- 1Key Laboratory of Microbial Resources Exploitation and Application of Gansu Province, Institute of Biology, Gansu Academy of Sciences, Lanzhou, China
- 2State Key Laboratory of Aridland Crop Science, Gansu Agricultural University, Lanzhou, China
Introduction: Heparan sulfate (HS), a linear anionic polysaccharide, participates in many physiological processes and exhibits many pharmacological activities. D-glucuronyl C5-epimerase (Glce) is one of the key enzymes in the biosynthesis of heparan sulfate proteoglycans. However, the recombinant Glce protein exhibits reduced catalytic activity and production yield, which substantially impedes the development of enzymatic methods for producing pharmaceutical-grade heparin.
Methods: In this experiment, we established a valid method for heterologous expression in Escherichia coli (E. coli) and subsequent purification of two N-terminal truncated Glce proteins using the SUMO-fused expression system. Characterization of human Glce167-617 was described by dynamic light scattering size-exclusion chromatography, and X-ray crystallographic.
Results: In the present study, we successfully overexpressed and purified human Glce167-617 protein in E. coli. Subsequently, the recombinant Glce167-617 was found to exist as a dimer in solution. X-ray crystallographic result further confirmed its dimeric assembly while maintaining the integrity of the catalytic domain.
Discussion: In summary, this study successfully overexpressed and purified human Glce protein in E. coli. The purified Glce protein will be applied to chemoenzymatic synthesis of heparin and heparan sulfates in vitro, which facilitating the future bioengineering of pharmaceutical heparins.
1 Introduction
Heparan sulfate, a highly sulfated polysaccharide, constitutes a fundamental component of cell surface and extracellular matrix structures (Préchoux et al., 2015). This glycosaminoglycan involves in a variety of biological activities including cell growth, blood coagulation, embryonic development, inflammatory response and tumor metastasis (Bartolini et al., 2020; Bendersky et al., 2020). HS biosynthesis is accomplished through multi-step enzymatic reactions following the polymerization of its precursor disaccharide units. The HS chain precursor is composed of GlcA-GlcNAc repeating disaccharide units. N-deacetylation/N-sulfation of glucosamine is catalyzed by N-deacetylase/N-sulfotransferase (NDST), followed by the epimerization of glucuronic acid (GlcA) unit to L-iduronic acid (IdoA) by Glce protein. The side chain of the polysaccharide further undergoes sulfation modification by 2-O-sulfatetransferase (2OST), 6-O-sulfatetransferase (6OST) and 3-O-sulfate transferase (3OST) (El Masri et al., 2017; Préchoux et al., 2015). In vertebrates, there are multiple isoforms of sulfotransferases in HS biosynthetic activities, while Glce and 2OST each have only a single isoform (Tarja et al., 2005).
At present, pharmaceutical grade heparan sulfate is mainly derived from animal tissues. However, these preparations inevitably include trace contaminants including structurally similar glycosaminoglycans, as well as tissue-derived viruses and prions (Liu et al., 2009). To address these limitations, a chemoenzymatic bioengineered approach has emerged as a promising alternative to animal-sourced heparan sulfate (Fu et al., 2016). This methodology commences with the microbial fermentation of Escherichia coli K5 capsular polysaccharide, follows by sequential enzymatic modifications to generate well-defined heparan sulfate structures. Among these steps, Glce protein catalyzes the epimerization of GlcA residues to L-IdoA, a reaction that constitutes a critical step in determining the characteristic structural heterogeneity of heparan sulfate.
Glce is one of the key enzymes in the biosynthesis of heparan sulfate. Targeted disruption of Glce gene in mice leads to a structurally aberrant HS lacking of L-IdoA residues, which correlates with a lethal phenotype characterized by renal agenesis, lung defects, and skeletal malformations (Li et al., 2003). Functional investigations demonstrate that Glce may affect the proliferation, angiogenesis and metastasis of Ewing sarcoma by altering heparan sulfate biosynthesis and is a potential prognostic indicator in Ewing sarcoma (Wen et al., 2023). Notably, many studies have found that Glce gene expression is associated with many tumorigenesis and virus infections (Marques et al., 2021). For example, Glce significantly suppresses the proliferative activity of breast cancer cells in vitro and tumor growth in vivo (Grigorieva et al., 2011; Prudnikova et al., 2014). Cell-surface Glce binds to the porcine deltacoronavirus spike protein, facilitating viral entry (Xiao et al., 2024). Currently, the crystal structure of human Glce protein was reported, these structures elucidate its catalytic mechanism with heparin sulfate. Structure analysis found that the N-terminal β-hairpin and C-terminal α-barrel were major domains to form the dimer structure (Debarnot et al., 2019). However, the molecular basis of N-terminal β-hairpin involvement in functional dimer formation remains to be established.
At present, there is still lack an effective strategy for producing soluble and high-yield Glce protein. Mammalian cells serve as the primary protein expression system to obtain human Glce protein (Cui et al., 2023; Debarnot et al., 2019), but their high operational costs and limited production capacities render large-scale manufacturing technically challenging. In contrast, the E. coli expression system offers simplicity, low cost, and high yield, making it an important platform for heterologous expression of membrane proteins and glycosylation modification enzymes. To address this challenge, we utilized the E. coli SUMO-fused expression system to express and purify two N-terminal truncated Glce proteins. Static- light and dynamic-light scattering experiments revealed that Glce167-617 is consistent with a homogeneous and stable dimer. Additionally, crystallography experiments further demonstrated that Glce167-617 protein is dimeric assembly and retains its glycosylation substrate binding sites. This study established a protocol for efficient expression and purification of recombination human Glce167-617 in E. coli, while confirming its catalytic activity through structural characterization. The crystallographic study was specially designed to compare substrate-binding mechanisms between human and zebrafish Glce orthologs. These founding may facilitate industrial production of Glce protein for the chemoenzymatic synthesize of specific heparin sulfate.
2 Materials and methods
2.1 Construction of two Glce truncation expression plasmids
The Glce gene was chemically synthesized according to the codon preference of Escherichia coli. The Glce (G102-N617) and Glce (Y167-N617) gene fragments were amplified from full-length Glce cDNA with a 5′ overhang containing the EcoRI site and a 3′ overhang containing the XhoI site. The PCR products were subsequently inserted into the pET-15b-SUMO vector (Invitrogen) using the EcoRI and XhoI restriction sites, producing a SUMO and His6 tag at the N-terminus. The pET-15b-SUMO-Glce102-617 and pET-15b-SUMO-Glce167-617 constructs were confirmed by double restriction enzyme digestion and sequencing.
2.2 Expression of Glce truncated proteins in Escherichia coli
The recombinant plasmids were transformed into C2566H E. coli competent cells (New England Biolabs) for expression of SUMO-Glce102-617 and SUMO-Glce167-617 fusion proteins. Cells containing the Glce gene expression plasmid were initially grown in LA (Luria-Bertani Agar) medium with 100 μg/mL ampicillin at 37°C. Selected colonies were inoculated into Luria Bertani (LB) medium containing 100 μg/mL ampicillin and incubated at 37°C, 180 rpm for overnight. The seed culture was subsequently transferred to LB medium (0.1% inoculation volume) and grown under identical conditions until the cell density reached an OD600 of 0.8–1.2. Protein induction was initiated by reducing the temperature was lowered to 20°C, followed by addition of 0.15 mM IPTG and incubation for 16 h at 18°C, 180 rpm.
2.3 Purification of Glce truncated proteins
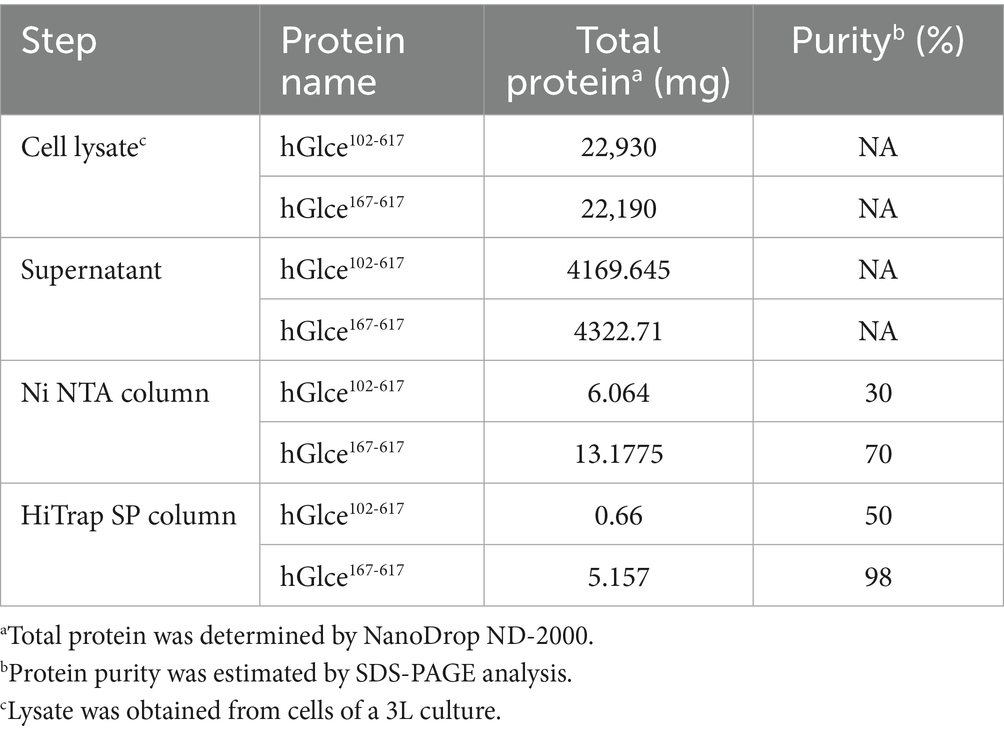
Bacterial cells were collected by centrifugation and resuspended in lysis buffer containing 20 mM Tris–HCl, pH 8.0, 500 mM NaCl, 5 mM imidazole and 5% glycerol (v/v). Following sonication, the lysate was centrifuged at 12,000 rpm for 1 h. The supernatant was loaded onto a Ni2+ charged immobilized metal affinity chromatography (IMAC) column (GE Healthcare), followed by sequential washing with lysis buffer and wash buffer [20 mM Tris-HCl, pH 8.0, 500 mM NaCl, 20 mM imidazole and 5% glycerol (v/v)]. Target proteins were eluted with elution buffer [20 mM Tris-HCl, pH 8.0, 500 mM NaCl, 200 mM imidazole and 5% glycerol (v/v)]. Proteolytic cleavage of the N-terminal SUMO and His6 tag was removed by incubation the target proteins with SUMO protease at a molar ratio of 250:1 for overnight at 4°C. The tag-free target proteins were diluted in buffer [20 mM Tris–HCl, pH 8.0, 5% glycerol (v/v), 1 mM 1, 4-dithioerythritol (DTT)] and loaded onto a 1 mL HiTrap SP column (GE Healthcare) at 18°C. Bound proteins were eluted using a linear NaCl gradient to 1 M, and the target protein-containing fractions were subsequently diluted in buffer (20 mM Tris-HCl, pH 8.0, 200 mM NaCl, 1 mM DTT). Final the target protein-containing fractions were concentrated to 8 mg/mL using ultrafiltration devices and stored at −80°C.
2.4 Static- and dynamic-light scattering
The aggregation state of Glce167-617 protein in solution was analyzed using static- and dynamic-light scattering. For static light scattering experiment, which was performed on a Superdex 200 10/300 GL column (GE Healthcare) using a fast protein liquid chromatography (FPLC) system (ÄKTA Purifier, GE Healthcare) at 18°C. The column was equilibrated with buffer (20 mM Tris-HCl, pH 8.0, 200 mM NaCl, 1 mM DTT) and calibrated using gel filtration standard (Bio-Rad) at a flow-rate of 0.2 mL/min. Subsequently, 100 μg of Glce167-617 was loaded onto the column, and elution profiles were monitored via UV absorbance at 280 nm. Molecular weight determination of Glce167-617 protein was calculated with the fitted standard curve of ln(Mw) = −8.4755Kav + 15.145 (R2 = 0.9841).
Dynamic light scattering (DLS) was performed at room temperature using a DynaPro NanoStar instrument (Wyatt Technology Corporation, United States) with a 20 μL micro-cuvette (Liu et al., 2015). All buffers and samples were filtered with 0.22 μm filter membranes and centrifuged at 12,000 rpm for 30 min. A preparation 2 μM Glce167-617 protein was used for the DLS experiment. Following the instrument equilibration, data acquisition durations were maintained between 3 and 5 min, and the collected data were analyzed using Dynamics 7.0 software.
2.5 Crystallization
Crystallization experiments were conducted using commercially available crystallization screens with the sitting-drop vapour diffusion method at 20°C. During preliminary screening, crystals of human Glce were grown in drops containing 0.25 μL protein solution (8 mg/mL) and 0.25 μL reservoir solution composed of 0.2 M ammonium sulfate, 0.1 M Tris, pH 6.5–8.5, and 25% (w/v) polyethylene glycol 3,350. Following optimization, rod-shaped crystals were successfully obtained with a reservoir solution containing 0.2 M ammonium sulfate, 0.1 M Tris, pH 8.5, and 22% (w/v) polyethylene glycol 3,350.
2.6 Date collection and structure determination
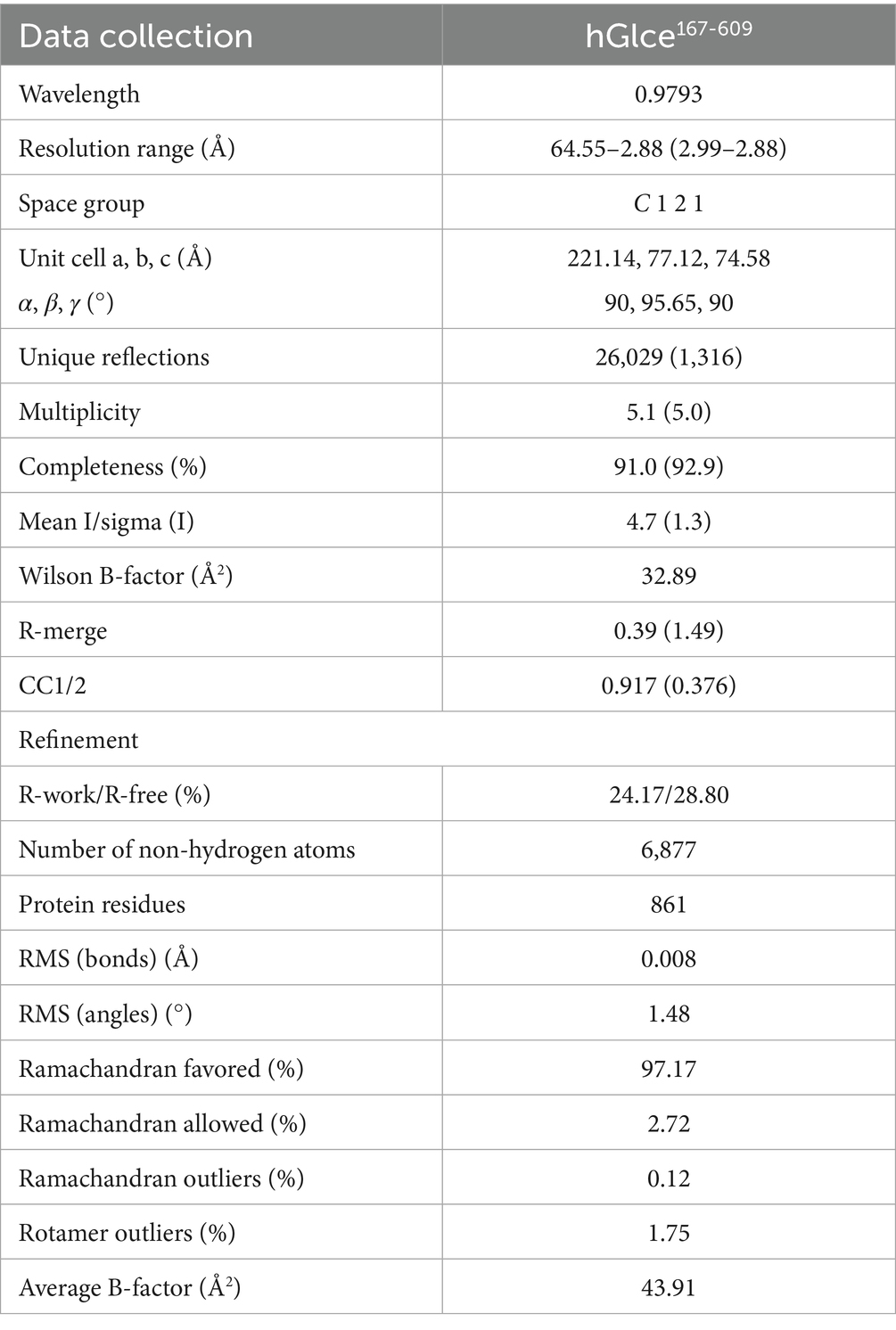
All X-ray diffraction data were collected at beamline BL19U1 of the Shanghai Synchrotron Radiation Facility (SSRF, Shanghai, China) using a Pilatus 6M detector (Dectris) and processed using X-ray Detector Software (XDS) (Kabsch, 2010). Date collection and processing statistics are summarized in Table 1.
3 Results
3.1 Plasmid construction and test expression of SUMO-fused recombinant proteins Glce102-617 and Glce167-617
To enhance the soluble fragment of human Glce protein and simplify the purification steps, the coding sequence was fused to the C-terminus of the SUMO tag. This fusion provides a simple and rapid purification strategy (Zhou et al., 2020). The recombination protein bearing the SUMO tag could be cleaved by SUMO protease. In addition, a His6 tag was incorporated at the N-terminus of the SUMO tag, thereby enabling protein purification via Ni2+-IMAC affinity chromatography.
Multiple sequence alignment was conducted to demonstrate the sequence homology among human, mouse, zebrafish and Drosophila melanogaster. The alignment illustrated that Glce possesses a highly conserved protein sequence across species (Figure 1). AlphaFold structure prediction showed that the N-terminus of Glce composes one α-helical domain and disordered regions (Supplementary Figure 1). To achieve enhanced expression of Glce protein, two truncated constructs differing in N-terminal truncation positions were generated. In summary, the Glce102-617 and Glce167-617 gene fragments were cloned into the pET-15b-SUMO vector (Figures 2A,D), with construct validation performed through double restriction enzyme and Sanger sequencing (Figures 2B,E).
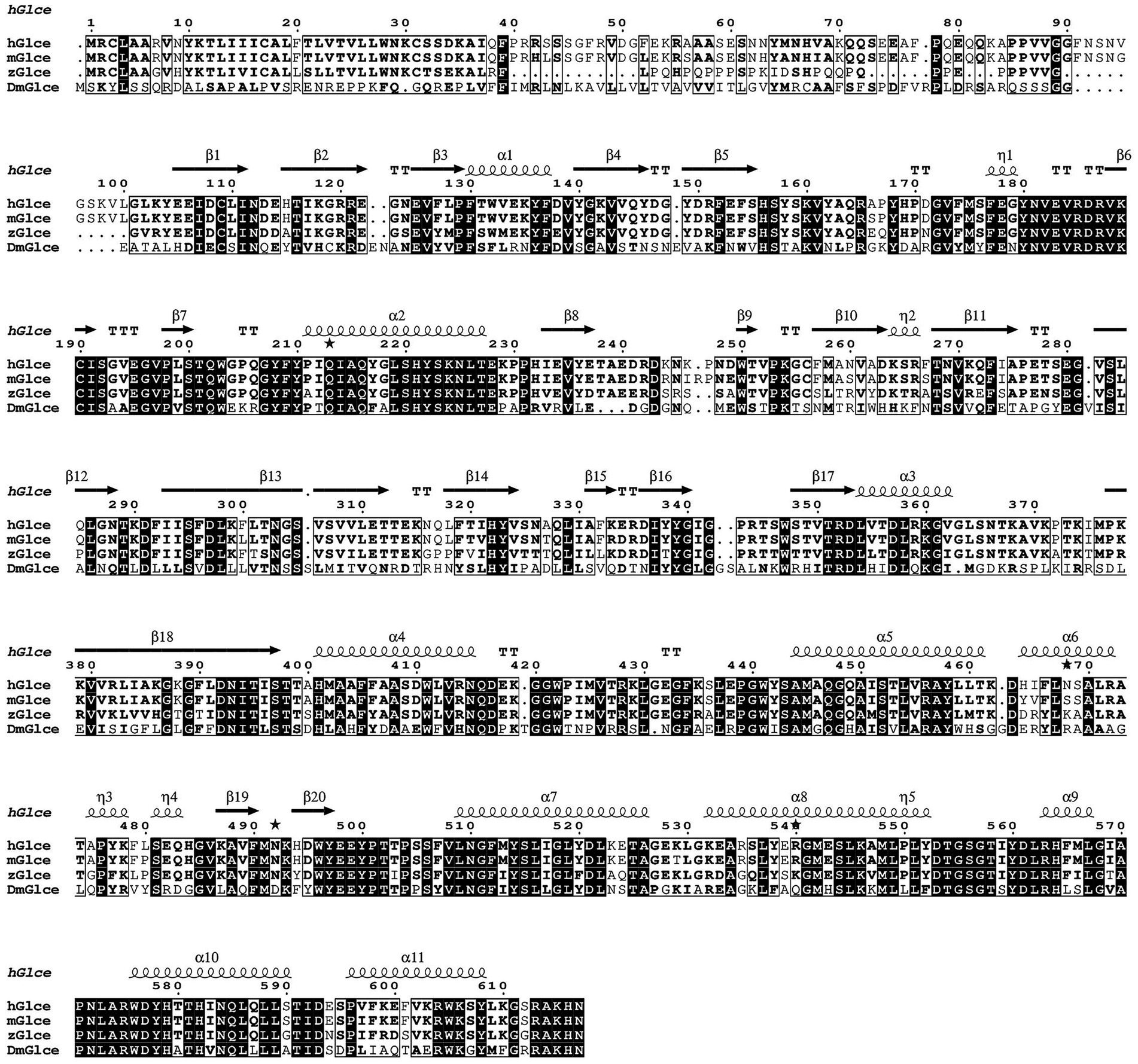
Figure 1. Multiple sequence alignment of hGlce, its close homologs from Mus musculus (mGlce) and Zebrafish (zGlce), as well as Drosophila melanogaster (DmGlce), was constructed using ClustalW and visualized via ESPript. The identical residues are showed with white color, whereas similar residues are enclosed in boxes. Secondary structure elements of Glce are indicated above the alignment result.
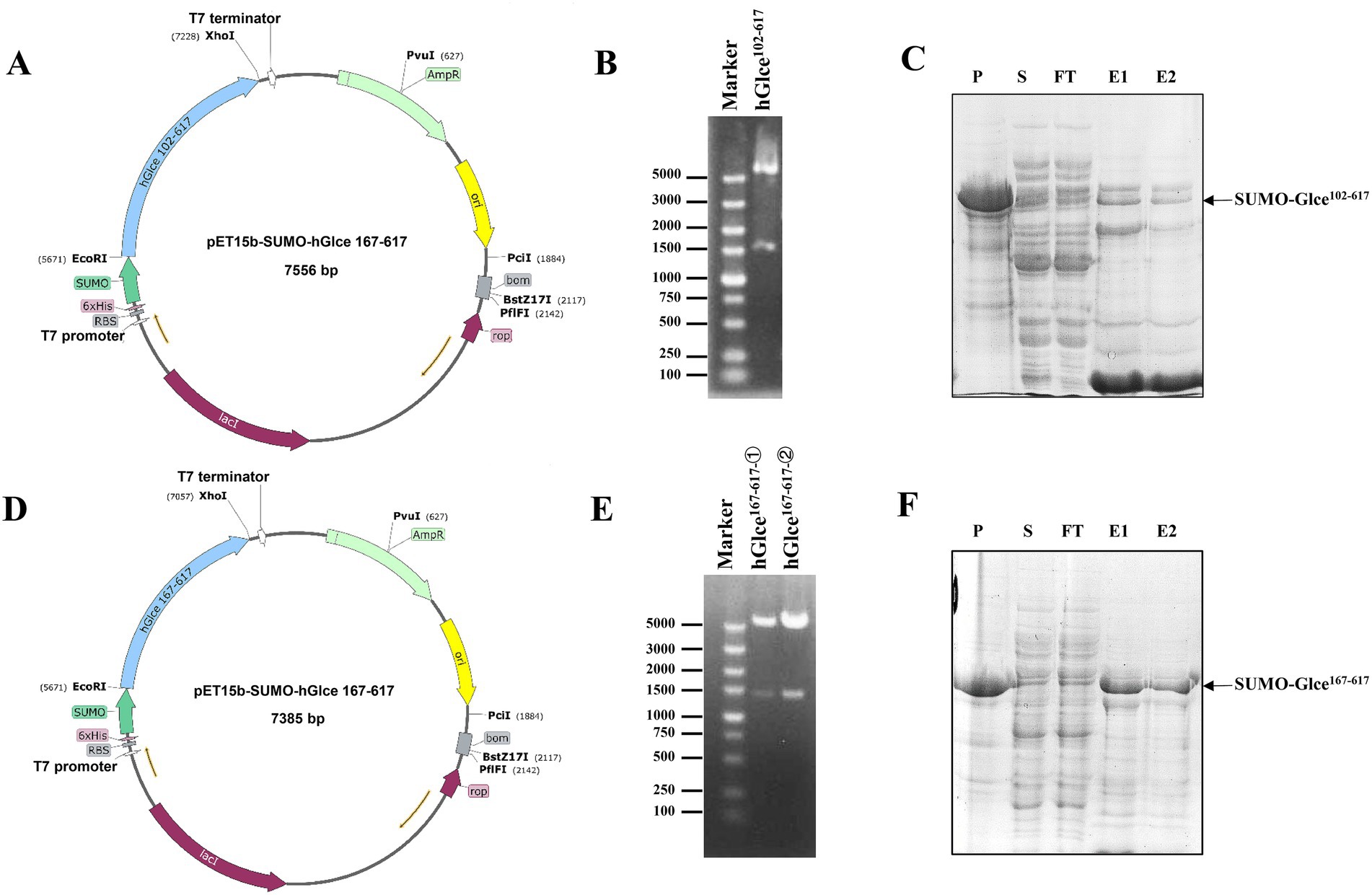
Figure 2. Recombinant plasmids construction and expression of human Glce102-617 and Glce167-617 fusion protein. (A) Schematic of prokaryote expression vector pET-15b-SUMO-Glce102-617. (B) Enzymatic digestion of the recombinant plasmid pET-15b-SUMO-Glce102-617. (C) The SDS-PAGE analysis of SUMO-Glce102-617 fusion protein. Arrow shows SUMO-Glce102-617 fusion protein. P, precipitate; S, supernatant; FT, flow-through fraction; E1, the first tube of eluted protein; E2, the second tube of eluted protein. (D) Schematic of pET-15b-SUMO-Glce167-617 plasmid. (E) Enzymatic digestion of the recombinant plasmid pET-15b-SUMO-Glce167-617. (F) The SDS-PAGE analysis of SUMO-Glce167-617 fusion protein. Arrow shows SUMO-Glce167-617 fusion protein.
The E. coli C2566H strain was employed as the expression host for recombinant glucuronyl C5-epimerase. An engineered E. coli strain harboring Glce102-617 or Glce167-617 gene was cultured at 37°C to reach an OD600 value of approximately 0.85. Following induction with 0.15 mM IPTG, SUMO-Glce102-617 and SUMO-Glce167-617 proteins were induced for efficient overexpression. SDS-PAGE analysis demonstrated that SUMO-Glce167-617 protein exhibited a significantly higher expression level compared to SUMO-Glce102-617 (Figures 2C,F).
3.2 Purification of human Glce102-617 and Glce167-617 proteins
An efficient purification protocol for the hGlce protein was established, demonstrating suitability for biochemical characterization and crystallization. The expressed Glce102-617 and Glce167-617 proteins underwent purification via a two-step chromatography method comprising Ni2+-IMAC column purification followed by HiTrap SP ion exchange chromatography. The first purification step using Ni2+-IMAC column eluted SUMO-Glce102-617 or SUMO-Glce167-617 fused proteins (Figures 3A,C). Subsequently, the eluted proteins underwent SUMO tag cleavage using SUMO protease at 4°C for overnight. The second purification step comprised HiTrap SP ion exchange column with elution conducted through a linear NaCl gradient (0.05–0.5 M) (Figures 3B,D). The purity of purified Glce167-617 was exceeded 98%, whereas Glce102-617 displayed a purity of approximately 50% (Figure 3E). SDS-PAGE analysis indicated that both proteins migrated with apparent molecular weights slightly smaller than their theoretical molecular weight (Mw). The purified Glce102-617 (theoretical Mw: 58.9 kDa) migrated as ~55 kDa protein as determined by SDS-PAGE. Similarly, the purified Glce167-617 (theoretical Mw: 52.1 kDa) migrated as ~50 kDa protein by SDS-PAGE. Purification procedures are detailed in Table 2. The yield of Glce167-617 was observably higher than that of Glce102-617.
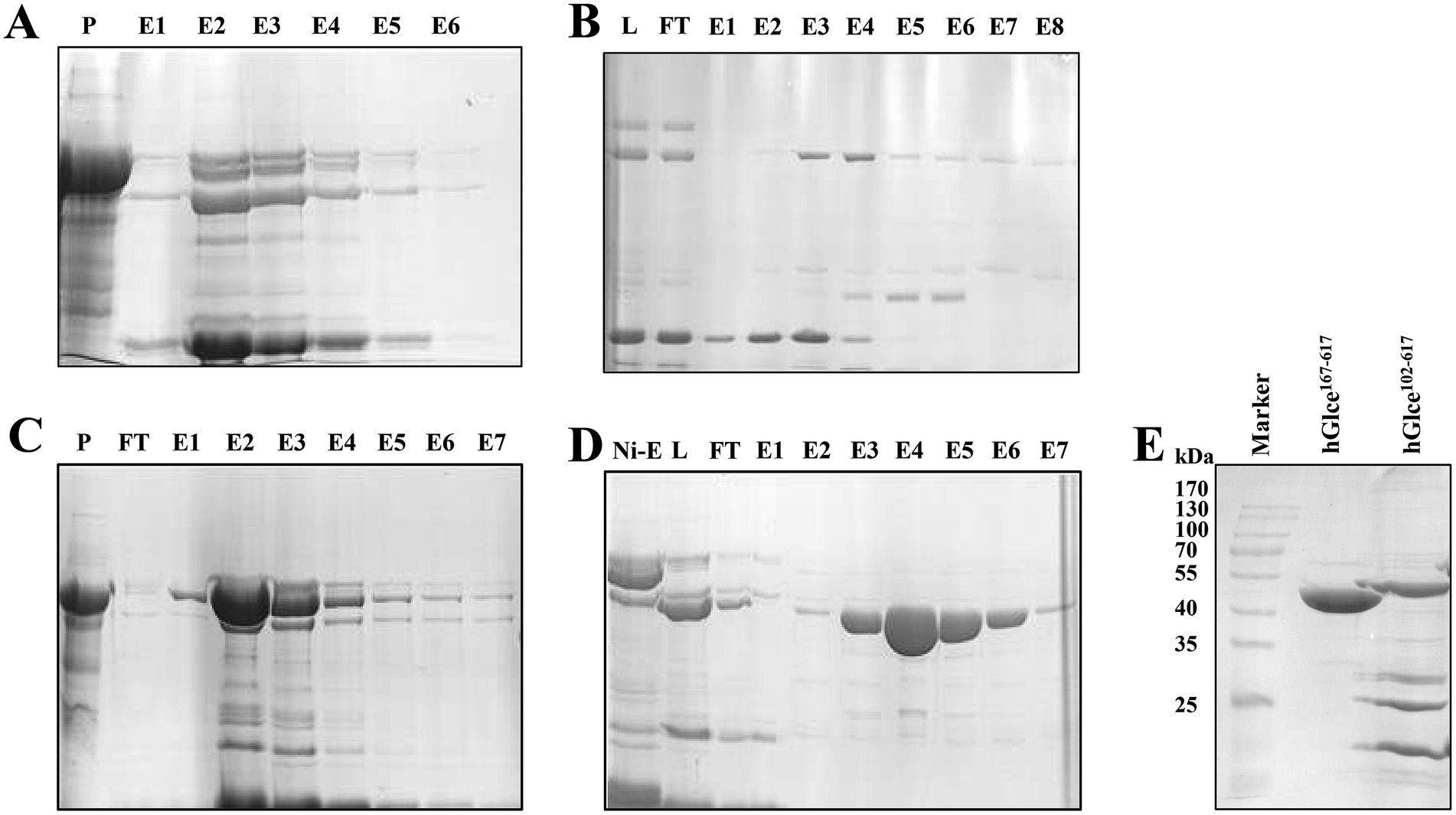
Figure 3. Purification of Glce protein using affinity and ion exchange chromatography. (A) SDS-PAGE analysis of SUMO-Glce102-617 fusion protein after Ni2+ affinity chromatography. P, precipitate; E, the eluted protein. (B) Purification of Glce102-617 protein after ion exchange chromatography. L: SUMO-Glce102-617 was treated with SUMO protease to load onto ion exchange chromatography. (C) SDS-PAGE analysis of SUMO-Glce167-617 fusion protein after Ni2+-IMAC affinity chromatography. (D) Purification of Glce167-617 protein after ion exchange chromatography. (E) The SDS-PAGE analysis of the purified Glce102-617 and Glce167-617 proteins.
3.3 Glce167-617 protein act as dimer in solution
The aggregation state of Glce167-617 may be disrupted by denaturing conditions during SDS-PAGE. To further determine the homogenicity and aggregation state of Glce167-617 in solution, DLS and size exclusion chromatography (SEC) analyses were conducted for Glce167-617 characterization. DLS analysis revealed a hydrodynamic radius of 4.715 nm for Glce167-617, which corresponded to an estimated molecular weight of 127 kDa (Figure 4A). These results demonstrate that Glce167-617 exists as a dimer in solution. Similarly, the SEC analysis revealed a single peak at Kav = 0.425, yielding an apparent molecular weight of 102.5 kDa based on the calibration equation (Figure 4B).
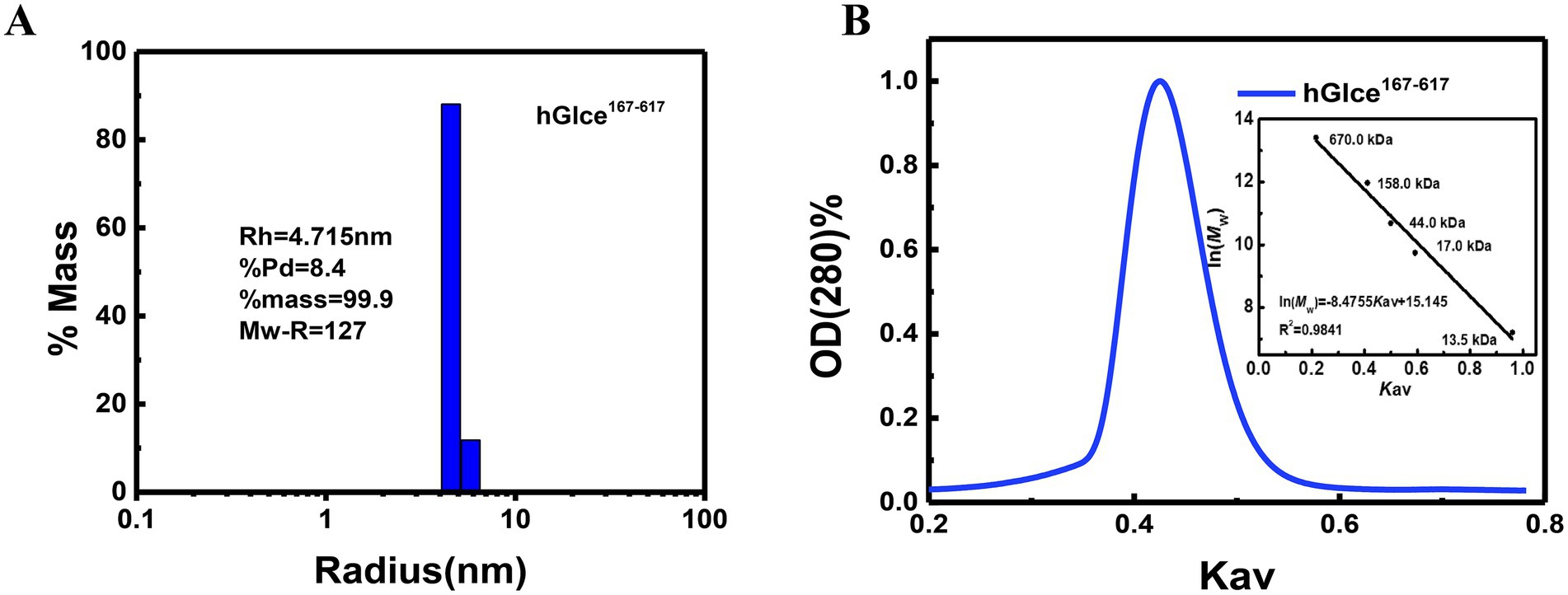
Figure 4. The aggregation state of Glce167-617 protein in solution. (A) Size distribution histogram of Glce167-617 measured by DLS. (B) Size exclusion chromatography analysis of Glce167-617 by Superdex 200 10/300 GL column. The insert graph displays elution profiles of calibration kit protein on the same column.
3.4 Crystallization and dimer structure of Glce167-617
The pure and homogenous Glce167-617 protein was utilized for crystallization trails with different commercial screening kits. We succeeded in screening crystallization conditions, which grew a few single crystals with lamellar structures (Figure 5A). The crystal structure of Glce167-617 was determined by molecular replacement and refined to 2.88 Å resolution with R-work/R-free values of 0.24/0.28, respectively. Comprehensive statistics for structure determination and refinement are summarized in Table 1.
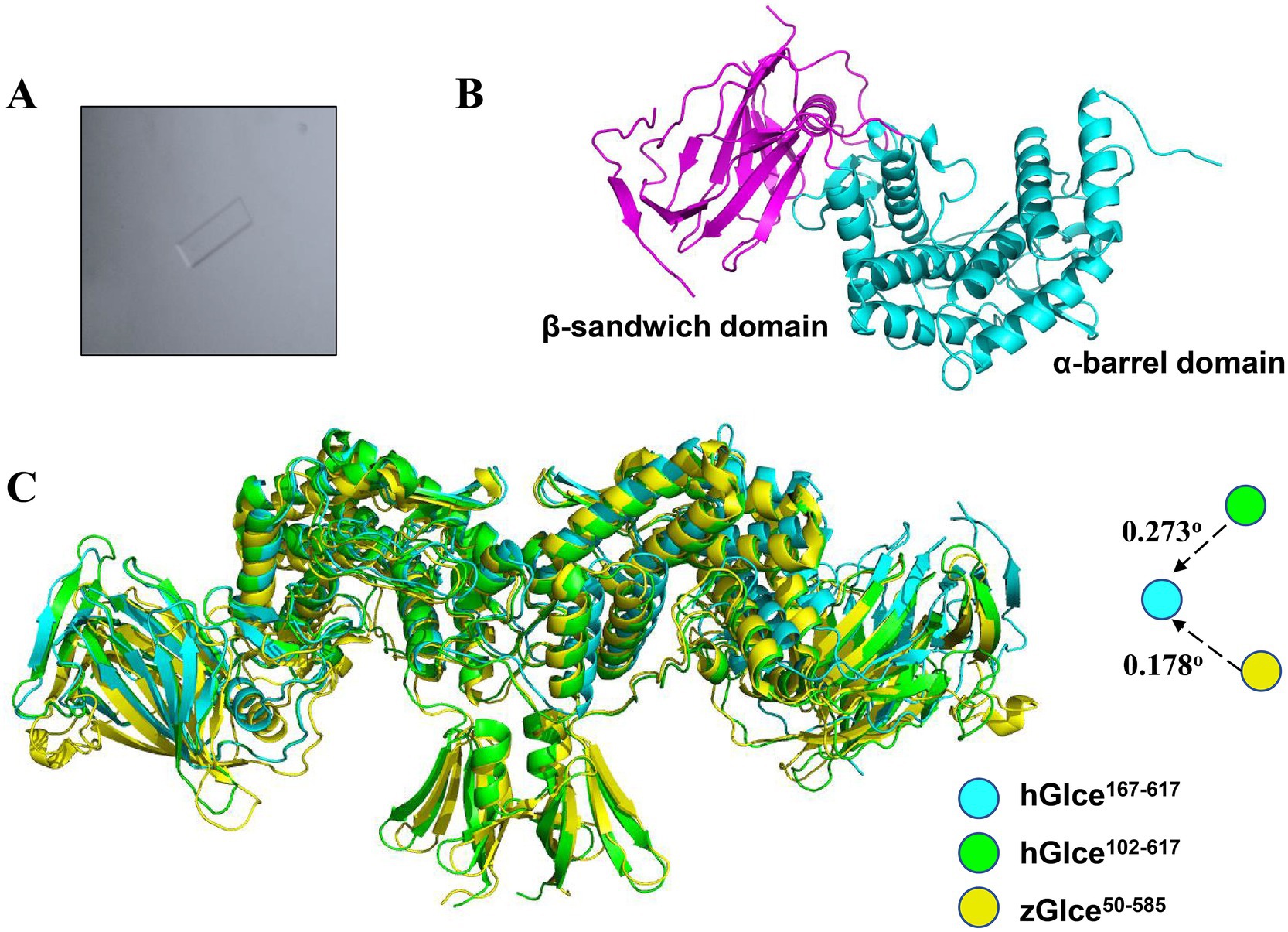
Figure 5. Structure of hGlce167-617. (A) The micrograph shows typical lamellar crystal of hGlce167-617. (B) Overall structure of the Glce167-617 monomer. Each subunit comprising of β-sandwich and α-barrel domain are colored in magentas and cyan. (C) Structure alignment of hGlce167-617 to the structure of hGlce102-617 and zGlce50-585. hGlce167-617, hGlce102-617 and zGlce50-585 are colored in cyan, green, and yellow, respectively.
Glce167-617 crystallized in space group C 1 2 1 with two molecules in the asymmetric unit. The Glce167-617 structure comprises two domains: an α-barrel domain (residues P167-I233 and D353-N617) and a β-sandwich domain (residues E234-R352). The overall dimeric structure adopts a butterfly-like shape (Figure 5B). Structure analysis reveals a high degree of structural conservation between Glce167-617 and both human Glce102-617 and zebrafish Glce50-585. Structural superposition of hGlce167-617 with hGlce102-617 and zGlce50-585 yielded root-mean-square deviations (RMSD) of 0.273 Å and 0.178 Å, respectively (Figure 5C). As previously reported, dimer formation in Glce involves N-terminal β-hairpin and C-terminal α-barrel domains (Debarnot et al., 2019; Qin et al., 2015). Our results found that deletion of the N-terminal β-hairpin did not disrupt the dimeric architecture in Glce. At the dimerization interface, the C-terminal α-barrel domain forms hydrophobic packing interactions with residues F490, M491, A547, L551, T554, and F565 (Supplementary Figure 2).
3.5 hGlce167–617 contains conserved substrate binding sites
To identify the substrate-binding pocket, structural superposition was performed among three conformations: hGlce167-617 apo, hGlce102-617 in complex with (GlcA-GlcNS)5, and zGlce50-585 in complex with heparin. Structural superposition of hGlce167-617 apo and the hGlce102-617-(GlcA-GlcNS)5 complex through alignment of α-barrel domain revealed that the (GlcA-GlcNS)5 substrate occupies a cleft between the α-barrel and β-sandwich domains in hGlce167-617 (Figure 6A). Similarly, superposition of hGlce167-617 apo with zGlce50-585-heparin complex via α-barrel domain alignment demonstrated that heparin substrate occupies the same cleft between α-barrel and β-sandwich domains in hGlce167-617 (Figure 6B). Structural analysis suggests that hGlce167-617 retains evolutionarily conserved substrate-binding sites.
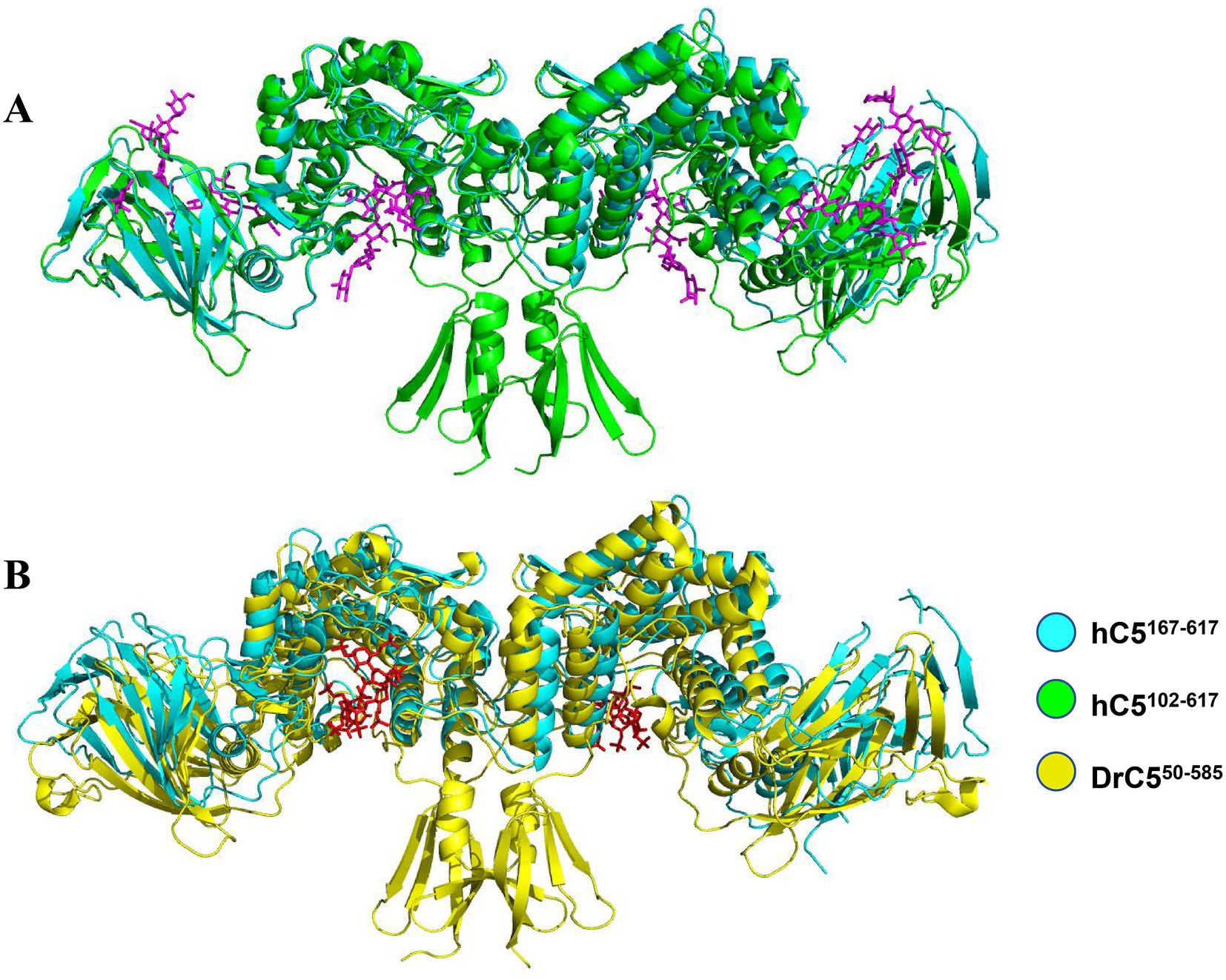
Figure 6. Structure of hGlce167-617 contain the conserved substrate binding sites. (A) Overall view of substrate binding sites in hGlce167-617 and hGlce102-617. (B) Overall view of (GlcA-GlcNS)5 substrate binding sites in hGlce167-617 and zGlce50-585.
4 Discussion
Currently, bioengineered heparin synthesis represents a cost-effective method that has been proposed as an alternative to animal-sourced heparin (Vaidyanathan et al., 2020). Glucuronyl C5-epimerase serves as a key enzyme in the biosynthesis of heparin and heparin sulfate, which plays a vital role in cell–cell and cell–matrix interaction and signaling (Cui et al., 2021; Ori et al., 2008). Human glucuronyl C5-epimerase has been the most extensively characterized because of its functional involvement in tumor suppression (Belyavskaya et al., 2017; Wen et al., 2023; Zhu et al., 2021). However, the recombinant Glce protein exhibits reduced catalytic activity and production yield, which substantially impedes the development of enzymatic methods for producing pharmaceutical-grade heparin.
The E. coli expression system was used for the overexpressed human Glce protein in Origami-B cell through the pMalc2x vector, demonstrating epimerase functionality (Li et al., 2010; Préchoux et al., 2015). Although the recombinant Glce protein was successfully expressed and purified, it remained fused to maltose-binding protein (MBP) and contained some GroEL contaminant (Vaidyanathan et al., 2020). Moreover, mammalian HEK293 and HEK-EBNA cells were utilized for human Glce protein expression, resulting in enhanced catalytic activity and production yield (Cui et al., 2023; Debarnot et al., 2019). However, ekaryotic expression systems exhibit high operational costs and low expression efficiency, rendering them unsuitable for large-scale protein expression production. In summary, the E. coli expression system provides a streamlined, cost-effective, and scalable platform for high-yield production of target proteins.
In this study, hGlce gene was successfully cloned and overexpressed in the host E. coli C2566H strain with the pET-15b-SUMO expression vector. It was previously reported that deletion of 203 amino acids from the N-terminus of the Glce protein resulted complete loss of epimerase activity (Li et al., 2010). In order to enhance Glce protein expression efficiency, two N-terminally truncated constructs (Glce102-617 and Glce167-617) were generated. The overall expression level and solution fraction of Glce167-617 protein demonstrated a remarkable increase compared to full-length Glce, thereby facilitating high-throughput screen of crystallization conditions. Notably, the expression level of Glce167-617 protein was considerably improved, which is compared with other different truncations (Li et al., 2010). Hence, this optimization established a feasible strategy for recombinant Glce protein production.
For protein purification, affinity chromatography represents a prevalently adopted technique for target proteins purification, thereby enabling high-yield recovery and exceptional purity from complex protein mixtures (Gomari et al., 2020). The SUMO tag strategy was employed to promote the solubility of Glce protein (Nazir et al., 2024). The purification method of Glce protein involves a two-step chromatographic process that contains affinity purification and ion exchange purification, resulting in highly purified target protein.
The structure and function of Glce are highly conserved across species. Current structural investigations of human and zebrafish Glce have achieved the most comprehensive characterization. Studies have reported that human and zebrafish Glce exist as dimer and function in dimeric form (Debarnot et al., 2019; Qin et al., 2015). Furthermore, Bermanella marisrubri Glce adopts dimeric and tetrameric states, with aggregate formation maintaining functional activity (Raedts et al., 2013). Structure analysis found that human and zebrafish Glce protein predominantly adopt tight dimeric configurations through hydrophobic packing interaction and ionic bonds between N-terminal β-hairpin and C-terminal α-barrel domains (Debarnot et al., 2019; Qin et al., 2015). In this study, it was investigated whether Glce dimer remained stable following N-terminal β-hairpin deletion. Initially, DLS and SEC analyses confirmed the dimeric state of Glce167-617 protein in solution. Subsequently, crystallographic analysis of Glce167-617 showed that the dimeric architecture was preserved after N-terminal β-hairpin removal. Collectively, these findings identify the C-terminal α-barrel domain as the primary dimerization interface. Each Glce dimer contains two active sites that facilitate heparin sulfate modifications.
5 Conclusion
In the present study, an Escherichia coli expression system has been developed to facilitate efficient expression and purification of Glce protein. SEC and DLS analyses confirmed the stable dimeric configuration of Glce167-617 with homogeneous oligomeric state. Moreover, the crystal structure of Glce167-617 was resolved at 2.88 Å resolution, revealing a complete dimeric architecture that preserves the GlcA epimerization site. Collectively, these findings establish a simple and efficient approach for producing active Glce protein, thereby facilitating the biosynthesis of heparin and heparan sulfate in vitro.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Ethics statement
The manuscript presents research on animals that do not require ethical approval for their study.
Author contributions
Q-xS: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. L-jG: Data curation, Formal analysis, Methodology, Project administration, Conceptualization, Writing – original draft.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the Gansu Province Youth Science and Technology Fund program, China (No. 25JRRA403); Youth Science and Technology Talents Project of Lanzhou city, China (No. 2024-QN-130); The Special Fund for Talent Introduction and Scientific Research of Gansu Academy of Sciences, China (No. QD2024-02).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1641598/full#supplementary-material
References
Bartolini, B., Caravà, E., Caon, I., Parnigoni, A., Moretto, P., Passi, A., et al. (2020). Heparan sulfate in the tumor microenvironment. Adv. Exp. Med. Biol. 1245, 147–161. doi: 10.1007/978-3-030-40146-7_7
Belyavskaya, V. A., Prudnikova, T. Y., Domanitskaya, N. V., Litviakov, N. V., Maksimov, V. N., Cherdyntseva, N. V., et al. (2017). GLCE rs3865014 (Val597Ile) polymorphism is associated with breast cancer susceptibility and triple-negative breast cancer in Siberian population. Gene 628, 224–229. doi: 10.1016/j.gene.2017.07.054
Bendersky, V., Yang, Y., and Brennan, T. V. (2020). Immunomodulatory activities of the heparan sulfate mimetic PG545. Adv. Exp. Med. Biol. 1221, 461–470. doi: 10.1007/978-3-030-34521-1_18
Cui, H., Li, Q., Wang, P., and Fang, J. (2023). Characterization and expression of highly active recombinant human glucuronyl C5-epimerase in mammalian cells. Glycobiology 33, 432–440. doi: 10.1093/glycob/cwad021
Cui, H., Wang, Z., Zhang, T., Li, J. P., and Fang, J. (2021). Re-expression of glucuronyl C5-epimerase in the mutant MEF cells increases heparan sulfate epimerization but has no influence on the Golgi localization and enzymatic activity of 2-O-sulfotransferase. Glycobiology 31, 1018–1025. doi: 10.1093/glycob/cwab019
Debarnot, C., Monneau, Y. R., Roig-Zamboni, V., Delauzun, V., Le Narvor, C., Richard, E., et al. (2019). Substrate binding mode and catalytic mechanism of human heparan sulfate d-glucuronyl C5 epimerase. Proc. Natl. Acad. Sci. U.S.A. 116, 6760–6765. doi: 10.1073/pnas.1818333116
El Masri, R., Seffouh, A., Lortat-Jacob, H., and Vives, R. R. (2017). The “in and out” of glucosamine 6-O-sulfation: the 6th sense of heparan sulfate. Glycoconj. J. 34, 285–298. doi: 10.1007/s10719-016-9736-5
Fu, L., Suflita, M., and Linhardt, R. J. (2016). Bioengineered heparins and heparan sulfates. Adv. Drug Deliv. Rev. 97, 237–249. doi: 10.1016/j.addr.2015.11.002
Gomari, M. M., Afshari, S. A., Farsimadan, F., Rostami, N., Aghamiri, S., and Farajollahi, M. M. (2020). Opportunities and challenges of the tag-assisted protein purification techniques: applications in the pharmaceutical industry. Biotechnol. Adv. 45:107653. doi: 10.1016/j.biotechadv.2020.107653
Grigorieva, E. V., Prudnikova, T. Y., Domanitskaya, N. V., Mostovich, L. A., Pavlova, T. V., Kashuba, V. I., et al. (2011). D-glucuronyl C5-epimerase suppresses small-cell lung cancer cell proliferation in vitro and tumour growth in vivo. Br. J. Cancer 105, 74–82. doi: 10.1038/bjc.2011.170
Li, K., Bethea, H. N., and Liu, J. (2010). Using engineered 2-O-sulfotransferase to determine the activity of heparan Sulfate C5-epimerase and its mutants. J. Biol. Chem. 285, 11106–11113. doi: 10.1074/jbc.M109.081059
Li, J. P., Gong, F., Asa, H. M., Erik, F., Magnus, A., Robert, K., et al. (2003). Targeted disruption of a murine glucuronyl C5-epimerase gene results in heparan sulfate lacking L-iduronic acid and in neonatal lethality. J. Biol. Chem. 278, 28363–28366. doi: 10.1074/jbc.C300219200
Liu, N. N., Duan, X. L., Ai, X., Yang, Y. T., Li, M., Dou, S. X., et al. (2015). The Bacteroides sp. 3_1_23 Pif1 protein is a multifunctional helicase. Nucleic Acids Res. 43, 8942–8954. doi: 10.1093/nar/gkv916
Liu, H., Zhang, Z., and Linhardt, R. J. (2009). Lessons learned from the contamination of heparin. Nat. Prod. Rep. 26, 313–321. doi: 10.1039/b819896a
Marques, C., Reis, C. A., Vivès, R. R., and Magalhães, A. (2021). Heparan sulfate biosynthesis and sulfation profiles as modulators of cancer signalling and progression. Front. Oncol. 11:778752. doi: 10.3389/fonc.2021.778752
Nazir, A., Shad, M., and Rehman, N. S. M. (2024). Application of sumo fusion technology for the enhancement of stability and activity of lysophospholipase from pyrococcus abyssi. World J. Microbiol. Biotechnol. 40:183. doi: 10.1007/s11274-024-03998-w
Ori, A., Wilkinson, M. C., and Fernig, D. G. (2008). The heparanome and regulation of cell function: structures, functions and challenges. Front. Biosci. 13, 4309–4338. doi: 10.2741/3007
Préchoux, A., Halimi, C., Simorre, J. P., Lortat-Jacob, H., and Laguri, C. (2015). C5-epimerase and 2-O-sulfotransferase associate in vitro to generate contiguous epimerized and 2-O-sulfated heparan sulfate domains. ACS Chem. Biol. 10, 1064–1071. doi: 10.1021/cb501037a
Prudnikova, T. Y., Mostovich, L. A., Kashuba, V. I., Ernberg, I., Zabarovsky, E. R., and Grigorieva, E. V. (2014). miRNA-218 contributes to the regulation of D-glucuronyl C5-epimerase expression in normal and tumor breast tissues. Epigenetics 7, 1109–1114. doi: 10.4161/epi.22103
Qin, Y., Ke, J., Gu, X., Fang, J., Wang, W., Cong, Q., et al. (2015). Structural and functional study of d-glucuronyl C5-epimerase. J. Biol. Chem. 290, 4620–4630. doi: 10.1074/jbc.M114.602201
Raedts, J., Lundgren, M., Kengen, S. W. M., Li, J. P., and van der Oost, J. (2013). A novel bacterial enzyme with d-glucuronyl C5-epimerase activity. J. Biol. Chem. 288, 24332–24339. doi: 10.1074/jbc.M113.476440
Tarja, K., Zebo, H., Joanne, T., Gatdula, M. M., Brown, J. R., Esko, J. D., et al. (2005). Heparan 2-O-sulfotransferase, hst-2, is essential for normal cell migration in Caenorhabditis elegans. Proc. Natl. Acad. Sci. U.S.A. 102, 1507–1512. doi: 10.1073/pnas.0401591102
Vaidyanathan, D., Paskaleva, E., Vargason, T., Ke, X., McCallum, S. A., Linhardt, R. J., et al. (2020). Elucidating the unusual reaction kinetics of D-glucuronyl C5-epimerase. Glycobiology 30, 847–858. doi: 10.1093/glycob/cwaa035
Wen, J., Yi, L., Wan, L., and Dong, X. (2023). Prognostic value of GLCE and infiltrating immune cells in Ewing sarcoma. Heliyon 9:e19357. doi: 10.1016/j.heliyon.2023.e19357
Xiao, W., Chen, C., Xia, S., Li, Z., Ding, T., Zhou, J., et al. (2024). Cell-surface d-glucuronyl C5-epimerase binds to porcine deltacoronavirus spike protein facilitating viral entry. J. Virol. 98:e0088024. doi: 10.1128/jvi.00880-24
Zhou, X. F., Zhang, C. L., Gao, X. P., Wang, W. L., He, Z. F., Jiang, F. Y., et al. (2020). A simple and rapid protein purification method based on cell-surface display of SUMO-fused recombinant protein and Ulp1 protease. AMB Express 10:65. doi: 10.1186/s13568-020-00999-4
Keywords: D-glucuronyl C5-epimerase, heparan sulfate biosynthesis, Escherichia coli protein expression, two-step protein purification, dimer structure
Citation: Song Q-x and Guo L-j (2025) A high effective expression of human D-glucuronyl C5-epimerase with dimer structure in Escherichia coli. Front. Microbiol. 16:1641598. doi: 10.3389/fmicb.2025.1641598
Edited by:
Deniz Yildirim, Çukurova University, TürkiyeCopyright © 2025 Song and Guo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qin-xia Song, c29uZ3F4QG53YWZ1LmVkdS5jbg==
 Qin-xia Song
Qin-xia Song Li-jian Guo2
Li-jian Guo2