- 1College of Marine Sciences, South China Agricultural University, Guangzhou, China
- 2Nansha-South China Agricultural University Fishery Research Institute, Guangzhou, China
The disease caused by Largemouth bass virus (LMBV) is one of the most serious viral diseases that threaten the development of largemouth bass industry. Point-of-Care testing of LMBV infection is crucial to prevent and control disease. Here, we designed a simple and sensitive aptamer-based lateral flow biosensor (LFB) method for the rapid detection of LMBV. In this method, two different aptamers (LA38s and LA13s) specific for LMBV particles were used for target capture and initiation of isothermal strand displacement amplification (SDA), respectively. After adding the reaction product to LFB, the color change of the control zone (C-line) and the test zone (T-line) can be observed by naked eyes within 5 min. The LFB possesses high specificity and accuracy for LMBV detection in vivo and in vitro. The detection limit of LFB is as low as 8 × 101/mL LMBV infected cells, with the detection time of 1 h. Thus, this novel LFB provides a new detection tool, which can be used for rapid detection of LMBV in field.
1 Introduction
Largemouth bass (Micropterus salmoides) is one of the important economic fish species in China, and the production has reached 888,030 metric tons in 2023. However, with the increase of breeding density and the expansion of breeding scale, the outbreak of viral diseases is becoming more and more frequent.
Iridovirus is one of the large nucleoplasmic DNA viruses, which not only brings huge economic losses to the aquaculture industry, but also poses a serious threat to biodiversity (Whittington et al., 2010). Largemouth bass virus (LMBV) was initially isolated from diseased largemouth bass and characterized as genus Ranavirus, family Iridoviridae (Zhang et al., 2022a). It is the main viral pathogen seriously threatening the largemouth bass culture and causing huge economic losses. In addition, LMBV not only infects fish such as bluegill sunfish, red-breasted sunfish, striped bass, spotted bass, smallmouth bass and largemouth bass, but also amphibians such as salamanders and bullfrogs (George et al., 2015; Piaskoski et al., 1999). Largemouth bass infected with LMBV often showed different symptoms, include lethargy, abnormal movement, surface ulcers, hemorrhage, and spleen enlargement (Deng et al., 2011; Zhao et al., 2020; Liu et al., 2023). Hence, it is difficult to diagnose by symptoms of disease caused by LMBV.
Early diagnosis is crucial for virus control and prevention. Presently, several detection methods of LMBV have been developed, mainly including polymerase chain reaction (PCR), real-time quantitative PCR (qRT-PCR), loop-mediated isothermal amplification (LAMP) and isothermal recombinase polymerase amplification assay (RPA), as well as CRISPR/Cas based methods (Zhang et al., 2022b; Zhu et al., 2020, 2022; Guang et al., 2024; Li et al., 2025). Thereinto, conventional PCR and qRT-PCR are the most commonly used. Generally, these detection methods possess high sensitivity and reliability, but require special equipment, well-trained operator and complicated operation in the laboratory (Luo et al., 2014). Thus, an accurate and rapid method for LMBV on-site detection is still needed to ensure the healthy aquaculture of largemouth bass.
Point-of-Care testing (POCT) refers to a rapid and easy-to-use test occurring close the patient rather than in traditional laboratory, which is an attractive pattern for the on-site monitoring pathogen (Ferreira et al., 2018). Lateral flow biosensors (LFBs), also known as test strips, are a representative method in the POCT analysis, due to the noteworthy advantages of rapid, low-cost, user-friendly, high sensitivity and easy interpretation of results (Udhani et al., 2025). LFBs have been widely applied in multiple areas such as disease diagnosis, food safety and environmental assessments (Jauset-Rubio et al., 2017; Majdinasab et al., 2019; Xiao et al., 2021; Wang et al., 2024). Traditionally, most LFBs were designed based on antibodies. In recent years, aptamers have attracted more and more attention and have gradually become a new choice for bio-recognition element.
Aptamers are single-stranded oligonucleotide sequences (DNA or RNA) that can recognize various targets such as proteins, viruses, and bacteria, toxin and heavy metal ions (Zou et al., 2019; Santarpia and Carnes, 2024). In addition, aptamers also have the advantages of high sensitivity and specificity, easy modification, low cost, strong stability and short synthesis cycle, making them suitable as precision tools for diagnosing and treating diseases (Sujith et al., 2024; Majdinasab et al., 2022). Based on aptamers, a variety of LFBs have been developed for detecting various pathogens, such as Escherichia coli, Salmonella, Singapore grouper iridovirus (SGIV), red-spotted grouper nervous necrosis virus (RGNNV), and Mycoplasma hyopneumoniae (Fang et al., 2014; Liu et al., 2020, 2021; Ren et al., 2021; Zhang et al., 2022c).
In previous study, three DNA aptamers targeting LMBV particles were selected, and showed high sensitivity and specificity (Zhang et al., 2022a). From this, we further developed an aptamer based LFB combined with strand displacement amplification (SDA) and gold nanoparticle (AuNPs), for rapid detection of LMBV. This LFB can effectively detect different samples including LMBV particles, LMBV-infected cells, and LMBV-infected tissues. The detection limit reach as low as 8 × 101 cells/mL, and the detection process can be completed within 1 h. Therefore, this novel LFB is a promising and effective detection method for on-site detection of LMBV.
2 Materials and methods
2.1 Reagents and materials
The bovine serum albumin (BSA) (A1933), trisodium citrate (S1804), HAuCl4 (254169), sucrose (S5016), Tween-20 (P9416), and Triton X-100 (93443) were purchased from Sigma-Aldrich (St. Louis, MO, USA). The streptavidin (SA) magnetic beads (Dynabeads™ MyOne™ Streptavidin C1)(65001) were purchased from Invitrogen (Carlsbad, CA, USA). The Nt.BbvCI (R0632L) and the Klenow Fragment exo-DNA polymerase (M0212L) were purchased from New England Biolabs (Ipswich, MA, USA). The LA Taq® (RR02MA) and deoxynucleoside triphosphates (dNTPs)(639125) were purchased from Takara (Shiga, Japan). The nitrocellulose membranes (SHF1350425) were purchased from Millipore (Burlington, MA, USA). The glass fiber (FH8975) and absorbent pads (FH-DB-8025-2) were purchased from Allway Biotech (Guangzhou, China).
2.2 Cells, virus, and aptamers
The fathead minnow (FHM) cells, grouper spleen (GS) cells and grouper brain (GB) cells used in this experiment were cultured in Leibovitz’s L15 (Gibco) medium containing 10% FBS and maintained in 28 °C (Huang et al., 2009, 2011).
LMBV isolated from diseased largemouth bass, was propagated in FHM cells and stored at −80 °C in our laboratory (Huang et al., 2014). Both SGIV and RGNNV were isolated from diseased grouper, propagated in GS and GB cells, respectively, and stored at −80 °C (Qin et al., 2001, 2003).
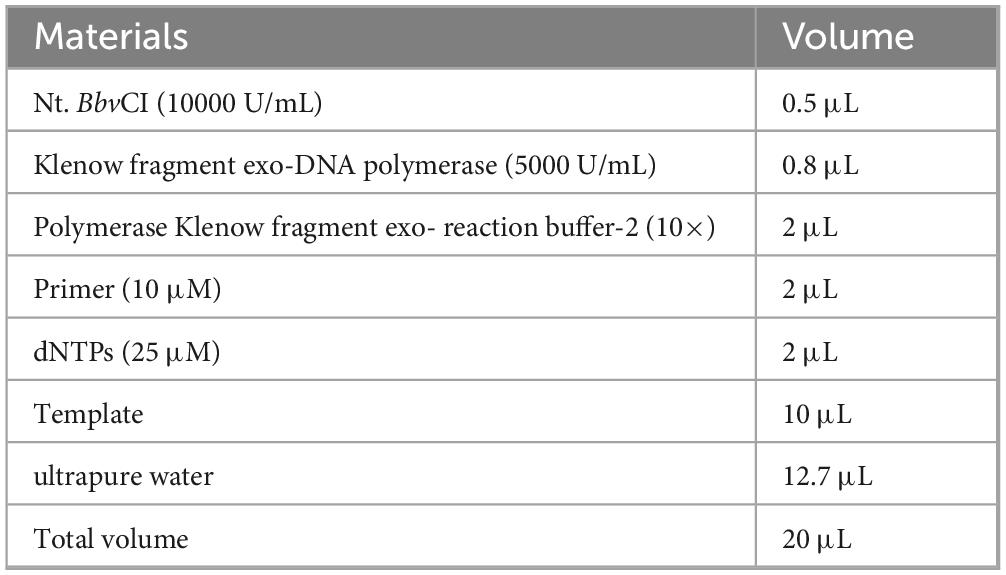
LA38 and LA13 are two specific aptamers targeting to LMBV particles. Through ELASA experiments, their Kd values were calculated to be 5.09 nM and 5.43 nM, respectively (Zhang et al., 2022a). To enhance the performance of these aptamers, the fixed primer sequences at both ends were truncated, and the truncated aptamers were named LA38s and LA13s, respectively. Compared to the original aptamers, the truncated aptamers demonstrated higher binding affinity to LMBV virions, with Kd values of 3.42 nM and 2.34 nM, respectively (Zhang et al., 2022b). In this study, LA38s were modified with biotin (Bio) and used as capture-aptamer. As an amplification aptamer, two specific sequences were added to LA13s for the SDA reaction. One sequence serves as the target site for the Nt.BbvCI (GCTGAGG), while the other sequence acts as the binding site for the primers used in the SDA reaction (TGGACACGGTGGCTTAGT). All aptamers, probes and primer were listed in Table 1, and synthesized by Sangon Biotech.
2.3 Preparation of AuNPs and AuNPs-probe conjugates
The synthesis procedure of AuNPs was carried out as previously described, with some modifications (Fang et al., 2014; Liu et al., 2021). Firstly, 200 mL HAuCl4 (0.01%) was added into a round bottom flask and placed on a magnetic stirrer for heating and stirring. Once the solution boiled, 4 mL of trisodium citrate (1%) was immediately added and stirred, and then the solution was continued to be heated and stirred until it turned wine red. Then, the solution was boiled for 5 min and cooled to room temperature with stirring. AuNPs were obtained, collected and stored in 4 °C with dark environment.
To prepare AuNPs-probe conjugates, AuNPs needs to be condensed. Briefly, 6 mL of AuNPs was centrifuged at 12,000 × g for 20 min, and the precipitate was collected by 900 μL PBS. Then, the thiol modified DNA (AuNPs probe, 10 μM, 100 μL) was added and gently shaken at 4 °C for 12 h. The solution was blocked by 10% BSA for 3 h, followed by the addition of 1.5 M NaCl and 1% sodium dodecyl sulfate (final concentrations of 150 mM and 0.01%, respectively). The solution was gently shaken at 4 °C for 12 h, and centrifuged at 12,000 × g for 20 min at 4 °C. The precipitate was gently resuspended in 1 mL of rinsing buffer (20 mM Na3PO4, 5% BSA, 0.25% Tween-20 and 10% sucrose). Finally, the solution was repeatedly centrifuged twice, and the precipitate was resuspended in 100 μL of rinsing buffer. The AuNP-probe solution was stored at 4 °C until use.
2.4 Construction of the LFB
As shown in Figure 1, the LFB consisted of an adhesive backing, a sample pad, an AuNPs-probe conjugate pad, a nitrocellulose membrane, and an absorbent pad. Firstly, the sample pad was immersed in the sample pad buffer (0.5% Triton, 2% sucrose, 1% BSA, 50 mM boric acid, pH 8.0) for 2 h, then dried overnight at 37 °C for 12 h and stored at room temperature. Subsequently, the sample pad, conjugate pad, nitrocellulose membrane, and absorbent pad were adhered to the adhesive backing in turn, with an overlap of approximately 1.5 mm between adjacent parts. The T-line (T-line probe, 100 μM, 40 μL) and C-line (C-line probe, 100 μM, 40 μL) were dispensed onto the nitrocellulose membrane using the three-dimensional reciprocating gold sprayer (HM3260; Shanghai Kinbio, China) at a dispensing rate of 0.7 mL/cm, creating the C-line and T-line, respectively. The distance between the two probes was approximately 7 mm. Finally, the strip was cut into a size of 6 cm in length and 3.5 mm in width by a guillotine cutting machine (ZQ2002; Shanghai Kinbio, China) and stored at room temperature with a dry environment.
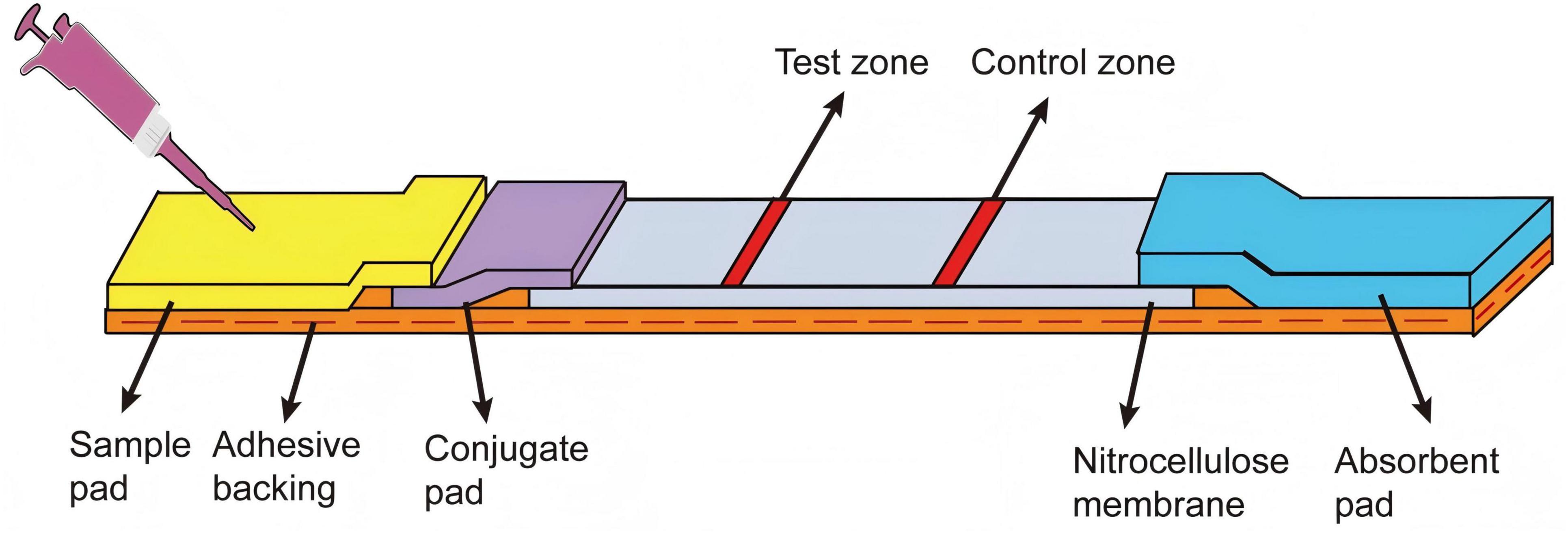
Figure 1. Configuration of the LFB. The sample pad, AuNPs-probe conjugate pad, nitrocellulose membrane and absorbent pad were assembled on the adhesive backing. The test zone (T-line) and control zone (C-line) were marked on the nitrocellulose membrane.
2.5 Virus-infected cell samples
To obtain cell samples for testing, cells (FHM cells, GS cells and GB cells) were cultured in a 24 well plates for 18 h. Then the virus (LMBV, SGIV and RGNNV) at a multiplicity of infection (MOI) of 0.5 was added, and the cytopathic effect was observed by optical microscope at 96 h post-infection. The virus-infected cell suspension was collected, centrifuged at 900 × g for 10 min, and lysed in Pierce IP™ Lysis buffer (Thermo Fisher Scientific, United States). Then they were washed with phosphate buffered saline (PBS; 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, pH 7.4) and resuspended in 1 mL PBS. Uninfected cells were used as the control group. The cell samples were stored at −80 °C until use.
2.6 Virus-infected tissue samples
Largemouth bass (8–10 cm) were purchased from a farm in Foshan, Guangdong Province, China, and used for LMBV challenge experiments. They were maintained in a circulating water system at 28 ± 1 °C and fed twice daily. Before the challenge experiment, fish were randomly selected for virus detection to ensure that they did not infect LMBV. As previously described, largemouth bass were infected with 100 μL LMBV (104.7TCID50/mL) by intraperitoneal injection. At 4 days post-infection, liver, spleen and kidney tissues were collected from diseased fish. After washing with PBS, the tissues were cut into small pieces and lysed in 300 μL of Pierce IP™ Lysis buffer (Thermo Fisher Scientific, United States) at 4 °C for 10 min. Then, the lysates were filtered through medical gauze, and the supernatants were collected for LFB detection.
2.7 Procedure for LMBV detection
The detection process of the LFB was illustrated in Figure 2. Firstly, the capture-aptamer (500 nM) and amplification-aptamer (500 nM) were incubated with LMBV in 100 μL PBS for 1 h. Subsequently, 1.5 μL of SA-modified magnetic beads were added, and the mixture was shaken at 4 °C for 30 min. The capture-aptamer-LMBV-amplification-aptamer-magnetic beads complex was collected by a magnetic separator. After washing three times of PBST (PBS supplied with 0.1% Tween-20), the complex was resuspended in 10 μL of ultrapure water and used as a template for the SDA reaction. As shown in Table 2, the SDA reaction system mainly consisted of primer, Nt.BbvCI enzyme and Klenow Fragment exo-DNA polymerase, performed at 37 °C for 30 min. The amplification-aptamers were modified with Nt.BbvCI recognition sites. The reaction starts with the hybridization of the primer and primer binding sequence, followed by DNA polymerase extension. The newly formed complementary chain was cleaved at the Nt.BbvCI cleavage site, and then the extension reaction continued at the cleavage site. This process was repeated, and a large amount of ssDNA was amplified. Finally, the ssDNA were added to the sample pad of LFB. After rinsing with 30 μL of saline-sodium citrate (0.6 M NaCl, 0.06 M sodium citrate, pH = 7.0), the detection results of LFB can be observed within 5 min.
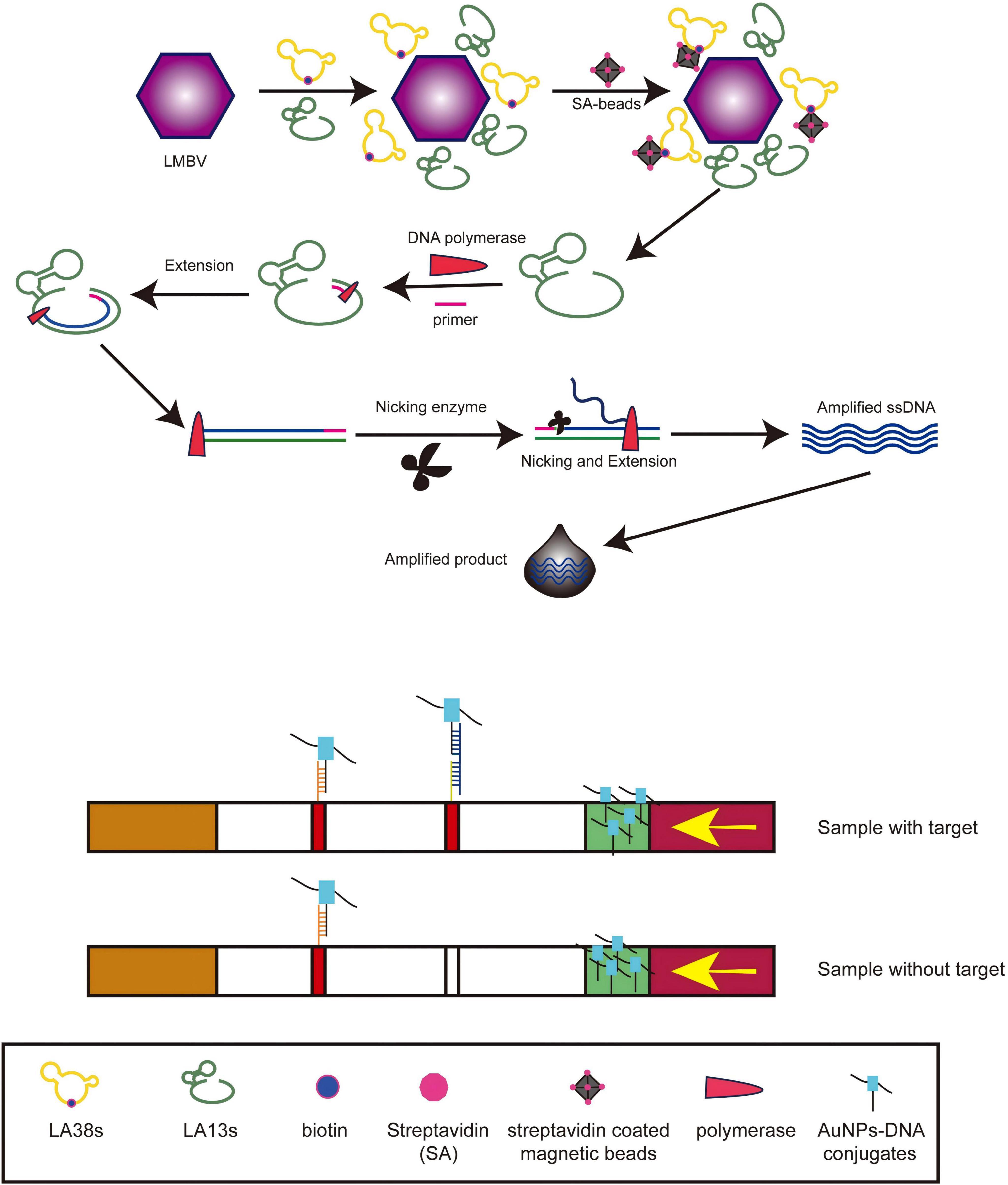
Figure 2. The procedure for LMBV detection by LFB. After incubation of LA38s and LA13s with LMBV, SA-modified magnetic beads were added, and the capture-aptamer-LMBV-amplification-aptamer-magnetic beads complex was collected through a magnetic separator. After SDA reaction, the product was added to the sample pad for detection.
2.8 The optimum working condition
Briefly, LMBV-infected FHM cells at a concentration of 8 × 107/mL were used as the test samples, while uninfected FHM cells served as the control. In order to obtain the optimal temperature of the SDA reaction, we set the temperature gradient (25 °C, 37 °C, 42 °C, 50 °C, and 65 °C) for testing. In addition, in order to shorten the detection time, we first changed the incubation time of the sample, capture-aptamer and amplification-aptamer (2 min, 5 min, 10 min, 20 min, and 30 min). Subsequently, we varied the incubation time of the the capture-aptamer-sample-amplification-aptamer complex and magnetic beads (2 min, 5 min, 10 min, 20 min, and 30 min) to test the assay performance of the LFB.
2.9 Sensitivity analysis of LFB
Briefly, LMBV-infected FHM cells were diluted to various concentrations: 8 × 107,8 × 106, 8 × 105, 8 × 104, 8 × 103, 8 × 102 and 8 × 101/mL. Uninfected FHM cells were used as the control group. The cells were collected, lysed and tested for the LFB as described above. In order to detect accurately, LFB was scanned by a portable test strip reader (Hangzhou Allsheng, Zhuantang Town, China). It can record the optical intensity of the C-line and the T-line.
2.10 Sample preparation for LFB and PCR detection
Thirty largemouth bass (10 ± 5 cm in length) with suspected LMBV infection were collected from farms in Guangdong Province, China. These fish were dissected and the spleen tissue were divided into two parts, one for LFB detection and the other part was used to extract DNA for PCR detection.
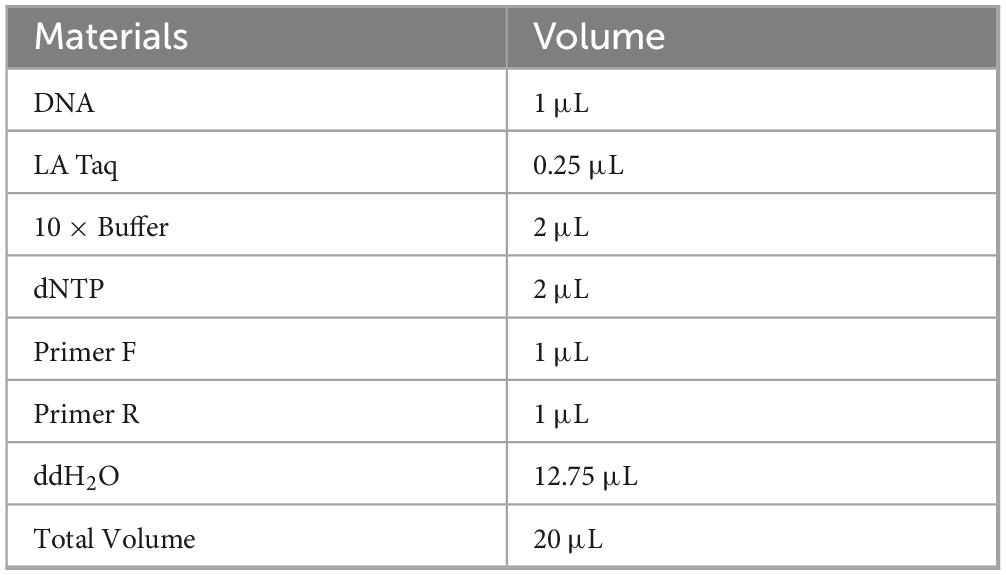
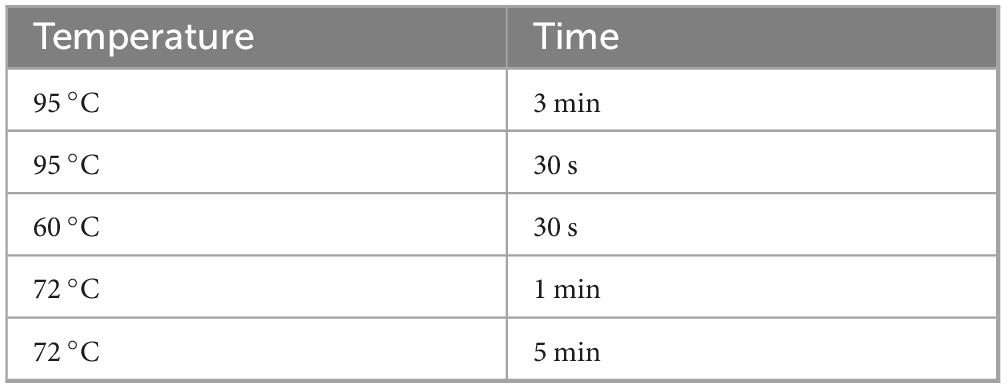
Sample preparation and testing procedures for LFB are as described above. DNA from spleen tissue was extracted using the TIANGEN DNA extraction kit. The PCR system was prepared using the LA Taq (TAKARA) kit as shown in Table 3. Primers (forward: ATTATCCCGTGGGTTGGTT and reverse: CGATGGGCTTGACTTCTCC) specific for the gene (ORF579) encoding the LMBV MCP protein were used for PCR detection. According to the reaction conditions shown in Table 4, 35 cycles were performed by the PCR instrument to amplify the product. Subsequently, 7 μL PCR product was diluted with 10-fold loading buffer (Takara, Shiga, Japan) and coated on 1% (w/v) agarose gel containing ethidium bromide. The agarose gel was electrophoresed in 1 × TAE buffer at 170 V constant voltage for 15 min. Finally, it was placed in a gel imager for imaging analysis.
3 Results
3.1 Characterization of synthesized AuNPs- probe conjugates
As shown in Figure 3, the AuNPs solution had a transparent red with no precipitation. After 6 times of concentration, concentrated AuNPs turned to wine red. The 6 × AuNPs was coupled with the probe, blocked and aged to obtain AuNPs-probe conjugate. The conjugate was dark red, stable, clear and transparent.
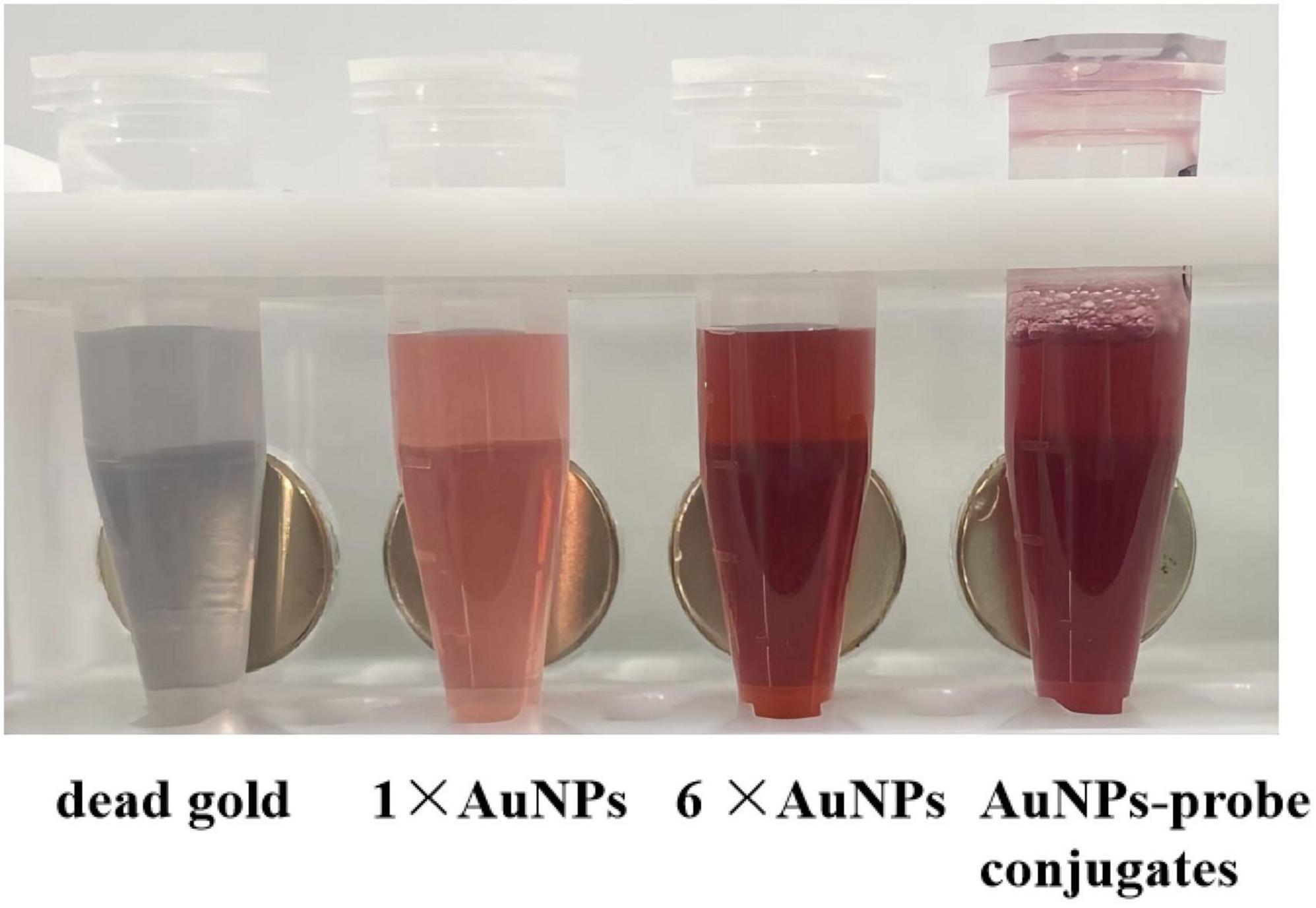
Figure 3. The preparation of AuNPs-probe conjugates. From left to right: dead gold caused by excessive NaCl, AuNPs solution, concentrated 6 times of AuNPs solution, AuNPs-probe conjugates solution.
3.2 Validation of the biosensor
After assembling the LFB, we designed four group of experiments to verify the biosensor for LMBV detection. In the experimental group, LMBV (MOI = 10) was incubated with both the amplification-aptamer (LA13s) and the capture-aptamer (LA38s). In control group 1, LMBV was incubated with only capture-aptamer. In control group 2, LMBV was incubated with only amplification-aptamer. In control group 3, PBS was incubated with both the amplification-aptamer and the capture-aptamer. The detection results were shown in Figure 4. Only when LMBV coexists with the amplification-aptamer and the capture-aptamer, the T-line and C-line of LFB can become red. In the three control groups, only the C-line showed red color, with no color change on the T-line.
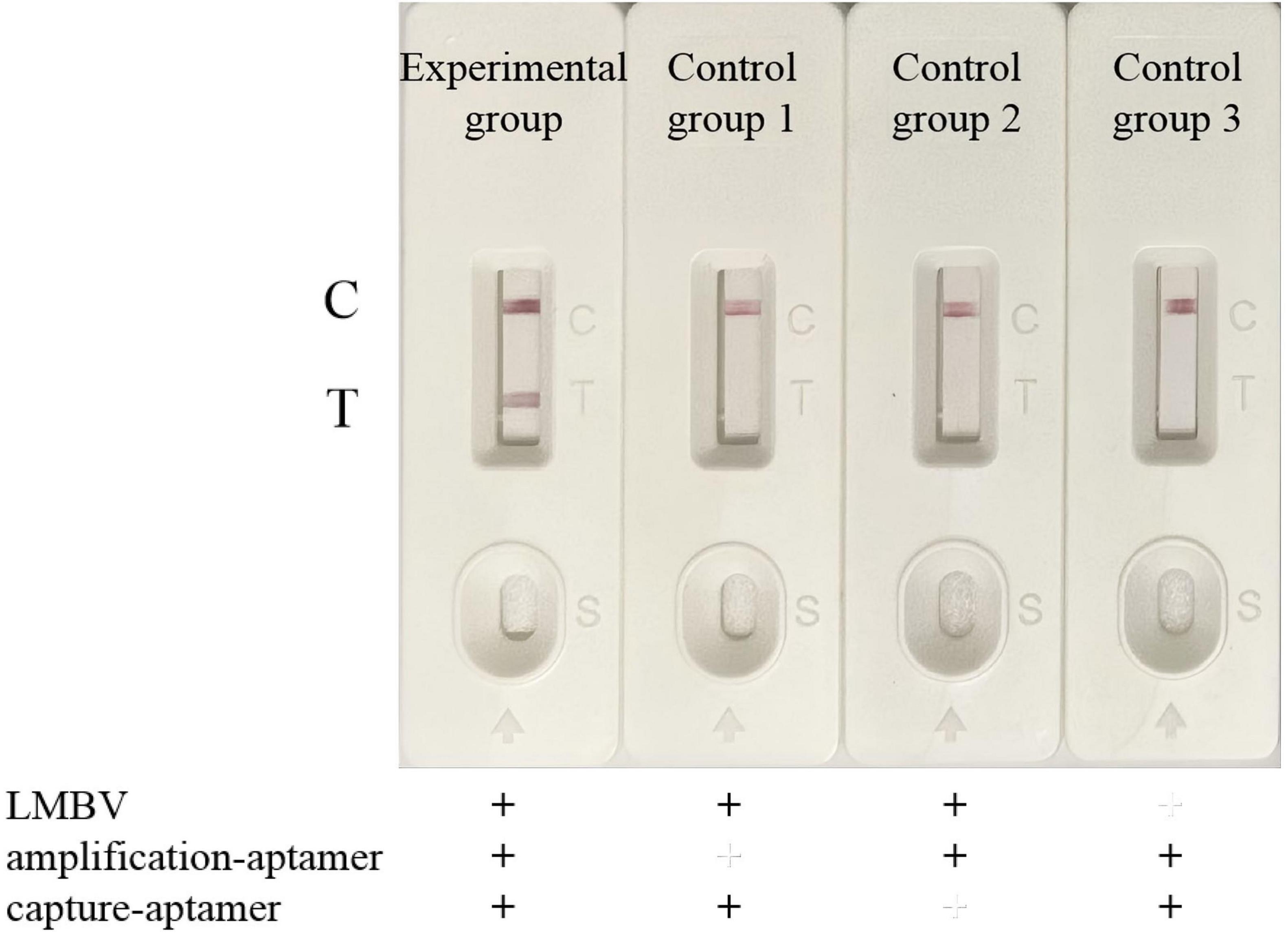
Figure 4. Verification of the LFB for LMBV detection. From left to right: Experimental group, LMBV was co-incubated with amplification-aptamer and capture-aptamer. Control group 1, LMBV was incubated with capture-aptamer. Control group 2, LMBV was incubated with the amplification-aptamer. Control group 3, PBS was incubated with amplification-aptamer and capture-aptamer. T-line could only be observed obviously in the presence of LMBV, amplification-aptamer and capture-aptamer. In the absence of LMBV, capture-aptamer or amplification-aptamer, LFB only showed C-line, and no positive T line appeared. The experiment was performed in triplicates.
The capture-aptamer was biotinylated, allowing it to non-covalently bind to SA-modified magnetic beads, thereby enriching the captured target in the magnetic beads. The amplification-aptamer has an enzyme cleavage site, which is a necessary condition for the SDA reaction. Only when the capture-aptamer, amplification-aptamer and target exist, the capture-aptamer-LMBV-amplification-aptamer-magnetic bead complex can be formed, and ssDNA can be amplified by SDA reaction. The ssDNA, which is complementary to the sequence of T-line probe and the sequence of AuNPs-probe, produced a red strip in the T-line region. Additionally, the sequence of AuNPs-probe was also complementary to the sequence of C-line probe. Under the action of capillary force, the excessive AuNPs-probe conjugate complex continued to migrate, and color reaction occured in the C-line region. When there is no LMBV in the detection system, the complex cannot be formed. In the absence of capture-aptamers or amplification-aptamers, the target cannot be enriched by magnetic beads or SDA reaction cannot be performed. Therefore, only when the LMBV, capture-aptamer and amplification-aptamer are present, the positive T line can be seen. In the absence of target, capture-aptamer or amplification-aptamer, the positive T-line could not be observed. The above results showed that the LFB was able to effectively detect LMBV.
3.3 Specificity of the LFB
To evaluate the specificity of LFB, different purified viruses, virus-infected cell and virus-infected tissue were used as samples to be tested. The PBS or uninfected tissue were used as the control groups. As shown in Figure 5A, LMBV particle samples showed a clear and visible T-line, while SGIV particle samples and control group only showed C-line, without positive T-line. As shown in Figure 5B, only LMBV-infected FHM cell samples were detected positive, while SGIV-infected GS cell samples and RGNNV-infected GB cells samples did not show positive T lines. Furthermore, biosensors were used to detect the liver, spleen and body kidney tissue samples of the infected largemouth bass. As shown in Figure 5C, the three tissue samples all displayed the visible T-line. These results demonstrated that the LFB had high specificity for LMBV detection, which can effectively detect LMBV particles, LMBV-infected cells and tissue samples.
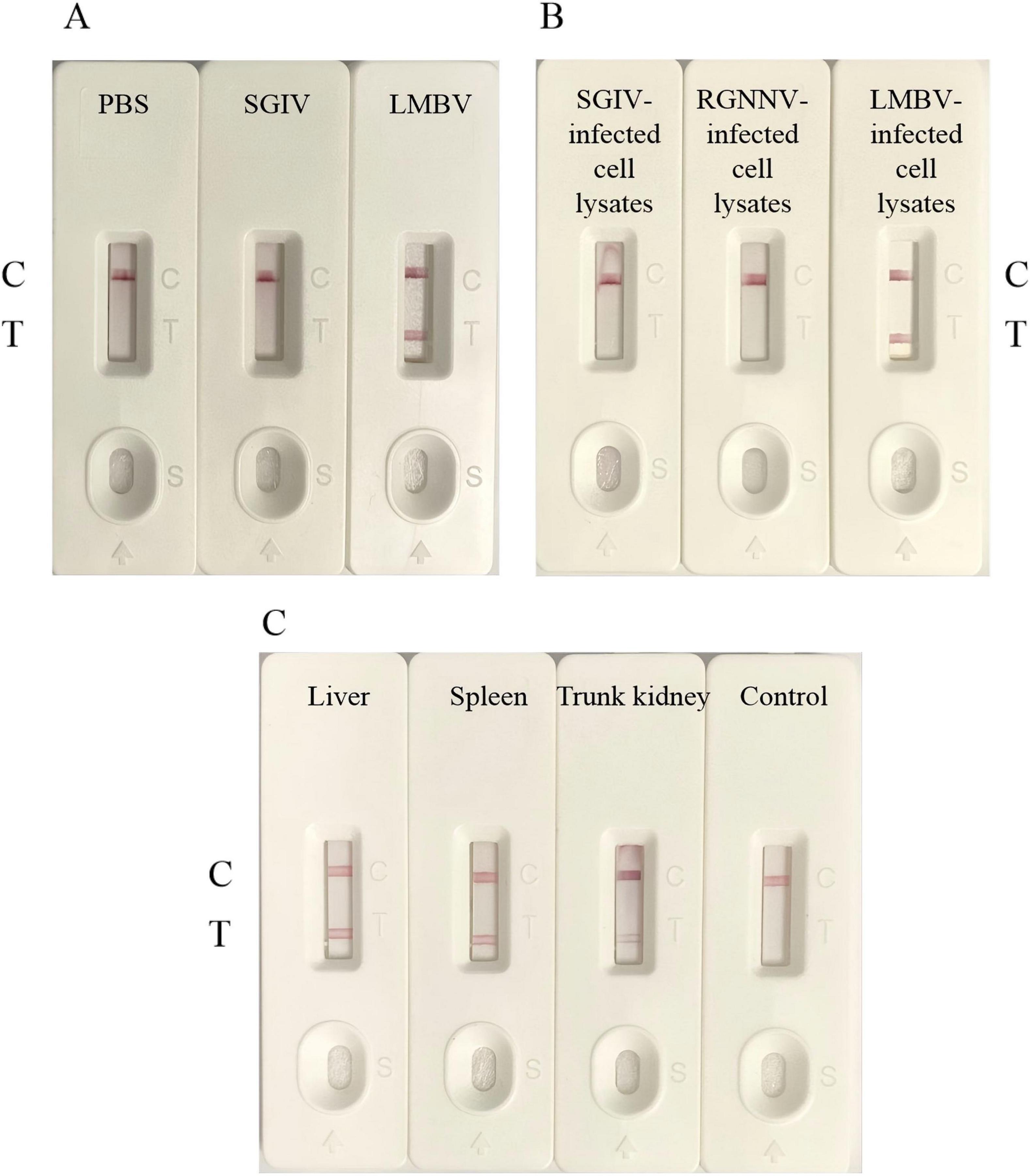
Figure 5. The specificity of LFB. (A) LFB could specifically detect LMBV particles. (B) LFB could specifically detect LMBV infected cell lysates. (C) LFB could specifically detect the liver, spleen and kidney tissues of largemouth bass infected with LMBV. The experiment was performed in triplicates.
3.4 Optimization of experimental conditions
In order to further improve the detection efficiency of LFB, we optimized some detection steps. We collected and lysed 8 × 107 LMBV-infected FHM cells for LFB detection. Uninfected FHM cells were used as the control. Firstly, different temperatures (25 °C, 37 °C, 42 °C, 50 °C, and 65 °C) were set to be applied to SDA. When the reaction temperature was as low as 25 °C or as high as 65 °C, the SDA reaction could not proceed properly, and the LFB displayed only a C-line without T-line (Figure 6A). When the reaction temperature was 37 °C, 42 °C and 50 °C, the SDA reaction can proceed normally, and the LFB showed a clear red detection line. It is worth noting that the T-line band of the 50 °C experimental group was significantly darker than the other two groups (Figure 6A). This indicated that the SDA reaction has higher amplification efficiency under this condition, so that the biosensor could capture more targets. Subsequently, we reduced the incubation time of the sample and two aptamers to determine the effect of incubation time. Even though the incubation time was reduced to 2 min, the LFB could still showed a light red T-line (Figure 6B). In addition, we also changed the incubation time of capture-aptamer-sample-amplification-aptamer complex and magnetic beads. As shown in Figure 6C, when the incubation time was 2 min, 5 min and 10 min, LFB all showed a red T-line, but the color of the T-line became darker with the increase of incubation time. We believed that this phenomenon was due to the fact that the binding reaction between the magnetic beads and the capture-aptamer had not yet reached the upper limit. There was almost no difference in the color of the detection line between the 20 min experimental group and the 30 min experimental group. Therefore, the experimental results showed that 20 min could fully combine the magnetic beads with the capture-aptamer-sample-amplification-aptamer complex. The above results showed that the optimal reaction temperature of SDA was 50 °C. After optimizing the LFB detection procedure, the whole detection process could be completed within 1 h.
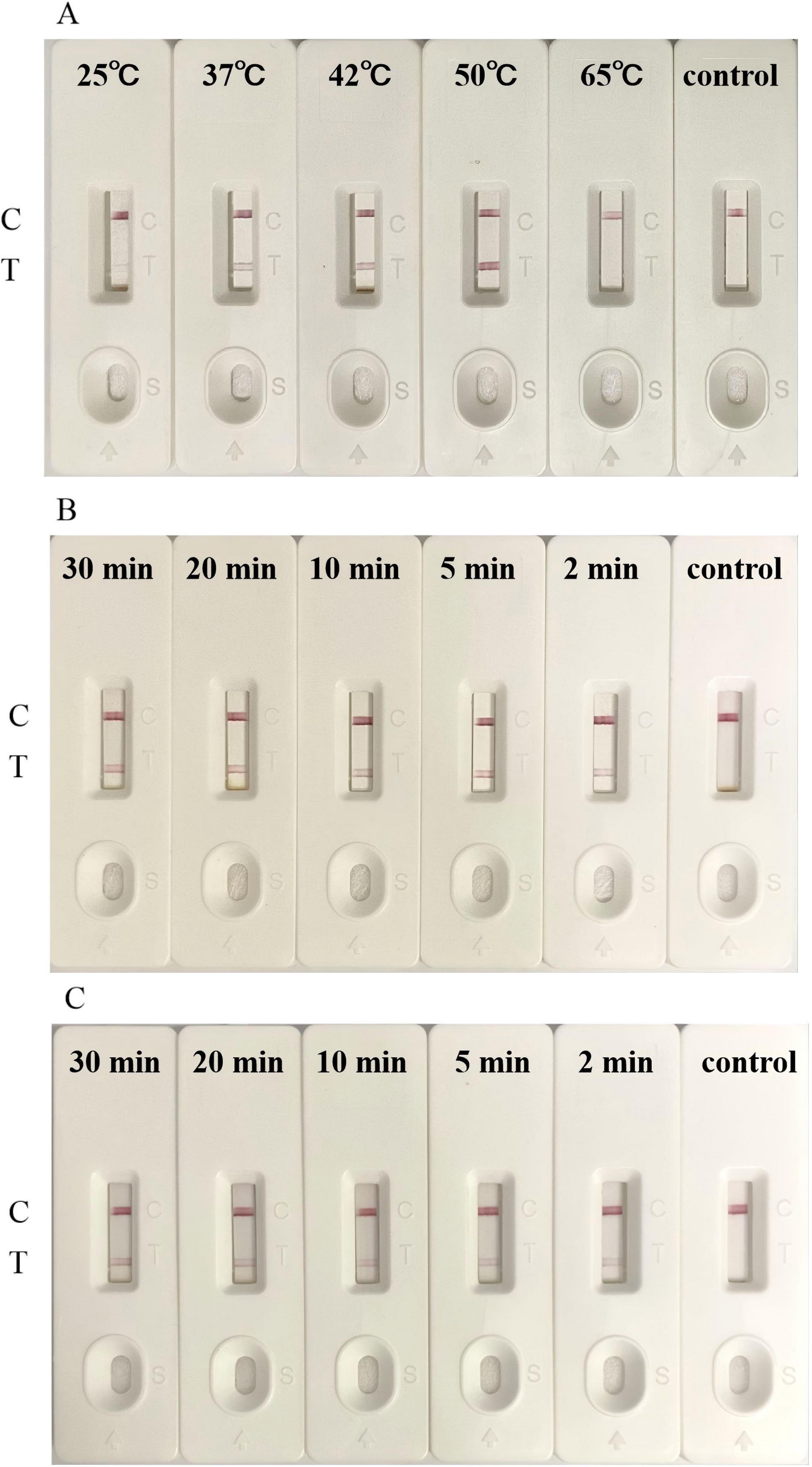
Figure 6. Optimization of LFB for detection. (A) The optimal SDA temperature for the detection of LMBV infection by LFB was 50 °C. (B) The LFB detected LMBV infection with an incubation time for sample and two aptamers as short as 2 min. (C) The optimal incubation time between the capture-aptamer-sample-amplification-aptamer complex and magnetic beads was 20 min. Uninfected FHM cells were used as the control. The experiment was performed in triplicates.
3.5 Sensitivity of the LFB
The sensitivity of LFB was verified by various concentrations of LMBV-infected FHM cells. As shown in Figure 7, in the control group, no positive T-line was observed. When LMBV-infected FHM cells were as low as 8 × 101/mL, their lysates could still be detected by LFB. At the same time, the peaks corresponding to the T-line and the C-line were observed by the strip reader. The peak area of the T-line decreased with the decrease of the concentration of LMBV-infected cells. Thus, these results suggested that the detection limit of the LFB was 8 × 101/mL.
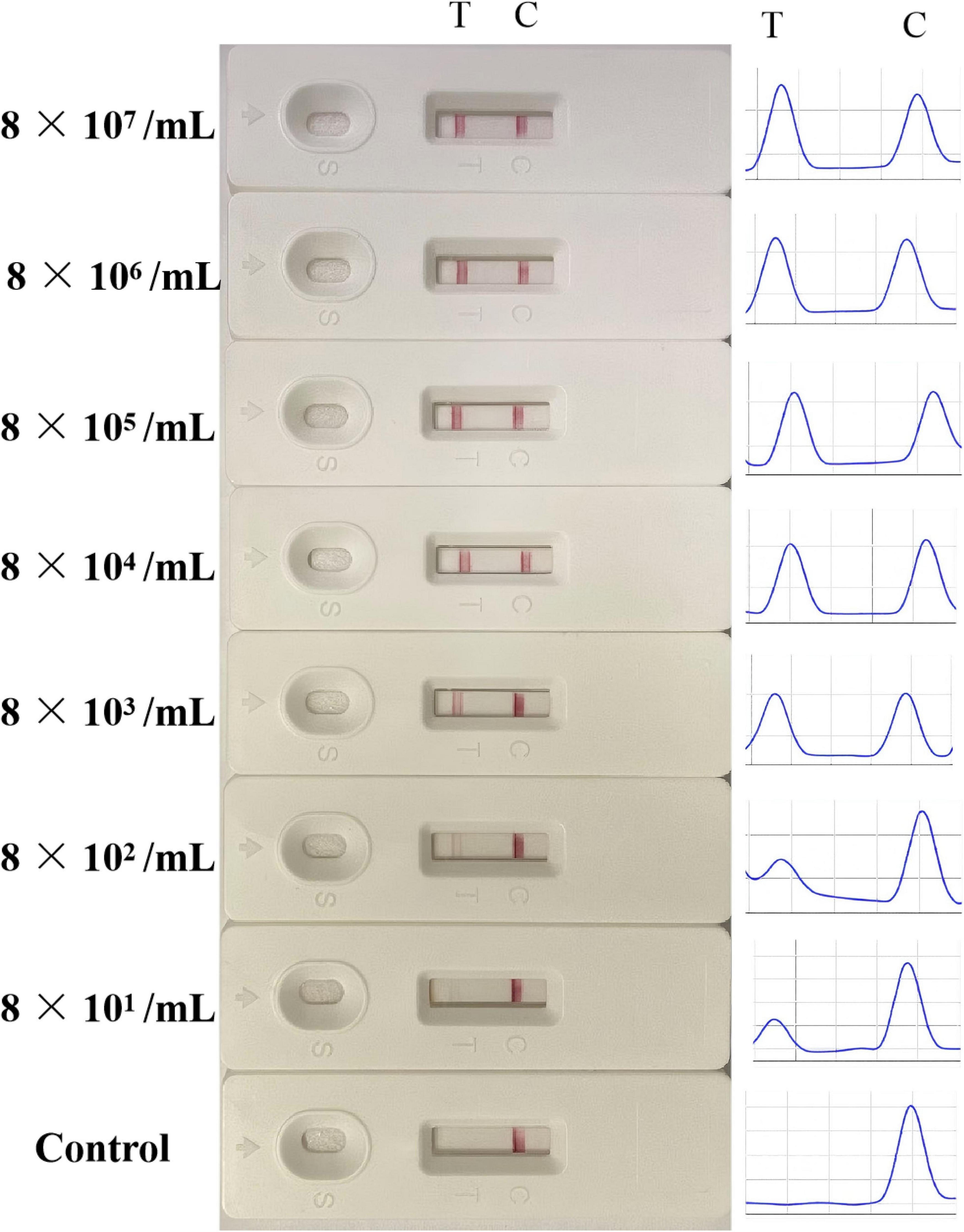
Figure 7. The sensitivity analysis of the LFB. Various concentrations of LMBV-infected FHM cells (8 × 107, 8 × 106, 8 × 105, 8 × 104, 8 × 103, 8 × 102 and 8 × 101/mL) were prepared, and uninfected FHM cells were used as control group. The experiment was performed in triplicates.
3.6 Sample for LFB and PCR detection
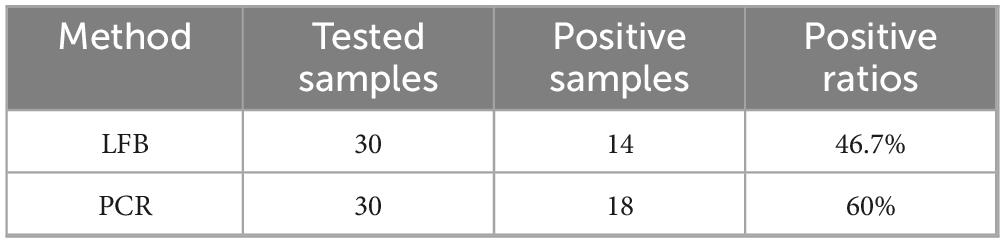
The existence of LMBV in 30 largemouth bass collected from farms was detected by both LFB and PCR. The results of LFB showed that 14 (46.7%) samples were positive of LMBV and 16 (60%) samples were negative of LMBV (Figure 8A). Among them, 18 were confirmed positive by the PCR (Figure 8B). This means that 4 (13.3%) samples were positive by PCR, but LFB were negative (Table 5). Overall, the accuracy of LFB was slightly lower than that of PCR, but LFB was able to obtain the results within 1 h.
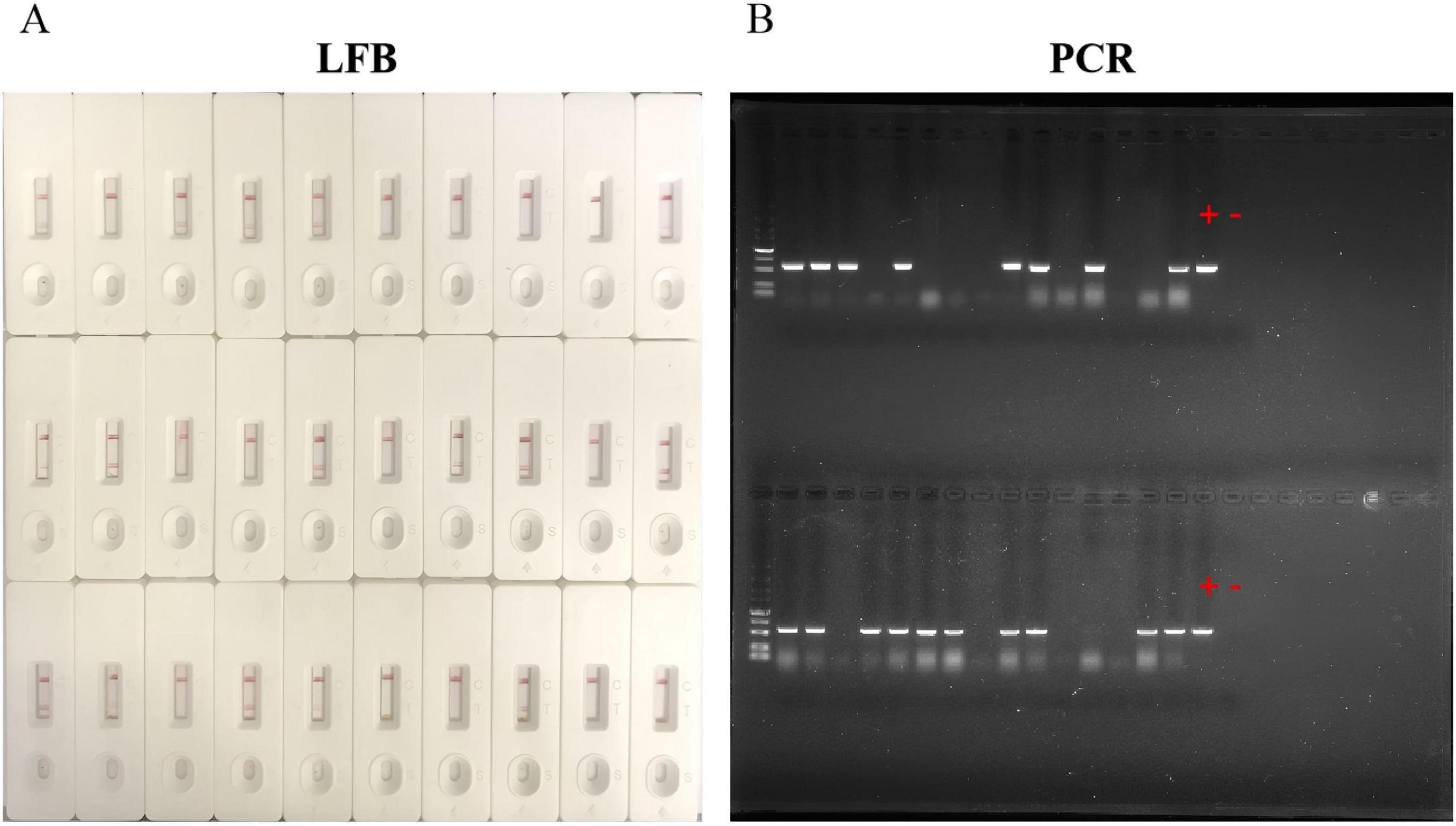
Figure 8. Detection of largemouth bass. (A) 14 positive samples were detected by LFB. (B) 18 positive samples were detected by PCR. “+” represents the positive control sample, “–” represents the negative control sample.
4 Discussion
The frequent outbreaks of disease caused by LMBV have severely interfered the healthy development of the largemouth bass industry. The clinical symptoms of LMBV infection are variable and complicated, making it difficult to diagnose by symptom signs (Jin et al., 2022). This poses new challenges for detection. The results of clinical symptoms and pathological observations can only be used as auxiliary judgments. Accurate diagnose is essentially important for preventing and controlling disease. Hence, rapid and on-site detection methods for LMBV are necessary.
For the detection of LMBV, multiple nucleic acid amplification techniques have been developed, such as droplet digital PCR (ddPCR), qRT-PCR, LAMP, and RPA. The lowest detection limit of RT-qPCR for LMBV was 5.8 copies/μL, with a 100% positive detection rate (Guo et al., 2022). Jiang et al. (2023) developed a ddPCR method that detect as low as 2 copies/μL. Guang et al. (2024) employed recombinase-aided amplification technology coupled with CRISPR and Cas13a for LMBV detection, achieving a detection limit as low as 3.1 × 101 copies/μL. Similarly, a detection method based on RPA-CRISPR/Cas12 was established, achieving a sensitivity of 50 copies/reaction within 40 min (Li et al., 2025). The LAMP assay targeting the LMBV MCP gene is highly specific for LMBV diagnosis, as it shows no cross-reactivity with large yellow croaker iridovirus or SGIV (Zhu et al., 2020). Additionally, an RPA assay targeting the LMBV MCP gene was developed for LMBV detection, with a detection limit of 89 copies/μL (Zhu et al., 2022). In this study, the LFB demonstrated high detection specificity for LMBV without cross-reactivity with RGNNV or SGIV. This method achieved a lower detection limit of 8 × 101/mL LMBV-infected cell. However, LFB does not target specific viral genes and eliminates the need for nucleic acid extraction steps.
To date, various aptamer-based LFBs have been reported, which are increasingly recognized as user-friendly detection tools. They have been applied in areas such as pathogens, hormones, and disease-associated biomarkers. Based on aptamer (Np-A48), an ultrasensitive electrochemical aptamer-based aptasensor for SARS-CoV-2 detection was developed using CRISPR/Cas12a, achieving a detection limit of 16.5 pg/mL (Han et al., 2022). Xu et al. (2025) developed an ultrasensitive surface-enhanced Raman spectroscopy technique using aptamer-antibody dual-recognition elements for rapid detection of intraoperative parathyroid hormone. There is also an LFB that utilizes aptamers to capture targets for the detection of human osteopontin protein (OPN), achieving qualitative and semi-quantitative detection of OPN in the serum of cancer patients at clinical cutoff levels (Mukama et al., 2020).
In field testing, LFBs demonstrated a detection rate of 46.7%, slightly lower than PCR. Nevertheless, they maintain significant advantages in operational simplicity and rapid detection. Furthermore, sensitivity of LFBs can be further improved through optimization of both aptamers and the detection system. Aptamers are the key recognition element of the LFB method, so their properties largely determine the detection sensitivity. Aptamers can be further optimized in different ways such as truncation, chemical modification, mutation of nucleic acid base, and construction of multivalent aptamers. For example, the truncation of aptamers against Gonyautoxin 1/4 obviously increase the binding affinity, with a higher Kd of 17.7 nM (Gao et al., 2016). The aptamers used in this study were also truncate, and showed higher affinity compared with the original aptamers.
Several rapid detection methods have been developed based on aptamer-designed detection systems combined with SDA. For example, an aptamer-mediated double SDA-microchip electrophoresis method achieved a detection limit as low as 6 CFU/mL for Salmonella Typhimurium (Lu et al., 2024). Lu et al. (2025) developed an A-LKS-mediated SDA-Cas12a signal cascade biosensor for ultrasensitive aflatoxin B1 detection. Islam et al. (2021) developed a label-free method for detecting miR-215 in saliva and serum using aptamers, thioflavin T dye, and SDA. In this study, LA38s and LA13s with high affinity and specificity of LMBV particles were used to capture target and initiate subsequent SDA reaction, respectively. SDA reaction could be conducted under isothermal condition, and reaction product further amplified detection signal (Zhang et al., 2021). Combined with aptamers and SDA, a novel LFB was developed for visual detecting of LMBV, without requiring special equipment and laboratory conditions. After optimization, the detection for LMBV could be completed in 1 h, with detection limit of 8 × 101/mL LMBV infected cells. These two aptamers had been generated and applied in developing aptamer-based sandwich enzyme-linked apta-sorbent assay (ELASA) for LMBV detection. The detection limit of ELASA was 1.25 × 102/mL LMBV-infected cells, and the detection time required approximately 4 h (Zhang et al., 2022b). Obviously, the LFB showed higher sensitivity and shorter detection time, compared with that of ELASA.
Currently, the development of aptamer-based LFB for aquatic animal virus detection remains at an early stage. Building upon the RGNNV-specific aptamer, Liu et al. (2020) developed a sensitive LFB capable of detecting RGNNV-CP protein at concentrations as low as 5 ng/mL. In another study, researchers developed an LFB for rapid SGIV detection by employing two SGIV-specific aptamers, achieving detection within 90 min (Liu et al., 2021). Several studies have integrated nucleic acid amplification techniques with LFB to achieve visual detection (Preena et al., 2022). For instance, rapid detection LFBs have been developed targeting specific viral genes, such as the SS N gene of Micropterus salmoides rhabdovirus (MSRV), as well as the MCP and ORF 007 genes of infectious spleen and kidney necrosis virus (ISKNV; Feng et al., 2022; Li et al., 2020). In this study, two LMBV-specific aptamers were combined with SDA, and LFB was utilized to achieve visual detection. The detection limit of LFB is as low as 8 × 101/mL LMBV infected cells, and the color change of the LFB can be observed by the naked eye within 5 min.
5 Conclusion
In conclusion, we developed a novel LFB based on specific aptamers, with high sensitivity, specificity and stability. It was time-saving, easy to handle, visualization of results, and did not need special equipment, thus providing a valuable option for rapid detection for LMBV in field. Thus, LFBs contribute to sustainable aquaculture by ensuring fish health, optimizing therapeutic interventions, mitigating disease-related financial risks, and enhancing production yields.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics statement
The animal study was approved by the Animal Care and Use Committee of College of Marine Sciences, South China Agricultural University. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
XZ: Writing – review & editing, Writing – original draft, Resources, Methodology. LG: Software, Writing – original draft, Formal analysis. ZX: Investigation, Writing – review & editing. HW: Validation, Writing – review & editing. XW: Writing – review & editing, Visualization. MY: Data curation, Investigation, Writing – review & editing, Conceptualization. QQ: Writing – review & editing, Project administration, Conceptualization, Supervision. SW: Funding acquisition, Writing – review & editing, Validation.
Funding
The author(s) declare that financial support was received for the research and/or publication of this article. This work was supported by the National Key Research and Development Program of China (2023YFD2402300) and the Seed Industry vitalizatio project in Rural Vitalization Strategy of Guangdong, China (2024SPY00019).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Generative AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Deng, G. C., Li, S. J., Xie, J., Bai, J. J., Chen, K. C., Ma, D. M., et al. (2011). Characterization of a ranavirus isolated from cultured largemouth bass (Micropterus salmoides) in China. Aquaculture 312, 198–204. doi: 10.1016/j.aquaculture.2010.12.032
Fang, Z., Wu, W., Lu, X., and Zeng, L. (2014). Lateral flow biosensor for DNA extraction-free detection of salmonella. Biosens. Bioelectron. 192–197. doi: 10.1016/j.bios.2014.01.015
Feng, Z., Chu, X., Han, M., Yu, C., Jiang, Y., Wang, H., et al. (2022). Rapid visual detection of Micropterus salmoides rhabdovirus using recombinase polymerase amplification combined with lateral flow dipsticks. J. Fish. Dis. 45, 461–469. doi: 10.1111/jfd.13575
Ferreira, C., Guerra, J., Slhessarenko, N., Scartezini, M., França, C., Colombini, M., et al. (2018). Point-of-care testing: General aspects. Clin. Lab. 64, 1–9. doi: 10.7754/Clin.Lab.2017.170730
Gao, S., Hu, B., Zheng, X., Cao, Y., Liu, D., Sun, M., et al. (2016). Gonyautoxin 1/4 aptamers with high-affinity and high-specificity: From efficient selection to aptasensor application. Biosens. Bioelectron. 79, 938–944. doi: 10.1016/j.bios.2016.01.032
George, M. R., John, K. R., Mansoor, M. M., Saravanakumar, R., Sundar, P., and Pradeep, V. (2015). Isolation and characterization of a ranavirus from koi, Cyprinus carpio L., experiencing mass mortalities in India. J. Fish. Dis. 38, 389–403. doi: 10.1111/jfd.12246
Guang, M., Zhang, Q., Chen, R. G., Li, H. M., Xu, M. R., Wu, X. M., et al. (2024). Rapid and facile detection of largemouth bass ranavirus with CRISPR/Cas13a. Fish. Shellfish. Immun. 148:e109517. doi: 10.1016/j.fsi.2024.109517
Guo, Y. M., Wang, Y. H., Fan, Z. B., Zhao, X. L., Bergmann, S. M., Dong, H. X., et al. (2022). Establishment and evaluation of qPCR and real-time recombinase-aided amplification assays for detection of largemouth bass ranavirus. J. Fish. Dis. 45, 1033–1043. doi: 10.1111/jfd.13627
Han, C., Li, W., Li, Q., Xing, W., Luo, H., Ji, H., et al. (2022). CRISPR/Cas12a-Derived electrochemical aptasensor for ultrasensitive detection of COVID-19 nucleocapsid protein. Biosens. Bioelectron. 200:113922. doi: 10.1016/j.bios.2021.113922
Huang, X. H., Huang, Y. H., Ouyang, Z. L., and Qin, Q. W. (2011). Establishment of a cell line from the brain of grouper (Epinephelus akaara) for cytotoxicity testing and virus pathogenesis. Aquaculture 311, 65–73. doi: 10.1016/j.aquaculture.2010.11.037
Huang, X. H., Huang, Y. H., Sun, J. J., Han, X., and Qin, Q. W. (2009). Characterization of two grouper Epinephelus akaara cell lines: Application to studies of Singapore grouper iridovirus (SGIV) propagation and virus-host interaction. Aquaculture 292, 172–179. doi: 10.1016/j.aquaculture.2009.04.019
Huang, X. H., Wang, W., Huang, Y. H., Xu, L., and Qin, Q. W. (2014). Involvement of the PI3K and ERK signaling pathways in largemouth bass virus-induced apoptosis and viral replication. Fish. Shellfish. Immun. 41, 371–379. doi: 10.1016/j.fsi.2014.09.010
Islam, M. M., Ghielmetti, V. M., and Allen, P. B. (2021). Graphene oxide assisted light-up aptamer selection against Thioflavin T for label-free detection of microRNA. Sci. Rep. 11:4291. doi: 10.1038/s41598-021-83640-z
Jauset-Rubio, M., El-Shahawi, M. S., Bashammakh, A. S., Alyoubi, A. O., and O’Sullivan, C. K. (2017). Advances in aptamers-based lateral flow assays. Trac-trend Anal. Chem. 97, 385–398. doi: 10.1016/j.trac.2017.10.010
Jiang, N., Shen, J. Y., Zhou, Y., Liu, W. Z., Meng, Y., Li, Y. Q., et al. (2023). Development of a droplet digital PCR method for the sensitive detection and quantification of largemouth bass ranavirus. J. Fish. Dis. 46, 91–98. doi: 10.1111/jfd.13721
Jin, Y. Q., Bergmann, S. M., Mai, Q. Y., Yang, Y., Liu, W. Q., Sun, D. L., et al. (2022). Simultaneous isolation and identification of largemouth bass virus and rhabdovirus from moribund largemouth bass (Micropterus salmoides). Viruses-Basel 14:1643. doi: 10.3390/v14081643
Li, H. X., Yuan, G. L., Luo, Y. Z., Yu, Y. Z., Ai, T. S., and Su, J. G. (2020). Rapid and sensitive detection of infectious spleen and kidney necrosis virus by recombinase polymerase amplification combined with lateral flow dipsticks. Aquaculture 519:734926. doi: 10.1016/j.aquaculture.2020.734926
Li, N., Dai, C., Mao, Y., Zhang, Y., Yan, H., Wang, H., et al. (2025). Development of a rapid on-site detection method for largemouth bass virus based on RPA-CRISPR/Cas12a system. Front. Microbiol. 16:1599006. doi: 10.3389/fmicb.2025.1599006
Liu, J. X., Qin, Q. W., Zhang, X. Y., Li, C., Yu, Y. P., Huang, X. H., et al. (2020). Development of a novel lateral flow biosensor combined with aptamer-based isolation: Application for rapid detection of grouper nervous necrosis virus. Front. Microbiol. 11:886. doi: 10.3389/fmicb.2020.00886
Liu, J. X., Zhang, X. Y., Zheng, J. Y., Yu, Y. P., Huang, X. H., Wei, J. G., et al. (2021). A lateral flow biosensor for rapid detection of singapore grouper iridovirus (SGIV). Aquaculture 541:e736756. doi: 10.1016/j.aquaculture.2021.736756
Liu, X. D., Zhang, Y. B., Zhang, Z. L., An, Z. H., Zhang, X. J., Vakharia, V. N., et al. (2023). Isolation, identification and the pathogenicity characterization of a santee-cooper ranavirus and its activation on immune responses in juvenile largemouth bass (Micropterus salmoides). Fish. Shellfish. Immun. 135:108641. doi: 10.1016/j.fsi.2023.108641
Lu, B., Lin, S., Lang, Z., Yin, Q., and Cao, H. (2025). Aptamer probe-assisted strand displacement-CRISPR/Cas12a biosensor for ultrasensitive detection of AFB1. J. Agric. Food Chem. 31, 20500–20507. doi: 10.1021/acs.jafc.5c05775
Lu, Y., Xie, Q., Chen, J., Chu, Z., Zhang, F., and Wang, Q. (2024). Aptamer-mediated double strand displacement amplification with microchip electrophoresis for ultrasensitive detection of Salmonella typhimurium. Talanta 273:125875. doi: 10.1016/j.talanta.2024.125875
Luo, R., Li, Y., Lin, X., Dong, F., Zhang, W., Yan, L., et al. (2014). A colorimetric assay method for invA gene of Salmonella using DNAzyme probe self-assembled gold nanoparticles as single tag. Sensor. Actuat. B-Chem. 198, 87–93. doi: 10.1016/j.snb.2014.02.104
Majdinasab, M., Badea, M., and Marty, J. L. (2022). Aptamer-based lateral flow assays: Current trends in clinical diagnostic rapid tests. Pharmaceuticals-Base 15:90. doi: 10.3390/ph15010090
Majdinasab, M., Zareian, M., Zhang, Q., and Li, P. W. (2019). Development of a new format of competitive immunochromatographic assay using secondary antibody-europium nanoparticle conjugates for ultrasensitive and quantitative determination of ochratoxin. Food Chem. 275, 721–729. doi: 10.1016/j.foodchem.2018.09.112
Mukama, O., Wu, W., Wu, J. H., Lu, X. W., Liu, Y. M., Liu, Y. J., et al. (2020). A highly sensitive and specific lateral flow aptasensor for the detection of human osteopontin. Talanta 210:120624. doi: 10.1016/j.talanta.2019.120624
Piaskoski, T. O., Plumb, J. A., and Roberts, S. R. (1999). Characterization of the largemouth bass virus in cell culture. J. Aquat. Anim. Health 11, 45–51. doi: 10.1577/1548-86671999011
Preena, P. G., Kumar, T. V., Johny, T. K., Dharmaratnam, A., and Swaminathan, T. R. (2022). Quick hassle-free detection of cyprinid herpesvirus 2 (CyHV-2) in goldfish using recombinase polymerase amplification-lateral flow dipstick (RPA-LFD) assay. Aquac. Int. 30, 1211–1220. doi: 10.1007/s10499-021-00806-2
Qin, Q. W., Chang, S. F., Ngoh-Lim, G. H., Gibson-Kueh, S., Shi, C., and Lam, T. J. (2003). Characterization of a novel ranavirus isolated from grouper Epinephelus tauvina. Dis. Aquat. Organ. 53, 1–9. doi: 10.3354/dao053001
Qin, Q. W., Lam, T. J., Sin, Y. M., Shen, H., Chang, S. F., Ngoh, G. H., et al. (2001). Electron microscopic observations of a marine fish iridovirus isolated from brown-spotted grouper, Epinephelus tauvina. J. Virol. Methods 98, 17–24. doi: 10.1016/s0166-0934(01)00350-0
Ren, Y. W., Gao, P. P., Song, Y., Yang, X. Y., Yang, T., Chen, S. H., et al. (2021). An aptamer-exonuclease iii (exo iii)-assisted amplification-based lateral flow assay for sensitive detection of escherichia coli. J. Dairy Sci. 104, 8517–8529. doi: 10.3168/jds.2020-19939
Santarpia, G., and Carnes, E. (2024). Therapeutic applications of aptamers. Int. J. Mol. Sci. 25:6742. doi: 10.3390/ijms25126742
Sujith, S., Naresh, R., Srivisanth, B. U., Sajeevan, A., Rajaramon, S., David, H., et al. (2024). Aptamers: Precision tools for diagnosing and trea ting infectious diseases. Front. Cell. Infect. Microbiol. 14:1402932. doi: 10.3389/fcimb.2024.1402932
Udhani, R., Kothari, C., and Kumar, S. (2025). Biosensors and lateral flow immunoassays: Current state and future prospects. Clin. Chim. Acta 574:120272. doi: 10.1016/j.cca.2025.120272
Wang, S., Zhou, Z., Cao, M., Pan, Y., Zhang, Y., Fang, Y., et al. (2024). A comprehensive review of aptamer screening and application for lateral flow strip: Current status and future perspectives. Talanta 275:126181. doi: 10.1016/j.talanta.2024.126181
Whittington, R. J., Becker, J. A., and Dennis, M. M. (2010). Iridovirus infections in finfish-critical review with emphasis on ranaviruses. J. Fish. Dis. 32, 95–122. doi: 10.1111/j.1365-2761.2009.01110.x
Xiao, X. Y., Hu, S., Lai, X. C., Peng, J., and Lai, W. H. (2021). Developmental trend of immunoassays for monitoring hazards in food samples: A review. Trends. Food Sci. Tech. 111, 68–88. doi: 10.1016/j.tifs.2021.02.045
Xu, S., Wang, S., Wu, J. J., Guo, X. M., Zhai, J. M., Hu, G. Y., et al. (2025). Aptamer-antibody birecognized sandwich SERRS biosensor in accurate and rapid identification of intraoperative parathyroid hormone. Anal. Chem. 97, 3117–3124. doi: 10.1021/acs.analchem.4c06377
Zhang, X. Y., Wang, L. Q., Liu, J. X., Zhang, Z. M., Zhou, L. L., Huang, X. H., et al. (2022a). Generation and identification of novel DNA aptamers with antiviral activities against largemouth bass virus (LMBV). Aquaculture 547:e737478. doi: 10.1016/j.aquaculture.2021.737478
Zhang, X. Y., Zhang, Z. M., Li, J. R., Huang, X. H., Wei, J. G., Yang, J. H., et al. (2022b). A novel sandwich elasa based on aptamer for detection of largemouth bass virus (LMBV). Viruses-Basel 14:945. doi: 10.3390/v14050945
Zhang, Y. B., Dai, J., Yang, Y., Guo, J. X., Cao, L. Q., and Ye, M. (2022c). Lateral flow strip assay for detection of Mycoplasma hyorhinis based on a pair of sandwich-type aptamers. J. Biomed. Nanotechnol. 18, 166–174. doi: 10.1166/jbn.2022.3230
Zhang, Y., Li, S. T., Tian, J. J., Li, K., Du, Z. H., and Xu, W. T. (2021). Universal linker polymerase chain reaction-triggered strand displacement amplification visual biosensor for ultra-sensitive detection of salmonella. Talanta 222:121575. doi: 10.1016/j.talanta.2020.121575
Zhao, R. X., Geng, Y., Qin, Z. Y., Wang, K. Y., Ouyang, P., Chen, D. F., et al. (2020). A new ranavirus of the santee-cooper group invades largemouth bass (Micropterus salmoides) culture in southwest China. Aquaculture 526:e735363. doi: 10.1016/j.aquaculture.2020.735363
Zhu, Q., Wang, Y., and Feng, J. (2020). Rapid diagnosis of largemouth bass ranavirus in fish samples using the loop-mediated isothermal amplification method. Mol. Cell. Probes 52:e101569. doi: 10.1016/j.mcp.2020.101569
Zhu, Q., Wu, Y., Song, G., and Feng, J. (2022). Onsite and visual detection of ranavirus in largemouth bass (Micropterus salmoides) by an isothermal recombinase polymerase amplification method. Aquaculture 551:e737907. doi: 10.1016/j.aquaculture.2022.737907
Keywords: largemouth bass, LMBV, aptamer, lateral flow biosensor, rapid detection
Citation: Zhang X, Guan L, Xu Z, Wang H, Wei X, Yang M, Qin Q and Wang S (2025) Aptamer based lateral flow biosensor for rapid detection of largemouth bass virus. Front. Microbiol. 16:1643764. doi: 10.3389/fmicb.2025.1643764
Received: 09 June 2025; Accepted: 19 September 2025;
Published: 01 October 2025.
Edited by:
Eleonora Cella, University of Central Florida, United StatesReviewed by:
Sumi Mukhopadhyay, Calcutta School of Tropical Medicine, IndiaYong Zhou, Chinese Academy of Fishery Sciences (CAFS), China
Copyright © 2025 Zhang, Guan, Xu, Wang, Wei, Yang, Qin and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qiwei Qin, cWlucXdAc2NhdS5lZHUuY24=; Shaowen Wang, d2FuZ3N3QHNjYXUuZWR1LmNu
 Xinyue Zhang1,2
Xinyue Zhang1,2 Min Yang
Min Yang Qiwei Qin
Qiwei Qin Shaowen Wang
Shaowen Wang