- 1Guangxi Key Laboratory of Aquatic Genetic Breeding and Healthy Aquaculture, Guangxi Academy of Fishery Science, Nanning, Guangxi, China
- 2Key Laboratory of Comprehensive Development and Utilization of Aquatic Germplasm Resources of China (Guangxi) and ASEAN (Co-Construction by Ministry and Province), Ministry of Agriculture and Rural Affairs, Nanning, Guangxi, China
- 3Guangdong Provincial Key Laboratory of Aquatic Animal Disease Control and Healthy Culture, Fisheries College of Guangdong Ocean University, Shenzhen Institute of Guangdong Ocean University, Guangdong, China
A bacterial strain (No. 20230510) was isolated from the kidneys of diseased Siniperca chuatsi in Guangxi, China, since 2023. Artificial infection experiments demonstrated that this strain caused the observed disease in S. chuatsi. The isolate underwent morphological, pathological, genomic (whole-genome sequencing, WGS), and antibiotic susceptibility analyses. Infection trials revealed 100% mortality in high-concentration groups, with an LD50 of 3.89 × 104 CFU/mL, indicating high virulence. WGS results showed a circular chromosome of 8,123,106 bp (GC content: 68.14%), containing 7,638 CDSs, 72 tRNAs, and 12 rRNAs. Phylogenomic analysis revealed that strain 20230510 (CP130742) clusters with three N. seriolae strains with 98% bootstrap supporting, confirming its identification as N. seriolae. Further analysis identified 403 potential virulence genes linked to nutrient metabolism, regulatory factors, immune modulation, effector delivery systems, and exotoxins. Chromosomal comparisons also detected multiple antibiotic resistance genes. Susceptibility testing confirmed sensitivity to nine antibiotics, including enrofloxacin, doxycycline, florfenicol, and sulfamethoxazole. Histopathology revealed chronic granulomatous lesions, most severe in the kidneys, with similar but milder damage in the liver, spleen, gills, and intestines. These results confirm N. seriolae strain 20230510 as the pathogenic agent behind S. chuatsi mortality, offering key insights for developing control strategies.
1 Introduction
Siniperca chuatsi, a commercially important fish species in China, is prized for its tender meat and high nutritional value, driving substantial consumer demand. Official statistics report annual aquaculture production exceeding 400,000 tons (Ministry of Agriculture and Rural Affairs Fisheries Administration Bureau et al., 2023). However, intensified farming practices have led to rising disease prevalence. Documented pathogens include infectious spleen and kidney necrosis virus, iridovirus, Aeromonas hydrophila, A. veronii, Streptococcus uberis, Flavobacterium columnare, and A. salmonicida (Chen et al., 2018; Lin et al., 2020; Liu et al., 2018; Luo et al., 2017; Liu et al., 2020; Zhou et al., 2015; Zhu et al., 2023). Notably, no documented cases of N. seriolae infection in S. chuatsi have been reported to date, despite its known capability to induce mass mortality and severe economic losses in aquaculture systems.
N. seriolae is an environmentally ubiquitous opportunistic pathogen, with 113 Nocardia species identified across diverse niches, including soil, aquatic systems, rhizospheres, insects, fish, and human clinical specimens (Han et al., 2019). Nocardiosis, a chronic systemic disease characterized by high fatality rates (Zhang et al., 2022), primarily presents as cutaneous ulcers and visceral granulomatous lesions (Chen et al., 2000; Huang et al., 2021; Kim et al., 2018; Liao et al., 2021; Shimahara et al., 2008). First isolated from Japanese amberjack in 1968 (Kariya et al., 1968), Nocardia spp. have since been documented in numerous aquatic species, including Channa maculata, Micropterus salmoides (Chen, 1992), Crassostrea gigas (Friedman et al., 1998), Oncorhynchus nerka (Isik et al., 1999), Terapon jarbua (Wang et al., 2009), Trachinotus blochii (Vu-Khac et al., 2016), Anguilla japonica (Kim et al., 2018), Sciaenops ocellatus (Rio-Rodriguez et al., 2021), hybrid snakehead (Zhang et al., 2022), Oreochromis niloticus, Chanos chanos, and Lates calcarifer (Nazareth et al., 2024). Among these, N. seriolae constitutes the predominant pathogen (Kim et al., 2018; Nazareth et al., 2024; Rio-Rodriguez et al., 2021; Vu-Khac et al., 2016; Wang et al., 2009; Zhang et al., 2022), alongside N. asteroides (Chen, 1992), N. crassostrea (Friedman et al., 1998), and N. salmonicida (Isik et al., 1999). Aquacultural outbreaks caused by N. seriolae exhibit cumulative mortality rates ranging from 0.86 to 70%, posing substantial economic risks (Chen et al., 2000; Huang et al., 2021; Kim et al., 2018; Liao et al., 2021; Matsumoto et al., 2017; Rio-Rodriguez et al., 2021; Wang et al., 2009).
The advancement of whole-genome sequencing (WGS) technologies has enabled comprehensive analyses of bacterial virulence mechanisms, host-pathogen-environment interactions, and strain-specific traits (Strauss and Falkow, 1997). As a pivotal tool for virulence factor research, WGS has been applied to characterize N. seriolae genomes, including virulence determinants, antimicrobial resistance genes (Han et al., 2019; Kim et al., 2021; Umeda et al., 2023; Yasuike et al., 2017), and draft assemblies (Xia et al., 2015).
In May, 2023, a disease outbreak causing significant mortality in S. chuatsi was documented at an aquaculture facility in Guangxi, China. Laboratory investigations identified N. seriolae as the causative agent through (1) histopathological concordance with nocardial infections, and (2) 16S rRNA gene sequencing (>99% identity with N. seriolae type strain). To our knowledge, no previous studies have reported N. seriolae infections in this fish species. In this study, we performed histopathological examinations and whole-genome sequencing (WGS) analysis of the isolated bacterium, comparing it with previously reported N. seriolae genomes. Using multiple databases, we classified and annotated the genome sequence of this strain, predicted gene functions, and identified virulence and resistance-related genes. These findings provide foundational data for understanding N. seriolae's pathogenic mechanisms in S. chuatsi and support the development of disease prevention strategies.
2 Materials and methods
2.1 Ethical statement
The research adhered to China's regulations on the ethical treatment and utilization of laboratory animals. The experimental protocols were reviewed and authorized by the Animal Ethics Committee of Guangxi Academy of Fisheries Sciences (Approval No. GAFS2021001).
2.2 Experimental materials
Diseased mandarin fish (S. chuatsi) specimens (mean ± SD = 76 ± 10.46 g) were collected from a commercial aquaculture facility in Nanning, Guangxi Province. Healthy specimens (50 ± 8.12 g) were procured from the Guangxi Aquatic Science Research Institute's Wuming experimental base, undergoing a 7-day acclimation period before experimentation.
2.3 Isolation and identification of bacteria from diseased S. chuatsi
Following sterile protocols, S. chuatsi specimens were surface-sterilized using 75% ethanol and aseptically dissected to expose muscle tissue. Bacterial isolation involved streaking tissue samples onto blood agar plates (10% sheep blood agar plates, Beijing Luqiao Technology Co., Ltd.) with subsequent 25°C incubation for 3–5 days. Morphologically distinct colonies were isolated through serial purification, yielding strain 20230510. This strain was then cultured in tryptic soy broth (25°C, 120 rpm, 5–7 days). An aliquot was subjected to Gram staining per manufacturer's protocol (Gram Stain Kit, Beijing Solarbio Science & Technology Co., Ltd.) for morphological analysis, while the remainder was allocated for virulence assessment and cryopreservation (−86°C in 20% glycerol medium).
2.4 Pathogenicity testing of isolated strain on S. chuatsi
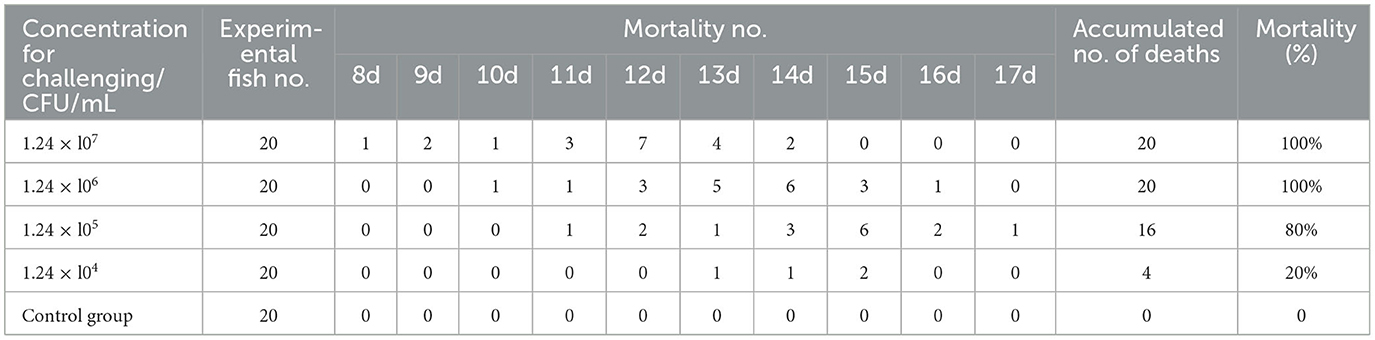
Healthy mandarin fish (S. chuatsi) were acclimatized in controlled aquarium systems for 1 week prior to experimental procedures. Infection challenges were implemented in 200-L tanks across a 20-day exposure period, with test subjects stratified into five cohorts (4 experimental + 1 control; n = 20/group). Quadruplicate bacterial suspensions (1.24 × 107 CFU/mL) prepared in sterile saline were administered via intraperitoneal (IP) injection (0.2 mL/fish), while controls received equivalent volumes of saline. Aquaria maintenance included semiweekly 50% water renewal with continuous aeration, temperature regulation (20–28°C), and scheduled feeding (2% BW/day). Daily monitoring documented clinical signs and mortality events, with moribund specimens undergoing pathogen recovery. LD50 values with 95% confidence intervals were determined by probit regression (GraphPad Prism 9.5), with supplementary verification using the classical Reed-Muench method.
2.5 Whole-genome sequencing and analysis
2.5.1 Molecular identification of isolated strain
Genomic DNA was extracted using the DNAzol™ Reagent DNA extraction kit (Hangzhou BoRi Technology Co., Ltd.). PCR reactions and subsequent sequencing were performed according to the methods described in reference (Heuer et al., 1997). The universal primers for 16S rRNA gene amplification and PCR reagents were all supplied by Takara Biotechnology (Dalian) Co., Ltd. The sequencing results were subjected to BLAST comparison in the NCBI database.
2.5.2 Sequencing
The target strain (20230510) was cultured in tryptic soy broth (TSB) at 25 °C for 7 days under standard conditions. Genomic DNA was extracted using DNAzol™ Reagent, resuspended in 100 μL of ultrapure water, and treated with RNase A (20 μg/mL) to remove residual RNA. DNA quality was evaluated through multiple approaches, including visual inspection for particulate contamination, agarose gel electrophoresis to assess integrity, NanoDrop™ spectrophotometry for purity (A260/A280 ratio), and Qubit™ fluorometry for accurate quantification.
For Oxford Nanopore sequencing, high-quality DNA was subjected to size selection using magnetic beads. The selected DNA was then end-repaired and purified, followed by barcoding using the EXP-NBD104/114 kit. A second round of purification was performed before ligating sequencing adapters with the SQK-LSK109 kit. The final library was quantified using a Qubit™ fluorometer and loaded onto a PromethION flow cell for single-molecule real-time sequencing.
For DNBSEQ-T7 sequencing, DNA was fragmented using a Covaris ultrasonicator to produce fragments between 150 and 300 bp. The library construction included terminal polishing, addition of A-overhangs, and ligation of adapters, followed by PCR amplification. After passing quality control, the amplified libraries were denatured and circularized. DNA nanoballs (DNBs) were generated through rolling circle amplification and loaded onto patterned nanoarrays for 150 bp paired-end sequencing using the DNBSEQ-T7 platform.
2.5.3 Genome assembly
Oxford Nanopore sequencing datasets underwent initial quality filtering through LongQC v1.2.0c, implementing a Q-score threshold of ≥7 for read retention. Concurrently, Illumina short-read data were processed via fastp v0.23.2 with standard parameters. De novo genome assembly was conducted using Flye v2.8, followed by iterative polishing cycles integrating both sequencing platforms (ONT + Illumina) to achieve high-fidelity consensus sequences.
2.5.4 Gene annotation and functional clustering analysis
Genome annotation was executed with Prokka v1.14.5 for structural prediction and primary functional characterization. Functional profiling included: (1) NCBI NR database queries through protein homology searches; (2) COG functional classification via eggNOG-mapper (evolutionary genealogy framework); (3) KEGG pathway mapping using orthology-based prediction. Pathogenicity assessment involved BLASTp alignment against the Virulence Factor Database (VFDB), while antimicrobial resistance determinants were identified through CARD database interrogation using DIAMOND alignment (E-value < 1e-5).
2.5.5 Phylogenomic tree construction
The genome sequence data was uploaded to the Type (Strain) Genome Server (TYGS) for a whole genome-based phylogenomic tree construction (Meier-Kolthoff and Göker, 2019). All pairwise comparisons among a set of Nocardia genomes were conducted using GBDP and accurate intergenomic distances inferred under the algorithm “trimming” and distance formula d5 (Meier-Kolthoff et al., 2013). Hundred distance replicates were calculated each. The resulting intergenomic distances were used to infer a balanced minimum evolution tree with branch support via FASTME 2.1.6.1 including SPR postprocessing (Lefort et al., 2015). Branch support was inferred from 100 pseudo-bootstrap replicates each. The tree was rooted at the midpoint (Farris, 1972) and visualized with PhyD3 (Kreft et al., 2017).
2.6 Histopathological observations
Pathological specimens (liver, spleen, kidney, gill, intestine) from infected mandarin fish underwent 24-h fixation in 4% paraformaldehyde (PFA). Standard histological processing included graded ethanol dehydration, paraffin embedding, and microtome sectioning (5 μm thickness). H&E-stained sections were mounted with DPX resin for bright-field microscopy analysis (Nikon Eclipse Ci-L system) with digital image acquisition via NIS-Elements software (v5.02.03).
2.7 Antibiotic susceptibility testing
Antimicrobial susceptibility profiling was conducted via standardized Kirby-Bauer assay. Bacterial lawns were prepared by aseptic inoculation of 200 μL adjusted suspension (0.5 McFarland) onto blood agar surfaces, followed by precision placement of antibiotic discs (Hangzhou Tianhe Microbial Reagents Co., Ltd.) using sterile forceps. Plates were subjected to standardized incubation parameters (25°C, 7 days) with subsequent quantitative assessment of inhibition zones using digital caliper measurements (Mitutoyo 500-196-30), determining N. seriolae 2023050's resistance profile.
3 Results
3.1 Strain isolation
Isolate 20230510 demonstrated extended lag-phase growth (3–5 days) on blood agar, forming pale yellow, granular colonies with irregular margins and rugose surface topography (Figure 1B). Microscopic analysis revealed Gram-positive staining with characteristic filamentous branching morphology (Figure 1C).
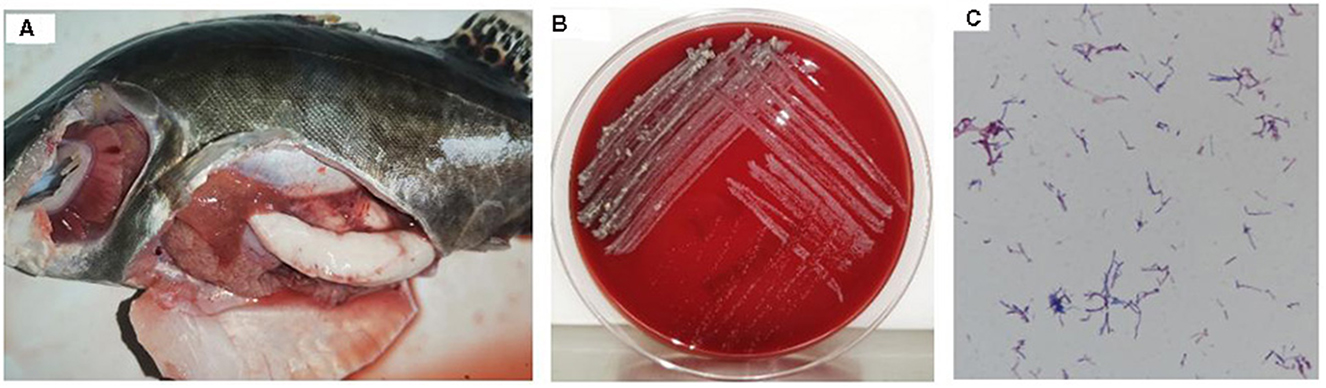
Figure 1. Diseased mandarin fish and bacterial morphology of strain 20230510. Diseased mandarin fish (A); colony morphology on the blood plates (B); Gram staining of strain 20230510 [(C), 1000×].
3.2 Pathogenicity testing
Healthy S. chuatsi were challenged with N. seriolae strain 20230510. High-dose groups (1.24 × 106−1.24 × 107 CFU/fish) exhibited behavioral abnormalities (lethargy, anorexia, delayed responsiveness) by day 3 post-infection, with mortality initiating on day 8 (Table 1). Comparatively, groups receiving 1.24 × 104−1.24 × 106 CFU showed delayed mortality (days 10–13). By day 16, all high-dose fish succumbed, while only four mortalities occurred in the low-dose group (1.24 × 104 CFU/fish) by day 20. No mortality was observed in controls over 21 days. The calculated LD50 was 3.89 × 104 CFU/mL.
Affected fish lacked skin ulcers but displayed fin congestion/ulceration (dorsal, pelvic, anal). Necropsy identified abdominal distension, ascites, and posterior kidney enlargement (Figure 1A), mirroring natural infection phenotypes. Control specimens remained asymptomatic. Reisolated bacteria from experimental mortalities matched natural infection isolates in purity, colony morphology, and growth kinetics.
3.3 WGS results of strain 20230510
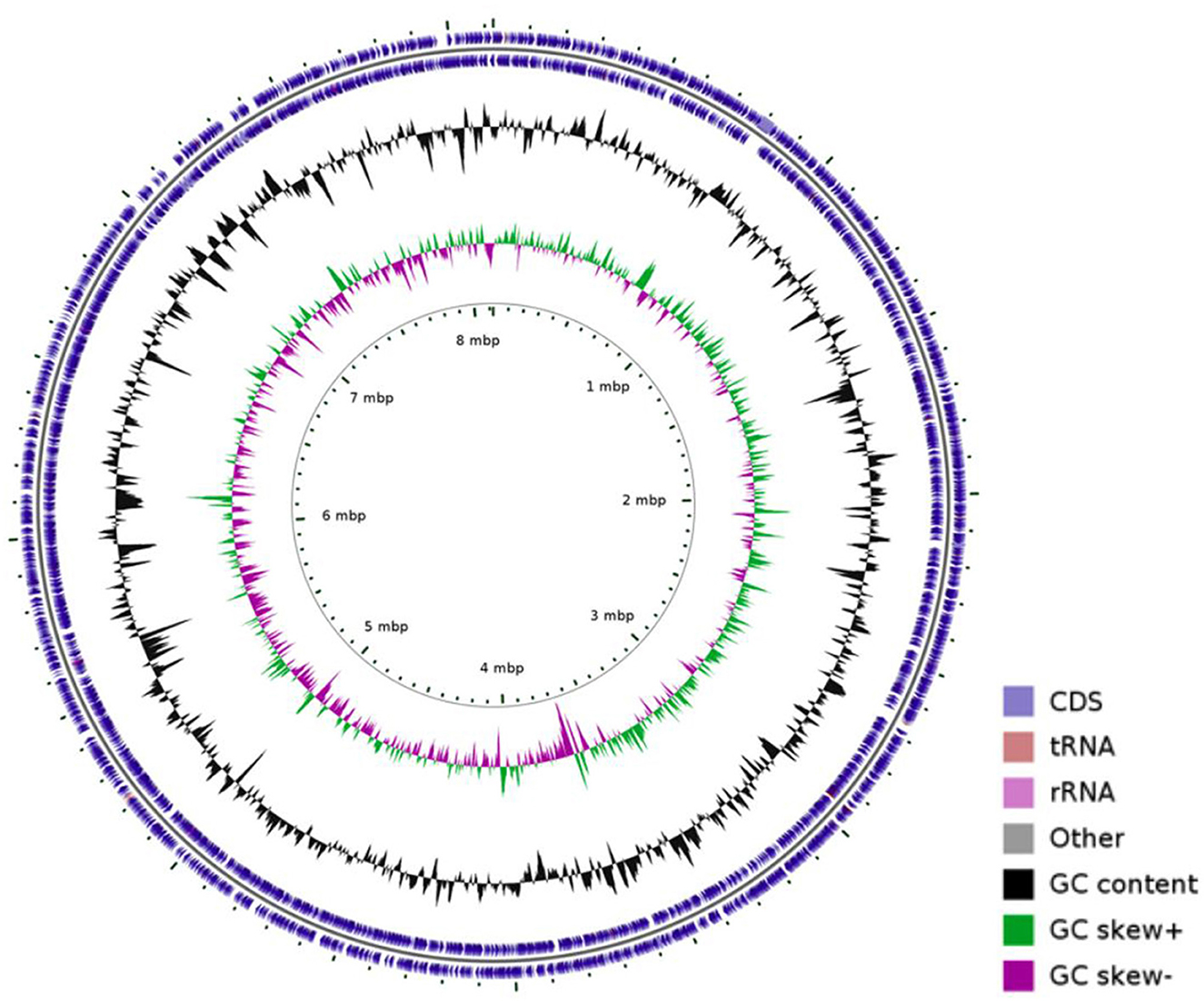
Whole-genome sequencing of N. seriolae strain 20230510 revealed a circular chromosome (8,123,106 bp; GC 68.14%) encoding 7,638 CDSs, 12 rRNAs, 72 tRNAs, and 1 tmRNA (Figure 2). The genome (GenBank: CP130742) was compared with seven reference strains, showing conserved GC content (~68.1%) despite size variations (7.70–8.37 Mb; Table 2).
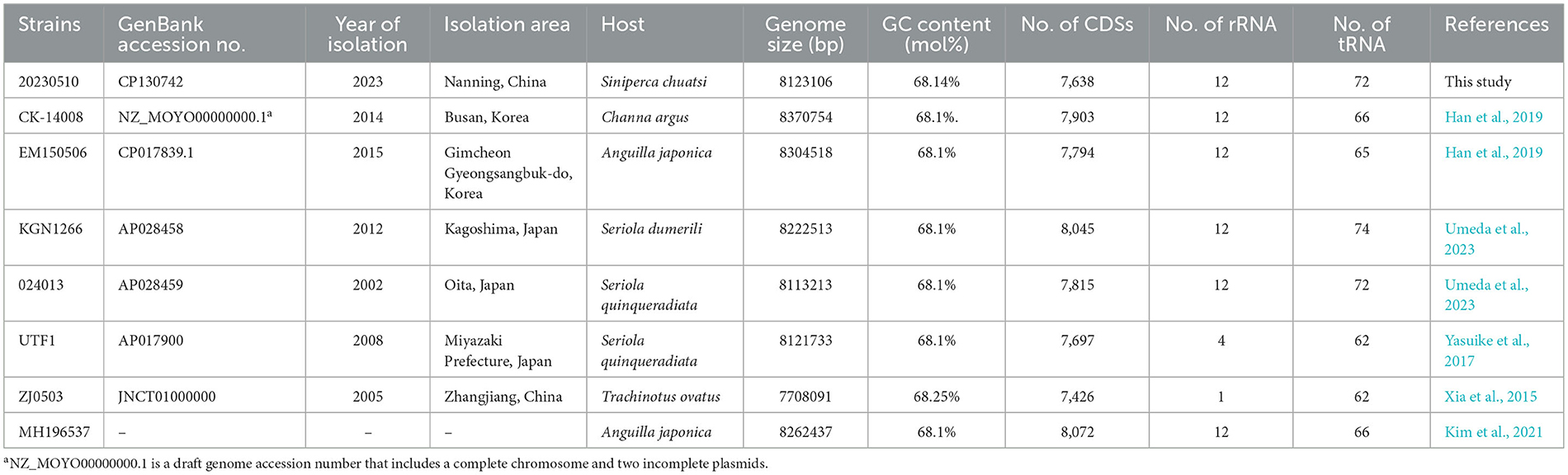
Table 2. Genomic features of the N. seriolae strain 20230510 genome and comparison with genomes of other N. seriolae strains.
COG functional annotation classified 7,108 CDSs into 23 categories (Figure 3), with dominant functional groups including transcription (Class K, 861), replication/repair (L, 681), amino acid transport (E, 534), lipid metabolism (I, 454), secondary structures (Q, 433), and energy production (C, 412). Additionally, 1,462 genes lacked functional assignments.
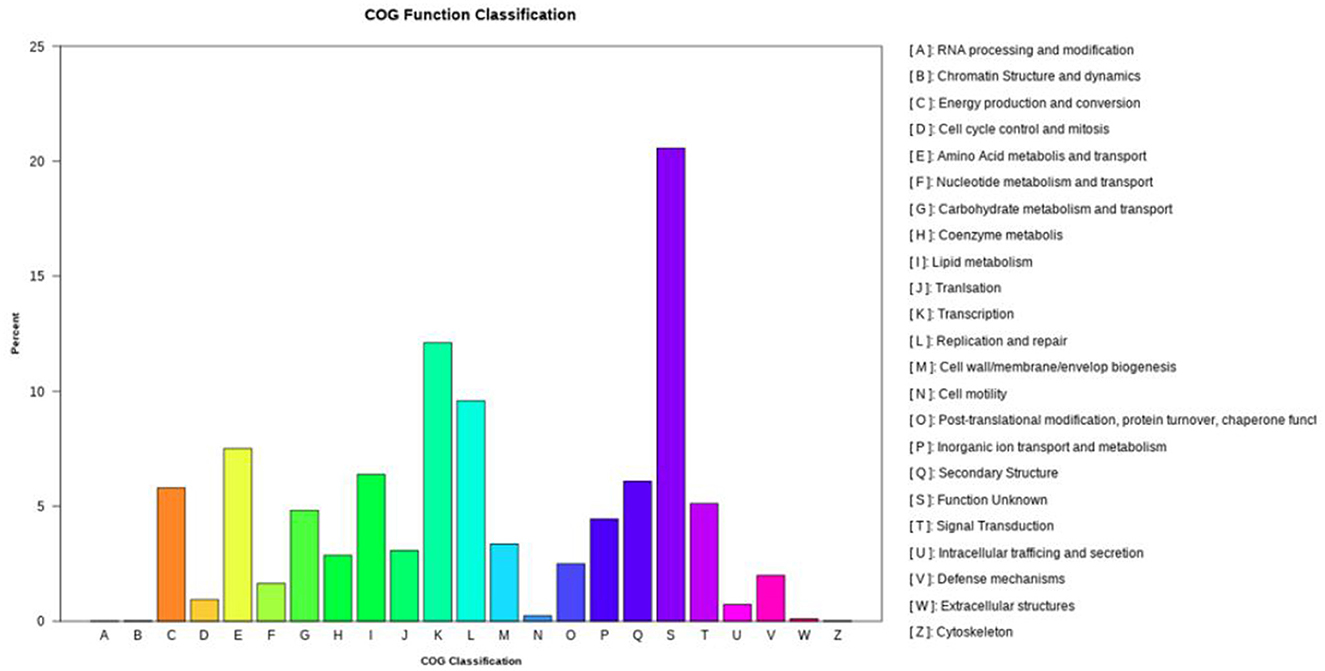
Figure 3. The clusters of orthologous genes (COG) functional annotation in the whole genome of N. seriolae strain 20230510.
KEGG pathway analysis mapped 6,272 genes to 252 metabolic processes (Figure 4), predominantly involved in core metabolism (68.3%), environmental signal transduction (15.1%), and genetic information processing (11.6%).
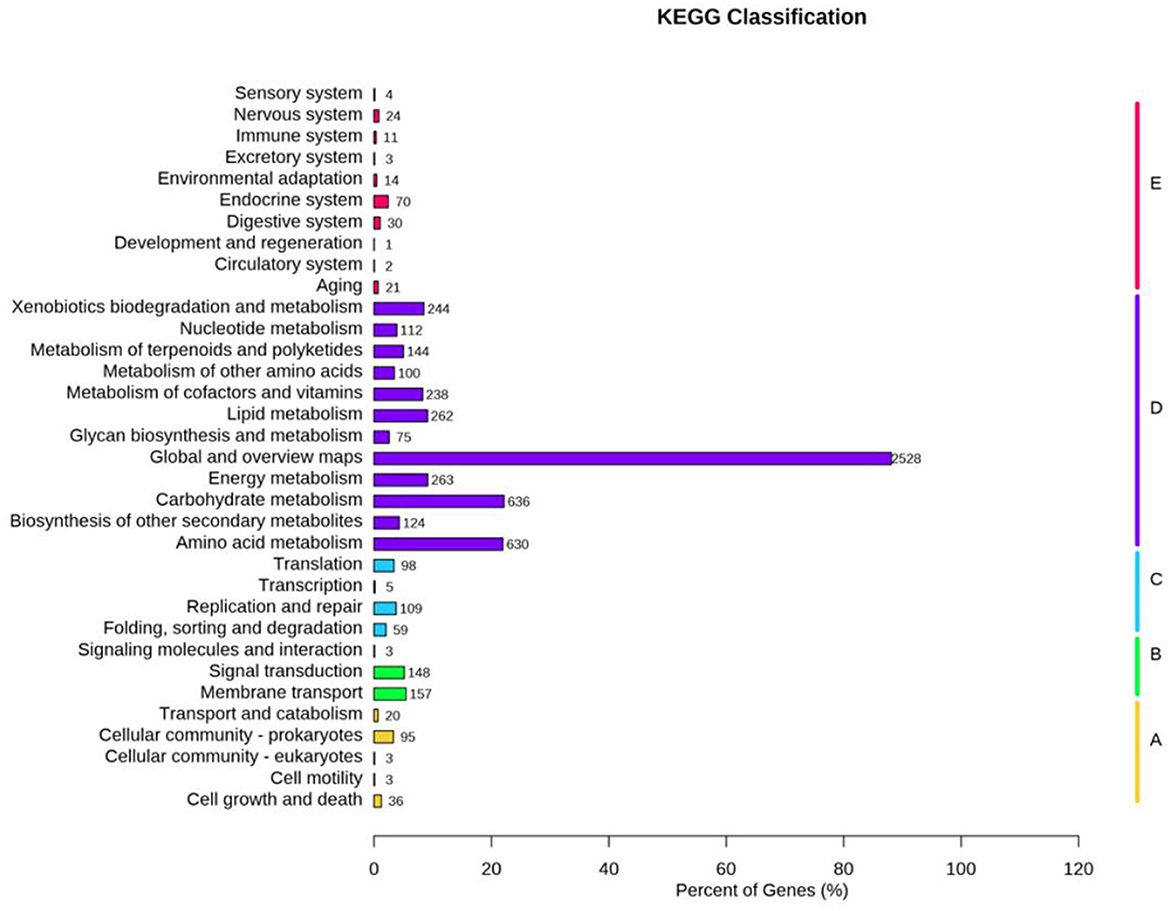
Figure 4. The Kyoto encyclopedia of genes and genomes (KEGG) functional annotation of N. seriolae strain 20230510.
3.4 Phylogenomic tree analysis
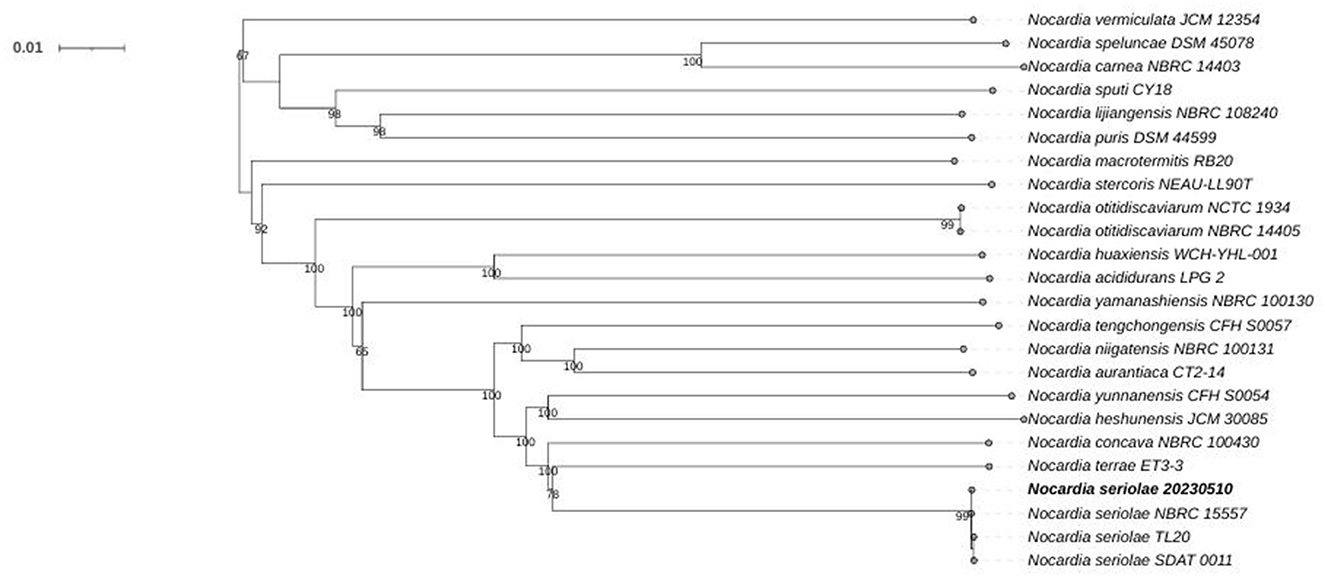
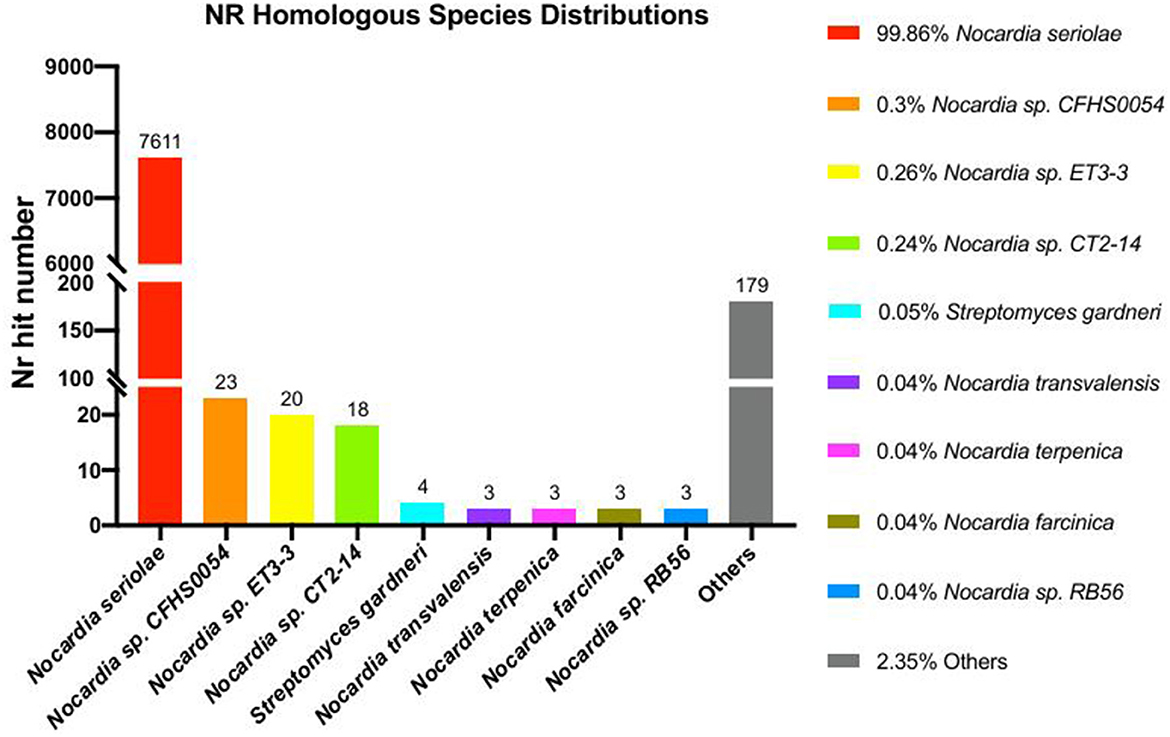
Phylogenomic analysis of 24 Nocardia species revealed that strain 20230510 clusters within the N. seriolae clade, with 99% bootstrap supporting (Figure 5). NR database annotations further confirmed taxonomic alignment, with 99.96% genus-level (Nocardia) and 99.86% species-level (N. seriolae) sequence homology (Figure 6), conclusively identifying the isolate as N. seriolae.
3.5 Virulence factor analysis
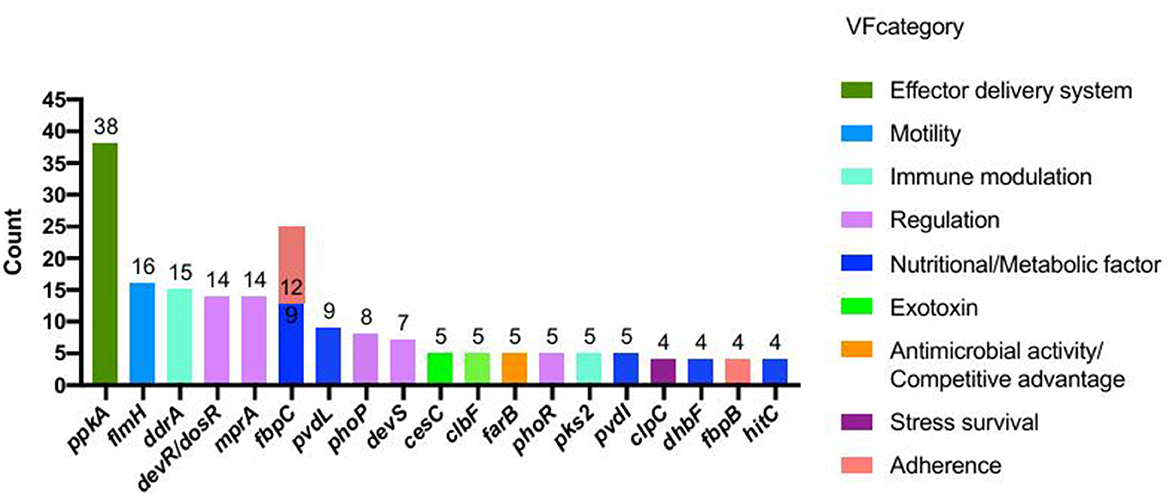
VFDB analysis identified 403 putative virulence-associated CDSs in strain 20230510 (Supplementary Table 1). These genes were functionally annotated into 12 categories, with the most prevalent involving nutrient metabolism (115 genes, e.g., fbpC, pvdL, dhbF, hitC), regulatory systems (76 genes; devR/dosR, mprA, phoP), immune modulation (63 genes; ddrA, pks2, cpsA/uppS), effector delivery (54 genes; ppkA, cdsN, eccA1-C3), and exotoxin production (28 genes; cesC, clbF/D, cyaB). The top 20 virulence factors by gene count are illustrated in Figure 7.
3.6 Antibiotic resistance gene analysis
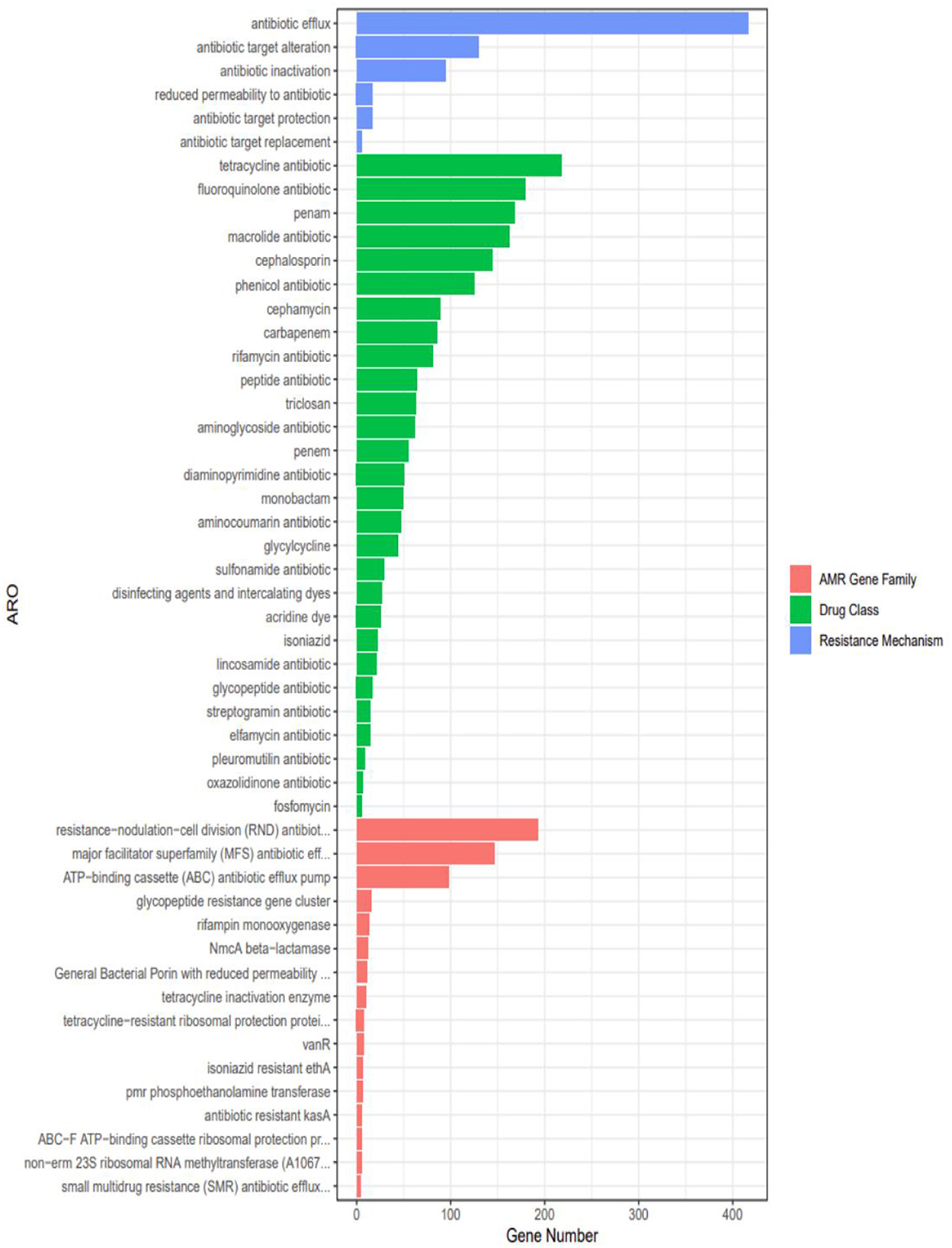
Genomic analysis identified 28 classes of antimicrobial resistance (AMR) genes in strain 20230510, spanning tetracyclines, fluoroquinolones, β-lactams (penams, cephalosporins, cephamycins, carbapenems), macrolides, chloramphenicol, and rifamycins (Figure 8). Quantitative profiling revealed predominant resistance to tetracyclines (218 genes), followed by fluoroquinolone-(179 genes) and penam-targeting mechanisms (168 genes).
3.7 Histopathological observations
Histopathological assessment of infected S. chuatsi demonstrated systemic granulomatous inflammation, with acid-fast stained N. seriolae (blue) present in all examined organs (Figure 9). Organ-specific manifestations were as follows:
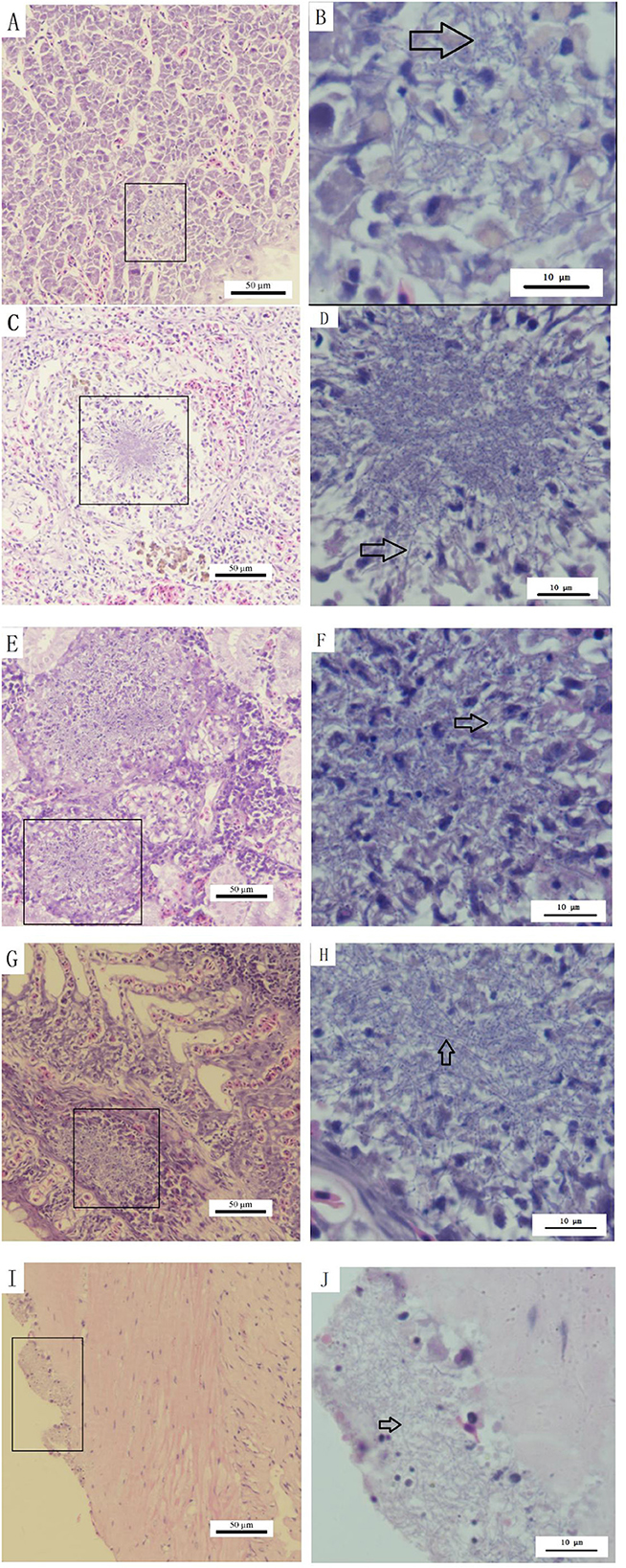
Figure 9. Histopathological observations of diseased S. chuatsi. (A, C, E, G, I) Show granulomas or lesion areas in or on the surface of the liver, spleen, kidneys, gills, and intestines, respectively; (B, D, F, H, J) provide magnified views of selected granulomas or lesion areas of (A, C, E, G, I), respectively. Within these images, N. seriolae are indicated by arrows, and the lesion or granulomatous areas are marked by squares.
Liver: Hepatocellular degeneration/necrosis accompanied by developing granulomas (Figure 9A), with intralesional bacterial colonization (Figure 9B).
Spleen: Connective tissue hyperplasia and lymphocyte depletion (Figure 9C), featuring necrotic foci with epithelioid cell aggregates, neutrophil infiltration, and marked macrophage proliferation (Figure 9D).
Kidneys: Most severe granulomatous involvement, showing large-diameter lesions (Figure 9E) with interstitial necrosis and dense macrophage/neutrophil infiltration (Figure 9F).
Gills: Granulomatous infiltration by macrophages/lymphocytes (Figures 9G, H).
Intestines: Structurally intact but with surface-adherent pathogens (Figures 9I, J).
3.8 Antibiotic susceptibility testing
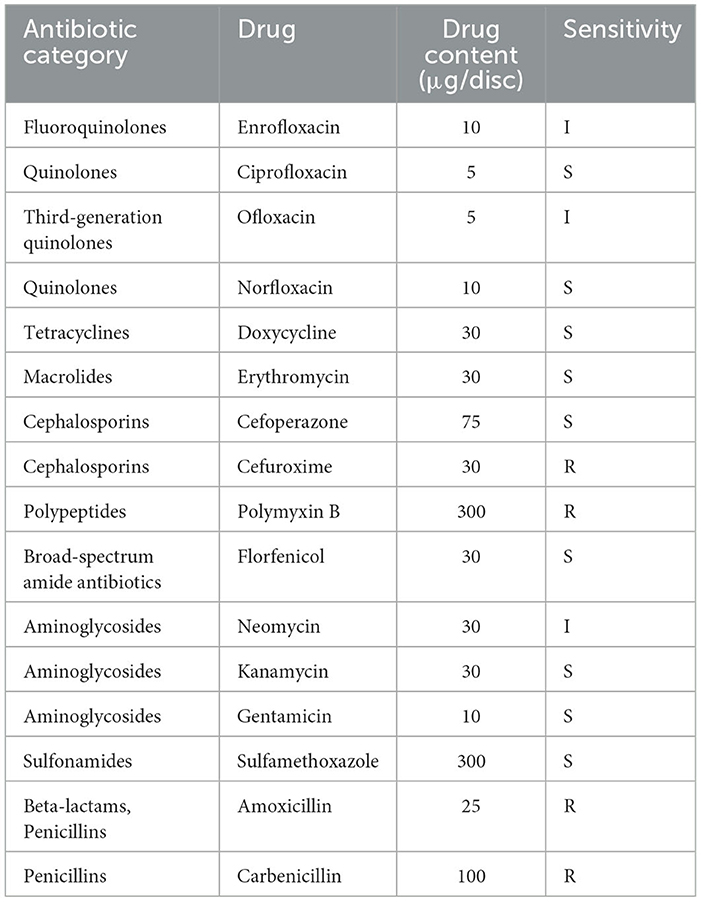
Antibiotic susceptibility profiling of strain 2023510 against 16 agents (Table 3) identified sensitivity to nine antimicrobials spanning five classes: quinolones (ciprofloxacin, norfloxacin), tetracyclines (doxycycline), macrolides (erythromycin), cephalosporins (cefoperazone), sulfonamides (florfenicol, sulfamethoxazole), and aminoglycosides (kanamycin, gentamicin). The strain demonstrated resistance to cefuroxime, polymyxin B, amoxicillin, and carbenicillin.
Notably, four sensitive agents—enrofloxacin, doxycycline, florfenicol, and sulfamethoxazole—are approved by China's Ministry of Agriculture and Rural Affairs under the Aquaculture Medication Guidelines (2024 Documents 2). These compounds, along with ciprofloxacin, erythromycin, cefoperazone, kanamycin, and gentamicin, constitute viable therapeutic options for managing N. seriolae infections in S. chuatsi.
4 Discussion
Experimental infection of S. chuatsi with N. seriolae isolated from naturally diseased fish reproduced clinical signs consistent with natural infections, including fin congestion, ulceration, and ascites, though ulcerative skin lesions were absent. Histopathological analysis revealed systemic granulomatous inflammation in the liver, spleen, kidneys, intestines, and gills, with visible clusters of N. seriolae in affected tissues. These findings align with previous reports in other species but exhibit notable variations. For instance, Huang et al. (2021) described granulomatous necrosis and lymphoid depletion in largemouth bass, while Kim et al. (2018) documented nodular lesions in eels with myocardial necrosis and hepatic congestion. Conversely, tilapia and milkfish failed to develop granulomas (Nazareth et al., 2024), and East Asian four-finger threadfins exhibited acute necrotic lesions with purulent fluid (Liao et al., 2021). The pathological changes observed in this study do not completely align with the typical characteristics of N. seriolae infections in other fish species. This discrepancy may be attributed to different responses of various fish species to N. seriolae or the varying pathogenicity of different strains. Further comparative studies are needed to elucidate the mechanisms underlying these interspecies differences.
Traditional bacterial identification relies on a polyphasic approach combining morphological, physiological, and molecular techniques (16S rRNA and multilocus sequence analysis) to ensure taxonomic accuracy (Friedman et al., 1998; Isik et al., 1999; Rio-Rodriguez et al., 2021; Vu-Khac et al., 2016; Wang et al., 2009). However, advancements in next-generation sequencing have revolutionized microbial identification by enabling rapid whole-genome characterization. In the present study, strain 20230510 was isolated from diseased S. chuatsi in Guangxi aquaculture facilities. Comprehensive genomic analysis confirmed its identity as N. seriolae through: (1) 16S rRNA sequencing (>99% homology with N. seriolae), and (2) whole-genome sequencing (99.86% average nucleotide identity with N. seriolae reference strains). The complete genome assembly revealed a typical Nocardia genomic architecture—a single circular chromosome spanning 8.12 Mb with 68.14% GC content and 7,638 coding sequences. These genomic features align well with previous reports documenting N. seriolae genomes ranging 7.70–8.37 Mb with ~68.1% GC content and 7,426–8,072 CDSs (Han et al., 2019; Kim et al., 2021; Strauss and Falkow, 1997; Umeda et al., 2023; Yasuike et al., 2017). The minor variations observed in genome size and gene content likely reflect natural strain-to-strain genomic diversity within the species.
Vaccination is considered an effective method to control bacterial infections, and scholars have been working toward developing vaccines against N. seriolae (Chang et al., 2016; Chen et al., 2019; Matsumoto et al., 2017; Nayak et al., 2014; Shimahara et al., 2010; Wang et al., 2022). However, no vaccine for N. seriolae has yet been approved for use, and antibiotics remain the primary treatment for N. seriolae infections in aquaculture production. In Japan, only sulfamonomethoxine and sulfisozole sodium are licensed for the treatment of N. seriolae infections (Imajoh et al., 2016; Ismail et al., 2012). Liao et al. (2021) found that N. seriolae strains NM107152 and NM108007 were susceptible to oxytetracycline, doxycycline, lincomycin, erythromycin, and florfenicol. Chang et al. (2016) discovered that oxytetracycline, trimethoprim, erythromycin, florfenicol, and thiamphenicol were effective drugs for treating N. seriolae infections in fish. Imajoh et al. (2016) identified that the antibiotic resistance genes of the N. seriolae U-1 strain included one gene for vancomycin resistance, two for fluoroquinolone resistance, and 10 for β-lactamase resistance, indicating that the draft genome of U-1 lacks genes responsible for resistance to macrolides and tetracyclines. Yasuike et al. (2017) found that the N. seriolae UTF1 strain was resistant to oxytetracycline (a tetracycline antibiotic) but susceptible to erythromycin (a macrolide antibiotic). Our experimental results do not fully align with the above studies. Moreover, the antibiotic susceptibility testing results of this strain were not fully consistent with the resistance genes. These discrepancies may be due to variations in strains isolated from different regions, leading to different results in antibiotic susceptibility testing or resistance gene identification. Additionally, the discrepancies could be due to the different principles of the two testing methods: resistance genes are detected in the genome sequence as mutations of one or several drug resistance-related genes, while antibiotic susceptibility testing is a phenotypic test that measures the in vitro sensitivity of the bacterium to a specific drug. Furthermore, antibiotic resistance genes do not necessarily translate into an antibiotic-resistant phenotype (Zhu et al., 2023). As suggested above, the mechanisms underlying the inconsistency between resistance genes and antibiotic susceptibility results for N. seriolae require further in-depth investigation.
Antimicrobial susceptibility testing demonstrated that strain 20230510 exhibited sensitivity to multiple antibiotic classes, including quinolones, tetracyclines, macrolides, cephalosporins, sulfonamides, and aminoglycosides. These findings provide valuable insights for clinical management of N. seriolae infections in aquaculture. In accordance with the antimicrobial susceptibility profile and considering the approved veterinary drugs listed by the Ministry of Agriculture and Rural Affairs of China, we recommend the following treatment protocol for N. seriolae infections in S. chuatsi: primary options is enrofloxacin (quinolone) and doxycycline (tetracycline), and alternative choices is florfenicol (phenicol) and sulfamethoxazole (sulfonamide). This therapeutic strategy is proposed based on: demonstrated in vitro efficacy against the isolated strain; compliance with national regulations on veterinary drug use; practical considerations for aquaculture applications. The broad susceptibility pattern observed suggests these antimicrobials remain effective for controlling N. seriolae outbreaks in mandarin fish populations, though continuous monitoring of resistance development is advised.
The results of the present study indicate that N. seriolae is pathogenic to Mandarin fish; however, further research on the source, infection pathways, pathogenicity, and immune mechanism of the pathogen is required.
Data availability statement
The original contributions presented in the study are publicly available. This data can be found in here: https://www.ncbi.nlm.nih.gov/genbank/, CP130742.
Ethics statement
The animal study was approved by Guangxi Academy of Fishery Sciences. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
LC: Writing – original draft, Formal analysis, Data curation. XY: Data curation, Writing – original draft, Formal analysis. YL: Funding acquisition, Formal analysis, Visualization, Writing – original draft. ZL: Investigation, Writing – original draft, Methodology. XW: Methodology, Investigation, Writing – original draft. ZT: Methodology, Investigation, Writing – original draft. LX: Methodology, Formal analysis, Validation, Writing – original draft. FC: Writing – original draft, Methodology, Formal analysis. ML: Methodology, Formal analysis, Writing – original draft. ZG: Writing – original draft, Formal analysis, Methodology. ZH: Writing – original draft, Formal analysis. TH: Supervision, Writing – review & editing, Funding acquisition, Validation, Project administration.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by Guangxi Key Research and Development Program (Project No. Guike nong AB2506910041), Construction Project of National Modern Agricultural Industrial Technology System (CARS-46), Guangxi project of Key Research and development (GuikeAB No.21220017), National Modern Agricultural Industrial Technology System Guangxi Characteristic Freshwater Fish Innovation Team Disease Prevention and Control post Functional Expert Project (nycytxgxcxtd-2021-08-02), and National Modern Agricultural Industry Technology System Guangxi Characteristic Freshwater Fish Innovation Team Ecological Breeding Position Functional Expert (nycytxgxcxtd-2021-08-03).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Gen AI was used in the creation of this manuscript.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1623741/full#supplementary-material
References
Chang, H. Y., Lai, J. M., and Wang, J. H. (2016). The minimum inhibitory concentration of antibiotics against Nocardia seriolae isolation from diseased fish in Taiwan. Taiwan Vet. J. 42, 81–84. doi: 10.1142/S1682648515720087
Chen, J., Wang, W., Hou, S., Fu, W., Cai, J., Xia, L., et al. (2019). Comparison of protective efficacy between two DNA vaccines encoding DnaK and GroEL against fish nocardiosis. Fish Shellfish Immunol. 95, 128–139. doi: 10.1016/j.fsi.2019.10.024
Chen, N., Jiang, J., Gao, X., Li, X., Zhang, Y., Liu, X., et al. (2018). Histopathological analysis and the immune related gene expression profiles of mandarin fish (Siniperca chuatsi) infected with Aeromonas hydrophila. Fish Shellfish Immunol. 83, 410–415. doi: 10.1016/j.fsi.2018.09.023
Chen, S. C. (1992). Study on the pathogenicity of Nocardia asteroides to the Formosa snakehead, Channa maculata (Lacepède), and largemouth bass, Micropterus salmoides (Lacepède). J. Fish Dis. 15, 47–53. doi: 10.1111/j.1365-2761.1992.tb00635.x
Chen, S. C., Lee, J. L., Lai, C. C., Gu, Y. W., Wang, C. T., Chang, H. Y., et al. (2000). Nocardiosis in sea bass, Lateolabrax japonicus, in Taiwan. J. Fish Dis. 23, 299–307. doi: 10.1046/j.1365-2761.2000.00217.x
Farris, J. S. (1972). Estimating phylogenetic trees from distance matrices. Am. Nat. 106, 645–668. doi: 10.1086/282802
Friedman, C. S., Beaman, B. L., Chun, J., Goodfellow, M., Gee, A., and Hedrick, R. P. (1998). Nocardia crassostreae sp. nov., the causal agent of nocardiosis in Pacific oysters. Int. J. Syst. Evol. Microbiol. 48, 237–246. doi: 10.1099/00207713-48-1-237
Han, H. J., Kwak, M. J., Ha, S. M., Yang, S. J., Kim, J. D., Cho, K. H., et al. (2019). Genomic characterization of Nocardia seriolae strains isolated from diseased fish. Microbiologyopen 8:e00656. doi: 10.1002/mbo3.656
Heuer, H., Krsek, M., Baker, P., Smalla, K., and Wellington, E. (1997). Analysis of actinomycete communities by specific amplification of genes encoding 16S rRNA and gel-electrophoretic separation in denaturing gradients. Appl. Environ. Microbiol. 63, 3233–3241. doi: 10.1128/aem.63.8.3233-3241.1997
Huang, X., Liu, S., Chen, X., Zhang, H., Yao, J., Geng, Y., et al. (2021). Comparative pathological description of nocardiosis in largemouth bass (Micropterus salmoides) and other Perciformes. Aquaculture 534:736193. doi: 10.1016/j.aquaculture.2020.736193
Imajoh, M., Sukeda, M., Shimizu, M., Yamane, J., Ohnishi, K., and Oshima, S. I. (2016). Draft genome sequence of erythromycin-and oxytetracycline-sensitive Nocardia seriolae strain U-1 (NBRC 110359). Genome Announc. 4, 10–1128. doi: 10.1128/genomeA.01606-15
Isik, K., Chun, J., Hah, Y. C., and Goodfellow, M. (1999). Nocardia salmonicida nom. rev., a fish pathogen. Int. J. Syst. Evol. Microbiol. 49, 833–837. doi: 10.1099/00207713-49-2-833
Ismail, T. F., Nakamura, A., Nakanishi, K., Minami, T., Murase, T., Yanagi, S., et al. (2012). Modified resazurin microtiter assay for in vitro and in vivo assessment of sulfamonomethoxine activity against the fish pathogen Nocardia seriolae. Fish. Sci. 78, 351–357. doi: 10.1007/s12562-011-0450-8
Kariya, T., Kubota, S., Nakamura, Y., and Kira, K. (1968). Nocardial infection in cultured yellowtails (Seriola quinqueruiata and S. purpurascens)—I Bacteriological study. Fish Pathol. 3, 16–23. doi: 10.3147/jsfp.3.16
Kim, B. S., Huh, M. D., and Roh, H. (2021). The complete genome of Nocardia seriolae MH196537 and intra-species level as analyzed by comparative genomics based on random Forest algorithm. Curr. Microbiol. 78, 2391–2399. doi: 10.1007/s00284-021-02490-0
Kim, J. D., Lee, N. S., Do, J. W., Kim, M. S., Seo, H. G., Cho, M., et al. (2018). Nocardia seriolae infection in the cultured eel Anguilla japonica in Korea. J. Fish Dis. 41, 1745–1750. doi: 10.1111/jfd.12881
Kreft, Ł., Botzki, A., Coppens, F., Vandepoele, K., and Van Bel, M. (2017). PhyD3: a phylogenetic tree viewer with extended phyloXML support for functional genomics data visualization. Bioinformatics 33, 2946–2947. doi: 10.1093/bioinformatics/btx324
Lefort, V., Desper, R., and Gascuel, O. (2015). FastME 2.0: a comprehensive, accurate, and fast distance-based phylogeny inference program. Mol. Biol. Evol. 32, 2798–2800. doi: 10.1093/molbev/msv150
Liao, P. C., Tsai, M. A., See, M. S., Wang, P. C., and Chen, S. C. (2021). Isolation and characterization of Nocardia seriolae, a causative agent of systematic granuloma in cultured East Asian four finger threadfin, Eleutheronema rhadinum, and red snapper, Lutjanus erythropterus. Aquac. Res. 52, 763–770. doi: 10.1111/are.14932
Lin, Q., Li, J., Fu, X., Liu, L., Liang, H., Niu, Y., et al. (2020). Hemorrhagic gill disease of Chinese perch caused by Aeromonas salmonicida subsp. salmonicida in China. Aquaculture 519:734775. doi: 10.1016/j.aquaculture.2019.734775
Liu, X., Chen, N., Gao, X., Zhang, Y., Li, X., Zhang, Y., et al. (2018). The infection of red seabream iridovirus in mandarin fish (Siniperca chuatsi) and the host immune related gene expression profiles. Fish Shellfish Immunol. 74, 474–484. doi: 10.1016/j.fsi.2018.01.020
Liu, X., Sun, W., Zhang, Y., Zhou, Y., Xu, J., Gao, X., et al. (2020). Impact of Aeromonas hydrophila and infectious spleen and kidney necrosis virus infections on susceptibility and host immune response in Chinese perch (Siniperca chuatsi). Fish Shellfish Immunol. 105, 117–125. doi: 10.1016/j.fsi.2020.07.012
Luo, X., Fu, X., Liao, G., Chang, O., Huang, Z., and Li, N. (2017). Isolation, pathogenicity and characterization of a novel bacterial pathogen Streptococcus uberis from diseased mandarin fish Siniperca chuatsi. Microb. Pathog. 107, 380–389. doi: 10.1016/j.micpath.2017.03.049
Matsumoto, M., Araki, K., Hayashi, K., Takeuchi, Y., Shiozaki, K., Suetake, H., et al. (2017). Adjuvant effect of recombinant interleukin-12 in the Nocardiosis formalin-killed vaccine of the amberjack Seriola dumerili. Fish Shellfish Immunol. 67, 263–269. doi: 10.1016/j.fsi.2017.06.025
Meier-Kolthoff, J. P., Auch, A. F., Klenk, H. P., and Göker, M. (2013). Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinf. 14, 1–14. doi: 10.1186/1471-2105-14-60
Meier-Kolthoff, J. P., and Göker, M. (2019). TYGS is an automated high-throughput platform for state-of-the-art genome-based taxonomy. Nat. Commun. 10:2182. doi: 10.1038/s41467-019-10210-3
Ministry of Agriculture and Rural Affairs Fisheries Administration Bureau, National Aquatic Product Technology Extension Station, and Fisheries, C. S. o. (2023). 2023 China Fishery Statistical Yearbook. Beijing: Agricultural Press of China.
Nayak, S. K., Shibasaki, Y., and Nakanishi, T. (2014). Immune responses to live and inactivated Nocardia seriolae and protective effect of recombinant interferon gamma (rIFN γ) against nocardiosis in ginbuna crucian carp, Carassius auratus langsdorfii. Fish Shellfish Immunol. 39, 354–364. doi: 10.1016/j.fsi.2014.05.015
Nazareth, S. C., Cheng, L. W., Wang, P. C., and Chen, S. C. (2024). Comparative pathogenicity of Nocardia seriolae in Nile tilapia (Oreochromis niloticus), milkfish (Chanos chanos) and Asian seabass (Lates calcarifer). J. Fish Dis. 47:e13947. doi: 10.1111/jfd.13947
Rio-Rodriguez, R. E. D., Ramirez-Paredes, J. G., Soto-Rodriguez, S. A., Shapira, Y., Huchin-Cortes, M. D. J., Ruiz-Hernandez, J., et al. (2021). First evidence of fish nocardiosis in Mexico caused by Nocardia seriolae in farmed red drum (Sciaenops ocellatus, Linnaeus). J. Fish Dis. 44, 1117–1130. doi: 10.1111/jfd.13373
Shimahara, Y., Huang, Y. F., Tsai, M. A., Wang, P. C., and Chen, S. C. (2010). Immune response of largemouth bass, Micropterus salmoides, to whole cells of different Nocardia seriolae strains. Fish. Sci. 76, 489–494.
Shimahara, Y., Nakamura, A., Nomoto, R., Itami, T., Chen, S. C., and Yoshida, T. (2008). Genetic and phenotypic comparison of Nocardia seriolae isolated from fish in Japan. J. Fish Dis. 31, 481–488. doi: 10.1111/j.1365-2761.2008.00920.x
Strauss, E. J., and Falkow, S. (1997). Microbial pathogenesis: genomics and beyond. Science 276, 707–712. doi: 10.1126/science.276.5313.707
Umeda, K., Matsuura, Y., Shimahara, Y., Takano, T., and Matsuyama, T. (2023). Complete genome sequences of α-glucosidase-positive and-negative strains of Nocardia seriolae from Seriola species in Japan. Microbiol. Resource Announce. 12:e00712-23. doi: 10.1128/MRA.00712-23
Vu-Khac, H., Chen, S. C., Pham, T. H., Nguyen, T. T. G., and Trinh, T. T. H. (2016). Isolation and genetic characterization of Nocardia seriolae from snubnose pompano Trachinotus blochii in Vietnam. Dis. Aquat. Org. 120, 173–177. doi: 10.3354/dao03023
Wang, P. C., Chen, S. D., Tsai, M. A., Weng, Y. J., Chu, S. Y., Chern, R. S., et al. (2009). Nocardia seriolae infection in the three striped tigerfish, Terapon jarbua (Forsskål). J. Fish Dis. 32, 301–310. doi: 10.1111/j.1365-2761.2008.00991.x
Wang, W., Hou, S., Chen, J., Xia, L., and Lu, Y. (2022). Construction of an attenuated glutamyl endopeptidase deletion strain of Nocardia seriolae. Fish Shellfish Immunol. 129, 161–169. doi: 10.1016/j.fsi.2022.08.044
Xia, L., Cai, J., Wang, B., Huang, Y., Jian, J., and Lu, Y. (2015). Draft genome sequence of Nocardia seriolae ZJ0503, a fish pathogen isolated from Trachinotus ovatus in China. Genome Announce. 3:e01223-14. doi: 10.1128/genomeA.01223-14
Yasuike, M., Nishiki, I., Iwasaki, Y., Nakamura, Y., Fujiwara, A., Shimahara, Y., et al. (2017). Analysis of the complete genome sequence of Nocardia seriolae UTF1, the causative agent of fish nocardiosis: The first reference genome sequence of the fish pathogenic Nocardia species. PLoS ONE 12:e0173198. doi: 10.1371/journal.pone.0173198
Zhang, N., Zhang, H., Dong, Z., and Wang, W. (2022). Molecular identification of Nocardia seriolae and comparative analysis of spleen transcriptomes of hybrid snakehead (Channa maculata female × Channa argus male) with nocardiosis disease. Front. Immunol. 13:778915. doi: 10.3389/fimmu.2022.778915
Zhou, W., Zhang, Y., Wen, Y., Ji, W., Zhou, Y., Ji, Y., et al. (2015). Analysis of the transcriptomic profilings of Mandarin fish (Siniperca chuatsi) infected with Flavobacterium columnare with an emphasis on immune responses. Fish Shellfish Immunol. 43, 111–119. doi: 10.1016/j.fsi.2014.12.006
Keywords: Siniperca chuatsi, Nocardia seriolae, pathogenicity, whole-genome analysis, antibiotic susceptibility testing
Citation: Chen L, Yan X, Luo Y, Lu Z, Wei X, Tang Z, Xia L, Chen F, Li M, Guo Z, He Z and Huang T (2025) Pathogenicity and whole-genome analysis of a Siniperca chuatsi-derived Nocardia seriolae strain. Front. Microbiol. 16:1623741. doi: 10.3389/fmicb.2025.1623741
Received: 07 May 2025; Accepted: 23 July 2025;
Published: 25 August 2025.
Edited by:
Angel Isidro Campa, Centro de Investigación Biológica del Noroeste (CIBNOR), MexicoReviewed by:
Jaime Eugenio Figueroa, Austral University of Chile, ChileAmelia Cristina Montoya Martinez, Instituto Tecnológico de Sonora (ITSON), Mexico
Copyright © 2025 Chen, Yan, Luo, Lu, Wei, Tang, Xia, Chen, Li, Guo, He and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ting Huang, aHR3aXNoQDE2My5jb20=
†These authors have contributed equally to this work
 Liting Chen
Liting Chen Xin Yan1,2†
Xin Yan1,2† Liqun Xia
Liqun Xia Ting Huang
Ting Huang