- 1Allied Health Sciences Department, College of Health and Sport Sciences, University of Bahrain, Manama, Bahrain
- 2Center of Environmental and Biological Studies, Arabian Gulf University, Manama, Bahrain
- 3Biodiversity Research Center, Academia Sinica, Taipei, Taiwan
- 4Department of Pharmaceutical Chemistry, Faculty of Pharmacy, Alexandria University, Alexandria, Egypt
Steroid estrogens, including the naturally occurring hormones estrone (E1), estradiol (E2), and estriol (E3), as well as the synthetic estrogen ethinylestradiol (EE2), play essential physiological roles in the regulation of the reproductive systems and development of secondary sex characteristics in humans and animals. Environmental pollution with steroid estrogens is gaining rising concerns worldwide due to their endocrine-disrupting and carcinogenic properties, which can harm humans and aquatic organisms. Hence, efficient removal of these compounds, particularly from wastewater, is deemed key to prevent environmental pollution with estrogens. Although several physicochemical treatments contribute to estrogen elimination from wastewater treatment plants (WWTPs), biological treatment via microbial biodegradation remains the most efficient estrogen removal approach. Several estrogen-degrading/transforming bacteria were isolated mainly from activated sludge samples collected from WWTPs. Moreover, biochemical, and molecular aspects for estrogen degradation pathways were revealed recently for estrone and estradiol. On the contrary, less knowledge is currently available for E3 and EE2 biodegradation pathways. Despite high structural similarity among steroid estrogens, they can be degraded via a diversity of biodegradation and biotransformation pathways. Nonetheless, these pathways exhibit common as well as unique biochemical and molecular features. Moreover, steroid estrogens are interconvertible, which can affect their environmental concentrations, and hence, their persistence/biodegradability. In this review, we present and discuss the various steroid estrogen biodegradation and biotransformation pathways, with a focus on the biochemical aspects. Furthermore, we highlight some of the known abiotic estrogen reactions and the recent discoveries on microbial estrogenesis and envisage how they can affect estrogen susceptibility to microbial degradation.
1 Introduction
Environmental pollution by hazardous chemicals is one of the most critical global challenges. Some of these chemical pollutants can interfere with the natural functions of hormones in humans and animals, and hence are termed endocrine-disrupting chemicals (EDCs) (Zhang et al., 2015; Chiang et al., 2020; López-Velázquez et al., 2023; Lerdsuwanrut et al., 2025). The latter were defined by the World Health Organization (WHO) as “an exogenous substance or a mixture that alters function(s) of the endocrine system and consequently causes adverse health effects in an intact organism, or its progeny, or (sub) populations” (Bergman et al., 2012; Gonsioroski et al., 2020).
In the same context, scarcity of freshwater resources has become a global challenge that threatens sustainable development in many countries. This issue will escalate in the future due to the increasing global human population and urbanization and will be compounded by the negative impacts of climate change (Stikker, 1998; Stefanakis, 2022). Hence, reuse and recycling of wastewater from municipal, industrial, and agricultural sectors are being adopted as counter measures to increase the water resource base. However, wastewater streams require efficient treatment and remediation processes to remove hazardous pollutants, like EDCs, and ensure safe reuse (Stefanakis, 2018).
Estrogens are steroid hormones produced by humans and animals where they play key physiological roles in the regulation of the reproductive systems and development of secondary sex characteristics (Bilal et al., 2021). Although estrogens are necessary for humans and other vertebrates, chronic exposure to even trace amounts of estrogens (sub-nanomolar levels) can disrupt the endocrine system and sexual development (Kolodziej et al., 2003; Lee et al., 2006; Lambert et al., 2015; Regnault et al., 2018; Predieri et al., 2022). Hence, estrogens are considered EDCs and were classified by the WHO as group-1 carcinogens, and some catechol metabolites of estrogens were reported as carcinogens (Liehr et al., 1986; Fernandez et al., 2006; http://monographs.iarc.fr/ENG/Classification/latest_classif.php). These hazardous properties raised substantial concerns on environmental pollution with estrogens.
Steroid estrogens are represented by three naturally occurring hormones, including estrone (E1), estradiol (E2), and estriol (E3), as well as the synthetic estrogen ethinylestradiol (EE2) (Pratush et al., 2020a). Estrogens are found in aquatic environments and raise significant concerns on water and wastewater quality (Roudbari and Rezakazemi, 2018). They enter the environment via different routes, but mainly through discharge of urine and feces reaching wastewater treatment plants (WWTPs) (Adeel et al., 2017). Estrogens are commonly present in wastewater in trace amounts, ranging from a few ng/L to several μg/L, yet they are associated with adverse ecological impact and influence marine organisms in the impacted area, accumulating in the ecosystem and causing acute and chronic toxicity to organisms, as well as loss of habitat and biodiversity (Adeel et al., 2017; Chiang et al., 2020).
Estrogens accumulate in the tissues of fish and other marine organisms (Ros et al., 2016) and are associated with increased liver size, increased plasma glucose concentration, smaller gonad size, and less mature gametes. This may adversely disrupt the reproductive potential of marine organisms, reducing their numbers and thus causing decline in a sustainable food source for the exposed populations (Elliott et al., 2017). Likewise, the accumulation of estrogens seriously threatens humans and the food chain. Eventually, they build up in the human body and trigger health concerns, such as breast cancer in women and prostate cancer in men (Adeel et al., 2017). Consequently, effective elimination of estrogens and other EDCs from the environment has become an active research area.
Efficient removal of estrogens during wastewater treatment is essential to avoid environmental pollution. Physical removal methods such as reverse osmosis and adsorption on activated carbon do not eliminate the pollutants, albeit only transfer them from one phase to another. Moreover, chemical oxidation is not economically viable, and chemicals are reactive, and the process is associated with by-products with unknown effects (Koh et al., 2008; Bolong et al., 2009; Liu et al., 2009a). While several techniques, such as electrochemical oxidation, adsorption, photocatalysis, and membrane filtration, have been proposed for the removal of EDCs from the aquatic environment, yet they can be associated with high cost, operational complexity, and environmental impact (Ben et al., 2017).
On the contrary, microbial biodegradation is a green and economic process which plays a key role in estrogen removal from WWTPs (de Lorenzo, 2017). Several estrogen-degrading bacteria have been isolated mainly from activated sludge collected from WWTPs (Chiang et al., 2020). Despite the high structural similarity among the steroid estrogens, the reported biotransformation reactions and biodegradation pathways are very versatile. In this review, we first give a brief account on estrogens, their characteristics, how they reach and impact the environment and public health. Then, we explore and discuss the status quo of what is known about the microbial biodegradation of estrogens in terms of microbiology and diversity of the biodegradation pathways, highlighting knowledge gaps and proposing future research directions. We included early as well as recent studies reporting proposed biotransformation reactions and biodegradation pathways with more focus on bacteria. While we focus on the degradation and transformation metabolites and the flow of the pathways, we refer to involved key enzymes and genes whenever relevant. For more information on the genes and enzymes of estrogen degradation we refer the readers to several reviews (Olivera and Luengo, 2019; Chiang et al., 2020; Pratush et al., 2020a).
2 Estrogens: properties and physiological roles
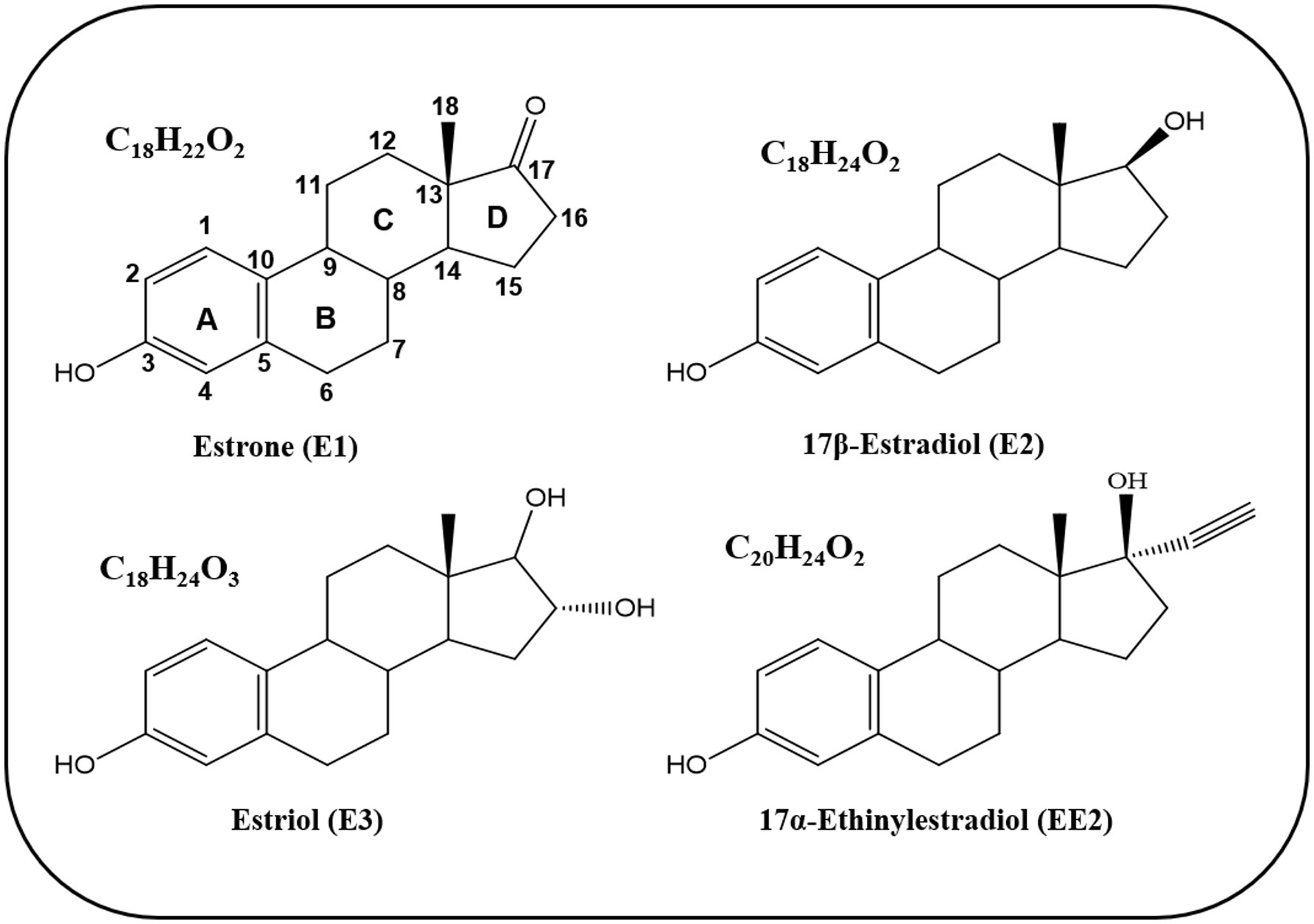
Estrogens are steroid hormones that function as signaling molecules, including natural estrogens such as estrone (E1), 17β-estradiol (E2), estriol (E3), as well as the synthetic estrogen 17α-ethinylestradiol (EE2) (Bilal et al., 2021). The core structure of estrogens comprises 18 carbon atoms and shares the same core molecular structure of sterane, tetracyclic framework, consisting of four fused rings (A, B, C, and D) (Figure 1) (Bilal et al., 2021). The A-ring is phenolic, B- and C-rings are cyclohexane, whereas the D-ring is a cyclopentane. The structural differences among estrogens lie in the configuration of the D-ring, specifically in carbons at positions 16 and 17.
The precursor of natural estrogens is dietary cholesterol (C27H46O), particularly the low-density lipoprotein (LDL) (Olivera and Luengo, 2019). In steroidogenesis, cholesterol is converted into sterane, a C18 steroid derivative, and the leading site of estrogen biosynthesis is the granulosa cells of the ovaries (Vazakidou et al., 2024). Yet, the translocation of cholesterol occurs in the inner membrane of the mitochondria and is regulated by a steroidogenic acute regulatory protein (StAR) (Lennartz et al., 2024). In the mitochondrial membrane, cholesterol is transformed to pregnenolone by the enzyme P450scc, which is further converted to androstenedione by the enzymes CYP17A1 and 3β-hydroxysteroid dehydrogenase (3β-HSD) (Cui et al., 2013). Androstenedione disperses to the granulosa cells across the basal lamina, where it produces E1 and E2 by the enzymes CYP19A1 (aromatase) and 17β-hydroxysteroid dehydrogenase (17β-HSD), respectively (Cui et al., 2013).
In addition to the well-known aromatase-dependent biosynthesis of C18 estrogens from C19 androgens, a very recent study showed that androgens can be transformed to estrogens in a Peptococcaceae bacterium isolated from the gut of the great, blue-spotted mudskipper (Boleophthalmus pectinirostris) (Wang et al., 2025). This bacterium, Phosphitispora sp. strain TUW77, has a novel testosterone fermentation pathway which can transform testosterone (androgen) into estrogens and androstanediol under anaerobic conditions. Upon growth on testosterone, the strain TUW77 strongly expresses a polycistronic gene cluster abeABC (anaerobic bacterial estrogenesis), encoding components of a classic cobalamin-dependent methyltransferase system, which enables the removal of the C19 methyl group of testosterone. The strain TUW77 then utilizes the removed methyl group as a carbon and electron source by oxidation to CO2 via the oxidative Wood-Ljungdahl pathway. Accordingly, Wang et al. (2025) suggested that the Wood-Ljungdahl pathway may have had a role in the emergence of estrogens in the early biosphere. These findings suggest that bacterial estrogenesis may have existed in nature prior to the O2-dependent production in vertebrates, which challenges the legacy that estrogen biosynthesis is restricted to aromatase-containing vertebrates. They also open new venues for research addressing the occurrence of this unusual phenotype within the microbial world, particularly in obligately anaerobic bacteria.
Natural estrogens are essential sex hormones that play important physiological roles in the development of the female reproductive system and secondary sexual characteristics (Fuentes and Silveyra, 2019) in addition to male external normal genitalia (Baskin et al., 2022). It is known that estrogens have roles in regulating immunity through their effect on the immune cells to prevent atherosclerosis and endothelial dysfunction by maintaining the endothelial cells and promoting angiogenesis (Trenti et al., 2018). The synthetic estrogen EE2 is commonly used in contraceptives. It is one of the most common medications for humans and animals and has raised various health issues and ecological concerns (Laurenson et al., 2014). It is a highly potent oral contraceptive and ranked among the top 15 US active ingredients in terms of its daily use (Kostich and Lazorchak, 2008).
3 Environmental pollution with estrogens
Humans and animals excrete steroid hormones in different amounts depending on age, state of health, diet, or pregnancy. Natural and synthetic estrogens are considered among the most significant pollutants in different environments, and the main sources of estrogens pollution are WWTPs, hospitals, pharmaceutical industry, agricultural manure, and effluents from livestock feedlots (Adeel et al., 2017; Chiang et al., 2020; López-Velázquez et al., 2021; Torres et al., 2021) (Table 1). Natural estrogens are free or conjugated with sulfate and/or glucuronide groups. Free estrogens are more hydrophobic with lower solubility than conjugated estrogens, even though they pose an estrogenic potential (Yu et al., 2019). After being metabolized in the body via conjugation with sulfate and hydroxylation reactions, the inactive forms of estrogens are excreted into the environment in urine and feces, which end up in WWTPs (Table 2).
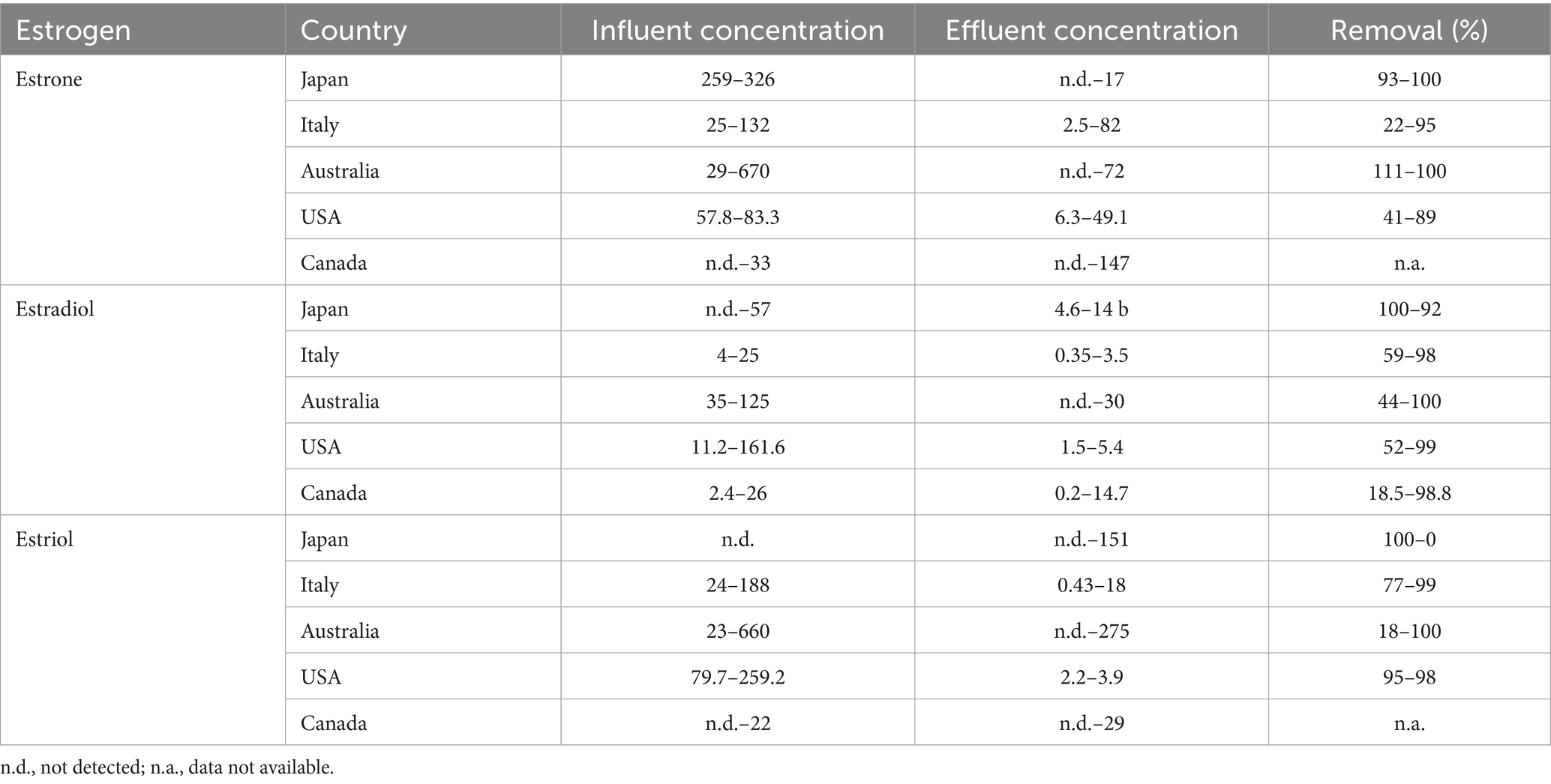
Table 1. Steroid estrogen concentrations (ng/L) in influents and effluents of WWTPs and their removal percentage (Liu et al., 2009b, 2009c; Pratush et al., 2020a).
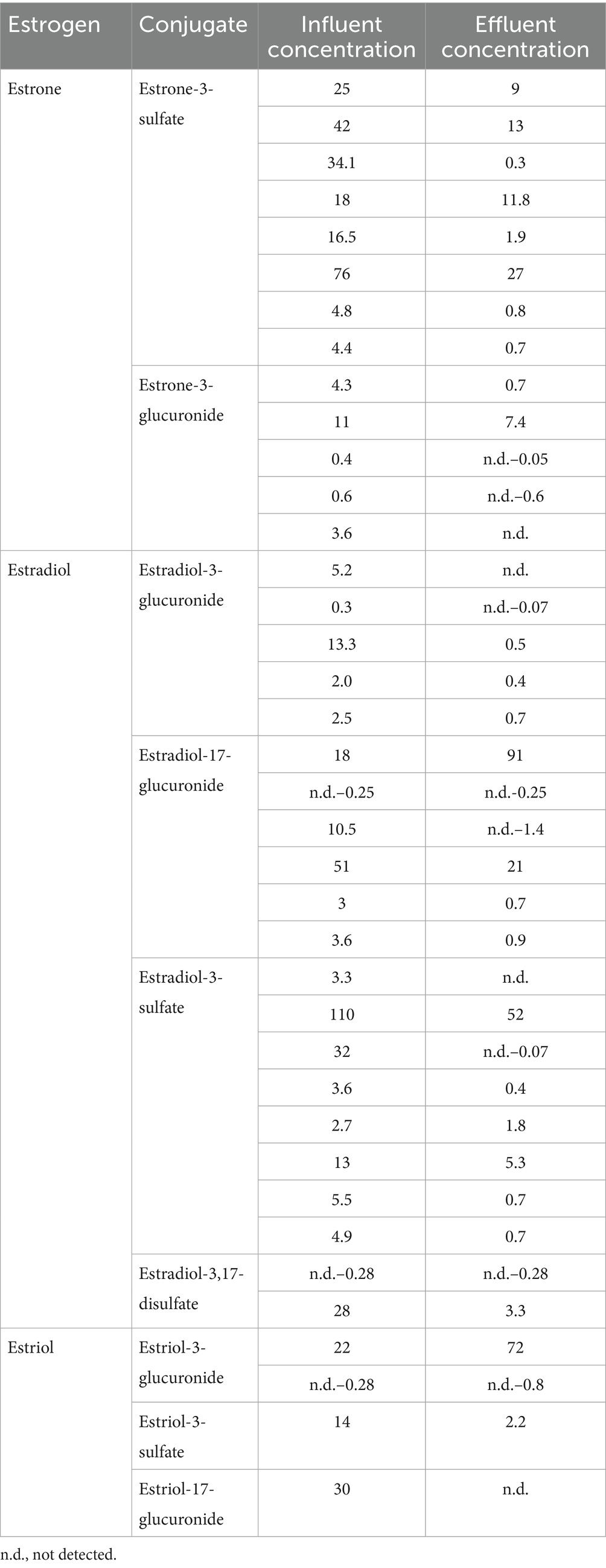
Table 2. Examples of concentrations (ng/L) of estrogen conjugates detected in WWTPs (Yu et al., 2019; Pratush et al., 2020a).
In WWTPs, estrogens’ conjugates revert to free estrogens (active forms) through enzymatic reactions performed by microbes existing in the WWTPs (Palme et al., 1996; Liu et al., 2009c; Zhang et al., 2024). The presence of estrogens in aquatic ecosystems is hazardous to aquatic life due to their endocrine-disrupting activity, and the hazards may include synthesis and secretion of vitellogenin (a female specific protein) in male fish, development of intersex characteristics and/or failure in the development of normal secondary characteristics (Racz and Goel, 2010; Ying et al., 2002) which leads to changes in reproduction. In addition to their endocrine-disrupting activity, estrogens were listed by the WHO as “group 1″ carcinogens.
Moreover, some catechol metabolites of estrogens, such as 4-hydroxyestrone, were reported to exhibit carcinogenic activity (Liehr et al., 1986; Fernandez et al., 2006). Due to their biological activity at very low concentrations, EDCs can threaten the polluted environment with acute and chronic toxicity to organisms, loss of habitat and biodiversity, and a range of potential adverse effects on environmental and ecological health (Torres et al., 2021; Zheng et al., 2023).
In humans, studies have reported a close association between exposure to estrogens and disorders of the male and female reproductive systems. Estrogens can disrupt the hormone-dependent pathways via direct interaction with the receptors or epigenetic changes. Some documented disorders include decline in fertility, congenital malformation of gonads development, and increased incidence of reproductive diseases and cancer (Adeel et al., 2017; Rashtian et al., 2019).
The increasing worldwide concern about environmental exposure to estrogens has prompted programs to understand and improve their removal from WWTPs, which are the main source of environmental pollution with estrogens (Leech et al., 2009; Yu and Chu, 2009). Some research studies also addressed estrogens removal from drinking water treatment plants, constructed wetlands, sewer, manure, poultry litter and diary disposal systems (Yu et al., 2013).
Removal of steroid estrogens is challenging due to their recalcitrant sterane structure and low aqueous solubility, which were found to be as low as 30, 3.6, 441, and 116 mg/L, for E1, E2, E3, and EE2, respectively (Zhang et al., 2015). While WWTPs are essential for eliminating estrogens; they are not designed to eliminate them completely (López-Velázquez et al., 2021). Thus, estrogens are continuously discharged into surface water (López-Velázquez et al., 2025; Alfonso Murillo-Tovar et al., 2025). Various studies have evaluated estrogens removal from WWTPs using microfiltration, activated carbon adsorption, chemical degradation, and advanced treatment such as reverse osmosis. However, despite the relative efficiency of the physical and chemical techniques, biological methods, predominantly the use of biofilms, are emerging (Gadupudi et al., 2021).
Biological removal of estrogens in WWTPs includes deconjugation, complete degradation by heterotrophic microorganisms, or co-metabolism (Fernández et al., 2017; Ke et al., 2007; Liu et al., 2016; Zhang et al., 2024). However, the removal efficiency varies from one plant to another and depends on several factors such as operating conditions, geological location, estrogen concentration, and the biological system type (Yu et al., 2013). Estrogen elimination is more efficient in WWTPs equipped with biological treatment systems with long solid and hydraulic retention times. The latter enable longer contact time for better estrogen adsorption and sufficient time for the growth of slow-growing estrogen degraders (Maeng et al., 2013).
In all cases, it is generally acknowledged that estrogens are challenging substrates for microbial utilization due to their physicochemical properties such as low aqueous solubility, low number of functional groups, the presence of four alicyclic rings, and the presence of two quaternary carbon atoms (Olivera and Luengo, 2019), which may contribute to the various removal efficiencies reported in the literature, ranging between 14–94%, 76–92%, and 83–87% for E1, E2, and EE2, respectively (Baronti et al., 2000). It is also worth noting that steroid estrogens exhibit different susceptibilities to microbial degradation, following the descending order E3 > E2 > E1 > EE2 (Coello-Garcia et al., 2019).
Many studies addressed estrogen biodegradation in the environment, with more focus on WWTPs, which revealed a diversity of estrogen-degrading microorganisms and biodegradation/biotransformation mechanisms. Several factors may affect the type of estrogen metabolic mode in different bacteria. However, the key determinants appear to be the bacterial species and the applied growth conditions, which are discussed and analyzed in the following sections.
4 Microbial biodegradation and biotransformation of estrogens
4.1 Estrogen-degrading microorganisms
Microbial biodegradation was reported as the major mechanism which removes estrogens from engineered and natural polluted environments (Yu et al., 2013; Huang et al., 2014; Liyanage et al., 2024). Despite many studies on estrogens biodegradation, there are still many knowledge gaps regarding the catabolic pathways. Moreover, the involved enzymes, genes, and their regulation mechanisms are not fully characterized yet. Degradation of estrogens by aerobic bacteria has been reported, and many estrogens–degrading bacteria have been isolated from various environments (Yu et al., 2013; Zhang et al., 2015; Olivera and Luengo, 2019; Chiang et al., 2020; Pratush et al., 2020a).
Estrogens, like other steroids, are carbon-rich and highly reduced compounds that are abundant and ubiquitous in the environment. Therefore, they represent a precious carbon and energy source for some microorganisms (Chiang et al., 2020). In fact, some microorganisms (including bacteria, yeasts, fungi, and microalgae) can perform biotransformation reactions on estrogens (Pratush et al., 2020a). However, only dedicated bacteria have the ability to mineralize estrogens (complete degradation to CO2) (Bergstrand et al., 2016; Holert et al., 2018). Recent studies identified steroid-degrading bacteria and elucidated the biodegradation mechanisms using both culture-dependent and culture-independent approaches.
Bacteria that can degrade estrogens under aerobic conditions were isolated mainly from activated sludge, soil, compost, sandy aquifers, and very little from the marine environment. Although the isolates belong to different phyla, most estrogen-degrading bacteria reported to date belong mainly to the Actinomycetota (Actinobacteria) and Pseudomonadota (Proteobacteria) (Supplementary Tables S1, S2) (Bergstrand et al., 2016; Chiang et al., 2020; Pratush et al., 2020a; Shah et al., 2024).
Key enzymes involved in aerobic estrogen biodegradation and biotransformation are classified in different categories including dehydrogenases, hydroxylases, ring-cleavage dioxygenases, isomerases, monooxygenases, hydratases, and demethylases, as well as cytochrome P450 (Chiang et al., 2020; Pratush et al., 2020a).
4.2 Initial degradation and transformation reactions
Various estrogen biotransformation reactions were reported, including hydroxylation, dehydrogenation, and ring cleavage reactions (Figure 2). Most of the studies have focused on the aerobic biodegradation E1 and E2 as model substrates. On the contrary, E3 and EE2 have not been less thoroughly investigated. Moreover, the biodegradation of estrogens under anaerobic conditions was reported only recently (Wang et al., 2020).
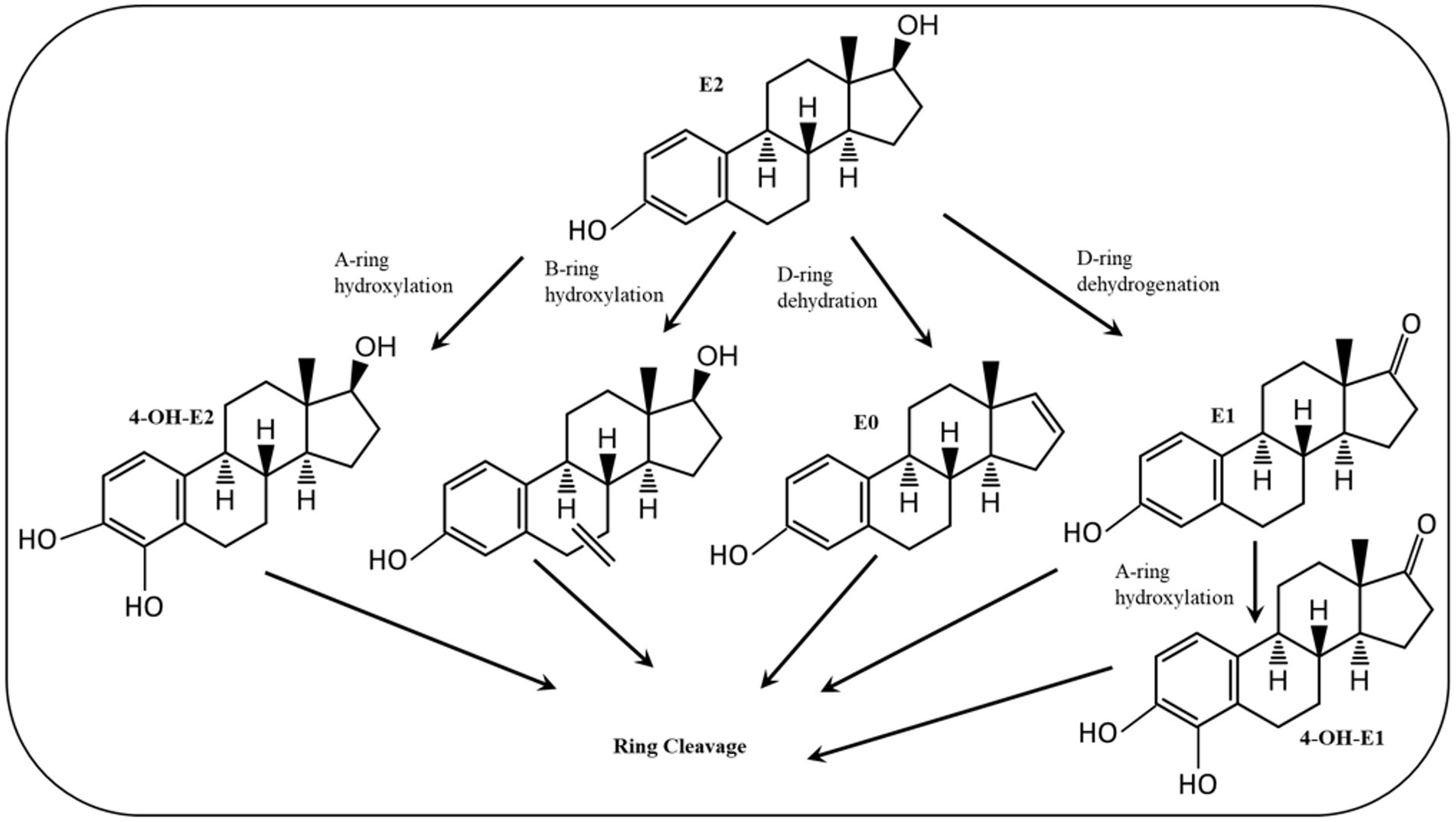
Figure 2. Initial reactions of major E2 degradation pathways (Mensitieri et al., 2023).
Despite the high structural similarity among estrogens, a diversity of biotransformation reactions and degradation pathways have been reported in the literature. Some of those reactions/pathways were proposed based on the detection and identification of metabolic intermediates. Another important feature of estrogen biodegradation is the fact that estrogens are interconvertible, i.e., one estrogen can be transformed to another during biodegradation or biotransformation. To date, three major catabolic pathways for steroid biodegradation have been reported. Under aerobic conditions, sterols and androgens are degraded via the 9,10-seco pathway, while estrogens are commonly degraded via the 4, 5-seco pathway.
On the contrary, under anaerobic conditions sterols and androgens are catabolized via the 2,3-seco pathway, while the anaerobic degradation of estrogens has not been studied in detail (Chiang et al., 2020; Jacoby et al., 2025). Regardless of the oxygen availability, the three pathways merge at a common key intermediate, namely HIP (3aα-H-4α(3′-propanoate)-7aβ-methylhexahydro-1,5-indanedione), which is subsequently degraded via a common central pathway to produce metabolites that are further degraded through central metabolic networks (Figure 3). In the following, different estrogen biodegradation and biotransformation mechanisms will be presented. For clarity, E2 and E1 will be discussed together because their degradation pathways are linked. Subsequently, EE2 and E3 degradation pathways will be presented separately.
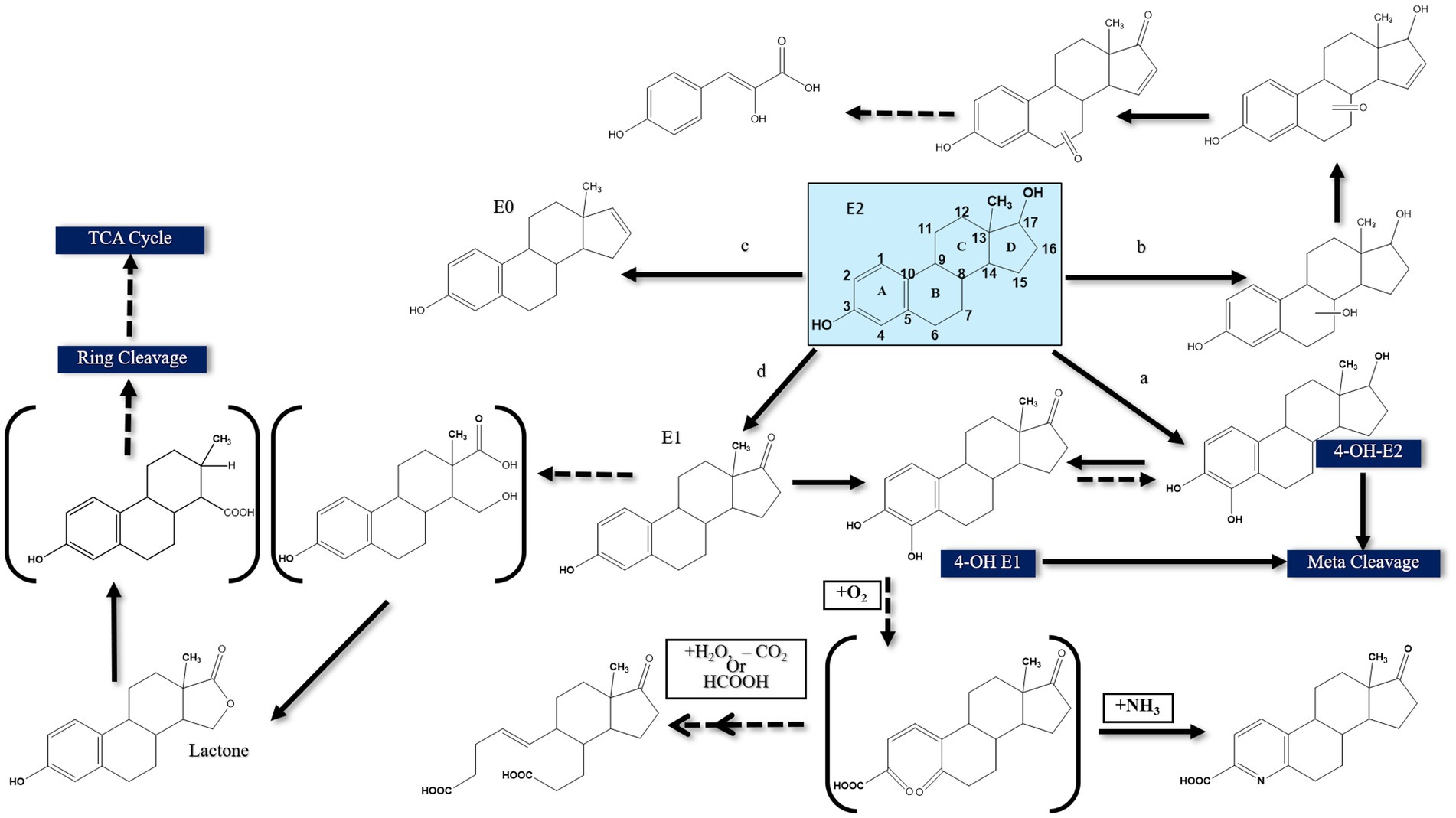
Figure 3. Proposed E2 degradation pathways by aerobic bacteria (Solid lines indicate confirmed pathways. Dashed lines indicate uncertain pathways) [modified from Yu et al., 2013].
4.3 Aerobic degradation of E2 and E1
The first E2-degrading bacterium was isolated from activated sludge and identified as Novosphingobium tardaugens sp. nov. (Fujii et al., 2002), and Coombre et al. (1966) proposed the first pathway for E1 degradation by the soil bacterium Nocardia sp. E110. It was proposed that a dioxygenase catalyzes the cleavage of the A-ring of E1 and this pathway was included as a part of E2 degradation pathways reported in subsequent studies, proposing that E2 degradation can be initiated by one of the following reactions: (a) hydroxylation of the A-ring at C4, (b) hydroxylation of a saturated ring, (c) dehydration of the D-ring at C17, and (d) dehydrogenation of the D-ring at C17 (Yu et al., 2013) (Figure 3).
The first mode of degradation was reported by Kurisu et al. (2010) who detected 4-OH-E2 during degradation by the soil isolate Sphingomonas sp. ED8, which suggests that E2 was hydroxylated at C4. The authors proposed that degradation of this hydroxylated product proceeds via meta cleavage. Kurisu et al. (2010) detected other products of E2 degradation by the ED8 strain, including hydroxy-E2, keto-E2, keto-E1, and 3-(4-hydroxyphenyl)-2-hydroxyprop-2-enoic acid. This latter product suggested that E2 degradation was initiated at a saturated ring. However, the authors did not further investigate how the saturated ring was cleaved. In another study, Nakai et al. (2011) used Nitrosomonas europeae to degrade E2 and detected a metabolite identified as estra-1, 3, 5 (10), 16 tetraen-3-ol (estratetra enol, or E0), which results from dehydration at C17 of the D-ring. Nitrosomonas europeae further degraded E0 to non-estrogenic compounds.
Transformation of E2 to E1 via dehydrogenation of C17 at the D-ring was also reported (Yu et al., 2007). Estrone can be further transformed to 4-OH-E1, followed by further degradation via meta cleavage (Kurisu et al., 2010). Lee and Liu (2002) suggested another degradation pathway for E2 when utilized by mixed sewage bacteria based on the detection of a new metabolite designated XI, which contains a lactone structure at the D-ring. All those previous studies proposed some biotransformation reactions, albeit complete pathway for estrogen degradation was not elucidated in terms of metabolic intermediates, genes, and enzymes, thus rendering the assessment of the fate and biodegradation potential of estrogens in the polluted environments challenging. Furthermore, the structural elucidations were mostly based on mass-spectrometry and no ring-cleave products were reported.
4.3.1 Elucidation of the 4,5-seco pathway
To address these knowledge gaps, Chen et al. (2017) adopted functional genomic analyses and enzyme characterization to study E2 degradation by Sphingomonas sp. KC8. They showed that E2 degradation by the KC8 strain starts by oxidation of the C17 hydroxyl group leading to E1, followed by hydroxylation at C4 of the A-ring to produce 4-OH-E1. The A-ring is then opened via meta cleavage. These reactions constitute the initial steps of the so called 4,5-seco pathway for aerobic bacterial degradation of E2/E1 (Figure 4). A key feature of this pathway is the abiotic reaction of the A-ring cleavage product with NH4+ to produce pyridinestrone acid (PEA), a typical side product and functional marker of the 4,5-seco pathway. Further characterization of 17β-E2 dehydrogenase (OecA), which catalyzes the transformation of E2 to E1, and 4-OH-E1 4,5-dioxygenase, which catalyzes the meta cleavage of the A-ring, was conducted by Chen et al. (2017). Moreover, the genes encoding these two enzymes were identified in the genome of the KC8 strain.
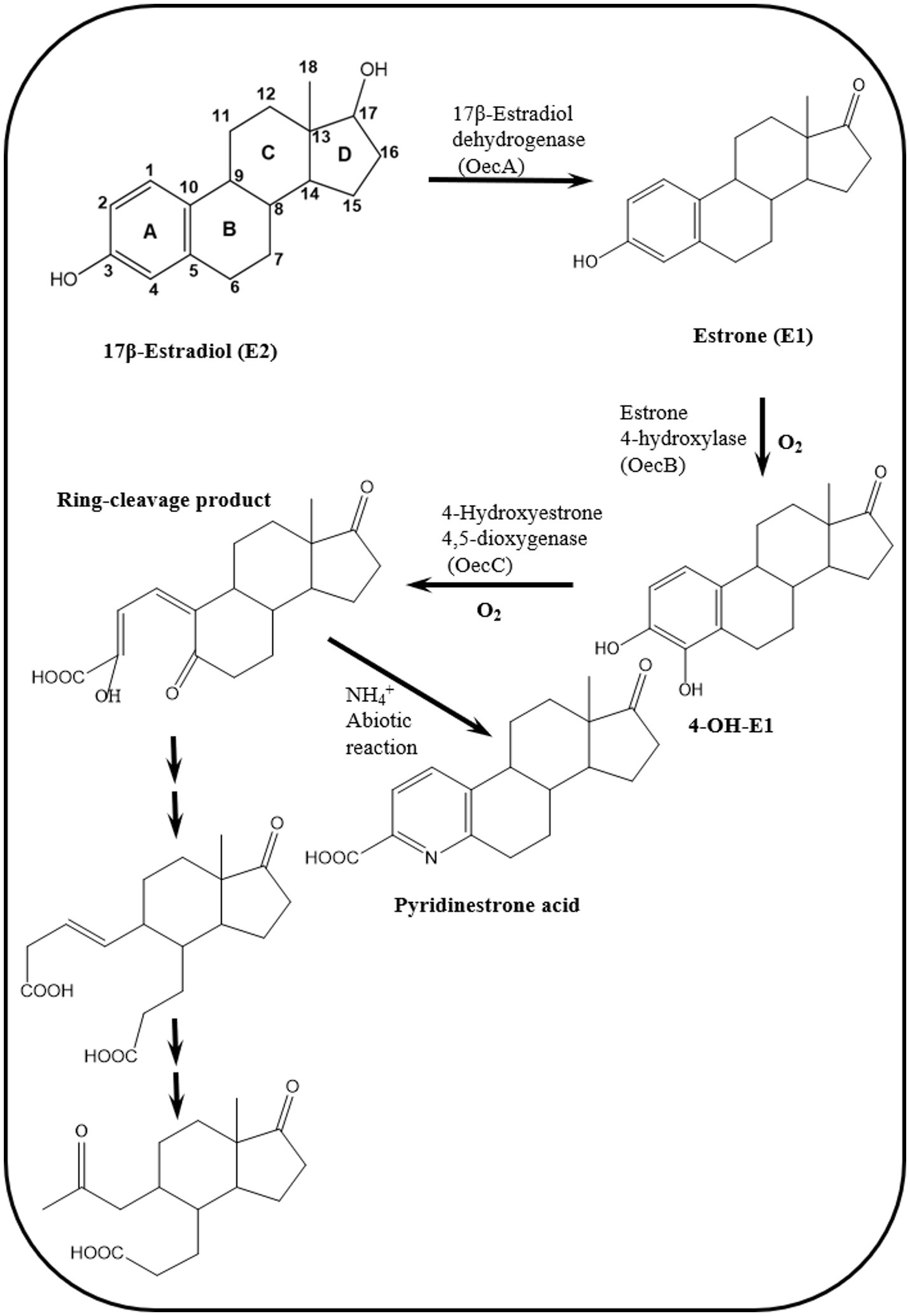
Figure 4. Reactions of the 4,5-seco pathway of aerobic biodegradation of estrogens in activated sludge and Novosphingobium sp. SLCC, showing intermediates downstream of the A-ring cleavage product (Chen et al., 2017, 2018).
Two gene clusters were specifically expressed during growth on E2 compared to cultures grown on testosterone. In one of these gene clusters, Chen et al. (2017) identified a gene (oecB), proposed to encode a putative flavin-dependent monooxygenase, which catalyzes hydroxylation of E1 at C4 of the A-ring. However, this enzyme was not further characterized. Enzymes encoded by gene cluster II were proposed to catalyze the A/B rings degradation. The authors also identified gene cluster III, which was similarly expressed in E2- and testosterone-grown cells. The gene products of this gene cluster exhibit high identity to β-oxidation enzymes and were proposed to be involved in the degradation of the C/D-rings of E2. Detection of PEA in [3,4C-13C]E1-spiked river water and activated sludge samples from a WWTP suggested that the 4,5-seco pathway might play a role in estrogen degradation in the environment (Chen et al., 2017).
Further biochemical and microbiological studies were performed by Chen et al. (2018) to detect additional downstream intermediates of the 4,5-seco pathway and to assess the ecological relevance of estrogen-degrading bacteria. In activated sludge samples incubated with El, PEA and two A/B-rings cleavage products were detected (Figure 4). Furthermore, PCR detected homologs of the alphaproteobacterial oecC, a functional marker of the 4,5-seco pathway. Metagenomics analysis revealed that Novosphingobium spp. are key estrogen degraders in activated sludge, which was corroborated by the isolation of Novosphingobium sp. SLCC strain from the E1-amended activated sludge. In batch cultures, this strain degraded E1 and produced the A/B-rings cleavage products detected in the activated sludge, thus confirming their role in E1 degradation.
Further investigations on the 4,5-seco pathway were reported by Wu et al. (2019) using Sphingomonas sp. KC8 as a model organism and applying NMR and UPLC-HRMS (Ultraperformance Liquid Chromatography-High Resolution Mass Spectroscopy) to identify metabolic intermediates of E1 degradation. In E1 cultures, the authors could detect and identify 4-norestrogen-5(10)-en-3-oyl-coenzyme A (CoA) and the corresponding non-conjugate (4-norestrogenic acid), in addition to the common metabolite of microbial steroid metabolism (HIP). Based on these findings, they proposed that after A-ring cleavage, C4 is removed by oxidative decarboxylation by a 2-oxoacid oxidoreductase to produce 4-norestrogen-5(10)-en-3-oyl-CoA. Then the B-ring is opened by hydrolysis and β-oxidation reactions transform the A/B-ring cleavage product into the common metabolite HIP (Table 3; Figure 5).
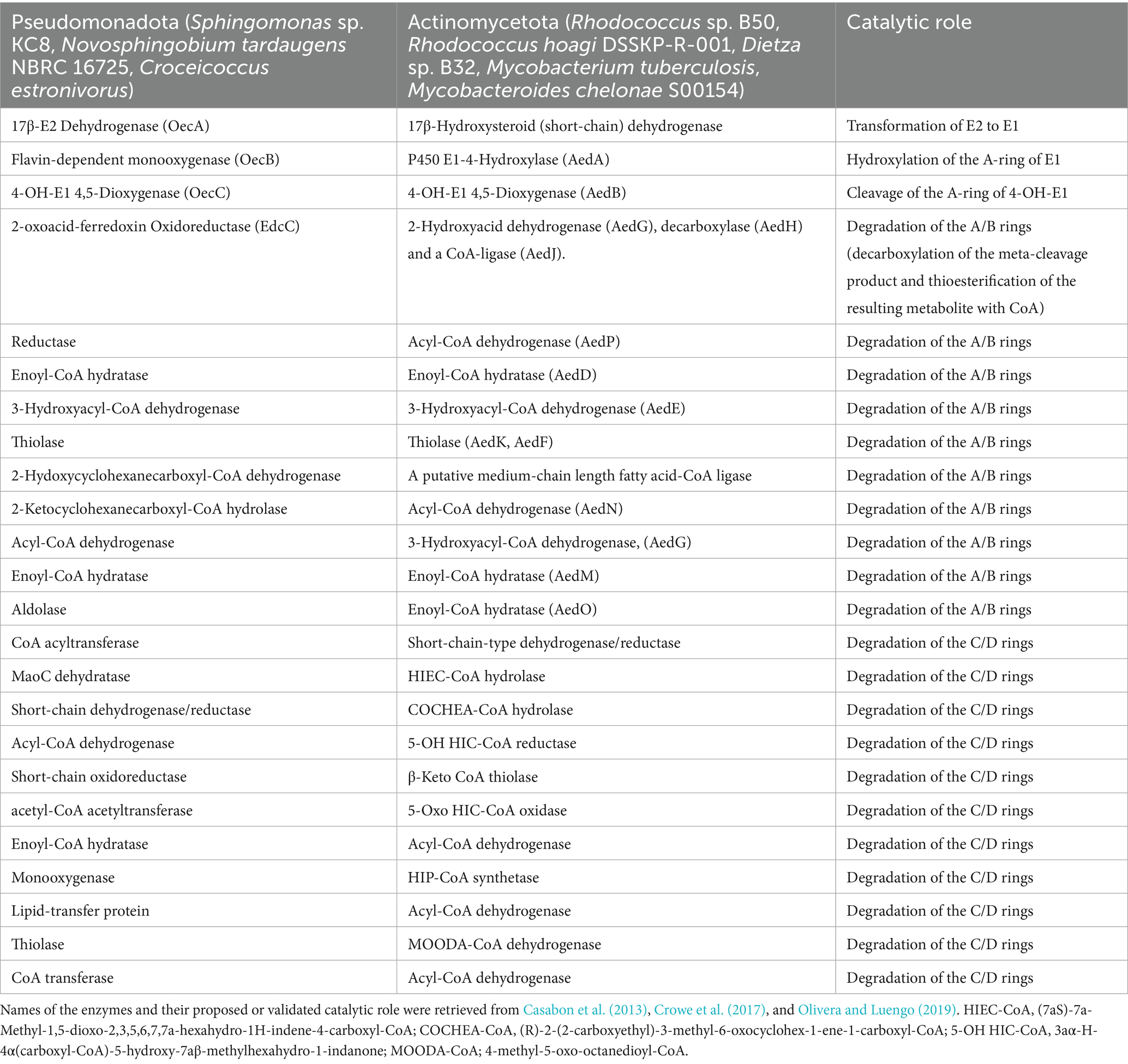
Table 3. Key enzymes of bacterial aerobic estrogen degradation via the 4,5-seco pathway and the HIP pathway.
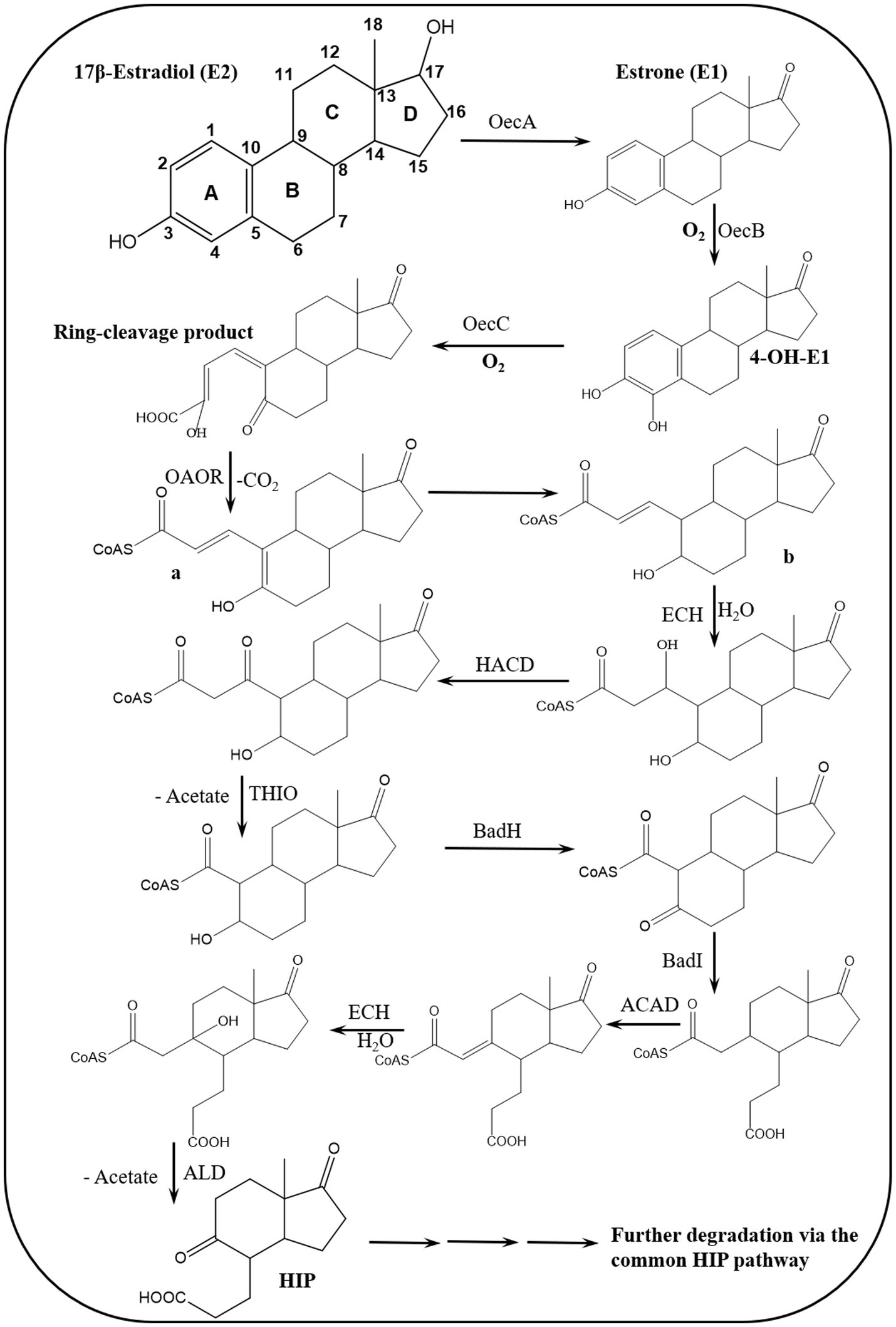
Figure 5. Proposed bacterial degradation pathway of natural estrogens. ACAD, acyl-CoA dehydrogenase; ALD, aldolase; BadH, 2-hydroxycyclohexanecarboxyl-CoA dehydrogenase; BadI, 2-ketocyclohexanecarboxyl-CoA hydrolase; ECH, enoyl-CoA hydratase; HACD, β-hydroxyacyl-CoA dehydrogenase; OAOR, 2-oxoacid oxidoreductase; THIO, thiolase (Wu et al., 2019).
Although PEA was commonly regarded as a dead-end product, it may undergo further reactions such as ring cleavage as reported recently in Microbacterium proteolyticum ZJSK01 (Chen et al., 2025). Furthermore, E2 degradation can be initiated at the saturated rings after hydroxylation of the A-ring to produce 4-OH-E2 (Chen et al., 2025). Consolidated cleavage of both the A- and B-ring of E2 after transformation to 4-OH-E2 was enhanced in the presence of Fe(III) which promoted the expression of the involved monooxygenase and extradiol dioxygenase (Li et al., 2024).
A diversity of estrogen biotransformation reactions was reported by Zhu et al. (2024), who identified 18 transformation products of E2 in a culture medium inoculated with the human intestinal fungus Aspergillus niger RG13B1. Hydroxylation, oxidation, dehydrogenation, and alkylation were the key transformation reactions. While hydroxylation reactions were the most common and occurred at various positions on the E2 structure, oxidation and alkylation reactions were mostly on the D- and B-rings. Notably, no ring-cleavage products were detected, further confirming that the metabolic mode is biotransformation not complete degradation.
Although the majority of research reported on E1 and E2 degradation focused on members of the Pseudomonadota and Actinomycetota, less information is available regarding the involved genes, enzymes and pathways for actinobacterial estrogen degraders, and Hsiao et al. performed a series of detailed studies to fill these knowledge gaps. In the first study, Hsiao et al. (2021) applied metabolomics and gene disruption studies using a soil isolate Rhodococcus sp. B50 as a model estrogen-degrading strain which utilized E1, E2, E3, testosterone, or cholesterol as a sole carbon and energy source. In E1-fed resting cell assays, UPLC-APCI-HRMS analysis revealed key metabolites of the 4,5-seco pathway, such as PEA and HIP.
Comparative genomic analysis revealed a putative estrogen degradation gene cluster carried on a mega plasmid and encodes a cytochrome P450 E1-4-hydroxylase (aedA) and a meta cleavage dioxygenase (aedB) involved in estrogen degradation via the 4,5-seco pathway. The function of these two enzymes was validated via gene disruption and phenotype analysis, showing functional similarity with the corresponding homologs from Pseudomonadota, EdcA and EdcB (Ibero et al., 2020) and OecA and OecB (Chen et al., 2017).
Despite this functional similarity, the amino acid sequence identity between the actinobacterial and proteobacterial homologs is relatively low (<40%). Subsequently, the authors used the extracellular metabolites PEA and HIP in addition to primers specific to the actinobacterial 4-hydroxyestrone 4,5-dioxygenase gene (aedB) as functional biomarkers to study estrogen biodegradation in urban estuarine sediments. Using E1-spiked mesocosms and PCR-based functional assays, it was concluded that estrogen biodegradation in the estuarine sediments proceeds via the 4,5-seco pathway, and Actinomycetota are active estrogen degraders in this ecosystem.
In a follow up study, Hsiao et al. (2022) applied comparative transcriptomics, gene disruption, and metabolite analysis to unravel genes and intermediates of downstream degradation of the estrogen A/B rings using Rhodococcus sp. B50 as a model. Analysis of the transcriptomes of B50 cultures grown on E1 compared to control cultures on cholesterol or testosterone showed that genes involved in C/D-rings degradation were similarly expressed in all three cultures. On the contrary, the aed gene cluster, which was identified by Hsiao et al. (2021) was significantly upregulated in the E1-grown culture. This gene cluster carries genes encoding AedA (E1 hydroxylase), AedB (meta cleavage dioxygenase), a putative medium-chain length fatty acid-CoA ligase, genes encoding two sets of β-oxidation enzymes including acyl-CoA dehydrogenase (AedN, AedP), enoyl-CoA hydratase (AedD, AedM, AedO), 3-hydroxyacyl-CoA dehydrogenase, (AedE, AedG) and thiolase (AedF, AedK). The authors then constructed knockouts for the genes AedF and AedK and validated their function via phenotype and metabolite analysis. Based on their findings, an estrogen degradation pathway for Actinomycetota was established. In addition to the common metabolites of the 4,5-seco pathway, three novel metabolites were identified including 4,5-seco-estrogenic acid, 5-oxo-4-norestogenic acid, 2, 3, 4-trinorestrogenic acid.
Obviously, estrogen degradation in Actinomycetota proceeds via the 4, 5-seco pathway as reported for Pseudomonadota. However, there are still some differences that distinguish both bacterial groups. For instance, actinobacterial estrogen metabolites bear a 5-oxo group, whereas the corresponding metabolites in Pseudomonadota have a hydroxy group at C5. Moreover, the ring-A cleavage products in Pseudomonadota possess 3 unsaturated bonds (at C1, C3, and C5), while in Actinomycetota the corresponding 4,5-seco-estrogenic acid has only two double bonds (at C1 and C5) and is less likely recyclized. Oxidation of the A-ring-cleaved products also reveals some differences. In Sphingomonas sp. KC8, oxidative decarboxylation after opening of the A-ring is catalyzed by a 2-oxoacid oxidoreductase (EdcC), a member of the indolepyruvate ferredoxin oxidoreductase family, to produce 4-norestrogen-5 (10)-en-3-oyl-CoA (Wu et al., 2019; Ibero et al., 2020). In Rhodococcus sp. B50, an EdcC homolog is lacking in the genome. Hence, the EdcC function may be carried out by a putative 2-hydroxyacid dehydrogenase (AedG), a decarboxylase (AedH) and a CoA-ligase (AedJ).
Hsiao et al. (2023) continued their investigations on the topic and conducted further comparative genomics, gene disruption, and metabolite analysis studies to validate the proposed function for the actinobacterial enzymes AedGHJ. They first revealed that the genes aedGHJ are found exclusively in the genomes of estrogen-degrading Actinomycetota. In contrast, the α-oxoacid-ferredoxin oxidoreductase EdcC is unique to estrogen-degrading Pseudomonadota. These findings suggested the presence of phylum-specific functional markers that delineate estrogen degradation mechanisms. Gene disruption experiments and metabolite analysis validated the proposed functions for AedGHJ proteins (Table 3; Figure 6). The authors further detected a C17 metabolite (1-hydroxy-5-oxo-4-norestrogenic acid) that accumulated in the wild type.
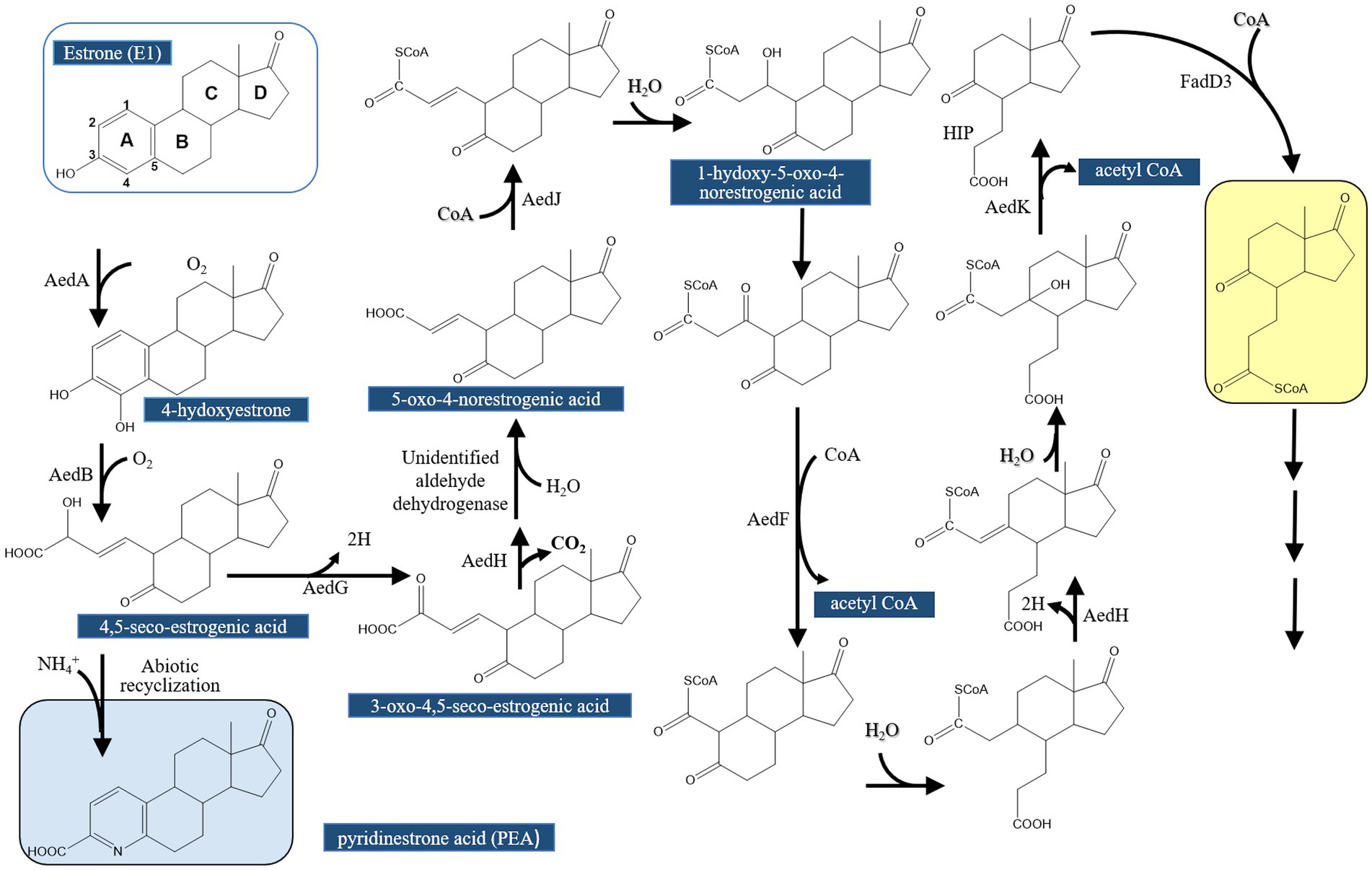
Figure 6. Proposed estrogen degradation pathway in Actinobacteria (Actinomycetota). AedG, AedH, and AedJ were functionally characterized in this study [modified from Hsiao et al., 2023].
Based on these findings, it appears that activation and degradation of the estrogenic A-ring proceeds via different strategies. In Pseudomonadota, a single oxidative decarboxylation reaction catalyzed by an α-oxoacid-ferredoxin oxidoreductase EdcC removes the C4 of E1 and produces a CoA-ester C17 metabolite (Ibero et al., 2020; Wu et al., 2019). Homologs of EdcC are lacking in estrogen-degrading members of the Actinomycetota, which depend on the enzymes AedGHJ to degrade the A-ring. Accordingly, these phylum-specific genes were used as biomarkers to study estrogen degradation in the environment (Table 3; Figure 7) using degenerate specific primers for the aedJ gene and edcC genes and phylum-specific 16S rRNA gene primers.
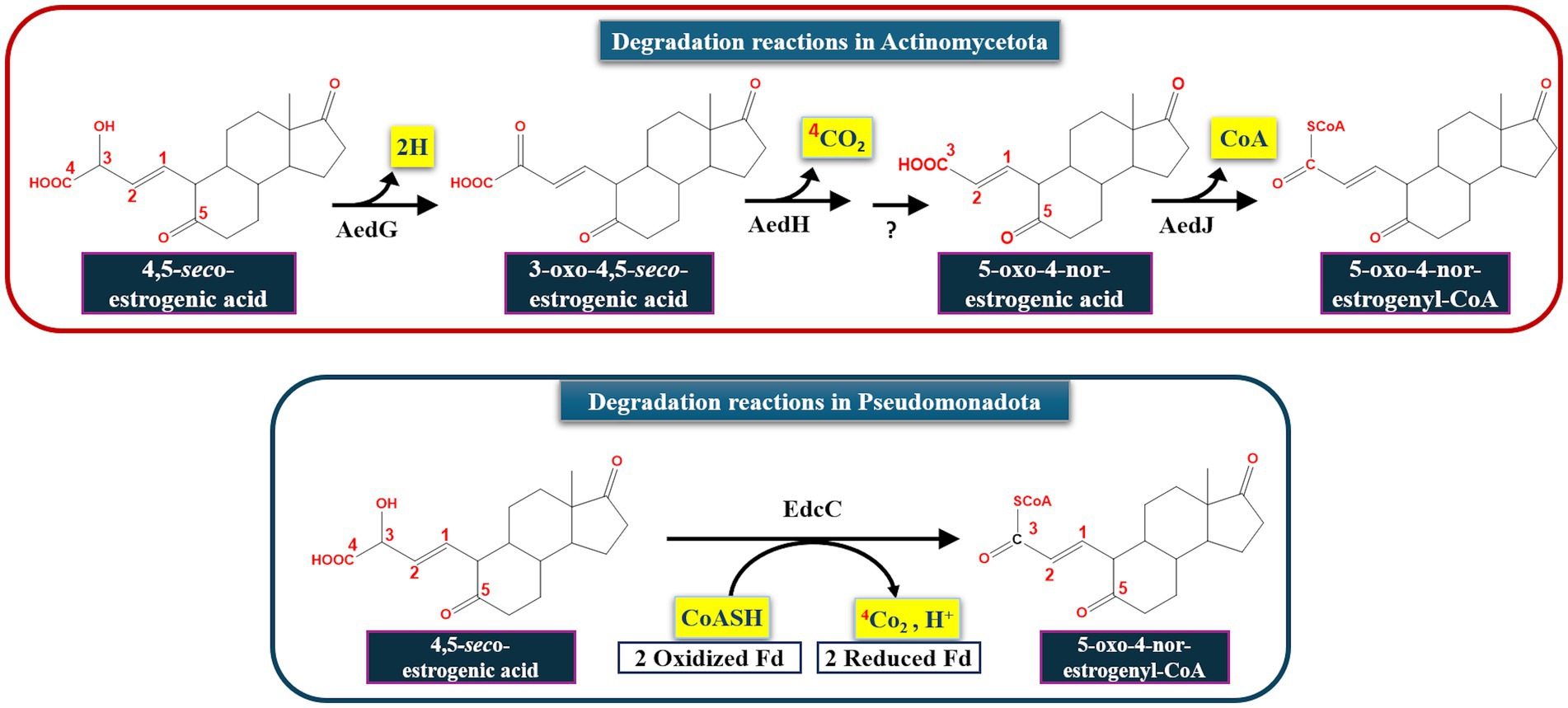
Figure 7. Bacterial taxa-specific enzymes involved in aerobic estrogen degradation reactions. Fd, ferredoxin (Hsiao et al., 2023).
Quantitative PCR assays using DNA from various estrogen-contaminated ecosystems in Taiwan revealed that γ-Proteobacteria were most abundant in the examined environmental samples. However, the actinobacterial gene aedJ was more abundant than the proteobacterial gene edcC in most of the examined samples, particularly those collected from aquatic ecosystems, suggesting that the aedJ gene can be adopted as a specific biomarker to probe for estrogen-degrading Actinomycetota in the environment due to the following reasons: (a) the biochemical function of the AedJ enzyme was characterized and its substrate and product were identified, (b) an aedJ homolog is lacking in estrogen-degrading Pseudomonadota and non-estrogen-degrading Actinomycetota, (c) the long sequence of aedJ (1,500 bp) enables the design of specific primers, and (d) aedJ is highly expressed only in the presence of estrogens (Hsiao et al., 2022). Similarly, the gene encoding α-oxoacid-ferredoxin oxidoreductase edcC can be used as a specific biomarker for estrogen-degrading Pseudomonadota.
While estrogen degradation via the 4,5-seco pathway by other Rhodococcus spp. was proposed (Tian et al., 2022), it appears that alternative pathways could exist in different actinobacterial members. This could be inferred from a study on a mangrove sediment isolate, Rhodococcus spp., which utilized E1 as the sole carbon and energy source and produced two non-accumulating E1 metabolites, namely, 3-hydroxyandrosta-5,7,9(11)-trien-17-one and androsta-1,4,6-triene-3,17-dione (Pratush et al., 2020b). Genomic analysis revealed that the strain BH2-1 has 46 genes belonging to 6 major steroid degradation gene classes including cholesterol oxidase, steroid Δ-isomerase, cytochrome P450 monooxygenase, 3δ-steroid-1 dehydrogenase, 3-steroid-9α-hydroxylase, and 3α,20β-hydroxysteroid dehydrogenase.
Based on LC-MS analysis, E1, E3, and dehydroepiandrosterone, androsta-1,4-diene-3,17-dione, homovanillic acid, and vanillylmandelic acid, were detected as E2 metabolites in Microbacterium resistens MZT7, isolated from farm-derived activated sludge (Hao et al., 2022). Transformation of E2 to E1 in Microbacterium sp. MZT7 is catalyzed by a 17β-hydroxysteroid (short-chain) dehydrogenase which is active even in synthetic livestock wastewater (Hao et al., 2023). A similar study identified E1, E3, and dehydroepiandrosterone, androsta-1,4-diene-3,17-dione, etiocholanolone, 2-methoxyestradiol, homovanillic acid, and 3,4-dehydroxyphenylpropionic acid as E2 metabolites of Rhodococcus sp. RCBS9 strain isolated from soil of a dairy farm (Hao et al., 2024). Accordingly, several potential E2 degradation pathways were proposed by Hao et al. (2022, 2024), although no quantification data was reported to properly assess the relative significance of the different proposed pathways. The physiological or ecological significance of having several biodegradation pathways for the same compound was not discussed. Moreover, the possibility that some of the detected metabolites could result from unspecific biotransformation reactions was not highlighted. In those studies, the detection of androgenic metabolites is of particular interest because conversion of estrogens into androgens is thermodynamically challenging and has only been reported recently as a key step in anaerobic estrogen degradation (Wang et al., 2020), thus contrasting with the findings of Hao et al. (2022, 2024), which were reported for aerobic E2 degradation. It was not reported in those studies whether the genomes of the studied bacteria (Rhodococcus sp. RCBS9 and Microbacterium sp. MZT7) carry genes encoding homologs of the cobalamin-dependent methyltransferases which catalyze the estrogen retroconversion to androgens (Wang et al., 2020).
Gordonia sp. strain R9, isolated from an enrichment culture of chicken leachate, degraded E2, E1, E3, and EE2 as a sole carbon and energy source. E2 degradation by the strain R9 produced several metabolic intermediates including E1, E3, (3Z)-3-(3-hydroxy-3a-methyl-7-oxododecahydro-6H-cyclopenta[a]naphthalene-6-ylidene) propanoic acid, and 3-hydroxy-3a-methyl-7-oxododecahydro-1H-cyclopenta[a]naphthalene-6-carboxylic acid. The latter two metabolites suggest that E2 degradation proceeds via cleavage of the A-ring. The authors also reasoned that detection of E1 and E3 in the E2-degrading culture may indicate the presence of alternative estrogen degradation pathways in the R9 strain (Liu et al., 2020). It is worth noting that no CoA-bound intermediates were reported in this study, in contrast to the studies of Wu et al. (2019) which could be differences in degradation mechanisms by actinobacterial and proteobacterial members.
Estradiol biodegradation was studied by Li et al. (2020) using a Novosphingobium sp. ES2-1 that was isolated from activated sludge in a domestic sewage treatment plant. Using 13C labeling, NMR and HRMS, and as reported by Chen et al. (2017, 2018) and Wu et al. (2019), the authors proposed that E2 is transformed to E1 then to 4-OH-E1 where the A-ring is opened, and PEA is produced. However, ring cleavage appeared to occur by successive monooxygenation reactions leading to long-chain ketonic structures. The authors also observed the production of several pyridine derivatives.
Degradation of E2/E1 via the 4,5-seco pathway was also reported in Novosphingobium tardaugens NBRC 16725 (Ibero et al., 2020). Transcript analysis in this bacterium revealed a gene cluster, designated edc, that encodes estrogen degradation proteins. Comamonas testosteroni JLU460ET is another Proteobacterium in which E2 was proposed to be degraded via the 4, 5-seco pathway based on the transcriptomic data. This was shown by Wang et al. (2023) via comparative transcriptomics, which revealed a 100 kb steroid degradation gene cluster including genes for sensing substrates in the environment, steroid uptake, ring-cleavage, and β-oxidation enzymes. However, no degradation metabolites were reported from E2-grown cultures.
4.3.2 Alternative E1 and E2 degradation pathways
Some other estrogen degradation pathways, which are different from the 4,5-seco pathway, were reported in cyanobacteria, yeast, and Gram-positive bacteria, further attesting taxon-dependent variations in biodegradation pathways. However, those pathways were proposed solely based on metabolite detection. For instance, Sami et al. (2023) studied E1 biodegradation by the cyanobacterium Spirulina CPCC-695 and showed that it utilizes E1 as a carbon and energy source for growth without addition of co-substrates. The degradation intermediates included fatty alcohols, alkanes, and a degradation pathway was proposed which starts with the reduction of E1 to E2. Detection of these intermediates suggested E1 metabolism may include hydrogenation, dehydrogenation, oxidation–reduction, hydroxylation, and side chain cleavage reactions. Moreover, E1 metabolism was proposed to start by reduction of the C17 keto group to produce E2, and the D-ring is cleaved first after formation of E1-formate and E1-propionate. After ring opening, esterification, methylation, and hydrogenation reactions produce diones/diols such as 3-phenylundecane, cyclo-tetradecane, and tetradecane.
After methylation, 4, 6-dimethyldodecane undergoes dehydrogenation to form lauryl alcohol. Further degradation via oxidation reactions produces lauric acid that condenses with CoA. The resulting fatty acyl-CoA is further degraded to acetyl-CoA (Sami et al., 2023). This proposed pathway is completely different from the commonly reported 4, 5-seco pathway and does not converge at HIP. Although these findings suggest the presence of different estrogen degradation pathways in cyanobacteria, the authors did not present biochemical evidence at the enzyme level. Furthermore, the culture medium contained several other carbon sources in addition to E1, such as acetone (the solvent of E1), citric acid, ferric ammonium citrate, Na-EDTA, and Na-CO3, which questions the origin of at least some of the detected intermediates.
Various pathways were proposed for E2 degradation by Candida utilis CU-2 and Lactobacillus casei LC-1 in the presence of additional carbon sources (Ge et al., 2021). Using LC-MS/MS analysis, 12 degradation products were detected in E2 biodegradation experiments and accordingly proposed five degradation pathways (Figure 8). In pathway I, degradation proceeds via a dehydration reaction at C17 of the D-ring to produce E0 as proposed earlier by Nakai et al. (2011). In pathway II, E2 is hydroxylated at the D-ring and then methylated. No ring cleavage products were reported for pathways I and II. As reported by several studies, E2 is first transformed to E1 in the other three alternative pathways reported by Ge et al. (2021), and E1 is then oxidized at the C-ring via dehydrogenation, or at the D-ring via oxygen incorporation as reported by Wang et al. (2019b), or at C4 of the A-ring to produce 4-OH-E1. However, further degradation of the intermediate proceeds via opening of the D-ring. Enzymes that could catalyze the proposed hydroxylation, methylation, and dehydrogenation reactions were not reported in this study.
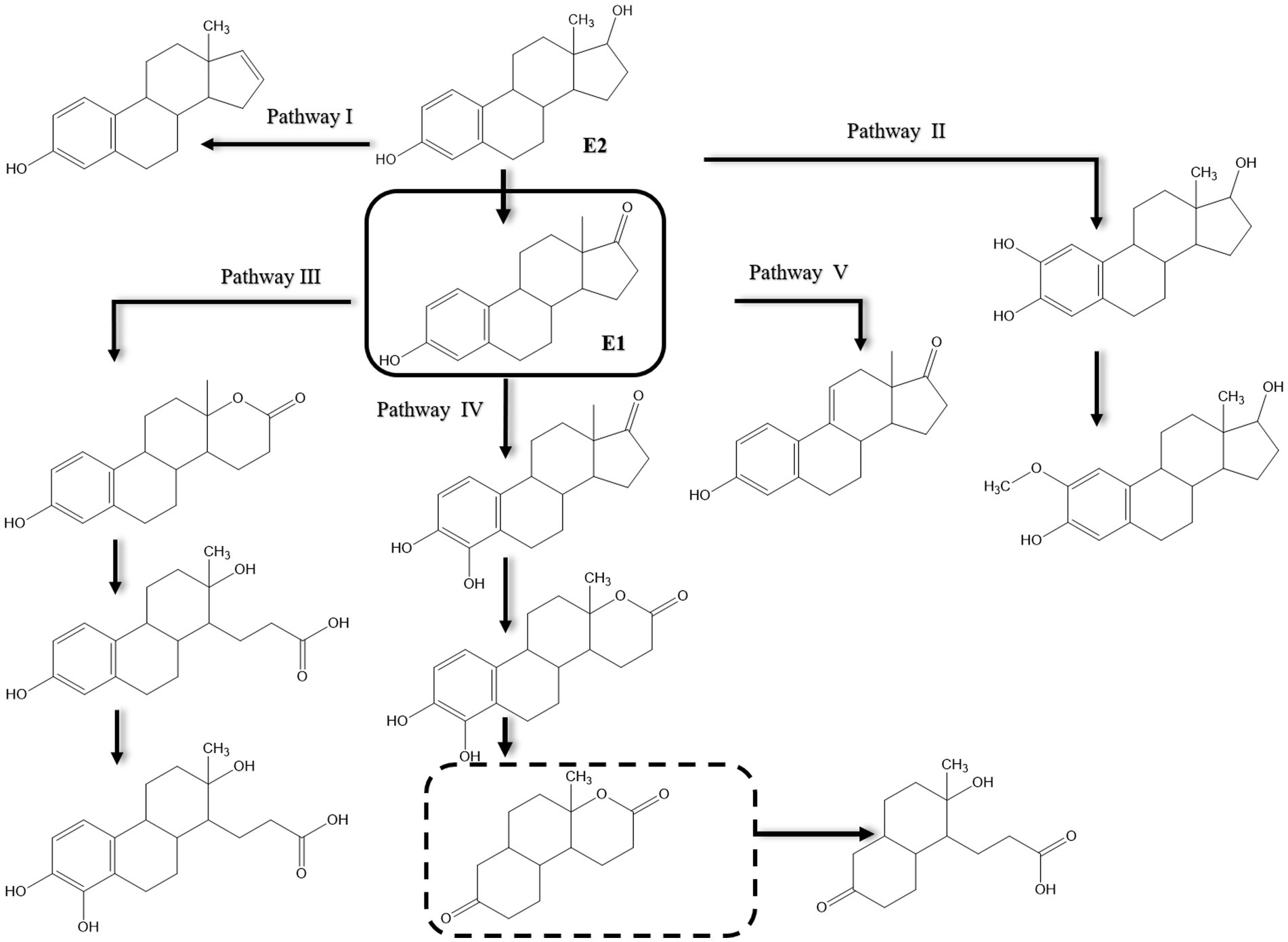
Figure 8. Proposed pathways of E2 biodegradation in Candida utilis CU-2 and Lactobacillus casei LC-1. The metabolite within the dashed rectangle was not detected by liquid chromatography–tandem mass spectrometry (LC-MS/MS) [modified from Ge et al., 2021].
4.3.3 Estrogen degradation by bacterial consortia
Despite the known advantages of applying microbial consortia for degradation of environmental pollutants (Zhang and Zhang, 2022), degradation of estrogens by bacterial consortia has not been often reported. Mixed bacterial cultures were isolated from activated sludge and degraded estrogen in a minimal medium, however, the degradation pathways were not elucidated (Weber et al., 2005; Muller et al., 2010a). Estrogen degradation can be more efficient by mixed cultures compared to axenic cultures as shown by Wang et al. (2019a) for E2 degradation by a mixed culture of Rhodococcus equi DSSKP-R-001 and Comamonas testosteroni QYYZ0150409. Moreover, concerted metabolic capabilities of members of bacterial consortia could lead to novel biotransformations and/or degradation pathways that are lacking in pure-estrogen degrading bacteria. This was also shown by Wang et al. (2019a) who identified three unique E2 metabolites in the co-culture of Rhodococcus equi DSSKP-R-001 and Comamonas testosteroni QYYZ0150409 that were lacking in the pure cultures of these bacteria. These findings suggested E2 degradation via alternative pathways, all of which start by transformation of E2 to El. In one of the proposed pathways E1 is transformed to 4-hydroxyestrone, which is further hydroxylated at C-16 to give a metabolite that is further degraded via the TCA cycle. In the second pathway, the A-ring of 4-hydroxyestrone is cleaved between C-4 and C-5 and C-5 is oxidized to a carboxyl group, and the C-9–C-10 bond is then cleaved. In the third pathway, the B-ring of E1 is first cleaved by hydroxylation at C-9α, followed by hydroxylation of the A-ring at C-4 and cleavage between C-4 and C-5. Eventually, the C-5–C-6 bond is cleaved. However, neither pyridine derivatives or CoA thioesters were reported, and the involved enzymes and genes were not identified.
Estrone biodegradation was studied in E1-spiked activated sludge microcosms from a major WWTP in Bahrain, and the key metabolites of the 4,5-seco pathway, namely, 4-OH-E1 and PEA were detected, indicating that E1 was degraded via the 4,5-seco pathway (AlDhafiri et al., 2022). The alphaproteobacterial genera Novosphingobium and Sphingoaurantiacus were significantly enriched in the E1-spiked microcosms after 2 days, and the authors enriched a mixed culture from the activated sludge which degraded E1 in batch cultures. From this consortium, an alphaproteobacterial strain, identified as Sphingobium estrogenivorans, was isolated and degraded E1 in batch cultures, revealing the role of Alphaproteobacteria in E1 degradation.
4.4 Aerobic EE2 degradation
Biodegradation of EE2 has been less frequently reported, and most of the available studies were conducted with microbial cultures that can utilize/transform EE2 in the presence of co-substrates, thus following a cometabolic mode of degradation (O’Grady et al., 2009; Pauwels et al., 2008). This could be attributed to recalcitrance of EE2 compared to the natural estrogens (Zhang et al., 2015; Olivera and Luengo, 2019). Co-substrates may strengthen central metabolism, promote cell growth, and supply reducing power, and can even induce the expression of some enzymes that play a role in EE2 metabolism (Gröning et al., 2007; Tran et al., 2013; Fischer and Majewsky, 2014). Currently, no complete EE2 biodegradation pathway is available and knowledge of the involved genes, enzymes are largely lacking.
Only a few bacteria can degrade EE2 as a sole carbon and energy source such as Sphingobacterium sp. JCR5 (Haiyan et al., 2007) and Rhodococcus equi (Yoshimoto et al., 2004). The number of reported microorganisms that can transform or cometabolize EE2 is very limited including Sphingomonas spp., Pseudomonas spp., Nitrosomonas europaea, the cyanobacterium Microcystis novacekii, the microalgal strain Selenastrum capricornutum, and the fungi Phoma sp. and Pleurotus ostreatus (Flint, 2019; Pratush et al., 2020a). Degradation of EE2 occurs under aerobic and anaerobic conditions, albeit higher degradation rates were reported under aerobic conditions (Aris et al., 2014; Cajthaml et al., 2009). EE2 degradation was slow in marine sediments (Ying and Kookana, 2003) and aquifer material (Ying et al., 2003), while in agricultural soils (Colucci and Topp, 2001) and nitrifying activated sludge (Vader et al., 2000) it was more rapidly degraded. Earlier studies reported biotransformation of EE2 by some bacteria and microalgae (Haiyan et al., 2007; Della Greca et al., 2008) including Sphingobium sp. JCR5, Rhodococcus zopfii, and Nitrosomonas europaea EE2 (Shi et al., 2002; Yoshimoto et al., 2004; Haiyan et al., 2007; Aris et al., 2014). One of the possible degradation pathways starts by transforming EE2 to E1, then E3, but no further degradation was reported (Li et al., 2013a, 2013b), or transformation of EE2 to E1, followed by ring cleavage (Cajthaml et al., 2009). At low concentrations, EE2 could be co-metabolized with E1, E2, or E3 in sludge and wastewater; however, no further metabolites were detected (Cajthaml et al., 2009; Writer et al., 2012; Rhee et al., 2014).
Several studies reported biotransformation reactions for EE2 without proposing complete degradation pathways (Figure 9). Yi and Harper (2007) detected 2-OH-EE2 during their studies on EE2 degradation by a nitrifying culture and showed that the A-ring was cleaved before the other rings, probably due to the higher electron density of the A-ring carbon units. These findings suggested that EE2 can be cometabolically transformed under nitrification conditions.
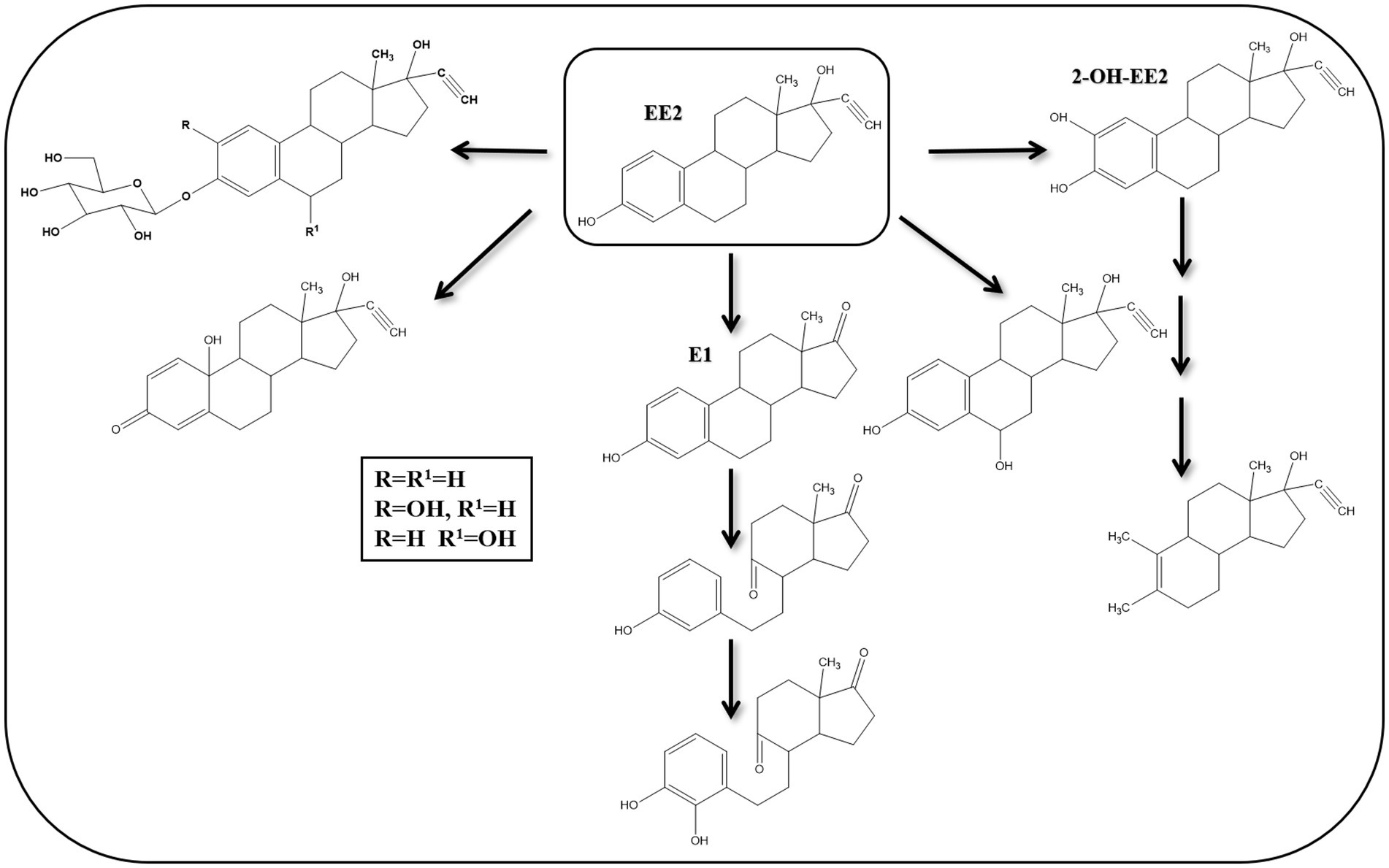
Figure 9. Proposed EE2 degradation pathways in bacteria and algae [modified from Yu et al. (2013)].
Some algal species can transform EE2 into EE2-glucoside, 3-β-D-glucopyranosyl-2-hydroxy-EE2, 3-β-D-glucopyranoside-6β-hydroxy-EE2, 17α-ethinyl-1,4, estradien-10,17β-diol-3-one, and 6-α-OH-EE2 (Della Greca et al., 2008). Thus, hydroxylation, oxidation, and glycosylation reactions were the main biotransformation reactions. In another study, Haiyan et al. (2007) isolated Sphingobacterium sp. JCR5 strain from activated sludge and showed that it can grow on EE2 as a sole carbon and energy source. Mass spectrometry analysis revealed that EE2 is first transformed to E1, followed by hydroxylation and oxidation reactions at C9, leading to B-ring cleavage. The A-ring is then hydroxylated at C4.
Co-metabolism of EE2 was studied with several bacteria isolated from compost. The isolated bacteria degraded E1, E2, and E3. However, EE2 was not degraded when used as a sole carbon source, albeit it was degraded in the presence of E2, and in this case E1 was detected as a metabolite. No other metabolites were detected, and EE2 removal started after complete removal of E2, and it increased as the initial E2 concentration increased (Pauwels et al., 2008). R. erythropolis and R. equi removed EE2 from minimal medium in the presence of co-substrates (adipic acid and glucose). Two metabolites were identified as phenol and a derivative of EE2 with a higher molecular mass (331 atomic mass unit) where the ethinyl group was absent (O’Grady et al., 2009).
In a study by Ma et al. (2018), lab-scale percolation columns were used to investigate EE2 biodegradation during groundwater recharge with reclaimed water. Based on metabolite analysis, different pathways for EE2 degradation were proposed depending on groundwater recharge mode. The dominant pathway in wetting and drying alternative recharge mode started by oxidation of C17 at the D-ring, which is then hydroxylated and cleaved. Thus, EE2 is transformed to E1 → E3 → 7-hydroxy-1-hydroxymethyl-2-methyl-1,2,3,4,4a,9.10,10a-octahydrophenanathrene-2-carboxylic acid → 2-hydroxy-2,4-diene-1, 6-dioic acid. On the contrary, in a continual recharge mode, EE2 metabolism started by hydroxylation of C4 on the A-ring and then the A- or B-ring is cleaved. This transition in the degradation mechanism was proposed to be related to dissolved oxygen and the structure of the involved microbial community.
Recent studies on EE2 degradation in synthetic wastewater by microalgae proposed hydroxylation reactions at C2 or C4, followed by ring opening (Wang et al., 2019b) (Figure 10). However, enzymes and genes of the proposed biochemical reactions were not reported. Peng et al. (2023) studied EE2 degradation by Pseudomonas citronellolis SJTE-3 and showed that it can grow on and degrade EE2 only in the presence of co-substrates like ethanol and glucose. The authors also identified two new degradation metabolites. Metabolism of EE2 was inducible, and 8 genes were significantly upregulated in the presence of EE2, including genes encoding a short chain dehydrogenase, a membrane transporter, and a cytochrome P450 hydroxylase, which were crucial for EE2 metabolism.
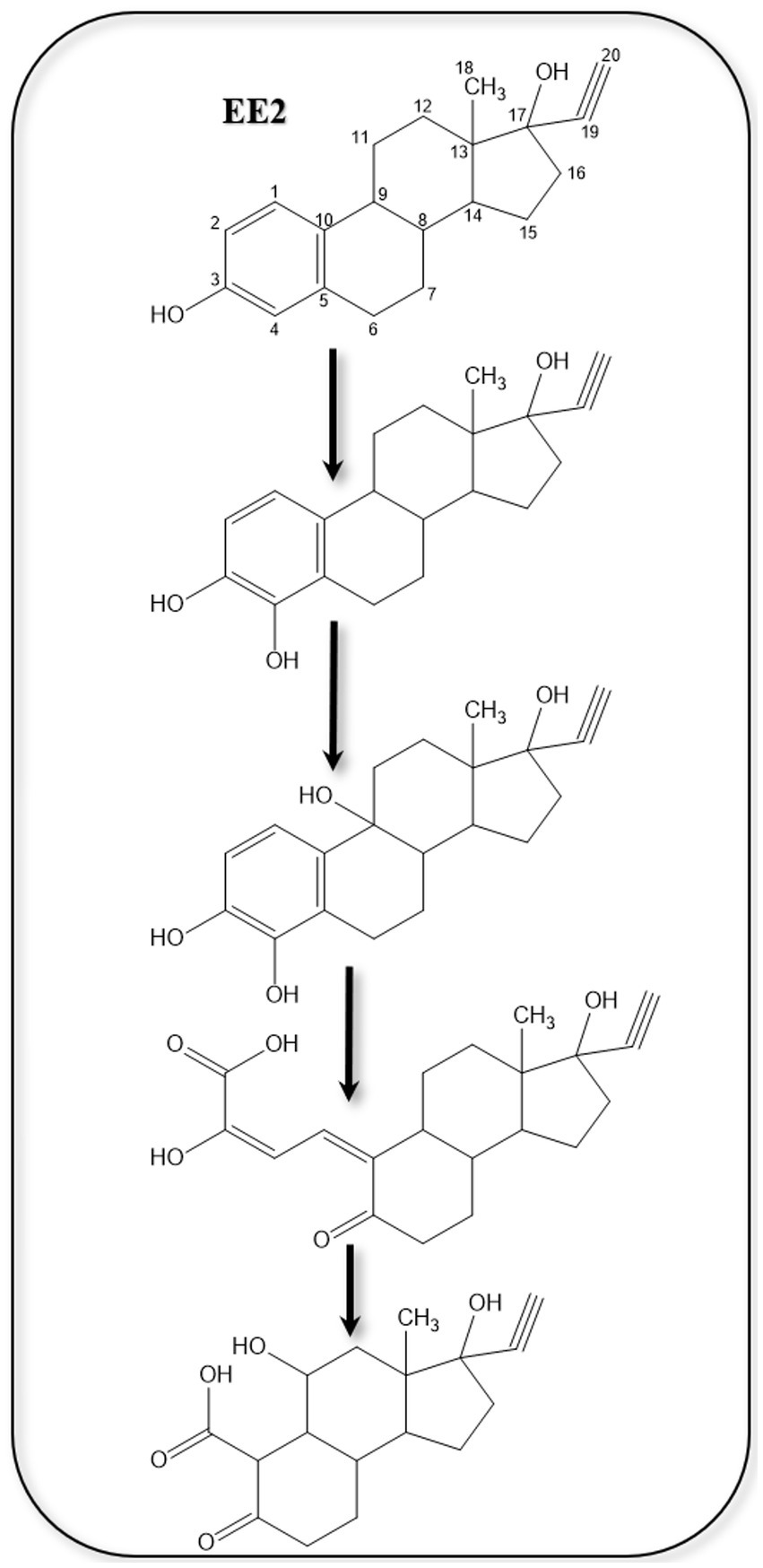
Figure 10. Proposed EE2 metabolites (Wang et al., 2019b).
Guo et al. (2021) studied the effect of rhamnolipid biosurfactants produced by Pseudomonas aeruginosa MIG-N146 on the biodegradation of EE2 in shake flasks containing sediment/water systems. The authors reported enhanced biodegradation of EE2 as the rhamnolipid concentration increased, which was attributed to better mass transfer and stimulation of the indigenous microbial consortium. Three metabolic intermediates were detected including a dehydrogenation product of EE2 at C15-C16 of the D-ring, and a carboxylation product of EE2. This study did not report any data on the involved enzymes or microbial community.
Recently, bacteria affiliated with the genera Aeromonas, Rhizobium, and Paraburkholderia isolated from an acid mine drainage degraded EE2 in a minimal medium and produced several degradation metabolites which were mainly ring cleavage products (Palma and Costa, 2024). The authors mentioned that EE2 was the sole carbon source in the cultures. However, EE2 was added from a methanol stock solution, and it was not clear whether it was evaporated before inoculation. Furthermore, while the aim of the study was to isolate EE2-degrading bacterial consortia, all the enrichment and isolation procedures were performed in a rich medium (LB medium) in the presence of paracetamol.
4.5 Aerobic E3 degradation
Compared to E1, E2 and EE2, a limited number of studies addressed E3 biodegradation. In most of those studies, E3 was mentioned in the context of substrate spectrum experiments together with other steroid estrogens. Moreover, those studies reported only E3 biotransformation reactions, albeit a complete E3 mineralization pathway is lacking. For instance, Ke et al. (2007) isolated three bacteria from marine sand in microcosms using E1, E2, E3, and EE2 as sole carbon sources. The isolates were identified as members of the genera Sphingomonas, Agromyces, and Acinetobacter. These bacteria, individually or as a consortium, had different substrate spectra in terms of estrogen degradation. The strain identified as Acinetobacter sp. degraded E3 in aerobic cultures and transiently produced 16α-OH-E1, which was reported as a metabolite of E2 degradation by sewage bacteria (Lee and Liu, 2002) and of E1 degradation by intestinal bacteria (Järvenpää et al., 1980). Moreover, Staphylococcus aureus can reduce 16α-OH-E1 to E3 under anaerobic conditions (Järvenpää et al., 1980). Nitrifying activated sludge and ammonia-oxidizing bacterium Nitrosomonas europaea degraded E1, E2, E3 and EE2 in minimal medium (Shi et al., 2004). However, no further information was reported on the degradation mechanisms. Ueda et al. (2012) isolated and characterized a laccase from the mushroom Pleurotus eryngii and showed that it can remove E3 from aqueous solutions. A Comamonas testosteroni strain isolated from animal waste grew on E3 as a sole carbon and energy source (Liu et al., 2021). Nothing was reported in this study on the specific E3 degradation products, enzymes, or genes. However, genomic analysis revealed the presence of 22 steroid degradation genes. The authors also identified a testosterone degradation gene cluster and a gene encoding 17β-hydroxysteroid dehydrogenase, which catalyzes the transformation of E2 to E1. In the same context, Rhodococcus sp. PI4, a marine isolate, utilized E2, E3, and testosterone as sole carbon sources and induced the expression of a 17β-hydroxysteroid dehydrogenase which transforms E3 to 16α-OH-E1 (Ye et al., 2019).
A Gordonia sp. strain R9 isolated from chicken leachates utilized E3 and other steroid estrogens as sole carbon sources (Liu et al., 2020). Although no information on E3 degradation was reported, the authors detected E3 as a metabolite of E2. Estriol was also reported as a product of E2 degradation by Microbacterium resistens MZT7 (Hao et al., 2022) and Rhodococcus sp. RCBS9 (Hao et al., 2024), and it was not clear how E3 is further metabolized.
A study by Hsiao et al. (2021) showed that a Rhodococcus sp. strain B50 can grow on E3 as a sole carbon source. However, no further information on the mechanism of E3 degradation was reported. Wang P. et al. (2019) isolated a Pseudomonas putida strain SJTE-1 from activated sludge and showed that it can degrade E1, E2, E3 and testosterone as sole carbon sources. Nothing was reported in this study on the E3 degradation mechanisms.
Evidence supporting the conclusion that E3 is degraded by some bacteria via the 4,5-seco pathway was reported in recent studies. We showed that a bacterial consortium obtained from raw domestic sewage transformed E3 to E1 potentially via transient formation of 16α-OH-E1. Further degradation of E1 proceeded via the 4,5-seco pathway and the community structure of the consortium shifted strongly toward Croceicoccus estronivorus which dominated the community after 10 days of incubation (Hashem et al., 2025). Liu et al. (2024) reported the degradation of E3 by Novosphingobium sp. ES2-1 and proposed two alternative pathways for E3 degradation based on metabolite analysis. The first proposed pathway proceeds via E3 transformation to E1, which is further degraded via the 4,5-seco pathway. However, the authors did not clarify how E3 was transformed to E1. They proposed oxidation and dehydration reactions, albeit no biochemical evidence was presented. The other pathway proposed by Liu et al. (2024) also proceeds by transforming E3 to E1, then to 4-OH-E1, which undergoes B-ring cleavage, resembling the 9,10-seco pathway. However, nothing was reported on the genes and enzymes involved in this later pathway. Moreover, the mechanism of the B-ring cleavage was not discussed. The various detected metabolites were not quantified, which is essential to reflect on the relative significance or dominance of the two proposed pathways. Degradation of E3 via the 9,10-seco pathway was also proposed by Thathola et al. (2024) using a psychrotolerant Pseudomonas proteolytica strain based on the detection of E1, 16-OH-E1, in addition to two proposed ring cleavage downstream products: 2-hydroxy2-4-dienevaleric acid, and 2-hydroxy-2,4-diene-1,6-dioic acid. These products were identified only using mass spectrometry and no further biochemical evidence was presented.
4.6 Anaerobic degradation of estrogens
In contrast to the aerobic estrogen degradation, biodegradation of estrogens under anaerobic conditions has been very rarely investigated. The low aqueous solubility (1.5 mg/L at room temperature) of estrogens and stability of the aromatic A-ring render estrogens challenging substrates for microbial utilization (Shareef et al., 2006; Wang et al., 2020). Therefore, under aerobic conditions, bacteria depend on oxygenases and oxygen to activate and cleave the aromatic A-ring through the 4,5-seco pathway (Chen et al., 2017; Chiang et al., 2020). Lack of oxygen in anaerobic environments, such as river and marine sediments, prohibits estrogen degradation and makes the process slower compared to aerobic degradation. Hence, anaerobic environments are considered as major reservoirs for estrogens (Hanselman et al., 2003; Czajka and Londry, 2006). Probably, slow degradation rates and difficulties in isolating anaerobic estrogen-degrading bacteria explain why anaerobic estrogen degradation has been less investigated compared to aerobic degradation. Only two bacterial species are known to degrade estrogens under anaerobic conditions, namely, Denitratisoma oestradiolicum and Steroidobacter denitrificans (Fahrbach et al., 2006, 2008).
To elucidate the biochemical mechanisms of anaerobic estrogen degradation, Wang et al. (2020) isolated a denitrifying bacterium, Denitratisoma sp. strain DHT3, from a municipal WWTP and showed that it can efficiently degrade estrogens under denitrifying conditions. The authors sequenced the genome of this strain and performed comparative transcriptomics to identify potential genes involved in anaerobic estrogen degradation using E2- and testosterone-grown cultures. Genes involved in the anaerobic 2,3-seco pathway were expressed at similar levels in both cultures. On the contrary, genes of transport, salvage, and reductive activation of cobalamin were upregulated in the E2 culture. In addition, a gene cluster encoding a putative methyltransferase (emtABCD) was also upregulated in the E2 culture. Gene disruption experiments confirmed that emtA plays a role in anaerobic estrogen degradation by the DHT3 strain.
Interestingly, the emtABCD gene cluster is present in the genome of estrogen-degrading anaerobes, but lacking in steroid-degrading bacteria that cannot utilize estrogens (Wang et al., 2020). Moreover, metabolite analysis of E1-grown cultures of the DHT3 strain revealed several androgenic metabolites including two ring-cleavage products which are characteristic of the 2,3-seco pathway, i.e., 17β-hydroxy-1-oxo-2,3-seco-androstan-3-oic acid (2,3-SAOA) and HIP. These findings suggested that anaerobic estrogen degradation in the DHT3 strain is initiated by conversion of the C18 estrogens to C19 androgens which are further degraded via the 2,3-seco pathway. The key step is the conversion of estrogens to androgens via a methylation reaction catalyzed by a cobalamin-dependent methyltransferase (emtABCD) (Figure 11).
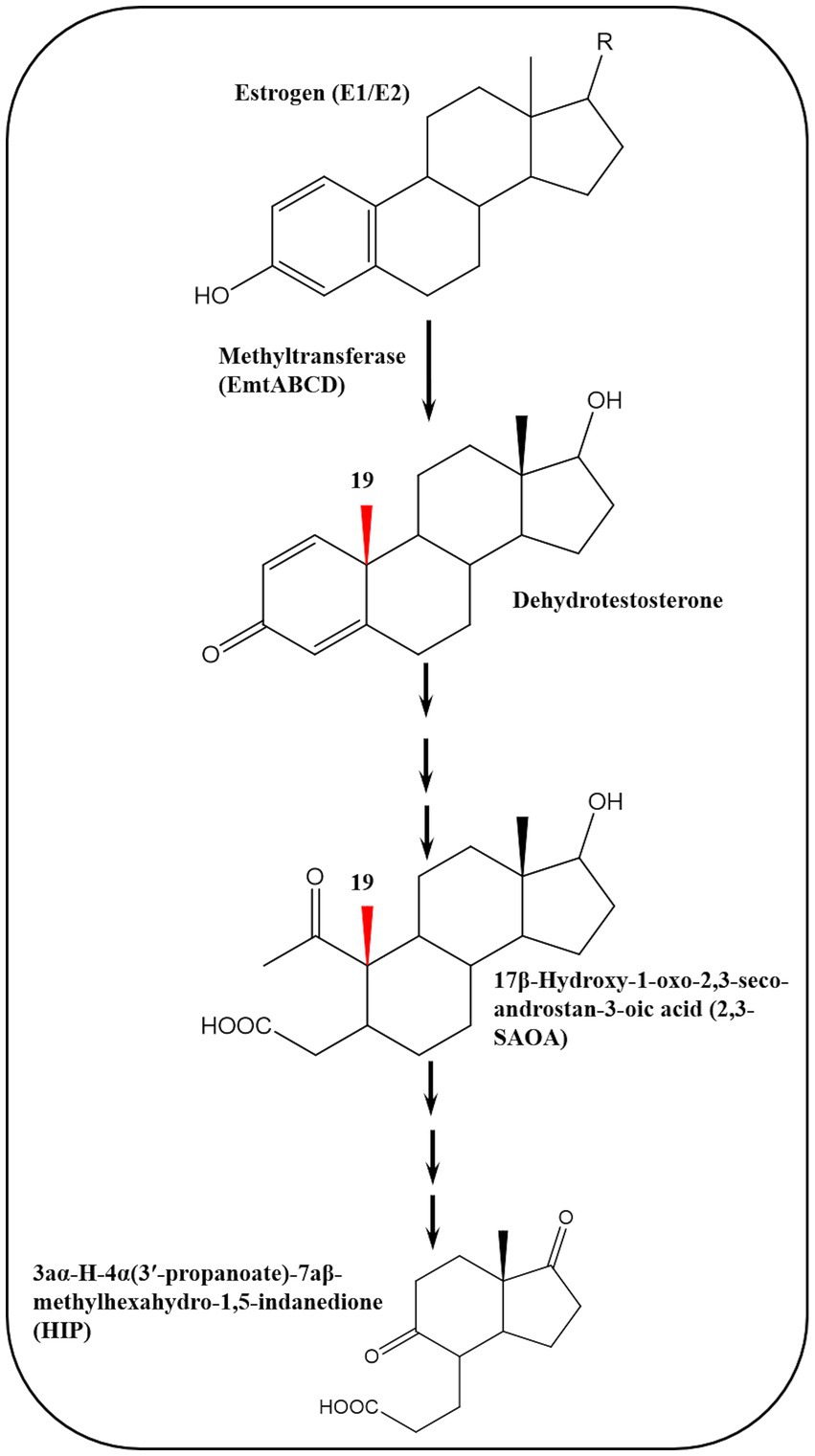
Figure 11. Cobalamin-dependent retroconversion of estrogens into androgens during anaerobic estrogen degradation by Denitratisoma sp. strain DHT3 (Wang et al., 2020).
These findings highlight microbe-host metabolic relationships mediated through retroconversion of estrogens into androgens based on the role of sex steroids in bidirectional metabolic interactions between bacteria and their eukaryotic hosts (Karavolos et al., 2013; Markle et al., 2013; vom Steeg and Klein, 2017). Conversion of estrogens into androgens represents the reverse process of estrogen (C18) biosynthesis from androgens (C19) via removal of the C19 angular methyl group, leading to formation of the aromatic A-ring (Miyairi and Fishman, 1985), and it is a thermodynamically challenging reaction which has not been reported in any organism (Wang et al., 2020).
The reverse reaction, namely, the conversion of C19 androgens into C18 estrogens, was recently discovered in strict anaerobic bacteria (Wang et al., 2025). This aromatase-independent estrogenesis is catalyzed by a cobalamin-dependent methyltransferase (AbeABC), which is 57% identical to the EmtAB methyltransferase identified in the denitrifying Denitratisoma spp. Homologs of AbeAB were identified in uncultivated bacteria from marine sediments, suggesting widespread occurrence of anaerobic estrogenesis in anoxic ecosystems. Based on those findings, some ecological implications can be envisaged. For instance, the reversible conversion of estrogens into androgens in anaerobic environments (animal guts and sediments) could potentially affect steroid hormone cycling and fate in these habitats. Furthermore, the unique presence of the abeAB genes in microbial genomes render them a functional biomarker that enables monitoring of ecosystem health and tracking steroid pollution (Wang et al., 2025). A recent study showed that EE2 was transformed to E1 and E2 in an electrochemical anaerobic membrane bioreactor (Lei et al., 2025), suggesting that anaerobic degradation of EE2 may proceed via E2 or E1 conversion to androgen. The discovery of this unprecedented steroid microbial bioconversion paves the way for potential diverse applications in environmental monitoring and bioremediation.
The recent studies and findings on anaerobic estrogen biodegradation reflect an increasing interest in this topic whose understanding has been lagging compared to the aerobic degradation processes. Three overlapping drivers can be envisaged to have motivated researchers to investigate this topic further. The first is purely scientific curiosity to understand how bacteria deal with challenging substrates like steroid estrogens in the absence of oxygen or under oxygen fluctuation conditions. Recalling the unprecedented biochemical aspects revealed by extensive studies on the anaerobic degradation of aromatic compounds (Ismail and Gescher, 2012; Boll and Estelmann, 2020; Boll et al., 2020a, 2020b), enables us to reconcile the importance of this driver. The second driver could be the rising global concerns on steroid estrogens as emerging pollutants and the need to better understand their fate and biodegradation mechanisms particularly in anaerobic environments where they tend to accumulate (Hanselman et al., 2003; Czajka and Londry, 2006). While earlier and recent studies addressed the fate and kinetics of estrogen degradation in anaerobic ecosystems (Muller et al., 2010b; Zheng et al., 2012; Luo et al., 2024), unraveling the biochemical mechanisms is pivotal for the development of efficient bioremediation technologies for polluted anaerobic environments due to the recalcitrance of estrogens in the (Pratush et al., 2020a). The third driver is related to the overwhelming recent interest in understanding the functional relevance of the gut microbiome in health and pathogenesis, which is instrumental toward the development of novel therapies and probiotics (Li et al., 2022; Diviccaro et al., 2025). In this context, it is known that sex steroids mediate bidirectional interactions between bacteria and their eukaryotic hosts (Kverka and Stepan, 2024; Nieto et al., 2025). Moreover, bacteria can alter a host’s sex steroid profile through interconversions (biotransformations), utilization as a carbon and electron source, and affecting endogenous estrogen metabolism in postmenopausal women (Horinouchi et al., 2012; Markle et al., 2013; Ridlon et al., 2013; Fuhrman et al., 2014; Zhang et al., 2025). While the three proposed drivers guide research in different directions with different objectives, they share better understanding of the subject matter as a key enabler and will continue to fuel research on estrogen microbial metabolism and associated applications in health care and environmental biotechnology.
4.7 Central pathway of bacterial estrogen degradation
A C13 metabolite (HIP), containing the C/D rings of the steroid substrate, was commonly identified in all bacterial pathways for steroid catabolism. Hence, it is degraded through a common central catabolic pathway which targets the C/D rings (Table 3; Figure 12). After activation by a specific acyl-CoA synthetase (FadD3), the remaining carbons of the steroid B-ring are removed through β-oxidation. This is followed by hydrolytic cleavage the D-ring by an enoyl-CoA hydratase (EchA20), and another hydrolase (IpdAB) opens the C-ring. Interestingly, gene mining studies revealed the presence of conserved gene clusters for HIP degradation in the majority of experimentally verified steroid-degrading bacteria (Chiang et al., 2020).
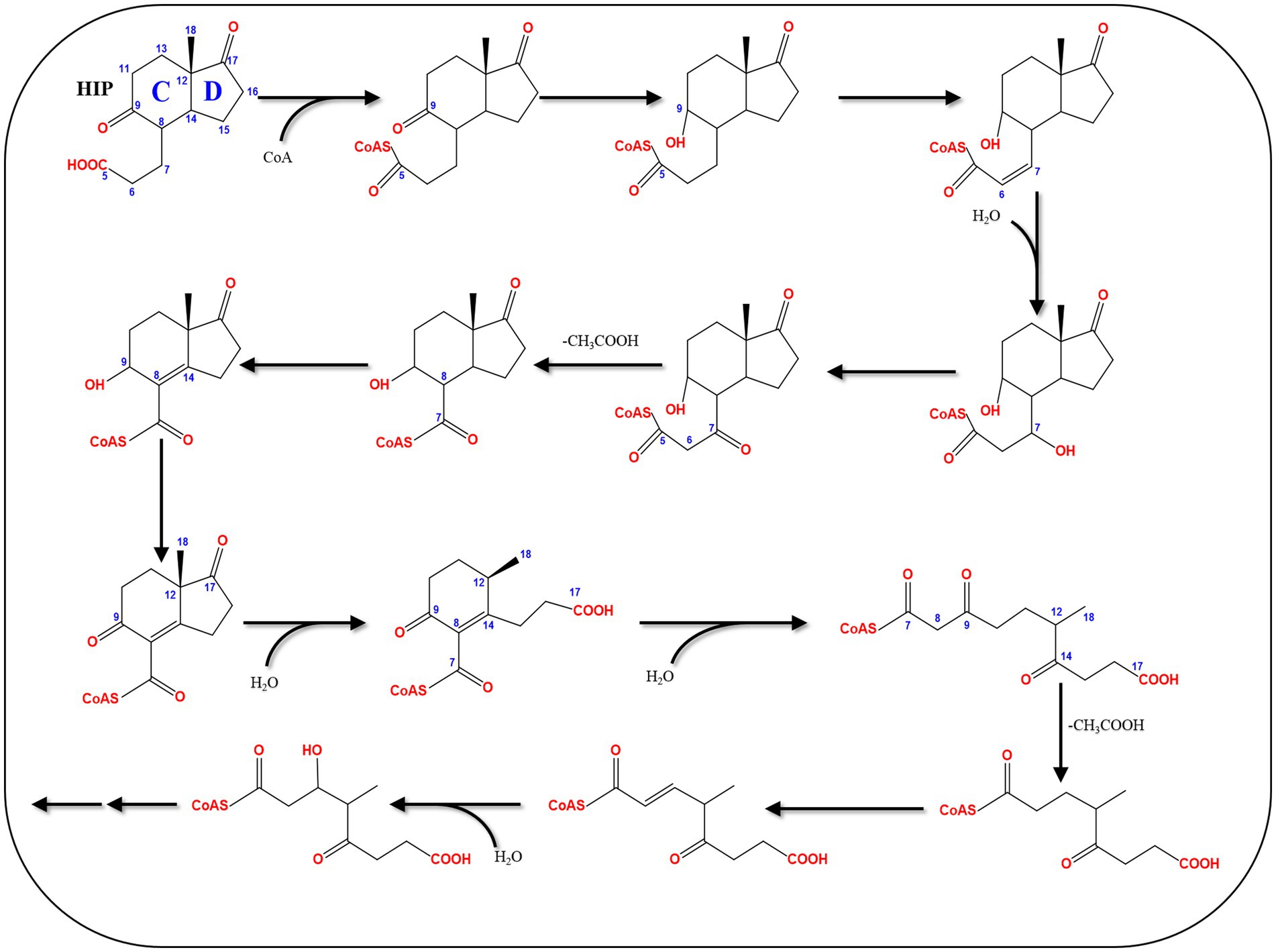
Figure 12. The HIP degradation pathway (Chiang et al., 2020).
4.8 Abiotic degradation and transformation of estrogens
In addition to biological degradation and transformation, estrogens are exposed to several abiotic factors in the environment, which can lead to some degradation and transformation reactions, including chemical and photodegradation/transformation. These abiotic reactions may affect the fate of estrogens in the environment and can also directly interfere with the susceptibility of estrogens to biodegradation and biotransformation (Griffith et al., 2023). This depends on the nature of the products of the abiotic reactions, which could be more prone to biodegradation or more recalcitrant than the estrogen substrate. For instance, chlorine is commonly applied as a disinfectant during water treatment due to its strong oxidation capability, continuous disinfection, and low cost (Deborde and von Gunten, 2008). However, chlorine often produces undesirable intermediate byproducts and potentially carcinogenic halogenated byproducts (Richardson and Kimura, 2016).
Estrogen degradation during chlorination was reported mostly in lab-scale studies, showing the formation of intermediate products and disinfection byproducts (Deborde et al., 2004; Lee and von Gunten, 2009). At an initial concentration of 50 mg/L, E2 was completely degraded after 10 min during chlorination at pH 7.5 (Hu et al., 2003). In batch tests, E3 degradation during chlorination followed a second-order kinetic (Zhao et al., 2014). Moreover, E1 degradation with free chlorine produced chlorinated estrone by-products (Nakamura et al., 2007). The kinetics and products of E2 chlorination were also studied in a pilot-scale water distribution system (He et al., 2016; Li et al., 2017). Further studies by Li et al. (2017) on the kinetics and mechanism of E2 chlorination in a pilot-scale water distribution system showed that chlorination degraded E2 in three stages: (1) halogenation of the aromatic ring, (2) cleavage of the benzene moiety and chlorine substitution, (3) production of trihalomethanes and halogenated acetic acids from the phenolic intermediates.
During E3 chlorination in a pilot-scale water distribution system, E2, E1, 16α-OH-E1, and chlorinated E3, in addition to four types of disinfection by-products including trihalomethanes, haloacetonitriles, haloketones, and chloral hydrate were identified as intermediate products (Dong et al., 2019). Based on the identified products, three possible pathways for E3 chlorination were proposed (Figure 13). During water chlorination, EE2 was transformed into 4-chloro-EE2 and 2,4-dichloro-EE2. In the presence of Br−, the bromo-analogs of the chloro-EE2 derivatives were additionally produced (Lee and von Gunten, 2009). In drinking water, chlorination of E2 produced several transformation products including 2,4-dichloro-E2, monochloro-E1, 2,4-dichloro-E1, and by-product such as 4-[2-(2,6-dichloro-3-hydroxyphenyl)ethyl]-7α-methyloctahydroinden-5-one (Hu et al., 2003).
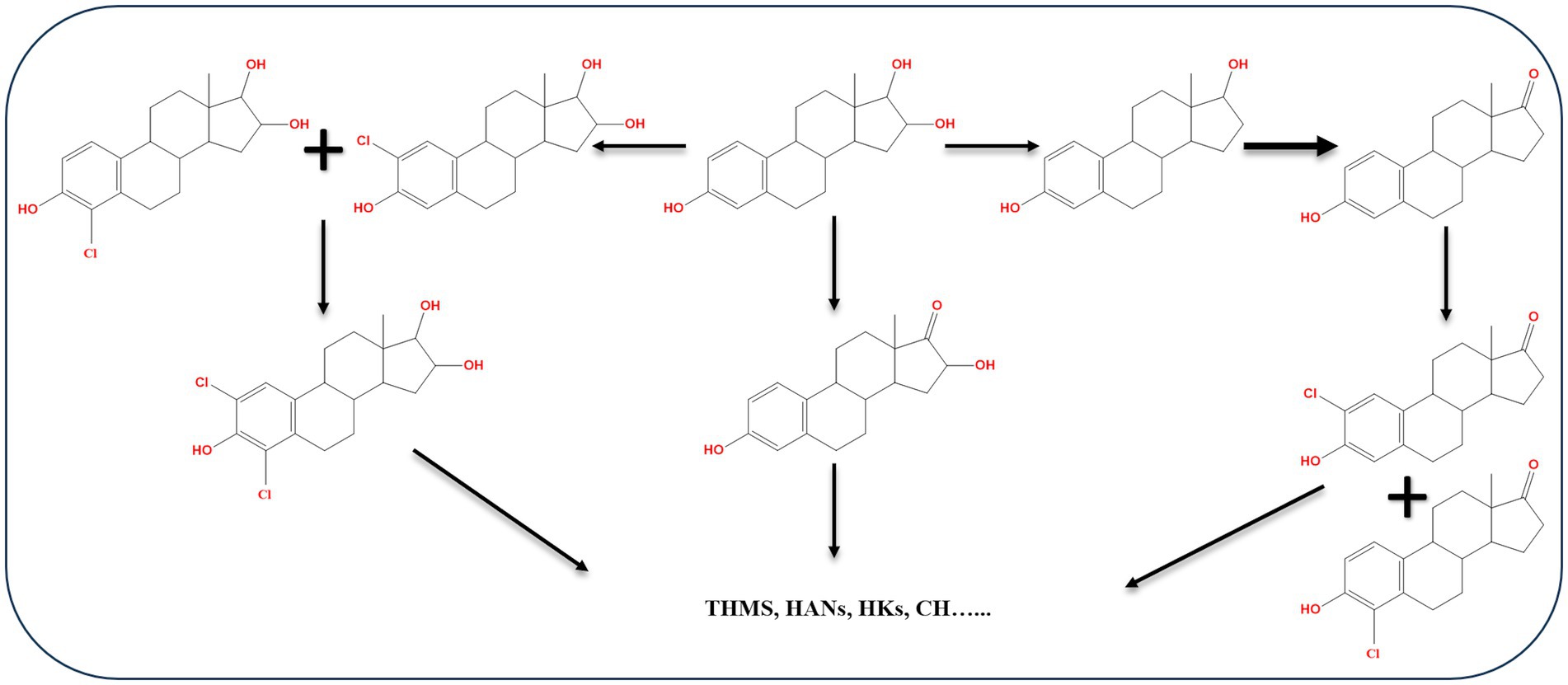
Figure 13. Proposed E3 chlorination pathways. THMs, trihalomethane; HANs, haloacetonitrile; HKs, haloketone; CH, chloral hydrate (Dong et al., 2019).
Chlorinated E1, such as 2-chloro-E1, 4-chloro-E1, 2,4-dichloro-E1, were detected in a sewage treatment plant effluent in Japan. Moreover, in a buffer of pH 7 and 9 combining E1 with sodium hypochlorite, the same chlorinated products were produced in addition to 1,4-estradiene-3,17-dione derivatives resulting from the reaction of 2,4-dichloro-E1 and sodium hypochlorite (Nakamura et al., 2007). Ten oxidation products of E2 were identified during treatment with chloramine in a water distribution system. The identified products were formed by addition or loss of a functional group at different positions. The proposed reactions include electrophilic substitution of the ortho-position of the phenol moiety, elimination reaction between C9 and C11, and a hydrogenation reaction on the benzene ring (He et al., 2016). In general, biodegradation studies on halogenated estrogens are very scarce. Recently, the findings of Griffith et al. (2023) on biodegradation kinetics of free and halogenated estrogens in river water-sediment microcosms suggested that halogenated estrogens degrade more slowly than the corresponding free estrogens. Halogenated E1 derivatives were detected as transformation products of halogenated E2. While detection of halogenated E1 derivatives suggests subsequent metabolism via the most common 4,5-seco pathway, further studies are needed to unravel how dehalogenation of the A-ring occurs before hydroxylation and ring cleavage. Moreover, biodegradation kinetics and pathways in wastewater microcosms containing a mixture of free and halogenated estrogens is worth investigating.
Beside chlorination, advanced oxidation processes are being investigated as a wastewater treatment to eliminate endocrine-disrupting chemicals including estrogens. These processes include ozonation, photocatalysis, Fenton and Fenton-like processes, and they depend on the production of highly reactive hydroxyl radicals to oxidize organic pollutants (Wang and Wang, 2016; Dewil et al., 2017). There is also an interest in the development of chemical-free electrochemical oxidation processes (Dewil et al., 2017). As these processes depend on very strong oxidizing agents, namely hydroxyl radicals, they can lead to several hydroxylated and ring-cleavage transformation products. Even the ethinyl group of EE2 can be removed (Reis et al., 2023).
In the environment, estrogens are prone to direct and indirect photo-transformation reactions, which can play a role in estrogen removal (Chen et al., 2013; Adeel et al., 2017). The most common photo-transformation products of estrogen are hydroxylated derivatives bearing hydroxy groups at various positions (Mazellier et al., 2008; Li Puma et al., 2010; Caupos et al., 2011; Chen et al., 2013; Zhao et al., 2019). In addition, some ring cleavage products were also reported during estrogen photodegradation (Perondi et al., 2020). Susceptibility of estrogens to photodegradation can be exploited to develop concerted photocatalysis-biocatalysis technologies that can enhance estrogens removal, as shown for conjugated E2. Photocatalysis can contribute to deconjugation transformations which make free estrogens available for microbial degradation (Ding et al., 2024).
5 Concluding remarks and future research directions
Steroid estrogens are endocrine-disrupting chemicals that can be harmful to public health and the aquatic environment at extremely low concentrations. It has been recognized that effluents from WWTPs are major source of environmental pollution with estrogens. Hence, proper treatments of wastewater are instrumental to avoid environmental pollution with these hazardous compounds. Microbial biodegradation has been studied as a key process for the elimination of estrogens from wastewater. Despite the very low aqueous solubility and relatively inert chemical structure, several bacteria are endowed with metabolic capabilities to utilize these challenging substrates as a carbon and energy source for growth, while others can only perform specific biotransformations on estrogens. Estrogen-degrading bacteria were isolated from engineered and natural environments such as activated sludge, soil, compost, and sandy aquifers, and the majority of them belong to Actinomycetota and Pseudomonadota. Most studies focused on the aerobic degradation of E1 and E2, and the involved enzymes, genes and pathways were investigated in some detail. On the contrary, EE2 was much less studied and even fewer studies are available for E3. This may be due to the very high structural similarity among the different estrogens, which could lead to the perception that E3 and EE2 biodegradation may proceed similarly to E1and E2. Furthermore, the relatively high recalcitrance of EE2, due to the ethinyl group, rendered the isolation of EE2-degrading microbes a challenge. However, the large diversity of the reported biotransformation reactions and biodegradation pathways justifies more in-depth studies to unravel specific biodegradation mechanisms and key degrading microbes for E3 and EE2.
Despite the high structural similarity between steroid estrogens, diverse estrogen degradation pathways have been reported. A common feature of those pathways is the start by modification of a certain ring of the estrogen substrate followed by cleavage of that ring. However, for most of the proposed degradation pathways or biotransformation reactions, biochemical and molecular evidence at the level of the involved enzymes and genes is lacking. Exceptionally, only the 4,5-seco pathway of aerobic estrogen degradation has been thoroughly investigated. Interestingly, biochemical and molecular studies on the 4,5-seco pathway revealed some differences between Actinomycetota and Pseudomonadota, which can be exploited as functional biomarkers for environmental and ecological assessment of estrogen biodegradation. Anaerobic estrogen degradation has been rarely studied, and key mechanisms have been only recently elucidated, indicating that anaerobic estrogen degradation proceeds through the 2,3-seco pathway after a unique initial retro conversion of estrogens into androgens.
Regardless of oxygen availability, both the aerobic and anaerobic pathways converge at a common central metabolite of steroid catabolism by bacteria; namely, HIP. The latter is then degraded through a common central metabolic pathway. One of the characteristic features of estrogen biodegradation is the interconversion of one compound to another as shown in Figure 14. This feature might have some implications on the environmental fate of estrogens due to changes in their concentrations and potential substrate preference differences among the involved microbial communities, particularly in the presence of estrogen mixtures. In addition, abiotic degradation of estrogens occurs in the environment via photochemical reactions which can potentially impact their susceptibility to biodegradation/biotransformation process. Nonetheless, biodegradation of the products of abiotic estrogen degradation and transformation have been scarcely investigated.
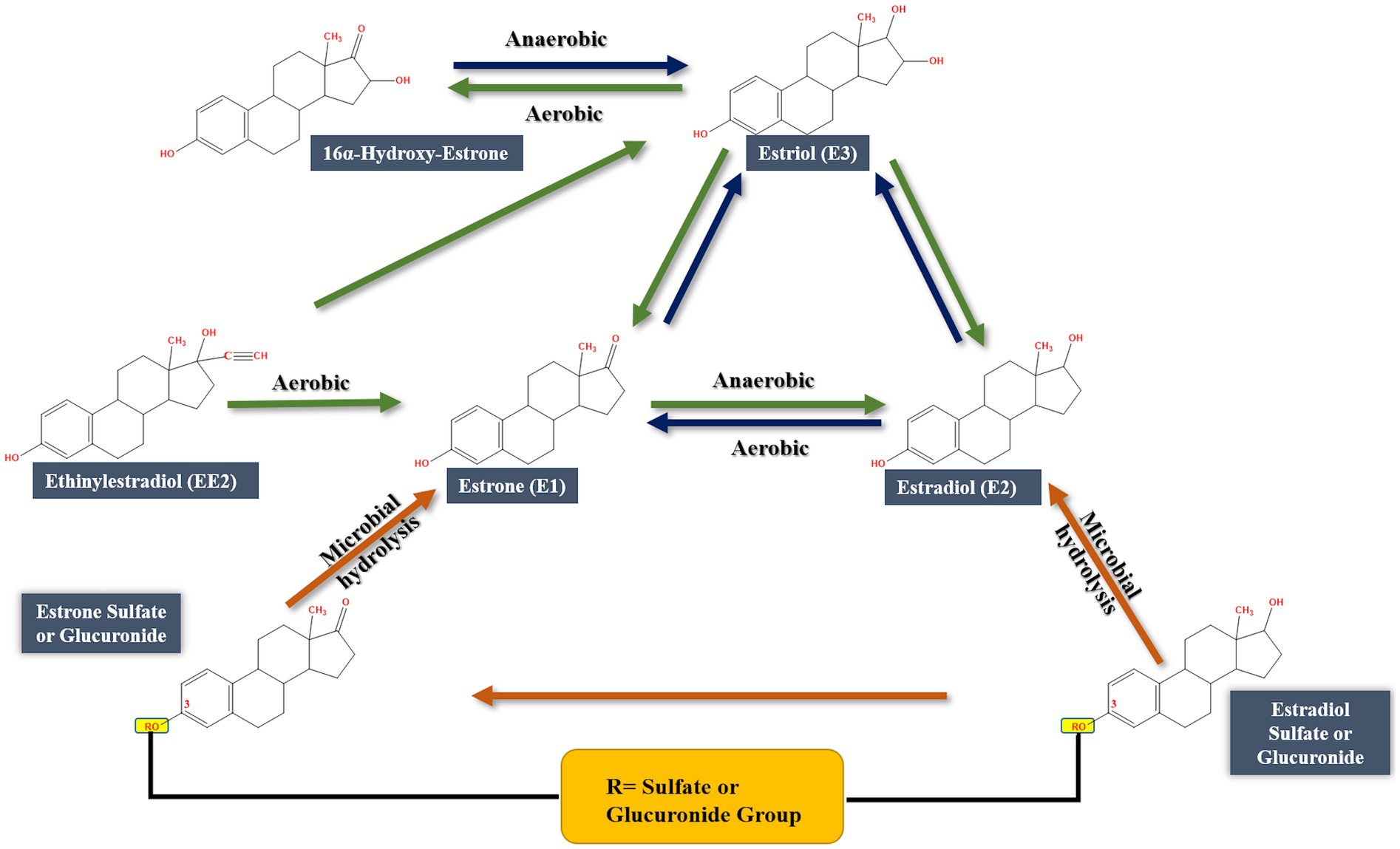
Figure 14. Interconversions between different steroid estrogens (Adeel et al., 2017; Yu et al., 2019).
Studies on estrogen biodegradation applied mostly pure cultures and single estrogen substrates, which is not necessarily environmentally relevant where there is a mixture of estrogens and other carbon sources, and microbes exist as complex and heterogeneous communities. Moreover, there are still many knowledge gaps on the mechanisms and microbes involved in E3 and EE2 biodegradation. It was clear that E3 and EE2 need further studies to unravel their degradation mechanisms and the involved microbial communities. Furthermore, future research should focus more on microbial consortia as they are prevailing in natural and engineered ecosystems. However, it is crucial to investigate the functionality of isolated microbial consortia in polluted environmental matrices such as raw sewage and activated sludge. Moreover, stability of estrogen-degrading microbial consortia in terms of functional resilience/resistance needs to be properly addressed. In addition, more work is needed to elucidate estrogen biodegradation process in environmental matrices to gain more realistic and conclusive data for in-situ applications. A holistic or systems approach, encompassing proteomics, transcriptomics, metabolomics, comparative genomics, metagenomics, and metaproteomics, can be instrumental in future studies. It is also crucial to unravel how estrogen uptake occurs in estrogen-degrading microbes as knowledge of estrogen uptake systems is largely lacking. The impact of abiotic estrogen degradation on the biological treatment of estrogen-polluted environments deserves proper attention as it might augment or prohibit the biodegradation/biotransformation processes. Some limitations of most of the available studies need to be rectified in future. The use of more environmentally relevant estrogen concentrations, the use of estrogen mixtures (with and without other steroids), and the effect of non-steroid co-substrates are of particular interest.
Despite active research on estrogens–degrading bacteria and accumulation of knowledge during recent years, it is obvious that we have just scratched the surface of this field. Further intensive research is needed on different aspects of estrogens biodegradation. This is a pre-requisite to comprehensively understand the underlying mechanisms and elucidate the structural and functional diversity of estrogens-degrading microbial communities, which should enable efficient estrogens removal from engineered and natural polluted environments via implementation of well-designed bioremediation technologies.
Author contributions
JH: Writing – original draft, Writing – review & editing, Investigation. WI: Writing – original draft, Project administration, Writing – review & editing, Supervision, Conceptualization. Y-RC: Writing – review & editing, Conceptualization, Writing – original draft. AB: Writing – review & editing, Writing – original draft, Conceptualization.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Acknowledgments
Our studies on estrogen biodegradation were supported by the Arabian Gulf University and Sheikh Zayed bin Sultan Al Nahyan Chair for Environmental Sciences, hosted by Arabian Gulf University (Bahrain). The authors also wish to thank University of Bahrain and the Ministry of Science and Technology of Taiwan for generous support.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1630636/full#supplementary-material
References
Adeel, M., Song, X., Wang, Y., Francis, D., and Yang, Y. (2017). Environmental impact of estrogens on human, animal and plant life: a critical review. Environ. Int. 99, 107–119. doi: 10.1016/j.envint.2016.12.010
AlDhafiri, S., Chiang, Y. R., EL Nayal, A. M., Abed, R. M. M., Abotalib, N., and Ismail, W. (2022). Temporal compositional shifts in an activated sludge microbiome during estrone biodegradation. Environ. Sci. Pollut. Res. 29, 32702–32716. doi: 10.1007/s11356-021-18185-9
Alfonso Murillo-Tovar, M., Vergara-Sánchez, J., Albeiro Saldarriaga-Noreña, H., Luisa García-Betancourt, M., Torres-Segundo, C., López-Velázquez, K., et al. (2025). Endocrine-disrupting compound in Mexican aquatic environments. In: Advances in Water Resources Science, eds. Tarhouni, J., Farrukh, M. A., Ali, S. London: IntechOpen. doi: 10.5772/intechopen.1010168
Aris, A. Z., Shamsuddin, A. S., and Praveena, S. M. (2014). Occurrence of 17α-ethynylestradiol (EE2) in the environment and effect on exposed biota: a review. Environ. Int. 69, 104–119. doi: 10.1016/j.envint.2014.04.011
Baronti, C., Curini, R., D’Ascenzo, G., Di Corcia, A., Gentili, A., and Samperi, R. (2000). Monitoring natural and synthetic estrogens at activated sludge sewage treatment plants and in a receiving river water. Environ. Sci. Technol. 34, 5059–5066. doi: 10.1021/es001359q
Baskin, L., Sinclair, A., Derpinghaus, A., Cao, M., Li, Y., Overland, M., et al. (2022). Estrogens and development of the mouse and human external genitalia. Differentiation 119, 82–106. doi: 10.1016/j.diff.2020.09.004
Ben, W., Zhu, B., Yuan, X., Zhang, Y., Yang, M., and Qiang, Z. (2017). Transformation and fate of natural estrogens and their conjugates in wastewater treatment plants: influence of operational parameters and removal pathways. Water Res. 124, 244–250. doi: 10.1016/j.watres.2017.07.065
Bergman, Å., Heindel, J., Jobling, S., Kidd, K., and Zoeller, R. T. (2012). State-of-the-science of endocrine disrupting chemicals, 2012. Toxicol. Lett. 211:S3. doi: 10.1016/j.toxlet.2012.03.020
Bergstrand, L. H., Cardenas, E., Holert, J., van Hamme, J. D., and Mohn, W. W. (2016). Delineation of steroid-degrading microorganisms through comparative genomic analysis. mBio 7, e00166–e00116. doi: 10.1128/mBio.00166-16
Bilal, M., Barceló, D., and Iqbal, H. M. N. (2021). Occurrence, environmental fate, ecological issues, and redefining of endocrine disruptive estrogens in water resources. Sci. Total Environ. 800:149635. doi: 10.1016/j.scitotenv.2021.149635
Boll, M., and Estelmann, S. (2020). “Catabolic pathways and enzymes involved in the anaerobic degradation of polycyclic aromatic hydrocarbons” in Anaerobic utilization of hydrocarbons, oils, and lipids. Handbook of hydrocarbon and lipid microbiology. ed. M. Boll (Cham: Springer).
Boll, M., Estelmann, S., and Heider, J. (2020a). “Anaerobic degradation of hydrocarbons: mechanisms of hydrocarbon activation in the absence of oxygen” in Anaerobic utilization of hydrocarbons, oils, and lipids. Handbook of hydrocarbon and lipid microbiology. ed. M. Boll (Cham: Springer).
Boll, M., Estelmann, S., and Heider, J. (2020b). “Catabolic pathways and enzymes involved in the anaerobic degradation of monocyclic aromatic compounds” in Anaerobic utilization of hydrocarbons, oils, and lipids. Handbook of hydrocarbon and lipid microbiology. ed. M. Boll (Cham: Springer).
Bolong, N., Ismail, A. F., Salim, M. R., and Matsuura, T. (2009). A review of the effects of emerging contaminants in wastewater and options for their removal. Desalination 239, 229–246. doi: 10.1016/j.desal.2008.03.020
Cajthaml, T., Kresinová, Z., Svobodová, K., Sigler, K., and Rezanka, T. (2009). Microbial transformation of synthetic estrogen 17 α-ethinylestradiol. Environ. Pollut. 157, 3325–3335. doi: 10.1016/j.envpol.2009.06.027
Casabon, I., Zhu, S. H., Otani, H., Liu, J., Mohn, W. W., and Eltis, L. D. (2013). Regulation of the KstR2 regulon of Mycobacterium tuberculosis by a cholesterol catabolite. Mol. Microbiol. 89, 1201–1212. doi: 10.1111/mmi.12340
Caupos, E., Mazellier, P., and Croue, J. P. (2011). Photodegradation of estrone enhanced by dissolved organic matter under simulated sunlight. Water Res. 45, 3341–3350. doi: 10.1016/j.watres.2011.03.047
Chen, Y., Chen, T., Xu, W., Yu, X., Tang, X., Wang, Y., et al. (2025). Characterization of a rapid 17β-estradiol-degrading strain, Microbacterium proteolyticum ZJSU01. Appl. Microbiol. Biotechnol. 109:112. doi: 10.1007/s00253-025-13480-8
Chen, Y. L., Fu, H. Y., Lee, T. H., Shih, C. J., Huang, L., Wang, Y. S., et al. (2018). Estrogen degraders and estrogen degradation pathway identified in an activated sludge. Appl. Environ. Microbiol. 84, e00001–e00018. doi: 10.1128/aem.00001-18
Chen, Y. L., Yu, C. P., Lee, T. H., Goh, K. S., Chu, K. H., Wang, P. H., et al. (2017). Biochemical mechanisms and catabolic enzymes involved in bacterial estrogen degradation pathways. Cell Chem. Biol. 24, 712–724.e7. doi: 10.1016/j.chembiol.2017.05.012
Chen, Y., Zhang, K., and Zuo, Y. (2013). Direct and indirect photodegradation of estriol in the presence of humic acid, nitrate and iron complexes in water solutions. Sci. Total Environ. 463–464, 802–809. doi: 10.1016/j.scitotenv.2013.06.026
Chiang, Y. R., Wei, S. T., Wang, P. H., Wu, P. H., and Yu, C. P. (2020). Microbial degradation of steroid sex hormones: implications for environmental and ecological studies. Microb. Biotechnol. 13, 926–949. doi: 10.1111/1751-7915.13504
Coello-Garcia, T., Curtis, T. P., Mrozik, W., and Davenport, R. J. (2019). Enhanced estrogen removal in activated sludge processes through the optimization of the hydraulic flow pattern. Water Res. 164:114905. doi: 10.1016/j.watres.2019.114905
Colucci, M. S., and Topp, E. (2001). Persistence of estrogenic hormones in agricultural soils: II. 17α-ethynylestradiol. J. Environ. Qual. 30, 2077–2080. doi: 10.2134/jeq2001.2077
Coombre, R. G., Tsong, Y. Y., Hamilton, P. B., and Sih, C. J. (1966). Mechanisms of steroid oxidation by microorganisms. X. Oxidative cleavage of estrone. J. Biol. Chem. 241, 1587–1595. doi: 10.1016/s0021-9258(18)96753-0
Crowe, A. M., Casabon, I., Brown, K. L., Liu, J., Lian, J., Rogalski, J. C., et al. (2017). Catabolism of the last two steroid rings in mycobacterium tuberculosis and other bacteria. mBio 8, e00321–e00317. doi: 10.1128/mBio.00321-17
Cui, J., Shen, Y., and Li, R. (2013). Estrogen synthesis and signaling pathways during aging: from periphery to brain. Trends Mol. Med. 19, 197–209. doi: 10.1016/j.molmed.2012.12.007
Czajka, C. P., and Londry, K. L. (2006). Anaerobic biotransformation of estrogens. Sci. Total Environ. 367, 932–941. doi: 10.1016/j.scitotenv.2006.01.021
de Lorenzo, V. (2017). Seven microbial bio-processes to help the planet. Microb. Biotechnol. 10, 995–998. doi: 10.1111/1751-7915.12816
Deborde, M., Rabouan, S., Gallard, H., and Legube, B. (2004). Aqueous chlorination kinetics of some endocrine disruptors. Environ. Sci. Technol. 38, 5577–5583. doi: 10.1021/es040006e
Deborde, M., and von Gunten, U. (2008). Reactions of chlorine with inorganic and organic compounds during water treatment-kinetics and mechanisms: a critical review. Water Res. 42, 13–51. doi: 10.1016/j.watres.2007.07.025
Della Greca, M., Pinto, G., Pistillo, P., Pollio, A., Previtera, L., and Temussi, F. (2008). Biotransformation of ethinylestradiol by microalgae. Chemosphere 70, 2047–2053. doi: 10.1016/j.chemosphere.2007.09.011
Dewil, R., Mantzavinos, D., Poulios, I., and Rodrigo, M. A. (2017). New perspectives for advanced oxidation processes. J. Environ. Manag. 195, 93–99. doi: 10.1016/j.jenvman.2017.04.010
Ding, X., Yu, Q., Ren, H., and Geng, J. (2024). Degradation of conjugated estrogen in visible light-driven intimately coupled photocatalysis and biodegradation system. Bioresour. Technol. 406:131045. doi: 10.1016/j.biortech.2024.131045
Diviccaro, S., Giatti, S., Cioffi, L., Chrostek, G., and Melcangi, R. C. (2025). The gut-microbiota-brain axis: focus on gut steroids. J. Neuroendocrinol. 37:e13471. doi: 10.1111/jne.13471
Dong, F., Li, C., Crittenden, J., Zhang, T., Lin, Q., He, G., et al. (2019). Sulfadiazine destruction by chlorination in a pilot-scale water distribution system: kinetics, pathway, and bacterial community structure. J. Environ. Manag. 366, 88–97. doi: 10.1016/j.jhazmat.2018.11.096
Elliott, S. M., Brigham, M. E., Lee, K. E., Banda, J. A., Choy, S. J., Gefell, D. J., et al. (2017). Contaminants of emerging concern in tributaries to the Laurentian Great Lakes: I Patterns of occurrence. PLoS One 12:e0182868. doi: 10.1371/journal.pone.0182868
Fahrbach, M., Kuever, J., Meinke, R., Kämpfer, P., and Hollender, J. (2006). Denitratisoma oestradiolicum gen. nov., sp. nov., a 17 β-oestradiol-degrading, denitrifying betaproteobacterium. Int. J. Syst. Evol. Microbiol. 56, 1547–1552. doi: 10.1099/ijs.0.63672-0
Fahrbach, M., Kuever, J., Remesch, M., Huber, B. E., Kämpfer, P., Dott, W., et al. (2008). Steroidobacter denitrificans gen. Nov., sp. nov., a steroidal hormone-degrading gammaproteobacterium. Int. J. Syst. Evol. Microbiol. 58, 2215–2223. doi: 10.1099/ijs.0.65342-0
Fernández, L., Louvado, A., Esteves, V. I., Gomes, N. C. M., Almeida, A., and Cunha, Â. (2017). Biodegradation of 17β-estradiol by bacteria isolated from deep sea sediments in aerobic and anaerobic media. J. Environ. Manag. 323, 359–366. doi: 10.1016/j.jhazmat.2016.05.029
Fernandez, S. V., Russo, I. H., and Russo, J. (2006). Estradiol and its metabolites 4-hydroxyestradiol and 2-hydroxyestradiol induce mutations in human breast epithelial cells. Int. J. Cancer 118, 1862–1868. doi: 10.1002/ijc.21590
Fischer, K., and Majewsky, M. (2014). Cometabolic degradation of organic wastewater micropollutants by activated sludge and sludge-inherent microorganisms. Appl. Microbiol. Biotechnol. 98, 6583–6597. doi: 10.1007/s00253-014-5826-0
Flint, S. L. (2019). The genomic evaluation of estrogen degradation by Rhodococcus equi ATCC 13557. PhD Thesis, Newcastle University.
Fuentes, N., and Silveyra, P. (2019). Estrogen receptor signaling mechanisms. Adv. Protein Chem. Struct. Biol. 116, 135–170. doi: 10.1016/bs.apcsb.2019.01.001
Fuhrman, B. J., Feigelson, H. S., Flores, R., Gail, M. H., Xu, X., Ravel, J., et al. (2014). Associations of the fecal microbiome with urinary estrogens and estrogen metabolites in postmenopausal women. J. Clin. Endocrinol. Metab. 99, 4632–4640. doi: 10.1210/jc.2014-2222
Fujii, K., Kikuchi, S., Satomi, M., Ushio-Sata, N., and Morita, N. (2002). Degradation of 17β-estradiol by a gram-negative bacterium isolated from activated sludge in a sewage treatment plant in Tokyo, Japan. Appl. Microbiol. Biotechnol. 68, 2057–2060. doi: 10.1128/AEM.68.4.2057-2060.2002
Gadupudi, C. K., Rice, L., Xiao, L., and Kantamaneni, K. (2021). Endocrine disrupting compounds removal methods from wastewater in the United Kingdom: a review. Science 3, 1–10. doi: 10.3390/sci3010011
Ge, H., Yang, L., Li, B., Feng, Y., Wang, S., Zheng, Y., et al. (2021). A comparative study on the biodegradation of 17β-estradiol by Candida utilis CU-2 and Lactobacillus casei LC-1. Front. Energy Res. 9:850. doi: 10.3389/fenrg.2021.661850
Gonsioroski, A., Mourikes, V. E., and Flaws, J. A. (2020). Endocrine disruptors in water and their effects on the reproductive system. Int. J. Mol. Sci. 21:929. doi: 10.3390/ijms21061929
Griffith, D. R., Carolan, M., Gutierrez, M. M., Romig, A., Garcia-Diaz, N., Hutchinson, C. P., et al. (2023). Microbial degradation of free and halogenated estrogens in river water-sediment microcosms. Environ. Sci. Technol. 57, 10782–10791. doi: 10.1021/acs.est.3c00801
Gröning, J., Held, C., Garten, C., Claußnitzer, U., Kaschabek, S. R., and Schlömann, M. (2007). Transformation of diclofenac by the indigenous microflora of river sediments and identification of a major intermediate. Chemosphere 69, 509–516. doi: 10.1016/j.chemosphere.2007.03.037
Guo, Y., Guan, Z., Lin, H., and Ou, X. (2021). Enhanced biodegradation of 17α-ethinylestradiol by rhamnolipids in sediment/water systems. Environ. Chem. 18, 300–310. doi: 10.1071/EN20175
Haiyan, R., Shulan, J., Ud Din Ahmad, N., Dao, W., and And Chengwu, C. (2007). Degradation characteristics and metabolic pathway of 17α-ethynylestradiol by Sphingobacterium sp. JCR5. Chemosphere 66, 340–346. doi: 10.1016/j.chemosphere.2006.04.064
Hanselman, T. A., Graetz, D. A., and Wilkie, A. C. (2003). Manure-borne estrogens as potential environmental contaminants: a review. Environ. Sci. Technol. 37, 5471–5478. doi: 10.1021/es034410+
Hao, P., Lv, Z., Pan, H., Zhang, J., Wang, L., Zhu, Y., et al. (2024). Characterization and low-temperature biodegradation mechanism of 17β-estradiol-degrading bacterial strain Rhodococcus sp. RCBS9. Environ. Res. 240:117513. doi: 10.1016/j.envres.2023.117513
Hao, P., Pan, H., Lv, Z., Zhang, J., Wang, L., Zhu, Y., et al. (2023). Characterization of 17β-estradiol-degrading enzyme from Microbacterium sp. MZT7 and its function on E2 biodegradation in wastewater. Microb. Cell Factories 22:116. doi: 10.1186/s12934-023-02119-w
Hao, P., Wu, S., Zhang, X., Gou, C., Wang, Y., Wang, L., et al. (2022). Characterization and degradation pathways of Microbacterium resistens MZT7, a novel 17β-estradiol-degrading bacterium. Int. J. Environ. Res. Public Health 19:11097. doi: 10.3390/ijerph191711097
Hashem, J. S., Ismail, W., Chiang, Y. R., Sangal, V., Hentati, D., Abotalib, N., et al. (2025). A native bacterial consortium degrades estriol in domestic sewage and activated sludge via the 4,5-seco pathway and requires estriol to retain its biodegradation phenotype. Microbiol. Spectr 26:e0074125. doi: 10.1128/spectrum.00741-25
He, G., Li, C., Dong, F., Zhang, T., Chen, L., Cizmas, L., et al. (2016). Chloramines in a pilot-scale water distribution system: transformation of 17β-estradiol and formation of disinfection byproducts. Water Res. 106, 41–50. doi: 10.1016/j.watres.2016.09.047
Holert, J., Cardenas, E., Bergstrand, L. H., Zaikova, E., Hahn, A. S., Hallam, S. J., et al. (2018). Metagenomes reveal global distribution of bacterial steroid catabolism in natural, engineered, and host environments. mBio 9:17. doi: 10.1128/mBio.02345-17
Horinouchi, M., Hayashi, T., and Kudo, T. (2012). Steroid degradation in Comamonas testosteroni. J. Steroid Biochem. Mol. Biol. 129, 4–14. doi: 10.1016/j.jsbmb.2010.10.008
Hsiao, T., Chen, Y., Meng, M., Chuang, M., Horinouchi, M., Hayashi, T., et al. (2021). Mechanistic and phylogenetic insights into actinobacteria-mediated oestrogen biodegradation in urban estuarine sediments. Microb. Biotechnol. 14, 1212–1227. doi: 10.1111/1751-7915.13798
Hsiao, T. H., Chen, P. H., Wang, P. H., Brandon-Mong, G. J., Li, C. W., Horinouchi, M., et al. (2023). Harnessing microbial phylum-specific molecular markers for assessment of environmental estrogen degradation. Sci. Total Environ. 896:165152. doi: 10.1016/j.scitotenv.2023.165152
Hsiao, T. H., Lee, T. H., Chuang, M. R., Wang, P. H., Meng, M., Horinouchi, M., et al. (2022). Identification of essential β-oxidation genes and corresponding metabolites for oestrogen degradation by actinobacteria. Microb. Biotechnol. 15, 949–966. doi: 10.1111/1751-7915.13921
Hu, J., Cheng, S., Aizawa, T., Terao, Y., and Kunikane, S. (2003). Products of aqueous chlorination of 17beta-estradiol and their estrogenic activities. Environ. Sci. Technol. 37, 5665–5670. doi: 10.1021/es034324+
Huang, S.-L., Chen, H., Hu, A., Tuan, N. N., and Yu, C.-P. (2014). Draft genome sequence of Pseudomonas nitroreducens strain TX1, which degrades nonionic surfactants and estrogen-like alkylphenols. Genome Announc. 2, e01262–e01213. doi: 10.1128/genomeA.01262-13
Ibero, J., Galán, B., Rivero-Buceta, V., and García, J. L. (2020). Unraveling the 17β-estradiol degradation pathway in Novosphingobium tardaugens NBRC 16725. Front. Microbiol. 11:588300. doi: 10.3389/fmicb.2020.588300
Ismail, W., and Gescher, J. (2012). Epoxy coenzyme a thioester pathways for degradation of aromatic compounds. Appl. Environ. Microbiol. 78, 5043–5051. doi: 10.1128/AEM.00633-12
Jacoby, C., Peller, L., Wenzler, J., Luttermann, M., Seiche, W., Breit, B., et al. (2025). Ring a cleaving beta-diketone hydrolase is a key enzyme of steroid degradation in anaerobic bacteria. Environ. Microbiol. 27:e70034. doi: 10.1111/1462-2920.70034
Järvenpää, P., Kosunen, T., Fotsis, T., and Adlercreutz, H. (1980). In vitro metabolism of estrogens by isolated intestinal microorganisms and by human faecal microflora. J. Steroid Biochem. 13, 345–349. doi: 10.1016/0022-4731(80)90014-x
Karavolos, M. H., Winzer, K., Williams, P., and Khan, C. M. A. (2013). Pathogen espionage: multiple bacterial adrenergic sensors eavesdrop on host communication systems. Mol. Microbiol. 87, 455–465. doi: 10.1111/mmi.12110
Ke, J., Zhuang, W., Gin, K. Y. H., Reinhard, M., Hoon, L. T., and Tay, J. H. (2007). Characterization of estrogen-degrading bacteria isolated from an artificial sandy aquifer with ultrafiltered secondary effluent as the medium. Appl. Microbiol. Biotechnol. 75, 1163–1171. doi: 10.1007/s00253-007-0923-y
Koh, Y. K. K., Chiu, T. Y., Boobis, A., Cartmell, E., Scrimshaw, M. D., and Lester, J. N. (2008). Treatment and removal strategies for estrogens from wastewater. Environ. Technol. 29, 245–267. doi: 10.1080/09593330802099122
Kolodziej, E. P., Gray, J. L., and Sedlak, D. L. (2003). Quantification of steroid hormones with pheromonal properties in municipal wastewater effluent. Environ. Toxicol. Chem. 22, 2622–2629. doi: 10.1897/03-42
Kostich, M. S., and Lazorchak, J. M. (2008). Risks to aquatic organisms posed by human pharmaceutical use. Sci. Total Environ. 389, 329–339. doi: 10.1016/j.scitotenv.2007.09.008
Kurisu, F., Ogura, M., Saitoh, S., Yamazoe, A., and Yagi, O. (2010). Degradation of natural estrogen and identification of the metabolites produced by soil isolates of Rhodococcus sp. and Sphingomonas sp. J. Biosci. Bioeng. 109, 576–582. doi: 10.1016/j.jbiosc.2009.11.006
Kverka, M., and Stepan, J. J. (2024). Associations among estrogens, the gut microbiome and osteoporosis. Curr. Osteoporos. Rep. 23:2. doi: 10.1007/s11914-024-00896-w
Lambert, M. R., Giller, G. S. J., Barber, L. B., Fitzgerald, K. C., and Skelly, D. K. (2015). Suburbanization, estrogen contamination, and sex ratio in wild amphibian populations. Proc. Natl. Acad. Sci. USA 112, 11881–11886. doi: 10.1073/pnas.1501065112
Laurenson, J. P., Bloom, R. A., Page, S., and Sadrieh, N. (2014). Ethinyl estradiol and other human pharmaceutical estrogens in the aquatic environment: a review of recent risk assessment data. AAPS J. 16, 299–310. doi: 10.1208/s12248-014-9561-3
Lee, H. B., and Liu, D. (2002). Degradation of 17β-estradiol and its metabolites by sewage bacteria. Water Air Soil Pollut. 134, 351–366. doi: 10.1023/A:1014117329403
Lee, Y., and von Gunten, U. (2009). Transformation of 17alpha-ethinylestradiol during water chlorination: effects of bromide on kinetics, products, and transformation pathways. Environ. Sci. Technol. 43, 480–487. doi: 10.1021/es8023989
Lee, Y. C., Wang, L. M., Xue, Y. H., Ge, N. C., Yang, X. M., and Chen, G. H. (2006). Natural estrogens in the surface water of Shenzhen and the sewage discharge of Hong Kong. Hum. Ecol. Risk. Assess. 12, 301–312. doi: 10.1080/10807030500533394
Leech, D. M., Snyder, M. T., and Wetzel, R. G. (2009). Natural organic matter and sunlight accelerate the degradation of 17β-estradiol in water. Sci. Total Environ. 407, 2087–2092. doi: 10.1016/J.SCITOTENV.2008.11.018
Lei, Z., Zheng, J., Luo, H., Li, Y. Y., and Chen, R. (2025). Anaerobic electrochemical membrane bioreactors strengthen steroid estrogen removal: the pivotal role of planktonic sludge. Environ. Sci. Technol. 59, 14518–14527. doi: 10.1021/acs.est.5c01995
Lennartz, M., Amezada, D., Höflmayer, D., Dwertmann Rico, S., von Bargen, C., and Kind, S. (2024). Steroidogenic acute regulatory protein is a useful marker for sex-cord-stroma tumors and normal and neoplastic adrenocortical tissue. Arch. Pathol. Lab Med. 148:1327. doi: 10.5858/arpa.2023-0281-oa
Lerdsuwanrut, N., Zamani, R., and Akrami, M. (2025). Environmental and human health risks of estrogenic compounds: a critical review of sustainable management practices. Sustainability 17:491. doi: 10.3390/su17020491
Li, C., Dong, F., Crittenden, J. C., Luo, F., Chen, X., and Zhao, T. (2017). Kinetics and mechanism of 17β-estradiol chlorination in a pilot-scale water distribution systems. Chemosphere 178, 73–79. doi: 10.1016/j.chemosphere.2017.03.039
Li, J., Fu, J., Zhang, H., Li, Z., Ma, Y., and Wu, M. (2013a). Spatial and seasonal variations of occurrences and concentrations of endocrine disrupting chemicals in unconfined and confined aquifers recharged by reclaimed water: a field study along the Chaobai River, Beijing. Sci. Total Environ. 450–451, 162–168. doi: 10.1016/j.scitotenv.2013.01.089
Li, J., Jiang, L., Liu, X., and Lv, J. (2013b). Adsorption and aerobic biodegradation of four selected endocrine disrupting chemicals in soil–water system. Int. Biodeterior. Biodegrad. 76, 3–7. doi: 10.1016/j.ibiod.2012.06.004
Li, S., Liu, J., Sun, K., Yang, Z., and Ling, W. (2020). Degradation of 17β-estradiol by Novosphingobium sp. ES2-1 in aqueous solution contaminated with tetracyclines. Environ. Pollut. 260:114063. doi: 10.1016/j.envpol.2020.114063
Li, D., Liu, R., Wang, M., Peng, R., Fu, S., and Fu, A. (2022). 3β-hydroxysteroid dehydrogenase expressed by gut microbes degrades testosterone and is linked to depression in males. Cell Host Microbe 30, 329–339.e5. doi: 10.1016/j.chom.2022.01.001
Li Puma, G., Puddu, V., Tsang, H. K., Gora, A., and Toepfer, B. (2010). Photocatalytic oxidation of multicomponent mixtures of estrogens (estrone (E1), 17β-estradiol (E2), 17α-ethynylestradiol (EE2) and estriol (E3)) under UVA and UVC radiation: photon absorption, quantum yields and rate constants independent of photon absorption. Appl Catal B 99, 388–397. doi: 10.1016/j.apcatb.2010.05.015
Li, S., Wang, Y., Sun, K., Li, Y., Lu, C., and Gao, Y. (2024). Fe(III)-aided Novosphingobium sp. ES2-1 regulates molecular mechanisms of 17β-estradiol biodegradation. Environ. Sci. Technol. 58, 22245–22256. doi: 10.1021/acs.est.4c08818
Liehr, J. G., Wan-Fen, F., Sirbasku, D. A., and Ari-Ulubelen, A. (1986). Carcinogenicity of catechol estrogens in Syrian hamsters. J. Steroid Biochem. 24, 353–356. doi: 10.1016/0022-4731(86)90080-4
Liu, Z. H., Ito, M., Kanjo, Y., and Yamamoto, A. (2009b). Profile and removal of endocrine disrupting chemicals (EDCs) in two municipal wastewater treatment plants by using an ER(AR) competitive ligand binding assay and chemical analyses. J. Environ. Sci. 21, 900–906. doi: 10.1016/S1001-0742(08)62356-6
Liu, Z. H., Kanjo, Y., and Mizutani, S. (2009a). Removal mechanisms for endocrine disrupting compounds (EDCs) in wastewater treatment—physical means, biodegradation, and chemical advanced oxidation: a review. Sci. Total Environ. 407, 731–748. doi: 10.1016/j.scitotenv.2008.08.039
Liu, Z. H., Kanjo, Y., and Mizutani, S. (2009c). Urinary excretion rates of natural estrogens and androgens from humans, and their occurrence and fate in the environment: a review. Sci. Total Environ. 407, 4975–4985. doi: 10.1016/j.scitotenv.2009.06.001
Liu, J., Liu, J., Xu, D., Ling, W., Li, S., and Chen, M. (2016). Isolation, immobilization, and degradation performance of the 17β-estradiol-degrading bacterium Rhodococcus sp. JX-2. Water Air Soil Pollut. 227, 1–13. doi: 10.1007/s11270-016-3122-6
Liu, N., Shi, Y. E., Li, J., Zhu, M., and Zhang, T. (2020). Isolation and characterization of a new highly effective 17β-estradiol-degrading Gordonia sp. Strain R9. 3 Biotech 10, 1–11. doi: 10.1007/s13205-020-2156-z
Liu, N., Shi, Y. E., Li, J., Zhu, M., and Zhang, T. (2021). Identification and genome analysis of Comamonas testosteroni strain JLU460ET, a novel steroid-degrading bacterium. 3 Biotech 11, 1–12. doi: 10.1007/s13205-021-02949-8
Liu, X., Wang, Z., Wang, X., Liu, J., and Waigi, M. G. (2024). Conversion of estriol to estrone: a bacterial strategy for the catabolism of estriol. Ecotoxicol. Environ. Saf. 280:116564. doi: 10.1016/j.ecoenv.2024.116564
Liyanage, S., Lay, M., Glasgow, G., Tanner, C., Craggs, R., and Northcott, G. (2024). Nature-based solutions for removal of steroid estrogens in wastewater. Front. Microbiol. 15:1437795. doi: 10.3389/fmicb.2024.1437795
López-Velázquez, K., Guzmán-Mar, J. L., Saldarriaga-Noreña, H. A., Murillo-Tovar, M. A., Hinojosa-Reyes, L., and Villanueva-Rodríguez, M. (2021). Occurrence and seasonal distribution of five selected endocrine-disrupting compounds in wastewater treatment plants of the metropolitan area of Monterrey, Mexico: the role of water quality parameters. Environ. Pollut. 269:116223. doi: 10.1016/j.envpol.2020.116223
López-Velázquez, K., Guzmán-Mar, J. L., Saldarriaga-Noreña, H. A., Murillo-Tovar, M. A., and Villanueva-Rodríguez, M. (2023). Ecological risk assessment associated with five endocrine-disrupting compounds in wastewater treatment plants of Northeast Mexico. Environ. Sci. Pollut. Res. 30, 30714–30726. doi: 10.1007/s11356-022-24322-9
López-Velázquez, K., Ronderos-Lara, J. G., Saldarriaga-Noreña, H. A., Murillo-Tovar, M. A., Villanueva-Rodríguez, M., Guzmán-Mar, J. L., et al. (2025). Endocrine-disrupting compounds in urban rivers of the southern border of Mexico: occurrence and ecological risk assessment. Emerg Contam. 11:100456. doi: 10.1016/j.emcon.2024.100456
Luo, X., Zhao, B., Yao, J., Peng, M., Mao, L., Zhang, W., et al. (2024). Degradation performance of estrogen during anaerobic digestion of pig manure. Waste Biomass Valor. 15, 2625–2635. doi: 10.1007/s12649-023-02286-2
Ma, W., Sun, J., Li, Y., Lun, X., Shan, D., Nie, C., et al. (2018). 17α-ethynylestradiol biodegradation in different river-based groundwater recharge modes with reclaimed water and degradation-associated community structure of bacteria and archaea. J. Environ. Sci. (China) 64, 51–61. doi: 10.1016/j.jes.2016.11.022
Maeng, S. K., Choi, B. G., Lee, K. T., and Song, K. G. (2013). Influences of solid retention time, nitrification and microbial activity on the attenuation of pharmaceuticals and estrogens in membrane bioreactors. Water Res. 47, 3151–3162. doi: 10.1016/j.watres.2013.03.014
Markle, J. G. M., Frank, D. N., Mortin-Toth, S., Robertson, C. E., Feazel, L. M., Rolle-Kampczyk, U., et al. (2013). Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science 339, 1084–1088. doi: 10.1126/science.1233521
Mazellier, P., Méité, L., and De Laat, J. (2008). Photodegradation of the steroid hormones 17beta-estradiol (E2) and 17alpha-ethinylestradiol (EE2) in dilute aqueous solution. Chemosphere 73, 1216–1223. doi: 10.1016/j.chemosphere.2008.07.046
Mensitieri, F., Bosso, A., Dal Piaz, F., Charlier, B., Notomista, E., Izzo, V., et al. (2023). Bioconversion of 4-hydroxyestradiol by extradiol ring-cleavage dioxygenases from Novosphingobium sp. PP1Y. Sci. Rep. 13, 1–13. doi: 10.1038/s41598-023-28908-2
Miyairi, S., and Fishman, J. (1985). Radiometric analysis of oxidative reactions in aromatization by placental microsomes: presence of differential isotope effects. J. Biol. Chem. 260, 320–325. doi: 10.1016/s0021-9258(18)89734-4
Muller, M., Combalbert, S., Delgenès, N., Bergheaud, V., Rocher, V., Benoît, P., et al. (2010b). Occurrence of estrogens in sewage sludge and their fate during plant-scale anaerobic digestion. Chemosphere 81, 65–71. doi: 10.1016/j.chemosphere.2010.06.062
Muller, M., Patureau, D., Godon, J. J., Delgenès, J. P., and Hernandez-Raquet, G. (2010a). Molecular and kinetic characterization of mixed cultures degrading natural and synthetic estrogens. Appl. Microbiol. Biotechnol. 85, 691–701. doi: 10.1007/S00253-009-2160-Z
Nakai, S., Yamamura, A., Tanaka, S., Shi, J., Nishikawa, M., Nakashimada, Y., et al. (2011). Pathway of 17β-estradiol degradation by Nitrosomonas europaea and reduction in 17β-estradiol-derived estrogenic activity. Environ. Chem. Lett. 9, 1–6. doi: 10.1007/s10311-010-0308-9
Nakamura, H., Kuruto-Niwa, R., Uchida, M., and Terao, Y. (2007). Formation of chlorinated estrones via hypochlorous disinfection of wastewater effluent containing estrone. Chemosphere 66, 1441–1448. doi: 10.1016/j.chemosphere.2006.09.011
Nieto, M. R., Rus, M. J., Areal-Quecuty, V., Lupian Lopez, D., and Simon-Soro, A. (2025). Menopausal shift on women’s health and microbial niches. NPJ Womens Health 3:50. doi: 10.1038/s44294-024-00050-y
O’Grady, D., Evangelista, S., and Yargeau, V. (2009). Removal of aqueous 17α-ethinylestradiol by Rhodococcus species. Environ. Eng. Sci. 26, 1393–1400. doi: 10.1089/ees.2008.0272
Olivera, E. R., and Luengo, J. M. (2019). Steroids as environmental compounds recalcitrant to degradation: genetic mechanisms of bacterial biodegradation pathways. Genes 10, 1–12. doi: 10.3390/genes10070512
Palma, T. L., and Costa, M. C. (2024). Biodegradation of 17α-ethinylestradiol by strains of Aeromonas genus isolated from acid mine drainage. Clean Technol. 6, 1–24. doi: 10.3390/cleantechnol6010008
Palme, R., Fischer, P., Schildorfer, H., and Ismail, M. N. (1996). Excretion of infused 14C-steroid hormones via faeces and urine in domestic livestock. Anim. Reprod. Sci. 43, 43–63. doi: 10.1016/0378-4320(95)01458-6
Pauwels, B., Wille, K., Noppe, H., De Brabander, H., Van De Wiele, T., Verstraete, W., et al. (2008). 17α-ethinylestradiol cometabolism by bacteria degrading estrone, 17β-estradiol and estriol. Biodegradation 19, 683–693. doi: 10.1007/s10532-007-9173-z
Peng, W., Lin, S., Deng, Z., and Liang, R. (2023). Bioaugmentation removal and microbiome analysis of the synthetic estrogen 17α-ethynylestradiol by Pseudomonas citronellolis SJTE-3 under hostile conditions and environmental samples. Chemosphere 317:137893. doi: 10.1016/j.chemosphere.2023.137893
Perondi, T., Michelon, W., Basso, A., Bohrer, J. K., Viancelli, A., Fonseca, T. G., et al. (2020). Degradation of estriol (E3) and transformation pathways after applying photochemical removal processes in natural surface water. Water Sci. Technol. 82, 1445–1453. doi: 10.2166/wst.2020.411
Pratush, A., Yang, Q., Peng, T., Huang, T., and Hu, Z. (2020b). Identification of non-accumulating intermediate compounds during estrone (E1) metabolism by a newly isolated microbial strain BH2-1 from mangrove sediments of the South China Sea. Environ. Sci. Pollut. Res. 27, 5097–5107. doi: 10.1007/s11356-019-06894-1
Pratush, A., Ye, X., Yang, Q., Kan, J., Peng, T., Wang, H., et al. (2020a). Biotransformation strategies for steroid estrogen and androgen pollution. Appl. Microbiol. Biotechnol. 104, 2385–2409. doi: 10.1007/s00253-020-10374-9
Predieri, B., Alves, C. A. D., and Iughetti, L. (2022). New insights on the effects of endocrine-disrupting chemicals on children. J. Pediatr. 98, S73–S85. doi: 10.1016/j.jped.2021.11.003
Racz, L., and Goel, R. K. (2010). Fate and removal of estrogens in municipal wastewater. J. Environ. Monit. 12, 58–70. doi: 10.1039/b917298J
Rashtian, J., Chavkin, D. E., and Merhi, Z. (2019). Water and soil pollution as determinant of water and food quality/contamination and its impact on female fertility. Reprod. Biol. Endocrinol. 17:5. doi: 10.1186/s12958-018-0448-5
Regnault, C., Usal, M., Veyrenc, S., Couturier, K., Batandier, C., Bulteau, A. L., et al. (2018). Unexpected metabolic disorders induced by endocrine disruptors in Xenopus tropicalis provide new lead for understanding amphibian decline. Proc. Natl. Acad. Sci. USA 115, E4416–E4425. doi: 10.1073/pnas.1721267115
Reis, R., Dhawle, R., Du Pasquier, D., Tindall, A. J., Frontistis, Z., Mantzavinos, D., et al. (2023). Electrochemical degradation of 17α-ethinylestradiol: transformation products, degradation pathways and in vivo assessment of estrogenic activity. Environ. Int. 176:107992. doi: 10.1016/j.envint.2023.107992
Rhee, J. S., Kim, B. M., Lee, B. Y., Hwang, U. K., Lee, Y. S., and Lee, J. S. (2014). Cloning of circadian rhythmic pathway genes and perturbation of oscillation patterns in endocrine disrupting chemicals (EDCs)-exposed mangrove killifish Kryptolebias marmoratus. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 164, 11–20. doi: 10.1016/j.cbpc.2014.04.001
Richardson, S. D., and Kimura, S. Y. (2016). Water analysis: emerging contaminants and current issues. Anal. Chem. 88, 546–582. doi: 10.1021/acs.analchem.5b04493
Ridlon, J. M., Ikegawa, S., Alves, J. M., Zhou, B., Kobayashi, A., Iida, T., et al. (2013). Clostridium scindens: a human gut microbe with a high potential to convert glucocorticoids into androgens. J. Lipid Res. 54, 2437–2449. doi: 10.1194/jlr.M038869
Ros, O., Vallejo, A., Olivares, M., Etxebarria, N., and Prieto, A. (2016). Determination of endocrine disrupting compounds in fish liver, brain, and muscle using focused ultrasound solid–liquid extraction and dispersive solid phase extraction as clean-up strategy. Anal. Bioanal. Chem. 408, 5689–5700. doi: 10.1007/s00216-016-9697-3
Roudbari, A., and Rezakazemi, M. (2018). Hormones removal from municipal wastewater using ultrasound. AMB Express 8:91. doi: 10.1186/s13568-018-0621-4
Sami, N., Afzal, B., Rehmanji, M., Naaz, H., Yasin, D., Jutur, P. P., et al. (2023). Insight into the mechanism of estrone biodegradation by Spirulina CPCC-695. Environ. Dev. Sustain. 27:739. doi: 10.1007/s10668-023-03873-y
Shah, B. A., Malhotra, H., Papade, S. E., Dhamale, T., Ingale, O. P., Kasarlawar, S. T., et al. (2024). Microbial degradation of contaminants of emerging concern: metabolic, genetic and omics insights for enhanced bioremediation. Front. Bioeng. Biotechnol. 12:1470522. doi: 10.3389/fbioe.2024.1470522
Shareef, A., Angove, M. J., Wells, J. D., and Johnson, B. B. (2006). Aqueous solubilities of estrone, 17β-estradiol, 17α-ethynylestradiol, and bisphenol a. J. Chem. Eng. Data 51, 879–881. doi: 10.1021/je050318c
Shi, J., Fujisawa, S., Nakai, S., and Hosomi, M. (2004). Biodegradation of natural and synthetic estrogens by nitrifying activated sludge and ammonia-oxidizing bacterium Nitrosomonas europaea. Water Res. 38, 2323–2330. doi: 10.1016/j.watres.2004.02.022
Shi, J. H., Suzuki, Y., Lee, B. D., Nakai, S., and Hosomi, M. (2002). Isolation and characterization of the ethynylestradiol-biodegrading microorganism Fusarium proliferatum strain HNS-1. Water Sci. Technol. 45, 175–179. doi: 10.2166/wst.2002.0424
Stefanakis, A. I. (2018). “Introduction to constructed wetland technology” in Constructed wetlands for industrial wastewater treatment. ed. A. I. Stefanakis (Chichester: Wiley), 1–21.
Stefanakis, A. (2022). Constructed wetlands for wastewater treatment in hot and arid climates. Wetlands: Ecology, Conservation and Management, vol 7. Cham: Springer.
Stikker, A. (1998). Water today and tomorrow: prospects for overcoming scarcity. Futures 30, 43–62. doi: 10.1016/S0016-3287(98)00005-6
Thathola, P., Agnihotri, V., Pandey, A., and Upadhyay, S. K. (2024). Biodegradation of steroid hormone estriol by Pseudomonas proteolytica GBPI_Hb61, a psychrotolerant Himalayan bacterium. Water Air Soil Pollut. 235:289. doi: 10.1007/s11270-024-07079-4
Tian, K., Meng, Q., Li, S., Chang, M., Meng, F., Yu, Y., et al. (2022). Mechanism of 17β-estradiol degradation by Rhodococcus equi via the 4,5-seco pathway and its key genes. Environ. Pollut. 312:120021. doi: 10.1016/j.envpol.2022.120021
Torres, N. H., Santos, G., De, O. S., Romanholo Ferreira, L. F., Américo-Pinheiro, J. H. P., Eguiluz, K. I. B., et al. (2021). Environmental aspects of hormones estriol, 17β-estradiol and 17α-ethinylestradiol: electrochemical processes as next-generation technologies for their removal in water matrices. Chemosphere 267:888. doi: 10.1016/j.chemosphere.2020.128888
Tran, N. H., Urase, T., Ngo, H. H., Hu, J., and Ong, S. L. (2013). Insight into metabolic and cometabolic activities of autotrophic and heterotrophic microorganisms in the biodegradation of emerging trace organic contaminants. Bioresour. Technol. 146, 721–731. doi: 10.1016/j.biortech.2013.07.083
Trenti, A., Tedesco, S., Boscaro, C., Trevisi, L., Bolego, C., and Cignarella, A. (2018). Estrogen, angiogenesis, immunity and cell metabolism: solving the puzzle. Int. J. Mol. Sci. 19:859. doi: 10.3390/ijms19030859
Ueda, M., Shintani, K., Nakanishi-Anjyuin, A., Nakazawa, M., Kusuda, M., Nakatani, F., et al. (2012). A protein from Pleurotus eryngii var. tuoliensis C.J. Mou with strong removal activity against the natural steroid hormone, estriol: purification, characterization, and identification as a laccase. Enzym. Microb. Technol. 51, 402–407. doi: 10.1016/j.enzmictec.2012.08.010
Vader, J. S., van Ginkel, C. G., Sperling, F. M. G. M., de Jong, J., de Boer, W., de Graaf, J. S., et al. (2000). Degradation of ethinylestradiol by nitrifying activated sludge. Chemosphere 41, 1239–1243. doi: 10.1016/s0045-6535(99)00556-1
Vazakidou, P., Evangelista, S., Li, T., Lecante, L. L., Rosenberg, K., Koekkoek, J., et al. (2024). The profile of steroid hormones in human fetal and adult ovaries. Reprod. Biol. Endocrinol. 22, 1–14. doi: 10.1186/s12958-024-01233-7
vom Steeg, L. G., and Klein, S. L. (2017). Sex steroids mediate bidirectional interactions between hosts and microbes. Horm. Behav. 88, 45–51. doi: 10.1016/j.yhbeh.2016.10.016
Wang, Z., Chen, M., Liu, N., Zhao, Y., Ru, J., Qin, C., et al. (2023). Common and unique testosterone and 17β-estradiol degradation mechanisms in Comamonas testosteroni JLU460ET by transcriptome analysis. Front. Microbiol. 14:1238855. doi: 10.3389/fmicb.2023.1238855
Wang, P. H., Chen, Y. L., Wei, S. T. S., Wu, K., Lee, T. H., Wu, T. Y., et al. (2020). Retroconversion of estrogens into androgens by bacteria via a cobalamin-mediated methylation. Proc. Natl. Acad. Sci. USA 117, 1395–1403. doi: 10.1073/pnas.1914380117
Wang, Y., Shao, H., Zhu, S., Tian, K., Qiu, Q., and Huo, H. (2019a). Degradation of 17β-estradiol and products by a mixed culture of Rhodococcus equi DSSKP-R-001 and Comamonas testosteroni QYY20150409. Biotechnol. Biotechnol. Equip. 33, 268–277. doi: 10.1080/13102818.2019.1568913
Wang, Y., Sun, Q., Li, Y., Wang, H., Wu, K., and Yu, C. P. (2019b). Biotransformation of estrone, 17β-estradiol, and 17α-ethynylestradiol by four species of microalgae. Ecotoxicol. Environ. Saf. 180, 723–732. doi: 10.1016/j.ecoenv.2019.05.061
Wang, J., and Wang, S. (2016). Removal of pharmaceuticals and personal care products (PPCPs) from wastewater: a review. J. Environ. Manag. 182, 620–640. doi: 10.1016/j.jenvman.2016.07.049
Wang, P. H., Wu, T. Y., Chen, Y. L., Gicana, R. G., Lee, T. H., Chen, M. J., et al. (2025). Bacterial estrogenesis without oxygen: wood-Ljungdahl pathway likely contributed to the emergence of estrogens in the biosphere. Proc. Natl. Acad. Sci. USA 122:e2422930122. doi: 10.1073/pnas.2422930122
Wang, P., Zheng, D., and Liang, R. (2019). Isolation and characterization of an estrogen-degrading Pseudomonas putida strain SJTE-1. 3 Biotech 9, 1–11. doi: 10.1007/s13205-018-1537-z
Weber, S., Leuschner, P., Kämpfer, P., Dott, W., and Hollender, J. (2005). Degradation of estradiol and ethinyl estradiol by activated sludge and by a defined mixed culture. Appl. Microbiol. Biotechnol. 67, 106–112. doi: 10.1007/s00253-004-1693-4
Writer, J. H., Ryan, J. N., Keefe, S. H., and Barber, L. B. (2012). Fate of 4-nonylphenol and 17β-estradiol in the Redwood River of Minnesota. Environ. Sci. Technol. 46, 860–868. doi: 10.1021/es2031664
Wu, K., Lee, T. H., Chen, Y. L., Wang, Y. S., Wang, P. H., Yu, C. P., et al. (2019). Metabolites involved in aerobic degradation of the a and B rings of estrogen. Appl. Microbiol. Biotechnol. 85, 123–135. doi: 10.1128/AEM.02223-18
Ye, X., Peng, T., Feng, J., Yang, Q., Pratush, A., Xiong, G., et al. (2019). A novel dehydrogenase 17β-HSDx from Rhodococcus sp. P14 with potential application in bioremediation of steroids-contaminated environment. J. Environ. Manag. 362, 170–177. doi: 10.1016/j.jhazmat.2018.09.023
Yi, T., and Harper, W. F. (2007). The link between nitrification and biotransformation of 17α-ethinylestradiol. Environ. Sci. Technol. 41, 4311–4316. doi: 10.1021/es070102q
Ying, G. G., and Kookana, R. S. (2003). Degradation of five selected endocrine-disrupting chemicals in seawater and marine sediment. Environ. Sci. Technol. 37, 1256–1260. doi: 10.1021/es0262232
Ying, G. G., Kookana, R. S., and Dillon, P. (2003). Sorption and degradation of selected endocrine-disrupting chemicals in aquifer material. Water Res. 37, 3785–3791. doi: 10.1016/s0043-1354(03)00261-6
Ying, G. G., Kookana, R. S., and Ru, Y. J. (2002). Occurrence and fate of hormone steroids in the environment. Environ. Int. 28, 545–551. doi: 10.1016/S0160-4120(02)00075-2
Yoshimoto, T., Nagai, F., Fujimoto, J., Watanabe, K., Mizukoshi, H., Makino, T., et al. (2004). Degradation of estrogens by Rhodococcus zopfii and Rhodococcus equi isolates from activated sludge in wastewater treatment plants. Appl. Microbiol. Biotechnol. 70, 5283–5289. doi: 10.1128/aem.70.9.5283-5289.2004
Yu, C. P., and Chu, K. H. (2009). Occurrence of pharmaceuticals and personal care products along the west prong little Pigeon River in East Tennessee, USA. Chemosphere 75, 1281–1286. doi: 10.1016/j.chemosphere.2009.03.043
Yu, C. P., Deeb, R. A., and Chu, K. H. (2013). Microbial degradation of steroidal estrogens. Chemosphere 91, 1225–1235. doi: 10.1016/j.chemosphere.2013.01.112
Yu, W., Du, B., Yang, L., Zhang, Z., Yang, C., Yuan, S., et al. (2019). Occurrence, sorption, and transformation of free and conjugated natural steroid estrogens in the environment. Environ. Sci. Pollut. Res. 26, 9443–9468. doi: 10.1007/s11356-019-04402-z
Yu, C. P., Roh, H., and Chu, K. H. (2007). 17β-estradiol-degrading bacteria isolated from activated sludge. Environ. Sci. Technol. 41, 486–492. doi: 10.1021/es060923f
Zhang, W., Jia, J., Yang, Y., Ye, D., Li, Y., Li, D., et al. (2025). Estradiol metabolism by gut microbiota in women's depression pathogenesis: inspiration from nature. Front. Psych. 16:1505991. doi: 10.3389/fpsyt.2025.1505991
Zhang, C., Li, Y., Wang, C., Niu, L., and Cai, W. (2015). Occurrence of endocrine-disrupting compounds in the aqueous environment and their bacterial degradation: a review. Crit. Rev. Environ. Sci. Technol. 46, 1–59. doi: 10.1080/10643389.2015.1061881
Zhang, W., Yu, Q., and Geng, J. (2024). Biodegradation of conjugated estrogens in wastewater treatment: species, mechanisms, and influencing factors. Biotechnol. Environ. 1:15. doi: 10.1186/s44314-024-00014-1
Zhang, T., and Zhang, H. (2022). Microbial consortia are needed to degrade soil pollutants. Microorganisms 10:261. doi: 10.3390/microorganisms10020261
Zhao, X., Grimes, K. L., Colosi, L. M., and Lung, W. S. (2019). Attenuation, transport, and management of estrogens: a review. Chemosphere 230, 462–478. doi: 10.1016/j.chemosphere.2019.05.086
Zhao, T. T., Li, C., and Yu, T. C. (2014). Chlorination degradation of estriol under various pH values in aqueous solution. Appl. Mech. Mater. 535, 713–717. doi: 10.4028/www.scientific.net/AMM.535.713
Zheng, W., Li, X., Yates, S. R., and Bradford, S. A. (2012). Anaerobic transformation kinetics and mechanism of steroid estrogenic hormones in dairy lagoon water. Environ. Sci. Technol. 46, 5471–5478. doi: 10.1021/es301551h
Zheng, R., Zhang, Y., Cheng, S., and Xiao, T. (2023). Environmental estrogens shape disease susceptibility. Int. J. Hyg. Environ. Health 249:125. doi: 10.1016/j.ijheh.2023.114125
Zhu, Q. M., Wang, C., Liu, J. W., Zhang, R., Xin, X. L., Zhang, J., et al. (2024). Degradation profile of environmental pollutant 17β-estradiol by human intestinal fungus Aspergillus niger RG13B1 and characterization of genes involved in its degradation. J. Hazard. Mater. 461:132617. doi: 10.1016/j.jhazmat.2023.132617
Keywords: environment, wastewater, estradiol, pollution, endocrine-disrupting chemicals, 4,5-seco pathway
Citation: Hashem JS, Ismail W, Chiang Y-R and Bekhit AA (2025) Diversity of estrogen biodegradation pathways and application in environmental bioremediation. Front. Microbiol. 16:1630636. doi: 10.3389/fmicb.2025.1630636
Edited by:
Yating Luo, Zhejiang Agriculture and Forestry University, ChinaReviewed by:
Khirbet López Velázquez, Polytechnic University of Tapachula, MexicoWanli Peng, Shanghai Jiao Tong University, China
Copyright © 2025 Hashem, Ismail, Chiang and Bekhit. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wael Ismail, d2FlbGFtZUBhZ3UuZWR1LmJo
 Jaleela S. Hashem1
Jaleela S. Hashem1 Wael Ismail
Wael Ismail Yin-Ru Chiang
Yin-Ru Chiang Adnan A. Bekhit
Adnan A. Bekhit