- Center of Clinical Laboratory Medicine, The Fourth Affiliated Hospital of Soochow University (Suzhou Dushu Lake Hospital), Suzhou, Jiangsu, China
Microascus spp., globally distributed fungi, are increasingly recognized as causative agents of rare and refractory invasive infections in immunocompromised populations, with approximately 50 cases reported worldwide. This study aimed to characterize the epidemiological features, antifungal resistance profiles, and identification strategies for Microascus-associated pulmonary infections. Ten Microascus isolates were collected from respiratory specimens of patients with pulmonary infections (2021–2024). Clinical characteristics were analyzed, and antifungal susceptibility testing (AFST) was performed following CLSI M38-A3 guidelines. Taxonomic identification integrated MALDI-TOF mass spectrometry, and multilocus phylogenetic analysis (ITS/EF-1α/TUB). Nine of ten cases occurred in hematopoietic stem cell transplant (HSCT) recipients, with concurrent infections by cytomegalovirus (7/9), Pseudomonas aeruginosa (5/9), Corynebacterium striatum (5/9), or Aspergillus spp. (5/9). Three patients succumbed to refractory infections. Morphologically, colonies exhibited olive-to-black concentric rings, floccose hyphae with dark granules, and basally swollen conidiophores producing oval or pear-shaped conidia in chains. Nine isolates were M. gracilis, and one was M. cirrosus. All strains demonstrated resistance to fluconazole, amphotericin B, and flucytosine (MIC >64 μg/mL) but high sensitivity to terbinafine (MIC ≤0.125 μg/mL). MALDI-TOF accurately identified M. gracilis (100%), while M. cirrosus required sequencing for confirmation. Multilocus sequence typing revealed a monophyletic cluster among M. gracilis isolates. Microascus spp. represent underdiagnosed pathogens in HSCT-associated fungal pneumonia, often complicated by polymicrobial infections. Terbinafine demonstrates promising in vitro efficacy against multidrug-resistant strains. A multimodal diagnostic approach combining morphology, MALDI-TOF, and sequencing is essential for species-level identification.
Introduction
Microascus spp., classified within the family Microascaceae (order Microascales, class Sordariomycetes), represent emerging opportunistic pathogens associated with life-threatening infections globally. These fungi predominantly afflict immunocompromised hosts, including hematopoietic stem cell transplant (HSCT) recipients, critically ill patients, and individuals undergoing prolonged immunosuppressive therapy (Schoeppler et al., 2015). High mortality rates persist due to diagnostic delays, intrinsic antifungal resistance, and limited therapeutic options. Clinical manifestations range from localized cutaneous abscesses to disseminated infections involving lungs, sinuses, cardiac valves, and the central nervous system, often demonstrating poor prognosis despite aggressive interventions (Sandoval-Denis et al., 2016). Recent years have witnessed increased reports of severe pulmonary, hematogenous, and deep-tissue infections, paralleling the growing population of transplant recipients (Baddley et al., 2000; Miossec et al., 2011; Ding et al., 2020; Brasch et al., 2018). However, systematic analyses of clinical features and microbiological characteristics remain scarce, with current literature predominantly limited to case reports.
Ecologically diverse, Microascus spp. typically exhibit saprophytic or weakly parasitic lifestyles, colonizing air, soil, animal feces, plant debris, and humid environments. M. Sandoval-Denis et al. conducted a study on 141 clinical and environmental isolates by integrating morphological, physiological, and multigene sequence analyses of ITS, LSU, EF-1α, and β-tubulin genes. Based on these analyses and applying the Genealogical Concordance Phylogenetic Species Recognition (GCPSR) criterion, they revealed that Microascus and Scopulariopsis belong to two distinct evolutionary lineages. Additionally, they analyzed 54 species from 12 genera within the family Microascaceae, as well as one species from the family Graphiaceae. The results demonstrated that Microascus and Scopulariopsis are polyphyletic genera, with their species distributed across multiple distantly related clades (Sandoval-Denis et al., 2016). According to MycoBank (as of September 2025), 98 species names are currently accepted in Microascus; however, some of these taxa are based on lost specimens or invalid names. To better delineate species boundaries and elucidate evolutionary relationships within the genus, Wei et al. (2024) conducted multi-locus phylogenetic analyses using a combined dataset of four gene regions (ITS, LSU, tef1, and tub2), incorporating expanded taxon sampling and morphological assessments. Their results resolved 49 phylogenetic species in Microascus with high statistical support. Differentiated by ascus morphology (peridium structure) and ascospore characteristics (size, symmetry, surface ornamentation) [drfungus.org/Microascus Species]. Critical gaps persist in morphological identification criteria, antifungal susceptibility patterns, and genomic characterization, contributing to diagnostic and therapeutic challenges.
Our team recently reported China’s first fatal M. cirrosus-associated invasive pulmonary infection in an HSCT recipient, with whole-genome sequencing data deposited in NCBI (PRJNA835605) (Cheng et al., 2023). Subsequent clinical surveillance identified nine additional M. gracilis infections. This study systematically analyzes the clinical and microbiological profiles of Microascus-related severe pneumonia to inform improved management strategies.
Materials and methods
Strain collection and clinical data
Ten Microascus isolates (preliminarily identified by Bruker MALDI-TOF MS and confirmed via Sanger sequencing) were collected from patients at the Fourth Affiliated Hospital of Soochow University (September 2021–September 2024). Clinical data included: (1) Demographics (sex, age, Underlying disease); (2) Time to infection post-transplantation; (3) Antifungal prophylaxis post-transplantation; (4) Specimen type; (5) Imaging findings; (6) Co-pathogens; (7) Antimicrobial treatment strategy; and (8) outcomes (improvement or deterioration).
Case data collection from the literature: Case reports published between 1990 and 2025 were retrieved from the PubMed database using the keywords “Microascus,” “infection,” and “human.” Information including patient age, sex, pathogen species, infection site, underlying condition/transplant type, treatment regimen, and clinical outcome was extracted from each eligible report.
Instruments and reagents
Bruker MALDI-TOF MS (Bruker Daltonics, Germany), SLAN-96P real-time PCR system (Hongshi Medical, China). Reagents: Sabouraud dextrose agar (Autobio, China), lactophenol cotton blue stain (Besio Biotechnology), antifungal agents (amphotericin B, caspofungin acetate, terbinafine, fluconazole, flucytosine; BBI Solutions), and fungal DNA extraction kit (Sangon Biotech).
Strain isolation and culture
Sputum/bronchoalveolar lavage fluid (BALF) specimens were inoculated onto Sabouraud dextrose agar (SDA) and incubated at 35 °C or 28 °C for 3–5 days. Brown colonies (<1 cm diameter) were subcultured on fresh SDA for morphological characterization. Extended incubation (≤14 days) enabled assessment of colony morphology and microscopic structures. The morphological characteristics were observed under an Olympus CX-33 optical microscope (400× and 1,000× magnification) using lactophenol cotton blue staining, and images were captured with the MShot Image Analysis System software.
MALDI-TOF identification
According to the filamentous fungi identification SOP recommended by Bruker Daltonics, fungal colonies were liquid-cultured for 48 h, harvested by centrifugation (10,000 ×g, 10 min), and washed twice with 1 mL deionized water (vortexed, then centrifuged at 10,000 ×g for 10 min). The pellet was resuspended in 300 μL deionized water and 900 μL absolute ethanol, vortexed, and centrifuged (10,000 ×g, 10 min). After air-drying, the pellet was treated with 50 μL 70% formic acid (thoroughly mixed), followed by addition of 50 μL acetonitrile and vigorous mixing. The final suspension was centrifuged (10,000 ×g, 10 min), and 1 μL supernatant was spotted onto a MALDI target plate, air-dried at room temperature, and overlaid with 1 μL matrix solution (α-cyano-4-hydroxycinnamic acid in 50% acetonitrile/2.5% trifluoroacetic acid). MALDI-TOF MS analysis was performed using the Bruker Biotyper system (Bruker Daltonics) according to the manufacturer’s protocol. Protein profiles were acquired using flexControl 3.4 (Bruker) and analyzed against the MBT Filamentous Fungi Library v2.0.
DNA extraction and multilocus sequencing
Genomic DNA was extracted using Ezup Column Fungal DNA Kit (Sangon Biotech) from mechanically disrupted hyphae. Three loci—ITS, EF-1α, and β-tubulin—were amplified (50 μL reaction: 95 °C/5 min; 40 cycles of 95 °C/5 s, 52–58 °C/30 s, 72 °C/30 s). Sanger sequencing (Sangon Biotech) results were aligned with GenBank references via Clustal W in MEGA-12. Phylogenetic trees were reconstructed using maximum likelihood (ML) method.
Antifungal susceptibility testing
Broth microdilution assays were performed per CLSI M38-A2 guidelines. Tested agents included caspofungin acetate (0.125–16 μg/mL), terbinafine (0.125–16 μg/mL), amphotericin B (0.5–64 μg/mL), fluconazole (0.5–64 μg/mL), and flucytosine (0.5–64 μg/mL).
Environmental monitoring and surface sampling
Environmental and surface samplings were performed according to the WS/T 592-2018 standard (Standard Terminology for Healthcare-associated Infection Management). Air sampling was conducted in the bronchoscopy procedure room using the sedimentation method at five sampling points and one negative control. The exposure time was 15 min per plate. Surface sampling was performed in the bronchoscopy procedure room on three operating table surfaces, and in the respiratory ward on twelve hospital bed surfaces, including bed rails and mattresses. All samples were incubated at 37 °C for 7 days with daily examinations for filamentous fungal growth. Fungal isolates were identified by Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry (MALDI-TOF MS) (Bruker Daltonics) following standard protocols.
Results
Microascus spp. as emerging pathogens in HSCT recipients
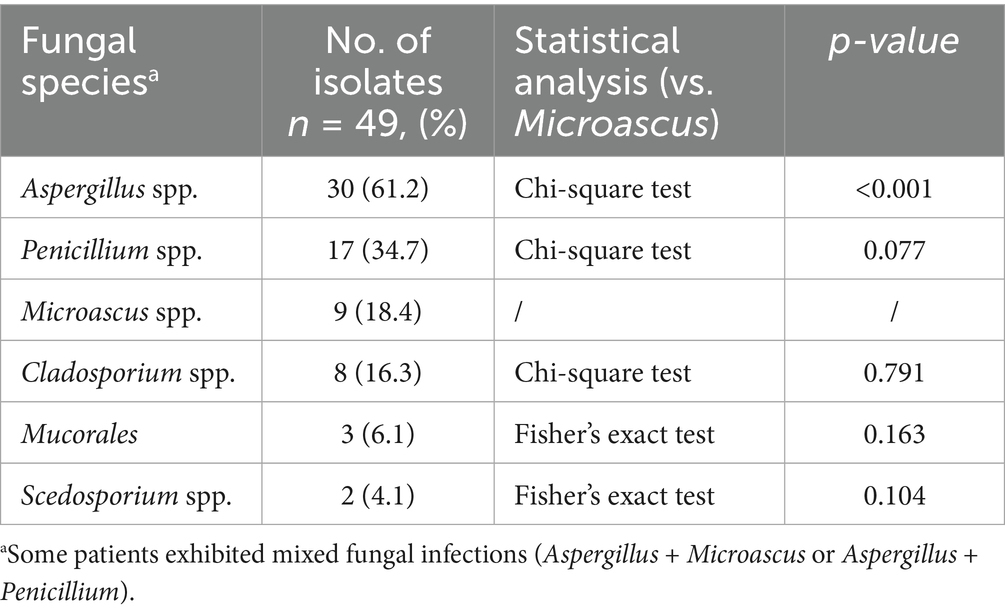
Among 49 filamentous fungal isolates from HSCT patients (January 2021–December 2024), Microascus spp. accounted for 18.4% (9/49), ranking after Aspergillus (61.2%, 30/49) and preceding Penicillium (34.7%, 17/49). All nine Microascus-positive cases (90%) were HSCT recipients in stable remission (median post-transplant interval: 31 months), while one case involved a COPD exacerbation patient (Table 1).
Clinical profiles and therapeutic outcomes
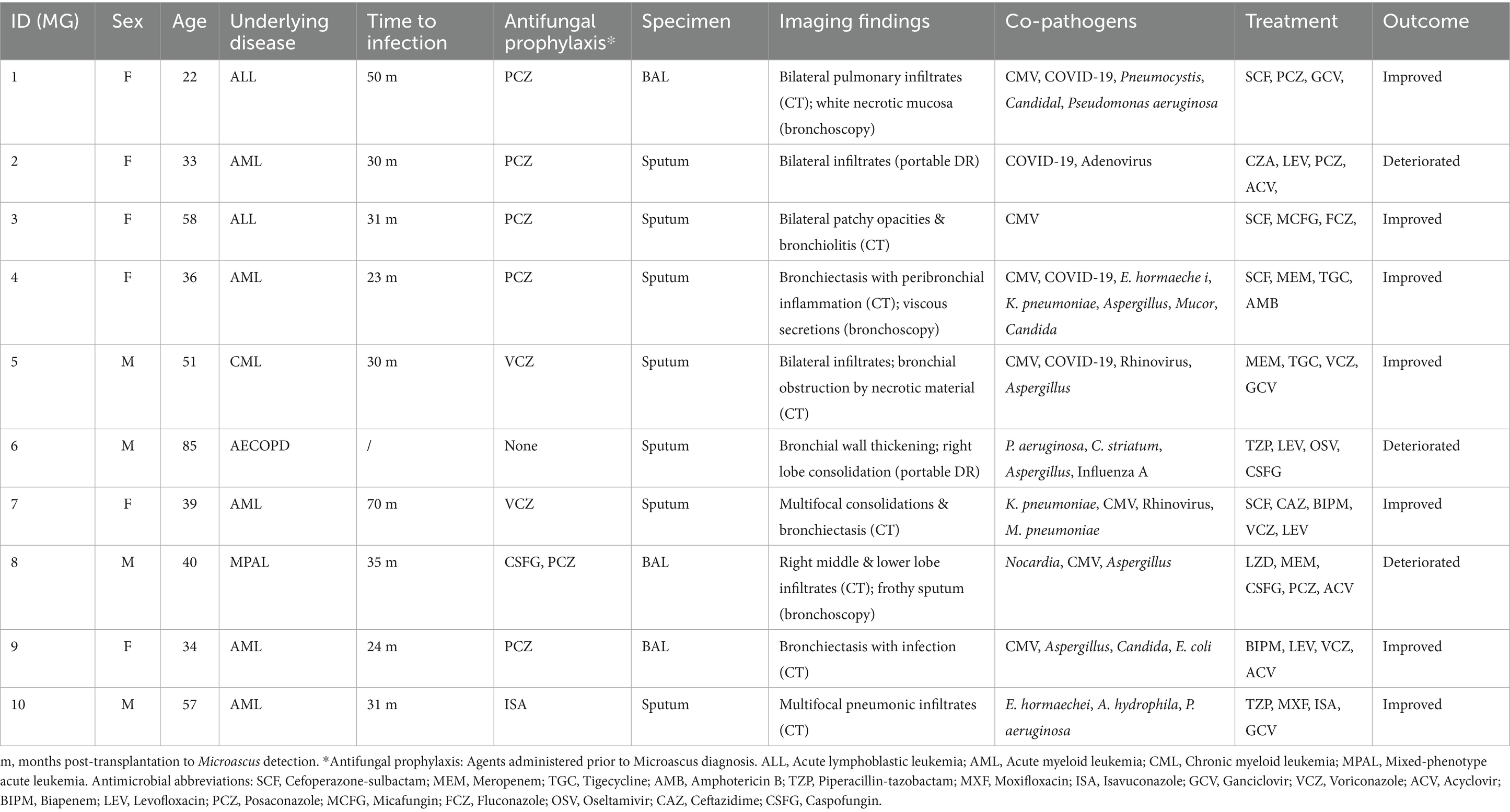
Ten patients (MG1-MG10; male:female = 4:6; median age = 39 years) exhibited bronchiolitis obliterans on High-Resolution Computed Tomography (HRCT, 100%), with characteristic radiologic findings including bronchial wall thickening (8/10), patchy infiltrates (6/10), and cavitation (2/10). Co-infections were predominantly caused by viral pathogens (CMV, Rhinovirus, 2019-nCoV, Adenovirus, Influenza A) and Aspergillus spp. (5/10), as shown in Table 2. Despite receiving multimodal therapy (broad-spectrum antibiotics + antifungals ± antivirals), three patients developed clinical deterioration.
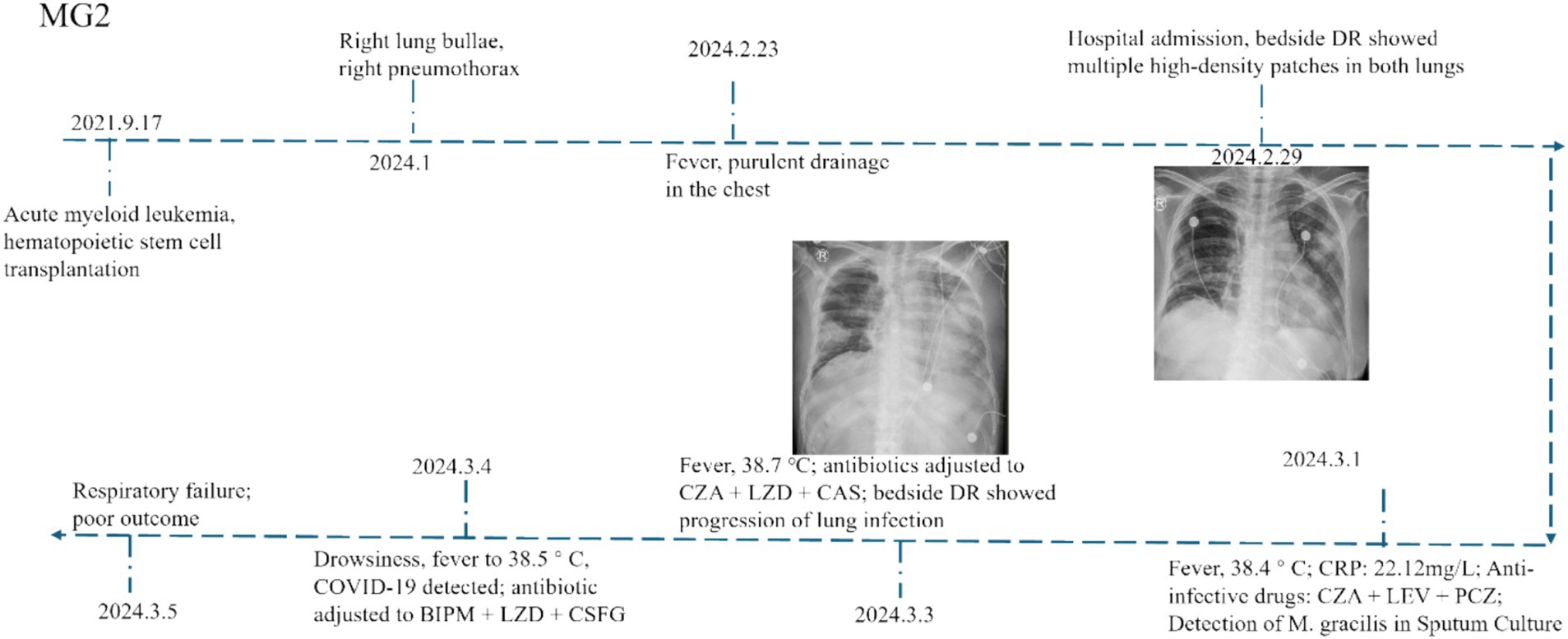
Compared with other cases, MG2 showed both typical disease characteristics and unique clinical features, we selected one representative case (MG2) for detailed analysis. The patient was initially diagnosed with M. gracilis infection during hospitalization, followed by concurrent 2019-nCoV infection. The patient’s post-transplantation status combined with mixed infection likely caused the poor clinical outcome. The disease progression then unfolded as follows. Patient MG2 who had undergone allogeneic HSCT 2 years prior developed pulmonary complications in January 2024, manifesting as right-side pneumatoceles with concomitant pleural effusion and radiologically confirmed pulmonary infection. On February 23, 2024, the patient presented with recurrent fever and purulent transformation of pleural effusion. Moreover, chest X-ray demonstrated multiple patchy consolidations with air bronchograms in bilateral lung fields (Figure 1). The patient developed persistent fever during hospitalization. Microbiological analysis of sputum cultures identified M. gracilis, with concurrent COVID-19 coinfection. Despite aggressive treatment with broad-spectrum antibiotics (bipenem, linezolid, micafungin) and molnupiravir, the patient’s condition deteriorated. The patient subsequently developed acute respiratory distress syndrome (ARDS) requiring mechanical ventilation.
Comparison of patients infected with Microascus spp. in the literature and our study
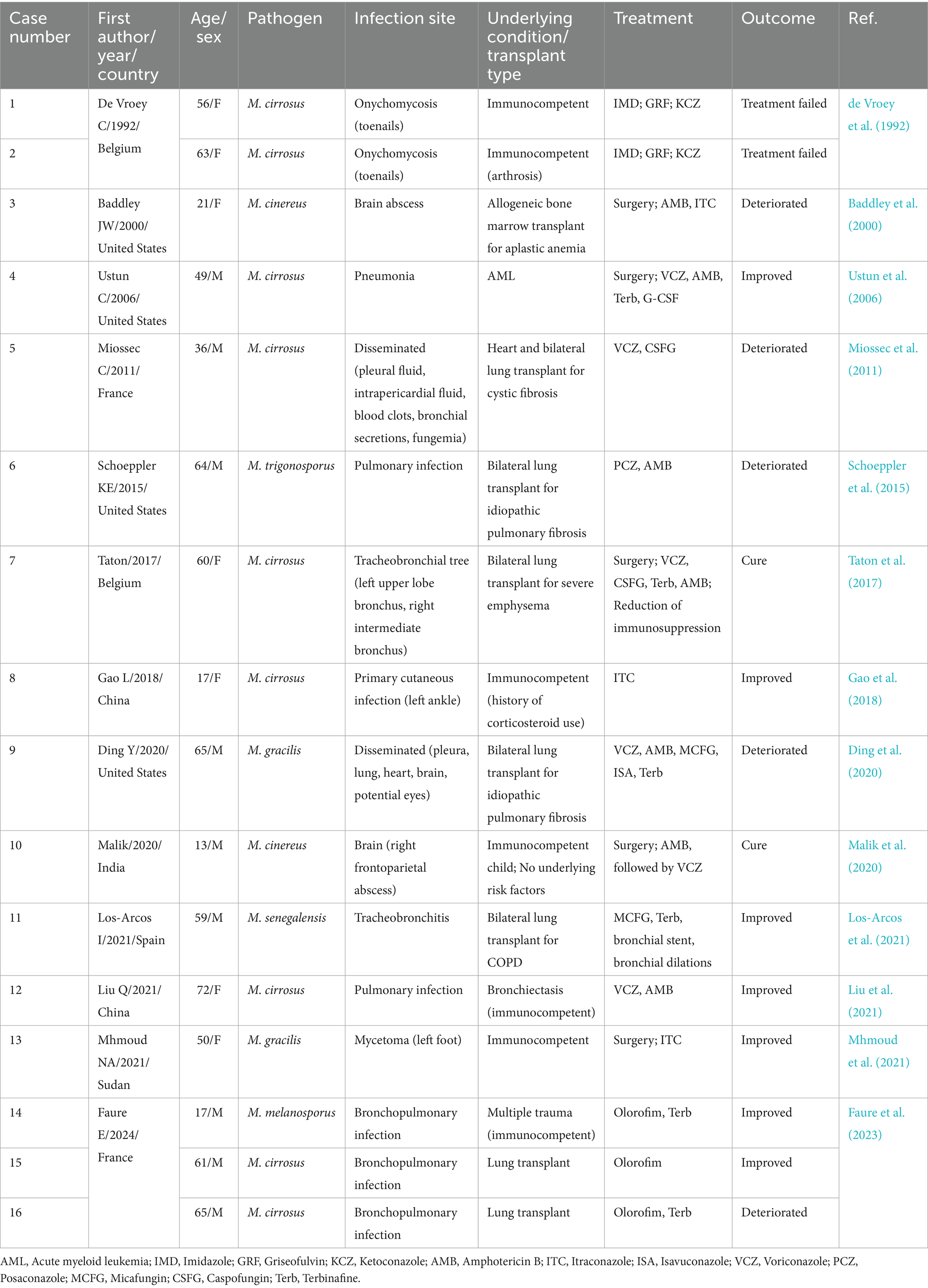
Between 1992 and 2023, 16 cases of Microascus infection were reported in studies indexed in PubMed, with a median age of 56 years and no clear sex predominance. Among these cases, immunocompromised patients accounted for the majority (9/16), and disseminated infections-particularly in this group-were associated with high mortality (5/9), as shown in Table 3. Microascus often demonstrates reduced in vitro susceptibility to common antifungals, underscoring the need for susceptibility-guided therapy. Comparison with our cohort of 10 patients reinforces that pulmonary infections occur predominantly in severely immunocompromised individuals, such as patients with hematologic malignancies or lung transplant recipients, and often present as mixed infections. Breakthrough infections despite posaconazole prophylaxis highlight the importance of comprehensive microbiologic testing-including BALF metagenomic next-generation sequencing (mNGS) and culture - and tailored antifungal coverage.
Morphological characterization
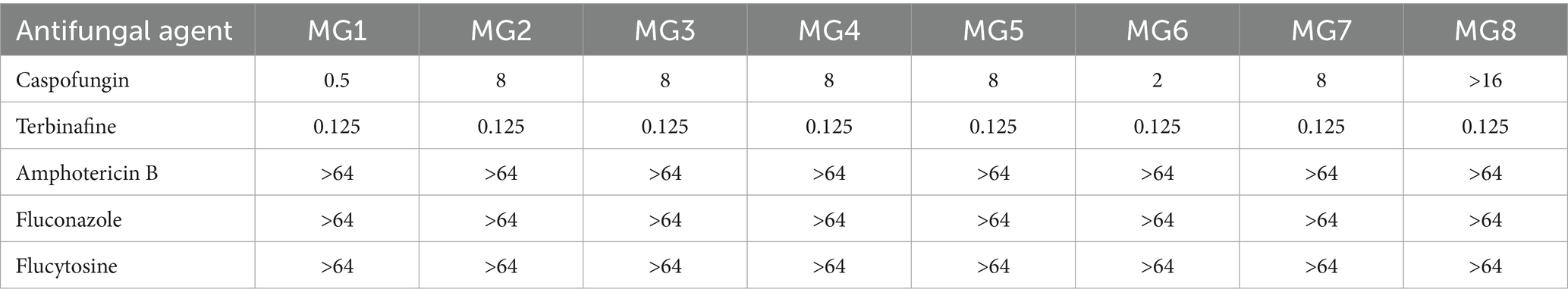
Seven M. gracilis and one M. cirrosus isolate formed concentric olive-black colonies with floccose hyphae and dark granules after 14 days of SDA incubation (Figures 2A–D). Lactophenol cotton blue staining revealed septate hyphae, basally swollen conidiophores, and chain-forming oval/pear-shaped conidia (Figures 2E,F). Cleistothecia containing smooth reniform ascospores were observed in M. cirrosus (Figures 2G,H) (Cheng et al., 2023).
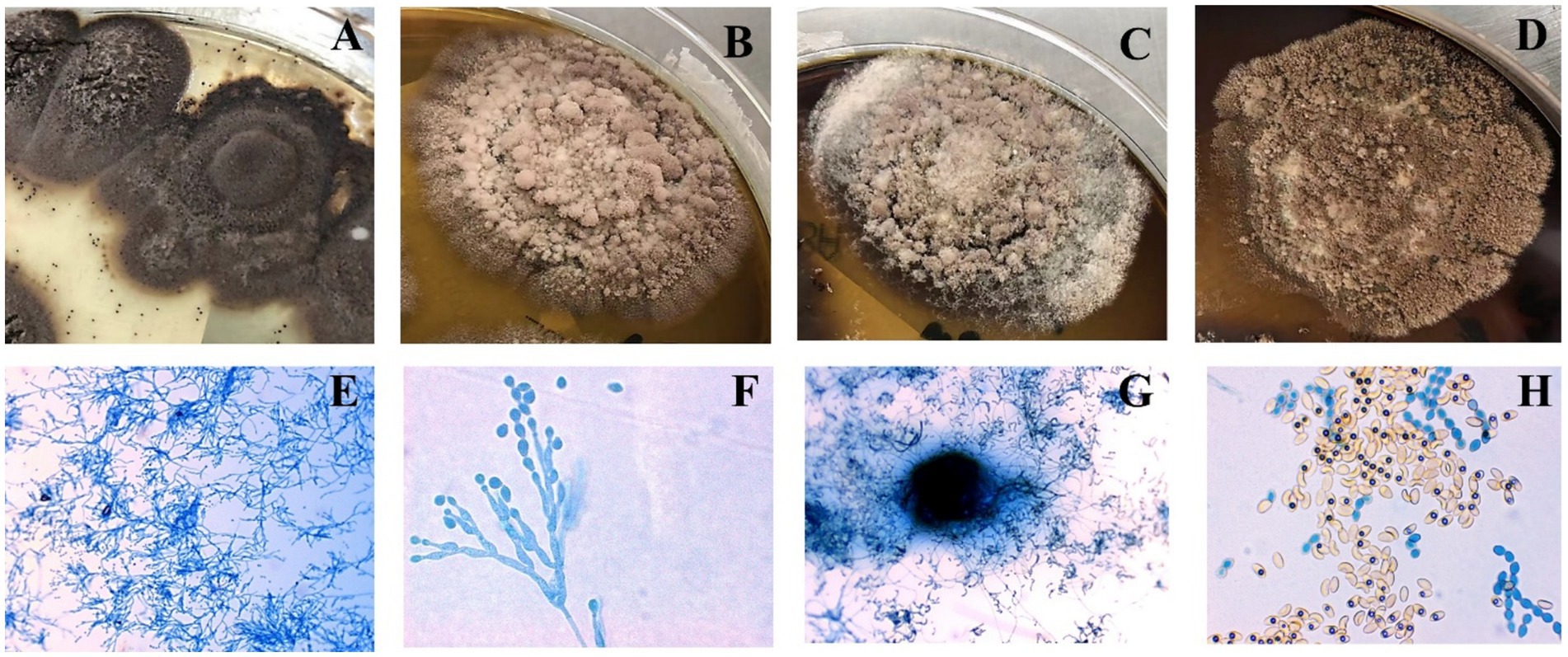
Figure 2. Colonial morphology (A–D) and lactophenol cotton blue-stained microscopic features (E–H) of Microascus spp. (E,G x400; F,H x1000).
Antifungal susceptibility patterns
All isolates exhibited elevated MIC to amphotericin B, fluconazole, and flucytosine (MIC >64 μg/mL) but were highly susceptible to terbinafine (MIC ≤0.125 μg/mL) (Table 4). Caspofungin demonstrated strain-dependent activity (MIC = 0.5–16 μg/mL). Among the nine patients who underwent HSCT, antifungal prophylaxis/therapy regimens included posaconazole (n = 6), voriconazole (n = 2), and isavuconazole (n = 1). Following Microascus infection diagnosis, all patients maintained their initial triazole antifungal therapy. Of these, two received adjunctive micafungin, four received caspofungin (of whom three exhibited disease progression and clinical deterioration).
MALDI-TOF profiling and strain typing
Protein fingerprinting distinguished two clusters: MG1-MG7 shared conserved peaks (3–4 high-intensity signals at 3400–3600 m/z; 1–2 low-intensity peaks at 2000–2200 m/z), while MG8 exhibited a unique 6,186 m/z signature (Figure 3A). Principal component analysis (PCA) and hierarchical clustering confirmed the taxonomic divergence between these clusters (Figures 3B,C).
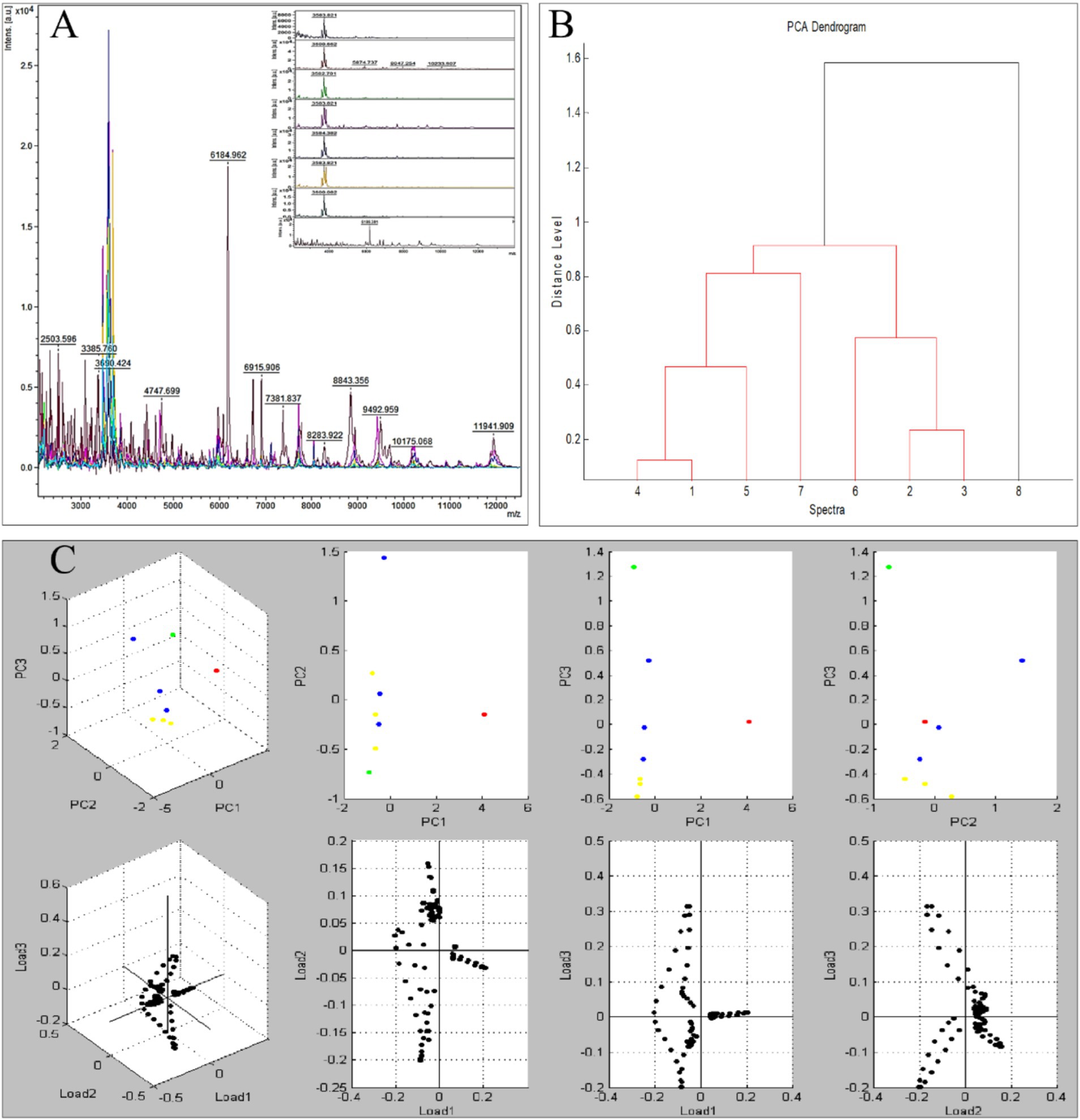
Figure 3. Protein fingerprinting and principal component analysis of eight Microascus strains. (A: protein fingerprint patterns of eight Microascus strains; B: cluster analysis of eight Microascus strains; C: principal component analysis of eight Microascus strains).
Phylogenetic relationships and species delineation
The multilocus phylogenetic analysis based on ITS, EF-1α, and TUB loci revealed remarkable interspecific diversity within Microascus. Notably, a highly supported monophyletic clade comprising reference strains of M. gracilis and clinical strains MG1/MG3–7 validated their taxonomic classification (Figure 4). Strain MG2 occupied a distinct phylogenetic position between the M. gracilis and M. cinereus clades, necessitating whole-genome sequencing for definitive classification. Furthermore, strain MG8 showed close phylogenetic affinity with the M. cirrosus reference strain despite minor branch length variations, which did not affect its taxonomic assignment.
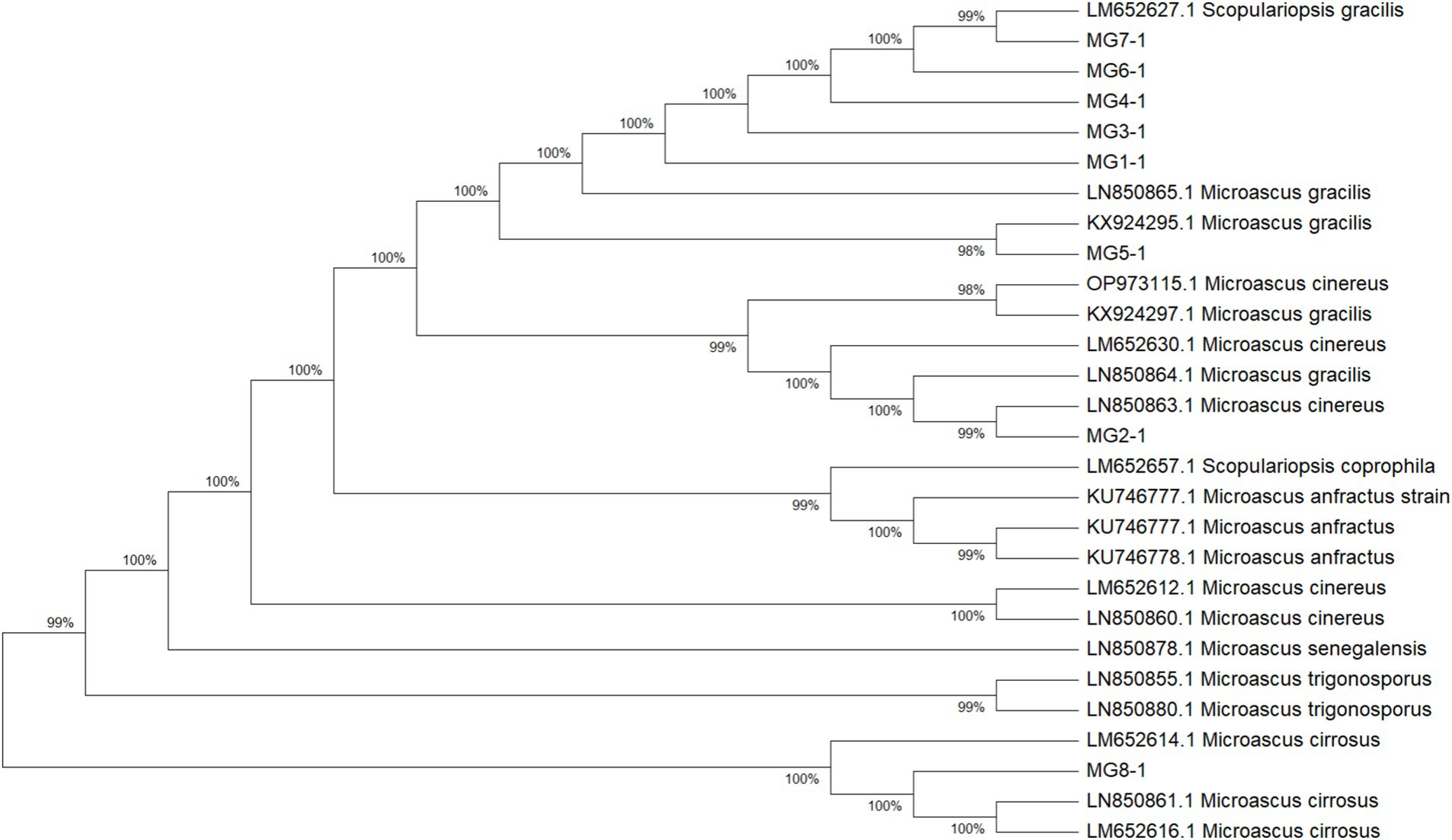
Figure 4. Phylogenetic analysis of the 8 Microascus isolates. (MG1-MG8: Microascus strains isolated in the present study; other strains: reference sequences obtained from GenBank http://www.ncbi.nlm.nih.gov/genbank/).
Environmental sampling of air and surfaces tested negative for Microascus spp
We performed targeted environmental sampling of air and surfaces in bronchoscopy suites and patient wards. Culture results showed no detectable Microascus spp. in the following samples: bronchoscopy suite air samples (Figure 5A), surfaces of bronchoscopy procedure tables (Figure 5B), and high-touch surfaces in patient wards (Figure 5C).
Figure 5. Targeted environmental sampling of air and surfaces in bronchoscopy suites and patient wards. (A: Settling bacterial monitoring of the air in the bronchoscopy operating room; B: surface monitoring of the bronchoscopy operating table; C: surface monitoring of the patient wards).
Discussion
The occurrence of severe pneumonia during the stable phase of hematopoietic stem cell transplantation (HSCT) represents a clinically significant challenge. Current evidence implicates multiple risk factors, including environmental distribution of pathogens, host immunocompromised status, and prior antifungal exposure. In our cohort, Microascus ranked third most prevalent fungal genus in HSCT recipients, following Aspergillus and Penicillium spp. While existing literature predominantly documents Microascus infections as sporadic cases or reviews, this pathogen demonstrates concerning clinical versatility, capable of causing cutaneous infections or invasive dissemination to critical organs (heart, lungs, kidneys, and brain) with fatal outcomes (Miossec et al., 2011; Patterson et al., 2016; Eisman and Sinclair, 2014; Skiada et al., 2018; Baddley et al., 2010). A comparative analysis of 26 patients-16 from the literature and 10 from our study-regarding clinical features (age, sex, underlying diseases, infection site), treatment strategies, and outcomes revealed that Microascus infections can occur in immunocompetent individuals, typically presenting as localized infections of the skin, nails, or foot. However, these infections are more prevalent in immunocompromised patients, particularly those with solid organ transplants, mainly lung, or hematopoietic stem cell transplant recipients, and are frequently associated with pulmonary or disseminated infections that lead to high mortality. Both infection site and baseline immune status were identified as critical prognostic factors. Early intervention, including prompt diagnosis and treatment initiation, and individualized therapy are essential. Localized infections may be managed with surgical treatment combined with antifungal therapy, whereas disseminated or mixed fungal infections require aggressive combination antifungal regimens and, where applicable, reduction of immunosuppressive therapy.
Of particular relevance to our cohort, HSCT recipients were maintained on long-term cyclosporine-based immunosuppression for graft-versus-host disease prophylaxis alongside routine posaconazole/voriconazole prophylaxis. This therapeutic landscape likely selects for resistant Microascus strains, potentially explaining the observed fulminant fungal pneumonias. Radiological findings consistently revealed characteristic infectious patterns, including bronchovascular bundle thickening and cavitary lesions. Meanwhile, we evaluated the impact of the timing of antifungal therapy initiation on patient outcomes. Among the 10 patients studied, 9 were HSCT recipients, of whom 7 received prolonged prophylactic antifungal therapy initiated pre-admission or at admission, resulting in favorable outcomes. Conversely, the three cases with clinical deterioration comprised two HSCT patients who began antifungal treatment within 24 h post-admission and one non-HSCT patient with delayed initiation at 7 days post-admission. This suggests that early or prophylactic initiation correlates with improved outcomes, aligning with causal inference principles regarding exposure and outcome relationships, although this association does not confirm causation.
Notably, Microascus was detected within 10 days of admission in all cases, frequently alongside cytomegalovirus, Pseudomonas aeruginosa, or Aspergillus spp. The three fatal cases demonstrated synergistic morbidity with influenza A virus, 2019-nCoV, or Nocardia co-infections. The poor outcomes observed were primarily influenced by profound immunosuppression due to HSCT-related immune reconstitution failure and chronic comorbidities impairing antifungal efficacy in all three cases; additionally, one patient experienced COPD exacerbations compromising pulmonary defenses, and viral coinfections such as COVID-19 or influenza A triggered cytokine storms and multi-organ failure, further amplifying pathophysiological deterioration. This aligns with evidence that viral pathogens in immunocompromised hosts exacerbate immune dysfunction, creating a permissive environment for polymicrobial infections and poor outcomes (Vanderbeke et al., 2018; Arastehfar et al., 2020; Uhde et al., 2010).
Despite standard posaconazole/voriconazole prophylaxis, breakthrough Microascus infections occurred in our hematopoietic stem cell transplantation (HSCT) cohort, suggesting that conventional azole-based prophylactic regimens may be inadequate against this pathogen. All eight isolates exhibited elevated MICs (>64 μg/mL) to azoles, polyenes, and flucytosine, indicating potential intrinsic resistance mechanisms. In contrast, terbinafine demonstrated excellent in vitro activity (MIC ≤0.125 μg/mL across all strains). Heterogeneous susceptibility to echinocandins was observed, as reflected by variable MIC values to caspofungin among the isolates. These findings are consistent with those reported by Skóra et al. (2015), who also documented strong resistance of Microascus to single antifungal agents such as terbinafine, caspofungin, and posaconazole, with generally high MICs. Notably, their study revealed significant synergistic effects with triple combination therapy (terbinafine, caspofungin, and posaconazole) in vitro. Our clinical observations further support these results: a dose-escalated voriconazole regimen combined with caspofungin showed efficacy in partial responders, underscoring the potential of combination therapy to overcome monotherapeutic resistance. However, suboptimal responses were still observed in some cases even with susceptible agents, which may be attributed to concurrent viral or Aspergillus co-infections. This complexity underscores the need for comprehensive therapeutic strategies that integrate host immune status, co-infecting pathogens, and antifungal resistance profiles. Further genomic studies are warranted to elucidate species-specific resistance determinants in Microascus.
Current fungal infection diagnostics still rely heavily on morphological identification. These methods are time-consuming and often inaccurate, which may result in missed or incorrect pathogen detection. However, the slow growth kinetics of Microascus and technical challenges in rapidly obtaining observable diagnostic structures significantly impede timely detection. Notably, six next-generation sequencing (NGS) analyses in our cohort failed to detect Microascus species, highlighting the technical limitations of metagenomics for rare fungi detection, especially when pathogen-specific genomic signatures are underrepresented in reference databases. These diagnostic difficulties enhance the clinical significance of our findings, as better characterization of Microascus epidemiology and resistance patterns directly guides therapeutic decision-making for HSCT recipients during the stabilization phase. Our data underscore the necessity for developing rapid molecular diagnostics targeting conserved genomic regions.
The Microascus isolates in our study exhibited grayish-brown to black pigmentation with conidial morphological features consistent with established descriptions of the Microascus morphology (Liu et al., 2021). The observed morphological plasticity (melanized conidia and cleistothecial structures) suggests evolutionary adaptations related to environmental persistence or pathogenic regulation, though the underlying mechanisms remain unclear (Skiada et al., 2018). MALDI-TOF mass spectrometry proved highly effective for fungal identification, showing 100% concordance (7/7 strains) in genus-level classification based on characteristic protein fingerprints (3400–3,600 m/z spectral peaks) (Patel, 2015; Wei et al., 2022). However, species-level discrimination required supplementary phylogenetic analysis using multilocus sequence data. This highlights the necessity of combined approaches, whose diagnostic complexity warrants careful consideration. Notably, the phenotypic similarities among Microascus, Aspergillus, and Penicillium species necessitate implementing a tiered diagnostic algorithm in clinical laboratories. We propose a hierarchical identification framework incorporating: Morphological screening + MALDI-TOF + Multilocus sequence typing (MLST).
Our data suggest Microascus spp. as emerging opportunistic pathogens in post-hematopoietic stem cell transplantation (HSCT) invasive fungal infections, demonstrating a secondary prevalence rate to Aspergillus and Penicillium spp. This epidemiological pattern warrants heightened clinical vigilance, particularly because therapeutic complexities emerge from both frequent polymicrobial coinfections and intrinsic antifungal resistance. The technical challenges in species identification necessitate a multidimensional diagnostic protocol: (1) Prolonged incubation (5–14 days) for cleistothecia development; (2) MALDI-TOF MS spectral analysis focusing on the characteristic range; (3) Multilocus sequence typing (MLST) targeting β-tubulin, ITS, and CAL loci for definitive speciation. Antifungal susceptibility profiles revealed universal resistance to triazoles and echinocandins, while terbinafine showed the lowest MIC values (≤0.125 μg/mL). These data underscore the urgency to establish species-specific interpretive criteria for Microascus, distinct from existing guidelines for Aspergillus or Candida.
To investigate potential nosocomial sources of Microascus spp., targeted environmental sampling of air and surfaces in bronchoscopy suites and patient wards was performed. Although these fungi are ubiquitously distributed in natural environments—including soil, decaying plant materials, animal feces, and moist habitats—all environmental specimens tested negative for Microascus spp. by culture. This finding suggests an absence of detectable contamination within the examined healthcare settings during the study period. However, temporal and spatial variations in fungal load may occur, and the inherent limitations of conventional culturing in detecting low-abundance spores must be considered. These negative culture results, while not fully defining transmission dynamics, support excluding hospital-based environmental exposure as a source of these infections. If consecutive cases of Microascus spp. detection occur in the same clinical unit, surveillance involving multiple sampling locations and time points in high-risk areas, combined with molecular genotyping techniques for source attribution, should be prioritized.
The novel contributions of this study include the development of a tiered diagnostic strategy integrating morphological, MALDI-TOF MS, and MLST for accurate identification of Microascus; evidence-based recommendations for prophylaxis adjustment informed by in vitro susceptibility and literature review; and environmental surveillance data supporting targeted infection control. These findings provide critical insights for early detection and tailored management of Microascus infections in high-risk populations. Nevertheless, our study has three main limitations. First, its single-center design and small sample size (10 patients) limit the generalizability of the findings, particularly to non-HSCT populations. Second, the lack of molecular resistance data (e.g., ERG11 mutations, efflux pump expression) constrains the mechanistic interpretation of antifungal resistance. Third, the absence of fungal burden quantification and autopsy data impedes accurate correlation of pathogen load with clinical outcomes and precise attribution of mortality. To address these issues, future studies should incorporate molecular profiling (e.g., whole-genome sequencing), standardize specimen processing, and implement fungal quantification methods (e.g., qPCR). Furthermore, enhanced autopsy efforts with ethical compliance are needed to clarify mortality causes. Prioritized research directions include developing rapid molecular diagnostics for high-risk groups, establishing multicenter surveillance to monitor resistance, and exploring combination therapies that leverage unique antifungal mechanisms such as those of terbinafine.
The increasing incidence of severe pulmonary Microascus infections, particularly among immunocompromised patients, is accompanied by high treatment failure rates and significant mortality. This is further compounded by the pathogen’s pan-resistance to conventional antifungal agents, underscoring an urgent need for improved early detection, elucidation of resistance mechanisms, and development of novel therapeutic strategies. In response, our future perspectives emphasize several key directions: First, diagnostic accuracy must be enhanced through the integration of Microascus-specific protein fingerprints into MALDI-TOF-MS databases and standardization of mNGS reporting protocols to facilitate early and reliable identification. Second, due to the high in vitro resistance observed, although resistance patterns are not fully defined, empirical antifungal susceptibility testing remains critical for tailoring targeted therapy. This is especially important for high-risk groups, such as transplant recipients receiving prophylaxis with azoles (e.g., posaconazole, voriconazole). Clinical vigilance should be heightened in these populations, incorporating multimodal data to differentiate colonization from invasive infection. Finally, strengthened environmental surveillance in high-risk hospital areas is essential to control potential nosocomial transmission. Collectively, these strategies aim to improve clinical outcomes through earlier intervention, individualized treatment, and evidence-based infection control in the face of this emerging and challenging pathogen.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, PRJNA835605.
Ethics statement
The studies were conducted in accordance with the local legislation and institutional requirements. Informed consent was obtained from all subjects involved in the study.
Author contributions
TZ: Conceptualization, Formal analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing. J-JC: Investigation, Methodology, Writing – original draft. R-RB: Investigation, Methodology, Writing – original draft. J-MZ: Investigation, Methodology, Writing – original draft. L-TZ: Formal analysis, Investigation, Writing – original draft. YC: Formal analysis, Investigation, Writing – original draft. Q-ZH: Conceptualization, Funding acquisition, Investigation, Methodology, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was funded by the Suzhou Science and Technology Agency Project (SKY2022091), and Suzhou Science and Technology Plan Project (SZM 2024019), and Suzhou Industrial Park Healthcare Innovation Research Project (CXYJ2024A08), and Jiangsu Province International Joint Laboratory for Regeneration Medicine. The corresponding author, Qingzhen Han, was supported by the Health Talents Project of Suzhou Industrial Park.
Acknowledgments
The authors would like to thank the Suzhou Industrial Park Healthcare In-novation and Jiangsu Province International Joint Laboratory for Regeneration Medicine for their financial support to this research.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Arastehfar, A., Carvalho, A., Nguyen, M. H., Hedayati, M. T., Netea, M. G., Perlin, D. S., et al. (2020). COVID-19-associated candidiasis (CAC): an underestimated complication in the absence of immunological predispositions? J. Fungi (Basel) 6:211. doi: 10.3390/jof6040211
Baddley, J. W., Andes, D. R., Marr, K. A., Kontoyiannis, D. P., Alexander, B. D., Kauffman, C. A., et al. (2010). Factors associated with mortality in transplant patients with invasive aspergillosis. Clin. Infect. Dis. 50, 1559–1567. doi: 10.1086/652768
Baddley, J. W., Moser, S. A., Sutton, D. A., and Pappas, P. G. (2000). Microascus cinereus (anamorph scopulariopsis) brain abscess in a bone marrow transplant recipient. J. Clin. Microbiol. 38, 395–397. doi: 10.1128/JCM.38.1.395-397.2000
Brasch, J., Beck-Jendroschek, V., Iturrieta-González, I., Voss, K., and Gené, J. (2018). A human subcutaneous infection by Microascus ennothomasiorum Sp. Nov. Mycoses 62, 157–164. doi: 10.1111/myc.12861
Cheng, J., Zeng, D., Zhang, T., Zhang, L., Han, X., Zhou, P., et al. (2023). Microascus cirrosus SZ 2021: a potentially new genotype of Microascus cirrosus, which can cause fatal pulmonary infection in patients with acute leukemia following haplo-HSCT. Exp. Ther. Med. 26:404. doi: 10.3892/etm.2023.12103
de Vroey, C., Lasagni, A., Tosi, E., Schroeder, F., and Song, M. (1992). Onychomycoses due to Microascus cirrosus (syn. M. desmosporus). Mycoses 35, 193–196. doi: 10.1111/j.1439-0507.1992.tb00845.x
Ding, Y., Steed, L. L., and Batalis, N. (2020). First reported case of disseminated Microascus gracilis infection in a lung transplant patient. IDCases :e00984:22. doi: 10.1016/j.idcr.2020.e00984
Eisman, S., and Sinclair, R. (2014). Fungal nail infection: diagnosis and management. BMJ 348:g1800. doi: 10.1136/bmj.g1800
Faure, E., Brugière, O., de Verdiere, S. C., Vuotto, F., Limousin, L., Cardot, E., et al. (2023). Refractory Microascus bronchopulmonary infection treated with Olorofim, France. Emerg. Infect. Dis. 29, 2401–2403. doi: 10.3201/eid2911.230984
Gao, L., Chen, J., Gao, D., and Li, M. (2018). Primary cutaneous infection due to Microascus cirrosus: a case report. BMC Infect. Dis. 18:604. doi: 10.1186/s12879-018-3535-5
Liu, Q., Kong, L., Hua, L., and Xu, S. (2021). Pulmonary Microascus cirrosus infection in an immunocompetent patient with bronchiectasis: a case report. Respir. Med. Case. Rep. 34:101484. doi: 10.1016/j.rmcr.2021.101484
Los-Arcos, I., Berastegui, C., Martín-Gómez, M. T., Grau, S., Campany-Herrero, D., Deu, M., et al. (2021). Nebulized micafungin treatment for Scopulariopsis/Microascus tracheobronchitis in lung transplant recipients. Antimicrob. Agents Chemother. 65, e02174–e02120. doi: 10.1128/AAC.02174-20
Malik, S., Bajpai, V., Betai, S., Pal, L., and Marak, R. S. (2020). An unusual case of Microascus brain abscess in an immunocompetent child and a review of the literature. J. Family Med. Prim. Care 9, 1244–1247. doi: 10.4103/jfmpc.jfmpc_1038_19
Mhmoud, N. A., Siddig, E. E., Nyuykonge, B., Bakhiet, S. M., van de Sande, W. W. J., and Fahal, A. H. (2021). Mycetoma caused by Microascus gracilis: a novel agent of human eumycetoma in Sudan. Trans. R. Soc. Trop. Med. Hyg. 115, 426–430. doi: 10.1093/trstmh/trab010
Miossec, C., Morio, F., Lepoivre, T., le Pape, P., Garcia-Hermoso, D., Gay-Andrieu, F., et al. (2011). Fatal invasive infection with fungemia due to Microascus cirrosus after heart and lung transplantation in a patient with cystic fibrosis. J. Clin. Microbiol. 49, 2743–2747. doi: 10.1128/JCM.00127-11
Patel, R. (2015). MALDI-TOF MS for the diagnosis of infectious diseases. Clin. Chem. 61, 100–111. doi: 10.1373/clinchem.2014.221770
Patterson, T. F., Thompson, G. R. III, Denning, D. W., Fishman, J. A., Hadley, S., Herbrecht, R., et al. (2016). Practice guidelines for the diagnosis and management of aspergillosis: 2016 update by the Infectious Diseases Society of America. Clin. Infect. Dis. 63, e1–e60. doi: 10.1093/cid/ciw326
Sandoval-Denis, M., Gené, J., Sutton, D. A., Cano-Lira, J. F., de Hoog, G. S., Decock, C. A., et al. (2016). Redefining Microascus, Scopulariopsis and allied genera. Persoonia 36, 1–36. doi: 10.3767/003158516X688027
Schoeppler, K. E., Zamora, M. R., Northcutt, N. M., Barber, G. R., O’Malley-Schroeder, G., and Lyu, D. M. (2015). Invasive Microascus trigonosporus species complex pulmonary infection in a lung transplant recipient. Case Rep. Transplant. 2015:745638. doi: 10.1155/2015/745638
Skiada, A., Lass-Floerl, C., Klimko, N., Ibrahim, A., Roilides, E., and Petrikkos, G. (2018). Challenges in the diagnosis and treatment of mucormycosis. Med. Mycol. 56, S93–S101. doi: 10.1093/mmy/myx101
Skóra, M., Bulanda, M., and Jagielski, T. (2015). In vitro activities of a wide panel of antifungal drugs against various Scopulariopsis and Microascus species. Antimicrob. Agents Chemother. 59, 5827–5829. doi: 10.1128/AAC.00978-15
Taton, O., Bernier, B., Etienne, I., Bondue, B., Lecomte, S., Knoop, C., et al. (2017). Necrotizing Microascus tracheobronchitis in a bilateral lung transplant recipient. Transpl. Infect. Dis. 20:e12806. doi: 10.1111/tid.12806
Uhde, K. B., Pathak, S., McCullum, I. Jr., Jannat-Khah, D. P., Shadomy, S. V., Dykewicz, C. A., et al. (2010). Antimicrobial-resistant nocardia isolates, United States, 1995–2004. Clin. Infect. Dis. 51, 1445–1448. doi: 10.1086/657399
Ustun, C., Huls, G., Stewart, M., and Marr, K. A. (2006). Resistant Microascus cirrosus pneumonia can be treated with a combination of surgery, multiple anti-fungal agents and a growth factor. Mycopathologia 162, 299–302. doi: 10.1007/s11046-006-0067-0
Vanderbeke, L., Spriet, I., Breynaert, C., Rijnders, B. J. A., Verweij, P. E., and Wauters, J. (2018). Invasive pulmonary aspergillosis complicating severe influenza: epidemiology, diagnosis and treatment. Curr. Opin. Infect. Dis. 31, 471–480. doi: 10.1097/QCO.0000000000000504
Wei, L., Shao, J., Song, Y., Wan, Z., Yao, L., Wang, H., et al. (2022). Performance of matrix-assisted laser desorption ionization-time of flight mass spectrometry for identification of Scedosporium, Acremonium-like, Scopulariopsis, and Microascus species. Front. Microbiol. 13:841286. doi: 10.3389/fmicb.2022.841286
Keywords: Microascus spp., hematopoietic stem cell transplantation, antifungal susceptibility testing, MALDI-TOF, multilocus sequence typing, environmental monitoring
Citation: Zhang T, Cheng J-J, Bi R-R, Zhu J-M, Zhou L-T, Chen Y and Han Q-Z (2025) Clinical characterization and etiological insights: a small cohort study of pulmonary infections caused by Microascus spp. in critically ill patients. Front. Microbiol. 16:1632354. doi: 10.3389/fmicb.2025.1632354
Edited by:
Hector Mora Montes, University of Guanajuato, MexicoReviewed by:
Husam Eldin Salah Ismail Mohamed, Hamad Medical Corporation, QatarTuhin Mukherjee, Birla Institute of Technology, Mesra, India
Copyright © 2025 Zhang, Cheng, Bi, Zhu, Zhou, Chen and Han. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qing-Zhen Han, Z3locXoyMDIxQDE2My5jb20=
†ORCID: Qing-Zhen Han, https://orcid.org/0000-0001-7373-891X
†These authors have contributed equally to this work
 Ting Zhang
Ting Zhang Jian-Jun Cheng†
Jian-Jun Cheng† Li-Ting Zhou
Li-Ting Zhou Qing-Zhen Han
Qing-Zhen Han