- 1Laboratory of Deep-Sea Microbial Cell Biology, Institute of Deep-Sea Science and Engineering, Chinese Academy of Sciences, Sanya, China
- 2College of Marine Sciences, University of Chinese Academy of Sciences, Beijing, China
- 3International Associated Laboratory of Evolution and Development of Magnetotactic Multicellular Organisms, CNRS-Marseille/CAS, Sanya, China
- 4Hainan Deep-Sea Technology Laboratory, Institution of Deep-Sea Life Sciences, IDSSE-BGI, Sanya, Hainan, China
- 5Laboratory of Marine Viruses and Molecular Biology, Institute of Deep-Sea Science and Engineering, Chinese Academy of Sciences, Sanya, China
- 6State Key Laboratory of Proteomics, Beijing Proteome Research Center, National Center for Protein Sciences Beijing, Beijing Institute of Lifeomics, Beijing, China
- 7CNRS, LCB, IMM, IM2B, Aix Marseille University, Marseille, France
Thermococcus species are ubiquitously distributed across both shallow and deep-sea hydrothermal vent ecosystems. Elemental sulfur (S°) reduction plays a pivotal role in their energy metabolism. While extensive characterization of the MBS and MBH pathways, along with their SurR-dependent regulatory network, has been established in shallow-water model strains, understanding of the high hydrostatic pressure (HHP) and sulfur-responsive regulation of these pathways in deep-sea Thermococcus lineages remains limited. In this study, we investigated the effects of HHP on both growth and S° reduction in the deep-sea SY113 strain, as well as its regulatory impact on mbs and mbh expression. Our results demonstrate that HHP enhances both S° reduction and growth in SY113 strain, independent of the general regulator SurR. Genetic disruption of mbsL significantly impaired H2S production and growth under HHP conditions, establishing the essential role of S° reduction in HHP adaptation. Furthermore, disrupted mbhL1 gene confirmed that a single MBS complex is sufficient to maintain pressure-stimulated growth. The gene expression analysis revealed that the expression of mbsL gene is primarily promoted by S°, while the expression of mbhL1 gene is induced by HHP. Moreover, the expression of these genes exhibits correlation. Additionally, we found that the expression of mbsL gene, mbhL1 gene, and mbhL2 gene in SY113 strain is not only regulated by SurR, and HHP also plays a role in modulating the expression of these genes. Overall, the sulfur responsive regulation of gene expression in SY113 strain distinguishes from that in the shallow model strains, which implies an adaptive strategy for Thermococcus species used to dwell in the deep-sea hydrothermal vent.
1 Introduction
The order Thermococcales is widely distributed across geothermal habitats, including shallow and deep-sea hydrothermal vents, as well as high-temperature oil reservoirs (Schut et al., 2014). It consists of three genera, Pyrococcus (Fiala and Stetter, 1986), Thermococcus (Zillig et al., 1983) and Paleococcus (Takai et al., 2000), these microorganisms represent deep evolutionary branches within the domain Archaea (Stetter, 2006; Canganella and Wiegel, 2014). Their phylogenetic position makes them particularly valuable for understanding early stages of unicellular life evolution.
Members of the Thermococcales order are hyperthermophilic, anaerobic heterotrophs that utilize peptides, amino acids, and oligosaccharides as carbon sources, while employing protons or S° as terminal electron acceptors to produce molecular hydrogen (H2) or hydrogen sulfide (H2S) (Leigh et al., 2011; Schut et al., 2014). While S0 generally enhances growth and is essential for certain Thermococcales species, it's extremely low solubility necessitates conversion to biologically accessible forms. Cyclooctasulfur is the main form of S0 and its S8 ring reacts readily with H2S to form soluble polysulfide (Wu et al., 2018). These polysulfides serve as substrates for the membrane-bound sulfane reductase (MBS), first characterized in Pyrococcus furiosus (Schut et al., 2007). The MBS complex catalyzes the reduction of organic and anionic polysulfides using reduced ferredoxins as electron donors, ultimately yielding H2S through spontaneous disproportionation of the resulting small anionic polysulfides (Wu et al., 2018).
MBS is encoded by 13 genes (mbsABCDEGHH'MJKLN) and composed of a transmembrane MbsABCDEGHH'M complex and a cytoplasmic MbsJKLN module. MbsM anchors the cytoplasmic module to the transmembrane module. Among them, the MbsJKLN module transfers the electrons from the ferredoxin to the substrate polysulfide with the MbsL as the catalytic subunit (Schut et al., 2013; Wu et al., 2018; Yu et al., 2020, 2021). In the transmembrane module, both MbsH and MbsH' subunits function as H+ pumps that transport protons out of the membrane and creates a proton gradient. The MbsABCDEG transmembrane sub-complex contains some H+/Na+-translocating units that generate a sodium ion gradient across the cell membrane using the proton gradient. The Na+-driven ATP synthase uses the created sodium ion gradient to produce ATP (Silva et al., 2000; Sapra et al., 2003; Wu et al., 2018). In Thermococcales genome, there is also a 14-gene cluster termed membrane-bound hydrogenase complex (MBH), which is highly homologous to MBS (Schut et al., 2013; Yu et al., 2018, 2021). The most obvious difference between them is MBH has additional cysteine residues in the [NiFe] binding motif of its catalytic subunit MbhL, and could use reduced ferredoxins to reduce proton, following H2 production (Schut et al., 2013; Yu et al., 2018). During this reaction, reducing equivalents are disposed of as H2, coupled with the generation of a sodium ion gradient to produce ATP (Sapra et al., 2003; Kanai et al., 2011; Schut et al., 2013). In addition, MBH contains only a single proton pump (MbhH), which means that MBH may conserve less energy than MBS.
Studies on shallow water model strains P. furiosus and Thermococcus kodakarensis demonstrated that the MBS and MBH were antagonistically regulated by the addition of S0 through a sulfur-dependent redox switch SurR (Lipscomb et al., 2009; Yang et al., 2010; Santangelo et al., 2011; Lipscomb et al., 2017). The DNA binding activity of SurR is dependent on the redox status of a CxxC motif located in its N-terminal region. In the absence of S0, SurR with reduced CxxC motif could bind to the target DNA sequence “GTTn3AAC” (Yang et al., 2010). This DNA motif is present in the promoter regions of both mbs and mbh. It is located upstream of the BRE/TATA element in the mbh promoter region, but overlap with the BRE/TATA element of mbs (Hidese et al., 2017). Consequently, SurR binding to the “GTTn3AAC” motif activates transcription of the mbh but prevents RNA polymerase recruitment and access of the transcription apparatus to the promoter, thus inhibit the transcription of mbs. In the presence of S0, the cysteine residues of the CxxC motif are oxidized and the oxidized SurR loses its capacity to bind the “GTTn3AAC” motif. Therefore, transcription of mbh is repressed, while mbs is de-repressed (Lipscomb et al., 2009; Yang et al., 2010).
At the time of writing, the order Thermococcales comprises 48 species with valid published names (https://lpsn.dsmz.de/order/thermococcales). Among them, 39 species belong to the genus Thermococcus, including 27 species isolated from deep-sea hydrothermal vents. Although most Thermococcus strains have been isolated from deep-sea hydrothermal vents, very little is known about how they adapt to the HHP. Studies of deep-sea strains of Pyrococcus yayanosii (Michoud and Jebbar, 2016), Thermococcus barophilus (Vannier et al., 2015) and Thermococcus piezophilus (Moalic et al., 2021) showed that HHP regulated the expression of genes associated with some metabolic pathways, especially pathways involved in energy production. Moreover, recent studies on T. barophilus have revealed that SurR-mediated regulation of energy metabolism genes is influenced by both sulfur availability and HHP (Moalic et al., 2025). In this study, we selected Thermococcus aciditolerans SY113, isolated from a deep-sea hydrothermal vent chimney on the Southwest Indian Ridge at a depth of 2,770 m (Li et al., 2021), as a representative deep-sea Thermococcus strain. We focused on the S0 reduction in connection with MBS and MBH, and the HHP regulation of the expression of these enzyme complexes. Here we show that HHP improves the S0 reduction and growth of SY113 strain. Disruption of mbsL gene impairs the H2S production and growth of SY113 strain under HHP, which indicates that S0 reduction plays an important role in the HHP tolerance. Furthermore, disruption of the mbhL1 gene resulted in increased growth rate, maximum biomass, and H2S production under HHP in the mutant strain compared to the wild type, further supporting the role of elemental sulfur reduction in enhancing HHP tolerance. Gene expression analysis revealed that SurR regulates mbs and mbh operons in SY113 strain in a manner distinct from shallow-water model strains. Notably, this SurR-mediated regulation exhibits conserved features with the deep-sea model strain T. barophilus MP (Moalic et al., 2025). Furthermore, HHP was identified as an additional regulatory factor influencing these operons. Collectively, these results reveal the adaptive strategy of strain SY113 to deep-sea hydrothermal vent environments, expanding our understanding of the regulatory mechanisms governing elemental sulfur metabolism in deep-sea Thermococcus species.
2 Materials and methods
2.1 Strains and growth conditions
T. aciditolerans SY113 was cultivated under anaerobic conditions in TRM medium at 85 °C (Zeng et al., 2009; Li et al., 2021). To investigate the growth of SY113 strain, culture was grown in TRM medium supplemented with S0 until the stationary phase, with OD600nm values ranging from 0.5 to 0.6. Then the culture was centrifuged at 2,000 × g and 4°C for 10 min to remove sulfur powder, transfer the supernatant liquid to a new tube and centrifuged again at 5,000 × g and 4°C for 15 min. The cell pellet was collected and washed three times with fresh anoxic TRM medium in an anaerobic chamber. The cells were then resuspended in anaerobic TRM medium to an OD600nm of ~1.0. The resuspended cells were inoculated into TRM medium at 1% (v/v), then dispensed into 11 ml serum bottles in an anaerobic chamber with complete liquid filling (no headspace). For sulfur-supplemented culture conditions, elemental sulfur was added at a final concentration of 2 g/l. High-pressure cultivation at 27 MPa was performed in a stainless steel pressure vessel (Nantong Feiyu Petroleum Technology Development Co., Ltd., China) equipped with a pressure gauge and an automated water-pumping system for pressurization control. For colony selection, the TRM medium was solidified with 1% (w/v) Gelzan CM (Sigma-Aldrich, CAS 71010-52-1), and polysulfide (10 g Na2S·9H2O was dissolved in 15 ml deionized water, followed by addition of 3 g sublimed sulfur powder) was used as a substitute for elemental sulfur at a final concentration of 10 μl/ml. Under these conditions, visible colonies typically developed within 2–3 days at 85 °C.
2.2 RNA extraction, cDNA synthesis and quantitative real-time PCR
For RNA extraction, ~3 × 109 cells in the mid-exponential growth phase were harvested by centrifugation at 8,000 × g for 20 min. Total RNA was extracted with a TRI reagent-RNA/DNA/protein isolation kit (Molecular Research Center, Inc.) according to the manufacturer's instructions, yielding ~20–30 μg of RNA per sample. For RNA purification and cDNA synthesis, the PrimeScriptTM II 1st strand cDNA synthesis kit (TAKARA, Shiga, Japan) was used. The RT-PCR was performed as described previously (Li et al., 2018). The expression levels of target genes were normalized to the reference gene por (pyruvate/ketoisovalerate ferredoxin oxidoreductase subunit gamma, POR, FPV09_09760). Each experiment was performed in triplicate for each sample, and a mean value and standard deviation were calculated for the relative RNA expression levels.
The absolute copy number of target genes was quantified by correlating the cycle threshold (Ct) values with a standard curve. The genes mbsL, mbhL1, and mbhL2 from SY113 strain were cloned into the pMD™19-T vector (TAKARA, Shiga, Japan). The recombinant plasmids were transformed into E. coli DH5α competent cells and verified by DNA sequencing. Plasmids containing mbsL, mbhL1, and mbhL2 were extracted and quantified using a NanoDrop 2000/2000c spectrophotometer (Thermo Scientific, Shanghai, China). Real-time quantitative PCR (qPCR) was conducted on a QuantStudio 3 system (Applied Biosystems). Gradient-diluted plasmids were used as templates to generate Ct values for the construction of standard curves, which were subsequently used to calculate the absolute copy numbers of the target genes.
2.3 Construction of gene deletion mutant
To overcome the limitations of available genetic markers, gene deletions were performed by replacing the target gene with the 3-hydroxy-3-methylglutaryl coenzyme A (HMG-CoA) reductase gene, which confers simvastatin resistance (SimR) as a positive selection marker. The SimR gene cassette was derived by fusing the promoter regions of glutamate dehydrogenase (PF1602) from P. furiosus DSM 3638 and HMG-CoA reductase (PYCH_01180) from P. yayanosii CH1 (Li et al., 2014). For counter-selection, an HHP-responsive toxin-antitoxin cassette (TAC) was utilized. The toxin gene is controlled by a HHP-inducible promoter, while the antitoxin gene is constitutively expressed under the Pgdh promoter to minimize basal toxin activity and eliminate background growth (Song et al., 2019).
Using the SY113 genome as a template, ~1 kb upstream and downstream regions of the mbsL, mbhL1, and surR genes were amplified using the primer pairs 04690-up-F/R, 03665-up-F/R, and 08560-up-F/R, respectively (Supplementary Table 1). The SimR cassette was amplified from the pUSW01 plasmid using primers Pgdh-F and HMG-CoA-R, and the HHP-TAC cassette was amplified using primers hhp-tac-F/R (Supplementary Table 1). The upstream and downstream fragments, along with the SimR and HHP-TAC cassette, were assembled into a linearized pUC18 vector containing 15 bp overlapping sequences using the ClonExpress® Ultra One Step Cloning Kit (Vazyme). The resulting suicide plasmids-pUS04690, pUS03665, and pUS08560-were introduced into strain SY113 via CaCl2-mediated transformation method (Li et al., 2014), with a transformation efficiency of ~3.3 × 103 CFU per μg of plasmid DNA. Transformants were initially plated on solid TRM medium supplemented with 10 μM simvastatin and incubated at 85 °C for 2–3 days. Single colonies were then transferred to 5 ml of liquid TRM medium and incubated at 85 °C for 12–16 h. Take 500 μl of the culture and inoculate it into 50 ml of TRM medium. After incubating at 85 °C for 12–16 h, harvest the cells for genomic DNA extraction. Subsequently, verify the transformants by PCR amplification. Verified transformants were then inoculated into syringes and incubated in TRM under 27 MPa for 12 h to facilitate mutant selection via secondary homologous recombination. Following high-pressure incubation, cultures were plated onto solid TRM medium supplemented 10 μM simvastatin to isolate mutants resulting from secondary homologous recombination. The Hungate tube were incubated at 85 °C for 2–3 days. For each mutant strain, we typically selected 30–50 single colonies. These were subjected to liquid culture, followed by genomic DNA extraction, PCR amplification, and restriction enzyme digestion verification to obtain the gene knockout mutants. Overall, it takes ~3 weeks from vector construction to finally obtaining the mutant strain.
2.4 Quantification of H2S and H2
For quantitative determination of H2S, 100 μl of culture from the stationary phase was mixed with 1 ml of H2S absorption solution, and centrifuged at 12,000 × g for 10 min at 4 °C to remove the supernatant. Subsequently, 375 μl of N, N-dimethyl-p-phenylenediamine, and 375 μl ferric ammonium sulfate solutions were added, thoroughly mixed, and incubated at room temperature for 10 min. The absorbance was measured at 665 nm using a UV spectrophotometer. The specific H2S concentration measurement followed the protocol of the H2S assay kit (Solarbio, BC2050). The H2S production yield was determined by normalizing to OD600nm-measured biomass, using the formula: [H2S] sample—[H2S] blank control/OD600nm, where [H2S] represents the H2S concentration measured by methylene blue method.
For quantitative determination of H2, freshly prepared TRM medium was inoculated to an initial OD600nm of ~0.01. Then, 5 ml of inoculated culture was dispensed into 15 ml Hungate anaerobic tubes with N2 as the headspace gas. The tubes were incubated at 85 °C until the stationary phase was reached. Headspace gas samples were collected from the top of the sealed anaerobic tubes to quantify H2 production. Gas analysis was performed using a gas chromatograph (Trace GC Ultra, Thermo Scientific) equipped with a thermal conductivity detector (TCD) set at 250 °C. Chromatographic separation was achieved using a CBP-01 column (6 ft [1.83 m] × 1/8 inch), with the injector and column oven temperatures set at 200 and 50 °C, respectively. Nitrogen served as the carrier gas at a flow rate of 20 ml/min, and the injection volume was 0.1 ml. The H2 production yield was determined by normalizing to OD600nm-measured biomass.
3 Results
3.1 HHP enhance the growth and S0 reduction of SY113 strain
To investigate the effects of HHP on the growth and S0 metabolism, SY113 strain was cultured at 0.1 MPa and 27 MPa, both with and without S0 supplementation. As shown in Figure 1A1, in the absence of S0, the SY113 strain exhibited only limited growth under both 0.1 MPa and 27 MPa (corresponding to the strain's isolation depth). The addition of S0 significantly promoted the growth of the SY113 strain at atmospheric pressure. Notably, application of HHP further improved the growth. At 0.1 MPa, S0 supplementation increased the maximum biomass of the SY113 strain by ~6.5-fold (OD600nm: 0.26 ± 0.01 with S0 vs. 0.04 ± 0.02 without S0). At 27 MPa, the biomass increased by about 11.6-fold (OD600nm: 0.58 ± 0.04 with S0 vs. 0.05 ± 0.02 without S0). Notably, under HHP conditions with S0, not only enhanced the growth rate (0.32 ± 0.04 h−1 at 0.1 MPa vs. 0.47 ± 0.06 h−1 at 27 MPa), but also increased the H2S concentrations from 10,062.03 ± 461.81 mmol/OD600nm at 0.1 MPa to 13,118.21 ± 582.30 mmol/OD600nm at 27 MPa (Figure 1B1). These results suggest that HHP promotes the growth of SY113 strain by enhancing H2S production.
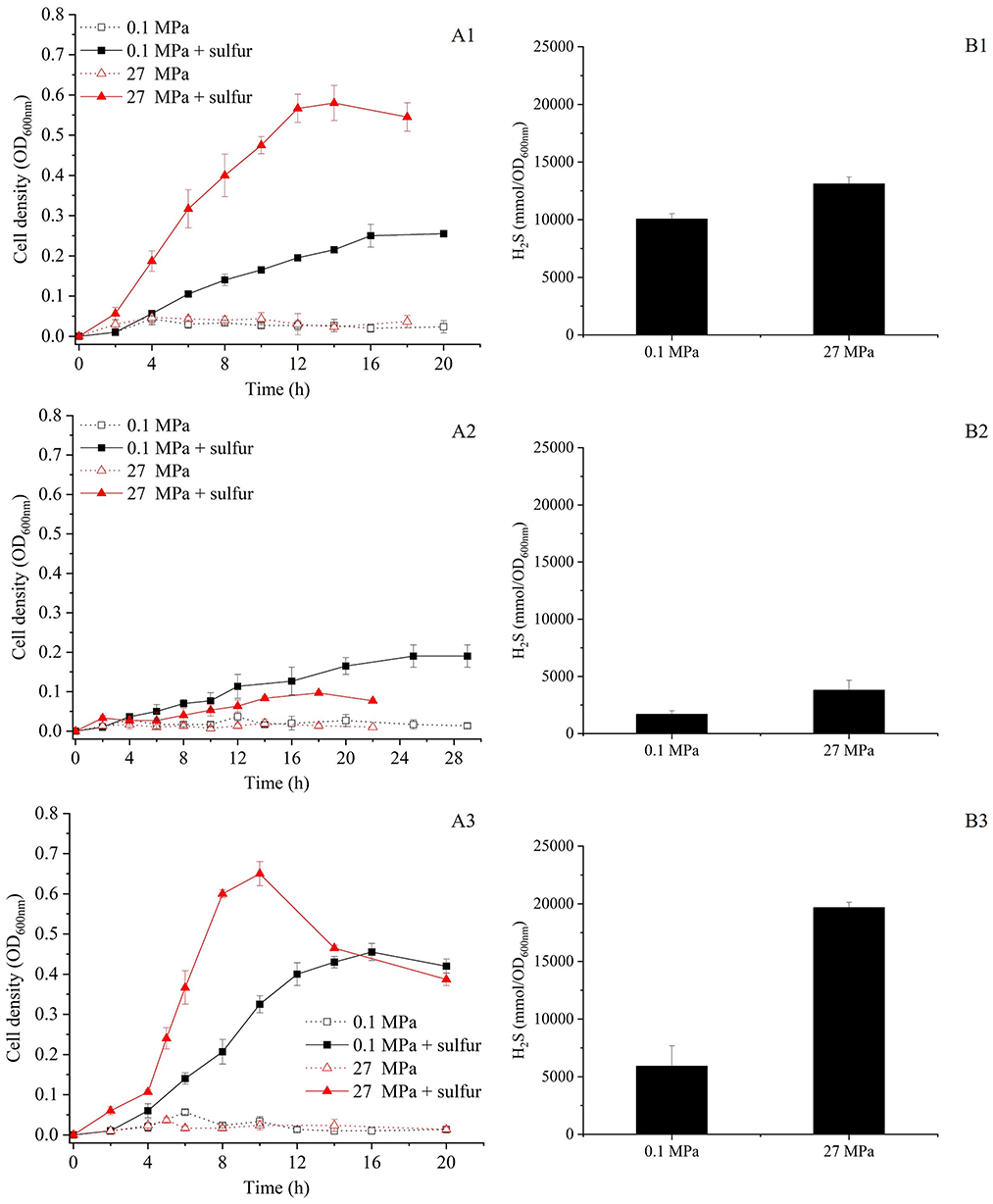
Figure 1. Growth curve and H2S production of the SY113 strain and its ΔmbsL and ΔmbhL1 mutants. (A1–A3) Growth curves of SY113 (A1), ΔmbsL (A2), and ΔmbhL1 (A3) cultured at 0.1 MPa and 27 MPa in the presence or absence of elemental sulfur, measured by OD600nm over time. (B1–B3) Corresponding H2S concentrations for the SY113 strain (B1), ΔmbsL (B2), and ΔmbhL1 (B3) measured at stationary phase. Black bars represent cultures with sulfur supplementation, error bars represent the standard deviation of three biological replicates. No H2S was produced in WT, ΔmbsL and ΔmbhL1 mutants in the absence of elemental sulfur.
3.2 HHP and S0 regulated gene expression of mbsL and mbhL
To further evaluate the impact of HHP and S0 on the gene expression of SY113 strain, we first analyzed the S0 reduction related genes in its genome. As shown in Figure 2, SY113 strain harbors a well conserved mbs gene cluster (FPV09_03610 to FPV09_03670), with gene composition and arrangement identical to those of shallow-water model strains. All corresponding subunits exhibited high similarity (average 85.6%) to those of the shallow-sea strain T. kodakarensis KOD1 (Supplementary Table 2). In addition, SY113 strain had two copies of mbh gene clusters, which were located next to each other on the chromosome. One copy, named as MBH1 (FPV09_08505 to FPV09_08570), showed a high sequence identity (average 73.3%) with shallow-sea strain KOD1, while the other copy, MBH2 (FPV09_08575 to FPV09_08645), possessed a relatively low sequence identity (average 44.5%) (Supplementary Table 2).
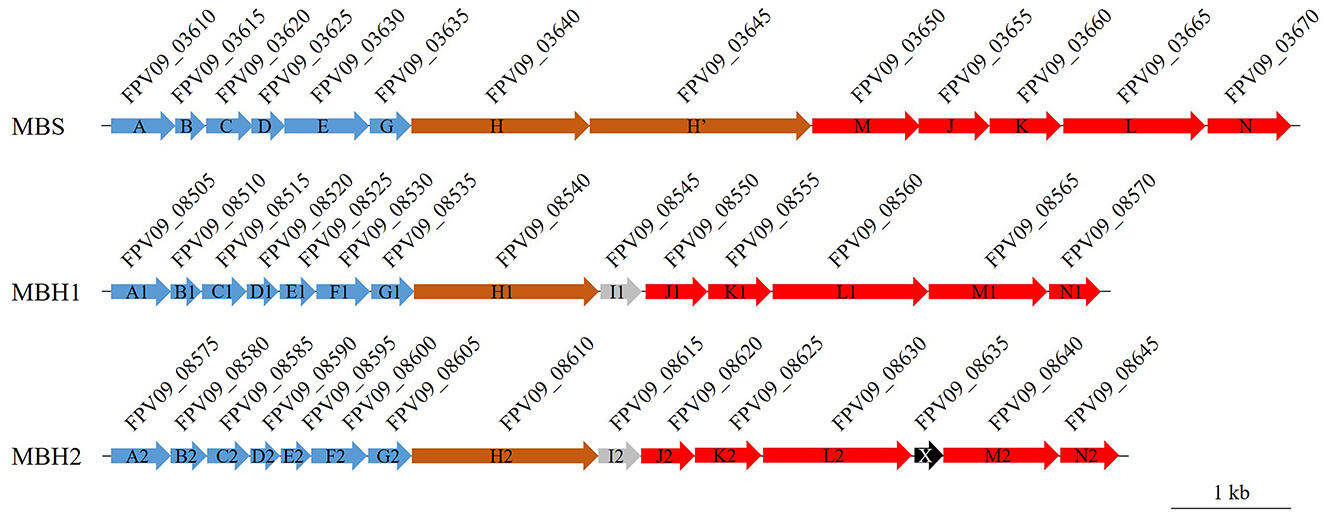
Figure 2. Gene cluster organization of the MBS, MBH1, and MBH2 complexes in SY113 strain. MBS, MBH1, and MBH2 are membrane-bound complexes involved in elemental sulfur or proton reduction. Arrows indicate gene order and transcriptional direction. Genes are color-coded based on function: blue (Na+/H+ antiporters), brown (proton pumps), red (ferredoxin-dependent oxidation–sulfur reduction modules), and gray/black (proteins of unknown function). The locus tags of genes are displayed above each corresponding gene. Scale bar = 1 kb.
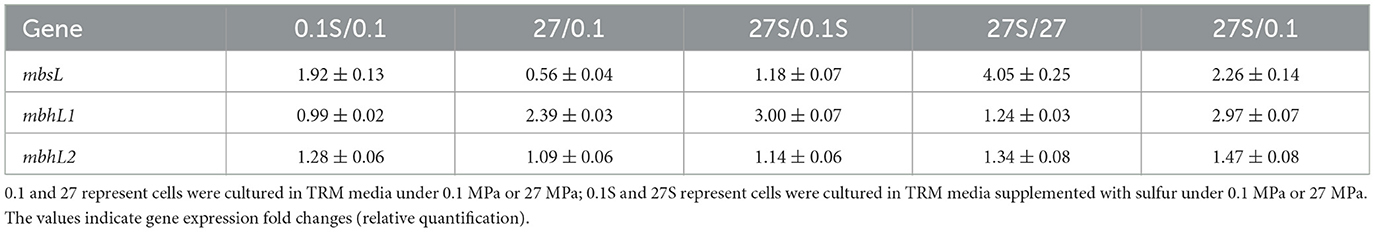
Using the catalytic subunit encoding genes mbsL (FPV09_03665), mbhL1 (FPV09_08560) and mbhL2 (FPV09_08630) as reference, we analyzed the effects of HHP and S0 on the transcriptional levels of these gene clusters. As shown in Table 1, the expression of mbsL was primarily enhanced by S0, while HHP showed no significant regulatory effect. The addition of S0 induced the expression of mbsL gene at both 0.1 MPa (0.1S/0.1, RQ: 1.92 ± 0.13) and 27 MPa (27S/27, RQ: 4.05 ± 0.25). However, when HHP was applied, no significant expression changes in mbsL gene were observed in either S0-supplemented (27S/0.1S, RQ: 1.18 ± 0.07) or S0-free (27/0.1, RQ: 0.56 ± 0.04) conditions. Notably, combination of S0 and HHP produced a synergistic effect, resulting in a 2.26-fold upregulation of mbsL gene expression compared to control conditions (27S/0.1, RQ: 2.26 ± 0.14). On the contrary, HHP markedly enhanced the mbhL1 gene expression irrespective of S0 availability (27/0.1, RQ: 2.39 ± 0.03 and 27S/0.1S, RQ: 3.00 ± 0.07). In contrast, S0 had no effect on mbhL1 gene expression (0.1S/0.1, RQ: 0.99 ± 0.02 and 27S/27, RQ: 1.24 ± 0.03). Strikingly, the combined application of S0 and HHP synergistically enhanced mbhL1 gene expression by ~3-fold (27S/0.1, RQ: 2.97 ± 0.03). For another copy of mbhL2 gene, neither S0 nor HHP treatment exerted any significant regulatory effects.
We further analyzed the transcriptional abundance of these genes under different conditions by means of absolute quantitative RT-PCR. As shown in Supplementary Figure 1, the transcript abundance of mbhL1 gene at 0.1 MPa was modestly elevated relative to mbsL gene (1.4-fold and 1.9-fold with or without S0, respectively). At 27 MPa, this difference became more pronounced, with mbhL1 gene expression exceeding mbsL gene by 3.2-fold and 7.1-fold under S0-supplemented and S0-free conditions, respectively. In contrast to both mbsL gene and mbhL1 gene, mbhL2 gene exhibited significantly low transcriptional abundance (10-fold lower) in all tested conditions. Collectively, these findings demonstrate distinct regulatory responses among the three gene clusters, MBS expression was predominantly induced by S0, and MBH1 was primarily activated by HHP, while MBH2 cluster remained transcriptionally inert under all tested conditions.
3.3 MBS is essential for HHP-promoted growth of SY113 strain
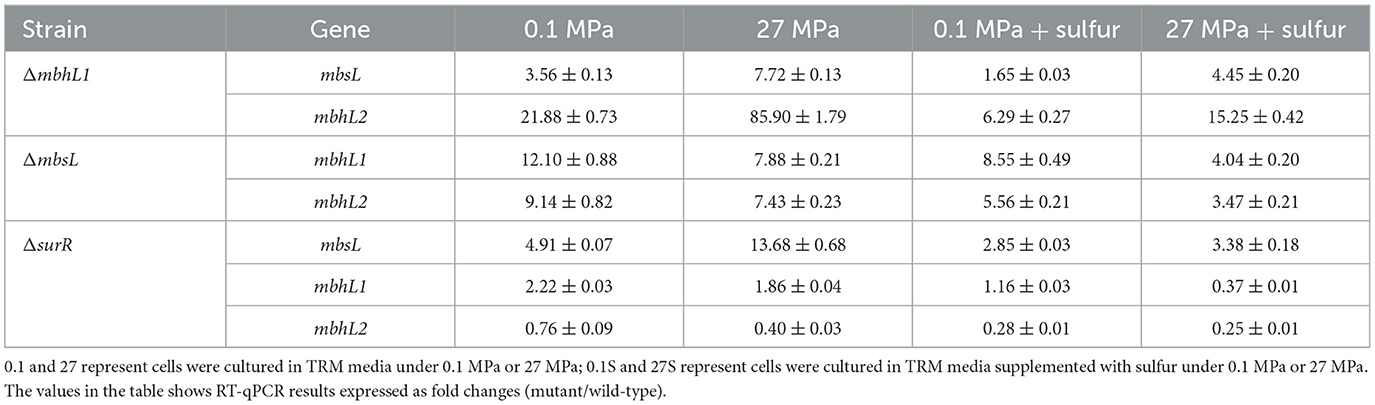
To further investigate the roles of MBS and MBH1 in HHP-enhanced growth, we first inactivated the mbsL gene (Supplementary Figure 2A). As illustrated in Figure 1A2, the disruption of the mbsL gene significantly impaired the growth of the SY113 strain under S0-supplemented conditions, with particularly pronounced inhibition observed at 27 MPa. At 0.1 MPa, the maximum biomass decreased from OD600nm 0.26 to 0.19 and the growth rate decreased from 0.32 h−1 to 0.18 h−1. While at 27 MPa, the maximum biomass decreased by 5.8 times (OD600nm: 0.58 ± 0.04 for wild-type vs. 0.10 ± 0.01 for ΔmbsL) and the growth rate decreased by 3.4 times (0.47 ± 0.06 h−1 for wild-type vs. 0.14 ± 0.04 h−1 for ΔmbsL). Thus, the strain's growth phenotype shifted from piezophilic to piezosensitive. Notably, under S0-supplemented conditions, the ΔmbsL mutant exhibited a pronounced reduction in H2S production at both 0.1 MPa and 27 MPa compared to the wild-type strain. Specifically, H2S yields decreased from 10,062.03 ± 461.81 mmol/OD600nm (wild-type) to 1,685.89 ± 307.28 nmol/OD600nm (ΔmbsL) at 0.1 MPa, and from 13,118.21 ± 582.30 mmol/OD600nm to 3,790.13 ± 865.36 nmol/OD600nm at 27 MPa, representing 76.5% and 68.2% reductions, respectively (Figures 1B1, B2).
Moreover, we further disrupted the mbhL1 gene to investigate the growth and metabolic profiles of the strain when only the MBS was functional (Supplementary Figure 2B). Surprisingly, compared to the wild-type strain, the ΔmbhL1 strain exhibited enhanced growth in the presence of S0 (Figure 1A3). At 0.1 MPa, the maximum biomass increased by 1.8 times (OD600nm: 0.46 ± 0.02 for ΔmbhL1 vs. 0.26 ± 0.01 for wild-type) and the growth rate increased by 1.6 times (0.50 ± 0.05 h−1 for ΔmbhL1 vs. 0.32 ± 0.04 h−1 for wild-type). While at 27 MPa, the maximum biomass increased from 0.58 to 0.65 and the growth rate increased by 1.8 times (0.86 ± 0.1 h−1 for ΔmbhL1 vs. 0.47 ± 0.06 h−1 for wild-type). The ΔmbhL1 strain produced H2S concentrations of 5,912.02 ± 1,775.23 mmol/OD600nm and 19,676.19 ± 455.25 mmol/OD600nm at 0.1 MPa and 27 MPa (Figure 1B3), respectively, which were comparable to those of the wild-type strain. These results indicate that MBS alone is sufficient for HHP-enhanced growth.
3.4 The expression of MBS and MBH in SY113 strain is correlated
Although HHP stimulated mbhL1 gene expression, disruption of this gene unexpectedly enhanced the growth of SY113 strain. To resolve this discrepancy, we analyzed the expression patterns of mbsL gene and mbhL2 gene in the ΔmbhL1 mutant. As shown in Table 2, compared to the wild-type strain, mbhL1 gene disruption led to significantly elevated mbsL gene expression under all tested conditions, except for only a 1.65-fold increase at 0.1 MPa with S0 supplementation. Notably, mbhL2 gene expression was also markedly upregulated in the ΔmbhL1 mutant, particularly under HHP conditions (Table 2). These results suggest a regulatory link between mbhL1 gene and the expression of mbsL gene and mbhL2 gene. Furthermore, we examined mbhL1 gene and mbhL2 gene expression in the ΔmbsL mutant. Similarly, mbsL gene disruption resulted in significant upregulation of both mbhL1 gene and mbhL2 gene across all tested conditions (Table 2). Collectively, these findings demonstrate that the expression of MBS and MBH genes is interdependent.
3.5 The HHP-promoted growth of SY113 strain is independent of SurR regulator
In shallow-water model strains, sulfur promotes growth by altering the conformation of SurR, which subsequently enhances MBS expression while repressing MBH expression (Lipscomb et al., 2009; Yang et al., 2010). However, how HHP interacts with SurR in SY113 strain is yet to be elucidated. Analysis of the SY113 genome identified a SurR homologous gene (FPV09_04690) flanked by glutaredoxin and ferredoxin, a syntenic region characteristic of Thermococcales. To investigate the potential role of SurR in HHP response, we generated a ΔsurR mutant (Supplementary Figure 2C). Surprisingly, surR gene disruption did not impair the HHP-promoted growth. On the contrary, the ΔsurR mutant demonstrated superior growth performance under HHP compared to the wild-type strain (Figure 3A). At 0.1 MPa, the ΔsurR mutant achieved a maximum biomass of OD600 = 0.36 ± 0.03 with a growth rate of 0.38 ± 0.04 h−1, whereas at 27 MPa it showed increased maximum biomass (OD600nm = 0.73 ± 0.06) and significantly enhanced growth rate (1.13 ± 0.16 h−1). Consistently, H2S production under sulfur-supplemented conditions reached 7,561.41 ± 1,448.27 mmol/OD600nm at 0.1 MPa and 15,350.00 ± 1,024.52 mmol/OD600nm at 27 MPa (Figure 3B).
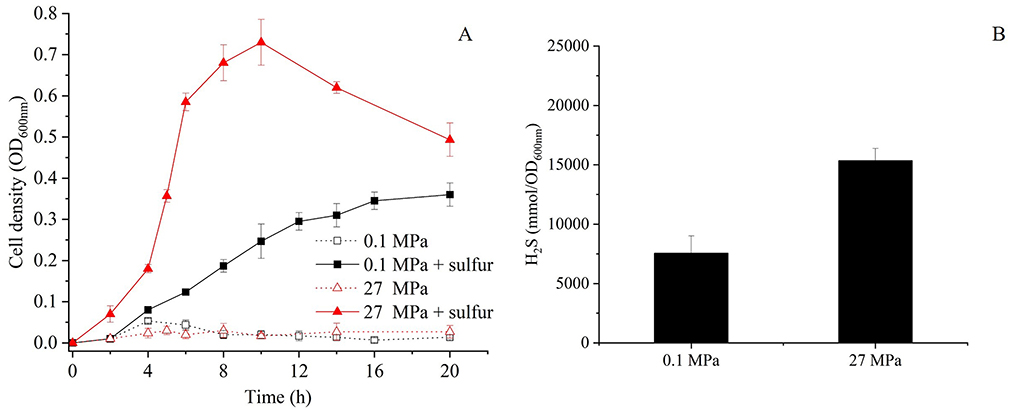
Figure 3. Growth curve and H2S production of the ΔsurR mutants at 0.1 MPa and 27 MPa with or without elemental sulfur. (A) Growth curves of ΔsurR cultured at 0.1 MPa and 27 MPa in the presence or absence of elemental sulfur, measured by OD600nm over time. (B) Corresponding H2S concentrations for ΔsurR strain measured after incubation under the same conditions. Black bars represent cultures with sulfur supplementation, error bars represent the standard deviation of three biological replicates. No H2S was produced in ΔsurR mutant in the absence of elemental sulfur.
We further analyzed the transcriptional profiles of mbsL gene, mbhL1 gene and mbhL2 gene following surR gene disruption. As shown in Table 2, surR gene disruption resulted in upregulated expression of mbsL gene under all tested conditions, particularly under HHP. In contrast, mbhL2 gene exhibited significant expression downregulation in S0-supplemented conditions upon surR gene disruption. Disruption of the surR gene also resulted in a slight expression upregulation of mbhL1 gene at 0.1 MPa (RQ: 2.22 ± 0.03), while causing a slight downregulation under 27 MPa with S0 supplementation (RQ: 0.37 ± 0.01). No significant changes in expression were observed under the other two tested conditions. These findings demonstrate that SurR does not directly regulate either HHP-promoted growth or elemental sulfur reduction.
4 Discussion
S0 reduction for ATP generation represents a crucial energy metabolism pathway in Thermococcus. While studies on shallow-water model strains have described their S0 reduction pathway and regulatory mechanisms, the corresponding pathways and regulatory networks in deep-sea Thermococcus strain remain poorly characterized. Members of the Thermococcus genus typically possess two energy metabolic complexes MBH and MBS. The former reduces protons to generate H2, while the latter reduces elemental sulfur to produce H2S. In this study, we observed strikingly different HHP responses between these two metabolic pathways (Figure 1). The ΔmbsL mutant exhibited significantly reduced H2S production but markedly increased H2 production, accompanied by impaired growth under HHP (Figure 1, Supplementary Figure 3). Conversely, the ΔmbhL1 mutant nearly ceased H2 production while showing enhanced H2S generation, with improved HHP growth performance (Figure 1, Supplementary Figure 3). These findings demonstrate that sulfur reduction confers superior pressure tolerance. Further analysis revealed that the MBS complex contains two proton-pumping subunits (MbsH and MbsH'), compared to just one (MbhH) in MBH (Figure 2), theoretically doubling the energy production efficiency of MBS relative to MBH (Yu et al., 2021). This may explain why the ΔmbsL mutant displayed greater pressure sensitivity than the ΔmbhL1 mutant.
Moreover, for the ΔmbhL1 mutant, there was no significant difference in H2S production compared to the wild-type strain at 0.1 MPa (P > 0.05), but it was significantly higher than that of the wild-type strain at 27 MPa (P < 0.001). Correspondingly, in the presence of S0, the expression level of mbsL gene in the ΔmbhL1 strain showed no significant difference at 0.1 MPa compared to the wild-type strain (RQ: 1.65 ± 0.03), but was significantly upregulated at 27 MPa (RQ: 4.45 ± 0.20). However, the mechanism by which HHP enhances mbsL gene expression remains unclear. Similarly, in the ΔsurR strain, mbsL gene expression was significantly upregulated at 27 MPa with S0 supplementation, indicating that SurR is not involved in the HHP regulation. These findings suggest that other regulatory factors may be responsible for HHP-dependent mbsL gene expression regulation.
The gene expression profile of SY113 strain revealed that the addition of S0 did not significantly alter the transcriptional levels of either mbsL gene (0.1S/0.1, RQ: 1.92 ± 0.13) or mbhL1 gene (0.1S/0.1, RQ: 0.99 ± 0.02) at 0.1 MPa. This regulatory pattern is significantly different from that in the extensively studied shallow strains P. furiosus (Schut et al., 2007) and T. kodakarensis (Santangelo et al., 2011). S0 significantly enhanced H2S production of the SY113 wild-type strain, while the transcription of mbsL gene did not show obvious upregulation. This indicates that the expression of mbsL gene is not regulated by SurR and remains at a high expression level even in the absence of S0. Analysis of the promoter region of mbs cluster in the SY113 strain revealed that, although it contains two SurR-binding consensus sequence (SBS) like the KOD1 strain, the middle three bases of the SBS differ (Supplementary Figure 4). Therefore, we hypothesize that one possible explanation is that the variation in the SBS bases prevents SurR from binding, while another possibility is that the SurR protein sequence in the SY113 strain differs from that in the KOD1 strain, rendering it unable to bind these SBS. Both scenarios would result in the high expression of mbs even without S0 supplementation. Further comparison of SurR sequences across different Thermococcus strains revealed that their N-terminal region and C-terminal region are relatively conserved, whereas the linker region exhibits low homology (Supplementary Figure 5). Differences in the linker region may alter the structure of SurR in the SY113 strain compared to KOD1 strain, affecting its binding to SBS.
As for mbhL1 gene, its expression abundance was consistently higher than that of mbsL gene regardless of the presence of S0 (Supplementary Figure 1), suggesting that S0 cannot regulate its gene expression through SurR. This regulatory mechanism is distinct from the SurR-dependent regulation observed in shallow-sea strains. In addition, the transcriptional disparity between mbhL1 gene and mbsL gene was more pronounced under high-pressure conditions, suggesting that high pressure also contributes to their transcriptional regulation. However, disrupting surR gene significantly suppressed mbhL1 gene transcription under HHP in the presence of elemental sulfur and completely inhibited H2 production (Supplementary Figure 3), indicating that expression of the mbhL1 gene depends on SurR regulation. A similar phenomenon was observed in the T. barophilus MP strain, where the expression level of the mbh1 was significantly suppressed after the knockout of the surR gene (Moalic et al., 2025). Analysis showed that the mbh1 promoter region contains two SBS, one located at upstream of the BRE/TATA and the other downstream (Supplementary Figure 4). We speculate that SurR binds to the upstream SBS to activate transcription regardless of the presence of S0 but cannot bind to the downstream SBS, resulting in the consistently high transcriptional level of mbhL1 gene. This regulatory pattern resembles the SurR-mediated regulation of ferredoxin: NADP+ oxidoreductases 2 (FNOR2) gene expression in T. kodakarensis. It showed that a proportion of reduced form of SurR persists even when S0 is supplemented in the culture medium. The reduced form of SurR maintains the ability to bind SBS in the FNOR2 promoter region, thereby activating its gene expression (Hidese et al., 2017).
Additionally, the SY113 strain harbors another copy of MBH. Disruption of mbhL1 gene led to a significant upregulation of mbhL2 gene, suggesting functional complementarity between the two systems. Therefore, we hypothesize that possessing dual MBH systems may enhance the strain's adaptability to deep-sea environments. Deep-sea microbial adaptation to HHP frequently involves metabolic pathway redundancy. A notable example is the deep-sea Shewanella piezotolerant WP3, which maintains duplicate periplasmic nitrate reductases systems (Li et al., 2018) and dimethyl sulfoxide reductase systems (Xiong et al., 2016), conferring exceptional pressure tolerance. We further analyzed 29 published Thermococcus genomes and found that 13 strains possess dual MBH systems (Supplementary Figure 6). Among these 13 strains, 12 clustered into two distinct evolutionary clades. One clade comprised 7 strains, all isolated from deep-sea hydrothermal vents. The other clade contained 5 strains: 2 from deep-sea hydrothermal vent, 2 from high temperature and high-pressure oil reservoirs, and was derived from a shallow submarine thermal spring. These results suggest that possessing dual MBH systems may facilitate these strains' adaptation to extreme high-temperature and/or high-pressure environments. It is noteworthy that HHP affects multiple aspects of cells, such as membrane fluidity, biomacromolecule function, and energy metabolic pathways (Oger and Jebbar, 2010). Correspondingly, microbial adaptation to HHP environments is not determined by a single factor but rather results from the combined effects of multiple factors.
5 Conclusion
Our findings revealed that the MBS-mediated elemental sulfur reduction pathway is important for the SY113 strain's adaptation to deep-sea HHP environments. Moreover, we found significant differences in the regulatory patterns of MBS and MBH between the deep-sea strain SY113 and shallow-water strains in response to elemental sulfur. Notably, HHP also modulates the expression of MBS and MBH, mirroring observations in other deep-sea piezophiles such as T. piezophilus and T. barophilus. This research uncovers the diversity of sulfur metabolism regulation in deep-sea Thermococcus species and expands our understanding of their HHP adaptation mechanisms.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
Z-XJ: Formal analysis, Writing – review & editing, Methodology, Writing – original draft, Investigation. X-GL: Funding acquisition, Writing – review & editing, Supervision, Writing – original draft, Conceptualization, Formal analysis. W-JZ: Supervision, Writing – review & editing, Conceptualization. G-YZ: Methodology, Investigation, Writing – review & editing. S-JB: Formal analysis, Writing – review & editing. LF: Writing – review & editing, Formal analysis, Conceptualization. L-FW: Supervision, Writing – review & editing, Conceptualization.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by National Key R&D Program of China (Grant No. 2023YFC2812800), National Natural Science Foundation of China (Grant Nos. NSFC 42376123 and 32270126), and Natural Science Foundation of Hainan Province (Grant No. 321RC616).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The author(s) declare that Gen AI was used in the creation of this manuscript. For English correction.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1643593/full#supplementary-material
References
Canganella, F., and Wiegel, J. (2014). Anaerobic thermophiles. Life 4, 77–104. doi: 10.3390/life4010077
Fiala, G., and Stetter, K. O. (1986). Pyrococcus furiosus sp. nov. represents a novel genus of marine heterotrophic archaebacteria growing optimally at 100 C. Arch. Microbiol. 145, 56–61. doi: 10.1007/BF00413027
Hidese, R., Yamashita, K., Kawazuma, K., Kanai, T., Atomi, H., Imanaka, T., et al. (2017). Gene regulation of two ferredoxin:NADP(+) oxidoreductases by the redox-responsive regulator SurR in Thermococcus kodakarensis. Extremophiles 21, 903–917. doi: 10.1007/s00792-017-0952-0
Kanai, T., Matsuoka, R., Beppu, H., Nakajima, A., Okada, Y., Atomi, H., et al. (2011). Distinct physiological roles of the three [NiFe]-hydrogenase orthologs in the hyperthermophilic archaeon Thermococcus kodakarensis. J. Bacteriol. 193, 3109–3116. doi: 10.1128/JB.01072-10
Leigh, J. A., Albers, S. V., Atomi, H., and Allers, T. (2011). Model organisms for genetics in the domain Archaea: methanogens, halophiles, Thermococcales and Sulfolobales. FEMS Microbiol. Rev. 35, 577–608. doi: 10.1111/j.1574-6976.2011.00265.x
Li, X., Fu, L., Li, Z., Ma, X., Xiao, X., Xu, J., et al. (2014). Genetic tools for the piezophilic hyperthermophilic archaeon Pyrococcus yayanosii. Extremophiles 19, 59–67. doi: 10.1007/s00792-014-0705-2
Li, X. G., Tang, H. Z., Zhang, W. J., Qi, X. Q., Qu, Z. G., Xu, J., et al. (2021). Thermococcus aciditolerans sp. nov., a piezotolerant, hyperthermophilic archaeon isolated from a deep-sea hydrothermal vent chimney in the Southwest Indian Ridge. Int. J. Syst. Evol. Microbiol. 71. doi: 10.1099/ijsem.0.004934
Li, X. G., Zhang, W. J., Xiao, X., Jian, H. H., Jiang, T., Tang, H. Z., et al. (2018). Pressure-regulated gene expression and enzymatic activity of the two periplasmic nitrate reductases in the deep-sea bacterium Shewanella piezotolerans WP3. Front. Microbiol. 9:3173. doi: 10.3389/fmicb.2018.03173
Lipscomb, G. L., Keese, A. M., Cowart, D. M., Schut, G. J., Thomm, M., Adams, M. W., et al. (2009). SurR: a transcriptional activator and repressor controlling hydrogen and elemental sulphur metabolism in Pyrococcus furiosus. Mol. Microbiol. 71, 332–349. doi: 10.1111/j.1365-2958.2008.06525.x
Lipscomb, G. L., Schut, G. J., Scott, R. A., and Adams, M. W. W. (2017). SurR is a master regulator of the primary electron flow pathways in the order Thermococcales. Mol. Microbiol. 104, 869–881. doi: 10.1111/mmi.13668
Michoud, G., and Jebbar, M. (2016). High hydrostatic pressure adaptive strategies in an obligate piezophile Pyrococcus yayanosii. Sci. Rep. 6:27289. doi: 10.1038/srep27289
Moalic, Y., Hartunians, J., Dalmasso, C., Courtine, D., Georges, M., Oger, P., et al. (2021). The piezo-hyperthermophilic archaeon thermococcus piezophilus regulates its energy efficiency system to cope with large hydrostatic pressure variations. Front. Microbiol. 12:2021. doi: 10.3389/fmicb.2021.730231
Moalic, Y., Nguyen, T. B. H., Hartunians, J., Birien, T., Thiel, A., Jebbar, M., et al. (2025). Regulation of gene expression under high hydrostatic pressure: the versatile role of the master regulator SurR in energy metabolism. Front. Microbiol. 16:1593936. doi: 10.3389/fmicb.2025.1593936
Oger, P. M., and Jebbar, M. (2010). The many ways of coping with pressure. Res. Microbiol. 161, 799–809. doi: 10.1016/j.resmic.2010.09.017
Santangelo, T. J., Cubonova, L., and Reeve, J. N. (2011). Deletion of alternative pathways for reductant recycling in Thermococcus kodakarensis increases hydrogen production. Mol. Microbiol. 81, 897–911. doi: 10.1111/j.1365-2958.2011.07734.x
Sapra, R., Bagramyan, K., and Adams, M. W. (2003). A simple energy-conserving system: proton reduction coupled to proton translocation. Proc. Natl. Acad. Sci. 100, 7545–7550. doi: 10.1073/pnas.1331436100
Schut, G., Lipscomb, G., Han, Y., Notey, J., Kelly, R., Adams, M., et al. (2014). “The order Thermococcales and the family Thermococcaceae,” in The Prokaryotes, eds. E. Rosenberg, E. F. DeLong, S. Lory, E. Stackebrandt, and F. Thompson (Berlin, Heidelberg: Springer), 363–383. doi: 10.1007/978-3-642-38954-2_324
Schut, G. J., Boyd, E. S., Peters, J. W., and Adams, M. W. (2013). The modular respiratory complexes involved in hydrogen and sulfur metabolism by heterotrophic hyperthermophilic archaea and their evolutionary implications. FEMS Microbiol. Rev. 37, 182–203. doi: 10.1111/j.1574-6976.2012.00346.x
Schut, G. J., Bridger, S. L., and Adams, M. W. W. (2007). Insights into the metabolism of elemental sulfur by the hyperthermophilic archaeon Pyrococcus furiosus: characterization of a coenzyme A-dependent NAD(P)H sulfur oxidoreductase. J. Bacteriol. 189, 4431–4441. doi: 10.1128/JB.00031-07
Silva, P. J., van den Ban, E. C., Wassink, H., Haaker, H., de Castro, B., and Robb, T. (2000). Enzymes of hydrogen metabolism in Pyrococcus furiosus. Eur. J. Biochem. 267, 6541–6551. doi: 10.1046/j.1432-1327.2000.01745.x
Song, Q., Li, Z., Chen, R., Ma, X., Xiao, X., Xu, J., et al. (2019). Induction of a toxin-antitoxin gene cassette under high hydrostatic pressure enables markerless gene disruption in the hyperthermophilic Archaeon Pyrococcus yayanosii. Appl. Environ. Microbiol. 85, e02662–18. doi: 10.1128/AEM.02662-18
Stetter, K. O. (2006). History of discovery of the first hyperthermophiles. Extremophiles 10, 357–362. doi: 10.1007/s00792-006-0012-7
Takai, K., Sugai, A., Itoh, T., and Horikoshi, K. (2000). Palaeococcus ferrophilus gen. nov., sp. nov., a barophilic, hyperthermophilic archaeon from a deep-sea hydrothermal vent chimney. Int. J. Syst. Evol. Microbiol. 50, 489–500. doi: 10.1099/00207713-50-2-489
Vannier, P., Michoud, G., Oger, P., þór Marteinsson, V., and Jebbar, M. (2015). Genome expression of Thermococcus barophilus and Thermococcus kodakarensis in response to different hydrostatic pressure conditions. Res. Microbiol. 166, 717–725. doi: 10.1016/j.resmic.2015.07.006
Wu, C. H., Schut, G. J., Poole, F. L. 2nd, Haja, D. K., and Adams, M. W. W. (2018). Characterization of membrane-bound sulfane reductase: a missing link in the evolution of modern day respiratory complexes. J. Biol. Chem. 293, 16687–16696. doi: 10.1074/jbc.RA118.005092
Xiong, L., Jian, H., Zhang, Y., and Xiao, X. (2016). The two sets of DMSO respiratory systems of Shewanella piezotolerans WP3 are involved in deep sea environmental adaptation. Front. Microbiol. 7:1418. doi: 10.3389/fmicb.2016.01418
Yang, H., Lipscomb, G. L., Keese, A. M., Schut, G. J., Thomm, M., Adams, M. W., et al. (2010). SurR regulates hydrogen production in Pyrococcus furiosus by a sulfur-dependent redox switch. Mol. Microbiol. 77, 1111–1122. doi: 10.1111/j.1365-2958.2010.07275.x
Yu, H., Wu, C. H., Schut, G. J., Haja, D. K., Zhao, G., Peters, J. W., et al. (2018). Structure of an ancient respiratory system. Cell 173, 1636–1649. e1616. doi: 10.1016/j.cell.2018.03.071
Yu, H. J., Haja, D. K., Schut, G. J., Wu, C. H., Meng, X., Zhao, G. P., et al. (2020). Structure of the respiratory MBS complex reveals iron-sulfur cluster catalyzed sulfane sulfur reduction in ancient life. Nat. Commun. 11:5953. doi: 10.1038/s41467-020-19697-7
Yu, H. J., Schut, G. J., Haja, D. K., Adams, M. W. W., and Li, H. L. (2021). Evolution of complex I-like respiratory complexes. J. Biol. Chem. 296:100740. doi: 10.1016/j.jbc.2021.100740
Zeng, X., Birrien, J. L., Fouquet, Y., Cherkashov, G., Jebbar, M., Querellou, J., et al. (2009). Pyrococcus CH1, an obligate piezophilic hyperthermophile: extending the upper pressure-temperature limits for life. ISME J. 3, 873–876. doi: 10.1038/ismej.2009.21
Keywords: Thermococcus aciditolerans SY113, high hydrostatic pressure, elemental sulfur reduction, membrane-bound sulfane reductase, membrane-bound hydrogenase, SurR
Citation: Jiao Z-X, Li X-G, Zhang W-J, Zhang G-Y, Bai S-J, Fu L and Wu L-F (2025) High hydrostatic pressure enhanced the growth of deep-sea Thermococcus aciditolerans by promoting the reduction of elemental sulfur. Front. Microbiol. 16:1643593. doi: 10.3389/fmicb.2025.1643593
Received: 09 June 2025; Accepted: 25 July 2025;
Published: 18 August 2025.
Edited by:
Philippe M. Oger, UMR5240 Microbiologie, Adaptation et Pathogenie (MAP), FranceReviewed by:
Doug Bartlett, University of California, San Diego, United StatesYann Moalic, ISEN Yncréa Ouest, France
Copyright © 2025 Jiao, Li, Zhang, Zhang, Bai, Fu and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xue-Gong Li, eHVlZ29uZ2xpQGlkc3NlLmFjLmNu
 Ze-Xi Jiao
Ze-Xi Jiao Xue-Gong Li
Xue-Gong Li Wei-Jia Zhang
Wei-Jia Zhang Guan-Yuan Zhang
Guan-Yuan Zhang Shi-Jie Bai
Shi-Jie Bai Ling Fu
Ling Fu Long-Fei Wu
Long-Fei Wu