- 1Department of Infectious Diseases, Zhongnan Hospital of Wuhan University, Wuhan, China
- 2AIDS Research Center, Wuhan University, Wuhan, China
- 3Vision Medicals Co. Ltd., Guangzhou, China
- 4Department of Clinical Laboratory, Zhongnan Hospital of Wuhan University, Wuhan, China
Background: Probe-Capture Metagenomics is a newly developed method for detecting infectious pathogens. However, its application in HIV-infected patients with pulmonary infection remains limited. This study utilized Probe-Capture Metagenomics to analyze lung microbiomes and Drug Resistance Mutations of HIV and bacteria in people living with HIV (PLWH) with pneumonia.
Methods: We retrospectively investigated lung microorganisms in PLWH hospitalized at Zhongnan Hospital of Wuhan University. A combination of bronchoalveolar lavage fluid Probe-Capture Metagenomics and conventional microbiological tests were performed in all patients.
Results: A total of 91 patients were included in the study. Excluding the EB and Torque teno virus, at least two organisms were identified in 85 patients using Probe-Capture Metagenomics combined with conventional microbiological tests. The top six detected organisms were CMV, Pneumocystis jirovecii, Mycobacterium tuberculosis complex, HHV-7, Candida albicans and Aspergillus. For specific organisms, the detection rate of CMV and Candida albicans by Probe-Capture Metagenomics was significantly higher than that of conventional microbiological tests (p < 0.0001). Moreover, the detection rates of CMV (p = 0.0167) and Pneumocystis jirovecii (p = 0.04) in patients with CD4+T count ≥ 200 cells/μL were higher than that with CD4+T count < 200 cells/μL. Importantly, Probe-Capture Metagenomics can uncover potentially clinically relevant drug-resistance mutations linked to HIV and bacteria.
Conclusion: Probe-Capture Metagenomics provides a promising method of detecting suspected opportunistic infections in PLWH with pneumonia, especially for mixed infections and rare microorganisms. In addition, Probe-Capture Metagenomics was a potential valuable tool for genotyping resistance testing of HIV and bacteria.
1 Introduction
Lower respiratory tract infections can be caused by a variety of microbes including bacteria, fungi and viruses, which was one of the main causes of morbidity and mortality among people living with HIV (PLWH), especially in low-income countries countries (Zarrinfar et al., 2012; Zarrinfar et al., 2014; Zarrinfar et al., 2016). Pneumonia diagnoses in PLWH with immune dysfunction pose an additional challenge as these patients are susceptible to mixed respiratory pathogens (Di Pasquale et al., 2019; Wang et al., 2019). Patients with mixed pulmonary infection may exhibit a more severe and diverse clinical manifestations compared to those with a single pathogen infection. In addition, conventional microbiological tests (CMTs) of detecting microorganisms have several limitations, including low positivity rates, long culture times, susceptibility to interference from empirical antibiotic treatment, and difficulties in detecting uncommon or emerging pathogens (Xu et al., 2022). Each of these causes contributes to the complexity of the diagnosis. Therefore, timely and precise identification of multi-pathogenic pneumonia is crucial for optimizing treatment strategies, improving the prognosis, and reducing mortality rates.
In recent years, metagenomic high-throughputhigh-throughput sequencing (mHTS) has been around for over two decades now (since the onset of the 2nd gen sequencing with 454), gradually gaining utility in clinical diagnosis expanded to the field of infectious diseases due to its rapid, unbiased and highly sensitive for identification of pathogens (Gu et al., 2019; Wüthrich et al., 2016; Shi et al., 2022). It can simultaneously detect various infectious pathogens, which provides additional valuable information for the detection of pathogens (Gu et al., 2019; Wüthrich et al., 2016; Shi et al., 2022). However, the large proportion of host DNA in human cells may mask other nucleic acids and affect the detection of certain microbial populations. DNA and RNA need to be processed separately, and the application of mHTS has mainly detected DNA microorganisms previously. Currently, RNA viruses, such as influenza virus, are also part of the important causes of lung infections. We developed a Probe-Capture Metagenomics assay for detecting infectious pathogens that integrates metagenomics techniques with magnetic bead capture technology (Sun et al., 2025). It employs a hybridization capture-based approach using pathogen-specific probes to enrich microbial genomic material while simultaneously depleting host DNA, and DNA/RNA library detection could be performed simultaneously. Thus, in addition to the advantages of mHTS, Probe-Capture Metagenomics can also detect RNA viruses and microbial resistance genes. However, the application of Probe-Capture Metagenomics in PLWH with pulmonary infection remains limited.
Therefore, the aim of this study was to explore the microbiome spectrum in PLWH with pneumonia using Probe-Capture Metagenomics, and to initially investigate the resistance gene phenotypes of respiratory pathogens and HIV in PLWH with pneumonia.
2 Materials and methods
2.1 Study design and participants
Bronchoalveolar Lavage Fluid (BALF) samples from PLWH with pneumonia were submitted to Probe-Capture Metagenomics, which were retrospectively reviewed from the Zhongnan Hospital of Wuhan University from 1 may 2022 to 30 December 2022. Patients who met the following criteria were considered as pneumonia: (1) exhibited at least one of the following manifestations: fever; cough, expectoration, dyspnea or other respiratory symptoms; (2) chest imaging suggested pneumonia; (3) determined by the consensus of two experienced senior clinicians based on the clinical manifestation, laboratory tests, chest imaging and response to antimicrobial treatment. The exclusion criteria were as follows: (1) Probe-Capture Metagenomics of BALF were not performed; (2) age < 18 years old; (3) Hospital stay less than 24 h (Figure 1). At the same time, CMTs, including chest imaging, smear and culture of bacteria and fungi, 1,3-β-D glucan (G) assay (G test) (β-D-glucan detection kit, Shanghai Fuxing Changzheng Medical Science Co., Ltd., China) and galactomannan (GM) assay (Platelia™ Aspergillus Ag, Bio-Rad, United States), polymerase chain reaction (PCR) test [including cytomegalovirus (CMV), EB virus, Pneumocystis jirovecii (P. jirovecii), Mycobacterium tuberculosis complex (MTB)], tuberculin skin test (TST), interferon-gamma release assay (T-SPOT.TB kit, TB.300, Oxford Immunotec, UK), acid-fast stain (TB Auramine-Rhodamine kit, K12345, Thermo Fisher, United States) and GeneXpert MTB/RIF (Xpert) (Cepheid. Xpert® MTB/RIF Assay, Cepheid, United States), were performed for pulmonary infection. The sequence and conditions of the primer pairs used for P. jirovecii PCR was described in one previous study (Chen et al., 2024). The primers for CMV PCR were as follows: Forward: 5’-CGCGACTTGACCTGGACTAC-3′, Reverse: 5’-TGGTGGCAGTGTACCCGTAA-3′. The conditions used were: 45 cycles of denaturation for 5 min at 95°C; annealing for 15 s at 95°C; and extension for 30 s at 60°C. The primers for EB PCR were as follows: Forward: 5’-GCCAGAGGTAAGTGGACTTT-3′, Reverse: 5’-TCTTGGTGAGCGGTAGTAAA-3′. The conditions used were: 40 cycles of denaturation for 3 min at 95°C; annealing for 10 s at 95°C; and extension for 30 s at 58°C. The primers for MTB PCR were as follows: Forward: 5’-CCTGCGAGCGTAGGCGTCGG-3′, Reverse: 5’-CTCGTCCAGCGCCGCTTCGG-3′. The conditions used were: 50 cycles of denaturation for 10 min at 95°C; annealing for 15 s at 95°C; and extension for 60 s at 62°C. The date of collection, symptoms and test results were included in the clinical data. All data was anonymized in our study. These patient clinical data was included as Supplementary Material. Every fungi/bacteria/virus evaluated in CMTs were present in the databases of Probe-Capture Metagenomics. The diagnosis was made based on clinical symptoms, laboratory findings, microbiologic examination, chest radiology, and the response to treatment. This study was approved by the institutional ethics committee of Zhongnan hospital of Wuhan university (2022101). All subjects have voluntarily taken part in the research and signed the informed consent form. The Declaration of Helsinki’s standards and any applicable laws were followed when conducting the study.
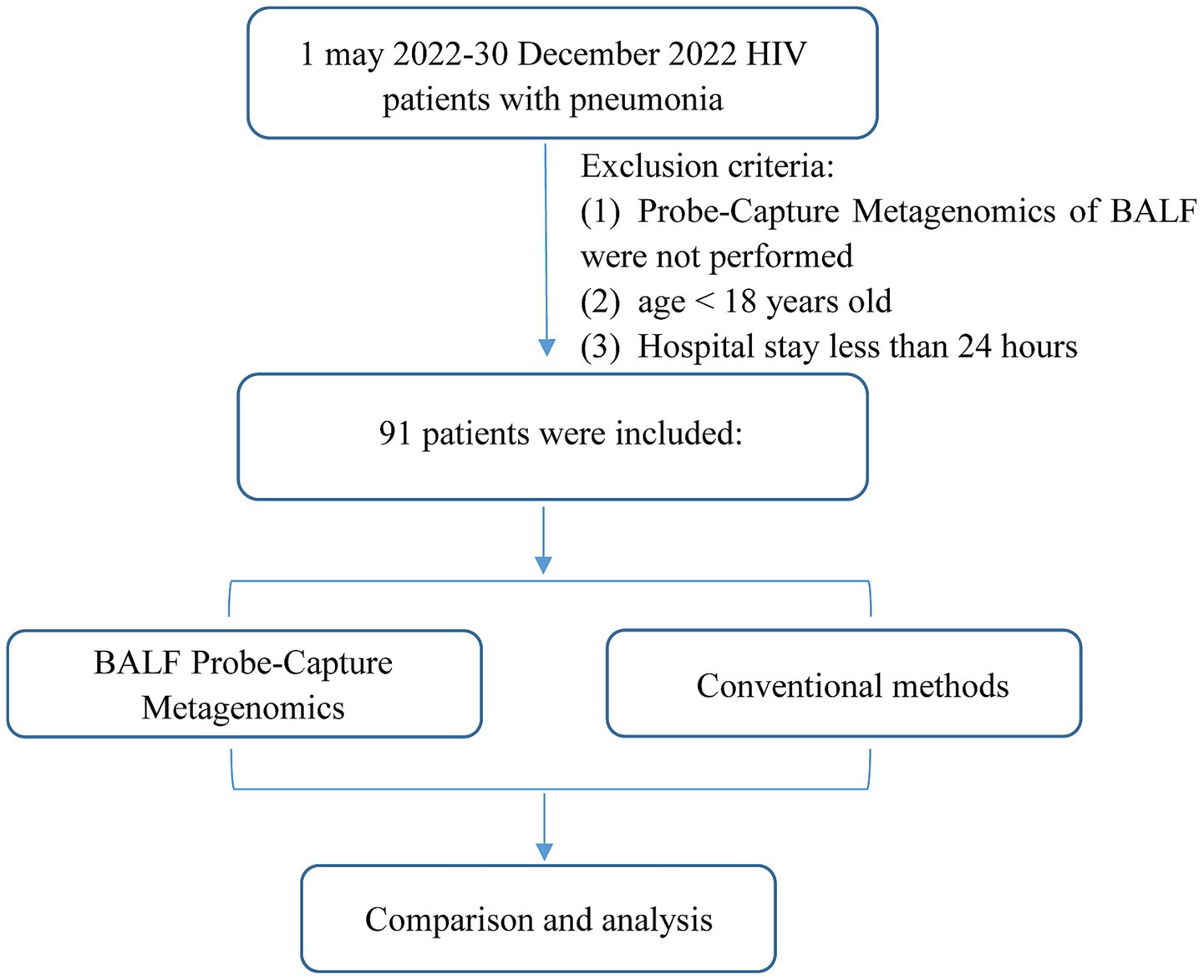
Figure 1. Flowchart of case selection. A total of 91 HIV-infected cases in the infection department were selected for further analysis.
2.2 Data collection
Clinical data were retrospectively obtained from medical records. These following data were extracted: age, gender, HIV infection status, antiretroviral therapy (ART), antibiotic use within 3 months, lymphocyte count, CD4+ T lymphocyte count (CD4 count), HIV viral load, C-reactive protein (CRP), Interleukin-6 (IL-6), procalcitonin (PCT), erythrocyte sedimentation rate (ESR), D-dimer, lactate dehydrogenase (LDH), alanine aminotransferase (ALT), albumin, creatinine, results of CMTs, chest imaging, and Probe-Capture Metagenomics tests.
2.3 Probe design for pathogen detection
To optimize probe design, we employed HUBDesign, a bioinformatics pipeline that generates probes from representative sequences at multiple taxonomic levels. This method allows for precise targeting and enrichment of specific clades, facilitating the detection of both known and novel organisms. The default parameters of HUBDesign were applied in this study. We designed a comprehensive probe library targeting up to 167 clinically relevant pathogenic microorganisms. And the pathogen-specific probe set includes 62 bacteria and 41 antimicrobial resistance genes, 16 fungi, 14 parasites, 15 DNA viruses, and 60 RNA viruses, collectively covering >95% of pathogens detected in clinical samples. A total of 640 probes were designed including 517 probes targeting pathogens and 183 probes targeting resistance genes. Pathogens used for probe design are provided in Supplementary Materials. Leveraging sequence homology and DNA hybridization, this pipeline has been successfully demonstrated in designing and validating probe sets for two key applications: (1) comprehensive detection of all sequenced coronaviruses, and (2) identification of bacterial pathogens associated with sepsis. The Probe-Capture assay developed in this work has been granted an invention patent by the China National Intellectual Property Administration (patent name: Group, method, kit and application of Probe Capture for detecting pathogenic microorganisms, patent No. ZL2020116367800).
2.4 Nucleic acid extraction, library preparation, probe hybridization and sequencing
DNA was extracted from all samples using a QIAamp® UCP Pathogen DNA Kit (Qiagen) following the manufacturer’s instructions. Human DNA was removed using Benzonase (Qiagen) and Tween20 (Sigma). Using a Qubit dsDNA HS Assay Kit (Invitrogen, United States), the isolated DNA was quantified. Total RNA was extracted with a QIAamp® Viral RNA Kit (Qiagen) and ribosomal RNA was removed by a Ribo-Zero rRNA Removal Kit (Illumina). cDNA was generated using reverse transcriptase and dNTPs (Thermo Fisher). Libraries were constructed for the DNA and cDNA samples using a Nextera XT DNA Library Prep Kit (Illumina, San Diego, CA). An aliquot of 750-ng library from each sample was used for hybrid capture-based enrichment of microbial probe one rounds of hybridization (SeqCap EZ Library, Roche, United States). Purification and size selection were carried out following the double-sided bead purification procedure. Library was quality assessed by Qubit dsDNA HS Assay kit followed by High Sensitivity DNA kit (Agilent) on an Agilent 2,100 Bioanalyzer. Library pools were then loaded onto an Illumina Nextseq CN500 sequencer for 75 cycles of single-end sequencing to generate approximately 20 million reads for each library. For negative controls, we also prepared sterile deionized water in parallel with each batch to serve as non-template controls (NTC), using the same protocol.
2.5 Bioinformatics analyses
Raw reads were adapter trimmed and quality filtered using Trimmomatic (v0.39). All reads longer than 40 base pairs with a minimum phred quality score of 30 were kept for further analysis (Bolger et al., 2014). Low-complexity reads were removed by fastp (v0.23.4) default settings (Chen et al., 2018). The human sequence data were identified and eliminated by mapping them to the hg38 reference genome utilizing Burrows-Wheeler Aligner software (BWA-0.7.17) (Li and Durbin, 2009). Taxonomic classification of sequencing reads was performed using Kraken2 (v2.1.3) with default parameters, including a confidence threshold of 0.2 (-confidence 0.2) and 40 threads (-threads 40). The Kraken2 reference database was constructed using the kraken2-build –standard pipeline, incorporating complete genomes and assemblies from the NCBI RefSeq archaea, bacteria, viruses, plasmids, human (GRCh38), UniVec_Core, protozoa, fungi and plants, all downloaded on December 28, 2024. To enhance microbial representation, genomes of Pneumocystis jirovecii (GenBank accession no. GCA_001477535.1) were manually incorporated into the database, as detailed in Supplementary Table 1. To mitigate spurious alignments arising from PCR duplicates or low complexity regions, microbial reads were subsequently aligned against the same reference database using the Burrows–Wheeler Aligner (BWA, v0.7.17) with default parameters. Only alignments exhibiting a nucleotide identity of ≥96% to the reference genome were considered valid. Furthermore, reads aligning to multiple loci within the same genus were excluded from downstream analyses to prevent ambiguous taxonomic assignments. To remove the mistakes brought on by different sequencing depths between samples, we normalized the sequencing reads using reads per million (RPM). The clinical reportable range (CRR) for pathogens was established according to the following three references indicated in a previous study (Jing et al., 2021): I. Johns Hopkins ABX Guide,1 II. Manual of Clinical Microbiology, and III. clinical case reports or research articles published in peer-reviewed journals.
The clinical reportable range (CRR) for pathogens was established according to a previous study (Sun et al., 2025). The suspected pathogens were determined according to the following rules (Wilson et al., 2019): A virus was considered positively detected if RPM ≥ 3 covered at least three non-overlapping regions in the genome. For identification of bacteria, fungi, and parasites, we developed a RPM ratio metric, defined as RPM-r = RPM sample/RPMNTC, or RPM ratio = RPM sample if RPMNTC = 0. A positive detection of a bacterium, fungus, or parasite required an RPM ratio ≥10. The results can be considered positive when they are consistent with the pathogenicity of positive microorganisms, clinical characteristics, and therapeutic efficacy.
2.6 Drug resistance mutations analysis
The Stanford HIV Genotypic Resistance Interpretation Algorithm2 was used to identify mutations associated with HIV drug resistance. HIVDR mutations linked with integrase strand transfer inhibitor (INSTI), reverse-transcriptase inhibitor (RT), and protease inhibitor (PI) were reported with threshold of mutation detection threshold (MDT) ≥ 1%, and divided into three categories: major, accessory, and other. The estimated drug resistance based on the total point score was designated: susceptible, potential low-level resistance, low-level resistance, intermediate resistance, and high-level resistance.
To identify clinically important antimicrobial resistance bacteria, such as carbapenem-resistant Enterobacterales, carbapenem-resistant Acinetobacter baumannii, carbapenem-resistant Pesudomonas aerugeinosa, third-generation cephalosporin-resistant Enterobacterales, extended-spectrum β-lactamases (ESBL) producing Enterobacteriaceae, vancomycin-resistant Enterococcus faecium, methicillin-resistant Staphylococcus aureus, and so on, the specimen’s sequence was screened and aligned with sequences of antibiotic resistance genes in the Comprehensive Antibiotic Resistance Database (CARD; https://card.mcmaster.ca) (Jia et al., 2017). This analysis was only performed when the targeted bacterial sequence surpassed a threshold of 1,000. And the relationship between antibiotic resistance genes and the detected bacteria was evaluated using the CARD database (Alcock et al., 2023).
2.7 Statistical analysis
SPSS 25.0 software (IBM Corp., Armonk, NY, United States) was used for statistical analysis. The numerical variables were presented as medians and interquartile ranges and nominal variables as counts and percentages. Comprehensive clinical diagnosis and determination of microbial aetiology were used as reference standards. The McNemar test was used to compare the diagnostic performance of CMTs and Probe-Capture Metagenomics. Fisher’s exact test was used to test for association of pathogen with the categorical variables of CD4 count (≥200 vs. < 200 cells per μL). All tests were two-tailed, and the p value of <0.05 was considered statistically significant.
3 Results
3.1 Patient characteristics
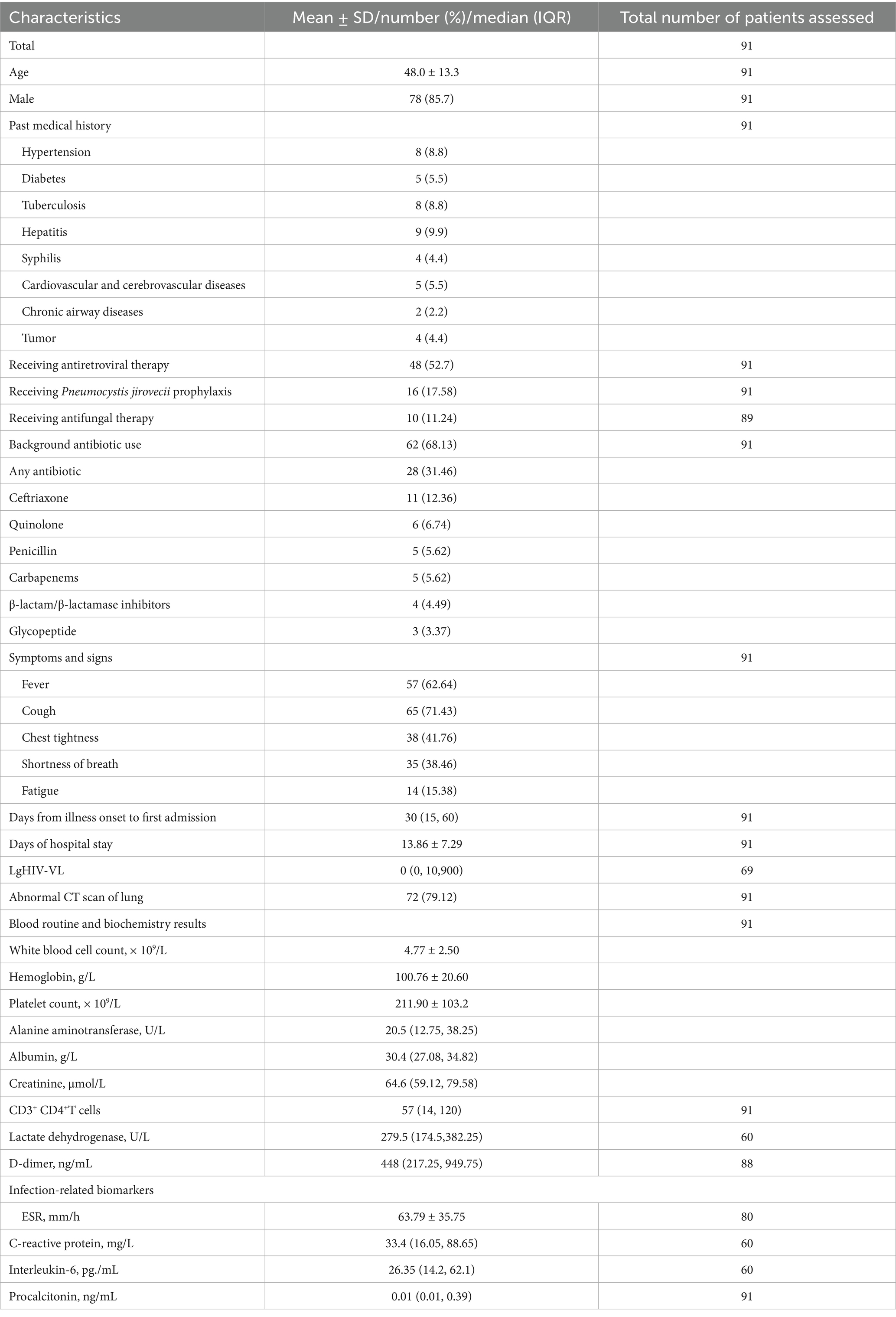
During the study period, a total of 91 HIV patients (mean age: 48 years) including 78 men (85.7%) met the inclusion criteria. Upon admission to hospital, the median CD4 count of 91 patients was 57 cells/μL. 48 (52.7%) were taking antiretroviral therapy, and 16 (17.58%) had been receiving P. jirovecii pneumonia prophylaxis. 10 (11.24%) out of 89 patients were receiving empirical antifungal therapy, and 62 (68.13%) out of 91 patients were receiving empirical antibiotic therapy. 57 patients (62.64%) had fever, and 72 patients (79.12%) of CT scan of the lung were abnormal. Detailed clinical characteristics are shown in Table 1.
3.2 Distribution of pathogens
Of 91 samples, excluding the EB and Torque teno virus, 6 had single pathogen and 85 had at least two microorganism (Figure 2A) detected by Probe-Capture Metagenomics combined with CMTs. In addition, the top six organisms were CMV, P. jirovecii, MTB, HHV-7, Candida albicans (C. albicans) and Aspergillus (Figures 2B–E).
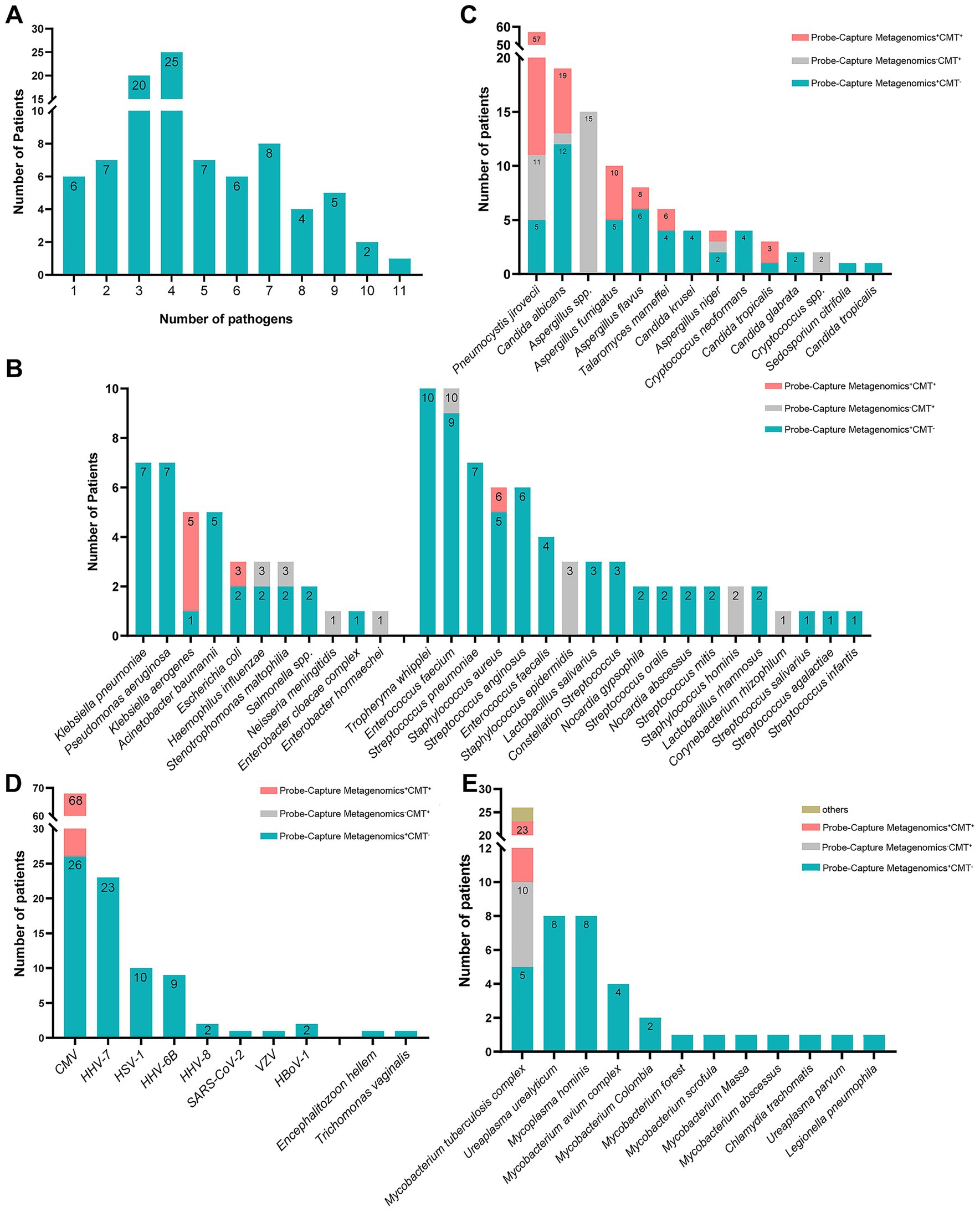
Figure 2. The organism spectrums of pulmonary detected by probe-capture metagenomics and CMTs in PLWH with pneumonia. (A) number of patients in different amounts of detected organisms; (B) number of patients in different amounts of detected bacterial species; (C) number of patients in different amounts of detected fungus species; (D) number of patients in different amounts of detected virus species; (E) number of patients in different amounts of detected other atypical pathogen species. PLWH, people living with HIV; CMTs, conventional microbiological tests; CMV, cytomegalovirus; HHV-7, human herpes virus 7; HSV-1, herpes simplex virus 1; HHV-6B, human herpes virus 6B; HHV-8, human herpes virus 8; VZV, varicella-zoster virus; HBoV-1, human bocavirus 1.
As shown in Figure 2B, Tropheryma whipplei (T. whipplei), Enterococcus faecium, Klebsiella pneumoniae, Pseudomonas aeruginosa and Streptococcus pneumoniae were more prevalent in the detected bacteria spectrum among PLWH with pulmonary infection.
As shown in Figure 2C, P. jirovecii and C. albicans were more common in the detected fungus spectrum. Among the detected fungus, two kinds (Aspergillus and Cryptococcus) of them were detected by CMTs only, five kinds (Candida krusei, Cryptococcus neoformans, Candida glabrata, Sedosporium citrifolia and Candida tropicalis) of them were detected by Probe-Capture Metagenomics only. In the cases of P. jirovecii detection, 5 cases were diagnosed by Probe-Capture Metagenomics alone, and 6 cases were diagnosed by CMTs alone. In the cases of C. albicans detection, 12 cases were diagnosed by Probe-Capture Metagenomics alone, and 1 case was diagnosed by CMTs alone. In the cases of Aspergillus fumigatus detection, 5 cases were diagnosed by Probe-Capture Metagenomics alone. In the cases of Aspergillus flavus detection, 6 cases were diagnosed by Probe-Capture Metagenomics alone and 2 cases were diagnosed both by Probe-Capture Metagenomics and CMTs. In the cases of T. marneffei detection, 4 cases were diagnosed by Probe-Capture Metagenomics alone. In the cases of Aspergillus niger detection, 2 cases were diagnosed by Probe-Capture Metagenomics alone, and 1 case was diagnosed by CMTs alone. In the cases of Candida tropicalis detection, 1 case was diagnosed by Probe-Capture Metagenomics alone.
As shown in Figure 2D, CMV and HHV-7 were more common in the detected virus spectrum. In the cases of CMV detection, 26 case was diagnosed by Probe-Capture Metagenomics alone. In addition, except for CMV, the remaining viruses were detected only by Probe-Capture Metagenomics.
As shown in Figure 2E, MTB and Mycobacterium avium complex (MAC) were more common in the detected mycobacterium spectrum. In the cases of MTB detection, 5 case was diagnosed by Probe-Capture Metagenomics alone, 13 cases were diagnosed both by Probe-Capture Metagenomics and CMTs, and 5 cases were diagnosed by CMTs alone. In addition, all of the nontuberculous mycobacteria (NTM), Mycoplasma hominis, Chlamydia trachomatis, Ureaplasma and Legionella pneumophila were detected only by Probe-Capture Metagenomics.
3.3 Comparison of diagnostic performance among probe-capture metagenomics and CMTs in PLWH with pneumonia
As shown in Table 2 and Supplementary Table 2, Probe-Capture Metagenomics demonstrated superior detection performance compared to CMTs for CMV and C. albicans infections in PLWH with pulmonary infections. Using the McNemar test to compare paired binary outcomes (positive/negative) from Probe-Capture Metagenomics and CMTs, Probe-Capture Metagenomics showed significantly higher detection rates for CMV (p < 0.001) and C. albicans (p < 0.001). For CMV viremia, Probe-Capture Metagenomics achieved a sensitivity of 100% (95% CI, 94.65–100%) compared to 67.65% for CMTs (95% CI, 55.84–77.56). Similarly, for C. albicans infection, Probe-Capture Metagenomics sensitivity was 94.74% (95% CI, 75.36–99.73), significantly higher than CMTs at 31.58% (95% CI, 15.36–53.99; p < 0.001). In contrast, BALF Probe-Capture Metagenomics showed comparable detection performance to CMTs for P. jirovecii, MTB, and Aspergillus infections (p > 0.05, McNemar test).
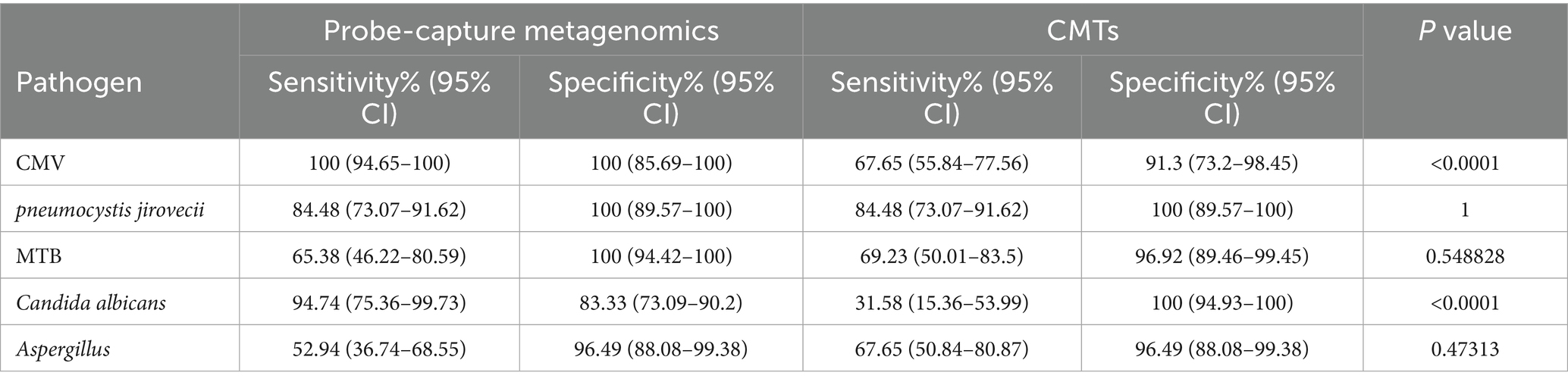
Table 2. Comparison of diagnostic performance among probe-capture metagenomics and CMTs in PLWH with pulmonary infection.
3.4 Comparison analysis of lung microorganisms in patients with CD4+T count < 200 cells/μL and those with a count ≥ 200 cells/μL
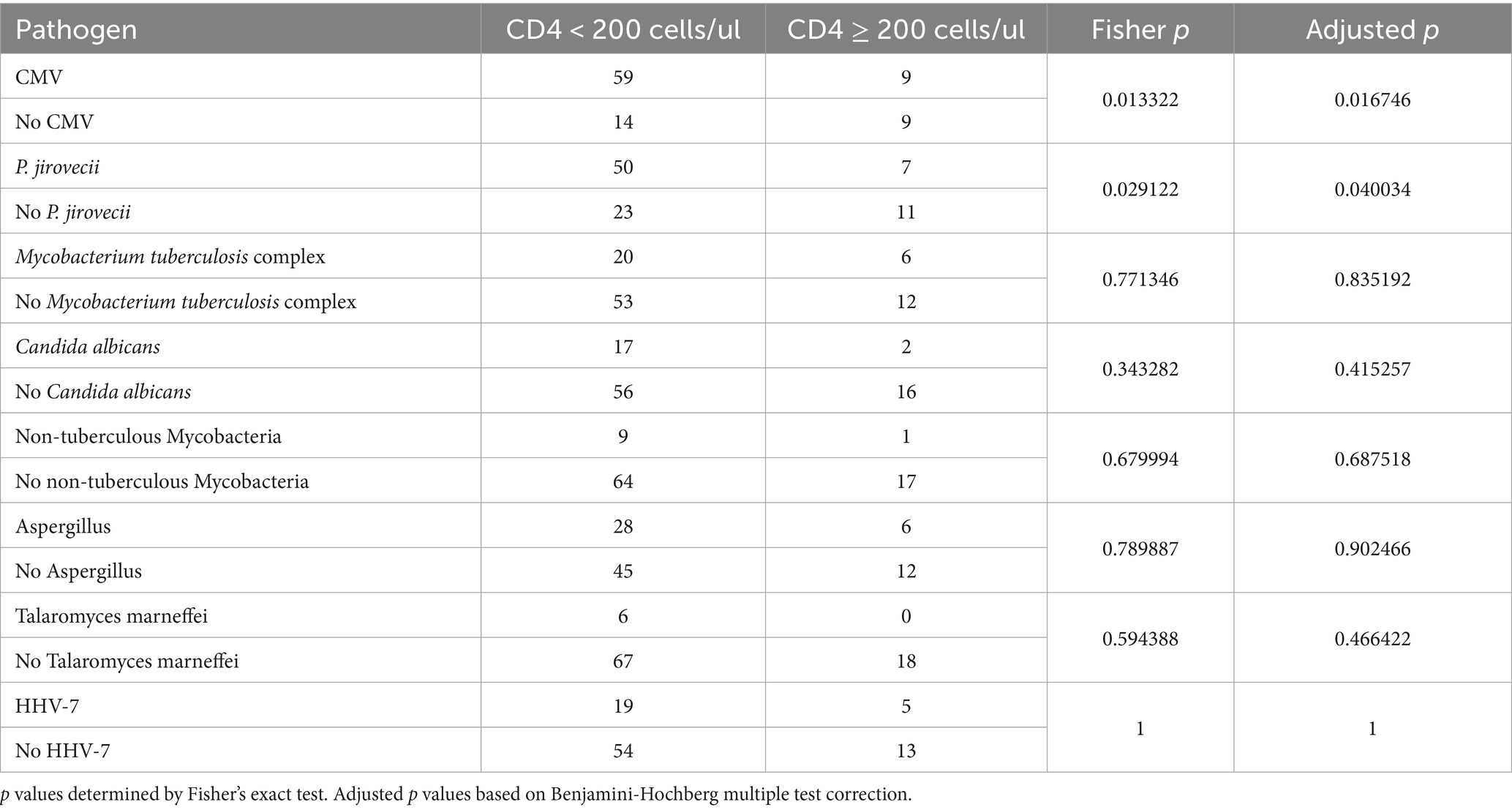
We further compared the lung microorganisms of PLWH with pulmonary infection between patients with CD4+T count < 200 cells/μL to those with a count ≥ 200 cells/μL. As shown in Table 3, the detection ratios of CMV and P. jirovecii in patients with CD4+T count < 200 cells/μL were significantly higher than that of CD4+T count ≥ 200 cells/μL (p < 0.05). However, there are no significant differences of the detection ratios of MTB, C. albicans, NTM, Aspergillus, T. marneffei, HHV-7 and EBV between patients with CD4+T count < 200 cells/μL and CD4+T count ≥ 200 cells/μL (p > 0.05).
3.5 The drug-resistance mutations (DRMs) associated with HIV and bacteria detected by probe-capture metagenomics in PLWH with pneumonia
Furthermore, 91 samples were successfully sequenced to detect DRMs using Probe-Capture Metagenomics. HIV sequences were detected in 26 BALF samples. Detailed results are presented in the Supplementary Materials. As shown in Figure 3, DRMs (V179E, V179D) for nonnucleoside reverse transcriptase inhibitors (NNRTI) were found in the BALF of 3 patients. The scores of DRMs associated with HIV resistance were all 10 in each cases, indicating that HIV in these three patients had potentially low-level resistance to NNRTI (Figure 3B). In addition, the BALF of 27 patients exhibited the presence of antimicrobial resistance genes (ErmB, blaTEM, ErmC, mecA, blaCTX-M, blaSHV), which were identified by Probe-Capture Metagenomics (Table 4).
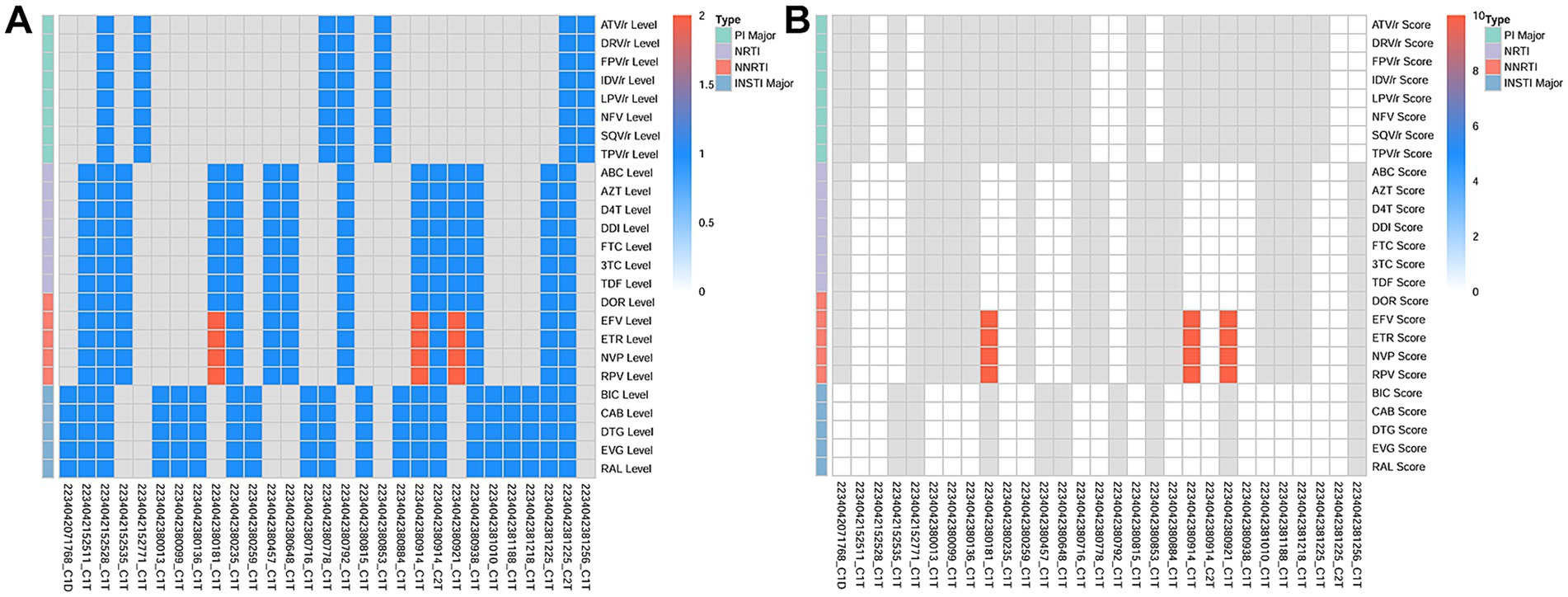
Figure 3. The DRMs associated with HIV detected in BALF by probe-capture metagenomics in PLWH with pneumonia. (A) The HIV resistance level. 1 represents susceptible. 2 represents potential low-level resistance. (B) The HIV resistance score of drug resistance analysis. A score of 0–9 is susceptible and a score of 10–14 is potential low-level resistance. The gray color indicates that no detection sequence is covered. DRMs, drug-resistance mutations; BALF, Bronchoalveolar Lavage Fluid; PLWH, people living with HIV; PIs, protease inhibitors; NRTI, nucleotide reverse transcriptase inhibitor; NNRTI, non-nucleoside reverse transcriptase inhibitors; INSTI, integrase inhibitor; ATV/r, Atazanavir/Ritonavir; DRV/r, Darunavir/Ritonavir; FPV/r, Fosamprenavir/Ritonavir; IDV/r, Indinavir/Ritonavir; LPV/r, Lopinavir/Ritonavir; NFV/r, Nelfinavir/Ritonavir; SQV/r, Saquinavir/Ritonavir; TPV/r, Tipranavir/Ritonavir; ABC, Abacavir; AZT, Zidovudine; D4T, Stavudine; DDI, Didanosine; FTC, Emtricitabine; 3TC, Lamivudine; TDF, Tenofovir Disoproxil Fumarate; DOR, Doravirine; EFV, Efavirenz; ETR, Etravirine; NVP, Nevirapine; RPV, Rilpivirine; BIC, Bictegravir; CAB, Cabotegravir; DTG, Dolutegravir; EVG, Elvitegravir; RAL, Raltegravir.
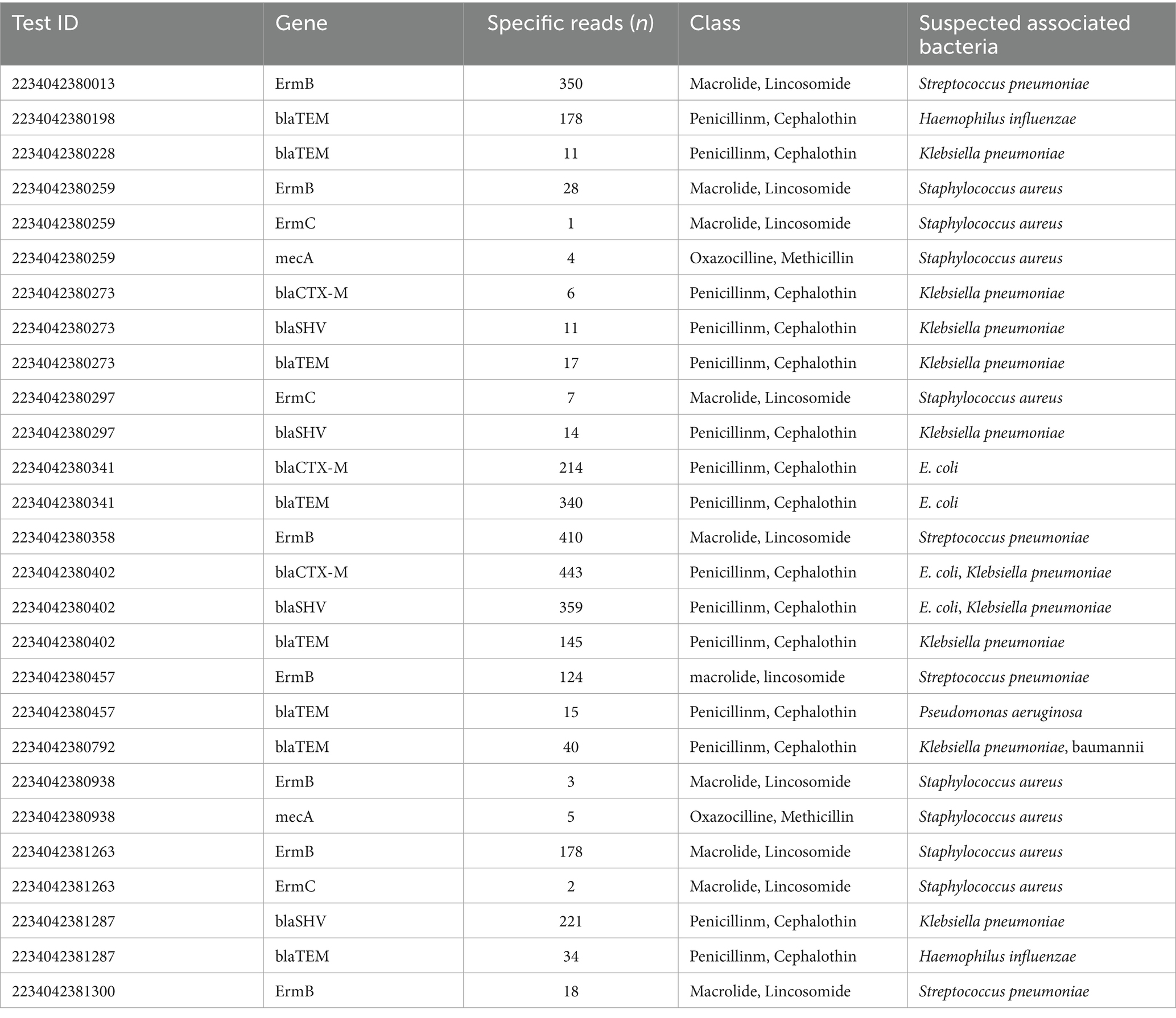
Table 4. Lower respiratory tract antimicrobial resistance genes identified by probe-capture metagenomics in 27 patients.
4 Discussion
Various infections or frequent co-infections, including bacterial, fungal, viral and other atypical pathogens, are common in PLWH with severely immunosuppression (Chen et al., 2024). By utilizing Probe-Capture Metagenomics technology based on DNA/RNA library detection simultaneously, we facilitated the understanding of pneumonia microorganisms in the unique vulnerable population of PLWH. In addition to clinical manifestations and CMTs, the results of Probe-Capture Metagenomics can also provide potential reference for the clinical diagnosis and treatment of PLWH with pneumonia. Furthermore, its application in detecting drug-resistance mutations of HIV and bacteria was also initially explored for PLWH with pneumonia.
Currently, CMTs used in the clinic include the detection of specific antigens or antibodies, PCR, microscopy observation and culture. And culture and PCR in CMTs remain the primary detection methods, known as the gold standard for confirming microbial pathogen infection. However, there are certain limitations of CMTs, including low sensitivity, time-consuming procedures, difficulties in simultaneous detection of multiple pathogens (Jain et al., 2015; Labelle et al., 2010). In addition, there are numerous pathogens that cannot be effectively cultured or cultured for too long time within hospital settings (Wilson et al., 2014; Bateman et al., 2020; Sawatpanich et al., 2022; Getahun et al., 2007; Xie et al., 2018), such as P. jirovecii, NTM and MTB. Therefore, prompt appropriate diagnosis and treatment of pneumonia with co-infections in PLWH remains challenging. Probe-Capture Metagenomics is an emerging pathogen detection method based on molecular biology. This approach enables the targeting and enrichment of specific clades for the identification of both known and previously unidentified organisms. In our study, the Probe-Capture Metagenomics comprised a total of 640 probes, including 517 pathogen-specific probes and 183 antimicrobial resistance gene-targeting probes, which could cover >95% of pathogens detected in clinical samples. The top six detected organisms were CMV, P. jirovecii, MTB, HHV-7, C. albicans and Aspergillus using BALF Probe-Capture Metagenomics combined with CMTs. Furthermore, the sensitivity of Probe-Capture Metagenomics for the detection of CMV and C. albicans was markedly higher than that of CMTs. However, the sensitivity of BALF Probe-Capture Metagenomics was comparable to CMTs for diagnosing the infections of P. jirovecii, MTB, and Aspergillus in PLWH with pulmonary infection. While Probe-Capture Metagenomics offers technological advantages, its high cost currently limits practical application, especially in low- and middle-income countries. Therefore, the CMTs, such as PCR and Xpert, may be more appropriate for diagnosing P. jirovecii, Aspergillus and MTB in low- and middle-income areas. Furthermore, future development of additional CMTs with enhanced pathogen detection capabilities remains imperative. Importantly, more than 40 pathogens were detected solely by Probe-Capture Metagenomics, including all of the NTM, T. whipplei. Studies have shown that compared with HIV-uninfected patients, significantly higher burden of T. whipplei colonization and NTM infection in the lungs have been observed in HIV-infected patients (Tan et al., 2023; Lozupone et al., 2013). Therefore, Probe-Capture Metagenomics showed obvious advantages in the detection of NTM and T. whipplei compared with CMTs. However, pulmonary infection caused by T. whipplei is rarely reported in PLWH (Yan et al., 2021; Stein et al., 2013). Whether T. whipplei could cause pathological changes in the lung should be further explored. Other commonly pathogens, such as Aspergillus, T. marneffei, could also be detected with high sensitivity. Moreover, the results can be obtained within 24 h, which was shorter than multiple CMTs. These results suggested that Probe-Capture Metagenomics combines the advantages of the speed, unbiased, high sensitivity and specificity of mHTS to detect multiple pathogens simultaneously (Han et al., 2019). Therefore, Probe-Capture Metagenomics which represent a valuable adjunct to CMTs can be indicated in the following clinical scenarios: (1) when CMTs fail to identify suspected opportunistic pathogens, especially in immunocompromised hosts, atypical presentations, or situations requiring rapid diagnosis; (2) for suspected polymicrobial co-infections; and (3) when CMTs has limited coverage, such as for emerging viruses or atypical bacteria. Notably, multiple organisms were diagnosed by Probe-Capture Metagenomics or CMTs alone in our study, which showed that it is still impossible for Probe-Capture Metagenomics to completely replace CMTs in clinical application. And detection from Probe-Capture Metagenomics cannot be considered the same as confirming it as an infectious agent, just that its genetic material could be found in the samples. Thus, Probe-Capture Metagenomics alone cannot distinguish pathogens between colonization and infection.
Through our study, a deeper understanding of the microbial spectrums of pulmonary was gained in PLWH with pneumonia. T. whipplei, E. faecium, K. pneumoniae, P. aeruginosa and S. pneumoniae in bacteria spectrum, P. jirovecii and C. albicans in fungus spectrum, CMV and HHV-7 in virus spectrum, MTB and MAC in mycobacterium spectrum were more commonly detected among PLWH with pulmonary infection. These results were partly similar with the study of Tan et al., which also showed that MTB and MAC were more common in the mycobacterium spectrum (Tan et al., 2023). And the detection ratios of CMV and P. jirovecii in PLWH with CD4+T count < 200 cells/μL were significantly higher than that with CD4+T count ≥ 200 cells/μL, which indicated that the pathogen spectrum of pulmonary infection varies in PLWH according to their immune conditions (Xu et al., 2022; Tan et al., 2023). In addition, HIV or other RNA viruses such as the SARS-CoV-2 can also be detected by Probe-Capture Metagenomics, which was superior to conventional mHTS method mainly detecting DNA viruses. Therefore, our findings could help physicians early recognize the pathogens of pulmonary infection in PLWH, which facilitates rapid and correctly empirical treatment for favourable prognoses of opportunistic infections. Notably, the detection rates of SARS-CoV-2 and influenza virus were low in our study. Since the onset of Omicron, all variants are from the same lineage and one of these have a higher affinity against cells in the upper respiratory tract, so lung cells are secondary (contrary to Delta and earlier variants). While Delta relies on ACE2-TMPRSS2-mediated entry in pulmonary tissues, the Omicron variants do not induce cell syncytia in vitro and favoured a TMPRSS2-independent endosomal entry pathway (Meng et al., 2022; Willett et al., 2022).
Currently, although effective ART has significantly improved survival in PLWH (Poorolajal et al., 2016), it only control the HIV level in PWLH below detection threshold and can not achieve a complete cure for HIV (Ryom et al., 2022; Chun et al., 1997; Lu et al., 2018). One important reason is that ART cannot eliminate the latent HIV reservoir in multiple cellular at diverse anatomical sites which can undergo clonal expansion and persist for many years (Ryom et al., 2022; Chun et al., 1997; Lu et al., 2018). Therefore, a comprehensive understanding of HIV reservoirs and their kinetics would be crucial in formulating efficacious to achieve a cure for HIV infection. Some sequencing methods to measure and characterize the HIV reservoir are also gradually emerging (Moar et al., 2023). But one single approach cannot be universally used due to its own advantages and limitations. Nowadays, researchers have widely recognized that in tissues, potential HIV reservoir has also been found in seminal vesicles, the urethra, adipose, liver, lung, bone marrow in femoral head as well as the central nervous system (Chen et al., 2022). Researches have demonstrated that the lung also serves as an HIV reservoir. HIV primarily infects CD4+T cells, while also capable of infecting lung-resident immune cells such as alveolar macrophages and dendritic cells (Cribbs et al., 2020). HIV-related lung injury and disease can occur secondary to a number of mechanisms including altered pulmonary and systemic inflammatory pathways, viral persistence in the lung, oxidative stress with additive effects of smoke exposure, microbial translocation, and alterations in the lung and gut microbiome (Cribbs et al., 2020). Our results suggested that Probe-Capture Metagenomics can detect HIV in the BALF of PLWH with pneumonia, providing further evidence that the lungs act as an important HIV reservoir, and Probe-Capture Metagenomics provides a supplement detection method for finding reservoirs of latent HIV.
HIV drug resistance (HIVDR) will impair the efficacy of ART, leading to virologic failure, and hinder the progress in HIV/AIDS treatment (Zhu et al., 2024). HIVDR assays are usually performed through Sanger sequencing, which could detect the frequency of minority variants at a 15–20% in PWLH (Casadellà and Paredes, 2017). In recent years, studies have shown that HTS was able to detect more of the minority drug resistance variants than Sanger (Li et al., 2021). Furthermore, although serum HIV drug resistance testing (HIVDR) are common, HIVDR in BALF still holds greater clinical significance. Different resistance mutations can be detected simultaneously in the blood and the lung of HIV-1 infected individuals on antiretroviral therapy (White et al., 2004). HIV strains in the lung may evolve independently from those in blood, developing unique drug resistance mutation profiles (White et al., 2004; Shahid et al., 2022). Consequently, even when blood viral load is well-controlled, latent virus in the lungs may still lead to localized viral replication and treatment failure. In our study, we discovered DRMs for NNRTI in 3 patients by Probe-Capture Metagenomics, which was targeted towards the HIV genome. This suggested that Probe-Capture Metagenomics could be used as a valuable additional tool to gather further data on HIVDR. However, the detection rate was not high in our study. In the future, as the gene probes for HIV resistance increase in Probe-Capture Metagenomics, its application value will improve in PLWH.
Furthermore, the antimicrobial resistance genes were detected in the BALF of 27 patients through Probe-Capture Metagenomics. In one previous study, by the direct detection of key gene features associated with carbapenem resistance of P. aeruginosa, the carbapenem resistance of P. aeruginosa was directly predicted from cultured isolates by HTS, which could assist the timely treatment and surveillance of carbapenem-resistant P. aeruginosa (Liu et al., 2023). This demonstrated that Probe-Capture Metagenomics is capable of detecting antimicrobial resistance genes in bacterial strain and can offer improved guidance for clinical medication. But the results of Probe-Capture Metagenomics were not completely consistent with BALF bacterial culture. Thus, Probe-Capture Metagenomics cannot completely replace culture in terms of antimicrobial resistance genes detection. In the future, it is still necessary to further optimize the sensitivity of probes in Probe-Capture Metagenomics, and then improve the application value in the detection of antimicrobial resistance genes.
Some limitations also exist in our study. Firstly, while RPM normalization accounts for sequencing depth, it does not adjust for genome size or nucleic acid type, which could bias cross-species abundance comparisons. However, as our analysis focused on pathogen detection within patient samples using a fixed RPM threshold, these factors did not impact our conclusions. Secondly, Probe-Capture Metagenomics does not detect actual Virus-like particles or organisms but rather their nucleic acids. Thus, the organisms detected by Probe-Capture Metagenomics may not be actual pathogenic microorganisms. Therefore, it is crucial to remove any contaminating and/or colonizing bacteria, taking into consideration the patient’s clinical history and other results from CMTs. Thirdly, the duration of antibiotic therapy prior to sampling was not considered in our study. Future research is needed to investigate the impact of antibiotic usage duration on the pneumonia microorganisms in the unique vulnerable population of PLWH. Finally, our study was limited by a small sample size, which was confined to a single center. The reliability of Probe-Capture Metagenomics needs to be verified with a larger sample size. Thus, the impact of Probe-Capture Metagenomics on regime adjustment of anti-infection and ART needs further investigation.
5 Conclusion
Probe-Capture Metagenomics provides a promising method for detecting suspected opportunistic infections in PLWH with pneumonia, especially for mixed infections and rare microorganisms. And the lungs may be a reservoir for HIV. In addition, Probe-Capture Metagenomics was a potential valuable tool for detecting HIVDR and antimicrobial resistance genes in bacteria.
Data availability statement
The original contributions presented in the study are publicly available. This data can be found here: https://www.ncbi.nlm.nih.gov/, PRJNA1254612.
Ethics statement
The studies involving humans were approved by the institutional ethics committee of Zhongnan hospital of Wuhan university. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
QC: Writing – review & editing, Methodology, Writing – original draft, Formal analysis, Visualization, Data curation. PM: Formal analysis, Resources, Writing – review & editing, Validation, Writing – original draft, Investigation. RY: Visualization, Validation, Software, Writing – original draft. HX: Methodology, Formal analysis, Software, Writing – review & editing. HL: Data curation, Writing – review & editing, Software, Methodology. ZZ: Resources, Data curation, Writing – review & editing. QD: Writing – review & editing, Investigation, Formal analysis. QJ: Writing – review & editing, Investigation, Visualization. QG: Writing – review & editing, Methodology, Resources, Formal analysis. LC: Formal analysis, Methodology, Data curation, Writing – review & editing. YZ: Validation, Investigation, Project administration, Writing – review & editing, Writing – original draft. YX: Resources, Writing – review & editing, Funding acquisition, Project administration, Writing – original draft, Investigation. LD: Conceptualization, Project administration, Funding acquisition, Writing – original draft, Writing – review & editing, Resources, Formal analysis.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by Center for AIDS Research, Wuhan University (PTPP2023002) and Discipline Cultivation Funding, Zhongnan Hospital of Wuhan University (ZNXKPY2021021).
Conflict of interest
HX and HL were employed by Vision Medicals Co. Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1616937/full#supplementary-material
Footnotes
References
Alcock, B. P., Huynh, W., Chalil, R., Smith, K. W., Raphenya, A. R., Wlodarski, M. A., et al. (2023). CARD 2023: expanded curation, support for machine learning, and resistome prediction at the comprehensive antibiotic resistance database. Nucleic Acids Res. 51, D690–D699. doi: 10.1093/nar/gkac920
Bateman, M., Oladele, R., and Kolls, J. K. (2020). Diagnosing pneumocystis jirovecii pneumonia: a review of current methods and novel approaches. Med. Mycol. 58, 1015–1028. doi: 10.1093/mmy/myaa024
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Casadellà, M., and Paredes, R. (2017). Deep sequencing for HIV-1 clinical management. Virus Res. 239, 69–81. doi: 10.1016/j.virusres.2016.10.019
Chen, Q., Chen, X., Mo, P., Chen, L., Du, Q., Hu, W., et al. (2024). Diagnostic values of BALF metagenomic next-generation sequencing, BALF real-time PCR and serum BDG for pneumocystis jirovecii pneumonia in HIV-infected patients. Front. Microbiol. 15:1421660. doi: 10.3389/fmicb.2024.1421660
Chen, S., Zhou, Y., Chen, Y., and Gu, J. (2018). Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. doi: 10.1093/bioinformatics/bty560
Chen, J., Zhou, T., Zhang, Y., Luo, S., Chen, H., Chen, D., et al. (2022). The reservoir of latent HIV. Front. Cell. Infect. Microbiol. 12:945956. doi: 10.3389/fcimb.2022.945956
Chun, T. W., Stuyver, L., Mizell, S. B., Ehler, L. A., Mican, J. A., Baseler, M., et al. (1997). Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc. Natl. Acad. Sci. USA 94, 13193–13197. doi: 10.1073/pnas.94.24.13193
Cribbs, S. K., Crothers, K., and Morris, A. (2020). Pathogenesis of HIV-related lung disease: immunity, infection, and inflammation. Physiol. Rev. 100, 603–632. doi: 10.1152/physrev.00039.2018
Di Pasquale, M. F., Sotgiu, G., Gramegna, A., Radovanovic, D., Terraneo, S., Reyes, L. F., et al. (2019). Prevalence and etiology of community-acquired pneumonia in immunocompromised patients. Clin. Infect. Dis. 68, 1482–1493. doi: 10.1093/cid/ciy723
Getahun, H., Harrington, M., O'Brien, R., and Nunn, P. (2007). Diagnosis of smear-negative pulmonary tuberculosis in people with HIV infection or AIDS in resource-constrained settings: informing urgent policy changes. Lancet 369, 2042–2049. doi: 10.1016/S0140-6736(07)60284-0
Gu, W., Miller, S., and Chiu, C. Y. (2019). Clinical metagenomic next-generation sequencing for pathogen detection. Annu. Rev. Pathol. 14, 319–338. doi: 10.1146/annurev-pathmechdis-012418-012751
Han, D., Li, Z., Li, R., Tan, P., Zhang, R., and Li, J. (2019). mNGS in clinical microbiology laboratories: on the road to maturity. Crit. Rev. Microbiol. 45, 668–685. doi: 10.1080/1040841X.2019.1681933
Jain, S., Self, W. H., Wunderink, R. G., Fakhran, S., Balk, R., Bramley, A. M., et al. (2015). Community-acquired pneumonia requiring hospitalization among U.S. adults. N. Engl. J. Med. 373, 415–427. doi: 10.1056/NEJMoa1500245
Jia, B., Raphenya, A. R., Alcock, B., Waglechner, N., Guo, P., Tsang, K. K., et al. (2017). CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 45, D566–D573. doi: 10.1093/nar/gkw1004
Jing, C., Chen, H., Liang, Y., Zhong, Y., Wang, Q., Li, L., et al. (2021). Clinical evaluation of an improved metagenomic next-generation sequencing test for the diagnosis of bloodstream infections. Clin. Chem. 67, 1133–1143. doi: 10.1093/clinchem/hvab061
Labelle, A. J., Arnold, H., Reichley, R. M., Micek, S. T., and Kollef, M. H. (2010). A comparison of culture-positive and culture-negative health-care-associated pneumonia. Chest 137, 1130–1137. doi: 10.1378/chest.09-1652
Li, H., and Durbin, R. (2009). Fast and accurate short read alignment with burrows-Wheeler transform. Bioinformatics 25, 1754–1760. doi: 10.1093/bioinformatics/btp324
Li, M., Liang, S., Zhou, C., Chen, M., Liang, S., Liu, C., et al. (2021). HIV drug resistance mutations detection by next-generation sequencing during antiretroviral therapy interruption in China. Pathogens 10:264. doi: 10.3390/pathogens10030264
Liu, B., Gao, J., Liu, X. F., Rao, G., Luo, J., Han, P., et al. (2023). Direct prediction of carbapenem resistance in Pseudomonas aeruginosa by whole genome sequencing and metagenomic sequencing. J. Clin. Microbiol. 61:e0061723. doi: 10.1128/jcm.00617-23
Lozupone, C., Cota-Gomez, A., Palmer, B. E., Linderman, D. J., Charlson, E. S., Sodergren, E., et al. (2013). Widespread colonization of the lung by Tropheryma whipplei in HIV infection. Am. J. Respir. Crit. Care Med. 187, 1110–1117. doi: 10.1164/rccm.201211-2145OC
Lu, D. Y., Wu, H. Y., Yarla, N. S., Xu, B., Ding, J., and Lu, T. R. (2018). HAART in HIV/AIDS treatments: future trends. Infect. Disord. Drug Targets 18, 15–22. doi: 10.2174/1871526517666170505122800
Meng, B., Abdullahi, A., Ferreira, I. A. T. M., Goonawardane, N., Saito, A., Kimura, I., et al. (2022). Altered TMPRSS2 usage by SARS-CoV-2 omicron impacts tropism and fusogenicity. Nature 603, 706–714. doi: 10.1038/s41586-022-04474-x
Moar, P., Premeaux, T. A., Atkins, A., and Ndhlovu, L. C. (2023). The latent HIV reservoir: current advances in genetic sequencing approaches. MBio 14:e0134423. doi: 10.1128/mbio.01344-23
Poorolajal, J., Hooshmand, E., Mahjub, H., Esmailnasab, N., and Jenabi, E. (2016). Survival rate of AIDS disease and mortality in HIV-infected patients: a meta-analysis. Public Health 139, 3–12. doi: 10.1016/j.puhe.2016.05.004
Ryom, L., De Miguel, R., Cotter, A. G., Podlekareva, D., Beguelin, C., Waalewijn, H., et al. (2022). Major revision version 11.0 of the European AIDS clinical society guidelines 2021. HIV Med. 23, 849–858. doi: 10.1111/hiv.13268
Sawatpanich, A., Petsong, S., Tumwasorn, S., and Rotcheewaphan, S. (2022). Diagnostic performance of the Anyplex MTB/NTM real-time PCR in detection of Mycobacterium tuberculosis complex and nontuberculous mycobacteria from pulmonary and extrapulmonary specimens. Heliyon 8:e11935. doi: 10.1016/j.heliyon.2022.e11935
Shahid, A., Jones, B. R., Yang, J. S. W., Dong, W., Shaipanich, T., Donohoe, K., et al. (2022). HIV proviral genetic diversity, compartmentalization and inferred dynamics in lung and blood during long-term suppressive antiretroviral therapy. PLoS Pathog. 18:e1010613. doi: 10.1371/journal.ppat.1010613
Shi, Y., Peng, J. M., Qin, H. Y., and Du, B. (2022). Metagenomic next-generation sequencing: a promising tool for diagnosis and treatment of suspected pneumonia in rheumatic patients with acute respiratory failure: retrospective cohort study. Front. Cell. Infect. Microbiol. 12:941930. doi: 10.3389/fcimb.2022.941930
Stein, A., Doutchi, M., Fenollar, F., and Raoult, D. (2013). Tropheryma whipplei pneumonia in a patient with HIV-2 infection. Am. J. Respir. Crit. Care Med. 188, 1036–1037. doi: 10.1164/rccm.201304-0692LE
Sun, Q., Teng, R., Shi, Q., Liu, Y., Cai, X., Yang, B., et al. (2025). Clinical implement of probe-capture metagenomics in sepsis patients: a multicentre and prospective study. Clin. Transl. Med. 15:e70297. doi: 10.1002/ctm2.70297
Tan, Y., Chen, Z., Zeng, Z., Wu, S., Liu, J., Zou, S., et al. (2023). Microbiomes detected by Bronchoalveolar lavage fluid metagenomic next-generation sequencing among HIV-infected and uninfected patients with pulmonary infection. Microbiol Spectr. 11:e0000523. doi: 10.1128/spectrum.00005-23
Wang, J., Han, Y., and Feng, J. (2019). Metagenomic next-generation sequencing for mixed pulmonary infection diagnosis. BMC Pulm. Med. 19:252. doi: 10.1186/s12890-019-1022-4
White, N. C., Israel-Biet, D., Coker, R. J., Mitchell, D. M., Weber, J. N., and Clarke, J. R. (2004). Different resistance mutations can be detected simultaneously in the blood and the lung of HIV-1 infected individuals on antiretroviral therapy. J. Med. Virol. 72, 352–357. doi: 10.1002/jmv.20010
Willett, B. J., Grove, J., Mac Lean, O. A., Wilkie, C., De Lorenzo, G., Furnon, W., et al. (2022). SARS-CoV-2 omicron is an immune escape variant with an altered cell entry pathway. Nat. Microbiol. 7, 1161–1179. doi: 10.1038/s41564-022-01143-7
Wilson, M. R., Naccache, S. N., Samayoa, E., Biagtan, M., Bashir, H., Yu, G., et al. (2014). Actionable diagnosis of neuroleptospirosis by next-generation sequencing. N. Engl. J. Med. 370, 2408–2417. doi: 10.1056/NEJMoa1401268
Wilson, M. R., Sample, H. A., Zorn, K. C., Arevalo, S., Yu, G., Neuhaus, J., et al. (2019). Clinical metagenomic sequencing for diagnosis of meningitis and encephalitis. N. Engl. J. Med. 380, 2327–2340. doi: 10.1056/NEJMoa1803396
Wüthrich, D., Boujon, C. L., Truchet, L., Selimovic-Hamza, S., Oevermann, A., Bouzalas, I. G., et al. (2016). Exploring the virome of cattle with non-suppurative encephalitis of unknown etiology by metagenomics. Virology 493, 22–30. doi: 10.1016/j.virol.2016.03.009
Xie, Y. L., Cronin, W. A., Proschan, M., Oatis, R., Cohn, S., Curry, S. R., et al. (2018). Transmission of Mycobacterium tuberculosis from patients who are nucleic acid amplification test negative. Clin. Infect. Dis. 67, 1653–1659. doi: 10.1093/cid/ciy365
Xu, J., Huang, Q., Yu, J., Liu, S., Yang, Z., Wang, F., et al. (2022). Metagenomic next-generation sequencing for the diagnosis of suspected opportunistic infections in people living with HIV. Infect Drug Resist. 15, 1767–1775. doi: 10.2147/IDR.S350047
Yan, J., Zhang, B., Zhang, Z., Shi, J., Liu, S., Qi, J., et al. (2021). Case report: Tropheryma whipplei Hide in an AIDS patient with pneumocystis pneumonia. Front. Public Health 9:663093. doi: 10.3389/fpubh.2021.663093
Zarrinfar, H., Kaboli, S., Dolatabadi, S., and Mohammadi, R. (2016). Rapid detection of Candida species in bronchoalveolar lavage fluid from patients with pulmonary symptoms. Braz. J. Microbiol. 47, 172–176. doi: 10.1016/j.bjm.2015.02.001
Zarrinfar, H., Mirhendi, H., Fata, A., Khodadadi, H., and Kordbacheh, P. (2014). Detection of aspergillus flavus and A. fumigatus in bronchoalveolar lavage specimens of hematopoietic stem cell transplants and hematological malignancies patients by real-time polymerase chain reaction, nested PCR and mycological assays. Jundishapur J. Microbiol. 8:e13744. doi: 10.5812/jjm.13744
Zarrinfar, H., Saber, S., Kordbacheh, P., Makimura, K., Fata, A., Geramishoar, M., et al. (2012). Mycological microscopic and culture examination of 400 bronchoalveolar lavage (BAL) samples. Iran. J. Public Health 41, 70–76
Zhu, M., Sun, Z., Zhang, X., Luo, W., Wu, S., Ye, L., et al. (2024). Epidemiological dynamics and molecular characterization of HIV drug resistance in eastern China from 2020 to 2023. Front. Microbiol. 15:1475548. doi: 10.3389/fmicb.2024.1475548
Glossary
PLWH - people living with HIV
CMTs - conventional microbiological tests
mHTS - metagenomic high-throughput sequencing
BALF - Bronchoalveolar Lavage Fluid
G test - 1,3-β-D glucan (G) assay
GM - galactomannan
PCR - polymerase chain reaction
CMV - cytomegalovirus
P. jirovecii - Pneumocystis jirovecii
MTB - Mycobacterium tuberculosis complex
TST - tuberculin skin test
Xpert - GeneXpert MTB/RIF
ART - antiretroviral therapy
CRP - C-reactive protein
IL-6 - Interleukin-6
PCT - procalcitonin
ESR - erythrocyte sedimentation rate
LDH - lactate dehydrogenase
ALT - alanine aminotransferase
NTC - non-template controls
RPM - reads per million
CRR - clinical reportable range
INSTI - integrase strand transfer inhibitor
RT - reverse-transcriptase inhibitor
PI - protease inhibitor
MDT - mutation detection threshold
ESBL - extended-spectrum β-lactamases
C. albicans - Candida albicans
T. whipplei - Tropheryma whipplei
T. marneffei - Talaromyces marneffei
MAC - Mycobacterium avium complex
NTM - nontuberculous mycobacteria
DRMs - drug-resistance mutations
NNRTI - nonnucleoside reverse transcriptase inhibitors
VOA - virus outgrowth assay
HIVDR - HIV drug resistance
Keywords: microorganisms, probe-capture metagenomics, HIV, pneumonia, drug resistance
Citation: Chen Q, Mo P, Yang R, Xu H, Liu H, Zhang Z, Du Q, Jiang Q, Guo Q, Chen L, Zhang Y, Xiong Y and Deng L (2025) Analysis of microorganisms and drug-resistance mutations detected by probe-capture metagenomics among HIV-infected patients with pneumonia. Front. Microbiol. 16:1616937. doi: 10.3389/fmicb.2025.1616937
Edited by:
Ana Elena Pérez-Cobas, Ramón y Cajal Institute for Health Research, SpainReviewed by:
Rodrigo Garcia-Lopez, National Autonomous University of Mexico, MexicoVeronica Ueckermann, University of Pretoria, South Africa
Copyright © 2025 Chen, Mo, Yang, Xu, Liu, Zhang, Du, Jiang, Guo, Chen, Zhang, Xiong and Deng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liping Deng, ZGVuZ2xpcGluZ0B3aHUuZWR1LmNu; Yongxi Zhang, em5hY3QxOTM2QDEyNi5jb20=; Yong Xiong, eW9uZ3hpb25nNjRAMTYzLmNvbQ==
†These authors have contributed equally to this work
‡These authors have contributed equally to this work and share first authorship
 Qianhui Chen
Qianhui Chen Pingzheng Mo
Pingzheng Mo Rongrong Yang
Rongrong Yang Huan Xu
Huan Xu Huifang Liu
Huifang Liu Zhongwei Zhang
Zhongwei Zhang Qian Du1,2
Qian Du1,2 Qunqun Jiang
Qunqun Jiang Liangjun Chen
Liangjun Chen Yongxi Zhang
Yongxi Zhang Yong Xiong
Yong Xiong