- 1Microbial Pathogen and Anti-Infection Research Group, School of Basic Medicine and Forensic Medicine, Henan University of Science and Technology, Luoyang, China
- 2State Key Laboratory of Antiviral Drugs, Pingyuan Laboratory, National Medical Products Administration (NMPA) Key Laboratory for Research and Evaluation of Innovative Drug, School of Chemistry and Chemical Engineering, Henan Normal University, Xinxiang, China
Introduction: Pseudomonas aeruginosa is an important opportunistic and foodborne disease-related bacterium, and the increasing antibiotic resistance of the pathogen leads to the urgent exploration of new and effective antibacterial agents. In this study, a scorpion peptide derivative HTP2 was designed.
Methods: The in vitro anti-P. aeruginosa activity was evaluated using a broth microdilution assay. A mouse model of P. aeruginosa skin subcutaneous infection was used to evaluate the in vivo anti-P. aeruginosa activity of HTP2. The antibacterial mechanism and influence on pathogenic factors of P. aeruginosa of HTP2 were also investigated.
Results: HTP2 could effectively inhibit the growth of P. aeruginosa cells with low hemolytic activity. HTP2 killed P. aeruginosa in a concentration-dependent manner, and could damage the membrane, induce ROS accumulation, and interact with nucleic acids. HTP2 could also inhibit biofilm formation, motility, pyocyanin production, and elastase activity of P. aeruginosa. In the mouse subcutaneous infection model, HTP2 significantly reduced the bacterial load of P. aeruginosa cells and inhibited inflammatory infiltration in the infection area.
Conclusion: HTP2 could effectively kill P. aeruginosa in vitro and in vivo, and had the potential as an anti-P. aeruginosa agent.
1 Introduction
Pseudomonas aeruginosa is a Gram-negative bacterium that is widespread in nature, which can be found in water, soil, plant, animal, and human. It is not only a vital food-related microorganism, but also an opportunistic pathogen. As a vital food-related microorganism, P. aeruginosa causes a variety of contamination and spoilage of foods (such as dairy, meat, aquatic products, fresh vegetables, etc.), drinking water, and fruit juices, which can lead to intoxications or infections associated with foodborne diseases (Wu et al., 2016; Lee et al., 2020; Nahar et al., 2021; Schalli et al., 2023). As an opportunistic pathogen, P. aeruginosa mainly colonizes the skin and intestines of humans, and can cause skin infections, pneumonia, urinary tract infections, and sometimes result in serious systemic infections, especially in immunosuppressed individuals (Quiñones-Vico et al., 2024). Nowadays, P. aeruginosa can also be isolated from hospital environment, clinical instruments, medical products, and cosmetics. Thus, effective removal and killing of P. aeruginosa is necessary to prevent foodborne infections and opportunistic infections caused by the pathogen. However, P. aeruginosa can form biofilm on various surfaces, which can protect the pathogen from antibiotics and biocides (Hauser et al., 2011; Maurice et al., 2018). Moreover, antibiotic resistance also increases the difficulty of P. aeruginosa clearance (Kunz Coyne et al., 2022; Liao et al., 2022). Nowadays, concerns about synthetic preservatives and antibiotic-resistant food pathogens have increased significantly. Thus, developing new and effective antimicrobial agents with no side effects and food pollution potential is urgently necessary.
Antimicrobial peptides (AMPs) are usually composed of 10 to 50 amino acids, which can be identified in bacteria, fungi, plants, vertebrates, and invertebrates (Verma et al., 2024). They usually have a positive net charge and a significant proportion of hydrophobic residues (Bin Hafeez et al., 2021). Nowadays, lots of AMPs have been artificially designed based on their characteristics (Lourenço et al., 2023). According to the Collection of Anti-Microbial Peptides (CAMP), 24,243 AMPs have been recorded: 11827 natural AMPs and 12,416 synthetic AMPs. They show activities against a broad spectrum of microorganisms, including antibiotic-resistant strains, via targeting multiple sites, especially damaging the cell membrane, leading to rapid cell death (Gagat et al., 2024). Due to the specific action modes different from traditional antibiotics, AMPs exhibit a low propensity for inducing bacterial resistance (Fjell et al., 2011; Nguyen et al., 2011). Moreover, they also have the potential to inhibit biofilm formation (Shahrour et al., 2019). Currently, they have shown potential as drugs to inhibit infectious disease, and also receive special attention in food safety.
In this study, we designed several peptide derivatives based on the peptide (FWSFLAKIATKALPALFGSRKKSSSR, renamed as Helepsin in this study) from the scorpion Hemiscorpius lepturus (Kazemi-Lomedasht et al., 2017), among which a peptide derivative HTP2 with improved antibacterial activity against P. aeruginosa and decreased hemolysis was obtained. A mouse model of P. aeruginosa skin subcutaneous infection was used to evaluate the potential application of HTP2 as an antibacterial agent. The antibacterial mechanism and influence on pathogenic factors of P. aeruginosa of HTP2 were also investigated.
2 Materials and methods
2.1 Peptides and bacterial strains
The peptides used in the study were synthesized by GL Biochem (Shanghai) Ltd. with an amidated C-terminus, and the purity of the peptides was more than 95%. P. aeruginosa PAO1, P. aeruginosa ATCC27853, P. aeruginosa ATCC9027, and P. aeruginosa CCTCC93066 used in study were cultured using Luria-Bertani (LB) broth medium at 37°C.
2.2 Antimicrobial activity
The minimum inhibitory concentration (MIC) of the peptides against P. aeruginosa was measured using a broth microdilution assay recommended by the Clinical and Laboratory Standards Institute guidelines (CLSI, 2019). Briefly, exponential-phase P. aeruginosa cells were diluted in LB medium to 105–106 cfu/mL, and the peptides were dissolved and serially diluted in 0.9% saline. Then, the bacterial dilution (80 μL) and peptide dilution (20 μL) were added into sterile 96-well cell culture plates. The final concentration of each peptide was 3.13 μg/mL, 6.25 μg/mL, 12.5 μg/mL, 25 μg/mL, 50 μg/mL, and 100 μg/mL, respectively. Each concentration was conducted in triplicate. Thereafter, the plates were incubated with continuous shaking at 200 rpm for 18–24 h at 37°C. The lowest peptide concentration with no bacterial growth was determined as the MIC.
2.3 Hemolytic activity
Hemolytic activity was used to evaluate the in vitro toxicity of the peptides, using the method described previously (Yuan et al., 2022). Briefly, fresh mouse red blood cells (mRBCs) were washed with and then resuspended in 0.9% saline to 2% (v/v), and the peptides were dissolved and serially diluted in 0.9% saline. Then, the mRBCs suspension (100 μL) and peptide dilution (100 μL) were added into sterile 96-well cell culture plates. The final concentration of each peptide was 25 μg/mL, 50 μg/mL, and 100 μg/mL, respectively. Each concentration was conducted in triplicate, and 1% Triton X-100 and no peptide (0.9% saline) treatment were set as the positive and negative controls, respectively. Thereafter, the plates were incubated with continuous shaking at 200 rpm for 1 h at 37°C. After incubation, the plates were centrifuged at 1,000 g for 10 min, and 100 μL of the supernatant was transferred to a new 96-well plate, and the absorbance was measured at 490 nm. Hemolytic activity was evaluated using the following equation: Hemolysis% = (Hsample − Hnegative)/(Hpositive − Hnegative) × 100%. H: absorbance at 490 nm.
2.4 Stability assay
The stability of the peptide was determined using the method described previously (Mirzaei et al., 2022; Yuan et al., 2022). For the thermal stability assay, the peptide was incubated at 60°C for 24 h. Then, the MIC of the peptide against P. aeruginosa PAO1 was measured. For salt stability assay, the exponential phase P. aeruginosa PAO1 cells were washed with and diluted in NaCl-free LB medium to 105–106 cfu/mL with specific amounts of NaCl, CaCl2, MgCl2, or KCl, respectively. The peptide was dissolved and serially diluted in sterile water. Then, the bacterial dilution (80 μL) and peptide dilution (20 μL) were added into sterile 96-well cell culture plates. The final concentration of each peptide was 3.13 μg/mL, 6.25 μg/mL, 12.5 μg/mL, 25 μg/mL, 50 μg/mL, and 100 μg/mL, respectively. Each concentration was conducted in triplicate. The final concentration of NaCl, CaCl2, MgCl2, and KCl was 150 mM/300 mM, 1 mM/2 mM, 1 mM/2 mM, and 2 mM/4 mM, respectively. Thereafter, the MIC of the peptide against P. aeruginosa PAO1 was measured.
2.5 Time-killing kinetics
The time-killing kinetics assay of the peptide against P. aeruginosa PAO1 was performed using the method described previously (Li et al., 2020). Briefly, exponential-phase P. aeruginosa PAO1 cells were diluted in LB medium to 105–106 cfu/mL, and the peptide was dissolved and serially diluted in 0.9% saline. Then, the bacterial dilution (400 μL) was incubated with peptide dilution (100 μL) at 37°C with continuous shaking at 200 rpm. The final concentration of the peptide was 12.5 μg/mL, 25 μg/mL, and 50 μg/mL, respectively. No peptide (0.9% saline) treatment served as a negative control. Aliquots were collected at 0 min, 15 min, and 30 min, serially diluted in 0.9% saline, and plated on LB agar plates. The plates were incubated at 37°C for 18–24 h, and the CFU was counted.
2.6 Confocal laser-scanning microscopy
The action site of the peptide on P. aeruginosa was determined using the method described previously (Li et al., 2014). Briefly, exponential-phase P. aeruginosa PAO1 cells were diluted in LB medium to 106–107 cfu/mL, and the FITC-labeled peptide was dissolved in 0.9% saline. Then, the bacterial dilution was incubated with the FITC-labeled peptide dilution at a volume ratio of 4:1. The final concentration of the FITC-labeled peptide was 10 μg/mL. After incubation for 20 min at 37°C, the cells were washed with and resuspended in PBS, and then immobilized on a glass slide. The cells were observed using a confocal laser-scanning microscope.
2.7 LPS competition binding assay
The interaction of the peptide with LPS was evaluated using the method described previously (Cao et al., 2012). Briefly, exponential-phase P. aeruginosa PAO1 cells were diluted in LB medium to 105–106 cfu/mL, and the peptides were dissolved and serially diluted in 0.9% saline containing LPS. Then, the bacterial dilution (80 μL) and peptide dilution (20 μL) were added into sterile 96-well cell culture plates. The final concentration of each peptide was 3.13 μg/mL, 6.25 μg/mL, 12.5 μg/mL, 25 μg/mL, 50 μg/mL, and 100 μg/mL, respectively. Each concentration was conducted in triplicate. The final concentration of LPS was 100 μg/mL, 200 μg/mL, or 500 μg/mL, respectively. Thereafter, the plates were incubated with continuous shaking at 200 rpm for 18–24 h at 37°C. The MIC of the peptide in the presence of LPS was determined.
2.8 Membrane permeability assay
The influence of the peptide on the membrane integrity of P. aeruginosa cells was evaluated using the method described previously (Li et al., 2016). Briefly, exponential-phase P. aeruginosa PAO1 cells were washed with and diluted in 0.9% saline to 106–107 cfu/mL, and then incubated with SYTOX green (final concentration: 5 μM) in the dark for 10 min at 37°C. The peptide was dissolved and serially diluted in 0.9% saline. Then, the bacterial dilution (50 μL) and peptide dilution (50 μL) were added into a Costar 96-well flat-bottom black plate. The final concentration of the peptide was 6.25 μg/mL, 12.5 μg/mL, and 25 μg/mL, respectively. Each concentration was conducted in triplicate, and no peptide (0.9% saline) treatment was served as a negative control. Thereafter, the fluorescence was measured every 2 min at the excitation and emission wavelengths of 488 and 525 nm, respectively.
2.9 Membrane potential assay
The influence of the peptide on the membrane potential of P. aeruginosa cells was evaluated using the method described previously (Lee and Lee, 2014). Briefly, exponential-phase P. aeruginosa PAO1 cells were washed with and diluted in 0.9% saline to 106–107 cfu/mL, and then incubated with DiBAC4(3) (final concentration: 10 μM) in the dark for 10 min at 37°C. The peptide was dissolved and serially diluted in 0.9% saline. Then, the bacterial dilution (50 μL) and peptide dilution (50 μL) were added into a Costar 96-well flat-bottom black plate. The final concentration of the peptide was 6.25 μg/mL, 12.5 μg/mL, and 25 μg/mL, respectively. Each concentration was conducted in triplicate, and no peptide (0.9% saline) treatment was served as a negative control. Thereafter, the fluorescence was measured every 2 min at the excitation and emission wavelengths of 488 and 525 nm, respectively.
2.10 ROS measurements
The influence of the peptide on ROS generation of P. aeruginosa cells was evaluated using the method described previously (Shi et al., 2021). Briefly, exponential-phase P. aeruginosa PAO1 cells were washed with and diluted in 0.9% saline to 106–107 cfu/mL, and then incubated with DCFH-DA (final concentration: 10 μM) in the dark for 20 min at 37°C. The peptide was dissolved and serially diluted in 0.9% saline. Then, the bacterial dilution (50 μL) and peptide dilution (50 μL) were added into a Costar 96-well flat-bottom black plate. The final concentration of the peptide was 6.25 μg/mL, 12.5 μg/mL, and 25 μg/mL, respectively. Each concentration was conducted in triplicate, and no peptide (0.9% saline) treatment was served as a negative control. Thereafter, the fluorescence was measured every 10 min at the excitation and emission wavelengths of 488 and 525 nm, respectively.
2.11 Nucleic acid binding assay
The interaction of the peptide with nucleic acids was evaluated using the method described previously (Li Z. et al., 2022). Briefly, approximately 300 ng of DNA (plasmid pET-28a) or RNA (P. aeruginosa PAO1 RNA) was incubated with varying concentrations of the peptide for 10 min at room temperature. Then, the mixtures were electrophoresed in a 1% agarose gel. The migration of nucleic acids in the gel was visualized using a Bio-Rad Gel Documentation system.
2.12 Effects on biofilm formation
The influence of the peptide on biofilm formation of P. aeruginosa was evaluated using the method described previously (Artini et al., 2022). Briefly, exponential-phase P. aeruginosa PAO1 cells were diluted in LB medium to 105–106 cfu/mL, and 200 μL of the bacterial dilution was added into sterile 96-well cell culture plates. After incubation for 4 h at 37°C, the wells were washed with PBS to remove the non-adherent cells. Then, 200 μL of LB medium containing varying concentrations of the peptide was added to the wells. The final concentration of the peptide was 6.25 μg/mL, 12.5 μg/mL, and 25 μg/mL, respectively. Each concentration was conducted in triplicate, and no peptide (0.9% saline) treatment was served as a negative control. After static cultivation for another 24 h at 37°C, biomasses were quantified using a crystal violet staining assay.
2.13 Motility assay
The influence of the peptide on the motility of P. aeruginosa was evaluated using the method described previously (She et al., 2018; Chen et al., 2020). Briefly, exponential-phase P. aeruginosa PAO1 cells were diluted in LB medium to 106–107 cfu/mL. To investigate swimming motility, 0.3% LB agar medium (1 mL) supplemented with varying concentrations of the peptide was poured into sterile 24-well cell culture plates. Then, 2 μL of the bacterial dilution was spotted in the center of the agar plate. After incubation for 18–24 h at 37°C, the swimming zone was observed and pictured with a camera. To investigate swarming motility, 0.5% LB agar medium (1 mL) supplemented with varying concentrations of the peptide was poured into sterile 24-well cell culture plates. Then, 2 μL of the bacterial dilution was spotted in the center of the agar plate. After incubation for 18–24 h at 37°C, the swarming zone was observed and pictured with a camera.
2.14 Pyocyanin assay
The influence of the peptide on the pyocyanin production of P. aeruginosa was evaluated using the method described previously (Pan et al., 2023). Briefly, exponential-phase P. aeruginosa PAO1 cells were diluted in LB medium to 106–107 cfu/mL, and the peptides were dissolved and serially diluted in 0.9% saline. Then, the bacterial dilution was incubated with the peptide dilution at a volume ratio of 4:1. The final concentration of each peptide was 6.25 μg/mL and 12.5 μg/mL, respectively. Each concentration was conducted in triplicate, and no peptide (0.9% saline) treatment was served as a negative control. After incubation for 16 h at 37°C with continuous shaking at 200 rpm, pyocyanin was extracted using 0.6 mL of chloroform from 1 mL supernatant of the bacterial culture. Then, the organic layer was extracted using 0.2 mL of HCl (0.2 N), and the absorbance of the upper layer was measured at 520 nm.
2.15 Elastase assay
The influence of the peptide on the elastase of P. aeruginosa was evaluated using the method described previously (Musthafa et al., 2012; Xu et al., 2015). Briefly, exponential-phase P. aeruginosa PAO1 cells were diluted in LB medium to 106–107 cfu/mL, and the peptides were dissolved and serially diluted in 0.9% saline. Then, the bacterial dilution was incubated with the peptide dilution at a volume ratio of 4:1. The final concentration of each peptide was 6.25 μg/mL and 12.5 μg/mL, respectively. Each concentration was conducted in triplicate, and no peptide (0.9% saline) treatment was served as a negative control. After incubation for 6 h at 37°C with continuous shaking at 200 rpm, the cultures were centrifuged at 12,000 r/min for 10 min at 4°C. Then, the supernatant was filtered using a 0.22 μm syringe filter. Thereafter, the filtered supernatant (100 μL) was incubated with ECR solution (900 μL: ECR 20 mg/mL, 0.1 M Tris–HCl, 1 mM CaCl2, pH 7.2) for 4 h at 37°C with continuous shaking at 200 rpm. After incubation, the reaction mixture was incubated on ice for 10 min, and then centrifuged at 12,000 r/min for 10 min at 4°C. The absorbance of the supernatant was measured at 520 nm.
2.16 Animals and subcutaneous infection model
Male BALB/c mice (20–30 g), obtained from Henan SKBEX Biology Co., Ltd., were maintained under a standard condition of humidity (50 ± 5%), temperature (25 ± 2°C), and dark–light cycles (12 h each) with free access to food and water. All the animal-associated experiments were conducted following the Animal Care and Ethics guidelines with protocols approved by the Animal Care and Use Committee of Henan University of Technology and Science. At the end of the experiments, all the mice were humanely euthanized by intraperitoneal injection of excessive pentobarbital.
A mouse subcutaneous infection model was used to evaluate the in vivo anti-P. aeruginosa activity of the peptide according to the method previously described (Li S. et al., 2022). Briefly, exponential-phase P. aeruginosa PAO1 cells were washed with and resuspended in 0.9% saline to approximately 109 CFU/mL. Each mouse was subcutaneously injected with 50 μL of the bacterial suspension at the back near the tail. Then, the mice were randomly divided into three groups (six mice per group). One hour after infection, 50 μL of 0.9% saline (negative control), peptide solution (500 μg/mL in 0.9% saline), or ciprofloxacin solution (positive control, 500 μg/mL in 0.9% saline) was subcutaneously injected into the infected area of each mouse in the corresponding group. All the treatments were administered once a day continuously for 3 days. On the 4th day, the mice were humanely euthanized, and the infected area of each mouse was sterilized using 10% povidone/iodine solution and 70% ethyl alcohol after shaving the fur. Thereafter, half of the skin abscesses were excised and homogenized in 0.9% saline. After being serially diluted in 0.9% saline, the homogeneous samples were cultured on LB agar at 37°C for 18–24 h. The number of colony-forming units (CFU) per gram of the skin abscess was subsequently calculated. Another half of the samples were used for hematoxylin and eosin staining, and the infiltration of inflammatory cells was observed.
2.17 Statistical analysis
The data were expressed as mean ± standard error of the mean (SEM) and analyzed using the GraphPad Prism 6 software. Differences between groups were analyzed using one-way ANOVA.
3 Results
3.1 Peptides and in vitro antibacterial activity
Based on the peptide Helepsin, three peptide derivatives (HTP, HTP1, and HTP2) were designed (Figure 1A). HTP was a truncated peptide from Helepsin; HTP1 and HTP2 were designed based on HTP by amino acid substitution with lysine. As shown in Figure 1B, the MICs of Helepsin against the tested P. aeruginosa strains were all 17.3 μM, while HTP had no inhibitory effects against the tested P. aeruginosa strains at 66.9 μM. The MICs of HTP1 against the tested P. aeruginosa strains were close to that of Helepsin, which were 16–32 μM. HTP2 displayed an improved activity against the tested P. aeruginosa strains compared to that of Helepsin with the MICs all 7.7 μM.
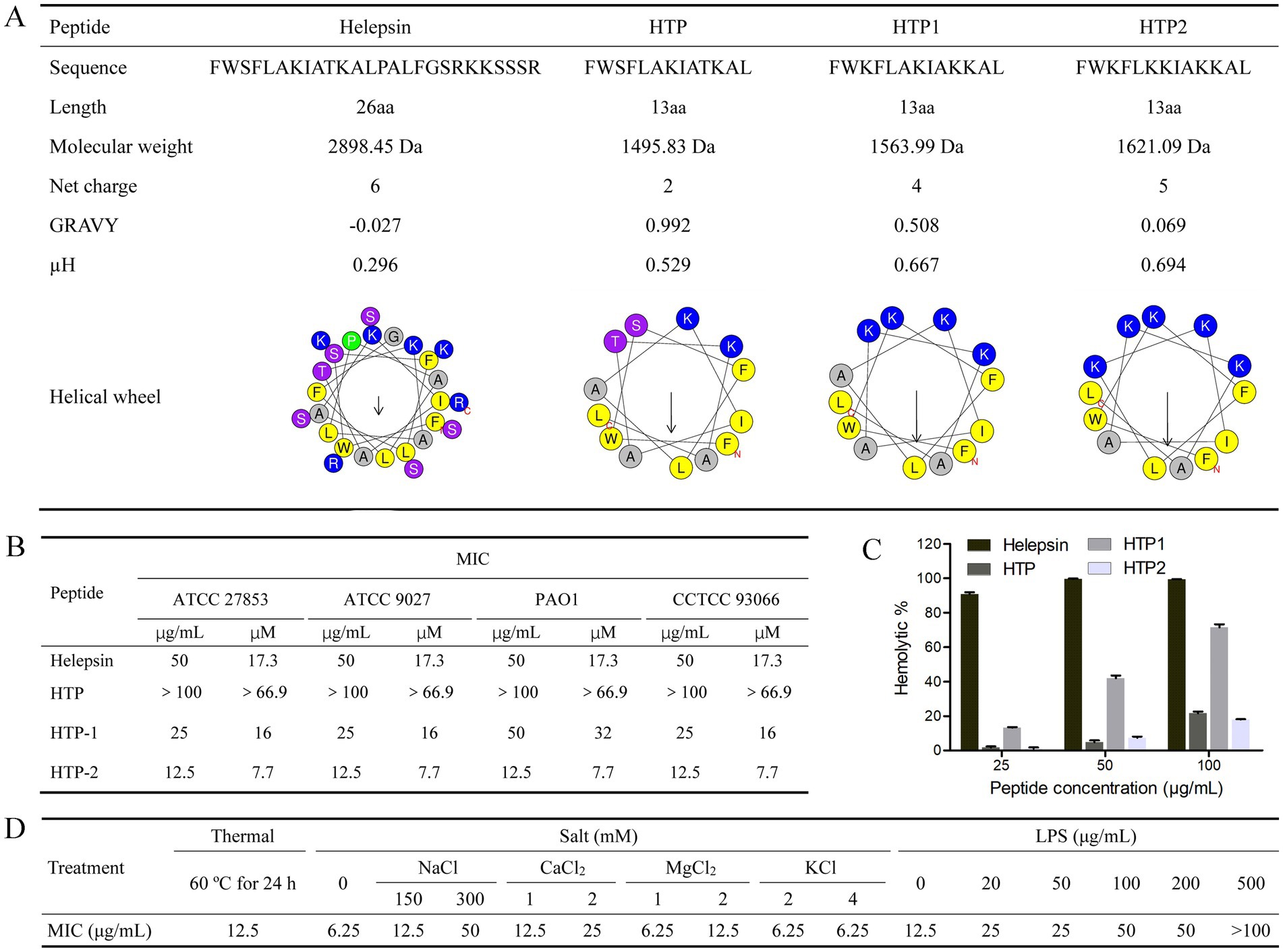
Figure 1. Peptides and in vitro activities. (A) Characterization and helical wheel diagram. (B) Anti-P. aeruginosa activity. (C) Hemolytic activity. (D) Stability and competition binding assay. GRAVY (Grand average of hydropathicity): Determined by ProtParam (https://web.expasy.org/protparam/). μH (Hydrophobic moment) and helical wheel diagram: Determined by the Heliquest (https://heliquest.ipmc.cnrs.fr/cgi-bin/ComputParams.py).
3.2 Hemolytic activity of the peptides
Hemolytic activity was used to evaluate the in vitro toxicity of the peptides. As shown in Figure 1C, Helepsin showed about 91% hemolysis at the concentration of 25 μg/mL (8.6 μM), while HTP showed only about 22% hemolysis at the concentration of 100 μg/mL (66.9 μM), and HTP2 showed only about 18% hemolysis at the concentration of 100 μg/mL (61.7 μM). Although HTP1 only showed about 13% hemolysis at the concentration of 25 μg/mL (16 μM), it showed about 72% hemolysis at the concentration of 100 μg/mL (63.9 μM).
3.3 Peptide stability of HTP2
According to the antibacterial activity and hemolytic activity, HTP2 was selected, and its stability was evaluated. As shown in Figure 1D, there are no changes in the MICs for HTP2 against P. aeruginosa after incubation at 60°C for 24 h. Thus, HTP2 had good thermal stability. When treated with HTP2 with different salts at varying concentrations, MICs showed varying degrees of changes. Low concentration monovalent ions (2 mM KCl, 4 mM KCl) had little effect on the antibacterial activity of HTP2, but high concentration monovalent ions (300 mM NaCl) had a greater impact on the antibacterial activity of HTP2. And low concentrations of divalent cations (1 mM/2 mM CaCl2, 2 mM MgCl2) have a significant impact on activity. Therefore, the impact of different ions, especially cations, on the activity of HTP2 varies.
3.4 Interaction between LPS and HTP2
Electrostatic interaction mediates the activity of AMPs, and LPS is the main anionic component on the surface of P. aeruginosa cells. Thus, the interaction between HTP2 and LPS was determined. As shown in Figure 1D, the MICs of HTP2 increased in the presence of additional LPS, indicating a decrease in the activity of HTP2 against P. aeruginosa. These results indicated that HTP2 could interact with LPS.
3.5 Time-killing kinetics of HTP2
To gain insights into the killing mode of HTP2 against P. aeruginosa, the time-killing kinetics were performed. As shown in Figure 2A, the amount of P. aeruginosa PAO1 cells did not show any changes after being treated with HTP2 at a concentration of 12.5 μg/mL for 30 min. When treated with HTP2 for 15 min at the concentrations of 25 μg/mL and 50 μg/mL, the amount of P. aeruginosa PAO1 cells decreased by about 1-log and about 3-log, respectively. Thus, HTP2 killed P. aeruginosa cells in a concentration-dependent manner.
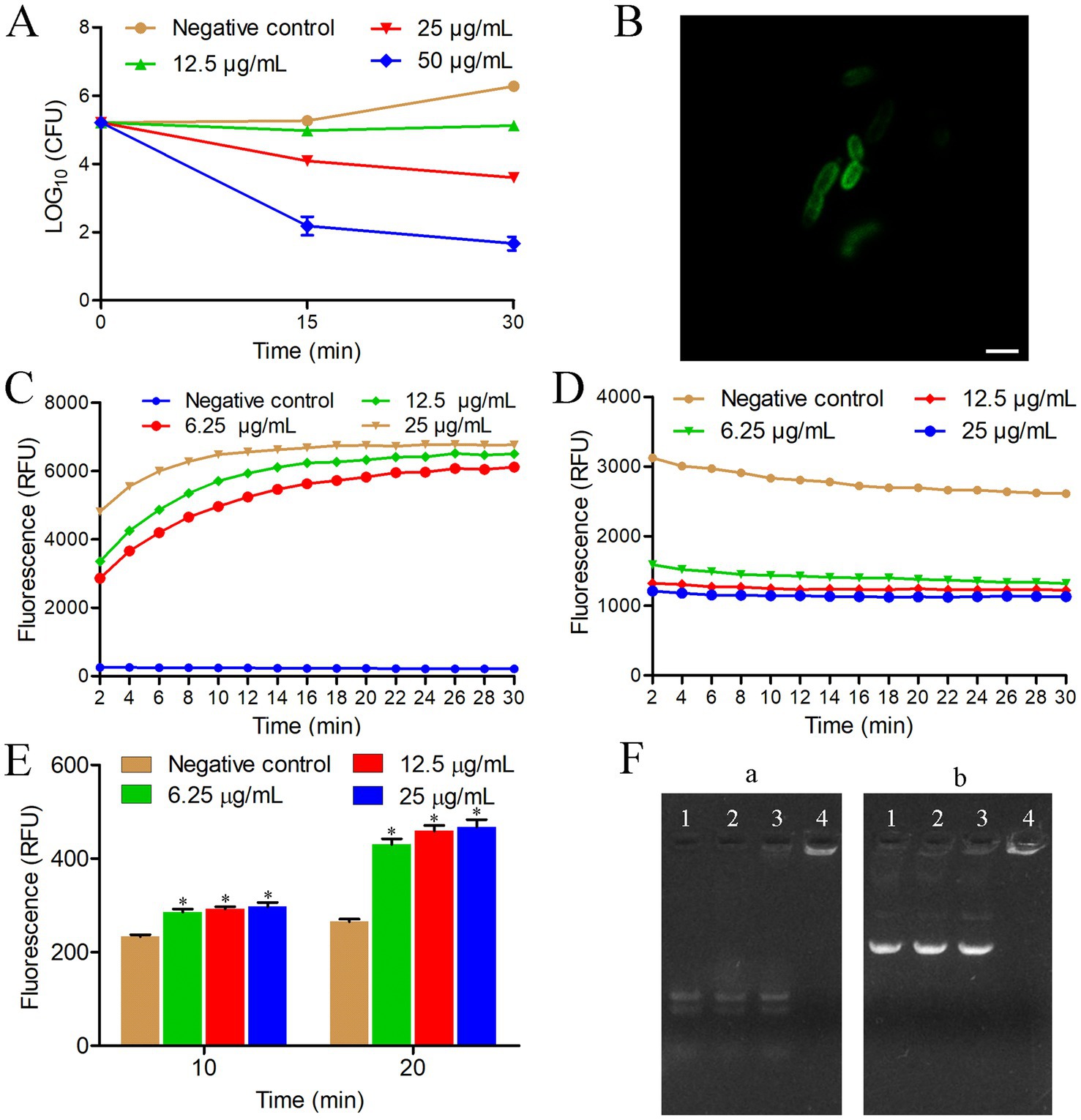
Figure 2. Anti-P. aeruginosa mechanism of HTP2. (A) Time-killing kinetics. Negative control group, 0.9% saline. (B) Confocal fluorescence microscopic images of P. aeruginosa cells treated with FITC-HTP2. (C) Membrane integrity measurement. Negative control group, 0.9% saline. RFU: Relative fluorescence unit. (D) Membrane potential measurement. Negative control group, 0.9% saline. RFU: Relative fluorescence unit. (E) ROS measurement. Negative control group, 0.9% saline. RFU: Relative fluorescence unit. *p < 0.05. (F) Nucleic acids binding assay. (a) P. aeruginosa RNA; (b) pET-28a; Ratio of peptide/nucleic acids: line 1 (0:1), line 2 (5:1), line 3 (10:1), line 4 (20:1).
3.6 Action sites of HTP2
To determine the action sites of HTP2, P. aeruginosa PAO1 cells were treated with FITC-HTP2. As shown in Figure 2B, a strong fluorescence distribution appeared on the cell surface, while there was a weak fluorescence distribution inside the cell. These results indicated that FITC-HTP2 mainly bound to the surface of P. aeruginosa PAO1 cells, and it could also act inside the cell.
3.7 Effects of HTP2 on the membrane integrity of Pseudomonas aeruginosa
To evaluate the influence of HTP2 on the membrane integrity of P. aeruginosa cells, the membrane permeability assay was performed using the fluorescence dye SYTOX green. As shown in Figure 2C, there is a significant increase in fluorescence intensity within a few minutes after treating P. aeruginosa PAO1 cells with HTP2 at the concentration of 6.25 μg/mL, 12.5 μg/mL, or 25 μg/mL, respectively. These results indicated that the membrane integrity of P. aeruginosa cells was disrupted by HTP2.
3.8 Effects of HTP2 on the membrane potential of Pseudomonas aeruginosa
To evaluate the influence of HTP2 on the membrane potential of P. aeruginosa cells, the membrane potential assay was performed using the fluorescence dye DiBAC4(3). As shown in Figure 2D, there is a rapid decrease in fluorescence intensity after treating P. aeruginosa PAO1 cells with HTP2 at the concentration of 6.25 μg/mL, 12.5 μg/mL, or 25 μg/mL, respectively. These results indicated that the membrane potential of P. aeruginosa cells was disrupted by HTP2.
3.9 Effects of HTP2 on ROS production of Pseudomonas aeruginosa
To evaluate the influence of HTP2 on the production of ROS in P. aeruginosa cells, the amount of ROS was detected using the fluorescence dye DCFH-DA. As shown in Figure 2E, there is a significant increase in fluorescence intensity after treating P. aeruginosa PAO1 cells with HTP2 at the concentration of 6.25 μg/mL, 12.5 μg/mL, or 25 μg/mL for 10 min or 20 min, respectively. These results indicated that HTP2 induced the production of ROS in P. aeruginosa cells.
3.10 Interaction between nucleic acid and HTP2
To evaluate whether HTP2 could interact with nucleic acids, the migration of nucleic acid in gel in the presence of HTP2 was tested. As shown in Figure 2F, the migration of RNA (Figure 2Fa) and plasmid DNA (Figure 2Fb) is retarded by HTP2. These results indicated that HTP2 could interact with different types of nucleic acids.
3.11 Effects of HTP2 on biofilm formation of Pseudomonas aeruginosa
Biofilm is an important pathogenic factor for P. aeruginosa, therefore, the influence of HTP2 on biofilm formation of P. aeruginosa was evaluated. As shown in Figure 3A, after treatment with HTP2 at the concentration of 6.25 μg/mL, 12.5 μg/mL, and 25 μg/mL, the biomass of P. aeruginosa biofilms was reduced by about 24.1% (p < 0.05), 41% (p < 0.05), and 71.7% (p < 0.05), respectively. Thus, HTP2 could significantly inhibit the biofilm formation of P. aeruginosa.
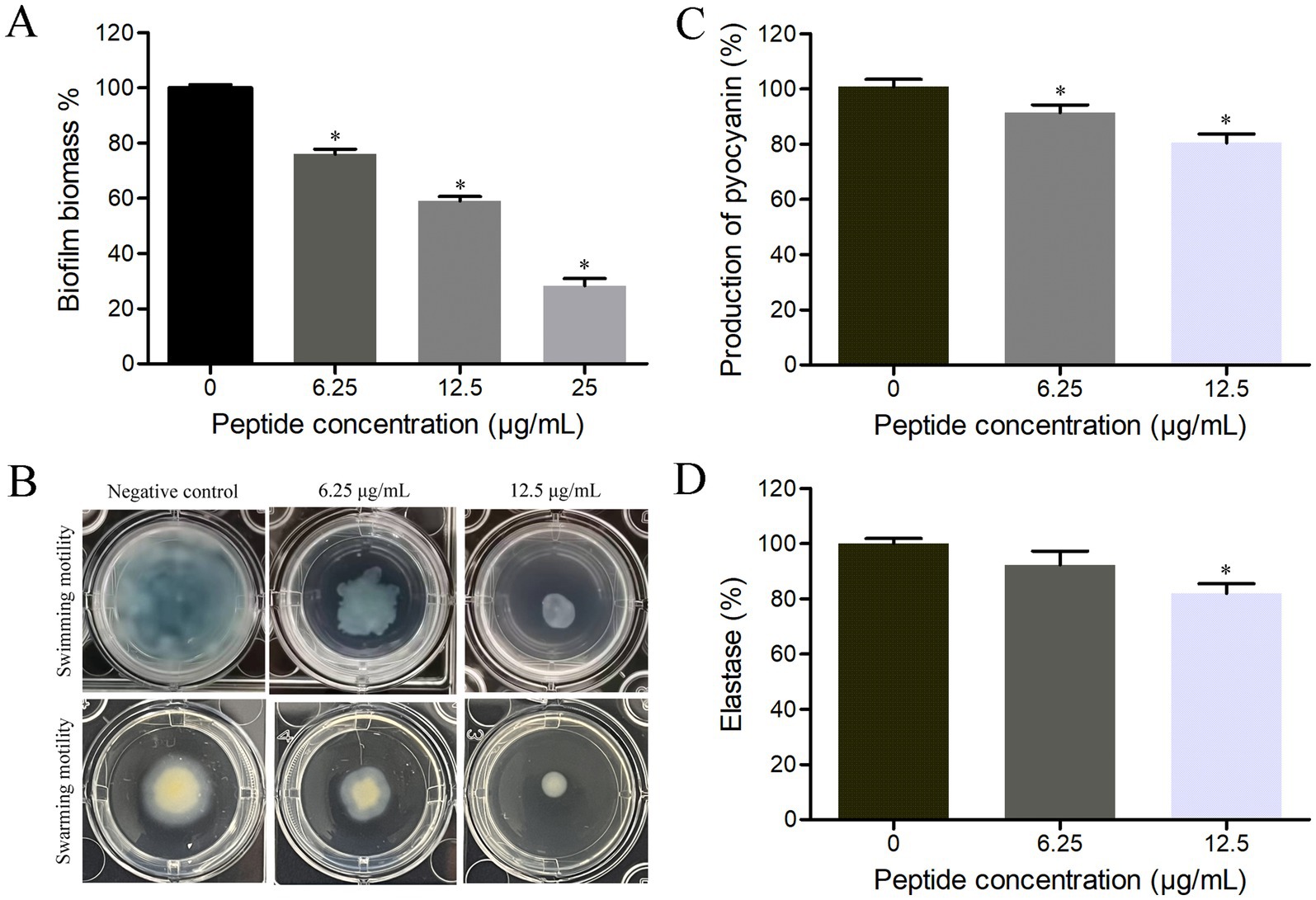
Figure 3. Influence of HTP2 on pathogenic factors of P. aeruginosa. (A) Biofilm formation. *p < 0.05. (B) Swimming motility assay. (C) Pyocyanin production. *p < 0.05. (D) Elastase activity. *p < 0.05.
3.12 Effects of HTP2 on motility of Pseudomonas aeruginosa
Motility ability plays a crucial role in the adhesion, colonization, and invasion processes of bacteria with flagella, therefore, the influence of HTP2 on the motility ability of P. aeruginosa was evaluated. As shown in Figure 3B, after treatment with HTP2 at the concentrations of 6.25 μg/mL and 12.5 μg/mL, the size of the bacterial lawn area was significantly smaller than that of the negative control (non-HTP2) treatment, respectively. Thus, HTP2 could significantly inhibit the motility ability of P. aeruginosa.
3.13 Effects of HTP2 on pyocyanin production of Pseudomonas aeruginosa
Pyocyanin can enhance the pathogenicity of P. aeruginosa through multiple modes, therefore, the influence of HTP2 on pyocyanin production of P. aeruginosa was evaluated. As shown in Figure 3C, after treatment with HTP2 at the concentrations of 6.25 μg/mL and 12.5 μg/mL, the production of pyocyanin decreased by about 9.6% (p < 0.05) and 19.6% (p < 0.05), respectively. Thus, HTP2 could significantly inhibit the pyocyanin production of P. aeruginosa.
3.14 Effects of HTP2 on elastase of Pseudomonas aeruginosa
Elastase plays an important role in the invasiveness of P. aeruginosa, therefore, the influence of HTP2 on the elastase production/activity of P. aeruginosa was evaluated. As shown in Figure 3D, after treatment with HTP2 at the concentrations of 6.25 μg/mL and 12.5 μg/mL, the elastase production/activity decreased by about 7.7% (p > 0.05) and 18.1% (p < 0.05), respectively. Thus, HTP2 could inhibit the production/activity of the elastase of P. aeruginosa.
3.15 In vivo anti-Pseudomonas aeruginosa activity of HTP2
A mouse subcutaneous infection model was used to evaluate the activity of HTP2 against P. aeruginosa under physiological conditions. As shown in Figure 4D, the bacterial load of P. aeruginosa from the infection area of HTP2-treated mice and Ciprofloxacin-treated mice was significantly decreased (p < 0.05) compared to that from the negative control group. And there was no significant difference between the HTP2-treated group and the Ciprofloxacin-treated group. Moreover, HTP2 significantly inhibited inflammatory infiltration (Figure 4B) in the infection area compared to that of the negative control group (Figure 4A). Thus, HTP2 had good anti-P. aeruginosa activity under physiological conditions.
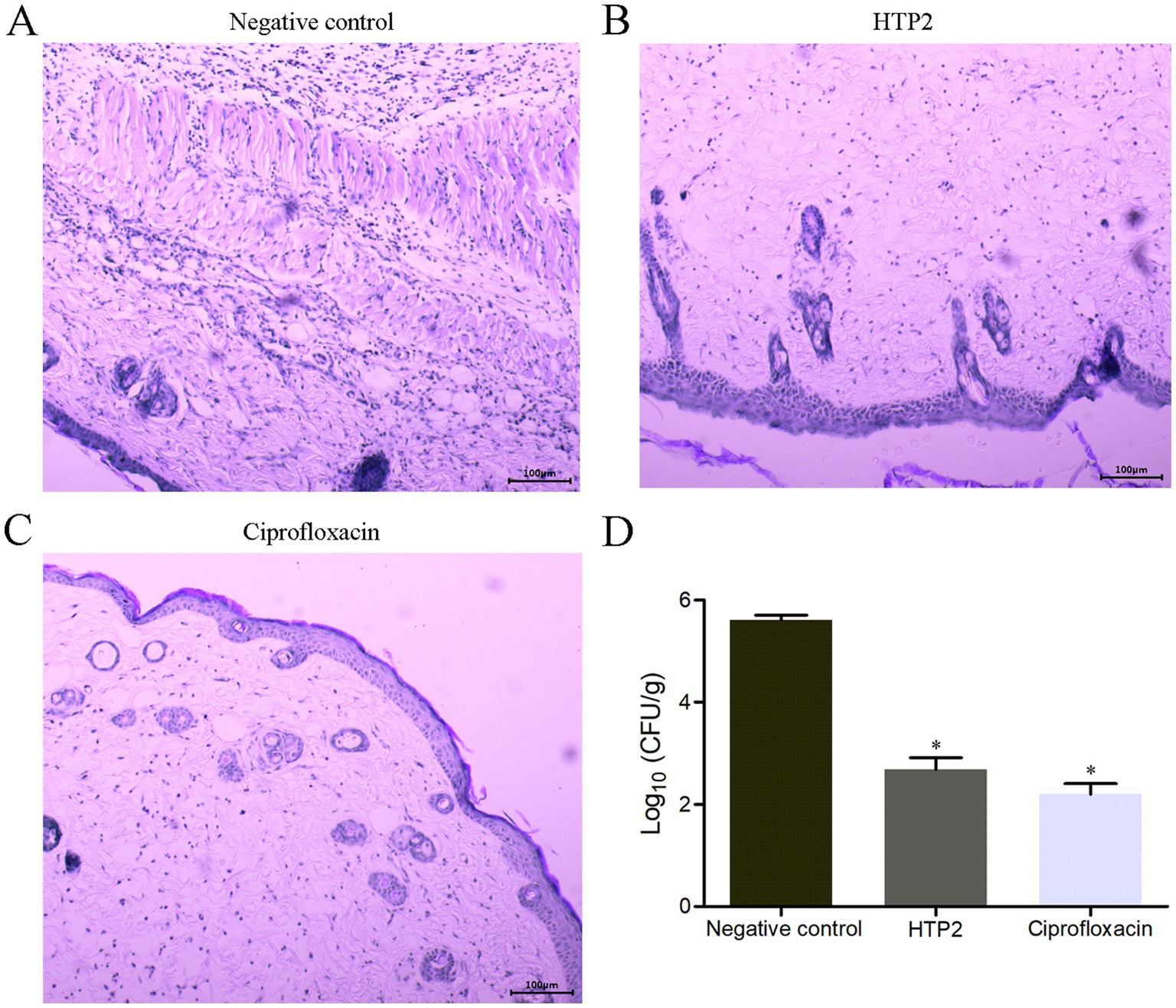
Figure 4. In vivo anti-P. aeruginosa activity of HTP2. (A) Hematoxylin–Eosin staining for the negative control group. (B) Hematoxylin–Eosin staining for the HTP2 treatment group. (C) Hematoxylin–Eosin staining for the Ciprofloxacin treatment group. (D) CFU per gram of the tissue. Negative control group, 0.9% saline. *p < 0.05.
4 Discussion
As one of the widely distributed bacteria, P. aeruginosa can cause food contamination, opportunistic infections, and foodborne diseases (Nahar et al., 2021; Li X. et al., 2022; Quiñones-Vico et al., 2024). The increasing antibiotic resistance of P. aeruginosa has made its impact more severe and its elimination more difficult (Reynolds and Kollef, 2021; Antimicrobial Resistance Collaborators, 2022). Thus, the development of novel antimicrobial agents that are safe, effective, and environmentally friendly is both critically important and urgently needed. Among these candidates, AMPs have attracted great attention.
In this study, the scorpion peptide Helepsin showed anti-P. aeruginosa activity with the MICs of 17.3 μM (Figure 1B), but it had a high hemolytic activity (90.8% hemolysis) at the concentration of 8.7 μM (25 μg/mL) (Figure 1C). To decrease the hemolytic activity, maintain/increase the anti-P. aeruginosa activity and reduce the cost, a truncated peptide HTP was designed based on Helepsin, and then two derivatives (HTP1 and HTP2) were designed based on HTP using amino acid substitution with lysine (Figure 1A). Previous studies have shown that an increase in the proportion of hydrophobic amino acids, hydrophobic moment, or net positive charge can enhance the antibacterial activity of AMPs (Brogden, 2005). Although HTP had a lower hemolytic activity compared to Helepsin (Figure 1C), it did not show anti-P. aeruginosa activity at the concentration of 66.9 μM (Figure 1B). It was mainly due to the decrease in percentage of hydrophobic amino acids (a higher GRAVY) and net positive charge, even with a higher hydrophobic moment (Figure 1A). Both HTP1 and HTP2 had an increased anti-P. aeruginosa activity, which was mainly due to the increase in proportion of hydrophobic amino acids (decrease in GRAVY), hydrophobic moment, and net positive charge (Figure 1A) compared to HTP. Moreover, HTP2 also had a very low hemolytic activity among the four peptides (Figure 1C). Thus, HTP2 was the optimal peptide in this study, which was selected for the in vivo study. In the mouse cutaneous infection model, HTP2 could significantly (p < 0.05) reduce the bacterial load of P. aeruginosa cells (Figure 4D) and inhibit inflammatory infiltration (Figure 4B) in the infection area. Thus, HTP2 had the potential as an antibacterial agent or a sanitizer against P. aeruginosa.
To investigate the mechanism of action of HTP2 against P. aeruginosa, P. aeruginosa PAO1 was selected as the model bacterial strain. Our results showed that HTP2 killed P. aeruginosa cells in a dose-dependent way (Figure 2A) and mainly bound to the surface of the cells (Figure 2B). It’s well known that cationic AMPs can disrupt the cytoplasmic membrane after binding to the surface of the bacterial cell, and the binding is mediated by electrostatic interactions between the peptide and anions on the cell surface (Lohner, 2009; Fjell et al., 2011). To verify whether the binding of HTP2 to P. aeruginosa cells was related to electrostatic attraction, the cation stability assay and LPS competition binding assay were performed. Our results showed that external cations (especially multivalent cations) and external LPS could decrease the anti-P. aeruginosa of HTP2 (Figure 1D), indicating that electrostatic interactions played an important role in the anti-P. aeruginosa activity of HTP2, and HTP2 interacted with LPS (the major anionic components on the cell surface of P. aeruginosa). To further verify whether HTP2 affected the integrity and function of cell membranes, the membrane permeability assay and membrane potential assay were performed. Our results showed that HTP2 could disrupt the integrity (Figure 2C) and potential (Figure 2D) of P. aeruginosa membrane, which would finally disrupt the normal physiological functions of the cell membranes. Besides acting on the cell membrane, AMPs can interfere with intracellular physiological regulation in various ways, such as interacting with intracellular targets and inducing ROS accumulation (Nicolas, 2009). Our results showed that HTP2 could induce the accumulation of ROS (Figure 2E), which might damage DNA, RNA, proteins, and membrane lipids (Borisov et al., 2021). HTP2 could also interact with nucleic acids (Figure 2F), which might inhibit the replication, transcription, and translation of nucleic acids. Thus, HTP2 killed P. aeruginosa cells via a multi-mode manner.
Many virulence factors of P. aeruginosa play important roles during the pathogenic processes, or are beneficial for contaminating food, or make it difficult to be eliminated. P. aeruginosa can form biofilm on the surface of tissue, food, or objects, which protects the pathogen from being killed by the host immune system and/or antibacterial agents (Li X. et al., 2022; Yin et al., 2022). Motility can promote bacterial adhesion and invasion of P. aeruginosa, help the pathogen migrate to favorable environments, and escape from harmful environments (Matilla et al., 2021; Muggeo et al., 2023). Pyocyanin, an extracellular secreted redox-active phenazine secondary metabolite produced by P. aeruginosa, can induce the generation of reactive oxygen species and activate pro-inflammatory signaling pathways of host cells, and can disrupt macrophage phagocytic function (Lew et al., 2025). Elastase is an extracellular secreted metalloprotease, which can promote host invasion and immune evasion of P. aeruginosa by cleaving the host substrates and immune system components, and can activate pathogenicity-associated proteins/proteases/exotoxins (Camberlein et al., 2022). Our results showed that HTP2 could inhibit the biofilm formation (Figure 3A), motility (Figure 3B), pyocyanin production (Figure 3C), and elastase activity (Figure 3D) of P. aeruginosa. Thus, HTP2 could prevent the pathogenic process of opportunistic infections and foodborne diseases caused by P. aeruginosa via inhibiting various pathogenic factors.
5 Conclusion
In conclusion, the peptide HTP2 effectively inhibited the growth of P. aeruginosa in vitro and in vivo. HTP2 killed P. aeruginosa cells via a multi-manner, including damaging the membrane, inducing ROS accumulation, and interacting with nucleic acids. HTP2 could also inhibit biofilm formation, motility, pyocyanin production, and elastase activity of P. aeruginosa. Taken together, HTP2 had the potential as an antibacterial agent or a sanitizer against P. aeruginosa.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The animal study was approved by Animal Care and Use Committee of Henan University of Technology and Science. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
ZL: Data curation, Validation, Methodology, Project administration, Investigation, Funding acquisition, Writing – review & editing, Resources, Formal analysis, Writing – original draft. JZ: Formal analysis, Visualization, Resources, Methodology, Writing – review & editing, Investigation. YL: Resources, Writing – review & editing, Validation. QD: Validation, Writing – review & editing, Resources. SL: Formal analysis, Validation, Methodology, Writing – review & editing, Resources. BD: Validation, Resources, Writing – review & editing. PW: Validation, Resources, Writing – review & editing. WL: Resources, Validation, Writing – review & editing. YD: Validation, Writing – review & editing, Resources. PX: Supervision, Funding acquisition, Resources, Writing – review & editing. WZ: Writing – review & editing, Funding acquisition, Resources, Supervision, Data curation.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was funded by Young Backbone Teachers of Henan Province Colleges and Universities (2024GGJS054), Key Research and Development Program of Henan Province (242102310253), Key Scientific Research Projects of Henan Province Higher Education Institutions (25A310024), National Natural Science Foundation of China (22271079), and Research Project from Pingyuan Laboratory (2023PY-ZZ-0201).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Antimicrobial Resistance Collaborators (2022). Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet 399, 629–655. doi: 10.1016/S0140-6736(21)02724-0
Artini, M., Imperlini, E., Buonocore, F., Relucenti, M., Porcelli, F., Donfrancesco, O., et al. (2022). Anti-virulence potential of a Chionodracine-derived peptide against multidrug-resistant Pseudomonas aeruginosa clinical isolates from cystic fibrosis patients. Int. J. Mol. Sci. 23:13494. doi: 10.3390/ijms232113494
Bin Hafeez, A., Jiang, X., Bergen, P. J., and Zhu, Y. (2021). Antimicrobial peptides: an update on classifications and databases. Int. J. Mol. Sci. 22:11691. doi: 10.3390/ijms222111691
Borisov, V. B., Siletsky, S. A., Nastasi, M. R., and Forte, E. (2021). ROS defense systems and terminal oxidases in Bacteria. Antioxidants (Basel) 10:839. doi: 10.3390/antiox10060839
Brogden, K. A. (2005). Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat. Rev. Microbiol. 3, 238–250. doi: 10.1038/nrmicro1098
Camberlein, V., Jézéquel, G., Haupenthal, J., and Hirsch, A. K. H. (2022). The structures and binding modes of small-molecule inhibitors of Pseudomonas aeruginosa elastase LasB. Antibiotics (Basel) 11:1060. doi: 10.3390/antibiotics11081060
Cao, L., Dai, C., Li, Z., Fan, Z., Song, Y., Wu, Y., et al. (2012). Antibacterial activity and mechanism of a scorpion venom peptide derivative in vitro and in vivo. PLoS One 7:e40135. doi: 10.1371/journal.pone.0040135
Chen, T., Xu, Y., Xu, W., Liao, W., Xu, C., Zhang, X., et al. (2020). Hypertonic glucose inhibits growth and attenuates virulence factors of multidrug-resistant Pseudomonas aeruginosa. BMC Microbiol. 20:203. doi: 10.1186/s12866-020-01889-2
CLSI (2019). Performance standards for antimicrobial susceptibility testing. 29th Edn: CLSI. Available at: https://clsi.org/
Fjell, C. D., Hiss, J. A., Hancock, R. E., and Schneider, G. (2011). Designing antimicrobial peptides: form follows function. Nat. Rev. Drug Discov. 11, 37–51. doi: 10.1038/nrd3591
Gagat, P., Ostrówka, M., Duda-Madej, A., and Mackiewicz, P. (2024). Enhancing antimicrobial peptide activity through modifications of charge, hydrophobicity, and structure. Int. J. Mol. Sci. 25:10821. doi: 10.3390/ijms251910821
Hauser, A. R., Jain, M., Bar-Meir, M., and McColley, S. A. (2011). Clinical significance of microbial infection and adaptation in cystic fibrosis. Clin. Microbiol. Rev. 24, 29–70. doi: 10.1128/Cmr.00036-10
Kazemi-Lomedasht, F., Khalaj, V., Bagheri, K. P., Behdani, M., and Shahbazzadeh, D. (2017). The first report on transcriptome analysis of the venom gland of Iranian scorpion, Hemiscorpius lepturus. Toxicon 125, 123–130. doi: 10.1016/j.toxicon.2016.11.261
Kunz Coyne, A. J., el Ghali, A., Holger, D., Rebold, N., and Rybak, M. J. (2022). Therapeutic strategies for emerging multidrug-resistant Pseudomonas aeruginosa. Infect. Dis. Ther. 11, 661–682. doi: 10.1007/s40121-022-00591-2
Lee, D. H., Kim, B. S., and Kang, S. S. (2020). Bacteriocin of Pediococcus acidilactici HW01 inhibits biofilm formation and virulence factor production by Pseudomonas aeruginosa. Probiotics Antimicrob. Proteins 12, 73–81. doi: 10.1007/s12602-019-09623-9
Lee, W., and Lee, D. G. (2014). Magainin 2 induces bacterial cell death showing apoptotic properties. Curr. Microbiol. 69, 794–801. doi: 10.1007/s00284-014-0657-x
Lew, S. Q., Chong, S. Y., and Lau, G. W. (2025). Modulation of pulmonary immune functions by the Pseudomonas aeruginosa secondary metabolite pyocyanin. Front. Immunol. 16:1550724. doi: 10.3389/fimmu.2025.1550724
Li, X., Gu, N., Huang, T. Y., Zhong, F., and Peng, G. (2022). Pseudomonas aeruginosa: a typical biofilm forming pathogen and an emerging but underestimated pathogen in food processing. Front. Microbiol. 13:1114199. doi: 10.3389/fmicb.2022.1114199
Li, Z., Jing, X., Yuan, Y., Shui, Y., Li, S., Zhao, Z., et al. (2022). In vitro and in vivo activity of Phibilin against Candida albicans. Front. Microbiol. 13:862834. doi: 10.3389/fmicb.2022.862834
Li, Z., Liu, G., Meng, L., Yu, W., Xu, X., Li, W., et al. (2016). K1K8: an Hp1404-derived antibacterial peptide. Appl. Microbiol. Biotechnol. 100, 5069–5077. doi: 10.1007/s00253-016-7395-x
Li, S., Shui, Y., Ma, J., Yuan, Y., Jiang, W., Xu, C., et al. (2022). Antimicrobial activity of CT-K3K7, a modified peptide by lysine substitutions from ctry2459 - a Chaerilus tryznai scorpion venom peptide. Toxicon 218, 88–98. doi: 10.1016/j.toxicon.2022.09.004
Li, Z., Xu, X., Meng, L., Zhang, Q., Cao, L., Li, W., et al. (2014). Hp1404, a new antimicrobial peptide from the scorpion Heterometrus petersii. PLoS One 9:e97539. doi: 10.1371/journal.pone.0097539
Li, Z., Yuan, Y., Li, S., Deng, B., and Wang, Y. (2020). Antibacterial activity of a scorpion-derived peptide and its derivatives in vitro and in vivo. Toxicon 186, 35–41. doi: 10.1016/j.toxicon.2020.07.028
Liao, C., Huang, X., Wang, Q., Yao, D., and Lu, W. (2022). Virulence factors of pseudomonas aeruginosa and Antivirulence strategies to combat its drug resistance. Front. Cell. Infect. Microbiol. 12:926758. doi: 10.3389/fcimb.2022.926758
Lohner, K. (2009). New strategies for novel antibiotics: peptides targeting bacterial cell membranes. Gen. Physiol. Biophys. 28, 105–116. doi: 10.4149/gpb_2009_02_105
Lourenço, A. L. P., Rios, T. B., da Silva, Á. P., Franco, O. L., and Ramada, M. H. S. (2023). Peptide stapling applied to antimicrobial peptides. Antibiotics (Basel) 12:1400. doi: 10.3390/antibiotics12091400
Matilla, M. A., Martín-Mora, D., Gavira, J. A., and Krell, T. (2021). Pseudomonas aeruginosa as a model to study chemosensory pathway signaling. Microbiol. Mol. Biol. Rev. 85:e00151-20. doi: 10.1128/MMBR.00151-20
Maurice, N. M., Bedi, B., and Sadikot, R. T. (2018). Pseudomonas aeruginosa biofilms: host response and clinical implications in lung infections. Am. J. Respir. Cell Mol. Biol. 58, 428–439. doi: 10.1165/rcmb.2017-0321TR
Mirzaei, R., Alikhani, M. Y., Arciola, C. R., Sedighi, I., Irajian, G. R., Jamasbi, E., et al. (2022). Highly synergistic effects of Melittin with vancomycin and rifampin against vancomycin and rifampin resistant Staphylococcus epidermidis. Front. Microbiol. 13:869650. doi: 10.3389/fmicb.2022.869650
Muggeo, A., Coraux, C., and Guillard, T. (2023). Current concepts on Pseudomonas aeruginosa interaction with human airway epithelium. PLoS Pathog. 19:e1011221. doi: 10.1371/journal.ppat.1011221
Musthafa, K. S., Balamurugan, K., Pandian, S. K., and Ravi, A. V. (2012). 2,5-piperazinedione inhibits quorum sensing-dependent factor production in Pseudomonas aeruginosa PAO1. J. Basic Microbiol. 52, 679–686. doi: 10.1002/jobm.201100292
Nahar, S., Ha, A. J. W., Byun, K. H., Hossain, M. I., Mizan, M. F. R., and Ha, S. D. (2021). Efficacy of flavourzyme against Salmonella Typhimurium, Escherichia coli, and Pseudomonas aeruginosa biofilms on food-contact surfaces. Int. J. Food Microbiol. 336:108897. doi: 10.1016/j.ijfoodmicro.2020.108897
Nguyen, L. T., Haney, E. F., and Vogel, H. J. (2011). The expanding scope of antimicrobial peptide structures and their modes of action. Trends Biotechnol. 29, 464–472. doi: 10.1016/j.tibtech.2011.05.001
Nicolas, P. (2009). Multifunctional host defense peptides: intracellular-targeting antimicrobial peptides. FEBS J. 276, 6483–6496. doi: 10.1111/j.1742-4658.2009.07359.x
Pan, X., Liang, H., Zhao, X., Zhang, Q., Chen, L., Yue, Z., et al. (2023). Regulatory and structural mechanisms of PvrA-mediated regulation of the PQS quorum-sensing system and PHA biosynthesis in Pseudomonas aeruginosa. Nucleic Acids Res. 51, 2691–2708. doi: 10.1093/nar/gkad059
Quiñones-Vico, M. I., Ubago-Rodríguez, A., Fernández-González, A., Sanabria-de la Torre, R., Sierra-Sánchez, Á., Montero-Vilchez, T., et al. (2024). Antibiotic nanoparticles-loaded wound dressings against Pseudomonas aeruginosa's skin infection: a systematic review. Int. J. Nanomedicine 19, 7895–7926. doi: 10.2147/IJN.S469724
Reynolds, D., and Kollef, M. (2021). The epidemiology and pathogenesis and treatment of Pseudomonas aeruginosa infections: an update. Drugs 81, 2117–2131. doi: 10.1007/s40265-021-01635-6
Schalli, M., Platzer, S., Haas, D., and Reinthaler, F. F. (2023). The behaviour of Escherichia coli and Pseudomonas aeruginosa in bottled mineral water. Heliyon 9:e21634. doi: 10.1016/j.heliyon.2023.e21634
Shahrour, H., Ferrer-Espada, R., Dandache, I., Bárcena-Varela, S., Sánchez-Gómez, S., Chokr, A., et al. (2019). AMPs as anti-biofilm agents for human therapy and prophylaxis. Adv. Exp. Med. Biol. 1117, 257–279. doi: 10.1007/978-981-13-3588-4_14
She, P., Wang, Y., Luo, Z., Chen, L., Tan, R., Wang, Y., et al. (2018). Meloxicam inhibits biofilm formation and enhances antimicrobial agents efficacy by Pseudomonas aeruginosa. Microbiology 7:e00545. doi: 10.1002/mbo3.545
Shi, J., Chen, C., Wang, D., Tong, Z., Wang, Z., and Liu, Y. (2021). Amphipathic peptide antibiotics with potent activity against multidrug-resistant pathogens. Pharmaceutics 13:438. doi: 10.3390/pharmaceutics13040438
Verma, D. P., Tripathi, A. K., and Thakur, A. K. (2024). Innovative strategies and methodologies in antimicrobial peptide design. J Funct Biomater 15:320. doi: 10.3390/jfb15110320
Wu, S., Liu, G., Jin, W., Xiu, P., and Sun, C. (2016). Antibiofilm and anti-infection of a marine bacterial exopolysaccharide against Pseudomonas aeruginosa. Front. Microbiol. 7:102. doi: 10.3389/fmicb.2016.00102
Xu, Z. G., Gao, Y., He, J. G., Xu, W. F., Jiang, M., and Jin, H. S. (2015). Effects of azithromycin on Pseudomonas aeruginosa isolates from catheter-associated urinary tract infection. Exp. Ther. Med. 9, 569–572. doi: 10.3892/etm.2014.2120
Yin, R., Cheng, J., Wang, J., Li, P., and Lin, J. (2022). Treatment of Pseudomonas aeruginosa infectious biofilms: challenges and strategies. Front. Microbiol. 13:955286. doi: 10.3389/fmicb.2022.955286
Keywords: Pseudomonas aeruginosa , skin infection, food contamination, scorpion, antimicrobial peptide
Citation: Li Z, Zhang J, Liu Y, Dai Q, Li S, Deng B, Wu P, Li W, Dong Y, Xin P and Zhang W (2025) In vitro and in vivo anti-Pseudomonas aeruginosa activity of a scorpion peptide derivative. Front. Microbiol. 16:1622282. doi: 10.3389/fmicb.2025.1622282
Edited by:
Aravind Madhavan, Amrita Vishwa Vidyapeetham University, IndiaReviewed by:
Jhon Carlos Castaño, University of Quindío, ColombiaSatya Deo Pandey, University of Louisville, United States
Copyright © 2025 Li, Zhang, Liu, Dai, Li, Deng, Wu, Li, Dong, Xin and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenlu Zhang, ejkwNzczMzI3MEAxNjMuY29t; Pengyang Xin, cHl4aW4yN0AxNjMuY29t
 Zhongjie Li
Zhongjie Li Jiao Zhang1
Jiao Zhang1 Pengfei Wu
Pengfei Wu Wanwu Li
Wanwu Li Pengyang Xin
Pengyang Xin