- 1National Reference Centre for Botulism, Department of Food Safety, Nutrition and Veterinary Public Health, Istituto Superiore di Sanità, Rome, Italy
- 2Department of Bioscience and Technology for Food, Agriculture and Environment, University of Teramo, Teramo, Italy
Essential oils (EOs) hold significant potential as antimicrobials in food, due to their high concentration of active phenolic compounds. These compounds can target bacterial cells through various mechanisms, such as membrane disruption, quorum sensing inhibition, and interference in virulence factors, affecting microorganisms at a genomic level. Bacillus cereus and Bacillus subtilis are key foodborne bacteria that could be managed using these natural preservatives. The present study investigated the effects of stress induced by applying Thymus vulgaris and Origanum vulgare subsp. hirtum EOs on genetic modifications in B. cereus 11 and B. subtilis 58C strains isolated from shelf-stable gnocchi, through their gene expression analysis by quantitative real-time RT-PCR. Sublethal EO concentrations were tested, at increasing time intervals (6, 12, 18, 24, and 48 h). Most of the genes were downregulated at 6 h, indicating that the stressful situation prolonged the lag phase. Only spo0A for both B. cereus and B. subtilis, and pbpF and sigB for B. subtilis were upregulated after 6 h, suggesting an attempt to restore cellular communication and repair membrane damage. The pbpF gene was the most significant in the stress response of B. subtilis. Conversely, B. cereus responded through different mechanisms, primarily driven by the plcR and nheB genes, illustrating the role of virulence mechanisms in its stress response. In both strains, the genes were generally more upregulated at a higher concentration of EO (0.58 mg/mL), which was more stimulating than at 0.29 mg/mL. Moreover, the two EOs elicited variable stress responses, which implies different cellular mechanisms and genes in the same microorganism. Therefore, the outcomes of this study suggest that the action of the two EOs mainly influenced cell membrane integrity and quorum sensing mechanisms, with differences in the genes involved for the two species and the two EOs.
1 Introduction
Bacillus cereus and Bacillus subtilis are spore-forming, Gram-positive, rod-shaped bacteria that are commonly found in the environment, particularly in soil, sediments, dust, plants, and foods. These two species are of interest for a variety of foods, particularly starchy and potato-based products, where they are frequently detected (Del Torre et al., 2001; Purgatorio et al., 2024). Due to their ability to form spores, Bacillus spp. pose a concern for food products, even if thermally treated. Indeed, sporulation confers resistance to various stressful situations, including those induced by heat, high hydrostatic pressure, acids, antibiotics, or other antimicrobials (Stenfors Arnesen et al., 2008; Chaves López et al., 2009; Jin et al., 2021).
B. cereus is well recognised as a spoilage agent and a pathogen in foods. This microorganism is responsible for two types of foodborne diseases: the emetic syndrome, which is caused by consuming foods contaminated with cereulide, and diarrhoea, along with abdominal pain, due to toxins produced by enteropathogenic strains in the small intestine. For the latter disease, the infectious dose is estimated between 105 and 108 CFU/g viable cells or spores (Stenfors Arnesen et al., 2008). B. cereus-associated gastroenteric diseases are mostly mild and self-limiting; however, some fatal cases have been reported (Hoffmaster et al., 2008).
B. subtilis is a particularly competitive microorganism that adapts well to different environmental conditions due to its ease of genetic modification. In the food industry, B. subtilis can be used as a starter for fermentation (Kovács, 2019) and for other technological applications, such as chitosan production (Sini et al., 2007) or the antifungal activity of its volatile compounds (Chaves López et al., 2015). Additionally, while possessing probiotic and functional properties, B. subtilis strains can also lead to food spoilage, characterised by red discoloration, slime formation, and a sticky texture (Purgatorio et al., 2024).
Purgatorio et al. (2024) have recently investigated the occurrence of various species of Bacillus spp. in ambient gnocchi. B. subtilis and B. cereus were the two most frequently isolated species in the samples formulated without the organic acids traditionally used as preservatives. The removal of conventional additives in food formulations is becoming increasingly popular, and research is shifting towards the use of natural substitutes. This trend is connected to the rising phenomenon of antimicrobial resistance and the growing interest in clean label foods, to create alternative and eco-friendly products. Essential oils (EOs) are particularly appealing among these natural alternatives for their well-documented antimicrobial properties (D’Amato et al., 2018).
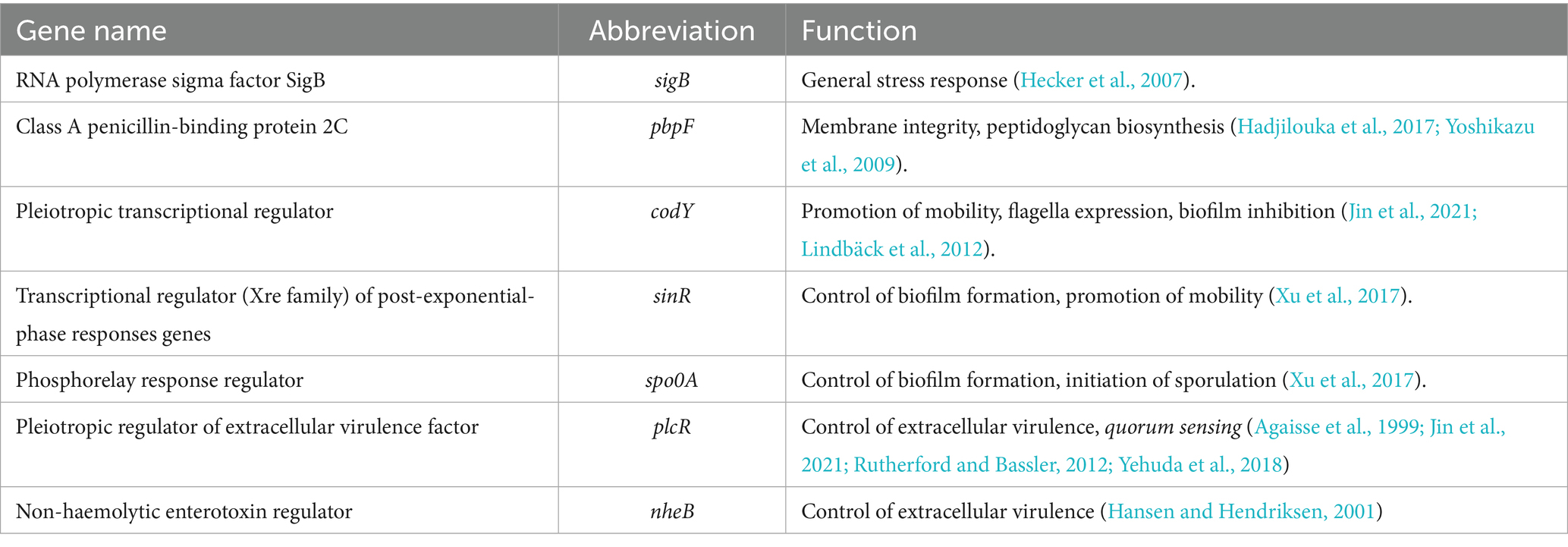
In fact, EOs can act against target cells through several mechanisms, including the destabilisation of cytoplasmic membrane phospholipids, leakage of cellular material, loss of ions, protein denaturation, interference with quorum sensing mechanisms, sporulation, and the expression of virulence factors (Purgatorio et al., 2022; Rossi et al., 2022). Therefore, the contact with EOs represents a stressful event for the cells, which attempt to adapt and survive by regulating the expression of a wide range of genes. The molecular targets that may be affected include those involved in quorum sensing and virulence (e.g., plcR, nhe genes) (Jin et al., 2021; Rutherford and Bassler, 2012). As previously demonstrated in B. cereus, applying sublethal concentrations of biopreservatives can hinder vital communication between bacteria, resulting in reduced expression of virulence factors (Jin et al., 2021). The alteration of communication within the bacterial population can also influence their biofilm production and the gene expression involved in its formation (e.g., codY, sinR, spo0A) (Lindbäck et al., 2012; Jin et al., 2021; Xu et al., 2017; Zhao et al., 2021). Other genes that may be affected by biopreservative-induced stress include those involved in cellular metabolism and growth control (Jin et al., 2021) and the maintenance of membrane integrity (e.g., pbpF) (Chowdhury et al., 2021).
Thymus vulgaris and Origanum vulgare subsp. hirtum EOs have been extensively studied for their antimicrobial properties, which are associated with the high level of phenolic compounds, such as thymol, carvacrol, p-cymene, γ-terpinene, and linalool (D’Amato et al., 2024; Kosakowska et al., 2024; Pellegrini et al., 2018; Tardugno et al., 2022).
This study aims to evaluate the impact of treatments with sublethal concentrations of Thymus vulgaris and Origanum vulgare subsp. hirtum EOs on the expression of the genes involved in the stress response in B. cereus and B. subtilis.
2 Materials and methods
2.1 Bacterial strains and culture conditions
B. cereus strain 11 and B. subtilis strain 58C, isolated and characterized in a previous study (Purgatorio et al., 2024), were used in the design of experiments. B. cereus strain 11 was isolated from ambient gnocchi prepared without preservatives, packed in a modified atmosphere (MAP) and stored at room temperature (~25°C) for 5 days. The pathogenicity of this strain was underlined by the fact that it encodes the cesC gene, which is involved in the biosynthesis of the cereulide toxin, responsible for B. cereus emetic syndrome. B. subtilis strain 58C was isolated from ambient gnocchi containing lactic acid as a preservative, packed in MAP and stored under thermal abuse (30°C) for 7 days. The details of the isolation, identification, and molecular characterisation of the strains are reported in the previous study (Purgatorio et al., 2024). The strains were stored at −80°C in cryovials containing Brain Heart Infusion broth (BHI) (Oxoid, UK) and 20% (w/v) glycerol.
Each strain was streaked onto BHI agar and incubated at 37°C for 24–48 h. A single colony from each strain was then inoculated into BHI broth and incubated at 37°C for 18 h to obtain a fresh working culture.
2.2 Essential oils
Commercial and food-grade Thymus vulgaris thymol chemotype EO was kindly provided by Flora S.r.l. (Pisa, Italy), while Origanum vulgare subsp. hirtum carvacrol chemotype EO was kindly supplied by Exentiae S.r.l. Soc. Agricola (Catania, Italy). EOs were prepared at a concentration of 36.0 mg/mL, by adding PBS (Phosphate Buffer Saline) and Tween 80 (10.0 μL/mL). Homogeneous emulsions, obtained through vortexing, were subsequently sterilised using a 0.22 μm polytetrafluoroethylene (PTFE) Minisart syringe filter (Sartorius, Göttingen, Germany).
2.3 Primers design and SYBR green real-time PCR optimisation
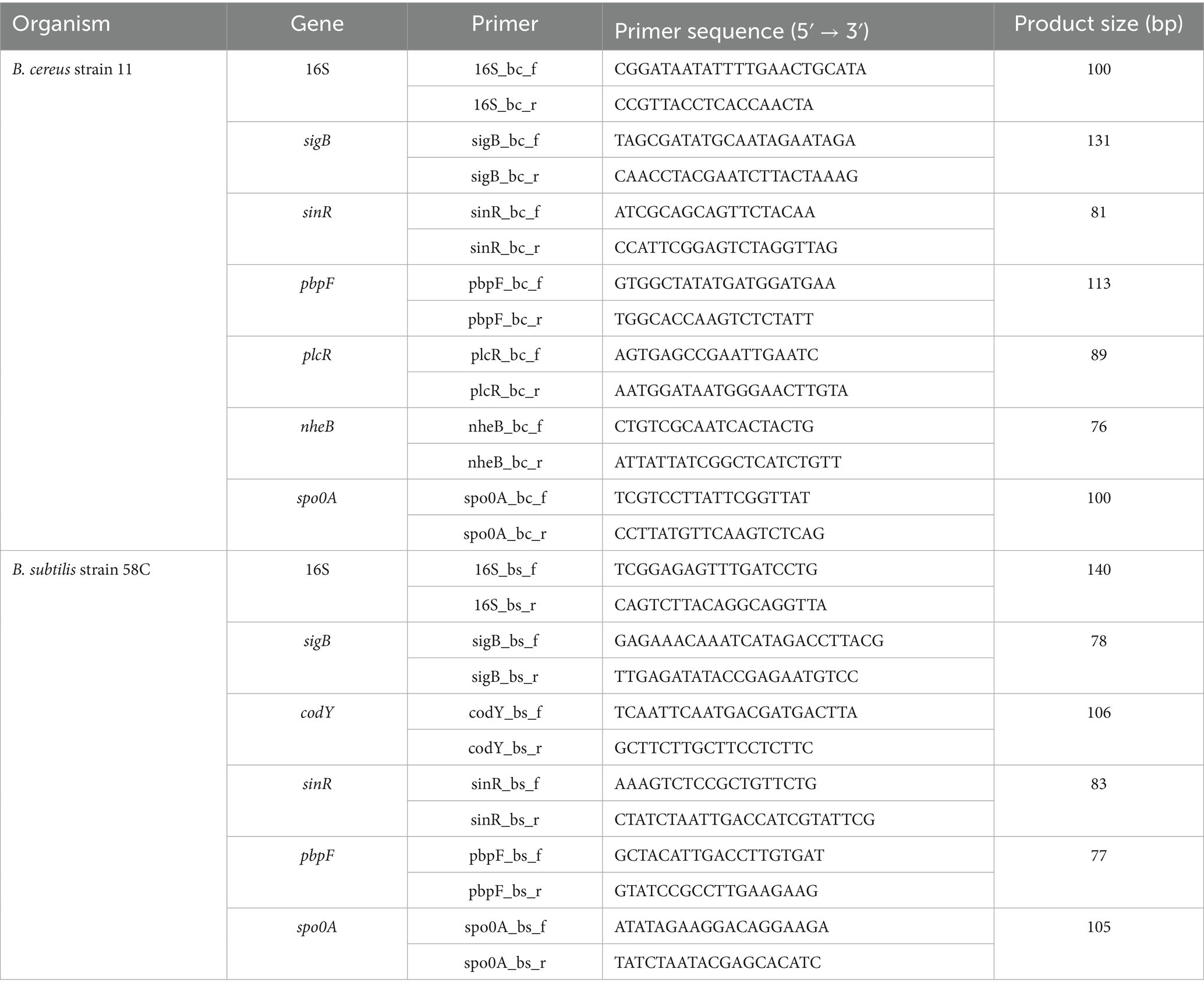
The genes used in this study, presented in Table 1, were retrieved from the literature (Fouet et al., 2000; Hecker et al., 2007; Jin et al., 2021). Their functions are summarised in Table 2. The nucleotide sequence of each gene was extracted from publicly available reference genomes at accession numbers CP020383 and CP34551. These sequences were aligned with those of B. cereus strain 11 and B. subtilis strain 58C using the Clustal Omega algorithm.1 The consensus sequences obtained for each gene harboured by both B. cereus and B. subtilis generated by the alignment study were used to design primers through Beacon Design version 7.91 (Premier Biosoft International, USA).
The specificity of each primer was assessed through a basic local alignment search on BLASTn.2 The selectivity study (inclusivity and exclusivity) was performed in silico by running the freely available PCR amplification tool at the website http://insilico.ehu.es/, using the most permissive PCR conditions, against all available Bacillus species. Additional selectivity studies were conducted, testing each primer couple against the following strains: Bacillus cereus ATTC 11778, Bacillus cereus ATCC 27884, Bacillus coagulans ATCC 7050, Bacillus subtilis ATCC 6633, Citrobacter freundii ATCC 8090, Clostridium botulinum ATCC19397, Clostridium butyricum ATCC19398, Enterococcus faecalis ATCC 29212, Escherichia coli ATCC 25922, Listeria innocua ATCC 33090, Listeria monocytogenes ATCC 7644, Pseudomonas aeruginosa ATCC 9027, Salmonella enterica ser. Enteritidis ATCC 13076, Staphylococcus aureus ATCC 13565, Streptococcus thermophilus ATCC 19258 and Rhodococcus equi ATCC 6939, along with 13 wild strains isolated from ambient gnocchi (6 B. subtilis, 6 B. cereus, and 1 B. atrophaeus). All real-time PCR runs assessing selectivity were conducted in a Rotor-Gene Q (Qiagen, Hilden, Germany) real-time PCR machine using QuantiNova SYBR Green master mix (Qiagen, Hilden, Germany – cat. No 208056) according to the conditions suggested by the manufacturer. Inclusivity was defined as the ability of the PCR method to detect the target analyte from a wide range of strains. Exclusivity was defined as the lack of interference from a relevant range of nontarget strains in the PCR methods (Malorny et al., 2003).
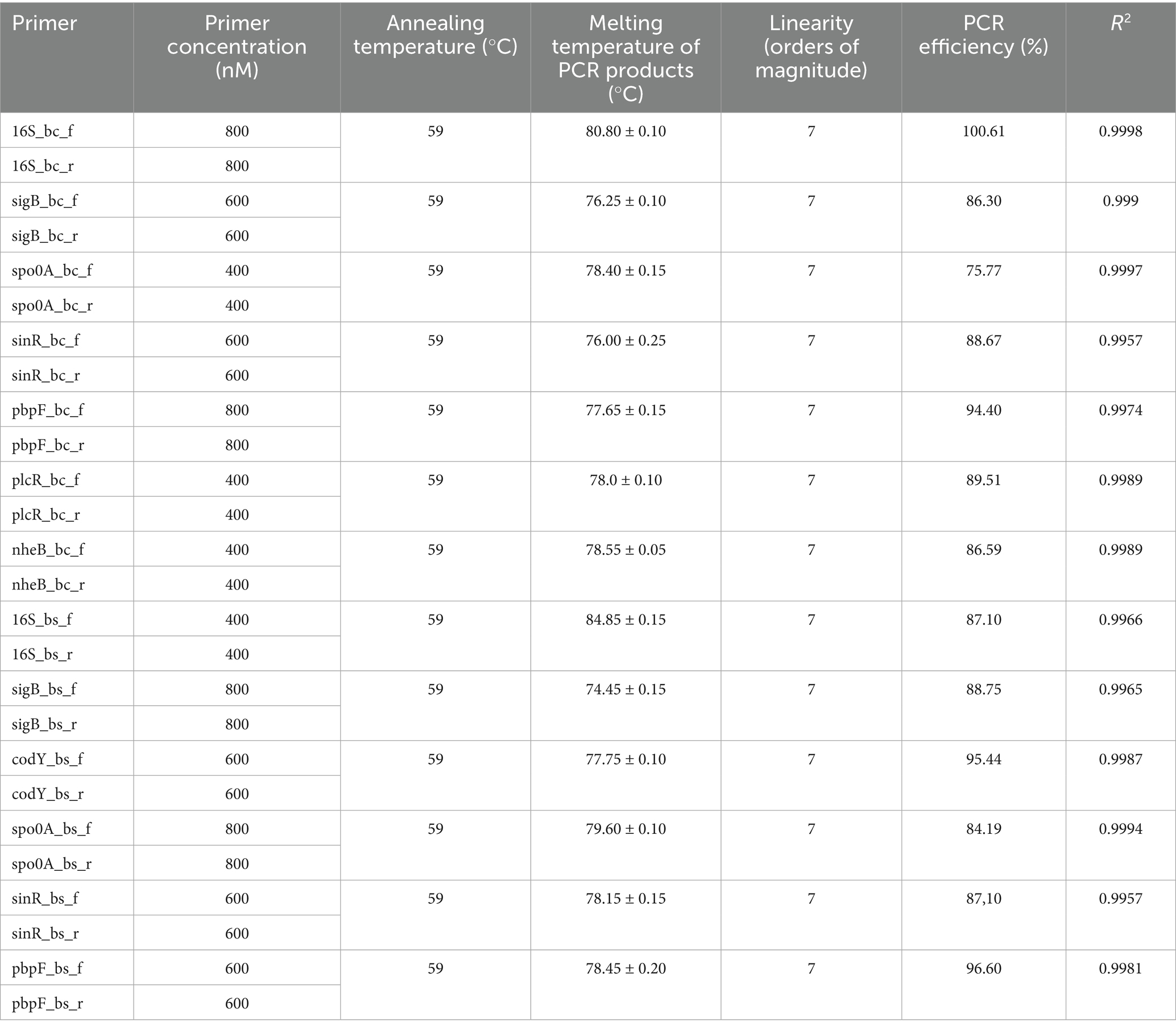
Further optimisation involved identifying the optimal annealing temperature for each primer couple (range 55°C–59°C) and the best primers concentration (400, 600, 700, and 800 nM). Lastly, the dynamic range of linearity and amplification efficiency was assessed for each primer couple, testing a known quantity of DNA extracted from B. cereus strain 11 and B. subtilis strain 58C in triplicate. To evaluate amplification efficiency, a standard curve was plotted with the log of the number of DNA copies (x-axis) extracted from B. cereus strain 11 and B. subtilis strain 58C against the threshold cycle (Ct) for these copies (y-axis) (data not shown). The amplification efficiency (E) for the various targets was calculated using the following equation:
DNA copies were calculated using the following equation:
2.4 Treatment with Thymus vulgaris and Origanum vulgare subsp. hirtum essential oils
B. cereus strain 11 and B. subtilis strain 58C were treated with T. vulgaris and O. vulgare EOs using the broth microdilution method described by Clinical and Laboratory Standards Institute guidelines (CLSI, 2020). The EOs emulsions prepared at a concentration of 36.0 mg/mL were two-fold diluted in BHI broth in 2-mL Eppendorf tubes, to obtain concentrations ranging from 18.0 to 0.58 mg/mL. Cells from the 18 h culture were collected by centrifugation at 13,000 rpm (Eppendorf-Centrifuge 5415D, Hamburg, Germany) for 5 min and washed three times with PBS. The inoculum was standardised using a Jenway 6305 spectrophotometer at 5 × 105 CFU/mL and tested in duplicate. Positive controls (BHI broth and inoculum) and negative controls (T. vulgaris or O. vulgare EOs and BHI broth) were also tested. Tubes were incubated at 37°C for 6, 12, 18, 24, and 48 h to assess the effect of EOs on B. cereus strain 11 and B. subtilis strain 58C at various exposure times. The lowest concentration of EOs that inhibited microbial growth after incubation at 37°C for 48 h was considered the Minimum Inhibitory Concentration (MIC). The analyses were carried out in triplicate.
2.5 RNA extraction
B. cereus strain 11 and B. subtilis strain 58C RNA were extracted from the tubes with EOs treatments. The concentrations to be tested were selected based on the MIC results at 48 h. Two sub-inhibitory concentrations were considered: 0.29 mg/mL (1/4 MIC) and 0.58 mg/mL (1/2 MIC) for both Bacillus species. The extractions were carried out by RNeasy Mini Kit (Qiagen, Germany) with some modifications. Briefly, 1 mL of each culture was chilled on ice for 30 min prior to extraction to limit metabolic and enzymatic activity. Cells were recovered by centrifugation at 13,000 rpm for 3 min and then washed four times with PBS. The pellet was resuspended in 500 μL of TE buffer, and 2 μL of lysozyme (20 mg/mL) was added. Samples were subsequently incubated at 37°C for 60 min to allow for cell rupture. Then, 700 μL of RLT buffer and 500 μL of 70% ethanol were added to the suspension, homogenising gently with the micropipette without vortexing. Then, 600 μL of the samples were transferred to a RNeasy Mini spin column and were centrifuged at 10,000 rpm for 1 min. The supernatant was discarded, and the process was repeated twice with the remaining sample. DNA digestion was performed by incubating the spin columns at 37°C for 30 min, in which 20 μL of DNase I solution and 140 μL of RDD buffer were deposited. After DNA digestion, 500 μL of buffer RPE was added to the spin columns and centrifuged at 10,000 rpm for 2 min. RNA was eluted in 100 μL of RNase-free water.
The RNA purity was assessed by a Nanodrop spectrometer (Thermo Fisher Scientific, United States), considering the A260/A280 ratio acceptable between 1.8 and 2.1. The RNA integrity was evaluated with the Agilent Technologies 2,100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). The absence of DNA was confirmed by running 3 μL of each RNA sample as a template for real-time PCR, using the optimised protocols described above (no amplification demonstrated the absence of DNA traces).
2.6 Quantitative real-time RT-PCR SYBR green
RT-PCR SYBR Green reactions were conducted in Rotor-Gene Q (Qiagen, Hilden, Germany) using the QuantiNova SYBR Green RT-PCR kit (Qiagen, Hilden, Germany – cat. No 208154). Each assay was performed with 2 μL of template RNA, 10 μL of SYBR Green RT-PCR Master Mix, 0.2 μL of QuantiNova SYBR Green RT-Mix, variable volumes of each primer, according to the concentration selected during optimisation (see Table 3), along with RNase-free water to reach a total volume of 20 μL. The RT-PCR conditions consisted of an initial RT-step at 50°C for 10 min to allow cDNA production, followed by 40 cycles of PCR initial heat activation at 95°C for 2 min (2-step cycling) and denaturation at 95°C for 5 s and followed by combined annealing/extension for 10 s at the optimised temperature. No-RT and no-template controls were included in each run to check for contamination of reagents. Gene expression studies were carried out according to the MIQE guidelines.3
2.7 Statistical analysis
All the real-time PCR experiments were conducted in triplicate. Arithmetic means and standard deviations of melting temperature (Tm) values were calculated to define positive results. A positive result was assigned to an assay that generated a Ct value at the expected Tm for the target gene. RT-qPCR runs were performed in triplicate using 16S rrn as the reference gene. Normalised data were converted to relative expression as described by Pfaffl (2001) and log2-values (fold change) according to Kubista (2007) for further analysis with one-way ANOVA. p < 0.05 was considered significant. Data visualisation and statistical analysis were performed using Microsoft Excel and Prism 9.5.1 software (GraphPad, Boston, MA, USA).
3 Results
3.1 SYBR green real-time PCR protocols and their performance parameters
Each primer pair was designed to produce a specific signal for B. cereus strain 11 and B. subtilis strain 58C. Table 3 shows the PCR conditions optimised for each set of primers using the QuantiNova master mix in the Rotor-Gene Q thermal cycler. The melting temperature (Tm) of PCR products, obtained by running positive control DNAs, was used to distinguish between positive and negative results. The selectivity study provided 100% inclusivity and 100% exclusivity (data not shown).
As reported in Table 3, all primer sets produced a linear response over seven orders of magnitude with PCR efficiency ranging from 84.19 to 100.61%.
3.2 Treatment with Thymus vulgaris and Origanum vulgare subsp. hirtum essential oils
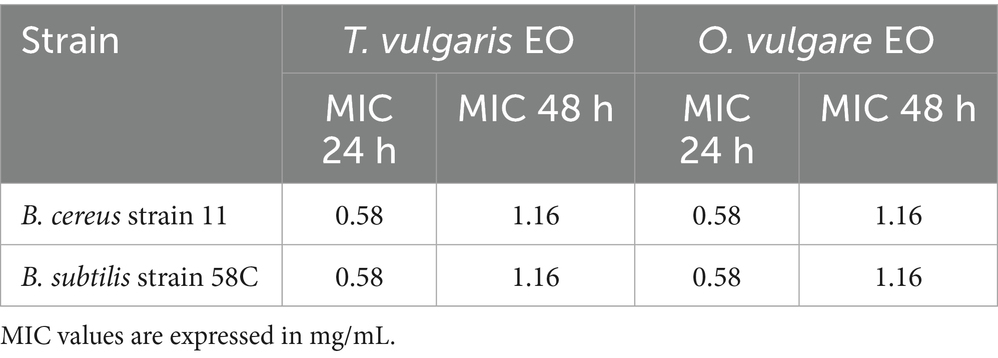
MIC values of T. vulgaris and O. vulgare EOs against B. cereus strain 11 and B. subtilis strain 58C after 24 h and 48 h of exposure are summarised in Table 4.
Based on the obtained MIC values, cultures of the mentioned strains exposed to sub-inhibitory concentrations of 0.29 mg/mL (1/4 MIC for 48 h) and 0.58 mg/mL (1/2 MIC for 48 h) were subjected to RNA extraction and gene expression analysis.
3.3 Effect of Thymus vulgaris and Origanum vulgare subsp. hirtum essential oils on gene expression
Results of relative gene expression are presented in Tables 5, 6, and Figures 1–8.
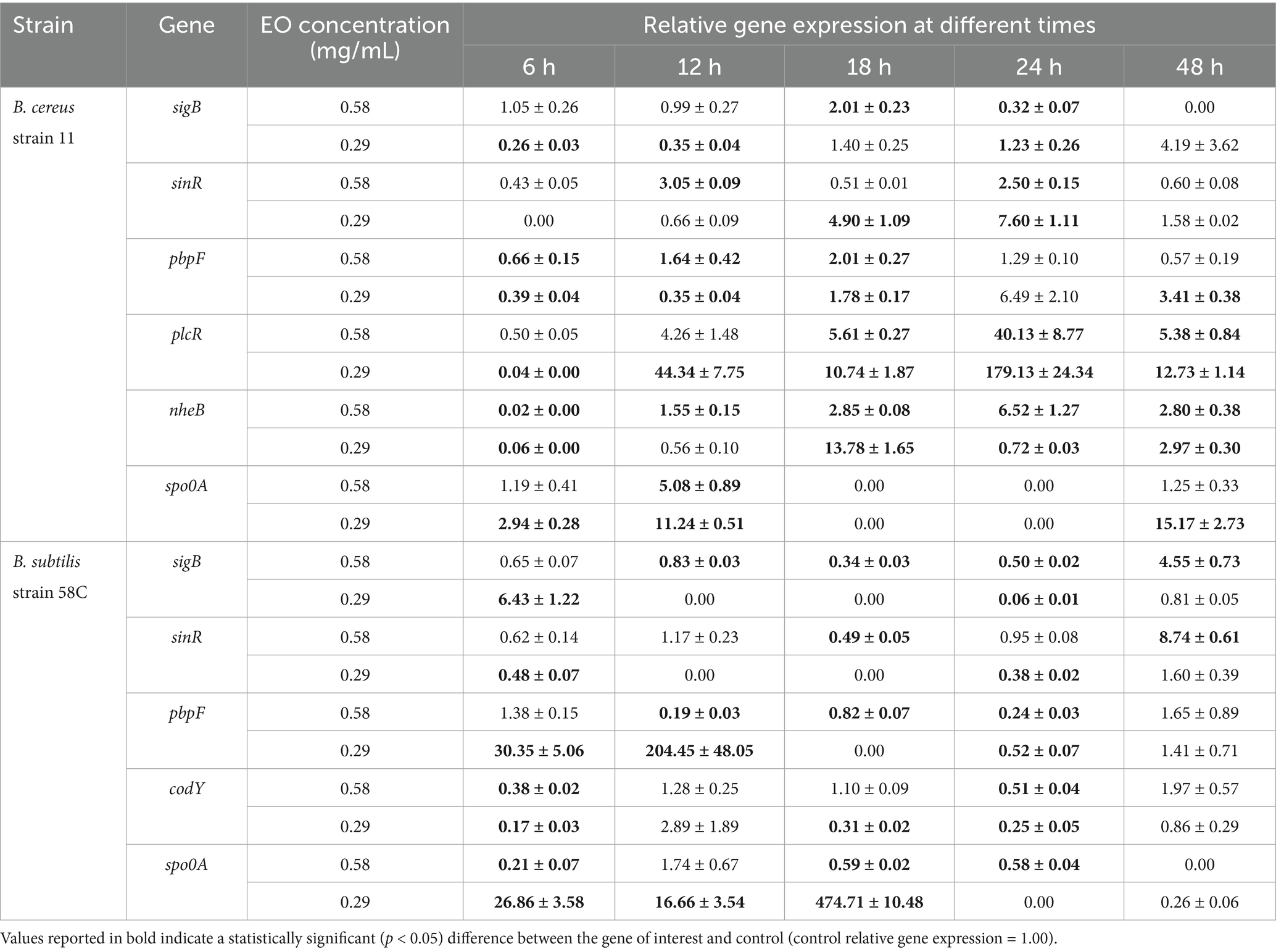
Table 5. Effect of Thymus vulgaris EO on B. cereus strain 11 and B. subtilis strain 58C at different exposure times.
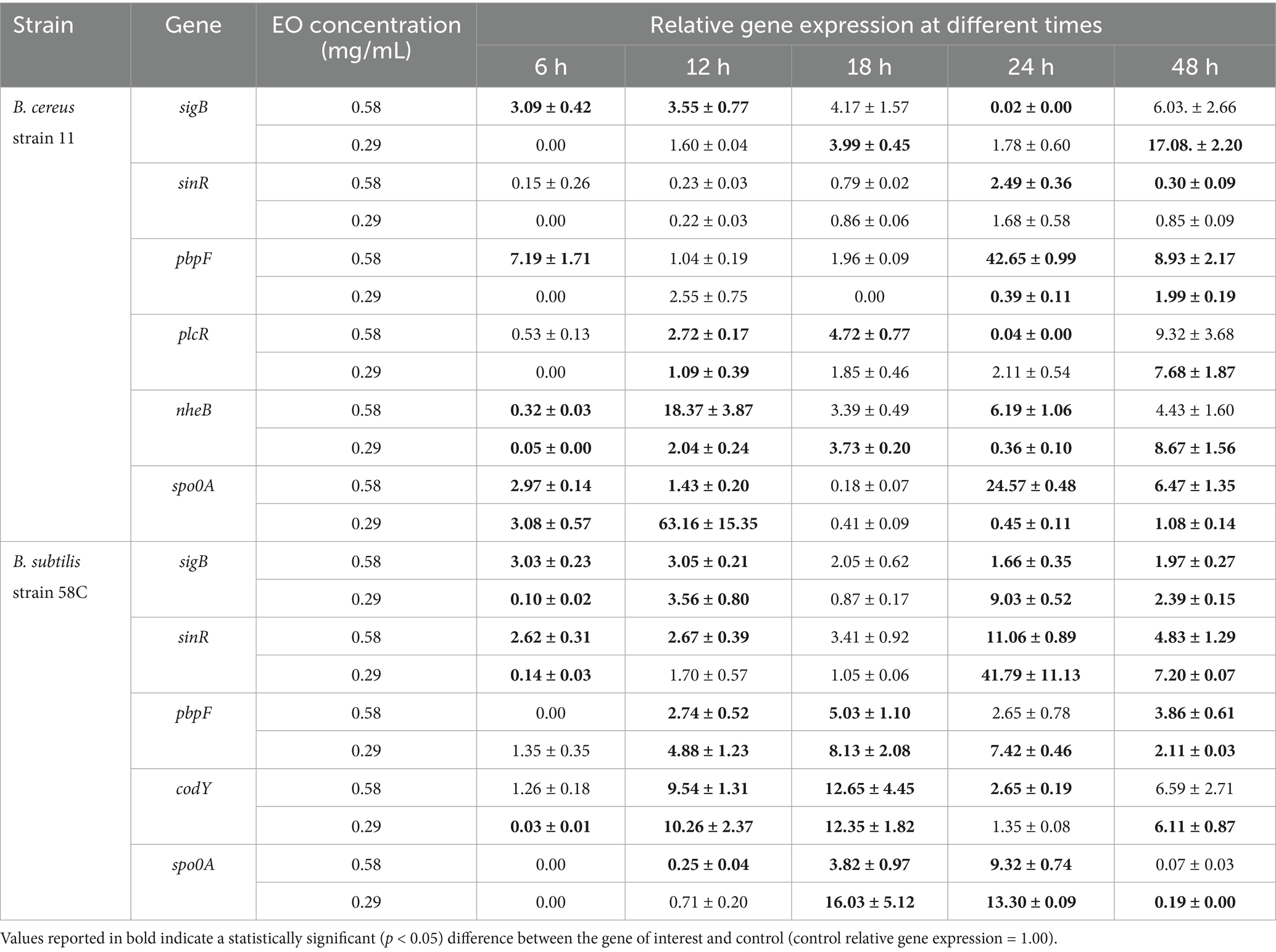
Table 6. Effect of Origanum vulgare EO on B. cereus strain 11 and B. subtilis strain 58C at different exposure times.
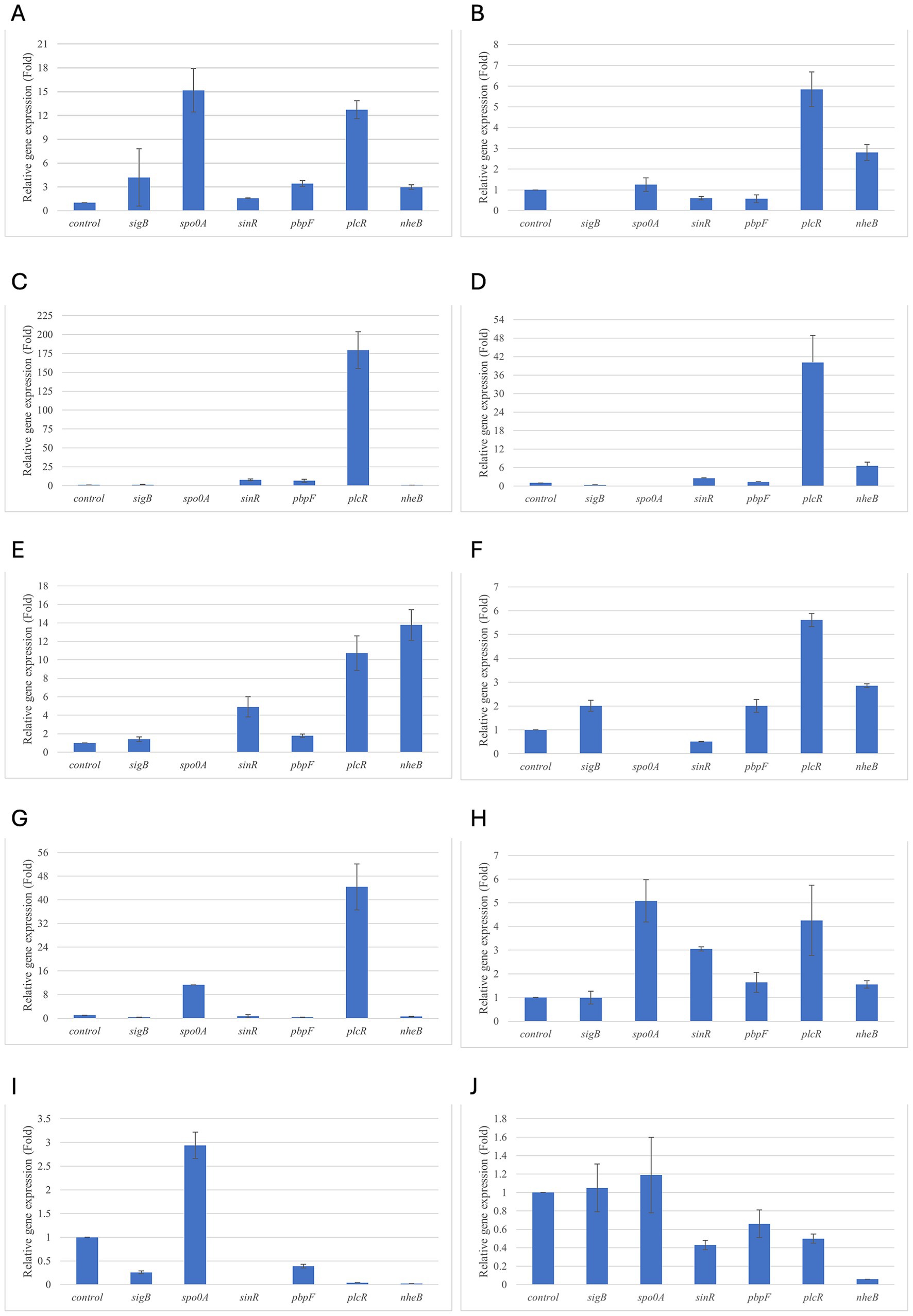
Figure 1. Relative gene expression of B. cereus strain 11 at various exposure durations to Thymus vulgaris EO. (A) 0.58 mg/mL for 48 h, (B) 0.29 mg/mL for 48 h, (C) 0.58 mg/mL for 24 h, (D) 0.29 mg/mL for 24 h, (E) 0.58 mg/mL for 18 h, (F) 0.29 mg/mL for 18 h, (G) 0.58 mg/mL for 12 h, (H) 0.29 mg/mL for 12 h, (I) 0.58 mg/mL for 6 h, (J) 0.29 mg/mL for 6 h. Bars depict the mean of three replicates, with error bars showing standard deviation.
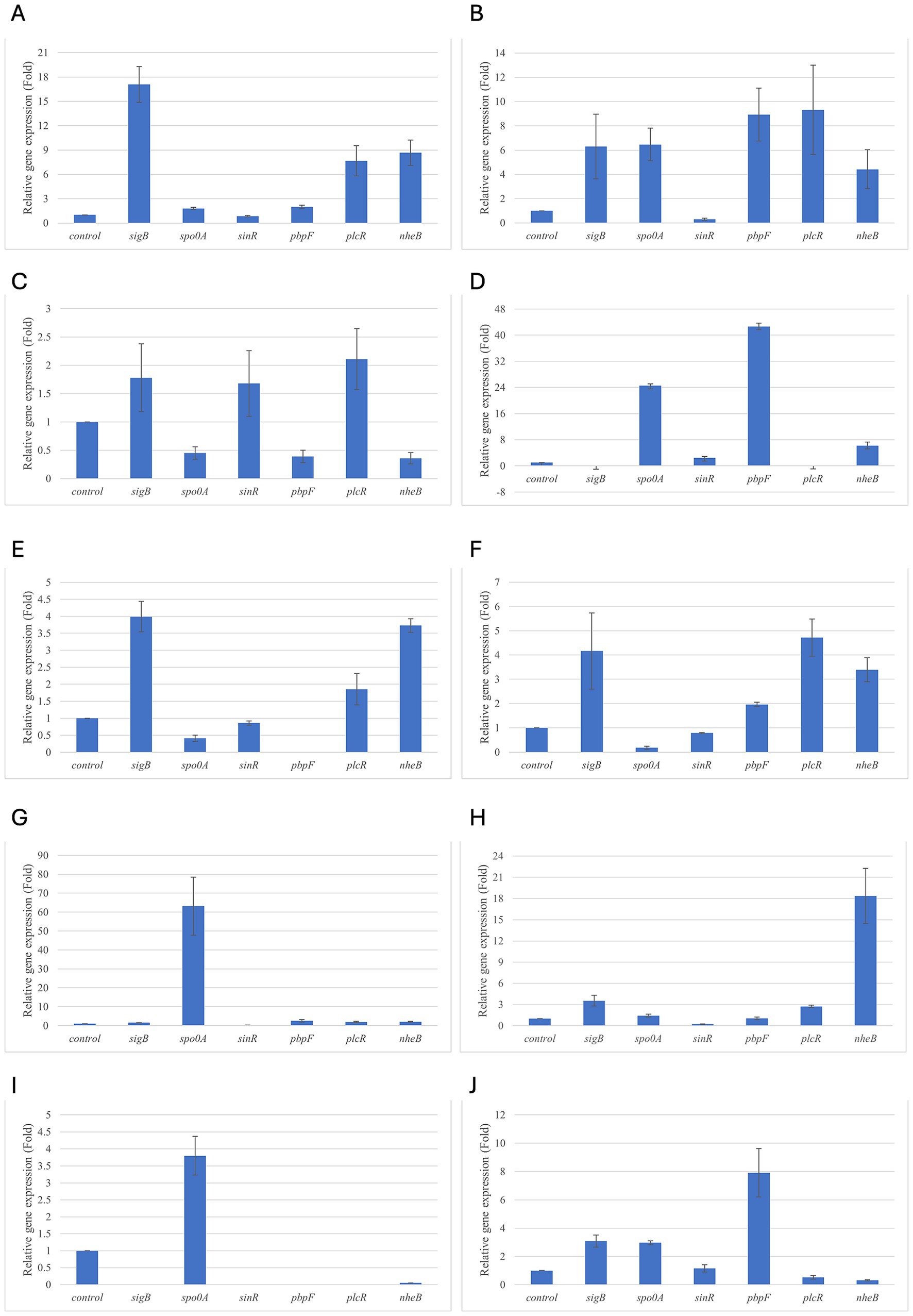
Figure 2. Relative gene expression of B. cereus strain 11 at various exposure durations to Origanum vulgare EO. (A,B) 48 h, (C,D) 24 h, (E,F) 18 h, (G,H) 12 h, (I,J) 6 h. Bars depict the mean of three replicates, with error bars showing standard deviation.
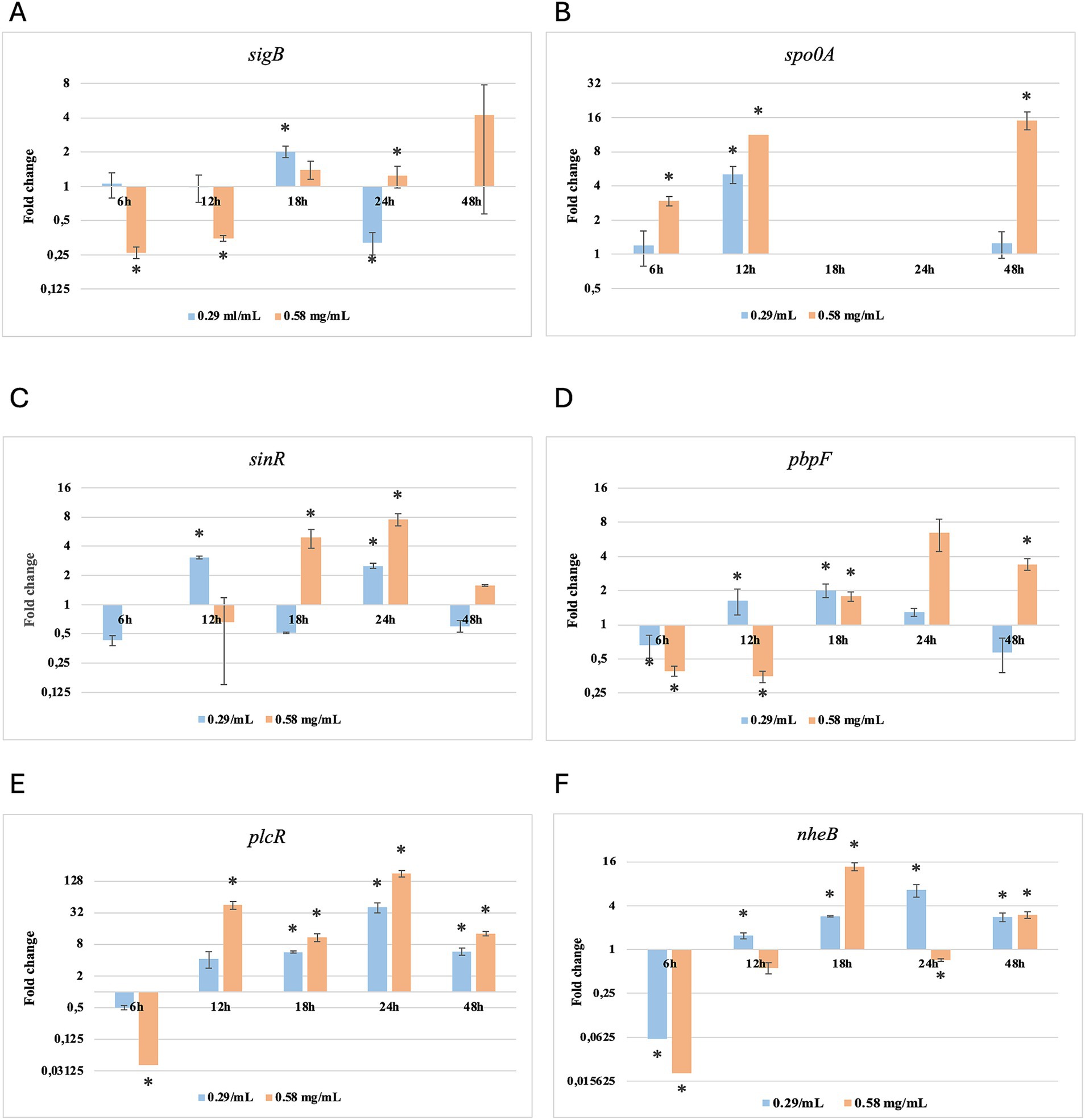
Figure 3. Log2 fold change of B. cereus strain 11 at various exposure durations to Thymus vulgaris EO. Bars depict the mean of three replicates, with error bars showing standard deviation. Asterisks (*) indicate significant differences between treatments and control (p < 0.05).
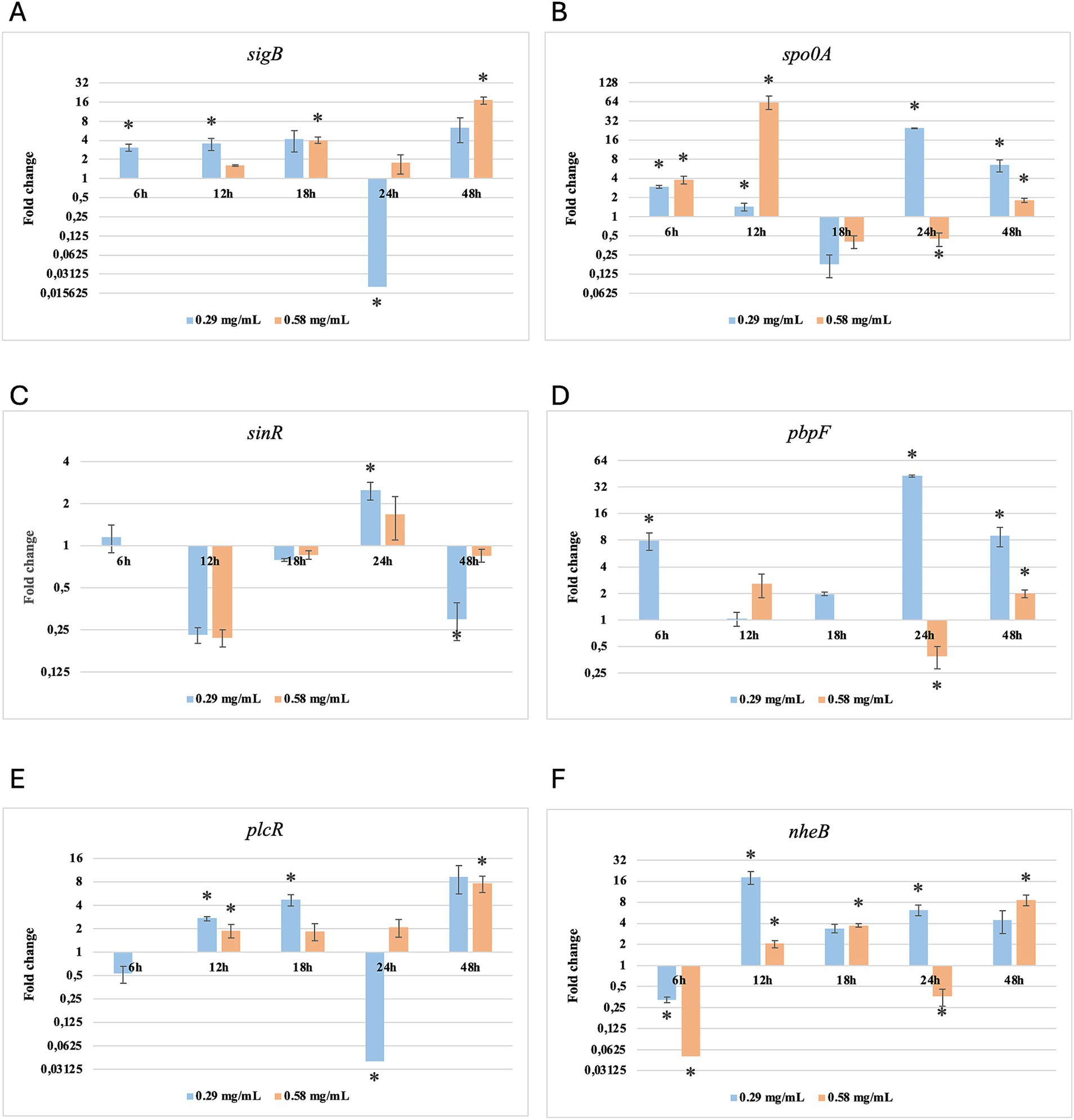
Figure 4. Log2 fold change of B. cereus strain 11 at various exposure durations to Origanum vulgare EO. Bars depict the mean of three replicates, with error bars showing standard deviation. Asterisks (*) indicate significant differences between treatments and control (p < 0.05).
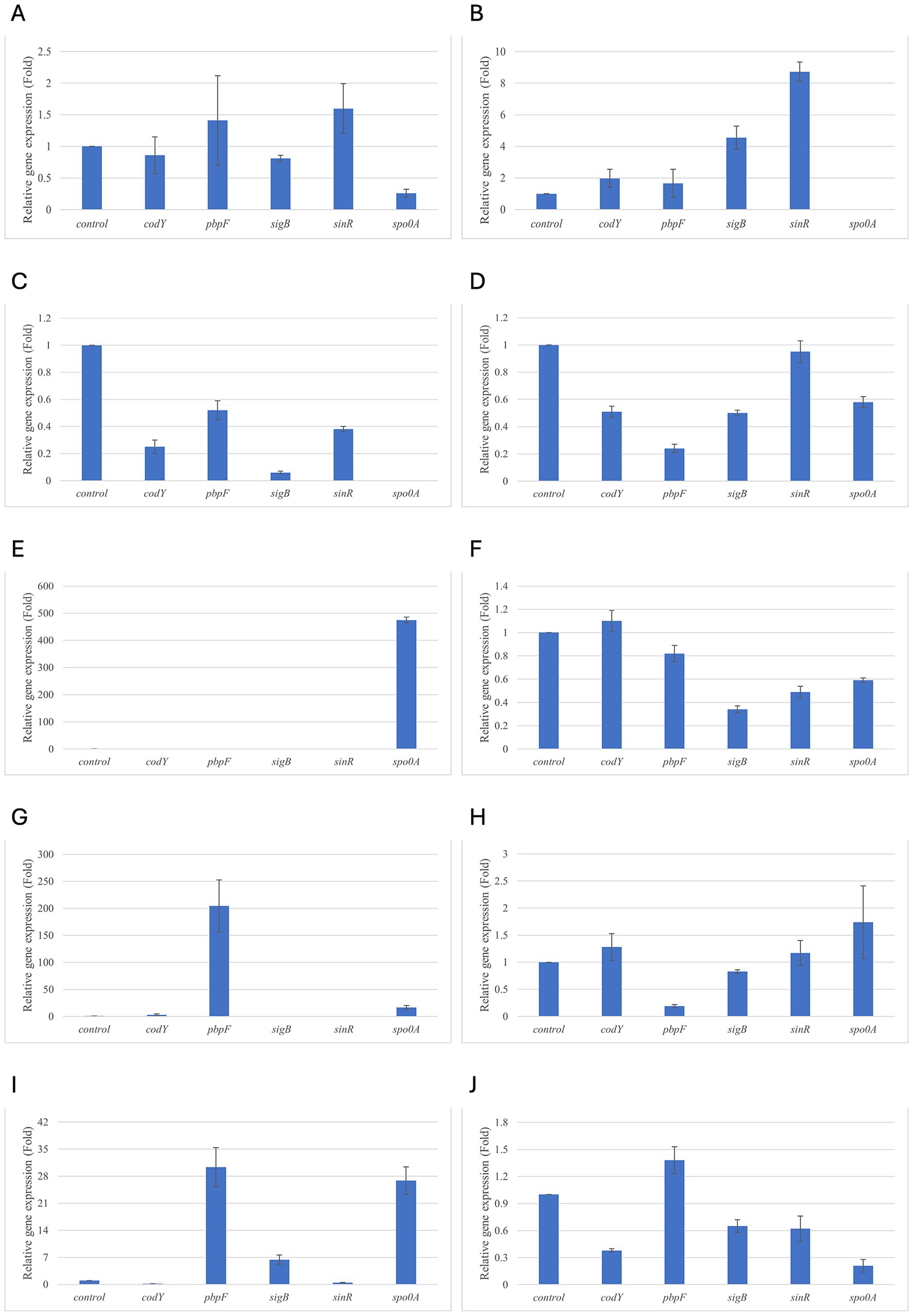
Figure 5. Relative gene expression of B. subtilis strain 58C at various exposure durations to Thymus vulgaris EO. (A) 0.58 mg/mL for 48 h, (B) 0.29 mg/mL for 48 h, (C) 0.58 mg/mL for 24 h, (D) 0.29 mg/mL for 24 h, (E) 0.58 mg/mL for 18 h, (F) 0.29 mg/mL for 18 h, (G) 0.58 mg/mL for 12 h, (H) 0.29 mg/mL for 12 h, (I) 0.58 mg/mL for 6 h, (J) 0.29 mg/mL for 6 h. Bars depict the mean of three replicates, with error bars showing standard deviation.
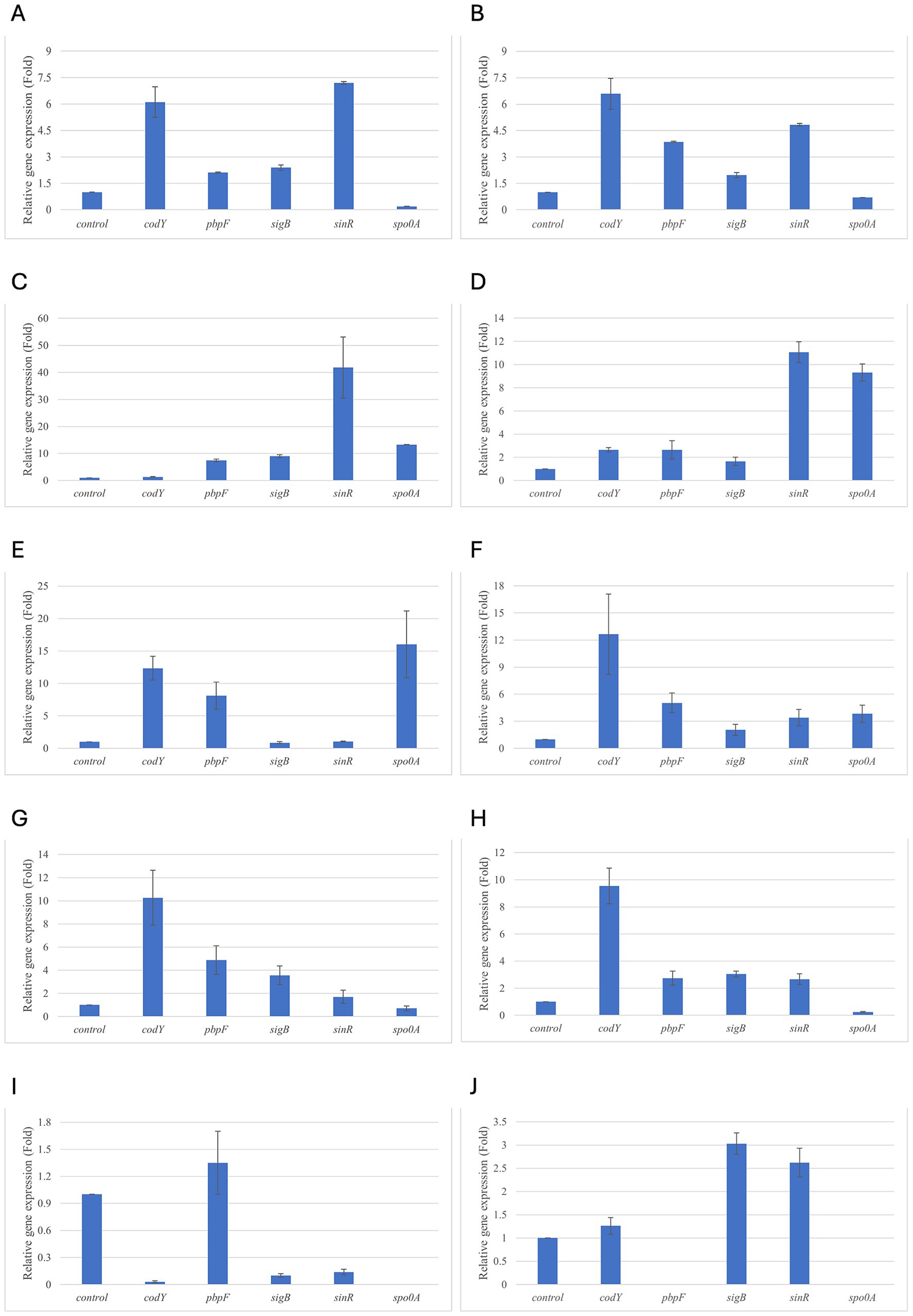
Figure 6. Relative gene expression of B. subtilis strain 58C at various exposure durations to Origanum vulgare EO. (A) 0.58 mg/mL for 48 h, (B) 0.29 mg/mL for 48 h, (C) 0.58 mg/mL for 24 h, (D) 0.29 mg/mL for 24 h, (E) 0.58 mg/mL for 18 h, (F) 0.29 mg/mL for 18 h, (G) 0.58 mg/mL for 12 h, (H) 0.29 mg/mL for 12 h, (I) 0.58 mg/mL for 6 h, (J) 0.29 mg/mL for 6 h. Bars depict the mean of three replicates, with error bars showing standard deviation.
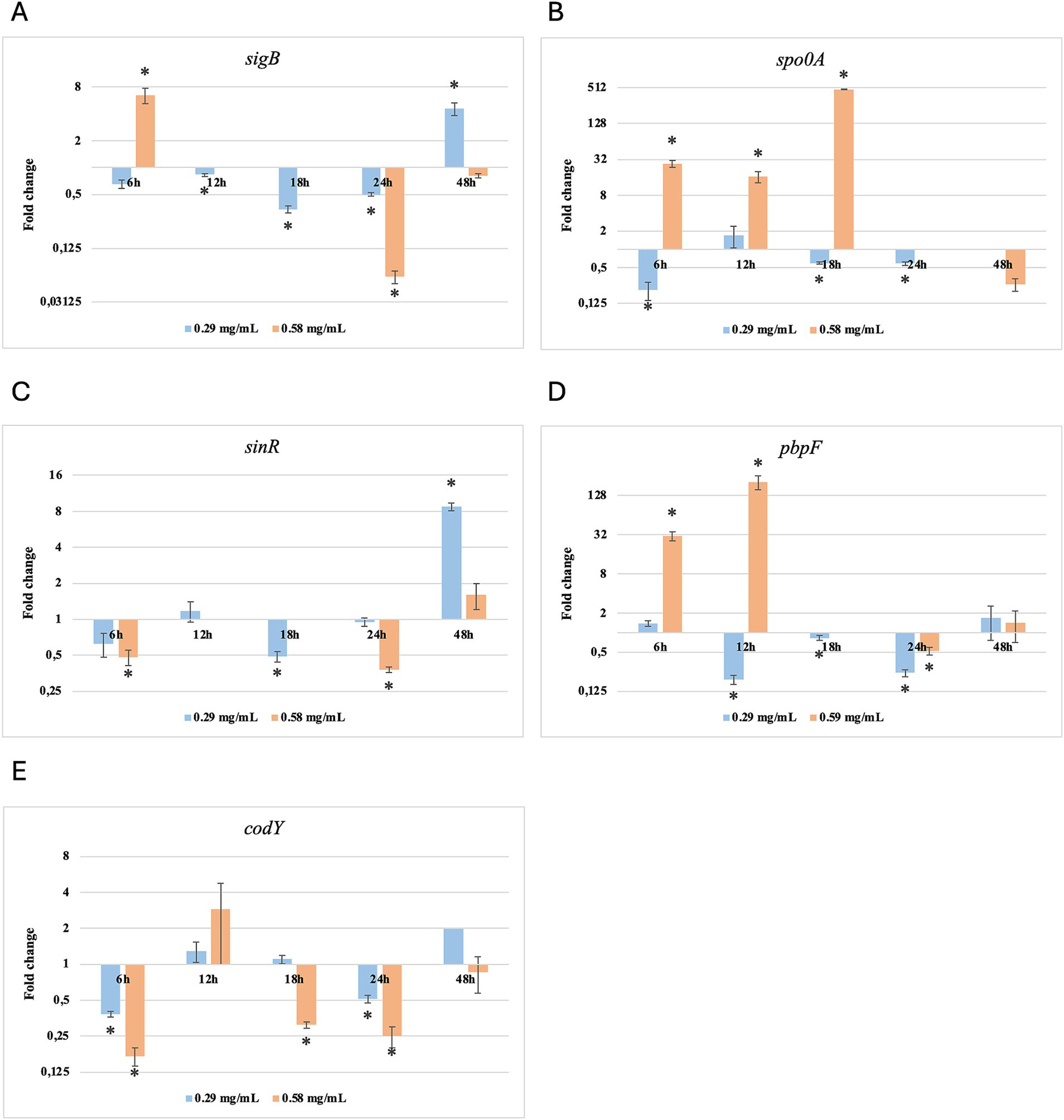
Figure 7. Log2 fold change of B. subtilis strain 58C at various exposure durations to Thymus vulgaris EO. Bars depict the mean of three replicates, with error bars showing standard deviation. Asterisks (*) indicate significant differences between treatments and control (p < 0.05).
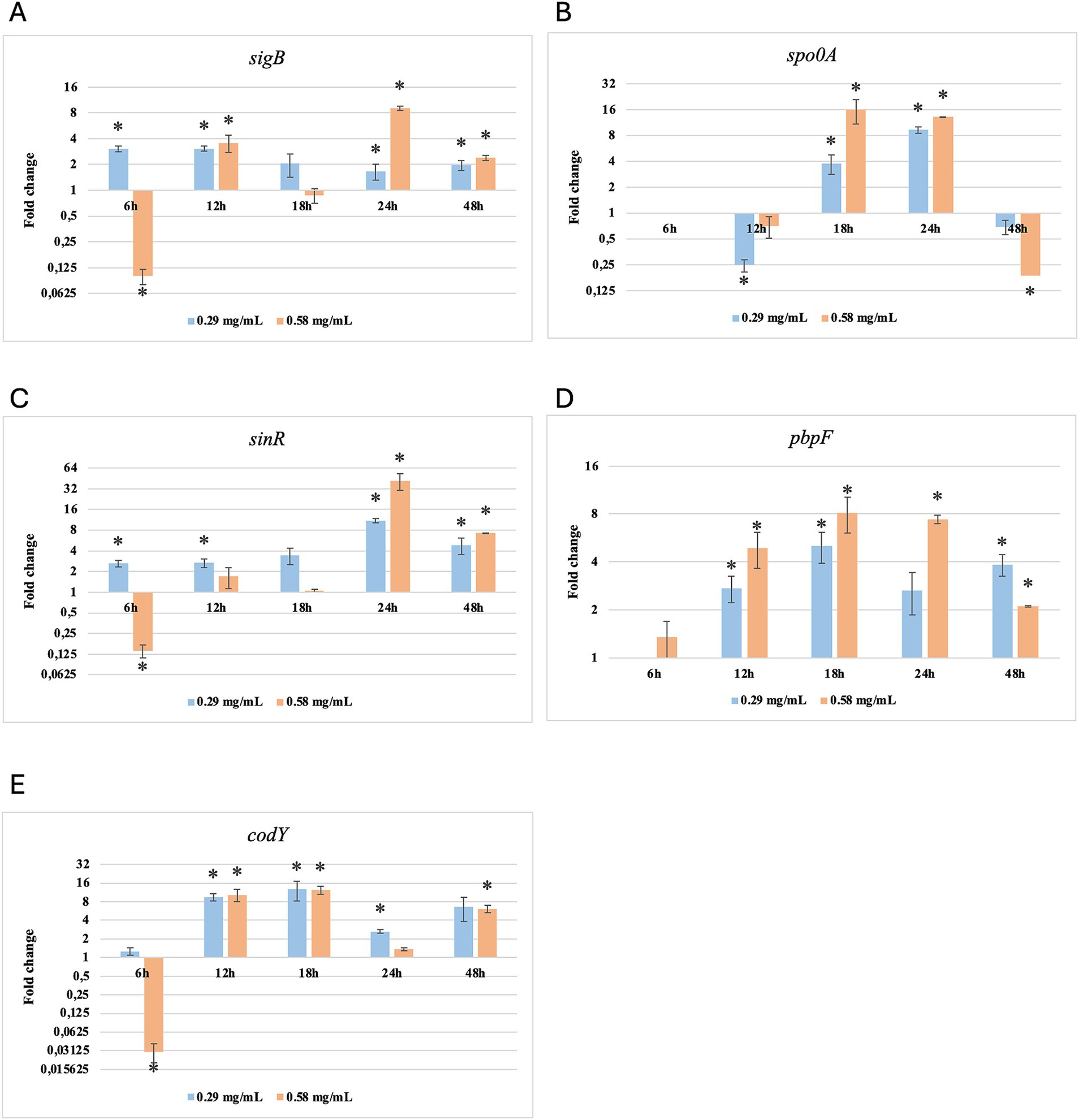
Figure 8. Log2 fold change of B. subtilis strain 58C at different exposure times to Origanum vulgare EO. Bars depict the mean of three replicates, with error bars showing standard deviation. Asterisks (*) indicate significant differences between treatments and control (p < 0.05).
After 6 h of exposure to both EOs, most of the genes in the two tested strains exhibited a downregulation, with some exceptions in which an upregulation was observed (T. vulgaris: B. cereus spo0A; B. subtilis sigB, spo0A, pbpF; O. vulgare: B. cereus sigB, spo0A, pbpF; B. subtilis: sigB, sinR) (Tables 5, 6; Figures 3B, 4A,B,D, 7A,B,D, 8A,C). The sigma factor B (sigB) was the most significantly upregulated gene (p < 0.05) at 6 h.
3.3.1 Bacillus cereus: gene expression and comparison of exposure times
For B. cereus, after 12 h of exposure, almost all genes exhibited increased regulation levels compared to those seen at 6 h, likely because the strains began to organise their cellular functions to counteract the antimicrobial activity exerted by the EOs (Figures 1,2G,H). At this time, the genes that primarily increased their expression in B. cereus were plcR and spo0A for both EOs. For O. vulgare EO, nheB was significantly overexpressed at 12 h, while an evident upregulation began at 18 h for T. vulgaris. Generally, there was an upregulation at 18 h, but no high peaks were seen for 12 h (Figures 1,2E,F). This is likely because the cells, at the start of their stationary phase, had already implemented their stress response, thus maintaining these functions. Expression values exceeding 10.00 were observed for plcR and nheB (treatment with T. vulgaris), highlighting how the reaction to stressful situations involves pathogenicity factors of the microorganism. This trend was also evident at 24 h and 48 h, when genes associated with virulence continued to be overexpressed (Figures 1,2A–D). The highest values were recorded at 24 h for plcR (between approximately 40.00 and 180.00, T. vulgaris treatment), at which point the gene reached its peak level. Unexpectedly, spo0A subjected to Thymus vulgaris at 18 h and 24 h showed no expression levels.
O. vulgare treatment also influenced other genes at 24 h and 48 h, particularly sigB, pbpF, and spo0A, which were involved to a lesser extent in the response to stress caused by T. vulgaris. In particular, pbpF was significantly upregulated to values higher than 42.00 after O. vulgare EO exposure. This difference indicates that the cells could respond variably to the different EOs, because of their composition of active molecules. The sinR gene, for both EOs, was only modestly upregulated at 18–24 h, while it was downregulated at the other analysis times.
While for the most significant part of the genes there was no evident difference between the expression at the two concentrations tested, for other genes (plcR, spo0A, sinR) T. vulgaris EO determined a more significant overexpression at the concentration of 0.58 mg/mL, as indicated by the fold change (Figures 3B,C,E). In contrast, O. vulgare EO was more stimulating at 0.29 mg/mL for the other genes, particularly pbpF (Figure 4D).
3.3.2 Bacillus subtilis: gene expression and comparison of exposure times
For B. subtilis, expression at 6 h was limited, as for B. cereus. The sigB gene was among the most upregulated initially, but its expression decreased during exposure to T. vulgaris EO, while it continued, with a peak at 24 h, during exposure to O. vulgare EO (Figures 7,8A). For B. cereus, gene expressions were generally higher at 12 h, when cells began to activate their defense mechanisms (Figures 5,6G,H). The pbpF gene was significantly expressed for both EOs, particularly for T. vulgaris, which peaked at approximately 205.00 at 12 h, following a high expression level at 6 h (about 30.35). This indicates that the pbpF gene is significantly involved in the gene expression of B. subtilis, especially during the first hours of exposure to T. vulgaris (Figure 7D). The same gene remained more consistently expressed up to 48 h for O. vulgare (Figure 8D). The codY gene was overexpressed at 12 h for both EOs, to a greater extent for O. vulgare (values around 10.00) (Figures 7,8E). The spo0A gene, instead, showed a significant role in the first hours of exposure only for T. vulgaris, while it was downregulated for O. vulgare. However, at 18 h, there was upregulation for both EOs, lower for O. vulgare (around 16.00 at a concentration of 0.58 mg/mL) and higher for T. vulgaris, with a peak of almost 475.00 at 0.58 mg/mL (Figures 7,8B).
At 18, 24, and 48 h, almost all the genes in B. subtilis treated with T. vulgaris EO showed significant downregulation, or slight non-significant upregulation (Figures 5A–F). This indicates that the active compounds in this EO trigger a response from the microorganism particularly in the early stages of exposure, especially at the highest concentration tested, when the stress was greater. Conversely, after exposure to O. vulgare for 18, 24, or 48 h, the level of overexpression remained relatively constant, and almost always significant for all tested genes (Figures 6A–F). This indicates that the molecules present in the O. vulgare EO act continuously, prompting the cell to implement numerous cellular functions. Furthermore, for this EO, the expression was similar for the two concentrations tested, with some exceptions where the higher concentration was more stimulating, particularly sinR at 24 h (0.29 mg/mL: about 11.00; 0.58 mg/mL: about 42.00).
4 Discussion
The multiple properties of EOs have been exploited since ancient times. Among EOs, those from the Thymus and Origanum genera are now widely used as herbal teas, tonics, carminatives, antitussives, and antiseptics. The species T. vulgaris and O. vulgare subsp. hirtum exhibit antibacterial and antifungal properties primarily due to their phenolic compounds content. It has been proposed that the minor components may be essential for the antimicrobial activity of EOs, because of their ability to establish synergistic effects (Gutierrez et al., 2008; Tardugno et al., 2022). Based on the profiles of aromatic compounds, T. vulgaris and O. vulgare have been classified into various chemotypes (Raal et al., 2024). In this study, the effect of two common chemotypes, T. vulgaris chemotype thymol and O. vulgare chemotype carvacrol, on the transcription of genes involved in the stress response in Bacillus spp. has been evaluated.
The aromatic compounds of EOs are “generally recognised as safe” (GRAS) by health food authorities (FDA (Food and Drug Administration), 2009). Due to their antimicrobial activity, they may represent promising alternatives to food preservatives and even antibiotics (Prakash et al., 2024). However, their sensory impact and the elevated cost may influence their practical application in the food industry. For this reason, the use of sublethal concentrations of EOs could be of great interest. This study aims to evaluate how Bacillus spp. strains respond at the genetic level to concentrations of EOs that are lower than the inhibitory dose. The genes sigB, sinR, pbpF, spo0A, plcR, and nheB were monitored in B. cereus strain 11, while sigB, sinR, pbpF, spo0A, and codY were observed in B. subtilis strain 58C. The effects of sub-inhibitory concentrations of EOs on these two strains were evaluated after 6, 12, 18, 24, and 48 h of exposure.
At the beginning of exposure (6 h), most of the tested genes were downregulated. Similarly, other authors have noted a delay in the lag phase in strains subjected to sub-inhibitory concentrations of EOs (Burt, 2004; Mazzarrino et al., 2015). During this period, cells organise their functions to respond to the stress they experience. At 6 h, one of the most upregulated genes was sigma factor B (sigB). It is a general transcription factor activated by various cellular stresses, including the pressure experienced upon entering the stationary growth phase. Harms et al. (2024) describe sigB as an “emergency system” that cells utilise under stressful conditions. Consequently, it is a central control mechanism for numerous stress responses. This factor appears capable of regulating the expression of over 150 genes, including those with the sigB operon (Hecker et al., 2007; Yeak et al., 2023). The sigB operon exhibits similar clusters across various Bacillus spp. species, with some differences (e.g., three genes for the B. anthracis sigB operon and eight genes for the B. subtilis sigB operon) (Fouet et al., 2000). Being integral to the general stress response, it was upregulated from the initial hours of exposure of B. cereus and B. subtilis to T. vulgaris and O. vulgare EOs. However, this gene did not appear to be mainly involved in the stress induced by the EO, compared to other genes, especially for T. vulgaris treatment. Indeed, sigB is widely recognised for its regulatory activity under contrasting stress conditions, primarily due to different types of stress, such as acid or thermal shocks (Hecker et al., 2007).
The genes plcR and nheB, involved in the virulence of B. cereus, were identified in the genome of B. cereus strain 11 in a previous study conducted by Purgatorio et al. (2024). plcR was particularly significant in B. cereus response. According to Gohar et al. (2008), plcR regulates genes encoding for proteins that are either secreted or situated at the cell wall, forming the interface between the bacterial cell and its environment, as well as genes related to sporulation, biofilm formation, and the synthesis of extracellular enzymes and toxins. The authors noted that plcR transcription was auto-induced just before the onset of the stationary phase, and that the action of spo0A repressed its expression (Gohar et al., 2008). Our results confirmed the initiation of plcR transcription after 12 h, with expression increasing up to 48 h, despite the concurrent upregulation of spo0A. In B. cereus, plcR also serves a crucial role as a virulence regulator in controlling gene transcription for enterotoxins. Hadjilouka et al. (2017) consistently observed the expression of numerous virulence genes following the treatment of L. monocytogenes with lemongrass EO. The nheB gene was also overexpressed, albeit less than plcR, particularly following T. vulgaris EO treatment. It regulates extracellular virulence, contributing to the production of non-haemolytic enterotoxin (Hansen and Hendriksen, 2001; Li et al., 2016). Both nheB and pbpF, a gene associated with peptidoglycan biosynthesis, were significantly upregulated to their maximum levels after 18 h of T. vulgaris EO exposure.
The spo0A gene is well-known as a sporulation factor that contributes to biofilm formation and regulates the transition phase of growth. It influences the expression of hundreds of genes and can prevent the initiation of DNA replication by binding to the origins of DNA replication. Although this gene is widely studied in the Bacillus and Clostridium genera, to the best of our knowledge, the behaviour we observed in our study (no expression at 18 h and 24 h under treatment with Thymus vulgaris) has never been documented in the literature. Further research is required to confirm this phenomenon in other B. cereus strains using different Thymus vulgaris EOs; however, this finding likely represents the most significant discovery of our work, at least regarding the use of EO for B. cereus control in foods. By controlling sporulation, spo0A promotes the transcription of an enzyme that antagonises sinR. In this study, sinR was expressed in most cases after 18 h of exposure. In Bacillus thuringiensis, sinR regulates genes involved in detoxification processes, sugar metabolism, DNA recombination and degradation, peptidoglycan turnover, and energy production. Additionally, this gene represses biofilm formation and is necessary for swimming motility (Fagerlund et al., 2014).
B. subtilis strain 58C responded to the stress induced by exposure to T. vulgaris EO quite differently from B. cereus strain 11. This finding aligns with other authors who reported essential differences between the two species in several regulatory pathways, including those involved in stress response (Fagerlund et al., 2014; Gohar et al., 2008). At the early stages of exposure to T. vulgaris EO, B. subtilis showed a significant improvement in gene expression, particularly for pbpF, sigB, and spo0A. According to Harms et al. (2024), sigB, spo0A, and sinR are involved in the adaptive response of B. subtilis to unfavourable conditions. Our results demonstrate that sinR was upregulated but only in the final hours of exposure, indicating that this is not the first gene activated in response to the presence of EOs.
The codY gene regulates over two hundred B. subtilis genes (Barbieri et al., 2015). During our observation, codY and sinR remained predominantly down-regulated (p < 0.05), and began to be significantly upregulated after 24 h for O. vulgare EO and 48 h for T. vulgaris EO treatments. These genes regulate the promotion of mobility, flagella expression, and biofilm control (Jin et al., 2021). Some of these functions overlap with those of the spo0A gene; however, the latter is more involved in the initiation of sporulation, which appears to play a significant role in stress response (Xu et al., 2017).
B. subtilis did not use the same main genetic repair mechanisms as B. cereus, favouring the transcription of the gene pbpF encoding for a protein family known as penicillin-binding proteins. This observation agrees with Yoshikazu et al. (2009), who described the regulation of cell wall morphogenesis in B. subtilis by recruiting PBP1 proteins. The involvement of genes that encode for membrane integrity and peptidoglycan biosynthesis is also reported by Hadjilouka et al. (2017) for L. monocytogenes.
The higher sublethal concentration (0.58 mg/mL) generally resulted in more significant gene overexpression than the lower concentration (0.29 mg/mL), likely because the cells, experiencing more severe damage, activate emergency mechanisms more vigorously. For some genes, O. vulgare EO resulted in higher expression at 0.29 mg/mL, particularly against B. cereus. These differences, along with the fact that for B. cereus and B. subtilis the two EOs determine slightly different gene expression profiles, are closely related to the composition of EOs. In fact, even substances present in minimal quantities can influence the mechanisms of action against the target cell and how it genetically reacts to the external agent. T. vulgaris EO is the thymol chemotype and contains significant amounts of γ-terpinene, p-cymene, linalool, and carvacrol. In contrast, O. vulgare subsp. hirtum EO is the carvacrol chemotype and also includes γ-terpinene, p-cymene, and (E)-caryophyllene as its main components. These substances interact with one another and those present at very low concentrations, resulting in effects that could vary significantly between them. Even a phenolic compound present in what might seem like an irrelevant concentration can substantially influence the impact on the cell (D’Amato et al., 2024). Further studies on how the interaction between the different components of antimicrobial substances may affect gene expression would be beneficial.
5 Conclusion
The present study provides a first overview of how two of the most common EOs act at the molecular level on B. cereus and B. subtilis strains. Although further studies are required, the results improve understanding of the effects of EOs on these species, as only a few gene expression studies have focused on these microorganisms and their responses to EOs, especially at sublethal treatments.
The higher sublethal concentration (0.58 mg/mL) was generally more stimulating. Moreover, in most cases, significant upregulation started at 12 h, and continued differently depending on the gene, while at the beginning of the exposure (6 h), most genes were downregulated. These findings also suggest that a better understanding of the molecular mechanisms involved in the repression of these genes could provide a foundation for new research in the field of natural substances used as antimicrobials to replace conventional food preservatives or to substitute or synergise with antibiotics to counteract the rampant phenomenon of antimicrobial resistance. In fact, elucidating the cellular mechanisms implicated in controlling food pathogenic and spoilage microorganisms, along with the concentrations and exposure time at which EOs can exert their activity, would allow for the optimisation of their application to foods. Although some of the genes studied in this work are well-known, further exploration is needed to investigate the complex regulatory pathways triggered by the stress induced by T. vulgaris, O. vulgare subsp. hirtum and other commonly used EOs, considering the significant variability of phenolic compounds and interaction between them, which can influence gene expression profiles.
Data availability statement
The original contributions presented in the study are included in the article, further inquiries can be directed to the corresponding author.
Author contributions
FA: Writing – review & editing, Funding acquisition, Conceptualization, Writing – original draft. CP: Writing – review & editing, Writing – original draft, Investigation, Conceptualization. AS: Data curation, Validation, Software, Writing – review & editing, Formal analysis. CS: Methodology, Investigation, Writing – review & editing, Data curation, Formal analysis. ST: Investigation, Data curation, Formal analysis, Writing – review & editing, Methodology. AP: Writing – review & editing, Supervision, Funding acquisition, Validation, Conceptualization.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work has been funded by the European Union-NextGenerationEU, Mission 4, Component 1, under the Italian Ministry of University and Research (MUR) National Innovation Ecosystem grant ECS00000041-VITALITY-CUP: C43C22000380007.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
References
Agaisse, H., Gominet, M., Økstad, O. A., Kolstø, A. B., and Lereclus, D. (1999). PlcR is a pleiotropic regulator of extracellular virulence factor gene expression in Bacillus thuringiensis. Mol. Microbiol. 32, 1043–1053. doi: 10.1046/j.1365-2958.1999.01419.x
Barbieri, G., Voigt, B., Albrecht, D., Hecker, M., Albertini, A. M., Sonenshein, A. L., et al. (2015). Cody regulates expression of Bacillus subtilis extracellular protease Vpr and Mpr. J. Bacteriol. 197, 1423–1432. doi: 10.1128/jb.02588-14
Burt, S. (2004). Essential oils: their antibacterial properties and potential applications in foods—a review. Int. J. Food Microbiol. 94, 223–253. doi: 10.1016/j.ijfoodmicro.2004.03.022
Chaves López, C., Lanciotti, R., Serio, A., Paparella, A., Guerzoni, M. E., and Suzzi, G. (2009). Effect of high pressure homogenization applied individually or in combination with other mild physical or chemical stresses on Bacillus cereus and Bacillus subtilis spore viability. Food Control 20, 691–695. doi: 10.1016/j.foodcont.2008.09.001
Chaves López, C., Serio, A., Gianotti, A., Sacchetti, G., Ndagijimana, M., Ciccarone, C., et al. (2015). Diversity of foodborne Bacillus volatile compounds and influence on fungal growth. J. Appl. Microbiol. 119, 487–499. doi: 10.1111/jam.12847
Chowdhury, N., Goswami, G., Boro, R. C., and Barooah, M. (2021). A pH-dependent gene expression enables Bacillus amyloliquefaciens MBNC to adapt to acid stress. Curr. Microbiol. 78, 3104–3114.
CLSI (2020). Performance standards for antimicrobial susceptibility testing. 30th Edn, CLSI 447 Supplement M100S. Wayne, PA: CLSI.
D’Amato, S., Rossi, C., Maggio, F., Valbonetti, L., Savini, V., Paparella, A., et al. (2024). Antilisterial effectiveness of Origanum vulgare var. hirtum and Coridothymus capitatus essential oils and hydrolates alone and in combination. Foods 13:860. doi: 10.3390/foods13060860
D’Amato, S., Serio, A., López, C. C., and Paparella, A. (2018). Hydrosols: biological activity and potential as antimicrobials for food applications. Food Control 86, 126–137. doi: 10.1016/j.foodcont.2017.10.030
Del Torre, M., Della Corte, M., and Stecchini, M. L. (2001). Prevalence and behaviour of Bacillus cereus in a REPFED of Italian origin. Int. J. Food Microbiol. 63, 199–207. doi: 10.1016/s0168-1605(00)00421-9
Fagerlund, A., Dubois, T., Økstad, O. A., Verplaetse, E., Gilois, N., Bennaceur, I., et al. (2014). SinR control enterotoxins expression in Bacillus thuringiensis biofilms. PLoS One 9:e87532. doi: 10.1371/journal.pone.0087532
FDA (Food and Drug Administration) (2009). Food generally, recognized as safe (GRAS). Available online at: https://www.fda.gov/food/food-ingredients-packaging/generallyrecognized-safe-gras (Accessed February 4, 2023).
Fouet, A., Namy, O., and Lambert, G. (2000). Characterization of the operon encoding the alternative ςB factor from Bacillus anthracis and its role in virulence. J. Bacteriol. 182, 5036–5045. doi: 10.1128/jb.182.18.5036-5045.2000
Gohar, M., Faegri, K., Perchat, S., Ravnum, S., Økstad, O. A., Gominet, M., et al. (2008). The PlcR virulence regulon of Bacillus cereus. PLoS One 3:e2793. doi: 10.1371/journal.pone.0002793 B., Lereclus, D
Gutierrez, J., Rodriguez, G., Barry-Ryan, C., and Bourke, P. (2008). Efficacy of plant essential oils against foodborne pathogens and spoilage bacteria associated with ready-to-eat vegetables: antimicrobial and sensory screening. J. Food Prot. 71, 1846–1854. doi: 10.4315/0362-028x-71.9.1846
Hadjilouka, A., Mavrogiannis, G., Mallouchos, A., Paramithiotis, S., Mataragas, M., and Drosinos, E. H. (2017). Effect of lemongrass essential oil on Listeria monocytogenes gene expression. LWT 77, 510–516. doi: 10.1016/j.lwt.2016.11.080
Hansen, B. M., and Hendriksen, N. B. (2001). Detection of enterotoxic Bacillus cereus and Bacillus thuringiensis strains by PCR analysis. Appl. Environ. Microbiol. 67, 185–189. doi: 10.1128/AEM.67.1.185-189.2001
Harms, M., Michalik, S., Hildebrandt, P., Schaffer, M., Gesell Salazar, M., Gerth, U., et al. (2024). Activation of the general stress response sigma factor SigB prevents competence development in Bacillus subtilis. MBio 15:e02274-24. doi: 10.1128/mbio.02274-24
Hecker, M., Pané-Farré, J., and Uwe, V. (2007). Sigb-dependent general stress response in Bacillus subtilis and related gram-positive bacteria. Ann. Rev. Microbiol. 61, 215–236. doi: 10.1146/annurev.micro.61.080706.093445
Hoffmaster, A. R., Novak, R. T., Marston, C. K., Gee, J. E., Helsel, L., Pruckler, J. M., et al. (2008). Genetic diversity of clinical isolates of Bacillus cereus using multilocus sequence typing. BMC Microbiol. 8, 191–199. doi: 10.1186/1471-2180-8-191
Jin, Z., Li, L., Zheng, Y., and An, P. (2021). Diallyl disulfide, the antibacterial component of garlic essential oil, inhibits the toxicity of Bacillus cereus ATCC 14579 at sub-inhibitory concentrations. Food Control 126:108090. doi: 10.1016/j.foodcont.2021.108090
Kosakowska, O., Węglarz, Z., Styczyńska, S., Synowiec, A., Gniewosz, M., and Bączek, K. (2024). Activity of common thyme (Thymus vulgaris L.), Greek oregano (Origanum vulgare L. ssp. hirtum), and common oregano (Origanum vulgare L. ssp. vulgare) essential oils against selected phytopathogens. Molecules 29:4617. doi: 10.3390/molecules29194617
Kovács, Á. T. (2019). Bacillus subtilis. Trends Microbiol. 27, 724–725. doi: 10.1016/j.tim.2019.03.008
Kubista, M. (2007). The prime technique real-time PCR data analysis. GIT Lab. J. Europe 11:33. doi: 10.1002/bmb.21552
Li, F., Zuo, S., Yu, P., Zhou, B., Wang, L., Liu, C., et al. (2016). Distribution and expression of the enterotoxin genes of Bacillus cereus in food products from Jiangxi Province, China. Food Control 67, 155–162. doi: 10.1016/j.foodcont.2016.02.049
Lindbäck, T., Mols, M., Basset, C., Granum, P. E., Kuipers, O. P., and Kovács, Á. T. (2012). CodY, a pleiotropic regulator, influences multicellular behaviour and efficient production of virulence factors in Bacillus cereus. Environ. Microbiol. 14, 2233–2246. doi: 10.1111/j.1462-2920.2012.02766.x
Malorny, B., Hoorfar, H., Bunge, C., and Helmuth, R. (2003). Multicenter validation of the analyticia accuracy of Salmonella PCR: towards an international standard. Appl. Environ. Microbiol. 69, 290–296. doi: 10.1128/AEM.69.1.290-296.2003
Mazzarrino, G., Paparella, A., Chaves-López, C., Faberi, A., Sergi, M., Sigismondi, C., et al. (2015). Salmonella enterica and Listeria monocytogenes inactivation dynamics after treatment with selected essential oils. Food Control 50, 794–803. doi: 10.1016/j.foodcont.2014.10.029
Pellegrini, M., Ricci, A., Serio, A., Chaves-López, C., Mazzarrino, G., D’Amato, S., et al. (2018). Characterization of essential oils obtained from Abruzzo autochthonous plants: antioxidant and antimicrobial activities assessment for food application. Foods 7:19. doi: 10.3390/foods7020019
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. doi: 10.1093/nar/29.9.e45
Popham, D. L., and Setlow, P. (1993). Cloning, nucleotide sequence, and regulation of the Bacillus subtilis pbpF gene, which codes for a putative class a high-molecular-weight penicillin-binding protein. J. Bacteriol. 175, 4870–4876. doi: 10.1128/jb.175.15.4870-4876.1993
Prakash, B., Singh, P. P., Gupta, V., and Raghuvanshi, T. S. (2024). Essential oils as green promising alternatives to chemical preservatives for agri-food products: new insight into molecular mechanism, toxicity assessment, and safety profile. Food Chem. Toxicol. 183:114241. doi: 10.1016/j.fct.2023.114241
Purgatorio, C., Anniballi, F., Scalfaro, C., Serio, A., and Paparella, A. (2024). Occurrence and molecular characterization of Bacillus spp. strains isolated from gnocchi ingredients and ambient gnocchi stored at different temperatures. LWT 192:115703. doi: 10.1016/j.lwt.2023.115703
Purgatorio, C., Serio, A., Chaves-López, C., Rossi, C., and Paparella, A. (2022). An overview of the natural antimicrobial alternatives for sheep meat preservation. Compr. Rev. Food Sci. Food Saf. 21, 4210–4250. doi: 10.1111/1541-4337.13004
Raal, A., Gontova, T., Ivask, A., Orav, A., and Koshovyi, O. (2024). Yield, composition, and chemotypes of essential oils from Origanum vulgare L. aerial parts cultivated in different European countries. Agronomy 14:3046. doi: 10.3390/agronomy14123046
Rossi, C., Chaves-López, C., Serio, A., Casaccia, M., Maggio, F., and Paparella, A. (2022). Effectiveness and mechanisms of essential oils for biofilm control on food-contact surfaces: an updated review. Crit. Rev. Food Sci. Nutr. 62, 2172–2191. doi: 10.1080/10408398.2020.1851169
Rutherford, S. T., and Bassler, B. L. (2012). Bacterial quorum sensing: its role in virulence and possibilities for its control. Cold Spring Harb. Perspect. Med. 2:a012427. doi: 10.1101/cshperspect.a012427
Sini, T. K., Santhosh, S., and Mathew, P. T. (2007). Study on the production of chitin and chitosan from shrimp shell by using Bacillus subtilis fermentation. Carbohydr. Res. 342, 2423–2429. doi: 10.1016/j.carres.2007.06.028
Stenfors Arnesen, L. P., Fagerlund, A., and Granum, P. E. (2008). From soil to gut: Bacillus cereus and its food poisoning toxins. FEMS Microbiol. Rev. 32, 579–606. doi: 10.1111/j.1574-6976.2008.00112.x
Tardugno, R., Serio, A., Purgatorio, C., Savini, V., Paparella, A., and Benvenuti, S. (2022). Thymus vulgaris L. essential oils from Emilia Romagna Apennines (Italy): phytochemical composition and antimicrobial activity on food-borne pathogens. Nat. Prod. Res. 36, 837–842. doi: 10.1080/14786419.2020.1798666
Xu, S., Yang, N., Zheng, S., Yan, F., Jiang, C., Yu, Y., et al. (2017). The spo0A-sinI-sinR regulatory circuit plays an essential role in biofilm formation, nematicidal activities, and plant protection in Bacillus cereus AR156. Mol. Plant-Microbe Interact. 30, 603–619. doi: 10.1094/MPMI-02-17-0042-R
Yeak, K. Y. C., Boekhorst, J., Wels, M., Abee, T., and Wells-Bennik, M. H. (2023). Prediction and validation of novel SigB regulon members in Bacillus subtilis and regulon structure comparison to Bacillales members. BMC Microbiol. 23, 1–23. doi: 10.1186/s12866-022-02700-0
Yehuda, A., Slamti, L., Bochnik-Tamir, R., Malach, E., Lereclus, D., and Hayouka, Z. (2018). Turning off Bacillus cereus quorum sensing system with peptidic analogs. Chem. Comm. 54, 9777–9780. doi: 10.1039/C8CC05496G
Yoshikazu, K., Daniel, R. A., and Errington, J. (2009). Regulation of cell wall morphogenesis in B. subtilis by recruiting PBP1 to the MreB helix. Mol. Microbiol. 71, 1131–1144. doi: 10.1111/j.1365-2958.2009.06601.x
Zhao, L., Duan, F., Gong, M., Tian, X., Guo, Y., Jia, L., et al. (2021). (+)-Terpinen-4-ol inhibits Bacillus cereus biofilm formation by upregulating the interspecies quorum sensing signals diketopiperazines and diffusing signalling factors. J. Agric. Food Chem. 69, 3496–3510. doi: 10.1021/acs.jafc.0c07826
Keywords: Bacillus cereus , Bacillus subtilis , Thymus vulgaris , Origanum vulgare subsp. hirtum, essential oils, gene expression Bacillus cereus, gene expression Bacillus subtilis
Citation: Anniballi F, Purgatorio C, Serio A, Scalfaro C, Taglieri S and Paparella A (2025) Gene expression dynamics in Bacillus cereus and Bacillus subtilis treated with Thymus vulgaris and Origanum vulgare subsp. hirtum essential oils. Front. Microbiol. 16:1643608. doi: 10.3389/fmicb.2025.1643608
Edited by:
Emilia Ghelardi, University of Pisa, ItalyReviewed by:
Supriyo Ray, Bowie State University, United StatesKashif Shamim, University of Mississippi, United States
Copyright © 2025 Anniballi, Purgatorio, Serio, Scalfaro, Taglieri and Paparella. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Fabrizio Anniballi, ZmFicml6aW8uYW5uaWJhbGxpQGlzcy5pdA==
†These authors have contributed equally to this work and share first authorship
 Fabrizio Anniballi
Fabrizio Anniballi Chiara Purgatorio2†
Chiara Purgatorio2† Annalisa Serio
Annalisa Serio Concetta Scalfaro
Concetta Scalfaro Antonello Paparella
Antonello Paparella