- 1Department of Plant Protection, Faculty of Agriculture, Azarbaijan Shahid Madani University, Tabriz, Iran
- 2East Azarbaijan Agricultural and Natural Resources Research and Education Centre, Plant Protection Research Department, Agricultural Research, Education and Extension Organization (AREEO), Tabriz, Iran
- 3Plankton and Microbial Ecology, Leibniz Institute for Freshwater Ecology and Inland Fisheries (IGB), Berlin, Germany
- 4Bermuda Institute of Ocean Sciences, St. George's, Bermuda
- 5Institute of Biochemistry and Biology, University of Potsdam, Potsdam, Germany
During the study of oomycete biodiversity in aquatic environments of northwestern Iran (East Azarbaijan), four Globisporangium isolates were recovered from a river and irrigation canal. These isolates were identified based on multi-locus phylogenetic analyses (ITS, cox1, and cox2 genomic regions) and morphological features. As a result, two novel species were described, namely Globisporangium parvizense sp. nov. and G. sarabense sp. nov., both exhibiting unique sporangial structures and growth patterns. Pathogenicity assays on cucumber seedlings confirmed strains' high potential to cause root and crown rot. This research highlights the diversity of Globisporangium in Iranian freshwater habitats, providing insights into its taxonomy and phylogenetic relationships. Detailed morphological descriptions and illustrations are provided for these novel species.
1 Introduction
Pythium Pringsheim, nom. cons., sensu lato (s.l.) is a diverse and ecologically important genus within the phylum Oomycota (commonly known as water molds), comprising over 200 species (Uzuhashi et al., 2010; Derevnina et al., 2016). As a member of the kingdom Straminipila which also includes diatoms, brown algae, and slime molds (Mueller, 2011; Watkinson et al., 2015), Pythium s.l. shares phylogenetic affinities with other important plant pathogenic genera such as Albugo Roussel, Aphanomyces de Bary, Bremia Regel, Peronospora Corda, Phytophthora de Bary, Plasmopara Schroet., and Saprolegnia Nees. This genus is particularly notable for its wide distribution across both terrestrial and aquatic habitats and for its significant impact on agriculture, natural ecosystems, and even animal health (Robideau et al., 2011; Shreves et al., 2024; Masigol et al., 2025).
Pythium s.l. causes economic losses in vegetables, fruits, and ornamentals, resulting in damping-off, root and crown rot (Martin and Loper, 1999; Uzuhashi et al., 2010; Nzungize et al., 2012; Nguyen et al., 2022). Thriving in soil and aquatic agroecosystems, members of Pythium s.l. are frequently encountered in agricultural fields, nurseries, and greenhouses, where they persist in soil, water, plant debris, and even snow (Al-Sheikh and Abdelzaher, 2012; Chenari Bouket et al., 2015). Although capable of surviving in both aquatic and dryland environments (Barton, 1958; Abdelzaher and Kageyama, 2020), their motile zoospores, equipped with flagella, favor water for dispersal, which enhances their infectivity in moist conditions. Some species have even adapted to saline environments, further expanding their ecological range (Al-Sheikh and Abdelzaher, 2012).
Additionally, Pythium s.l. could be a potential threat to aquatic ecosystems, particularly when introduced into water by human activities (Abdelzaher and Kageyama, 2020). In fact, certain water-dwelling species such as Pythium insidiosum, could be responsible for severe infections in humans and dogs, causing “pythiosis,” a disease that affects cutaneous and vascular tissues (Hamlin et al., 2024). The ability of Pythium s.l. to infect multiple hosts across different ecosystems highlights its adaptability and pathogenic potential.
Given the ecological significance of Pythium, delineating its species boundaries is essential, as it lays the foundation for subsequent ecological and functional studies. In recent years, the taxonomy of Pythium s.l. has undergone significant transformations, primarily driven by advancements in molecular techniques (Villa et al., 2006). Molecular phylogenetic analyses revealed the paraphyletic nature of Pythium s.l., leading to proposals for its division into several distinct lineages. Initially, Lévesque and De Cock (2004) classified Pythium s.l. into 11 clades (A–K) based on phylogenetic data derived from the nuclear rDNA internal transcribed spacer region (ITS1–5.8S–ITS2) and the D1–D3 domains of the 28S rDNA. These clades were later reassessed through multigene phylogenetic analyses, which resulted in a revised classification that consolidated the original clades into 10 groups, with clade K reassigned to the newly established genus Phytopythium (Villa et al., 2006; Bala et al., 2010). Furthermore, Uzuhashi et al. (2015) proposed a major revision, restructuring Pythium s.l. into five distinct genera: Pythium sensu stricto (clades A–D), Globisporangium (clades E–G, I, and J), Elongisporangium (clade H), Ovatisporangium (clade K = Phytopythium), and Pilasporangium, which stands apart from the previously defined clades.
Several studies have focused on the isolation and identification of Pythium s.l. associated with aquatic ecosystems. In one of the earliest studies, Abdelzaher et al. (1995) reported Pythium group F from aquatic environments. Later, P. myriotylum, P. ultimum var. sporangiferum (Kageyama, 2010), and P. rishiriense (Rahman et al., 2015) were reported from water, soil and floating water from Rishiri Island, Japan. More novel Pythium species have been recently reported from different aquatic ecosystems in Japan (Uzuhashi et al., 2015; Abdelzaher and Kageyama, 2020), South Korea (Nam and Choi, 2019), and China (Chen and Zheng, 2019).
Although oomycetes have been studied in Iran for decades, the focus has traditionally been on plant pathogenic species in Peronosporomycetes (e.g., Pythium, Mostowfizadeh-Ghalamfarsa, 2015; Salmaninezhad and Mostowfizadeh-Ghalamfarsa, 2017, 2019, 2022, 2023; Salmaninezhad et al., 2022). Only recently, Saprolegniomycetes (Ghiasi et al., 2010; Masigol et al., 2018, 2019, 2020, 2022, 2023, 2025; Mirmazloomi et al., 2022) and, to a lesser extent, Peronosporomycetes (Ahadi et al., 2024) have been investigated taxonomically and ecologically in aquatic ecosystems. However, as research on aquatic oomycetes is still limited and geographically specific, this study aims to identify Globisporangium isolates from agroecosystems in East Azarbaijan province, Iran, using a combination of morphological analysis and multigene phylogeny, based on the nuclear rDNA ITS1-5.8S-ITS2 region and partial cytochrome C oxidase subunit I (cox1) and subunit II (cox2) sequences. Additionally, the potential pathogenicity of these isolates on cucumber, a common host plant of oomycetes, was evaluated.
2 Materials and methods
2.1 Sample collection and isolation
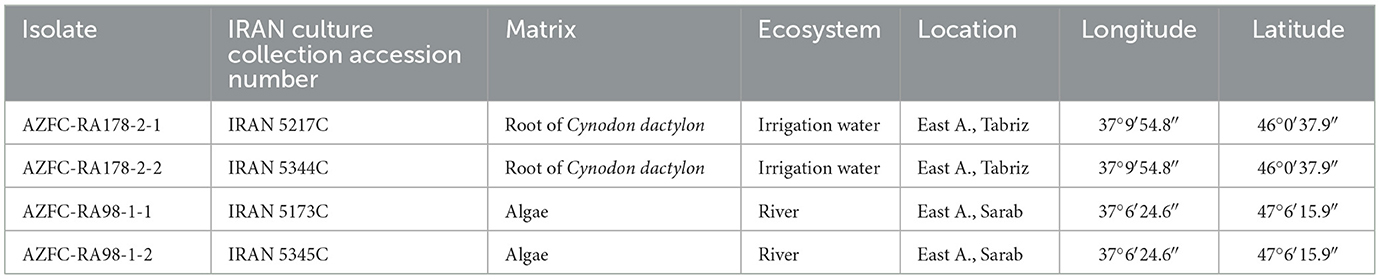
Sampling was conducted in the aquatic environments of East Azerbaijan Province, Iran (Table 1). Algae and roots of the grass species Cynodon dactylon were collected from a river and irrigation canal in 50 mL Falcon tubes and stored at 4°C prior to processing in the laboratory. Following surface sterilization for 1 min, tissue samples were cultured on NARF medium (Morita and Tojo, 2007) and incubated at 15°C for 5 days. Upon hyphal growth, a portion of the culture was transferred to WA medium (20.0 g/L agar) for purification using the hyphal tip method (Goh, 1999). Purified isolates were preserved on CMA medium in McCarthy vials at 10°C.
2.2 Morphological analysis
Growth patterns of the isolates were observed 2 weeks after inoculation on various agar media, including corn meal agar (CMA) (MIRMEDIA, Iran), potato dextrose agar (PDA) (Sigma Aldrich, Germany), malt extract agar (MEA) (DIFCO, USA), potato carrot agar (PCA) (Sigma Aldrich, Germany), and V8-juice agar (SIGMA, Germany) at 25°C. Morphological evaluations were performed on sexual and asexual structures produced on autoclaved hemp seeds and ryegrass pieces floating in sterile water from different sources (pond water, distilled water, tap water) (Martin, 1992). Twenty measurements were taken for each structure. Microscopic structures were photographed using a Nikon Eclipse Ti2 microscope with a digital camera system (Nikon, Japan). All purified cultures were deposited in the fungal culture collection of Azarbaijan Shahid Madani University, Tabriz, Iran (AZFC) and Iranian Research Institute of Plant Protection, Tehran, Iran (IRAN). Type specimens were also deposited in the herbarium of the Iranian Research Institute of Plant Protection.
The critical temperatures for growth were determined by incubating the strains on potato carrot agar (PCA) at 0, 2, 5, 10, 15, 20, 25, 30, 35, and 40°C. Descriptions were provided based on ex-type strains, and additional data for strains showing distinct morphological differences were included.
2.3 Phylogenetic analyses
Genomic DNA was extracted from 5-day-old CMA cultures using a modified manual procedure (Möller et al., 1992). Three genomic regions were targeted for amplification: the ITS-rDNA region, which contains the 5.8S nuclear ribosomal RNA gene and the internal transcribed spacers ITS1 and ITS2, and partial sequences of the mitochondrial cox1 and cox2 genes. These regions were amplified using specific primer pairs: ITS5 (GGAAGTAAAAGTCGTAACAAGG) and ITS4 (TCCTCCGCTTATTGATATGC) (White et al., 1990) for the ITS-rDNA region, FM55 (GGCATACCAGCTAAACCTAA) and FM52R (TTAGAATGGAATTAGCACAAC) (Martin, 2000) for the cox1 gene, and FM58 (CCACAAATTTCACTACATTGA) and FM66 (TAGGATTTCAAGATCCTGC) (Villa et al., 2006) for the cox2 gene. All reactions were conducted in a total volume of 50 μL, consisting of 25 μL of ready-to-use PCR Master mix (SinaClon, Iran), 1.2 μM of each primer, 18.6 μL of DNase-free water, and 10 ng of DNA. Amplifications were carried out using a PeqStar 96X universal thermal cycler with the following conditions: 95°C for 5 min, followed by 30 cycles of denaturation at 95°C for 30 s, annealing at 55°C for 30 s, and extension at 72°C for 1 min, with a final extension step at 72°C for 7 min for ITS-rDNA (White et al., 1990), and 94°C for 5 min, followed by 40 cycles of denaturation at 94°C for 30 s, annealing at 54°C for 30 s, and extension at 72°C for 1 min, with a final extension step at 72°C for 7 min for cox1 and cox2 genes (Martin, 2000; Chenari Bouket et al., 2015).
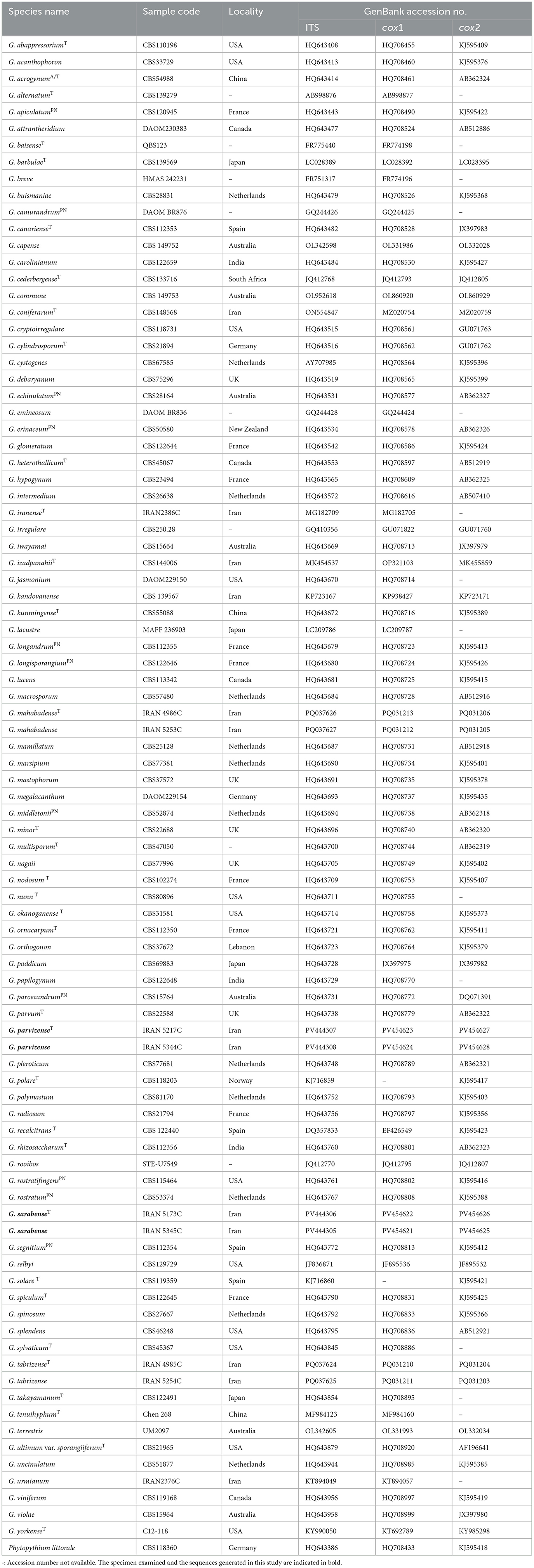
PCR amplicons were sequenced by Macrogen (Amsterdam, the Netherlands). Raw sequences were manually edited using SeqManII® (DNA STAR) and MEGA v. 6 (Tamura et al., 2013). Sequence data from ex-type and reference strains of known Globisporangium species were obtained from NCBI GenBank (www.ncbi.nlm.nih.gov/genbank/) (Table 2). The retrieved sequences were assembled using Geneious (version 5.6) and aligned using the Q-INS-I algorithm in MAFFT (latest version), available on the MAFFT web server (Katoh and Standley, 2013; Katoh et al., 2019), separately for each of the genomic regions. Subsequently, after the removal of gaps, phylogenetic analyses were performed on the TrEase webserver (Mishra et al., 2023) for the individual genes using FastTree2 (Price et al., 2010) for Minimum Evolution, RAxML (Stamatakis, 2014) for Maximum Likelihood, and MrBayes (Ronquist et al., 2012) for Bayesian inference. For the Bayesian analysis, a GTR model was selected, and the analyses were run on random trees for 1,000,000 generations, discarding 30% of the first trees as burn-in steps of the analysis to determine posterior probabilities from the remaining trees. RAxML and FastTree2 trees were generated using the GTRGAMMA and GTR algorithms, respectively, and the reliability of the inferred trees was assessed through bootstrap analysis with 1,000 replications. After ensuring that there were no conflicting topologies in the phylogeny of the individual loci, they were concatenated, with the borders marked to ensure independent modeling of substitution rates for each partition. Multigene phylogenies (ITS, cox1, and cox2) with support values were calculated in the same manner as mentioned above, using three different approaches to assess the robustness of the inferred phylogenies. The sequences obtained in this study were deposited in GenBank, and their accession numbers have been provided in Table 2.
2.4 Pathogenicity assays
Pathogenicity tests were conducted using a single isolate of each species (IRAN 5217C and IRAN 5173C, respectively). Cucumber (Cucumis sativus L.), a known host for a wide range of oomycetes (Ben-Yephet and Nelson, 1999; Lévesque and De Cock, 2004; Roberts et al., 2016; Sigillo et al., 2020; Zhang et al., 2023), was selected as the test plant. The inoculum was prepared according to the methods of Ahadi et al. (2024) with minor modifications. A sterile substrate containing sandy loam soil, wheat seed, and distilled water was inoculated with five mycelial plugs (5 mm3) from three-day-old PDA cultures of the target isolates. After 9 days of incubation at 25°C, cucumber seeds were sown and grown in a greenhouse at 25 ± 2°C with a 16-h photoperiod for 14 days. Positive controls consisted of plants grown in soil inoculated with each of the test isolates, while negative controls were grown in non-inoculated soil. Symptoms including wilting, crown and root rot, and stem discoloration and deterioration were assessed daily for 14 days. Symptomatic plants were further analyzed to isolate and identify potential pathogens using previously described isolation methods. A randomized complete block design with six replicates per treatment was employed.
3 Results
3.1 Phylogeny
In the analysis of multi-locus alignment (Globisporangium species), which included the gene boundaries of ITS (1–1,066), cox1 (1,067–1,560), and cox2 (1,561–2,069), a total of 89 isolates belonging to Globisporangium, along with an outgroup, were examined. The combined dataset (ITS + cox1 + cox2) comprised 2,069 characters, including alignment gaps, with 958 variable characters (636 for ITS-rDNA, 169 for cox1, and 153 for cox2) and 1,002 constant characters (the sequence lengths were 333 bp for ITS-rDNA, 320 bp for cox1, and 349 bp for cox2).
Bayesian analysis confirmed the tree topology obtained from minimum evolution (ME) and maximum likelihood (ML) methods. While most Bayesian posterior probability values were consistent with bootstrap supports, the Bayesian tree was selected to represent the phylogeny, with bootstrap values from ME and ML methods incorporated for comparison (Figure 1). In all three-locus phylogenetic trees (Bayesian, ML, and ME), the four Globisporangium isolates were placed into two distinct, well-supported clades, each representing a separate species. Specifically, the isolates IRAN 5217C and IRAN 5344C formed a robust monophyletic group, with maximum support from Bayesian posterior probability (1) and bootstrap values (100% for both ME and ML). Similarly, isolates IRAN 5173C and IRAN 5345C were clustered together in a separate, strongly supported clade [support values: 1.00 (Bayesian posterior probability), 83% (ME), and 67% (ML)]. In the multi-locus analysis, the topologies of the single-locus phylogenies did not conflict with the respective three-locus phylogenies, confirming the reliability and accuracy of the inferred relationships among these isolates.
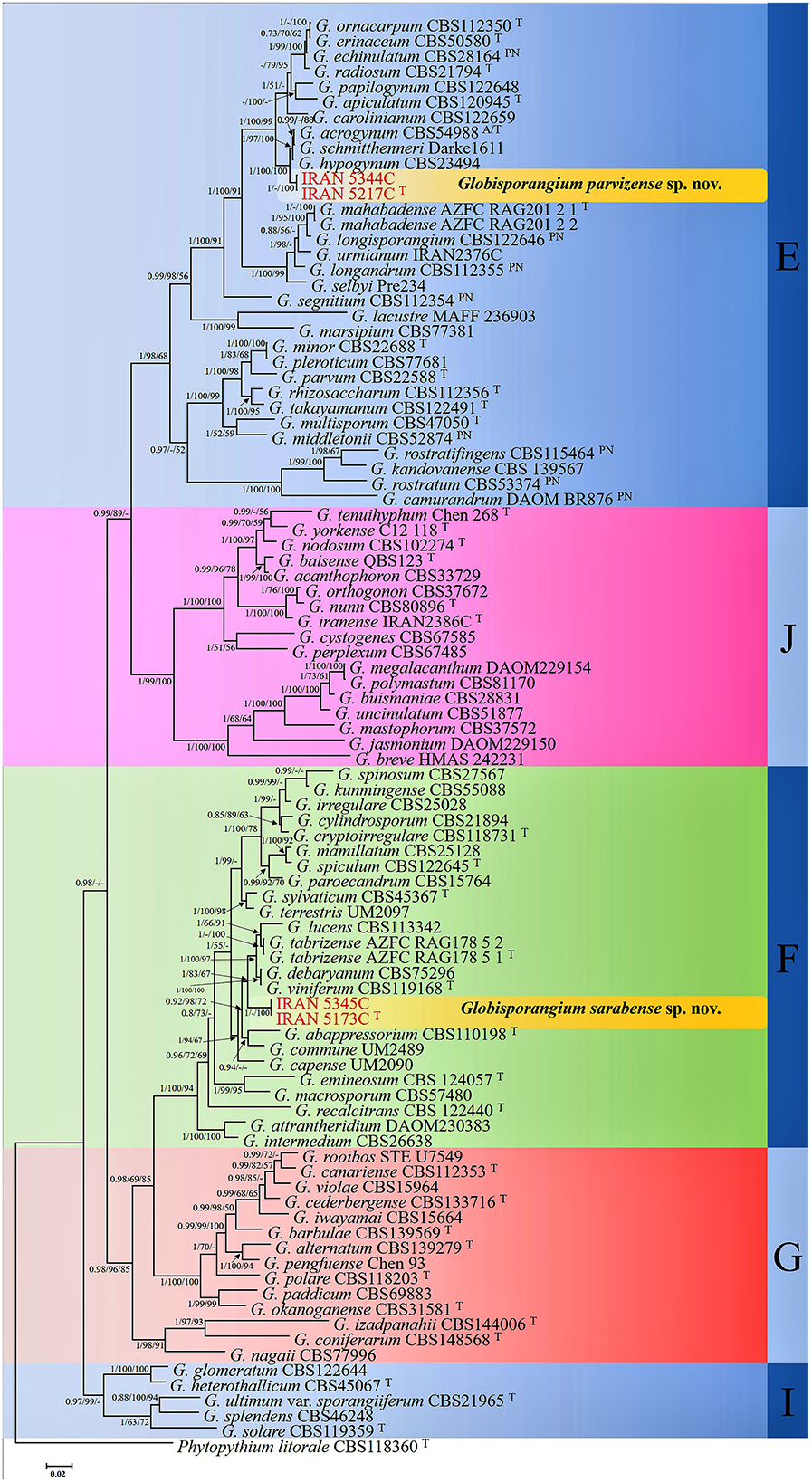
Figure 1. Phylogram generated from Bayesian inference analysis based on ITS-rDNA, cox1, and cox2 sequence data for four examined strains and 89 reference strains of Globisporangium. Numbers on the branches represent posterior probabilities from Bayesian Inference, as well as bootstrap support from Minimum Evolution (ME) and Maximum Likelihood (ML), in the order 0.7/50%/50%. A dash indicates lower support for the presented topology or the possibility of an alternative topology. Phytopythium litorale type strain CBS118360 was used as the outgroup. Clades E–G, I, and J, which were identified by Lévesque and De Cock (2004) within the Pythium sensu lato, are depicted on the right side of the figure. Strains obtained in this study are shown in red. “T” denotes ex-type strains, and “PN” indicates authentic strains used for description in the monograph by Van der Plaats-Niterink (1981).
3.2 Taxonomy
This study identified two Globisporangium species from various aquatic environments in Iran. Four isolates, including two from the roots of the grass species Cynodon dactylon growing in an irrigation canal (IRAN 5217C and IRAN 5344C), and two from water surface algae in the Sarab River in Sarab City (IRAN 5173C and IRAN 5345C) were reported as new species: G. parvizense sp. nov. and G. sarabense sp. nov.
Globisporangium parvizense Ahadi, Alizadeh, Chenari Bouket sp. nov. (Figure 2).
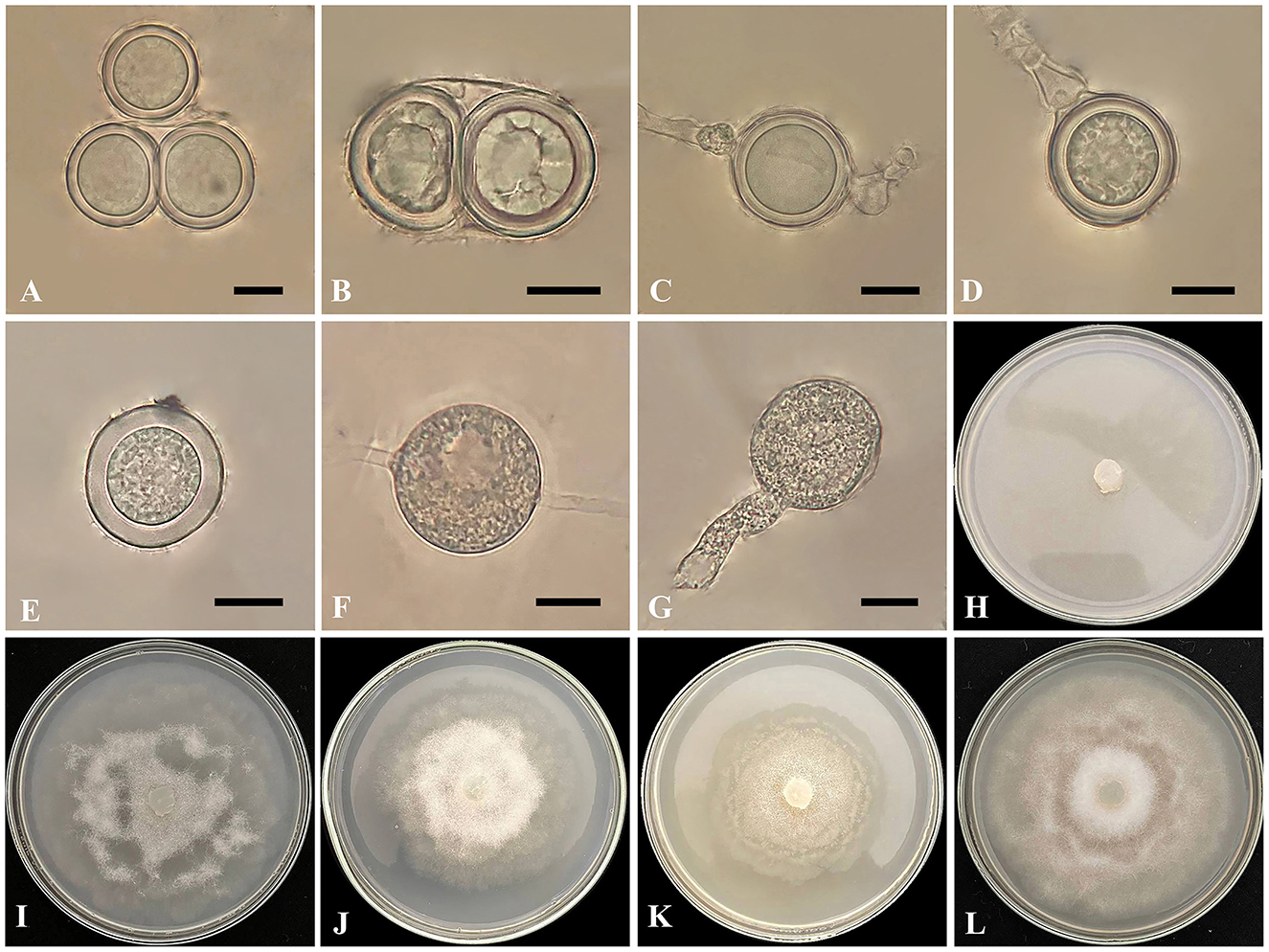
Figure 2. Globisporangium parvizense isolate IRAN 5217C. (A, B) Two oospores per oogonium. (C) Intercalary oogonium with one antheridium. (D) Terminal oogonium with one antheridium. (E) Plerotic oospore. (F) Intercalary sporangium. (G) Sporangium with discharge tubes. (H) Corn Meal Agar (CMA). (I) Potato Carrot Agar (PCA). (J) Potato Dextrose Agar (PDA). (K) Malt Extract Agar (MEA). (L) V8 Juice Agar (V8A). Scale bars: 10 μm.
MycoBank: 857765
Typification: IRAN. East Azarbaijan: Tabriz (Varanaq), from roots of the grass species Cynodon dactylon, growing within the agricultural irrigation canals, Oct 2022, R. Ahadi (holotype IRAN 18632F). Ex-holotype culture IRAN 5217C = AZFC-RA178-2-1. GenBank: ITS = PV444307; cox1 = PV454623; cox2 = PV454627.
Etymology: The species is named in honor of the first author's father (Parviz), whose assistance was invaluable in the collection of the species.
Morphology: The colony pattern on MEA and PCA is a semi-rosette pattern, pattern on CMA is entirely within the growth medium and pattern on PDA and V8A appears submerged to stellate, with a daily growth rate of 9 mm at 25°C on PCA. The cardinal temperatures are at a minimum of 5°C, an optimum of 25°C, and a maximum of 30°C on PCA. The main hyphae are hyaline, aseptate, and range from 2.4 to 6 μm in width. The globose sporangia appear intercalary or terminal, measuring 11–32 μm in diameter (mean 23 μm). Globose hyphal swelling, terminal or intercalary, 8.5–16.5 μm (mean 10 μm) in diameter. Discharge tube arising from sporangia, short to long, one and rarely two per sporangia, and thick, 13–41 μm (mean 19.5 μm) in length and 7–8.5 μm (mean 8 μm) in width. Zoospores are not observed. Oogonia are globose and smooth, appearing intercalary or terminal, with a diameter range of 14–23 μm (mean 20.5 μm) and produced in single cultures. Antheridia are typically one, occasionally two per oogonium, diclinous. Oospores are plerotic, with one, rarely two per oogonium, measuring 15–23 μm in diameter (mean 20 μm). The wall thickness ranged from 1.8 to 3.6 μm.
Additional specimen examined: IRAN. East Azarbaijan: Tabriz (Varanaq), from roots of the grass species Cynodon dactylon, growing within the agricultural irrigation canals, Oct 2022, R. Ahadi. culture IRAN 5344C = AZFC-RA178-2-2. GenBank: ITS = PV444308; cox1 = PV454624; cox2 = PV454628.
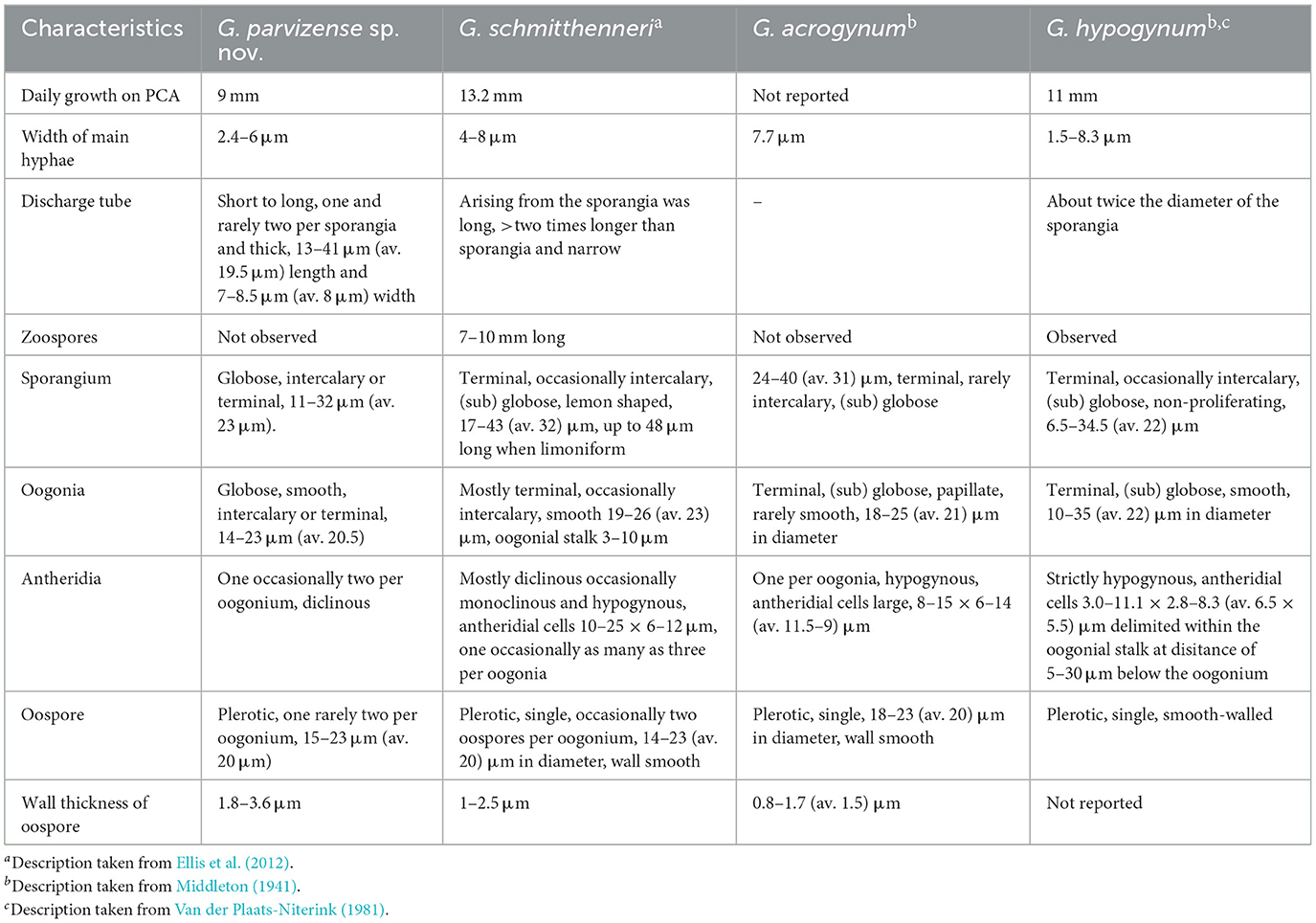
Notes: Phylogenetic analysis revealed a close relationship between Globisporangium parvizense and G. hypogynum, followed by a slightly less close relationship with G. acrogynum and G. schmitthenneri. These species, as well as all other Globisporangium species, can be differentiated from G. parvizense based on ITS, cox1, and cox2 sequence data. Blastn searches on NCBI GenBank revealed that the ITS sequence of the G. parvizense ex-type strain (IRAN 5217C = AZFC-RA178-2-1) exhibited 99% identity with the G. hypogynum type strain CBS23494 (HQ643565), G. schmitthenneri type strain Darke1611 (JF836869), and G. acrogynum isolate CBS54988 (HQ643414) (with two, three, and three nucleotide differences, respectively). Similarly, the cox1 sequence of the ex-type strain shared 99% identity, respectively (with four nucleotide differences for both), with the G. hypogynum type strain CBS23494 (HQ708609), G. acrogynum isolate CBS54988 (HQ708461), and G. schmitthenneri type strain Darke1611 (JF895534). Comparative analysis of G. parvizense with the cox2 sequence showed 98% identity, with seven nucleotide differences compared to G. hypogynum type strain CBS23494 (AB362325), G. acrogynum isolate CBS54988 (AB362324), and G. schmitthenneri type strain Darke1611 (JF895530). The phylogenetic data robustly establish G. parvizense as a phylogenetically isolated species within the genus Globisporangium.
Globisporangium parvizense is morphologically distinct from its phylogenetically close species based on several morphological characteristics. A detailed comparison of G. parvizense, G. hypogynum, G. acrogynum, and G. schmitthenneri reveals several distinct features. G. parvizense is characterized by narrower hyphae, up to 6 μm in width, compared to 7.7, 8, and 8.3 μm in G. acrogynum, G. schmitthenneri, and G. hypogynum, respectively. While G. parvizense exclusively forms globose sporangia, G. schmitthenneri also produces lemon-shaped sporangia. Globisporangium parvizense also has smaller oogonia (up to 23 μm) compared to G. hypogynum (up to 35 μm), and consistently smooth oogonia, unlike G. acrogynum. Globisporangium parvizense typically has one, occasionally two, antheridia per oogonium, while G. schmitthenneri can have up to three, and G. acrogynum has only one antheridium per oogonium. Globisporangium parvizense is uniquely characterized by the consistent formation of two spherical oospores per oogonium, while G. acrogynum and G. hypogynum each produce a single oospore. These morphological characteristics, including narrower hyphae, simpler sporangial shapes, consistent oospore formation, and unique antheridial structures, clearly differentiate G. parvizense from its closely related species (Table 3).
Globisporangium sarabense Ahadi, Alizadeh, Chenari Bouket sp. nov. (Figure 3).
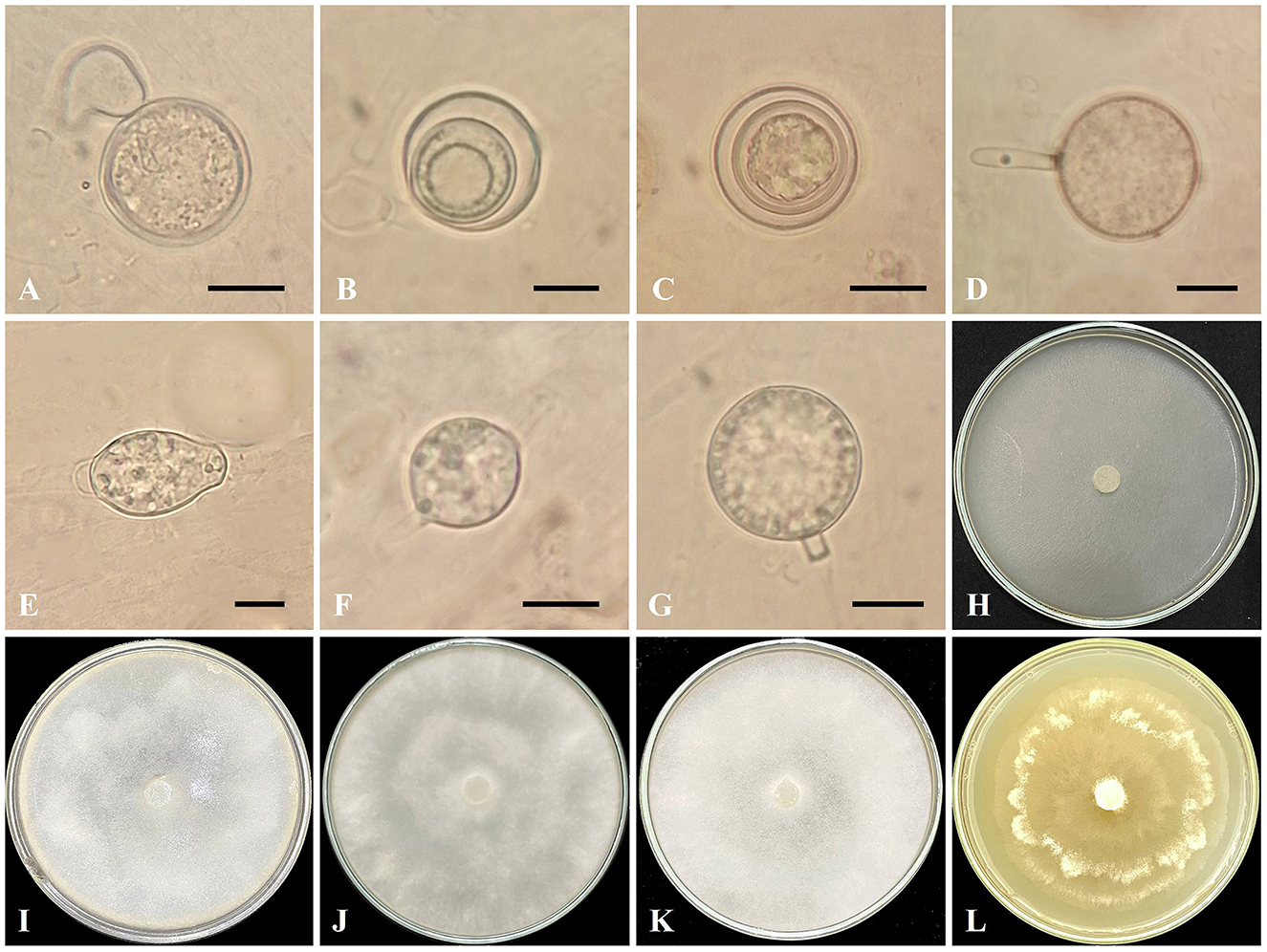
Figure 3. Globisporangium sarabense isolate IRAN 5173C. (A) Oogonium with one antheridium. (B, C) Aplerotic oospores. (D) Sporangium with discharge tubes. (E, F) Intercalary sporangium. (G) Terminal sporangium. (H) Corn Meal Agar (CMA). (I) Potato Carrot Agar (PCA). (J) Potato Dextrose Agar (PDA). (K) V8 Juice Agar (V8A). (L) Malt Extract Agar (MEA). Scale bars: 10 μm.
MycoBank: 857791
Typification: IRAN. East Azarbaijan: Sarab, from water surface algae in Sarab River, Oct 2022, R. Ahadi (holotype IRAN 18544F). Ex-holotype culture IRAN 5173C = AZFC-RA98-1-1. GenBank: ITS = PV444306; cox1 = PV454622; cox2 = PV454626.
Etymology: Referring to the type location, Sarab City.
Morphology: The colony pattern on MEA is a rosette pattern, whereas the pattern on CMA, PDA, PCA, and V8A are submerged to stellate. Daily growth at 25°C on PCA is 20 mm. Cardinal temperatures are a minimum of 5°C, an optimum of 25°C, and a maximum of 35°C on Potato Carrot Agar. Main hyphae are hyaline, aseptate, and 2.4–6 μm (mean 3.1 μm) wide. Sporangia are globose or subglobose, intercalary or terminal, 17–26.8 μm (mean 21.6 μm) in diameter, and lemon-shaped sporangia are intercalary or terminal, 23.1–31.7 μm (mean 27.4 μm) in length and 17–26.8 μm (mean 21.7 μm) in width. The discharge tube is 12.8–10.3 μm (mean 11.5 μm) in length and 3–3.6 μm (mean 3.3 μm) in width. Zoospores are not observed. Oogonia are rarely formed, mostly terminal, smooth, globose, 18.3–23.1 μm (mean 20 μm) in diameter, and produced in a single culture. Antheridia are one per oogonium, mostly diclinous. Oospores are globose, aplerotic, one per oogonium, measuring 12.2–23.3 μm (mean 15.8 μm) in diameter, with wall thickness ranging from 0.6 to 2.4 μm.
Additional specimen examined: IRAN. East Azarbaijan, Sarab, from water surface algae in Sarab River, Oct 2022, R. Ahadi. culture AZFC-RA98-1-2. GenBank: ITS = PV444305; cox1 = PV454621; cox2 = PV454625.
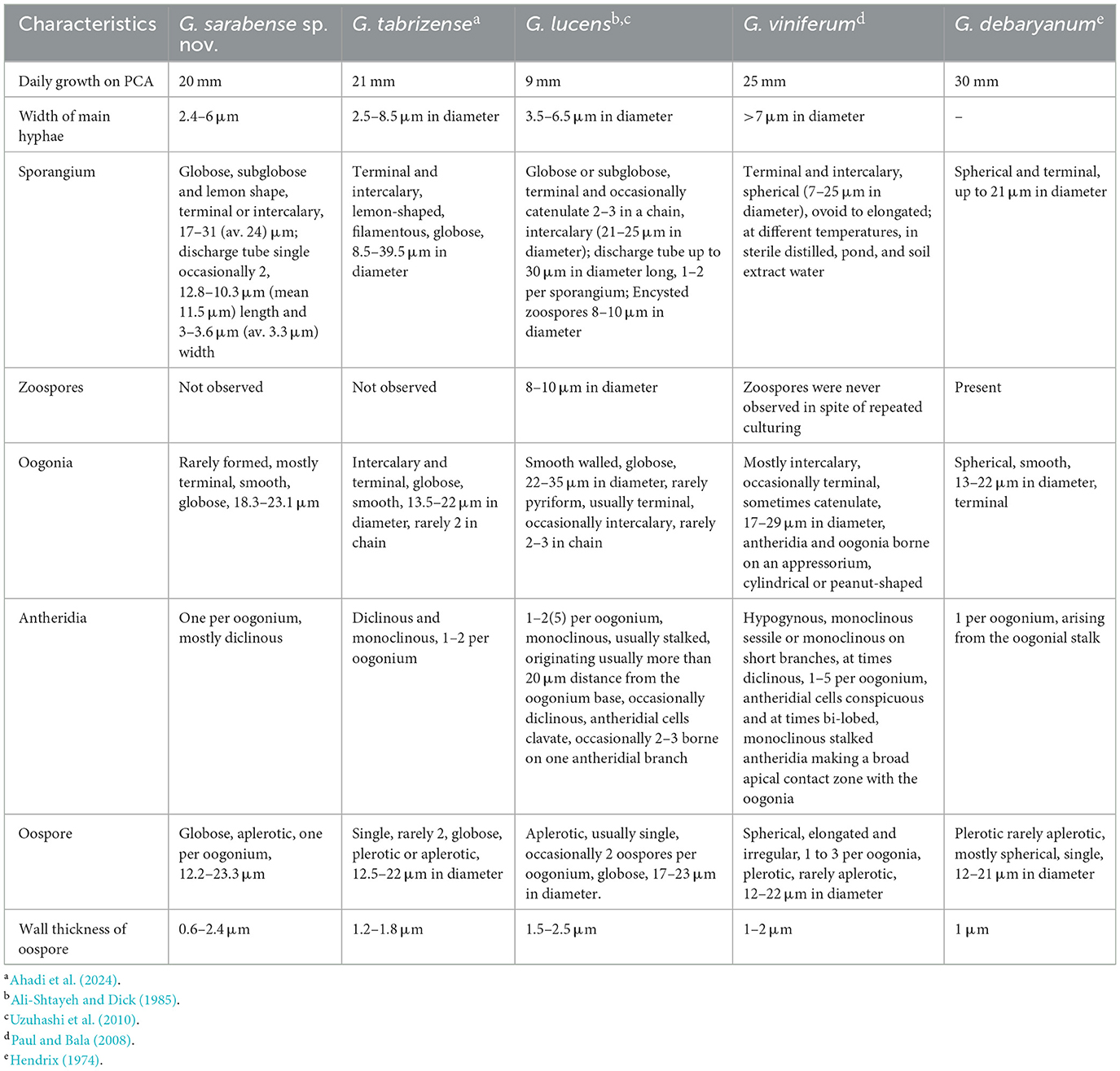
Notes: Phylogenetic analysis revealed a close phylogenetic relationship between G. sarabense, G. lucens, and G. tabrizense, followed by a slightly less close relationship with G. viniferum and G. debaryanum. The ex-type strain of G. sarabense (IRAN 5173C = AZFC-RA98-1-1) exhibited 94% ITS identity with the G. lucens strain CBS113342, 96% identity in the cox1 gene, and <96% identity in the cox2 gene, as well as <94% ITS identity with the G. tabrizense strain IRAN 18502F. It showed 96.62% identity in the cox1 gene and <97% identity in the cox2 gene. However, it also displayed 21 and 15 nucleotide differences in ITS, 18 and 17 in cox1, and 10 and 12 in cox2, respectively, compared to this reference strain. Blastn searches on NCBI GenBank indicated that the ITS sequence of G. sarabense shared the highest similarity (97% identity, with 18 nucleotide differences and 5 gaps) with the G. viniferum isolate Tr-Sm01 (MT251151). Its cox1 sequence was most similar (97%) to Pythium sp. isolate Kb003 (ON202818), and the cox2 sequence was most similar (98%) to G. intermedium (MG256947). The ex-type strain of G. sarabense shared 96% identity in ITS, <95% in cox1 and cox2 genes with the G. viniferum voucher CBS119168, differing by 20 nucleotides in ITS, 16 in cox1, and 12 in cox2. Additionally, it shared 97% ITS identity, 96% cox1 identity, and 97% cox2 identity with G. debaryanum CBS75296, but exhibited 19 nucleotide differences in ITS, 16 in cox1, and 12 in cox2. The examined loci consistently placed G. sarabense in a separate phylogenetic clade, confirming its status as a distinct species within Globisporangium.
Globisporangium sarabense shares some morphological similarities with G. lucens, G. tabrizense, G. viniferum, and G. debaryanum. However, distinct morphological features differentiate G. sarabense from its congeners. Compared to G. lucens, G. sarabense lacks zoospores, while G. lucens produces zoospores. Furthermore, G. sarabense has single oospores, whereas G. lucens occasionally has two oospores per oogonium. Oogonia chains were not identified in G. sarabense, but G. lucens exhibits the formation of oogonia chains. In terms of sporangium morphology, G. sarabense exhibits greater diversity, including lemon-shaped, globose, or subglobose forms, while G. lucens primarily has globose or subglobose sporangia. Globisporangium sarabense possesses a single antheridium per oogonium, whereas G. lucens may have as many as five antheridia.
In contrast to G. tabrizense, G. sarabense is characterized by the absence of filamentous sporangia and the possession of a discharge tube, which further distinguishes it from G. tabrizense. Furthermore, in G. sarabense, the formation of oogonia is infrequent, and in contrast to G. tabrizense, the development of oogonia chains does not occur. In G. tabrizense, as many as two antheridia are produced, whereas in G. sarabense, only a single antheridium is formed. Oospores in G. tabrizense produce both plerotic and aplerotic types, with two oospores per oogonium. In contrast, G. sarabense only produces a single aplerotic oospore. Moreover, G. viniferum has up to three plerotic oospores per oogonium, but G. sarabense has a single, aplerotic oospore.
Differentiating G. sarabense from G. debaryanum is primarily based on the absence of zoospores and the production of only aplerotic oospores in G. sarabense. At the same time, G. debaryanum produces mostly plerotic and rarely aplerotic oospores. Additionally, in G. sarabense, the wall thickness of the oospore is 1.5 μm wider than in G. debaryanum.
In summary, G. sarabense is morphologically distinct from its congeners due to its unique combination of sporangia types, absence of zoospores, oospore characteristics, and number of antheridia. These morphological differences highlight the taxonomic distinctiveness of G. sarabense within the genus Globisporangium (Table 4 and Figure 3).
4 Pathogenicity assay
Pathogenicity experiments confirmed the pathogenicity of all examined isolates, including G. parvizense sp. nov. (IRAN 5217C) and G. sarabense sp. nov. (IRAN 5173C), resulting in crown and root rot and subsequent seedling death (Figure 4). All inoculated seedlings developed characteristic water-soaked lesions at the crown, leading to crown and root rot and eventually plant collapse within 2 weeks.
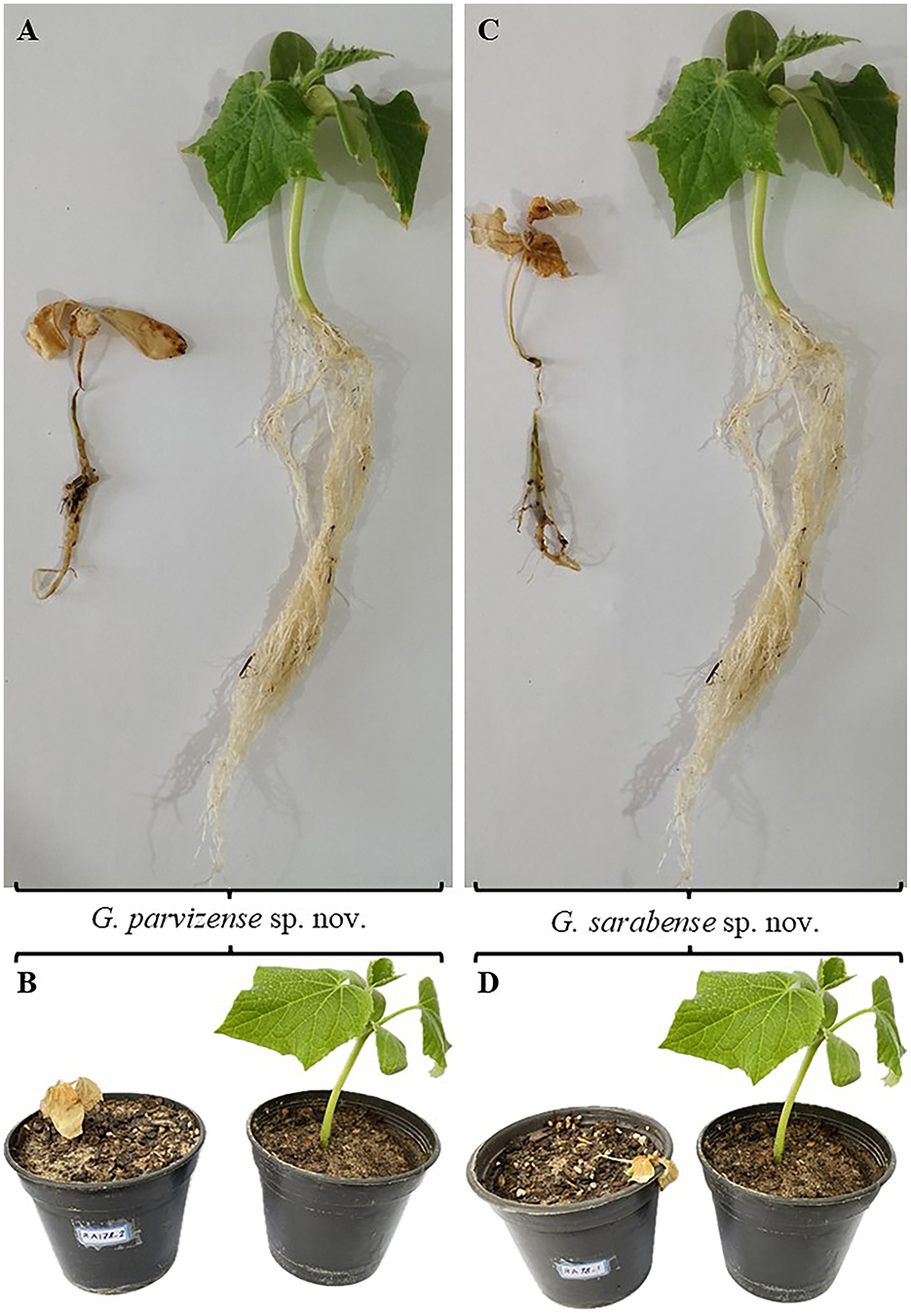
Figure 4. Pathogenicity test results. (A, B) Plants inoculated with Globisporangium parvizense sp. nov. (IRAN 5217C) on the left; corresponding negative control plants on the right. (C, D) Plants inoculated with G. sarabense sp. nov. (IRAN 5173C) on the left; corresponding negative control plants on the right.
Seedlings emerged normally 3 days post-sowing (dps). However, by 5 dps, initial symptoms—water-soaked lesions on the crown accompanied by incipient wilting—became apparent. These symptoms were intensified by 7 dps, with lesion coalescence, stem browning, and tissue decay extending into the root system. Control plants (negative control) remained symptomless throughout the experiment and exhibited significantly greater root and stem development compared to the inoculated treatments. Microscopical examination of infected tissues revealed the presence of sporangia and oospores in all three tested isolates. Furthermore, successful re-isolation of the original isolates from infected plants (100%) and their subsequent identification through detailed microscopy and morphometry fulfilled Koch's postulates, unequivocally confirming their pathogenicity.
5 Discussion
This study was part of a broader research project aimed at investigating the diversity of Pythium s.l. populations from Iranian aquatic environments. The primary focus of this survey was to study the diversity of Globisporangium species in non-agricultural environments in Iran. We described two new species of Globisporangium, namely G. parvizense sp. nov. and G. sarabense sp. nov., using a multi-gene phylogenetic framework that combined nuclear rDNA ITS sequences with partial sequences of cytochrome c oxidase subunits I and II (cox1 and cox2), supported by morphological assessments. The multi-gene phylogenetic approach enabled a comparative evaluation of the discriminatory robustness of these three genetic markers—ITS, cox1, and cox2—based on their nucleotide-level variation among the newly described species.
The newly described species can be clearly differentiated from previously described species in Globisporangium based on their morphological characteristics. Morphologically, G. parvizense differs from its close relatives in several ways: it lacks zoospores (unlike G. schmitthenneri and G. hypogynum), has a discharge tube (unlike G. acrogynum), possesses one to two antheridia per oogonium (unlike G. schmitthenneri and G. acrogynum), and forms two oospores per oogonium (unlike G. acrogynum and G. hypogynum). Similarly, G. sarabense can be distinguished by its uniquely shaped sporangia, absence of zoospores (unlike G. lucens and G. debaryanum), lack of oogonium chains (unlike G. tabrizense and G. lucens), and presence of a single antheridium and oospore per oogonium (in contrast to G. tabrizense, G. lucens, and G. viniferum).
For G. sarabense, the ITS region exhibited 21 and 15 nucleotide differences compared to G. lucens and G. tabrizense, respectively, making it the most informative marker for distinguishing these species. In contrast, cox1 showed 18 and 17 differences, while cox2 showed 10 and 12, respectively. Thus, while all three loci contributed to species delimitation, ITS provided the highest resolution in this case. Conversely, in G. parvizense, ITS showed only two to three nucleotide differences compared to G. hypogynum, G. schmitthenneri, and G. acrogynum, indicating limited discriminatory power. Cox1 showed four differences, while cox2 showed seven, making cox2 the most informative marker in this lineage.
These findings underscore that the effectiveness of genetic markers can vary across lineages. While ITS proved most effective for G. sarabense, the mitochondrial genes (cox1 and cox2) offered superior resolution for G. parvizense. This highlights (I) the limitations of relying solely on ITS—commonly used as a DNA barcode for oomycetes—which may lack sufficient resolution for some taxa, and (II) the importance of a multi-locus phylogenetic approach for robust species delimitation, providing a deeper understanding of the genetic diversity and evolutionary relationships within Pythium s.l.
The isolation of two novel Globisporangium species from irrigation systems and their positive pathogenicity toward cucumber plants might suggest that such organisms are not merely transient inhabitants but may actively exploit freshwater systems as a means of dispersal, raising concerns about their potential role in introducing plant diseases into agricultural settings. This is particularly important in Iran as it is one of the top cucumber producers globally and grows it widely near rivers and other natural water bodies (Nikolaou et al., 2021).
Moreover, the confirmed pathogenicity of these species on cucumber raises important ecological questions about the ability of aquatic-associated oomycetes to transition from water-associated habitats to terrestrial plant hosts. Although cucumber is not an aquatic plant, its constant exposure to irrigation water, flooding, and high soil moisture creates an ecological bridge that allows aquatic-origin oomycetes to colonize terrestrial crops. This highlights the importance of future research into the environmental adaptability of these pathogens and their potential infection under natural field conditions (Redekar et al., 2020).
The findings of this study highlight the urgent need for systematic monitoring and risk assessment of oomycetes in irrigation systems. If left unchecked, these pathogens could pose a serious threat to food security and sustainable agriculture. Future work should aim to monitor oomycete populations in agricultural water, identify high-risk species, and develop targeted disease management strategies to prevent future outbreaks.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Ethics statement
The pathogenicity experiment on cucumber was complied with relevant institutional, national, and international guidelines and legislation. We confirm that all methods were performed in accordance with the relevant guidelines.
Author contributions
RA: Funding acquisition, Resources, Writing – original draft, Formal analysis, Conceptualization, Investigation. AA: Project administration, Formal analysis, Supervision, Conceptualization, Writing – review & editing, Resources, Funding acquisition, Investigation, Writing – original draft. AC: Writing – original draft, Resources, Conceptualization, Supervision, Writing – review & editing, Funding acquisition, Investigation. HM: Investigation, Writing – review & editing, Writing – original draft. H-PG: Funding acquisition, Writing – review & editing, Supervision.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Acknowledgments
We would like to express our sincere gratitude to the Research Deputy of Azarbaijan Shahid Madani University, Tabriz, Iran, for providing financial support for this project. We also extend our thanks to the East Azerbaijan Agriculture and Natural Resources Research and Education Center for their valuable cooperation throughout the study. We are deeply appreciative of the IGB Leibniz-Institute of Freshwater Ecology and Inland Fisheries for not only providing essential facilities for this project but also facilitating the first author's participation at the institute, which enabled a portion of the research to be conducted there. Finally, we wish to thank Mr. Parviz Ahadi, the father of the first author, for his invaluable assistance during the sampling process. A special thanks to the Research Deputy of Azarbaijan Shahid Madani University, Tabriz, Iran and East Azerbaijan Agriculture and Natural Resources Research and Education Center for funding this project, and IGB Leibniz-Institute of Freshwater Ecology and Inland Fisheries Institute for providing financial support for this study under The German Science Foundation (DFG) Pycnotrap project (GR1540/37-1) given to H-PG. HM gratefully acknowledges support from the Simon's Foundation International as part of the BIOSSCOPE program (http://scope.bios.edu/).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that Gen AI was used in the creation of this manuscript. In the preparation of this manuscript, ChatGPT (OpenAI, 2024) was used exclusively for language editing and improving the clarity of the text. The tool was not used to generate or analyze scientific content. All ChatGPT-assisted edits were manually reviewed and verified for correctness. OpenAI. (2024, March 25). ChatGPT (Mar 25 version) [Large language model; https://chat.openai.com].
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abdelzaher, H. M. A., and Kageyama, K. (2020). Diversity of aquatic Pythium and Phytopythium spp. from rivers and a pond of Gifu city, Japan. Nov. Res. Microbiol. J. 4, 1029–1044. doi: 10.21608/nrmj.2020.130851
Abdelzaher, H. M. A., Morikawa, T., Ichitan, T., and Elnaghy, M. A. (1995). Classification of Pythium ‘group F'based on mycelial protein and isozyme patterns. Mycoscience 36, 45–49. doi: 10.1007/BF02268572
Ahadi, R., Bouket, A. C., Alizadeh, A., Masigol, H., and Grossart, H. P. (2024). Globisporangium tabrizense sp. nov., Globisporangium mahabadense sp. nov., and Pythium bostanabadense sp. nov. (Oomycota), three new species from Iranian aquatic environments. Sci. Rep. 14:31701. doi: 10.1038/s41598-024-81651-0
Ali-Shtayeh, M. S., and Dick, M. W. (1985). Five new species of Pythium (Peronosporomycetidae). Bot. J. Linn. Soc. 91, 297–317.
Al-Sheikh, H., and Abdelzaher, H. M. A. (2012). Occurrence, identification and pathogenicity of Pythium aphanidermatum, P. diclinum, P. dissotocum and Pythium “group P” isolated from Dawmat Al-Jandal Lake, Saudi Arabia. Res. J. Environ. Sci. 6 :196. doi: 10.3923/rjes.2012.196.209
Bala, K., Robideau, G. P., Désaulniers, N., De Cock, A., and Lévesque, C. A. (2010). Taxonomy, DNA barcoding and phylogeny of three new species of Pythium from Canada. Persoonia Mol. Phylog. Evol. Fungi 25, 22–31. doi: 10.3767/003158510X524754
Barton, R. (1958). Occurrence and establishment of Pythium in soils. Trans. Br. Mycol. Soc. 41, 207–222. doi: 10.1016/S0007-1536(58)80033-9
Ben-Yephet, Y., and Nelson, E. B. (1999). Differential suppression of damping-off caused by Pythium aphanidermatum, P. irregulare, and P. myriotylum in composts at different temperatures. Plant Dis. 83, 356–360. doi: 10.1094/PDIS.1999.83.4.356
Chen, J. J., and Zheng, X. B. (2019). Pythium subutonaiense, a new aquatic oomycete from southern china based on morphological and molecular characters. Mycobiology 47, 273–279. doi: 10.1080/12298093.2019.1642700
Chenari Bouket, A., Arzanlou, M., Tojo, M., and Babai-Ahari, A. (2015). Pythium kandovanense sp. nov., a fungus-like eukaryotic micro-organism (Stramenopila, Pythiales) isolated from snow-covered ryegrass leaves. Int. J. Syst. Evol. Microbiol. 65, 2500–2506. doi: 10.1099/ijs.0.000291
Derevnina, L., Petre, B., Kellner, R., Dagdas, Y. F., Sarowar, M. N., Giannakopoulou, A., et al. (2016). Emerging oomycete threats to plants and animals. Philos. Trans. R. Soc. B Biol. Sci. 371:20150459. doi: 10.1098/rstb.2015.0459
Ellis, M. L., Paul, P. A., Dorrance, A. E., and Broders, K. D. (2012). Two new species of Pythium, P. schmitthenneri and P. selbyi, pathogens of corn and soybean in Ohio. Mycologia 104, 477–487. doi: 10.3852/11-162
Ghiasi, M., Khosravi, A. R., Soltani, M., Binaii, M., Shokri, H., Tootian, Z., et al. (2010). Characterization of Saprolegnia isolates from Persian sturgeon (Acipencer persicus) eggs based on physiological and molecular data. J. Mycol. Med. 20, 1–7. doi: 10.1016/j.mycmed.2009.11.005
Hamlin, A. N., Locker, S., Huguet, E., Berry, C. R., Cole, R., Iv, J. F. G., et al. (2024). Computed tomographic characteristics of confirmed and presumed noncutaneous pythiosis in 25 dogs. Vet. Radiol. Ultrasound 65, 87–98. doi: 10.1111/vru.13326
Hendrix, J. r. F. F. (1974). Taxonomy and genetics of Pythium. Proc. Am. Phytopathol. Soc. 30, 200–207. doi: 10.5555/19751634442
Kageyama, K. (2010). Mycobiota in the subtropical and cool temperate areas in Japan. IFO Res. Commun. Nov. Res. Microbiol. J. 24:117.
Katoh, K., Rozewicki, A. j., and Yamada, K. D. (2019). MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinformatics 20, 1160–1166. doi: 10.1093/bib/bbx108
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Lévesque, C. A., and De Cock, A. W. A. M. (2004). Molecular phylogeny and taxonomy of the genus Pythium. Mycol. Res. 108, 1363–1383. doi: 10.1017/S0953756204001431
Martin, F. N. (2000). Phylogenetic relationships among some Pythium species inferred from sequence analysis of the mitochondrially encoded cytochrome oxidase II gene. Mycologia 92, 711–727. doi: 10.1080/00275514.2000.12061211
Martin, F. N., and Loper, J. E. (1999). Soilborne plant diseases caused by Pythium spp.: ecology, epidemiology, and prospects for biological control. CRC. Crit. Rev. Plant Sci. 18, 111–181. doi: 10.1080/07352689991309216
Martin, J. (1992). Cultures in Organizations: Three Perspectives. New York, NY: Oxford University Press. doi: 10.1093/oso/9780195071634.001.0001
Masigol, H., Khodaparast, S. A., Mostowfizadeh-Ghalamfarsa, R., Mousanejad, S., Rojas-Jimenez, K., and Grossart, H. P. (2018). Notes on Dictyuchus species (Stramenopila, Oomycetes) from Anzali lagoon, Iran. Mycol. Iran 5, 79–89. doi: 10.22043/mi.2018.120353
Masigol, H., Khodaparast, S. A., Mostowfizadeh-Ghalamfarsa, R., Rojas-Jimenez, K., Woodhouse, J. N., Neubauer, D., et al. (2020). Taxonomical and functional diversity of Saprolegniales in Anzali lagoon, Iran. Aquat. Ecol. 54, 323–336. doi: 10.1007/s10452-019-09745-w
Masigol, H., Khodaparast, S. A., Woodhouse, J. N., Rojas-Jimenez, K., Fonvielle, J., Rezakhani, F., et al. (2019). The contrasting roles of aquatic fungi and oomycetes in the degradation and transformation of polymeric organic matter. L&O 64, 2662–2678. doi: 10.1002/lno.11242
Masigol, H., Mostowfizadeh-Ghalamfarsa, R., and Grossart, H. P. (2022). The current status of Saprolegniales in Iran: calling mycologists for better taxonomic and ecological resolutions. Mycol. Iran 8, 1–13. doi: 10.22043/MI.2022.358121.1212
Masigol, H., Solbach, M. D., Pourmoghaddam, M. J., Ahadi, R., Mostowfizadeh-Ghalamfarsa, R., Taheri, S. R., et al. (2025). A glimpse into Oomycota diversity in freshwater lakes and adjacent forests using a metabarcoding approach. Sci. Rep. 15:19124. doi: 10.1038/s41598-025-01727-3
Masigol, H., van West, P., Taheri, S. R., Fregeneda-Grandes, J. M., Pârvulescu, L., McLaggan, D., et al. (2023). Advancements, deficiencies, and future necessities of studying Saprolegniales: A semi-quantitative review of 1073 published papers. Fungal Biol. Rev. 46:100319. doi: 10.1016/j.fbr.2023.100319
Middleton, J. T. (1941). Boot rot of barley caused by Pythium hypogynum n. sp. Phytopathology 31:863.
Mirmazloomi, S., Ghiasi, M., Shirangi, S. A., Lashgari, S. N., and Khosravi, A. R. (2022). Morphological and molecular characterization of two species of Saprolegnia isolated from a rainbow trout (Oncorhynchus mykiss) hatchery in Iran. Int. Aquat. Res. 14:223. doi: 10.22034/iar.2022.1959159.1282
Mishra, A., Shukla, A., Rana, N. P., Currie, W. L., and Dwivedi, Y. K. (2023). Re-examining post-acceptance model of information systems continuance: a revised theoretical model using MASEM approach. Int. J. Inf. Manag. 68:102571. doi: 10.1016/j.ijinfomgt.2022.102571
Möller, E. M., Bahnweg, G., Sandermann, H., and Geiger, H. H. (1992). A simple and efficient protocol for isolation of high molecular weight DNA from filamentous fungi, fruit bodies, and infected plant tissues. Nucleic Acids Res. 20:6115. doi: 10.1093/nar/20.22.6115
Morita, Y., and Tojo, M. (2007). Modifications of PARP medium using fluazinam, miconazole, and nystatin for detection of Pythium spp. in soil. Plant Dis. 91, 1591–1599. doi: 10.1094/PDIS-91-12-1591
Mostowfizadeh-Ghalamfarsa, R. (2015). The current status of Pythium species in Iran: challenges in taxonomy. Mycol. Iran 2, 79–87. doi: 10.22043/mi.2015.19901
Mueller, G. M. (2011). Biodiversity of Fungi: Inventory and Monitoring Methods. Amsterdam: Elsevier.
Nam, B., and Choi, Y.-J. (2019). Phytopythium and Pythium species (Oomycota) isolated from freshwater environments of Korea. Mycobiology 47, 261–272. doi: 10.1080/12298093.2019.1625174
Nguyen, H. D. T., Dodge, A., Dadej, K., Rintoul, T. L., Ponomareva, E., Martin, F. N., et al. (2022). Whole genome sequencing and phylogenomic analysis show support for the splitting of genus Pythium. Mycologia 114, 501–515. doi: 10.1080/00275514.2022.2045116
Nikolaou, G., Neocleous, D., Christou, A., Polycarpou, P., Kitta, E., and Katsoulas, N. (2021). Energy and water related parameters in tomato and cucumber greenhouse crops in semiarid mediterranean regions. A review, part II: irrigation and fertigation. Horticulturae 7:548. doi: 10.3390/horticulturae7120548
Nzungize, J. R., Lyumugabe, F., Busogoro, J. P., and Baudoin, J. P. (2012). Pythium root rot of common bean: biology and control methods. A review. Biotechnol. Agron. Soc. Environ. 16, 405–413.
Paul, B., and Bala, K. (2008). A new species of Pythium with inflated sporangia and coiled antheridia, isolated from India. FEMS Microbiol. Lett. 282, 251–257. doi: 10.1111/j.1574-6968.2008.01138.x
Price, M. N., Dehal, P. S., and Arkin, A. P. (2010). FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS ONE 5:e9490. doi: 10.1371/journal.pone.0009490
Rahman, M. Z., Abdelzaher, H. M. A., Mingzhu, L., Motohashi, K., Suga, H., and Kageyama, K. (2015). Pythium rishiriense sp. nov. from water and P. alternatum sp. nov. from soil, two new species from Japan. FEMS Microbiol. Lett. 362:fnv086. doi: 10.1093/femsle/fnv086
Redekar, N. R., Bourret, T. B., Eberhart, J. L., Johnson, G. E., Pitton, B. J. L., Haver, D. L., et al. (2020). The population of oomycetes in a recycled irrigation water system at a horticultural nursery in southern California. Water Res. 183:116050. doi: 10.1016/j.watres.2020.116050
Roberts, D. P., Lakshman, D. K., McKenna, L. F., Emche, S. E., Maul, J. E., and Bauchan, G. (2016). Seed treatment with ethanol extract of Serratia marcescens is compatible with Trichoderma isolates for control of damping-off of cucumber caused by Pythium ultimum. Plant Dis. 100, 1278–1287. doi: 10.1094/PDIS-09-15-1039-RE
Robideau, G. P., De Cock, A. W. A. M., Coffey, M. D., Voglmayr, H., Brouwer, H., Bala, K., et al. (2011). DNA barcoding of oomycetes with cytochrome c oxidase subunit I and internal transcribed spacer. Mol. Ecol. Resour. 11, 1002–1011. doi: 10.1111/j.1755-0998.2011.03041.x
Ronquist, F., Teslenko, M., Van Der Mark, P., Ayres, D. L., Darling, A., Höhna, S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Salmaninezhad, F., Aloi, F., Pane, A., Mostowfizadeh-Ghalamfarsa, R., and Cacciola, S. O. (2022). Globisporangium coniferarum sp. nov., associated with conifers and Quercus spp. Fungal Syst. Evol. 10, 127–137. doi: 10.3114/fuse.2022.10.05
Salmaninezhad, F., and Mostowfizadeh-Ghalamfarsa, R. (2017). Taxonomy, phylogeny and pathogenicity of Pythium species in rice paddy fields of Fars Province. Iran. J. Plant Pathol. 53, 31–50.
Salmaninezhad, F., and Mostowfizadeh-Ghalamfarsa, R. (2019). Three new Pythium species from rice paddy fields. Mycologia 111, 274–290. doi: 10.1080/00275514.2018.1543486
Salmaninezhad, F., and Mostowfizadeh-Ghalamfarsa, R. (2022). The current status of Globisporangium species in Iran: from Pythium sensu lato to Newly Described Species. Mycol. Iran. 9, 1–14. doi: 10.22092/MI.2022.359205.1221
Salmaninezhad, F., and Mostowfizadeh-Ghalamfarsa, R. (2023). The current status of Phytopythium species in Iran: challenges in identification of an intermediate taxon. Mycol. Iran 10, 1–12. doi: 10.3390/jof10060405
Shreves, K. V., Saraiva, M., Ruba, T., Miller, C., Scott, E. M., McLaggan, D., et al. (2024). Specific phylotypes of Saprolegnia parasitica associated with atlantic salmon freshwater aquaculture. J. Fungi 10:57. doi: 10.3390/jof10010057
Sigillo, L., Pane, C., Garaguso, I., Luongo, L., Galli, M., Valente, M. T., et al. (2020). First report of Pythium spinosum as a causal agent of crown and root rot in greenhouse cucumber cultivation in Italy. Plant Dis. 104:3269. doi: 10.1094/PDIS-02-20-0305-PDN
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Uzuhashi, S., Kakishima, M., and Tojo, M. (2010). Phylogeny of the genus Pythium and description of new genera. Mycoscience 51, 337–365. doi: 10.1007/S10267-010-0046-7
Uzuhashi, S., Okada, G., and Ohkuma, M. (2015). Four new Pythium species from aquatic environments in Japan. Antonie van Leeuwenhoek. Int. J. Gen. Mol. Microbiol. 107, 375–391. doi: 10.1007/s10482-014-0336-8
Villa, N. O., Kageyama, K., Asano, T., and Suga, H. (2006). Phylogenetic relationships of Pythium and Phytophthora species based on ITS rDNA, cytochrome oxidase II and -tubulin gene sequences. Mycologia 98, 410–422. doi: 10.3852/mycologia.98.3.410
White, T. J., Bruns, T., Lee, S., and Taylor, J. (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protoc. Guid. Methods Appl. 18, 315–322. doi: 10.1016/B978-0-12-372180-8.50042-1
Keywords: aquatic oomycetes, plant parasites, Pythium sensu lato, systematics, taxonomy
Citation: Ahadi R, Alizadeh A, Chenari Bouket A, Masigol H and Grossart H-P (2025) Multigene phylogeny, morphology, and pathogenicity uncover two novel Globisporangium species (Oomycota) from freshwater habitats in northwestern Iran. Front. Microbiol. 16:1615096. doi: 10.3389/fmicb.2025.1615096
Received: 20 April 2025; Accepted: 30 June 2025;
Published: 06 August 2025.
Edited by:
Samantha Chandranath Karunarathna, Qujing Normal University, ChinaReviewed by:
Danfeng Bao, Mae Fah Luang University, ThailandJian Ma, Mae Fah Luang University, Thailand
Copyright © 2025 Ahadi, Alizadeh, Chenari Bouket, Masigol and Grossart. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alireza Alizadeh, YWxpcmV6YS5hbGl6YWRlaEBhemFydW5pdi5hYy5pcg==; Hans-Peter Grossart, aGFuc3BldGVyLmdyb3NzYXJ0QGlnYi1iZXJsaW4uZGU=
†These authors have contributed equally to this work
 Reza Ahadi
Reza Ahadi Alireza Alizadeh
Alireza Alizadeh Ali Chenari Bouket
Ali Chenari Bouket Hossein Masigol
Hossein Masigol Hans-Peter Grossart
Hans-Peter Grossart