- 1College of Agriculture and Animal Husbandry, Qinghai University, Xining, China
- 2Key Laboratory of Medicinal Animal and Plant Resources of Qinghai Tibetan Plateau, Qinghai Normal University, Xining, China
- 3Academy of Plateau Science and Sustainability, Qinghai Normal University, Xining, China
Diarrhea poses a significant challenge to the growth of the livestock industry by decreasing the productivity and increasing mortality rates in animals. Several factors such as bacteria, viruses, parasites, and stress have been identified as potential contributors to diarrhea. The gut microbiota, a complex micro-ecosystem consisting of trillions of microorganisms such as bacteria, fungi, and viruses, plays a key role in host metabolism, immunity, and nutrient absorption. The gut microbial homeostasis is essential for the intestine to perform physiological functions that maintain the host health. Conversely, gut microbial dysbiosis can lead to the development of various diseases. Recent research has highlighted that gut microbial dysbiosis is a driving factor in the animal diarrhea. Consequently, maintaining the gut microbial homeostasis has become a key focus for the prevention and treatment of diarrhea. This review examines the composition, metabolites and functions of gut microbiota as well as the causes of diarrhea and the alterations in gut microbiota during diarrhea. Furthermore, this review provides insights for future research in this field, especially for alleviating animal diarrhea from gut microbial perspective.
Introduction
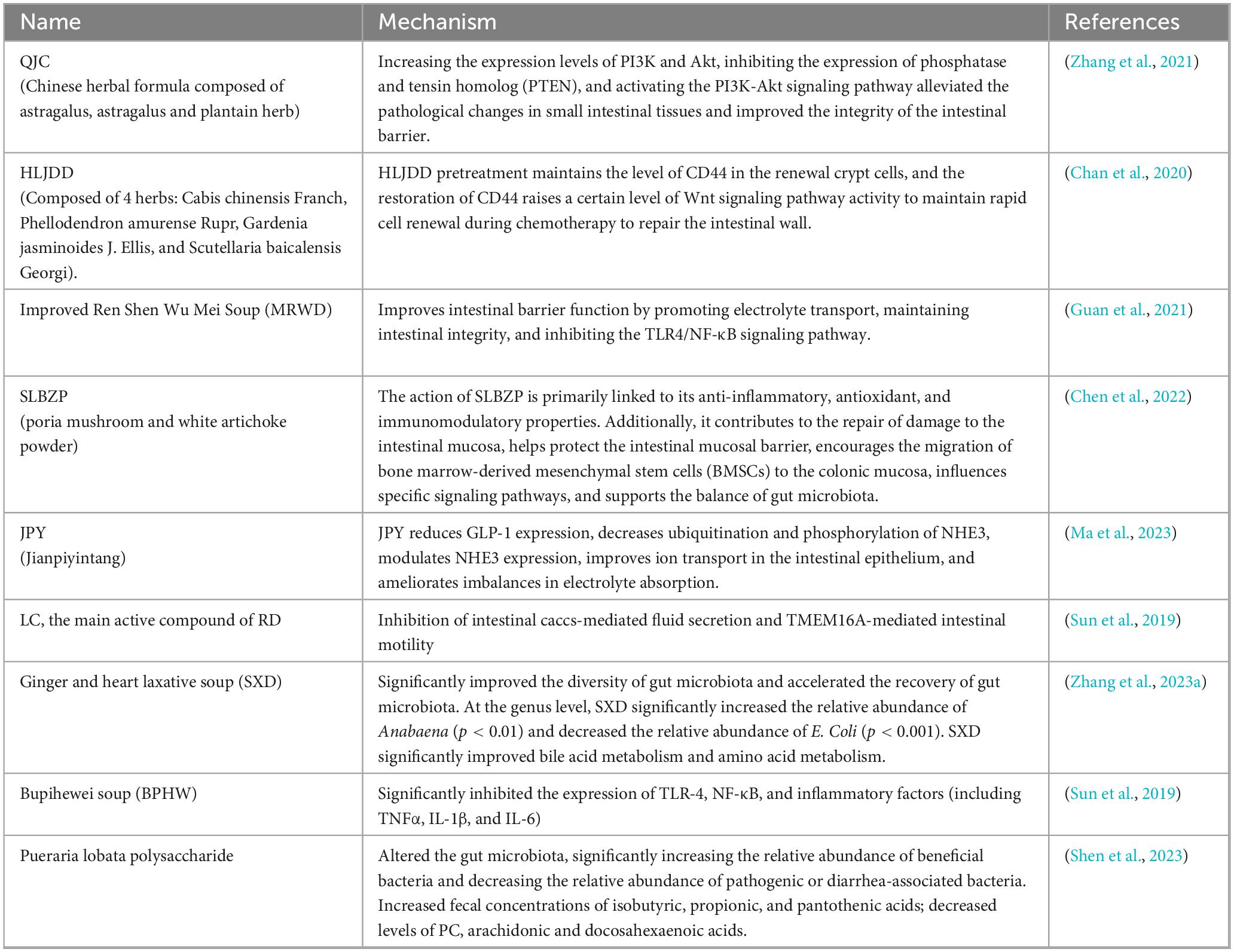
Intestine is the largest immune organ in the body, and the gut microbial balance is crucial for overall health. These microorganisms, with a significant portion inhabiting the gastrointestinal tract, exert pivotal role in the regulation of physiological functions (Wei et al., 2025). Numerous factors contribute to the imbalance of gut microbiota, including management practices, environmental conditions, diet, additives, and host-related factors (Figure 1). Developing a mutually beneficial connection between the host and gut microorganisms is essential for mucosal immunity and thwarting pathogen colonization (Du et al., 2023).
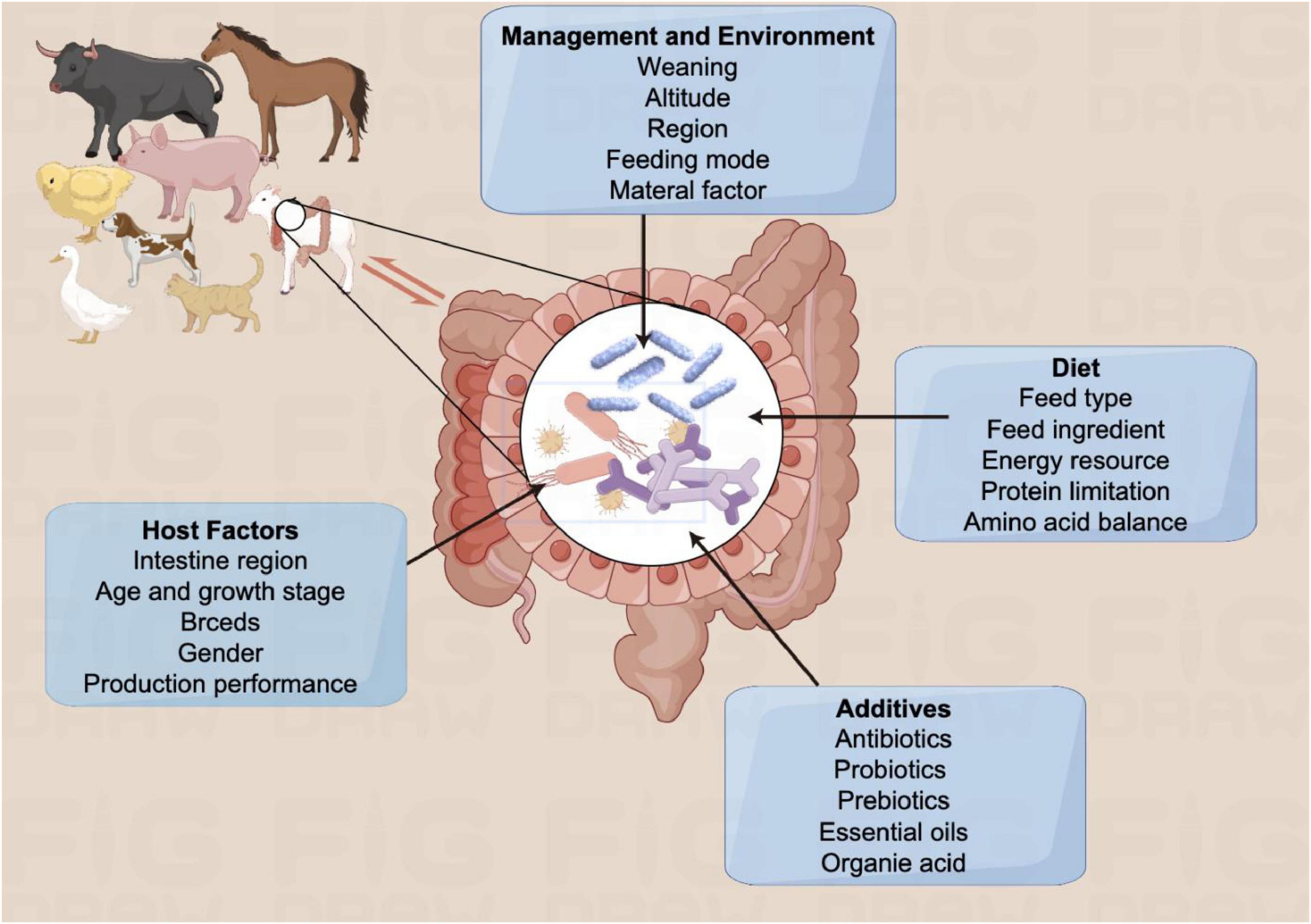
Figure 1. Bidirectional effects between animal and gut microbiota. Gut is an important immune organ of the animal organism, in which gut microbiota can be affected by various factors, such as host factors, additives, diet, management and environment etc.
Previous studies showed the importance of early manipulation of gut microbiota in young animals, especially during the postnatal and early weaning stages, to enhance immunity. Moreover, the prompt colonization of gut microbiota plays a vital roles in developing intestinal barrier function and the immune system maturation (Torow and Hornef, 2017). However, gut microbial dysbiosis may cause various diseases such as neurological, cardiovascular, gastrointestinal issues, and even cancer can arise. Diarrhea is a common consequence of gut microbial dysbiosis, affecting animal health. Therefore, effective preventive measures to reduce diarrhea incidence in animals are crucial in practical production.
Animal diarrhea is a prevalent issue in modern large-scale animal husbandry, affecting various species, particularly young animals, and posing a significant threat to animal health and productivity. Research indicated that young animals such as calves, piglets, and lambs are more susceptible to diarrhea because of their immature immune systems and low resistance levels. In large-scale pig farms, piglet diarrhea rates can exceed 50%, causing a mortality rate of at least 15%. Research conducted by the National Dairy Animal Health Surveillance Program indicates that diarrhea accounts for 57% of deaths among weaned calves, leading to a 38% reduction in net income (Zhang et al., 2022).
Diarrhea in newborn animals can hinder growth and decrease reproductive and milk production in later stages (Du et al., 2023). To address this issue, practitioners employ various strategies to prevent and manage diarrhea. However, China has implemented a policy banning the use of antibiotics in animal feed. However, the complexity of the pathogenesis of diarrhea poses a challenge for effective prevention and control of diarrhea outbreaks. Diarrhea could be divided into two main categories: infectious and non-infectious. Non-infectious factors encompass causes such as stress from weaning, environmental pressures, and mycotoxicosis. Conversely, infectious factors are linked to pathogenic microorganisms, including bacteria, viruses, and parasites.
Presently, research on the causes and mechanisms of diarrhea has demonstrated the potential for reducing incidence through early interventions such as probiotics, prebiotics, herbal additives, and polysaccharides to maintain a balanced gut microbiota (Saha et al., 2024; Yang et al., 2024). Furthermore, fecal microbiota transplantation (FMT) has demonstrated effectiveness in alleviating diarrhea. This review provides a comprehensive analysis of various etiologies and mechanisms of diarrhea, as well as the strategies for modulating gut microbiota and their potential mechanisms, aiming to offer valuable insights for prevention and control of diarrhea.
Gut microbiota
Composition of the gut microbiota
The gut microbiota, a diverse microbial community consisting of bacteria, fungi, archaea, viruses, and protozoa, plays a key roles in maintaining the intestinal structural integrity, immune function, and metabolic balance. Microbial density and diversity increase along the gastrointestinal tract due to varying levels of gastric acid, nutrients, oxygen, and antimicrobial peptides. Recent research indicates a 1:1 ratio of microorganisms to human cells in the body, with the majority residing in the colon (Jordan et al., 2019). Studies on ducks, broilers, laying hens, and piglets have indicated that livestock and poultry gut microbiota are primarily composed of Phylum Firmicutes and Bacteroidetes, with unique variations at the genus level (Wei et al., 2025; Altintas et al., 2025). Monogastric animals like pigs, chickens, and rabbits typically have dominant genera such as Anabacterium, Clostridium, Bifidobacterium, Lactobacillus, and others, with species abundance influenced by factors such as geography, diet, and nutrition (Ley et al., 2008; Sergeant et al., 2014).
Mechanism of gut microbiota in maintaining intestinal homeostasis
The intestinal barrier consists of four primary components: the microbial barrier, mechanical barrier, chemical barrier, and immune barrier (Figure 2). The microbial barrier, primarily composed of anaerobic bacteria, plays a crucial role in resisting the colonization and growth of harmful pathogens within the intestine. Beneficial gut bacteria enhance the microbial barrier by tightly binding to intestinal epithelial cells, outcompeting pathogenic bacteria for nutrients, and producing protective substances like organic acids and antimicrobial peptides. The mechanical barrier, formed by intact intestinal mucosal epithelial cells and tight junctions, acts as a physical defense against bacterial invasion. Beneficial gut microbiota supports the mechanical barrier by promoting epithelial cell development, aiding in mucosal repair, and facilitating mucin synthesis and secretion. Mucins, large glycoproteins secreted by intestinal cells, create a protective barrier that hinders the passage of irritants, toxins, and pathogens, safeguarding intestinal epithelial cells from damage. Bifidobacterium and Lactobacillus are particularly important for maintaining the integrity of the intestinal mucosal barrier, a key component of the body’s defense system against external antigens. Compromise of this barrier can lead to systemic inflammation. Various components, including gastric acid, bile, enzymes, and organic acids produced by normal gut microbiota, contribute to the chemical barrier. The immune barrier involves intestinal epithelial cells, immune cells, and molecules distributed throughout the mucosa and gut-associated lymphoid tissue (GALT). These barriers work together to maintain intestinal homeostasis. Within the intestinal mucosa, Peyer’s patches, innate and adaptive immune cells, dendritic cells, macrophages, antimicrobial peptides, and immunoglobulin A (IgA) are involved in immune defense mechanisms. Intestinal lymphoid tissues can produce cytokines and antimicrobial peptides to protect against microbial invasion. Additionally, some microbial metabolites have been shown to inhibit the virulence factors of pathogens such as Salmonella, thereby contributing to the maintenance of a healthy intestinal ecology (Antunes et al., 2014).
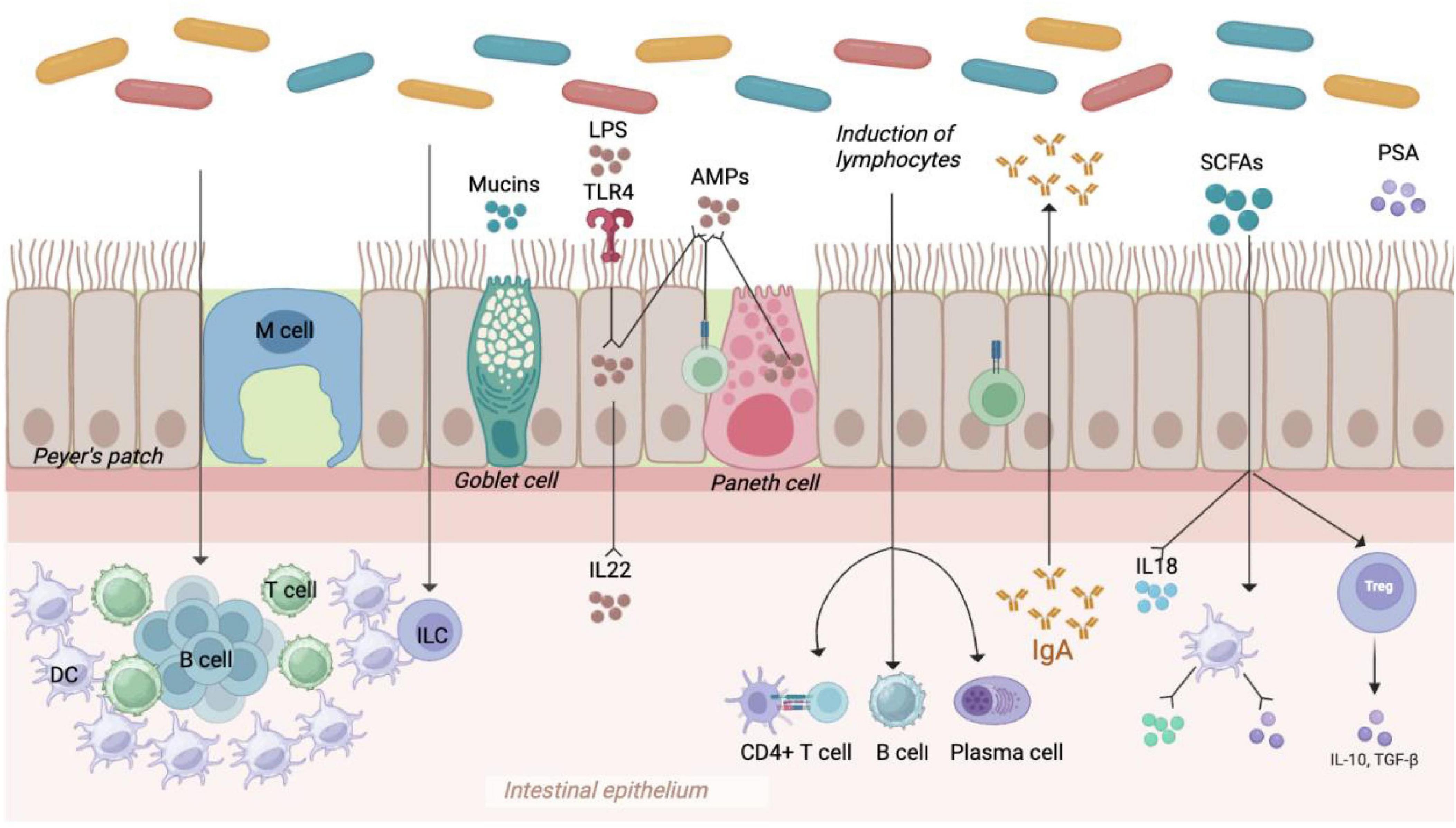
Figure 2. Gut microbiota in the intestinal barrier. Gut barriers include microbial barriers (composed of abundant intestinal flora), mechanical barriers (composed of multifunctional intestinal epithelial cells), and immune barriers (composed of immune cells and cytokines, etc.), which work together to maintain intestinal homeostasis.
Gut microbiota are essential for maintaining the intestinal barrier and promoting lipolysis (Round et al., 2010; Olszak et al., 2012). Moreover, they are also key players in the development and function of both innate and adaptive immune systems. Notably, gut microbiota has also been shown to be closely related to the intestinal immune maturation. Commensal bacteria in the gastrointestinal tract contribute significantly to the development of the mucosal immune system, which helps maintain the integrity of the intestinal barrier and triggers a natural immune response by recognizing specific receptors. The interaction between innate immunity and gut microbiota is crucial for shaping adaptive immunity (Lundin et al., 2008; Watanabe et al., 2012). Studies indicated that gut microbiota influences the function of dendritic cells (DCs), which are crucial for activating T cells and promoting immune responses (Tan and O’Neill, 2005). Intestinal DCs can drive the differentiation of T- helper cells into subsets that secrete specific cytokines, thereby activating other immune cells like B cells or macrophages. Several research has revealed the symbiotic relationship between microbial communities and intestinal immune cells. Gut microbial dysbiosis can impact the activation of Toll-like receptors (TLRs), decrease the secretion of soluble immunoglobulin A, and modify mucosal innate immunity. Gut microbial dysbiosis can also hinder the differentiation of T cell subtypes and the production of intestinal epithelial cytokines, leading to reduced adaptive immune responses in the intestinal mucosa (Watanabe et al., 2012). The gut microbiota plays a vital role in stimulating both innate and adaptive immunity, such as the production of cytokine IL-22 that enhances the production of antimicrobial peptide AMP by acting on epithelial cells. Furthermore, pattern recognition receptors (PRRs) are a key component of innate immunity, which could recognize and response to various microbial components, such as peptidoglycan, lipopolysaccharide, and formylated peptides (Rooks and Garrett, 2016). The initiation of pattern recognition receptors (PRRs) leads to the secretion of chemokines, cytokines, and apoptotic factors via a signaling cascade involved in disease mechanisms. For instance, the microbial polysaccharide PSA is capable of directly entering the circulatory system by the host intestinal epithelial cells (Watanabe et al., 2012).
In the circulatory system, PSA facilitates the interaction between dendritic cells (DCs) and T cells through MHC class II molecules and T cell receptors (TCRs) to inhibit inflammation. Within the host’s genito-urinary tract, host defense peptides (HDPs) enhance the release of pro-inflammatory cytokines by immune cells in the host via the TIFA/TRAF6/NF-κB signaling pathway. Additionally, formyl peptides modulate the inflammatory response of neutrophils by binding to formyl peptide receptor 1 (FPR1), which is expressed on neutrophils (Blyth et al., 2020).
Diarrhea induces alterations in the gut microbial composition
Gut microbial dysbiosis poses a threat to the host’s health, potentially leading to systemic diseases like obesity, diabetes, and cancer in addition to diarrhea. Previous studies indicated that diarrhea can significantly reduce the gut microbial diversity and alter their taxonomic composition, causing a decrease in beneficial bacteria. Various studies have explored the specific changes in gut microbiota during animal diarrhea, offering insights for preventing and treating diarrhea by targeting the gut microbiota (Li et al., 2022). In piglets, the most common genera in the gut microbiota include Lactobacillus spp., Shigella spp., Enterococcus spp., Bacteroides spp., and Clostridium spp. Healthy piglets typically exhibit a predominance of Lactobacillus, while diarrheic pigs show increased levels of E. coli-Shigella and Enterococcus, and decreased levels of Bacteroides. The composition of the microbiome varies across different parts of the intestine, with Lactobacillus being most prevalent in the ileum, and Clostridium and Bacteroides being more common in the rectum (Gryaznova et al., 2022). At the phylum level, healthy calf herds exhibit high abundances of Firmicutes, Bacteroides, Aspergillus, Clostridium, and Actinobacteria. Diarrhea is associated with an increase in Aspergillus and Clostridium, and a decrease in Bacteroides and Actinobacterium (Shen et al., 2023). Furthermore, significant alterations in the alpha and beta diversities of the gut fungal community have been observed in diarrheic horses, leading to marked changes in taxonomic composition. Although healthy and diarrheic horses share major fungal phyla like Neocallimastigaceae, Ascomycetes, and Basidiomycota, the relative abundance of these phyla differs between the two groups. The composition of dominant fungal genera in diarrheic horses differed significantly from that of healthy horses. Metastats analysis revealed a significant decrease in various fungal phyla during the period of diarrhea. Furthermore, a total of 175 fungal genera were identified in the gut fungal community of healthy and diarrheic horses, with 4 fungal genera showing a significant increase and 171 bacterial genera showing a significant decrease during diarrhea. Among the reduced bacteria, 74 fungal genera were completely absent from the intestine (Li et al., 2022).
Diarrhea
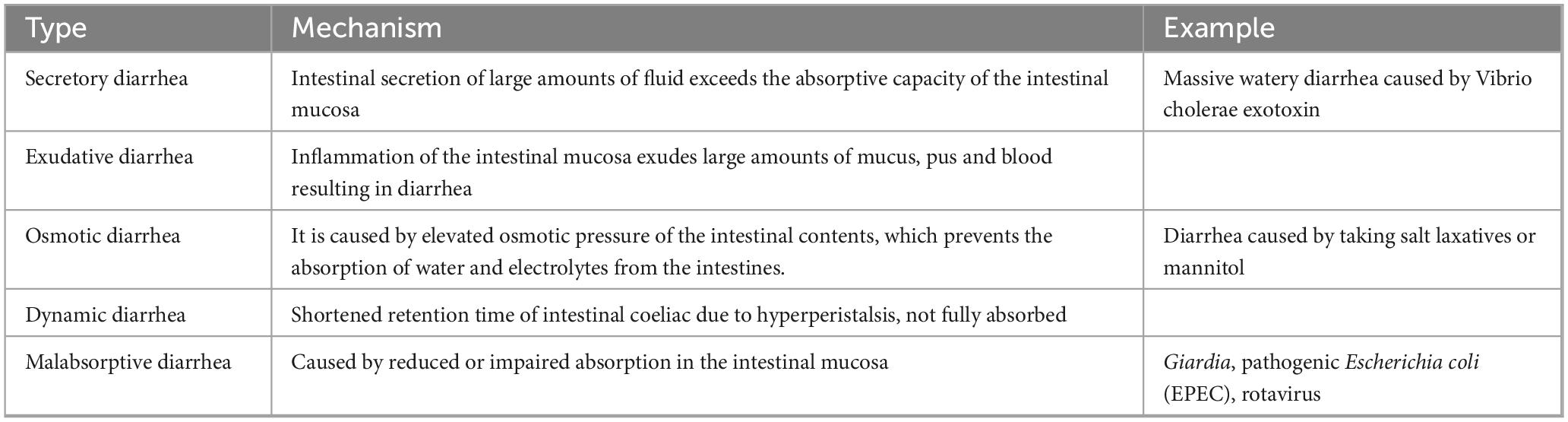
The etiology of diarrhea can be classified into infectious and non-infectious diarrhea. Moreover, it can be classified as secretory, exudative, hyperactive, osmotic, and malabsorptive diarrhea depending on the underlying mechanism (Table 1).
Non-infectious diarrhea
In animal production and management, various factors including malnutrition, environmental changes, autoimmune diseases, and management practices cause stress in animals in addition to the infections of pathogenic bacteria. This stress can disrupt intestinal homeostasis and cuase diarrhea. For instance, weaning stress in piglets can suppress their immune response, affect digestive physiology, and reduce the activity of digestive enzymes, leading to diarrhea. Other causes of diarrhea include metabolic issues due to an imbalanced nutritional ratio in feed and the presence of mycotoxins. Young animals, characterized by underdeveloped body functions, weak digestive capabilities, poor gastrointestinal function, and low resistance, are particularly susceptible to bacterial, viral, and parasitic infections that can induce diarrhea. Secretory diarrhea, a prevalent type of diarrhea, occurs when there is an imbalance between the absorptive and secretory functions of the intestines. This imbalance results in excessive fluid secretion, primarily driven by Cl– secretion, which exceeds the absorptive capacity of the intestines, leading to increased water content in the feces. Secretory diarrhea commonly arises from infections caused by pathogenic bacteria and viruses, exposure to allergens, disruption of bile acid homeostasis, or as a side effect of various medications (Keely and Barrett, 2022).
Infectious diarrhea
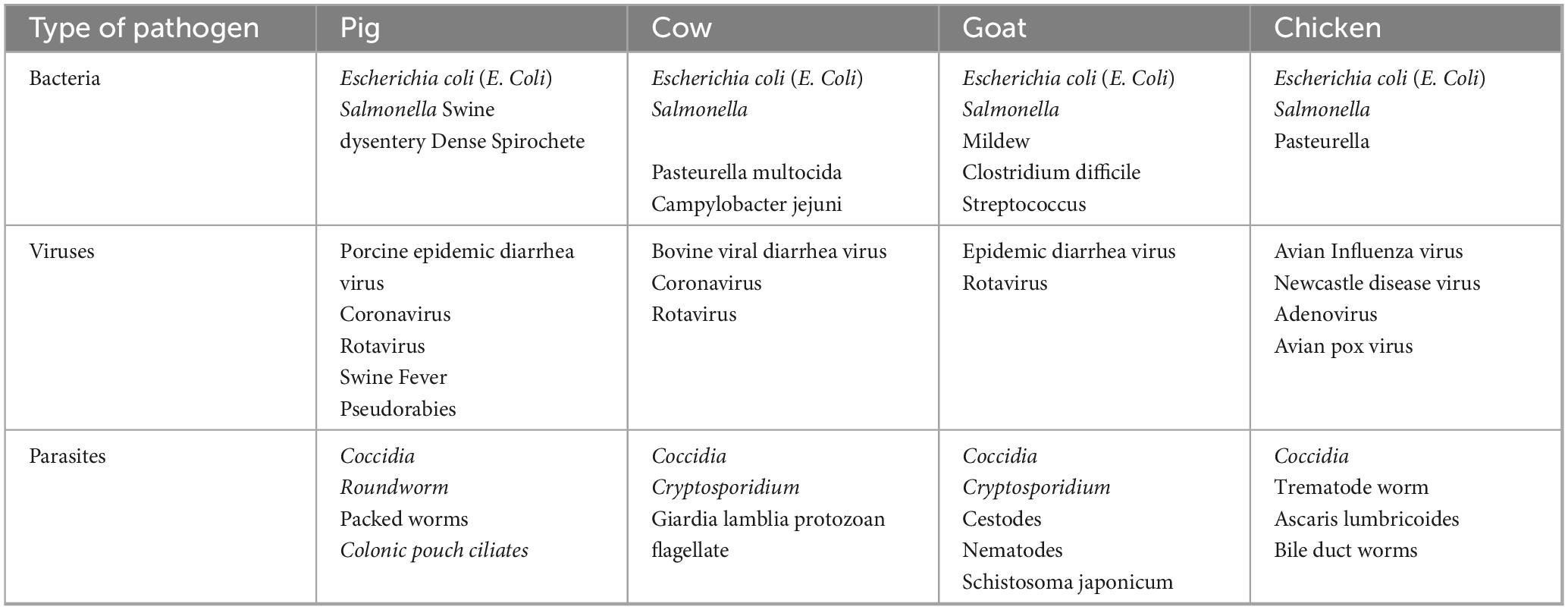
The etiology of infectious diarrhea is diverse, typically involving bacteria, fungi, viruses, and parasites. These intestinal pathogens can directly impact epithelial ion transport processes and barrier function, causing diarrhea. Moreover, they may indirectly cause intestinal dysfunction by inflammation, neuropeptides, or loss of absorptive surfaces in the gut, causing diarrhea. For instance, pathogens like the intestinal parasite Giardia flagellata can result in the loss of the absorptive surface of the brush border and diffuse shortening of the villi. Similarly, enteropathogenic Escherichia coli (EPEC) can lead to the loss of intestinal microvilli, reducing the surface area for nutrient absorption and causing increased osmotic pressure and malabsorption of intestinal contents. Various bacteria, viruses, and parasites can induce diarrhea in animals, as illustrated in Table 2. Notably, different pathogens exhibit varying levels of pathogenicity in animals, resulting in a diverse range of clinical symptoms. For example, E. coli is known to trigger diarrhea in pigs, cattle, and sheep, whereas chickens infected with E. coli typically display symptoms such as hepato-pancreatitis and pericarditis.
Bacteria and diarrhea
Escherichia coli, Shigella, Salmonella, Campylobacter, Clostridium difficile, and Aeromonas are the main pathogens that cause diarrhea. Previous studies indicated that the rapid onset of Enteropathogenic Escherichia coli (EPEC)-induced diarrhea may be attributed to a direct impact on intestinal epithelial ion transport processes (Croxen et al., 2013). Invasive pathogens like Shigella and Salmonella species lead to inflammatory diarrhea characterized by fever and the presence of polymorphonuclear cells (PMN) in feces. PMN could regulate absorption through cytokine secretion and induce diarrhea by secreting adenosine precursors, which activate CFTR (cystic fibrosis transmembrane conductance regulator). Additionally, Clostridium difficile and Clostridium rotundum infections indirectly affect ion transport through cytokine secretion and by stimulating enteric nerves via neuropeptides. Escherichia coli are common opportunistic pathogens in the large intestine and are typically non-pathogenic. However, the acquisition of virulence-encoding genes can render them pathogenic to the host. Pathogenic E. coli strains can be categorized as enterotoxin-producing (ETEC), enteropathogenic (EPEC), adherent-invasive (AIEC), entero-invasive (EIEC), diffuse adherent (DAEC), enteroaggregative (EAEC), and Shiga toxin-producing (STEC) strains, including enterohemorrhagic (EHEC). Some of these types of E. coli can cause diarrhea in various animals, such as ETEC in piglets, calves, and lambs, EPEC in rabbits, dogs, cats, pigs, calves, and lambs, and Shiga toxin-producing E. coli in calves and piglets (Aydin et al., 2022). So far, EAEC, EIEC, and DAEC have not been identified as pathogens causing animal diarrhea (Croxen et al., 2013). Piglets infected with E. coli-associated diarrhea typically exhibit feces that are white-gray or yellow, often characterized by yellow bubbles and a fishy odor. Yellow diarrhea is observed in piglets during the first week of life, referred to as early onset, while white diarrhea is more common in piglets aged between 1 and 4 weeks, known as late onset. Infection with enterotoxin-producing Escherichia coli (ETEC) F4 + disrupts the intestinal epithelial barrier, decreases the number of goblet cells, and lowers the total short-chain fatty acid (SCFA) levels in the colon (López-Colom et al., 2020). In calves, E. coli infection induces diarrhea by disrupting tight junctional proteins in the intestines and releasing cell-damaging toxins that harm the small intestinal mucosa, leading to mucosal inflammation (Wu et al., 2022). Invasion of pathogenic bacteria triggers the disruption of tight junction proteins and activation of inflammatory signaling pathways, such as I-kappa-B-alpha, mitogen-activated protein kinase, and nuclear factor-κB. The invasion additionally prompts small intestinal epithelial cells to produce IL-8, IL-6, and TNF-α, which causes an inflammatory response in calves (Gandhar et al., 2022).
Furthermore, weaning stress can increase serum cortisol levels, potentially compromising cellular immunity and suppressing various non-specific immune responses, thereby increasing susceptibility to diseases (Kim et al., 2020). This stress also leads to elevated blood levels of neutrophils, TNF-α, IL-6, and IL-1β, alongside increased expression of apoptotic and pro-inflammatory factors, including IL-1β, IL-8, interferon-gamma, TNF-α, and TLR-4. Collectively, these factors trigger an inflammatory response in the calf intestine (O’Loughlin et al., 2011). Infection of lambs with E. coli NMGCF-19 results in severe diarrhea and neurological disorders, research on mice models showed a significant decrease in ZO-1 and occludin expression in mice brain tissue. Furthermore, NMGCF-19 infection induces upregulation of pro-inflammatory cytokines TNF-α, IL-6, IL-1β, and IL-18, higher expression levels of HMGB1, and activation of the TLR2/TLR4/MyD88 and NLRP3 inflammasome pathways (Wang et al., 2020).
Salmonella, a gram-negative intracellular pathogen, is classified based on its lipopolysaccharide (LPS) O antigen and flagellar H antigen, with over 2,600 serotypes identified to date (Lamichhane et al., 2024). It is a significant cause of infectious diarrhea and a prominent zoonotic pathogen worldwide, posing significant public health risks and economic losses in livestock (Quach et al., 2022). Salmonella infections typically spread through fecal-oral contamination among livestock, often originating from contaminated feed, water, equipment, and inadequate hygiene practices among farm workers. Furthermore, certain serotypes, such as Salmonella Dublin, can be transmitted via aerosols in confined captive calves. Studies indicate that Salmonella-induced diarrhea may result from the inhibition of the Notch signaling pathway, which leads to a transition in intestinal cells from absorptive to secretory types, ultimately diminishing cellular absorption and resulting in diarrhea (Diepold and Armitage, 2015). In piglets, Salmonella infection is characterized by persistent watery feces, which are often yellowish-green or dark brown, and contain necrotic intestinal mucosal tissue, emitting a foul odor. This condition can lead to growth retardation and, in severe cases, mortality. In sheep, Salmonella infections are frequently associated with sheep enterotoxemia, exhibiting the highest infection and mortality rates in lambs, along with severe diarrhea in pregnant ewes. The Salmonella type III protein secretion system (T3SS) plays an important role in bacterial invasion of host cells and subsequent replication (Diepold and Armitage, 2015). The invasion process involves the translocation of effector proteins such as SipA, SipB, SipC, and SopB into the host cell, leading to cytoskeletal remodeling that facilitates Salmonella cell invasion (Lou et al., 2019). SipA is the key protein involved in bacterial invasion, with SipB forming protein complexes with SipC to promote host cell invasion by S. typhimurium (Myeni et al., 2013). Structural components of T3SS-1 are essential for Salmonella invasion of bovine and porcine ileal mucosa, with specific effector proteins (SipA, SipC, SopE/E2, SopB, SopD, and SptP) playing roles in manipulating actin dynamics (Shi et al., 2025). These effector proteins can mimic eukaryotic guanine nucleotide exchange factors (GEFs) or produce secondary messengers to activate host GEFs and Rho family GTPases, ultimately affecting actin assembly and cellular structure (Singh et al., 2020). Additionally, SptP acts as a mimic of host GTPase-activating proteins to deactivate Cdc42 and Rac1, reversing signaling pathways initiated by SopB, SopE, and SopE2 (Ly and Cyert, 2017). Salmonella infections occur through fecal-oral transmission, overcoming various intestinal barriers to interact with the intestinal epithelium and penetrate host tissues (Gopinath et al., 2012; Barbara et al., 2021). Previous studies showed that Salmonella attaches to epithelial tissues and disrupts cellular junctions, ultimately causing intestinal inflammation and diarrhea (Wang et al., 2022; Zhang et al., 2012). Therefore, preserving the integrity of epithelial tissue could potentially reduce the pathogenicity and severity of Salmonella infection.
The transmission of Clostridium difficile (C. difficile) occurs via the fecal-oral route, leading to a spectrum of symptoms ranging from asymptomatic cases to severe diarrhea, which can culminate in life-threatening conditions. The normal gut microbiota serves as a protective barrier against C. difficile infection (CDI). Disruption of this balance allows C. difficile to colonize the large intestine and become predominant. CDI is increasingly recognized as a significant cause of enteritis in neonatal piglets, potentially resulting in growth retardation, delayed weaning, and mortality in swine. Additionally, it can cause colitis in large birds such as ostriches. The primary virulence factors are two toxins, A (TcdA) and B (TcdB). Research has indicated that toxin A primarily drives the pathogenic process, with toxin B exerting its effects following the tissue damage caused by toxin A. These toxins disrupt the intestinal epithelial cytoskeleton and tight junctions, increase fluid secretion, promote neutrophil adhesion, and ultimately compromise the integrity and function of the intestinal barrier (Czepiel et al., 2019). Most C. difficile strains isolated from animals also produce a binary toxin (C. difficile transferase) that enhances virulence. Toxins are transported to the cytoplasm where they inactivate the Rho family of GTPases. Rho proteins are crucial for actin polymerization and maintaining cytoskeleton stability. When Rho proteins are inactivated, cytoskeletal stability diminishes, exacerbating the inflammatory process. In severe cases, piglets infected with C. difficile may develop watery, yellow, pasty, non-hemorrhagic diarrhea, with micro-ulcers and pseudomembranes on the intestinal mucosa.
Campylobacter jejuni, a gram-negative bacterium with a spiral or rod-shaped form, is recognized for causing fever, bloody diarrhea, and inflammatory diarrhea, similar to Salmonella and Shigella (Giallourou et al., 2018). Aeromonas, a genus of anaerobic gram-negative bacillus, produces oxidizing enzymes, catalase, and other exoenzymes. Clostridium perfringens infection in piglets leads to pink and reddish-brown feces, known as necrotizing enteritis, with high mortality rates in 1-week-old piglets. Swine dysentery and Lawsonella intracellularis can also cause diarrhea in pigs (Burrough, 2017; Campillo et al., 2021).
Parasitic infestation and diarrhea
Intestinal parasites represent a significant cause of diarrhea, often underestimated in terms of their public health implications. These parasites can profoundly affect the intestinal physiology and immune system of the host, leading to intestinal damage and disrupting the relationship between gut microbiota and host immunity. Common parasites, including Coccidia, Cryptosporidium, Roundworms, Whipworms, and Colonic pouch ciliates, can induce diarrhea by compromising the intestinal mucus and epithelial barriers. Furthermore, they impact both the innate and adaptive immunity of the host while altering the gut microbiota, thereby affecting the overall intestinal environment.
For instance, unicellular protozoa of the phylum Apicomplexa, such as Eimeria spp., cause severe infections in livestock, particularly in poultry and cattle (Morgoglione et al., 2020). Avian coccidiosis alone has caused over $3 billion in economic losses globally to the poultry industry (Balta et al., 2021). These protozoa not only affect gastrointestinal function and resident microbial composition but also contribute to mucosal damage, which can predispose individuals to necrotizing enterocolitis and subsequent bacterial infections like Salmonella typhimurium and Clostridium perfringens (Macdonald et al., 2019; McKnight et al., 2019). In calves, Eimeria bovis and Eimeria kewi are common and highly pathogenic, with Eimeria bovis inhibiting NF-κB activation, impairing gene expression of immunomodulatory molecules, regulating apoptosis and cholesterol metabolism, and reducing cytoskeletal integrity. These effects lead to acute hemorrhagic diarrhea, dehydration, weight loss, and a significant decrease in growth rate (Velásquez et al., 2021).
Following infection with Cryptosporidium, epithelial cells release pro-inflammatory cytokines and chemokines to the infection site, potentially causing increased epithelial permeability, impaired intestinal absorption, and increased secretion. In cases of intestinal cryptosporidiosis, villous atrophy and crypt hyperplasia have been identified as factors contributing to impaired monosaccharide and glucose-Na absorption (Certad et al., 2017). It has been proposed that the down-regulation of specific key components of tight junctions (TJ) and adherents junctions (AJ) to disrupt barrier function may play a significant role in diarrhea resulting from Cryptosporidium infection (Kumar et al., 2018). Cryptosporidium is commonly found in conjunction with other pathogens, such as Cryptosporidium minis, Cryptosporidium bovis, Cryptosporidium anserineum, and Cryptosporidium ruber. Cryptosporidium infection induces cytoskeletal changes that control actin reorganization and the insertion of channel/transporter proteins (Meremikwu et al., 2012). Numerous studies have indicated that the infection of intestinal and biliary epithelial cells necessitates host cell actin polymerization and cytoskeletal remodeling (Färnert et al., 2014). This polymerization involves the actin branching and nucleation mechanisms of the Arp2/3 complex protein (Snow et al., 2015). Multiple signaling pathways have been demonstrated that regulate actin reorganization and the internalization of Cryptosporidium, including PI3K, guanine exchange factor, and CDC42- and c-Src-dependent activation of corticosterone (Meremikwu et al., 2012). Cryptosporidium activates NF-κB to inhibit apoptosis in biliary epithelial cells, which is crucial for cell survival. NF-κB plays a role in activating various intracellular survival signals, including the C-myc oncogene (Carneiro et al., 2010). A study conducted on Cryptosporidium-infected human (Caco-2) and bovine (MDBK and NBL-1) epithelial cells in vitro revealed disruption of occluded ZO-1 and fragmentation of the nucleus during infection (Roca-Feltrer et al., 2010).
Giardia infection disrupts, reduces expression, and relocates tight junctions and cytoskeletal proteins such as ZO-1, claudin-1, F-actinin, and α-actinin, resulting in increased intestinal permeability and decreased trans-epithelial resistance. This disruption can lead to paracellular leakage and exudative diarrhea. Giardia flagellates interfere with tight junctions without penetrating the epithelium, causing secretory diarrhea characterized by elevated chloride ion concentrations, loss of absorptive function (including microvilli atrophy and increased cell death), heightened secretion (due to epithelial barrier disruption), and inadequate disaccharidase activity and nutrient malabsorption stemming from the loss of the microvillar brush border and villi atrophy, ultimately resulting in osmotic diarrhea.
Viruses and diarrhea
Common clinical viruses that can cause diarrhea in animals include rotavirus, coronavirus, and epidemic diarrhea virus. The mechanisms underlying viral diarrhea are primarily secretory and exudative. Rotavirus predominantly affects the small intestine of pigs, damaging intestinal villi and resulting in acute diarrhea, dehydration, and mortality in piglets (Chepngeno et al., 2020). In calves, bovine rotavirus (BRV) can induce acute diarrhea within 12–24 h post-infection, with substantial amounts of the virus excreted in feces, thereby facilitating transmission (Jang et al., 2019). The infection mechanism involves viral replication in small intestinal epithelial cells, leading to destruction of mature cells, activation of the enteric nervous system, and secretion of viral enterotoxin (Kayasaki et al., 2021). Additionally, rotavirus infection affects the diversity, homogeneity, and abundance of gut microbiota. Sheep and alpacas are also susceptible to rotavirus infection, which can result in diarrhea (Rojas et al., 2016; Shabana et al., 2017).
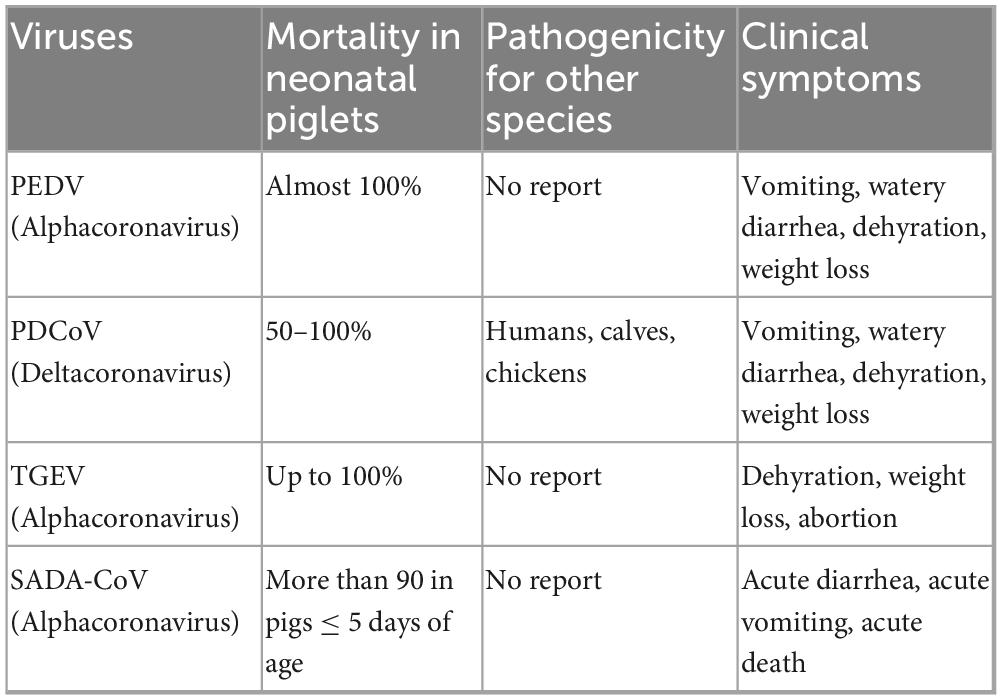
Porcine enteric coronaviruses (PECs) such as PEDV, PDCoV, TGEV, and SADS-CoVare known to cause severe diarrhea in young pigs, posing a significant threat to the global swine industry (Table 3). These viruses primarily target the digestive system of piglets, resulting in symptoms such as weight loss, lethargy, vomiting, loss of appetite, watery diarrhea, and, in severe cases, death. The pathological effects include necrosis and detachment of enterocytes, as well as damage to intestinal villi (Pan et al., 2017; Suzuki et al., 2018).
Studies have indicated that PEDV S1 facilitates viral invasion through interaction with EGFR, triggering the EGFR-regulated downstream EGFR/ERK signaling pathway. This cascade results in decreased NHE3 expression, impaired NHE3 migration at the plasma membrane, and ultimately reduced NHE3 activity (Zhang et al., 2023c). The downregulation of NHE3 expression in intestinal epithelial cells is believed to play a significant role in the development of PEDV-induced diarrhea in neonatal piglets. Infection with porcine intestinal coronavirus SeCoV induces autophagy, apoptosis, and innate immune responses, with the intricate interplay between these responses potentially impacting viral replication or modifying specific signaling pathways to induce pathological damage in SeCoV-infected hosts. Coronaviruses can also infect cattle, with bovine coronavirus (BCV) causing calf enteritis in dairy and beef cattle. Animals typically affected are usually aged between 1 day to 3 months and diarrhea occurs within 1–2 weeks after birth, reaching its peak between days 7 and 10. Calves can become infected with BCV via their oral and respiratory routes, exhibiting clinical symptoms around 2 days after infection, which typically persist for a duration of 3–6 days (Hodnik et al., 2020).
Calves, particularly those that do not receive colostrum, are at an increased risk of severe diarrhea due to secondary coronavirus infections. These infections typically originate in the upper portion of the small intestine and subsequently spread throughout the entire small intestine (Vlasova and Saif, 2021). The virus utilizes spiny projections and hemagglutinin glycoproteins to attach to the epithelial cells of the intestine, leading to the fusion of the viral envelope with either the host cell membrane or intracellular vesicles. Following this, the virus is released to replicate and induce cell death via normal secretory mechanisms, ultimately resulting in diarrhea. Bovine viral diarrhea virus (BVDV) is recognized as a causative agent of diarrhea in cattle, with 21 subtypes A (BVDV 1a-u), 4 subtypes B (BVDV 2a-d), and 4 subtypes H (BVDV 3a-d) identified. BVDV has the capability to cross the placental barrier during early gestation, which can lead to persistent infections in calves. Managing these infected calves poses significant challenges, as they continuously harbor and shed the virus throughout their lives, becoming the primary reservoir for BVDV infection. In addition to the aforementioned pathogens, other viruses such as bovine astrovirus, bovine circovirus, bovine norovirus, bovine enterovirus, and bovine microvirus may also contribute to diarrhea in calves. Moreover, BCoV and bovine-like CoV have been identified in various domestic and wild ruminants, as well as in dogs and humans (Vlasova and Saif, 2021).
Improving the gut microbiota alleviates symptoms of diarrhea
Diarrhea ranks among the foremost reasons for death in young animals, such as calves, piglets, and lambs. Due to their immature immune systems, these young animals are more vulnerable to deterimental factors such as bacteria, viruses, and stress, which may disrupt the gut microbial balance and result in diarrhea. Therefore, interventions such as dietary supplementation with probiotics, prebiotics, herbal additives, and polysaccharides are effective in maintaining gut microbial balance and alleviating diarrhea. Furthermore, fecal microbiota transplantation (FMT) has also shown promise in reducing the incidence of diarrhea.
Probiotics and their metabolites
Probiotics play a key role in regulating the gut microbial homeostasis, enhancing resistance to intestinal pathogens by maintaining a healthy gut microbial balance (Figure 3). They produce metabolites that can inhibit pathogenic bacteria, interfere with microbial virulence gene expression, and regulate host gene expression (Önlü et al., 2025). Additionally, probiotics modulate intestinal immune function and effectively treat pathogen-induced diarrhea. The active ingredients of probiotics include antimicrobials, vitamins, peptides, biosurfactants, extracellular polysaccharides, and enzymes. The structural features on the surfaces of probiotics, including flagella, hyphae, surface layer proteins (SLP), podocarp polysaccharides (CPS), lipophosphatidic acids, and lipopolysaccharides, function as MAMPs that interact with PRRs, such as NLRs and TLRs (Lebeer et al., 2018; Liu et al., 2020).
Figure 3. Protective mechanisms of probiotics. Probiotics can maintain intestinal microbial homeostasis in multiple ways: (1) Increasing the relative abundance of beneficial bacteria; (2) regulating T cells differentiation to relieve inflammatory reaction and immunologic derangement; (3) maintaining the intestinal barrier by regulating tight junction proteins; (4) enhancing humoral immune response and inducing anti-inflammatory effects; (5) promoting fluid absorption via the exchange of Na+ and H+ in epithelial cells, reducing the risk of antibiotic-associated diarrhea.
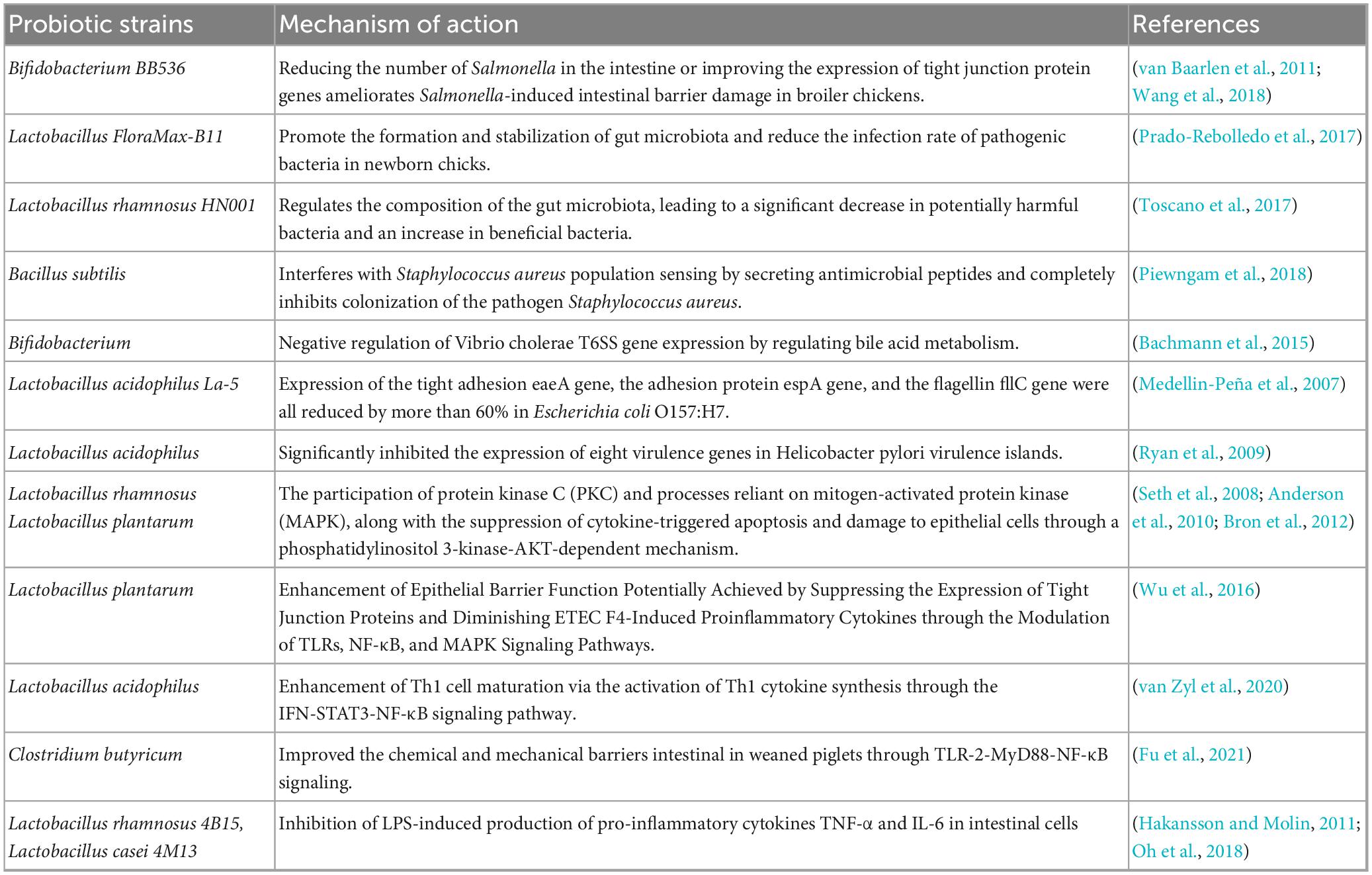
Probiotics may prevent or treat diarrhea by outcompeting pathogens for nutrients, producing antimicrobial compounds, and inhibiting harmful bacterial colonization (Endt et al., 2010). Research has demonstrated that Lactobacillus XLTG11 can reduce intestinal pathology by decreasing abundance of LPS, D-LA, and DAO, as well as improving intestinal permeability. This probiotic additionally promotes the expression of proteins involved in water channels and tight junctions, inhibits the TLR4/NF-κB signaling pathway, elevates the levels of anti-inflammatory cytokines, and diminishes the levels of pro-inflammatory cytokines. The content of acetic acid, propionic acid, butyric acid, and total short-chain fatty acids significantly increased after treatment with Lactobacillus XLTG11. Compared to the MC group, Lactobacillus XLTG11 enhanced the gut microbial abundance and diversity and induced changes in the gut microbial composition (Xu et al., 2022). Moreover, the rest of the probiotics listed in Table 4 show some effectiveness in relieving diarrhea and regulating gut microbiota.
Probiotics are capable of producing beneficial metabolites, including prebiotics such as oligofructose, inulin, and pectin oligosaccharides. These metabolites function as soluble decoy receptors, which help prevent pathogen colonization and facilitate the elimination of pathogens from the intestine (Rigo-Adrover et al., 2017). Furthermore, metabolites like secreted proteins, indoles, and short-chain fatty acids produced by probiotics play a crucial role in protecting the intestinal barrier. Notably, butyrate synthesized by Clostridium butyricum improves the immune function and intestinal barrier by increasing the levels of hypoxia-inducible factor (HIF-1α), mucins, antimicrobial peptides, and IL-22 in the intestines (Pral et al., 2021). Indole-3-lactic acid produced by Bifidobacterium infantis and Lactobacillus rhamnosus activates aryl hydrocarbon receptors (AhRs), which enhances the expression of CYP1A1. This process ultimately promotes the transcription of IL-22 and the expression of antimicrobial peptides (Ehrlich et al., 2020; Hou et al., 2021). Additionally, the soluble proteins P40 and p75 derived from Lactobacillus rhamnosus activate EGFR, promoting IgA secretion and preserving intestinal homeostasis via the EGFR-PIK3-Akt signaling pathway (Wang et al., 2017). The proliferation of short-chain fatty acid-producing microorganisms in the hindgut of weaned piglets reduces intestinal pro-inflammatory factors and enhances the intestinal immune barrier through the MyD88-NF-κB signaling pathway (Tian et al., 2022).
Prebiotics and postbiotics
Oligofructose, oligogalactose, oligo xylose, oligo isomaltose, soybean oligosaccharides, and inulin are commonly used prebiotics. Certain microalgae, such as spirulina and other algae, can also function as prebiotics. Additionally, polysaccharides (e.g., Yunzhi polysaccharides, carrageenan polysaccharides with nitrogen), protein hydrolyzates (e.g., casein hydrolyzates, alpha lactalbumin, lactoferrin), and natural plants from vegetables, herbs, and wild plants can be utilized as prebiotics. Postbiotics have garnered significant attention due to their potential applications in functional foods and pharmaceuticals. The International Society for the Science of Probiotics and Prebiotics (ISAPP) has established a standardized consensus definition for postbiotics. According to ISAPP, postbiotics are preparations of non-living microorganisms or their constituents that offer health benefits to the host. These postbiotics can include fully or partially inactivated bacteria, with or without metabolic byproducts. Various terms have been employed in published studies to refer to postbiotics, including inactive probiotic, butylated probiotic, heat-killed probiotic, cell lysate, parabiotic, ghost probiotic, and postbiotic (Salminen et al., 2021). Postbiotic formulations often include microbially produced components such as metabolites, peptides, enzymes, proteins, vitamins, and extracellular polysaccharides (EPS), which contribute to the overall health benefits of postbiotics (Deshpande et al., 2018). Postbiotic forms of probiotics can be produced through optimal application of heat, ultraviolet light, and sonication. Live probiotics may encounter difficulties adhering to the intestinal mucosa due to the barrier posed by the mucosal layer, which limits direct bacterial contact. In contrast, postbiotics can easily penetrate the mucosal layer, posing no risk of infection or transfer of antibiotic resistance genes in vulnerable individuals. This enhances the convenience of dosage maintenance, transport, standardization, and storage of postbiotics (Deshpande et al., 2018). Therefore, utilizing inactivated bacteria as a supplement may present a safer alternative to live probiotics for susceptible populations, such as neonates, potentially aiding in the treatment of gastrointestinal diseases and immune disorders (Ou et al., 2011; De Marco et al., 2018). In their inactivated form, strains like Lactobacillus and Bifidobacterium exhibit immunomodulatory effects by stimulating innate and adaptive immunity, as well as enhancing the membrane integrity of intestinal epithelial cells (Hirose et al., 2006; Adams, 2010; Sugahara et al., 2017). These effects are mediated through various signaling receptors, including TLR in dendritic cells, intestinal epithelial cells, and other immune cells (Soares et al., 2010).
FMT and diarrhea
FMT involves transferring functional microbiota from healthy donors to diseased individuals to restore normal gut microbiota and treat intestinal diseases. Early FMT intervention has been demonstrated to increase growth performance and intestinal barrier function in piglets by promoting symbiotic bacterial colonization and increasing intestinal oxygen concentration (Geng et al., 2018). Previous research indicated that feeding neonatal piglets FMT suspensions from healthy adult pigs can significantly improve growth performance and intestinal barrier function (Zeng et al., 2016). Additionally, probiotics play a key role in maintaining intestinal homeostasis, regulating gut microbiota, and reducing diarrhea. Combining early FMT intervention with probiotics has been demonstrated to enhance growth performance, reduce diarrhea, and improve intestinal barrier function by increasing the abundance of beneficial bacteria in the intestine (Xiang et al., 2020). FMT has also been linked to an increase in beneficial bacterial species and a decrease in pro-inflammatory cytokines and markers of inflammation such as CRP and fecal calreticulin (Konturek et al., 2017).
Traditional Chinese medicine
Recently, traditional Chinese medicine (TCM) has shown promising results in treating diarrhea and is widely utilized in clinical practice. The ancient theoretical framework of Chinese medicine, which dates back over 2,000 years, has demonstrated both clinical and scientific efficacy in treating various diseases. Through extensive clinical practice, TCM has developed numerous effective and safe formulas, grounded in the principles of “Jun”-“Chen”-“Zuo”-“Envoy” (Yang et al., 2023). Due to the advantages of low side effects, long-lasting efficacy, and wide range of targets of action, the composition and mechanism of action of TCM have been extensively studied in modern times (Zhang et al., 2023b; Zhu et al., 2025). Herbal formulations and monomers in TCM can alleviate diarrhea symptoms through their anti-inflammatory, antioxidant, antimicrobial, and immune-boosting properties. The inflammatory signaling pathways associated with intestinal inflammation, along with the mechanisms of action of TCM formulas and monomers in addressing this inflammation, underscore the potential application of TCM in treating diarrheal diseases in animals (Table 5).
Conclusion
Diarrhea is closely associated with alterations in gut microbiota. Therefore, it is crucial to investigate how microbial communities detect pathogen infections to enhance our understanding of the pathogenesis of diarrhea. Furthermore, given that various probiotics possess distinct properties, future research should focus on further elucidating the specific mechanisms by which probiotics contribute to the rehabilitation of beneficial microbial colonies, thereby facilitating the clearance of diarrhea-causing pathogens.
Author contributions
YZ: Writing – original draft. YM: Writing – review and editing. YQ: Writing – review and editing, Writing – original draft.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was funded by the Natural Science Foundation of Qinghai Province (2023-ZJ-964Q).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Adams, C. (2010). The probiotic paradox: Live and dead cells are biological response modifiers. Nutr. Res. Rev. 23, 37–46. doi: 10.1017/S0954422410000090
Altintas, T., Ceylani, T., Önlü, H., Sağır, E., Yılmaz, M., Bora, A., et al. (2025). Targeting gut microbiota health in aged rats through the potent strategy of probiotics supplementation during intermittent fasting. Pak. Vet. J. 45, 286–294. doi: 10.1016/j.tifs.2023.06.013
Anderson, R., Cookson, A., McNabb, W., Park, Z., McCann, M., Kelly, W., et al. (2010). Lactobacillus plantarum MB452 enhances the function of the intestinal barrier by increasing the expression levels of genes involved in tight junction formation. BMC Microbiol. 10:316. doi: 10.1186/1471-2180-10-316
Antunes, L., McDonald, J., Schroeter, K., Carlucci, C., Ferreira, R., Wang, M., et al. (2014). Antivirulence activity of the human gut metabolome. mBio 5:e01183-14. doi: 10.1128/mBio.01183-14
Aydin, O., Ulas, N., Genc, A., Baysal, S., Kandemir, O., and Aktas, M. (2022). Investigation of hemogram, oxidative stress, and some inflammatory marker levels in neonatal calves with Escherichia coli and coronavirus diarrhea. Microb Pathog. 173(Pt A):105802. doi: 10.1016/j.micpath.2022.105802
Bachmann, V., Kostiuk, B., Unterweger, D., Diaz-Satizabal, L., Ogg, S., and Pukatzki, S. (2015). Bile salts modulate the mucin-activated type VI secretion system of pandemic Vibrio cholerae. PLoS Negl. Trop. Dis. 9:e0004031. doi: 10.1371/journal.pntd.0004031
Balta, I., Marcu, A., Linton, M., Kelly, C., Stef, L., Pet, I., et al. (2021). The in vitro and in vivo anti-virulent effect of organic acid mixtures against Eimeria tenella and Eimeria bovis. Sci. Rep. 11:16202. doi: 10.1038/s41598-021-95459-9
Barbara, G., Barbaro, M., Fuschi, D., Palombo, M., Falangone, F., Cremon, C., et al. (2021). Inflammatory and microbiota-related regulation of the intestinal epithelial barrier. Front. Nutr. 8:718356. doi: 10.3389/fnut.2021.718356
Blyth, G., Connors, L., Fodor, C., and Cobo, E. (2020). The network of colonic host defense peptides as an innate immune defense against enteropathogenic bacteria. Front. Immunol. 11:965. doi: 10.3389/fimmu.2020.00965
Bron, P., van Baarlen, P., and Kleerebezem, M. (2012). Emerging molecular insights into the interaction between probiotics and the host intestinal mucosa. Nat. Rev. Microbiol. 10, 66–78. doi: 10.1038/nrmicro2690
Campillo, M., Smith, S., Gally, D., and Opriessnig, T. (2021). Review of methods for the detection of Lawsonia intracellularis infection in pigs. J. Vet. Diagn. Invest. 33, 621–631. doi: 10.1177/10406387211003551
Carneiro, I., Roca-Feltrer, A., Griffin, J., Smith, L., Tanner, M., Schellenberg, J., et al. (2010). Age-patterns of malaria vary with severity, transmission intensity and seasonality in sub-Saharan Africa: A systematic review and pooled analysis. PLoS One 5:e8988. doi: 10.1371/journal.pone.0008988
Certad, G., Viscogliosi, E., Chabé, M., and Cacciò, S. (2017). Pathogenic mechanisms of cryptosporidium and giardia. Trends Parasitol. 33, 561–576. doi: 10.1016/j.pt.2017.02.006
Chan, Y., Cheung, F., Zhang, C., Fu, B., Tan, H., Norimoto, H., et al. (2020). Ancient Chinese medicine herbal formula huanglian jiedu decoction as a neoadjuvant treatment of chemotherapy by improving diarrhea and tumor response. Front. Pharmacol. 11:252. doi: 10.3389/fphar.2020.00252
Chen, J., Shen, B., and Jiang, Z. (2022). Traditional Chinese medicine prescription Shenling BaiZhu powder to treat ulcerative colitis: Clinical evidence and potential mechanisms. Front. Pharmacol. 13:978558. doi: 10.3389/fphar.2022.978558
Chepngeno, J., Takanashi, S., Diaz, A., Michael, H., Paim, F., Rahe, M., et al. (2020). Comparative sequence analysis of historic and current porcine rotavirus C strains and their pathogenesis in 3-Day-Old and 3-week-old piglets. Front. Microbiol. 11:780. doi: 10.3389/fmicb.2020.00780
Croxen, M., Law, R., Scholz, R., Keeney, K., Wlodarska, M., and Finlay, B. (2013). Recent advances in understanding enteric pathogenic Escherichia coli. Clin. Microbiol. Rev. 26, 822–880. doi: 10.1128/CMR.00022-13
Czepiel, J., Dróżd, M., Pituch, H., Kuijper, E. J., Perucki, W., Mielimonka, A., et al. (2019). Clostridium difficile infection: Review. Eur. J. Clin. Microbiol. Infect. Dis. 38, 1211–1221. doi: 10.1007/s10096-019-03539-6
De Marco, S., Sichetti, M., Muradyan, D., Piccioni, M., Traina, G., Pagiotti, R., et al. (2018). Probiotic cell-free supernatants exhibited anti-inflammatory and antioxidant activity on human gut epithelial cells and macrophages stimulated with LPS. Evid. Based Complement. Alternat. Med. 2018:1756308. doi: 10.1155/2018/1756308
Deshpande, G., Athalye-Jape, G., and Patole, S. (2018). Para-probiotics for preterm neonates-the next frontier. Nutrients 10:871. doi: 10.3390/nu10070871
Diepold, A., and Armitage, J. (2015). Type III secretion systems: The bacterial flagellum and the injectisome. Philos. Trans. R Soc. Lond. B Biol. Sci. 370:20150020. doi: 10.1098/rstb.2015.0020
Du, W., Wang, X., Hu, M., Hou, J., Du, Y., Si, W., et al. (2023). Modulating gastrointestinal microbiota to alleviate diarrhea in calves. Front. Microbiol. 14:1181545. doi: 10.3389/fmicb.2023.1181545
Ehrlich, A., Pacheco, A., Henrick, B., Taft, D., Xu, G., Huda, M., et al. (2020). Indole-3-lactic acid associated with Bifidobacterium-dominated microbiota significantly decreases inflammation in intestinal epithelial cells. BMC Microbiol. 20:357. doi: 10.1186/s12866-020-02023-y
Endt, K., Stecher, B., Chaffron, S., Slack, E., Tchitchek, N., Benecke, A., et al. (2010). The microbiota mediates pathogen clearance from the gut lumen after non-typhoidal Salmonella diarrhea. PLoS Pathog. 6:e1001097. doi: 10.1371/journal.ppat.1001097
Färnert, A., Yman, V., Homann, M., Wandell, G., Mhoja, L., Johansson, M., et al. (2014). Epidemiology of malaria in a village in the Rufiji River Delta, Tanzania: Declining transmission over 25 years revealed by different parasitological metrics. Malar J. 13:459. doi: 10.1186/1475-2875-13-459
Fu, J., Wang, T., Xiao, X., Cheng, Y., Wang, F., Jin, M., et al. (2021). Clostridium butyricum ZJU-F1 benefits the intestinal barrier function and immune response associated with its modulation of gut microbiota in weaned piglets. Cells 10:527. doi: 10.3390/cells10030527
Gandhar, J., De, U., Kala, A., Malik, Y., Yadav, S., Paul, B., et al. (2022). Efficacy of microencapsulated probiotic as adjunct therapy on resolution of diarrhea, copper-zinc homeostasis, immunoglobulins, and inflammatory markers in serum of spontaneous rotavirus-infected diarrhoetic calves. Probiotics Antimicrob Proteins 14, 1054–1066. doi: 10.1007/s12602-021-09862-9
Geng, S., Cheng, S., Li, Y., Wen, Z., Ma, X., Jiang, X., et al. (2018). Faecal microbiota transplantation reduces susceptibility to epithelial injury and modulates tryptophan metabolism of the microbial community in a piglet model. J. Crohns Colitis. 12, 1359–1374. doi: 10.1093/ecco-jcc/jjy103
Giallourou, N., Medlock, G., Bolick, D., Medeiros, P., Ledwaba, S., Kolling, G., et al. (2018). A novel mouse model of Campylobacter jejuni enteropathy and diarrhea. PLoS Pathog. 14:e1007083. doi: 10.1371/journal.ppat.1007083
Gopinath, S., Carden, S., and Monack, D. (2012). Shedding light on Salmonella carriers. Trends Microbiol. 20, 320–327. doi: 10.1016/j.tim.2012.04.004
Gryaznova, M., Dvoretskaya, Y., Syromyatnikov, M., Shabunin, S., Parshin, P., Mikhaylov, E., et al. (2022). Changes in the microbiome profile in different parts of the intestine in piglets with diarrhea. Animals 12:320. doi: 10.3390/ani12030320
Guan, Z., Zhao, Q., Huang, Q., Zhao, Z., Zhou, H., He, Y., et al. (2021). Modified Renshen Wumei decoction alleviates intestinal barrier destruction in rats with diarrhea. J. Microbiol. Biotechnol. 31, 1295–1304. doi: 10.4014/jmb.2106.06037
Hakansson, A., and Molin, G. (2011). Gut microbiota and inflammation. Nutrients 3, 637–682. doi: 10.3390/nu3060637
Hirose, Y., Murosaki, S., Yamamoto, Y., Yoshikai, Y., and Tsuru, T. (2006). Daily intake of heat-killed Lactobacillus plantarum L-137 augments acquired immunity in healthy adults. J. Nutr. 136, 3069–3073. doi: 10.1093/jn/136.12.3069
Hodnik, J., Ježek, J., and Starič, J. (2020). Coronaviruses in cattle. Trop. Anim. Health Prod. 52, 2809–2816. doi: 10.1007/s11250-020-02354-y
Hou, Q., Ye, L., Liu, H., Huang, L., Yang, Q., Turner, J., et al. (2021). Correction: Lactobacillus accelerates ISCs regeneration to protect the integrity of intestinal mucosa through activation of STAT3 signaling pathway induced by LPLs secretion of IL-22. Cell Death Differ. 28, 2025–2027. doi: 10.1038/s41418-020-00630-w
Jang, J., Kim, S., Kwon, M., Lee, J., Yu, D., Song, R., et al. (2019). Rotavirus-mediated alteration of gut microbiota and its correlation with physiological characteristics in neonatal calves. J. Microbiol. 57, 113–121. doi: 10.1007/s12275-019-8549-1
Jordan, J., Moeller, R., Chakraborty, S., Vijay-Kumar, M., and Joe, B. (2019). Pressure from the bugs within. Hypertension 73, 977–979. doi: 10.1161/HYPERTENSIONAHA.119.12685
Kayasaki, F., Okagawa, T., Konnai, S., Kohara, J., Sajiki, Y., Watari, K., et al. (2021). Direct evidence of the preventive effect of milk replacer-based probiotic feeding in calves against severe diarrhea. Vet. Microbiol. 254:108976. doi: 10.1016/j.vetmic.2020.108976
Keely, S., and Barrett, K. (2022). Intestinal secretory mechanisms and diarrhea. Am. J. Physiol. Gastrointest. Liver Physiol. 322, G405–G420. doi: 10.1152/ajpgi.00316.2021
Kim, E., Lee, H., Kim, D., Son, J., Kim, B., Joo, S., et al. (2020). Hydrolyzed yeast supplementation in calf starter promotes innate immune responses in holstein calves under weaning stress condition. Animals 10:1468. doi: 10.3390/ani10091468
Konturek, P., Koziel, J., Dieterich, W., Haziri, D., Wirtz, S., Glowczyk, I., et al. (2017). Successful therapy of Clostridium difficile infection with fecal microbiota transplantation. J. Physiol. Pharmacol. 67, 859–866.
Kumar, A., Chatterjee, I., Anbazhagan, A., Jayawardena, D., Priyamvada, S., Alrefai, W., et al. (2018). Cryptosporidium parvum disrupts intestinal epithelial barrier function via altering expression of key tight junction and adherens junction proteins. Cell Microbiol. 20:e12830. doi: 10.1111/cmi.12830
Lamichhane, B., Mawad, A., Saleh, M., Kelley, W., Harrington, P., Lovestad, C., et al. (2024). Salmonellosis: An overview of epidemiology, pathogenesis, and innovative approaches to mitigate the antimicrobial resistant infections. Antibiotics 13:76. doi: 10.3390/antibiotics13010076
Lebeer, S., Bron, P., Marco, M., Van Pijkeren, J., O’Connell Motherway, M., Hill, C., et al. (2018). Identification of probiotic effector molecules: Present state and future perspectives. Curr. Opin. Biotechnol. 49, 217–223. doi: 10.1016/j.copbio.2017.10.007
Ley, R., Lozupone, C., Hamady, M., Knight, R., and Gordon, J. (2008). Worlds within worlds: Evolution of the vertebrate gut microbiota. Nat. Rev. Microbiol. 6, 776–788. doi: 10.1038/nrmicro1978
Li, Y., Lan, Y., Zhang, S., and Wang, X. (2022). Comparative analysis of gut microbiota between healthy and diarrheic horses. Front. Vet. Sci. 9:882423. doi: 10.3389/fvets.2022.882423
Liu, Q., Yu, Z., Tian, F., Zhao, J., Zhang, H., Zhai, Q., et al. (2020). Surface components and metabolites of probiotics for regulation of intestinal epithelial barrier. Microb Cell Fact. 19:23. doi: 10.1186/s12934-020-1289-4
López-Colom, P., Castillejos, L., Rodríguez-Sorrento, A., Puyalto, M., Mallo, J., and Martín-Orúe, S. (2020). Impact of in-feed sodium butyrate or sodium heptanoate protected with medium-chain fatty acids on gut health in weaned piglets challenged with Escherichia coli F4. Arch. Anim. Nutr. 74, 271–295. doi: 10.1080/1745039X.2020.1726719
Lou, L., Zhang, P., Piao, R., and Wang, Y. (2019). Salmonella pathogenicity Island 1 (SPI-1) and its complex regulatory network. Front. Cell Infect. Microbiol. 9:270. doi: 10.3389/fcimb.2019.00270
Lundin, A., Bok, C., Aronsson, L., Björkholm, B., Gustafsson, J., Pott, S., et al. (2008). Gut flora, Toll-like receptors and nuclear receptors: A tripartite communication that tunes innate immunity in large intestine. Cell Microbiol. 10, 1093–1103. doi: 10.1111/j.1462-5822.2007.01108.x
Ly, N., and Cyert, M. (2017). Calcineurin, the Ca2+-dependent phosphatase, regulates Rga2, a Cdc42 GTPase-activating protein, to modulate pheromone signaling. Mol. Biol. Cell 28, 576–586. doi: 10.1091/mbc.E16-06-0432
Ma, J., Chen, T., Xue, H., Zhang, M., Li, Z., Li, X., et al. (2023). Jian-Pi-Yin decoction attenuates lactose-induced chronic diarrhea in rats by regulating GLP-1 and reducing NHE3 ubiquitination and phosphorylation. Heliyon 9:e17444. doi: 10.1016/j.heliyon.2023.e17444
Macdonald, S., van Diemen, P., Martineau, H., Stevens, M., Tomley, F., Stabler, R., et al. (2019). Impact of Eimeria tenella coinfection on Campylobacter jejuni colonization of the chicken. Infect. Immun. 87:e00772-18. doi: 10.1128/IAI.00772-18
McKnight, L., Peppler, W., Wright, D., Page, G., and Han, Y. (2019). A blend of fatty acids, organic acids, and phytochemicals induced changes in intestinal morphology and inflammatory gene expression in coccidiosis-vaccinated broiler chickens. Poult. Sci. 98, 4901–4908. doi: 10.3382/ps/pez241
Medellin-Peña, M., Wang, H., Johnson, R., Anand, S., and Griffiths, M. (2007). Probiotics affect virulence-related gene expression in Escherichia coli O157:h7. Appl. Environ. Microbiol. 73, 4259–4267. doi: 10.1128/AEM.00159-07
Meremikwu, M., Donegan, S., Sinclair, D., Esu, E., and Oringanje, C. (2012). Intermittent preventive treatment for malaria in children living in areas with seasonal transmission. Cochrane Database Syst. Rev. 2012:CD003756. doi: 10.1002/14651858.CD003756.pub4
Morgoglione, M., Bosco, A., Maurelli, M., Alves, L., Saralli, G., Bruni, G., et al. (2020). A 10-year surveillance of Eimeria spp. in cattle and buffaloes in a mediterranean area. Front. Vet. Sci. 7:410. doi: 10.3389/fvets.2020.00410
Myeni, S., Wang, L., and Zhou, D. (2013). SipB-SipC complex is essential for translocon formation. PLoS One 8:e60499. doi: 10.1371/journal.pone.0060499
Oh, N., Joung, J., Lee, J., and Kim, Y. (2018). Probiotic and anti-inflammatory potential of Lactobacillus rhamnosus 4B15 and Lactobacillus gasseri 4M13 isolated from infant feces. PLoS One 13:e0192021. doi: 10.1371/journal.pone.0192021
O’Loughlin, A., McGee, M., Waters, S., Doyle, S., and Earley, B. (2011). Examination of the bovine leukocyte environment using immunogenetic biomarkers to assess immunocompetence following exposure to weaning stress. BMC Vet. Res. 7:45. doi: 10.1186/1746-6148-7-45
Olszak, T., An, D., Zeissig, S., Vera, M., Richter, J., Franke, A., et al. (2012). Microbial exposure during early life has persistent effects on natural killer T cell function. Science 336, 489–493. doi: 10.1126/science.1219328
Önlü, H., Teker, H., Keskin, S., Genc, A., Allahverdi, H., Elarslan, A., et al. (2025). Role of the probiotic supplementation on intestinal inflammation and structural integrity in wistar rats subjected to a cafeteria diet during development. Pak. Vet. J. 45, 226–235. doi: 10.29261/pakvetj/2025.002
Ou, C., Lin, S., Tsai, J., and Lin, M. (2011). Heat-killed lactic acid bacteria enhance immunomodulatory potential by skewing the immune response toward Th1 polarization. J. Food Sci. 76, M260–M267. doi: 10.1111/j.1750-3841.2011.02161.x
Pan, Y., Tian, X., Qin, P., Wang, B., Zhao, P., Yang, Y., et al. (2017). Discovery of a novel swine enteric alphacoronavirus (SeACoV) in southern China. Vet. Microbiol. 211, 15–21. doi: 10.1016/j.vetmic.2017.09.020
Piewngam, P., Zheng, Y., Nguyen, T., Dickey, S., Joo, H., Villaruz, A., et al. (2018). Pathogen elimination by probiotic Bacillus via signalling interference. Nature 562, 532–537. doi: 10.1038/s41586-018-0616-y
Prado-Rebolledo, O., Delgado-Machuca, J., Macedo-Barragan, R., Garcia-Márquez, L., Morales-Barrera, J., Latorre, J., et al. (2017). Evaluation of a selected lactic acid bacteria-based probiotic on Salmonella enterica serovar Enteritidis colonization and intestinal permeability in broiler chickens. Avian Pathol. 46, 90–94. doi: 10.1080/03079457.2016.1222808
Pral, L., Fachi, J., Corrêa, R., Colonna, M., and Vinolo, M. (2021). Hypoxia and HIF-1 as key regulators of gut microbiota and host interactions. Trends Immunol. 42, 604–621. doi: 10.1016/j.it.2021.05.004
Quach, A., Jayaratne, R., Lee, B., Ibeawuchi, S., Lim, E., Das, S., et al. (2022). Diarrhoeal pathogenesis in Salmonella infection may result from an imbalance in intestinal epithelial differentiation through reduced Notch signalling. J. Physiol. 600, 1851–1865. doi: 10.1113/JP282585
Rigo-Adrover, M., Pérez-Berezo, T., Ramos-Romero, S., van Limpt, K., Knipping, K., Garssen, J., et al. (2017). A fermented milk concentrate and a combination of short-chain galacto-oligosaccharides/long-chain fructo-oligosaccharides/pectin-derived acidic oligosaccharides protect suckling rats from rotavirus gastroenteritis. Br. J. Nutr. 117, 209–217. doi: 10.1017/S0007114516004566
Roca-Feltrer, A., Carneiro, I., Smith, L., Schellenberg, J., Greenwood, B., and Schellenberg, D. (2010). The age patterns of severe malaria syndromes in sub-Saharan Africa across a range of transmission intensities and seasonality settings. Malar J. 9:282. doi: 10.1186/1475-2875-9-282
Rojas, M., Manchego, A., Rocha, C., Fornells, L., Silva, R., Mendes, G., et al. (2016). Outbreak of diarrhea among preweaning alpacas (Vicugna pacos) in the southern Peruvian highland. J. Infect. Dev. Ctries 10, 269–274. doi: 10.3855/jidc.7398
Rooks, M., and Garrett, W. (2016). Gut microbiota, metabolites and host immunity. Nat. Rev. Immunol. 16, 341–352. doi: 10.1038/nri.2016.42
Round, J., O’Connell, R., and Mazmanian, S. (2010). Coordination of tolerogenic immune responses by the commensal microbiota. J. Autoimmun. 34, J220–J225. doi: 10.1016/j.jaut.2009.11.007
Ryan, K., O’Hara, A., van Pijkeren, J., Douillard, F., and O’Toole, P. (2009). Lactobacillus salivarius modulates cytokine induction and virulence factor gene expression in Helicobacter pylori. J. Med. Microbiol. 58(Pt 8), 996–1005. doi: 10.1099/jmm.0.009407-0
Saha, S., Namai, F., Nishiyama, K., Villena, J., and Kitazawa, H. (2024). Role of immunomodulatory probiotics in alleviating bacterial diarrhea in piglets: A systematic review. J. Anim. Sci. Biotechnol. 15:112. doi: 10.1186/s40104-024-01070-z
Salminen, S., Collado, M., Endo, A., Hill, C., Lebeer, S., Quigley, E., et al. (2021). The International scientific association of probiotics and prebiotics (ISAPP) consensus statement on the definition and scope of postbiotics. Nat. Rev. Gastroenterol. Hepatol. 18, 649–667. doi: 10.1038/s41575-021-00440-6
Sergeant, M., Constantinidou, C., Cogan, T., Bedford, M., Penn, C., and Pallen, M. (2014). Extensive microbial and functional diversity within the chicken cecal microbiome. PLoS One 9:e91941. doi: 10.1371/journal.pone.0091941
Seth, A., Yan, F., Polk, D., and Rao, R. (2008). Probiotics ameliorate the hydrogen peroxide-induced epithelial barrier disruption by a PKC- and MAP kinase-dependent mechanism. Am. J. Physiol. Gastrointest Liver Physiol. 294, G1060–G1069. doi: 10.1152/ajpgi.00202.2007
Shabana, I., Bouqellah, N., and Zaraket, H. (2017). Investigation of viral and bacterial enteropathogens of diarrheic sheep and goats in Medina, Saudi Arabia. Trop. Biomed. 34, 944–955.
Shen, L., Shen, Y., You, L., Zhang, Y., Su, Z., Peng, G., et al. (2023). Pueraria lobata polysaccharides alleviate neonatal calf diarrhea by modulating gut microbiota and metabolites. Front. Vet. Sci. 9:1024392. doi: 10.3389/fvets.2022.1024392
Shi, Z., Guo, Z., Li, S., Jiang, C., Wang, J., Deng, X., et al. (2025). Purpurin suppresses Salmonella invasion of host cells by reducing the secretion of T3SS-1 effector proteins. Sci. Rep. 15:4507. doi: 10.1038/s41598-025-86822-1
Singh, V., Davidson, A., Hume, P., and Koronakis, V. (2020). Arf6 can trigger wave regulatory complex-dependent actin assembly independent of Arno. Int. J. Mol. Sci. 21:2457. doi: 10.3390/ijms21072457
Snow, R., Kibuchi, E., Karuri, S., Sang, G., Gitonga, C., Mwandawiro, C., et al. (2015). Changing Malaria prevalence on the Kenyan Coast since 1974: Climate, drugs and vector control. PLoS One 10:e0128792. doi: 10.1371/journal.pone.0128792
Soares, J., Pimentel-Nunes, P., Roncon-Albuquerque, R., and Leite-Moreira, A. (2010). The role of lipopolysaccharide/toll-like receptor 4 signaling in chronic liver diseases. Hepatol. Int. 4, 659–672. doi: 10.1007/s12072-010-9219-x
Sugahara, H., Yao, R., Odamaki, T., and Xiao, J. (2017). Differences between live and heat-killed bifidobacteria in the regulation of immune function and the intestinal environment. Benef. Microbes 8, 463–472. doi: 10.3920/BM2016.0158
Sun, Z., Hu, Y., Wang, Y., Feng, J., and Dou, Y. (2019). BuPiHeWei decoction ameliorates 5-Fu-induced intestinal mucosal injury in the rats by regulating the TLR-4/NF- κ B signaling pathway. Evid. Based Complement. Alternat. Med. 2019:5673272. doi: 10.1155/2019/5673272
Suzuki, T., Shibahara, T., Imai, N., Yamamoto, T., and Ohashi, S. (2018). Genetic characterization and pathogenicity of Japanese porcine deltacoronavirus. Infect. Genet. Evol. 61, 176–182. doi: 10.1016/j.meegid.2018.03.030
Tan, J., and O’Neill, H. (2005). Maturation requirements for dendritic cells in T cell stimulation leading to tolerance versus immunity. J. Leukoc Biol. 78, 319–324. doi: 10.1189/jlb.1104664
Tian, S., Wang, J., Wang, J., and Zhu, W. (2022). Differential effects of early-life and postweaning galacto-oligosaccharide intervention on colonic bacterial composition and function in weaning piglets. Appl. Environ. Microbiol. 88:e0131821. doi: 10.1128/AEM.01318-21
Torow, N., and Hornef, M. (2017). The neonatal window of opportunity: Setting the stage for life-long host-microbial interaction and immune homeostasis. J. Immunol. 198, 557–563. doi: 10.4049/jimmunol.1601253
Toscano, M., De Grandi, R., Stronati, L., De Vecchi, E., and Drago, L. (2017). Effect of Lactobacillus rhamnosus HN001 and Bifidobacterium longum BB536 on the healthy gut microbiota composition at phyla and species level: A preliminary study. World J. Gastroenterol. 23, 2696–2704. doi: 10.3748/wjg.v23.i15.2696
van Baarlen, P., Troost, F., van der Meer, C., Hooiveld, G., Boekschoten, M., Brummer, R., et al. (2011). Human mucosal in vivo transcriptome responses to three lactobacilli indicate how probiotics may modulate human cellular pathways. Proc. Natl. Acad. Sci. U. S. A. 108(Suppl. 1), 4562–4569. doi: 10.1073/pnas.1000079107
van Zyl, W., Deane, S., and Dicks, L. (2020). Molecular insights into probiotic mechanisms of action employed against intestinal pathogenic bacteria. Gut Microbes 12:1831339. doi: 10.1080/19490976.2020.1831339
Velásquez, Z., López-Osorio, S., Waiger, D., Manosalva, C., Pervizaj-Oruqaj, L., Herold, S., et al. (2021). Eimeria bovis infections induce G1 cell cycle arrest and a senescence-like phenotype in endothelial host cells. Parasitology 148, 341–353. doi: 10.1017/S0031182020002097
Vlasova, A., and Saif, L. (2021). Bovine coronavirus and the associated diseases. Front. Vet. Sci. 8:643220. doi: 10.3389/fvets.2021.643220
Wang, L., Li, L., Lv, Y., Chen, Q., Feng, J., and Zhao, X. (2018). Lactobacillus plantarum restores intestinal permeability disrupted by Salmonella infection in newly-hatched chicks. Sci. Rep. 8:2229. doi: 10.1038/s41598-018-20752-z
Wang, W., Cai, M., Hu, J., Zhang, Z., Wang, X., Chang, X., et al. (2020). Mechanism of blood-brain barrier disruption by an Escherichia coli from lambs with severe diarrhea and meningoencephalitis. Microb Pathog. 147:104288. doi: 10.1016/j.micpath.2020.104288
Wang, W., Wang, Y., Lu, Y., Zhu, J., Tian, X., Wu, B., et al. (2022). Reg4 protects against Salmonella infection-associated intestinal inflammation via adopting a calcium-dependent lectin-like domain. Int. Immunopharmacol. 113(Pt A), 109310. doi: 10.1016/j.intimp.2022.109310
Wang, Y., Liu, L., Moore, D., Shen, X., Peek, R., Acra, S., et al. (2017). An LGG-derived protein promotes IgA production through upregulation of APRIL expression in intestinal epithelial cells. Mucosal Immunol. 10, 373–384. doi: 10.1038/mi.2016.57
Watanabe, T., Yamashita, K., Fujikawa, S., Sakurai, T., Kudo, M., Shiokawa, M., et al. (2012). Involvement of activation of toll-like receptors and nucleotide-binding oligomerization domain-like receptors in enhanced IgG4 responses in autoimmune pancreatitis. Arthritis Rheum. 64, 914–924. doi: 10.1002/art.33386
Wei, M., Han, Z., Lu, S., Gao, Q., Liu, J., Yang, Y., et al. (2025). Effect of high-density rearing of pregnant ewes on the intestinal microbiota of their offsprings. Pak. Vet. J. 45, 236–245.
Wu, Y., Nie, C., Luo, R., Qi, F., Bai, X., Chen, H., et al. (2022). Effects of multispecies probiotic on intestinal microbiota and mucosal barrier function of neonatal calves infected with E. coli K99. Front. Microbiol. 12:813245. doi: 10.3389/fmicb.2021.813245
Wu, Y., Zhu, C., Chen, Z., Chen, Z., Zhang, W., Ma, X., et al. (2016). Protective effects of Lactobacillus plantarum on epithelial barrier disruption caused by enterotoxigenic Escherichia coli in intestinal porcine epithelial cells. Vet. Immunol. Immunopathol. 172, 55–63. doi: 10.1016/j.vetimm.2016.03.005
Xiang, Q., Wu, X., Pan, Y., Wang, L., Guo, Y., Cui, C., et al. (2020). Early intervention using fecal microbiota transplantation combined with probiotics influence the growth performance, diarrhea, and intestinal barrier function of piglets. Appl. Sci. 10:2. doi: 10.3390/app10020568
Xu, B., Liang, S., Zhao, J., Li, X., Guo, J., Xin, B., et al. (2022). Bifidobacterium animalis subsp. Lactis XLTG11 improves antibiotic-related diarrhea by alleviating inflammation, enhancing intestinal barrier function and regulating intestinal flora. Food Funct. 13, 6404–6418. doi: 10.1039/d1fo04305f
Yang, H., Fan, X., Mao, X., Yu, B., He, J., Yan, H., et al. (2024). The protective role of prebiotics and probiotics on diarrhea and gut damage in the rotavirus-infected piglets. J. Anim. Sci. Biotechnol. 15:15. doi: 10.1186/s40104-024-01018-3
Yang, Y., Xiao, G., Cheng, P., Zeng, J., and Liu, Y. (2023). Protective application of chinese herbal compounds and formulae in intestinal inflammation in humans and animals. Molecules 28:6811. doi: 10.3390/molecules28196811
Zeng, M., Cisalpino, D., Varadarajan, S., Hellman, J., Warren, H., Cascalho, M., et al. (2016). Gut microbiota-induced immunoglobulin g controls systemic infection by symbiotic bacteria and pathogens. Immunity 44, 647–658. doi: 10.1016/j.immuni.2016.02.006
Zhang, B., Shao, Y., Liu, D., Yin, P., Guo, Y., and Yuan, J. (2012). Zinc prevents Salmonella enterica serovar Typhimurium-induced loss of intestinal mucosal barrier function in broiler chickens. Avian Pathol. 41, 361–367. doi: 10.1080/03079457.2012.692155
Zhang, C., Yu, X., Cui, Y., Wang, H., Chen, X., Ma, X., et al. (2023b). Shengjiang Xiexin Decoction ameliorates antibiotic-associated diarrhea by altering the gut microbiota and intestinal metabolic homeostasis. Phytomedicine 113:154737. doi: 10.1016/j.phymed.2023.154737
Zhang, H., Wang, K., An, T., Zhu, L., Chang, Y., Lou, W., et al. (2022). Genetic parameters for dairy calf and replacement heifer wellness traits and their association with cow longevity and health indicators in Holstein cattle. J. Dairy Sci. 105, 6749–6759. doi: 10.3168/jds.2021-21450
Zhang, Y., Wang, Y., Yan, K., Li, H., Zhang, X., Essola, J., et al. (2023a). Traditional Chinese medicine formulae QY305 reducing cutaneous adverse reaction and diarrhea by its nanostructure. Adv. Sci. 11:e2306140. doi: 10.1002/advs.202306140
Zhang, Y., Yu, F., Hao, J., Nsabimana, E., Wei, Y., Chang, X., et al. (2021). Study on the effective material basis and mechanism of traditional Chinese medicine prescription (QJC) against stress diarrhea in mice. Front. Vet. Sci. 8:724491. doi: 10.3389/fvets.2021.724491
Zhang, Y., Zhang, S., Sun, Z., Liu, X., Liao, G., Niu, Z., et al. (2023c). Porcine epidemic diarrhea virus causes diarrhea by activating EGFR to regulates NHE3 activity and mobility on plasma membrane. Front. Microbiol. 14:1237913. doi: 10.3389/fmicb.2023.1237913
Zhu, Y., Lu, S., Cidan, Y., Ali, M., Zhang, X., Pubu, P., et al. (2025). Protective effects of traditional Chinese herbal medicine formulas (TCHMFs) Via influencing anti-oxidative capacity, inflammatory mediators, and gut microbiota in Weaned Yaks. Pak. Vet. J. 45, 184–194. doi: 10.29261/pakvetj/2025.114
Keywords: diarrhea, gut microbiota, metabolites, treatment, prevention
Citation: Zhang Y, Ma Y and Qi Y (2025) Potential relationship between gut microbiota and animal diarrhea: a systematic review. Front. Microbiol. 16:1637331. doi: 10.3389/fmicb.2025.1637331
Received: 29 May 2025; Accepted: 09 July 2025;
Published: 24 July 2025.
Edited by:
Houqiang Luo, Wenzhou Vocational College of Science and Technology, ChinaReviewed by:
Shuai Guo, Huazhong Agricultural University, ChinaFazul Nabi, Sindh Agriculture University, Pakistan
Copyright © 2025 Zhang, Ma and Qi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Youchao Qi, MjAyMDk5MDAxM0BxaHUuZWR1LmNu
 Yuxin Zhang
Yuxin Zhang Yonggui Ma2,3
Yonggui Ma2,3 Youchao Qi
Youchao Qi