. Strain PPS120 exhibited 98.56% similarity to Erwinia tasmaniensis Et/199. Strain PPS68 shared 99.47% similarity with Arthrobacter luteolus CF-25. Strain PPS72 exhibited 99.93% similarity to Paenarthrobacter nitroguajacolicus JCM 14115.
The GTDB-Tk ani_rep command was used to compare query genomes against a comprehensive reference dataset of 113,104 bacterial genomes, providing detailed assessments of genomic similarity through Average Nucleotide Identity (ANI) and alignment fractions. For strain PPS68, high ANI matches were observed and identified as A. koreensis (GCF_009193255.1) since ANI was 98.7% and 0.9317 alignment fraction. Strain PPS72 shows highest matches (88.65 and 88.26%) with Paenarthrobacter nitroguajacolicus (GCF_001375615.1) and Arthrobacter sp. (GCF_000281065.1). Strain PPS120 showed a closest match to Erwinia sp. (GCF_900068895.1) with an ANI of 94.1% and an alignment fraction of 0.8363, though it did not meet the threshold for a close match. For strain PPS117, ANI analysis found no significant matches, despite a 16S rRNA similarity of 99.0% with M. zhadangensis ZD1-4, indicating the potential for a novel species. However, according to FastANI, the closest match for the strain PPS117 was with M. zhadangensis (82.9%, GCF_014636455.1) (Table 4; Supplementary Table 3).
3.2 Comparative genomics of 4 selected cleanroom strains
The functional analysis identified 1,990 genes, observed across four bacterial species, which were classified into various function-based categories according to RAST (Figure 1A; Supplementary Table 4). In this dataset, 522 roles of genes were found to be conserved across four bacterial species, constituting ~26% of the total dataset. These conserved genes form part of the core genome shared by four species analyzed (Figure 1B; Supplementary Table 5). The conserved genes have functional annotations predictive of their role in regulating essential pathways like, carbohydrate metabolism, amino acid biosynthesis, protein processing, and cofactor and vitamin metabolism. Moreover, these genes are also related to the processes, nucleotide synthesis, DNA and RNA metabolism, and respiration which are consistently maintained, underscoring the fundamental requirements for cellular maintenance and energy generation. Additionally, several genes linked to membrane transport, lipid biosynthesis, and stress defense mechanisms were conserved, reflecting their important role in maintaining structural integrity and adaptive potential under dynamic environmental conditions.
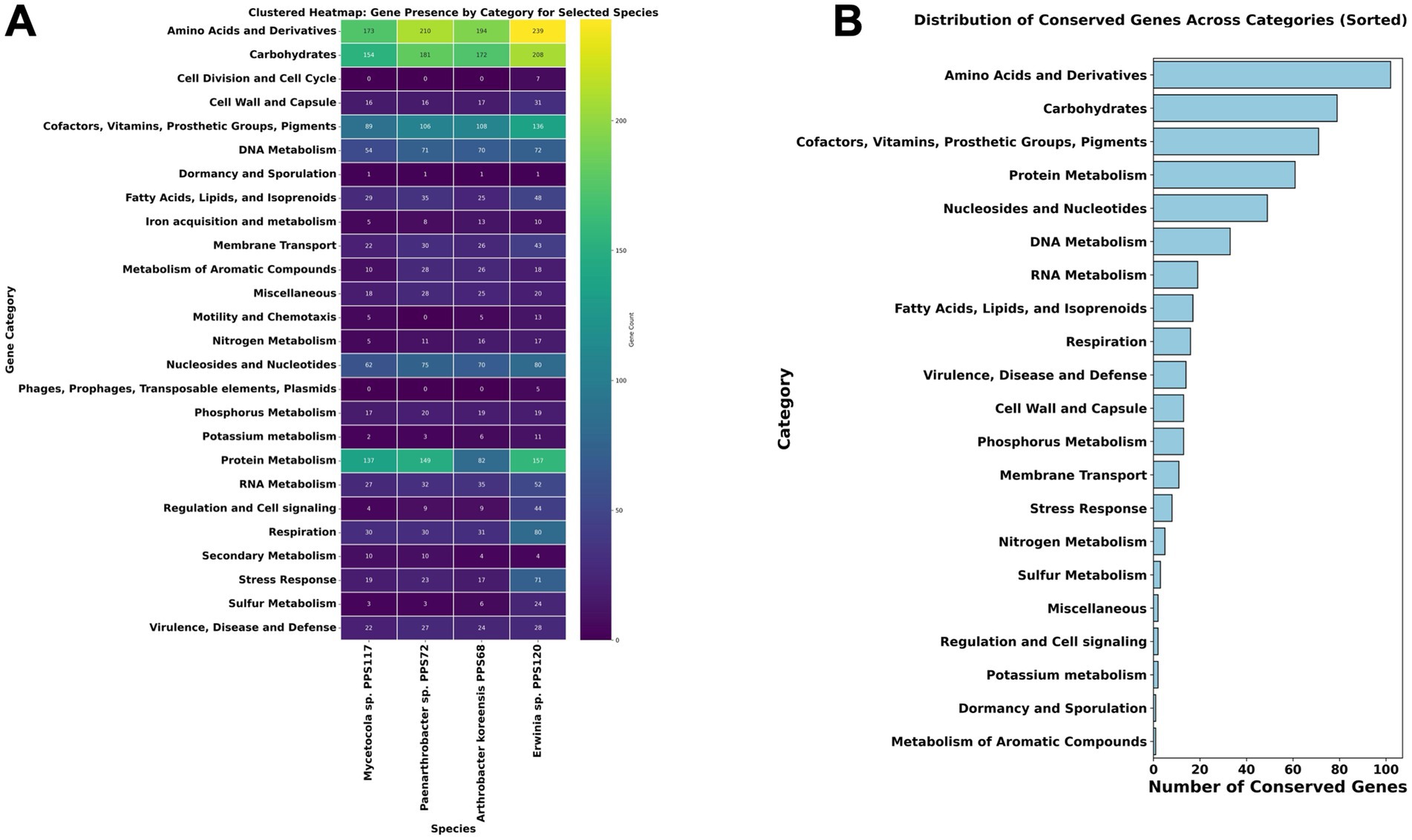
Figure 1. (A) Clustered heatmap of gene presence across functional categories in four bacterial species. This heatmap illustrates the distribution of gene counts across 25 functional categories for four bacterial species: Arthrobacter koreensis PPS68, Erwinia sp. PPS120, Mycetocola manganoxydans PPS117, and Paenarthrobacter nitroguajacolicus PPS72. Gene categories include pathways related to metabolism, stress response, dormancy, and more. The color gradient reflects gene abundance, with higher counts represented by lighter colors. This visualization highlights both shared and distinct genetic characteristics among the species, shedding light on their potential functional adaptations to various environmental conditions. (B) The bar chart represents the distribution of conserved genes across different functional categories identified in the core genome shared by the analyzed bacterial species. Categories such as Amino Acids and Derivatives, Carbohydrates, and Cofactors, Vitamins, Prosthetic Groups, and Pigments exhibit the highest conservation levels, indicating their essential roles in fundamental metabolic and cellular processes. The chart provides insights into the functional significance of conserved pathways and their contribution to bacterial survival and adaptation in varied environments. The heatmap uses a color gradient to indicate gene count, ranging from dark blue (low) to green (high) and yellow (highest).
3.3 Variation in pathway specialization in selected 4 cleanroom strains
Despite the conservation of core genes, variations in specific metabolic pathways were observed. Notably, carbohydrate metabolism genes showed significant diversity in these 4 organisms (Supplementary Table 4). Erwinia sp. PPS120 exhibited a broader set of genes associated with sugar utilization, indicating metabolic flexibility and the ability to exploit diverse carbon sources. Conversely, A. koreensis sp. PPS68 and Paenarthrobacter sp. PPS72 had more specialized genes in some pathways (Supplementary Table 4), which may reflect adaptations to specific environments with limited substrate diversity.
3.4 Stress response functions in selected 4 cleanroom strains
Around 90 genes were directly reported in the stress response category in RAST annotations, with an additional 463 genes identified as possible stress-response related. Out of these 553 functional entries, 21% were conserved across all four species and were mainly associated with stress response, membrane permeases, transporters, efflux pumps, resistance genes, antimicrobial genes, heat shock proteins and DNA repair genes.
3.5 Survival in harsh conditions
All species carry genes related to oxidative stress responses and detoxification, that might help them enhance survival in challenging environments. The presence of glutathione-dependent detoxification pathways in Erwinia sp. PPS120, for instance, may highlight its adaptability to oxidative environments. Compared to the other 3 cleanroom strains, A. koreensis PPS68 also displayed a robust set of stress response genes, including those for oxidative damage management and membrane stability, which may be contributing to its resilience to desiccation, radiation, and extreme temperatures. To cross-validate the genes that might be contributing to the radiation resistance in A. koreensis PPS68, we conducted further comparisons of these cleanroom strains with literature searched radioresistant and radiosensitive strains predominantly belonging to the phylum Actinobacteria.
3.6 Comparison of predicted resistome of A. koreensis PPS68 to radioresistant and radiosensitive strains
Building on the above comparative overview of stress-response related functional annotations, a resistomics analysis was conducted by selecting literature searched 11 radioresistant strains of phylum Actinobacteria and 3 probable radiosensitive strains which includes a strain PAMC 25486 of Arthrobacter sp. (Figure 2; Supplementary Table 4). Further, by using Pan-ResistomeFinder, we have identified 220 PPS68 roles present in at least one additional radio-resistant strain (Figure 3). These functional roles include multiple oxidoreductases, membrane-associated efflux pumps, and DNA-repair enzymes, underscoring a conserved molecular machinery that may underpin the high radiation and desiccation tolerance shared across the resistant panel. Conversely, only 26 PPS68 roles/genes remained after all genes found in the sensitive controls were subtracted; this exclusive subset pinpoints candidate determinants that are retained by resistant strains yet absent from radiation-sensitive taxa, and therefore may contribute most directly to the extreme-tolerance phenotype observed in A. koreensis PPS68 (Supplementary Table 6). This analysis supports the prediction that PPS68 possesses an expanded DNA repair capacity, characterized by an augmented base-excision repair (BER) module operating in tandem with non-homologous end-joining (NHEJ) ligases (LigC/LigD) and accessory polymerases. Together, these components constitute a multi-layered strategy for resolving radiation-induced DNA damages. Concurrently, osmoprotection and redox balance in PPS68 appear to be regulated by dual glycine-betaine uptake and synthesis systems, thiamin-precursor importers, and folate-cycling enzymes Supplementary Table 7. In parallel, detoxification and adaptive signaling may be supported by multidrug efflux pumps, a c-di-GMP-modulating diguanylate cyclase/phosphodiesterase, and nitrite/phosphate regulatory elements (Supplementary Table 6).
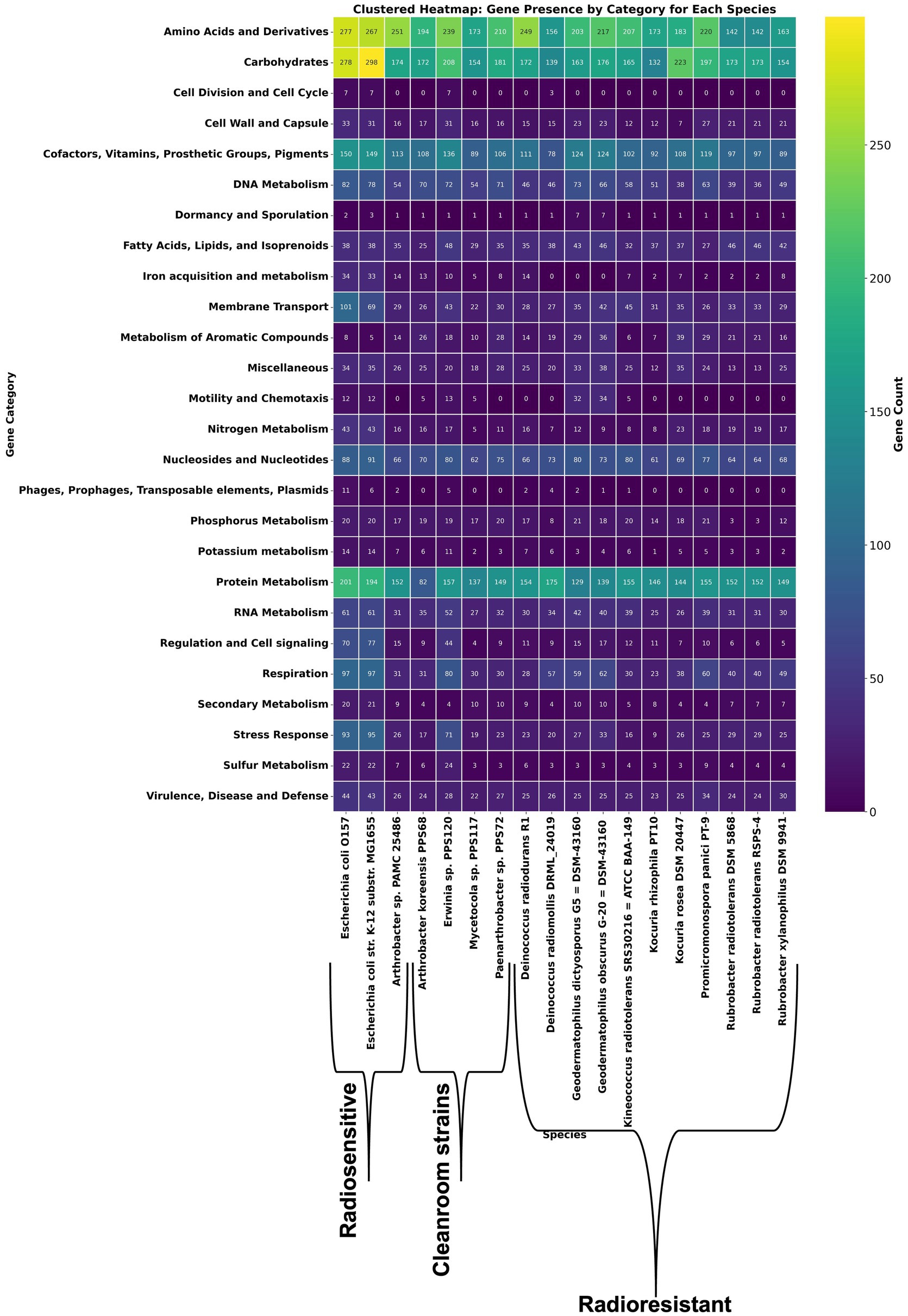
Figure 2. Clustered heat-map of gene counts in 26 functional categories across literature based radioresistant, radiosensitive representatives and clean-room isolates. Rows represent functional gene categories; columns represent individual bacterial genomes ordered as radiosensitive strains, clean-room isolates (including Arthrobacter sp. PPS68) and reference radioresistant taxa. Each cell contains the number of genes assigned to the corresponding category in the indicated genome. The heatmap uses a color gradient to indicate gene count, ranging from dark blue (low) to green (high) and yellow (highest).
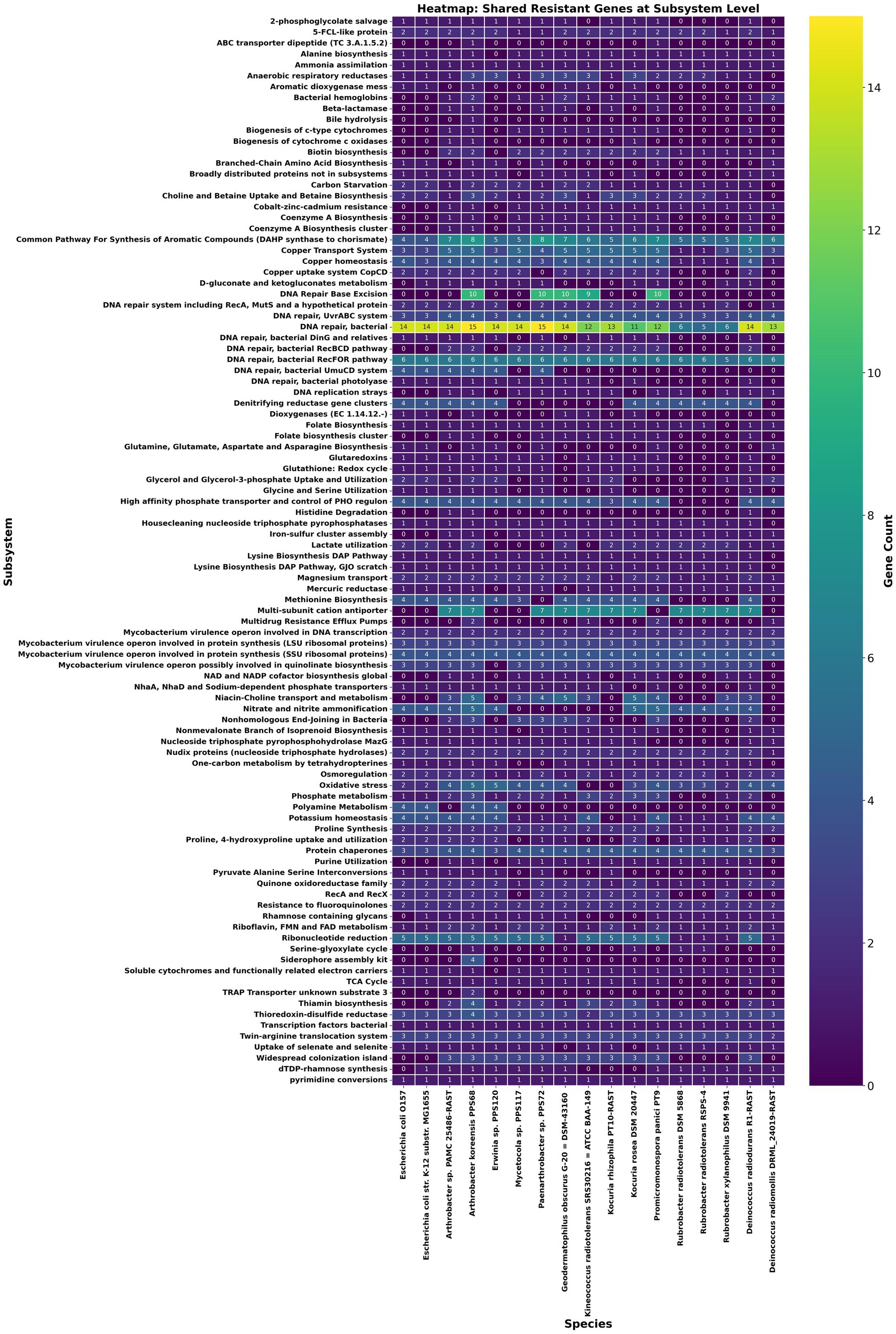
Figure 3. Heat-map of resistance-associated functions shared between Arthrobacter sp. PPS68 and at least one radioresistant comparator. Rows list individual resistance-related subsystems; columns list 11 radioresistant and 3 radiosensitive actinobacterial genomes together with Arthrobacter sp. PPS68 and other cleanroom strains. The heatmap used a color gradient to indicate gene count for each function, ranging from dark blue (low) to green (high) and yellow (highest).
4 Discussion
Bacterial and fungal isolates (n = 82) were recovered from built environments of various cleanliness levels and a subset was screened using increasingly harsh conditions mimicking some features of spaceflight and identified using sequencing of ribosomal RNA genes. A modular method was adapted from the literature to expose dried microbes to proton radiation in a clean and efficient manner to rapidly screen microbial isolates. The method used to expose microorganisms to proton radiation in this study was adapted from formerly published methods of exposing dried microbes to proton radiation in a relatively clean and efficient manner (Schuerger et al., 2003; Moeller et al., 2012; Cortesão et al., 2019; Hase et al., 2021). It can be modified to change many different features of experimental conditions, including what microbes are tested, the physical substrate the microbes are applied to, the density of the microbial population, the exposure/stressor employed, and the quantification method.
Using this platform, we have identified under-studied, non-sporulating isolates with resistance to ionizing radiation. Four isolates with the greatest tolerance for radiation were further characterized using WGS and comparative genomics analysis. Many of the genera detected in our study are consistent with taxa that have been detected in other NASA cleanrooms studies (Mahnert et al., 2015; Dworkin et al., 2017; Regberg et al., 2020; Hendrickson et al., 2021). Of note are microorganisms from the genera Acinetobacter, Bacillus, Brevibacillus, Erwinia, Paenibacillus, Micrococcus, Staphylococcus, Pseudomonas, Cladosporium, and Penicillium. Although prior studies have primarily analyzed cleanroom microbial communities using cultivation-independent molecular methods, the prevalence of these genera across multiple detection methods indicates that they are prevalent within cleanroom environments, irrespective of the location within the United States, are more easily sampled than other contaminants, or both.
Since microbes were isolated primarily from floors and surfaces of human-used, built environments, it is unsurprising that many microbes identified were associated with soil or human skin (Flowers and Grice, 2020; Philippot et al., 2023). Although many of these microbes have been poorly characterized, some of them have been previously identified as extremophiles. Specifically, species of the genus Brevundimonas and Kocuria have demonstrated the ability to survive in simulated Martian conditions (Dartnell et al., 2010; Vallalar, 2012). Furthermore, some genera, including Deinococcus and Janibacter, have been associated with resistance to certain stressors like ultraviolet radiation within the space environment (Shivaji et al., 2009; Ho et al., 2016). Finally, some of the genera identified, including those from the genera Alkalihalobacillus, Bacillus, Brevibacillus, Calidifontibacillus, Fictibacillus, Lederbergia, Paenibacillus, Priestia, Psychrobacillus, Pseudogracilibacillus, and Streptomyces, are known to form endospores which are capable of withstanding extreme environmental conditions and would likely be in the recoverable population from the NSA (Dietz and Mathews, 1971; Ash et al., 1993; Shida et al., 1996; Logan et al., 2009; Park et al., 2018; Adiguzel et al., 2020; Gupta et al., 2020; Rodríguez et al., 2020) Likely these documented, spore forming microbes did not demonstrate survival in our studies because they were exposed in a vegetative state. These conclusions further support previous findings that cleanrooms house many microbes that could be of potential risk to NASA missions as they relate to PP (Stieglmeier et al., 2009; Ghosh et al., 2010). GTDB-Tk ani_rep shows PPS68 at 98.7% ANI to Arthrobacter koreensis DSM 16760, the alkalitolerant soil type-species from Daejeon, Korea (Lee et al., 2003); its actual FastANI hit, strain Gar NS 3, was taken from bovine skin swabs in Guwahati, India and carries no irradiation data (NCBI BioSample: SAMN29049796). The strain PPS72 shares just 88.7% ANI with Paenarthrobacter nitroguajacolicus strain HG (GCF_001375615.1), a soil bacterium for which the exact source of isolation was not reported, but which is characterized by its ability to utilize papaverine an opium alkaloid antispasmodic drug as the sole carbon source (NCBI BioSample: SAMEA3310068). PPS120’s closest genome (94.1% ANI) is Erwinia sp. ErVv1 (GCF_900068895.1), a grape-vine (Vitis vinifera) endophyte from Italy (NCBI BioSample: SAMEA3216243). For PPS117, FastANI recovered only 82.9% identity to Mycetocola zhadangensis CGMCC 1.12042 (BMEK00000000.1), the type strain isolated from snow on the high-UV Zhadang Glacier, Tibetan Plateau, China (Shen et al., 2013), and no published radiation assays exist for this species.
The conserved genes in the cleanroom resistant strains are part of the core genome and can be involved in several functions like DNA repair and stress tolerance. For instance, DNA repair genes like RecA and the UvrABC excinuclease system provide protection against UV and oxidative DNA damage (Lenhart et al., 2012; Sinha et al., 2020; Kalogiannis and Eyre-Walker, 2024). Heat shock proteins, such as the 16 kDa heat shock protein A, stabilize and refold proteins during thermal stress, ensuring cellular functionality (Trilling et al., 2011; Srivastava et al., 2013). Furthermore, genes involved in oxidative stress and metal resistance, such as Alkyl hydroperoxide reductase and CopD, enhance resilience against reactive oxygen species and metal toxicity were present, facilitating adaptation to diverse environments (Hu and Zhao, 2007; Krishnamurthy et al., 2024; Ramnarine et al., 2024). Erwinia sp. PPS120 had an extensive array of stress response genes, particularly those involved in glutathione-dependent detoxification pathways, such as S-(hydroxymethyl)glutathione dehydrogenase and S-formylglutathione hydrolase (Chen et al., 2016; Osman et al., 2016). This predicts enhanced oxidative stress management, providing a survival advantage in environments prone to high oxidative damage. Additionally, all four species possessed the DedA protein for selenate and selenite transport, predictive of adaptations to environments with elevated selenium levels (Chen et al., 2016).
Specifically, A. koreensis PPS68 has reported notable resistance to both proton radiation and UV radiation. It is difficult to compare across studies where Bacillus spores have been exposed to protons (Moeller et al., 2012), but results from Run 3 preliminarily indicated that A. koreensis PPS68 was less resistant to radiation than B. atrophaeus spores (data not shown). While little is known about this strain of A. koreensis, previous literature suggests tolerance of both desiccation and alkaline conditions, and these features make it an organism of PP concern with the potential to survive the extreme conditions of space environments (Lee et al., 2003; Manzanera et al., 2015). A. koreensis PPS68 had several unique genes which were absent in the other cleanroom species genomically compared. Key genes include Thiol peroxidase-Tpx-type (EC 1.11.1.15), essential for detoxifying peroxides and protecting against oxidative damage induced by radiation (Lushchak, 2001). Multidrug resistance efflux pumps, such as the Acriflavin resistance protein and Multi antimicrobial extrusion protein, are known for exporting harmful compounds, enhancing oxidative stress management (Bogomolnaya et al., 2013). Thioredoxin-disulfide reductase maintains protein stability and supports recovery under stress (Kern et al., 2003). Additional genes like Sialidase (EC 3.2.1.18) may contribute to membrane stability (Kim et al., 2011), while the Histidine transport permease ensures nutrient acquisition under limited conditions (Zhang et al., 2015; Beetham et al., 2024). Stress response regulators with GGDEF and EAL domains modulate biofilm formation and stress responses, supporting survival in extreme environments (Rajeev et al., 2014). Siderophore biosynthesis and transport proteins (e.g., DesA, DesB, DesC, DesD) might also have roles in stress management because siderophores scavenge iron, which is crucial for many cellular processes, including those involved in stress responses. Siderophores play a critical role in mitigating radiation-induced oxidative damage by limiting free intracellular iron and preventing Fenton chemistry, a mechanism observed in extremophiles like Deinococcus radiodurans and potentially mirrored in cleanroom isolates (Daly, 2009; Krisko and Radman, 2013). In contrast, the presence of unique radiation and stress tolerance related genes does not confirm that they can be the contributing factor for their resistance until supported by further wetlab analysis. Thus, we tried to further validate these genomic predictions for their importance in radiation resistance through detailed comparative genomic analysis by including literature proven radiation resistant strains of phylum Actinomycetota.
Excluding genes shared with E. coli and a radio‑sensitive Arthrobacter strain revealed a distinct subset of genes in PPS68 that is present in at least one radiotolerant reference and absent from all three sensitive comparators (Supplementary Table 6). This genomic complement encodes a PhoU homolog that may regulate phosphate uptake during γ‑irradiation (diCenzo et al., 2017); the NHEJ ligase LigC, which may couple with LigD to re‑seal double‑strand breaks (Bhattarai et al., 2014); and a formate/nitrite exporter that mitigates nitrosative stress (Maeda et al., 2015). Osmoprotection and redox balance might be reinforced by dual glycine‑betaine uptake/synthesis genes (OpuD and betaine aldehyde dehydrogenase) (Boch et al., 1996; Kappes et al., 1996), whereas a Na+‑coupled MATE antiporter and an Acriflavin protein can remove toxic metabolites (Omote et al., 2006; Fischer and Kandt, 2013). An expanded base‑excision repair arsenal including polymerase‑like MT3142, Ku, LigD, LigC, polymerase I, and multiple glycosylases and endonucleases might be capable to facilitate rapid repair of clustered DNA breaks, while quinate/shikimate‑5‑dehydrogenase supports antioxidant and folate biosynthesis (Krokan and Bjørås, 2013; Marienhagen, 2025). Stress signalling in the strain may be modulated by a PAS‑domain GGDEF/EAL diguanylate cyclase/phosphodiesterase (Rajeev et al., 2014) whereas Vanillate O-demethylase oxidoreductase may dissipate reactive species and regenerate NAD+; and the ABC dipeptide transporter DppD could be responsible for acquisition of amino acids and peptides needed for DNA repair‑protein synthesis (Xu et al., 2021; Hu et al., 2024) in the presence of radiation stress conditions. Collectively, these functions absent from all sensitive genomes constitute an integrated network of DNA repair, osmotic and redox homeostasis, detoxification, and adaptive signalling that underpins their probable role in PPS68’s exceptional radiotolerance than rest of the strains. It is interesting to note that Paenarthrobacter sp. PPS72 also has 17 of these genes common whereas other two cleanroom strains share 2 for each.
Although this study has focused on a single type of radiation exposure (protons), there are other stressors that space-faring microbes will be exposed to, including: temperature extremes, broad spectrum ultraviolet (UV) radiation, microgravity, chemical assault, and starvation. Much of the benefit of this study was developing protocols and setting baseline exposures to better prepare for more stringent future studies. In addition, the UV experiment we performed was simple and used a common lab UV crosslinker which has a much higher intensity, perhaps as much as two orders of magnitude higher, than the intensity of 254 nm wavelength light in space. Therefore, future work from this proposal would include tightening the space-relevance of experimental parameters and performing stringent individual and combination studies to understand the ability of these microbes to survive the effects of multiple stressors (Smith et al., 2009; Schuerger, 2024).
The results of this study emphasize the necessity to modernize PP policies by shifting from spore burden analysis to a broader evaluation of microbes, including non-spore-formers, to address their ability to survive and adapt to space-related stressors. Molecular methods enhance the understanding of microbial diversity, especially non-culturable organisms. However, characterizing cultivable non-spore-forming microorganisms is essential for refining decontamination strategies. Investigations into the effects of desiccation, vacuum, and proton radiation on non-sporulating cleanroom isolates highlight critical gaps in knowledge regarding microbial survival, adaptation, and resilience in oligotrophic environments like spacecraft assembly facilities and cleanrooms. This information is vital for mitigating microbial risks in robotic and crewed missions beyond low Earth orbit, where extreme conditions may impact microbial behavior, crew health, and life support systems. Furthermore, genomic and functional analyses of high-risk strains provide insights to develop targeted mitigation strategies. These findings have broader implications for astrobiology, space biology, and environmental monitoring, ensuring PP, mission success, and the preservation of scientific objectives.
This study highlights the necessity of modernizing PP policies to include non-spore-forming microbes in bioburden assessments, offering broader applications beyond spacecraft. Insights into microbial survival and adaptation to extreme conditions are relevant to contamination control in pharmaceutical, medical, and semiconductor industries, enhancing sterility, product integrity, and operational safety.
Data availability statement
The sequencing data generated in this study have been deposited in the NCBI Sequence Read Archive (SRA) under BioProject accession PRJNA1211674. The raw sequencing reads for the four microbial genomes are available under the following SRA accessions: Strain PPS72: SRX27599488; Strain PPS68: SRX27599487; Strain PPS120: SRX27599486, Strain PPS117: SRX27599485.
Author contributions
CC: Writing – original draft, Funding acquisition, Writing – review & editing, Resources, Investigation, Methodology, Supervision, Data curation, Project administration, Conceptualization. AC: Writing – original draft, Writing – review & editing, Formal analysis, Methodology, Conceptualization, Investigation, Data curation. JV: Supervision, Writing – review & editing, Methodology, Investigation, Resources. KK: Formal analysis, Methodology, Writing – review & editing, Data curation, Investigation. SG: Formal analysis, Methodology, Supervision, Project administration, Conceptualization, Investigation, Writing – review & editing, Resources. KV: Conceptualization, Writing – review & editing, Supervision, Methodology, Investigation, Writing – original draft, Project administration, Funding acquisition, Resources. PB: Conceptualization, Methodology, Investigation, Resources, Writing – review & editing. CB: Data curation, Investigation, Writing – review & editing. SM: Writing – review & editing, Data curation, Investigation. HM: Investigation, Data curation, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. The research described in this manuscript was partially funded by Amentum Space Exploration Division Technology Innovation and Process Improvement grants: C803-Molecular Identification of Microbial Contaminants and Innovation Lab Microbiology Equipment grant and C903-Space Environmental Effects on Clean Room Microbes grant to CDC. It was also partially funded by the NASA ROSES grant NNH18ZDA001N-PPR award 18-PPR18-0011 to KV.
Acknowledgments
The authors would like to thank the Innovation Grant committee at Amentum Space Exploration Division for their support of this work, specifically Roland Bruyns, Cyndi Russell, and Jeff Haars. We would also like to thank the Office of Planetary Protection at NASA Headquarters for their funding to support travel and sequencing efforts at JSC, specifically Nick Benardini, Elaine Seasly, and Andy Spry. We would like to thank the group at Johnson Space Center who planned and assisted us with our sequencing efforts, including Sarah Wallace, Aaron Regberg, Hang Nguyen, Sarah Rommel, and Christian Castro. We would also like to thank Todd Schneider, Erin Hayward, and Jarvis Caffrey, Mary Nehls, and Lee Allen at NASA MSFC for their support, suggestions, and guidance on radiation experiments. Finally, we thank Surabhi Naik from Rush University for her contributions to the FAST ANI analysis.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1600106/full#supplementary-material
References
Adiguzel, A., Ay, H., Baltaci, M. O., Akbulut, S., Albayrak, S., and Omeroglu, M. A. (2020). Genome-based classification of Calidifontibacillus erzurumensis gen. Nov., sp. nov., isolated from a hot spring in Turkey, with reclassification of Bacillus azotoformans as Calidifontibacillus azotoformans comb. nov. and Bacillus oryziterrae as Calidifontibacillus oryziterrae comb. nov. Int. J. Syst. Evol. Microbiol. 70, 6418–6427. doi: 10.1099/IJSEM.0.004549
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. doi: 10.1093/NAR/25.17.3389
Asgarani, E., Soudi, M. R., Borzooee, F., and Dabbagh, R. (2012). Radio-resistance in psychrotrophic Kocuria sp. ASB 107 isolated from ab-e-Siah radioactive spring. J. Environ. Radioact. 113, 171–176. doi: 10.1016/J.JENVRAD.2012.04.009
Ash, C., Priest, F. G., and Collins, M. D. (1993). Molecular identification of rRNA group 3 bacilli (Ash, farrow, Wallbanks and Collins) using a PCR probe test. Proposal for the creation of a new genus Paenibacillus. Antonie Van Leeuwenhoek 64, 253–260. doi: 10.1007/BF00873085
Bagwell, C. E., Bhat, S., Hawkins, G. M., Smith, B. W., Biswas, T., Hoover, T. R., et al. (2008). Survival in nuclear waste, extreme resistance, and potential applications gleaned from the genome sequence of Kineococcus radiotolerans SRS30216. PLoS One 3:e3878. doi: 10.1371/JOURNAL.PONE.0003878
Beblo-Vranesevic, K., Galinski, E. A., Rachel, R., Huber, H., and Rettberg, P. (2017). Influence of osmotic stress on desiccation and irradiation tolerance of (hyper)-thermophilic microorganisms. Arch. Microbiol. 199, 17–28. doi: 10.1007/S00203-016-1269-6
Beetham, C. M., Schuster, C. F., Kviatkovski, I., Santiago, M., Walker, S., and Gründling, A. (2024). Histidine transport is essential for the growth of Staphylococcus aureus at low pH. PLoS Pathog. 20:e1011927. doi: 10.1371/JOURNAL.PPAT.1011927
Bhattarai, H., Gupta, R., and Glickman, M. S. (2014). DNA ligase C1 mediates the LigD-independent nonhomologous end-joining pathway of Mycobacterium smegmatis. J Bacteriol 196, 3366–3376. doi: 10.1128/JB.01832-14/SUPPL_FILE/ZJB999093276SO1.PDF
Boch, J., Kempf, B., Schmid, R., and Bremer, E. (1996). Synthesis of the osmoprotectant glycine betaine in Bacillus subtilis: characterization of the gbsAB genes. J Bacteriol 178, 5121–5129. doi: 10.1128/JB.178.17.5121-5129.1996
Bogomolnaya, L. M., Andrews, K. D., Talamantes, M., Maple, A., Ragoza, Y., Vazquez-Torres, A., et al. (2013). The ABC-type efflux pump MacAB protects Salmonella enterica serovar typhimurium from oxidative stress. MBio 4, e630–13. doi: 10.1128/mBio.00630-13
Bruckbauer, S. T., Minkoff, B. B., Yu, D., Cryns, V. L., Cox, M. M., and Sussman, M. R. (2020). Ionizing radiation-induced proteomic oxidation in Escherichia coli. Mol. Cell. Proteomics 19, 1375–1395. doi: 10.1074/MCP.RA120.002092
Callegan, R. P., Noble, M. F., McTernan, P. M., Battista, J. R., Navarro-González, R., McKay, C. P., et al. (2008). Description of four novel psychrophilic, ionizing radiation-sensitive Deinococcus species from alpine environments. Int. J. Syst. Evol. Microbiol. 58, 1252–1258. doi: 10.1099/IJS.0.65405-0
Cao, G., Zhang, M., Miao, J., Li, W., Wang, J., Lu, D., et al. (2015). Effects of X-ray and carbon ion beam irradiation on membrane permeability and integrity in Saccharomyces cerevisiae cells. J. Radiat. Res. 56, 294–304. doi: 10.1093/JRR/RRU114
Cengel, K. A., Diffenderfer, E. S., Avery, S., Kennedy, A. R., and McDonough, J. (2010). Using electron beam radiation to simulate the dose distribution for whole body solar particle event proton exposure. Radiat. Environ. Biophys. 49, 715–721. doi: 10.1007/S00411-010-0315-Z
Chander, A. M., Nair, R. G., Kaur, G., Kochhar, R., Dhawan, D. K., Bhadada, S. K., et al. (2017). Genome insight and comparative Pathogenomic analysis of Nesterenkonia jeotgali strain CD08_7 isolated from duodenal mucosa of celiac disease patient. Front. Microbiol. 8:129. doi: 10.3389/FMICB.2017.00129
Chen, N. H., Djoko, K. Y., Veyrier, F. J., and McEwan, A. G. (2016). Formaldehyde stress responses in bacterial pathogens. Front. Microbiol. 7:257. doi: 10.3389/FMICB.2016.00257
Cortesão, M., de Haas, A., Unterbusch, R., Fujimori, A., Schütze, T., Meyer, V., et al. (2020). Aspergillus niger spores are highly resistant to space radiation. Front. Microbiol. 11:560. doi: 10.3389/FMICB.2020.00560
Cortesão, M., Fuchs, F. M., Commichau, F. M., Eichenberger, P., Schuerger, A. C., Nicholson, W. L., et al. (2019). Bacillus subtilis spore resistance to simulated mars surface conditions. Front. Microbiol. 10:333. doi: 10.3389/FMICB.2019.00333
Daly, M. J. (2009). A new perspective on radiation resistance based on Deinococcus radiodurans. Nat. Rev. Microbiol. 7, 237–245. doi: 10.1038/nrmicro2073
Dance, A. (2025). Microbes in space: how bacteria could help sustain long-distance space travel. Nature 638, 282–284. doi: 10.1038/D41586-025-00319-5
Dartnell, L. R., Hunter, S. J., Lovell, K. V., Coates, A. J., and Ward, J. M. (2010). Low-temperature ionizing radiation resistance of Deinococcus radiodurans and Antarctic Dry Valley bacteria. Astrobiology 10, 717–732. doi: 10.1089/AST.2009.0439
Deng, A., Wang, T., Wang, J., Li, L., Wang, X., Liu, L., et al. (2023). Adaptive mechanisms of Bacillus to near space extreme environments. Sci. Total Environ. 886:163952. doi: 10.1016/J.SCITOTENV.2023.163952
diCenzo, G. C., Sharthiya, H., Nanda, A., Zamani, M., and Finan, T. M. (2017). PhoU allows rapid adaptation to high phosphate concentrations by modulating PstSCAB transport rate in Sinorhizobium meliloti. J Bacteriol 199, e00143–17. doi: 10.1128/JB.00143-17
Dietz, A., and Mathews, J. (1971). Classification of Streptomyces spore surfaces into five groups. Appl. Microbiol. 21, 527–533. doi: 10.1128/AM.21.3.527-533.1971
Dworkin, J. P., Adelman, L. A., Ajluni, T., Andronikov, A. V., Aponte, J. C., Bartels, A. E., et al. (2017). OSIRIS-REx contamination control strategy and implementation. Space Sci. Rev. 214:19. doi: 10.1007/S11214-017-0439-4
Ferreira, A. C., Nobre, M. F., Moore, E., Rainey, F. A., Battista, J. R., and Da Costa, M. S. (1999). Characterization and radiation resistance of new isolates of Rubrobacter radiotolerans and Rubrobacter xylanophilus. Extremophiles 3, 235–238. doi: 10.1007/S007920050121
Fischer, N., and Kandt, C. (2013). Porter domain opening and closing motions in the multi-drug efflux transporter AcrB. Biochimica et Biophysica Acta (BBA) – Biomembranes 1828, 632–641. doi: 10.1016/J.BBAMEM.2012.10.016
Flowers, L., and Grice, E. A. (2020). The skin microbiota: balancing risk and reward. Cell Host Microbe 28, 190–200. doi: 10.1016/J.CHOM.2020.06.017
Ghosh, S., Osman, S., Vaishampayan, P., and Venkateswaran, K. (2010). Recurrent isolation of extremotolerant bacteria from the clean room where Phoenix spacecraft components were assembled. Astrobiology 10, 325–335. doi: 10.1089/AST.2009.0396
Guesmi, S., Nouioui, I., Pujic, P., Dubost, A., Najjari, A., Ghedira, K., et al. (2021a). Draft genome sequence of Promicromonospora panici sp. nov., a novel ionizing-radiation-resistant actinobacterium isolated from roots of the desert plant Panicum turgidum. Extremophiles 25, 25–38. doi: 10.1007/S00792-020-01207-8
Guesmi, S., Pujic, P., Nouioui, I., Dubost, A., Najjari, A., Ghedira, K., et al. (2021b). Ionizing-radiation-resistant Kocuria rhizophila PT10 isolated from the Tunisian Sahara xerophyte Panicum turgidum: Polyphasic characterization and proteogenomic arsenal. Genomics 113, 317–330. doi: 10.1016/J.YGENO.2020.11.029
Gupta, R. S., Patel, S., Saini, N., and Chen, S. (2020). Robust demarcation of 17 distinct Bacillus species clades, proposed as novel Bacillaceae genera, by phylogenomics and comparative genomic analyses: description of Robertmurraya kyonggiensis sp. nov. and proposal for an emended genus Bacillus limiting it only to the members of the subtilis and Cereus clades of species. Int. J. Syst. Evol. Microbiol. 70, 5753–5798. doi: 10.1099/IJSEM.0.004475
Hase, Y., Satoh, K., Chiba, A., Hirano, Y., Moribayashi, K., and Narumi, K. (2021). Proton-cluster-beam lethality and mutagenicity in Bacillus subtilis spores. Quantum Beam Sci. 5:25. doi: 10.3390/QUBS5030025/S1
Hendrickson, R., Urbaniak, C., Minich, J. J., Aronson, H. S., Martino, C., Stepanauskas, R., et al. (2021). Clean room microbiome complexity impacts planetary protection bioburden. Microbiome 9, 1–17. doi: 10.1186/S40168-021-01159-X
Ho, J., Adeolu, M., Khadka, B., and Gupta, R. S. (2016). Identification of distinctive molecular traits that are characteristic of the phylum “Deinococcus-Thermus” and distinguish its main constituent groups. Syst. Appl. Microbiol. 39, 453–463. doi: 10.1016/J.SYAPM.2016.07.003
Hu, N., and Zhao, B. (2007). Key genes involved in heavy-metal resistance in Pseudomonas putida CD2. FEMS Microbiol. Lett. 267, 17–22. doi: 10.1111/J.1574-6968.2006.00505.X
Hu, T., Yang, X., Zhu, Y., Liu, F., Yang, X., Xiong, Z., et al. (2024). Molecular basis for substrate transport of Mycobacterium tuberculosis ABC importer DppABCD. Sci Adv 10, 8521. doi: 10.1126/SCIADV.ADK8521/SUPPL_FILE/SCIADV.ADK8521_MOVIE_S1.ZIP
Jain, C., Rodriguez-R, L. M., Phillippy, A. M., Konstantinidis, K. T., and Aluru, S. (2018). High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 9:5114. doi: 10.1038/S41467-018-07641-9
Kalogiannis, G., and Eyre-Walker, A. (2024). The effect of the presence and absence of DNA repair genes on the rate and pattern of mutation in Bacteria. Genome Biol. Evol. 16:evae216. doi: 10.1093/GBE/EVAE216
Kappes, R. M., Kempf, B., and Bremer, E. (1996). Three transport systems for the osmoprotectant glycine betaine operate in Bacillus subtilis: characterization of OpuD. J Bacteriol 178, 5071–5079. doi: 10.1128/JB.178.17.5071-5079.1996
Kern, R., Malki, A., Holmgren, A., and Richarme, G. (2003). Chaperone properties of Escherichia coli thioredoxin and thioredoxin reductase. Biochemical Journal 371, 965. doi: 10.1042/BJ20030093
Khodadad, C. L., Wong, G. M., James, L. M., Thakrar, P. J., Lane, M. A., Catechis, J. A., et al. (2017). Stratosphere conditions inactivate bacterial endospores from a mars spacecraft assembly facility. Astrobiology 17, 337–350. doi: 10.1089/AST.2016.1549
Kim, S., Oh, D. B., Kang, H. A., and Kwon, O. (2011). Features and applications of bacterial sialidases. Appl. Microbiol. Biotechnol. 91, 1–15. doi: 10.1007/S00253-011-3307-2
Kimura, S., Ishikawa, S., Hayashi, N., Fujita, K., Inatomi, Y., and Suzuki, S. (2023). Bacterial and fungal bioburden reduction on material surfaces using various sterilization techniques suitable for spacecraft decontamination. Front. Microbiol. 14:1253436. doi: 10.3389/FMICB.2023.1253436/FULL
KOPRI Repository: Investigation of the Susceptibility of Arctic Arthrobacter sp.PAMC 25486 to Mutagens (n.d.). Available online at: https://repository.kopri.re.kr/handle/201206/3810 (Accessed June 12, 2025).
Krishnamurthy, H. K., Rajavelu, I., Pereira, M., Jayaraman, V., Krishna, K., Wang, T., et al. (2024). Inside the genome: understanding genetic influences on oxidative stress. Front. Genet. 15:1397352. doi: 10.3389/FGENE.2024.1397352
Krisko, A., and Radman, M. (2013). Biology of extreme radiation resistance: the way of Deinococcus radiodurans. Cold Spring Harb. Perspect. Biol. 5:a012765. doi: 10.1101/CSHPERSPECT.A012765
Krokan, H. E., and Bjørås, M. (2013). Base Excision Repair. Cold Spring Harb Perspect Biol 5, a012583. doi: 10.1101/CSHPERSPECT.A012583
La Duc, M. T., Dekas, A., Osman, S., Moissl, C., Newcombe, D., and Venkateswaran, K. (2007). Isolation and characterization of bacteria capable of tolerating the extreme conditions of clean room environments. Appl. Environ. Microbiol. 73, 2600–2611. doi: 10.1128/AEM.03007-06
Lee, J. S., Lee, K. C., Pyun, Y. R., and Bae, K. S. (2003). Arthrobacter koreensis sp. nov., a novel alkalitolerant bacterium from soil. Int. J. Syst. Evol. Microbiol. 53, 1277–1280. doi: 10.1099/IJS.0.02492-0
Lenhart, J. S., Schroeder, J. W., Walsh, B. W., and Simmons, L. A. (2012). DNA repair and genome maintenance in Bacillus subtilis. Microbiol. Mol. Biol. Rev. 76, 530–564. doi: 10.1128/MMBR.05020-11
Logan, N. A., Berge, O., Bishop, A. H., Busse, H. J., De Vos, P., Fritze, D., et al. (2009). Proposed minimal standards for describing new taxa of aerobic, endospore-forming bacteria. Int. J. Syst. Evol. Microbiol. 59, 2114–2121. doi: 10.1099/IJS.0.013649-0
Lushchak, V. I. (2001). Oxidative stress and mechanisms of protection against it in bacteria. Biochemistry (Mosc) 66, 476–489. doi: 10.1023/A:1010294415625
Maeda, S. I., Murakami, A., Ito, H., Tanaka, A., and Omata, T. (2015). Functional characterization of the FNT family Nitrite transporter of marine Picocyanobacteria. Life, 5, 432–446. doi: 10.3390/LIFE5010432
Mahnert, A., Vaishampayan, P., Probst, A. J., Auerbach, A., Moissl-Eichinger, C., Venkateswaran, K., et al. (2015). Cleanroom maintenance significantly reduces abundance but not diversity of indoor microbiomes. PLoS One 10:e0134848. doi: 10.1371/JOURNAL.PONE.0134848
Manzanera, M., Narváez-Reinaldo, J. J., García-Fontana, C., Vílchez, J. I., and González-López, J. (2015). Genome sequence of Arthrobacter koreensis 5J12A, a plant growth-promoting and desiccation-tolerant strain. Genome Announc. 3:e00648-15. doi: 10.1128/GENOMEA.00648-15
Marienhagen, J. (2025). Engineering of Corynebacterium glutamicum for the synthesis of aromatic compounds. Applied Microbiology and Biotechnology 2025, 1–22. doi: 10.1007/S00253-025-13520-3
Mattimore, V., and Battista, J. R. (1996). Radioresistance of Deinococcus radiodurans: functions necessary to survive ionizing radiation are also necessary to survive prolonged desiccation. J. Bacteriol. 178, 633–637. doi: 10.1128/jb.178.3.633-637.1996
Moeller, R., Reitz, G., Li, Z., Klein, S., and Nicholson, W. L. (2012). Multifactorial resistance of Bacillus subtilis spores to high-energy proton radiation: role of spore structural components and the homologous recombination and non-homologous end joining DNA repair pathways. Astrobiology 12, 1069–1077. doi: 10.1089/AST.2012.0890
Moissl-Eichinger, C., Rettberg, P., and Pukall, R. (2012). The first collection of spacecraft-associated microorganisms: a public source for extremotolerant microorganisms from spacecraft assembly clean rooms. Astrobiology 12, 1024–1034. doi: 10.1089/AST.2012.0906
Montero-Calasanz, M. d. C., Hezbri, K., Göker, M., Sghaier, H., Rohde, M., Spröer, C., et al. (2015). Description of gamma radiation-resistant Geodermatophilus dictyosporus sp. nov. to accommodate the not validly named Geodermatophilus obscurus subsp. dictyosporus (Luedemann, 1968). Extremophiles 19, 77–85. doi: 10.1007/S00792-014-0708-Z
Olsson-Francis, K., Doran, P. T., Ilyin, V., Raulin, F., Rettberg, P., Kminek, G., et al. (2023). The COSPAR planetary protection policy for robotic missions to Mars: a review of current scientific knowledge and future perspectives. Life Sci. Space Res. 36, 27–35. doi: 10.1016/J.LSSR.2022.12.001
Omote, H., Hiasa, M., Matsumoto, T., Otsuka, M., and Moriyama, Y. (2006). The MATE proteins as fundamental transporters of metabolic and xenobiotic organic cations. Trends Pharmacol Sci 27, 587–593. doi: 10.1016/j.tips.2006.09.001
Osman, S., Peeters, Z., La Duc, M. T., Mancinelli, R., Ehrenfreund, P., and Venkateswaran, K. (2007). Effect of shadowing on survival of bacteria under conditions simulating the Martian atmosphere and UV radiation. Appl. Environ. Microbiol. 74:959. doi: 10.1128/AEM.01973-07
Osman, D., Piergentili, C., Chen, J., Sayer, L. N., Usón, I., Huggins, T. G., et al. (2016). The effectors and sensory sites of formaldehyde-responsive regulator FrmR and metal-sensing variant. J. Biol. Chem. 291, 19502–19516. doi: 10.1074/JBC.M116.745174
Park, J., Kim, M. K., Yun, B. R., Han, J. H., and Kim, S. B. (2018). Pseudogracilibacillus endophyticus sp. nov., a moderately thermophilic and halophilic species isolated from plant root. Int. J. Syst. Evol. Microbiol. 68, 165–169. doi: 10.1099/IJSEM.0.002475
Philippot, L., Chenu, C., Kappler, A., Rillig, M. C., and Fierer, N. (2023). The interplay between microbial communities and soil properties. Nat. Rev. Microbiol. 22, 226–239. doi: 10.1038/s41579-023-00980-5
Popovich, K. J., Green, S. J., Okamoto, K., Rhee, Y., Hayden, M. K., Schoeny, M., et al. (2021). MRSA transmission in intensive care units: genomic analysis of patients, their environments, and healthcare workers. Clin. Infect. Dis. 72, 1879–1887. doi: 10.1093/CID/CIAA731
Rajeev, L., Luning, E. G., Altenburg, S., Zane, G. M., Baidoo, E. E. K., Catena, M., et al. (2014). Identification of a cyclic-di-GMP-modulating response regulator that impacts biofilm formation in a model sulfate reducing bacterium. Front. Microbiol. 5:382. doi: 10.3389/FMICB.2014.00382
Ramnarine, S. D. B., Ali, O., Jayaraman, J., and Ramsubhag, A. (2024). Early transcriptional changes of heavy metal resistance and multiple efflux genes in Xanthomonas campestris pv. Campestris under copper and heavy metal ion stress. BMC Microbiol. 24:81. doi: 10.1186/S12866-024-03206-7
Regberg, A. B., Castro, C. L., Connolly, H. C., Davis, R. E., Dworkin, J. P., Lauretta, D. S., et al. (2020). Prokaryotic and fungal characterization of the facilities used to assemble, test, and launch the OSIRIS-REx spacecraft. Front. Microbiol. 11:530661. doi: 10.3389/FMICB.2020.530661
Rodríguez, M., Reina, J. C., Béjar, V., and Llamas, I. (2020). Psychrobacillus vulpis sp. nov., a new species isolated from faeces of a red fox in Spain. Int. J. Syst. Evol. Microbiol. 70, 882–888. doi: 10.1099/IJSEM.0.003840
Schoch, C. L., Seifert, K. A., Huhndorf, S., Robert, V., Spouge, J. L., Levesque, C. A., et al. (2012). Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc. Natl. Acad. Sci. USA 109, 6241–6246. doi: 10.1073/PNAS.1117018109
Schuerger, A. C. (2024). Synergistic interactions among vacuum, solar heating, and UV irradiation enhance the lethality of interplanetary space. Microorganisms 12:1976. doi: 10.3390/MICROORGANISMS12101976
Schuerger, A. C., Mancinelli, R. L., Kern, R. G., Rothschild, L. J., and McKay, C. P. (2003). Survival of endospores of Bacillus subtilis on spacecraft surfaces under simulated martian environments: implications for the forward contamination of Mars. Icarus 165, 253–276. doi: 10.1016/S0019-1035(03)00200-8
Schuerger, A. C., and Moores, J. E. (2023). UV reflectance of spacecraft materials and analog soils: implications for bioburden reductions on the undersides of Mars rovers. J. Geophys. Res. Planets 128:e2023JE007975. doi: 10.1029/2023JE007975
Shen, L., Liu, Y., Yao, T., Kang, S., Wang, Y., Jiao, N., et al. (2013). Mycetocola zhadangensis sp. nov., isolated from snow. Int. J. Syst. Evol. Microbiol. 63, 3375–3378. doi: 10.1099/IJS.0.047159-0
Shida, O., Takagi, H., Kadowaki, K., and Komagata, K. (1996). Proposal for two new genera, Brevibacillus gen. Nov. and Aneurinibacillus gen. Nov. Int. J. Syst. Bacteriol. 46, 939–946. doi: 10.1099/00207713-46-4-939
Shivaji, S., Chaturvedi, P., Begum, Z., Pindi, P. K., Manorama, R., Padmanaban, D. A., et al. (2009). Janibacter hoylei sp. nov., Bacillus isronensis sp. nov. and Bacillus aryabhattai sp. nov., isolated from cryotubes used for collecting air from the upper atmosphere. Int. J. Syst. Evol. Microbiol. 59, 2977–2986. doi: 10.1099/IJS.0.002527-0
Sinha, A. K., Løbner-Olesen, A., and Riber, L. (2020). Bacterial chromosome replication and DNA repair during the stringent response. Front. Microbiol. 11:582113. doi: 10.3389/FMICB.2020.582113
Smith, D. J., Schuerger, A. C., Davidson, M. M., Pacala, S. W., Bakermans, C., and Onstott, T. C. (2009). Survivability of Psychrobacter cryohalolentis K5 under simulated martian surface conditions. Astrobiology 9, 221–228. doi: 10.1089/AST.2007.0231
Srivastava, S. K., Ruigrok, V. J. B., Thompson, N. J., Trilling, A. K., Heck, A. J. R., van Rijn, C., et al. (2013). 16 kDa heat shock protein from heat-inactivated Mycobacterium tuberculosis is a homodimer - suitability for diagnostic applications with specific llama VHH monoclonals. PLoS One 8:e64040. doi: 10.1371/JOURNAL.PONE.0064040
Stieglmeier, M., Wirth, R., Kminek, G., and Moissl-Eichinger, C. (2009). Cultivation of anaerobic and facultatively anaerobic bacteria from spacecraft-associated clean rooms. Appl. Environ. Microbiol. 75, 3484–3491. doi: 10.1128/AEM.02565-08
Tarpley, W., Ilavsky, J., Manowitz, B., and Horrigan, R. V. (1953). Radiation Sterilization I.: the effect of high energy gamma radiation from kilocurie radioactive sources on Bacteria. J. Bacteriol. 65, 305–309. doi: 10.1128/JB.65.3.305-309.1953
Trilling, A. K., de Ronde, H., Noteboom, L., van Houwelingen, A., Roelse, M., Srivastava, S. K., et al. (2011). A broad set of different llama antibodies specific for a 16 kDa heat shock protein of Mycobacterium tuberculosis. PLoS One 6:e26754. doi: 10.1371/JOURNAL.PONE.0026754
Vaishampayan, P., Moissl-Eichinger, C., Pukall, R., Schumann, P., Sproöer, C., Augustus, A., et al. (2013). Description of Tersicoccus phoenicis gen. nov., sp. nov. isolated from spacecraft assembly clean room environments. Int. J. Syst. Evol. Microbiol. 63, 2463–2471. doi: 10.1099/IJS.0.047134-0
Vaishampayan, P., Roberts, A. H., Augustus, A., Pukall, R., Schumann, P., Schwendner, P., et al. (2014). Deinococcus phoenicis sp. nov., an extreme ionizing-radiation-resistant bacterium isolated from the Phoenix Lander assembly facility. Int. J. Syst. Evol. Microbiol. 64, 3441–3446. doi: 10.1099/IJS.0.063107-0
Vallalar, B. (2012). Investigation of the growth and survival of bacteria from Mars analog environments when exposed to Mars-like conditions : Louisiana State University Master’s Thesis.
Verbeelen, T., Fernandez, C. A., Nguyen, T. H., Gupta, S., Leroy, B., Wattiez, R., et al. (2024). Radiotolerance of N-cycle bacteria and their transcriptomic response to low-dose space-analogue ionizing irradiation. iScience 27:109596. doi: 10.1016/J.ISCI.2024.109596
Xu, X., Chen, J., Huang, X., Feng, S., Zhang, X., She, F., et al. (2021). The role of a Dipeptide transporter in the virulence of human pathogen, Helicobacter pylori. Front Microbiol 12, 633166. doi: 10.3389/FMICB.2021.633166/BIBTEX
Keywords: whole genome sequencing, radiation, planetary protection, non-spore-forming bacteria, Pan-Resistomics
Citation: Cassilly CD, Chander AM, Vaughn JA, Kunstman KJ, Green SJ, Venkateswaran K, Bertone PF, Bahr CW, Marcella SA and Morris HC (2025) Effects of simulated space environmental conditions on cleanroom microbes. Front. Microbiol. 16:1600106. doi: 10.3389/fmicb.2025.1600106
Edited by:
Wolfram Weckwerth, University of Vienna, AustriaReviewed by:
Rubens Tadeu Delgado Duarte, Federal University of Santa Catarina, BrazilBruno Fosso, University of Bari Aldo Moro, Italy
Copyright © 2025 Cassilly, Chander, Vaughn, Kunstman, Green, Venkateswaran, Bertone, Bahr, Marcella and Morris. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chelsi D. Cassilly, Q2hlbHNpLmQuY2Fzc2lsbHlAbmFzYS5nb3Y=
‡ORCID: Chelsi D. Cassilly, orcid.org/0000-0001-9611-7438
Atul M. Chander, orcid.org/0000-0001-9582-6638
Kasthuri Venkateswaran, orcid.org/0000-0002-6742-0873
†These authors have contributed equally to this work and share first authorship
 Chelsi D. Cassilly
Chelsi D. Cassilly Atul M. Chander2†‡
Atul M. Chander2†‡ Stefan J. Green
Stefan J. Green