- 1Department of Gastroenterology, Lanzhou University Second Hospital, Lanzhou, China
- 2Department of Gastroenterology, Key Laboratory of Digestive Diseases of Lanzhou University Second Hospital, Lanzhou, China
- 3Department of Respiratory, Gansu Second People’s Hospital, Lanzhou, China
- 4The First People’s Hospital of Jiande City, Hangzhou, China
- 5Department of Oncology, Gansu Provincial Central Hospital, Lanzhou, China
Helicobacter pylori is a gram-negative bacterium that associated with diseases such as gastritis, peptic ulcer and gastric cancer. In recent years, various treatment options have been evaluated, such as bismuth-containing quadruple therapy, high-dose dual therapy, and the use of acid-suppressing drugs such as Vonoprazan, however, the effectiveness of H. pylori eradication treatment is still dramatically decreasing due to the rising antibiotic resistance rate, and successful eradication of H. pylori has become a major public health problem. Therefore a promising strategy against drug-resistant H. pylori is to individualize treatment based on the outcome of antibiotic resistance. This article reviews the antibiotic resistance situation in recent years in various regions. The advantages and disadvantages of novel antibiotic resistance detection methods are examined, and the therapeutic efficacy of individualized therapy under different detection methods is evaluated. Molecular methods have developed rapidly in recent years, and non-invasive methods can quickly and accurately determine the presence of drug resistance. Clinical application of antibiotic resistance test results to guide medication use needs to be used as early as possible. Customized therapies based on antibiotic drug sensitivity testing and individualized therapies guided by personal medication history can contribute to future therapeutic strategies.
1 Introduction
With drug resistance rates of Helicobacter pylori (H. pylori) climbing and eradication rates declining globally this year, new breakthroughs need to be found in the eradication treatment of H. pylori. Unfortunately, curing this infection remains challenging for clinicians, as there are no available empirical treatment regimens capable of eradicating the bacteria in all treated patients. The application of drug susceptibility testing not only enables precise treatment but also prevents antimicrobial drug resistance. The 2022 Chinese Guidelines for the Treatment of Helicobacter pylori Infections propose individualized treatment guided by bacterial culture and antimicrobial drug susceptibility testing for patients with refractory H. pylori infections when available (Zhou et al., 2022), but there is still a lack of definitive evidence to support individualized rather than empirical treatment. There are various detection methods for drug resistance testing, and those with high specificity and sensitivity are still being explored. Literature published was systematically identified in PubMed from 2018 to 2025 using the following search terms: (Helicobacter pylori) AND (antibiotic OR antimicrobial OR antimicrobial OR antibacterial OR anti-bacterial OR drug) AND (resistance OR resistant*). Each protein was searched individually to reduce the rate of missed detection. This article compares the existing methods for drug resistance testing with the aim of clarifying the direction for future resistance testing investigations.
2 Detection of Helicobacter pylori antibiotic resistance
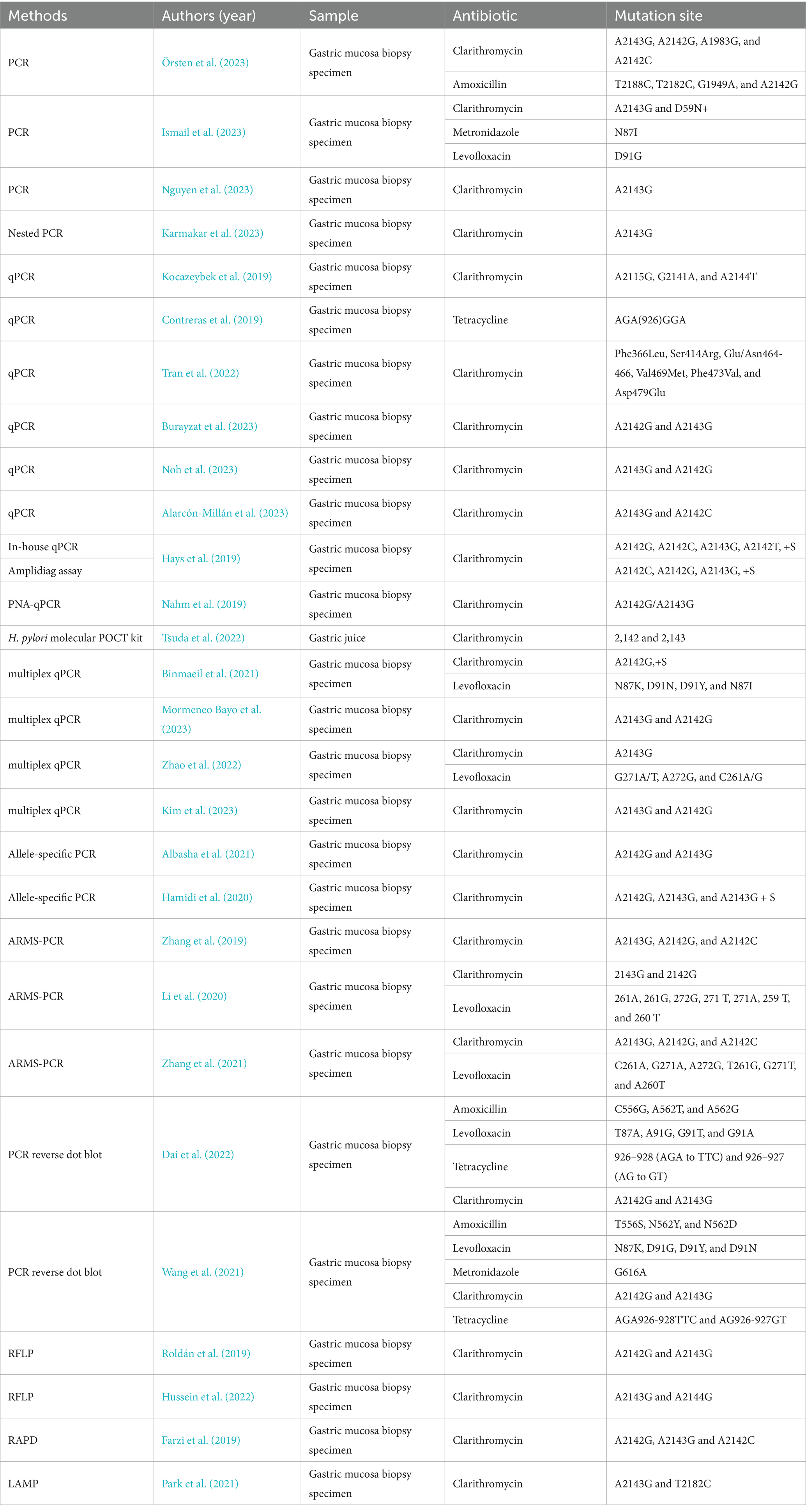
There are two different approaches for testing antimicrobial resistance in H. pylori: cultures with antimicrobial susceptibility testing (AST) and molecular testing. The AST are phenotypic identification methods used to isolate H. pylori from gastroscopic biopsy specimens and determine antibiotic susceptibility by agar dilution, disk diffusion, broth microdilution, or E-test. Molecular-based methods primarily examine mutations for specific antimicrobials, such as 23S rRNA and gyrA genes. These methods are mainly based on polymerase chain reaction (PCR), real-time PCR (qPCR), and others (Table 1). Next-generation sequencing (NGS), a technology capable of massively parallel sequencing, is a powerful tool for examining mutations in multiple regions. In addition to gastric mucosal biopsies specimens, studies on the use of non-invasive samples such as gastric juices and stool have obtaining promising results.
2.1 Culture-based method
The culture method can be divided into broth microdilution, disk diffusion, agar dilution, and E-test. The Clinical and Laboratory Standards Institute (CLSI) recommends agar dilution as the “gold standard” (Shakir et al., 2022). This method requires multiple fresh blood agar plates, which is time-consuming and laborious. Therefore, it is less commonly used for routine antibiotic sensitivity testing of clinical isolates. Disk diffusion and broth microdilution are the two most commonly used methods for clinical pathogens. E-test is a simple method that can be utilized to determine the Minimum Inhibitory Concentration (MIC) value of a single clinical isolate by using different concentration gradients on a single strip of paper. The studies showed good agreement between clarithromycin and levofloxacin when compared to broth microdilution and agar dilution methods (Tang et al., 2022). E-test and agar dilution methods compared with levofloxacin, metronidazole, tetracycline and clarithromycin showed good agreement and amoxicillin correlation was poor (Miftahussurur et al., 2020). The disk diffusion method compared with E-test showed good agreement for levofloxacin, clarithromycin and metronidazole and poor correlation for amoxicillin and tetracycline (Tang et al., 2020). E-test can be used as an alternative to agar dilution as a common clinical test for H. pylori drug sensitivity.
2.2 Molecular-based methods
The mechanism of H. pylori resistance to key antibiotics is well understood. Clinical resistance is caused by mutations in the nucleotide and amino acid sequences of H. pylori at the binding site to antibiotics. The molecular method can detect allelic mutation sequence sites, which is easier, faster, and more successful than the culture method. Additionally, it can detect heterogeneous resistance (Hays et al., 2019). Molecular methods can examine mutations for clarithromycin and levofloxacin. Clarithromycin resistance is mainly due to the point mutations especially at the A2142 and A2143 of 23S rRNA. The mutation sites of fluoroquinolones leading to resistance are in positions 86, 87, 88, and 91 of the genes gyrA and gyrB in DNA gyrase (Egli et al., 2020). However, molecular methods currently have limitations in detecting resistance to tetracycline (16S rRNA), rifampicin (rpoB), and metronidazole. In recent years, novel methods for detecting resistance to H. pylori have emerged.
Real-time quantitative fluorescence PCR (qPCR) allows for the direct amplification and analysis of gastric mucosal specimens, including formalin-fixed, paraffin-embedded samples, without the requirement for incubation and agar gel electrophoresis. qPCR is now more widely used in clinical practice because it enables easy and accurate interpretation of mutations directly through the signal of fluorescence melting peaks. Several modified qPCR methods and commercial kits have been developed.
A peptide nucleotide acid (PNA) probe-based qPCR test is a method with a PNA probe that allows for perfect hybridization, partial hybridization, and mismatch hybridization with template alleles. The genotypes of the alleles were obtained by analyzing the quenching temperature. PNA qPCR and dual priming oligonucleotide (DPO)-based multiplex PCR were used to analyze the A142G and A2143G mutation types, and the results were compared with those obtained using conventional PCR. The sensitivity and specificity of PNA qPCR were 100 and 100%, whereas the sensitivity and specificity of DPO-based PCR were 92.9 and 100%. DPO-based PCR involves two steps, PCR and agarose gel electrophoresis, whereas PNA qPCR only requires a single step of the real-time fluorescence PCR instrument, making it simpler and more accurate (Bénéjat et al., 2021).
The ready-to-use PCR microtiter plate is based on the principle of qPCR with optimized premix and emitter probe fluorophores to detect mutations at clarithromycin A2142G, A2143G, and A2142C sites. The time consumption, sensitivity, and specificity of this test strip need further exploration (Nahm et al., 2019).
To simultaneously detect resistance mutation sites in clarithromycin and levofloxacin, the researchers developed a multiplexed qPCR assay for the detection of mutations in clarithromycin 23S rRNA (A2142 and A2143) and levofloxacin gyrA (N87K and N87I, as well as D91N and D91Y). qPCR targeting the 23S rRNA showed a sensitivity of 100% and a specificity of 98.7%, while that targeting the gyrA gene demonstrated a sensitivity of 100% and a specificity of 99.8%. The method has high performance in detecting resistance to levofloxacin and is in good agreement with the E-test (Binmaeil et al., 2021).
Allplex™ is a quantitative multiplex PCR for the detection of clarithromycin A2142G, A2143G, and A2142C mutation sites. It is more sensitive (97.6% vs. 88.3%) and specific (96.0% vs. 82.0%) than DPO-PCR. Allplex™ showed higher accuracy in detecting the A2142G mutation site, which has a lower mutation rate (Kim et al., 2023).
The amount of H. pylori DNA template extracted from gastric mucosal specimens was small. To enhance the detection rate and quickly determine drug resistance directly from biopsy samples, a nested-allele specific primer-polymerase chain reaction (nested-ASP-PCR) was developed. Nested-ASP-PCR involves two steps. The first step includes amplifying 23S rRNA using DNA templates from biopsy samples. In the second step, the amplification product from the first step is used as a template and amplified using allele-specific primers for clarithromycin -sensitive or drug-resistant phenotypes. Results were in high concordance with Sanger sequencing. Nested-ASP-PCR only detects the A2143G mutation and not any other single nucleotide polymorphisms (SNPs) on the 23S rRNA. Multi-step PCR can be contaminated, leading to a high rate of false positives (Karmakar et al., 2023).
The amplification refractory mutation system combined with quantitative real-time PCR (ARMS-PCR) is based on designing specific ARMS primers that target mismatches at the 3′ end of the main chain, enhancing specificity and sensitivity. The ARMS-PCR method involves only requires 2 steps: nucleic acid extraction from gastric mucosal specimens and PCR amplification, which can be completed within 2 h. APMS-PCR has higher sensitivity than sequencing when the mutation rate is low. High concordance with the E-test method (97.1%, p > 0.05) was observed. ARMS-PCR requires optimization of the primers and fluorescent probes to target as many different sites as possible to prevent a decrease in the detection rate (Zhang et al., 2019). Multiplex ARMS-PCR in a single tube enables the detection of clarithromycin and levofloxacin resistance mutation sites, optimized for simplicity and speed. Compared with Sanger sequencing, multiplex ARMS-PCR is highly sensitive and specific, especially for detecting clarithromycin (Li et al., 2020).
PCR-RLFP can detect point mutations at common mutation sites in clarithromycin. Different restriction endonucleases can cut different point mutations into DNA fragments of different sizes. The fragments can be identified by separating and digesting them on an agar gel. PCR-RFLP does not require sequencing and is highly cost-effective. Detection time can be shortened compared with culture method, but it is necessary to isolate gastric mucosal H. pylori strains to extract nucleic acids, which may appear to reduce the chance of detection due to culture difficulties. Some less common mutations cannot be detected by RFLP (Hussein et al., 2022).
Allele-specific PCR is the use of multiple pairs of primers to amplify strain DNA templates, where nucleotides with different point mutations will bind to different primers, yielding PCR products of different lengths. Allele-specific PCR extracts DNA templates directly from the gastric mucosa without the need for isolation and culture. Alleles can detect mixed infections (Hamidi et al., 2020). Allele-specific PCR targeted only two common mutations. Allele-specific PCR was less sensitive compared to sequencing (allele PCR results: 9/53 A2142G, no A2143G, sequencing results: 1/25A2142G, 5/25A2143G) (Albasha et al., 2021).
Loop-mediated isothermal amplification (LAMP) amplifies DNA at 60–65°C without the need for a thermal cycler, and DNA amplification can be visualized by dye staining after 30 min. Park CG et al. detected two mutation sites, 23S rRNA A2143G and T2182C mutation sites. DNA was extracted from biopsy tissues. LAMP can detect H. pylori infection and drug resistance mutation sites, and the results are in high concordance with RUT and PCR sequencing results (Park et al., 2021).
Hybridization, washing, and color development of the amplified product, the mutation point can be changed to blue color by naked eye (Dai et al., 2022). The time-consuming and accuracy of the method still needs to be further explored.
Due to increased sensitivity and specificity, non-invasive assays have recently been developed for fecal and gastric fluids, etc. Smart Gene™'s Helicobacter pylori Molecular Evening Test (POCT) kit detects the H. pylori Clarithromycin 23S rRNA mutations at positions 2,142 and 2,143 in gastric fluids. Nucleic acid extraction, amplification and detection are automated. The test can be completed in approximately 1 h. all mutations at position 2,142 can be detected by the POCT kit, but only mutations at position 2,143 with a mutation rate of 15% or higher can be detected. The sensitivity is 91.7% and the specificity is 100% compared to the broth dilution method (Tsuda et al., 2022). Pichon M et al. investigated H. pylori clarithromycin resistance in fecal samples using qPCR with a sensitivity and specificity of 96.3 and 98.7%, respectively (Pichon et al., 2022). Meta-analysis by Gong, R. J. et al. showed that PCR-based analysis of fecal samples had high diagnostic accuracy for detecting clarithromycin resistance in patients with H. pylori infection. The combined sensitivity was 0.91 (95% CI: 0.83 ~ 0.95) and the combined specificity was 0.97 (95% CI: 0.62 ~ 1.00) (Gong et al., 2021).
Compared with the culture method, most commercial systematic molecular methods are only capable of detecting two antibiotics, clarithromycin and levofloxacin, and a single specific mutation site in the target gene. Therefore, both methods are necessary for the study of drug resistance, and the mechanism of antibiotic resistance needs to be further explored, with a view to discovering assays with higher clinical utility.
3 Tailored therapy guided by antibiotic susceptibility testing
Pre-medication resistance testing can prevent resistance-related treatment failure and the emergence of resistance caused by antibiotic misuse, but due to the high cost and workload of the H. pylori culture method, molecular-based AST is an emerging method for obtaining H. pylori resistance through mutations in genes associated with resistance, but due to genetic diversity, the clinical efficacy of molecular-based AST for individualized treatment needs further study. Moreover, the eradication rate is related to the host’s own factors such as PPI-associated CYP2C19 polymorphisms, and some authors have suggested that empirical treatment based on patients’ medication histories and local resistance rates can be an alternative to pharmacovigilance-guided individualized testing (Ishibashi et al., 2023) (Table 2).
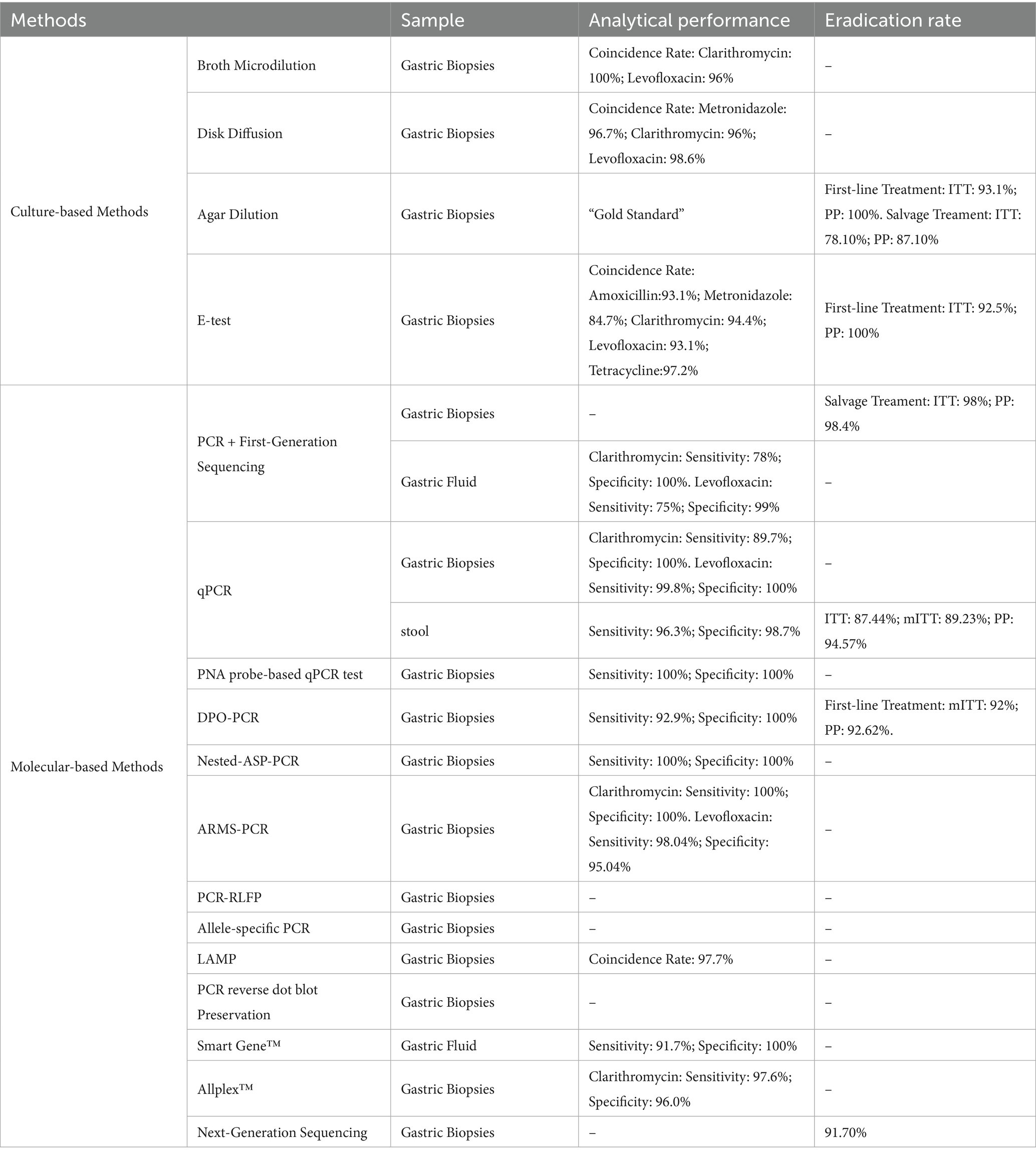
Table 2. Effectiveness of H. pylori drug resistance testing methods for detection and individualized treatment.
3.1 Personalized treatment guided by culture detection
In the first-line treatment, the individualized treatment group had a significantly higher eradication rate than empirical treatment and a lower incidence of adverse events (Perkovic et al., 2021). Meta-analysis by Francesco, V. et al. showed higher eradication rates with individualized treatment than with empirical treatment (89.7% vs. 77.6%). Individualized treatment prior to third-line therapy was significantly more effective than empirical treatment in achieving optimal eradication rates (>90%) (Francesco et al., 2022). For second-line and above-treated patients, AST and personal medication history-guided therapy (PMH) eradication rates were comparable (intention-to-treat analysis:78.10% vs. 74.29%, p = 0.42; protocol analysis:87.10% vs. 88.64%, p = 0.80), and the healthcare costs of pmh-guided therapy were lower (Ji et al., 2020).
3.2 Personalized treatment guided by molecular detection
In recent years, there have been numerous studies on individualized treatment for clarithromycin-related conditions using primer-based dual-start oligonucleotide primer (DPO) technology-based multiplex PCR. However, the results have been inconsistent, with variations in control group therapies and treatment durations. Most studies have administered a bismuth-containing quadruple therapy regimen without clarithromycin to patients with clarithromycin resistance in the individualized treatment group. While sensitive patients were given a three-drug regimen containing clarithromycin. In the empirical treatment group, different studies administered varying treatment regimens to patients (including three-drug regimens containing clarithromycin, three-drug regimens without clarithromycin, four-drug regimens containing clarithromycin, or four-drug regimens without clarithromycin), and the comparison of eradication rates across groups yielded inconsistent results. Compared with the clarithromycin-containing triple therapy, the eradication rate in the individualized therapy group was significantly higher than that in the empirical therapy group (Cho et al., 2019); compared with the clarithromycin-free triple therapy, even the eradication rate in the empirical therapy group was significantly higher than that in the individualized therapy group (Choi et al., 2021); compared with the clarithromycin-containing quadruple therapy, the eradication rate in the individualized therapy group was significantly higher than that in the empirical therapy group (Kim et al., 2022); when compared with the quadruple therapy regimen without clarithromycin but containing bismuth, there was no significant difference in eradication rates between the two groups (Cho et al., 2022). A recent study proposed a treatment regimen selection strategy: in empirical therapy, a combination of three antibiotics (clarithromycin, amoxicillin, and metronidazole) with a proton pump inhibitor (PPI), in individualized therapy, a regimen of clarithromycin, amoxicillin, and PPI for clarithromycin-sensitive patients, and a quadruple therapy regimen of oxytetracycline, metronidazole, PPI, and bismuth for clarithromycin-resistant patients. The results showed that the eradication rate in the individualized therapy group was significantly higher than that in the empirical therapy group, and adverse reactions were significantly lower than those in the empirical therapy group (Cho et al., 2025). All of the above detection methods are based on primer-double-start oligonucleotide primer (DPO technology) multiplex PCR to detect clarithromycin resistance by detecting mutations at the 23S RNA A2142G and A2143G sites. DPO-based molecular diagnosis may not only be used for the selection of sensitive antibiotics but also has guiding value in the application of bismuth-containing quadruple therapy.
Most published studies have used clarithromycin testing for first-line therapy. Choi et al. (2019) reported that in second-line therapy, the individualized treatment group (resistant group: PPI + tetracycline + metronidazole + bismuth compound, sensitive group: PPI + amoxicillin + clarithromycin) had a higher eradication rate than empirical therapy (PPI + tetracycline + metronidazole + bismuth compound) (90.1% vs. 96.0%, p < 0.001), and adverse reactions were significantly lower than those in empirical therapy. A randomized clinical trial in Taiwan evaluated the combined resistance of clarithromycin and levofloxacin in patients with refractory Hp infection. The results showed that individualized treatment based on resistance gene testing did not demonstrate a significant advantage over individualized treatment based on medication history (ITT analysis: 78% vs. 72.2%, p = 0.170; PP analysis: 78.4% vs. 74.4%, p = 0.346) (Liou et al., 2018). This cannot rule out the influence of factors such as the selection of treatment regimens and the effectiveness of conventional PCR testing, so further prospective studies are needed to determine the optimal salvage regimen for such patients.
New H. pylori resistance detection technologies developed in recent years, as mentioned earlier, have higher sensitivity and specificity than DPO PCR and conventional PCR, so it is necessary to explore the clinical value of these new technologies. Additionally, regarding the selection of resistant gene mutation sites, in a study by Cho et al. (2023) administering 500 mg of clarithromycin and extending the treatment duration to more than 7 days for patients negative for the A2142G and A2143G mutation sites did not improve the eradication rate. Although the common clarithromycin resistance mutation sites are A2142G and A2143G, we have reason to suspect that these two sites may not fully reflect Hp’s sensitivity to clarithromycin, and more sites are needed.
Due to the invasive nature of gastric mucosal samples and the difficulty in obtaining them, PCR techniques based on fecal samples have gained attention in recent years due to their non-invasive, convenient, rapid, and low-cost characteristics. Currently, PCR methods used for detecting fecal samples include nested PCR, real-time fluorescent quantitative PCR, and conventional PCR. A meta-analysis showed that molecular methods for detecting H. pylori resistance in feces have high sensitivity (0.97) and specificity (0.98) (Al Qady et al., 2025). In recent years, randomized controlled studies have been conducted to detect H. pylori resistance to clarithromycin and levofloxacin by identifying point mutations in the 23S rRNA and gyrA genes in fecal samples, with the aim of guiding clinical drug use based on resistance profiles. The results showed that compared to empirical treatment and individualized treatment regimens based on clarithromycin usage history, individualized treatment based on fecal resistance gene detection results achieved higher eradication rates (Yu et al., 2025). In addition to stool samples, gastric fluid is another non-invasive sample that can be obtained. Studies have shown that genetic testing of clarithromycin and levofloxacin from gastric aspirates has high accuracy rates of 97 and 95%, respectively (Vasapolli et al., 2025). However, there are currently no reported studies on the use of gastric fluid samples to guide clinical drug use.
3.3 Personalized treatment guided by NGS detection
Guo et al. (2022) detected H. pylori resistance to clarithromycin, levofloxacin, amoxicillin, and metronidazole in refractory patients using NGS and agar dilution, with a concordance rate higher than 90%, and eradication therapy was performed in this patient based on the results of the drug sensitivity test, and the eradication rates based on AST, PCR, and WGS were 90.9% (10/11), 83.3% (10/12), and 91.7% (11/12), which suggests that genotypic resistance-guided therapy may achieve satisfactory results with phenotypic resistance-guided therapy.
3.4 Comparison of genotype- and phenotype-guided individualized treatment
Chen et al. (2023) tested for resistance to clarithromycin and levofloxacin using PCR direct sequencing and agarose dilution methods in patients on first- and third-line treatment, respectively, and gave different eradication regimens based on the results. The results showed that molecular assay-guided therapy in first-line treatment had similar results to culture assay-guided therapy (eradication rate: 86% vs. 87%, p = 0.81, rate of difference: −0.7%), and molecular assay-guided therapy in third-line treatment was not inferior to culture assay-guided therapy results (eradication rate: 88% vs. 87%, p = 0.74, rate of difference: 1.3%).
4 Conclusion
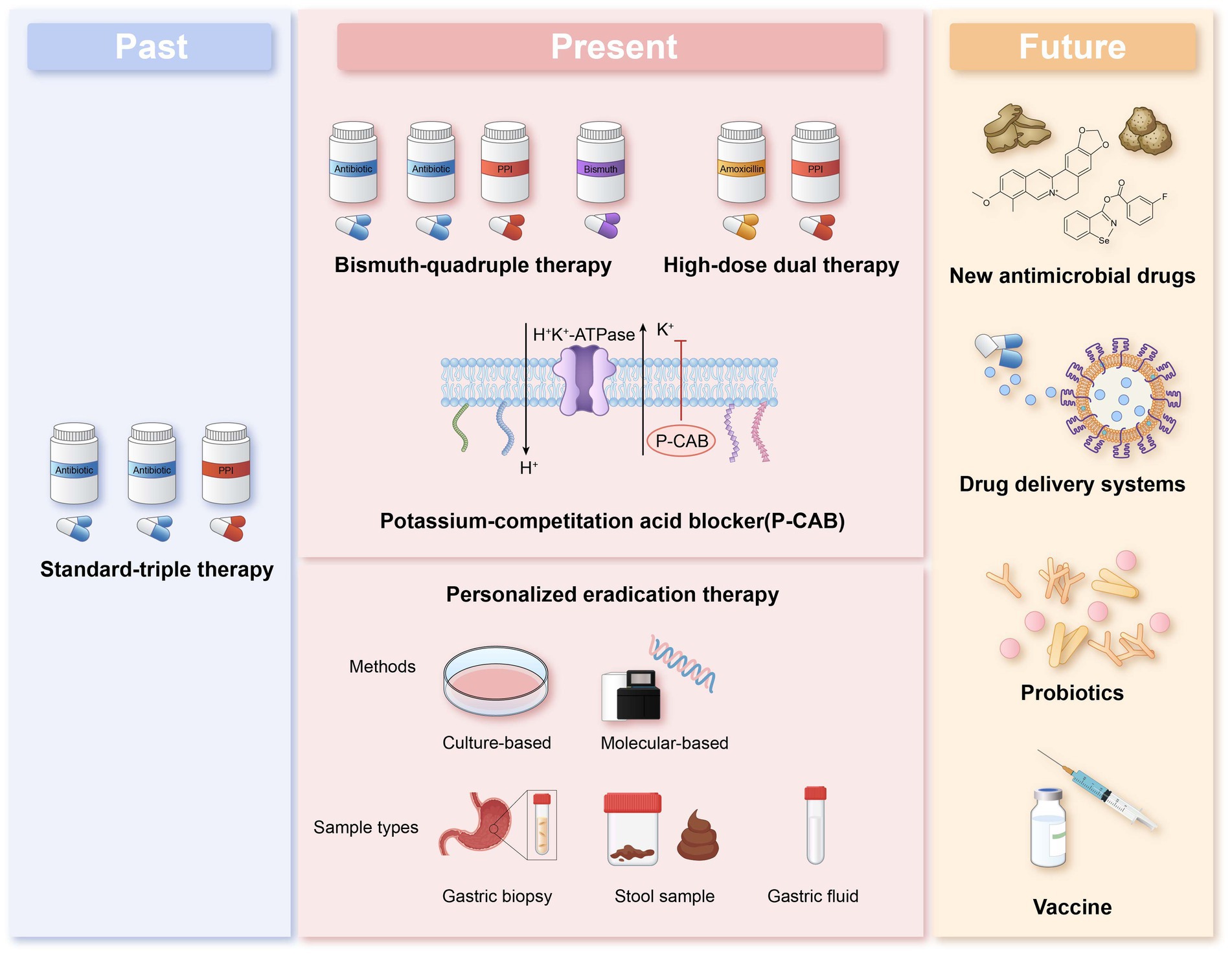
This article reviews the current status of Helicobacter pylori antibiotic resistance, commonly used resistance detection methods, and the efficacy of eradication therapy guided by different methods. It is worth noting that the current situation of antibiotic resistance in Helicobacter pylori remains severe. Non-invasive molecular detection methods show significant potential for future development. In the future, empirical treatment should be replaced by individualized treatment. However, there is currently limited clinical evidence to suggest that individualized treatment guided by antibiotic susceptibility testing is superior to treatment guided by medication history. In regions with poorer economic conditions, individualized treatment guided by personal medication history could be considered as an alternative to antibiotic resistance testing. Additionally, more accurate, rapid, and patient-compliant detection methods need to be developed. The introduction of nanomaterials in recent years has made Hp diagnosis and treatment more precise and efficient, and this may represent another significant opportunity in the exploration of antibiotic resistance detection methods (Figure 1).
Author contributions
LG: Writing – original draft, Writing – review & editing. ZM: Conceptualization, Writing – original draft. MH: Conceptualization, Writing – original draft. WT: Conceptualization, Writing – original draft. XC: Conceptualization, Writing – original draft. XK: Conceptualization, Writing – original draft. DZ: Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This study was supported by the Natural Science Foundation of Gansu Province, China (no. 24ZDFA006), the Natural Science Foundation of Gansu Province, China (no. 23JRRA1501), and the Cuiying Science and Technology Innovation, Outstanding Doctorate Fund (no. CY2024-YB-B03).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Alarcón-Millán, J., Bonilla-Delgado, J., Fernández-Tilapa, G., Nieto-Velázquez, N. G., Sierra-Martínez, M., Alvarado-Castro, V. M., et al. (2023). Helicobacter pylori Virulence Factors and Clarithromycin Resistance-Associated Mutations in Mexican Patients. Pathogens 12.
Al Qady, A., Aldhaleei, W., Salih, M., Ali, M., Menakuru, S., Nayar, K. D., et al. (2025). Accuracy of fecal polymerase chain reaction testing in clarithromycin-resistant Helicobacter pylori: a systematic review and Meta-analysis. Clin. Transl. Gastroenterol. 16:e00792. doi: 10.14309/ctg.0000000000000792
Albasha, A. M., Elnosh, M. M., Osman, E. H., Zeinalabdin, D. M., Fadl, A. A. M., Ali, M. A., et al. (2021). Helicobacter pylori 23S rRNA gene A2142G, A2143G, T2182C, and C2195T mutations associated with clarithromycin resistance detected in Sudanese patients. BMC Microbiol. 21:38. doi: 10.1186/s12866-021-02096-3
Bénéjat, L., Ducournau, A., Domingues-Martins, C., Lecoeur, M., Blosse, A., Mégraud, F., et al. (2021). Adaptation of an in-house PCR for the detection of Helicobacter pylori and the mutations associated with macrolide resistance into ready-to-use PCR microwell strips. Helicobacter 26:e12855. doi: 10.1111/hel.12855
Binmaeil, H., Hanafiah, A., Mohamed Rose, I., and Raja Ali, R. A. (2021). Development and validation of multiplex quantitative PCR assay for detection of Helicobacter pylori and mutations conferring resistance to clarithromycin and levofloxacin in gastric biopsy. Infect. Drug Resist. 14, 4129–4145. doi: 10.2147/IDR.S325056
Burayzat, S., Al-Tamimi, M., Barqawi, M., Massadi, M. S., Abu-Raideh, J., Albalawi, H., et al. (2023). Antimicrobial Resistance Molecular Mechanisms of Helicobacter pylori in Jordanian Children: A Cross-Sectional Observational Study. Antibiotics (Basel) 12.
Chen, M. J., Chen, P. Y., Fang, Y. J., Bair, M. J., Chen, C. C., Chen, C. C., et al. (2023). Molecular testing-guided therapy versus susceptibility testing-guided therapy in first-line and third-line Helicobacter pylori eradication: two multicentre, open-label, randomised controlled, non-inferiority trials. Lancet Gastroenterol. Hepatol. 8, 623–634. doi: 10.1016/S2468-1253(23)00097-3
Cho, J. H., Jeon, S. R., Kim, H. G., Jin, S. Y., and Park, S. (2019). Cost-effectiveness of a tailored Helicobacter pylori eradication strategy based on the presence of a 23S ribosomal RNA point mutation that causes clarithromycin resistance in Korean patients. J. Gastroenterol. Hepatol. 34, 700–706. doi: 10.1111/jgh.14383
Cho, J. H., Jin, S. Y., and Park, S. (2022). Comparison of tailored Helicobacter pylori eradication versus modified bismuth quadruple therapy in Korea: a randomized controlled trial. Expert Rev. Anti-Infect. Ther. 20, 923–929. doi: 10.1080/14787210.2022.2017280
Cho, Y. S., Kim, S. M., Kang, S. H., Moon, H. S., Sung, J. K., Bang, K. B., et al. (2025). Comparison of therapeutic outcomes between concomitant therapy and tailored therapy for Helicobacter pylori: a multicenter, prospective, and randomized study. Helicobacter 30:e70040. doi: 10.1111/hel.70040
Cho, S. H., Park, M. S., Park, S. Y., Kim, D. H., You, H. S., and Kim, H. S. (2023). Effectiveness of 7-day triple therapy with half-dose clarithromycin for the eradication of Helicobacter pylori without the A2143G and A2142G point mutations of the 23S rRNA gene in a high clarithromycin resistance area. Front. Med. (Lausanne) 10:1150396. doi: 10.3389/fmed.2023.1150396
Choi, Y. I., Chung, J. W., Kim, K. O., Kwon, K. A., Kim, Y. J., Kim, J. H., et al. (2021). Tailored eradication strategy vs concomitant therapy for Helicobacter pylori eradication treatment in Korean patients. World J. Gastroenterol. 27, 5247–5258. doi: 10.3748/wjg.v27.i31.5247
Choi, Y. I., Chung, J. W., Park, D. K., Kim, K. O., Kwon, K. A., Kim, Y. J., et al. (2019). Tailored eradication vs empirical bismuth-containing quadruple therapy for first-line Helicobacter pylori eradication: a comparative, open trial. World J. Gastroenterol. 25, 6743–6751. doi: 10.3748/wjg.v25.i46.6743
Contreras, M., Benejat, L., Mujica, H., Peña, J., García-Amado, M. A., Michelangeli, F., et al. (2019). Real-time PCR detection of a 16S rRNA single mutation of Helicobacter pylori isolates associated with reduced susceptibility and resistance to tetracycline in the gastroesophageal mucosa of individual hosts. J Med Microbiol 68, 1287–1291.
Dai, J., Zhao, J., Mao, L., Hu, Y., and Lv, B. (2022). Study on the value of antibiotic-resistant gene detection in Helicobacter pylori in China. Exp. Ther. Med. 23:228. doi: 10.3892/etm.2022.11153
Egli, K., Wagner, K., Keller, P. M., Risch, L., Risch, M., and Bodmer, T. (2020). Comparison of the diagnostic performance of qPCR, sanger sequencing, and whole-genome sequencing in determining clarithromycin and levofloxacin resistance in Helicobacter pylori. Front. Cell. Infect. Microbiol. 10:596371. doi: 10.3389/fcimb.2020.596371
Farzi, N., Behzad, C., Hasani, Z., Alebouyeh, M., Zojaji, H., and Zali, M. R. (2019). Characterization of clarithromycin heteroresistance among Helicobacter pylori strains isolated from the antrum and corpus of the stomach. Folia Microbiol (Praha) 64, 143–151.
Francesco, V., Zullo, A., Manta, R., Satriano, A., Fiorini, G., Pavoni, M., et al. (2022). Culture-based antibiotic susceptibility testing for Helicobacter pylori infection: a systematic review. Ann. Gastroenterol. 35, 127–134. doi: 10.20524/aog.2022.0689
Gong, R. J., Xu, C. X., Li, H., and Liu, X. M. (2021). Polymerase chain reaction-based tests for detecting Helicobacter pylori clarithromycin resistance in stool samples: a meta-analysis. World J. Clin. Cases 9, 133–147. doi: 10.12998/wjcc.v9.i1.133
Guo, Z., Tian, S., Wang, W., Zhang, Y., Li, J., and Lin, R. (2022). Correlation analysis among genotype resistance, phenotype resistance, and eradication effect after resistance-guided quadruple therapies in refractory Helicobacter pylori infections. Front. Microbiol. 13:861626. doi: 10.3389/fmicb.2022.861626
Hamidi, S., Badmasti, F., Sadeghpour Heravi, F., Safapoor, M. H., Mohammad Ali Tabrizi, A., Ghorbani, M., et al. (2020). Antibiotic resistance and clonal relatedness of Helicobacter pylori strains isolated from stomach biopsy specimens in northeast of Iran. Helicobacter 25:e12684. doi: 10.1111/hel.12684
Hays, C., Delerue, T., Lamarque, D., Burucoa, C., Collobert, G., Billöet, A., et al. (2019). Molecular diagnosis of Helicobacter pylori infection in gastric biopsies: evaluation of the Amplidiag(®) H. pylori + ClariR assay. Helicobacter 24:e12560. doi: 10.1111/hel.12560
Hussein, R. A., Al-Ouqaili, M. T. S., and Majeed, Y. H. (2022). Detection of clarithromycin resistance and 23SrRNA point mutations in clinical isolates of Helicobacter pylori isolates: phenotypic and molecular methods. Saudi J Biol Sci 29, 513–520. doi: 10.1016/j.sjbs.2021.09.024
Ishibashi, F., Suzuki, S., Nagai, M., Mochida, K., and Morishita, T. (2023). Optimizing Helicobacter pylori treatment: an updated review of empirical and susceptibility test-based treatments. Gut Liver 17, 684–697. doi: 10.5009/gnl220429
Ismail, M., Majaliwa, N. D., Vale, F. F., Cumbana, R., Sumbana, J. J., Muchongo, A., et al. (2023). Molecular detection of Helicobacter pylori and its genotypic antimicrobial resistance patterns in dyspeptic Mozambican patients. Helicobacter 28:e13000.
Ji, C. R., Liu, J., Li, Y. Y., Qiao, C., Qu, J. Y., Hu, J. N., et al. (2020). Susceptibility-guided quadruple therapy is not superior to medication history-guided therapy for the rescue treatment of Helicobacter pylori infection: a randomized controlled trial. J. Dig. Dis. 21, 549–557. doi: 10.1111/1751-2980.12934
Karmakar, B. C., Paul, S., Basak, S., Ghosh, M., Mukherjee, P., Das, R., et al. (2023). Development and evaluation of a simple PCR assay and nested PCR for rapid detection of clarithromycin-resistant Helicobacter pylori from culture and directly from the biopsy samples in India. Gut Pathog 15:7. doi: 10.1186/s13099-023-00530-7
Kim, S. J., Jee, S. R., Park, M. I., Jung, K., Kim, G. H., Lee, M. W., et al. (2022). A randomized controlled trial to compare Helicobacter pylori eradication rates between the empirical concomitant therapy and tailored therapy based on 23S rRNA point mutations. Medicine (Baltimore) 101:e30069. doi: 10.1097/MD.0000000000030069
Kim, I., Maeng, L. S., Kim, J. S., Kim, B. W., Cheung, D. Y., Kim, J. I., et al. (2023). Quantitative multiplex real-time polymerase chain reaction assay for the detection of Helicobacter pylori and clarithromycin resistance. BMC Microbiol. 23:155. doi: 10.1186/s12866-023-02868-z
Kocazeybek, B., Sakli, M. K., Yuksel, P., Demirci, M., Caliskan, R., Sarp, T. Z., et al. (2019). Comparison of new and classical point mutations associated with clarithromycin resistance in Helicobacter pylori strains isolated from dyspeptic patients and their effects on phenotypic clarithromycin resistance. J Med Microbiol 68, 566–573.
Li, Y., Lv, T., He, C., Wang, H., Cram, D. S., Zhou, L., et al. (2020). Evaluation of multiplex ARMS-PCR for detection of Helicobacter pylori mutations conferring resistance to clarithromycin and levofloxacin. Gut Pathog 12:35. doi: 10.1186/s13099-020-00373-6
Liou, J. M., Chen, P. Y., Luo, J. C., Lee, J. Y., Chen, C. C., Fang, Y. J., et al. (2018). Efficacies of genotypic resistance-guided vs empirical therapy for refractory Helicobacter pylori infection. Gastroenterology 155, 1109–1119. doi: 10.1053/j.gastro.2018.06.047
Miftahussurur, M., Fauzia, K. A., Nusi, I. A., Setiawan, P. B., Syam, A. F., Waskito, L. A., et al. (2020). E-test versus agar dilution for antibiotic susceptibility testing of Helicobacter pylori: a comparison study. BMC. Res. Notes 13:22. doi: 10.1186/s13104-019-4877-9
Mormeneo Bayo, S., Bellés Bellés, A., Vázquez Gómez, D., Planella de Rubinat, M., Bayas Pastor, D. C., Morales Portillo, A., et al. (2023). Antibiotic Susceptibility and Clarithromycin Resistance Determinants in Helicobacter pylori in the Northeast of Spain: A One-Year Prospective Study. Antibiotics (Basel) 12.
Nahm, J. H., Kim, W. K., Kwon, Y., and Kim, H. (2019). Detection of Helicobacter pylori with clarithromycin resistance-associated mutations using peptide nucleic acid probe-based melting point analysis. Helicobacter 24:e12634. doi: 10.1111/hel.12634
Nguyen, T. C., Le, G. K. N., Pham, D. T. H., Pham, B. V., Nguyen, L. T. H., Che, T. H., et al. (2023). Antibiotic resistance and heteroresistance in Helicobacter pylori isolates from symptomatic Vietnamese children: A prospective multicenter study. Helicobacter 28:e13009.
Noh, J. H., Ahn, J. Y., Choi, J., Park, Y. S., Na, H. K., Lee, J. H., et al. (2023). Real-Time Polymerase Chain Reaction for the Detection of Helicobacter pylori and Clarithromycin Resistance. Gut Liver 17, 375–381.
Örsten, S., Yılmaz, E., and Akyön, Y. (2023). Molecular Characterization of Clarithromycin Resistance in Helicobacter pylori Strains. Turk J Gastroenterol 34, 427–432.
Park, C. G., Kim, S., Jeon, H. S., and Han, S. (2021). Validation of loop-mediated isothermal amplification to detect Helicobacter pylori and 23S rRNA mutations: a prospective, observational clinical cohort study. J. Clin. Lab. Anal. 35:e23563. doi: 10.1002/jcla.23563
Perkovic, N., Mestrovic, A., Bozic, J., Ivelja, M. P., Vukovic, J., Kardum, G., et al. (2021). Randomized clinical trial comparing concomitant and tailored therapy for eradication of Helicobacter pylori infection. J Pers Med 11:534. doi: 10.3390/jpm11060534
Pichon, M., Freche, B., and Burucoa, C. (2022). New strategy for the detection and treatment of Helicobacter pylori infections in primary care guided by a non-invasive PCR in stool: protocol of the French HepyPrim study. J. Clin. Med. 11:1151. doi: 10.3390/jcm11051151
Roldán, I. J., Castaño, R., and Navas, M. C. (2019). Mutations in the Helicobacter pylori 23S rRNA gene associated with clarithromycin resistance in patients at an endoscopy unit in Medellín, Colombia. Biomedica 39, 117–129.
Shakir, S. M., Otiso, J., Keller, G., Heule, H. V., Osborn, L. J., Cole, N., et al. (2022). Multicenter evaluation of a gradient diffusion method for antimicrobial susceptibility testing of Helicobacter pylori. Microbiol. Spectr. 10:e0211121. doi: 10.1128/spectrum.02111-21
Tang, X., Shen, Y., Hu, R., Yang, T., Benghezal, M., Li, H., et al. (2020). Re-assessment of the disk diffusion technique for routine antimicrobial susceptibility testing for Helicobacter pylori. Helicobacter 25:e12703. doi: 10.1111/hel.12703
Tang, X., Shen, Y., Song, X., Benghezal, M., Marshall, B. J., Tang, H., et al. (2022). Reassessment of the broth microdilution method for susceptibility testing of Helicobacter pylori. J. Infect. Dis. 226, S486–s492. doi: 10.1093/infdis/jiac389
Tran, T. T., Nguyen, A. T., Quach, D. T., Pham, D. T., Cao, N. M., Nguyen, U. T., et al. (2022). Emergence of amoxicillin resistance and identification of novel mutations of the pbp1A gene in Helicobacter pylori in Vietnam. BMC Microbiol 22:41.
Tsuda, M., Watanabe, Y., Oikawa, R., Watanabe, R., Higashino, M., Kubo, K., et al. (2022). Clinical evaluation of a novel molecular diagnosis kit for detecting Helicobacter pylori and clarithromycin-resistant using intragastric fluid. Helicobacter 27:e12933. doi: 10.1111/hel.12933
Vasapolli, R., Ailloud, F., Spießberger, B., Malfertheiner, P., Suerbaum, S., and Schulz, C. (2025). Real-time assessment of H. pylori infection to guide molecular antibiotic resistance testing: a combined endoscopy-gastric juice analysis approach. Aliment. Pharmacol. Ther. 61, 465–471. doi: 10.1111/apt.18378
Wang, L., Zhang, J., Hu, M., and Pang, X. (2021). Comparison of Drug Resistance of Helicobacter pylori Between Children and Adults in Jilin, China. Turk J Gastroenterol 32, 1012–1018.
Yu, Z., Liu, X., Qiao, J., Shen, W., Mao, X., Lou, G., et al. (2025). Is tailored bismuth quadruple therapies (with clarithromycin or Furazolidone) based on fecal molecular susceptibility testing in first-line Helicobacter pylori eradication treatment more effective? A three-arm, multicenter randomized clinical trial. Helicobacter 30:e70018. doi: 10.1111/hel.70018
Zhang, C., Cao, M., Lv, T., Wang, H., Liu, X., Xie, Y., et al. (2021). Molecular testing for H. pylori clarithromycin and quinolone resistance: a prospective Chinese study. Eur J Clin Microbiol Infect Dis 40, 1599–1608.
Zhang, X. Y., Shen, W. X., Chen, C. F., Sheng, H. H., Cheng, H., Li, J., et al. (2019). Detection of the clarithromycin resistance of Helicobacter pylori in gastric mucosa by the amplification refractory mutation system combined with quantitative real-time PCR. Cancer Med. 8, 1633–1640. doi: 10.1002/cam4.1986
Zhao, Y., Li, Y., Luan, Z., Ma, C., Yang, L., Zhang, W., et al. (2022). Establishment of a TaqMan-MGB probe multiplex real-time PCR system for one-step levofloxacin and clarithromycin resistant Helicobacter pylori detection. J Microbiol Methods 192:106393.
Keywords: Helicobacter pylori, antibiotics resistance, eradication treatment, culture-based methods, molecular-based methods
Citation: Gou L, Ma Z, Han M, Tian W, Chen X, Kang X and Zhang D (2025) Antimicrobial drug susceptibility testing for the management of Helicobacter pylori infection in personalized eradication therapy. Front. Microbiol. 16:1626930. doi: 10.3389/fmicb.2025.1626930
Edited by:
Santi M. Mandal, Indian Institute of Technology Kharagpur, IndiaReviewed by:
Pankaj Chaudhary, Indian Institute of Technology Roorkee, IndiaNusrat Tanni, Dhaka Medical College and Hospital, Bangladesh
Copyright © 2025 Gou, Ma, Han, Tian, Chen, Kang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dekui Zhang, emhhbmdkazg2MTZAMTI2LmNvbQ==
 Lingzhu Gou
Lingzhu Gou Zenghui Ma
Zenghui Ma Mengyu Han4
Mengyu Han4 Xiaojuan Kang
Xiaojuan Kang Dekui Zhang
Dekui Zhang