- 1School of Public Health, Lanzhou University, Lanzhou, China
- 2Department of Medical Administration, Gansu Provincial Maternity and Child-Care Hospital, Lanzhou, China
- 3Department of Respiratory and Critical Care Medicine, The Second Hospital of Lanzhou City, Lanzhou, China
Introduction: Exposure to heavy metals is a growing environmental concern. Although exposure to individual metals is associated with pulmonary damage, real-world exposure typically involves multiple metals simultaneously. This study hypothesizes that combined exposure to nickel, copper, and arsenic induces lung injury through disruption of the bronchoalveolar lavage fluid (BALF) microbial ecosystem in rats. The primary objective of this study was to verify the hypothesis.
Methods: Thirty-two male Sprague–Dawley (SD) rats were randomly assigned to four groups: one control group and three exposure groups (low, medium, and high doses). The exposed groups received mixed heavy metal aerosols containing nickel (Ni), copper (Cu), and arsenic (As) at low (Ni: 210.9 ng/m3, Cu: 108.4 ng/m3, As: 104.6 ng/m3), medium (5×low), and high (10×low) concentrations. Exposure occurred via inhalation twice daily for 1 h over 90 days. Lung function was assessed non-invasively, and histological examinations, 16S ribosomal DNA (16S rDNA) sequencing, and microbial functional predictions were performed to evaluate exposure effects. We measured heavy metal concentrations in lung tissues and assessed the associations with microbial changes. Microbial community structure and function were further analyzed using LEfSe, PICRUSt2, and ecological network analysis.
Results: Compared exposure to Ni, Cu, and As induced dose-dependent lung damage, including inflammation, alveolar deformation, and bronchial thickening, accompanied by significant declines in lung function, including a 21.2% reduction in tidal volume and a 34.5% increase in airway resistance in the high-dose group (P < 0.05). Microbial diversity and phylogenetic richness were significantly reduced (Chao1, Richness, ACE, P < 0.05), with taxonomic shifts characterized by the enrichment of metal-resistant genera (Pseudomonas, Burkholderia) and depletion of sensitive taxa (Ralstonia, Achromobacter). Functional prediction suggested impairments in xenobiotic metabolism and amino acid biosynthesis. Ecological network complexity declined with increasing exposure dose. Microbiota dysbiosis is strongly associated with lung function impairments.
Conclusions: Combined exposure to Ni, Cu, and As disrupts respiratory microbiota and impairs lung function in rats. These findings highlight a critical link between environmental heavy metal exposure and respiratory health, underscoring the need for stricter regulation of metal pollutants and further research into microbiota-related lung injury mechanisms.
1 Introduction
Chronic respiratory disease refers to a group of conditions that persistently impair respiratory system function, including chronic obstructive pulmonary disease (COPD), asthma, and others. These diseases are characterized by a prolonged course, slow recovery, and frequent relapse (Lin et al., 2018), representing a major global health burden. Lung function, a key indicator for assessing chronic lung disease, is widely used to evaluate the impact of environmental pollution, particularly to quantify the effects of air pollution on the respiratory tract (Duprez and Jacobs, 2018; Zheng et al., 2023).
Heavy metals are commonly defined as elements with an atomic number exceeding 20 and a density >5 g/cm3, including metalloids like arsenic (As) due to their similar chemical properties and environmental behavior (Tchounwou et al., 2012; Gautam et al., 2016; Jacob et al., 2018). In recent years, intensified industrial activities, fossil fuel combustion, and heavy fertilizer use have led to rising levels of heavy metals in the environment (Long et al., 2021; Lavanya et al., 2024). This issue is particularly pronounced in China and other developing countries (Jacob et al., 2018; Liu et al., 2022). Inhaled heavy metal particles typically deposited in the bronchioles and alveoli, causing severe lung damage. Moreover, these pollutants may enter the blood circulation through pathways such as ingestion or absorption through the skin, triggering systemic inflammation that further exacerbates lung damage (Balali-Mood et al., 2021; Hashem et al., 2021).
Although numerous studies have confirmed the adverse effects of heavy metal exposure on lung function, they have predominantly focused on individual metals and mechanisms such as inflammation, oxidative stress, and apoptosis (Wu et al., 2022), or on the gastrointestinal microbiota under metal exposure (Li X. et al., 2019). However, real-world exposure involves complex metal mixtures, and their combined effects, particularly on the lower respiratory tract microbiome, a key interface for immune defense and pollutant interaction, remain poorly understood (Xue et al., 2020).
Historically, healthy lungs were thought to be sterile environments (Chambers et al., 2014; Taylor et al., 2015). However, recent studies have revealed a complex microbial ecosystem within the airways and lung tissues that serves as a “gatekeeper” of respiratory health (Morris et al., 2013; Venkataraman et al., 2015). Imbalances in these microbiota are associated with various respiratory diseases, including COPD, asthma, and cystic fibrosis (Huang et al., 2015; Li et al., 2017), with changes in microbial communities correlating with reduced lung function (Wang et al., 2019; Zhang et al., 2023). However, research on microbiome changes induced by heavy metal exposure has primarily focused on the gut flora, with limited studies exploring the interaction between respiratory microbiota and heavy metal exposure.
Based on the above, we hypothesize that combined exposure to nickel, copper, and arsenic induces dose-dependent lung injury through disruption of the respiratory microbiota. Such exposure is expected to alter the composition and function of airway microbial communities, leading to reduced microbial diversity and the enrichment of pollutant-tolerant taxa. These microbial shifts may contribute to or mediate the observed decline in lung function, suggesting a potential link between microbiota dysbiosis and metal-induced pulmonary toxicity.
In this study, we evaluated lung structure and functional parameters and profiled the respiratory microbiota in rats exposed to varying concentrations of mixed heavy metal aerosols. The objective was to investigate the impact of combined heavy metal exposure on pulmonary injury and to elucidate the potential role of BALF microbiota dysbiosis in this process. This study will provide a novel perspective on pollution-induced respiratory dysfunction and support future efforts toward microbiota-informed environmental health policies.
2 Materials and methods
2.1 Animal exposure programs
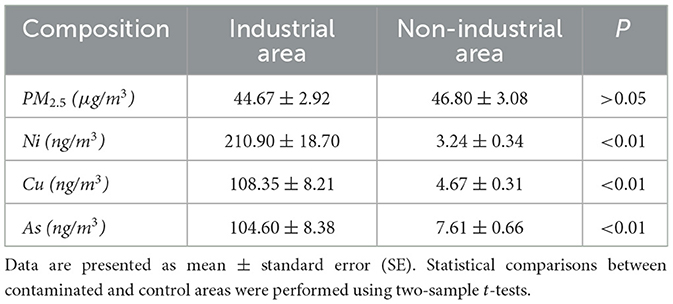
Our research team has long focused on the health impacts of heavy metal pollution in an industrial region northwest of China. Based on our atmospheric monitoring data (Table 1), nickel (Ni), copper (Cu), and arsenic (As) were identified as the predominant metals in atmospheric PM2.5 from the industrial zone, with average concentrations of 210.90 ± 18.70 ng/m3 for Ni, 108.35 ± 8.21 ng/m3 for Cu, and 104.60 ± 8.38 ng/m3 for As. These values were significantly higher than those observed in a nearby non-industrial area (3.24 ± 0.34 ng/m3 for Ni, 4.67 ± 0.31 ng/m3 for Cu, and 7.61 ± 0.66 ng/m3 for As) (P < 0.01).
Based on these findings, we designed three exposure groups, using the concentrations of Ni, Cu, and As detected in the ambient PM2.5 of the industrial region as the reference for the low-dose group (Table 1). The medium and high doses were set at 5 and 10 times the low-dose level, respectively. To replicate environmental exposure conditions, an aerosol mixture was prepared using nickel sulfate hexahydrate (NiSO4·6H2O), copper sulfate pentahydrate (CuSO4·5H2O), and sodium arsenite (NaAsO2), dissolved in deionized water and aerosolized with a nebulizer for controlled inhalation delivery.
Thirty-two SPF-grade SD male rats, aged 6–8 weeks and weighing between 180–220 g, were purchased from the Laboratory Animal Center of Lanzhou University. The rats were randomly assigned to four groups (n = 8 per group): control group (Group C), low-dose group (Group L), medium-dose group (Group M), and high-dose group (Group H). Group C was housed in a conventional laboratory environment without aerosol atomization exposure, while Groups L, M, and H were respectively placed in isolated chambers within a transparent exposure box for aerosol exposure to solutions of varying concentration gradients (Figure 1). The transparent exposure box measured 0.5 m × 0.3 m × 0.35 m (length × width × height), and contained a mesh fence to separate the box into isolated chambers to prevent rats from aggregating and burying their mouths and noses into each other's hairs (similar to “wearing a mask”), which would affect the effect of the exposure to the aerosol. The right side of the chamber was connected to a nebulizer serving as the inlet of the heavy metal aerosol mixture, while the left side was connected to a particle sampler to monitor the aerosol concentration in the exposure chamber.
The exposure was conducted twice daily, with each session lasting 1 h. After each exposure, the rats were returned to their cages. The exposure protocol spanned 90 days and was performed in a fume hood connected to handling and collection units. The ambient temperature was maintained at 18–26°C, and the relative humidity at 40%−70%.
2.2 Non-invasive lung function assessment in rats
Within 24 h of completing exposure, lung function was assessed non-invasively in all rats using the NAM system (DSI BUXCO, USA). Eight rats per group were individually placed in the cavity for 10 min to acclimate, after which relevant parameters were recorded, including respiratory rate (f), tidal volume (Tv), specific airway resistance (sRaw), airway resistance (Raw), maximal inspiratory flow rate (PIF), maximal expiratory flow rate (PEF), functional residual air volume (Frc), and expiratory flow rate at mid-expiration (EF50).
2.3 Collection of rat bronchoalveolar lavage fluid and lung tissue
Bronchoalveolar lavage fluid (BALF) was collected from one lung of each rat, with the contralateral lung harvested for tissue analysis. Rats were anesthetized with an intraperitoneal injection of 0.3 ml/100 g chloral hydrate, and the trachea and lungs were exposed. The epidermis of the animals was sterilized with 75% ethanol, and sterilized grade surgical instruments were used throughout. Under sterile conditions, the thoracic cavity was opened, and one lung was ligated at the bronchus for isolation. A tracheal cannula was inserted into each rat, and 10 ml of phosphate-buffered saline (PBS) was slowly injected into the lungs, with a dwell time of 30–60 seconds instillation. The lavage was repeated 2–3 times, and the recovered BALF was transferred to a sterile test tube, centrifuged to separate the cells and supernatant, and the supernatant was stored in a −80°C refrigerator for subsequent analysis. Concurrently, lung tissues were washed with saline and divided into two parts. One portion was fixed in 4% paraformaldehyde solution for hematoxylin and eosin (H&E) staining, while the other was snap-frozen in liquid nitrogen and stored at −80°C for heavy metal analysis.
2.4 Lung histopathology
Fixed rat lung tissues were dehydrated with alcohol and xylene, embedded in paraffin, and sectioned into 3–5 μm thin slices using a machine. The sections were stained with hematoxylin and eosin (H&E), coverslipped, and examined microscopically for histopathological analysis.
2.5 Determination of heavy metals in rat lung tissue
Heavy metal concentrations in lung tissue were determined using inductively coupled plasma mass spectrometry (ICP-MS) technique. Approximately 20–30 mg of lung tissue was digested in 10 ml of concentrated nitric acid using a microwave digestor for 1 h. After cooling and acid evaporation, the digestate was diluted, filtered, and analyzed for Ni, Cu, and As concentrations by ICP-MS.
2.6 16S rDNA sequencing of rat alveolar lavage fluid
Bacterial genomic DNA was extracted in BALF using the Mabiote Bacterial DNA Mini Kit, and the concentration and quality of the extracted genomic DNA were assessed with a Thermo Scientific Nanodrop One (Thermo Fisher Scientific, MA, USA). For each group, only BALF samples with qualified bacterial DNA were selected for 16S rRNA gene sequencing, while those with insufficient DNA quality or quantity were excluded to reduce contamination risks and ensure data reliability (Saladié et al., 2020; Baker et al., 2021). The V3-V4 region of 16S rDNA was amplified using the Illumina HiSeq platform with amplification primers 338F (5′-ACTCCTACGGGGAGGCAGCA- 3′) and 806R (5′-GGACTACHVGGGTWTCTAAT- 3′). Each amplification reaction consisted of denaturation at 98°C for 30 seconds, followed by 30 cycles of amplification (10 seconds at 98°C, annealing at 50°C for 30 seconds, and extension at 72°C for 1 min), concluding with a final extension at 72°C for 2 min. Amplicons were purified PCR amplicons from agarose gels and sequenced. Raw sequencing data were processed using QIIME2 (version 2020.11.0), including trimming of low-quality sequences and quality filtering with Cutadapt. Paired-end reads were assembled by FLASH 1.2.11. The Raw Tags sequence quality is filtered by using fastp (an ultra-fast all-in-one FASTQ preprocessor, version 0.14.1). Performs sliding window quality clipping (-W 4 -M 20) on Raw Tags data to obtain valid splicing fragments (Clean Tags).
For each sample, we used Deblur to process sequences, identify amplicon sequence variants (ASVs), and assign ASVs to taxonomic taxa (Amir et al., 2017).
2.7 Statistical analysis
We conducted dilution curve analysis and calculated multiple alpha diversity indices, including Chao1, ACE, Shannon_e, Simpson, Richness, Jost, dominance, Good's-coverage, and PD_whole_tree indices. The Kruskal-Wallis rank-sum test was applied to compare alpha diversity differences across groups.
Beta diversity analysis was employed to assess differences in species composition and structural organization among various biomes. A distance matrix was constructed to calculate the distances between biological communities for principal coordinate analysis (PCoA). β diversity was visualized using PCoA based on the ASV abundance table, utilizing the vegan package1 in R. The Bray-Curtis, weighted-UniFrac, and unweighted-UniFrac distance algorithms were applied.
Differences in bacterial community composition were assessed using PCoA at the ASV level, with Bray-Curtis, weighted and unweighted UniFrac distance metrics. Statistical significance was assessed by analysis of similarity (ANOSIM). Linear discriminant analysis of effect size (LEfSe) was applied to identify biomarkers that could lead to differences between the four groups, with LDA scores >3.5 as the threshold for distinguishing significantly different taxa. Concentration of Ni, Cu, and As in rat lung tissue was quantified, and their associations with lung function and respiratory microbial communities were investigated. To assess functional differences in metabolic pathways, microbial functional genes were predicted using the KEGG database.
All statistical analyses were performed using SPSS (version 26.0) and R (version 4.2.2). The Mantel test2 implemented in R was used to calculate correlation coefficients between the environmental factor matrix and community composition matrix, assessing significant correlations existed and the influence of environmental factors on community structure. Distance matrix calculations employed the Manhattan, Euclidean, and Bray-Curtis distance algorithms.
2.8 Microbial ecological networks analysis
Bacterial molecular ecological networks (MENs) in the BALF were constructed using the Integrated Network Analysis Pipeline (iNAP) (Feng et al., 2022), an online tool for ecological network analysis. Networks were inferred using the Meinschausen-Buhlmann (MB) neighborhood selection method, implemented through the SParse InversE Covariance Estimation (SPEIC-EASI) algorithm (Kurtz et al., 2015). The resulting networks were further explored and visualized with Gephi (Bastian et al., 2009), an interactive platform, utilizing the Fruchterman-Reingold layout, which provides an intuitive representation of network structures.
During network construction, only ASVs detected in >80% of samples were retained to ensure reliability and minimize noise. Key topological parameters and node scores were calculated using Gephi,3 including average degree (number of edges per node, reflecting connectivity), average path length (shortest distance between two nodes), network diameter (longest path between nodes), and clustering coefficient (degree of interconnection among a node's neighbors, indicating localized density). Community structure was identified by optimizing modularity, which quantified the strength of division into distinct clusters or modules. Networks with higher modularity exhibited dense intra-cluster connections and sparse inter-cluster connections, signifying distinct functional or ecological groupings.
Keystone taxa were identified based on Gephi-calculated parameters: degree >7, harmonic closeness centrality >0.28, and betweenness centrality <0.18 (Banerjee et al., 2019). The degree distribution was wide, with the highest degree (up to 18) representing <1% of nodes, and approximately 20% of nodes had degrees >7. Consequently, the degree threshold was adjusted from 10 to 7 to capture relevant taxa.
Microbial functions were predicted using DESeq2 for differential abundance analysis (McCarthy et al., 2012) and the Kyoto Encyclopaedia of Genes and Genomes (KEGG) database for metabolic pathway analysis (Kanehisa et al., 2017). KEGG enrichment analysis was performed using the clusterProfiler R package (Wu et al., 2021).
The correlation network map, correlation heat map, KEGG enrichment analysis and functional prediction volcano map among microbiota were generated using ChiPlot4 (accessed January 2025). For clarity, all abbreviations used in this manuscript are provided in Supplementary Table S1.
3 Results
3.1 General status and weight
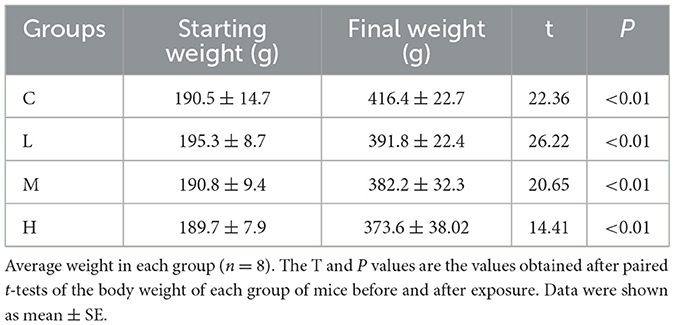
Rats in Group L displayed normal behavior. Group M showed dull fur, reduced mobility, and occasional sneezing. Group H experienced fur thinning, nasal bleeding, respiratory distress, lethargy, and decreased food and water intake, worsening over time. Although all groups of rats gained weight (Table 2), the control group began to gain more weight than the other groups after 6 weeks of exposure, which may indicate that heavy metal exposure slowed down the rate of body weight gain. However, differences in body weight gain among groups were not statistically significant (P > 0.05).
3.2 Levels of Ni, Cu, and As in rat lung tissue
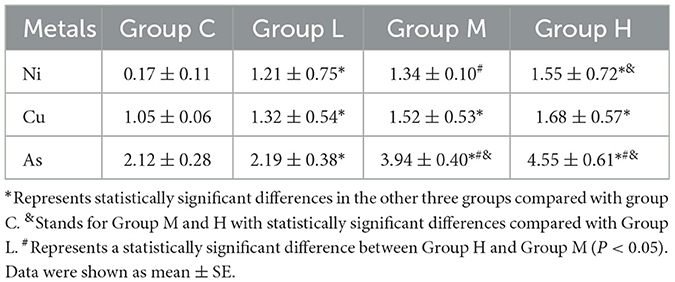
As shown in Table 3, the concentrations of Ni and As in rat lung tissues exhibited significant increases with escalating exposure doses of the mixed aerosol, culminating in the highest levels in the H group (P < 0.01). In contrast, although the overall comparison across all groups did not reveal significant differences in Cu levels (P = 0.352), pairwise comparisons between each experimental group and the control group unveiled significant elevations in Cu concentrations in the experimental groups (P < 0.05). These results underscore that varying doses of Ni and As aerosol exposure significantly augment the accumulation of these heavy metals in rat lungs.
3.3 Lung inflammation and lung function differences
In Group C, the respiratory tract and alveolar structures remained intact with no abnormal changes. Group L rats exhibited mild inflammation and vasodilation. Group M rats displayed enlarged alveoli, thickened bronchial walls, epithelial cell detachment, and moderate inflammation. Group H rats showed severe lung and bronchial damage, with disorganized alveolar and bronchial structures and abundant inflammatory cell infiltration. The number of dust cells increased progressively with higher exposure doses (Figure 2).
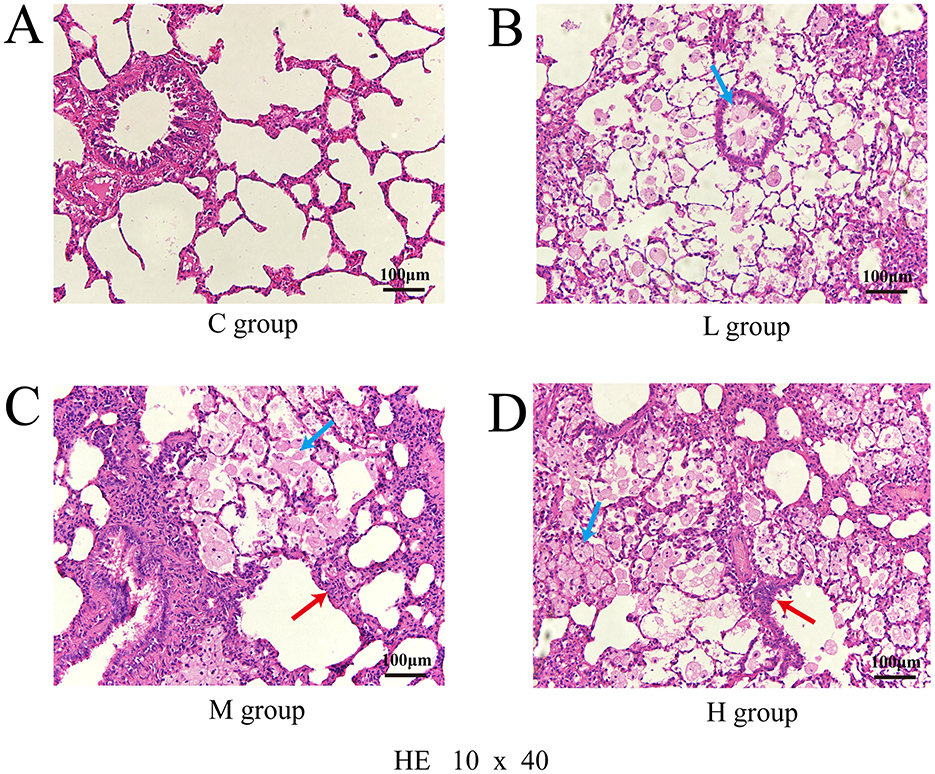
Figure 2. Pathological section of lung injury stained with hematoxylin and eosin (HE), observed under a light microscope at 10 × 40 magnification; Control group (A); Low-dose group (B); Medium-dose group (C); High-dose group (D); Blue arrows point to dust cells and red arrows to inflammatory cells.
With increasing heavy metal exposure, respiratory frequency (f), airway resistance (sRaw, Raw), and functional residual capacity (FRC) tended to increase, whereas tidal volume (Tv), peak inspiratory flow rate (PIF), peak expiratory flow rate (PEF), and flow rate at 50% expiration (EF50) decreased (Figure 3).
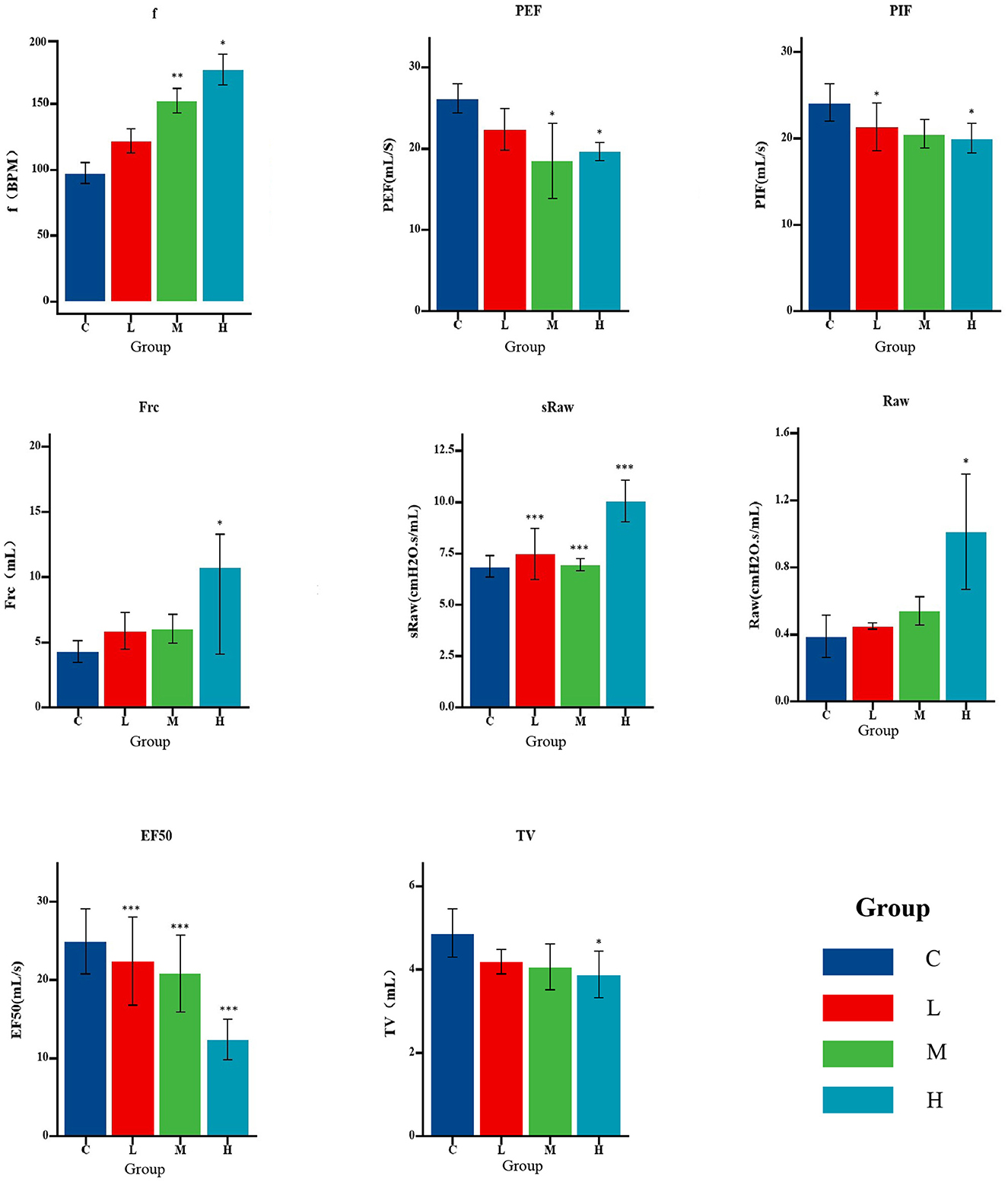
Figure 3. Pulmonary function parameters of rats in each exposure group (n = 8 per group). Bar graphs show mean ± standard error (SE) for each index. Statistical analysis was performed using a one-way ANOVA test; *P < 0.05, **P < 0.01, ***P < 0.001 compared to control (Group C).
FRC was positively correlated with lung arsenic (As) concentration, and f, sRaw, and Raw were positively correlated with Ni and As concentrations. Tv was negatively correlated with As concentration, and PIF, PEF, and EF50 were negatively correlated with Ni and As concentrations. The correlation between As and lung function was stronger than that with Ni, while copper (Cu) concentrations showed no significant correlations with lung function indices (Figure 4).
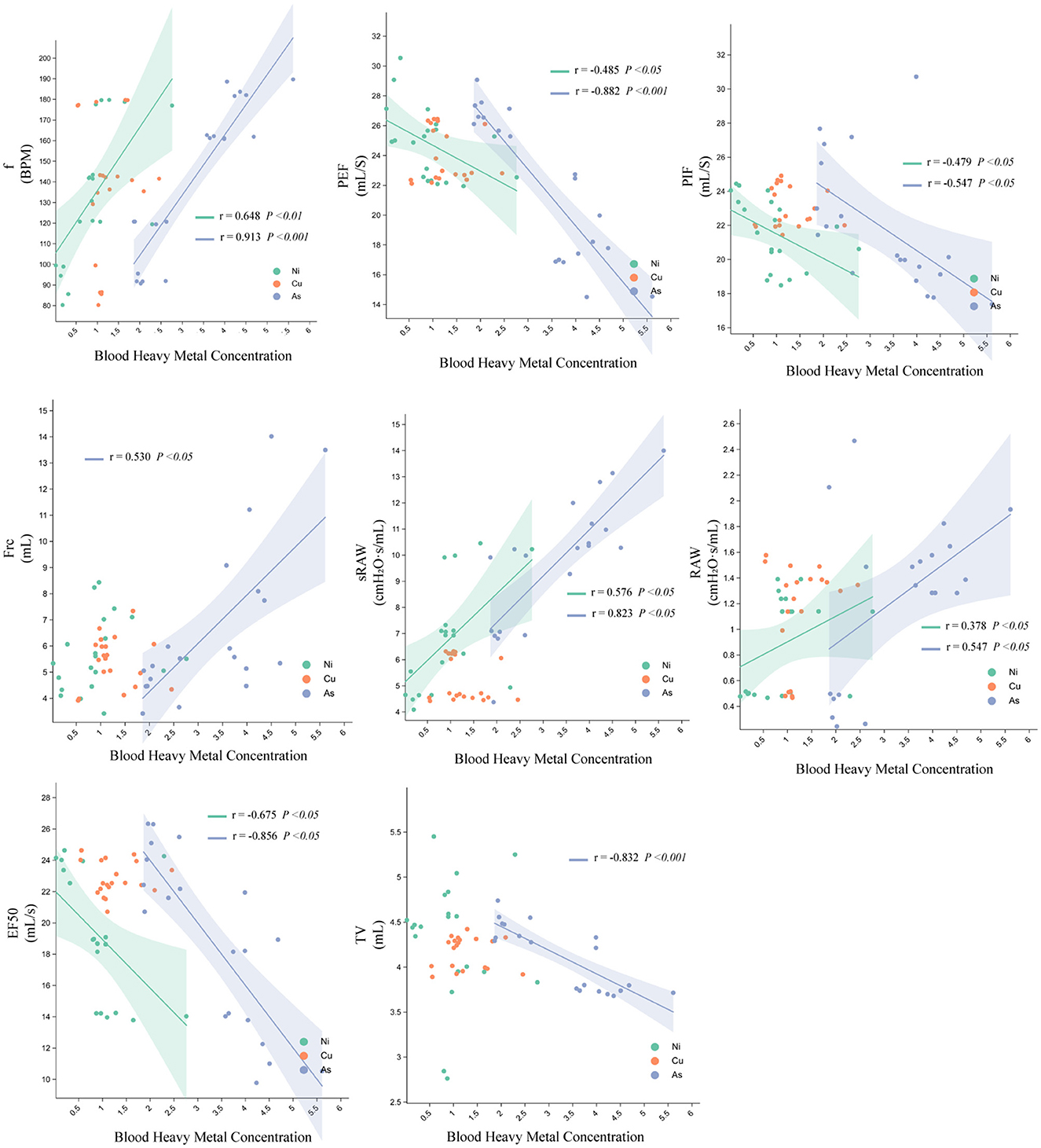
Figure 4. Relationship between Ni, Cu, and As heavy metal concentrations and pulmonary function parameters in rats.
3.4 Bacterial population structure in alveolar lavage fluid of rats
The abundance clustering heatmap (Figure 5A) visually depicts microbial community abundances. Darker regions indicate higher microbial abundance, while lighter regions signify lower abundance. In Group C, microbial abundance showed a relatively uniform distribution, reflecting the maintenance of typical microbial communities in animals without pollutant exposure. In Group L, microbial abundances were relatively balanced, with minor increases in certain ASVs compared to the control group but minimal overall changes. Group M exhibited greater variability in microbial abundance, with a marked increase in specific ASVs, such as ASV_10 and ASV_7. Group H demonstrated a more concentrated distribution, with significant increases in specific ASVs like ASV_201 and ASV_6. Overall, the microbial communities in the M and H groups underwent significant restructuring, with certain groups showing pronounced increases in abundance while others decreased or maintained low abundance.
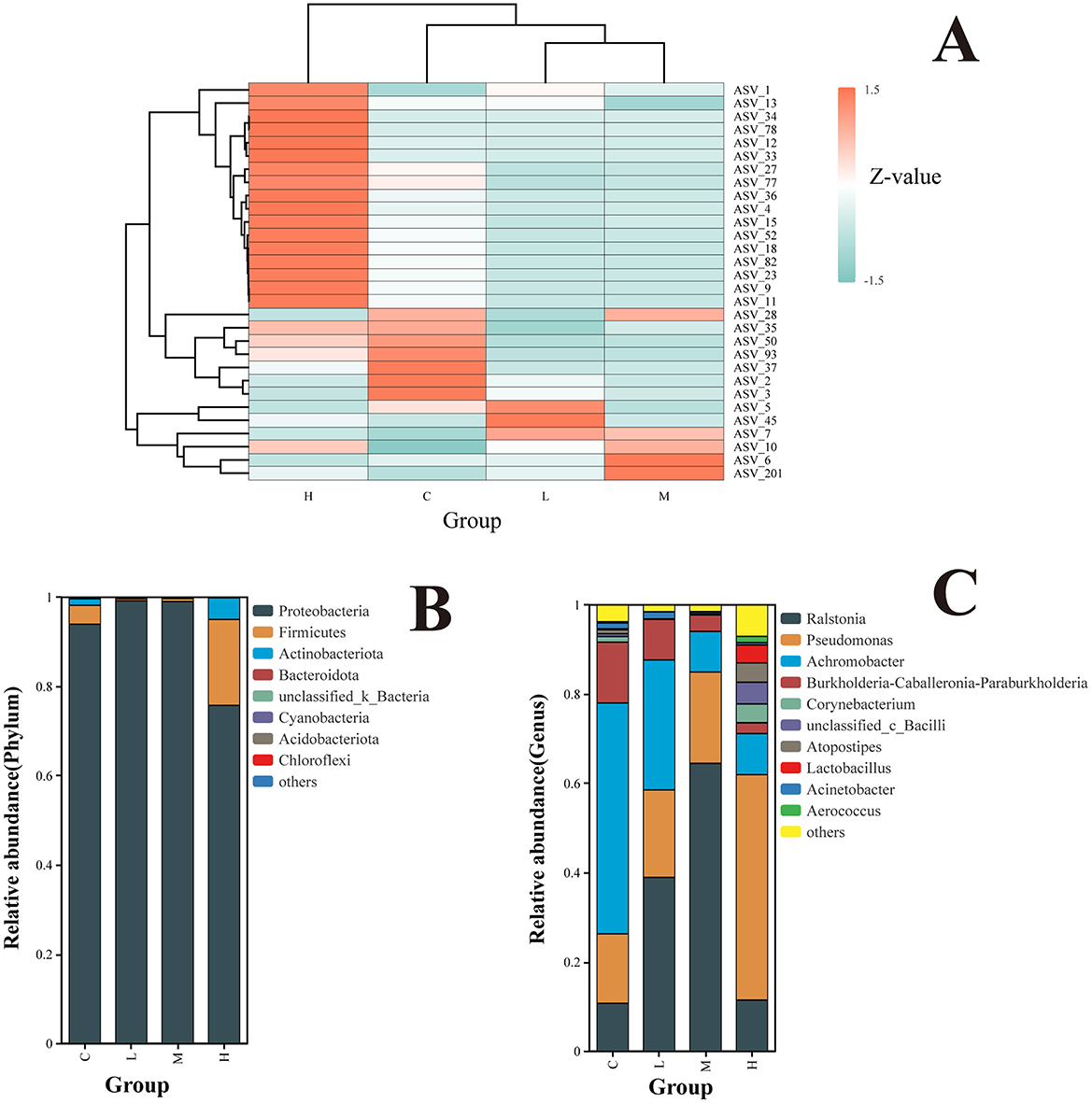
Figure 5. Clustering of species relative abundance at the ASV level (A). Z-scores represent species abundance deviations from the mean across samples, with red indicating higher abundance and blue indicating lower abundance. Hierarchical clustering trees show species and sample similarities; Relative abundance of the rat lower respiratory tract microbiota at the level of phyla (B); relative abundance of rat lower respiratory tract microbiota at the genus level (C). The top 10 phyla/genera are shown.
At the phylum level (Figure 5B), Proteobacteria, Firmicutes, and Actinobacteriota were identified as the dominant phyla across all groups. The abundance of Proteobacteria significantly increased in Groups L and M but decreased significantly in Group H. In contrast, Firmicutes and Actinobacteriota exhibited the opposite trend, showing significant decreases in Groups L and M and significant increases in Group H.
At the genus level, the top 10 genera in terms of relative abundance in the lower respiratory microbial community were identified, with the top five genera being Ralstonia, Pseudomonas, Achromobacter, Burkholderia-Caballeronia-Paraburkholderia, and Corynebacterium.
Across dose groups, the relative abundance of genera exhibited distinct patterns. Between Group C and Groups L and M, a gradual increase in the relative abundance of Ralstonia and Pseudomonas was observed with increasing exposure doses. Conversely, the relative abundance of Achromobacter and Burkholderia-Caballeronia-Paraburkholderia showed a decreasing trend. Upon further dose escalation, Group H demonstrated diverse changes in community structure. The relative abundance of Pseudomonas increased, while Ralstonia decreased. Additionally, less abundant genera, such as Lactobacillus, Corynebacterium, and Atopostipes, exhibited increased relative abundance, reflecting altered community diversity (Figure 5C).
LEfSe analyses based on relative abundance data were performed to identify bacterial taxa represented by differences between groups. The LDA bar plot results (Figure 6A) revealed significant bacterial taxa associated with each group (LDA threshold ≥ 3.5). In Group C, Bacteroidota, Clostridiales, Bacteroides, Weissella, and Nosocomiicoccus were significantly enriched, suggesting a dominance of anaerobic and gut-associated taxa. Group L showed increased abundance of Proteobacteria, particularly Sphingomonadales and Sphingomonadaceae. Group M was defined by Burkholderiales, Burkholderiaceae, and Ralstonia. In contrast, Group H exhibited a broader range of differential taxa, including Firmicutes, Bacilli, Lactobacillales, Actinobacteriota, and genera such as Atopostipes, Aerococcus, Psychrobacter, and Enteractinococcus.
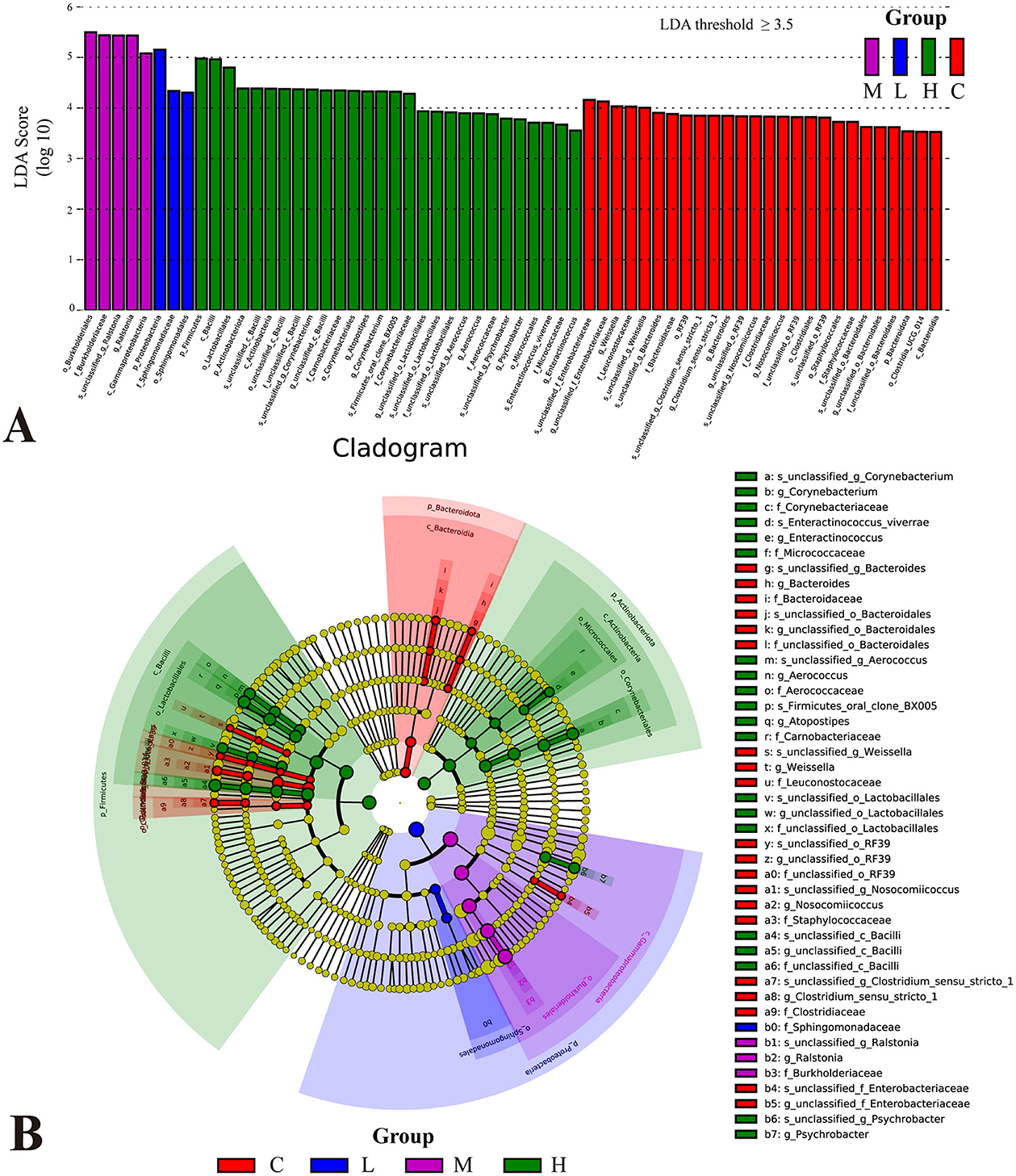
Figure 6. Differential analysis of bacterial flora in the lower respiratory tract of rats based on LEfSe analysis. Histogram of linear discriminant analysis (LDA) scores distribution (A). Taxa with LDA scores >3.5 are considered significantly enriched. Phylogenetic tree illustrating the distribution of significantly different microbial taxa across groups (B). The letters before the underscores represent, in order: phylum(p_), class(c_), order(o_), family(f_), genus(g_), and species(s_).
The cladogram (Figure 6B) illustrates the taxonomic distribution of significantly different microbial taxa across groups. The node colors corresponded to the colors in the LDA bar plot, with each color representing significant taxa for a specific group. The central part of the cladogram demonstrated differences among the groups at higher taxonomic levels, such as phylum, class, and order. For example, in Group C, Bacteroidota, Clostridiales, and Staphylococcales dominated. However, Firmicutes (Bacilli and Lactobacillales) and Actinobacteriota exhibited higher abundance in Group H, while Proteobacteria (Gammaproteobacteria, Burkholderiales, and Sphingomonadales) were significantly enriched in Group L and Group M, indicating notable variations in the proportions of these major taxa across exposure groups. The peripheral part of the cladogram highlighted differences at lower taxonomic levels, such as genus and species.
3.5 Bacterial diversity in alveolar lavage fluid of rats
Alpha diversity analysis revealed significant differences in species richness across groups. The Richness, ACE, and Chao1 indices (Figures 7A–C) were significantly higher in Group C compared to Groups L and M (P < 0.05 for all), indicating a reduction in microbial richness following metal exposure. Similarly, the PD_Whole_Tree index, reflecting phylogenetic diversity, was significantly lower in the exposure groups relative to Group C (P < 0.05), suggesting that inhaled heavy metals decreased phylogenetic breadth (Figure 7D).
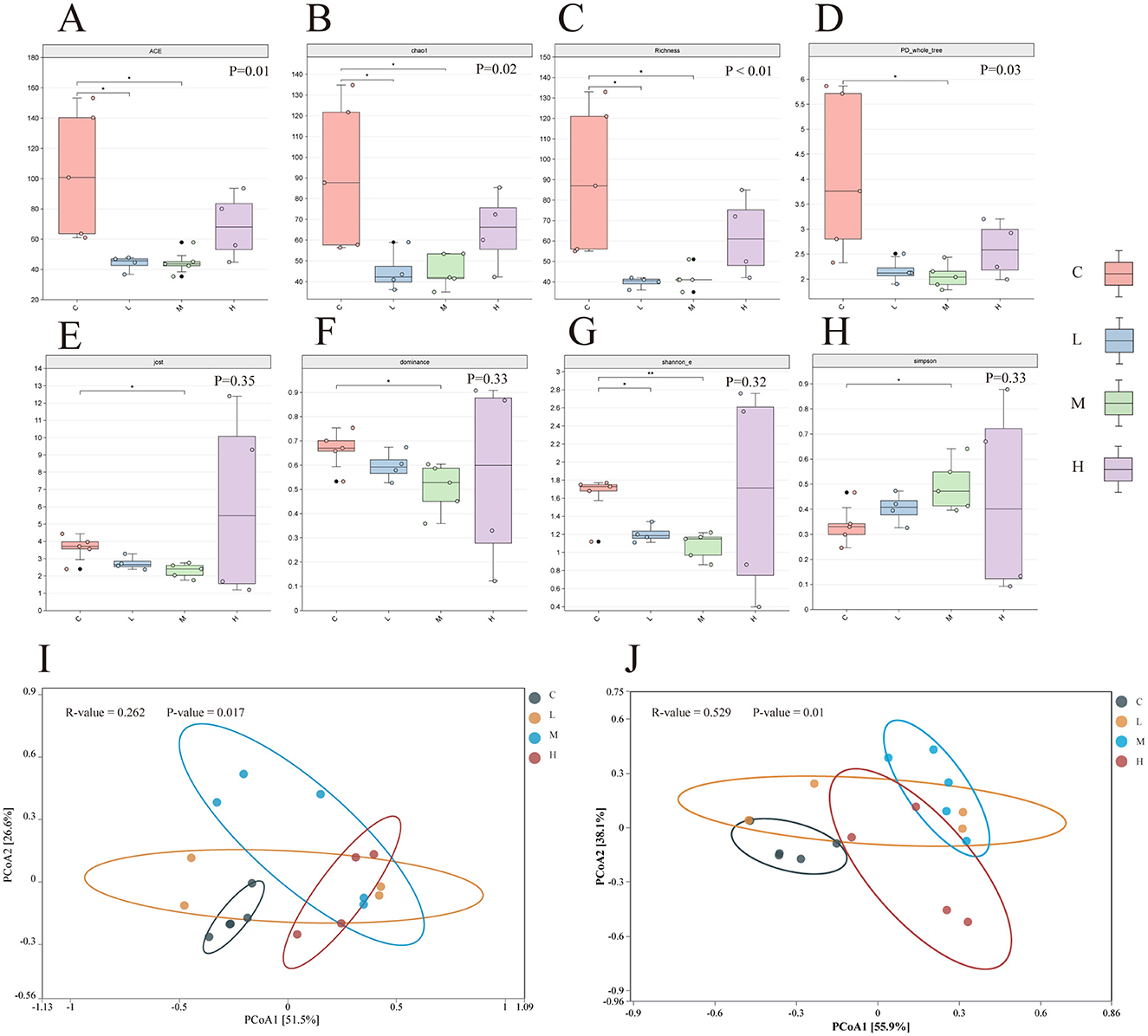
Figure 7. Alpha diversity index of lower respiratory tract microbiota in four groups of rats (A–H). Significance of differences between two groups was determined using the Wilcoxon rank-sum test, while differences among multiple groups were assessed using the Kruskal-Wallis test. *P ≤ 0.05; **P ≤ 0.01. PCoA analysis of lower respiratory tract microbiota in rats under the Bray-Curtis distance algorithm (I, J). ASV level (I) and Genus level (J). The ANOSIM (Analysis of Similarities) test was employed to evaluate the significant differences in bacterial β-diversity across multiple groups. C-Group C; L-Group L; M-Group M; H-Group H.
In contrast, community evenness and dominance showed no significant group differences. Although the Dominance index suggested a higher proportion of dominant species in Group H and the Jost index indicated slightly greater evenness in this group, neither reached statistical significance (P > 0.05; Figures 7E, F).
The Shannon_E and Simpson indices, which account for both richness and evenness, showed a trend of decreasing diversity from Group C to Group H. However, these changes were not statistically significant (Shannon_E: P = 0.32; Simpson: P = 0.33; Figures 7G, H).
Beta diversity was assessed using Principal Coordinates Analysis (PCoA) based on Bray–Curtis dissimilarity. At the ASV level (Figure 7I), the first two principal coordinates explained 77% of the total variance (PCoA1: 51.5%; PCoA2: 25.5%). ANOSIM results (R = 0.262, P = 0.017) indicated a moderate but significant separation among the four groups, with Group C microbiota clearly distinct from Group H, while Groups L and M showed partial overlap.
At the genus level (Figure 7J), PCoA1 and PCoA2 explained 55.5% and 38.9% of the variance, respectively, accounting for 94.4% of total variation. Group separation was more pronounced at this taxonomic resolution, as supported by a higher ANOSIM R-value (R = 0.529, P = 0.01), with clear divergence observed between the control and exposure groups, especially between Group C and Group H.
3.6 Correlation of lower respiratory flora with heavy metals and lung function
As shown in Figure 8, the network diagram represents microbial genera and environmental/lung function parameters as gray nodes. Green edges indicate significant correlations with moderate strength (0.01 <P ≤ 0.05), while orange edges denote stronger significant correlations (0.001 <P ≤ 0.01). Gray edges represent statistically non-significant correlations (P > 0.05). The type of correlation is further indicated by the line style: solid lines represent significant positive correlations, whereas dashed lines indicate significant negative correlations. The line thickness reflects the strength of the correlation (Mantel's r), with thicker lines indicating stronger correlations.
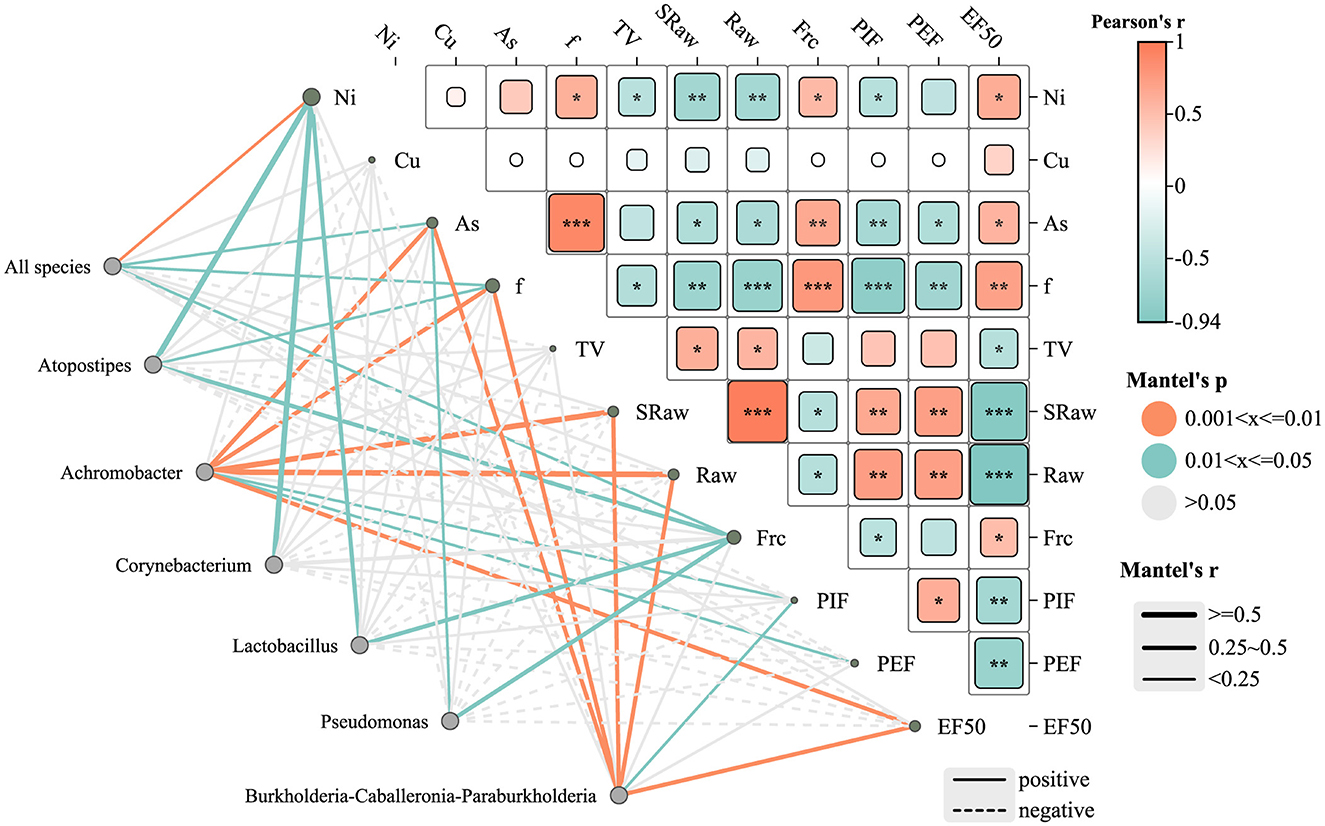
Figure 8. Heat map of the correlation network between lower respiratory tract microbiota, heavy metals, and lung function parameters in rats. *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001. Edge color denotes significance level: green = P ≤ 0.05; orange = P ≤ 0.01; gray = not significant (P > 0.05). Edge style indicates correlation direction: solid = positive, dashed = negative. Edge thickness reflects Mantel correlation strength.
Several genera, including Achromobacter, Burkholderia-Caballeronia-Paraburkholderia, Pseudomonas, Lactobacillus, and Atopostipes, were notably correlated with heavy metal concentrations (e.g., Ni and As) and lung function indicators. Among them, Burkholderia-Caballeronia-Paraburkholderia exhibited significant positive correlations with EF50 and PEF (P ≤ 0.01), suggesting a potential role in preserving lung function under metal exposure. Pseudomonas showed a moderate positive correlation with airway resistance (Raw) (P ≤ 0.05). In contrast, Atopostipes showed a negative correlation with Ni and As exposure, although these associations were not statistically significant (P > 0.05), implying a possible sensitivity of this genus to metal stress.
The heatmap further illustrates Pearson correlation coefficients (r) between lung function parameters and metal concentrations. Arsenic (As) levels were negatively associated with multiple lung function indicators, with a significant inverse correlation observed for PEF (**P < 0.01), suggesting As exposure may impair airflow capacity. Nickel (Ni) showed strong negative correlations with EF50 and Raw (***P < 0.001), indicating substantial impairment of lung function at higher Ni concentrations. Conversely, copper (Cu) exhibited moderate positive correlations with EF50 and PEF (*P < 0.05), suggesting a potential stimulatory effect on lung function, possibly through metabolic activation mechanisms.
The “all species” node, representing the overall microbial community, was significantly positively correlated with Ni (P ≤ 0.01) and As (P ≤ 0.05), suggesting a community-wide response to these metals. No significant correlation was found between Cu levels and the overall community structure. Regarding lung function, the “all species” node showed positive correlations with breathing frequency (f) and functional residual capacity (Frc) (P < 0.05), indicating that microbial community composition may influence specific pulmonary parameters.
3.7 Functional prediction of respiratory tract communities
Functional gene profiling revealed significant differences among exposure groups. Kruskal–Wallis tests indicated that pathways involved in xenobiotic biodegradation and metabolism varied significantly across groups (P < 0.05), whereas core metabolic pathways remained largely conserved (P > 0.05).
PICRUSt2 analysis based on KEGG annotations demonstrated that metabolism dominated KEGG Level 1 functions across all groups, accounting for approximately 80%−83% of the total functional capacity. However, a slight reduction in metabolic activity was observed in Group H, suggesting disruption of key microbial functions. Genetic information processing (8%−9%) and cellular processes (~5%) showed mild but consistent dose-dependent alterations.
At the KEGG Level 2 tier, although no pathways reached statistical significance (P > 0.05), minor decreases in amino acid metabolism and xenobiotic degradation were noted in the high-dose group. More distinct patterns emerged at KEGG Level 3, where five pathways—including ketone body synthesis, amino acid biosynthesis, and fatty acid degradation—showed significant differences (P < 0.05). The low-dose group (L) displayed elevated gene abundances in several pathways, possibly reflecting adaptive compensation, whereas the high-dose group (H) exhibited reduced functional gene expression, indicative of impaired microbial metabolic capacity.
KEGG enrichment analysis (Figure 9D) highlighted significant upregulation in pathways related to biosynthesis of cofactors, carbon metabolism, and amino acid biosynthesis, with high –log10 (Q-value) scores indicating robust enrichment. Additional enrichment was observed in glycolysis/gluconeogenesis, fructose and mannose metabolism, and amino sugar and nucleotide sugar metabolism, possibly suggesting that heavy metal exposure may trigger metabolic reprogramming to maintain energy homeostasis and structural integrity. Enrichment of the phosphotransferase system (PTS) and bacterial secretion system pathways points to possible microbial adaptation mechanisms involving nutrient uptake and intercellular communication.
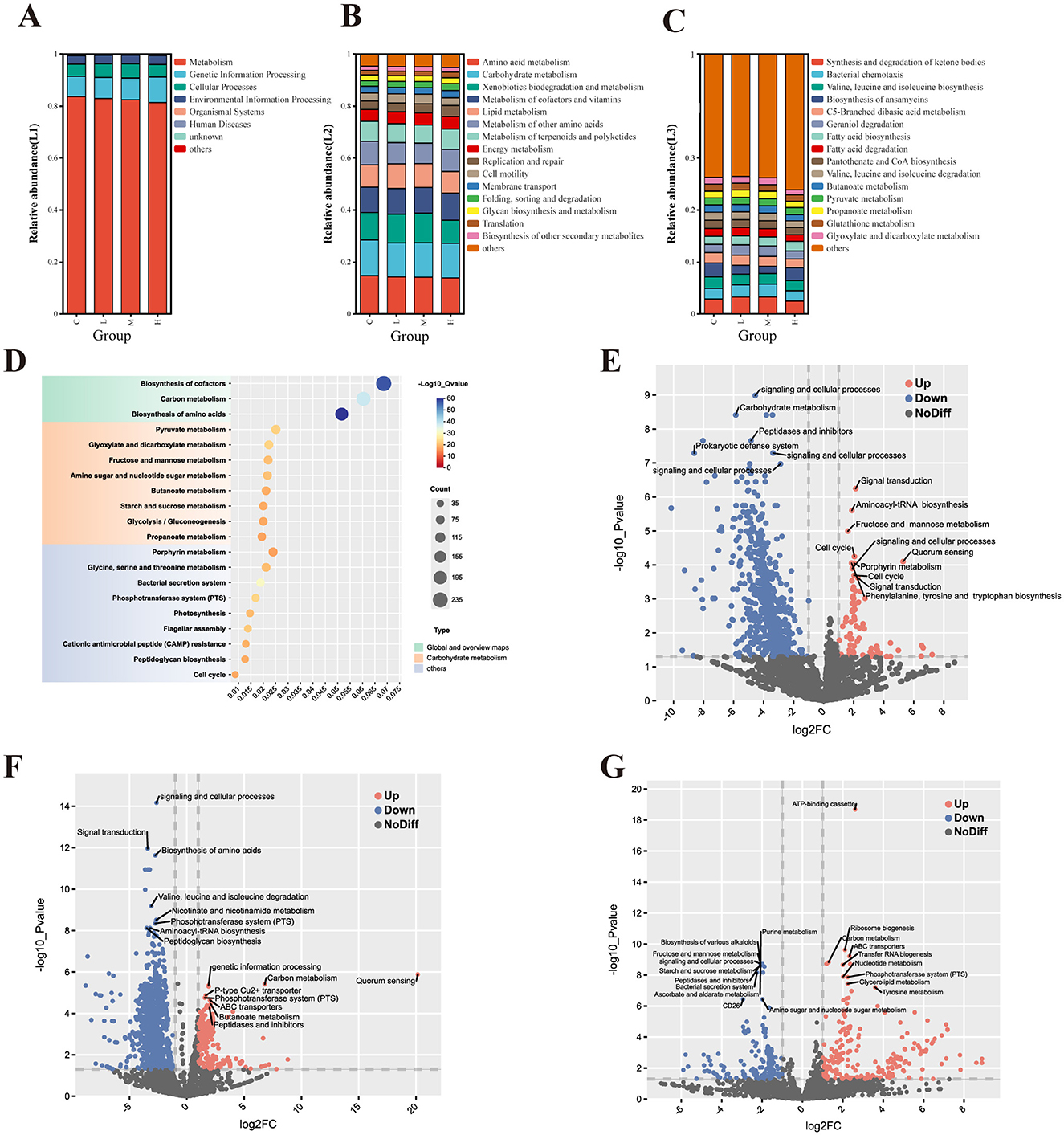
Figure 9. Functional prediction of lower respiratory tract microbiota based on PICRUSt2 analysis with KEGG annotation. Analysis of the pathway composition of the lower respiratory tract microbiota in rats (A–C). Level 1 pathway (A); Level 2 pathway (B); and Level 3 pathway (C). KEGG enrichment analysis of BALF microorganisms in the experimental group and control group (D). KEGG function prediction volcano map of each experimental group compared with the control group (E-G). Group L vs. Group C (E); Group M vs. Group C (F); and Group H vs. Group C (G). Significantly upregulated (red) and downregulated (blue); non-significant (gray).
Figures 9E–G display volcano plots of KEGG Orthology (KO) terms under different exposure levels. In both low-dose (Figure 9E) and medium-dose (Figure 9F) groups, downregulation was observed in pathways associated with carbohydrate metabolism, signal transduction, and prokaryotic defense systems. Concurrently, upregulation occurred in signal transduction, quorum sensing, and aminoacyl-tRNA biosynthesis pathways, probably suggesting early microbial responses to environmental stress. The medium-dose group showed more pronounced downregulation in amino acid degradation pathways such as valine, leucine, and isoleucine metabolism, as well as in PTS and peptidase-related functions, while upregulation was observed in amino acid biosynthesis and peptidoglycan biosynthesis.
High-dose exposure (Figure 9G) resulted in the most pronounced functional shifts. Notable upregulation was observed in purine metabolism, ATP-binding cassette (ABC) transporters, and ribosome biogenesis pathways, which may reflect enhanced microbial activity associated with protein synthesis and cellular stress responses. In contrast, pathways related to carbohydrate metabolism, amino sugar metabolism, and quorum sensing were significantly downregulated, suggesting potential impairments in core metabolic and communication functions within the microbial community.
3.8 Microbial ecological networks analysis of bacterial interactions in BALF
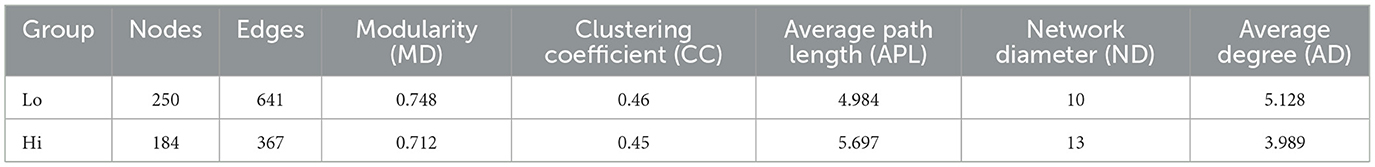
To further explore the impact of Ni and As heavy metal concentrations on microbial interactions and relationships, ecological network analyses were conducted by ranking samples based on heavy metal levels. The samples were divided into low-dose (Lo, Figure 10A) and high-dose (Hi, Figure 10B) groups at the 50% threshold. Nodes in the networks were annotated with their corresponding phyla.
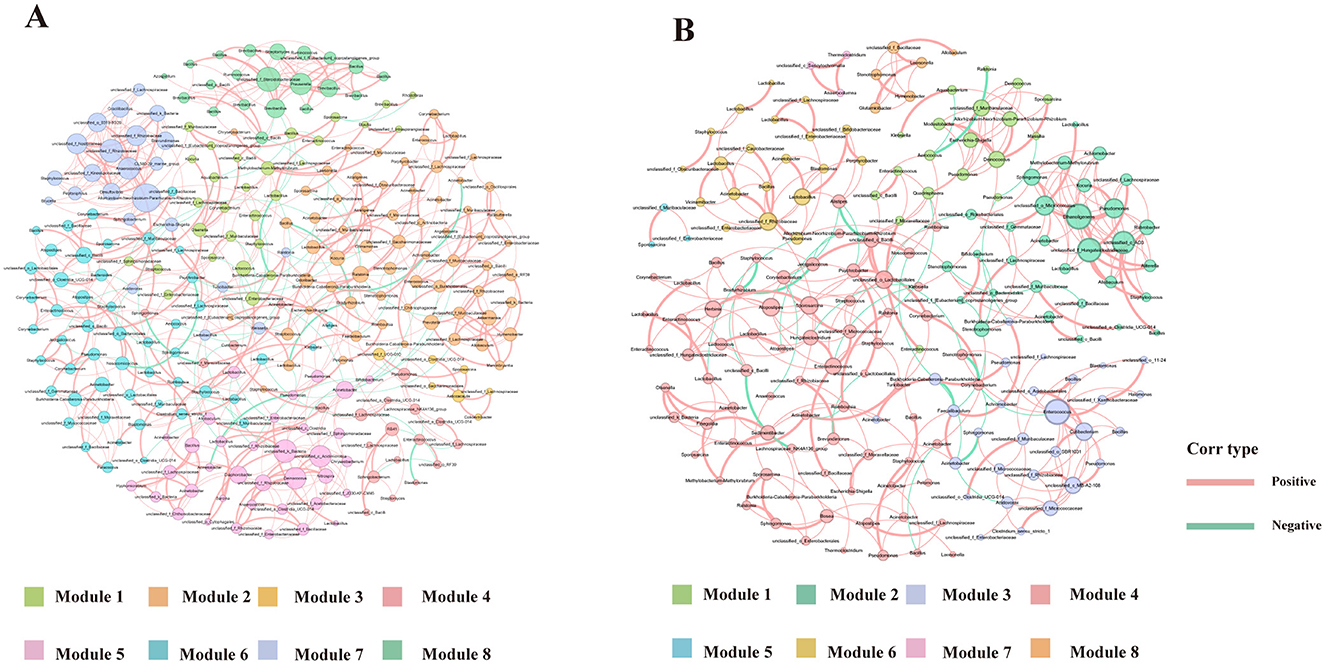
Figure 10. Ecological networks genera illustrating bacterial interactions in the Group Lo (A) and Group H (B) were constructed based on correlations among bacterial ASVs. Each connection indicates a strong (Spearman's r > 0.6) and significant (P < 0.01) correlation. The size of each node is proportional to the number of connections, and the edge thickness is proportional to the weight of each correlation.
Network complexity differed significantly between the two groups. Compared to the Lo group, the Hi group exhibited a less complex network, characterized by fewer edges (367 vs. 521), fewer nodes (184 vs. 251), a lower average degree (3.989 vs. 4.719), and reduced modularity (0.712 vs. 0.823) (Table 4). Keystone genera also differed between the groups. In the Hi group, Atopostipes and Sporosarcina were identified as keystone taxa, whereas in the Lo group, the keystones included Bacill, Gammaproteobacteria, Alphaproteobacteria, and Acidimicrobiia. While direct links between these genera and lung function or damage are limited, the differences in keystone taxa suggest notable shifts in microbial interactions between low-dose and high-dose exposure groups.
Further analysis focused on the largest module in each network, defined by the highest number of nodes. In the Lo group, Module 2 represented the largest module (25.6% of the network), while in the Hi group, Module 3 dominated (39.67%). The dominant genera in these modules varied as well, with the Lo group primarily comprising genera such as Ralstonia, Lactobacillus, and Prevotella, while the other group was predominantly composed of genera like Atopostipes, Enteractinococcus, and Sporosarcina.
The network diagrams reveal that most relationships between microbial taxa were positive, with only a small fraction displaying negative correlations. Notably, negative relationships involved genera such as Lactobacillus and Burkholderia-Caballeronia-Paraburkholderia in their interactions with other taxa. These changes in modularity and relationships suggest a reorganization of microbial network structures under heavy metal exposure, potentially reflecting the effects of environmental stress on community functionality.
4 Discussion
This study investigated, for the first time, the effects of mixed aerosol exposure to nickel (Ni), copper (Cu), and arsenic (As) on rat lungs using high-throughput sequencing technology, and analyzed the correlation with the respiratory microbiota. Although the effects of combined heavy metal exposure on lung tissues are an emerging field in air pollution research, the underlying mechanisms remain unclear. The findings demonstrated that sub-chronic combined exposure to mixed aerosols of Ni, Cu, and As resulted in significant lung damage and reduced lung function in rats. Additionally, combined exposure to these heavy metals altered the composition of the respiratory microbiota. Importantly, a significant association was found between changes in several respiratory bacterial communities and decreased lung function. Overall, this study reveals the potential role of respiratory microbiota in lung function reduction induced by combined heavy metal exposure and provides new insights into the mechanisms underlying heavy metal-induced lung function impairment.
Exposure to heavy metals may lead to reduced lung function (Wu et al., 2022). After exposure to a mixture of three heavy metals, rats in the M and H groups exhibited significant inflammatory cell infiltration, thickening of alveolar walls, and deformation of alveoli. These pulmonary injuries may impair airway patency and affect lung ventilation function. Compared to the control group, the airway resistance indicators (sRaw and Raw) in the heavy metal exposure groups were significantly elevated. A reduction in tidal volume can trigger a compensatory increase in respiratory frequency in rats to maintain adequate ventilation (Liu, 2022), consistent with our results. With escalating levels of heavy metal exposure, compensatory pulmonary overexpansion was observed, as indicated by a significant increase in functional residual capacity (FRC) in the H group. FRC serves as an indicator of lung overinflation (Laveneziana and Palange, 2012; Aalstad et al., 2018); The PEF values in the exposure groups were all lower than those in the control group, indicating that combined heavy metal exposure exacerbates airway obstruction. Peak inspiratory flow (PIF), which is influenced primarily by respiratory muscle strength and airway patency, was also reduced in exposed rats. This decline may be attributed to weakened respiratory muscle function (Liu, 2022). Changes in muscle strength and endurance should be interpreted dependent on the density of slow muscle fibers (Fuso et al., 2012). Notably, microvascular lesions primarily affect slow muscle fibers, thereby impacting endurance. Our study shows that PIF decreases with increasing heavy metal concentration, indicating that the decline in lung function in rats due to heavy metals may be related not only to increased airway resistance but also to weakened respiratory muscle strength and complications in lung microvasculature.
High-throughput sequencing confirmed sufficient data depth. At the phylum level, Groups L and M showed increased Proteobacteria and decreased Firmicutes and Actinobacteria, consistent with findings in PM2.5 and ventilation-exposed animal models, and patients with respiratory diseases like asthma and COPD (Hilty et al., 2010; Chen, 2022). In contrast, Group H had lower Proteobacteria and higher Firmicutes, suggesting that high heavy metal levels may suppress sensitive taxa while favoring resistant ones. This pattern aligns with studies reporting Proteobacteria decline and Firmicutes enrichment under heavy metal stress (Duan et al., 2024; Jin et al., 2024). Increased Firmicutes in Group H may reflect ecological adaptation or selective pressure. Minor phyla remained stable, indicating limited responsiveness.
At the genus level, Ralstonia, Pseudomonas, and Achromobacter dominated. In Group H, Pseudomonas increased while Ralstonia declined. Rare genera such as Lactobacillus, Corynebacterium, and Aerococcus also increased, possibly indicating adaptive or protective roles under metal stress. Then, the high relative abundance of Group H for Pseudomonas may reflect selective environmental pressure. If certain metals or pollutants favor Pseudomonas growth and inhibit Ralstonia, Pseudomonas' relative abundance would rise (Choudhary and Sar, 2016). This could be due to Pseudomonas' stronger adaptability and more efficient metal-resistance mechanisms (Adhikary et al., 2019).
Alpha diversity indices (ACE, Chao1, PD) showed significant reductions in richness and phylogenetic diversity in exposure groups, especially under high-dose exposure, in line with previous research on cadmium and air pollution (Tao et al., 2020; Laiman et al., 2022). Shannon_E and Simpson indices indicated lower diversity and evenness, with a dose-dependent decline. However, changes in dominance were not statistically significant, though trends pointed to enrichment of resistant genera like Burkholderia and Pseudomonas (Awais et al., 2024). Beta diversity (PCoA) showed clear group separation. The high-dose group had the greatest dispersion, indicating disrupted microbial stability and increased heterogeneity, likely due to selective pressure favoring metal-resistant taxa and suppressing sensitive ones. Genus-level resolution revealed stronger group differences than ASV-level, highlighting taxonomic restructuring. These results echo NMDS-based studies on polluted soils showing increased heterogeneity under heavy metal stress (Wen et al., 2024).
The LDA analysis (Figure 6A) revealed dose-dependent community shifts. In Group C, Bacteroidota and Bacilli (e.g., Weissella) predominated, consistent with a stable, anaerobe-dominated community, suggesting a relatively stable microbiota. Low-dose exposure caused minimal structural changes, while medium-dose exposure enriched resistant taxa like Proteobacteria. In the high-dose group, enrichment broadened to include multiple phyla (Firmicutes, Actinobacteriota) and opportunistic genera such as Atopostipes, Aerococcus, and Psychrobacter, suggesting a possible shift toward dysbiosis and colonization by opportunistic or environmental bacteria. The cladogram (Figure 6B) further confirmed phylogenetic divergence across groups, highlighting the distinct microbial signatures induced by increasing metal burden. These patterns align with previous findings that heavy metal exposure can reduce microbial diversity, disrupt metabolic function, and promote inflammatory responses (Giambò et al., 2021). Consistent with our observations, study reported that rising Ni, Cu, and As sharply lower α-diversity and favor metal-tolerant generalists (e.g., Gammaproteobacteria, Bacillus) as keystone generalists, simplifying microbial networks under heavy-metal stress (Qi et al., 2022). Together, these findings suggest that heavy metal exposure-associated alterations in the lower airway microbiome might contribute to compromised mucosal immunity and increased susceptibility to lung injury.
The Mantel Test (Figure 8) provided critical insights into the relationships between microbial genera, environmental factors, and lung function parameters, highlighting the complex interplay between microbiota and heavy metal exposure. Genera such as Achromobacter, Burkholderia-Caballeronia-Paraburkholderia, and Pseudomonas were significantly positively correlated with heavy metal concentrations like Ni and As. Notably, Burkholderia was linked to better respiratory parameters, possibly due to its robust oxidative stress defenses and detoxification systems (Eissa, 2024), which have been shown to enhance bacterial survival and support host adaptability. Pseudomonas was moderately correlated with Raw, reflecting its adaptive capacity in polluted environments. Conversely, Atopostipes showed a non-significant negative correlation with Ni and As, hinting at suppression under metal stress, aligning with reports of metal-induced microbial imbalance and inflammation (Popov Aleksandrov et al., 2021). However, while Pseudomonas may help maintain ecological stability, its pathogenic potential, particularly in chronic lung diseases such as COPD, cannot be overlooked (Jingsheng et al., 2023). Correlation analysis with the three heavy metals Ni, Cu, and As revealed that Lactobacillus, Corynebacterium, and Atopostipes were positively correlated with Ni, while Achromobacter and Pseudomonas were positively correlated with As (Figure 8). Lactobacillus, known for its immunomodulatory effects (Huang et al., 2014), may support lung health through the gut-lung axis (Heeney et al., 2018). In contrast, Corynebacterium and Actinobacteria—frequently found in respiratory infections—may indicate potential pathogenicity under environmental stress (Sze et al., 2015; Li H. et al., 2019).
Functional prediction using PICRUSt2 (Figures 9A–C) revealed that core metabolic pathways, including carbohydrate metabolism, amino acid metabolism, and lipid metabolism, were largely conserved across all groups (P > 0.05). Heavy metal exposure can impair the metabolic potential of microbial communities, thereby disrupting host nutrient absorption, energy metabolism, and respiratory health (He et al., 2020; Liu et al., 2024). In our study, compositional differences in the BALF microbiota across exposure groups suggested that microbial community structure is sensitive to heavy metal stress, necessitating further investigation into functional gene diversity.
The KEGG enrichment analysis (Figure 9D) demonstrates a significant perturbation in metabolic pathways, particularly those associated with carbohydrate metabolism, amino acid biosynthesis, and energy production processes. The most significantly enriched pathways included biosynthesis of amino acids, carbon metabolism, and pyruvate metabolism, aligning with previous studies that indicate microbial communities respond to heavy metal stress by enhancing essential metabolic pathways to maintain cellular stability and energy homeostasis (Prabhakaran et al., 2016; Ayangbenro and Babalola, 2017). The enrichment of pathways related to bacterial secretion systems and cationic antimicrobial peptide (cAMP) resistance suggests that microbial communities are actively deploying defense mechanisms against environmental stress, particularly under toxic metal exposure conditions (Patil et al., 2024).
In the differential expression analysis (Figures 9E–G), significant differences were observed across experimental groups. Low-dose exposure predominantly affected genes involved in signal transduction, prokaryotic defense systems, and carbohydrate metabolism. At medium doses, genes related to amino acid biosynthesis, branched-chain amino acid degradation (valine, leucine, isoleucine), and membrane transport via the phosphotransferase system (PTS) were enriched. In contrast, high-dose exposure triggered a pronounced microbial stress response, with upregulation of purine metabolism, ABC transporters, and ribosome biogenesis, indicating a shift toward protein synthesis and cellular repair mechanisms, typical of environmental stress adaptation. These findings align with previous reports that metals such as mercury (Hg), arsenic (As), and cadmium (Cd) can activate detoxification pathways, where metal-glutathione complexes are expelled through ABC transporters to alleviate toxicity (Pearson and Cowan, 2021). Study demonstrated that microbial systems often trade off metabolic efficiency for stress survival, reallocating resources toward protective pathways under adverse conditions (Escoll and Buchrieser, 2019). Such adaptive responses may have broader implications for microbial ecology and environmental resilience.
Ecological network analysis revealed that microbial interactions in the high-dose exposure group (Hi) were significantly simplified, as evidenced by a reduction in network nodes and edges. This suggests that high concentrations of nickel and arsenic suppress sensitive microbial taxa, thereby reducing microbial diversity and weakening overall community connectivity. Such network simplification is indicative of ecological stress and aligns with prior findings showing that heavy metal exposure exerts strong selective pressure, favoring metal-tolerant species while inhibiting sensitive groups. Similar patterns have been observed in other environmental studies, where pollutants such as polycyclic aromatic hydrocarbons (PAHs) and heavy metals led to reduced microbial network complexity and stability, ultimately making ecosystems more vulnerable (Wang et al., 2024). These changes underscore a shift in community structure, with cooperative interactions being replaced by competitive or stress-adaptive dynamics. For instance, the decline of sensitive genera like Acidobacteria and Verrucomicrobia has been previously reported under heavy metal stress, contributing to lower modularity and increased inter-species competition (Ma et al., 2020). In our study, this trend is further reflected in the shift of keystone genera from Lactobacillus and Ralstonia in the low-dose group to stress-adaptive genera such as Atopostipes and Sporosarcina in the high-dose group. These findings highlight how heavy metal exposure not only simplifies microbial networks but also reorganizes keystone taxa, potentially altering key ecological functions such as nutrient cycling and immune regulation.
This study also has several limitations that warrant consideration. First, the lack of single-metal exposure groups prevents the differentiation of independent effects and interactions among the heavy metals in mixed exposure. Future studies should include single-metal exposure groups to clarify these contributions and provide more comprehensive insights. Second, Sample sizes (n = 5 for each group) may limit the generalizability of our conclusions, we plan to increase sample sizes in future experiments to enhance the robustness and reproducibility of our results. Third, while 16S rDNA sequencing offered valuable functional predictions of the lower respiratory microbiota, the absence of direct validation through metabolomics, transcriptomics, or functional assays limits the ability to confirm the predicted microbial functions and their relevance to host physiology. Incorporating high-throughput techniques and metabolite detection in future studies could enhance the understanding of microbial functional changes under heavy metal stress. Additionally, the study relies on endpoint data without capturing the temporal progression of microbial dysbiosis and its link to lung injury, highlighting the need for longitudinal analyses.
5 Implications and future perspectives of this study
This study reveals that exposure to heavy metals such as Ni, Cu, and As induces specific disruptions in the respiratory microbiota of rat BALF, including the enrichment of Pseudomonas and Burkholderia and depletion of Ralstonia, which may act as early biomarkers of dysbiosis linked to lung dysfunction. Unlike previous work focused on gut microbiota or general respiratory inflammation, our study highlights for the first time the distinct compositional and predicted functional alterations of the lower-airway microbiota under co-exposure to multiple metals. Although based on 16S rRNA data, our findings provide new insights into potential microbial symbioses and their association with host injury, offering a basis for hypothesis-driven mechanistic research. Future studies applying multi-omics and longitudinal models are essential to establish causality and delineate microbial contributions to lung injury. These results also support the integration of microbiota-sensitive endpoints into air pollution toxicology and regulatory frameworks, emphasizing the need for strengthened emission controls, metal-specific PM2.5 monitoring, and early-warning strategies tailored to vulnerable populations.
6 Conclusion
This study demonstrates that mixed exposure to nickel (Ni), copper (Cu), and arsenic (As) induces significant, dose-dependent lung injury and alters the composition and diversity of the lower respiratory tract microbiota in rats. The exposure led to microbial dysbiosis, characterized by enrichment of metal-tolerant genera such as Pseudomonas and Burkholderia, and depletion of sensitive taxa including Ralstonia and Actinobacteria. These microbial changes were closely associated with impaired lung function, suggesting a potential mechanistic link. Ecological network analysis further revealed simplified microbial interactions and a shift in keystone taxa under high-dose exposure. Although the exact mechanisms remain to be clarified, these findings highlight the impact of heavy metal exposure on respiratory microecology and support the need for stricter environmental control, particularly of Ni and As emissions, as well as further studies to explore microbiota-based strategies for mitigating lung injury.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA1253313.
Ethics statement
The animal study was approved by the Ethical Committees of the School of Public Health, Lanzhou University. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
PD: Writing – review & editing, Writing – original draft, Data curation. XW: Funding acquisition, Writing – review & editing, Writing – original draft. NL: Writing – review & editing, Data curation, Writing – original draft. SY: Data curation, Writing – review & editing. YZ: Writing – review & editing, Data curation. JY: Data curation, Writing – review & editing. TT: Writing – review & editing. RC: Writing – review & editing. BL: Writing – review & editing. LM: Writing – review & editing, Conceptualization, Funding acquisition. RZ: Writing – review & editing, Conceptualization.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was funded by the National Training Program of Innovation and Entrepreneurship for Undergraduates (202410730173), the Fundamental Research Funds for the Central Universities of China (lzujbky-2024-1) and the Scientific Research Project of Health Industry in Gansu Province (GSWSKY2023-15).
Acknowledgments
The authors thank Professor Aimin Yang (Department of Medicine and Therapeutics, The Chinese University of Hong Kong) for English modification.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Gen AI was used in the creation of this manuscript.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1615130/full#supplementary-material
Footnotes
References
Aalstad, L. T., Hardie, J. A., Espehaug, B., Thorsen, E., Bakke, P. S., Eagan, T. M. L., et al. (2018). Lung hyperinflation and functional exercise capacity in patients with COPD - a three-year longitudinal study. BMC Pulm. Med. 18:187. doi: 10.1186/s12890-018-0747-9
Adhikary, A., Kumar, R., Pandir, R., Bhardwaj, P., Wusirika, R., and Kumar, S. (2019). Pseudomonas citronellolis; a multi-metal resistant and potential plant growth promoter against arsenic (V) stress in chickpea. Plant Physiol. Biochem. 142, 179–192. doi: 10.1016/j.plaphy.2019.07.006
Amir, A., McDonald, D., Navas-Molina, J. A., Kopylova, E., Morton, J. T., Zech Xu, Z., et al. (2017). Deblur rapidly resolves single-nucleotide community sequence patterns. mSystems 2:00191-16. doi: 10.1128/mSystems.00191-16
Awais, M., Xiang, Y., Shah, N., Bilal, H., Yang, D., Hu, H., et al. (2024). Unraveling the role of contaminants reshaping the microflora in zea mays seeds from heavy metal-contaminated and pristine environment. Microb. Ecol. 87:133. doi: 10.1007/s00248-024-02445-5
Ayangbenro, A. S., and Babalola, O. O. (2017). A new strategy for heavy metal polluted environments: a review of microbial biosorbents. Int. J. Environ. Res. Public Health 14:94. doi: 10.3390/ijerph14010094
Baker, J. M., Hinkle, K. J., McDonald, R. A., Brown, C. A., Falkowski, N. R., Huffnagle, G. B., et al. (2021). Whole lung tissue is the preferred sampling method for amplicon-based characterization of murine lung microbiota. Microbiome 9:99. doi: 10.1186/s40168-021-01055-4
Balali-Mood, M., Naseri, K., Tahergorabi, Z., Khazdair, M. R., and Sadeghi, M. (2021). Toxic mechanisms of five heavy metals: mercury, lead, chromium, cadmium, and arsenic. Front. Pharmacol. 12:643972. doi: 10.3389/fphar.2021.643972
Banerjee, S., Walder, F., Büchi, L., Meyer, M., Held, A. Y., Gattinger, A., et al. (2019). Agricultural intensification reduces microbial network complexity and the abundance of keystone taxa in roots. ISME J. 13, 1722–1736. doi: 10.1038/s41396-019-0383-2
Bastian, M., Heymann, S., and Jacomy, M. (2009). “Gephi: an open source software for exploring and manipulating networks,” in Proceedings of the International AAAI Conference on Web and Social Media, 361–362. doi: 10.1609/icwsm.v3i1.13937
Chambers, D. C., Gellatly, S. L., Hugenholtz, P., and Hansbro, P. M. (2014). JTD special edition ‘Hot Topics in COPD'-The microbiome in COPD. J. Thorac. Dis. 6, 1525–1531. doi: 10.3978/j.issn.2072-1439.2014.11.08
Chen, X. (2022). Correlation between changes of lower respiratory tract flora and lung injury in rats after mechanical ventilation. Master thesis, Southern Medical University.
Choudhary, S., and Sar, P. (2016). Real-time PCR based analysis of metal resistance genes in metal resistant Pseudomonas aeruginosa strain J007. J. Basic Microbiol. 56, 688–697. doi: 10.1002/jobm.201500364
Duan, X., Li, J., Li, Y., Xu, Y., Chao, S., and Shi, Y. (2024). Accumulation of typical persistent organic pollutants and heavy metals in bioretention facilities: Distribution, risk assessment, and microbial community impact. Environ. Res. 252:119107. doi: 10.1016/j.envres.2024.119107
Duprez, D. A., and Jacobs, D. R Jr. (2018). Lung function decline and increased cardiovascular risk: quo vadis. J. Am. Coll. Cardiol. 72, 1123–1125. doi: 10.1016/j.jacc.2018.07.015
Eissa, M. (2024). Genus burkholderia: a double-edged sword with widespread implications for human health, agriculture, and the environment. J. Biol. Res. Rev. 1:79. doi: 10.5455/JBRR.20240516055714
Escoll, P., and Buchrieser, C. (2019). Metabolic reprogramming: an innate cellular defence mechanism against intracellular bacteria? Curr. Opin. Immunol. 60, 117–123. doi: 10.1016/j.coi.2019.05.009
Feng, K., Peng, X., Zhang, Z., Gu, S., He, Q., Shen, W., et al. (2022). iNAP: an integrated network analysis pipeline for microbiome studies. imeta 1:e13. doi: 10.1002/imt2.13
Fuso, L., Pitocco, D., Longobardi, A., Zaccardi, F., Contu, C., Pozzuto, C., et al. (2012). Reduced respiratory muscle strength and endurance in type 2 diabetes mellitus. Diabetes Metab. Res. Rev. 28, 370–375. doi: 10.1002/dmrr.2284
Gautam, P. K., Gautam, R. K., Banerjee, S., Chattopadhyaya, M., and Pandey, J. J. N. S.P. (2016). “Heavy metals in the environment: fate, transport, toxicity and remediation technologies,” in Heavy Metals: Sources Toxicity And Remediation Techniques, 101–130.
Giambò, F., Italia, S., Teodoro, M., Briguglio, G., Furnari, N., Catanoso, R., et al. (2021). Influence of toxic metal exposure on the gut microbiota (Review). World Acad. Sci. J. 3:19. doi: 10.3892/wasj.2021.90
Hashem, M. A., Hasan, M. A., Nayan, A. H., Payel, S., Hasan, M., and Sahen, M. S. (2021). The environmental impacts of heavy metals in soil, certain plants and wastewater near industrial area of Brahmanbaria, Bangladesh. Environ. Monit. Assess. 193:688. doi: 10.1007/s10661-021-09497-x
He, X., Qi, Z., Hou, H., Qian, L., Gao, J., and Zhang, X. X. (2020). Structural and functional alterations of gut microbiome in mice induced by chronic cadmium exposure. Chemosphere 246:125747. doi: 10.1016/j.chemosphere.2019.125747
Heeney, D. D., Gareau, M. G., and Marco, M. L. (2018). Intestinal Lactobacillus in health and disease, a driver or just along for the ride? Curr. Opin. Biotechnol. 49, 140–147. doi: 10.1016/j.copbio.2017.08.004
Hilty, M., Burke, C., Pedro, H., Cardenas, P., Bush, A., Bossley, C., et al. (2010). Disordered microbial communities in asthmatic airways. PLoS ONE 5:e8578. doi: 10.1371/journal.pone.0008578
Huang, Y. J., Nariya, S., Harris, J. M., Lynch, S. V., Choy, D. F., Arron, J. R., et al. (2015). The airway microbiome in patients with severe asthma: associations with disease features and severity. J. Allergy Clin. Immunol. 136, 874–884. doi: 10.1016/j.jaci.2015.05.044
Huang, Y. J., Sethi, S., Murphy, T., Nariya, S., Boushey, H. A., and Lynch, S. V. (2014). Airway microbiome dynamics in exacerbations of chronic obstructive pulmonary disease. J. Clin. Microbiol. 52, 2813–2823. doi: 10.1128/JCM.00035-14
Jacob, J. M., Karthik, C., Saratale, R. G., Kumar, S. S., Prabakar, D., Kadirvelu, K., et al. (2018). Biological approaches to tackle heavy metal pollution: a survey of literature. J. Environ. Manage. 217, 56–70. doi: 10.1016/j.jenvman.2018.03.077
Jin, J., Wang, X., Sha, Y., Wang, F., Huang, X., Zong, H., et al. (2024). Changes in soil properties and microbial activity unveil the distinct impact of polyethylene and biodegradable microplastics on chromium uptake by peanuts. Environ. Sci. Pollut. Res. Int. 31, 53369–53380. doi: 10.1007/s11356-024-34743-3
Jingsheng, H., shuxin, Z., Weichao, K., Zhijie, D., Yunmei, Y., chengzhi, L., et al. (2023). Study on antibacterial peptide BNBDS regulating the innate immune response of lung against actinobacillus pleuropneumoniae [in Chinese]. China Animal Husb. Veter. Med. 50, 4261–4269. doi: 10.16431/j.cnki.1671-7236.2023.10.040
Kanehisa, M., Furumichi, M., Tanabe, M., Sato, Y., and Morishima, K. (2017). KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 45, D353–d361. doi: 10.1093/nar/gkw1092
Kurtz, Z. D., Müller, C. L., Miraldi, E. R., Littman, D. R., Blaser, M. J., and Bonneau, R. A. (2015). Sparse and compositionally robust inference of microbial ecological networks. PLoS Comput. Biol. 11:e1004226. doi: 10.1371/journal.pcbi.1004226
Laiman, V., Lo, Y.-C., Chen, H.-C., Yuan, T.-H., Hsiao, T.-C., Chen, J.-K., et al. (2022). Effects of antibiotics and metals on lung and intestinal microbiome dysbiosis after sub-chronic lower-level exposure of air pollution in ageing rats. Ecotoxicol. Environ. Saf. 246:114164. doi: 10.1016/j.ecoenv.2022.114164
Lavanya, M. B., Viswanath, D. S., and Sivapullaiah, P. V. (2024). Phytoremediation: an eco-friendly approach for remediation of heavy metal-contaminated soils-A comprehensive review. Environ. Nanotechnol. Monitor. Manag. 22:100975. doi: 10.1016/j.enmm.2024.100975
Laveneziana, P., and Palange, P. (2012). Physical activity, nutritional status and systemic inflammation in COPD. Eur. Respir. J. 40, 522–529. doi: 10.1183/09031936.00041212
Li, H., Hua, J., Li, L., and Wu, Q. (2019). Case analysis and literature review of corynebacterium striatum pulmonary infection [in Chinese]. Chinese J. Urban Rural Enter. Hygiene 34, 33–37. doi: 10.16286/j.1003-5052.2019.10.013
Li, N., Qiu, R., Yang, Z., Li, J., Chung, K. F., Zhong, N., et al. (2017). Sputum microbiota in severe asthma patients: Relationship to eosinophilic inflammation. Respir. Med. 131, 192–198. doi: 10.1016/j.rmed.2017.08.016
Li, X., Brejnrod, A. D., Ernst, M., Rykær, M., Herschend, J., Olsen, N. M. C., et al. (2019). Heavy metal exposure causes changes in the metabolic health-associated gut microbiome and metabolites. Environ. Int. 126, 454–467. doi: 10.1016/j.envint.2019.02.048
Lin, J., Wang, W., Chen, P., Zhou, X., Wan, H., Yin, K., et al. (2018). Prevalence and risk factors of asthma in mainland China: the CARE study. Respir. Med. 137, 48–54. doi: 10.1016/j.rmed.2018.02.010
Liu, T., Zheng, X., Zhang, Q., Wang, Y., and Tang, G. (2024). The effects of Cd2+ and Cu2+ on characteristics of soluble microbial products from activated sludge via altering expression of cellular metabolism pathways. J. Water Proc. Eng. 66:106050. doi: 10.1016/j.jwpe.2024.106050
Liu, X. (2022). The effects oflong-term exposure to cadmium in drinking water on pulmonary impairment in diabetic rats. Master thesis, Soochow University.
Liu, Y., Yu, L., Ji, H., Zhu, M., Liu, Y., Fu, Y., et al. (2022). Association between urinary metal levels and slow vital capacity in Chinese preschoolers. Hum. Ecol. Risk Assess. 28, 621–634. doi: 10.1080/10807039.2022.2084029
Long, Z., Huang, Y., Zhang, W., Shi, Z., Yu, D., Chen, Y., et al. (2021). Effect of different industrial activities on soil heavy metal pollution, ecological risk, and health risk. Environ. Monit. Assess. 193:20. doi: 10.1007/s10661-020-08807-z
Ma, J., Lu, Y., Chen, F., Li, X., Xiao, D., and Wang, H. (2020). Molecular ecological network complexity drives stand resilience of soil bacteria to mining disturbances among typical damaged ecosystems in China. Microorganisms 8:433. doi: 10.3390/microorganisms8030433
McCarthy, D. J., Chen, Y., and Smyth, G. K. (2012). Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 40, 4288–4297. doi: 10.1093/nar/gks042
Morris, A., Beck, J. M., Schloss, P. D., Campbell, T. B., Crothers, K., Curtis, J. L., et al. (2013). Comparison of the respiratory microbiome in healthy nonsmokers and smokers. Am. J. Respir. Crit. Care Med. 187, 1067–1075. doi: 10.1164/rccm.201210-1913OC
Patil, A., Chakraborty, S., Yadav, Y., Sharma, B., Singh, S., and Arya, M. (2024). Bioremediation strategies and mechanisms of bacteria for resistance against heavy metals: a review. Bioremediat. J. 2024, 1–33. doi: 10.1080/10889868.2024.2375204
Pearson, S. A., and Cowan, J. A. (2021). Glutathione-coordinated metal complexes as substrates for cellular transporters. Metallomics 13:mfab015. doi: 10.1093/mtomcs/mfab015
Popov Aleksandrov, A., Mirkov, I., Tucovic, D., Kulas, J., Zeljkovic, M., Popovic, D., et al. (2021). Immunomodulation by heavy metals as a contributing factor to inflammatory diseases and autoimmune reactions: Cadmium as an example. Immunol. Lett. 240, 106–122. doi: 10.1016/j.imlet.2021.10.003
Prabhakaran, P., Ashraf, M. A., and Aqma, W. S. (2016). Microbial stress response to heavy metals in the environment. RSC Adv. 6, 109862–109877. doi: 10.1039/C6RA10966G
Qi, Q., Hu, C., Lin, J., Wang, X., Tang, C., Dai, Z., et al. (2022). Contamination with multiple heavy metals decreases microbial diversity and favors generalists as the keystones in microbial occurrence networks. Environ. Pollut. 306:119406. doi: 10.1016/j.envpol.2022.119406
Saladié, M., Caparrós-Martín, J. A., Agudelo-Romero, P., Wark, P. A. B., Stick, S. M., and O'Gara, F. (2020). Microbiomic analysis on low abundant respiratory biomass samples; improved recovery of microbial DNA from bronchoalveolar lavage fluid. Front. Microbiol. 11:572504. doi: 10.3389/fmicb.2020.572504
Sze, M. A., Utokaparch, S., Elliott, W. M., Hogg, J. C., and Hegele, R. G. (2015). Loss of GD1-positive Lactobacillus correlates with inflammation in human lungs with COPD. BMJ Open 5:e006677. doi: 10.1136/bmjopen-2014-006677
Tao, C., Pei, Y., Zhang, L., and Zhang, Y. (2020). Microbial communities respond to microenvironments in lungs of mice under simulated exposure to cadmium aerosols. Sci. Total Environ. 710:136300. doi: 10.1016/j.scitotenv.2019.136300
Taylor, S. L., Rogers, G. B., Chen, A. C. H., Burr, L. D., McGuckin, M. A., and Serisier, D. J. (2015). Matrix metalloproteinases vary with airway microbiota composition and lung function in non-cystic fibrosis bronchiectasis. Ann. Am. Thorac. Soc. 12, 701–707. doi: 10.1513/AnnalsATS.201411-513OC
Tchounwou, P. B., Yedjou, C. G., Patlolla, A. K., and Sutton, D. J. (2012). Heavy metal toxicity and the environment. Exper. Suppl. 101, 133–164. doi: 10.1007/978-3-7643-8340-4_6
Venkataraman, A., Bassis, C. M., Beck, J. M., Young, V. B., Curtis, J. L., Huffnagle, G. B., et al. (2015). Application of a neutral community model to assess structuring of the human lung microbiome. MBio 6:02284-14. doi: 10.1128/mBio.02284-14
Wang, C., Wu, H., Zhao, W., Zhu, B., and Yang, J. (2024). Effects of polycyclic aromatic hydrocarbons on soil bacterial and fungal communities in soils. Diversity 16, 675. doi: 10.3390/d16110675
Wang, L., Cheng, H., Wang, D., Zhao, B., Zhang, J., Cheng, L., et al. (2019). Airway microbiome is associated with respiratory functions and responses to ambient particulate matter exposure. Ecotoxicol. Environ. Saf. 167, 269–277. doi: 10.1016/j.ecoenv.2018.09.079
Wen, P., Feng, S. W., Liang, J.-L., Jia, P., Liao, B., Shu, W. S., et al. (2024). Heavy metal pollution in farmland soils surrounding mining areas in China and the response of soil microbial communities. Soil Secur. 17:100173. doi: 10.1016/j.soisec.2024.100173
Wu, L., Cui, F., Ma, J., Huang, Z., Zhang, S., Xiao, Z., et al. (2022). Associations of multiple metals with lung function in welders by four statistical models. Chemosphere 298:134202. doi: 10.1016/j.chemosphere.2022.134202
Wu, T., Hu, E., Xu, S., Chen, M., Guo, P., Dai, Z., et al. (2021). clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation 2:100141. doi: 10.1016/j.xinn.2021.100141
Xue, Y., Chu, J., Li, Y., and Kong, X. (2020). The influence of air pollution on respiratory microbiome: a link to respiratory disease. Toxicol. Lett. 334, 14–20. doi: 10.1016/j.toxlet.2020.09.007
Zhang, J., Cheng, H., Di Narzo, A., Zhu, Y., Xie, S., Shao, X., et al. (2023). Profiling microbiota from multiple sites in the respiratory tract to identify a biomarker for PM2.5 nitrate exposure-induced pulmonary damages. Environ. Sci. Technol. 57, 7346–7357. doi: 10.1021/acs.est.2c08807
Keywords: heavy metal, exposure, respiratory microbiota, microbial dysbiosis, 16S rDNA sequencing, host-microbiota interaction
Citation: Ding P, Wang X, Li N, Yu S, Zhang Y, Yu J, Tian T, Chen R, Luo B, Ma L and Zhang R (2025) Effects of combined exposure to heavy metals on lower respiratory flora and its role of lung injury in rats. Front. Microbiol. 16:1615130. doi: 10.3389/fmicb.2025.1615130
Received: 20 April 2025; Accepted: 22 May 2025;
Published: 13 June 2025.
Edited by:
Yuanzhao Ding, University of Oxford, United KingdomReviewed by:
SsuJu Li, National Yang Ming Chiao Tung University, TaiwanBasanta Kumar Biswal, National University of Singapore, Singapore
Shan Chen, Nanyang Technological University, Singapore
Copyright © 2025 Ding, Wang, Li, Yu, Zhang, Yu, Tian, Chen, Luo, Ma and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Li Ma, bWxAbHp1LmVkdS5jbg==; Rongxuan Zhang, Mzc1MTQyMzAyQHFxLmNvbQ==
†These authors have contributed equally to this work
 Ping Ding
Ping Ding Xiaoxia Wang
Xiaoxia Wang Nan Li1†
Nan Li1† Yiwen Zhang
Yiwen Zhang Junpu Yu
Junpu Yu Tian Tian
Tian Tian Bin Luo
Bin Luo Li Ma
Li Ma