- 1Department of Food Science & Technology, Chungnam National University, Daejeon, Republic of Korea
- 2Department of Biotechnology, The Catholic University of Korea, Bucheon, Republic of Korea
Listeria monocytogenes is a foodborne pathogen that causes listeriosis, a disease with a mortality rate of 20 ~ 30%. This bacterium enters the human body through contaminated food or ingredients and encounters primary innate defense systems, including gastric acid, bile salts, and gut microbiota. These systems play a critical role in preventing pathogen colonization and infection. However, interactions with pathogens can also alter the gut microbiota profile. This study aimed to investigate the host’s defense mechanisms against L. monocytogenes and the changes in the gut microbiota profile following infection. L. monocytogenes ATCC 7644 showed the greatest reduction (7.6 log CFU), followed by ATCC 19111 (5.71 log), F2365 (5.02 log), ATCC 19113 (3.96 log), and NCCP 14714 (3.38 log), while the pooled cocktail exhibited a 3.49 log CFU reduction. Notably, the clinical isolates NCCP 14714 and F2365 exhibited greater resistance to the simulated digestive process compared to the food isolate ATCC 7644. L. monocytogenes infection induced notable shifts in specific bacterial groups, including Bacteroides, Bifidobacterium, and the Mediterraneibacter gnavus group, as well as an increase in ethanol levels. These alterations may contribute to gut barrier disruption and the upregulation of immune responses, ultimately promoting the pathogenesis of L. monocytogenes infection. The findings from this study provide valuable insights into the interaction between L. monocytogenes and the human gut microbiota, offering a comparative reference for the interpretation of future research.
1 Introduction
Listeria monocytogenes is a Gram-positive, foodborne pathogen renowned for its capacity to cause severe infections, termed listeriosis (Osek et al., 2022). Listeriosis is recognized as one of the top five foodborne illnesses, with a mortality rate of 20 ~ 30% (Koopmans et al., 2023; Osek et al., 2022). The illness primarily occurs in pregnant women, newborns, elderly, and immunocompromised individuals, with pregnant women being over 100 times more likely to develop the infection compared to women with reproductive capability (Centers for Disease Control and Prevention, 2024; Koopmans et al., 2023). The incidence of foodborne outbreaks caused by L. monocytogenes has decreased through the implementation of regulations, practices, and screening methods such as HACCP (Koopmans et al., 2023). However, the costs associated with listeriosis still reached up to 22 billion dollars in North America, serving as a significant public health concern worldwide (Koopmans et al., 2023).
Listeria monocytogenes can primarily be transmitted through contaminated food ingredients or food products, such as meat, fish, ready-to-eat products, sliced vegetables, juice, and salad (Buchanan et al., 2017; Je et al., 2024). L. monocytogenes is introduced into the digestive system along with these contaminated foods, where it encounters primary defense mechanisms in the human body: gastric acid and bile salts (Koopmans et al., 2023). These harsh conditions of the digestive system can structurally damage the bacterial surface, increasing surface irregularities and potentially causing dissolution during passage through the gastrointestinal tract (Zou et al., 2024). Bile hinders microbial growth, and its toxicity against bacteria leads to heightened DNA damage, formation of secondary RNA structures, and instability in cellular membranes (Dowd et al., 2011). However, some L. monocytogenes can overcome acidic and enzymatic stress through the glutamate decarboxylase system, acid tolerance response, bile acid deconjugation, and the upregulation of multidrug efflux pumps (Koopmans et al., 2023; Zou et al., 2024). The σB operon is also considered a critical genetic element in adapting to acidic stress and surviving in the gastrointestinal tract (Gahan and Hill, 2014; Guerreiro et al., 2022). L. monocytogenes, which survives the digestive process, reaches the intestine, where it adheres to or invades the intestinal epithelium, leading to infection in the host (Koopmans et al., 2023).
Gut microbiota, comprising Bacteroidota (formerly Bacteroidetes), Bacillota (formerly Firmicutes), Pseudomonadota (formerly Proteobacteria), and Actinomycetota (formerly Actinobacteria), colonize the human intestine and play a crucial role in maintaining homeostasis (Park and Im, 2020). They are resistant against pathogens by competing with limited resources, altering the gut environment, and secreting antimicrobial substances such as short-chain fatty acids (SCFAs) and bacteriocins (Kamada et al., 2013). Some Bacteroides and lactic acid bacteria exhibit direct colonization resistance and infection resistance against pathogens such as Citrobacter rodentium, Escherichia coli, Clostridium difficile, and Salmonella (Buffie and Pamer, 2013). SCFAs decrease pH-sensitive pathogens, such as Enterobacteriaceae and Clostridia, and reduce the pathogenicity of Campylobacter jejuni and Staphylococcus aureus (Alva-Murillo et al., 2012; Sittipo et al., 2019; Van Deun et al., 2008). Pathogens can also lead to the death of anaerobic microbes and modify the gut microbiota profile by inducing secretion of antibacterial compounds, such as reactive oxygen species and reactive nitrogen species (Bäumler and Sperandio, 2016; Rogers et al., 2021). These imbalances reduce the diversity of gut microbiota and metabolite production, which aid pathogens in evading the microbial defense mechanisms (Rogers et al., 2021).
Various studies have explored pathogenic susceptibility, survival rates, and dynamics of intestinal bacteria in animal models (Becattini et al., 2017; Las Heras et al., 2019; Wolter et al., 2021). However, significant biological differences in intestinal structures, immune systems, and particularly in gut microbiota profiles between animals and humans can lead to results that may not accurately reflect human conditions (Park and Im, 2020). An in vitro fecal fermentation model, which utilizes human fecal inoculation and maintains a stable gut microbiota composition over extended periods, offers a more relevant platform for studying bacteria-to-bacteria interactions between human gut microbiota and L. monocytogenes (Li et al., 2019). Additionally, in vitro studies offer several advantages, including speed, cost-effectiveness, reduced labor demands, the ability to process multiple samples simultaneously, and no ethical concerns (Li et al., 2019; Minekus et al., 2014).
The aim of this study was to investigate the host’s defense strategy against L. monocytogenes and the modulation of the gut microbiota profile following listerial infection. The survival of L. monocytogenes during digestion was assessed in vitro, and the changes in gut microbiota and their metabolites induced by the pathogen were explored using a fecal fermentation model (Li et al., 2019; Minekus et al., 2014). Ultimately, understanding the interactions between L. monocytogenes and the gut microbiota is crucial for developing effective strategies to enhance the host’s defense mechanisms. Insights from this study may enhance our understanding of the interactions between gut microbiota and L. monocytogenes, aiming to mitigate the impact of L. monocytogenes infection and thereby promoting overall gut health and food safety.
2 Materials and methods
2.1 Selection of L. monocytogenes strains and growth conditions
Five L. monocytogenes strains [L. monocytogenes F2365 (4b), ATCC 19111 (1/2a), ATCC 19113 (3a), ATCC 7644 (1/2c), and NCCP 14714 (1/2b)] were selected from the culture collection at the Food Safety Lab of Chungnam National University, South Korea. These strains were chosen to represent a diverse range of serotypes commonly associated with foodborne outbreaks and environmental isolates (Koopmans et al., 2023). All strains were stored at −80°C. For each experiment, the strains were incubated on brain heart infusion (BHI; Kisan Bio, Seoul, South Korea) agar at 37°C for 24 h. Subsequently, a single colony of each L. monocytogenes was inoculated into BHI broth and cultured overnight at 37°C. A pooled L. monocytogenes culture was prepared by combining equal proportions of each strain incubated in broth.
2.2 In vitro digestion model
The survival of L. monocytogenes during gastrointestinal transit was assessed using an in vitro digestion model, with modifications based on the protocol of Pettersen et al. (2019). The five L. monocytogenes strains described above were tested with pooled cocktail to represent the genetic and phenotypic diversity in nature and compared the result with each single-strain. Overnight cultures of L. monocytogenes strains were centrifuged, and the pellets were resuspended in 0.1% peptone water (Kisan Bio). The bacterial suspension was exposed to simulated gastric fluid (SGF) containing porcine pepsin (2,000 U/mL; Sigma-Aldrich, St. Louis, MO, United States) in a 1:1 ratio. The SGF composition included 6.9 mM KCl (Junsei, Tokyo, Japan), 0.9 mM KH2PO4 (Sigma-Aldrich), 25 mM NaHCO3 (Daejung, Siheung, South Korea), 47.2 mM NaCl (Daejung), 0.1 mM MgCl2·7H2O (Daejung), 0.5 mM (NH4)2·CO3 (Junsei), and 0.15 mM CaCl2·2H2O (Daejung). The pH was adjusted to either 2.0 or 5.5 using 1 M HCl (Daejung) to mimic fasted or fed states (AquaSearcher AB23PH; Ohaus Corporation, Parsippany, NJ, United States). The gastric mixtures were incubated at 37°C in a shaking water bath (Maxturdy-18, Daihan Scientific, South Korea) at 100 rpm for 40 min. For the intestinal phase, the gastric mixture was combined with a simulated intestinal fluid (SIF), supplemented with 10 mM bovine bile (Sigma-Aldrich), and 100 U/mL porcine pancreatin (Sigma-Aldrich) at a 1:1 ratio. The SIF composition included 6.8 mM KCl, 0.8 mM KH2PO4, 85 mM NaHCO3, 38.4 mM NaCl, 0.33 mM MgCl2·6H2O, and 0.6 mM CaCl2·2H2O. The pH was adjusted to 7.0 using 1 M NaOH (Daejung) and incubated at 37°C in a shaking water bath at 100 rpm for 2 h. At each step, L. monocytogenes was enumerated by serial dilution in 0.1% peptone water and plating onto Oxford agar (Kisan Bio).
2.3 Fecal sample collection
Fecal samples (~5 g) were collected from three healthy donors using stool containers (SPL Life Sciences, Pocheon, South Korea). The samples were stored at 4°C and processed within 6 h. The donors were selected based on the absence of congenital or chronic diseases, no medication, and no antibiotic treatment within 4 weeks prior to collection. The ethical approval for fecal sample collection was obtained from the Institutional Review Board (IRB, 202209-BR-125-01).
2.4 In vitro fecal fermentation model
An in vitro fecal fermentation was conducted, based on Li et al. (2019) with slight modifications. Fresh fecal samples (2 g) were homogenized using a vortexer in 20 mL of sterile phosphate-buffered saline (PBS; Gibco BRL, Paisley, Scotland, UK) with 0.5% (w/v) L-cysteine hydrochloride (Junsei). The homogenized fecal mixture was filtered through sterile gauze. All experiments were conducted in a Coy Anaerobic Vinyl chamber (Coy Laboratory Product, MI, United States) under an anaerobic atmosphere (5% H2, 5% CO2, and 90% N2) at 37°C.
MiPro medium was prepared with the following components: 2.0 g/L peptone water, 2.0 g/L yeast extract (Kisan Bio), 0.5 g/L L-cysteine hydrochloride, 2 mL/L Tween 80 (Daejung), 5 mg/L hemin bovine (Sigma-Aldrich), 10 μL/L vitamin K1 (Sigma-Aldrich), 0.4 g/L K2HPO4 (Sigma-Aldrich), 0.4 g/L KH2PO4, 0.1 g/L MgSO4·7H2O, 0.1 g/L CaCl2·2H2O, 4.0 g/L NaHCO3, 4.0 g/L porcine gastric mucin (Sigma-Aldrich), and 0.5 g/L bile salts (Merck, Darmstadt, Germany) (Li et al., 2019). The sterile medium was placed in the anaerobic chamber a day prior to the experiment to ensure anaerobic conditions. Overnight cultures of L. monocytogenes were centrifuged and resuspended in fresh MiPro medium. The L. monocytogenes cocktail and the gut microbiota inocula were adjusted to reach approximately 8 log gene copies (GC)/mL. Fermentation was conducted in 96-deep well plates (Eppendorf, Hamburg, Germany) covered with silicone mats punctured for sampling, and incubated at 37°C. The samples were collected at 6 h, 12 h, and 24 h, and pH was measured at each time point. The survival of L. monocytogenes was verified by plating onto Oxford agar. The pellets were collected via centrifugation at 8,000 rpm for 10 min and stored at −80°C for subsequent analysis. The supernatants were filtered using a 0.22 μm syringe filter (13 mm; polytetrafluoroethylene) (Advantec, Tokyo, Japan) and stored at −80°C for SCFA analysis.
2.5 Quantitative PCR, and 16S rRNA amplicon sequencing
DNA was extracted using a NucleoSpin DNA Stool kit (Macherey-Nagel, Duren, Germany). Quantitative PCR (qPCR) was conducted on a CFX Connect Real-Time System (Bio-Rad, Hercules, CA, United States), using iQ SYBR-Green supermix (Bio-Rad) with 0.5 μM forward and reverse primers and template DNA. Universal primers targeting the 16S rRNA gene (forward: 5`-GTG STG CAY GGY YGT CGT CA-3`; reverse: 5`-ACG TCR TCC MCN CCT TCC TC-3`) were used following cycling conditions: 95°C for 3 min followed by 40 cycles of 95°C for 5 s, and 60°C for 30 s (Fuller et al., 2007; Reichardt et al., 2018). The 16S rRNA gene of Bifidobacterium miconisargentati 82T25 (accession ID: NR_181805.1, 148 bp) was used as the standard with concentrations ranging from 104 to 109 GC/ng (Macrogen Co. Ltd., Seoul, South Korea).
The 16S rRNA amplicon sequencing of the gut microbiota was conducted by Sanigen Co. Ltd. (Anyang, South Korea). In short, PCR was conducted with 341F and 806R primers to amplify the V3-V4 region of bacterial 16S rRNA genes. Then, the amplicon purification was carried out employing AMPure XP beads (Beckman Coulter, CA, United States). Library preparation for sequencing was carried out using the Nextera XT library prep kit (Illumina, San Diego, CA, United States). Sequencing was performed on the Illumina MiSeq platform (Illumina), generating 2 × 300 bp paired-end reads. Sequence processing, including trimming of low-quality reads, error correction of noisy reads, and elimination of chimeric sequences, was performed with Trimmomatic v0.39 (Bolger et al., 2014) and DADA2-QIIME2 (Callahan et al., 2016). Taxonomic classification was conducted using the SILVA 138 reference database (Quast et al., 2012), and diversity analyses were performed with QIIME2 (Bolyen et al., 2019). Metadata from this study is publicly available through the NCBI Sequence Read Archive under BioProject accession number PRJNA1254141, with associated BioSample accessions SAMN48096620 to SAMN48096647. To clearly understand the effects of L. monocytogenes infection, the OTU value of L. monocytogenes was excluded prior to analyzing diversity and profile abundance.
2.6 Quantitative analysis of SCFAs
Short-chain fatty acids concentrations in the fermentation supernatants were determined using the Nexera series high-performance liquid chromatography (HPLC) system (Shimadzu, Kyoto, Japan) installed with a RID-20A detector (Shimadzu) and an Aminex HPX-87H column (300 × 7.8 mm; Bio-Rad). The mobile phase consisted of 0.008 N sulfuric acid (Sigma-Aldrich), maintained at a constant flow rate of 0.6 mL/min at 35°C. Sample 20 μL was injected, and the analytes were detected over a 25 min run time. All samples were treated under the same conditions.
2.7 Statistical analysis
All experiments were conducted in triplicate to ensure data reproducibility and reliability. Statistical analysis was performed using one-way analysis of variance (ANOVA), followed by Tukey’s multiple comparisons test. A p-value of <0.05 was considered statistically significant for all analyses.
3 Results
3.1 Survival of L. monocytogenes during digestion
Five individual L. monocytogenes strains and their cocktail were subjected to an in vitro digestion model. During the gastric phase at pH 2.0, the five individual strains exhibited an average reduction of 5.13 log CFU, while the pooled L. monocytogenes cocktail demonstrated a 3.49 log CFU reduction (Figure 1A). Log reduction rates varied across individual strains, ranging from 3.38 to 7.6. L. monocytogenes ATCC 7644 showed the highest reduction at 7.6 log, followed by ATCC 19111 with 5.71 log, F2365 with 5.02 log, ATCC 19113 with 3.96 log, and ATCC 14714 with 3.38 log reduction. In the gastric phase at pH 5.5, all strains showed no significant differences in survival (Figure 1B). In the subsequent intestinal phase, regardless of initial pH, survival of all strains remained consistent within a ± 1 log range, indicating stable resistance throughout this phase (Figure 1).
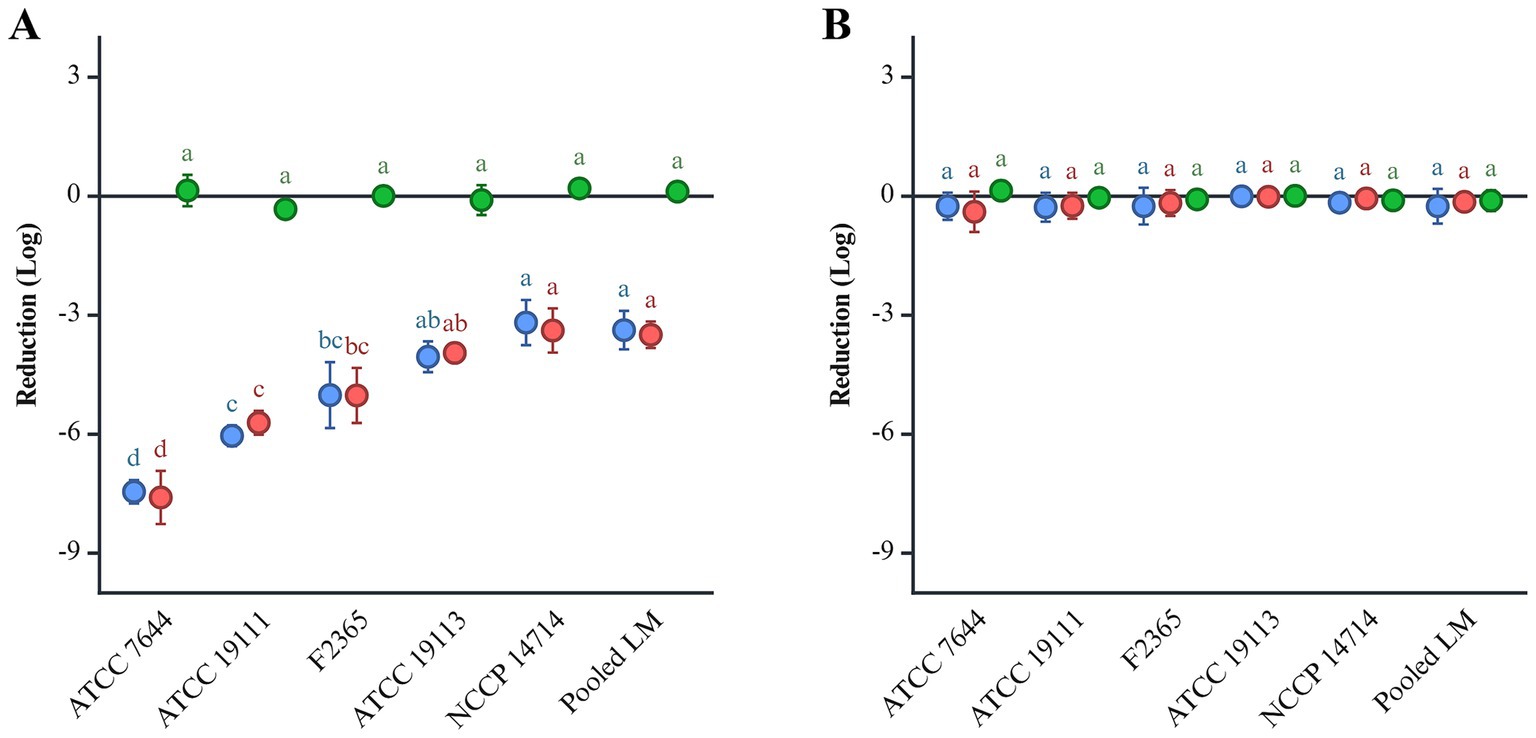
Figure 1. The reduction of L. monocytogenes at pH 2.0 (A) and 5.5 (B) during in vitro gastric digestion. L. monocytogenes was inoculated at a final concentration of 9 log CFU/mL. Gastric and intestinal digestions are indicated in red and green, respectively. The overall reduction is shown in blue. Significant differences (p < 0.05) are indicated by alphabets.
3.2 Survival of gut microbiota and L. monocytogenes during fecal fermentation
Gut microbiota in the L. monocytogenes-infected and non-infected samples reached 10.62 and 10.25 log GC/mL at 6 h, respectively (Supplementary Figure S1). The levels further increased to 11.39 and 11.46 log GC/mL at 12 h, then stabilized at 11.61 and 11.65 log GC/mL at 24 h. The L. monocytogenes cocktail, 7.34 log CFU/mL, increased to 8.58 log CFU/mL at 6 h, stabilized at 8.55 log CFU/mL at 12 h, and slightly declined to 8.17 log CFU/mL by 24 h. When cultured in MiPro medium alone, L. monocytogenes reached 8.41 log CFU/mL at 6 h, rose to 8.61 log CFU/mL at 12 h, and decreased slightly to 8.47 log CFU/mL by 24 h. Initial pH was 7.6, which decreased to 6.98 at 6 h, remained steady until 12 h, and then slightly increased to 7.2 by 24 h.
3.3 Dynamics of gut microbiota profile
At 0 h, the dominant phyla in fecal samples were Bacillota (51.98%), Bacteroidota (38.21%), Pseudomonadota (6.89%), and Fusobacteriota (0.11%) comprising over 95% of the microbial community (Figure 2). At 6 h, there was a significant shift in the microbial composition: Pseudomonadota and Fusobacteriota increased to 49.72 and 4.47%, respectively, while Bacteroidota and Bacillota decreased to 28.43 and 16.46%, respectively. Pseudomonadota reduced by 22.12% at 12 h and by 16.40% at 24 h, while Bacteroidota and Bacillota recovered to 35.90 and 25.26% at 12 h, respectively, with further stabilization at 24 h. Fusobacteriota increased to 15.39% at 12 h and remained relatively stable at 12.66% at 24 h. Notably, the abundance of Fusobacteriota in single feces 3 (SF3) decreased to below 0.05% after 12 h.
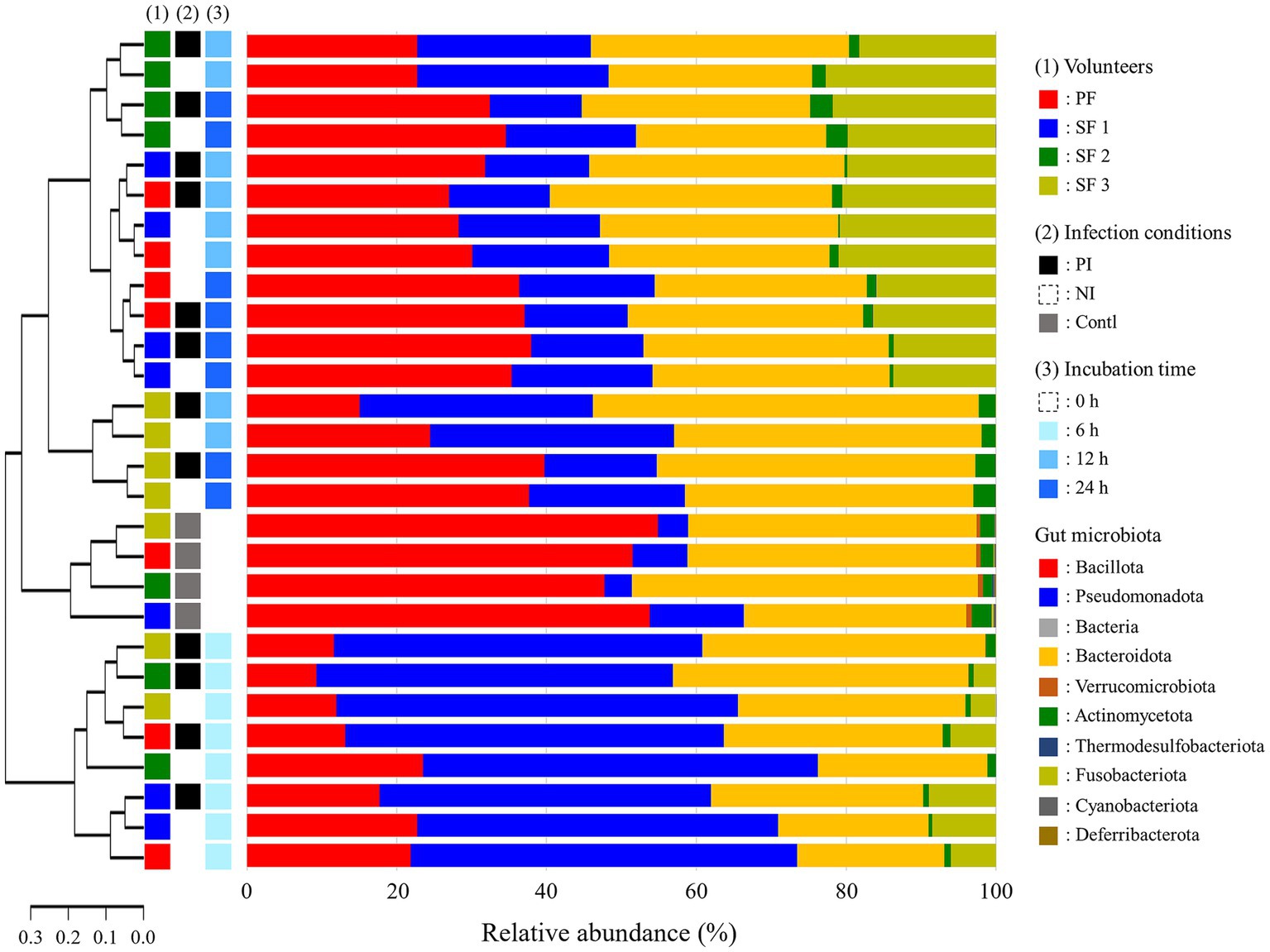
Figure 2. Phylogenetic tree and relative abundance of gut microbiota at the phylum level after L. monocytogenes infection during fecal fermentation, categorized by (1) volunteers, (2) in vitro infection conditions, and (3) incubation time. The phylogenetic analysis was performed using weighted distance values of gut microbiota. PF, pooled feces; SF, single feces; PI, post-infected; NI, non-infected; Contl, control.
Alpha and beta diversity analyses indicated that infection with L. monocytogenes reduced richness while increased evenness, though the changes were not statistically significant (Figures 3A,B). The diversity was significantly decreased at 6 h, recovered at 12 h, with richness and evenness continuing to increase through 24 h (Figures 3C,D). Weighted distance analysis displayed clustering differences based on infection conditions, with incubation time exerting a stronger influence on cluster patterns (Figure 3E). Notably, 0 h samples were distinct from latter time points, while 12 h and 24 h samples exhibited greater proximity, clustering more closely together, which aligns with phylogenetic results (Figure 2).
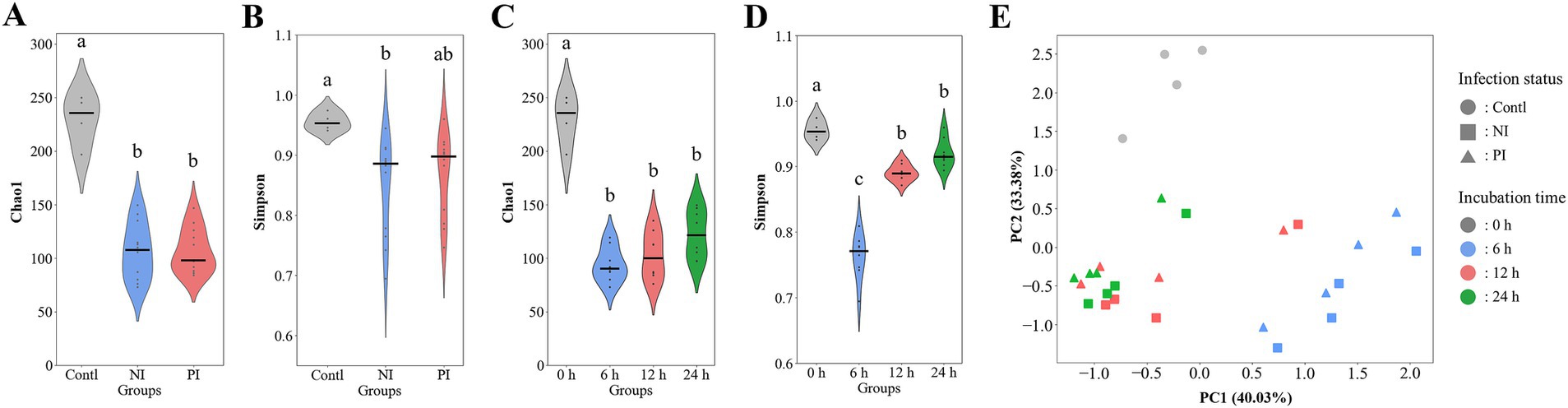
Figure 3. Violin plots and principal coordinate analysis (PCA) comparing the alpha and beta diversity of gut microbiota during fecal fermentation based on infection status (A,B) and incubation time (C,D). The plots include the species richness as measured by the chao1 index (A,C) as well as the species evenness as measured by the Simpson diversity index (B,D). PCA is presented by weighted Bray–Curtis distances (E). Significant differences (p < 0.05) are indicated by alphabets. Contl, control; NI, non-infected; PI, post-infected.
3.4 Gut microbiota profile in the phylum, family, and genus level
Pseudomonadota was more dominant in the non-infected (NI) group (18.77 ~ 51.54%) than post-infected (PI) groups (14.03 ~ 47.90%), while Bacteroidota was more abundant in PI (33.69 ~ 39.43%) compared to NI (23.17 ~ 32.37%) groups (Figure 4). This pattern remained consistent across all fecal samples and time points. Bacillota showed higher levels in NI (20.00%) than PI (12.93%) at 6 h, with similar levels thereafter. L. monocytogenes infection delayed the recovery of Bacillota at 6 h in the pooled feces (PF) group and SF1 and 2 groups and 12 h in the SF3 group (data not shown). At the family level, PI groups consistently exhibited lower Erysipelotrichaceae but higher Bacteroidaceae, Lachnospiraceae, and Bifidobacteriaceae abundances than NI at all time points (Supplementary Figure S2). Notably, both Bacteroidaceae and Lachnospiraceae showed a marked increase in response to L. monocytogenes infection across all fecal groups (data not shown). Eight genera within these families exhibited abundance patterns consistent with trends observed at the family level (Figure 5). Bacteroides (Bacteroidaceae) and Bifidobacterium (Bifidobacteriaceae) were more abundant in PI (30.55 ~ 34.90% and 0.10 ~ 0.74%) than NI (20.80 ~ 28.74% and 0.12 ~ 0.61%). Faecalitalea, belonging to Erysipelotrichaceae, was more prevalent in NI (7.77 ~ 12.72%) than PI (4.12 ~ 11.19%). Blautia and Mediterraneibacter gnavus group of Lachnospiraceae exhibited higher abundances in PI (0.14 ~ 1.17% and 1.04 ~ 3.41%) than NI (0.14 ~ 0.94% and 0.83 ~ 3.32%).
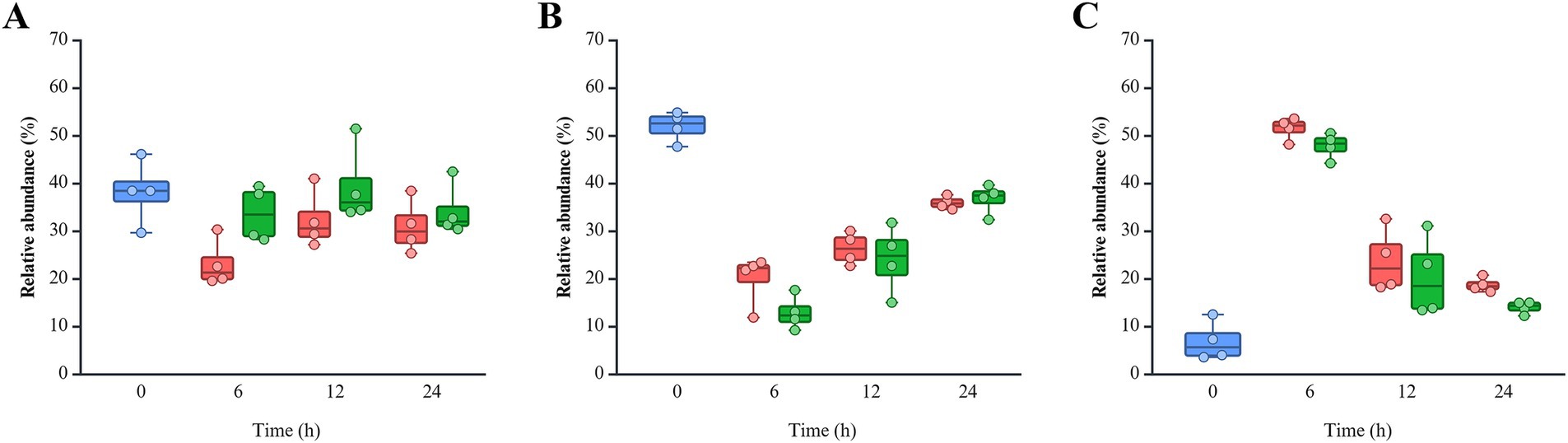
Figure 4. Relative abundance of three phyla of interest: (A) Bacteroidota, (B) Bacillota, and (C) Pseudomonadota. The color of each bar chart represents different infection conditions: control (blue), non-infected (red), and post-infected (green). Each dot represents the value for each sample.
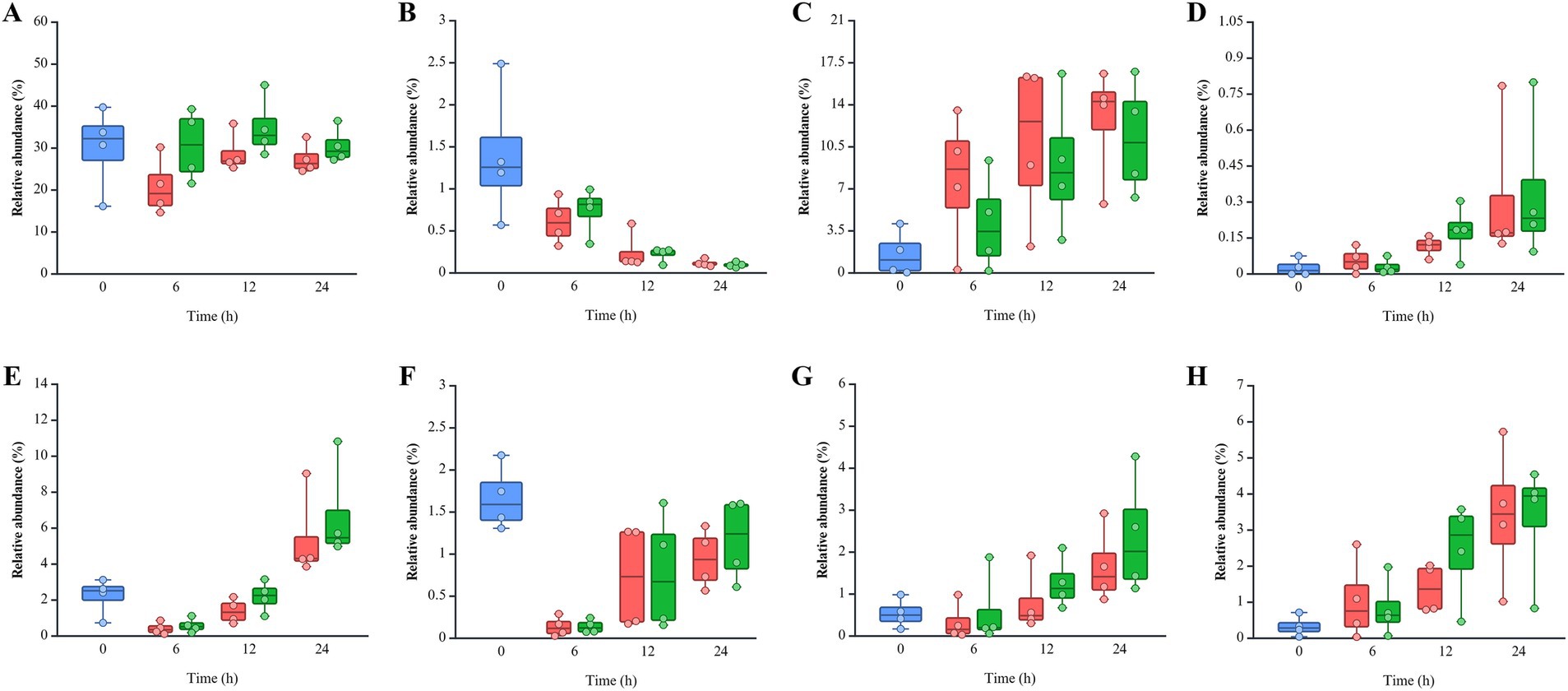
Figure 5. Relative abundance of eight genera belonging to Bacteroidaceae, Bifidobacteriaceae, Erysipelotrichaceae, and Lachnospiraceae: (A) Bacteroides, (B) Bifidobacterium, (C) Faecalitalea, (D) Clostridium innocuum group, (E) Lachnoclostridium, (F) Blautia, (G) Ruminococcus torques group, and (H) Mediterraneibacter gnavus group. The color of each bar chart represents different infection conditions: control (blue), non-infected (red), and post-infected (green). Each dot represents the value for each sample.
3.5 Changes in SCFAs and ethanol
Acetate and propionate concentrations were comparable between the NI and PI groups, with acetate ranging from 0 to 30.14 mM in NI and 0 to 30.47 mM in PI, and propionate ranging from 0 to 11.74 mM in NI and 0 to 11.88 mM in PI (Figure 6). Butyrate levels were slightly higher in NI (0 ~ 15.55 mM) compared to PI (0 ~ 14.87 mM). In contrast, ethanol production was elevated in the PI group, ranging from 39.07 to 63.82 mM, compared to 27.78 to 55.13 mM in the NI group. Over time, the concentration of both SCFAs and ethanol increased progressively throughout the incubation period.
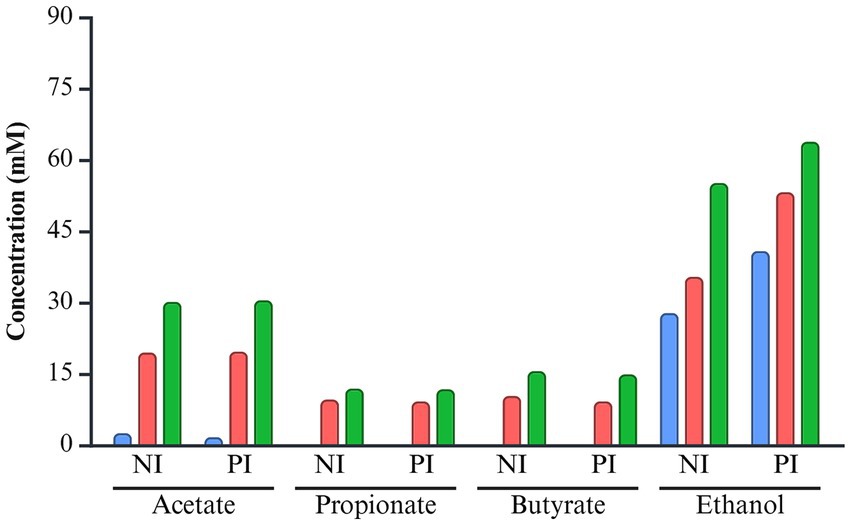
Figure 6. The concentration of short-chain fatty acids and ethanol of L. monocytogenes infected and non-infected pooled fecal samples during fermentation. The color of each bar chart represents the different incubation times: 6 h (blue), 12 h (red), and 24 h (green). NI, non-infected; PI, post-infected.
4 Discussion
Listeria monocytogenes is a significant public health and food safety concern because of its virulence and resistance against various environmental stresses (Muchaamba et al., 2022). Its infectious dose can be influenced by the human innate immune system, which plays a crucial role in determining the severity of infection (Guo et al., 2023; Koopmans et al., 2023). During the gastrointestinal passage, L. monocytogenes must endure the stressful environment, with its survival capability varying by strain, isolate origin, and serotype (Ramalheira et al., 2010; Zou et al., 2024). Survived L. monocytogenes that bypass host’s defense systems must encounter gut microbiota, which competes for energy sources and secretes antibacterial compounds (Kamada et al., 2013). In immunocompromised individuals, L. monocytogenes can establish prolonged colonization in the cecum and colon, thereby altering the gut microbiota profile (Koopmans et al., 2023). Although several studies have explored the changes in gut microbiota due to L. monocytogenes infection, specific interactions between bacteria remain poorly understood (Becattini et al., 2017; Guo et al., 2023; Wolter et al., 2021). In this study, we aimed to investigate the survival of L. monocytogenes during digestion, assess its pathogenic risk, and determine its impact on gut microbiota and metabolite profiles.
Among the 14 serotypes of L. monocytogenes, 92 ~ 95% of the clinical isolates belong to serotypes 4b, 1/2a, and 1/2b (Koopmans et al., 2023). Serotypes 4b and 1/2a exhibit higher resistance to environmental stresses, including acidity, than serotypes 4a and 1/2c (Chakravarty et al., 2021; Jiang et al., 2010; Zou et al., 2024). Additionally, clinical isolates exhibited greater resistance to acidic and bile stresses compared to food isolates (Ramalheira et al., 2010). L. monocytogenes F2365 (serotype 4b) and NCCP 14714 (serotype 1/2b) were clinical isolates, while ATCC 7644 (serotype 1/2c) was isolated from food (Koopmans et al., 2023). L. monocytogenes F2365 and NCCP 14714 showed significantly higher resistance in the simulated gastric phase (pH 2.0) than ATCC 7644. Acidic stress adaption is regulated by the stress response factor σB, encoded by the rsbRSTUVWX and sigB genes (Gahan and Hill, 2014; Guerreiro et al., 2022). The rsbS gene plays a critical role in stress signaling, as it mediates the phosphorylation and activation of the stressosome (Guerreiro et al., 2022). Notably, a ΔrsbS mutant strain exhibited reduced acid tolerance, falling below detection limits after just 15 min of exposure to pH 5.0 (Guerreiro et al., 2022). L. monocytogenes ATCC 7644 has an adenine deletion in the rsbS gene with a premature stop codon, resulting in reduced survival under acidic conditions. This characteristic suggests the need for further research to explore the mechanisms underlying this phenomenon. Previous studies have documented the presence of multiple L. monocytogenes strains in a single food sample, as well as multi-strain involvement in listeriosis outbreaks (Zilelidou and Skandamis, 2018). In addition to the individual strains, a five-strain cocktail was evaluated to reflect potential real-world contamination scenarios involving multiple serotypes. The cocktail exhibited greater resistance than the more acid-sensitive individual strains, such as ATCC 7644, but showed lower resistance than F2365 and NCCP 14714. Although inter-strain interactions among L. monocytogenes, such as quorum sensing, bacteriocin production, or biofilm cooperation, may enhance virulence or survival, the overall survival observed in the cocktail can be attributed primarily to the intrinsic resistance of the more tolerant strains (Zilelidou and Skandamis, 2018).
The human colon contains nearly 11 to 12 log CFU/g of bacteria, comprising 500 to 1,000 species (Rizzatti et al., 2017). Factors such as diet, illness, and antibiotic treatment can significantly impact microbial diversity and often result in gut dysbiosis (Rogers et al., 2021). In the present work, alpha and beta diversity of the gut microbiota showed three distinct phases regardless of infection status. A significant decline in the diversity was observed within the first 6 h, followed by a gradual recovery, highlighting a rebalance within the microbial ecosystem and the resilience of the gut microbiota after exposure to stress. A decline can be attributed to dysbiosis induced by the initial fecal processing step, resulting in unfavorable conditions (Rogers et al., 2021). Pseudomonadota decreased over time, which includes various human pathogens, such as Shigella, Escherichia, Salmonella, Yersinia, and Helicobacter (Rizzatti et al., 2017). Conversely, Fusobacteriota showed a continuous increase, suggesting that the gut microbiota had reached an alternate stable state which was different from its baseline status.
Gut dysbiosis can increase susceptibility to L. monocytogenes infection. Once established, listerial infections may exacerbate the condition by further disrupting the microbial community structure (Bäumler and Sperandio, 2016; Becattini et al., 2017). In C57BL/6 mice, L. monocytogenes significantly decreased Chao1 and Shannon indices, indicating a reduction in microbial diversity (Alam et al., 2021; Keane et al., 2023). Similarly, a decrease in evenness was found in the macaque monkey, although the difference was insignificant (Hugon et al., 2023). In this study, L. monocytogenes infection resulted in decreased richness and increased evenness, consistent with a reduction in specific taxa and the reorganization of community structure. Furthermore, PCA analysis of weighted distances also revealed distinct clustering between non-infected and infected groups, highlighting differences in the microbial community composition. The observed differences in gut microbiota diversity can be, in part, attributed to bacteriocins produced by L. monocytogenes, such as listeriolysin S (LLS) and Lmo2776 (Lee, 2020). L. monocytogenes ATCC 7644 and F2365 have the Lmo2776 synthesis operon and Lmo2776 expressed under in vitro conditions (Rolhion et al., 2019). Lmo2776 specifically targeted Prevotella spp. and Segatella copri (formerly Prevotella copri) (Rolhion et al., 2019). L. monocytogenes NCCP 14714 and F2365 contain the LLS synthesis cluster. LLS is expressed in gastrointestinal tracts in vivo and decreases the abundance of Allobaculum and Alloprevotella in mice (Quereda et al., 2016; Quereda et al., 2017). The antimicrobial activities of these L. monocytogenes-derived bacteriocins may suppress the richness of specific microbial taxa. Conversely, the absence of such bacteriocins could facilitate the proliferation of other microbial species by relieving competitive pressures within the gut environment.
In this study, L. monocytogenes infection led to a decrease in Bacillota, while Bacteroidota was increased, consistent with the findings from the BALB/c mice study by Guo et al. (2023), though opposite results were reported in C57BL/6 mice model (Alam et al., 2021). Bacteroidota has been linked to inflammation, suggesting that L. monocytogenes can modulate inflammation by altering the gut microbial composition (Guo et al., 2023; Rogers et al., 2021). The reduction and delayed recovery of Bacillota may weaken gut barrier function and impair immune regulation, heightening susceptibility to pathogens and promoting inflammation (Rogers et al., 2021). L. monocytogenes infection led to an increase in Pseudomonadota in the BALB/c mice, which contradicts the findings of this study (Guo et al., 2023). Elevated levels of Pseudomonadota was reported in pregnant individuals, who are at the highest risk for listeriosis (Becattini et al., 2017; Koopmans et al., 2023). However, little research has been conducted on the specific relationship between Pseudomonadota and L. monocytogenes.
Several bacterial groups known for their protective roles against intestinal pathogens showed increased abundance following L. monocytogenes infection in this study. Bacteroides spp. contribute to intestinal homeostasis by degrading mucin and supporting nutrient availability for other microbes (Zafar and Saier Jr, 2021). They also interact with the host immune system and compete with pathogens (Wexler, 2007; Zafar and Saier Jr, 2021). In murine models, L. monocytogenes infection altered Bacteroides populations: B. caccae increased while B. ovatus decreased in BALB/c mice, and in C57BL/6 mice, B. uniformis and Bacteroidaceae decreased while B. acidifaciens increased (Alam et al., 2021; Guo et al., 2023; Las Heras et al., 2019; Wolter et al., 2021). The observed increase in Bacteroides may have contributed to the reduction of Pseudomonadota in our study, potentially through colonization resistance mechanisms. In particular, B. thetaiotaomicron has been shown to directly inhibit the colonization of E. coli and Salmonella, both members of Pseudomonadota, supporting the proposed role of Bacteroides in colonization resistance (Buffie and Pamer, 2013). Bifidobacterium spp. also play a well-established role in inhibiting pathogen colonization. They improve host outcomes in infections by E. coli O157: H7 and Clostridium perfringens, and can inhibit L. monocytogenes EGDe invasion by 60–90% through secreted proteinaceous factors (Corr et al., 2007; O'Callaghan and Van Sinderen, 2016). The increased abundance of Bifidobacterium following listerial infection may be related to L. monocytogenes mitigating oxidative stress on B. bifidum through neutralization of reactive oxygen species (Yu et al., 2020). Lachnospiraceae, also known as Clostridium cluster XIVa, are fiber-degrading bacteria that produce SCFAs and help maintain gut barrier function (Vacca et al., 2020). Within this family, Blautia producta has demonstrated anti-listerial activity by inhibiting L. monocytogenes propagation (Becattini et al., 2017). Increased abundance of Lachnospiraceae has been observed in infected C57BL/6 mice fed a high-fat diet and in aged C57BL/6 mice (Alam et al., 2021; Las Heras et al., 2019), and a positive correlation (ρ = 0.33) was reported between Lachnospiraceae and L. monocytogenes abundance in human listeriosis patients (Hafner et al., 2021). These findings align with our observation of elevated Lachnospiraceae following infection. The increased abundance of Bacteroides, Bifidobacterium, and Lachnospiraceae, which are known to protect against L. monocytogenes, following infection may reflect an intrinsic compensatory mechanism of the human gut microbiota to restore microbial balance in response to pathogenic challenge (Becattini et al., 2017; Buffie and Pamer, 2013; Corr et al., 2007). This observation supports the hypothesis that individuals with a diverse and resilient gut microbiota are better protected against listeriosis (Guo et al., 2023; Hafner et al., 2021; Wolter et al., 2021).
The direct impact of SCFA on L. monocytogenes has been unknown, however, SCFAs can regulate the host’s immune system and enhance gut barrier function by strengthening tight junctions (Becattini et al., 2017; Rogers et al., 2021). Acetate and propionate exhibited a synergistic effect with nisin, produced by Lactococcus lactis, potentially contributing to shifts in the bacterial profile (Rodpan et al., 2022). SCFAs produced by beneficial microbes such as Bifidobacterium could lower intestinal pH, thereby creating an unfavorable environment for opportunistic pathogens (O'Callaghan and Van Sinderen, 2016). The pH in this study decreased to 6.98 due to the buffering capacity of MiPro medium, limiting the ability to assess microbial changes directly related to pH fluctuations.
Acetate, propionate, and butyrate are detected in the human colon and stool at a 3:1:1 ratio, with total SCFA concentrations ranging from 20 to 70 mM in the distal colon (Den Besten et al., 2013). Comparable levels were observed in this study, with ~30.14 mM acetate, ~11.88 mM propionate, and ~15.55 mM butyrate. Although aformentioned bacteria such as Bacteroides, Bifidobacterium, and Lachnospiraceae are known contributors to SCFA production, no significant differences in SCFA levels were observed between infected and non-infected groups (O'Callaghan and Van Sinderen, 2016; Vacca et al., 2020). Listeria infection increased ethanol levels, a primary metabolite associated with bacteria such as B. thetaiotaomicron, Bifidobacterium, and M. gnavus group (Crost et al., 2018; Elshaghabee et al., 2016; Yamaguchi et al., 2018). Although many gut microbiota are capable of producing ethanol, its elevated levels during the infected state are linked to an increased abundance of these bacteria. Ethanol can disrupt epithelial cells and weaken tight junctions. This “leaky gut” condition allows harmful substances, such as endotoxins, to penetrate the bloodstream, triggering systemic inflammation and potentially exacerbating Listeria infection (Chen et al., 2022). Since most gut microbiota cannot metabolize ethanol, it is predominantly converted to acetate through host metabolic pathways (Martino et al., 2022). Ethanol consumption increases Bacteroidetes more through elevated acetate levels, an ethanol metabolite, than through ethanol itself (Martino et al., 2022). As this in vitro model excludes host interactions, predicting acetate accumulation from ethanol metabolism and its impact on the gut microbiota remains challenging. Therefore, the observed increase in Bacteroidetes cannot be solely attributed to ethanol.
This study highlights the intricate interactions between L. monocytogenes and the gut microbiota, emphasizing the pathogen’s ability to survive and adapt within the gastrointestinal environment. Our study demonstrates that clinical L. monocytogenes strains exhibit greater resistance to the human digestive process than food-derived strains. L. monocytogenes was observed to cause minimal to no significant changes in the gut microbiota diversity, consistent with previous studies (Becattini et al., 2017; Hugon et al., 2023; Keane et al., 2023; Las Heras et al., 2019; Li et al., 2019). Hugon et al. (2023) reported that L. monocytogenes alone did not induce dysbiosis; however, listeriosis may contribute to gut microbiota alterations under specific conditions, such as pregnancy. Comparative studies using macaque monkeys, BALB/c mice, and C57BL/6 mice have reported varying results, highlighting discrepancies between animal models and human microbiome data (Alam et al., 2021; Guo et al., 2023; Hafner et al., 2021; Hugon et al., 2023). These differences may stem from defense mechanisms against pathogens and variations in the host’s immune system. The administration of Akkermansia muciniphila to mice has been shown to decrease susceptibility to L. monocytogenes (Keane et al., 2023). This protective effect was not directly attributed to changes in gut microbiota composition or the bacterium itself but rather through interactions with the host’s immune system, indicating that the inhibition of L. monocytogenes by gut microbiota may be more intricate than previously understood (Keane et al., 2023). LLS expressed by L. monocytogenes has been found to have no effect on human eukaryotic cells but exhibits antimicrobial activity against prokaryotes, suggesting that Listeria can directly influence gut microbiota composition (Quereda et al., 2017). This study primarily focused on the interplay between human gut microbiota and Listeria and did not account for the potential involvement of the immune system. The findings provide valuable insights into understanding complex mechanisms underlying gut microbiota-pathogen interactions. Notably, Listeria infection was associated with an increased abundance of bacteria such as Bacteroides, Bifidobacterium, and the M. gnavus group, which are known ethanol producers. Elevated ethanol levels may compromise epithelial barrier integrity to a leaky gut that facilitates the translocation of harmful substances, exacerbates inflammatory responses, and further complicates conditions like listeriosis. Although ethanol has not been previously recognized as a metabolic biomarker in Listeria infection studies, the unexpected increase observed in this study highlights the need for further investigation into its role in gut function and host health. Due to the use of pooled samples, correlation analyses between specific taxa and ethanol production were not feasible, limiting direct functional inference. Future studies with larger sample sizes and individual-level measurements will be required to validate these observations.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found: https://www.ncbi.nlm.nih.gov/, PRJNA1254141.
Ethics statement
The studies involving humans were approved by Chungnam National University Institutional Review Board. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
DK: Data curation, Visualization, Validation, Investigation, Writing – review & editing, Formal analysis, Writing – original draft. SS: Writing – review & editing, Investigation, Validation. UK: Writing – review & editing, Investigation. SA: Writing – review & editing, Investigation. HJ: Writing – review & editing. DL: Validation, Investigation, Writing – review & editing. EY: Methodology, Supervision, Resources, Writing – review & editing. OK: Supervision, Conceptualization, Writing – review & editing, Resources, Project administration, Funding acquisition.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was supported by the National Research Foundation of Korea (RS-2023-00242749 and RS-2024-00396978).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1616720/full#supplementary-material
References
Alam, M. S., Gangiredla, J., Hasan, N. A., Barnaba, T., and Tartera, C. (2021). Aging-induced dysbiosis of gut microbiota as a risk factor for increased Listeria monocytogenes infection. Front. Immunol. 12:672353. doi: 10.3389/fimmu.2021.672353
Alva-Murillo, N., Ochoa-Zarzosa, A., and López-Meza, J. E. (2012). Short chain fatty acids (propionic and hexanoic) decrease Staphylococcus aureus internalization into bovine mammary epithelial cells and modulate antimicrobial peptide expression. Vet. Microbiol. 155, 324–331. doi: 10.1016/j.vetmic.2011.08.025
Bäumler, A. J., and Sperandio, V. (2016). Interactions between the microbiota and pathogenic bacteria in the gut. Nature 535, 85–93. doi: 10.1038/nature18849
Becattini, S., Littmann, E. R., Carter, R. A., Kim, S. G., Morjaria, S. M., Ling, L., et al. (2017). Commensal microbes provide first line defense against Listeria monocytogenes infection. J. Exp. Med. 214, 1973–1989. doi: 10.1084/jem.20170495
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9
Buchanan, R. L., Gorris, L. G., Hayman, M. M., Jackson, T. C., and Whiting, R. C. (2017). A review of Listeria monocytogenes: an update on outbreaks, virulence, dose-response, ecology, and risk assessments. Food Control 75, 1–13. doi: 10.1016/j.foodcont.2016.12.016
Buffie, C. G., and Pamer, E. G. (2013). Microbiota-mediated colonization resistance against intestinal pathogens. Nat. Rev. Immunol. 13, 790–801. doi: 10.1038/nri3535
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016). DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nMeth.3869
Centers for Disease Control and Prevention. (2024). Listeria infection (listeriosis). Available online at: https://www.cdc.gov/ (accessed April 10, 2025)
Chakravarty, D., Sahukhal, G., Arick, M., Davis, M. L., and Donaldson, J. R. (2021). Transcriptomic analysis of Listeria monocytogenes in response to bile under aerobic and anaerobic conditions. Front. Microbiol. 12:754748. doi: 10.3389/fmicb.2021.754748
Chen, G., Shi, F., Yin, W., Guo, Y., Liu, A., Shuai, J., et al. (2022). Gut microbiota dysbiosis: the potential mechanisms by which alcohol disrupts gut and brain functions. Front. Microbiol. 13:916765. doi: 10.3389/fmicb.2022.916765
Corr, S. C., Gahan, C. G., and Hill, C. (2007). Impact of selected Lactobacillus and Bifidobacterium species on Listeria monocytogenes infection and the mucosal immune response. FEMS Immunol. Med. Microbiol. 50, 380–388. doi: 10.1111/j.1574-695X.2007.00264.x
Crost, E. H., Le Gall, G., Laverde-Gomez, J. A., Mukhopadhya, I., Flint, H. J., and Juge, N. (2018). Mechanistic insights into the cross-feeding of Ruminococcus gnavus and Ruminococcus bromii on host and dietary carbohydrates. Front. Microbiol. 9:2558. doi: 10.3389/fmicb.2018.02558
Den Besten, G., Van Eunen, K., Groen, A. K., Venema, K., Reijngoud, D. J., and Bakker, B. M. (2013). The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J. Lipid Res. 54, 2325–2340. doi: 10.1194/jlr.R036012
Dowd, G. C., Joyce, S. A., Hill, C., and Gahan, C. G. (2011). Investigation of the mechanisms by which Listeria monocytogenes grows in porcine gallbladder bile. Infect. Immun. 79, 369–379. doi: 10.1128/IAI.00330-10
Elshaghabee, F. M., Bockelmann, W., Meske, D., De Vrese, M., Walte, H. G., Schrezenmeir, J., et al. (2016). Ethanol production by selected intestinal microorganisms and lactic acid bacteria growing under different nutritional conditions. Front. Microbiol. 7:47. doi: 10.3389/fmicb.2016.00047
Fuller, Z., Louis, P., Mihajlovski, A., Rungapamestry, V., Ratcliffe, B., and Duncan, A. J. (2007). Influence of cabbage processing methods and prebiotic manipulationof colonic microflora on glucosinolate breakdown in man. Br. J. Nutr. 98, 364–372. doi: 10.1017/S0007114507709091
Gahan, C. G., and Hill, C. (2014). Listeria monocytogenes: survival and adaptation in the gastrointestinal tract. Front. Cell. Infect. Microbiol. 4:9. doi: 10.3389/fcimb.2014.00009
Guerreiro, D. N., Pucciarelli, M. G., Tiensuu, T., Gudynaite, D., Boyd, A., Johansson, J., et al. (2022). Acid stress signals are integrated into the σB-dependent general stress response pathway via the stressosome in the food-borne pathogen Listeria monocytogenes. PLoS Pathog. 18:e1010213. doi: 10.1371/journal.ppat.1010213
Guo, L., Yin, X., and Liu, Q. (2023). Fecal microbiota transplantation reduces mouse mortality from Listeria monocytogenes infection. Microb. Pathog. 178:106036. doi: 10.1016/j.micpath.2023.106036
Hafner, L., Pichon, M., Burucoa, C., Nusser, S. H., Moura, A., Garcia-Garcera, M., et al. (2021). Listeria monocytogenes faecal carriage is common and depends on the gut microbiota. Nat. Commun. 12:6826. doi: 10.1038/s41467-021-27069-y
Hugon, A. M., Deblois, C. L., Simmons, H. A., Mejia, A., Schotzo, M. L., Czuprynski, C. J., et al. (2023). Listeria monocytogenes infection in pregnant macaques alters the maternal gut microbiome. Biol. Reprod. 109, 618–634. doi: 10.1093/biolre/ioad104
Je, H. J., Kim, U. I., and Koo, O. K. (2024). A comprehensive systematic review and meta-analysis of Listeria monocytogenes prevalence in food products in South Korea. Int. J. Food Microbiol. 415:110655. doi: 10.1016/j.ijfoodmicro.2024.110655
Jiang, L., Olesen, I., Andersen, T., Fang, W., and Jespersen, L. (2010). Survival of Listeria monocytogenes in simulated gastrointestinal system and transcriptional profiling of stress-and adhesion-related genes. Foodborne Pathog. Dis. 7, 267–274. doi: 10.1089/fpd.2009.0361
Kamada, N., Chen, G. Y., Inohara, N., and Núñez, G. (2013). Control of pathogens and pathobionts by the gut microbiota. Nat. Immunol. 14, 685–690. doi: 10.1038/ni.2608
Keane, J. M., Las Heras, V., Pinheiro, J., FitzGerald, J. A., Núñez-Sánchez, M. A., Hueston, C. M., et al. (2023). Akkermansia muciniphila reduces susceptibility to Listeria monocytogenes infection in mice fed a high-fat diet. Gut Microbes 15:2229948. doi: 10.1080/19490976.2023.2229948
Koopmans, M. M., Brouwer, M. C., Vázquez-Boland, J. A., and van de Beek, D. (2023). Human listeriosis. Clin. Microbiol. Rev. 36, e0006019–e0000019. doi: 10.1128/cmr.00060-19
Las Heras, V., Clooney, A. G., Ryan, F. J., Cabrera-Rubio, R., Casey, P. G., Hueston, C. M., et al. (2019). Short-term consumption of a high-fat diet increases host susceptibility to Listeria monocytogenes infection. Microbiome 7, 7–12. doi: 10.1186/s40168-019-0621-x
Lee, S. (2020). Bacteriocins of Listeria monocytogenes and their potential as a virulence factor. Toxins 12:103. doi: 10.3390/toxins12020103
Li, L., Abou-Samra, E., Ning, Z., Zhang, X., Mayne, J., Wang, J., et al. (2019). An in vitro model maintaining taxon-specific functional activities of the gut microbiome. Nat. Commun. 10:4146. doi: 10.1038/s41467-019-12087-8
Martino, C., Zaramela, L. S., Gao, B., Embree, M., Tarasova, J., Parker, S. J., et al. (2022). Acetate reprograms gut microbiota during alcohol consumption. Nat. Commun. 13:4630. doi: 10.1038/s41467-022-31973-2
Minekus, M., Alminger, M., Alvito, P., Ballance, S., Bohn, T., Bourlieu, C., et al. (2014). A standardised static in vitro digestion method suitable for food–an international consensus. Food Funct. 5, 1113–1124. doi: 10.1039/c3fo60702j
Muchaamba, F., Eshwar, A. K., Stevens, M. J., Stephan, R., and Tasara, T. (2022). Different shades of Listeria monocytogenes: strain, serotype, and lineage-based variability in virulence and stress tolerance profiles. Front. Microbiol. 12:792162. doi: 10.3389/fmicb.2021.792162
O'Callaghan, A., and Van Sinderen, D. (2016). Bifidobacteria and their role as members of the human gut microbiota. Front. Microbiol. 7:206360. doi: 10.3389/fmicb.2016.00925
Osek, J., Lachtara, B., and Wieczorek, K. (2022). Listeria monocytogenes–how this pathogen survives in food-production environments? Front. Microbiol. 13:866462. doi: 10.3389/fmicb.2022.866462
Park, J. C., and Im, S. H. (2020). Of men in mice: the development and application of a humanized gnotobiotic mouse model for microbiome therapeutics. Exp. Mol. Med. 52, 1383–1396. doi: 10.1038/s12276-020-0473-2
Pettersen, K. S., Skjerdal, T., Wasteson, Y., Lindbäck, T., Vegarud, G., Comi, I., et al. (2019). Survival of Listeria monocytogenes during in vitro gastrointestinal digestion after exposure to 5 and 0.5% sodium chloride. Food Microbiol. 77, 78–84. doi: 10.1016/j.fm.2018.08.010
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2012). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Quereda, J. J., Dussurget, O., Nahori, M. A., Ghozlane, A., Volant, S., Dillies, M. A., et al. (2016). Bacteriocin from epidemic Listeria strains alters the host intestinal microbiota to favor infection. Proc. Natl. Acad. Sci. 113, 5706–5711. doi: 10.1073/pnas.1523899113
Quereda, J. J., Nahori, M. A., Meza-Torres, J., Sachse, M., Titos-Jiménez, P., Gomez-Laguna, J., et al. (2017). Listeriolysin S is a streptolysin S-like virulence factor that targets exclusively prokaryotic cells in vivo. MBio 8:10-1128. doi: 10.1128/mbio.00259-17
Ramalheira, R., Almeida, M., Azeredo, J., Brandao, T. R., Almeida, G., Silva, J., et al. (2010). Survival of clinical and food isolates of Listeria monocytogenes through simulated gastrointestinal tract conditions. Foodborne Pathog. Dis. 7, 121–128. doi: 10.1089/fpd.2009.0319
Reichardt, N., Vollmer, M., Holtrop, G., Farquharson, F. M., Wefers, D., Bunzel, M., et al. (2018). Specific substrate-driven changes in human faecal microbiota composition contrast with functional redundancy in short-chain fatty acid production. ISME J. 12, 610–622. doi: 10.1038/ismej.2017.196
Rizzatti, G., Lopetuso, L., Gibiino, G., Binda, C., and Gasbarrini, A. (2017). Proteobacteria: a common factor in human diseases. Biomed. Res. Int. 2017:9351507. doi: 10.1155/2017/9351507
Rodpan, S., Usman, J. N., Koga, Y., and Jongruja, N. (2022). Synergistic effect of nisin with acetic and propionic acids inactivates Bacillus subtilis on meat and potato. Biocatal. Agric. Biotechnol. 41:102317. doi: 10.1016/j.bcab.2022.102317
Rogers, A. W., Tsolis, R. M., and Bäumler, A. J. (2021). Salmonella versus the microbiome. Microbiol. Mol. Biol. Rev. 85, 10–1128. doi: 10.1128/MMBR.00027-19
Rolhion, N., Chassaing, B., Nahori, M. A., De Bodt, J., Moura, A., Lecuit, M., et al. (2019). A Listeria monocytogenes bacteriocin can target the commensal Prevotella copri and modulate intestinal infection. Cell Host Microbe 26, 691–701.e5. doi: 10.1016/j.chom.2019.10.016
Sittipo, P., Shim, J. W., and Lee, Y. K. (2019). Microbial metabolites determine host health and the status of some diseases. Int. J. Mol. Sci. 20:5296. doi: 10.3390/ijms20215296
Vacca, M., Celano, G., Calabrese, F. M., Portincasa, P., Gobbetti, M., and De Angelis, M. (2020). The controversial role of human gut lachnospiraceae. Microorganisms 8:573. doi: 10.3390/microorganisms8040573
Van Deun, K., Pasmans, F., Van Immerseel, F., Ducatelle, R., and Haesebrouck, F. (2008). Butyrate protects Caco-2 cells from Campylobacter jejuni invasion and translocation. Br. J. Nutr. 100, 480–484. doi: 10.1017/S0007114508921693
Wexler, H. M. (2007). Bacteroides: the good, the bad, and the nitty-gritty. Clin. Microbiol. Rev. 20, 593–621. doi: 10.1128/CMR.00008-07
Wolter, M., Steimle, A., Parrish, A., Zimmer, J., and Desai, M. S. (2021). Dietary modulation alters susceptibility to Listeria monocytogenes and Salmonella typhimurium with or without a gut microbiota. mSystems 6:e00717-00721. doi: 10.1128/mSystems.00717-21
Yamaguchi, M., Yang, Y., Ando, M., Kumrungsee, T., Kato, N., and Okazaki, Y. (2018). Increased intestinal ethanol following consumption of fructooligosaccharides in rats. Biomed. Rep. 9, 427–432. doi: 10.3892/br.2018.1150
Yu, X., Wu, X., Shah, N. P., and Xu, F. (2020). Interaction between Bifidobacterium bifidum and Listeria monocytogenes enhances antioxidant activity through oxidoreductase system. LWT 127:109209. doi: 10.1016/j.lwt.2020.109209
Zafar, H., and Saier, M. H. Jr. (2021). Gut Bacteroides species in health and disease. Gut Microbes 13, 1–20. doi: 10.1080/19490976.2020.1848158
Zilelidou, E. A., and Skandamis, P. N. (2018). Growth, detection and virulence of Listeria monocytogenes in the presence of other microorganisms: microbial interactions from species to strain level. Int. J. Food Microbiol. 277, 10–25. doi: 10.1016/j.ijfoodmicro.2018.04.011
Keywords: Listeria monocytogenes , gut microbiota, in vitro digestion, fecal fermentation, microbial interactions
Citation: Kim DW, Singh S, Kim UI, An SH, Je HJ, Lee DY, Yun EJ and Koo OK (2025) Exploring the fate of Listeria monocytogenes in an in vitro digestion and fecal fermentation model: insights into survival during digestion and interaction with gut microbiota. Front. Microbiol. 16:1616720. doi: 10.3389/fmicb.2025.1616720
Edited by:
Junling Shi, Northwestern Polytechnical University, ChinaReviewed by:
Rishi Drolia, Old Dominion University, United StatesCarolina Matto, Ministerio de Ganadería, Agricultura y Pesca, Uruguay
Copyright © 2025 Kim, Singh, Kim, An, Je, Lee, Yun and Koo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ok Kyung Koo, b2tvb0BjbnUuYWMua3I=
 Dong Woo Kim
Dong Woo Kim Saloni Singh1
Saloni Singh1 Dong Young Lee
Dong Young Lee Ok Kyung Koo
Ok Kyung Koo