- Institute for Open and Transdisciplinary Research Initiatives (OTRI), The University of Osaka, Osaka, Japan
Alkaliphilic Bacillaceae strains likely utilize a limited number of free H+, producing ATP through an H+-based electrochemical membrane potential more efficiently than neutralophiles do. One possible mechanism responsible for this involves a structure that accumulates H+ through a hydrogen-bonding network formed by water molecules and the acidic, amido-, and hydroxyl- groups of amino acids located at the N-terminal site of membrane-bound cytochromes c, which are specifically found in obligate alkaliphiles. The segment of cytochromes c facilitates the formation of an H+-capacitor at the outer membrane surface. The H+-capacitor would produce an additional unbalanced vertical force to drive F1F0-ATP synthase via H+ concentrations and electrical charges across the membrane. Accumulated H+ ions are transferred from cytochrome c to the H+ influx gate of the a-subunit of F1F0-ATP synthase. However, the relative abundance of protonable basic amino acids at this site is low, suggesting that H+ transfer occurs via a membrane-bound protein containing the DUF2759 domain. This protein exposes basic amino acids that outnumber the deprotonatable acidic amino acids, effectively recruiting H+ from cytochrome c near the H+ influx gate of F1F0-ATP synthase. The disparity in abundance between acidic and basic amino acids within the H+ carrier segment may play a crucial role in determining H+ transfer efficiency. In alkaliphiles, significant gaps in H+ release or acceptance exist between the outer membrane and the intracellular side of F1F0-ATP synthase. This indicates that the hydrophilic segments involved in H+ transfer are specifically designed to enhance the performance of F1F0-ATP synthase. This hypothetical mechanism for the effective transportation of accumulated H+ to the N-terminal region of the cytochrome c amino acid sequence is essential for ATP production in obligate alkaliphilic Bacillaceae. The unique bioenergetic configuration of these alkaliphiles is evident in their high maximum ATP production rates. Maximizing the activity of F1F0-ATP synthase can be achieved through efficient H+ transport and a high transmembrane electrical potential (ΔΨ), particularly in environments where H+ availability is limited.
1 Introduction
Microorganisms thrive in various environments on Earth. This is due to their wide distribution and high adaptability to natural environments. Thus, studying the physiology of microorganisms is key to understanding the relationship between biological systems and their environments. Microorganisms that can adapt to extreme environments, such as low or high temperatures and pH levels, UV radiation, and high salinity, are called extremophiles (Satyanarayana et al., 2005; Rampelotto, 2013; Coker, 2023; Cowan et al., 2024). Studying organisms that live in extreme environments can improve our understanding of the hidden biological and biochemical mechanisms employed by microorganisms that live in normal environments (Goto et al., 2022). For instance, the physiological features observed during environmental adaptation in alkaliphiles may be present to a different extent in neutralophiles. We can gain a more comprehensive understanding of parameters sustaining biological systems by exploring these aspects in the microorganisms.
The mainstream view is that mutations favorable for environmental adaptation occur randomly and rarely at the same time, with adaptation then progressing through natural selection (Orr, 2005). However, microorganisms alter various cellular components and processes to adapt to alkaline environments. Given the phylogenetic proximity and synthesis of similar molecular features of proteins between facultative alkaliphiles and neutrophiles, the alkaliphilicity of facultative alkaliphiles may stem from physiological (e.g., transcriptional) gene regulation and transposable elements (Lebre and Cowan, 2020). Transposable elements are thought to have played an important role in evolution not only in facilitating horizontal gene transfer but also in gene rearrangements within genomes (Takami et al., 2001). Transposable elements in alkaliphilic bacteria may play diverse roles, such as enhancing genetic diversity, facilitating the transmission of resistance genes, regulating gene expression, and acquiring new functions for the corresponding environment (Siguier et al., 2014). This allows microorganisms to adapt and proliferate in alkaline environments. Over a long interaction period with environmental pH changes in a niche where the pH fluctuates between 7 and 10, facultative alkaliphiles with superior gene regulatory functions or post-translational modifications that facilitate adaptation to alkaline environments may have diverged.
Obligate alkaliphiles are often phylogenetically distant from that of neutralophiles, and they produce proteins with specialized molecular features for that support growth under high pH. These features may have originated from transposable elements and been retained through natural selection in persistently alkaline environments. Transposable elements and horizontal gene transfer are thought to contribute to the acquisition and diversification of alkaline-adaptive functions. These mechanisms may explain the coordinated changes in multiple cellular functions observed in alkaliphiles, contrasting with more gradual adaptation in neutralophiles.
Obligately alkaliphilic Bacillaceae within the phylum Bacillota (formerly Firmicutes) and class Bacilli, have been studied as model organisms for investigating bioenergetic adaptations to high-pH environments. On the other hand, other phylogenetically distinct obligately alkaliphilic bacteria also thrive in microbial communities adapted to high-pH serpentinizing systems by employing distinct physiological mechanisms to overcome the same energetic constraints. For example, Alkaliphilus metalliredigens, a strict anaerobe belonging to the phylum Bacillota, class Clostridia, could be thought to produce energy through metal reduction and potentially fermentative pathways (Ye et al., 2004). In contrast, aerobic hydrogen-oxidizing bacteria of the genus Serpentinomonas, classified within the phylum Pseudomonadota (formerly Proteobacteria), class Gammaproteobacteria, could be thought to generate ATP under alkaline conditions through hydrogen oxidation (Suzuki et al., 2014). In both cases, it is plausible that specific mechanisms exist to minimize H+ expenditure. These distinct metabolic strategies underscore the evolutionary adaptability of obligate alkaliphiles and emphasize the value of comparative studies across taxonomic lineages.
A key adaptation is the ability to maintain intracellular pH near neutrality, enabling survival in extreme alkaline conditions. Therefore, maintaining physiological mechanisms that regulate intracellular pH is essential for optimal metabolic activity within cells. Caldalkalibacillus thermarum strain TA2. A1, an alkaliphilic member of the family Bacillaceae, utilizes H+ ions to generate an electrochemical gradient that drives F₁F₀-ATP synthase (Dimroth and Cook, 2004). A Na+ potential could possibly be used across the membrane to drive F1F0-ATP synthase; in reality, alkaliphilic Bacillaceae, use H+ to produce ATP. However, anaerobic alkaliphilic Alkaliphilus spp. and the alkaliphilic, halotolerant cyanobacterium Aphanothece halophytica, use a Na+ potential across their membranes to drive ATP synthase (Hicks et al., 2010; Soontharapirakkul and Incharoensakdi, 2010). The reason why alkaliphilic Bacillaceae strains utilize H+ to drive F1F0-ATP synthase in H+-deficient environments remains unclear. However, using H+ for ATP production may be advantageous in some alkaliphiles (Banciu and Sorokin, 2013). For example, making more efficient use of scarce H+ would likely not require significant alterations to the physiological functions of the organism. Alkaliphilic Bacillaceae strains use the transmembrane Na+ potential (sodium motive force; SMF) for solute transport and flagellar rotation. The Na+/H+ antiporter, Mrp (Sha), plays a critical role in generating an SMF by translating the H+-based electrochemical potential generated by respiration into the transmembrane Na+ potential (Hamamoto et al., 1994; Krulwich et al., 2001). Through this translation, alkaliphilic Bacillaceae can dramatically reduce the frequency of H+ utilization. This key strategy uses limited H+ only to drive F1F0-ATP synthase. At the same time, the exchange of intracellular Na+ for extracellular H+ reduces the intracellular pH. Thus, the Na+/H+ antiporter plays a crucial role in conserving H+ for metabolism and in lowering intracellular pH levels (Krulwich et al., 2001).
In addition to utilizing ions, alkaliphiles protect themselves by displaying acidic substances on their cell surfaces. These negatively charged secondary cell walls repel OH− under alkaline conditions, protecting cells from the environment. For instance, Halalkalibacterium halodurans (formerly Bacillus halodurans) C-125 exhibits teichuronopeptide and teichuronic acid on its cell surface as secondary cell walls (Aono and Horikoshi, 1983; Aono et al., 1993, 1995, 1999), whereas Alkalihalobacillus pseudofirmus OF4 exhibits cell surface layer (S-layer) protein A (SlaA) for this purpose (Gilmore et al., 2000). Although the substances displayed on the cell surface differ by species; they all play a crucial role in protecting cells from the harsh extracellular environment. Additionally, alkaliphilic Bacillaceae strains have been reported to produce acids to avoid direct interaction with harsh pH levels (Mano, 2020). Extracellular acid production mitigates the surrounding environment more effectively when bacterial cells exist as a group or live in a confined space. Thus, for alkaliphilic Bacillaceae strains, protection via the secondary cell wall and mitigation of the harsh environment around cells are important adaptation strategies to alkaline environments.
In addition to the previously described alkaline adaptation strategies, the membrane lipids that protect intracellular spaces differ between alkaliphilic Bacillaceae strains and their neutralophilic counterparts (e.g., Bacillus subtilis). These membrane lipid characteristics may play an important role in conferring the bioenergetic properties required for adaptation to alkaline environments. When alkaliphilic Bacillaceae strains use the respiratory chain to produce ATP by forming a H+-capacitor, the membrane lipid involved acts as a dielectric, providing insulation between the electrode plates of an electrical capacitor. Obligate alkaliphilic strains have 1.5 times more total membrane lipids than neutralophilic B. subtilis does (Clejan et al., 1986). The ratio of neutral to polar lipids is greater than ≥80% in obligate alkaliphiles, compared with the 33–54 and 43% in facultative alkaliphiles and B. subtilis, respectively. These results suggest that obligate alkaliphilic bacteria have fundamentally different physiological mechanisms than facultative alkaliphilic and neutralophilic bacteria do. Obligate and facultative alkaliphiles contain higher ratios of squalene, C40 isoprenoid, and negatively charged cardiolipin than B. subtilis does (Clejan et al., 1986). Cardiolipin may contribute to the negative charge of the membrane surface; however, the functions of squalene and C40 isoprenoid remain unknown.
Microorganisms require H+ to produce ATP. However, alkaliphiles have adapted to survive in conditions with one-thousandth of the H+ concentration found in neutralophilic conditions through use of the multiple physiologically supportive mechanisms described above. In addition to these features, alkaliphiles possess unique mechanisms for managing H+ through respiratory components. Due to the presence of a high transmembrane electrical potential (ΔΨ) across the membrane, the ability to retain H+ on the outer surface of the membrane (Yoshimune et al., 2010), and the H+-capacitor mechanism (Goto et al., 2022), alkaline environments are not harsh for alkaliphiles to survive in. The H+-capacitor mechanism is based on the Asn-rich N-terminal segment, which is specifically observed in the cytochrome c of obligately alkaliphilic Evansella clarkii (H+ carrier segment; Goto et al., 2022). The H+-capacitor function is enhanced under air-limited conditions by increased cytochrome c expression (Goto et al., 2022). This review summarizes and discusses the formation of an H+-capacitor and how H+ are transferred from the outer surface membrane to the intracellular membrane via F1F0-ATP synthase in alkaliphilic Bacillaceae for adapting to alkaline conditions.
2 Taxonomic background of alkaliphilic Bacillaceae
Since Vedder first isolated Alkalihalobacillus alcalophilus (formerly Bacillus alcalophilus) from the feces of healthy individuals in 1934, numerous alkaliphilic Bacillaceae strains have been discovered in various environments, including soil, animal manure, and ocean and lake water. Strains belonging to A. alcalophilus are obligate alkaliphiles, meaning that they cannot grow at a neutral pH. Facultative alkaliphiles, however, can grow at nearly the same intensity in both alkaline and neutral pH environments. The reason why many alkaliphiles have been isolated from normal environments remains unclear. Ammonia is naturally produced from the decomposition of proteins in animal urine. Ammonia could be released during the decomposition of proteins from dead plants and animals. Therefore, localized alkaline environments may be ubiquitous in nature, and alkaliphilic Bacillaceae may originate in these environments and later diffuse into normal environments. Facultative alkaliphiles are more frequently isolated than obligate alkaliphiles are from the normal soil samples used to isolate alkaliphilic Bacillaceae strains (Yumoto et al., 1997). A phylogenetic tree was constructed based on the 16S rRNA gene sequence (Figure 1). Each clade in the phylogeny exhibits characteristics of pH adaptation. For instance, the large clade comprising the genera Halalkalibacterium, Alkalihalobacillus, and Shouchella is facultatively alkaliphilic. In contrast, the clade containing the genera Salisediminibacterium, Salipaludibacillus, and Evansella is obligately alkaliphilic. However, the Alkalibacillus haloalkaliphilus strain isolated from these clades shows a wide growth pH range of 7.5–13. Thus, the strategies used for adapting to high pH levels are likely different in each class of alkaliphiles. Remarkably, we can infer that species exhibiting different growth characteristics for a pH range are not phylogenetically distant. The alkaliphilic species used thus far in bioenergetic studies to investigate alkaline adaptation mechanisms are as follows: Halalkali. halodurans (Hamamoto et al., 1994; Aono et al., 1996), A. alcalophilus (Lewis et al., 1980; Hoffmann and Dimroth, 1991), Alkalihalophilus pseudofirmus (formerly Bacillus pseudofirmus) (Lewis et al., 1980; Guffanti et al., 1986; Guffanti and Krulwich, 1994). Sutcliffeiella cohnii (formerly Bacillus cohnii) (Yumoto et al., 1991, 1993); E. clarkii (formerly Bacillus clarkii) (Ogami et al., 2009; Goto et al., 2022), and Caldalkalibacillus thermarum (formerly thermoalkaliphilic Bacillus sp.) (Meier et al., 2007; McMillan et al., 2009; Ferguson et al., 2016). Most of these strains are facultative alkaliphiles; only two, A. alcalophilus and E. clarkii, are obligate alkaliphiles. While each species employ their own strategy for adapting to high pH levels, each clade of alkaliphiles may contain common strategies to facilitate these adaptations.

Figure 1. Maximum-Likelihood phylogenetic tree derived from the 16S rRNA gene sequences of obligate alkaliphilic Evansella clarkii and other related alkaliphilic and neutralophilic Bacillaceae. The evolutionary history was inferred by using the maximum-likelihood method and General Time Reversible model (Nei and Kumar, 2000). The tree with the highest log likelihood (−10998.89) is shown. The percentage of trees in which the associated taxa clustered together is shown alongside the branches. Initial tree(s) for the heuristic search were automatically obtained by applying the Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the Maximum Composite Likelihood approach and thereafter selecting the topology with a superior log likelihood value. A discrete Gamma distribution was used to model differences in evolutionary rate among sites (five categories [+G, parameter = 0.1885]). The rate variation model allowed for some sites to be evolutionarily invariable ([+I], 34.77% sites). The strains used in physiological studies for alkaline adaptation are indicated in bold text. The neutralophilic or alkaliphilic groups are categorized as follows: FA, facultative alkaliphiles; OB/FA, differ in obligate or facultative alkaliphiles depending on the strain; NA, neutralophilic; W, facultative alkaliphile that grows in a wider range than that of other facultative alkaliphiles; OB, obligate alkaliphiles; OB > FA, most of the species are obligate alkaliphiles, whereas a few strains are facultative alkaliphiles. FA (pH 10 > 7), facultative alkaliphiles, which grow much better in alkaline conditions than in neutralophilic conditions. The bootstrap values (>50%) based on 1,000 replications are shown in the branch node. Paenibacillus polymyxa IAM 13419T was used as an outgroup. Bar, 0.05 substitutions per nucleotide position. Evolutionary analyses were conducted in MEGA11 (Tamura et al., 2021).
3 Metal acquisition in alkaliphilic Bacillaceae
Alkaliphilic Bacillaceae face significant challenges in metal ion utilization for survival. Therefore, these organisms must possess efficient mechanisms for capturing and utilizing scarce metals. In most aqueous environments, ferric iron (Fe3+) tends to precipitate as insoluble Fe(OH)₃, significantly reducing its bioavailability above pH 6. This tendency becomes even worse in alkaline conditions. The concentration of iron available to microorganisms at pH 10 is estimated to be approximately 10−23 M (Drechsel and Jung, 1998), which is substantially lower than the 10−18 M typically found at pH 7 (McMillan et al., 2010). Limited iron availability impairs electron transfer efficiency, as respiratory chain components such as cytochromes and copper-dependent oxidases require precise metallation with Fe2+ and Cu2+ at the membrane interface, mediated by dedicated transport systems.
Although Fe2+ remains relatively soluble, it is highly susceptible to oxidation. In oxidative environments, Fe2+ is rapidly converted to Fe3+, further limiting its availability to microorganisms. If Fe3+ accumulates, it can lead to the production of toxic reactive oxygen species, such as oxygen free radicals. To circumvent this issue, alkaliphilic bacteria may adopt strategies to suppress oxidative conditions, thereby avoiding direct interactions with Fe3+. Alkaliphilic Bacillaceae strains encounter a shortage of Fe2+ and oxidative toxicity resulting from Fe3+.
To prevent iron deficiency and mitigate the toxicity of Fe3+, microorganisms secrete small molecules known as siderophores, that strongly bind to Fe3+ and facilitate its transport (Drechsel and Jung, 1998; Andrews et al., 2003). Approximately 500 types of siderophores have currently been reported, and in addition to iron ions, they can bind to other metal ions (e.g., Zn, Cu, Mo) (Ahmed and Holmström, 2014). Structural analyses of novel siderophores from alkaliphilic Bacillaceae, such as Caldalkalibacillus thermarum, have been documented (McMillan et al., 2010).
The genome of A. pseudofirmus OF4 reveals a six-gene cluster (asbABCDEF) responsible for synthesizing a catecholate siderophore (Janto et al., 2011). This siderophore is similar to Peribactin produced by the Bacillus cereus group. Adjacent to this gene cluster are four additional genes (FhuGB and FhuRD) that are predicted to encode an ABC-type siderophore transporter and its regulator, which is one of six ABC transporters anticipated to transport ferric siderophores. The genome also contains genes for two siderophore ferric reductases necessary for releasing iron from siderophore complexes through reduction. One reductase is located near a ferric dicitrate ABC uptake system, whereas the second reductase shows significant homology with proteins in H. halodurans and Caldalkalibacillus thermarum.
Another potential strategy used by alkaliphilic bacteria to prevent iron deficiency and mitigate oxidative conditions is the retention of metals by acidic compounds on the cell surface, such as polyglutamate capsules, teichuronopeptides, and teichuronic acid, which serve as secondary cell walls (Aono and Horikoshi, 1983; Aono et al., 1993, 1995, 1999), as well as S-layer protein A (SlaA) (Gilmore et al., 2000). Extracellular acidic polymers can accumulate deficient iron by adsorbing iron oxides, thereby reducing the direct toxic effects of Fe3+ on cells. These polymers may also alter the reactivity of Fe3+ by adjusting the surrounding pH and concentrations of other chemicals.
In some cases, alkaliphilic Bacillaceae species, such as A. pseudofirmus, exhibit metal ion-reducing abilities (Ma et al., 2012). Additionally, other Bacillaceae such as S. cohnii (Aino et al., 2018) and Halalkalibacter hemicellulosilyticus (Lopes et al., 2021) may also possess the ability to reduce indigo. The extracellular electron transfer capabilities of these organisms can reduce Fe2+, thereby detoxifying reactive oxygen species and enhancing the utilization of iron for their metabolic processes, particularly within their respiratory systems.
4 Bioenergetics in alkaliphilic Bacillaceae
According to Mitchell’s chemiosmotic theory (Mitchell, 1961), the H+ motive force (Δp) that drives F1F0-ATP synthase consists of the electrical potential difference (Δψ) across the membrane (ψin – ψout) and the transmembrane pH gradient (ΔpH: pHin – pHout).
where R = gas constant (8.315 J/K/mol), T = absolute temperature, and F = Faraday constant (96.485 kJ/mol/V). The constant holds for charge z = +1 |e| (i.e., H+).
Both the bulk-based bioenergetic parameters and growth rate of the facultatively alkaliphilic A. pseudofirmus OF4 on malate-containing medium were estimated at different culture pH levels under pH-controlled culture conditions (Sturr et al., 1994). The strain exhibited a specific growth rate and Δp potential of 1.10 h−1 and −26 mV, respectively, at pH 10.6. At pH 7.5, the specific growth rate and Δp were 0.77 h−1 and −140 mV, respectively. These results suggested that the bulk-based Δp did not reflect a real bioenergetic parameter. A similar observation was made in an experiment using right-side-out membrane vesicles (Guffanti and Krulwich, 1994). On ascorbate plus phenazine methosulfate energized membrane vesicles prepared from cells of the OF4 strain grown at pH 10.5, the Δp was measured at −36 mV (including bulk base ΔpH), and ATP production was 2.6 nmoles • mg protein−1 • 30 min−1. At pH 7.5, the energized membrane vesicles of cells generated a Δp and ATP production of −166 mV (including bulk base ΔpH) and 1.4 nmoles • mg protein−1 • 30 min−1, respectively. In addition, an artificially imposed K+ diffusion potential equivalent to the respiration-derived Δψ failed to synthesize ATP, whereas α-aminoisobutyric acid (AIB) uptake occurred. These results suggest that ATP production can be realized by the H+-based Δψ produced by the respiratory chain. If H+ are abundant in the extracellular space, ATP could be produced by the imposed K+-based diffusion potential. At the same time, alkaliphilic bacteria could transport the limited H+ translocated by the respiratory chain to the F1F0-ATPase in a unique way. Therefore, Guffanti and Krulwich (1994) hypothesized that H+ translocated across the membrane via respiratory complexes are directly transferred to the H+ influx gate of F1F0-ATP synthase.
This review discusses the H+-capacitor attributed to the Δψ and H+-transferable amino acids associated with water molecules. The H+-capacitor concept was first reported by Lee in 2012 (Lee, 2012, 2021). Based on the protonic capacitor concept, the ideal transmembrane electrostatically localized H+ (TELP) concentration [HL+]0 (mol/L = M) is given by the following equation:
where C/S is the specific membrane capacitance per unit surface area: C the capacitance in coul (electric charge)/V (voltage) (i.e., farads); S the area of the plates (m2); l the thickness (m) of the layer of transmembrane-electrostatically localized H+; and F the Faraday constant (96,486 coul/mol). In this review, configuration of the bioenergetic mechanisms of obligate alkaliphilic E. clarkii in alkaline conditions is mainly forced, and the system can be considered to show an enhanced version of the H+-capacitor compared with that of other organisms. Facultatively alkaliphilic Bacillaceae strains, while lacking the corresponding configuration of obligate alkaliphiles, can thrive in environments similar to those wherein obligate alkaliphiles grow. The TELP theory may explain the growth of facultative alkaliphilic strains in alkaline conditions.
5 Structure and function of cytochromes c
Cytochrome c is an electron-transfer protein that acts as a component of the respiratory chain by transferring electrons from the cytochrome bc1 complex (complex III) to cytochrome c oxidase. Compared with those in gram-positive bacteria, cytochromes c in gram-negative bacteria have been intensively studied (Yamanaka, 1992). One reason for this disparity is that cytochrome c in gram-negative bacteria is soluble, whereas in gram-positive bacteria, it is membrane-bound (Sone and Toh, 1994). Cytochromes c in gram-negative bacteria are thought to exist in the periplasmic space. However, cytochrome c in gram-positive bacteria is membrane-bound and has the same function as that of cytochrome c in gram-negative bacteria. As cytochrome c is membrane-bound in gram-positive bacteria, its movement is restricted; thus, fulfilling physiological functions that require accumulated cytochrome c is possible. Below, we discuss the molecular adaptation strategies of cytochromes c in alkaline environments by comparing them in neutralophilic, facultative, and obligate alkaliphilic bacteria.
5.1 Neutralophilic cytochromes c
Few examples of studies on membrane-bound cytochrome c in Bacillaceae can be found; a typical example from neutralophilic bacteria is that of the B. subtilis cytochrome c. As B. subtilis exhibits strong protease activity, its cytochrome c has been reported as a soluble protein (Miki and Okunuki, 1969a,b). Two different membrane anchor types of cytochrome c (c-550 and c-551) from B. subtilis 168 were purified and characterized. Cytochrome c-551, which is equivalent to the cytochrome c-550 of E. clarkii discussed below, is composed of 112 amino acid residues, including a signal peptide sequence of 20 residues (Table 1; Bengtsson et al., 1999). The processed form, which is dissociated from the signal peptide, consists of 92 amino acids, including 18 acidic and 14 basic amino acids, and binds to the membrane via a diacylglyceryl moiety (the acyl chain length of which is C15) through its N-terminal cysteine. The redox potential of this cytochrome c was reported to be higher than +100 mV. The phylogenetic position of the amino acid sequence of B. subtilis cytochrome c-551 was found to be similar to that of Bacillus licheniformis, which is distantly related to the clade of obligate alkaliphilic strains (Figure 2). The other cytochrome c, cytochrome c-550, consists of approximately 120 amino acids, including a single α-helical transmembrane segment of approximately 30 hydrophobic amino acids, which serves as a membrane anchor (von Wachenfeldt and Hederstedt, 1993; David et al., 2000). These cytochromes c may likely have a function in electron transfer reactions between the b6c complex and cytochrome c oxidase (cytochrome caa3). An almost pure b6c complex associated with cytochromes c-550 and c-551 has been isolated previously (Picón Garrido et al., 2022). The estimated molecular masses of cytochromes c-550 and c-551 from B. subtilis were not reported via gel filtration. However, cytochrome c-551 from Geobacillus sp. PS3, which is equivalent to the cytochrome c-551 found in B. subtilis, was reported as a trimer via gel filtration (Table 1; Sone et al., 1989; Fujiwara et al., 1993). According to the amino acid sequence alignment of membrane-bound cytochromes c, B. subtilis cytochrome c-551, Geobacillus sp. PS3 cytochrome c-551, and B. licheniformis cytochrome c are classified as neutralophilic. The B. subtilis cytochrome c-551 is in the same clade as B. licheniformis cytochrome c, whereas Geobacillus sp. PS3 cytochrome c-551 is in the same clade with the facultatively alkaliphilic Sutcliffiella horikoshii cytochrome c. This may be reflected in the phylogenetic distance observed between the genera Geobacillus and Bacillus.
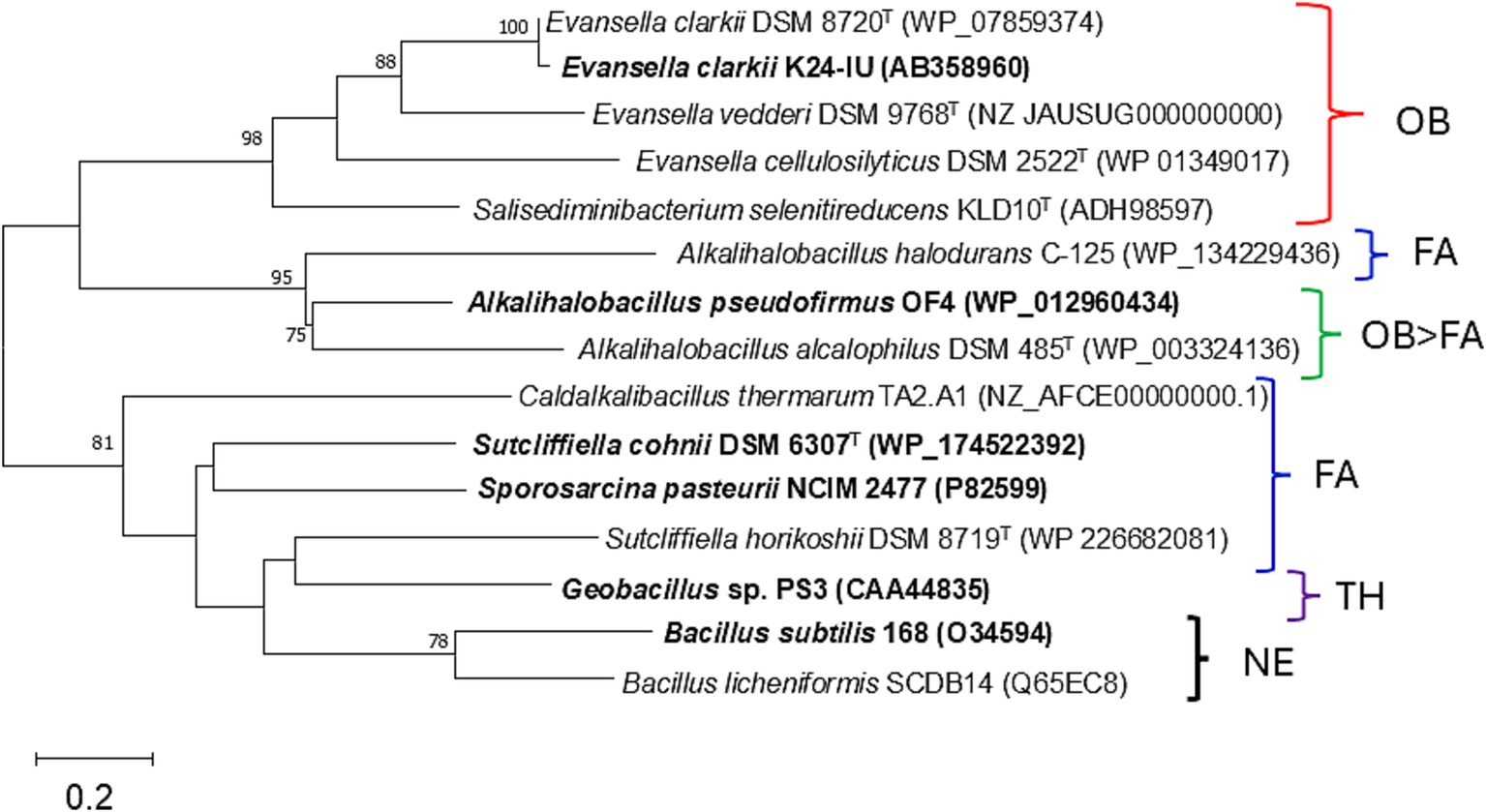
Figure 2. Maximum-likelihood phylogenetic tree of the membrane bound cytochromes c from Evansella clarkii and other alkaliphilic and neutralophilic Bacillaceae. Evolutionary history was inferred by using the Whelan and Goldman model (Whelan and Goldman, 2001). The tree with the highest log likelihood (−2807.82) is shown. The percentage of trees in which the associated taxa clustered together is shown alongside the branches. Initial tree(s) for the heuristic search were automatically obtained by applying the Neighbor-Join and BioNJ algorithms to a matrix of pairwise distances estimated using the JTT model and then selecting the topology with a superior log likelihood value. A discrete Gamma distribution was used to model differences in evolutionary rate among sites (five categories [+G, parameter = 4.1748]). The rate variation model allowed for some sites to be evolutionarily invariable ([+I], 8.25% sites). Strains or species for which protein studies have actually been conducted are indicated in bold. Bar, 0.2 substitutions per nucleotide position. Evolutionary analyses were conducted in MEGA11 (Tamura et al., 2021). The neutralophilic or alkalophilic strains were categorized as follows: OB, obligate alkaliphiles; FA, facultative alkaliphiles; OB > FA, most of the strains are obligate alkaliphiles, whereas a few are facultative alkaliphiles; NA, neutralophilic; TH, thermophile.
5.2 Facultative alkaliphilic cytochromes c
The abundance of cytochrome c was found to be higher when grown at pH 10 than at pH 7 in reduced minus oxidized forms of the membrane of the facultatively alkaliphilic S. cohnii YN-2000 (Yumoto et al., 1991). The elution profile obtained after anion-exchange chromatography of the loaded crude membrane extract showed that an increase in cytochrome c-553 was the main reason for the observed increase in cytochrome c abundance. Therefore, cytochrome c-553 is likely responsible for adaptation to high pH levels in the strain YN-2000. Cytochrome c-553 has a molecular mass of 10.5 kDa (according to SDS-PAGE results), and the native molecular mass determined using gel filtration showed that it forms a tetramer. The phylogenetic position of the amino acid sequence of S. cohnii DSM 6307T was similar to that of other facultative alkaliphilic species (Figure 2). It consists of 93 amino acids, including 14 acidic and 7 basic amino acids (Table 1; Figure 3). The redox potential was +87 mV in the pH range of 6–8. Cytochrome c-553 reacted strongly with its physiological electron acceptor, cytochrome c oxidase (cytochrome aco3), in the presence of poly-L-lysine. This suggests that positively charged molecules are necessary for effective binding between cytochromes c-553 and aco3 (Yumoto et al., 1993).

Figure 3. Amino acid sequence alignment of the membrane-bound cytochrome c-550 from Evansella clarkii and cytochromes c from other alkaliphilic and neutralophilic Bacillaceae. Obligate and facultative alkaliphilic strains are indicated by red and black, respectively, whereas neutralophilic strains are shown by blue. Asn (N)-rich segment: the Asn (N)4–Asn (N)27 sequence in the processed protein base (Asn [N]21–Asn [N]44 in the entire sequence base) in the E. clarkii cytochromes c and corresponding positions in other strains are indicated by a red box. The equivalent sequences were observed in obligate alkaliphilic strains ([1]–[7]). The N-terminal amino acid residue in processed cytochrome c (Cys [C]) is indicated by a blue marker. Amino acids representing the heme-binding site (C) and axical ligands (H and M) are indicated by a light blue marker. Acidic (D, E) and basic (H, K and R) residues are indicated by blue and red letters, respectively; amino acids representing amido (N, Q) or hydroxyl (S, T) group side-chains are indicated by black letters; and hydrophobic amino acids are shown in pale green. Sequences showing a trend from neutral to alkaliphilic (D + E) + (N + Q + T + S) > (H + K + R) are indicated by blue boxes. While the A. pseudofirmus OF4 is a facultative alkaliphile, most strains belonging to A. pseudofirmus are obligate alkaliphiles (Nielsen et al., 1995), and the cytochrome c sequence presented is classified as obligate alkaliphile.
Both the biochemical properties and three-dimensional structure of cytochrome c-553 from Sporosarcina pasteurii (formerly Bacillus pasteurii) has been reported previously (Benini et al., 2000). Cytochrome c-553 consists of 92 amino acids in the mature protein, with an N-terminal cysteine. Therefore, it is thought to consist of 92 amino acids, including 13 acidic and 6 basic amino acids. This cytochrome also has a low redox potential of +47 mV (Benini et al., 1998, 2000). Its structural features indicate that most of the charges are localized to the opposite side of the heme edge suggesting that most of the charges are localized between the protein molecule and outer membrane surface. This may be related to the accumulation and transfer of H+ localized between the cytochrome c molecule and outer membrane surface. Analysis of physicochemical parameters showed that the solvent accessibility of heme c correlated with entropy (ΔS: disorder) in the relationship between enthalpy (ΔH: heat content) and ΔS in the Gibbs free energy equation (ΔG = ΔH – TΔS; T: absolute temperature [in Kelvin]). This suggests a direct relationship between the main determinant of the electrochemical reduction potential (ΔS) and a structural parameter (heme c solvent exposure). The low midpoint redox potential of cytochrome c-553 from S. pasteurii could be explained by the reduction of ΔS via water-molecule extrusion, which increases solvent accessibility. The water molecules are produced from the hydration shell in the cytochrome molecule during heme c reduction.
In conclusion, the particular characteristics of cytochromes c from facultative alkaliphiles include a low redox potential (<+90 mV) and lower content of basic amino acids (<8 residues) in the mature protein. Phylogenetic positions of the cytochromes c of S. cohnii and S. pasteurii are relatively close to that of B. subtilis cytochrome c-551 when compared with those of the other alkaliphilic cytochromes c. As the S. cohnii cytochrome c is relatively similar to that of B. subtilis, a higher amount of cytochrome c-553 may be expressed for adaptation under high pH conditions. Cytochrome c expression intensity may depend on the requirement of an H+-retaining function during environmental adaptation.
5.3 Cytochromes c from obligate alkaliphiles
The first cytochrome c identified from an obligate alkaliphilic Bacillaceae strain was cytochrome c-552 from A. pseudofirmus RAB (formerly Bacillus firmus RAB) (Davidson et al., 1988), which was purified as a soluble protein and had a molecular mass of 16.5 kDa. This size is larger than expected for common Bacillaceae based on its corresponding gene sequence length. The mean redox potential of cytochrome c-552 is +66 mV at pH 7, which decreases at pH > 8.3 at a rate of −54 mV/pH. For example, its redox potential at pH 9.25 is +8 mV. According to resonance Raman spectroscopy, this change in midpoint redox potential is attributed to a switch in the sixth ligand of heme c from methionine to histidine when oxidized cytochrome c is reduced (Larsen et al., 1990). Although it is unclear whether the A. pseudofirmus RAB cytochrome c-552 is equivalent to the cytochrome c of A. pseudofirmus OF4, the methionine could not have been replaced by histidine in the fifth ligand of heme c based on the amino acid sequence (Figure 3). The A. pseudofirmus RAB cytochrome c-552 is autoxidizable and has been isolated in a soluble form; these characteristics suggest that this protein is partially digested by intrinsic proteases during purification.
As cytochromes c in Bacillaceae have been shown to be sensitive to intrinsic protease activity, an attempt was made to isolate obligate alkaliphilic Bacillaceae strains that exhibit weak protease activity. Consequently, cytochrome c-550 was purified and characterized from an E. clarkii K24-1 U (Ogami et al., 2009). The purified cytochrome c-550 exhibited dimeric (23 kDa) and tetrameric (40 kDa) states in blue native PAGE and gel filtration results, respectively. Cytochrome c-550 binds to fatty acids with carbon lengths of C15–C17 via glycerol-Cys18 as a membrane anchor. The molecular species of the fatty acid corresponded to its membrane fatty acid, suggesting that when cytochrome c-550 expression fluctuates with culture conditions, the fatty acid composition of the membrane is not strongly affected. The redox potential of cytochrome c-550 determined via redox titration was +83 mV (pH 7–9), whereas that determined through cyclic voltametric measurements was +7 mV (pH 6.8). A possible explanation for this difference is the reorientation of the protein backbone that likely occurs due to the effect that the electric field of the electrode has on cytochrome c-550. Electric field-induced redox potential shifts have been reported to occur due to Coulombic forces from an electrode (Rivas et al., 2005). The phylogenetic position of cytochromes c from obligate alkaliphilic bacteria is distant from that of neutralophilic B. subtilis cytochromes c-551 when compared with those from facultative alkaliphilic bacteria (Figure 2). This reflects that cytochromes c in obligate alkaliphiles have evolved to be even more specialized for functioning under alkaline environments. Two clades comprising the genera Evancella and Alkalihalobacillus are present in the cytochrome c phylogeny of obligate alkaliphiles. In this sense, cytochromes c of the A. pseudofirmus OF4 and E. clarkii K24-1 U belong to different clades.
In conclusion, the particular characteristics of cytochromes c from obligate alkaliphiles include a low redox potential (< +90 mV) and lower content of basic amino acids (< 3 residues) in the mature protein. Phylogenetic positions of the cytochromes c of A. pseudofirmus OF4 and E. clarkii are more distant from that of B. subtilis cytochrome c-551 when compared with those of the other alkaliphilic cytochromes c. Since the cytochrome c of S. cohnii is relatively similar to that of B. subtilis, compared to A. pseudofirmus OF4 and E. clarkii, a higher level of cytochrome c expression may contribute to adaptation under high-pH conditions. The intensity of cytochrome c expression may depend on the necessity of the H+-retaining function.
6 Comparison of amino acid sequences in cytochrome c and F1F0-ATP synthase
6.1 Cytochrome c
The amino acid sequence of Asn4-Asn37 in the mature protein base in E. clarkii cytochromes c is quite unique among obligate alkaliphiles compared with those of facultative and neutralophilic membrane-bound cytochromes c (Figure 3; Table 1). The Asn4-Asn37 sequence is located between the first N-terminal α-helix in the main body of the protein and the membrane anchor, based on the structural data of S. pasteurii (Benini et al., 2000). Between Asn4-Asn37, 10 acidic amino acids (glutamic acid [Glu (E)] or aspartic acid [Asp (D)]), nine amido group side-chains possessing asparagine [Asn (N)] and glutamine [Gln (Q)], and one hydroxyl group side-chain (possessing threonine [Thr (T)] and serine [Ser (S)]) can be found in E. clarkii cytochrome c. Glu (E) and Asp (D) are acidic amino acids with a H+-transferable carboxyl group on their side-chains (Voet et al., 2016). Asn (N), Gln (Q), Thr (T), and Ser (S) can act as hydrogen-bond acceptors and contribute to hydrogen-bond formation (Schoneich et al., 1994; Doukov et al., 2007; Vennelakanti et al., 2021). This structural feature contributes to the hydrogen-bond network in the space between the main body of the cytochrome c-550 molecule and outer membrane surface. The tetrameric structure of this cytochrome c can further enhance the advantages of its H+-transferring properties. The segment of the cytochromes c lead the formation of an H+-capacitor at the outer membrane surface. The H+-capacitor would produce an additional unbalanced vertical force to drive F1F0-ATP synthase via H+ concentrations and electrical charges across the membrane.
Alignment of the amino acid sequence of cytochromes c from obligate and facultative alkaliphilic and neutralophilic strains with Bacillaceae showed obvious difference among them (Figure 3). The pronounced differences between cytochromes c from obligate alkaliphilic strains and those of others are the paucity of basic amino acids and presence of long sequences between the main body of the cytochrome c molecule and three amino acids at the N-terminus. These membrane-bound cytochromes c can potentially regulate H+-transferability via the acid ratio and amino acid composition of the sequence adjacent to the N-terminal sequence. Acidic amino acids, such as Asp (D) and Glu (E), can deprotonate their carboxyl groups and release H+ under high pH, whereas basic amino acids, such as Lys (K) and Arg (R), can protonate their amino side-chains under low pH and thus become H+-receivers (Voet et al., 2016). Given that H+ tend to accumulate at the cell membrane surface, resulting in a local pH assumed to be below 8. Both arginine (Arg) and lysine (Lys) are expected to accept protons due to their high pKa values (~12.5 and ~10.5, respectively). Therefore, the feature of the amino acid sequence of obligate alkaliphiles precedes H+-transfer to the H+-gate in F1F0-ATP synthase. However, the presence of amido groups on the side-chain possessing Asn (N), which act as proton carries, can accumulate H+ at the outer membrane surface. In addition to the N-terminal sequence, the C-terminal sequence and presence of two α-helices tend to decrease the frequency of basic amino acids and increase that of acidic amino acids.
6.2 Cytochrome c moiety in complexes III and IV
In addition to independent cytochromes c, such as the cytochrome c-550 of E. clarkii, cytochrome c components can exist in complexes, such as complexes III and IV. The subunit II of complex IV (cytochrome c oxidase in S. cohnii and Shouchella lehensis) has been reported to lack basic amino acids in its alkaliphilic form (Denda et al., 2001; Noor et al., 2014). Cytochromes c are attached to complexes III and IV in obligate alkaliphiles; a relatively higher acidic amino acid abundance plus the amino acids contributing to H+ transfer and a lower basic amino acid abundance could be present in areas exposed to the outer membrane surface (Supplementary Figures S1, S2). However, the pronounced trend in the abundance of Asn (N) observed in E. clarkii cytochrome c-550 and the other obligate alkaliphilic cytochromes c is not intense in the cytochrome c subunits in complexes III and IV.
In conclusion, all cytochromes c, including the cytochrome c moiety in complexes III and IV, are believed to cooperatively and efficiently transport the H+ that are pumped to the outer membrane surface via the respiratory chain to F1F0-ATP synthase and retain them on the outer membrane surface in conjunction in obligate alkaliphiles. However, independent cytochrome c contains more acidic and fewer basic amino acids than the cytochrome c moieties in complexes III and IV. Specificity of the additional N-terminal sequences may have higher efficiencies for H+ retention and transfer functions than those of the other moiety in the respiratory system. The location of the additional N-terminal sequence moiety in obligate alkaliphiles, which is found between the main cytochrome c molecule and membrane anchor moiety, may have a central role in coupling the redox properties of cytochrome c.
6.3 DUF2759 domain-containing protein and F1F0-ATP synthase: a- and c-subunits
The increase in the abundance of acidic amino acids, paucity of basic amino acids, and increase in the amino acids contributing to H+ transfer via the hydrogen-bonding network (Marx et al., 1999; Marx, 2006) for cytochromes c observed in obligate alkaliphiles are higher than those in the other categorized members. Those differences are not as intense as those in the respiratory complexes. To draw the accumulated H+ from cytochrome c to the H+ influx gate in the a-subunit of F1F0-ATP synthase, a relative abundance of protonable basic amino acids should be present at the site. A multiple alignment was performed for the a-subunit of F1F0-ATP synthase because it serves as an H+ gate for the production of ATP (Vik et al., 2000). The most pronounced differences between the a-subunit of obligate alkaliphilic and the other categories of strains (facultative alkaliphiles and neutralophiles) are the N-terminal sequences that localize to the outer membrane surface (Figure 4). However, deprotonatable acidic amino acids at the N-terminal sequences that localize to the outer membrane surface in the a-subunit slightly outnumber basic ones. Therefore, H+ are likely transferred from cytochrome c to F1F0-ATP synthase via a small membrane-bound protein (BpOF4_01690) found in A. pseudofirmus OF4 (Figure 5; Takahashi et al., 2018). This protein exposes four or three basic amino acids and one acidic amino acid, may recruit H+ at the vicinity of the H+ influx gate of F1F0-ATP synthase. This protein has been reported to play a crucial role in oxidative phosphorylation (Takahashi et al., 2018). The difference in abundance of acidic and basic amino acids in an H+ carrier segment may indicate its ability to release or accept H+. Acidic minus basic amino acid residue numbers in the H+-transferring segments indicate a H+ releasing ability with a negative value, indicating H+-release or acceptability. The number of acidic minus basic amino acids in the amino acid sequence corresponding to the N-terminal domain of Asn4-Asn37 in cytochrome c, DUF2759 domain-containing protein (equivalent to BpOF4_01690), a-subunit of the H+ influx gate, and intracellular side of F1F0-ATP synthase in E. clarkii are +10, −2, +1, and −6, respectively (Table 2; Figure 6). Therefore, significant gaps could be present in the H+-releasing and -accepting abilities between the outer and intracellular membrane sides of F1F0-ATP synthase in alkaliphiles. Such acidic minus basic amino acid residue-related number gaps could also be observed between the N-terminal site of the cytochrome c and H+-recruiting small membrane-bound protein amino acid sequences. The above-described H+ might facilitate H+ transfer from the H+-capacitor in cytochrome c to the intracellular side of F1F0-ATP synthase. The higher abundance of protonatable amino acids in the low pH environment at the intracellular side of the F1F0-ATP synthase may be of some importance in driving H+ from the outer membrane surface to the intracellular space through F1F0-ATP synthase.
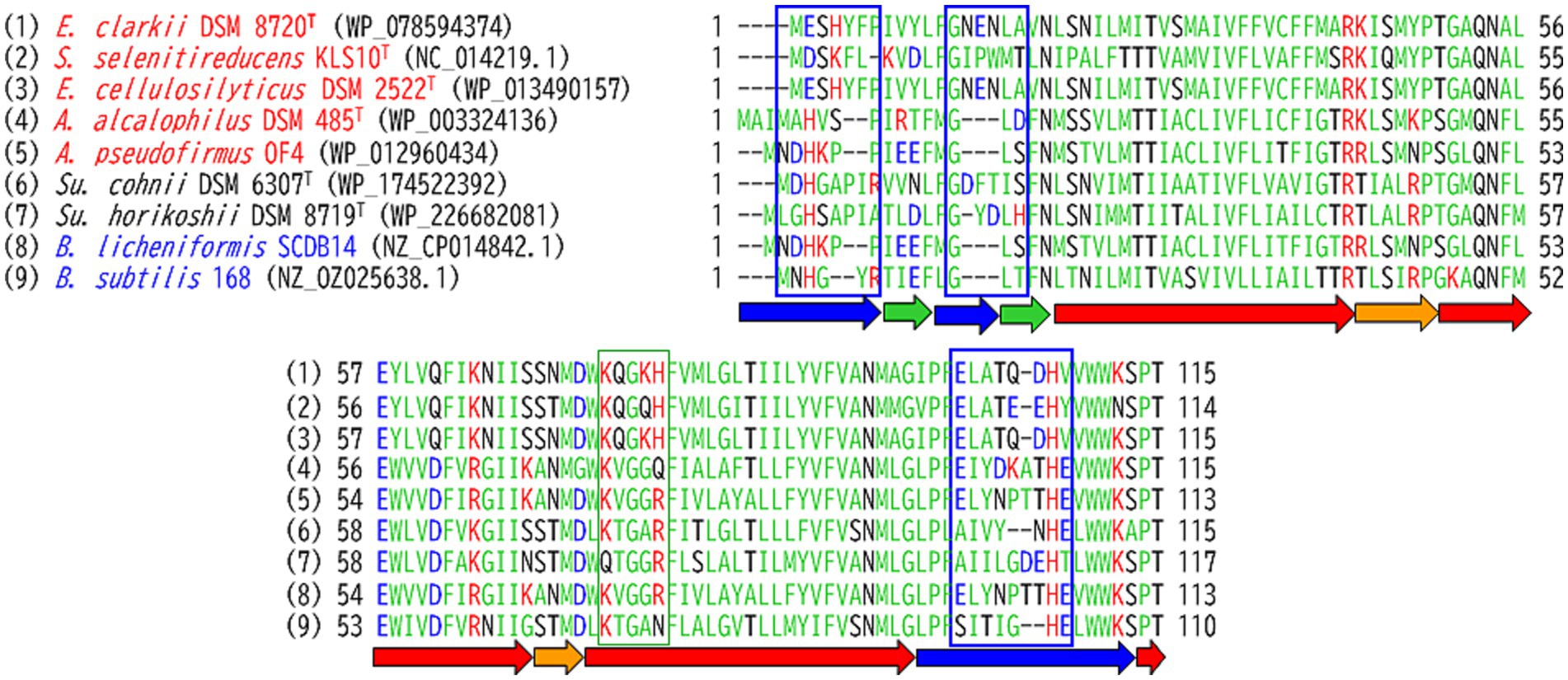
Figure 4. Amino acid sequence alignment of the a-subunit of F1F0-ATP synthase from Evancella clarkii DSM 8720T and other alkaline and neutral hydrophilic Bacillaceae. Obligate and facultative alkaliphilic strains are indicated by red and black, respectively, and neutralophilic strains shown by blue. Green and red arrows indicate β-sheet and α-helix structures, respectively. Blue and orange arrows highlight the outer and inner membrane sides of the loop structures, respectively. Acidic and basic residues are indicated by blue and red letters, respectively; amino acids representing amido or hydroxyl group side-chains indicated by black letters; and hydrophobic amino acids shown as pale green. Sequences showing a trend from neutral to alkaliphilic (D + E) + (N + T + S) > (H + K + R) are indicated by blue boxes, and those showing a trend from neutral to alkaliphilic (D + E) + (N + T + S) < (H + K + R) indicated by green boxes.

Figure 5. Amino acid sequence alignment of the small membrane-bound protein (DUF2759) equivalent to BpOF4_01690 (Takahashi et al., 2018) from Evansella clarkii DSM 8720T and other alkaline and neutral hydrophilic Bacillaceae. Obligate and facultative alkaliphilic strains are indicated by red and black, respectively, and neutralophilic strains are shown by blue. Acidic and basic residues are indicated with blue and red letters, respectively; amino acids representing amido or hydroxyl group side-chains indicated by black letters; hydrophobic amino acids presented as pale green.
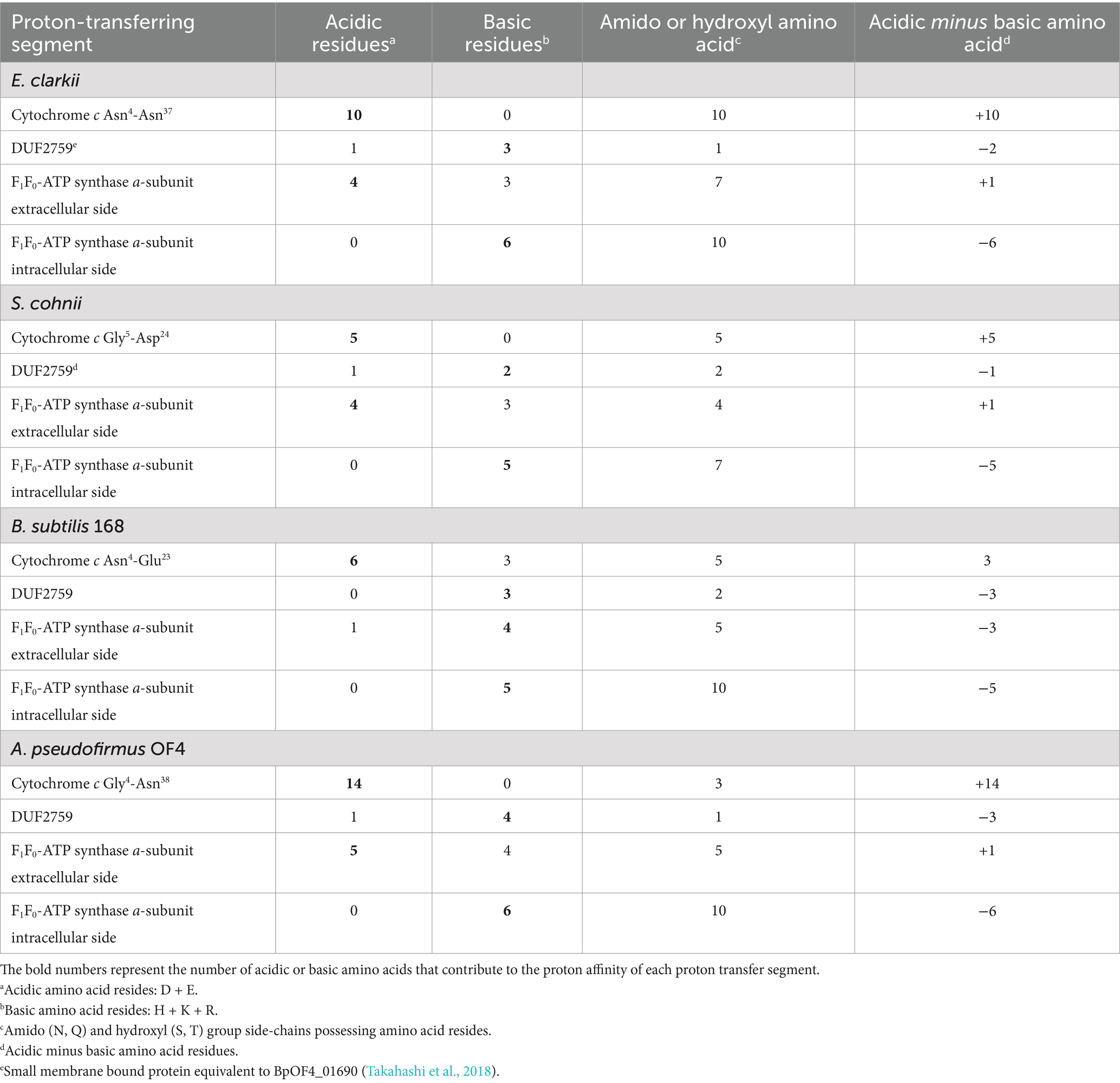
Table 2. Number of amino acids (acidic, basic and H+ transfer assist) that associated H+ transfer from cytochrome c to at the outer-surface membrane of obligate alkaliphilic Evansella clarkii, facultative alkaliphilic Sutcliffiella cohnii, and neutralophilic Bacillus subtilis and Alkalihalophilus pseudofirmus.
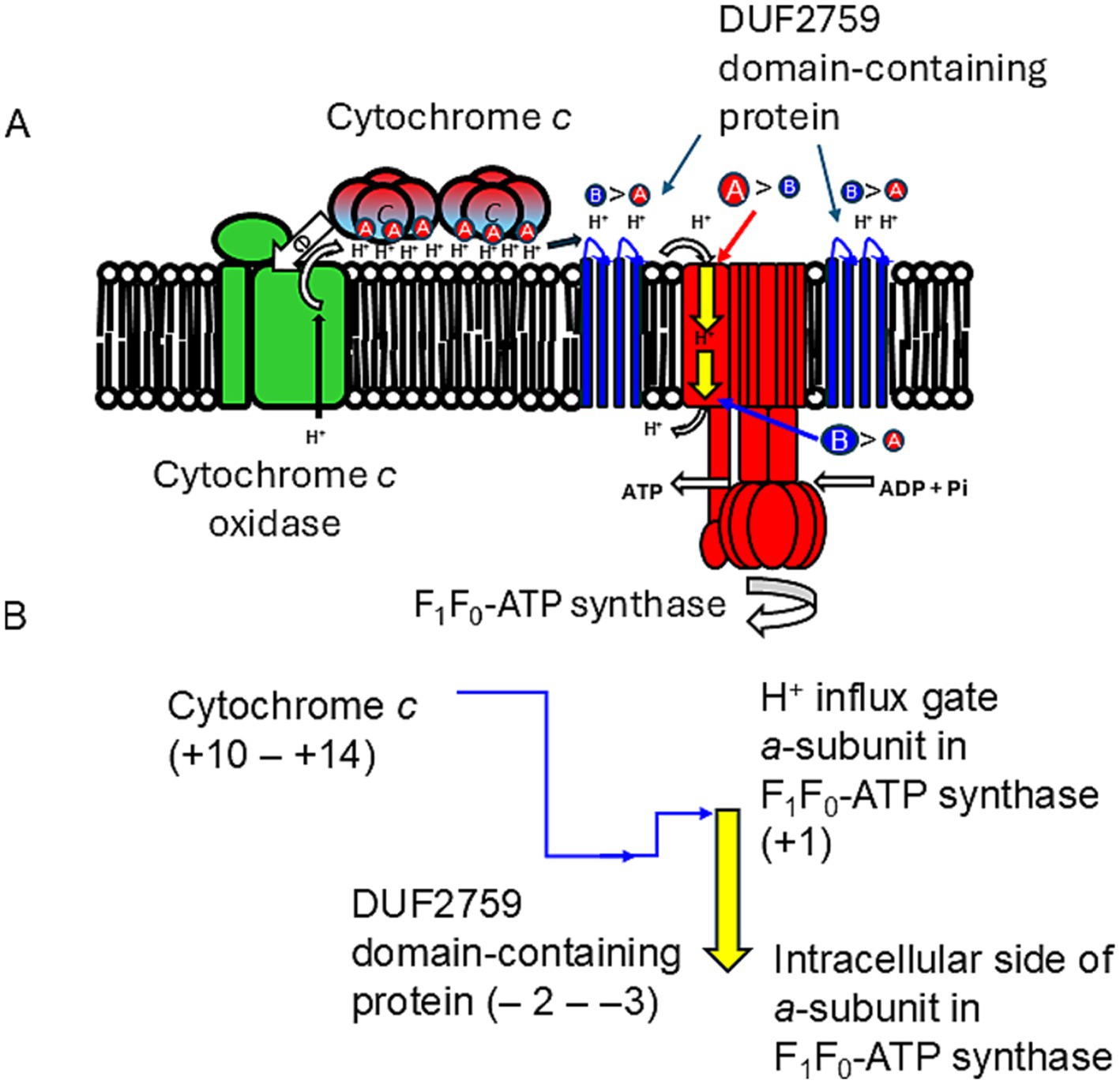
Figure 6. High efficiency in H+ localization on the outer surface of the membrane is achieved through a hydrogen-bonded network formed by the membrane-bound cytochrome c N-terminal segment (corresponding to Asn4-Asn27 in E. clarkii cytochrome c) in obligate alkaliphiles. (A) H+ capacitor formation and H+ transfer in the outer surface of membrane and F1F0-ATP synthase. The membrane-bound cytochrome c acts as an H+ capacitor due to its N-terminal segment. This segment is rich in acidic, amido-, and hydroxyl groups of amino acids, which, along with its tetrameric structure, play a crucial role in the accumulation and transfer of H+ on the outer membrane surface. H+ is transferred from the H+-capacitor by cytochrome c to F1F0-ATP synthase via a DUF2759 domain-containing protein (illustrated in blue). This protein features four or three basic amino acids and one acidic amino acid, which facilitate H+ recruitment near the H+ influx gate of the a-subunit of F1F0-ATP synthase. The disparity in the abundance of acidic (red A) versus basic (blue B) amino acids in the H+ carrier segment suggests its H+ release (refer to Table 2). The data indicate significant differences in H+ releasing and accepting capabilities between the outer membrane side (red arrow) and the intracellular side (blue arrow) of F1F0-ATP synthase in alkaliphiles. (B) Changes in amino acid composition in H+ transfer segments. This illustrates the changes in the abundance of acidic to basic amino acids in H+ carrier segments, highlighting the H+ transfer cascade from cytochrome c to F1F0-ATP synthase. It also presents a schematic representation of the high performance of F1F0-ATP synthase. Notably, a substantial gap exists between cytochrome c and the DUF2759 domain-containing protein that recruit H+. This larger gap is essential for overcoming the reversal difference between the DUF2759 domain-containing protein and the H+ influx gate of the a-subunit of F1F0-ATP synthase.
Alkaliphilic Bacillaceae strains have been reported to harbor specific amino acid sequences in the c-subunit of F1F0-ATP synthase compared with those of neutralophilic strains (Liu et al., 2011). A multiple alignment was performed for the c-subunit of F1F0-ATP synthase in obligate and facultative alkaliphilic and neutralophilic Bacillaceae (Supplementary Figure S3). The outer membrane surface-exposed amino acid sequence of obligate alkaliphilic E. clarkii contains a higher frequency of amino acids that contribute to the hydrogen-bonding network and a lower frequency of basic amino acids compared with those of facultative alkaliphilic and neutralophilic sequences. Three Glu (E) residues were observed in neutrophilic B. subtilis, whereas in the three obligate alkaliphilic species, E. clarkii, Salisediminibacterium selenitireducens, and E. cellulosilytica, only one Glu (E) residue was observed among the 20 residues on the outer-surface side. These results suggest that the outer-surface side of the c-subunit residues in F1F0-ATP synthase is also designed to balance H+-acceptance and release.
7 Bioenergetic parameter and capacitance formation
From the amino acid alignment and characteristics of the amino acid sequence, we can predict the physiological role of E. clarkii cytochrome c-550. In addition, the measurements of bioenergetic parameters (cytochrome c content in the membrane, oxygen consumption rate during respiration, ψ, and ratio of H+ extrusion to oxygen consumption [H+/O]) will be necessary to refer to the outcome (ATP production rate). From the oxygen consumption rate and H+/O, we can predict the H+ extrusion frequency•min−1. By considering estimations of the H+ extrusion frequency•min−1 and ATP production•min−1, we can predict the actual potential for H+ extruded at the outer membrane for ATP production. Therefore, we could possibly verify the physiological role of cytochrome c-550 from E. clarkii and the value of H+ transfer by comparing bioenergetic parameters and the outcomes.
7.1 H+ translocation frequencies
The oxygen consumption rates of the obligate alkaliphile E. clarkii and the facultative alkaliphile S. cohnii are lower than that of the neutralophile B. subtilis. This difference may be attributed to the higher membrane electrical potential (Δψ) observed in alkaliphilic species. The H+-translocation by the respiratory complex is coupled to electron transfer to molecular oxygen. The translocation of positively charged protons (H+) from the intracellular to the extracellular side is impeded by this Δψ, which tends to attract cations back toward the cytoplasmic side of the membrane (Table 3).

Table 3. Summary of the bioenergetic parameters of obligate alkaliphilic Evansella clarkii, facultative alkaliphilic Sutcliffiella cohnii and neutralophilic Bacillus subtilis high aeration conditions*.
Additionally, the H+/O ratio—representing the number of protons extruded per oxygen molecule consumed—is lower in alkaliphiles than in B. subtilis (Table 3). One possible reason is the accumulation of H+ at the outer membrane surface, facilitated by cytochrome c, which may act as a limiting factor for efficient proton extrusion. This localized proton crowding could generate a back-pressure effect, impeding further H+ translocation by the respiratory chain. As a result, the rate of H+ export mediated by the respiratory chain in alkaliphilic strains is likely much lower than in B. subtilis.
It has been reported that both the membrane electrical potential (Δψ) and the transmembrane pH gradient (ΔpH) act to hinder proton translocation simultaneously in the respiratory chain, thereby limiting the effective generation of membrane potential (Gregory and Ferguson-Miller, 1989). This inhibitory effect is more pronounced in alkaliphiles than in neutralophiles. Nevertheless, once protons are successfully translocated across the membrane despite these barriers, they exhibit a remarkably high energetic potential. Consequently, the driving force per proton (Δp) for ATP synthesis via the F₁F₀-ATP synthase is substantially greater in alkaliphilic species (Table 3). However, the retention of protons at the outer membrane surface—mediated by Δψ and surface-associated cytochrome c—may impede their lateral transfer to ATP synthase. Therefore, enhancing proton transfer mechanisms at the membrane surface, potentially through strategically positioned amino acid residues that facilitate proton mobility, could be critical for optimizing ATP synthesis efficiency in alkaliphilic bacteria.
7.2 Transmembrane electrical potential
Δψ is the voltage difference across a biological membrane, which is usually between the inside and outside of a cell or organelle is a key component of the electrochemical gradient that drives many cellular processes. In bacteria, the inside of the membrane (cytoplasmic side) is generally negatively charged, whereas the outside (external environment side) is positively charged. This potential difference is mainly caused by differences in ion concentration and membrane permeability. For measurements of the bacterial Δψ, cationic probes permeable to the membrane have often been used and according to the distribution across the membrane Δψ (Vm), can be calculated via the Nernst equation:
where Vm is the equilibrium potential of the ion (=Δψ), z represents the number of moles of electrons transferred in the electrochemical reaction and Cout and Cin are the ion concentrations outside and inside the membrane, respectively. One of the main sources of the bacterial Δψ is the Donnan potential (Donnan, 1924). This occurs when negatively charged molecules that are impermeable to the membrane are present on the intracellular side of the membrane (Figure 7). Intracellular DNA was shown to be naturally charged. In addition, negatively charged proteins corresponding to the intracellular pH are thought to contribute to the Donnan potential. Acidic proteins with a low isoelectric point (pI) will exhibit the intracellular pH of alkaliphilic bacteria (pH 8.1–8.4 when the extracellular pH is approximately 10). Considering the Donnan potential that contributes to the Δψ, the intrinsic non-energized Δψ (ΔψNE) can be assumed to be attributable to the initial ion balance across the membrane and existence of negatively charged intracellular molecules. However, H+ are translocated through respiratory activity of the energized Δψ (ΔψE), which affects ATP production via F1F0-ATP synthase. Intracellular DNA and acidic proteins contribute to increases in the Donnan potential, which subsequently contributes to the ΔψNE. Analysis of the predicted protein pI derived from whole genome analyses indicates that the obligate alkaliphile, E. clarkii, has a higher preference for acidic amino acids than the facultative alkaliphile, S. cohnii, and neutralophilic B. subtilis do (Figure 8). Lebre and Cowan (2020) also reported the preference of acidic proteins (pI 4.01–5) in alkaliphiles compared with that in neutralophiles, and acidophiles. The results of comparative gene expressions that correspond to culture conditions show that obligatory alkaliphilic E. clarkii has a predisposition to exhibit a higher ΔψNE, which is attributed by the Donnan potential, than that of S. cohnii and B. subtilis. A negative ion capacity was estimated in E. clarkii and S. cohnii grown at pH 10 and B. subtilis grown at pH 7 (Goto et al., 2016). In addition, the Donnan potential can be predicted by the negative ion capacity of intracellular constituents. The capacity in two alkaliphilic strains increased as the studied pH increased in the range of pH 6–8 and remained at high values in the range of pH 8–10, whereas the values hardly changed at pH 6–10 in B. subtilis. As the ΔψNE attracts H+ to the intracellular site, it hinders the transport of H+ via the respiratory chain. However, once translocated to the outer membrane surface, H+ will have a higher potential for Δp than in the case of a moderate ΔψNE. To overcome the difficulty of H+ translocation, cytochrome c and its moieties of respiratory complexes exhibit a lower redox potential compared with that in neutralophiles to produce a larger gap between the electron donor (cytochrome c) and acceptor (cytochrome a) in complex IV (Lewis et al., 1981; Orii et al., 1991).

Figure 7. In alkaliphilic bacteria, the abundance of acidic proteins and nucleic acids in the intracellular space makes the intracellular side more likely to be negatively charged. This generates a strong membrane potential (Δψ), or high inward electrical potential for H+, between the intracellular space and the high-pH external environment. It is thought that the Donnan effect contributes to the formation of this potential. The Donnan potential is an electrical potential difference across a membrane caused by charged molecules (e.g., acidic proteins within cells) that cannot diffuse across a semi-permeable barrier (e.g., a cell membrane), resulting in a distribution bias of diffusible ions (e.g., Na+, Cl−).
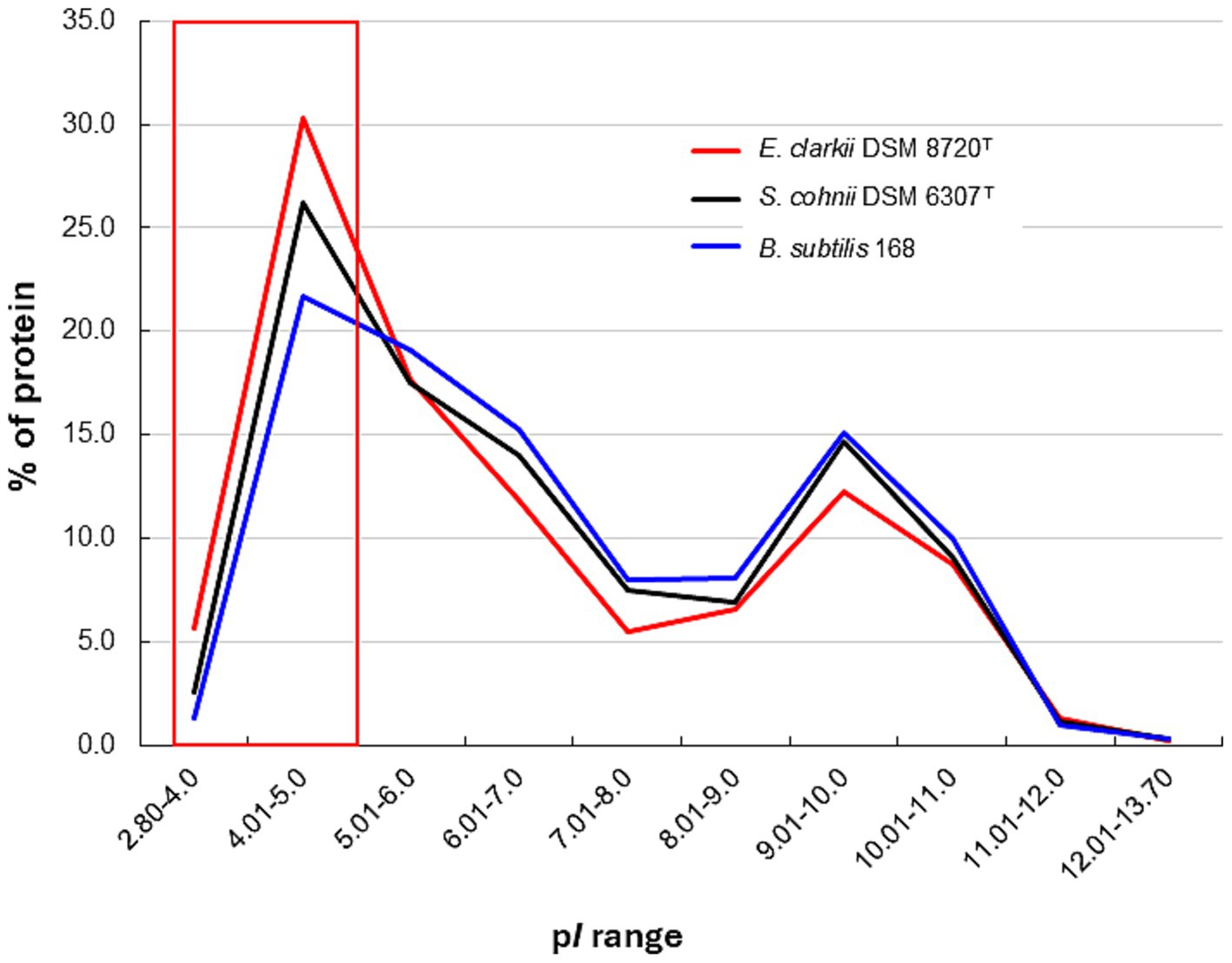
Figure 8. Protein isoelectric point (pI) distribution in obligate alkaliphilic Evansella clarkii DSM 8720T, facultative alkaliphilic Sutcliffiella cohnii DSM 6306T and neutralophilic Bacillus subtilis 168. The red line indicates E. clarkii DSM 8720T, black line indicates S. cohnii DSM 6306T, and blue line indicates B. subtilis 168. The red boxes indicate that the pI of proteins from obligate alkaliphilic E. clarkii DSM 8720T is biased toward the low-pH side compared with that of facultative alkaliphilic S. cohnii DSM 6306T and neutralophilic B. subtilis 168. The pI values of each genome were calculated using the Sequence Manipulation Suite server (Stothard, 2000).
Thus, the produced congenial high ΔψNE can lead to a high ΔψE. The effect of the ψΕ retention of extruded H+ via the respiratory chain has been reported previously (Yoshimune et al., 2010). The rate of H+ extrusion during respiratory activity in E. clarkii is slower than that in B. subtilis. This suggests that H+ extruded through respiration in E. clarkii are retained at the outer membrane surface. However, the retained H+ are rapidly released into the bulk phase after the introduction of valinomycin, which disrupts the ΔψΕ. However, the retardation of H+ at the outer membrane surface was enhanced after the introduction of monensin, which acts as a Na+/H+ exchanger. This suggests that H+ accumulating at the outer membrane surface were exchanged for intercellular Na+.
7.3 Efficiency of ATP production
To estimate the ATP production efficiency of each strain (E. clarkii, S. cohnii, and B. subtilis), the rate of maximum ATP synthesis per H+ translocated by the respiratory chain was estimated (Table 3). This experimental system used endogenous substrates. Less than 20% of ATP can be assumed to be derived from substrate-level phosphorylation (Russell and Cook, 1995). The maximum ATP synthesis efficiency per H+ extrusion rate of E. clarkii was approximately 3.4 times higher than that of S. cohnii, whereas it was approximately 130-fold higher than that of B. subtilis. This high efficiency cannot be explained by the high ΔψΕ alone; the H+-capacitor function is likely responsible for this, which involves the retention of accumulated cytochrome c and H+ on the outer membrane surface and their transfer to the F1F0-ATP synthase. This configuration creates an additional imbalance across the membrane in the electrochemical Η+ potential that exists along the outer membrane surface for driving F1F0-ATP synthase. Thus, alkaliphiles strategically employ even more effective bioenergetic mechanics to reverse the harsh alkaline environment.
8 Conclusion and perspective
Obligate alkaliphilic E. clarkii and facultative alkaliphilic S. cohnii exhibit a phylogenetic position near that of neutralophilic B. subtilis; however, these species are able to grow vigorously under harsh environments at pH 10 in which ordinary neutralophiles are unable to grow. This review focused on the H+-transferable amino acids at the N-terminal side of membrane-bound cytochromes c that can effectively accumulate H+ on the outer membrane surface, which forms a H+-capacitor. Thus, the produced H+-capacitor would bring an additional unbalanced vertical force to drive F1F0-ATP synthase via H+ concentrations and electrical charges across the membrane. Furthermore, the maximal driving performance of proton translocation and ATP synthesis in E. clarkii is likely achieved through a steep proton affinity gradient between the efficient H+ transfer across the outer membrane surface and the H+ entry site of the F₁F₀-ATP synthase on the cytoplasmic side. The proton transfer mechanism is governed by the differential abundance of acidic and basic amino acid residues within key protein segments. Specifically, the net number of acidic minus basic residues in the amino acid sequences corresponding to the N-terminal domain of cytochrome c, the DUF2759 domain-containing protein, the a-subunit of the H+ influx gate, and the intracellular side of the F₁F₀-ATP synthase are +10, −2, +1, and −6, respectively. This pronounced cascade of proton affinity differences exemplifies a unique bioenergetic adaptation characteristic of alkaliphilic bacteria. Taken together, these findings suggest that the H+-capacitor mechanism involving cytochrome c plays a contributory role in driving ATP synthesis. In the future, verifying the physiological role of the N-terminal sequence of cytochrome c-550 using various mutant proteins, and clarifying the relationship between the amino acid sequence and H+ retention ability would be crucial.
Formation of a high ΔψE is indispensable for the high performance of an H+-capacitor and physiological functioning in alkaliphilic Bacillaceae. The total bacterial transmembrane potential consists of charge separation driven by heterologous permeable ions across the membrane, impermeable substances across the membrane (Donnan potential), and chemical modifications to the membrane surface (e.g., membrane lipid head group) (Benarroch and Asally, 2020). Among them, charge separation driven by the existence of intracellular, impermeable substances with strong negative charges are intrinsically advantageous to produce large amounts of ATP in the obligate alkaliphilic E. clarkii. The experimentally confirmed intracellular negative charge (Goto et al., 2016) may be largely attributable to the presence of negatively charged proteins (Figure 6). However, these proteins and their biochemical processes have yet to be identified. These negative charges use the intracellular pH of alkaliphiles, which are approximately one unit higher (i.e., ca. pH 8) than that in neutralophilic bacterial strains. The intracellular negative ion capacity decreases under low aeration conditions in both E. clarkii and S. cohnii (Goto et al., 2016). Thus, these alkaliphiles can regulate the expression of negatively charged substances by sensing external environmental conditions. Identification of substances contributing to the intracellular negative ion capacity and expression of associated regulatory mechanisms should be clarified in future studies.
Alkaliphilic bacteria have a lower respiratory rate than that of neutrophiles. This is not only due to the lack of H+, but also to the inhibition of transfer through the Δψ that attribute to an intrinsic, large, intracellularly negative ion capacity. Alkaliphilic bacteria strategically produce an even higher membrane potential than neutralophiles do because they must maintain the function of an even more complicated solute transport system. Under high pH conditions, alkaliphilic bacteria have thus adopted the H+-capacitor system. This mechanism increases the driving force of F1F0-ATP synthase by allowing H+ to accumulate on the outer membrane surface, thereby driving F1F0-ATP synthase more efficiently. In addition, as the positive charge of H+ accumulates on the outer membrane surface, this likely creates an attractive force between the positive and negative charges present, which is equivalent to the electrostatic attraction observed in an actual electrical capacitor (opposite charge), providing the driving force for F1F0-ATP synthase propulsion. The presence of a factor equivalent to the electrostatic attraction should be verified in future studies.
Author contributions
IY: Writing – original draft, Writing – review & editing.
Funding
The author declares that no financial support was received for the research and/or publication of this article.
Acknowledgments
We thank Editage (www.editage.com) for the English language editing.
Conflict of interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The author declares that no Gen AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1637315/full#supplementary-material
References
Ahmed, E., and Holmström, S. (2014). Siderophores in environmental research. Microb. Biotechnol. 7, 196–208. doi: 10.1111/1751-7915.12117
Aino, K., Hirota, K., Okamoto, T., Tu, Z., Matsuyama, H., and Yumoto, I. (2018). Microbial communities associated with indigo fermentation that thrive in anaerobic alkaline environments. Front. Microbiol. 9:2196. doi: 10.3389/fmicb.2018.02196
Andrews, S. C., Robinson, A. K., and Rodriguez-Quinones, F. (2003). Bacterial iron homeostasis. FEMS Microbiol. Rev. 27, 215–237. doi: 10.1016/S0168-6445(03)00055-X
Aono, R., and Horikoshi, K. (1983). Chemical composition of cell walls of alkalophilic strains of alkalophilic strains of Bacillus. J. Gen. Microbiol. 129, 1083–1087.
Aono, R., Ito, M., and Horikoshi, K. (1993). Occurrence of teichurono peptide in cell walls of group 2 alkaliphilic Bacillus sp. J. Gen. Microbiol. 139, 2738–2744.
Aono, R., Ito, M., Joblin, K. N., and Horikoshi, K. (1995). A high cell wall negative charge is necessary for the growth of alkaliphile Bacillus lentus C−125 at elevated pH. Microbiology 141, 2955–2964.
Aono, R., Ito, M., and Machida, T. (1999). Contribution of the cell wall component teichuronopeptide to pH homeostasis and alkaliphily in the alkaliphilic Bacillus lentus C-125. J. Bacteriol. 181, 6600–6606. doi: 10.1128/JB.181.21.6600-6606.1999
Aono, R., Kaneko, H., and Horikoshi, K. (1996). Alkaline growth pH-dependent increase of respiratory and NADH-oxidation activities of the facultatively alkaliphilic strain Bacillus lentus C-125. Biosci. Biotechnol. Biochem. 60, 1243–1247. doi: 10.1271/bbb.60.1243
Banciu, H. L., and Sorokin, D. Y. (2013). “Adaptation in haloalkaliphiles and natronophilic bacteria” in Polyextremophiles (Dordrecht: Springer), 121–178.
Benarroch, J. M., and Asally, M. (2020). The microbiologist's guide to membrane potential dynamics. Trends Microbiol. 28, 304–314. doi: 10.1016/j.tim.2019.12.008
Bengtsson, J., Rivolta, C., Hederstedt, L., and Karamata, D. (1999). Bacillus subtilis contains two small c-type cytochromes with homologous heme domains but different types of membrane anchors. J. Biol. Chem. 274, 26179–26184. doi: 10.1074/jbc.274.37.26179
Benini, S., Borsari, M., Ciurli, S., Dikiy, A., and Lamborghini, M. (1998). Modulation of Bacillus pasteurii cytochrome c553 reduction potential by structural and solution-parameters. J. Biol. Inorg. Chem. 3, 371–382. doi: 10.1007/s007750050247
Benini, S., González, A., Rypniewski, W. R., Wilson, K. S., Van Beeumen, J. J., and Ciurli, S. (2000). Crystal structure of oxidized Bacillus pasteurii cytochrome c553 at 0.97-Å resolution. Biochemistry 39, 13115–13126.
Clejan, S., Krulwich, T. A., Mondrus, K. R., and Seto-Yung, D. (1986). Membrane lipid composition of obligately and facultatively alkaliphilic strains of Bacillus spp. J. Bacteriol. 168, 334–340. doi: 10.1128/jb.168.1.334-340.1986
Cowan, D. A., Albers, S. V., Antranikian, G., Atomi, H., Averhoff, B., Basen, M., et al. (2024). Extremophiles in a changing world. Extremophiles 28:26. doi: 10.1007/s00792-024-01341-7
David, P. S., Dutt, P. S., Wathen, B., Jia, Z., and Hill, B. C. (2000). Characterization of structural model of membrane bound cytochrome c−550 from Bacillus subtilis. Arch. Biochem. Biophys. 377, 22–30.
Davidson, M. W., Gray, K. A., Knaff, D. B., and Krulwich, T. A. (1988). Purification and characterization of two soluble cytochromes from the alkalophile Bacillus firmus RAB. Biochim. Biophys. Acta 933, 470–477. doi: 10.1016/0005-2728(88)90082-5
Denda, K., Oshima, A., and Fukumori, Y. (2001). Structural analyses of the deduced amino acid sequences of a novel type heme-copper terminal oxidase, cytochrome aco3, from alkalophilic Bacillus YN-2000. Can. J. Microbiol. 47, 1075–1081. doi: 10.1139/cjm-47-12-1075
Dimroth, P., and Cook, G. M. (2004). Bacterial Na+− or H+-coupled ATP synthases operating a low electrical potential. Adv. Microb. Physiol. 49, 175–218. doi: 10.1016/S0065-2911(04)49004-3
Donnan, F. G. (1924). The theory of membrane equilibria. Chem. Rev. 1, 73–90. doi: 10.1021/cr60001a003
Doukov, T. I., Hemmi, H., Drenman, C. L., and Ragsdale, S. W. (2007). Structural and kinetic evidence for extended hydrogen-bonding network in catalysis of methyl group transfer. Role of active site asparagine residue in activation of methyl transfer by methyltransferase. J. Biol. Chem. 282, 6609–6618. doi: 10.1074/jbc.M609828200
Drechsel, H., and Jung, G. (1998). Peptide siderophores. J. Pept. Sci. 4, 147–181. doi: 10.1002/(SICI)1099-1387(199805)4:3%3C147::AID-PSC136%3E3.0.CO;2-C
Ferguson, S. A., Cook, G. M., Montgomery, M. G., Leslie, A. G., and Walker, J. E. (2016). Regulation of the thermoalkaliphilic F1-ATPase from Caldalkalibacillus thermarum. Proc. Natl. Acad. Sci. USA 113, 10860–10865. doi: 10.1073/pnas.1612035113
Fujiwara, Y., Oka, M., Hamamoto, T., and Sone, N. (1993). Cytochrome c-551 of the thermophilic bacterium PS3, DNA sequence and analysis of the mature cytochrome. Biochim. Biophys. Acta 1144, 213–219. doi: 10.1016/0005-2728(93)90175-F
Gilmore, R., Messner, P., Guffanti, A. A., Kent, R., Scheberl, A., Kendrick, N., et al. (2000). Two-dimensional gel electrophoresis of pH-dependent protein expression in facultatively alkaliphilic Bacillus pseudofirmus OF4 lead to characterization OF an S-layer protein with a role in alkaliphily. J. Bacteriol. 182, 5969–5981. doi: 10.1128/JB.182.21.5969–5981.2000
Goto, T., Hirabayashi, T., Morimoto, H., Yamazaki, K., Inoue, N., Matsuyama, H., et al. (2016). Contribution of intracellular negative ion capacity to Donnan effect across the membrane in alkaliphilic Bacillus spp. J. Bioenerg. Biomembr. 48, 87–96. doi: 10.1007/s10863-015-9641-9
Goto, T., Ogami, S., Yoshimume, K., and Yumoto, I. (2022). Differences in bioenergetic metabolism of obligately alkaliphilic Bacillaceae under high pH depend on the aeration conditions. Front. Microbiol. 13:842785. doi: 10.3389/fmicb.2022.842785
Gregory, L., and Ferguson-Miller, S. (1989). Independent control of respiration in cytochrome c oxidase vesicles by pH and electrical gradients. Biochemistry 28, 2655–2662. doi: 10.1021/bi00432a044
Guffanti, A. A., Finkelthal, O., Hicks, D. B., Falk, L., Sidhu, A., Garro, A., et al. (1986). Isolation and characterization of new facultatively alkalophilic strains of Bacillus species. J. Bacteriol. 167, 766–773. doi: 10.1128/jb.167.3.766-773.1986
Guffanti, A. A., and Krulwich, T. A. (1994). Oxidative phosphorylation by ADP + pi-loaded membrane vesicles of alkaliphilic Bacillus firmus OF4. J. Biol. Chem. 269, 21576–21582. doi: 10.1016/S0021-9258(17)31843-4
Hamamoto, T., Hashimoto, M., Hino, M., Kitada, M., Seto, Y., Kudo, T., et al. (1994). Characterization of a gene responsible for the Na+/H+ antiporter system of alkalophilic Bacillus species strain C-125. Mol. Microbiol. 14, 939–946. doi: 10.1111/j.1365-2958.1994.tb01329.x
Hicks, D. B., Liu, J., Fujisawa, M., and Krulwich, T. A. (2010). F1F0-ATP synthases of alkaliphilic bacteria: lessons from their adaptations. Biochim. Biophys. Acta 1797, 1362–1377. doi: 10.1016/j.bbabio.2010.02.028
Hoffmann, A., and Dimroth, P. (1991). The electrochemical proton potential of Bacillus alcalophilus. Eur. J. Biochem. 201, 467–473. doi: 10.1111/j.1432-1033.1991.tb16304.x
Janto, B., Ahmed, A., Ito, M., Liu, J., Hicks, D. B., Pagni, S., et al. (2011). Genome of alkaliphilic Bacillus pseudofirmus OF4 reveals adaptations that support the ability to grow in an external pH range from 7.5 to 11.4. Environ. Microbiol. 13, 3289–3309. doi: 10.1111/j.1462-2920.2011.02591.x
Krulwich, T. A., Ito, M., and Guffanti, A. A. (2001). The Na+-dependence of alkaliphily in Bacillus. Biochim. Biophys. Acta 1505, 158–168. doi: 10.1016/S0005-2728(00)00285-1
Larsen, R. W., Chavez, M. D., Nunez, D. J., Davidson, M. W., Knaff, D. B., Krulwich, T. A., et al. (1990). Resonance Raman investigation of a soluble cytochrome c552 from alkaliphilic Bacillus firmus RAB. Arch. Biochem. Biophys. 283, 266–270. doi: 10.1016/0003-9861(90)90641-b
Lebre, P. H., and Cowan, D. A. (2020). Genomics of alkaliphiles. Adv. Biochem. Eng. Biotechnol. 172, 135–155. doi: 10.1007/10_2018_83
Lee, J. W. (2012). Proton-electrostatics hypothesis for localization proton coupling bioenergetics. Bioenergetics 1, 1–8. doi: 10.4172/2167-7662.1000104
Lee, J. W. (2021). Mitochondrial energetics with transmembrane electrostatically localized protons: do we have a thermotropic feature? Sci. Rep. 11, 1–13. doi: 10.1038/s41598-021-93853-x
Lewis, R. J., Belkina, S., and Krulwich, T. A. (1980). Alkalophiles have much higher cytochrome contents than conventional bacteria and then their own nonalkaliphilic mutant derivatives. Biochem. Biophys. Res. Commun. 95, 857–863. doi: 10.1016/0006-291x(80)90866-9
Lewis, R. J., Prince, R. C., Dutton, P. L., Knaff, D. B., and Krulwich, T. A. (1981). The respiratory chain of Bacillus alcalophilus and its nonalkalophilic mutant derivative. J. Biol. Chem. 256, 10543–10549. doi: 10.1016/S0021-9258(19)68656-4
Liu, J., Fackelmayer, O. J., Hicks, D. B., Preiss, L., Meier, T., Sobie, E. A., et al. (2011). Mutations in a helix-1 motif of the ATP synthase c-subunit of Bacillus pseudofirmus OF4 cause functional deficits and changes in the c-ring stability and mobility on sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Biochemistry 50, 5497–5506. doi: 10.1021/bi2005009
Lopes, H. F. S., Tu, Z., Sumi, H., Furukawa, H., and Yumoto, I. (2021). Indigofera tinctoria leaf powder as a promising additive to improve indigo fermentation prepared with Sukumo (composted Polygonum tinctorium leaves). World J. Microbiol. Biotechnol. 37:179. doi: 10.1007/s11274-021-03142-y
Ma, C., Zhuang, L., Zhou, S. G., Yang, G. Q., Yuan, Y., and Xu, R. X. (2012). Alkaline extracellular reduction: isolation and characterization of an alkaliphilic and halotolerant bacterium, Bacillus pseudofirmus MC02. J. Appl. Microbiol. 112, 883–891. doi: 10.1111/j.1365-2672.2012.05276.x
Mano, G. (2020). Challenges and adaptation of life in alkaline habitats. Adv. Biochem. Eng. Biotechnol. 172, 85–134. doi: 10.1007/10_2019_97
Marx, D. (2006). Proton transfer 200 years after von Grotthuss: insights from ab initio simulations. ChemPhysChem 7, 1848–1870. doi: 10.1002/cphc.200600128
Marx, D., Tuckerman, M. E., Hutter, J., and Parrinello, M. (1999). The nature of the hydrated excess proton in water. Nature 397, 601–604. doi: 10.1038/17579
McMillan, D. G., Keis, S., Berney, M., and Cook, G. M. (2009). Nonfermentative thermoalkaliphilic growth is restricted to alkaline environments. Appl. Environ. Microbiol. 75, 7649–7654. doi: 10.1128/AEM.01639-09
McMillan, D. G. G., Velasquez, I., Nunn, B. L., Goodlett, D. R., Hunter, K. A., Lamont, I., et al. (2010). Acquisition of iron by alkaliphilic Bacillus species. Appl. Environ. Microbiol. 76, 6955–6961. doi: 10.1128/AEM.01393-10
Meier, T., Morgner, N., Matthies, D., Pogoryelov, D., Keis, S., Cook, G. M., et al. (2007). A tridecameric c ring of the adenosine triphosphate (ATP) synthase from the thermoalkaliphilic Bacillus sp. strain TA2.A1 facilitates ATP synthesis at low electrochemical proton potential. Mol. Microbiol. 65, 1181–1192. doi: 10.1111/j.1365-2958.2007.05857.x
Miki, K., and Okunuki, K. (1969a). Cytochrome of Bacillus subtilis II. Purification and spectral properties of cytochrome c-550 and c-554. J. Biochem. 66, 831–843. doi: 10.1093/oxfordjournals.jbchem.a129214
Mitchell, P. (1961). Coupling of phosphorylation to electron and hydrogen transfer by a chemi-osmotic type of mechanism. Nature 191, 144–148. doi: 10.1038/191144a0
Nei, M., and Kumar, S. (2000). Molecular evolution and phylogenetics. New York, NY: Oxford University Press.
Nielsen, P., Fritze, D., and Priest, F. G. (1995). Phenetic diversity of alkaliphilic Bacillus strains: proposal for nine new species. Microbiology 141, 1745–1761. doi: 10.1099/13500872-141-7-1745
Noor, Y. M., Samsulrizal, N. H., Jema'on, N. A., Low, K. O., Ramli, A. N., Alias, N. I., et al. (2014). A comparative genomic analysis of the alkalitolerant soil bacterium Bacillus lehensis G1. Gene 545, 253–261. doi: 10.1016/j.gene.2014.05.012
Ogami, S., Higikata, S., Tsukahara, T., Mie, Y., Matsuno, T., Morita, N., et al. (2009). A novel membrane-anchored cytochrome c-550 of alkaliphilic Bacillus clarkii K24-1U: expression, molecular features and properties of redox potential. Extremophiles 13, 491–504. doi: 10.1007/s00792-009-0234-6
Orii, Y., Yumoto, I., Fukumori, Y., and Yamanaka, T. (1991). Stopped-flow and rapid-scan studies of the redox behavior of cytochrome aco from facultative alkalophilic Bacillus. J. Biol. Chem. 266, 14310–14316. doi: 10.1016/S0021-9258(18)98685-0
Orr, H. A. (2005). The genetic theory of adaptation: a brief history. Nat. Rev. Genet. 6, 119–127. doi: 10.1038/nrg1523
Picón Garrido, G. I., García García, A. P., González de la Vara, L., Chagolla-López, A., Gómez-Lojero, C., and Gutiérrez-Cirlos, E. B. (2022). Separation and analysis of Bacillus subtilis respiratory chain complexes. J. Bioenerg. Biomembr. 54, 251–271. doi: 10.1007/s10863-022-09951-6
Rampelotto, P. H. (2013). Extremophiles and extreme environments. Life (Basel) 3, 482–485. doi: 10.3390/life3030482
Rivas, L., Soares, C. M., Baptista, A. M., Simaan, J., Di Paola, R. E., Murgida, D., et al. (2005). Electric-field-induced redox potential shifts of tetraheme cytcochromes c3 immobilized on self assembled monolayers: surface-enhanced resonance spectroscopy and simulation studies. Biophys. J. 88, 4188–4199. doi: 10.1529/biophysj.104.057232
Russell, J. B., and Cook, G. M. (1995). Energetics of bacterial growth: balance of anabolic and catabolic reactions. Microbiol. Rev. 59, 48–62. doi: 10.1128/mr.59.1.48-62.1995
Satyanarayana, T., Raghukumar, C., and Sisinthy, S. (2005). Extremophilic microbes: diversity and perspectives. Curr. Sci. 89, 78–90.
Schoneich, C., Zhao, F., Madden, K. P., and Bobrowski, K. (1994). Side chain fragmentation of N-terminal threonine or serine residue induced through intramolecular proton transfer to hydroxy sulfuranyl radical formed at neighboring methionine in dipeptides. J. Am. Chem. Soc. 116, 4641–4652. doi: 10.1021/ja00090a012
Siguier, P., Gourbeyre, E., and Chandler, M. (2014). Bacterial insertion sequences: their genomic impact and diversity. FEMS Microbiol. Rev. 38, 865–891. doi: 10.1111/1574-6976.12067
Sone, N., Kutoh, E., and Yanagita, Y. (1989). Cytochrome c-551 from the thermophilic bacterium PS3 grown under air-limited conditions. Biochim. Biophys. Acta 977, 329–334. doi: 10.1016/S0005-2728(89)80088-X
Sone, N., and Toh, H. (1994). Membrane-bound Bacillus cytochromes c and their phylogenetic position among bacterial class I cytochromes c. FEMS Microbiol. Lett. 122, 203–210. doi: 10.1111/j.1574-6968.1994.tb07168.x
Soontharapirakkul, K., and Incharoensakdi, A. (2010). A Na+ −stimulated ATPase of alkaliphilic halotolerant cyanobacterium Aphanothece halophytica translocates Na+ into proteoliposomes via Na+ uniport mechanism. BMC Biochem. 11:30. doi: 10.1186/1471-2091-11-30
Stothard, P. (2000). The sequence manipulation suite: javascript programs for analysing and formatting protein and DNA sequences. BioTechniques 28, 1102–1104. doi: 10.2144/00286ir01
Sturr, M. G., Guffanti, A. A., and Krulwich, T. A. (1994). Growth and bioenergetics of alkaliphilic Bacillus firmus OF4 in continuous culture at high pH. J. Bacteriol. 176, 3111–3116. doi: 10.1128/jb.176.11.3111-3116.1994
Suzuki, S., Kuenen, J. G., Schipper, K., van der Velde, S., Ishii, S., Wu, A., et al. (2014). Physiological and genomic features of highly alkaliphilic hydrogen-utilizing Betaproteobacteria from a continental serpentinizing site. Nat. Commun. 5:3900. doi: 10.1038/ncomms4900
Takahashi, T., Krulwich, T. A., and Ito, M. (2018). A hydrophobic small protein, BpOF4_01690, is critical for alkaliphily of alkaliphilic Bacillus pseudofirmus OF4. Front. Microbiol. 9:01994. doi: 10.3389/fmicb.2018.01994
Takami, H., Han, C. G., Takaki, Y., and Ohtsubo, E. (2001). Identification and distribution of new insertion sequences in the genome of alkaliphilic Bacillus halodurans C-125. J. Bacteriol. 183, 4345–4356. doi: 10.1128/JB.183.14.4345-4356.2001
Tamura, K., Stecher, G., and Kumar, S. (2021). MEGA 11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Vennelakanti, V., Qi, H. W., Mehmood, R., and Kulik, H. J. (2021). When are two hydrogen bonds better than one? Accurate first-principles models explain the balance of hydrogen bond donors and acceptors found in proteins. Chem. Sci. 12, 1147–1162. doi: 10.1039/d0sc05084a
Vik, S. B., Long, J. C., Wada, T., and Zhang, D. (2000). A model for the structure of subunit a of the Escherichia coli ATP synthase and its role in proton translocation. Biochim. Biophys. Acta 1458, 457–466. doi: 10.1016/S0005-2728(00)00094-3
Voet, D., Voet, J. G., and Pratt, C. W. (2016). “Fundamentals of biochemistry: Life at the molecular level” in Life at the molecular level. 5th ed (New York, NY: Wiley Global Education).
von Wachenfeldt, C., and Hederstedt, L. (1993). Physico-chemical characterization of membrane-bound and water-soluble forms of Bacillus subtilis cytochrome c−550. Eur. J. Biochem. 212, 499–509. doi: 10.1111/j.1432-1033.1993.tb17687.x
Whelan, S., and Goldman, N. (2001). A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol. Biol. Evol. 18, 691–699. doi: 10.1093/oxfordjournals.molbev.a003851
Yamanaka, T. (1992). The biochemistry of bacterial cytochromes. Tokyo: Japan Scientific Society Press.
Ye, Q., Roh, Y., Carroll, S. L., Blair, B., Zhou, J., Zhang, C. L., et al. (2004). Alkaline anaerobic respiration: isolation and characterization of a novel alkaliphilic and metal-reducing bacterium. Appl. Environ. Microbiol. 70, 5595–5602. doi: 10.1128/AEM.70.9.5595-5602.2004
Yoshimune, K., Morimoto, H., Hirano, Y., Sakamoto, J., Matsuyama, H., and Yumoto, I. (2010). The obligate alkaliphile Bacillus clarkii K24-1U retains extruded protons at the beginning of respiration. J. Bioenerg. Biomembr. 42, 111–116. doi: 10.1007/s10863-010-9278-7
Yumoto, I., Fukumori, Y., and Yamanaka, T. (1991). Purification and characterization of two membrane-bound c-type cytochromes from a facultative alkalophilic Bacillus. J. Biochem. 110, 267–273. doi: 10.1093/oxfordjournals.jbchem.a123569
Yumoto, I., Nakajima, K., and Ikeda, K. (1997). Comparative study on cytochrome content of alkaliphilic Bacillus strains. J. Ferment. Bioeng. 83, 466–469. doi: 10.1016/S0922-338X(97)83002-4
Keywords: membrane-bound cytochrome c, hydrogen-bonding network, H+ transfer amino acids, H+-capacitor, F1F0-ATP synthase
Citation: Yumoto I (2025) H+-capacitor and ATP production in obligate alkaliphilic Bacillaceae: insights into cytochrome c and H+ transport mechanisms. Front. Microbiol. 16:1637315. doi: 10.3389/fmicb.2025.1637315
Edited by:
Masahiro Ito, Toyo University, JapanReviewed by:
Doug Bartlett, University of California, San Diego, United StatesGiovanni La Penna, National Research Council (CNR), Italy
Copyright © 2025 Yumoto. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Isao Yumoto, eXVtb3RvLmlzYW8uYXRyQG9zYWthLXUuYWMuanA=
 Isao Yumoto
Isao Yumoto