- 1Institute of Vegetable, Hangzhou Academy of Agricultural Sciences, Hangzhou, China
- 2Ningbo Jiangbei District Agricultural Technology Extension Service Station, Ningbo, China
- 3Department of Biology, College of Science, United Arab Emirates University, Abu Dhabi, United Arab Emirates
- 4State Key Laboratory of Rice Biology and Breeding, Ministry of Agriculture Key Laboratory of Molecular Biology of Crop Pathogens and Insects, Zhejiang Key Laboratory of Biology and Ecological Regulation of Crop Pathogens and Insects, Institute of Biotechnology, Zhejiang University, Hangzhou, China
- 5Department of Life Sciences, Western Caspian University, Baku, Azerbaijan
Background: In order to ensure food security, China is actively carrying out conversion of nongrain cultivated land to paddy field. Therefore, it is very necessary to investigate the influence of this conversion on soil health, which has been well known to play an important role in crop growth.
Methods: A combined analysis of soil physicochemical properties, bacterial community structure, and metabolite was conducted on 72 soil samples, which were collected in this study from the converted paddy fields and the corresponding non-grain cultivated lands including loquat garden, mulberry field, blueberry garden, vineyard, bamboo garden and nursery stock base.
Results: In this study, conversion of non-grain cultivated land to paddy field significantly influenced physicochemical properties, bacterial community structure, and metabolite of root-zone soil with 8.08–43.85%, 8.90–64.14%, 24.98–91.97%, 38.74–92.52%, and 5.12–32.99% reduction in soil organic matter content (SOM), alkaline hydrolysis nitrogen (AHN), available phosphorus (AP), available potassium (AK), and microbial biomass carbon (MBC), respectively; 0.81–3.08 fold, 1.26–21.50 fold, and 4.29–14.54 fold increase in relative abundance (RAs) of Chloroflexi, Desulfobacterota, and Nitrospirota, respectively; and 2,204 differentially expressed metabolite (DEMs) belonging to amino acids and derivatives, benzene and substituted derivatives, flavonoids, lipids, organic acids, terpenoids. Furthermore, correlation analysis indicated that these DEMs were significantly correlated with some specific bacteria, thereby helping in coordinating the root-zone soil community during conversion, while these bacteria were also correlated with soil properties.
Conclusion: Overall, this study highlights the importance of bacterial communities during conversion of non-grain cultivated land to paddy field, which provided a scientific basis and supporting evidence for the renovation of non-grain cultivated land.
1 Background
Cultivated land is the major vehicle of food production, and demonstrates a central role in guaranteeing national food security, social stability and sustainable development (Lai et al., 2020; Lu et al., 2024). As the most populous country in the world, China feeds about 20% of the world population with just 7% of the world farmland (Cui and Shoemaker, 2018), thus China’s bumper grain plays a central role in maintaining global food security (Wu et al., 2023). However, due to the acceleration of urbanization, continuous upgrading of national food consumption demand from grain to pluralism, society’s blind pursuit of economic interests considering that comparative economic benefit gap between grain crops and economic forest fruits, and agricultural industrial policies, a significant tendency of using cultivated lands for non-grain production (such as bamboo shoots, perennial fruits, tea, vegetables, flowers) has widely occurred in recent years, in turn severely restricting local grain productivity, and leading to decline of cultivated land quantity and quality (Su et al., 2014; Yuan et al., 2019; Boulanger et al., 2022; Zhu et al., 2022; Chen et al., 2023; Deng et al., 2024).
To solve above-mentioned problems, the Chinese government has issued the “Opinions on Preventing the Non-Grain Production on Cultivated Land and Stabilizing Grain Production” on November 17, 2020, clearly understanding the profound urgency of preventing non-grain production on cultivated land and stabilizing grain production, earnestly grasping the national food security initiative, and establishing the reporting system of non-grain production on cultivated land so as to keep it strictly controlled (State Council of China, 2020). Since then, the Delta area of China has been actively carrying out conversion of non-grain cultivated land to paddy field, including consolidating non-grain paddy fields and restoring grain production functions to ensure food production, and improving the quality of cultivated land, which has significantly increased the scale of grain cultivation and thereby partly resolved issues caused by non-grainization (Shi et al., 2018; Liang and Geng, 2023). Therefore, it is of great significant to further solving non-grain related issues during the conversation of non-grain cultivated land to paddy field.
Over the past few decades, scholars have conducted lots of investigations from different perspectives on land use changes (Yu and Zhang, 2019; Meng et al., 2022; Hao et al., 2024). For example, different types of land use (agricultural, industrial, recreational, coastal, and residential areas) influenced soil physiochemical properties, the abundance of nitrifying bacteria, and microbial interactions in tropical urban soil (Das, 2023; Medriano et al., 2023). Land-use change from natural grasslands to shrub plantations, tree plantations, and arable lands altered patterns of soil biodiversity in arid lands of northwestern China (Li et al., 2018). The changes of typical six land-use types (forest, open forest, shrub, grassland, corn field and abandoned farmland) significantly affected soil phytolith-occluded organic carbon accumulation in Southwest China (Wang et al., 2023b). Conversion of upland crop to paddy field significantly changed soil water moisture and organic carbon contents, with increased bacterial diversity and changed bacterial community composition (Sun et al., 2021). Land use changes have been reported to be associated with the growth of different plant species, which shift the soil physiochemical properties and microbial community (Li et al., 2007; Jiang et al., 2016; Legrand et al., 2018; Lin et al., 2019; Sun et al., 2021). Farmland ecosystems exhibit distinct microclimates shaped by crops and land use changes by human intervention, which profoundly influence the composition and function of soil microbes (Deng et al., 2022; Agyekum et al., 2023).
Soil bacteria, the most prevalent microbes in soil, are vital for maintaining soil fertility and crop production by driving lots of soil ecosystem functions (including organic matter decomposition, humus formation, nutrients transformation, and suppression of soil-borne disease) (Ahmed et al., 2023; Chen et al., 2024). Simultaneously, soil nutrition gives rise to a vast diversity of soil bacteria (Wang X. et al., 2024), while soil metabolites strongly affect bacterial community structure and function (Bi et al., 2022). Obviously, these studies clearly revealed that soil bacteria exhibited important theoretical and practical implications for sustainable agricultural development by improving physiochemical properties and metabolites of soil. However, little attention has been paid on role of soil bacterial community structure and their ecological function during conversation of different types of non-grain production lands to paddy fields.
The objective of this study was to evaluate the impact of conversion of land use from non-grain cultivated land to paddy field on soil bacterial community structure, metabolite, and physicochemical properties by collecting soil samples from six types of non-grain cultivated land and the corresponding converted paddy fields, which will provide a scientific basis for the renovation of non-grain cultivated land, thus ensuring food security.
2 Methods
2.1 Soil sampling
On November 15, 2023, 72 soil samples (5–20 cm depth) were collected according to the method of (Butler et al., 2003) by mixing a total of nine random soil cores, which were picked up at the drip line around the crown of crops (including non-grain plants, and rice planted in the surrounding paddy fields converted from the corresponding non-grain lands), located in Jiande (loquat), Chun’an (mulberry), Tonglu (blueberry plant), Fuyang (grapevine), Lin’an (bamboo tree), and Yuhang (nursery stock), Hangzhou city (experiencing a subtropical monsoon climate with an average annual temperature of 17.8°C and precipitation of 1,454 mm), Zhejiang province, China (Table 1). Each treatment had six replicates. After passing through a 2 mm sieve, individual sample was partitioned into three: (a) air-dried at room temperature and passed through a 0.45 mm gauze for analysis of soil pH, soil organic matter (SOM), alkaline hydrolysis nitrogen (AHN), available phosphorus (AP), and available potassium (AK), (b) stored at 4°C for microbial biomass carbon (MBC) analysis, and (c) stored at −70°C for genome sequencing and metabolomic profiling analyses.
2.2 Analyses of soil characteristics and MBC
Air-dried soil samples were used for analysis of soil characteristics. In detail, soil pH was estimated within 1: 5 soil suspension (soil: water, w/v) via pH meter (FE28, Met tlerToledo, Zurich, Switzerland) (Rathje, 1959). SOM was determined using K2Cr2O7 oxidation heating method (Nelson and Sommers, 1996). AHN was measured by conductometric titration (Chen et al., 2016). AP and AK were extracted with ammonium lactate solution and then analyzed using spectrophotometry and flame photometry, respectively (Tian et al., 2021). In contrast, fresh soil samples were used to determine MBC, which was performed by employing chloroform fumigation-extraction method (Vance et al., 1987).
2.3 Soil genome sequencing
Following the extraction of DNAs using the E.Z.N.ATM Mag-Bind Soil DNA Kit (OMEGA, USA), soil bacterial diversity were determined by amplifing the V3–V4 region of the bacterial 16S rRNA gene using two universal primers 341F (5′-CCTACGGGNGGCWGCAG-3′) and 805R (5′-GACTACHVGGGTATCTAATCC-3′) (Wu et al., 2015), which was carried out as described by Li et al. (2024). The reaction mixture of PCR contained 2 × Hieff® Robust PCR Master Mix (15 μL), 10 μM primer 341F (1 μL), 10 μM primer 805R (1 μL), ddH2O (12 μL), and DNA (1 μL). PCR was run at 95°C for 3 min; 95°C for 30 s, 45°C for 30 s, 72°C for 30 s, 5 cycles; 95°C for 30 s, 55°C for 30 s, 72°C for 30 s, 20 cycles; 72°C for 5 min. The final amplicon was detected by 2% agarose gel, purified by Hieff NGS™ DNA selection beads (Yeasen, China), quantified using a Qubit 4.0 (Thermo, USA), and subsequently pair-end (2 × 250 bp) sequenced on an Illumina MiSeq platform (Sangon BioTech, Shanghai, China).
After the sequencing process, the primers were cut off using Cutadapt (v3.5) (Martin, 2011). The short Illumina reads were assembled adopting PEAR (v0.9.8), and then the reads with Phred33 score of less than 20 were removed via Trimmomatic (v0.39) to ensure data integrity (Bolger et al., 2014). After, raw reads were further filtered, denoised, and concatenated by DADA2 (v1.14.0) (Callahan et al., 2016), the chimera was then clustered into operational taxonomic units (OTUs) using Usearch (v11.0.667) with a 97% similarity cutoff. After selection of the representative read of each cluster using QIIME (v2020.06), taxonomic classification of each OTU was performed with Silva (v138.1) using the RDP classifier (v2.12) (Wang et al., 2007; Quast et al., 2012; Wang et al., 2023a).
2.4 Soil metabolomics assay
Metabolic assay was carried out as described by Li et al. (2024) using liquid chromatography-mass spectrometry (LC–MS) system (Vanquish, Thermo), in which LC coupled to an Orbitrap Exploris 120 mass spectrometer (Orbitrap MS, Thermo). To assess the quality and reproducibility of data, pooled quality control samples were included by adding equal amount of all sample supernatants. The original data obtained via LC–MS was changed into mzXML format by ProteoWizard. Peak extraction, peak alignment, and time retention correction were, respectively, performed by XCMS. The peak area was corrected by SVR, and the peaks with detection rate lower than 50% in each group of samples were discarded. Afterwards, metabolite annotation was executed against an in-house MS2 database (Sangon BioTech, Shanghai, China).
2.5 Statistical analysis
The comparative analysis were performed on each land type between the non-grain cultivated lands and the corresponding converted paddy fields, while the conversation of each land type from non-grain cultivated land to paddy field was arranged in a county, ensuring consistency and minimize variability across sampling locations. One-way analysis of variance (ANOVA) tests were adopted to analyze variance using SPSS (v16.0) (Chicago, USA). To assess bacterial abundance and α-diversity, the OTU richness and α-diversity indexes (including Chao1, Shannon, and Simpson indexes) were visualized via Origin (v2022) (Hampton, USA) after normalizing data by Usearch (v11) (California, USA). To assess changes in the bacterial community structure, principal component analysis (PCA) was performed using Bray–Curtis dissimilarity matrix (Ramette, 2007). The significant differences between groups were tested by permutational multivariate ANOVA (PERMANOVA), with 999 permutations used to calculate p-values (Dixon, 2003). In visualizing bacterial community composition, relative abundances (RAs) and heat maps of dominant bacteria taxa were conducted using Origin (v2022). To evaluate the influence of biomarkers on different groups, linear discriminant analysis (LDA) effect size (LEfSe) was carried out using LEfSe Galaxy based on the LDA score (Segata et al., 2011). To investigate how the conversion processes affect soil bacterial co-occurrence patterns, co-occurrence networks were constructed using a SparCC correlation matrix, based on RAs (>1%) and statistically significant correlations of RAs (p < 0.01, SparCC’s coefficient N > 0.22 or < −0.22) among OTUs, and visualized via Gephi (v0.9.2) (Friedman and Alm, 2012; Weiss et al., 2016; Gloor et al., 2017; Peschel et al., 2021; Yu et al., 2022). To explore the differences in metabolites under different groups, orthogonal projections to latent structures discriminant analysis (OPLS-DA), volcano plots, pathway enrichment analysis of differential metabolites were adopted with the MetaboAnalyst 4.0 platform. To gain a better understanding of the potential association between differentially expressed metabolites (DEMs) and bacteria, the correlation heat maps were clustered as described by Hollander et al. (2015) based on the Spearman’s rank correlation coefficient among the top 20 RA of root-zone soil bacteria and significant DEMs with largest variable importance in projection (VIP). Meanwhile, redundancy discriminant analysis (RDA) was performed to investigate the impact of different environmental factors (such as pH and nutrition) on microbial community structure by Origin (v2022, Hampton, MA, USA).
3 Results
3.1 Impacts on soil pH and chemical properties
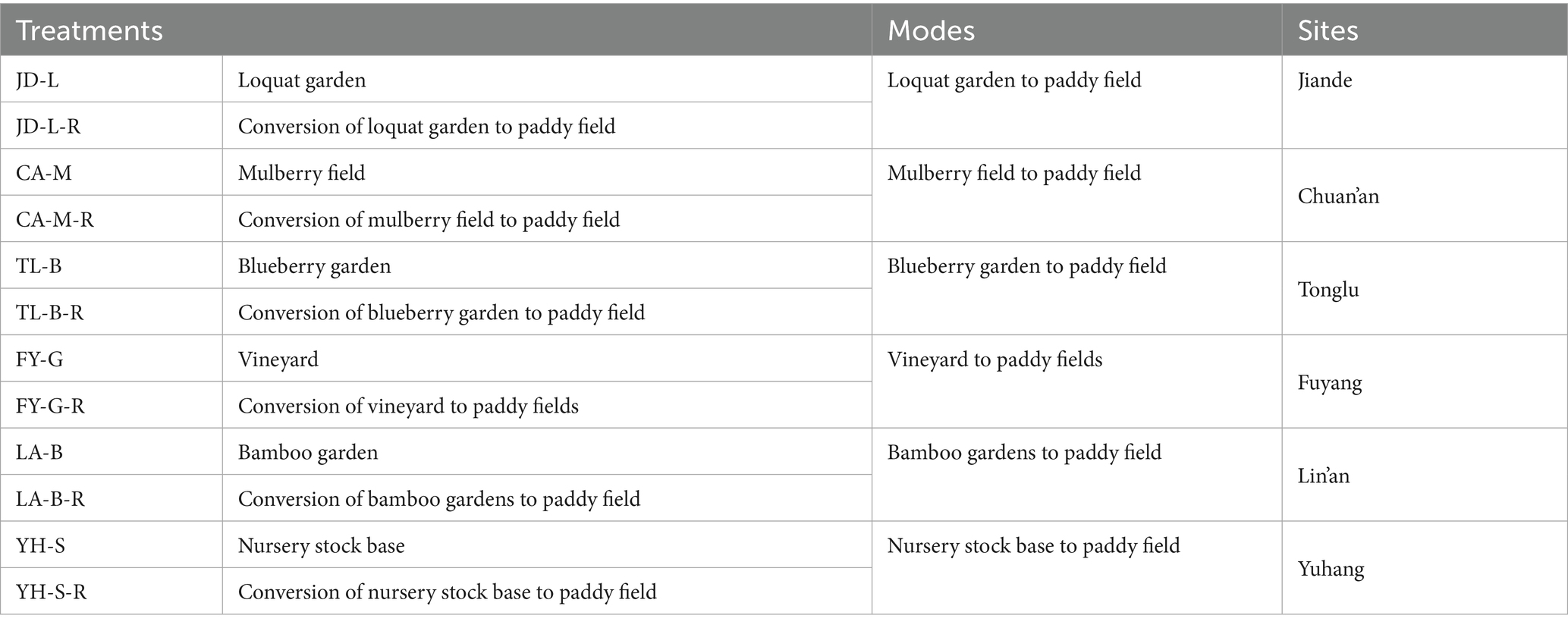
Results from this study showed that soil physicochemical properties were differentially affected by conversion of six non-grain cultivated lands to paddy field, while the effect depends on both the type of non-grain cultivated lands and the kind of soil parameters. Indeed, the soil pH was significantly (p < 0.05) reduced by conversion of loquat garden, mulberry field, blueberry garden to paddy field (5.30–16.97%), but increased by conversion of vineyard, bamboo garden and nursery stock base to paddy field (10.04–22.62%). However, conversion of six non-grain cultivated lands to paddy field significantly (p < 0.05) decreased the SOM (8.08–43.85%), AHN (8.90–64.14%), AP (24.98–91.97%), AK (38.74–92.52%), and MBC (5.12–32.99%), except a slight increase in the SOM (0.96%) and AHN (1.63%) by conversion of blueberry garden to paddy field, a significant (p < 0.05) increase in the AP (372.11%) and AK (25.50%) by conversion of nursery stock base to paddy field, and in the MBC (10.20%) by conversion of vineyard to paddy field (Figure 1; Table 2).
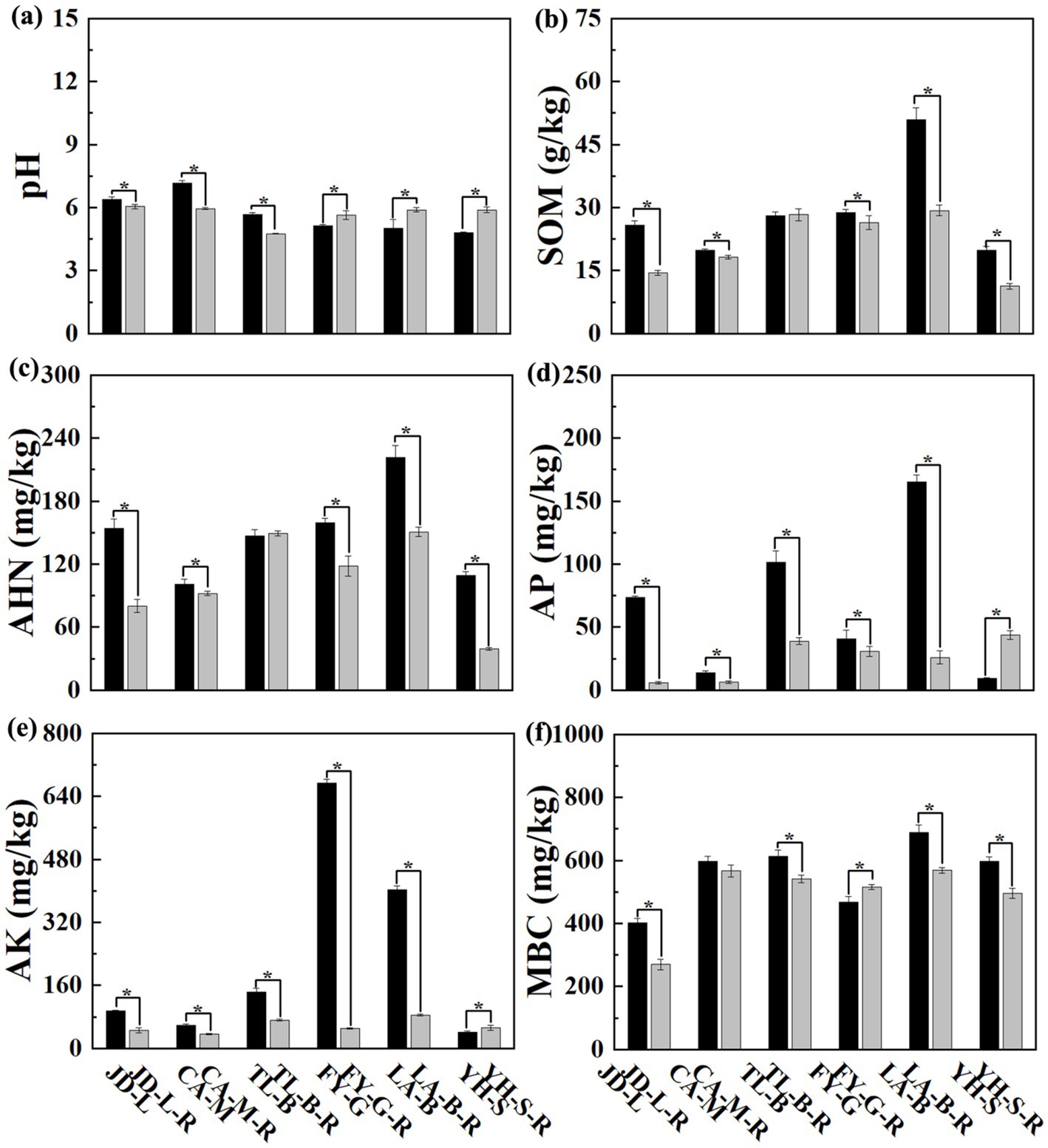
Figure 1. The pH and chemical properties of the root-zone soil between non-grain cultivated land and paddy field under different groups.
3.2 Impacts on soil bacterial community characteristics
3.2.1 Soil bacterial community diversity
In six diverse conversion modes from non-grain cultivated lands to paddy field (Figure 2a), a total of 363,752 OTUs from 21 bacterial phyla were identified, while the distribution of OTUs across all treatments was shown in Figure 2b. Generally, the conversion of non-grain cultivated land to paddy field significantly changed the richness and diversity of bacteria in the root-zone soils. In detail, the number of bacterial OTUs in the converted paddy fields was increased by 5.67–45.26% except a significant 17.64% reduction by conversion of blueberry garden to paddy field. The α-diversity analysis was chosen to evaluate the bacterial community (Figures 2c,d), while the trend of bacterial Chao1 index was basically same as OTUs, with increases of 3.62–45.22% in the converted paddy fields, and a significant reduction of 15.48% by conversion of blueberry garden to paddy field. Whereas, the Shannon index was increased by 3.17–16.27% by conversion of mulberry field, vineyard and bamboo garden to paddy fields, but decreased by 0.82–10.11% by conversion of the other three non-grain cultivated land to paddy field.
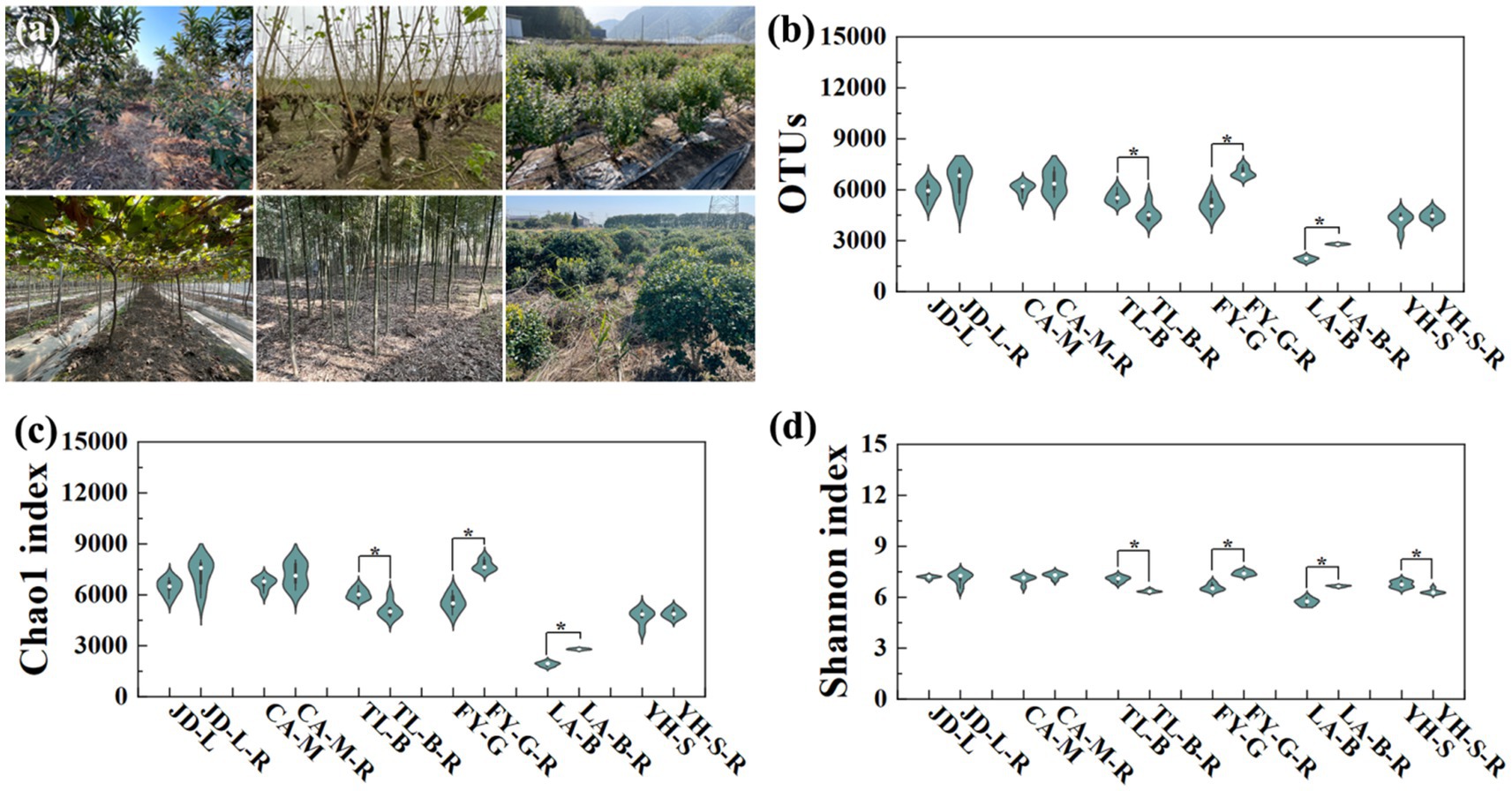
Figure 2. Six diverse conversion modes from non-grain cultivated lands to paddy field (a), and the bacterial OTU distribution (b), Chao1 (c), and Shannon (d) index under different groups. “*” above columns indicate statistical significant differences (p < 0.05). A total of 363,752 OTUs (5,079–7,294 for Jiande, 5,454–7,333 for Chun’an, 4,138–6,135 for Tonglu, 4,399–7,486 for Fuyang, 1,750–2,853 for Lin’an, and 3,339–4,899 for Yuhang) from 21 bacterial phyla were identified.
To further examine the effect of conversion of non-grain cultivated land to paddy field on root-zone soil bacterial communities, the PCA analysis at the OTU level was carried out based on the Bray-Curtis distance (Figure 3). Results from this study indicated that the conversion of non-grain cultivated land to paddy field significantly changed the bacterial community structure of the root-zone soil, while the effect depends on the type of conversion. Indeed, the root-zone soil bacterial communities of paddy fields converted from six non-grain cultivated lands formed two significantly different groups, and all groups were well separated from each other. Furthermore, the PCA1 and PCA2 revealed 49.45–68.56% and 18.67–39.51% of the variability in the bacterial communities, respectively, while the results of PERMANOVA indicated that the type of non-grain cultivated land explained 99.3–100% of the variation (p = 0.002–0.006).
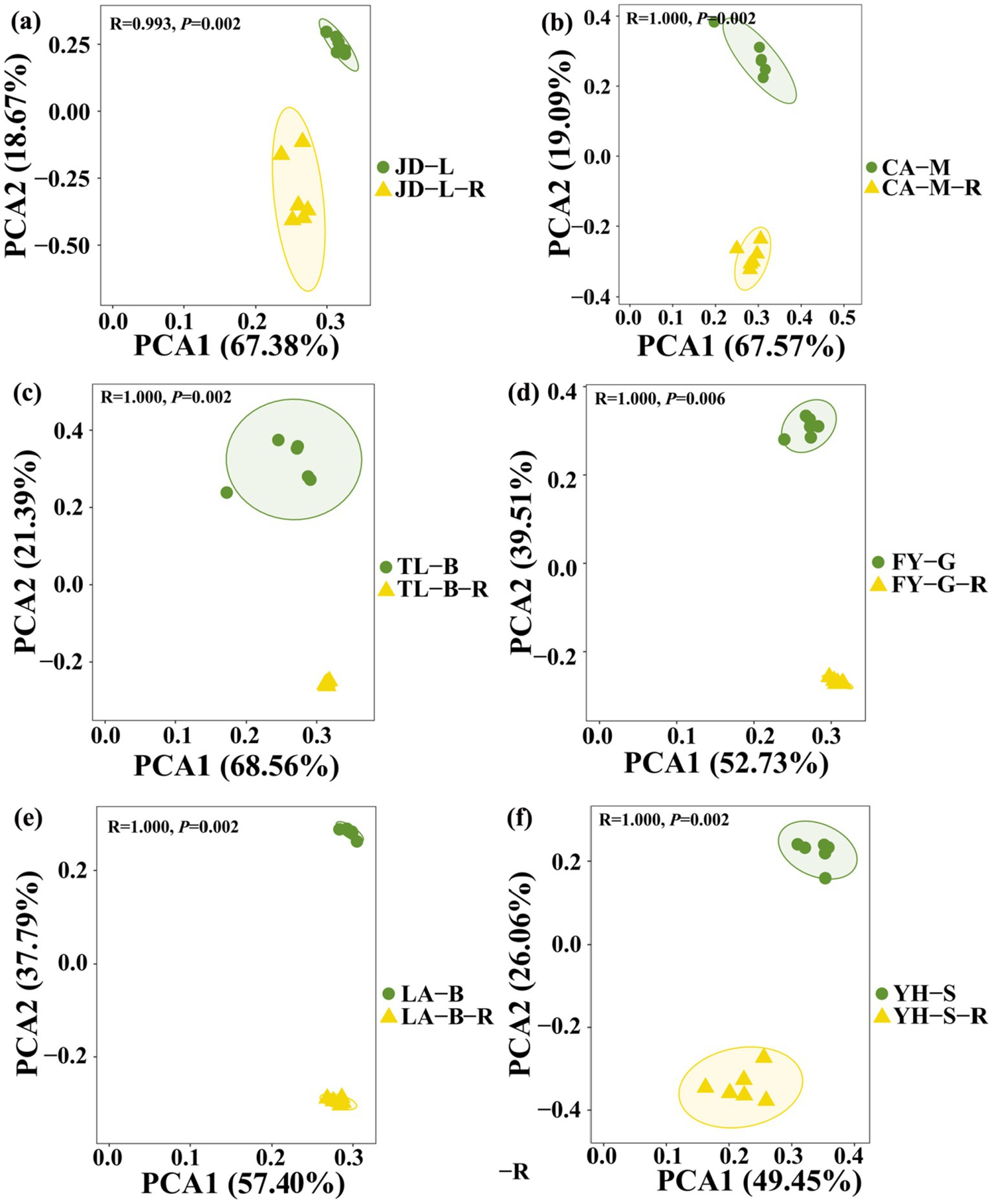
Figure 3. Principal component analysis (PCA) of the soil root-zone bacterial communities at the OTU level under different groups. The conversion of loquat garden to paddy field (a), conversion of mulberry field to paddy field (b), conversion of blueberry garden to paddy field (c), conversion of vineyard to paddy field (d), conversion of bamboo garden to paddy field (e), and conversion of nursery stock base to paddy field (f). Ellipses have been drawn for each treatment with a confidence limit of 0.95.
3.2.2 Soil bacterial community structure
Results indicated that conversion of non-grain cultivated land to paddy field led to significant changes in the bacterial community composition of the root-zone soil at the phylum level, while the relative abundance of the top 10 bacterial phyla was noted across all soil samples (Figure 4). In details, conversion of loquat garden to paddy field significantly increased Desulfobacterota (12.43 fold), Nitrospirota (4.29 fold), Chloroflexi (1.97 fold), and Myxococcota (0.92 fold), but significantly decreased Actinobacteriota (0.62 fold) (Figure 4a). Conversion of mulberry field to paddy field significantly increased Patescibacteria (1.84 fold), Desulfobacterota (1.26 fold), and Chloroflexi (0.81 fold) (Figure 4b). Conversion of blueberry garden to paddy field significantly increased Desulfobacterota (15.00 fold) and Acidobacteriota (0.55 fold), but significantly decreased Firmicutes (0.85 fold) and Bacteroidota (0.50 fold) (Figure 4c). Conversion of vineyard to paddy field significantly increased Desulfobacterota (21.50 fold), Nitrospirota (14.54 fold), Chloroflexi (2.41 fold), Verrucomicrobiota (0.97 fold) and Planctomycetota (0.82 fold), but significantly decreased Actinobacteriota (0.68 fold) and Proteobacteria (0.58 fold) (Figure 4d). Conversion of bamboo garden to paddy field significantly increased Nitrospirota (7.53 fold), Chloroflexi (3.08 fold), and Myxococcota (2.55 fold), but significantly decreased Actinobacteriota (0.53 fold) (Figure 4e). Conversion of nursery stock base to paddy field significantly increased Desulfobacterota (4.33 fold), Bacteroidota (2.35 fold), Chloroflexi (1.48 fold), and Actinobacteriota (0.69 fold) (Figure 4f).
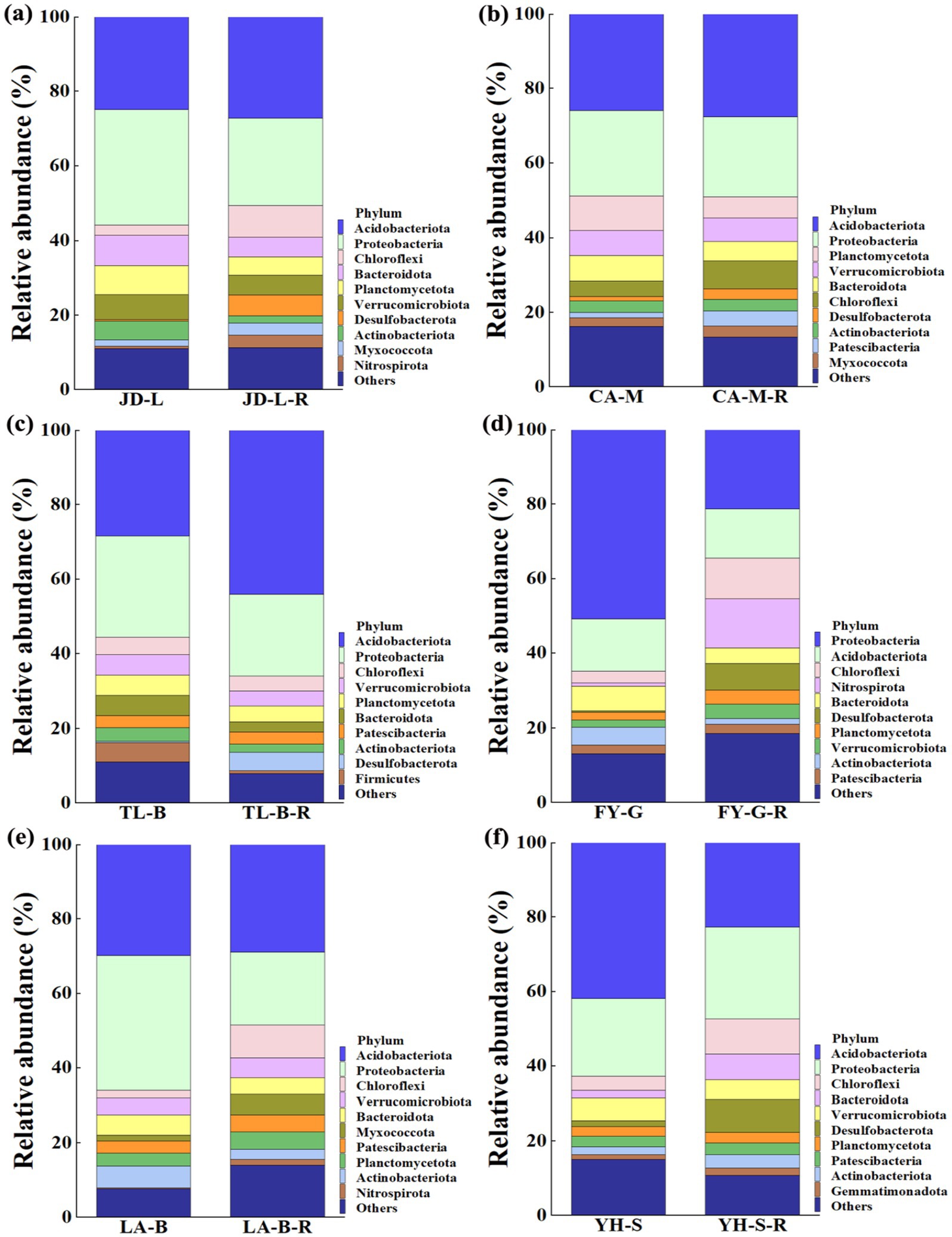
Figure 4. Relative abundance (RA) of the top 10 dominant bacteria at the phlyum level under different groups. The conversion of loquat garden to paddy field (a), conversion of mulberry field to paddy field (b), conversion of blueberry garden to paddy field (c), conversion of vineyard to paddy field (d), conversion of bamboo garden to paddy field (e), and conversion of nursery stock base to paddy field (f).
Furthermore, the difference in relative abundance composition of soil bacterial community among all treatments at the family level was further visually illustrated through heat maps (Figure 5). In detail, the converted paddy fields were enriched with Anaerolineaceae, Bryobacteraceae, Comamonadaceae, Gallionellaceae, Geobacteraceae, Haliangiaceae, Hydrogenophilaceae, Koribacteraceae, Ktedonobacteraceae, MBNT15, Nitrosomonadaceae, Pedosphaeraceae, Solibacteraceae, Subgroup_7, Subgroup_18, Sva0485, 4–29-1, Thermodesulfovibrionia, and WD2101, but were reduced with Acetobacteraceae, Chthoniobacteraceae, Elsterales, Gammaproteobacteria, Gemmatimonadaceae, KF-JG30-C25, Pirellulaceae, Rhodanobacteraceae, Sphingomonadaceae, Subgroup_2, and Vicinamibacteraceae. The results demonstrate that rice cultivation can increase or reduce the presence of certain species, accounting in a change in the root-zone soil bacterial community structure. Notably, these bacteria may have significant potential in colonizing and altering soil bacterial communities during conversion of non-grain cultivated land to paddy field.
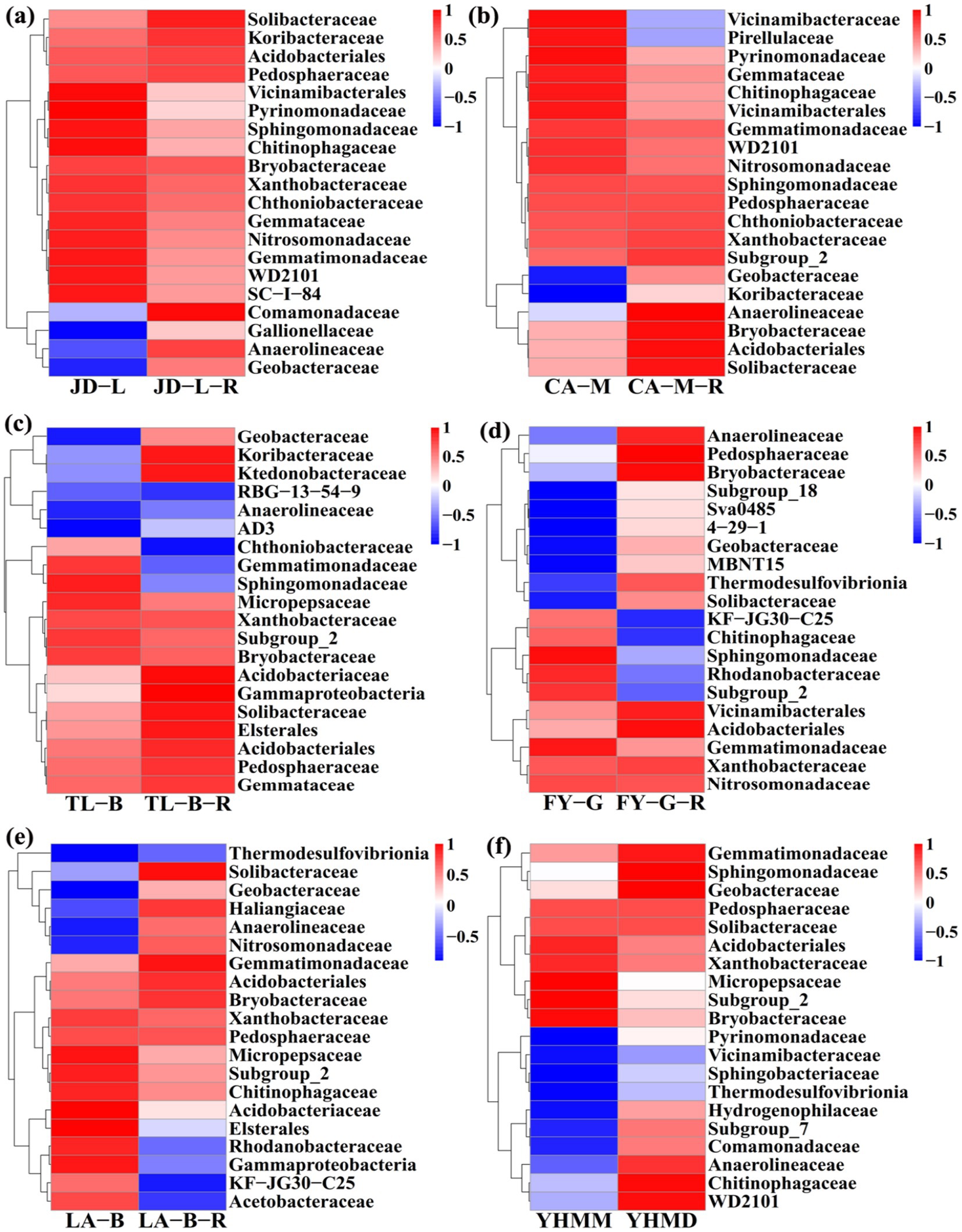
Figure 5. Heat map at the family level. The conversion of loquat garden to paddy field (a), conversion of mulberry field to paddy field (b), conversion of blueberry garden to paddy field (c), conversion of vineyard to paddy field (d), conversion of bamboo garden to paddy field (e), and conversion of nursery stock base to paddy field (f). The tree plot represents a clustering analysis of the top 20 bacteria at the family level according to their Person correlation coefficient matrix and relative abundance.
3.2.3 Soil microbiome and biomarker
LEfSe was performed to identify the biomarkers with the most difference in the root-zone soil bacterial communities between non-grain cultivated land and the converted paddy fields (Figure 6). Results showed that a total of 48 bacteria biomarkers (LDA > 4.5, p < 0.05) were found in all groups. Indeed, the loquat garden was enriched with Alphaproteobacteria, Proteobacteria, Sphingomonadaceae, and Sphingomonadales; the paddy field converted from mulberry field was enriched with Acidobacteriales and Acidobacteriae; the paddy field converted from blueberry field was enriched with three types of Acidobacteriales, Acidobacteriae, Acidobacteriota, Candidatus_Solibacter, Solibacteraceae, and Solibacterales; the vineyard was enriched with Anaerolineae, Chloroflexi, Desulfobacterota, four types of Thermodesulfovibrionia, and Nitrospirota, while the converted paddy field was enriched with Alphaproteobacteria, Chujaibacter, Gammaproteobacteria, three types of KF_JG30_C25, Proteobacteria, Rhodanobacteraceae, and Xanthomonadales; the bamboo garden was enriched with Acidobacteriaceae, Alphaproteobacteria, Gammaproteobacteria, and Proteobacteria, while the converted paddy field was enriched with Chloroflexi; the nursery stock base was enriched with three types of Acidobacteriales, Acidobacteriae, Acidobacteriota, Anaerolineae, and three types of Subgroup_2, while the converted paddy field was enriched with Burkholderiales, Desulfobacterota, and Gammaproteobacteria. As stated above, 25 and 23 bacteria biomarkers were associated with six kinds of non-grain cultivated land and the converted paddy fields, respectively.
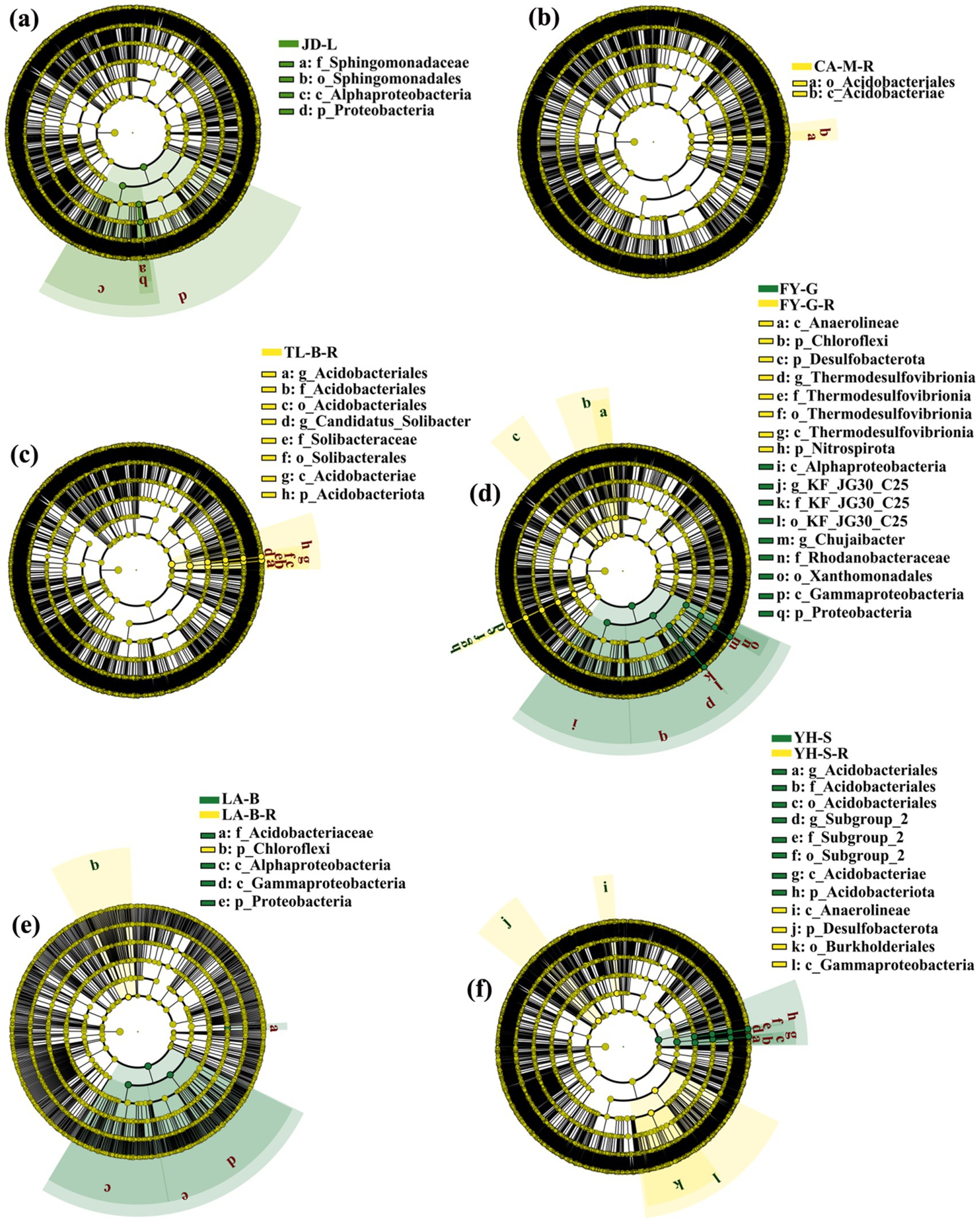
Figure 6. Linear discriminant analysis (LDA) effect size (LEfSe) of the root-zone soil bacterial taxa. The conversion of loquat garden to paddy field (a), conversion of mulberry field to paddy field (b), conversion of blueberry garden to paddy field (c), conversion of vineyard to paddy field (d), conversion of bamboo garden to paddy field (e), and conversion of nursery stock base to paddy field (f).
3.2.4 Co-occurrence networks of root-zone soil bacteria
To visualize the complexity and stability of soil bacterial community responses to conversion of non-grain cultivated land to paddy field in each group, co-occurrence networks were built, and then the topological properties were estimated to characterize differences between different groups (Figure 7; Table 3). Nodes represent microbes derived from OTUs, edges (positive/negative links between nodes) correspond to potential associations between nodes, while modularity indicates the presence of dense cluster of related nodes embedded within the network. In other word, fewer nodes or edges signify a less interconnected community and higher modularity represent higher structural stability of network (Freundt, 2021; Ma et al., 2021; Yang et al., 2022). Results showed that the nodes, edges and average degree was decreased by 32.17, 54.46, and 32.86%, respectively, in the paddy fields converted from mulberry field, decreased by 71.64, 76.47, and 17.00%, respectively, in the paddy fields converted from blueberry garden, increased by 1.23% and decreased by 8.00 and 9.13%, respectively, in the paddy fields converted from vineyard, decreased by 69.82, 84.36, and 48.19%, respectively, in the paddy fields converted from bamboo garden, increased by 29.63, 45.10, and 11.99%, respectively, in the paddy fields converted from loquat garden, increased by 48.67, 68.18, and 13.10%, respectively, in the paddy fields converted from nursery stock base. Whereas conversion of non-grain cultivated land to paddy field resulted in 4.38–65.17% increase in the modularity, suggesting that the bacterial community structure became more stable in the converted paddy field.
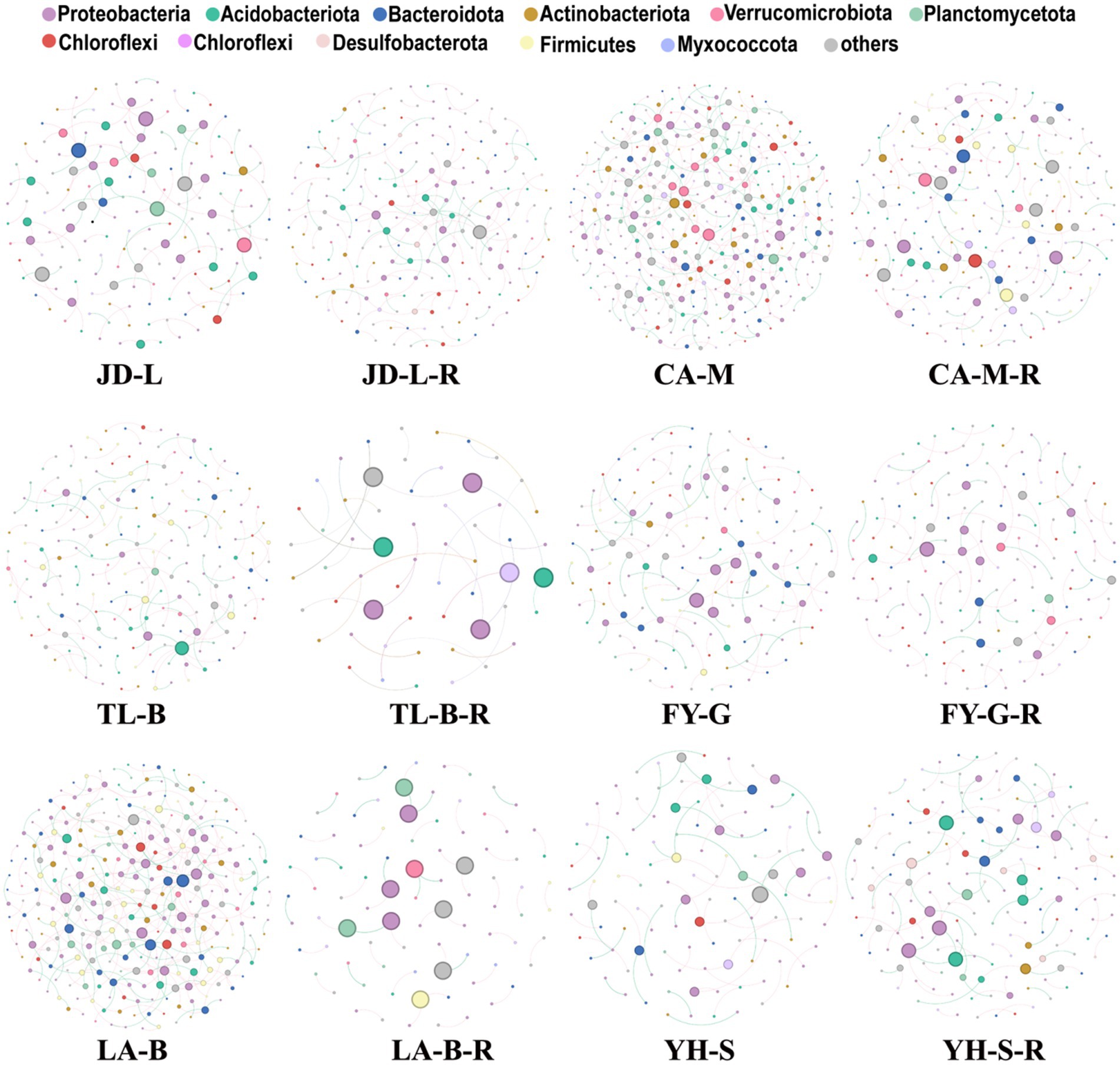
Figure 7. The differences on the co-occurrence patterns of soil bacterial communities. Networks were constructed at the OTU level. The size of the nodes (OTUs) represented the relative abundance (RA) of the bacteria, and the nodes were colored according to the phylum.
3.3 Soil metabolomics
To analyze and compare the changes in the soil metabolites in the paddy fields converted from non-grain cultivated lands, LC–MS analysis was performed, while OPLS-DA was used to construct a score map of metabolites to identify variables differed between different treatments. Results revealed that the distribution of soil metabolites could be effectively distinguished between the non-grain cultivated lands and the converted paddy fields, as reflected by the sample distributions of each group in the positive and negative directions of t[1], with corresponding model values of R2X (cum) = 0.446–0.648, R2Y (cum) = 0.999–1.000, and Q2 (cum) = 0.951–0.991. Furthermore, this inference could also be confirmed by volcano plot (based on VIP > 1 and p < 0.05) (Figures 8–13). A total of 5,827 metabolites were obtained from all groups, which mainly referred to amino acids and derivatives (9.76–13.78%), benzene and substituted derivatives (9.27–14.14%), flavonoids (3.45–8.54%), lipids (3.02–9.22%), organic acids (12.12–16.30%), terpenoids (4.27–9.58%), and so on. Furthermore, enrichment analysis of the KEGG pathway indicated that these DEMs might be associated with ABC transporters, biosynthesis of cofactors, biosynthesis of secondary metabolites, fructose and mannose metabolism, metabolic pathways, microbial metabolism in diverse environments, nucleotide metabolism, phosphotransferase system, purine metabolism, starch and sucrose metabolism, consequently playing a significant role during conversation of non-grain cultivated land to paddy field.
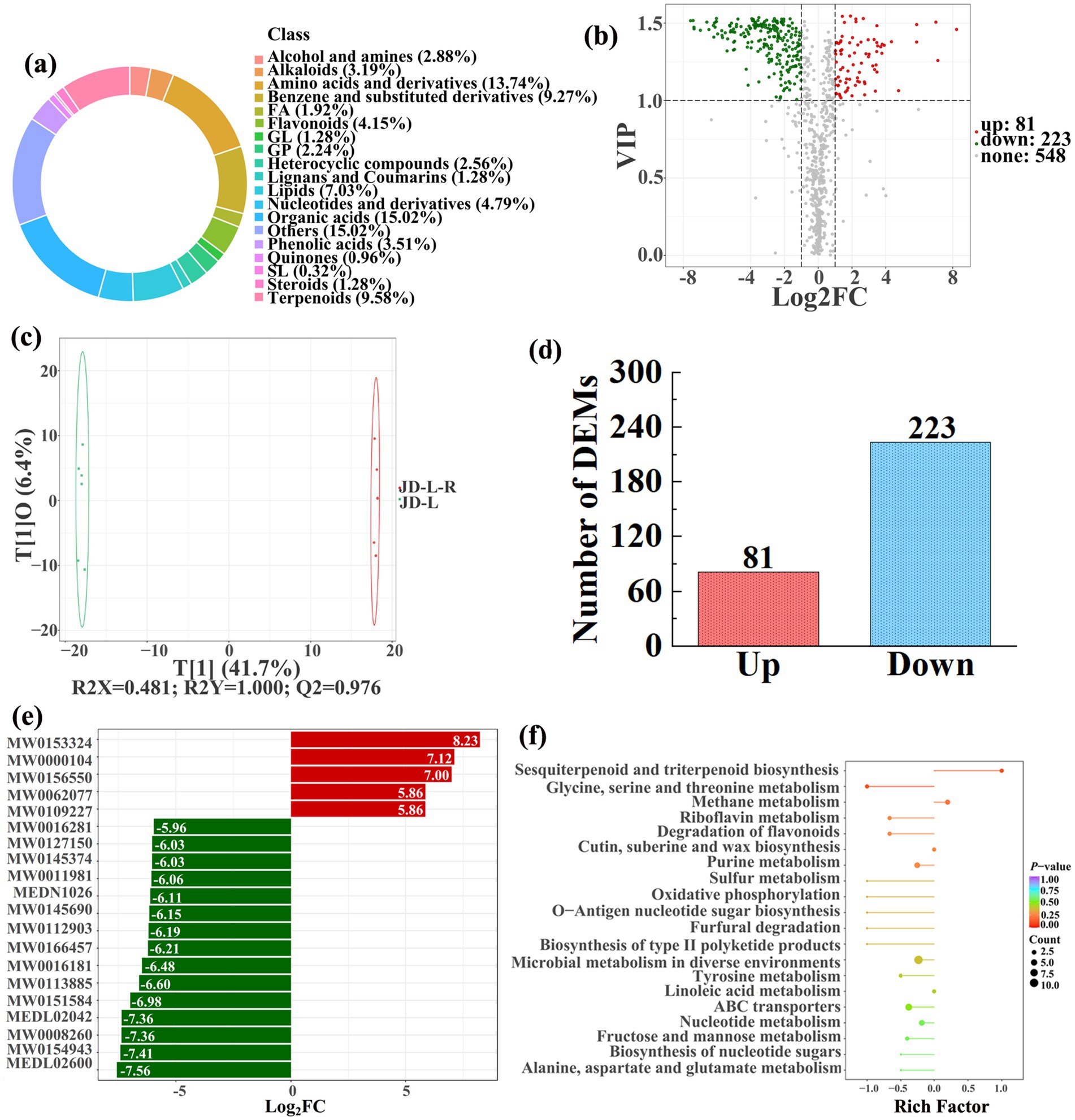
Figure 8. Donut plots showing metabolite classification and proportion (a), volcano plot (b), orthogonal projection to latent structures-discriminant analysis (OPLS-DA) score map (c), the number of DEMs (d), top 20 DEMs with largest VIP (e), KEGG enrichment analysis of differential soil metabolites in conversion of loquat garden to paddy field (JD-L-R) vs loquat garden (JD-L) (f).
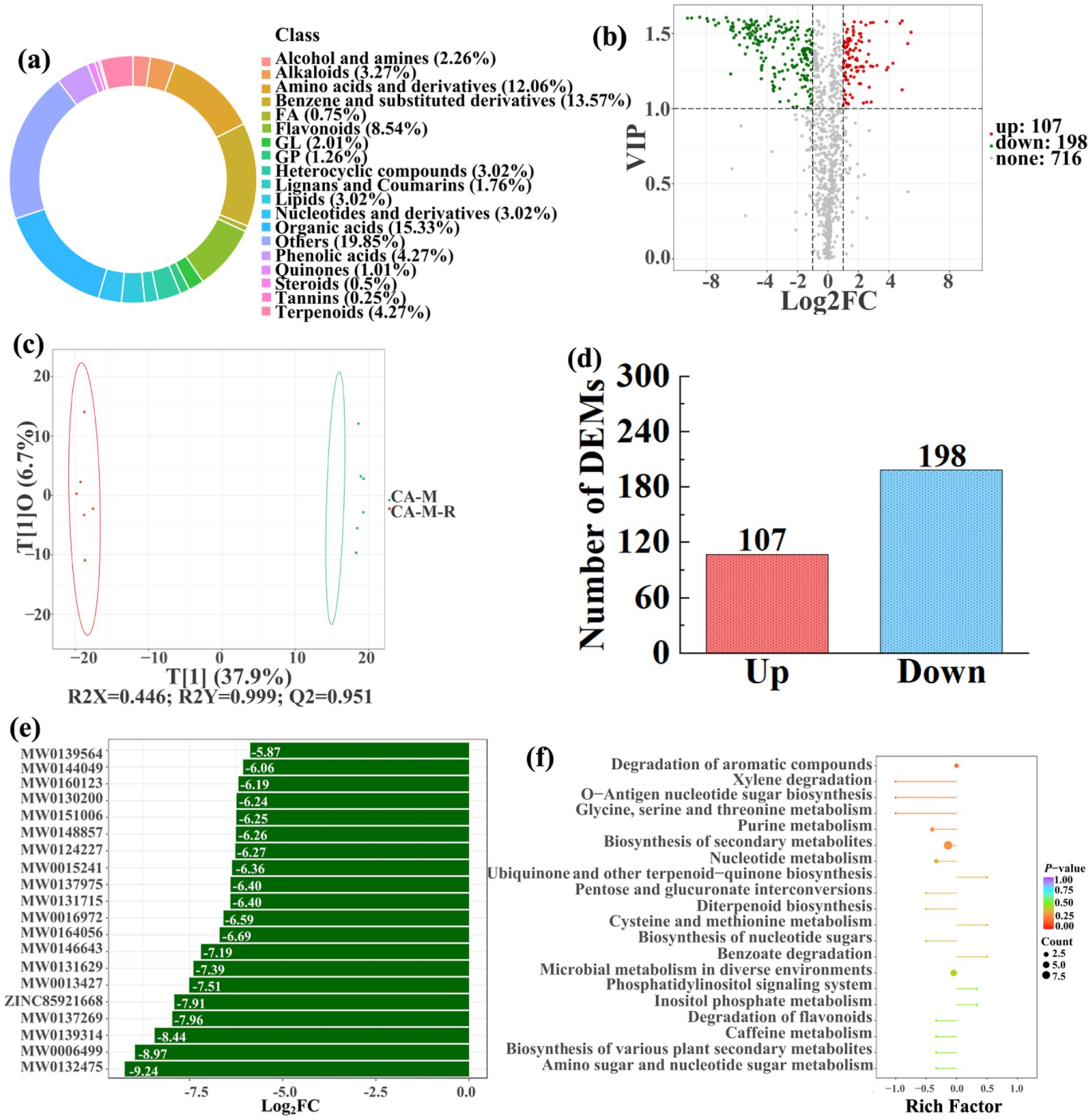
Figure 9. Donut plots showing metabolite classification and proportion (a), volcano plot (b), orthogonal projection to latent structures-discriminant analysis (OPLS-DA) score map (c), the number of DEMs (d), top 20 DEMs with largest VIP (e), KEGG enrichment analysis of differential soil metabolites in conversion of mulberry field to paddy field (CA-M-R) vs mulberry field (CA-M) (f).
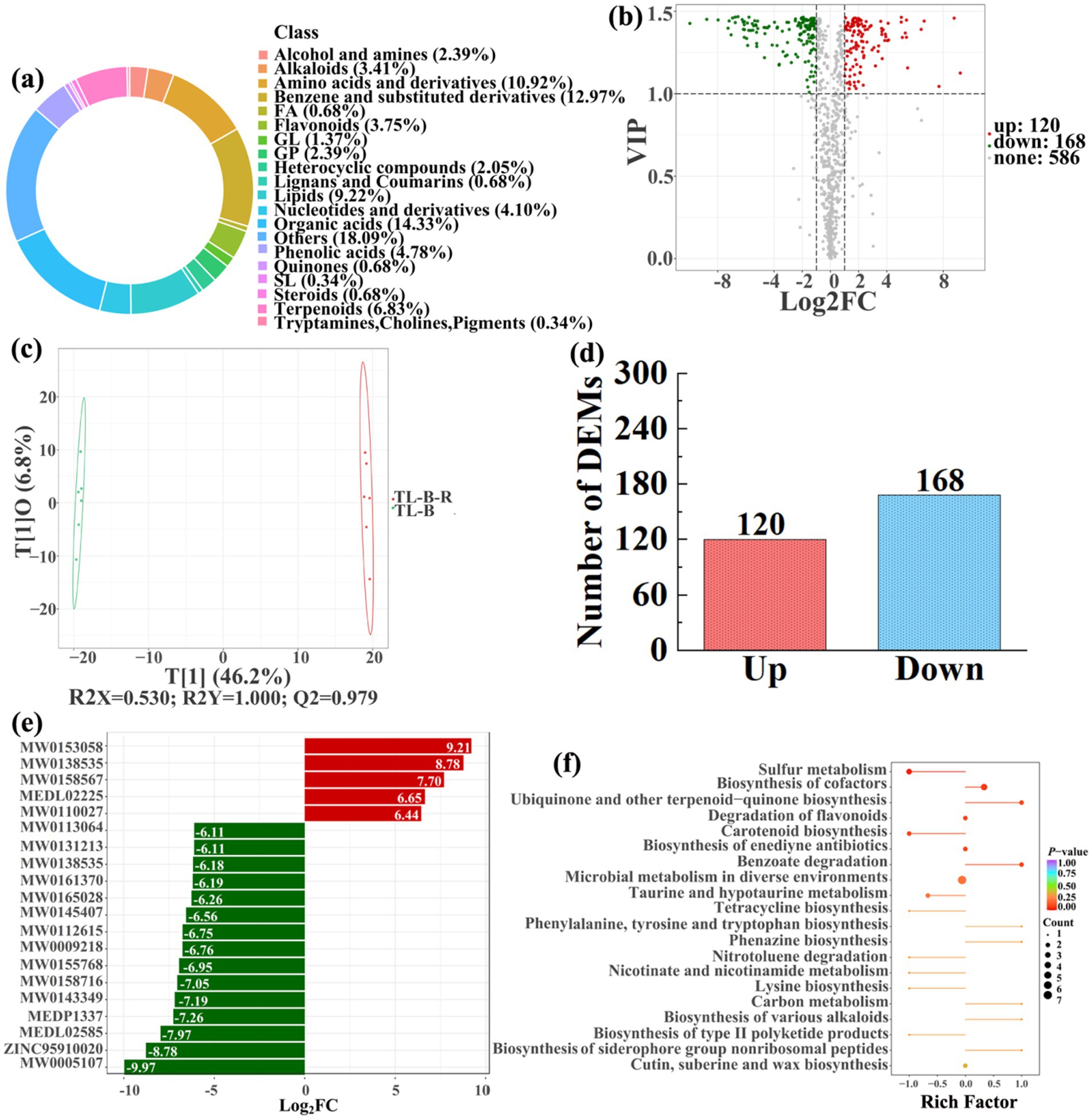
Figure 10. Donut plots showing metabolite classification and proportion (a), volcano plot (b), orthogonal projection to latent structures-discriminant analysis (OPLS-DA) score map (c), the number of DEMs (d), top 20 DEMs with largest VIP (e), KEGG enrichment analysis of differential soil metabolites in conversion of blueberry garden to paddy field (TL-B-R) vs blueberry garden (TL-B) (f).
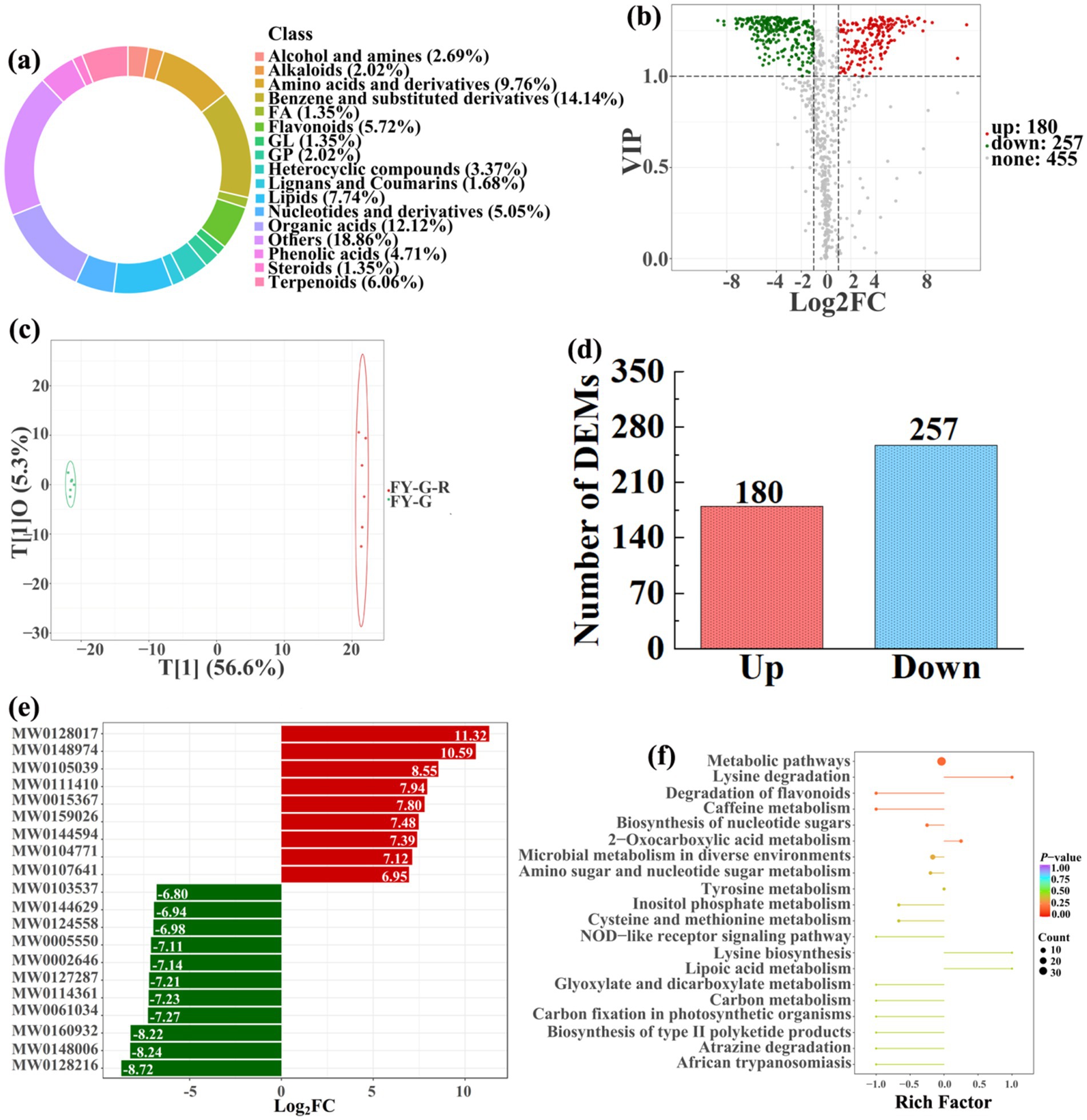
Figure 11. Donut plots showing metabolite classification and proportion (a), volcano plot (b), orthogonal projection to latent structures-discriminant analysis (OPLS-DA) score map (c), the number of DEMs (d), top 20 DEMs with largest VIP (e), KEGG enrichment analysis of differential soil metabolites in conversion of vineyard to paddy field (FY-G-R) vs vineyard (FY-G) (f).
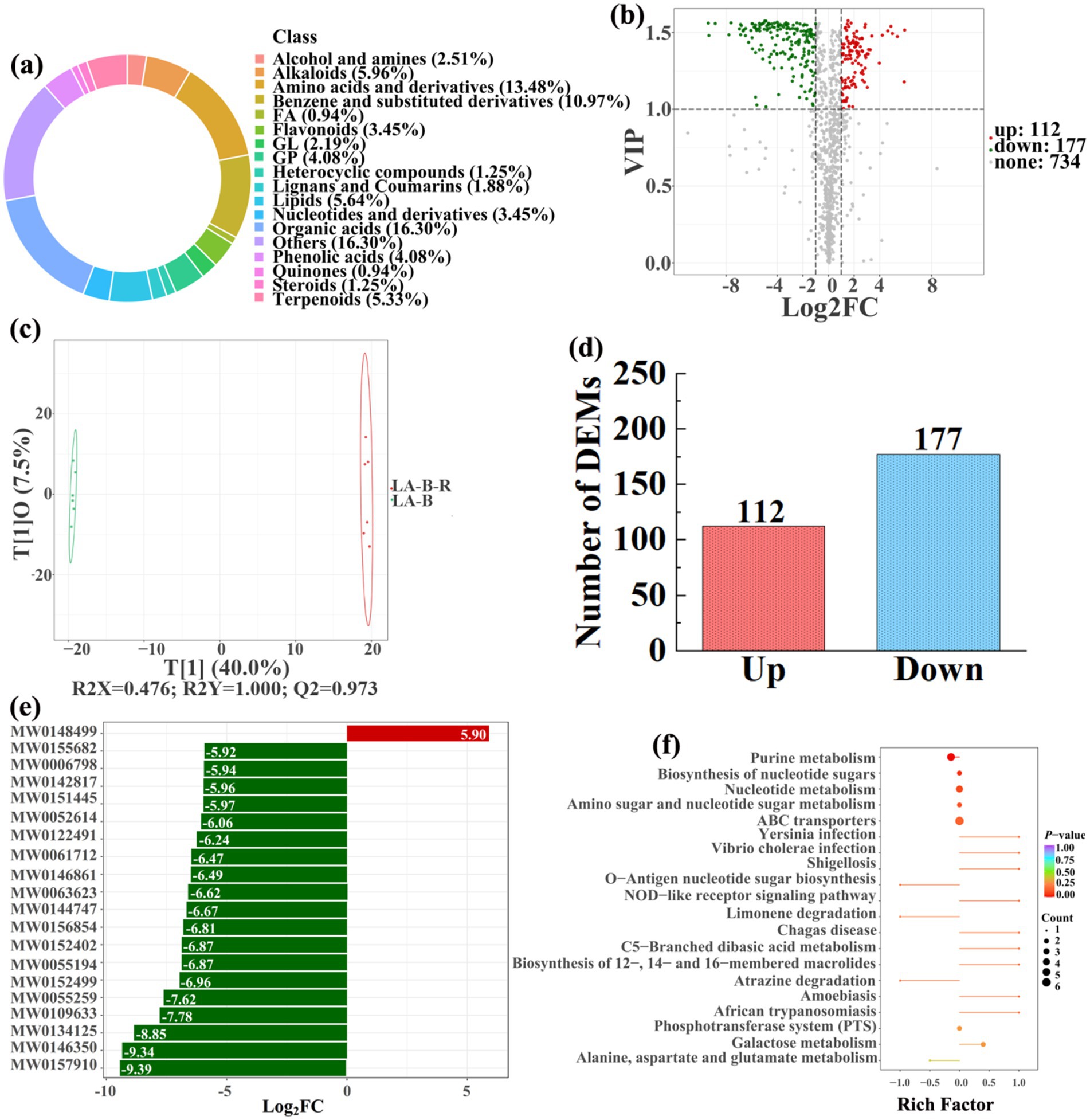
Figure 12. Donut plots showing metabolite classification and proportion (a), volcano plot (b), orthogonal projection to latent structures-discriminant analysis (OPLS-DA) score map (c), the number of DEMs (d), top 20 DEMs with largest VIP (e), KEGG enrichment analysis of differential soil metabolites in conversion of bamboo garden to paddy field (LA-B-R) vs bamboo garden (LA-B) (f).
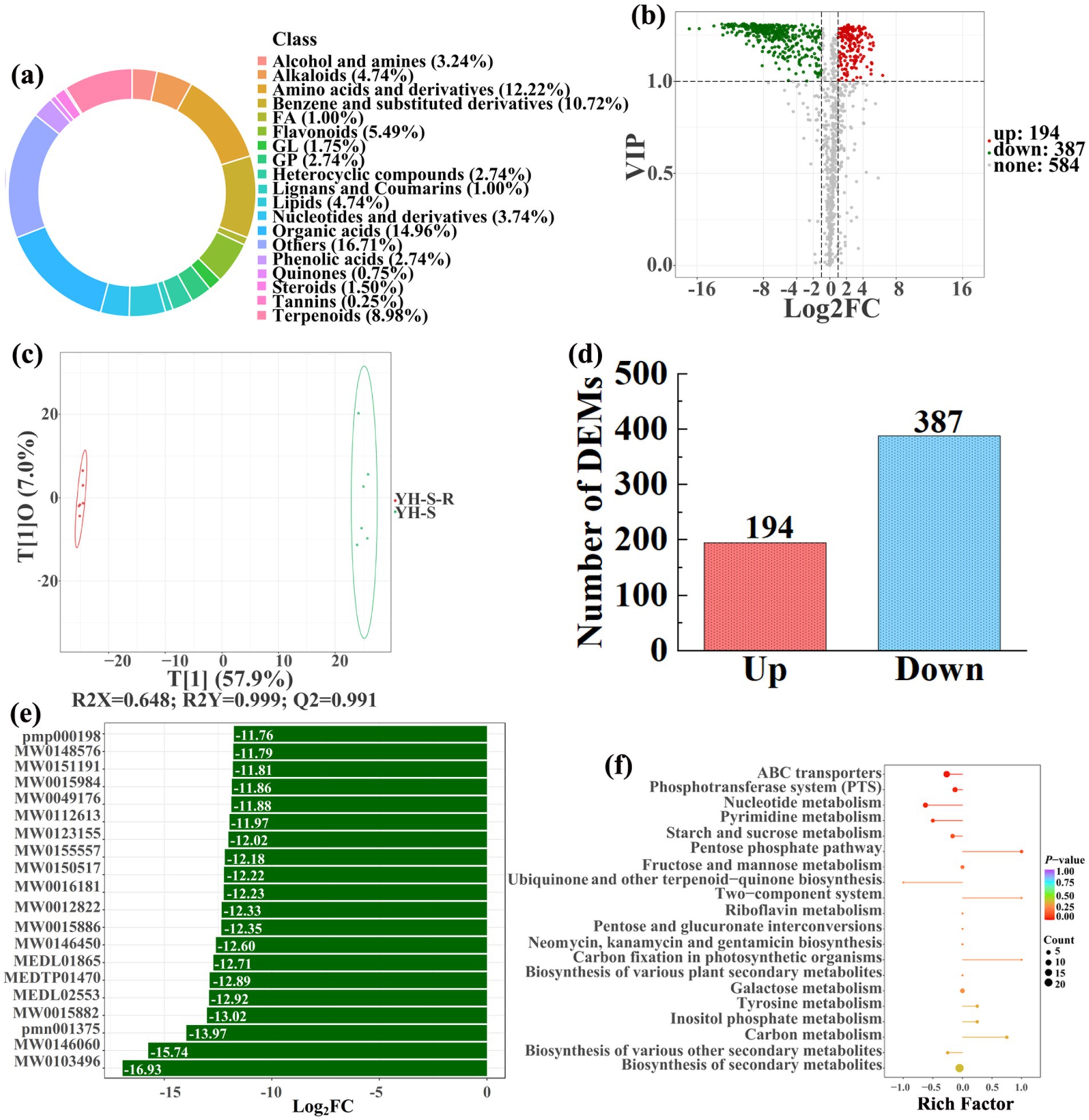
Figure 13. Donut plots showing metabolite classification and proportion (a), volcano plot (b), orthogonal projection to latent structures-discriminant analysis (OPLS-DA) score map (c), the number of DEMs (d), top 20 DEMs with largest VIP (e), KEGG enrichment analysis of differential soil metabolites in conversion of nursery stock base to paddy field (YH-S-R) vs nursery stock base (YH-S) (f).
In detail, 304 out of 852 identified metabolites were differentially expressed by conversion of loquat garden to paddy field, while the top 20 DEMs (5 upregulation and 15 downregulation) with largest VIP were visualized by bar chart (Figure 8), 305 out of 1,021 identified metabolites were differentially expressed by conversion of mulberry field to paddy field, while the top 20 DEMs with largest VIP were all downregulated (Figure 9), 288 out of 874 identified metabolites were differentially expressed by conversion of blueberry garden to paddy field, while 5 upregulation and 15 downregulation were found in the top 20 DEMs with largest VIP (Figure 10), 437 out of identified 892 metabolites were differentially expressed by conversion of vineyard to paddy field, while 9 upregulation and 11 downregulation were found in the top 20 DEMs with largest VIP (Figure 11), 289 out of identified 1,023 metabolites were differentially expressed by conversion of bamboo garden to paddy field, while 1 upregulation and 19 downregulation were found in the top 20 DEMs with largest VIP (Figure 12), 581 out of 1,165 identified metabolites were differentially expressed by conversion of nursery stock base to paddy field, while top 20 DEMs with largest VIP were all downregulated (Figure 13). Across all six different land-use conversions, amino acids and derivatives, benzene and substituted derivatives, heterocyclic compounds, organic acids, and terpenoids were the most consistently differentially expressed metabolite classes (Table 4). The functions of these DEMs were determined by the KEGG pathway analysis (Figures 8–13f). In contrast with non-grain cultivated land, the distinct metabolites mostly pertain to “microbial metabolism in diverse environments” and “ABC transporters” in paddy field converted from loquat garden; “biosynthesis of secondary metabolites” and “microbial metabolism in diverse environments” in paddy field converted from mulberry field; “microbial metabolism in diverse environments” and “biosynthesis of cofactors” in paddy field converted from blueberry garden; “metabolic pathways” and “microbial metabolism in diverse environments” in paddy field converted from vineyard; “ABC transporters,” “purine metabolism,” and “nucleotide metabolism” in paddy field converted from bamboo garden; “biosynthesis of secondary metabolites” and “ABC transporters” in paddy field converted from nursery stock base. An overlapping enrichment of “microbial metabolism in diverse environments” and “ABC transporters” in most paddy field converted from non-grain cultivated land suggests that the two specific metabolic pathways are vital for their survival and ecological niche establishment.
3.4 Correlations among soil properties, bacteria, and metabolites
RDA based on family-level abundances was carried out to examine the correlation between environmental factors and soil bacterial communities, and the results showed that a total of 56.42–88.61% of the cumulative variance of bacterial community-factor correction occurred at the family level (Figure 14; Table 5). In detail, the contributions of the 6 variables were AK (F = 73.54–97.78%, p = 0.001–0.04), AHN (F = 70.70–96.49%, p = 0.001–0.008), pH (F = 65.39–97.31%, p = 0.001–0.017), AP (F = 53.27–99.02%, p = 0.001–0.037), MBC (F = 27.12–82.86%, p = 0.002–0.229), and SOM (F = 4.75–94.83%, p = 0.001–0.818) during conversion of non-grain cultivated land to paddy field. Thus, it can be inferred that AK, AHN, pH, and AP were the primary factors associated with bacterial community variation, suggesting that soil nutrient elements clearly affected bacterial family-level distributions.
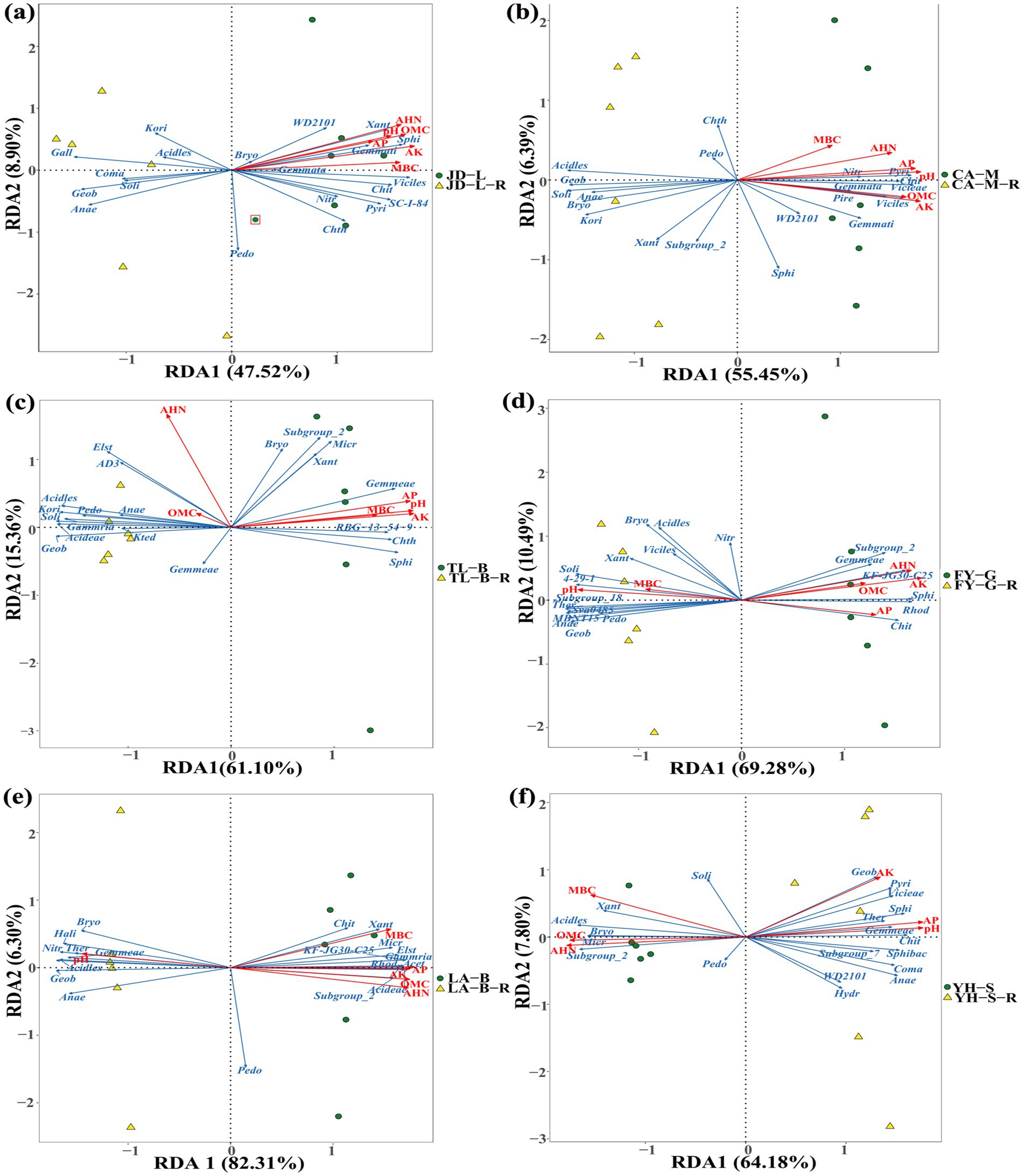
Figure 14. Redundancy discriminant analysis (RDA) of the root-zone soil bacterial community compositions at the family level with soil properties. The conversion of loquat garden to paddy field (a), conversion of mulberry field to paddy field (b), conversion of blueberry garden to paddy field (c), conversion of vineyard to paddy field (d), conversion of bamboo garden to paddy field (e), and conversion of nursery stock base to paddy field (f). Acet, Acetobacteraceae; Acideae, Acidobacteriaceae; Acidles, Acidobacteriales; Anae, Anaerolineaceae; Bryo, Bryobacteraceae; Chit, Chitinophagaceae; Chth, Chthoniobacteraceae; Coma, Comamonadaceae; Elst, Elsterales; Gall, Gallionellaceae; Geob, Geobacteraceae; Gemmata, Gemmataceae; Gemmati, Gemmatimonadaceae; Gammria, Gammaproteobacteria; Hali, Haliangiaceae; Hydr, Hydrogenophilaceae; Kori, Koribacteraceae; Kted, Ktedonobacteraceae; Micr, Micropepsaceae; Nitr, Nitrosomonadaceae; Pedo, Pedosphaeraceae; Pire, Pirellulaceae; Pyri, Pyrinomonadaceae; Rhod, Rhodanobacteraceae; Soli, Solibacteraceae; Sphi, Sphingomonadaceae; Sphibac, Sphingobacteriaceae; Ther, Thermodesulfovibrionia; Vicieae, Vicinamibacteraceae; Viciles, Vicinamibacterales; Xant, Xanthobacteraceae. SOM, organic matter contains; AHN, alkaline hydrolysis N; AP, available P; AK, available K; MBC, microbial biomass carbon. Arrows indicate the direction and magnitude of soil properties (pH, SOM, AHN, AP, AK, and MBC) associated with the different bacteria.
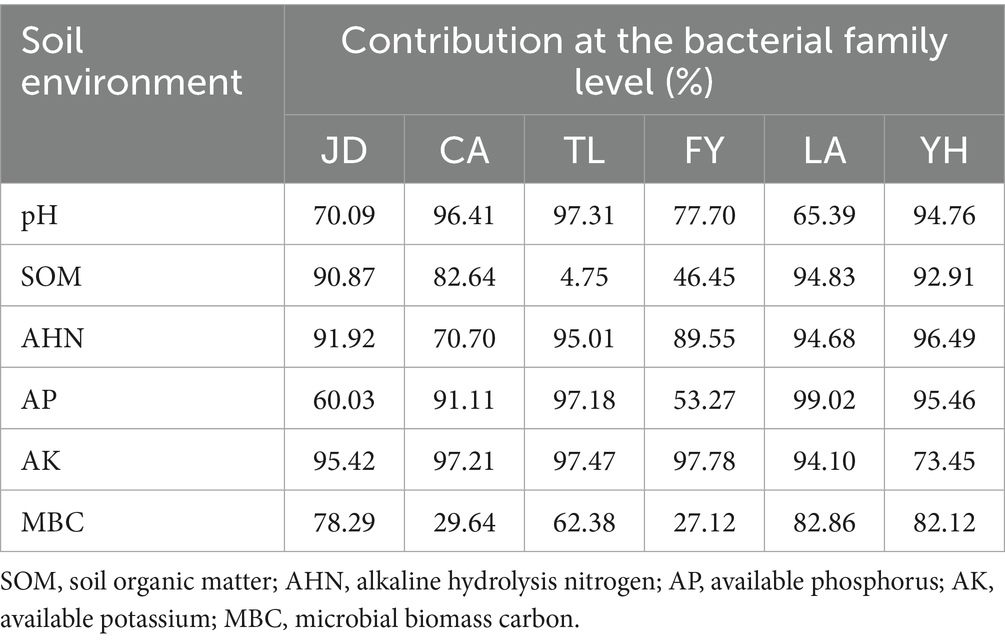
Table 5. Contribution of the soil environment to bacterial taxa at the family level under different groups.
To further analyze the correlation relationship between bacteria and metabolites, the clustering heat map was drawn based on bacterial top 20 family and 20 DEMs with largest VIP (Figure 15). Indeed, the top 20 upregulated DEMs were significantly correlated with 15 bacteria (except Acidobacteriales, Gemmataceae, Koribacteraceae, Nitrosomonadaceae, and Pedosphaeraceae) in the paddy fields converted from loquat garden, 17 bacteria (except Chthoniobacteraceae, Pedosphaeraceae and Sphingomonadaceae) in the paddy fields converted from mulberry field, 19 bacteria (except Gemmataceae) in the paddy fields converted from blueberry garden, 18 bacteria (except Nitrosomonadaceae and Vicinamibacterales) in the paddy fields converted from vineyard, 19 bacteria (except Pedosphaeraceae) in the paddy fields converted from bamboo garden, 18 bacteria (except Pedosphaeraceae and Solibacteraceae) in the paddy fields converted from nursery stock base, respectively (Table 4).
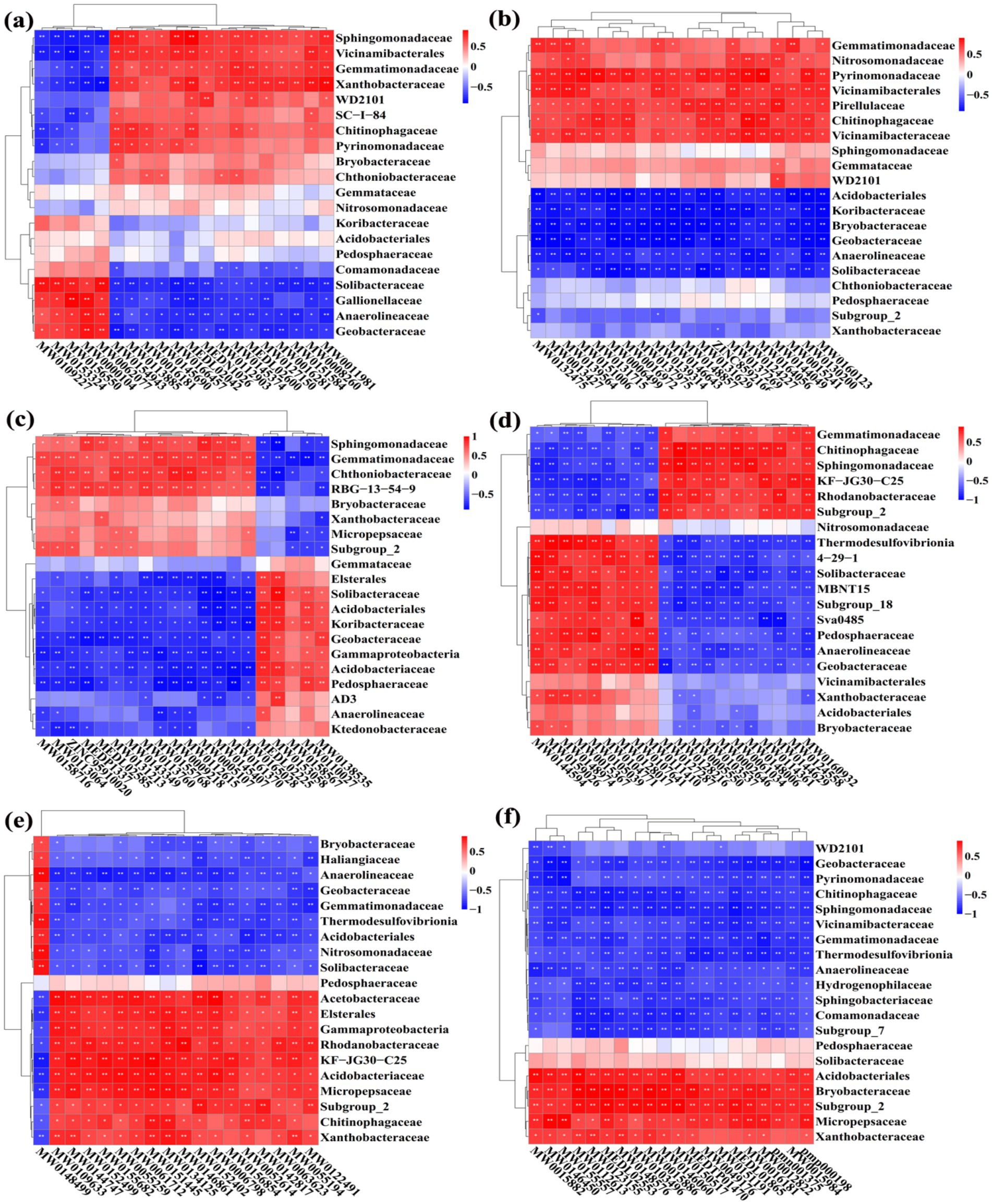
Figure 15. Correlation heat map between the top 20 family of bacteria and significant DEMs under different groups. The conversion of loquat garden to paddy field (a), conversion of mulberry field to paddy field (b), conversion of blueberry garden to paddy field (c), conversion of vineyard to paddy field (d), conversion of bamboo garden to paddy field (e), and conversion of nursery stock base to paddy field (f). *Indicated a significant correlation at p < 0.05, **indicated a significant correlation at p < 0.01.
Taken together, the DEMs (amino acids and derivatives, organic acids, benzene and substituted derivatives, heterocyclic compounds, and terpenoids) of root-zone soils could significantly alter the soil bacteria. Especially, they were significantly negative correlated with the two key families of Anaerolineaceae and Geobacteraceae, while the two families were also negative correlated with AP and AK. In other word, the lower AP, AK, and DEMs thereby could help in the coordination of the root-zone bacteria during conversion of non-grain cultivated land to paddy field. Previous research reported that soil moisture regimes influenced bacterial community structure both directly and indirectly by changing nutrient availability and oxygen concentrations, and flooding with less oxygen availability promoted the growth of facultative anaerobic bacteria, such as Anaerolineaceae (with the ability to providing organic acid such as acetate to other microbes) and Geobacteraceae (the main ferric reducers in anaerobic environments) (Liang et al., 2015; Wang et al., 2020).
4 Discussion
Cultivated land is paramount for grain production, serving as a vital strategic resource worldwide. However, due to the pursuit for economic benefits, numerous cultivated lands have been used for non-grain production, thus leading to significant impacts on the arable land-use structure and ecosystem function (Su et al., 2020). Soil microbes mainly including bacteria, fungi, archaea play an essential role in soil ecosystem by driving soil functional process including nutrient cycling, organic matter transformation, plant disease control, and plant productivity promotion (Philippot et al., 2013; Ling et al., 2014; Finzi et al., 2015; Pieterse et al., 2016). Among them, soil bacteria are essential members of soil microbial community (He et al., 2023), and presented larger environmental niche breadths than fungi and archaea (Malard et al., 2022). However, soil interference (such as changes in land use, vegetation, soil fertility, pH) can significantly affect soil bacteria in their taxonomy and functionality (Liang and Geng, 2023; Cordovez et al., 2019; Samaddar et al., 2019). In addition, soil bacteria are related to soil metabolites (Song et al., 2020). Here, the combined analyses of the soil physicochemical properties, bacterial community structure, and metabolite were able to explore the influence mechanism of non-grain production on cultivated land, which will in guiding the conversion of non-grain cultivated land to paddy field.
The soil properties in the non-grain cultivated land were significantly affected by the conversion to paddy field, and the impact depended on the soil parameters and the type of non-grain crops. In general, conversion of non-grain cultivated land to paddy field caused a remarkable decrease in the concentration of soil nutrients, while there were some recovery trajectories of soil nutrients in the vineyard or nursery stock conversions. Indeed, results revealed that the soil pH can be significantly reduced in the paddy fields converted from loquat garden, mulberry field, and blueberry garden, but improved from vineyard, bamboo garden, and nursery stock base. The SOM, AHN, AP (except nursery stock base), AK (except nursery stock base), and MBC (except vineyard) can be significantly reduced by the conversion of non-grain cultivated land to paddy fields, which may be due to the excessive use of fertilizer in non-grain cultivated lands. For example, the quantity of fertilizer usage on flowers and fruit trees was 3.85 fold and 2.45 fold of grain crops, respectively (Wang et al., 2018). Vegetable production systems had higher N (264.3 kg/ha) and P (101.0 kg/ha) fertilizer input than rice cultivation (Wang et al., 2019). Broadly, farmers were inclined to apply more fertilizer on dry land than in paddy fields (Jiao et al., 2017), while fertilization with N and P could increase soil organic carbon, N, and P concentrations (Wang N. et al., 2024). However, long-term overuse of chemical fertilizers could alter the soil pH, increase acidification, decrease soil quality (Pahalvi et al., 2021), while conversion of upland crop cultivation to paddy rice is an essential methodology to improving ecology (Lü et al., 2017). Therefore, it can be inferred that it is beneficial for soil sustainable ecological function restoration by conversion of non-grain cultivated land to paddy field.
Following the measurement of the bacterial community diversity in all soil samples using 16S rRNA gene high-throughput sequencing, we found that the bacterial OTUs number and Chao1 index was increased by conversion of non-grain cultivated land to paddy field (except blueberry garden). These results suggest that conversion of non-grain cultivated land to paddy field had the distinct impact in increasing the robustness of bacterial communities. Further PCA assay also revealed that there were significant differences in the root-zone soil bacterial communities between non-grain cultivated lands and the paddy fields converted from the corresponding non-grain cultivated lands. Liang and Geng (2023) showed that non-grainization consolidation by conversion of dryland to paddy field enhanced the α-diversity content (including Ace, Chao1, Coverage, and Shannon indices) in terms of soil bacterial community diversity. Indeed, agricultural land consolidation that widely applied in farmland improvement has been found to be able to significantly impact soil microbial community diversity and composition (Lin et al., 2020). In agreement with previous reports, conversion of non-grain cultivated land to paddy field also caused the alteration of some certain microbes, thus reshaping the root-zone soil bacterial community of rice field converted from non-grain cultivated land.
At the phylum level, Chloroflexi, Desulfobacterota, and Nitrospirota were significantly increased in most of the paddy fields converted from non-grain cultivated lands. At the family level, conversion of non-grain cultivated land to paddy field could significantly enrich Anaerolineaceae, Bryobacteraceae, Comamonadaceae, Gallionellaceae, Geobacteraceae, Haliangiaceae, Hydrogenophilaceae, Koribacteraceae, Ktedonobacteraceae, MBNT15, Nitrosomonadaceae, Pedosphaeraceae, Solibacteraceae, Subgroup_7, Subgroup_18, Sva0485, 4-29-1, Thermodesulfovibrionia, and WD2101. Furthermore, LEfSe also obtained 48 bacterial biomarkers in all treatments between different groups. In particular, conversion of six non-grain cultivated lands to paddy field in this study resulted in enrichment of some bacteria including Desulfobacterota (1.26–21.50 fold), Nitrospirota (4.29–14.54 fold), and Chloroflexi (0.81–3.08 fold), which might have important functional implications on rice growth, and could be used as potential biomarkers for successful land restoration.
In agreement with the result of this study, Chloroflexi and Nitrospirota play important roles in the nitrification process in the soil nitrogen cycle by oxidizing nitrite to nitrate (Prabhu et al., 2022). Desulfobacterales plays an important role in nitrogen cycling and contributes 12% of the genes of nitrogen pathways on average (Nie et al., 2021). Anaerolineaceae can be used as abundant primary fermenters in anaerobic digesters, and has the ability of providing organic acid such as acetate to other microbes (Liang et al., 2015; Mcllroy et al., 2017). Geobacteraceae is an important dissimilatory Fe(III) reducer that affects the cycles of multiple elements, while dissimilatory iron reduction mediated by the Geobacteraceae may influence the rice yields by affecting the biogeochemical cycles of nutrient elements (Li et al., 2020). Nitrosomonadaceae plays a key role in the nitrogen cycle (Prosser et al., 2014). Pedosphaeraceae can detoxify arsenic and antimony (Huang et al., 2014). Obviously, these bacteria may have significant potential in colonizing and altering soil fertility in paddy fields converted from non-grain cultivated land by enhancing nutrient uptake, improving soil conditions and increasing bioavailability. On the other hand, this study focuses on bacteria due to it is the most major kingdom in soil, however, it will capture a more holistic soil microbiome profile by including fungal or archaeal community analyses.
Moreover, network assay can reveal the co-occurrence patterns between soil microbial members and complex associations within soil microbial communities (Wang J. et al., 2022). The co-occurrence networks constructed in this study indicated that the number of network nodes and edges were higher in non-grain cultivated lands (except loquat garden and nursery stock base) compared to the paddy fields converted from the corresponding non-grain cultivated land. Conversely, the modularity of networks was higher in the converted paddy fields than that of from the corresponding non-grain cultivated land. Due to more nodes and edges indicating a more complex network structure, while high modularity representing high structural stability of network (Freundt, 2021; Ma et al., 2021), thus, it can be inferred that conversion of non-grain cultivated land to paddy fields is good for stability of soil bacterial community. Align with this study, Cornell et al. (2023) also showed that land use conversion in a temperate grassland increased network complexity and stability of soil microbial communities, which have been reported to be highly associated with soil ecosystem function.
The variability within each treatment was the main challenges in comparing metabolomic profiles across such ecologically distinct conversion types. In order to address the issue, soil sample in this study was collected by mixing a total of nine random soil cores, while the comparative analysis of metabolites was performed on each land type between the non-grain cultivated lands and the corresponding converted paddy fields. Generally, a sum of 5,827 metabolites were identified from all different groups, which were composed mainly of amino acids and derivatives, benzene and substituted derivatives, flavonoids, lipids, organic acids, terpenoids, with 794 upregulated and 1,410 downregulated metabolites. The OPLS-DA, volcano plot, and KEGG enrichment analysis showed that there was significant difference in the metabolite compositions between non-grain cultivated lands and the paddy fields converted from the corresponding non-grain cultivated land. Previous researches indicated that the differentially expressed metabolites were involved in different bio-activities and many bio-chemical activities in relation to rice growth and enhancement of soil fertility. For example, flavonoids play important roles in plant development and plant-environmental interactions (Dong and Lin, 2021), while lipids affect plant growth and development by involving cell membrane remodeling, anther fertility, seed formation, and response to adverse stresses (Wan et al., 2020).
Previous study also indicated that some secondary metabolism (such as flavonoids, terpenoids, strigolactones, and coumarins) could regulate the assembly of specific microbial taxa in the rhizosphere, while soil microbes can also enhance the promotion or inhibition of soil metabolites accumulation (Cheng et al., 2022; Wang L. et al., 2022). Finally, the regulation of key metabolites can increase crop yields in agroecosystems (Zhao et al., 2022). For example, flavoniods could regulate rhizosphere bacterial community structure (with a significant increase in the RA of Micrococcaceae and Nocardioidaceae) to enhance organic P mineralization, thereby facilitating P uptake and plant growth (Wang S. et al., 2024). In agreement with previous reports, the result of this study showed that amino acids and derivatives, organic acids, and terpenoids could recruit beneficial microbes, such as Anaerolineaceae and Geobacteraceae, to improve plant fitness and help plants cope with environmental changes during conversation non-grain cultivated land to paddy field. In other word, soil metabolites might play important roles in maintenance of soil ecosystem functions and crop yields during this conversation.
The correlation relationship between bacteria and DEMs in different group was determined by drawing the clustering he during conversation non-grain cultivated land to paddy field at map, which showed that the DEMs (amino acids and derivatives, organic acids, benzene and substituted derivatives, and other secondary metabolites) of root-zone soils were significantly positively or negatively correlated with the relative abundances of some bacteria, thereby helping in coordinating the root-zone bacteria during conversion of non-grain cultivated lands to paddy fields. Meanwhile, RDA in this study also indicated that soil pH, AK, AHN, and AP were the primary factors associated with bacterial community variation, showing the strongest predictive value for bacterial community shifts. For example, the two key families of Anaerolineaceae and Geobacteraceae in paddy field were negatively correlated with AP and AK. Therefore, these specific soil nutrients should be considered for land rehabilitation from non-grain land to paddy land. In agreement with the result of our study, Wan et al. (2023) and Li et al. (2024) also indicated that the growth of soil bacteria was often impacted by various environmental factors. Taken overall, this study revealed that good teamwork occurs among soil properties, bacteria, and metabolites during conversation of non-grain cultivated land to paddy field.
5 Conclusion
In conclusion, conversion of non-grain field to paddy field changed the soil properties, bacterial communities, and metabolites. Specifically, a higher OTUs number, a more diversity and stable bacterial community was obtained in paddy fields converted from non-grain fields than the corresponding non-grain fields. Furthermore, 48 bacterial biomarkers were identified across all different groups, with enriched abundances of Chloroflexi, Desulfobacterota, Nitrospirota, Anaerolineaceae, Geobacteraceae, Nitrosomonadaceae, and Pedosphaeraceae in converted paddy fields converted from non-grain cultivated lands, while 5,827 metabolites were identified in converted paddy fields and non-grain cultivated lands, with 794 of upregulation and 1,410 of downregulation. In addition, DEMs of root-zone soils were significantly correlated with bacteria, thereby helping in coordinating the root-zone bacteria during conversion of non-grain cultivated land to paddy field, while soil environmental properties (especially soil pH, AK, AHN, and AP) were related to variations in bacterial community composition. Overall, the result of this study indicated that the paddy fields converted from non-grain cultivated lands can be characterized by more richness, diversity and stable bacterial community structure, specific bacteria and metabolites, lower nutrition. Overall, this study provides a scientific basis and supporting evidence to explain the mechanism of conversion from non-grain cultivated lands to paddy fields, thus ensuring national food security.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/, PRJNA1192420; https://db.cngb.org/, CNP0006336.
Author contributions
XL: Software, Resources, Formal analysis, Writing – original draft, Methodology, Visualization, Investigation, Validation, Conceptualization. HC: Methodology, Writing – original draft, Conceptualization, Resources. XW: Writing – original draft, Resources, Formal analysis, Investigation, Conceptualization, Methodology, Validation. QA: Writing – original draft, Methodology, Conceptualization, Supervision, Data curation, Funding acquisition, Project administration, Writing – review & editing. LL: Conceptualization, Writing – original draft, Methodology, Software. TZ: Methodology, Supervision, Investigation, Writing – review & editing, Project administration, Data curation. MI: Data curation, Project administration, Methodology, Writing – review & editing, Supervision. TA: Data curation, Supervision, Software, Writing – review & editing, Project administration. JY: Conceptualization, Formal analysis, Visualization, Writing – review & editing, Supervision, Investigation, Writing – original draft, Funding acquisition, Validation. BL: Software, Supervision, Writing – review & editing, Conceptualization, Funding acquisition, Writing – original draft, Visualization, Project administration, Resources, Data curation.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This research was funded by Hangzhou Science and Technology Development Plan Project (20231203A05), Science and Technology Innovation and Promotion Demonstration Project of Hangzhou Academy of Agricultural Sciences (2025HNCT-09), Hangzhou City Agricultural Science and Technology Collaboration and Innovation Project (202409SX16), Zhejiang Province Key Research and Development Program of China (2019C02035). Additionally, we thank United Arab Emirates University for providing a postdoctoral grant on climate action to Qurban Ali (#12S140).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that Gen AI was used in the creation of this manuscript. Declaration of Generative AI and AI-assisted technologies in the writing process during the preparation of this work the author(s) used ChatGpt tool to improve language and readability. After using this tool, the author(s) reviewed and edited the content as needed and take(s) full responsibility for the content of the publication.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Agyekum, D. V. A., Kobayashi, T., Dastogeer, K. M. G., Yasuda, M., Sarkodee-Addo, E., Ratu, S. T. N., et al. (2023). Diversity and function of soybean rhizosphere microbiome under nature farming. Front. Microbiol. 14:1130969. doi: 10.3389/fmicb.2023.1130969
Ahmed, T., Luo, J., Noman, M., Ijaz, M., Wang, X., Masood, H. A., et al. (2023). Microbe-mediated nanoparticle intervention for the management of plant diseases. Crop Health 1:3. doi: 10.1007/s44297-023-00006-9
Bi, B., Yuan, Y., Zhang, H., Wu, Z., Wang, Y., and Han, F. (2022). Rehizosphere soil metabolites mediated microbial community changes of Pinus sylvestris var. mongolica across stand ages in the Mu Us Desert. Appl. Soil Ecol. 169:104222. doi: 10.1016/j.apsoil.2021.104222
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Boulanger, P., Dudu, H., Ferrari, E., Mainar-Causapé, A. J., and Ramos, M. P. (2022). Effectiveness of fertilizer policy reforms to enhance food security in Kenya: a macro-micro simulation analysis. Appl. Econ. 54, 841–861. doi: 10.1080/00036846.2020.1808180
Butler, J. L., Williams, M. A., Bottomley, P. J., and Myrold, D. D. (2003). Microbial community dynamics associated with rhizosphere carbon flow. Appl. Environ. Microbiol. 69, 6793–6800. doi: 10.1128/AEM.69.11.6793-6800.2003
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016). DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Chen, Q., Song, Y., An, Y., Lu, Y., and Zhong, G. (2024). Soil microorganisms: their role in enhancing crop nutrition and health. Diversity 16:734. doi: 10.3390/d16120734
Chen, B., Yang, H., Song, W., Liu, C., Xu, J., Zhao, W., et al. (2016). Effect of N fertilization rate on soil alkali-hydrolyzable N, subtending leaf N concentration, fiber yield, and quality of cotton. Crop J 4, 323–330. doi: 10.1016/j.cj.2016.03.006
Chen, S., Zhang, Y., Zhu, Y., Ma, L., and Zhao, J. (2023). The battle of crops: unveiling the shift from grain to non-grain use of farmland in China? Int. J. Agric. Sustain. 21:2262752. doi: 10.1080/14735903.2023.2262752
Cheng, H., Yuan, M., Tang, L., Shen, Y., Yu, Q., and Li, S. (2022). Integrated microbiology and metabolomics analysis reveal responses of soil microorganisms and metabolic functions to phosphorus fertilizer on semiarid farm. Sci. Total Environ. 817:152878. doi: 10.1016/j.scitotenv.2021.152878
Cordovez, V., Dini-Andreote, F., Carrión, V. J., and Raaijmakers, J. M. (2019). Ecology and evolution of plant microbiomes. Ann. Rev. Microbiol. 73, 69–88. doi: 10.1146/annurev-micro-090817-062524
Cornell, C. R., Zhang, Y., Ning, D., Xiao, N., Wagle, P., Xiao, X., et al. (2023). Land use conversion increases network complexity and stability of soil microbial communities in a temperate grassland. ISME J. 17, 2210–2220. doi: 10.1038/s41396-023-01521-x
Cui, K., and Shoemaker, S. P. (2018). A look at food security in China. NPJ Sci. Food 2:4. doi: 10.1038/s41538-018-0012-x
Das, S. K. (2023). Stability of organic carbon pools and sequestration potential as affected under different agroforestry systems. Crop Health 1:14. doi: 10.1007/s44297-023-00016-7
Deng, O., Ran, J., Huang, S., Duan, J., Reis, S., Zhang, J., et al. (2024). Managing fragmented croplands for environmental and economic benefits in China. Nat. Food 5, 230–240. doi: 10.1038/s43016-024-00938-7
Deng, F. B., Wang, H. J., Xie, H. T., Bao, X. L., He, H. B., Zhang, X. D., et al. (2022). Low-disturbance farming regenerates healthy deep soil toward sustainable agriculture—evidence from long-term no-tillage with Stover mulching in Mollisols. Sci. Total Environ. 825:153929. doi: 10.1016/j.scitotenv.2022.153929
Dixon, P. (2003). VEGAN, a package of R functions for community ecology. J. Veg. Sci. 14, 927–930. doi: 10.1111/j.1654-1103.2003.tb02228.x
Dong, N., and Lin, H. (2021). Contribution of phenylpropanoid metabolism to plant development and plant-environment interactions. J. Integr. Plant Biol. 63, 180–209. doi: 10.1111/jipb.13054
Finzi, A. C., Abramoff, R. Z., Spiller, K. S., Brzostek, E. R., Darby, B. A., Kramer, M. A., et al. (2015). Rhizosphere processes are quantitatively important components of terrestrial carbon and nutrient cycles. Glob. Change Biol. 21, 2082–2094. doi: 10.1111/gcb.12816
Freundt, S. (2021). Emergence in a complex network with two types of directed edges – a numerical investigation. Results Phys. 30:104819. doi: 10.1016/j.rinp.2021.104819
Friedman, J., and Alm, E. J. (2012). Inferring correlation networks from genomic survey data. PLoS Comput. Biol. 8:e1002687. doi: 10.1371/journal.pcbi.1002687
Gloor, G. B., Macklaim, J. M., Pawlowsky-Glahn, V., and Egozcue, J. J. (2017). Microbiome datasets are compositional: and this is not optional. Front. Microbiol. 8:2224. doi: 10.3389/fmicb.2017.02224
Hao, Q., Zhang, T., Cheng, X., He, P., Zhu, X., and Chen, Y. (2024). GIS-based non-grain cultivated land susceptibility prediction using data mining methods. Sci. Rep. 14:4433. doi: 10.1038/s41598-024-55002-y
He, C., Li, K., Wen, C., Li, J., Fan, P., Ruan, Y., et al. (2023). Changes in physicochemical properties and bacterial communities of tropical soil in China under different soil utilization types. Agronomy 13:1897. doi: 10.3390/agronomy13071897
Hollander, M., Wolfe, D. A., and Chicken, E. (2015). “Nonparametric statistical methods” in Biostatistics and microbiology: a survival manual. ed. S. P. Daryl (New York, NY: Springer), 121–162.
Huang, L., Chakrabarti, S., Cooper, J., Perez, A., John, S. M., Daroub, S. H., et al. (2014). Ammonia-oxidizing archaea are integral to nitrogen cycling in a highly fertile agricultural soil. ISME Commun. 1:19. doi: 10.1038/s43705-021-00020-4
Jiang, N., Wei, K., and Chen, L. (2016). Long-term chronological shifts in bacterial communities and hydrolytic extracellular enzyme activities in the forty years following a land-use change from upland fields to paddy fields. Pedobiologia 59, 17–26. doi: 10.1016/j.pedobi.2015.12.002
Jiao, W., Min, Q., and Fuller, A. M. (2017). Converting rice paddy to dry land farming in the tai Lake Basin, China: toward an understanding of environmental and economic impacts. Paddy Water Environ. 15, 171–179. doi: 10.1007/s10333-016-0538-y
Lai, Z., Chen, M., and Liu, T. (2020). Changes in and prospects for cultivated land use since the reform and opening up in China. Land Use Pol. 97:104781. doi: 10.1016/j.landusepol.2020.104781
Legrand, F., Picot, A., Cobo-Díaz, J. F., Carof, M., Chen, W., and Le Floch, G. (2018). Effect of tillage and static abiotic soil properties on microbial diversity. Appl. Soil Ecol. 132, 135–145. doi: 10.1016/j.apsoil.2018.08.016
Li, X., Ding, L., Li, X., and Zhu, Y. (2020). Abundance, diversity, and structure of Geobacteraceae community in paddy soil under long-term fertilization practices. Appl. Soil Ecol. 153:103577. doi: 10.1016/j.apsoil.2020.103577
Li, F., Liu, J., Ren, W., and Liu, L. (2018). Land-use change alters patterns of soil biodiversity in arid lands of northwestern China. Plant Soil 428, 371–388. doi: 10.1007/s11104-018-3673-y
Li, X., Ren, X., Su, Y., Zhou, X., Wang, Y., Ruan, S., et al. (2024). Differential effects of winter cold stress on soil bacterial communities, metabolites, and physicochemical properties in two varieties of Tetrastigma hemsleyanum Diels & Gilg in reclaimed land. Microbiol. Spectr. 12:e02425. doi: 10.1128/spectrum.02425-23
Li, Z., Wu, X., and Chen, B. (2007). Changes in transformation of soil organic C and functional diversity of soil microbial community under different land uses. Agr. Sci. China 6, 1235–1245. doi: 10.1016/S1671-2927(07)60168-0
Liang, Y., and Geng, B. (2023). Effects of paddy field non-grainization consolidation on sustainable eco-functions protection of soil bacterial: empirical evidence from Zhejiang province, China. Front. Environ. Sci. 11:1130234. doi: 10.3389/fenvs.2023.1130234
Liang, B., Wang, L., Mbadinga, S. M., Liu, J., Yang, S., Gu, J., et al. (2015). Anaerolineaceae and Methanosaeta turned to be the dominant microorganisms in alkanes-dependent methanogenic culture after long-term of incubation. AMB Expr. 5:37. doi: 10.1186/s13568-015-0117-4
Lin, Y., Ye, Y., Wu, C., Hu, Y., and Shi, H. (2020). Changes in microbial community structure under land consolidatio n in paddy soils: a case study in eastern China. Ecol. Eng. 145:105696. doi: 10.1016/j.ecoleng.2019.105696
Lin, Y., Ye, Y., Wu, C., Yang, J., Hu, Y., and Shi, H. (2019). Comprehensive assessment of paddy soil quality under land consolidation: a novel perspective of microbiology. Peer J. 7:e7351. doi: 10.7717/peerj.7351
Ling, N., Deng, K., Song, Y., Wu, Y., Zhao, J., Raza, W., et al. (2014). Variation of rhizosphere bacterial community in watermelon continuous mono-cropping soil by long-term application of a novel bioorganic fertilizer. Microbiol. Res. 169, 570–578. doi: 10.1016/j.micres.2013.10.004
Lü, Y., Bai, W., Wang, X., Cai, Q., and Liang, W. (2017). Responses of soil micro-food web to land use change from upland to paddy fields with different years of rice cultivation. Pedosphere 27, 155–164. doi: 10.1016/S1002-0160(15)60102-3
Lu, D., Wang, Z., Su, K., Zhou, Y., Li, X., and Lin, A. (2024). Understanding the impact of cultivated land-use changes on China's grain production potential and policy mplications: a perspective of non-agriculturalization, non-grainization, and marginalization. J. Clean. Prod. 436:140647. doi: 10.1016/j.jclepro.2024.140647
Malard, L. A., Mod, H. K., Guex, N., Broennimann, O., Yashiro, E., Lara, E., et al. (2022). Comparative analysis of diversity and environmental niches of soil bacterial, archaeal, fungal and protist communities reveal niche divergences along environmental gradients in the Alps. Soil Biol. Biochem. 169:108674. doi: 10.1016/j.soilbio.2022.108674
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12. doi: 10.14806/ej.17.1.200
Ma, W., Yang, Z., Liang, L., Ma, Q., Wang, G., and Zhao, T. (2021). Characteristics of the fungal communities and co-occurrence networks in hazelnut tree root endospheres and rhizosphere soil. Front. Plant Sci. 12:749871.doi: 10.3389/fpls.2021.749871
Mcllroy, S. J., Kirkegaard, R. H., Dueholm, M. S., Fernando, E., Karst, S. M., Albertsen, M., et al. (2017). Culture-independent analyses reveal novel Anaerolineaceae as abundant primary fermenters in anaerobic digesters treating waste activated sludge. Front. Microbiol. 8:1134. doi: 10.3389/fmicb.2017.01134
Medriano, C. A., Chan, A., Sotto, R. D., and Bae, S. (2023). Different types of land use influence soil physiochemical properties, the abundance of nitrifying bacteria, and microbial interactions in tropical urban soil. Sci. Total Environ. 869:161722. doi: 10.1016/j.scitotenv.2023.161722
Meng, F., Tan, F., Chen, H., and Xiong, W. (2022). Spatial-temporal evolution patterns and influencing factors of “non-grain” utilization, of cultivated land in China. China Land Sci. 6, 97–106. (In Chinese). doi: 10.11994/zgtdkx.20211206.092159
Nelson, D. W., and Sommers, L. E. (1996). “Total carbon, organic carbon, and organic matter” in Methods of soil analysis: part 3: chemical methods, SSSA and ASA, Madison, vol. 34, 961–1010.
Nie, S., Zhang, Z., Mo, S., Li, J., He, S., Kashif, M., et al. (2021). Desulfobacterales stimulates nitrate reduction in the mangrove ecosystem of a subtropical gulf. Sci. Total Environ. 769:144562. doi: 10.1016/j.scitotenv.2020.144562
Pahalvi, H. N., Rafiya, L., Rashid, S., Nisar, B., and Kamili, A. N. (2021). “Chemical fertilizers and their impact on soil health” in Microbiota and biofertilizers. eds. G. H. Dar, R. A. Bhat, M. A. Mehmood, and K. R. Hakeem, vol. 2 (Cham: Springer).
Peschel, S., Müller, C. L., von Mutius, E., Boulesteix, A. L., and Depner, M. (2021). NetCoMi: network construction and comparison for microbiome data in R. Brief. Bioinform. 22:bbaa290. doi: 10.1093/bib/bbaa290
Philippot, L., Raaijmakers, J. M., Lemanceau, P., and van der Putten, W. H. (2013). Going back to the roots: the microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 11, 789–799. doi: 10.1038/nrmicro3109
Pieterse, C. M. J., de Jonge, R., and Berendsen, R. L. (2016). The soil-borne supremacy. Trends Plant Sci. 21, 171–173. doi: 10.1016/j.tplants.2016.01.018
Prabhu, D. M., Rao, N., Luo, Z., Li, Q., Liu, B., Guo, S., et al. (2022). Metagenomic analysis further extends the role of Chloroflexi in fundamental biogeochemical cycles. Envron. Res. 207:112888. doi: 10.1016/j.envres.2022.112888
Prosser, J. I., Head, I. M., and Stein, L. Y. (2014). “The family nitrosomonadaceae” in The prokaryotes. eds. E. Rosenberg, E. F. DeLong, S. Lory, E. Stackebrandt, and F. Thompson (Berlin, Heidelberg: Springer).
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2012). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Ramette, A. (2007). Multivariate analyses in microbial ecology. FEMS Microbiol. Ecol. 62, 142–160. doi: 10.1111/j.1574-6941.2007.00375.x
Rathje, M. L. (1959). Jackson, M. L: Soil chemical analysis. Verlag: prentice Hall, Inc., Englewood Cliffs, NJ. 1958, 498 S. DM 39.40. J. Plant Nutr. Soil Sci. 85, 251–252. doi: 10.1002/jpln.19590850311
Samaddar, S., Chatterjee, P., Truu, J., Anandham, R., Kim, S., and Sa, T. (2019). Long-term phosphorus limitation changes the bacterial community structure and functioning in paddy soils. Appl. Soil Ecol. 134, 111–115. doi: 10.1016/j.apsoil.2018.10.016
Segata, N., Izard, J., Waldron, L., Gevers, D., Miropolsky, L., Garrett, W., et al. (2011). Metagenomic biomarker discovery and explanation. Genome Biol. 12:R60. doi: 10.1186/gb-2011-12-6-r60
Shi, Y., Cao, X., Fu, D., and Wang, Y. (2018). Comprehensive value discovery of land consolidation projects: An empirical analysis of Shanghai, China. Sustainability 10:2039. doi: 10.3390/su10062039
Song, Y., Li, X., Yao, S., Yang, X., and Jiang, X. (2020). Correlations between soil metabolomics and bacterial community structures in the pepper rhizosphere under plastic greenhouse cultivation. Sci. Total Environ. 728:138439. doi: 10.1016/j.scitotenv.2020.138439
State Council of China. (2020). Available at: https://language.chinadaily.com.cn/a/202006/01/WS5ed46379a310a8b241159ce5.html
Su, Y., Qian, K., Lin, L., Wang, K., Guan, T., and Gan, M. (2020). Identifying the driving forces of non-grain production expansion in rural China and its implications for policies on cultivated land protection. Land Use Pol. 92:104435. doi: 10.1016/j.landusepol.2019.104435
Su, S., Yang, C., Hu, Y., Luo, F., and Wang, Y. (2014). Progressive landscape fragmentation in relation to cash crop cultivation. Appl. Geogr. 53, 20–31. doi: 10.1016/j.apgeog.2014.06.002
Sun, M., Li, T., Li, D., Zhao, Y., Gao, F., Sun, L., et al. (2021). Conversion of land use from upland to paddy field changes soil bacterial community structure in Mollisols of Northeast China. Microb. Ecol. 81, 1018–1028. doi: 10.1007/s00248-020-01632-4
Tian, H., Qiao, J., Zhu, Y., Jia, X., and Shao, M. (2021). Vertical distribution of soil available phosphorus and soil available potassium in the critical zone on the loess plateau, China. Sci. Rep. 11:3159. doi: 10.1038/s41598-021-82677-4
Vance, E. D., Brookes, P. C., and Jenkinson, D. S. (1987). An extraction method for measuring soil microbial biomass C. Soil Biol. Biochem. 19, 703–707. doi: 10.1016/0038-0717(87)90052-6
Wan, Q., Li, L., Liu, B., Zhang, Z., Liu, Y., and Xie, M. (2023). Different and unified responses of soil bacterial and fungal community composition and predicted functional potential to 3 years’ drought stress in a semiarid alpine grassland. Front. Microbiol. 14:1104944. doi: 10.3389/fmicb.2023.1104944
Wan, X., Wu, S., Li, Z., An, X., and Tian, Y. (2020). Lipid metabolism: critical roles in male fertility and other aspects of reproductive development in plants. Mol. Plant 13, 955–983. doi: 10.1016/j.molp.2020.05.009
Wang, N., Ai, Z., Zhang, Q., Leng, P., Qiao, Y., Li, Z., et al. (2024). Impacts of nitrogen (N), phosphorus (P), and potassium (K) fertilizers on maize yields, nutrient use efficiency, and soil nutrient balance: insights from a long-term diverse NPK omission experiment in the North China plain. Field Crop Res. 318:109616. doi: 10.1016/j.fcr.2024.109616
Wang, M., Chen, S., Chen, L., and Wang, D. (2020). Microbial mechanisms responsible for the variation of soil cd availability under different pe+pH environments. Ecotoxicol. Environ. Saf. 206:111057. doi: 10.1016/j.ecoenv2020.111057
Wang, L., Chen, M., Lam, P., Dini-Andreote, F., Dai, L., and Wei, Z. (2022). Multifaceted roles of flavonoids mediating plant-microbe interactions. Microbiome 10:233. doi: 10.1186/s40168-022-01420-x
Wang, X., Chi, Y., and Song, S. (2024). Important soil microbiota’s effects on plants and soils: a comprehensive 30-year systematic literature review. Front. Microbiol. 15:1347745. doi: 10.3389/fmicb.2024.1347745
Wang, S., Duan, S., George, T. S., Feng, G., and Zhang, L. (2024). Adding plant metabolites improve plant phosphorus uptake by altering the rhizosphere bacterial community structure. Plant Soil 497, 503–522. doi: 10.1007/s11104-023-06409-5
Wang, L., Fang, H., Xue, Z., De, J., and Guo, X. (2023b). Agrochemical exposure-induced seed microbiome response in barley. Crop Health 1:16. doi: 10.1007/s44297-023-00013-w
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2007). Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microb. 73, 5261–5267. doi: 10.1128/AEM.00062-07
Wang, J., Liao, L., Ye, Z., Liu, H., Zhang, C., Zhang, L., et al. (2022). Different bacterial co-occurrence patterns and community assembly between rhizosphere and bulk soils under N addition in the plant–soil system. Plant Soil 471, 697–713. doi: 10.1007/s11104-021-05214-2
Wang, R., Min, J., Kronzucker, H. J., Li, Y., and Shi, W. (2019). N and P runoff losses in China’s vegetable production systems: loss characteristics, impact, and management practices. Sci. Total Environ. 663, 971–979. doi: 10.1016/j.scitotenv.2019.01.368
Wang, L., Wang, K., and Sheng, M. (2023a). Changes in land use are associated with the accumulated with the accumulation of soil pehytoluth-occluded organic carbon. Ecol. Indic. 151:110300. doi: 10.1016/j.ecolind.2023.110300
Wang, J., Zhang, Z., and Liu, Y. (2018). Spatial shifts in grain production increases in China and implications for food security. Land Use Pol. 74, 204–213. doi: 10.1016/j.landusepol.2017.11.037
Weiss, S., Van Treuren, W., Lozupone, C., Faust, K., Friedman, J., Deng, Y., et al. (2016). Correlation detection strategies in microbial data sets vary widely in sensitivity and precision. ISME J. 10, 1669–1681. doi: 10.1038/ismej.2015.235
Wu, L., Wen, C., Qin, Y., Yin, H., Tu, Q., Van Nostrand, J. D., et al. (2015). Phasing amplicon sequencing on Illumina Miseq for robust environmental microbial community analysis. BMC Microbiol. 15:125. doi: 10.1186/s12866-015-0450-4
Wu, Y., Yuan, C., Liu, Z., Wu, H., and Wei, X. (2023). Decoupling relationship between the non-grain production and intensification of cultivated land in China based on Tapio decoupling model. J. Clean. Prod. 424:8800. doi: 10.1016/j.jclepro.2023.138800
Yang, Y., Shi, Y., Fang, J., Chu, H., and Adams, J. M. (2022). Soil icrobial network complexity varies with pH as a continuum, not a threshold across the North China plain. Front. Microbiol. 13:895687. doi: 10.3389/fmicb.2022.895687
Yu, F., Jayawardena, R. S., Thongklang, N., Lv, M., Zhu, X., and Zhao, Q. (2022). Morel production associated with soil nitrogen-fixing and nitrifying microorganisms. J. Fungi 8:299. doi: 10.3390/jof8030299
Yu, W., and Zhang, P. (2019). Study on the spatial-temporal differentiation characteristics and influencing factors of agricultural development resilience in China. Geogr. Geo Inf. Sci. 35, 102–108. (In Chinese). doi: 10.3969/j.issn.1672-0504.2019.01.016
Yuan, M., Seale, J. L. Jr., Wahl, T., and Bai, J. (2019). The changing dietary patterns and health issues in China. China Agr. Econ. Rev. 11, 143–159. doi: 10.1108/CAER-12-2017-0254
Zhao, G., Wu, K., An, T., Wen, L., Zi, S., Fan, Z., et al. (2022). Integrated analysis of changes in soil microbiota and metabolites following long-term fertilization in a subtropical maize-wheat agroecosystem. Pedosphere 33, 521–533. doi: 10.1016/j.pedsph.2022.06.055
Keywords: non-grain cultivated land, converted paddy fields, soil bacterial community structure, soil metabolite, soil physicochemical properties
Citation: Li X, Chen H, Wang X, Ali Q, Lv L, Zhou T, Ijaz M, Ahmed T, Yan J and Li B (2025) Reshaping of soil properties and microbial community by the conversion from non-grain cultivated land to paddy field. Front. Microbiol. 16:1643144. doi: 10.3389/fmicb.2025.1643144
Edited by:
Decai Jin, Chinese Academy of Sciences (CAS), ChinaCopyright © 2025 Li, Chen, Wang, Ali, Lv, Zhou, Ijaz, Ahmed, Yan and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianli Yan, eWFuamlhbmxpMDBAZ21haWwuY29t; Qurban Ali, cmF0dGFycXVyYmFuQHVhZXUuYWMuYWU=
†These authors have contributed equally to this work and share first authorship
 Xuqing Li
Xuqing Li Han Chen1†
Han Chen1† Qurban Ali
Qurban Ali Temoor Ahmed
Temoor Ahmed Bin Li
Bin Li