- 1State Key Laboratory of Microbial Diversity and Innovative Utilization, Institute of Microbiology, Chinese Academy of Sciences, Beijing, China
- 2State Key Laboratory of Biopharmaceutical Preparation and Delivery, Chinese Academy of Sciences, Beijing, China
- 3State Key Laboratory of Microbial Technology, Shandong University, Qingdao, China
- 4Department of Rheumatology and Clinical Immunology, Peking Union Medical College Hospital, Chinese Academy of Medica Sciences & Peking Union Medical College, Beijing, China
Autism spectrum disorders (ASD), a group of neurodevelopmental disorders characterized by the core symptoms of impaired social communication and stereotyped behaviors, is strongly associated with dysregulated microbiota-gut-brain axis. Emerging evidence suggests that Faecalibacterium, which showed reduced abundance in ASD cohorts, holds therapeutic potential, though its interaction with host remain unexplored. Here, we investigated the efficacy and molecular basis of Faecalibacterium hominis 4P-15 (4P-15) in BTBR T+ Itpr3tf/J (BTBR) mice, an idiopathic ASD mouse model. Oral administration of 4P-15 significantly reduced the intestinal levels of indole, indole-3-propionic acid (IPA), and indole-3-acetic acid (IAA), as well as the level of IPA in brain. Furthermore, the decreased levels of IPA in brain contributed to the attenuated aryl hydrocarbon receptor (AhR) signaling characterized by increased expression of downstream elements, including glutamate transporters and GABA receptors. Ultimately, this modulation led to the restoration of excitatory/inhibitory imbalance, a typical pathophysiological feature of ASD, and thereby alleviated ASD core behavioral symptoms. Our findings underscore Faecalibacterium-mediated AhR modulation as a promising therapeutic strategy for ASD, highlighting the dual potential of Faecalibacterium-based probiotics and targeted interventions against indole-AhR signaling to address neurodevelopmental disorders.
Introduction
Autism spectrum disorders (ASD), a group of neurodevelopmental disorders characterized by core symptoms of impaired social communication (e.g., reduced social reciprocity and atypical eye contact) and stereotyped behaviors such as stereotypical motor movements and insistence on sameness, has increasingly been linked to dysregulated gut-brain axis (Association, 2013). Emerging evidence highlights the gut microbiome as a pivotal modulator of neurodevelopment, with microbiota dysbiosis now recognized as a hallmark of ASD. Studies consistently report altered gut microbial composition in ASD patients and animal models (Liu et al., 2019; Lou et al., 2022; Chen et al., 2019). Notably, microbial signatures in ASD significantly correlate with the core symptoms, underlining the potential clinical applicability of the fecal microbiome for aiding in the diagnosis of ASD (Su et al., 2024; Wan et al., 2024; Manghi et al., 2024). Furthermore, therapeutic strategies targeting the gut microbiome, such as fecal microbiota transplantation, probiotic colonization, and prebiotic and dietary interventions, have shown promise in alleviating ASD symptoms (Li et al., 2024; Sgritta et al., 2019; Kang et al., 2017; Yu et al., 2025). These advances position gut microbiota remodeling as a viable therapeutic frontier in ASD.
Emerging evidence highlights the gut microbiome as a pivotal regulator of neurodevelopment by modulating synaptic plasticity and neurotransmitter synthesis, primarily via microbial metabolites, immune activation, and vagal nerve signaling. In recent years, bacteria-derived metabolites have garnered significant attention as key mediators in the microbiota-gut-brain axis, influencing how commensal gut microbes shape host neural activity. Indole and its derivatives, produced through the metabolism of tryptophan by gut microbes, have emerged as crucial elements influencing neurological function (Agus et al., 2018; Zhou et al., 2023). Cohort studies have indicated a correlation between indoles and abnormalities in the nervous system. Elevated levels of several indoles, such as indole, indole-3-acetic acid, and 3-methylindole, have been observed in the feces and urine of children with ASD (Khan et al., 2022), suggesting the potentially detrimental effect of indole and its derivatives. As the receptor for indole and its derivatives, the aryl hydrocarbon receptor (AhR) plays a critical role in regulating the transcription of various downstream genes, some of which are responsible for neurotransmission. Furthermore, AhR signaling pathway is dysregulated in the brain of ASD mice. However, the mechanisms through which gut microbiota impact the AhR signaling pathway in ASD and how this interference affects ASD-related phenotypes via the gut-brain axis remain poorly understood.
Faecalibacterium, emerging as a next-generation probiotic candidate, has drawn significant interest due to its anti-inflammatory properties and ability to enhance intestinal barrier integrity (Sokol et al., 2008; Lapiere et al., 2020). Notably, a reduced abundance of Faecalibacterium has been observed in multiple cohorts of individuals with ASD (Kang et al., 2018; Zhao et al., 2023; Pang et al., 2023; De Angelis et al., 2013; Ding et al., 2020; Wan et al., 2022) and the abundance of Faecalibacterium was negatively associated with the severity of ASD (Ding et al., 2020), yet its therapeutic potential and underlying mechanisms in ASD remain largely unexplored. In this study, we focused on Faecalibacterium hominis 4P-15 (4P-15), a new species isolated by our team from the feces of a typically developing individual (Liu et al., 2021), to assess its efficacy and investigate its underlying mechanisms using the BTBR T+ Itpr3tf/J (BTBR) mice, a widely recognized idiopathic model of ASD. The BTBR mice exhibit pronounced social deficits, as evidenced by their avoidance of social interaction with unfamiliar mice in three-chamber social interaction test and stereotypic behaviors supported by excessive self-grooming in self-grooming test. Our investigation identifies that 4P-15 normalizes abnormal levels of indole and its derivatives in the intestine and brain, and corrects the downstream of AhR signaling pathway associated with excitatory/inhibitory imbalance in ASD, thereby alleviating core behavioral deficits of ASD. Our investigation underscores the significance of intestinal commensal bacteria in regulating ASD and highlights potential mechanisms, laying a theoretical groundwork for probiotic intervention and indicating a fresh perspective for the clinical management of ASD by targeting indole and its derivatives and AhR signaling.
Results
Faecalibacterium hominis 4P-15 has been isolated and cultured from fecal samples of the typically developing individual
We observed that the abundance of Faecalibacterium reduced in ASD individuals compared to typically developing (TD) individuals in several cohorts (Kang et al., 2018; Zhao et al., 2023; Pang et al., 2023; De Angelis et al., 2013; Ding et al., 2020), suggesting that Faecalibacterium may play a protective role in the context of ASD (Figure 1a and Supplementary Figure 1a). To obtain the microbial strains belonging to Faecalibacterium, we isolated and cultured the gut microbes derived from the fecal samples of TD and ASD individuals. We harvested 183 bacterial strains belonging to 65 species (Figure 1b). Focusing on Faecalibacterium, two Faecalibacterium prausnitzii strains and four Faecalibacterium hominis strains were obtained (Figure 1b). Notably, F. hominis was identified as a new species, and all strains of this species were isolated from the TD sample (Figure 1c). Consequently, we selected the strain F. hominis 4P-15 (4P-15) for further functional research (Figure 1b, the red triangle).
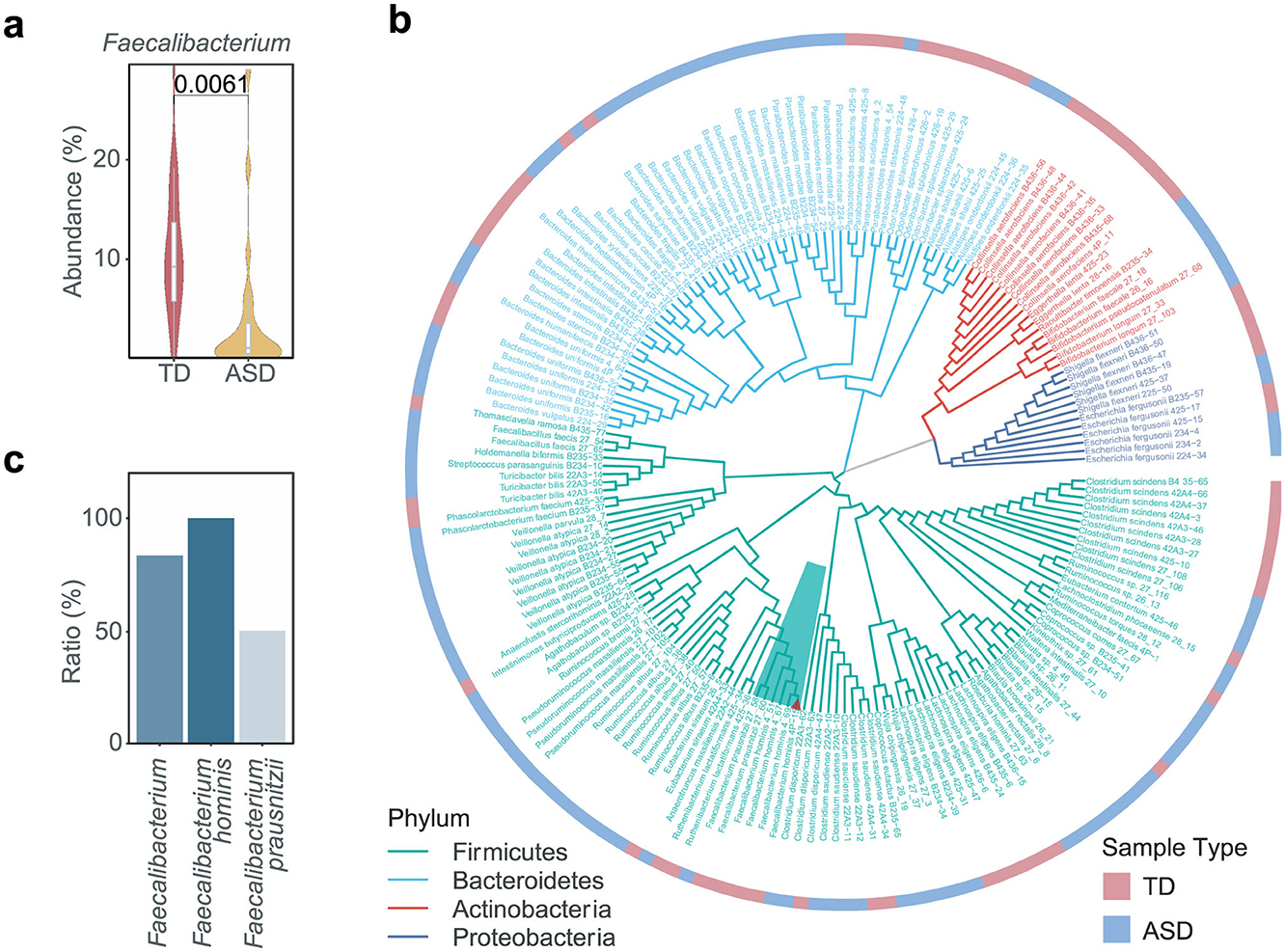
Figure 1. The isolation and culture of Faecalibacterium from fecal samples. (a) The relative abundance of Faecalibacterium between TD and ASD individuals was shown. The figure was based on the original data of a selected ASD cohort (ID number 11169 in the database “Qiita”) (Kang et al., 2018). TD, typically developing children. (b) The phylogenetic tree depicts the isolated gut microbial strains from TD and ASD samples. The branches are color-coded to represent four distinct phyla, with the dark green background specifically highlighting the genus Faecalibacterium. The red triangle denotes the strain Faecalibacterium hominis 4P-15. The heatmap surrounding the phylogenetic tree indicates the type of sample from which each strain was isolated. (c) The ratio of Faecalibacterium strains isolated from fecal samples of the TD individual compared to those isolated from fecal samples of both TD and ASD individuals. Quantitative data are shown as the mean ± SEM (a). The P-value was determined by the two-sided Wilcoxon rank-sum test and the adjusted P-value was evaluated using the false discovery rate (FDR) correction of P-value (a), and the significance was indicated by adjusted P-value (a).
4P-15 has shown potential in alleviating the core deficits of ASD in BTBR mice
To explore whether 4P-15 could play a protective role in ASD by alleviating the ASD-like phenotypes in ASD animal model, we gavaged weaned BTBR mice daily for 4 weeks (Figure 2a). Subsequently, we evaluated ASD-relevant behaviors using the three-chamber test for social interaction deficits and the self-grooming test for stereotyped behavior. The results indicated that 4P-15 restored the stereotyped behavior in the self-grooming test (Figure 2b) and the impaired social interaction in the social ability session of the three-chamber test (Figure 2c and Supplementary Figure 2a), the core deficits of ASD. Furthermore, we conducted the open-field test for anxiety and the sucrose preference test for depression to assess the impact of 4P-15 on these behaviors. We did not observe any significant improvement of 4P-15 on anxiety or depression (Supplementary Figures 2b, c). Furthermore, we analyzed the correlation between the abundance of F. hominis in the feces and behavioral outcomes, and found a significant negative correlation between F. hominis abundance and stereotypic behavior in the F. hominis-gavaged BTBR mice (Supplementary Figures 2d, e).
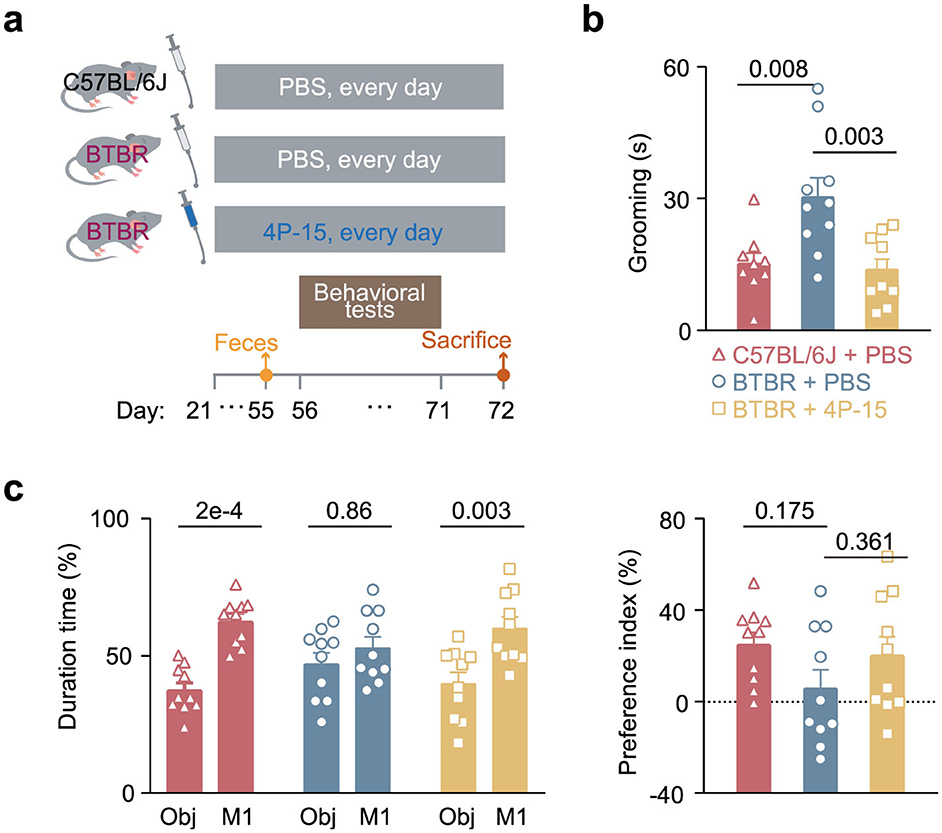
Figure 2. The restoration of core ASD-like behaviors in BTBR mice by F. hominis 4P-15. (a) Schematic diagram of the study design. (b) Grooming time in the self-grooming test. n = 10 mice for each group. c, Percentage of duration time (left panel) and the preference index (right panel) in the social ability session of the three-chamber social interaction test. Ob, object. n = 10 mice for each group. Quantitative data are shown as the mean ± SEM. The adjusted P-values were determined by one-way ANOVA with two-tailed Tukey's multiple comparison tests [(b) and (c) (right panel)] and two-way ANOVA with two-tailed Turkey's test for multiple comparisons (c, left panel). Significance was indicated by adjusted P-values (b, c).
4P-15 does not affect inflammatory cytokines and SCFAs in the intestine and neurotransmitters in the brain
Next, we explored the alterations associated with the mechanisms of the gut-brain axis in both the intestine and the brain in response to 4P-15 administration (Yu, 2021). Given that the genome of 4P-15 contained a microbial anti-inflammatory molecule showing 96.56% similarity (https://blast.ncbi.nlm.nih.gov/Blast.cgi) to F. prausnitzii (Auger et al., 2022) (Figure 3a), we hypothesized the anti-inflammatory properties of 4P-15 and the anti-inflammatory effect might mediate the protective effect of 4P-15 on ASD-like phenotypes. We analyzed six inflammatory cytokines in the cerebral cortex following gavage and found that 4P-15 did not affect the levels of these cytokines (Figure 3b). Then, we examined bacteria-derived compounds such as SCFAs and neurotransmitters that were previously reported to have altered levels in BTBR mice and participate in the modulation of CNS activity by gut microbes (Liu et al., 2024; Bove et al., 2024). Our results showed no significant changes in the levels of SCFAs in the feces or neurotransmitters in the brain after 4P-15 treatment (Supplementary Figures 2f, g).
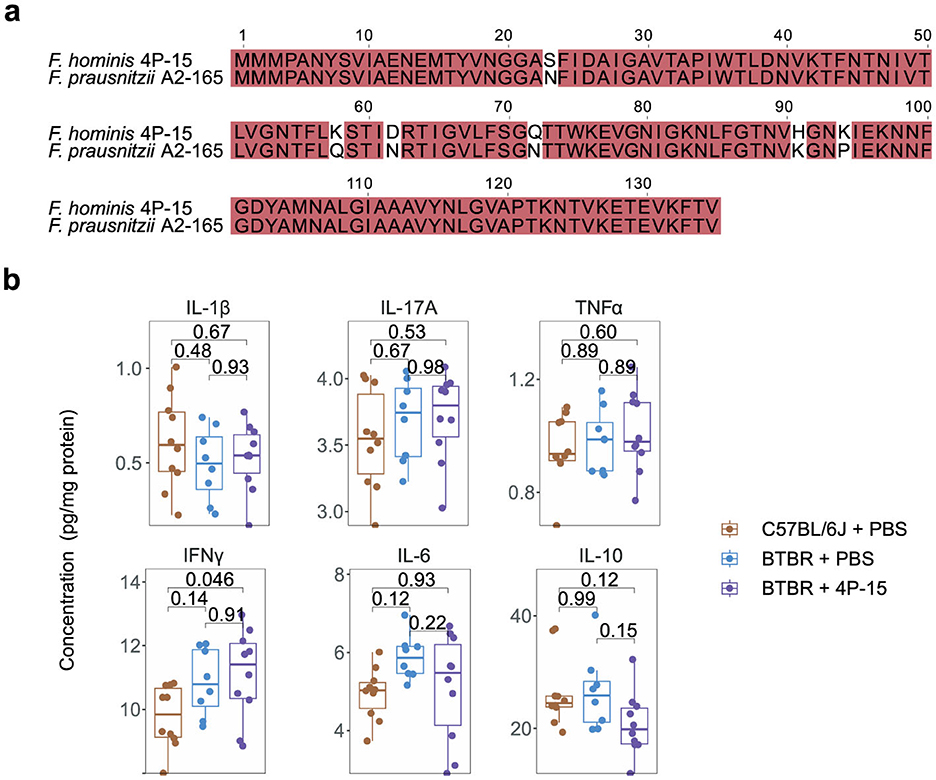
Figure 3. The non-effect of F. hominis 4P-15 on inflammatory cytokines, neurotransmitters, or SCFAs in BTBR mice. (a) The microbial anti-inflammation molecule (MAM) amino acid sequence alignment between F. hominis 4P-15 and Faecalibacterium prausnitzii A2-165. The identical sequences were highlighted in red. (b) The concentrations of inflammatory cytokines in the cerebral cortex of the brain. n = 10, 8 and 10 mice, respectively. Quantitative data are shown as the mean ± SEM. The adjusted P-values were determined by one-way ANOVA with two-tailed Tukey's multiple comparison tests (b). Significance was indicated by adjusted P-values (b).
4P-15 restores the abnormal levels of intestinal indole and its derivatives by modulating the potential indole-producing bacteria
Next, we explore the functional mechanisms via the gut-brain axis following the 4P-15 intervention, focusing on the intrinsic defects of the ASD mouse model we utilized. Previous studies reported excessive activation of the AhR signaling pathway in BTBR mice, characterized by increased expression of AhR (Uddin et al., 2023). Hyperactivation of AhR may lead to neurodevelopmental toxicity, contributing to the pathogenesis of ASD (Kimura et al., 2017; Siddiqui et al., 2021; Martin et al., 2022). In our investigation, we sought to determine whether 4P-15 could alleviate the hyperactivation of AhR by reducing the levels of AhR ligands, such as indole and its derivatives, which are primarily produced by gut bacteria and function mainly through AhR activation. We analyzed levels of indole and six indole derivatives in fecal samples (Figure 4a). Our results revealed elevated levels of indole and four derivatives, indole-3-acetic acid (IAA), indole-3-propionic acid (IPA), indole-3-lactic acid (ILA) and indole-3-aldehyde (IAId) in the feces of BTBR mice compared to C57BL/6J mice (Figure 4b). Notably, administration of 4P-15 led to a reduction in the levels of indole, IPA, and IAA (Figure 4b).
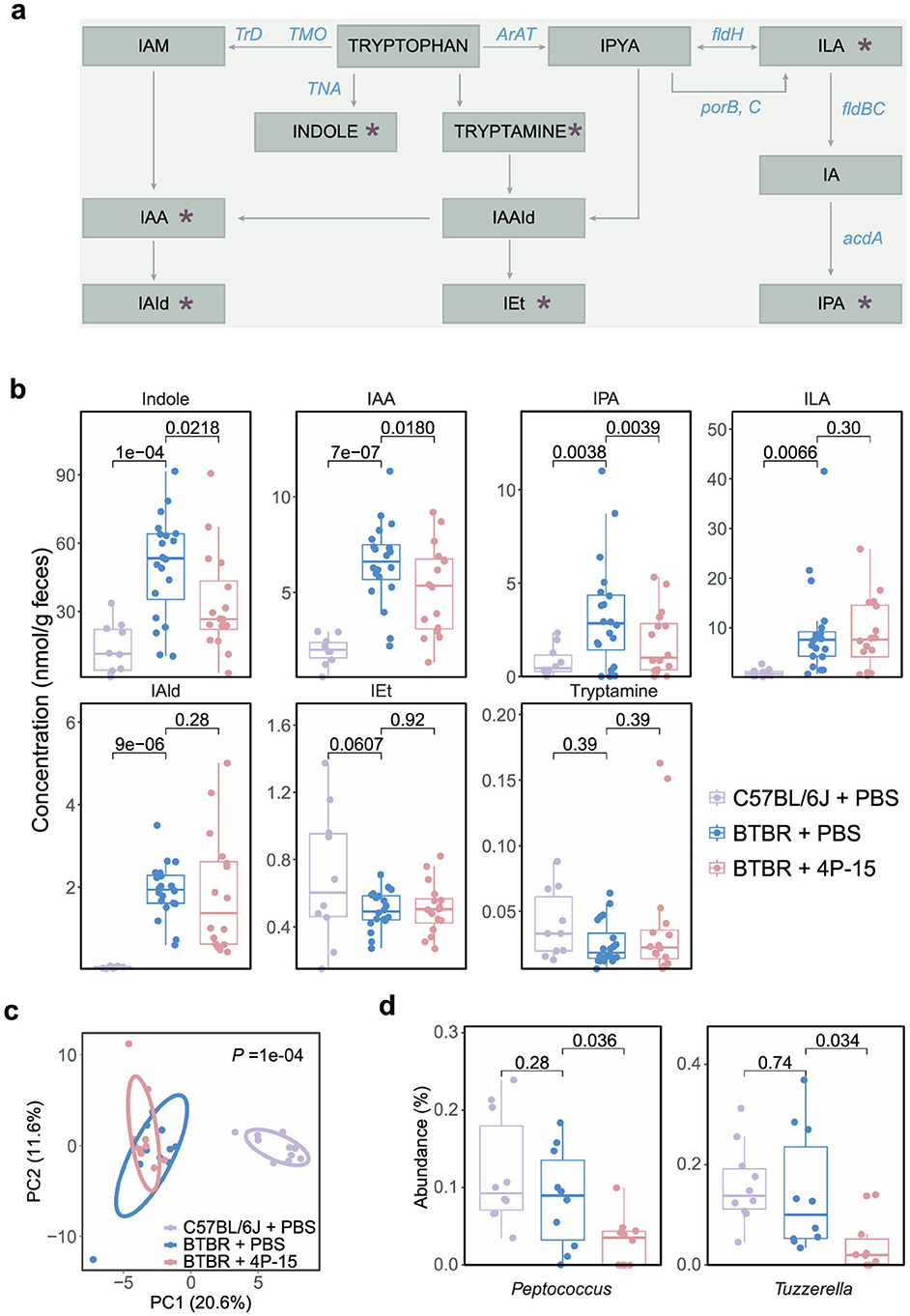
Figure 4. The modulation of intestinal indole and its derivatives, as well as gut microbes by F. hominis 4P-15 in BTBR mice. (a) The metabolites and enzymes in the indole pathway of tryptophan metabolism. The metabolites analyzed in this study were marked by red asterisks. IAM, indole-3-acetamide. IAA, indole-3-acetic acid. IAId, indole-3-aldehyde. IAAId, indole-3-acetaldehyde. IEt, indole-3-ethanol. IPYA, indole-3-pyruvate. ILA, indole-3-lactic acid. IPA, indole-3-propionic acid. TrD, tryptophan decarboxylase. TMO, tryptophan 2-monooxygenase. TNA, tryptophanase. ArAT, aromatic amino acid aminotransferase. fldH, phenylacetate dehydrogenase. porB, C, pyruvate: ferredoxin oxidoreductase B and C. fldBC, phenylacetate dehydratase. acdA, acyl-CoA dehydrogenase. (b) Box plots demonstrating the concentrations of indole and its six derivatives in the feces of C57BL/6J mice gavaged with PBS, BTBR mice gavaged with PBS, and BTBR mice gavaged with 4P-15. n = 10, 20, 16 mice, respectively. The adjusted P-values were calculated using one-way ANOVA with multiple comparisons by controlling FDR. The significant difference was defined as the adjusted P-value < 0.05. (c, d) A 16S rRNA sequencing analysis of gut microbiota from C57BL/6J mice treated with PBS, BTBR mice treated with PBS, and BTBR mice treated with 4P-15. n = 10 mice for each group. (c) PCA plots of the fecal microbiota composition at the genus level (PERMANOVA). (d) The relative abundance of genera significantly different between BTBR treated with 4P-15 and PBS. The significantly different genera were defined as the adjusted P-value < 0.05, and the adjusted P-values were evaluated using the FDR correction of P-value (one-way ANOVA). Quantitative data are shown as the mean ± SEM (b, d). Significance was indicated by adjusted P-values (b, d).
Subsequently, we delved into understanding how the administration of 4P-15 reduces the levels of intestinal indole and its derivatives. We first examined whether the genome of 4P-15 contained the enzymes capable of indole biotransformation and did not identify any functional elements (Ma et al., 2018) (Supplementary Figure 3a). We then assessed whether 4P-15 had the potential to decrease the concentrations of indole, indole-3-acetic acid (IAA), and indole-3-propionic acid (IPA) in vitro. By comparing the concentrations of these three compounds in the fermentation broth samples of 4P-15 compared with its YCFA medium, we observed no significant difference in IAA levels (Supplementary Figure 3b). Since the concentrations of indole and IPA were below the detection threshold in YCFA medium, we supplemented the YCFA medium with indole and IPA, respectively. Our findings indicated that 4P-15 failed to reduce the concentrations of both indole and IPA in the supplemented YCFA medium (Supplementary Figures 3c, d).
Next, our focus shifted to the influence of 4P-15 on the gut microbiota, particularly the abundance of indole-producing bacteria. Utilizing 16S rRNA gene sequencing of cecal contents, we found that 4P-15 administration did not significantly alter either alpha or beta diversity (Figure 4c and Supplementary Figures 3e–h). Subsequent analysis revealed a significant reduction in the abundance of two genera following 4P-15 treatment (Figure 4d). Notably, one of these genera, Peptococcus, is identified as a potential indole-producing bacterium (Lanigan, 1976; Schwan, 1979). These results suggest that 4P-15 might decrease intestinal indole concentrations by reducing the abundance of potential indole-producing bacteria.
4P-15 restores the abnormal levels of indole derivatives and AhR downstream pathways related to ASD pathology in BTBR mice
We next examined whether 4P-15 gavage-induced reductions in intestinal indole and its derivatives influenced brain metabolite concentrations. 4P-15 administration significantly decreased IPA levels and modestly reduced indole concentrations in the brain (Figure 5a). To assess the potential neuroprotective effects of these metabolic changes, we prioritized AhR-regulated pathways directly relevant to ASD pathophysiology. While AhR modulates numerous downstream targets, we focused on glutamate transporters (Slc1a1, Slc1a2, and Slc1a3) and GABA receptors (Gabra1, Gabrb2, Gabrg2, and Gabbr1), given their dual rationale: (1) prior studies demonstrate AhR suppresses glutamate transporter expression (Silva-Parra et al., 2024) and GABA receptor transcription (Dever et al., 2016), and (2) ASD pathogenesis involves excitatory/inhibitory imbalance driven by glutamate-GABA signaling dysregulation. While 4P-15 administration did not significantly change Ahr mRNA level (Figure 5b and Supplementary Figure 4a), it notably upregulated the expression of glutamate transporters Slc1a2 and GABA receptors Gabra1, Gabrb2, and Gabbr1 (Figures 5c, d)
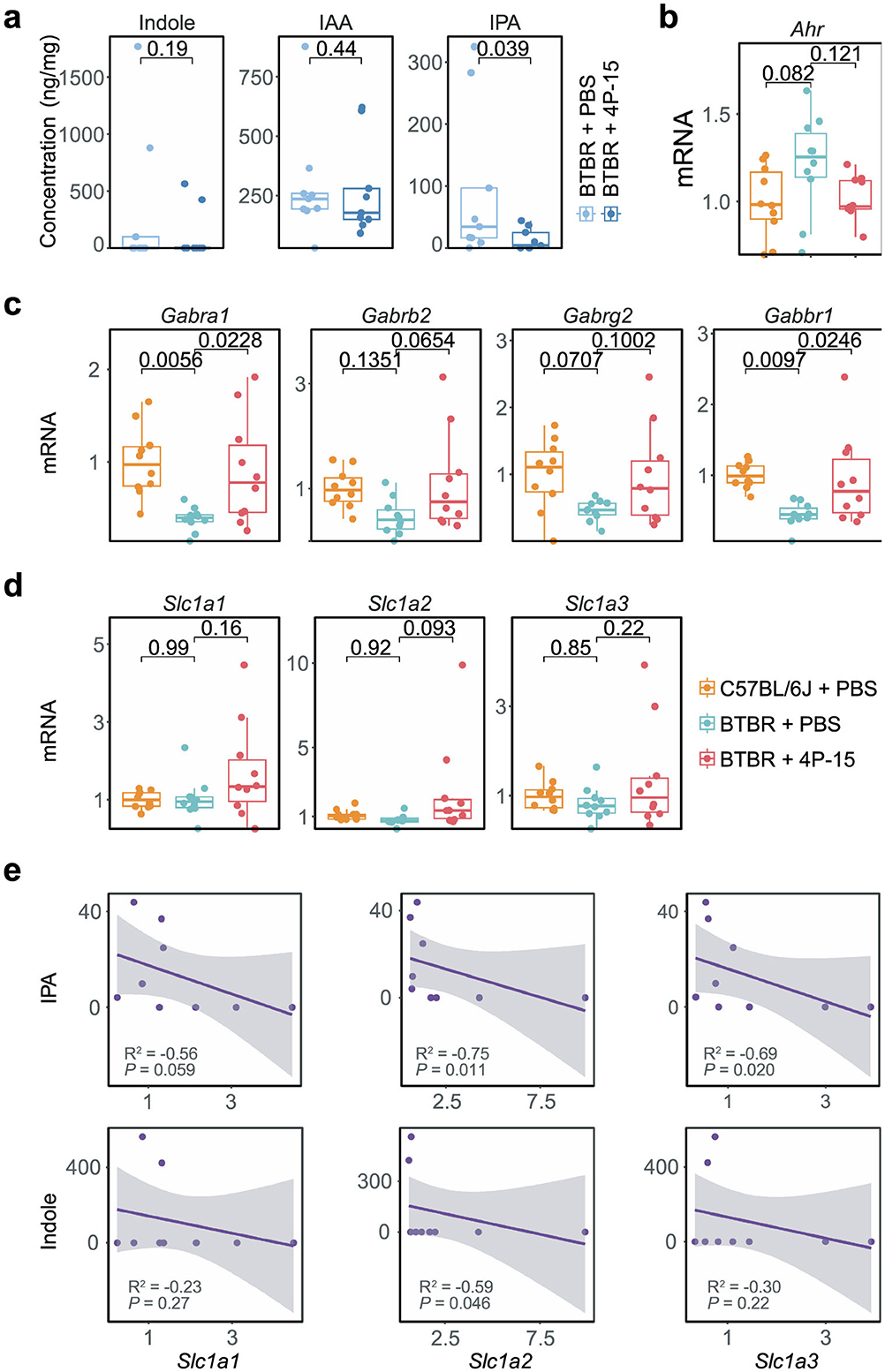
Figure 5. The modulation of indoles and AhR signaling by F. hominis 4P-15 in the brain of BTBR mice. (a) Box plots showing the concentration of indole, IAA, and IPA in the cerebellum of BTBR mice gavaged with PBS and 4P-15. n = 9 mice for each group. The P-values were calculated by the one-sided Student's t-test. The significant difference was defined as the P-value < 0.05. (b–d) Box plots showing the qPCR analysis in the cerebral cortex of C57BL/6J mice gavaged with PBS, BTBR mice with PBS, and BTBR mice with 4P-15. n = 10 mice for each group. The adjusted P-values were calculated using one-way ANOVA with Tukey's multiple comparisons test. The significant difference was defined as the adjusted P-value < 0.1. (e) Scatter plots showing the association between the mRNA levels of AhR downstream genes (X-axis) and the concentrations of AhR ligands (Y-axis) in BTBR mice gavaged with 4P-15. n = 8 mice for each group. Statistical significances (P-value) and correlation coefficients (R2) were determined by the function cor.test (method = “spearman”) in R. The significant difference was defined as the P-value < 0.05. Quantitative data are shown as the mean ± SEM (a–d). Significance was indicated by P-value (a, e) and adjusted P-value (b–d).
To investigate how AhR ligand dynamics influence its downstream targets, specifically glutamate transporters and GABA receptors, we analyzed correlations between brain indole/IPA levels and AhR pathway gene expression in BTBR mice gavaged with 4P-15. Notably, IPA concentrations exhibited significant inverse associations with Slc1a2/Slc1a3 expression, while indole levels showed a significant negative correlation specifically with Slc1a2 (Figure 5e and Supplementary Figure 4b). Together, these data suggest that 4P-15 derepresses glutamate transporters and GABA receptors through diminished AhR signaling by reducing indole-related AhR ligands, thereby alleviating excitatory-inhibitory imbalance.
Discussion
Our study identifies Faecalibacterium hominis 4P-15 as a pioneering microbial therapeutic avenue in ameliorating ASD pathophysiology in BTBR mice through gut-brain axis modulation. By reducing indole and its derivatives in both the gut and brain, 4P-15 rectifies dysregulated AhR signaling pathways linked to excitatory-inhibitory imbalance in ASD, and thus ameliorates core behavioral deficits (Figure 6). These findings establish a new therapeutic paradigm for ASD by targeting indole-AhR signaling pathways, offering clinically translatable strategies.
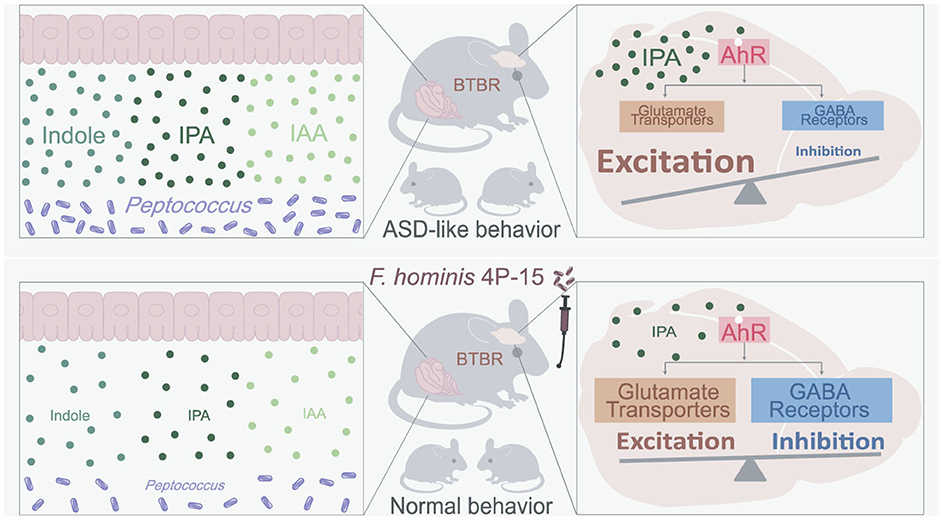
Figure 6. The summary of how 4P-15 alleviates the ASD-like phenotypes in BTBR mice via the gut-brain axis. Oral administration of 4P-15 attenuated the concentrations of intestinal indole and its derivatives IPA and IAA in BTBR mice possibly by reducing the abundance of Peptococcus, potential indole-producing bacteria. Furthermore, 4P-15 decreased the levels of indoles, especially IPA in the brain and increased the expression of glutamate transporter and GABA receptors, which were downstream elements of the AhR signaling and were negatively regulated by it. Ultimately, this led to the restoration of excitation/inhibition imbalance and ASD-like behaviors.
Our prior work demonstrated that Bacteroides uniformis ameliorates ASD-associated excitatory/inhibitory imbalance in CHD8 haploinsufficient mice by attenuating intestinal amino acid transport, thereby decreasing glutamate precursor availability for excitatory neurotransmission (Yu et al., 2022b; Ji et al., 2025). Intriguingly, while both B. uniformis and F. hominis 4P-15 alleviate ASD-like behaviors, they operate through distinct yet complementary mechanisms: 4P-15 reduces microbial indoles to suppress AhR-driven suppression of glutamate/GABA synaptic transmission regulators, whereas B. uniformis directly reduces glutamate synthesis via amino acid metabolic rewiring. This mechanistic divergence underscores the potential for personalized microbiota-based therapies tailored to individual ASD subpopulations. For instance, 4P-15 may preferentially benefit patients with elevated fecal indole levels and AhR overactivation, whereas B. uniformis could target those exhibiting dysregulated amino acid metabolism. Such stratification aligns with the growing recognition of ASD as a disorder of heterogeneous etiology, where microbiome-directed interventions must align with host-specific metabolic signatures to optimize efficacy.
Our research demonstrates that 4P-15 decreases the expression of genes downstream of the indole-AhR signaling pathway, which is linked to the regulation of excitatory-inhibitory imbalance in ASD. While prior investigations into AhR's role in ASD have predominantly focused on its canonical activation of cytochrome P450 enzymes and associated oxidative stress and inflammation pathways (Dhulkifle et al., 2021; Ahmed et al., 2022), the connection between microbial AhR ligands and synaptic excitatory-inhibitory dysregulation has remained unexplored. Notably, we demonstrate that 4P-15 administration reduces bacterial-derived neuroactive metabolites, particularly indole and its derivatives, which are potent AhR agonists. This reduction attenuates AhR overactivation, resulting in increased expression of glutamate transporters and GABA receptors, critical regulators of neurotransmission equilibrium. By shifting the focus from AhR's traditional metabolic roles to its synaptic regulatory functions, our findings provide distinctive mechanistic insights into how microbial modulation of the indole-AhR axis may rectify excitatory-inhibitory imbalance in ASD.
Our findings suggest that indole and its derivatives, including IPA and IAA, may exert neurotoxic effects in ASD, as evidenced by their association with AhR overactivation and disrupted excitatory-inhibitory balance. However, this contrasts with reports in other neurological contexts where indole derivatives exhibit neuroprotective properties (Hwang et al., 2009; Serger et al., 2022; Yin et al., 2023). These divergent roles may stem from context-dependent factors such as metabolite concentration, tissue-specific AhR activation thresholds, or disease-specific neuroinflammatory microenvironments. In ASD, where chronic neuroinflammation and glutamate excitotoxicity are hallmarks, indole derivatives might act as double-edged swords: at physiological levels, they could maintain gut-brain homeostasis, but their pathological accumulation, as observed in our study, may tip the balance toward synaptic dysfunction. Resolving this paradox will require delineating dose-dependent effects, AhR isoform specificity, and interactions with comorbid factors in future studies.
Our study revealed that oral administration of 4P-15 reduced the abundance of Peptococcus in the gut microbiota, though the precise inhibitory mechanisms remain uncharacterized. Interbacterial suppression within microbial communities commonly involves resource competition, metabolite-mediated inhibition, immune modulation, or niche occupation (Jiang et al., 2025; Ikryannikova et al., 2020). Carbon source utilization analysis demonstrated distinct substrate preferences between the Faecalibacterium and Peptococcus: Faecalibacterium primarily metabolizes fermentable sugars (e.g., fructose, glucose, lactose) (Liou et al., 2024; Sakamoto et al., 2022), whereas Peptococcus specializes in proteolytic metabolism, relying on protein-derived substrates such as peptones, amino acids, and polypeptides (Reece et al., 1976; Wilkins et al., 1975; Shkoporov et al., 2016). This divergence in nutritional niches suggests limited direct competition for carbon sources under typical dietary conditions. Instead, metabolite-mediated inhibition emerged as a plausible mechanism. Faecalibacterium generates multiple organic acids, including SCFAs (e.g., butyrate), lactate, and formate (Sakamoto et al., 2022), which collectively lower intestinal luminal pH. This pH reduction likely creates a hostile microenvironment that selectively inhibits the growth of pH-sensitive Peptococcus (Lanigan, 1976).
While our study highlights the role of 4P-15 in alleviating ASD-like behaviors through the indole-AhR-glutamate/GABA axis, additional mechanisms may contribute to its neuromodulatory effects. First, although we observed reduced indole levels in the brain and prior studies suggest indoles can cross the blood-brain barrier (Guo et al., 2023; Pappolla et al., 2021; Kunevičius et al., 2024), we cannot exclude indirect effects mediated by peripheral AhR activation. Intestinal epithelia and immune cells express AhR (Madison et al., 2023; Rothhammer and Quintana, 2019), whose activation may modulate gut-brain communication via skewed T cell differentiation or intestinal barrier function (Hao and Whitelaw, 2013; Stockinger et al., 2021). Second, the upregulation of glutamate transporters and GABA receptors by 4P-15 may involve AhR-independent pathways. For instance, indole and its derivatives can act as agonists or antagonists of serotonin receptors and tropomyosin receptor kinases (TrKs) (Tammiku-Taul et al., 2016; van Niel et al., 1999; Bermudez et al., 1990; Swain et al., 1991; Hogendorf et al., 2019), which regulate synaptic plasticity and are implicated in neurodevelopmental disorders (Stoleru et al., 2013). Additionally, indole and its derivatives are predicted to interact with GABAA receptor-associated proteins, potentially enhancing receptor surface expression (Chen et al., 2005; Leil et al., 2004). These putative mechanisms underscore the pleiotropic nature of microbial metabolites in neuroregulation and warrant further investigation.
While our study primarily focused on male BTBR mice to investigate the effects and underlying mechanisms of 4P-15, we acknowledge that clinical data demonstrate a depletion of Faecalibacterium in both male (Kang et al., 2018) and female ASD cohorts (PRJEB42687). This raises the possibility that females with ASD could also benefit from the 4P-15 intervention. However, it remains uncertain whether the underlying mechanisms, particularly indole-AhR modulation, operate similarly across sexes. Emerging evidence highlights significant sex differences in the regulation of the gut-brain axis. It is plausible that hormonal-immune crosstalk may contribute to differential responses to microbiota interventions between males and females. Therefore, while we believe that the core mechanism of 4P-15 is likely applicable to both sexes, it is crucial to validate these findings in sex-stratified ASD models to identify sex-specific nuances. Future research in this area could clarify the necessity for personalized treatment regimens, such as hormone-adjusted probiotic dosing, to optimize therapeutic outcomes in females with ASD.
Methods
Sample collection and gut microbiome isolation
The fecal samples from the typically developing (TD) individual were obtained as previously described (Liu et al., 2021). The fecal samples from the ASD individual used for gut microbial isolation were collected with approval from the Ethics Committee of Peking Union Medical College Hospital under the ethical approval number ZS-1393. The samples intended for bacterial isolation were kept fresh and transported to an anaerobic workstation (AW500, Electrotek, UK) within 2 hours of collection. The samples were suspended in anaerobic PBS and filtered through a 40 μm cell strainer. The resulting suspensions were divided into two portions: one portion was directly cultured on modified YCFA agar plates (YCFA agar medium supplemented with glucose at 1 g/L, hemin at 0.01 g/L, and L-cysteine at 1 g/L) and BHI agar plates supplemented with 5% defibrinated sheep blood. The other portion underwent alcohol pretreatment before being cultured on a different set of modified YCFA agar plates (YCFA agar medium supplemented with glucose at 1 g/L, hemin at 0.01 g/L, L-cysteine at 1 g/L, and sodium taurocholate at 0.1%) (Browne et al., 2016).
Colony isolation and identification were carried out as described in our previous study (Liu et al., 2021). All colonies on the plates were picked after cultivation periods of 24 h, 3 days, and 7 days, followed by incubation at 37°C under anaerobic conditions in the corresponding media for a duration of 1–7 days. Subsequently, 500 μl of the culture media were collected, and the bacterial pellets were harvested for PCR-based amplification of the 16S rRNA gene sequences using primers 27F and 1492R. The sequences of the PCR products were identified by aligning them against the NCBI 16S rRNA sequence database and the EZBioCloud database (updated on August 30, 2024). The threshold for 16S rRNA gene sequence identity for novel species was set at 98.7% (Liu et al., 2021). The phylogenetic tree of isolated gut microbiome strains was constructed by MEGA (Version 11.0.13) using the Neighbour-Joining Tree method. The 16S rRNA gene sequences of the isolated bacteria are provided in Supplementary Table 1. Isolated bacteria from the TD sample have been preserved in hGMB (Liu et al., 2021).
Animals
All animal experiments complied with the National Institute of Health Guide for the Care and Use of Laboratory Animals. The permission for animal experiment procedures was granted by the Animal Ethics Committee at the Institute of Microbiology, Chinese Academy of Sciences.
Mice were maintained on a 12 h/12 h dark/light cycle (lights on at 7:00 am) in the animal facility at the Institute of Microbiology, Chinese Academy of Sciences, under specific pathogen-free conditions. Male and female BTBR T+ Itpr3tf/J (BTBR) mice purchased from Jackson Lab (America) were crossed to obtain BTBR mice offspring. After weaning, BTBR mice were reared separately by gender. C57BL/6J mice were purchased from SPF (Beijing) Biotechnology Co., Ltd (Beijing, China). After weaning, the mice were group-housed in cages containing two to five mice per cage. Mice from different experimental groups (C57BL/6J mice gavaged with PBS, BTBR mice gavaged with PBS, and BTBR mice gavaged with 4P-15) were housed separately. Male mice and samples from male mice were used in this study.
Animal numbers: The number of animals used in the behavioral tests (Figures 2b, c and Supplementary Figures 2a–c) were 10 for C57BL/6J mice gavaged with PBS, BTBR mice gavaged with PBS, and BTBR mice gavaged with 4P-15. The number of animals used for the cytokine analysis (Figure 3b) were 10, 8, and 10 for C57BL/6J mice gavaged with PBS, BTBR mice gavaged with PBS, and BTBR mice gavaged with 4P-15, respectively. The number of animals used in the 16S rRNA sequencing (Figures 4c, d and Supplementary Figures 3e–h) were 10 for C57BL/6J mice gavaged with PBS, BTBR mice gavaged with PBS, and BTBR mice gavaged with 4P-15. The number of animals used for intestinal indole detection (Figure 4b) were 10, 20, and 16 for C57BL/6J mice gavaged with PBS, BTBR mice gavaged with PBS, and BTBR mice gavaged with 4P-15, respectively. The number of animals used for the expression analysis of AhR, glutamate transporters and GABA receptors (Figures 5b–d) were 10 for C57BL/6J mice gavaged with PBS, BTBR mice gavaged with PBS, and BTBR mice gavaged with 4P-15. The number of animals used for neurotransmitter analysis (Supplementary Figure 2f) were 10 for BTBR mice gavaged with PBS and BTBR mice gavaged with 4P-15. The number of animals used for SCFA analysis (Supplementary Figure 2g) were 9 and 10 for BTBR mice gavaged with PBS and BTBR mice gavaged with 4P-15, respectively.
F. hominis 4P-15 administration
F. hominis 4P-15 was grown anaerobically in a modified YCFA medium supplemented with 3 mM acetate (1M acetate-sodium acetate buffer) to stimulate bacterial growth (Duncan et al., 2002). The dosage of F. hominis 4P-15 chosen for colonization was based on previous publications (Yu et al., 2022b; Martín et al., 2015; Miquel et al., 2015), which reported effective dosages ranging from 1 × 108 to 1 × 109 colony-forming units (CFU). For each mouse in the F. hominis 4P-15 administration group, 4 × 108 CFU were suspended in 200 μl PBS and delivered by gavage immediately. For the control group, an equal volume of PBS was given. The wean C57BL/6J or BTBR mice at 4 weeks old were gavaged with PBS or 4P-15 for a duration of 4 weeks, after which fecal samples were collected. The mice underwent behavioral tests at 8 weeks of age or were sacrificed for brain tissue collection. The 11-week-old mice that completed behavioral tests were sacrificed. The gavage regimen began when the mice were 4 weeks old and continued until they were sacrificed. The numbers of mice for gavage were 10, 20, and 20 for C57BL/6J mice gavaged with PBS, BTBR mice gavaged with PBS, and BTBR mice gavaged with 4P-15, respectively.
Behavioral tests
The test mice were acclimated by handling them for three consecutive days before the experiments. Behavioral tests were carried out in the behavioral test room between 8:00 a.m. and 7:00 p.m., with the mice being transferred at least an hour in advance for acclimation (Yu et al., 2022b).
Self-grooming
The self-grooming test was conducted following the methodology outlined in a previous study (Gkogkas et al., 2013). Specifically, a test mouse was introduced into a clean arena and allowed to explore freely for 20 min. Self-grooming behavior (including genital or tail grooming, body grooming, paw or leg licking, and head washing) was then observed and recorded during the second 10-min interval, with the initial 10 min as the habituation phase.
Three-chamber social interaction test
The three-chamber sociability test was performed in a white plastic arena [60 cm × 15 cm] divided into three equal-sized chambers by clear movable plastic dividers. Three chambers were named ZONE 2, ZONE 1, and ZONE 3 from left to right. In the first session, the test mouse was introduced to the center of ZONE 1 and moved freely for 10 min. In the second session (social ability session), an age-, sex- and strain-matched stranger mouse (M1) previously habituated to the cylinder was placed in the center of ZONE 2, while an object in another cylinder (Object) was placed in the center of ZONE 3. The test mouse was then allowed to explore freely for 10 min. The movement was recorded by a camera over the arena. The duration time in ZONE 2 (M1) and ZONE 3 (Object) were analyzed using ANY-maze software. The percentage of duration time was calculated as duration time/600, and the preference index was calculated as (duration time in ZONE 2 – duration time in ZONE 3)/(duration time in ZONE 2 + duration time in ZONE 3). For the third session (social novelty preference session), another age-, sex- and strain-matched stranger mouse (M2) previously habituated to the cylinder was placed in the center of ZONE 3, while M1 was placed in the center of ZONE 2. The test mouse was then allowed to explore freely for 10 min. The movement was recorded by a camera over the arena. The duration time in ZONE 2 (M1) and ZONE 3 (M2) were analyzed using ANY-maze software. The percentage of duration time was calculated as duration time/600, and the preference index was calculated as (duration time in ZONE 3 – duration time in ZONE 2)/(duration time in ZONE 3 + duration time in ZONE 2) (Kaidanovich-Beilin et al., 2011; Yang et al., 2011).
Open-field test
The mouse was introduced to the center of the arena [40 cm (L) × 40 cm (W) × 50 cm (H)] and allowed to explore for 10 min. The video was recorded by a ceiling camera. Total distance traveled and time in the center (20 cm × 20 cm) were analyzed by Xeye Aba (3.2 version, Beijing Macroambition S&T Development, China) (Deng et al., 2019; Qiao et al., 2017).
Sucrose preference test
Mice were individually housed in standard cages for 72 h prior to experimentation to eliminate social interference. During the training phase, animals were acclimated to a two-bottle choice paradigm over two consecutive days: on Day 1, both bottles contained plain water, whereas on Day 2, both bottles were replaced with 1% (w/v) sucrose solution. Following a 24-h food deprivation period, the test session commenced with the simultaneous presentation of two identical bottles containing sucrose solution and plain water, respectively. To minimize position preference effects, bottle positions were systematically rotated every 12 h during the 24-h testing period. The weight of bottles was measured before and after the test, and the consumed weight was calculated. The preference index was calculated as sucrose solution consumed/(sucrose solution consumed + plain water consumed) (Yu et al., 2022a).
Targeted metabolomics for indole and its derivatives
Sample preparation
Brain samples were taken out and subjected to rapid freezing in liquid nitrogen, subsequently stored at −80°C until later utilization. After thawing the the brain samples on ice, the cerebellum was isolated and homogenized in ice-cold methanol (MeOH) (5 μl/mg). After centrifugation at 14,000 rcf for 20 min at 4°C, 100 μl supernatant was transferred to a fresh tube and dried using a speed vacuum at room temperature. The pellets were dissolved in 10 μl of MeOH: H2O (1:1, v/v) and then subjected to centrifugation at 14,000 rcf for 20 min at 4°C. The supernatant was utilized for metabolomic analysis (Yu et al., 2022b).
Fecal samples were collected, immediately frozen in liquid nitrogen, and stored at −80°C until use. Frozen feces were thawed on ice and dispersed in ice-cold MeOH (5 μl/mg). The mixture was centrifuged at 14,000 rcf for 20 min at 4°C, and the supernatant was used for metabolomic assay (Yu et al., 2022b).
The fermentation broth samples of 4P-15 were collected after the strains had been grown anaerobically for 48 h and then centrifuged at 14,000 rcf for 20 min at 4°C. Three hundred microliter YCFA medium or supernatant of the broths were added to 1.5 ml ice-cold MeOH. The mixture was centrifuged at 14,000 rcf for 20 min at 4°C, and 1.2 ml of the supernatant was transferred to a new tube and then dried in a speed vacuum at room temperature. The pellets were re-suspended in 200 μl of MeOH: H2O (1:1, v/v) and then centrifuged at 14,000 rcf for 20 min at 4°C. The resulting supernatant was used for metabolomic analysis (Yu et al., 2022b).
Metabolomic assay
Metabolomics assays were conducted on a 6500 QTRAP triple quadrupole mass spectrometer (Applied Biosystems/Sciex) that was coupled to an ExionLC LC system (Applied Biosystems/Sciex). Chromatographic separations were accomplished on a Phenomenex Gemini® 3 μm NX-C18 100 Å LC column (50 × 2 mm) (Yu et al., 2022b). Indole, indole-3-acetic acid, indole-3-aldehyde, indole-3-ethanol, indole-3-lactic acid, indole-3-propionic acid, tryptamine, kynurenine, and tryptophan were detected using multiple reaction monitoring modes with specific parent-daughter ion transitions at m/z ratios for each compound. Compounds were identified by comparing parent-daughter ion transitions and the retention time of reference standards: indole (118.0/91.0), indole-3-acetic acid (176.1/130.0), indole-3-aldehyde (146.1/91.0), indole-3-ethanol (162.1/144.1), indole-3-lactic acid (204.0/158.0), indole-3-propionic acid (190.0/130.1), and tryptamine (161.1/144.0) (Huang et al., 2021). The various concentrations of standards were employed for quantification purposes.
Measurement of short-chain fatty acids (SCFAs)
For testing the SCFAs, including acetic acid, propionic acid, butyric acid, pentanoic acid, and valeric acid in feces, 0.1 g feces of each sample were freeze-dried and extracted in 1 ml methanol by 10-min ultrasound bathing in iced water. After the centrifuge, the supernatant liquid was prepared for GC-MS analysis. GC-MS analysis was performed on a GCMS-QP2010 Ultra with an autosampler (SHIMADZU) and the DB-wax capillary column (30 m, 0.25 mm i.d., 0.25 μm film thickness; SHIMADZU). The temperature of the oven was programmed from 80°C to 140°C at a 20°C/min gradient, with a 1 min hold; to 290°C at 3.5°C/min, with a 15 min hold. Injection of a 1 μl sample was performed at 280°C. The carrier gas, helium, flowed at 1.2 ml/min. The electronic impact was recorded at 70 eV. Referenced to standard curves, the products from feces were calculated by peak area (Li et al., 2022).
Enzyme-linked immunosorbent assay (ELISA)
Neurotransmitters
The entire mouse brain including the cerebellum was homogenized by pulverization in cold PBS using a tissue homogenizer (60 Hz, 2 min, twice) at a ratio of 0.2 g brain tissue to 100 μl PBS and centrifuged at 3,000 rpm for 10 min. The supernatant was collected for ELISA using a Mouse 5-Hydroxytryptamine ELISA kit and a Mouse dopamine ELISA kit (QiSong, Beijing, China) according to the manufacturer's protocols.
Inflammatory cytokines
The right cerebral cortex of the brain was homogenized in lysis buffer containing a protease inhibitor cocktail (P0013B, Beyotime, China) and then centrifuged (4°C, 14,000 rcf, 20 min). The supernatant was utilized for ELISA analysis according to the manufacturer's guidelines. ELISA kits were used for IL-1β, IL-17A, TNFα, and IFNγ detection (Thermo Fisher, USA) and for IL-6 and IL-10 analysis (SinoBestBio, Shanghai, China). The concentrations of the inflammatory cytokines in the supernatant were normalized to the concentrations of total proteins (Yu et al., 2022b).
16S rRNA gene sequencing and data analysis
Genomic DNA was extracted from mouse cecal contents or human feces by QIAamp PowerFecal® Pro DNA kit (Qiagen, United States). The V3-V4 region of the 16S rRNA gene was amplified using the primer set 338F (5′-CCTACGGGNGGCWGCAG-3′) and 806R (5′-GACTACHVGGGTWTCTAAT-3′). PCR reactions were amplified using PremixTaq (TaKaRa, China), DNA products were purified by gel electrophoresis, and a sequencing adapter was connected to build the library followed by the standard protocol of NEBNext®UltraTM DNALibraryPrepKitforIllumina®. The 16S rRNA gene amplicon sequencing was conducted on Illumina novaseq 6000 PE250 platform.
The raw sequencing data of 16S rRNA gene sequences were processed using QIIME2 (v2021.4.0) (Bolyen et al., 2019). Sequences were trimmed to remove primer and adapter sequences. The trimmed raw reads were then analyzed with DADA2 (Callahan et al., 2016) to perform quality trimming, denoising, error-correction, paired-end read merging, chimera removal, and dereplication. The produced amplicon sequence variants (ASVs) were taxonomically classified according to the SILVA database (release 138) using “classify-sklearn,” after training the classifier on the S database (v.2022.10) (McDonald et al., 2024) with 341F/806R primers. Subsequently, the microbial profile table was exported for downstream analyses.
For diversity analysis, the microbial table was first normalized by total sum reads. Alpha diversity was assessed using the “diversity” function in the R package vegan, with statistical significance determined by the “aov” function. Then the “p.adjust” function (method = “bonferroni”) in R. Beta diversity analysis involved conducting principal component analysis (PCA) using the “prcomp” function (center = T, scale = T) in R and visualization with the ggbioplot R package. Multivariate analysis of variance in beta diversity analysis using distance matrices (PERMANOVA) was carried out with the “adonis2” function (permutations = 9999, method = “euclidean”) in the vegan package.
The metabolic gene content was estimated from 16S rRNA gene data using Phylogenetic Investigation of Communities by Reconstruction of Unobserved States 2 (PICRUSt2) pipeline with default parameters. ASVs with nearest-sequenced taxon index (NSTI) values >2 were filtered from the analysis. KEGG orthology (KO), and enzyme Commission number (EC number) metagenomes, and MetaCyc pathway abundances were predicted with PICRUSt2 (Renga et al., 2023).
qPCR analysis
Total RNA extracted from the left cerebral cortex of mice was used for cDNA preparation using a Hifair® AdvanceFast One-step RT-gDNA Digestion SuperMix for qPCR (11151ES60, Yeasen, Shanghai, China). Real-time PCR analysis was performed using Hieff® qPCR SYBR Green Master Mix (11203ES08, Yeasen, Shanghai, China) to determine the expression levels of glutamate transporters (Slc1a1, Slc1a2, Slc1a3), GABA receptors (Gabra1, Gabrb2, Gabrg2, Gabbr1), AhR (Ahr), cytochrome P450 (CYP) enzymes (Cyp1b1), and the Gapdh gene. The sequences of primers were provided in Supplementary Table 2.
Statistical analysis
False discovery rate (FDR)-adjusted P-values determined by two-sided Wilcoxon rank-sum tests was used for analyzing Feacalibacterium abundance differences in published cohorts (Figure 1a and Supplementary Figure 1). Mann-Whitney (Supplementary Figures 2b, c, f, g) and Student's t-tests (Figure 5a and Supplementary Figures 3b–d) were used for comparing data between two groups. One-way ANOVA with Tukey's multiple comparison tests [Figures 2b, c (right panel), Figures 3b, 5b–d] and one-way ANOVA with multiple comparisons by controlling FDR (Figures 4b, d) were used for multiple comparisons across three groups. Two-way ANOVA with Tukey's multiple comparison tests were used for comparing the effects of two factors on a response variable [Figure 2c (lefe panel) and Supplementary Figure 2a (left panel)]. Spearman correlation tests (Figure 5e and Supplementary Figure 4b) and Pearson correlation tests (Supplementary Figures 2d, e) in R were used to quantify associations. PERMANOVA with the adonis2 function (vegan package) using Euclidean distances was used for beta diversity analysis (Figure 4c).
Data availability statement
The assembled genome of 4P-15 was available at NODE with the project accession OEP001106 (https://www.biosino.org/node/project/detail/OEP001106) (Liu et al., 2021). The 16S rRNA gene sequence data have been deposited in the the Genome Sequence Archive (GSA) in National Genomics Data Center, Beijing Institute of Genomics (China National Center for Bioinformation), Chinese Academy of Sciences under the accession code PRJCA038009 (https://ngdc.cncb.ac.cn/search/specific?db=biosample&q=PRJCA038009).
Ethics statement
The studies involving humans were approved by The fecal sample from ASD used for gut microbiome isolation and 16S rRNA gene sequencing was collected with approval from the Ethics Committee of Peking Union Medical College Hospital under the ethical approval number ZS-1393. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation in this study was provided by the participants' legal guardians/next of kin. The animal study was approved by All procedures were approved by the Animal Ethics Committee at the Institute of Microbiology, Chinese Academy of Sciences (APIMCAS2023026). The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
YY: Conceptualization, Writing – original draft, Writing – review & editing. YujW: Formal analysis, Writing – original draft. JZ: Formal analysis, Writing – original draft. SL: Formal analysis, Writing – original draft. YulW: Formal analysis, Writing – original draft. XY: Resources, Writing – original draft. XC: Formal analysis, Writing – original draft. MD: Formal analysis, Writing – original draft. LX: Formal analysis, Writing – original draft. S-JL: Conceptualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by grants from National Natural Science Foundation of China (32300094).
Acknowledgments
We thank Dr. Weilin Li (Institutional Center for Shared Technologies and Facilities of Institute of Microbiology, Chinese Academy of Sciences), for their work of targeted metabolomics analysis for Indole and its derivatives via a 6500 QTRAP triple quadrupole mass spectrometer (Applied Biosystems/Sciex) in this manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Gen AI was used in the creation of this manuscript.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1640149/full#supplementary-material
References
Agus, A., Planchais, J., and Sokol, H. (2018). Gut microbiota regulation of tryptophan metabolism in health and disease. Cell Host Microbe 23, 716–724. doi: 10.1016/j.chom.2018.05.003
Ahmed, H., Leyrolle, Q., Koistinen, V., Kärkkäinen, O., Layé, S., Delzenne, N., et al. (2022). Microbiota-derived metabolites as drivers of gut-brain communication. Gut Microbes 14:2102878. doi: 10.1080/19490976.2022.2102878
Association, A. P. (2013). Diagnostic and Statistical Manual of Mental Disorders (DSM-5®). Arlington, VA: American Psychiatric Pub.
Auger, S., Kropp, C., Borras-Nogues, E., Chanput, W., Andre-Leroux, G., Gitton-Quent, O., et al. (2022). Intraspecific diversity of microbial anti-inflammatory molecule (MAM) from Faecalibacterium prausnitzii. Int. J. Mol. Sci. 23:1705. doi: 10.3390/ijms23031705
Bermudez, J., Fake, C. S., Joiner, G. F., Joiner, K. A., King, F. D., Miner, W. D., et al. (1990). 5-Hydroxytryptamine (5-HT3) receptor antagonists. 1. Indazole and indolizine-3-carboxylic acid derivatives. J. Med. Chem. 33, 1924–1929. doi: 10.1021/jm00169a016
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9
Bove, M., Palmieri, M. A., Santoro, M., Agosti, L. P., Gaetani, S., Romano, A., et al. (2024). Amygdalar neurotransmission alterations in the BTBR mice model of idiopathic autism. Transl. Psychiatry 14:193. doi: 10.1038/s41398-024-02905-z
Browne, H. P., Forster, S. C., Anonye, B. O., Kumar, N., Neville, B. A., Stares, M. D., et al. (2016). Culturing of ‘unculturable' human microbiota reveals novel taxa and extensive sporulation. Nature 533, 543–546. doi: 10.1038/nature17645
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J., and Holmes, S. P. (2016). DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Chen, K., Luan, X., Liu, Q., Wang, J., Chang, X., Snijders, A. M., et al. (2019). Drosophila histone demethylase KDM5 regulates social behavior through immune control and gut microbiota maintenance. Cell Host Microbe 25, 537–552.e8. doi: 10.1016/j.chom.2019.02.003
Chen, Z. W., Chang, C. S., Leil, T. A., Olcese, R., and Olsen, R. W. (2005). GABAA receptor-associated protein regulates GABAA receptor cell-surface number in Xenopus laevis oocytes. Mol. Pharmacol. 68, 152–159. doi: 10.1124/mol.104.009878
De Angelis, M., Piccolo, M., Vannini, L., Siragusa, S., De Giacomo, A., Serrazzanetti, D. I., et al. (2013). Fecal microbiota and metabolome of children with autism and pervasive developmental disorder not otherwise specified. PLoS ONE 8:e76993. doi: 10.1371/journal.pone.0076993
Deng, X., Gu, L., Sui, N., Guo, J., and Liang, J. (2019). Parvalbumin interneuron in the ventral hippocampus functions as a discriminator in social memory. Proc. Natl. Acad. Sci. U. S. A. 116, 16583–16592. doi: 10.1073/pnas.1819133116
Dever, D. P., Adham, Z. O., Thompson, B., Genestine, M., Cherry, J., Olschowka, J. A., et al. (2016). Aryl hydrocarbon receptor deletion in cerebellar granule neuron precursors impairs neurogenesis. Dev. Neurobiol. 76, 533–550. doi: 10.1002/dneu.22330
Dhulkifle, H., Agouni, A., Zeidan, A., Al-Kuwari, M. S., Parray, A., Tolefat, M., et al. (2021). Influence of the aryl hydrocarbon receptor activating environmental pollutants on autism spectrum disorder. Int. J. Mol. Sci. 22:9258. doi: 10.3390/ijms22179258
Ding, X., Xu, Y., Zhang, X., Zhang, L., Duan, G., Song, C., et al. (2020). Gut microbiota changes in patients with autism spectrum disorders. J. Psychiatr. Res. 129, 149–159. doi: 10.1016/j.jpsychires.2020.06.032
Duncan, S. H., Hold, G. L., Harmsen, H. J. M., Stewart, C. S., and Flint, H. J. (2002). Growth requirements and fermentation products of Fusobacterium prausnitzii, and a proposal to reclassify it as Faecalibacterium prausnitzii gen. nov., comb. nov. Int. J. Syst. Evol. Microbiol. 52(Pt 6), 2141–2146. doi: 10.1099/00207713-52-6-2141
Gkogkas, C. G., Khoutorsky, A., Ran, I., Rampakakis, E., Nevarko, T., Weatherill, D. B., et al. (2013). Autism-related deficits via dysregulated eIF4E-dependent translational control. Nature 493, 371–377. doi: 10.1038/nature11628
Guo, X., Li, C., Zhang, J., Sun, M., Xu, J., Xu, C., et al. (2023). Chiral nanoparticle-remodeled gut microbiota alleviates neurodegeneration via the gut-brain axis. Nat. Aging 3, 1415–1429. doi: 10.1038/s43587-023-00516-9
Hao, N., and Whitelaw, M. L. (2013). The emerging roles of AhR in physiology and immunity. Biochem. Pharmacol. 86, 561–570. doi: 10.1016/j.bcp.2013.07.004
Hogendorf, A. S., Hogendorf, A., Popiołek-Barczyk, K., Ciechanowska, A., Mika, J., Satała, G., et al. (2019). Fluorinated indole-imidazole conjugates: selective orally bioavailable 5-HT(7) receptor low-basicity agonists, potential neuropathic painkillers. Eur. J. Med. Chem. 170, 261–275. doi: 10.1016/j.ejmech.2019.03.017
Huang, Z., Schoones, T., Wells, J. M., Fogliano, V., and Capuano, E. (2021). Substrate-driven differences in tryptophan catabolism by gut microbiota and aryl hydrocarbon receptor activation. Mol. Nutr. Food Res. 65:e2100092. doi: 10.1002/mnfr.202100092
Hwang, I. K., Yoo, K. Y., Li, H., Park, O. K., Lee, C. H., Choi, J. H., et al. (2009). Indole-3-propionic acid attenuates neuronal damage and oxidative stress in the ischemic hippocampus. J. Neurosci. Res. 87, 2126–2137. doi: 10.1002/jnr.22030
Ikryannikova, L. N., Kurbatov, L. K., Gorokhovets, N. V., and Zamyatnin, A. A. (2020). Contact-dependent growth inhibition in bacteria: do not get too close! Int. J. Mol. Sci. 21:7990. doi: 10.3390/ijms21217990
Ji, P., Wang, N., Yu, Y., Zhu, J., Zuo, Z., Zhang, B., et al. (2025). Single-cell delineation of the microbiota-gut-brain axis: probiotic intervention in Chd8 haploinsufficient mice. Cell Genom 5:100768. doi: 10.1016/j.xgen.2025.100768
Jiang, K., Pang, X., Li, W., Xu, X., Yang, Y., Shang, C., et al. (2025). Interbacterial warfare in the human gut: insights from Bacteroidales' perspective. Gut Microbes 17:2473522. doi: 10.1080/19490976.2025.2473522
Kaidanovich-Beilin, O., Lipina, T., Vukobradovic, I., Roder, J., and Woodgett, J. R. (2011). Assessment of social interaction behaviors. J. Vis. Exp. 25:2473. doi: 10.3791/2473
Kang, D. W., Adams, J. B., Gregory, A. C., Borody, T., Chittick, L., Fasano, A., et al. (2017). Microbiota transfer therapy alters gut ecosystem and improves gastrointestinal and autism symptoms: an open-label study. Microbiome 5:10. doi: 10.1186/s40168-016-0225-7
Kang, D. W., Ilhan, Z. E., Isern, N. G., Hoyt, D. W., Howsmon, D. P., Shaffer, M., et al. (2018). Differences in fecal microbial metabolites and microbiota of children with autism spectrum disorders. Anaerobe 49, 121–131. doi: 10.1016/j.anaerobe.2017.12.007
Khan, Z. U. N., Chand, P., Majid, H., Ahmed, S., Khan, A. H., Jamil, A., et al. (2022). Urinary metabolomics using gas chromatography-mass spectrometry: potential biomarkers for autism spectrum disorder. BMC Neurol. 22:101. doi: 10.1186/s12883-022-02630-4
Kimura, E., Kubo, K. I., Endo, T., Nakajima, K., Kakeyama, M., and Tohyama, C. (2017). Excessive activation of AhR signaling disrupts neuronal migration in the hippocampal CA1 region in the developing mouse. J. Toxicol. Sci. 42, 25–30. doi: 10.2131/jts.42.25
Kunevičius, A., Sadauskas, M., Raudyte, J., Meškys, R., and Burokas, A. (2024). Unraveling the dynamics of host-microbiota indole metabolism: an investigation of indole, indolin-2-one, isatin, and 3-hydroxyindolin-2-one. Molecules 29:993. doi: 10.3390/molecules29050993
Lanigan, G. W. (1976). Peptococcus heliotrinreducans, sp. nov., a cytochrome-producing anaerobe which metabolizes pyrrolizidine alkaloids. J. Gen. Microbiol. 94, 1–10. doi: 10.1099/00221287-94-1-1
Lapiere, A., Geiger, M., Robert, V., Demarquay, C., Auger, S., Chadi, S., et al. (2020). Prophylactic Faecalibacterium prausnitzii treatment prevents the acute breakdown of colonic epithelial barrier in a preclinical model of pelvic radiation disease. Gut Microbes 12, 1–15. doi: 10.1080/19490976.2020.1812867
Leil, T. A., Chen, Z. W., Chang, C. S., and Olsen, R. W. (2004). GABAA receptor-associated protein traffics GABAA receptors to the plasma membrane in neurons. J. Neurosci. 24, 11429–11438. doi: 10.1523/JNEUROSCI.3355-04.2004
Li, D. H., Abuduaini, R., Du, M. X., Wang, Y. J., Chen, H. H., Zhou, N., et al. (2022). Alkaliphilus flagellatus sp. nov., Butyricicoccus intestinisimiae sp. nov., Clostridium mobile sp. nov., Clostridium simiarum sp. nov., Dysosmobacter acutus sp. nov., Paenibacillus brevis sp. nov., Peptoniphilus ovalis sp. nov. and Tissierella simiarum sp. nov., isolated from monkey faeces. Int. J. Syst. Evol. Microbiol. 72:005276. doi: 10.1099/ijsem.0.005276
Li, Y., Hu, W., Lin, B., Ma, T., Zhang, Z., Hu, W., et al. (2024). Omic characterizing and targeting gut dysbiosis in children with autism spectrum disorder: symptom alleviation through combined probiotic and medium-carbohydrate diet intervention - a pilot study. Gut Microbes 16:2434675. doi: 10.1080/19490976.2024.2434675
Liou, J. S., Zhang, W. L., Hsu, L. W., Chen, C. C., Wang, Y. T., Mori, K., et al. (2024). Faecalibacterium taiwanense sp. nov., isolated from human faeces. Int. J. Syst. Evol. Microbiol. 74:006413. doi: 10.1099/ijsem.0.006413
Liu, C., Du, M. X., Abuduaini, R., Yu, H. Y., Li, D. H., Wang, Y. J., et al. (2021). Enlightening the taxonomy darkness of human gut microbiomes with a cultured biobank. Microbiome 9:119. doi: 10.1186/s40168-021-01064-3
Liu, F., Li, J., Wu, F., Zheng, H., Peng, Q., and Zhou, H. (2019). Altered composition and function of intestinal microbiota in autism spectrum disorders: a systematic review. Transl. Psychiatry 9:43. doi: 10.1038/s41398-019-0389-6
Liu, S., Xi, H., Xue, X., Sun, X., Huang, H., Fu, D., et al. (2024). Clostridium butyricum regulates intestinal barrier function via trek1 to improve behavioral abnormalities in mice with autism spectrum disorder. Cell Biosci. 14:95. doi: 10.1186/s13578-024-01278-6
Lou, M., Cao, A., Jin, C., Mi, K., Xiong, X., Zeng, Z., et al. (2022). Deviated and early unsustainable stunted development of gut microbiota in children with autism spectrum disorder. Gut 71, 1588–1599. doi: 10.1136/gutjnl-2021-325115
Ma, Q., Zhang, X., and Qu, Y. (2018). Biodegradation and biotransformation of indole: advances and perspectives. Front. Microbiol. 9:2625. doi: 10.3389/fmicb.2018.02625
Madison, C. A., Hillbrick, L., Kuempel, J., Albrecht, G. L., Landrock, K. K., Safe, S., et al. (2023). Intestinal epithelium aryl hydrocarbon receptor is involved in stress sensitivity and maintaining depressive symptoms. Behav. Brain Res. 440:114256. doi: 10.1016/j.bbr.2022.114256
Manghi, P., Filosi, M., Zolfo, M., Casten, L. G., Garcia-Valiente, A., Mattevi, S., et al. (2024). Large-scale metagenomic analysis of oral microbiomes reveals markers for autism spectrum disorders. Nat. Commun. 15:9743. doi: 10.1038/s41467-024-53934-7
Martin, N. R., Patel, R., Kossack, M. E., Tian, L., Camarillo, M. A., Cintrón-Rivera, L. G., et al. (2022). Proper modulation of AHR signaling is necessary for establishing neural connectivity and oligodendrocyte precursor cell development in the embryonic zebrafish brain. Front. Mol. Neurosci. 15:1032302. doi: 10.3389/fnmol.2022.1032302
Martín, R., Miquel, S., Chain, F., Natividad, J. M., Jury, J., Lu, J., et al. (2015). Faecalibacterium prausnitzii prevents physiological damages in a chronic low-grade inflammation murine model. BMC Microbiol. 15:67. doi: 10.1186/s12866-015-0400-1
McDonald, D., Jiang, Y., Balaban, M., Cantrell, K., Zhu, Q., Gonzalez, A., et al. (2024). Greengenes2 unifies microbial data in a single reference tree. Nat. Biotechnol. 42, 715–718. doi: 10.1038/s41587-023-01845-1
Miquel, S., Leclerc, M., Martin, R., Chain, F., Lenoir, M., Raguideau, S., et al. (2015). Identification of metabolic signatures linked to anti-inflammatory effects of Faecalibacterium prausnitzii. mBio 6:e00300-15. doi: 10.1128/mBio.00300-15
Pang, X., Zhang, Q., Wang, Y., Zhan, Y., Guo, M., Chen, B., et al. (2023). Characteristics of the gut microbiota in young adults with autism spectrum disorder. J. Integr. Neurosci. 22:141. doi: 10.31083/j.jin2206141
Pappolla, M. A., Perry, G., Fang, X., Zagorski, M., Sambamurti, K., and Poeggeler, B. (2021). Indoles as essential mediators in the gut-brain axis. Their role in Alzheimer's disease. Neurobiol. Dis. 156:105403. doi: 10.1016/j.nbd.2021.105403
Qiao, Y., Wang, X., Ma, L., Li, S., and Liang, J. (2017). Functional inactivation of dorsal medial striatum alters behavioral flexibility and recognition process in mice. Physiol. Behav. 179, 467–477. doi: 10.1016/j.physbeh.2017.07.026
Reece, P., Toth, D., and Dawes, E. A. (1976). Fermentation of purines and their effect on the adenylate energy charge and viability of starved Peptococcus prevotii. J. Gen. Microbiol. 97, 63–71. doi: 10.1099/00221287-97-1-63
Renga, G., D'Onofrio, F., Pariano, M., Galarini, R., Barola, C., Stincardini, C., et al. (2023). Bridging of host-microbiota tryptophan partitioning by the serotonin pathway in fungal pneumonia. Nat. Commun. 14:5753. doi: 10.1038/s41467-023-41536-8
Rothhammer, V., and Quintana, F. J. (2019). The aryl hydrocarbon receptor: an environmental sensor integrating immune responses in health and disease. Nat. Rev. Immunol. 19, 184–197. doi: 10.1038/s41577-019-0125-8
Sakamoto, M., Sakurai, N., Tanno, H., Iino, T., Ohkuma, M., and Endo, A. (2022). Genome-based, phenotypic and chemotaxonomic classification of Faecalibacterium strains: proposal of three novel species Faecalibacterium duncaniae sp. nov., Faecalibacterium hattorii sp. nov. and Faecalibacterium gallinarum sp. nov. Int. J. Syst. Evol. Microbiol. 72:e005379. doi: 10.1099/ijsem.0.005379
Schwan, O. (1979). Biochemical, enzymatic, and serological differentiation of Peptococcus indolicus (Christiansen) Sorensen from Peptococcus asaccharolyticus (Distaso) Douglas. J. Clin. Microbiol. 9, 157–162. doi: 10.1128/jcm.9.2.157-162.1979
Serger, E., Luengo-Gutierrez, L., Chadwick, J. S., Kong, G., Zhou, L., Crawford, G., et al. (2022). The gut metabolite indole-3 propionate promotes nerve regeneration and repair. Nature 607, 585–592. doi: 10.1038/s41586-022-04884-x
Sgritta, M., Dooling, S. W., Buffington, S. A., Momin, E. N., Francis, M. B., Britton, R. A., et al. (2019). Mechanisms underlying microbial-mediated changes in social behavior in mouse models of autism spectrum disorder. Neuron 101, 246–259.e6. doi: 10.1016/j.neuron.2018.11.018
Shkoporov, A. N., Efimov, B. A., Kondova, I., Ouwerling, B., Chaplin, A. V., Shcherbakova, V. A., et al. (2016). Peptococcus simiae sp. nov., isolated from rhesus macaque faeces and emended description of the genus Peptococcus. Int. J. Syst. Evol. Microbiol. 66, 5187–5191. doi: 10.1099/ijsem.0.001494
Siddiqui, T., Bhattarai, P., Popova, S., Cosacak, M. I., Sariya, S., Zhang, Y., et al. (2021). KYNA/Ahr signaling suppresses neural stem cell plasticity and neurogenesis in adult zebrafish model of alzheimer's disease. Cells 10:2748. doi: 10.3390/cells10102748
Silva-Parra, J., Ramírez-Martínez, L., Palafox-Gómez, C., Sandu, C., López-Bayghen, E., Vega, L., et al. (2024). Aryl hydrocarbon receptor involvement in the sodium-dependent glutamate/aspartate transporter regulation in cerebellar bergmann glia cells. ACS Chem. Neurosci. 15, 1276–1285. doi: 10.1021/acschemneuro.4c00046
Sokol, H., Pigneur, B., Watterlot, L., Lakhdari, O., Bermúdez-Humarán, L. G., Gratadoux, J. J., et al. (2008). Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl. Acad. Sci. U. S. A. 105, 16731–16736. doi: 10.1073/pnas.0804812105
Stockinger, B., Shah, K., and Wincent, E. (2021). AHR in the intestinal microenvironment: safeguarding barrier function. Nat. Rev. Gastroenterol. Hepatol. 18, 559–570. doi: 10.1038/s41575-021-00430-8
Stoleru, B., Popescu, A. M., Tache, D. E., Neamtu, O. M., Emami, G., Tataranu, L. G., et al. (2013). Tropomyosin-receptor-kinases signaling in the nervous system. Maedica 8, 43–48.
Su, Q., Wong, O. W. H., Lu, W., Wan, Y., Zhang, L., Xu, W., et al. (2024). Multikingdom and functional gut microbiota markers for autism spectrum disorder. Nat. Microbiol. 9, 2344–2355. doi: 10.1038/s41564-024-01739-1
Swain, C. J., Baker, R., Kneen, C., Moseley, J., Saunders, J., Seward, E. M., et al. (1991). Novel 5-HT3 antagonists. Indole oxadiazoles. J. Med. Chem. 34, 140–151. doi: 10.1021/jm00105a021
Tammiku-Taul, J., Park, R., Jaanson, K., Luberg, K., Dobchev, D. A., Kananovich, D., et al. (2016). Indole-like Trk receptor antagonists. Eur. J. Med. Chem. 121, 541–552. doi: 10.1016/j.ejmech.2016.06.003
Uddin, M. N., Mondal, T., Yao, Y., Manley, K., and Lawrence, D. A. (2023). Oxidative stress and neuroimmune proteins in a mouse model of autism. Cell Stress Chaperones 28, 201–217. doi: 10.1007/s12192-023-01331-2
van Niel, M. B., Collins, I., Beer, M. S., Broughton, H. B., Cheng, S. K., Goodacre, S. C., et al. (1999). Fluorination of 3-(3-(piperidin-1-yl)propyl)indoles and 3-(3-(piperazin-1-yl)propyl)indoles gives selective human 5-HT1D receptor ligands with improved pharmacokinetic profiles. J. Med. Chem. 42, 2087–2104. doi: 10.1021/jm981133m
Wan, Y., Wong, O. W. H., Tun, H. M., Su, Q., Xu, Z., Tang, W., et al. (2024). Fecal microbial marker panel for aiding diagnosis of autism spectrum disorders. Gut Microbes 16:2418984. doi: 10.1080/19490976.2024.2418984
Wan, Y., Zuo, T., Xu, Z., Zhang, F., Zhan, H., Chan, D., et al. (2022). Underdevelopment of the gut microbiota and bacteria species as non-invasive markers of prediction in children with autism spectrum disorder. Gut 71, 910–918. doi: 10.1136/gutjnl-2020-324015
Wilkins, T. D., Moore, W. E. C., West, S. E. H., and Holdeman, L. V. (1975). Peptococcus niger (Hall) Kluyver and van Niel 1936: emendation of description and designation of neotype strain. Int. J. Syst. Evol. Microbiol. 25, 47–49. doi: 10.1099/00207713-25-1-47
Yang, M., Silverman, J. L., and Crawley, J. N. (2011). Automated three-chambered social approach task for mice. Curr. Protoc. Neurosci. doi: 10.1002/0471142301.ns0826s56
Yin, J., Zhang, Y., Liu, X., Li, W., Hu, Y., Zhang, B., et al. (2023). Gut microbiota-derived indole derivatives alleviate neurodegeneration in aging through activating GPR30/AMPK/SIRT1 pathway. Mol. Nutr. Food Res. 67:e2200739. doi: 10.1002/mnfr.202200739
Yu, H., Chen, L., Lei, H., Pi, G., Xiong, R., Jiang, T., et al. (2022a). Infralimbic medial prefrontal cortex signalling to calbindin 1 positive neurons in posterior basolateral amygdala suppresses anxiety- and depression-like behaviours. Nat. Commun. 13:5462. doi: 10.1038/s41467-022-33139-6
Yu, Y., Zhang, B., Ji, P., Zuo, Z., Huang, Y., Wang, N., et al. (2022b). Changes to gut amino acid transporters and microbiome associated with increased E/I ratio in Chd8(+/-) mouse model of ASD-like behavior. Nat. Commun. 13:1151. doi: 10.1038/s41467-022-28746-2
Yu, Y., Zhang, B., Wang, N., Zuo, Z., Ji, P., and Zhao, F. (2025). Unravelling lipidomic disruptions across multiple tissues in Chd8-mutant ASD mice through integration of lipidomics and single-cell transcriptomics. Gut 74, 684–686. doi: 10.1136/gutjnl-2024-332972
Yu, Y., and Zhao, F. (2021). Microbiota-gut-brain axis in autism spectrum disorder. J. Genet. Genomics 48, 755–762. doi: 10.1016/j.jgg.2021.07.001
Zhao, Y., Wang, Y., Meng, F., Chen, X., Chang, T., Huang, H., et al. (2023). Altered Gut microbiota as potential biomarkers for autism spectrum disorder in early childhood. Neuroscience 523, 118–131. doi: 10.1016/j.neuroscience.2023.04.029
Keywords: the gut-brain axis, autism spectrum disorders (ASD), microbiome, indole, aryl hydrocarbon receptor (AhR), BTBR
Citation: Yu Y, Wang Y, Zhang J, Li S, Wang Y, You X, Chen X, Du M, Xie L and Liu S-J (2025) The gut commensal Faecalibacterium hominis attenuates indole—AhR signaling and restores ASD—like behaviors with BTBR mice. Front. Microbiol. 16:1640149. doi: 10.3389/fmicb.2025.1640149
Received: 03 June 2025; Accepted: 28 July 2025;
Published: 22 August 2025.
Edited by:
Julio Plaza-Diaz, Children's Hospital of Eastern Ontario (CHEO), CanadaReviewed by:
Lorena Coretti, University of Naples Federico II, ItalyKiesha Wilson, University of South Carolina, United States
Copyright © 2025 Yu, Wang, Zhang, Li, Wang, You, Chen, Du, Xie and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: You Yu, eXV5b3UyMDE0QHNpbmEuY29t; Shuang-Jiang Liu, bGl1c2pAaW0uYWMuY24=
†These authors have contributed equally to this work
 You Yu
You Yu Yujing Wang
Yujing Wang Jie Zhang1
Jie Zhang1 Yulin Wang
Yulin Wang Xin You
Xin You Xue Chen
Xue Chen Shuang-Jiang Liu
Shuang-Jiang Liu