- School of Bio Sciences and Technology, Vellore Institute of Technology, Vellore, Tamil Nadu, India
Introduction: Methicillin-resistant Staphylococcus aureus (MRSA) colonization in the nasal nares significantly contributes to hospital-acquired infections. This poses a global health concern due to its resistance to β-lactam antibiotics and its virulence mediated by quorum sensing (QS). This study investigates the antimicrobial efficacy of two natural bioactive compounds (BACs)—curcumin and eugenol—against clinical MRSA isolates, specifically focusing on their impact on mecA and agrA gene expression.
Methods: Nasal swab samples from 150 patients yielded 53 staphylococcal isolates, among which 25 were identified as MRSA. Three highly resistant isolates (VITKV25, VITKV32, and VITKV39), identified as S. aureus through 16S rRNA gene sequencing, were selected for further analysis. The antibacterial efficacy of curcumin, eugenol, and gallic acid was assessed through agar well diffusion and minimum inhibitory concentration (MIC) assays. The presence of mecA and agrA genes was detected using polymerase chain reaction (PCR). Additionally, the impact of these compounds on mecA and agrA gene expression was assessed using Quantitative real-time PCR (qRT-PCR).
Results and discussion: The isolates exhibited resistance to multiple antibiotics, including ampicillin, oxacillin, vancomycin, cefoxitin, and erythromycin. Curcumin and eugenol distinctly downregulated mecA and agrA expression levels. In particular, curcumin completely suppressed mecA gene expression in VITKV39 (0.0) and agrA expression in VITKV25 and MRSA (ATCC 43300) (0.0 and 0.1, respectively). Eugenol reduced both genes, mecA and agrA, in VITKV32 to 0.01 and 0.02, respectively. These findings highlight the therapeutic potential of curcumin and eugenol as adjuncts to conventional antibiotics by targeting both resistance and virulence pathways in MRSA.
1 Introduction
Staphylococci are opportunistic pathogens that can cause infections in specific conditions, such as when the host's immune system is compromised or the skin or mucosal barriers are breached (Jaradat et al., 2020). Although many strains of Staphylococci are harmless and also part of the human body's normal flora, some strains may cause various infections, including skin diseases (boils, impetigo, and cellulitis), respiratory diseases, or indeed serious conditions like septicaemia, endocarditis, and pneumonia (Al-Dmour et al., 2023). The virulence arsenal and increasing multidrug resistance have been responsible for its adaptability, persisting in hospital and community settings. The nasal cavity is a critical site of both asymptomatic carriage and subsequent transmission of the organism, particularly in hospitals. Studies revealed that the nasal carriage rate can be as high as 60–70% among patients in hospitals and health care workers, significantly indicating the dynamic aspects of nosocomial infections (Rongpharpi et al., 2013). The S. aureus was the first organism to develop resistance to penicillin shortly after its introduction (Lowy, 2003).
The semi-synthetic β-lactam antibiotic, methicillin, was developed in 1959 as an alternative to penicillin, but MRSA was reported within 2 years. The emergence and widespread occurrence of MRSA strains have impelled the global antimicrobial resistance crisis. The mecA gene, which encodes penicillin-binding protein 2a (PBP2a), decreases the binding efficacy of β-lactam antibiotics to the target, leading to resistance in MRSA organisms. The mecA gene is located on the staphylococcal cassette chromosome mec (SCCmec), which contains distinct resistance determinants (Deurenberg et al., 2007; Urushibara et al., 2020). These features contribute to MRSA's multidrug-resistant phenotype and limit the number of therapeutic and management approaches for MRSA infections (Idrees et al., 2023). In addition to antibiotic resistance, S. aureus pathogenicity is strongly regulated by QS systems, particularly the accessory gene regulator (agr) locus, which governs key virulence factors such as adhesins, exotoxins, and degradative enzymes (Fuqua et al., 1994; Vinodhini and Kavitha, 2024). This operon is organized around two promoters, P2 and P3, that produce two transcripts, RNAII and RNAIII, respectively. The RNAII encodes AgrB, AgrD, AgrC, and AgrA—the core elements for autoinducing peptide (AIP) production and signal transduction. AgrC-AgrA represents a typical two-component system, where AgrC phosphorylates AgrA in response to AIP binding, thereby stimulating the transcription of RNAIII and virulence gene expression (Thoendel et al., 2011). Downregulation of AgrA disrupts this cascade, significantly reducing pathogenicity. Furthermore, numerous in vivo and in vitro studies have demonstrated that the inhibition of AgrA drastically attenuated virulence factors. Therefore, it may be possible to inhibit or reduce virulence by targeting AgrA, which would increase the pathogen's susceptibility to antibiotics. Additionally, the QS inhibitor would gain great significance if it receives FDA approval, as it might be a convenient adjunct to current antibiotics (Palaniappan et al., 2021). Consequently, targeting both mecA and agrA provides a promising strategy to simultaneously suppress resistance and virulence.
Bioactive compounds (BACs) of biological origin are known for their medicinal properties and role in preventing infectious diseases. The BACs play a key role in the production of novel synthetic drugs, ranging from anticancer treatments to antibiotics. They offer several advantages over the synthetic compounds, such as fewer side effects, a broader spectrum of action, lower cost, and abundant natural availability (Stan et al., 2021; Nandhini et al., 2023). BACs are mostly certain secondary metabolites, exhibit antimicrobial, immunomodulating, inflammatory, and antioxidant properties (Sytar and Smetanska, 2022). Secondary metabolites possessing remarkable activity against MRSA have been taken into consideration and investigated for their inhibitory activity. BACs have been selected in this study according to recent research outcomes and potential effectiveness against MRSA. All five compounds are secondary metabolite classes, including polyphenols, alkaloids, and terpenes. Curcumin is a polyphenolic compound derived from the rhizome of Curcuma longa L. and has been known to possess a wide range of medicinal properties, including antimicrobial activity (Batista de Andrade Neto et al., 2021). Gallic acid, a phenolic compound present in the bark, leaves, and roots of berries, nuts, and tea leaves, exhibits a wide range of biological activities, such as antibacterial, anti-inflammatory, anti-cancer, and antioxidant properties (Jiamboonsri et al., 2023). Piperine, a naturally occurring alkaloid in black pepper (Piper nigrum) has antioxidant, antiproliferative, antibacterial and immunomodulatory effects. Its antibiofilm effect against MRSA has been studied, highlighting its potential for developing novel antibiofilm therapies (Das et al., 2024). Eugenol (4-ally-2-methoxyphenol), a key component of clove oil, is widely used as a flavoring agent in culinary and cosmetic l. Studies have shown its antibacterial, antioxidant, anti-inflammatory, and antispasmodic properties (El-Far et al., 2021). α-Terpineol is a monocyclic terpene alcohol widely used in the pharmaceutical industry, is extracted from the essence of Citrus aurantium (neroli), Melaleuca alternifolia. Apart from possessing a well-established bactericidal action against both Gram-positive and Gram-negative bacteria, α-terpineol has analgesic, anti-inflammatory, antioxidant, and antiproliferative properties. And incredibly, it is effective against drug-resistant bacteria (Johansen et al., 2022).
To our knowledge, no studies have evaluated the effectiveness of the aforementioned BACs on the transcription levels of the mecA and agrA genes in MRSA isolates. In this context, the present study aimed to investigate the antimicrobial activity of curcumin, eugenol, and gallic acid against multidrug-resistant clinical MRSA isolates. Particularly, this study focused on their inhibitory effect on mecA and agrA gene expression by qRT-PCR. By examining phenotypic as well as molecular endpoints, this study affords intuitive insights into the potential of these natural compounds as supplemental therapeutics against resistant and virulent strains of S. aureus.
2 Materials and methods
2.1 Study design and sample collection
A cross-sectional comparative study was performed to determine prevalence rates of nasal carriage of S. aureus, especially methicillin-resistant strains. Totally 150 nasal swab samples were collected from patients at Sri Bala Nursing Home, Chennai, Tamil Nadu, India, irrespective of age, sex, or occupation. Samples were collected with sterile cotton swabs and placed into the anterior nares, where the swabs were gently rotated twice. The swabs were placed in sterile transport containers and processed within 6 h under aseptic conditions. Ethical clearance for this study has been obtained from the Institutional Ethical Committee for Human Studies, Vellore Institute of Technology, Vellore, India (Approval No. VIT/IECH/XI/2021/11). All participants gave written informed consent, and confidentiality was maintained throughout the study.
2.2 Isolation and preliminary identification of staphylococcal isolates
All nasal swab samples were streaked onto mannitol salt agar (MSA; HiMedia, India) and incubated at 37 °C for 24–48 h. Plates with yellow-colored colonies indicating mannitol fermentation were selected as presumptive staphylococcal isolates. Distinct colonies were sub cultured and subjected to preliminary identification through standard microbiological and biochemical methods. The cell morphology was determined by Gram staining. Catalase activity was tested with 3% hydrogen peroxide, while coagulase production was assessed with the tube coagulase test using rabbit plasma (HiMedia). Oxidase activity was determined using oxidase reagent discs (HiMedia). Deoxyribose production was evaluated in DNase medium (10g—Pancreatic digest of casein, 10g—yeast extract, 2g—deoxyribonucleic acid, 5g—NaCl, 15g—agar, pH 7.5), which was then flooded with 1N HCl solution to visualize the zone of clearance (Kateete et al., 2010). Isolates showing Gram-positive cocci in clusters, catalase-positive, coagulase-positive, oxidase-negative, and DNase-positive profiles were presumptively considered as S. aureus. Final confirmation was performed by 16S rRNA gene sequencing, as described in a subsequent section.
2.3 Antibiotic susceptibility test
The antibiotic susceptibility of the Staphylococcal isolates was determined by following the Kirby-Bauer disc diffusion method on Mueller-Hinton agar (MHA; HiMedia, India), by following Clinical and Laboratory Standards Institute (CLSI) guidelines (Beargie et al., 1965; Clinical and Laboratory Standards Institute, 2018). The isolates were inoculated in Luria-Bertani broth (HiMedia, India) and incubated at 37 °C overnight, the density of which matched the turbidity of 0.5 McFarland standard (~1.5 × 106 CFU/mL). The bacterial suspension was uniformly swabbed onto the surface of MHA plates. Octo discs (HiMedia, India) containing ampicillin (10 μg), levofloxacin (5 μg), vancomycin (30 μg), gentamicin (10 μg), oxacillin (1 μg), clindamycin (2 μg), erythromycin (15 μg) was carefully placed at the center of the plates. The diameters of inhibition zones were determined after 24 h of incubation at 37 °C and interpreted using the CLSI-recommended breakpoints to categorize isolates as susceptible, intermediate, or resistant. For methicillin resistance screening, additional discs of methicillin (5 μg) and cefoxitin (30 μg) were used under the same conditions. Isolates showing resistance to either disc were considered presumptive MRSA and were further confirmed by a molecular approach. S. aureus ATCC 43300 and ATCC 25923 were used as MRSA and MSSA quality control strains, respectively, in all the experiments.
2.4 Molecular confirmation by 16S rRNA gene sequencing
For species-level confirmation, three staphylococcal isolates labeled as VITKV25, VITKV32, and VITKV39, exhibiting the maximum antibiotic resistance profiles, were selected. The strains were subjected to 16S rRNA gene sequencing using agarose gel electrophoresis and PCR analysis by the Rajiv Gandhi Centre of Biotechnology (RGCB), Kerala. The primers used for this process were 16S-RS-F 5′CAGGCCTAACACATGCAAGTC3′ as forward primer and 16S-RS-R 5′GGGCGGWGTGTACAAGGC3′ as reverse primer. The resulting sequences were compared with known bacterial taxa using the NCBI BLASTn tool. Phylogenetic relationships were further evaluated using MEGA11 software using the neighbor-joining method (Sada et al., 2024).
2.5 Biofilm formation assay
The biofilm-forming capacity of confirmed S. aureus isolates (VITKV25, VITKV32, and VITKV39) was evaluated using the crystal violet staining method in a 96-well microtiter plate. Each isolate was streaked onto trypticase soy agar (TSA; HiMedia, India) and kept at 37 °C for overnight incubation. A single colony from the TSA plate was inoculated onto 5 mL of trypticase soy broth (TSB; HiMedia, India) and incubated overnight at 37 °C. The overnight cultures were diluted 1:100 in fresh TSB, and 200 μL of the diluted suspension (adjusted to an OD600 of 0.5) was dispensed into a sterile, flat-bottom 96-well polystyrene microtiter plate (HiMedia, India). The plates were then incubated under static conditions at 37 °C for 16 h. After incubation, wells were washed three times with sterile distilled water to remove non-adherent cells, and plates were allowed to dry in blotting paper. Then, 200 μL of 0.1% crystal violet solution was added to the wells and incubated at room temperature for 30 min. The wells were blotted dry and washed with distilled water, and air-dried. To quantify the retained stain, 200 μL of 95% ethanol was added to each well to solubilize the bound dye, and spectrophotometric absorbance was taken at 540 nm using a microplate reader (Bio-Rad, USA). The results were an average of the three experiments (Shukla and Rao, 2013; Turk et al., 2013). Biofilm formation was categorized according to the mean OD values: OD <0.120 as non-biofilm producers, 0.120 – 0.240 as moderate biofilm producers, and >0.240 as strong biofilm producers (Kala et al., 2012). S. aureus ATCC 43300 (MRSA reference strain) and ATCC 25923 (MSSA reference strain) were included as strong and moderate biofilm-producing positive controls, respectively.
2.6 Agar well diffusion assay and MIC
The antibacterial activity of selected BACs—curcumin, piperine, eugenol, gallic acid and α-terpineol—was evaluated against VITKV25, VITKV32, and VITKV39 using agar well diffusion and minimum inhibitory concentration (MIC) assay. The selected bacterial strains were grown on Nutrient broth and the turbidity of the bacterial suspension was adjusted to 0.5 MacFarland's standard. The MHA plates were spread with 100 μl of the bacterial suspension. Using a sterile borer, 6 mm diameter wells were made in the inoculated agar. BACs, curcumin, piperine (HiMedia, India), eugenol, gallic acid and α-terpineol (Sigma-Aldrich, USA) were used to prepare stock solutions at a concentration of 100 mg/mL. The 50μl of each stock solution was loaded in the respective wells and the same volume of solvent (5% DMSO) was used as a negative control. The plates were incubated at 37 °C for 24 h and the zone of inhibition in diameter were measured in mm (Hossain et al., 2022).
The MIC of S. aureus was determined for three compounds—curcumin, eugenol and gallic acid which showed activity against the isolates, using micro-broth dilution method in 96 well plate following CLSI guidelines. In the 96 well plate, the compounds were serially diluted to obtain final concentrations of 10, 5, 2.5, 1.25, 0.62, and 0.31 mg/mL. Then, 90 μL of double strength MHB was transferred to each well, followed be the addition of 100 μL of the respective compound from each dilution. Finally, 10 μL of the respective bacterial culture (adjusted to 0.5 McFarland) was added to each well, and the microtiter plate was incubated at 37 °C for 24 h and bacterial growth was assessed by measuring absorbance at 600 nm using a microplate reader (Bio-Rad, USA). The MIC was defined as the lowest concentration of the compound that completely inhibited visible growth (Moses et al., 2020). MRSA and MSSA reference strains were used (ATCC 43300 & ATCC 25923) for both assays.
2.7 DNA extraction and detection of mecA and agrA genes by PCR
Genomic DNA was extracted from the isolates VITKV25, VITKV32, and VITKV39 using the phenol-chloroform method. Overnight cultures were grown in Luria-Bertani (LB) broth at 37 °C and centrifuged at 10,000 rpm for 5 min to pellet the cells. The pellet was resuspended in TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8.0), followed by cell lysis using lysozyme (10 mg/mL) and proteinase K (20 mg/mL). after incubation at 37 °C for 1 h, DNA was extracted with an equal volume of phenol: chloroform: isoamyl alcohol (25:24:1) and precipitated using cold isopropanol. The resulting DNA pellet was washed with 70% ethanol, air-dried, and resuspended in nuclease-free water. DNA quality and concentration were assessed using 1% agarose gel electrophoresis and spectrophotometric quantification (Nanodrop, Thermo Fisher Scientific, USA) (Divyakolu et al., 2021).
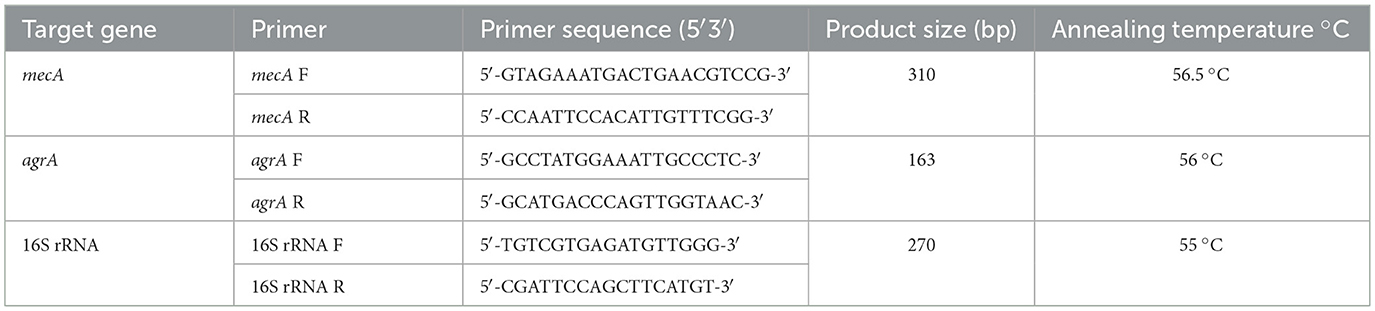
The extracted DNA was used as a template DNA in PCR to detect the presence of the mecA and agrA genes. The effective primer pairs were designed from the FASTA sequences retrieved from the NCBI database, thereby validating their melting temperature, GC content, and other required specifications. The primer sequences (Eurofins, Bangalore) used in this study have been included in Table 1. The reaction mixture (10 μl) contained 2 μL template DNA, 1 μL forward primer (FP), 1 μL reverse primer (RP), 2 μL of Taq DNA polymerase master mix (Ampliqon, Denmark), and 4 μl of nuclease-free water for both genes. Amplification of mecA gene were performed for 30 cycles under the following cyclic conditions: initial denaturation at 95 °C for 3 min, denaturation at 95 °C for 30 s, followed by annealing at 56.5 °C for 30 s and elongation at 72 °C for 1 min with the final step of extension at 72 °C for 5 min. Similarly, amplification of the agrA gene was performed for 30 cycles with the same conditions, except that the annealing temperature was set at 56 °C. (Divyakolu et al., 2021). PCR amplicons were resolved by electrophoresis on 1.5% agarose gels stained with ethidium bromide and visualized under UV illumination using a gel documentation system (Bio-Rad, USA). MRSA and MSSA reference strains were used as positive and negative controls, respectively.
2.8 Assessment of bacterial viability
To confirm the viability of bacterial cells following treatment with BACs at MIC concentrations, a colony-forming unit (CFU) assay was performed. MRSA (ATCC 43300) and MSSA (ATCC 25923) strains were grown overnight and then treated with curcumin and eugenol at their respective MIC concentrations for 24 h at 37 °C. Following incubation, cultures were serially diluted from 10−1 to 10−5 in sterile PBS. From each dilution, 100 μL was plated on Mueller-Hinton Agar (MHA) plates in triplicate and incubated at 37 °C for 24 hours. Only plates from the 10−3 dilution were selected for colony enumeration and comparative visualization, as they consistently yielded distinct, countable colonies across all treatment groups. The presence of visible colonies was interpreted as evidence of bacterial viability after BACs exposure (Rahman and Butzin, 2024).
2.9 RNA extraction and cDNA synthesis
The total RNA was extracted from treated and untreated cells using RNAiso Plus reagent (Takara Bio Inc., Japan), following the manufacturer's protocol. For the untreated batch, the strains (VITKV25, VITKV32, VITKV39) and MRSA, MSSA controls were inoculated in Nutrient broth and incubated overnight at 37 °C. For the treated batch, the overnight cultures of the isolates (VITKV25, VITKV32, VITKV39) and MRSA, MSSA controls were treated with curcumin, eugenol, and gallic acid at their respective MIC values for 24 h. The rationale for using MIC concentrations was to capture a transcriptional snapshot under growth-inhibited conditions, allowing us to examine how exposure to these BACs affects the gene expression. Despite inhibition of visible growth, sufficient intact bacterial cells remained for successful RNA extraction and cDNA synthesis. Then both the untreated and treated strains were taken to extract total RNA using RNAiso Plus. Briefly, the bacterial cells were harvested by centrifugation at 10,000 rpm for 10 min. the pellet was resuspended in RNAiso Plus reagent and subjected to mechanical disruption using sterile glass beads with intermittent vortexing to ensure efficient cell lysis. Phase separation was carried out by chloroform addition and centrifugation, followed by RNA precipitation using isopropanol. The RNA pellet was washed with 75% ethanol, air-dried, and resuspended in RNase-free water. RNA concentration and purity were determined using a NanoDrop spectrophotometer (Thermo Fisher Scientific, USA) before cDNA synthesis. First-strand cDNA synthesis was performed using the ABScript II cDNA first-strand synthesis kit (Abclonal, China) according to the manufacturer's protocol. The cDNA samples were stored at −20 °C until further use in qRT-PCR analysis.
2.10 Gene expression analysis by qRT-PCR
The qRT-PCR was carried out using Genious 2X SYBR Green Fast qPCR mix (Abclonal, China) following the manufacturer's instructions. The primers used in this study targeted mecA, agrA and 16S rRNA (housekeeping gene as an internal control) (Divyakolu et al., 2021). Amplification was performed using a Bio-Rad real-time PCR system at 95 °C for 5 min, by the subsequent steps: 35 cycles of 95 °C for 15 s, 56 °C for 30 s and 60 °C for 1 min. All samples were analyzed in triplicate. Expression levels of mecA and agrA were normalized to the 16S rRNA housekeeping gene, and relative expression was calculated using the 2∧–ΔΔCT technique. MRSA, MSSA reference strains were used as controls. Also, to confirm the absence of genomic DNA contamination, no-reverse-transcriptase (–RT) control reactions were performed, and melt peak analysis showed no specific amplification (see Supplementary Figure S4). Primer efficiencies were determined using five 10-fold serial dilutions of cDNA, and all primer pairs (mecA, agrA, 16S rRNA) demonstrated amplification efficiencies within the acceptable range (96.8–98.7%) and high linearity (R2 ≥ 0.996), as shown in Supplementary Table S1.
2.11 Statistical analysis
All experiments were conducted with two independent biological replicates. The CV assay, MIC, and qRT-PCR were each performed in triplicate. Results are presented as mean ± standard deviation (SD). For all graphical representations, optical density (OD) and percentage values are indicated above the bars. Statistical analysis was performed using one-way analysis of variance (ANOVA) followed by Tukey's multiple comparison test. A p-value of <0.001 (p < 0.001) was considered statistically significant. All statistical analysis and data visualizations were performed using Origin 2024b software (OriginLab Corporation, USA).
3 Results
3.1 S. aureus prevalence among patients
In this study, a total of 150 nasal swab samples were equally collected from male (n = 75) and female (n = 75) patients. Out of these, 53 isolates (35.3%) were identified as S. aureus isolates based on morphological and biochemical characterization. The proportion of S. aureus positive isolates was higher among females (45.3%) compared to males (25.3%), and this difference was statistically significant (χ2 = 6.56, p = 0.01). On MSA, the presumptive isolates formed yellow-colored colonies with surrounding yellow zones. Gram staining revealed Gram-positive cocci in clusters. The isolates also tested positive for catalase (bubble formation), tube coagulase (visible clot formation), and DNase (zone of clearance on DNase agar). The oxidase test was negative, as indicated by no color change on the oxidase disc. A summary of the phenotypic test results is presented in Table 2.
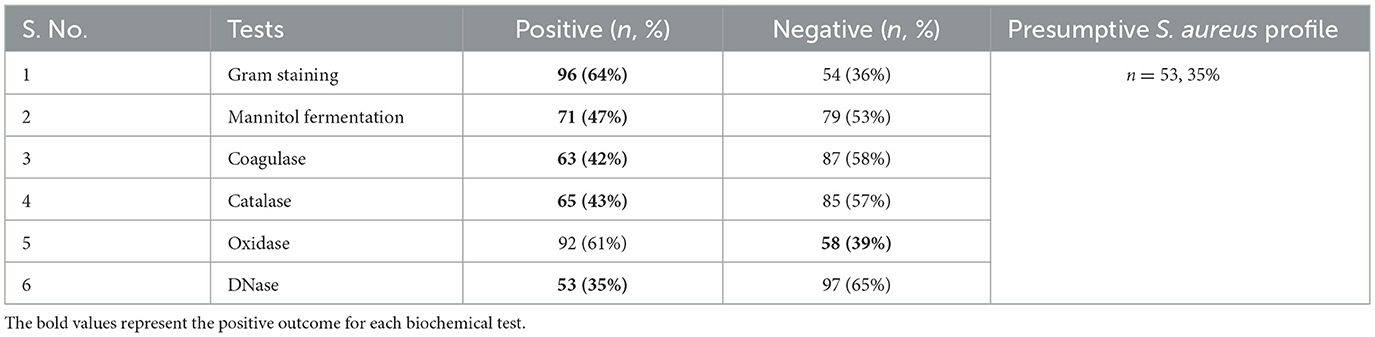
Table 2. Morphological and biochemical characterization of presumptive S. aureus isolates (n = 150).
3.2 Antibiotic susceptibility profiling of S. aureus isolates
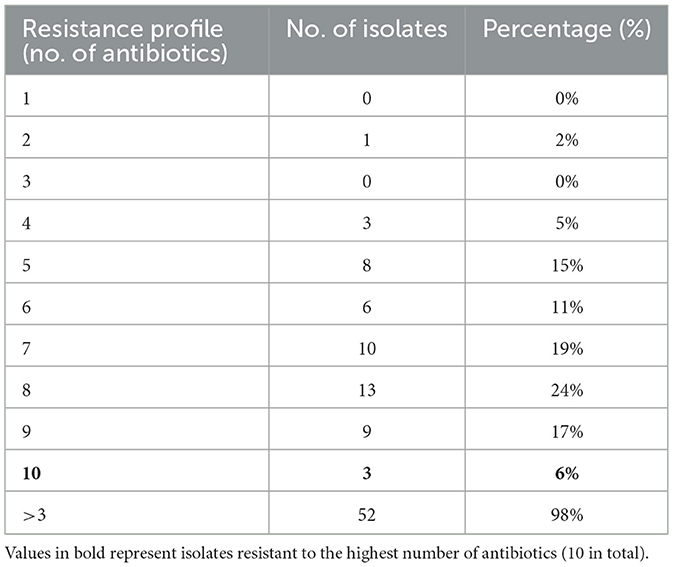
Antibiotic susceptibility testing was performed for all 53 presumptively confirmed S. aureus isolates using the disc diffusion method. The isolates exhibited a high level of resistance to multiple antibiotics. Resistance to ampicillin (100%) was most significant, followed by oxacillin (98%) and vancomycin (94%). In contrast, lower resistance was observed against clindamycin (30%). Methicillin resistance was assessed using cefoxitin disc diffusion, which identified 62% (25/53) of the isolates as MRSA. Figure 1 presents the resistance profile of all isolates based on the disc diffusion assay, showing the percentage of resistance, intermediate susceptibility, and sensitivity across the tested antibiotics. Representative disc diffusion results are shown in Figure 2, the distribution of resistance levels among the isolates is summarized in Table 3, and the three isolates (VITKV25, VITKV32, and VITKV39) that demonstrated resistance to all 10 antibiotics tested were highlighted. These multidrug-resistant strains were selected for further analysis involving compound treatment and gene expression profiling.
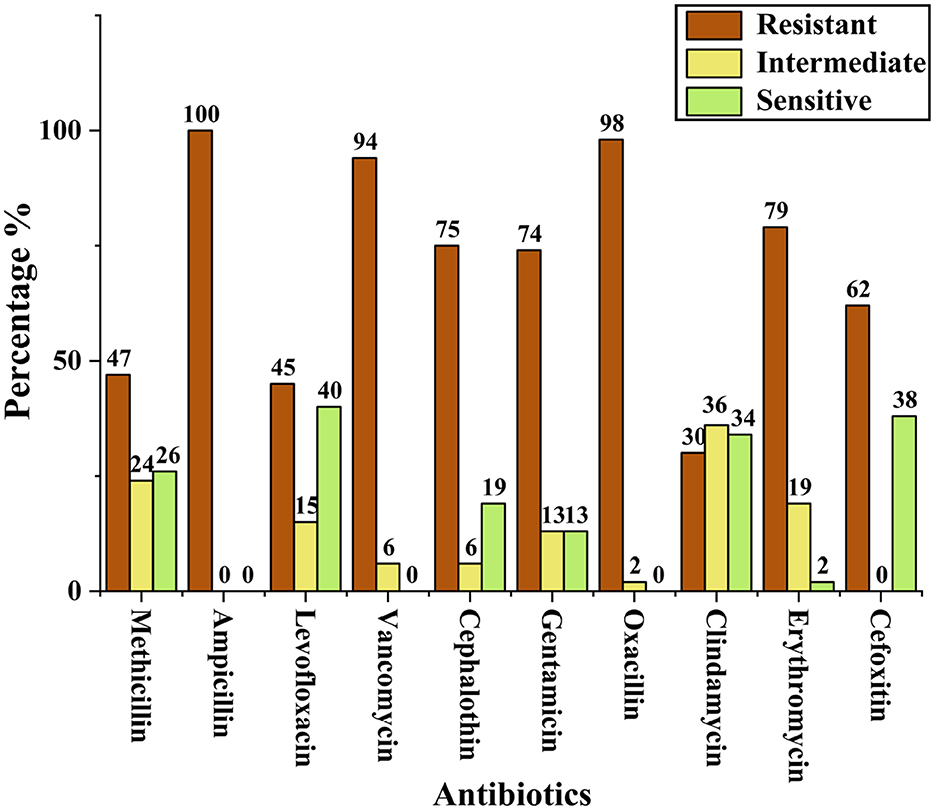
Figure 1. Bar graph representing the percentage of presumptive S. aureus isolates categorized as resistant, intermediate, or susceptible to each antibiotic tested by the antibiotic disc diffusion method.
Figure 2. Antibiotic sensitivity pattern of presumptive S. aureus isolates using Octo disc in MHA by the antibiotic disc diffusion method. The clear inhibition zones around antibiotic discs indicate varying degrees of susceptibility. Images shown are representative and do not correspond to a specific strain.
3.3 Molecular identification of S. aureus isolates through 16S rRNA sequencing
Three multidrug-resistant isolates (VITKV25, VITKV32, and VITKV39), which demonstrated resistance to all 10 antibiotics tested were selected for molecular identification. The isolates were subjected to 16S RNA gene sequencing, and the results sequences were analyzed using the NCBI BLASTn tool. All three isolates showed high sequence similarity to S. aureus strains in the GenBank database. The isolates were confirmed as S. aureus and the corresponding nucleotide sequences were submitted to GenBank with accession numbers PQ215935 (VITKV25), PQ302583 (VITKV32), and PQ303245 (VITKV39). A phylogenetic tree was constructed using the neighbor-joining method in MEGA11 to assess the evolutionary relationships of the sequenced isolates with reference Staphylococcus species. As shown in Figure 3, all three isolates clustered closely with known S. aureus sequences, further supporting their taxonomic classification.
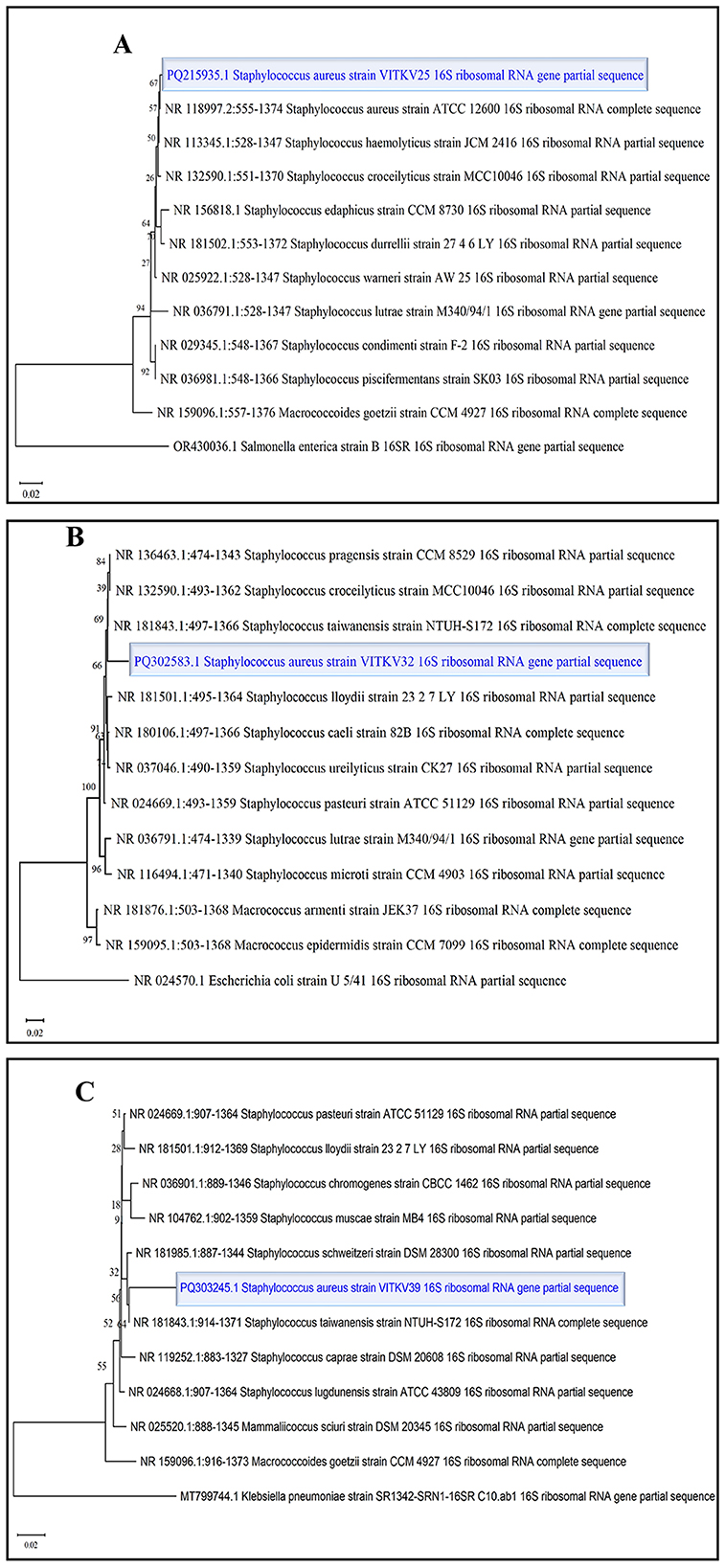
Figure 3. Phylogenetic tree analysis of S. aureus isolates. Neighbor-joining phylogenetic trees were constructed for clinical isolates (A) VITKV25, (B) VITKV32, and (C) VITKV39 using partial 16S rRNA gene sequences—each isolate (highlighted) clustered closely with S. aureus reference strains from GenBank, confirming species-level identity.
3.4 Biofilm formation assay
The crystal violet assay results confirmed that all three multidrug-resistant S. aureus isolates were biofilm producers. Absorbance values at 540 nm were used to classify biofilm strength. Isolate VITKV32 exhibited strong biofilm formation with a mean absorbance of 0.34, while VITKV25 (0.14) and VITKV39 (0.22) were categorized as moderate biofilm producers. The positive control strains MRSA and MSSA reference strains, showed strong (0.32) and moderate (0.15) biofilm formation, respectively. The negative control (TSB only) exhibited negligible absorbance (0.03), confirming assay specificity. As shown in Figure 4, biofilm formation among the clinical isolates was comparable to that of standard control strains. The results are expressed as the mean of three biological replicates, with error bars representing standard error of the mean (SEM).
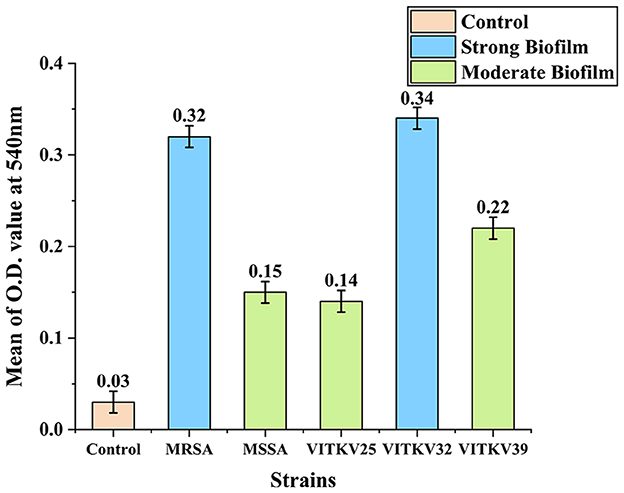
Figure 4. Biofilm formation in clinical and reference S. aureus strains. Bar chart representing the mean absorbance values at 540 nm (±SEM) from the crystal violet assay for clinical isolates VITKV25, VITKV32, and VITKV39 and control strains (S. aureus ATCC4300 and ATCC 25923). The TSB-only control showed negligible biofilm formation. VITKV32 and MRSA control strain were categorized as strong biofilm producers, while VITKV25, VITKV39, and MSSA control strain demonstrated moderate biofilm formation. Data represent the mean of three independent replicates.
3.5 Antibacterial activity of BACs
The selected BACs—curcumin, eugenol, piperine, gallic acid, and α-terpineol—were chosen to represent distinct classes of bioactive secondary metabolites, including polyphenols (gallic acid), diarylheptanoids (curcumin), phenolic monoterpenes (eugenol), alkaloids (piperine), and monoterpenoids (α-terpineol). This chemical diversity was intentional, allowing us to evaluate the broad-spectrum potential of structurally different phytocompounds against S. aureus. These BACs were evaluated against the multidrug-resistant S. aureus isolates using the agar well diffusion method. The inhibitory effect was evaluated based on the diameter of the zone of inhibition. Among the tested compounds, curcumin, eugenol, and gallic acid exhibited measurable zones of inhibition against all three isolates, indicating effective antibacterial activity. Meanwhile, piperine and α-terpineol did not produce any visible zones at the tested concentrations and were considered inactive under these conditions. The results were summarized in Figure 5, which denotes the zone of inhibition produced by each compound against the clinical isolates. Based on these findings, curcumin, eugenol, and gallic acid were selected for further evaluation through MIC assays.
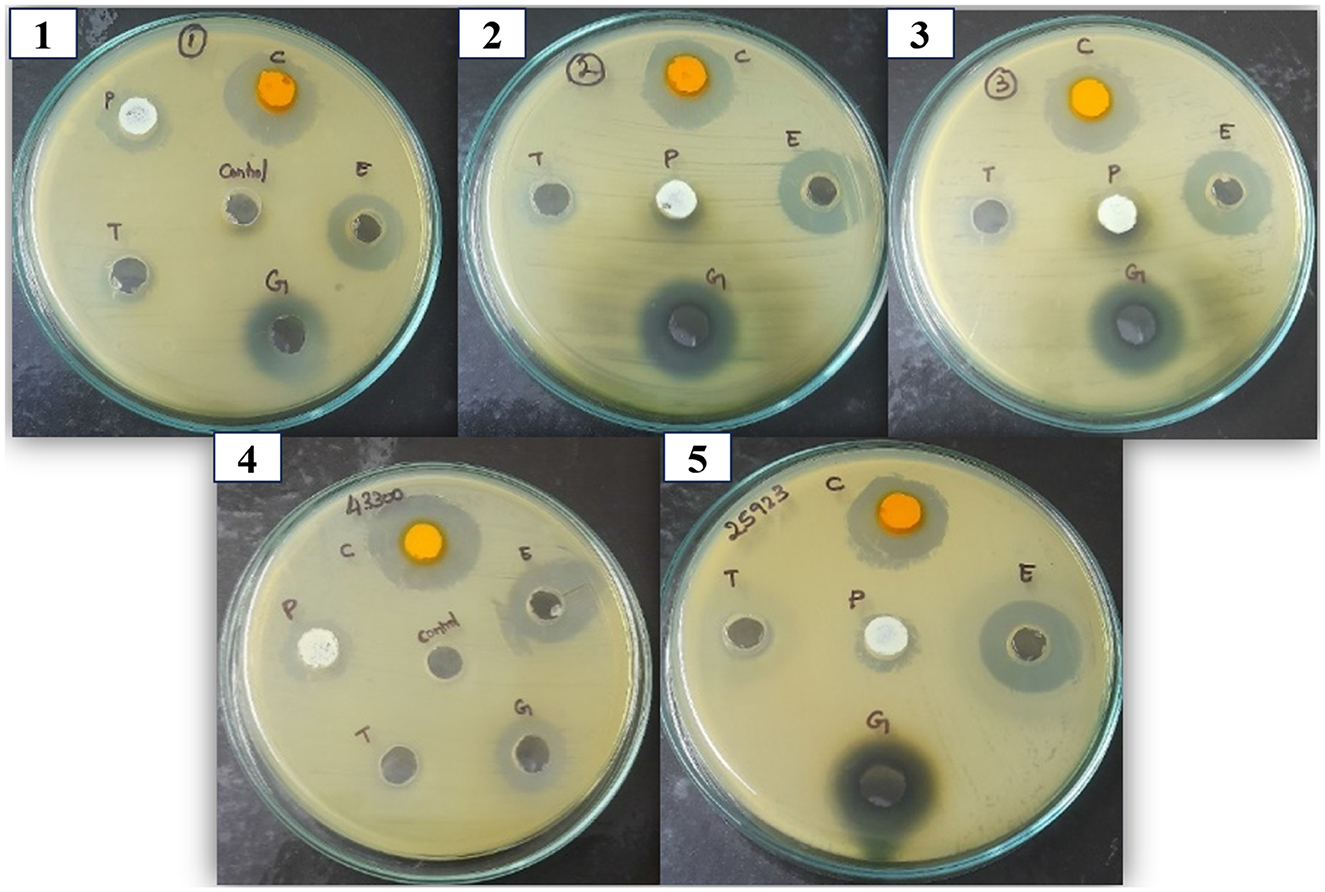
Figure 5. Antibacterial activity of the BACs against clinical and control S. aureus isolates. The zone of inhibition denotes the antibacterial activity of compounds against the S. aureus strains. 1,2, and 3 correspond to VITKV25, VITKV32, and VITKV39, respectively. 4 and 5: MRSA and MSSA reference strains respectively. C, curcumin; E, Eugenol; G, Gallic acid; T, α-terpineol; P, Piperine. DMSO was used as the negative control.
3.6 MIC determination of BACs
Based on the results of the agar well diffusion assay, the MICs of curcumin, eugenol, and gallic acid were determined by the broth microdilution method. All three compounds exhibited inhibitory effects against the isolates and reference strains, with compound-specific and strain-specific variability. Curcumin showed the highest potency with MIC values of 2.5 mg/mL against MRSA, MSSA, VITKV25, and VITKV39 and the lowest MIC of 1.25 mg/mL against VITKV32. Eugenol also demonstrated strong activity with MICs of 1.25 mg/mL for MRSA and VITKV32 and 2.5 mg/mL for all other strains. In contrast, gallic acid exhibited comparatively lower efficacy, with MICs of 10 mg/mL for MRSA, VITKV32, and VITKV39 and 5 mg/mL for MSSA and VITKV25. The comparative MIC values are graphically represented in Figure 6. These findings indicate that curcumin and eugenol are more effective than gallic acid in inhibiting the growth of multidrug-resistant S. aureus strains.
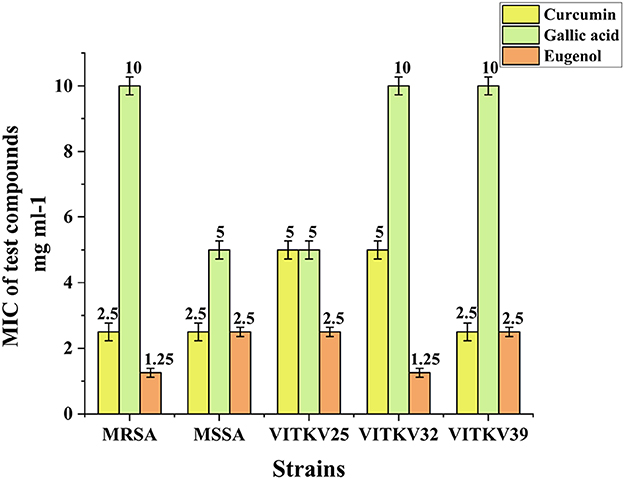
Figure 6. Bar graph representing the MIC values (mg/mL) of curcumin, eugenol, and gallic acid against reference strains (MRSA & MSSA) clinical S. aureus isolates (VITKV25, VITKV32, VITKV39). Data represent mean ± standard error of triplicate experiments.
3.7 Molecular detection of mecA and agrA genes by PCR
Conventional PCR was performed to detect the presence of the mecA and agrA genes in the isolates. Amplification of the mecA gene yielded a product of 310bp, which was observed in all three clinical isolates and the MRSA reference strain. No amplification was detected in the MSSA control, confirming its mecA-negative phenotype. Subsequently, amplification of the agrA gene resulted in a 163bp product in all tested strains, which includes three clinical isolates, MRSA and MSSA controls, indicating the presence of a functional agr quorum sensing system across both resistant and susceptible strains. The PCR amplicons were visualized using 1.5% agarose gel electrophoresis, and the results are illustrated in Figure 7.
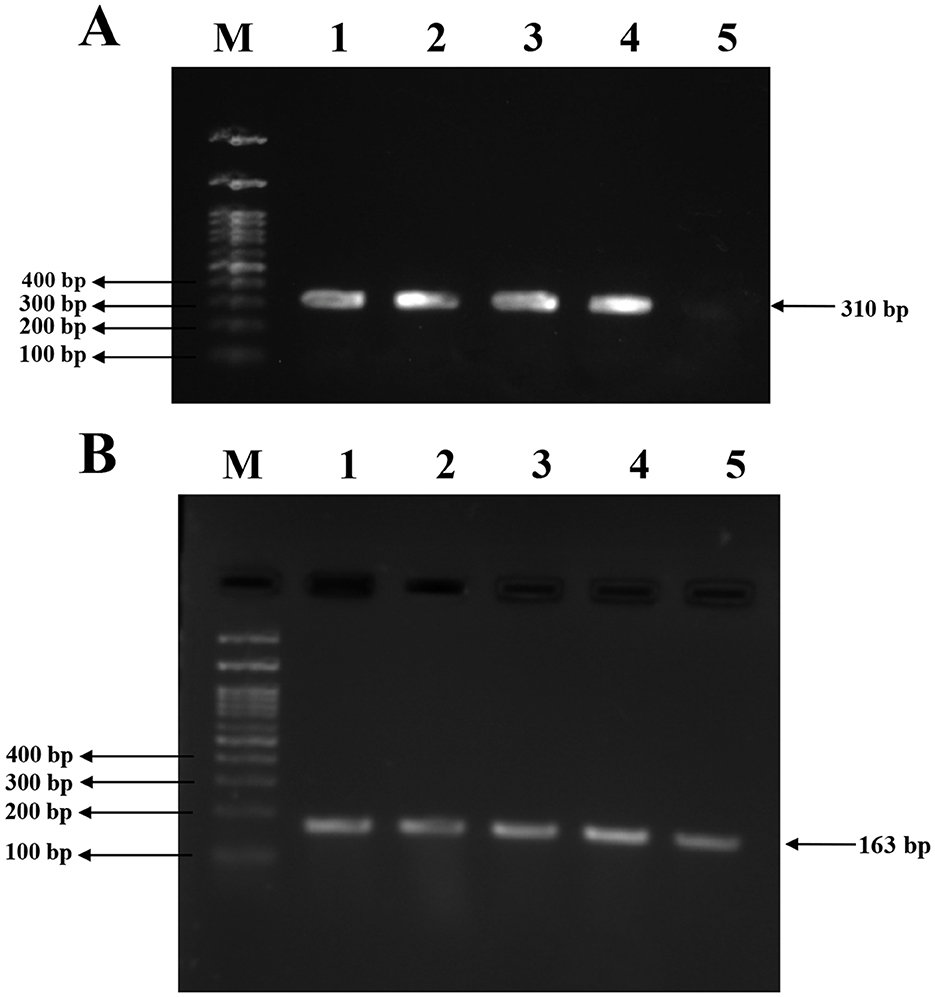
Figure 7. PCR amplification for mecA and agrA genes in clinical and reference S. aureus strains. (A) Amplification of the mecA gene (310bp) in S. aureus isolates. Lane 1, 2, 3: clinical S. aureus isolates VITKV25, VITKV32, VITKV39, respectively; Lane 4: positive control (MRSA control strain); Lane 5: negative control (MSSA control strain); Lane M: 100 bp DNA ladder. (B) Amplification of the agrA gene (163 bp) across isolates. Lane 1,2,3: VITKV25, VITKV32, and VITKV39; Lanes 4 and 5: MRSA and MSSA control strains, respectively; Lane M: 100 bp DNA ladder. All isolates tested positive for agrA, while only MRSA strains showed mecA amplification.
3.8 Viability assessment
The bacterial cells remained viable following exposure to curcumin and eugenol at MIC levels, a CFU assay was conducted on MRSA and MSSA strains. Cultures were serially diluted and plated after 24 h of treatment. As shown in Supplementary Figure S1, the 10−3 dilution consistently produced visible, countable colonies in both treated and untreated groups. The presence of colonies in curcumin- and eugenol-treated cultures confirmed that growth was inhibited but not eliminated, supporting the feasibility of RNA extraction and downstream qRT-PCR analysis.
3.9 Gene expression analysis of mecA and agrA by qRT-PCR
The relative expression levels of the mecA and agrA genes in the multidrug-resistant S. aureus isolates after treatment with BACs were evaluated using qRT-PCR. The expression levels were normalized to the 16S rRNA housekeeping gene, and it was assessed by comparing treated cultures to untreated controls using the 2∧–ΔΔCt method. Both mecA and agrA gene expressions varied across the treated S. aureus isolates. Curcumin treatment significantly downregulated both genes in multiple strains (p < 0.001). Notably, mecA expression in VITKV39 was completely suppressed (0.00 ± 0.00), and agrA expression in VITKV25 and MRSA was also strongly reduced (0.00 ± 0.01, respectively). Eugenol produced a moderate but statistically significant decrease in gene expression, particularly in VITKV32 (mecA: 0.01 ± 0.00; agrA: 0.02 ± 0.01; p < 0.001). In contrast, gallic acid had no significant effect on mecA gene expression and only a modest effect on agrA expression (p < 0.05). The expression values were normalized to untreated controls, which were set at 1.0. The results are represented as mean ± SEM from three individual experiments. The Supplementary Figure S5 demonstrates the amplification of the reference gene 16S rRNA by PCR. The fold changes in mecA and agrA expression following compound treatment are summarized in Figure 8. Furthermore, PCR analysis was performed to study the downregulated strains, and the results are presented in Figure 9. In addition to transcriptional modulation, an antibiotic synergy assay was performed to evaluate whether curcumin and eugenol enhance the efficacy of β-lactam antibiotics against MRSA. Combination treatments using sub-inhibitory concentrations (½ MIC) of BACs with methicillin and ampicillin were assessed via agar well diffusion. As shown in Supplementary Figure S2, the combination treatments resulted in larger zones of inhibition compared to antibiotics alone, suggesting a potential synergistic or additive effect of BACs in re-sensitizing MRSA to β-lactam antibiotics. The effect of BACs on MRSA biofilm formation was assessed using the crystal violet assay. Treatment with curcumin, and eugenol resulted in a marked reduction in biofilm formation across all three clinical MRSA isolates. Based on OD540 values, treated strains showed a shift from strong to moderate or non-biofilm producers, suggesting a functional impact on biofilm development. These findings are presented in Supplementary Figure S3.
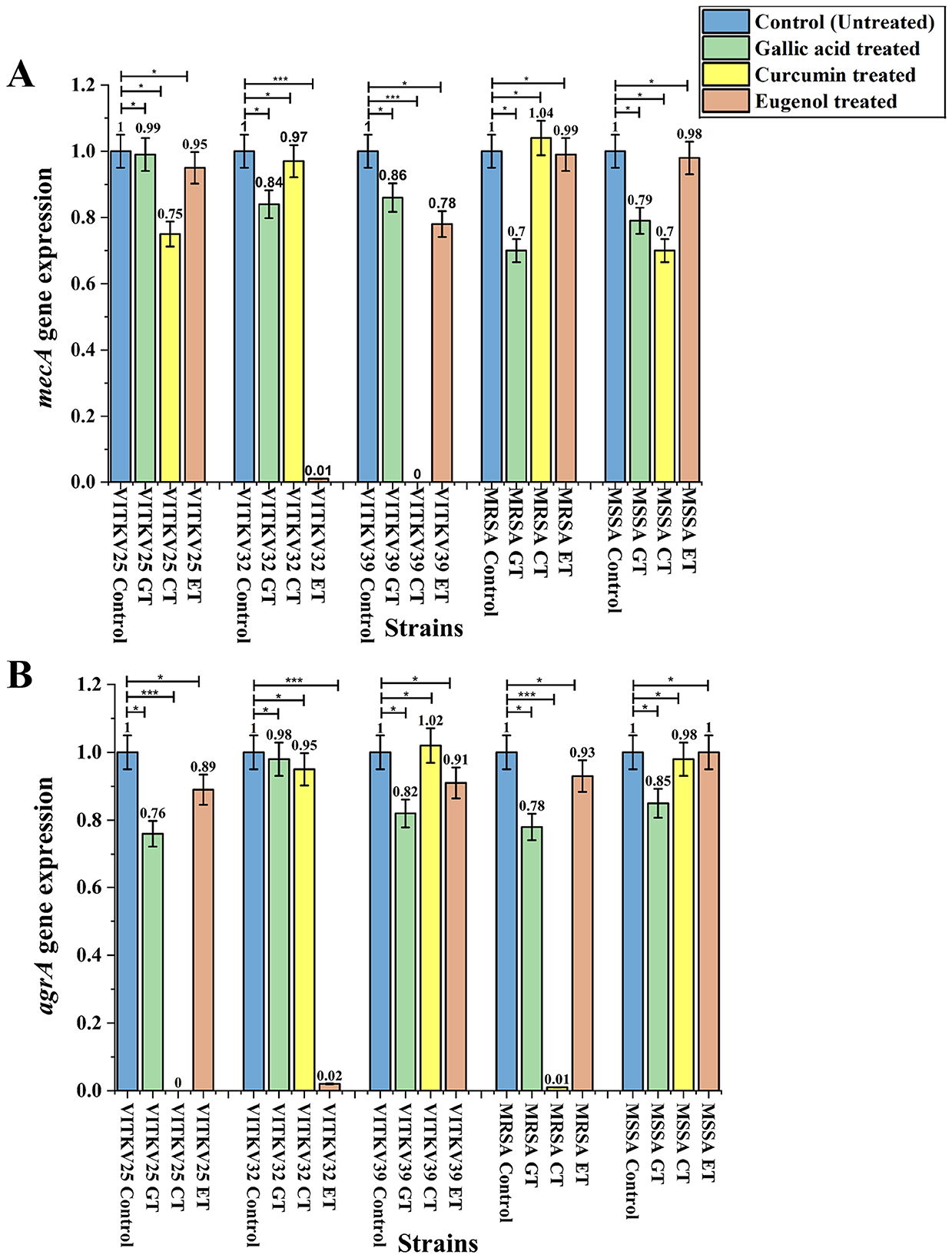
Figure 8. Relative gene expression analysis of mecA (A) and agrA (B) gene expression in S. aureus isolates treated with gallic acid (GT), curcumin (CT), and eugenol (ET). Expression levels were normalized to 16S rRNA and are represented relative to untreated controls (set as 1.0). Significant downregulation of mecA was observed in VITKV39 (CT) and VITKV32 (ET), while agrA gene expression was effectively decreased in VITKV25 (CT), MRSA (CT), and VITKV32 (ET). Data represent the mean ± SEM from three independent experiments. ***p < 0.001, *p < 0.05.
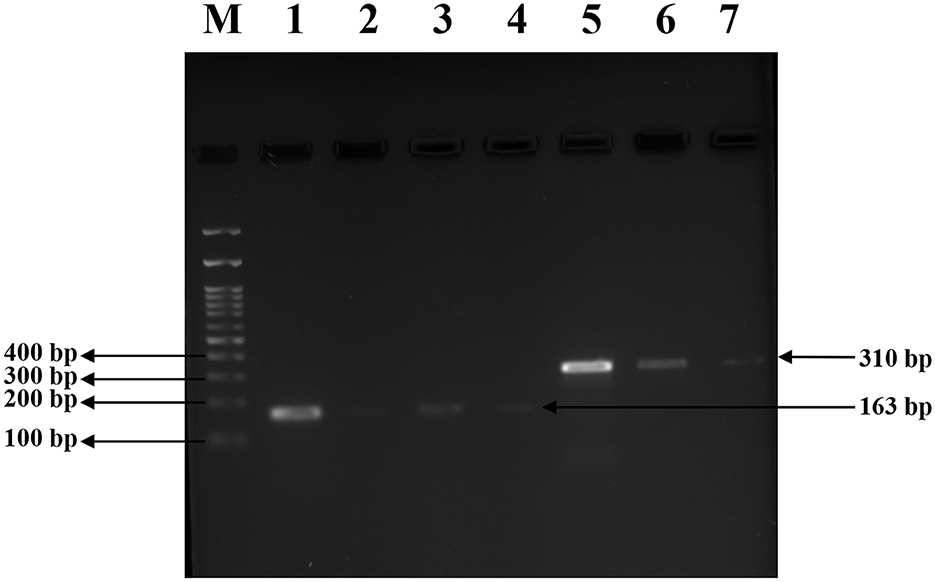
Figure 9. Agarose gel electrophoresis for visualizing the downregulated gene expression following BACs treatment. Gel image indicating PCR products of mecA (310 bp) and agrA (163 bp) amplified from treated S. aureus strains. Lane M: 100 bp DNA ladder; Lane 1: positive control for agrA (ATCC43300); Lanes 2, 3 and 4: agrA amplification from VITKV25 (CT), and MRSA (CT), and VITKV32 (ET), respectively; Lane 5: positive control for mecA (ATCC 43300); Lane 6,7: mecA amplification from VITKV39 (CT) and VITKV32 (ET), respectively. Reduced band intensity in treated samples indicates transcriptional suppression of target genes.
4 Discussion
MRSA infections have emerged as a major pathogen in both the general public and clinical settings, contributing significantly to antimicrobial resistance challenges. MRSA isolates typically carry the mecA gene, which encodes the PBP2a and confers resistance to β-lactam antibiotics, particularly cefoxitin and penicillin. However, some MRSA strains may remain sensitive to other classes of antibiotics depending on the local resistance ecology and clinical exposure history (Idrees et al., 2023). In the present study, the prevalence of MRSA among nasal S. aureus isolates was 16% (25 out of 150), which aligns with previous reports of nasal MRSA carriage in healthcare environments in India, where rates range from 12% to 20% (Wolde et al., 2023). The prevalence of S. aureus was higher in female subjects, with an overall carriage rate of 35.3%, and these results highlight the importance of routine screening and decontamination techniques, particularly in hospital settings, to prevent infection and control the spread of MRSA (Aravind et al., 2000).
The phenotypic and biochemical assays, including cefoxitin resistance, catalase, coagulase, DNase, and mannitol fermentation tests, were employed to identify S. aureus strains. Initial screening and identification of S. aureus isolates in this study were performed using a combination of standard phenotypic assays. Gram-staining facilitated the preliminary classification of Gram-positive cocci. Subsequent growth on MSA, characterized by yellow colonies due to mannitol fermentation in high-salt conditions, served as a selective and differential indicator for presumptive S. aureus (Thakur et al., 2017). Further, these observations were supported by positive results in the coagulase test, which remains a gold standard for differentiating S. aureus from other coagulase-negative Staphylococcus species (Sperber and Tatini, 1975). The catalase test helped differentiate Staphylococci spp. from catalase-negative Streptococci spp. (Mustafa, 2014). The oxidase test, which detects the presence of cytochrome c oxidase, was negative in all presumptive S. aureus isolates, consistent with the known oxidase-negative profile of Staphylococcal species and helping to exclude oxidase-positive genera such as Neisseria and Pseudomonas (Arjyal et al., 2020). The DNase test determines whether an organism can produce the DNase enzyme, which degrades DNA into smaller fragments that the cell can absorb. This test can distinguish S. aureus from other Staphylococcus species. Gram-positive, catalase-positive cocci that produce DNase can potentially be identified as S. aureus (Kateete et al., 2010). Altogether, these assays provided a comprehensive and reliable phenotypic profile. Out of 150 total isolates, 53 were confirmed as presumptive S. aureus using this phenotypic strategy, establishing the basis for downstream antimicrobial resistance and molecular characterizations.
The antibiotic susceptibility testing using the Kirby-Bauer disc diffusion method remains a standard phenotypic approach for evaluating bacterial resistance. In this study, cefoxitin, methicillin, and oxacillin were among the β-lactam agents used to determine resistance profiles, with interpretive breakpoints referenced from CLSI guidelines (Jorgensen and Ferraro, 2009). The zone of inhibition diameters provided a reliable estimate of antibiotic efficacy, and the consistent resistance observed to cefoxitin helped to confirm MRSA phenotypes among the selected isolates. Three isolates (VITKV25, VITKV32, and VITKV39) exhibited complete resistance to all ten antibiotics tested. These extensively drug-resistant profiles prompted their selection for molecular identification and further analysis. The molecular characterization by 16S rRNA sequencing results confirmed all three strains as S. aureus, with high sequence similarity to reference strains in GenBank, as validated by phylogenetic tree analysis.
The QS system in S. aureus, predominantly mediated by the agr operon, coordinates population-dependent gene regulation related to virulence, adhesion, motility, and notable biofilm formation. In MRSA, biofilm development plays a crucial role in persistent infections by enhancing bacterial survival on host tissues and medical devices, impeding phagocytosis, and restricting antibiotic penetration (Yamazaki et al., 2024). In this study, the crystal violet biofilm assay revealed that VITKV32 was a strong biofilm producer, while VITKV25 and VITKV39 exhibited moderate biofilm-forming capabilities, relative to MRSA and MSSA reference strains. These findings are consistent with recent reports, including those by B. Chaudhary et al., which indicate that biofilm production among clinical MRSA isolates varies widely, with stronger biofilm phenotypes frequently associated with co-expression of resistance determinants and virulence factors (Chaudhary et al., 2021). These insights reinforce the clinical significance of assessing biofilm potential alongside resistance profiling, particularly in multidrug-resistant strains, where biofilm formation increases the risk of chronic infection in addition to antimicrobial resistance.
The antibacterial activity of selected compounds was first evaluated through agar well diffusion, which allowed for a preliminary assessment of efficacy against the clinical isolates. Compounds were chosen based on their natural sources and recent research highlighting their antibacterial effects against S. aureus. Among all five tested compounds, curcumin, eugenol, and gallic acid produced clear inhibition zones and were selected for further analysis. In contrast, piperine and α-terpineol showed no measurable activity under the tested conditions. MIC assays were performed to determine the potency of these compounds. The MIC values revealed that curcumin exhibited the highest inhibitory activity, followed by eugenol, while gallic acid showed relatively lower activity. The outcomes are consistent with previous studies, where curcumin demonstrated strong antibacterial activity against S. aureus isolates as reported by Teow et al. (2016), the notable efficacy of eugenol is demonstrated by Li et al. (2024) and gallic acid exhibited moderate activity in line with observations by Jiamboonsri et al. (2023). These results, identifying curcumin and eugenol as the most potent inhibitors, provided a rational basis for selecting these compounds to investigate their effects on gene expression at their respective MICs.
Followed by the MIC profiling, the PCR amplification confirmed the presence of the mecA and agrA genes in all three isolates, with MRSA and MSSA reference strains used as positive and negative controls, respectively. Methicillin resistance was molecularly confirmed by the identification of mecA, which was consistent with the previously noted phenotypic cefoxitin resistance. The detection of mecA justify the classification of these strains as MRSA as it is dependable diagnostic marker in clinical microbiology (Dhungel et al., 2021). A functional QS system that supports the regulatory control of virulence was confirmed by the presence of agrA. Also, its structural susceptibility to small-molecule inhibition, agrA is increasingly recognized as a promising ant virulence target (Podkowik et al., 2023). These findings are reinforced by recent clinical surveillance data in which mecA was found in 95% of MRSA strains and agrA in 85% of the same isolates, according to a study that examined S. aureus from hospital settings (Eidaroos et al., 2025). Therefore, these molecular confirmations established the basis for evaluating the transcriptional modulation of virulence and resistance genes in response to BACs treatment.
The expression of mecA and agrA relative to the 16S rRNA housekeeping gene was evaluated by qRT-PCR to investigate the transcriptional effects of BACs treatment on the clinical isolates. Remarkably, mecA gene expression was downregulated in curcumin-treated VITKV39 and eugenol-treated VITKV32, whereas agrA was suppressed in curcumin-treated VITKV25 and the MRSA reference strain, as well as in eugenol-treated VITKV32. These findings depicted that both curcumin and eugenol have gene-specific inhibitory effects on significant resistance and virulence factors in S. aureus. Systematically, the transcriptional suppression of the mecA and agrA genes may involve interference with upstream regulatory elements. Our study results are consistent with the previous studies that have demonstrated curcumin's ability to downregulate mecA gene expression, leading to a decrease in PBP2a levels and restoring the efficacy of β-lactam antibiotics against MRSA strains (Teow et al., 2016). Additionally, curcumin has been reported to inhibit the agrA gene, impacting the QS system, attenuating virulence, and biofilm formation (Kashi et al., 2024). Curcumin's antimicrobial activity has also been attributed to its ability to compromise bacterial cell wall integrity. Previous studies have shown that curcumin increases membrane permeability, alters lipid composition, and causes structural disintegration of the cell wall, ultimately leading to cytoplasmic leakage and cell death (Mun et al., 2014). Such membrane damage may activate stress-responsive regulatory pathways in S. aureus, which could contribute to the observed downregulation of mecA and agrA in our study. Eugenol has also demonstrated significant anti-biofilm activity and QS disruption in S. aureus, which may indirectly affect mecA expression and enhance susceptibility to antibiotics (El-Far et al., 2021). Also, eugenol is known to disrupt membrane integrity and may alter signal transduction through sensor kinase like AgrC, indirectly suppressing AgrA activation. Although this study focused on mecA and agrA as key targets, ongoing work includes the transcriptional analysis of additional regulators and virulence genes such as SarA, blaZ, icaA, and hla. The role of diadenylate cyclase in biofilm regulation is also acknowledged and will be explored in future transcriptomic studies.
Antimicrobial-induced transcriptional changes are often attributed to indirect effects arising from cellular stress, rather than direct modulation of gene expression. As outlined by Kashi et al. (2024), compounds may alter transcriptional profiles through disruption of membrane integrity or metabolic processes without specifically targeting transcription factors. In this context, the observed downregulation of mecA and agrA in response to BACs may reflect secondary stress responses. Further mechanistic studies are required to determine whether these effects result from direct regulatory interference (Kashi et al., 2024).
5 Conclusion
This study confirms the prevalence of MRSA and multidrug-resistant S. aureus in nasal carriage within a clinical environment, with a prevalence rate of 16% (25/150). Among the BACs tested, curcumin and eugenol exhibited significant antibacterial activity and also downregulated key resistance and virulence genes at their MIC concentrations. Curcumin effectively inhibited mecA in VITKV39 and agrA in both VITKV25 and MRSA, while eugenol inhibited mecA and agrA in VITKV32. While gene expression modulation may be influenced in part by growth inhibition, the consistent transcriptional downregulation observed suggests an additional regulatory effect of these compounds. The study outcomes underscore the potential of curcumin and eugenol as promising phytocompounds by targeting both the mecA gene for antimicrobial resistance and the agrA gene for QS-mediated virulence pathways in MRSA. Such mechanisms could help reduce pathogenicity and may enhance the efficacy of β-lactam antibiotics against resistant strains. This dual-action approach can help limit the risk of chronic infection and the development of multidrug-resistant strains. Future studies exploring quorum quenching, anti-virulence mechanisms, and synergy with antibiotics are crucial to advance these compounds toward clinical application.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Ethics statement
The studies involving humans were approved by Institutional Ethical Committee for studies on human subjects (IECH) of Vellore Institute of Technology, Vellore. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
MK: Project administration, Formal analysis, Methodology, Supervision, Writing – original draft, Conceptualization, Writing – review & editing. VV: Data curation, Writing – original draft, Conceptualization, Investigation, Validation, Writing – review & editing, Formal analysis, Methodology.
Funding
The author(s) declare that no financial support was received for the research and/or publication of this article.
Acknowledgments
The authors would like to acknowledge the Vellore Institute of Technology for granting the required facilities to conduct this research.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Gen AI was used in the creation of this manuscript.
Any alternative text (alt text) provided alongside figures in this article has been generated by Frontiers with the support of artificial intelligence and reasonable efforts have been made to ensure accuracy, including review by the authors wherever possible. If you identify any issues, please contact us.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1643774/full#supplementary-material
Abbreviations
MRSA, Methicillin-resistant Staphylococcus aureus; MSSA, Methicillin-sensitive Staphylococcus aureus; QS, Quorum sensing; Agr, Accessory gene regulator; Mec, Methicillin resistance; PBP2a, Penicillin-binding protein 2a; PCR, Polymerase chain reaction; MIC, Minimum inhibitory concentration; qRT-PCR, Quantitative real-time reverse transcriptase-polymerase chain reaction; FDA, Food and Drug Administration.
References
Al-Dmour, O., Al-Groom, R., Alsheikh, A., Mahmoud, S., Amawi, K., Yousef, I., et al. (2023). Genetic identification of methicillin-resistant Staphylococcus aureus nasal carriage and its antibiogram among kidney dialysis patients at a tertiary care hospital in AL-Karak, Jordan. Int. J. Microbiol. 2023:9217014. doi: 10.1155/2023/9217014
Aravind, P., Unny Krishnan, P., Srinivasa, H., and Joseph, V. (2000). Screening of burns unit staff of a tertiary care hospital for methicillin-resistant Staphylococcus aureus colonisation. McGill J. Med. 5, 80–84. doi: 10.26443/mjm.v5i2.701
Arjyal, C., Kc, J., and Neupane, S. (2020). Prevalence of methicillin-resistant Staphylococcus aureus in shrines. Int. J. Microbiol. 2020. doi: 10.1155/2020/7981648
Batista de Andrade Neto, J., Pessoa de Farias Cabral, V., Brito Nogueira, L. F., Rocha da Silva, C., Gurgel do Amaral Valente Sá, L., Ramos da Silva, A., et al. (2021). Anti-MRSA activity of curcumin in planktonic cells and biofilms and determination of possible action mechanisms. Microb. Pathog. 155:104892. doi: 10.1016/j.micpath.2021.104892
Beargie, R. A., Bracken, E. C., and Riley, H. D. (1965). Micromethod (spot-plate) determination of in vitro antibiotic. Appl. Microbiol. 13, 279–280. doi: 10.1128/am.13.2.279-280.1965
Chaudhary, B. L., Bisht, D., and Faujdar, S. S. (2021). Biofilm formation and its association with antibiotic susceptibility pattern in methicillin-resistant Staphylococcus aureus isolates. J. Pure Appl. Microbiol. 15, 2041–2049. doi: 10.22207/JPAM.15.4.26
Clinical and Laboratory Standards Institute (2018). Institute CaLS. Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from animals CLSI Supplement VET08. Wayne: CLSI.
Das, S., Malik, M., Dastidar, D. G., Roy, R., Paul, P., Sarkar, S., et al. (2024). Piperine, a phytochemical prevents the biofilm city of methicillin-resistant Staphylococcus aureus: a biochemical approach to understand the underlying mechanism. Microb. Pathog. 189:106601. doi: 10.1016/j.micpath.2024.106601
Deurenberg, R. H., Vink, C., Kalenic, S., Friedrich, A. W., Bruggeman, C. A., and Stobberingh, E. E. (2007). The molecular evolution of methicillin-resistant Staphylococcus aureus. Clin. Microbiol. Infect. 13, 222–235. doi: 10.1111/j.1469-0691.2006.01573.x
Dhungel, S., Rijal, K. R., Yadav, B., Dhungel, B., Adhikari, N., Shrestha, U. T., et al. (2021). Methicillin-Resistant Staphylococcus aureus (MRSA): prevalence, antimicrobial susceptibility pattern, and detection of mec a gene among cardiac patients from a tertiary care heart center in Kathmandu, Nepal. Infect. Dis. Res. Treat. 14:117863372110373. doi: 10.1177/11786337211037355
Divyakolu, S., Chikkala, R., Kamaraju, S., and Sritharan, V. (2021). Quorum quenching as a strategy for treating methicillin resistant S. Aureus (MRSA)-effect of ε-polylysine, ethanolic extracts of guava leaves and mango seed kernel. Indian J. Biochem. Biophys. 58, 171–177. doi: 10.56042/ijbb.v58i2.35248
Eidaroos, N. H., Algammal, A. M., Mohamaden, W. I., Alenzi, A. M., Alghamdi, S., Kabrah, A., et al. (2025). Virulence traits, agr typing, multidrug resistance patterns, and biofilm ability of MDR Staphylococcus aureus recovered from clinical and subclinical mastitis in dairy cows. BMC Microbiol. 25:3. doi: 10.1186/s12866-025-03870-3
El-Far, A., Samir, S., El-Gebaly, E., Taha, N. Y., Fahmy, E. M., Diab, T. M., et al. (2021). Assessment of eugenol inhibitory effect on biofilm formation and biofilm gene expression in methicillin resistant Staphylococcus aureus clinical isolates in Egypt. Infect. Genet. Evol. 89:104722. doi: 10.1016/j.meegid.2021.104722
Fuqua, W. C., Winans, S. C., and Greenberg, E. P. (1994). Quorum sensing in bacteria: the LuxR-LuxI family of cell density- responsive transcriptional regulators. J. Bacteriol. 176, 269–275. doi: 10.1128/jb.176.2.269-275.1994
Hossain, M. L., Lim, L. Y., Hammer, K., Hettiarachchi, D., and Locher, C. (2022). A review of commonly used methodologies for assessing the antibacterial activity of honey and honey products. Antibiotics 11. doi: 10.3390/antibiotics11070975
Idrees, M. M., Saeed, K., Shahid, M. A., Akhtar, M., Qammar, K., Hassan, J., et al. (2023). Prevalence of mecA- and mecC-associated methicillin-resistant Staphylococcus aureus in clinical specimens, Punjab, Pakistan. Biomedicines 11, 1–11. doi: 10.3390/biomedicines11030878
Jaradat, Z. W., Ababneh, Q. O., Sha'aban, S. T., Alkofahi, A. A., Assaleh, D., and Al Shara, A. (2020). Methicillin resistant Staphylococcus aureus and public fomites: a review. Pathog. Glob. Health 114, 426–450. doi: 10.1080/20477724.2020.1824112
Jiamboonsri, P., Eurtivong, C., and Wanwong, S. (2023). Assessing the potential of gallic acid and methyl gallate to enhance the efficacy of β-lactam antibiotics against methicillin-resistant Staphylococcus aureus by targeting β-lactamase: in silico and in vitro studies. Antibiotics 12:1622. doi: 10.3390/antibiotics12111622
Johansen, B., Duval, R. E., and Sergere, J. C. (2022). First evidence of a combination of terpinen-4-ol and α-terpineol as a promising tool against ESKAPE pathogens. Molecules 27, 1–18. doi: 10.3390/molecules27217472
Jorgensen, J. H., and Ferraro, M. J. (2009). Antimicrobial susceptibility testing: a review of general principles and contemporary practices. Clin. Infect. Dis. 49, 1749–1755. doi: 10.1086/647952
Kala, R., Chauhan, H., Rajput, A., and Kutty, R. (2012). Biofilm characterization and quorum quenching in pathogenic strains Staphylococcus aureus and Pseudomonas Aeruginosa. Int. J. Adv. Biotechnol. Res. 3, 515–522.
Kashi, M., Noei, M., Chegini, Z., and Shariati, A. (2024). Natural compounds in the fight against Staphylococcus aureus biofilms: a review of antibiofilm strategies. Front. Pharmacol. 15:1491363. doi: 10.3389/fphar.2024.1491363
Kateete, D. P., Kimani, C. N., Katabazi, F. A., Okeng, A., Okee, M. S., Nanteza, A., et al. (2010). Identification of Staphylococcus aureus: DNase and Mannitol salt agar improve the efficiency of the tube coagulase test. Ann. Clin. Microbiol. Antimicrob. 9, 1–7. doi: 10.1186/1476-0711-9-23
Li, H., Li, C., Shi, C., Alharbi, M., Cui, H., and Lin, L. (2024). Phosphoproteomics analysis reveals the anti-bacterial and anti-virulence mechanism of eugenol against Staphylococcus aureus and its application in meat products. Int. J. Food Microbiol. 414, 110621. doi: 10.1016/j.ijfoodmicro.2024.110621
Lowy, F. D. (2003). Antimicrobial resistance: the example of Staphylococcus aureus. J. Clin. Invest. 111, 1265–1273. doi: 10.1172/JCI18535
Moses, V. K., Kandi, V., and Rao, S. K. D. (2020). Minimum inhibitory concentrations of vancomycin and daptomycin against methicillin-resistant Staphylococcus aureus isolated from various clinical specimens: a study from South India. Cureus 12, 1–8. doi: 10.7759/cureus.6749
Mun, S. H., Kim, S. B., Kong, R., Choi, J. G., Kim, Y. C., Shin, D. W., et al. (2014). Curcumin reverse methicillin resistance in Staphylococcus aureus. Molecules 19, 18283–18295. doi: 10.3390/molecules191118283
Mustafa, H. S. I. (2014). Staphylococcus aureus can produce catalase enzyme when adding to human WBCs as a source of H2O2 productions in human plasma or serum in the laboratory. Open J. Med. Microbiol. 04, 249–251. doi: 10.4236/ojmm.2014.44028
Nandhini, P., Gupta, P. K., Mahapatra, A. K., Das, A. P., Agarwal, S. M., Mickymaray, S., et al. (2023). In-Silico molecular screening of natural compounds as a potential therapeutic inhibitor for Methicillin-resistant Staphylococcus aureus inhibition. Chem. Biol. Interact. 374:110383. doi: 10.1016/j.cbi.2023.110383
Palaniappan, B., Solomon, A. P., and C, D. R. (2021). Targeting AgrA quorum sensing regulator by bumetanide attenuates virulence in Staphylococcus aureus – a drug repurposing approach. Life Sci. 273:119306. doi: 10.1016/j.lfs.2021.119306
Podkowik, M., Perault, A. I., Putzel, G., Pountain, A., Kim, J., Dumont, A. L., et al. (2023). Quorum-sensing agr system of Staphylococcus aureus primes gene expression for protection from lethal oxidative stress. Elife 12, 1–52. doi: 10.7554/eLife.89098.4
Rahman, K. M. T., and Butzin, N. C. (2024). Counter-on-chip for bacterial cell quantification, growth, and live-dead estimations. Sci. Rep. 14, 1–14. doi: 10.1038/s41598-023-51014-2
Rongpharpi, S. R., Hazarika, N. K., and Kalita, H. (2013). The prevalence of nasal carriage of Staphylococcus aureus among healthcare workers at a tertiary care hospital in Assam with special reference to MRSA. J. Clin. Diagnostic Res. 7, 257–260. doi: 10.7860/JCDR/2013/4320.2741
Sada, T. S., Alemayehu, D. H., Tafese, K. M., and Tessema, T. S. (2024). Genomic analysis and characterization of lytic bacteriophages that target antimicrobial resistant Escherichia coli in Addis Ababa, Ethiopia. Heliyon 10:e40342. doi: 10.1016/j.heliyon.2024.e40342
Shukla, S. K., and Rao, T. S. (2013). Effect of calcium on Staphylococcus aureus biofilm architecture: a confocal laser scanning microscopic study. Colloids Surf. B Biointerfaces 103, 448–454. doi: 10.1016/j.colsurfb.2012.11.003
Sperber, W. H., and Tatini, S. R. (1975). Interpretation of the tube coagulase test for identification of Staphylococcus aureus. Appl. Microbiol. 29, 502–505. doi: 10.1128/am.29.4.502-505.1975
Stan, D., Enciu, A. M., Mateescu, A. L., Ion, A. C., Brezeanu, A. C., Stan, D., et al. (2021). Natural compounds with antimicrobial and antiviral effect and nanocarriers used for their transportation. Front. Pharmacol. 12, 1–25. doi: 10.3389/fphar.2021.723233
Sytar, O., and Smetanska, I. (2022). Special issue “bioactive compounds from natural sources (2020, 2021).” Molecules 27, 2020–2023. doi: 10.3390/molecules27061929
Teow, S. Y., Liew, K., Ali, S. A., Khoo, A. S. B., and Peh, S. C. (2016). Antibacterial action of curcumin against Staphylococcus aureus: a brief review. J. Trop. Med. 2016. doi: 10.1155/2016/2853045
Thakur, P., Nayyar, C., Tak, V., and Saigal, K. (2017). Mannitol-fermenting and tube coagulase-negative staphylococcal isolates: unraveling the diagnostic dilemma. J. Lab. Physicians 9, 064–065. doi: 10.4103/0974-2727.187926
Thoendel, M., Kavanaugh, J. S., Flack, C. E., and Horswill, A. R. (2011). Peptide signaling in the Staphylococci. Chem. Rev. 111, 117–151. doi: 10.1021/cr100370n
Turk, R., Singh, A., Rousseau, J., and Weese, J. S. (2013). In vitro evaluation of DispersinB on methicillin-resistant Staphylococcus pseudintermedius biofilm. Vet. Microbiol. 166, 576–579. doi: 10.1016/j.vetmic.2013.07.011
Urushibara, N., Aung, M. S., Kawaguchiya, M., and Kobayashi, N. (2020). Novel staphylococcal cassette chromosome mec (SCCmec) type XIV (5A) and a truncated SCCmec element in SCC composite islands carrying speG in ST5 MRSA in Japan. J. Antimicrob. Chemother. 75, 46–50. doi: 10.1093/jac/dkz406
Vinodhini, V., and Kavitha, M. (2024). Deciphering agr quorum sensing in Staphylococcus aureus: insights and therapeutic prospects. Mol. Biol. Rep. 51. doi: 10.1007/s11033-023-08930-3
Wolde, W., Mitiku, H., Sarkar, R., and Shume, T. (2023). Nasal carriage rate of Staphylococcus aureus, its associated factors, and antimicrobial susceptibility pattern among health care workers in public hospitals, Harar, Eastern Ethiopia. Infect. Drug Resist. 16, 3477–3486. doi: 10.2147/IDR.S396570
Keywords: antibiotic resistance, bioactive compounds, methicillin-resistant Staphylococcus aureus, quorum sensing, mecA gene, agrA gene
Citation: Vinodhini V and Kavitha M (2025) Targeted inhibition of mecA and agrA genes in clinical MRSA isolates by natural bioactive compounds. Front. Microbiol. 16:1643774. doi: 10.3389/fmicb.2025.1643774
Received: 09 June 2025; Accepted: 11 August 2025;
Published: 26 August 2025.
Edited by:
Amira Zairi, University of Sousse, TunisiaReviewed by:
Rakesh Sikdar, University of Minnesota Twin Cities, United StatesDenny Chin, Concerto Biosciences, United States
Copyright © 2025 Vinodhini and Kavitha. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: M. Kavitha, bWthdml0aGExOTcyQGdtYWlsLmNvbQ==
†ORCID: M. Kavitha orcid.org/0000-0002-1862-8037
 V. Vinodhini
V. Vinodhini M. Kavitha
M. Kavitha