- 1College of Life Sciences and Technology, Tarim University, Alar, China
- 2College of Life Science and Agriculture Forestry, Qiqihar University, Qiqihar, China
- 3Key Laboratory of Tarim Animal Husbandry Science and Technology, College of Animal Science and Technology, Tarim University, Alar, China
In recent years, the role of the respiratory tract microbiota in respiratory tract infections has attracted considerable attention. Respiratory microbiota have important effects on respiratory physiology, immune regulation, and the occurrence and development of various respiratory viral infectious diseases. The microbial composition in the different parts of the respiratory tract, such as the nose, oropharynx, and lower respiratory tract, varies. Under physiological conditions, the respiratory microbiota remains relatively stable; however, when this homeostatic balance is disrupted, respiratory microbiota imbalance occurs, increasing the risk of infection. An increasing number of studies have revealed the complex relationship between bacterial dysregulation and respiratory viral infections. Dysregulation of the respiratory tract microbiota plays an important role in both innate and adaptive immune responses. In this study, changes in respiratory microbes and their interactions with host immunity, respiratory viral infections and malignant tumors were reviewed. Future studies should further explore the interaction mechanism between respiratory microbiota and host immunity, develop new diagnostic and therapeutic strategies, and improve the current level of clinical treatment for respiratory diseases.
1 Introduction
Microbiota are found in all parts of the respiratory tract. Respiratory microbiota participate in the maturation of respiratory physiology and local immune regulation and are closely related to the occurrence and progression of various respiratory diseases. The human respiratory tract is anatomically divided into the upper and lower respiratory tracts. The upper respiratory tract includes the mouth, pharynx, and larynx, whereas the lower respiratory tract includes the trachea, bronchi, and lungs. Different anatomical parts of the respiratory tract have different microbial compositions (Zhou et al., 2024). When the microbiota structure deviates from the physiological state, microbiota dysregulation occurs, which manifests as a decrease in the abundance of probiotics and symbiotic bacteria and an increase in the abundance of potentially pathogenic bacteria (Natalini et al., 2023). Microbial dysregulation is closely related to the occurrence and development of various respiratory diseases, especially chronic airway diseases, such as chronic obstructive pulmonary disease and bronchiectasis (Chellappan et al., 2019). In recent years, increasing attention has been paid to respiratory tract infections, and outstanding progress has been made in research on the role of respiratory tract microbiota in respiratory tract infections. This paper reviews the recent findings on the relationship between the respiratory microbiota and host immunity, respiratory viral infections and malignant tumors, respectively.
2 Main distribution of respiratory microbiota
The adult respiratory microbiota has a different structure in different respiratory regions. The main microbes in the nasal cavity include Staphylococcus, Propionibacterium, Corynebacterium, Moraxella, and Streptococcus. The microbiota of the nasopharynx is similar to that of the nasal cavity, with dominant bacteria including Moraxella, Staphylococcus, Corynebacterium, and Streptococcus, and other major microbiota including Dolosigranulum and Hemophilus. Microbes in the oropharynx are very different from those in the nasal cavity and nasopharynx. In addition to Streptococcus, the genera Prevotella, Veillonella, Rothia, Leptotrichia, Neisseria, and Fusobacterium are present (Natalini et al., 2023; Man et al., 2017). The microbiota of the lower respiratory tract is relatively simple, with a low load, and is mainly composed of oral symbiotic bacteria, such as Streptococcus, Veronella, and Prevotella. The microbiota of the oropharynx can enter the lower respiratory tract through microinhalation or other means, thereby affecting respiratory health. The spread of oral microbiota to the lungs is heterogeneous, and enrichment in the lungs is associated with decreased lung function and increased lung proinflammatory cytokine levels (Zhang et al., 2022). The composition of the lung microbiota is tightly controlled by airway clearance mechanisms such as cough, mucociliary transport, and the innate immune system to prevent colonization and infection by pathogens. It is generally believed that nasal microbiota are the main source of healthy lung bacteria. The oral cavity is structurally adjacent to the respiratory tract and alveoli, and the oral microbiota directly affects the composition of the lung microbiota. Multiple studies have highlighted differences in microbial diversity, biomass, and community structure between the upper and lower respiratory tracts (Charlson et al., 2011; Morris et al., 2013; Li et al., 2024).
3 The effect on innate and adaptive immune responses
3.1 Innate immune response
Host-microbial interactions affect different aspects of the immune system development, promoting immune maturation, immune tolerance, and immune response (Hu et al., 2020) (Figure 1). Lung epithelial cells, macrophages, and dendritic cells (DC) have different receptors to sense microbiota; these microbial pattern recognition receptors (PRR) include Toll-like receptors (TLRS) and NOD-like receptors (NLRs) (Uehara et al., 2007). Epithelial cells are involved in multiple mechanisms by which the microbiota in the lung interact and act as a permeable barrier, sensing microbiota and responding to their presence (Evans et al., 2010). By providing a strong barrier in the lower respiratory tract, the airway epithelium is the primary line of defense against potentially harmful environmental irritants. It is the first site to interact with inhaled compounds and is designed to promote the efficient removal of particles and microbiota by the mucous cilia (Leiva-Juarez et al., 2018). In chronic lung disease, the increased mucus produced by epithelial cells promotes bacterial growth and leads to low oxygen concentrations and areas of high temperature, which promote the selectivity and stability of specific bacteria (Invernizzi et al., 2020).
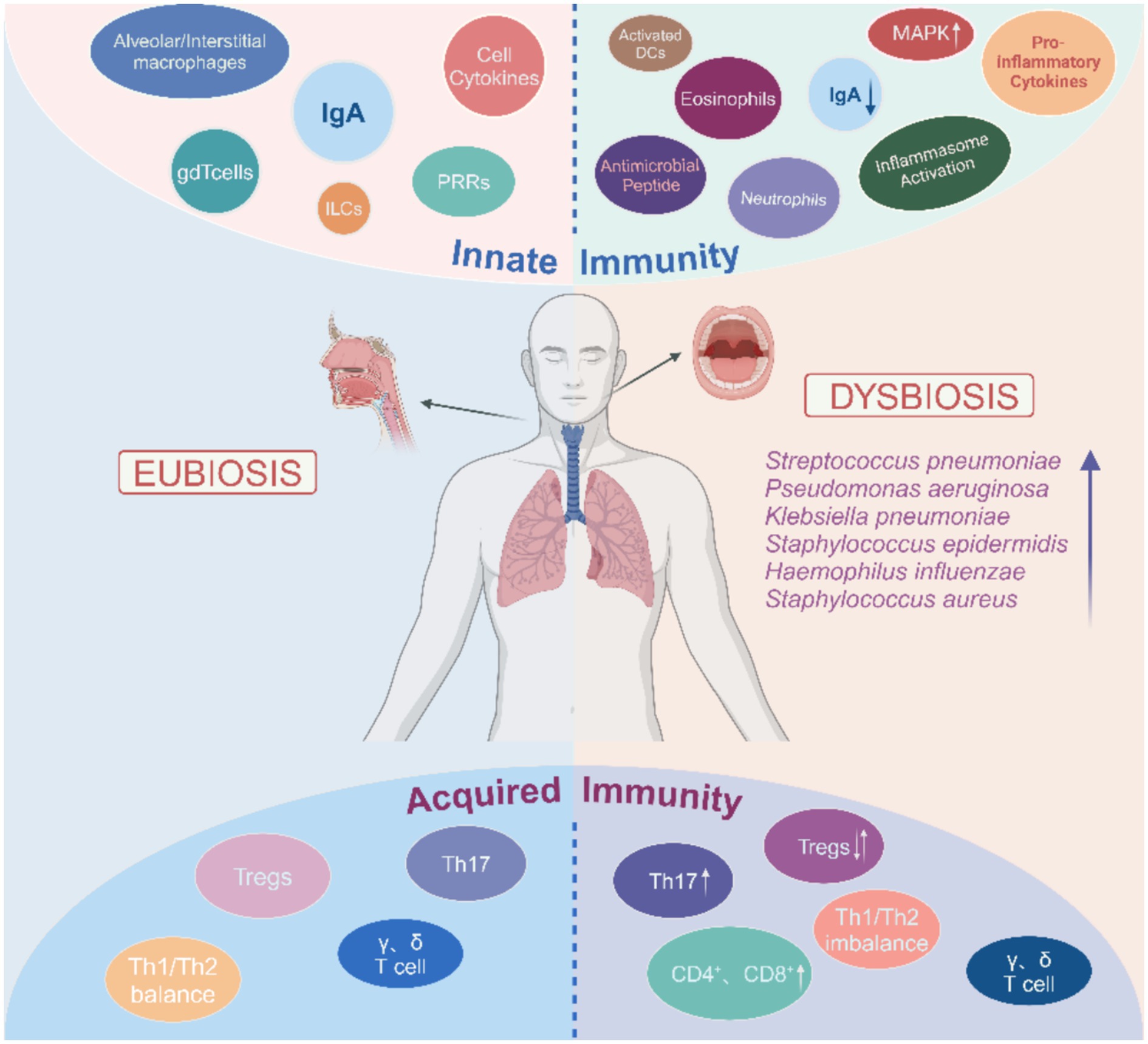
Figure 1. Overview of the respiratory tract microbiota dysbiosis and immunity. Respiratory tract microbiota dysbiosis leads to activation of immune cells. The immune cells then migrate into the tissue and produce pro-inflammatory cytokines, which ultimately contribute to the local inflam-matory response. In addition, changes in the cytokine environment promote pathological fibrotic remodeling, NETosis, and apoptosis. However, this also maintains the pulmonary immune balance through the differentiation of Tregs and Th17 cells. The arrows indicate changes in the relative abundance of immune cell subsets and bacterial species.
Human and mouse models have demonstrated the important role of innate immune responses in regulating microbiota variation, composition, and individual differences (Levy et al., 2015). In healthy mice, IL-1α is a key cytokine in anti-bacterial innate immune activity in the lungs and is negatively correlated with the diversity of bacterial communities and the presence of pathogenic bacteria. In addition, lung concentrations of this inflammatory cytokine is more closely associated with changes in the lung microbiota than with distal intestinal or oral concentrations (Dickson et al., 2018). Upper respiratory tract symbiotic bacteria defend against influenza virus infection in mice through polarization of M2 macrophages and secretion of anti-inflammatory mediators such as IL-10 and transforming growth factor-β (TGF-β) (Iwasaki and Medzhitov, 2015). Microbiota also produce metabolites associated with immune responses. For example, oral commensals generate short-chain fatty acids (SCFAs) that regulate the action of Treg or other immune cells and the expression of pro-inflammatory factors. These effects on immunity and inflammation can be direct or indirect, but all serve to maintain microbial homeostasis (Furusawa et al., 2013; Smith et al., 2013; Oh et al., 2019). Long-chain fatty acids (LCFA) also play a role in immunity but cannot be synthesized directly by mammals. Instead, microbial taxa (e.g., Prevotella, Lactobacillus, and Alistipes) in the gastrointestinal tract catabolize food to yield LCFAs (Zhao et al., 2018). In a cohort study, antibiotic treatment of pulmonary malignant transformation in cystic fibrosis led to significant LCFA upregulation, implying a link between LCFA and pulmonary inflammation (Hsu et al., 2015). Other major immunity-related metabolites include D-phenyllactic acid (D-PLA), produced by lactic acid bacteria and the most potent natural agonist of human hydroxycarboxylic acid receptor 3 (HCA 3). The latter is expressed in immune cells such as macrophages, neutrophils, monocytes, and adipose tissue (Zhou et al., 2023).
After transplanting respiratory microbiota from mice infected with chronic pneumonia into germ-free mice, lung inflammation increased the secretion of interleukin-17A (IL-17A) in germ free mice (Yadava et al., 2016). After nasal drops of bacteria rich liquid were administered to germ-free mice, the level of IL-17 increased. Lung inflammation of the model mice was significantly relieved after neutralizing IL-17, indicating that the increase in lung bacterial load mediates the increase in the level of the inflammatory factor IL-17 and aggravates lung inflammation (Mannion et al., 2023). Yang et al. (2019) analyzed and compared the bacterial community composition of normal lung tissue and pathological tissue of pulmonary fibrosis and found that outer membrane vesicles (OMVs) were secreted after the abundance of Prevotella and Bacteroides increased. OMVs carry lipopolysaccharides (LPS) and lipoproteins by acting on Toll-like receptors of alveolar macrophages. PRRs further activate the downstream myeloid differentiation factor 88 (MyD88) signaling pathway, thereby promoting the expression of interleukin-17B (IL-17B). IL-17B acts directly on the lung epithelial cells and causes pulmonary fibrosis by recruiting neutrophils and promoting the differentiation of Th17 cells (Tsai et al., 2013). Singanayagam et al.(Singanayagam et al., 2019) found that the disturbance of the pulmonary microbiota in patients with chronic obstructive pulmonary disease (COPD) was closely related to the decreased expression of cathelicidin, and increasing exogenous antimicrobial peptides inhibited the expansion of Streptococcus pneumoniae and reduced the total number of bacteria in the lung tissue. These results indicate that antimicrobial peptides may affect immune homeostasis by altering the structure of lung microbiota. In lung diseases, pathologic changes of lung structure and impaired mucous clearance mechanism may lead to microecological imbalance, and microbial imbalance may promote the occurrence and development of the disease by upregulating inflammatory signals such as NF-κB, Ras, IL-17, and PI3K, or inhibiting the production of TNF and IFN-γ in response to pathogens in the lower respiratory tract (Faniyi et al., 2022).
Certain microbiota, such as S. pneumoniae, promote a wide range of intrinsic responses in the respiratory tract, support the clearance of pathogens, and improve host survival during infection via IL-17 axis and granulocyte macrophage colony stimulating factor signaling (Brown et al., 2017). S. pneumoniae and Hemophilus influenzae can be activated by the mitogen-activated protein kinase (MAPK) signaling pathway, which causes inflammation in the lungs (Lynch, 2016). Klebsiella pneumoniae ST258 can change the metabolic response of the host, enhance glutamine metabolism and fatty acid β oxidation after glucose depletion, lead to an increase in lung active oxide content, recruit immunosuppressive cells, promote the secretion of anti-inflammatory factors, and prolong the survival time of pathogens (Dominguez-Andres et al., 2019).
When germ-free mice are inoculated with respiratory symbiotic microbes such as Staphylococcus aureus and Staphylococcus epidermidis, the granulocyte macrophage colony stimulating factor (GM-CSF) signaling pathway and NLRs are activated, and the NLRs signaling pathway improves resistance to infection (Brown et al., 2017). The lung microbiota of patients with lung cancer is significantly different from that of healthy people, and pathogens such as Mycobacterium tuberculosis can produce in-flammatory cytokines (such as IL-1β and IL-23) by activating the MyD88 signaling pathway, further stimulating lung γδT cells to produce IL-17 and other effector molecules to promote tumor cell proliferation (Mao et al., 2018; Jin et al., 2019).
3.2 Adaptive immune response
The pulmonary microbiota mainly induces the differentiation of Tregs and Th17 cells by producing PRRs to maintain the pulmonary immune balance. During the fetal period, the immune response is mainly dominated by Th2 cells because the immune system is immature. About two weeks after birth, the main members of the lung microbiota gradually change from Proteobacteria and Firmicutes to Bacteroides, promoting natural T cells from Th2 type immune response to Th1 type immune response, which can enhance resistance to asthma and allergic diseases (Wang et al., 2017). The enrichment of pulmonary oropharyngeal microbiota, such as Veillonella and Prevotella, has been associated with inflammatory phenotypes, including elevated Th17 lymphocyte levels, increased expression of inflammatory cytokines, and decreased expression of the inflammatory cytokine TLR4 in alveolar macrophages (Segal et al., 2016). In mice, neutrophil infiltration, high levels of IL-6 and TNF-α, and moderate levels of CD4+ T-cell-derived IFN-γ and IL-17 were associated with Proteobacterium catarrhalis infection (Alnahas et al., 2017). Inhalation of symbiotic bacteria induces a long-term immune response in healthy mice. This includes CD4+ and CD8+ T cell activation, Th17 and γδT cell recruitment, and other anti-regulatory immune responses, such as increases in Tregs and immune checkpoint inhibitor markers on T cells (Wu et al., 2021). Certain lung microbes, including Staphylococcus, produce short chain fatty acids that regulate changes in oral microbes (Remot et al., 2017). In the epithelial lining of patients in an immunocompromised condition, short chain fatty acids production was associated with elevated levels of Tregs induced by M. tuberculosis antigen (Remot et al., 2017; Segal et al., 2017).
The exact outcome of changes in microbial diversity in patients with lung cancer has not been elucidated; however, previous studies have shown that increased α-diversity is generally associated with improved survival and treatment outcomes in several cancers, such as cervical cancer, because α-diversity increases tumor invasion of CD4+ lymphocytes and expression of ki67+ and CD69+ (Sims et al., 2021). Lung tumor growth is associated with an increase in the number of bacteria in the airway and changes in bacterial composition; for example, a dysregulated protomicrobiome triggers MyD88 dependent IL-1 and IL-23 production, and induces the activation and proliferation of lung-resident Vγ6 + Vδ1+γδT cells (Jin et al., 2019).
The microbiota diversity in patients with pneumonia with acquired immune deficiency syndrome (AIDS) is higher than that in patients without AIDS (Twigg et al., 2016). In 60 Ugandan patients with human immunodeficiency virus (HIV) infections in whom pneumonia was treated with antimicrobial agents, patients with reduced airway bacterial diversity showed increased bacterial load and increased expression of matrix metalloproteinase (MMP)-9 and pro-inflammatory TNF-α (Iwai et al., 2014). In addition, the differences in the lower respiratory tract of patients with advanced HIV infection were much greater than those in healthy individuals. These studies suggest that the composition of the respiratory microbiota is correlated with immune response (Lozupone et al., 2013).
4 Respiratory microbiota and respiratory viral infections
The respiratory microbiota can interact with respiratory viruses at multiple levels. On the one hand, respiratory microbiota and its metabolites can promote the proliferation of respiratory viruses and enhance their infectivity. Patients with cystic fibrosis are often susceptible to respiratory syncytial virus infection following infection with pathogenic Pseudomonas aeruginosa, influenza virus, rhinoviruses, and adenovirus (Kiedrowski and Bomberger, 2018). On the other hand, respiratory viruses can also cause secondary bacterial infections by damaging the host mucosal barrier, affecting immune function, and causing changes in the abundance and diversity of respiratory microbiota (Mazel-Sanchez et al., 2019) (Figure 2).
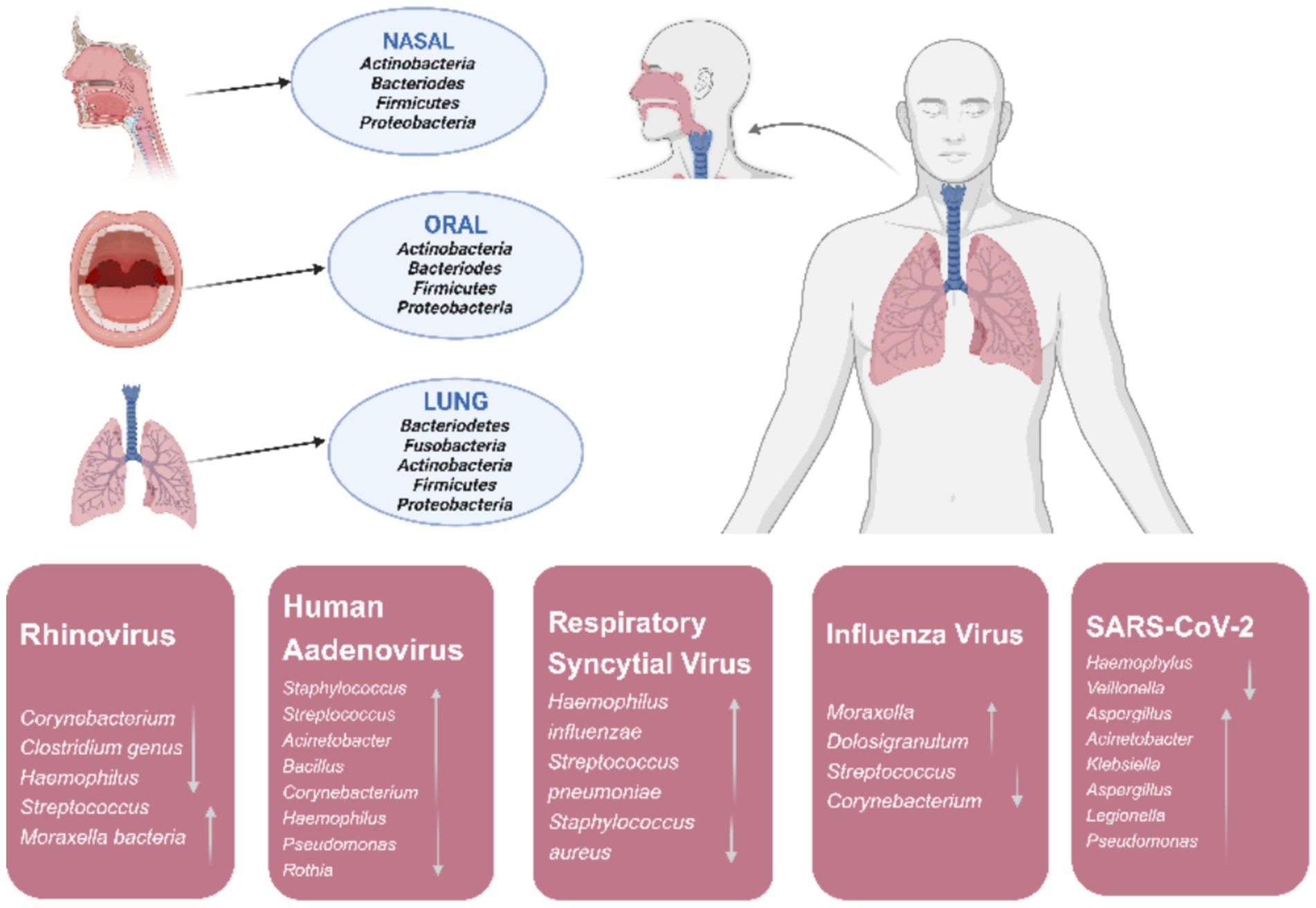
Figure 2. In a healthy state major phyla originating from the microbiota of nasal, oral, and lung in the human respiratory tract and changes of respiratory tract microbiota during respiratory viral infections.
The novel coronavirus disease has triggered a global pandemic that has infected hundreds of millions of people and killed millions of people, with profound consequences for economies, societies, and public health systems worldwide. The severity of the coronavirus disease 2019 (COVID-19) is strongly correlated with the respiratory microbiome. As the severity of COVID-19 increases, oropharyngeal α-diversity tends to decrease in patients (Hernandez-Teran et al., 2021; Rueca et al., 2021; Bradley et al., 2022). The composition and dynamics of oropharyngeal microbiota were significantly associated with COVID-19 mortality. In patients with severe COVID-19, the relative abundances of Hemophilus and Neisseria decreased, which is consistent with previous studies (de Castilhos et al., 2022; Liu et al., 2015). Veillonella, especially Veillonella parvula, may be a potential biomarker of COVID-19 (Ma et al., 2021). In addition, the higher the abundance of Streptococcus at admission, the better the prognosis (Ren et al., 2021). The relative abundance of opportunistic pathogens (including the aforementioned bacterial species) in the nasopharynx of patients with severe COVID-19 was lower than that in patients with mild cases (de Castilhos et al., 2022). Patients with severe COVID-19 are more likely to have reduced α-diversity and increased bacterial load in their lungs than healthy individuals and patients with mild COVID-19 (Merenstein et al., 2021). A metatranscriptomics and metagenomics analysis of bronchoalveolar lavage fluid and sputum samples from 116 COVID-19 patients investigated the relationship between microbial composition and disease severity (mild, severe, and critical). It is indicated that disease severity increased with the Chao index. However, Streptococcus and Rothia abundance both decreased with disease progression from mild to critical. Firmicutes was the dominant microbial taxon in the mild group, while Bacteroidetes abundance increased in the severe group. Finally, the critical group was characterized by higher Actinobacteria and Proteobacteria abundance (Wang et al., 2025). These results suggest that SARS CoV-2 infection disrupts both the oropharyngeal microbiome and the microbiome of the lower respiratory tract. This microbial imbalance potentially increases the risk of secondary pulmonary infections (Wang et al., 2025).
Approximately 250,000–500,000 people die from influenza annually, placing a huge burden on public health (Javanian et al., 2021). Changes in the respiratory microbiome may affect susceptibility to influenza (Tsang et al., 2020), which is related to age (Huffnagle et al., 2017). In a mouse model, after infection with influenza A virus (IAV), the threshold of bacterial invasion decreased, which was mainly caused by the change in bacterial composition in the respiratory tract, but had limited association with the change in overall bacterial abundance. Symbiotic bacteria such as Streptococcus infantis and Streptococcus mitis may play a key role in resisting the overgrowth of these pathogens (Li et al., 2023).
Ding et al. (2019) found differences in the composition of the oropharyngeal microbiota in acute respiratory infections caused by different IAV subtypes. In patients with influenza A, the main species were Atopobium and Prevotella, whereas in patients with influenza B, Bergeyella and Prevotella dominated (Li et al., 2023). In addition, vaccination was also found to correlate with microbiota composition in cohorts of influenza A and B, which could mean that influenza vaccination not only prevents influenza infection but may also play a role in controlling secondary bacterial infections (Ding et al., 2019).
Rhinovirus (RV) infection has also been associated with changes in respiratory microbiome (Nakagome et al., 2014; Lehtinen et al., 2018). With an increase in RV load, the abundance of Corynebacterium and Dolosigranulum decreased, while the abundance of Hemophilus increased. In addition, the abundance of Streptococcus and Moraxella increased and decreased, respectively, with changes in the RV replication level. The enrichment of Corynebacterium and Guignococcus may help individuals with infection maintain normal respiratory physiological functions during rhinovirus infection, thereby reducing or preventing respiratory symptoms (Kloepfer et al., 2017).
Respiratory syncytial virus (RSV) infection is closely associated with nasopharyngeal microbiota. The main manifestation includes an increase in H. influenzae and S. pneumoniae. Additionally, there appeared to be an interaction between RSV and these two bacteria (de Steenhuijsen Piters et al., 2016). Kanmani et al. (2017) found that the injection Corynebacterium pseudodiphtheriticum, one of the components of the upper respiratory tract microbiota in healthy people, into the nasal cavity of young mice enhanced their resistance to RSV and pneumococcal secondary infection.
Human adenovirus (HAdV) is an important infectious respiratory tract pathogen in children, accounting for 4–10% of community-acquired pediatric pneumonia cases (Jobran et al., 2018). Adenoviruses can cause an increase in Staphylococcus, Streptococcus, Acinetobacter, Bacillus, and Corynebacterium, and a decrease in Hemophilus, Pseudomonas, and Rothia. Adenoviral infection is often accompanied by mycoplasma co-infection (Zhong and Dong, 2021). Children with HAdV infection have higher microbial diversity in their lungs than patients with Mycoplasma pneumoniae infection. Compared with patients with M. pneumoniae infection alone, patients with HAdV infection had decreased BALF microbial richness and increased β-diversity (Zhou et al., 2022). The reason may be that M. pneumoniae inhibits the growth of other bacteria by directly competing with or activating bacterial clearance mechanisms, whereas HAdV infection damages lung epithelial cells and suppresses the immune response, thus promoting bacterial growth (Wang et al., 2019; Yang et al., 2004).
5 Respiratory microbiota and malignant tumors
The oral microbiota plays a major role in oral health and systemic diseases, including malignancies. A study on oral microbiota and lung cancer risk in low-income populations in the southeastern United States analyzed 156 cases of lung cancer (73 European Americans and 83 non-European Americans) and 156 individually matched controls in a cohort study using 16S rRNA gene sequencing to analyze oral microbiota in oral rinse samples and evaluate the association between individual bacterial abundance or prevalence and lung cancer risk. Oral microbiota may play a role in the development of lung cancer (Shi et al., 2021). Jin et al. (2019) analyzed the BALF of 91 patients with lung cancer, 29 patients with non-malignant lung disease, and 30 healthy subjects using metagenomics and found that compared to healthy subjects, the airway microbial abundance of patients with lung cancer was reduced, and the microbiome of patients with non-malignant lung disease was similar to that of patients with lung cancer. Bradyrhizobium japonicum was found only in patients with lung cancer, whereas Acidovorax was found in patients with lung cancer and non-malignant lung diseases. This study showed reduced microbiome abundance in patients with lung cancer and that microbiome specific biomarkers may help diagnose lung cancer in patients when lung biopsy is not feasible. Zhang et al. (2020) screened for colorectal cancer by detecting the oral microbiota and found that the microecological imbalance of oral pathogens, such as Clostridium, Prevotella, and Porphyromonas may be the main risk factor for colorectal cancer. The detection of oral microbial markers may serve as an effective, non-invasive method for colorectal cancer screening. Patients with pancreatic cancer have oral microbiota dysbiosis. DNA microarrays were used to compare the oral salivary microbiota of patients with pancreatic cancer, chronic pancreatitis, and healthy controls. The abundance of Neisseria elongatus and S. mitis decreased significantly in patients with pancreatic cancer. The abundance of Granulicatella adiacens was significantly higher in patients with chronic pancreatitis, and significantly lower than S. mitis (Vogtmann et al., 2020).
6 Administration of antibiotics and probiotics
The effects of antibiotics, the routine clinical treatment of respiratory infections, on the respiratory microbiota are not always favorable. For example, long-term use of macrolides can significantly alter the composition of the microbiota in the lower respiratory tract (Taylor et al., 2019; Ritchie and Singanayagam, 2020), and mice treated with neomycin were more likely to be susceptible to influenza than the control group (Ichinohe et al., 2011). In addition, the use of antibiotics can lead to selection pressure. One study compared the oropharyngeal microbial composition and related functional changes between healthy individuals and patients with moderate to severe COVID-19 (Wu et al., 2023). The results showed that disease-causing pathogens enriched in patients with COVID-19 had higher virulence and resistance, suggesting that antibiotic use may make the disease more difficult to control. Probiotics play an important role in microbial therapy as a common means of regulating microbiota. The use of gut-lung or gut-respiratory axis interactions to regulate the airway response by oral probiotics has become a mainstream strategy (Dang and Marsland, 2019). Oral microbiological therapy with lactic acid bacteria can help reduce the risk of respiratory failure, as shown in a small clinical study on patients COVID-19 (d'Ettorre et al., 2020). Furthermore, ClinicalTri-als.gov has registered several clinical trials investigating the effectiveness of probiotics (such as Lactobacillus or Bifidobacterium) in preventing or treating respiratory viral infections. These clinical trials not only reflect the recognition of the potential clinical value of probiotics by the scientific community but also indicate the significant clinical and economic benefits that may be brought about in the future.
Compared with the indirect effect of oral probiotics on respiratory health through the entero-lung axis, the direct application of microbiota to the respiratory tract to exert activity at the site of infection may be more effective in regulating the susceptibility to infection and its course. Previous studies demonstrated the effectiveness of local application. For example, a single prophylactic fogging treatment with H. influenzae lysate successfully prevented the occurrence of influenza pneumonia (Tuvim et al., 2009), whereas an intranasal injection of fermentative Lactobacillus CJL-112 L promoted the production of specific protective IgA and enhanced immune defense in a mouse model of influenza infection (Yeo et al., 2014).
Lactobacillus is considered a promising candidate strain owing to its various probiotic properties and potential for local application in the airway (De Boeck et al., 2021; De Boeck et al., 2020), Several studies have demonstrated the feasibility and safety of its airway administration. However, determining the optimal probiotic strains, mixtures, dosages, and formulations remains challenging in clinical practice.
In addition, the metabolites of the microbiota, as key signals for the microbiota-host interaction, also show considerable potential as a treatment for lung infections (Montassier et al., 2023). The discovery of bacteria derived host isoenzymes in the gut provides a new therapeutic approach for metabolic diseases (Wang et al., 2023), suggesting that similar mechanisms may also exist in the respiratory tract; however, their specific existence and mechanism of action need to be further explored.
7 Conclusion
In recent years, important progress has been made in the study of respiratory microbiota, and the main microbiota types in each anatomical region of the upper and lower respiratory tracts have been identified. Respiratory infections are often accompanied by a decline in microbial diversity, an increase in microbial load, and an increase in the abundance of opportunistic pathogens, especially in patients in intensive care unit (ICU), where the abundance of multiple bacteria is correlated with patient outcomes. Recent studies on respiratory microbiota in disease states have identified microbial signatures associated with differential diagnosis, disease severity, and prognosis. However, the lack of consistency among the results of different studies makes it challenging to clearly define respiratory microbiota dysbiosis.
Even within the course of a single disease, multiple forms of dysregulation may occur. The phenotypes of dysregulation of the respiratory microbiota are inconsistent across cohorts, infection types, and disease progression, making the discovery of common markers of the respiratory microbiota challenging. Future studies should shift from crosssectional designs and correlation studies to longitudinal sampling methods, consider geographical characteristics and experimental conditions, and establish animal models that can closely simulate the human respiratory microbiota to better control the microbiota and host conditions to verify the mechanism of microbial action.
At present, clear evidence for the local use of probiotics in the respiratory tract to regulate respiratory microbiota and prevent respiratory infections is lacking. Although intermittent drops of oral symbiotic bacteria in the lower respiratory tract altered the host susceptibility to respiratory pathogens in multiple animal experiments (Jin et al., 2019; Zhang et al., 2020), there is still a lack of institutional studies and feasible implementation plans in this field. Conducting safe and meaningful clinical trials at present is challenging.
Author contributions
XJ: Writing – original draft. SZ: Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was funded by the Natural Science Foundation of Heilongjiang Province (PL2024C038); Heilongjiang Province Education Department Fundamental Scientific Research Funds (145309510).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Correction note
This article has been corrected with minor changes. These changes do not impact the scientific content of the article.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Alnahas, S., Hagner, S., Raifer, H., Kilic, A., Gasteiger, G., Mutters, R., et al. (2017). IL-17 and TNF-alpha are key mediators of Moraxella catarrhalis triggered exacerbation of allergic airway inflammation. Front. Immunol. 8:1562.
Bradley, E. S., Zeamer, A. L., Bucci, V., Cincotta, L., Salive, M. C., Dutta, P., et al. (2022). Oropharyngeal microbiome profiled at admission is predictive of the need for respiratory support among COVID-19 patients. Front. Microbiol. 13:1009440. doi: 10.3389/fmicb.2022.1009440
Brown, R. L., Sequeira, R. P., and Clarke, T. B. (2017). The microbiota protects against respiratory infection via GM-CSF signaling. Nat. Commun. 8:1512. doi: 10.1038/s41467-017-01803-x
Charlson, E. S., Bittinger, K., Haas, A. R., Fitzgerald, A. S., Frank, I., Yadav, A., et al. (2011). Topographical continuity of bacterial populations in the healthy human respiratory tract. Am. J. Respir. Crit. Care Med. 184, 957–963. doi: 10.1164/rccm.201104-0655OC
Chellappan, D. K., Sze Ning, Q. L., Su Min, S. K., Bin, S. Y., Chern, P. J., Shi, T. P., et al. (2019). Interactions between microbiome and lungs: paving new paths for microbiome based bio-engineered drug delivery systems in chronic respiratory diseases. Chem. Biol. Interact. 310:108732. doi: 10.1016/j.cbi.2019.108732
Dang, A. T., and Marsland, B. J. (2019). Microbes, metabolites, and the gut-lung axis. Mucosal Immunol. 12, 843–850. doi: 10.1038/s41385-019-0160-6
De Boeck, I., Spacova, I., Vanderveken, O. M., and Lebeer, S. (2021). Lactic acid bacteria as probiotics for the nose? Microb. Biotechnol. 14, 859–869. doi: 10.1111/1751-7915.13759
De Boeck, I., van den Broek, M. F. L., Allonsius, C. N., Spacova, I., Wittouck, S., Martens, K., et al. (2020). Lactobacilli have a niche in the human nose. Cell Rep. 31:107674. doi: 10.1016/j.celrep.2020.107674
de Castilhos, J., Zamir, E., Hippchen, T., Rohrbach, R., Schmidt, S., Hengler, S., et al. (2022). Severe Dysbiosis and specific Haemophilus and Neisseria signatures as hallmarks of the oropharyngeal microbiome in critically ill coronavirus disease 2019 (COVID-19) patients. Clin. Infect. Dis. 75, e1063–e1071. doi: 10.1093/cid/ciab902
de Steenhuijsen Piters, W. A., Heinonen, S., Hasrat, R., Bunsow, E., Smith, B., Suarez-Arrabal, M. C., et al. (2016). Nasopharyngeal microbiota, host transcriptome, and disease severity in children with respiratory syncytial virus infection. Am. J. Respir. Crit. Care Med. 194, 1104–1115. doi: 10.1164/rccm.201602-0220OC
d'Ettorre, G., Ceccarelli, G., Marazzato, M., Campagna, G., Pinacchio, C., Alessandri, F., et al. (2020). Challenges in the management of SARS-CoV2 infection: the role of Oral Bacteriotherapy as complementary therapeutic strategy to avoid the progression of COVID-19. Front Med 7:389. doi: 10.3389/fmed.2020.00389
Dickson, R. P., Erb-Downward, J. R., Falkowski, N. R., Hunter, E. M., Ashley, S. L., and Huffnagle, G. B. (2018). The lung microbiota of healthy mice are highly variable, cluster by environment, and reflect variation in baseline lung innate immunity. Am. J. Respir. Crit. Care Med. 198, 497–508. doi: 10.1164/rccm.201711-2180OC
Ding, T., Song, T., Zhou, B., Geber, A., Ma, Y., Zhang, L., et al. (2019). Microbial composition of the human nasopharynx varies according to influenza virus type and vaccination status. MBio 10:296. doi: 10.1128/mBio.01296-19
Dominguez-Andres, J., Novakovic, B., Li, Y., Scicluna, B. P., Gresnigt, M. S., Arts, R. J. W., et al. (2019). The Itaconate pathway is a central regulatory node linking innate immune tolerance and trained immunity. Cell Metab. 29, 211–220.e5. doi: 10.1016/j.cmet.2018.09.003
Evans, S. E., Xu, Y., Tuvim, M. J., and Dickey, B. F. (2010). Inducible innate resistance of lung epithelium to infection. Annu. Rev. Physiol. 72, 413–435. doi: 10.1146/annurev-physiol-021909-135909
Faniyi, A. A., Hughes, M. J., Scott, A., Belchamber, K. B. R., and Sapey, E. (2022). Inflammation, ageing and diseases of the lung: potential therapeutic strategies from shared biological pathways. Br. J. Pharmacol. 179, 1790–1807. doi: 10.1111/bph.15759
Furusawa, Y., Obata, Y., Fukuda, S., Endo, T. A., Nakato, G., Takahashi, D., et al. (2013). Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 504, 446–450. doi: 10.1038/nature12721
Hernandez-Teran, A., Mejia-Nepomuceno, F., Herrera, M. T., Barreto, O., Garcia, E., Castillejos, M., et al. (2021). Dysbiosis and structural disruption of the respiratory microbiota in COVID-19 patients with severe and fatal outcomes. Sci. Rep. 11:21297. doi: 10.1038/s41598-021-00851-0
Hsu, A. C., Starkey, M. R., Hanish, I., Parsons, K., Haw, T. J., Howland, L. J., et al. (2015). Targeting PI3K-p110alpha suppresses influenza virus infection in chronic obstructive pulmonary disease. Am. J. Respir. Crit. Care Med. 191, 1012–1023.
Hu, X., Wu, M., Ma, T., Zhang, Y., Zou, C., Wang, R., et al. (2020). Single-cell transcriptomics reveals distinct cell response between acute and chronic pulmonary infection of Pseudomonas aeruginosa. Medcomm 3:e193.
Huffnagle, G. B., Dickson, R. P., and Lukacs, N. W. (2017). The respiratory tract microbiome and lung inflammation: a two-way street. Mucosal Immunol. 10, 299–306. doi: 10.1038/mi.2016.108
Ichinohe, T., Pang, I. K., Kumamoto, Y., Peaper, D. R., Ho, J. H., Murray, T. S., et al. (2011). Microbiota regulates immune defense against respiratory tract influenza a virus infection. Proc. Natl. Acad. Sci. USA 108, 5354–5359. doi: 10.1073/pnas.1019378108
Invernizzi, R., Lloyd, C. M., and Molyneaux, P. L. (2020). Respiratory microbiome and epithelial interactions shape immunity in the lungs. Immunology 160, 171–182. doi: 10.1111/imm.13195
Iwai, S., Huang, D., Fong, S., Jarlsberg, L. G., Worodria, W., Yoo, S., et al. (2014). The lung microbiome of Ugandan HIV-infected pneumonia patients is compositionally and functionally distinct from that of san Franciscan patients. PLoS One 9:e95726. doi: 10.1371/journal.pone.0095726
Iwasaki, A., and Medzhitov, R. (2015). Control of adaptive immunity by the innate immune system. Nat. Immunol. 16, 343–353. doi: 10.1038/ni.3123
Javanian, M., Barary, M., Ghebrehewet, S., Koppolu, V., Vasigala, V., and Ebrahimpour, S. (2021). A brief review of influenza virus infection. J. Med. Virol. 93, 4638–4646. doi: 10.1002/jmv.26990
Jin, J., Gan, Y., Liu, H., Wang, Z., Yuan, J., Deng, T., et al. (2019). Diminishing microbiome richness and distinction in the lower respiratory tract of lung cancer patients: a multiple comparative study design with independent validation. Lung Cancer 136, 129–135. doi: 10.1016/j.lungcan.2019.08.022
Jin, C., Lagoudas, G. K., Zhao, C., Bullman, S., Bhutkar, A., Hu, B., et al. (2019). Commensal microbiota promote lung cancer development via gammadelta T cells. Cell 176:e16, 998–1013.
Jobran, S., Kattan, R., Shamaa, J., Marzouqa, H., and Hindiyeh, M. (2018). Adenovirus respiratory tract infections in infants: a retrospective chart-review study. Lancet 391:S43. doi: 10.1016/S0140-6736(18)30409-4
Kanmani, P., Clua, P., Vizoso-Pinto, M. G., Rodriguez, C., Alvarez, S., Melnikov, V., et al. (2017). Respiratory commensal Bacteria Corynebacterium pseudodiphtheriticum improves resistance of infant mice to respiratory syncytial virus and Streptococcus pneumoniae superinfection. Front. Microbiol. 8:1613. doi: 10.3389/fmicb.2017.01613
Kiedrowski, M. R., and Bomberger, J. M. (2018). Viral-bacterial co-infections in the cystic fibrosis respiratory tract. Front. Immunol. 9:3067. doi: 10.3389/fimmu.2018.03067
Kloepfer, K. M., Sarsani, V. K., Poroyko, V., Lee, W. M., Pappas, T. E., Kang, T., et al. (2017). Community-acquired rhinovirus infection is associated with changes in the airway microbiome. J. Allergy Clin. Immunol. 140, 312–315. doi: 10.1016/j.jaci.2017.01.038
Lehtinen, M. J., Hibberd, A. A., Mannikko, S., Yeung, N., Kauko, T., Forssten, S., et al. (2018). Nasal microbiota clusters associate with inflammatory response, viral load, and symptom severity in experimental rhinovirus challenge. Sci. Rep. 8:11411. doi: 10.1038/s41598-018-29793-w
Leiva-Juarez, M. M., Kolls, J. K., and Evans, S. E. (2018). Lung epithelial cells: therapeutically inducible effectors of antimicrobial defense. Mucosal Immunol. 11, 21–34. doi: 10.1038/mi.2017.71
Levy, M., Thaiss, C. A., and Elinav, E. (2015). Metagenomic cross-talk: the regulatory interplay between immunogenomics and the microbiome. Genome Med. 7:120. doi: 10.1186/s13073-015-0249-9
Li, R., Li, J., and Zhou, X. (2024). Lung microbiome: new insights into the pathogenesis of respiratory diseases. Signal Transduct. Target. Ther. 9:19. doi: 10.1038/s41392-023-01722-y
Li, H., Wu, X., Zeng, H., Chang, B., Cui, Y., Zhang, J., et al. (2023). Unique microbial landscape in the human oropharynx during different types of acute respiratory tract infections. Microbiome. 11:157. doi: 10.1186/s40168-023-01597-9
Liu, G., Tang, C. M., and Exley, R. M. (2015). Non-pathogenic Neisseria: members of an abundant, multi-habitat, diverse genus. Microbiology 161, 1297–1312. doi: 10.1099/mic.0.000086
Lozupone, C., Cota-Gomez, A., Palmer, B. E., Linderman, D. J., Charlson, E. S., Sodergren, E., et al. (2013). Widespread colonization of the lung by Tropheryma whipplei in HIV infection. Am. J. Respir. Crit. Care Med. 187, 1110–1117. doi: 10.1164/rccm.201211-2145OC
Lynch, S. V. (2016). The lung microbiome and airway disease. Ann. Am. Thorac. Soc. 13, S462–S465. doi: 10.1513/AnnalsATS.201605-356AW
Ma, S., Zhang, F., Zhou, F., Li, H., Ge, W., Gan, R., et al. (2021). Metagenomic analysis reveals oropharyngeal microbiota alterations in patients with COVID-19. Signal Transduct. Target. Ther. 6:191. doi: 10.1038/s41392-021-00614-3
Man, W. H., de Steenhuijsen Piters, W. A., and Bogaert, D. (2017). The microbiota of the respiratory tract: gatekeeper to respiratory health. Nat. Rev. Microbiol. 15, 259–270. doi: 10.1038/nrmicro.2017.14
Mannion, J. M., McLoughlin, R. M., and Lalor, S. J. (2023). The airway microbiome-IL-17 Axis: a critical regulator of chronic inflammatory disease. Clin. Rev. Allergy Immunol. 64, 161–178. doi: 10.1007/s12016-022-08928-y
Mao, Q., Jiang, F., Yin, R., Wang, J., Xia, W., Dong, G., et al. (2018). Interplay between the lung microbiome and lung cancer. Cancer Lett. 415, 40–48. doi: 10.1016/j.canlet.2017.11.036
Mazel-Sanchez, B., Yildiz, S., and Schmolke, M. (2019). Menage a trois: virus, host, and microbiota in experimental infection models. Trends Microbiol. 27, 440–452.
Merenstein, C., Liang, G., Whiteside, S. A., Cobian-Guemes, A. G., Merlino, M. S., Taylor, L. J., et al. (2021). Signatures of COVID-19 severity and immune response in the respiratory tract microbiome. MBio 12:e0177721. doi: 10.1128/mBio.01777-21
Montassier, E., Kitsios, G. D., Radder, J. E., Le Bastard, Q., Kelly, B. J., Panzer, A., et al. (2023). Robust airway microbiome signatures in acute respiratory failure and hospital-acquired pneumonia. Nat. Med. 29, 2793–2804. doi: 10.1038/s41591-023-02617-9
Morris, A., Beck, J. M., Schloss, P. D., Campbell, T. B., Crothers, K., Curtis, J. L., et al. (2013). Comparison of the respiratory microbiome in healthy nonsmokers and smokers. Am. J. Respir. Crit. Care Med. 187, 1067–1075. doi: 10.1164/rccm.201210-1913OC
Nakagome, K., Bochkov, Y. A., Ashraf, S., Brockman-Schneider, R. A., Evans, M. D., Pasic, T. R., et al. (2014). Effects of rhinovirus species on viral replication and cytokine production. J. Allergy Clin. Immunol. 134, 332–341. doi: 10.1016/j.jaci.2014.01.029
Natalini, J. G., Singh, S., and Segal, L. N. (2023). The dynamic lung microbiome in health and disease. Nat. Rev. Microbiol. 21, 222–235. doi: 10.1038/s41579-022-00821-x
Oh, J. H., Alexander, L. M., Pan, M., Schueler, K. L., Keller, M. P., Attie, A. D., et al. (2019). Dietary fructose and microbiota-derived short-chain fatty acids promote bacteriophage production in the gut symbiont Lactobacillus reuteri. Cell Host Microbe 25:e6, 273–284.
Remot, A., Descamps, D., Noordine, M. L., Boukadiri, A., Mathieu, E., Robert, V., et al. (2017). Bacteria isolated from lung modulate asthma susceptibility in mice. ISME J. 11, 1061–1074. doi: 10.1038/ismej.2016.181
Ren, L., Wang, Y., Zhong, J., Li, X., Xiao, Y., Li, J., et al. (2021). Dynamics of the upper respiratory tract microbiota and its association with mortality in COVID-19. Am. J. Respir. Crit. Care Med. 204, 1379–1390. doi: 10.1164/rccm.202103-0814OC
Ritchie, A. I., and Singanayagam, A. (2020). Metagenomic characterization of the respiratory microbiome. A piece de resistance. Am. J. Respir. Crit. Care Med. 202, 321–322.
Rueca, M., Fontana, A., Bartolini, B., Piselli, P., Mazzarelli, A., Copetti, M., et al. (2021). Investigation of nasal/oropharyngeal microbial community of COVID-19 patients by 16S rDNA sequencing. Int. J. Environ. Res. Public Health 18:174. doi: 10.3390/ijerph18042174
Segal, L. N., Clemente, J. C., Li, Y., Ruan, C., Cao, J., Danckers, M., et al. (2017). Anaerobic bacterial fermentation products increase tuberculosis risk in antiretroviral-drug-treated HIV patients. Cell Host Microbe 21, 530–537. doi: 10.1016/j.chom.2017.03.003
Segal, L. N., Clemente, J. C., Tsay, J. C., Koralov, S. B., Keller, B. C., Wu, B. G., et al. (2016). Enrichment of the lung microbiome with oral taxa is associated with lung inflammation of a Th17 phenotype. Nat. Microbiol. 1:16031. doi: 10.1038/nmicrobiol.2016.31
Shi, J., Yang, Y., Xie, H., Wang, X., Wu, J., Long, J., et al. (2021). Association of oral microbiota with lung cancer risk in a low-income population in the southeastern USA. Cancer Causes Control 32, 1423–1432. doi: 10.1007/s10552-021-01490-6
Sims, T. T., El Alam, M. B., Karpinets, T. V., Dorta-Estremera, S., Hegde, V. L., Nookala, S., et al. (2021). Gut microbiome diversity is an independent predictor of survival in cervical cancer patients receiving chemoradiation. Commun Biol. 4:237. doi: 10.1038/s42003-021-01741-x
Singanayagam, A., Glanville, N., Cuthbertson, L., Bartlett, N. W., Finney, L. J., Turek, E., et al. (2019). Inhaled corticosteroid suppression of cathelicidin drives dysbiosis and bacterial infection in chronic obstructive pulmonary disease. Sci. Transl. Med. 11:507. doi: 10.1126/scitranslmed.aav3879
Smith, P. M., Howitt, M. R., Panikov, N., Michaud, M., Gallini, C. A., Bohlooly, Y. M., et al. (2013). The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 341, 569–573.
Taylor, S. L., Leong, L. E. X., Mobegi, F. M., Choo, J. M., Wesselingh, S., Yang, I. A., et al. (2019). Long-term azithromycin reduces Haemophilus influenzae and increases antibiotic resistance in severe asthma. Am. J. Respir. Crit. Care Med. 200, 309–317. doi: 10.1164/rccm.201809-1739OC
Tsai, H. C., Velichko, S., Hung, L. Y., and Wu, R. (2013). IL-17A and Th17 cells in lung inflammation: an update on the role of Th17 cell differentiation and IL-17R signaling in host defense against infection. Clin. Dev. Immunol. 2013:267971, 1–12. doi: 10.1155/2013/267971
Tsang, T. K., Lee, K. H., Foxman, B., Balmaseda, A., Gresh, L., Sanchez, N., et al. (2020). Association between the respiratory microbiome and susceptibility to influenza virus infection. Clin. Infect. Dis. 71, 1195–1203. doi: 10.1093/cid/ciz968
Tuvim, M. J., Evans, S. E., Clement, C. G., Dickey, B. F., and Gilbert, B. E. (2009). Augmented lung inflammation protects against influenza a pneumonia. PLoS One 4:e4176. doi: 10.1371/journal.pone.0004176
Twigg, H. L., Knox, K. S., Zhou, J., Crothers, K. A., Nelson, D. E., Toh, E., et al. (2016). Effect of advanced HIV infection on the respiratory microbiome. Am. J. Respir. Crit. Care Med. 194, 226–235. doi: 10.1164/rccm.201509-1875OC
Uehara, A., Fujimoto, Y., Fukase, K., and Takada, H. (2007). Various human epithelial cells express functional toll-like receptors, NOD1 and NOD2 to produce anti-microbial peptides, but not proinflammatory cytokines. Mol. Immunol. 44, 3100–3111. doi: 10.1016/j.molimm.2007.02.007
Vogtmann, E., Han, Y., Caporaso, J. G., Bokulich, N., Mohamadkhani, A., Moayyedkazemi, A., et al. (2020). Oral microbial community composition is associated with pancreatic cancer: a case-control study in Iran. Cancer Med. 9, 797–806. doi: 10.1002/cam4.2660
Wang, D., Duan, Y., He, L., Jiang, J., Xian, J., Yuan, K., et al. (2025). Altered microbiota of the lower respiratory tract and its association with COVID-19 severity analysed by metagenomics and metatranscriptomics. Commun Biol. 8:804. doi: 10.1038/s42003-025-08234-1
Wang, J., Li, F., and Tian, Z. (2017). Role of microbiota on lung homeostasis and diseases. Sci. China Life Sci. 60, 1407–1415. doi: 10.1007/s11427-017-9151-1
Wang, K., Zhang, Z., Hang, J., Liu, J., Guo, F., Ding, Y., et al. (2023). Microbial-host-isozyme analyses reveal microbial DPP4 as a potential antidiabetic target. Science 381:eadd5787.
Wang, H., Zhou, Q., Dai, W., Feng, X., Lu, Z., Yang, Z., et al. (2019). Lung microbiota and pulmonary inflammatory cytokines expression vary in children with Tracheomalacia and adenoviral or Mycoplasma pneumoniae pneumonia. Front. Pediatr. 7:265. doi: 10.3389/fped.2019.00265
Wu, J., Liu, W., Zhu, L., Li, N., Luo, G., Gu, M., et al. (2023). Dysbiosis of oropharyngeal microbiome and antibiotic resistance in hospitalized COVID-19 patients. J. Med. Virol. 95:e28727. doi: 10.1002/jmv.28727
Wu, B. G., Sulaiman, I., Tsay, J. J., Perez, L., Franca, B., Li, Y., et al. (2021). Episodic aspiration with oral commensals induces a MyD88-dependent, pulmonary T-helper cell type 17 response that mitigates susceptibility to Streptococcus pneumoniae. Am. J. Respir. Crit. Care Med. 203, 1099–1111. doi: 10.1164/rccm.202005-1596OC
Yadava, K., Pattaroni, C., Sichelstiel, A. K., Trompette, A., Gollwitzer, E. S., Salami, O., et al. (2016). Microbiota promotes chronic pulmonary inflammation by enhancing IL-17A and autoantibodies. Am. J. Respir. Crit. Care Med. 193, 975–987. doi: 10.1164/rccm.201504-0779OC
Yang, D., Chen, X., Wang, J., Lou, Q., Lou, Y., Li, L., et al. (2019). Dysregulated lung commensal Bacteria drive interleukin-17B production to promote pulmonary fibrosis through their outer membrane vesicles. Immunity 50, 692–706.e7. doi: 10.1016/j.immuni.2019.02.001
Yang, J., Hooper, W. C., Phillips, D. J., and Talkington, D. F. (2004). Cytokines in Mycoplasma pneumoniae infections. Cytokine Growth Factor Rev. 15, 157–168. doi: 10.1016/j.cytogfr.2004.01.001
Yeo, J. M., Lee, H. J., Kim, J. W., Lee, J. B., Park, S. Y., Choi, I. S., et al. (2014). Lactobacillus fermentum CJL-112 protects mice against influenza virus infection by activating T-helper 1 and eliciting a protective immune response. Int. Immunopharmacol. 18, 50–54. doi: 10.1016/j.intimp.2013.10.020
Zhang, S., Kong, C., Yang, Y., Cai, S., Li, X., Cai, G., et al. (2020). Human oral microbiome dysbiosis as a novel non-invasive biomarker in detection of colorectal cancer. Theranostics 10, 11595–11606. doi: 10.7150/thno.49515
Zhang, J., Wu, Y., Liu, J., Yang, Y., Li, H., Wu, X., et al. (2022). Differential Oral microbial input determines two microbiota Pneumo-types associated with health status. Adv Sci (Weinh). 9:e2203115. doi: 10.1002/advs.202203115
Zhao, L., Huang, Y., Lu, L., Yang, W., Huang, T., Lin, Z., et al. (2018). Saturated long-chain fatty acid-producing bacteria contribute to enhanced colonic motility in rats. Microbiome. 6:107. doi: 10.1186/s40168-018-0492-6
Zhong, H., and Dong, X. (2021). Analysis of clinical characteristics and risk factors of severe adenovirus pneumonia in children. Front. Pediatr. 9:566797. doi: 10.3389/fped.2021.566797
Zhou, W., Chen, J., Xi, Z., Shi, Y., Wang, L., and Lu, A. (2022). Characteristics of lung microbiota in children's refractory Mycoplasma pneumoniae pneumonia Coinfected with human adenovirus B. Can. J. Infect. Dis. Med. Microbiol. 2022, 1–8. doi: 10.1155/2022/7065890
Zhou, Y., Liu, M., Liu, K., Wu, G., and Tan, Y. (2023). Lung microbiota and potential treatment of respiratory diseases. Microb. Pathog. 181:106197. doi: 10.1016/j.micpath.2023.106197
Keywords: respiratory tract microbiota, viral infection, airway dysbiosis, immune, interaction
Citation: Jiang X and Zhang S (2025) Respiratory microbiota, host immunity, respiratory viral infections and malignant tumors. Front. Microbiol. 16:1626077. doi: 10.3389/fmicb.2025.1626077
Edited by:
Svetlana Khaiboullina, University of Nevada, Reno, United StatesReviewed by:
Salvatore Walter Papasergi, National Research Council (CNR), ItalyIngrid Gisell Bustos Moya, Universidad de La Sabana, Colombia
Copyright © 2025 Jiang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sujiang Zhang, enNqZGt5QDEyNi5jb20=
 Xinjie Jiang
Xinjie Jiang Sujiang Zhang1,3*
Sujiang Zhang1,3*