- 1Department of Microbiology, Graduate School of Medical and Dental Sciences, Kagoshima University, Kagoshima, Japan
- 2Division of Microbiology, National Institute of Health Sciences, Kawasaki, Kanagawa, Japan
- 3Department of Bacteriology I, National Institute of Infectious Diseases, Tokyo, Japan
- 4Department of Bacteriology, Faculty of Medical Sciences, Kyushu University, Fukuoka, Japan
- 5Advanced Genomics Center, National Institute of Genetics, Shizuoka, Japan
- 6Department of Microbiology, Hoshi University, Tokyo, Japan
Introduction: Enteroaggregative Escherichia coli heat-stable enterotoxin 1 (EAST1) encoded by the astA gene was first identified in an enteroaggregative E. coli strain isolated from a patient with persistent diarrhea. While astA-positive strains sometimes cause large food poisoning outbreaks, the significance of EAST1 as a virulence factor remains unclear. Additionally, although the prototype and seven variants of the astA gene have been identified, the biological significance of these genetic variations remains undefined. This study aimed to elucidate the characteristics of the A gene by investigating its distribution and sequence diversity within the evolutionary lineages of Escherichia coli.
Methods: We conducted PCR screening for the astA gene in 2,726 E. coli strains isolated from children with diarrhea in Kagoshima, Japan, and blastn search of the astA gene was conducted on 9,065 publicly available finished E. coli genomes. The astA gene identified were subjected to analysis of sequence variation and comparison of their flanking genomic regions. In addition, the phylogenetic distribution of astA gene variants in E. coli lineage was also investigated.
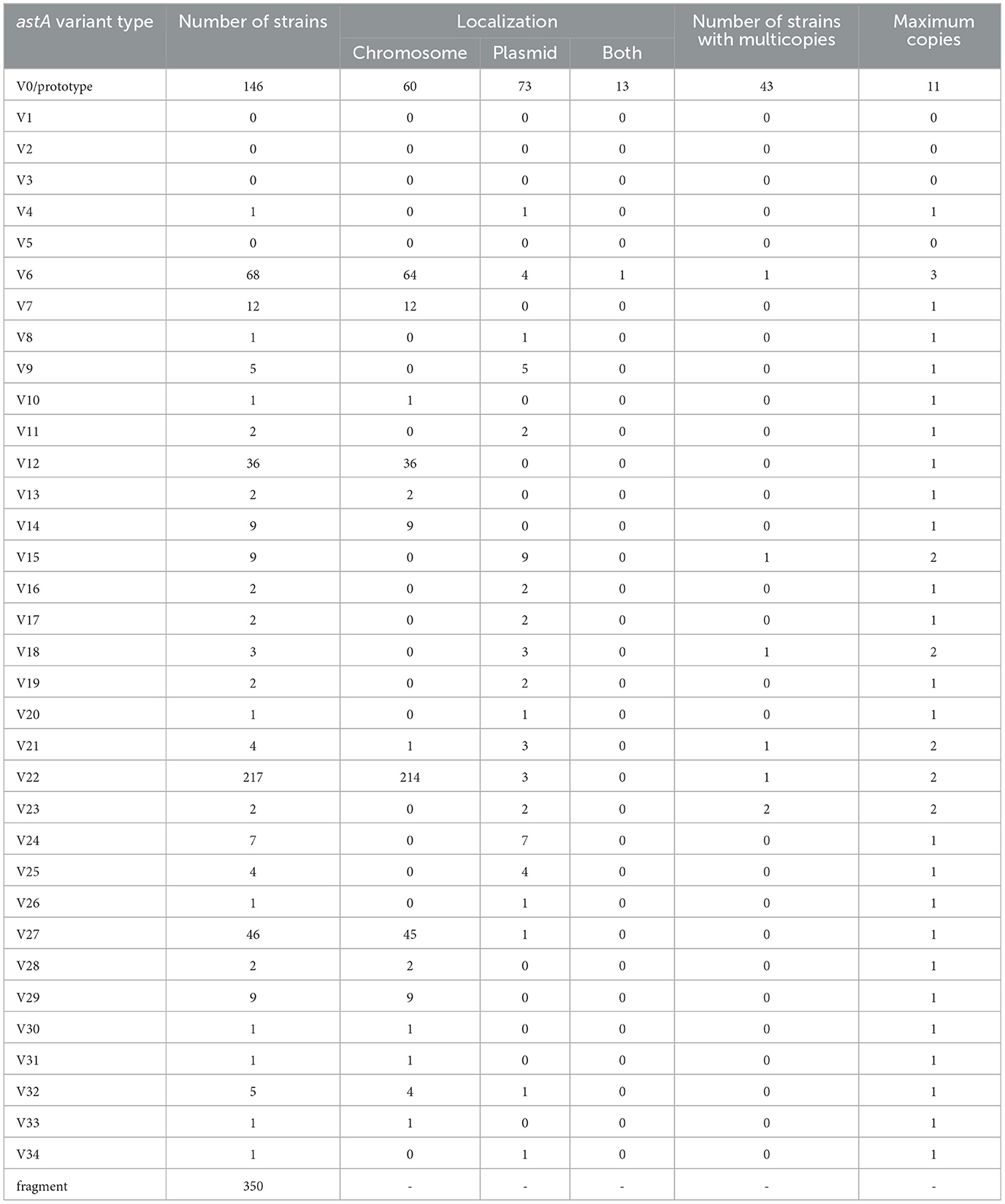
Results and discussion: The results showed that 185 (6.8%) of the Kagoshima strains and 690 (7.6%) of the database strains had similar possession rates. We identified 31 sequence variations (four known and 27 new variants [V8-34]) which were widely distributed in the E. coli lineages. Detailed sequence analyses revealed that 31 of the 35 astA gene types are intact and encode 23 types of EAST1 peptides. Although all 35 types were associated with IS1414, only three (prototype, V30, and V31) of the 31 intact astA gene types were encoded in the intact IS1414. A notable number of prototype-bearing strains (43/146 strains) possessed multiple copies (two to 11 copies) of this type of astA gene, indicating that the amplification has predominantly occurred in the prototype, which was driven by IS1414 amplification. However, given that the IS1414 associated with V30 and V31 also remain structurally intact, it is plausible that similar amplification events may occur in these variants in the future. These results provide an important basis to investigate the virulence of the astA-positive strains and the role of EAST1 as a virulence factor.
Introduction
Enteroaggregative Escherichia coli heat-stable enterotoxin1 (EAST1), a small peptide (38 amino acids of 4.1 kDa) encoded by the astA gene, was first discovered in an enteroaggregative E. coli (EAggEC) strain 17-2 (Savarino et al., 1991; Ménard et al., 2004). The astA gene is known to be widely distributed in a variety of pathogenic E. coli strains (Yamamoto and Echeverria, 1996; Savarino et al., 1996; Paiva de Sousa and Dubreuil, 2001; Ménard and Dubreuil, 2002; Beutin et al., 2008; Maluta et al., 2017; Paniagua-Contreras et al., 2017). EAST1 shares 50% amino acid sequence identity with the enterotoxic domain of heat-stable enterotoxin (STa) and is proposed to exhibit similar mechanism of action to that of STa, which elicits an increase in cGMP on intestinal epithelial cells and subsequent fluid secretion (Dubreuil, 2019). Functional studies using Ussing chamber assays in rabbit ileal mucosa, as well as human T84 epithelial cell monolayers, have demonstrated that EAST1 can stimulate chloride ion secretion, evidenced by sustained increases in short-circuit current (Savarino et al., 1991; Veilleux et al., 2008). These findings support its potential role in the onset of diarrhea. On the other hand, the role of EAST1 in diarrhea in vivo is still questioned because some volunteers challenged with EAST1-producing EAEC strains did not develop diarrhea, even when the strains effectively colonized the intestine (Nataro et al., 1995). However, large-scale food poisoning outbreaks occurred in Japan by E. coli strains of serotypes O7:H4 and O166:H5, in which only the astA gene was detected as a potential virulence-related gene (Zhou et al., 2002; Kashima et al., 2021), suggesting that a possible contribution of EAST1 to the onset of diarrhea.
A notable sequence variation in the astA gene sequence has also been detected. Besides the prototype (referred to as V0 in this manuscript), seven variants (named V1–V7) have been identified to date (Yamamoto et al., 1997; Yamamoto and Taneike, 2000; Savarino et al., 1993; Zhou et al., 2002; Silva et al., 2014; Maluta et al., 2017). However, the biological significance of this genetic variation, the actual sequence diversity, and the prevalence of the variants have not yet been elucidated. A unique feature of the astA gene is that it is embedded within a transposase gene (tnp) of an insertion sequence (IS) IS1414 but in its−1 reading frame (Figure 1A) (McVeigh et al., 2000). Although this was shown for the prototype astA gene in an enterotoxigenic E. coli strain 27D (McVeigh et al., 2000), it is unknown how the astA and its variants are associated with IS1414 in other E. coli strains.
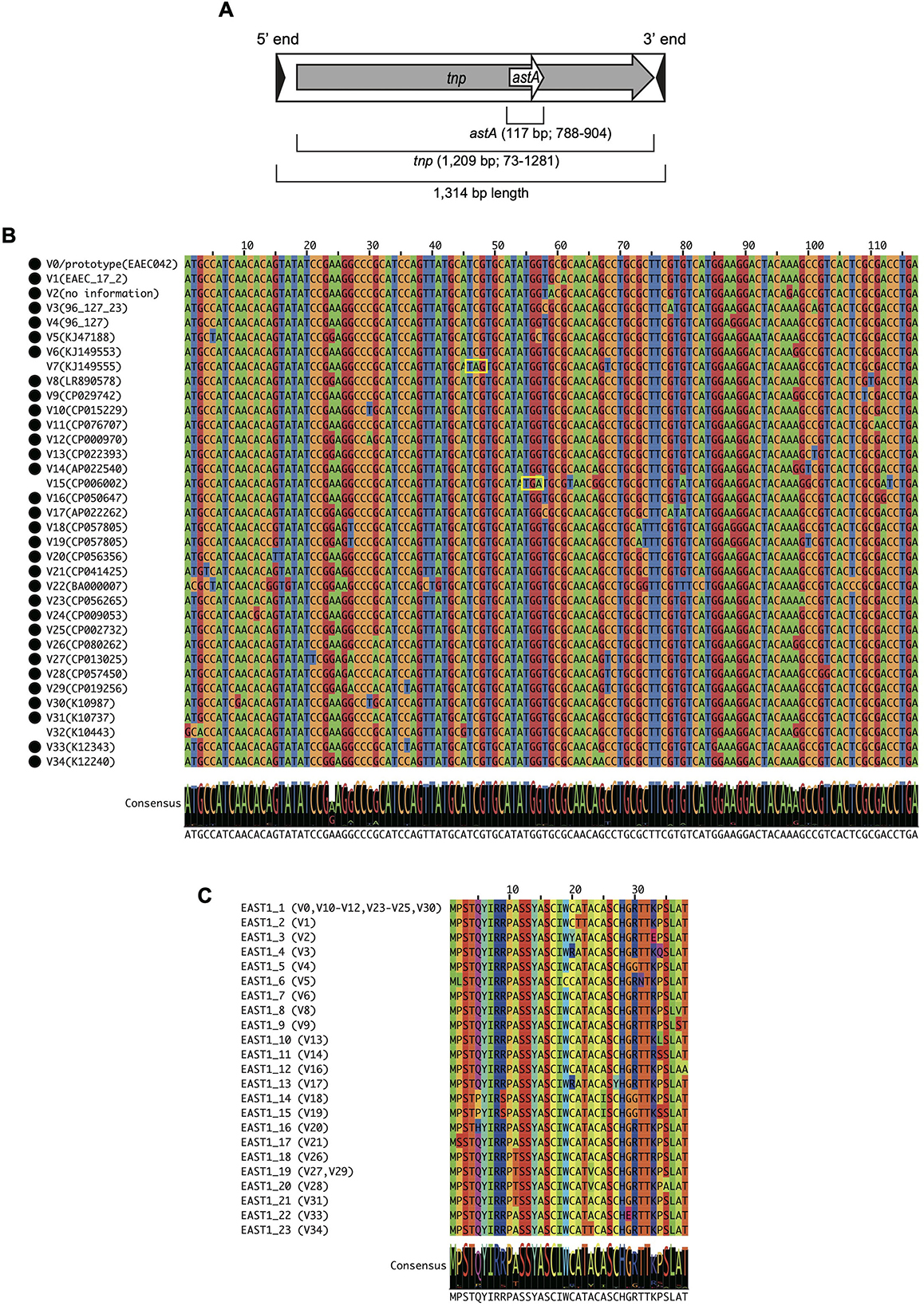
Figure 1. The structure of IS1414 encoding astA gene (A). Multiple nucleotide (B) and amino-acid (C) sequence alignments of the astA and EAST1 variants. Consensus sequences were shown below each sequence alignment in B and C. (B) The stop codon is indicated by yellow boxes and intact astA genes are marked with filled circles. (C) Variants with the same amino acid sequence are shown in parenthesis.
In this study, to better understand the prevalence, sequence variation, and IS association of the astA gene, we conducted PCR screening for the astA gene in 2,726 E. coli strains isolated from children with diarrhea in Kagoshima, Japan, and blastn search in 9,065 publicly available finished E. coli genomes. Using the astA-positive strains and astA genes identified through these analyses, we examined the sequence variation and IS association of astA and the phylogenetic distribution of the astA and its variants in the entire E. coli lineage. In addition, we report the amplification of astA genes associated with intact IS1414 in multiple E. coli strains.
Materials and methods
Bacterial strains and genomic DNA preparation
For PCR screening of the astA gene, 2,726 E. coli strains isolated from stool specimens of diarrheal children who visited clinics in Kagoshima, Japan, from 2013 to 2020 (referred to as Kagoshima strains; listed in Supplementary Table 1) were used. For the blastn search of astA, 9,065 finished E. coli genomes retrieved from the NCBI database (accessed on 10 May 2022; listed in Supplementary Table 2) were used.
Template genomic DNA for PCR screening and multi-locus sequence typing (MLST) analysis was prepared by the alkaline boiling method from a 1 ml culture grown at 37°C in Lysogeny broth (nacalai tesque). Genomic DNA for whole genome sequencing was purified from a 2 ml overnight culture using the NucleoBond HMW DNA (MACHEREY-NAGEL) according to the manufacturer's instructions.
PCR screening of the astA gene
Detection of the astA gene by PCR was performed using a primer pair (EAST1S (5′-GCCATCAACACAGTATATCC-3′) and EAST1AS (5′-GAGTGACGGCTTTGTAGTCC-3′) (Yatsuyanagi et al., 2002) and KAPATaq EXtra PCR Kit (NIPPON Genetics, Tokyo, Japan). Each reaction mixture (15 μl) contained 1 μl of template DNA, 4.5 μM of each primer and 0.3 U of polymerase. PCR was conducted with initial denaturation for 2 min at 94°C, followed by 30 cycles of 30 s at 94°C, 30 s at 55°C and 30 s at 72°C. PCR products were analyzed by agarose gel electrophoresis using 2 % Agarose S (Nippon Gene).
Randomly amplified polymorphic DNA -PCR
RAPD-PCR was performed as described by Pacheco et al. (1997) using P1252 (5′-GCGGAAATAG-3′), P1254 (5′-CCGCAGCCAA-3′), and P1290 (5′-GTGGATGCGA-3′) primers and the KAPATaq EXtra PCR Kit (NIPPON Genetics Co., Ltd.). Each reaction mixture (25 μl) contained 5 μl of 5x KAPATaq Extra buffer, 2.5 mM MgCl2, 300 mM dNTPs, 1 μl of template DNA, 0.4 μM primer and 0.6 U of KAPATaq DNA polymerase. The PCR amplification steps employed were as follows: 4 cycles of 94°C for 5 min, 37°C for 5 min, and 72°C for 5 min, followed by 30 cycles of 94°C for 1 min, 37°C for 1 min and 72°C for 2 min and a final extension step at 72°C for 10 min. After PCR amplification, 10 μl of each PCR product was analyzed by agarose gel electrophoresis using 1.2% agarose S (Nippon Gene).
Multi-locus sequence typing
MLST was performed by PCR amplification and sequencing of seven housekeeping genes (adk, fumC, gyrB, icd, mdh, purA, and recA) as previously described (Ooka et al., 2012). Alleles of each gene and the sequence types (STs) and clonal complexes (CCs) were assigned using the PubMLST (https://pubmlst.org/organisms/escherichia-spp) (Jolley et al., 2018) and the EnteroBase E. coli/Shigella MLST website (https://enterobase.warwick.ac.uk/) (Zhou et al., 2015), respectively.
Whole-genome sequencing
Short-read sequencing libraries were prepared using the Nextera XT DNA Sample Prep Kit (Illumina) to obtain paired-end sequences (300 bp × 2) on the Illumina MiSeq platform. Long-read sequencing libraries were prepared using a Rapid Barcoding Kit (Oxford Nanopore Technologies) and sequenced using an R9.4.1 flow cell. After base-calling and demultiplexed using Guppy GPU v3.4.5 (Oxford Nanopore Technologies), long raw reads were filtered based on quality cut-off score of 10, and minimum length of 2,000 bp and trimmed 100 nucleotides from the start of the read using NanoFilt. A hybrid assembly was performed using microPIPE (Murigneux et al., 2021) with long and short reads with default parameters. The complete and draft genome sequences obtained in this study have been deposited in the GenBank/EMBL/DDBJ database under Bioproject no. PRJDB18009 (see Supplementary Table 2 for the list of sequenced strains and their sequencing statuses and accession numbers).
Blastn search of the astA gene and assignment of new variants
The astA gene in the E. coli genomes was identified by blastn search using the known sequences of astA (V0–V7) (Supplementary Table 3) as queries with cutoff values of 90% nucleotide sequence identity and 60% length match. The BLAST+ source code was downloaded from the NCBI website (https://ftp.ncbi.nlm.nih.gov/blast/executables/blast+/LATEST/). New astA variants were defined if an identified astA gene showed one or more nucleotide sequence differences compared with all of the eight known sequences.
Sequence comparison and characterization of astA genes, their encoding peptides, and astA-flanking regions
The nucleotide sequences of astA genes and their 1,000-bp upstream and downstream flanking regions were extracted from the genome sequences listed in Supplementary Table 2. The multiple nucleotide sequence alignments of astA genes and their 1,000 bp upstream and downstream regions and the amino-acid sequence alignment of astA gene products (EAST1 peptides) were constructed using the ClustalW function of MEGAX (Kumar et al., 2018) with default parameters. Multiple sequence alignments were visualized using JalView (V2.11.5.0) (Waterhouse et al., 2009).
Core-gene based phylogenetic analyses, in silico phylo-typing, and serotyping of strains belonging to phylogroup E
The genome assemblies of 720 strains, including those of 690 strains from the NCBI database and those of 30 strains obtained in this study (18 finished and 12 draft sequences), were annotated using Prokka (Seemann, 2014), and core genes (n = 1,632) were identified using Roary v3.13.0 (Page et al., 2015) with 90% amino acid sequence identity cut-off. Core gene single nucleotide polymorphisms (SNPs) (n = 51,418) were extracted using the core gene alignment tool in Roary and used as inputs for maximum-likelihood (ML) inference with RAxML v8 (Stamatakis, 2014). The ML tree was displayed and annotated using iTOL v6 (https://itol.embl.de) (Letunic and Bork, 2016). The tree was mid-point rooted and the confidence value of each branch were estimated by bootstrap with 200 replications. Genomes showing no core gene SNPs (14 strains forming nine pairs or groups) were deduplicated (strains excluded are indicated in Supplementary Table 2). Strains used as the references for E. coli phylogroup assignment are also indicated in Supplementary Table 2. Serotypes of strains belonging to phylogroup E were determined by SerotypeFinder 2.0 (Joensen et al., 2015).
Identification of other virulence-related genes in the astA-positive Kagoshima strains
To identify potential virulence-associated genes in each genome sequenced Kagoshima strain, we retrieved the core data set of protein sequences from the Virulence Factor Database (VFDB) website (https://www.mgc.ac.cn/VFs/). A blastx search was performed for the genome sequences using the amino acid sequences of the core dataset, with the following parameters: minimum identity of 60%, minimum coverage of 90%, and E-value cutoff of 0.01.
Ethical approval
This study was conducted with the approval of the Ethics Committee for Epidemiological Research, Graduate School of Medical and Dental Sciences, Kagoshima University (#190105).
Results
Prevalence of astA genes in two strain sets
Of the 2,726 Kagoshima strains isolated from children with diarrhea, 185 (6.8%) were positive in the PCR screening for the astA gene (listed in Supplementary Table 1). In a blastn search of the astA gene in 9,065 finished E. coli genomes obtained from NCBI, 690 (7.6%) were found to possess one or more astA genes.
MLST analysis of the astA-positive Kagoshima strains
Prior to the MLST analysis of the 185 astA-positive strains, we performed a RAPD analysis to identify genetically related strains among them. As 39 groups of strains with epidemiological links showed identical amplification patterns in each group (data not shown), one strain was selected from each group and used for the MLST analysis. Thus, 139 strains were subjected to the MLST analysis. This analysis revealed that they belonged to diverse STs: 63 STs were identified with ST6196 being as the largest group containing 14 strains (Supplementary Table 1). This result suggests a wide distribution of astA-positive Kagoshima strains in the entire E. coli lineage. Among the 139 astA-positive strains, 30 strains were selected so that they represented the phylogenetic diversity of astA-positive Kagoshima strains as much as possible and subjected to genome sequencing and following whole genome sequence (WGS)-based analyses.
Sequence variation in the astA gene, its gene product and genomic location
By analyzing the astA genes in the genomes of 720 astA-positive E. coli strains (those of the 30 Kagoshima strains sequenced in this study and the 690 finished genomes obtained from the NCBI database), we identified 31 nucleotide sequence types. They included four of the eight known sequences (V1, V2, V3, and V5 were not detected in this strain set) and 27 newly identified types which were named V8–V34. In addition, various lengths of astA fragments that encode only C-terminal parts of EAST1 were detected in 350 genomes.
As shown in Figure 1B, while many of the variants (23/34 variants) showed 1- or 2-bp difference compared with the V0/prototype, the remaining 11 variants showed 3–12 bp difference. Of these variants, four variants were inactivated by premature stop codons (V7 and V15) or base-changes in the start codon (V22 and V32). Amino-acid sequence alignment of the 31 astA genes that encode full-length EAST1 (Figure 1C) revealed that seven variants encoded EAST1 identical to that encoded by the V0/prototype. In addition, two variant astA genes encoded an identical EAST1 peptide. Thus, 23 variants of EAST1 were identified. We named them EAST1_1 - EAST1_23, of which EAST1_1 corresponds to the EAST1 encoded by the V0/prototype astA gene. Although three EAST1 types contained three, four or five amino-acid substitution compared to the EAST1_1, the remaining types showed one or two amino-acid differences. Of the 38 amino acid residues, 22 were fully conserved, including two of the four cysteine residues present in EAST1_1. These four cysteine residues (Cys-17, 20, 24, and 27) are involved in the formation of two disulfide bridges responsible for the heat stability and, especially, the Cys-17 residue is important for a disulfide bridge integrity for toxicity expression (Uzzau and Fasano, 2000). Of these four cysteine residues, Cys-17 is conserved in all EAST1 types, but in EAST1_3, 4, and 13, one or two amino-acid substitutions occurred at Cys-20 and Cys-27.
Among the 720 strains analyzed, the most dominant nucleotide sequence type was V22 (217 strains; 30.1%), followed by V0/prototype (146 strains; 20.3%), V6 (68 strains; 9.4%), V27 (46 strains; 6.4%), and V12 (36 strains; 5.0%). The other types were detected in less than 2% of the 720 strains (Table 1). Of the top five types, V22 has been inactivated as mentioned above. V12 encoded the EAST1 identical to that encoded by V0/prototype (EAST1_1). As additional seven variants encoded EAST1_1, the most predominant EAST1 type was EAST1_1, which can be potentially produced by 199 strains.
As for the genomic locations of the astA genes identified in the 720 strains, they were located on chromosome or plasmid. While the locations of the astA genes other than V0/prototype showed some bias toward either chromosome or plasmid, V0/prototype was located almost evenly on chromosome and plasmids and 12 strains carried it on both chromosome and plasmid (Table 1). As astA-bearing plasmids were 28-380 kb in size (data not shown), they are likely single- or low-copy plasmids and many of them are probably transmissible or were previously transmissible.
Strains possessing multiple types of astA genes and multiple copies of the V0/prototype gene
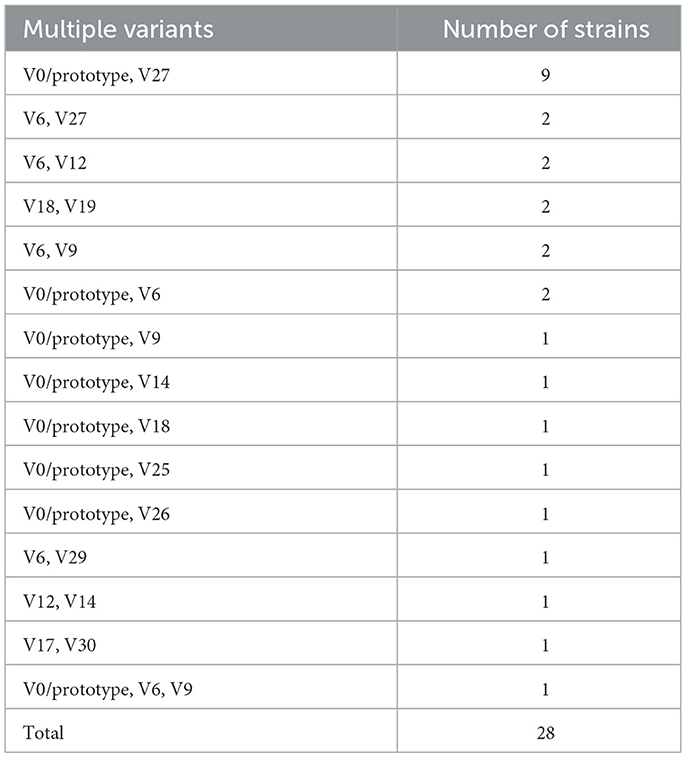
Interestingly, 28 strains harbored two or three types of potentially active astA genes (encoding a full length EAST1) with various combinations: two types in 27 strains and three types in one strain (Table 2). Of the 15 combinations detected, V0/prototype involved in eight combinations, which was an expected finding from its wide distribution. A more interesting and important finding was that a notable number of V0/prototype-containing strains (43/146) possessed multiple copies of this type of astA gene (Table 1). Of these strains, while 22 strains possessed two copies and seven strains possessed three copies, 14 strains contained five or more copies (up to 11 copies). This was in sharp contrast to other types of astA genes: only six variants, of which two were inactivated ones (V15 and V22), appeared twice or three times in the genomes of only eight strains. These findings indicate that the amplification of astA occurred almost specifically for the V0/prototype.
IS1414-association of astA genes
To investigate how various types of astA identified in the 720 strains are associated with IS1414, we analyzed the sequences flanking each astA gene (1,000-bp sequences upstream and downstream of astA). This analysis revealed that while all were associated with IS1414, many of the associated IS1414 have been decayed by deletion or inactivated by mutations in the transposase gene (Supplementary Figures 1, 2). However, most V0/prototype genes (203/270) were associated with intact IS1414, thus they are transposable as a part of IS element (Supplementary Table 2). The V30 and V31 genes were also associated with intact IS1414, but they were harbored by only one strain, respectively.
Phylogenetic view of the 720 astA-positive E. coli strains and the distribution of V0/prototype and variant astA genes
We constructed an ML phylogenetic tree of the 720 astA-positive strains based on their core gene sequences to investigate the phylogenetic relationship of the 720 strains and the distribution of the V0/prototype and variant astA genes and astA fragments in this strain set. As shown in Figure 2, the astA genes were distributed in all E. coli phylogroups. Notably, most of V22 (the most prevalent but inactivated variant) were distributed in phylogroup E: 94.5% of V22-positive strains belonged to this phylogroup (Supplementary Table 4). However, this bias was apparently introduced by the presence of a large number of O157:H7 strains in the strain set analyzed: it included 258 O157:H7 strains, of which 202 contained the V22 variant (Supplementary Table 5). Except for this bias of V22, there was no clear association between the astA type and the lineage (phylogroup) of strains. For example, the V0/prototype astA gene was distributed in all phylogroups and the strains harboring multiple copies of V0/prototype were also found in all phylogroups.
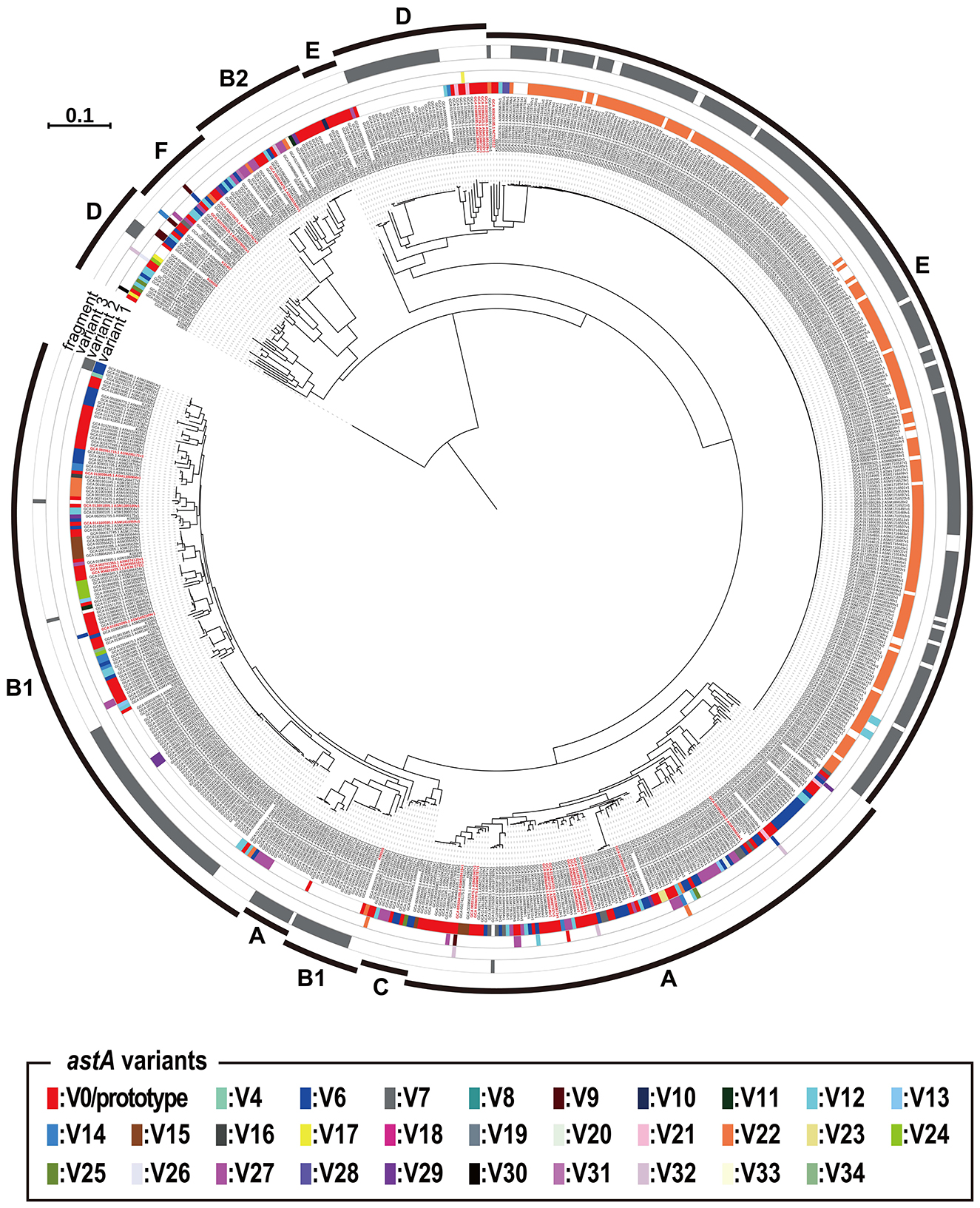
Figure 2. Phylogenetic view of the 720 astA-positive E. coli strains that have been genome sequenced and the distribution of the 35 astA variants and the astA fragment of in these strains. The types of variants carried by each strain are indicated as variant 1, variant 2, and variant 3. For example, strains carrying three types such as V0, V3, and V5 have each type shown under variant 1, 2, and 3, respectively. Strains that possess short astA gene fragments, which are insufficient to determine the variant type, are indicated in gray as “fragment”. The strains possessing multiple copies of the V0/prototype astA gene are indicated in red.
Correlation between the type and copy number of the astA gene and the severity of diarrhea
We investigated the relationship between clinical symptoms and astA gene variants in 30 Escherichia coli isolates collected in Kagoshima Prefecture that were subjected to whole-genome sequencing in this study. Detailed epidemiological data were available for 14 of the 30 patients. However, isolates of Campylobacter jejuni or norovirus were also detected in 8 of these cases, suggesting that E. coli was not the primary causative agent; thus, these cases were excluded from further analysis. In the remaining 6 cases, only astA-positive E. coli strains were isolated. Among these, five isolates—excluding strain K9291—harbored intact astA variants (V0/prototype or V33), while K9291 carried the inactive V15 variant. Furthermore, a virulence factor search against the VFDB database revealed that only strain K12343 did not possess any potential virulence factors other than the astA gene. The remaining strains carried additional virulence factors, including senB (encoding enterotoxin), the cfa operon (encoding CFA/I fimbriae), or genes encoding effector proteins (EspL, EspR, EspX, EspY) secreted via the locus of enterocyte effacement-encoded type III secretion system (LEE-T3SS). However, none of the five strains possessed the LEE region itself in their genomes, suggesting that the LEE-T3SS is absent and therefore the effector proteins are unlikely to be secreted (Supplementary Table 6).
Discussion
The results of our analyses of two sets of E. coli genomes indicate that the frequency of astA-positive E. coli strains is about 7% both in the 2,726 strains isolated from children with diarrhea in Kagoshima, Japan, and the 9,065 finished E. coli genomes obtained from NCBI, most of which were the genomes of non-Japanese strains. In previous studies, the detection rate of the astA gene in animal-, healthy human-, and diarrheal patient-derived E. coli strains was 20.7–86.8%, 2.4–20.5%, and 4.8–11.2%, respectively, although the number of samples and detection methods were different between the studies (Fujihara et al., 2009; Wang et al., 2017; Sukkua et al., 2017; Awad et al., 2020). Thus, the detection rate in the Kagoshima strains (diarrheal patient-derived E. coli strains) was in a range similar to those of the previous studies.
Our analysis of the astA genes identified in this study added 27 novel sequence variants of astA (V8–V34) to the previous list of astA sequences (V0/prototype and variants V1–V7), revealing the notable sequence diversity of astA (Figure 1B). Importantly, four of the 35 types of astA have been inactivated by premature stop codons or mutations in start codon (Figure 1B). In particular, one of the four variants was the most prevalent astA type, V22, which was found in 217 strains out of the 720 strains analyzed (Table 1). Moreover, astA fragments of various lengths were found in as many as 350 strains. These findings indicate the need of distinguishing strains harboring functional and non-functional astA genes when considering the contribution of EAST1 as a virulence factor. As PCR protocols currently used cannot distinguish them (Yamamoto and Echeverria, 1996; Yatsuyanagi et al., 2002), novel methods that specifically detect functional astA genes detect need to be developed.
At the amino-acid sequence level, 23 types of EAST1 (EAST1_1 to EAST1_23) were identified. EAST1_1 encoded by V0/prototype (second most prevalent) and seven variant astA genes were most predominant. Most of other EAST1 types contained one or two amino-acid substitutions. However, in four EAST1 types, substitutions occurred at one or two cysteine residues which may be important for the function as a heat-stable enterotoxin. The functions of these EAST1 types as well as other EAST1 types containing any amino-acid substitution(s) also need to be examined to understand the contribution of EAST1 as a virulence factor.
There are several studies showing conflicting results regarding the correlation between the copy number of the astA gene and toxicity (McVeigh et al., 2000; Ruan et al., 2012), but a clear conclusion has yet to be established. Although the correlation between the copy number of the astA gene and toxicity remains unclear, it is well established that increases in the copy number of virulence-related genes can enhance pathogenicity in other bacterial species. In Vibrio cholerae, the ctx operon encoding cholera toxin forms tandem repeats through gene duplication, and strains with higher numbers of repeats exhibit increased virulence (Mekalanos, 1983). Additionally, in Yersinia enterocolitica, it is known that pathogenicity is enhanced during infection by increasing the copy number of a plasmid encoding a type III secretion system (Wang et al., 2016). Although the further functional analyses are required to understand a potential of virulence of the astA gene, if the copy number of astA gene correlates positively with virulence, then isolates carrying multiple copies of V0/prototype are of particular importance because they included strains carrying more than five copies (up to 11 copies) (Table 2).
The emergence of these strains carrying multiple copies of V0/prototype astA gene is apparently linked to the fact that most of the V0/prototype are associated with intact IS1414 (Supplementary Table 2), and thus, they can be amplified on the genome upon the transposition of IS1414. It should also be emphasized that all types of astA genes are associated with IS1414, but the IS1414 elements associated with the astA genes other than V0/prototype and two minor variants (V30 and V31) are currently inactive due to various deletions or the mutations in their transpose genes. A notable number of prototype-bearing strains (43/146 strains) possessed multiple copies (two to 11 copies) of this type of astA gene, indicating that the amplification has predominantly occurred in the prototype, which was driven by IS1414 amplification. However, given that the IS1414 associated with V30 and V31 also remain structurally intact, it is plausible that similar amplification events may occur in these variants in the future. Frequent structural alterations of IS1414, such as deletion and point mutations, are likely responsible for the generation of astA gene fragments of different lengths, found in as many as 350 strains.
Since all astA variants are encoded within IS1414, it is suggested that IS1414 may be involved in the wide distribution of astA genes in almost all the E. coli lineages (Figure 2). It also resulted in the variable genomic locations of astA, either or both of chromosome and plasmids. As the IS1414-bearing plasmids are large plasmids, many of them are probably transmissible (or were transmissible before). Thus, these plasmids also likely contributed to the spread of astA.
Based on the analysis of six cases in which only E. coli strains harboring the astA gene were isolated, we found that strain K12343, which carries an intact V33 variant, did not possess any additional virulence-associated genes. This suggests the possibility that EAST1 may be directly involved in the diarrheal symptom in this case. In contrast, the four strains (K9228, K9910, K11627, and K12196) carrying the V0/prototype type also harbored other virulence-associated factors, making it difficult to conclude that the V0/prototype type alone was directly responsible for the diarrheal symptoms.
In conclusion, our screening of two large E. coli strain sets, followed by detailed analyses of the astA gene and astA-positive strains, revealed several important findings: (i) notable sequence diversity in astA gene and its gene product, EAST1; (ii) the presence of several non-functional astA variants; (iii) widespread distribution of astA gene fragments; (iv) a strong association between the V0/prototype astA genes and intact IS1414, which have led to amplification of this type of astA gene in some strains; and (v) broad dissemination of the astA gene in almost all the E. coli lineages, likely driven by IS1414-mediated transposition. Furthermore, since the IS1414 associated with variants V30 and V31 also remain structurally intact, it is plausible that similar amplification events could occur in these variants in the future. These findings will be an important basis to investigate the virulence of astA-positive strains and the role of EAST1 as a virulence factor.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
TO: Writing – original draft, Investigation, Writing – review & editing, Visualization, Funding acquisition, Validation, Data curation, Formal analysis, Project administration, Conceptualization. SA: Writing – review & editing, Validation, Visualization. KL: Writing – review & editing, Validation, Visualization. YG: Visualization, Investigation, Validation, Writing – review & editing. AK: Visualization, Validation, Writing – review & editing. NI: Resources, Writing – review & editing. YH-K: Validation, Funding acquisition, Writing – review & editing, Visualization. SI: Validation, Writing – review & editing, Visualization. TH: Visualization, Writing – review & editing, Data curation, Validation. JN: Writing – review & editing, Resources, Funding acquisition, Visualization, Validation.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by JSPS KAKENHI (Grant Numbers 17K10118 to JN) and a Health Labour Sciences Research Grant (21KA0701, 24KA0501, and 24KA1002).
Acknowledgments
The authors thank K. Saito and F. Funakura for their technical assistance. We would like to thank Editage (www.editage.jp) for English Language Editing.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Gen AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1635769/full#supplementary-material
References
Awad, W. S., El-Sayed, A. A., Mohammed, F. F., Bakry, N. M., Abdou, N. M. I., and Kamel, M. S. (2020). Molecular characterization of pathogenic Escherichia coli isolated from diarrheic and in-contact cattle and buffalo calves. Trop. Anim. Health Prod. 52, 3173–3185. doi: 10.1007/s11250-020-02343-1
Beutin, L., Krüger, U., Krause, G., Miko, A., Martin, A., and Strauch, E. (2008). Evaluation of major types of Shiga toxin 2E-producing Escherichia coli bacteria present in food, pigs, and the environment as potential pathogens for humans. Appl. Environ. Microbiol. 74, 4806–4816. doi: 10.1128/AEM.00623-08
Dubreuil, J. D. (2019). EAST1 toxin: an enigmatic molecule associated with sporadic episodes of diarrhea in humans and animals. J. Microbiol. 57, 541–549. doi: 10.1007/s12275-019-8651-4
Fujihara, S., Arikawa, K., Aota, T., Tanaka, H., Nakamura, H., Wada, T., et al. (2009). Prevalence and properties of diarrheagenic Escherichia coli among healthy individuals in Osaka City, Japan. Jpn. J. Infect. Dis. 62, 318–323. doi: 10.7883/yoken.JJID.2009.318
Joensen, K. G., Tetzschner, A. M., Iguchi, A., Aarestrup, F. M., and Scheutz, F. (2015). Rapid and easy in silico serotyping of Escherichia coli isolates by use of whole-genome sequencing data. J. Clin. Microbiol. 53, 2410–2426. doi: 10.1128/JCM.00008-15
Jolley, K. A., Bray, J. E., and Maiden, M. C. J. (2018). Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 3:124. doi: 10.12688/wellcomeopenres.14826.1
Kashima, K., Sato, M., Osaka, Y., Sakakida, N., Kando, S., Ohtsuka, K., et al. (2021). An outbreak of food poisoning due to Escherichia coli serotype O7:H4 carrying astA for enteroaggregative E. coli heat-stable enterotoxin1 (EAST1). Epidemiol. Infect. 149:e244. doi: 10.1017/S0950268821002338
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Letunic, I., and Bork, P. (2016). Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44, W242–W245. doi: 10.1093/nar/gkw290
Maluta, R. P., Leite, J. L., Rojas, T. C. G., Scaletsky, I. C. A., Guastalli, E. A. L., Ramos, M. C., et al. (2017). Variants of astA gene among extra-intestinal Escherichia coli of human and avian origin. FEMS Microbiol. Lett. 364. doi: 10.1093/femsle/fnw285
McVeigh, A., Fasano, A., Scott, D. A., Jelacic, S., Moseley, S. L., Robertson, D. C., et al. (2000). IS1414, an Escherichia coli insertion sequence with a heat-stable enterotoxin gene embedded in a transposase-like gene. Infect. Immun. 68, 5710–5715. doi: 10.1128/IAI.68.10.5710-5715.2000
Mekalanos, J. J. (1983). Duplication and amplification of toxin genes in Vibrio cholerae. Cell 35, 253–263. doi: 10.1016/0092-8674(83)90228-3
Ménard, L. P., and Dubreuil, J. D. (2002). Enteroaggregative Escherichia coli heat-stable enterotoxin 1 (EAST1): a new toxin with an old twist. Crit. Rev. Microbiol. 28, 43–60. doi: 10.1080/1040-840291046687
Ménard, L. P., Lussier, J. G., Lépine, F., Paiva de Sousa, C., and Dubreuil, J. D. (2004). Expression, purification, and biochemical characterization of enteroaggregative Escherichia coli heat-stable enterotoxin 1. Protein Expr. Purif. 33, 223–231. doi: 10.1016/j.pep.2003.09.008
Murigneux, V., Roberts, L. W., Forde, B. M., Phan, M. D., Nhu, N. T. K., Irwin, A. D., et al. (2021). MicroPIPE: validating an end-to-end workflow for high-quality complete bacterial genome construction. BMC Genomics 22:474. doi: 10.1186/s12864-021-07767-z
Nataro, J. P., Deng, Y., Cookson, S., Cravioto, A., Savarino, S. J., Guers, L. D., et al. (1995). Heterogeneity of enteroaggregative Escherichia coli virulence demonstrated in volunteers. J. Infect. Dis. 171, 465–468. doi: 10.1093/infdis/171.2.465
Ooka, T., Seto, K., Kawano, K., Kobayashi, H., Etoh, Y., Ichihara, S., et al. (2012). Clinical significance of Escherichia albertii. Emerg. Infect. Dis. 18, 488–492. doi: 10.3201/eid1803.111401
Pacheco, A. B., Guth, B. E., Soares, K. C., Nishimura, L., de Almeida, D. F., and Ferreira, L. C. (1997). Random amplification of polymorphic DNA reveals serotype-specific clonal clusters among enterotoxigenic Escherichia coli strains isolated from humans. J. Clin. Microbiol. 35, 1521–1525. doi: 10.1128/jcm.35.6.1521-1525.1997
Page, A. J., Cummins, C. A., Hunt, M., Wong, V. K., Reuter, S., Holden, M. T., et al. (2015). Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31, 3691–3693. doi: 10.1093/bioinformatics/btv421
Paiva de Sousa, C., and Dubreuil, J. D. (2001). Distribution and expression of the astA gene (EAST1 toxin) in Escherichia coli and Salmonella. Int. J. Med. Microbiol. 291, 15–20. doi: 10.1078/1438-4221-00097
Paniagua-Contreras, G. L., Hernández-Jaimes, T., Monroy-Pérez, E., Vaca-Paniagua, F., Díaz-Velásquez, C., Uribe-García, A., et al. (2017). Comprehensive expression analysis of pathogenicity genes in uropathogenic Escherichia coli strains. Microb. Pathog. 103, 1–7. doi: 10.1016/j.micpath.2016.12.008
Ruan, X., Crupper, S. S., Schultz, B. D., Robertson, D. C., and Zhang, W. (2012). Escherichia coli expressing EAST1 toxin did not cause an increase of cAMP or cGMP levels in cells, and no diarrhea in 5-day old gnotobiotic pigs. PLoS One 7:e43203. doi: 10.1371/journal.pone.0043203
Savarino, S. J., Fasano, A., Robertson, D. C., and Levine, M. M. (1991). Enteroaggregative Escherichiacoli elaborate a heat-stable enterotoxin demonstrable in an in vitro rabbit intestinal model. J. Clin. Invest. 87, 1450–1455. doi: 10.1172/JCI115151
Savarino, S. J., Fasano, A., Watson, J., Martin, B. M., Levine, M. M., Guandalini, S., et al. (1993). Enteroaggregative Escherichia coli heat-stable enterotoxin 1 represents another subfamily of E. coli heat-stable toxin. Proc. Natl. Acad. Sci. U.S.A. 90, 3093–3097. doi: 10.1073/pnas.90.7.3093
Savarino, S. J., McVeigh, A., Watson, J., Cravioto, A., Molina, J., Echeverria, P., et al. (1996). Enteroaggregative Escherichia coli heat-stable enterotoxin is not restricted to enteroaggregative E. coli. J. Infect. Dis. 173, 1019–1022. doi: 10.1093/infdis/173.4.1019
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Silva, L. E., Souza, T. B., Silva, N. P., and Scaletsky, I. C. (2014). Detection and genetic analysis of the enteroaggregative Escherichia coli heat-stable enterotoxin (EAST1) gene in clinical isolates of enteropathogenic Escherichia coli (EPEC) strains. BMC Microbiol. 14:135. doi: 10.1186/1471-2180-14-135
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Sukkua, K., Manothong, S., and Sukhumungoon, P. (2017). Seroprevalence and molecular epidemiology of EAST1 gene-carrying Escherichia coli from diarrheal patients and raw meats. J. Infect. Dev. Ctries. 11, 220–227. doi: 10.3855/jidc.6865
Uzzau, S., and Fasano, A. (2000). Cross-talk between enteric pathogens and the intestine. Cell. Microbiol. 2, 83–89. doi: 10.1046/j.1462-5822.2000.00041.x
Veilleux, S., Holt, N., Schultz, B. D., and Dubreuil, J. D. (2008). Escherichia coli EAST1 toxin toxicity of variants 17-2 and O 42. Comp. Immunol. Microbiol. Infect. Dis. 31, 567–578. doi: 10.1016/j.cimid.2007.10.003
Wang, H., Avican, K., Fahlgren, A., Erttmann, S. F., Nuss, A. M., Dersch, P., et al. (2016). Increased plasmid copy number is essential for Yersinia T3SS function and virulence. Science 353, 492–495. doi: 10.1126/science.aaf7501
Wang, L., Zhang, S., Zheng, D., Fujihara, S., Wakabayashi, A., Okahata, K., et al. (2017). Prevalence of diarrheagenic Escherichia coli in foods and fecal specimens obtained from cattle, pigs, chickens, asymptomatic carriers, and patients in Osaka and Hyogo, Japan. Jpn. J. Infect. Dis. 70, 464–469. doi: 10.7883/yoken.JJID.2016.486
Waterhouse, A. M., Procter, J. B., Martin, D. M. A., Clamp, M., and Barton, G. J. (2009). Jalview Version 2–a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191. doi: 10.1093/bioinformatics/btp033
Yamamoto, T., and Echeverria, P. (1996). Detection of the enteroaggregative Escherichia coli heat-stable enterotoxin 1 gene sequences in enterotoxigenic E. coli strains pathogenic for humans. Infect. Immun. 64, 1441–1445. doi: 10.1128/iai.64.4.1441-1445.1996
Yamamoto, T., and Taneike, I. (2000). The sequences of enterohemorrhagic Escherichia coli and Yersinia pestis that are homologous to the enteroaggregative E. coli heat-stable enterotoxin gene: cross-species transfer in evolution. FEBS Lett. 472, 22–26. doi: 10.1016/S0014-5793(00)01414-9
Yamamoto, T., Wakisaka, N., Sato, F., and Kato, A. (1997). Comparison of the nucleotide sequence of enteroaggregative Escherichia coli heat-stable enterotoxin 1 genes among diarrhea-associated Escherichia coli. FEMS Microbiol. Lett. 147, 89–95. doi: 10.1111/j.1574-6968.1997.tb10225.x
Yatsuyanagi, J., Saito, S., Sato, H., Miyajima, Y., Amano, K., and Enomoto, K. (2002). Characterization of enteropathogenic and enteroaggregative Escherichia coli isolated from diarrheal outbreaks. J. Clin. Microbiol. 40, 294–297. doi: 10.1128/JCM.40.1.294-297.2002
Zhou, Z., Alikhan, N. F., Mohamed, K., the Agama Study Group, and Achtman, M. (2015). The User's Guide to Comparative Genomics with EnteroBase. Three Case Studies: Micro-Clades Within Salmonella enterica Serovar Agama, Ancient and Modern Populations of Yersinia pestis, and Core Genomic Diversity of All Escherichia. bioRxiv [Preprint]. doi: 10.1101/613554
Zhou, Z., Ogasawara, J., Nishikawa, Y., Seto, Y., Helander, A., Hase, A., et al. (2002). An outbreak of gastroenteritis in Osaka, Japan due to Escherichia coli serogroup O166:H15 that had a coding gene for enteroaggregative E. coli heat-stable enterotoxin 1 (EAST1). Epidemiol. Infect. 128, 363–371. doi: 10.1017/S0950268802006994
Keywords: Escherichia coli, enterotoxin EAST1, astA variant, IS1414, genotyping, pathogenesis
Citation: Ooka T, Arai S, Lee K, Gotoh Y, Kubomura A, Imuta N, Hara-Kudo Y, Iyoda S, Hayashi T and Nishi J (2025) Prevalence, sequence diversity, and amplification of an IS-associated enterotoxin gene, astA, in Escherichia coli. Front. Microbiol. 16:1635769. doi: 10.3389/fmicb.2025.1635769
Received: 28 May 2025; Accepted: 03 October 2025;
Published: 22 October 2025.
Edited by:
Ilya V. Kublanov, Hebrew University of Jerusalem, IsraelReviewed by:
Tales Fernando da Silva, Universidade Federal de Minas Gerais, BrazilHege Smith Tunsjø, Oslo Metropolitan University, Norway
Copyright © 2025 Ooka, Arai, Lee, Gotoh, Kubomura, Imuta, Hara-Kudo, Iyoda, Hayashi and Nishi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tadasuke Ooka, dGFvaG9rYTFAbS5rdWZtLmthZ29zaGltYS11LmFjLmpw
 Tadasuke Ooka
Tadasuke Ooka Sakura Arai2
Sakura Arai2 Kenichi Lee
Kenichi Lee Yasuhiro Gotoh
Yasuhiro Gotoh Naoko Imuta
Naoko Imuta Sunao Iyoda
Sunao Iyoda Tetsuya Hayashi
Tetsuya Hayashi