- 1NUBAD, LLC, Greenville, SC, United States
- 2Laboratory of Medicinal Chemistry, Department of Chemistry, Clemson University, Clemson, SC, United States
Globally, it is predicted that by 2050, 10 million people will die annually because of infections with drug-resistant bacteria. Since antibacterial agents with novel mechanisms of action have not been developed in the past 30 years, there has been a surge of interest in combination therapies using existing drugs. The combination of aminoglycosides and colistin is often used to treat pneumonia caused by multidrug-resistant bacteria. The goal of this study is to investigate the relationship between the antibacterial activity of a peptide-neomycin library and polymyxin B in extensively drug-resistant and pandrug-resistant bacteria. The peptide-neomycin library contained conjugates with one or two amino acids linked to neomycin, rendering them unsuitable substrates for aminoglycoside-modifying enzymes. Neomycin- susceptible and neomycin-resistant members of Acinetobacter baumannii, Klebsiella pneumoniae, and Pseudomonas aeruginosa were screened for synergy with polymyxin B using two-way checkerboard and time-kill methods. Most A. baumannii strains are resistant to amikacin, gentamicin, tobramycin, and plazomicin, and approximately half are susceptible to neomycin. P. aeruginosa strains have a similar resistance profile but was more susceptible to plazomicin. K. pneumoniae strains are most susceptible to a wide variety of aminoglycosides. Bacteria challenged with a combination of neomycin, other aminoglycosides, and polymyxin B exhibited an additive to indifferent relationship, whereas synergy was found with several neomycin-peptide conjugates containing cysteine, arginine, or tryptophan, lowering the minimal inhibitory concentration for the peptide-neomycin conjugate by 8-64-fold and polymyxin B by 2-8-fold. Cysteine, arginine, or tryptophan conjugates were the most effective against A. baumannii and K. pneumoniae carrying a 16S rRNA methyltransferase gene and a pandrug-resistant P. aeruginosa strain. Resistance to the combination of R-, C-, or RC-NEO conjugates and PB did not develop over a 14-day period in neomycin-susceptible strains of A. baumannii, K. pneumoniae, and P. aeruginosa. Based on this survey of the peptide-neomycin library, circumvention of aminoglycoside-modifying enzymes and alluding to bacterial resistance is an important step toward the design and development of peptide aminoglycoside-based motifs for antimicrobial drug development.
1 Introduction
The global burden associated with antibacterial resistance has resulted in an estimated 4.71 million deaths reported for 2021 (Murray et al., 2022; World Health Organization, 2024; Naghavi et al., 2024). The World Health Organization's (WHO) most recent update of the list of pathogens in rank order of critical and high-priority pathogens includes, carbapenem-resistant Klebsiella pneumoniae, third-generation cephalosporin-resistant Escherichia coli, carbapenem-resistant Acinetobacter baumannii, rifampicin-resistant Mycobacterium tuberculosis fluoroquinolone-resistant Salmonella Typhi, fluoroquinolone-resistant Shigella spp., vancomycin-resistant Enterococcus faecium, and carbapenem-resistant Pseudomonas aeruginosa (Sati et al., 2025). The natural habitats of A. baumannii, Mycobacterium spp., and P. aeruginosa are soil and aquatic environments, whereas E. coli, K. pneumoniae, and other members of the order Enterobacterales are part of the normal human intestinal microbial flora. These opportunistic pathogens comprise the ESKAPE opportunistic pathogens, including Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter spp., which are identified as being highly virulent and antibiotic resistant and are the leading cause of hospital-acquired infections (De Oliveira et al., 2020). In particular, A. baumannii has been identified as having high rates of acquired pandrug resistance, defined as resistance to all agents within all classes of antibiotics (Falagas and Bliziotis, 2007), because of its versatility in the upregulation of intrinsic resistance determinants and the rapid acquisition of multiple antibiotic resistance genes under the selective pressure of antibiotic use. Among the well-characterized antibiotic resistance mechanisms, aminoglycoside-modifying enzymes (AMEs) are the most abundant and diverse in nature, with >85 resistance determinants identified (Ramirez and Tolmasky, 2010, 2017; Aishwarya et al., 2020; Fernanda et al., 2022). Aminoglycoside resistance mechanisms fall into three categories: (1) enzymatic modification of antibiotics by AMEs (acquired resistance), (2) alteration or upregulation of intrinsic factors (outer membrane proteins, efflux pumps, and penicillin-binding proteins), and (3) methylation of the 16S ribosomal RNA amino-acyl site (acquired resistance). In particular, A. baumannii strains can acquire resistance genes at an alarming rate compared with other groups of bacteria. Chromosomal analysis of extensively drug-resistant A. baumannii strains has identified large resistance islands on the chromosome with up to 50 resistance genes (Imperi et al., 2011). These resistance islands are flanked by insertion sequences and encode transposase genes, indicating their recent origin from multiple genera of Gram-negative bacteria, including P. aeruginosa, K. pneumoniae, and E. coli. The acquisition of highly adaptable resistance determinants in the face of antibiotic-induced selective pressure necessitates the development of new therapies to treat infections caused by opportunistic bacterial pathogens.
Considering that no new approved antibacterial with a novel mechanism of action has been developed in the past 30 years, there has been an increase in the use of combination therapy with standard care drugs and the development of new drugs. There is a rapid rate of resistance to clinically important aminoglycosides (amikacin, gentamicin, tobramycin, and plazomicin) (Golkar et al., 2021; Bassenden et al., 2021). The occurrence of toxicity associated with aminoglycosides can narrow the window for their clinical use (Arya, 2007). However, in light of combination drug therapy, there is renewed interest. Combination therapy using tobramycin or amikacin and the last resort drug colistin is often necessary for treating invasive lung infections caused by Gram-negative bacteria harboring multidrug-resistant genes (Ramsey et al., 1999; Hodson et al., 2002; Fulnecky et al., 2005). Polymyxin B and colistin (polymyxin E) are both used in the clinical setting for treating bacterial infections, and it is recommended that colistin be administered in combination with one or more active antimicrobial agents, such as tobramycin. Polymyxins are composed of polycationic lipopeptides that interact with lipopolysaccharides of the Gram-negative bacterial outer membrane by binding divalent cations (Mg2+ and Ca2+), resulting in the loss of outer membrane integrity, eventually leading to cell lysis and death (Newton, 1956; Hancock, 1997; Evans et al., 1999; Hermsen et al., 2003). Aminoglycosides, such as tobramycin and neomycin, exert antibacterial activity by binding to the amino-acyl site (A-site) of the 16S rRNA component of the 23S ribosomal subunit, which results in the inhibition of protein synthesis and ultimately cell death (Davis, 1987; Tevyashova and Shapovalova, 2021). It is thought that the mechanism of action of the aminoglycoside/colistin combination is because of an accelerated influx of tobramycin caused by cell membrane damage and heightened permeabilization of the outer membrane (OM), which induces more rapid cell death than either alone (Hurdle et al., 2011; Zou et al., 2018). Generally, the interaction between aminoglycosides and polymyxins, as used clinically, has an additive effect that can lower the concentrations required for each drug, thereby reducing toxicity (Almutairi, 2022; Bayatinejad et al., 2023; Wang et al., 2022; Güzel and Gerçeker, 2008; Zhu et al., 2022).
Several studies have demonstrated that conjugation of aminoglycosides with small molecules improves target binding and/or biological activity (Aradi et al., 2020; Arya, 2005; Charles and Arya, 2005; Jiang et al., 2015; Jin et al., 2016; Watkins et al., 2017; Ghosh et al., 2018; Kellish et al., 2014; Ranjan and Arya, 2019; McFarland et al., 2024; Kumar et al., 2013; Ranjan et al., 2020; Story et al., 2019; Watkins et al., 2019; Chandrika et al., 2018; Ramos et al., 2017; Degtyareva et al., 2017; Ranjan and Arya, 2016; Nahar et al., 2015; Ranjan and Arya, 2013; Willis and Arya, 2009), allowing the action of two (Xue et al., 2002) or more (Willis and Arya, 2009, 2010) parts of the conjugate (for example, the aminoglycoside and amino acid, peptide, or aromatic unit attached) to function together and present a synergistic effect against the target binding site. Positively charged amino acids can form hydrogen bonds with unpaired RNA bases, have strong electrostatic interactions with the negatively charged RNA backbone, and interact with the outer membrane (OM) of Gram-negative bacteria. Basic amino acids, such as arginine and lysine, when conjugated to aminoglycosides, kanamycin or gentamicin, have been shown to have increased affinity and selectivity for RNA targets (Jiang et al., 2015; Litovchick et al., 1999; Lapidot et al., 2004; Tevyashova and Shapovalova, 2021; Jin et al., 2016). Additionally, more than one mechanism of action was found for lysine-neomycin conjugates, allowing increased penetration (membrane action) and inhibition of efflux (Bera et al., 2011). Another example of molecules with multiple modes of action is conjugates of membrane-acting compounds with tobramycin, allowing both membrane action and inhibition of protein synthesis (Dhondikubeer et al., 2012; Du et al., 2015; Rebizant, 2022; Gambato et al., 2023). In our previous studies, mono- and di-amino acid conjugates of neomycin or kanamycin were synthesized with the goal of overcoming the mechanisms of antibiotic resistance via enzymatic inactivation and drug efflux through improvement of the affinity and selectivity toward the bacterial 16S rRNA A-site. Several conjugates of the 215-member peptide-neomycin (P-NEO) library had higher affinity for A-site rRNA than neomycin (NEO). P-NEO conjugates that were more effective than NEO contained cysteine, tryptophan, lysine, and/or arginine residues. However, in aminoglycoside-resistant and/or intrinsically polymyxin-resistant bacteria, several tryptophan-NEO conjugates are ineffective (Jiang et al., 2015; Jin et al., 2016; Kukielski et al., 2018).
We hypothesized that the OM of Gram-negative bacteria had limited permeability to some P-NEO conjugates and that increasing the permeability of the OM with a sub-minimal inhibitory concentration (sub-MIC) of polymyxin B (PB) would act synergistically with the P-NEO conjugates and improve their bactericidal effect. In this study, the antibacterial activity of a P-NEO conjugate library (Jiang et al., 2015) in combination with PB was assayed against several strains of aminoglycoside-resistant A. baumannii, K. pneumoniae, and P. aeruginosa. These include extensive pandrug-resistant bacteria harboring 16S rRNA methyltransferases. A significant synergy between P-NEO and PB was observed across bacterial genera. However, an additive to an indifferent effect was observed with the parent compound, NEO, and other aminoglycosides. The MICs for the top P-NEO and PB combinations were below the maximum concentration that could be achieved in the serum and below the minimal inhibitory concentration of the parent compound NEO. This synergistic action improves the bactericidal effect of P-NEO to achieve an increased immunity against antibiotic resistance, such that the use of much lower drug concentrations can potentially reduce possible aminoglycoside- and polymyxin-based nephrotoxicities.
2 Materials and methods
2.1 Bacterial strains and test compounds
Genotypic and phenotypic antibiotic resistance profiles of the bacterial strains used in this study are given in Table 1. The majority of the bacterial strains used in this study were obtained from the Centers for Disease Control (CDC) and Prevention's Antimicrobial Isolate Resistance Bank. For each bacterial isolate, the complete antibiotic susceptibility profiles and antibiotic resistance genes present can be found on the CDC website: cdc.gov/ARIsolateBank/Panel/AllIsolate. Other bacterial isolates were received or purchased from BEI Resources, the Antimicrobial Resistance Leadership Group (ARLG), and the American Type Culture Collection (ATCC). Polymyxin B and aminoglycosides were purchased as sulfate salts from various vendors. Concentrated stocks of control aminoglycosides, PB, and P-NEO conjugates were prepared in water, aliquoted in cryovials, and stored at −80°C until use. P-NEO conjugates were synthesized in our laboratory, and their purity and other characteristics have been reported previously (Jiang et al., 2015). Bacterial stocks were stored at −80°C until use. Isolates were maintained on Luria-Bertani agar plates and routinely checked for purity and identity verification.
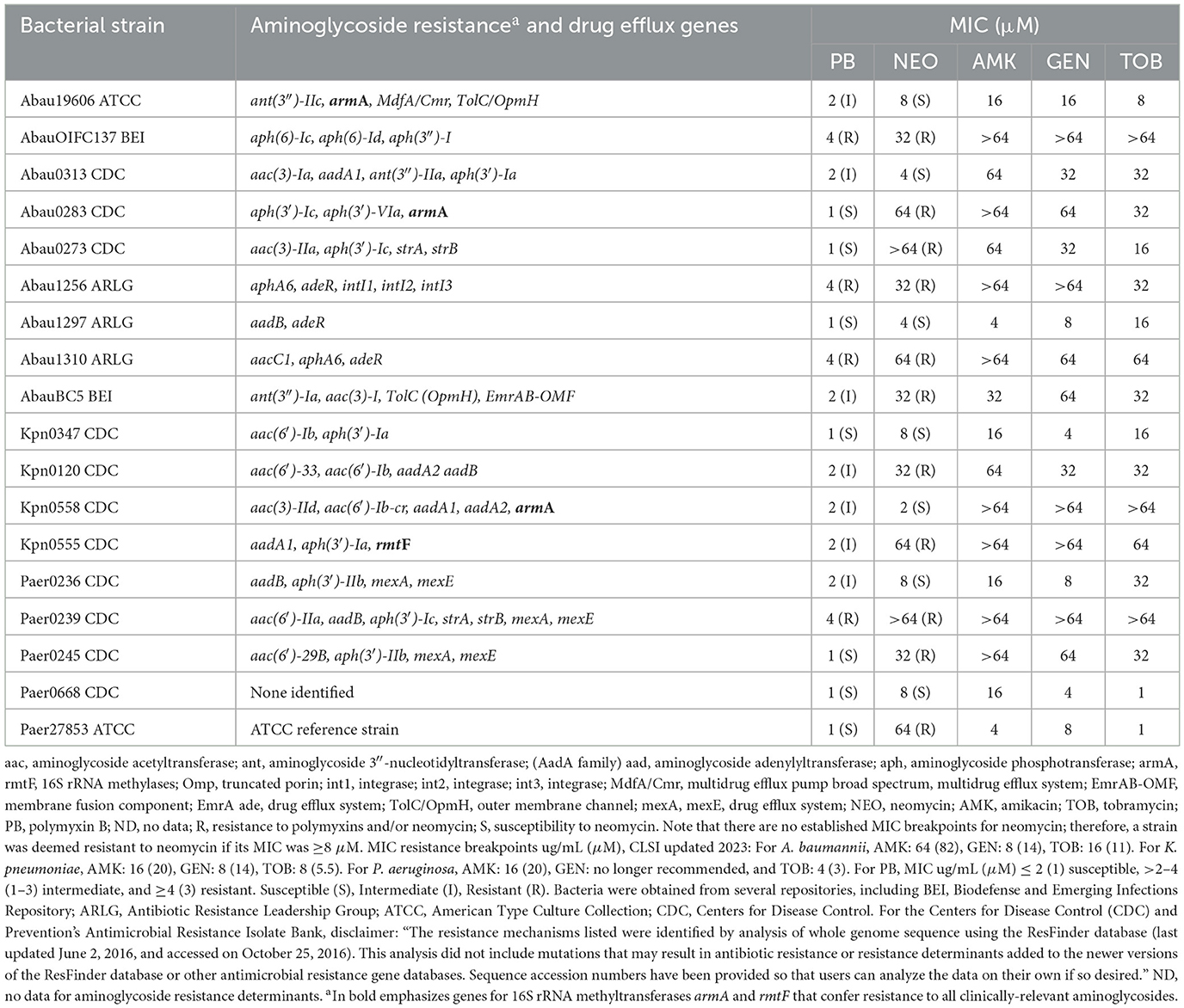
Table 1. Relevant genotypic and phenotypic profiles of aminoglycoside-resistant bacterial strains were chosen to screen an NEO-peptide library.
2.2 Minimal inhibitory concentration determination
MIC assays were conducted according to the Clinical Laboratory Standards Institute (CLSI) guidelines (document M100-M07). Briefly, the MICs of control aminoglycosides and P-NEO conjugates were determined in triplicate using the broth microdilution method in Mueller–Hinton II cation-adjusted (MHII-CA) medium. To determine the MICs, the concentration range of each drug (0.019–64 μM) was prepared by serial dilution in MHII-CA broth. To identify the sub-MIC of each test compound, the IC50 and IC25 were determined from the MICs. Fetal bovine serum (20%) was used to determine the effect of serum proteins on the MIC for the selected drug and drug combinations. Microtiter 96-well polystyrene plates containing 10 μL of 10X stock of each drug were inoculated with 90 μl of each bacterial strain (final cell concentration adjusted to ~5 × 105 cells/mL) and incubated at 37°C for 16–20 h. MHII-CA broth with and without bacterial inoculum was used as the positive and negative controls, respectively. Optical density at 600 nm was measured using a plate reader. The MIC was defined as the lowest concentration that showed ≥90% growth inhibition in the MHII-CA medium. The percent growth inhibition was determined using the following equation:
2.3 Single-point concentration synergy screening
Bacterial strains were used in a high-throughput single-point concentration screening assay to determine the efficacy of the additive effect of the drug combination of the sub-MIC of PB (PB) and each P-NEO conjugate at 5 μM. The sub-MIC PB for each strain varied (0.25–1 μM) depending on the PB susceptibility of the bacterial strain. The single-point assay was performed as follows: a 10X solution (50 μM) of each P-NEO conjugate was prepared from 1 mM stock, and 10 uL of each compound was aliquoted into the wells of a 96-well styrene plate using an automated liquid handler. Bacteria were grown to the early exponential phase in tryptic soy broth and diluted 10,000-fold to a final cell concentration of ~ 5 × 105 cells/mL in an appropriate volume of MH II broth containing sub-MIC PB. The bacterial suspension was manually dispensed in 90 μL aliquots to the wells of the P-NEO test plates and mixed immediately using a multichannel pipettor to reduce the localized action of the concentrated P-NEO solution at the bottom of the plate. Controls included sterile water (sterility control), bacterial suspension without drug (growth control), sub-MIC PB, 5 μM NEO, sub-MIC PB + NEO, and minimal inhibitory concentration of NEO or PB that killed ≥ 90% of the bacterial culture (positive controls). Plates were incubated for 24–48 h in a humidified chamber at 37°C and bacterial growth was monitored by light absorbance (OD600nm) using a plate reader. The results were averaged from triplicate samples, and the variation was calculated as the standard deviation from the mean.
2.4 Checkerboard titrations
The two-way dilution checkerboard assay was conducted in MHII-CA broth for each P-NEO conjugate that exhibited 70%−100% growth inhibition with sub-MIC PB in the single-point concentration synergy screen (SPCSS) described above. Six 2-fold dilutions for each test compound stock in MHII-CA were performed to obtain a 20X P-NEO conjugate with a concentration range from 31.25 μM to 100 μM and a concentration range of 0.625–40 μM for PB. The first drug component dilution (PB) was added to the top row of a 96-well polystyrene plate and diluted. The second drug dilution was added to the first column (P-NEO) and diluted to produce a 10X checkerboard plate. Then, ten microliters from each well of the stock checkerboard plate were transferred using an automated liquid handler to 96-well assay plates. A 90 μL volume of exponential-phase bacteria at a cell density of 5 x 105 CFU mL−1 was added to each well of the assay plate, and the plates were incubated at 37°C in a humidified chamber for 18–24 h. After the incubation period, the absorbance at OD600 was measured using a plate reader. The MIC and MIC in combination with ≥90% growth inhibition were used to determine the fractional inhibitory concentration index (FICI), which is defined as follows:
where FIC is the fractional inhibitory concentration in combination and MIC is the minimal inhibitory concentration.
2.5 Time kill analysis
Time-kill assays were performed on the combinations found to be “synergistic” or “additive” using the checkerboard method described above. Time-kill analysis was performed according to CLSI document M26-A guidelines. For each test compound and test combination, three polypropylene 50 mL tubes containing 10 mL of MHII-CA broth containing the drug or drug combination were inoculated with a mid-log-phase aliquot of the test strain to a density of ~5 × 105 CFU mL−1 in a final volume of 10 ml and incubated in a shaking incubator at 37°C in ambient air. Aliquots were removed at 0, 2, 4, 6, 8, and 24 h post-inoculation and serially diluted in sterile 0.85% sodium chloride solution to determine the culturable counts. At each time point and for each of the three replicate assay tubes, 100 μL aliquots were spread-plated on 80 mm diameter nutrient agar plates in duplicate trypticase soy agar plates using a spiral plater. Total culturable bacteria (LOG10 CFU mL−1) were determined after 24 h of incubation at 37°C.
Synergy was defined as a ≥ 2- LOG10 decrease in colony count at 6, 8, or 24 h with the antimicrobial combination compared to the most active single agent. Indifference was defined as a <2-LOG10 increase or decrease in colony count at 6, 8, or 24 h with the combination compared with the most active drug alone. Antagonism was defined as a ≥2-LOG10 increase in colony count at 6, 8, or 24 h with the combination compared with that of the most active drug alone. Three time-kill assay tubes were used: untreated growth control, sterile control (MHII-CA broth without drug or bacterial inoculum), sub-MIC PB alone or sub-MIC NEO alone, MIC for PB or NEO, and 0.5×, 1× , and 2× the MIC for the combination found in the checkerboard synergy assay.
2.6 Microtiter plate assay for biofilm quantification of A. baumannii strains
For biofilm inhibition, 100 μl of 1 × 105 cells mL−1 in Mueller Hinton II cation-adjusted (MHII-CA) broth was added to the wells of a high-binding polystyrene microtiter plate, and 100 μL of test solution was added to the cell suspension and mixed well by pipetting using a multichannel pipettor. Test solutions were as follows: MHII-CA (growth control), 0.4 μM PB, 4 μM NEO & 0.4 μM PB or 4 μM P-NEO & 0.4 μM PB. Assay plates were incubated for 16 h at 37°C. After incubation, planktonic cells were gently aspirated from the biofilm layer, and the biofilm was washed twice with phosphate-buffered saline (PBS; pH 7.4). After incubating the assay plates, biofilms were fixed with methanol for 15 min at room temperature. Fixed biofilms were washed twice with PBS, and 200 μl of 0.2% crystal violet solution was added to each well. After 5 min, excess crystal violet was removed by aspiration, the cells were washed twice with PBS, and they were air-dried. Cell-bound crystal violet was dissolved in 33% acetic acid, and the biofilm mass was quantified by spectroscopy (OD570) using a microplate reader (SPARK, Tecan).
Biofilm eradication: Biofilms were pre-formed onto 96-well flat-bottom polystyrene microtiter plates as follows: 100 μl of exponential phase cells in MHII-CA with an OD600 of 0.1 was inoculated into each well, and the plate was incubated for 16 h at 37°C to allow attachment of cells and biofilm formation. After incubation, the biofilm was washed as described above and treated with 100 μL of test solutions containing MHII-CA (growth control), 0.8 μM PB, 8 μM NEO, and 0.8 μM PB or 8 μM P-NEO & 0.8 μM PB. Biofilms were incubated for 16 h at 37°C. After incubation, the procedures for crystal violet staining of biofilms outlined in the previous paragraph were performed.
2.7 Resistance development
A 14-day resistance development assay was performed using neomycin-susceptible strains A. baumannii 19606, K. pneumoniae 0558, and P. aeruginosa 0668. To prepare for the drug challenge, bacteria were grown overnight in MHII non-cation-adjusted (MHII) broth. After overnight incubation, the cell densities were adjusted to 5 × 105 CFU mL−1, and 90 μL was dispensed into the wells of a 96-well non-binding microtiter plate containing 10X solutions of the drug or a combination over the dilution range above and below the MICs and incubated at 37°C for 24 h. The cell suspension was quickly mixed with the drug solution using a multichannel pipettor to avoid the localized effects of the drugs. For subsequent challenge days, the wells at the subinhibitory MIC were sampled and diluted to achieve an OD600 reading of 0.08 and then diluted again by mixing 2 μL of the cell suspension with 5 mL MHII broth. After adjusting for cell density, 90 μL of the cell suspension was dispensed into drug challenge plates. Growth was monitored by measuring absorbance at OD600 over a period of 14 days. Purity checks were performed every 2 days to identify whether cross-contamination occurred by spread plating from wells with the most diluted drug exposure. The minimum bactericidal concentration was determined on day 14 using the spread plate method. When resistance development was observed, the stability of resistance was determined by taking the day 14 contents at the MIC and suspending them in 2 mL MHII, taking the contents at the sub-MIC, suspending them in 10 mL MHII, and incubating overnight. After incubation, 5 × 105 CFU mL−1 was dispensed onto the drug challenge plate and incubated for 24 h.
2.8 Statistical analysis
The primary single-point synergy screen data were expressed as the mean and standard deviation of triplicate data. The paired two-tailed Student's t-test was used to determine the differences between the untreated and treated bacteria in the biofilm assays. Two-way ANOVA with repeated measures and post-hoc Kruskal–Wallis and Mann–Whitney tests were performed for the time-kill data. Data were analyzed using OriginPro software.
3 Results
3.1 Extensive aminoglycoside resistance among clinical isolates of A. baumannii, K. pneumoniae, and P. aeruginosa
A list of P-NEO conjugates and their descriptions is provided in Supplementary Table S1 (refer to Figure 1 for representative P-NEO conjugates). Prior to the selection of bacteria for screening of the P-NEO library (Jiang et al., 2015), a panel of ~30 strains of clinical isolates of A. baumannii, K. pneumoniae, and P. aeruginosa was profiled for their susceptibility to a wide range of aminoglycosides, including NEO (Figures 2a–c). Clinical isolates were obtained from the Centers for Disease Control and Prevention's Antibiotic Resistance Isolate Bank, which has a large repository of bacteria collected from healthcare and food industries and communities across the United States. The aminoglycoside susceptibility profile differed between the three groups of bacteria with respect to NEO compared to plazomicin, the newest generation aminoglycoside. Many of these strains carried at least one gene encoding AME, and several strains carried three or more genes. Among the A. baumannii strains, 44% carried armA, compared to K. pneumoniae (23%) and P. aeruginosa (6%), which carried either armA or rmtA-G (Supplementary Table S2). Among the A. baumannii strains, approximately half were susceptible to NEO, and approximately one-third were susceptible to plazomicin (Figure 2a). For other aminoglycosides such as amikacin and tobramycin, A. baumannii susceptibility rates were low, with <30% susceptibility. K. pneumoniae strains showed greater susceptibility to most of the aminoglycosides tested (Figure 2b). The P. aeruginosa panel showed greater resistance to NEO but greater susceptibility to plazomicin (Figure 2c).
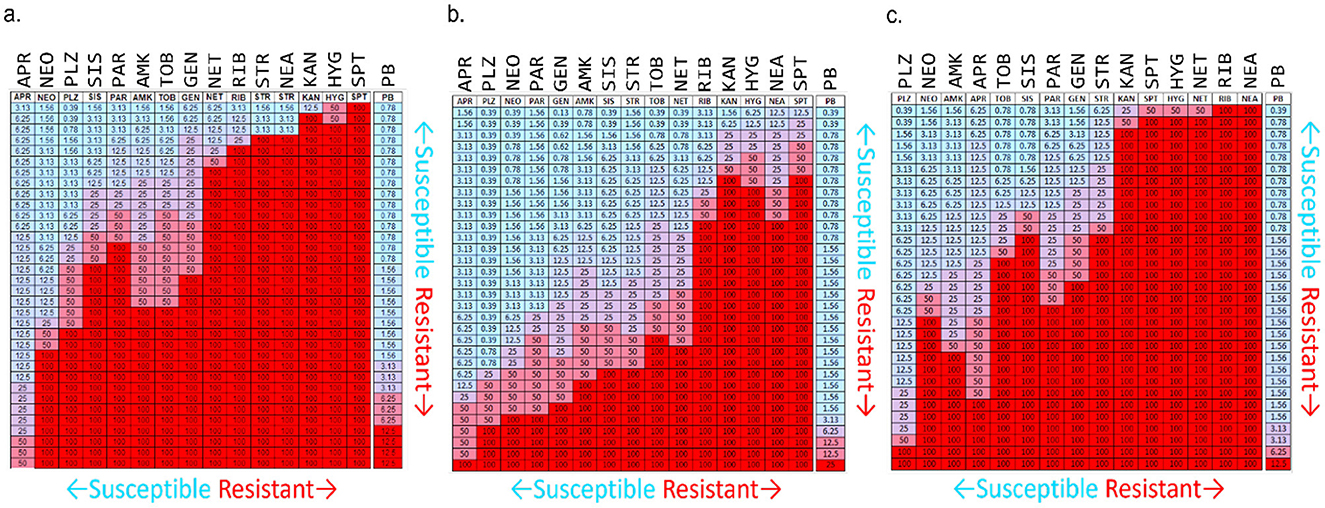
Figure 2. Susceptibility/resistance profiles for aminoglycosides and polymyxin B in ~30 clinical isolates of (a) A. baumannii, (b) K. pneumoniae, and (c) P. aeruginosa. Heat map interpretation: Blue to lavender = susceptible (MIC range 0.39–12.5 μM), purple to red = resistant (MIC range 25 to ≥100 μM). Numbers reflect the MIC for each aminoglycoside. The highest concentration used was 50 μM (purple cells); if an MIC was not found, 100 μM was administered instead (red cells). APR, apramycin; NEO, neomycin; PLZ, plazomicin; SIS, sisomicin; PAR, paromomycin; AMK, amikacin; TOB, tobramycin; GEN, gentamicin; NET, netilmicin; RIB, ribostamycin; STR, streptomycin; NEA, neamine; KAN, kanamycin; HYG, hygromycin; SPT, spectinomycin; PB, polymyxin B. Bacterial strains were acquired from the Centers for Infectious Disease Control and Prevention's Antimicrobial Resistance Bank, American Type Culture Collection, Antimicrobial Resistance Leadership Group, and BEI Resources. For a list of bacteria represented in this figure for aminoglycoside susceptibility profiling and their aminoglycoside resistance determinants (see Supplementary Table S2).
Based on the aminoglycoside susceptibility profiles shown in Figure 2, strains of A. baumannii, K. pneumoniae, and P. aeruginosa were selected for screening of the P-NEO library. Those that were chosen varied in susceptibility to NEO and PB and were mostly resistant to amikacin (AMK), gentamicin (GEN), and tobramycin (TOB) (Table 1, Supplementary Table S2). Selection was also based on the types of AME genes and 16S rRNA methyltransferases (Jouybari et al., 2021; Galimand et al., 2005, 2012; Yang and Hu, 2022) those carrying drug efflux genes and multidrug-resistant integrons (Vaillancourt et al., 2021; Richmond et al., 2016; Shetty et al., 2024), and susceptibility to PB. Both neomycin-resistant and sensitive strains are present in the selected panel, and 7 of the 18 strains were susceptible to NEO. Of the 18 strains, 7 were susceptible, 7 were intermediate, and 4 were resistant to PB. The majority of bacteria in the panel were also resistant to clinically relevant aminoglycosides, amikacin, gentamicin, and tobramycin.
3.2 Primary screening of the P-NEO library reveals P-NEO conjugates with arginine, cysteine, tyrosine, and/or tryptophan are effective against aminoglycoside-resistant A. baumannii
Previously, we observed little growth inhibition with P-NEO conjugates alone when screened against aminoglycoside-resistant bacteria, although several P-NEO conjugates demonstrated good binding affinity to 16S bacterial ribosomal A-site RNA using our fluorescent-neomycin binding displacement probe (Jiang et al., 2015; Watkins et al., 2013; Story and Arya, 2024; Watkins et al., 2015a; King et al., 2013). Therefore, a primary screen using a single-point concentration synergy screen (SPCSS) with 5 μM P-NEO and the specific sub-MIC PB for a particular strain was used to screen the P-NEO library. The P-NEO library is represented by conjugates of one or two L-form amino acids linked to NEO. The primary SPCSS began with NEO-susceptible (NEOS) and NEO-resistant strains (NEOR) of A. baumannii (Figure 3). Abau19606 had a NEO MIC of 8 μM, and AbauOIFC137 had a NEO MIC of 32 μM, respectively. These two A. baumannii strains also differed in their susceptibility to PB, with Abau19606 being susceptible and AbauOIFC137 being resistant. A comparison of the activity of the P-NEO conjugates between the NEOS and NEOR strains may indicate better target binding to the 16S rRNA A-site (Story and Arya, 2024), rather than better permeabilization facilitated by PB.
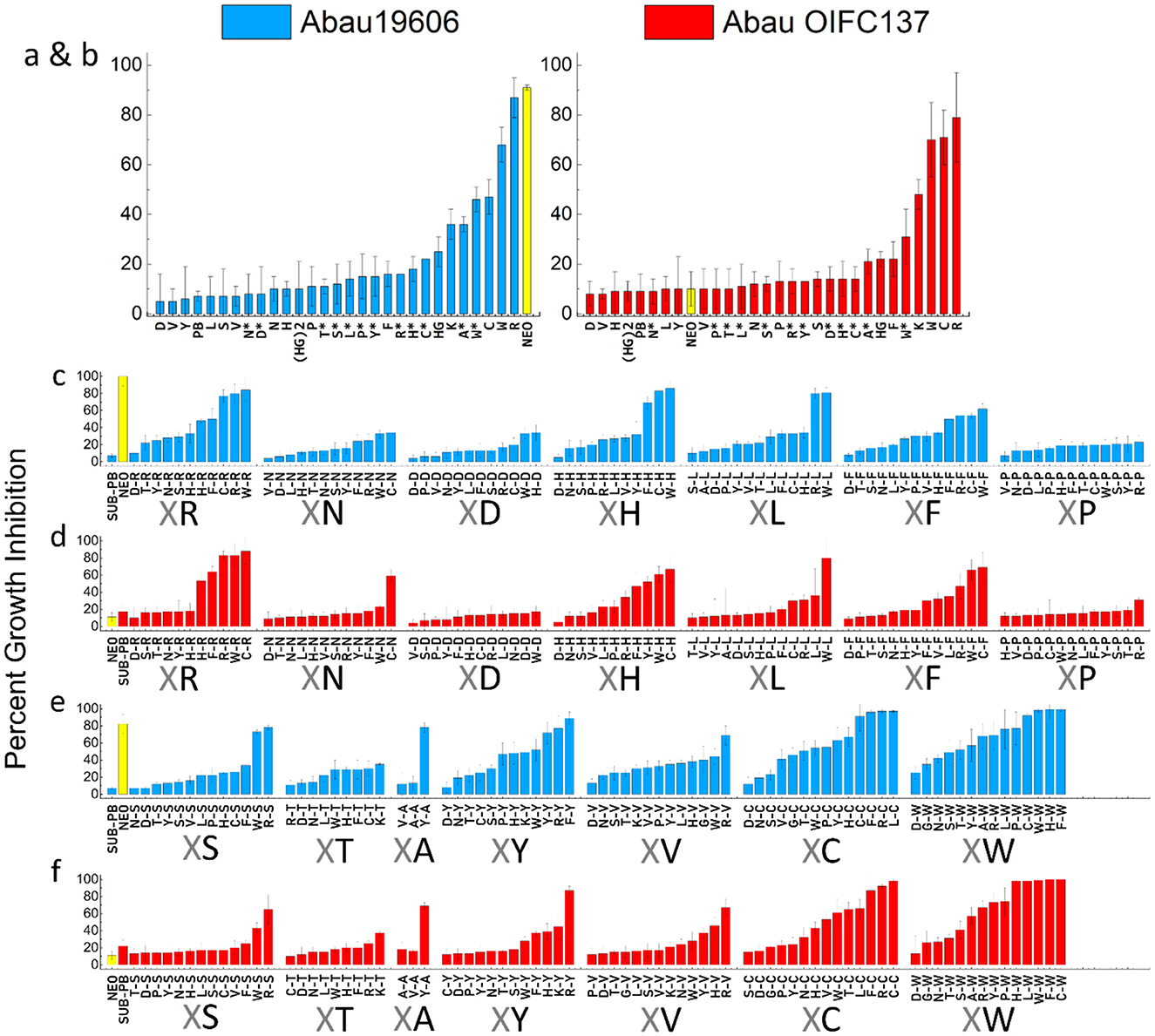
Figure 3. Percentage growth inhibition in a single-point synergy screen (SPCSS) of a P-NEO library (5 μM) in combination with sub-MIC of PB (PB) against A. baumannii strains. NEO-susceptible Abau19606 (blue = NEOS, MIC 4–8 μM), and NEO-resistant AbauOIFC137 (red = NEOR, MIC 32 μM). The sub-MIC of PB did not significantly affect growth after 24 h of incubation (≤ 20% growth inhibition). Neo linked with one amino acid (a, b) and with a β-alanine end group (marked with an asterisk*). NEO linked to two amino acids (c–f), where X is any of the 18 amino acids, followed by the common amino acid linked to NEO (e.g., X-R-NEO, X-N-NEO and so on). For the common amino acid adjacent to the X amino acid and linked to NEO: R, Arginine; N, Asparagine; D, Aspartic acid; H, Histidine; L, Leucine; F, Phenylalanine; P, Proline; S, Serine; T, Threonine; A, Alanine; Y, Tyrosine; V, Valine; C, Cysteine; W, Tryptophan. Error bars represent the standard deviation of the mean of three replicates. The percent growth inhibition values are relative to growth control without PB, ≤ 20% growth inhibition is considered insignificant. Neomycin (NEO) was screened at 5 μM (yellow bar) and served as a control. Conjugates that inhibited growth by ≥70% in combination with PB were used in the checkerboard assays (refer to Supplementary Figures S2–S4 for an expanded presentation of this figure).
The primary SPCSS with P-NEO conjugates containing one L-form amino acid for NEOS and NEOR A. baumannii strains showed ≥70 growth inhibition for P-NEO conjugates with arginine (R), cysteine (C), and tryptophan (W) (Figures 3a, b). R*, C*, and W* had an additional β-alanine end group, which was not as effective (20%−50% growth inhibition). Next, P-NEO conjugates with two L-form amino acids were compared (Figures 3c, d), where the end amino acid (X) is any of the 18 amino acids followed by a common amino acid that is linked to NEO (i.e., XR, XN, XD, and so on). For the diamino acid NEO group, we found that any combination of amino acids R, C, W, F, Y, S, and H was the most effective (≥ 80% growth inhibition) in NEOS and NEOR A. baumannii strains. Overall, NEOR AbauOIF137 had a lower susceptibility to the P-NEO and PB combination than NEOS Abau19606. The least effective P-NEO conjugates were those containing amino acids N, D, L, P, T, and A linked to NEO. Across NeoS and NEOR strains, the most effective P-NEO conjugates (with ≥80% growth inhibition) found in the SPCSS had a combination of R, W, H, F, and S.
3.3 Checkerboard analysis of P-NEO conjugates supports a synergistic interaction between P-NEO conjugates and PB
Although the SPCSS screen for the combination of P-NEO and PB against several NEOS and NEOR strains has provided some insight into which P-NEO conjugates may be most effective, SPCSS was susceptible to a high false-negative hit rate. Based on the primary screening, several P-NEO conjugates that included both negative and positive hits were selected for checkerboard analysis to determine the relationship between the interaction of P-NEO and PB (Figure 4). For this screening, the assay was extended to other bacteria with different aminoglycoside resistance mechanisms, including NEOS, NEOR, A. baumannii, K. pneumoniae, and P. aeruginosa. The checkerboard analysis included NEO as the control, and for all strains tested, an additive to indifferent effect was observed. The NEO MICs for the NEOS strains Abau0313, Kpn0347, and Paer0236 were 4 μM, and those for the NEOR strains Abau0283, Kpn0120, and Pear0239 were 64, 32, and 64 μM, respectively. MICs for P-NEO conjugates ranged from 16 to >64 μM across NEOS and NEOR strains, with NEOS strains generally having lower MIC values (Supplementary Table S3). MICs for PB were in the intermediate range (1 μM) to the intermediate-resistant range (2–4 μM). For bacterial strains in which synergy was observed (Figure 4), the MIC in combination with P-NEO conjugates was reduced by 4–64-fold below its MIC (Supplementary Table S3). In contrast, the MIC for PB in combination was much narrower, being reduced by 2–4-fold. RC-NEO, CW-NEO, and HW-NEO demonstrated significant synergy with PB and P. aeruginosa strains, and the additive effects of the combination were primarily observed for other P-NEO conjugates.
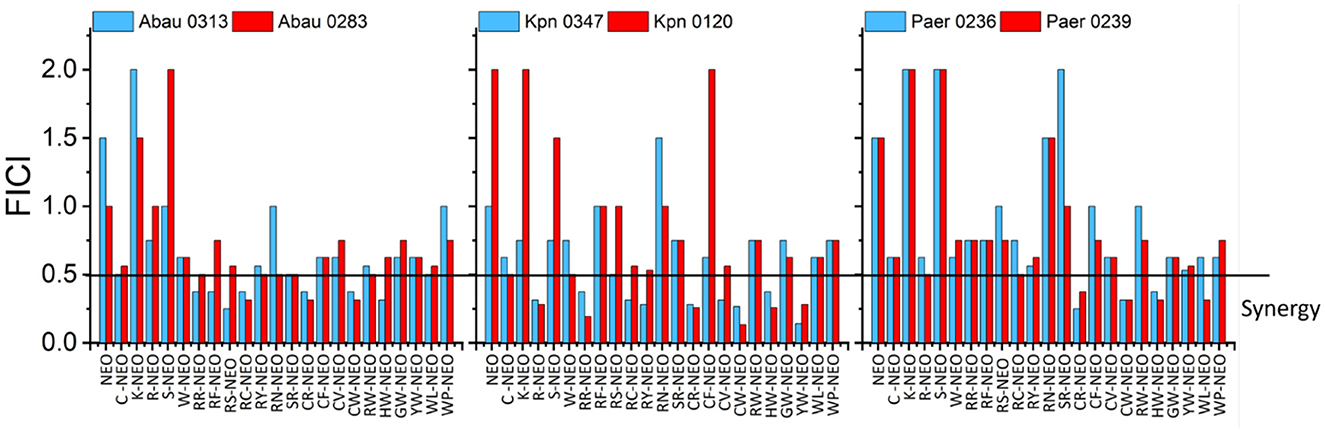
Figure 4. Comparison of fractional inhibitory concentration indices (FICI) derived from two-way checkerboard dilutions for the combination of the selected L-form amino acids P-NEO and PB in one NEOS (blue) and one NEOR (red) for A. baumannii (Abau0313, Abau0283), K. pneumoniae (Kpn0347, Kpn0120), and P. aeruginosa (Paer0236, Paer0239). FICI was calculated as follows: FICI = (FICP − NEO/MICP − NEO + FICPB/MICPB). FICI interpretation: ≤ 0.5 indicates a synergistic interaction, >0.5–1.0 indicates an additive interaction, >1–4 indicates an indifferent interaction, and >4 indicates an antagonistic interaction. NEO, neomycin; C, cysteine; K, lysine; R, arginine; S, serine; W, tryptophan; F, phenylalanine; Y, tyrosine; N, asparagine; V, valine; H, histidine; G, glycine; P, proline. Because it was impractical to find the P-NEO MIC for some of the highly resistant strains, the FICI value was calculated using 64 μM as the cutoff. See Supplementary Table S3 for the MIC and FIC values from which FICI calculations were derived.
3.4 Time-kills demonstrate synergy for the RC-NEO and PB combination
For the time-kill assay, synergy was defined as a 2-log10 decrease in colony count by the combination compared with that by the most active single agent (PB), the most active combination (NEO+PB), or as a 2-log10 decrease in colony count compared with the starting inoculum. NEOR strains of A. baumannii (Abau 0283, extensively drug-resistant), K. pneumoniae (Kpn 0120, extensively drug-resistant), and P. aeruginosa (Paer 0239, pandrug-resistant) were used. Colony-forming units per milliliter (CFU mL−1) were monitored for 24 h, and the following drug conditions were compared to the untreated control: sub-MIC for PB, sub-MIC for the combination of NEO+PB, and 0.5, 1, and 2 times the MICs found for the combination of RC-NEO and PB in the checkerboard analyses for each strain (Supplementary Table S3). Synergy was assessed in the 4–8 h range because of the resumption of growth by 24 h (Figure 5). For all strains, a bactericidal effect on PB was observed with a 3-fold reduction in the CFU mL−1 by 6 h and little to no recovery by 24 h, and NEO at MIC was bacteriostatic, with little change in the CFU/mL over 4 h and a gradual increase in growth by 24 h (data not shown). Some growth inhibition with sub-MIC of PB and NEO+PB was observed for most strains, with complete or near-complete recovery of growth after 24 h. A dose-dependent effect was observed for the combination of RC-NEO and PB at 0.5, 1, and 2 times the MIC. For A. baumannii, there was a significant difference between the combination of PB+NEO and PB+RC-NEO using twice the MIC at 4 h and 8 h, where a ≥2-fold LOG CFU mL−1 reduction with PB+RC-NEO was observed (p = 0.003). Additionally, a significant difference between 1 and 2 times the combination for RC-NEO+PB was found (p = 0.028) (Supplementary Tables S5–S10). Two-fold log reductions in CFU mL−1 for PB+RC-NEO compared to PB+NEO were found to be statistically significant for K. pneumoniae and P. aeruginosa using 1 and 2 times the RC-NEO+PB combination (Supplementary Tables S11–S22). Because of rebound growth after 8 h, a second administration of drugs between 6 and 8 h may extend the period of growth inhibition. The results from the time-kill assays support the SPCSS and checkerboard observations for the RC-NEO.
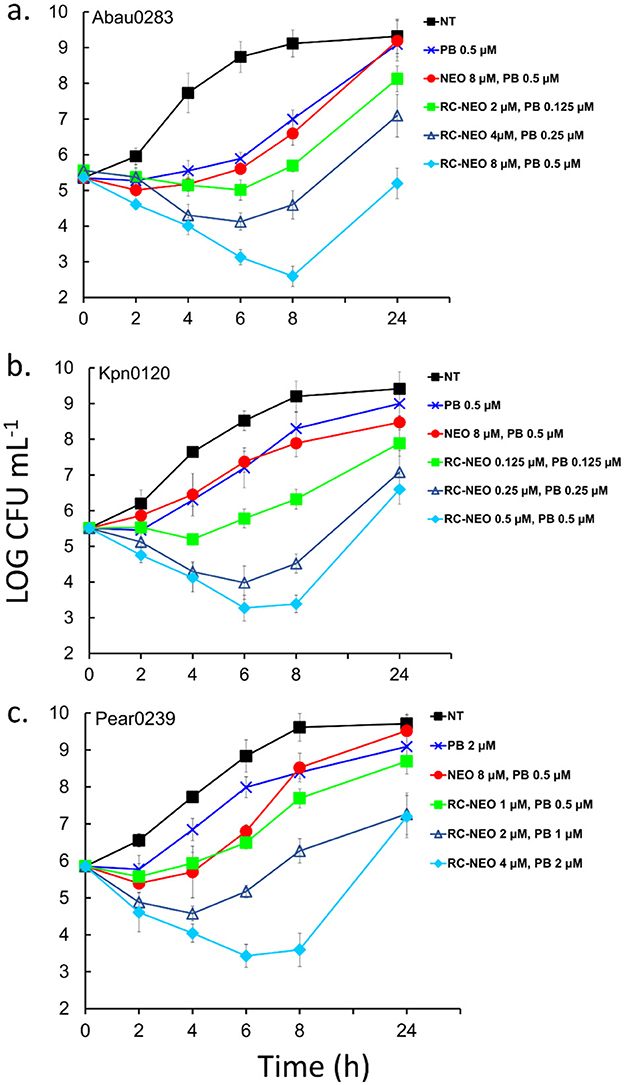
Figure 5. Time-kill curves of NEO-resistant strains of (a) extensively drug-resistant A. baumannii strain 0283, (b) extensively drug-resistant K. pneumoniae strain 0120, and (c) pandrug-resistant P. aeruginosa strain 0239 for RC-NEO and control NEO in combination with PB. LOG10 colony-forming units per milliliter (CFU mL−1) were monitored for 24 h. The concentrations used for the drugs alone and in combination reflect the concentrations used for each bacterial strain and were determined using the checkerboard synergy assay (Supplementary Table S3). Synergy was defined as a ≥2-LOG10 decrease in colony count at 6 or 8 h with the combination compared to the most active single agent (PB). Indifference was defined as a <2-LOG10 increase or decrease in colony count at 6, 8, or 24 h with the combination compared with the most active component. Antagonism was defined as a ≥2-LOG10 increase in colony count at 6, 8, or 24 h with the combination compared with that of the most active component alone. The NEO and PB MICs for A. baumannii (64 and 1 μM), K. pneumoniae (64 and 1 μM), and P. aeruginosa (>64 and 4 μM), respectively. Error bars represent the standard deviation from the mean of three replicate assay vials, with duplicate plating from each assay vial. Tests for significance: Two-way ANOVA with repeated measures was followed by post-hoc Bonferroni tests and additional non-parametric Kruskal–Wallis and Mann–Whitney tests for significance at p = 0.005 and p = 0.001. Data tables for the statistical metrics are presented in Supplementary Tables S5–S22.
The effect of serum on the drug MIC and drug MIC in combination was evaluated in the neomycin-susceptible strains of A. baumannii ATCC 19606, K. pneumoniae CDC 0558, and P. aeruginosa CDC 0668 (Table 2). For PB and NEO, the MIC was generally unchanged, whereas the MICs for R-NEO, C-NEO, and RC-NEO increased by 2-fold. For the MICs in combination, no change was observed for K. pneumoniae and P. aeruginosa, whereas a 2–4-fold decrease was observed for the combination of PB and R-NEO, C-NEO, and RC-NEO with A. baumannii. Brightfield microscopy demonstrated changes in cell morphology with different treatments, where elongation of the cells and/or long chains of cells were observed with RC-NEO alone or in combination with PB, lysis of cells with PB, and very small cells with NEO (Supplementary Figure S6).
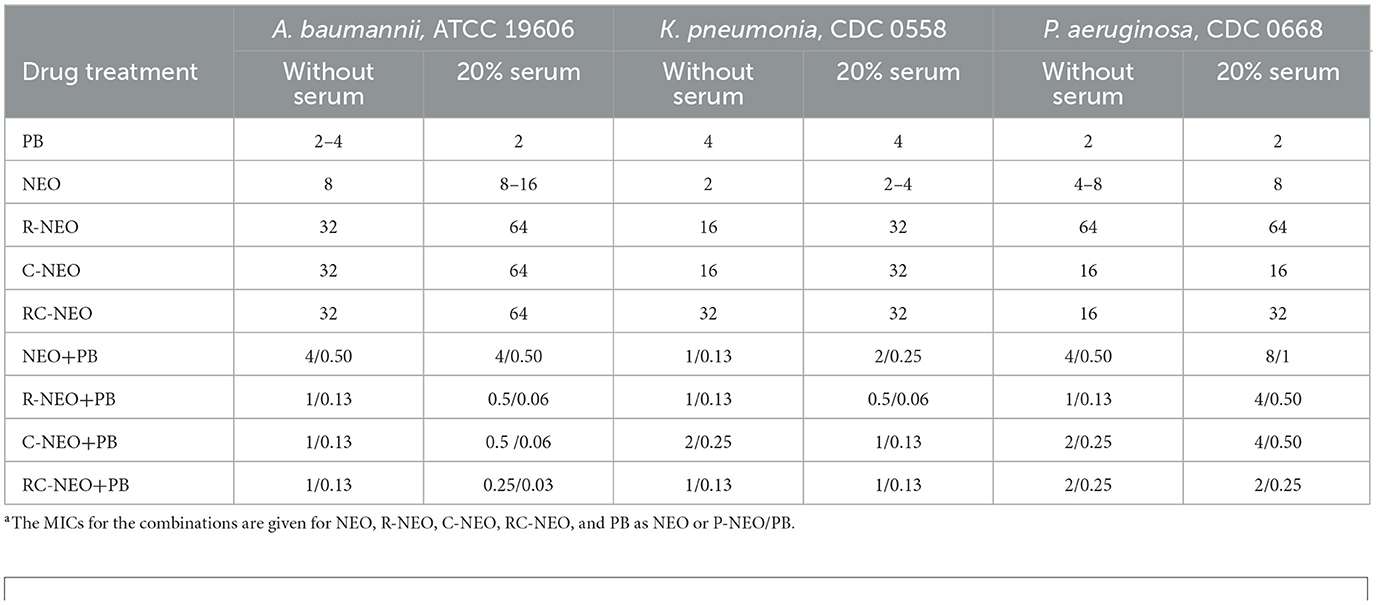
Table 2. Effect of serum on the minimal inhibitory concentrationa (MIC) of drugs alone and in combination with neomycin-susceptible bacteria.
3.5 Inhibition of A. baumannii biofilm formation
The time-kill curves demonstrated a synergistic interaction between P-NEO and PB. The time-kill assay specifically tested the effect of the drugs on cells in the planktonic state, which is much more susceptible to bactericidal and bacteriostatic actions of the drugs alone and in combination than in the biofilm-associated cell state. Using the same drug combinations as in the time-kill assay, biofilm formation and eradication were evaluated for four A. baumannii strains with different levels of NEO susceptibility and aminoglycoside resistance determinants (Figure 6). The biofilm formation for the RC-NEO & PB combination against A. baumannii strains relative to the untreated control was 21% for Abau19606, 37% for Abau0273, 39% for Abau0283, and 17% for Abau0313 (Figure 6a). The percentage of biofilm remaining for established A. baumannii biofilms by the combination of RC-NEO and PB for Abau19606, Abau0273, Abau0283, and Abau0313 was 52%, 63%, 77%, and 39% of the untreated growth control, respectively. Significant reductions in the RC-NEO & PB combination relative to the NEO & PB combination were observed for Abau19606, Abau0273, and Abau0283 (Figure 6b).
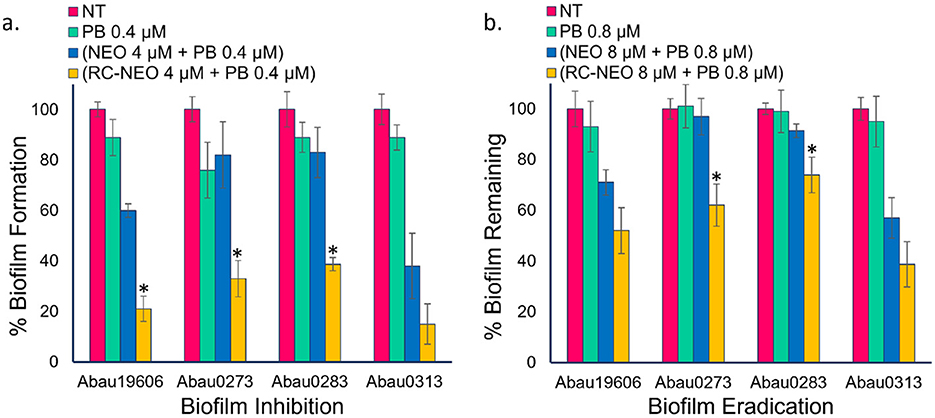
Figure 6. Biofilm inhibition and eradication indicate the percentage of biofilms formed or remaining in comparing four A. baumannii strains. (a) NT: no treatment growth control, PB: treatment with 0.4 μM PB alone, treatment with 4 μM NEO or RC-NEO in combination with 0.4 μM PB, (b) NT, no treatment growth control; PB, treatment with 0.8 μM PB alone, treatment with 8 μM NEO or RC-NEO in combination with 0.8 μM PB. Data are expressed as the mean ± standard deviation of triplicate data. The percent biofilm for the treatment groups was normalized to the biofilm formed or remaining for the untreated growth control, based on the crystal violet retained. Error bars represent the standard deviation of the mean of three replicates. A two-tailed Student's t-test was used to determine the differences in biofilm formation between the control (NEO+PB) and test P-NEO (RC-NEO+PB) combinations, where *p ≤ 0.001.
3.6 Lack of resistance development to peptide-neomycin conjugates
A 14-day resistance development assay was performed using the neomycin-susceptible strains A. baumannii 19606, K. pneumoniae 0558, and P. aeruginosa 0668 (Figure 7). Resistance development was not observed for the P-NEO conjugates R-NEO, C-NEO, and RC-NEO, alone or in combination with PB (Supplementary Tables S23–S25). In A. baumannii and K. pneumoniae, the MICs for the combinations demonstrated a narrow range of MICs for combinations of 1–2 μM, and for P. aeruginosa, the combination MIC range was 2–4 μM. By day 9, A. baumannii began to develop resistance to PB, with an 8-fold increase in MIC by day 14. However, this resistance was transient once the PB was removed (Figure 7). Resistance to NEO was not observed in A. baumannii and P. aeruginosa during this period. In contrast, K. pneumoniae developed significant resistance to both PB (8-fold increase in the MIC) and NEO (16-fold increase in the MIC) when administered alone, and this resistance appeared to be stable. Additionally, the MIC for the NEO+PB combination increased 4-fold in K. pneumoniae. It appeared that P. aeruginosa developed resistance to PB toward the end of the assay period, with a 4-fold increase in the MIC after the drug was removed for 24 h.
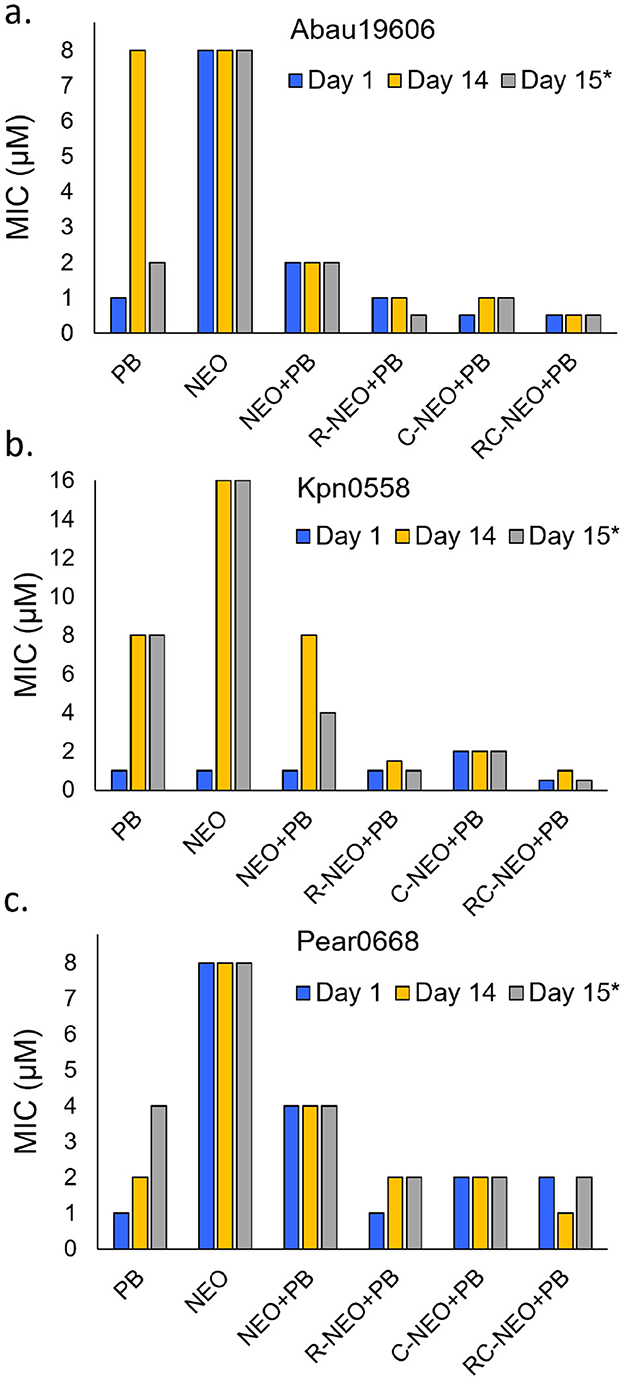
Figure 7. Test for resistance development comparing drug challenge on day 1 and day 14 as reflected in changes in the MIC for drugs alone or in combination. Day 15*: reflects the MICs after a 24 h removal of the drug or drug combination from day 14 cultures. (a) NEO susceptible A. baumannii 19606, (b) NEO susceptible K. pneumoniae 0558, (c) NEO susceptible P. aeruginosa 0668. The concentration range for PB administered alone was 0.125–8 μM in the combinations was 0.0625–4 μM. The MICs are given for NEO, R-NEO, C-NEO and RC-NEO in the combination with PB. The median MIC is presented for Day 14 where a trend in resistance was not observed over 14 days (Supplementary Tables S23–S25).
3.7 Additive to indifferent effect for the combination of aminoglycosides and polymyxin B
The significant synergy observed between the P-NEO conjugates and PB was in stark contrast to that observed with NEO and PB, where an additive to indifferent relationship was found. To identify whether this lack of synergy with PB was unique to NEO or was a general rule for all aminoglycosides, ten other aminoglycosides in combination with PB were assessed with several strains of A. baumannii (Table 3). Generally, for strains exhibiting susceptibility, an additive effect was observed, and for those highly resistant, indifference to antagonism was found. One strain, Acinetobacter radioresistens, which is part of the normal flora of human skin, although susceptible to carbapenems and other antibiotics, is thought to be a source of carbapenem resistance and acquisition of other antibiotic resistance determinants in A. baumannii (Poirel et al., 2008; Lazarev et al., 2022). A. radioresistens is significantly more susceptible to aminoglycosides. A synergistic relationship with PB was observed with NEO, AMK, TOB, GEN, KAN, and PAR, for which the lowest MICs were observed. However, an additive to the indifferent relationship was observed for APR, SPT, STR, and RIB, where higher MICs were observed. Bacteria of other genera, such as Klebsiella, Escherichia, Salmonella, and Shigella, showed a similar pattern for the interaction of PB and the aminoglycosides amikacin and the latest-generation aminoglycoside plazomicin, which depended on the degree of susceptibility to the aminoglycoside (Supplementary Table S4). It is believed that both specific and indirect synergistic interactions occur. Notably, for highly susceptible strains, although the MIC for aminoglycosides in combination was markedly reduced (up to 16-fold), the reduction in the MIC for PB was at most 4-fold.
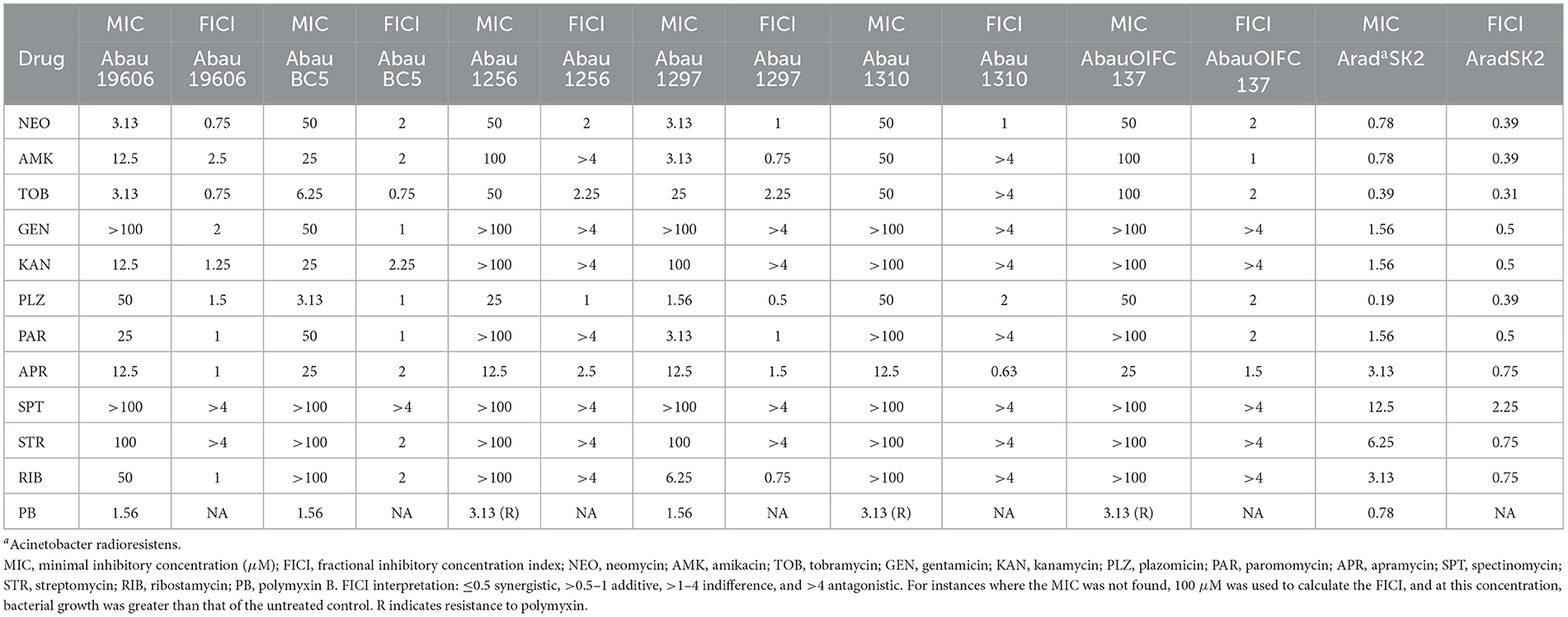
Table 3. Minimal inhibitory concentrations and fractional inhibitory concentration indices for the combination of aminoglycosides and PB in A. baumannii strains.
4 Discussion
We tested the growth inhibitory effect of a 191-member P-NEO library in combination with PB for extensively aminoglycoside-resistant and susceptible Gram-negative opportunistic pathogenic strains of A. baumannii, K. pneumoniae, and P. aeruginosa. The library was designed to compare the P-NEO conjugates with one or two amino acids. Each bacterial group in this study poses a unique challenge for drug treatment options because of differences in both acquired and intrinsic resistance factors. Previously, we identified members of the P-NEO library that did not serve as substrates for AMEs (Jiang et al., 2015). Besides AME genes, other acquired and intrinsic factors contribute to resistance to aminoglycosides. For example, there is a high prevalence of A. baumannii strains carrying armA, an acquired resistance determinant that confers resistance to clinically relevant aminoglycosides. Intrinsic colistin heteroresistance is common among A. baumannii strains. K. pneumoniae produces a thick extracellular capsular material that is important for biofilm formation and supports intrinsic aminoglycoside tolerance and resistance. Consistently, clinical isolates of K. pneumoniae had a greater diversity of AME resistance determinants than either A. baumannii or P. aeruginosa. P. aeruginosa strains are especially well-equipped to upregulate their intrinsic drug efflux systems and employ quorum sensing in the establishment of biofilms and drug tolerance.
The combination of PB with specific P-NEO conjugates in aminoglycoside-resistant bacteria appeared not to serve as substrates for AMEs and bypass other resistance determinants as follows: (1) Previously, we found that P-NEO conjugates were not modified by purified AMEs (Jiang et al., 2015; Jin et al., 2016), and for the present study, all bacteria tested carried some variation of AME resistance determinants for aminoglycoside acetyltransferases and/or aminoglycoside phosphotransferases. (2) P-NEO conjugates are effective in bacteria carrying 16S rRNA methyltransferases such as armA and rmt genes, which included A. baumannii 0283 and K. pneumoniae strains 0558 and 0555. (3) The combination with PB allows uptake of P-NEO conjugates that alone cannot pass the outer membrane of Gram-negative bacteria and also allows uptake in capsulated bacteria (all K. pneumoniae tested). (4) It appears that drug efflux mechanisms are not sufficient for P-NEO export (all P. aeruginosa tested carried mexA and mexE drug efflux determinants). An additional factor that affects treatment options is the prevalence of colistin resistance among A. baumannii. In addition to the colistin resistance determinant mcr-1, A. baumannii can regulate the composition of the outer membrane lipid A or lose the lipid A portion through the expression of intrinsic factors, resulting in heteroresistance and complete resistance to colistin (Jean ShioShin and Hsueh PoRen, 2011; Moffatt et al., 2011; López-Rojas et al., 2011; Ko et al., 2007). However, the loss of the lipid A component of the outer membrane can increase A. baumannii susceptibility to drugs it is resistant to, including aminoglycosides (Moffatt et al., 2011, 2010; Mu et al., 2016; Carretero-Ledesma et al., 2018). Resistance did not develop with R-, C-, or RC-NEO conjugates alone or in combination with PB in the NEO-susceptible strains of A. baumannii 19606, K. pneumoniae 0558, and P. aeruginosa 0668. A. baumannii 19606 developed transient resistance to PB, K. pneumoniae 0558 developed a stable resistance to both PB and NEO, and P. aeruginosa 0668 developed resistance to PB toward the end of the drug-challenge time period. These bacteria host various acquired and intrinsic resistance factors. Resistance to polymyxins in vitro can develop via heteroresistance or chromosomal mutations, causing a transient or stable modification of lipid A and/or polysaccharides of the outer membrane (Andersson et al., 2019), and has been reported to occur rapidly among the populations of A. baumannii, K. pneumoniae, and P. aeruginosa strains in vitro (Thi Khanh Nhu et al., 2016; Janssen et al., 2020; Dößelmann et al., 2017). Surprisingly, the K. pneumoniae strain 0558 developed stable resistance to both PB and NEO. This strain has multiple AME genes and carries 16S rRNA methyltransferase armA. With the exception of NEO and apramycin, this strain was resistant to all clinically relevant aminoglycosides, including plazomicin (Supplementary Table S27 for the full aminoglycoside resistance profile and antibiotic resistance determinants).
P-NEO conjugates with cysteine (C), arginine (R), tryptophan (W), or tyrosine (Y) were the most effective, consistent with our previous study that analyzed the structure-activity relationship for P-NEO binding to the bacterial 16S rRNA A-site and bacterial growth inhibition. Previous studies (Jiang et al., 2015; Watkins et al., 2015b; Jin et al., 2016) demonstrated that the top P-NEO 16S rRNA A-site binders have a combination of W, Y, R, K, S, C, R, and H. The lowest MIC for E. coli was found with R-NEO (5 μM), followed by RN-NEO, RH-NEO, RS-NEO, RY-NEO, RV-NEO, RC-NEO, and RW-NEO (20 μM), well above the MIC for NEO (<1 μM). In contrast, the top P-NEOs for P. aeruginosa were WX-NEO, CX-NEO, XS-NEO, and XW-NEO (where X is any other amino acid). In the present study, the most effective P-NEO conjugates had the amino acid residues R, C, Y, and W, or combinations thereof. Notably, several strains carrying 16S rRNA methyltransferases were susceptible. Arginine carries a very stable positive charge among amino acids. The ionic interactions of arginine and PB with the outer membrane likely contributed to the synergistic relationship observed for A. baumannii and K. pneumoniae and the additive effects observed with P. aeruginosa. In bacteria, cysteine is the least abundant residue in structural proteins and is primarily found as a functional site in proteins (Marino and Gladyshev, 2012). Cysteine is cytotoxic to bacteria at low concentrations; thus, its intracellular concentration is highly regulated through its degradation and efflux as a part of sulfur assimilation (Takumi and Nonaka, 2016). Cysteine at low levels induces amino acid starvation, inhibits isoleucine synthesis, promotes reactive oxygen species, and elicits sulfide production, all contributing to cysteine toxicity (Sørensen and Pedersen, 1991; Korshunov et al., 2020; Marino and Gladyshev, 2012). It is unknown whether NEO conjugates with cysteine residues exhibit the same toxic properties as cysteine. C-NEO conjugates in combination with PB may result in the accumulation of C-NEO in the cytoplasm and may be resistant to intracellular degradation, in addition to the synergistic effects observed with PB. In addition to cysteine- and arginine-NEO conjugates, P-NEO with tryptophan and tyrosine residues appeared to be more effective against P. aeruginosa strains. Previous studies have demonstrated that both D- and L-isomers of tryptophan inhibit P. aeruginosa biofilm formation and disrupt quorum sensing (Brandenburg et al., 2013). The combination of amikacin and tyrosine inhibits P. aeruginosa biofilms (She et al., 2015). Another study found that the end modification of short antimicrobial peptides with one tryptophan residue enhanced the growth inhibitory activity of the peptide against P. aeruginosa, and additional tryptophan residues further improved the anti-pseudomonad activity. These tryptophan-enriched peptides were proposed to induce killing through cell wall and vesicle damage and increased binding to the LPS of the outer membrane (Pasupuleti et al., 2009). Our W-NEO conjugates, in combination with PB, may also inhibit biofilm formation in P. aeruginosa by binding to outer membrane LPS, in addition to binding to the RNA A-site.
For extensively drug-resistant Gram-negative bacteria, combination drug therapy is often deployed to treat infection. Colistin (polymyxin E) is considered a last-resort drug for the treatment of multidrug-resistant infections and is used in combination with amikacin or tobramycin, which have been shown to be effective in treating lung infections caused by multidrug-resistant bacteria (Tappenden et al., 2013; Herrmann et al., 2010; Taccetti et al., 2021; Buendía et al., 2024). Generally, the effect of the colistin/aminoglycoside combination has been shown to have an additive to indifferent relationship in vitro (Almutairi, 2022; Bayatinejad et al., 2023; Wang et al., 2022; Güzel and Gerçeker, 2008; Zhu et al., 2022). A synergistic relationship was found between P-NEO and PB, but at best, an additive effect was observed with NEO and other aminoglycosides for aminoglycoside-resistant bacteria and most of the susceptible bacteria. Based on the mechanism of action of aminoglycosides compared to PB, the data given in Table 3 and Supplementary Table S4 indicate both specific synergism and indirect synergism (Cokol et al., 2011). When the strain is highly aminoglycoside resistant because of aminoglycoside-modifying enzymes, drug efflux, and/or 16S rRNA methylases and polymyxin resistance factors, an antagonistic relationship prevails where the combination promotes growth, as observed in extensively drug-resistant A. baumannii strains (Ocampo et al., 2014). In highly aminoglycoside-susceptible strains, more extensive membrane damage results from both aminoglycoside and PB binding to the OM of Gram-negative cells. Initially, destabilization of the OM by PB would give aminoglycosides a “push” in passage across the cell wall and increased accumulation in the periplasmic space. Aminoglycoside entry into the cytoplasm is facilitated by the proton-motive force of the inner membrane. Once in the cell, irreversible binding to the target 16S rRNA A-site of the 30S ribosomal subunit induces misreading of the mRNA code and production of aberrant proteins. Misfolded truncated proteins cause further damage to the cell membrane, resulting in a surge of aminoglycosides in the cytoplasm, ultimately leading to cell death.
Bacterial biofilms pose a significant challenge in treating chronic infections, as biofilm cells are more tolerant to antibiotic treatment and grow rapidly on surfaces such as medical devices. The RC-NEO and PB combination was very effective in inhibiting biofilm growth but less effective in reducing pre-established A. baumannii biofilms, as it required four times more of the combination to observe a significant reduction.
5 Conclusions
In conclusion, we report a novel finding of a synergistic relationship between P-NEO and PB in both extensively drug- and pandrug-resistant bacteria, including those carrying 16S rRNA methyltransferase genes. In contrast, an additive to the indifferent relationship between aminoglycosides and PB was consistently observed in both aminoglycoside-sensitive and -resistant strains. P-NEO conjugates containing cysteine, arginine, or tryptophan residues were the most effective in synergy with PB, significantly lowering the MIC for P-NEO several-fold, with a smaller reduction in the MIC for PB. Amino acid-linked NEO can evade modification by AMEs and facilitate the “ESKAPE” of resistance development. Given that peptide modifications can be rapidly achieved using solution- or solid-phase chemistries, these findings provide promising new and rapidly tunable tools to mitigate the growth of drug-resistant pathogens using peptide-linked aminoglycosides.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
SS: Data curation, Formal analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing. LJ: Data curation, Writing – review & editing. AL: Data curation, Formal analysis, Writing – review & editing. DA: Conceptualization, Funding acquisition, Project administration, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by National Institutes of Health (NIH) grants AI126874 and AI114114 to DA.
Conflict of interest
SS and AL were employed by NUBAD LLC. DA has ownership interest in NUBAD LLC.
The remaining author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Gen AI was used in the creation of this manuscript.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1605813/full#supplementary-material
References
Aishwarya, K. V. L., Geetha, P. V., Eswaran, S., Mariappan, S., and Sekar, U. (2020). Spectrum of aminoglycoside modifying enzymes in gram-negative bacteria causing human infections. J. Lab. Physicians 12, 27–31. doi: 10.1055/s-0040-1713687
Almutairi, M. M. (2022). Synergistic activities of colistin combined with other antimicrobial agents against colistin-resistant Acinetobacter baumannii clinical isolates. PLoS ONE 17:e0270908. doi: 10.1371/journal.pone.0270908
Andersson, D. I., Nicoloff, H., and Hjort, K. (2019). Mechanisms and clinical relevance of bacterial heteroresistance. Nat. Rev. Microbiol. 17, 479–496. doi: 10.1038/s41579-019-0218-1
Aradi, K. D. I., Giorgio, A., and Duca, M. (2020). Aminoglycoside conjugation for RNA targeting: antimicrobials and beyond. Chem. Eur. J. 26, 12273–12309. doi: 10.1002/chem.202002258
Arya, D. P. (2005). Aminoglycoside–nucleic acid interactions: the case for neomycin. DNA Binders Relat. Sub. 253, 149–178. doi: 10.1007/b100446
Arya, D. P. (2007). Aminoglycoside Antibiotics: From Chemical Biology to Drug Discovery. Hoboken, NJ: John Wiley and Sons. doi: 10.1002/9780470149676
Bassenden, A. V., Dumalo, L., Park, J., Blanchet, J., Maiti, K., Arya, D. P., et al. (2021). Structural and phylogenetic analyses of resistance to next-generation aminoglycosides conferred by AAC(2′) enzymes. Sci. Rep. 11:11614. doi: 10.1038/s41598-021-89446-3
Bayatinejad, G., Salehi, M., Beigverdi, R., Halimi, S., Emaneini, M., and Jabalameli, F. (2023). In Vitro antibiotic combinations of Colistin, Meropenem, Amikacin, and Amoxicillin/clavulanate against multidrug-resistant Klebsiella pneumonia isolated from patients with ventilator-associated pneumonia. BMC Microbiol. 23:298. doi: 10.1186/s12866-023-03039-w
Bera, S., Zhanel, G. G., and Schweizer, F. (2011). Synthesis and antibacterial activity of amphiphilic lysine-ligated neomycin B conjugates. Carbohydr. Res. 346, 560–568. doi: 10.1016/j.carres.2011.01.015
Brandenburg, K. S., Rodriguez, K. J., Mcanulty, J. F., Murphy, C. J., Abbott, N. L., Schurr, M. J., et al. (2013). Tryptophan inhibits biofilm formation by Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 57, 1921–1925. doi: 10.1128/AAC.00007-13
Buendía, J. A., Guerrero Patiño, D., and Zuluaga Salazar, A. F. (2024). Efficacy of adjunctive inhaled colistin and tobramycin for ventilator-associated pneumonia: systematic review and meta-analysis. BMC Pulm. Med. 24:213. doi: 10.1186/s12890-024-03032-7
Carretero-Ledesma, M., García-Quintanilla, M., Martín-Peña, R., Pulido, M. R., Pachón, J., and Mcconnell, M. J. (2018). Phenotypic changes associated with Colistin resistance due to Lipopolysaccharide loss in Acinetobacter baumannii. Virulence 9, 930–942. doi: 10.1080/21505594.2018.1460187
Chandrika, N. T., Shrestha, S. K., Ranjan, N., Sharma, A., Arya, D. P., and Garneau-Tsodikova, S. (2018). New application of neomycin B-bisbenzimidazole hybrids as antifungal agents. ACS Infect. Dis. 4, 196–207. doi: 10.1021/acsinfecdis.7b00254
Charles, I., and Arya, D. P. (2005). Synthesis of neomycin-DNA/peptide nucleic acid conjugates. J. Carbohydr. Chem. 24, 145–160. doi: 10.1081/CAR-200059973
Cokol, M., Chua, H. N., Tasan, M., Mutlu, B., Weinstein, Z. B., Suzuki, Y., et al. (2011). Systematic exploration of synergistic drug pairs. Mol. Syst. Biol. 7:544. doi: 10.1038/msb.2011.71
Davis, B. D. (1987). Mechanism of bactericidal action of aminoglycosides. Microbiol. Rev. 51, 341–350. doi: 10.1128/mr.51.3.341-350.1987
De Oliveira, D. M., Forde, B. M., Kidd, T. J., Harris, P. N., Schembri, M. A., Beatson, S. A., et al. (2020). Antimicrobial resistance in ESKAPE pathogens. Clin. Microbiol. Rev. 33:e00181-1. doi: 10.1128/CMR.00181-19
Degtyareva, N. N., Gong, C., Story, S., Levinson, N. S., Oyelere, A. K., Green, K. D., et al. (2017). Antimicrobial activity, AME resistance, and A-site binding studies of anthraquinone–neomycin conjugates. ACS Infect. Dis. 3, 206–215. doi: 10.1021/acsinfecdis.6b00176
Dhondikubeer, R., Bera, S., Zhanel, G. G., and Schweizer, F. (2012). Antibacterial activity of amphiphilic tobramycin. J. Antibiot. 65, 495–498. doi: 10.1038/ja.2012.59
Dößelmann, B., Willmann, M., Steglich, M., Bunk, B., Nübel, U., Peter, S., et al. (2017). Rapid and consistent evolution of colistin resistance in extensively drug-resistant Pseudomonas aeruginosa during morbidostat culture. Antimicrob. Agents Chemother. 61:e00043-17. doi: 10.1128/AAC.00043-17
Du, J., Bandara, H., Du, P., Huang, H., Hoang, K., Nguyen, D., et al. (2015). Improved biofilm antimicrobial activity of polyethylene glycol conjugated tobramycin compared to tobramycin in Pseudomonas aeruginosa biofilms. Mol. Pharm. 12, 1544–1553. doi: 10.1021/mp500846u
Evans, M. E., Feola, D. J., and Rapp, R. P. (1999). Polymyxin B sulfate and colistin: old antibiotics for emerging multiresistant gram-negative bacteria. Ann. Pharmacother. 33, 960–967. doi: 10.1345/aph.18426
Falagas, M. E., and Bliziotis, I. A. (2007). Pandrug-resistant gram-negative bacteria: the dawn of the post-antibiotic era? Int. J. Antimicrob. Agents 29, 630–636. doi: 10.1016/j.ijantimicag.2006.12.012
Fernanda, P. A., Liu, S., Yuan, T., Ramalingam, B., Lu, J., and Sekar, R. (2022). Diversity and abundance of antibiotic resistance genes and their relationship with nutrients and land use of the inflow rivers of Taihu Lake. Front. Microbiol. 13:1009297. doi: 10.3389/fmicb.2022.1009297
Fulnecky, E. J., Wright, D., scheld, W. M., Kanawati, L., and Shoham, S. (2005). Amikacin and colistin for treatment of Acinetobacter baumannii meningitis. J. Infect. 51, e249–e251. doi: 10.1016/j.jinf.2005.04.003
Galimand, M., Courvalin, P., and Lambert, T. (2012). RmtF, a new member of the aminoglycoside resistance 16S rRNA N7 G1405 methyltransferase family. Antimicrob. Agents Chemother. 56, 3960–3962. doi: 10.1128/AAC.00660-12
Galimand, M., Sabtcheva, S., Courvalin, P., and Lambert, T. (2005). Worldwide disseminated armA aminoglycoside resistance methylase gene is borne by composite transposon Tn1548 Antimicrob. Agents Chemother. 49, 2949–2953. doi: 10.1128/AAC.49.7.2949-2953.2005
Gambato, S., Bellotto, O., Mardirossian, M., Di Stasi, A., Gennaro, R., Pacor, S., et al. (2023). Designing new hybrid antibiotics: proline-rich antimicrobial peptides conjugated to the aminoglycoside tobramycin. Bioconjug. Chem. 34, 1212–1220. doi: 10.1021/acs.bioconjchem.2c00467
Ghosh, A., Degyatoreva, N., Kukielski, C., Story, S., Bhaduri, S., Maiti, K., et al. (2018). Targeting miRNA by tunable small molecule binders: peptidic aminosugar mediated interference in miR-21 biogenesis reverts epithelial to mesenchymal transition. Medchemcomm 9, 1147–1154. doi: 10.1039/C8MD00092A
Golkar, T., Bassenden, A. V., Maiti, K., Arya, D. P., Schmeing, T. M., and Berghuis, A. M. (2021). Structural basis for plazomicin antibiotic action and resistance. Commun. Biol. 4:729. doi: 10.1038/s42003-021-02261-4
Güzel, Ç. B., and Gerçeker, A. A. (2008). In vitro activities of various antibiotics, alone and in combination with colistin methanesulfonate, against Pseudomonas aeruginosa strains isolated from cystic fibrosis patients. Chemotherapy 54, 147–151. doi: 10.1159/000119741
Hermsen, E. D., Sullivan, C. J., and Rotschafer, J. C. (2003). Polymyxins:: Pharmacology, pharmacokinetics, pharmacodynamics, and clinical applications. Infect. Dis. Clin. 17, 545–562. doi: 10.1016/S0891-5520(03)00058-8
Herrmann, G., Yang, L., Wu, H., Song, Z., Wang, H., Høiby, N., et al. (2010). Colistin-tobramycin combinations are superior to monotherapy concerning the killing of biofilm Pseudomonas aeruginosa. J. Infect. Dis. 202, 1585–1592. doi: 10.1086/656788
Hodson, M., Gallagher, C., and Govan, J. (2002). A randomised clinical trial of nebulised tobramycin or colistin in cystic fibrosis. Eur. Respir. J. 20, 658–664. doi: 10.1183/09031936.02.00248102
Hurdle, J. G., O'Neill, A. J., Chopra, I., and Lee, R. E. (2011). Targeting bacterial membrane function: an underexploited mechanism for treating persistent infections. Nat. Rev. Microbiol. 9, 62–75. doi: 10.1038/nrmicro2474
Imperi, F., Antunes, L. C., Blom, J., Villa, L., Iacono, M., Visca, P., et al. (2011). The genomics of Acinetobacter baumannii: insights into genome plasticity, antimicrobial resistance and pathogenicity. IUBMB Life 63, 1068–1074. doi: 10.1002/iub.531
Janssen, A. B., Doorduijn, D. J., Mills, G., Rogers, M. R., Bonten, M. J., Rooijakkers, S. H., et al. (2020). Evolution of colistin resistance in the Klebsiella pneumoniae complex follows multiple evolutionary trajectories with variable effects on fitness and virulence characteristics. Antimicrob. Agents Chemother. 65:e01958-20. doi: 10.1128/AAC.01958-20
Jean ShioShin, J. S., and Hsueh PoRen, H. P. (2011). High burden of antimicrobial resistance in Asia. Int. J. Antimicrob. Agents 37(4):291-5. doi: 10.1016/j.ijantimicag.2011.01.009
Jiang, L., Watkins, D., Jin, Y., Gong, C., King, A., Washington, A. Z., et al. (2015). Rapid synthesis, RNA binding, and antibacterial screening of a peptidic-aminosugar (PA) library. ACS Chem. Biol. 10, 1278–1289. doi: 10.1021/cb5010367
Jin, Y., Watkins, D., Degtyareva, N. N., Green, K. D., Spano, M. N., Garneau-Tsodikova, S., et al. (2016). Arginine-linked neomycin B dimers: synthesis, rRNA binding, and resistance enzyme activity. Medchemcomm 7, 164–169. doi: 10.1039/C5MD00427F
Jouybari, M. A., Ahanjan, M., Mirzaei, B., and Goli, H. R. (2021). Role of aminoglycoside-modifying enzymes and 16S rRNA methylase (ArmA) in resistance of Acinetobacter baumannii clinical isolates against aminoglycosides. Rev. Soc. Bras. Med. Trop. 54:e05992020. doi: 10.1590/0037-8682-0599-2020
Kellish, P. C., Kumar, S., Mack, T. S., Spano, M. N., Hennig, M., and Arya, D. P. (2014). Multivalent amino sugars to recognize different TAR RNA conformations. Medchemcomm 5, 1235–1246. doi: 10.1039/C4MD00165F
King, A., Watkins, D., Kumar, S., Ranjan, N., Gong, C., Whitlock, J., et al. (2013). Characterization of ribosomal binding and antibacterial activities using two orthogonal high-throughput screens. Antimicrob. Agents Chemother. 57, 4717–4726. doi: 10.1128/AAC.00671-13
Ko, K. S., Suh, J. Y., Kwon, K. T., Jung, S.-I., Park, K.-H., Kang, C. I., et al. (2007). High rates of resistance to colistin and polymyxin B in subgroups of Acinetobacter baumannii isolates from Korea. J. Antimicrob. Chemother. 60, 1163–1167. doi: 10.1093/jac/dkm305
Korshunov, S., Imlay, K. R. C., and Imlay, J. A. (2020). Cystine import is a valuable but risky process whose hazards Escherichia coli minimizes by inducing a cysteine exporter. Mol. Microbiol. 113, 22–39. doi: 10.1111/mmi.14403
Kukielski, C., Maiti, K., Bhaduri, S., Story, S., and Arya, D. P. (2018). Rapid solid-phase syntheses of a peptidic-aminoglycoside library. Tetrahedron 74, 4418–4428. doi: 10.1016/j.tet.2018.07.012
Kumar, S., Kellish, P., Robinson, J. R. W. E., Wang, D., Appella, D. H., et al. (2013). Correction to click dimers to target HIV TAR RNA conformation. Biochemistry 52, 7159–7159. doi: 10.1021/bi401088n
Lapidot, A., Vijayabaskar, V., Litovchick, A., Yu, J., and James, T. L. (2004). Structure–activity relationships of aminoglycoside-arginine conjugates that bind HIV-1 RNAs as determined by fluorescence and NMR spectroscopy. FEBS Lett. 577, 415–421. doi: 10.1016/j.febslet.2004.10.038
Lazarev, A., Hyun, J., Sanchez, J. L., and Verda, L. (2022). Community-acquired Acinetobacter radioresistens bacteremia in an immunocompetent host. Cureus 14:e29650. doi: 10.7759/cureus.29650
Litovchick, A., Evdokimov, A. G., and Lapidot, A. (1999). Arginine-aminoglycoside conjugates that bind to HIV transactivation responsive element RNA in vitro. FEBS Lett. 445, 73–79. doi: 10.1016/S0014-5793(99)00092-7
López-Rojas, R., Jiménez-Mejías, M. E., Lepe, J. A., and Pachón, J. (2011). Acinetobacter baumannii resistant to colistin alters its antibiotic resistance profile: a case report from Spain. J. Infect. Dis. 204, 1147–1148. doi: 10.1093/infdis/jir476
Marino, S. M., and Gladyshev, V. N. (2012). Analysis and functional prediction of reactive cysteine residues. J. Biol. Chem. 287, 4419–4425. doi: 10.1074/jbc.R111.275578
McFarland, A. W. Jr., Fernando, L. P., Kellish, P., Story, S. P., Schober, G. B., KUMAR, S., et al. (2024). Nucleic acid specificity, cellular localization and reduced toxicities of thiazole orange-neomycin conjugates. Chem. Open 14:e202400189. doi: 10.1002/open.202400189
Moffatt, J. H., Harper, M., Adler, B., Nation, R. L., Li, J., and Boyce, J. D. (2011). Insertion sequence IS Aba11 is involved in colistin resistance and loss of lipopolysaccharide in Acinetobacter baumannii. Antimicrob. Agents Chemother. 55, 3022–3024. doi: 10.1128/AAC.01732-10
Moffatt, J. H., Harper, M., Harrison, P., Hale, J. D., Vinogradov, E., Seemann, T., et al. (2010). Colistin resistance in Acinetobacter baumannii is mediated by complete loss of lipopolysaccharide production. Antimicrob. Agents Chemother. 54, 4971–4977. doi: 10.1128/AAC.00834-10
Mu, X., Wang, N., Li, X., Shi, K., Zhou, Z., Yu, Y., et al. (2016). The effect of colistin resistance-associated mutations on the fitness of Acinetobacter baumannii. Front. Microbiol. 7:1715. doi: 10.3389/fmicb.2016.01715
Murray, C. J., Ikuta, K. S., Sharara, F., Swetschinski, L., Aguilar, G. R., Gray, A., et al. (2022). Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 399, 629–655. doi: 10.1016/S0140-6736(21)02724-0
Naghavi, M., Vollset, S. E., Ikuta, K. S., Swetschinski, L. R., Gray, A. P., Wool, E. E., et al. (2024). Global burden of bacterial antimicrobial resistance 1990–2021: a systematic analysis with forecasts to 2050. Lancet 404, 1199–1226. doi: 10.1016/S0140-6736(24)01867-1
Nahar, S., Ranjan, N., Ray, A., Arya, D. P., and Maiti, S. (2015). Potent inhibition of miR-27a by neomycin–bisbenzimidazole conjugates. Chem. Sci. 6, 5837–5846. doi: 10.1039/C5SC01969A
Newton, B. (1956). The properties and mode of action of the polymyxins. Bacteriol. Rev. 20, 14–27. doi: 10.1128/br.20.1.14-27.1956
Ocampo, P. S., Lázár, V., Papp, B., Arnoldini, M., Abel Zur Wiesch, P., Busa-Fekete, R., et al. (2014). Antagonism between bacteriostatic and bactericidal antibiotics is prevalent. Antimicrob. Agents Chemother. 58, 4573–4582. doi: 10.1128/AAC.02463-14
Pasupuleti, M., Chalupka, A., Mörgelin, M., Schmidtchen, A., and Malmsten, M. (2009). Tryptophan end-tagging of antimicrobial peptides for increased potency against Pseudomonas aeruginosa. Biochim. Biophys. Acta 1790, 800–808. doi: 10.1016/j.bbagen.2009.03.029
Poirel, L., Figueiredo, S., Cattoir, V., Carattoli, A., and Nordmann, P. (2008). Acinetobacter radioresistens as a silent source of carbapenem resistance for Acinetobacter spp. Antimicrob. Agents Chemother. 52, 1252–1256. doi: 10.1128/AAC.01304-07
Ramirez, M. S., and Tolmasky, M. E. (2010). Aminoglycoside modifying enzymes. Drug Resist. Updates 13, 151–171. doi: 10.1016/j.drup.2010.08.003
Ramirez, M. S., and Tolmasky, M. E. (2017). Amikacin: uses, resistance, and prospects for inhibition. Molecules 22:2267. doi: 10.3390/molecules22122267
Ramos, L. M., Degtyareva, N. N., Kovacs, N. A., Holguin, S. Y., Jiang, L., Petrov, A. S., et al. (2017). Eukaryotic ribosomal expansion segments as antimicrobial targets. Biochemistry 56, 5288–5299. doi: 10.1021/acs.biochem.7b00703
Ramsey, B. W., Pepe, M. S., Quan, J. M., Otto, K. L., Montgomery, A. B., Williams-Warren, J., et al. (1999). Intermittent administration of inhaled tobramycin in patients with cystic fibrosis. N. Engl. J. Med. 340, 23–30. doi: 10.1056/NEJM199901073400104
Ranjan, N., Andreasen, K. F., Arora, Y., Xue, L., and Arya, D. P. (2020). Surface dependent dual recognition of a G-quadruplex DNA with neomycin-intercalator conjugates. Front. Chem. 8:60. doi: 10.3389/fchem.2020.00060
Ranjan, N., and Arya, D. P. (2013). Targeting C-myc G-quadruplex: dual recognition by aminosugar-bisbenzimidazoles with varying linker lengths. Molecules 18, 14228–14240. doi: 10.3390/molecules181114228
Ranjan, N., and Arya, D. P. (2016). Linker dependent intercalation of bisbenzimidazole-aminosugars in an RNA duplex; selectivity in RNA vs. DNA binding. Bioorg. Med. Chem. Lett. 26, 5989–5994. doi: 10.1016/j.bmcl.2016.10.076
Ranjan, N., and Arya, D. P. (2019). A fluorescent aminosugar to rapidly screen and study RNA binders. Meth. Enzymol. 623, 291–314. doi: 10.1016/bs.mie.2019.05.027
Rebizant, J. (2022). The design, synthesis, and microbiological investigation of tobramycin catecholate conjugates (Thesis). Winnipeg, MB: University of Manitoba.
Richmond, G. E., Evans, L. P., Anderson, M. J., Wand, M. E., Bonney, L. C., Ivens, A., et al. (2016). The Acinetobacter baumannii two-component system AdeRS regulates genes required for multidrug efflux, biofilm formation, and virulence in a strain-specific manner. MBio 7:00430-16. doi: 10.1128/mBio.00430-16
Sati, H., Carrara, E., Savoldi, A., Hansen, P., Garlasco, J., Campagnaro, E., et al. (2025). The WHO bacterial priority pathogens list 2024: a prioritisation study to guide research, development, and public health strategies against antimicrobial resistance. Lancet Infect. Dis. Open Access. 11. doi: 10.1016/S1473-3099(25)00118-5
She, P., Chen, L., Liu, H., Zou, Y., Luo, Z., Koronfel, A., et al. (2015). The effects of D-tyrosine combined with amikacin on the biofilms of Pseudomonas aeruginosa. Microb. Pathog. 86, 38–44. doi: 10.1016/j.micpath.2015.07.009
Shetty, V. P., Rodrigues, C., and Deekshit, V. K. (2024). Detection of multidrug-resistant integrons associated with Acinetobacter baumannii Isolated from clinical and environmental samples. J. Pure Appl. Microbiol. 18:9013. doi: 10.22207/JPAM.18.1.44
Sørensen, M., and Pedersen, S. (1991). Cysteine, even in low concentrations, induces transient amino acid starvation in Escherichia coli. J. Bacteriol. 173, 5244–5246. doi: 10.1128/jb.173.16.5244-5246.1991
Story, S., and Arya, D. P. (2024). A cell-based screening assay for rRNA-targeted drug discovery. ACS Infect. Dis. 10, 4194–4207. doi: 10.1021/acsinfecdis.4c00446
Story, S., Skriba, M. J., Maiti, K., Ranjan, N., Degtyareya, N., Green, K. D., et al. (2019). Synthesis, antimicrobial activity, attenuation of aminoglycoside resistance in MRSA, and ribosomal A-site binding of pyrene-neomycin conjugates. Eur. J. Med. Chem. 163, 381–393. doi: 10.1016/j.ejmech.2018.11.022
Taccetti, G., Francalanci, M., Pizzamiglio, G., Messore, B., Carnovale, V., Cimino, G., et al. (2021). Cystic fibrosis: recent insights into inhaled antibiotic treatment and future perspectives. Antibiotics 10:338. doi: 10.3390/antibiotics10030338
Takumi, K., and Nonaka, G. (2016). Bacterial cysteine-inducible cysteine resistance systems. J. Bacteriol. 198, 1384–1392. doi: 10.1128/JB.01039-15
Tappenden, P., Harnan, S., Uttley, L., Mildred, M., Carroll, C., and Cantrell, A. (2013). Colistimethate sodium powder and tobramycin powder for inhalation for the treatment of chronic Pseudomonas aeruginosa lung infection in cystic fibrosis: systematic review and economic model. Health Technol. Assess. 17, v–xvii, 1–81. doi: 10.3310/hta17560
Tevyashova, A., and Shapovalova, K. (2021). Potential for the development of a new generation of aminoglycoside antibiotics. Pharm. Chem. J. 55, 860–875. doi: 10.1007/s11094-021-02510-0
Thi Khanh Nhu, N., Riordan, D. W., Do Hoang Nhu, T., Thanh, D. P., Thwaites, G., Huong Lan, N. P., et al. (2016). The induction and identification of novel Colistin resistance mutations in Acinetobacter baumannii and their implications. Sci. Rep. 6:28291. doi: 10.1038/srep28291
Vaillancourt, M., Limsuwannarot, S. P., Bresee, C., Poopalarajah, R., and Jorth, P. (2021). Pseudomonas aeruginosa mexR and mexEF antibiotic efflux pump variants exhibit increased virulence. Antibiotics 10:1164. doi: 10.3390/antibiotics10101164
Wang, Y., Li, C., Wang, J., Bai, N., Zhang, H., Chi, Y., et al. (2022). The efficacy of Colistin combined with amikacin or levofloxacin against Pseudomonas aeruginosa biofilm infection. Microbiol. Spectr. 10, e01468–e01422. doi: 10.1128/spectrum.01468-22
Watkins, D., Gong, C., Kellish, P., and Arya, D. P. (2017). Probing A –form DNA: a fluorescent aminosugar probe and dual recognition by anthraquinone-neomycin conjugates. Bioorg. Med. Chem. 25, 1309–1319. doi: 10.1016/j.bmc.2016.11.003
Watkins, D., Jiang, L., Nahar, S., Maiti, S., and Arya, D. P. (2015a). A pH sensitive high-throughput assay for miRNA binding of a peptide-aminoglycoside (PA) library. PLoS ONE 10:e0144251. doi: 10.1371/journal.pone.0144251
Watkins, D., Kumar, S., Green, K. D., Arya, D. P., and Garneau-Tsodikova, S. (2015b). Influence of linker length and composition on enzymatic activity and ribosomal binding of neomycin dimers. Antimicrob. Agents Chemother. 59, 3899–3905. doi: 10.1128/AAC.00861-15
Watkins, D., Maiti, K., and Arya, D. P. (2019). Aminoglycoside functionalization as a tool for targeting nucleic acids. Methods Mol. Biol. 1973, 147–162. doi: 10.1007/978-1-4939-9216-4_9
Watkins, D., Norris, F., Kumar, S., and Arya, D. P. (2013). A fluorescence-based screen for ribosome binding antibiotics. Anal. Biochem. 434, 300–307. doi: 10.1016/j.ab.2012.12.003
Willis, B., and Arya, D. P. (2009). Triple recognition of B-DNA. Bioorg. Med. Chem. Lett. 19, 4974–4979. doi: 10.1016/j.bmcl.2009.07.079
Willis, B., and Arya, D. P. (2010). Triple recognition of B-DNA by a neomycin–hoechst 33258–pyrene conjugate. Biochemistry 49, 452–469. doi: 10.1021/bi9016796
World Health Organization (2024). WHO Updates List of Drug-Resistant Bacteria Most Threatening to Human Health. Geneva: World Health Organization.
Xue, L., Charles, I., and Arya, D. P. (2002). Pyrene–neomycin conjugate: dual recognition of a DNA triple helix. Chem. Commun. 1, 70–71. doi: 10.1039/b108171c
Yang, W., and Hu, F. (2022). Research updates of plasmid-mediated aminoglycoside resistance 16S rRNA methyltransferase. Antibiotics 11:906. doi: 10.3390/antibiotics11070906
Zhu, S., Song, C., Zhang, J., Diao, S., Heinrichs, T. M., Martins, F. S., et al. (2022). Effects of amikacin, polymyxin-B, and sulbactam combination on the pharmacodynamic indices of mutant selection against multi-drug resistant Acinetobacter baumannii. Front. Microbiol. 13:1013939. doi: 10.3389/fmicb.2022.1013939
Keywords: aminoglycosides, peptide neomycin conjugates, polymyxins, synergy, drug-resistant bacteria, amino acids
Citation: Story S, Jiang L, Leutou AS and Arya DP (2025) Synergistic action between peptide-neomycin conjugates and polymyxin B against multidrug-resistant gram-negative pathogens. Front. Microbiol. 16:1605813. doi: 10.3389/fmicb.2025.1605813
Received: 04 April 2025; Accepted: 25 June 2025;
Published: 07 August 2025.
Edited by:
Peter E. Nielsen, University of Copenhagen, DenmarkReviewed by:
Kavita De, Colorado State University, United StatesLise Goltermann, University of Copenhagen, Denmark
Bouke Boekema, Association of Dutch Burn Centres, Netherlands
Copyright © 2025 Story, Jiang, Leutou and Arya. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dev P. Arya, ZHBhcnlhQGNsZW1zb24uZWR1
 Sandra Story1
Sandra Story1 Alain S. Leutou
Alain S. Leutou Dev P. Arya
Dev P. Arya