- 1Jiaxing Center for Disease Control and Prevention, Jiaxing, China
- 2Jiashan County Center for Disease Control and Prevention, Jiaxing, China
Introduction: Salmonella is an important cause of foodborne diarrheal diseases worldwide. The emergence of blaNDM-positive carbapenem-resistant Salmonella enterica isolates in recent years poses a huge public health challenge.
Methods: In this study, two clinical S. enterica isolates carrying blaNDM–5: a serotype 4,[5],12:i:- strain (2023JX045) and a serovar Stanley strain (2024–406) were analyzed using antimicrobial susceptibility testing and whole genome sequencing.
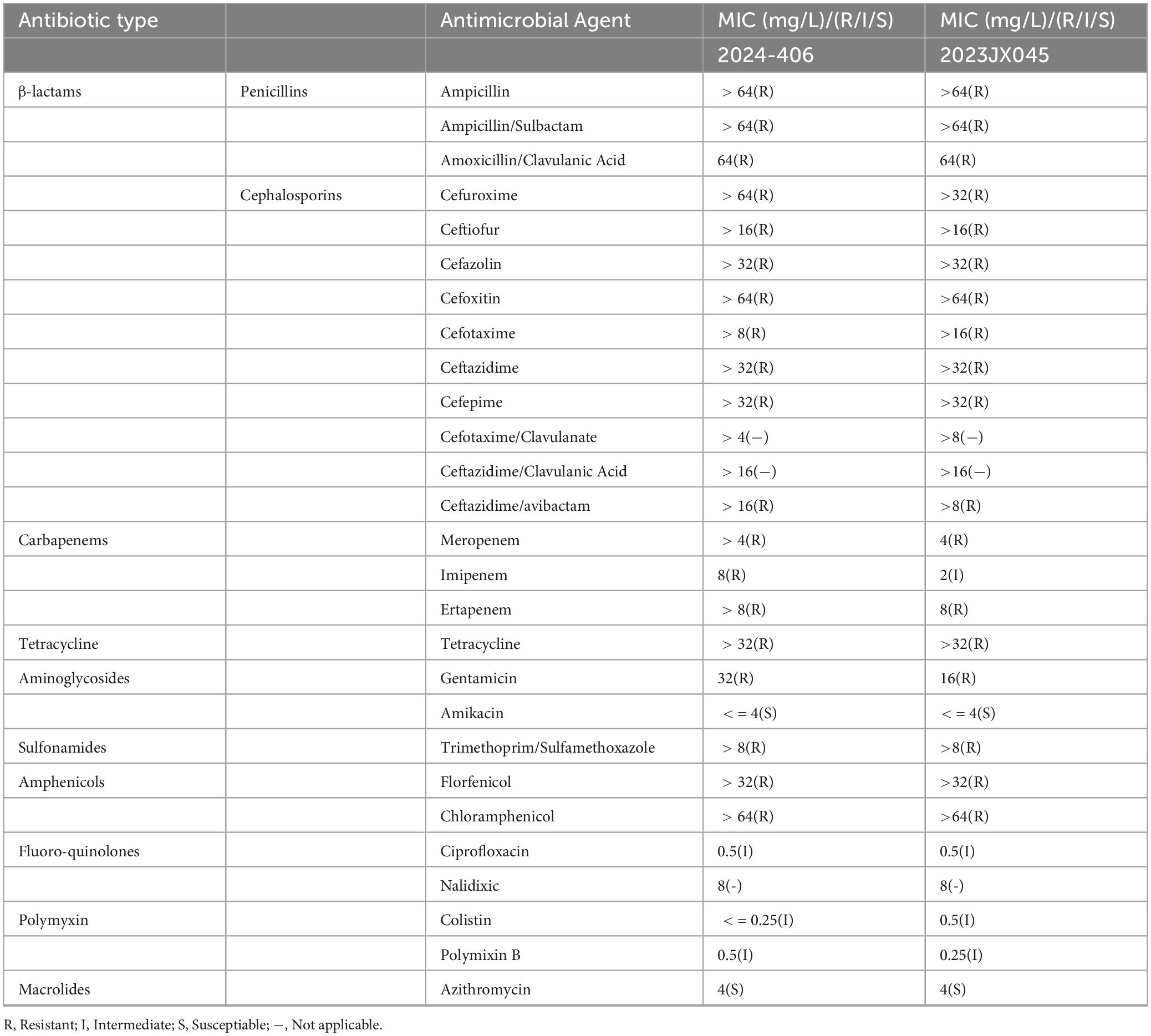
Results: Both isolates were multidrug resistant, with insusceptibility to ampicillin, ampicillin/sulbactam, amoxicillin/clavulanic acid, cefuroxime, ceftiofur, cefazolin, cefoxitin, cefotaxime, ceftazidime, cefepime, ceftazidime/avibactam, meropenem, imipenem, ertapenem, tetracycline, gentamicin, trimethoprim/sulfamethoxazole, florfenicol, chloramphenicol, ciprofloxacin, colistin, and polymixin B. The blaNDM–5-carrying plasmids in 2023JX045 and 2024–406 were named p23045-NDM5 and p2024406-NDM5, respectively, with both belonging to the incompatibility (Inc)HI2/IncHI2A group and sequence type ST3. In 2023JX045, blaNDM–5 was flanked by two same-oriented copies of IS26 elements (IS26-dsbC-trpF-bleMBL-blaNDM–5-IS5-ΔIS3000-ΔISKox3-umuC-umuD-IS26). In 2024–406, p2024406-NDM5 was found to carry two copies of blaNDM–5, possibly resulting from duplication of IS26-dsbC-trpF-bleMBL-blaNDM–5-IS5-ΔISAba125-ΔIS3000-ΔISKox3-umuC-umuD-IS26 and interrupted by mobile element IS1 at the upstream region of ΔIS3000.
Discussion: This is the first report to describe the presence of two blaNDM–5 copies on an IncHI2/IncHI2A plasmid carried by serovar Stanley, as well as the dissemination of blaNDM–5 in Salmonella in Jiaxing City, China. IS26-flanked composite transposons appeared to play an important role in the formation of this region. The dissemination of blaNDM in Salmonella isolates and the complexity of the blaNDM–5 region highlight the urgent need to monitor carbapenem-resistant S. enterica.
1 Introduction
Non-typhoidal Salmonella (NTS) is one of the most prevalent foodborne pathogens, consistently causing gastrointestinal infections. More than 2,600 serovars of NTS have been identified, among which S. Typhimurium is one of the most common (Lamichhane et al., 2024). Previous studies have indicated that Salmonella enterica may act as a reservoir for carbapenemase genes, contributing to the transmission of carbapenem resistance via the food chain (Day et al., 2015). Whole-genome sequencing (WGS) data of global carbapenem-resistant S. enterica (CRSE) isolates revealed S. Typhimurium (21.8%) to be the most prevalent serotype of CRSE worldwide (Wu et al., 2023). Additionally, Typhimurium (25.8%) and Senftenberg (19.4%) were the most prevalent serovars of global CRSE isolates harboring the New Delhi metallo-β-lactamase (blaNDM) gene (Zhao et al., 2025). In recent years, the blaNDM gene has been identified in S. Stanley isolated from clinical and environmental samples (Deng et al., 2024, Huang et al., 2013).
The worldwide spread of multidrug-resistant (MDR) Enterobacteriaceae strains, particularly carbapenem-resistant Enterobacteriaceae (CRE), has become an increasing public health threat (Nordmann et al., 2012). Mobile resistance elements carrying blaNDM–1 have contributed to the dramatic increase in the prevalence of CRE in clinical settings (Huang et al., 2016). Twenty-nine NDM protein variants have been identified since 2009 (Mojica et al., 2022). The blaNDM-5 gene was first identified in a clinical Escherichia coli strain (EC045) from India in 2011, and was later commonly identified among strains of E. coli (Yang et al., 2014, Sassi et al., 2014), Klebsiella pneumoniae (Bathoorn et al., 2015), and Morganella morganii (Guo et al., 2019). In China, a report on blaNDM–1 in Acinetobacter baumannii isolates appeared in early 2011 (Chen et al., 2011). Among Salmonella spp., blaNDM–1 was first identified in a Senftenberg isolate in 2011, and was located on an incompatibility (Inc)L/M group plasmid (Savard et al., 2011, Rasheed et al., 2013). A 2012 report described the identification of a blaNDM–1-bearing strain of Salmonella Stanley isolated from the feces of an 11-month-old girl (Huang et al., 2013). The first blaNDM–5-positive IncFII plasmid, isolated from an S. Typhimurium sequence type (ST) 34 isolate in China, was reported in 2015 (Li et al., 2017). Compared with blaNDM–1, blaNDM–5 has two amino acid substitutions (Val88Leu and Met154Leu) and confers a high level of resistance to carbapenems and broad-spectrum cephalosporins (Hornsey et al., 2011).
Here, we aimed to better understand the antimicrobial resistance determinants and transmission risk of blaNDM-positive Salmonella in Jiaxing City, China by conducting a comprehensive investigation of two blaNDM–5-carrying carbapenem-resistant Salmonella isolates that were recovered from clinical samples. To the best of our knowledge, this is the first report of Salmonella isolates carrying blaNDM in this part of China. Notably, it is also the first report of an IncHI2/IncHI2A plasmid co-carrying two copies of blaNDM–5 in S. Stanley. Jiaxing is located in the Yangtze River Delta region, with well-developed water and land transportation and a continuously growing population. It is an important economic and population aggregation area in Zhejiang Province. These findings further complicate the challenges of establishing effective treatment modalities and management strategies.
2 Materials and methods
2.1 Bacterial collection and characterization
Fecal samples from patients with acute clinical diarrhea were collected to isolate Salmonella spp. Within 4 h of collection, undiluted samples were streaked onto Columbia Blood Agar plates (CHROMagar, Shanghai, China) and cultured overnight at 37°C. Suspected Salmonella spp. colonies were analyzed using matrix-assisted laser desorption/ionization–time of flight mass spectrometry. Serotyping was conducted using the slide agglutination method to detect somatic (O) antigen and flagellar (H) antigens (phase 1 and 2) following the White–Kaufmann–Le Minor Scheme. Salmonella Serotyping by Whole Genome Sequencing was confirmed using the Sequence query tool implemented in SeqSero2/SeqSero2S.1
2.2 Antimicrobial susceptibility testing
AST of the following antimicrobial agents was performed to determine the minimum inhibitory concentration (MIC) of each using the microdilution method: ampicillin, ampicillin/ sulbactam, amoxicillin/clavulanic acid, cefuroxime, ceftiofur, cefazolin, cefoxitin, cefotaxime, ceftazidime, cefepime, cefotaxime/ clavulanate, ceftazidime/clavulanic acid, ceftazidime/avibactam, meropenem, imipenem, ertapenem, tetracycline, gentamicin, amikacin, trimethoprim/sulfamethoxazole, florfenicol, chloramphenicol, ciprofloxacin, nalidixic acid, colistin, polymixin, and azithromycin. The resistance breakpoints of ampicillin, ceftiofur, imipenem, meropenem, ertapenem, azithromycin, tetracycline, ciprofloxacin, trimethoprim/sulfamethoxazole, and chloramphenicol were determined in accordance with the principles outlined in relevant documents from the Clinical and Laboratory Standards Institute (CLSI) (M100-S32, M45-A3). Amoxicillin/clavulanic acid, ampicillin/sulbactam, cefazolin, cefepime, cefotaxime, cefoxitin, ceftazidime, cefuroxime, ceftazidime/avibactam, gentamicin, and amikacin were determined in accordance with the European Committee on Antimicrobial Susceptibility Testing (EUCAST). Colistin, Polymixin B, and florfenicol were interpreted in accordance with the “National Food Contamination and Hazardous Factor Risk Monitoring Work Manual 2024” (China National Center for Food Safety Risk Assessment, 2024). Escherichia coli ATCC 25922, Enterococcus faecalis ATCC29212, Pseudomonas aeruginosa ATCC27853, and Staphylococcus aureus ATCC29213 was used as a quality control strains for AST.
2.3 Genomic DNA extraction and WGS
Total genomic DNA was extracted from overnight (16–18 h) cultures of strains 2023JX045 and 2024-406 using the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions. WGS was performed on the two strains using both the long-read Nanopore MinION (Nanopore, Oxford, United Kingdom) and the short-read NextSeq 550 (Illumina, San Diego, CA, United States) platforms. The derived short reads and long reads were assembled using SPAde v.3.6 software.
2.4 Bioinformatic analysis
The ST of Salmonella isolates were determined using multilocus sequence typing software.2 Antimicrobial-resistant genes and plasmid profiles were analyzed used ResFinder3 and PlasmidFinder.4 Annotation of mobile elements was carried out using online databases, such as ISfinder.5 Plasmid sequence alignment was performed using BRIG v0.95 (Alikhan et al., 2011) and Easyfig v2.2.5.6
To investigate the epidemic characteristics of blaNDM-carrying plasmids in Salmonella, we obtained 31 blaNDM-positive plasmids from the GenBank core nucleotide database (last accessed on 6th January, 2025. Plasmids from Salmonella isolates (taxid: 590) were selected). For the input sequences, multiple sequence alignment (MSA) was performed with the Multiple Alignment Using Fast Fourier Transform in auto mode. The resulting MSA was then entered into ModelTest using default parameters to estimate the best model for constructing the evolutionary tree. Subsequently, the MSA and the selected model were used as input for RAxML-NG with the arguments –all –seed 12345 –bs-trees 1000) to generate the final evolutionary tree. Visualization and annotation of the phylogenetic tree were performed using iTOL v7.7
2.5 Nucleotide sequence accession number
The sequences of plasmids p23045-NDM5 and p2024406-NDM5 were submitted to the GenBank database and assigned accession numbers OR497833 and PQ844496, respectively.
3 Results
3.1 Characterization of two carbapenem-resistant Salmonella strains
S. enterica strains 2023JX045 and 2024-406 were isolated from clinical diarrhea samples collected from a 20-month-old boy and 57-year-old woman, respectively. The main symptoms of case 1 were a fever of 39.3°C and watery diarrhea 10 times per day. No history of suspected food exposure was found. Case 2 did not have a fever. The digestive system symptoms were abdominal pain and watery diarrhea five times a day. It is suspected that it might be related to the consumption of bulk fruits and their products. The patients and his/her family had not traveled to any country in recent 7 days, and no family members were affected. Strain 2023JX045, identified as 4,[5],12:i:-, a ST34 monophasic variant of S. enterica serovar Typhimurium, was found to carry the following antimicrobial resistance genes: aph(4)-Ia, aph(3’)-Ia, aac(3)-IV, aadA2b, blaNDM–5, blaOXA–10, blaTEM–1B, lnu(F), qnrS1, and sul3. Strain 2024-406 was identified as belonging to ST29 S. enterica serovar Stanley (4,12:d:2). The antimicrobial resistance genes identified in this isolate included the following: aph(4)-Ia, aph(6)-Id, aph(3’)-Ia, aph(3”)-Ib, aadA1, aadA1, aadA2b, aac(3)-IV, blaTEM–1B, blaNDM–5, blaNDM–5, blaOXA–10, cmlA1, cmlA1, floR, qnrS1, ARR-2, sul3, tet(A), tet(A), and dfrA14. Furthermore, 2024-406 was shown to possess a single point mutation in the quinolone resistance-determining region of parC (T57S). While a sole plasmid carrying IncHI2/IncHI2A replicons was identified in 2024-406, 2023JX045 was found to carry multiple plasmid replicons, namely Col (pHAD28), IncHI2/IncHI2A, IncI2 (Delta), IncQ1, and p0111 (Table 1).
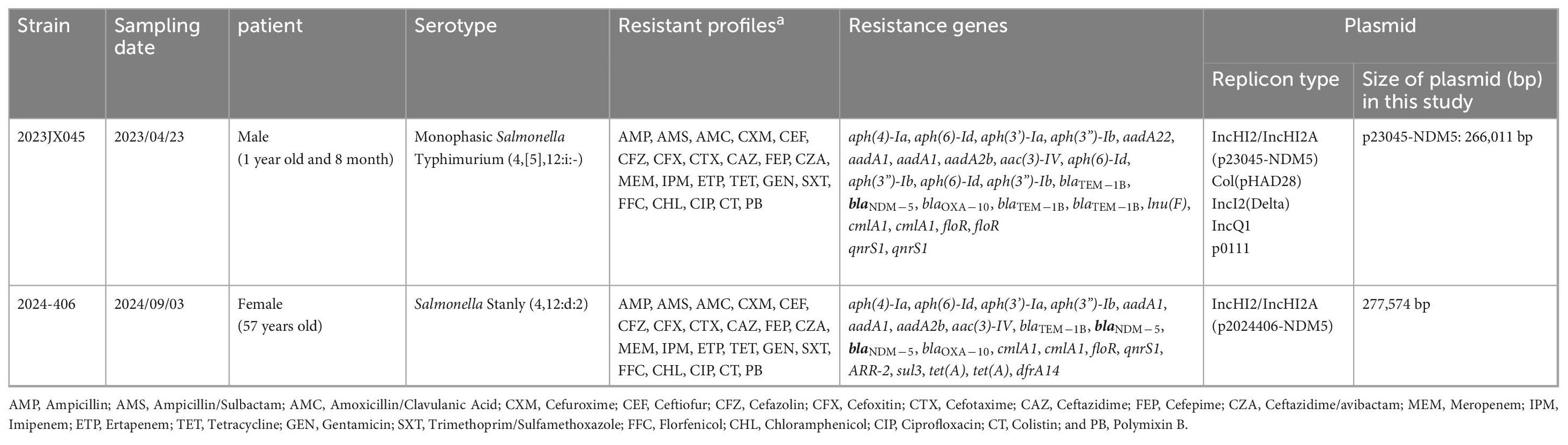
Table 1. Information about the NDM-5-harboring Salmonella strains 2023JX045 and 2024-406 identified in this study and its plasmids.
The AST analysis showed that these two isolates were MDR to β-lactams, including penicillins (ampicillin, ampicillin/sulbactam, and amoxicillin/clavulanic acid) and cephalosporins (cefuroxime, ceftiofur, cefazolin, cefoxitin, cefotaxime, ceftazidime, cefepime, and ceftazidime/avibactam), carbapenems (meropenem, imipenem, and ertapenem), tetracycline (tetracycline), aminoglycosides (gentamicin), sulfonamides (trimethoprim/sulfamethoxazole), amphenicols (florfenicol and chloramphenicol), fluoroquinolones (ciprofloxacin), and polymyxins (colistin and polymixin B), but remained susceptible to amikacin and azithromycin (Table 2).
3.2 Plasmid characterization
The carbapenemase-encoding gene blaNDM–5 was found on a 266,011-bp plasmid (p23045-NDM5) with 47.1% GC content in strain 2023JX045. Strain 2024-406 was found to carry a 277,574-bp plasmid (p2024406-NDM5) with 47.3% GC content. Both p23045-NDM5 and p2024406-NDM5 were classified as IncHI2/IncHI2A and ST3 plasmids. Exhibiting 99% coverage and 100% identity with p23045-NDM5, p2024406-NDM5 is distinguished by numerous rearrangements and inversions within accessory regions.
The resistance genes in p23045-NDM5 were found to be arranged in three regions. The first region contains △tet(A), qnrS1, aadA22, lnu(F), aph(3”)-Ib, aph(6)-Id, aph(3’)-Ia, aph(4)-Ia, aac(3)-Iva, sul3, aadA1, cmlA1, and aadA2b, with the major antimicrobial resistance genes arranged within two class 1 integrons (IntI1). The blaNDM–5-region carries resistance genes including floR, tet(A), dfrA14, aadA1, blaOXA–10, cmlA, ARR-2, bleMBL, and blaNDM–5. This plasmid also carries blaTEM–1 within a truncated Tn2.
In p2024406-NDM5, we found the resistance genes aph(4)-Ia, aph(6)-Id, aph(3’)-Ia, aph(3”)-Ib, aadA1, aadA1, aadA2b, aac(3)-IV, blaTEM–1B, blaOXA–10, cmlA1, cmlA1, floR, qnrS1, ARR-2, sul3, tet(A), tet(A), dfrA14, along with two copies of blaNDM–5 are clustered in a complicated accessory region. The blaOXA–10-region containing tet(A), qnrS1, △ blaTEM–1B, tet(A), floR, and a class 1 integron carrying ARR-2, cmlA1, blaOXA–10, aadA1, and dfrA14 resistant genes.
Compared with the genetic background of the typical blaNDM5–IncX3 plasmid pNDM_MGR194 (IS3000-△ ISAba125-IS5-blaNDM–5-ble-trpF-dsbC-IS26), the blaNDM–5 gene on plasmid p23045-NDM5 was found to be flanked by two same-oriented copies of IS26 elements (IS26-umuD-umuC-△ ISKox3-△ IS3000-IS5-blaNDM–5-bleMBL-trpF-dsbC-IS26). Note the absence of the △ ISAba125 feature in this region. We identified more complex genetic arrangements in p2024406-NDM5, formed by duplication of the IS26-umuD-umuC-△ ISKox3-△ IS3000-△ ISAba125-IS5-blaNDM–5-bleMBL-trpF-dsbC-IS26 unit. Additionally, mobile element IS1 was found upstream of △ IS3000, which may have led to the loss of IS26-umuD-umuC-△ ISKox3 (Figure 1). A direct repeat was not detected immediately upstream or downstream of those two IS26-flanked regions.
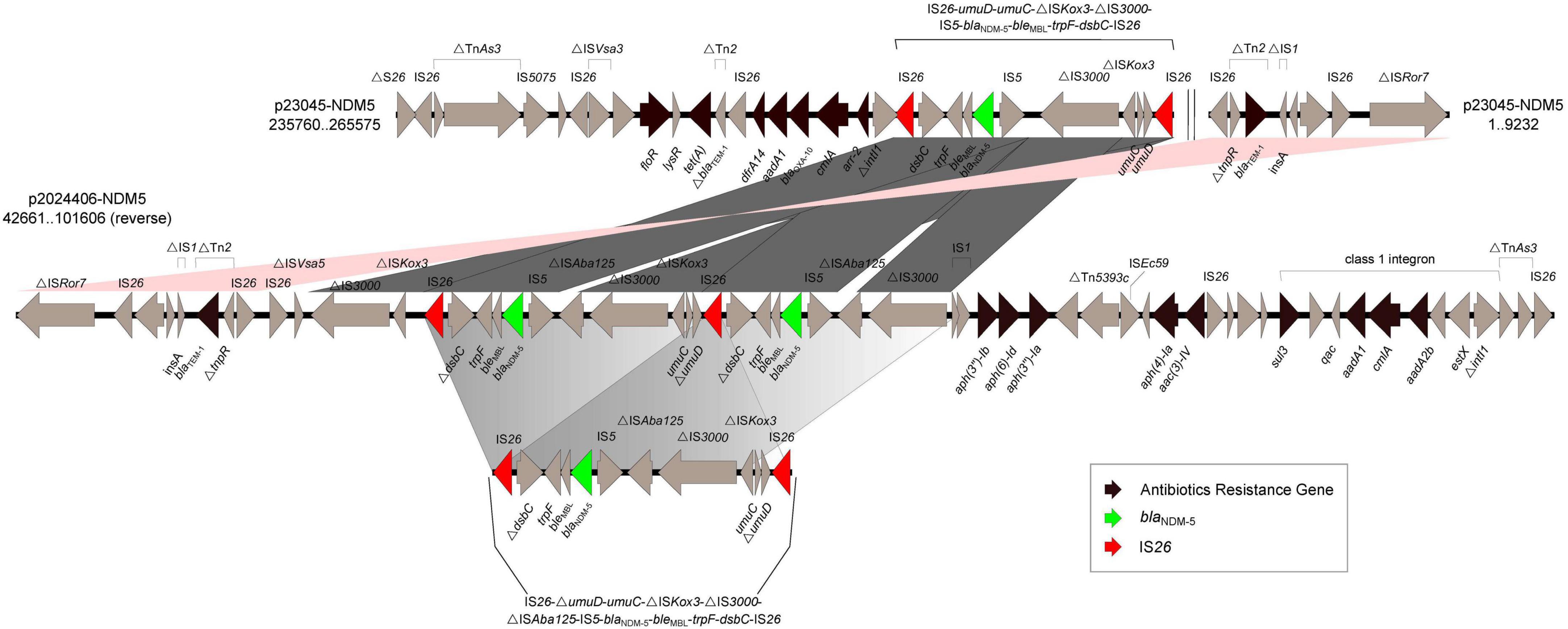
Figure 1. Schematic representation and comparison of the genetic environments of the blaNDM-flanking region in p23045-NDM5 and p2024406-NDM5 recovered from this study. Arrows indicate the direction of transcription of each gene. Numbers in brackets indicate nucleotide positions within corresponding plasmid sequences.
3.3 Comparative study with other NDM-positive plasmids in S. enterica strains already identified
A total of 31 S. enterica strains with positivity for blaNDM plasmids were obtained from the GenBank core nucleotide database (Table 3). The serovars of these strains, mostly isolated from Homo sapiens, were Senftenberg (n = 2), Bareilly (n = 1), London (n = 2), Lomita (n = 1), Enteritidis (n = 1), Mbandaka (n = 2), Rissen (n = 1), 1,4,[5],12:i:- (n = 11), Stanley (n = 2), Corvallis (n = 2), Kottbus (n = 1), 1,4,[5],12:i:2 (n = 2), Typhimurium (n = 1), and Indiana (n = 1). The most prevalent blaNDM gene was blaNDM–1, followed by blaNDM–5, which was the major blaNDM gene in 1,4,[5],12:i:- isolates. Two S. Stanley strains carried a blaNDM gene, with one isolate harboring blaNDM–1 and the other blaNDM–5. Despite the presence of blaNDM, other β-lactamase genes (e.g., blaCMY, blaOXA, and blaTEM) were also detected in some of these strains.
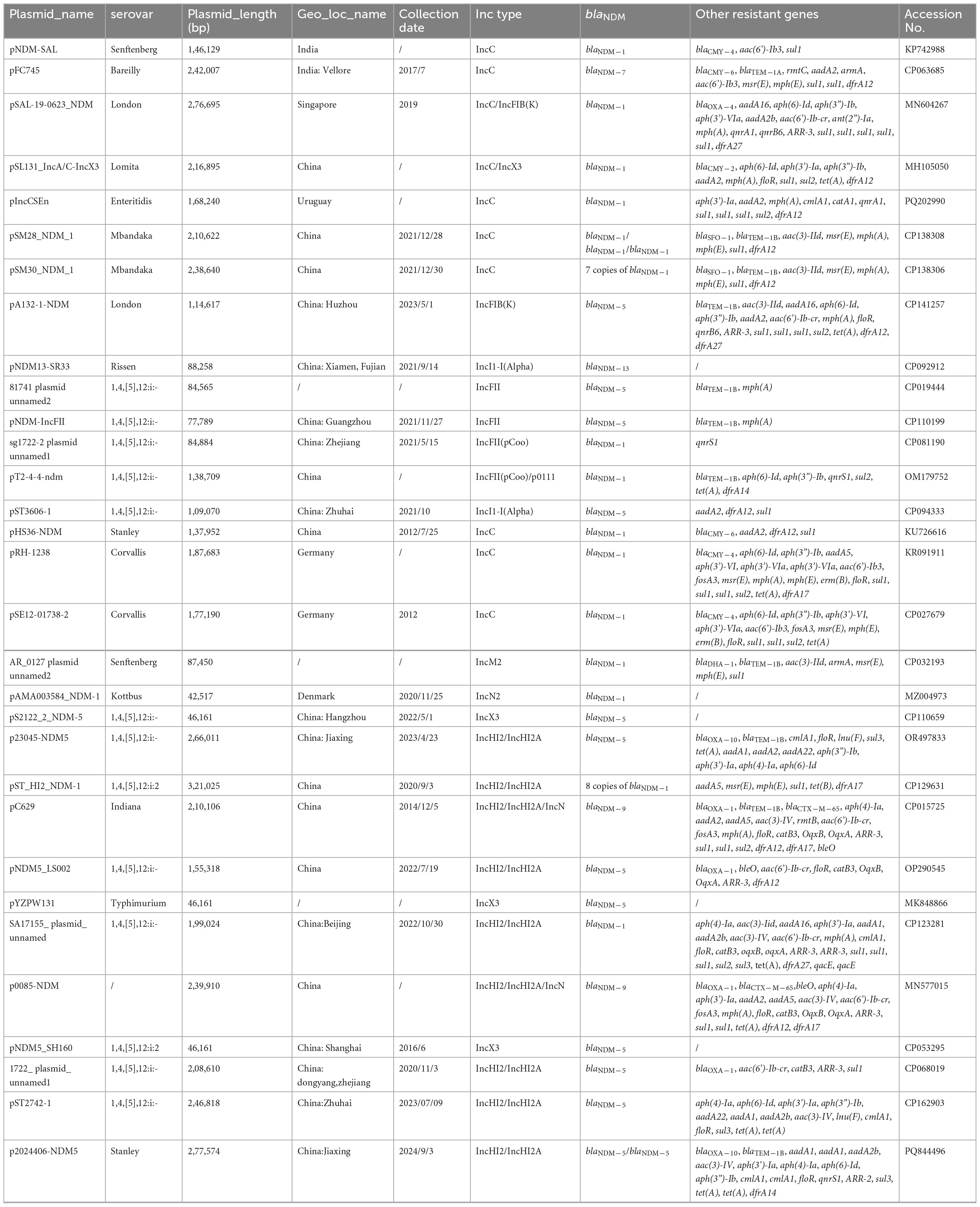
Table 3. 31 S. enterica strains with positivity for blaNDM plasmids obtained from the GenBank core nucleotide database.
Plasmids of different Inc groups, including IncC, IncFII, IncM, IncX3, and IncHI2/IncHI2A, were also found to carry blaNDM genes (Table 3), with IncC and IncHI2/IncHI2A being the most common. Plasmids in S. Stanley strains, namely pHS36-NDM and p2024406-NDM5 (this study), belong to IncC and IncHI2/IncHI2A, respectively. Multiple copies of blaNDM on a single plasmid were found in pSM28_NDM_1 (three copies of blaNDM–1), pSM30_NDM_1 (seven copies of blaNDM–1), and pST_HI2_NDM-1 (eight copies of blaNDM–1). Both pSM28_NDM_1 and pSM30_NDM_1 are IncC-type plasmids, hosted by S. Mbandaka. The IncHI2/IncHI2A-type plasmid pST_HI2_NDM-1 was isolated from a 1,4,[5],12:i:2 strain in 2020. Two copies of blaNDM–5 were found in p2024406-NDM5. Overall, IncHI2/IncHI2A and IncC plasmids carry more resistant genes than IncX3, IncFII, and IncN2 plasmids.
Phylogenetic analysis showed similarity between p23045-NDM5 and pST_HI2_NDM-1 carried by 1,4,[5],12:i:2. Eight copies of blaNDM–1, in addition to 14 other resistance genes, were found in pST_HI2_NDM-1. The highest homology was between p2024406-NDM5 and pST2742-1, which harbors blaNDM–5 and was isolated from a 1,4,[5],12:i:- strain. IncHI2/IncHI2A- and IncX3-type plasmids were more similar than IncC- and IncFII-type NDM-positive plasmids in Salmonella isolates (Figure 2).
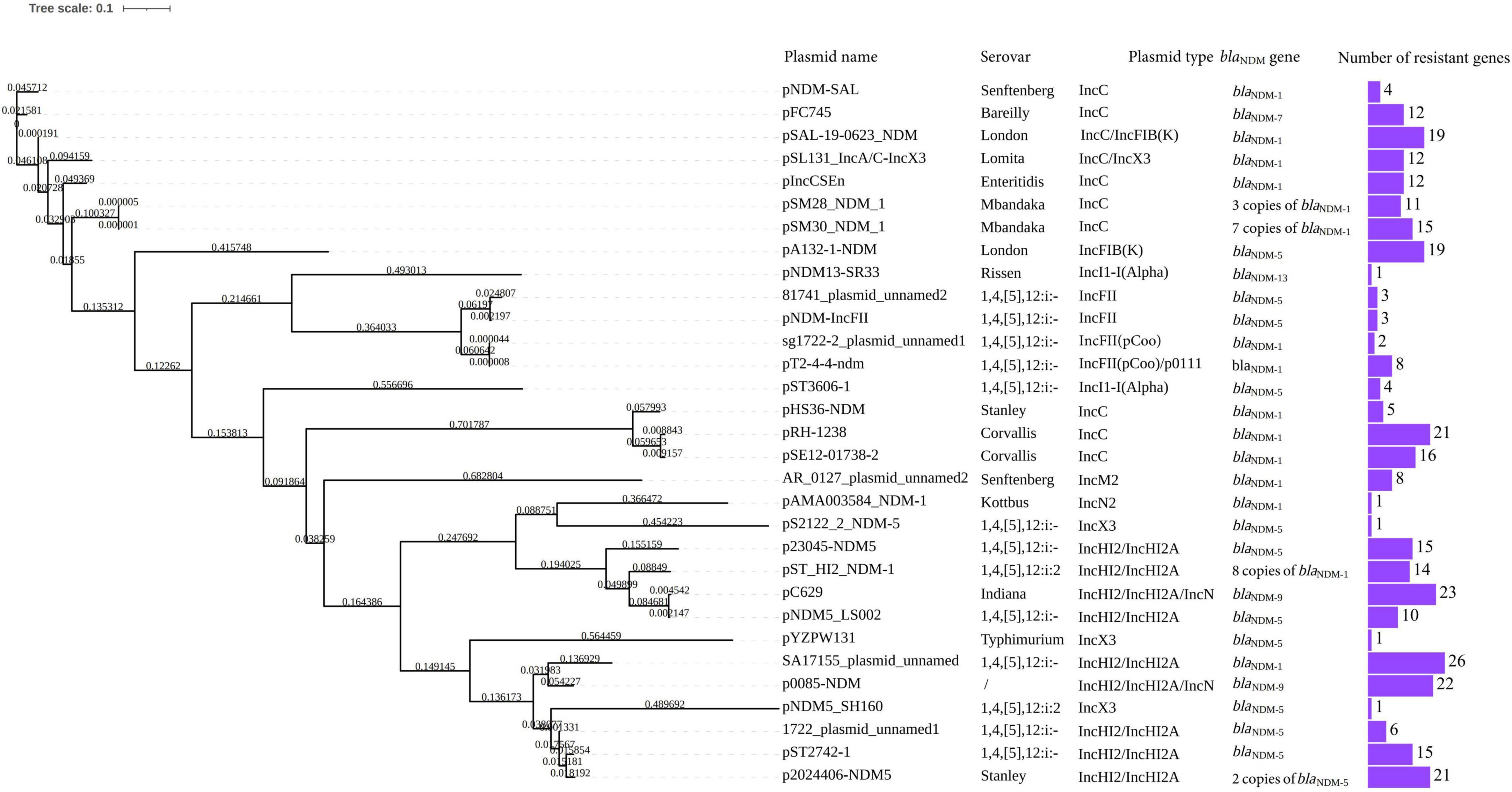
Figure 2. Phylogenetic tree of p23045-NDM5 and p2024406-NDM5 and 29 screened blaNDM-carrying plasmid of Salmonella isolates from GenBank.
4 Discussion
Carbapenems are last-resort antimicrobial agents against infections caused by MDR Gram-negative bacteria. Infection with CRE has become an urgent and continuous threat to public health worldwide (Temkin et al., 2014). Resistance to carbapenems among Salmonella isolates is primarily attributed to the presence of mobile genetic elements encoding various classes of β-lactamases. These include carbapenemase, temoneira, NDM, oxacillinase, imipenemase, and Verona integron-encoded metallo-β-lactamase. The carrier isolates with a single copy gene have high minimum inhibitory concentration (MIC) values for all β-lactams (Han et al., 2020, Miao et al., 2018).
In China, the most prevalent carbapenemase gene among blaNDM-positive isolates is blaNDM–1, followed by blaNDM–5 and blaNDM–3 (Hu et al., 2017). In recent years, blaNDM genes have also been identified in rare Salmonella serotypes, such as S. Kottbus, S. Corvallis, and S. Lomita (Nielsen et al., 2021, Villa et al., 2015, Li et al., 2020). ST34 S. Typhimurium is often characterized by MDR expressed through several resistance genes, including mcr-1, blaCTX–M–55, and qnrS. A previous report characterized an ST34 S. Typhimurium isolate carrying blaNDM–5 and the clonal dissemination of blaNDM–1-positive ST34 S. Typhimurium in South China (Deng et al., 2024). Here, we have described the first identification of Salmonella isolates carrying blaNDM in Jiaxing City. Our isolation of two unrelated clinical Salmonella isolates of different serovars, both carrying blaNDM–5, indicates that the major NDM type in Jiaxing is NDM-5.
Horizontal transmission mediated by various Inc groups of plasmids constitute the major route for the ongoing spread of carbapenem resistance, and include IncC, IncC/IncFIB(K), IncC/IncX3, IncFIB(K), IncI1-I(Alpha), IncFII, IncFII(pCoo), IncFII(pCoo)/p0111, IncM2, IncN2, IncX3, IncHI2/IncHI2A, and IncHI2/IncHI2A/IncN. All of these were isolated from different serotypes across various countries from 2012 to 2024, highlighting the global burden of blaNDM-positive plasmid in S. enterica. Most IncHI2 plasmids found in ST34 S. Typhimurium strains shared a similar backbone, with the capture of blaNDM–1 through an IncHI2/ST3 plasmid (Deng et al., 2024). Although IncX3 has been deemed the primary vehicle for blaNDM transmission worldwide in Enterobacteriaceae (Guo et al., 2019), IncHI2/ST3 plasmids have replaced IncX3 plasmids as the primary plasmid vector for blaNDM–5 transmission on some farms (He et al., 2023).
ST3-IncHI2 plasmids exhibit high sequence conservation in backbones, but possess highly genetic plasticity in accessory regions, allowing for the acquisition of numerous antibiotic resistance genes through mobile elements (Fang et al., 2018). Many mobile elements have played crucial roles in the dissemination of blaNDM, including IS26, ISAba125, IS5, ISCR1, Tn3, Tn125, and Tn3000 (Feng et al., 2018, Zhao et al., 2021, Li et al., 2021). A novel IS26-flanked composite transposon (Tn7540) in the chromosome of an S. Indiana isolate was found to carry blaNDM–9 and fosA3 (Sun et al., 2023). An IS15DIV-flanked composite transposon also contributed to the dissemination of blaNDM–5 in S. Typhimurium (Zhao et al., 2025). NDM-positive isolates consistently carry either a complete or fragmented ISAba125, providing a promoter region for blaNDM and playing a critical role in the horizontal transmission of blaNDM–5 and other resistance determinants (Zhao et al., 2025). Our comparative plasmid analysis showed that the deletion of ISAba125 may have been occurred late in the evolution of p23045-NDM5. Up to eight tandem copies of an ISCR1 unit (ISCR1-dsbD-trpF-ble-blaNDM–1-ΔISAba125) were found on an HI2 plasmid in S. Typhimurium (Song et al., 2023). Although plasmid-borne blaNDM–5 is usually found as a single copy, we previously identified two non-tandem copies of blaNDM–5 on a 144,225-bp IncF plasmid from a carbapenem-resistant clinical isolate of E. coli (Feng et al., 2018). The coexistence of two blaNDM–5 genes was attributed to duplication of an IS26-bracketed region containing ISCR1. In the present study, the two blaNDM–5 regions within one IncHI2/IncHI2A plasmid carried by S. Stanley may have resulted from the duplication of a unit comprising IS26-umuD-umuC-△ ISKox3-△ IS3000-△ ISAba125-IS5-blaNDM–5-bleMBL-trpF-dsbC-IS26 that was subsequently interrupted by IS1 upstream of △ IS3000. No ISCR1 sequences were found in p23045-NDM5 or p2024406-NDM5.
5 Conclusion
In conclusion, we described two MDR Salmonella strains carrying blaNDM–5 that were isolated in Jiaxing City, China, specifically 2023JX045 (4,[5],12:i:-) and 2024-406 (S. Stanley). Each strain was shown to harbor a blaNDM–5-positive IncHI2/IncHI2A plasmid (p23045-NDM5 in 2023JX045 and p2024406-NDM5 in 2024-406), exhibiting signs of multiple evolutionary events that contributed to the diversity of the blaNDM–5-region. IS26-flanked composite transposons appeared to play an important role in the formation of this region. The complex diversity of the blaNDM–5 region is one explanation for the common development of MDR host strains. To the best of our knowledge, this is the first report of a blaNDM gene carried by Salmonella, a major foodborne pathogen, in this region of China. Importantly, this is also the first report of a single IncHI2/IncHI2A plasmid carrying two copies of blaNDM–5 in an S. Stanley host. The identification of CRSE isolates harboring blaNDM and the expanding diversity of blaNDM–5-positive plasmids indicate the potential for widespread dissemination.
This study has several limitations. Since only two isolates were analyzed in this study, the transmission and evolution mechanism of NDM in Salmonella has not been fully explained. The sources of infection of the two cases were also not successfully identified. Therefore, we recommend heightened vigilance and international cooperation to mitigate the public health impact of these pathogens.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Author contributions
PL: Formal Analysis, Writing – original draft, Writing – review & editing. YYu: Data curation, Formal Analysis, Investigation, Visualization, Writing – review & editing. YYa: Software, Writing – review & editing. MJ: Software, Writing – review & editing. LG: Software, Writing – review & editing. XL: Conceptualization, Writing – review & editing. YS: Conceptualization, Visualization, Writing – original draft, Writing – review & editing. GZ: Methodology, Writing – review & editing. ZC: Data curation, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the National Key Research and Development Program of China (2023YFC260510401 and 2023YFC2605100), the Medical Science and Technology Project of Zhejiang Province (2024KY1697) and the Science and Technology Program of Jiaxing City (2023AY31028 and 2023AY11037).
Acknowledgments
We thank Michelle Kahmeyer-Gabbe, PhD, from Liwen Bianji (Edanz) (www.liwenbianji.cn) for editing the English text of a draft of this manuscript.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
1. ^http://denglab.info/SeqSero2
3. ^https://cge.food.dtu.dk/services/ResFinder/
4. ^https://cge.food.dtu.dk/services/PlasmidFinder/
References
Alikhan, N., Petty, N., Ben Zakour, N., and Beatson, S. (2011). BLAST Ring Image Generator (BRIG): Simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402
Bathoorn, E., Rossen, J., Lokate, M., Friedrich, A., and Hammerum, A. (2015). Isolation of an NDM-5-producing ST16 Klebsiella pneumoniae from a Dutch patient without travel history abroad, August 2015. Euro Surveill. 20:30040. doi: 10.2807/1560-7917.ES.2015.20.41.30040
Chen, Y., Zhou, Z., Jiang, Y., and Yu, Y. (2011). Emergence of NDM-1-producing Acinetobacter baumannii in China. J. Antimicrob Chemother. 66, 1255–1259. doi: 10.1093/jac/dkr082
China National Center for Food Safety Risk Assessment (2024). National foodborne disease surveillance manual in 2024. Beijing: China National Center for Food Safety Risk Assessment.
Day, M., Meunier, D., Doumith, M., de Pinna, E., Woodford, N., and Hopkins, K. (2015). Carbapenemase-producing Salmonella enterica isolates in the UK. J. Antimicrob Chemother. 70, 2165–2167. doi: 10.1093/jac/dkv075
Deng, L., Lv, L., Tu, J., Yue, C., Bai, Y., He, X., et al. (2024). Clonal spread of blaNDM-1-carrying Salmonella enterica serovar Typhimurium clone ST34 and wide spread of IncHI2/ST3-blaNDM-5 plasmid in China. J. Antimicrob Chemother. 79, 1900–1909. doi: 10.1093/jac/dkae178
Fang, L., Li, X., Deng, G., Li, S., Yang, R., Wu, Z., et al. (2018). High genetic plasticity in multidrug-resistant sequence type 3-IncHI2 plasmids revealed by sequence comparison and phylogenetic analysis. Antimicrob Agents Chemother. 62:e02068-17. doi: 10.1128/AAC.02068-17.
Feng, Y., Liu, L., McNally, A., and Zong, Z. (2018). Coexistence of two blaNDM-5 genes on an IncF plasmid as revealed by nanopore sequencing. Antimicrob Agents Chemother. 62, e110–e118. doi: 10.1128/AAC.00110-18
Guo, X., Rao, Y., Guo, L., Xu, H., Lv, T., Yu, X., et al. (2019). Detection and genomic characterization of a Morganella morganii isolate from China that produces NDM-5. Front. Microbiol. 10:1156. doi: 10.3389/fmicb.2019.01156
Han, R., Shi, Q., Wu, S., Yin, D., Peng, M., Dong, D., et al. (2020). Dissemination of Carbapenemases (KPC, NDM, OXA-48, IMP, and VIM) among carbapenem-resistant Enterobacteriaceae isolated from adult and children patients in China. Front. Cell Infect. Microbiol. 10:314. doi: 10.3389/fcimb.2020.00314
He, W., Gao, M., Lv, L., Wang, J., Cai, Z., Bai, Y., et al. (2023). Persistence and molecular epidemiology of blaNDM-positive Gram-negative bacteria in three broiler farms: A longitudinal study (2015-2021). J. Hazard Mater. 446:130725. doi: 10.1016/j.jhazmat.2023.130725
Hornsey, M., Phee, L., and Wareham, D. (2011). A novel variant, NDM-5, of the New Delhi metallo-β-lactamase in a multidrug-resistant Escherichia coli ST648 isolate recovered from a patient in the United Kingdom. Antimicrob Agents Chemother. 55, 5952–5954. doi: 10.1128/AAC.05108-11
Hu, X., Xu, X., Wang, X., Xue, W., Zhou, H., Zhang, L., et al. (2017). Diversity of New Delhi metallo-beta-lactamase-producing bacteria in China. Int. J. Infect. Dis. 55, 92–95. doi: 10.1016/j.ijid.2017.01.011
Huang, J., Wang, M., Ding, H., Ye, M., Hu, F., Guo, Q., et al. (2013). New Delhi metallo-β-lactamase-1 in carbapenem-resistant Salmonella strain, China. Emerg. Infect. Dis. 19, 2049–2051. doi: 10.3201/eid1912.130051
Huang, Y., Yu, X., Xie, M., Wang, X., Liao, K., Xue, W., et al. (2016). Widespread dissemination of carbapenem-resistant Escherichia coli sequence type 167 strains harboring blaNDM-5 in clinical settings in China. Antimicrob Agents Chemother. 60, 4364–4368. doi: 10.1128/AAC.00859-16
Lamichhane, B., Mawad, A., Saleh, M., Kelley, W., Harrington, P., Lovestad, C., et al. (2024). Salmonellosis: An overview of epidemiology, pathogenesis, and innovative approaches to mitigate the antimicrobial resistant infections. Antibiotics. 13:10. doi: 10.3390/antibiotics13010076
Li, R., Xie, M., Liu, L., Huang, Y., Wu, X., Wang, Z., et al. (2020). Characterisation of a cointegrate plasmid harbouring blaNDM-1 in a clinical Salmonella Lomita strain. Int. J. Antimicrob Agents 55:105817. doi: 10.1016/j.ijantimicag.2019.09.021
Li, X., He, J., Jiang, Y., Peng, M., Yu, Y., and Fu, Y. (2021). Genetic characterization and passage instability of a hybrid plasmid Co-Harboring blaIMP-4 and blaNDM-1 reveal the contribution of insertion sequences during plasmid formation and evolution. Microbiol. Spectr. 9:e0157721. doi: 10.1128/Spectrum.01577-21
Li, X., Jiang, Y., Wu, K., Zhou, Y., Liu, R., Cao, Y., et al. (2017). Whole-genome sequencing identification of a multidrug-resistant Salmonella enterica serovar Typhimurium strain carrying blaNDM-5 from Guangdong. China. Infect Genet Evol. 55, 195–198. doi: 10.1016/j.meegid.2017.09.005
Miao, M., Wen, H., Xu, P., Niu, S., Lv, J., Xie, X., et al. (2018). Genetic Diversity of Carbapenem-Resistant Enterobacteriaceae (CRE) clinical isolates from a tertiary hospital in Eastern China. Front. Microbiol. 9:3341. doi: 10.3389/fmicb.2018.03341
Mojica, M., Rossi, M., Vila, A., and Bonomo, R. (2022). The urgent need for metallo-β-lactamase inhibitors: An unattended global threat. Lancet Infect Dis. 22, e28–e34. doi: 10.1016/S1473-3099(20)30868-9
Nielsen, H., Thomsen, P., Litrup, E., Torpdahl, M., Overballe-Petersen, S., Hansen, F., et al. (2021). A case of bla NDM-1-positive Salmonella Kottbus, Denmark, November 2021. Euro Surveill. 26:2100569. doi: 10.2807/1560-7917.ES.2021.26.26.2100569
Nordmann, P., Dortet, L., and Poirel, L. (2012). Carbapenem resistance in Enterobacteriaceae: Here is the storm! Trends Mol. Med. 18, 263–272. doi: 10.1016/j.molmed.2012.03.003
Rasheed, J., Kitchel, B., Zhu, W., Anderson, K., Clark, N., Ferraro, M., et al. (2013). New Delhi metallo-β-lactamase-producing Enterobacteriaceae United States. Emerg. Infect. Dis. 19, 870–878. doi: 10.3201/eid1906.121515
Sassi, A., Loucif, L., Gupta, S., Dekhil, M., Chettibi, H., and Rolain, J. (2014). NDM-5 carbapenemase-encoding gene in multidrug-resistant clinical isolates of Escherichia coli from Algeria. Antimicrob Agents Chemother. 58, 5606–5608. doi: 10.1128/AAC.02818-13
Savard, P., Gopinath, R., Zhu, W., Kitchel, B., Rasheed, J., Tekle, T., et al. (2011). First NDM-positive Salmonella sp. strain identified in the United States. Antimicrob Agents Chemother. 55, 5957–5958. doi: 10.1128/AAC.05719-11
Song, H., Zou, S., Huang, Y., Jian, C., Liu, W., Tian, L., et al. (2023). Salmonella Typhimurium with Eight Tandem Copies of blaNDM-1 on a HI2 Plasmid. Microorganisms 12:10. doi: 10.3390/microorganisms12010020
Sun, Y., Han, Y., Qian, C., Zhang, Q., Yao, Z., Zeng, W., et al. (2023). A novel transposon Tn7540 carrying blaNDM-9 and fosA3 in chromosome of a pathogenic multidrug-resistant Salmonella enterica serovar Indiana isolated from human faeces. J. Glob. Antimicrob Resist. 33, 72–77. doi: 10.1016/j.jgar.2023.01.013
Temkin, E., Adler, A., Lerner, A., and Carmeli, Y. (2014). Carbapenem-resistant Enterobacteriaceae: Biology, epidemiology, and management. Ann. N. Y. Acad. Sci. 1323, 22–42. doi: 10.1111/nyas.12537
Villa, L., Guerra, B., Schmoger, S., Fischer, J., Helmuth, R., Zong, Z., et al. (2015). IncA/C plasmid carrying bla(NDM-1), bla(CMY-16), and fosA3 in a Salmonella enterica serovar corvallis strain isolated from a migratory wild bird in Germany. Antimicrob Agents Chemother. 59, 6597–6600. doi: 10.1128/AAC.00944-15
Wu, Y., Jiang, T., Bao, D., Yue, M., Jia, H., Wu, J., et al. (2023). Global population structure and genomic surveillance framework of carbapenem-resistant Salmonella enterica. Drug Resist. Updat. 68:100953. doi: 10.1016/j.drup.2023.100953
Yang, P., Xie, Y., Feng, P., and Zong, Z. (2014). blaNDM-5 carried by an IncX3 plasmid in Escherichia coli sequence type 167. Antimicrob Agents Chemother. 58, 7548–7552. doi: 10.1128/AAC.03911-14
Zhao, K., Jin, J., Liao, Y., Liu, A., Liu, W., and Wu, W. (2025). IS 15DIV-flanked composite transposon harboring bla NDM-5 in multidrug-resistant Salmonella typhimurium. iScience 28:111720. doi: 10.1016/j.isci.2024.111720
Keywords: non-typhoidal Salmonella, carbapenem-resistant, IncHI2/IncHI2A, IS26 unit, blaNDM–5
Citation: Li P, Yuan Y, Yan Y, Jia M, Gao L, Liu X, Sun Y, Zhu G and Chen Z (2025) Characterization of blaNDM–5-carrying plasmids in two clinical Salmonella isolates from Jiaxing city, China. Front. Microbiol. 16:1620907. doi: 10.3389/fmicb.2025.1620907
Received: 30 April 2025; Accepted: 05 June 2025;
Published: 26 June 2025.
Edited by:
Zhangnv Yang, Zhejiang Center for Disease Control and Prevention (Zhejiang CDC), ChinaReviewed by:
Steven L. Foley, National Center for Toxicological Research (FDA), United StatesZhang Sheng Wei, Beijing University of Chinese Medicine, China
Keke Liu, Shandong Provincial Hospital, China
Gianluigi Ferri, University of Teramo, Italy
Copyright © 2025 Li, Yuan, Yan, Jia, Gao, Liu, Sun, Zhu and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guoying Zhu, anhjZGN6aHVndW95aW5nQDE2My5jb20=; Zhongwen Chen, Y3p3MjAwN0Bzb2h1LmNvbQ==
†These authors have contributed equally to this work
 Ping Li
Ping Li Yongjuan Yuan2†
Yongjuan Yuan2†