- 1The Provincial and Ministerial Co-founded Collaborative Innovation Center for R&D in Xizang Characteristic Agricultural and Animal Husbandry Resources, Xizang Agriculture and Animal Husbandry University, Linzhi, China
- 2Key Laboratory of Traditional zang's Medicine Resources Conservation and Utilization of Xizang Autonomous Region, Xizang Agriculture and Animal Husbandry University, Linzhi, China
- 3Resources and Environment College, Xizang Agriculture and Animal Husbandry University, Linzhi, China
Gymnadenia conopsea has high economic value and can be used as a medicinal and ornamental plant. Owing to its low natural reproduction rate and overexploitation, the risk of extinction of this plant is gradually increasing. Endophytic fungi play crucial roles in host growth and development; however, the characteristics of the endophytic fungal community in various tissues of G. conopsea have not been fully characterized. Illumina MiSeq high-throughput sequencing technology was employed to sequence the fungal ITS (A non-coding DNA sequence located between the 18S rRNA gene and the 5.8S rRNA gene within the ribosomal RNA gene cluster) region, thereby characterizing the community structure and assembly processes of endophytic fungi in roots, stems, leaves, and fruits. A total of 7,371 OTUs were obtained from all the samples and were dominated by Ascomycota and Basidiomycota. The richness indices of various tissues were significantly different, whereas the diversity indices were not significantly different. The composition of the dominant genera differed; overall, the compositions of the endophytic fungal communities were similar among the leaf, stem, and fruit tissues. The relative abundances of Ceratobasidium, Cadophora, and Mortierella were significantly higher in root tissues than in other tissues, G. conopsea roots should be prioritized as the material for isolating growth-promoting endophytic fungi. Cooccurrence network analysis revealed that endophytic fungi in different tissues presented typical modular structures and the network was mainly positive. The assembly processes in different tissues were affected mainly by deterministic factors. The proportions of pathotrophs, saprotrophs, and symbiotrophs were different among various tissues, and the proportion of pathotrophs was greater than those of saprotrophs and symbiotrophs. In conclusion, tissue type affects the composition of endophytic fungi. Furthermore, by dissecting the composition and functions of the fungal community associated with G. conopsea, this study aims to provide a reference for exploring the interaction mechanisms between endophytic fungi and G. conopsea.
1 Introduction
Gymnadenia conopsea (L.) R. Br, which is widely distributed in Asia and Europe, is a perennial and terrestrial herb of the Orchidaceae family (Lin et al., 2020; Shi et al., 2023). In Xizang of China, G. conopsea grows mainly in forests, grasslands, and waterlogged meadows at altitudes of 1,265–4,700 m (Shang et al., 2017). G. conopsea is a valuable traditional medicine and edible plant, and its tubers have multiple benefits, including antifatigue (Zhao and Liu, 2011), antioxidative (Morikawa et al., 2006), sedative and hypnotic (Lin, 2009), immunoregulatory (Li et al., 2006), antiaging (Si and Liu, 2013), and antihyperlipidemic (Zhang et al., 2013) activities. In recent years, tubers of G. conopsea have also been used as an ingredient and tonic added to food to strengthen bodies and prevent illness (Shang et al., 2017). Owing to overexploitation, habitat destruction, and its low natural reproductive capacity, the G. conopsea populations are shrinking rapidly (Lin et al., 2020; Shang et al., 2017; Gao et al., 2020). G. conopsea has been listed as a grade II endangered species by the Convention on International Trade in Endangered Species of Wild Fauna and Flora (CITES) (Lin et al., 2020).
Fungal endophytes are a group of nonpathogenic fungal organisms that can enter plants through the root cortex, wounds, or stomata and reside in different tissues or organs without causing any harm to the host plants, with benefits for both the host and fungus (Tan and Zou, 2001; Schulz and Boyle, 2005; Kusari et al., 2013). Previous studies have indicated that endophytic fungi can be considered the second genome of the plant, playing a crucial role in nutrient cycling and plant growth and development, for example, by conferring resistance to various biotic and abiotic stresses (Seleiman et al., 2021), producing biocontrol agents and phytohormones (Gong et al., 2020; Firáková et al., 2007), secreting extracellular enzymes to promote nutrient cycling (Chathurdevi and Gowrie, 2016), and producing bioactive properties (Omomowo et al., 2023). Owing to the tiny seeds of orchids lacking endosperm and sufficient nutrients, seed germination under natural conditions requires mycorrhizal fungal colonization for nutrient supply (Lin et al., 2020). Previous studies have shown that mycorrhizal fungi can facilitate seed germination, protocorm growth, and seedling establishment in orchids (Wang et al., 2024; Huynh et al., 2009; Bonnardeaux et al., 2007; Sathiyadash et al., 2012). In comparison to soil-inhabiting fungi, dominant endophytic fungal taxa exhibit greater stability, and these may represent key fungal symbionts influencing orchid growth and development (Chen et al., 2019). A culturable orchid mycorrhizal (OM) fungal strain of Epulorhiza sp. isolated from the rhizome significantly promoted seed germination in Dendrobium officinale and seedling growth in Epidendrum secundum orchids (Wang et al., 2024). The mycorrhizal fungus Ceratobasidium GS2, isolated from G. conopsea, produces several steroid compounds that significantly promote the growth and differentiation of its protocorms (Shi et al., 2023). These findings suggest that symbiotic fungi may play a crucial role in the symbiotic germination of G. conopsea seeds via metabolite-mediated interactions. In recent years, endophytic fungi have garnered significant attention due to their potential applications in pharmacology, ecology, and agroindustry (Jha et al., 2023). However, the composition and diversity of endophytic fungal communities within different tissues of G. conopsea remain largely unexplored, underscoring the necessity of this study.
Microbial community assembly, based on the niche and neutral theories, is a crucial determinant of microbial diversity and ecological function, which are shaped by both deterministic and stochastic processes (Zhang et al., 2024). The deterministic processes are based on the niche theory, which posits that environmental filtering, interspecies interactions and species characteristics govern microbial community structure (Vellend, 2010). The neutral theory emphasizes that stochastic processes, such as gene and ecological drift, birth and death, determine microbial community dynamics (Ning et al., 2019). Many studies have shown that microbial community assembly is influenced by environmental factors, host species and tissue type (Kivlin et al., 2022; Zheng and Gong, 2019; Carper et al., 2018). Moreover, host metabolites, such as salicylic acid, triterpenoids and benzoxazines, also affect community assembly (Pang et al., 2021; Lebeis et al., 2015). However, the process of endophytic fungal community assembly in different tissues of G. conopsea remains unclear. In this study, high-throughput sequencing technology was used to investigate the diversity and composition of the endophytic fungal community in different tissues of G. conopsea. Subsequently, correlation network analysis, fungal community assembly and functional prediction were conducted to elucidate endophytic fungal community dynamics and function.
2 Materials and methods
2.1 Sample collection and treatment
In August 2019, the roots, stems, leaves, and fruit of G. conopsea plants (5 years old) were collected from Linzhi city, Xizang, China (geographical coordinates: 94°41′51.90′′E and 29°37′ 0.85′′N), and the altitude of the sampling site ranged from 3,244 m to 4,513 m, with a highland, temperate, semihumid climate, an annual average temperature of −0.73 °C, and an annual average precipitation of 1,134 mm. Fifteen biological replicates were collected for each tissue type, resulting in a total of 60 samples (15 replicates × 4 tissue types). Each replicate was sampled from 45 individual 5-year-old G. conopsea plants exhibiting uniform growth and no signs of disease or pest infestation. Each sample will undergo independent sequencing following subsequent processing. GcR represents the tuber samples, GcL represents the leaf samples, GcS represents the stem samples, and GcF represents the fruit samples of G. conopsea. The collected samples were randomly packed in sterile plastic bags and brought to the laboratory. Tissue samples were rinsed thoroughly with ultrapure water, followed by surface washing of G. conopsea roots, stems, leaves, and fruits with sterile water in a laminar flow hood. Surface moisture was blotted dry using sterile filter paper. Samples were then immersed in 75% ethanol for 30 s, rinsed 2–3 times with sterile water, and subsequently soaked with agitation in 10% sodium hypochlorite solution for 3–5 min. After rinsing 2–3 times with sterile water and drying with sterile filter paper, the samples were stored for further use. If surface sterilization of root and fruit tissues remained insufficient, the entire procedure was repeated up to multiple times until sterility was confirmed. The sterile water from the final rinse was inoculated onto PDA (Potato Dextrose Agar) medium as a control and incubated at 28 °C for 2–3 days. Surface sterilization was deemed complete if no microbial growth was observed.
2.2 DNA extraction, PCR amplification, and sequencing
Total DNA was extracted from all the samples using fungal genomic DNA extraction kits (Solarbio, D2300). Afterward, the quality of extracted DNA was assessed by 1% agarose gel electrophoresis, and the concentration and purity were determined with a NanoDrop 2000 UV–vis spectrophotometer (Thermo Scientific, Wilmington, USA). The primers ITS1F(F) (5′-CTTGGTCATTTAGAGGAAGTAA3′) and ITS2(R) (5′-GCTGCGTTCTTCATCGATGC3′) were used to amplify the ITS1 region from all the fungal samples (Chen et al., 2019; Liang et al., 2022), and PCR amplification was performed using a 20 μL reaction system (5 × TransStart FastPfu buffer, 4 μL; 2.5 mM dNTPs, 2 μL; 5 μM primer ITSIF, 0.8 μL; 5 μM primer ITS2R, 0.8 μL; TransStart FastPfu DNA Polymerase, 0.4 μL; template DNA, 10 ng; and ddH2O up to 20 μL). The PCR procedure was as follows: initial denaturation at 95 °C for 3 min; 27 cycles of denaturation at 95 °C for 30 s, annealing at 55 °C for 30 s and extension at 72 °C for 45 s; and a single extension at 72 °C for 10 min, followed by holding at 4 °C. The PCR products were detected by 2% agarose gel electrophoresis. An AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, USA) was used for purification of the products. The purified products were sent to Shanghai Majorbio Biopharm Technology Co., Ltd., for sequencing, and the sequences were deposited into the NCBI Sequence Read Archive (SRA) database (accession number: PRJNA699676).
2.3 Statistical analysis
The raw gene sequencing reads were quality filtered and merged by FLASH (Magoc and Salzberg, 2011). Operational taxonomic units (OTUs) with a 97% similarity cutoff were clustered using UPARSE (version 7.0.1), and chimeric sequences were removed (Edgar, 2013). The taxonomy of each OTU representative sequence was analyzed by RDP Classifier (version 2.2) (Zhao et al., 2018) against the UNITE database (version 8.0) (Kõljalg et al., 2013) with a confidence threshold of 70%. Alpha diversity indices, including the Sobs, Chao, Shannon, and coverage indices, were determined for all the samples using Mothur (version 1.30.2) to analyze the richness, diversity, and coverage of the microbial community. A Venn diagram, which could intuitively show the composition similarities and overlap of species, was used to analyze the number of common and unique OTU in multiple groups (Wang et al., 2019). Analysis of fungal community composition among all samples was performed with the R package at different taxonomic levels. Principal coordinate analysis (PCoA) was implemented with QIIME (v1.9.1) (Yuan et al., 2018). The functions of the endophytic fungi were predicted using the FUNGuild database (Nguyen et al., 2016). Cooccurrence network analysis was conducted on the basis of Spearman's correlation to explore the interactions among fungal genera with absolute correlation coefficients ≥0.7 and p < 0.05, and the results were visualized using Gephi (0.9.7) (Chen et al., 2022). A neutral community model (NCM) was used to assess the microbial community assembly process (Li et al., 2024). Differences were analyzed using Duncan's multiple comparison tests and one-way analysis of variance (ANOVA) by SPSS 26.0 software at a significance level of 0.05.
3 Results
3.1 Diversity of endophytic fungi in different tissues
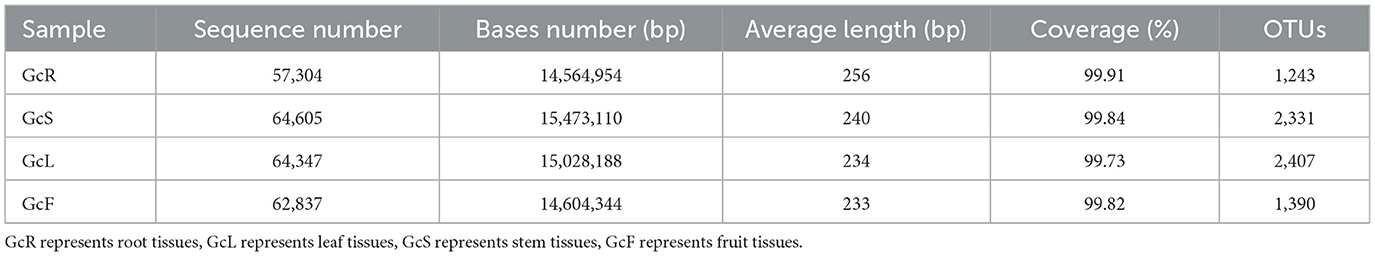
After filtering the sequencing data, a total of 249,093 valid sequences (with numbers ranging from 57,304 to 64,605 valid sequences in each sample) were obtained from 60 samples, with an average length of 241 bp. The fungal dataset yielded 7,371 OTUs. Among them, the number of OTUs in GcL was highest, whereas that in GcR was lowest, the lowest number of OTUs in root samples may be due to the fact that some dominant mycorrhizal fungi in the roots of photosynthetic orchids were not amplified due to mismatches with the primers used (Table 1). The rarefaction curves, combined with the coverage index analysis results, indicated that the sequencing depth was appropriate, and the sequencing results reflected the endophytic fungal diversity in all the samples (Supplementary Figure S1, Table 1). The alpha diversity analysis revealed significant differences in the Chao1 and Sobs indices among the different tissues (P < 0.05), the richness of GcL and GcS was highest, and that of GcR was lowest; Similarly, significant differences in Shannon and Simpson indices were observed among different tissues (P < 0.05) (Figure 1).
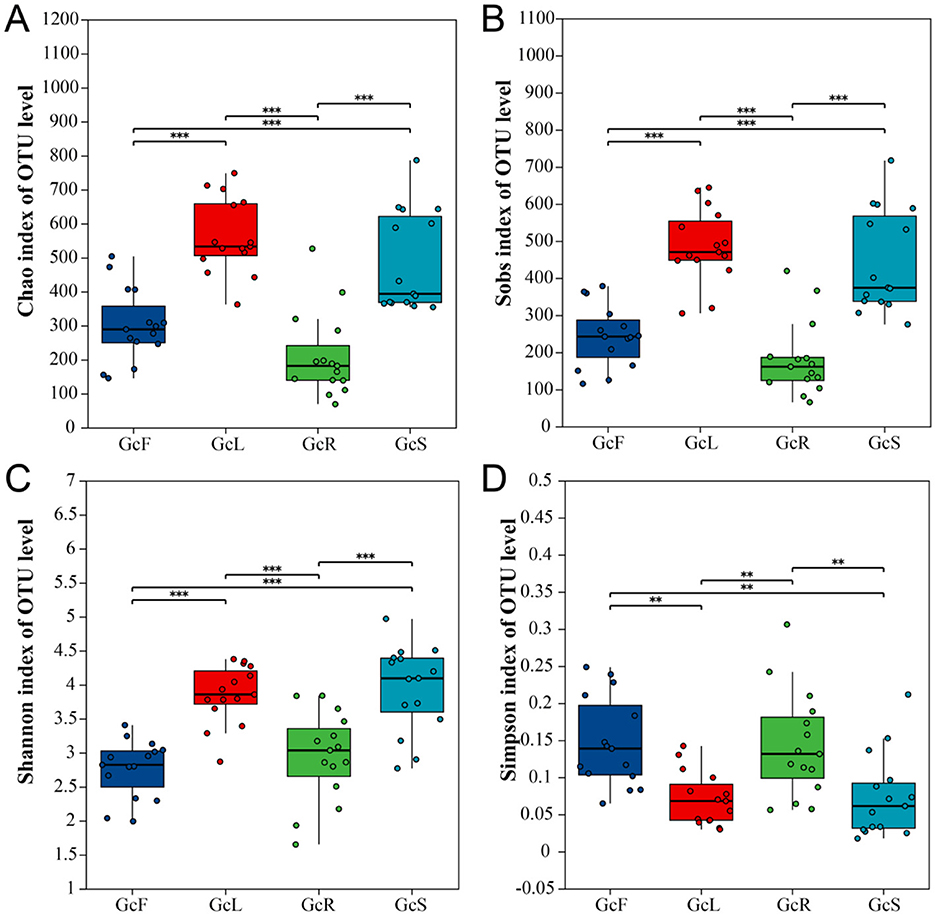
Figure 1. Alpha diversity analysis in different tissues of G. conopsea. (A) Chao1 index; (B) Sobs index; (C) Shannon index; (D) Simpson index. GcR represents root tissues, GcL represents leaf tissues, GcS represents stem tissues, GcF represents fruit tissues. Significance levels were presented as follows: **P < 0.01 and ***P < 0.001.
According to the OTU cluster analysis, there were 433 shared OTUs among the different tissues of G. conopsea, accounting for 11.11%, and the percentages of unique OTUs of GcR, GcL, GcS, and GcF were 375 (9.62%), 699 (17.93%), 641 (16.44%), and 246 (6.31%), respectively (Figure 2A). To explore the effects of different tissues on the endophytic fungal community, PCoA was performed at the OTU level on the basis of the Bray–Curtis distance. The results suggested that the distance between root tissues and other tissues of G. conopsea was relatively large, and the different contribution rates of PC1 and PC2 were 13.07 and 10.59%, respectively, suggesting that the endophytic fungal community in root tissues significantly differed from that in other tissues (Figure 2B). The samples from stem, leaf, and fruit tissues were relatively clustered, suggesting that the endophytic fungal communities were similar.
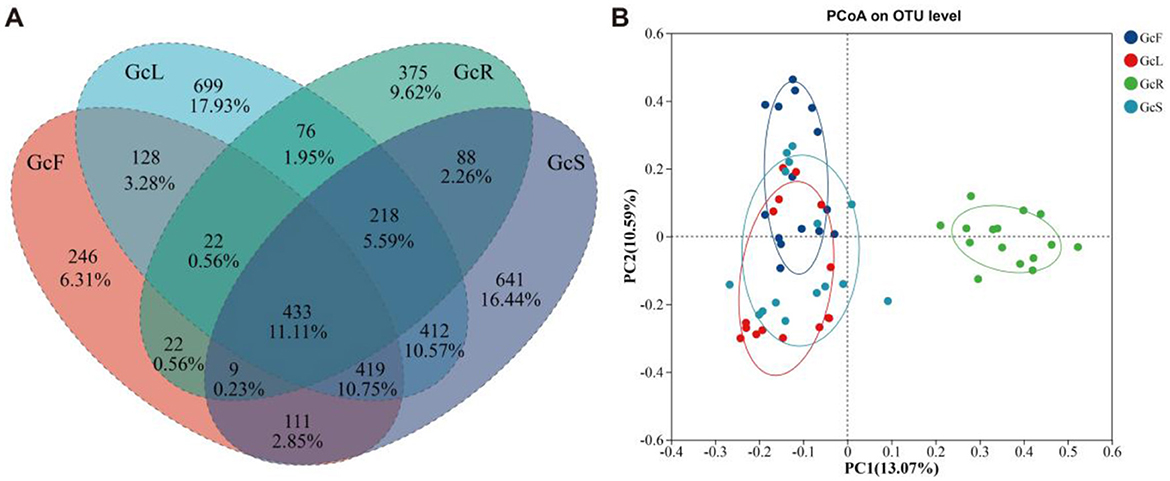
Figure 2. Venn diagram and PCoA analysis of endophytic fungal community among different tissues of G. conopsea. (A) Venn diagram; (B) PCoA analysis. GcR represents root tissues, GcL represents leaf tissues, GcS represents stem tissues, GcF represents fruit tissues.
3.2 Analysis of endophytic fungal community composition
The ITS sequences obtained from 60 tissue samples were classified into 12 phyla, 37 classes, 113 orders, 294 families, and 621 genera at the 97% similarity level. The endophytic fungal community composition and relative abundance varied among different tissues of G. conopsea (Supplementary Table S1, Figure 3). As shown in Figure 3A, the dominant fungal phyla (relative abundance >1%) were Ascomycota, Basidiomycota, and Mucoromycota across all the samples. Ascomycota was the dominant phylum, with a relative abundance ranging from 56.96% to 63.83%, followed by Basidiomycota, with a relative abundance ranging from 22.87% to 39.67%, whereas the relative abundance of Mucoromycota in root tissues was greater than that in other tissues, at 10.13%, and that in leaf, stem, and fruit tissues was less than 1%. The composition of the endophytic fungal community at the genus level and clustering of the top 20 genera in terms of relative abundance were shown in Supplementary Figure S2 and Figure 3B. The distributions of the dominant genera significantly differed among the different tissues. In the GcR samples, the dominant genera (relative abundance >1%) were Cadophora (16.45%), Mortierella (10.08%), Cladophialophora (9.41%), Tetracladium (4.03%), Ceratobasidium (2.63%), Russula (2.52%), Exophiala (2.30%), Hyaloscypha (2.01%), Trichocladium (1.02%), Vishniacozyma (1.13%), Ilyonectria (1.93%), and Phialocephala (1.20%). In the GcS samples, the dominant genera (relative abundance >1%) were Sistotrema (5.41%), Vishniacozyma (5.09%), Cadophora (2.78%), Protomyces (2.69%), Cladosporium (2.48%), Heterocephalacria (2.10%), Leucosporidium (1.81%), Gibberella (1.71%), Genolevuria (1.71%), Rachicladosporium (1.49%), Exophiala (1.37%), Trichomerium (1.37%), Knufia (1.24%), Zymoseptoria (1.15%), Plenodomus (1.14%), Mycocentrospora (1.09%), and Filobasidium (1.00%). In the GcL samples, the dominant genera (relative abundance >1%) were Vishniacozyma (12.31%), Zymoseptoria (5.93%), Gibberella (4.46%), Rachicladosporium (2.69%), Protomyces (2.29%), Podospora (1.84%), Ramularia (1.56%), Cladosporium (1.54%), Trichomerium (1.28%), Leucosporidium (1.17%), Knufia (1.13%), Heterocephalacria (1.06%), and Septoria (1.01%). In the GcF samples, the dominant genera (relative abundance >1%) were Cladosporium (16.17%), Rachicladosporium (9.20%), Vishniacozyma (4.39%), Epicoccum (3.08%), Leucosporidium (2.58%), Genolevuria (2.59%), Naganishia (2.97%), Curvibasidium (2.82%), Papiliotrema (1.87%), Heterocephalacria (1.64%), Protomyces (1.59%), and Plenodomus (1.03%). The clustering of dominant genera analysis shown that the composition of endophytic fungal community was similar between stem and leaf tissues, and varied between roots and the other two tissues.
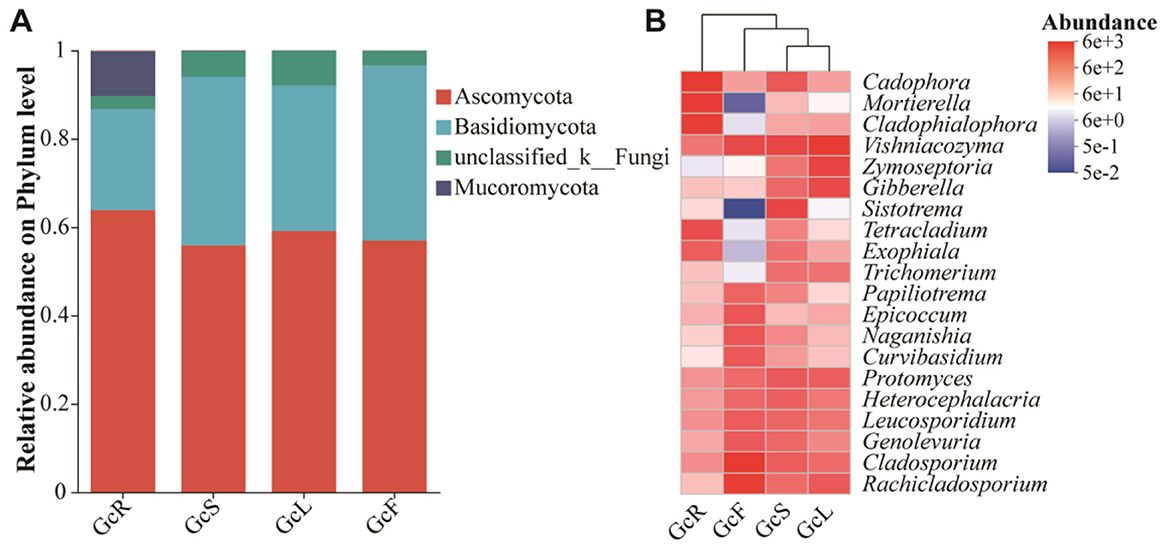
Figure 3. Relative abundance of endophytic fungi in different tissues of G. conopsea at the phylum level (A) and the genus level (B). GcR represents root tissues, GcL represents leaf tissues, GcS represents stem tissues, GcF represents fruit tissues.
3.3 Genus-level differential analysis across distinct tissue types
The distribution of the top 20 genera in different tissues were compared (Figure 4A). The relative abundances of Cadophora, Mortierella, Cladophialophora, Tetracladium, Exophiala, and Ceratobasidium were significantly greater in root tissues than in other tissues (p < 0.05). The relative abundances of Protomyces, Sistotrema, Heterocephalacria, Knufia, and Plenodomus were significantly greater in stem tissues than in other tissues (p < 0.05). The relative abundances of Vishniacozyma, Zymoseptoria, and Gibberella were significantly greater in the leaf tissues than in other tissues (p < 0.05). The relative abundances of Cladosporium, Rachicladosporium, Leucosporidium, Genolevuria, Epicoccum, and Curvibasidium were significantly greater in fruit tissues than in other tissues (p < 0.05). Overall, the relative abundance of endophytic fungi significantly differed among different tissues.
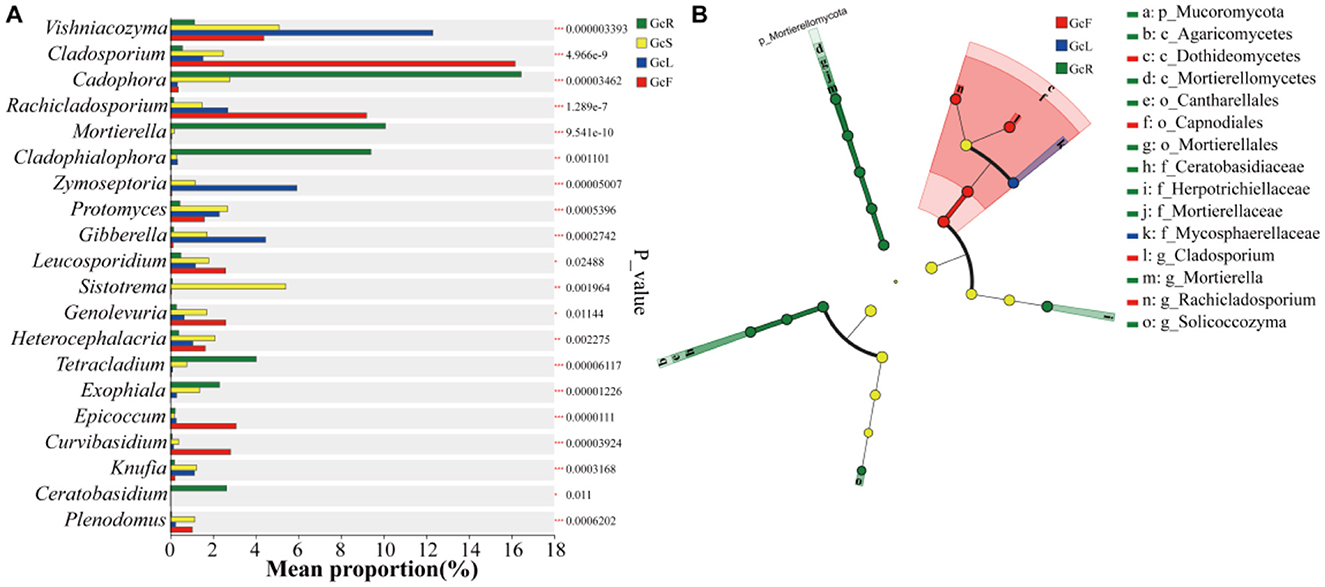
Figure 4. Statistical comparison of relative abundance based on the Kruskal-Wallis rank sum test (A) and LEfSe differential analysis (B) among different tissues of G. conopsea. The phylogenetic tree diagram illustrates taxonomic differences across hierarchical levels, providing a visual representation of differentially abundant taxa identified between groups at various taxonomic ranks. Nodes with distinct colors indicate microbial taxa that are significantly enriched in corresponding groups and exert a significant impact on inter-group differences; pale yellow nodes represent taxa with no significant differential abundance across groups or no substantial contribution to inter-group distinctions. When the number of significantly differential taxa is ≤ 50, the legend on the right is displayed in a single column; when >50, the legend on the right is presented in two columns.
Linear discriminant analysis Effect Size (LEfSe) enables multi-level taxonomic differential analysis (spanning phylum, class, order, family, genus, and species ranks) to test for differential taxa across multiple hierarchical levels. It quantifies the impact of each taxon on the observed differences using the Linear Discriminant Analysis (LDA) score, indicating potential key roles of these taxa in response to environmental changes. LEfSe (Figure 4B) revealed 15 fungal biomarkers at the phylum, class, order, family and genus levels on the basis of the criterion LDA>4. Among these, p_Mucoromycota, c_Mortierellomycetes, c_Agaricomycetes, o_Cantharellales, o_Mortierellales, f_Ceratobasidiaceae, f_Herpotrichiellaceae, g_Mortierella, and g_Solicoccozyma were identified in the root tissues; c_Dothideomycetes, o_Capnodiales, g_Cladosporium, and g_Cladosporium were identified in the fruit tissues; and f_Mycosphaerellaceae was identified in the leaf tissues. However, no fungal biomarkers were detected in the stem tissues.
3.4 Cooccurrence network analysis of the endophytic fungal community in different tissues
To explore the potential interactions within endophytic fungi among different tissues, co-occurrence network analysis was performed at the genus level (Figure 5). The correlation network analysis revealed that the positive proportions were greater than the negative proportions in the GcR, GcS, GcL, and GcF tissues; the positive proportions ranged from 94.77% to 99.52%. In the GcR tissues, a total of 204 nodes and 826 edges were significantly correlated (P < 0.05, |rho| > 0.7), with an average degree of 8.098 and clustering coefficient of 0.677; the nodes were mainly affiliated with Ascomycota (60.29%), Basidiomycota (37.25%), Glomeromycota (1.47%), and Mucoromycota (0.98%). In the GcS tissues, a total of 304 nodes and 1,148 edges were significantly correlated (P < 0.05, |rho| > 0.7), with an average degree of 7.553 and clustering coefficient of 0.620; the nodes were mainly affiliated with Ascomycota (58.88%), Basidiomycota (39.14%), Mucoromycota (0.99%), Glomeromycota (0.33%), Chytridiomycota (0.33%), and Olpidiomycota (0.33%). In the GcL tissues, a total of 328 nodes and 1,476 edges were significantly correlated (P < 0.05, |rho| > 0.7), with an average degree of 9.000 and clustering coefficient of 0.637; the nodes were mainly affiliated with Ascomycota (57.93%), Basidiomycota (40.55%), Mucoromycota (0.61%), Glomeromycota (0.3%), Olpidiomycota (0.3%), Chytridiomycota (0.3%). In the GcF tissues, a total of 219 nodes and 968 edges were significantly correlated (P < 0.05, |rho| > 0.7), with an average degree of 8.440 and clustering coefficient of 0.712; the nodes were mainly affiliated with Ascomycota (60.27%), Basidiomycota (39.27%), and Mucoromycota (0.46%). The results revealed that the complexity of endophytic fungi significantly differed among different tissues.
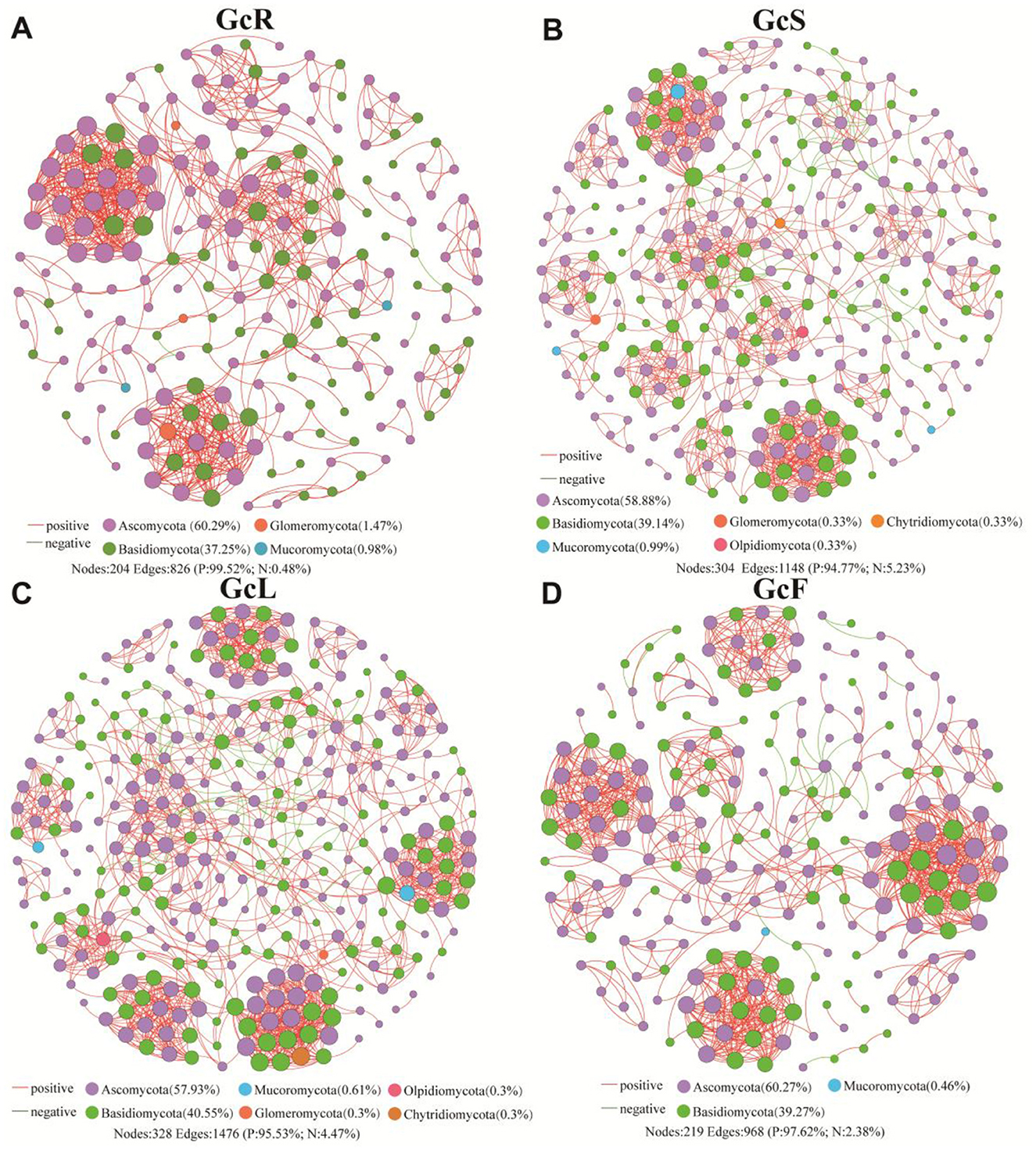
Figure 5. Co-occurrence network analysis of endophytic fungi in different tissues. (A) root tissues, (B) stem tissues, (C) leaf tissues, (D) fruit tissues.
3.5 Assembly processes of the endophytic fungal community
NCM-based analysis revealed distinct assembly dynamics of endophytic fungal communities across different tissues (Figure 6). The goodness of fit (R2) for the endophytic fungal communities in the GcS sample (R2 = 0.4458) was greater than that in the GcR sample (R2 = 0.1298), GcL sample (R2 = 0.3704), and GcF sample (R2 = 0.3427), showing an initial rapid increase followed by a gradual decrease in the explanatory rate for G. conopsea from root tissues to fruit tissues. Therefore, the endophytic fungal community assembly in root tissues was more strongly influenced by deterministic processes, and the deterministic processes in the GcS, GcL, and GcF samples became less important. These results indicated that the degree of diffusion for endophytic fungal communities in the GcR sample was limited, while that in other tissues gradually increased.
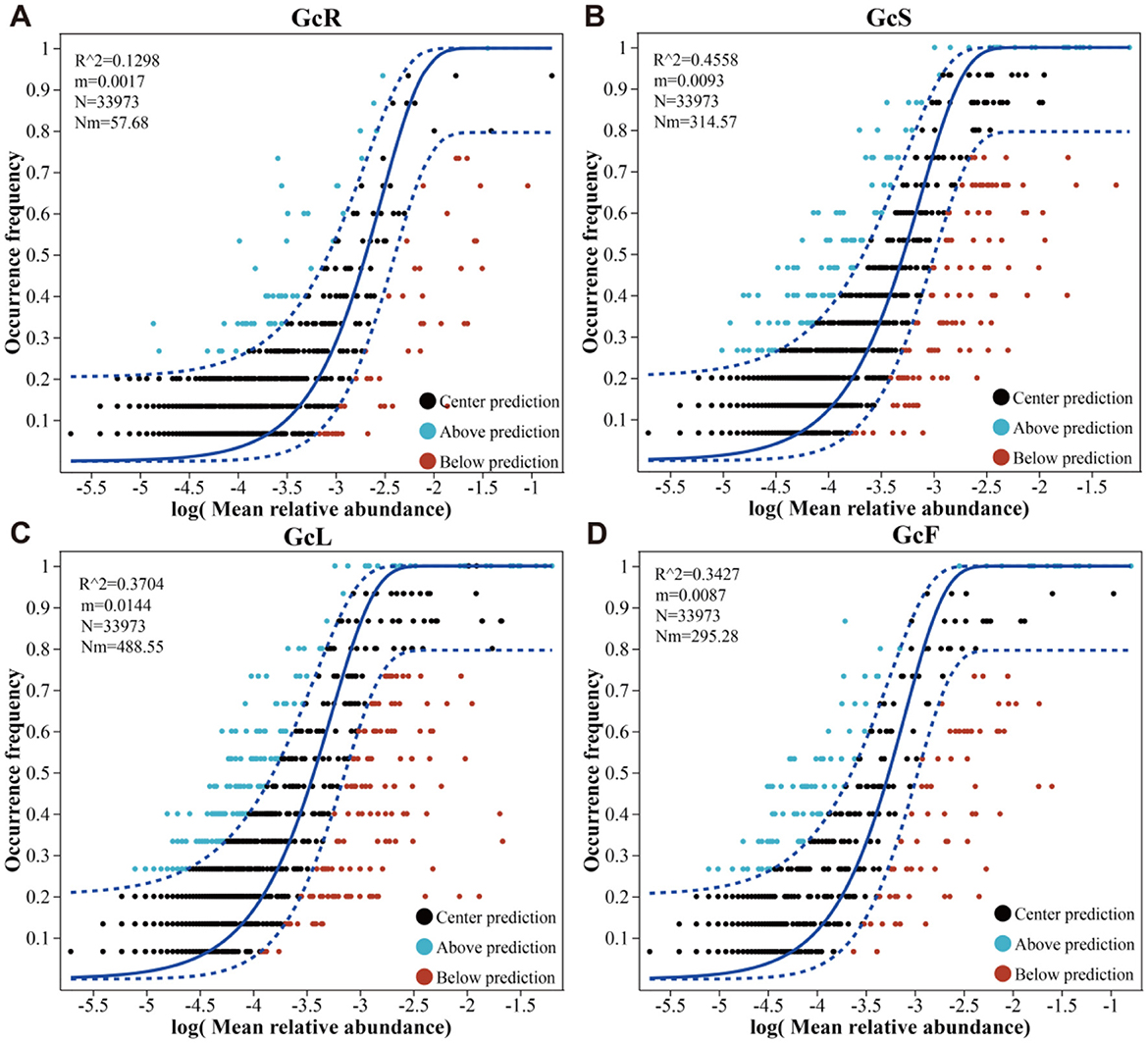
Figure 6. Endophytic fungal community assembly in different tissues of G. conopsea. (A) root tissues, (B) stem tissues, (C) leaf tissues, (D) fruit tissues.
3.6 Functional prediction for endophytic fungi in different tissues
The FUNGuild database was used to predict the potential functions of endophytic fungi in different tissues of G. conopsea on the basis of ecological guilds. Fungal OTUs were classified into three ecologically relevant trophic modes (pathotrophs, saprotrophs, and symbiotrophs). Of these, the relative abundance of pathotrophs (28.5%−88.1%) gradually increased from the root to fruit tissues, while that of saprotrophs (44.7%−11.2%) and symbiotrophs (26.8%−0.7%) gradually decreased from root to fruit tissues (Figure 7A). At the guild level, endophytes (34.9%), undefined saprotrophs (25.4%), and ectomycorrhizal fungi (20.7%) were the top three high abundance guilds in the root tissues; plant pathogens (26.8%), fungal parasites (19.2%), animal pathogens (15.4%), and ectomycorrhizal fungi (13.7%) were the top four high-abundance guilds in the stem tissues; and plant pathogens (35.5%), fungal parasites (28.0%), and animal pathogens (13.4%) were the top three high-abundance guilds in the leaf tissues. Animal pathogens (46.3%), plant pathogens (22.8%), and fungal parasites (18.0%) were the top three high-abundance guilds in the fruit tissues (Figure 7B). The growth morphology in the root tissues was associated with microfungi (71.3%) and facultative yeasts (17.8%), whereas that in the stem tissues was associated with microfungi (59.2%), tremelloid fungi (21.4%), and corticioid fungi (9.9%), and that in the leaf and fruit tissues was associated with microfungi (60.6 and 79.2%) and tremelloid fungi (31.8 and 18.1%) (Figure 7C). These results indicated that the relative abundance of endophytic fungi in different tissues significantly differed at the three functional levels.
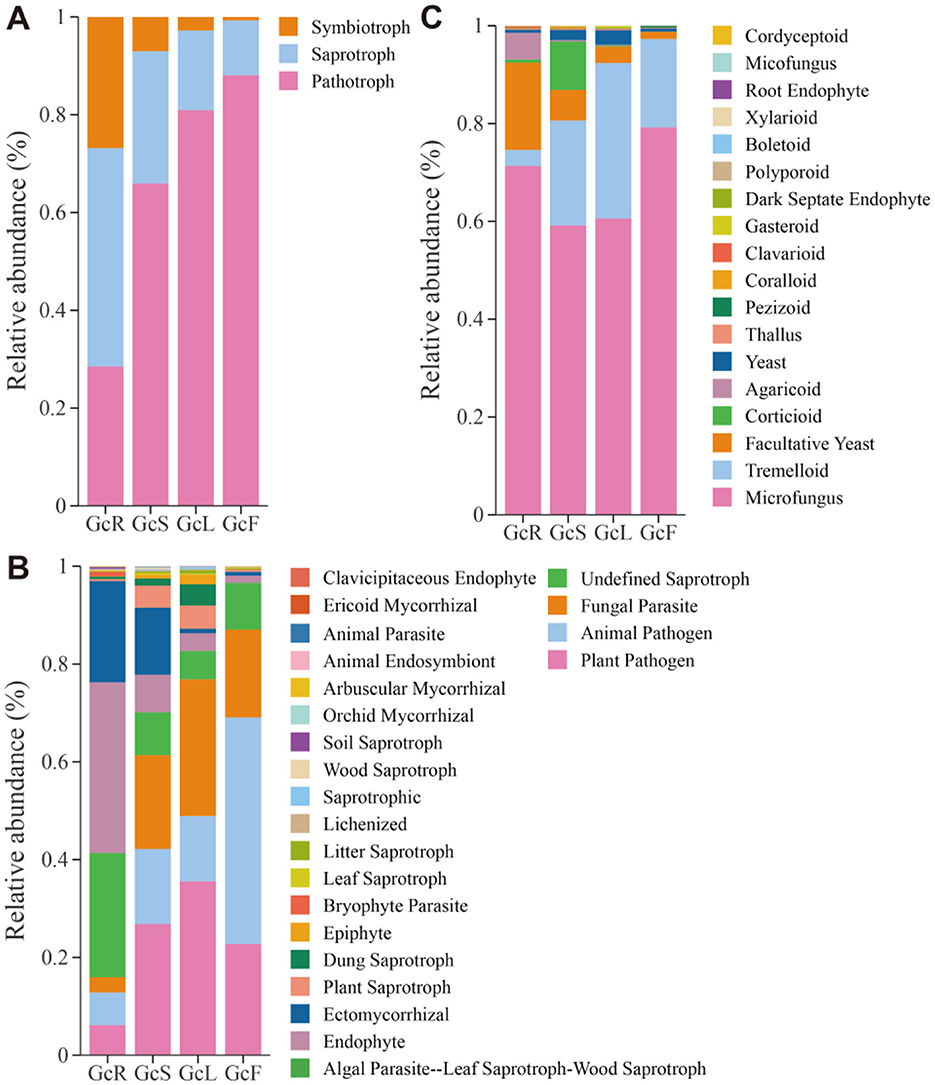
Figure 7. Functional prediction of endophytic fungi in different tissues of G. conopsea. (A) At the trophic mode level. (B) At the guilds level. (C) At the growth morphology level. GcR, root tissues, GcL, leaf tissues, GcS, stem tissues, GcF, fruit tissues.
4 Discussion
Extensive studies on endophytic plant interactions have focused mainly on endophytic microbes because of their potential functions in pharmaceuticals, ecology, industry and agriculture (Jha et al., 2023). In this study, we explored the endophytic fungal community structure and assembly in different tissues of G. conopsea. Prior to conducting relevant analyses, the sample sequences were first rarefied according to the minimum number of sequences per sample. The present study revealed that the endophytic fungal communities in the GcL and GcS samples presented the greatest number of OTUs, followed by those in the GcF and GcR samples. The richness indices of the GcS and GcL samples were significantly greater than those of the GcF and GcR samples. The richness index of the GcR samples was the lowest; however, there were no significant differences in the diversity indices among the different tissues. Previous studies have shown that endophytes have unique tissue specificity and are affected by various factors, such as climate, geographic location, genotype, growth stage, seasonality, and metabolites (Zhang et al., 2023; Yang et al., 2023; Li et al., 2025; Wang et al., 2022). Saha et al. reported that the diversity of endophytic fungi in Chromolaena odorata was highest in stems, followed by roots and leaves (Saha, 2024). Pooja et al. (2025) reported that the diversity in Lagenandra toxicaria root samples was greater than that in leaf samples. These results indicated that the endophytic diversity pattern was not universal.
The endophytic fungi in different tissues of G. conopsea were classified into 12 phyla, 37 classes, 113 orders, 294 families, and 621 genera. Ascomycota, Basidiomycota, and Mucoromycota were the dominant phyla; however, the relative abundance of Mucoromycota in root tissues, at 10.13%, was greater than that in other tissues, at less than 1% in leaf, stem, and fruit tissues. These findings corroborate previous studies showing that Ascomycota, Basidiomycota, and Mucoromycota were the predominant fungal phyla in the root tissues of G. conopsea (Lin et al., 2020). Previous studies have shown that endophytic fungi from different tissues belong to Ascomycota, Basidiomycota, and Zoopagomycota (Saha, 2024). Some species of Ascomycota and Basidiomycota participate in the carbon cycle by degrading organic matter (Unterseher et al., 2013), whereas Mucoromycota species have various agricultural benefits (Li et al., 2020), and the relative abundance of Mucoromycota is related to altitude and soil substrate (SOM, N, P, and K) (Ren et al., 2024). At the genus level, the dominant endophytic fungal community in different tissues differed, which might be related to tissue type and metabolites that varied among different tissues (Li et al., 2025; Wang et al., 2022). The germination of Orchidaceae seeds depends on mycorrhizal fungal colonization under natural conditions (Li et al., 2020). Previous studies have shown that endophytic fungi, such as Gibberella (Silva et al., 2018), Leptosphaerulina (Favre-Godal et al., 2020), Trichoderma (Zhang et al., 2016), Cladosporium (Favre-Godal et al., 2020), and Cadophora (Berthelot et al., 2016) species, can produce phytohormones. The mycorrhizal fungus Ceratobasidium GS2, isolated from G. conopsea, produces several steroid compounds that significantly promote the growth and differentiation of its protocorms (Shi et al., 2023). This study revealed that the relative abundances of Ceratobasidium, Cadophora, and Mortierella were significantly higher in root tissues than in other tissues. Furthermore, Ceratobasidium is classified under Basidiomycota, which was the dominant phylum across the four tissue types. These results indicate that roots harbor a relatively high abundance of growth-promoting fungi, suggesting that G. conopsea roots should be prioritized as the material for isolating growth-promoting endophytic fungi. In the GcR samples, the dominant genus Ceratobasidium had a relative abundance of 2.63%, which is relatively low for a genus composed of mycorrhizal fungi. We hypothesize two potential explanations for this phenomenon: First, the low abundance of Ceratobasidium may be one of the factors contributing to the endangered status of G. conopsea, though this remains merely a preliminary speculation. Second, other dominant mycorrhizal fungi may have colonized the roots of G. conopsea but were not amplified by the ITS primers used in this study. This speculation could be further validated through microscopic observation of the mycorrhizal morphology in G. conopsea roots in subsequent research. Mortierella species have potential functions in biocontrol, phosphorus absorption, and phytohormone production (Yakun, 2024). Cadophora (16.45%) and Mortierella (10.08%) were significantly enriched in the root tissues of G. conopsea, Cladosporium (1.54%−16.17%) was the dominant genus in the stem, leaf, and fruit tissues, and Gibberella was significantly enriched in the stem and leaf tissues. These genera might play crucial roles in the growth of G. conopsea.
A co-occurrence network not only can reveal more microbial complex interactions among different microbial taxa but is also a useful tool for identifying the key species that play crucial roles in maintaining microbial community stability (Xu et al., 2024). These highly complex microbial networks contribute to resisting biotic and abiotic stress to maintain host health (Qiao et al., 2024), which can be reflected by the number of edges, nodes, modularity index, and other topological parameters (Fan et al., 2022). In this study, the numbers of nodes and edges in the leaf (328, 1476) and stem (304, 1148) tissues were greater than those in the fruit (219, 968) and root (204, 804) tissues, and the modularity indices of the different tissues were greater than 0.4, indicating a stable modular structure. The smallest number of nodes and edges in root samples may be attributed to the failure of amplification of some dominant mycorrhizal fungi in the roots of G. conopsea due to mismatching with the primers used. The results corroborated those of a previous study showing that the endophytic fungal co-occurrence network was more complex in the leaf tissues than in the root tissues of medicinal plants (He et al., 2023). Moreover, these indices suggested that the endophytic fungal community in various tissues of G. conopsea exhibited complex network relationships. The G. conopsea samples were collected from high-altitude areas that were subject to multiple environmental pressures, such as intense UV-B radiation, low temperatures, and strong winds. Endophytic fungi associated with G. conopsea may assist their host in resisting abiotic stresses through highly intricate interactions, thereby endeavoring to maintain host health. In addition, the network analysis revealed that the network structure was mainly positive (≥94.77%). This result was similar to that of previous studies showing that the endophytic fungal co-occurrence network in Sophora alopecuroides tissues was mainly positive (96.6%) (Ju et al., 2024).
Microbial community assembly processes play crucial roles in shaping microbial diversity, composition, and ecological function (Zhang et al., 2024). In our study, we found that deterministic processes play crucial roles in the endophytic fungal community of G. conopsea. The endophyte community assembly in Eucommia ulmoides (Tang et al., 2024) and Rhododendron (Zhang et al., 2024) was mainly affected by stochastic processes, whereas the assembly processes of the root endophytic community in alpine grasslands shift under different climate and precipitation conditions (Wei et al., 2024). In addition, He et al. revealed that endophytic fungal community assembly is more strongly affected by the host than by the season (He et al., 2023). These results indicate that microbial community assembly processes are influenced by multiple biotic and abiotic factors; therefore, species identities, tissue niches and environmental factors might determine endophytic fungal community assembly processes in various tissues of G. conopsea. In this study, FUNGuild analysis revealed that the abundances of saprotrophs (44.7%−11.2%) and symbiotrophs (26.8%−0.7%) gradually decreased from root to fruit tissues. The function of saprotrophic fungi is mainly to degrade plant cells (non-host plant cells) to provide essential nutrients for the host, and research has shown that Ascomycota members constitute a large group of saprophytic species that promote nutrient cycling (He et al., 2023). Currently, we lack experimental evidence to support this hypothesis, as this study did not investigate the structure and diversity of the fungal community in the rhizospheric soil and bulk soil of G. conopsea. Therefore, additional subsequent experimental evidence is required to verify this hypothesis. Mucoromycota is a widely prevalent fungal phylum in nature, and its members are important lower saprophytic fungi, such as Mortierella (Yakun, 2024; Zhou et al., 2024); therefore, the relatively high abundance of Ascomycota and Mucoromycota in the root tissues of G. conopsea might be associated with nutrient cycling.
5 Conclusions
The various tissues of G. conopsea were rich in endophytic fungi, dominated by Ascomycota and Basidiomycota. The richness indices of various tissues were significantly different; however, the diversity was not significantly different, and that in the root tissues being relatively low. The compositions of the endophytic fungal communities in leaf, stem, and fruit tissues were similar, while that in root tissues differed from those in other tissues. The relative abundances of Ceratobasidium, Cadophora, and Mortierella were significantly higher in root tissues than in other tissues, G. conopsea roots should be prioritized as the material for isolating growth-promoting endophytic fungi. Co-occurrence network analysis revealed that endophytic fungi in different tissues presented typical modular structures and were mainly positive. Deterministic processes play crucial roles in the endophytic fungal community of G. conopsea. The proportions of pathotrophs, saprotrophs, and symbiotrophs were different among various tissues, and the proportion of pathotrophs was greater than those of saprotrophs and symbiotrophs. Overall, tissue type affects the composition and function of endophytic fungi.
Data availability statement
The datasets presented in this study are publicly available. This data can be found here: https://www.ncbi.nlm.nih.gov/sra, accession number PRJNA699676.
Author contributions
XY: Data curation, Formal analysis, Investigation, Writing – original draft. YL: Data curation, Formal analysis, Investigation, Writing – original draft. HQ: Data curation, Formal analysis, Writing – original draft. EZ: Data curation, Formal analysis, Writing – review & editing. ZW: Data curation, Formal analysis, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was supported financially by the selection and cultivation of High-Quality Orchid Varieties in Linzhi City (XDHZ-2025-04), the Ministry of Education's Experimental Teaching and Laboratory Construction Research Project (SYJX2024-195), Xizang Autonomous Region Major Special Science and Technology (grant no. XZ201901-GA-04), Xizang Autonomous Region Major Special Science and Technology (Nos. XZ202101ZD0023G), Base and Talent Fundation of Science and Technology Department of Xizang Autonomous Region (XZ202501JD0026), The Forth National Survey of Traditional Chinese Medicine Resources, and Chinese or Xizang Medicinal Resources Investigation in Xizang Autonomous Region (State Administration of Chinese Traditional Medicine (20200501-542301), Xizang Agriculture and Animal Husbandry University Doctoral Program in Forestry (Phase I) (533325001).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Gen AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1640133/full#supplementary-material
References
Berthelot, C., Leyval, C., Foulon, J., Chalot, M., and Blaudez, D. (2016). Plant growth promotion, metabolite production and metal tolerance of dark septate endophytes isolated from metal-polluted poplar phytomanagement sites. FEMS Microbiol. Ecol. 92:fiw144. doi: 10.1093/femsec/fiw144
Bonnardeaux, Y., Brundrett, M., Batty, A., Dixon, K., Koch, J., and Sivasithamparam, K. (2007). Diversity of mycorrhizal fungi of terrestrial orchids: compatibility webs, brief encounters, lasting relationships and alien invasions. Mycol. Res. 111, 51–61. doi: 10.1016/j.mycres.2006.11.006
Carper, D. L., Carrell, A. A., Kueppers, L. M., and Frank, A. C. (2018). Bacterial endophyte communities in Pinus flexilis are structured by host age, tissue type, and environmental factors. Plant Soil 428, 335–352. doi: 10.1007/s11104-018-3682-x
Chathurdevi, G., and Gowrie, S. U. (2016). Endophytic fungi isolated from medical plant—a promising source of potential bioactive metabolites. Int. J. Curr. Pharm. Res. 8 50–56.
Chen, C. J., Guo, G., Li, M., Liang, X. Y., and Gu, Y. Y. (2022). Diversity of endophytic bacteria of mulberry (Morus L.) under cold conditions. Front. Microbiol. 13:923162. doi: 10.3389/fmicb.2022.923162
Chen, L., Wang, Y. C., Qin, L. Y., He, H. Y., Yu, X. L., Yang, M. Z., et al. (2019). Dynamics of fungal communities during Gastrodia elata growth. BMC Microbial. 19:158. doi: 10.1186/s12866-019-1501-z
Edgar, R. C. (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nature Met. 10, 996–998. doi: 10.1038/nmeth.2604
Fan, Q., Xie, K., Cui, X., Zhang, G., Zheng, H., Chang, S., et al. (2022). Microecosystem of yak rumen on the Qinghai-Tibetan Plateau is stable and is unaffected by soil or grass microbiota. Environ. Microbiol. 24, 5760–5773. doi: 10.1111/1462-2920.16236
Favre-Godal, Q., Gourguillon, L., Lordel-Madeleine, S., Gindro, K., and Choisy, P. (2020). Orchids and their mycorrhizal fungi: an insufficiently explored relationship. Mycorrhiza 30, 5–22. doi: 10.1007/s00572-020-00934-2
Firáková, S., Šturdíková, M., and Múčková, M. (2007). Bioactive secondary metabolites produced by microorganisms associated with plants. Biologia 62, 251–257. doi: 10.2478/s11756-007-0044-1
Gao, Y., Zhao, Z., Li, J., Liu, N., Jacquemyn, H., Guo, S., et al. (2020). Do fungal associates of co-occurring orchids promote seed germination of the widespread orchid species Gymnadenia conopsea. Mycorrhiza 30, 221–228. doi: 10.1007/s00572-020-00943-1
Gong, Z., Xiong, L., Shi, H., Yang, S., Herrera-Estrella, L. R., Chao, D. Y., et al. (2020). Plant abiotic stress response and nutrient use efficiency. Sci. China Life Sci. 63, 635–674. doi: 10.1007/s11427-020-1683-x
He, C., Meng, D., Li, W., Li, X., and He, X. (2023). Dynamics of endophytic fungal communities associated with cultivated medicinal plants in farmland ecosystem. J. Fungi 9:1165. doi: 10.3390/jof9121165
Huynh, T. T., Thomson, R., Mclean, C. B., and Lawrie, A. C. (2009). Functional and genetic diversity of mycorrhizal fungi from single plants of Caladenia formosa (Orchidaceae). Ann. Botany 104, 757–765. doi: 10.1093/aob/mcp153
Jha, P., Kaur, T., Chhabra, I., Panja, A., Paul, S., Kumar, V., et al. (2023). Endophytic fungi: hidden treasure chest of antimicrobial metabolites interrelationship of endophytes and metabolites. Front. Microbiol. 14:1227830. doi: 10.3389/fmicb.2023.1227830
Ju, M., Zhang, Q., Wang, R., Yan, S., Zhang, Q., Li, P., et al. (2024). Community ecological succession of endophytic fungi associates with medicinal compound accumulation in Sophora alopecuroides. Microbiol. Spect. 12, e03076–e03023. doi: 10.1128/spectrum.03076-23
Kivlin, S. N., Mann, M. A., Lynn, J. S., Kazenel, M. R., Taylor, D. L., and Rudgers, J. A. (2022). Grass species identity shapes communities of root and leaf fungi more than elevation. ISME Commun. 2:25. doi: 10.1038/s43705-022-00107-6
Kõljalg, U., Nilsson, R. H., Abarenkov, K., Tedersoo, L., Taylor, A. F. S., Bates, S. T., et al. (2013). Towards a unified paradigm for sequence-based identification of fungi. Mol. Ecol. 22, 5271–5277. doi: 10.1111/mec.12481
Kusari, S., Pandey, S. P., and Spiteller, M. (2013). Untapped mutualistic paradigms linking host plant and endophytic fungal production of similar bioactive secondary metabolites. Phytochemistry 91, 81–87. doi: 10.1016/j.phytochem.2012.07.021
Lebeis, S. L., Paredes, S. H., Lundberg, D. S., Breakfield, N., Gehring, J., McDonald, M., et al. (2015). Salicylic acid modulates colonization of the root microbiome by specific bacterial taxa. Science 349, 860–864. doi: 10.1126/science.aaa8764
Li, F., Zhang, S., Wang, Y., Li, Y., and Han, Y. (2020). Rare fungus, mortierella capitata, promotes crop growth by stimulating primary metabolisms related genes and reshaping rhizosphere bacterial community. Soil Biol. Biochemist. 151:108017. doi: 10.1016/j.soilbio.2020.108017
Li, L., Zheng, R., Wang, Z., Li, H., Shi, Y., Pan, Z., et al. (2024). Leaf health status regulates endophytic microbial community structure, network complexity, and assembly processes in the leaves of the rare and endangered plant species Abies fanjingshanensis. Microorganisms 12:1254. doi: 10.3390/microorganisms12071254
Li, M., Wang, C., and Guo, S. (2006). Advances in studies on chemical constituents and pharmacological activities for plants of Gymnadenia R. Br. Chin. Trad. Herb. Drug. 37:1264. (in Chinese)
Li, S., Liu, Y., Yang, X., Yang, Y., Peng, J., Xu, Y., et al. (2025). Spatiotemporal composition and diversity of endophyte communities in Dracaena cambodiana on Hainan Island. Front. Microbiol. 16:1540669. doi: 10.3389/fmicb.2025.1540669
Liang, J., Zou, R., Huang, Y., Qin, H., Tang, J., Wei, X., et al. (2022). Structure and diversity of mycorrhizal fungi communities of different part of Bulbophyllum tianguii in three terrestrial environments. Front. Plant Sci. 13:992184. doi: 10.3389/fpls.2022.992184
Lin, M., Xiong, H., Xiang, X., Zhou, Z., Liang, L., and Mei, Z. (2020). The effect of plant geographical location and developmental stage on root-associated microbiomes of Gymnadenia conopsea. Front. Microbiol. 11:1257. doi: 10.3389/fmicb.2020.01257
Lin, Z. C. (2009). The pharmacology of Gymnadenia conopsea. Dissertation, Guangzhou University of Chinese Medicine, 10–23. (in Chinese)
Magoc, T., and Salzberg, S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
Morikawa, T., Xie, H., Matsuda, H., Wang, T., and Yoshikawa, M. (2006). Bioactive constituents from Chinese natural medicines. XVII. Constituents with radical scavenging effect and new glucosyloxybenzyl 2-isobutylmalates from Gymnadenia conopsea. Chem. Pharmaceut. Bull. 54, 506–513. doi: 10.1248/cpb.54.506
Nguyen, N. H., Song, Z., Bates, S. T., Branco, S., Tedersoo, L., Menke, J., et al. (2016). FUNGuild: an open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 20, 241–248. doi: 10.1016/j.funeco.2015.06.006
Ning, D., Deng, Y., Tiedje, J. M., and Zhou, J. (2019). A general framework for quantitatively assessing ecological stochasticity. Proceed. Nat. Acad. Sci. 116, 16892–16898. doi: 10.1073/pnas.1904623116
Omomowo, I. O., Amao, J. A., Abubakar, A., Ogundola, A. F., Ezediuno, L. O., and Bamigboye, C. O. (2023). A review on the trends of endophytic fungi bioactivities. Sci. African 20:e01594. doi: 10.1016/j.sciaf.2023.e01594
Pang, Z., Chen, J., Wang, T., Gao, C., Li, Z., Guo, L., et al. (2021). Linking plant secondary metabolites and plant microbiomes: a review. Front. Plant Sci. 12:621276. doi: 10.3389/fpls.2021.621276
Pooja, P., Edison, L. K., and Pradeep, N. S. (2025). Ecotype variations and endophytic fungal diversity of aquatic angiosperm A case study with Lagenandra toxicaria Dalz. Horizon 12, 1–12. doi: 10.14719/pst.5200
Qiao, Y., Wang, T., Huang, Q., Guo, H., Zhang, H., Xu, Q., et al. (2024). Core species impact plant health by enhancing soil microbial cooperation and network complexity during community coalescence. Soil Biol. Biochemist. 188:109231. doi: 10.1016/j.soilbio.2023.109231
Ren, Y. L., Cao, Q. B., Lu, M., and Li, C. (2024). Differentiated contributions of plant diversity and soil parameters to shifts in fungal taxonomic composition across three altitudinal transects. Microbiology 93, 640–653. doi: 10.1134/S0026261723600404
Saha, K. (2024). Seasonal variation and diversity of endophytic fungi in Chromolaena odorata (L.) King and Robinson, an invasive alien weed of Tripura, Northeast, India. Asian J. Pharmacy Pharmacol. 10, 1–12. doi: 10.31024/ajpp.2024.10.1.1
Sathiyadash, K., Muthukumar, T., Uma, E., and Pandey, R. R. (2012). Mycorrhizal association and morphology in orchids. J. Plant Interact. 7, 238–247. doi: 10.1080/17429145.2012.699105
Schulz, B., and Boyle, C. (2005). The endophytic continuum. Mycol. Res. 109, 661–686. doi: 10.1017/S095375620500273X
Seleiman, M. F., Al-Suhaibani, N., Ali, N., Akmal, M., Alotaibi, M., Refay, Y., et al. (2021). Drought stress impacts on plants and different approaches to alleviate its adverse effects. Plants 10:259. doi: 10.3390/plants10020259
Shang, X., Guo, X., Liu, Y., Pan, H., Miao, X., and Zhang, J. (2017). Gymnadenia conopsea (L.) R. Br.: a systemic review of the ethnobotany, phytochemistry, and pharmacology of an important Asian folk medicine. Front. Pharmacol. 8:24. doi: 10.3389/fphar.2017.00024
Shi, L., Zhao, Z., Yang, L., Ding, G., and Xing, X. (2023). Bioactive steroids from seed germination supporting fungus (Ceratobasidium GS2) of the terrestrial orchid Gymnadenia conopsea. Mycology 14, 371–380. doi: 10.1080/21501203.2023.2254893
Si, Q., and Liu, T. H. (2013). Anti-aging effect and mechanism of Shouzhangshen-37 Pill from Mongolian medicine on subacute aging mice. Chin. J. Exper. Trad. Med. Formul. 19, 194–197. (in Chinese)
Silva, F. D. A., Liotti, R. G., Boleti, A. P. D. A., Reis, É. D. M., Passos, M. B. S., Dos Santos, E. L., et al. (2018). Diversity of cultivable fungal endophytes in Paullinia cupana (Mart.) Ducke and bioactivity of their secondary metabolites. PloS ONE 13:e0195874. doi: 10.1371/journal.pone.0195874
Tan, R. X., and Zou, W. X. (2001). Endophytes: a rich source of functional metabolites. Nat. Prod. Rep. 18, 448–459. doi: 10.1039/b100918o
Tang, Y., Tian, C., Yao, D., Yang, S., Shi, L., Yi, L., et al. (2024). Community assembly and potential function analysis of the endophyte in eucommia ulmoides. BMC Microbiol. 24:460. doi: 10.1186/s12866-024-03601-0
Unterseher, M., Peršoh, D., and Schnittler, M. (2013). Leaf-inhabiting endophytic fungi of European Beech (Fagus sylvatica L.) co-occur in leaf litter but are rare on decaying wood of the same host. Fungal Divers. 60, 43–54. doi: 10.1007/s13225-013-0222-0
Vellend, M. (2010). Conceptual synthesis in community ecology. Quart. Rev. Biol. 85, 183–206. doi: 10.1086/652373
Wang, L. X., Ren, L. L., Li, C. C., Gao, C. L., Liu, X. B., Wang, M., et al. (2019). Effects of endophytic fungi diversity in different coniferous species on the colonization of Sirex noctilio (Hymenoptera: Siricidae). Sci Rep. 9:5077. doi: 10.1038/s41598-019-41419-3
Wang, T., Chi, M., Chen, J., Liang, L., Wang, Y., Chen, Y., et al. (2024). The diversity and growth-promoting potential of the endophytic fungi of Neuwiedia singapureana (Orchidaceae) in China. Diversity 16:34. doi: 10.3390/d16010034
Wang, Y., Tong, Y., Adejobi, O. I., Wang, Y., and Liu, A. (2022). Research advances in multi-omics on the traditional Chinese herb Dendrobium officinale. Front. Plant Sci. 12:808228. doi: 10.3389/fpls.2021.808228
Wei, X., Han, B., Zhang, J., and Shao, X. (2024). Shifts in structure and assembly processes of root endophytic community caused by climate warming and precipitation increase in alpine grassland. Microorganisms 12:1780. doi: 10.3390/microorganisms12091780
Xu, Q., Jiang, D., Zhou, N., Kang, Y., Li, M., Yang, C., et al. (2024). Community structure of soil microorganisms and endophytes of honeysuckle at different ecological niche specificities. BMC Microbiol. 24:367. doi: 10.1186/s12866-024-03518-8
Yakun, S. I. (2024). Mechanism and application of Mortierella fungi mediating the formation of fluvo-aquic soil aggregates and phosphorus turnover [D], Henan Agricultural University. In Chinese.
Yang, H. J., Ye, W. W., Yu, Z., Shen, W. L., Li, S. Z., Wang, X., et al. (2023). Host niche, genotype, and field location shape the diversity and composition of the soybean microbiome. J. Integr. Agric. 2095–3119. doi: 10.1016/j.jia.2023.01.006
Yuan, X. L., Cao, M., Liu, X. M., Du, Y. M., Shen, G. M., et al. (2018). Composition and genetic diversity of the Nicotiana tabacum microbiome in different topographic areas and growth periods. Int. J. Mol. Sci. 19:3421. doi: 10.3390/ijms19113421
Zhang, L., Hu, H., Gui, T., Gao, X., Xu, Q., and Cai, J. (2023). Diversity of endophytic bacterial community in different tissues of Pyracantha fortuneana. Guihaia 43, 1193–200. doi: 10.11931/guihaia.gxzw202206031
Zhang, R., Zhou, X. L., Yang, L., Long, B., and Shen, S. K. (2024). Composition and assembly of the endophytic fungal community of alpine rhododendron hosts along elevation gradients. Phytobiom. J. 8, 236–247. doi: 10.1094/PBIOMES-02-24-0015-R
Zhang, S., Gan, Y., and Xu, B. (2016). Application of plant-growth-promoting fungi Trichoderma longibrachiatum T6 enhances tolerance of wheat to salt stress through improvement of antioxidative defense system and gene expression. Front. Plant Sci. 7:1405. doi: 10.3389/fpls.2016.01405
Zhang, T. E., Chen, C. Y., Li, S. H., Chen, C., Liu, W. W., and Yan, Z. Y. (2013). Effect of the extract of Gymnadenia conopsea on the blood lipid and liver function in experimental hyperlipidemia rats. Lishizhen Med. Mater. Med. Res. 24, 865–867. (in Chinese)
Zhao, L., and Liu, G. Q. (2011). Experimental study of Shouzhang Shen liquids on anti-fatigue effects in mice. Clin. J. Chin. Med. 22:17. (in Chinese)
Zhao, Y., Xiong, Z., Wu, G. L., Bai, W. X., Zhu, Z. Q., Gao, Y. H., et al. (2018). Fungal endophytic communities of two wild Rosa varieties with different powdery mildew susceptibilities. Front. Microbiol. 9:2462. doi: 10.3389/fmicb.2018.02462
Zheng, Y., and Gong, X. (2019). Niche differentiation rather than biogeography shapes the diversity and composition of microbiome of Cycas panzhihuaensis. Microbiome 7, 1–19. doi: 10.1186/s40168-019-0770-y
Keywords: Gymnadenia conopsea, endophytic fungi, community structure, network, assembly, tissue type
Citation: Yin X, Lu Y, Quan H, Zhang E and Wang Z (2025) Exploring the fungal community structure and assembly in different tissues of Gymnadenia conopsea. Front. Microbiol. 16:1640133. doi: 10.3389/fmicb.2025.1640133
Received: 03 June 2025; Accepted: 18 September 2025;
Published: 10 October 2025.
Edited by:
Mariusz Cycoń, Medical University of Silesia, PolandReviewed by:
Chaoyuan Zheng, Fujian Agriculture and Forestry University, ChinaMin Yang, Kunming University, China
Monika Malicka, University of Silesia in Katowice, Poland
Shu Wang, Southwest Forestry University, China
Copyright © 2025 Yin, Lu, Quan, Zhang and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Erhao Zhang, emhhbmdlcmhhb0B4emEuZWR1LmNu; Zhongbin Wang, MDgyM3l1YW4xMDIwQDE2My5jb20=
 Xiu Yin
Xiu Yin Yazhou Lu
Yazhou Lu Hong Quan1,2
Hong Quan1,2 Erhao Zhang
Erhao Zhang Zhongbin Wang
Zhongbin Wang