- 1Cellular and Molecular Diagnostics (Molecular Biology Group), ICMR-National Institute of Cancer Prevention and Research, Noida, India
- 2Symbiosis School of Biological Sciences, Symbiosis International (Deemed University) (SIU), Pune, India
- 3Department of Bioscience, Jamia Millia Islamia, New Delhi, India
- 4Laboratory Oncology Unit, Dr. BRA-IRCH, All India Institute of Medical Sciences, New Delhi, India
- 5Department of Pathology, Government Institute of Medical Sciences, Greater Noida, India
Cervical cancer is a one of the leading causes of mortality in women, and WHO’s initiative to eliminate cervical cancer by 2030 needs to explore several emerging research areas for its elimination such as epigenetics which could play a crucial important role in the cervical cancer pathogenesis driven by persistent high-risk-human papillomavirus infection. Understanding the molecular and epigenetic mechanisms underlying HPV infection and its progression to cancer is critical for advancing prevention, diagnosis, and treatment strategies, which may play a crucial role in eliminating cervical cancer. Persistent infection of human Papillomavirus is intricately linked to the initiation and progression of cervical cancer with different molecular mechanisms, pathways, viral genes, and proteins. HPV-mediated alterations in the host epigenome play a pivotal role in driving oncogenic transformation by modulating gene expression, chromatin dynamics, and DNA methylation patterns, ultimately disrupting normal cellular functions. The relationship between HPV-induced epigenetic changes and cancer progression underscores the virus’s ability to bypass conventional gene-silencing mechanisms. By altering critical regulatory pathways, HPV not only fosters cancerous growth but also influences patient responses to existing treatments, posing challenges to effective disease management. In this current review, we have discussed the role of epigenetic disruptions caused by HPV, which provided a unique opportunity to identify novel therapeutic targets and biomarkers. Epigenetic factors, being reversible and independent of direct genetic manipulation, offer promising avenues for innovative drug delivery strategies. Such approaches could enhance disease management by advancing therapeutic strategies and diagnostics for improving patient outcomes.
1 Introduction
Cervical cancer (CC) remains one of the leading causes of cancer-related deaths, especially in low-and middle-income countries (LMICs), and also 4th most common cancer in women globally. It is well-established that cervical cancer occurs due to persistent infection of high-risk human papillomavirus (HR-HPV); among all HR-HPV, HPV 16 and 18 are responsible for most of the cervical cancers (Khan et al., 2022). Although HPV infection usually goes away on its own, in some cases, it stays in the body and can lead to cancer due to other cellular, molecular mechanisms, and genetic and epigenetic alterations, which are crucial contributors to cervical carcinogenesis (Neyaz et al., 2008).
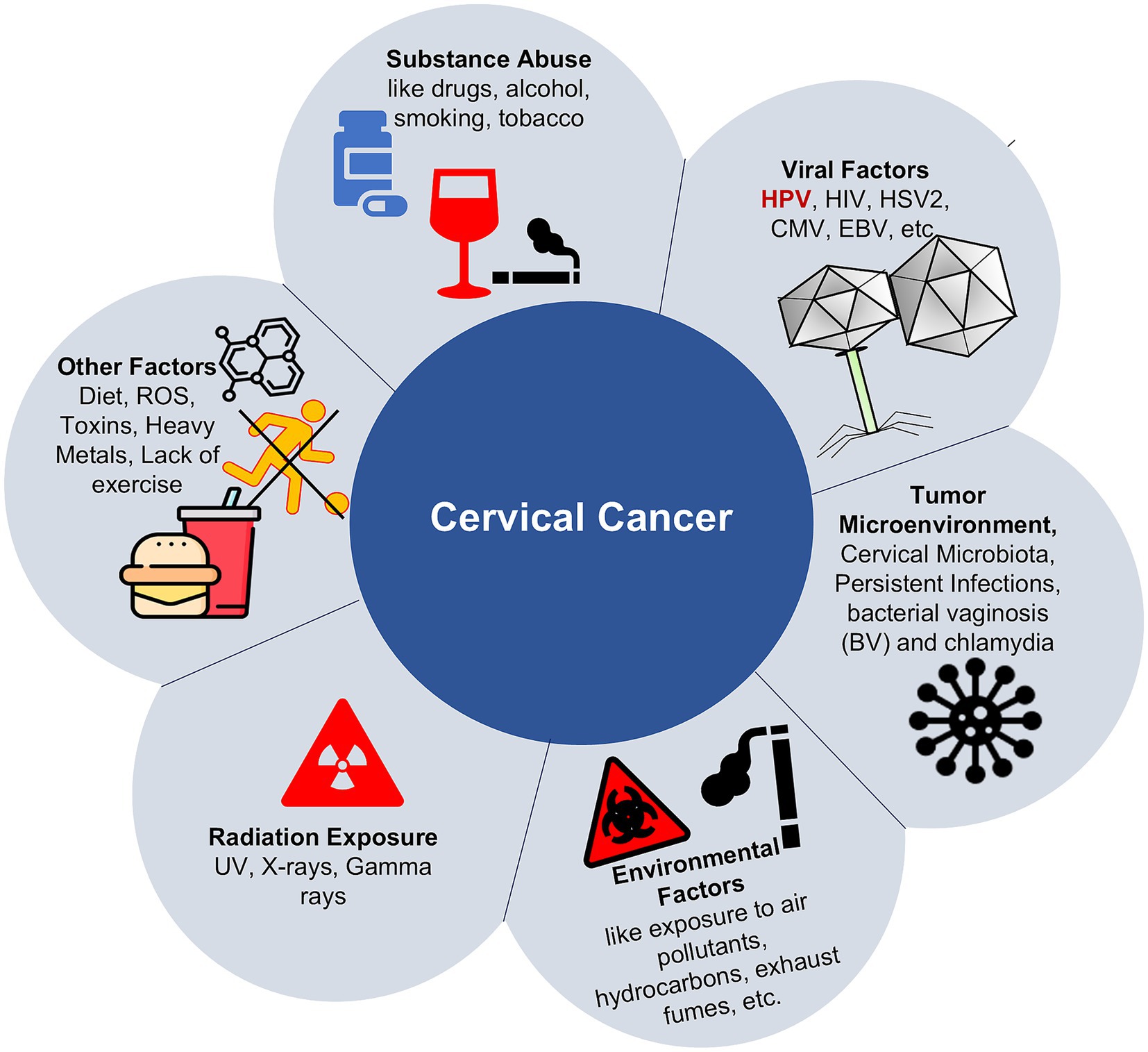
Epigenetics involves the regulation of gene expression through heritable modifications that do not alter the underlying DNA sequence (Hamilton, 2011; Gibney and Nolan, 2010; Dasari et al., 2015; Hanahan, 2022). There are several factors that influence epigenetic changes, such as diet, exposure to toxins, psychological stress, and smoking (Figure 1). This involves several epigenetic mechanisms, such as DNA methylation, histone modifications, chromatin remodeling, and regulation by non-coding RNAs, all of which are essential for controlling gene expression and preserving genomic stability. When these processes become dysregulated, these alterations can result in abnormal gene expression, contributing significantly to the initiation and progression of cancer (Hussain et al., 2022; Sobti et al., 2011).
Epigenetic alterations, including global DNA hypomethylation, tumor suppressor gene hypermethylation, and histone modifications, occur throughout cervical carcinogenesis in both HPV and host genomes. Establishment of HPV shows that it exploits the host’s epigenetic machinery to promote viral persistence, evade immune surveillance, and drive oncogenic transformation. The viral oncoproteins E6 and E7 are central players in this process, as they not only disrupt tumor suppressor pathways, notably p53 and Rb, but also modulate the host epigenetic landscape to favor malignancy (Da Silva et al., 2021).
Aberrant DNA methylation is among the earliest and most extensively investigated epigenetic modifications in HPV-associated cervical cancer. This modification involves attaching a methyl group to the 5-carbon position of cytosine bases located within CpG dinucleotide sequences, particularly in promoter regions, often resulting in transcriptional repression of the associated genes. In cervical cancer, hypermethylation of tumor suppressor gene promoters such as CDKN2A, DAPK1, RASSF1A, CADM1, and MAL has been consistently observed. These alterations result in the silencing of critical genes involved in apoptosis, cell cycle regulation, and cell adhesion, thereby promoting carcinogenesis (Zhu et al., 2020). Simultaneously, global hypomethylation, especially in repetitive elements and oncogenes, contributes to genomic instability and aberrant activation of proto-oncogenes. The E7 oncoprotein has been shown to upregulate DNA methyltransferases (DNMTs), particularly DNMT1 and DNMT3b, which mediate these methylation changes. E6, through degradation of p53, also indirectly influences DNA methylation and histone modification pathways, underscoring the cooperative roles of these viral proteins in epigenetic reprogramming (Sen et al., 2018).
An important epigenetic mechanism centers on histone proteins, which forms the structural framework around which the DNA is wrapped. These histones are subject to a range of post-translational modifications, such as acetylation, methylation, phosphorylation, and ubiquitination, that collectively regulate chromatin architecture and gene expression (Figure 2). These changes can either stimulate or suppress gene transcription depending on the nature and position of the modifications. In cervical cancer, aberrant histone modification patterns are frequently observed and are often mediated by the deregulation of histone-modifying enzymes (Zhao and Shilatifard, 2019). HPV oncoproteins have been found to interact with histone acetyltransferases (HATs) and histone deacetylases (HDACs), leading to an imbalance in acetylation patterns that influence gene expression. For example, E7 has been shown to inhibit HDAC activity, thereby promoting the expression of cell cycle-related genes. Additionally, the HPV E6 oncoprotein has been shown to interact with the histone methyltransferase EZH2, leading to increased trimethylation of histone H3 at lysine 27 (H3K27me3), a repressive epigenetic mark linked to gene silencing. This mechanism contributes to the epigenetic inactivation of crucial tumor suppressor genes and regulatory pathways, thereby promoting the progression of cervical carcinogenesis (Lourenco de Freitas et al., 2020).
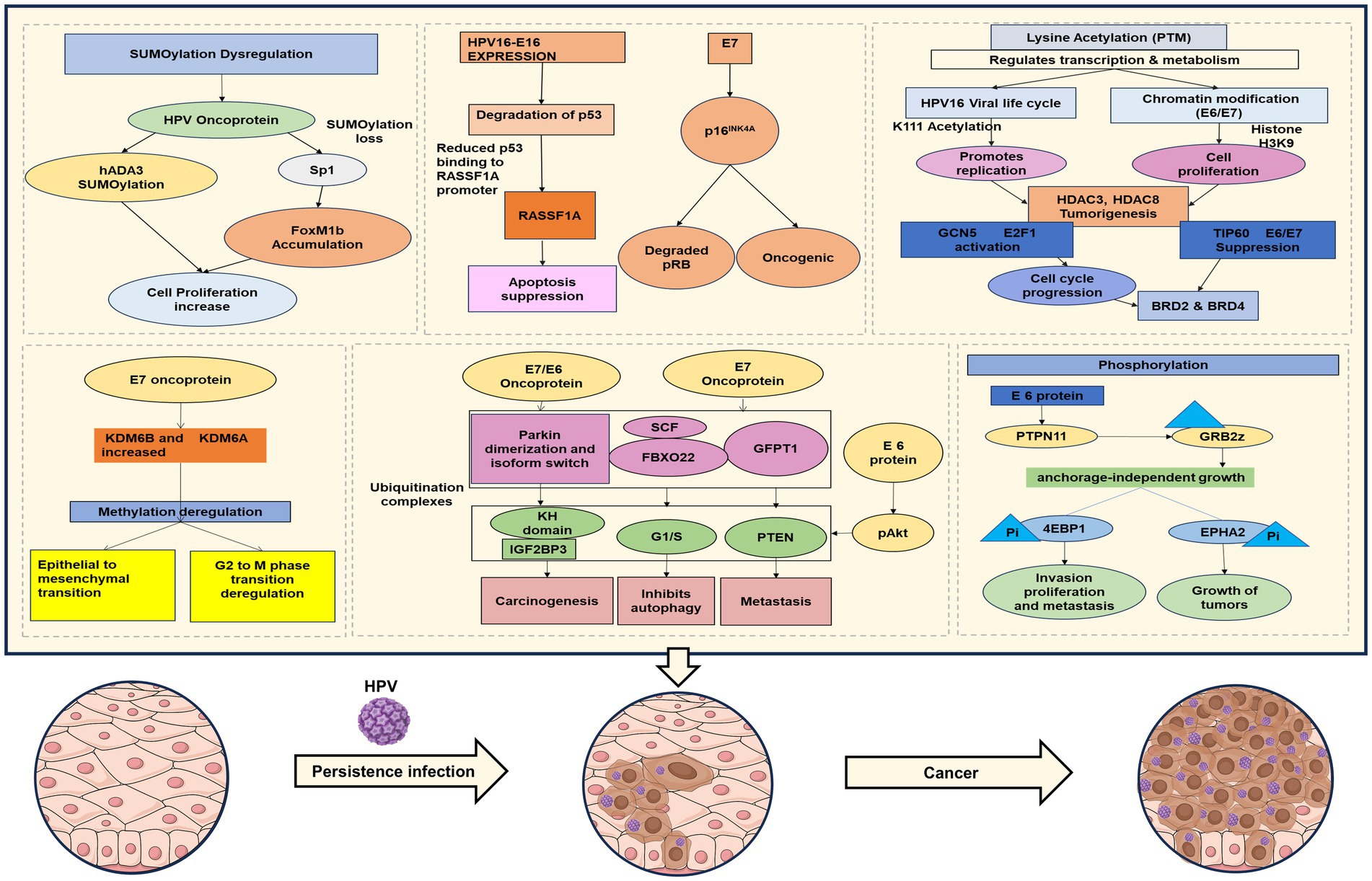
Figure 2. Overview of epigenetic changes/factors contributing to HPV-associated cervical cancer pathogenesis.
Non-coding RNAs (ncRNAs), particularly microRNAs (miRNAs) and long non-coding RNAs (lncRNAs), are increasingly recognized as crucial epigenetic regulators in the development and progression of cervical cancer. These RNAs do not encode proteins but regulate gene expression post-transcriptionally and via chromatin remodeling complexes. HPV infection alters the expression of several ncRNAs, which in turn modulate pathways critical to cell proliferation, apoptosis, and immune response (Hussain et al., 2022; Sharma and Munger, 2020). MiRNAs such as miR-34a, miR-203, and miR-375 are often downregulated in HPV-positive cervical cancers, either through direct targeting by HPV oncoproteins or through promoter hypermethylation. Conversely, oncogenic miRNAs like miR-21 are upregulated and contribute to tumor progression LncRNAs, such as HOTAIR, MALAT1, and PVT1, are also dysregulated in HPV-associated cervical cancer and function by recruiting epigenetic modifiers to specific genomic loci, thereby altering chromatin states and gene expression (Gomez-Gomez et al., 2013; Jiang et al., 2014; Zhou et al., 2021; Cho et al., 2018).
The interplay between HPV infection and host epigenetic machinery is central to the development and progression of cervical cancer. HPV-induced epigenetic reprogramming drives the silencing of tumor suppressor genes, activation of oncogenes, and disruption of normal cellular pathways, culminating in malignant transformation, and understanding these changes is crucial for future cervical cancer management. The aim of this current review is to discuss the role of epigenetics and its clinical implications in HPV-mediated cervical cancer, focusing on molecular epigenetic modifications, diagnostics/screening, and therapeutic strategies in cervical cancer and its future prospects.
2 Epigenetic mechanism and an overview of HPV mediated cervical cancer
Over the past few decades, extensive research into the natural history of cervical cancer has established that persistent infection with certain high-risk human papillomavirus (HPV) types is the primary etiological factor driving the disease’s development (Chan et al., 2019). Since the early 1980s, a strong and well-documented association has been established between human papillomavirus (HPV) infection and cervical squamous cell carcinoma. Notably, the strength of this association surpasses that observed between tobacco use and lung cancer (Chan et al., 2019). Around 30 sexually transmitted HPV types have been identified that can primarily infect the cervix, vagina, vulva, penis, and anus. Notably, one or more of these types are found in 99.7% of cervical squamous cell carcinoma cases, highlighting their crucial role in the pathogenesis of the disease (Chan et al., 2019; Walboomers et al., 1999).
High-risk human papillomavirus (HPV) types, especially HPV16 and HPV18, play a pivotal role in the development of cervical cancer, primarily through the oncogenic functions of their E6 and E7 proteins. HPV comprises a group of genetically related viruses, each classified based on nucleotide sequence variations and assigned a type number corresponding to the order of identification. More than 200 HPV types are known to exist (de Villiers et al., 2004) and Among all, approximately 15 are strongly linked to the development of cervical cancer. Genital HPV types are broadly categorized into high-risk (oncogenic) and low-risk (non-oncogenic) groups based on their potential to cause cervical malignancies and related precursor lesions. Low-risk types, such as HPV 6, 11, 42, 43, and 44, are typically associated with benign conditions like genital warts, whereas high-risk types—including HPV 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 68, 73, and 82—are implicated in cervical cancer and its premalignant stages (Walboomers et al., 1999; Unger and Barr, 2004). Although low-risk HPV subtypes are primarily associated with benign lesions, they have occasionally been detected in cervical carcinoma cases. HPV typically targets the mucocutaneous epithelium, where it replicates in differentiated epithelial cells. This infection can interfere with normal cell-cycle regulation, promoting unrestrained cell proliferation and contributing to the accumulation of genetic alterations (Unger and Barr, 2004; Okunade, 2020). HPV infection has been implicated in nearly 90% of cervical adenocarcinomas occurring in women under the age of 40, whereas its presence drops significantly to approximately 43% in adenocarcinoma cases among women aged 60 and above (Burd, 2003).
Nonetheless, several additional factors can influence an individual’s capacity to eliminate HPV infection. These include genetic susceptibility, such as polymorphisms in major histocompatibility complex (MHC) genes and specific variants in the p53 gene implicated in HPV persistence and clearance, as well as genetic diversity among HPV types, co-infection with multiple HPV strains, reinfection frequency, hormonal influences, and the host immune response. Thus, while the presence of high-risk HPV is a critical prerequisite, it alone may not be sufficient for cervical cancer development. The progression to malignancy depends on the interplay between high-risk HPV types and various cofactors that modulate viral oncogenic activity. These modifiers include: Suppressed primary immune response, long-term use of oral contraceptives, cigarette smoking, and increasing parity (Okunade, 2020). Apart from this, persistent infection with high-risk HPV types plays a crucial role in cervical carcinogenesis by triggering epigenetic modifications in both the viral and host genomes. These alterations, including changes in DNA methylation patterns, histone modifications, and dysregulation of non-coding RNA (ncRNA) expression, result in the activation of oncogenes or the silencing of tumor suppressor genes, thereby facilitating the progression toward cervical cancer (Figueroa-Angulo et al., 2025).
2.1 Methylation
DNA methylation is a widely researched epigenetic modification in mammals. In healthy cells, it plays a key role in regulating gene expression and maintaining stable gene silencing. It works in conjunction with histone modifications, and their interaction is vital for genome regulation by altering chromatin structure (Kulis and Esteller, 2010). Methylation involves the enzymatic addition of a methyl group to the 5′ positions of cytosine, using S-adenosyl-methionine as the methyl donor, a process catalyzed by DNA methyltransferases DNMT1, DNMT3a, and DNMT3b (Kulis and Esteller, 2010; Tsai and Baylin, 2011). In the mammalian genome, methylation predominantly occurs at cytosines within CpG dinucleotides, with a small percentage of these dinucleotides clustered in regions known as CpG islands. These islands are often associated with gene promoters, transcription start sites, and/or first exons. In cancer, two major altered methylation patterns are observed: global DNA hypomethylation and promoter DNA hypermethylation (Tsai and Baylin, 2011). Hypermethylation, observed across nearly all tumor types, is often confined to CpG islands in gene regions, whereas hypomethylation frequently affects repeated DNA sequences, retrotransposons, endogenous retroviral elements, and unique transcription control sequences, suggesting its independent role in cancer development and progression (Esteller, 2002; Ehrlich, 2002). Epigenetic silencing of genes impacts key pathways involved in cancer initiation, progression, invasion, and metastasis, including cervical cancer. One of the pioneering works done by F Rösl, A. Arab, et al., suggested that DNA methylation is a stringent regulatory pathway for modulation of HPV gene expression (Rosl et al., 1993).
2.2 Histone modification and chromatin remodeling
Within cells, DNA is organized into chromatin, a highly dynamic structure in which the nucleosome serves as the fundamental unit of packaging. Histones constitute the core of the nucleosome, which is composed of an octamer containing the four core histone proteins—H3, H4, H2A, and H2B—around which approximately 147 base pairs of DNA are tightly wrapped. Each of these core histones possesses an N-terminal tail that is abundant in positively charged lysine and arginine residues. These tails are subject to various covalent post-translational modifications (PTMs), which play a coordinated role in regulating chromatin structure and function. Some modifications can influence the electrostatic interactions between histones and DNA, thereby affecting chromatin compaction and gene transcription. Additionally, certain PTMs act as docking sites for chromatin-binding proteins, which may further mediate changes in chromatin dynamics and gene regulation. Dysregulation of these chromatin-associated factors has been implicated in multiple cancers, including cervical cancer, where aberrant histone modifications and the malfunction of components like the SWI/SNF complex subunits BAF47 and BAF250A have been associated with tumorigenesis (Tsai and Baylin, 2011).
Histone modifications influence chromatin compaction, nucleosome dynamics, and transcription by altering DNA accessibility and facilitating the recruitment of DNA-binding proteins, thereby regulating gene expression. Post-translational modifications (PTMs) of histones serve as a diverse and flexible set of epigenetic marks, playing crucial roles not only in dynamic cellular processes like transcription regulation and DNA repair, but also in the long-term maintenance of repressive chromatin states (Audia and Campbell, 2016). This intricate regulatory system is coordinated by three main groups of proteins: “writers,” which add histone modifications; “readers,” which recognize and interpret these marks; and “erasers,” which remove them. The reversibility of these modifications contributes to the plasticity of the genome, enabling dynamic control of gene expression (Gomez-Gomez et al., 2013). Histone deacetylase inhibitors (HDACis) function by inhibiting the removal of acetyl groups from histones, thereby enhancing histone acetylation levels. This loosens the interaction between DNA and histones, facilitating a more relaxed chromatin structure, and as a result, contributes to the regulation of gene expression (de Villiers et al., 2004).
There are several types of modifications that occur, such as SUMOylation, a type of post-translational modification categorized by the covalent bonds of Small Ubiquitin-like Modifier (SUMO) proteins to specific target proteins, which has gained recognition as a key regulatory mechanism in cancer. It influences the activity of cell cycle regulators, oncogenes, and tumor suppressor genes, thereby impacting tumor development and progression. The dynamic changes in SUMOylation throughout the cell cycle are maintained by precise spatial and temporal control of the SUMO machinery. Moreover, SUMOylation works in concert with other post-translational modifications to support proper cell cycle advancement. Disruption of SUMOylation or deSUMOylation enzymes can lead to significant impairments in cell proliferation, genome stability (Eifler and Vertegaal, 2015) DNA damage response (Jackson and Durocher, 2013) and protein trafficking (Melchior et al., 2003).
Acetylation is a post-translational modification and is reversible in nature. It targets lysine residues across a broad range of proteins, which play a crucial role in modulating cellular plasticity. While approximately 80% of human proteins can undergo N-terminal acetylation, its most profound effects are seen in histone modifications, where it directly impacts gene regulation (Lopez-Banuelos and Vega, 2022). While acetylation was initially identified as a major post-translational modification (PTM) in histones, more recent research has shown that acetylation of non-histone proteins also plays a critical role in regulating protein–protein and protein-DNA interactions, influencing protein localization, stability, and functional activation (Narita et al., 2019). The acetylation of non-histone proteins plays a vital role in various cellular functions, including gene expression, metabolism, and the processes of DNA replication and repair. It also governs cell cycle progression by influencing nutrient uptake, initiating signaling pathways, activating transcription factors, and regulating checkpoints that determine whether the cell cycle continues or is arrested (Lopez-Banuelos and Vega, 2022; Narita et al., 2019).
Functionally, acetylation of lysine residues on histone tails reduces chromatin compaction by neutralizing the positive charge of lysine. This weakens the electrostatic attraction between the negatively charged DNA and histones, resulting in a more open chromatin structure. Histone acetylation is closely linked to transcriptional activation, particularly at enhancers, promoters, and within gene bodies, where it facilitates access to transcriptional machinery (Di Cerbo and Schneider, 2013). Altered histone acetylation levels, particularly at lysine 16 on histone H4, are linked to cancer phenotypes. Hyperacetylation of proto-oncogenes may lead to their activation or altered regulation of gene expression, whereas hypoacetylation of tumor suppressor genes, particularly at promoter regions, is often associated with DNA methylation, contributing to the transcriptional repression process. Enzymes known as lysine acetyltransferases (KATs), also referred to as histone acetyltransferases (HATs), catalyze the addition of acetyl groups to both histone and non-histone proteins such as p53, Rb, and MYC. In contrast, histone deacetylases (HDACs) remove these acetyl groups, thereby influencing gene expression and chromatin structure. Both HAT and HDAC activities are crucial for proper gene expression regulation, and any changes in their expression or function can impact chromatin regulation.
Apart from this, phosphorylation and ubiquitination are also a part of these histone/chromatin modifications. Phosphorylation is one of the key post-translational modifications in which phosphate groups are added to proteins by specific kinases, playing an essential role in controlling various cellular processes such as cell cycle progression, apoptosis, and signal transduction. The ubiquitination process involves the attachment of ubiquitin molecules to substrate proteins, which plays a central role in maintaining cellular homeostasis by regulating protein degradation, trafficking, DNA repair, and signal transduction.
2.3 Non-coding RNAs
The discovery of ncRNA dates back to the 1950s and has been an evolving field of research since then (Chen and Kim, 2024). ncRNAs, especially long non-coding RNAs (lncRNAs) and microRNAs (miRNAs), are crucial and important regulators of gene expression and epigenetic changes in both normal physiology and cancer biology. LncRNAs can interact with DNA, particularly purine-rich double-stranded regions, through Hoogsteen base pairing to form RNA–DNA triplex structures, functioning as either transcriptional repressors or activators depending on the required context or function. This structure enables lncRNAs to recruit chromatin modifiers to specific loci, contributing to epigenetic changes and transcriptional regulation (Li, 2023). Moreover, various ncRNAs, including miRNAs and small nucleolar RNAs (snoRNAs), operate as internal molecular signals that regulate key aspects and functions of genes, such as chromatin organization, transcription, RNA splicing, RNA editing, translation, and mRNA degradation, thus shaping cellular differentiation and disease progression (Mattick and Makunin, 2006).
In summary, epigenetic mechanisms comprising DNA methylation, histone modifications, chromatin remodeling, and non-coding RNAs form a complex and interconnected regulatory network that governs gene expression and cellular identity. Disruption of these pathways plays a pivotal role in the initiation and progression of various cancers, including HPV-associated malignancies such as cervical cancer (Da Silva et al., 2021). A deeper understanding of these processes is crucial for future research regarding HPV mediated cancers, including cervical cancer.
3 HPV-epigenome interaction
Epigenetics refers to the study of genome regulation through mechanisms beyond the DNA sequence itself, leading to stable changes in gene expression. The landmark discovery was the association of histone complexes with Human Papilloma Virus DNA, as first described by Favre and colleagues in Favre et al. (1977). In recent years, it has been well-established that epigenetics significantly influences various biological processes, including embryonic development, cancer progression, and immune responses. Among the most extensively researched HPV influenced epigenetic changes or modifications are E6/E7-mediated epigenetic disruption, DNA methylation, histone modifications, and non-coding RNAs, which may be influenced by HPV infection (Herceg, 2016; Figure 2).
3.1 E6/E7-mediated epigenetic disruption
The high-risk HPV oncoproteins E6 and E7 drive epigenetic reprogramming in host cells to facilitate persistent infection, cellular immortalization, and oncogenic progression. E6 promotes degradation of the tumor suppressor p53, disrupting genomic surveillance and indirectly promoting DNMT upregulation, leading to aberrant DNA methylation patterns. Meanwhile, E7 inactivates pRB, freeing E2F transcription factors to activate targets including EZH2, the catalytic core of the Polycomb Repressive Complex 2 (PRC2), elevating its expression in HPV-infected cells and HPV-positive lesions (Ghiani and Chiocca, 2022). Recent research indicates that E7 binds to and inactivates E2F6 protein, which is associated with polycomb complexes, resulting in epigenetic alterations associated with gene silencing. E7 CD2 domain’s LXCXE motif is seen to be required for pRB inactivation, which is also required for the downregulation of p107 and p130 (Tomaic, 2016).
Despite of the fact that, EZH2, a histone methyltransferase, is overexpressed, in HPV infected cells, H3K27me3 levels, is paradoxically reduced in E6/E7 expressing keratinocytes and cervical precancerous lesions. This unforeseen observation is attributed to the simultaneous induction of the demethylases KDM6A and KDM6B by E7, which remove H3K27me3 and derepress loci such as HOX and p16-INK4A, driving epigenetic reprogramming toward a stem-like, proliferative phenotype. Indeed, depletion of either demethylase in HPV-positive cervical cancer cells inhibits cell proliferation, highlighting their oncogenic relevance (McLaughlin-Drubin et al., 2011).
Additional dysregulation includes phosphorylation of EZH2 (at Ser21) via Akt activation and downregulation of PRC1 component BMI1, further contributes to altered chromatin landscapes and loss of H3K27me3 in E6/E7-expressing cells (Hyland et al., 2011). Transcriptomic data from over 800 human tumors (cervical and head-and-neck) in TCGA confirm elevated expression of EZH2, KDM6A, and KDM6B in HPV-positive cases, along with altered methylation in the CDKN2A locus, reinforcing the mechanistic model in clinical samples (Gameiro et al., 2017).
In sum, the E6/E7 oncoproteins remodel the host epigenome through a coordinated dysregulation of methyl writers and erasers, establishing a unique epigenetic signature characterized by high EZH2 yet low H3K27me3, which helps transcriptional deregulation, loss of differentiation, and malignant transformation. This interaction reveals potential vulnerabilities, as targeting KDM6 demethylases or EZH2 activity may reverse epigenetic reprogramming and inhibit HPV-driven oncogenesis.
3.2 DNA methylation alterations
During persistent high-risk HPV (HR-HPV) infection, abnormal DNA methylation events occur in both the host cervical squamous epithelial genome and the HPV viral genome. Increased levels of DNA methylation have been found with histological grades in cervical cancer, and also been found that methylation may increase the cervical intraepithelial neoplasia (CIN) grade I to invasive cervical cancer, while being undetectable in normal cervical epithelium (Laengsri et al., 2018). Epigenetic changes in both the host genome and the viral genome contribute to oncogenesis. In host cells, promoter hypermethylation of tumor suppressor genes such as p16INK4a, RASSF1A, CADM1, DAPK1, and ZNF582 leads to their silencing and promotes uncontrolled cellular proliferation and survival (Laengsri et al., 2018; Boscolo-Rizzo et al., 2017). Concurrently, methylation of the HPV genome, particularly within the long control region (LCR) and E2 binding sites, may alter viral gene expression, including upregulation of E6 and E7 oncoproteins, which in turn inactivate p53 and pRb tumor suppressors (Clarke et al., 2012; Moody and Laimins, 2010; Castro-Oropeza and Pina-Sanchez, 2022). It has been observed that methylation of the long control region (LCR) is more common in uterine cervical cancer (UCC) compared to cervical intraepithelial neoplasia (CIN). Moreover, the hypermethylation of CpG islands within the LCR of HPV16 increases progressively with lesion severity, showing significantly higher levels in invasive cervical cancer (Wang et al., 2017).
Moreover, due to methylation, dysregulations of several genes occur, which affect the immune cell development and their response in the tumor microenvironment (TME) and also affect the prognosis of cervical cancer patients. This may happen due to the methylation of immune-related genes, which affects their expression and dysregulation (Liu et al., 2022). For example, methylation of ERBB3 is oncogenic as RNA and DNA hypermethylation at transcription site genes may change the immune response in cervical cancer (Yang et al., 2022).
HPV16 E7 has been shown to interact with both histone acetyltransferases (HATs) and histone deacetylases (HDACs), thereby influencing chromatin structure and transcriptional activity. In a study by Burgers et al. (2007), E7 was found to interact directly with DNA methyltransferase 1 (DNMT1), enhancing its methyltransferase activity and contributing to aberrant DNA methylation in host cells (Burgers et al., 2007). Additionally, E7 is associated with increased expression of histone H3 lysine 27 (H3K27) demethylases, KDM6A and KDM6B, which remove the repressive H3K27me3 mark, leading to transcriptional activation of oncogenic and stemness-related genes (Basukala and Banks, 2021). Furthermore, recent evidence suggests that E7 enhances the post-translational modification of SETD2, a histone methyltransferase responsible for H3K36me3 deposition, thereby influencing transcriptional regulation and genome integrity (Castro-Oropeza and Pina-Sanchez, 2022). These documented changes in methylation suggest its utility as a biomarker for early detection, triage, and prognosis of cervical cancer.
3.3 Histone and chromatin alterations
Several genes were shown to be histone modification markers linked to cervical cancer in a study by Laengsri et al. Although the precise uses of the tumor suppressor gene p53 were not stated, it was examined in cell lines. Histone H3 acetyl K9 (H3K9ac) and Histone H3 trimethyl K4 (H3K4) were analyzed in tissue samples for chromatin remodeling, and their functions were hypothesized to be connected to the prognosis of disease. Retinoic acid receptor beta 2 (RARB2) and p21Cip1/WAF1 were found in cell lines to regulate apoptosis, suggesting possible uses for therapy and treatment monitoring (Laengsri et al., 2018). Without altering the expression of viral oncoproteins E6 and E7, valproate, a histone deacetylase (HDAC) inhibitor, demonstrated an inhibitory effect on cervical cancer cell lines and primary tumors by causing hyperacetylation of p53. Protease inhibitors like bortezomib and HDAC inhibitors like vorinostat or trichostatin A (TSA) also showed a synergistic effect when combined, efficiently eliminating HPV-positive cervical cancer cell lines. These results demonstrate how HDAC inhibitors may be used as epigenetic treatment approaches to treat cervical cancer (Liu et al., 2019).
Through epigenetic regulation, E6 also consecutively expresses hTERT (human telomerase reverse transcriptase, the catalytic unit of the telomerase enzyme). It also regulates methylases and demethylases, raising the H3K4Me3 activation mark and lowering the H3K9Me2 repressive mark (Pal and Kundu, 2019). NFX1-91, a transcriptional repressor, binds to the X box element of the hTERT promoter and also interacts with mSin3A, which is a transcriptional co-repressor that recruits HDACs. E6/E6AP complex alters chromatin accessibility at the hTERT by altering histone acetyltransferase (HAT) and histone deacetylase (HDAC) recruitment to the hTERT promoter. E6/E6AP also degrades NFX1-91, which leads to loss of HDAC activity at the promoter, whereas the activity of HAT is increased, which results in an increase in histone acetylation. HPV infection alters the DNA methylation pattern at the hTERT promoter. E6 and E7 of high-risk HPV are also hypermethylated and hypomethylated in certain promoter regions (Katzenellenbogen, 2017).
Moreover, HPV16 E7 also interacts with p300/CBP-associated factor (PCAF) histone acetyltransferase, which in turn downregulates IL-8, which helps the cells infected with HPV evade immune response (Huang and McCance, 2002). Mi2β protein in the presence of HPV E7 influences HDAC1/HDAC2, which regulates cell cycle deregulation and immune evasion through the histone modification process and transcription (Basukala and Banks, 2021). There are several other processes also altered, such as SUMOylation, Acetylation, Phosphorylation, and Ubiquitination. Recent research/literature is discussed below in detail.
3.4 SUMOylation
One of the early findings with respect to SUMOylation and HPV comes from the work of D. Rangasamy and K Woytek, where they listed that SUMOylation plays a key role in nuclear transport in turn, regulating E1 replication function, by regulating nuclear replication domains (Rangasamy et al., 2000). Apart from this, recent research has established that SUMOylation has a role in ferroptosis, a controlled type of cell death. The tumor microenvironment frequently contains hypoxia-like circumstances, which greatly increase cervical cancer cells’ resistance to ferroptosis. Hypoxia-like conditions decrease the deSUMOylation enzyme SENP1 while increasing the amounts of KDM4A, SUMO1, and Ubc9. As a result, KDM4A is SUMOylated at the K471 locus, which facilitates its interaction with SUMO1 in the nucleus. These epigenetic modifications increase the expression of GPX4 and SLC7A11, strengthening cervical cancer cells’ resistance to ferroptosis (Xiong et al., 2024).
By influencing protein stability, localization, and activity, SUMOylation plays a pivotal role in HPV-mediated oncogenesis. Sabtani and colleagues showed that SUMOylation, the UBC9/SUMO pathway, facilitates E-cadherin cleavage, which enhances metastasis in HPV associated head and neck cancer (Sabatini et al., 2022). Through the analysis of clinical samples and bioinformatic data, this study demonstrated that SUMOylation of E-cadherin promotes the degradation of its cleaved fragments (Chen et al., 2023). These findings provide new perspectives on the mechanisms underlying E-cadherin downregulation in HPV-positive head and neck cancers, identifying potential avenues for therapeutic intervention. Moreover, the study highlighted the UBC9/SUMO pathway as a key mediator of E-cadherin cleavage in these cancers, underscoring its critical role in HPV-associated tumor progression (Sabatini et al., 2022; Chen et al., 2023).
Chand and team demonstrated that degradation of hADA3, a transcriptional coactivator protein that HPV16 E6 targets, demonstrates a strong correlation between SUMOylation and HPV oncoproteins. The E6AP ubiquitin ligase mediates this connection and promotes the ubiquitin-mediated degradation of hADA3. Interestingly, HPV16 E6 speeds up hADA3’s SUMOylation, destabilizing the protein and preparing it for ubiquitination later on. While overexpression of hADA3 inhibits SiHa cell migration and proliferation, depletion of Ubc9, the only SUMO-conjugating enzyme, stops hADA3 from degrading quickly. These outcomes establish a clear relation in SUMOylation and HPV-mediated carcinogenesis by highlighting the interaction between SUMOylation and ubiquitination in controlling hADA3 stability (Chand et al., 2014).
Moreover, SUMOylation destabilizes the proteins and promotes the nucleocytoplasmic shuttling of the oncogenic transcription factor Forkhead box M1 b (FoxM1 b), a crucial cell cycle regulator. FoxM1b accumulates and overexpresses because of decreased SUMOylation in HPV16 E7-expressing cells. This change highlights the part SUMOylation dysregulation plays in the development of cervical cancer and is ascribed to HPV16 E7’s disruption of the interaction between FoxM1b and Ubc9 (Jaiswal et al., 2015). HPV E6 targets Ubc9 for proteasomal degradation, which has further consequences on the SUMOylation mechanism. Normal cellular processes are hampered when Ubc9 levels are decreased because it alters the host cells’ SUMOylation profile. These modifications imply that changes in SUMOylation brought on by HPV E6 are essential for viral replication and the development of cervical cancer (Heaton et al., 2011). Another level of intricacy is added by the complex interaction between autophagy and SUMOylation in HPV-mediated oncogenesis. An upregulation of Ubc9, an enzyme essential for SUMOylation, accompanies early HPV-driven transformation. Ubc9 breakdown is prevented by HPV E6/E7 proteins, which limit autophagosome-lysosome fusion, even though autophagy normally controls Ubc9 levels. As a result, Ubc9 builds up, strengthening host cells’ resistance to apoptosis, a sign of cancer growth. These results suggest that Ubc9 may be a therapeutic target for cancers linked to HPV (Mattoscio et al., 2017). When taken as a whole, these investigations demonstrate that SUMOylation is an essential regulatory mechanism in cervical cancer. HPV oncoproteins use SUMOylation to promote carcinogenesis by altering important proteins like KDM4A, Sp1, hADA3, and FoxM1b, as well as the SUMOylation mechanism itself. Gaining insight into these processes opens up new approaches that may target SUMOylation in cervical cancer.
3.5 Acetylation
In 2018, Elliot J. Androphy and his team highlighted the regulation of HPV replication, mentioning acetylation at the conserved lysine residue of E2 protein (Thomas and Androphy, 2018). Acetylation of lysine residues on host proteins is also essential for advancing the viral life cycle in the context of Human Papillomavirus (HPV) 16. Important regulatory sites are the di-lysines at positions 111 and 112, which are substantially conserved across papillomaviruses. The delicate balance between acetylation and viral replication is demonstrated by the fact that acetylation at K111 promotes viral replication, whereas deacetylation at the same location limits the helicase activity of E1, through P300 protein and by affecting topo 1 recruitment, hence stops replication (Thomas and Androphy, 2018; Thomas and Androphy, 2019). Chromatin alteration is also influenced by HPV genes, including the E6 and E7 oncogenes. Acetylation of histone H3 at lysine 9 (H3K9), a change linked to the stimulation of cell proliferation, survival, and carcinogenesis, results from the activation of E6 and E7 (Feng et al., 2016). Cervical cancer may develop as a result of these oncogenes’ effects on cell proliferation, division, and death. Lysine acetyltransferase (KAT) enzymes catalyze lysine acetylation. By altering histones and non-histones, these enzymes affect gene expression and, consequently, cellular activities via PTMs (Liu et al., 2019).
Histone deacetylases (HDACs), including HDAC2, HDAC3, and HDAC8, also significantly impact various cellular processes. HDAC2 and HDAC3 influence spindle assembly checkpoint signaling, crucial for mitotic regulation. Additionally, HDAC3’s interaction with PIWIL2 affects its role in epigenetic regulation and cell proliferation in cancer. HDAC3’s ability to inhibit the tumor suppressor proteins p53 and p27 further underscores its importance in cellular transformation and tumorigenesis. HDAC8, expressed both in the cytoplasm and nucleolus of HeLa cells, functions as a tubulin deacetylase, with its inhibition stabilizing microtubules and preventing cell migration and mitotic progression. In contrast, the inhibition of HDAC6 leads to the accumulation of connexin 32 (Cx32) in HeLa cells, where the acetylation of lysines in the C-terminal region of Cx32 plays a critical role in regulating cellular proliferation (Alaei et al., 2018; Zhang et al., 2018).
The role of lysine acetyltransferase GCN5 in HPV-induced cell proliferation has also been highlighted. In cells expressing HPV E7 protein, GCN5 is upregulated and promotes acetylation of histones at the E2F1 promoter. This increase in acetylation stabilizes the transcription factor E2F1, facilitating the G1/S transition and promoting cell cycle progression. Knockout of GCN5 disrupts this process, leading to G1-phase arrest. These findings reveal a novel mechanism by which GCN5 contributes to the pathogenesis of cervical cancer by promoting HPV E7-induced proliferation.
In addition, the bromodomain protein BRD2 plays a critical role in acetylation-mediated chromatin remodeling. By binding to acetylated lysines, BRD2 protects acetylated chromatin from HDACs and promotes acetylation spread, thereby enhancing transcriptional activity. BRD4, a member of the same protein family, has been shown to regulate HPV infection, further emphasizing the importance of acetylation in viral gene expression and cellular transformation.
Collectively, these findings underscore the pivotal role of acetylation in cervical cancer biology, particularly in the regulation of histone acetyltransferases, chromatin remodeling, cell proliferation, and viral oncogene expression. The ongoing exploration of acetylation pathways provides valuable insights into potential therapeutic strategies for targeting cervical cancer and other malignancies.
3.6 Phosphorylation
Aberrations in phosphorylation signaling pathways are commonly observed in numerous cancers, including cervical cancer, and are often associated with uncontrolled cell growth and survival (Singh et al., 2017), where Human Papillomavirus (HPV) infection is a primary etiological factor. A crucial aspect express oncoproteins E6 and E7 of this process involves phosphorylation-mediated modulation of both viral and host proteins, which alters their activity, stability, and interactions, thereby driving carcinogenesis. HPV E7, for instance, is phosphorylated by host kinases such as casein kinase II (CK2), and this modification regulates its ability to bind and inactivate the retinoblastoma tumor suppressor protein (pRb). Inactivation of pRb releases E2F transcription factors, promoting uncontrolled cell cycle progression. Moreover, phosphorylation affects the stability and oncogenic potential of E7, highlighting phosphorylation as a regulatory switch in HPV-mediated tumorigenesis (Bhattacharjee et al., 2022). Similarly, the E6 oncoprotein targets the p53 tumor suppressor for proteasomal degradation. The phosphorylation status of p53 is crucial for its stability and activation; HPV infection leads to altered phosphorylation patterns that destabilize p53, impairing its role in DNA damage response and apoptosis (Zhang et al., 2015). Beyond direct modification of viral proteins and tumor suppressors, HPV also manipulates host signaling pathways governed by phosphorylation cascades. The phosphatidylinositol 3-kinase (PI3K)/Akt pathway, which controls cell survival and proliferation, is frequently activated in HPV-positive cervical cancer cells through phosphorylation events induced by E7. This activation confers resistance to apoptosis and enhances cellular proliferation, contributing to tumor progression (Wang et al., 2018). Also, USF1/USF2 suppress the activity of hTERT, but when E6 is present, it enhances hTERT transcription by promoting serine 2 phosphorylation of RNA polymerase (McMurray and McCance, 2003). Additionally, mitogen-activated protein kinase (MAPK)/ERK pathway, often modulated via phosphorylation, is also influenced by HPV oncoproteins to promote oncogenic signaling. NF-κB, a key transcription factor involved in inflammation and immune response, is activated by phosphorylation-mediated signaling in HPV-infected cells, facilitating viral persistence and immune evasion (Moody and Laimins, 2010). These phosphorylation-driven alterations in signaling not only enable HPV to bypass cellular checkpoints but also create a microenvironment conducive to malignant transformation. Furthermore, phosphorylation influences HPV’s ability to evade the host immune system by modulating molecules involved in antigen presentation and inflammatory signaling. The crosstalk between HPV oncoproteins and host kinases creates a complex regulatory network that disrupts normal phosphorylation homeostasis, driving cervical carcinogenesis. Understanding these phosphorylation events offers potential therapeutic targets; for example, kinase inhibitors targeting CK2 or PI3K/Akt pathways may counteract HPV-mediated oncogenic signaling. In summary, phosphorylation acts as a critical molecular mechanism by which HPV oncoproteins regulate both viral functions and host cellular pathways, facilitating cervical cancer development. These insights underscore the importance of phosphorylation in HPV-induced carcinogenesis and provide avenues for developing targeted treatments against cervical cancer.
3.7 Ubiquitination
The ubiquitin-proteasome system (UPS) is critical in controlling protein turnover and is frequently hijacked by viruses, including Human Papillomavirus (HPV), to facilitate infection, persistence, and oncogenic transformation. In the context of cervical cancer, which is predominantly caused by high-risk HPV types such as HPV16 and HPV18, the viral oncoproteins E6 and E7 manipulate the ubiquitination machinery to degrade tumor suppressor proteins and deregulate cell cycle control. One of the most well-characterized mechanisms involves the HPV E6 protein, which forms a trimeric complex with the cellular E3 ubiquitin ligase E6-AP (UBE3A) and the tumor suppressor protein p53, targeting p53 for ubiquitin-mediated degradation via the proteasome. Loss of functional p53 disrupts cell cycle arrest and apoptosis, allowing cells with DNA damage to proliferate uncontrollably (Scheffner et al., 1993). Similarly, the E7 oncoprotein promotes the ubiquitin-mediated degradation of retinoblastoma protein (pRb), either directly or through induction of specific E3 ligases like cullin 2-containing complexes. This leads to the release of E2F transcription factors, promoting entry into the S-phase of the cell cycle and unregulated cellular proliferation (Huh et al., 2007). Beyond degrading key tumor suppressors, HPV also exploits the UPS to modulate immune responses, enhancing its persistence in host cells. For example, E6 and E7 downregulate components of the antigen presentation machinery by promoting the degradation of MHC class I molecules, reducing immune recognition (Huh et al., 2007).
In addition to targeting host proteins, HPV oncoproteins are themselves regulated by ubiquitination. The stability of E6 and E7 can be modulated by cellular ubiquitin ligases such as HERC1 and UBR4, which fine-tune the levels of viral proteins and influence the progression from precancerous lesions to invasive cancer. Aberrations in the UPS have been observed in HPV-positive cervical cancers, with overexpression of specific deubiquitinating enzymes (DUBs) and E3 ligases contributing to malignant phenotypes and therapy resistance. Therapeutically, the UPS represents a promising target; proteasome inhibitors like bortezomib have shown potential in sensitizing HPV-positive cancer cells to apoptosis by restoring p53 levels and reducing E6/E7 stability (Rampias et al., 2010).
4 Non-coding RNA-mediated regulation and alterations
In the HPV-associated carcinogenesis, ncRNAs can play dual roles as oncogenic or tumor-suppressive behavior. Strategies to modulate ncRNA activity, either by inhibiting oncogenic ncRNAs or overexpressing tumor-suppressive ncRNAs, have been proposed as potential therapeutic interventions (Li, 2023). For instance, miR-106a and miR-27a increase radiosensitivity in HPV-positive head and neck squamous cell carcinoma (HNSCC) by targeting RUNX3 and SMG1, respectively, whereas overexpression of miR-125b in HPV-negative cells reduces radiosensitivity by downregulating ICAM2. Additionally, circular RNAs (circRNAs) act as miRNA sponges, modulating the availability of tumor-related miRNAs. For example, circPVT1 suppresses miR-497-5p, promoting cell proliferation in HPV-negative oral squamous cell carcinoma (OSCC) (Bonelli et al., 2021).
HPV oncoproteins E6 and E7 have been shown to upregulate miR-18a, which enhances proliferation and invasion via suppression of STK4, a key tumor suppressor in the Hippo pathway (Morgan et al., 2020). Several tumor suppressor miRNAs (tsmiRs) such as miR-125 and miR-375 inhibit cervical cancer progression by targeting VEGF and MELK, respectively, impairing proliferation, migration, and invasion while promoting apoptosis (Chauhan et al., 2024; Gui et al., 2021). Interestingly, miRNAs like miR-9-5p exhibit dual roles: acting as an oncomiR in HPV16 + SCC cell lines (CaSki, SiHa) by repressing TWIST1 and CDH1, thus promoting epithelial–mesenchymal transition (EMT), but displaying a tumor-suppressive profile in HPV18 + adenocarcinoma cells (HeLa) due to differential regulation of CDH2 (Banuelos-Villegas et al., 2021).
Other oncogenic miRNAs, such as miR-499, enhance tumor aggressiveness by downregulating SOX6 (Chen et al., 2020), while miR-21, a microRNA, fosters the growth, proliferation, invasion, and spread of tumor cells. It accomplishes this by suppressing the tumor suppressor PTEN, which in turn triggers the abnormal activation of the PI3K/Akt signaling pathway. This activation is characterized by an increase in phosphorylated Akt (p-Akt) (Wang et al., 2017; Silvia Lima et al., 2024). Moreover, miR-29a regulates HSP47, a collagen-processing protein, with its downregulation linked to increased cancer cell migration and invasion, implying a role in cervical cancer metastasis. Collectively, these findings underscore the multifaceted roles of ncRNAs in HPV-driven cervical cancer and highlight their potential as diagnostic biomarkers and therapeutic targets, especially for enhancing the efficacy of chemo-radiotherapy (Mattick and Makunin, 2006).
5 Clinical aspects of HPV mediated epigenetics in cervical cancer
HPV infection induces several epigenetic modifications in the host tumor suppressor genes and oncogenes, which may serve as biomarkers for cervical cancer screening, triage, and the development of therapeutic targets. Some of the important clinical implications are mentioned below.
5.1 Screening potential
Recently, DNA methylation status has been suggested for a triage strategy for HPV cases and for atypical squamous cells of undetermined significance (ASCUS) cases (Burdier et al., 2024). It is well established that HPV infection can develop cervical cancer, and there is a long window for early detection and treatment. So, if any case is found HPV positive, it should go through a second test triage to differentiate the precursor lesions, which may be through persistent infection, and those that do not have infection. This triage strategy to check the DNA methylation of cellular and viral genes has potential for diagnostics for HPV positive patients (Castro-Oropeza and Pina-Sanchez, 2022).
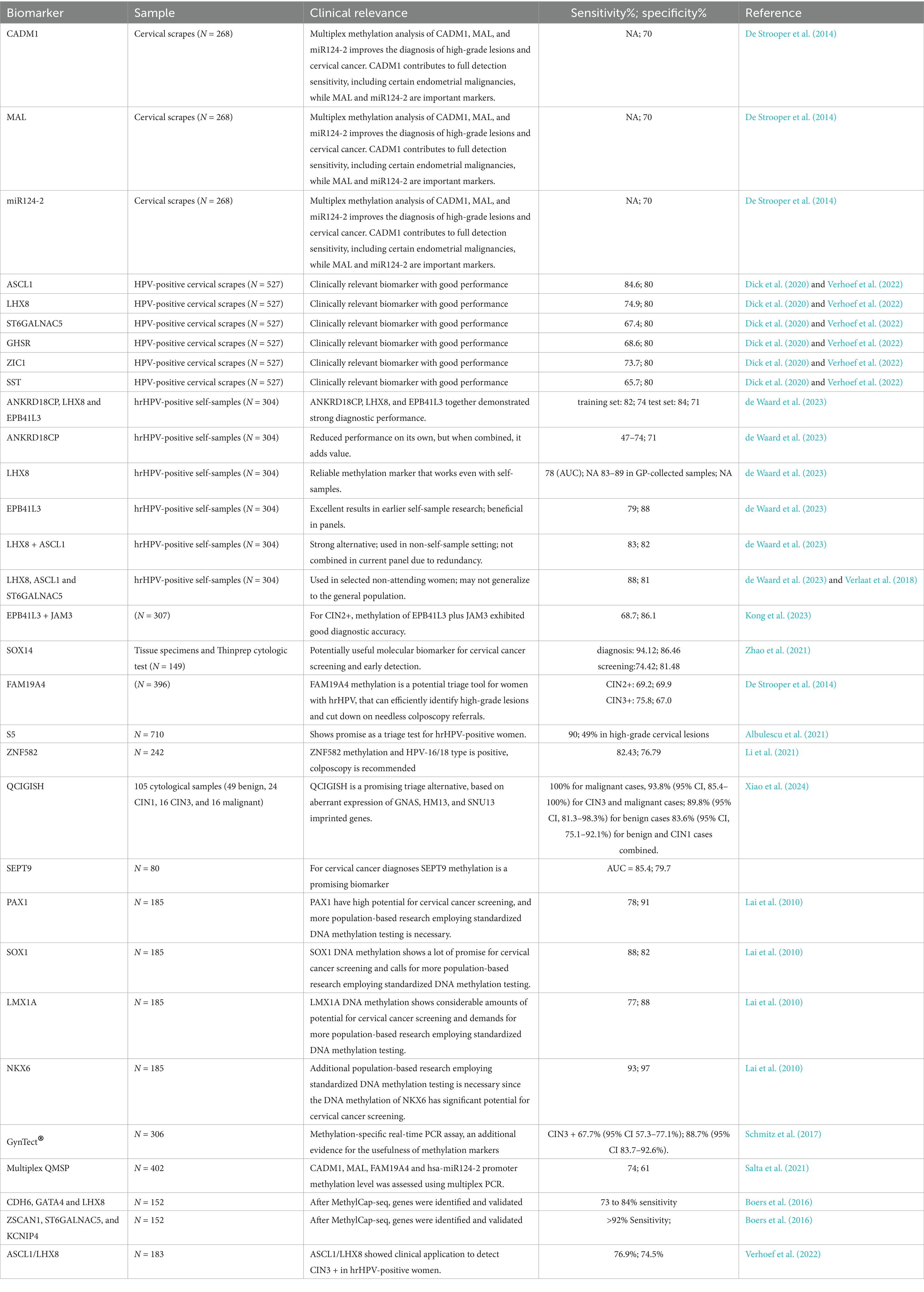
Previous investigations suggested that methylation of the CADM1 and MAL genes may serve as effective biomarkers for identifying cervical intraepithelial neoplasia grades 2 and 3 (CIN2/3), and demonstrating higher sensitivity than traditional cytology-based triage methods (Verhoef et al., 2015). Additionally, epigenetic changes in the form of methylation of the transcription factor ZNF582 have been linked to CIN3 in patients with ASCUS (Burdier et al., 2024), and when it is combined with PAX1, ZNF582 methylation markers have demonstrated potential for detecting high-grade squamous intraepithelial lesions (HSIL) with high sensitivity and specificity (Tian et al., 2017). Apart from this, various genes have been identified with methylation patterns that may serve as promising biomarkers for detecting cervical precursor lesions, such as POUF4 (Castro-Oropeza and Pina-Sanchez, 2022; Pun et al., 2015; Kocsis et al., 2017), ASCL1, LHX8, and ST6GALNAC5 (Verlaat et al., 2018).
Some clinical validations on methylation profiles as diagnostic biomarkers for CIN2, CIN3, and cervical cancer (CC), analyze the methylation status of EPB41L3 and the late regions of HPV types 16, 18, 31, and 33, reporting a sensitivity of 74% and a specificity of 90% (Lorincz et al., 2016; Cook et al., 2019). Recently, a multicenter study reported an impressive 99.8% sensitivity for identifying CIN3 and invasive cervical cancer (Banila et al., 2022). Beyond cervical applications, a panel targeting EPB41L3 along with L1, L2, and E2 regions of HPV16 has been explored for the early detection of oropharyngeal carcinoma, demonstrating a 70% sensitivity and 91% specificity using a non-invasive oral gargle sample (Giuliano et al., 2020).
There are several commercial assays for detecting malignant neoplasms associated with high-risk HPV (HPV-HR), including GynTect® and QIAsure®. GynTect® is a molecular test specifically designed to diagnose cervical carcinoma by analyzing the methylation profiles of six biomarkers: ASTN1, ZNF671, DLX1, ITGA4, RXFP3, and SOX17 (Schmitz et al., 2018; Schmitz et al., 2017). In contrast, QIAsure® uses multiplex real-time methylation-specific PCR (qMSP) to detect hypermethylation of FAM19A4 and miR124-2 (Floore et al., 2019). This assay consistently detects cervical cancer across various histological types, including HPV HR-negative cases (Vink et al., 2020). A comparative studies indicate that GynTect® has superior specificity, identifying CIN2 + at 87% versus QIAsure® at 67%, and CIN3 at 84% compared to QIAsure®'s 68% (Dippmann et al., 2020). This suggests that GynTect® may be a more reliable choice for early detection of precancerous lesions. In view of these studies, we may use epigenetic changes to enhance the screening methods for early detection of cervical cancer. Some biomarkers are discussed in Table 1.
5.2 Therapeutic potential
The reversible nature of epigenetic alterations gives crucial therapeutic windows for cervical cancer as well as HPV-driven malignancies. Unlike genetic mutations, epigenetic changes such as DNA methylation and histone modifications are dynamic and potentially reversible, making them suitable targets for therapeutic intervention. In HPV-positive cervical cancers, the viral oncoproteins E6 and E7 induce epigenetic silencing of tumor suppressor genes and reprogram host transcription, thus creating a dependency on the altered epigenome that can be exploited therapeutically.
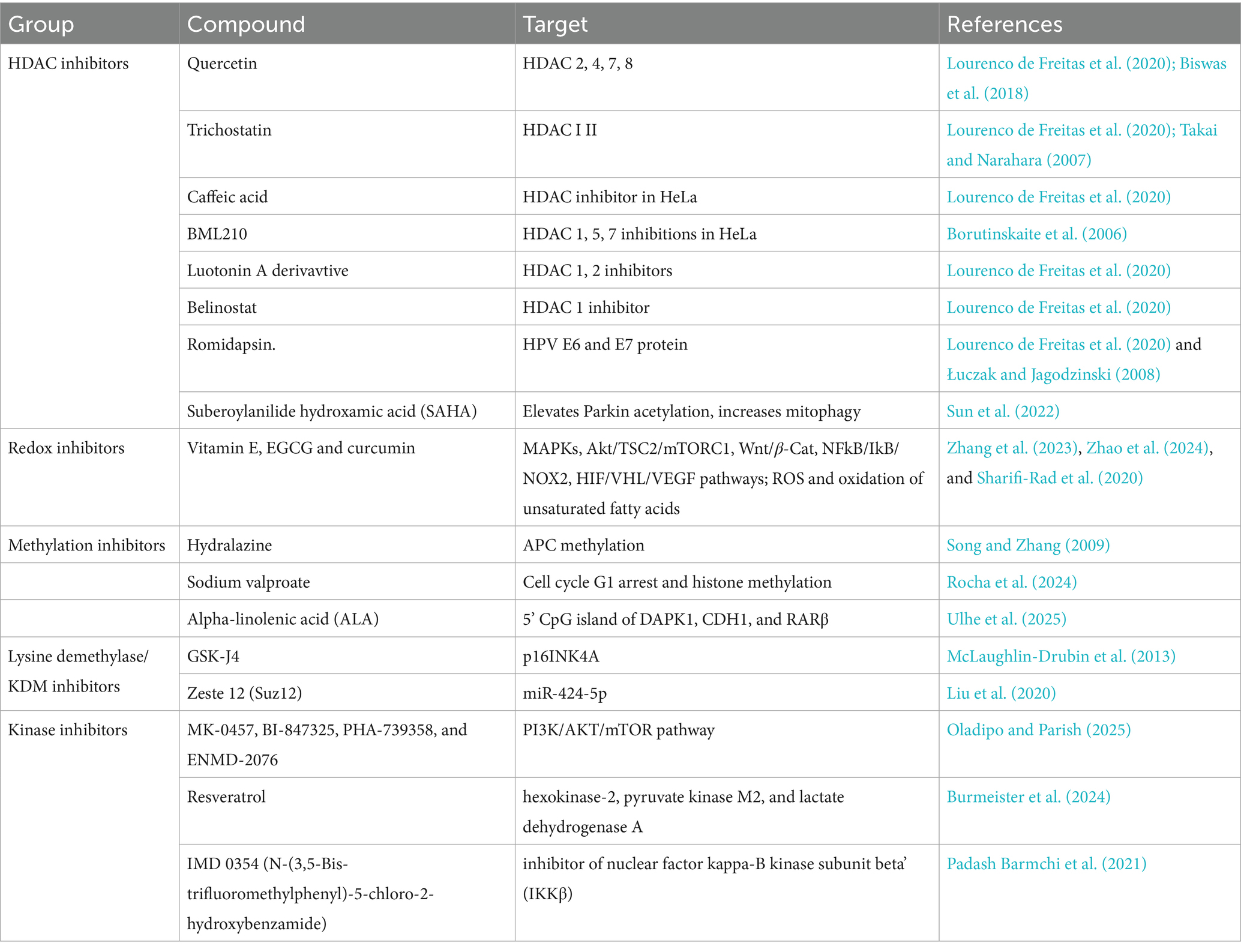
Drugs such as decitabine and azacitidine, which inhibit DNA methyltransferases (DNMTs), have been extensively studied for their ability to demethylate and reactivate silenced tumor suppressor genes. These agents incorporate into DNA and trap DNMTs, leading to hypomethylation and re-expression of epigenetically silenced genes, including those involved in cell cycle regulation and apoptosis (e.g., p16INK4a, DAPK1) (Hu et al., 2021; Chiappinelli et al., 2015). HDAC inhibitors such as vorinostat (SAHA), belinostat, and romidepsin function by increasing histone acetylation, leading to a more relaxed chromatin structure and transcriptional activation of silenced genes. In cervical cancer, HDAC inhibitors can induce apoptosis and cell cycle arrest, downregulate HPV E6/E7 expression, upregulate tumor suppressor genes (e.g., p53, p21), and enhance radiosensitivity and chemosensitivity of cervical cancer cells (Lourenco de Freitas et al., 2020; Cheng et al., 2019).
Apart from this, recent studies suggest that epigenetic regulation plays a crucial role in shaping host immune responses during viral infections, including those caused by human papillomavirus (HPV). Studies have shown that viral infections can induce epigenetic alterations in key immune response genes such as AIM2, BST2, BTN3A3, and IL12RB1 (Gasperov et al., 2014). HPV oncoproteins E6, E7, and E5 are known to manipulate the immune system by targeting multiple immune-modulatory pathways. E6 interferes with type I interferon signaling by preventing STAT1 and STAT2 phosphorylation through its interaction with TYK2, impairing IFN-α-mediated innate immunity (Castro-Munoz et al., 2022). E7, in turn, interacts with IRF-1 and the Nucleosome Remodeling and Deacetylase (NuRD) complex containing HDAC3, suppressing transcriptional activation of IFN-β and other interferon-stimulated genes. Furthermore, E5 and E7 downregulate major histocompatibility complex (MHC) class I expression, hampering antigen presentation to cytotoxic T lymphocytes (Westrich et al., 2017).
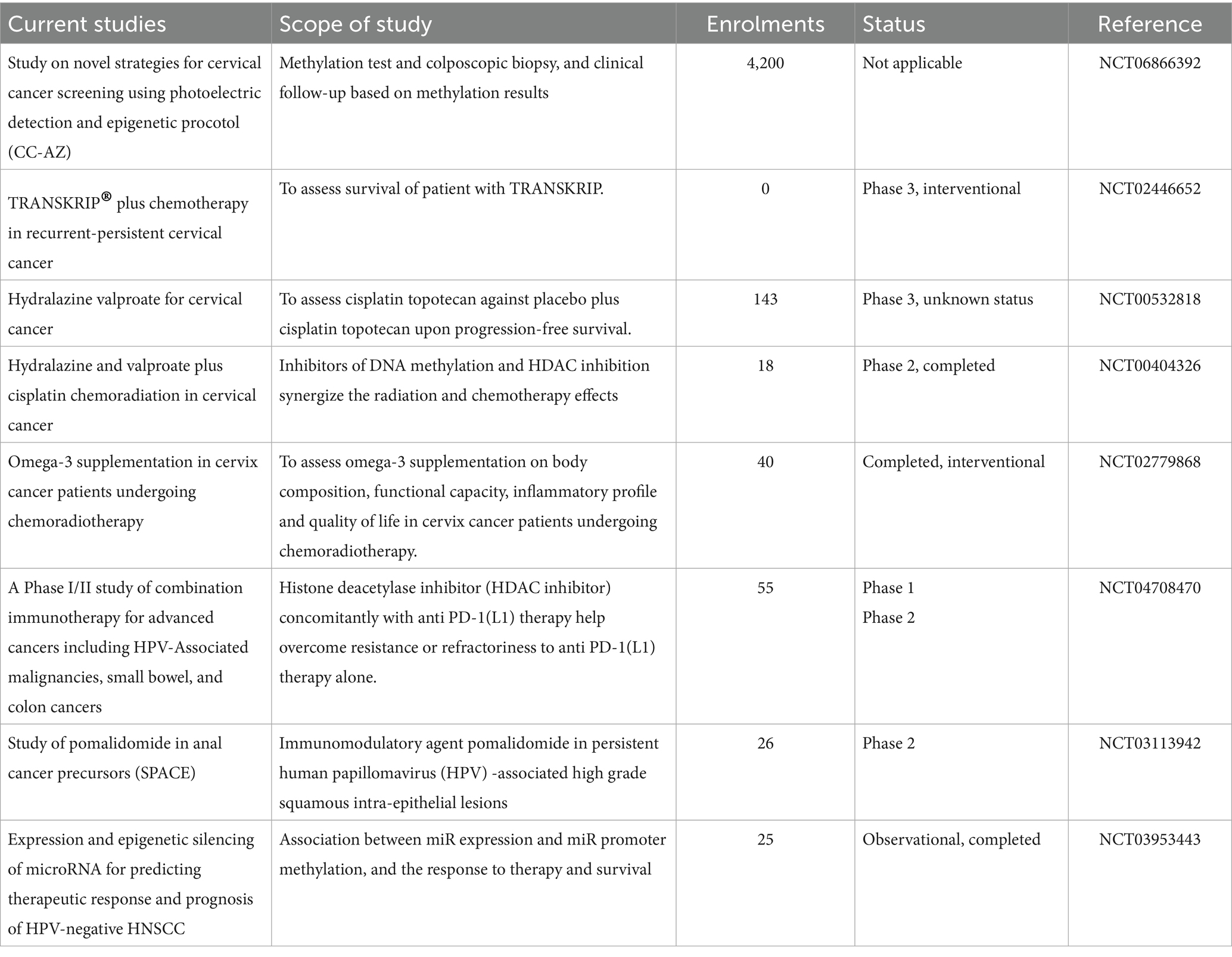
There is growing interest in combining epigenetic drugs with immunotherapy. Epigenetic modulation can enhance expression of tumor-associated antigens, upregulate MHC class I molecules, and induce type I interferon responses, thereby increasing immune recognition of HPV-transformed cells. Early-phase clinical trials are exploring the combination of DNMT inhibitors and HDAC inhibitors with PD-1/PD-L1 checkpoint blockade in HPV-associated malignancies, including cervical cancer (Zhou et al., 2021; Chen et al., 2022). Some therapeutic targets and inhibitors and ongoing trials have been listed in Tables 2, 3, respectively.
5.3 Treatment resistance
Substantial evidences are available for the therapeutic potential of epigenetic modulation in mitigating treatment resistance including those associated with high-risk human papillomavirus (HPV). Several recent studies have emphasized how targeting epigenetic mechanisms such as DNA methylation and histone modifications can reverse chemoresistance and radio resistance (Wang et al., 2023; Folliero et al., 2023). In cervical cancer, in vitro demethylation has been shown to restore the expression of p73, a crucial pro-apoptotic gene often silenced in radioresistant cells, thereby sensitizing them to radiation therapy (Liu et al., 2004). Also, Inhibition of histone deacetylases (HDACs) has been reported to enhance the chemosensitivity of HPV-positive tumor cells to agents like cisplatin and taxol by reactivating silenced apoptotic pathways (Ozaki et al., 2008).
Specifically, the combination of DNA methyltransferase inhibitors (DNMTis) with standard chemotherapy has shown synergistic effects, as DNMTis modulate the epigenetic landscape, allowing previously silenced tumor suppressor genes to re-express and respond to cytotoxic stress (Gravina et al., 2010). Moreover, carbon-ion irradiation has been shown to overcome radio resistance induced by disruption of the HPV E2 gene in cervical keratinocytes, suggesting that epigenetic changes in the tumor microenvironment significantly influence therapeutic outcomes (Arians et al., 2019).
These recent findings emphasize on the concept that epigenetic mechanisms not only drive resistance in HPV-associated cancers but also represent actionable therapeutic targets. The reversibility of these changes highlights the translational potential of epigenetic drugs in improving treatment responses and combating resistance, particularly in tumors with HPV etiology.
6 Conclusion and future prospects
Understanding cervical cancer and its epigenetic alterations driven by HPV infection is crucial for its management. DNA methylation, histone modifications, and non-coding RNA regulation are reversible and dynamic changes; these changes help the virus persist and contribute to oncogenic transformation. They also offer promising opportunities for diagnosis and treatment. However, more research is needed. In particular, identifying and validating epigenetic signatures at different stages of HPV-associated cervical cancer is essential. Previously, therapeutic agents such as DNA methyltransferase inhibitors (e.g., 5-azacytidine) and histone deacetylase inhibitors (e.g., vorinostat) showed promising results in restoring normal gene function and expression. In this review, we have highlighted how HPV-mediated epigenetic reprogramming affects the function of tumor suppressors and oncogenes and modulates immune evasion mechanisms. These epigenetic changes contribute to disease progression. However, their reversible nature provides a unique therapeutic window. They also offer opportunities to develop biomarkers that can complement current HPV screening and treatment strategies. Such biomarkers may enable early detection, better prognostication, and personalized treatment. Integrating epigenetic biomarkers into cervical cancer care could improve outcomes and support global health efforts. Exploration of HPV-epigenome interactions holds the potential to transform cervical cancer prevention, biomarker discoveries and therapeutic scope.
Author contributions
SS: Conceptualization, Writing – review & editing, Data curation, Visualization, Writing – original draft. PS: Visualization, Writing – review & editing. TJ: Visualization, Writing – review & editing. MA: Writing – review & editing. NF: Writing – review & editing. AK: Writing – review & editing. NM: Writing – review & editing, Supervision. NJ: Writing – review & editing. PT: Writing – review & editing. VG: Writing – review & editing. SH: Conceptualization, Supervision, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Acknowledgments
We would like to acknowledge Nazneen Arif and Ekta Gupta for their support during the manuscript writing. Also, would like to acknowledge the support of the Indian Council of Medical Research, New Delhi.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Alaei, S. R., Abrams, C. K., Bulinski, J. C., Hertzberg, E. L., and Freidin, M. M. (2018). Acetylation of C-terminal lysines modulates protein turnover and stability of Connexin-32. BMC Cell Biol. 19:22. doi: 10.1186/s12860-018-0173-0
Albulescu, A., Plesa, A., Fudulu, A., Iancu, I. V., Anton, G., and Botezatu, A. (2021). Epigenetic approaches for cervical neoplasia screening (review). Exp. Ther. Med. 22:1481. doi: 10.3892/etm.2021.10916
Arians, N., Nicolay, N. H., Brons, S., Koerber, S. A., Jaschke, C., Vercruysse, M., et al. (2019). Carbon-ion irradiation overcomes HPV-integration/E2 gene-disruption induced radioresistance of cervical keratinocytes. J. Radiat. Res. 60, 564–572. doi: 10.1093/jrr/rrz048
Audia, J. E., and Campbell, R. M. (2016). Histone modifications and cancer. Cold Spring Harb. Perspect. Biol. 8:a019521. doi: 10.1101/cshperspect.a019521
Banila, C., Lorincz, A. T., Scibior-Bentkowska, D., Clifford, G. M., Kumbi, B., Beyene, D., et al. (2022). Clinical performance of methylation as a biomarker for cervical carcinoma in situ and cancer diagnosis: A worldwide study. Int. J. Cancer. 150, 290–302. doi: 10.1002/ijc.33815
Banuelos-Villegas, E. G., Perez-yPerez, M. F., and Alvarez-Salas, L. M. (2021). Cervical cancer, papillomavirus, and miRNA dysfunction. Front. Mol. Biosci. 8:758337. doi: 10.3389/fmolb.2021.758337
Basukala, O., and Banks, L. (2021). The not-so-good, the bad and the ugly: HPV E5, E6 and E7 oncoproteins in the orchestration of carcinogenesis. Viruses 13:1892. doi: 10.3390/v13101892
Bhattacharjee, R., Das, S. S., Biswal, S. S., Nath, A., Das, D., Basu, A., et al. (2022). Mechanistic role of HPV-associated early proteins in cervical cancer: molecular pathways and targeted therapeutic strategies. Crit. Rev. Oncol. Hematol. 174:103675. doi: 10.1016/j.critrevonc.2022.103675
Biswas, S., Reddy, N. D., Jayashree, B. S., and Rao, C. M. (2018). Evaluation of novel 3-Hydroxyflavone analogues as HDAC inhibitors against colorectal cancer. Adv. Pharmacol. Sci. 2018, 1–14. doi: 10.1155/2018/4751806
Boers, A., Wang, R., van Leeuwen, R. W., Klip, H. G., de Bock, G. H., Hollema, H., et al. (2016). Discovery of new methylation markers to improve screening for cervical intraepithelial neoplasia grade 2/3. Clin. Epigenetics 8:29. doi: 10.1186/s13148-016-0196-3
Bonelli, P., Borrelli, A., Tuccillo, F. M., Buonaguro, F. M., and Tornesello, M. L. (2021). The role of circ RNAs in human papillomavirus (HPV)-associated cancers. Cancers 13:1173. doi: 10.3390/cancers13051173
Borutinskaite, V. V., Navakauskiene, R., and Magnusson, K. E. (2006). Retinoic acid and histone deacetylase inhibitor BML-210 inhibit proliferation of human cervical cancer HeLa cells. Ann. N. Y. Acad. Sci. 1091, 346–355. doi: 10.1196/annals.1378.079
Boscolo-Rizzo, P., Furlan, C., Lupato, V., Polesel, J., and Fratta, E. (2017). Novel insights into epigenetic drivers of oropharyngeal squamous cell carcinoma: role of HPV and lifestyle factors. Clin. Epigenetics 9:124. doi: 10.1186/s13148-017-0424-5
Burd, E. M. (2003). Human papillomavirus and cervical cancer. Clin. Microbiol. Rev. 16, 1–17. doi: 10.1128/CMR.16.1.1-17.2003
Burdier, F. R., Waheed, D. E., Nedjai, B., Steenbergen, R. D. M., Poljak, M., Baay, M., et al. (2024). DNA methylation as a triage tool for cervical cancer screening - a meeting report. Prev. Med. Rep. 41:102678. doi: 10.1016/j.pmedr.2024.102678
Burgers, W. A., Blanchon, L., Pradhan, S., de Launoit, Y., Kouzarides, T., and Fuks, F. (2007). Viral oncoproteins target the DNA methyltransferases. Oncogene 26, 1650–1655. doi: 10.1038/sj.onc.1209950
Burmeister, C. A., Khan, S. F., and Prince, S. (2024). Drugs and drug targets for the treatment of HPV-positive cervical cancer. Tumour Virus Res. 19:200309. doi: 10.1016/j.tvr.2024.200309
Castro-Munoz, L. J., Rocha-Zavaleta, L., Lizano, M., Ramirez-Alcantara, K. M., Madrid-Marina, V., and Manzo-Merino, J. (2022). Alteration of the IFN-pathway by human papillomavirus proteins: antiviral immune response evasion mechanism. Biomedicine 10:2965. doi: 10.3390/biomedicines10112965
Castro-Oropeza, R., and Pina-Sanchez, P. (2022). Epigenetic and transcriptomic regulation landscape in HPV+ cancers: biological and clinical implications. Front. Genet. 13:886613. doi: 10.3389/fgene.2022.886613
Chan, C. K., Aimagambetova, G., Ukybassova, T., Kongrtay, K., and Azizan, A. (2019). Human papillomavirus infection and cervical Cancer: epidemiology, screening, and vaccination-review of current perspectives. J. Oncol. 2019, 1–11. doi: 10.1155/2019/3257939
Chand, V., John, R., Jaiswal, N., Johar, S. S., and Nag, A. (2014). High-risk HPV16E6 stimulates hADA3 degradation by enhancing its SUMOylation. Carcinogenesis 35, 1830–1839. doi: 10.1093/carcin/bgu104
Chauhan, P., Pramodh, S., Hussain, A., Elsori, D., Lakhanpal, S., Kumar, R., et al. (2024). Understanding the role of miRNAs in cervical cancer pathogenesis and therapeutic responses. Front. Cell Dev. Biol. 12:1397945. doi: 10.3389/fcell.2024.1397945
Chen, L. L., and Kim, V. N. (2024). Small and long non-coding RNAs: past, present, and future. Cell 187, 6451–6485. doi: 10.1016/j.cell.2024.10.024
Chen, Y., Qiu, X., Wang, W., Li, D., Wu, A., Hong, Z., et al. (2020). Human papillomavirus infection and cervical intraepithelial neoplasia progression are associated with increased vaginal microbiome diversity in a Chinese cohort. BMC Infect. Dis. 20:629. doi: 10.1186/s12879-020-05324-9
Chen, X., Smaldone, G., Piccaluga, P. P., and Qi, Y. (2023). Editorial: role of the SUMOylation in cancer regulation. Front. Mol. Biosci. 10:1236230. doi: 10.3389/fmolb.2023.1236230
Chen, S., Xie, P., Cowan, M., Huang, H., Cardenas, H., Keathley, R., et al. (2022). Epigenetic priming enhances antitumor immunity in platinum-resistant ovarian cancer. J. Clin. Invest. 132:132. doi: 10.1172/JCI158800
Cheng, Y., He, C., Wang, M., Ma, X., Mo, F., Yang, S., et al. (2019). Targeting epigenetic regulators for cancer therapy: mechanisms and advances in clinical trials. Signal Transduct. Target. Ther. 4:62. doi: 10.1038/s41392-019-0095-0
Chiappinelli, K. B., Strissel, P. L., Desrichard, A., Li, H., Henke, C., Akman, B., et al. (2015). Inhibiting DNA methylation causes an interferon response in cancer via ds RNA including endogenous retroviruses. Cell 162, 974–986. doi: 10.1016/j.cell.2015.07.011
Cho, S. W., Xu, J., Sun, R., Mumbach, M. R., Carter, A. C., Chen, Y. G., et al. (2018). Promoter of lncRNA gene PVT1 is a tumor-suppressor DNA boundary element. Cell 173, 1398–1412.e22. doi: 10.1016/j.cell.2018.03.068
Clarke, M. A., Wentzensen, N., Mirabello, L., Ghosh, A., Wacholder, S., Harari, A., et al. (2012). Human papillomavirus DNA methylation as a potential biomarker for cervical cancer. Cancer Epidemiol. Biomarkers Prev. 21, 2125–2137. doi: 10.1158/1055-9965.EPI-12-0905
Cook, D. A., Krajden, M., Brentnall, A. R., Gondara, L., Chan, T., Law, J. H., et al. (2019). Evaluation of a validated methylation triage signature for human papillomavirus positive women in the HPV FOCAL cervical cancer screening trial. Int. J. Cancer 144, 2587–2595. doi: 10.1002/ijc.31976
Da Silva, M. L. R., De Albuquerque, B., Allyrio, T., De Almeida, V. D., Cobucci, R. N. O., Bezerra, F. L., et al. (2021). The role of HPV-induced epigenetic changes in cervical carcinogenesis (review). Biomed Rep. 15:60. doi: 10.3892/br.2021.1436
Dasari, S., Wudayagiri, R., and Valluru, L. (2015). Cervical cancer: biomarkers for diagnosis and treatment. Clin. Chim. Acta 445, 7–11. doi: 10.1016/j.cca.2015.03.005
De Strooper, L. M., Meijer, C. J., Berkhof, J., Hesselink, A. T., Snijders, P. J., Steenbergen, R. D., et al. (2014). Methylation analysis of the FAM19A4 gene in cervical scrapes is highly efficient in detecting cervical carcinomas and advanced CIN2/3 lesions. Cancer Prev. Res. 7, 1251–1257. doi: 10.1158/1940-6207.CAPR-14-0237
De Strooper, L. M., van Zummeren, M., Steenbergen, R. D., Bleeker, M. C., Hesselink, A. T., Wisman, G. B., et al. (2014). CADM1, MAL and miR124-2 methylation analysis in cervical scrapes to detect cervical and endometrial cancer. J. Clin. Pathol. 67, 1067–1071. doi: 10.1136/jclinpath-2014-202616
de Villiers, E. M., Fauquet, C., Broker, T. R., Bernard, H. U., and zur Hausen, H. (2004). Classification of papillomaviruses. Virology 324, 17–27. doi: 10.1016/j.virol.2004.03.033
de Waard, J., Bhattacharya, A., de Boer, M. T., van Hemel, B. M., Esajas, M. D., Vermeulen, K. M., et al. (2023). Identification of a methylation panel as an alternative triage to detect CIN3+ in hr HPV-positive self-samples from the population-based cervical cancer screening programme. Clin. Epigenetics 15:103. doi: 10.1186/s13148-023-01517-6
Di Cerbo, V., and Schneider, R. (2013). Cancers with wrong HATs: the impact of acetylation. Brief. Funct. Genomics 12, 231–243. doi: 10.1093/bfgp/els065
Dick, S., Verhoef, L., De Strooper, L. M., Ciocanea-Teodorescu, I., Wisman, G. B. A., Meijer, C. J., et al. (2020). Evaluation of six methylation markers derived from genome-wide screens for detection of cervical precancer and cancer. Epigenomics. 12, 1569–1578. doi: 10.2217/epi-2019-0331
Dippmann, C., Schmitz, M., Wunsch, K., Schutze, S., Beer, K., Greinke, C., et al. (2020). Triage of hr HPV-positive women: comparison of two commercial methylation-specific PCR assays. Clin. Epigenetics 12:171. doi: 10.1186/s13148-020-00963-w
Ehrlich, M. (2002). DNA methylation in cancer: too much, but also too little. Oncogene 21, 5400–5413. doi: 10.1038/sj.onc.1205651
Eifler, K., and Vertegaal, A. C. O. (2015). SUMOylation-mediated regulation of cell cycle progression and cancer. Trends Biochem. Sci. 40, 779–793. doi: 10.1016/j.tibs.2015.09.006
Esteller, M. (2002). CpG island hypermethylation and tumor suppressor genes: a booming present, a brighter future. Oncogene 21, 5427–5440. doi: 10.1038/sj.onc.1205600
Favre, M., Breitburd, F., Croissant, O., and Orth, G. (1977). Chromatin-like structures obtained after alkaline disruption of bovine and human papillomaviruses. J. Virol. 21, 1205–1209. doi: 10.1128/jvi.21.3.1205-1209.1977
Feng, D., Yan, K., Zhou, Y., Liang, H., Liang, J., Zhao, W., et al. (2016). Piwil 2 is reactivated by HPV oncoproteins and initiates cell reprogramming via epigenetic regulation during cervical cancer tumorigenesis. Oncotarget 7, 64575–64588. doi: 10.18632/oncotarget.11810
Figueroa-Angulo, E. E., Puente-Rivera, J., Perez-Navarro, Y. F., Condado, E. M., and Alvarez-Sanchez, M. E. (2025). Epigenetic alteration in cervical cancer induced by human papillomavirus. Int. Rev. Cell Mol. Biol. 390, 25–66. doi: 10.1016/bs.ircmb.2024.09.001
Floore, A., Hesselink, A., Ostrbenk, A., Alcaniz, E., Rothe, B., Pedersen, H., et al. (2019). Intra-and inter-laboratory agreement of the FAM19A4/mir 124-2 methylation test: results from an international study. J. Clin. Lab. Anal. 33:e22854. doi: 10.1002/jcla.22854
Folliero, V., Dell'Annunziata, F., Chianese, A., Morone, M. V., Mensitieri, F., Di Spirito, F., et al. (2023). Epigenetic and genetic keys to fight HPV-related cancers. Cancers 15:5583. doi: 10.3390/cancers15235583
Gameiro, S. F., Kolendowski, B., Zhang, A., Barrett, J. W., Nichols, A. C., Torchia, J., et al. (2017). Human papillomavirus dysregulates the cellular apparatus controlling the methylation status of H3K27 in different human cancers to consistently alter gene expression regardless of tissue of origin. Oncotarget 8, 72564–72576. doi: 10.18632/oncotarget.19885
Gasperov, M., Farkas, S. A., Nilsson, T. K., and Grce, M. (2014). Epigenetic activation of immune genes in cervical cancer. Immunol. Lett. 162, 256–257. doi: 10.1016/j.imlet.2014.09.019
Ghiani, L., and Chiocca, S. (2022). High risk-human papillomavirus in HNSCC: present and future challenges for epigenetic therapies. Int. J. Mol. Sci. 23:3483. doi: 10.3390/ijms23073483
Gibney, E. R., and Nolan, C. M. (2010). Epigenetics and gene expression. Heredity 105, 4–13. doi: 10.1038/hdy.2010.54
Giuliano, A. R., Nedjai, B., Lorincz, A. T., Schell, M. J., Rahman, S., Banwait, R., et al. (2020). Methylation of HPV 16 and EPB41L3 in oral gargles: associations with oropharyngeal cancer detection and tumor characteristics. Int. J. Cancer 146, 1018–1030. doi: 10.1002/ijc.32570
Gomez-Gomez, Y., Organista-Nava, J., and Gariglio, P. (2013). Deregulation of the mi RNAs expression in cervical cancer: human papillomavirus implications. Biomed. Res. Int. 2013:407052. doi: 10.1155/2013/407052
Gravina, G. L., Festuccia, C., Marampon, F., Popov, V. M., Pestell, R. G., Zani, B. M., et al. (2010). Biological rationale for the use of DNA methyltransferase inhibitors as new strategy for modulation of tumor response to chemotherapy and radiation. Mol. Cancer 9:305. doi: 10.1186/1476-4598-9-305
Gui, Y., Wang, L., and Huang, Z. (2021). MiR-137 inhibits cervical cancer progression via down-modulating notch 1 and inhibiting the PI3K/AKT/mTOR signaling pathway. Transl. Cancer Res. 10, 3748–3756. doi: 10.21037/tcr-21-1049
Hamilton, J. P. (2011). Epigenetics: principles and practice. Dig. Dis. 29, 130–135. doi: 10.1159/000323874
Hanahan, D. (2022). Hallmarks of cancer: new dimensions. Cancer Discov. 12, 31–46. doi: 10.1158/2159-8290.CD-21-1059
Heaton, P. R., Deyrieux, A. F., Bian, X. L., and Wilson, V. G. (2011). HPV E6 proteins target Ubc 9, the SUMO conjugating enzyme. Virus Res. 158, 199–208. doi: 10.1016/j.virusres.2011.04.001
Herceg, Z. (2016). Epigenetic mechanisms as an Interface between the environment and genome. Adv. Exp. Med. Biol. 903, 3–15. doi: 10.1007/978-1-4899-7678-9_1
Hu, C., Liu, X., Zeng, Y., Liu, J., and Wu, F. (2021). DNA methyltransferase inhibitors combination therapy for the treatment of solid tumor: mechanism and clinical application. Clin. Epigenetics 13:166. doi: 10.1186/s13148-021-01154-x
Huang, S. M., and McCance, D. J. (2002). Down regulation of the interleukin-8 promoter by human papillomavirus type 16 E6 and E7 through effects on CREB binding protein/p 300 and P/CAF. J. Virol. 76, 8710–8721. doi: 10.1128/JVI.76.17.8710-8721.2002
Huh, K., Zhou, X., Hayakawa, H., Cho, J. Y., Libermann, T. A., Jin, J., et al. (2007). Human papillomavirus type 16 E7 oncoprotein associates with the cullin 2 ubiquitin ligase complex, which contributes to degradation of the retinoblastoma tumor suppressor. J. Virol. 81, 9737–9747. doi: 10.1128/JVI.00881-07
Hussain, S., Tulsyan, S., Dar, S. A., Sisodiya, S., Abiha, U., Kumar, R., et al. (2022). Role of epigenetics in carcinogenesis: recent advancements in anticancer therapy. Semin. Cancer Biol. 83, 441–451. doi: 10.1016/j.semcancer.2021.06.023
Hyland, P. L., McDade, S. S., McCloskey, R., Dickson, G. J., Arthur, K., McCance, D. J., et al. (2011). Evidence for alteration of EZH2, BMI1, and KDM6A and epigenetic reprogramming in human papillomavirus type 16 E6/E7-expressing keratinocytes. J. Virol. 85, 10999–11006. doi: 10.1128/JVI.00160-11
Jackson, S. P., and Durocher, D. (2013). Regulation of DNA damage responses by ubiquitin and SUMO. Mol. Cell 49, 795–807. doi: 10.1016/j.molcel.2013.01.017
Jaiswal, N., John, R., Chand, V., and Nag, A. (2015). Oncogenic human papillomavirus 16E7 modulates SUMOylation of fox M1b. Int. J. Biochem. Cell Biol. 58, 28–36. doi: 10.1016/j.biocel.2014.11.002
Jiang, Y., Li, Y., Fang, S., Jiang, B., Qin, C., Xie, P., et al. (2014). The role of MALAT1 correlates with HPV in cervical cancer. Oncol. Lett. 7, 2135–2141. doi: 10.3892/ol.2014.1996
Katzenellenbogen, R. (2017). Telomerase induction in HPV infection and oncogenesis. Viruses 9:180. doi: 10.3390/v9070180
Khan, A., Das, B. C., Abiha, U., Sisodiya, S., Chikara, A., Nazir, S. U., et al. (2022). Insights into the role of complement regulatory proteins in HPV mediated cervical carcinogenesis. Semin. Cancer Biol. 86, 583–589. doi: 10.1016/j.semcancer.2021.05.031
Kocsis, A., Takacs, T., Jeney, C., Schaff, Z., Koiss, R., Jaray, B., et al. (2017). Performance of a new HPV and biomarker assay in the management of hr HPV positive women: subanalysis of the ongoing multicenter TRACE clinical trial (n > 6, 000) to evaluate POU4F3 methylation as a potential biomarker of cervical precancer and cancer. Int. J. Cancer 140, 1119–1133. doi: 10.1002/ijc.30534
Kong, L., Wang, L., Wang, Z., Xiao, X., You, Y., Wu, H., et al. (2023). Cytological DNA methylation for cervical cancer screening: a validation set. Front. Oncol. 13:1181982. doi: 10.3389/fonc.2023.1181982
Kulis, M., and Esteller, M. (2010). DNA methylation and cancer. Adv. Genet. 70, 27–56. doi: 10.1016/B978-0-12-380866-0.60002-2
Laengsri, V., Kerdpin, U., Plabplueng, C., Treeratanapiboon, L., and Nuchnoi, P. (2018). Cervical cancer markers: epigenetics and micro RNAs. Lab. Med. 49, 97–111. doi: 10.1093/labmed/lmx080
Lai, H. C., Lin, Y. W., Huang, R. L., Chung, M. T., Wang, H. C., Liao, Y. P., et al. (2010). Quantitative DNA methylation analysis detects cervical intraepithelial neoplasms type 3 and worse. Cancer 116, 4266–4274. doi: 10.1002/cncr.25252
Li, Y. (2023). Non-coding RNA performs its biological function by interacting with macromolecules. Int. J. Mol. Sci. 24:16246. doi: 10.3390/ijms242216246
Li, S. Y., Yu, Y., Zhen, S., Xing, Z., and Gai, X. (2021). Clinical study on the effect of whole course nursing intervention on critically ill patients and the occurrence of complications. Indian J. Pharm. Sci. 83, 73–76. doi: 10.36468/pharmaceutical-sciences.spl.254
Liu, S., Chang, W., Jin, Y., Feng, C., Wu, S., He, J., et al. (2019). The function of histone acetylation in cervical cancer development. Biosci. Rep. 39:BSR20190527. doi: 10.1042/BSR20190527
Liu, S. S., Leung, R. C., Chan, K. Y., Chiu, P. M., Cheung, A. N., Tam, K. F., et al. (2004). P 73 expression is associated with the cellular radiosensitivity in cervical cancer after radiotherapy. Clin. Cancer Res. 10, 3309–3316. doi: 10.1158/1078-0432.CCR-03-0119
Liu, B., Zhai, J., Wang, W., Liu, T., Liu, C., Zhu, X., et al. (2022). Identification of tumor microenvironment and DNA methylation-related prognostic signature for predicting clinical outcomes and therapeutic responses in cervical cancer. Front. Mol. Biosci. 9:872932. doi: 10.3389/fmolb.2022.872932
Liu, J., Zhao, H., Zhang, Q., Shi, Z., Zhang, Y., Zhao, L., et al. (2020). Human papillomavirus type 16 E7 oncoprotein-induced upregulation of lysine-specific demethylase 5A promotes cervical cancer progression by regulating the micro RNA-424-5p/suppressor of zeste 12 pathway. Exp. Cell Res. 396:112277. doi: 10.1016/j.yexcr.2020.112277
Lopez-Banuelos, L., and Vega, L. (2022). Inhibition of acetylation, is it enough to fight cancer? Crit. Rev. Oncol. Hematol. 176:103752. doi: 10.1016/j.critrevonc.2022.103752
Lorincz, A. T., Brentnall, A. R., Scibior-Bentkowska, D., Reuter, C., Banwait, R., Cadman, L., et al. (2016). Validation of a DNA methylation HPV triage classifier in a screening sample. Int. J. Cancer 138, 2745–2751. doi: 10.1002/ijc.30008
Lourenco de Freitas, N., Deberaldini, M. G., Gomes, D., Pavan, A. R., Sousa, A., Dos Santos, J. L., et al. (2020). Histone deacetylase inhibitors as therapeutic interventions on cervical cancer induced by human papillomavirus. Front cell. Dev. Biol. 8:592868. doi: 10.3389/fcell.2020.592868
Łuczak, M. W., and Jagodzinski, P. P. (2008). Apicidin down-regulates human papillomavirus type 16 E6 and E7 transcripts and proteins in SiHa cervical cancer cells. Cancer Lett. 272, 53–60. doi: 10.1016/j.canlet.2008.06.030
Mattick, J. S., and Makunin, I. V. (2006). Non-coding RNA. Hum. Mol. Genet. 15, R17–R29. doi: 10.1093/hmg/ddl046
Mattoscio, D., Casadio, C., Miccolo, C., Maffini, F., Raimondi, A., Tacchetti, C., et al. (2017). Autophagy regulates UBC9 levels during viral-mediated tumorigenesis. PLoS Pathog. 13:e1006262. doi: 10.1371/journal.ppat.1006262
McLaughlin-Drubin, M. E., Crum, C. P., and Munger, K. (2011). Human papillomavirus E7 oncoprotein induces KDM6A and KDM6B histone demethylase expression and causes epigenetic reprogramming. Proc. Natl. Acad. Sci. USA 108, 2130–2135. doi: 10.1073/pnas.1009933108
McLaughlin-Drubin, M. E., Park, D., and Munger, K. (2013). Tumor suppressor p16INK4A is necessary for survival of cervical carcinoma cell lines. Proc. Natl. Acad. Sci. USA 110, 16175–16180. doi: 10.1073/pnas.1310432110
McMurray, H. R., and McCance, D. J. (2003). Human papillomavirus type 16 E6 activates TERT gene transcription through induction of c-Myc and release of USF-mediated repression. J. Virol. 77, 9852–9861. doi: 10.1128/jvi.77.18.9852-9861.2003
Melchior, F., Schergaut, M., and Pichler, A. (2003). SUMO: ligases, isopeptidases and nuclear pores. Trends Biochem. Sci. 28, 612–618. doi: 10.1016/j.tibs.2003.09.002
Moody, C. A., and Laimins, L. A. (2010). Human papillomavirus oncoproteins: pathways to transformation. Nat. Rev. Cancer 10, 550–560. doi: 10.1038/nrc2886
Morgan, E. L., Patterson, M. R., Ryder, E. L., Lee, S. Y., Wasson, C. W., Harper, K. L., et al. (2020). Micro RNA-18a targeting of the STK4/MST1 tumour suppressor is necessary for transformation in HPV positive cervical cancer. PLoS Pathog. 16:e1008624. doi: 10.1371/journal.ppat.1008624
Narita, T., Weinert, B. T., and Choudhary, C. (2019). Functions and mechanisms of non-histone protein acetylation. Nat. Rev. Mol. Cell Biol. 20, 156–174. doi: 10.1038/s41580-018-0081-3
Neyaz, M. K., Kumar, R. S., Hussain, S., Naqvi, S. H., Kohaar, I., Thakur, N., et al. (2008). Effect of aberrant promoter methylation of FHIT and RASSF1A genes on susceptibility to cervical cancer in a north Indian population. Biomarkers 13, 597–606. doi: 10.1080/13547500802078859
Okunade, K. S. (2020). Human papillomavirus and cervical cancer. J. Obstet. Gynaecol. 40, 602–608. doi: 10.1080/01443615.2019.1634030
Oladipo, K. H., and Parish, J. L. (2025). De-regulation of aurora kinases by oncogenic HPV; implications in cancer development and treatment. Tumour Virus Res. 19:200314. doi: 10.1016/j.tvr.2025.200314
Ozaki, K., Kishikawa, F., Tanaka, M., Sakamoto, T., Tanimura, S., and Kohno, M. (2008). Histone deacetylase inhibitors enhance the chemosensitivity of tumor cells with cross-resistance to a wide range of DNA-damaging drugs. Cancer Sci. 99, 376–384. doi: 10.1111/j.1349-7006.2007.00669.x
Padash Barmchi, M., Thomas, M., Thatte, J. V., Vats, A., Zhang, B., Cagan, R. L., et al. (2021). Inhibition of kinase IKKbeta suppresses cellular abnormalities induced by the human papillomavirus oncoprotein HPV 18E6. Sci. Rep. 11:1111. doi: 10.1038/s41598-020-80193-5
Pal, A., and Kundu, R. (2019). Human papillomavirus E6 and E7: the cervical cancer hallmarks and targets for therapy. Front. Microbiol. 10:3116. doi: 10.3389/fmicb.2019.03116
Pun, P. B., Liao, Y. P., Su, P. H., Wang, H. C., Chen, Y. C., Hsu, Y. W., et al. (2015). Triage of high-risk human papillomavirus-positive women by methylated POU4F3. Clin. Epigenetics 7:85. doi: 10.1186/s13148-015-0122-0
Rampias, T., Boutati, E., Pectasides, E., Sasaki, C., Kountourakis, P., Weinberger, P., et al. (2010). Activation of Wnt signaling pathway by human papillomavirus E6 and E7 oncogenes in HPV16-positive oropharyngeal squamous carcinoma cells. Mol. Cancer Res. 8, 433–443. doi: 10.1158/1541-7786.MCR-09-0345
Rangasamy, D., Woytek, K., Khan, S. A., and Wilson, V. G. (2000). SUMO-1 modification of bovine papillomavirus E1 protein is required for intranuclear accumulation. J. Biol. Chem. 275, 37999–38004. doi: 10.1074/jbc.M007777200
Rocha, M. A., Cardoso, A. L., Martins, C., and Mello, M. L. S. (2024). Sodium valproate affects the expression of p 16 (INK4a) and p 21 (WAFI/Cip 1) cyclin-dependent kinase inhibitors in HeLa cells. Oncol. Lett. 28:432. doi: 10.3892/ol.2024.14563
Rosl, F., Arab, A., Klevenz, B., and zur Hausen, H. (1993). The effect of DNA methylation on gene regulation of human papillomaviruses. J. Gen. Virol. 74, 791–801.
Sabatini, M. E., Compagnoni, M., Maffini, F., Miccolo, C., Pagni, F., Lombardi, M., et al. (2022). The UBC9/SUMO pathway affects E-cadherin cleavage in HPV-positive head and neck cancer. Front. Mol. Biosci. 9:940449. doi: 10.3389/fmolb.2022.940449
Salta, S., Maia-Moco, L., Estevao-Pereira, H., Sequeira, J. P., Vieira, R., Bartosch, C., et al. (2021). Performance of DNA methylation-based biomarkers in the cervical cancer screening program of northern Portugal: a feasibility study. Int. J. Cancer 149, 1916–1925. doi: 10.1002/ijc.33778
Scheffner, M., Huibregtse, J. M., Vierstra, R. D., and Howley, P. M. (1993). The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p 53. Cell 75, 495–505. doi: 10.1016/0092-8674(93)90384-3
Schmitz, M., Eichelkraut, K., Schmidt, D., Zeiser, I., Hilal, Z., Tettenborn, Z., et al. (2018). Performance of a DNA methylation marker panel using liquid-based cervical scrapes to detect cervical cancer and its precancerous stages. BMC Cancer 18:1197. doi: 10.1186/s12885-018-5125-8
Schmitz, M., Wunsch, K., Hoyer, H., Scheungraber, C., Runnebaum, I. B., Hansel, A., et al. (2017). Performance of a methylation specific real-time PCR assay as a triage test for HPV-positive women. Clin. Epigenetics 9:118. doi: 10.1186/s13148-017-0419-2
Sen, P., Ganguly, P., and Ganguly, N. (2018). Modulation of DNA methylation by human papillomavirus E6 and E7 oncoproteins in cervical cancer. Oncol. Lett. 15, 11–22. doi: 10.3892/ol.2017.7292
Sharifi-Rad, J., Rayess, Y. E., Rizk, A. A., Sadaka, C., Zgheib, R., Zam, W., et al. (2020). Turmeric and its major compound curcumin on health: bioactive effects and safety profiles for food, pharmaceutical, biotechnological and medicinal applications. Front. Pharmacol. 11:01021. doi: 10.3389/fphar.2020.01021
Sharma, S., and Munger, K. (2020). The role of long noncoding RNAs in human papillomavirus-associated pathogenesis. Pathogens. 9:289. doi: 10.3390/pathogens9040289
Silvia Lima, R. Q., Vasconcelos, C. F. M., Gomes, J. P. A., de Menezes EdS, B., de Oliveira Silva, B., Montenegro, C., et al. (2024). miRNA-21, an oncomi R that regulates cell proliferation, migration, invasion and therapy response in lung cancer. Pathol. Res. Pract. 263:155601. doi: 10.1016/j.prp.2024.155601
Singh, V., Ram, M., Kumar, R., Prasad, R., Roy, B. K., and Singh, K. K. (2017). Phosphorylation: implications in cancer. Protein J. 36, 1–6. doi: 10.1007/s10930-017-9696-z
Sobti, R. C., Singh, N., Hussain, S., Suri, V., Nijhawan, R., Bharti, A. C., et al. (2011). Aberrant promoter methylation and loss of suppressor of cytokine signalling-1 gene expression in the development of uterine cervical carcinogenesis. Cell. Oncol. 34, 533–543. doi: 10.1007/s13402-011-0056-2
Song, Y., and Zhang, C. (2009). Hydralazine inhibits human cervical cancer cell growth in vitro in association with APC demethylation and re-expression. Cancer Chemother. Pharmacol. 63, 605–613. doi: 10.1007/s00280-008-0773-z
Sun, X., Shu, Y., Ye, G., Wu, C., Xu, M., Gao, R., et al. (2022). Histone deacetylase inhibitors inhibit cervical cancer growth through Parkin acetylation-mediated mitophagy. Acta Pharm. Sin. B 12, 838–852. doi: 10.1016/j.apsb.2021.07.003
Takai, N., and Narahara, H. (2007). Human endometrial and ovarian cancer cells: histone deacetylase inhibitors exhibit antiproliferative activity, potently induce cell cycle arrest, and stimulate apoptosis. Curr. Med. Chem. 14, 2548–2553. doi: 10.2174/092986707782023299
Thomas, Y., and Androphy, E. J. (2018). Human papillomavirus replication regulation by acetylation of a conserved lysine in the E2 protein. J. Virol. 92:jvi.01912-17. doi: 10.1128/JVI.01912-17
Thomas, Y., and Androphy, E. J. (2019). Acetylation of E2 by P 300 mediates topoisomerase entry at the papillomavirus replicon. J. Virol. 93:10.1128/jvi.02224-18. doi: 10.1128/JVI.02224-18
Tian, Y., Yuan Wu, N. Y., Liou, Y. L., Yeh, C. T., Cao, L., Kang, Y. N., et al. (2017). Utility of gene methylation analysis, cytological examination, and HPV-16/18 genotyping in triage of high-risk human papilloma virus-positive women. Oncotarget 8, 62274–62285. doi: 10.18632/oncotarget.19459
Tomaic, V. (2016). Functional roles of E6 and E7 Oncoproteins in HPV-induced malignancies at diverse anatomical sites. Cancers 8:95. doi: 10.3390/cancers8100095
Tsai, H. C., and Baylin, S. B. (2011). Cancer epigenetics: linking basic biology to clinical medicine. Cell Res. 21, 502–517. doi: 10.1038/cr.2011.24
Ulhe, A., Raina, P., Chaudhary, A., and Kaul-Ghanekar, R. (2025). Alpha-linolenic acid-mediated epigenetic reprogramming of cervical cancer cell lines. Epigenetics 20:2451551. doi: 10.1080/15592294.2025.2451551
Unger, E. R., and Barr, E. (2004). Human papillomavirus and cervical cancer. Emerg. Infect. Dis. 10, 2031–2032. doi: 10.3201/eid1011.040623_09
Verhoef, L., Bleeker, M. C. G., Polman, N., Steenbergen, R. D. M., Meijer, C., Melchers, W. J. G., et al. (2022). Performance of DNA methylation analysis of ASCL1, LHX8, ST6GALNAC5, GHSR, ZIC1 and SST for the triage of HPV-positive women: results from a Dutch primary HPV-based screening cohort. Int. J. Cancer 150, 440–449. doi: 10.1002/ijc.33820
Verhoef, V. M., van Kemenade, F. J., Rozendaal, L., Heideman, D. A., Bosgraaf, R. P., Hesselink, A. T., et al. (2015). Follow-up of high-risk HPV positive women by combined cytology and bi-marker CADM1/MAL methylation analysis on cervical scrapes. Gynecol. Oncol. 137, 55–59. doi: 10.1016/j.ygyno.2015.01.550
Verlaat, W., Snoek, B. C., Heideman, D. A. M., Wilting, S. M., Snijders, P. J. F., Novianti, P. W., et al. (2018). Identification and validation of a 3-gene methylation classifier for HPV-based cervical screening on self-samples. Clin. Cancer Res. 24, 3456–3464. doi: 10.1158/1078-0432.CCR-17-3615
Vink, F. J., Meijer, C., Clifford, G. M., Poljak, M., Ostrbenk, A., Petry, K. U., et al. (2020). FAM19A4/miR124-2 methylation in invasive cervical cancer: a retrospective cross-sectional worldwide study. Int. J. Cancer 147, 1215–1221. doi: 10.1002/ijc.32614
Walboomers, J. M., Jacobs, M. V., Manos, M. M., Bosch, F. X., Kummer, J. A., Shah, K. V., et al. (1999). Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J. Pathol. 189, 12–19. doi: 10.1002/(SICI)1096-9896(199909)189:1<>3.0.CO;2-F
Wang, P., Guan, Q., Zhou, D., Yu, Z., Song, Y., and Qiu, W. (2017). Mir-21 inhibitors modulate biological functions of gastric cancer cells via PTEN/PI3K/mTOR pathway. DNA Cell Biol. 37, 38–45. doi: 10.1089/dna.2017.3922
Wang, X., Huang, X., and Zhang, Y. (2018). Involvement of human papillomaviruses in cervical Cancer. Front. Microbiol. 9:2896. doi: 10.3389/fmicb.2018.02896
Wang, N., Ma, T., and Yu, B. (2023). Targeting epigenetic regulators to overcome drug resistance in cancers. Signal Transduct. Target. Ther. 8:69. doi: 10.1038/s41392-023-01341-7
Wang, W., Sun, Z., Liu, J., Wang, G., Lu, Z., Zhou, W., et al. (2017). Increased methylation of human papillomavirus type 16 DNA is associated with the severity of cervical lesions in infected females from Northeast China. Oncol. Lett. 13, 3809–3816. doi: 10.3892/ol.2017.5903
Westrich, J. A., Warren, C. J., and Pyeon, D. (2017). Evasion of host immune defenses by human papillomavirus. Virus Res. 231, 21–33. doi: 10.1016/j.virusres.2016.11.023
Xiao, X., Wang, W., Bai, P., Chen, Y., Qin, Z., Cheng, T., et al. (2024). Genomic imprinting biomarkers for cervical cancer risk stratification. Cancer Commun. 44, 1385–1390. doi: 10.1002/cac2.12617
Xiong, J., Chen, P., He, L., Chai, X., Zhang, Y., and Sun, S. (2024). Functional mechanism of hypoxia-like conditions mediating resistance to ferroptosis in cervical cancer cells by regulating KDM4A SUMOylation and the SLC7A11/GPX4 pathway. Environ. Toxicol. 39, 4207–4220. doi: 10.1002/tox.24304
Yang, X., Chen, Y., Li, M., and Zhu, W. (2022). ERBB3 methylation and immune infiltration in tumor microenvironment of cervical cancer. Sci. Rep. 12:8112. doi: 10.1038/s41598-022-11415-1
Zhang, L., Wu, J., Ling, M. T., Zhao, L., and Zhao, K. N. (2015). The role of the PI3K/Akt/mTOR signalling pathway in human cancers induced by infection with human papillomaviruses. Mol. Cancer 14:87. doi: 10.1186/s12943-015-0361-x
Zhang, Y., Zheng, X., Tan, H., Lu, Y., Tao, D., Liu, Y., et al. (2018). PIWIL2 suppresses Siah 2-mediated degradation of HDAC3 and facilitates CK2alpha-mediated HDAC3 phosphorylation. Cell Death Dis. 9:423. doi: 10.1038/s41419-018-0462-8
Zhang, X., Zhu, L., Wang, X., Zhang, H., Wang, L., and Xia, L. (2023). Basic research on curcumin in cervical cancer: progress and perspectives. Biomed. Pharmacother. 162:114590. doi: 10.1016/j.biopha.2023.114590
Zhao, J., Cao, H., Zhang, W., Fan, Y., Shi, S., and Wang, R. (2021). SOX14 hypermethylation as a tumour biomarker in cervical cancer. BMC Cancer 21:675. doi: 10.1186/s12885-021-08406-2
Zhao, Z., and Shilatifard, A. (2019). Epigenetic modifications of histones in cancer. Genome Biol. 20:245. doi: 10.1186/s13059-019-1870-5
Zhao, X., Zhang, R., Song, Z., Yang, K., He, H., Jin, L., et al. (2024). Curcumin suppressed the proliferation and apoptosis of HPV-positive cervical cancer cells by directly targeting the E6 protein. Phytother. Res. 38, 4967–4981. doi: 10.1002/ptr.7868
Zhou, Y., Wang, Y., Lin, M., Wu, D., and Zhao, M. (2021). LncRNA HOTAIR promotes proliferation and inhibits apoptosis by sponging miR-214-3p in HPV16 positive cervical cancer cells. Cancer Cell Int. 21:400. doi: 10.1186/s12935-021-02103-7
Zhou, L., Xu, N., Shibata, H., Saloura, V., and Uppaluri, R. (2021). Epigenetic modulation of immunotherapy and implications in head and neck cancer. Cancer Metastasis Rev. 40, 141–152. doi: 10.1007/s10555-020-09944-0
Keywords: cervical cancer, human papillomavirus, epigenetics, screening, biomarker
Citation: Sisodiya S, Singh P, Joshi T, Aftab M, Firdausi N, Khan A, Mishra N, Jamil Khan N, Tanwar P, Gupta V and Hussain S (2025) Human papillomavirus-mediated cervical cancer: epigenetic interplay and clinical implications. Front. Microbiol. 16:1633283. doi: 10.3389/fmicb.2025.1633283
Edited by:
Gurdeep Singh, University of New Mexico School of Medicine, United StatesReviewed by:
Deepak Parashar, Medical College of Wisconsin, United StatesJonas Wolf, Moinhos de Vento Hospital, Brazil
Copyright © 2025 Sisodiya, Singh, Joshi, Aftab, Firdausi, Khan, Mishra, Jamil Khan, Tanwar, Gupta and Hussain. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Showket Hussain, c2hvd2tldC5odXNzYWluQGdvdi5pbg==
†These authors have contributed equally to this work
 Sandeep Sisodiya
Sandeep Sisodiya Payal Singh
Payal Singh Tannu Joshi
Tannu Joshi Mehreen Aftab
Mehreen Aftab Nasera Firdausi
Nasera Firdausi Asiya Khan
Asiya Khan Neetu Mishra
Neetu Mishra Nida Jamil Khan
Nida Jamil Khan Pranay Tanwar
Pranay Tanwar Vivek Gupta
Vivek Gupta Showket Hussain
Showket Hussain