- 1Shanghai Veterinary Research Institute, Chinese Academy of Agricultural Sciences, Shanghai, China
- 2College of Animal Science and Technology, Anhui Agricultural University, Hefei, China
Introduction: Ticks are the primary vectors of Babesia sp, with the midgut as the initial site of pathogen invasion following blood feeding. Elucidating the molecular interactions between tick midguts and Babesia is essential for developing targeted strategies to control tick-borne babesiosis. However, studies in this field remain limited.
Methods: To investigate tick-pathogen interactions, we employed RNA-seq to profile gene expression, and qRT-PCR served to validate key findings. Apoptosis and autophagy were assessed via TUNEL staining and Transmission Electron Microscopy (TEM). Furthermore, RNA interference (RNAi) and pharmacological modulation were employed to evaluate the impact of ticks on pathogen load.
Results: Our RNA-seq analysis identified 540 and 569 Differentially Expressed Genes (DEGs) in infected midguts at 0 and 4 d post-engorgement, respectively. These DEGs were enriched in pathways related to metabolic processes, immunity, and cellular processes. To clarify the functional relevance of these findings, the roles of apoptosis and autophagy during infection were further evaluated. Quantitative Real-Time PCR (qRT-PCR) analysis revealed significant upregulation of apoptosis-related genes (caspase-7, caspase-8, and caspase-9) and autophagy genes (ATG5, ATG8, and ATG12) in response to B. microti infection. TUNEL assay and Transmission Electron Microscopy (TEM) analysis demonstrated that B. microti infection significantly induced apoptosis and autophagosome formation in tick midgut tissues. Functional assays demonstrated that RNA interference (RNAi)-mediated knockdown of caspase-7, caspase-9, and ATG5 significantly reduced the burden of B. microti. Conversely, pharmacological induction of autophagy using rapamycin increased B. microti load, whereas inhibition with 3-methyladenine (3-MA) decreased B. microti load.
Discussion: These findings underscore the critical roles of apoptosis and autophagy in facilitating B. microti infection within tick midguts, highlighting these pathways as potential molecular targets for disrupting the transmission of tick-borne Babesia infections.
1 Introduction
The Asian longhorned tick (Haemaphysalis longicornis) is an invasive ectoparasitic arthropod of significant concern to public health and agriculture. Originally endemic to East Asia, Australia, and New Zealand, this highly adaptable species has successfully colonized persistent, self-sustaining populations in at least nineteen states across the eastern United States, underscoring its notable ecological plasticity. This adaptability is largely attributed to biological traits, such as parthenogenetic reproduction and a broad thermal tolerance range (-4 to 40 C) (Beard et al., 2018; Rainey et al., 2018; Rochlin, 2019; Tanne, 2018). As a generalist hematophagous vector, H. longicornis exhibits a broad host specificity, parasitizing over forty species of mammals and birds. It serves as a competent vector for numerous pathogens, including Severe Fever With Thrombocytopenia Syndrome Virus (SFTSV), Tick-Borne Encephalitis Virus (TBEV), Anaplasma phagocytophilum, Borrelia burgdorferi, Babesia spp., Theileria orientalis, Rickettsia spp., and Ehrlichia spp (Dobler et al., 2012; Kang et al., 2016; Rosenberg et al., 2018; Zhao et al., 2021; Zhuang et al., 2018). The combination of its biological characteristics—cold resistance, asexual reproduction, and broad host specificity—facilitates the rapid establishment of populations and supports the enzootic maintenance of pathogen transmission cycles in newly colonized regions, thus posing significant threats to public health and veterinary disease management (Yu et al., 2023).
Among Babesia species, B. microti is the most prevalent zoonotic pathogen. Although approximately 2,000 annual cases are reported, epidemiological evidence indicates the true incidence is significantly higher (Bloch et al., 2019; Diuk-Wasser et al., 2014; Krause et al., 2003). Although Ixodes spp. are recognized as the primary vectors ofB. microti, recent epidemiological surveillance has detected B. microti DNA in H. longicornis, suggesting a possible vectorial role (Zhang et al., 2017). Experimental transmission models have demonstrated that H. longicornis can acquire B. microti from infected murine hosts during blood feeding and subsequently transmit the parasite to naïve mice, thereby establishing its competence as an alternative transmission vector (Wu et al., 2017). Upon infection, H. longicornis initiates complex innate immune responses mediated by several effector molecules, including antimicrobial peptides (such as defensin, microplusin, and hebraein), protease regulators (Kunitz domain-containing proteins), transport molecules (lipocalins), and enzymatic regulators (proteases) (Antunes et al., 2017; De la Fuente et al., 2017; Wikel, 1999). However, these responses are often countered by the parasite's ability to exploit host-derived factors, thereby enhancing its colonization and transmission efficiency. While these vector-parasite interactions have been partially elucidated in other Babesia-tick systems, the precise molecular mechanisms underlying B. microti infection in H. longicornis remain poorly understood.
Unlike vertebrates, ticks lack adaptive immunity and rely solely on their innate immune mechanisms for defense against pathogens (Brossard and Wikel, 2004; Hart and Thangamani, 2021). This defense system comprises various immune cells and signaling molecules capable of pathogen recognition and elimination (Fogaça et al., 2021; Yuan et al., 2020). Programmed Cell Death (PCD), including apoptosis, autophagy, and ferroptosis, is a fundamental component of innate immunity and plays a crucial role in cellular homeostasis and developmental processes in eukaryotes (Jorgensen et al., 2017; Nagata and Tanaka, 2017). Notably, several tick-borne pathogens have evolved mechanisms to modulate host PCD pathways to facilitate their survival and transmission (Chen et al., 2021). For instance, Rickettsia rickettsii inhibits apoptosis in infected tick cells by suppressing caspase-3 activity, thereby enhancing the growth and proliferation of the bacteria within the host cells (Martins et al., 2020). Similarly, A. phagocytophilum promotes intracellular survival by downregulating Porin expression, which decreases mitochondrial cytochrome C release and impairs apoptosis (Ayllón et al., 2015). Although bacterial modulation of autophagy pathways has been extensively studied in mammalian hosts infected with Anaplasmataceae, the role of autophagy in tick-pathogen interactions remains elusive (Lin et al., 2016; Niu et al., 2012). Our previous study demonstrated that B. microti infection upregulates the expression of Hemolymph-Related Factor (HRF) in the midgut of H. longicornis, inducing ferroptosis and promoting parasite colonization (Chen et al., 2025). However, the involvement of apoptosis and autophagy in tick responses to B. microti infection has not been fully elucidated, warranting further investigations.
In this study, a B. microti—mouse—H. longicornis infection model was established to investigate early-stage molecular interactions between B. microti and the midguts of H. longicornis. Dissected midgut tissues from engorged H. longicornis nymphs were subjected to RNA Sequencing (RNA-Seq) to assess transcriptomic changes associated with B. microti infection. Comparative analysis of infected and uninfected ticks identified Differentially Expressed Genes (DEGs) associated with apoptosis and autophagy. Functional validation using RNA interference (RNAi) demonstrated that silencing of caspase-7, caspase-9, and ATG5 significantly decreased B. microti burden, indicating the parasite's dependence on these host cellular pathways for successful colonization. Furthermore, pharmacological modulation of autophagy with rapamycin (an autophagy activator) and 3-methyladenine (an autophagy inhibitor) demonstrated that B. microti modulates host cell PCD mechanisms to promote its survival. These findings provide novel insights into Babesia-ticks interactions and highlight potential molecular targets for transmission-blocking interventions against tick-borne babesiosis.
2 Materials and methods
2.1 Ethics statement
All experimental protocols were approved by the Institutional Animal Care and Use Committee and the Animal Ethics Committee of the Shanghai Veterinary Research Institute (Approval Nos. SHVRI-SZ-202008026-01, SHVRI-SV-20230616-03, and SHVRI-20230602-01).
2.2 Babesia, tick, and animal models
B. microti strains (ATCC PRA-99™; Manassas, VA, U.S.A.) were maintained in the laboratory through serial intraperitoneal passages in BALB/c mice. Female BALB/c mice (5–6 weeks old, 18–20 g) were obtained from Suzhou Sibifu Biotechnology Co., Ltd. (Suzhou, China) for parasite propagation and tick infection studies. Laboratory colonies of H. longicornis were maintained under controlled environmental conditions (25 °C, 60% relative humidity, complete darkness) and fed on New Zealand White rabbits supplied by the Shanghai Laboratory Animal Center (Chinese Academy of Sciences).
2.3 microti infection in H. longicornis
Tick infection with B. microti was conducted following previously established protocols (Wu et al., 2017). Cryopreserved B. microti strains (ATCC PRA-99™) were rapidly thawed in a 37 °C water bath, and 500 μL of the suspension was administered intraperitoneally into specific pathogen-free BALB/c mice. B. microti was monitored daily through microscopic examination of thin peripheral blood smears stained with 10% Giemsa solution (pH 7.2). B. microti infection was typically confirmed within 5-−7 d post-inoculation. Upon reaching a B. microti level of 50%, blood was collected into EDTA-coated tubes, and 200 μL aliquots were used to infect naïve, age-matched immunocompetent mice to maintain the infection cycle. For tick exposure, 60 H. longicornis nymphs were applied to the shaved dorsal skin of each B. microti-infected mouse (10−15%) and allowed to feed to repletion. This method was used for all groups. This time point was selected to synchronize the rapid engorgement phase of ticks with peak B. microti, thereby optimizing the efficiency of pathogen acquisition.
3 Quantitative detection of B. microti
Quantification of B. microti burden was performed using a TaqMan probe-based quantitative PCR (qPCR) assay following established protocols (Persing et al., 1992; Rollend et al., 2013). A 429-bp fragment of the B. microti 18S rDNA (GenBank accession no. AB190435.1) was cloned into a pMD18-T vector (TaKaRa Bio, Japan) to generate a standard curve using serial ten-fold dilutions (101 – 108 copies/μL). qPCR analysis was conducted in triplicate on a QuantStudio 5 Real-Time PCR System (Applied Biosystems, U.S.A.). Each 20 μL reaction contained 10 μL 2 × Premix Ex Taq (Hot Start DNA polymerase), 0.6 μL of each primer (10 μM), 0.3 μL of FAM/BHQ1-labeled probe (10 μM), and 3 μL of DNA template. The thermal cycling conditions included an initial denaturation at 95°C for 30 s, followed by 40 amplification cycles of 95°C for 5 s and 60°C for 34 s. Fluorescence signals were recorded at the end of each extension phase. Primer and probe sequences are provided in Supplementary Table 1. Each qPCR run included negative controls (no template) and inter-run calibrators to ensure the specificity, sensitivity, and reproducibility of the assay.
3.1 Midgut collection from H. longicornis
Engorged H. longicornis nymphs were collected from both B. microti-infected and uninfected groups at two critical time points: 0 day post-engorgement and 4 d post-engorgement. Each biological replicate consisted of a pooled sample of thirty ticks, with three replicates per group (n = 3). Ticks were surface-sterilized by immersion in 70% ethanol with gentle agitation (100 rpm) for 90 s, followed by three sequential washes in sterile phosphate-buffered saline (PBS, pH 7.4) to remove residual ethanol. Following meticulous dissection with fine forceps to isolate midguts and prevent contamination from neighboring tissues (e.g., salivary glands and reproductive organs), samples were immediately transferred to pre-cooled PBS. Each midgut was then incised using sterile surgical blades and subjected to three sequential washes with PBS to ensure the complete removal of residual hemolymph components. Finally, samples were promptly flash-frozen in 500 μL of RNAlater Stabilization Solution (Thermo Fisher Scientific) to preserve RNA integrity.
3.2 RNA extraction and qPCR
Total RNA was extracted from the dissected midgut tissues using TRIzol reagent (Invitrogen), according to the manufacturer's protocol. Frozen samples were thawed on ice and homogenized in 1 mL TRIzol reagent per 50-−100 mg of tissue. Following a 5 min incubation at room temperature, 0.2 mL chloroform was added per 1 mL of TRIzol, and the mixture was vigorously shaken for 15 s before centrifugation at 12,000 × g for 15 min at 4 °C. The aqueous phase, containing the RNA, was carefully transferred to a new RNase-free tube and mixed with an equal volume of isopropanol to precipitate RNA. After centrifugation at 12,000 × g for 10 min at 4 °C, the supernatant was discarded, and the resulting RNA pellet was washed twice with 75% ethanol, air-dried, and resuspended in RNase-free water. RNA concentration and purity were assessed using a NanoDrop spectrophotometer (Thermo Scientific), and the integrity was verified by agarose gel electrophoresis, ensuring a RIN > 8.0. RNA Aliquots were stored at −80 °C until further use. All procedures were performed under RNase-free conditions using DEPC-treated materials to minimize RNA degradation.
The RNA was converted to first-strand cDNA using a HiScript III RT SuperMix for qPCR (gDNA wiper) kit (Vazyme Biotech, China). The cDNA was used to analyze the relative quantitative changes in gene expression (Supplementary Table 2). Samples were subjected to qRT-PCR using ChamQ Universal SYBR qPCR Master Mix (Q711, Vazyme) in a QuantStudio™5 Real-Time PCR System (Applied Biosystems™, New York, U.S.A.), and all samples were analyzed with three replicates. Elongation factor-1 (ELF1A, GenBank registry number AB836665) is an internal control for relative gene expression (following the 2−ΔΔCt method) (Nijhof et al., 2009).
3.3 RNA-seq and transcriptomic analysis
High-quality total RNA samples were submitted to Omicsmart (China) for transcriptomic sequencing. Ribosomal RNA (rRNA) was depleted from the RNA samples using the Ribo-Zero Globin kit (Illumina, San Diego, CA, USA), and the enriched mRNA was fragmented and reverse-transcribed into first-strand cDNA using random hexamer primers. Second-strand cDNA synthesis was performed using a reaction mixture containing buffer, dNTPs (substituting dUTP for dTTP), RNase H, and DNA polymerase I. The resulting double-stranded cDNA was purified using a QiaQuick PCR purification kit (Qiagen) and subjected to end repair, adenine (A)-tailing, and adapter ligation to generate sequencing libraries. Second-strand cDNA was selectively degraded using Uracil-N-Glycosylase (UNG) to ensure strand specificity during sequencing. Library fragments were size-selected using agarose gel electrophoresis and amplified by PCR.
Sequencing was performed on the Illumina HiSeqTM 4000 platform, generating 150 bp paired-end reads. Raw sequencing reads were quality-filtered to obtain clean reads, which were subsequently aligned to the reference genome of H. longicornis using HISAT2 v2.1.0 (http://daehwankimlab.github.io/hisat2/). Transcript assembly and quantification were performed using StringTie v1.3.4 (https://ccb.jhu.edu/software/stringtie/index.shtml), enabling the identification of both annotated and novel transcripts. Gene expression levels were quantified across all samples based on the HISAT2 alignments. Differential gene expression analysis was conducted using the edgeR 3.12.1 (http://www.bioconductor.org/packages/release/bioc/html/edgeR.html). Read counts were normalized, and statistical significance was evaluated using negative binomial models, with False Discovery Rate (FDR) correction for multiple comparisons. Differentially Expressed Genes (DEGs) were defined based on the thresholds of FDR < 0.05 and |log2FC| > 1. Functional enrichment analyses were performed by mapping identified DEGs to the Gene Ontology (GO) (https://www.bioconductor.org/packages/release/data/annotation/html/GO.db.html) and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases (http://www.kegg.jp). Significantly enriched GO terms and KEGG pathways (P < 0.05) were identified, providing mechanistic insights into the transcriptomic responses of H. longicornis midgut tissue to B. microti infection and highlighting key host-pathogen interactions.
3.4 TUNEL assay
Midguts from engorged H. longicornis nymphs were dissected in PBS, fixed in 4% paraformaldehyde (PFA) at 4 °C for 24 h, dehydrated, and embedded in paraffin, and sectioned at a thickness of 5 μm. Tissue sections were deparaffinized, rehydrated, and subjected to proteinase K digestion (20 μg/mL, 37 °C, 30 min) to facilitate antigen retrieval, followed by permeabilization with 0.1% Triton X-100 on ice for 10 min. TUNEL staining was performed using a commercially available kit (Roche) following the manufacturer's instructions. Sections were incubated with a terminal deoxynucleotidyl transferase (TdT)/FITC-dUTP labeling mixture at 37 °C for 1 h. Negative (TdT) and positive (+DNase I) controls were included to confirm assay specificity. Nuclei were counterstained with DAPI and visualized using a fluorescence microscope equipped with appropriate filter sets. Apoptotic rates were calculated by TUNEL?/DAPI? percentages across three randomly selected fields.
3.5 Transmission electron microscopy (TEM)
This study used the same H. longicornis midgut epithelial cell samples and experimental methods as in our previous publication to observe the autophagosome (Magnification,7,000 × ) following B. microti infection (Chen et al., 2025). Specifically, dissected midguts were washed thrice with Phosphate-Buffered Saline (PBS) and fixed overnight at 4 °C in 2.5% glutaraldehyde. Subsequent post-fixation was performed using 1% osmium tetroxide (OsO4) in 0.1 M phosphate buffer (PBS; pH 7.4) for 2 h in the dark. After three PBS rinses (15 min), samples were dehydrated in a graded ethanol series (30−100%, 20 min), acetone-embedded, and polymerized (37°C overnight). Ultrathin sections (60-−80 nm) were mounted on copper grids and double-stained with 2% uranyl acetate and 2.6% lead citrate for 8 min in a CO2-free environment. Sections were air-dried and imaged using a HITACHI transmission electron microscope.
3.6 RNAi
Ticks were treated with gene-RNA interference (RNAi) according to previously published methods using the primers listed in Supplementary Table 3. Gene-specific RNA interference primers were designed against caspase-7, caspase-9, and ATG5 sequences from our transcriptome database, each incorporating a5′-T7 RNA polymerase promoter sequence, with Luciferase serving as the normalization control. Double-stranded RNA (dsRNA) was synthesized using the T7 RiboMAX™ Express RNAi System (Promega, Madison, WI, U.S.A.) following the manufacturer's protocol. Briefly, target-specific DNA fragments (200−500 bp) flanked by T7 promoters were transcribed at 37°C for 4 h, followed by thermal denaturation and controlled annealing to generate dsRNA. The product was treated with DNase I (15 min, 37°C) to eliminate template DNA, purified by ethanol precipitation, and quantified spectrophotometrically. For tick RNAi, 23 nL (10 μg/μL) of synthesized dsRNA was precisely injected into the root of the last pair of legs of the nymphs using a microinjector (Drummond Scientific, U.S.A.). Interference-treated ticks were left to stand for 12 h (n = 50) and then fed simultaneously with controls (n = 50) on the same mice infected with B. microti. Two engorged nymphs were assigned to each group, with at least five biological replicates included, followed by DNA extracted from the ticks for B. microti detection. One limitation of this study is that the susceptibility of each mouse to B. microti varies, which resulted in inter-batch and inter-group variations.
3.7 Rapamycin and 3-methyladenine treatment
The effects of autophagy on B. microti infection were investigated using the autophagy inducer Rapamycin (Beyotime # S1842) and the inhibitor 3-methyladenine (Solarbio #IM0190). Those were microinjected into engorged nymphs infected with B. microti at a volume of 69 nL (Feng et al., 2021). DMSO was used as a control, and samples were collected 3 d after injection.
3.8 Data analysis
Statistical analyses were performed using GraphPad Prism 6 software (GraphPad Software Inc., San Diego, CA, U.S.A.). Quantitative data were expressed as mean ± Standard Deviation (SD). Intergroup comparisons were conducted using the two-tailed Mann–Whitney U test, unpaired Student's t test, or one-way analysis of variance (ANOVA), as appropriate. A P value < 0.05 was considered statistically significant.
4 Results
4.1 RNA-seq of B. microti-infected ticks
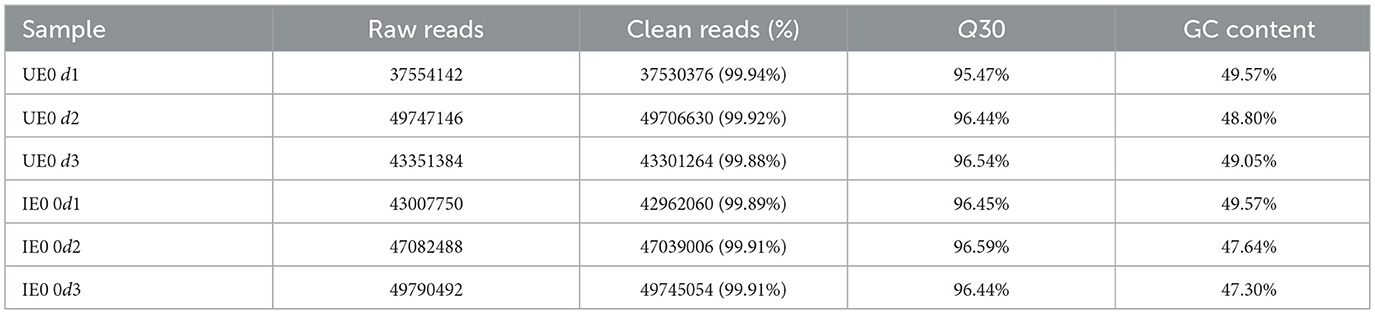
B. microti invade the tick midgut, differentiate into gametes, form a syncytium that migrates to the hemolymph and salivary glands (Jalovecka et al., 2019). We quantified B. microti in the midguts of engorged nymphal ticks by qPCR (n = 10 per group, with three biological replicates). The analysis demonstrated that parasite load peaked immediately after engorgement. Subsequently, a significant decline in parasite numbers was observed between d 1 and 3 post-engorgement. A transient rebound in parasite load occurred on day 4, followed by a further reduction on d 5 and 6 (Figure 1A). Principal Component Analysis (PCA) was conducted using the gmodels package in R to assess the variance in gene expression data. The resulting PCA plot revealed a strong tendency for biological replicates within each experimental group to cluster tightly, suggesting a high degree of reproducibility and reliability in the experimental data (Figure 1B). RNA-seq was performed on midgut tissues from H. longicornis nymphs at 0 and 4 d post-engorgement following B. microti infection based on quantitative detection of B. microti in engorged nymphal midguts (n = 10, 3 replicates). Raw sequencing reads from infected and uninfected midgut underwent stringent quality assessment before bioinformatic analysis at 0 and 4 d post-engorgement. Each sample generated over 4 GB of clean reads, with >99% read retention. The GC content ranged between 48% and 50%, and Q30 scores consistently exceeded 95%, meeting established quality thresholds for transcriptomic analyses. Clean reads were aligned to the H. longicornis reference genome (ASM966319v1) using HISAT2, achieving alignment rates greater than 50% across all samples. While this moderate mapping efficiency likely reflects genomic divergence between the reference bisexual strain and the parthenogenetic colony used in this study, the high Q30 scores (>93%) and stable GC content confirm the reliability of the dataset for downstream transcriptomic profiling (Table 1).
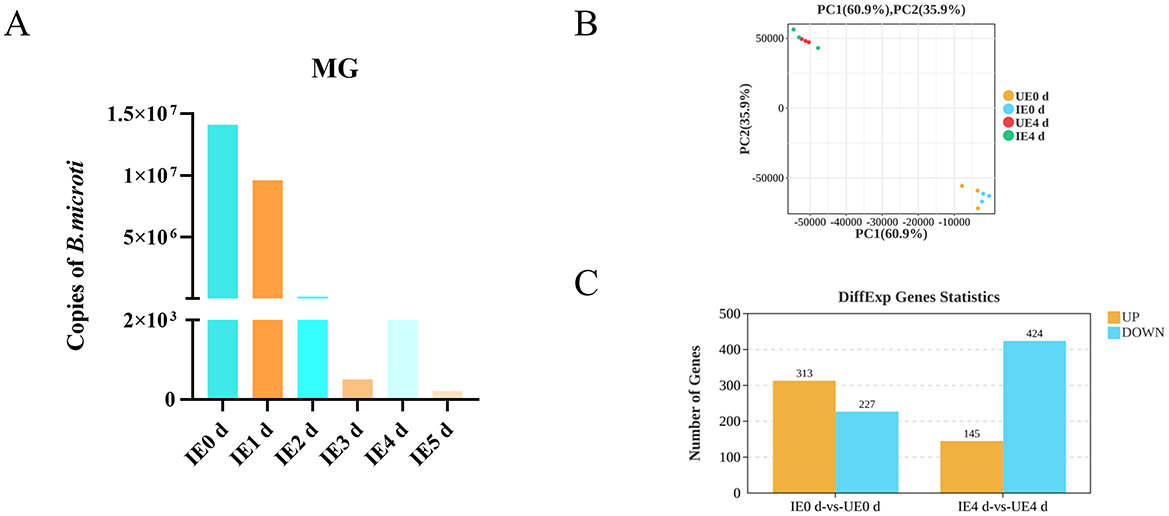
Figure 1. B. microti presence and differential gene expression in B. microti-infected tick midguts. (A) B. microti presence in the engorged nymph midgut (n = 3). (B) Principal component analysis (PCA) of gene expression profiles. (C) Upregulated and downregulated genes are indicated in red and blue, respectively. IE0 d: Infected B. microti at 0 day post-engorgement; IE1 d: Infected B. microti at 1 d post-engorgement; IE4 d: Uninfected B. microti at 4 d post-engorgement; UE0 d: Uninfected B. microti at 0 day post-engorgement; UE4 d: Uninfected B. microti at 4 d post-engorgement.
Comparative transcriptomic analysis revealed significant temporal changes in midgut gene expression in response to B. microti infection. At 0 d post-engorgement, 540 DEGs were identified, comprising three hundred and thirteen upregulated and two hundred and twenty seven downregulated transcripts (FDR < 0.05). By 4 d post-engorgement, 569 DEGs were identified, including one hundres and forty five upregulated and four hundred and twenty four downregulated, indicating a shift toward global transcriptional suppression (Figure 1C, Supplementary Tables 4, 5).
4.2 GO annotation
GO enrichment analysis of DEGs revealed temporally distinct functional responses in the midgut during B. microti infection. At 0 d post-engorgement, fifty eight significantly enriched GO terms (FDR < 0.05) were identified, comprising twenty four biological processes, fifteen molecular functions, and nineteen cellular components (Figure 2A). At 4 d post-engorgement, fifty five significantly enriched GO terms were identified, including twenty three biological processes, twelve molecular functions, and twenty cellular components, indicating persistent but restructured transcriptional activity (Figure 2B). Enriched biological processes included key cellular and metabolic processes, responses to external stimuli, developmental regulation, and cellular localization. Molecular functions were significantly associated with protein binding domains (particularly receptor-ligand interactions), enzymatic activity, and membrane transport. Enriched cellular components were related to plasma membrane structures, supramolecular complexes, and organelle luminal compartments (Figure 2).

Figure 2. GO enrichment analysis of DEGs in tick midguts at 0 (A) and 4 days (B) post-engorgement. Bar plots show significantly enriched GO terms across three functional categories: Biological Process (BP); Molecular Function (MF); and Cellular Component (CC). The x-axis represents the number of DEGs associated with each GO term, while the y-axis indicates the corresponding GO terms. Upregulated terms are shown in yellow, while downregulated terms are in blue. IE0 d: Infected B. microti at 0 day post-engorgement; IE4 d: Infected B. microti at 4 d post-engorgement; UE0 d: Uninfected B. microti at 0 day post-engorgement; UE4 d: Uninfected B. microti at 4 d post-engorgement.
4.3 KEGG pathway enrichment analysis
KEGG pathway analysis of DEGs revealed significant enrichment in six functional categories in response to B. microti infection: human disease, organismal systems, metabolism, genetic information processing, and cellular processes.
At 0 d post-engorgement, B. microti infection significantly perturbed several key biological pathways in H. longicornis (Supplementary Figure 1A). Pathway enrichment analysis identified significant alterations (Q < 0.05) in immune system processes, including antigen processing and presentation; metabolic pathways, such as steroid hormone biosynthesis, and linoleic acid metabolism; and digestive system functions, including protein digestion and absorption, pancreatic juice secretion, and mineral absorption. Furthermore, pathways associated with specific diseases, including Legionellosis, Toxoplasmosis, and Measles, were also significantly enriched. Although not reaching statistical significance (Q > 0.05), the cellular processes of apoptosis and autophagy exhibited a trend toward enrichment. These findings collectively suggest that B. microti infection elicits broad effects on a range of physiological functions in the tick host, encompassing immune responses, metabolic regulation, and nutrient absorption, even at the early stages of infection.
Although pathway enrichment analysis at 4 d post-engorgement did not identify statistically significant results (Q > 0.05), examining the top thirty pathways demonstrating trend changes revealed potentially relevant regulatory shifts (Supplementary Figure 1B). These pathways encompass processes related to the immune system, such as complement and coagulation cascades; the digestive system, including protein digestion and absorption; and cellular processes, including lysosome, apoptosis, and autophagy. While these pathways did not meet the threshold for statistical significance, they warrant further investigation as potential targets of regulation following engorgement.
4.4 Validation of RNA-seq findings by qRT-PCR
Given the important role of cellular processes in host-pathogen interactions, pathway enrichment analysis suggests that apoptosis and autophagy pathways may be involved in B. microti infection processes (Chen et al., 2021). To validate the transcriptomic results, the expression levels of key apoptosis-related genes (caspase-7, caspase-8, and caspase-9) and autophagy-related genes (ATG5, ATG6, ATG8, and ATG12) were analyzed by qRT-PCR at 0 d post-engorgement. These genes were selected based on their significant upregulation (P < 0.05) in the RNA-seq dataset and their established roles in cellular stress response, particularly apoptosis and autophagy. Our analysis revealed significant upregulation of key apoptosis-related genes, including caspase-7 (P = 0.022), caspase-8 (P = 0.0003), and caspase-9 (P = 0.0003), in response to infection (Figure 3A). Similarly, autophagy-related genes ATG5 (P = 0.029), ATG8 (P<0.0001), and ATG12 (P = 0.0045) showed marked transcriptional activation, while ATG6 (P = 0.28) and expression remained unchanged (Figure 3B). The qRT-PCR results corroborated the RNA-seq findings, demonstrating consistent and statistically significant upregulation of most selected transcripts. These results validated the reliability of the RNA-seq data, highlighting the activation of apoptotic and autophagic pathways in tick midgut following B. microti infection.
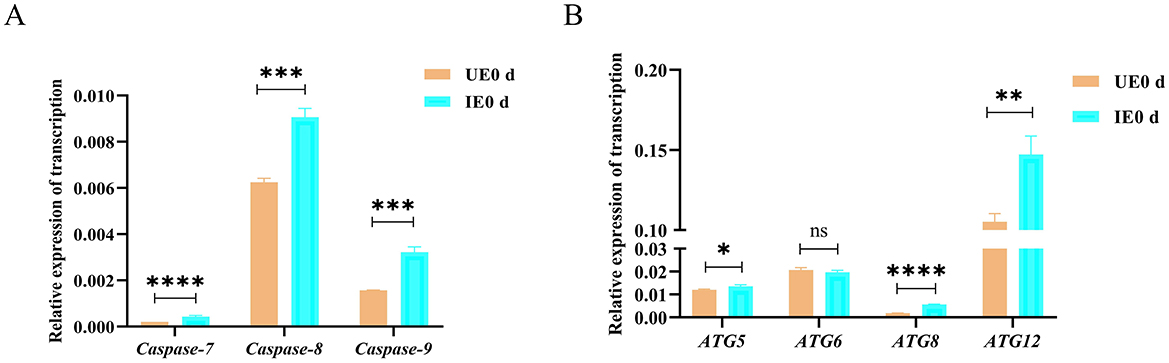
Figure 3. Transcriptional activation of apoptosis and autophagy pathways in tick midguts following B. microti infection (A) qRT-PCR analysis of apoptosis-related genes (caspase-7, caspase-8, and caspase-9) and in H. longicornis nymphs at day 0 post-engorgement (n = 3). (B) autophagy-related genes (ATG5, ATG6, ATG8, ATG12) in H. longicornis nymphs at day 0 post-engorgement (n = 3). B. microti-infected midguts (green) exhibited significant upregulation of apoptosis- and autophagy-related genes compared with uninfected controls (red). Data are presented as the mean ± standard error. P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001, differential gene expression analysis determined using Student's t test.
4.5 B. microti infection induces autophagy and apoptosis in tick midgut
Apoptosis and autophagy are tightly regulated cellular processes essential for maintaining tissue homeostasis and modulating host responses to pathogen invasion. Apoptosis is characterized by distinct nuclear morphological changes, including chromatin condensation, nuclear fragmentation, and karyolysis (Jiang et al., 2000). In contrast, autophagy involves the sequestration of damaged organelles or misfolded proteins within double-membrane autophagosomes, which subsequently fuse with lysosomes for degradation. To evaluate the impact of B. microti infection on apoptosis, TUNEL staining was performed on the tick midgut at 0 d post-engorgement. The results revealed a significant increase in fluorescence signal intensity in the midgut tissues of the infected group compared with the control (P < 0.05), indicating elevated apoptotic activity (Figure 4A). Complementary ultrastructural analysis by TEM revealed characteristic autophagic structures, including double- and multi-membrane-bound vesicles, within the midgut epithelial cells of infected ticks at 0 day post-engorgement, suggesting enhanced autophagic activity during early infection (Figure 4B, Supplementary Figure 2). These findings collectively indicate that B. microti infection concurrently activates apoptotic and autophagic pathways in tick midgut cells, highlighting their potential synergistic role in host-pathogen interactions.
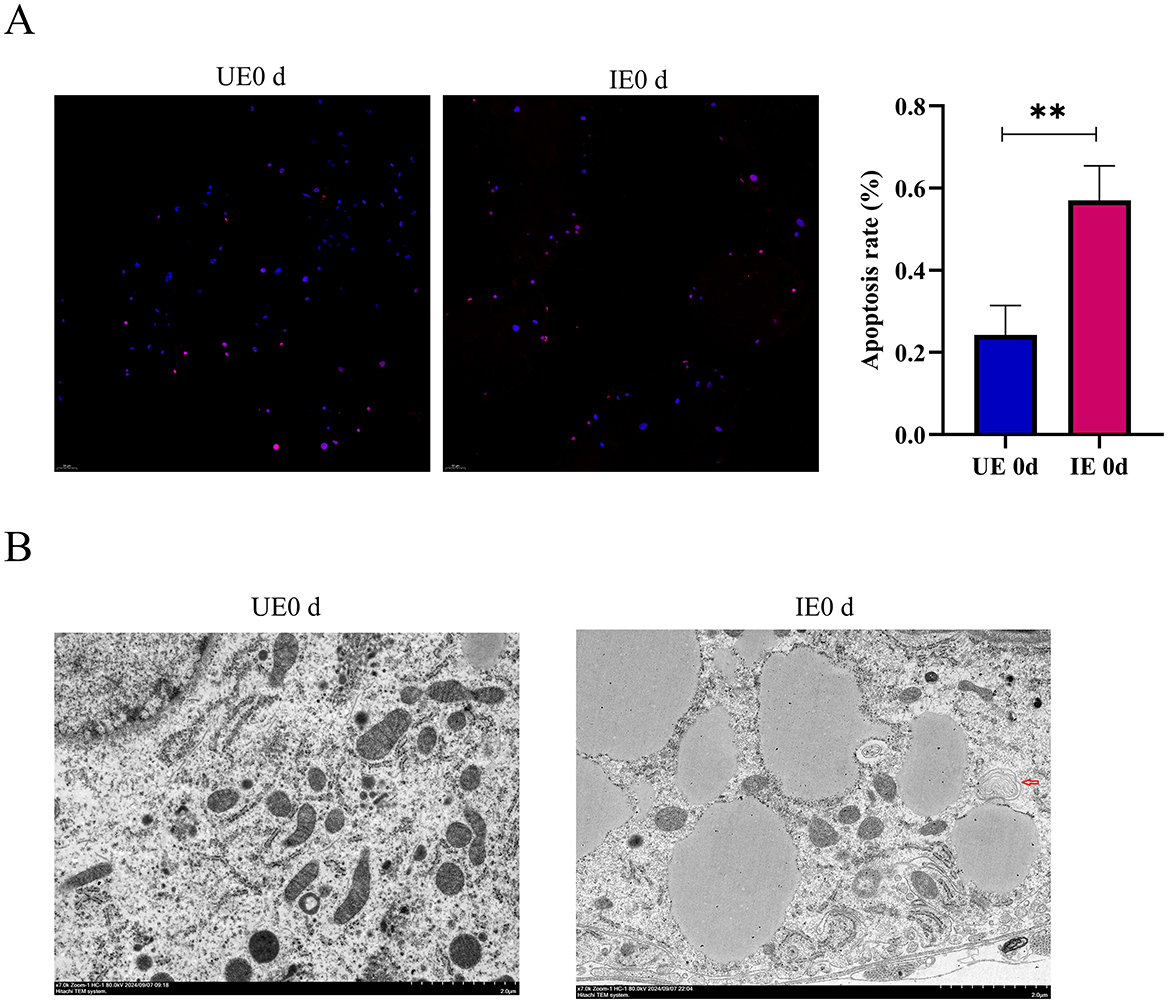
Figure 4. B. microti infection induces apoptosis and autophagy in tick midguts. (A) Apoptosis assessment in midgut tissues of engorged ticks by TUNEL staining (n = 3). Nuclei were counterstained with DAPI (blue) and apoptotic cells were labeled with TUNEL (red). Quantitative analysis of fluorescence intensity (right) showed significantly elevated apoptosis levels in B. microti-infected ticks. (B) TEM revealed autophagy activation in B. microti-infected ticks, indicated by characteristic double- or multi-membrane vesicles enclosing cytoplasmic contents (arrows, n = 3). This experiment was conducted concurrently with the mitochondrial observations reported by our previous publication, utilizing the same biological samples for autophagosome observation (Chen et al., 2025). UE0 d: Uninfected B. microti at 0 day post-engorgement; IE0 d: infected B. microti at 0 day post-engorgement. Data are presented as the mean ± standard error. P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001. differential gene expression analysis determined using Student's t test.
4.6 Caspase-7 and caspase-9 regulate tick acquisition of B. microti
To evaluate the functional role of apoptosis in B. microti acquisition, RNAi was used to silence the apoptotic regulators caspase-7 and caspase-9 in H. longicornis nymphs. Caspase-7 functions as an executioner caspase mediating the terminal phase of apoptosis in mammalian systems, whereas caspase-9 serves as an initiator caspase in the intrinsic (mitochondrial) apoptotic pathway (An et al., 2020; Ghazavi et al., 2022). Quantitative reverse transcription PCR (qRT-PCR) analysis confirmed the successful knockdown of caspase-7 (P = 0.0007) and caspase-9 (P < 0.0001) expression in nymphal ticks achieved through RNAi (Figure 5A). Gene knockdown of both caspases resulted in a significant reduction in B. microti load in infected ticks compared with the control at 0 d post-engorgement, indicating that these caspases are essential for efficient B. microti establishment in the ticks (Figure 5B). The observed variation in 18S rRNA copy number among control samples reflects biological differences in parasite acquisition, rather than technical or sampling errors.
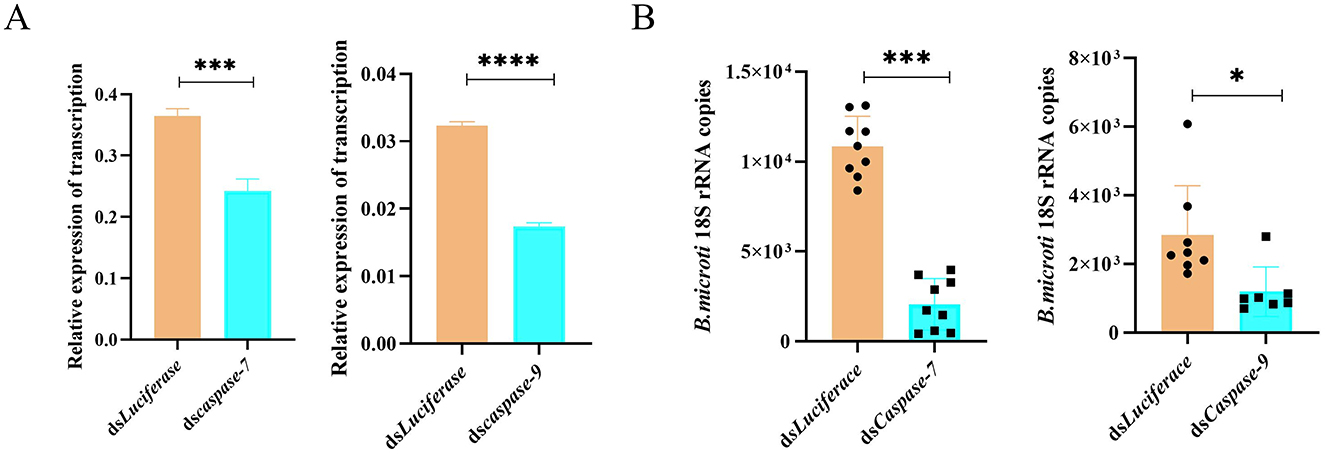
Figure 5. RNAi of caspase-7 and caspase-9 reduces B. microti infection in H. longicornis. (A) qPCR analysis of RNAi efficiency for caspase-7 and caspase-9 in engorged nymphs (n = 3). (B) qRT-PCR analysis demonstrated a significant reduction in B. microti load following caspase-7 (n = 9) and caspase-9 (n = 6) gene silencing in H. longicornis nymphs compared with luciferase dsRNA controls (n = 9 for caspase-7 comparison; n = 8 for caspase-9 comparison). Data are presented as the mean ± standard error. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001, differential gene expression analysis determined using Student's t test; B. microti load analysis determined using two-tailed Mann-Whitney U test.
4.7 Autophagy enhances tick susceptibility to B. microti infection
To further investigate the role of autophagy in B. microti infection, pharmacological modulation of autophagy was performed in infected ticks. Rapamycin (10 mM), an autophagy activator, and 3-MA (5 mM), an autophagy inhibitor, were microinjected into B. microti-infected ticks post-engorgement. Parasite burden was assessed 3 d post-treatment for each group, respectively. Rapamycin treatment significantly increased the B. microti load, whereas 3-MA significantly decreased the parasite burden, indicating that enhanced autophagic activity promotes B. microti survival (Figure 6A). To confirm the genetic basis of this observation, RNAi was conducted to silence ATG5 (P = 0.0001), a critical gene implicated in autophagosome formation and cross-regulation with apoptosis (Figure 6B). ATG5 knockdown significantly reduced B. microti load in tick midgut tissues, corroborating the pharmacological results (Figure 6C). These findings suggest that autophagy facilitates B. microti infection in H. longicornis, enhancing tick susceptibility to the parasite. Therefore, targeting autophagy-related pathways may represent a novel strategy to reduce vector competence and limit transmission of tick-borne babesiosis.
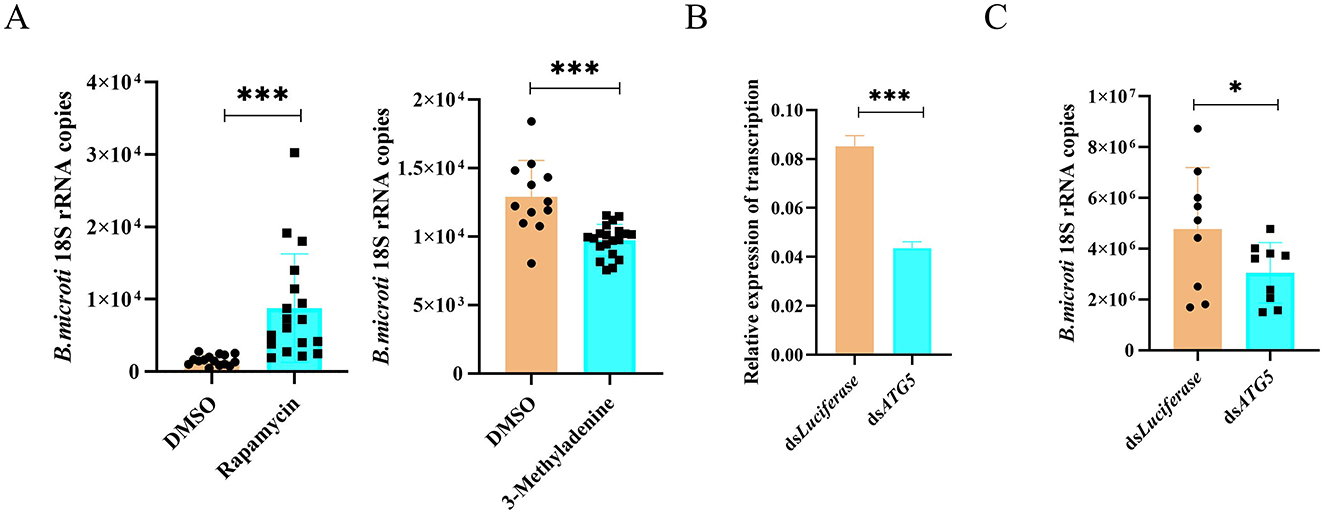
Figure 6. Autophagy promotes B. microti infection in H. longicornis. (A) Differential analysis of B. microti load in ticks treated with rapamycin (autophagy inducer, n = 18), 3-methyladenine (autophagy inhibitor, n = 20) and DMSO (n = 15 for rapamycin comparison; n = 12 for 3-methyladenine comparison). (B) qPCR analysis assessing RNAi efficiency for ATG5 genes in engorged tick nymphs (n = 3). (C) qRT-PCR analysis revealed that ATG5 knockdown significantly reduced B. microti acquisition in H. longicornis nymphs (n = 9). Data are presented as the mean ± standard error. *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001, differential gene expression analysis determined using Student's t test; B. microti load analysis determined using two-tailed Mann-Whitney U test.
5 Discussion
The transmission dynamics of B. microti primarily involve horizontal acquisition by tick larvae and nymphs during blood feeding, rather than transovarial transmission (Florin-Christensen et al., 2021; Wu et al., 2017). Consequently, the tick midgut and salivary glands serve as the principal sites for B. microti acquisition, replication, and transmission to vertebrate hosts (Gray et al., 2002; Rudzinska et al., 1983). Despite the crucial role of the midgut in vector competence, the specific molecular mechanisms underlying the complex interactions between Babesia spp. and their tick vectors in this tissue remain poorly understood. This study characterized tick midgut transcriptional responses to B. microti infection at 0 and 4 d post-engorgement. RNA-seq revealed significant autophagy and apoptosis pathway activation, validated by qRT-PCR and histology. The coordinated induction of these processes suggests B. microti manipulates host cell death pathways to enhance midgut infection, providing new insights into vector-pathogen adaptation.
Comparative transcriptomic analyses of B. microti-infected ticks during the early stages of infection revealed significant enrichment of metabolic and cellular processes, particularly those associated with nutrient transport, energy metabolism, cell proliferation, and cell death. Unlike previous transcriptomic studies that analyzed whole engorged nymphs, this study specifically focused on the tick midgut—the primary site for B. microti invasion and establishment, thus providing tissue-specific insights into tick-pathogen interactions at this critical interface (Feng et al., 2025). The whole transcriptome analysis identified a total of 1,051 DEGs, with a predominance of upregulated genes. Similarly, this study demonstrated a significant increase in upregulated genes following B. microti infection in the midgut of H. longicornis. This trend aligns with findings in other tick species, such as Amblyomma aureolatum, where Rickettsia rickettsii infection resulted in a significantly increased number of upregulated genes within the midgut compared with down regulated genes (Martins et al., 2017). The preferential upregulation of gene expression levels in response to B. microti infection suggests a selective activation of host immune and metabolic pathways rather than global suppression of host transcription. Given the midgut's critical role as the primary site for B. microti colonization and as the initial immunologic and physiologic barrier to pathogen invasion, the observed transcriptional changes likely reflect key host-pathogen interactions, particularly within pathways related to immunity, digestion, lipid metabolism, infectious disease, and cell growth (Cabezas-Cruz et al., 2019; De la Fuente et al., 2017). The predominance of upregulated genes in the midgut transcriptome indicates the tick's active counter-response aimed at limiting parasite proliferation through mechanisms such as antimicrobial peptide synthesis, oxidative stress induction, or metabolic reprogramming—a hypothesis requiring further functional validation.
Apoptosis and autophagy emerged as the predominant cell death pathways significantly enriched among upregulated Coding Sequences (CDSs) in the B. microti-infected H. longicornis nymphal midgut, highlighting their critical roles in modulating tick-pathogen interactions. Apoptosis, a highly regulated form of PCD, has been increasingly recognized for its role in modulating tick-pathogen interactions. Several studies have explored the potential mechanisms underlying apoptosis in tick cells and how pathogens manipulate these processes to enhance their survival and replication within the tick vector (Wang and Cull, 2022). While typically activated as a defense mechanism in response to cellular damage or infection, several tick-borne pathogens have evolved strategies to modulate apoptotic signaling to enhance their survival. For instance, Anaplasma phagocytophilum infection in I. scapularis induces host cell apoptosis, which paradoxically restricts bacterial proliferation by eliminating infected cells (Ayllón et al., 2015). However, under certain conditions, apoptosis may facilitate pathogen invasion and colonization. Specifically, the apoptosis-associated protein porin in ticks has been shown to promote Babesia infection during the early stages of tick blood engorgement (Zheng et al., 2020). These context-dependent effects of apoptosis suggest that, in some cases, this pathway may support pathogen infection and transmission. The present study supports this hypothesis, demonstrating increased apoptotic activity in tick midgut tissues following B. microti infection, ultimately enhancing parasite burden, as evidenced by RNAi-mediated silencing of caspase-7, caspase-9. Notably, Babesia infection exhibits tissue-specific modulation of host cell death pathways in ticks. Transcriptomic analyses of B. bovis and B. bigemina-infected Rhipicephalus microplus hemolymph reveal significant suppression of apoptotic pathways (Vimonish et al., 2025). This study provides the first experimental evidence that the apoptotic machinery of H. longicornis contributes to B. microti acquisition, highlighting caspase-7 and caspase-9 as potential molecular targets for transmission-blocking intervention against tick-borne babesiosis.
Autophagy, a conserved cellular degradation process, mediates lysosomal degradation of damaged organelles, misfolded proteins, and intracellular pathogens through the formation of double-membrane autophagosomes (Siqueira et al., 2018). Although autophagy typically functions as a host defense mechanism, certain tick-borne pathogens can exploit this process to enhance their intracellular survival. For instance, A. phagocytophilum secretes the effector protein Ats-1, which interacts with Beclin-1 to induce autophagosome formation, creating a nutrient-rich niche favorable to pathogen replication (Niu et al., 2012). Similarly, Rickettsia buchneri activates autophagy in tick cells to enhance its proliferation (Wang et al., 2024). In contrast, Plasmodium vivax infections induce autophagy in Anopheles aquasalis mosquitoes, leading to a reduction in parasite load and transmission potential (Santana et al., 2019). RNAi-mediated knockdown of the autophagy gene ATG5 significantly reduced parasite burden, whereas pharmacological induction of autophagy promoted parasite proliferation. These findings underscore the complex interplay between autophagy and tick-borne pathogens, highlighting its dual role as an immune effector and as a resource exploited by invading pathogens to support their survival and colonization within tick midguts.
Emerging evidence indicates that some pathogens concurrently induce autophagy and apoptosis in host cells to enhance their survival, replication, and transmission. For instance, Toxoplasma gondii secretes effector proteins such as ROP16 and ROP18, which modulate autophagosome formation, leading to the encapsulation of the parasite within a protective endo-vesicular structure, thereby facilitating immune evasion during early infection stages (Cheng et al., 2022). Additionally, T. gondii activates host caspase pathways, particularly caspase-3, which plays a critical role in inducing cell apoptosis, facilitating parasite egress and propagation (Payne et al., 2003). Similarly, Plasmodium spp. promotes parasite proliferation by activating autophagic pathways, enabling the acquisition of essential nutrients such as amino acids and lipids from host cells (Thieleke-Matos et al., 2016). It also induces apoptosis in erythrocytes to promote parasite release and systemic spread of the infection. Consistent with these findings, the present study demonstrated that Babesia infection induces both autophagy and apoptosis in ticks to increase infectivity. Functional validation through RNAi confirmed that silencing of caspase-7, caspase-9, and ATG5 genes significantly reduced B. microti loads, reinforcing the hypothesis that these pathways are co-opted by the parasite to enhance its proliferation and survival within the tick midgut. While transcriptional changes in apoptosis (caspase-7, caspase-9) and autophagy (ATG5, ATG8) genes were observed, protein-level validation (cleaved caspases, LC3B-II) is required to confirm pathway activation. Future studies should quantify caspase cleavage and LC3-I/II conversion via Western blot to elucidate their roles in Babesia-tick interactions. Future investigations should examine the incidence of these processes in both infected and adjacent uninfected cells, characterize their cell-type specificity (e.g., differentiating between immune and non-immune populations), and determine the relative contributions of apoptosis and autophagy to the course of B. microti infection. Clarifying these spatial and cellular dynamics will elucidate how Babesia modulates host pathways, informing novel intervention strategies. Subsequent studies are warranted to elucidate the precise molecular mechanism underlying the modulatory role of B. microti on autophagy and apoptosis in ticks and to provide crucial insights into the cellular pathogenesis of Babesia infection, establishing a theoretical foundation for novel intervention strategies.
GO and KEGG enrichment analyses suggest that B. microti infection may modulate autophagy and apoptosis pathways in the tick midgut. Subsequent experimental validation confirmed that both processes facilitate B. microti infection. Collectively, these findings provide robust molecular evidence that B. microti actively modulates tick autophagy and apoptosis pathways to promote its survival and vector competence, offering novel insights into the complex interactions between B. microti and its tick vector, H. longicornis.
6 Conclusion
This study demonstrated that B. microti infection significantly upregulated genes associated with PCD pathways, particularly apoptosis and autophagy, in H. longicornis. Functional experiments demonstrated that RNAi-mediated knockdown of caspase-7, caspase-9, and ATG5 genes effectively suppressed parasite proliferation, highlighting the pro-parasitic roles of apoptosis and autophagy in B. microti-infected ticks. These findings strongly suggest that B. microti modulates host PCD mechanisms to enhance its survival and transmission potential in tick midguts. Furthermore, this study provides a theoretical foundation for future investigations into the precise molecular mechanisms underlying Babesia-tick interactions and highlights the potential of targeting apoptosis and autophagy pathways as transmission-blocking strategies against tick-borne babesiosis.
Data availability statement
The raw sequencing data supporting the findings of this study have been deposited in the NCBI Sequence Read Archive (SRA) under the BioProject accession number PRJNA1328440.
Ethics statement
The animal study was approved by the Institutional Animal Care and Use Committee and the Animal Ethics Committee of the Shanghai Veterinary Research Institute. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
SC: Methodology, Validation, Data curation, Formal analysis, Writing – review & editing, Software, Writing – original draft, Resources, Investigation. SH: Data curation, Methodology, Writing – review & editing, Resources. FG: Methodology, Writing – review & editing, Formal analysis. HZhu: Writing – review & editing, Methodology, Formal analysis. YZ: Writing – review & editing, Investigation. JC: Investigation, Writing – review & editing. HZha: Visualization, Project administration, Conceptualization, Software, Writing – review & editing, Investigation. YW: Conceptualization, Writing – review & editing, Resources. JZ: Funding acquisition, Resources, Conceptualization, Validation, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This study was supported by the National Key Research and Development Program of China [grant number 2022YFD1800200].
Acknowledgments
We thank LetPub (www.letpub.com.cn) for its linguistic assistance during the preparation of this manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Gen AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1632974/full#supplementary-material
References
An, H. K., Chung, K. M., Park, H., Hong, J., Gim, J. E., Choi, H., et al. (2020). CASP9 (caspase 9) is essential for autophagosome maturation through regulation of mitochondrial homeostasis. Autophagy 16, 1598–1617. doi: 10.1080/15548627.2019.1695398
Antunes, S., Rosa, C., Couto, J., Ferrolho, J., and Domingos, A. (2017). Deciphering Babesia-Vector Interactions. Front. Cell. Infect. Microbiol. 7:429. doi: 10.3389/fcimb.2017.00429
Ayllón, N., Villar, M., Galindo, R. C., Kocan, K. M., Šíma, R., López, J.A., et al. (2015). Systems biology of tissue-specific response to Anaplasma phagocytophilum reveals differentiated apoptosis in the tick vector Ixodes scapularis. PLoS Genet. 11:e1005120. doi: 10.1371/journal.pgen.1005120
Beard, C. B., Occi, J., Bonilla, D. L., Egizi, A. M., Fonseca, D. M., Mertins, J. W., et al. (2018). Multistate infestation with the exotic disease-vector tick Haemaphysalis longicornis- United States, August 2017-September 2018. MMWR Morb. Mortal. Wkly. Rep. 67, 1310–1313. doi: 10.15585/mmwr.mm6747a3
Bloch, E. M., Kumar, S., and Krause, P. J. (2019). Persistence of Babesia microti Infection in Humans. Pathogens 8:102. doi: 10.3390/pathogens8030102
Brossard, M., and Wikel, S. K. (2004). Tick immunobiology. Parasitology 129, S161–S176. doi: 10.1017/S0031182004004834
Cabezas-Cruz, A., Espinosa, P., Alberdi, P., and de la Fuente, J. (2019). Tick-pathogen interactions: the metabolic perspective. Trends Parasitol. 35, 316–328. doi: 10.1016/j.pt.2019.01.006
Chen, S., Hu, S., Zhou, Y., Cao, J., Zhang, H., Wang, Y., et al. (2025). Tick HRF-dependent ferroptosis pathway to promote tick acquisition of Babesia microti. Front. Cell. Infect. Microbiol. 15:1560152. doi: 10.3389/fcimb.2025.1560152
Chen, X., Zeh, H. J., Kang, R., Kroemer, G., and Tang, D. (2021). Cell death in pancreatic cancer: from pathogenesis to therapy. Nat. Rev. Gastroenterol. Hepatol. 18, 804–823. doi: 10.1038/s41575-021-00486-6
Cheng, A., Zhang, H., Chen, B., Zheng, S., Wang, H., Shi, Y., et al. (2022). Modulation of autophagy as a therapeutic strategy for Toxoplasma gondii infection. Front. Cell. Infect. Microbiol. 12:902428. doi: 10.3389/fcimb.2022.902428
De la Fuente, J., Antunes, S., Bonnet, S., Cabezas-Cruz, A., Domingos, A. G., Estrada-Peña, A., et al. (2017). Tick-pathogen interactions and vector competence: identification of molecular drivers for tick-borne diseases. Front. Cell. Infect. Microbiol. 7:114. doi: 10.3389/fcimb.2017.00114
Diuk-Wasser, M. A., Liu, Y., Steeves, T. K., Folsom-O'Keefe, C., Dardick, K. R., Lepore, T., et al. (2014). Monitoring human babesiosis emergence through vector surveillance New England, USA. Emerg. Infect. Dis. 20, 225–231. doi: 10.3201/eid1302/130644
Dobler, G., Gniel, D., Petermann, R., and Pfeffer, M. (2012). Epidemiology and distribution of tick-borne encephalitis. Wien. Med. Wochenschr. 162, 230–238. doi: 10.1007/s10354-012-0100-5
Feng, T., Tong, H., Zhang, F., Zhang, Q., Zhang, H., Zhou, X., et al. (2025). Transcriptome study reveals tick immune genes restrict Babesia microti infection. Insect. Sci. 32, 457–470. doi: 10.1111/1744-7917.13384
Feng, Y., Chen, L., Gao, L., Dong, L., Wen, H., Song, X., et al. (2021). Rapamycin inhibits pathogen transmission in mosquitoes by promoting immune activation. PLoS Pathog. 17:e1009353. doi: 10.1371/journal.ppat.1009353
Florin-Christensen, M., Wieser, S. N., Suarez, C. E., and Schnittger, L. (2021). In silico survey and characterization of Babesia microti functional and non-functional proteases. Pathogens 10:1457. doi: 10.3390/pathogens10111457
Fogaça, A. C., Sousa, G., Pavanelo, D. B., Esteves, E., Martins, L. A., Urbanová, V., et al. (2021). Tick immune system: what is known, the interconnections, the gaps, and the challenges. Front. Immunol. 12:628054. doi: 10.3389/fimmu.2021.628054
Ghazavi, F., Huysentruyt, J., De Coninck, J., Kourula, S., Martens, S., Hassannia, B., et al. (2022). Executioner caspases 3 and 7 are dispensable for intestinal epithelium turnover and homeostasis at steady state. Proc. Nat. Acad. Sci. U S A. 119:e2024508119. doi: 10.1073/pnas.2024508119
Gray, J., von Stedingk, L. V., Gürtelschmid, M., and Granström, M. (2002). Transmission studies of Babesia microti in Ixodes ricinus ticks and gerbils. J. Clin. Microbiol. 40, 1259–1263. doi: 10.1128/JCM.40.4.1259-1263.2002
Hart, C. E., and Thangamani, S. (2021). Tick-virus interactions: current understanding and future perspectives. Parasite Immunol. 43:e12815. doi: 10.1111/pim.12815
Jalovecka, M., Sojka, D., Ascencio, M., and Schnittger, L. (2019). Babesia Life cycle—When Phylogeny Meets Biology. Trends Parasitol. 35, 356–368. doi: 10.1016/j.pt.2019.01.007
Jiang, Z. F., Zhao, Y., Hong, X., and Zhai, Z. H. (2000). Nuclear apoptosis induced by isolated mitochondria. Cell Res. 10, 221–232. doi: 10.1038/sj.cr.7290051
Jorgensen, I., Rayamajhi, M., and Miao, E. A. (2017). Programmed cell death as a defence against infection. Nat. Rev. Immunol. 17, 151–164. doi: 10.1038/nri.2016.147
Kang, J. G., Ko, S., Smith, W. B., Kim, H. C., Lee, I. Y., and Chae, J. S. (2016). Prevalence of Anaplasma, Bartonella and Borrelia Species in Haemaphysalis longicornis collected from goats in North Korea. J. Vet. Sci. 17, 207–216. doi: 10.4142/jvs.2016.17.2.207
Krause, P. J., McKay, K., Gadbaw, J., Christianson, D., Closter, L., Lepore, T., et al. (2003). Increasing health burden of human babesiosis in endemic sites. Am. J. Trop. Med. Hyg. 68, 431–436. doi: 10.4269/ajtmh.2003.68.431
Lin, M., Liu, H., Xiong, Q., Niu, H., Cheng, Z., Yamamoto, A., et al. (2016). Ehrlichia secretes Etf-1 to induce autophagy and capture nutrients for its growth through RAB5 and class III phosphatidylinositol 3-kinase. Autophagy 12, 2145–2166. doi: 10.1080/15548627.2016.1217369
Martins, L. A., Galletti, M., Ribeiro, J. M., Fujita, A., Costa, F. B., Labruna, M. B., et al. (2017). The distinct transcriptional response of the midgut of amblyomma sculptum and Amblyomma aureolatum ticks to Rickettsia rickettsii correlates to their differences in susceptibility to infection. Front. Cell. Infect. Microbiol. 7:129. doi: 10.3389/fcimb.2017.00129
Martins, L. A., Palmisano, G., Cortez, M., Kawahara, R., de Freitas Balanco, J. M., Fujita, A., et al. (2020). The intracellular bacterium Rickettsia rickettsii exerts an inhibitory effect on the apoptosis of tick cells. Parasit Vectors 13:603. doi: 10.1186/s13071-020-04477-5
Nagata, S., and Tanaka, M. (2017). Programmed cell death and the immune system. Nat. Rev. Immunol. 17, 333–340. doi: 10.1038/nri.2016.153
Nijhof, A. M., Balk, J. A., Postigo, M., and Jongejan, F. (2009). Selection of reference genes for quantitative RT-PCR studies in Rhipicephalus (Boophilus) microplus and Rhipicephalus appendiculatus ticks and determination of the expression profile of Bm86. BMC Mol. Biol. 10:112. doi: 10.1186/1471-2199-10-112
Niu, H., Xiong, Q., Yamamoto, A., Hayashi-Nishino, M., and Rikihisa, Y. (2012). Autophagosomes induced by a bacterial Beclin 1 binding protein facilitate obligatory intracellular infection. Proc. Nat. Acad. Sci. U S A. 109, 20800–20807. doi: 10.1073/pnas.1218674109
Payne, T. M., Molestina, R. E., and Sinai, A. P. (2003). Inhibition of caspase activation and a requirement for NF-kappaB function in the Toxoplasma gondii-mediated blockade of host apoptosis. J. Cell. Sci. 116, 4345–4358. doi: 10.1242/jcs.00756
Persing, D. H., Mathiesen, D., Marshall, W. F., Telford, S. R., Spielman, A., Thomford, J. W., et al. (1992). Detection of Babesia microti by polymerase chain reaction. J. Clin. Microbiol. 30, 2097–2103. doi: 10.1128/jcm.30.8.2097-2103.1992
Rainey, T., Occi, J. L., Robbins, R. G., and Egizi, A. (2018). Discovery of Haemaphysalis longicornis (Ixodida: Ixodidae) parasitizing a sheep in New Jersey, United States. J. Med. Entomol. 55, 757–759. doi: 10.1093/jme/tjy006
Rochlin, I. (2019). Modeling the Asian longhorned tick (Acari: Ixodidae) suitable habitat in north America. J. Med. Entomol. 56, 384–391. doi: 10.1093/jme/tjy210
Rollend, L., Bent, S. J., Krause, P. J., Usmani-Brown, S., Steeves, T. K., States, S. L., et al. (2013). Quantitative PCR for detection of Babesia microti in Ixodes scapularis ticks and in human blood. VBZD 13, 784–790. doi: 10.1089/vbz.2011.0935
Rosenberg, R., Lindsey, N. P., Fischer, M., Gregory, C. J., Hinckley, A. F., Mead, P. S., et al. (2018). Vital signs: trends in reported vectorborne disease cases—United States and Territories, 2004-2016. MMWR. 67, 496–501. doi: 10.15585/mmwr.mm6717e1
Rudzinska, M. A., Spielman, A., Lewengrub, S., Trager, W., and Piesman, J. (1983). Sexuality in piroplasms as revealed by electron microscopy in Babesia microti. Proc. Nat. Acad. Sci. U S A. 80, 2966–2970. doi: 10.1073/pnas.80.10.2966
Santana, R. A. G., Oliveira, M. C., Cabral, I., Junior, R., de Sousa, D. R. T., Ferreira, L., et al. (2019). Anopheles aquasalis transcriptome reveals autophagic responses to Plasmodium vivax midgut invasion. Parasit Vectors 12:261. doi: 10.1186/s13071-019-3506-8
Siqueira, M. D. S., Ribeiro, R. M., and Travassos, L. H. (2018). Autophagy and its interaction with intracellular bacterial pathogens. Front. Immunol. 9:935. doi: 10.3389/fimmu.2018.00935
Tanne, J. H. (2018). New tick seen in nine US states is an emerging disease threat, warns CDC. BMJ 36:k5191. doi: 10.1136/bmj.k5191
Thieleke-Matos, C., Lopes da Silva, M., Cabrita-Santos, L., Portal, M. D., Rodrigues, I. P., Zuzarte-Luis, V., et al. (2016). Host cell autophagy contributes to Plasmodium liver development. Cell. Microbiol. 18, 437–450. doi: 10.1111/cmi.12524
Vimonish, R., Capelli-Peixoto, J., Johnson, W., Kappmeyer, L., Saelao, P., Taus, N., et al. (2025). Transcriptomic analysis of Rhipicephalus microplus hemocytes from female ticks infected with Babesia bovis or Babesia bigemina. Parasit Vectors 18:37. doi: 10.1186/s13071-025-06662-w
Wang, X. R., and Cull, B. (2022). Apoptosis and autophagy: current understanding in tick-pathogen interactions. Front. Cell. Infect. Microbiol. 12:784430. doi: 10.3389/fcimb.2022.784430
Wang, X. R., Cull, B., Oliver, J. D., Kurtti, T. J., and Munderloh, U. G. (2024). The role of autophagy in tick-endosymbiont interactions: insights from Ixodes scapularis and Rickettsia buchneri. Microbiol. Spectr. 12:e0108623. doi: 10.1128/spectrum.01086-23
Wikel, S. K. (1999). Tick modulation of host immunity: an important factor in pathogen transmission. Int. J. Parasitol. 29, 851–859. doi: 10.1016/S0020-7519(99)00042-9
Wu, J., Cao, J., Zhou, Y., Zhang, H., Gong, H., and Zhou, J. (2017). Evaluation on infectivity of Babesia microti to domestic animals and ticks outside the Ixodes Genus. Front. Microbiol. 8:1915. doi: 10.3389/fmicb.2017.01915
Yu, Z., Wang, R., Zhang, T., Wang, T., Nwanade, C. F., Pei, T., et al. (2023). The genome-wide characterization and associated cold-tolerance function of the superoxide dismutase in the cold response of the tick Haemaphysalis longicornis. Pestic. Biochem. Physiol. 195:105573. doi: 10.1016/j.pestbp.2023.105573
Yuan, C., Wu, J., Peng, Y., Li, Y., Shen, S., Deng, F., et al. (2020). Transcriptome analysis of the innate immune system of Hyalomma asiaticum. J. Invertebr. Pathol. 177:107481. doi: 10.1016/j.jip.2020.107481
Zhang, H., Sun, Y., Jiang, H., and Huo, X. (2017). Prevalence of severe febrile and thrombocytopenic syndrome virus, Anaplasma spp. and Babesia microti in hard ticks (Acari: Ixodidae) from Jiaodong Peninsula, Shandong Province. Vector Borne Zoonotic Dis. 17, 134–140. doi: 10.1089/vbz.2016.1978
Zhao, G. P., Wang, Y. X., Fan, Z. W., Ji, Y., Liu, M. J., Zhang, W. H., et al. (2021). Mapping ticks and tick-borne pathogens in China. Nat. Commun. 12:1075. doi: 10.1038/s41467-021-21375-1
Zheng, W., Umemiya-Shirafuji, R., Zhang, Q., Okado, K., Adjou Moumouni, P. F., Suzuki, H., et al. (2020). Porin expression profiles in Haemaphysalis longicornis infected with Babesia microti. Front. Physiol. 11:502. doi: 10.3389/fphys.2020.00502
Keywords: B. microti, tick, midgut, RNA-seq, apoptosis, autophagy
Citation: Chen S, Hu S, Gong F, Zhu H, Zhou Y, Cao J, Zhang H, Wang Y and Zhou J (2025) Apoptosis and autophagy promote Babesia microti infection in tick midguts: insights from transcriptomic and functional RNAi studies. Front. Microbiol. 16:1632974. doi: 10.3389/fmicb.2025.1632974
Received: 22 May 2025; Accepted: 20 August 2025;
Published: 19 September 2025.
Edited by:
Mubasher Hussain, Guangdong Pest Control Technology Group, ChinaReviewed by:
Mirinda Van Kleef, Agricultural Research Council of South Africa (ARC-SA), South AfricaElisa Azuara-Liceaga, Universidad Autónoma de la Ciudad de México, Mexico
Wang Fangfang, Hebei University of Engineering, China
Vipin Rana, University of Maryland, United States
Prasanna Babu Araveti, Boston University, United States
Abdul Qadeer, Central South University, China
Venkatesh Kumaresan, University of Texas at San Antonio, United States
Dávid Hargitai, Eötvös Loránd University, Hungary
Copyright © 2025 Chen, Hu, Gong, Zhu, Zhou, Cao, Zhang, Wang and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jinlin Zhou, amlubGluemhvdUBzaHZyaS5hYy5jbg==
 Songqin Chen1
Songqin Chen1 Yongzhi Zhou
Yongzhi Zhou Jie Cao
Jie Cao Houshuang Zhang
Houshuang Zhang Yanan Wang
Yanan Wang Jinlin Zhou
Jinlin Zhou