- 1College of Plant Protection, Northeast Agricultural University, Harbin, China
- 2State Key Laboratory of Green Pesticide, Key Laboratory of Green Pesticide and Agricultural Bioengineering, Ministry of Education, Guizhou University, Guiyang, China
The Potyviridae family is one of the most economically significant groups of plant RNA viruses, causing severe yield losses in agriculturally important crops. Among the viral proteins encoded by potyviruses, the 6-kilodalton peptide 1 (6K1) has emerged as a critical, albeit poorly understood player in viral pathogenesis. Despite its small size, 6K1 exhibits diverse functions, including facilitating the assembly of viral replication complex (VRC), altering host membrane permeability as a viroporin, and interacting with host factors to promote infection. This review synthesizes current knowledge on 6K1, focusing on its structural characteristics, evolutionary conservation, molecular interactions, and potential as a target for antiviral strategies. We further discuss unresolved questions surrounding its putative ion channel activity, polyprotein processing dynamics, and functional parallels with animal virus viroporins. Understanding 6K1’s multifunctionality provides new insights into viral infection mechanisms and opens avenues for novel disease control approaches.
1 Introduction
Viral diseases represent a major threat to global food security and sustainable agriculture. The Potyviridae family is the largest family of plant RNA viruses, comprising at least 228 species grouped into 12 genera (Martínez-Turiño and García, 2020; Yang et al., 2021; Jaramillo-Mesa and Rakotondrafara, 2023). Viruses in the largest genus, Potyvirus, such as potato virus Y (PVY; Potyvirus yituberosi), soybean mosaic virus (SMV; P. glycitessellati), tobacco etch virus (TEV; P. nicotianainsculpentis), bean common mosaic virus (BCMV; P. phaseoli), maize dwarf mosaic virus (MDMV; P. zaenanus), turnip mosaic virus (TuMV; P. rapae), plum pox virus (PPV; P. plumpoxi), sweet potato feathery mottle virus (SPFMV; P. batataplumei), and sugarcane mosaic virus (SCMV; P. sacchari), which cause significant economic yield losses in crops of the families Solanaceae, Leguminosae, and Chenopodiaceae worldwide. For instance, PVY alone can reduce potato yields by 20–80% during severe outbreaks (Yang et al., 2021; Wani et al., 2023; Liu et al., 2023; Belabess et al., 2024; Kumar and Dasgupta, 2024; Mardanova et al., 2024).
Members of the Potyviridae family (potyvirids) induce a range of symptoms that affect plant growth and yield, including leaf shrinkage, necrosis, mottling, yellowing, and plant stunting. They also reduce the quality of fruits and tubers and increase susceptibility to other phytopathogens (Yang et al., 2021; Luan et al., 2024; Qin et al., 2024; Yang et al., 2024; Dupuis et al., 2024; Kamran et al., 2025). In addition, virus infection compromises the storage quality of potatoes, imposing an economic burden on farmers (Yang et al., 2021; Kamran et al., 2025; Dupuis et al., 2024). The severity of potyviral infection is exacerbated by their diverse transmission modes, including aphids, mechanical fiction, fungus, and seed, which complicates efforts for disease prevention and control (Crosslin, 2013; Gadhave et al., 2020; Bhoi et al., 2022). Therefore, understanding the biological characteristics and mechanisms of their pathogenesis are crucial to developing effective prevention and control strategies.
The genome of potyvirids consists of one or two positive-sense single-stranded RNAs (+ssRNA) of approximately 10 kilobases (kb) in length in total, that is covalently linked to VPg at the 5′ end and polyadenylated at the 3′ end (Yang et al., 2021). Each genomic +ssRNA encodes a long open reading frame (ORF) that is translated into a polypeptide. Typically, potyvirids encode 10 multifunctional proteins, namely, First Protein (P1), Helper Component-Proteinase (HC-Pro), Third Protein (P3), 6-kilodalton peptide 1 (6K1), Cylindrical Inclusion (CI), 6-kilodalton peptide 1 (6K2), Viral Protein Genome-linked (VPg), Nuclear Inclusion a-Protease (NIa-Pro), Nuclear Inclusion b (NIb), and Coat Protein (CP) (Yang et al., 2021; Hýsková et al., 2024). In addition, all potyvirids contain an RNA polymerase slippage motif within P3 cistron, enabling the expression of an additional polypeptide that is cleaved into P1, HC-Pro, and P3 N-terminal fused with Pretty Interesting Potyviridae ORF (P3N-PIPO). A few sweet-potato-infecting potyvirids also possess a similar slippage motif within the P1 cistron, leading to the translation of an additional protein, P1 N-terminus fused with the Pretty Interesting Sweet Potato potyviral ORF (P1N-PISPO) (Yang et al., 2021; Jaramillo-Mesa and Rakotondrafara, 2023; Valli et al., 2024). Emerging evidence suggests that the complementary RNA strand of Potyviridae has protein-coding capacity, challenging the traditional view of their unidirectional genomic expression and enriching our understanding of viral coding potential and pathogenicity (Gong et al., 2021; Gong et al., 2023; Li et al., 2024; Gong et al., 2025). Among these proteins, 6K1 remains one of the least understood viral proteins. It is highly conserved in potyvirids and located between P3 and CI cistrons (Cui and Wang, 2016; Hu et al., 2023a,b). Recent studies reveal that 6K1 functions as a viroporin, which plays a critical role in viral infection. This discovery provides new insights into the molecular mechanisms of potyviral infection (Chai et al., 2024). This review summarized the structural and functional properties of 6K1, discussed its role in viral replication and membrane remodeling, explored interactions with host factors and immune evasion strategies, evaluated evolutionary adaptations across Potyviridae, and assessed potential antiviral strategies targeting 6K1.
2 Structural and biochemical properties of 6K1
2.1 Protein characteristics and subcellular localization
The 6K1 protein is located between the P3 and CI cistrons in all known potyvirids with sizes ranging from 6 to 7 kilodalton (kDa). Bioinformatics analyses suggest that 6K1 contains two transmembrane helices (TMH1 and TMH2) (Chai et al., 2024). TMH1 is highly hydrophobic, and is primarily composed of the amino acids Ser, Glu, Ile, Phe, Val, and Phe, while TMH2 is less hydrophobic and contains several highly conserved basic residues, e.g., His and Lys, which comprises a so-called K/R-rich motif (Chai et al., 2024; Hu et al., 2025). Alphafold-assistant modeling suggests that the two transmembrane helices adopt a helix-turn-helix structure. The cleavage site between P3 and 6K1 in many, if not all potyvirids, is not favor for NIa-Pro cleavage, leading to the coexistence of free 6K1 and P3-6K1 fusion proteins in virus-infected cells, which highlights the functional versatility of 6K1 in viral infection (Riechmann et al., 1995; Cui and Wang, 2016).
It is widely accepted that 6K1, together with P3 and 6K2, constitutes three potyviral membrane-associated proteins (Cui and Wang, 2016; Bera et al., 2022; Hu et al., 2025). Immunogold labeling using polyclonal antiserum revealed that soybean mosaic virus (SMV)-encoded 6K1 predominately localizes at the cell periphery (Hu et al., 2023a,b). Transient expression of 6K1 from TuMV and PPV as C-terminal YFP-tagged recombinant proteins in Nicotiana benthamiana ephemeral cell showed cytosolic and nuclear localization, while N-terminal YFP-tagged TuMV 6K1 displayed membrane-associated subcellular distribution and colocalized with the endoplasmic reticulum (ER) network. By contrast, P3-6K1 fusion protein appeared as small granules localized to the ER network (Riechmann et al., 1995; Cui and Wang, 2016; Chai et al., 2024; Hu et al., 2025). In the context of virus infection, the subcellular localized of C-terminal YFP-tagged PPV 6K1 was analyzed by expressing an additional copy between P1 and HC-Pro (Riechmann et al., 1995; Cui and Wang, 2016; Chai et al., 2024; Hu et al., 2025). Results showed that PPV 6K1 forms punctuate membrane-associated granules and colocalizes with viral replication complexes (VRCs) adjacent to chloroplasts. These findings underscore the multifunctional role of 6K1 in viral life cycle and suggest that it serve as critical regulatory hubs for coordinating viral processes (Cui and Wang, 2016).
2.2 Function as a viroporin
Alphafold–assisted structure modeling and biochemical assays suggest that TuMV and PVY 6K1 forms pentamers with a central hydrophobic cavity, resembling viroporins, a specialized group of virus-encoded small ion channels (Chai et al., 2024). Decades ago, researchers observed increased permeability to ions and small molecules of cells infected with animal viruses, but it was not until 1995 that Louis Carrasco coined the term “viroporins” to describe viral proteins with ion channel activity (Carrasco, 1995). According to the classification criteria proposed by Devantier et al. (2024a,b) and Nieva et al. (2012), viroporins are divided into two major classes (I and II) based on the number of TMHs) (Figure 1a). Furthermore, depending on the cytoplasmic and organellar accessibility of their N- and C-terminal domains, viroporins are further divided into two subtypes A and B (Pielak and Chou, 2011; Nieva et al., 2012; Devantier et al., 2024a,b; Gebert et al., 2024). Recently, a class III viroporin category has been proposed (Pielak and Chou, 2011; Nieva et al., 2012; Devantier et al., 2024a,b; Gebert et al., 2024). Structural predictions of the TuMV 6K1 pentamer indicate that 6K1 adopts a type II viroporins classification, characterized by two TMHs. In addition, a topology assay demonstrated that the N-terminus of 6K1 is exposed to the cytosol (Chai et al., 2024), confirming that 6K1 belongs to the class IIB subtype viroporins.
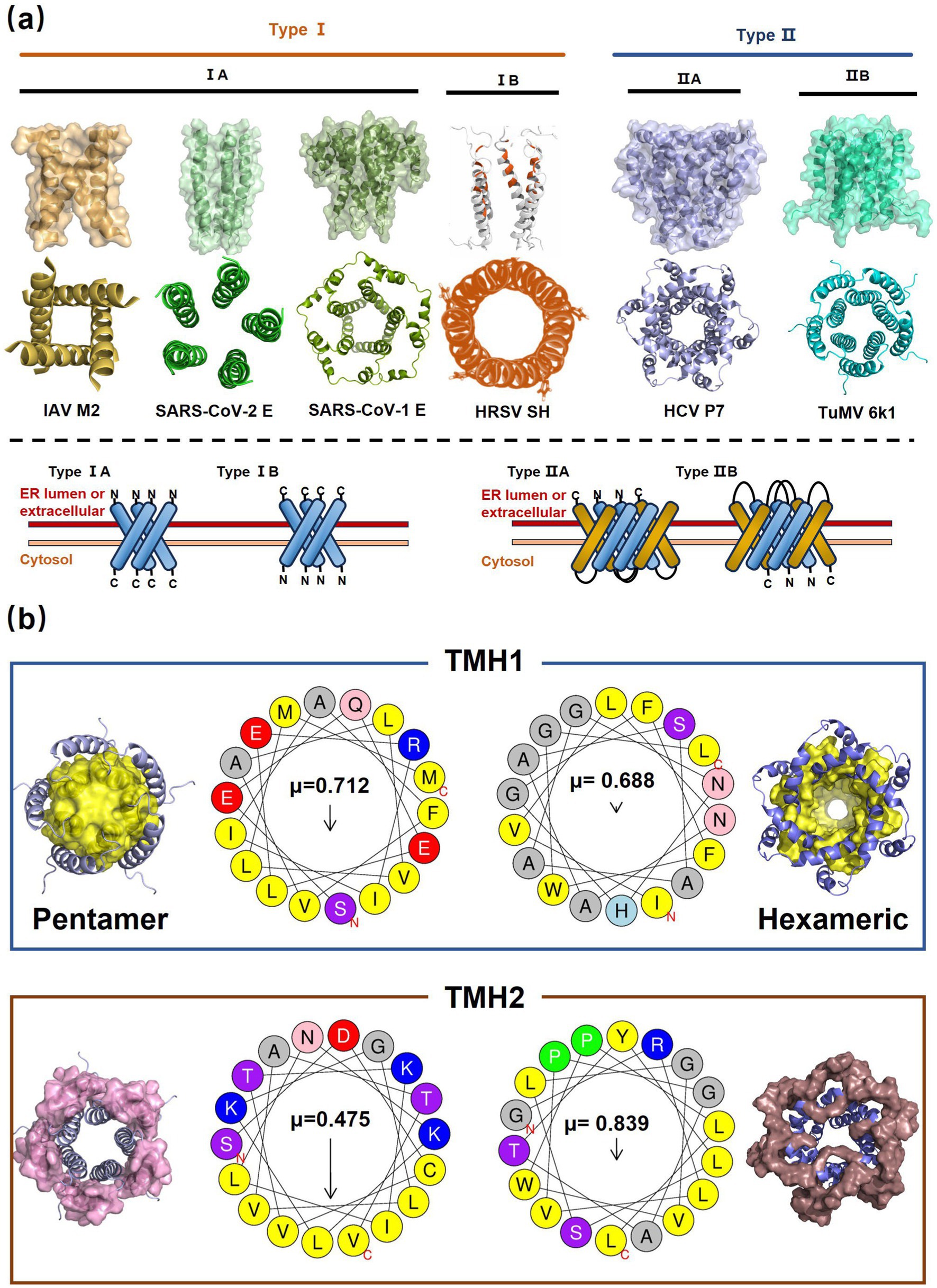
Figure 1. The structure of viroporins. (a) Schematic classification of viroporins. IA and IB viroporins contain one TMH with the C terminus in the cytosol and endoplasmic reticulum (ER) lumen or extracellular, respectively; IIA and IIB viroporins contain two TMHs with both N- and C-termini in the cytosol and ER lumen or extracellular, respectively. Type III viroporins contain three TMHs and are not illustrated due to lack of structure; Influenza virus A M2 (IAV M2; PDB ID: 4QKM); Syndrome coronavirus 2 E peptide (SARS-COV-2 E; PDB ID: 8SUZ); Syndrome coronavirus 1 E peptide (SARS-COV-1 E; PDB ID: 5 × 29);Human respiratory syncytial virus SH (HRSV SH); Hepatitis C virus P7 (HCV P7; PDB ID: 2M6X); (b) Structural comparison of TuMV 6K1 and HCV P7. The helical wheels showing TMH 1 (top panel) and TMH 2 (top panel) of the putative pentamer of TuMV 6K1 and HCV P7 hexamer, generated using HeliQuest (numbers indicate the hydrophobic moment). The cartoon views of the TMH 1 (top panel) and TMH 2 (top panel) of the putative pentamer of TuMV 6K1 and HCV P7 hexamer are also shown.
TuMV 6K1 forms oligomers via self-interaction on the ER and can enhances membrane permeability in Escherichia coli and N. benthamiana. Additionally, it complements the growth of yeast mutants deficient in potassium ion channels on low-potassium medium (Chai et al., 2024; Gao et al., 2024). In addition to TuMV 6K1, other potyviral 6K1 proteins and their cognate 7K counterparts similarly alter membrane permeability, exhibiting toxicity in E. coli (Chai et al., 2024; Gao et al., 2024). Notably, both TuMV 6K1 and hepatitis C virus (HCV) P7 facilitate the uptake of the macromolecular dye Sytox green when expressed in N. benthamiana leaves (Figure 2a) (Chai et al., 2024; Gao et al., 2024), confirming their role in modulating membrane permeability. Viroporins exhibit significant diversity in amino acid composition and ionic selectivity. For instance, severe acute respiratory syndrome coronavirus-1 (SARS-CoV-1) E, HCV P7, human immunodeficiency virus-1 Vpu (HIV1 Vpu), and ross river virus 6K (RRV 6K) display high selectivity for Na+ and K+, but low affinity for Cl− (Melton et al., 2002; Premkumar et al., 2004; Wilson et al., 2004; Surya and Torres, 2022; Surya et al., 2023). SARS-CoV-1 E shows a 5- to 10-fold preference for Na+ over K+ and an 10-fold preference for K+ over Cl− (Surya et al., 2023; Surya and Torres, 2022; Wilson et al., 2004; Melton et al., 2002). IAV M2 is proton-selective, mediating virion acidification, viral envelope-organelle membrane fusion, and cytoplasmic virion release (Lakadamyali et al., 2003; Xia et al., 2022). TuMV 6K1 and barley yellow striate mosaic virus (BYSMV; Cytorhabdovirus hordei) P7 also exhibit K+-selective permeability (Chai et al., 2024; Gao et al., 2024). Despite these functional parallels, potyviral 6K1 proteins share no detectable amino acid sequence homology with known viroporins. The specific ion selectivity of potyviral 6K1 remain unresolved, primarily due to the absence of electrophysiological evidence.
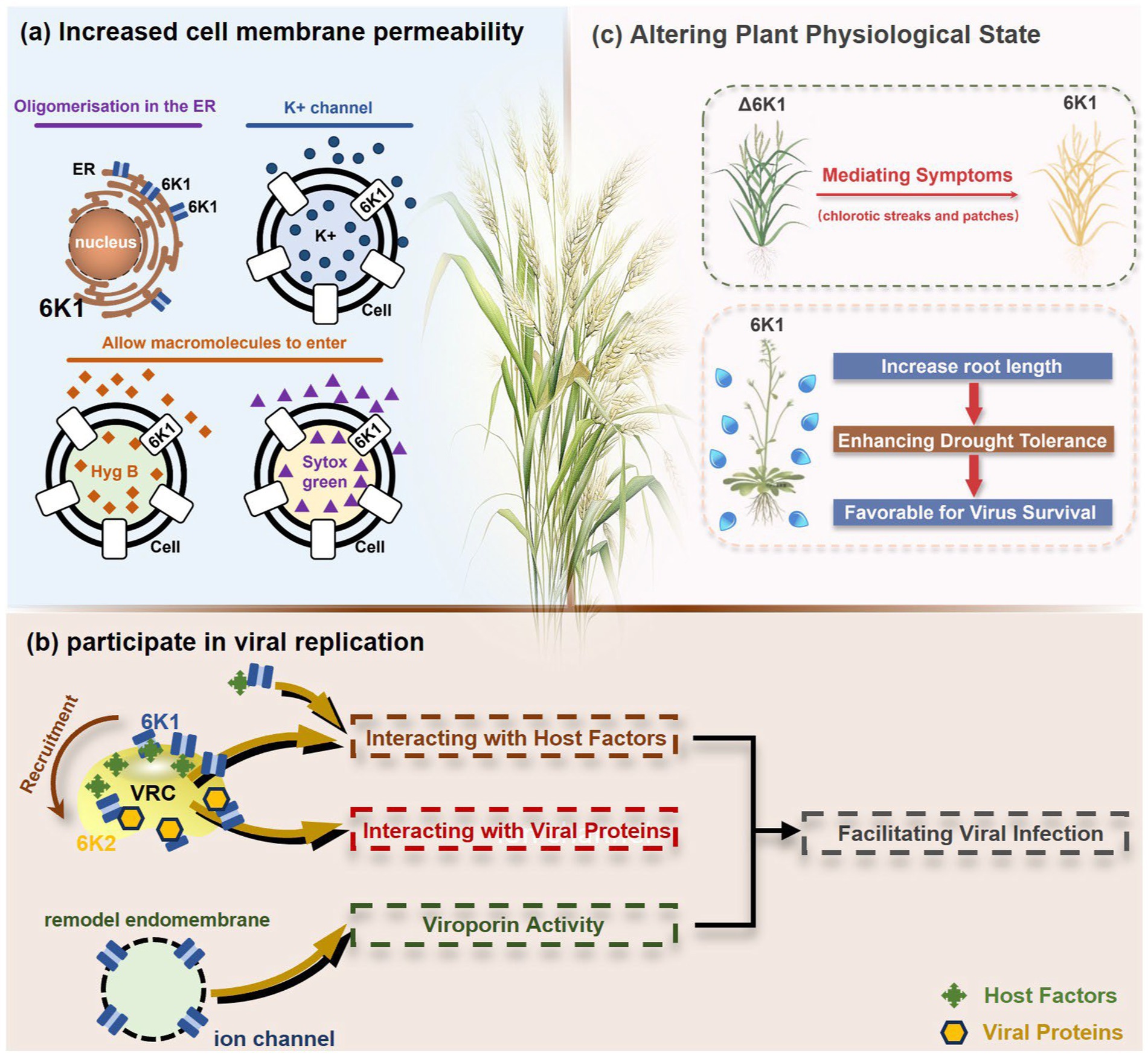
Figure 2. The illustration of 6K1 function during viral infection. (a) 6K1 localizes to ER and form oligomers. It increases membrane permeability, enabling the uptake of Hygromycin B and Sytox Green and enhances potassium ion flux. (b) 6K1 is recruited to VRCs to facilitate viral infection possibly via interacting with host and viral proteins and its viroporin activity. (c) 6K1 adds in viral symptom development, promotes root elongation and improves drought tolerance.
3 Functional roles of 6K1 in viral infection
3.1 Coordinates VRC assembly and participates in replication
6K1 plays a central role in viral replication (Figure 2b) (Chai et al., 2024; Cui and Wang, 2016; Cui and Wang, 2019). In the early stages of viral infection, 6K1 of PPV forms punctate structures and colocalizes with VRCs induced by 6K2 from ER (Cui and Wang, 2016). During tobacco vein banding mosaic virus (TVBMV) infection, 6K2 recruits 6K1 to the VRCs near the chloroplasts, where it colocalizes with NIb, the viral RNA-dependent RNA polymerase (RdRp) (Geng et al., 2017). Deletion or functional defects of pepper veinal mottle virus (PVMV; P. capsivenae)-encoded 6K1 result in a substantial reduction in viral replication (Hu et al., 2025). Recent studies on PPV infection have found that 6K1, 6K2, and NIb colocalize and jointly coordinate the assembly of VRCs during the early stages of infection (Cui and Wang, 2016). Importantly, disruption of the N- or C-terminal cleavage sites of 6K1 prevents its proteolytic release from the viral polyprotein precursor, thereby leading to partial or complete inhibition of viral replication, suggesting the crucial role of 6K1 in the viral life cycle (Cui and Wang, 2016). Moreover, a series of deletion analyses of PPV 6K1 have demonstrated that the removal of any internal short sequence, truncation of the extension region (even when the conserved cleavage sites are retained), or complete deletion of the 6K1-encoding sequence results in attenuated viral replication. These findings suggest that 6K1 has stringent spatiotemporal requirements for coordinating viral replication dynamics (Cui and Wang, 2016). However, the precise role of 6K1 in viral replication remains poorly understood. One possibility is that 6K1 is directly involved in the assembly of VRCs (Bera et al., 2022; Chai et al., 2024; Cui and Wang, 2016; Fang et al., 2024). Alternatively, 6K1 may affect the spatial organization or stability of VRCs. Additionally, 6K1 can interact with other viral proteins, which may affect VRCs assembly and viral replication (Fang et al., 2024; Morozov and Solovyev, 2020; Wei and Wang, 2008; Xue et al., 2023). Another hypothesis is that the function of 6K1 viroporin activity may contribute to viral replication by modulating the pH or ion homeostasis in VRCs. Nevertheless, further investigations are needed to fully illustrate these possibilities.
3.2 Viroporin activity modulates viral replication
Many viral membrane proteins remodel endomembrane for viral replication or movement. For instance, 6K2 can remodel ER into vesicles to host viral replication (Bera et al., 2022; Cui and Wang, 2016; Hu et al., 2023a,b). Similarly, 6K1 also exhibits endomembrane-remodeling activity: transiently expressed TuMV 6K1 forms granules of various sizes on the ER (Chai et al., 2024). Confocal microscopy observations revealed that PPV 6K1 forms punctate inclusion during viral infection when expressed as a 6K1-GFP fusion protein between the P1 and HC-Pro cistron (Cui and Wang, 2016). Notably, the P3-6K1 fusion protein, along with other potential 6K1-containing complexes, may play distinct yet complementary roles in viral replication, membrane remodeling, and intracellular trafficking, thereby contributing to the establishment of VRCs and systemic viral spread.
3.3 Autophagy-mediated degradation orchestrates viral replication
The accumulation of 6K1 is extremely low in late viral infection stages, which is closely linked to its targeted degradation by the host autophagy system (Hu et al., 2025). For instance, treatment with the E-64d inhibitor or silencing of the autophagy gene N. benthamiana Autophagy-related Protein 7 (NbATG7) has been shown to enhance the stability of 6K1 while concomitantly delaying systemic viral infection (Hu et al., 2025). This paradoxical phenomenon suggests that the degradation of 6K1 may represent an active viral strategy to co-degrade host antiviral factors, thereby indirectly promoting viral replication. Alanine scanning mutagenesis has revealed that the K/R-rich motif is critical for its autophagy-mediated degradation of 6K1: mutants (e.g., V32A or K34A) that evade autophagy recognition exhibit delayed viral spread in the host (Hu et al., 2025). A similar mechanism has also been demonstrated in TuMV, where 6K1 degradation depends on its interaction with the autophagy receptor ATG8; disruption of this process significantly reduces viral replication (Bera et al., 2022; Hu et al., 2025). These finding suggest that the 6K1 orchestrates viral replication through a dual mechanism: firstly, it serves as a structural component of the replication complex to directly facilitate viral RNA synthesis; secondly, it may simultaneously coordinate the elimination of host defense factors through dynamic regulation of its own abundance through autophagy-mediated degradation. This sophisticated “synthesis-degradation” regulatory paradigm not only optimizes viral resource utilization but also underscores the evolutionary adaptability of 6K1 in host-pathogen interactions, revealing new therapeutic targets for antiviral intervention.
4 Host-virus interactions
4.1 Subversion of host defense
In recent years, accumulating evidence has demonstrated that 6K1 establishes dynamic interaction networks with both host factors and viral proteins to optimize the microenvironment for viral replication and facilitate viral proliferation (Figure 2b) (Cui and Wang, 2016; Bera et al., 2022; Hu et al., 2023a,b; Tatineni et al., 2023; Fang et al., 2024; Hu et al., 2025). Soybean 40S ribosomal protein S8 (GmRPS8), an essential ribosomal component, interacts with 6K1 in yeast-two-hybrid (Y2H) and bimolecular fluorescence complementation (BiFC) assays, play a crucial role in SMV infection (Hu et al., 2023a,b). During TVBMV infection, 6K2 recruits 6K1 to VRCs, where 6K1 interacts with both 6K2 and NbPsbO1, a component of the PSII oxygen-evolving complex, to participate in viral replication (Geng et al., 2017). 6K1 competitively binds to Nb14-3-3 h, a key plant defense protein, disrupting its interaction with N. benthamiana Translationally Controlled Tumor Protein (NbTCTP) to promote PVY infection (Fang et al., 2024). When ectopic expression of TuMV 6K1 in N. benthamiana, it reduces transcripts related to jasmonic acid biosynthesis and cysteine proteinase inhibitors while enhancing TuMV accumulation in systemic leaves (Bera et al., 2022). However, whether 6K1 helps viruses evade host immune system by suppressing RNA silencing or interfering with other antiviral defense mechanisms, and whether 6K1 modulates host immune responses through interactions with host immune factors (such as products of R genes), require further investigations.
4.2 Symptom development
Studies have shown that co-expression of 6K1, 6K2, and NIa-Pro in A. thaliana significantly improves drought tolerance, and A. thaliana overexpressing 6K1 shows notably greater root length compared to those expressing NIa-Pro or 6K2 (Prakash et al., 2023). This drought tolerance mechanism benefits both virus and host by increasing host survival rate under drought conditions and therefore extending the viral replication and transmission window (Prakash et al., 2023). Heterologous expression of wheat streak mosaic virus (WSMV; Tritimovirus tritici) induces characteristic viral symptoms, including severe chlorotic streaks and leaf spotting, which mirrors symptoms seen during natural WSWV infection (Tatineni et al., 2023). This dual functionality of 6K1, enhancing host stress tolerance while promoting viral pathogenesis, highlights its sophisticated role in virus-host interactions. The physiological modifications induced by 6K1 create an environment that simultaneously sustains host viability while facilitating viral proliferation and symptom development (Figure 2c).
5 Evolutionary adaptations
5.1 Evolution of 6K1: dual-driven by both host and vector pressures
Phylogenetic analyses reveal both vector-driven clustering patterns and host-specific lineage diversification of potyviral 6K1 and its 7K homologs. For instance, aphid-transmitted genera (Potyvirus and Macluravirus) and eriophyid mite-transmitted genus (Tritimovirus) form well-supported distinct monophyletic clades. Rymovirus members (e.g., parthenium mottle virus, ryegrass mosaic virus, and agropyron mosaic virus) exhibit evolutionary convergence with potyviruses, while sweet potato mild mottle virus (Potyvirus) shows atypical clustering with ipomoviruses (e.g., Squash vein yellowing virus), suggesting ecological niche adaptation. Poacevirus and Tritimovirus (Poaceae specialists) share close genetic relationship, whereas Arepavirus (Arecaceae specialists) and Bymovirus (Poaceae specialists) form discrete host-adapted clusters. Potyvirus, the largest genus with broad host range spanning monocots and dicots, displays extensive intragenus differentiation, reflecting clear patterns of adaptive radiation across monocot/dicot hosts. For instance, bermuda grass southern mosaic virus clusters with Amaranthaceae and Poaceae-infecting lineages infecting. These complex evolutionary patterns of potyviral 6K1 suggesting selective pressures from both hosts and transmission vectors, such as host antiviral RNA silencing, crop-specific protein adaptations, and vector transmission efficiency optimization. Such pressures result in a unique evolutionary balance between strict conservation of replication-critical domains and adaptive plasticity in host/vector-interacting regions. This evolutionary framework highlights 6K1’s dual evolutionary strategy - maintaining core functional elements while acquiring specialized adaptations to diverse ecological niches (Figure 3) (Xie et al., 2023).
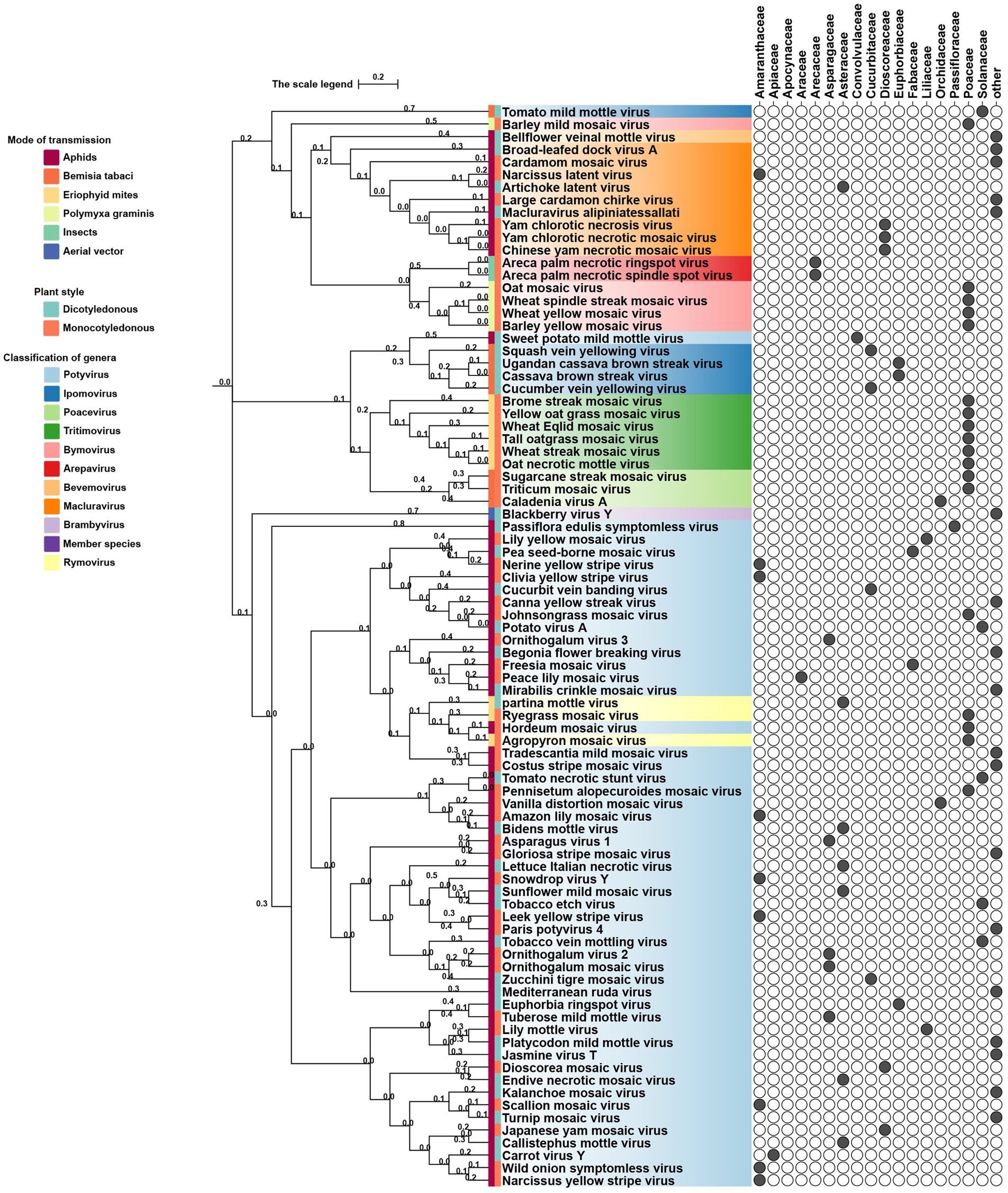
Figure 3. Phylogeny of potyviral 6K1 and its 7K homolog proteins. The tree was constructed using the Neighbor-Joiningmethod and the numbers on branches indicate the bootstrap supporting values. The transmission manner, host specificity, and taxonomy of each virus were indicated. Full information of the tree can be found in the Supplementary Figure 1.
5.2 Core residue stability vs. vector-specific adaptation
All known potyvirids contain 6K1 or its 7 K homolog between the P3 and CI cistrons. Bioinformatics analyses reveal two conserved TMHs in all 6K1 proteins, and the helix-turn-helix hairpin topology also is also consistently in 6K1 across potyvirids (Chai et al., 2024). Moreover, the three highly conserved Lys residues in 6K1 (Lys33, Lys37, and Lys39) that are critical for oligomerization are also functional conserved (Figure 4) (Chai et al., 2024; Cui and Wang, 2016). The “RSD” motif, which is highly conserved in potyviruses, have been replaced by “IAE” “LAL,” and “TAN” in viruses of the genera Macluravirus, Bymovirus, and Tritimovirus, respectively. These data highlight the crucial role of 6K1 in viral infection process with universal structural conservation, genus-specific functional adaptation, and essential conserved residues for oligomerization (Figure 4).

Figure 4. Multiple sequence alignment of potyviral 6K1 and its 7K homologs. The alignment was produced using the Clustal X software and presented using ESPript. The completely and functional conserved residues are shown in red and yellow background, respectively. The full information of this figure can be found in the Supplementary Figure 2.
6K1 also display significant amino acid sequence and size diversity across potyvirids. The length of 6K1 proteins is typically 6 kDa in most potyvirids, while expands to 7 kDa in certain viruses of the genera Macluravirus and Bymovirus, which have been designated as 7K. Sequence alignment suggest that the 7K variants contain an 8–10 residues extension at the TMH2 C-terminus. The function of these amino acids is unknown at the present, although these residues have been proposed in interacting with viral or host proteins or regulating viroporin activity. Besides, the N- and C-terminal residues of 6K1 also show high variability. These two parts comprising the NIa-Pro recognition and cleavage site and thus likely co-evolved with NIa-Pro protease. These findings indicate that the 6K1 protein follows a dual evolutionary strategy to balance functional constraints and ecological adaptation. 6K1 of viruses infecting dicot plants tend to enrich acidic residues (Asp/Glu), while those of viruses infecting monocot plants prefer basic residues (Lys/Arg). Additionally, the 6K1 of dicot-infecting viruses maintain a hydrophobic transmembrane core (Leu/Val/Ile > 85%) and conserved Phe/Leu residues, while those of monocot-infecting viruses in genera Tritimovirus and Bymovirus accumulate Ser → Pro/Lys → Glu mutations in their transmembrane regions (Figure 4).
The 6K1 protein also exhibits evolutionary adaptations corresponding to different transmission pathways within the Potyviridae family. For instance, 6K1 of aphid-transmitted viruses exhibit C-terminal Pro/Gly enrichment and contain highly conserved Asp/Glu residues (mutation rate <3%), while that of mite-transmitted viruses show a higher frequency of Lys → Glu mutations in the transmembrane region (22%) and an increased Phe/Leu residue proportion, 6K1 of whitefly-transmitted viruses possess GRREESF sequence insertions and Val → Leu mutations, and that of fungus-transmitted viruses exhibit a “LNKLK” to “LNHV” motif alteration. These transmission-specific adaptations likely demonstrate how 6K1 has evolved unique molecular strategies to optimize vector-specific requirements, highlighting its remarkable evolutionary plasticity while maintaining core functions.
6 Antiviral strategies targeting 6K1
6.1 Small-molecule inhibitors
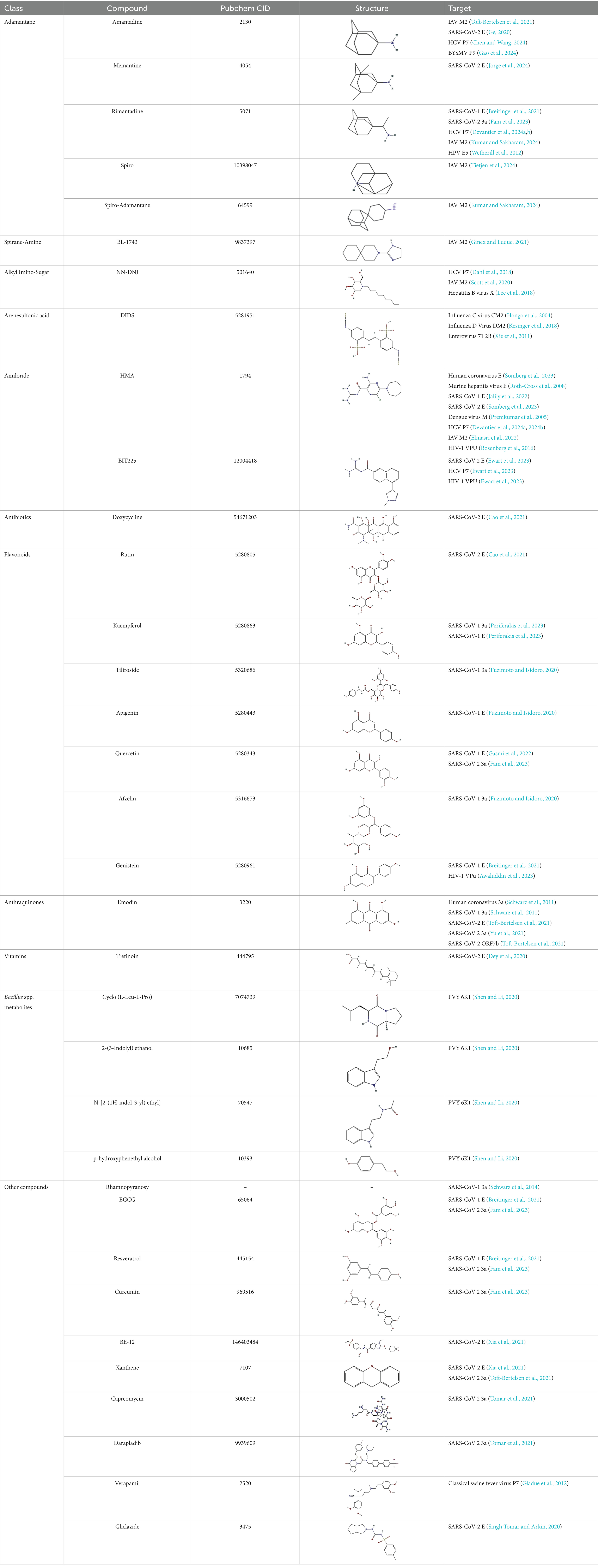
Traditional inhibitors primarily target viral RdRp or helicase, such as ribavirin, which disrupts the catalytic core of TSWV RdRp (Li et al., 2025; Wang et al., 2025), Z9 effectively inhibits co-condensate formation between TSWV nucleoprotein (N) and viral RNA, reducing viral proliferation (Zan et al., 2025). Recently, pH-sensitive (GPS) polymer nanoparticles have been found to selectively bind to infected cell membranes or viral envelopes, even envelope disruption (Sun et al., 2022). This leverages pH changes in infected cells as a therapeutic target. For viroporins, most inhibitors act via either physical block the channel or disrupt their oligomerisation. Although 6K1 have recently been confirmed function as a viroporin, research on specific inhibitors remains lacking. Despite lacking sequence homology, 6K1 adopts spatial folding and conformations analogous to IAV M2, HCV P7, SARS-CoV E1/E2 and HIV Vpu with a high hydrophobic central cavity (Figure 1b) (Devantier et al., 2024a,b; Elmasri et al., 2022; Surya et al., 2023; Surya and Torres, 2022). Therefore, it is possible that these inhibitors may also function for 6K1. For example, amantadine also inhibits BYSMV P9, another recently identified plant virus-encoded Class I viroporin (Gao et al., 2024). Table 1 listed the major viroporin inhibitors of viroporins. Among these inhibitors, Adamantane and Amiloride have significant inhibitory effects on IAV M2 and HCV P7, while Flavonoids, Emodin, and Tretinoin target SARS-CoV E1/E2. Thus, these compounds may function against 6K1 as well. Nevertheless, it is necessary to test the existing viroporin inhibitors against using the membrane permeability and electrophysiological assays and then rational optimize these compounds for higher specificity and potency against 6K1.
6.2 Biocontrol agents
Several species of the genus Bacillus, such as Bacillus subtilis and B. licheniformis, have demonstrated efficacy in controlling potyviral diseases (Miljaković et al., 2020; Xuan et al., 2024). These bacteria can inhibit the accumulation and spread of viruses through multiple mechanisms, including activation of the plant defense systems, secretion of antimicrobial compounds, and nutrient competition with other pathogens (Veselova et al., 2022; Amin et al., 2023; Xuan et al., 2024). Recent studies reveal that some Bacillus species secret compounds directly target potyviruses. For instance, nine organic compounds extracted from B. pumilus E303035 fermentation showed strong inhibition against PVY, among which cyclo (L-Leu-L-Pro), 2-(3-Indolyl) ethanol, N-[2-(1H-indol-3-yl) ethyl] and phydroxyphenethyl alcohol significantly inhibit the activity of PVY 6K1 (Shen and Li, 2020). Furthermore, these compounds also affect the expression of viral genes involving in viral proliferation (Shen and Li, 2020). In addition, Bacillus can also produce other metabolites, such as surfactin, iturin, foenomycin, hydrolases, and volatile organic compounds (Wang et al., 2018; Rajaofera et al., 2019; Jin et al., 2020). These compounds exhibit antiviral activities against various plant viruses, such as cucumber mosaic virus (CMV; Cucumovirus CMV), tomato spotted wilt virus (TSWV; Orthotospovirus tomatomaculae), pepper mild mottle virus (PMMoV; Tobamovirus capsici), and tomato yellow leaf curl virus (TYLCV; Begomovirus coheni) (Ongena and Jacques, 2008; Kong et al., 2018). In the future, it is necessary to identify the precise target of these active compounds and structurally optimize their structures for enhanced specificity. In conclusion, the secondary metabolites produced valuable resources for identifying potential 6K1 inhibitor.
7 Unresolved questions and future directions
Despite significant advances concerning the critical functions of 6K1 including increasing cell membrane permeability, facilitating viral replication, and interacting with host factors, several key knowledge gaps remained. For instance, the precise atomic structure of 6K1, its ion selectivity of 6K1, and its interaction network with viral and host proteins. In addition, investigation of the evolutionary pressures shaping 6K1 diversity across potyviruses and potential role of 6K1 in virus-vector interaction will underscore its functional diversity. Finally, screen small molecule antiviral agents that targets 6K1 will significantly benefit the control of potyviral diseases. In conclusion, 6K1 is a multifunctional protein represents the central hub in potyviral infection, bridging replication, membrane dynamics, and host manipulation and the discovery of 6K1 as a plant viroporin not only enhances our understanding of the pathogenesis of potyvirids but also provides an important target for developing antiviral agents.
Author contributions
LY: Investigation, Resources, Writing – original draft, Writing – review & editing. XK: Validation, Visualization, Writing – original draft. ZW: Resources, Validation, Writing – original draft. SC: Methodology, Validation, Writing – original draft. XJ: Data curation, Formal analysis, Resources, Writing – review & editing. XW: Project administration, Supervision, Writing – review & editing. XC: Conceptualization, Funding acquisition, Project administration, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was financial supported by the National Key Research and Development Program of China (2023YFD1700701-03) and the Open Fund of State Key Laboratory of Green Pesticide (QianJiaoJi[2023]115).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1605199/full#supplementary-material
References
Amin, H. A., El Kammar, H. F., Saied, S. M., and Soliman, A. M. (2023). Effect of Bacillus subtilis on potato virus Y (PVY) disease resistance and growth promotion in potato plants. Eur. J. Plant Pathol. 167, 743–758. doi: 10.1007/s10658-023-02774-0
Awaluddin, F., Batubara, I., and Wahyudi, S. T. (2023). Virtual screening of natural compounds against six protein receptors coded by the SARS-CoV-2 genome. Mol. Ther. 18, 147–158. doi: 10.20884/1.jm.2023.18.1.7884
Belabess, Z., Tahiri, A., and Lahlali, R. (2024). Contemporary perspectives on the global evolution of potato virus Y pathogen. Indian Phytopathol. 77, 13–34. doi: 10.1007/s42360-024-00709-1
Bera, S., Arena, G. D., Ray, S., Flannigan, S., and Casteel, C. L. (2022). The potyviral protein 6K1 reduces plant proteases activity during turnip mosaic virus infection. Viruses 14:1341. doi: 10.3390/v14061341
Bhoi, T. K., Samal, I., Majhi, P. K., Komal, J., Mahanta, D. K., Pradhan, A. K., et al. (2022). Insight into aphid mediated potato virus Y transmission: a molecular to bioinformatics prospective. Front. Microbiol. 13:1001454. doi: 10.3389/fmicb.2022.1001454
Breitinger, U., Ali, N. K. M., Sticht, H., and Breitinger, H. G. (2021). Inhibition of SARS CoV envelope protein by flavonoids and classical viroporin inhibitors. Front. Microbiol. 12:692423. doi: 10.3389/fmicb.2021.692423
Cao, Y., Yang, R., Lee, I., Zhang, W., Sun, J., Wang, W., et al. (2021). Characterization of the SARS-CoV-2 E protein: sequence, structure, viroporin, and inhibitors. Protein Sci. 30, 1114–1130. doi: 10.1002/pro.4075
Carrasco, L. (1995). Modification of membrane permeability by animal viruses. Advances in virus research. Adv. Virus Res. 45, 61–112. doi: 10.1016/s0065-3527(08)60058-5
Chai, M., Li, L., Li, Y., Yang, Y., Wang, Y., Jiang, X., et al. (2024). The 6-kilodalton peptide 1 in plant viruses of the family Potyviridae is a viroporin. Proc. Natl. Acad. Sci. USA 121:e2401748121. doi: 10.1073/pnas.2401748121
Chen, X., and Wang, X. (2024). Computational investigation in inhibitory effects of amantadine on classical swine fever virus p7 ion channel activity. Sci. Rep. 14:20387. doi: 10.1038/s41598-024-71477-1
Crosslin, J. M. (2013). PVY: an old enemy and a continuing challenge. Am. J. Potato Res. 90, 2–6. doi: 10.1007/s12230-012-9286-8
Cui, H., and Wang, A. (2016). The plum pox virus 6K1 protein is required for viral replication and targets the viral replication complex at the early infection stage. J. Virol. 90, 5119–5131. doi: 10.1128/jvi.00024-16
Cui, H., and Wang, A. (2019). The biological impact of the hypervariable N-terminal region of potyviral genomes. Annu. Rev. Virol. 6, 255–274. doi: 10.1146/annurev-virology-092818-015843
Dahl, S. L., Kalita, M. M., and Fischer, W. B. (2018). Interaction of antivirals with a heptameric bundle model of the p7 protein of hepatitis C virus. Chem. Biol. Drug Des. 91, 942–950. doi: 10.1111/cbdd.13162
Devantier, K., Kjær, V. M., Griffin, S., Kragelund, B. B., and Rosenkilde, M. M. (2024a). Advancing the field of viroporins—structure, function and pharmacology: IUPHAR review X. Br. J. Pharmacol. 181, 4450–4490. doi: 10.1111/bph.17317
Devantier, K., Toft-Bertelsen, T. L., Prestel, A., Kjaer, V. M., Sahin, C., Giulini, M., et al. (2024b). The SH protein of mumps virus is a druggable pentameric viroporin. bioRxiv. doi: 10.1101/2024.08.09.607002
Dey, D., Borkotoky, S., and Banerjee, M. (2020). In silico identification of tretinoin as a SARS-CoV-2 envelope (E) protein ion channel inhibitor. Comput. Biol. Med. 127:104063. doi: 10.1016/j.compbiomed.2020.104063
Dupuis, B., Nkuriyingoma, P., and Ballmer, T. (2024). Economic impact of potato virus Y (PVY) in Europe. Potato Res. 67, 55–72. doi: 10.1007/s11540-023-09623-x
Elmasri, Z., Negi, V., Kuhn, R. J., and Jose, J. (2022). Requirement of a functional ion channel for Sindbis virus glycoprotein transport, CPV-II formation, and efficient virus budding. PLoS Pathog. 18:e1010892. doi: 10.1371/journal.ppat.1010892
Ewart, G., Bobardt, M., Bentzen, B. H., Yan, Y., Thomson, A., Klumpp, K., et al. (2023). Post-infection treatment with the E protein inhibitor BIT225 reduces disease severity and increases survival of K18-hACE2 transgenic mice infected with a lethal dose of SARS-CoV-2. PLoS Pathog. 19:e1011328. doi: 10.1371/journal.ppat.1011328
Fam, M. S., Sedky, C. A., Turky, N. O., Breitinger, H. G., and Breitinger, U. (2023). Channel activity of SARS-CoV-2 viroporin ORF3a inhibited by adamantanes and phenolic plant metabolites. Sci. Rep. 13:5328. doi: 10.1038/s41598-023-31764-9
Fang, L., Geng, C., Wei, X. Y., Dong, C. C., Pang, J. P., Yan, Z. Y., et al. (2024). Potato virus Y viral protein 6K1 inhibits the interaction between defense proteins during virus infection. Plant Physiol. 194, 1447–1466. doi: 10.1093/plphys/kiad612
Fuzimoto, A. D., and Isidoro, C. (2020). The antiviral and coronavirus-host protein pathways inhibiting properties of herbs and natural compounds-additional weapons in the fight against the COVID-19 pandemic? J. Tradit. Complement. Med. 10, 405–419. doi: 10.1016/j.jtcme.2020.05.003
Gadhave, K. R., Gautam, S., Rasmussen, D. A., and Srinivasan, R. (2020). Aphid transmission of potyvirus: the largest plant-infecting RNA virus genus. Viruses 12:773. doi: 10.3390/v12070773
Gao, Q., Zang, Y., Qiao, J. H., Zhang, Z. Y., Wang, Y., Han, C. G., et al. (2024). The plant rhabdovirus viroporin P9 facilitates insect-mediated virus transmission in barley. Plant Cell 36, 3483–3497. doi: 10.1093/plcell/koae162
Gasmi, A., Mujawdiya, P. K., Lysiuk, R., Shanaida, M., Peana, M., Gasmi, B. A., et al. (2022). Quercetin in the prevention and treatment of coronavirus infections: a focus on SARS-CoV-2. Pharmaceuticals 15:1049. doi: 10.3390/ph15091049
Ge, A. A. (2020). The Sars-Cov-2 Viroporin E and its interaction with amantadine an analysis. J. Pharm. Pharmacol. 8:S200003. doi: 10.13188/2327-204X
Gebert, J. T., Scribano, F. J., Engevik, K. A., Philip, A. A., Kawagishi, T., Greenberg, H. B., et al. (2024). Viroporin activity from rotavirus nonstructural protein 4 induces intercellular calcium waves that contribute to pathogenesis. bioRxiv. doi: 10.1101/2024.05.07.592929
Geng, C., Yan, Z. Y., Cheng, D. J., Liu, J., Tian, Y. P., Zhu, C. X., et al. (2017). Tobacco vein banding mosaic virus 6K2 protein hijacks NbPsbO1 for virus replication. Sci. Rep. 7:43455. doi: 10.1038/srep43455
Ginex, T., and Luque, F. J. (2021). Searching for effective antiviral small molecules against influenza a virus: a patent review. Expert Opin. Ther. Pat. 31, 53–66. doi: 10.1080/13543776.2020
Gladue, D. P., Holinka, L. G., Largo, E., Fernandez Sainz, I., Carrillo, C., O’Donnell, V., et al. (2012). Classical swine fever virus p7 protein is a viroporin involved in virulence in swine. J. Virol. 86, 6778–6791. doi: 10.1128/jvi.00560-12
Gong, P., Gao, M., Chen, Y., Zhang, M., Huang, Y., Hu, X., et al. (2025). Cucumber green mottle mosaic virus encodes additional small proteins with specific subcellular localizations and virulence function. Sci. China Life Sci., 1674–7305. doi: 10.1007/s11427-024-2892-1
Gong, P., Shen, Q., Zhang, M., Qiao, R., Jiang, J., Su, L., et al. (2023). Plant and animal positive-sense single-stranded RNA viruses encode small proteins important for viral infection in their negative-sense strand. Mol. Plant 16, 1794–1810. doi: 10.1016/j.molp.2023.09.020
Gong, P., Tan, H., Zhao, S., Li, H., Liu, H., Ma, Y., et al. (2021). Geminiviruses encode additional small proteins with specific subcellular localizations and virulence function. Nat. Commun. 12:4278. doi: 10.1038/s41467-021-24617-4
Hongo, S., Ishii, K., Mori, K., Takashita, E., Muraki, Y., Matsuzaki, Y., et al. (2004). Detection of ion channel activity in Xenopus laevis oocytes expressing influenza C virus CM2 protein. Arch. Virol. 149, 35–50. doi: 10.1007/s00705-003-0209-3
Hu, W., Deng, C., Qin, L., Liu, P., Wang, L., Wang, X., et al. (2025). A conserved lysine/arginine-rich motif is essential for the autophagic degradation of potyviral 6K1 protein and virus infection. J. Virol. 99:e0218324. doi: 10.1128/jvi.02183-24
Hu, T., Guo, D., Li, B., Wang, L., Liu, H., Yin, J., et al. (2023a). Soybean 40S ribosomal protein S8 (GmRPS8) interacts with 6K1 protein and contributes to soybean susceptibility to soybean mosaic virus. Viruses 15:2362. doi: 10.3390/v15122362
Hu, T., Luan, H., Wang, L., Ren, R., Sun, L., Yin, J., et al. (2023b). Soybean mosaic virus 6K1 interactors screening and GmPR4 and GmBI1 function characterization. Int. J. Mol. Sci. 24:5304. doi: 10.3390/ijms24065304
Hýsková, V., Bělonožníková, K., Chmelík, J., Hoffmeisterová, H., Čeřovská, N., Moravec, T., et al. (2024). Potyviral helper-component protease: multifaced functions and interactions with host proteins. Plan. Theory 13:1236. doi: 10.3390/plants13091236
Jalily, P. H., Jalily Hasani, H., and Fedida, D. (2022). In silico evaluation of hexamethylene amiloride derivatives as potential luminal inhibitors of SARS-CoV-2 E protein. Int. J. Mol. Sci. 23:10647. doi: 10.3390/ijms231810647
Jaramillo-Mesa, H., and Rakotondrafara, A. M. (2023). All eggs in one basket: how potyvirus infection is controlled at a single cap-independent translation event. Semin. Cell Dev. Biol. 148-149, 51–61. doi: 10.1016/j.semcdb.2022.12.011
Jin, P., Wang, H., Tan, Z., Xuan, Z., Dahar, G. Y., Li, Q. X., et al. (2020). Antifungal mechanism of bacillomycin D from Bacillus velezensis HN-2 against Colletotrichum gloeosporioides Penz. Pestic. Biochem. Physiol. 163, 102–107. doi: 10.1016/j.pestbp.2019.11.004
Jorge, L. M., Juan, H. M., and Garduño-Juárez, R. (2024). In silico screening, ADMET analysis, MD simulations, and MM/PBSA binding free energy identify new inhibitor molecules for Viropiron E. Authorea 24, 1–29. doi: 10.22541/au.172716199.92346731
Kamran, A., Hussain, M. D., Farooq, T., Li, F., Khan, M., Li, X., et al. (2025). Deciphering intricate plant-virus interactions: Potyvirids orchestrate protein posttranslational modifications to regulate pathogenicity. Microbiol. Res. 290:127940. doi: 10.1016/j.micres.2024.127940
Kesinger, E., Liu, J., Jensen, A., Chia, C. P., Demers, A., and Moriyama, H. (2018). Influenza D virus M2 protein exhibits ion channel activity in Xenopus laevis oocytes. PLoS One 13:e0199227. doi: 10.1371/journal.pone.0199227
Kong, H. G., Shin, T. S., Kim, T. H., and Ryu, C.-M. (2018). Stereoisomers of the bacterial volatile compound 2, 3-butanediol differently elicit systemic defense responses of pepper against multiple viruses in the field. Front. Plant Sci. 9:90. doi: 10.3389/fpls.2018.00090
Kumar, R., and Dasgupta, I. (2024). Abscisic acid: an emerging player in plant-virus interactions. Plant Physiol. Biochem. 215:109046. doi: 10.1016/j.plaphy.2024.109046
Kumar, G., and Sakharam, K. A. (2024). Tackling influenza a virus by M2 ion channel blockers: latest progress and limitations. Eur. J. Med. Chem. 267:116172. doi: 10.1016/j.ejmech.2024.116172
Lakadamyali, M., Rust, M. J., Babcock, H. P., and Zhuang, X. (2003). Visualizing infection of individual influenza viruses. Proc. Natl. Acad. Sci. USA 100, 9280–9285. doi: 10.1073/pnas.0832269100
Lee, H. R., Cho, Y. Y., Lee, G. Y., You, D. G., Yoo, Y. D., and Kim, Y. J. (2018). A direct role for hepatitis B virus X protein in inducing mitochondrial membrane permeabilization. J. Viral Hepat. 25, 412–420. doi: 10.1111/jvh.12831
Li, J., Cao, L., Zhao, Y., Shen, J., Wang, L., Feng, M., et al. (2025). Structural basis for the activation of plant bunyavirus replication machinery and its dual-targeted inhibition by ribavirin. Nat. Plant. 11, 518–530. doi: 10.1038/s41477-025-01940-y
Li, F., Jia, M., and Wang, A. (2024). Hidden viral proteins: how powerful are they? PLoS Pathog. 20:e1011905. doi: 10.1371/journal.ppat.1011905
Liu, J., Wu, X., Fang, Y., Liu, Y., Bello, E. O., Li, Y., et al. (2023). A plant RNA virus inhibits NPR1 sumoylation and subverts NPR1-mediated plant immunity. Nat. Commun. 14:3580. doi: 10.1038/s41467-023-39254-2
Luan, Y., Yang, S., Wang, Y., Zhao, Y., Wu, X., Chen, Q., et al. (2024). Genome-wide association analysis of cowpea mild mottle virus resistance in soybean germplasms from Northeast China. Agronomy 14:489. doi: 10.3390/agronomy14030489
Mardanova, E. S., Vasyagin, E. A., and Ravin, N. V. (2024). Virus-like particles produced in plants: a promising platform for recombinant vaccine development. Plan. Theory 13:3564. doi: 10.3390/plants13243564
Martínez-Turiño, S., and García, J. A. (2020). Chapter five-potyviral coat protein and genomic RNA: a striking partnership leading virion assembly and more. Adv. Virus Res. 108, 165–211. doi: 10.1016/bs.aivir.2020.09.001
Melton, J. V., Ewart, G. D., Weir, R. C., Board, P. G., Lee, E., and Gage, P. W. (2002). Alphavirus 6K proteins form ion channels. J. Biol. Chem. 277, 46923–46931. doi: 10.1074/jbc.M207847200
Miljaković, D., Marinković, J., and Balešević-Tubić, S. (2020). The significance of bacillus spp. in disease suppression and growth promotion of field and vegetable crops. Microorganisms 8:1037. doi: 10.3390/microorganisms8071037
Morozov, S. Y., and Solovyev, A. G. (2020). Small hydrophobic viral proteins involved in intercellular movement of diverse plant virus genomes. AIMS Microbiol. 6, 305–329. doi: 10.3934/microbiol.2020019
Nieva, J. L., Madan, V., and Carrasco, L. (2012). Viroporins: structure and biological functions. Nat. Rev. Microbiol. 10, 563–574. doi: 10.1038/nrmicro2820
Ongena, M., and Jacques, P. (2008). Bacillus lipopeptides: versatile weapons for plant disease biocontrol. Trends Microbiol. 16, 115–125. doi: 10.1016/j.tim.2007.12.009
Periferakis, A., Periferakis, A. T., Troumpata, L., Periferakis, K., Scheau, A. E., Savulescu-Fiedler, I., et al. (2023). Kaempferol: a review of current evidence of its antiviral potential. Int. J. Mol. Sci. 24:16299. doi: 10.3390/ijms242216299
Pielak, R. M., and Chou, J. J. (2011). Influenza M2 proton channels. Biochim. Biophys. Acta 1808, 522–529. doi: 10.1016/j.bbamem.2010.04.015
Prakash, V., Nihranz, C. T., and Casteel, C. L. (2023). The potyviral protein 6K2 from turnip mosaic virus increases plant resilience to drought. Mol. Plant-Microbe Interact. 36, 189–197. doi: 10.1094/MPMI-09-22-0183-R
Premkumar, A., Horan, C., and Gage, P. (2005). Dengue virus M protein C-terminal peptide (DVM-C) forms ion channels. J. Membr. Biol. 204, 33–38. doi: 10.1007/s00232-005-0744-9
Premkumar, A., Wilson, L., Ewart, G., and Gage, P. (2004). Cation-selective ion channels formed by p7 of hepatitis C virus are blocked by hexamethylene amiloride. FEBS Lett. 557, 99–103. doi: 10.1016/s0014-5793(03)01453-4
Qin, L., Liu, H. J., Liu, P. L., Jiang, L., Cheng, X. F., Li, F. F., et al. (2024). Rubisco small subunit (RbCS) is co-opted by potyvirids as the scaffold protein in assembling a complex for viral intercellular movement. PLoS Pathog. 20:e1012064. doi: 10.1371/journal.ppat.1012064
Rajaofera, M. J. N., Wang, Y., Dahar, G. Y., Jin, P., Fan, L., Xu, L., et al. (2019). Volatile organic compounds of Bacillus atrophaeus HAB-5 inhibit the growth of Colletotrichum gloeosporioides. Pestic. Biochem. Physiol. 156, 170–176. doi: 10.1016/j.pestbp.2019.02.019
Riechmann, J. L., Cervera, M. T., and García, J. A. (1995). Processing of the plum pox virus polyprotein at the P3-6K1 junction is not required for virus viability. J. Gen. Virol. 76, 951–956. doi: 10.1099/0022-1317-76-4-951
Rosenberg, M. R., Weaver, L. M., and Casarotto, M. G. (2016). Probing interactions of Vpu from HIV-1 with amiloride-based compounds. Biochim. Biophys. Acta 1858, 733–739. doi: 10.1016/j.bbamem.2015.12.028
Roth-Cross, J. K., Bender, S. J., and Weiss, S. R. (2008). Murine coronavirus mouse hepatitis virus is recognized by MDA5 and induces type I interferon in brain macrophages/microglia. J. Virol. 82, 9829–9838. doi: 10.1128/JVI.01199-08
Schwarz, S., Sauter, D., Wang, K., Zhang, R., Sun, B., Karioti, A., et al. (2014). Kaempferol derivatives as antiviral drugs against the 3a channel protein of coronavirus. Planta Med. 80, 177–182. doi: 10.1055/s-0033-1360277
Schwarz, S., Wang, K., Yu, W., Sun, B., and Schwarz, W. (2011). Emodin inhibits current through SARS-associated coronavirus 3a protein. Antivir. Res. 90, 64–69. doi: 10.1016/j.antiviral.2011.02.008
Scott, C., Kankanala, J., Foster, T. L., Goldhill, D. H., Bao, P., Simmons, K., et al. (2020). Site-directed M2 proton channel inhibitors enable synergistic combination therapy for rimantadine-resistant pandemic influenza. PLoS Pathog. 16:e1008716. doi: 10.1371/journal.ppat.1008716
Shen, S., and Li, W. (2020). The inhibitory effects of metabolites from Bacillus pumilus on potato virus Y and the induction of early response genes in Nicotiana tabacum. AMB Express 10, 152–112. doi: 10.1186/s13568-020-01089-1
Singh Tomar, P. P., and Arkin, I. T. (2020). SARS-CoV-2 E protein is a potential ion channel that can be inhibited by Gliclazide and Memantine. Biochem. Biophys. Res. Commun. 530, 10–14. doi: 10.1016/j.bbrc.2020.05.206
Somberg, N. H., Medeiros-Silva, J., Jo, H., Wang, J., DeGrado, W. F., and Hong, M. (2023). Hexamethylene amiloride binds the SARS-CoV-2 envelope protein at the protein–lipid interface. Protein Sci. 32:e4755. doi: 10.1002/pro.4755
Sun, Y., Gong, L., Yin, Y., Zhang, L., Sun, Q., Feng, K., et al. (2022). A gradient pH-sensitive polymer-based antiviral strategy via viroporin-induced membrane acidification. Adv. Mater. 34:e2109580. doi: 10.1002/adma.202109580
Surya, W., Tavares-Neto, E., Sanchis, A., Queralt-Martín, M., Alcaraz, A., Torres, J., et al. (2023). The complex proteolipidic behavior of the SARS-CoV-2 envelope protein channel: weak selectivity and heterogeneous oligomerization. Int. J. Mol. Sci. 24:12454. doi: 10.3390/ijms241512454
Surya, W., and Torres, J. (2022). Oligomerization-dependent Beta-structure formation in SARS-CoV-2 envelope protein. Int. J. Mol. Sci. 23:13285. doi: 10.3390/ijms232113285
Tatineni, S., Alexander, J., and Nunna, H. (2023). 6K1, NIa-VPg, NIa-pro, and CP of wheat streak mosaic virus are collective determinants of wheat streak mosaic disease in wheat. Phytopathology 113, 1115–1127. doi: 10.1094/PHYTO-10-22-0401-R
Tietjen, I., Kwan, D. C., Petrich, A., Zell, R., Antoniadou, I. T., Gavriilidou, A., et al. (2024). Antiviral mechanisms and preclinical evaluation of amantadine analogs that continue to inhibit influenza a viruses with M2 S31N-based drug resistance. Antivir. Res. 236:106104. doi: 10.1016/j.antiviral.2025.106104
Toft-Bertelsen, T. L., Jeppesen, M. G., Tzortzini, E., Xue, K., Giller, K., Becker, S., et al. (2021). Amantadine inhibits known and novel ion channels encoded by SARS-CoV-2 in vitro. Commun. Biol. 4:1347. doi: 10.1038/s42003-021-02866-9
Tomar, P. P. S., Krugliak, M., and Arkin, I. T. (2021). Blockers of the SARS-CoV-2 3a channel identified by targeted drug repurposing. Viruses 13:532. doi: 10.3390/v13030532
Valli, A. A., Domingo-Calap, M. L., González de Prádena, A., García, J. A., Cui, H., Desbiez, C., et al. (2024). Reconceptualizing transcriptional slippage in plant RNA viruses. MBio 15:e0212024. doi: 10.1128/mbio.02120-24
Veselova, S., Sorokan, A., Burkhanova, G., Rumyantsev, S., Cherepanova, E., Alekseev, V., et al. (2022). By modulating the hormonal balance and ribonuclease activity of tomato plants Bacillus subtilis induces defense response against potato virus X and potato virus Y. Biomol. Ther. 12:288. doi: 10.3390/biom12020288
Wang, X., Yang, W., Wu, S., Jin, F., Shen, Z., Li, X., et al. (2025). Carbene-catalyzed phthalide ether functionalization for discovering chiral phytovirucide that specifically targets viral NIa protein to inhibit proliferation. Research 8:0637. doi: 10.34133/research.0637
Wang, X., Zhao, D., Shen, L., Jing, C., and Zhang, C. (2018). “Chapter one: application and mechanisms of Bacillus subtilis in biological control of plant disease” in Role of Rhizospheric Microbes in Soil, vol. 1, 225–250.
Wani, S., Nisa, Q., Fayaz, T., Naziya, N., Aasiya, N., Lateef, I., et al. (2023). An overview of major bean diseases and current scenario of common bean resistance. Dis. Legume Crops 3358, 99–123. doi: 10.1007/978-981-99-3358-7_5
Wei, T., and Wang, A. (2008). Biogenesis of cytoplasmic membranous vesicles for plant potyvirus replication occurs at endoplasmic reticulum exit sites in a COPI-and COPII-dependent manner. J. Virol. 82, 12252–12264. doi: 10.1128/JVI.01329-08
Wetherill, L. F., Holmes, K. K., Verow, M., Müller, M., Howell, G., Harris, M., et al. (2012). High-risk human papillomavirus E5 oncoprotein displays channel-forming activity sensitive to small-molecule inhibitors. J. Virol. 86, 5341–5351. doi: 10.1128/jvi.06243-11
Wilson, L., Mckinlay, C., Gage, P., and Ewart, G. (2004). SARS coronavirus E protein forms cation-selective ion channels. Virology 330, 322–331. doi: 10.1016/j.virol.2004.09.033
Xia, X., Cheng, A., Wang, M., Ou, X., Sun, D., Mao, S., et al. (2022). Functions of viroporins in the viral life cycle and their regulation of host cell responses. Front. Immunol. 13:890549. doi: 10.3389/fimmu.2022.890549
Xia, B., Shen, X., He, Y., Pan, X., Liu, F. L., Wang, Y., et al. (2021). SARS-CoV-2 envelope protein causes acute respiratory distress syndrome (ARDS)-like pathological damages and constitutes an antiviral target. Cell Res. 31, 847–860. doi: 10.1038/s41422-021-00519-4
Xie, J., Chen, Y., Cai, G., Cai, R., Hu, Z., and Wang, H. (2023). Tree visualization by one table (tvBOT): a web application for visualizing, modifying and annotating phylogenetic trees. Nucleic Acids Res. 51, W587–W592. doi: 10.1093/nar/gkad359
Xie, S., Wang, K., Yu, W., Lu, W., Xu, K., Wang, J., et al. (2011). DIDS blocks a chloride-dependent current that is mediated by the 2B protein of enterovirus 71. Cell Res. 21, 1271–1275. doi: 10.1038/cr.2011.112
Xuan, Z., Wang, Y., Shen, Y., Pan, X., Wang, J., Liu, W., et al. (2024). Bacillus velezensis HN-2: a potent antiviral agent against pepper veinal mottle virus. Front. Plant Sci. 15:1403202. doi: 10.3389/fpls.2024.1403202
Xue, M., Arvy, N., and German-Retana, S. (2023). The mystery remains: how do potyviruses move within and between cells? Mol. Plant Pathol. 24, 1560–1574. doi: 10.1111/mpp.13383
Yang, X. L., Li, Y. Z., and Wang, A. M. (2021). Research advances in potyviruses: from the laboratory bench to the field. Annu. Rev. Phytopathol. 59, 1–29. doi: 10.1146/annurev-phyto-020620-114550
Yang, T. Q., Zhao, X. Y., Bai, J. J., Lv, W. X., Chen, Q., Hu, J., et al. (2024). Transcriptome analysis of genes involved in the pathogenesis mechanism of potato virus Y in potato cultivar You Jin. Front. Microbiol. 15:1353814. doi: 10.3389/fmicb.2024.1353814
Yu, H. G., Sizemore, G., Smoot, K., and Perrotta, P. (2021). Detecting SARS-CoV-2 Orf3a and E ion channel activity in COVID-19 blood samples. J. Clin. Transl. Sci. 5:e196. doi: 10.1017/cts.2021.856
Keywords: 6K1, Potyviridae, viroporin, viral replication complex, membrane remodeling, antiviral targets
Citation: Yu L, Korxeelor X, Wang Z, Chang S, Jiang X, Wu X and Cheng X (2025) The 6-kilodalton peptide 1 of the family Potyviridae: small in size but powerful in function. Front. Microbiol. 16:1605199. doi: 10.3389/fmicb.2025.1605199
Edited by:
Qingfa Wu, University of Science and Technology of China, ChinaReviewed by:
Ibukun A. Akinyemi, University of Florida, United StatesWentao Shen, Chinese Academy of Tropical Agricultural Sciences, China
Copyright © 2025 Yu, Korxeelor, Wang, Chang, Jiang, Wu and Cheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaofei Cheng, eGZjaGVuZ0BuZWF1LmVkdS5jbg==
†These authors have contributed equally to this work
 Liansheng Yu1†
Liansheng Yu1† Xiaofei Cheng
Xiaofei Cheng